Abstract
Lipid nanoparticles (LNPs) have proven effective in mRNA delivery, as evidenced by COVID-19 vaccines. Its key ingredient, ionizable lipids, is traditionally optimized by inefficient and costly experimental screening. This study leverages artificial intelligence (AI) and virtual screening to facilitate the rational design of ionizable lipids by predicting two key properties of LNPs, apparent pKa and mRNA delivery efficiency. Nearly 20 million ionizable lipids were evaluated through two iterations of AI-driven generation and screening, yielding three and six new molecules, respectively. In mouse test validation, one lipid from the initial iteration, featuring a benzene ring, demonstrated performance comparable to the control DLin-MC3-DMA (MC3). Notably, all six lipids from the second iteration equaled or outperformed MC3, with one exhibiting efficacy akin to a superior control lipid SM-102. Furthermore, the AI model is interpretable in structure-activity relationships.
Subject terms: Computational models, Virtual drug screening
Ionizable lipid optimization is essential for mRNA therapy via lipid nanoparticle, but experimental screening is investment-intensive. Here, authors developed AI models achieving the rational design of lipid molecules and fast high-throughput screening.
Introduction
The success of mRNA vaccines against COVID-191,2 has firmly established lipid nanoparticles (LNPs) as the foremost method for mRNA delivery. Additionally, the growing adoption of LNPs as a delivery system for potential mRNA therapies targeting diverse infectious diseases, cancers, and genetic disorders3,4 further underscores its promising potential. In the late 1990s, it was found that the addition of positively charged lipids to liposomes significantly enhanced their efficacy in delivering nucleic acids, resulting in LNPs. These positively charged lipids have a strong propensity to interact with the negatively charged phosphoric acid backbones of nucleic acids5,6. Subsequently, various analogous lipids, either permanently charged (known as cationic lipids) or conditionally charged (referred to as ionizable lipids), were designed and gradually assumed a more significant role in LNP formulations. Over time, the usage of ionizable lipids has dominated7–10 due to their desirable safety and pharmacokinetic characteristics5,11.
A typical LNP formulation generally is composed of four types of lipids: ionizable lipids, helper lipids, cholesterol, and polyethylene glycol (PEG) lipids3. At the heart of LNPs lie the ionizable lipids, which play a pivotal role. These lipids possess positively chargeable head groups with amine functionalities, capable of protonation under acidic conditions due to their distinct apparent pKa (often around 6.5 when formulated in LNPs)8,12. The positively charged ionizable lipids serve multiple purposes: entrapping mRNA during LNP formation and interacting with anionic endosomal membranes, thereby facilitating mRNA release from endosomes into the cytoplasm9. Once within the neutral environment of the bloodstream, ionizable lipids are discharged, preventing rapid clearance and extending systemic circulation13,14. Moreover, the chemical structures of ionizable lipids have been observed to impact the overall mRNA expression and the distribution of loaded mRNA within tissues and organs.
The intricate influence of each component of an ionizable lipid on its function presents a challenge in their precise design. For instance, the head groups influence pKa values11 and hydrogen bonding strengths with mRNA15,16, linker groups impact biodegradability17, and lipid tails influence pKa18 as well as membrane stability and lipid fluidity7. These characteristics collectively shape mRNA delivery efficiency.
Historically, identifying optimal ionizable lipid structures relied on screening tests through trial-and-error experiments. This approach, however, is beset by limitations. Extensive screening entails substantial time, significant quantities of materials, considerable animal use, and cutting-edge equipment (e.g., combinatorial chemistry and high-throughput technologies)19–21. Even with these extensive resources, experimental efficiency and success rates remain very low given the expansive chemical space of ionizable lipids. Conversely, relying solely on human intuition for lipid design is restricted by personal experience and limited capacity to fully exploit accumulated data.
The challenges are promising to be addressed through the integration of artificial intelligence (AI) models. AI excels in discerning underlying relationships within big data and extrapolating these relationships to predict new cases. In the field of new drug molecule discovery, AI has achieved remarkable strides22,23. Moreover, AI models have been developed to predict multiple drug features in various dosage forms, including solid dispersions, cyclodextrin complexes, and nanoparticles24,25. Previously, our group developed an AI model to predict IgG titers induced by mRNA vaccines in a previous study related to formulation development26. The involved features included structures of ionizable lipids, compositions, vaccination schedules, and animal types. Optimizing the IgG titer profile could lead to the selection of the desired formulation, especially the ionizable lipid type. This was a proof-of-concept study of AI application to the design of mRNA-LNP delivery systems. The latest research has introduced AI models in association with high-throughput synthesis to screen a class of ionizable lipids (synthesized from amine, isocyanide, aldehyde and carboxylic acid) to optimize mRNA delivery27.
In this work, we meticulously gathered various structures of ionizable lipids from literature sources13,15,18,20,28–32 and patents33–41 aiming to develop AI models. The chemical structures and LNP formulations of some collected ionizable lipids are shown in Supplementary Table 1. The AI models predict apparent pKa values and mRNA delivery efficiency of LNPs. They provided insights for lipid generation and were applied to predict their properties, accelerating the screening work (Fig. 1). As a result of this approach, several ionizable lipids are successfully identified and demonstrate robust performance upon experimental validation.
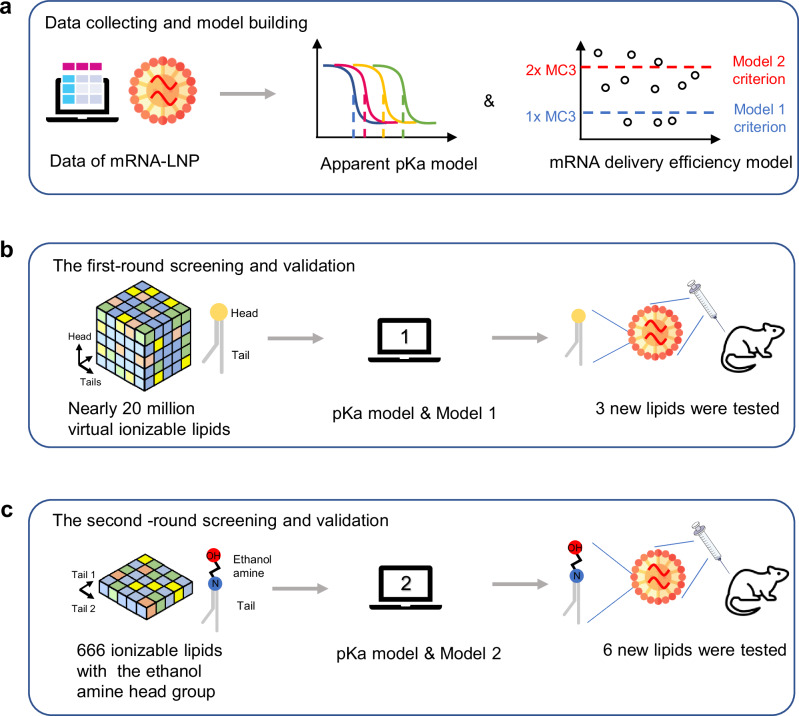
Fig. 1. Overview of AI-driven rational design of ionizable lipids for mRNA lipid nanoparticles.
a The collected data was used to build models predicting the apparent pKa and mRNA delivery efficiency of LNPs. The 1- and 2-fold of MC3 mRNA delivery efficiency was used in Models 1 and 2 as the criterion in the model classifying ionizable lipids with positive delivery efficiency. b The first-round of virtual screening of ionizable lipids and validation based on pKa model and Model 1. c The second-round of virtual screening of ionizable lipids and validation based on pKa model and Model 2. AI artificial intelligence, LNP lipid nanoparticle, MC3, DLin-MC3-DMA.
Results
Overview of the first-round lipid virtual screening
In this study, the lipid virtual screening was carried out in two sequential stages, ultimately resulting in the synthesis and evaluation of two separate batches of lipids. Figure 2 shows the workflow of the first-round of screening. Initially, the AI model was built to predict mRNA delivery efficiency (Model 1) and the apparent pKa of LNPs containing ionizable lipids. This model informed significant substructures that should be highlighted during lipid generation. With the built models, each lipid could be predicted, and all substructures were ranked. This ranking information helped in selecting lipids for experimental testing, eventually identifying three lipids.
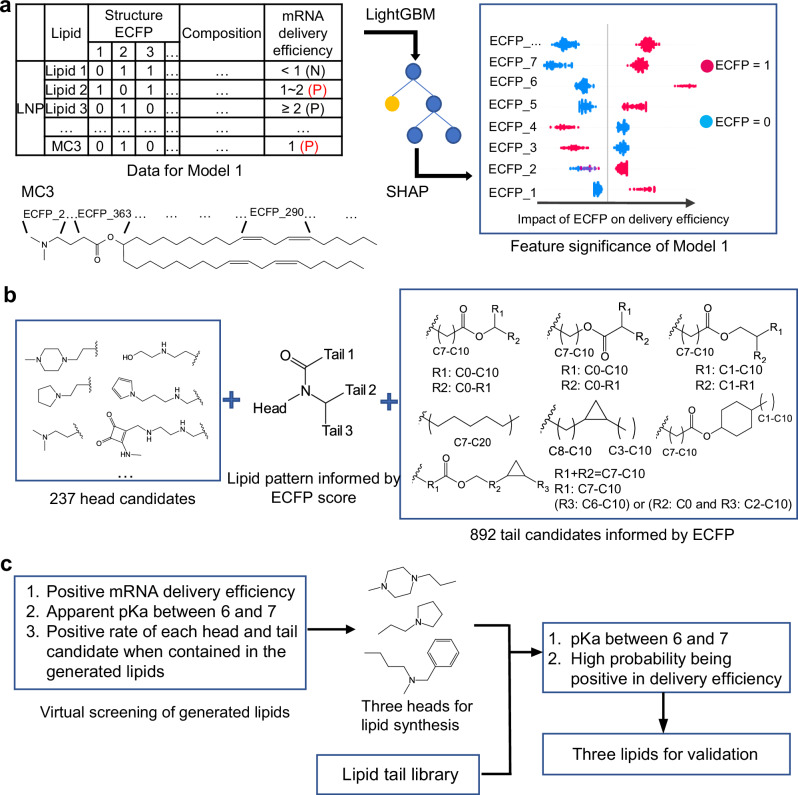
Fig. 2. Overview of the first-round lipid virtual screening.
a Data representation of the AI model (Model 1), and the methods of model training (LightGBM) and feature significance calculation (SHAP). Some typical ECFP bits and their corresponding substructure of MC3 are shown as an example. P, positive; N, negative. b Significant substructures informed by the model and used for lipid generation. c The method of lipid virtual screening and three lipids were selected for experimental validation. ECFP extended connectivity fingerprints, LightGBM Light Gradient Boosting Machine, SHAP SHapley Additive exPlanations, MC3 DLin-MC3-DMA, AI artificial intelligence.
