Abstract
Human gut microbial species are crucial for dietary metabolism and biosynthesis of micronutrients. Digested products are utilised by the host as well as several gut bacterial species. These species are influenced by various factors such as diet, age, geographical location, and ethnicity. India is home to the largest human population in the world. It is spread across diverse ecological and geographical locations. With variable dietary habits and lifestyles, Indians have unique gut microbial composition. This review captures contrasting and common trends of gut bacterial community establishment in infants (born through different modes of delivery), and how that bacterial community manifests itself along infancy, through old age between Indian and global populations. Because dysbiosis of the gut community structure is associated with various diseases, this review also highlights the common and unique bacterial species associated with various communicable as well as noncommunicable diseases such as diarrhoea, amoebiasis, malnutrition, type 2 diabetes, obesity, colorectal cancer, inflammatory bowel disease, and gut inflammation and damage to the brain in the global and Indian population.
Keywords: human gut microbiome, gut microbiome development, diet and lifestyle, dysbiosis, communicable and noncommunicable diseases
Introduction
The human microbiome is a complex microbial community structure that resides at different body sites, namely skin, oral cavity, gastrointestinal tract (GIT), respiratory tract, and vagina. However, microbial diversity and richness vary across all body sites (Costello et al., 2009; Human Microbiome Project Consortium, 2012) The community belongs to several domains of life, that is, bacteria, viruses, fungi, archaea, and protists (Shreiner et al., 2015; Sender et al., 2016). Unlike bacterial species, others have been poorly studied for their role in human physiology (Matijašić et al., 2020). The extensively researched gut bacterial species outnumbers human body cells and genes by 10 and 100 times, respectively (Bull and Plummer, 2014). Its role in breakdown of complex carbohydrates into short-chain fatty acids (SCFAs) such as acetate, propionate, and butyrate, branched-chain amino acids, hydrolysis of polyphenols, and biosynthesis of Vitamin K and water-soluble B-vitamins is well explored (Magnúsdóttir et al., 2015; Rowland et al., 2018; Sharma et al., 2019; Chandel et al., 2023).
The microbiome composition varies across different parts of the GIT with distinct community structures along the mucosal-lumen axis (Bäckhed et al., 2012; Ruan et al., 2020), in different development stages of a particular individual (Rinninella et al., 2019), and among individuals (Human Microbiome Project Consortium, 2012; Rinninella et al., 2019). A healthy human gut microbiome is a stable community composed of a defined set of microbial species, which resist change or return to an equilibrium state following perturbation (Bäckhed et al., 2012). It consists of a few phyla with a relatively higher abundance (Bacillota, Bacteroidota, Actinomycetota, and Pseudomonadota) as compared to several others (Fusobacteriota, Tenericutes, Spirochaetes, Cyanobacteria, Verrucomicrobia, and TM7) (Human Microbiome Project Consortium, 2012). Some of the highly abundant and/or prevalent genera include Bacteroides, Eubacterium, Faecalibacterium, Alistipes, Ruminococcus, Clostridium, Prevotella, Roseburia, and Blautia, and highly abundant species include Faecalibacterium prausnitzii, Oscillospira guillermondii, and Blautia obeum (Arumugam et al., 2011; Piquer-Esteban et al., 2022; Qin et al., 2010; Ruan et al., 2020). They are also the core taxa of a healthy individual (Qin et al., 2010). However, there is little consensus about how the taxonomic core microbiome should be quantified, as different researchers use different quantification metrics (Neu, 2021). For instance, with 90% and 0.01% threshold of prevalence and relative abundance, respectively, only Faecalibacterium prausnitzii was observed as the core microbiome across Indian cohorts from multiple locations (Chandel et al., 2023). Moreover, the studies on inferring core gut microbiome have not fully captured the variability in microbiome composition due to various factors like geographical location, race, diet, lifestyle, and age.
Large-scale studies on human gut microbiomes have largely been from the U.S. and European countries (Human Microbiome Project Consortium, 2012). But if we look at India, it has the largest human population and is spread across six different physiographic regions, and has a huge diversity in habitat, lifestyle, ethnicity, and dietary habits, which makes the Indian gut microbiota an interesting community to study. While population-specific variations in gut microbial composition have earlier been reported (Yatsunenko et al., 2012), a recent study captured the uniqueness of the Indian gut microbiome (Dhakan et al., 2019). Not only a substantially large number (943,395) of unique genes were observed in Indian samples, but a few species belonging to genera Prevotella, Mitsuokella, Dialister, Megasphaera, and Lactobacillus were also found highly associated with the Indian population (Dhakan et al., 2019).
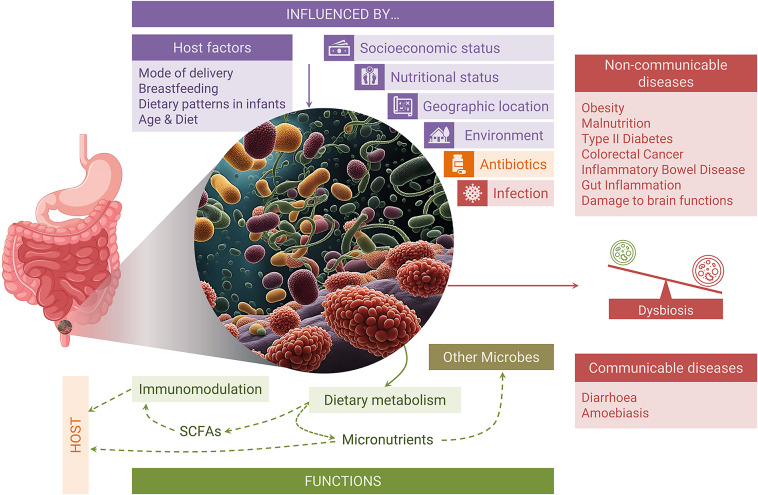
Pulipati et al. (2020) recently analysed the features, and determinants of Indian gut microbiota and compared it with worldwide data (Pulipati et al., 2020). However, the association of gut microbiota with human health and various infectious/noninfectious diseases in the Indian population has not been systematically reviewed. This review provides Indian population-specific characteristics of the gut microbiome at different developmental stages of life, discusses the factors that shape the gut microbiome, and their association with noninfectious and infectious diseases while comparing them with the findings or trends in global populations (Figure 1).
Figure 1.
Pictorial representation of the key aspects discussed in this review article.
Establishment of gut microbiome
Pregnancy, birth, and infancy
The sterile womb hypothesis and microbial community acquisition from the external environment (Mackie et al., 1999) were challenged when microbes were identified in the placenta, amniotic fluid, and meconium (Perez-Muñoz et al., 2017). It was further supported by the presence of phyla Bacillota, Pseudomonadota, and Bacteroidota and genera Enterococcus and Staphylococcus, in the meconium microbiome, which was majorly affected by maternal rather than perinatal factors (Jiménez et al., 2008; Perez-Muñoz et al., 2017; Tapiainen et al., 2018). The similarity of the placental microbial community with the oral (Walker et al., 2017), and a higher dissimilarity with the vaginal and stool microbiome, were highly unlikely the result of contamination (Wassenaar and Panigrahi, 2014; Walker et al., 2017; Cariño et al., 2021).
A Finland-based study reported highly variable gut microbiota in T3 (third trimester of pregnancy) as compared to T1, resembling a rather disease-associated dysbiosis. The T3 stage also had a lower abundance of Faecalibacterium (butyrate producer) and a higher abundance of phyla Actinomycetota and Pseudomonadota. The Pseudomonadota has often been associated with inflammation-associated dysbiosis (Koren et al., 2012) (Figure 2). In contrast, there were no significant changes in the gut community structure of the Indian population between T1 and T3; although Pseudomonadota showed a higher abundance during T3, however, this difference was not statistically significant (Kumbhare et al., 2020). There were no reported adverse effects of higher Pseudomonadota in T3 on infants’ health. The difference in the findings was attributed to either a difference in data analysis or a smaller sample size of the Indian cohort (Kumbhare et al., 2020).
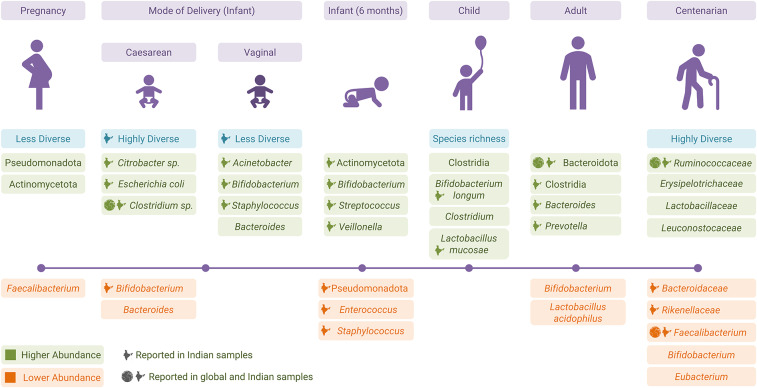
Figure 2.
Changes in the gut microbiota from pregnancy to old age.
Mode of delivery, that is, caesarean section delivery (CS) and vaginal delivery (VD), has a strong influence on infants’ gut community. CS infants from Finland and the United States showed a delay in gut microbial community colonisation and reported a lower Bacteroides abundance as compared to VD infants (Grönlund et al., 1999; Mitchell et al., 2020). The inverse correlation of Bacteroides with Streptococcus or Haemophilus in CS was the result of direct competition between the two species (Mitchell et al., 2020). Early colonisation of Bifidobacterium-like and Lactobacillus-like beneficial bacteria was seen in the VD children (Grönlund et al., 1999). Corroborating the findings from Western countries, an Indian study reported higher Bifidobacterium – a primary coloniser in VD children along with Acinetobacter sp., Staphylococcus sp. (Pandey et al., 2012). The absence of Bifidobacterium and a higher abundance of opportunistic bacteria (Citrobacter, Clostridium difficile, and E. coli) were seen in Indian CS infants (Pandey et al., 2012) (Figure 2). The exposure of CS infants to environmental microbes makes them susceptible to colonisation of undesired microbes, which results in higher microbiome diversity (Pandey et al., 2012).
Studies from Italy and the United States showed that the maternal microbiome from all body sites was the main source of the infant’s gut microbiome; however, the gut microbiome was more persistent compared to other body sites (Ferretti et al., 2018; Mitchell et al., 2020). Indian infants at 6 months of age had a higher abundance of phylum Actinomycetota, genera Bifidobacterium, Streptococcus, and Veillonella, and a lower abundance of phylum Pseudomonadota, genera Staphylococcus, and Enterococcus as compared to the birth stage (Kumbhare et al., 2020). Bifidobacterium and Streptococcus are one of the most abundant and core bacterial members, respectively, of an infant’s gut (Jost et al., 2013; Underwood et al., 2015). The role of Veillonella in infancy is poorly understood (Ferretti et al., 2018; Kumbhare et al., 2020) (Figure 2). There was a similarity between Indian infants’ and their mothers’ microbiomes, but the results were not significant.
Childhood
Three studies from Norway, Sweden, and Finland were compared with the ones available for Indian cohorts. A Norwegian study showed that a certain bacterial species pool is shared between mother and infant. Mother-associated operational taxonomic units start depleting after 3 months of age. Over the period, microbiota gets enriched with class Bacteroidia and Clostridia (Avershina et al., 2016) and species Bifidobacterium breve (Agans et al., 2011; Avershina et al., 2016; Roswall et al., 2021). Bifidobacterium breve acts as an inhibitor or is negatively associated with late-appearing microbes (Avershina et al., 2016). The first 5 years of the developmental trajectory in the Swedish population showed a higher abundance of lactic acid bacteria (Enterococcus, Streptococcus, and Lactobacillus) and gamma-Proteobacteria (Enterobacteriaceae, Citrobacter, and Serratia) along with Bifidobacterium in the first few months. At the age of 1 year, adult-associated genera such as Akkermansia, Faecalibacterium, Prevotella, Roseburia (Roswall et al., 2021) and Ruminococcus (Agans et al., 2011) become highly prevalent, and their abundance increases as they grow older (Roswall et al., 2021).
Healthy children from the south Indian slum had a higher abundance of the genera Prevotella, Bifidobacterium, and Escherichia-Shigella (Shivakumar et al., 2021). Partially in line with the Swedish population, children from southern India showed a higher abundance of Lactobacillus, Bifidobacterium, Eubacterium rectale, and Faecalibacterium prausnitzii (Balamurugan et al., 2008). A comparison of Indian and Finnish children’s microbiomes showed enrichment of Prevotella and Megasphaera in Indian children (Kumbhare et al., 2017) (Figure 2). A higher prevalence of Prevotella indicates enterotype 2 in the Indian population, which is well established in other studies as well (Dhakan et al., 2019; Kaur et al., 2020)
Adult
The Norwegian data showed that Bifidobacterium breve had a higher prevalence in the first year of life and was negatively associated with a range of adult-like species. Its disappearance suggestively drives (at least partially) the transition from infant to adult-associated gut microbiome (Avershina et al., 2016). According to a study from the Netherlands, the adult gut microbiome is stable and highly diverse compared to children, with the dominance of Blautia and Bacteroides in the former and latter groups, respectively (Radjabzadeh et al., 2020). On the contrary, data from Ohio, USA showed that it was relative abundance, not the presence–absence of specific genera that differentiated the two groups (Agans et al., 2011). The western adult gut microbiome is dominated by phyla Bacillota, Bacteroidota, Actinomycetota, and Pseudomonadota with carbohydrate metabolism remaining the dominant pathway (Human Microbiome Project Consortium, 2012).
Comparison of the Indian with Chinese populations showed no difference in diversity; however, composition and relative abundance differed (Jain et al., 2018). Both the populations were enriched with Bacillota and Actinomycetota, with fewer Bacteroides. Differences in dietary patterns led to a significantly higher abundance of Bacteroidota and Prevotella in Indians in contrast to Chinese (Jain et al., 2018). Bacterial succession from childhood to adulthood in Indians showed a decline in Bifidobacterium and Lactobacillus. Contrary to Radjabzadeh et al. (2020) and Jain et al. (2018), a higher abundance of Bacteroides during late adolescence and adulthood, and a sharp decline of Eubacterium rectale and F. prausnitzii in Indian adults were reported (Balamurugan et al., 2008; Jain et al., 2018; Radjabzadeh et al., 2020). Similar to the western microbial profile at the phylum level, Indian communities are also dominated by Bacillota, Bacteroidota, Actinomycetota, and Pseudomonadota (Figure 2) (Ramakrishna, 2013; Das et al., 2018).
