Abstract
The crosstalk of light signaling pathways with other signaling cascades has just started to be revealed. Here, we report the identification and functional characterization of a Z-box binding factor (ZBF1) in light signaling pathways. Arabidopsis thaliana ZBF1 encodes AtMYC2/JIN1, a basic helix-loop-helix transcription factor, which has recently been shown to be involved in abscisic acid (ABA), jasmonic acid (JA), and jasmonate-ethylene signaling pathways. We demonstrate that AtMYC2 interacts with the Z- and G-box light-responsive elements of minimal light–regulated promoters. AtMYC2 is expressed in various light-grown seedlings, including in red, far red, and blue light. Genetic analyses suggest that AtMYC2 acts as a negative regulator of blue light–mediated photomorphogenic growth and blue and far-red-light–regulated gene expression; however, it functions as a positive regulator of lateral root formation. Our results further demonstrate that atmyc2 mutants have compromised sensitivity to ABA- and JA-mediated responses. Taken together, these results demonstrate that AtMYC2 is a common transcription factor of light, ABA, and JA signaling pathways in Arabidopsis.
INTRODUCTION
Light is one of the most important environmental stimuli for plant growth and development (Kendrick and Kronenberg, 1994; Deng and Quail, 1999; Neff et al., 2000; Quail, 2002). Light is perceived by several photoreceptors: far-red and red light by phytochromes (phyA to phyE) and blue and UV-A light by cryptochromes (cry1 and cry2) (Ahmad and Cashmore, 1993; Furuya, 1993; Neff et al., 2000; Lin, 2002; Quail, 2002). Whereas cytosolic phytochromes are translocated into the nucleus upon light-mediated activation, cryptochromes are localized in the nucleus (Cashmore et al., 1999; Guo et al., 1999; Kircher et al., 1999; Yamaguchi et al., 1999; Quail, 2002; Schepens et al., 2004). Significant progress has been made in understanding the functions of photoreceptors and in the identification of early signaling components of light signaling pathways. However, the connection of photoperception to transcription is still largely unclear (Deng and Quail, 1999; Martinez-Garcia et al., 2000; Nagy and Schafer, 2002; Yadav et al., 2002). Additionally, information about crosstalk of light signaling pathways with other signaling cascades is still at its infancy.
Arabidopsis thaliana seedling development follows two distinct pathways: skotomorphogenesis or etiolation in the dark and photomorphogenesis or deetiolation in the light. The shift from skotomorphogenic to photomorphogenic development leads to a change in expression of approximately one-third of the total genes in Arabidopsis (Ma et al., 2001; Tepperman et al., 2001). Several transcription factors in light signaling pathways have been reported that are involved in photomorphogenic development. HY5 is a bZIP transcription factor in light signaling pathways (Oyama et al., 1997; Ang et al., 1998; Chattopadhyay et al., 1998a). The hy5 mutant seedlings show a partially etiolated phenotype in red, far-red, or blue light and have more lateral roots as compared with wild-type plants (Koornneef et al., 1980; Oyama et al., 1997). Recently, a homolog of HY5, HYH, has been reported, mutation in which results in blue light–specific partial etiolation (Holm et al., 2002). Mutations in bHLH protein HFR1/REP1/RSF1 lead to an etiolated phenotype in the far-red light (Fairchild et al., 2000; Soh et al., 2000; Spiegelman et al., 2000). Two other bHLH proteins, PIF3 and PIF4, have been shown to be involved in phytochrome-mediated transcriptional regulation. Furthermore, it has been demonstrated that phyB interacts with PIF3, which is bound to DNA (Ni et al., 1998; Huq and Quail, 2002). Mutational studies have recently shown that PIF3 negatively regulates phyB-mediated inhibition in hypocotyl elongation (Kim et al., 2003). LAF1, a MYB protein, has been shown to be involved in far-red light signaling (Ballesteros et al., 2001). Two other MYB proteins, LHY and CCA1, are involved in circadian rhythm (Wang and Tobin, 1998; Mizoguchi et al., 2002).
A group of 11 different repressors of photomorphogenesis, COP/DET/FUS, acting downstream to photoreceptors has been identified and demonstrated to be downregulating the expression of several light-inducible genes in the darkness (Miséra et al., 1994; Deng and Quail, 1999; Wei and Deng, 1999). Among these, COP1 has been studied in detail. The cop1 mutant seedlings show photomorphogenic growth in dark and develop less lateral roots as compared with wild-type plants (Deng et al., 1991; Deng and Quail, 1999). COP1 acts as a ubiquitin ligase and helps in the degradation of HY5, HYH, and LAF1 in the dark (Ang et al., 1998; Osterlund et al., 2000; Holm et al., 2002; Seo et al., 2003). SPA1 acts as a negative regulator of far-red light signaling. Recent studies have shown that COP1 interacts with SPA1, and this interaction modulates the proteasome-mediated degradation of HY5 and LAF1 (Hoecker et al., 1998; Saijo et al., 2003; Seo et al., 2003; Laubinger et al., 2004).
Regulation of transcription of specific genes is an important mechanism by which light regulates plant growth and development (Tobin and Kehoe, 1994; Terzaghi and Cashmore, 1995; Millar and Kay, 1996). CAB, RBCS, and CHS are well-studied genes that are upregulated by light (Ha and An, 1988; Donald and Cashmore, 1990; Sun and Tobin, 1990; Gilmartin et al., 1992). Investigations of the promoters of the light-inducible genes, including CAB, RBCS, and CHS, have led to identification of four commonly found light-responsive elements (LREs): G, GATA, GT1, and Z-box, which have been demonstrated to be essential for light-mediated transcriptional activity (Terzaghi and Cashmore, 1995; Puente et al., 1996; Yadav et al., 2002). Several LRE-specific transacting factors have been identified, and in some cases, their functions in light signaling pathways have been investigated (Tobin and Kehoe, 1994; Terzaghi and Cashmore, 1995; Wang et al., 1997).
The existence of crosstalk among various signaling pathways in plants has just started to be revealed. The Arabidopsis DEAD-box RNA helicase mutant los4 is chilling sensitive and impaired in the cold-regulated expression of CBF genes (Gong et al., 2002). Phytochrome-mediated light signaling has been shown to be involved in the regulation of TOP2, one of the components of DNA replication and cell cycle machinery (Hettiarachchi et al., 2003). Interestingly, a promoter determinant, C/DRE, which is known to respond to low temperature, has been shown to be involved in phyB-mediated light signaling to cold-induced gene expression (Kim et al., 2002). Using studies with Arabidopsis mutants in light perception, it was recently shown that phytochrome signaling interacts with salicylic acid signal transduction (Genoud et al., 2002). Weatherwax (1996) earlier demonstrated an interaction of light and abscisic acid (ABA) in the regulation of plant gene expression in Lemna gibba.
ABA plays an important role in the regulation of plant water balance and osmotic stress tolerance (Leung and Giraudat, 1998; Finkelstein and Lynch, 2000; Shinozaki and Yamaguchi-Shinozaki, 2000). AtMYC2 is a basic helix-loop-helix (bHLH) transcription factor, which has been shown to be functioning in ABA signaling pathways (Abe et al., 2003). Additionally, very recently AtMYC2/JIN1 has been shown to be acting as a transcription factor in jasmonic acid (JA) and JA-ethylene signaling pathways (Anderson et al., 2004; Boter et al., 2004; Lorenzo et al., 2004). In this article, we further demonstrate that AtMYC2/JIN1 is involved in light-regulated gene expression and photomorphogenic growth in Arabidopsis. It was previously shown by DNA–protein interaction studies that Z-box binding activity was present in Arabidopsis (Yadav et al., 2002). We have performed ligand binding screening to screen an Arabidopsis cDNA expression library for Z-box binding factors (ZBFs) and have identified several such factors. We have investigated the functional relevance to light-regulated gene expression and photomorphogenic growth of one such factor, ZBF1 (AtMYC2), in this study.
