Summary
Background
The APOE-ε4 genotype is the highest genetic risk factor for Alzheimer's disease (AD), and exercise training can reduce the risk of AD. Two early pathologies of AD are degradation of tight junctions between brain microvascular endothelial cells (BMEC) and brain glucose hypometabolism. Therefore, the objective of this work was to determine how the APOE-ε4 genotype and serum from exercise trained individuals impacts BMEC barrier function and metabolism.
Methods
iPSC homozygous for the APOE-ε3 and APOE-ε4 alleles were differentiated to BMEC-like cells and used to measure barrier function and metabolism. To investigate exercise effects, serum was collected from older adults pre- and post- 6 months of exercise training (n = 9 participants per genotype). APOE-ε3 and APOE-ε4 BMEC were treated with genotype-matched serum, and then barrier function and metabolism were measured.
Findings
APOE-ε4 genotype impaired BMEC barrier function and metabolism by reducing sirtuin 1 (SIRT1) levels by 27% (p = 0.0188) and baseline insulin signalling by 37% (p = 0.0186) compared to APOE-ε3 BMEC. Exercise-trained serum increased SIRT1 by 33% (p = 0.0043) in APOE-ε3 BMEC but decreased SIRT1 by 22% (p = 0.0004) in APOE ε4 BMEC.
Interpretation
APOE-ε4 directly impairs glucose metabolism and barrier function. Serum from exercise trained individuals alters SIRT1 in a genotype-dependent manner but may require additional cues from exercise to decrease AD pathologies.
Funding
Brain and Behaviour Initiative at the University of Maryland through the Seed Grant Program, NSF-GRFP DGE 1840340, Fischell Fellowship in Biomedical Engineering, NSF CBET-2211966 and DGE-1632976, National Niemann-Pick Disease Foundation, University of Maryland ASPIRE Program, NIH R01HL165193, R01HL140239-01, and R01AG057552.
Keywords: Blood-brain barrier, Alzheimer's disease, Glucose metabolism, APOE genotype, Exercise training
Research in context.
Evidence before this study
We reviewed the literature using traditional (e.g., PubMed) sources. The APOE-ε4 genotype decreases SIRT1 and insulin signalling in neurons and decreases brain microvascular endothelial cell (BMEC) in mice. Exercise training increases brain glucose metabolism in mice and humans. The impacts of APOE-ε4 and exercise on BMEC are not well established.
Added value of this study
We found the APOE-ε4 genotype reduces BMEC barrier function by reducing SIRT1 and decreases BMEC glucose metabolism by impairing insulin signalling. Finally, we found serum from exercise-trained individuals has genotype-dependent dimorphic effects on SIRT1 but does not alter BMEC barrier function or metabolism.
Implications of all the available evidence
This manuscript demonstrates two mechanisms through which the APOE-ε4 genotype directly impairs BMEC metabolism and function which can be leveraged to develop therapeutics against AD and calls for further studies of the mechanisms behind exercise benefits in prevention of AD.
Introduction
The APOE-ε4 genotype is the strongest genetic risk factor for AD and also increases the risk of vascular disease.1 Individuals homozygous for the APOE-ε4 allele have a ten-fold higher risk for AD and develop clinically detectable AD manifestations 7–10 years before individuals homozygous for the APOE-ε3 allele.2,3 Even so, not all APOE-ε4 allele carriers develop AD, suggesting vulnerability to AD may be modifiable through lifestyle behaviours. Consistent exercise training is estimated to reduce AD risk by 45%,4 though it is unknown if individuals with the APOE-ε3 and -ε4 genotypes receive equal benefit from exercise training.
Brain microvascular endothelial cells (BMEC) line the cerebral blood vessels and have specialised tight junctions designed to strictly regulate nutrient and waste transfer between the blood and the brain. Two early indicators of AD development are breakdown of the tight junctions and whole brain glucose hypometabolism. In humans, the APOE-ε4 genotype is correlated with many AD biomarkers, including increased blood–brain barrier (BBB) permeability,5 decreased brain glucose metabolism,6 and increased amyloid-β (Aβ) and tau accumulation.7 In mice, the APOE-ε4 genotype reduced BMEC barrier function8, 9, 10 and glycolysis9 compared to the APOE-ε3 genotype. In vitro, APOE-ε4 astrocytes had decreased glycolysis but increased pentose phosphate pathway (PPP) activity as compared to APOE-ε3 astrocytes, leading to increased lipid and nucleotide biosynthesis.11 Notably, none of these studies examined mechanisms through which the APOE genotype itself regulates BMEC barrier function or cellular metabolism.
Exercise training in healthy older adults increases cerebral blood flow,12 reduces hippocampal atrophy,13 and increases brain glucose metabolism,14 each of which reduces AD risk. Exercise training also improves Aβ clearance in rodents15 and may reduce humans plasma Aβ levels as well.16 From a mechanistic standpoint, exercise elevates circulating irisin, lactate, and cathepsin B, which are all hypothesised to increase brain derived neurotrophic factor (BDNF),17, 18, 19, 20 a molecule that supports neuronal growth, synaptic plasticity, learning, and memory.21,22 Past studies primarily focused on how exercise training improves neuronal signalling to improve cognition. However, little is known of how exercise alters BMEC function, which is of particular importance because BMEC are directly in contact with the circulation and therefore act as first responders to the benefits of exercise training.
The goal of this study was to investigate APOE-ε4 induced mechanisms of BMEC barrier and metabolic dysfunction, and the role of exercise training in counteracting these deficits. We used iPSC-derived BMEC-like cells to investigate the role of APOE genotype alone in BMEC barrier function and metabolism and then added human serum from healthy older adults (ages 65–80) collected pre- and post-6 months exercise training before re-evaluating barrier function and metabolism. Here, we outline two mechanisms through which APOE-ε4 regulates barrier function and glucose metabolism: reduced sirtuin 1 (SIRT1) and reduced insulin signalling. We then investigate how human serum collected pre- and post- 6 months of exercise training alters these pathways in BMEC. We used CRISPR/Cas9 to generate induced pluripotent stem cells (iPSC) homozygous for the APOE-ε3 and -ε4 alleles. We then differentiated the iPSC into BMEC (hiBMEC), which have better barrier function and similar glucose metabolism to primary BMEC.23 Using the APOE-ε3 and -ε4 hiBMEC, we studied how the APOE-ε4 genotype leads to barrier and glycolytic deficits. Finally, using serum collected from older adults pre- and post-6 months of exercise training, we assessed how genotype-matched exercise-trained serum impacts APOE-ε3 and -ε4 hiBMEC barrier function and metabolism.
Methods
Ethics
This work was conducted in line with the ARRIVE reporting guidelines.24 All experimental procedures were approved by the Institutional Review Board of University of Maryland, College Park and human serum was collected in this study in accordance with the approved clinical trial NCT03727360. All donors provided written informed consent prior to participation.
iPSC culture
IMR90 iPSC (RRID: CVCL_C437)25 were cultured on Matrigel (Corning, 354230) and maintained in mTeSR–Plus medium (STEMCELL Technologies, 100-0276). iPSC were passaged after reaching 70% confluence using Versene (Thermo Fisher, 15040066).
CRISPR/Cas9
IMR90 iPSC are homozygous for the APOE-ε3 allele.26 Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology was used to induce a point mutation and alter the iPSC to be homozygous for the APOE-ε4 allele based on previously established protocols.27 Small guide RNA (sgRNA; Table 1) targeting rs429358 (IDT) was resuspended to 100 μM in Resuspension Buffer R (Thermo Fisher, MPK10096). Single-stranded oligodeoxynucleotides (ssODN) were also resuspended to 200 μM in Resuspension Buffer R. iPSC at 80% confluence were dissociated using TrypLE (Thermo Fisher, 12605010), centrifuged at 300×g for 5 min, then resuspended at 13.5 × 106 cells/mL in 100 μL of Resuspension Buffer R. 3 μL sgRNA, 2 μL ssODN, 3 μL Alt-R Cas9 Nuclease V3 (IDT), and 7 μL cell suspension were combined and electroporated using a Neon Transfection Instrument (Thermo Fisher) at 1100 V, with 2 pulses of 30 ms each. Following electroporation cells were replated on Matrigel coated plates in mTeSR–Plus medium supplemented with 10 μM Y-27632 (Tocris, 1254) and 1:1000 Alt-R HDR Enhancer (IDT, 1081072).
Table 1.
CRISPR/Cas9 and genotyping sequences.
| Item | Sequence |
|---|---|
| sgRNA | GCGGACATGGAGGACGTGTGCGG |
| ssODN | AGGAGCTGCAGGCGGCGCAGGCCCGGCTGGGCGCGGACATGGAGGATGTACGTGGCCGCCTGGTGCAGTACCGCGGCGAGGTGCAGGCCATGCTCGGCCAG |
| APOE-ε3 Forward | GTAAAACGACGGCCAGTGTCTCTGTCTCCTTCTCTC |
| APOE-ε3 Reverse | AGGTCATCGGCATCGCGGA |
| APOE-ε4 Forward | GTAAAACGACGGCCAGACGAGACCATGAAGGAGTTG |
| APOE-ε4 Reverse | ACACTGCCAGGCGCTTCTG |
iPSC expansion and genotyping
Following CRISPR editing, iPSC were cultured to 70% confluence, passaged using Accutase (Thermo Fisher, A1110501), replated at 15,000 cells/well, and allowed to grow from single cells into colonies in which each cell was a replicate of a single cell. Individual colonies were expanded, and DNA was sequenced to find successfully edited colonies. DNA was isolated using a Quick-DNA Miniprep Plus Kit (Zymo Research; D4068) following manufacturer's protocol. PCR reactions were run using AmpliTaq Gold 360 Master Mix (Thermo Fisher; 4398881) and associated protocols. Samples were thermally cycled using a Pro Flex Thermal Cycler (Thermo Fisher) with an initial 10-min activation at 95 °C followed by 40 cycles of 30 s denaturing at 95 °C, 30 s of annealing at 70 °C, and 60 s/kb at 72 °C. A final extension was performed at 72 °C for 7 min before cooling back to 4 °C. Samples were then sent to Genewiz (Azenta Life Sciences) for sequencing. Sequencing results were processed through the Synthego ICE CRISPR Analysis tool and successful homozygous APOE-ε3 (isogenic controls) and -ε4 (edited samples) were expanded and frozen in liquid nitrogen for future use.
hiBMEC differentiation
hiBMEC were differentiated following established protocols.28,29 iPSC at 70% confluence were passaged using Accutase onto Matrigel coated plates at 150,000 cells/well in mTeSR–Plus medium containing 10 μM Y-27632. Over the next four days media was replaced daily with E6 Medium (STEMCELL Technologies, 05946). On the fifth day, media was replaced with Neurobasal medium (Thermo Fisher, 2110304) supplemented with 10 mM glucose (Millipore Sigma, G5767), 4.5 mM glutamine (Fisher Scientific, 25-030-081), 2% B27 (Thermo Fisher, 17504001), 20 ng/mL basic fibroblast growth factor (bFGF; Peprotech, 100-18B), and 10 μM retinoic acid (RA; Millipore Sigma, R2625-50MG). Media was not changed on the sixth day. On the seventh day, cells were dissociated using Accutase and re-plated at 1 × 106 cells/cm2 on 0.4 μm Transwell Filters (Corning, 3460), 12-well plates (CELLTREAT, 229112), or 96-well plates (Cellvis, P96-1.5P) coated with extracellular matrix (ECM) containing 0.4 mg/mL collagen IV (Millipore Sigma, C7521) and 0.1 mg/mL fibronectin (Millipore Sigma, F2006). For all experiments, cells were cultured for 2 days following subculture in Neurobasal medium with 2% B27, then treated with any applicable treatments for 24 h before collecting endpoint experimental outputs. In experiments comparing APOE-ε3 and -ε4 genotypes, biological replicates were collected from separate cell culture wells.
Exercise training
Adults between the ages of 60 and 80 years old were recruited in compliance with clinical trial NCT03727360. Participants included in the trial were screened for signs of mild cognitive impairment, dementia, and neurological illness, and were deemed cognitively healthy. Inclusion in the trial required participants to be physically inactive, meaning they did not partake in physical activity more than 2 days a week over the past 6 months. Participants were excluded from the trial if they had history or evidence of cardiovascular disease, cerebrovascular disease, or pulmonary disease. During the 6-month clinical trial, each participant was randomly assigned to either a low or a moderate intensity exercise training intervention. At the start of the trial, the peak rate of oxygen consumption (O2peak) was measured in each participant to quantify their cardiorespiratory fitness. To minimise potential confounders, all low and moderate intensity participants began each workout with the same 20-min warmup. Following the warmup, the moderate intensity group completed a moderate intensity aerobic exercise regimen while the participants in the low intensity group completed a set of mild resistance exercises. For both the low and moderate intensity groups, exercise duration and frequency increased over the course of the study, starting with 10 min of exercise one day a week and finishing with 60 min of exercise four days per week. O2peak testing was then repeated at the end of the 6-month trial (Table 2).
Table 2.
