Abstract
Background
Asthma is a global chronic respiratory disease with complex pathogenesis. While current therapies offer some relief, they often fall short in effectively managing symptoms and preventing exacerbations for numerous patients. Thus, understanding its mechanisms and discovering new drug targets remains a pressing need for better treatment.
Methods
Using the GEO dataset, we screened differentially expressed genes (DEGs) in asthma patients' blood. Employing Summary Data-based Mendelian Randomization (SMR) and Two-Sample Mendelian Randomization (TSMR), we pinpointed asthma causal genes, causal DNA methylation sites, and methylation sites affecting gene expression, cross validated with at least 2 large-scale GWAS from each source. We utilized colocalization for genetic associations, meta-analysis for data integration, two-step MR for methylation-gene-asthma mediation mechanism. Druggability was evaluated using Open Target, virtual screening, and docking.
Results
Among the 954 DEGs found in asthma patients' blood, increased expression of CEP95 (discovery, OR_SMR = 0.94, 95% CI: 0.91–0.97), RBM6 (discovery, OR_SMR = 0.97, 95% CI: 0.95–0.99), and ITPKB (discovery, OR_SMR = 0.82, 95% CI: 0.74–0.92) in the blood decreased the risk of asthma, higher levels of HOXB-AS1 (discovery, OR_SMR = 1.05, 95% CI: 1.03–1.07), ETS1 (discovery, OR_SMR = 1.62, 95% CI: 1.29–2.04), and JAK2 (discovery, OR_SMR = 1.13, 95% CI: 1.06–1.21) in the blood increased the risk of asthma. Additionally, a total of 8 methylation sites on ITPKB, ETS1, and JAK2 were identified to influence asthma. An increase in methylation at site cg16265553 raised the risk of asthma partially by suppressing ITPKB expression. Similarly, increased methylation at cg13661497 reduced the asthma risk totally by suppressing JAK2 expression. The impact of CEP95, HOXB-AS1, and RBM6 expressions on asthma was further confirmed in lung tissues. Except for HOXB-AS1, all the other genes were potential druggable targets.
Conclusion
Our study highlighted that specific gene expressions and methylation sites significantly influence asthma risk and revealed a potential methylation-to-gene-to-asthma mechanism. This provided pivotal evidence for future targeted functional studies and the development of preventive and treatment strategies.
Keywords: Asthma, Mendelian randomization, Gene expression, DNA methylation, Mediation analysis
Introduction
Asthma is a common lung disease that affects approximately 262.4 million people worldwide.1 It results in over 400,000 deaths annually. The pathogenesis of asthma is complex, being influenced by both genetic and environmental factors.2 The exact causes remain not fully understood. While glucocorticoids are the primary treatment for asthma, many patients develop hormone dependence or resistance. Besides, prolonged use of these drugs can lead to side effects, including osteoporosis.3 Therefore, a deeper understanding of the precise causes and underlying mechanisms of asthma is essential for developing more effective treatment strategies.
Transcriptome studies play a pivotal role in disease studies.4 This approach provides a profound understanding of gene expression. It is recognized that diseases often coincide with changes in gene expression patterns. Such analyses are instrumental in deciphering disease mechanisms and identifying diagnostic markers. In conditions such as asthma, transcriptome studies become invaluable for elucidating the disease's molecular intricacies and proposing potential therapeutic avenues.5
While transcriptomics provides valuable insights, it alone cannot establish the influence of genes on diseases. Current randomized controlled trials (RCTs) and observational studies are not equipped to rapidly assess causal genes for diseases due to the extensive time, manpower, financial resources, and ethical considerations involved. Fortunately, developments in molecular genetics have provided new opportunities. Mendelian Randomization (MR), a prominent epidemiological method, leverages genetic variations as instrumental variables to elucidate causal relationships between exposures and outcomes, almost as credible as RCTs.6 Compared with traditional clinical research, MR has several benefits: It minimizes biases since genetic variations are randomly assigned, avoids reverse causality issues, is ethical without participant interventions, and quickly assesses exposure-outcome causal relationship using existing GWAS data, laying the groundwork for further clinical research.7 By incorporating expression quantitative trait loci (eQTL), which represent associations between genotypes and gene expression levels, into MR studies, we can effectively clarify the causal genes for diseases.
DNA methylation, a common chemical modification of DNA, plays a significant role in asthma.8 Research has shown that DNA methylation is both heritable and reversible.9 Consequently, investigating it is valuable for developing drugs and diagnostic tools that target sites associated with asthma. Meanwhile, DNA methylation can alter gene expression, with increased methylation typically resulting in reduced gene expression.10 Thus, comprehending DNA methylation patterns is vital. Including DNA methylation quantitative trait loci (mQTL) in our research, which link genotypes to DNA methylation levels, further advances our understanding of DNA methylation's impact on asthma and the factors driving changes in gene expression.
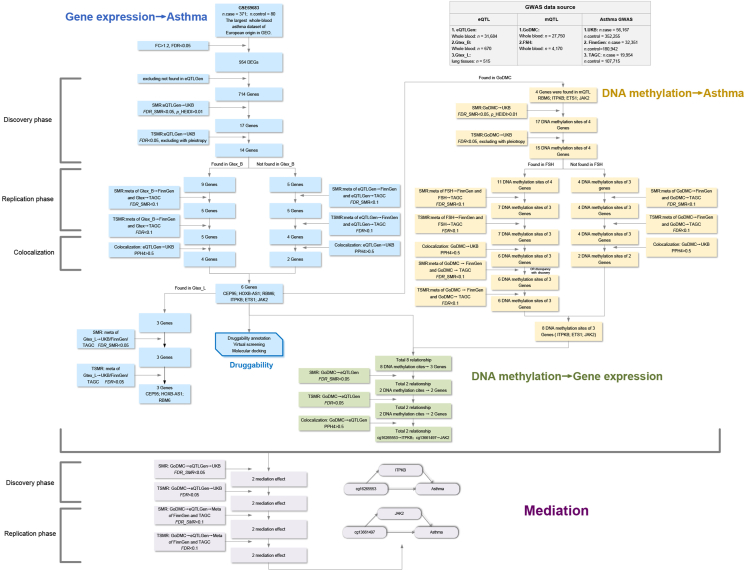
In our study, we aimed to identify potential causal genes and DNA methylation sites associated with asthma and elucidate the underlying mechanisms, as illustrated in Fig. 1. Initially, we utilized the blood GEO dataset from asthma patients to analyze differentially expressed genes (DEGs). We then employed Summary Data-based Mendelian Randomization (SMR) and Two-Sample Mendelian Randomization (TSMR) to investigate the causalities between circulating gene expression or circulating DNA methylation and asthma, as well as DNA methylation and gene expression. These analyses were carried out in both the discovery and replication phases using large-scale GWAS data. We also utilized meta-analysis and colocalization methods to enhance credibility. Some of these causal genes were validated in lung tissue. Regarding the mechanistic aspects, we proceeded to explore which methylation sites could influence asthma by suppressing gene expression through a two-step mediation analysis. Furthermore, we explored whether these identified targets could potentially serve as viable therapeutic drug targets for asthma.
Fig. 1.
Study design. FC, fold change; DEGs, differentially expressed genes; SMR, Summary Data-based Mendelian Randomization; TSMR: Two-Sample Mendelian Randomization; UKB, the UK Biobank; TAGC, Trans-National Asthma Genetic Consortium; eQTL, expression quantitative trait loci; mQTL, DNA methylation quantitative trait loci.
Materials and methods
Data resource
For transcriptomic data, we analyzed the whole-blood gene expression profile (GSE69683) from the GEO database consisting of 371 asthma patients and 80 controls of European descent. Using R packages "GEOquery" for data extraction, "limma" for DEG analysis, and "ggplot2" for visualization, we identified DEGs based on criteria: FC > 1.2 and FDR<0.05, referencing 2 criteria: 1 with Fold Change (FC) > 1.211,12 and the other based on |logFC|>[mean(|logFC|)+2sd(|logFC|)],13 which resulted in an FC > 1.21 after calculation.