Performance of the AI model in the initial screening round
During the construction of the model predicting mRNA delivery efficiency, an initial attempt with a regression model revealed unsatisfactory performance due to potential data source impacts (Supplementary Table 2). Consequently, a classification model was adopted based on the criterion of delivery efficiency compared to DLin-MC3-DMA (MC3), distinguishing between lipids that outperformed MC3 (positive) and those that did not (negative). The model performance from two training algorithms, Random Forest and LightGBM (Light Gradient Boosting Machine), was compared (Supplementary Table 3). The LightGBM model exhibited superior scores in terms of Recall, ACC, and F1, and was thus chosen for subsequent use.
Conversely, the apparent pKa model was built as a regression using the LightGBM algorithm. The performance of the pKa model is shown in Supplementary Table 4. For the test set, RMSE and MAE were calculated as 0.25 and 0.19, respectively, with R2 around 0.59. While the initial impression may not seem entirely satisfactory, a closer examination of the scatter plot in (Supplementary Fig. 1a) revealed close alignment between predicted and actual pKa values in the range of 6 to 7. Outside this range, there is a severe deviation. Notably, the pKa range of 6 to 7 encapsulated most ionizable lipids in our dataset, especially those with superior mRNA delivery efficiency relative to MC3 (Supplementary Fig. 1b). Consequently, considering the primary objective of the AI model is to screen ionizable lipids with exceptional delivery efficiency, the existing pKa prediction model aligned well with this purpose.
In addition to validating our model’s performance using our collected dataset, we also subjected the model to validation using an external dataset. Briefly, the mRNA delivery efficiency of 14 ionizable lipids (Supplementary Fig. 2) was predicted and subsequently compared to experimental data (Supplementary Table 5). Besides the values predicted by the model presented above (“Prediction”), values predicted by the model trained on the whole collected data after the determination of hyperparameters (“Prediction_all”) are also shown. The prediction from the model trained on the whole data showed high accuracy, with a correct rate achieving around 0.78.
Similarly, the apparent pKa of nine LNPs containing different ionizable lipids in the external dataset was also compared to the predicted values, which are shown in Supplementary Fig. 3. Seven out of the nine samples exhibited close alignment between predicted and experimental data. The two outliers, which exhibited less accurate predictions contain hydroxyl groups in their tails – a feature scarcely represented in our collected dataset.
The first-round ionizable lipids generation and virtual screening
After validation, the model served as a guide in the design of ionizable lipids, effectively steering lipid generation and expediting virtual screening. In this model, the structure of ionizable lipids was denoted by extended connectivity fingerprints42 (ECFP), with each ECFP bit corresponding to a substructure within a lipid. The question of which substructure to emphasize in lipid generation was addressed by using the SHAP (SHapley Additive exPlanations) algorithm43. The contribution of all ECFP bits in each ionizable lipid could be quantified as SHAP value (Supplementary Fig. 4a). This further allowed for the calculation of ECFP scores for every lipid using Equation 5 and 6. Surprisingly, the ECFP score was found to be correlated with mRNA delivery efficiency (Supplementary Fig. 4b). Next, 40 lipids with the highest ECFP scores and their molecular similarities to other lipids were visualized in Supplementary Fig. 5. In these lipids, tails containing cyclopropyl and cyclohexyl were distinct in structure, and a joint containing an amide bond linking the head and three tails often appeared in top-performing lipids. Thus, they were worthy of exploration for molecule generation (Fig. 2b). Besides, commonly seen ester bond-containing tails and single carbon chains were also considered. As for head groups, all heads in the collected data were included. After introducing some variance in the chosen tail and head segments, 892 tail and 237 head candidates (Fig. 2b) were constructed. The tails and head candidates were combined according to the structure pattern to generate virtual ionizable lipids exhaustively, but Tail 2 and Tail 3 (Fig. 2b) were kept the same for simplicity.
Through a comprehensive permutation, a pool of nearly 20 million lipids was generated. Employing the workflow depicted in Fig. 2c, these lipids’ pKa and mRNA delivery efficiencies were predicted. Here, lipids were deemed positive if they showed positive mRNA delivery efficiency and a desired pKa range (6.0–7.0). Since each head or tail candidate was used in various lipids and may lead to disparate mRNA delivery efficiency, the positive rate (Equation 7) could be calculated and ranked for each segment candidate (Supplementary Fig. 6). Consistently, an overview of good lipids was illustrated in Supplementary Fig. 7, where advantageous tails and the top 32 head groups were collectively presented. However, the decision of which lipids to synthesize for further exploration was informed by practical considerations. Lipids chosen for synthesis should be feasible at this stage, and the lipid structures, especially head groups, should exhibit diversity to explore a broader chemical space. Guided by these principles, three heads ranked 10, 12, and 32 in Supplementary Fig. 7 were selected. The choice of lipid tails was influenced by our synthesis capabilities. Our tail library allowed the synthesis of 666 possible lipids for each head type. They were predicted for pKa and delivery efficiency, and the probability of being positive in delivery efficiency was output by the model. Consequently, lipids with desired pKa, high probability, and positive mRNA delivery efficiency were chosen for synthesis: LQ085, LQ086, and LQ087 (Fig. 3a). Their predicted pKa and positive probability are listed in Supplementary Table 6.
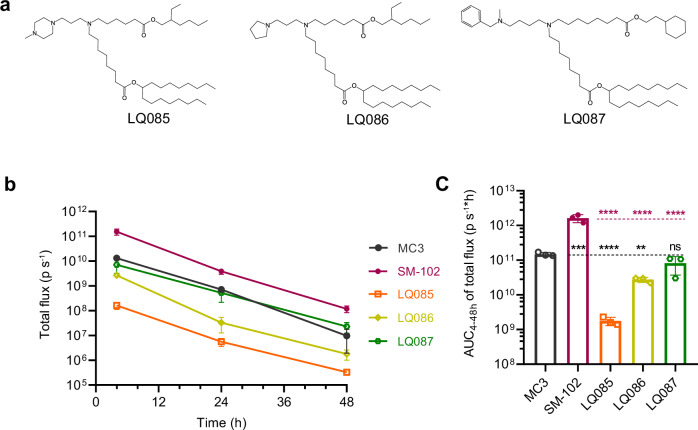
Fig. 3. Experimental validation of the three ionizable lipids resulted from the first-round of virtual screening.
a The structure of the three ionizable lipids selected from the first round of virtual screening. b Female BALB/c mice (6-8 weeks old) were intravenously injected with LNPs loaded with luciferase mRNA at a dose of 5 μg per mouse, and at certain time points, total luminescence was detected after injection of D-luciferin. c The AUC of total luminescence. All data are presented as the mean ± SD (n = 3). Statistical significance was analyzed by one-way ANOVA (ns, not significant; *p < 0.0332; **p < 0.0021; ***p < 0.0002; ****p < 0.0001. The P makers in black are results of the comparisons were with MC3, and those in red are with SM-102. Source data are provided as a Source Data file.). MC3 DLin-MC3-DMA, AUC area under curve, LNP lipid nanoparticle, SD standard deviation, ANOVA Analysis of Variance.
Experimental validation of the first-round of screening
LQ085, LQ086, and LQ087 were formulated into LNPs encapsulating luciferase mRNA and compared to two positive control lipids, MC3 and SM-102. SM-102 was used in Moderna’s COVID-19 vaccines44 and often shows even higher capacity in delivering luciferase and hEPO mRNA in rodents12. The basic characteristics of these LNPs, including particle size, polydispersity index (PDI), and encapsulation efficiency (EE), were measured (Supplementary Table 6). The apparent pKa of LNPs generated from LQ087, SM-102, and MC3 lay within the desired range of 6.0 to 7.0, while the pKa of LQ085 and LQ086 exceeded 7.0.
The LNPs loaded with luciferase mRNA were intravenously administered to mice. After LNP injection, luminescence signals were detected at 4, 24, and 48 hours, following the administration of the substrate, D-luciferin (Fig. 3b and c, Supplementary Fig. 8). At the starting point, LNP containing LQ087 and LQ086 induced similar luminescence which was higher than that of LQ085 by 10 to 100 fold. Later, the luminescence of LQ086 decreased faster than that of LQ087, getting close to that of LQ085. The luminescence-time curve and AUC showed that LQ087 was the best of the three ionizable lipids we proposed. Compared to the positive controls of SM-102 and MC3, LQ087 matched MC3 but still performed worse than SM-102. This experimental result should be robust (Supplementary Fig. 9). The gender factor made little difference to the performance of the lipids, and the data collected at the three-time points were sufficient to measure the AUC of the administration.
The second-round lipid virtual screening
The relatively modest performance of the three lipids prompted a reevaluation. One possible explanation was that the selected head groups were underrepresented in the dataset, potentially biasing the model. As a result, in the second round of lipid screening, we focused on lipids containing the ethanolamine head group, a component tested most frequently in our collected dataset. Besides, the synthesis capability was considered at the beginning of this round. These factors shrank the pool of candidate lipids. To pick competent lipids from the narrow domain, a stricter classifier was preferred to the above method which required statistical analysis of a large number of virtual lipids.
Given these considerations, Model 2 was built, increasing the criterion that an ionizable lipid was judged as positive to 2-fold the delivery capability of MC3 (Fig. 4a). The pKa prediction model remained unaltered. Compared to Model 1, the performance of Model 2 was defective in validation using the collected data (Supplementary Table 7). This defective performance was also evidenced by the external validation, in which the number of wrong predictions increased from three to six (Fig. 4b). However, all the mistakes happened to be that truly positive lipids were falsely predicted as negative, while truly negative lipids were predicted correctly. In other words, Model 2 showed a stricter criterion when assessing mRNA delivery efficiency, which was also reflected by the increasing precision index if validated against the original data (Supplementary Table 8). Higher precision means fewer false positive predictions.
