Abstract
Bacterial infections commonly complicate cutaneous leishmaniasis (CL), worsening the disease and delaying healing. Despite this, there is a gap in research concerning the characteristics of pathogenic microorganisms associated in CL patients. This study aims to identify bacterial isolates and drug susceptibility patterns in CL patients. A purposive cross-sectional study was conducted among CL patients attending the ALERT hospital from January 2021 to June 2021. Standardized questionnaires were used to collect socio-demographic and clinical data. Each patient’s lesion was aseptically sampled with two swabs. The swabs were aseptically inoculated onto blood agar plates (BAP) and Mac Conkey agar (MAC) and cultured following standard protocols. The isolates were identified by gram staining, colony morphology, and biochemical testing. Antimicrobial susceptibility patterns were done using the disk diffusion technique according to 2021 CLSI guidelines. SPSS version 20 was used to enter and analyze data. In this study 384 CL patients (66.9% male), aged 2–85 years were enrolled. 390 pathogenic bacteria were isolated from CL lesions, with 58.0% and 42.0% Gram-positive and Gram-negative bacteria, respectively. S aureus (41.5%), coagulase-negative Staphylococci (14.4%), Citrobacter spp. (10.8%), Klebsiella spp. (9.9%), and Proteus spp. (7.9%) were the most commonly identified bacteria. Over 80% of the isolates demonstrated multi-drug resistance to two or more antibiotics. S. aureus showed high resistance to penicillin (86.4%), methicillin (83.9%), and tetracycline (47.5%). These findings highlight the critical needs of microbial diagnostics and antibiotic susceptibility testing in CL patients due to the rising prevalence of drug-resistance, including the multi-drug resistant bacteria.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-024-10409-w.
Keywords: Cutaneous leishmaniasis, Bacterial isolates, Drug susceptibility, Ethiopia
Introduction
Leishmaniasis is a disease caused by the bite of Leishmania-infected female phlebotomine sandflies [1]. It is characterized by cutaneous, mucocutaneous, or visceral clinical manifestations.
depending on the species of parasite and the immune response of the host [2, 3]. Cutaneous leishmaniasis (CL) is the most common leishmaniasis, including in Ethiopia. In Ethiopia, Leishmania aethiopica is the primary causative species for CL [1]. CL is known for its diverse clinical presentations, ranging from small nodules on the skin to gross mucosal tissue damage [2]. The severity and type of lesions are closely tied to the host’s immune response. The initial immune response to the parasite includes a rapid influx of neutrophils and inflammatory monocytes. Neutrophils can play dual roles, being both protective and potentially harmful, while inflammatory monocytes actively combat the Leishmania parasites and differentiate into monocyte-derived dendritic cells. These dendritic cells are crucial for facilitating the development of protective CD4 + T helper 1 (TH1) cells [4]. The ulcers of CL are usually slow to heal and can persist for months or years without the recommended first-line anti-parasitic drug, pentavalent antimony [5, 6]. This chronicity of the ulcers, the presence of skin lesions primarily on exposed parts of the body, and poor lesion site hygiene due to the proximity of lesions in the lower extremities, which are most affected, to the ground promote secondary bacterial infection, which is frequently observed in local CL (LCL) patients [7–9].
Secondary bacterial infection is the major complication of CL. The Leishmania parasite which was inserted into the cutaneous part of the skin by the sand flies bite damages the tissue of the skin and facilitates the bacterial invasion. During normal circumstances, a typical CL lesion usually presents a painless, ulcerative skin with a rounded or oval shape, an infiltrated base, a well-demarcated raised erythematous edge, a reddish background with rough granulation, and discharge [10].The presence of a bacterial infection causes localized pain and produces serious and purulent exudates that completely or partially reconstitute the lesion, which then dries into crusts and may delay the healing process and resulting scarring [1, 4].
The presence of complex bacterial colonization in lesions of chronic wounds can cause adverse effects on the healing process. The incidence of secondary bacterial infection in CL lesions ranges from 23.6–81% [7, 8, 11]. In LCL lesions, individuals with secondary infections with purulent discharge and concomitant infection can delay the healing process. The most frequently isolated bacteria from ulcers of skin and soft-tissue infections are Staphylococcus aureus, Streptococcus pyogenes, Staphylococcus epidermidis, Pseudomonas aeruginosa, Morganella morganii, Klebsiella pneumoniae, Corynebacterium diphtheria and Enterococcus durans [7, 8, 12]. CL ulcer infected with secondary bacterial infection need to follow antimicrobial stewardship during ulcer management, which was recently proposed for all chronic [12]. Studies suggest that bacterial pyogenic infections should be eliminated from patients with CL ulcers by appropriate treatment before starting anti-parasitic antimony [13].
Antibiotics have proven useful in treating bacterial infections. However, it is important to note that nearly all pathogenic bacteria emerge to develop acquired antimicrobial resistance over time, while some bacteria may also possess inherent resistant to newly developed antimicrobials. The rise of antimicrobial resistance has become a major public health problem, affecting healthcare outcome and posing a serious threats to economic and social development, thereby complicating the treatment of infectious diseases [14]. The overuse of antibiotics is the primary driver of this problem, as it creates selective pressure and promotes the emergence of resistant bacterial strains [15].
