Abstract
Reln mRNA and protein levels are reduced by ∼50% in various cortical structures of post-mortem brain from patients diagnosed with schizophrenia or bipolar illness with psychosis. To study mechanisms responsible for this down-regulation, we have analyzed the promoter of the human reelin gene. We show that the reelin promoter directs expression of a reporter construct in multiple human cell types: neuroblastoma cells (SHSY5Y), neuronal precursor cells (NT2), differentiated neurons (hNT) and hepatoma cells (HepG2). Deletion constructs confirmed the presence of multiple elements regulating Reln expression, although the promoter activity is promiscuous, i.e. activity did not correlate with expression of the endogenous gene as reflected in terms of reelin mRNA levels. Co-transfection of the –514 bp human reelin promoter with either Sp1 or Tbr1 demonstrated that these transcription factors activate reporter expression by 6- and 8.5-fold, respectively. Within 400 bp of the RNA start site there are 100 potential CpG targets for DNA methylation. Retinoic acid (RA)-induced differentiation of NT2 cells to hNT neurons was accompanied by increased reelin expression and by the appearance of three DNase I hypersensitive sites 5′ to the RNA start site. RA-induced differentiation was also associated with demethylation of the reelin promoter. To test if methylation silenced reelin expression, we methylated the promoter in vitro prior to transfection. In addition, we treated NT2 cells with the methylation inhibitor aza-2′-deoxycytidine and observed a 60-fold increase in reelin mRNA levels. The histone deacetylase inhibitors trichostatin A (TSA) and valproic acid also induced expression of the endogenous reelin promoter, although TSA was considerably more potent. These findings indicate that one determinant responsible for regulating reelin expression is the methylation status of the promoter. Our data also raise the interesting possibility that the down-regulation of reelin expression documented in psychiatric patients might be the consequence of inappropriate promoter hypermethylation.
INTRODUCTION
To date, in all post-mortem brain areas obtained from patients diagnosed with schizophrenia, reelin and glutamate decarboxylase 67 mRNA and protein levels are reduced by ∼50% (1). This includes multiple cortical regions, hippocampi and cerebella (reviewed in 2). These findings were replicated in the Stanley Foundation cohort of 60 brains, and similar decreases in both parameters were obtained in the prefrontal cortices and cerebella of schizophrenia and bipolar patients with psychosis but not in patients with unipolar depression (3). Since there were no changes in the levels of GAD65 immunoreactivity, which is expressed in the same neurons that express GAD67, and the changes persisted after adjusting for the presence of a specific neuronal marker, these results cannot be explained by a neuronal loss related to the illness. Interestingly, a reduction in the size of the neuropil (4) was detected that could be related to a decrease in the dendritic spines of cortical pyramidal neurons associated with reduced reelin levels. Another report reproduced the decreased reelin levels in patients with schizophrenia and also noted a decrease in CA4 areas of subjects with bipolar disorder and a non-significant decrease in this same region in patients with major depression (5). The decrease observed in the hippocampus of unipolar depressed patients is interesting, but the significance remains unclear. Nevertheless, there is considerable interest in understanding mechanisms underlying the regulation of human reelin expression and it is our goal to provide the requisite framework for this understanding to allow for more detailed studies in the context of psychiatric illness.
Reelin is an extracellular matrix protein (6) that is expressed in the developing brain and continues to be expressed during the lifespan of GABAergic neurons of the cortex and hippocampus, in glutamatergic granule cells of the cerebella, in neostriatal medium spiny neurons of adult rodents (7,8), in non-human primates (9) and in many of these structures in adult humans (1,3,10). Very little is currently known with respect to how the expression of reelin is regulated in mature neurons. The human and mouse cDNAs share 88% nucleotide identity and show a high degree of sequence similarity surrounding the start site of transcription (11). Sequences flanking the human RNA start site are very GC-rich (75%) and form a large CpG island. This suggested to us that the reelin promoter might be epigenetically regulated through changes in DNA cytosine methylation (12–15). In contrast to the vast majority of CpG dinucleotides in the genome which are generally methylated, CpG islands tend to be undermethylated and this pattern can vary in both a temporal and spatially selective manner (14,16). This provides one mechanism by which promoters embedded in CpG islands may be differentially regulated.
In addition, it has been proposed that there may be an interplay between transcription factor binding and DNA methylation of promoters regulated epigenetically through alterations in their patterns of methylation (17). The interaction of Sp1, for example, with its recognition site in the adenine phosphoribosyltransferase promoter has been suggested as a signal for keeping the CpG island upstream of the adenine phosphoribosyltransferase gene free of methylation in mouse embryonic stem cells (18). Equally important in this context is the interplay between methylation, chromatin structure and histone deacetylaton. Transcription factors and methyl-CpG-binding proteins interact with various classes of histone deacetylases (HDACs) in complexes that repress transcription and perhaps induce demethylation (14). This would provide a means by which genes could be turned on and off through epigenetic switches that ultimately regulate local chromatin structure and gene expression. While this type of mechanism has not been shown to operate in regulating genes expressed in post-mitotic cells, the role of methylation/demethylation as a determinant dynamically regulating gene activity in neurons is largely unexplored.
In the present study, we examined the human reelin promoter using transient transfection assays to identify regions that participate in targeting expression to both reelin mRNA-expressing and non-expressing cell lines. We also demonstrate that the non-methylatable cytosine analog 5-aza-deoxycytidine (AzAdc), which prevents methylation, activates expression of the endogenous reelin gene, while in vitro methylation of the reelin promoter silences expression. In addition, we present data that show that the increased expression of the reelin gene following retinoic acid (RA)-induced differentiation of neural progenitor cells in vitro is accompanied by a reduced methylation of the reelin promoter. Finally, we examined the effect of altering the pattern of histone acetylation of neural progenitor (NT2) cells on reelin expression. The HDAC inhibitor trichostatin A (TSA) increased reelin mRNA levels. Of particular interest in the area of psychiatric pharmacology, the mood-stabilizing drug valproic acid (VPA) has been shown to act as an HDAC inhibitor (19,20). VPA moderately increased reelin expression, although it was considerably less potent than TSA. These studies provide a framework for the pursuit of hypotheses which propose that alterations in epigenetic gene regulation may be relevant to neuropsychiatric disease.
MATERIALS AND METHODS
Cell culture
SHSY5Y cells were maintained in a 1:1 mixture of Eagle’s MEM/F12K (Life Technologies) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin, streptomycin and glutamine. HepG2 cells were grown in HAM supplemented with 10% FBS and 1% penicillin/streptomycin. NT2 cells were maintained in DMEM/F12, 10% FBS and 1% penicillin, streptomycin and glutamine. hNT neurons were induced from cultures of NT2 cells by treating with RA for up to 6 weeks. Low density cultures of NT2 neural progenitors were treated with AzAdC at several concentrations (1, 5 and 10 µM) and TSA (0.2, 1 and 5 µM) for various times and cells were harvested for total RNA isolation. In parallel, NT2 cells were treated with various concentrations of VPA (0.2, 2, 5 and 10 mM) and the inactive VPA amide valpromide (VPM) (5 mM) for 40 h and cells were harvested for RNA analysis. RNA was isolated following ultracentrifugation through CsCl as previously described (21).
