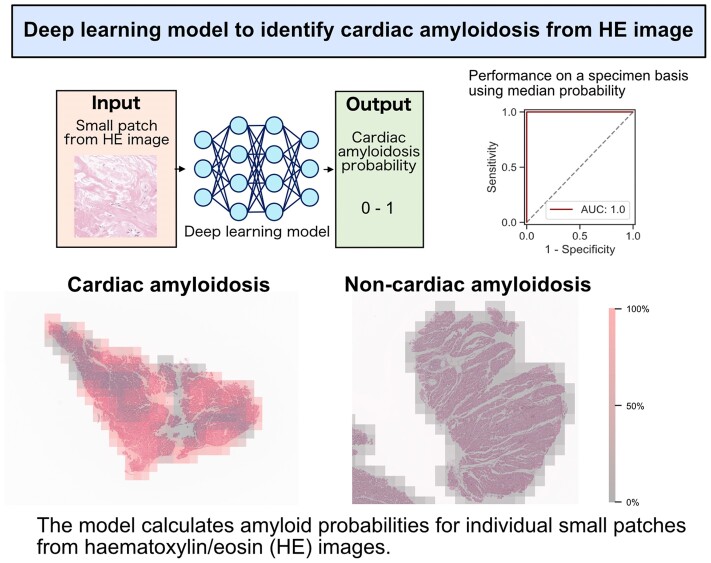
Abstract
Aims
Amyloid deposition in myocardial tissue is a definitive feature for diagnosing cardiac amyloidosis, though less invasive imaging modalities such as bone tracer cardiac scintigraphy and cardiac magnetic resonance imaging have been established as first steps for its diagnosis. This study aimed to develop a deep learning model to support the diagnosis of cardiac amyloidosis from haematoxylin/eosin (HE)-stained myocardial tissue.
Methods and results
This single-centre retrospective observational study enrolled 166 patients who underwent myocardial biopsies between 2008 and 2022, including 76 patients diagnosed with cardiac amyloidosis and 90 with other diagnoses. A deep learning model was developed to output the probabilities of cardiac amyloidosis for all the small patches cutout from each myocardial specimen. The developed model highlighted the area in the stained images as highly suspicious, corresponding to where Dylon staining marked amyloid deposition, and discriminated the patches in the evaluation dataset with an area under the curve of 0.965. Provided that the diagnostic criterion for cardiac amyloidosis was defined as a median probability of cardiac amyloidosis >50% in all patches, the model achieved perfect performance in discriminating patients with cardiac amyloidosis from those without it, with an area under the curve of 1.0.
Conclusion
A deep learning model was developed to diagnose cardiac amyloidosis from HE-stained myocardial tissue accurately. Although further prospective validation of this model using HE-stained myocardial tissues from multiple centres is needed, it may help minimize the risk of missing cardiac amyloidosis and maximize the utility of histological diagnosis in clinical practice.
Keywords: myocardial biopsy, cardiac amyloidosis, deep learning, haematoxylin–eosin stain, computer-aided diagnosis
Graphical Abstract
Graphical Abstract.
Introduction
The clinical importance of endomyocardial biopsy has been increasingly recognized, with the pathological classification of cardiomyopathies becoming more detailed.1 Although it is clinically meaningful to extract as much diagnostic information as possible from biopsy specimens, routinely performing various special staining on all samples, which is expected to increase in the future, will become challenging in terms of both time- and cost-effectiveness, thus necessitating the technological innovations for differentiating specific cardiomyopathies through histological analysis.
Cardiac amyloidosis (CA), in which amyloid deposits are observed in the myocardial tissue, is one of cardiomyopathies with a poor prognosis, resulting in refractory heart failure.2 Recent reports suggest that it is more common in older patients with heart failure than previously thought.3,4 Diagnostic advances in non-invasive modalities, such as echocardiography, bone tracer cardiac scintigraphy, and cardiac magnetic resonance imaging, have made screening for CA increasingly feasible.5,6 However, these tests are not entirely reliable for excluding CA, and even in the absence of characteristic findings on echocardiography or scintigraphy, there is still a possibility of reaching a diagnosis of amyloidosis.7,8 Therefore, once a myocardial biopsy is performed, it is clinically necessary to exclude amyloidosis as a part of the diagnostic process, given that CA is currently a treatable disease.9–13
Histologic diagnosis of CA by endomyocardial biopsy requires specific staining with Dylon or Congo Red to detect amyloid deposition. However, staining with Dylon or Congo Red is not routinely performed in all cases, possibly resulting in missed opportunities to diagnose CA when it is not suspected. This may occur particularly in atypical CA cases where dilated cardiomyopathy is suspected based on the cardiac morphology14,15 or cases of patients with heart transplantation,16,17 but amyloidosis is also present as a comorbidity. In contrast, haematoxylin/eosin (HE) staining is a basic procedure for histological analysis performed for most myocardial tissue biopsies. Therefore, it is beneficial to develop a new approach for the diagnosis of CA from HE-stained myocardial tissue.
Artificial intelligence is an innovative solution to address a variety of challenging tasks across all fields. Currently, deep learning applications for medical images, such as computed tomography, skin images, and histological images, have been developed for computer-aided diagnosis.18–20 Histological images are digitally available in high resolution as whole slide images; therefore, deep learning may be a good application for histological analysis.21 Indeed, deep learning models have focused on histological diagnosis,22,23 and image segmentation on specimens.24–26 We thus hypothesized that a deep learning model could accurately diagnose CA from HE-stained specimens.
This study aimed to develop a deep learning model to assess the likelihood of CA in HE-stained myocardial tissue to aid in diagnosis from individual histological images.