Elderly
The transition from a stable and diverse bacterial community in adults to a less diverse one in the elderly population was compared between four global studies (China, Italy, Ireland, and Japan) and available Indian studies. An increase in Pseudomonadota species was reported in several studies (Rampelli et al., 2013; Kumar et al., 2016; Kong et al., 2018). An Ireland-based study reported significantly higher dominance of Prevotella and Ruminococcus in the adults and Alistipes and Oscillibacter in the elderly group (Claesson et al., 2012). The study done on the same cohort showed Bacteroides, Alistipes, Parabacteroides, Faecalibacterium, and Ruminococcus as the core genera in the elderly population (Jeffery et al., 2015). An overall decrease in SCFAs production, shift from proteolytic to saccharolytic fermentation, loss of organisms such as Eubacterium, Bifidobacterium, and Faecalibacterium, and increased abundance of pathogens such as Escherichia-Shigella were considered as functions of the ageing process (Kumar et al., 2016; Kong et al., 2018).
In line with the results from other countries, an Indian study done by Tuikhar et al. (2019) also reported a higher diversity in the Ruminococcaceae family in centenarians (~100 years old). Direct comparison with samples from Italy, Japan, and China in the same study also showed similar results. A decrease in the abundance of Faecalibacterium was also observed in the Indian population. Species from genera Akkermansia, Alistipes, and Ruminococcoaceae D16 were reported as signatures of longevity in all four populations. Akkermansia was reported to be associated with health and anti-inflammatory activity. The unclassified species Ruminococcoaceae D16 was reported to be a butyrate producer in herbivorous and omnivorous animals (Figure 2) (Tuikhar et al., 2019; Badal et al., 2020).
Factors affecting gut microbiome composition
Diet
Trends from three studies done on global cohorts (the United States, Japan, Europe, and Africa) were compared with available data on Indian cohorts. The long-term effect of diet has a huge impact on microbial community structure; however, short-term (5 days) consumption of entirely plant-based or animal-based foods has also rapidly changed the gut community structure (David et al., 2013). Animal-based diet showed a higher abundance of bile-tolerant bacteria such as Bacteroides, Alistipes, and Bilophila (David et al., 2013; Pareek et al., 2019), whereas the higher abundance of Bacillota that metabolise plant polysaccharides such as Roseburia, Eubacterium rectale, and Ruminococcus bromii reported in plant-based diet consuming individuals (David et al., 2013). Another study done by De Filippo et al. (2010) on European and African children, consuming western and rural diets, respectively, showed partial overlapping patterns. A higher abundance of phylum Bacteroidota (Prevotella) and SCFAs, and depletion of phylum Bacillota and family Enterobacteriaceae (Shigella and Escherichia) reported in Africans (De Filippo et al., 2010). In line with the above results, the Indian population consuming a plant-based diet had a higher abundance of Prevotella (Dhakan et al., 2019; Jain et al., 2018; Kaur et al., 2020). It was also reported to have higher lipopolysaccharide pathway genes and serum BCAA levels; Latter is because of the presence of fewer in-ward transporters in bacteria; hence; they get absorbed in serum (Dhakan et al., 2019). In contrast, the omnivorous group showed higher bacterial BCAA transporters and hence their high abundance in faecal matter (Dhakan et al., 2019). Partially overlapping results on the association of omnivorous diet with butyrate-producing bacteria such as Roseburia–E. Rectale (Kabeerdoss et al., 2012), Bacteroides, Ruminococcus, and Faecalibacterium, and enrichment of SCFAs biosynthesis pathways were also observed (Dhakan et al., 2019). Another Indian study by Bamola et al. (2017), however, presented a completely different picture, reporting a higher Bacteroidota to Bacillota ratio in the non-vegetarian group as compared to vegetarians. It was not clearly explained if the abundance profile comparison of taxa between the vegetarian and omnivorous groups was statistically significant (sequence data involved just 96 sequences per group) (Bamola et al., 2017).
Lifestyle
Despite being crucial in maintaining health, little is known to what extent modernisation has impacted gut microbiota structure. Less affected tribal populations still use traditional ways to survive (Shetty et al., 2013). Here, the comparison of Indian studies was made with data from Tanzania, America, Malawi, Mongolia, and Italy. Yanomami, who live a hunter-gatherer lifestyle similar to human ancestors, not exposed to antibiotics, were first contacted in ~1960 in Venezuela. Their gut composition showed significantly huge diversity than the U.S. population, with high Prevotella and low Bacteroides abundance, similar to that in African hunter-gatherers, Guahibo Amerindians, and Malawians (Clemente et al., 2015). They also showed high functional diversity, gene prevalence, and less intragroup variation as compared to the United States (Clemente et al., 2015). An interesting pattern of seasonal variation in community structure emerged in Hadza hunter-gatherers of Tanzania. This seasonal variation was based on food acquisition activities which were affected by the local environment and type of food availability in two different seasons. Bacillota, for instance, remained stable in both dry (May–October) and wet (November–April) seasons; however, the abundance of family Prevotellace significantly declined during the wet season compared to the dry season (Smits et al., 2017). Surprisingly, seasonally volatile taxa in Hadza differentiated this traditional population from the industrialised one, indicating a decrease in the prevalence and abundance of some taxa in modernised populations (Smits et al., 2017). Prevotella was the dominant genus in Mongolian, Amerindian, and Malawian groups, while Faecalibacterium was in the American, Italian, and Hadza populations (Dehingia et al., 2015). India, with six major physiographic divisions, namely The Himalayan mountains, Northern plains, Peninsular plateau, Indian desert, Coastal plains, and Islands along with multiple ethnic groups living in each division, have many distinct dietary habits and lifestyles (urban, rural, tribals from forests, hills, hot deserts, cold deserts, remote islands, mangroves, etc.). While there are multiple studies on tribal populations, no proper study has been done on Indian ethnic groups. Similar to the trends mentioned above, gut bacterial profiles of tribal populations from four different geographical locations, namely Assam, Telangana, Manipur, and Sikkim, showed the dominance of Prevotella. Likewise, a comparison of three different tribes from Mongoloid (Ladakh), Caucasoid (Jaisalmer), and Australoid (Khargone) ancestry revealed that despite the differences in ethnicity and geographical locations, genera Prevotella, Bifidobacterium, Bacteroides, Eubacterium, and Faecalibacterium were abundant in overall populations (Kaur et al., 2020; Hazarika et al., 2022). A small cohort size study in Tamil Nadu, India, revealed a higher Bacillota/Bacteroidota ratio and higher Actinomycetota abundance in the rural population than in tribal (Ramadass et al., 2017). A study on the Nicobarese community, one of the six tribal communities of Andaman and Nicobar Islands, revealed that their lifestyle has a profound impact on the gut bacterial composition, where the remote subset of the community had Bacteroides–Prevotella–Porphyromonas as the dominant bacterial group, while the rural and urban subsets had Clostridium coccoides, Eubacterium rectale, and Bifidobacterium as the predominant bacterial groups, respectively (Anwesh et al., 2016).
Antibiotic usage
The benefits of antibiotic usage in humans as well as livestock come at a cost with the inevitable evolution of antibiotic-resistant variants and the collateral damaging effect of antibiotics on commensal bacteria (Blaser, 2016). A longitudinal study conducted on 12 individuals in Denmark observed that antibiotic usage reduces microbial diversity, especially that of butyrate-producing species with a restoration period of 1.5 months to obtain the baseline composition (Palleja et al., 2018) A similar restoration period of 1 month was observed in a study which included 39 children from Finland (Yassour et al., 2016). However, Palleja et al. (2018) observed that several common species were not restored even after 1.5 months and until the end of their study period which was 180 days. Moreover, disruptions in the balance of gut microbial species lead to an increase in pathobionts such as Clostridium difficile (Buffie and Pamer 2013). Another study conducted on 21 participants from Spain, who were treated with broad-spectrum antibiotics indicated a reduction in bacterial diversity due to the elimination of antibiotic-susceptible bacteria and an increase in the overall microbial load due to the replacement and rapid multiplication of antibiotic-resistant bacterial species (Panda et al., 2014). Studies conducted across Canada and the United States provide increasing evidence that early antibiotic exposure in life is associated with obesity, diabetes, inflammatory bowel diseases (IBDs), allergies, and asthma (Arrieta et al., 2015; Azad et al., 2014; Bokulich et al., 2016) in the later stages of life. Whereas, the short-term and medium-term consequences include antibiotic-associated diarrhoea, C. difficile infections, and H. pylori-related gut dysbiosis (Ramirez et al., 2020).
In the Indian context, a study from southern India, which included 120 infants, revealed that azithromycin has a moderate impact on their gut microbiota (Parker et al., 2017). This study indicated a decrease in the microbial diversity and abundance during antibiotic intake; however, no effect was observed on the maturity of the microbiota. Although studies depicting the direct effect of antibiotic usage on the gut microbiota may be rare in India, the other major concern of gut microbiota acting as a reservoir for antibiotic resistance genes has been reported in various studies. Antibiotic abuse is a common phenomenon in low- and middle-income countries. In India, the usage of antibiotics has increased from 3.2 billion defined daily doses in 2000 to 6.5 billion in 2015, an increase of 103% (Klein et al., 2018). In such situations, the human gut microbiome acts as a reservoir of antibiotic-resistance genes, capable of transferring the genes rapidly to transient pathogens within the holobiont through horizontal gene transfer (Sitaraman 2018; Groussin et al., 2021). An insightful gut microbiome study among 18 Swedish students who travelled to India on an exchange programme showed that 12 of the students acquired ESBL-producing E. coli, even without taking antibiotics (Bengtsson-Palme et al., 2015). Another study on 122 travellers from the Netherlands to India revealed increased acquisition rates of beta-lactam and quinolone resistance genes (von Wintersdorff et al., 2014). This emphasises the potential for antibiotic resistance transmission in regions with heightened antibiotic use. Furthermore, a study conducted in 2019 among 207 healthy individuals from Chandigarh, India, reported that 70.5% of the stool samples had antibiotic-resistant isolates of which 2.4% were multi-drug resistant and the most common genes identified were β-lactamases (Gupta et al. 2019b). Similarly, a high prevalence of β-lactamases was observed in the rectal swabs collected from neonates and mothers in India (Carvalho et al. 2022). A study on 25 healthy individuals from Kolkata, India, reported that all the samples carried aminoglycoside resistance markers and most of them showed resistance to tetC and sul-2 genes (De et al. 2023).
Gut microbiome association with health and diseases
Gut microbiota has a crucial role in regulating gut homeostasis, maintaining intestinal barrier and immunity by metabolising complex dietary substrates, and synthesising micronutrients. The microbial community dysbiosis or modulation could lead to or associate with various noncommunicable and communicable diseases. Studies across the globe and from India have suggested their role/association in malnourishment, diabetes, obesity, inflammatory diseases, neurological disorders, diarrhoea, amoebiasis, and so forth.
Noncommunicable diseases
Malnourishment
Excess, deficiency, and/or imbalanced micronutrients and energy intake lead to malnutrition. The various forms of malnutrition include undernutrition, micronutrient-related malnutrition, overweight, obesity, and other diet-related diseases. Around 45% of children’s deaths are caused by malnutrition globally (Fact Sheets – Malnutrition, n.d.).
A comparison of four global studies from Indonesia, Mexico, Bangladesh, South Africa, Guatemala, and Malawi with Indian studies provides evidence that gut microbiota dysbiosis could also predispose to various forms of malnutrition. A study from Indonesia reported low Bacteroidota and high Bacillota in stunted children of 3–5 years (Surono et al., 2021), which was also true in undernourished and obese children from Mexico (Méndez-Salazar et al., 2018). High species richness and diversity along with significant enrichment of Prevotella 9 in healthy children correlated with their height and high dietary fibre intake (Méndez-Salazar et al., 2018; Surono et al., 2021). However, it has not been confirmed if this species could revert the malnutrition. Malnourished and poorly growing Bangladeshi children had a higher abundance of Pseudomonadota species such as Klebsiella, Escherichia/Shigella, and a lower abundance of Prevotella, compared to healthy controls (Monira et al., 2011, Perin et al., 2020) (Table 1). The gastrointestinal infection caused by these pathogenic species could lead to nutrient malabsorption (Monira et al., 2011), likely by dissolution of the brush border membrane and loss of microvilli structure due to lesions induced by adherence of pathogens to the intestine (Neto and Scaletsky, 2000). These pathogens are also associated with poor growth, and inflammation and can also detoxify nitric oxide, which is produced by colonic epithelial cells as an inflammatory response (Perin et al., 2020). Million et al., 2017 also reviewed the link between malnutrition and gut microbiota in studies from countries including South Africa, Guatemala, Bangladesh, Malawi, and India, and reported early depletion of Bifidobacterium longum as the first step in severe acute malnutrition.
Table 1.