V̇O2peak of individuals pre- and post-exercise training.
| Genotype | Training condition | Time point | V̇O2peak | ΔV̇O2peak |
|---|---|---|---|---|
| APOE ε3 | Low Intensity | pre | 17.23 (±1.90) | 0.63 (±1.60) |
| post | 17.33 (±0.46) | |||
| Moderate Intensity | pre | 15.96 (±4.38) | 0.10 (±1.81) | |
| post | 17.86 (±4.96) | |||
| APOE ε4 | Low Intensity | pre | 18.35 (±6.21) | 1.67 (±4.05) |
| post | 20.02 (±4.14) | |||
| Moderate Intensity | pre | 12.80 (±3.88) | 2.00 (±3.85) | |
| post | 17.73 (±3.85) |
Human serum collection and use
Human serum was collected during clinical trial NCT03727360 and banked for this ancillary study. At the start and finish of the trial, 6 mL of whole blood was drawn from each participant in Vacutainer Serum Separator Tubes (SST; BD, 367989), and deidentified for downstream analyses. To isolate serum, whole blood in SST tubes was left undisturbed at room temperature for 20 min, then centrifuged at 2000×g for 10 min at 4 °C. Serum was then aliquoted and stored at −80 °C until use. Sample size was determined by the number of APOE-ε4 serum samples collected during the duration of this trial. Of the 40 serum donors included in the trial, 9 had at least one copy of the APOE-ε4 allele, matching with estimated population allele frequencies.30 Researchers responsible for the collection and preparation of serum samples were blinded to the APOE genotype of the donors, and all serum samples were deidentified before being transferred to the researchers performing cell culture experiments. Researchers performing experiments were not blinded to APOE genotype to ensure even numbers of each group. To ensure equal sample sizes in each group, 9 APOE-ε3 homozygous donors were selected at random for downstream experiments. For all experiments, APOE-ε3 and -ε4 hiBMEC were treated with 20% genotype-matched human serum for 24 h before downstream analysis. Serum donor demographics are reported (Table 3). In experiments with serum treatment, biological replicates were cells treated with serum from donors of the same genotype.
Table 3.
Serum donor demographics.
| APOE-ε3 (n = 9) | APOE-ε4 (n = 9) | |
|---|---|---|
| Donor sex | ||
| Male | 0 (0%) | 3 (33%) |
| Female | 9 (100%) | 6 (66%) |
| Donor genotype | ||
| Homozygous | 9 (100%) | 1 (11%) |
| Heterozygous | 0 (0%) | ε2/ε4: 2 (22%) ε3/ε4: 6 (66%) |
| Donor race | ||
| Undisclosed | 0 (0%) | 1 (11%) |
| White | 6 (67%) | 5 (56%) |
| Black/African American | 2 (22%) | 3 (33%) |
| Asian | 1 (11%) | 0 (0%) |
| Age at consent | ||
| Average (±SD) | 65.3 ± 5.0 | 65.1 ± 3.9 |
ELISA
Enzyme-linked immunosorbent assays (ELISAs) were used to quantify SIRT1 (Thermo Fisher, EH427RB) and APOE (Thermo Fisher, EHAPOE). For both assays aliquots of serum were thawed on ice, and assays were run following manufacturer's protocols. Absorbances were read at 450 nm using a Tecan Spark Multimode Microplate Reader, and concentrations were calculated based on standards included in the assay kits.
TEER
To measure transendothelial electrical resistance (TEER), hiBMEC were subcultured onto 0.4 μm ECM-coated Transwell filters in Neurobasal medium with 2% B27, 20 ng/mL bFGF, and 10 μM RA. Every day post-subculture, TEER was measured in triplicate using STX2-Plus electrodes and the Epithelial Volt/Ohm Meter 3 (EVOM3; World Precision Instruments).
Immunostaining and junction analyser program
hiBMEC in 96-well plates were fixed with either 4% paraformaldehyde (PFA; Millipore Sigma, P6148) (GLUT1, ZO-1, occludin) or ice-cold 100% methanol (Millipore Sigma, 646377) (claudin-5) for 20 min. Cells were then blocked and permeabilised for 1 h at room temperature in 5% normal goat serum (Millipore Sigma, S26) in PBS supplemented with 0.2% Triton X-100 (Alfa Aesar, A16046), then incubated overnight at 4 °C in primary antibodies (Table 4). The next day, cells were incubated in either Alexa Fluor 594 goat anti-rabbit or Alexa Fluor 488 goat anti-mouse secondary antibodies (1:1000) and Hoechst (1:2000) for 1 h at room temperature. Z-stacks of the junctions were captured on an Eclipse Ti2 spinning disk confocal microscope (Nikon) at 60× magnification. 3 images were taken per well, and 9 wells were imaged per treatment group. Maximum intensity projections were created in Fiji of each image. The Junction Analyser Program (JAnaP)31 was then used to characterise the percent of continuous junctions surrounding each cell. Continuous junctions were classified as areas along the traced edge of the cell with >15 pixels of continuous labelling. 5 cells were quantified per image.
Table 4.
Antibodies.
| Antibody | Manufacturer | Catalogue number | Dilution | RRID |
|---|---|---|---|---|
| GLUT1 | Thermo Fisher Scientific | 21829-1-AP | 1:100 (Immunostaining) 1:4000 (Western Blot) |
AB_10837075 |
| HK1 | Santa Cruz Biotechnology | sc-46695 | 1:1000 (Western Blot) | AB_627721 |
| HK2 | Santa Cruz Biotechnology | sc-374,091 | 1:1000 (Western Blot) | AB_10917915 |
| PFKFB3 | Cell Signaling Technology | 13123S | 1:1000 (Western Blot) | AB_2617178 |
| GAPDH | Thermo Fisher Scientific | 437,000 | 1:1000 (Western Blot) | AB_10374327 |
| LDH | Santa Cruz Biotechnology | sc-133,123 | 1:1000 (Western Blot) | AB_2134964 |
| β-Actin | Santa Cruz Biotechnology | sc-47778 | 1:1000 (Western Blot) | AB_626632 |
| INSR | Santa Cruz Biotechnology | sc-57342 | 1:1000 (Western Blot) | AB_784102 |
| SIRT1 | Cell Signaling Technology | 9475S | 1:1000 (Western Blot) | AB_2617130 |
| APOE | Thermo Fisher Scientific | MA516146 | 1:1000 (Western Blot) | AB_11157884 |
| GFAT | Cell Signaling Technology | 5322S | 1:1000 (Western Blot) | AB_10699031 |
| G6PD | Cell Signaling Technology | 12263S | 1:1000 (Western Blot) | AB_2797861 |
| Na/K ATPase | Santa Cruz Biotechnology | sc-28800 | 1:1000 (Western Blot) | AB_290063 |
| Akt | Cell Signaling Technology | 9272S | 1:1000 (Western Blot) | AB_329827 |
| p-Akt | Cell Signaling Technology | 9271S | 1:1000 (Western Blot) | AB_329825 |
| ZO-1 | Cell Signaling Technology | 13663S | 1:100 (Immunostaining) | AB_2798287 |
| Occludin | Cell Signaling Technology | 91131S | 1:100 (Immunostaining) | AB_2934013 |
| Claudin 5 | Thermo Fisher Scientific | 35–2500 | 1:100 (Immunostaining) | AB_2533200 |
| Anti-mouse IgG (H + L), HRP Conjugate | Promega | W4021 | 1:2000 (Western Blot) | AB_430834 |
| Anti-rabbit IgG (H + L), HRP Conjugate | Promega | W4011 | 1:2000 (Western Blot) | AB_430833 |
| Goat anti-rabbit IgG (H + L) 594 | Thermo Fisher Scientific | A-11012 | 1:1000 (Immunostaining) | AB_2534079 |
| Goat anti-mouse IgG (H + L) 488 | Thermo Fisher Scientific | A-11001 | 1:1000 (Immunostaining) | AB_2534069 |
| Hoechst | Thermo Fisher Scientific | PI62249 | 1:2000 (Immunostaining) | AB_2651133 |
Western Blot
hiBMEC were lysed in RIPA Buffer (Thermo Fisher, 89901) containing Halt Protease and Phosphatase Inhibitor (Fisher Scientific, PI78440) and Deacetylation Inhibition Cocktail (Santa Cruz Biotechnology, sc-362323). Protein was quantified using a bicinchoninic acid assay (Thermo Fisher, 23225), and 35 μg protein was combined with 7.5 μL sample buffer (Thermo Fisher, NP0008) and 3 μL reducing agent (Thermo Fisher, NP0009). Samples were either warmed to 37 °C (for glucose transporter 1 (GLUT1) only) or boiled at 70 °C (for all other proteins) for 5 min and loaded into 4–12% Bis Tris gels (Thermo Fisher, NP0323). Protein was transferred to either polyvinylidene fluoride (for GLUT1; Thermo Fisher, IB23001) or nitrocellulose (for all other proteins, Thermo Fisher, IB24001) membranes using an iBlot2 (Thermo Fisher, IBL21001). Polyvinylidene fluoride membranes were blocked in 5% milk in tris buffered saline (TBS; Fisher Scientific, BP24711) with 0.5% Tween 20 (Thermo Fisher, 85113) for 1 h, incubated in primary antibodies (Table 4) overnight at 4 °C in 1% bovine serum albumin (BSA) in TBS with 0.5% Tween 20, and then incubated for 2 h at room temperature in secondary antibody in 1% BSA in TBS with 0.5% Tween 20. Membranes for all other proteins were blocked for 1 h at room temperature in 5% bovine serum albumin (BSA; Millipore Sigma, 126609) in PBS with 0.5% Tween 20, then incubated in primary antibodies (Table 4) overnight at 4 °C in 1% BSA in PBS with 0.5% Tween 20, and secondary antibodies for 2 h at room temperature in 1% BSA in PBS with 0.5% Tween 20. Membranes were imaged using an Alpha Inotech Fluorchem Imager (Protein Simple) and analysed using AlphaView.
SIRT1 activity assay
SIRT1 activity was quantified using a fluorometric SIRT1 activity assay (abcam, ab156065) run according to manufacturer's protocols. 2 technical replicates were run for each sample. Protein concentrations from APOE-ε3 and -ε4 hiBMEC were quantified using a BCA assay (Thermo Fisher, 23225). SIRT1 fluorescence readings were normalised to total protein levels for each biological replicate.
Extracellular metabolite flux measurements
Glucose and glutamine uptake and lactate and glutamate secretion, as well as transport of each metabolite, were measured using a Yellow Springs Instruments (YSI) 2950 (Yellow Springs Instruments, 527690). Media samples were collected from either 12 well plates or both the apical and basolateral chambers of Transwell inserts at 0- and 24-h. Glucose, lactate, glutamine, and glutamate measurements were measured on the YSI. Metabolite uptake was calculated by subtracting metabolite concentrations at 24 h from the initial concentrations. Metabolite secretion was calculated by subtracting initial concentrations from the 24-h metabolite concentrations.
Seahorse glycolytic rate assay
A Seahorse glycolytic rate assay (GRA; Agilent, 103344-100) was used to evaluate rates of glycolysis. hiBMEC were seeded at 50,000 cells/well in 96-well Seahorse assay plates. On the day of the assay, cells were washed once with fresh Seahorse DMEM, and then incubated for 1 h in Seahorse DMEM supplemented with 10 mM glucose, 2 mM glutamine, 1 mM pyruvate, and 2% B27 before running the GRA in a Seahorse XFe96 following manufacturer's instructions.
Membrane fractionation
To quantify GLUT1 levels in the cell membrane of APOE-ε3 and -ε4 hiBMEC, a Mem-PER Plus Membrane Protein Extraction Kit (Thermo Fisher, 89842) was used. APOE-ε3 and -ε4 hiBMEC cultured in 12-well plates were lysed in membrane permeabilisation buffer supplemented with Halt Protease and Phosphatase Inhibitor. Samples were agitated for 10 min at 4 °C and then centrifuged at 17,000×g for 10 min at 4 °C. Supernatant containing the cytosolic proteins was removed and stored on ice. The pellet was then resuspended in membrane solubilisation buffer, agitated for 30 min at 4 °C, and then centrifuged at 17,000×g for 30 min at 4 °C. Protein was quantified, and western blot was run following previously described protocols.
Mass spectrometry
APOE-ε3 and -ε4 hiBMEC were cultured in Neurobasal medium supplemented with 8.5 mM glucose, 4.5 mM glutamine, and 2% B27 for 2 days following subculture. Cells were then incubated for 24 h in Neurobasal medium (±20% human serum) supplemented with 4.5 mM glutamine and 8.5 mM U-13C6-glucose (Fisher Scientific, NC9207695). To extract metabolites, cells were incubated in 80:20 methanol:water for 15 min at −80 °C and then centrifuged at 16,000 g for 10 min to pellet debris. Mass spectrometry was performed by the University of Colorado School of Medicine Metabolomics Core. Samples were randomised and a Vanquish ultra-high performance liquid chromatograph (UHPLC; Thermo Fisher) was used to inject 8 μL of each sample into a Q Exactive Mass Spectrometer (MS).32 Eluent was introduced to the MS via electrospray ionisation. The MS scanned 2 μscans over 65–950 m/z. Maven (Princeton University) and the KEGG database were used to manually annotate the metabolites, and peak quality was determined using technical mixes, blanks, and 13C natural abundances.33 The IsoCor Python package was used to correct isotope labelling,34 and metabolite pool sizes were analysed using Metaboanalyst 6.0.35 Metaboanalyst was used to generate partial least squares discriminant analysis (PLS-DA) plots wherein principal component 1 (PC1) on the x-axis is derived from the variables contributing to the greatest amount of variance in the dataset, while PC2 on the y-axis is derived from the variables contributing to greatest amount of variance once those in PC1 are removed from the dataset. The percent variance accounted for by each PC is noted on the axis.
