Abstract
We describe the design and detailed characterization of 6-hydroxy-nicotine (6HNic)-adjustable transgene expression (NICE) systems engineered for lentiviral transduction and in vivo modulation of angiogenic responses. Arthrobacter nicotinovorans pAO1 encodes a unique catabolic machinery on its plasmid pAO1, which enables this Gram-positive soil bacterium to use the tobacco alkaloid nicotine as the exclusive carbon source. The 6HNic-responsive repressor-operator (HdnoR-ONIC) interaction, controlling 6HNic oxidase production in A.nicotinovorans pAO1, was engineered for generic 6HNic-adjustable transgene expression in mammalian cells. HdnoR fused to different transactivation domains retained its ONIC-binding capacity in mammalian cells and reversibly adjusted transgene transcription from chimeric ONIC-containing promoters (PNIC; ONIC fused to a minimal eukaryotic promoter [Pmin]) in a 6HNic-responsive manner. The combination of transactivators containing various transactivation domains with promoters differing in the number of operator modules as well as in their relative inter-ONIC and/or ONIC-Pmin spacing revealed steric constraints influencing overall NICE regulation performance in mammalian cells. Mice implanted with microencapsulated cells engineered for NICE-controlled expression of the human glycoprotein secreted placental alkaline phosphatase (SEAP) showed high SEAP serum levels in the absence of regulating 6HNic. 6HNic was unable to modulate SEAP expression, suggesting that this nicotine derivative exhibits control-incompatible pharmacokinetics in mice. However, chicken embryos transduced with HIV-1-derived self-inactivating lentiviral particles transgenic for NICE-adjustable expression of the human vascular endothelial growth factor 121 (VEGF121) showed graded 6HNic response following administration of different 6HNic concentrations. Owing to the clinically inert and highly water-soluble compound 6HNic, NICE-adjustable transgene control systems may become a welcome alternative to available drug-responsive homologs in basic research, therapeutic cell engineering and biopharmaceutical manufacturing.
INTRODUCTION
The Gram-positive soil bacterium Arthrobacter nicotinovorans pAO1 acquired the metabolic capacity to metabolize the tobacco alkaloid nicotine as an exclusive carbon source. Utilization is initiated by hydroxylation of nicotine's pyridine ring at position C6 followed by oxidation mediated by the 6-hydroxy-nicotine oxidase (6HNO). Recent sequence analysis of the catabolic plasmid pAO1 revealed the 6HNO gene repressor HdnoR, which controls 6HNO expression in the presence of 6-hydroxy-nicotine (6HNic) (1). 6HNic modulates HdnoR's allosteric conformation in a way that prevents further binding and repression of the 6HNO promoter, thereby resulting in induction of follow-up nicotine-specific metabolic pathways. 6HNic produced by A.nicotinovorans pAO1 came into the limelight as a bio-catalytic bulk product, which can be employed in conventional esterification processes or as an educt for the production of particular special chemicals, including 6-alkoxynicotine derivatives known to exhibit antibacterial and antifungal activities (2). In addition to being an intermediate product in (bio-) chemical production scenarios, 6HNic is also a by-product of pioneering efforts to denicotinize tobacco for the production of ‘mild’ or ‘light’ cigarettes characterized by a low nicotine content (3,4). Besides its involvement in the aforementioned processes, knowledge of 6HNic's physiologic impact and pharmacokinetics is limited or non-existing. However, unlike nicotine itself, 6HNic is expected to be more soluble and unable to trigger nicotine-specific receptor responses because of its hydroxylated aromatic ring (5).
HdnoR has been reported to belong to the TetR family of bacterial response regulators, which repress target genes in a physiologic compound-responsive manner (1). In particular, TetR dissociates from a cognate PTetA promoter in the presence of tetracycline antibiotics and so induces the expression of the tetracycline resistance gene tetA (6). Pioneering efforts in using bacterial response regulators for mammalian transgene expression fine-tuning have resulted in the design of the tetracycline-responsive expression system [the TET system(s)], which consists of its generic configuration of TetR fused to a Herpes simplex-derived VP16 transactivation domain (tTA) and a heptameric TetR-specific tetA promoter-derived operator module (tetO7) functionally linked to the minimal version of the human cytomegalovirus immediate early promoter (tetO7-PhCMVmin; PhCMV*−1). tTA binding to PhCMV*−1 induced desired transgene transcription in the absence of tetracycline. However, tetracycline switched tTA's allosteric conformation to a PhCMV*−1 binding-incompetent state, which resulted in dose-dependent transgene repression (7).