In Africa, empirical antimicrobial therapy is an important therapeutic modality due to the relative lack of adequate laboratory equipment for bacterial culture and susceptibility testing in many healthcare settings [16]. In institutions that have microbiology laboratory facilities, swab samples from ulcer of CL showed multidrug resistant bacterial growth. For instance, from the ulcers of CL patients in Ghana more than ten pathogenic bacterial infections were identified, with the majority of the isolates were resistant to third-generation cephalosporins and beta-lactam antibiotics. Remarkably, 92.3% of S. aureus isolates were ampicillin resistant, 69.2% were only moderately sensitive to erythromycin, and 84.6% of isolates were resistant to both methicillin and ciprofloxacin [16]. This highlights a significant concern regarding antibiotic resistance in managing infections associated with CL. In Ethiopia, secondary bacterial infections of CL patients range from 4 to 44%, including diffuse (DCL) [17–19]. Based on a combined incidence of bacterial isolates with multidrug resistance that ranged from 64.9 to 76.1% of antibiotic resistance is a rising hazard to public health [20]. Among the pathogenic bacteria isolated from wound infections in patients with lower extremity lymphedema, 52.4% multiple-microbial isolates, 47.6% single bacterial isolates, and 5.3% multiple Gram-positive isolates were identified [21]. In Ethiopia, culture tests are only available in a few hospitals and research institutions which are not fully functional due to inadequate supplies and microbiology laboratory facilities. However, to our knowledge, there is no or limited information on the spread of pathogenic bacteria and their antimicrobial resistance patterns in CL-infected patients. This study aimed to identify bacterial isolates and drug susceptibility patterns in infected lesions of CL patients at ALERT Hospital, Addis Ababa, Ethiopia.
Materials and methods
Study settings and period
This study was conducted in the ALERT specialized hospital, Addis Ababa, Ethiopia, from 01 January 2021 to 30 June 2021. The hospital is one of the specialized tertiary hospitals in the country, located in the south west parts of the capital, Addis Ababa. The hospital currently provides wide range of services in various departments, including the departments of dermatology, ophthalmology, general surgery, and gynecology for patients with more than 400 beds and serves 350,000- 400,000 patients per year.
Study design and sample size determination
A health-institution-based cross-sectional study design was employed to identify pathogenic bacteria isolates and drug sensitivity pattern among CL patients attending ALERT specialized hospital, Addis Ababa, Ethiopia. Sample size was estimated by considering 50% as baseline for the sample size calculation due to difficulty of getting similar previous studies. For unknown number of population and if there is no previous similar study, the appropriate sample size calculation is assuming 50% or doing pilot study is the recommended method. Doing pilot study to get P value in resource limited area is not feasible. So, we used p as 50% and the calculated sample size by using p as 50% is well represent the study population. As a result, based on the formula for sample size (n) estimation = (Z95%) 2 p (1-p)/ (w) 2 at 95% confidence level and5% degree of accuracy [17], the estimated sample size was 384 respondents.
Study participants and data collection
The study populations were patients who were suspected of having a Lieshmania parasite infection and were referred to ALERT specialized hospital. After a dermatologist ruled out the patients’ condition as CL and patients’ skin lesion microscopic investigation revealed a positive Leishmania parasite infection patients were selected as study population. Then patients diagnosed with CL and ulcerative lesions were included in the study. All patients with ulcerated lesions of parasitologically diagnosed as CL-positive in the hospital, and willingly provided written informed consent/ assent to participate in the study during of the study period were purposely enrolled in the study. A structured questionnaire was prepared for data collection purpose after intensive literature reviews. The questionnaire contains socio-demographic characteristics and other information’s. Respondents were interviewed using a structured questionnaire to collect socio-demographics data such as gender, age, and educational status, prior leishmaniasis treatment history, and anatomical sites of infection such as arms, legs, feet, face and abdomen. With trained professional laboratory personnel, the lesion and surrounding skin were thoroughly cleaned with moistened sterile gauze and sterile normal saline solution. The indurate margins of the lesions were scraped using a flame-sterilized vaccinostyle.
The scratched area and the center of the lesion are thoroughly scraped with a sterile swab and double swab samples were collected at a time from each lesion to reduce contamination. To ensure that the same patients’ results can be repeated, it’s crucial to take two swabs from the wound. It is unlikely to be contamination if the two swabs from the same patient that were cultured showed identical bacteria growth. It might be contamination if the second swab sample culture didn’t show growth whereas the first did. The swab samples were inoculated onto blood agar plate (BAP) and Mac Conkey(MAC) agar to culture, isolate and to further identify drug susceptibility patterns of the isolated bacteria in microbiology department of the ALERT specialized hospital laboratory.
Culture and identification of bacteria
A swab samples were applied to a small area of the plate and streaked out to the rest of the surface with a sterile wire loop on BAP and MAC aseptically in a bio-safety cabinet (BSC) level II. The plates were kept in a 5% CO2 jar for BAP and in aerobic for MAC (oxoid) and incubated for 18–24 h at 37 °C. The growth organisms were preliminary identified based on colony characteristics like haemolysis on blood agar, and changes in physical appearance in differential MAC media of the organisms. Gram staining was performed to differentiate gram negative and gram positive bacteria and series of biochemical tests (oxoid) were performed on colonies from primary cultures for identification of the isolates. A series of Catalase, coagulase, bacitracin, novobiocin, optochin, pyrrolidonylarylamidase (PYRase) and Christie–Atkins–Munch-Peterson (CAMP) tests were performed for Gram-positive bacteria identification, and a series of triple sugar iron agar, indole production test, urease test, Hydrogen sulfide production test, oxidase test, Gas Glucose production, Citrate utilization test, motility test, and Lysine decarboxylase test for Gram-negative pathogenic bacteria species identifications.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was carried out using the disc diffusion method on Muller Hinton agar (oxoid) based on the 2021 CLSI guideline. The inoculums were prepared by picking parts of similar test organisms with a sterile wire loop were transferred into a tube containing sterile physiological saline and mixed gently until the suspension became turbid. The suspension was adjusted by comparison with the opacity standard on McFarland 0.5 Barium sulfate solution. The suspension was uniformly swabbed into Muller Hinton agar surface and left at room temperature for 15 min and then exposed to appropriately impregnated but diffuse radial manner antibiotic disk and incubated at 37 °C for 16–18 h. The following antibiotics were selected based on the availability and prescription frequency of these drugs in the study area. Antibiotics like doxycycline(30 µg), gentamicine(10 µg), ciprofloxacin(5 µg), penicillin(10 IU), trimethoprim-sulfamethoxazole (1.25/23.75 µg), erythromycin(15 µg) and cefoxitin (30 µg) were applied to assess the antimicrobial susceptibility pattern of Gram-positive bacteria. Using the cefoxitin (30 g) antibiotic disk, the methicillin resistance pattern of S. aureus and CoNS was identified. Methicillin-resistant S. aureus and CoNS were reported when the zone of inhibition was ≤ 21 and ≤ 24 mm, respectively, and methicillin-sensitive when the zone of inhibition was ≥ 22 and ≥ 25 mm. Similarly, antibiotics including ampicillin(10 µg), tetracycline(30 µg), cefuroxime(30 µg), meropenem(30 µg), ciprofloxacin(5 µg), amikacin(30 µg), chloramphenicol (30 µg), gentamicin(10 µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), ceftriaxone(30 µg), amoxicillin-clavulanate(30 µg) and imipenem(10 µg)were used for gram negative isolates. The antimicrobial agent immediately diffuses from the disk into the surrounding agar, where it may inhibit the growth of the microorganism. If growth is inhibited, a clear zone of “no growth” appears around the paper disk after incubation. The presence or absence of this zone serves as a measure of the microorganism’s susceptibility to the tested antimicrobial agent. The diameters of the zone of inhibition around the discs was measured to the nearest millimeter using graduated ruler to classify as sensitive, intermediate, and resistant the isolates were classified as sensitive, intermediate, and resistant to the tested drugs based on CLSI 2021 guideline [18].