Generation of the reelin promoter/reporter constructs
The 5′ portion of the human mRNA and upstream sequence maps to human BAC clone RG126M09, which contains 163 kb of human genomic DNA. We subcloned a 3′ terminal 4.2 kb EcoRI fragment from this BAC DNA which contained the entire first exon, 255 bp of the first intron and 3.7 kb of 5′ flanking DNA (Fig. 1A). To generate the reelin –514 bp minimal promoter construct, we designed primers to PCR amplify this region from the BAC DNA. The sequences of the primers were: –514 bp, 5′-AAA AAC AGG GCA CAC TGA CGG CCA-3′; +76 bp, 5′-CGG AGA GAA GGC GAG AAG AAG GCG-3′. Following PCR, the amplicon was subcloned into the pGL3-Basic luciferase vector (Promega/Fisher). The –514 bp promoter/reporter was used to introduce additional upstream sequences using the restriction sites indicated in Figure 1A and the downstream AatII site. The –514 bp promoter/reporter was also used to generate a series of Bal31 deletions as previously described (22). Sequences between –303 and –137 were cloned upstream of the SV40 promoter using the pGL-3-Promoter parent vector (Promega/Fisher) in both orientations (sense and antisense). We assessed activity by transient transfection of numerous cell lines.
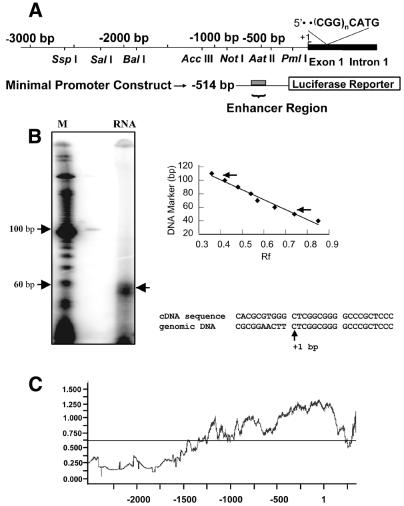
Figure 1.
Reelin promoter structure and transient transfections. (A) Schematic representation of the cloned human reelin promoter. The top line shows a linear representation of sequences along the BAC DNA and associated restriction enzyme cleavage sites. The transcriptional start site is indicated (+1). The position of the CGG repeat present in the 5′ UTR of the first exon is shown where n varies from 4 to 24 repeats. The subcloned minimal promoter/luciferase reporter vector contains a region (reelin enhancer) that acts to enhance expression of the SV40 promoter. (B) Primer extension analysis. The products of reverse transcription of SHSY5Y RNA using the +60 bp primer is shown. To the right of the gel is the standard curve that was generated and the position of migration of this product and that corresponding to the +110 bp primer. The sequence alignment shows a comparison of the human reelin cDNA (GenBank accession no. NM_005045) and the genomic sequence (as shown in Fig. 4B). Similar results were obtained using RNA from HepG2 cells (data not shown). (C) CpG islands plot of the human reelin promoter and first exon. The CpG islands plot was generated using GeneTool software (BioTools) that is based on an algorithm that plots the observed number of CpG sites divided by the expected number of CpG sites along the length of the DNA fragment. The numbering is based on the RNA start site being equal to +1. y-axis values above the horizontal line indicate regions likely to be CpG islands (28).
The RNA start site was mapped using a primer extension assay (as previously described; 22). In brief, the reelin +60 primer (reverse complement sequence underlined in Fig. 4B) was end-labeled using T4 polynucleotide kinase. The specific activity of the primer was 300 000 c.p.m./ng. The primer (5 ng) and 10 µg cellular RNA (SHSY5Y or HepG2) were denatured in 15 µl of hybridization buffer (10 mM Tris–HCl pH 8.3, 150 mM KCl, 1 mM EDTA) at 85°C for 5 min and hybridized at 50°C for 30 min. Primer extension was carried out by adding an aliquot of the primer extension mix (to 20 mM Tris–HCl pH 8.3, 10 mM MgCl2, 10 mM DTT, 1.6 mM dNTP, with 10 U AMV reverse transcriptase). The reaction was incubated at 42°C for 1 h. RNase reaction mix (100 µg/ml salmon sperm DNA and 20 µg/ml RNase A) was added to the sample and incubated at 37°C for 30 min. Following phenol/chloroform extraction and ethanol precipitation, the reaction products were analyzed on a 10% denaturing TBE–urea gel (Invitrogen). The gel was dried and exposed to a phosphorimager screen. The image was scanned using a Storm PhosphorImager (Molecular Dynamics) to visualize the reaction products. The 10 bp ladder DNA marker was used for sizing the products (Invitrogen).
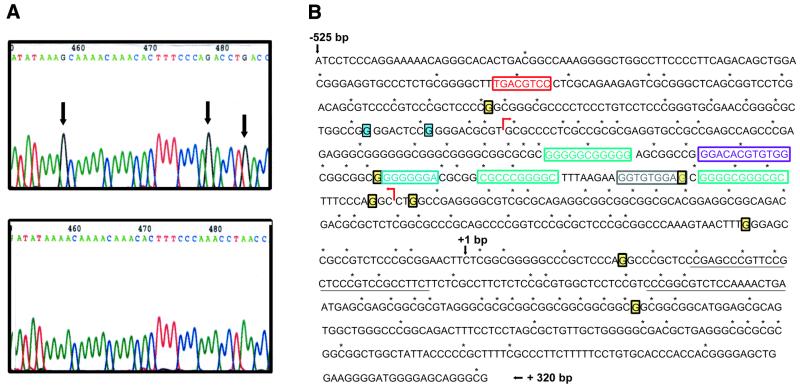
Figure 4.
Methylation status of the reelin promoter. (A) Genomic DNA was isolated from NT2 cells (top) or RA-differentiated hNT neurons (bottom) and modified using the bisulfite reaction. The DNA was amplified and subcloned. Subsequent sequencing showed differences indicated by the arrows in which the presence of a G residue indicates a methylated cytosine on the opposite strand. The numbering refers to the position in the sequencing chromatogram. A total of eight residues were modified in NT2 cells while two different G residues were methylated in the hNT neurons. (B) Data obtained from the methylation studies are summarized. Individual G residues shaded yellow were methylated in NT2 cells, while those that are boxed and shaded blue were methylated in hNT neurons. The red arrows show the boundaries of the enhancer region. The boxed base pairs indicate putative binding sites for a variety of transcription factors, including Sp1 (aqua), CREB (red), N-myc (purple), Mzf1 (light blue) and Tbr1 (gray). The middle Sp1 site is also a putative AP-2 element. Sequences underlined downstream of +1 indicate the position of the primer used in the primer extension analysis (Fig. 1B).
DNase I hypersensitive site analysis
Nuclei were isolated from either NT2 neuroprogenitor cells or from hNT neurons as previously described (23). Isolated nuclei from either group were treated with increasing amounts of DNase I (0, 10, 20, 30, 40, 60, 80 and 100 U DNase I, FPLC pure; Pharmacia) for 10 min at 37°C. Following treatment, the reaction was stopped by the addition of 50 mM Tris–HCl pH 7.5, 100 mM NaCl and 1% SDS. Genomic DNA was extracted, purified and digested with EcoRI and subjected to agarose gel electrophoresis and Southern blotting. The blot was hybridized with random primed, 32P-labeled probe corresponding to the 5′ most portion of the subcloned genomic EcoRI fragment (shown in Fig. 1A). The specific activity of the probe used was 1 × 109 d.p.m./µg. The washed blots were exposed to phosphorimager screens for 12–16 h and the image was obtained by scanning the screen with a Storm PhosphorImager (Molecular Dynamics).
