Abstract
Mental health disorders emerge from complex interactions among neurobiological processes across multiple scales, which poses challenges in uncovering pathological pathways from molecular dysfunction to neuroimaging changes. Here, we proposed a multiscale fusion (mFusion) method to evaluate the relevance of each gene to the neuroimaging traits of mental health disorders. We combined gene-neuroimaging associations with gene-positron emission tomography (PET) and PET-neuroimaging associations using protein-protein interaction networks, where various genes traced by PET maps are involved in neurotransmission. Compared with previous methods, the proposed algorithm identified more disease genes on both simulated and empirical data sets. Applying mFusion to eight mental health disorders, we found that these disorders formed three clusters with distinct associated genes. In summary, mFusion is a promising tool of prioritizing genes for mental health disorders by establishing gene-PET-neuroimaging pathways.
Subject terms: Computational models, Gene expression
We introduced mFusion, a method that integrates gene and neuroimaging data to identify disease-related genes in mental disorders. By analyzing gene interactions and PET data, mFusion successfully clusters disorders and highlight critical gene pathways.
Introduction
Mental health disorders, constituting 16% of the global burden of diseases, rank among the leading causes of disability worldwide1. In severe cases, they can diminish life expectancy by 10 to 20 years2. Despite substantial progresses in understanding molecular mechanisms of brain functions in animal models, the rate of successful clinical translations to humans remains notably low3. The primary obstacle lies in the current knowledge gap between molecular processes4 and psychiatric symptoms. There exist many complex interactions across multiple scales from genes, through neurotransmitters, to neural networks. This complexity is compounded by the challenge of concurrently collecting multiscale data within the human brain. As human brain data rapidly accumulate but separately at various scales, there is an urgent need for dedicated analytic method to integrate these data comprehensively, enabling the discovery of insights into mental health disorders.
At present, some public collection databases can identify disease-related genes, such as DisGeNET5 and CTD (Comparative Toxicogenomics Database)6, but they lack the capacity to establish connections with neurotransmitter systems or pathways. Both gene differential expression analysis and Genome-wide association study (GWAS) analysis fall short in addressing this challenge7, with limited coverage of disease phenotypes. Partial Least Squares (PLS) regression analysis can establish associations between genes and imaging phenotypes based on spatial molecular distribution patterns in the brain8,9. However, it can only perform pairwise correlation analysis, necessitating a method to facilitate the establishment of cross-scale pathway associations.
Neuroimaging studies have identified various alterations in neuroimaging features of human brains associated with mental health disorders, i.e., spatial distributions of alterations across different brain regions in psychiatric patients compared with healthy controls10. Leveraging transcriptomic data from postmortem brain tissues11, researchers have initiated efforts to correlate neuroimaging features with gene expressions, prioritizing relevant genes and molecular pathways12. In this way, genes associated with neurodevelopment, neuroplasticity, and neurotransmission have been implicated in autism spectrum disorder (ASD)9 and schizophrenia (SCZ)13. Despite these progresses, a significant knowledge gap persists between gene expressions and neuroimaging traits. Recently, positron emission tomography (PET) studies have started to reveal spatial associations between neurotransmitter receptors/transporters and structural/functional traits of mental health disorders in the human brain14,15. Leveraging neurotransmissions revealed by PET images, this study aims to establish biological bridges for the gap between gene expressions and neuroimaging traits for mental disorders. The disease related genes are defined by 4 curated disease gene databases listed in Table 1.
Table 1.
Four gene-disease databases
| Database | # of SCZ risk genes | # of ASD risk genes | Collection date | URL |
|---|---|---|---|---|
| DisGeNet | 2872 (score > 0) | 1071 (score > 0) | June, 2020 (v7.0) | https://www.disgenet.org/ |
| CTD | 2875 (score > 15.28) | 1071 (score > 29) | June 30, 2023 (17123) | https://ctdbase.org/ |
| DISEASES | 1548 (Z > 3) | 211 (Z > 3) | March, 2015 | https://diseases.jensenlab.org/Downloads |
| PGC-GWAS | 380 (p < 5e-8) | 56 (p < 5e-4) |
SCZ:202257/ ASD:201953 |
https://pgc.unc.edu/for-researchers/download-results/ |
This study proposes a multiscale fusion (mFusion) method to bridge genes to mental disorders through establishing links between gene expressions in brain tissues, neurotransmissions, and neuroimaging traits of these disorders. Leveraging the knowledge in the protein-protein interaction (PPI) network made available by the STRING database16, mFusion provides a tool for integrating 15,408 gene expression maps from the Allen Human Brain Atlas (AHBA)17,18, 45 PET maps across various neurotransmitter systems14,19,20, and neuroimaging traits associated with mental disorders. Performances of mFusion were first evaluated by numerical simulations, and then demonstrated by applying to neuroimaging traits of two mental disorders (i.e., autism9 and schizophrenia13). The ENIGMA (Enhancing NeuroImaging Genetics through Meta-Analysis) consortium has reported neuroimaging traits for mental disorders by analyzing thousands of neuroimaging scans21. Using these neuroimaging traits, mFusion enabled us to reveal the clustering structure for eight major mental disorders.
Results
Overview of mFusion framework
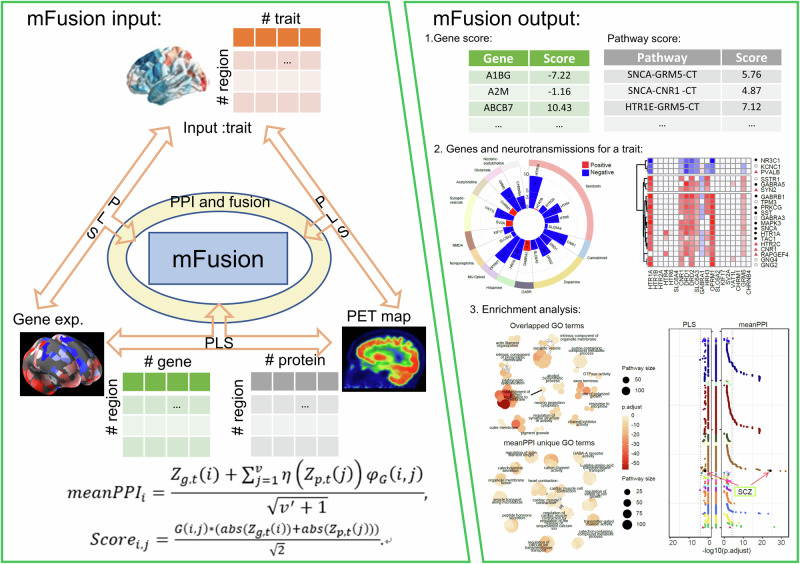
In this study, the mFusion integrated gene expressions in brain tissues and PET maps for specific proteins (related to the receptors, transporters, or release of neurotransmitters) within a PPI network, to link neuroimaging traits to genes (Fig. 1; Additional file 1: Fig. S1) through proteins (measured by PET maps; Table 2; Supplementary Table S1). First, we examined Z-scores value of genes or proteins from three types of (PLS) associations independently, including gene-trait, PET-trait, and gene-PET associations. Second, we utilized the Z-transform test, also referred to as the “Stouffer’s method”22, to combine multiscale Z-scores of a gene. Meanwhile, the neighboring information of PPI network from STRING database was used to boost the ability of identifying disease related genes. Finally, disease category5 and Gene Ontology (GO)23 term enrichment analysis was conducted on the top-ranked genes, which were determined by the mFusion methods, to identify important biomolecular pathways or processes that relate to candidate genes. Further details are provided in Methods, and Supplementary Fig. S1.
Fig. 1. The framework and working interface of the “mFusion” method.
By using partial least square association to integrate spatial correlations of gene expressions in the human brain with information about neurotransmission and neuroimaging, the mFusion method yields a relevance score for each gene and pathway associated with a mental disorder, facilitating the identification of top-ranked genes and pathways. This fusion method additively provided the potential reasons for neurochemical architectures (neurotransmissions) in PET images influencing gene scores. Subsequent enrichment analysis of top genes identifies biological process and pathways relate to the mental disorder.
Table 2.
