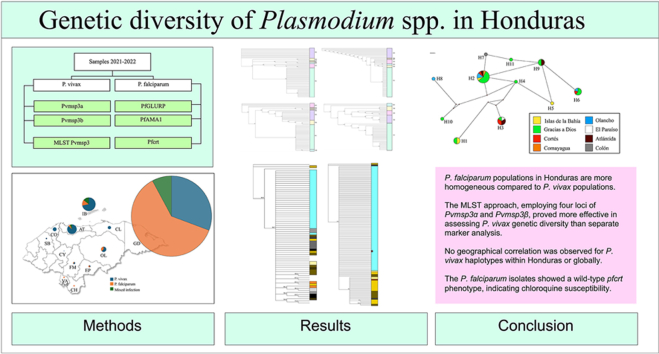
Abstract
Malaria continues to be a major threat to public health in tropical regions, primarily affecting sub-Saharan Africa but also Asia, the Middle East, and Latin America. Malaria cases in Honduras have seen a significant decline and the country aims to eliminate the disease by 2030. This study examines the genetic diversity of Plasmodium falciparum and Plasmodium vivax in Honduras using four molecular markers (Pfama1, Pfglurp, Pvmsp3α, and Pvmsp3β), and the chloroquine resistance marker pfcrt in the context of the elimination phase. Our findings indicate that P. falciparum populations in Honduras are more homogeneous compared to P. vivax. The multilocus sequence typing (MLST) approach, using four loci from Pvmsp3α and Pvmsp3β, proved more effective in assessing the genetic diversity of P. vivax than individual marker analyses. No geographical clustering was observed for P. vivax haplotypes, either within Honduras or globally. In Honduras, P. falciparum appears to be under more effective control, while P. vivax presents a greater challenge due to its higher genetic diversity. This requires enhanced surveillance, targeted control strategies, and measures to prevent the reintroduction of variants. The isolates of P. falciparum also displayed a wild-type Pfcrt phenotype, suggesting susceptibility to chloroquine.
Keywords: Plasmodium vivax, Plasmodium falciparum, Genetic diversity, Honduras
Graphical abstract
Highlights
-
•
Our findings suggest that P. falciparum populations in Honduras are more homogeneous compared to P. vivax populations.
-
•
An MLST approach using four Pvmsp3α and Pvmsp3β loci better assessed P. vivax diversity than separate markers.
-
•
No geographical clustering was observed for P. vivax haplotypes within Honduras or globally.
-
•
P. falciparum isolates showed a wild-type pfcrt phenotype, indicating chloroquine susceptibility.
1. Introduction
Malaria is a significant global health concern, causing 249 million cases and 608,000 deaths in 2022, particularly among young children in sub-Saharan Africa (WHO, 2023). Malaria exerts an important socioeconomic burden on affected countries leading to decreased productivity as a result of sickness and mortality, elevated healthcare expenses, and hindered economic progress perpetuating a cycle of poverty in endemic regions (Sarma et al., 2019). Although sub-Saharan Africa suffers the majority of malaria cases and deaths, the disease is also prevalent in Latin America and other tropical regions (WHO, 2023).
Continued efforts to fight malaria have resulted in remarkable advances in global health programmes. Honduras, together with other countries in Central America and the island of Hispaniola, has announced a commitment to eliminate malaria by the year 2030 (WHO, 2015a). According to National reports (Secretaría de Salud de Honduras, 2024), the occurrence of malaria in Honduras has dropped by a factor of 15 between 2000 and 2023. As of epidemiological week 25 of 2024, Honduras has registered 1093 indigenous and 49 imported cases (Secretaría de Salud de Honduras, 2024). Less than 10% of malaria cases are attributed to Plasmodium falciparum malaria, whereas the remaining 90% are caused by Plasmodium vivax. Furthermore, 90% of the cases are concentrated in the Department Gracias a Dios.
While progress has been made in reducing the malaria burden, several challenges remain to be addressed, including tackling drug and insecticide resistance (Hancock et al., 2024; Schafer et al., 2024), detecting asymptomatic reservoirs (Matamoros et al., 2023), reaching remote or conflict-affected regions (Mertens, 2024), and developing effective vaccines (Laurenson and Laurens, 2024). Several obstacles hinder the advancement of malaria vaccine development including the complex parasite life cycle, its ability to evade the immune system, the complex host immunological response, and the parasites’ antigenic diversity (Stanisic et al., 2013).
Several molecular strategies are commonly used to evaluate the genetic diversity and population structure of Plasmodium spp., such as microsatellites (Larranaga et al., 2013; Li et al., 2023a) and whole genome sequencing (Kattenberg et al., 2023). Additional genes are used to explore genetic diversity, particularly in the context of vaccine development. These genes exhibit a high level of polymorphism, such as the merozoite surface proteins (MSPs) (Huang et al., 2018), glutamate-rich protein (PfGLURP) (Paul et al., 2024), circumsporozoite proteins (CSP) and apical membrane antigens (AMA1) (Kusi et al., 2024). These markers are also essential tools in the study of malaria epidemiology, contributing to our understanding of parasite biology, transmission patterns, and the development of control strategies. Four molecular markers are used in this study to evaluate the genetic diversity of the two species of Plasmodium (Pfama1, Pfglurp, Pvmsp3α, and Pvmsp3β). These loci encode proteins that are important for the interaction between the parasite and the human immune system. These genes are widely used to gain insight into the variability in parasite antigens that could affect the immune response and the effectiveness of potential vaccines.
The current epidemiological characteristics of malaria in Honduras require an assessment of the genetic diversity of Plasmodium spp. This study aimed to assess the effectiveness of four molecular markers (Pfama1, Pfglurp, Pvmsp3α, and Pvmsp3β), applied for the first time in Honduras, to analyze the genetic diversity of the two species of Plasmodium circulating in the country.
2. Materials and methods
2.1. Sample collection, malaria diagnosis, and DNA extraction
Blood samples were obtained from febrile patients who sought medical care in health establishments in 12 departments of Honduras (Atlántida, Choluteca, Comayagua, Colón, Cortés, El Paraíso, Francisco Morazán, Gracias a Dios, Islas de la Bahía, Olancho, Santa Bárbara, and Valle) between 2020 and 2022. The patients were diagnosed with malaria using either thick smear microscopy or rapid diagnostic tests (RDTs). Simultaneously, the Ministry of Health collected a drop of blood on Whatman® FTA filter paper (Merck, Darmstadt, Germany) for molecular analyses. These analyses involved studying parasite genes associated with susceptibility to antimalarial drugs and served as part of a routine diagnostic quality control system.
The blood samples on filter paper were placed in separate sealed plastic bags with desiccant and then sent to the National Malaria Surveillance Laboratory in Tegucigalpa. Two circles (2 mm in diameter) were cut from paper impregnated with blood for DNA extraction. Genomic DNA was isolated using the Extracta® DNA Prep for PCR kit (QuantaBio, Beverly, MA, USA) following the manufacturer’s instructions and kept at −20 °C for further analysis.
2.2. PCR amplification and sequencing
2.2.1. Molecular diagnosis of malaria
Each sample was amplified in duplicate using specific primers for the genus Plasmodium by PET-PCR (Lucchi et al., 2013, 2014; Matamoros et al., 2023). Positive specimens were subjected to amplification using two separate pairs of primers targeting the two parasite species found in Honduras, P. falciparum and P. vivax.
Amplification reactions were carried out in a volume of 20 μl containing 10 μl Go Taq® Probe qPCR master mix (Promega Corp. Madison, WI, USA), 0.5 μl of each primer (10 μM) (Table 1), 4 μl of nuclease-free water, and 5 μl of DNA (∼40 ng/μl). Reactions were run on a Mic qPCR Cycler (Bio Molecular Systems, Brisbane, Australia) and the results were visualized in the Mic qPCR Cycler Software v.2.10.1. The amplification conditions for the diagnosis of genus and species were 95 °C for 15 min, 45 cycles at 95 °C for 20 s, 63 °C for 40 s, and 72 °C for 30 s. The fluorescence channel was selected for each labelled primer (6FAM for genus and HEX for species). Each experiment comprised both positive and negative controls. A cycle threshold (Ct) of 42 or below was used to consider samples as positive.
Table 1.