RESULTS
DNA-Ligand Binding Screening Leads to Molecular Cloning of ZBF1 (AtMYC2)
A DNA-ligand binding screening was set up to identify and clone ZBF(s). We screened ∼2 × 106 clones of a cDNA expression library, made of 5-d-old constant white light–grown seedlings, using a dimeric Z-box LRE as probe (Figure 1A). Thus far, three genes have been identified and cloned from this screening, the products of which showed specific interactions with the Z-box. One such gene, ZBF1 (AtMYC2), represented by four independent cDNA clones, was selected here for further study. To determine the binding specificity of the clone (AtMYC2) obtained from tertiary screening, we blotted the plaques onto the membrane and cut the membrane into two halves: one half was probed with the Z-box and the other half was probed with either the GT1 or GATA LRE. Whereas a strong binding activity was found with the Z-box, no such binding activity was detected with the GATA or GT1 LRE (Figure 1A; data not shown), suggesting that AtMYC2 specifically interacts with the Z-box.
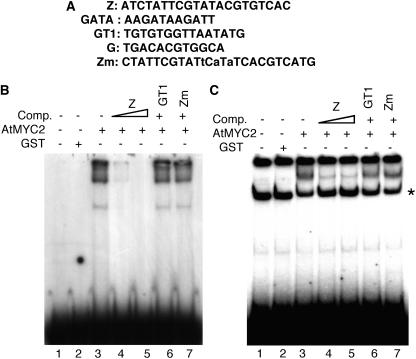
Figure 1.
AtMYC2 Interacts with the Z-Box LRE of Light-Responsive Promoters.
(A) The DNA sequences of various LREs used in this study (Puente et al., 1996; Yadav et al., 2002).
(B) Gel shift assays using GST-AtMYC2 and the consensus dimeric Z-box LRE as probe. Approximately 200 ng of recombinant protein was added (lanes 3 to 7) to the radioactively labeled Z-box. No protein was added in lane 1, and 500 ng of GST protein was added in lane 2. The DNA–protein complexes were resolved on 8% native polyacrylamide gel. The triangle indicates increasing concentrations of the competitors (Comp.), and the plus and minus signs indicate the presence or absence of competitors, respectively. A tetrameric GT1 LRE was used as a nonspecific competitor (Puente et al., 1996). Zm is a mutated version of the Z-box LRE (Yadav et al., 2002) that was also used as nonspecific competitor.
(C) Gel shift assays using GST-AtMYC2 and the CAB1 minimal light-responsive promoter as probe. Approximately 200 ng of recombinant protein was added (lanes 3 to 7) to radioactively labeled, 189-bp DNA fragment of the CAB1 minimal promoter. No protein was added in lane 1, and 500 ng of GST protein was added in lane 2. The DNA–protein complexes were resolved on a 6% native polyacrylamide gel. A tetrameric GT1 LRE was used as a nonspecific competitor (Puente et al., 1996). Zm is a mutated version of the Z-box (Yadav et al., 2002) that has also been used as nonspecific competitor. The triangle indicates increasing concentrations of the competitors (Comp.), and the plus and minus signs indicate the presence and absence of competitor DNA, respectively. The asterisk indicates a spurious band present in all the lanes.
The coding sequence of AtMYC2 cDNA isolated from the ligand binding screening appeared to be a full-length cDNA (At1g32640). It codes for a protein of 623 amino acids (predicted molecular mass of 68 kD) with a bHLH domain. Previously, the same protein was identified from ligand binding screenings by two independent groups and designated as RAP1 (de Pater et al., 1997), and AtMYC2 (Abe et al., 1997, 2003). Studies with RAP1 revealed that the protein interacted with the G-box (CACNTG) motif in pea (Pisum sativum) lectin promoter (de Pater et al., 1997). On the other hand, studies with AtMYC2 demonstrated that the protein interacted with the CACATG sequence, a dehydration-responsive cis-acting element in rd22 promoter (Abe et al., 1997). Boter et al. (2004) have very recently demonstrated that JAMYC2, a functional homolog of AtMYC2, recognizes the AAACGTG element.
Deletion analyses of Arabidopsis CAB1 promoter have demonstrated that the Z-box is essential for the light-dependent developmental expression of CAB1 (Ha and An, 1988). Furthermore, combinatorial interactions of Z-box with other LREs have revealed that the Z-box containing synthetic as well as native promoters are regulated by several components of the light signaling pathways (Puente et al., 1996; Yadav et al., 2002). In general, the bHLH proteins are demonstrated to be interacting with the hexameric DNA sequence referred to as E-box (CANNTG). Depending on the phylogenetic analysis, bHLH proteins have been divided into four monophyletic groups (Ledent and Vervoort, 2001). One such group binds to the ACGTG core sequence, which is included in the Z-box (ATACGTGT).
AtMYC2 Interacts with the Z- and G-Box LREs Commonly Found in Minimal Light–Responsive Promoters
To further test whether AtMYC2 specifically interacts with the Z-box, we used purified glutathione S-transferase–AtMYC2 (GST-AtMYC2) fusion protein and dimeric Z-box DNA as probe in electrophoretic mobility shift (gel shift) assays. A high affinity DNA–protein complex was detected along with the free probe, as shown in Figure 1B (lane 3). Whereas this DNA binding activity was competed out with 50 or 100 molar excess of unlabeled Z-box DNA (Figure 1B, lanes 4 and 5), no competition was observed with 100 molar excess of GT1 or Zm, a mutated version of the Z-box (Figure 1B, lanes 6 and 7).
We then tested the ability of AtMYC2 to interact with the Z-box of native light-regulated CAB1 minimal promoter. We used the 189-bp, light-responsive minimal promoter region of Arabidopsis CAB1 for gel shift assays. As shown in Figure 1C, GST alone did not show any binding activity; however, a strong low mobility DNA–protein complex was formed with GST-AtMYC2 fusion protein (lanes 2 and 3). This DNA–protein complex was efficiently competed out with 50 and 100 molar excess of unlabeled Z-box (Figure 1C, lanes 4 and 5) but not with 100 molar excess of GT1 or Zm (Figure 1C, lanes 6 and 7). Taken together, these results suggest that AtMYC2 specifically interacts with Z-box LRE.
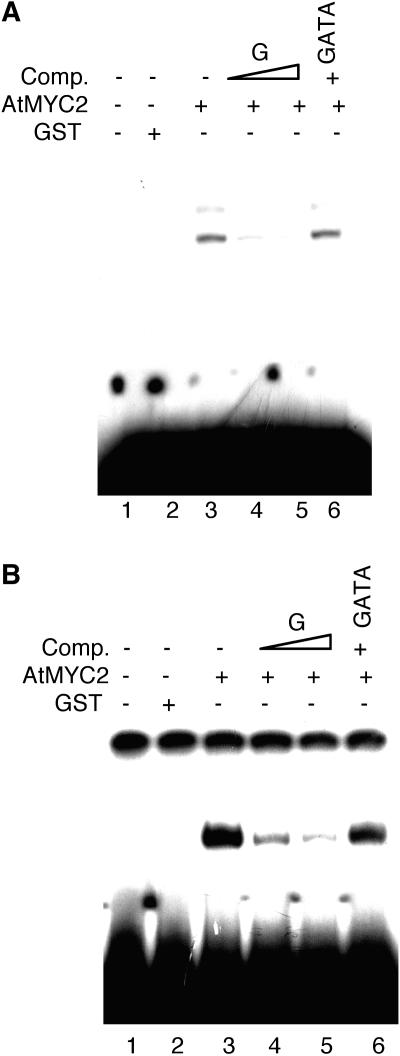
To test whether the bHLH protein AtMYC2 is also able to interact with the G-box (which includes the E-box) of light-regulated promoters, we performed gel shift assays using purified GST-AtMYC2 fusion protein and a consensus tetrameric G-box LRE as probe (Chattopadhyay et al., 1998a). As shown in Figure 2A, a low mobility DNA–protein complex was formed that was competed out by 80 and 150 molar excess of unlabeled G-box but not with 150 molar excess of unlabeled GATA LRE (Figure 2A, lanes 3 to 6).