Materials and methods
Data source and participants
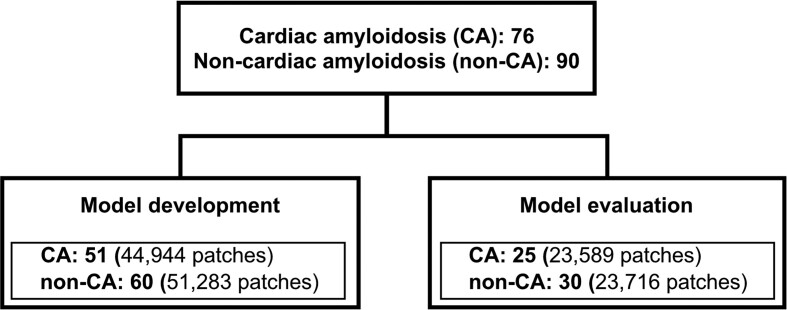
This retrospective observational study included patients who underwent endomyocardial biopsy from the septum of the right ventricular at the Kyushu University Hospital between January 2008 and July 2022. Seventy-six cases diagnosed with CA were included, and 90 patients with other diagnoses (non-CA) were randomly selected. The patients were allowed to opt-out. This study was approved by the Institutional Review Board of Kyushu University Hospital (approval numbers: 22175 and 23209) and was conducted in accordance with the Declaration of Helsinki. This study follows the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE)27 and the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis + AI (TRIPOD + AI) statements.28 The 166 patients were randomly assigned to two datasets: a dataset for model development (51 patients with CA and 60 without) and another dataset for model evaluation (25 with CA and 30 without) (Figure 1).
Figure 1.
Patient flowchart. Patients with and without cardiac amyloidosis were divided into 2:1 for the model development and evaluation.
Outcome
The proposed model was designed to calculate the probability of CA for patch images (224 × 224 px) cutout from each specimen. In addition, we diagnosed each specimen-based on the probabilities of all the patches. We then evaluated the performance of the model based on all patches and specimens respectively. More than two pathologists determined the ground truths of pathological diagnoses on each specimen. CA was diagnosed based on amyloid deposition detected by Dylon and/or Congo Red staining.
Preparation of cardiac specimens and images for digital analysis
The input images for the deep learning model were small patches cutout from whole slide images of HE-stained myocardial specimens. Originally, specimens were fixed in 10% buffered formalin and embedded in paraffin. Each block was sliced into 4-μm thick sections. All slides were digitally converted to whole slide images using a virtual slide scanner (Aperio GT450, Leica) at 40× magnification (0.23 µm/px).
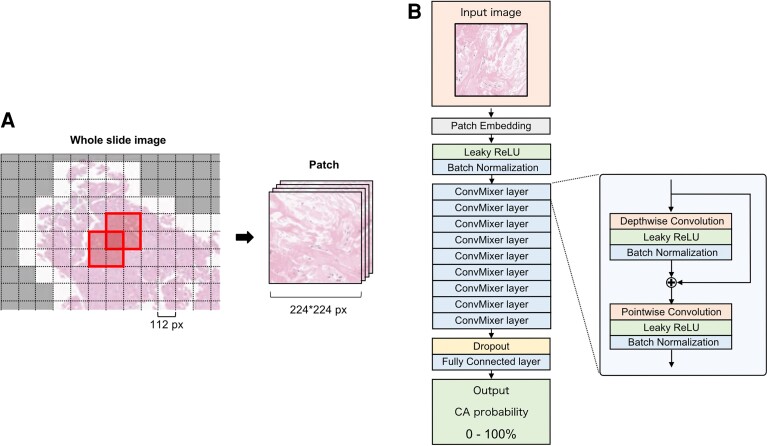
As shown in Figure 2A, each whole slide image specimen was cropped into multiple small patches at a size of 224 × 224 px at level-1 resolution (OpenSlide Python ver. 1.3.1) while being shifted by half. During this process, patches without myocardial tissue were removed. All patches in each specimen with amyloid deposition were labelled CA, and those without were labelled non-CA. Therefore, the CA group included some patches with no apparent amyloid deposition.
Figure 2.
(A) Each whole slide image of the specimen was cropped into multiple small patches of 224 × 224 px size and shifted by half (a patch is an area bordered by a bold line). Patches without myocardial tissue were removed (the white area was used for analysis, and the gray area was excluded). (B) The architecture of the deep learning model is based on ConvMixer. The input image is a patch (224 × 224 px) cropped from the whole slide image of myocardial specimens. And the output is the probability of CA. To improve the flexibility of the model and mitigate overfitting, the Leaky ReLU was used as the activation function, and a dropout layer was inserted at the end of the model. The hyperparameters of this model were determined using GS as follows: Kernel size = 5 (GS: 3, 5, 7, 9), patch size = 4 (GS: 2, 4, 8), number of filters = 128 (GS: 32, 64, 128, 256). GS, grid search.
The patches were allocated to the model development and evaluation datasets at a 2:1 ratio on a specimen basis; 80% of the model development dataset was used to train the model, and the remaining 20% was used to determine the hyperparameters.
The architecture of the deep learning model
The architecture of the deep learning model was based on ConvMixer (Figure 2B).29 The hyperparameters of the model were determined using a grid search. To improve the flexibility of the model and mitigate overfitting, the Leaky ReLU was used as the activation function, and a dropout layer was inserted at the end of the model.30,31 The model was trained directly on the training data, which was augmented by randomly flipping left/right and up/down, changing brightness, and using the cutout method.32
Statistics
The discrimination performance of the developed model was evaluated using the receiver operating characteristic curve, area under the curve, and confusion matrix for all patches in the evaluation dataset. By setting the threshold with the median amyloid-positive probability on each specimen at 50%, the performance of the developed model was evaluated with specimen-based receiver operating characteristic curves, areas under the curves, and confusion matrices. The analysis was performed using Python 3.11.0 and TensorFlow 2.15.