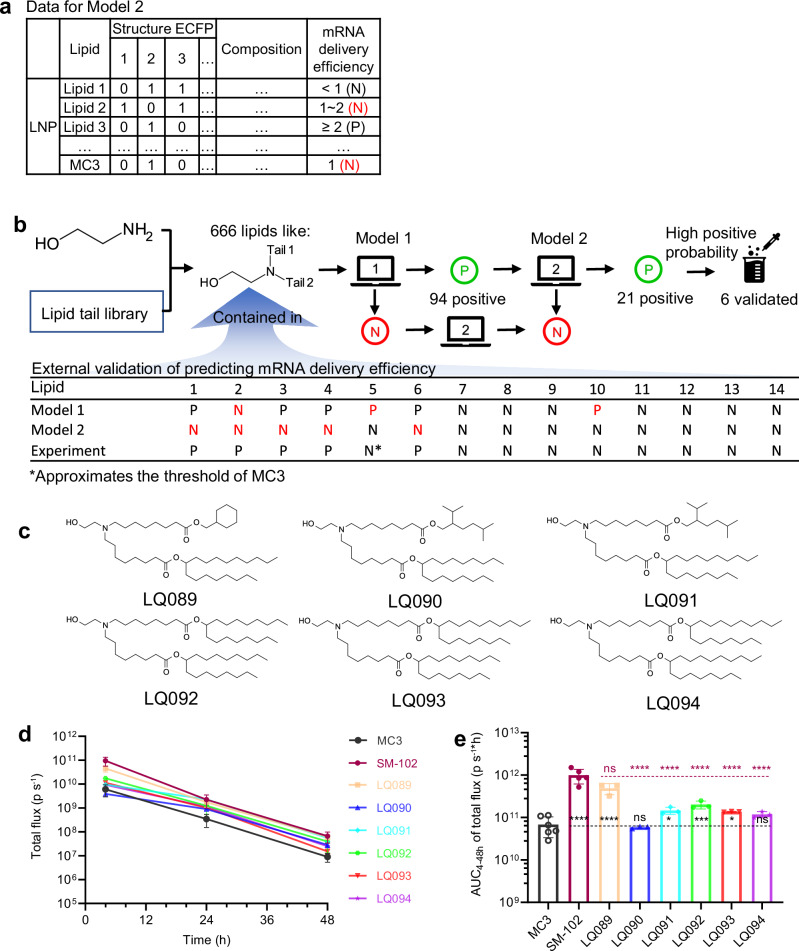
Fig. 4. Overview of the second-round lipid virtual screening and experimental validation.
a Data representation of Model 2 predicting mRNA delivery efficiency. Compared to the representation method of Model 1, the positive criterion was set as 2-fold the delivery efficiency of the standard MC3 LNP. b External validation of Model 2, and associating Model 1 and 2 to screen the generated ionizable lipids. c The six lipids selected for experimental validation. d Female BALB/c mice (6–8 weeks old) were intravenously injected with LNPs loaded with luciferase mRNA at a dose of 5 μg per mouse, and at certain time points, total luminescence was detected after injection of D-luciferin. Time courses of the total flux of the screened six lipids. e The AUC of total luminescence of the screened six lipids. For MC3, SM-102, and other groups, n = 6, 5, 3, respectively. All data are presented as the mean ± SD. Statistical significance was analyzed by one-way ANOVA (ns, not significant; *p < 0.0332; **p < 0.0021; ***p < 0.0002; ****p < 0.0001. The P makers in black are results of the comparisons with MC3, and those in red are with SM-102. Source data are provided as a Source Data file). MC3 DLin-MC3-DMA, AUC area under curve, LNP lipid nanoparticle, SD standard deviation, ANOVA Analysis of Variance.
Combining the ethanolamine head group and our tail library, 666 lipids were constructed, which included the molecules in the external validation set. Among the 666 lipids, Model 1 predicted 94 positive lipids, from which Model 2 predicted 21 positive lipids (Supplementary Fig. 10), while the other 645 molecules were negative in delivery (Fig. 4b). Therefore, the 21 lipids were more likely to have better delivery efficiency and worth exploring. Like the first-round, lipids with desired properties and diverse tail structures, such as two long branches, dendritic branches, and cyclohexyl groups, were preferred. Eventually, six of them (Fig. 4c) were selected for synthesis and evaluation. Their predicted pKa and probability of being positive in mRNA delivery efficiency were reported in Supplementary Table 9.
Experimental validation of the second-round screening
Analogously, the positive controls (SM-102, MC3) and the newly designed (LQ089-094) ionizable lipids were formulated into LNPs with luciferase mRNA. The particle size, PDI, apparent pKa, and EE of these LNPs were measured (Supplementary Table 9). The pKa values of all lipids were well-contained within the 6.0 to 7.0 range.
Following intravenous administration of LNPs loaded with luciferase mRNA, luminescence signals stemming from luciferase activity were detected at 4, 24, and 48 hours, subsequent to the administration of D-luciferin (Fig. 4d and Supplementary Fig. 8). Impressively, all new lipids performed well in terms of mRNA delivery efficiency, among which LQ089 and LQ091-LQ093 exhibited significantly higher efficacy than MC3. Notably, LQ089 surpassed the performance of all the previously tested lipids. Its luminescence signal approached SM-102, showing no significant difference from SM-102 in the area under curves (AUC) of luminescence signals (Fig. 4e).
Structure-activity relationship in hydroxyl-containing lipids
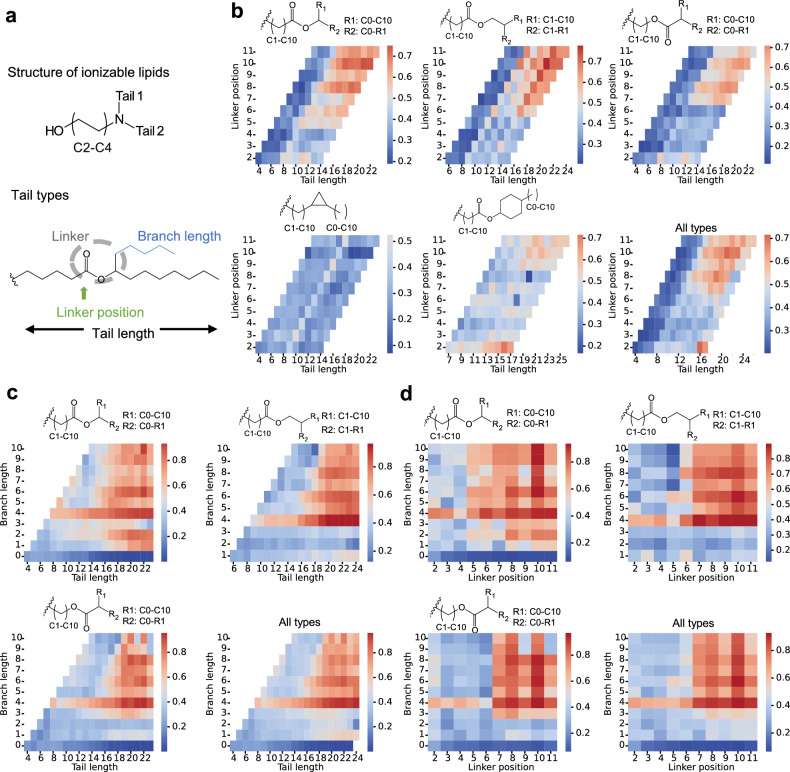
For the commonly synthesized ionizable lipids where the head group contains hydroxyl and two tails directly linked to the nitrogen atom (Fig. 5a), a structure-activity relationship was inferred with the AI model. The tail types analyzed covered those used for virtual screening. Variables such as tail length, linker position, and branch length and their effect on the positive rate of lipid molecules were analyzed to show the structure-activity relationship.
Fig. 5. Structure analysis of ionizable lipids informed by AI models.
a Ionizable lipid pattern and tail types for analysis. b–d Heatmap of positive rates for specific tail types. All types of tails and heads containing hydroxyl were combined to form ionizable lipids in an exhaustive manner. Their mRNA delivery efficiency and apparent pKa were predicted with Model 1. For each type of tail, the number of resulting lipids containing the tail and among them the number of positive lipids (efficiency higher than the standard MC3 formulation and pKa between 6.0 and 7.0) were used to calculate the positive rate. The structure-activity relationship is shown as the influence of tail length and linker position (b) tail length and branch length (c), and the branch length and linker position (d) on the positive rate. AI, artificial intelligence.
Figure 5b shows tails containing ester linkers are more likely to show better performance (positive rate more than 0.5) than those with a single linker of cyclopropyl. The tail length more than 10 and linker position is more than 5 or 6 seem to be a safe space. Meanwhile, linkers should be located in the mid-area of the tail, remaining a moderate carbon chain length before and after it. For different types of tails, the threshold of tail length and linker position is different. But tail length exceeding 20 and linker position more than 10 seem to bring about defectiveness in performance, especially in lipids with inverse ester bond as the linker. In contrast, constraints in length and linker position are not as strict in tails with the linker of ester bond and cyclohexyl, where no clear thresholds were drawn. For tails with the linker of a single ester bond, the influence of branch length can be analyzed. Branch length of more than 3 increases the possibility of achieving better performance (Fig. 5c). Since the branch length is restricted to being not longer than the main chain, a long branch length almost does not lower performance if only the chain length before the linker is guaranteed (Fig. 5d).
Comprehensive in vivo study of the newly generated lipids
The screening method above evaluated the general performance of new lipids. However, distribution in different organs and the expression of mRNA via different routes of administration are also of great importance for lipid molecule assessment. In order to evaluate the newly generated lipids more comprehensively, qualified LNPs (Supplementary Fig. 11) loaded with luciferase mRNA were administered by intravenous injection (Fig. 6 and Supplementary Fig. 12 and 13) and intramuscular injection (Fig. 7 and Supplementary Fig. 14 and 15) at a dosage of 5 μg mRNA per mouse, respectively. In vivo luminescence signals were detected at 4 hours post-administration, subsequent to the administration of D-luciferin. Then, the mice were euthanized and the organs were isolated for the detection of organ-distributed luminescence ex vivo.