Following the generic design principle of the TET system, a wide variety of bacterial response regulators have been adapted for use as mammalian gene regulation systems, including those responsive to (i) tetracycline derivatives (7,8), (ii) streptogramin (9), (iii) macrolide (10) and (iv) coumermycin (11) antibiotics, (v) immunosuppressive rapamycin (12), hormones such as (vi) estrogen (13), (vii) progesterone (14) and (viii) ecdysone (15), (ix) temperature (16), (x) quorum-sensing molecules (17,18), (xi) the terpene cumate (http://www.qbiogene.com/products/gene-expression/qmateslideshow/index.htm), (xii) the type-2 diabetes drug roziglitazone (19) and (xiii) gaseous acetaldehyde (20).
Most transgene regulation modalities were conceived or have been used as stand-alone systems [one-gene control system modulating a single (set of) transgene(s)] for targeted molecular interventions in complex regulatory networks, including (i) prototype gene therapy and tissue engineering scenarios (12,21), (ii) drug discovery (22,23), (iii) biopharmaceutical manufacturing (24,25) and (iv) gene-function analysis (26). Clinically licensed antibiotics (macrolide, streptogramin, coumermycin and tetracycline), immunosuppressive agents (rapamycin), hormones (mifepristone) and PPAR-γ (peroxisome proliferator-activated receptor-γ) agonists (roziglitazone) seem to be ideal candidates for gene therapy-based conditional transgene interventions. However, some of these drugs may elicit side effects following long-term administration at regulation-effective concentrations (27–29). Furthermore, drug-based inducers are less suited for transgene modulation of biotechnologically relevant production cell lines, since preparation of inducer-free product formulations remains a costly downstream processing challenge.
Availability of different compatible transgene regulation systems enabled their functional interconnection to produce synthetic mammalian networks with unprecedented signal integration. The most prominent mammalian cell-embedded synthetic regulatory networks include: (i) an artificial regulatory cascade consisting of three heterologous transcription control units interconnected in a linear manner to produce discrete multilevel expression control of a terminally encoded transgene in response to clinical doses of different antibiotics (30), (ii) an epigenetic circuitry able to switch between two stable transgene expression states after transient administration of two alternate drugs (31) and BioLogic gates providing transgene expression integration reminiscent of digital electronics (32).
We have designed a novel gene regulation system [6HNic-adjustable transgene expression (NICE)] responsive to the non-toxic nicotine derivative 6HNic. NICE technology enabled fine-tuning of transgene expression in mammalian cells, was compatible with state-of-the-art lentiviral transduction and provided precise control of angiogenic responses in chicken embryos. Since NICE systems are responsive to a clinically inert, highly water-soluble nicotine derivative, we believe it will foster advances in basic research as well as in biopharmaceutical manufacturing.
MATERIALS AND METHODS
Plasmid construction
All plasmids used in this study are listed in Table 1; detailed information on their construction is also provided.
Table 1.
Plasmids used and designed in this study
| Plasmid | Description and cloning strategy | Reference or source |
|---|---|---|