Common and/or unique trends observed between gut microbiome of Indian and global populations in noncommunicable and communicable diseases
| Phenotype | Country | Sample size | Age group | Sequenced region | Sequencing platform | High | Low | References |
|---|---|---|---|---|---|---|---|---|
| Malnutrition | Indonesia | Healthy = 53, Stunted = 78 | 3–5 years | V3–V4 | Illumina Miseq | p–Bacillota | p–Bacteroidota, g–Prevotella 9 | Surono et al. (2021) |
| Mexico | Healthy = 12, Undernourished = 12, Obese = 12 | 9–11 years | V3–V4 | Illumina Miseq | p–Pseudomonadota | alpha diversity, p–Bacteroidota | Méndez–Salazar et al. (2018) | |
| Bangladesh | Healthy = 7, Malnourished = 7 | 2–3 years | V5–V6 | 454 parallel sequencing | p–Pseudomonadota, g–Klebsiella, Escherichia, Neisseria | p–Bacteroidota | Monira et al. (2011) | |
| Bangladesh | Cases and controls = 68 | 6–31 months | V1–V3 | Illumina Miseq | p–Pseudomonadota, g–Escherichia/Shigella | g–Prevotella | Perin et al. (2020) | |
| India | Stunted, wasted, and underweight = 41 | 18–12 months | V3–V4 | llumina HiSeq2500 | g–Prevotella 9, Bifidobacterium, Escherichia–Shigella | Shivakumar et al. (2021) | ||
| India | Control = 10, Stunted = 10 | Birth to 2 years | V4 | Illumina MiSeq | g–Desulfovibrio, o–Campylobacterales | s–Bifidobacterium longum, Lactobacillus mucosae | Dinh et al. (2016) | |
| India | Undernourished = 53 | 10–18 months | V3–V4 | Illumina MiSeq | p–Pseudomonadota, o–Aeromonadales, g–Enterococcus, g– Anaerococcus, g–Vibrio | Huey et al. (2020) | ||
| Obesity | Finland | Normal–weight women = 36, Overweight women = 18 | ~30 years | fluorescent in situ hybridisation coupled with flow cytometry (FCM–FISH) and by quantitative real–time polymerase chain reaction (qPCR) | g–Bacteroides, g–Staphylococcus | g–Bifidobacterium | Collado et al. (2008) | |
| European countries (Cyprus, Estonia, Germany, Hungary, and Sweden) | 70 subjects (2 time points), Time point 0: Normal = 70, Time point 1: Normal = 34, Obese = 36 | 2–9 years | V3–V4 | Illumina MiSeq | p–Pseudomonadota, f–Bacteroidaceae | diversity, f–Clostridiaceae, f–Ruminococcaceae, f–Prevotellaceae | Rampelli et al. (2018) | |
| Germany | Normal weight = 30, Overweight = 35, Obese = 33 | 14–74 years | qPCR to detect a group of commensals | p–Bacteroidota, g–Bacteroides | g–Bifidobacterium, s–Ruminococcus flavefaciens | Schwiertz et al. (2010) | ||
| India | 20 (5 lean, 5 normal, 5 obese, 5 surgically treated obese) | 21–62 years | 900 bases amplicon | BigDye™ Terminator Cycle Sequencing Ready Reaction Kit v3.1 in an automated 3730 DNA analyser | g–Bacteroides | Ppatil et al. (2012) | ||
| India | Normal = 13, Obese = 15 | 11–14 years | 16S rRNA | qPCR | s–F. prausnitzii | Balamurugan et al. (2010) | ||
| India | Normal = 10, Obese = 10 | NA | V3 | Denaturing Gradient Gel Electrophoresis analysed in Gel Compar II version 6.6 software (Sequencing platform was not mentioned) | s–Collinsella aerofaciens, g–Dialister, g– Eubacterium, g–Mitsuokella, g–Victivallis | Diversity | Bahadur et al. (2021) | |
| Type 2 diabetes | West Africa | Controls = 193, Cases = 98 | 57 years (mean) | V4 | Illumina MiSeq | s–Desulfovibrio piger, g–Prevotella, g–Peptostreptococcus, g–Eubacterium | f–Clostridiaceae, f–Peptostreptococcaceaea | Doumatey et al. (2020) |
| China | Normal glucose tolerance = 97, Prediabetese patients = 80, Newly diagnosed treatment naive T2D patient = 77 | 62.53 years (mean) | WGS | Combinatorial probe–anchor synthesis (cPAS)–based BGISEQ–500 sequencing | s–Dialister invisus, s–Roseburia hominis | Zhong et al. (2019) | ||
| Denmark and India | Indian non–diabetics = 137, Danish non–diabetics = 138, Indian T2D patients = 157, Danish diabetic patients = 141 | 35–74 years | V1–V5 | 454 GS FLX+ pyrosequencer platform | f–Lachnospiraceae | g–Subdoligranulum and Butyricicoccus | Alvarez–Silva et al. (2021) | |
| Meta–analysis (Denmark, Sweden, China) | Danish non–diabetic = 277, Swedish non–diabetic = 92, Chinese non–diabetic = 185, Danish T2D = 75, T1D = 31, Swedish T2D = 52, Chinese T2D = 71 | 35–75 years | WGS + 16S rRNA | Illumina shotgun sequencing | metformin untreated: s–Roseburia spp., Subdoligranulum spp | Forslund et al. (2015) | ||
| China | Non–diabetic = 185, Diabetic = 183 | 13–86 years | WGS | Illumin aHiSeq 2000 | s–Bacteroides caccae, Clostridium hathewayi, Clostridium ramosum, Clostridium symbiosum, Eggerthella lenta, and Escherichia coli | s–Clostridiales sp. SS3/4, Eubacterium rectale, Faecalibacterium prausnitzii, Roseburia intestinalis, and Roseburia inulinivorans | Wang et al. (2012) | |
| India | Healthy = 19, New diabetic patients = 14, Known diabetic patients = 16 | 49.37 years (mean) | V3 | Ion Torrent | g–Lactobacillus, p–Bacillota | s–P. copri, s–Faecalibacterium prausnitzii, f–Ruminococcaceae, Lachnospiraceae | Bhute et al. (2017) | |
| India | Healthy = 9, T1D = 8, T2D = 10, T3cD = 17 | 18–60 years (Healthy), patient’s age was not mentioned | V3–V4 | Illumina MiSeq | Diversity, g–Fecalibacterium, Eubacterium, and Ruminococcus | Talukdar et al. (2021) | ||
| India | Healthy = 30, T2D and no diabetic retinopathy (DR) = 25, T2D + DR = 28 | 54.86 years (mean) | V3–V4 | Illumina HiSeq | g–Escherichia, Enterobacter, Methanobrevibacter, and Treponema | g–Roseburia, Lachnospira, Sutterella, Coprococcus, Phascolarctobacterium, Haemophilus, Blautia, Comamonas, Anaerostipes, and Turicibacter | Das et al. (2021) | |
| Colorectal cancer | China | Healthy = 56, Patients = 46 | 40–77 years | V3 | 454 pyrosequencing | s–Bacteroides fragilis, g–Escherichia/Shigella, Klebsiella, Streptococcus, Enterococcus, Peptostreptococcus, Eggerthella, Fusobacterium | s–Bacteroides uniformis, Roseburia spp. and Eubacterium spp. | T. Wang et al. (2012) |
| China | Healthy = 130, Patients = 130 | 59.1 years (mean) | V3–V4 | Illumina MiSeq | s–Peptostreptococcus stomatis, Fusobacterium nucleatum, etc. | s–Roseburia faecis, Ruminococcus lactaris, Eubacterium desmolans, Streptococcus salivarius, etc. | Zhang et al. (2018) | |
| China | Patients = 23 (tumour tissue and surrounding healthy tissue) (early and late stages) | 49–70 years | V4 | Illumina MiSeq | late stage: g–Akkermansia, Fusobacterium, Peptostreptococcus, Streptococcus, and Ruminococcus | Pan et al. (2020) | ||
| USA | Healthy = 52, Patients = 52 | 61 years (mean) | WGS | Illumina HiSeq 2000/2500 | g–Fusobacterium, Porphyromonas | Vogtmann et al. (2016) | ||
| India | Healthy = 30, Patients = 30 | Not mentioned | WGS | Illumina NextSeq 500 | Diversity, g–Bacteroides, s–Flavonifractor plautii | Gupta et al. (2019a) | ||
| India | Patients = 5 (healthy tissue = 5, tumour tissue = 5) | 40–83 years | V3–V4 | Ion 520 OT2 | s–Bacteroides massiliensis, Alistipes sp. Alistipes onderdonkii, Bifidobacterium pseudocatenulatum, Corynebacterium appendicis, and Acidiphilium sp. | s–Bacillus sp., Veillonella atypica, etc. | Hasan et al. (2022) | |
| Inflammatory bowel diseases | USA | Non–IBD = 27, UC = 38, CD = 67 | 27.5 years (mean) | WGS | Illumina HiSeq2500 | s–E. coli, Ruminococcus torques and Ruminococcus gnavus | Faecalibacterium prausnitzii, and Roseburia hominis | Lloyd–Price et al. (2019) |
| USA and Netherlands | Non–IBD = 34, UC = 53, CD = 68 | >18 years | WGS | Illumina HiSeq2500 | g–Unclassified Roseburia | s–Roseburia hominis, Dorea formicigenerans and Ruminococcus obeum | Franzosa et al. (2019) | |
| China | Healthy = 30, IBD patients = 18 | 37 years (mean) | V3–V4 | Illumina MiSeq | p–Pseudomonadota, Fusobacteriota, g–Escherichia_Shigella | s–Eubacterium coprostanoligenes, Eubacterium hallii group | T. Wang et al. (2022) | |
| India | Health control = 17, CD = 20, UC = 22 | 33.6 years (mean) | 16S rRNA gene sequences specific to C. leptum group | Not mentioned | s–Faecalibacterium prausnitzii, C. leptum group | Kabeerdoss et al. (2013) | ||
| India | Control individuals (haemorrhoid patients only) = 14, UC patients (severe: n = 12, moderate: n = 6, remission: n = 8) = 26 | 36 years (mean) | Clostridium cluster population targeted by 16S rRNA gene | Not mentioned | s–Faecalibacterium prausnitzii, R. intestinalis, a member of the C. coccoides group, reduced SCFA | Kumari et al. (2013) | ||
| India | Control = 65, UC = 72, CD = 12 | 38 years (mean) | Real–time analysis using 16S rRNA | g–Eubacterium, Peptostreptococcus | g–Lactobacillus, Ruminococcus, and Bifidobacterium, C. leptum group | Verma et al. (2010) | ||
| Gut inflammation and damage to the brain function | ||||||||
| ASD | Italy | Healthy control = 14, ASD patients = 11 | 35 months (mean) | V3–V4 | Illumina Miseq | p–Bacteroidota, Proteobacteria, s–F. prausnitzii, B. uniformis and B. vulgatus and P. distasonis, f–Enterobacteriaceae and Pasteurellaceae | p–Actinomycetota, s–Bifidobacterium longum and Eggerthella lenta | Coretti et al. (2018) |
| China | Healthy control = 48, ASD patients = 48 | 2–7 years | V3–V4 | Illumina Miseq | s–P. copri, Bacteroides coprocola, B. vulgatus, Eubacterium eligens, Roseburia faecis | s–A. muciniphila, Dialister invisus, Escherichia coli, B. fragilis, Haemophilus parainfluenzae, Flavonifractor plautii | Zou et al. (2020) | |
| China | Healthy Control = 18, ASD patients = 71 | 3–6 years | V1–V2 | Illumina Miseq | Eisenbergiella, Klebsiella, Faecalibacterium, and Blautia | Escherichia, Shigella, Veillonella, Akkermansia, Provindencia, Dialister, Bifidobacterium, Streptococcus | Ye et al. (2021) | |
| India | Family–matched healthy = 24, ASD children = 30 | 3–16 years | V3 | Illumin NextSeq500 | p–Bacillota, g–Lactobacillus (f–Lactobacillaceae), Bifidobacterium (f–Bifidobacteraceae), Megasphaera, and Mitsuokella (f–Veillonellaceae) | f–Prevotellaceae, g–Faecalibacterium and Roseburia | Pulikkan et al. (2018) | |
| PD | China | Healthy control = 114, ASD patients = 106 (early stage = 48, advanced stage = 58) | 67.6 years (mean) | V3–V4 | Illumina Miseq | In advanced PD patients: p–Desulfobacterota, f–Lachnospiraceae, Desulfovibrionaceae, g–Parasutterella | In advanced PD patients: g–Subdoligranulum | Zhang et al. (2022) |
| Luxembourg | Healthy control = 162, PD patients = 147 | 66.3 years (mean) | V3–V4 | Illumina Miseq | Akkermansia muciniphila, Biolophila, Christensenella, Lactobacillus, Christensenella, and Lactobacillus | Turicibacter | Baldini et al. (2020) | |
| Germany | Healthy control = 25, PD patients = 34 | V4–V5 | Ion Torrent PGM | Clostridiales family XI, Peptoniphilus | Faecalibacterium and Fusicatenibacter | Weis et al. (2019) | ||
| Alzheimer’s disease | Italy | No brain amyloidosis and no cognitive impairment = 10, cognitively impaired patients with amyloidosis = 40, cognitively impaired patients with NO brain amyloidosis = 33 | 69.6 years (mean) | Selected bacterial DNA quantification using the Microbial DNA qPCR Assay Kit | Escherichia/Shigella | E. rectale | Cattaneo et al. (2017) | |
| USA | Non–demented individuals = 25, Dementia due to AD = 25 | 70.3 years (mean) | V4 | Illumina Miseq | p–Bacteroidota, g–Bacteroides, Blautia, Phascolarctobacterium, Alistipes, Bilophila | alpha diversity, –p Bacillota, Actinomycetota, g–Bifidobacterium, Adlercreutzia, SMB53, Dialister, Clostridium, Turicibacter, and cc115 | Vogt et al. (2017) | |
| Diarrhoea | Bangladesh | Time–series metagenomic study with 7 patients, 50 healthy children, 12 healthy adult males | NA | V4 | Illumina Miseq | s–R. obeum restricts V. cholerae colonisation | Hsiao et al. (2014) | |
| Bangladesh | Patients’ household members who shared a cooking pot were defined as contacts (n = 27), cholera cohort 1 = 13, cholera cohort 2 = 10 | ≥6 months | 16S rRNA gene (V4) and WGS sequencing | Illumina HiSeq | Microbial succession follows secretory diarrheal illness in humans | David et al. (2015) | ||
| India | Healthy control = 0, Patients = 20 | 8 months to 56 years | V3–V4, WGS of 5 samples | Illumina MiSeq | p–Bacillota, Presence of s–V. cholerae, Helicobacter pylori, Eschericia sp. | p–Bacteroidota, significantly negative correlation between f–Enterobacteriaceae and Lachnospiraceae and Enterobacteriaceae and Ruminococcaceae | De et al. (2020) | |
| India | 46 children during an episode of acute diarrhoea, immediately after recovery from diarrhoea, and 3 months after recovery | 3 months to 5 years | 16srRNA gene (rDNA) sequences of specific bacterial group | qPCR | Bacteroides–Prevotella–Porphyromonas group, s–Eubacterium rectale, Faecalibacterium prauznitzii significantly less abundant during or immediately after diarrhoea than during normal health | Balamurugan et al. (2008) | ||
| India | Healthy infant = 1, diarrhoea infected infants = 3 | 3–18 months | V3 | Illumina MiSeq | p–Pseudomonadota, g–Klebsiella, Haemophilus, Rothia, Granulicatella, Chelonobacter and Vibrio species were identified as key pathogenic lineages in diarrheal samples | p–Bacillota, Bacteroidota | Thakur et al. (2018) | |
| India | 105 Central Indian participants comprising 35 rural (12 with diarrhoea) and 70 urban (46 with diarrhoea) | 38.8 years (mean) | WGS | Illumina | Rural habitants have g–Prevotella–dominant microbiome compared with the urban population. Urbanisation is associated with functional enrichment of genes involved in xenobiotic and lipid metabolism, have a much higher burden of AMR overall. | Monaghan et al. (2020) | ||
| Amoebiasis | Bangladesh | Uninfected = 85, Infected = 307 | Birth to 2 years | qPCR | Prevotella copri | Gilchrist et al. (2016) | ||
| Japan | Asymptomatic infection = 13, Symptomatic infection = 51 | 43 years (mean) | V3–V4 | Illumina Miseq | f–Streptococcaceae | f–Ruminococcaceae, Coriobacteriaceae, and Clostridiaceae, s–Collinsella aerofaciens | Yanagawa et al. (2021) | |
| India | Healthy = 22, chronic/acute diarrheal patients = 550 | 21–40 years | 16S rRNA | qPCR | g–Bifidobacterium | g–Bacteroides, Eubacterium, C. leptum subgroup, C. coccoides, Lactobacillus | Verma et al. (2012) | |
| India | Healthy = 29, E. histolytica positive patients = 14 | 15–69 years | V1–V5 | Illumina HiSeq 2500 | g–Escherichia, Klebsiella, and Ruminococcus | g–Prevotella, Sutterella, and Collinsella | Iyer et al. (2023) | |
An Indian study showed enrichment of bacterial genera Prevotella 7, Prevotella 9, and Sutterella, and depletion of Clostridiaceae 1 family, Intestinibacter and Fusicatenibacter genera and Bifidobacterium longum subsp longum species in stunted children compared to non-stunted children (Shivakumar et al., 2021). This conflicting trend (of Prevotella genera in malnourished children) in Shivakumar et al. (2021), which was also observed in Kristensen et al. (2016), could be either due to the difference in the age group of children being compared (<2 years vs. 3–5 years) or due to dietary differences between the cohorts, which needs further examination (Kristensen et al. (2016); Shivakumar et al., 2021). However, a higher abundance of pathogenic genera Escherichia/Shigella was in sync with the global trend (Shivakumar et al., 2021; Surono et al., 2021). A longitudinal study on persistently stunted children from south India showed an increase in diversity in both groups (stunted and healthy controls) with age. Partially in line with Shivakumar et al. (2021), stunted children at 12 months of age showed a higher abundance of Bacteroidota. Enrichment of inflammogenic taxa, that is, genus Desulfovibrio and order Campylobacterales, and lower abundance of probiotic species Bifidobacterium longum and Lactobacillus mucosae in stunted children were also observed (Dinh et al., 2016; Shivakumar et al., 2021). The gut microbiota of children living in Mumbai slums was enriched with Pseudomonadota and less Actinomycetota, representing the immaturity of the gut (Huey et al., 2020) (Table 1).