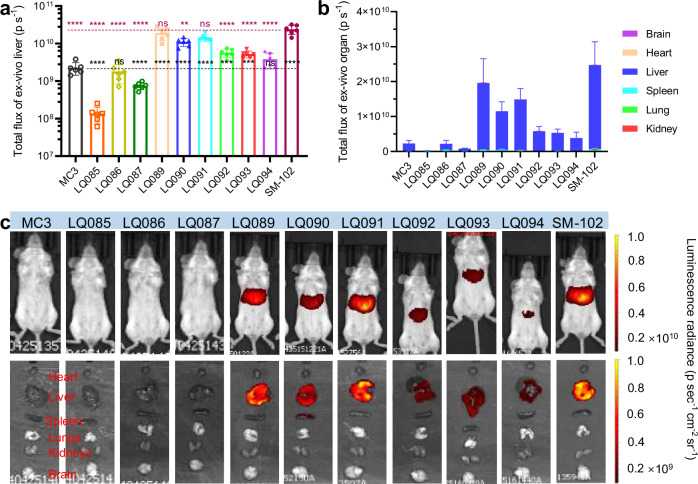
Fig. 6. Luminescence distribution and expression of luciferase mRNA loaded in different LNPs via intravenous administration.
a Liver luminescence of luciferase at 4 h. b Organ-distributed luminescence of luciferase at 4 h. (c) Representative images of the luminescence. Each group had three female BALB/c mice and three male ones. All data are presented as the mean ± SD. Statistical significance was analyzed by one-way ANOVA (ns, not significant; *p < 0.0332; **p < 0.0021; ***p < 0.0002; ****p < 0.0001. The P makers in black are the results of the comparisons with MC3, and those in red are with SM-102. Source data are provided as a Source Data file.). MC3 DLin-MC3-DMA, SD standard deviation, ANOVA Analysis of Variance.
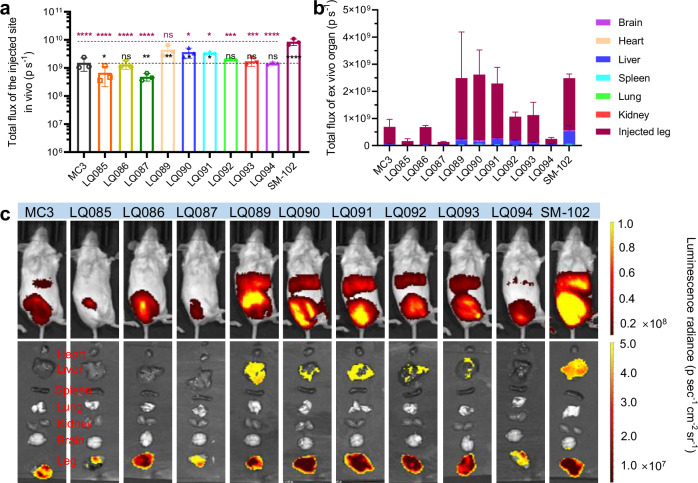
Fig. 7. Luminescence distribution and expression of luciferase mRNA loaded in different LNPs via intramuscular administration.
a Luminescence of the injection site at 4 h. b Organ-distributed luminescence of luciferase at 4 h. c Representative images of the luminescence. Each group had three female BALB/c mice. All data are presented as the mean ± SD. Statistical significance was analyzed by one-way ANOVA (ns, not significant; *p < 0.0332; **p < 0.0021; ***p < 0.0002; ****p < 0.0001. The P makers in black are results of the comparisons were with MC3, and those in red are with SM-102. Source data are provided as a Source Data file.). MC3 DLin-MC3-DMA, SD standard deviation, ANOVA Analysis of Variance.
As foreseen, the luminescence signal of new lipid groups was high in the liver which is the main targeted organ (Fig. 6a and b), due to their structural similarity to MC3 and SM-102, which were liver-targeted. LQ089 and LQ091 outperformed other tested lipids and showed no significant difference from SM-102. Furthermore, the isolated organs were homogenized to detect the concentration of Cy5-mRNA. The ratios of organ-distributed mRNA to administered mRNA were calculated and listed in Supplementary Fig. 13b. Although only a small amount of Cy5-mRNA was detected, the majority of it was in the liver, a result which matched well with the distribution of luminescence.
For the luminescence of the injection site detected in vivo in intramuscular groups, the order of luminescence intensity between each LNP was consistent with that of the intravenous route (Fig. 7a). It was worth noting that luminescence signals after intramuscular administration could also be detected in the liver, both in vivo (Fig. 7c, Supplementary Fig. 14, and Supplementary Fig. 15) and ex vivo (Fig. 7b and c, Supplementary Fig. 15).
Stability study on the LNPs containing newly generated lipids
Besides the efficient delivery of mRNA, proper LNPs must have long-term pharmaceutical stability and low toxicity. Taking account of both delivery efficiency and structural diversity, LQ086, LQ089 and LQ092 were selected as typical models to study the long-term storage stability and acute toxicity of the nine newly generated lipids.
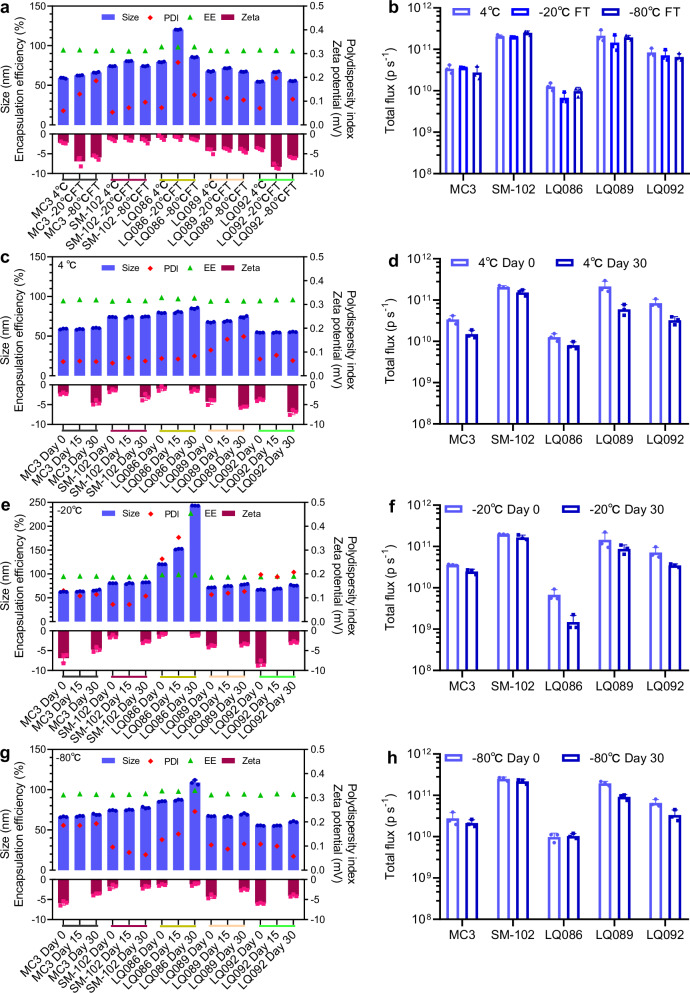
Due to the poor stability of mRNA, LNPs usually need to be frozen for storage, so LNPs must face the challenges of the freezing-thawing process on the pharmaceutical properties and in vivo effectiveness. Herein, LNPs loaded with luciferase mRNA were frozen at −20 °C and −80 °C and then melted at room temperature to examine the changes in particle size, PDI, potential, EE, and in vivo efficiency of mRNA expression before and after the freezing-thawing process (Figs. 8a and b). For LQ089, LQ092, MC3, and SM-102, the freezing-thawing process did not affect the particle characteristics of LNPs. The freezing-thawing process at -20 °C significantly increased the size of LQ086 LNP, while this change did not occur during the process at −80 °C. Besides, the freezing-thawing process at both −20 °C and −80 °C mildly impacted the in vivo efficiency of all groups.
Fig. 8. LNP stability during the freezing-thawing process and long-term storage.
a, b Changes in particle characteristics and in vivo efficiency before and after the freezing-thawing process. c, d Changes in particle characteristics and in vivo efficiency during long-term storage at 4 °C. e, f Changes in particle characteristics and in vivo efficiency during long-term storage at −20 °C. g, h Changes in particle characteristics and in vivo efficiency during long-term storage at −80 °C. The small colored lines beneath the X-axes in panels a, c, e and g are used to distinguish the lipid groups more clearly. For in vivo efficiency of each group, three female BALB/c mice were intravenously administered luciferase mRNA loaded in LNPs at a dosage of 5 μg per mouse and the luminescence signal of the whole body was detected 4 hours later post intraperitoneal administration of D-luciferin. All data are presented as the mean ± SD (n = 3). The analysis of change trends met the needs of the stability study, so significance analysis was not performed. Source data are provided as a Source Data file. MC3 DLin-MC3-DMA, PDI polydispersity index, EE encapsulation efficiency, LNP lipid nanoparticle, SD standard deviation.
For the long-term storage test, all samples were stored under three conditions, 4 °C, −20 °C and −80 °C (Fig. 8c–h). The particle size, PDI and EE were measured on Day 14 and Day 30, and the in vivo efficiency of mRNA expression and potential were measured at Day 30. Similar to the freezing-thawing process, storage at −20 °C seriously affected the stability of LQ086 LNP, while the condition of −80 °C had a lesser impact. LQ089 and LQ092 showed good pharmaceutical stability in all three conditions, which was comparable to that of MC3 and SM-102. For in vivo mRNA expression, all groups decreased after one month of storage, but SM-102 decreased less than other groups, which is possibly because it had the highest baseline. Interestingly, LNPs formed from the new lipids still had good particle characteristics and high mRNA expression levels after one month of storage at 4 °C, indicating that they had good stability.