For eQTL data, 3 datasets were utilized: a discovery blood eQTL dataset (n = 31,684) from the eQTLGen Consortium, a replication blood eQTL dataset (n = 670) from GTExV8, and an additional lung eQTL dataset (n = 515) from GTExV8. The lung tissues were primarily sampled from the inferior segment of the left upper lobe, 1 cm below the pleural surface, and can include airways, parenchyma, mesenchyme, etc.
For mQTL data, the study incorporates a discovery blood mQTL dataset (n = 27,750) from GoMDC, a replication blood mQTL dataset (n = 4170) from FSH.
For asthma GWAS summary data, the discovery set came from the UK Biobank (UKB) (ntotal = 408,422, ncases = 56,167, ncontrols = 352,255), while the replication sets were derived from FinnGen (ntotal = 213,293, ncases = 32,351, ncontrols = 180,942) and Trans-National Asthma Genetic Consortium (TAGC) (ntotal = 127,669, ncases = 19,954, ncontrols = 107,715).
In these datasets, only eQTLGen and GTEx originated from predominantly European populations, while the rest were from European populations.
Selection of instrumental variables (IVs)
Only QTLs that met the following criteria were included: 1) showed genome-wide significance level (p < 5 × 10−8); 2) positioned outside the major histocompatibility complex region;14 3) were cis-acting QTLs15 (SNP gene distance<1 Mb for eQTL;16 SNP DNAm site<1 Mb for mQTL17); 4) with a (MAF) minor allele frequency>0.01;18 5) with F > 10. An F, determined by (beta2/se2), assesses the potency of the SNPs;19,20 6) were both exist in exposure and outcome. Besides, for TSMR, the default setting was used, identifying independent associations through linkage disequilibrium (LD) clumping with r2 < 0.001 using the R package "ieugwasr".14,21 For SMR, the default configuration was used, excluding SNPs in very strong LD (r2 > 0.9) with the top associated QTL and removing SNPs in weak LD or not in LD (r2 < 0.05) with the top associated QTL.
Additionally, CEP95 had no eQTL in lung tissues with a p-value of 5 × 10−8, so the threshold was relaxed to 1 × 10−5. For the mQTL from FSH, only data with a p-value less than 2 × 10−11 was provided by the authors, so we chose these SNPs.
Summary data-based mendelian randomization (SMR) and two-sample MR (TSMR)
Mendelian Randomization (MR) is a reliable method for causal inference that uses genetic variants as instrumental variables to determine the causal relationship between exposures (such as genes/genomic sites) and outcomes (such as asthma). The instrumental variables, typically single nucleotide polymorphisms (SNPs), must be strongly associated with the exposure (p < 5e-8) and independent of the outcome. These SNPs should be present in both the exposure and outcome genome-wide association studies (GWAS). Therefore, these instrumental variables can represent the exposure, and by analyzing their results, we can determine whether there is a causal relationship between the exposure and the outcome.
SMR is an advanced method that builds on traditional Mendelian Randomization. By using SMR software, we investigated the causal relationships between gene expression, DNA methylation, and asthma. In the discovery phase, significance was set at FDR_SMR<0.05, while in replication, it was defined as FDR_SMR<0.1. Genes/genomic sites meeting these criteria are considered causally related in SMR.
TSMR is a traditional Mendelian Randomization method for causal analysis. In this study, TSMR was performed by the "TwoSampleMR" in R software. Referencing previous literature,14 when only 1 IV was present, the wald ratio was applied. If 2 or more IVs were available, the IVW method was adopted. In the discovery phase, significance was set at FDR<0.05, while in replication, it was defined as FDR<0.1. Genes/genomic sites that meet these criteria are regarded as having a causal relationship in TSMR.
Results were visually presented using forest plots by R package "forestploter".
Colocalization analysis
We used Bayesian colocalization to investigate genetic associations between exposure-outcome pairs within the corresponding genetic locus by R package "coloc".22,23
For significant MR results, we conducted co-localization analyses considering SNPs within ±100 kb for gene expression and asthma, methylation and asthma, methylation and gene expression. When the posterior probability for H4 (PPH4) >0.5,24 it suggests colocalization, reinforcing the MR findings.
Sensitivity analysis
We used the heterogeneity in dependent instruments (HEIDI) test by the SMR software to identify potential linkage heterogeneity, where a p < 0.0125 indicated their presence. For TSMR analysis, we employed Cochran's Q test to assess heterogeneity, with p < 0.05 indicating its presence. To evaluate pleiotropy, we utilized the MR-Egger regression test, considering a p < 0.05 as an indicator of pleiotropy.
We used the R package "phenoscanner" to perform phenotype scanning the SNPs related to asthma and its confounding factors for positive genes and sites in discovery phase. Significant SNPs met these criteria: they shared the same effect allele as our results, reached GWAS significance (p < 5 × 10−8), had an absolute effect size(β)>0.01, and originated from a European ancestry population.26
Druggability annotation, virtual screening, and molecular docking
Using Open Targets data (release 2023-06), we annotated genes with drug susceptibility information mainly as previously employed:27 1) Licensed drugs (bucket 1) for small molecules, antibodies, PROTAC, and others; 2) Drugs in clinical development (buckets 2 and 3) for small molecules, antibodies, PROTAC, and others; 3) Compounds in the preclinical phase (buckets 4 and 5) for small molecules; and 4) Predicted druggable (buckets 6 to 8) for small molecules, (buckets 4 and 5) for antibodies, and (buckets 4 to 6) for PROTAC.
We obtained protein structures from the AlphaFold Database and used the FDA-approved drug library from Selleckchem. After screening for bioavailability and physicochemical descriptors following Lipinski-Veber rules, we selected 1147 compounds. These, along with receptor molecules, were prepared using AutoDockTools. Blind docking screenings were conducted using AutoDock Vina, with automation through in-house scripts.28 Details of the pocket parameters can be found in Supplementary Table. S3.
Meta-analysis
In meta-analysis, 2 models were used: fixed-effect and random-effect. The choice depends on observed heterogeneity, assessed using Cochran's Q test and I2 statistic. If no significant heterogeneity (Q test p ≥ 0.05, I2 ≤ 50%), we used the fixed-effect model. Significant heterogeneity (Q test p < 0.05 or I2>50%) led to the random-effect model. We conducted meta-analysis using "meta" R package, with significance defined as FDR<0.05 in discovery and <0.1 in replication.
Mediation analysis
We used a two-step MR to explore the potential mediating role of gene expression in the causal effect of DNA methylation on asthma. This involves: 1) assessing the causal effect of DNA methylation on gene expression(β1), and 2) determining the causal effect of gene expression on asthma(β2). The mediation effect is the product of these 2 estimates(β1 × β2). We obtained the overall effect of DNA methylation on asthma(β3). To quantify the proportion of mediation, we divided the indirect effect by the total effect(β1 × β2/β3).29,30 To assess the mediation's significance, we used the Sobel test,31 and when FDR<0.05 in discovery and <0.1 in replication, we considered it significant.
Data details
All the detailed information of R package, software, and database, along with their relevant references, were provided in Supplementary Table. S1, and information of GEO, QTL, and asthma GWAS datasets, along with their relevant references, were provided in Supplementary Table. S2.