Results
Patient characteristics
Figure 1 and Table 1 show the flow chart of the study population and the patient backgrounds in the overall and divided datasets, respectively. The CA group included 54 patients with transthyretin amyloidosis (ATTR) (71.1%), 10 with light chain amyloidosis (AL) (13.2%), 9 with amyloid A amyloidosis (AA) (11.8%), and 3 with ATTR and AL (3.9%), all diagnosed based on immunohistochemistry. The non-CA group included 13 patients with hypertrophic cardiomyopathy, 12 with dilated cardiomyopathy, 12 with hypertensive heart disease, 10 with valvular heart disease, 9 post-transplant patients, 8 with cardiac sarcoidosis, and the remainder with other cardiac diseases. In the CA group, 56 out of 76 cases (73.7%) had a suspicion of amyloidosis as the primary indication for endocardial biopsy, whereas only 3 out of 90 cases (3.3%) had this suspicion in the non-CA group. In the model evaluation dataset, the median age was higher in the CA group {78 years [interquartile range (IQR): 64–82]} than in the non-CA group [60 years (IQR: 45–72)]. The left ventricular ejection fractions were slightly better in the CA group [50% (IQR: 42–66)] than in the non-CA group [43% (IQR: 32–58)]. The interventricular septum (IVS) in the CA group [14 mm (IQR: 10–16)] was thicker than in the non-CA group [10 mm (IQR: 9–10)].
Table 1.
Patient characteristics
| Total | Model development | Model evaluation | |||
|---|---|---|---|---|---|
| CA | Non-CA | CA | Non-CA | ||
| Subject, n | 166 | 51 | 60 | 25 | 30 |
| Age, years | 70 [58–79] | 78 [71–82] | 63 [53–70] | 78 [64–82] | 60 [45–72] |
| Female, n (%) | 48 (28.9) | 12 (24.0) | 20 (32.8) | 5 (19.2) | 11 (37.9) |
| Etiologies, n (%) | |||||
| Amyloidosis | 76 (45.8) | 50 (100.0) | 26 (100.0) | ||
| HCM | 13 (7.8) | 10 (16.4) | 3 (10.3) | ||
| DCM | 12 (7.2) | 8 (13.1) | 4 (13.8) | ||
| HHD | 12 (7.2) | 4 (6.6) | 8 (27.6) | ||
| VHD | 10 (6.0) | 5 (8.2) | 5 (17.2) | ||
| Post-transplant | 9 (5.4) | 8 (13.1) | 1 (3.4) | ||
| Cardiac sarcoidosis | 8 (4.8) | 6 (9.8) | 2 (6.9) | ||
| Others, undiagnosed | 26 (15.7) | 20 (32.8) | 6 (20.7) | ||
| Amyloid classification, n (%) | |||||
| ATTR | 54 (71.1) | 34 (68.0) | 20 (76.9) | ||
| AL | 10 (13.2) | 9 (18.0) | 1 (3.8) | ||
| AA | 9 (11.8) | 6 (12.0) | 3 (11.5) | ||
| ATTR and AL | 3 (3.9) | 1 (2.0) | 2 (7.7) | ||
| Prior HF hospitalization, n (%) | 53 (31.9) | 17 (34.0) | 22 (36.1) | 8 (30.8) | 6 (20.7) |
| PM, ICD, CRT, n (%) | 29 (17.5) | 4 (8.0) | 18 (29.5) | 4 (15.4) | 3 (10.3) |
| Moderate to severe AS, n (%) | 13 (7.8) | 3 (6.0) | 1 (1.6) | 3 (11.5) | 6 (20.7) |
| Rhythm, n (%) | |||||
| Sinus | 116 (69.9) | 29 (58.0) | 47 (77.0) | 15 (57.7) | 25 (86.2) |
| AF | 39 (23.5) | 18 (36.0) | 9 (14.8) | 8 (30.8) | 4 (13.8) |
| PM | 9 (5.4) | 2 (4.0) | 5 (8.2) | 2 (7.7) | 0 (0.0) |
| SBP, mm Hg | 113 [102–129] | 110 [100–121] | 122 [103–137] | 114 [103–127] | 115 [103–127] |
| DBP, mm Hg | 70 [60–76] | 62 [58–71] | 70 [62–85] | 70 [60–74] | 72 [66–80] |
| Heart rate, bpm | 70 [61–80] | 67 [62–75] | 69 [60–82] | 73 [61–78] | 78 [66–86] |
| BNP, pg/mL | 216 [104–483] | 288 [154–532] | 159 [58–440] | 216 [138–333] | 225 [125–694] |
| LVEF (%) | 49 [38–63] | 48 [41–61] | 49 [36–65] | 50 [42–66] | 43 [32–58] |
| IVST (mm) | 11 [9–15] | 15 [12–16] | 10 [8–11] | 14 [10–16] | 10 [9–11] |
| LVPWT (mm) | 11 [9–14] | 14 [12–16] | 9 [9–11] | 14 [10–16] | 10 [9–11] |
| LAD (mm) | 42 [36–48] | 44 [40–48] | 42 [35–46] | 43 [36–48] | 40 [34–48] |
Data are presented as n (%) or median [Q1–Q3].
HCM, hypertrophic cardiomyopathy; DCM, dilated cardiomyopathy; HHD, hypertensive heart disease; VHD, valvular heart disease; PM, pacemaker; ICD, implantable cardioverter defibrillator; CRT, cardiac resynchronization therapy; AS, aortic valve stenosis; AF, atrial fibrillation; SBP, systolic blood pressure; DBP, diastolic blood pressure; BNP, brain natriuretic peptide; LVEF, left ventricular ejection fraction; IVST, interventricular septum thickness; LVPWT, left ventricular posterior wall thickness; LAD, left atrial dimension.