Acute toxicity of LNPs containing newly generated lipids
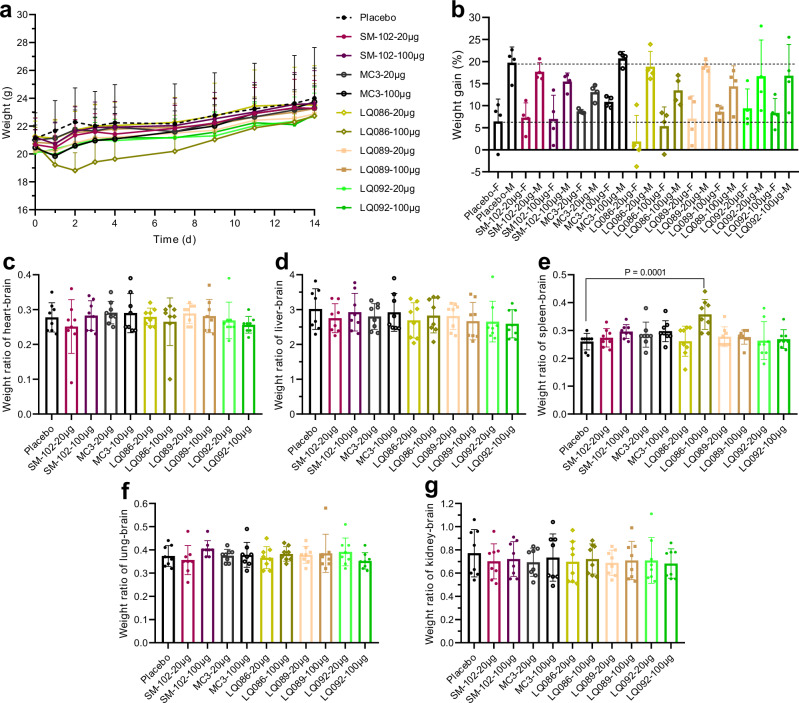
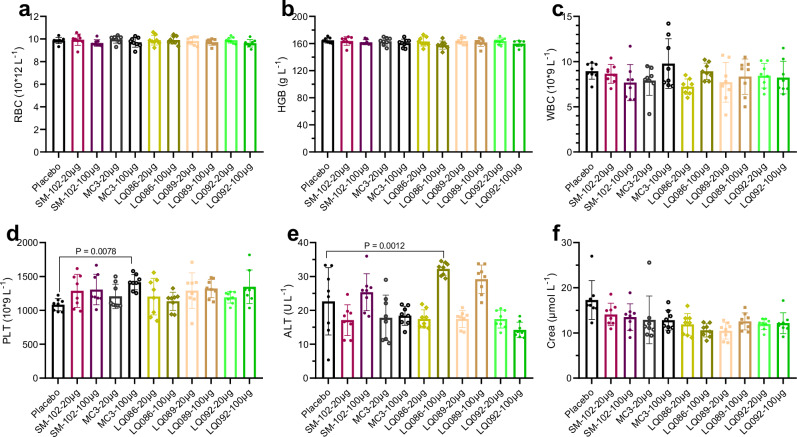
To preliminarily evaluate the in vivo safety of newly generated lipids, LNPs loaded with luciferase mRNA were intravenously administered to BALB/c mice at acute toxic dosages of 20 μg and 100 μg mRNA per mouse (1 mg kg-1 and 5 mg kg-1, respectively). The weight of mice was monitored on Days 1, 2, 3, 4, 7, 9, 11, 13, and 14 after administration (Fig. 9a and b). Mice were bled and euthanized to obtain organs on Day 14 for weighing (Fig. 9c–g and Supplementary Fig. 16). The whole blood was examined for blood cells and the serum was isolated to analyze the blood biochemistry (Fig. 10 and Supplementary Fig. 17). The group of LQ086-100 μg showed lower body weight gain and spleen enlargement, which might be caused by strong immunogenicity. The biochemistry analysis showed a slight rise in glutamic-pyruvic transaminase (ALT) in groups of LQ086-100 μg, LQ089-100 μg, and SM-102-100 μg, which was within the acceptable range. Meanwhile, no abnormality was found in the analysis of histopathology (Supplementary Fig. 18). In addition, the hemolysis test of LNP on rabbit red blood cells was also negative (Supplementary Fig. 19). Overall, the newly generated lipids exhibited excellent in vivo safety at an intravenous acute toxic dosage, supporting the AI-driven rational design of ionizable lipids to step forward.
Fig. 9. Weight monitoring and organ coefficient in the acute toxicity test.
a Weight-time curve. The placebo was saline. Each group had four female BALB/c mice and four male ones. b Weight gain at Day 14 compared to Day 0. F=female, M=male. c–g Coefficient of heart-brain, liver-brain, spleen-brain, lung-brain and kidney-brain, respectively. All data are presented as the mean ± SD (n = 8 for each dose, including four females and four males). Statistical significance was analyzed by one-way ANOVA (unmarked, not significant. Source data are provided as a Source Data file.). MC3 DLin-MC3-DMA, SD standard deviation, ANOVA Analysis of Variance.
Fig. 10. Hematological indices.
a–f Blood was obtained 14 days after intravenous administration. All data are presented as the mean ± SD (n = 8 for each dose, including four females and four males). Statistical significance was analyzed by one-way ANOVA. (Unmarked, not significant. Source data are provided as a Source Data file.). MC3 DLin-MC3-DMA, RBC red blood cell, HGB hemoglobin, WBC white blood cell, PLT platelet, ALT glutamic-pyruvic transaminase, Crea creatinine, SD standard deviation, ANOVA Analysis of Variance.
Discussion
The successful delivery of mRNA via LNPs heavily relies on the utilization of ionizable lipids, which govern both the encapsulation and release of mRNA. Consequently, the screening and design of effective ionizable lipids are pivotal in the development of mRNA-LNP delivery systems. This research aims to expedite the screening process by employing AI models to predict their critical properties of ionizable lipids. While mRNA delivery efficiency stands as the primary indicator for evaluating ionizable lipids, exploring intermediate indices becomes a logical step.
Previous studies have established the significance of the apparent pKa of LNPs8,12,18. The apparent LNP pKa is related to and found to be 2-3 units lower than the calculated pKa of the ionizable lipid molecule itself17,45. The LNP containing MC3 had an apparent pKa of 6.44, and LNPs with a pKa range of 6.2 to 6.5 exhibited optimal delivery efficiency for siRNA8. A similar pKa range of 6.2 to 6.8 was deemed advantageous for mRNA delivery18. However, for intramuscular administration and immunogenicity, the ideal pKa range leans towards 6.6 to 7.012. Additionally, Supplementary Fig. 1b indicates that an apparent pKa within the range of approximately 6.0 to 7.0 serves as a prerequisite for LNPs to exhibit positive mRNA delivery efficiency. Thus, predicting the apparent pKa emerges as another pivotal index.
The LightGBM algorithm was selected for model training. As a tree-based learning algorithms, LightGBM has outperformed well-known neural network frameworks in various pharmaceutical datasets46–49. Moreover, LightGBM is one of the fastest tree-based learning algorithms and is suitable for sparse feature datasets such as those using ECFP. Its feature grouping mechanism effectively consolidates sparse features while retaining essential information, mitigating the impact of high feature dimensionality and thus enhancing model performance.
Initially, regression was intended for predicting mRNA delivery efficiency. Nonetheless, this approach yielded varying performance outcomes within the sub-dataset (Supplementary Table 2). For example, validation on most lipid structures sourced from Acuitas37–41 and Protiva33 exhibited high performance (>0.7 in R2) likely due to multiple data points for each lipid conducted at different dosages. Conversely the validation on lipids from Moderna34,35, where they had a single test, yielded a suboptimal performance (~0.5 in R2). To better approximate the predictive capacity for genuinely novel lipids, the regression model was abandoned in favor of constructing a classification model. The model employed MC3 as a reference criterion, the ionizable lipid in the first approved LNP product (Onpattro®)50. Notably, the classification model exhibited an accuracy (ACC) of 0.82 and precision of 0.76 (Supplementary Table 3). ACC gauges the general predictivity, while precision indicates the likelihood of a predicted good ionizable lipid to genuinely excel in mRNA delivery—a primary concern. Regarding the apparent pKa, the regression model proved adept in the “optimal pKa range” (6.0–7.0).
Although the AI model was trained solely on data samples labeled with categories of mRNA delivery efficiency, it unexpectedly developed the ability to quantify this efficiency, which is a fortunate discovery. The efficiency is positively correlated with the newly defined ECFP score. Some lipids containing squaramide were reported to show remarkable delivery efficiency15, and they were also given high ECFP scores (Supplementary Fig. 5). The score is derived from lipid ECFP bits and their corresponding SHAP values. The SHAP algorithm, providing quantified assessments of feature contributions, has proven to be highly effective in explaining the outputs of AI models43,51. From this view, the model we have developed is interpretable in terms of its structure-activity relationship.
Considering both molecule performance and novelty in structure, some segments were picked up with the help of molecular similarity visualization to generate the lipid pool for the first-round of virtual screening. Design of new lipids was not conducted by straightforwardly maximizing ECFP score, because chemical structures cannot be derived from ECFP codes. After prediction on the generated lipids and statistical analysis, the segments were ranked based on their positive rate. Although heads featuring squaramide outperform other structures again (Supplementary Fig. 6), they were difficult to synthesize and had to be abandoned with regret.
The first-round of lipid virtual screening was culminated in the selection of three ionizable lipids for testing: LQ085, LQ086, and LQ087. Only LQ087 exhibited comparable performance to MC3, but it still fell notably short of SM-102. The specific reason for the underperformance of the three new lipids remains unclear, as the selected head groups were sparsely tested in the previous study, making mechanistic explanation challenging. The limitation in model generalization is a possible cause.
The second round of virtual screening focused on lipids containing the ethanol amine head and trained a stricter model (Model 2) to facilitate the screening. The evaluation process was actually an association of Model 1 and Model 2. Model 1 performed better in general prediction accuracy but still predicted too many lipids with positive mRNA delivery efficiency. Model 2 performed less well in general but showed high precision, making it effective in filtering out false positive predictions. This round of screening yielded six ionizable lipids, LQ089-094, all of which were equal to or superior to MC3, proving the validity of the screening strategy. Properly combining and leveraging the advantages of different models is important for AI applications.
This strategy yielded six ionizable lipids: LQ089-094. Among them, only LQ092 was previously reported18,35. LQ089-094 exhibited superior mRNA delivery efficiency compared to LQ085-087. Notably LQ089 showcased exceptional performance, comparable to SM-102. The distinguishing factor lies in the cyclohexane and branched alkane groups in their tails.
So far, all lipids were initially evaluated based on whole-body luminescence signals at three-time points. This reliable evaluation method is robust to gender differences and a number of mice and time points, with acceptable cost and satisfactory efficiency. After that, the lipids were comprehensively evaluated for organ-specific distribution. Similar to MC3 and SM-102, all new lipids led to high mRNA expression in the liver when administered both intravenously and intramuscularly. Storage stability and acute toxicity were tested for three representative lipids. LQ086 was less stable in the -20 °C storage condition, possibly due to its head structure, but LQ089 and LQ092 showed acceptable stability. Administration of these lipids was safe and no considerable acute toxicity was observed in the tests.
Lipids with head groups containing hydroxyl are commonly tested but show varied performance. The structure-activity relationship in this type of lipid has not been described in detail. However, with a well-trained AI model, this relationship can be comprehensively explored (Fig. 5), such as the influence of gradually extending the length of the carbon chain and moving the linker along the chain on delivery efficiency. It can be observed that, although the lipids, after ECFP transformation, produce high-dimensional and discontinuous features, the AI model output continuous trends in structure-activity relationships.