Neurotransmission-related PET maps included in analyses
| Protein | Neurotransmitter | Tracer | Measure | n | Age | Reference |
|---|---|---|---|---|---|---|
| HTR1A | Serotonin | [11C]CUMI-101 | BPND | 8 (5) | 28.4 ± 8.8 | Beliveau et al.75 |
| HTR1A | Serotonin | [11C]WAY-100635 | BPND | 35 (17) | 26.3 ± 5.2 | Savli et al.76 |
| HTR1B | Serotonin | [11C]AZ10419369 | BPND | 36 (12) | 27.8 ± 6.9 | Beliveau et al.75 |
| HTR1B | Serotonin | [11C]P943 | BPND | 23 (8) | 28.7 ± 7.0 | Savli et al.76 |
| HTR1B | Serotonin | [11C]P943 | BPND | 65 (16) | 33.7 ± 9.7 | Gallezot et al.77 |
| HTR2A | Serotonin | [18F]altanserin | BPND | 19 (8) | 28.2 ± 5.7 | Savli et al.76 |
| HTR2A | Serotonin | [11C]Cimbi-36 | BPND | 29 (14) | 22.6 ± 2.7 | Beliveau et al.75 |
| HTR2A | Serotonin | [11C]MDL100907 | BPND | 3 (1) | 35 ± 9 | Talbot et al.78 |
| HTR4 | Serotonin | [11C]SB207145 | BPND | 59 (18) | 25.9 ± 5.3 | Beliveau et al.75 |
| HTR6 | Serotonin | [11C]GSK215083 | BPND | 30 (0) | 36.6 ± 9.0 | Radhakrishnan et al.79 |
| SLC6A4 | Serotonin | [11C]DASB | BPND | 100 (71) | 25.1 ± 5.8 | Beliveau et al.75 |
| SLC6A4 | Serotonin | [11C]DASB | BPND | 18 (6) | 30.5 ± 9.5 | Savli et al.76 |
| SLC6A4 | Serotonin | [11C]MADAM | BPND | 10 (2) | range: 51–67 | Fazio et al.80 |
| SLC6A4 | Serotonin | [11C]MADAM | BPND | 16 (2) | range: 21–67 | Dukart et al.20 |
| CNR1 | Cannabinoid | [18F]FMPEP-d2 | VT | 22 (11) | male: 27 ± 6; female: 28 ± 10 | Laurikainen et al.81 |
| CNR1 | Cannabinoid | [11C]OMAR | VT | 77 (28) | 30.0 ± 8.9 | Normandin et al.82. |
| DRD1 | Dopamine | [11C]SCH23390 | BPND | 13 (7) | 33 ± 13 | Kaller et al.83. |
| DRD2 | Dopamine | [11C]FLB457 | BPND | 55 (29) | 32.5 ± 9.7 | Hansen et al.14. |
| DRD2 | Dopamine | [11C]FLB457 | BPND | 6 (2) | 39.5 ± 6.8 | Sandiego et al.84. |
| DRD2 | Dopamine | [18F]fallypride | BPND | 58 (22) | 18.5 ± 0.6 | Jaworska et al.85. |
| DRD2 | Dopamine | [11C]FLB457 | BPND | 37 (20) | 48.4 ± 16.9 | Smith et al.86. |
| DRD2 | Dopamine | [11C]raclopride | BPND | 7 (0) | 24 ± 2 | Alakurtti et al.87. |
| SLC6A3 | Dopamine | [123I]FP-CIT | SUVR | 174 (65) | 61 ± 11 | Dukart et al.88. |
| SLC6A3 | Dopamine | [123I]Ioflupano | SUVR | 26 (--) | range 35 ~ 65 | García-G et al.89. |
| SLC6A3 | Dopamine | [18F]FE-PE2I | SUVR | 10 (0) | 28.1 ± 6.9 | Sasaki et al.90. |
| GABRA1 | GABA | -- | -- | 26 (0) | 26 ± 5 | Dukart et al.88. |
| GABRA1 | GABA | [11C]flumazenil | Bmax | 16 (9) | 26.6 ± 8 | Nørgaard et al91. |
| HRH3 | Histamine | [11C]GSK189254 | VT | 8 (1) | 31.7 ± 9.0 | Gallezot et al.92. |
| OPRM1 | Opioid | [11C]carfentanil | BPND | 204 (72) | 32.3 ± 10.8 | Kantonen et al.93. |
| OPRM1 | Opioid | [11C]carfentanil | BPND | 39 (19) | 37.0 ± 4.9 | Turtonen et al.94. |
| SLC6A2 | Norepinephrine | [11C]MRB | BPND | 77 (27) | 33.4 ± 9.2 | Ding et al.95. |
| SLC6A2 | Norepinephrine | [11C]MRB | BPND | 20 (8) | 33.3 ± 10.0 | Hesse et al.96. |
| KIF17 | Glutamate | [18F]GE-179 | VT | 29 (8) | 40.9 ± 12.7 | Galovic et al.97. |
| SV2A* | -- | [11C]UCB-J | BPND | 10 (3) | 36 ± 10 | Finnema et al.98. |
| VAT1L | Acetylcholine | [18F]FEOBV | SUVR | 5 (4) | 68.4 ± 3.4 | Hansen et al.14. |
| VAT1L | Acetylcholine | [18F]FEOBV | SUVR | 6 (3) | 67.0 ± 11.1 | Aghourian et al.99. |
| VAT1L | Acetylcholine | [18F]FEOBV | SUVR | 4 (1) | 37 ± 10.2 | PI: Lauri Tuominen & Synthia Guimond |
| VAT1L | Acetylcholine | [18F]FEOBV | SUVR | 18 (13) | 66.8 ± 6.8 | Hansen et al.14. |
| VAT1L | Acetylcholine | [18F]FEOBV | SUVR | 5 (1) | 68.3 ± 3.1 | Bedard et al.100. |
| CHRM1 | Acetylcholine | [11C]LSN3172176 | BPND | 24 (11) | 40.5 ± 11.7 | Naganawa et al.101. |
| GRM5 | Glutamate | [11C]ABP688 | BPND | 22 (10) | 67.9 ± 9.6 | PI: Rosa-Neto, P. & Kobayashi, E. |
| GRM5 | Glutamate | [11C]ABP688 | BPND | 28 (13) | 33.1 ± 11.2 | DuBois et al.102. |
| GRM5 | Glutamate | [11C]ABP688 | BPND | 74 (49) | 20 ± 3.0 | Smart et al.103. |
| GRM5 | Glutamate | [11C]ABP688 | BPND | 22 (10) | 67.9 ± 9.6 | Hansen et al.14. |
| CHRNA4 | Acetylcholine | [18F]Flubatine | VT | 30 (10) | 33.5 ± 10.7 | Hillmer et al.104. |
The Protein column indicate the protein names in the STRING database. Supplementary Table S1 also includes more extensive methodological details, such as Excitatory/Inhibitory, Ionotropic/Metabotropic, and Source toolkit. Values in parentheses (under n) indicate the number of females.
BPND parametric and regional non-displaceable binding potential, Bmax density (pmol ml−1) converted from binding potential (5-HT) or distributional volume (GABA) using autoradiography-derived densities, VT tracer distribution volume, SUVR standardized uptake value ratio.
*The synaptic vesicle glycoprotein 2 A(SV2A) is targeted by PET imaging to quantify synaptic density in human brains98.
mFusion outperformed the traditional method on simulation data
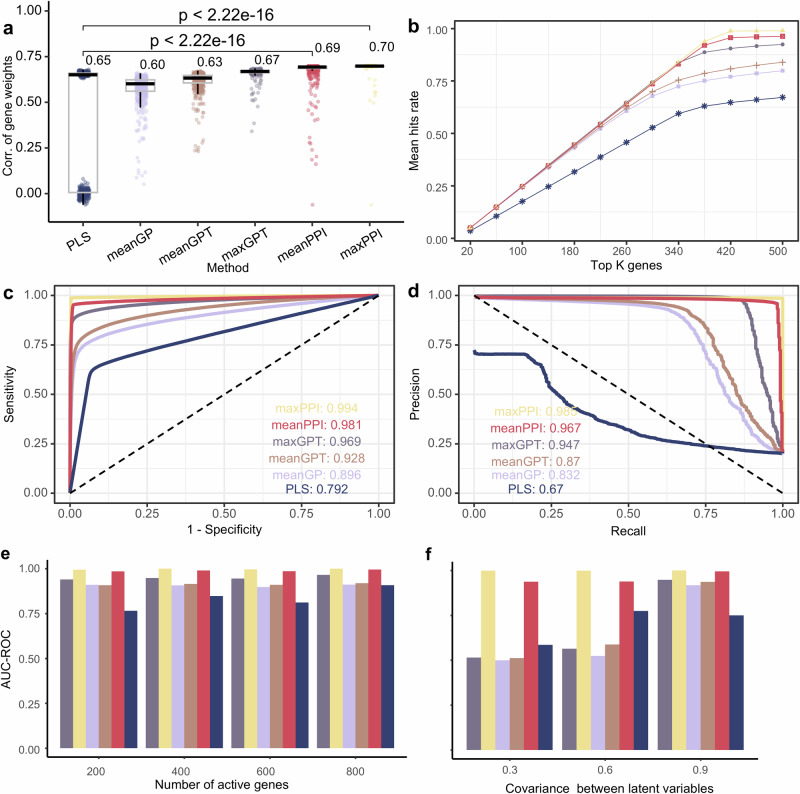
We compared performance on simulation data between the traditional partial least squares (PLS) association method, and five fusion methods proposed by this study (i.e., meanGP, meanGPT, meanPPI, maxGPT, and maxPPI, see “Methods”). Evaluation metrics included the correlation between estimated gene scores and real gene weights, the number (or rate) of hits, the area under curve (AUC) of receiver operating characteristic (ROC), AUC of precision-recall (PR) curve (see “Methods”).
Compared with other methods, we found that gene scores given by the meanPPI and maxPPI methods demonstrated higher correlation with real gene weights defined in the simulation model (Fig. 2a, unpaired Wilcoxon test, 500 times of simulations), higher hit rates of active genes in the simulation (Fig. 2b), and larger AUCs of both the ROC (Fig. 2c) and PR (Fig. 2d) curves, these curves were all generated by the mean value of 500 times of simulations.
Fig. 2. Evaluation of fusion methods from simulated datasets.
a The correlation between real gene weights and fusion weights measured by different fusion methods of 500 simulated experiments. The lower whisker extends from the first quartile (Q1) to the smallest data point that is within 1.5 * interquartile range (IQR) below Q1. The upper whisker extends from the third quartile (Q3) to the largest data point that is within 1.5 * IQR above Q3. The number next to bar represents the median of the population (using unpaired Wilcoxon test). b Average hit rates of genes in all 500 simulations. The hit rate was measured by the rate of really active genes in top K genes ranked by specific fusion method. c ROC (Receiver Operating Characteristic) curve of different fusion methods on simulation data. In simulation experiments, is completely accurate connection matrix, and this noiseless PPI information greatly improves the performance of maxPPI and meanPPI methods, so the AUC-ROC of maxPPI is 1. d PR (precision-recall) curve of different fusion methods on simulation data. e AUC-ROC value of different fusion method when number of active genes changed. f AUC-ROC value of different fusion method when covariance between latent variables changed.
We tested the performance of mFusion under different conditions as defined by both the sparsity in activate genes and the strength of the gene-PET covariance (Methods). The AUC-ROCs of both meanPPI and maxPPI outperformed the PLS method at different sparse levels of activate genes (Fig. 2e). Conversely, the results presented in Fig. 2f indicate that the two fusion methods, meanPPI and maxPPI, exhibited insensitivity to changes in the covariance between gene expression and neurotransmission PET maps.
And then, three kinds of perturbations were performed on the PPI networks to illustrated the influence of PPI information on the mFusion method for 500 repetitions: (1) randomly shuffle 30% of the elements within the adjacency matrix ; (2) set the minimum 30% of the elements in the adjacency matrix to be zero; (3) randomly shuffle 30% of the elements, and then set the minimum 30% of the elements in the adjacency matrix to be zero. We found that the meanPPI and maxPPI methods consistently outperformed their counterparts in all three conditions (Fig. S2).