Figure 2.
AtMYC2 Interacts with the G-Box LRE of Light-Regulated Promoters.
(A) Gel shift assays using GST-AtMYC2 and the consensus tetrameric G-box LRE. Approximately 300 ng of recombinant protein was added (lanes 3 to 6) to the radioactively labeled G-box. No protein was added in lane 1, and 500 ng of GST protein was added in lane 2. The DNA–protein complexes were resolved on a 7% native polyacrylamide gel. The triangle indicates increasing concentrations of the competitors (Comp.), and the plus and minus signs indicate the presence or absence of competitor DNA, respectively. A tetrameric GATA LRE was used as a nonspecific competitor (Puente et al., 1996).
(B) Gel shift assays using GST-AtMYC2 and the RBCS-1A minimal light-responsive promoter. Approximately 300 ng of recombinant protein was added (lanes 3 to 6) to radioactively labeled, 196-bp DNA fragment of the RBCS-1A minimal promoter. No protein was added in lane 1, and 500 ng of GST protein was added in lane 2. The DNA–protein complexes were resolved on a 6% native polyacrylamide gel. A tetrameric GATA LRE was used as a nonspecific competitor (Puente et al., 1996). The triangle indicates increasing concentrations of the competitors (Comp.), and the plus and minus signs indicate the presence or absence of competitors, respectively.
To further substantiate the above result, we used a 196-bp minimal promoter fragment of RBCS-1A for gel shift assays. The minimal promoter region of RBCS-1A contains a G-box LRE, which has been demonstrated to be critical for light-mediated activation of this promoter (Donald and Cashmore, 1990). This minimal promoter fragment contains three GT1 and two GATA (or I) LREs in addition to the G-box. AtMYC2 formed a strong DNA–protein complex (Figure 2B, lane 3), which was competed out by 80 and 150 molar excess of unlabeled 26-bp double-stranded oligonucleotide containing the native G-box of RBCS-1A promoter (Chattopadhyay et al., 1998a) but not with 150 molar excess of GATA (Figure 2B, lanes 4 to 6). Taken together, these results suggest that AtMYC2 interacts with both the Z- and G-box LREs of light-regulated promoters.
Isolation and Characterization of Mutations in AtMYC2
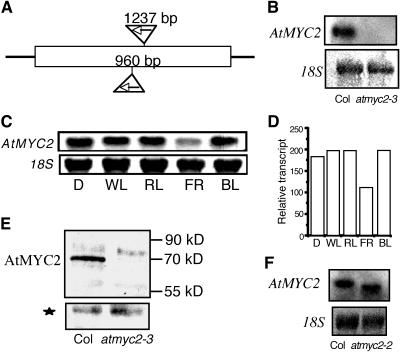
Because AtMYC2 interacts with the Z- and G-box LREs present in the light-regulated promoters of CAB1 and RBCS-1A, respectively, we ask whether AtMYC2 is involved in the regulation of photomorphogenic growth in Arabidopsis. To address this question, we searched for mutants in T-DNA knockout collections (Alonso et al., 2003). A mutant line with a T-DNA insertion at the 5′ end of AtMYC2 coding sequence (Salk_017005) was identified, and the corresponding allele was designated as atmyc2-3 (atmyc2-1 and atmyc2-2 alleles were already described to have less sensitivity to JA in Boter et al., 2004). Heterozygous T1 plants with the T-DNA insertion allele showed 3:1 segregation ratios with kanamycin resistance versus sensitive lines in T2 progeny, suggesting that one single T-DNA insertion locus is present in atmyc2-3 mutant plants. The junctions of T-DNA and AtMYC2 were amplified by PCR, and the DNA sequence analyses revealed that the T-DNA was inserted in nucleotide position 960 bp from the start codon (Figure 3A). RNA gel blot and protein gel blot analyses were unable to detect any transcript or protein encoded by AtMYC2 in atmyc2-3 mutant background (Figures 3B and 3E). Therefore, the T-DNA insertion in AtMYC2 likely caused instability of the corresponding transcript, resulting in a null mutant. A second mutant line (atmyc2-2) with a T-DNA insertion (Salk_083483) at the 5′ end of the AtMYC2 coding sequence was also identified where the T-DNA was inserted in nucleotide position 1237 bp from the start codon (Figures 3A and 3F) (Boter et al., 2004).
Figure 3.
Identification of T-DNA–Tagged Mutation in AtMYC2.
(A) The schematic diagram of the T-DNA insertion sites in AtMYC2. The inverted triangles show the T-DNA insertion sites after 960 or 1237 bp from the start codon.
(B) RNA gel blot of 20 μg of total RNA isolated from 6-d-old white light–grown wild-type (Columbia [Col]) and atmyc2-3 mutant seedlings. A 1.8-kb AtMYC2 DNA fragment was used as probe. The 18S rRNA has been shown as loading control.
(C) Light-regulated expression of AtMYC2. Six-day-old seedlings grown in constant dark (D), white light (WL), far-red light (FR), red light (RL), or blue light (BL) were used for RNA gel blot analyses. Twenty micrograms of total RNA was loaded onto each lane. A 1.8-kb AtMYC2 DNA fragment was used as probe. The 18S rRNA has been shown as loading control. A representative autorad from at least three independent experiments is shown.
(D) Quantification of the data in (C) by the Fluor-S-MultiImager (Bio-Rad, Hercules, CA).
(E) Protein gel blot of 20 μg of total protein extracts prepared from 6-d-old white light–grown wild-type (Col) and atmyc2-3 mutant seedlings. The AtMYC2 protein detected by AtMYC2 antibodies is indicated. The molecular weights of the protein bands are marked. The star marks a cross-reacting protein band in the same blot, indicating the loading control.
(F) RNA gel blot of 20 μg of total RNA isolated from 6-d-old white light–grown wild-type (Col) and atmyc2-2 mutant seedlings. A 1.8-kb AtMYC2 DNA fragment was used as probe. The 18S rRNA is shown as loading control.
To characterize the light regulation of AtMYC2 expression, we examined the relative levels of AtMYC2 expression in 6-d-old constant dark or various light-grown wild-type seedlings, including red light (RL), far-red light (FR), and blue light (BL). As shown in Figures 3C and 3D, AtMYC2 is expressed in dark and in all light conditions tested. The levels of expression were found to be almost similar in dark and various light-grown conditions with slightly lower level in FR (Figure 3D). These results suggest that AtMYC2 is constitutively expressed in dark- and light-grown Arabidopsis seedlings.
atmyc2 Mutants Exhibit BL-Specific Morphological Defects in Seedling Development
We measured the hypocotyl length of 6-d-old atmyc2 mutants and wild-type seedlings grown under constant dark or white light (WL) conditions. However, no significant difference in hypocotyl length was detected between wild-type and atmyc2 mutant seedlings grown in constant darkness or WL conditions (Figure 4A; data not shown). To determine whether the atmyc2 mutants have any altered morphology in a particular wavelength of light, we examined the growth of 6-d-old seedlings under various wavelengths of light, such as RL, FR, and BL. The enhanced inhibition in hypocotyl elongation of atmyc2 was observed in constant BL; however, no significant change in hypocotyl length was observed in constant FR or RL (Figures 4B to 4D, 4G, and 4H). Measurements of hypocotyl length revealed that 6-d-old BL-grown atmyc2 mutant seedlings had significantly shorter hypocotyls as compared with wild-type seedlings with no significant change in RL or FR at various fluences (Figures 5A to 5C). These results suggest that AtMYC2 acts as a negative regulator of BL-mediated photomorphogenic growth.