Transfection assays and reporter expression measurements
Cells were transfected using Lipofectamine 2000 (Life Technologies). We used the dual-luciferase reporter assay system (Promega/Fisher) and routinely used 2–4 µg each test DNA with 30 ng pRL-CMV vector (Renilla luciferase) for each well of a 6-well plate. Cell lysates were prepared 24–36 h after transfection and aliquots (∼20 µl) were used for determination of luciferase activity in a TD20/20 luminometer (Turner Design). Data (minimum of three to five transfections/construct) are expressed as a ratio to the signal obtained from the SV40 promoter/luciferase vector that was transfected in parallel (pGL3-Control vector; Promega/Fisher).
Competitive RT–PCR assay
We previously developed the use of competitive RT–PCR to make quantitative measurements of specific mRNAs in total cellular RNA with specific internal standards (24,25). The internal standard corresponding to reelin mRNA has been described (1). Following RT–PCR, amplicons were digested with BanI and electrophoresed on agarose gels. Gels were stained with Syber Gold (Molecular Probes) and scanned directly using a Storm PhosphorImager (Molecular Dynamics). The RNA analysis was performed three times per sample from a minimum of three different RNA isolations. The data were analyzed for significance using a one-way ANOVA.
In vitro DNA methylation reaction
Escherichia coli DNA methylases were used to methylate the –514 promoter/luciferase construct to test the effects of sequence-specific methylation on reporter activity. The following methylases were used, each of which is followed by its selectivity and number of sites present in the template: SssI (*CG), 122 sites in the –514 promoter construct; HpaII (C*CGG), 10 sites; MspI (*CCGG), 11 sites; HhaI (G*CGC), 21 sites; HaeIII (GG*CC), 10 sites. The * indicates the methylated cytosine. An aliquot of 10 µg –514 promoter/reporter was used per reaction with 25 U enzyme in 100 mM NaCl, 50 mM Tris–HCl pH 7.5, 5 mM 2-mercaptoethanol and 80 µM S-adenosylmethionine for 10 h at 30°C. The extent of methylation was examined by digesting aliquots of the reaction with the corresponding restriction enzyme. The effect of methylation on promoter activity was assessed by transfecting the various constructs as indicated above.
Bisulfite modification of genomic DNA
Genomic DNA from various samples (NT2 and hNT neurons) was isolated using the proteinase K/SDS method (26) and was digested with EcoRI and denatured by treating with NaOH. Sodium bisulfite (5 M) converts deoxycytosine but not 5-methylcytosine residues into uracil, and the reaction was performed as described (27). Following the modification, the strands (designated the A and B strands) are no longer complementary and different sets of primers were designed for each strand. Nested sets of primers were designed for both strands. For the B strand: outer primers, –790 bp, 5′-TTT AAA ATC CTC TAC AAA TAA AAC TCT ATC ACT-3′, and +350 bp, 5′-TGT TTG TAA TAT GTA GGG AAA TGA GTA TTT-3′; inner primers, –527 bp, 5′-ACA TCC TCC CAA AAA AAA CAA AAC ACA CTA A-3′, and +305 bp, 5′-TTT TTT TAG TTT TTT GTG GTG GGT GTA TAG GAA-3′. The reaction products of the B strand were amplified and subcloned. Multiple clones from NT2 neural prognitor cells and from hNT neurons were sequenced by the UIC sequencing facility using fluorescent dideoxy technology.
RESULTS
The murine reelin mRNA is encoded by a gene that contains 65 exons which span ∼450 kb of genomic DNA. The complete intron/exon structure of the rodent gene has been mapped (11). We have mapped the intron/exon structure of human Reln to various BAC clones and have found that the intron/exon boundaries are remarkably conserved, as are the sizes of the corresponding introns (D. R. Grayson, unpublished observations). In the course of making these comparisons we noticed a discrepancy between the 5′ end of the human mRNA (10) and the genomic sequence containing the remainder of the first exon (BAC clone 96012; Research Genetics). The difference resided in the first 10 bp of the cDNA, which were not present in the genomic sequence. To verify the location of the RNA start site within the genomic sequence of the BAC, we performed primer extension assays using primers that mapped to within 60 and 110 bp of the region that differed in the genomic DNA and the cDNA. As can be seen from the analysis (Fig. 1B), the assay yielded a single start site using RNA from SHSY5Y cells. Similar results were obtained with RNA obtained from HepG2 cells (data not shown). The size of the products obtained with each primer are indicated on the standard curve. This site corresponds to that predicted from the genomic sequence, indicating the possibility that the first 10 bp of the cDNA may be present due to a cloning artifact (Fig. 1B). The downstream primer yielded several minor bands much like the mouse primer extension, which showed multiple start sites (11). Because these additional products were not seen with both primers, we presume that the extra sites likely represent pausing artifacts due to the high GC content of the template.
Similar to the rodent promoter region (11), sequences flanking the human RNA start site are very GC-rich (75%) and, together with the first exon, form a large CpG island. A linear representation of the CpG islands plot corresponding to sequences flanking the human reelin RNA start site is shown in Figure 1C. The abscissa represents the observed number of CpG sites divided by the expected number of CpG sites assuming random sorting along the reelin promoter sequence. The reelin promoter and first exon sequences are shown along the ordinate, where the transcriptional start site is 1 and sequences 5′ to this are represented as negative numbers while downstream sequences are positive. A candidate CpG island gives a y-value of >0.6 on this CpG islands plot over an extended window size (28). Using these criteria, the reelin promoter CpG island extends from approximately –1200 to +200 bp relative to the transcription start site.
Transient transfection analysis of the human reelin promoter
To create promoter expression vectors, we PCR amplified and subcloned a region that extends from –514 to + 76 bp relative to the human RNA start site (minimal promoter, Fig. 1A). The region subcloned extends into the 5′ UTR but does not include the previously described CGG repeat region (29,30). This triplet repeat was excluded so as to avoid any translational effects that might be associated with variations in triplet repeat length. The reelin promoter constructs, along with the normalization control (pRLCMV; Promega) were transiently transfected into human neural progenitor cells (NT2, low reelin levels, 0.0076 ± 0.00074 pg Reln mRNA/µg total RNA), RA-differentiated NT2 cells (hNT, high reelin mRNA levels, 0.75 ± 0.046 pg Reln mRNA/µg total RNA), neuroblastoma cells (SHSY5Y) and hepatoma cells (HepG2). SHSY5Y and HepG2 cells express intermediate reelin mRNA levels. Following DNA transfection, cell protein lysates were prepared and the amount of luciferase activity was quantified. The data showed that the reelin promoter is actively expressed in each of the cell types (Fig. 2A) and that the reporter activity was independent of the endogenous reelin mRNA levels. For example, high reelin promoter expression was observed in NT2 cells which express only very low levels of the endogenous reelin transcript (Table 1). The results suggest that individual variations in the levels of expression arising from each reelin promoter construct likely reflect differences in transcription factor binding but that biologically accurate reelin expression requires additional information that likely occurs at the level of chromatin structure.