To obtain well-performing lipids, all chain segments in tails should have harmonious lengths. The linker position should be compatible with the whole tail length, and the length threshold is dependent on the linker type. Ionizable lipids like SM-102, ALC-031517, and our selected LQ092, LQ093, and LQ094 all belong to the area with a relatively high positive rate. The structure-activity relationship represented in this way is easy to understand and applicable to guide molecule design. However this visualization method is limited in some specific molecule design space.
This work solidifies the potential of AI methodology in ionizable lipid design by accelerating the screening process and summarizing the structure-activity relationship. This type of application can be improved in many aspects in the future. First, the size of the dataset is relatively modest. Despite comprehensive data collection from literature and patents, the majority of ionizable lipids were tested in the siRNA delivery system8,9,52,53 and had to be excluded. Additionally, rigorous data cleaning inevitably resulted in some data exclusion to ensure dataset consistency. As a result, the data size constrained the modeling approach. Expanding databases stands as a critical avenue for optimizing AI models, and high-throughput methods serve as a valuable complement to the AI approach54.
Secondly, generalizing the model to a broader formulation design space is challenging, like novel lipid structures and different mRNA sequences. In this work, predictions on LQ089-094 are more accurate than those on LQ085-087, and the former are closer to the majority of lipids in the dataset. In other AI modeling work, the newly designed molecules are also similar to their training data27,54. This limitation might be alleviated through data augmentation, introducing more diverse data, or adopting a pre-train and fine-tune model building workflow. Besides, mechanistic modeling is a promising way to break through the generalization limitation, such as molecular dynamic (MD) simulation. The simulated LNP and the interaction between RNA and lipids have been reported many times26,55–57, with customized ionizable lipid structures. MD simulation should also facilitate the understanding of the lipid specificity to different mRNA sequences. In our work, data of luciferase and hEPO mRNA were merged, but only nearly 10 lipids were tested using both mRNA. The delivery efficiencies for the two mRNA show a consistent trend, but using hEPO seems to be more likely to obtain positive results.
Lastly, the goal of this work is to construct lipids with generally high mRNA delivery ability, not specific for any organ, disease, or therapy. Therefore, only data of luciferase and hEPO mRNA delivery in mice were collected, as this is a basic screening method. However, models tailored according to therapeutic objectives or types of diseases are more appealing. For example, maximizing protein expression level is the priority in mRNA therapy supplementing missed proteins, but in mRNA vaccines against viruses, immunogenicity of the formulation needs additional consideration58. Developing models predicting immunogenicity is important for mRNA delivery. Likewise, another iteration direction of the model will be to screen out lipids with high-level expression of mRNA in organs other than the liver to meet the needs of a variety of diseases. Additionally, prediction in primates and even humans for specific diseases is profound for clinical translation. AI modeling methodology is still possible to handle these tasks only if data supports. However, other advanced modeling methods such as physiologically-based pharmacokinetics (PBPK) and quantitative systems pharmacology (QSP) models59–63 are very useful. PBPK is specialized in inferring the fate of drugs across different species. This inference is based on the properties of the drug and the physiological conditions of the subject, and therefore such extrapolation is mechanistically based. QSP is also mechanistic, predicting dynamic changes in signal pathways, biomarkers, and even therapeutic effects. For a complex system such as immune response, QSP is promising to address it64,65. Further, the association of the two models can integrate various in vitro and in vivo data, being able to quantify rates of critical processes in nucleic acid delivery such as RNA escape from endosomes66.
To summary, this study demonstrates that AI models predicting mRNA delivery efficiency and apparent pKa can expedite the screening and design of ionizable lipids for LNP formulation. Notably, among the screened lipids, one with a benzene ring in its head group demonstrated comparable performance to the MC3 control, while six others outperformed MC3. Notably, one of these even approached the performance of SM-102. This research underscores the significance of properly associating different AI models to leverage their merits, especially when working with limited data size. Additionally, this AI model shows explicit interpretability in structure. This methodology and insights gleaned from this study hold the potential to advance the development of mRNA-LNP delivery systems and offer valuable guidance to similar projects.
Methods
Data preparation
The data was collected from patents33–41 and articles13,15,18,20,28–32. The data was sourced from companies of Moderna, Acuitas, Protiva, and several academic institutions. The extracted information included the LNP formulations (including chemical structures of ionizable lipids, types of helper lipids, types of PEG-lipids, and molar ratios of lipids), apparent pKa of LNPs, particle sizes, EE, species of animals, routes of administrations, doses of administrations, and mRNA expression levels. Structures of ionizable lipids were extracted from resources with InDraw (6.1.0) AI chemical structure recognizer and transformed to SMILES string. Other data was extracted by manually copying and checked. N/P ratio or weight ratio of lipids and mRNA was not included in the data because many resources only provide a ratio range instead of a clear value, but most LNPs had a N/P ratio near to 6.
For the analysis of nanomedicine from multiple data sources, ensuring internal consistency within the data is crucial67. For AI models predicting the in vivo mRNA delivery efficiency of LNPs, subsequent data processing work was conducted to improve its internal consistence and maintain as large data as possible: (1) removed data that was not measured in mice; (2) removed data where the LNP was not administrated intravenously; (3) removed data of mRNA expression level which was not measured as the luminescence signal or concentration of the luciferase or the human erythropoietin (hEPO) induced by mRNA delivery; (4) removed data of the luminescence signal of luciferase that was not measured for whole-body of subject animals or livers; (5) maintained the data where the mRNA expression levels of LNPs could be transformed as the fold-change based on a standard LNP formulation. The standard LNP formulation was composed of MC3 (the ionizable lipid), DSPC (the helper lipid), cholesterol, and PEG2000-DMG (the PEG lipid) at the molar ratio of 50/10/38.5/1.5, which is commonly used as the control since it is the LNP formulation of the first approved siRNA drug68. The standard expression level of this formulation included: (1) luciferase concentration at 198 ng g-1 liver tissue at 4 h after administration of 0.3 mg kg-1 mRNA38; (2) luciferase luminescence flux at 2.57E + 9 p s-1 in livers at 6 h after administration of 0.5 mg kg-1 mRNA (for data from the institution of Moderna)35; (3) luciferase luminescence flux at 8.66E + 8 p s-1 in the whole-body at 6 h after administration of 0.5 mg kg-1 mRNA (for data from the institution of Tufts University)31; (4) plasma hEPO concentration at 1570, 1830, 810 ng mL-1 at 3, 6, 24 h respectively, after administration of 0.5 mg kg-1 mRNA35. The value of concentrations of expressed proteins was comparable among different institutions, while the value of luminescence flux was not since the measurement of the flux is the signal after amplification via the photomultiplier, which is dependent on the experimental instrument of the institute.
All the mRNA delivery efficiency of ionizable lipids was normalized to that of MC3. For the classification model, lipids with normalized efficiency equal to or larger than 1 (Model 1) or 2 (Model 2) were labeled as positive, while the others as negative. The delivery efficiency was also predicted based on LNP formulations (types of ionizable lipids, helper lipids, PEG-lipid, cholesterol, and their molar ratio in the formulation). The dataset contained 387 LNP formulations, with 370 different ionizable lipids.
In the work of predicting the apparent pKa, no particular processing work was conducted. The dataset contained 352 LNP formulations with 351 different ionizable lipids. The apparent pKa was predicted merely based on LNP formulations.
In this study, the ionizable lipid structure was represented by ECFP converted via the RDKit package (2023.9.1) in Python (3.11.4). The ECFP radius was set to 9, and the number of bits was set to 1024. Each ionizable lipid had a unique ECFP sequence. The involved three helper phospholipids, DSPC, DOPE, and DOPC were represented by two ‘0-1’ category variables (‘DS’ or ‘DO’, ‘PC’ or ‘PE’). The PEG-lipids were represented by a single multiple-category variable. Only one type of cholesterol lipid was included in our data, so it was not represented. Molar ratios of the four types of lipids in LNP were represented as numeric variables between 0 and 1.
Data splitting and hyperparameters
The following methods apply to the building of the classification model of mRNA delivery efficiency. The whole data set was split into the training set and the test set. The stratified sampling method was used to keep the category distribution (proportion of positive and negative lipids) and the source distribution (proportion of samples from each data source) in the separate data set the same as the original data. The stratified splitting strategy was implemented by the scikit-learn (sklearn 1.1.3) package. Finally, we obtained the training set and test set at a ratio of 4:1.
A random search was applied to tune the hyperparameters of the models. Briefly, 1000 hyperparameter combinations were randomly chosen from the hyperparameter space to train on the training set, and the results of the 5-fold cross-validation (5_CV) on the training set were used to finetune the hyperparameters to find the best model. For models built with the LightGBM algorithm (package version 3.3.5), the important hyperparameters of the best model were colsample_bynode = 0.8, colsample_bytree = 0.5, learning_rate = 0.1, max_depth = 3, num_leaves = 4, reg_alpha = 1, reg_lambda = 1, subsample = 0.7, subsample_freq = 3 (Model 1); and colsample_bytree = 0.5, learning_rate = 0.01, max_depth = 5, n_estimators=100, num_leaves = 11, subsample = 0.5 (Model 2). Meanwhile, the hyperparameters for Random Forest were class_weight = None, criterion = entropy, max_depth = 9, max_features = sqrt, max_leaf_nodes = 20, min_samples_split = 2, n_estimators = 50.
For the apparent pKa model, the dataset was divided into three subsets, namely the training, validation, and test sets, with the data size ratio at approximately 8:1:1. The following strategies were used to divide the data set. Uncommon molecules, defined as those whose head and tail structure appeared in the dataset less than three times, were forcibly included in the training set in order to make the model be trained in molecular structure space as broad as possible. The remaining data were divided using random stratified sampling based on pKa ( < 6, 6-7, 7-8, > 8). The model was trained on the training set, while its hyperparameters were adjusted on the validation set to obtain the optimal configuration. Ultimately, the performance of the model was evaluated on the testing set to assess its generalization ability.
This model was trained with the LightGBM algorithm to establish a regression model, using the sklearn library. Finetuning the model’s hyperparameters was conducted on the validation set based on a random search approach. Ultimately, the optimal hyperparameter configuration was: colsample_bytree=1, learning_rate=0.01, max_depth=40, n_estimators=700, num_leaves=45, objective=’regression’, subsample=0.8.