The majority of the microbiota-associated malnutrition reports are coming from countries with low socioeconomic status. Increasing poverty, poor hygiene, altered dietary habits, exposure to pollutants, and accumulation of environmental pathogens could make them more prone to long-term health problems such as malnutrition (Leocádio et al., 2021). Association of a higher abundance of pathogenic genera from phylum Pseudomonadota with malnutrition, and depletion of Bifidobacterium longum emerged as a common trend in both Indian and Global populations. However, the sample size, age group, and sequenced region of the 16S rRNA gene were different in the above comparisons.
Obesity
Excessive or abnormal accumulation of fat in the body that could impair health is termed obesity or overweight (Obesity and Overweight, n.d.). Nearly 650 million people around the globe and 135 million in India are affected by obesity. Changes in gut microbial composition also lead to excessive energy storage and a high risk of obesity. Four studies from Germany, Finland, the United States, and other European countries were compared with Indian studies. The gut bacterial-regulated low-grade inflammation was associated with obesity. For instance, inflammation associated Staphylococcus aureus was enriched in overweight mothers (Collado et al., 2008). The onset of obesity was associated with an increase in the Pseudomonadota phylum and a decrease in the family Clostridiaceae and Ruminococcaceae, as reported in a longitudinal study from Europe (Rampelli et al., 2018). The gut microbiota of obese individuals was reported to exhibit a lower abundance of the genus Bifidobacterium (Collado et al., 2008), Clostridium leptum group of phylum Bacillota (Schwiertz et al., 2010), and family Prevotellaceae (Rampelli et al., 2018). Additionally, enrichment of Bacteroides (Collado et al., 2008; Schwiertz et al., 2010; Rampelli et al., 2018) and faecal SCFAs concentrations, particularly propionate and butyrate, were also observed. The latter could be a result of factors like higher microbial production, changes in microbial cross-feeding patterns, and low absorption (Schwiertz et al., 2010). (Table 1).
A consistent pattern was observed while comparing the global (the United States, Germany, Finland, and six other European countries) results to the Indian gut microbiota, for instance, a higher abundance of Bacteroides and a higher level of faecal SCFAs in obese as compared to lean/normal individuals was reported. However, no difference in the distribution of Bacillota and Bacteroidota was observed (Ppatil et al., 2012). Faecalibacterium prausnitzii from the Clostridium leptum group was higher in obese south Indian children suggesting an increase in energy salvage from undigested/unabsorbed carbohydrates, which otherwise would be unavailable (Balamurugan et al., 2010) (Table 1). Inconsistent with both global as well as other Indian studies, Bahadur et al., 2021 reported bacterial composition with denaturing gradient gel electrophoresis technique. They detected Collinsella aerofaciens, Dialister, Eubacterium, Mitsuokella, Victivallis in obese, and Paraclostridium bifermentans in lean individuals (Bahadur et al., 2021). Obesity-related microbiota differences strongly influenced by geographical location, lifestyle, and diet as western individuals follow a low fibre and saturated fat-rich diet (Ecklu-Mensah et al., 2023). These could be the reasons for non-overlapping pattern between global and Indian studies. Inconsistency within Indian studies could be due to different methodologies used for taxonomy identification, different targeted regions of the 16S rRNA gene, and variable age groups (Table 1). However, the association of Bacteroides with obesity has been observed in both Indian and global data.
Type 2 diabetes
The condition of increased blood glucose level due to impaired insulin production by pancreatic beta-cells and the inability of body cells to utilise it (insulin resistance) is termed Type 2 diabetes (T2D). There are about 422 million cases across the globe and India harbours 77 million diabetic cases in adults with a prevalence rate of 8.3% (Members, n.d.). This metabolic disorder is caused by genetic, environmental, or both factors. Here, five studies from global cohorts (Africa, China, and Denmark) were compared with reports from India. A direct link between gut microbiome alteration and T2D comes from clinical studies reporting an increase in the incidence of T2D in total or partial colectomy (Jensen et al., 2018). The dysbiosis leading to a reduction in the Bacillota phylum, which is otherwise enriched in the healthy subjects, was observed in Africa and Denmark (Zhong et al., 2019; Doumatey et al., 2020). Differences in gut microbial profiles in healthy, pre-diabetic, and treatment-naive T2D were shown in Chinese cohorts. There was an insignificant difference in microbial gene-based diversity and richness among all three groups. However, the butyrate producers from class Clostridia (Dialister invisus and Roseburia hominis) were highly abundant in healthy compared to the other two groups. Treatment-naive T2D group had a higher abundance of Bacteroides spp and lower Akkermansia muciniphila compared to healthy and pre-diabetic groups (Zhong et al., 2019). Similarly, African, Danish, and Chinese T2D patients also showed a reduced abundance of butyrate producers (Collinsella, Ruminococcus lactaris, Anaerostipes, and Clostridium) (J. Wang et al., 2012; Forslund et al., 2015; Doumatey et al., 2020; Alvarez-Silva et al., 2021) (Table 1). In contrast to Zhong et al., microbial gene diversity increased upon treatment with metformin (Forslund et al., 2015). The high diversity and richness in urban African T2D patients could be due to different lifestyles (Doumatey et al., 2020).
Consistent with the above results, Indian T2D patients also showed a reduction in butyrate producers (family Ruminococcaceae and Lachnospiraceae, genera Prevotella, Fecalibacterium, Ruminococcus, Roseburia) (Bhute et al., 2017; Alvarez-Silva et al., 2021; Talukdar et al., 2021). Reduction in anti-inflammatory (Roseburia, Lachnospira, Coprococcus, Phascolarctobacterium, Blautia, Anaerostipes), pro-inflammatory (Sutterella), a few pathogens (Haemophilus, Comamonas), and enrichment of pathogenic (Escherichia, Enterobacter, Treponem), Pro-inflammatory (Methanobrevibacter), anti-inflammatory bacteria (Butyricimonas, Acidaminococcus, Weissella) was reported in Indian T2D patients (Das et al., 2021), indicating that a balance between anti-inflammatory and pro-inflammatory bacteria is crucial. Global studies were fairly different in their experimental design and sample size (Table 1). Taking together, it has been observed that T2D diseases could be associated with a decreased abundance of butyrate producers; however, butyrate-producing species can be different.
Colorectal cancer
Colorectal cancer (CRC), a digestive tract tumour, is a leading cause of morbidity and mortality in developed countries like Japan and the United States. Mutation in tumour repressor genes (p53, DPC4/Smad4, APC, MSH2, MLH1, and PMS2) and activation of oncogenes (beta-catenin, COX-2, and K-RAS) are the causes of CRC (Hisamuddin & Yang, 2006). In this section, four studies from China and the United States were compared with all available Indian ones.
Association studies of gut bacterial dysbiosis with CRC revealed the reduced abundance of butyrate producers (Roseburia spp., Eubacterium spp., E. hallii, E. hadrum, E. desmolans, Roseburia faecis, and Coprococcus comes) (T. Wang et al., 2012; Zhang et al., 2018) and a higher abundance of opportunistic pathogens (Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus, and Peptostreptococcus) in CRC patients of China. Species Bacteroides vulgatus and Bacteroides uniformis were enriched in healthy (T. Wang et al., 2012) (Table 1); however, species Bacteroides fragilis, reported to trigger cell proliferation, was enriched in CRC patients (T. Wang et al., 2012; Pan et al., 2020). The reduced abundance of butyrate producers was possibly due to a higher abundance of pathogens such as Fusobacterium nucleatum (Vogtmann et al., 2016; Zhang et al., 2018; Pan et al., 2020), Porphyromonas asaccharolytica, (Vogtmann et al., 2016; Zhang et al., 2018) Peptostreptococcus stomatis (Zhang et al., 2018; Pan et al., 2020), Parvimonas micra etc., which are oral periodontopathic bacteria (Zhang et al., 2018). Healthy and CRC tissue microbiota from Chinese showed no difference in diversity; however, a significant difference was observed while comparing different CRC stages. Cancer progression was marked by an increasing abundance of phyla Bacteroidota, Bacillota, Fusobacteriota, genera Fusobacterium, Peptostreptococcus, Streptococcus, and Ruminococcus, Verrucomicrobia, and a decreasing abundance of Pseudomonadota (Pan et al., 2020).
In accordance with global studies, Bacteroides fragilis, Peptostreptococcus stomatis, and Parvimonas micra were associated with Indian CRC patients (Table 1). Apart from them, species Akkermansia muciniphila, Bacteroides eggerthii, Escherichia coli, Odoribacter splanchnicus, and Parabacteroides distasonis were also associated with CRC (Gupta et al., 2019a). Species Flavonifractor plautii, a degrader of key flavonoids, was differentially abundant in Indian CRC samples and separated Indian from Austrian and Chinese samples (Gupta et al., 2019a). Differentially higher abundance of phylum Pseudomonadota and species Alistipes onderdonkii, Bacteroides massiliensis, Bifidobacterium pseudocatenulatum, and Corynebacterium appendicis was also reported by Hasan et al. (2022). The above comparisons revealed a common trend of higher abundance of genus Bacteroides in both Indian and Global CRC patients; however, species were different. A higher abundance of Fusobacterium in global and Flavonifractor in Indian CRC patients was the unique trend.
Inflammatory bowel diseases
IBDs consist of Crohn’s disease (CD) and ulcerative colitis (UC). The CD is an inflammatory disease affecting the GIT with abdominal pain, fever, diarrhoea with mucus or blood, or both (Baumgart & Sandborn, 2012). UC is also a relapsing inflammatory disease mainly affecting the inner linings of the large intestine and rectum (Gajendran et al., 2019). Two major hypotheses have emerged for the nature of the pathogenesis of IBDs. One is an excessive immunological response to the normal gut microbiome by dysregulation of the mucosal immune system and the second is dysbiosis in the gut microbiome that evokes an inflammatory response (Strober et al., 2007; Kabeerdoss et al., 2013). As the gut microbiome flourishes on dietary components, an anti-inflammatory microbiota could be nourished by specific food intake. High animal food intake, alcohol, soft drinks, sugar, and processed food could lead to gut inflammation, while plant-based foods are associated with low pathobiont abundance and high SCFA producers (Bolte et al., 2021). Three studies from the United States, Netherlands, and China were compared with the Indians.
A characteristic feature of IBD deduced in cohorts from the United States was an increase in facultative anaerobes with a decrease in obligate anaerobes (butyrate producers), specifically enrichment of E. coli and depletion of F. prausnitzii and Roseburia hominis in CD. The differential abundance of two prominent species in IBD, Ruminococcus torques and Ruminococcus gnavus in CD and UC, respectively, was also confirmed in this study (Lloyd-Price et al., 2019). Partially overlapping results from a study on the United States and Netherlands cohorts showed depletion of Roseburia hominis, Dorea formicigenerans, and Ruminococcus obeum and enrichment of unclassified Roseburia species in IBD patients. Symbiosis of Bifidobacterium breve and Clostridium symbiosum was uniquely abundant in UC, while species R. gnavus, E. coli, and Clostridium clostridioforme were in CD (Franzosa et al., 2019). Reduced diversity, low Bacillota, higher Pseudomonadota, and Fusobacteriota, in IBD patients, were also reported (Franzosa et al., 2019; T. Wang et al., 2022) (Table 1).
In comparison with the results from global studies, a higher abundance of Pseudomonadota, depletion of butyrate producers F. prausnitzii and Clostridial cluster IV & XIVa (Roseburia, Clostridium, Eubacterium, and Ruminococcus), was observed in UC and CD patients of India (Kabeerdoss et al., 2013; Kumari et al., 2013; Das et al., 2018). In contrast, Verma et al. (2010) reported a higher abundance of species from Clostridium cluster XIVa (Eubacterium and Peptostreptococcus) in CD but not in UC indicating their different roles in pathogenesis in both groups (Verma et al., 2010) (Table 1).
Low gut bacterial diversity and reduction in butyrate producers (Kabeerdoss et al., 2013; Lloyd-Price et al., 2019), which inhibit the gut inflammatory response in IBD patients, were observed in both Indian and global samples (Kabeerdoss et al., 2013; Lloyd-Price et al., 2019). All these results suggest that the nature of the pathogenesis of IBD could be explained by the second hypothesis, that dysbiosis in the gut microbiome evokes an inflammatory response.
Gut inflammation and damage to the brain function
The bidirectional communication between gut bacterial cells and the brain is called the gut-microbiota brain axis. The bacterial cells produce neurotransmitters, amino acids, and metabolites, which influence host immune systems, gut barrier integrity, and the brain. Gut barrier integrity also gets disturbed during stress, anxiety, autism spectrum disorders (ASDs), and Parkinson’s disease (PD) (Morais et al., 2020). An association study from the United Kingdom revealed a positive correlation of abundant Lactobacillus spp. with positive self-judgement, and an inverse relation of CRP (C-reactive protein), a pro-inflammatory molecule, with cognitive empathy (Heym et al., 2019).
ASDs are a group of complex neurodevelopmental disorders, and, unfortunately, the cause is still unclear (Geetha et al., 2019). However, an association of socioeconomic and environmental risk factors with ASD has suggested that family history of ASD, paternal age, nutrition during pregnancy, mode of delivery, breastfeeding, and NICU stay were statistically significant factors associated with ASDs (Geetha et al., 2019). Three gut microbial association studies with ASD, from Italy and China, were compared with an Indian study. A Chinese and Italian study reported an increased abundance of Bacteroidota in ASD children (Coretti et al., 2018; Zou et al., 2020); however, the opposite trend was reported other Chinese data (Ye et al., 2021). High bacterial diversity (Zou et al., 2020; Ye et al., 2021), a significant increase in BCAAs synthesising species (B. vulgatus and P. copri), a reduction in butyrate-producing genera clusters Clostridium clusters IV and XIVa, probiotic bacteria like B. fragilis and A. muciniphila in ASD children compared to normal controls in China (Zou et al., 2020). Depletion of the dominant infant gut bacterium Bifidobacterium longum (Coretti et al., 2018; Ye et al., 2021) an increase in Faecalibacterium prausnitzii, a significant butyrate producer and late coloniser of the healthy gut, was also reported (Coretti et al., 2018; Ye et al., 2021) (Table 1).