Development of deep learning model
Each specimen image was cut into small patches, and the median number of patches per specimen was 816 (IQR: 573–1089) (Figure 2A). The deep learning model was developed with the patches from 111 patients (CA: 44 944 patches; non-CA: 51 283 patches) based on ConvMixer (Figure 2B).
Model performance on all patches
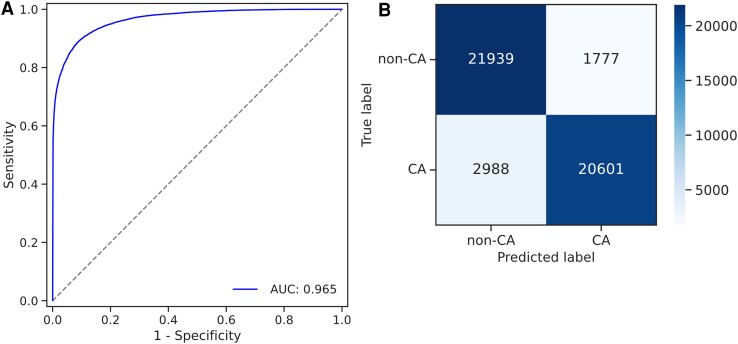
The developed deep learning model was evaluated using an evaluation dataset of 55 patients (CA: 23 589 patches; non-CA: 23 716 patches). Figure 3A shows the receiver operating characteristic curve for the discrimination of all patches; the area under the curve for this model was 0.965. Figure 3B shows the confusion matrix of the developed model, with an accuracy of 0.899, a sensitivity of 0.873, a specificity of 0.925, and a positive predictive value of 0.921.
Figure 3.
(A) The receiver operating characteristic curve of the developed deep learning model in all patches of the evaluation dataset. (B) Patch-based confusion matrix of the deep learning model.
Patch-based amyloid probabilities by the model in CA and non-CA specimens
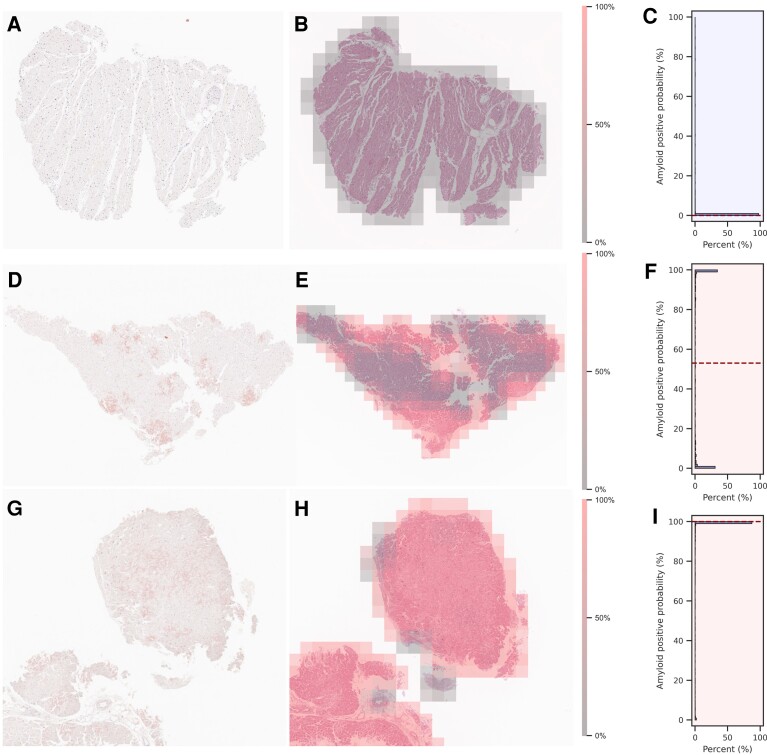
Figure 4 shows the representative operations of the deep learning model in non-CA (A to C) and CA cases (D to F and G to I). In the HE-stained specimens of the non-CA case (Figure 4B), the deep learning model scarcely detected amyloid-suspicious areas. Conversely, in the CA case with sparse amyloid deposition in the myocardial specimen (Figure 4E), the model specifically detected the areas corresponding to Dylon-positive amyloid deposition. Not surprisingly, in the CA case with dense amyloid deposition (Figure 4H), the model identified most patches as highly amyloid-suspicious areas. For the distribution of amyloid probability for all patches, in the non-CA case, most of the amyloid-positive probabilities were distributed close to 0% (Figure 4C); in the CA case with sparse amyloid deposition, the amyloid-positive probabilities had a biphasic distribution with 0 and 100% (Figure 4F). Understandably, in the CA case with dense amyloid deposition, most of the amyloid-positive probabilities were distributed close to 100% (Figure 4I).
Figure 4.
Sets of dylon staining (A, D, G), HE staining overlayed with the amyloid-positive area calculated by the model (B, E, H), and the distributions of amyloid-positive probabilities on a specimen basis (C, F, I). The set of A, B, and C is a representative case of non-cardiac amyloid. The set of D, E, and F is the case of cardiac amyloid with sparse amyloid deposition. The set of G, H, and I is the case of cardiac amyloid with dense amyloid deposition. In HE-stained images, high-probability areas of amyloid are shown in translucent red, and low-probability areas are shown in translucent gray. Since the patches were cropped while being shifted by half, the probabilities of overlapping areas are given as the average probability. The distributions are described by histograms, and the median probabilities are described by dotted lines.