Quality control
Once the necessary variables were determined and numerous related publications had been reviewed, the initial draft of the questionnaire was written in English and translated into Amharic, the national language, and then again translated back to English to ensure uniformity. The completed surveys were checked for completeness daily. Before starting each test, the quality of the culture media and functionality of the reagents were examined using QC strains of both gram positive and gram negative bacteria. BSC level II and autoclave were used to create a culture medium aseptically, and 5% of each batch was tested for sterility overnight at 37 °C. QC strain has been utilized to examine the quality of culture media, biochemical assays, and similar procedures. The quality of culture media and antimicrobial susceptibility testing was checked by using quality control standard strains of E. coli ATCC 25,922, S. aureus ATCC 25,923, E. faecalis ATCC 29,212, and K. pneumoniae ATCC 700,603, S. pyogen ATCC 19,615, S. agalactiae ATCC 12,386, P. mirabilis ATCC 35,659, P. aeruginosa ATCC 27,853.
Data analysis
Data was edited, cleaned, entered and analyzed using statistical package for social science (SPSS) version 20. Descriptive analysis such as frequencies and mean were used to express socio-demographic data, wound location and history of prior CL treatment. P-value of < 0.05 was considered to indicate statistically significant differences.
Results
Socio-demographic characteristics
A total of 384 CL patients (male, 66.9%), whose an age range from 2 to 85 years with median 26 and IQR 15 was participated in the study, with the majority, 187 (48.7%), between 16 and 30 years old. Most, 348 (90.6%), had no history of CL treatment, and 262 9(68.2%) had an education level of writing and reading or above. The sites of lesions were frequently found in decreasing order: ear, 120 (31.3%); hand, 118 (30.7%); face, 110 (28.6%); leg, 28 (7.3%); and nose, 8 (2.1%)(Table 1).
Table 1.
Socio-demographic characteristics and sites of the CL lesion of the patients
| Variables | Frequency (N) | Percentage (%) | |
|---|---|---|---|
| Sex of the patients | Male | 257 | 66.9 |
| Female | 127 | 33.1 | |
| Ages of the patients | 2–15 | 45 | 11.7 |
| 16–30 | 187 | 48.7 | |
| 31–45 | 95 | 27.7 | |
| 46–60 | 28 | 7.3 | |
| > 60 | 29 | 7.6 | |
| PriorCL treatment history | Yes | 36 | 9.4 |
| No | 348 | 90.6 | |
| Educational level | Write-read and above | 262 | 68.2 |
| Illiterate | 122 | 31.8 | |
| Sites of the lesion | Nose | 120 | 31.3 |
| Hand | 118 | 30.7 | |
| Face | 110 | 28.6 | |
| Leg | 28 | 7.3 | |
| Ear | 8 | 2.1 | |
Magnitude of bacterial etiology isolated from CL lesions
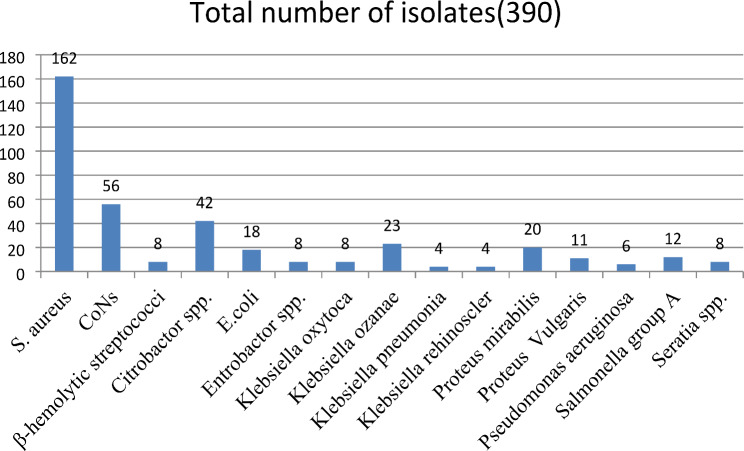
A total of 390 pathogenic bacteria were isolated from 384 CL patients’ lesions. While a single bacteria isolate was recovered from 350 (91.3%) patients, multiple bacterial isolates were isolated from 34 (8.7%) patients. Some bacteria grew with insignificant colony count and were ruled out as contaminants and excluded after re-subculture from primary inoculated media. After the isolation and identification process, a total of 390 pathogenic bacteria were further processed for drug susceptibility patterns. Most frequently, the isolates were isolated from the lesions found on the nose 125 (32.1%), hand 122 (31.3%), and face 108 (27.7%). Similarly, the isolated bacteria were isolated most frequently from patients whose ages ranged from 16 to 30, 178 (45.6%), followed by the 31–45, 102 (26.1%)-year-old age group. The majority of the isolates were gram-positive (226; 57.9%) versus gram-negative (164; 42.1%). Most of the gram-positives were isolated from noses (78, 34.5%), hands (67, 29.6%), and faces (62, 27.4%), and the gram-negatives were isolated from noses (47, 28.7%), hands (55, 33.5%), and faces (46, 28.0%). From the total isolates, the majority were Staphylococci spp. (218; 55.9%), particularly S. aureus and CoNS, which accounted for 162 (41.3%) and 56 (14.4%) of all isolates, respectively. Gram-negative organisms containing Citrobacter spp. 42 (10.8%), Klebsiella spp. (K. oxytoca, K. rhinoscleromatis, K. pneumoniae, and K. ozanae) 39 (9.9%), Proteus spp. (P. mirabilis and P. vulgaris) 22 (7.9%), and E. coli 18 (4.6%) were significantly isolated from lesions of study participants. Most of S. aureus 75 (46.3%) and CoNS 44 (78.6%) were isolated from lesions in patients whose ages ranged from 16 to 30. Most frequently, Gram-negatives were isolated among age groups of 31–45 (37.8%) and 16–30 (33.5%) (Table 2; Fig. 1).