Metabolic flux analysis
Isotopomer Network Compartmental Analysis (INCA) 2.2 was used to perform metabolic flux analysis (MFA).36,37 INCA is a user-friendly MATLAB based tool which uses experimental isotope datasets, extracellular fluxes, and a model of metabolic network reactions to estimate intracellular fluxes. INCA searches to identify the flux parameters for the intracellular fluxes to minimise the sum-of-squared residuals (SSR) between the experimental and computationally stimulated fluxes. The network model used here is based on similar models we developed for endothelial cells38 and includes reactions at isotopic and metabolic steady state for glycolysis, the PPP, the tricarboxylic acid (TCA) cycle, and amino acid metabolism. Briefly, to estimate the intracellular fluxes the 13C isotope distribution data for APOE-ε3 and -ε4 hiBMEC measured using mass spectrometry were corrected for the natural abundance of labelled and unlabelled atoms and input into the INCA software. The model was bounded by extracellular fluxes for glucose, glutamine, lactate, and glutamate measured using the YSI bioanalyser (converted to nmol/hr). The extracellular fluxes and isotope labelling data were fitted to the metabolic network map by generating a random initial flux guess and iterated until the best fit between the experimental and simulated fluxes was achieved. The fluxes were predicted using the Levenberg–Marquardt gradient descent algorithm37 and were repeated 100 times starting from random initial points to improve the statistical chance of finding a global optimum. Fit was based on n-p degrees of freedom, where n is the number of independent measurements and p is the number of fitted parameters. The final model contained 67 reactions. The number of fitted parameters differed between models, so the APOE-ε3 hiBMEC model had 79 degrees of freedom and the APOE-ε4 model had 76 degrees of freedom. The parameter continuation function was used by INCA to generate the confidence intervals.
Measurement error was set to a minimum of 1% based on previous recommendations.39 The distribution of residual error is expected to follow a normal distribution, and deviations from this suggest either inaccuracies in the model or gross measurement error. All isotopomers were scrutinised to identify poorly fitting measurements. Those that could not be explained by biochemically feasible reactions were rescaled to have an error of up to 5% to improve model fit and ensure a normal distribution of residual error. To minimise the SSR, the accepted error for some of the metabolites (GABA, ribose-5-phosphate, glutamate, citrate, and malate) was increased from 1% to 5%. For all other metabolites the accepted error was kept at 1%. To improve the accuracy of the flux predictions, experimental data from two different glucose labelling experiments were input into the model in parallel.
The final MFA output produced a set of possible flux predictions for each model. However, there is a degree of uncertainty in these predictions which allows each flux to take on a range of values. The parameter continuation function was used by INCA to generate 95% confidence intervals which estimate upper and lower boundaries for each metabolic reaction. Metabolic reactions without overlapping 95% confidence intervals were considered significantly different between the two genotypes, as is standard practice in MFA.38
Statistics
Statistics were analysed in GraphPad Prism. Data were not assumed to be normally distributed, so non-parametric statistical tests were used. Non-parametric Mann–Whitney tests were used to compare APOE-ε3 and -ε4 hiBMEC. Non-parametric Kruskal–Wallis tests were used to compare effects of insulin and wortmannin. Comparisons between APOE genotype and pre-and post-exercise training serum were analysed using repeated measures two-way ANOVA with Fisher's Least Significant Difference test. p-values <0.05 were considered statistically significant.
Role of funders
Funders were not involved in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
The APOE-ε4 genotype is associated with BBB breakdown and brain glucose hypometabolism. We therefore explored how the APOE-ε4 genotype impacts barrier strength and glucose metabolism in hiBMEC and how these metabolic changes propagate into systemic metabolic differences between APOE-ε3 and -ε4 hiBMEC. Then, we investigated how exercise training, as a preventative therapeutic against AD, changes hiBMEC barrier function and metabolism.
hiBMEC with the APOE-ε4 genotype had reduced barrier function
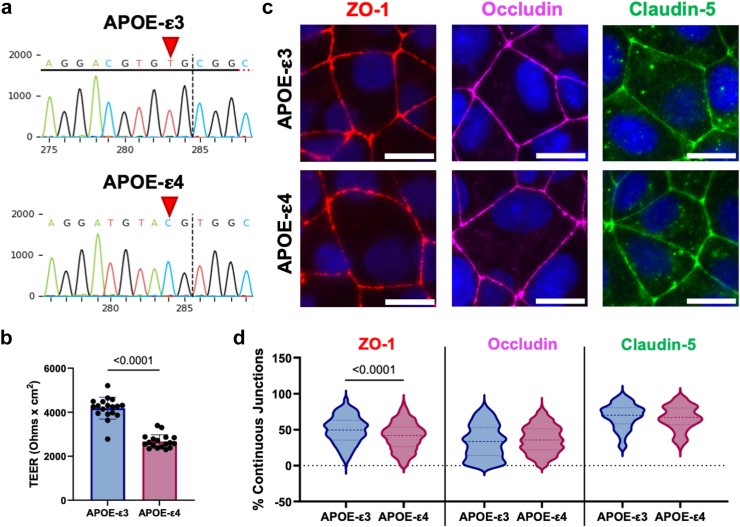
APOE-ε4 carriers have elevated BBB permeability relative to APOE-ε3 homozygotes.5 We began by determining if this phenotype also occurred in hiBMEC. CRISPR/Cas9 was used to insert a point mutation and thereby change iPSC homozygous for the APOE-ε3 allele to iPSC homozygous for the APOE-ε4 allele (Fig. 1a). These iPSC were then differentiated to hiBMEC,28,29 and barrier function was assessed using TEER and immunofluorescence. TEER was 36% lower in APOE-ε4 hiBMEC compared to -ε3 hiBMEC (p < 0.0001 [Mann–Whitney Test]; Fig. 1b). Tight junction protein ZO-1 continuity was 8.7% lower in APOE-ε4 hiBMEC compared to -ε3 hiBMEC (p < 0.0001 [Mann–Whitney Test]), suggesting that loss of ZO-1 may contribute to BBB breakdown in APOE-ε4 BMEC. Continuity of tight junction proteins occludin and claudin-5 did not significantly differ by genotype (Fig. 1c and d).
Fig. 1.
APOE-ε4 BMEC had reduced barrier function. (a) Sequencing electropherograms of iPSC edited with CRISPR/Cas9 targeting the APOE gene. Red arrows indicate the nucleotide that determines the APOE genotype in APOE-ε3 isogenic control iPSC and edited APOE-ε4 iPSC. (b) TEER measured on APOE-ε3 and -ε4 hiBMEC (n = 18 samples per genotype). (c) Immunofluorescent labelling of tight junction proteins in APOE-ε3 versus APOE-ε4 hiBMEC. ZO-1 (red), occludin (magenta), and claudin-5 (green). (d) Percent continuous junctions quantified via Junction Analyser Program. Scale bar = 20 μm (n = 176–327 cells from 16 biological replicates). Dotted lines indicate interquartile range. Data analysed using Mann–Whitney tests.
Reduced SIRT1 contributed to lower barrier function in APOE-ε4 compared to -ε3 hiBMEC
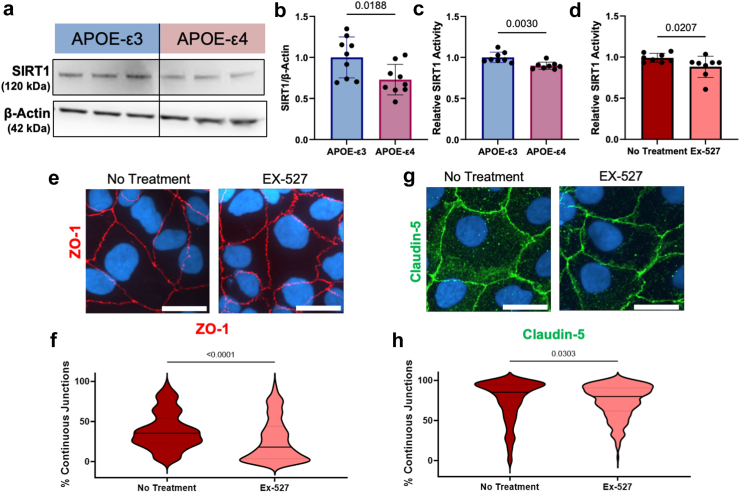
We next investigated mechanisms through which the APOE-ε4 genotype caused lower barrier function than the APOE-ε3 genotype. In neuroblastoma and glioblastoma cells, the APOE-ε4 protein directly suppresses transcription of SIRT1.40,41 Increasing SIRT1 decreases endothelial permeability through increased ZO-142 and claudin-5.43 Therefore, we hypothesised that APOE-ε4 hiBMEC have lower SIRT1 than APOE-ε3 hiBMEC, which impairs barrier function by decreasing ZO-1 junction continuity.
Western blots for SIRT1 demonstrated that APOE-ε4 hiBMEC had 27% less SIRT1 protein compared to APOE-ε3 hiBMEC (p = 0.0188 [Mann–Whitney Test]; Fig. 2a and b). A fluorometric SIRT1 activity assay showed SIRT1 activity was also 10% lower in APOE-ε4 compared to -ε3 hiBMEC (p = 0.0030 [Mann–Whitney Test]; Fig. 2c). To examine if the lower SIRT1 in the APOE-ε4 hiBMEC contributed to reduced barrier function, we modulated SIRT1 activity using a SIRT1 specific inhibitor, Ex-527. 10 μM Ex-527 decreased SIRT1 activity by 12% (p = 0.0570 [Mann–Whitney Test]; Fig. 2d). Then, we used immunofluorescent labelling and the Junction Analyser Program31 to examine how SIRT1 inhibition altered hiBMEC barrier function. Ex-527 decreased ZO-1 continuity by 26% (p < 0.0001 [Mann–Whitney Test]; Fig. 2e and f) but did not significantly alter claudin-5 continuity (Fig. 2g and h).
Fig. 2.
SIRT1 was lower in APOE-ε4 hiBMEC, and SIRT1 inhibition decreased barrier function in hiBMEC. (a) Representative Western blot and (b) quantification of SIRT1 relative to β-Actin in APOE-ε3 and -ε4 hiBMEC. Data were normalised to APOE-ε3 (n = 9 samples per genotype) (c) Relative SIRT1 activity measured using a SIRT1 activity assay and normalised to APOE-ε3 hiBMEC (n = 8 samples per genotype). (d) SIRT1 activity in hiBMEC treated with 10 μM EX-527 for 24 h relative to untreated hiBMEC (n = 8 samples per condition). Representative confocal microscopy images of tight junction proteins (e) ZO-1 (red) and (g) claudin-5 with (f, h) quantification of junction continuity by the Junction Analyser program (n = 225–251 cells per condition). Data analysed using a Mann–Whitney Test.
APOE-ε4 hiBMEC had reduced glucose metabolism relative to APOE-ε3 hiBMEC
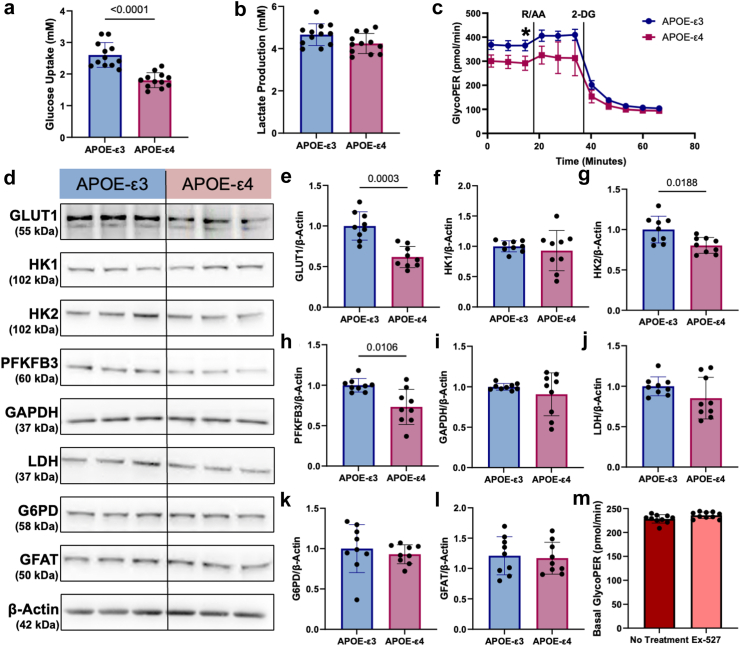
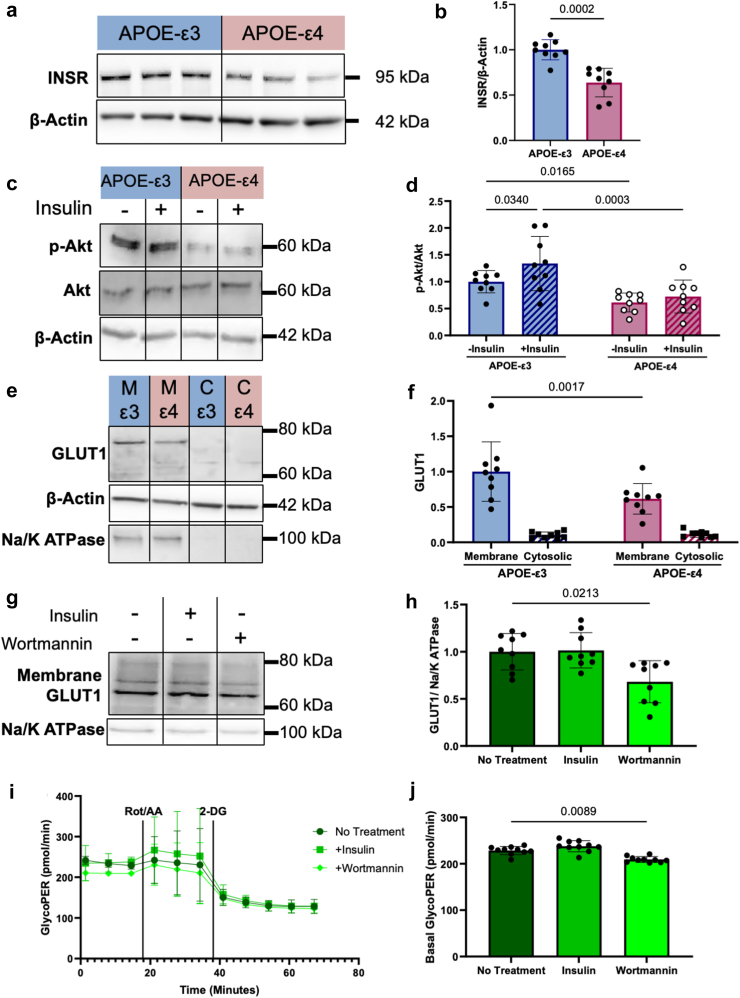
In addition to modulating barrier function, SIRT1 is known to regulate glycolysis. Glucose hypometabolism is a hallmark of AD, and low SIRT1 may decrease glycolysis through reduced GLUT144 and HK2.45 We used YSI and Seahorse Glycolytic Rate assays to assess if glycolytic rates were different between APOE-ε3 and -ε4 hiBMEC. APOE-ε4 hiBMEC had 31% lower glucose uptake than APOE-ε3 hiBMEC (p < 0.0001 [Mann–Whitney Test]; Fig. 3a), as measured by YSI. Lactate secretion was 9% lower in APOE-ε4 hiBMEC, although this was not statistically significant (p = 0.0780 [Mann–Whitney Test]; Fig. 3b). The basal glycolytic rate, measured by the Seahorse glycolytic proton efflux rate (GlycoPER), was 20% lower in APOE-ε4 hiBMEC than -ε3 hiBMEC (p < 0.0001 [Mann–Whitney Test]; Fig. 3c. Overall, these data indicate that APOE-ε4 hiBMEC have a decreased glycolytic rate compared to APOE-ε3 hiBMEC.