Specimen-based model performance
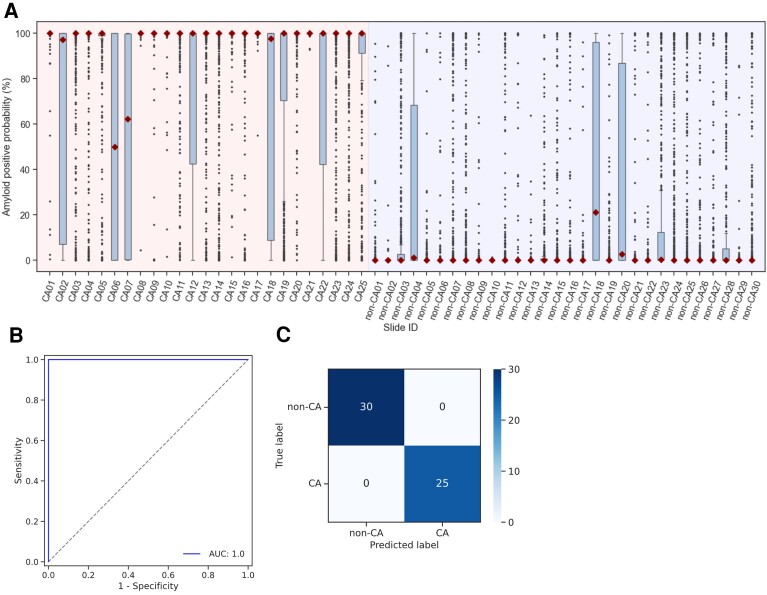
Figure 5A shows the box plots of the CA probabilities per specimen. By setting the threshold with the median amyloid-positive probability on each specimen at 50%, the developed deep learning model perfectly discriminated between CA and non-CA cases. As shown in Figures 5B and C, the model performance using a deep learning approach achieved a sufficient area under the curve, accuracy, and sensitivity and a positive predictive value of 1.0.
Figure 5.
(A) Box plots of amyloid-positive probabilities by specimens and the median probabilities are described by diamond symbols. (B) The specimen-based receiver operating characteristic curve evaluated with a threshold for the amyloid-positive probability of 50%. (C) Specimen-based confusion matrix of the deep learning model.
Discussion
CA, in which amyloid deposits are observed in the myocardial tissue, results in refractory heart failure with a poor prognosis.2 In recent years, CA has become a treatable disease with new therapeutic agents such as transthyretin stabilizers, RNAi, and monoclonal antibodies against TTR (NI006) for CA.9–13 Therefore, early and appropriate diagnosis is more important than ever. Although routine amyloid staining of endomyocardial biopsy specimens is commonly performed at our institution, particularly at high-volume centres, the anticipated increase in the number of cardiac specimens requiring differentiation among various cardiomyopathies presents a clinical challenge in efficiently screening all cases for CA. Here, we developed deep learning capable of accurately diagnosing CA from HE-stained specimens, which can help minimize the risk of missing amyloidosis in the analysis of valuable myocardial tissue.
Advances in imaging with non-invasive modalities, such as bone tracer cardiac scintigraphy and cardiac magnetic resonance imaging, have made the diagnosis of CA less invasive in patients with suspicious symptoms.5,6 However, histological evidence of amyloid deposition remains the definitive diagnostic feature.33 In addition, immunohistochemistry or laser microdissection with mass spectrometry of biopsied myocardial tissues can also identify the types of amyloid, including ATTR, AL, and AA, highlighting the clear merit of histological analysis.34 In this context, diagnosing CA is essential even in patients who do not show clinical suspicion but undergo endomyocardial biopsy, as CA is now a treatable disease.9–13 Retrospective analysis in this study revealed that 9 of 76 cases in the CA group were not suspected of CA prior to endomyocardial biopsy (see Supplementary data online, Table S1). Thus, the routine system for diagnosing CA from HE-stained specimens may be an innovative approach, as HE is standard staining applied to all myocardial biopsies.
In this study, we developed a deep learning model for diagnosing CA from HE-stained myocardial tissue. The developed model demonstrated remarkable accuracy in diagnosing CA using only HE-stained specimens. Previous deep learning models for histological analysis have focused on diagnosis for cancers,22,23 image segmentation,24–26 and image transformation.35 To the best of our knowledge, this is the first study to diagnose CA from images of HE-stained specimens without Dylon or Congo Red staining, although a few deep learning models that can quantify amyloid deposition in corneal or ligamentum flavum specimens stained with Congo Red have been reported.24,26 The present model can serve as the foundational basis for a system that routinely screens the possibility of CA from HE-stained specimens, thereby minimizing the risk of missing CA in CA-unsuspicious patients who are subject to endomyocardial biopsy.
Patient background in this study
In this study, cases of CA and non-CA were randomly selected from patients who underwent myocardial biopsy between 2008 and 2022. A model was developed using these myocardial biopsy specimens. Among the 76 cases of CA, 56 had amyloidosis as the primary indication for endomyocardial biopsy, whereas 11 had amyloidosis included as part of the differential diagnosis. The remaining 9 cases had no suspicion of amyloidosis prior to biopsy (see Supplementary data online, Table S1), and it was diagnosed clinically for the first time through the biopsy evaluation. This may be because of the approval of Tafamidis for use in Japan in 2019, which has since increased the inclusion of amyloidosis in the differential diagnosis. Prior to its approval, amyloidosis was less actively considered. However, approximately 10% of cases in this study still had amyloidosis diagnosed incidentally through endomyocardial biopsy. These findings highlight the importance of routine amyloid staining and the utility of the developed model as an alternative histological screening tool for CA in clinical practice.