Table 2.
Distribution of the isolates in ages and locations of the CL lesion of the participants
| Isolates | Anatomical location of the lesion from bacteria isolated (100%) | Ages of study participants (100%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ear | Face | Hand | Leg | Nose | 2–15 | 16–30 | 31–45 | 46–60 | > 60 | ||
| GP | S. aureus | 8(4.9) | 46(28.4) | 51(31.5) | 7(4.3) | 50(30.9) | 29(17.9) | 75(46.3) | 34(21.3) | 15(9.3) | 9 (5.6) |
| CoNs | - | 12(21.4) | 12(21.4) | 4(7.1) | 28(50.0) | - | 44(78.6) | 6 (10.7) | 2 (3.6) | 4 (7.1) | |
| β-hemolytic streptococci | - | 4(4.0) | 4(4.0) | - | - | - | 4 (50.0) | - | - | 4(50.0) | |
| Total GP | 8(3.5) | 62(27.4) | 67(29.6) | 11(4.9) | 78(34.5) | 29(12.8) | 123(54.4) | 40(17.7) | 17(7.5) | 17(7.5) | |
| GN | Citrobactor spp. | - | 6(14.3) | 20(47.6) | - | 16(38.1) | 8 (19.0) | 5(11.9) | 15(35.7) | 5(11.9) | 9(21.4) |
| E.coli | - | 8(44.4) | 4(22.2) | - | 6(33.3) | 2 (11.1) | - | 16(88.9) | - | - | |
| Entrobactor spp. | - | - | - | 4(50.0) | 4(50.0) | - | - | 7(87.5) | 1(12.5) | - | |
| K. oxytoca | - | - | 4(50.0) | 4(50.0) | - | - | 4 (50.0) | 4(50.0) | - | - | |
| K. ozanae | - | 8(34.8) | 7(30.4) | - | 8(34.8) | 4 (17.4) | 9(39.1) | 1(4.3) | 1(4.3) | 8(34.8) | |
| K. pneumonia | - | 4(100) | - | - | - | - | 4(100) | - | - | - | |
| K. rehinoscler | - | 4(100) | - | - | - | - | - | - | - | 4(100) | |
| P. mirabilis | - | 8(40.0) | 8(40.0) | 4(20.0) | - | - | 12(60.0) | 8(40.0) | - | - | |
| P. vulgaris | - | - | - | 4(36.4) | 7(63.6) | - | 7(63.6) | 3(27.3) | 1(9.1) | - | |
| P. aeruginosa | - | - | 4(66.7) | - | 2(33.3) | - | 2(33.3) | 4(66.7) | - | - | |
| Salmonella group A | - | 8(66.7) | - | - | 4(33.3) | - | 4(33.3) | 4(3.3) | 4(33.3) | - | |
| Seratia spp. | - | - | 8(100) | - | - | - | 8(100) | - | - | - | |
| Total GN | 46(28.0) | 55(33.5) | 16(9.8) | 47(28.7) | 14(8.5) | 55(33.5) | 62(37.8) | 12(7.3 | 21(12.8 | ||
| Total isolates | 8(2.1) | 108(27.7) | 122(31.3) | 28(7.2) | 125(32.1 | 43(11.0) | 178(45.6) | 102(26.1) | 28(7.2) | 38(9.7) | |
Fig. 1.
Etiology of bacterial isolated from swab samples collected from patients with CL infected lesions
Drug susceptibility patterns of gram positive bacteria
In this study, 226 gram-positive bacteria isolates were subjected to antimicrobial susceptibility pattern testing against eight antimicrobial agents. Among the isolates, the ranges of resistance to penicillin and methicillin were 50–86% and 50–83%, respectively. On the other hand, low levels of resistance were reported for doxycycline (8.4%), gentamycine (11.0%), trimethoprim sulfamethoxazole (21.2%), ciprofloxacin (19.9%), and erythromycin (35.4%). S. aureus showed high resistance to penicillin (86.4%), methicillin (83.9%), and tetracycline (47.5%). The most active antibiotics against S. aureus were doxycycline and gentamycin, each with 92.0% susceptibility, followed by trimethoprim-sulfamethoxazole (88.2%), ciprofloxacin (77.1%), and erythromycin (65.4%)(Table 3).
Table 3.