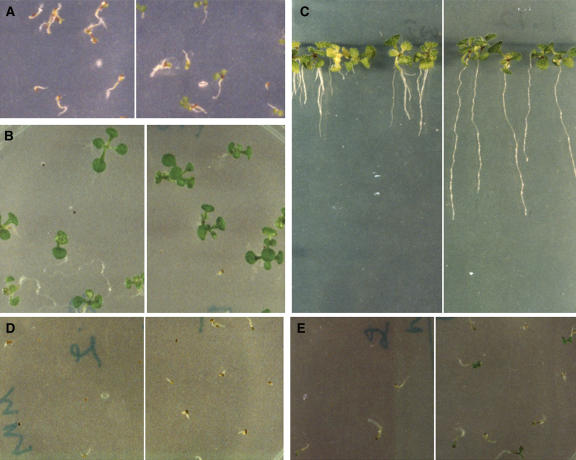
Figure 4.
The atmyc2 Mutants Show Multiple Phenotypes.
In each panel, segregated wild-type (Col) and atmyc2-3 ([A] to [F]) or atmyc2-2 ([G] and [H]) mutants are shown on the left and right sides, respectively.
(A) Six-day-old constant dark-grown seedlings.
(B) Six-day-old constant RL-grown (95 μmol/s/m2) seedlings.
(C) Six-day-old constant BL-grown (30 μmol/s/m2) seedlings.
(D) Six-day-old constant FR-grown (90 μmol/s/m2) seedlings. The arrowhead indicates the accumulation of anthocyanin.
(E) The root growth of 16-d-old wild-type and atmyc2-3 mutant plants grown in a long day cycle of 16 h of WL (100 μmol/s/m2) and 8 h of darkness.
(F) Adult plants (21 d old) grown in a long day cycle of 16 h of WL (100 μmol/s/m2) and 8 h of darkness.
(G) Six-day-old constant BL-grown (30 μmol/s/m2) seedlings.
(H) Six-day-old constant FR-grown (90 μmol/s/m2) seedlings.
(I) Six-day-old constant BL-grown (30 μmol/s/m2) wild-type (left) and complemented atmyc2-3 mutants with wild-type copy of AtMYC2 (right) are shown.
Figure 5.
atmyc2 Mutants Are Epistatic to cry1 and cry2.
(A) Hypocotyl length of 6-d-old constant RL-grown wild-type (Col) and atmyc2-3 mutant seedlings at various fluence rates.
(B) Hypocotyl length of 6-d-old constant FR-grown wild-type (Col) and atmyc2-3 mutant seedlings at various fluence rates.
(C) Hypocotyl length of 6-d-old constant BL-grown wild-type (Col), atmyc2-3, and atmyc2-2 mutant seedlings at various fluence rates.
(D) Hypocotyl lengths of 6-d-old constant BL-grown wild-type, atmyc2, cry1, and atmyc2 cry1 seedlings at various fluence rates.
(E) Hypocotyl lengths of 6-d-old constant BL-grown wild-type, atmyc2, cry2, and atmyc2 cry2 seedlings at various fluence rates.
(F) Hypocotyl lengths of 6-d-old constant BL-grown wild-type, atmyc2, phyA, and atmyc2 phyA seedlings at various fluence rates.
Although FR-grown atmyc2 mutants did not show any altered morphology, the mutant seedlings had higher accumulation of anthocyanin at the junction of hypocotyls and cotyledons (Figures 4D and 4H), a characteristic of hyperphotomorphogenic growth during early seedling development in Arabidopsis (Ang et al., 1998). Examination of root growth of atmyc2 mutant plants revealed that 16-d-old mutant plants developed significantly less lateral roots as compared with wild-type plants (Figure 4E). Furthermore, whereas atmyc2 mutant seedlings did not exhibit any altered morphology while grown in various fluences of WL, the mutant adult plants exhibited significantly short stature as compared with WL-grown wild-type plants (Figure 4F). Taken together, these results suggest that AtMYC2 acts as a negative regulator of photomorphogenesis and its effect is more pronounced under BL condition. These results further demonstrate that AtMYC2 acts as a positive regulator of lateral root formation.
A genomic fragment containing AtMYC2 and its upstream sequence of ∼1.5 kb was introduced into the atmyc2-3 mutant plants for a complementation test. The transgenic seedlings were unable to display a BL-specific hypersensitive response, suggesting that the observed phenotypes of atmyc2 mutants were caused by the loss of AtMYC2 function (Figure 4I; see Supplemental Figure 1 online). Because the loss of function of AtMYC2 leads to enhanced sensitivity to BL irradiation, we examined whether an increased level of AtMYC2 leads to reduced inhibition in hypocotyl elongation. However, the transgenic seedlings overexpressing AtMYC2 did not show significant change in sensitivity to WL or BL, although the transcript levels of AtMYC2 in these lines were dramatically elevated (see Supplemental Figure 2 online).
We performed epistasis analyses to determine the involvement of photoreceptors in AtMYC2 function. The atmyc2 cry1 and atmyc2 cry2 double mutants displayed similar hypocotyl lengths as atmyc2 mutant seedlings in BL (Figures 5D and 5E). However, atmyc2 phyA double mutants exhibited a hypocotyl length similar to phyA mutant seedlings in BL (Figure 5F). These results suggest that atmyc2 is epistatic to cry1 and cry2; however, phyA is likely to be epistatic to atmyc2 in BL.
atmyc2 Mutants Are Less Sensitive to ABA and JA Responsiveness
It was previously shown that mutation in AtMYC2 (generated by an Ac/Ds tagging system) caused Arabidopsis plants to be less sensitive to ABA (Abe et al., 2003). Furthermore, it has been recently demonstrated that jin1-1 mutants are less sensitive to JA (Lorenzo et al., 2004). To determine whether atmyc2 mutants respond to ABA and JA in a similar fashion, we monitored the effect of ABA and JA on atmyc2-3 mutant plants. Seeds of wild-type and mutant plants were plated on MS plates without or with various concentrations of ABA. Whereas 1 μM ABA reduced the rate of germination of wild-type seeds, the effect was significantly suppressed in atmyc2-3 mutants (Figure 6A). However, no noticeable effect of ABA on growth of the atmyc2-3 mutants, which were germinated in 1 μM ABA, was observed as compared with wild-type plants (Figure 6B).
Figure 6.
atmyc2-3 Mutants Are Less Responsive to ABA and JA.
In each panel, segregated wild-type (Col) and atmyc2-3 mutants are shown on the left and right sides, respectively.
(A) Six-day-old seedlings grown in constant WL (100 μmol/s/m2) in the presence of 1 μM ABA.
(B) Twelve-day-old constant WL-grown (100 μmol/s/m2) seedlings in the presence of 1 μM ABA.
(C) The root growth of 16-d-old constant WL-grown (100 μmol/s/m2) plants in the presence of 20 μM JA.
(D) Six-day-old seedlings grown in constant BL (30 μmol/s/m2) in the presence of 1 μM ABA.
(E) Six-day-old seedlings grown in constant RL (95 μmol/s/m2) in the presence of 1 μM ABA.
It has been reported recently that mutations in JIN1 result in less sensitivity to JA-mediated root growth retardation (Lorenzo et al., 2004). To determine the effect of JA on the root growth of atmyc2-3 mutant plants, we grew wild-type and atmyc2-3 mutant plants in the presence of 20 μM JA and monitored the root growth. JA caused severe root growth retardation in wild-type plants; however, the effect was drastically reduced in atmyc2-3 mutant plants (Figure 6C). These results altogether indicate that atmyc2-3 mutants are less sensitive to ABA- and JA-mediated responses. To determine whether the ABA- and JA-mediated effects are light specific, we performed the above experiments in various light conditions, including BL, where the effect of mutations in AtMYC2 is prominent. However, our results indicate that the less sensitivity of atmyc2-3 mutants to ABA and JA is not BL specific (Figures 6D and 6E; data not shown).