In this study, genetic testing was performed on 40 of the 57 TTR cases, including 3 cases with TTR and AL. The results revealed that 38 cases had wild-type TTR with no genetic mutations, whereas 2 cases had genetic mutations. One of these had a Lys61Glu (E61 K) mutation, and the other case had a Val30Met (V30 M) mutation (see Supplementary data online, Figure S1). As the case with E61 K was included in the development dataset, the case with V30 M assigned into the evaluation dataset was analysed in detail (see Supplementary data online, Figure S2 A–C); for reference, images of Dylon and HE staining from the case with E61 K are also shown in Supplementary data online, Figure S2 D and E. Although bone tracer scintigraphy has been reported to yield false negatives for certain variant TTRs, such as Phe84Leu (F84L) and Ser97Tyr (S97Y),36 which require myocardial biopsy for an accurate diagnosis, the analysis with this model demonstrated that it could accurately diagnose ATTRv (the V30M case corresponding to CA07 shown in Figure 5A and Supplementary data online, Figure S2C) and ATTRwt in the evaluation dataset (Figure 5A). Nevertheless, the median probability for the V30M case was lower than that of ATTRwt. This may be because some cases of ATTRv exhibit amyloid deposition primarily in the nervous system before cardiac symptoms and endomyocardial biopsy are present. Indeed, the patient with the V30M mutation had been diagnosed with amyloid polyneuropathy several years prior to the endomyocardial biopsy and was hospitalized for atrial fibrillation and bradyarrhythmia, which ultimately led to the biopsy. Despite the onset of arrhythmia, echocardiography revealed no evidence of hypertrophy. These findings were consistent with the median probability output by the model and the degree of amyloid deposition observed in Dylon staining. This case illustrates that ATTRv may follow a different diagnostic course compared with ATTRwt and that amyloidosis in other organs can precede the diagnosis of CA through myocardial biopsy. Consequently, endomyocardial biopsy in ATTRv may need to be performed at an earlier stage of CA, potentially resulting in lower median probability or a higher likelihood of false negatives because of sampling errors compared with ATTRwt. These concerns arise from the inherent limitations of myocardial biopsy in CA and are not directly related to the model development in this study. However, it should be noted that this model was carefully applied to cases of ATTRv, and further investigation into its predictive accuracy for ATTRv may be warranted, as the number of ATTRv cases was limited, and the majority of cases in this study were ATTRwt.
Deep learning modelling to diagnose CA from HE-stained specimen
In this study, we first developed a deep learning model to assess the probability of CA in each small patch cutout of an overall image. Then, we investigated whether using amyloidosis-positive probabilities in all patches from each specimen could accurately diagnose CA on a specimen basis. Figure 3 shows the accuracy of all patches in the evaluation dataset. Although not all small patches uniformly exhibited amyloid deposition, and thus, some patch labels in the CA cases could contain false positives, the developed models achieved good discrimination with an area under the curve of 0.965 in the evaluation dataset. To assess accuracy on a per-specimen basis, we plotted individual probability distributions for each specimen (Figure 5A) along with discrimination performance (Figures 5B and C). Most probabilities of patches in CA cases were mainly distributed close to 100%, and those in non-CA cases were distributed close to 0%. These distinct distributions did not require fine-tuning to set the threshold as the median value of the positive criterion, allowing us to discriminate effectively all specimens in the evaluation dataset by a crude threshold of 50% for median probability. This study mainly focused on developing a deep learning model to output the probability of cardiac amyloid based on individual small patches because available images were limited; however, with more data, the model could be extended to a more integrated end-to-end model for direct diagnosis from individual whole slide images using methods such as multiple instance learning.37
Identification of amyloid deposition by present deep learning model
The developed model almost did not detect any suspicious areas in the non-CA case shown in Figure 4B. In contrast, it marked the area in the digitally-stained images on HE-stained images as highly suspicious, corresponding to where Dylon staining showed amyloid deposition in the CA cases with sparsely and densely accumulated amyloid (Figures 4E and H). Due to the black-box nature of deep learning models, it is difficult to understand how the present deep learning model interprets images and exactly calculates the probability of CA. Since false positives were rare but present in the non-CA cases (Figure 3B), this model recognizes not only amyloid deposition but also other histological features of CA. In this context, we should carefully utilize the present model in clinical practice, as is always the case when using AI technology. Nevertheless, this study has suggested the clinical feasibility of the deep learning model in aiding in the diagnosis of CA from HE staining because the model achieved remarkable performance in specimen-based diagnosis.
Clinical perspectives
The present model, which outputs the possibility of CA based on images of HE-stained specimens, can provide two significant clinical benefits. First, the model accurately discriminated CA from non-CA even in cases where characteristic features of CA (such as myocardial hypertrophy) were not evident on the echocardiogram (Table 1). Retrospective analysis of cases in this study revealed that 9 of 76 cases in the CA group had no prior suspicion of CA before endomyocardial biopsy (see Supplementary data online, Table S1). This highlights the potential for underdiagnosis in cases of CA that could be missed without routine amyloid staining. The model can assess the possibility of CA in myocardial tissue that has not been stained with Dylon or Congo Red due to a lack of clinical suspicion of CA. Hence, this model could be used as a screening tool for cases not scheduled for specific CA staining as well as for re-evaluation of old specimens that were not specifically stained.
Second, staining with Dylon or Congo Red can affect diagnosis due to over-staining or over-depigmentation. This model can calculate the amyloid probabilities separately from Dylon or Congo Red staining, which could comprehensively enhance the diagnostic accuracy of histological analysis.