Antimicrobial susceptibility pattern of Gram- positive bacteria isolated from CL lesion
| Isolates (total number) | Antimicrobial agents (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| DA | GEN | SXT | CIP | TET | PEN | E | MET | ||
| S. aureus (162) | S | 149(92.0) | 149(92.0) | 143(88.2) | 125(77.1) | 84(51.9) | 21(12.9) | 106(65.4) | 25(15.4) |
| I | - | - | 3(1.9) | 1(0.6) | 1(0.6) | 1(0.6) | 1(0.6) | 1(0.6) | |
| R | 13(8.0) | 13 (8.0) | 16(9.9) | 36(22.2) | 77(47.5) | 140(86.4) | 55(33.9) | 136(83.9) | |
| CoNs (56) | S | 50(89.3) | 48 (85.5) | 28(50) | 47(83.9) | 20(35.7) | 9(16.0) | 31(55.4) | 16(28.6) |
| I | - | - | - | - | - | - | - | 2(3.6) | |
| R | 6(10.7) | 8(14.5) | 28(50) | 9(16.1) | 36(64.2) | 47(83.9) | 25(44.6) | 38(67.8) | |
| β-hem. streptococci (8) | S | 8(100) | 4 (50.0) | 4(50) | 8(100) | 8(100) | 4(50.0) | 8(100) | 4(50.0) |
| I | - | - | - | - | - | - | - | - | |
| R | - | 4 (50.0) | 4(50) | - | - | 4(50.0) | - | 4(50.0) | |
| Total (226) | S | 207(91.6) | 201(89.0) | 175(77.4) | 180(79.6) | 112(49.6) | 34(15) | 145(64.1) | 45(19.9) |
| I | - | - | 3(1.3) | 1(0.4) | 1(0.4) | 1(0.4) | 1(0.4) | 3(1.3) | |
| R | 19(8.4) | 25(11.0) | 48(21.2) | 45(19.9) | 113(50.0) | 191(84.5) | 80(35.4) | 178(78.8) | |
DA: doxycycline GEN: gentamycine SXT: trimethoprim-sulphamethoxazole CIP: ciprofloxacin TET: tetracycline PEN: penicillin E: erythromycin MET: methicillin CoNs: coagulase negative staphylococci S: Sensitive I: Intermediate R: Resistance
Antimicrobial susceptibilities of Gram-negative bacteria
All 164 gram-negative bacteria isolates were subjected to susceptibility tests against 12 antimicrobial agents. The isolates showed high drug resistance levels to cefuroxime (81.1%), ampicillin (70.3%), tetracycline (59.8%), and ceftriaxone (59.1%). Fortunately, the majority of gram-negative bacteria were susceptible to Amikacin (90.3%), Meropenem (87.2%), Imepinem (74.4%), Chloramphenicol (68.3%), Ciprofloxacin (63.4%), Gentamycin (63.4%), and Augmentin (55.5%), respectively. Citrobacter spp. was the predominant gram-negative bacteria found fully sensitive to amikacin (100%). However, a significant resistance rate was recorded to cefuroxime (97.6%), augmentine (64.3%), trimethoprim-sulfametoxazole (53.4%), ceftriaxone(52.4%), ciprofloxacillin(47.6%) and tetracycline (45.2%)(Table 4).
Table 4.
Antimicrobial susceptibility patterns of gram-negative bacterial isolates
| Isolates (N) | Antimicrobials agents No. (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | TE | CRX | MEM | CIP | AK | C | GEN | SXT | CRO | AMC | IMP | ||
|
Citrobactor spp. (42) |
S | 36(85.7) | 23(54.8) | 1(2.4) | 38(90.5) | 22(52.4) | 42(100) | 32(76.2) | 28(66.7) | 20(47.6) | 16(38.1) | 15(35.7) | 37(88.1) |
| I | - | - | - | - | - | - | - | - | - | 4(9.5) | - | 1(2.4) | |
| R | 6(14.3) | 19(45.2) | 41(97.6) | 4(9.5) | 20(47.6) | - | 10(23.8) | 14(33.3) | 22(53.4) | 22(52.4) | 27(64.3) | 4(9.4) | |
|
K. ozanae (23) |
S | - | 12(52.2) | 5(21.7) | 23(100) | 15(65.2) | 19(82.6) | 23(100) | 23(100) | 23(100) | 18(78.3) | 17(73.9) | 18(78.3) |
| I | 4(17.4) | - | 4(17.4) | - | 4(17.4) | - | - | - | - | - | - | 1(4.3) | |
| R | 19(82.6) | 11(47.8) | 14(60.9) | - | 4(17.4) | 4(17.4) | - | - | - | 5(21.7) | 6(26.1) | 4(17.4) | |
|
P. mirabilis (20) |
S | - | - | - | 20(100) | 4(20.0) | 12(60.0) | 4(20.0) | 8(40.0) | 4(20.0) | - | 12(60.0) | 12(60) |
| I | - | - | - | - | - | 4(20.0) | - | - | - | - | 4((20.0) | 4(20.0) | |
| R | 20(100) | 20(100) | 20(100) | - | 16(80.0) | 4(20.0) | 16(80.0) | 12(60.0) | 16(80.0) | 20(100) | 4(20.0) | 4(20) | |
|
E.coli (18) |
S | - | 2(11.1) | 2(11.1) | 18(100) | 18(100) | 18(100) | 18(100) | 18(100) | 6(33.3) | 10(55.6 | 18(100 | 14(77.8) |
| I | - | - | - | - | - | - | - | - | - | - | - | - | |
| R | 18(100) | 16(88.9) | 16(88.9) | - | - | - | - | - | 12(66.70 | 8(44.4) | - | 4(22.2) | |
|
S. group A (12) |
S | - | 4(33.3) | - | 8(66.7) | 12(100) | 8(66.7) | 4(33.3) | 8(66.7) | 12(100 | 4(33.3) | 4(33.3) | 12(100 |
| I | - | 4(33.3) | - | - | - | - | 4(33.3) | - | - | 4(33.3) | 4(33.3) | - | |
| R | 12(100) | 4(33.3) | 12(100) | 4(33.3) | - | 4(33.4) | 4(33.3) | 4(33.3) | - | 4(33.3) | 4(33.3) | - | |
|
P.vulgaris (11) |
S | - | 7(63.6) | 0 | 11(100) | 7(63.6) | 11(100) | 7(63.6) | 7(63.6 | 7(63.6) | 7(63.6) | 11(100 | 7(63.6) |
| I | - | - | 4(36.4) | - | - | - | - | - | - | - | - | 4(36.4) | |
| R | 11(100) | 4(36.4) | 7(63.4) | - | 4(36.4) | - | 4(36.4) | 4(36.4) | 4(36.4 | 4(36.4) | - | - | |
|
K.oxytoca (8) |