Mutation in AtMYC2 Results in a Higher Level of Chlorophyll and Anthocyanin Accumulation
Light signaling controls various physiological processes through the regulation of various light-responsive genes (Ma et al., 2001; Tepperman et al., 2001). The accumulation of chlorophyll and anthocyanin are two such important physiological responses. To determine whether AtMYC2 plays any role in chlorophyll or anthocyanin accumulation, we measured the chlorophyll and anthocyanin contents in wild-type and atmyc2 mutant seedlings under various wavelengths of light. As shown in Figures 7A and 7B, the chlorophyll and anthocyanin contents, respectively, were significantly higher in atmyc2-3 mutants as compared with wild-type seedlings in BL. Furthermore, the anthocyanin content of atmyc2-3 mutant seedlings was found to be significantly higher as compared with the wild type in FR (Figure 7C). While propagating atmyc2-3 mutant plants, we observed that atmyc2-3 mutation caused late flowering. Whereas long day–grown (16 h light/8 h dark) wild-type plants start flowering after the formation of approximately eight rosette leaves, the atmyc2-3 mutants flower after producing ∼13 rosette leaves (Figure 7D). However, the short day–grown (8 h light/16 h dark) atmyc2-3 mutant plants were unable to display such effects (Figure 7E).
Figure 7.
Characterization of atmyc2 Mutants.
(A) Accumulation of chlorophyll a/b in 6-d-old constant BL-grown (30 μmol/s/m2) wild-type and atmyc2-3 mutant seedlings.
(B) Accumulation of anthocyanin in 6-d-old constant BL-grown (30 μmol/s/m2) wild-type and atmyc2-3 mutant seedlings.
(C) Accumulation of anthocyanin in 6-d-old constant FR-grown (90 μmol/s/m2) wild-type (Col) and atmyc2-3 mutant seedlings.
(D) Number of rosette leaves formed at the time of bolting in wild-type (Col) and atmyc2-3 mutant plants grown in long-day conditions of 16 h of WL (100 μmol/s/m2) and 8 h of dark.
(E) Number of rosette leaves formed at the time of bolting in wild-type (Col) and atmyc2-3 mutant plants grown under short-day conditions of 8 h of WL (100 μmol/s/m2) and 16 h of dark.
AtMYC2 Negatively Regulates the Expression of Light-Inducible Genes
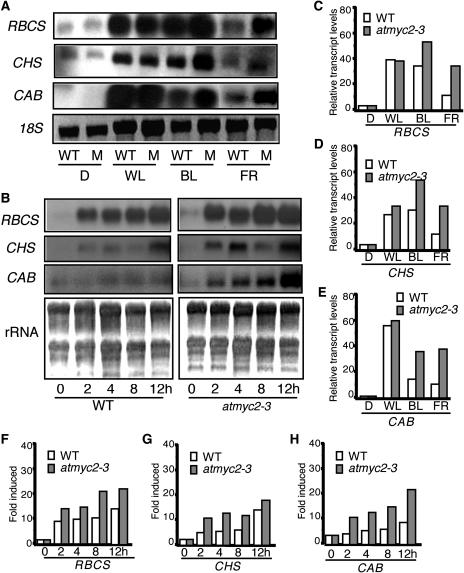
To determine the role of AtMYC2 in the regulation of light-inducible gene expression, we performed RNA gel blot analyses and measured the expression of CAB, RBCS, and CHS genes in 6-d-old various light-grown seedlings. As shown in Figure 8A, the expression of the light-inducible genes was significantly elevated in atmyc2-3 mutants as compared with wild-type seedlings in BL and FR. In the case of RBCS, whereas an approximately twofold increase in the transcript level was detected in BL, the expression of the gene was found to be more than threefold higher in the atmyc2-3 mutant background as compared with the wild type in FR (Figure 8C). Very little increase, if any, in the expression of CHS and CAB was detected in atmyc2-3 mutants in WL; however, an approximately twofold to threefold increase was detected in BL and FR as compared with wild-type background (Figures 8D and 8E). No significant change in expression of these genes was detected in the atmyc2-3 mutant in RL (data not shown). Taken together, these results suggest that AtMYC2 acts as a negative regulator of CAB, RBCS, and CHS in BL- and FR-meditated expression.
Figure 8.
Expression of Light-Inducible Genes in atmyc2 Mutant Background.
(A) Transcript level of RBCS, CHS, and CAB in 6-d-old wild-type and atmyc2-3 mutant (M) seedlings grown under constant dark (D), WL, BL, or FR conditions. Ten micrograms of total RNA was loaded onto each lane. The DNA fragments of CAB, RBCS, and CHS were used as probes (Deng et al., 1991). The 18S rRNA is shown as loading control.
(B) RNA gel blot analysis of RBCS, CHS, and CAB in wild-type and atmyc2-3 mutant seedlings. Four-day-old dark-grown seedlings were transferred to BL for 2, 4, 8, and 12 h, and total RNA was extracted from each sample for RNA gel blot analysis. Ten micrograms of total RNA was loaded onto each lane. Five-day-old dark-grown seedlings are shown as 0 h. rRNA has been shown as loading control.
(C) Quantification of the data in (A) of RBCS by the Fluor-S-MultiImager.
(D) Quantification of the data in (A) of CHS by the Fluor-S-MultiImager.
(E) Quantification of the data in (A) of CAB by the Fluor-S-MultiImager.
(F) Quantification of the data in (B) of RBCS by the Fluor-S-MultiImager.
(G) Quantification of the data in (B) of CHS by the Fluor-S-MultiImager.
(H) Quantification of the data in (B) of CAB by the Fluor-S-MultiImager.
To further examine the light-mediated induction of CAB, RBCS, and CHS in the atmyc2-3 mutant background, 4-d-old dark-grown seedlings were transferred to BL for 2, 4, 8, and 12 h, and the transcript levels were measured. As shown in Figure 8B, the level of induction of CAB, RBCS, and CHS genes was significantly elevated in atmyc2-3 mutants as compared with wild-type seedlings at various time points. Whereas a more than eightfold induction in RBCS expression was found in atmyc2-3 after 12 h, a less than fivefold induction was detected in the wild-type background (Figures 8B and 8F). In the case of CHS, an approximately sixfold induction was detected in atmyc2-3; however, an approximately fourfold induction was found in the wild-type background at 12 h (Figures 8B and 8G). Similarly, the expression of CAB was induced to approximately fivefold in atmyc2-3 mutants; however, an approximately twofold induction was detected in the wild-type background (Figures 8B and 8H). Taken together, these results suggest that AtMYC2 plays a negative regulatory role in the BL-mediated induction of CAB, RBCS, and CHS genes.
DISCUSSION
Several light-specific photomorphogenesis promoting factors have been reported in light signaling pathways. However, only a few repressors of photomorphogenesis have been reported that act in a light-specific manner (Hoecker et al., 1998, 1999; Dieterle et al., 2001; Guo et al., 2001). Here, we have reported a BL-specific repressor of photomorphogenic growth. Mutational studies with AtMYC2 highlight the existence of crosstalk among light, ABA, and JA signaling and thus establish a functional relationship among these signaling pathways.
AtMYC2 Interacts with the Z- and G-Box LREs of Light-Regulated Promoters
We have identified ZBF1 (AtMYC2) in a ligand binding screening using Z-box LRE as a probe. Our results with DNA–protein interaction studies provide several lines of evidence that AtMYC2 recognizes the Z- and G-box LREs of light-regulated promoters. Ligand binding and gel shift assays with individual LREs or light-regulated minimal promoter fragments of CAB1 and RBCS-1A clearly demonstrate that AtMYC2 specifically binds to the Z- or G-box LREs. Recognition of two different cis-acting elements by a specific transcription factor is not unprecedented. It has been demonstrated that ACGT-containing ABA responsive element and coupling element 3 are recognized by the same transcription factor, TRAB1 (Hobo et al., 1999). The functional equivalence of two or more elements is usually based on the sequence similarities and on being recognized by the same transcription factor. Therefore, the interaction of AtMYC2 transcription factor with G- and Z-box LREs [(C/T)ACGTG], which have been shown to be essential and sufficient for light-mediated induction of RBCS-1A and CAB1 promoters, respectively, probably indicate that these two LREs are functionally equivalent with respect to AtMYC2. It is worth mentioning here that at least one other ZBF that has been identified from the ligand binding screening is also able to recognize the G-box of light-inducible promoters as well (M. Chandrashekara and S. Chattopadhyay, unpublished data).