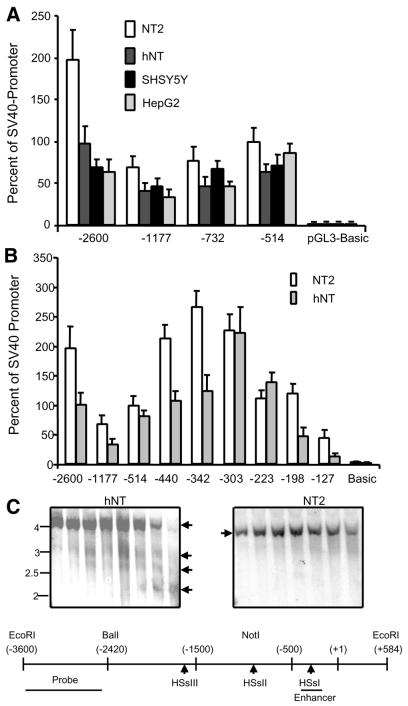
Figure 2.
Transfection analysis of the reelin promoter. (A) Transfection of the reelin promoter/luciferase reporter constructs into various cell lines. The restriction sites indicated in Figure 1A were used to generate the promoter fragments (–2600 contains 2.6 kb of 5′ flanking sequence; –1177, 1177 bp of 5′ flanking sequence; R-732, 732 bp of 5′ flanking sequence; R-514, 514 bp of 5′ flanking sequence). (B) Results obtained by transfection of various reelin promoter constructs and deletions into either NT2 neuroprogenitor cells or hNT neurons. The number below each pair of bars indicates the amount of 5′ flanking sequence present in the construct (relative to the RNA start site). Data represent the mean obtained from three measurements made from a minimum of three separate experiments after correcting for transfection efficiency. (C) DNase I hypersensitive sites are present in hNT neurons and not NT2 neuroprogenitor cells. NT2 cells were incubated with RA to induce differentiation as indicated in Materials and Methods. Nuclei were isolated from both cell types and incubated with DNase I. The genomic DNA was resolved by agarose gel electrophoresis and Southern blotted onto nitrocellulose membranes. The indicated probe was used to visualize the relevant bands. Arrows indicate sites that are hypersensitive. The genomic sequence is represented in the bottom portion of the panel with the approximate positions of the hypersensitive sites relative to the start site. The DNA probe used is indicated at the 5′-most portion of the promoter.
Table 1. Quantitative analysis of reelin mRNA levels following treatment of NT2 cells with a non-methylatable deoxycytosine analog and TSA.
| Cell | Treatment | Time (h) | Concentration (µM) | Amount of Reln RNAa (pg Reln RNA/µg RNA) | Fold increase |
|---|---|---|---|---|---|
| NT2 | Control | 0.0076 ± 0.00074 | |||
| 5′-AzAdC | 24 | 1 | 0.15 ± 0.021b | 21 ± 2.9 | |
| 5 | 0.17 ± 0.022b | 23 ± 2.9 | |||
| 10 | 0.14 ± 0.021b | 19 ± 2.8 | |||
| 72 | 1 | 0.40 ± 0.054b | 56 ± 6.0 | ||
| 5 | 0.47 ± 0.037b | 64 ± 5.1 | |||
| 10 | 0.43 ± 0.045b | 58 ± 6.1 | |||
| 5′-AzAdC (1 µM) + TSA (0.3 µM) | 24 + 24 | 0.21 ± 0.027b | 29 ± 3.7 | ||
| 48 + 24 | 0.35 ± 0.022b | 48 ± 3.1 | |||
| TSA | 24 | 0.2 | 0.14 ± 0.015b | 20 ± 2.1 | |
| 1 | 0.16 ± 0.018b | 21 ± 2.5 | |||
| 5 | 0.10 ± 0.014b | 14 ± 1.9 | |||
| hNT | 0.75 ± 0.046b | 103 ± 6.3 |
Values are means ± SEM.
aQuantitative measurements of mRNA three times using three independent RNA isolates.
bDenotes significantly different from NT2 control using one-way ANOVA (P < 0.01).
We also examined sequences smaller than the reelin minimal promoter (–514 bp). Multiple deletions were cloned that were evenly spaced between –514 and –127 bp of the reelin promoter. Figure 2B shows reporter activity results obtained following transient transfection of these deletions into neural progenitors (NT2) and differentiated neurons (hNT). NT2 cells express only low levels of endogenous reelin mRNA whereas hNT neurons express high levels of reelin mRNA. Results from these transfections showed that in both cell types there was a progressive decrease in high reporter activity as additional sequences were removed from the –303 reelin promoter construct.
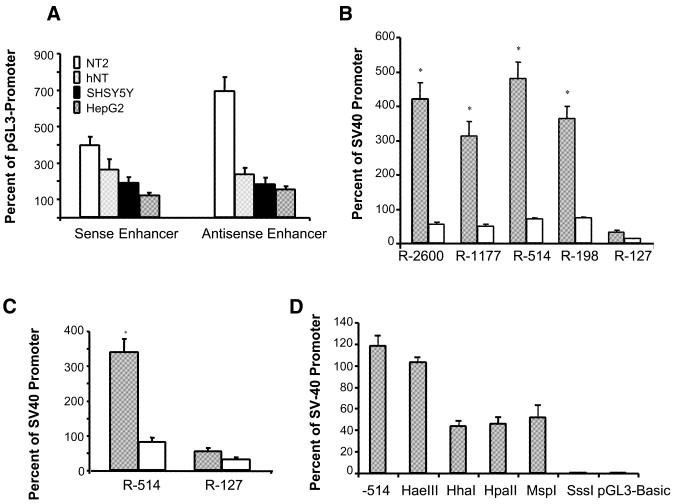
Sequences between –303 and –137 were cloned upstream of the SV40 promoter in both orientations to test for enhancer activity (22). These upstream sequences increased expression of the SV40 promoter independent of cell type (Fig. 3A). Embedded within this region are three binding sites for an Sp1-like transcription factor (31) and a site for the brain-specific T-box factor Tbr1 (32,33). We suspect that the presence of these three Sp1-like sites in the reelin enhancer/promoter may account for the observed promoter promiscuity. To test whether these factors were able to transactivate the reelin promoter, we co-transfected several reelin promoter deletions into NT2 cells along with expression vectors corresponding to Sp1 and Tbr1. As shown in Figure 3B, Tbr1 activates expression of the reelin promoter from 5- to 8-fold. This activation depends on the presence of the putative Tbr1 recognition site, which we have positioned at between –198 and –127 bp relative to the RNA start site (Fig. 4), based on both the transactivation data and the recognition sites of other T-box transcription factors. We also see a modest activation of the reelin promoter when we co-transfect a Sp1 expression cassette (Fig. 3C). There are three Sp1-like recognition sites located in the minimal reelin promoter (Fig. 4B) and when these sites are deleted, we no longer see Sp1-mediated promoter induction. The results imply a role for each of these transcription factors in modulating reelin expression (Fig. 3B and C) and provide an approximate location of their binding sites in the reelin promoter. While the extent of induction by each transcription factor is not extensive, NT2 cells endogenously express both Tbr1 and Sp1, which compromises the extent of transactivation (data not shown). The observation that these cells express both of these transcription factors but only express low levels of endogenous reelin mRNA suggests that an additional level of control is operative in regulating reelin promoter expression.