List of primers used for the amplification of five molecular markers of Plasmodium vivax and P. falciparum.
| Target gene | Reaction | Primer name | Primer sequence (5′-3′) | Reference |
|---|---|---|---|---|
| 18S rRNA gene | PET-PCR for Plasmodium spp. | Genus forward | GGCCTAACATGGCTATGACG | Lucchi et al. (2013); Matamoros et al. (2023) |
| Genus reverse | 6FAM-aggcgcatagcgcctggCTGCCTTCCTTAGATGTGGTAGCT | |||
| 18S rRNA gene | PET-PCR for P. falciparum | Falciparum forward | ACCCCTCGCCTGGTGTTTTT | Lucchi et al. (2013) |
| Falciparum reverse | HEX-aggcgcatagcgcctggTCGGGCCCCAAAAATAGGAA | |||
| 18S rRNA gene | PET-PCR for P. vivax | Vivax forward | ACTGACACTGATGATTTAGAACCCATTT | Kudyba et al. (2019) |
| Vivax reverse | HEX- aggcgcatagcgcctggTGGAGAGATCTTTCCATCCTAAACCT | |||
| Pfcrt | 1st round | AL6821 | AGCAAAAATGACGAGCGTTATAG | Griffing et al. (2010) |
| AL6822 | ATTGGTAGGTGGAATAGATTCTC | |||
| 2nd round | AL5631 | TTTTTCCCTTGTCGACCTTAAC | ||
| AL5632 | AGGAATAAACAATAAAGAACATAATCATAC | |||
| Pfama1 | 1st round | Pfama1F | GTACTTGTTATAAATTGTACA | Kang et al. (2018) |
| Pfama1R | TTTTAGCATAAAAGAGAAGC | |||
| 2nd round | Pfama1F1 | ACAAAAATGAGAAAATTATACTGC | ||
| Pfama1R1 | TTAATAGTATGGTTTTTCCATCAGAAC | |||
| Pfglurp | 1st round | GLURPOF | TGAATTTGAAGATGTTCACACTGAAC | Sathishkumar et al. (2022) |
| GLURPOR | GTGGAATTGCTTTTTCTTCAACACTAA | |||
| 2nd round | GLURPGNF | TGTTCACACTGAACAATTAGATTTAGATCA | ||
| GLURPOR | GTGGAATTGCTTTTTCTTCAACACTAA | |||
| Pvmsp3α | 1st round | Pvmsp3a P1 | CAGCAGACACCATTTAAGG | Thanapongpichat et al. (2019) |
| Pvmsp3a P2 | CCGTTTGTTGATTAGTTGC | |||
| 2nd round | Pvmsp3a N1 | GACCAGTGTGATACCATTAACC | ||
| Pvmsp3a N2 | ATACTGGTTCTTCGTCTTCAGG | |||
| Pvmsp3β | 1st round | Pvmsp3b P1 | GTATTCTTCGCAACACTC | Thanapongpichat et al. (2019) |
| Pvmsp3b P2 | CTTCTGATGTTATTTCCAG | |||
| 2nd round | Pvmsp3b N1 | CGAGGGGCGAAATTGTAAACC | ||
| Pvmsp3b N2 | GCTGCTTCTTTTGCAAAGG |
2.2.2. Pfcrt
A 264 bp fragment encoding amino acids 72–76 of exon 2 of the pfcrt gene, whose polymorphisms have been associated with antimalarial drug resistance in P. falciparum, was amplified and sequenced by nested PCR (nPCR) as described by Griffing et al. (2010). A reaction mixture was prepared for the first PCR reaction, containing 25 μl of KOD One™ PCR master mix (Toyobo Co., Ltd., Osaka, Japan), 2 μl of each primer at 10 μM (Table 1), 11 μl of nuclease-free water and 10 μl of template DNA. The amplification program consisted of 1 cycle at 98 °C for 10 s, 25 cycles at 98 °C for 10 s, 59 °C for 5 s, 68 °C for 5 s, and a final cycle at 68 °C for 30 s. The nested reaction was performed immediately after the first round of amplification, in a mixture containing 25 μl of 2× master mix, 2 μl of each primer at 10 μM (Table 1), 19 μl of nuclease-free water and 2 μl of the first PCR. The amplification program consisted of 1 cycle at 98 °C for 10 s, 25 cycles at 98 °C for 5 s, 56 °C for 5 s, 68 °C for 1 s, and a final cycle at 68 °C for 15 s. Amplicons were observed on 1% ethidium bromide agarose gels. PCR products were purified and sequenced at the Psomagen Inc. facilities (Rockville, MD, USA) (https://www.psomagen.com) using the primer AL5631 (Table 1). Geneious Prime v.2024.05 software (https://www.geneious.com) was used for sequence analysis and the residues at positions 72 to 76 of pfcrt exon-2 were evaluated.
2.2.3. Pfama1
A partial fragment of the Pfama1 gene was amplified by nested PCR. The first round of reactions was carried out in a volume of 25 μl, containing 12.5 μl of KOD One™ PCR master mix (Toyobo Co., Ltd., Osaka, Japan), 1 μl of each primer (10 μM) (Table 1), 9.5 μl of nuclease-free water, and 1 μl of genomic DNA. The amplification program consisted of one cycle at 98 °C for 30 s, 35 cycles at 98 °C for 10 s, 54 °C for 10 s, and 68 °C for 10 s, with a final cycle at 68 °C for 1 min. The second round of reactions used 12.5 μl of KOD One™ PCR master mix, 1 μl of each primer (10 μM) (Table 1), 9.5 μl of nuclease-free water, and 1 μl of the product from the first round of PCR. The amplification program was similar to that used in the first round of PCR except that instead of 35 cycles, 25 cycles were used, and the annealing temperature was changed from 54 °C to 55 °C for 5 s, and the extension cycle was 68 °C for 5 s.
2.2.4. Pfglurp
A semi-nested PCR was used to amplify a fragment of the P. falciparum glurp gene. The first round of reactions was carried out in a volume of 25 μl, containing 12.5 μl of KOD One™ PCR master mix (Toyobo Co., Ltd., Osaka, Japan), 1 μl of each primer (10 μM) (Table 1), 9.5 μl of nuclease-free water, and 1 μl of genomic DNA. The amplification program consisted of one cycle at 98 °C for 30 s, 35 cycles at 98 °C for 10 s, 62 °C for 10 s, and 68 °C for 10 s, with a final cycle at 68 °C for 1 min. The second round of reactions used 12.5 μl of KOD One™ PCR master mix, 1 μl of each primer (10 μM) (Table 1), 9.5 μl of nuclease-free water, and 1 μl of the product from the first PCR reaction. The amplification protocol was the same as the one employed in the initial round of PCR, with the only difference being the change in the number of cycles from 35 to 30.
2.2.5. Pvmsp3α and Pvmsp3β
Two molecular markers of P. vivax (Pvmsp3α and Pvmsp3β) were amplified. In both cases, nested PCR reactions contained 12.5 μl of 2× master mix (Promega Corp., Madison, WI, USA), 1 μl of each primer at 10 μM (Table 1), 8 μl of nuclease-free water and 2.5 μl of genomic DNA. The amplification program consisted of 1 cycle at 95 °C for 10 min, 35 cycles at 95 °C for 1 min, 54 °C for 70 s, 72 °C for 90 s, and a final cycle at 72 °C for 5 min. The nested reaction was performed in a reaction mixture containing 12.5 μl of 2× master mix, 1 μl of each primer at 10 μM (Table 1), 9.5 μl of nuclease-free water, and 1 μl of the first PCR. The amplification program was the same one used in the first reaction.
PCR products for all four markers described above were separated on 1% ethidium bromide agarose gels and subsequently were purified and sequenced on both sides at the Psomagen facilities. The same primers utilized for amplification were employed to sequence the fragments. Geneious Prime software was used for sequence analysis. The size of each amplicon was registered. Allele frequencies of Pvmsp3α and Pvmsp3β were calculated as the proportion of alleles observed for each gene. If there was more than one amplification band, it was interpreted as a polyclonal infection and the multiplicity of infection index (MOI) was calculated.
2.3. Data analysis
2.3.1. Sequences analyses
The sequences were edited using the “Trim ends” function of the Geneious Prime software. Afterward, each sequence was individually examined to assess its quality (HQ%) and length. All the electropherograms obtained showed only one peak in each position of the sequences. No double peaks were observed in any case. The number of identical sites between sequences of each marker was recorded as well as the pairwise percentage identity, following the default algorithm in the Geneious software. The forward and reverse sequences of each isolate were aligned to create consensus sequences for the Pfglurp gene. Nevertheless, the sequences obtained using the forward (N1) and reverse (N2) primers for Pfama1, Pvmsp3α, and Pvmsp3β were examined separately due to their lack of overlap.