Fig. 3.
Glycolysis and glycolytic enzymes were lower in APOE-ε4 hiBMEC, but this decrease was not caused by reduced SIRT1. YSI measurements of (a) glucose uptake and (b) lactate secretion over 24 h (n = 12 samples per genotype). (c) GlycoPER measured via Seahorse Glycolytic Rate Assay in APOE-ε3 versus -ε4 hiBMEC (n = 10 samples per genotype). (d) Representative Western blots with quantifications of (e) GLUT1 (f) hexokinase I (HK1), (g) hexokinase 2 (HK2), (h) 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3), (i) glyceraldehyde 3-phosphate dehydrogenase (GAPDH), (j) lactate dehydrogenase (LDH), (k) glucose-6-phosphate dehydrogenase (G6PD), and (l) glutamine fructose-6-phosphate amidotransferase (GFAT) relative to housekeeper protein β-Actin (n = 9 samples per genotype). Data were normalised to APOE-ε3 protein levels in each experiment. (m) Basal GlycoPER measured via Seahorse Glycolytic Rate Assay in hiBMEC treated with 10 μM EX-527 for 24 h (n = 10 samples per condition). Data were analysed using Mann–Whitney tests. ∗p < 0.05.
Next, we analysed glycolytic enzymes by Western blot to evaluate which glycolytic enzymes contributed to lowered glycolysis in APOE-ε4 hiBMEC (Fig. 3d) Glucose entry into BMEC primarily occurs through the glucose transporter GLUT1. GLUT1 levels were 38% lower in APOE-ε4 hiBMEC compared to -ε3 hiBMEC (p = 0.0003 [Mann–Whitney Test]; Fig. 3e). Glucose is then converted to glucose-6-phosphate by hexokinase (HK) 1 or 2. HK1 levels were similar between APOE-ε3 and -ε4 hiBMEC (Fig. 3F); however, HK2 was 20% lower in APOE-ε4 compared to -ε3 hiBMEC (p = 0.0188 [Mann–Whitney Test]; Fig. 3g). Downstream glycolytic rate limiting enzyme PFKFB3 was also 24% lower in APOE-ε4 compared to -ε3 hiBMEC (p = 0.0106 [Mann–Whitney Test]; Fig. 3h), while glycolytic enzymes GAPDH and LDH were unchanged between APOE-ε3 and -ε4 hiBMEC (Fig. 3i and j). Additionally, we measured levels of glucose-6-phosphate dehydrogenase (G6PD), the rate limiting enzyme for glucose entry into the PPP, and glutamine fructose-6-phosphate amidotransferase (GFAT), the rate limiting enzyme for glucose entry into the hexosamine biosynthesis pathway (HBP). Neither G6PD nor GFAT levels were different between APOE-ε3 and -ε4 hiBMEC (Fig. 3k and l).
Since lower levels of GLUT1 and HK2 in APOE-ε4 hiBMEC correlated with lower SIRT1, we next examined if using Ex-527 to inhibit SIRT1 would inhibit glycolysis. Interestingly, the Seahorse Glycolytic Rate Assay did not demonstrate reduced hiBMEC glycolysis in response to Ex-527 (Fig. 3m), indicating that SIRT1 either does not contribute to reduced glycolysis in the APOE-ε4 hiBMEC, or that SIRT1 only reduces glycolysis in conjunction with other metabolic regulators.
APOE-ε3 and -ε4 hiBMEC had similar metabolite transport and polarisation
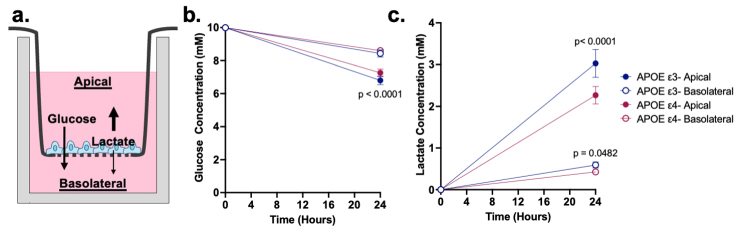
While glucose metabolism is important for energy production in BMEC themselves, BMEC are also responsible for transporting glucose from the blood into the brain for parenchymal brain cells to use as an energy source. To measure hiBMEC metabolite transport and polarisation, APOE-ε3 and -ε4 hiBMEC were seeded on Transwell inserts, and metabolite concentrations were measured on the apical and basolateral sides of the insert (Supplementary Fig. S1A). After 24 h, basolateral glucose concentrations were ∼8.5 mM glucose for both APOE-ε3 and -ε4 hiBMEC; however, the apical glucose concentration was 6.3% higher in APOE-ε4 hiBMEC compared to APOE-ε3 hiBMEC (p < 0.0001 [Two-Way ANOVA]), indicating reduced glucose uptake in APOE-ε4 hiBMEC (Supplementary Fig. S1B). Glucose was polarised in both -ε3 and APOE-ε4 hiBMEC, with higher glucose concentration in the basolateral compartment. Lactate was also polarised in both cell types, with higher lactate concentration in the apical compartment. APOE-ε4 hiBMEC secreted 25% less lactate in the apical compartment than APOE-ε3 hiBMEC, indicating decreased glycolysis (p < 0.0001 [Two-Way ANOVA]; Supplementary Fig. S1C).
APOE-ε4 genotype systemically changed hiBMEC metabolism
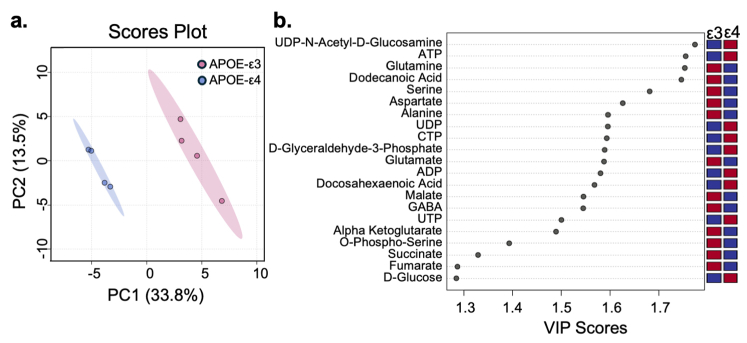
We next investigated if the APOE-ε4 genotype induced other metabolic changes in hiBMEC compared to APOE-ε3 hiBMEC using metabolomics. APOE-ε3 and -ε4 hiBMEC were labelled with 13C6-glucose for 24 h, and then metabolic mass spectrometry was used to identify changes in metabolite abundance and labelling patterns between the genotypes. Partial least squares-discriminant analysis (PLS-DA) indicated that APOE-ε3 and -ε4 hiBMEC separated primarily along component 1 (33.8%; Supplementary Fig. S2A). A variable importance plot (VIP) was then used to identify which metabolites most contributed to the PLS-DA separation (Supplementary Fig. S2B). The metabolite with the highest VIP score was UDP-N-Acetyl-d-Glucosamine (UDP-GlcNAc), the primary product of the HBP. Nucleotides including adenosine triphosphate (ATP), uridine diphosphate (UDP), cytidine triphosphate (CTP), adenosine diphosphate (ADP), and uridine triphosphate (UTP) were also elevated in APOE-ε4 hiBMEC. Amino acids glutamine, serine, aspartate, alanine, and glutamate were all elevated in APOE-ε3 hiBMEC, as were TCA cycle metabolites α-ketoglutarate, succinate, and fumarate.
13C metabolic flux analysis predicted lower glycolytic and higher malate shuttle and reductive carboxylation fluxes in APOE-ε4 hiBMEC
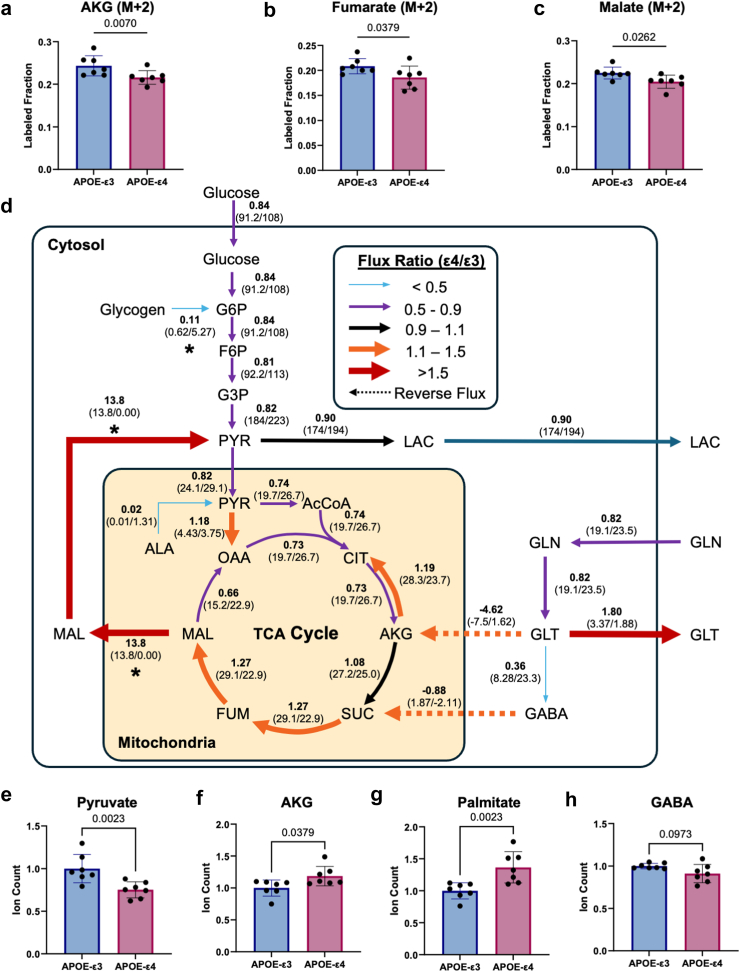
We then examined intracellular glucose metabolism by labelling APOE-ε3 and -ε4 hiBMEC with U-13C6-glucose and analysed the resulting mass isotopomers. Uniformly labelled glucose is metabolised into M+2 acetyl-CoA for entry into the TCA cycle. Notable, M+2 fractional enrichment decreased in APOE-ε4 hiBMEC for the TCA metabolites α-ketoglutarate (12% decrease, p = 0.0070 [Mann–Whitney Test]), fumarate (11% decrease, p = 0.0379 [Mann–Whitney Test]), and malate (9% decrease, p = 0.0262 [Mann–Whitney Test]) compared to APOE-ε3 hiBMEC (Fig. 4a–c).
Fig. 4.
Isotope labelling and13C metabolic flux analysis showed decreased glycolysis with redirection in the TCA cycle in APOE-ε4 hiBMEC. Fractional enrichment for M+2 isotopomers of (a) α-ketoglutarate (AKG), (b) fumarate, and (c) malate. Data pooled from two independent labelling experiments, n = 7. (d) Metabolic flux map showing ε4/ε3 flux ratios. Reversed fluxes are shown as dashed arrows. Fluxes without overlapping 95% confidence intervals shown with an asterisk under predicted flux ratio. Total ion counts for (e) pyruvate (f) AKG (g) palmitate and (h) GABA. All samples were normalised to mean of APOE-ε3 for the respective experiment. Data were pooled from two independent experiments (n = 7) and analysed with a Mann–Whitney test.