AtMYC2, a BL-Specific Repressor of Photomorphogenesis
The analyses of atmyc2 mutants clearly demonstrate that the short hypocotyl phenotype of atmyc2 seedlings is restricted to BL. These results suggest that although AtMYC2 is expressed in the dark-grown and various light-grown seedlings, it functions as a negative regulator of BL-mediated photomorphogenic growth. At least three downstream signaling components in BL, HYH, AtPP7, and SUB1, have been reported earlier. Whereas HYH and AtPP7 act as positive regulators of BL-mediated photomorphogenic growth, SUB1 acts as a negative regulator of BL- and FR-mediated signaling (Guo et al., 2001; Holm et al., 2002; Moller et al., 2003). SUB1 acts downstream to both cry1 and cry2 photoreceptors and is a point of crosstalk between phyA and cryptochrome signaling pathways (Guo et al., 2001). In this study, the epistasis analyses using atmyc2 cry1, atmyc2 cry2, or atmyc2 phyA double mutants indicate that AtMYC2 acts downstream to both cry1 and cry2 photoreceptors, and the increased sensitivity to BL caused by atmyc2 mutation also requires light perception by phyA. Therefore, AtMYC2-mediated inhibition may play an important role in negative feedback control of cryptochrome signaling. However, the function of phyA is likely to be independent of AtMYC2. Considering the altered light responsiveness of sub1 and atmyc2 mutants and the results of epistasis analyses, it could be envisioned that SUB1 and AtMYC2 might function closely in the same branch of the light signaling pathways.
Analyses of the light-regulated gene expression in atmyc2 mutants reveal that AtMYC2 represses the BL-mediated expression of CAB, RBCS, and CHS genes. Furthermore, although atmyc2 mutants do not exhibit any morphological defects in FR, the light-regulated genes are upregulated in FR in the atmyc2 mutant background. These results demonstrate that AtMYC2 plays a negative regulatory role in the expression of light-inducible genes in a BL- and FR-specific manner.
It has already been demonstrated that AtMYC2 (JIN1) acts as a transcriptional regulator in ABA and JA signaling pathways (Abe et al., 2003; Boter et al., 2004; Lorenzo et al., 2004). We have examined the ABA and JA responsiveness of atmyc2-3 mutants, and our results demonstrate that the atmyc2-3 mutant plants are partially insensitive to ABA and JA. However, the compromised sensitivity of atmyc2-3 mutants to ABA and JA is not specific to a particular wavelength of light. Abe et al. (2003) have reported that mutations in AtMYC2 result in better growth in the presence of ABA as compared with wild-type plants. However, our studies with atmyc2-3 mutants were unable to detect the corresponding effect. It is possible that this effect is more prominent at the adult stage (as found in Abe et al., 2003) rather than in 12-d-old plants. Studies with cop1 mutants have revealed that COP1, a master repressor of photomorphogenic growth in the darkness, acts as a positive regulator of lateral root formation (Ang et al., 1998). Analyses of atmyc2-3 mutants, in this study, have revealed that although AtMYC2 is a negative regulator of BL-mediated photomorphogenic growth, it is essential for optimum lateral root formation.
AtMYC2 Regulates Positive and Negative Responses of Light, ABA, and JA Signaling Pathways
Several light signaling components have been described previously, which function as positive as well as negative regulators of light responses (Deng et al., 1991; Wang et al., 1997; Chattopadhyay et al., 1998b; Liu et al., 2001). For example, PIF3, a phytochrome interacting bHLH protein, acts as a positive regulator for CHS induction but negatively regulates the inhibition of hypocotyl elongation, cotyledon opening, and expansion (Kim et al., 2003). We have demonstrated that AtMYC2 is a repressor of BL-mediated photomorphogenic growth and acts as a negative regulator of BL- and FR-regulated gene expression. However, it acts as a positive regulator of lateral root formation. Furthermore, whereas AtMYC2/JIN1 acts as a positive regulator of ABA signaling, it plays both positive and negative regulatory roles in JA signaling pathways (Boter et al., 2004; Lorenzo et al., 2004). However, the exact mechanism of AtMYC2/JIN1-mediated differential regulation is not known. A simple way to explain the differential regulation is to consider that AtMYC2 could function either as a transcriptional activator or repressor, depending on the specific promoter determinants of target genes. Alternatively, extensive heterodimerization of bHLH proteins has been reported (Robinson and Lopes, 2000; Ledent and Vervoort, 2001). Therefore, it could be envisioned that in vivo heterodimerization of AtMYC2 with other bHLH proteins might be a potential mechanism to generate positive and negative regulators, which in turn play opposite regulatory roles in signaling cascades.
Although the JA signaling pathway is poorly understood, ABA signaling has been studied in some detail. Furthermore, potential crosstalk between light and ABA signaling pathways has been reported. Potentially, light and ABA effects are antagonistic. For example, (1) suppression of seed germination by ABA is enhanced in the light (Fellner and Sawhney, 2002); (2) light-grown seedlings accumulate ABA when transferred to darkness and brief red light pulses decrease ABA amounts (Weatherwax et al., 1996); and (3) ABA mutants show altered responses to photoperiod and light quality (Rohde et al., 2000; Fellner and Sawhney, 2002). Plants have evolved the ability to integrate various signals and respond to them accordingly in a comprehensive manner. Demonstration of AtMYC2 as a common transcriptional regulator for light, ABA, and JA signaling establishes a functional relationship among these signaling cascades, which will help in deciphering the mechanism of integration of these signaling pathways in future studies.
METHODS
Plant Materials and Growth Conditions
Surface-sterilized seeds of Arabidopsis thaliana were sown on MS plates, kept at 4°C in darkness for 3 to 5 d, and transferred to specific light conditions at 22°C. The Arabidopsis growth conditions have been described by Yadav et al. (2002). The intensities of continuous light sources used in this study are as follows: WL (100, 30, 15, and 5 μmol/s/m2), BL (30, 20, 15, and 5 μmol/s/m2), RL (95, 30, 15, and 5 μmol/s/m2), and FR (90, 30, 15, and 5 μmol/s/m2). Unless otherwise mentioned, the highest light intensities were used for the experiments.
To obtain the homozygous atmyc2-3 or atmyc2-2 mutant line, plants heterozygous or homozygous for the atmyc2-3 or atmyc2-2 mutation were subjected to PCR genotyping analyses. Individual plants were examined by PCR using the left border specific primer LBP (5′-GCGTGGACCGCTTGCTGCACCT-3′) and the AtMYC2-specific primers LP2 (5′-GATCTGATTCTCCGGCGGTTT-3′) and RP2 (5′-GTTCGCCGCTTTCTACTC-3′) for atmyc2-3 and LP5 (5′-CGGCGAGCTCGAGTTTCACTT-3′) and RP5 (5′-AATTATCCGGGTCGGGTTGTG-3′) for atmyc2-2.