Thirdly, we conducted a simulation of brain maps at three distinct spatial resolutions. Specifically, the number of brain regions (n) was varied between 100, 200, and 500 (see “Methods” for further details), as delineated in Fig. S3. The results of this simulation demonstrated a positive correlation between the spatial resolution of the X, Y, and Z matrices and the efficacy of the methods in identifying activated genes. Notably, the meanPPI and maxPPI methodologies consistently exhibited superior performance compared to other methods, exhibiting a level of stability that highlights their robustness in high-resolution brain mapping analyses.
mFusion outperformed the traditional method on empirical data
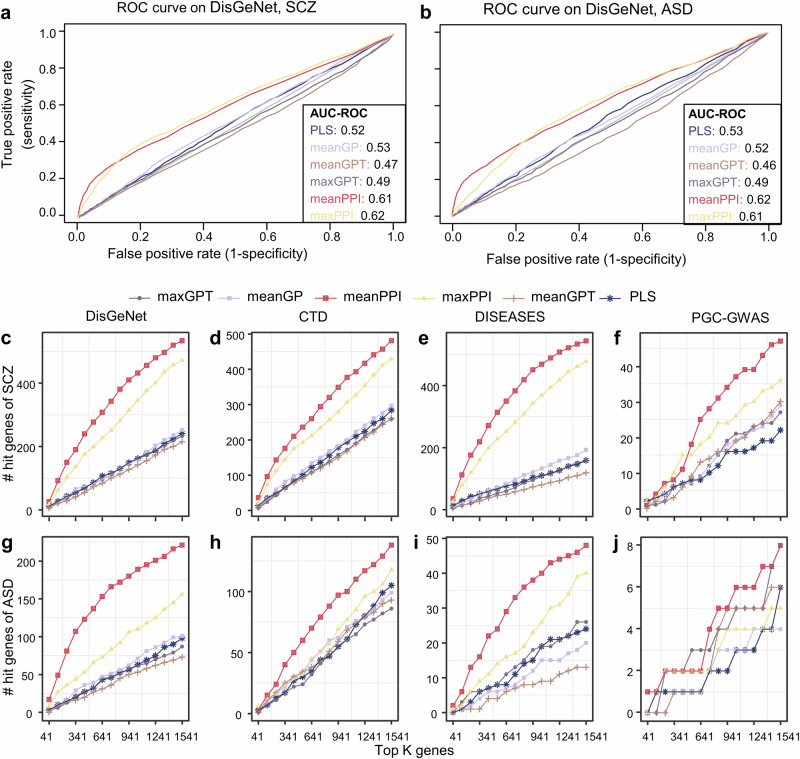
We used SCZ morphological similarity differences and ASD cortical thickness difference as the traits and get genes Z-scores from different fusion method, as described in Methods. Compared to the traditional PLS regression method and other fusion methods, the meanPPI and maxPPI method got a larger AUC on DisGeNet database (SCZ: Fig. 3a and Table S2; ASD: Fig. 3b and Table S3), which demonstrated superior identification of disorder-related genes. On the other hand, we compared the number of hits in the top K genes given by various methods. When we varied the parameter K from 41 to 1541, where 1541 was 10% of the total of 15,408 genes, we found that the proposed methods had consistently more hits as compared with the other algorithms (Fig. 3c–j). Notably, when referencing the DisGeNet database, the meanPPI method outperformed all the other methods in identifying SCZ-related hit genes significantly (Fig. 3c; p < 0.001, paired Wilcoxon test for meanPPI and PLS method. Gene scores refer to Supplementary Table S4). Among the ASD related genes in the DisGeNet database, the number of hit genes in the top K gene sets identified by the meanPPI method was also significantly greater than that identified by other five methods (Fig. 3g; p < 0.001, paired Wilcoxon test for meanPPI and PLS method. Gene scores refer to Supplementary Table S5). Furthermore, when compared to fusion methods lacking PPI information, such as meanGPT and maxGPT, their PPI-informed counterparts, meanPPI and maxPPI, consistently demonstrated superior performance across the board (Fig. 3c–j).
Fig. 3. Performance on SCZ and ASD disease of fusion methods under different disease databases.
a ROC curve of different fusion methods on DisGeNet database for SCZ. b ROC curve of different fusion methods on DisGeNet database for ASD. c–j Number of overlapped genes for SCZ (c–f) and ASD (g–j) in different standard datebases: DisGeNet, CTD, DISEASES, and PGC-GWAS datasets (corresponding to Table 1). Line types mean different fusion methods.
Sensitivity analysis on empirical data
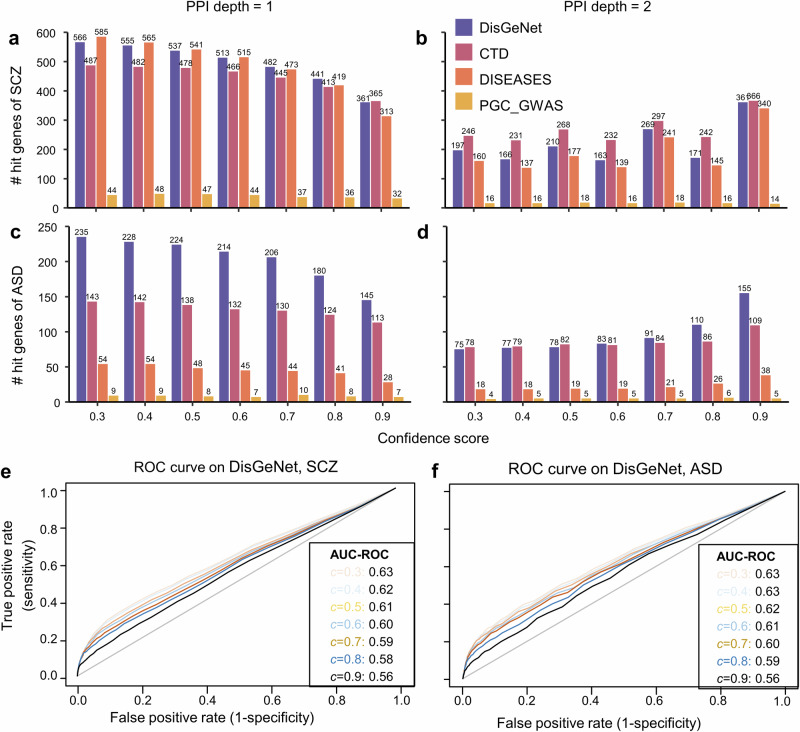
To identify optimal parameters for fusion methods, we compared performances of these methods with different network depths (d) and edge confidences (c) for the PPI. We observed that the meanPPI method exhibited superior performance (i.e., a larger number of hit genes, AUC-ROC value, or AUC-PR values) when its PPI depth d was set to 1 in comparison to 2 (Fig. 4 and Fig. S4). This trend was consistent across various edge confidence values ranging from 0.3 to 0.7. When the PPI depth was set as 2, meanPPI performed similarly to other methods (Fig. S5). Meanwhile, we noted that the meanPPI’s performance was less sensitive to the edge confidence of PPI when it varied from 0.3 to 0.7 (Fig. 4e, f). However, when it increased to 0.8 or 0.9, the meanPPI’s performance declined mainly owing to the fact that too few PPIs remained effective at such high confidence levels (Fig. 4 and Fig. S4). Using the physical subnetwork (i.e., with evidence of binding or forming a physical complex) instead of the full STRING PPI network, the meanPPI method exhibited a decrease in the number of hits. Nevertheless, it consistently outperformed other methods that did not incorporate the PPI information (Fig. S6). Consequently, we opted for d = 1 and c = 0.5 in subsequent analyses.
Fig. 4. Performance of meanPPI method on DisgeNet database with different threshold for pruning the PPI network.
a, b Number of hit genes for SCZ with different PPI depth d and confidence scores c, d = 1 in A and 2 in B, respectively. c, d Number of hit genes for ASD with different PPI depth and confidence scores, d = 1 in C and 2 in D, respectively. e ROC curve at different PPI confidence for SCZ. f ROC curve at different PPI confidence for ASD.
In order to evaluate the importance of PPIs in the context of the mFusion-meanPPI method, a comparative analysis was conducted on SCZ and ASD phenotypes separately. The analysis comprised a computational evaluation of 500 randomly generated PPIs for each disease (see “Methods”), with the resulting null distribution of the number of hit genes presented in Fig. S7A, B separately. The results demonstrated that the application of the meanPPI method using real PPI data markedly augmented the capacity to identify hit genes compared to the use of random PPI. In addition, a similar permutation was made for the 45 PET maps (see “Methods”) and reapplied to the analysis of the SCZ and ASD disease. The results in Figure S7C, D revealed a marked reduction in the ability of the meanPPI method in pinpointing disease-associated genes, thereby indicating that real PET maps are pivotal in the meanPPI method.
To assess the effect of the quality of PET maps on the results, the 45 redundant maps were synthesized and averaged into 20 unique maps (Fig. S8). Subsequently, the characteristics of SCZ and ASD were reanalyzed (Figs. S9, S10). The meanPPI method demonstrated remarkable consistency with the primary findings regarding the identification of disease risk genes, exhibiting a spearman correlation for gene scores of r = 0.97 (p < 2e-16) and r = 0.98 (p < 2e-16), respectively (Fig. S9). Furthermore, both the meanPPI and maxPPI methods emerged as the most effective approaches (Fig. S10).
Top-ranked genes enriched in the relevant diseases
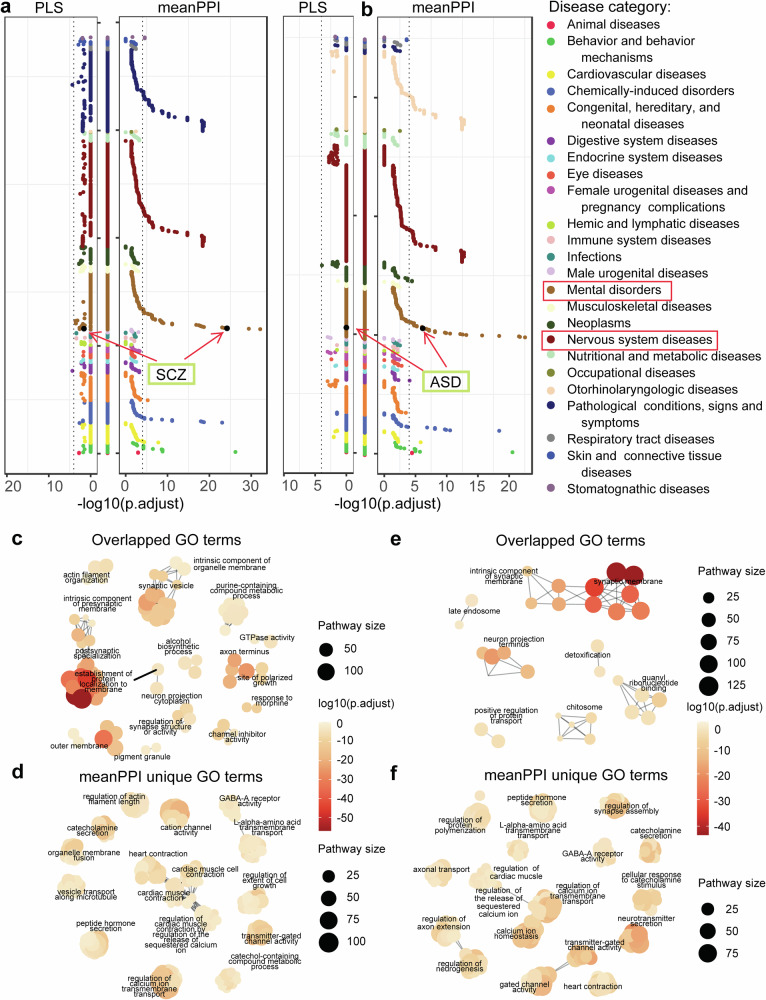
As an analysis module of mFusion analysis, we performed enrichment analysis for top 1541 (10% of 15,408) genes that had negative relevant scores to SCZ or ASD given by different methods (see “Methods”). Following the FDR correction among 30,170 diseases, traits, and phenotypes in the DisGeNet (Fig. 5a, b), genes prioritized by the meanPPI method for SCZ/ASD were enriched in the corresponding disease gene sets. In contrast, the top genes identified by the PLS method did not have such enrichments (Tables S8, S9).