S | - | - | - | 8(100) | 4(50.0) | 8(100) | 8(100) | 4(50.0) | 0 | - | 8(100) | 8(100) |
| I | - | - | - | - | - | - | - | - | - | - | - | - | |
| R | 8(100) | 8(100) | 8(100) | - | 4(50.0) | - | - | 4(50.0) | 8(100) | 8(100) | - | - | |
|
Seratia spp. (8) |
S | - | - | - | - | 4(50.0) | 4(50.0) | 4(50.0) | - | - | - | - | - |
| I | - | - | - | - | - | - | - | - | - | - | - | 4(50.0) | |
| R | 8(100) | 8(100) | 8(100) | 8(100) | 4(50.0) | 4(50.0) | 4(50.0) | 8(100) | 8(100) | 8(100) | 8(100) | 4(50.0) | |
|
*Other (22) |
S | 7(31.8) | 10(45.5) | 15(68.2) | 17(77.3) | 18(81.8 | 22(100) | 14(63.6 | 8(36.4) | 11(50.0 | 10(45.4) | 6(27.3) | 14(63.4) |
| I | 1(4.5) | 2(9.0) | - | 1(4.5) | - | - | 1(4.5) | 4(18.2) | 2(9.1) | 4(18.2) | 2(9.1) | 2(9.1) | |
| R | 14(63.6) | 8(36.4) | 7(31.8) | 4(18.2) | 4(18.2) | - | 7(31.8) | 10(45.4) | 8(36.4) | 8(36.4) | 14(63.4) | 6(27.3) | |
|
Total (164) |
S | 43(26.2) | 60(36.6) | 23(14.0) | 143(87.2) | 104(63.4) | 148(90.3 | 112(68.3) | 104(63.4 | 83(50.6) | 63(38.4) | 91(55.5) | 122(74.4) |
| I | 5(3.0) | 6(3.7) | 8(4.9) | 1(0.6) | 4(2.4) | 4((2.4) | 5(3.0) | 4(2.4) | 2(1.2) | 4(2.4) | 10(6.0) | 16(9.8) | |
| R | 116(70.3) | 98(59.8) | 133(81.1 | 20(12.2) | 56(34.2) | 12(7.3) | 45(27.4) | 56(34.1) | 79(48.2) | 97(59.1) | 63(38.4) | 26(15.8) | |
AMP: Ampicillin TE: tetracycline CRX: cefuroxime MEM: meropenem CIP: ciprofloxacillin AK: amikacin C: chloraphinicol GEN: gentamycine SXT: trimethoprim-sulphametoxazole CRO: ceftriaxone AMC: augmentine IMP: imipenem S: Sensitive I: Intermediate R: Resistance, *Enterobacter, spp., P.auruginosa, K. pneumoniae, and K.rhinoscleromatis
Discussion
Bacterial secondary infections on CL lesions are likely to be substantial difficulties in the care of CL patients [19, 20]. It hastens the beginning of the illness and delays recovery by increasing tissue damage and necrosis [15]. The purpose of this study was to isolate pathogenic bacteria from an infected lesion and assess their antimicrobial sensitivity pattern to antimicrobial medications.
In the current study, 390 pathogenic bacteria were recovered from 384 CL patient lesions, 350 (91.3%) CL patient lesions had mono pathogenic bacterium, and 34 (8.7%) patients had poly- bacteria growth. Some of the isolates with insignificant colony and ruled out as contaminant were not farther identified as pathogenic. Bacteria were most commonly isolated from lesions on the nose (32.1%), hand (31.3%), and face (27.7%), which contradicts a study reported by Fontes et al., 2023 [9] that indicated most isolates of microorganisms from the legs (47.6%), arms (19.0%), and face (14.3%). This discordance may differ from one location to the next, depending on the patient’s lifestyle and the body portions exposed to the environment, as well as their economic condition.
Of the isolates that were confirmed by culture, gram-positive bacteria accounted for 57.9% and gram-negative bacteria for 42.1%. From all isolates, S. aureus accounted for 41.7% and CONS for 24.8% of isolates, whereas from the gram-positive bacteria isolates, S. aureus accounted for 71.7% and CONS accounted for 14.4%. The outcome agrees with several related investigations that have been published elsewhere [9, 21–23]. Because pathogenic cocci can exist as commensalism on the skin [24], gram-positive cocci were the most frequently identified bacteria in our investigation. On the other hand Citrobacter spp. (10.8%) and Klebsiella spp. (9.9%) were the most prevalent gram-negative bacteria isolates in this investigation. The finding agrees with a previous similar study that found Proteus spp. and Klebsiella spp. to be the most common gram-negative etiological agents in CL lesions in Brazil [9], but this study disagrees with a study that reported E. coli to be the most common gram-negative isolate in India [21]. This could be due to infection prevention policies and level of health education of the countries. Lesion of CL is known to heal very slowly which exposes the lesion to microorganisms. It’s possible that germs that cause subsequent wound infections are resistant to antibiotics. Now a day, antibiotics are prescribed by any medical professionals and individuals in Ethiopia. Drug resistance is the result of this abuse of antibiotics. The majority of hospitals in Ethiopia lack a microbiology laboratory that can perform tests to determine antibiotic susceptibility and detect bacterial infections. The lesion caused by CL, which was highly contaminated with several bacteria, can result in serious sickness and require long time treatment.