Evaluation criteria
The performance of the prediction of the regression model was evaluated by mean absolute error (MAE), mean squared error (MSE), root mean squared error (RMSE), and determination coefficient (R2). The prediction performance of the classification model was evaluated by four metrics, including Accuracy (ACC), recall, precision, and F1_score (F1). These metrics are defined as follows:
| 1 |
| 2 |
| 3 |
| 4 |
where TP is the true positive, TN is the true negative, FP is the false positive, and FN is the false negative.
Calculation of lipid ECFP score
To inform lipid generation with the help of the built AI model, first, the SHAP algorithm43 was applied to the model to calculate feature importances of input parameters for the mRNA delivery efficiency prediction. Particularly, the SHAP value for each ECFP bit (indicating substructure) in each ionizable lipids in dataset was obtained. Then total contribution of each bit was calculated based on the sum of SHAP value in all lipids:
| 5 |
Where Coni is the contribution of the bit i, ILj is the ionizable lipid j, SHAPi,j is the SHAP value of bit i in lipid j, t_ECFPi,j is transformed value of ECFPi,j considering the contribution of the presence or absence of this bit in the lipid. Thus, Coni more than 0 means positively contributing bit and the other side means negatively contributing. Consequently, the ECFP score for lipids could be defined as the sum of the product of ECFP bit value and its contribution:
| 6 |
Where Scorej is the ECFP score for ionizable lipid j.
Molecular similarity analysis
The molecular similarity of ionizable lipids was calculated using RDKit script. To calculate the similarity of one ionizable lipid to a bundle of other lipids, the first step was to calculate Morgan fingerprint molecular similarity between the target lipid and each of the other lipids resulting a bundle of weight maps, then summed all weight maps to obtain the similarity of the target lipid and visualized it. From the similarity graph, distinct and typical substructures can be recognized.
Calculation of candidate segment positive rate
In virtual screening, the general performance of a candidate tail or head segment was judged by positive rate. A generated virtual ionizable lipid would be marked as positive if its predicted mRNA delivery efficiency was better than the standard MC3 LNP and apparent pKa was between 6.0 to 7.0. Since the lipids were generated by combining different head and tail segments, therefore, for each segment, the positive rate can be defined as:
| 7 |
Segments with high positive rate means they are more compatible in ionizable lipids to increase the LNP performance.
Ionizable lipids synthesis
Nine ionizable lipids (LQ085-087, 089-094) screened by AI models were synthesized for testing. The synthesis method of them is shown in the Supplementary Information. All synthesized lipids were chemically characterized in detail69, and their spectra of 1H NMR, 13C NMR, and MS are shown in Supplementary Figs. 20 to 28.
LNP formulation and characterization
The mRNA of firefly luciferase was synthesized in our lab. T7 RNA polymerase was used to mediate the transcription from a DNA template. Cap 1 was added to enhance the expression efficiency. MC3 was purchased from APExBIO. DSPC and cholesterol were purchased from Nippon Fine Chemical. DMG-PEG2000 was purchased from Zhejiang Guobang Pharmaceutical.
Appropriate amounts of ionizable lipids, cholesterol, DSPC, and DMG-PEG2000 were dissolved in ethanol to make stock solutions of each lipid. A mixed lipid solution was then prepared according to a molar ratio of ionizable lipid:DSPC:cholesterol:DMG-PEG2000 of 50:10:38.5:1.5, resulting in a final lipid concentration of 12.5 mM. Luciferase mRNA was dispersed in a citrate buffer to make an acidic mRNA solution. Using a PNI microfluidic device, the mRNA solution and the lipid ethanol solution (at a nitrogen-to-phosphorus ratio of 6:1) were mixed at a flow rate of 12 mL/min and a volume ratio of 3:1. The mixture was dialyzed against 0.01 M PBS for 12–24 hours to remove the ethanol. After dialysis, the LNP solution was concentrated by ultrafiltration (Amicon-Ultra, MWCO 10KDa) and sterilized by passing it through a 0.22 μm sterile filter. The particle size and PDI of the LNP were measured using a Malvern particle size analyzer; the EE of the LNP was determined using the Quant-it Ribogreen RNA assay kit; and the mRNA concentration was measured using a Stunner high-throughput concentration and particle size analyzer.
To measure the LNP apparent pKa, LNPs were incubated with TNS (2-(p-tolylamino)-6-naphthalenesulfonic acid) in different pH conditions. The negatively charged TNS interacted with cationic LNPs, emitting luminescence signals. The pH condition at which 50% of ionizable lipids are charged is defined as the apparent pKa. First, a series of buffers with a pH range from 2.0 to 12.0 was prepared by adding 2 M sodium hydroxide and 2 M hydrochloric acid to basal buffer (10 mM sodium phosphate, 10 mM sodium borate, 10 mM sodium citrate, 150 mM sodium chloride). Then, the series of pH buffer was added at 94 μL to a black-bottom, 96-well plate, followed by adding 4 μL the LNP solution (0.05 mg mL-1 mRNA, dissolved in PBS) and 2 μL TNS solution (300 μM, 10% DMSO). After the addition of the sample, the table was gently panned and shaken to mix well. Let the sample stand for 7 min at room temperature avoid light, and measure the luminescence intensity at 325 nm excitation wavelength and 435 nm emission wavelength using an enzyme marker. The luminescence intensity (Y-axis) was plotted against the pH of the assay buffer (X-axis), and the value of LogEC50 was considered as the pKa of the LNP to be measured.
AUC of the bioluminescence in vivo
This research complies with all relevant ethical regulations. Animal procedures were performed under the guidance of animal ethics and approved by the Institutional Animal Care and Use Committee of School of Life Sciences, Fudan University. Mice were housed in cages with six mice each, allowed unrestricted access to food and water, and kept in conditions with a temperature range of 20–26 °C, a relative humidity of 50–60%, and a 10 h/14 h light-dark cycle. For animal experiments, mice were randomly assigned to each experimental group and no data were excluded from the analyses. Female BALB/c mice (6–8 weeks old, Vital River) were used in verifying the performance of LNP. Mice were intravenously injected with LNPs loaded with luciferase mRNA at a dose of 5 μg per mouse. D-luciferin potassium salt was injected intraperitoneally at certain time points after administration, and the total luminescence in the mice was detected using an IVIS Spectrum small animal in vivo imaging system. The luminescence total flux of the lipids was first converted into logarithmic form and analyzed by One-Way ANOVA and followed by the Bonferroni test, α = 0.05. Prism 9 (GraphPad Software, San Diego, CA, USA) was used.
The Organ distribution of mRNA expression
BALB/c mice were intravenously or intramuscularly injected of LNPs loaded with luciferase mRNA at a dose of 5 μg mRNA per mouse. D-luciferin potassium salt was injected intraperitoneally a dose of 3 mg per mouse 4 hours after administration. The in vivo bioluminescence of whole body was detected and then the mice were euthanized to obtain hearts, livers, spleens, lungs, kidneys and brains. The bioluminescence of ex-vivo organs was detected to characterize the distribution of mRNA expression.
The Organ distribution of Cy5-labeled mRNA loaded in LNPs
BALB/c mice were intravenously injected of LNPs loaded with Cy5-labeled luciferase mRNA at a dose of 5 μg mRNA per mouse. The mice were euthanized and dissected to obtain the vital organs 4 hours after administration. Each organ was grinded with PBS buffer into 20% homogenate on ice and then the homogenate was centrifugated to get supernatant. The emitted light with a wavelength of 670 nm of homogenate supernatant was detected under the exciting light with a wavelength of 650 nm, which was the signal of Cy5. Tissue homogenates containing different gradient concentrations of Cy5 mRNA were prepared with blank mouse organs. For each organ, the standard curve of Cy5 luminescence intensity to concentration of Cy5 mRNA was drawn. Then, the concentration of Cy5 mRNA in homogenate and the ratio of organ-distributed Cy5 mRNA were calculated.
Acute toxicity test
BALB/c mice were intravenously injected of LNPs loaded with luciferase mRNA at acute toxic dosages of 20 μg and 100 μg mRNA per mouse (1 mg kg-1 and 5 mg kg-1, respectively). The weight of mice was monitored on Day 1, 2, 3, 4, 7, 9, 11, 13, and 14 after administration. Vital organs were obtained at Day 14, weighed, fixed in 4% buffered formalin for 3 days, embedded in paraffin, and cut into 5 μm thick slices. The slices were dewaxed in ethanol and xylene and then stained with hematoxylin-eosin (H&E). Pathology slides were scanned using a digital slide scanner (3DHISTECH). In the acute toxicity test, the whole blood was detected by an animal blood analyzer (HEMAVET). The glutamic-pyruvic transaminase (ALT) and creatinine (Crea) were detected using a biochemical analyzer (Rayto).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
This work was financially supported by the University of Macau Multi-Year Research Grant – Collaborative Research Grant (MYRG-CRG2022-00008-ICMS, China) to D.O., the Science and Technology Development Fund, Macau SAR (0071/2024/RIA1 and 005/2023/SKL) to D.O., the Shenzhen-Hong Kong-Macau Science and Technology Program (Category C) of Shenzhen Science and Technology Innovation Commission (SGDX20210823103802016) to D.O., Industry-university-research cooperation project and Zhuhai-Hong Kong-Macao cooperation project from Zhuhai Science and Technology Innovation Bureau (ZH22017002210010PWC) to D.O., National Natural Science Foundation of China (32301174) to K.C., and UM Postdoctoral Fellow of UM Talent Programme (ICMS/RTO/EP160/2023) to W.W. This work has received support from the Shanghai Zhangjiang mRNA Innovation and Translation Center and Fudan Center for mRNA Translational Research. We would like to express our gratitude to the Chemistry Department of RNACure for their assistance in lipid synthesis, as well as for providing their internal lipid library as an external dataset.
Author contributions
D.O. and J. Lin conceived and designed the study. W.W. collected the data, analyzed the AI modeling result, and wrote the article. K.C. designed the scheme of in vivo experiments, participated in the lipid synthesis, analyzed the data, and wrote the article. T.J. assisted with the preparation and characterization of LNPs, performed the evaluation LNPs in mice. Y.W. conducted the AI modeling of delivery efficiency. Z.W. conducted the AI modeling of apparent pKa. H.Ying and H.Yu assisted with optimization of models and algorithms for predicting lipids. D.O., J. Lin, and J. Lu supervised the study.