Fig. 5. Enrichment analysis of top-ranked genes related to SCZ and ASD traits.
a, b Disease enrichment results in DisGeNet diseases on top 1541 trait-related genes for SCZ (a) and ASD (b). The Y-axis lists disease with categories in alphabetical order. c–f Clusters of GO terms enrichment results on top 1541 genes for SCZ (overlapped terms in c, terms uniquely enriched by meanPPI method in d) and ASD (overlapped terms in (e), terms uniquely enriched by meanPPI method in (f). The size and color of the dots were proportional to the number of pathway genes and enrichment significance, respectively. The p-values were adjusted using Bonferroni correction. Clusters were generated from enriched GO terms by aPEAR (Advanced Pathway Enrichment Analysis Representation) package. It exploits the similarities between pathway gene sets and represents them as a network of interconnected clusters. Each cluster is assigned a meaningful name that highlights the main biological theme of the experiment.
Top-ranked genes enriched in more biological pathways
For SCZ, the meanPPI and PLS methods shared enrichment in 92 GO terms, while the meanPPI had enriched 837 new GO terms. The shared terms included the establishment of protein localization to the membrane (GO_BP:0090150), regulation of synapse structure or activity (GO_BP:0050803), channel inhibitor activity (GO_MF:0008200), etc. (Fig. 5c; Table S6). Newly enriched terms of meanPPI included the calcium ion transport (GO_BP:0060402), cation channel activity (GO_MF:0022843), GABA-A receptor activity (GO_CC:1902711), etc. (Fig. 5d). Importantly, these unique biological processes have been implicated in SCZ24,25.
For ASD, these two methods shared enrichment in 38 GO terms, including the synaptic membrane (GO_CC:0097060), neuron projection terminus (GO_CC:0044306), positive regulation of protein transport (GO_BP:0051222), etc. (Fig. 5e). In comparison to the PLS results, the meanPPI results introduced new enrichments in 795 GO terms, including the gated channel activity (GO_MF:0022836), neurotransmitter secretion (GO_BP:0001956), GABA-A receptor activity (GO_MF:0004890), etc. (Fig. 5f; Table S7).
Top-ranked genes had more hits in a disease-related gene database
To characterize differences between genes prioritized by the proposed method (i.e., mFusion-meanPPI) and the traditional PLS method, we compared the top 1541 (10% of 15,408) genes identified by different ranking methods (Fig. 6). By comparing gene scores with disease-related genes listed in the DisGeNet database, we observed that higher meanPPI fusion scores were associated with higher hit rates. Since the PLS-regression is essentially a multivariate approach, which is prone to overfitting, we found more false positives in the genes with high PLS-regression weights. In contrast, we demonstrated that the mFusion-meanPPI approach reduced the false positive rate by combining the information from multiscale. Among the top 10% genes, the meanPPI method identified 534 SCZ-related genes listed in the DisGeNet database, which was significantly more than the 235 genes identified by traditional PLS method (p < 2.2e-16, Chi-squared test; Fig. 6a; Tables S4, S6). Similarly, among the 1071 ASD risk genes listed in the DisGeNet database, the meanPPI method identified 221 of them within the top 10% genes, which was significantly more than the 98 genes identified by the PLS method (p = 5.42e-13, Chi-squared test; Fig. 6b; Tables S5, S7). Therefore, the proposed approach identified more genes that have already been implicated in mental disorders than the traditional PLS method did.
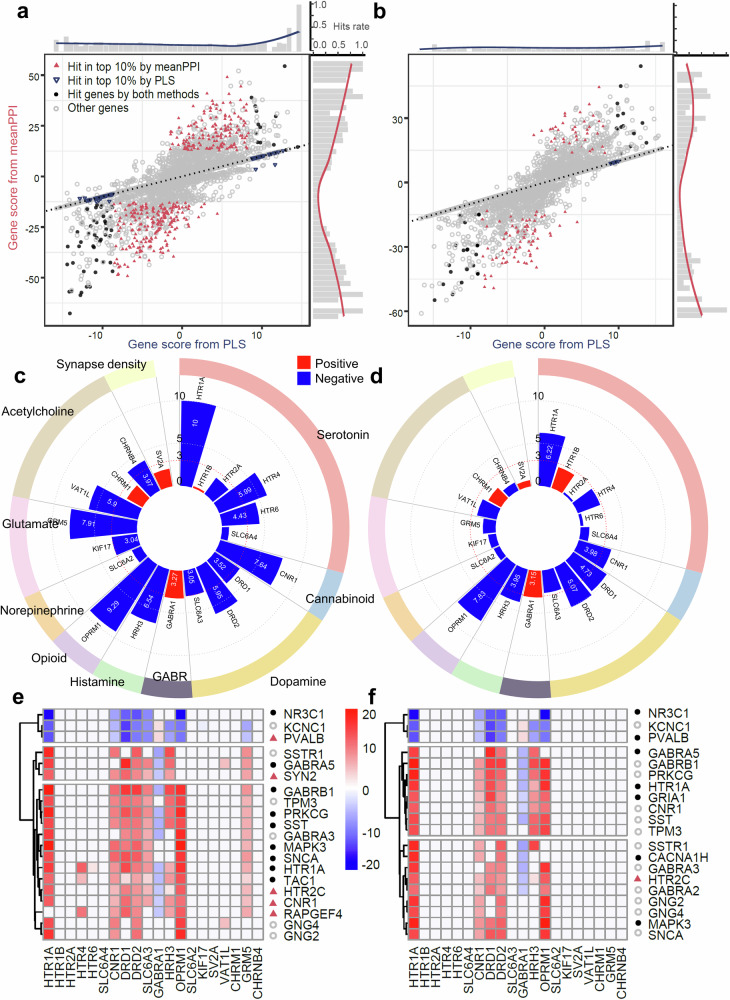
Fig. 6. Differential plot of genes by different fusion methods and neurotransmissions for SCZ and ASD.
a, b Gene scores from meanPPI method and PLS method. Black dots: genes overlapped among the genes from DisGeNet standard database, top 10% genes from meanPPI method, and top 10% genes from PLS method simultaneously. Blue triangles: genes overlapped between the genes from DisGeNet database and 10% genes from PLS method. Magenta triangles: genes overlapped between the genes from DisGeNet database and 10% genes from meanPPI methods. The bar chart at the edge shows the hit rates of these disease related genes. c, d Associations measured by PLS Z-score between all PET maps of various neurotransmission process and disease trait (c: SCZ; d: ASD). e, f Top 20 candidate genes identified by meanPPI method, and the gene-PET effects measured by PLS Z- score for SCZ (e) and ASD (f) disease trait. Point shapes of genes in (e–f) have the same meanings as in (a, b).
We examined the neurotransmissions-trait and gene-neurotransmissions association for SCZ and ASD (Fig. 6c, d). We found that the top 20 genes prioritized for SCZ by mFusion-meanPPI had two patterns of correlations with five neurotransmitter receptors, including 17 genes with positive correlations with HTR1A, CNR1, DRD1 DRD2, and OPRM1, and 3 genes with negative correlations with these receptors (Fig. 6e). Similar patterns were observed for ASD (Fig. 6f). Gene-neurotransmission PLS association analysis revealed that the majority of the top 20 genes were linked to these neurotransmissions (Fig. 6e, f). Specifically, 14 of the top 20 genes identified by the mFusion-meanPPI method were listed as SCZ-related genes in the DisGeNet database, and five of these 14 genes were not detected by the PLS method.
Comparison of correlations among multiple brain disorders
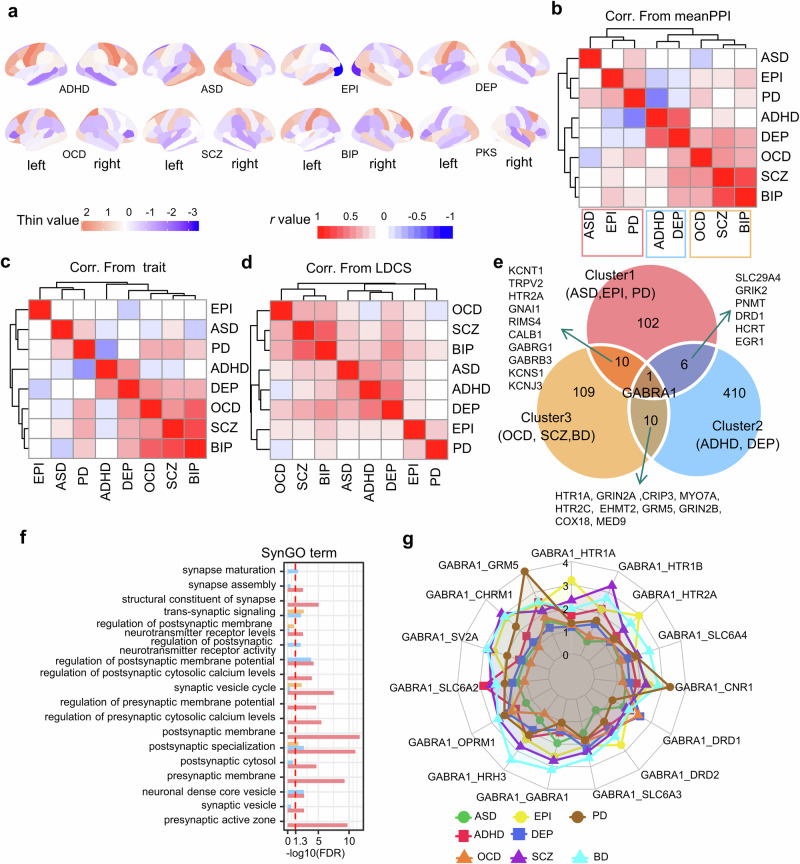
We applied the mFusion-meanPPI algorithm to neuroimaging traits of eight disorder cohorts separately (Fig. 7a, see “Methods”), and prioritized top 10% genes based on their Z-scores. Spearman correlation analysis of these genes was performed to assess the similarity between each pair of disorders. Following this, hierarchical clustering was applied to the spearman correlation coefficients among these diseases, resulting in the identification of three distinct clusters. These clusters reflected the expressional association among these diseases, as inferred from the gene Z-scores. The first cluster comprised the ASD, EPI, and PD, the second included the ADHD and DEP, and the third cluster encompassed the OCD, SCZ, and BIP (Fig. 7b). This clustering structure was supported by both morphological (Fig. 7c) and genetic (Fig. 7d,) correlations. Especially, the OCD-SCZ-BIP cluster and the EPI-PD cluster presented in all three clustering structures, which are supported by previous studies of the cross-disease similarity at different levels10,26,27. In the other two clusters, the EPI-PD correlation exhibited consistent stability. However, while genetically ASD showed more similarity to the DEP-ADHD cluster, neuroimaging traits placed it closer to the EPI-PD cluster. Simultaneously, the DEP-ADHD correlation was more pronounced genetically but less evident in terms of imaging trait correlation. Our identification of the clustering structure for eight major mental disorders unveiled a notable concordance of these disorders across multiple scales (Supplementary Table S8, Table S9, and Table S10).