Decreased fractional enrichment can occur due to decreased metabolism of labelled precursors, an influx of unlabelled carbons, or a combination of both scenarios. This greatly complicates labelling data interpretation, particularly in the TCA cycle, which has multiple carbon sources and several reversible metabolic steps. To better understand differences in metabolic activity between APOE-ε3 and -ε4 hiBMEC, we performed isotope-assisted MFA by integrating isotopomer data with extracellular flux measurements.46 Reactions used to generate the MFA maps, and net fluxes for the APOE-ε3 and -ε4 MFA are included in the supplemental information (Supplementary Tables S1–S3).
We produced a simplified flux map detailing estimated fluxes through some of the metabolic pathways that were predicted to differ between APOE-ε3 and -ε4 hiBMEC (Fig. 4d). Data are represented through the APOE-ε4/ε3 flux ratio, in which a flux ratio less than one indicates higher estimated flux in APOE-ε3 hiBMEC, and a flux ratio greater than one indicates higher estimated flux in APOE-ε4 hiBMEC. All glycolytic fluxes were reduced in APOE-ε4 hiBMEC (flux ratio 0.81–0.84) compared to APOE-ε3 hiBMEC up to the pyruvate branchpoint. At this metabolic junction, 13C-MFA estimated a major influx of malate-sourced pyruvate in APOE-ε4 hiBMEC, which was absent in APOE-ε3 hiBMEC. Despite these differences, it was estimated that both genotypes directed a similar percentage of pyruvate towards oxidative respiration in the mitochondria.
Within the mitochondria, the estimated pyruvate carboxylation to oxaloacetate was higher in APOE-ε4 hiBMEC (flux ratio = 1.18) while the estimated conversion of pyruvate dehydrogenase to acetyl-CoA was lower (flux ratio = 0.74) compared to APOE-ε3 hiBMEC. Citrate decarboxylation to α-ketoglutarate was also estimated to be lower in APOE-ε4 hiBMEC (flux ratio = 0.74). However, subsequent steps in the TCA cycle, including succinate, fumarate, and malate production, were estimated to be higher in the APOE-ε4 compared to APOE-ε3 hiBMEC (flux ratio 1.08–1.27).
Reductive carboxylation of α-ketoglutarate (α-ketoglutarate conversion to citrate) was estimated to be higher in APOE-ε4 hiBMEC (flux ratio = 1.19). The elevated reductive carboxylation in APOE-ε4 hiBMEC appeared to be fueled by glutamate conversion to α-ketoglutarate in the TCA cycle. In contrast, in APOE-ε3 hiBMEC α-ketoglutarate was estimated to be converted to glutamate (flux ratio = −4.62). APOE-ε4 hiBMEC also had an estimated lower glutamine influx and glutamine to glutamate flux (flux ratio = 0.82), and thus lower flux of glutamate to 4-aminobutanoate/GABA (flux ratio = 0.36). Interestingly, in APOE-ε4 hiBMEC, GABA was estimated to feed into succinate, whereas in APOE-ε3 hiBMEC the reaction was reversed (flux ratio = −0.88). Another major difference predicted by the 13C-MFA was in the malate shuttle (malate conversion to pyruvate), which was not predicted to be active in APOE-ε3 hiBMEC but was estimated to be highly active in APOE-ε4 hiBMEC (flux ratio = 13.8).
To corroborate the 13C-MFA, we examined the ion counts of key metabolites. Intracellular pyruvate ion count was 25% lower in APOE-ε4 hiBMEC (p = 0.0023 [Mann–Whitney Test]; Fig. 4e), while α-ketoglutarate was 19% higher in APOE-ε4 hiBMEC compared to APOE-ε3 hiBMEC (p = 0.0379 [Mann–Whitney Test]; Fig. 4f). Reductive carboxylation of α-ketoglutarate can also feed lipogenic pathways. We found palmitate to be 37% higher in APOE-ε4 hiBMEC (p = 0.0023; Fig. 4g). Finally, the 13C-MFA predicted lower GABA synthesis and higher GABA consumption in APOE-ε4 compared to APOE-ε3 hiBMEC. In accordance with the model, GABA ion counts were 9% lower in APOE-ε4 hiBMEC compared to APOE-ε3 hiBMEC (p = 0.0973 [Mann–Whitney Test]; Fig. 4h).
Impaired insulin signalling contributed to decreased glycolysis in APOE-ε4 hiBMEC
As decreased SIRT1 did not reduce hiBMEC glycolysis, we next investigated an alternative mechanism. The APOE-ε4 protein inhibits insulin receptor (INSR) recycling and glycolysis in neurons.47 We therefore hypothesised that APOE-ε4 hiBMEC would also have reduced INSR recycling, leading to decreased GLUT1 translocation to the cell membrane, and thereby decreasing glycolytic rate. We first investigated how INSR levels differed between APOE-ε3 and -ε4 hiBMEC by Western blot. APOE-ε4 hiBMEC had 36% lower INSR protein levels than APOE-ε3 hiBMEC (p = 0.0002 [Mann–Whitney Test]; Fig. 5a and b). Next, we determined how insulin signalling differed in APOE-ε3 and -ε4 hiBMEC by treating cells with insulin and probing for phosphorylated-Akt (p-Akt) by Western blot (Fig. 5c). APOE-ε4 hiBMEC had a 37% lower p-Akt:Akt ratio without insulin stimulation (p = 0.0186 [Two-Way ANOVA]), and an 82% lower p-Akt:Akt ratio with insulin stimulation (p = 0.0029 [Two-Way ANOVA]). While APOE-ε3 hiBMEC increased the p-Akt:Akt ratio by 30% following insulin stimulation (p = 0.0340 [Two-Way ANOVA]), APOE-ε4 hiBMEC did not significantly increase p-Akt:Akt ratio with insulin stimulation (Fig. 5d). These data suggest that insulin signalling is reduced in APOE-ε4 hiBMEC, possibly through reduced INSR levels.
Fig. 5.
Impaired insulin signalling may have contributed to reduced APOE-ε4 hiBMEC glycolysis. (a, b) Representative Western blots with quantification of insulin receptor (INSR) in APOE-ε3 and -ε4 hiBMEC (n = 9 samples per genotype). (c, d) Representative Western blots with quantification of p-Akt and Akt in APOE-ε3 and -ε4 hiBMEC treated with 10 μg/mL insulin for 30 min (n = 9 samples per condition). (e, f) Representative Western blots with quantification of membrane and cytosolic GLUT1 in APOE-ε3 and -ε4 hiBMEC (n = 9 samples per genotype). (g, h) Representative Western blots with quantification of membrane GLUT1 in hiBMEC treated with 10 μg/mL insulin and 1 μM Wortmannin for 30 min (n = 9 samples per condition). (i) Seahorse Glycolytic Rate Assay and (j) basal GlycoPER in hiBMEC following treatment with 10 μg/mL and 1 μM Wortmannin for 30 min (n = 10 samples per treatment group). Data were analysed using (b) Mann–Whitney test, (d, f) Two-way ANOVA with Fishers Least Significant Difference Test, and (h, j) Kruskall-Wallis with Dunn's multiple comparison test.
Next, we investigated how inhibition of insulin signalling impacted GLUT1 translocation to the membrane and glycolysis. Using membrane fractionation combined with Western blot, we showed that GLUT1 was 31% lower in APOE-ε4 hiBMEC relative to APOE-ε3 hiBMEC (p = 0.0017 [Two-Way ANOVA]; Fig. 5e and f). Then, we treated APOE-ε4 hiBMEC with 10 μg/mL insulin to stimulate insulin signalling and promote GLUT1 translocation to the membrane, or 1 μM Wortmannin to inhibit PI3K in the insulin signalling pathway and thus GLUT1 membrane translocation (Fig. 5g). Insulin treatment did not increase GLUT1 membrane translocation. Wortmannin, however, decreased membrane GLUT1 by 32% (p = 0.0213 [Kruskal–Wallis Test]; Fig. 5h), indicating that decreased insulin signalling can decrease membrane GLUT1. Finally, to determine how reduced insulin signalling and membrane GLUT1 corresponded with reduced hiBMEC glycolysis, we ran a Seahorse Glycolytic Rate Assay in which hiBMEC were pretreated with insulin or Wortmannin for 30 min (Fig. 5i). Basal GlycoPER was not significantly elevated by with insulin but decreased 8.4% with Wortmannin (p = 0.0089 [Kruskal–Wallis Test]; Fig. 5j).
Serum collected post-exercise training differentially regulated SIRT1, but not barrier function, in APOE-ε3 and -ε4 hiBMEC
Exercise training reduces hippocampal atrophy and increases cerebral glucose metabolism associated with AD.13,48 We therefore examined how serum from exercise trained individuals alters hiBMEC barrier function and metabolism to explore the molecular mechanisms by which exercise benefits brain health. Exercise training has been shown to increase cerebral lactate and AMP-activated protein kinase (AMPK), which in turn increase SIRT1.19,49,50 Therefore, we hypothesised hiBMEC treated with serum collected post-training would increase SIRT1, barrier function, and glucose metabolism compared to hiBMEC treated with serum collected pre-training.
First, we examined how cardiorespiratory fitness, measured using 2peak, changed in study participants from the start to the end of the trial (Table 2). Since the 2peak change was statistically similar between the low and moderate intensity exercise training groups, the data were combined. Overall, study participants did not have a significant change in O2peak following exercise training.
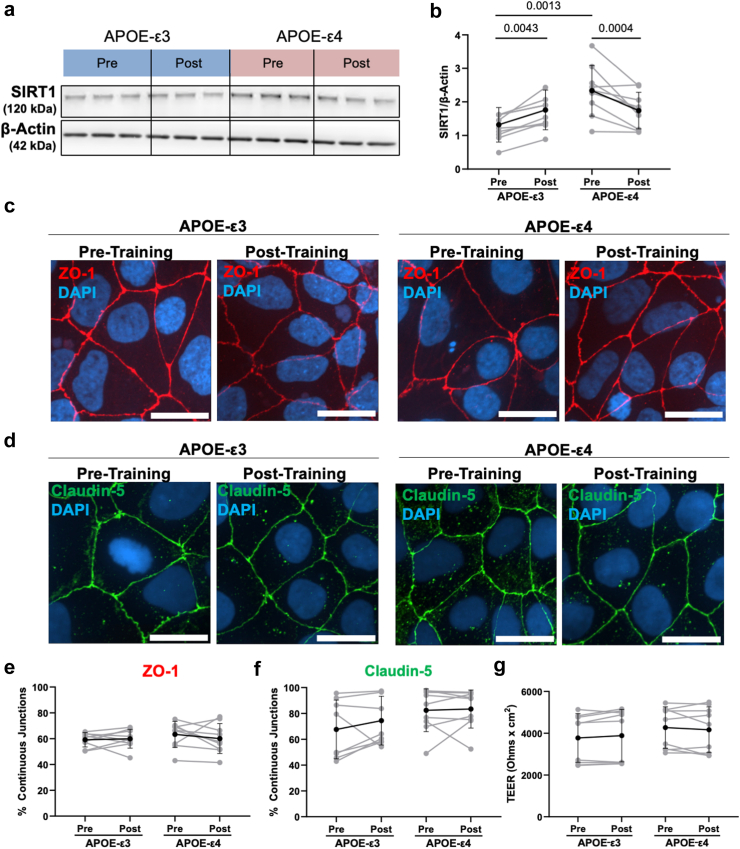
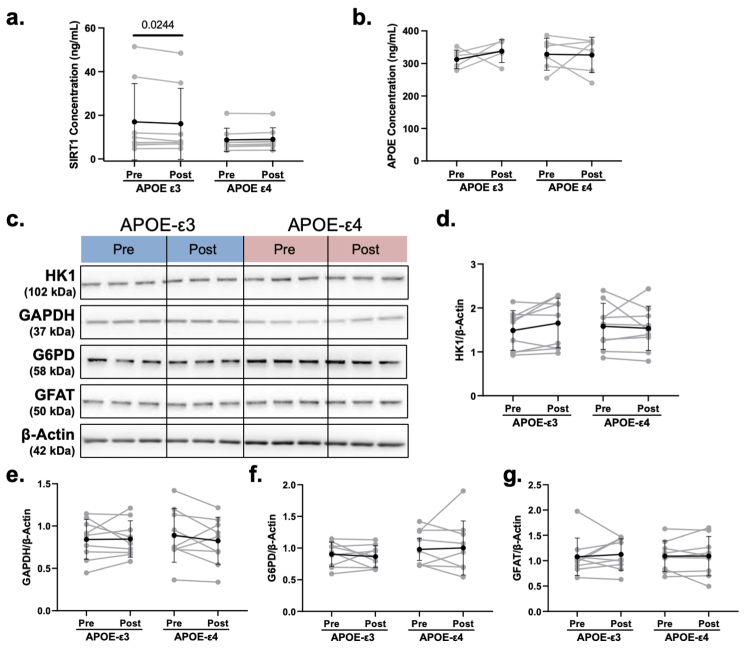
We then used Western blot to measure SIRT1 levels in APOE-ε3 and -ε4 hiBMEC treated with 20% serum collected pre- and post-training (Fig. 6a). Unexpectedly, SIRT1 levels were 71% higher in APOE-ε4 hiBMEC treated with pre-training serum compared to APOE-ε3 hiBMEC treated with pre-training serum (p = 0.0013 [Repeated Measures Two-Way ANOVA]). While SIRT1 levels in APOE-ε3 hiBMEC increased by 33% when treated with post-training serum (p = 0.0043 [Repeated Measures Two-Way ANOVA]), SIRT1 levels decreased by 22% in APOE-ε4 hiBMEC treated with post-training serum (p = 0.0004 [Repeated Measures Two-Way ANOVA]; Fig. 6b). There was no significant difference in SIRT1 levels between APOE-ε3 and -ε4 hiBMEC treated with post-training serum.