The results from Indian studies were not in line with the above global studies. However, a comparison done in the same study with ASD children from the United States showed an overlap. There was no difference in diversity between the control and ASD groups of Indian children. A higher relative abundance of families Lactobacillaceae (Lactobacillus), Bifidobacteraceae (Bifidobacterium), and Veillonellaceae (Megasphaera) was observed in ASD children. Despite the different diets of Indian ASD children (normal native diet) and the United States (gluten-free), the Lactobacillus genus was highly abundant compared to healthy. Support for this finding was also provided in the articles by Coretti et al. (2018) and Zou et al. (2020). However, it remains obscure whether the higher abundance of Lactobacillus is a cause or an effect of ASD (Pulikkan et al., 2018). Further metagenomic and metabolomic studies are needed to confirm this (Table 1).
The other common neurodegenerative disorders are PD and Alzheimer’s disease (AD). The former is caused by dead or impaired dopamine-producing basal ganglia cells, deposition of alpha-synuclein protein in the cells, and genetic or environmental factors (Parkinson’s Disease: Causes, Symptoms, and Treatments | National Institute on Aging, n.d.). The data from two studies from China and Germany were discussed here. Chinese study showed decreased levels of BCAAs (Leu, Ile, and Val) and Tyr in advanced as compared to early PD, which is probably due to increased energy expenditure which further accelerates amino acid consumption in advanced PD. It also showed a negative correlation between plasma BCAAs, aromatic amino acids, and microbial taxa such as Streptococcaceae, Streptococcus, and Lactobacillus, which consume or catabolise them (Zhang et al., 2022). The German study reported a decreased abundance of neuroprotective, health-promoting, anti-inflammatory species such as Faecalibacterium and Fusicatenibacter, enrichment of opportunistic pathogens, that is, Peptoniphilus and Finegoldia, higher level of calprotectin, a faecal inflammation marker in PD patients (Weis et al., 2019). Fang et al. (2020) reviewed several articles and revealed a higher abundance of Bifidobacterium, Lactobacillus, Akkermansia, and a lower abundance of Blautia, Coprococcus, and Prevotella in PD patients. The pro-inflammatory Bilophila species were associated with the progression of disease symptoms (Baldini et al., 2020) (Table 1). The burden of noncommunicable neurological disorders is increasing in India. There were 771,000 cases of PD in 2019 and 45,300 deaths reported in PD (Singh et al., 2021). The other noncommunicable disease is AD. It is a common type of dementia characterised by extracellular amyloid beta plaque and intracellular tau protein accumulation. In India, there were 3.69 million cases of AD or other dementias in 2019 (Singh et al., 2021).
Results from an Italian study showed a lower abundance of anti-inflammatory Eubacterium rectale and anti-inflammatory cytokines (IL-10), and a high abundance of pro-inflammatory Escherichia/Shigella in patients (cognitively impaired with and without brain amyloidosis) (Table 1). Both the studies from the United States and Italy showed more elevated pro-inflammatory cytokines (CXCL2, IL-1Beta, and NLRP3) in cognitively impaired patients with amyloidosis positively correlated with Escherichia/Shigella and negatively correlated with E. rectale (Cattaneo et al., 2017; Vogt et al., 2017) (Table 1). Despite increasing neurodegenerative cases in India, and their evident association with gut health in global studies, there are no studies done in India on gut microbial association with PD and AD.
Communicable diseases
Diarrhoea
Diarrhoea is one of the leading causes of mortality and is more prevalent in low- and middle-income countries (Naghavi et al., 2015). The common causes of diarrhoea are Vibrio cholera, Cryptosporidium sp., enterotoxigenic Escherichia coli, Clostridioides difficile, Rotavirus, and Shigella sp. infection (Guerrant et al., 1990; Monaghan et al., 2020). All the diarroeal studies compared with Indian ones were from Bangladesh.
Recovery from V. cholerae infection was characterised by the accumulation of a healthy gut microbial profile. For instance; upon infecting mice with the pathogen, the species Ruminococcus obeum consistently increased, which in turn restricted pathogens’ growth. The increased expression of autoinducer-2 synthase (luxS) in R. obeum repressed several colonisation factors of the pathogen (Table 1) (Hsiao et al., 2014). The recovery mechanism showed that infection or antibiotic treatment cleared both obligate and facultative anaerobes from the gut, followed by the accumulation of oxygen and dietary substrates in the gut. Recolonising facultative anaerobes majorly from dietary resources lowered the oxygen stress that enabled obligate anaerobes to colonise and utilise accumulated carbohydrates. Competition for the dietary substrates returned to the original state community (David et al., 2015). The disease-specific associations or changes in microbial composition revealed in a meta-analysis, where a higher abundance of Pseudomonadota and a low abundance of Bacteroidota and a few Bacillota, in particular, a reduction of butyrate producers from family Ruminococcaceae and Lachnospiraceae in diarrhoeal patients (Duvallet et al., 2017).
Similar to the above trends, Indian infants with acute and persistent diarrhoea showed the proliferation of facultative anaerobes of phylum Pseudomonadota (Chelonobacter, Granulicatella, Haemophilus, Klebsiella, Rothia, and Vibrio) and collapse of anaerobic bacteria (Bacillota, Bacteroides) (Thakur et al., 2018). However, the sample size was quite small in this study population. A high Bacillota to Bacteroidota ratio was associated with V. cholera infection (Thakur et al., 2018; De et al., 2020). A negative correlation between commensals of the family Bifidobacteriaceae and Lachnospiraceae and pathogenic families Enterobacteriaceae and Vibrionaceae, implying the obvious trend in diarrheal dysbiosis (De et al., 2020) (Table 1). The gut microbiome of acute diarrheal children from India showed a lower abundance of butyrate producers (E. rectale, F. prauznitzii, L. acidophilus), compared to after recovery microbiome (Balamurugan et al., 2008). Antibiotic-exposed urban diarrheal samples from central India were positive for Clostridioides difficile infection and were enriched with cephalosporins and carbapenem resistance genes (Monaghan et al., 2020). The observed differences between Indian and global studies are possible due to the experiment design, age of participants, and targeted region for the taxonomy profiling (Table 1).
Amoebiasis
Amoebiasis is caused by Entamoeba histolytica, and is the second most prevalent protozoan disease, especially in infants in developing countries (Gilchrist et al., 2016). Upon perturbation or host immune response compromisation, this can become virulent, and cause diarrhoea, and bloody stools. It can also invade other organs if left untreated (Sarjapuram et al., 2017; Yanagawa et al., 2021). Two studies on gut microbial association with amoebiasis from Bangladesh and Japan were compared with the Indian ones.
A report from Bangladesh showed a significantly higher parasitic load (E. histolytica) during the first year of life in symptomatic as compared to asymptomatics diarrheal infants and association of diarrheal onset with P. copri (Gilchrist et al., 2016). Japanese asymptomatic and symptomatic diarrheal children differed with significantly lower Streptococcaceae (Streptococcus salivarius and Streptococcus sinensis) and higher protective bacteria from Ruminococcaceae, Coriobacteriaceae, and Clostridiaceae families in former as compared to latter. However, there was no significant difference in the diversity (Yanagawa et al., 2021).
Real-time PCR quantification of E. histolytica infected gut microbiota of North Indians showed a significant decrease of predominant gut microbiome members (Bacteroides, Clostridium coccoides subgroup, Clostridium leptum subgroup, Campylobacter, Eubacterium, and Lactobacillus). An unusual rise in the Bifidobacterium population (SCFAs producer), which could also ferment mucin, in E. histolytica infected patients was reported (Verma et al., 2012). E. histolytica infection induces hypersecretion of mucus from goblet cells to counter adherence of pathogens, which in turn promotes Bifidobacterium growth (Verma et al., 2012; Cornick et al., 2017). Another study by Iyer et al. (2023) revealed a decreased abundance of Faecalibacterium, Prevotella, Sutterella, Subdoligranulum, and Colinsella and a higher abundance of Escherichia, Klebsiella, and Ruminococcus in the E. histolytica positive patients from Delhi, India. Association of high P. copri levels with diarrhoea was already reported; however, an opposite trend was observed in India (Gilchrist et al., 2016; Iyer et al., 2023) (Table 1). Another interesting finding was the preferential phagocytosis of beneficial bacteria from order Bifidobacteriales, Clostridales, Erysipelotrichales, and Lactobacillales cause dysbiosis which could help in the proliferation of pathogens (Iyer et al., 2019). Treatment of this protozoal disease with antiprotozoal drugs like Metronidazole could give rise to resistant E. histolytica. So efforts have been made to use LAB as probiotics to prevent this disease. The use of Saccharomyces boulardii strain and metronidazole in the clinical trial significantly reduced the duration of diarrhoea (Dinleyici et al. 2009). Co-culturing Lactobacillus casei and Enterococcus faecium with E. histolytica showed a significant reduction in parasite survival (Sarjapuram et al., 2017). The use of these probiotic strains could lead to amoebiasis treatment without using antibiotics.
Conclusion
This review provides insight into the establishment of the gut microbiome from pregnancy to birth, up till old age, and highlights the dynamics of gut microbiota upon perturbation during communicable and noncommunicable diseases. Gut metagenomic studies from diverse populations of Europe, North and South America, South Africa, and Asia were reviewed and the emerging global pattern of community composition, diversity, and abundance was compared with the Indian population. The differences start appearing right from the mode of delivery, where early colonisation of beneficial bacteria (Bifidobacterium and Lactobacillus) was seen in VD infants. The developmental trajectory from infant, child, and adult to elderly individuals from Indian and global studies showed overlapping as well as unique Indian-specific patterns. For instance, high diversity in the Ruminococcaceae family, and decreased abundance of Faecalibacterium in centenarians were reported in both global as well as Indian studies. On the other hand, a higher abundance of Bacteroides during late adolescence and adulthood, and a sharp decline of Eubacterium rectale and F. prausnitzii in adults were the unique features reported in Indians.
Among key factors influencing gut microbial composition, diet, lifestyle, antibiotic usage, and various diseased conditions have been discussed in depth. To the question of whether population affects these trends, both overlapping as well as unique trends were found, based on a limited number of populations. Since it was earlier reported that the major enterotypes are associated more with the diet rather than with the populations (Arumugam et al., 2011), so from where do the unique trends appear? Populations are known to have (a small set of) unique taxa (Dhakan et al., 2019), which may (at least partially) explain the observed unique trends. This review also highlighted that although reports on core gut microbiomes exist, they are highly limited in terms of capturing the variation present in populations across the globe. This hints towards the need for a systematic study that will prevent any bias associated with meta-analyses.
Studies within India and their comparison with global data also revealed contradictory/inconsistent patterns, which reflects the variability and complexity of metagenomic data. Apart from the various factors mentioned in the article, sampling, storage, DNA isolation methods, library preparation kits, sequencing techniques, and bioinformatic analysis could also influence the outcome of the metagenomic study (Szóstak et al., 2022). The majority of the Indian studies used amplicon-based different sequencing techniques such as Illumina, pyrosequencing, Ion-torrent, PCR quantification of specific anaerobes, denaturing gradient gel electrophoresis (DGGE), and only a few had used whole genome shotgun sequencing, suggesting a possible explanation for higher-level taxonomy resolution in most cases. Small sample size and lack of controls in comparative studies are other aspects that emerged while reviewing Indian studies. A smaller sample size does not represent a general population-based outcome and influences the significance of the results. As an example, a study done by De et al. (2020) on gut microbial signatures in diarrheal conditions has inferred the results without comparing them with healthy control. Another important limitation of several studies was their analysis’s ignorance of confounding factors, which might have added bias to the findings.
Lastly, dysbiosis linked with neurodevelopment and neurodegenerative disorders is an active area of research, yet there is only one study on ASD and none on AD and PD in the Indian population. Taken together, a large sample size across multiple geographical locations, analysed through the same robust pipeline, could give the true picture of the gut metagenome in healthy as well as diseased conditions.
Acknowledgements
We would like to thank Mr. Priyansh Patel, a JRF, and Mr. Angeo Saji, a PhD student, for their critical remarks on the manuscript.
Disclosure statement
The authors declare that there are no competing interests.
Author contribution
Conceptualisation: N.C., A.K.V., S.S., V.T.; Data curation: N.C.; Formal analysis: N.C., V.T.; Funding acquisition: V.T.; Supervision: V.T.; Writing – original draft: N.C., A.M.; Writing – review and editing: N.C., A.K.V., V.T.
Funding statement
N.C. was financially supported by funding from the University of Hyderabad’s Institution of Eminence (Grant No. UoH-IoE-RC2-21-023).