The curated sequences of each molecular marker were aligned using the “Geneious Alignment” tool with the default settings. The resulting alignments were utilized to create phylogenetic trees using the Tamura-Nei genetic distance model and the Neighbor-Joining tree construction method with 1000 bootstrap iterations (Geneious Prime v.2024.05 software). No sequences were included as outgroups in the analyses. The nucleotide sequences were translated using the correct open reading frame and the resulting amino acid sequences were aligned with each other to calculate the number of identical sites, the pairwise percentage identity, and the pairwise percentage positive. At least one sequence of each haplotype obtained in this study was deposited in GenBank under the accession numbers PP795732-PP795733 (Pfama1); PP681141, PP795719 (Pfglurp); PP795720-PP795721, PP913947-PP913970, PP934608 (Pvmsp3α); and P795722-PP795731, PP886053-PP886070 (Pvmsp3β).
Tajima’s D test (Tajima, 1989) and Fu and Li’s independent D∗ and F∗ tests (Fu and Li, 1993) were performed for the sequences of the four loci of the Pvmsp3α and Pvmsp3β genes.
2.3.2. Diversity analysis of Pvmsp3α and Pvmsp3β chimeric sequences with a multilocus approach
The number of haplotypes and genetic diversity indices were determined by creating chimeric sequences for each P. vivax isolate based on a multi-locus sequence typing (MLST) approach. These chimeric sequences were made by combining two segments of Pvmsp3α and two segments of Pvmsp3β (315 bp from the N1 primer, 571 bp from the N2 primer of Pvmsp3α; and 565 bp from the N1 primer and 561 bp from the N2 primer of Pvmsp3β, resulting in a composite sequence of 1919 bp).
Multilocus sequences were aligned using the “Geneious Alignment” tool. The number of polymorphisms, the number of segregating sites (S), the average nucleotide differences (k), the number of haplotypes (H), the haplotype diversity (Hd), and the nucleotide diversity (π) were calculated using DnaSP Software (version 6.12.03). Additionally, Tajima’s D test (Tajima, 1989) was performed in DnaSP to evaluate the neutrality theory of evolution, as well as with Fu and Li’s D∗ and F∗ tests (Fu and Li, 1993). Haplotype networks were generated using the median joining algorithm in Network 10.2.0.0.
2.3.3. Comparison with homologous sequences collected in other countries
To assess the similarity between the sequences obtained in this study and homologous sequences of parasites from different geographical regions, a minimum of 30 sequences per locus (for Pfama1, Pfglurp, Pvmsp3α, and Pvmsp3β) were downloaded from GenBank (Supplementary Table S1). The downloaded sequences were aligned together with the sequences obtained here and phylogenetic trees were constructed with Geneious Prime v.2024.05 software.
3. Results
3.1. Molecular diagnosis of Plasmodium species
PET-PCR results confirmed malaria in 197 samples collected from passive surveillance conducted by the Honduran public health system. Of these samples, 91 were positive for P. vivax, 92 were positive for P. falciparum and 14 represented mixed P. falciparum + P. vivax infections. Most of the samples were collected between 2021 and 2022 (Table 2).
Table 2.
Number and percentage of samples diagnosed with malaria by parasite species and year of collection.
| Year |
P. vivax |
P. falciparum |
Mixed infection |
Total |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | |
| 2020 | 0 (0) | 23 (11.7) | 1 (0.5) | 24 (12.2) |
| 2021 | 31 (15.7) | 56 (28.4) | 5 (2.5) | 92 (46.7) |
| 2022 | 60 (30.5) | 13 (6.6) | 8 (4.1) | 81 (41.1) |
| Total | 91 (46.2) | 92 (46.7) | 14 (7.1) | 197 (100) |
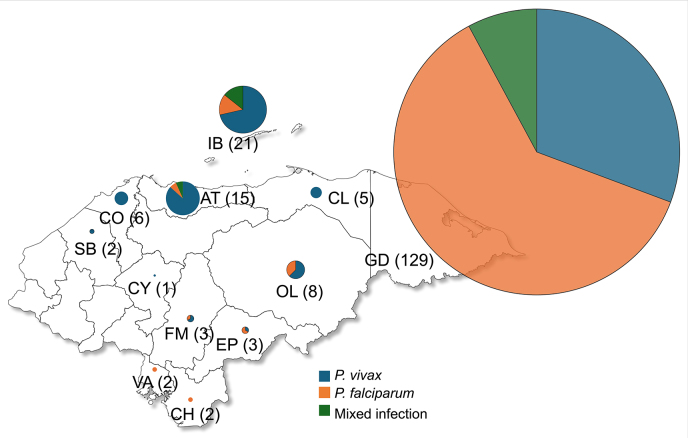
The Department Gracias a Dios (La Moskitia region) accounted for 65% of the samples, followed by Islas de la Bahía and Atlántida which contributed 10.7% and 7.6% of samples, respectively (Fig. 1). Due to the anonymization of the samples, patient demographics were not subjected to analysis.
Fig. 1.
Map of Honduras illustrating the number of samples collected by department, with indication of the parasite species. Circle size is proportional to the number of samples shown in parentheses. Department codes: GD, Gracias a Dios; IB, Islas de la Bahía; AT, Atlántida; OL, Olancho; CO, Cortés; CL, Colón; FM, Francisco Morazán; EP, El Paraíso; SB, Santa Bárbara; VA, Valle; CH, Choluteca; CY, Comayagua.
3.2. Pfcrt genotypes associated with susceptibility to chloroquine
Out of the 106 samples that tested positive for P. falciparum, 90 samples (84.9%) were genotyped for Pfcrt and analyzed. The selection was made based on the inclusion of specimens from all eight departments where P. falciparum was found, as well as from the three years of sample collection (2020–2022). All samples were found to have the wildtype genotype 72 CVMNK 76, which is associated with susceptibility to chloroquine.
3.3. Pfama1 and Pfglurp
The Pfama1 gene was amplified in 90 samples (84.9%) showing the absence of polyclonal infections. Sequencing was successful in 89 of the samples resulting in two segments obtained from the F1 and R1 primers with lengths of up to 763 bp and 757 bp, respectively. The sequences from each flank were aligned with each other but no polymorphisms were found. A sequence of each gene segment was deposited in the GenBank database under the accession numbers PP795732-PP795733.
Effective amplification of the Pfglurp gene was achieved in 89 samples (96.7%) showing bands that were classified into three groups according to their size range, as follows. The majority (98%) of the samples exhibited a size ranging from 550 to 600 bp, one sample had a size between 600 and 650 bp, and another sample - between 750 and 800 bp.
A 552 bp fragment of the Pfglurp gene was successfully sequenced in 89 P. falciparum samples and no polymorphisms were detected (GenBank: PP681141). However, one sample presented a 56 bp insertion (GenBank: PP795719).
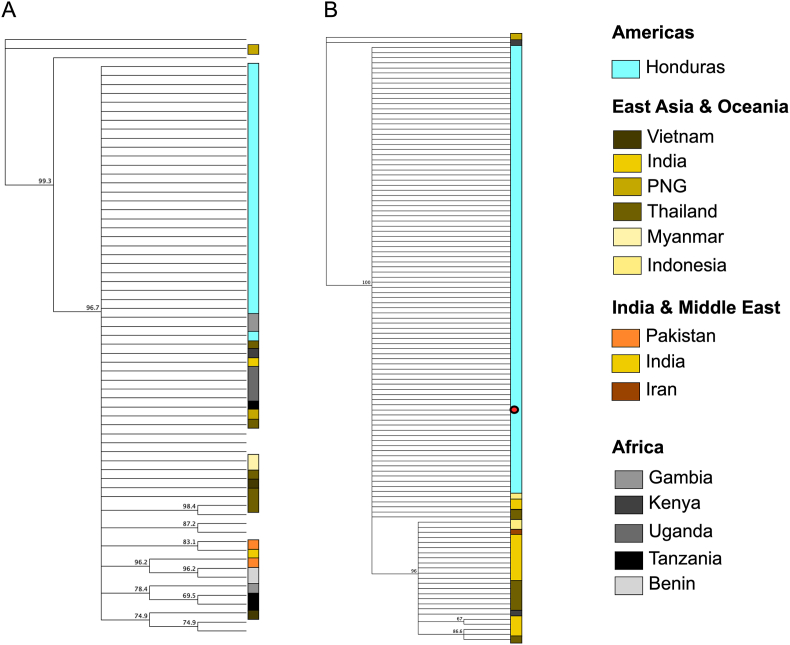
Geographical clustering analysis was carried out using the Pfama1 and Pfglurp sequences with homologous sequences from other countries downloaded from GenBank respectively (Supplementary Table S1). Our results show that the samples from Honduras appeared as a single population without geographical clustering using these genetic markers (Fig. 2).
Fig. 2.
Phylogenetic trees constructed with sequences of Pfama1 (A) and Pfglurp (B), obtained in this study, and homologous sequences available on GenBank. The red dot indicates the sequence from Honduras (GenBank: PP795719).