Figure 3.
Analysis of the reelin promoter proximal sequences. (A) Sequences between –303 and –137 activate expression of the SV40 promoter. This region of the reelin promoter was subcloned 5′ to the SV40 promoter in both sense and antisense oritentations to test for enhancer-like activity. These data are expressed relative to that obtained with the SV40 promoter containing vector alone (pGL3-Promoter). (B) The mouse Tbr1 cDNA was co-transfected with several reelin promoter/reporter constructs. The shaded bar indicates Tbr1 co-transfection while the unshaded bar indicates the same plasmid but without co-transfection of the Tbr1 sequence. The data are expressed relative to the SV40 promoter construct (pGL3-Promoter) transfected in parallel. (C) An Sp1 expression vector was co-transfected with either the –514 bp promoter or the –127 promoter/reporter. The shaded bar shows the Sp1 co-transfection while the unshaded bar shows results from the parent (no Sp1) plasmid co-transfection. (D) Bacterial methylases were used to methylate the reelin –514 bp promoter/reporter construct in vitro prior to transfection. The first bar (–514) represents the data obtained from transfecting the unmethylated –514 reelin promoter/reporter construct. The specificity of each indicated methylase is indicated in the text. The constructs were introduced into NT2 cells and reporter activity was measured. Except for the HaeIII methylase, results obtained for the modified constructs were different from the unmodified –514 bp promoter. All data are expressed relative to the mean signal obtained using the SV40 promoter/luciferase reporter transfections that were performed in parallel. *,P < 0.001.
RA-induced differentiation of neural progenitor cells is accompanied by changes in chromatin structure
The activity of the reelin promoter and enhancer constructs in NT2 cells was curious as expression of the endogenous transcript is minimal. We presume that, in addition to Tbr1 and Sp1, additional requisite transcription factors are present in these cells and that the endogenous gene is silenced through hypermethylation. As shown in Figure 2C, there are alterations in chromatin structure in the vicinity of the reelin promoter in cells that express the endogenous gene (hNT neurons) as indicated by the presence of three DNase I hypersensitive sites. These sites are not present in the undifferentiated NT2 neural progenitor cells. The highest molecular size site maps to within the reelin enhancer region, while the additional two sites are located further upstream. The data suggest that, upon neuronal differentiation, changes in the local chromatin structure occur, making the promoter more readily accessible to the transcriptional machinery. The transiently introduced promoter/reporter constructs are not confined by this restraint in the same way and expression is not silenced. These results indicate that an alteration in local chromatin structure likely mediated by changes in methylation status is an important epigenetic determinant modulating expression of the endogenous reelin gene.
Nucleotide analogs that prevent methylation and histone deacetylase inhibitors activate reelin expression in neural progenitor cells
We tested whether the non-methylatable cytosine analog AzAdC would activate expression of the previously silent endogenous reelin promoter. AzAdC was incubated with NT2 cells for 24 and 72 h. In parallel, NT2 cells were treated with the histone deacetylase inhibitor TSA (0.2–5 µM) for 24 h, either alone or with AzAdC (0.2 µM TSA, 1 µM AzAdC). RNA was isolated and the amount of the reelin mRNA present in each condition was quantified using competitive RT–PCR with an internal standard (1,25). The quantified amounts of reelin mRNA following each treatment are presented in Table 1 as pg reelin mRNA/µg total RNA. Incubating NT2 cells with the nucleotide analog AzAdC (1 µM) for 24 h increased reelin mRNA levels >20-fold (from 0.0076 ± 0.00074 to 0.17 ± 0.022 pg Reln RNA/µg total RNA). This increased to >50-fold after 72 h treatment. The HDAC inhibitor TSA also increased expression of the reelin mRNA some 20-fold after 24 h. Results obtained from including both inhibitors appeared to be time dependent and not necessarily additive (Table 1).
More recently, the anticonvulsant and mood-stabilizing drug VPA has been reported to act as a HDAC inhibitor causing hyperacetylation of histones in culture (19,20). We tested the action of VPA in increasing reelin expression by treating NT2 cells with different concentrations of the drug for 40 h. As shown in Table 2, VPA acted to increase reelin mRNA levels ∼20-fold over this time frame in a dose-dependent manner. In the same experiment, cultures were also treated with a low dose of TSA (300 nM) and with the inactive amide analog VPM. As can be seen by the data, VPA acts in a similar manner in activating reelin expression but is considerably less potent than TSA. VPM did not appreciably increase reelin mRNA levels.
Table 2. Increases in reelin mRNA levels after a 40 h treatment of NT2 neuroprogenitor cells with VPA, TSA and VPM.
| Treatment | Concentration | Amount of Reln RNA (pg Reln RNA/µg RNA) | Fold increase |
|---|---|---|---|
| Control | 0.0077 ± 0.0011 | ||
| VPA | 0.2 mM | 0.077 ± 0.018 | 10.0 ± 2.4 |
| 2.0 mM | 0.11 ± 0.021 | 15.0 ± 2.8 | |
| 5.0 mM | 0.16 ± 0.019 | 20.0 ± 2.5 | |
| 10.0 mM | 0.17 ± 0.027 | 22.0 ± 3.5 | |
| TSA | 0.3 µM | 0.17 ± 0.015 | 22.5 ± 2.0 |
| VPM | 5.0 mM | 0.0084 ± 0.0024 | 1.1 ± 0.3 |
NT2 neural progenitor cultures were treated with the indicated concentrations of drugs for 24 h and RNA was isolated. The amount of reelin mRNA was quantified using competitive RT–PCR (4) and is expressed relative to the amount present in the vehicle-treated cultures that were maintained in parallel. The RNA analysis was performed three times per dose on a minimum of three different culture preparations.
Methylation of the reelin promoter in vitro attenuates reporter activity
In the next series of experiments, we artificially methylated the –514 bp reelin promoter construct in vitro prior to transfection to assess the effects of differential methylation on reporter expression. The –514 construct contains 122 CpG sites which reside within the 5′ upstream region or downstream of the RNA start site. Each of these sites can be symmetrically methylated using the enzyme SssI methylase. We used additional methylases to specifically modify selected cytosines within the promoter region. For these experiments, we incubated the promoter-containing plasmid with the enzyme overnight and assessed the extent of methylation by restriction digestion. Following methylation, the modified constructs were transfected into NT2 cells to assess the effects on reporter activity (Fig. 3D). As can be seen, methylating all of the CpG sites in the promoter sequence inhibits expression completely (SssI). In contrast, methylating the first C in the GGCC sequence has very little effect on expression (HaeIII). The other recognition sites had intermediate effects with respect to expression of reporter activity. We predict that there are likely multiple motifs that are recognized by the mammalian DNA methylating enzymes. We would not have predicted that the methylation pattern generated using MspI methylase (CCGG) would reduce expression appreciably. However, recent studies have shown that non-CpG methylation is prevalent and appears to be mediated by Dnmt3a (34). These studies support the concept that methylation may be an important mode by which the reelin promoter is epigenetically regulated.