Results
Causal effect of gene expression on asthma
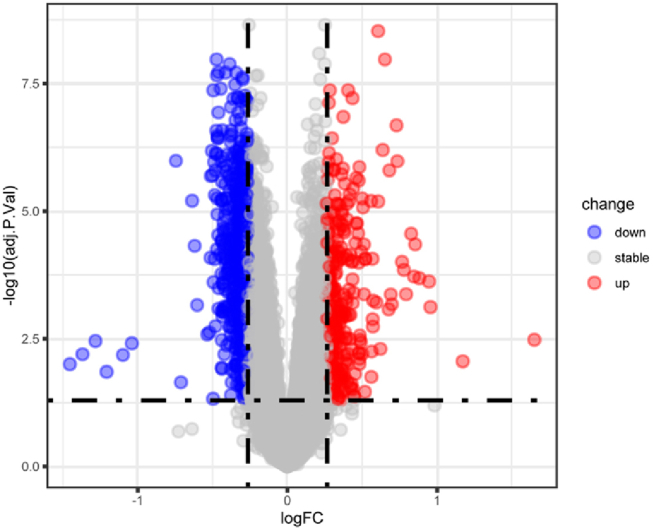
After analyzing the largest whole-blood GEO dataset with 371 European patients and 80 controls, we identified 954 DEGs (Fig. S1, Supplementary Table. S4). To identify causal DEGs in asthma, we utilized SMR, TSMR, and colocalization analyses, mutually validating them between the discovery and replication phases. Only genes meeting all these stringent criteria were considered as positive causal genes for asthma.
In the discovery phase, we utilized eQTL data from eQTLGen and asthma GWAS data from UKB. Initially, we conducted an SMR analysis and identified 17 potential causal DEGs for asthma (FDR_SMR<0.05, p_HEIDI>0.01) (Supplementary Table. S5). Subsequently, a TSMR analysis was conducted, confirming that 14 out of the initial 17 DEGs were potential causal genes (FDR<0.05, excluding with pleiotropy) (Supplementary Table. S6).
In the replication phase, we utilized eQTL data from GtexV8 and asthma GWAS data from both FinnGen and TAGC. However, we noticed disparities in the results between FinnGen and TAGC. To enhance the reliability of our findings, we conducted a meta-analysis by combining their results. Out of the 14 identified causal genes, only 9 were available in GtexV8. To ensure comprehensive coverage, we obtained eQTL data for the remaining 5 genes from eQTLGen. Among these 14 genes, 10 showed significant results in the meta-analysis (FDR_SMR<0.1) (Supplementary Table. S7), with 5 from GtexV8 and 5 from eQTLGen. In the subsequent TSMR analysis, 9 out of these 10 genes met the TSMR criteria (FDR<0.1) (Supplementary Table. S8).
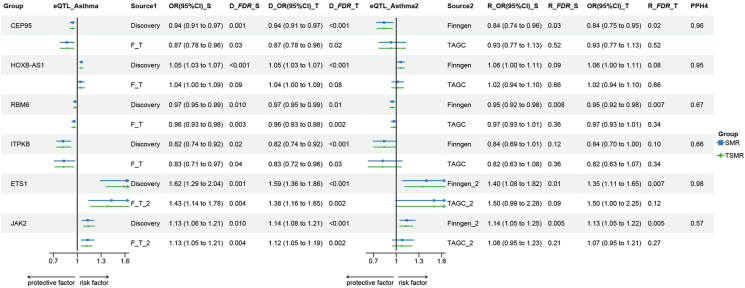
For colocalization analysis, 6 out of the 9 positive results met the criteria with PPH>0.5 (Supplementary Table. S9), including CEP95, HOXB-AS1, RBM6, ITPKB, ETS1, and JAK2 (Fig. 2). Since the results from SMR and TSMR were highly consistent, we have presented the SMR results below, detailed TSMR results are available in Fig. 2. Specifically, increased CEP95 (discovery, OR_SMR = 0.94, 95% CI: 0.91–0.97, FDR = 0.001; replication, OR_SMR = 0.87, 95% CI:0.78–0.96, FDR = 0.03; PPH4 = 0.96), RBM6 (discovery, OR_SMR = 0.97, 95% CI:0.95–0.99, FDR = 0.01; replication, OR_SMR = 0.96, 95% CI:0.93–0.98, FDR = 0.003; PPH4 = 0.67), ITPKB (discovery, OR_SMR = 0.82, 95% CI:0.74–0.92, FDR = 0.02; replication, OR_SMR = 0.83, 95% CI:0.71–0.97, FDR = 0.04; PPH4 = 0.66) decreased the risk of asthma, while elevated HOXB-AS1 (discovery, OR_SMR = 1.05, 95% CI:1.03–1.07, FDR<0.001; replication, OR_SMR = 1.04, 95% CI:1.00–1.09, FDR = 0.09; PPH4 = 0.95), ETS1 (discovery, OR_SMR = 1.62, 95% CI:1.29–2.04, FDR = 0.001; replication, OR_SMR = 1.43, 95% CI:1.14–1.78, FDR = 0.004; PPH4 = 0.98), JAK2 (discovery, OR_SMR = 1.13, 95% CI:1.06–1.21, FDR = 0.01; replication, OR_SMR = 1.13, 95% CI:1.05–1.21, FDR = 0.004; PPH4 = 0.57) increased the risk of asthma. Among these, ITPKB exhibited the strongest protective effect against asthma, while ETS1 displayed the most potent causative effect. Using PhenoScanner for the positive genes, we discovered that only the ETS1 eQTL rs55836957, used in both SMR and TSMR analyses, is associated with Eosinophil—a key component in asthma (Supplementary Table. S29). This suggests that this specific eQTL might directly impact asthma, affecting our conclusion's credibility. To address this, before performing the MR, we removed this eQTL associated with asthma and carried MR again. The results remained still significant with similar odds ratio values (Supplementary Table. S30).
Fig. 2.
Causal effect of gene expression in blood on asthma. SMR, Summary Data-based Mendelian Randomization; TSMR: Two-Sample Mendelian Randomization; eQTL, expression quantitative trait loci; OR, odds ratio; CI, confidence interval; _S, by using SMR; _T, by using TSMR; Discovery, exposure from eQTLGen, and outcome from UK Biobank (UKB); Finngen, exposure from Gtex and outcome from Finngen; TAGC, exposure from Gtex and outcome from Trans-National Asthma Genetic Consortium (TAGC); F_T, meta-analysis of results from Gtex→Finngen and Gtex→TAGC; Finngen_2, exposure from eQTLGen and outcome from Finngen; TAGC, exposure from eQTLGen and outcome from TAGC; F_T_2, meta-analysis of results from eQTLGen→Finngen and eQTLGen→TAGC.
Causal effect of DNA methylation on asthma
To explore the causal effect of DNA methylation on asthma, we employed the same approach we used for identifying positive genes, aiming to pinpoint positive causal methylation sites related to asthma.
Among the 6 positive genes, in the discovery phase, mQTL was derived from GoDMC, while asthma GWAS dataset came from UKB. Out of the 6 positive genes, 4 were identifiable within the mQTL dataset. Initially, we conducted an SMR analysis and identified 17 potential causal methylation sites of these 4 positive genes for asthma (FDR_SMR<0.05, p_HEIDI>0.01) (Supplementary Table. S10). Following this, we executed a TSMR analysis. Of the initial 17 sites, 15 potential sites of these the 4 positive genes had a significant association with asthma (FDR<0.05, excluding with pleiotropy) (Supplementary Table. S11).
In the replication phase, we selected mQTL from FSH and asthma data from both FinnGen and TAGC. Out of 15 identified causal sites, only 11 were present in FSH (group 1). To ensure comprehensive coverage, we sourced mQTL data for the remaining 4 sites from GoDMC (group 2). Of these 14 sites, 11 showed significant results in the meta-analysis (FDR_SMR<0.1), 7 were derived from FSH and 4 from GoDMC (Supplementary Table. S12); In the subsequent TSMR analysis, all 11 sites passed the TSMR criteria (FDR<0.1) (Supplementary Table. S13). In the colocalization analysis using mQTL data from GoDMC and asthma GWAS data from UKB, 8 sites met the required thresholds, 6 from FSH and 2 from GoDMC (PPH>0.5) (Supplementary Table. S14).