For the generation of overexpressor lines of AtMYC2, full-length cDNA was amplified by PCR using the primers (forward) 5′-GACTAGTAATCGTAGCTTTTGCAGCTTC-3′ and (reverse) 5′-GACTAGTATACAGACTCAAACATAGAGC-3′ and cloned into the BglII-SpeI site of the pCambia1303 vector. For the complementation test, a genomic fragment containing full-length AtMYC2 and ∼1.5-kb upstream DNA sequence was amplified by PCR using the primers (forward) 5′-TCCCCCGGGGAGTAATGGGACCATATTGGTG-3′ and (reverse) 5′-TCCCCCGGGTATCAATATATACAAGTTTACTC-3′ and cloned into the SmaI site of the pBI101.2 vector. The Agrobacterium tumefaciens strain GV3101 was transformed individually with each recombinant construct. The Arabidopsis wild-type (Wassilewskija) plants (for overexpression) or atmyc2-3 mutant plants (for complementation) were transformed with the recombinant plasmid or empty vector by the floral dip method, and transgenic plants were selected on 15-μg/mL hygromycin plates. Several transgenic lines homozygous for each transgene were generated for further studies. For ABA- or JA-responsive experiments, MS plates containing 0.5, 1, or 2 μM ABA or 20 μM JA were used for monitoring growth of atmyc2-3 mutant and wild-type plants.
For the generation of double mutants, such as atmyc2 cry1, atmyc2 cry2, and atmyc2 phyA, homozygous atmyc2-3 mutant plants were crossed individually with hy4-2.23N, cry2-1, and phyA-101 homozygous mutant lines. In the F2 generation, seedlings were grown in WL (60 μmol/s/m2) or FR (30 μmol/s/m2) for the identification of cry1, cry2, or phyA homozygous lines, and elongated seedlings were selected and transferred to soil. To determine the genotype at the AtMYC2 locus, ∼40 seedlings from each line were tested by genomic PCR. F3 progeny that are homozygous for atmyc2-3 mutant plants were further examined and considered as atmyc2 cry1, atmyc2 cry2, and atmyc2 phyA double mutants. Because atmyc2, cry1, cry2, and phyA were of different ecotype backgrounds, F2 seedlings, which were mutant for cry1, cry2, or phyA but homozygous for the wild-type AtMYC2, were used as control.
DNA-Ligand Binding Screening
Ligand binding screening was performed following the protocol of Singh et al. (1988) with some modifications. A cDNA expression library of 5-d-old constant light-grown Arabidopsis seedlings was constructed in λZapII vector (Stratagene, La Jolla, CA). Freshly prepared 150-mm NZY-agar plates (5 g NaCl, 2 g MgSO4, 5 g yeast extract, 10 g NZ amine [casein hydrolysate], and 15 g agar in one liter of water) were used for plating ∼10,000 pfu/plate and incubated for 4 to 6 h at 37°C. These plates were overlaid with nitrocellulose membrane (soaked in 10 mM isopropylthio-β-galactoside solution for 20 min, then dried briefly by keeping on Whatman filter paper [Clifton, NJ]) when the tiny plaques started to develop and incubated for 6 to 8 h at 37°C. These plates were then transferred from 37°C to 4°C for 15 min and marked. The membrane was then lifted off the plate and immersed in 50 mL of blocking solution per membrane. After incubation at room temperature for 1 h, the membrane was washed three times with 50 mL of TNE (15 mM Hepes, pH 7.5, 50 mM KCl, 1 mM EDTA, 1 mM DTT, 1 mM MgCl2, and 5% glycerol) for 5 min. The membrane was then incubated at room temperature with 3′ end–labeled Z-box and 250 μg of sonicated and denatured calf thymus DNA. The membrane was then washed three times with 50 mL of TNE for 10 min. The membrane was dried and autoradiographed. Putative positive plaques were picked up by aligning the autorad with the membrane and the plate. The clones were then subjected to further screening (secondary and tertiary) following the same procedure. The gene was cloned by plasmid rescue method (Stratagene).
RNA Gel Blot and Protein Gel Blot Analyses
Total RNA was extracted using the RNeasy plant minikit (Qiagen, Valencia, CA), and RNA gel blot analysis with 20 μg of total RNA for each sample was performed essentially as described by Hettiarachchi et al. (2003). The 1.8-kb AtMYC2 DNA fragment was used as probe after random prime labeling (Amersham Biosciences, Salem, OR). The DNA fragments of CAB, RBCS, and CHS genes were used for probes as described by Deng et al. (1991). The 18S rRNA was used as loading control. To quantify the RNA gel blot data, the intensity of each band was quantified by the Fluor-S-MultiImager, and ratios of the CAB, RBCS, or CHS gene versus its corresponding rRNA band were determined and plotted (Fluor-S-MultiImager). For protein gel blot analysis, affinity-purified AtMYC2 polyclonal antibodies were used. Protein extracts were prepared from 6-d-old constant WL-grown wild-type and atmyc2 mutant seedlings. Twenty micrograms of total protein was used for protein gel blot analysis. A cross-reacting band was used as a loading control.
Electrophoretic Mobility Shift (Gel Shift) Assays
GST-AtMYC2 was induced using 1 mM isopropylthio-β-galactoside and overexpressed in Escherichia coli. The overexpressed GST-AtMYC2 was affinity purified following the manufacturer's protocol (Amersham Biosciences). The DNA binding assays were performed at room temperature in a final volume of 30 μL with a binding buffer of 15 mM Hepes, pH 7.5, 35 mM KCl, 1 mM EDTA, 6% glycerol, 1 mM DTT, 1 mM MgCl2, and 2 μg of poly(dI-dC). The samples were incubated at room temperature for 15 min and then run on to 6 to 8% polyacrylamide gel at 12 to 15 mA. After drying, the gels were autoradiographed.
The 42-bp DNA fragment containing the Z-box dimer or 46-bp DNA fragments containing the tetrameric G-box cloned in pBluescript SK+ were digested with XhoI and HindIII, purified, and 3′ end labeled with [α-32P]dCTP (Chattopadhyay et al., 1998a; Yadav et al., 2002). The mutant Zm-box cloned in pBluescript was digested with EcoRI-BamHI and purified for competition studies (Yadav et al., 2002). The tetrameric GT1 or GATA elements were purified after digestion with HindIII-XhoI and used for competition reactions (Chattopadhyay et al., 1998a). The 189-bp DNA fragment of CAB1 minimal promoter region was cloned into pBluescript vector after PCR with primers (forward) 5′-CGGAATTCATAAGGATAGAGAGATCTATTC-3′ and (reverse) 5′-CGGGATCCTGAGGTTGCTATTGGCTAGTCAT-3′ using genomic DNA as template. The 189- and 196-bp fragments of native CAB1 and RBCS-1A promoters, respectively, were digested with EcoRI-BamHI, purified, and 3′ end labeled for use as probe for the DNA binding assays (Chattopadhyay et al., 1998a). One nanogram of labeled DNA was used for each binding reaction.
Chlorophyll and Anthocyanin Measurements
Chlorophyll and anthocyanin levels were measured following protocols as described by Holm et al. (2002).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AJ843256.
Supplementary Material
Acknowledgments
We thank Sushil Kumar for critically reading and commenting on this manuscript. We thank Prem Negi for helping with the DNA-ligand binding screening. This work is supported by Department of Biotechnology and Department of Science and Technology grants to S.C. V.Y., C.M., and S.N.G. are recipients of Council of Scientific and Industrial Research fellowships from the government of India.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sudip Chattopadhyay (sudipchatto@yahoo.com).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.032060.