Bisulfite mapping of methylated residues in non-expressing cells
We also present data to show that specific sites are methylated in NT2 cells that are not methylated in differentiated hNT neurons which express reelin mRNA (Fig. 4A). Genomic DNA was isolated from NT2 cells maintained in culture or from cultures treated with RA for 6 weeks and replated to enrich for hNT neurons. Both DNA samples were digested with EcoRI and treated with bisulfite under denaturing conditions (27,35). The region (from –527 to +322 bp) was subsequently amplified and cloned. A portion of the B strand sequence from NT2 (top) and hNT (bottom) neurons is shown (Fig. 4A). Representative sequences are shown and arrows point out specific bases that are methylated in the NT2 cells that are not methylated in the differentiated hNT neurons (methylated C residues shown as G residues in the B sequence). We have found that there are eight bases that are methylated in the non-expressing NT2 cells and these are not methylated in genomic DNA isolated from the reelin-positive hNT neurons. Instead, two additional bases are methylated in hNT neurons that are not methylated in NT2 cells (summarized in Fig. 4B). This is consistent with observations made by others which indicate that cytosines within CpG islands tend to be undermethylated and that selected cytosines serve as determinants that modulate gene expression (16).
DISCUSSION
The reelin promoter is competent in driving reporter expression in numerous cell types and in differentiated neurons in vitro. Sequences within the closed red arrows (Fig. 4B) correspond to the region of the promoter that also has enhancer-like activity (–304 to –137 bp). Interestingly, the mouse and human promoters show >92% sequence identity over a stretch of 125 bp upstream of the human RNA start site (–241 to –116 bp). Selected protein recognition sites are indicated in the boxed areas. Sp1 is one of many GC box-binding proteins (31) and is abundantly expressed in numerous cell types (36). There are at least three putative Sp1 binding sites in the reelin promoter/enhancer (Fig. 4B). Our data show that when Sp1 is co-transfected with the reelin promoter, reporter activity is significantly induced and that this induction is dependent on the presence of the GC boxes. Sp1 binding to its recognition sites has been shown to be a signal for demethylation and for preventing de novo methylation of specific sites upstream of the adenine ribosyltransferase gene (17). At the same time, cytosine methylation of an Sp1 site has been shown to contribute to cell-specific regulation of expression of the T1α gene (37). We suggest that the presence of multiple Sp1 sites in the reelin promoter contributes to the promiscuity of the promoter activity we observed in distinct cellular lineages.
We also provide evidence that Tbr1 is important in regulating reelin expression in the cell lines examined. Tbr1 is expressed at different levels in distinct bands of cells in the embryonic cortex and continues to be expressed in forebrain neurons in the adult (32). More recently, Tbr1 mutant mice were shown to exhibit a reeler-like cortical migration defect (38). The Cajal–Retzius cells of these mice showed reduced reelin expression, implicating Tbr1 as one molecular determinant regulating expression in these neurons. In another study, Tbr1 was shown to induce expression of reelin-driven reporter expression when co-transfected with the membrane-associated guanylate kinase, CASK (39). Induction of reelin promoter activity was dependent on the presence of both Tbr1 and CASK, as no induction was observed with Tbr1 alone. In contrast, we observed no interactions between Tbr1 and CASK in the cell lines we examined. The requisite T-box element was presumed to reside upstream of the reelin promoter. However, the position of the T-box element in the reelin promoter region remains uncharacterized (33). In our study, we provide more direct mapping of the T-box half palindrome in the reelin promoter which presumably lies within 190 bp of the RNA start site (Fig. 4B). This site is adjacent to one of the presumed Sp1-binding sites and, interestingly, one of the residues present in the putative site is methylated in non-expressing cells (Fig. 4B). This same base is not methylated in differentiated neurons that express the endogenous reelin promoter. Additional studies will be required to clarify whether methylation of this site interferes with transcription factor binding to its recognition site.
As indicated above, sequences flanking the human RNA start site together with the first exon form a large CpG island. In contrast to the vast majority of CpG dinucleotides in the genome, which are generally methylated, CpG islands tend to be undermethylated and this pattern can vary in both a temporal and spatially selective manner (14). There are 120 CpG sites within sequences proximal to the reelin minimal promoter which could be targets of the methylation machinery (see Fig. 4B). The presence of a CpG island and the observed promiscuity of the promoter suggested the possibility that the gene might be regulated through changes in methylation status. Addition of a non-methylatable deoxycytosine analog to neural progenitors (NT2 cells) in culture increased reelin expression >50-fold. Recent studies show that methylated DNA is recognized by a family of methylated DNA-binding proteins which recruit co-repressor complexes and HDACs (12,40). Consistent with this model, we show that HDAC inhibitors also induce expression of reelin independently of the methylation inhibitors. The combined use of the methylation and HDAC inhibitors did not increase expression to a greater extent than either treatment alone, which suggests that changes in local chromatin structure are likely mediated through either alterations in methylation status or the deacetylation of histone/DNA complexes. Interestingly, the anticonvulsant and mood-stabilizer VPA significantly increased reelin mRNA levels. The observed increase was dose-dependent, although it was considerably less potent than TSA in eliciting this response. Nevertheless, this is of considerable interest as VPA is widely prescribed and its use as an adjunct pharmacotherapy in the treatment of schizophrenia has tripled in recent years (41). While the gene targets of this drug have not been explored, it remains possible that its action on reelin and on GAD67 in vivo (L. Tremolizzo and A. Guidotti, unpublished data) could represent one aspect of its therapeutic value as these two genes are down-regulated in schizophrenia (2).
We took our analysis an additional step and mapped the location of the methylated residues in the non-expressing NT2 cells (yellow filled box, Fig. 4B). The sites methylated in RA-induced hNT neurons are also shown (blue filled box). The importance of those experiments that show that inhibition of methylation activates reelin expression is underscored by the change in the methylation profile of the promoter that accompanies these cells as they differentiate in culture. As tissue-specific and developmental expression patterns are associated with distinct alterations in chromatin structure and DNA methylation status (42), we suspect that methylation of critical residues within these sequences affects the ability of transcription factors to access these sites. While these data were obtained using a cell culture system, we suggest that methylation of the CpG island represents an epigenetic switch that is used to silence reelin promoter expression under appropriate conditions. The recent characterization of a DNA demethylase activity supports a model in which the pattern of methylation is maintained by a balance of methylation and demethylation activities and the local state of histone acetylation (43). Taken together with observations that high levels of Dnmt1 protein are expressed in post-mitotic neurons (44), one might speculate that methylation is a dynamic process used by neurons to regulate gene expression epigenetically. Recent literature suggests that the pattern of DNA methylation and local histone deacetylation states may be coordinated, establishing a relevant link between the action of these complementary regulatory processes.