3.4. Genetic diversity of Pvmsp3α and Pvmsp3β
Seventy-three of the 105 P. vivax-infected samples were effectively amplified for the Pvmsp3α gene. Four amplicon sizes were identified as type A (1700–2000 bp; 12%), type B (1500–1700 bp; 62.7%), type C (1200–1500 bp; 24%), and type D (400–500 bp; 1.3%). In addition, two samples with polyclonal infection were identified with two band sizes with a multiplicity of infection (MOI) value of 1.02. Regarding Pvmsp3β, 63 out of the 105 samples were effectively amplified, representing a success rate of 60%. The amplified samples exhibited three distinct band sizes classified as type A (1600–2000 bp; 24%), type B (1300–1500 bp; 13%), and type C (1100–1300 bp; 63%). Four samples exhibiting polyclonal infections were detected, with a MOI value of 1.06.
Pvmsp3α and Pvmsp3β genes were amplified from both sides. However, given the length of the genes, the sequences obtained were not overlapping, and consensus sequences were not constructed, so the sequences obtained with the forward primers (N1) and those obtained with the reverse primer (N2) were analyzed separately as four independent loci/markers.
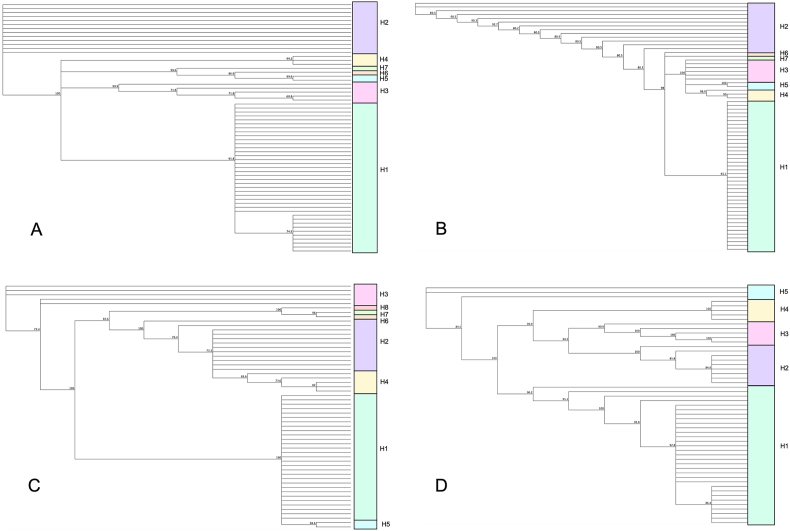
As shown in Table 3, a fraction of the total amplified samples was successfully sequenced. Sequence lengths ranged from 633 bp to 827 bp. The marker with the highest number of polymorphisms was Pvmsp3α-N1 followed by Pvmsp3β-N2, while the least diverse was Pvmsp3α-N2. In terms of haplotype analysis, also a fraction of samples was analyzed as sequences with shorter lengths had to be discarded (Supplementary Tables S2–S5). The highest number of haplotypes (n = 8) was found in Pvmsp3β-N1 and the lowest number of haplotypes in Pvmsp3β-N2 (n = 5). The two other markers (Pvmsp3α-N1 and Pvmsp3α-N2) produced a total of 7 distinct haplotypes each (Fig. 3). While the number of haplotypes varied among the four markers, individuals within each haplotype had a highly similar composition. Haplotype 1 was the most frequent allele, followed by Haplotype 2 (Supplementary Tables S2–S5).
Table 3.
Intraspecific genetic diversity indices for Pvmsp3α and Pvmsp3β.
| Pvmsp3α-N1 | Pvmsp3α-N2 | Pvmsp3β-N1 | Pvmsp3β-N2 | |
|---|---|---|---|---|
| No. of sequences | 65 | 66 | 56 | 53 |
| Length (bp) | Up to 827 | Up to 737 | Up to 824 | Up to 633 |
| Identical sites (%) | 453 (55%) | 701 (95.1%) | 577 (74.5%) | 352 (55.6%) |
| Pairwise % identity | 85.4 | 99 | 90.5 | 87.8 |
| No. of haplotypes (nt) | 7 | 7 | 8 | 5 |
| Polypeptide length (aa) | Up to 227 | Up to 244 | Up to 269 | Up to 205 |
| No. of haplotypes (aa) | 7 | 6 | 8 | 5 |
| Identical sites (%) | 71 (31.3%) | 227 (93%) | 130 (51.8%) | 82 (40.2%) |
| Pairwise % identity | 67.3 | 97.1 | 74.3 | 66.9 |
| Pairwise % positive | 76.2 | 98.1 | 82.8 | 77.2 |
| Tajima’s D test | 0.15863, P > 0.10 | 0.09266, P > 0.10 | 1.24308, P > 0.10 | 0.28050, P > 0.10 |
| Fu and Li’s D∗ test | 0.36392, P > 0.10 | 1.60341, P < 0.05∗ | 2.07742, P < 0.02∗ | 2.20871, P < 0.02∗ |
| Fu and Li’s F∗test | 0.33966, P > 0.10 | 1.24883, P > 0.10 | 2.10486, P < 0.02∗ | 1.76250, P < 0.05∗ |
| GenBank ID | PP795720; PP913947-PP913958 | PP795721; PP913959-PP913970; PP934608 | P795722-P795726; PP886053-PP886065 | PP795727-PP795731; PP886066-PP886070 |
Abbreviations: nt, nucleotides; aa, amino acids.
Fig. 3.
Dendrograms constructed with the sequences of Pvmsp3 genes obtained in this study from samples collected in Honduras. The colors of the blocks indicate the haplotypes based on nucleotide sequences for each marker: Pvmsp3α-N1 (A), Pvmsp3α-N2 (B), Pvmsp3β-N1 (C), Pvmsp3β-N2 (D).
Polymorphisms were also detected in the amino acid sequences in the four markers, revealing that Pvmsp3α-N1 exhibited the greatest diversity of residues, and Pvmsp3α-N2 was the most conserved region. The number of haplotypes generated with the amino acid sequences varied from 5 (Pvmsp3β-N2) to 8 (Pvmsp3β-N1) (Table 3). To explore the influence of natural selection on population structure, Tajima’s D test was performed on the four independent loci and Pvmsp3α/β MLST sequences. Positive values (ranging from 0.09266 to 1.24308) and non-significant P-values were observed. Fu and Li’s D∗ tests resulted in values ranging from 0.36 to 2.2, and three of them showed statistical significance except for Pvmsp3α-N1. Fu and Li’s F∗ tests resulted in values ranging from 0.33 to 2.1, and two loci (Pvmsp3β-N1 and Pvmsp3β-N2) showed statistical significance.
3.5. Haplotypes of MLST chimeric sequences
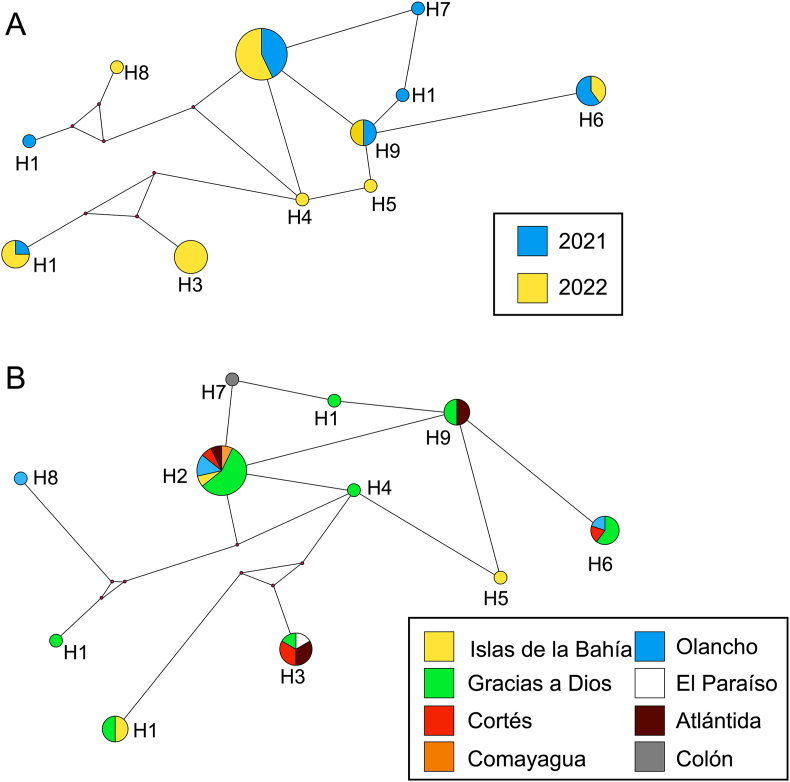
Chimera sequences were constructed using an MLST approach on 39 isolates for which high-quality sequences were obtained for the 4 markers: Pvmsp3α-N1, Pvmsp3α-N2, Pvmsp3β-N1, Pvmsp3β-N2. As indicated in Section 2.3.2, chimeric sequences were made by combining both segments of each gene. The number of segregating sites (S) was 344. The nucleotide diversity (π) was equal to 0.06107 (SD = 0.00723), and the average nucleotide differences (k) was 104.13. Eleven haplotypes (H) were found (Fig. 4), with a diversity index (Hd) of 0.8273. Out of all the MLST sequences, 84.7% were grouped into 5 haplotypes (Supplementary Table S6) whereas the six remaining haplotypes consisted of only one sequence each. Haplotype networks were constructed considering the year in which the blood samples were collected and the department of origin of the malaria case (Fig. 4). No association was observed between the circulating haplotypes of the parasite in either case.