References
- Agans R, Rigsbee L, Kenche H, Michail S, Khamis HJ and Paliy O (2011) Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiology Ecology 77(2), 404–412. 10.1111/J.1574-6941.2011.01120.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Silva C, Kashani A, Hansen TH, Pinna NK, Anjana RM, Dutta A, Saxena S, Støy J, Kampmann U, Nielsen T, Jørgensen T, Gnanaprakash V, Gnanavadivel R, Sukumaran A, Rani CSS, Færch K, Radha V, Balasubramanyam M, Nair GB, Das B, Vestergaard H, Hansen T, Mande SS, Mohan V, Arumugam M and Pedersen O (2021) Trans-ethnic gut microbiota signatures of type 2 diabetes in Denmark and India. Genome Medicine 13(1), 1–13. 10.1186/s13073-021-00856-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwesh M, Kumar KV, Nagarajan M, Chander MP, Kartick C and Paluru V (2016) Elucidating the richness of bacterial groups in the gut of Nicobarese tribal community – Perspective on their lifestyle transition. Anaerobe 39, 68–76. 10.1016/j.anaerobe.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Mohn WW, Turvey SE and Finlay BB (2015) Early infancy microbial and metabolic alterations affect risk of childhood asthma. Science Translational Medicine 7(307), 307ra152. 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Paslier DL, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium (additional members), Weissenbach J, Ehrlich SD and Bork P (2011) Enterotypes of the human gut microbiome. Nature 473(7346), 174–180. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avershina E, Lundgård K, Sekelja M, Dotterud C, Storrø O, Øien T, Johnsen R and Rudi K (2016) Transition from infant- to adult-like gut microbiota. Environmental Microbiology 18(7), 2226–2236. 10.1111/1462-2920.13248 [DOI] [PubMed] [Google Scholar]
- Azad MB, Bridgman SL, Becker AB and Kozyrskyj AL (2014) Infant antibiotic exposure and the development of childhood overweight and central adiposity. International Journal of Obesity 38(10), 1290–1298. 10.1038/ijo.2014.119 [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, Versalovic J, Young V and Finlay BB (2012) Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host & Microbe 12(5), 611–622. 10.1016/j.chom.2012.10.012 [DOI] [PubMed] [Google Scholar]
- Badal VD, Vaccariello ED, Murray ER, Yu KE, Knight R, Jeste DV and Nguyen TT (2020) The gut microbiome, aging, and longevity: A systematic review. Nutrients 12(12), 1–25. 10.3390/nu12123759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahadur T, Chaudhry R, Bamola VD, Chutani AM, Verma AK and Paul J (2021) Analysis of gut bacterial community composition in obese and lean Indian participants by denaturing gradient gel electrophoresis. Indian Journal of Health Sciences and Biomedical Research (KLEU) 14(1), 42–47. 10.4103/kleuhsj.kleuhsj_273_20 [DOI] [Google Scholar]
- Balamurugan R, George G, Kabeerdoss J, Hepsiba J, Chandragunasekaran AMS and Ramakrishna BS (2010) Quantitative differences in intestinal Faecalibacterium prausnitzii in obese Indian children. The British Journal of Nutrition 103(3), 335–338. 10.1017/S0007114509992182 [DOI] [PubMed] [Google Scholar]
- Balamurugan R, Janardhan HP, George S, Chittaranjan SP and Ramakrishna BS (2008) Bacterial succession in the colon during childhood and adolescence: Molecular studies in a southern Indian village. American Journal of Clinical Nutrition 88(6), 1643–1647. 10.3945/ajcn.2008.26511 [DOI] [PubMed] [Google Scholar]
- Balamurugan R, Janardhan HP, George S, Raghava MV, Muliyil J and Ramakrishna BS (2008) Molecular studies of fecal anaerobic commensal bacteria in acute diarrhea in children. Journal of Pediatric Gastroenterology and Nutrition 46(5), 514–519. 10.1097/MPG.0b013e31815ce599 [DOI] [PubMed] [Google Scholar]
- Baldini F, Hertel J, Sandt E, Thinnes CC, Neuberger-Castillo L, Pavelka L, Betsou F, Krüger R, Thiele I, NCER-PD Consortium (2020) Parkinson’s disease-associated alterations of the gut microbiome predict disease-relevant changes in metabolic functions. BMC Biology 18(1), 1–21. 10.1186/S12915-020-00775-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamola VD, Ghosh A, Kapardar RK, Lal B, Cheema S, Sarma P and Chaudhry R (2017) Gut microbial diversity in health and disease: Experience of healthy Indian subjects, and colon carcinoma and inflammatory bowel disease patients. Microbial Ecology in Health and Disease 28(1), 1322447. 10.1080/16512235.2017.1322447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgart DC and Sandborn WJ (2012) Crohn’s disease. The Lancet 380(9853), 1590–1605. 10.1016/S0140-6736(12)60026-9 [DOI] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Joakim Larsson DG and Johansson A (2015) The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy 59(10), 6551–6560. 10.1128/AAC.00933-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhute SS, Suryavanshi MV, Joshi SM, Yajnik CS, Shouche YS and Ghaskadbi SS (2017) Gut microbial diversity assessment of Indian type-2-diabetics reveals alterations in eubacteria, archaea, and eukaryotes. Frontiers in Microbiology 8, 214. 10.3389/fmicb.2017.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ (2016) Antibiotic use and its consequences for the normal microbiome. Science 352(6285), 544–545. 10.1126/science.aad9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, Lieber AD, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG and Blaser MJ (2016) Antibiotics, birth mode, and diet shape microbiome maturation during early life. Science Translational Medicine 8(343), 343ra82. 10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte LA, Vich Vila A, Imhann F, Collij V, Gacesa R, Peters V, Wijmenga C, Kurilshikov A, Campmans-Kuijpers MJE, Fu J, Dijkstra G, Zhernakova A and Weersma RK (2021) Long-term dietary patterns are associated with pro-inflammatory and anti-inflammatory features of the gut microbiome. Gut 70(7), 1287–1298. 10.1136/GUTJNL-2020-322670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG and Pamer EG (2013) Microbiota-mediated colonization resistance against intestinal pathogens. Nature Reviews. Immunology 13(11), 790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull MJ and Plummer NT (2014) Part 1: The human gut microbiome in health and disease. Integrative Medicine: A Clinician’s Journal 13(6), 17–22. [PMC free article] [PubMed] [Google Scholar]
- Cariño R, Takayasu L, Suda W, Masuoka H, Hirayama K, Konishi S and Umezaki M (2021) The search for aliens within us: A review of evidence and theory regarding the foetal microbiome. Critical Reviews in Microbiology 48(5), 611–623. 10.1080/1040841X.2021.1999903 [DOI] [PubMed] [Google Scholar]
- Carvalho MJ, Sands K, Thomson K, Portal E, Mathias J, Milton R, Gillespie D, Dyer C, Akpulu C, Boostrom I and Hogan P (2022) Antibiotic resistance genes in the gut microbiota of mothers and linked neonates with or without sepsis from low- and middle-income countries. Nature Microbiology 7(9), 1337–1347. 10.1038/s41564-022-01184-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, Ferrari C, Guerra UP, Paghera B, Muscio C, Bianchetti A, Volta GD, Turla M, Cotelli MS, Gennuso M, Prelle A, Zanetti O, Lussignoli G, Mirabile D, Bellandi D and Frisoni GB (2017) Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiology of Aging 49, 60–68. 10.1016/J.NEUROBIOLAGING.2016.08.019 [DOI] [PubMed] [Google Scholar]
- Chandel N, Somvanshi PR and Thakur V (2023) Characterisation of Indian gut microbiome for B-vitamin production and its comparison with Chinese cohort. British Journal of Nutrition 131(4), 686–697. 10.1017/S0007114523002179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, ’sullivan OO, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, Van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP and O’Toole PW (2012) Gut microbiota composition correlates with diet and health in the elderly. Nature 488(7410), 178–184. 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G and Dominguez-Bello MG (2015) The microbiome of uncontacted Amerindians. Science Advances 1(3). 10.1126/SCIADV.1500183/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Isolauri E, Laitinen K and Salminen S (2008) Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. The American Journal of Clinical Nutrition 88(4), 894–899. 10.1093/ajcn/88.4.894 [DOI] [PubMed] [Google Scholar]
- Coretti L, Paparo L, Riccio MP, Amato F, Cuomo M, Natale A, Borrelli L, Corrado G, Comegna M, Buommino E, Castaldo G, Bravaccio C, Chiariotti L, Canani RB and Lembo F (2018) Gut microbiota features in young children with autism spectrum disorders. Frontiers in Microbiology 9, 417648. 10.3389/FMICB.2018.03146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornick S, Moreau F, Gaisano HY and Chadee K (2017) Entamoeba histolytica-induced mucin exocytosis is mediated by VAMP8 and is critical in mucosal innate host defense. MBio 8(5), 1–14. 10.1128/mBio.01323-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI and Knight R (2009) Bacterial community variation in human body habitats across space and time. Science 326(5960), 1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Ghosh TS, Kedia S, Rampal R, Saxena S, Bag S, Mitra R, Dayal M, Mehta O, Surendranath A, Travis SPL, Tripathi P, Nair GB and Ahuja V (2018) Analysis of the gut microbiome of rural and urban healthy Indians living in sea level and high altitude areas. Scientific Reports 8(1), 1–15. 10.1038/s41598-018-28550-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Jayasudha R, Chakravarthy SK, Prashanthi GS, Bhargava A, Tyagi M, Rani PK, Pappuru RR, Sharma S and Shivaji S (2021) Alterations in the gut bacterial microbiome in people with type 2 diabetes mellitus and diabetic retinopathy. Scientific Reports 11(1), 1–15. 10.1038/s41598-021-82538-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ and Turnbaugh PJ (2013) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505(7484), 559–563. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Weil A, Ryan ET, Calderwood SB, Harris JB, Chowdhury F, Begum Y, Qadri F, LaRocque RC and Turnbaugh PJ (2015) Gut microbial succession follows acute secretory diarrhea in humans. MBio 6(3), 1–14. 10.1128/MBIO.00381-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G and Lionetti P (2010) Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America 107(33), 14691–14696. 10.1073/PNAS.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De R, Kanungo S, Mukhopadhyay AK and Dutta S (2023) The gut microbiome of the healthy population in Kolkata, India, is a reservoir of antimicrobial resistance genes emphasizing the need of enforcing antimicrobial stewardship. FEMS Microbiology Letters 370, fnad090. 10.1093/femsle/fnad090 [DOI] [PubMed] [Google Scholar]
- De R, Mukhopadhyay AK and Dutta S (2020) Metagenomic analysis of gut microbiome and resistome of diarrheal fecal samples from Kolkata, India, reveals the core and variable microbiota including signatures of microbial dark matter. Gut Pathogens 12(1), 1–48. 10.1186/s13099-020-00371-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehingia M, Devi KT, Talukdar NC, Talukdar R, Reddy N, Mande SS, Deka M and Khan MR (2015) Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Scientific Reports 5, 1–12. 10.1038/srep18563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakan DB, Maji A, Sharma AK, Saxena R, Pulikkan J, Grace T, Gomez A, Scaria J, Amato KR and Sharma VK (2019) The unique composition of Indian gut microbiome, gene catalogue, and associated fecal metabolome deciphered using multi-omics approaches. GigaScience 8(3), 1–20. 10.1093/gigascience/giz004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh DM, Ramadass B, Kattula D, Sarkar R, Braunstein P, Tai A, Wanke CA, Hassoun S, Kane AV, Naumova EN, Kang G and Ward HD (2016) Longitudinal analysis of the intestinal microbiota in persistently stunted young children in South India. PLoS One 11(5), 155405. 10.1371/JOURNAL.PONE.0155405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinleyici EC, Eren M, Yargic Z, Dogan N, Vandenplas Y (2009) Clinical efficacy of Saccharomyces boulardii and metronidazole compared to metronidazole alone in children with acute bloody diarrhea caused by amebiasis: a prospective, randomized, open label study. American Journal of Tropical Medicine and Hygiene, 80, 953–5. 10.4269/ajtmh.2009.80.953 [DOI] [PubMed] [Google Scholar]
- Doumatey AP, Adeyemo A, Zhou J, Lei L, Adebamowo SN, Adebamowo C and Rotimi CN (2020) Gut microbiome profiles are associated with type 2 diabetes in urban Africans. Frontiers in Cellular and Infection Microbiology 10, 63. 10.3389/FCIMB.2020.00063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C, Gibbons SM, Gurry T, Irizarry RA and Alm EJ (2017) Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nature Communications 8(1), 1–10. 10.1038/s41467-017-01973-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecklu-Mensah G, Choo-Kang C, Maseng MG, Donato S, Bovet P, Viswanathan B, Bedu-Addo K, Plange-Rhule J, Boateng PO, Forrester TE, Williams M, Lambert EV, Rae D, Sinyanya N, Luke A, Layden BT, O’Keefe S, Gilbert JA and Dugas LR (2023) Gut microbiota and fecal short chain fatty acids differ with adiposity and country of origin: The METS-microbiome study. Nature Communications 14, 5160. 10.1038/s41467-023-40874-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fact sheets – Malnutrition (n.d.) Malnutrition. Available at https://www.who.int/news-room/fact-sheets/detail/malnutrition (accessed 15 September 2023).