All isolated bacteria were challenged for antimicrobial susceptibility patterns to common antimicrobial agents. Tetracycline, erythromycin, chloramphenicol, gentamicin, ciprofloxacin, Trimethoprim-sulphamethoxazole, penicillin, ceftriaxone, norflaxocin, and amoxicillin were the most frequently used antibiotics in this study area. However, in the current study finding revealed that several isolates showed a significant level of resistance to various antimicrobial drugs. S. aureus showed a high multi drug resistance patterns (MDR) against penicillin (86.4%) and methicillin (83.9%) resistance. These finding is agreed with the resent studyreported,84.6% resistance to methicillin and 92.3% resistant to ampicillin in Ghana [25]. The findings are an assurance for the emerging an antibiotic-resistant pathogen, a well-recognizing public health challenge throughout the world [26]. S. aureus showed high susceptibility against doxycycline and gentamycin (each 92.0%), and against trimethoprim-sulfamethoxazole (88.2%). The finding is in line with the study reported 100% for gentamicin and 92.3% for trimethoprim-sulfamethoxazole susceptibilities in Ghana [25]. Cefuroxime resistance was found among 81.1% of gram-negative bacteria. It is consistent with recent studies that show significant rates of ampicillin and penicillin resistance [19, 27]. Bacteria isolates which showed high rate of antibiotics resistance can cause life threaten disease and long duration morbidity. The isolates demonstrated intermediate levels of resistance (50–80%) to ampicillin, tetracycline, and ceftriaxone, as well as low levels of resistance (< 50%) to meropenem, ciprofloxacin, amikacin, chloramphenicol, gentamycin, trimethoprim-sulfamethoxazole, augumentin, and imepinem, and these findings were supported by different studies [9, 25]. The prevalence of MDR with two or more routinely prescribed antimicrobials was found in more than 80% of the isolates. The high frequency of MDR might be attributed to the incoherent and inappropriate use of antimicrobial drugs in this study area, which as reasoned out previously somewhere, results in the formation of multidrug-resistant bacterial strains [28]. Based on our findings, most of the bacteria isolates were resistant to commonly prescribed antibiotics for patients diagnosed with bacterial infection. Gentamycin and trimethoprim-sulfamethoxazole were the commonly prescribed antibiotics with less resistant pattern for both gram positive and gram negative bacteria.
There are several limitations to this study. Because of the cross-sectional study design of this study, the influence of period fluctuations was not addressed. Furthermore, to supplement the phenotypic drug resistance approaches, genotypic characterization of antibiotic resistance and virulence genes might have supplied further information on the genetic profile of the isolates. In certain cases, fewer isolates were examined for the existence of drug resistance to specific antibiotics, which hampered generalization.
Conclusion
The research area has a high incidence of bacterial isolates in CL lesion, with the majority of gram-positive bacteria, S. aureus and CoNS were the most common isolates. Most of bacterial isolates tested positive for antibiotic resistance. As a result, before prescribing any antimicrobials, a drug susceptibility test is required. For the treatment of infected individuals with bacteria found in this finding, doxycycline and gentamicin for gram-positive isolates and meropenem and amikacin for gram-negative isolates can be effective drugs. Further studies with large sample sizes, covering other possibly associated factors and community practice, should be done. Factors like antibiotics usage history, sources of infection, knowledge on how to prevent infections, genetic makeup of the bacteria and the like should be done in large scale study group.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank study participants, ALERT hospital staff members, and Addis Ababa University.
Author contributions
All the authors contribute equally . GA, AA, MW, RD and BZ, all designed the research, wrote proposal, collect data, analyzed data, interprated data, drafted manuscript, read and agreed on the manuscript submission.
Funding
The study was financially supported by Addis Ababa University (AAU), Ethiopia. The funding body had no influence on the study at all points (i.e., during design, data collection, data analysis, interpretation, or preparation of manuscript).
Data availability
We used simple data. All data we used was summarized and included in this this manuscript.
Declarations
Ethical approval
of the study was obtained from the Department of Medical Laboratory Science, College of Health Sciences, Addis Ababa University Research Ethical Review Committee (DRERC/468/20/MLS). Permission for study work was obtained from the Addis Ababa City Administration Health Office and ALERT Hospital. Written informed consent was obtained from each of the study participants and their parents or guardians. Participants’ information sheet, which contains the objective of the study, inclusion/exclusion criteria, the required data, and methods of data collection as well as informed consent/assent document, were prepared in Amharic, the national language. The elements of the participants’ information sheet were described to each of the study participants or parents in the case of children under 18 years by trained local health personnel. Informed written consent was obtained from each participant or assent from children aged between 12 and 18. The Participant’s laboratory test results were maintained confidentially for the duration of the study.