Fig. 6.
SIRT1 increased with post-training serum in APOE-ε3 hiBMEC and decreased with post-training serum in APOE-ε4 hiBMEC, but this did not change barrier integrity. (a, b) Representative Western blot and quantification of SIRT1 in hiBMEC treated with 20% genotype-matched serum for 24 h (n = 9 donors per condition). Representative confocal microscopy images of tight junction proteins (c) ZO-1 and (d) claudin-5 following serum treatment and (e, f) quantified using the junction analyser program (45 cells analysed per donor per treatment group, n = 9 donors per genotype). (g) TEER measurements in APOE-ε3 and -ε4 hiBMEC following 24 h of treatment with 20% genotype-matched serum. Data analysed using repeated measures two-way ANOVA with Fisher's Least Significant Difference Test.
Since SIRT1 trends in APOE-ε3 and -ε4 hiBMEC were different in the presence of serum compared to those observed in serum-free culture, we next evaluated proteins in the serum that could differentially regulate SIRT1. When we quantified serum SIRT1 via ELISA, there were no significant differences in SIRT1 between APOE-ε3 and -ε4 serum samples (Supplementary Fig. S3A). Since APOE-ε4 proteins can regulate SIRT1 protein,40,41 we then quantified serum APOE via ELISA to see if that could induce the differential SIRT1regulation. APOE was also not significantly different between APOE-ε3 and -ε4 serum (Supplementary Fig. S3B). Therefore, there are likely other proteins in the serum that regulate SIRT1 in the APOE-ε3 and -ε4 hiBMEC.
We next used immunofluorescent imaging with the Junction Analyser Program and TEER to examine if the dimorphic changes in SIRT1 in APOE-ε3 and -ε4 hiBMEC with pre-and post-training serum correlated with barrier function changes (Fig. 6c and d). ZO-1 and claudin-5 immunostaining revealed no significant changes in tight junction protein continuity in the APOE-ε3 or -ε4 hiBMEC in response to pre- or post-training serum (Fig. 6e and f). There was also no difference in TEER between the APOE-ε3 and -ε4 hiBMEC pre- or post-training serum (Fig. 6g).
Post-training serum altered glycolytic enzymes but not glycolysis in APOE-ε3 and -ε4 hiBMEC
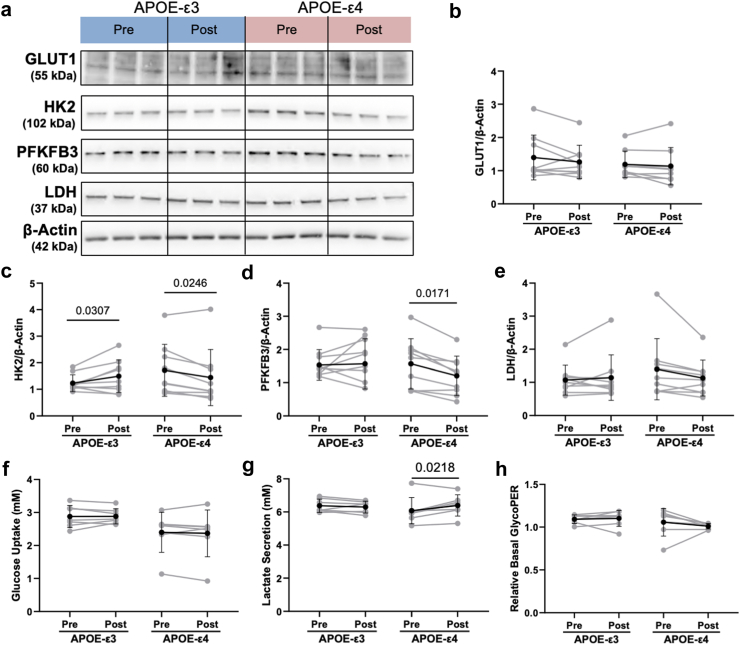
Since we previously showed changes in glycolytic proteins GLUT1, HK2, and PFKFB3 correlated with changes in SIRT1 in APOE-ε3 versus -ε4 hiBMEC, we next examined if these proteins matched the dimorphic changes in SIRT1 in APOE-ε3 and -ε4 hiBMEC treated with serum collected pre- and post-exercise training (Fig. 7a). GLUT1 protein levels were similar between APOE-ε3 and -ε4 hiBMEC treated with pre-versus post-exercise training serum (Fig. 7b). HK2 protein levels matched SIRT1 changes. HK2 increased by 27% with post-training serum in APOE-ε3 hiBMEC (p = 0.0307 [Repeated Measures Two-Way ANOVA]) but decreased by 15% in APOE-ε4 hiBMEC treated with post-training serum (p = 0.0246 [Repeated Measures Two-Way ANOVA]; Fig. 7c). PFKFB3 protein levels did not change consistently in APOE-ε3 hiBMEC treated with pre- or post-training serum but decreased by 23% with post-training serum in APOE-ε4 hiBMEC (p = 0.0171 [Repeated Measures Two-Way ANOVA]; Fig. 7d). Other glycolytic enzymes, including GAPDH, LDH, G6PD, and GFAT were not affected by genotype or serum (Supplementary Fig. S3C–G).
Fig. 7.
Serum from individuals post-training decreased rate limiting glycolytic enzymes HK2 and PFKFB3 in APOE-ε4 hiBMEC relative to serum collected pre-training. (a) Representative Western blots with quantification of (b) GLUT1, (c) HK2, (d) PFKFB3, and (e) LDH relative to housekeeper β-actin in APOE-ε3 and -ε4 hiBMEC following 24 h of treatment with 20% serum from genotype matched individuals pre- and post-exercise training. (n = 9 donors per genotype and condition). Glycolytic measures of (f) glucose uptake (YSI), (g) lactate secretion (YSI), and (h) basal GlycoPER (Seahorse Glycolytic Rate assay) in APOE-ε3 and -ε4 hiBMEC following 24 h of treatment with 20% serum from genotype matched individuals pre- and post-exercise training. (n = 9 donors per genotype). Data were analysed using repeated measures two-way ANOVA with Fisher's Least Significant Difference Test.
YSI and Seahorse Glycolytic Rate Assays were then used to determine if the changes in SIRT1 and glycolytic enzymes resulted in overall changes in glycolysis. Glucose uptake was unchanged between APOE-ε3 and -ε4 hiBMEC with pre- or post-training serum (Fig. 7f). Lactate secretion was 5% higher in APOE-ε4 hiBMEC treated with post-training serum compared to pre-training serum (p = 0.0218 [Repeated Measures Two-Way ANOVA], Fig. 7g) but was unchanged in APOE-ε3 hiBMEC. The Seahorse Glycolytic Rate Assay similarly showed no significant changes in basal glycolysis in APOE-ε3 or -ε4 hiBMEC treated with pre- or post-exercise training serum (Fig. 7h).
Serum systemically altered the hiBMEC metabolome, but exercise training did not
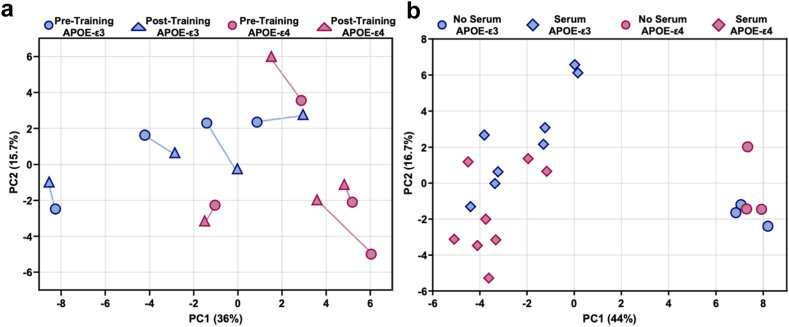
Our previous analysis revealed systemic metabolic differences between APOE-ε3 and -ε4 hiBMEC. We therefore used LC-MS to examine metabolomic changes in APOE-ε3 and -ε4 hiBMEC treated with serum collected pre- and post-exercise training. PLS-DA demonstrated no clear separation of APOE-ε3 and -ε4 hiBMEC treated with pre- or post-training serum (Fig. 8a). However, in 3/4 of the APOE-ε3 hiBMEC samples, the samples shifted rightward shift along PC1 with post-training serum. In contrast, post-training serum shifted all the APOE-ε4 hiBMEC samples leftward along PC1. Additionally, 3/4 of the APOE-ε4 hiBMEC samples treated with post-training serum shifted upwards along PC2 compared to those treated with pre-training serum.
Fig. 8.
Serum systemically altered the hiBMEC metabolome, but exercise training did not. (a) PLS-DA of the metabolite labelled fraction of APOE-ε3 (blue) and -ε4 (pink) hiBMEC treated for 24 h with 20% serum from genotype-matched individuals pre- (circle) and post- (triangle) exercise training and 13C6-glucose. A line connects hiBMEC treated with serum samples from the same donor (n = 4 donors per genotype). (b) PLS-DA of the metabolite labelled fraction of APOE-ε3 (blue) and -ε4 (pink) hiBMEC treated for 24 h with no serum (circle) or serum (diamond). Serum effects were analysed for combined pre- and post-training serum samples and compared to hiBMEC not treated with serum (n = 8 donors per genotype with serum, n = 3 donors per genotype without serum).
Finally, we analysed how APOE-ε3 and -ε4 hiBMEC metabolomes differed with serum addition. For this analysis, samples treated with serum collected pre- and post-training were combined and compared against serum-free APOE-ε3 and -ε4 hiBMEC. APOE-ε3 and -ε4 hiBMEC did not separate by genotype, as demonstrated by PLS-DA (Fig. 8b). However, serum separated hiBMEC along component 1 (PC1 = 44%). The metabolites that were the largest drivers of this separation are reported in Appendix 6 and include lactate, alanine, pyruvate, (4Z-7Z-10Z-13Z-16Z-19Z)-Docosahexaenoic acid, (9Z)-Octadecenoic acid, fructose 1,6-bisphosphate, and citrate, all of which decreased in hiBMEC treated with serum compared to serum-free hiBMEC culture. The metabolites with the largest contributions to the separation along component 1 largely belonged to glycolysis, fatty acid metabolism, and the TCA cycle.
Discussion
The APOE-ε4 genotype is one of the largest AD risk factors and is associated with reduced BMEC barrier function and whole brain glucose metabolism. Exercise training may counteract AD-related cognitive decline by increasing whole brain glucose metabolism.14,51 However, there are few, if any, identified mechanisms through which the APOE genotype alters barrier function and metabolism, or that investigate how exercise training may support BMEC barrier function and metabolism. Here, we demonstrated that APOE-ε4 hiBMEC have reduced barrier function and glucose metabolism compared to APOE-ε3 hiBMEC, and that reduced SIRT1 and insulin signalling may reduce barrier function and glycolysis in APOE-ε4 hiBMEC. Serum from exercise trained individuals did not significantly alter barrier function or glycolytic rates in hiBMEC but did have dimorphic effects on SIRT1 in APOE-ε3 and -ε4 hiBMEC. These findings indicate that the APOE-ε4 genotype induces BBB breakdown and glucose hypometabolism through SIRT1 and insulin signalling, and these changes cannot be counteracted by serum from exercise-trained individuals alone.
APOE-ε4 hiBMEC had reduced barrier function, as measured through TEER and ZO-1 continuity, relative to the APOE-ε3 hiBMEC. This agrees with past reports of reduced TEER in BMEC isolated from APOE-ε4 transgenic mice9; increased permeability with reduced ZO-1 and occludin in brain vessels of APOE-ε4 transgenic mice10; and increased BBB permeability in the hippocampus and parahippocampal gyrus of adults who are carriers of the APOE-ε4 allele.5 While APOE-ε4 may reduce barrier function through multiple mechanisms, APOE-ε4 cells had lower SIRT1. Previous studies have shown that decreasing SIRT1 reduces endothelial barrier integrity. For example, SIRT1 increased ZO-1 in human pulmonary microvascular endothelial cells,42 BMEC-specific SIRT1 knockout in mice increased BBB permeability,51 and SIRT1 siRNA knockdown increased mouse52 and primary human BMEC43 permeability by decreasing claudin-5. Although the mechanism has not been examined in BMEC, in ovarian cancer cells, SIRT1 deacetylates Krüppel-like factor 4 (KLF4) which promotes claudin-5 transcription.53 KLF4 can also interact with promotor regions on ZO-1 and occludin to increase their transcription.54 Therefore, it is possible that the low SIRT1 in APOE-ε4 hiBMEC increased KLF4 acetylation, which reduced ZO-1 transcription to reduce overall barrier function in APOE-ε4 hiBMEC.