Fig. 4.
Haplotype networks constructed for the samples obtained in this study according to the year the sample was collected (A), and the department of origin of the sample (B). Haplotypes were constructed with MLST chimera sequences comprising four gene regions: Pvmsp3α-N1, Pvmsp3α-N2, Pvmsp3β-N1, Pvmsp3β-N2. Each circle represents a different haplotype, and the size of the circles is indicative of the number of individuals within each haplotype.
To explore the influence of natural selection on population structure, Tajima’s D test was performed for the MLST chimeric sequences. A non-significant (P > 0.10) Tajima’s D value of 0.62954 was obtained. Fu and Li’s D∗ and F∗ tests resulted in a value of 1.50687 (P < 0.05) and 1.42040 (P > 0.10), respectively. Tajima’s D and Fu and Li’s D∗ and F∗ results did not show a significant influence of natural selection or demographic changes in the population based on the Pvmsp3α and Pvmsp3β markers. The result of Fu and Li’s for Pvmsp3α, which was statistically significant, suggests that there are rare alleles represented at low frequencies and that there could be a population structure or contraction in the studied population, although this signal is not sufficiently supported by the other tests. Although Fu and Li’s value was also positive for Pvmsp3β, it was not statistically significant. This indicates that there is no conclusive evidence in this test to confirm signals of selection or population structure, suggesting that these results could be due to chance.
3.6. Comparison with homologous Pvmsp3 sequences collected in other countries
Similarly to the approach used for Pfama1 and Pfglurp, we obtained 35 homologous sequences of Pvmsp3α and 46 homologous sequences of Pvmsp3β from four different geographical regions (Supplementary Table S1).
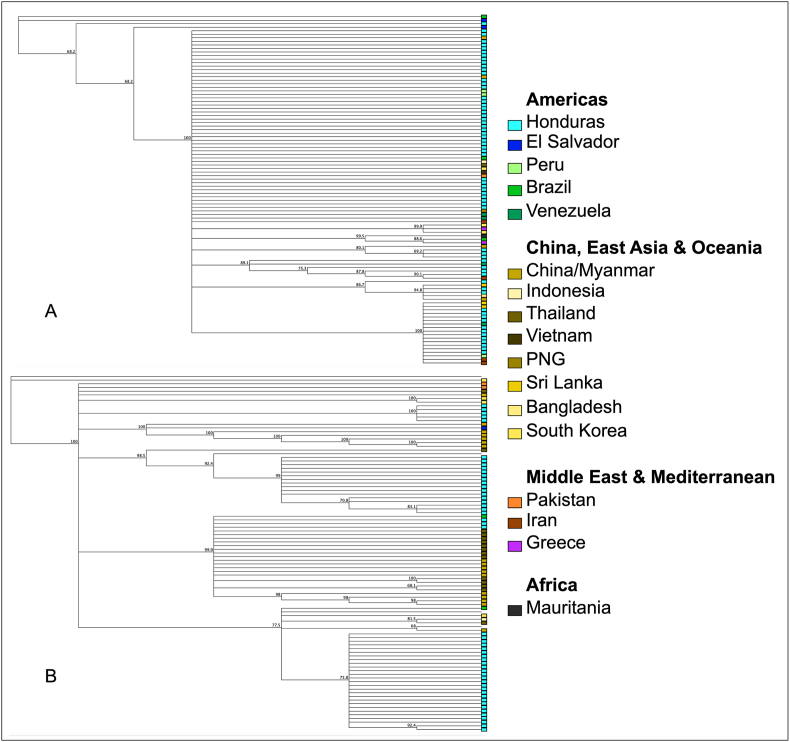
Based on the phylogenetic tree shown in Fig. 5, the haplotypes of the sequences obtained in this study did not form distinct clusters and there was no noticeable geographical clustering with strains of the parasite isolated in other countries. However, the small number of sequences isolated from countries outside Honduras is insufficient to categorically conclude that there is a lack of clustering.
Fig. 5.
Phylogenetic trees constructed with sequences of Pvmsp3α (A) and Pvmsp3β (B) obtained in this study and homologous sequences available on GenBank.
4. Discussion
This study assessed four markers of genetic diversity in P. falciparum and P. vivax, the two malaria-causing parasite species occurring in Honduras. Our results revealed a significant disparity in the intraspecific genetic diversity between the two species. The P. falciparum population seemed to be nearly uniform, whereas the P. vivax population exhibited significant variability. The minimal variability of the P. falciparum population in Honduras contrasts strongly with the considerable variation of Pfama1 and Pfglurp reported from Asia and Africa (Patgiri et al., 2019; Kang et al., 2021; Nirmolia et al., 2022; Olowe et al., 2023; Hawadak et al., 2024; Paul et al., 2024). The most likely explanation for this phenomenon is a bottleneck effect in Honduras, likely as a result of the decline in falciparum malaria cases from 2000 to 2014, with only 575 reported cases in 2014 (WHO, 2015b).
Recently, several studies have been carried out to evaluate the genetic diversity of Plasmodium spp. in Honduras. In 2012, a study analyzed the P. vivax population using Pvama1, Pvcsp, and Pvmsp1, as well as two P. falciparum markers: Pfmsp1 and Pfmsp2 (Lopez et al., 2012). These five molecular markers showed moderate genetic diversity and wide distribution across the country without geographical structuring. Samples of Lopez et al. (2012) were collected during 2010–2011 when the number of malaria cases in Honduras was around 13,000 per year (WHO, 2013) which is more than five times the number of cases (c.2500) reported in 2023 (Secretaría de Salud de Honduras, 2024).
In the present study, Pfama1 and Pfglurp showed that 100% and 98% of the isolates were identical and monoclonal, except for two samples for Pfglurp that showed size polymorphisms. In this regard, Haddad et al. (1999) published the first study of the genetic diversity of P. falciparum populations circulating in Honduras with 56 samples collected between 1995 and 1996 when the number of malaria cases in the country exceeded 35,000 per year (WHO, 2013). Haddad et al. (1999) genotyped four markers (Pfmsp1, Pfmsp2, Pfglurp, and Pf332) using nested PCR reactions to determine allelic families and size polymorphisms. These authors reported limited size polymorphism in all genes, with four and three variants for the Pfmsp1 and Pfmsp2 alleles, respectively, and two size variants for the Pfglurp and Pf332 genes, concluding that P. falciparum populations circulating in Honduras were genetically homogeneous.
More recently, an analysis of seven neutral STR markers in 77 P. falciparum field isolates collected between 2009 and 2012 in Honduras concluded that there was a low genetic diversity in the parasite population (Larranaga et al., 2013). Likewise, Pinto et al. (2021) conducted a study in which the polymorphic regions of Pfmsp1 and Pfmsp2 were sequenced, showing reduced genetic diversity of P. falciparum after a bottleneck effect between 2000 and 2014 followed by rapid expansion starting in 2015. In summary, our work supports earlier findings indicating that the genetic diversity of P. falciparum populations in Honduras is historically low, regardless of the specific molecular marker applied.
In contrast to the uniformity shown by P. falciparum, the two P. vivax markers assessed in the present study (Pvmsp3α and Pvmsp3β) exhibited high diversity indices with four and three alleles of different sizes, respectively. Pvmsp3α type B (1.7–2.0 kb) was the most frequent among field isolates from Honduras followed by type C (1.2–1.5 kb). This finding differs from studies carried out in Asia (Kim et al., 2006; Rungsihirunrat et al., 2011; Suphakhonchuwong et al., 2018; Thanapongpichat et al., 2019; Kuesap et al., 2022; Jalei et al., 2023) and South America (Cristiano et al., 2008) where type A (1.9 kb) was the most frequent; or from field isolates from China, in which type C (1.1–1.3 kb) was the most frequent (Li et al., 2022; Wang et al., 2023). Similar results were observed for Pvmsp3β, where the most common variant among the isolates from Honduras was type C (1.1–1.3 kb), but type A (1.8 kb) was more frequent in several countries (Zakeri et al., 2010; Rungsihirunrat et al., 2011; Suphakhonchuwong et al., 2018; Thanapongpichat et al., 2019; Jalei et al., 2023; Wang et al., 2023). These data confirm the significant variation observed in the size of this gene resulting from insertion/deletion mutations (Rayner et al., 2004).