Fig. 7. Correlation of eight brain disorders from multiple biomolecular levels.
a Cohen’s d maps of cortical thickness difference for eight disorders on Desikan–Killiany atlas regions. b Heatmap of expressional correlations across eight disorders (Spearman’s r value). c Heatmap of morphological correlations across eight disorders (Pearson r value). d Heatmap of genetic correlations across eight disorders (LDCS value). e The overlap of top10% genes among three disease clusters is shown in the Veen map. f GO:MF (molecular function) terms enrichment results for three groups of cluster-specific genes (Cluster1: 102; Cluster 2: 410; Cluster 3: 109). g GABRA1 related pathway scores across different neurotransmissions. ADHD Attention-deficit/hyperactivity disorder, ASD Autism spectrum disease, BIP Bipolar disorder, DEP Depression, EPI Epilepsy, OCD Obsessive-compulsive disorder, PD Parkinson’s disease, SCZ Schizophrenia.
Comparing among the top 10% genes for each disorder, we identified three cluster-specific gene sets including 102, 410 and 109 genes for three clusters, respectively (Fig. 7e; Table S11). Meanwhile, the genes related to cluster 1 were enriched in a wide range of pre- and post-synaptic functions, and the genes for cluster 2 enriched mainly in the postsynaptic functions (Fig. 7f). Notably, the “GABRA1” was the only gene associated with all eight disorders but with distinct gene-transmission pathways (Fig. 7g, Table S12). The GABRA1-GRM5 or -CNR1 pathway was prioritized for PD, while the GABRA1-HRH3 pathway was prioritized for OCD. This is consistent with the literature reporting that CNR1 agonists help relieve symptoms in PD patients28–30.
In total, all 43,126 gene-neurotransmissions-trait pathways among 15,408 genes, 20 neurotransmissions, and 29 disease traits were listed in a quadrable database (https://xomicsbio.shinyapps.io/mfusion_shiny/) and summarized in Supplementary Fig. S12.
Discussion
For making use of the human brain data, that have been rapidly accumulating but separately collected at various scales, this study proposed an analytical method, namely mFusion, to bridge neuroimaging traits and genes for mental disorders. Different from previous methods that examine pair-wise associations across two scales, mFusion establishes gene-neurotransmissions-trait pathways across three scales. The advantage of the mFusion method over the previous methods was demonstrated in both simulated and experimental datasets. Both well-known genes and new candidate genes were identified by this method for mental disorders. To our knowledge, it is the first method to prioritize cross-scale pathways for mental health disorders, providing a richer and more comprehensive perspective on disease exploration. In the current study, we demonstrated the performance of the proposed mFusion as a tool for finding gene hits in mental disorders using the PET maps, it is worth noting that the method could be applied to any brain maps, such as the functional MRI or magnetoencephalography, single-photon emission computed tomography, etc.
The proposed method, mFusion, also suggested new disease-related genes that have not been listed in the reference database (e.g., DisGeNet, Fig. 6E, F). For example, the gene CNR1 was prioritized for SCZ by mFution-meanPPI but not the traditional PLS method (Fig. 6E). The CNR1 (cannabinoid receptor 1) encodes cannabinoid receptors and is implicated in the pathophysiology of SCZ. In the literature, the decreased expression of this gene has been reported in the DLPFC of patients with schizophrenia31. The prioritization of this gene by the proposed method was contributed to by its gene-PET association with the DRD2, which is supported by its physical interaction with DRD2 to form CB1R–DRD2 heteromers32.
Another example is the gene KCNC1 (Potassium Voltage-Gated Channel Subfamily C Member 1, see Supplementary Fig. S11A for its PPI network), which is involved in the monoatomic ion channel activity and delayed rectifier potassium channel activity33. It was reported that the level of KCNC1 channels protein decreased in the neocortex of SCZ-infected mice compared with the control group34,35. Another example is GABRA3, which has already been associated with both dopamine transporter transcripts and the disinhibition of nigrostriatal dopamine neurotransmission in the literature36. A recent study using peripheral blood-mesenchymal stem cells has reported its transcriptomic association with ASD37.
Furthermore, for different disorders, gene-PET-trait pathways mediated by different neurotransmissions had great changes of influence (Fig. S11B, Table S12). For example, the neurotransmission GRM5 have strong effect on PD disease (average pathway score = 4.92, refer to Table S12) while not for SCZ (score = 1.64) and BD (score = 1.95) disease. When we refer to pathways in Table S12, the “SNCA” have stronger pathway scores mediated by neurotransmissions including GRM5 (score = 5.76), CHRNB4 (score = 5.00), and CNR1 (score = 4.87), compared with other disease (these pathways scores less than 3 all). The SNCA (alpha-synuclein gene) has been widely reported to be involved in the onset of Parkinson’s disease, especially in the formation of Lewy bodies38–40.
Nevertheless, the multiscale fusion analysis framework has its limitations. First, the currently available 45 PET maps of neurotransmissions cover only 9 neurotransmitter systems and the synaptic density, more PET maps of neurotransmitters remained exclusive due to numerous methodological and data-sharing challenges. The present study would be strengthened in future with advanced biomolecular imaging techniques. Second, the choice of processing parameters can influence the AHBA gene expression estimates41. To mitigate this challenge, we normalized the expression values and focused only on analyses related to the relative rank of genes as opposed to the absolute values. Third, the gene expression data within brain tissues is restricted to a finite set of samples. As additional data encompassing a broader range of genes becomes accessible in the future, the proposed method will be poised for application to these expanded datasets.
Conclusion
In this study, we proposed an analytical method to integrate information across multiple scales, including genes, neurotransmitters, and neuroimages. This method provides a neurotransmission bridge, bridging neuroimaging traits to genes in human brains for mental disorders. The mFusion method identified both well-known genes and new candidate genes of SCZ and ASD separately, demonstrating its advantages in mental disorder phenotypes. This novel method also prioritizes cross-scale pathways related to mental disorders, providing a richer and more comprehensive perspective on disease exploration.
Methods
Data preprocessing
Gene expression in human brain tissues
Microarray expression data for brain tissues were sourced from the Allen Human Brain Atlas (AHBA)11,17, featuring samples from six neurotypical donors aged between 26 to 54 years, with five males and one female. The database encompasses probe expressions from a total of 3702 samples, which have been normalized across all brains. Given the limited availability of right hemisphere samples from only two donors, our analysis focused on 2664 samples from the left hemisphere across all six donors. Following recommended preprocessing steps outlined by Arnatkevičiūtė et al. 18 and consistent with procedures detailed in our prior publication42, the data underwent re-annotation, intensity filtering, probe selection based on mean values, and normalization. This process yielded a matrix of gene expression comprising 2664 samples × 15,408 unique genes.
Neurotransmission images
PET imaging has proven invaluable for noninvasively mapping the in vivo spatial distributions of neurotransmissions within the human brain. In this study, we curated a comprehensive database comprising 45 neurotransmission-related PET maps for 9 neurotransmitter systems and synaptic density. Among them, 36 maps were provided in the neuromaps toolbox (https://netneurolab.github.io/neuromaps/index.html)19, 6 were available through the JuSpace toolbox (https://github.com/juryxy/JuSpace)20, and 3 were available at the PET imaging database provided by Hansen et al. 14 (https://github.com/netneurolab/hansen_receptors/tree/main/data/PET_nifti_images). These systems encompass serotonin, cannabinoid, dopamine, gamma-aminobutyric acid, histamine, mu-type opioid, norepinephrine, N-methyl-D-aspartate, synaptic vesicle membrane protein, acetylcholine, glutamate, and nicotinic-acetylcholine (Table 2 and Supplementary Table S1).
Protein-protein interaction (PPI) network
Recognizing the collaborative nature of proteins coded by genes in performing various functions43, our study employed the STRING Protein-Protein Interaction (PPI) network (Version 11.5, August 12, 2021)16. This repository stands as one of the largest and most widely utilized sources of PPI data, encompassing both direct (physical) and indirect (functional) interactions. These interactions are derived from a range of sources, including experimental data, gene co-expression, and text-mining. Within the PPI network, the strength of an edge is quantified by the confidence score (c), while the distance between two nodes is measured by the depth (d). Specifically, a larger c and a smaller d contribute to a PPI network that is substantiated by stronger evidence.
Brain traits of mental disorders using the Desikan–Killiany (DK) atlas
The ENIGMA consortium and ENIGMA toolbox (https://enigma-toolbox.readthedocs.io/en/latest/index.html#)21 have provided the structural case-control differences for eight mental disorders, including attention-deficit/hyperactivity disorder (ADHD)44, ASD45, bipolar disorder (BD)46, common epilepsy syndromes (EPI)47, depression (DEP)48, obsessive-compulsive disorder (OCD)49, Parkinson’s disease (PD)50, and SCZ51. In this study, we employed maps detailing case-control differences in cortical thicknesses, represented by inverted Cohen’s d values14 (this means, larger values represent greater cortical thinning), for 68 specific DK brain regions (Table S13).
Brain traits of mental disorders in the DK308 Atlas
In our investigation, we incorporated a brain map depicting case-control differences in morphological similarity, specifically the correlation of seven morphological parameters (i.e., gray matter volume, surface area, cortical thickness, Gaussian curvature, mean curvature, fractional anisotropy, and mean diffusivity) derived from MRI and diffusion-weighted imaging data, concerning schizophrenia. This map is defined by the Desikan–Killiany 308 atlas (DK308)13, an improved version of the DK atlas that maintains small-world properties of anatomical cortical networks while enhancing resolution with 308 regions8. We also employed another case-control differences map in cortical thickness for ASD illustrated by DK308 atlas9.
GWAS summary statistics for mental disorders
We compiled GWAS summary results for six mental disorders from published research, drawing from the Psychiatric Genomics Consortium (PGC) datasets for ADHD52, ASD53, BIP54, DEP55, OCD56, SCZ57. Additionally, we incorporated data from other relevant studies (EPI58, PD59). Table S14 offers comprehensive details on the individual GWAS samples, including references, sample sizes, and SNP numbers.