| pBM57 | HIV-1-derived lentiviral expression vector (5′LTR-ψ+-oriSV40-cPPT-RRE-EYFP-3′LTRΔU3) | (33) |
| pBM104 | Lentiviral expression vector encoding a PPIR8-driven VEGF121 expression unit (5′LTR-ψ+-oriSV40-cPPT-RRE-PPIR8-VEGF121-3′LTRΔU3) | (68) |
| pBM105 | Lentiviral expression vector encoding a PPIR8-driven SEAP expression unit (5′LTR-ψ+-oriSV40-cPPT-RRE-PPIR8-VEGF121-3′LTRΔU3) | (68) |
| pBP10 | Vector encoding a PETR5-driven SEAP expression unit (PETR5-SEAP-pA; PETR5, ETR-2bp-PhCMVmin) | (47) |
| pBP11 | Vector encoding a PETR6-driven SEAP expression unit (PETR6-SEAP-pA; PETR6, ETR-4bp-PhCMVmin) | (47) |
| pBP12 | Vector encoding a PETR7-driven SEAP expression unit (PETR7-SEAP-pA; PETR7, ETR-6bp-PhCMVmin) | (47) |
| pBP13 | Vector encoding a PETR8-driven SEAP expression unit (PETR8-SEAP-pA; PETR8, ETR-8bp-PhCMVmin) | (47) |
| pBP14 | Vector encoding a PETR9-driven SEAP expression unit (PETR9-SEAP-pA; PETR9, ETR-10bp-PhCMVmin) | (47) |
| pH6EX3-HdnoR | Vector encoding the A.nicotinovorans pAO16 repressor of the 6HNic oxidase (HdnoR) | (1) |
| pLM82 | Constitutive NT1 expression vector (PSV40-NT1-pA; NT1, HdnoR-VP16) | This work |
| HdnoR was PCR-amplified from pH6EX3-HdnoR using OLM83: 5-GTACgaattcCCACCatgcgtatttccacggtggatcg-3′ and OLM84: 5′-CTTATGgcgcgcGGCTGTACGCGGAtagtcctacccgatcgaggta-3′ (lower case, annealing sequence; lower case italics, restriction sites), restricted with EcoRI/BssHII and ligated into the corresponding sites (EcoRI/BssHII) of pWW35 | ||
| pLM83 | Vector encoding a PNIC1a-driven SEAP expression unit (PNIC1a-SEAP-pA; PNIC1a, ONIC-0bp-PhCMVmin) | This work |
| PhCMVmin was PCR-amplified from pRevTRE using OLM82: 5′-GATCgacgtcCCCCATTGACATGGACAGCTGTCCATGTATCAATAGGGTGcctgcaggtcgagctcggtacccgggtc-3′ and OWW22: 5′-GCTAgaattccgcggaggctggatcgg-3′ (lower case, annealing sequence; lower case italics, restriction sites; upper case, ONIC), restricted with AatII/EcoRI and ligated into the corresponding sites (AatII/EcoRI) of pMF111 | ||
| pLM101 | Constitutive NT2 expression vector (PSV40-NT2-pA; NT2, HdnoR-p65) | This work |
| The NF-κB-derived transactivation domain (p65) was excised from pWW42 using BssHII/BamHI and ligated into the corresponding sites (BssHII/BamHI) of pLM82 | ||
| pLM102 | Constitutive NT3 expression vector (PSV40-NT3-pA; NT3, HdnoR-E2F4) | This work |
| The E2F4-derived transactivation domain (E2F4) was excised from pWW64 using BssHII/BamHI and ligated into the corresponding sites (BssHII/BamHI) of pLM82 | ||
| pLM103 | Lentiviral NT1 expression vector (5′LTR-ψ+-oriSV40-cPPT-RRE-PhEF1α-NT1-3′LTRΔU3; NT1, HdnoR-VP16) | This work |
| NT1 was excised from pLM82 using NotI/XmaI and ligated into the corresponding sites (NotI/XmaI) of pMF391 | ||
| pLM104 | Vector encoding a PNIC1b-driven SEAP expression unit (PNIC1b-SEAP-pA; PNIC1b, ONIC-2bp-PhCMVmin) | This work |
| 2bp-PhCMVmin-SEAP was excised from pBP10 using SbfI/XhoI and ligated into the corresponding sites (SbfI/XhoI) of pLM83 | ||
| pLM105 | Vector encoding a PNIC1c-driven SEAP expression unit (PNIC1c-SEAP-pA; PNIC1c, ONIC-4bp-PhCMVmin) | This work |
| 4bp-PhCMVmin-SEAP was excised from pBP11 using SbfI/XhoI and ligated into the corresponding sites (SbfI/XhoI) of pLM83 | ||
| pLM106 | Vector encoding a PNIC1d-driven SEAP expression unit (PNIC1d-SEAP-pA; PNIC1d, ONIC-6bp-PhCMVmin) | This work |
| 6bp-PhCMVmin-SEAP was excised from pBP12 using SbfI/XhoI and ligated into the corresponding (SbfI/XhoI) sites of pLM83 | ||