Limitations
We acknowledge the following limitations. First, because the present model was developed based on single-centre data, it is unclear whether it can accurately diagnose CA when differences exist in the methods of HE staining or the quality of stained specimens, as these may vary by institution or pathologist. Further investigations using multi-centre data are needed to validate the accuracy and generalizability of the model for broader clinical use. Second, this model does not address the sampling error associated with endomyocardial biopsy. A biopsy samples only a small portion of the myocardium, and even in cases of CA, there is a risk that amyloid deposits may not be detected in the biopsied tissue. Therefore, it is important not to become overly reliant on pathological diagnosis alone. The diagnosis of CA should primarily follow the standard diagnostic algorithm, particularly for hypertrophic phenotypes, referencing T1/T2 mapping or late gadolinium enhancement on cardiac magnetic resonance imaging.38 Myocardial biopsy should be considered as an additional step. It should be emphasized that the developed system is intended as a complementary screening tool within the broader diagnostic process.
Conclusion
A deep learning model was developed to accurately diagnose CA from the images of HE-stained specimens. The present model may be effective in minimizing the risk of missing CA and maximizing the utility of histological diagnosis in clinical practice.
Supplementary data
Supplementary data are available at European Heart Journal - Imaging Methods and Practice online.
Supplementary Material
Acknowledgements
The authors thank Yukari Tanaka for her assistance.
Contributor Information
Takeshi Tohyama, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan; Center for Advanced Medical Open Innovation, Kyushu University, Fukuoka, Japan; Institute for Medical Engineering & Science, Massachusetts Institute of Technology, Cambridge, MA, USA.
Takeshi Iwasaki, Department of Anatomic Pathology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan.
Masataka Ikeda, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Masato Katsuki, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan; Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University Beppu Hospital, Beppu, Japan.
Tatsuya Watanabe, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Kayo Misumi, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Keisuke Shinohara, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Takeo Fujino, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Toru Hashimoto, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Shouji Matsushima, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Tomomi Ide, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Junji Kishimoto, Centre for Clinical and Translational Research of Kyushu University Hospital, Fukuoka, Japan.
Koji Todaka, Center for Advanced Medical Open Innovation, Kyushu University, Fukuoka, Japan; Centre for Clinical and Translational Research of Kyushu University Hospital, Fukuoka, Japan.
Yoshinao Oda, Department of Anatomic Pathology, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan.
Kohtaro Abe, Department of Cardiovascular Medicine, Faculty of Medical Sciences, Kyushu University, 3-1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan.
Data availability
The participants of this study did not give written consent for their data to be shared publicly, so due to the sensitive nature of the research supporting data are not available.
Funding
This work was supported by the Japan Foundation for Applied Enzymology (Cardiovascular Innovative Conference; CVIC) and Uehara Memorial Foundation Postdoctoral Fellowship (T.T.), JSPS KAKENHI (JP24K02449 and JP24K22274; M.I.), and a research grant from Pfizer (T.I. to K.A.). These funding agencies had no role in the design and conduct of the study; in the collection, management, analysis, and interpretation of the data; nor in the preparation, review, and submission of the manuscript for publication.
Lead author biography

Dr Takeshi Tohyama is a cardiologist and data scientist trained at Kyushu University. He is pursuing further advancements in AI within the medical field at Leo’s laboratory at the Massachusetts Institute of Technology, aiming for innovative breakthroughs.
References
- 1. Ammirati E, Frigerio M, Adler ED, Basso C, Birnie DH, Brambatti M et al. Management of acute myocarditis and chronic inflammatory cardiomyopathy: an expert consensus document. Circ Heart Fail 2020;13:e007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rapezzi C, Merlini G, Quarta CC, Riva L, Longhi S, Leone O et al. Systemic cardiac amyloidoses: disease profiles and clinical courses of the 3 main types. Circulation 2009;120:1203–12. [DOI] [PubMed] [Google Scholar]
- 3. AbouEzzeddine OF, Davies DR, Scott CG, Fayyaz AU, Askew JW, McKie PM et al. Prevalence of transthyretin amyloid cardiomyopathy in heart failure with preserved ejection fraction. JAMA Cardiol 2021;6:1267–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Castaño A, Narotsky DL, Hamid N, Khalique OK, Morgenstern R, DeLuca A et al. Unveiling transthyretin cardiac amyloidosis and its predictors among elderly patients with severe aortic stenosis undergoing transcatheter aortic valve replacement. Eur Heart J 2017;38:2879–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gillmore JD, Maurer MS, Falk RH, Merlini G, Damy T, Dispenzieri A et al. Nonbiopsy diagnosis of cardiac transthyretin amyloidosis. Circulation 2016;133:2404–12. [DOI] [PubMed] [Google Scholar]
- 6. Vogelsberg H, Mahrholdt H, Deluigi CC, Yilmaz A, Kispert EM, Greulich S et al. Cardiovascular magnetic resonance in clinically suspected cardiac amyloidosis: noninvasive imaging compared to endomyocardial biopsy. J Am Coll Cardiol 2008;51:1022–30. [DOI] [PubMed] [Google Scholar]