Most studies analyzing the genetic diversity of Pvmsp3 use a nested PCR followed by RFLP, either cutting with individual enzymes or combining two enzymes (Kim et al., 2006; Yang et al., 2006; Cristiano et al., 2008; Zakeri et al., 2010; Rungsihirunrat et al., 2011; Thanapongpichat et al., 2019; Kuesap et al., 2022; Jalei et al., 2023; Wang et al., 2023). This method exploits the extensive diversity largely present in the central domain of the PvMSP3 protein family (Jiang et al., 2013). However, in our study, the gene regions flanking the central domain of both markers, which have a lower degree of polymorphism, were sequenced. This approach could be more informative than PCR-RFLP to demonstrate genetic diversity and determine circulating haplotypes of the parasite (Rice et al., 2013).
The wide intraspecific diversity of P. vivax, as shown by size polymorphisms, was further confirmed through the sequencing of the four loci of Pvmsp3α and Pvmsp3β. This analysis revealed the presence of five to eight distinct haplotypes, based on both nucleotide and amino acid sequences. According to these results, more than half of the isolates were classified as Haplotype 1, and more than 20% were classified as Haplotype 2. This means that the other haplotypes are found at low frequencies in this population, as also confirmed by the Tajimaʼs D test.
While the independent analysis of the four loci appears to be sufficient for genotyping the parasite isolates, the MLST approach has proven to be a better technique for revealing the genetic diversity of the populations. One of the main limitations of the MLST approach is that it only evaluates a limited number of loci, which may underestimate overall genetic variability and may not adequately capture total diversity, especially in organisms with high recombination or in genomic regions outside those loci. Likewise, the MLST approach based on coding genes is a widely used strategy to classify bacteria (Azarsa et al., 2023; Li et al., 2023b; Rosa Rodrigues de Souza et al., 2023; Zhang et al., 2023) but very little used for eukaryotic parasites (Diosque et al., 2014; El Mazini et al., 2023; Hosseini et al., 2023). The combination of the four Pvmsp3 loci was more informative than PCR-RFLP, which neither detects mutations without a digestion target nor distinguishes between different haplotypes with equivalent band sizes (Rice et al., 2013).
On the other hand, phylogenetic analyses did not reveal any type of geographical clustering, neither within the country nor concerning P. vivax isolates collected on other continents. Only a few studies use Pvmsp3 to demonstrate population structure in P. vivax. In a study conducted by Kim et al. (2006), 151 isolates from Kolkata, India, were investigated and resulted in 37 Pvmsp3α alleles that were randomly distributed. Another study carried out in different geographical areas of Thailand showed differences in the allele frequencies of Pvmsp3α and Pvmsp3β when comparing two time periods, especially in isolates from the border area with Myanmar (Kuesap et al., 2022). Similarly, the diversity of the P. vivax population infecting local inhabitants of China was significantly more diverse than that of parasites infecting migrant labourers returning to China from 2012 to 2015 (Li et al., 2022). A larger study reported differences in Pvmsp3β diversity using PCR-RFLP among four Asian parasite populations representing both tropical and temperate strains (Yang et al., 2006). However, based on our findings, it appears that neither of the two Pvmsp3 markers can accurately determine the geographical origin on a regional or global level. Consequently, due to frequent insertion/deletion mutations and recurrent recombination between haplotypes (Rice et al., 2013), these markers do not seem suitable for tracking parasite populations in studies related to geography or extended periods.
Finally, this study using four molecular markers showed that P. falciparum populations in Honduras turned out to be much more homogeneous than those of P. vivax. These findings could have significant practical implications in the context of the malaria elimination phase in the country. The high homogeneity of P. falciparum populations suggests that the transmission of this parasite is probably more contained and that the population is dominated by a few genetic variants. This could imply that transmission is limited and that these are localized outbreaks or a small number of sources of infection. Consequently, from an elimination point of view, it is easier to track and control a more homogeneous population of the parasite, since the origin of infections is more likely to be localized to limited sources. On the other hand, the homogeneity in P. falciparum indicates that elimination efforts in Honduras are managing to reduce the genetic diversity of this parasite, which is a good indicator for short-term control and elimination. Therefore, surveillance should focus on monitoring these residual foci of transmission and avoiding reintroduction from external areas. Border control and surveillance of migratory movements or people who could be carriers of P. falciparum from other regions with greater genetic diversity must be a priority to avoid the reintroduction of more virulent variants.
In contrast, the greater genetic diversity of P. vivax suggests that there are multiple lineages of this parasite in circulation, which may indicate more active or persistent transmission compared to P. falciparum. This greater diversity can complicate elimination efforts, as there may be hidden reservoirs of the parasite and greater difficulty in identifying all sources of infection. For Honduras, during the malaria elimination phase, the study results suggest that P. falciparum is under more effective control and its elimination appears closer, while P. vivax represents a greater challenge due to its greater genetic diversity. This requires more intense surveillance, specific control strategies for P. vivax, and additional preventive measures to prevent the reintroduction of variants of both parasites from other regions.
To conclude, and in line with the low genetic diversity of P. falciparum, the Pfcrt gene sequencing results confirmed that the strains of the parasite circulating in Honduras continue to be susceptible to treatment with chloroquine, as previous publications have evidenced (Jovel et al., 2011; Mejia Torres et al., 2013; Fontecha et al., 2014, 2021). This result is encouraging as it facilitates the strategies used to eliminate malaria in Honduras.
5. Conclusions
This study evaluates the genetic diversity of the two parasite species that cause malaria in Honduras and demonstrates that the populations of P. falciparum are more homogeneous than P. vivax, based on the molecular markers evaluated. The MLST approach with four loci of Pvmsp3α and Pvmsp3β proved to be more informative in assessing the genetic diversity of P. vivax compared to individual markers. No geographical structuring was demonstrated within the country or on a global scale between the haplotypes of both species. In Honduras, P. falciparum appears to be under more effective control, while P. vivax poses a greater challenge due to its higher genetic diversity. This calls for intensified surveillance and targeted control strategies for P. vivax, and additional measures to prevent the reintroduction of parasite variants from other regions. Additionally, circulating strains of P. falciparum continue to show the wild phenotype of the Pfcrt gene associated with susceptibility to chloroquine.
CRediT authorship contribution statement
Alejandro Zamora: Investigation, Methodology, Formal analysis, Data curation, Writing – review & editing. Alejandra Pinto: Investigation, Methodology, Formal analysis, Data curation, Funding acquisition, Writing – review & editing. Denis Escobar: Investigation, Methodology, Formal analysis, Data curation, Writing – review & editing. Hugo O. Valdivia: Formal analysis, Funding acquisition, Writing – review & editing. Lesly Chaver: Investigation, Writing – review & editing. Gloria Ardón: Investigation, Writing – review & editing. Erick Carranza: Investigation, Writing – review & editing. Gustavo Fontecha: Investigation, Methodology, Writing – original draft, Writing – review & editing, Supervision, Visualization, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Ethical approval
The study made secondary use of biological specimens originally collected for malaria diagnosis as per standard of care in Honduras, to identify parasite species and analyze genes linked to drug resistance, following national regulations for routine malaria surveillance. The ethics committee CEI-MEIZ-UNAH reviewed and approved the study (03–2020 extended). The consent to participate was exempted based on: (a) The absence of personal information; (b) the studyʼs contribution to public health; and (c) the absence of any harm to the participants. The blood samples were anonymized and irrevocably stripped of direct identifiers, so future re-identification of individuals was not possible. The study protocol NAMRU6.2018.0002 was reviewed and approved by the Research Administration Programme of the Naval Medical Research Unit SOUTH (NAMRU SOUTH) in compliance with all applicable federal regulations governing the protection of human subjects.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary file. The newly generated sequences were submitted to the GenBank database under the accession numbers: PP795732-PP795733 (Pfama1); PP681141, PP795719 (Pfglurp); PP795720-PP795721, PP913947-PP913970, PP934608 (Pvmsp3α); and P795722-PP795731, PP886053-PP886070 (Pvmsp3β).
Disclaimer
One of the authors of this manuscript (H.O.V.) is an employee of the U.S. Government. This work was prepared as part of his official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this Title is not available for any work of the United States Government”. Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person's official duties. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the U.S. Government.