The APOE-ε4 genotype is associated with reduced glucose metabolism in transgenic mouse BMEC,9 astrocytes,11 and neurons.55 Here, we showed that these same changes occurred in hiBMEC. Our data suggest that reduced glycolysis may relate to INSR recycling. In neurons, APOE-ε4 similarly reduced endosomal INSR recycling, which in turn reduced insulin signalling and glycolysis.47 Neuronal glucose transport is primarily regulated by GLUT3,56 while hiBMEC glucose transport is primarily regulated by GLUT1. Increased INSR and insulin signalling can increase both GLUT1 and GLUT3 transcription and trafficking of the transporters to the cell membrane.57, 58, 59, 60 Interestingly, in our experiments insulin did not significantly increase membrane GLUT1 or glycolytic rate in hiBMEC. It is therefore possible that the cell membrane is already saturated with GLUT1, and thus GLUT1 translocation cannot increase with additional insulin.
Previous studies demonstrated potential links between glycolysis and barrier function. GLUT1 co-localises with tight junction protein ZO-1 in the ventromedial hypothalamus, indicating the function of the two proteins may be tightly linked.61 Reduced GLUT1 also increased vascular permeability in mouse models of AD.62 Finally, BMEC rely on oxidative phosphorylation for energy when glycolysis is inhibited. Oxidative phosphorylation leads to oxidative stress, which may then damage BBB integrity.63 Thus, a metabolic shift in APOE-ε4 BMEC from glycolysis to oxidative phosphorylation could reduce BBB barrier function through oxidative stress. Future studies should examine these mechanisms to identify how reduced glycolysis in APOE-ε4 hiBMEC reduces barrier function.
13C6-glucose metabolomics revealed systemic metabolomic differences between APOE-ε3 and -ε4 hiBMEC. To further delineate metabolic differences, we fit a 13C-MFA model and generated a metabolic flux map. The map unveiled several interesting changes in intracellular metabolism between APOE-ε3 and -ε4 hiBMEC, such as reduced GABA production, reversal of α-ketoglutarate-to-glutamate flux, and reversal of the GABA-succinate shuttle flux in APOE-ε4 hiBMEC. Additionally, APOE-ε4 hiBMEC have elevated estimated reductive carboxylation and pyruvate carboxylation compared to APOE-ε3 hiBMEC, both which are associated with lipogenesis.64,65 In microglia the APOE-ε4 genotype induces elevated triglyceride and lipid droplet formation, which are neurotoxic.66 The 13C-MFA predicts that increased fatty acid synthesis may also be occurring in the APOE-ε4 hiBMEC, which could contribute to neurodegeneration.
Interestingly, SIRT1 is associated with suppression of fatty acid synthase and lipogenesis in hepatocytes through an AMPK-mediated pathway.67 Lower SIRT1 in APOE-ε4 hiBMEC may therefore lead to increased activation of fatty acid synthase, which could further increase lipogenic pathways in the APOE-ε4 hiBMEC. Reductive carboxylation also generates NAD+,68 which can activate SIRT1 and other sirtuins. This may therefore suggest that reductive carboxylation is used to compensate for reduced SIRT1 levels by increasing activation of available SIRT1 and may help to explain why despite having 27% less SIRT1 protein, there is only 10% less SIRT1 activity in the APOE-ε4 compared to the APOE-ε3 hiBMEC.
Serum collected post-6 months of exercise training increased SIRT1 in APOE-ε3, but decreased SIRT1 in APOE-ε4 hiBMEC. A meta-analysis of SIRT1 changes with exercise training demonstrates that acute exercise elevates SIRT1 in skeletal muscle and consistent exercise can elevate circulating SIRT1.69 Here, we showed that in APOE-ε3 hiBMEC treated with pre-training serum, SIRT1 levels were significantly lower than in APOE-ε4 hiBMEC treated with pre-training serum, and that the addition of post-training serum normalised SIRT1 to the same level in APOE-ε3 and -ε4 hiBMEC. These data indicate exercise-trained serum may signal for homeostatic control of SIRT1, bringing them back to a general baseline. While SIRT1 levels were higher in APOE-ε3 hiBMEC compared to APOE-ε4 hiBMEC without serum treatment, this trend was not shown with serum treatment. Since neither SIRT1 nor APOE levels were significantly different between APOE-ε3 and -ε4 serum samples, other components of the serum likely regulate intracellular SIRT1 levels.
Since we previously showed SIRT1 regulates barrier function, we hypothesised that post-training serum would increase barrier function in APOE-ε3 hiBMEC and decrease barrier function in APOE-ε4 hiBMEC compared to serum collected pre-training. However, we did not see consistent changes in barrier function in response to serum from exercise trained individuals. Aerobic exercise increases cerebral blood flow,70 and shear stress can increase claudin-5 and ZO-1 levels in human BMEC.71,72 Thus, it is possible that changes in BBB function relate to increased shear stress rather than serum modifications in response to exercise.
Glycolytic enzyme HK2 followed a similar trajectory as SIRT1 with serum collected post-6 months of exercise training but did not change glycolysis. In leukemia cells, HK2 knockdown increased carbon flux into the PPP and TCA cycle but did not significantly impact glycolysis.73 Therefore, reduced HK2 in APOE-ε4 hiBMEC treated with exercise trained serum may alter PPP and TCA pathway activity more than glycolysis. Glycolytic rates are also modulated by many metabolites and proteins outside of the direct glycolytic pathway including ATP bioavailability,73 so other serum or intracellular factors may also override the changes in HK2 to modulate glycolytic rates.
PLS-DA revealed a clear separation of APOE-ε3 and -ε4 hiBMEC metabolites when the cells were cultured in serum-free conditions; however, this separation was no longer apparent when cells were cultured with human serum. Though this could relate to the inherent variability involved when using human serum samples, it is also possible that human serum reduced metabolomic differences between APOE-ε3 and -ε4 hiBMEC. Serum decreased intracellular fatty acid synthesis, which may be due to the presence of fatty acids in serum that are not generally present in serum-free cell culture experiments. Overall, the reduction in metabolomic separation between APOE-ε3 and -ε4 hiBMEC in the presence of serum reveals that despite intracellular differences, other cells in the body may secrete metabolites, proteins, and fatty acids to naturally regulate genotype-dependent differences in BMEC.
Although the data presented in this study are comprehensive, the study is not without limitations. The study was conducted in vitro using an entirely human model, which provides valuable insights into human physiology and pathology. However, in vivo animal models incorporate more holistic exercise effects, albeit in a different biological system. A prior in vivo analysis of exercise training in mice supports our results, showing that running increased gene expression associated with vascular integrity more in APOE-ε3/ε4 than APOE-ε4/ε4 mice.74 We used IMR90 iPSC in our in vitro model, which we differentiated into BMEC-like cells. IMR90 iPSC are a validated iPSC line homozygous for the APOE-ε3 genotype,25 and using one iPSC line enabled us to focus on human serum effects. iPSC-BMEC may also have an underlying epithelial transcriptome that could impact cell response,75 although we have previously validated that glucose metabolism is similar between hiBMEC and primary BMEC.23 Finally, this study lacks exercise training-induced physiological cues that are not in the human serum such as altered shear stress.
In conclusion, we demonstrated that the APOE-ε4 genotype reduces hiBMEC barrier function via SIRT1, and glucose metabolism via insulin signalling. We also showed exercise training may have different impacts on BMEC depending on APOE genotype. Future studies should build on these results to examine mechanisms of exercise benefits on brain health, stratified by APOE genotype, and develop genotype-dependent exercise recommendations.
Contributors
Conceptualization, C.M.W., B.M., J.C.S., and A.M.C.; Methodology, C.M.W., B.M., G.S.P., G.S.S., J.C.S., and A.M.C., Investigation, C.M.W., B.M., G.S.P., M.K., B.W., C.K., G.S.S.; Formal Analysis, C.M.W., B.M., M.K., B.W., C.K.; Data Curation, C.M.W., B.M., M.K.; Writing- Original Draft, C.M.W. and A.M.C.; Writing- Review & Editing, C.M.W., B.M., G.S.P., J.C.S., and A.M.C.; Visualization, C.M.W. and A.M.C.; Supervision, J.C.S. and A.M.C., Funding Acquisition, J.C.S. and A.M.C. All contributors have read and approved the final version of the manuscript.
Data sharing statement
All data are available in the main text or supplementary materials and can be made available by request to the corresponding author.
Declaration of interests
Authors declare they have no competing interests.
Acknowledgements
The authors would like to thank Sophia Zic, Deborah DiSilvestre, and Ivy Dick for valuable contributions to this research, and the University of Colorado School of Medicine Metabolomics Core for mass spectrometry analysis. AMC acknowledges funding from the Brain Behavior Initiative, NSF CBET 2211966, NIH R01HL165193 and NIH R01HL140239-01. CK and BW acknowledge funding from ASPIRE Program. BM acknowledges funding from Niemann-Pick Disease Foundation and NSF DGE 1632976. JCS acknowledges funding from the Brain Behavior Initiative and R01AG057552. CW acknowledges funding from NSF-GRFP DGE 1840340 and Fischell Fellowship.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105487.
Appendix A. Supplementary data
Supplemental Fig. S1.
Supplemental Fig. S2.
Supplemental Fig. S3.
References
- 1.Davignon J., Gregg R.E., Sing C.F. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.ATV.8.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Slooter A.J.C., Cruts M., Kalmijn S., et al. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the rotterdam study. Arch Neurol. 1998;55:964. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- 3.Fortea J., Pegueroles J., Alcolea D., et al. APOE4 homozygozity represents a distinct genetic form of Alzheimer's disease. Nat Med. 2024;30(5):1284–1291. doi: 10.1038/s41591-024-02931-w. [DOI] [PubMed] [Google Scholar]
- 4.Hamer M., Chida Y. Physical activity and risk of neurodegenerative disease: a systematic review of prospective evidence. Psychol Med. 2008;39:3–11. doi: 10.1017/S0033291708003681. [DOI] [PubMed] [Google Scholar]
- 5.Montagne A., Nation D.A., Sagare A.P., et al. APOE4 leads to blood–brain barrier dysfunction predicting cognitive decline. Nature. 2020;581:71–76. doi: 10.1038/s41586-020-2247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Small G.W., Ercoli L.M., Silverman D.H.S., et al. Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer's disease. Proc Natl Acad Sci U S A. 2000;97:6037–6042. doi: 10.1073/pnas.090106797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baek M.S., Cho H., Lee H.S., Lee J.H., Ryu Y.H., Lyoo C.H. Effect of APOE ε4 genotype on amyloid-β and tau accumulation in Alzheimer's disease. Alzheimers Res Ther. 2020;12:140. doi: 10.1186/s13195-020-00710-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishitsuji K., Hosono T., Nakamura T., Bu G., Michikawa M. Apolipoprotein E regulates the integrity of tight junctions in an isoform-dependent manner in an in vitro blood-brain barrier model. J Biol Chem. 2011;286:17536–17542. doi: 10.1074/jbc.M111.225532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marottoli F.M., Trevino T.N., Geng X., et al. Autocrine effects of brain endothelial cell-produced human apolipoprotein E on metabolism and inflammation in vitro. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.668296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montagne A., Nikolakopoulou A.M., Huuskonen M.T., et al. APOE4 accelerates advanced-stage vascular and neurodegenerative disorder in old Alzheimer's mice via cyclophilin A independently of amyloid-β. Nat Aging. 2021;1:506–520. doi: 10.1038/s43587-021-00073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams H.C., Farmer B.C., Piron M.A., et al. APOE alters glucose flux through central carbon pathways in astrocytes. Neurobiol Dis. 2020;136 doi: 10.1016/j.nbd.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J., Min L., Liu R., et al. The effect of exercise on cerebral blood flow and executive function among young adults: a double-blinded randomized controlled trial. Sci Rep. 2023;13:8269. doi: 10.1038/s41598-023-33063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J.C., Nielson K.A., Woodard J.L., et al. Physical activity reduces hippocampal atrophy in elders at genetic risk for Alzheimer's disease. Front Aging Neurosci. 2014;6 doi: 10.3389/fnagi.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dougherty R.J., Schultz S.A., Kirby T.K., et al. Moderate physical activity is associated with cerebral glucose metabolism in adults at risk for Alzheimer's disease. J Alzheimers Dis. 2017;58:1089–1097. doi: 10.3233/JAD-161067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He X., Liu D., Zhang Q., et al. Voluntary exercise promotes glymphatic clearance of amyloid beta and reduces the activation of astrocytes and microglia in aged mice. Front Mol Neurosci. 2017;10 doi: 10.3389/fnmol.2017.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker L.D., Frank L.L., Foster-Schubert K., et al. Effects of aerobic exercise on mild cognitive impairment. Arch Neurol. 2010;67 doi: 10.1001/archneurol.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nay K., Smiles W.J., Kaiser J., et al. Molecular mechanisms underlying the beneficial effects of exercise on brain function and neurological disorders. Int J Mol Sci. 2021;22:4052. doi: 10.3390/ijms22084052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lourenco M.V., Frozza R.L., de Freitas G.B., et al. Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer's models. Nat Med. 2019;25:165–175. doi: 10.1038/s41591-018-0275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Hayek L., Khalifeh M., Zibara V., et al. Lactate mediates the effects of exercise on learning and memory through SIRT1-dependent activation of hippocampal brain-derived neurotrophic factor (BDNF) J Neurosci. 2019;39:1661–1718. doi: 10.1523/JNEUROSCI.1661-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon H.Y., Becke A., Berron D., et al. Running-induced systemic cathepsin B secretion is associated with memory function. Cell Metab. 2016;24:332–340. doi: 10.1016/j.cmet.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaynman S., Ying Z., Gomez-Pinilla F. Hippocampal BDNF mediates the efficacy of exercise on synaptic plasticity and cognition. Eur J Neurosci. 2004;20:2580–2590. doi: 10.1111/j.1460-9568.2004.03720.x. [DOI] [PubMed] [Google Scholar]