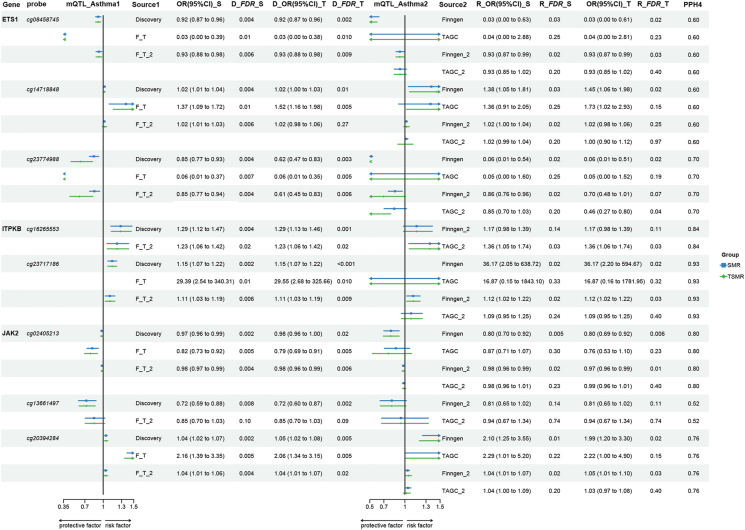
However, we observed notable discrepancies in the OR values when comparing the MR results when mQTL from GoDMC and FSH. We noted that when FSH was employed as the exposure database, several results exhibited large se, which may potentially compromise the reliability of the results. Given that GoDMC has a significantly larger sample size than FSH, we conducted another round of SMR and TSMR analyses for the 6 potential causal sites sourced from FSH. For this analysis, we used mQTL data from GoDMC and asthma data from FinnGen and TAGC. The findings affirmed the significance of these 6 sites, and their OR values closely aligned with those identified in the discovery phase (Supplementary Table. S12 and S13). Specifically, increased methylation levels of cg08458745 (ETS1, discovery, OR_SMR = 0.92, 95% CI:0.87–0.96, FDR = 0.004; replication, OR_SMR = 0.93, 95% CI:0.88–0.98, FDR = 0.006; PPH4 = 0.6), cg23774988 (ETS1, discovery, OR_SMR = 0.85, 95% CI:0.77–0.93, FDR = 0.004; replication, OR_SMR = 0.85, 95% CI:0.77–0.94, FDR = 0.004; PPH4 = 0.7), cg02405213 (JAK2, discovery, OR_SMR = 0.97, 95% CI:0.96–0.99, FDR = 0.002; replication, OR_SMR = 0.98, 95% CI:0.97–0.99, FDR = 0.004; PPH4 = 0.80); cg13661497 (JAK2, discovery, OR_SMR = 0.72, 95% CI:0.59–0.88, FDR = 0.008; replication, OR_SMR = 0.85, 95% CI:0.70–1.03, FDR = 0.10; PPH4 = 0.52) decreased the risk of asthma, whereas elevated methylation level of cg14718848 (ETS1, discovery, OR_SMR = 1.02, 95% CI:1.01–1.04, FDR = 0.004; replication, OR_SMR = 1.02, 95% CI: 1.01–1.03, FDR = 0.006; PPH4 = 0.60), cg16265553 (ITPKB, discovery, OR_SMR = 1.29, 95% CI: 1.12–1.47, FDR = 0.004; replication, OR_SMR = 1.23, 95% CI:1.06–1.42, FDR = 0.02; PPH4 = 0.84), cg23717186 (ITPKB, discovery, OR_SMR = 1.15, 95% CI:1.07–1.22, FDR = 0.002; replication, OR_SMR = 1.11, 95% CI:1.03–1.19, FDR = 0.006; PPH4 = 0.93), cg20394284 (JAK2, discovery, OR_SMR = 1.04, 95% CI:1.02–1.07, FDR = 0.002; replication, OR_SMR = 1.04, 95% CI:1.01–1.06, FDR = 0.004; PPH4 = 0.76) increased the risk of asthma (Fig. 3). Among these, cg13661497 of JAK2 exhibited the strongest protective effect against asthma, while cg16265553 of ITPKB displayed the most potent causative effect. Using PhenoScanner for the positively identified sites, we discovered that only the cg02405213 of JAK2 mQTL rs11789744, used in TSMR analyses, is associated with Eosinophil (Supplementary Table. S29). To address this, before performing the MR, we removed mQTLs associated with asthma and carried MR again. The results remained still significant with similar odds ratio values as before (Supplementary Table. S30).
Fig. 3.
Causal effect of DNA methylation in blood on asthma. SMR, Summary Data-based Mendelian Randomization; TSMR: Two-Sample Mendelian Randomization; eQTL, expression quantitative trait loci; OR, odds ratio; CI, confidence interval; _S, by using SMR; _T, by using TSMR; Discovery, exposure from GoDMC, and outcome from UK Biobank (UKB); Finngen, exposure from FSH and outcome from Finngen; TAGC, exposure from FSH and outcome from Trans-National Asthma Genetic Consortium (TAGC); F_T, meta-analysis of results from FSH→Finngen and FSH→TAGC; Finngen_2, exposure from GoDMC and outcome from Finngen; TAGC, exposure from GoDMC and outcome from TAGC; F_T_2, meta-analysis of results from GoDMC→Finngen and GoDMC→TAGC.
Besides, using the same method, we also identified 23 potential causal sites across 9 DEGs. Detailed information can be found in Supplementary Table. S18-S22.
Causal effect of DNA methylation on gene expression
To ascertain whether the 8 DNA methylation sites modulate the expression of 3 genes, we sourced mQTL data from GoDMC and eQTL data from eQTLGen. Our SMR analysis pinpointed 2 DNA methylation sites with notable impacts on their respective gene expression (FDR<0.05, p_HEIDI>0.01). Specifically, a rise in the methylation at site cg16265553 (SMR_β = −0.46, 95%CI: 0.63 to −0.28, FDR<0.001) decreased ITPKB expression. Likewise, an increase in methylation at site cg13661497 (SMR_β = −2.57, 95%CI: −3.27 to −1.91, FDR<0.001) decreased JAK2 expression (Supplementary Table. S15). Further TSMR analysis validated these findings, with both methylation sites meeting the established criteria (FDR<0.1), and the beta values being similar in magnitude (Supplementary Table. S16). Colocalization analysis further supported these findings (PPH>0.5) (Supplementary Table. S17). Collectively, our data implied that cg16265553 downregulated ITPKB expression, while cg13661497 inhibited JAK2 expression.
Mediation analysis
We used the Sobel method in a mediation analysis to study the effect of DNA methylation on asthma through gene expression levels. For exposure, we selected mQTL data from GoDMC, mediation data (eQTL) came from eQTLGen, and for the discovery phase, asthma GWAS dataset was from the UKB, with FinnGen and TAGC used in the replication phase.
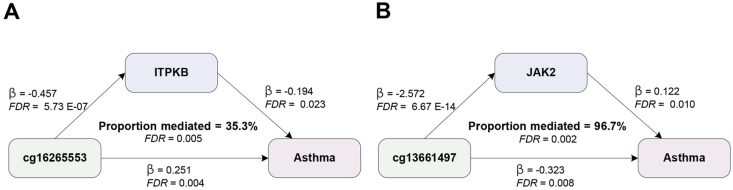
In both discovery and replication phases, our SMR results indicated that an increase in methylation at site cg16265553 raised the risk of asthma by suppressing ITPKB expression (discovery, mediation proportion = 35.3%, 95% CI:10.8%–59.8%, FDR = 0.005; replication, mediation proportion = 41.2%, 95% CI:3.3%–79.1%, FDR = 0.033). Similarly, increased methylation at cg13661497 reduced the asthma risk by suppressing JAK2 expression (discovery, mediation proportion = 96.7%, 95% CI:39.2%–154.3%, FDR = 0.002; replication, mediation proportion = 187.8%, 95% CI: 65.5%–310.2%, FDR = 0.005) (Fig. 4, Supplementary Table. S23 and S25). TSMR results were consistent with these findings, showing significant mediation effects for both sites (FDR<0.05) (details in Supplementary Table. S24 and S26). In summary, our data suggested that cg16265553 could raise asthma risk in part by suppressing ITPKB expression, while cg13661497 could reduce the risk nearly completely by inhibiting JAK2 expression.