Funding
Funding for this study was provided by the Genetic Research Center, CIG-UNAH, and by the Armed Forces Health Surveillance Division (AFHSD), Global Emerging Infections Surveillance (GEIS) Branch (PROMIS ID P0091_24_N6). The funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors wish to express their gratitude to the clinical staff of the Regions of the Ministry of Health of Honduras for the collaboration for the development of this project.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crpvbd.2024.100230.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- Azarsa M., Mosadegh M., Habibi Ghahfarokhi S., Pourmand M.R. Serotype distribution and multi locus sequence type (MLST) of erythromycin-resistant Streptococcus pneumoniae isolates in Tehran, Iran. Rep. Biochem. Mol. Biol. 2023;12:259–268. doi: 10.61186/rbmb.12.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristiano F.A., Perez M.A., Nicholls R.S., Guerra A.P. Polymorphism in the Plasmodium vivax msp 3 gene in field samples from Tierralta, Colombia. Mem. Inst. Oswaldo Cruz. 2008;103:493–496. doi: 10.1590/s0074-02762008000500015. [DOI] [PubMed] [Google Scholar]
- Diosque P., Tomasini N., Lauthier J.J., Messenger L.A., Monje Rumi M.M., Ragone P.G., et al. Optimized multilocus sequence typing (MLST) scheme for Trypanosoma cruzi. PLoS Negl. Trop. Dis. 2014;8 doi: 10.1371/journal.pntd.0003117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mazini S., Barhoumi M., Mhaidi I., Daoui O., Kbaich M.A., El Kacem S., et al. Genetic diversity and population structure of Leishmania infantum in Morocco as revealed by multilocus sequence typing (MLST) approach. Pathogens. 2023;12:785. doi: 10.3390/pathogens12060785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecha G., Pinto A., Archaga O., Betancourth S., Escober L., Henriquez J., et al. Assessment of Plasmodium falciparum anti-malarial drug resistance markers in pfcrt and pfmdr1 genes in isolates from Honduras and Nicaragua, 2018–2021. Malar. J. 2021;20:465. doi: 10.1186/s12936-021-03977-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecha G.A., Sanchez A.L., Mendoza M., Banegas E., Mejia-Torres R.E. A four-year surveillance program for detection of Plasmodium falciparum chloroquine resistance in Honduras. Mem. Inst. Oswaldo Cruz. 2014;109:492–493. doi: 10.1590/0074-0276140067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.X., Li W.H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing S., Syphard L., Sridaran S., McCollum A.M., Mixson-Hayden T., Vinayak S., et al. pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob. Agents Chemother. 2010;54:1572–1579. doi: 10.1128/AAC.01243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad D., Snounou G., Mattei D., Enamorado I.G., Figueroa J., Stahl S., Berzins K. Limited genetic diversity of Plasmodium falciparum in field isolates from Honduras. Am. J. Trop. Med. Hyg. 1999;60:30–34. doi: 10.4269/ajtmh.1999.60.30. [DOI] [PubMed] [Google Scholar]
- Hancock P.A., Ochomo E., Messenger L.A. Genetic surveillance of insecticide resistance in African Anopheles populations to inform malaria vector control. Trends Parasitol. 2024;40:604–618. doi: 10.1016/j.pt.2024.04.016. [DOI] [PubMed] [Google Scholar]
- Hawadak J., Kojom Foko L.P., Dongang Nana R.R., Yadav K., Pande V., Das A., Singh V. Genetic diversity and natural selection of apical membrane antigen-1 (ama-1) in Cameroonian Plasmodium falciparum isolates. Gene. 2024;894 doi: 10.1016/j.gene.2023.147956. [DOI] [PubMed] [Google Scholar]
- Hosseini S.A., Sharif M., Sarvi S., Mirzaei N., Abediankenari S., Arefkhah N., et al. Identification and multilocus genotyping of Toxoplasma gondii isolates from congenital infection in north of Iran. Parasitol. Res. 2023;122:177–184. doi: 10.1007/s00436-022-07714-1. [DOI] [PubMed] [Google Scholar]
- Huang B., Tuo F., Liang Y., Wu W., Wu G., Huang S., et al. Temporal changes in genetic diversity of msp-1, msp-2, and msp-3 in Plasmodium falciparum isolates from Grande Comore Island after introduction of ACT. Malar. J. 2018;17:83. doi: 10.1186/s12936-018-2227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalei A.A., Chaijaroenkul W., Na-Bangchang K. Genetic diversity of Plasmodium vivax field isolates from the Thai-Myanmar border during the period of 2006–2016. Trav. Med. Infect. Dis. 2023;8:210. doi: 10.3390/tropicalmed8040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Barnwell J.W., Meyer E.V., Galinski M.R. Plasmodium vivax merozoite surface protein-3 (PvMSP3): Expression of an 11 member multigene family in blood-stage parasites. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovel I.T., Mejia R.E., Banegas E., Piedade R., Alger J., Fontecha G., et al. Drug resistance associated genetic polymorphisms in Plasmodium falciparum and Plasmodium vivax collected in Honduras, Central America. Malar. J. 2011;10:376. doi: 10.1186/1475-2875-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.M., Le H.G., Vo T.C., Naw H., Yoo W.G., Sohn W.M., et al. Genetic polymorphism and natural selection of apical membrane antigen-1 in Plasmodium falciparum isolates from Vietnam. Genes. 2021;12:1903. doi: 10.3390/genes12121903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.M., Lee J., Moe M., Jun H., Le H.G., Kim T.I., et al. Population genetic structure and natural selection of Plasmodium falciparum apical membrane antigen-1 in Myanmar isolates. Malar. J. 2018;17:71. doi: 10.1186/s12936-018-2215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattenberg J.H., Fernandez-Minope C., van Dijk N.J., Llacsahuanga Allcca L., Guetens P., Valdivia H.O., et al. Malaria molecular surveillance in the Peruvian Amazon with a novel highly multiplexed Plasmodium falciparum AmpliSeq assay. Microbiol. Spectr. 2023;11 doi: 10.1128/spectrum.00960-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.R., Imwong M., Nandy A., Chotivanich K., Nontprasert A., Tonomsing N., et al. Genetic diversity of Plasmodium vivax in Kolkata, India. Malar. J. 2006;5:71. doi: 10.1186/1475-2875-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudyba H.M., Louzada J., Ljolje D., Kudyba K.A., Muralidharan V., Oliveira-Ferreira J., Lucchi N.W. Field evaluation of malaria malachite green loop-mediated isothermal amplification in health posts in Roraima State, Brazil. Malar. J. 2019;18:98. doi: 10.1186/s12936-019-2722-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuesap J., Rungsihirunrat K., Chaijaroenkul W., Mungthin M. Genetic diversity of Plasmodium vivax merozoite surface protein-3 alpha and beta from diverse geographic areas of Thailand. Jpn. J. Infect. Dis. 2022;75:241–248. doi: 10.7883/yoken.JJID.2021.457. [DOI] [PubMed] [Google Scholar]
- Kusi K.A., Amoah L.E., Acquah F.K., Ennuson N.A., Frempong A.F., Ofori E.A., et al. Plasmodium falciparum AMA1 and CSP antigen diversity in parasite isolates from southern Ghana. Front. Cell. Infect. Microbiol. 2024;14 doi: 10.3389/fcimb.2024.1375249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larranaga N., Mejia R.E., Hormaza J.I., Montoya A., Soto A., Fontecha G.A. Genetic structure of Plasmodium falciparum populations across the Honduras-Nicaragua border. Malar. J. 2013;12:354. doi: 10.1186/1475-2875-12-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenson A.J., Laurens M.B. A new landscape for malaria vaccine development. PLoS Pathog. 2024;20 doi: 10.1371/journal.ppat.1012309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Liu H., Tang L., Yang H., Bustos M.D.G., Tu H., Ringwald P. Genetic characteristics of P. falciparum parasites collected from 2012 to 2016 and anti-malaria resistance along the China-Myanmar border. PLoS One. 2023;18 doi: 10.1371/journal.pone.0293590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Lv R., Li X., Song C., Xingxin L., Zhang H. Antimicrobial resistance, serogroups, virulence gene profiles and MLST of Escherichia coli from giant panda. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1236227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Bai Y., Wu Y., Zeng W., Xiang Z., Zhao H., et al. PvMSP-3alpha and PvMSP-3beta genotyping reveals higher genetic diversity in Plasmodium vivax parasites from migrant workers than residents at the China-Myanmar border. Infect. Genet. Evol. 2022;106 doi: 10.1016/j.meegid.2022.105387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A.C., Ortiz A., Coello J., Sosa-Ochoa W., Torres R.E., Banegas E.I., et al. Genetic diversity of Plasmodium vivax and Plasmodium falciparum in Honduras. Malar. J. 2012;11:391. doi: 10.1186/1475-2875-11-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi N.W., Karell M.A., Journel I., Rogier E., Goldman I., Ljolje D., et al. PET-PCR method for the molecular detection of malaria parasites in a national malaria surveillance study in Haiti, 2011. Malar. J. 2014;13:462. doi: 10.1186/1475-2875-13-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchi N.W., Narayanan J., Karell M.A., Xayavong M., Kariuki S., DaSilva A.J., et al. Molecular diagnosis of malaria by photo-induced electron transfer fluorogenic primers: PET-PCR. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamoros G., Escobar D., Pinto A., Serrano D., Ksandrova E., Grimaldi N., et al. PET-PCR reveals low parasitaemia and submicroscopic malarial infections in Honduran Moskitia. Malar. J. 2023;22:110. doi: 10.1186/s12936-023-04538-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia Torres R.E., Banegas E.I., Mendoza M., Diaz C., Bucheli S.T., Fontecha G.A., et al. Efficacy of chloroquine for the treatment of uncomplicated Plasmodium falciparum malaria in Honduras. Am. J. Trop. Med. Hyg. 2013;88:850–854. doi: 10.4269/ajtmh.12-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J.E. A history of malaria and conflict. Parasitol. Res. 2024;123:165. doi: 10.1007/s00436-024-08167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nirmolia T., Ahmed M.A., Sathishkumar V., Sarma N.P., Bhattacharyya D.R., Mohapatra P.K., et al. Genetic diversity of Plasmodium falciparum AMA-1 antigen from the Northeast Indian state of Tripura and comparison with global sequences: Implications for vaccine development. Malar. J. 2022;21:62. doi: 10.1186/s12936-022-04081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olowe R.A., Ojo J.A., Funwei R.I., Oyedeji S.I., Olowe O.A., Thomas B.N., Ojurongbe O. Genetic diversity of Plasmodium falciparum among asymptomatic pregnant women on intermittent preventive treatment with sulfadoxine-pyrimethamine in Nigeria. Afr. Health Sci. 2023;23:765–773. doi: 10.4314/ahs.v23i1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patgiri S.J., Sarma K., Sarmah N., Bhattacharyya N., Sarma D.K., Nirmolia T., et al. Characterization of drug resistance and genetic diversity of Plasmodium falciparum parasites from Tripura, Northeast India. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-50152-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K.M.M., Simpson S.V., Nundu S.S., Arima H., Yamamoto T. Genetic diversity of glutamate-rich protein (GLURP) in Plasmodium falciparum isolates from school-age children in Kinshasa. DRC. Parasitol. Int. 2024;100 doi: 10.1016/j.parint.2024.102866. [DOI] [PubMed] [Google Scholar]
- Pinto A., Archaga O., Mejia A., Escober L., Henriquez J., Montoya A., et al. Evidence of a recent bottleneck in Plasmodium falciparum populations on the Honduran-Nicaraguan border. Pathogens. 2021;10:1432. doi: 10.3390/pathogens10111432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner J.C., Huber C.S., Feldman D., Ingravallo P., Galinski M.R., Barnwell J.W. Plasmodium vivax merozoite surface protein PvMSP-3 beta is radically polymorphic through mutation and large insertions and deletions. Infect. Genet. Evol. 2004;4:309–319. doi: 10.1016/j.meegid.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Rice B.L., Acosta M.M., Pacheco M.A., Escalante A.A. Merozoite surface protein-3 alpha as a genetic marker for epidemiologic studies in Plasmodium vivax: A cautionary note. Malar. J. 2013;12:288. doi: 10.1186/1475-2875-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa Rodrigues de Souza C., Bergis H., Ng P., Guillier L., Felix B., Leclercq A., Gnanou Besse N. Assessment of the relationship between the MLST genetic diversity of Listeria monocytogenes and growth under selective and non-selective conditions. Food Microbiol. 2023;114 doi: 10.1016/j.fm.2023.104303. [DOI] [PubMed] [Google Scholar]
- Rungsihirunrat K., Chaijaroenkul W., Siripoon N., Seugorn A., Na-Bangchang K. Genotyping of polymorphic marker (MSP3alpha and MSP3beta) genes of Plasmodium vivax field isolates from malaria endemic of Thailand. Trop. Med. Int. Health. 2011;16:794–801. doi: 10.1111/j.1365-3156.2011.02771.x. [DOI] [PubMed] [Google Scholar]
- Sarma N., Patouillard E., Cibulskis R.E., Arcand J.L. The economic burden of malaria: Revisiting the evidence. Am. J. Trop. Med. Hyg. 2019;101:1405–1415. doi: 10.4269/ajtmh.19-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathishkumar V., Nirmolia T., Bhattacharyya D.R., Patgiri S.J. Genetic polymorphism of Plasmodium falciparum msp-1, msp-2 and glurp vaccine candidate genes in pre-artemisinin era clinical isolates from Lakhimpur district in Assam, Northeast India. Access. Microbiol. 2022;4 doi: 10.1099/acmi.0.000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer T.M., Pessanha de Carvalho L., Inoue J., Kreidenweiss A., Held J. The problem of antimalarial resistance and its implications for drug discovery. Expet Opin. Drug Discov. 2024;19:209–224. doi: 10.1080/17460441.2023.2284820. [DOI] [PubMed] [Google Scholar]
- Secretaría de Salud de Honduras Boletín Epidemiológico Malaria, Equipo Técnico Nacional para la Eliminación de la Malaria. Tegucigalpa. 2024. https://sshome.salud.gob.hn/index.php/malaria#boletin
- Stanisic D.I., Barry A.E., Good M.F. Escaping the immune system: How the malaria parasite makes vaccine development a challenge. Trends Parasitol. 2013;29:612–622. doi: 10.1016/j.pt.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Suphakhonchuwong N., Chaijaroenkul W., Rungsihirunrat K., Na-Bangchang K., Kuesap J. Evaluation of Plasmodium vivax isolates in Thailand using polymorphic markers Plasmodium merozoite surface protein (PvMSP) 1 and PvMSP3. Parasitol. Res. 2018;117:3965–3978. doi: 10.1007/s00436-018-6106-1. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanapongpichat S., Khammanee T., Sawangjaroen N., Buncherd H., Tun A.W. Genetic diversity of Plasmodium vivax in clinical isolates from southern Thailand using PvMSP1, PvMSP3 (PvMSP3alpha, PvMSP3beta) genes and eight microsatellite markers. Kor. J. Parasitol. 2019;57:469–479. doi: 10.3347/kjp.2019.57.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Bai Y., Xiang Z., Zeng W., Wu Y., Zhao H., et al. Genetic diversity of Plasmodium vivax populations from the China-Myanmar border identified by genotyping merozoite surface protein markers. Trop. Med. Health. 2023;51:2. doi: 10.1186/s41182-022-00492-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; Geneva, Switzerland: 2013. World Malaria Report 2013.https://www.who.int/publications/i/item/9789241564694 [Google Scholar]
- WHO . World Health Organization; Geneva, Switzerland: 2015. Global Technical Strategy for Malaria 2016–2030. [Google Scholar]
- WHO . World Health Organization; Geneva, Switzerland: 2015. World Malaria Report 2015.https://www.who.int/publications/i/item/9789241565158 [Google Scholar]
- WHO . World Health Organization; Geneva, Switzerland: 2023. World Malaria Report 2023.https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2023 [Google Scholar]
- Yang Z., Miao J., Huang Y., Li X., Putaporntip C., Jongwutiwes S., et al. Genetic structures of geographically distinct Plasmodium vivax populations assessed by PCR/RFLP analysis of the merozoite surface protein 3beta gene. Acta Trop. 2006;100:205–212. doi: 10.1016/j.actatropica.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakeri S., Safi N., Afsharpad M., Butt W., Ghasemi F., Mehrizi A.A., et al. Genetic structure of Plasmodium vivax isolates from two malaria endemic areas in Afghanistan. Acta Trop. 2010;113:12–19. doi: 10.1016/j.actatropica.2009.08.025. [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., Tian B., Huang Y.H., Gu B., Ju P., Luo Y., Tang J., Wang L. Classification and prediction of Klebsiella pneumoniae strains with different MLST allelic profiles via SERS spectral analysis. PeerJ. 2023;11 doi: 10.7717/peerj.16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary file. The newly generated sequences were submitted to the GenBank database under the accession numbers: PP795732-PP795733 (Pfama1); PP681141, PP795719 (Pfglurp); PP795720-PP795721, PP913947-PP913970, PP934608 (Pvmsp3α); and P795722-PP795731, PP886053-PP886070 (Pvmsp3β).