References
- Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson, J.P., Badruzsaufari, E., Schenk, P.M., Manners, J.M., Desmond, O.J., Ehlert, C., Maclean, D.J., Ebert, P.R., and Kazan, K. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, L.-H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.-W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Ballesteros, M.L., Bolle, C., Lois, L.M., Moore, J.M., Vielle-Calzada, J.P., Grossniklaus, U., and Chua, N.H. (2001). LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15, 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter, M., Ruiz-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., Ang, L.-H., Puente, P., Deng, X.-W., and Wei, N. (1998. a). Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay, S., Puente, P., Deng, X.-W., and Wei, N. (1998. b). Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 15, 69–77. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., Caspar, T., and Quail, P.H. (1991). COP1: A regulatory gene, encodes a novel protein with both a Zn-binding motif and G homology domain. Cell 71, 791–801. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., and Quail, P.H. (1999). Signalling in light-controlled development. Semin. Cell Dev. Biol. 10, 121–129. [DOI] [PubMed] [Google Scholar]
- de Pater, S., Pham, K., Memelink, J., and Kijne, J. (1997). RAP-1 is an Arabidopsis MYC-like R protein homologue, that binds to G-box sequence motifs. Plant Mol. Biol. 34, 169–174. [DOI] [PubMed] [Google Scholar]
- Dieterle, M., Zhou, Y.-C., Schafer, E., Funk, M., and Kretsch, T. (2001). EID1, an E-box protein involved in phytochrome A-specific light signaling. Genes Dev. 15, 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald, R.G.K., and Cashmore, A.R. (1990). Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 9, 1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild, C.D., Schumaker, M.A., and Quail, P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14, 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fellner, M., and Sawhney, V.K. (2002). The 7B-1 mutant in tomato shows blue-light-specific resistance to osmotic stress and abscisic acid. Planta 214, 675–682. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya, M. (1993). Phytochromes: Their molecular species, gene families, and functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 617–646. [Google Scholar]
- Genoud, T., Buchala, A.J., Chua, N.H., and Metraux, J.P. (2002). Phytochrome signalling modulates the SA perceptive pathway in Arabidopsis. Plant J. 31, 87–95. [DOI] [PubMed] [Google Scholar]
- Gilmartin, P.M., Memelink, J., Hiratsuka, K., Kay, S.A., and Chua, N.-H. (1992). Characterization of a gene encoding a DNA-binding protein with specificity for a light-responsive element. Plant Cell 4, 839–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Z., Lee, H., Xiong, L., Jagendorf, A., Stevenson, B., and Zhu, J.K. (2002). RNA helicase like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99, 11507–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Duong, H., Ma, N., and Lin, C. (1999). The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J. 19, 279–287. [DOI] [PubMed] [Google Scholar]
- Guo, H., Mocker, T., Duong, H., and Lin, C. (2001). SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291, 487–490. [DOI] [PubMed] [Google Scholar]
- Ha, S.-B., and An, G. (1988). Identification of upstream regulatory elements involved in the developmental expression of the Arabidopsis thaliana cab1 gene. Proc. Natl. Acad. Sci. USA 85, 8017–8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettiarachchi, G.H.C.M., Yadav, V., Reddy, M.K., Chattopadhyay, S., and Sopory, S.K. (2003). Light mediated regulation defines a minimal promoter region of TOP2. Nucleic Acids Res. 31, 5256–5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo, T., Asada, M., Kowyama, Y., and Hattori, T. (1999). ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 19, 679–689. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A-specific signal transduction. Science 284, 496–499. [DOI] [PubMed] [Google Scholar]
- Hoecker, U., Xu, Y., and Quail, P.H. (1998). SPA1: A new genetic locus involved in phytochrome A-specific signal transduction. Plant Cell 10, 19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, M., Ma, L.-G., Qn, L.-J., and Deng, X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16, 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., and Quail, P. (2002). PIF4, a phytochrome interacting bHLH factor functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21, 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Kim, H.J., Kim, Y.R., Park, J.Y., and Kim, J. (2002). Light signaling mediated by phytochrome plays an important role in cold induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 6, 693–704. [DOI] [PubMed] [Google Scholar]
- Kim, J., Yi, H., Choi, G., Shin, B., Song, P.-S., and Choi, G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome mediated light signal transduction. Plant Cell 15, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana. Z. Pflanzenphysiol. 100, 147–160. [Google Scholar]
- Laubinger, S., Fittingoff, K., and Hoecker, U. (2004). The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledent, V., and Vervoort, M. (2001). The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 11, 754–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Lin, C. (2002). Blue light receptors and signal transduction. Plant Cell 14 (suppl.), S207–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.L., Covington, M.F., Fankhauser, C., Chory, J., and Wagner, D.R. (2001). ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis PHYB signal transduction pathway. Plant Cell 13, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo, O., Chico, J.M., Sanchez-Serrano, J.J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, L., Li, J., Qu, L., Hager, J., Chen, Z., Zhao, H., and Deng, X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13, 2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., and Kay, S.A. (1996). Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc. Natl. Acad. Sci. USA 93, 15491–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miséra, S., Müller, A.J., Weiland-Heidecker, U., and Jürgens, G. (1994). The FUSCA genes of Arabidopsis: Negative regulators of light responses. Mol. Gen. Genet. 244, 242–252. [DOI] [PubMed] [Google Scholar]
- Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.R., Carre, I.A., and Coupland, G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2, 629–641. [DOI] [PubMed] [Google Scholar]
- Moller, S.G., Kim, Y.-S., Kunkel, T., and Chua, N.-H. (2003). PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15, 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, F., and Schafer, E. (2002). Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 53, 329–355. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Fanhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Ni, M., Halliday, K.J., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente, P., Wei, N., and Deng, X.W. (1996). Combinational interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 15, 3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Photosensory perception and signaling in plant cells: New paradigms? Curr. Opin. Cell Biol. 14, 180–188. [DOI] [PubMed] [Google Scholar]
- Robinson, K.A., and Lopes, J.M. (2000). Saccharomyces cerevisiae basic helix-loop-helix proteins regulate diverse biological processes. Nucleic Acids Res. 28, 1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, A., DeRycke, R., Beeckman, T., Engler, G., Van Montagu, M., and Boerjan, W. (2000). ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12, 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo, Y., Sullivan, J.A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17, 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepens, I., Duek, P., and Fankhauser, C. (2004). Phytochrome-mediated light signalling in Arabidopsis. Curr. Opin. Plant Biol. 7, 564–569. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bole, C., Ballesteros, M.L., and Chua, N.-H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423, 995–999. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Singh, H., LeBowitz, J.H., Baldwin, A.S., Jr., and Sharp, P.A. (1988). Molecular cloning of an enhancer binding protein: Isolation by screening of an expression library with a recognizing site DNA. Cell 52, 414–423. [DOI] [PubMed] [Google Scholar]
- Soh, M.S., Kim, Y.-M., Han, S.J., and Song, P.S. (2000). REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12, 2061–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman, J.I., Mindrinos, M.N., Fankhauser, C., Richards, D., Lutes, J., Chory, J., and Oefner, P.J. (2000). Cloning of the Arabidopsis RSF1 gene by using a mapping strategy based on high-density DNA arrays and denaturing high-performance liquid chromatography. Plant Cell 12, 2485–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L., and Tobin, E.M. (1990). Phytochrome-regulated expression of genes encoding light-harvesting chlorophyll a/b-protein in two long hypocotyl mutants and wild type plants of Arabidopsis thaliana. Photochem. Photobiol. 15, 51–56. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription factor genes are early targets of phytochrome signaling. Proc. Natl. Acad. Sci. USA 98, 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi, W.B., and Cashmore, A.R. (1995). Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 445–474. [Google Scholar]
- Tobin, E.M., and Kehoe, D.M. (1994). Phytochrome regulated gene expression. Semin. Cell Biol. 5, 335–346. [DOI] [PubMed] [Google Scholar]
- Wang, Z.-Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.-Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Weatherwax, S.C., Ong, M.S., Degenhardt, J., Bray, E.A., and Tobin, E.M. (1996). The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 111, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.W. (1999). Making sense of the COP9 signalosome: A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 15, 98–103. [DOI] [PubMed] [Google Scholar]
- Yadav, V., Kundu, S., Chattopadhyay, D., Negi, P., Wei, N., Deng, X.W., and Chattopadhyay, S. (2002). Light regulated modulation of Z-box containing promoters by photoreceptors and downstream regulatory components, COP1 and HY5, in Arabidopsis. Plant J. 31, 741–753. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 145, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.