Fig. 4.
Mediation analysis. (A), ITPKB mediated the causal effect of cg16265553 on asthma risk using Summary Data-based Mendelian Randomization (SMR) method in discovery phase. (B), JAK2 mediated the causal effect of cg13661497 on asthma risk using SMR method in discovery phase
External validated in lung
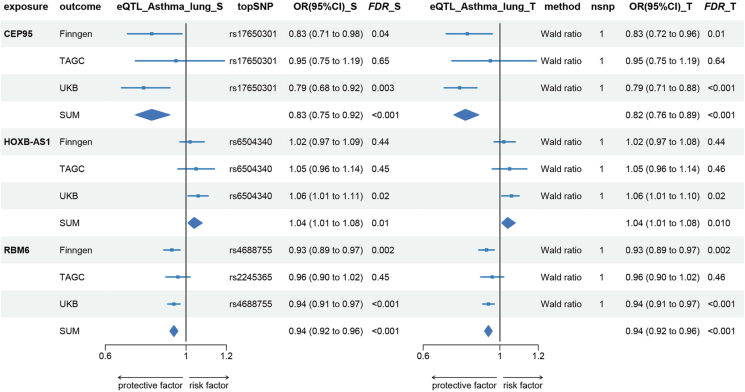
To further substantiate our conclusions, we used eQTL data in lung tissues obtained from GtexV8. Out of the 6 positive genes, only 3 were identifiable. The asthma GWAS datasets were sourced from UKB, FinnGen, and TAGC. We performed both SMR and TSMR analyses and consolidated the results using meta-analysis. Specifically, an increase in CEP95 (OR_SMR = 0.83, 95%CI:0.75–0.92, FDR<0.001) and RBM6 (OR_SMR = 0.94, 95%CI:0.92–0.96, FDR<0.001) in lung tissues decreased risk of asthma. While a rise in HOXB-AS1 (OR_SMR = 1.04, 95%CI:1.01–1.08, FDR = 0.01) in lung tissues increased asthma risk. In summary, our analysis revealed that in lung tissues, CEP95 and RBM6 functioned as protective factors for asthma, whereas HOXB-AS1 emerged as a risk factor (Fig. 5, Supplementary Table. S27 and S28).
Fig. 5.
Causal effect of gene expression in lung tissues on asthma. eQTL, expression quantitative trait loci; OR, odds ratio; CI, confidence interval; _S, by using Summary Data-based Mendelian Randomization (SMR); _T, by using Two-Sample Mendelian Randomization (TSMR).
Drug target analysis
Out of the 6 positive genes, 5 genes, namely JAK2, ITPKB, ETS1, RBM6 and CEP95, had the potential to be drug target by druggability annotation. Specifically, JAK2 had been identified as "licensed drugs". And ITPKB had been identified as "Compounds in preclinical". The rest 3 genes had been identified as "predicted druggable" (Supplementary Table. S31).
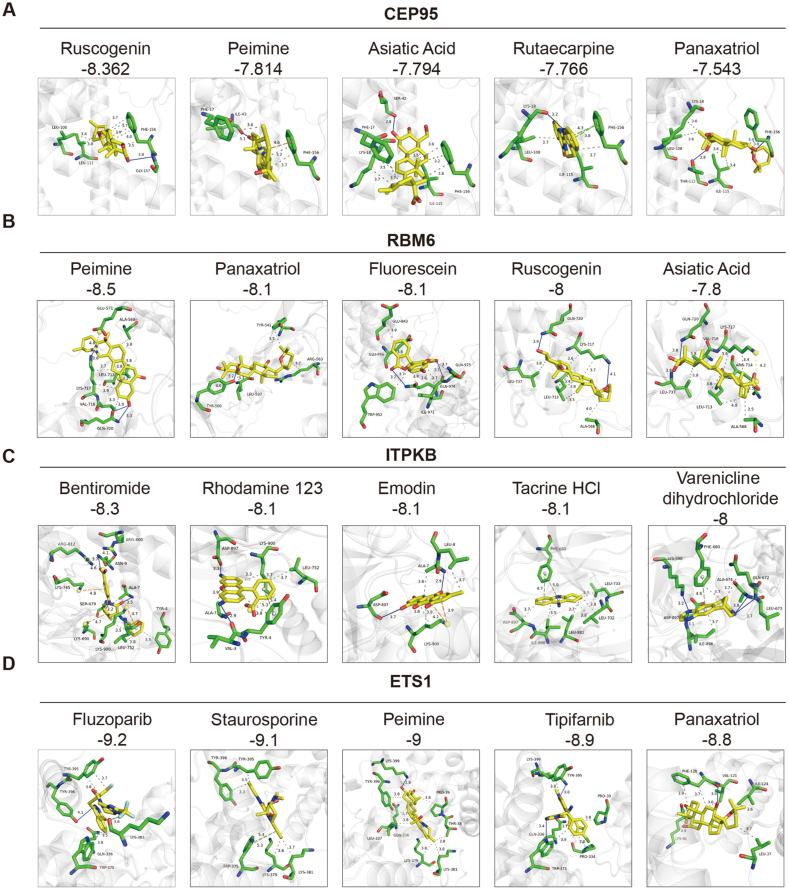
However, except JAK2, the other 4 candidates lacked existing target drug. To assist in drug discovery, we conducted virtual screening combined with molecular docking. We selected 1147 compounds from the FDA-approved drug library to investigate whether these drugs could potentially target these 4 candidates. Compounds with the docking score (binding energy) of less than -5 kcal/mol were considered more likely candidates. We displayed the top 5 compounds for each candidate based on their minimum binding energies (Fig. 6). Notably, Peimine and Panaxatriol both appeared among the top 5 compounds for CEP95, ETS1, and RBM6, suggesting they might influence asthma through these 3 candidates simultaneously. Ruscogenin and Asiatic Acid both ranked among the top 5 for CEP95 and RBM6, implying their potential influence on asthma through these 2 candidates. Besides, further research is required to validate these findings.
Fig. 6.
Virtual screening and molecular docking of potential positive genes. (A-D), Molecular docking of the top 5 compound in Virtual screening ranking by docking score. The number below the compound represents its docking score.
Discussion
To our knowledge, this is the first study integrating transcriptomic data, eQTL, and mQTL multi-omics to identify causal genes and methylation sites for asthma and their mechanisms. We based our data selection on European descent populations for consistency. Specifically, we utilized the largest available whole blood asthma GEO dataset of European origin and incorporated 3 eQTL, 2 mQTL, and 3 asthma GWAS datasets. For the design, we used both discovery and replication phase, with a meta-analysis to bolster our findings' reliability. For analysis, we applied SMR, TSMR, and co-localization. Only genes or sites that were validated by all 3 methods and remained consistent across both the discovery and replication phases were considered as positive target candidates.