The role that methylation plays in regulating gene expression in the nervous system is still underexplored. Mutations that occur in methyl-CpG-binding proteins have drastic consequences that occur post-natally. Rett syndrome is a neurodevelopmental disorder that results in mental retardation due to mutations in the methyl-CpG-binding protein MECP2 (45). It was first noted some 40 years ago that the administration of l-methionine to schizophrenia patients elicited a profound exacerbation of symptoms in 60–70% of patients (recently reviewed in 46). This exacerbation could not be explained by an increased accumulation of metabolites of catecholamines and has remained a mystery ever since. We would like to suggest that the mechanism of the methionine-induced psychosis could be related to increased levels of brain S-adenosylmethionine and subsequent alterations in the methylation status of selective promoters, including that corresponding to reelin. In support of this argument, we have preliminary data that show that the chronic injection of l-methionine to heterozygous reeler mice twice daily for 15 days further reduces the levels of reelin and reelin mRNA (46). These data are consistent with the hypothesis that the reelin promoter can be modulated through modifications in methylation and that a relationship may exist between the recrudescence of psychotic symptoms and the hypermethylation of sites within the reelin CpG island.
While the data presented here are based on experiments performed in cells maintained in culture, we anticipate that additional studies will provide insight into methylation and reelin promoter regulation in vivo. Ultimately, we are interested in assessing whether the reelin promoter is functionally compromised in patients diagnosed with psychosis (3). It seems plausible that alterations in reelin expression may be the consequence of inappropriate methylation that leads to altered chromatin structure and silenced expression (46). While we are presently attempting to substantiate our hypotheses with direct experiments, we would like to infer that the variable symptomatology associated with the schizophrenia spectrum of disorders might be the consequence of inappropriate methylation patterns resulting from the dysregulation of this fundamental regulatory mechanism. Interestingly, an analysis of genomic DNA from monozygotic twins discordant for schizophrenia showed discrepancies in the methylation patterns of certain genes, suggesting a possible epigenetic mechanism for differences between monozygotic twins (47). Current thinking also suggests that schizophrenia is a polygenic disorder. Biologically inappropriate expression patterns of multiple genes might result from changes that occur in the methylation status of specific promoters expressed in the nervous system, providing a potential clue to the polygenic nature of the disorder.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Mr David Gavin for his excellent technical assistance, Dr Genoveva Davidkova for providing reelin internal standard cRNA and Dr Jai Sung Noh for help with figure preparation. We would like to acknowledge Dr Robert Tjian (UC, Berkeley, CA) for providing the Sp1 expression vector and Dr Morgan Sheng (Massachusetts General Hospital and Harvard University, Cambridge, MA) for the Tbr1 expression vector. Finally, we would like to acknowledge Katwijk Chemie B.V. for the gift of valpromide that was used in these studies. This work was supported by 5R01MH062090-02 to E.C. and in part by 1R01MH062682-01A2 to D.R.G.
REFERENCES
- 1.Impagnatiello F., Guidotti,A.R., Pesold,C., Dwivedi,Y., Caruncho,H., Pisu,M.G., Uzunov,D.P., Smalheiser,N.R., Davis,J.M., Pandey,G.N., Pappas,G.D., Tueting,P., Sharma,R.P. and Costa,E. (1998) A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc. Natl Acad. Sci. USA, 95, 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa E., Davis,J., Grayson,D.R., Guidotti,A., Pappas,G.D. and Pesold,C. (2001) Dendritic spine hypoplasia and downregulation of reelin and GABAergic tone in schizophrenia vulnerability. Neurobiol. Dis., 8, 723–742. [DOI] [PubMed] [Google Scholar]
- 3.Guidotti A., Auta,J., Davis,J., Dwivedi,Y., Grayson,D.R., Impagnatiello,F., Pandey,G., Pesold,C., Sharma,R., Uzunov,D. and Costa,E. (2000) Reelin and GAD67 expression is decreased in postmortem brain of schizophrenia and bipolar disorder patients. Arch. Gen. Psychiatr., 57, 1061–1069. [DOI] [PubMed] [Google Scholar]
- 4.Selemon L.D. and Goldman-Rakic,P.S. (1999) The reduced neuropil hypothesis: a circuit based model of schizophrenia. Biol. Psychiatr., 45, 17–25. [DOI] [PubMed] [Google Scholar]
- 5.Fatemi S.H., Earle,J.A. and McMenomy,T. (2000) Reduction in reelin immunoreactivity in hippocampus of subjects with schizophrenia, bipolar disorder and major depression. Mol. Psychiatr., 5, 654–663. [DOI] [PubMed] [Google Scholar]
- 6.Rice D.S. and Curran,T. (2001) Role of the reelin signaling pathway in central nervous system development. Annu. Rev. Neurosci., 24, 1005–1039. [DOI] [PubMed] [Google Scholar]
- 7.Alcantara S., Ruiz,M., D’Arcangelo,G., Ezan,F., de Lecea,L., Curran,T., Sotelo,C. and Soriano,E. (1998) Regional and cellular patterns of reelin mRNA expression in the forebrain of the developing and adult mouse. J. Neurosci., 18, 7779–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pesold C., Impagnatiello,F., Pisu,M.G., Uzunov,D.P., Costa,E., Guidotti,A. and Caruncho,H.J. (1998) Reelin is preferentially expressed in neurons synthesizing γ-aminobutyric acid in cortex and hippocampus of adult rats. Proc. Natl Acad. Sci. USA, 95, 3221–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez M.A., Pesold,C., Liu,W.S., Kriho,V., Guidotti,A., Pappas,G.D. and Costa,E. (2000) Colocalization of integrin receptors and reelin in dendritic spine postsynaptic densities of adult nonhuman primate cortex. Proc. Natl Acad. Sci. USA, 97, 3550–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeSilva U., D’Arcangelo,G., Braden,V.V., Chen,J., Miao,G.G., Curran,T. and Green,E.D. (1997) The human reelin gene: isolation, sequencing, and mapping on chromosome 7. Genome Res., 7, 157–164. [DOI] [PubMed] [Google Scholar]
- 11.Royaux I., Lambert de Rouvroit,C., D’Arcangelo,G., Demirov,D. and Goffinet,A.M. (1997) Genomic organization of the mouse reelin gene. Genomics, 46, 240–250. [DOI] [PubMed] [Google Scholar]
- 12.Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression—belts, braces and chromatin. Cell, 9, 451–454. [DOI] [PubMed] [Google Scholar]
- 13.Lu R., Au,W.C., Yeow,W.S., Hageman,N. and Pitha,P.M. (2000) Regulation of the promoter activity of interferon regulatory-7 gene. Activation by interferon and silencing by hypermethylation. J. Biol. Chem., 275, 31805–31812. [DOI] [PubMed] [Google Scholar]
- 14.Newell-Price J., Clark,A.J. and King,P. (2000) DNA methylation and silencing of gene expression. Trends Endocrinol. Metab., 11, 142–148. [DOI] [PubMed] [Google Scholar]
- 15.Tucker K. (2001) Methylated cytosine and the brain: a new base for neuroscience. Neuron, 30, 649–652. [DOI] [PubMed] [Google Scholar]
- 16.Costello J.F. and Plass,C. (2001) Methylation matters. J. Med. Genet., 38, 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han L., Lin,I.G. and Hsieh,C.L. (2001) Protein binding protects sites on stable episomes and in the chromosome from de novo methylation. Mol. Cell. Biol., 21, 3416–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLeod D., Charlton,J., Mullins,J. and Bird,A. (1994) Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev., 8, 2282–2292. [DOI] [PubMed] [Google Scholar]
- 19.Phiel C.J., Zhang,F., Huang,E.Y., Guenther,M.G., Lazar,M.A. and Klein,P.S. (2001) Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem., 276, 36734–36741. [DOI] [PubMed] [Google Scholar]
- 20.Gottlicher M., Minucci,S., Zhu,P., Kramer,O.H., Schimpf,A., Giavara,S., Sleeman,J.P., Lo Coco,F., Nervi,C., Pelicci,P.G. and Heinzel,T. (2001) Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J., 20, 6969–6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szekely A.M., Costa,E. and Grayson,D.R. (1990) Transcriptional program coordination by NMDA-sensitive glutamate receptor stimulation in primary cultures of cerebellar neurons. Mol. Pharmacol., 38, 624–633. [PubMed] [Google Scholar]
- 22.Grayson D.R., Costa,R.H., Xanthopoulos,K.G. and Darnell,J.E.,Jr (1988) A cell-specific enhancer of the mouse α1-antitrypsin gene has multiple functional regions and corresponding protein binding sites. Mol. Cell. Biol., 8, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinha S., Degenstein,L., Copenhaver,C. and Fuchs,E. (2000) Defining the regulatory factors required for epidermal gene expression. Mol. Cell. Biol., 20, 2543–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bovolin P., Santi,M.R., Memo,M., Costa,E. and Grayson,D.R. (1992) Distinct developmental patterns of expression of the rat α1, α5, γ2S and γ2L GABAA receptor subunit mRNAs in vivo and in vitro. J. Neurochem., 59, 62–72. [DOI] [PubMed] [Google Scholar]
- 25.Grayson D.R. and Ikonomovic,S. (1998) Competitive RT-PCR to quantitate steady state mRNA levels. In Boulton,A.A., Baker,G.B. and Bateson,A. (eds), In Vitro Neurochemical Techniques: Neuromethods. Humana Press, Totowa, NJ, Vol. 34, pp. 127–151.