Peer review
Peer review information
Nature Communications thanks Hadi Valadi, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data generated in this study are provided in the Source Data file. Source data are provided with this paper and also deposited in figshare repository70 (https://figshare.com/s/ad928807e1b4795b9b5e). Source data of prediction result of the generated ionizable lipid library is available on request from the corresponding author D.O. Source data are provided with this paper.
Code availability
The codes that support the findings of this study are available on request from the corresponding author D.O.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Wei Wang, Kepan Chen, Ting Jiang, Yiyang Wu.
Contributor Information
Jinzhong Lin, Email: linjinzhong@fudan.edu.cn.
Defang Ouyang, Email: defangouyang@um.edu.mo.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-55072-6.
References
- 1.Baden Lindsey, R. et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med.384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polack, F. P. et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med.383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater.6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization. COVID-19 vaccine tracker and landscape. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (2022).
- 5.Semple, S. C. et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta BBA - Biomembr.1510, 152–166 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Maurer, N. et al. Spontaneous entrapment of polynucleotides upon electrostatic interaction with ethanol-destabilized cationic liposomes. Biophys. J.80, 2310–2326 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyes, J., Palmer, L., Bremner, K. & MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Rel.107, 276–287 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Jayaraman, M. et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl.51, 8529–8533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semple, S. C. et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol.28, 172–176 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Mui, B. L. et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol. Ther. Nucleic Acids2, e139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Y., Sun, C., Wang, C., Jankovic, K. E. & Dong, Y. Lipids and lipid derivatives for RNA delivery. Chem. Rev.121, 12181–12277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassett, K. J. et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. - Nucleic Acids15, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao, L. et al. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun.11, 2424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, S. et al. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Rel.235, 236–244 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Cornebise, M. et al. Discovery of a novel amino lipid that improves lipid nanoparticle performance through specific interactions with mRNA. Adv. Funct. Mater.32, 2106727 (2022). [Google Scholar]
- 16.Zhi, D. et al. The headgroup evolution of cationic lipids for gene delivery. Bioconjug. Chem.24, 487–519 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Eygeris, Y., Gupta, M., Kim, J. & Sahay, G. Chemistry of lipid nanoparticles for RNA delivery. Acc. Chem. Res. 55, 2–12 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Sabnis, S. et al. A novel amino lipid series for mRNA Delivery: Improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther.26, 1509–1519 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, B. et al. Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing. Nat. Biotechnol.41, 1410–1415 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao, L. et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol.37, 1174–1185 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Whitehead, K. A. et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun.5, 4277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pant, S. M. et al. Design, synthesis, and testing of potent, selective hepsin inhibitors via application of an automated closed-loop optimization platform. J. Med. Chem.61, 4335–4347 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Merk, D., Friedrich, L., Grisoni, F. & Schneider, G. De Novo design of bioactive small molecules by artificial intelligence. Mol. Inform.37, 1700153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bannigan, P. et al. Machine learning directed drug formulation development. Adv. Drug Deliv. Rev.175, 113806 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Wang, W., Ye, Z., Gao, H. & Ouyang, D. Computational pharmaceutics - A new paradigm of drug delivery. J. Control. Rel.338, 119–136 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Wang, W. et al. Prediction of lipid nanoparticles for mRNA vaccines by the machine learning algorithm. Acta Pharm. Sin. B12, 2950–2962 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, B. et al. Accelerating ionizable lipid discovery for mRNA delivery using machine learning and combinatorial chemistry. Nat. Mater.23, 1002–1008 (2024). [DOI] [PubMed] [Google Scholar]
- 28.Fenton, O. S. et al. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater.28, 2939–2943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajj, K. A. et al. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small15, 1805097 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Zhao, X. et al. Imidazole-based synthetic Lipidoids for in vivo mRNA delivery into primary T lymphocytes. Angew. Chem. Int. Ed. Engl.59, 20083–20089 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Qiu, M. et al. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc. Natl Acad. Sci.118, e2020401118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kauffman, K. J. et al. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett.15, 7300–7306 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Heyes, J. et al. Compositions and methods for delivering messenger RNA. WO2015011633Al. (2016).
- 34.Benenato, K. E. & Butcher, W. Compounds and compositions for intracellular delivery of agents. WO2017112865Al. (2017).
- 35.Benenato, K. E. Compounds and compositions for intracellular delivery of therapeutic agents. WO2017049245Al. (2018).
- 36.Benenato, K. E., Cornebise, M. & Hennessy, E. Compounds and compositions for intracellular delivery of therapeutic agents. WO2020061367A1. (2020).
- 37.Du, X. & Ansell, S. M. Lipids and lipid nanoparticle formulations for delivery of nucleic acids. US20160376224Al. (2017).
- 38.Du, X. Lipids for use in lipid nanoparticular formulations. WO2019036028A1. (2019).
- 39.Du, X. & Ansell, S. M. Novel carbonyl lipids and lipid nanoparticle formulations for delivery of nucleic acids. WO2018200943A1. (2018).
- 40.Ansell, S. & Du, X. Novel Lipids and Lipid Nanoparticle Formulations for Delivery of Nucleic Acids. WO2015199952Al. (2015).
- 41.Ansell, S. M. & Du, X. Novel lipids and lipid nanoparticle formulations for delivery of nucleic acids. WO2017075531A1. (2017).
- 42.Rogers, D. & Hahn, M. Extended-connectivity fingerprints. J. Chem. Inf. Model.50, 742–754 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Lundberg, S. M. & Lee, S.-I. A Unified Approach to Interpreting Model Predictions. in Advances in Neural Information Processing Systems vol. 30 (Curran Associates, Inc., 2017).
- 44.Verbeke, R., Lentacker, I., De Smedt, S. C. & Dewitte, H. The dawn of mRNA vaccines: The COVID-19 case. J. Control. Rel.333, 511–520 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carrasco, M. J. et al. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol.4, 1–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He, Y. et al. Can machine learning predict drug nanocrystals? J. Control. Rel.322, 274–285 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Deng, J. et al. Machine learning in accelerating microsphere formulation development. Drug Deliv. Transl. Res.13, 966–982 (2023). [DOI] [PubMed] [Google Scholar]
- 48.Zhao, Q., Ye, Z., Su, Y. & Ouyang, D. Predicting complexation performance between cyclodextrins and guest molecules by integrated machine learning and molecular modeling techniques. Acta Pharm. Sin. B9, 1241–1252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, J., Gao, H., Ye, Z., Deng, J. & Ouyang, D. In silico formulation prediction of drug/cyclodextrin/polymer ternary complexes by machine learning and molecular modeling techniques. Carbohydr. Polym.275, 118712 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Ledford, H. Gene-silencing technology gets first drug approval after 20-year wait. Nature560, 291–292 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Bannigan, P. et al. Machine learning models to accelerate the design of polymeric long-acting injectables. Nat. Commun.14, 35 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen, D. et al. Rapid discovery of Potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J. Am. Chem. Soc.134, 6948–6951 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Love, K. T. et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl Acad. Sci.107, 1864–1869 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, Y. et al. AGILE Platform: A Deep Learning-Powered Approach to Accelerate LNP Development for mRNA Delivery. 10.1101/2023.06.01.543345 (2023) [DOI] [PMC free article] [PubMed]
- 55.Rozmanov, D., Baoukina, S. & Peter Tieleman, D. Density based visualization for molecular simulation. Faraday Discuss.169, 225–243 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Paloncýová, M. et al. Atomistic insights into organization of RNA-loaded lipid nanoparticles. J. Phys. Chem. B127, 1158–1166 (2023). [DOI] [PubMed] [Google Scholar]
- 57.Rissanou, A. N., Ouranidis, A. & Karatasos, K. Complexation of single stranded RNA with an ionizable lipid: an all-atom molecular dynamics simulation study. Soft Matter16, 6993–7005 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Sahin, U., Karikó, K. & Türeci, Ö. mRNA-based therapeutics-developing a new class of drugs. Nat. Rev. Drug Discov.13, 759–780 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Parhiz, H. et al. Physiologically based modeling of LNP-mediated delivery of mRNA in the vascular system. Mol. Ther. - Nucleic Acids35, 1–11 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones, H. M. & Rowland-Yeo, K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacomet. Syst. Pharmacol.2, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeon, J. Y., Ayyar, V. S. & Mitra, A. Pharmacokinetic and pharmacodynamic modeling of siRNA therapeutics – a minireview. Pharm. Res.39, 1749–1759 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Apgar, J. F. et al. Quantitative systems pharmacology model of hUGT1A1-modRNA encoding for the UGT1A1 enzyme to treat Crigler-Najjar Syndrome Type 1. CPT Pharmacomet. Syst. Pharmacol.7, 404–412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, W., Deng, S., Lin, J. & Ouyang, D. Modeling on in vivo disposition and cellular transportation of RNA lipid nanoparticles via quantum mechanics/physiologically-based pharmacokinetic approaches. Acta Pharm. Sin. B14, 4591–4607 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruiz-Martinez, A. et al. Simulations of tumor growth and response to immunotherapy by coupling a spatial agent-based model with a whole-patient quantitative systems pharmacology model. PLOS Comput. Biol.18, e1010254 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bansal, L. et al. Mathematical modeling of complement pathway dynamics for target validation and selection of drug modalities for complement therapies. Front. Pharmacol. 13, 1–20 (2022). [DOI] [PMC free article] [PubMed]
- 66.Maugeri, M. et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun.10, 4333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mata Corral, M. Y., Alvarez, D. E. & Poon, W. Quantifying nanoparticle delivery: challenges, tools, and advances. Curr. Opin. Biotechnol.85, 103042 (2024). [DOI] [PubMed] [Google Scholar]
- 68.Schoenmaker, L. et al. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm.601, 120586 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dong, W. et al. Multicomponent synthesis of imidazole-based ionizable lipids for highly efficient and spleen-selective messenger RNA delivery. J. Am. Chem. Soc.146, 15085–15095 (2024). [DOI] [PubMed] [Google Scholar]
- 70.Wang, W. Dataset for artificial intelligence-driven rational design of ionizable lipids for mRNA delivery. figshare. 10.6084/m9.figshare.26379541.v1 (2024)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are provided in the Source Data file. Source data are provided with this paper and also deposited in figshare repository70 (https://figshare.com/s/ad928807e1b4795b9b5e). Source data of prediction result of the generated ionizable lipid library is available on request from the corresponding author D.O. Source data are provided with this paper.
The codes that support the findings of this study are available on request from the corresponding author D.O.