In this study, we identified 6 positive causal genes. JAK2 and ETS1 are already known in the asthma context, whereas CEP95, HOXB-AS1, RBM6, and ITPKB are novel targets, which have not been studied in asthma. JAK2 encodes a non-receptor tyrosine kinase, currently widely considered a risk molecule for asthma. Studies have shown that inhibiting JAK2 prevents antigen-induced eosinophil recruitment into the airways.32 Administration of JAK2 inhibitors has been proven to reduce inflammatory factors and oxidative stress levels in vitro, and to mitigate airway inflammation and enhance lung function in vivo33,34. In subsequent druggability annotation, we identified it as "licensed drug". However, current clinical studies on JAK inhibitors for asthma mainly emphasize pan-JAK inhibition.35 There is no direct clinical evidence for exclusive JAK2 research yet. Our MR analysis identified JAK2 as a risk factor for asthma using clinical evidence. Thus, targeting JAK2, either individually or combined with other molecules (like pan-JAK inhibition), may offer a promising therapeutic strategy for asthma.
ETS1, an ETS family transcription factor, has been reported to be a risk factor for asthma in vitro. Overexpressing ETS1 in CD4+ T cells induces TH2 cell polarization and elevates the expression of cytokines IL-5 and IL-13, thus aggravating asthma.36 In vitro, knockdown of ETS1 has been reported to reduce IL-6, VEGF,37 MMP9, and tenascin expression.38 However, compelling evidence is missing in animal and clinical trials. Our MR analysis identified ETS1 as a risk factor for asthma using clinical evidence. Subsequent druggability annotation categorizes ETS1 under "Predicted_druggable". This suggests its potential as a therapeutic target, even though a specific drug is yet to be developed.
Of the 4 newly identified target genes in this study, 3 demonstrate protective effects and 1 serves as a risk factor. The first gene, CEP95, belongs to the CCDC protein family. Research suggests that its expression is associated with immune infiltration levels,39 which are crucial in asthma. Through MR analysis, we determined CEP95 acts as a protective factor against asthma. Subsequently, in druggability annotation, it was categorized as "Predicted_druggable". The second gene, HOXB-AS1, is a long noncoding RNA (lncRNA). Although lncRNAs have a pivotal role in asthma, research into their involvement remains nascent. Through MR analysis, we identified HOXB-AS1 as a risk factor for asthma. Though HOXB-AS1 has not been designated as druggable, interventions can be applied via RNAi, CRISPR/Cas9, or antisense oligonucleotides. RBM6, the third gene, is a lesser-studied gene, involved in modulating gene expression and alternative splicing linked to diverse cellular processes.40 Through MR analysis, we found RBM6 to be a protective factor for asthma. Subsequent druggability annotation categorizes RBM6 under "Predicted_druggable". The fourth gene is ITPKB. Despite the absence of research focusing on ITPKB in asthma, its function appears intricately linked to the disease. On one hand, ITPKB can impede ER-to-mitochondria calcium transport, which reduces intracellular calcium levels41 — a mechanism tied to asthma symptom alleviation. On the other hand, ITPKB is pivotal in shaping the development and functionality of both T cells and B cells,42,43 entities fundamental to asthma's pathogenesis. In our study, MR analysis indicated ITPKB as a potential protective gene against asthma. Subsequent druggability annotation categorizes ITPKB under "Compounds_in_preclinical". Therefore, these 4 genes are causal genes for asthma and offer a new angle for understanding its pathogenesis and treatment.
Subsequently, we conducted virtual screening and molecular docking for ETS1, CEP95, RBM6, and ITPKB, identifying some potential target drugs, and further research is required to validate their function in asthma.
DNA methylation, a central facet of epigenetic regulation, plays a significant role in asthma. Changes in DNA methylation can be passed down to future generations, and it is reversible. Advancing our comprehension in this field offers potential new avenues for therapeutic targets, medication.10 Consequently, we concentrated on the methylation status of genes. We identified total 8 positive causal sites linked to asthma, including 3 sites in ETS1, 2 in ITPKB, and 3 in JAK2. These findings were never reported before. Targeting these methylation sites may provide a new therapeutic strategy for asthma.
It is well known that DNA methylation often typically inhibits gene expression. Therefore, we analyzed the 8 methylation sites for potential gene regulatory effects. We found that cg16265553 inhibited ITPKB expression, and cg13661497 suppressed JAK2. Within this framework, there are causal relationships between methylation and gene expression, gene expression and asthma, as well as methylation and asthma. We further explored if methylation could influence asthma through its impact on gene expression. Our results showed that cg16265553 could aggravate asthma via inhibiting ITPKB expression, with 35.3% mediation proportion, suggesting that other factors at this site might influence asthma or the site itself directly impacts the condition. On the other hand, cg13661497 could protect asthma through JAK2, with 96.7% mediation proportion, suggesting that the functionality of this site on asthma nearby entirely depends on JAK2. Hence, these results sheds light on the intricate mechanistic interplay between the methylation, gene expression and asthma, deepening our understanding of the pathogenesis.
Our study has several limitations. Firstly, we recognize that our study predominantly focuses on European populations, which may limit the generalizability of our findings to non-European populations. Despite our efforts to include data from other populations, we were unable to perform Mendelian randomization analysis due to the lack of suitable instrumental variables. Additionally, there have been no studies conducted on other racial groups to date. Our results still need to be explored in diverse populations in future research. Secondly, many prioritized genes and methylation sites exhibited a limited number of cis-acting SNPs. This limitation constrains the scope of our analyses, including heterogeneity tests and pleiotropy tests. This choice was made based on current research suggesting that utilizing cis-acting data might be preferable, as opposed to using a larger number of SNPs.14 Therefore, to enhance the reliability of our results, we opted to select only robust instrumental variables with an F-value greater than 10 as described before.14
Conclusion
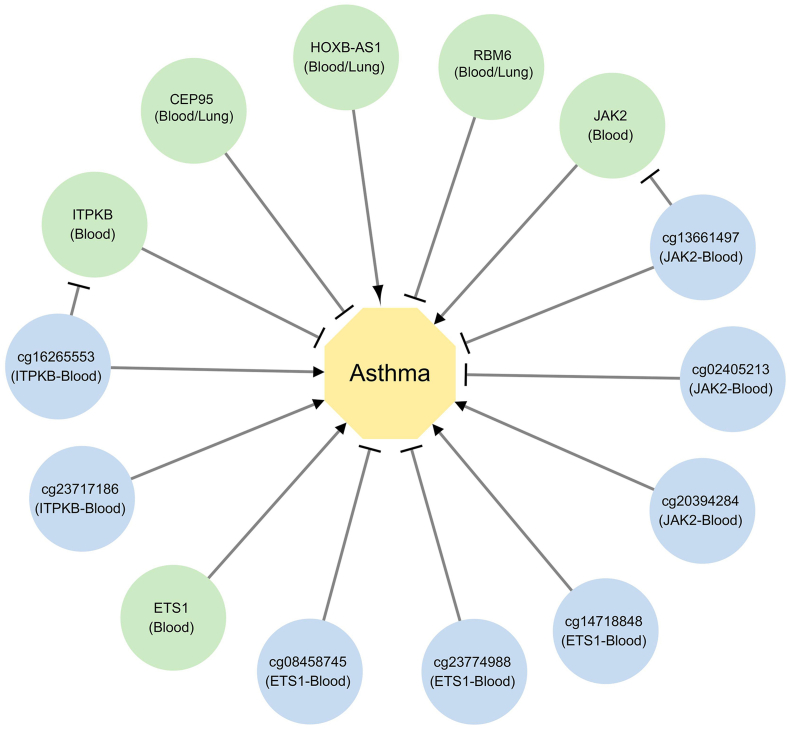
In summary, our integrative analysis reveals that the expression of 2 known asthma-related genes JAK2 and ETS1, 4 new genes CEP95, HOXB-AS1, RBM6, ITPKB and 8 specific DNA methylation sites is causally associated with asthma risk. We have identified 2 potential methylation-gene-asthma pathways (Fig. 7 and Table 1). Moreover, CEP95, RBM6, ITPKB, ETS1, and JAK2 may be prioritized as potential drug targets for asthma. Further research is needed to delve deeper into the roles these candidates play in asthma.
Fig. 7.
Causal genes, methylation sites, and corresponding pathways in Asthma.