Multiscale-fusion framework
Calculations of multiscale associations
We examined three types of associations, including gene-trait, PET-trait, and gene-PET associations. Each type of association was separately estimated by partial least squares (PLS) regression, a widely employed method for evaluating the association between a 1-dimention response variable () and a multidimensional predictor (), where n is the sample size and p is the dimension of predictors60–62. Let L be the rank of matrix X, the PLS regression iteratively computes L latent variables (i.e., components) for X by singular value decomposition (SVD). Suppose the data matrices have already been normalized, if not, we normalize (i.e., Z-score) them before entering the algorithm. At the initial step, the covariance matrix was decomposed by SVD as , where is an orthogonal vector and is the corresponding singular value. The first component of was X formed as . Therefore, U specifies the weight of each predictor for the first component (note as ). Next, data X is regressed on the first component and the residuals are used as the data for the next iteration. The second component is given by applying the SVD to . The iteration stops when L components are established as ().
For each component () established from X, the effect size of its association with can be estimated by the variance explained in the linear regression of on this component. The significance of this association was assessed by 1000 permutations, i.e., randomly shuffling the elements of to re-conduct the PLS regression and re-calculate the variance explained. Following the literature60, the estimation error in each weight is established empirically by 1000 bootstraps as the standard error in the bootstrapped estimations () and is adjusted for by The error-adjusted weights entered the following analyses. For brevity, the error-adjusted weight was referred to as the Z-score in the following texts.
For the gene-trait association, the regional gene expressions from AHBA (), n is the number of brain regions and p is the number of genes) served as predictors, while the neuroimage trait () was considered as the response variable. We selected the first component of PLS regression analysis if it had a p-value < 0.05 by permutation; otherwise, we selected the component that explained the most variance of Y. Subsequently, the weight of the ith gene was normalized and given by the Z-score of bootstrapped coefficients60, denoted as . These Z-scores was used to rank the relevance of genes to the trait in most previous studies.
The gene-PET associations were assessed by 45 PLS regressions separately for each of the 45 PET maps, and the regional gene expression matrix was the predictors. Therefore, for the j-th PET map (), the normalized Z-score was used to measure the association of the i-th gene to this PET map ().
Similarly, for the PET-trait association, we conducted the same PLS analysis using PET maps as the predictors and estimated the weight of the jth PET map. And then the weight was normalized by the Z-score from bootstraps as .
Combination of multiscale associations to rank genes
Here, we utilized the Z-transform test, also referred to as the “Stouffer’s method”22, to combine K independent Z-scores by
where the is a number drawn from a normal distribution with mean 0 and standard deviation 1. If the common null hypnosis is true, the has a standard normal distribution. Based on this test, we developed the following five fusion methods to rank genes according to their relevance to a given disorder, i.e., the larger the absolute value of the combined Z-score, the greater the relevance.
meanGP We combined the gene-trait association with gene-PET association for the ith gene by
meanGPT We first combined the PET-trait association with the gene-PET association by for the ith gene. Next, the was further combined with the gene-trait association by
meanPPI We applied a cut-off of high confidence (i.e., confidence threshold c) for edges of the STRING PPI network16, and focused exclusively on the closely interacted nodes within the network. The neighboring information is recorded in a binary matrix G(i, j) with 1 indicating a high-confidence interaction between genes i and j, and 0 indicates low-confidence interaction. To score the relevance of a pathway linking gene to trait via a neurotransmission, we combined the PET-trait and gene-PET associations by . This combination is especially informative when the PET-trait association is strong (i.e., above a threshold )63, so we defined a filtering function . The influence pathway of the ith gene through proteins (referred by specific PET maps) to trait were calculated by:
where maxGPT In biological systems, a strong signal can sometimes override several weaker signals to achieve a regulatory effect64,65. To mimic this phenome in the biological systems, we also introduced the following score:
where is defined as the operation to identify the element with the largest absolute value within a vector and then retain its original sign.
maxPPI To introduce the PPI information in the maxGPT score, we further defined:
Combination of multiscale associations to rank gene-neurotransmission-trait pathways Similar to gene ranking method above mentioned, we measure the associations between gene-PET pairs (i.e., gene i and PET map j) and disorders by the following method:
The larger the value of the combined Z-score, the more important the gene-PET pair.
Application of mFusion to numeric simulations
To assess the performance of mFusion, we adopted settings of numeric simulations in the literature66 to mimic properties of the real-world data. Specifically, we supposed to have 200 brain regions (n), 2000 genes ), 20 PET maps of neurotransmitters (), and 1 neuroimaging trait of a mental disorder . The trait and genes shared a latent variable , while the trait and PET maps shared another latent variable . The relationships between genes and PET maps were modeled by a multi-dimensional Gaussian distribution as , where is an identity matrix. Therefore, we generated the simulation data from the following model:
where the and are noises drawn independently from standard Gaussian distributions, and the and are weight vectors drawn from a uniform distribution . For sparsity, we retained 20% of these weights to be active and reset others to be zeros. We used the matrix to represent the interaction network between X and M (similar to the role of PPI in the real world, but not necessarily equivalent to the structure or characteristics of PPI). This simulation was repeated for 500 times. To test the performance of mFusion at different sparse levels, we varied the number of active genes from 200, 400, 600, to 800. To assess the impact of gene-PET covariance on fusion methods, we varied the covariance between latent variables within a range of 0.3, 0.6, and 0.9.
Furthermore, to evaluate the impact of Protein-Protein Interaction (PPI) networks on the mFusion method, we conducted a series of perturbations on the PPI network. Firstly, 30% of the elements within the adjacency matrix were randomly shuffled, thereby introducing a degree of artificiality to the PPI, akin to the inherent noise present in real-world PPI networks. Secondly, we set a minimum 30% of the elements in the adjacency matrix to zero, thereby simulating the incompleteness of PPI that is often encountered in biological systems. Thirdly, we changed the dimension of X, Y, and Z matrix (i.e., the brain regions n ranges in 100, 200, or 500) to investigate the influence of varying spatial resolutions of microarray-derived gene expression and PET maps to the mFusion method. In conclusion, we executed 500 repetitions of the mFusion method on these perturbed PPI matrices to evaluate the robustness and sensitivity to such perturbations.
The proposed fusion methods aim to discern active genes, indicated by non-zero elements of . We evaluate the performance through three metrics: (1) The Spearman correlation between true weights and their estimations, measured by the absolute values of gene scores provided by our method. (2) The AUC-ROC (Area under Curve of the receiver operating characteristic curve) captures the dynamic relationship between true positive rate and false positive rate in binary classification tasks. At each experiment, the first K genes with the highest score (the K was predefined) were predicted as active genes while others were deemed as inactive. A larger AUC-ROC signifies a more robust classification ability of the model. We used R package “multiROC” to compute the mean value of true positive and false positive genes at all simulated experiment. (3) The AUC-PR (Area under Curve of the precision-recall curve) illustrates the trade-off between recall rate and accuracy rate in the classification model. A higher AUC-PR denotes a stronger classification ability of the model.
Application of mFusion to two mental disorders
To assess the efficacy of the mFusion in real-world data, we conducted a comprehensive evaluation by comparing prioritized genes with four well-established gene-disease databases (Detailed information are in Table 1, and Tables S4, S5). The DisGeNet database stands as a comprehensive resource integrating data from expert curations, GWAS catalogues, animal models, and more, establishing gene-disease relationships across 21,671 genes and 30,170 diseases, traits, and phenotypes5. The CTD (Comparative Toxicogenomics Database) contains expert-curated and method-inferred gene–disease relationships drawn from published literature or the OMIM database6. The DISEASES, a weekly updated web resource, consolidates evidence on disease-gene associations from automatic text mining, manually curated literature, cancer mutation data, and genome-wide association studies67. Additionally, we applied fine-mapping analysis in MAGMA68 to derive GWAS-based gene sets from GWAS summary results provided by the SCZ and ASD working groups in the Psychiatric Genomics Consortium (PGC)53,57. A superior performance of a fusion method is indicated by a greater overlap of top-ranked genes generated by the method with those present in the gene-disease databases.
Sensitivity analysis
In above analyses, we employed a PPI confidence threshold (c) set at 0.5, and a PPI depth (d) set at 1. To assess the sensitivity of mFusion on different confidence (c) level and depth (d) in calculation, we conducted additional tests. Specifically, we explored a range of confidence values from 0.3 to 0.9 in increments of 0.1, while maintaining PPI depths at both 1 and 2.
To assess the influence of PPI information on the mFusion method, 500 times of shuffling were conducted on the nodes within the STRING PPI networks. This type of shift preserved the network properties of the original PPI and the characteristics of the node itself, while modifying the neighbor relationship of each node. The null distribution of results in random PPI network cases was obtained and compared with results from real STRING PPI networks. Similarly, we permutated the 45 PET maps 500 times (by shuffling the order of brain regions across 45 maps concurrently at each time) to evaluate their contributions to the mFusion methodology. If the number of hit genes significantly reduced, the molecular distributions given by the real PET maps contributed significantly to mFusion.
Furthermore, we averaged the PET maps measuring the same molecule and got 20 PET maps each of which measured a different molecule (Fig. S8). To test the potential bias introduced by the redundant PET maps for some molecules, we compared the gene scores given the mFusion using these 20 unique PET maps with the gene scores given by using all the 45 PET maps. If these scores are significantly correlated with each other, there is little evidence for the bias.
Enrichment analysis
For disease enrichment analysis, the R package “disgenet2r”5 was employed, and Gene Ontology (GO)23 term enrichment analysis was conducted using another R package, “clusterProfiler”69. Following the literature13, these analyses focused on the top 1541 genes, representing 10% of the total 15,408 genes70, exhibiting the most negative Z-scores as determined by the meanPPI method. To correct for multiple comparisons, a false discovery rate (FDR) correction (p.adjust < 0.05) was applied. The visualization was accomplished using the R package aPEAR (Advanced Pathway Enrichment Analysis Representation)71, which leverages similarities between pathways to represent them as an interconnected network of clusters.
Correlations between mental disorders assessed by mFusion
Applying the mFusion to neuroimaging traits of eight mental disorders given by the ENIGMA consortium as described above, we investigated the correlation between each pair of mental disorders. Spearman correlation coefficients were calculated between the fusion scores of the top 10% genes ranked by the meanPPI method. Subsequently, a hierarchical clustering analysis of these correlations was conducted, implementing complete-linkage clustering through the “hclust” function in R program72. To offer a comparative perspective, we investigated the clustering structure using both neuroimaging and genetic correlations. The former was established through Pearson correlation between neuroimaging traits for each pair of mental disorders. The latter was estimated using the Linkage Disequilibrium Score Regression (LDSC) method, employing default parameters for the European population63,73. To elucidate the clustering pattern of these disorders, we focused on the top 10% of disease-related genes within each disorder clusters and explored the cluster-specific gene sets. Subsequently, the study collected the gene-neurotransmissions-trait pathways of these diseases, and calculated the average scores of gene-neurotransmitter pairs across different neurotransmitter types, which provided the importance of pathways mediated by neurotransmissions in specific trait or disorder.