- Fagundes Neto U and Affonso Scaletsky IC (2000) Escherichia coli infections and malnutrition. The Lancet 356(9248), S27. 10.1016/S0140-6736(00)92013-0 [DOI] [PubMed] [Google Scholar]
- Fang P, Kazmi SA, Jameson KG and Hsiao EY (2020) The microbiome as a modifier of neurodegenerative disease risk. Cell Host & Microbe 28(2), 201–222. 10.1016/j.chom.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, Armanini F, Truong DT, Manara S, Zolfo M, Beghini F, Bertorelli R, De Sanctis V, Bariletti I, Canto R, Clementi R, Cologna M, Crifò T, Cusumano G, Gottardi S, Innamorati C, Masè C, Postai D, Savoi D, Duranti S, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Milani C, Mangifesta M, Anzalone R, Viappiani A, Yassour M, Vlamakis H, Xavier R, Collado CM, Koren O, Tateo S, Soffiati M, Pedrotti A, Ventura M, Huttenhower C, Bork P and Segata N (2018) Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host & Microbe 24(1), 133–145.e5. 10.1016/J.CHOM.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Krogh Pedersen H, Arumugam M, Kristiansen K, Yvonne Voigt A, Vestergaard H, Hercog R, Igor Costea P, Roat Kultima J, Li J, Jørgensen T, Levenez F, Dore J, MetaHIT Consortium, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P and Pedersen O (2015) Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528(7581), 262–266. 10.1038/nature15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, Sauk JS, Wilson RG, Stevens BW, Scott JM, Pierce K, Deik AA, Bullock K, Imhann F, Porter JA, Zhernakova A, Fu J, Weersma RK, Wijmenga C, Clish CB, Vlamakis H, Huttenhower C and Xavier RJ (2019) Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nature Microbiology 4(2), 293–305. 10.1038/s41564-018-0306-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajendran M, Loganathan P, Jimenez G, Catinella AP, Ng N, Umapathy C, Ziade N and Hashash JG (2019) A comprehensive review and update on ulcerative colitis. Disease-a-Month 65(12), 100851. 10.1016/J.DISAMONTH.2019.02.004 [DOI] [PubMed] [Google Scholar]
- Geetha B, Sukumar C, Dhivyadeepa E, Reddy JK and Balachandar V (2019) Autism in India: A case–control study to understand the association between socio-economic and environmental risk factors. Acta Neurologica Belgica 119(3), 393–401. 10.1007/S13760-018-01057-4 [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Petri SE, Schneider BN, Reichman DJ, Jiang N, Begum S, Watanabe K, Jansen CS, Elliott KP, Burgess SL, Ma JZ, Alam M, Kabir M, Haque R and Petri WA (2016) Role of the gut microbiota of children in diarrhea due to the protozoan parasite Entamoeba histolytica. The Journal of Infectious Diseases 213(10), 1579–1585. 10.1093/INFDIS/JIV772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönlund MM, Lehtonen OP, Eerola E and Kero P (1999) Fecal microflora in healthy infants born by different methods of delivery: Permanent changes in intestinal flora after cesarean delivery. Journal of Pediatric Gastroenterology and Nutrition 28(1), 19–25. 10.1097/00005176-199901000-00007 [DOI] [PubMed] [Google Scholar]
- Groussin M, Poyet M, Sistiaga A, Kearney SM, Moniz K, Noel M, Hooker J, Gibbons SM, Segurel L, Froment A, Mohamed RS, Fezeu A, Juimo VA, Lafosse S, Tabe FE, Girard C, Iqaluk D, Nguyen LTT, Shapiro BJ, Lehtimäki J, Ruokolainen L, Kettunen PP, Vatanen T, Sigwazi S, Mabulla A, Domínguez-Rodrigo M, Nartey YA, Agyei-Nkansah A, Duah A, Awuku YA, Valles KA, Asibey SO, Afihene MY, Roberts LR, Plymoth A, Onyekwere CA, Summons RE, Xavier RJ and Alm EJ (2021) Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184(8), 2053–2067.e18. 10.1016/J.CELL.2021.02.052 [DOI] [PubMed] [Google Scholar]
- Guerrant RL, Hughes JM, Lima NL and Crane J (1990) Diarrhea in developed and developing countries: Magnitude, special settings, and etiologies. Reviews of Infectious Diseases 12(Suppl 1), S41–S50. 10.1093/clinids/12.supplement_1.s41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Dhakan DB, Maji A, Saxena R, Vishnu Prasoodanan PK, Mahajan S, Pulikkan J, Kurian J, Gomez AM, Scaria J, Amato KR, Sharma AK and Sharma VK (2019a) Association of Flavonifractor plautii, a flavonoid-degrading bacterium, with the gut microbiome of colorectal cancer patients in India. mSystems, 4(6), e00438-19. 10.1128/MSYSTEMS.00438-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Didwal G, Bansal S, Kaushal K, Batra N, Gautam V and Ray P (2019b) Antibiotic-resistant Enterobacteriaceae in healthy gut flora: A report from north Indian semiurban community. The Indian Journal of Medical Research 149(2), 276–280. 10.4103/ijmr.IJMR_207_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan R, Bose S, Roy R, Paul D, Rawat S, Nilwe P, Chauhan NK and Choudhury S (2022) Tumor tissue-specific bacterial biomarker panel for colorectal cancer: Bacteroides massiliensis, Alistipes species, Alistipes onderdonkii, Bifidobacterium pseudocatenulatum, Corynebacterium appendicis. Archives of Microbiology 204(6), 1–10. 10.1007/S00203-022-02954-2 [DOI] [PubMed] [Google Scholar]
- Hazarika P, Chattopadhyay I, Umpo M, Choudhury Y and Sharma I (2022) Elucidating the gut microbiome alterations of tribal community of Arunachal Pradesh: Perspectives on their lifestyle or food habits. Scientific Reports 12(1), 1–12. 10.1038/s41598-022-23124-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heym N, Heasman BC, Hunter K, Blanco SR, Wang GY, Siegert R, Cleare A, Gibson GR, Kumari V and Sumich AL (2019) The role of microbiota and inflammation in self-judgement and empathy: Implications for understanding the brain-gut-microbiome axis in depression. Psychopharmacology 236(5), 1459–1470. 10.1007/S00213-019-05230-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamuddin IM and Yang VW (2006) Molecular genetics of colorectal cancer: An overview. Current Colorectal Cancer Reports 2(2), 53–59. 10.1007/S11888-006-0002-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA, Haque R, Ahmed T and Gordon JI (2014) Members of the human gut microbiota involved in recovery from vibrio cholerae infection. Nature 515(7527), 423–426. 10.1038/NATURE13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey SL, Jiang L, Fedarko MW, McDonald D, Martino C, Ali F, Russell DG, Udipi SA, Thorat A, Thakker V, Ghugre P, Potdar RD, Chopra H, Rajagopalan K, Haas JD, Finkelstein JL, Knight R and Mehta S (2020) Nutrition and the gut microbiota in 10- to 18-month-old children living in urban slums of Mumbai, India. mSphere 5(5), e00731-20. 10.1128/mSphere.00731-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486(7402), 207–214. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LR, Chandel N, Verma AK, Thakur V, Paul J, Mandal AK and Bhattacharya A (2023) Effect of Entamoeba histolytica infection on gut microbial diversity and composition in diarrheal patients from New Delhi. Parasitology Research 122(1), 285–298. 10.1007/s00436-022-07728-9 [DOI] [PubMed] [Google Scholar]
- Iyer LR, Verma AK, Paul J and Bhattacharya A (2019) Phagocytosis of gut bacteria by Entamoeba histolytica. Frontiers in Cellular and Infection Microbiology 9, 34. 10.3389/FCIMB.2019.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Li XH and Chen WN (2018) Similarities and differences in gut microbiome composition correlate with dietary patterns of Indian and Chinese adults. AMB Express 8(1), 1–12. 10.1186/s13568-018-0632-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery IB, Lynch DB and O’Toole PW (2015) Composition and temporal stability of the gut microbiota in older persons. The ISME Journal 10(1), 170–182. 10.1038/ismej.2015.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AB, Sørensen TIA, Pedersen O, Jess T, Brunak S and Allin KH (2018) Increase in clinically recorded type 2 diabetes after colectomy. eLife 7, e37420. 10.7554/ELIFE.37420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, Xaus J, Fernández L and Rodríguez JM (2008) Is meconium from healthy newborns actually sterile? Research in Microbiology 159(3), 187–193. 10.1016/J.RESMIC.2007.12.007 [DOI] [PubMed] [Google Scholar]
- Jost T, Lacroix C, Braegger C and Chassard C (2013) Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. British Journal of Nutrition 110(7), 1253–1262. 10.1017/S0007114513000597 [DOI] [PubMed] [Google Scholar]
- Kabeerdoss J, Sankaran V, Pugazhendhi S and Ramakrishna BS (2013) Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: A case–control study in India. BMC Gastroenterology 13(1), 20. 10.1186/1471-230X-13-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeerdoss J, Shobana Devi R, Regina Mary R and Ramakrishna BS (2012) Faecal microbiota composition in vegetarians: Comparison with omnivores in a cohort of young women in southern India. The British Journal of Nutrition 108(6), 953–957. 10.1017/S0007114511006362 [DOI] [PubMed] [Google Scholar]
- Kaur K, Khatri I, Akhtar A, Subramanian S and Ramya TNC (2020) Metagenomics analysis reveals features unique to Indian distal gut microbiota. PLoS One 15(4), e0231197. 10.1371/JOURNAL.PONE.0231197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H and Laxminarayan R (2018) Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proceedings of the National Academy of Sciences of the United States of America 115(15), E3463–E3470. 10.1073/PNAS.1717295115/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Deng F, Li Y and Zhao J (2018) Identification of gut microbiome signatures associated with longevity provides a promising modulation target for healthy aging. Gut Microbes 10(2), 210–215. 10.1080/19490976.2018.1494102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Kling Bäckhed H, Gonzalez A, Werner JJ, Angenent LT, Knight R, Bäckhed F, Isolauri E, Salminen S and Ley RE (2012) Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150(3), 470–480. 10.1016/J.CELL.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen KHS, Wiese M, Rytter MJH, Özçam M, Hansen LH, Namusoke H, Friis H and Nielsen DS (2016) Gut microbiota in children hospitalized with oedematous and non-oedematous severe acute malnutrition in Uganda. PLoS Neglected Tropical Diseases 10(1), 1–11. 10.1371/journal.pntd.0004369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Babaei P, Ji B and Nielsen J (2016) Human gut microbiota and healthy aging: Recent developments and future prospective. Nutrition and Healthy Aging 4(1), 3–16. 10.3233/NHA-150002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R, Ahuja V and Paul J (2013) Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World Journal of Gastroenterology 19(22), 3404–3414. 10.3748/WJG.V19.I22.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhare SV, Kumar H, Chowdhury SP, Dhotre DP, Endo A, Mättö J, Ouwehand AC, Rautava S, Joshi R, Patil NP, Patil RH, Isolauri E, Bavdekar AR, Salminen S and Shouche YS (2017) A cross-sectional comparative study of gut bacterial community of Indian and Finnish children. Scientific Reports 7(1), 1–13. 10.1038/s41598-017-11215-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbhare SV, Patangia DV, Mongad DS, Bora A, Bavdekar AR and Shouche YS (2020) Gut microbial diversity during pregnancy and early infancy: An exploratory study in the Indian population. FEMS Microbiology Letters 367(3), 1–7. 10.1093/femsle/fnaa022 [DOI] [PubMed] [Google Scholar]
- Leocádio PCL, Lopes SC, Dias RP, Alvarez-Leite JI, Guerrant RL, Malva JO and Oriá RB (2021) The transition from undernutrition to overnutrition under adverse environments and poverty: The risk for chronic diseases. Frontiers in Nutrition 8, 676044. 10.3389/FNUT.2021.676044/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA, IBDMDB Investigators, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, DPB McGovern, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ and Huttenhower C (2019) Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569(7758), 655–662. 10.1038/s41586-019-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie RI, Sghir A and Gaskins HR (1999) Developmental microbial ecology of the neonatal gastrointestinal tract. The American Journal of Clinical Nutrition 69(5), 1035S–1045S. 10.1093/AJCN/69.5.1035S [DOI] [PubMed] [Google Scholar]
- Magnúsdóttir S, Ravcheev D, De Crécy-Lagard V and Thiele I (2015) Systematic genome assessment of B-vitamin biosynthesis suggests cooperation among gut microbes. Frontiers in Genetics 6, 148. 10.3389/fgene.2015.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matijašić M, Meštrović T, Paljetak HČ, Perić M, Barešić A and Verbanac D (2020) Gut microbiota beyond bacteria – Mycobiome, virome, archaeome, and eukaryotic parasites in IBD. International Journal of Molecular Sciences 21(8), 2668. 10.3390/IJMS21082668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Members (n.d.) https://idf.org/our-network/regions-members/south-east-asia/members/94-india.html (accessed 15 October 2022).
- Méndez-Salazar EO, Ortiz-López MG, Granados-Silvestre MDLÁ, Palacios-González B and Menjivar M (2018) Altered gut microbiota and compositional changes in firmicutes and proteobacteria in Mexican undernourished and obese children. Frontiers in Microbiology 9, 1–11. 10.3389/fmicb.2018.02494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million M, Diallo A and Raoult D (2017) Gut microbiota and malnutrition. Microbial Pathogenesis 106, 127–138. 10.1016/j.micpath.2016.02.003 [DOI] [PubMed] [Google Scholar]
- Mitchell CM, Mazzoni C, Hogstrom L, Bryant A, Bergerat A, Cher A, Pochan S, Herman P, Carrigan M, Sharp K, Huttenhower C, Lander ES, Vlamakis H, Xavier RJ and Yassour M (2020) Delivery mode affects stability of early infant gut microbiota. Cell Reports Medicine 1(9), 100156. 10.1016/J.XCRM.2020.100156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan TM, Sloan TJ, Stockdale SR, Blanchard AM, Emes RD, Wilcox M, Biswas R, Nashine R, Manke S, Gandhi J, Jain P, Bhotmange S, Ambalkar S, Satav A, Draper LA, Hill C and Kashyap RS (2020) Metagenomics reveals impact of geography and acute diarrheal disease on the central Indian human gut microbiome. Gut Microbes 12(1), 1752605. 10.1080/19490976.2020.1752605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monira S, Nakamura S, Gotoh K, Izutsu K, Watanabe H, Alam NH, Endtz HP, Cravioto A, Ali SI, Nakaya T, Horii T, Iida T and Alam M (2011) Gut microbiota of healthy and malnourished children in Bangladesh. Frontiers in Microbiology 2, 228. 10.3389/FMICB.2011.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais LH, Schreiber HL and Mazmanian SK (2020) The gut microbiota–brain axis in behaviour and brain disorders. Nature Reviews Microbiology 19(4), 241–255. 10.1038/s41579-020-00460-0 [DOI] [PubMed] [Google Scholar]
- Naghavi M, Wang H, Lozano R, Davis A, Liang X, Zhou M, Vollset SE, Abbasoglu Ozgoren A, Abdalla S, Abd-Allah F, Abdel Aziz MI, GBD 2015 Mortality and Causes of Death Collaborators (2015) Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the global burden of disease study 2013. The Lancet 385(9963), 117–171. 10.1016/S0140-6736(14)61682-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu, A. T. (2021). Defining and quantifying the core microbiome: Challenges and prospects . Proceedings of the National Academy of Sciences of the United States of America 118(51), 1–10. 10.1073/pnas.2104429118/-/DCSupplemental.Published [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obesity and overweight (n.d.) https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed 13 October 2022).
- Palleja A, Mikkelsen KH, Forslund SK, Kashani A, Allin KH, Nielsen T, Hansen TH, Liang S, Feng Q, Zhang C, Pyl PT, Coelho LP, Yang H, Wang J, Typas A, Nielsen MF, Nielsen HB, Bork P, Wang J, Vilsbøll T, Hansen T, Knop FK, Arumugam M and Pedersen O (2018) Recovery of gut microbiota of healthy adults following antibiotic exposure. Nature Microbiology 3(11), 1255–1265. 10.1038/s41564-018-0257-9 [DOI] [PubMed] [Google Scholar]
- Pan HW, Du LT, Li W, Yang YM, Zhang Y and Wang CX (2020) Biodiversity and richness shifts of mucosa-associated gut microbiota with progression of colorectal cancer. Research in Microbiology 171(3–4), 107–114. 10.1016/J.RESMIC.2020.01.001 [DOI] [PubMed] [Google Scholar]
- Panda S, El Khader I, Casellas F, López Vivancos J, García Cors M, Santiago A, Cuenca S, Guarner F and Manichanh C (2014) Short-term effect of antibiotics on human gut microbiota. PLoS One 9(4), e95476. 10.1371/JOURNAL.PONE.0095476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey PK, Verma P, Kumar H, Bavdekar A, Patole MS and Shouche YS (2012) Comparative analysis of fecal microflora of healthy full-term Indian infants born with different methods of delivery (vaginal vs cesarean): Acinetobacter sp. prevalence in vaginally born infants. Journal of Biosciences 37(6), 989–998. 10.1007/S12038-012-9268-5 [DOI] [PubMed] [Google Scholar]
- Pareek S, Kurakawa T, Das B, Motooka D, Nakaya S, Rongsen-Chandola T, Goyal N, Kayama H, Dodd D, Okumura R, Maeda Y, Fujimoto K, Nii T, Ogawa T, Iida T, Bhandari N, Kida T, Nakamura S, Nair GB and Takeda K (2019) Comparison of Japanese and Indian intestinal microbiota shows diet-dependent interaction between bacteria and fungi. npj Biofilms and Microbiomes 5(1), 37. 10.1038/s41522-019-0110-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker EPK, Praharaj I, John J, Kaliappan SP, Kampmann B, Kang G and Grassly NC (2017) Changes in the intestinal microbiota following the administration of azithromycin in a randomised placebo-controlled trial among infants in South India. Scientific Reports 7(1), 9168. 10.1038/s41598-017-06862-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson’s Disease: Causes, Symptoms, and Treatments (n.d.) National Institute on Aging. https://www.nia.nih.gov/health/parkinsons-disease (accessed 13 October 2022).
- Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE and Walter J (2017) A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: Implications for research on the pioneer infant microbiome. Microbiome 5(1), 1–19. 10.1186/S40168-017-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin J, Burrowes V, Almeida M, Ahmed S, Haque R, Parvin T, Biswas S, Azmi IJ, Bhuyian SI, Talukder KA, Faruque AG, Stine OC and George CM (2020) A retrospective case–control study of the relationship between the gut microbiota, enteropathy, and child growth. American Journal of Tropical Medicine and Hygiene 103(1), 520–527. 10.4269/ajtmh.19-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquer-Esteban S, Ruiz-Ruiz S, Arnau V, Diaz W and Moya A (2022) Exploring the universal healthy human gut microbiota around the world. Computational and Structural Biotechnology Journal 20, 421–433. 10.1016/J.CSBJ.2021.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ppatil D, Pdhotre D, Gchavan S, Sultan A, Jain DS, Lanjekar VB, Gangawani J, Sshah P, Stodkar J, Shah S, Ranade DR, Patole MS and Shouche YS (2012) Molecular analysis of gut microbiota in obesity among Indian individuals. Journal of Biosciences 37(4), 647–657. 10.1007/S12038-012-9244-0 [DOI] [PubMed] [Google Scholar]
- Pulikkan J, Maji A, Dhakan DB, Saxena R, Mohan B, Anto MM, Agarwal N, Grace T and Sharma VK (2018) Gut microbial dysbiosis in Indian children with autism spectrum disorders. Microbial Ecology 76(4), 1102–1114. 10.1007/s00248-018-1176-2 [DOI] [PubMed] [Google Scholar]
- Pulipati P, Sarkar P, Jakkampudi A, Kaila V, Sarkar S, Unnisa M, Reddy DN, Khan M and Talukdar R (2020) The Indian gut microbiota-is it unique? Indian Journal of Gastroenterology: Official Journal of the Indian Society of Gastroenterology 39(2), 133–140. 10.1007/S12664-020-01037-8 [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD and Wang J (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464(7285), 59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radjabzadeh D, Boer CG, Beth SA, van der Wal P, Kiefte-De Jong JC, Jansen MAE, Konstantinov SR, Peppelenbosch MP, Hays JP, Jaddoe VWV, Ikram MA, Rivadeneira F, van Meurs JBJ, Uitterlinden AG, Medina-Gomez C, Moll HA and Kraaij R (2020) Diversity, compositional and functional differences between gut microbiota of children and adults. Scientific Reports 10(1), 1–13. 10.1038/s41598-020-57734-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadass B, Rani BS, Pugazhendhi S, John KR and Ramakrishna BS (2017) Faecal microbiota of healthy adults in South India: Comparison of a tribal & a rural population. The Indian Journal of Medical Research 145(2), 237–246. 10.4103/IJMR.IJMR_639_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna BS (2013) Role of the gut microbiota in human nutrition and metabolism. Journal of Gastroenterology and Hepatology 28(S4), 9–17. 10.1111/JGH.12294 [DOI] [PubMed] [Google Scholar]
- Ramirez J, Guarner F, Bustos Fernandez L, Maruy A, Sdepanian VL and Cohen H (2020) Antibiotics as major disruptors of gut microbiota. Frontiers in Cellular and Infection Microbiology 10, 572912. 10.3389/FCIMB.2020.572912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, O’Toole PW and Brigidi P (2013) Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging 5(12), 902–912. 10.18632/AGING.100623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S, Guenther K, Turroni S, Wolters M, Veidebaum T, Kourides Y, Molnár D, Lissner L, Benitez-Paez A, Sanz Y, Fraterman A, Michels N, Brigidi P, Candela M and Ahrens W (2018) Pre-obese children’s dysbiotic gut microbiome and unhealthy diets may predict the development of obesity. Communications Biology 1(1), 1–11. 10.1038/s42003-018-0221-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A and Mele MC (2019) What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 7(1), 14. 10.3390/microorganisms7010014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roswall J, Olsson LM, Kovatcheva-Datchary P, Nilsson S, Tremaroli V, Simon MC, Kiilerich P, Akrami R, Krämer M, Uhlén M, Gummesson A, Kristiansen K, Dahlgren J and Bäckhed F (2021) Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host and Microbe 29(5), 765–776.e3. 10.1016/j.chom.2021.02.021 [DOI] [PubMed] [Google Scholar]
- Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I and Tuohy K (2018) Gut microbiota functions: Metabolism of nutrients and other food components. European Journal of Nutrition 57, 1–24. 10.1007/S00394-017-1445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan W, Engevik MA, Spinler JK and Versalovic J (2020) Healthy human gastrointestinal microbiome: Composition and function after a decade of exploration. Digestive Diseases and Sciences 65(3), 695–705. 10.1007/S10620-020-06118-4 [DOI] [PubMed] [Google Scholar]
- Sarjapuram N, Mekala N, Singh M and Tatu U (2017) The potential of Lactobacillus casei and Entercoccus faecium combination as a preventive probiotic against Entamoeba. Probiotics and Antimicrobial Proteins 9(2), 142–149. 10.1007/S12602-016-9232-Z [DOI] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C and Hardt PD (2010) Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18(1), 190–195. 10.1038/OBY.2009.167 [DOI] [PubMed] [Google Scholar]
- Sender R, Fuchs S and Milo R (2016) Revised estimates for the number of human and bacteria cells in the body. PLoS Biology 14(8), e1002533. 10.1371/JOURNAL.PBIO.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Rodionov DA, Leyn SA, Tran D, Iablokov SN, Ding H, Peterson DA, Osterman AL and Peterson SN (2019) B-vitamin sharing promotes stability of gut microbial communities. Frontiers in Microbiology 10, 1485. 10.3389/fmicb.2019.01485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty SA, Marathe NP and Shouche YS (2013) Opportunities and challenges for gut microbiome studies in the Indian population. Microbiome 1(1), 1–12. 10.1186/2049-2618-1-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar N, Sivadas A, Devi S, Jahoor F, McLaughlin J, Smith CP, Kurpad AV and Mukhopadhyay A (2021) Gut microbiota profiles of young south Indian children: Child sex-specific relations with growth. PLoS One 16, 1–22. 10.1371/journal.pone.0251803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreiner AB, Kao JY and Young VB (2015) The gut microbiome in health and in disease. Current Opinion in Gastroenterology 31(1), 69–75. 10.1097/MOG.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G, Sharma M, Kumar GA, Rao NG, Prasad K, Mathur P, Pandian JD, Steinmetz JD, Biswas A, Pal PK, Prakash S, Sylaja PN, Nichols E, Dua T, Kaur H, Alladi S, Agarwal V, Aggarwal S, Ambekar A, Bagepally BS, Banerjee TK, Bender RG, Bhagwat S, Bhargava S, Bhatia R, Chakma JK, Chowdhary N, Dey S, Dirac MA, Feigin VL, Ganguli A, Golechha MJ, Gourie-Devi M, Goyal V, Gupta G, Gupta PC, Gupta R, Gururaj G, Hemalatha R, Jeemon P, Johnson CO, Joshi P, Kant R, Kataki AC, Khurana D, Krishnankutty RP, Kyu HH, Lim SS, Lodha R, Ma R, Malhotra R, Malhotra R, Mathai M, Mehrotra R, Misra UK, Mutreja P, Naghavi M, Naik N, Nguyen M, Pandey A, Parmar P, Perianayagam A, Prabhakaran D, Rath GK, Reinig N, Roth GA, Sagar R, Sankar MJ, Shaji KS, Sharma RS, Sharma S, Singh R, MVP Srivastava, Stark BA, Tandon N, Thakur JS, AS ThekkePurakkal, Thomas SV, Tripathi M, Vongpradith A, Wunrow HY, Xavier D, Shukla DK, Reddy KS, Panda S, Dandona R, CJL Murray, Vos T, Dhaliwal RS and Dandona L (2021) The burden of neurological disorders across the states of India: The global burden of disease study 1990–2019. The Lancet. Global Health 9(8), e1129–e1144. 10.1016/S2214-109X(21)00164-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman R (2018) Prokaryotic horizontal gene transfer within the human holobiont: Ecological-evolutionary inferences, implications and possibilities. Microbiome 6(1), 1–14. 10.1186/S40168-018-0551-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits SA, Leach J, Sonnenburg ED, Gonzalez CG, Lichtman JS, Reid G, Knight R, Manjurano A, Changalucha J, Elias JE, Dominguez-Bello MG and Sonnenburg JL (2017) Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357(6353), 802–806. 10.1126/science.aan4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober W, Fuss I and Mannon P (2007) The fundamental basis of inflammatory bowel disease. Journal of Clinical Investigation 117(3), 514–521. 10.1172/JCI30587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surono IS, Widiyanti D, Kusumo PD and Venema K (2021) Gut microbiota profile of Indonesian stunted children and children with normal nutritional status. PLoS One 16(1), e0245399. 10.1371/JOURNAL.PONE.0245399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szóstak N, Szymanek A, Havránek J, Tomela K, Rakoczy M, Samelak-Czajka A, Schmidt M, Figlerowicz M, Majta J, Milanowska-Zabel K, Handschuh L and Philips A (2022) The standardisation of the approach to metagenomic human gut analysis: From sample collection to microbiome profiling. Scientific Reports 12(1), 1–21. 10.1038/s41598-022-12037-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar R, Sarkar P, Jakkampudi A, Sarkar S, Aslam M, Jandhyala M, Deepika G, Unnisa M and Reddy DN (2021) The gut microbiome in pancreatogenic diabetes differs from that of type 1 and type 2 diabetes. Scientific Reports 11(1), 1–12. 10.1038/s41598-021-90024-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapiainen T, Paalanne N, Tejesvi MV, Koivusaari P, Korpela K, Pokka T, Salo J, Kaukola T, Pirttilä AM, Uhari M and Renko M (2018) Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatric Research 84(3), 371–379. 10.1038/PR.2018.29 [DOI] [PubMed] [Google Scholar]
- Thakur N, Changotra H, Grover N and Vashistt J (2018) Elucidation of bacterial species during childhood diarrhea through 16S rRNA Illumina Miseq approach. Meta Gene 16, 234–240. 10.1016/J.MGENE.2018.03.012 [DOI] [Google Scholar]
- Tuikhar N, Keisam S, Labala RK, Imrat, Ramakrishnan P, Arunkumar MC, Ahmed G, Biagi E and Jeyaram K (2019). Comparative analysis of the gut microbiota in centenarians and young adults shows a common signature across genotypically non-related populations. Mechanisms of Ageing and Development 179(February), 23–35. 10.1016/j.mad.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Underwood MA, German JB, Lebrilla CB and Mills DA (2015) Bifidobacterium longum subspecies infantis: Champion colonizer of the infant gut. Pediatric Research 77, 229–235. 10.1038/PR.2014.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Verma AK, Ahuja V and Paul J (2010) Real-time analysis of mucosal flora in patients with inflammatory bowel disease in India. Journal of Clinical Microbiology 48(11), 4279–4282. 10.1128/JCM.01360-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma AK, Verma R, Ahuja V and Paul J (2012) Real-time analysis of gut flora in Entamoeba histolytica infected patients of northern India. BMC Microbiology 12(1), 1–11. 10.1186/1471-2180-12-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, Carlsson CM, Asthana S, Zetterberg H, Blennow K, Bendlin BB and Rey FE (2017) Gut microbiome alterations in Alzheimer’s disease. Scientific Reports 7(1), 1–11. 10.1038/s41598-017-13601-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogtmann E, Hua X, Zeller G, Sunagawa S, Voigt AY, Hercog R, Goedert JJ, Shi J, Bork P and Sinha R (2016) Colorectal cancer and the human gut microbiome: Reproducibility with whole-genome shotgun sequencing. PLoS One 11(5), e0155362. 10.1371/JOURNAL.PONE.0155362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wintersdorff CJ, Penders J, Stobberingh EE, Oude Lashof AM, Hoebe CJ, Savelkoul PH and Wolffs PF (2014) High rates of antimicrobial drug resistance gene acquisition after international travel, the Netherlands. Emerging Infectious Diseases 20(4), 649–657. 10.3201/eid.2004.131718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker RW, Clemente JC, Peter I and Loos RJF (2017) The prenatal gut microbiome: Are we colonized with bacteria in utero? Pediatric Obesity 12(Suppl 1), 3–17. 10.1111/IJPO.12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S and Zhao L (2012) Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME Journal 6(2), 320–329. 10.1038/ISMEJ.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto J-M, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K and Wang J (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490(7418), 55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Wang T, Yu R, Zhu L, Wang X and Yang B (2022) Differences in the intestinal flora of patients with inflammatory bowel disease in Southwest China. Indian Journal of Microbiology 62(3), 384–392. 10.1007/S12088-022-01014-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM and Panigrahi P (2014) Is a foetus developing in a sterile environment? Letters in Applied Microbiology 59(6), 572–579. 10.1111/LAM.12334 [DOI] [PubMed] [Google Scholar]
- Weis S, Schwiertz A, Unger MM, Becker A, Faßbender K, Ratering S, Kohl M, Schnell S, Schäfer KH and Egert M (2019) Effect of Parkinson’s disease and related medications on the composition of the fecal bacterial microbiota. Npj Parkinson’s Disease 5(1), 1–9. 10.1038/s41531-019-0100-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa Y, Nagata N, Yagita K, Watanabe K, Okubo H, Kikuchi Y, Gatanaga H, Oka S and Watanabe K (2021) Clinical features and gut microbiome of asymptomatic Entamoeba histolytica infection. Clinical Infectious Diseases 73(9), e3163–e3171. 10.1093/CID/CIAA820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, Lander ES, Knip M and Xavier RJ (2016) Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Science Translational Medicine 8(343), 343ra81. 10.1126/SCITRANSLMED.AAD0917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight D and Gordon JI (2012) Human gut microbiome viewed across age and geography. Nature 486(7402), 222–227. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Gao X, Wang Z, Cao S, Liang G, He D, Lv Z, Wang L, Xu P and Zhang Q (2021) Comparison of gut microbiota in autism spectrum disorders and neurotypical boys in China: A case–control study. Synthetic and Systems Biotechnology 6(2), 120–126. 10.1016/J.SYNBIO.2021.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, He X, Qian Y, Xu S, Mo C, Yan Z, Yang X and Xiao Q (2022) Plasma branched-chain and aromatic amino acids correlate with the gut microbiota and severity of Parkinson’s disease. Npj Parkinson’s Disease 8(1), 1–10. 10.1038/s41531-022-00312-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yu X, Yu E, Wang N, Cai Q, Shuai Q, Yan F, Jiang L, Wang H, Liu J, Chen Y, Li Z and Jiang Q (2018) Changes in gut microbiota and plasma inflammatory factors across the stages of colorectal tumorigenesis: A case–control study. BMC Microbiology 18(1), 92. 10.1186/S12866-018-1232-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Ren H, Lu Y, Fang C, Hou G, Yang Z, Chen B, Yang F, Zhao Y, Shi Z, Zhou B, Wu J, Zou H, Zi J, Chen J, Bao X, Hu Y, Gao Y, Zhang J, Xu X, Hou Y, Yang H, Wang J, Liu S, Jia H, Madsen L, Brix S, Kristiansen K, Liu F and Li J (2019) Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naïve type 2 diabetics. eBioMedicine 47, 373–383. 10.1016/J.EBIOM.2019.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou R, Xu F, Wang Y, Duan M, Guo M, Zhang Q, Zhao H and Zheng H (2020) Changes in the gut microbiota of children with autism spectrum disorder. Autism Research 13(9), 1614–1625. 10.1002/AUR.2358 [DOI] [PubMed] [Google Scholar]