This figure illustrates 6 causal genes, 8 causal methylation sites, and 2 methylation-gene-asthma pathways. Green represents genes, blue represents methylation sites, and yellow represents asthma. Sharp arrows indicate promotion, while blunt arrows indicate inhibition.
Table 1.
Mendelian randomization analysis results for causal genes and methylation sites of asthma, along with corresponding pathways.
| Methylation sites (blood) → Asthma |
Genes (blood) → Asthma |
Methylation sites (blood) → Genes (blood) |
Methylation sites→Genes→Asthma |
Genes (lung)→Asthma |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Methylation sites | OR (95% CI) | FDR | Genes | OR (95% CI) | FDR | β (95%CI) | FDR | Proportion mediated | OR (95% CI) | FDR |
| – | – | – | CEP95 | 0.94 (0.91–0.97) |
0.001 | – | – | – | 0.83 (0.75–0.92) |
<0.001 |
| – | – | – | HOXB-AS1 | 1.05 (1.03–1.07) |
<0.001 | – | – | – | 1.04 (1.01–1.08) |
0.01 |
| – | – | – | RBM6 | 0.97 (0.95–0.99) |
0.01 | – | – | – | 0.94 (0.92–0.96) |
<0.001 |
| cg16265553 ITPKB |
1.29 (1.12–1.47) |
0.004 | ITPKB | 0.82 (0.74–0.92) |
0.02 | −0.457 (−0.63 to −0.28) |
<0.001 | 35.3% | – | – |
| cg23717186 ITPKB |
1.15 (1.07–1.22) |
0.002 | – | – | – | – | – | – | – | – |
| – | – | – | ETS1 | 1.62 (1.29–2.04) |
0.001 | – | – | – | – | – |
| cg08458745 ETS1 |
0.92 (0.87–0.96) |
0.004 | – | – | – | – | – | – | – | – |
| cg14718848 ETS1 |
1.02 (1.01–1.04) |
0.004 | – | – | – | – | – | – | – | – |
| cg23774988 ETS1 |
0.85 (0.77–0.93) |
0.004 | – | – | – | – | – | – | – | – |
| cg13661497 JAK2 |
0.72 (0.59–0.88) |
0.008 | JAK2 | 1.13 (1.06–1.21) |
0.01 | −2.572 (−3.24 to −1.91) |
<0.001 | 96.7% | – | – |
| cg02405213 JAK2 |
0.97 (0.96–0.99) |
0.002 | – | – | – | – | – | – | – | – |
| cg20394284 JAK2 |
1.04 (1.02–1.07) |
0.002 | – | – | – | – | – | – | – | – |
The table was based Summary Data-based Mendelian Randomization (SMR) method in discovery phase. OR, odds ratio; CI, confidence interval
Abbreviations
DEGs, Differentially expressed genes; eQTL, Expression quantitative trait loci; FC, Fold Change; HEIDI, heterogeneity in dependent instruments; IVs, Instrumental variables; LD, Linkage disequilibrium; lncRNA, long noncoding RNA; mQTL, DNA methylation quantitative trait loci; PPH4, posterior probability for H4; RCTs, Randomized controlled trials; SMR, Summary Data-based Mendelian Randomization; TAGC, Trans-National Asthma Genetic Consortium; TSMR, Two-Sample Mendelian Randomization; UKB, UK Biobank.
Credit authorship contribution statement
Jia Wang: Formal analysis, Investigation, Methodology, Software, Writing – review & editing. Jinxin Hu: Data curation, Writing – original draft, Writing – review & editing. Dan Qin: Formal analysis, Methodology, Software, Writing – review & editing. Dan Han: Writing – review & editing. Jiapeng Hu: Conceptualization, Data curation, Funding acquisition, Writing – review & editing.
Availability of data and materials
The datasets supporting the conclusions of this article are available in GEO (https://www.ncbi.nlm.nih.gov/geo/), eQTLGen Consortium (https://www.eqtlgen.org/cis-eqtls.html), GTExV8 (https://yanglab.westlake.edu.cn/software/smr/#DataResource), GoDMC (http://mqtldb.godmc.org.uk/index), FSH (https://ftp.ncbi.nlm.nih.gov/eqtl/), UK Biobank and TAGC (https://www.ebi.ac.uk/gwas/), FinnGen consortium R7 release data (https://www.finngen.fi/en). The detailed information of the repository/repositories and accession number(s) can be found in the Supplementary Table. S2.
Ethics statement
This research has been conducted using publicly available GWAS summary statistics. Ethical approval and participant consent were obtained in the original studies.
Authors’ consent for publication
All authors agreed to the publication of this work in the World Allergy Organization Journal.
Submission declaration
Authors confirm that this manuscript is original, has not been published before, is not currently being considered for publication elsewhere.
Funding
This work was supported by the National Natural Science Foundation of China [grant:82200036].
Declaration of competing interest
The authors have no conflict of interest relevant to this article to disclose.
Acknowledgments
The authors gratefully acknowledge the participants and investigators of eQTLGen, Gtex, GoDMC, FSH, UK Biobank, GWAS catalog, FinnGen, TAGC for providing the GWAS data, and thank the GEO, AlphaFold, and Open Targets Database. We also appreciate Selleckchem for providing the FDA-approved drug library.
Footnotes
Full list of author information is available at the end of the article.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.waojou.2024.101008.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Supplementary Fig. S1.
Differentially expressed genes (DEGs) analysis. Identification of 954 DEGs by analyzing the largest whole-blood GEO dataset (GSE69683) with 371 European patients and 80 controls
References
- 1.Global burden of chronic respiratory diseases and risk factors 1990-2019: an update from the global burden of disease study 2019. EClinicalMedicine. 2023;59 doi: 10.1016/j.eclinm.2023.101936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ntontsi P., Photiades A., Zervas E., et al. Genetics and epigenetics in asthma. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22052412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lane N.E. Glucocorticoid-induced osteoporosis: new insights into the pathophysiology and treatments. Curr Osteoporos Rep. 2019;17:1–7. doi: 10.1007/s11914-019-00498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casamassimi A., Federico A., Rienzo M., et al. Transcriptome profiling in human diseases: new advances and perspectives. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18081652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyler S.R., Bunyavanich S. Leveraging -omics for asthma endotyping. J Allergy Clin Immunol. 2019;144:13–23. doi: 10.1016/j.jaci.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birney E. Mendelian randomization. Cold Spring Harb Perspect Med. 2022;12 doi: 10.1101/cshperspect.a041302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekula P., Del Greco M.F., Pattaro C., et al. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016;27:3253–3265. doi: 10.1681/ASN.2016010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu C.J., Söderhäll C., Bustamante M., et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. 2018;6:379–388. doi: 10.1016/S2213-2600(18)30052-3. [DOI] [PubMed] [Google Scholar]
- 9.Sheikhpour M., Maleki M., Ebrahimi Vargoorani M., et al. A review of epigenetic changes in asthma: methylation and acetylation. Clin Epigenet. 2021;13:65. doi: 10.1186/s13148-021-01049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman A.R., Hu J.F. Directing DNA methylation to inhibit gene expression. Cell Mol Neurobiol. 2006;26:425–438. doi: 10.1007/s10571-006-9057-5. [DOI] [PubMed] [Google Scholar]