Consent for publication
Not applicable for our manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO. Leishmaniasis. Fact sheet [Internet]. World Health Organization. 2023 [cited 2023 Jan 30]. Available from: https://www.who.int/news-room/fact-sheets/detail/leishmaniasis
- 2.Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis. 2007;7(9):581–96. [DOI] [PubMed] [Google Scholar]
- 3.Mokni M. Leishmanioses cutanées. Annales de Dermatologie et de Vénéréologie. 2019;146(3):232–46. [DOI] [PubMed]
- 4.Scott P, Novais FO. Cutaneous leishmaniasis: immune responses in protection and pathogenesis. Nat Rev Immunol. 2016;16(9):581–92. [DOI] [PubMed] [Google Scholar]
- 5.Azeredo-Coutinho RBG, Mendonça SCF, Callahan H, Portal AC, Grögl M. Sensitivity of leishmania braziliensis promastigotes to meglumine antimoniate (glucantime) is higher than that of other leishmania species and correlates with response to therapy in American tegumentary leishmaniasis. J Parasitol. 2007;93(3):688–93. [DOI] [PubMed] [Google Scholar]
- 6.Salman S. Cutaneous leishmaniasis: clinical features and diagnosis. Clin Dermatol. 1999;17(3):291–6. [DOI] [PubMed] [Google Scholar]
- 7.Gonçalves E da G do R, Reis Filho SA dos, de Oliveira EG, Pareira ALN, da Silva AR, Costa JML. Infecção bacteriana na leishmaniose cutânea: padrão bacteriano e sensibilidade a antibióticos. Rev Soc Bras Med Trop. 2009;42(2):219–21. t. [DOI] [PubMed]
- 8.Vera LA, de Macedo JLS, Ciuffo IA, Santos CG, Santos JB. Sensibilidade antimicrobiana de bactérias aeróbicas isoladas de úlceras leishmanióticas, em Corte De Pedra, BA. Rev Soc Bras Med Trop. 2006;39(1):47–50. [DOI] [PubMed] [Google Scholar]
- 9.Fontes CO, Carvalho MAR, Nicoli JR, Hamdan JS, Mayrink W, Genaro O, et al. Identification and antimicrobial susceptibility of micro-organisms recovered from cutaneous lesions of human American tegumentary leishmaniasis in Minas Gerais, Brazil. J Med Microbiol. 2005;54(11):1071–6. [DOI] [PubMed] [Google Scholar]
- 10.Miró EM, Sánchez NP. Cutaneous manifestations of Infectious diseases. In: Sánchez NP, editor. Atlas Dermatol Int Med. New York, NY: Springer New York; 2012. pp. 77–119. [Google Scholar]
- 11.Sadeghian G, Ziaei H, Bidabadi LS, Baghbaderani AZ. Decreased effect of glucantime in cutaneous leishmaniasis complicated with secondary bacterial infection. Indian J Dermatol. 2011;56(1):37–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonio L, de Lyra F, Saheki MR, Schubach MN, de O A, Miranda L, de FC, Madeira M. Effect of secondary infection on epithelialisation and total healing of cutaneous leishmaniasis lesions. Mem Inst Oswaldo Cruz. 2017;112(9):640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isaac-Márquez AP, Lezama-Dávila CM. Detection of pathogenic bacteria in skin lesions of patients with chiclero’s ulcer: reluctant response to antimonial treatment. Mem Inst Oswaldo Cruz., Frimpong E, Donkor ES, Opintan JA, Asamoah-Adu A. Resistance to antimicrobial drugs in Ghana. Infect Drug Resist. 2011;4:215–20.
- 14.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277–83. [PMC free article] [PubMed] [Google Scholar]
- 15.Tadesse BT, Ashley EA, Ongarello S, Havumaki J, Wijegoonewardena M, González IJ, et al. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 2017;17(1):616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniel W. The Fisher exact test. Daniel WW Biostatistics: a foundation for analysis in the health sciences. 7th ed. New York: Wiley; 1999. pp. 606–11. [Google Scholar]
- 17.Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014 Dec;13(1):14. [DOI] [PMC free article] [PubMed]
- 18.Salgado VR, de Queiroz ATL, Sanabani SS, de Oliveira CI, Carvalho EM, Costa JML, et al. The microbiological signature of human cutaneous leishmaniasis lesions exhibits restricted bacterial diversity compared to healthy skin. Mem Inst Oswaldo Cruz. 2016;111(4):241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortes PR, Chiapello LS, Dib D, Herrero MV, Nuncira CT, De Petris C, et al. Coinfection of Leishmania (Viannia) braziliensis and Streptococcus pneumoniae in multiple cutaneous lesions. PLoS Negl Trop Dis. 2016;10(3):e0004388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziaei H, Sadeghian G, Hejazi SH. Distribution frequency of pathogenic bacteria isolated from cutaneus leishmaniasis lesions. Korean J Parasitol. 2008;46(3):191–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziaie H, Sadeghian G. Isolation of bacteria causing secondary bacterial infection in the lesions of cutaneous leishmaniasis. Indian J Dermatol. 2008;53(3):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Layegh P, Ghazvini K, Moghiman T, Hadian F, Zabolinejad N, Pezeshkpour F. Bacterial contamination in cutaneous leishmaniasis: its effect on the lesions’ healing course. Indian J Dermatol. 2015;60(2):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haile Z, Mengist HM, Dilnessa T. Bacterial isolates, their antimicrobial susceptibility pattern, and associated factors of external ocular infections among patients attending eye clinic at Debre Markos Comprehensive Specialized Hospital, Northwest Ethiopia. PLoS ONE. 2022;17(11):e0277230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeboaa C, Odoi H, Owusu Ntim R, Boakye YD, Kwakye-Nuako G, Agyare C, et al. Diversity and antibiograms of bacteria isolated from cutaneous leishmaniasis wounds in the Nkwanta South District of Ghana. Arch Microbiol. 2023;205(2):74. [DOI] [PubMed] [Google Scholar]
- 25.Zhou N, Cheng Z, Zhang X, Lv C, Guo C, Liu H, et al. Global antimicrobial resistance: a system-wide comprehensive investigation using the Global One Health Index. Infect Dis Poverty. 2022;11(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alkulaibi MM, Suleiman AM, Gasim Khalil EA, Al-Garadi MA. Prevalence of cutaneous leishmaniasis in Western Highlands in Yemen. J Trop Medic. 2019;2019:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolday D, Erge W. Increased incidence of resistance to antimicrobials by urinary pathogens isolated at Tikur Anbessa Hospital. Ethiop Med J. 1997;35(2):127–35. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We used simple data. All data we used was summarized and included in this this manuscript.