Statistics and reproducibility
Data manipulation and processing analyses were conducted using the packages MATLAB (version 2018b), R (version 4.1.0). We considered a p-value < 0.05 as significant unless otherwise stated. All relevant information of statistical tests and post hoc tests methods are included in the figure legends. The plots in our study are drawn by ggplot2 (version 3.5.1), aPEAR (version 1.0.0), and multiROC (version 1.1.1).
Ethics approval and consent to participate
All ethical regulations relevant to human research participants were followed. All the utilized data in our study were previously published and publicly available. The research conformed to the principles of the Helsinki Declaration.
Supplementary information
Description of Additional Supplementary File
Acknowledgements
This study was supported by grants from the National Key Research and Development Program of China (No. 2023YFE0109700), the National Natural Science Foundation of China (No. 82272079, 71834002), the Program of Shanghai Academic Research Leader (No. 23XD1423400), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01), University of Sydney – Fudan University BISA Flagship Research Program. This work was supported by the China National Postdoctoral Program for Innovative Talents [grant number BX20230229], the China Postdoctoral Science Foundation [grant number 2024M752038].
Author contributions
Q.L. conceived the study and designed experiments. L.C. implemented the methods, and analyzed the data. L.C., Z.W. and Z.Y. wrote the draft. Q.L., Z.Y. and Z.W. interpreted the results and revised the manuscript. L.C. and Q.L. made the visualization. All the authors have proofread and approved the manuscript. We are particularly thankful to Wenjie Hou for his significant contributions to the graphical representation and critical revision of the manuscript.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Jasmine Pan. A peer review file is available.
Data availability
Microarray expression data for brain tissues were sourced from the Allen Human Brain Atlas (AHBA), refer to https://atlas.brain-map.org/. This study employed the STRING Protein-Protein Interaction (PPI) network at https://version-11-5.string-db.org/ (Version 11.5, August 12, 2021). The disease-risk genes were acquired from the following websites: (1) DisGeNet database (https://www.disgenet.org/home/. Accessed 12 Dec 2023); (2) Comparative Toxicogenomics Database (https://ctdbase.org/. Accessed 12 Dec 2023); (3) DISEASES database (https://diseases.jensenlab.org/Search. Accessed 12 Dec 2023); (4) GWAS-based gene sets from GWAS summary results as described in Methods section. GWAS summary statistics of eight psychiatric disorders are available on the PGC website (https://www.med.unc.edu/pgc/results-and-downloads). All ethical regulations relevant to human research participants were followed in the public datasets used and approved by the respective boards and institutions. All Supplementary tables (Table S1–S12) are available at Supplementary Data 1.
Code availability
An open-source implementation of the mFusion method can be downloaded from https://github.com/CaoLuolong/XomicsEnrich. These codes are also publicly available in the Zenodo repository at 10.5281/zenodo.1437487974.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Luolong Cao, Zhenyi Wang.
These authors jointly supervised this work: Zhiyuan Yuan, Qiang Luo.
Contributor Information
Zhiyuan Yuan, Email: zhiyuan@fudan.edu.cn.
Qiang Luo, Email: qluo@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07404-x.
References
- 1.Arias, D., Saxena, S. & Verguet, S. Quantifying the global burden of mental disorders and their economic value. eClinicalMedicine54, 10.1016/j.eclinm.2022.101675 (2022). [DOI] [PMC free article] [PubMed]
- 2.World Health Organization. World mental health report: Transforming mental health for all. World Health Organization (2022).
- 3.Andersen, P. H. et al. Securing the future of drug discovery for central nervous system disorders. Nat. Rev. Drug Discov.13, 871–872, (2014). [DOI] [PubMed] [Google Scholar]
- 4.Wang, Z. et al. MarkovHC: Markov hierarchical clustering for the topological structure of high-dimensional single-cell omics data with transition pathway and critical point detection. Nucleic Acids Res.50, 46–56 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piñero, J. et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res.48, D845–D855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, A. P. et al. Comparative toxicogenomics database (CTD): update 2023. Nucleic Acids Res.51, D1257–d1262 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gandal, M. J. et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science359, 693–697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero-Garcia, R., Atienza, M., Clemmensen, L. H. & Cantero, J. L. Effects of network resolution on topological properties of human neocortex. NeuroImage59, 3522–3532 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Romero-Garcia, R., Warrier, V., Bullmore, E. T., Baron-Cohen, S. & Bethlehem, R. A. I. Synaptic and transcriptionally downregulated genes are associated with cortical thickness differences in autism. Mol. Psychiatry24, 1053–1064 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radonjić, N. V. et al. Structural brain imaging studies offer clues about the effects of the shared genetic etiology among neuropsychiatric disorders. Mol. Psychiatry26, 2101–2110 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawrylycz, M. J. et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richiardi, J. et al. Correlated gene expression supports synchronous activity in brain networks. Science348, 1241–1244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgan, S. E. et al. Cortical patterning of abnormal morphometric similarity in psychosis is associated with brain expression of schizophrenia-related genes. Proc. Natl Acad. Sci.116, 9604–9609 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen, J. Y. et al. Mapping neurotransmitter systems to the structural and functional organization of the human neocortex. Nat. Neurosci.25, 1569–1581 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hänisch, B. et al. Cerebral chemoarchitecture shares organizational traits with brain structure and function. eLife12, e83843 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res.49, D605–d612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen, E. H., Overly, C. C. & Jones, A. R. The Allen Human Brain Atlas: comprehensive gene expression mapping of the human brain. Trends Neurosci.35, 711–714 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Arnatkevic̆iūtė, A., Fulcher, B. D. & Fornito, A. A practical guide to linking brain-wide gene expression and neuroimaging data. NeuroImage189, 353–367 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Markello, R. D. et al. neuromaps: structural and functional interpretation of brain maps. Nat. Methods19, 1472–1479 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dukart, J. et al. JuSpace: a tool for spatial correlation analyses of magnetic resonance imaging data with nuclear imaging derived neurotransmitter maps. Hum. Brain Mapp.42, 555–566 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larivière, S. et al. The ENIGMA Toolbox: multiscale neural contextualization of multisite neuroimaging datasets. Nat. Methods18, 698–700 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitlock, M. C. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J. Evolut. Biol.18, 1368–1373 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marder, S. R. & Cannon, T. D. Schizophrenia. N. Engl. J. Med.381, 1753–1761 (2019). [DOI] [PubMed] [Google Scholar]
- 25.McCutcheon, R. A., Reis Marques, T. & Howes, O. D. Schizophrenia—an overview. JAMA Psychiatry77, 201–210 (2020). %J JAMA Psychiatry. [DOI] [PubMed] [Google Scholar]
- 26.Torsvik, A. et al. Patients with schizophrenia and bipolar disorder display a similar global gene expression signature in whole blood that reflects elevated proportion of immature neutrophil cells with association to lipid changes. Transl. Psychiatry13, 147 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hettwer, M. D. et al. Coordinated cortical thickness alterations across six neurodevelopmental and psychiatric disorders. Nat. Commun.13, 6851 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira, C., Almeida, C., Tenreiro, S. & Quintas, A. Neuroprotection or neurotoxicity of illicit drugs on Parkinson’s disease. Life10, 10.3390/life10060086 (2020). [DOI] [PMC free article] [PubMed]
- 29.Stampanoni Bassi, M., Sancesario, A., Morace, R., Centonze, D. & Iezzi, E. Cannabinoids in Parkinson’s Disease. Cannabis Cannabinoid Res.2, 21–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.More, S. V. & Choi, D. K. Promising cannabinoid-based therapies for Parkinson’s disease: motor symptoms to neuroprotection. Mol. Neurodegener.10, 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao, R. et al. Cannabinoid receptor CNR1 expression and DNA methylation in human prefrontal cortex, hippocampus and caudate in brain development and schizophrenia. Transl. Psychiatry10, 158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro, G. et al. Detection of receptor heteromers involving dopamine receptors by the sequential BRET-FRET technology. Methods Mol. Biol.964, 95–105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ried, T. et al. Localization of a highly conserved human potassium channel gene (NGK2-KV4; KCNC1) to chromosome 11p15. Genomics15, 405–411 (1993). [DOI] [PubMed] [Google Scholar]
- 34.Espinosa, F., Marks, G., Heintz, N. & Joho, R. H. Increased motor drive and sleep loss in mice lacking Kv3-type potassium channels. Genes Brain Behav.3, 90–100 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Joho, R. H., Ho, C. S. & Marks, G. A. Increased γ- and decreased δ-Oscillations in a mouse deficient for a potassium channel expressed in fast-spiking interneurons. J. Neurophysiol.82, 1855–1864 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Purves-Tyson, T. D., Brown, A. M., Weissleder, C., Rothmond, D. A. & Shannon Weickert, C. Reductions in midbrain GABAergic and dopamine neuron markers are linked in schizophrenia. Mol. Brain14, 96 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fazeli, Z., Ghaderian, S. M. H., Najmabadi, H. & Omrani, M. D. Understanding the molecular basis of fragile X syndrome using differentiated mesenchymal stem cells. Iran. J. Child Neurol.16, 85–95 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pan, H. et al. Genome-wide association study using whole-genome sequencing identifies risk loci for Parkinson’s disease in Chinese population. NPJ Parkinson’s Dis.9, 22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Shlomo, Y. et al. The epidemiology of Parkinson’s disease. Lancet403, 283–292 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kia, D. A. et al. Identification of candidate Parkinson disease genes by integrating genome-wide association study, expression, and epigenetic data sets. JAMA Neurol.78, 464–472, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Markello, R. D. et al. Standardizing workflows in imaging transcriptomics with the abagen toolbox. eLife10, e72129 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen, C. et al. What is the link between attention-deficit/hyperactivity disorder and sleep disturbance? A multimodal examination of longitudinal relationships and brain structure using large-scale population-based cohorts. Biol. Psychiatry88, 459–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baltoumas, F. A. et al. Biomolecule and bioentity interaction databases in systems biology: a comprehensive review. Biomolecules11, 10.3390/biom11081245 (2021). [DOI] [PMC free article] [PubMed]