- 22.Szuhany K.L., Bugatti M., Otto M.W. A meta-analytic review of the effects of exercise on brain-derived neurotrophic factor. J Psychiatr Res. 2015;60:56–64. doi: 10.1016/j.jpsychires.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber C.M., Moiz B., Zic S.M., Alpízar Vargas V., Li A., Clyne A.M. Induced pluripotent stem cell-derived cells model brain microvascular endothelial cell glucose metabolism. Fluids Barriers CNS. 2022;19:98. doi: 10.1186/s12987-022-00395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percie du Sert N., Hurst V., Ahluwalia A., et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J., Vodyanik M.A., Smuga-Otto K., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 26.Schaffer S., Lam V.Y.M., Ernst I.M.A., Huebbe P., Rimbach G., Halliwell B. Variability in APOE genotype status in human-derived cell lines: a cause for concern in cell culture studies? Genes Nutr. 2014;9:364. doi: 10.1007/s12263-013-0364-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid B., Prehn K.R., Nimsanor N., et al. Generation of a set of isogenic, gene-edited iPSC lines homozygous for all main APOE variants and an APOE knock-out line. Stem Cell Res. 2019;34 doi: 10.1016/j.scr.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Neal E.H., Marinelli N.A., Shi Y., et al. A simplified, fully defined differentiation scheme for producing blood-brain barrier endothelial cells from human iPSCs. Stem Cell Rep. 2019;12:1380–1388. doi: 10.1016/j.stemcr.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neal E.H., Katdare K.A., Shi Y., Marinelli N.A., Hagerla K.A., Lippmann E.S. Influence of basal media composition on barrier fidelity within human pluripotent stem cell-derived blood-brain barrier models. J Neurochem. 2021;159:980–991. doi: 10.1111/jnc.15532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corbo R.M., Scacchi R. Apolipoprotein E (APOE) allele distribution in the world. Is APOE∗4 a ‘thrifty’ allele? Ann Hum Genet. 1999;63:301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 31.Gray K.M., Jung J.W., Inglut C.T., Huang H.C., Stroka K.M. Quantitatively relating brain endothelial cell-cell junction phenotype to global and local barrier properties under varied culture conditions via the Junction Analyzer Program. Fluids Barriers CNS. 2020;17:1–20. doi: 10.1186/s12987-020-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemkov T., Hansen K.C., D'Alessandro A. A three-minute method for high-throughput quantitative metabolomics and quantitative tracing experiments of central carbon and nitrogen pathways. Rapid Commun Mass Spectrom. 2017;31:663–673. doi: 10.1002/rcm.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemkov T., D'Alessandro A., Hansen K.C. Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole orbitrap mass spectrometry. Amino Acids. 2015;47:2345–2357. doi: 10.1007/s00726-015-2019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Millard P., Delépine B., Guionnet M., Heuillet M., Bellvert F., Létisse F. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics. 2019;35:4484–4487. doi: 10.1093/bioinformatics/btz209. [DOI] [PubMed] [Google Scholar]
- 35.Pang Z., Zhou G., Ewald J., et al. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat Protoc. 2022;17:1735–1761. doi: 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- 36.Young J.D. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–1335. doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahim M., Ragavan M., Deja S., Merritt M.E., Burgess S.C., Young J.D. Inca 2.0: a tool for integrated, dynamic modeling of NMR-and MS-based isotopomer measurements and rigorous metabolic flux analysis. Metab Eng. 2022;69:1096–7176. doi: 10.1016/j.ymben.2021.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moiz B., Garcia J., Basehore S., et al. 13 C metabolic flux analysis indicates endothelial cells Attenuate metabolic perturbations by modulating tca activity. Metabolites. 2021;11 doi: 10.3390/metabo11040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antoniewicz M.R. A guide to 13C metabolic flux analysis for the cancer biologist. Exp Mol Med. 2018;50 doi: 10.1038/s12276-018-0060-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theendakara V., Peters-Libeu C.A., Spilman P., Poksay K.S., Bredesen D.E., Rao R.V. Direct transcriptional effects of apolipoprotein E. J Neurosci. 2016;36:685–700. doi: 10.1523/JNEUROSCI.3562-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theendakara V., Patent A., Libeu C.A.P., et al. Neuroprotective sirtuin ratio reversed by ApoE 4. Proc Natl Acad Sci U S A. 2013;110:18303–18308. doi: 10.1073/pnas.1314145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu C., Hao S., Xu X., et al. Activation of SIRT1 ameliorates LPS-induced lung injury in mice via decreasing endothelial tight junction permeability. Acta Pharmacol Sin. 2019;40:630–641. doi: 10.1038/s41401-018-0045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Cui G., Wang Y., Gong Y., Wang Y. SIRT1 activation alleviates brain microvascular endothelial dysfunction in peroxisomal disorders. Int J Mol Med. 2019;44(3):995–1005. doi: 10.3892/ijmm.2019.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J., Cao L., Li Z., Li Y. SIRT1 promotes GLUT1 expression and bladder cancer progression via regulation of glucose uptake. Hum Cell. 2019;32:193–201. doi: 10.1007/s13577-019-00237-5. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y., Yang H., Chen S., et al. SIRT1 regulated hexokinase-2 promoting glycolysis is involved in hydroquinone-enhanced malignant progression in human lymphoblastoid TK6 cells. Ecotoxicol Environ Saf. 2022;241 doi: 10.1016/j.ecoenv.2022.113757. [DOI] [PubMed] [Google Scholar]
- 46.Moiz B., Sriram G., Morss Clyne A. Interpreting metabolic complexity via isotope-assisted metabolic flux analysis Biochemical. Trends Biochem Sci. 2023;48 doi: 10.1016/j.tibs.2023.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao N., Liu C.-C., Van Ingelgom A.J., et al. Apolipoprotein E4 impairs neuronal insulin signaling by trapping insulin receptor in the endosomes. Neuron. 2017;96:115–129.e5. doi: 10.1016/j.neuron.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S., Zhu L., Peng Y., et al. Long-term running exercise improves cognitive function and promotes microglial glucose metabolism and morphological plasticity in the hippocampus of APP/PS1 mice. J Neuroinflammation. 2022;19:34. doi: 10.1186/s12974-022-02401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantó C., Gerhart-Hines Z., Feige J.N., et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng C.-K., Shang W., Liu J., et al. Activation of AMPK/miR-181b Axis alleviates endothelial dysfunction and vascular inflammation in diabetic mice. Antioxidants. 2022;11:1137. doi: 10.3390/antiox11061137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaitán J.M., Boots E.A., Dougherty R.J., et al. Brain glucose metabolism, cognition, and cardiorespiratory fitness following exercise training in adults at risk for Alzheimer's disease. Brain Plast. 2019;5:83–95. doi: 10.3233/BPL-190093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatovic S.M., Martinez-Revollar G., Hu A., Choi J., Keep R.F., Andjelkovic A.V. Decline in Sirtuin-1 expression and activity plays a critical role in blood-brain barrier permeability in aging. Neurobiol Dis. 2019;126:105–116. doi: 10.1016/j.nbd.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang X., Chen J., Sun L., Xu Y. SIRT1 deacetylates KLF4 to activate Claudin-5 transcription in ovarian cancer cells. J Cell Biochem. 2018;119:2418–2426. doi: 10.1002/jcb.26404. [DOI] [PubMed] [Google Scholar]
- 54.Ma J., Wang P., Liu Y., Zhao L., Li Z., Xue Y. Krüppel-like factor 4 regulates blood-tumor barrier permeability via ZO-1, occludin and claudin-5. J Cell Physiol. 2014;229:916–926. doi: 10.1002/jcp.24523. [DOI] [PubMed] [Google Scholar]
- 55.Zhang X., Wu L., Swerdlow R.H., Zhao L. Opposing effects of ApoE2 and ApoE4 on glycolytic metabolism in neuronal aging supports a warburg neuroprotective cascade against Alzheimer's disease. Cells. 2023;12:410. doi: 10.3390/cells12030410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagamatsu S., Kornhauser J.M., Burant C.F., Seino S., Mayo K.E., Bell G.I. Glucose transporter expression in brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose transporter isoform, and identification of sites of expression by in situ hybridization. J Biol Chem. 1992;267:467–472. doi: 10.1016/S0021-9258(18)48518-3. [DOI] [PubMed] [Google Scholar]
- 57.Taha C., Mitsumoto Y., Liu Z., Skolnik E.Y., Klip A. The insulin-dependent biosynthesis of GLUT1 and GLUT3 glucose transporters in L6 muscle cells is mediated by distinct pathways. J Biol Chem. 1995;270:24678–24681. doi: 10.1074/jbc.270.42.24678. [DOI] [PubMed] [Google Scholar]
- 58.Frazier H.N., Ghoweri A.O., Anderson K.L., et al. Elevating insulin signaling using a constitutively active insulin receptor increases glucose metabolism and expression of GLUT3 in hippocampal neurons. Front Neurosci. 2020;14 doi: 10.3389/fnins.2020.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riskin A., Nannegari V.H., Mond Y. Acute effectors of GLUT1 glucose transporter subcellular targeting in CIT3 mouse mammary epithelial cells. Pediatr Res. 2008;63:56–61. doi: 10.1203/PDR.0b013e31815b440b. [DOI] [PubMed] [Google Scholar]
- 60.Egert S., Nguyen N., Schwaiger M. Myocardial glucose transporter GLUT1: translocation induced by insulin and ischemia. J Mol Cell Cardiol. 1999;31:1337–1344. doi: 10.1006/jmcc.1999.0965. [DOI] [PubMed] [Google Scholar]
- 61.Ngarmukos C., Baur E.L., Kumagai A.K. Co-localization of GLUT1 and GLUT4 in the blood–brain barrier of the rat ventromedial hypothalamus. Brain Res. 2001;900:1–8. doi: 10.1016/S0006-8993(01)02184-9. [DOI] [PubMed] [Google Scholar]
- 62.Nguyen H.M., Mejia E.M., Chang W., et al. Reduction in cardiolipin decreases mitochondrial spare respiratory capacity and increases glucose transport into and across human brain cerebral microvascular endothelial cells. J Neurochem. 2016;139:68–80. doi: 10.1111/jnc.13753. [DOI] [PubMed] [Google Scholar]
- 63.Leung S.W.S., Shi Y. The glycolytic process in endothelial cells and its implications. Acta Pharmacol Sin. 2022;43:251–259. doi: 10.1038/s41401-021-00647-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metallo C.M., Gameiro P.A., Bell E.L., et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Patel M.S., Jomain-Baum M., Ballard F.J., Hanson R.W. Pathway of carbon flow during fatty acid synthesis from lactate and pyruvate in rat adipose tissue. J Lipid Res. 1971;12:179–191. doi: 10.1016/S0022-2275(20)39528-6. [DOI] [PubMed] [Google Scholar]
- 66.Haney M.S., Pálovics R., Munson C.N., et al. APOE4/4 is linked to damaging lipid droplets in Alzheimer's disease microglia. Nature. 2024;628:154–161. doi: 10.1038/s41586-024-07185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou X., Xu S., Maitland-Toolan K.A., et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283:20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cerutti R., Pirinen E., Lamperti C., et al. NAD+-Dependent activation of Sirt1 corrects the phenotype in a mouse model of mitochondrial disease. Cell Metab. 2014;19:1042–1049. doi: 10.1016/j.cmet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Juan C.G., Matchett K.B., Davison G.W. A systematic review and meta-analysis of the SIRT1 response to exercise. Sci Rep. 2023;13 doi: 10.1038/s41598-023-38843-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleinloog J.P.D., Mensink R.P., Ivanov D., Adam J.J., Uludağ K., Joris P.J. Aerobic exercise training improves cerebral blood flow and executive function: a randomized, controlled cross-over trial in sedentary older men. Front Aging Neurosci. 2019;11 doi: 10.3389/fnagi.2019.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cucullo L., Hossain M., Puvenna V., Marchi N., Janigro D. The role of shear stress in Blood-Brain Barrier endothelial physiology. BMC Neurosci. 2011;12 doi: 10.1186/1471-2202-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ranadewa D., Wu J., Subramanianbalachandar V.A., Steward R.L. Variable fluid flow regimes alter human brain microvascular endothelial cell–cell junctions and cytoskeletal structure. Cytoskeleton. 2021;78:323–334. doi: 10.1002/cm.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bennett N.K., Nguyen M.K., Darch M.A., et al. Defining the ATPome reveals cross-optimization of metabolic pathways. Nat Commun. 2020;11:4319. doi: 10.1038/s41467-020-18084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Foley K.E., Diemler C.A., Hewes A.A., Garceau D.T., Sasner M., Howell G.R. APOE ε4 and exercise interact in a sex-specific manner to modulate dementia risk factors. Alzheimers Dementia. 2022;8 doi: 10.1002/trc2.12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu T.M., Houghton S., Magdeldin T., et al. Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2016950118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.