- 7. Maurer MS, Elliott P, Comenzo R, Semigran M, Rapezzi C. Addressing common questions encountered in the diagnosis and management of cardiac amyloidosis. Circulation 2017;135:1357–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brownrigg J, Lorenzini M, Lumley M, Elliott P. Diagnostic performance of imaging investigations in detecting and differentiating cardiac amyloidosis: a systematic review and meta-analysis. ESC Heart Fail 2019;6:1041–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fontana M, Berk JL, Gillmore JD, Witteles RM, Grogan M, Drachman B et al. Vutrisiran in patients with transthyretin amyloidosis with cardiomyopathy. N Engl J Med 2024. Online ahead of print. [Google Scholar]
- 10. Maurer MS, Schwartz JH, Gundapaneni B, Elliott PM, Merlini G, Waddington-Cruz M et al. Tafamidis treatment for patients with transthyretin amyloid cardiomyopathy. N Engl J Med 2018;379:1007–16. [DOI] [PubMed] [Google Scholar]
- 11. Garcia-Pavia P, aus dem Siepen F, Donal E, Lairez O, van der Meer P, Kristen AV et al. Phase 1 trial of antibody NI006 for depletion of cardiac transthyretin amyloid. N Engl J Med 2023;389:239–50. [DOI] [PubMed] [Google Scholar]
- 12. Maurer MS, Kale P, Fontana M, Berk JL, Grogan M, Gustafsson F et al. Patisiran treatment in patients with transthyretin cardiac amyloidosis. N Engl J Med 2023;389:1553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gillmore JD, Judge DP, Cappelli F, Fontana M, Garcia-Pavia P, Gibbs S et al. Efficacy and safety of acoramidis in transthyretin amyloid cardiomyopathy. N Engl J Med 2024;390:132–42. [DOI] [PubMed] [Google Scholar]
- 14. Shimada S, Maekura S, Ino H, Matsuura M, Masunaga N, Matsumoto T et al. Unusual case of cardiac amyloidosis preceded by a twenty-year history of dilated cardiomyopathy and heart failure. Intern Med 2016;55:1109–15. [DOI] [PubMed] [Google Scholar]
- 15. GHy K, Ryu DR, Song PS, Song YB, Hahn JY, Choi SH et al. A case of a senile systemic amyloidosis patient presenting with angina pectoris and dilated cardiomyopathy. Korean Circ J 2011;41:209–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rushakoff JA, Kransdorf EP, Kittleson MM, Neyer JR, Luthringer D, Patel JK. Atypical cardiac amyloidosis phenotypes identified at transplant: a case series. Eur Heart J Case Rep 2023;7:ytad105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kintsler S, Jäkel J, Brandenburg V, Kersten K, Knuechel R, Röcken C. Cardiac amyloidosis in a heart transplant patient—a case report and retrospective analysis of amyloidosis evolution. Intractable Rare Dis Res 2015;4:60–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao W, Jiang W, Qiu X. Deep learning for COVID-19 detection based on CT images. Sci Rep 2021;11:14353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017;318:2199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nam S, Chong Y, Jung CK, Kwak TY, Lee JY, Park J et al. Introduction to digital pathology and computer-aided pathology. J Pathol Transl Med 2020;54:125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Byeon SJ, Park J, Cho YA, Cho BJ. Automated histological classification for digital pathology images of colonoscopy specimen via deep learning. Sci Rep 2022;12:12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sukegawa S, Ono S, Tanaka F, Inoue Y, Hara T, Yoshii K et al. Effectiveness of deep learning classifiers in histopathological diagnosis of oral squamous cell carcinoma by pathologists. Sci Rep 2023;13:11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kessel K, Mattila J, Linder N, Kivela T, Lundin J. Deep learning algorithms for corneal amyloid deposition quantitation in familial amyloidosis. Ocul Oncol Pathol 2020;6:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim HS, Yoon H, Thakur N, Hwang G, Lee EJ, Kim C et al. Deep learning-based histopathological segmentation for whole slide images of colorectal cancer in a compressed domain. Sci Rep 2021;11:22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang AY, Sharma V, Saini H, Tingen JN, Flores A, Liu D et al. Machine learning quantification of amyloid deposits in histological images of ligamentum Flavum. J Pathol Inform 2022;13:100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP et al. The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol 2008;61:344–9. [DOI] [PubMed] [Google Scholar]
- 28. Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD). Ann Intern Med 2015;162:735–6. [Google Scholar]
- 29. Trockman A, Kolter JZ. Patches are all you need? arXiv:2201.09792v1 [cs.CV], 10.48550/arXiv.2201.09792, 24 January 2022, preprint: not peer reviewed. [DOI]
- 30. Mass AL, Hannun AY, Ng AY. Rectifier nonlinearities improves neural network acoustic models. Proc Int Conf Mach Learn 2013;30:3. [Google Scholar]
- 31. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 2014;15:1929–58. [Google Scholar]
- 32. DeVries T, Taylor GW. Improved Regularization of Convolutional Neural Networks with Cutout. arXiv 1708.04552 [cs.CV], 10.48550/arXiv.1708.04552, 29 Nov 2017, preprint: not peer reviewed. [DOI]
- 33. Banypersad SM, Moon JC, Whelan C, Hawkins PN, Wechalekar AD. Updates in cardiac amyloidosis: a review. J Am Heart Assoc 2012;1:e000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nativi-Nicolau J, Maurer MS. Amyloidosis cardiomyopathy: update in the diagnosis and treatment of the most common types. Curr Opin Cardiol 2018;33:571–9. [DOI] [PubMed] [Google Scholar]
- 35. Runz M, Rusche D, Schmidt S, Weihrauch MR, Hesser J, Weis CA. Normalization of HE-stained histological images using cycle consistent generative adversarial networks. Diagn Pathol 2021;16:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garcia-Pavia P, Rapezzi C, Adler Y, Arad M, Basso C, Brucato A et al. Diagnosis and treatment of cardiac amyloidosis: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J 2021;42:1554–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ilse M, Tomczak JM, Welling M. Attention-based deep multiple instance learning. Proc 35th Int Conf Mach Learn 2018;80:2127–36. [Google Scholar]
- 38. Licordari R, Trimarchi G, Teresi L, Restelli D, Lofrumento F, Perna A et al. Cardiac magnetic resonance in HCM phenocopies: from diagnosis to risk stratification and therapeutic management. J Clin Med 2023;12:3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The participants of this study did not give written consent for their data to be shared publicly, so due to the sensitive nature of the research supporting data are not available.