- 44.Hoogman, M. et al. Brain imaging of the cortex in ADHD: a coordinated analysis of large-scale clinical and population-based samples. Am. J. Psychiatry176, 531–542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Rooij, D. et al. Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: results from the ENIGMA ASD Working Group. Am. J. Psychiatry175, 359–369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hibar, D. P. et al. Cortical abnormalities in bipolar disorder: an MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Mol. Psychiatry23, 932–942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whelan, C. D. et al. Structural brain abnormalities in the common epilepsies assessed in a worldwide ENIGMA study. Brain141, 391–408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmaal, L. et al. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA major depressive disorder working group. Mol. Psychiatry22, 900–909 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boedhoe, P. S. W. et al. Cortical abnormalities associated with pediatric and adult obsessive-compulsive disorder: findings from the ENIGMA obsessive-compulsive disorder working group. Am. J. Psychiatry175, 453–462 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laansma, M. A. et al. International multicenter analysis of brain structure across clinical stages of Parkinson’s disease. Mov. Disord.36, 2583–2594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Erp, T. G. M. et al. Cortical brain abnormalities in 4474 individuals with Schizophrenia and 5098 control subjects via the enhancing neuro imaging genetics through meta analysis (ENIGMA) consortium. Biol. Psychiatry84, 644–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Demontis, D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet.55, 198–208 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet.51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet.53, 817–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Giannakopoulou, O. et al. The genetic architecture of depression in individuals of East Asian ancestry: a genome-wide association study. JAMA Psychiatry78, 1258–1269, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Posthuma D. Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol. Psychiatry23, 1181–1188 (2018). [DOI] [PMC free article] [PubMed]
- 57.Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature604, 502–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dönertaş, H. M., Fabian, D. K., Valenzuela, M. F., Partridge, L. & Thornton, J. M. Common genetic associations between age-related diseases. Nat. Aging1, 400–412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet.53, 1415–1424 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Vértes, P. E. et al. Gene transcription profiles associated with inter-modular hubs and connection distance in human functional magnetic resonance imaging networks. Philos. Trans. R. Soc. Lond. Ser B Biol. Sci.371, 10.1098/rstb.2015.0362 (2016). [DOI] [PMC free article] [PubMed]
- 61.Krishnan, A., Williams, L. J., McIntosh, A. R. & Abdi, H. Partial least squares (PLS) methods for neuroimaging: a tutorial and review. NeuroImage56, 455–475 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Abdi, H. Partial least squares regression and projection on latent structure regression (PLS Regression). WIREs Comput. Stat.2, 97–106 (2010). [Google Scholar]
- 63.Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet.47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, R. et al. Winner-takes-all resource competition redirects cascading cell fate transitions. Nat. Commun.12, 853 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krijger, P. H. L. & de Laat, W. Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol.17, 771–782 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Mihalik, A. et al. Canonical correlation analysis and partial least squares for identifying brain–behavior associations: a tutorial and a comparative study. Biol. Psychiatry.: Cogn. Neurosci. Neuroimaging7, 1055–1067 (2022). [DOI] [PubMed] [Google Scholar]
- 67.Pletscher-Frankild, S., Pallejà, A., Tsafou, K., Binder, J. X. & Jensen, L. J. DISEASES: text mining and data integration of disease–gene associations. Methods74, 83–89 (2015). [DOI] [PubMed] [Google Scholar]
- 68.de Leeuw, C. A., Neale, B. M., Heskes, T. & Posthuma, D. The statistical properties of gene-set analysis. Nat. Rev. Genet.17, 353–364 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hawrylycz, M. et al. Canonical genetic signatures of the adult human brain. Nat. Neurosci.18, 1832–1844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kerseviciute, I. & Gordevicius, J. aPEAR: an R package for autonomous visualization of pathway enrichment networks. Bioinformatics39, 10.1093/bioinformatics/btad672 (2023). [DOI] [PMC free article] [PubMed]
- 72.Müllner, D. fastcluster: fast hierarchical, agglomerative clustering routines for R and python. J. Stat. Softw.53, 1–18 (2013). [Google Scholar]
- 73.Delaneau, O. et al. Integrating sequence and array data to create an improved 1000 genomes project haplotype reference panel. Nat. Commun.5, 3934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luolong, Cao et. al. Code for “mFusion: a multiscale fusion method bridging neuroimages to genes through neurotransmissions in mental health disorders” (v1.0.1). Zenodo, 10.5281/zenodo.14374879 (2024). [DOI] [PMC free article] [PubMed]
- 75.Beliveau, V. et al. A high-resolution in vivo atlas of the human brain’s serotonin system. J. Neurosci.37, 120–128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Savli, M. et al. Normative database of the serotonergic system in healthy subjects using multi-tracer PET. Neuroimage63, 447–459 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Gallezot, J. D. et al. Kinetic modeling of the serotonin 5-HT(1B) receptor radioligand [(11)C]P943 in humans. J. Cereb. Blood Flow. Metab.30, 196–210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Talbot, P. S. et al. Extended characterisation of the serotonin 2A (5-HT2A) receptor-selective PET radiotracer 11C-MDL100907 in humans: quantitative analysis, test-retest reproducibility, and vulnerability to endogenous 5-HT tone. Neuroimage59, 271–285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Radhakrishnan, R. et al. 2181 Age-related change in 5-HT6 receptor availability in healthy male volunteers measured with 11C-GSK215083 PET. J Clin Transl Sci. 2, 3–4. (2018). [DOI] [PMC free article] [PubMed]
- 80.Fazio, P. et al. Mapping the distribution of serotonin transporter in the human brainstem with high-resolution PET: validation using postmortem autoradiography data. Neuroimage133, 313–320 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Laurikainen, H. et al. Sex difference in brain CB1 receptor availability in man. Neuroimage184, 834–842 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Normandin, M. D. et al. Imaging the cannabinoid CB1 receptor in humans with [11C]OMAR: assessment of kinetic analysis methods, test-retest reproducibility, and gender differences. J. Cereb. Blood Flow. Metab.35, 1313–1322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaller, S. et al. Test-retest measurements of dopamine D(1)-type receptors using simultaneous PET/MRI imaging. Eur. J. Nucl. Med. Mol. Imaging44, 1025–1032 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Sandiego, C. M. et al. Reference region modeling approaches for amphetamine challenge studies with [11C]FLB 457 and PET. J. Cereb. Blood Flow. Metab.35, 623–629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jaworska, N. et al. Extra-striatal D2/3 receptor availability in youth at risk for addiction. Neuropsychopharmacology45, 1498–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith, C. T. et al. Partial-volume correction increases estimated dopamine D2-like receptor binding potential and reduces adult age differences. J. Cereb. Blood Flow. Metab.39, 822–833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alakurtti, K. et al. Long-term test-retest reliability of striatal and extrastriatal dopamine D2/3 receptor binding: study with [(11)C]raclopride and high-resolution PET. J. Cereb. Blood Flow. Metab.35, 1199–1205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dukart, J. et al. Cerebral blood flow predicts differential neurotransmitter activity. Sci. Rep.8, 4074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.García-Gómez, F. J. et al. Elaboration of the SPM template for the standardization of SPECT images with 123I-Ioflupane]. Rev. Esp. Med. Nucl. Imagen Mol.32, 350–356 (2013). [DOI] [PubMed] [Google Scholar]
- 90.Sasaki, T. et al. Quantification of dopamine transporter in human brain using PET with 18F-FE-PE2I. J. Nucl. Med.53, 1065–1073 (2012). [DOI] [PubMed] [Google Scholar]
- 91.Nørgaard, M. et al. A high-resolution in vivo atlas of the human brain’s benzodiazepine binding site of GABA(A) receptors. Neuroimage232, 117878 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gallezot, J. D. et al. Determination of receptor occupancy in the presence of mass dose: [(11)C]GSK189254 PET imaging of histamine H(3) receptor occupancy by PF-03654746. J. Cereb. Blood Flow. Metab.37, 1095–1107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kantonen, T. et al. Interindividual variability and lateralization of μ-opioid receptors in the human brain. Neuroimage217, 116922 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Turtonen, O. et al. Adult attachment system links with brain Mu opioid receptor availability in vivo. Biol. Psychiatry Cogn. Neurosci. Neuroimaging6, 360–369 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Ding, Y.-S. et al. PET imaging of the effects of age and cocaine on the norepinephrine transporter in the human brain using (S,S)-[11C]O-methylreboxetine and HRRT. Synapse64, 30–38 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hesse, S. et al. Central noradrenaline transporter availability in highly obese, non-depressed individuals. Eur. J. Nucl. Med Mol. Imaging44, 1056–1064 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Galovic, M. et al. Validation of a combined image derived input function and venous sampling approach for the quantification of [(18)F]GE-179 PET binding in the brain. Neuroimage237, 118194 (2021). [DOI] [PubMed] [Google Scholar]
- 98.Finnema, S. J. et al. Imaging synaptic density in the living human brain. Sci. Transl. Med.8, 348ra396–348ra396 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Aghourian, M. et al. Quantification of brain cholinergic denervation in Alzheimer’s disease using PET imaging with [(18)F]-FEOBV. Mol. Psychiatry22, 1531–1538 (2017). [DOI] [PubMed] [Google Scholar]
- 100.Bedard, M. A. et al. Brain cholinergic alterations in idiopathic REM sleep behaviour disorder: a PET imaging study with (18)F-FEOBV. Sleep. Med.58, 35–41 (2019). [DOI] [PubMed] [Google Scholar]
- 101.Naganawa, M. et al. First-in-human assessment of (11)C-LSN3172176, an M1 muscarinic acetylcholine receptor PET radiotracer. J. Nucl. Med.62, 553–560 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.DuBois, J. M. et al. Characterization of age/sex and the regional distribution of mGluR5 availability in the healthy human brain measured by high-resolution [(11)C]ABP688 PET. Eur. J. Nucl. Med. Mol. Imaging43, 152–162 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Smart, K. et al. Sex differences in [(11)C]ABP688 binding: a positron emission tomography study of mGlu5 receptors. Eur. J. Nucl. Med. Mol. Imaging46, 1179–1183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hillmer, A. T. et al. Imaging of cerebral α4β2* nicotinic acetylcholine receptors with (-)-[(18)F]Flubatine PET: implementation of bolus plus constant infusion and sensitivity to acetylcholine in human brain. Neuroimage141, 71–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary File
Data Availability Statement
Microarray expression data for brain tissues were sourced from the Allen Human Brain Atlas (AHBA), refer to https://atlas.brain-map.org/. This study employed the STRING Protein-Protein Interaction (PPI) network at https://version-11-5.string-db.org/ (Version 11.5, August 12, 2021). The disease-risk genes were acquired from the following websites: (1) DisGeNet database (https://www.disgenet.org/home/. Accessed 12 Dec 2023); (2) Comparative Toxicogenomics Database (https://ctdbase.org/. Accessed 12 Dec 2023); (3) DISEASES database (https://diseases.jensenlab.org/Search. Accessed 12 Dec 2023); (4) GWAS-based gene sets from GWAS summary results as described in Methods section. GWAS summary statistics of eight psychiatric disorders are available on the PGC website (https://www.med.unc.edu/pgc/results-and-downloads). All ethical regulations relevant to human research participants were followed in the public datasets used and approved by the respective boards and institutions. All Supplementary tables (Table S1–S12) are available at Supplementary Data 1.
An open-source implementation of the mFusion method can be downloaded from https://github.com/CaoLuolong/XomicsEnrich. These codes are also publicly available in the Zenodo repository at 10.5281/zenodo.1437487974.