- 26.Zuccotti M. and Monk,M. (1995) Methylation of the mouse Xist gene in sperm and eggs correlates with imprinted Xist expression and paternal X-inactivation. Nature Genet., 9, 316–320. [DOI] [PubMed] [Google Scholar]
- 27.Clark S.J., Harrison,J., Paul,C.L. and Frommer,M. (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res., 22, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardiner-Garden M. and Frommer,M. (1987) CpG islands in vertebrate genomes. J. Mol. Biol., 196, 261–282. [DOI] [PubMed] [Google Scholar]
- 29.Grayson D.R., Uzunov,D.P., Chen,Y., Siyanova,E., Raca,G., Sharma,R.P., Mirkin,S. and Costa,E. (2000) Studies of molecular mechanisms regulating reelin gene expression. Biol. Psychiatr., 47, 41S. [Google Scholar]
- 30.Persico A., D’Agruma,L., Maiorano,N., Totaro,A., Militerni,R., Bravaccio,C., Wassink,T., Schneider,C., Melmed,R., Trillo,S., Montecchi,F., Palermo,M., Pascucci,T., Puglisi-Allegra,S., Reichelt,K.-L., Conciatori,M., Marino,R., Quattrocchi,C., Baldi,A., Zelante,L., Gasparini,P. and Keller,F. (2001) Reelin gene alleles and haplotypes as a factor predisposing to autistic disorder. Mol. Psychiatr., 6, 150–159. [DOI] [PubMed] [Google Scholar]
- 31.Suske G. (1999) The Sp-family of transcription factors. Gene, 238, 291–300. [DOI] [PubMed] [Google Scholar]
- 32.Bulfone A., Smiga,S.M., Shimamura,K., Peterson,A., Puelles,L. and Rubenstein,J.L.R. (1995) T-brain-1: a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron, 15, 63–78. [DOI] [PubMed] [Google Scholar]
- 33.Tada M. and Smith,J.C. (2001) T-targets: clues to understanding the functions of T-box proteins. Dev. Growth Differ., 43, 1–11. [DOI] [PubMed] [Google Scholar]
- 34.Ramsahoye B.H., Biniszkiewicz,D., Lyko,F., Clark,V., Bird,A.P. and Jaenisch,R. (2000) Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl Acad. Sci. USA, 97, 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frommer M., McDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saffer J.D., Jackson,S.P. and Annarella,M.B. (1991) Developmental expression of Sp1 in the mouse. Mol. Cell. Biol., 11, 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao Y.X., Jean,J.C. and Williams,M.C. (2000) Cytosine methylation of an Sp1 site contributes to organ-specific and cell-specific regulation of expression of the lung epithelial gene T1α. Biochem. J., 350, 883–890. [PMC free article] [PubMed] [Google Scholar]
- 38.Hevner R.F., Shi,L., Justice,N., Hsueh,Y.P., Sheng,M., Smiga,S., Bulfone,A., Goffinet,A.M., Campagnoni,A.T. and Rubenstein,J.L.R. (2001) Tbr1 regulated differentiation of the preplate and layer 6. Neuron, 29, 353–366. [DOI] [PubMed] [Google Scholar]
- 39.Hsueh Y.P., Wang,T.F., Yang,F.C. and Sheng,M. (2000) Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature, 404, 298–302. [DOI] [PubMed] [Google Scholar]
- 40.Szyf M. (2001) Towards a pharmacology of DNA methylation. Trends Pharmacol. Sci., 22, 350–354. [DOI] [PubMed] [Google Scholar]
- 41.Citrome L., Levine,J. and Allingham,B. (2000) Changes in use of valproate and other mood stabilizers for patients with schizophrenia from 1994 to 1998. Psychiatr. Serv., 51, 634–638. [DOI] [PubMed] [Google Scholar]
- 42.Razin A. (1998) CpG methylation, chromatin structure and gene silencing—a three way connection. EMBO J., 17, 4905–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cervoni N. and Szyf,M. (2001) Demethylase activity is directed by histone acetylation. J. Biol. Chem., 276, 40778–40787. [DOI] [PubMed] [Google Scholar]
- 44.Inano K., Suetake,I., Ueda,T., Miyake,Y., Nakamura,M., Okada,M. and Tajima,S. (2000) Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and located in the cytoplasmic compartment. J. Biochem., 128, 315–321. [DOI] [PubMed] [Google Scholar]
- 45.Amir R.E., Van den Veyver,I.B., Wan,M., Tran,C.Q., Francke,U. and Zoghbi,H.Y. (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nature Genet., 23, 185–188. [DOI] [PubMed] [Google Scholar]
- 46.Costa E., Chen,Y., Davis,J., Dong,E., Noh,J.S., Tremolizzo,L., Veldic,M., Grayson,D.R. and Guidotti,A. (2002) Reelin and schizophrenia: a disease at the interface of the genome and epigenome. Mol. Intervent., 2, 47–57. [DOI] [PubMed] [Google Scholar]
- 47.Tsujita T., Niikawa,N., Yamashita,H., Imamura,A., Hamada,A., Nakane,Y. and Okazaki,Y. (1998) Genomic discordance between monozygotic twins discordant for schizophrenia. Am. J. Psychiatry, 155, 422–424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.