- 11.Gomez J.L., Diaz M.P., Nino G., et al. Impaired type I interferon regulation in the blood transcriptome of recurrent asthma exacerbations. BMC Med Genom. 2018;11:21. doi: 10.1186/s12920-018-0340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu Y., Huang D., Zhao Z., et al. Bioinformatic analysis identifies potential key genes of epilepsy. PLoS One. 2021;16 doi: 10.1371/journal.pone.0254326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia R., Li Z., Liang W., et al. Identification of key genes unique to the luminal a and basal-like breast cancer subtypes via bioinformatic analysis. World J Surg Oncol. 2020;18:268. doi: 10.1186/s12957-020-02042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J., Zhou J., Xu Y. Potential drug targets for multiple sclerosis identified through Mendelian randomization analysis. Brain. 2023;146:3364–3372. doi: 10.1093/brain/awad070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu S., Li X., Zhang S., et al. Oxidative stress gene expression, DNA methylation, and gut microbiota interaction trigger Crohn's disease: a multi-omics Mendelian randomization study. BMC Med. 2023;21:179. doi: 10.1186/s12916-023-02878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Võsa U., Claringbould A., Westra H.J., et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53:1300–1310. doi: 10.1038/s41588-021-00913-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min J.L., Hemani G., Hannon E., et al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet. 2021;53:1311–1321. doi: 10.1038/s41588-021-00923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P., Wang H., Guo L., et al. Association between gut microbiota and preeclampsia-eclampsia: a two-sample Mendelian randomization study. BMC Med. 2022;20:443. doi: 10.1186/s12916-022-02657-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie J., Huang H., Liu Z., et al. The associations between modifiable risk factors and nonalcoholic fatty liver disease: a comprehensive Mendelian randomization study. Hepatology. 2023;77:949–964. doi: 10.1002/hep.32728. [DOI] [PubMed] [Google Scholar]
- 20.de Klerk J.A., Beulens J.W.J., Mei H., et al. Altered blood gene expression in the obesity-related type 2 diabetes cluster may be causally involved in lipid metabolism: a Mendelian randomisation study. Diabetologia. 2023;66:1057–1070. doi: 10.1007/s00125-023-05886-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J., Cheng Y., Li M., et al. Genome-wide Mendelian randomization identifies actionable novel drug targets for psychiatric disorders. Neuropsychopharmacology. 2023;48:270–280. doi: 10.1038/s41386-022-01456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X., Xiao Q., Jiang F., et al. Dissecting the pathogenic effects of smoking and its hallmarks in blood DNA methylation on colorectal cancer risk. Br J Cancer. 2023;129:1306–1313. doi: 10.1038/s41416-023-02397-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battle A., Brown C.D., Engelhardt B.E., et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cronjé H.T., Karhunen V., Hovingh G.K., et al. Genetic evidence implicating natriuretic peptide receptor-3 in cardiovascular disease risk: a Mendelian randomization study. BMC Med. 2023;21:158. doi: 10.1186/s12916-023-02867-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W., Xiao J., Ji J., et al. Association of lipid-lowering drugs with COVID-19 outcomes from a Mendelian randomization study. Elife. 2021;10 doi: 10.7554/eLife.73873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y., Sundquist K., Zhang N., et al. Mitochondrial related genome-wide Mendelian randomization identifies putatively causal genes for multiple cancer types. EBioMedicine. 2023;88 doi: 10.1016/j.ebiom.2022.104432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasooly D., Peloso G.M., Pereira A.C., et al. Genome-wide association analysis and Mendelian randomization proteomics identify drug targets for heart failure. Nat Commun. 2023;14:3826. doi: 10.1038/s41467-023-39253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu R., Chen L., Lan R., et al. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter A.R., Sanderson E., Hammerton G., et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol. 2021;36:465–478. doi: 10.1007/s10654-021-00757-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai H., Hou T., Wang Q., et al. Causal relationships between the gut microbiome, blood lipids, and heart failure: a Mendelian randomization analysis. Eur J Prev Cardiol. 2023;30:1274–1282. doi: 10.1093/eurjpc/zwad171. [DOI] [PubMed] [Google Scholar]
- 31.Chong R.S., Li H., Cheong A.J.Y., et al. Mendelian randomization implicates bidirectional association between myopia and primary open-angle glaucoma or intraocular pressure. Ophthalmology. 2023;130:394–403. doi: 10.1016/j.ophtha.2022.11.030. [DOI] [PubMed] [Google Scholar]
- 32.Kumano K., Nakao A., Nakajima H., et al. Blockade of JAK2 by tyrphostin AG-490 inhibits antigen-induced eosinophil recruitment into the mouse airways. Biochem Biophys Res Commun. 2000;270:209–214. doi: 10.1006/bbrc.2000.2403. [DOI] [PubMed] [Google Scholar]
- 33.Ren Y., Yan Y., Zhen L., et al. Zhike Pingchuan Granule suppresses interleukin (IL)-6 or the medium of M2 macrophages induced apoptosis in human bronchial epithelial cells. Bioengineered. 2021;12:7694–7703. doi: 10.1080/21655979.2021.1982309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun W., Song S., Li G., et al. FcRn-targeting and ROS-responsive Fedratinib-incorporated nanoparticles alleviate asthma by inducing eosinophil apoptosis. Allergy. 2023;78:1659–1663. doi: 10.1111/all.15575. [DOI] [PubMed] [Google Scholar]
- 35.Georas S.N., Donohue P., Connolly M., et al. JAK inhibitors for asthma. J Allergy Clin Immunol. 2021;148:953–963. doi: 10.1016/j.jaci.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang T., Zhou Q., Shang Y. Downregulation of miRNA-451a promotes the differentiation of CD4+ T cells towards Th2 cells by upregulating ETS1 in childhood asthma. J Innate Immun. 2021;13:38–48. doi: 10.1159/000509714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian F., He S., Yang X., et al. Circular RNA DHTKD1 targets miR-338-3p/ETS1 axis to regulate the inflammatory response in human bronchial epithelial cells. Exp Ther Med. 2023;26:316. doi: 10.3892/etm.2023.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura Y., Esnault S., Maeda T., et al. Ets-1 regulates TNF-alpha-induced matrix metalloproteinase-9 and tenascin expression in primary bronchial fibroblasts. J Immunol. 2004;172:1945–1952. doi: 10.4049/jimmunol.172.3.1945. [DOI] [PubMed] [Google Scholar]
- 39.Liu S., Jia L., Quan B., et al. Coiled-coil domain-containing protein 45 is a potential prognostic biomarker and is associated with immune cell enrichment of hepatocellular carcinoma. Dis Markers. 2022;2022 doi: 10.1155/2022/7745315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machour F.E., Abu-Zhayia E.R., Awwad S.W., et al. RBM6 splicing factor promotes homologous recombination repair of double-strand breaks and modulates sensitivity to chemotherapeutic drugs. Nucleic Acids Res. 2021;49:11708–11727. doi: 10.1093/nar/gkab976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apicco D.J., Shlevkov E., Nezich C.L., et al. The Parkinson's disease-associated gene ITPKB protects against α-synuclein aggregation by regulating ER-to-mitochondria calcium release. Proc Natl Acad Sci USA. 2021;118 doi: 10.1073/pnas.2006476118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pouillon V., Hascakova-Bartova R., Pajak B., et al. Inositol 1,3,4,5-tetrakisphosphate is essential for T lymphocyte development. Nat Immunol. 2003;4:1136–1143. doi: 10.1038/ni980. [DOI] [PubMed] [Google Scholar]
- 43.Schurmans S., Pouillon V., Maréchal Y. Regulation of B cell survival, development and function by inositol 1,4,5-trisphosphate 3-kinase B (Itpkb) Adv Enzym Regul. 2011;51:66–73. doi: 10.1016/j.advenzreg.2010.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are available in GEO (https://www.ncbi.nlm.nih.gov/geo/), eQTLGen Consortium (https://www.eqtlgen.org/cis-eqtls.html), GTExV8 (https://yanglab.westlake.edu.cn/software/smr/#DataResource), GoDMC (http://mqtldb.godmc.org.uk/index), FSH (https://ftp.ncbi.nlm.nih.gov/eqtl/), UK Biobank and TAGC (https://www.ebi.ac.uk/gwas/), FinnGen consortium R7 release data (https://www.finngen.fi/en). The detailed information of the repository/repositories and accession number(s) can be found in the Supplementary Table. S2.