| pLM107 | Vector encoding a PNIC1e-driven SEAP expression unit (PNIC1e-SEAP-pA; PNIC1e, ONIC-8bp-PhCMVmin) | This work |
| 8bp-PhCMVmin-SEAP was excised from pBP13 using SbfI/XhoI and ligated into the corresponding (SbfI/XhoI) sites of pLM83 | ||
| pLM108 | Vector encoding a PNIC1f-driven SEAP expression unit (PNIC1f-SEAP-pA; PNIC1f, ONIC-10bp-PhCMVmin) | This work |
| 10bp-PhCMVmin-SEAP was excised from pBP14 using SbfI/XhoI and ligated into the corresponding (SbfI/XhoI) sites of pLM83 | ||
| pLM116 | BamHI-AscI-StuI-AatII-XbaI-ONIC-0bp-NheI-SbfI-EcoRI-EPO was PCR-amplified from pWW139 using OLM90 5′-CGggatccAggcgcgccAaggcctTTgacgtctctagaTACCCCATTGACATGGACAGCTGTCCATGTATCAATAGGGTGTgctagcTTcctgcagggaattccaccatgg-3′ and OWW51 5′-gcgcgcatcgattcacctgtcccctctcctgcag-3′, restricted with (BamHI/ClaI) and ligated into the corresponding sites (BamHI/ClaI) of pWW139 (lower case, annealing sequence; lower case italics, restriction sites; upper case, ONIC) | This work |
| pLM118 | ONIC-NheI-SbfI-EcoRI-EPO was excised from pLM116 using XbaI/ClaI and ligated into the compatible sites (XbaI/ClaI) of pLM116. PhEF1α-ONIC2-0bp-ONIC1-EPO-pA | This work |
| pLM119 | AscI-StuI-AatII-XbaI-ONIC-2bp-NheI-SbfI-EcoRI-EPO was PCR-amplified from pWW139 using OLM91 5′-CGggatccAggcgcgccAaggcctTTgacgtctctagaTACCCCATTGACATGGACAGCTGTCCATGTATCAATAGGGTGATTgctagcTTcctgcagggaattccaccatgg-3′ and OWW51 5′-gcgcgcatcgattcacctgtcccctctcctgcag-3′, restricted with AscI/ClaI and ligated into the corresponding sites (AscI/ClaI) of pLM116. PhEF1α-ONIC2-2bp-ONIC1-EPO-pA | This work |
| pLM120 | AscI-StuI-AatII-XbaI-ONIC-4bp-NheI-SbfI-EcoRI-EPO was PCR-amplified from pWW139 using OLM92 5′-CGggatccAggcgcgccAaggcctTTgacgtctctagaTACCCCATTGACATGGACAGCTGTCCATGTATCAATAGGGTGATCGTgctagcTTcctgcagggaattccaccatgg-3′ and OWW51 5′-gcgcgcatcgattcacctgtcccctctcctgcag-3′, restricted with AscI/ClaI and ligated into the corresponding sites (AscI/ClaI) of pLM116. PhEF1α-ONIC2-4bp-ONIC1-EPO-pA | This work |
| pLM121 | AscI-StuI-AatII-XbaI-ONIC-6bp-NheI-SbfI-EcoRI-EPO was PCR-amplified from pWW139 using OLM93 5′-CGggatccAggcgcgccAaggcctTTgacgtctctagaTACCCCATTGACATGGACAGCTGTCCATGTATCAATAGGGTGATCGTATgctagcTTcctgcagggaattccaccatgg-3′ and OWW51 5′-gcgcgcatcgattcacctgtcccctctcctgcag-3′, restricted with AscI/ClaI and ligated into the corresponding sites (AscI/ClaI) of pLM116. PhEF1α-ONIC2-6bp-ONIC1-EPO-pA | This work |
| pLM122 | AscI-StuI-AatII-XbaI-ONIC-8bp-NheI-SbfI-EcoRI-EPO was PCR-amplified from pWW139 using OLM94 5′-CGggatccAggcgcgccAaggcctTTgacgtctctagaTACCCCATTGACATGGACAGCTGTCCATGTATCAATAGGGTGATCGTAATTgctagcTTcctgcagggaattccaccatgg-3′ and OWW51 5′-gcgcgcatcgattcacctgtcccctctcctgcag-3′, restricted with AscI/ClaI and ligated into the corresponding sites (AscI/ClaI) of pLM116. PhEF1α-ONIC2-8bp-ONIC1-EPO-pA | This work |
| pLM123 | AscI-StuI-AatII-XbaI-ONIC-10bp-NheI-SbfI-EcoRI-EPO was PCR-amplified from pWW139 using OLM95 5′-CGggatccAggcgcgccAaggcctTTgacgtctctagaTACCCCATTGACATGGACAGCTGTCCATGTATCAATAGGGTGATCGTACGATTgctagcTTcctgcagggaattccaccatgg-3′ and OWW51 5′-gcgcgcatcgattcacctgtcccctctcctgcag-3′, restricted with AscI/ClaI and ligated into the corresponding sites (AscI/ClaI) of pLM116. PhEF1α-ONIC2-10bp-ONIC1-EPO-pA | This work |
| pLM124 | ONIC-NheI-SbfI-EcoRI-EPO was excised from pLM116 using XbaI/ClaI and ligated into the compatible sites (NheI/ClaI) of pLM119, thereby resulting in PhCMV-ONIC2-2bp-ONIC1-EPO-pA | This work |
| pLM125 | ONIC-NheI-SbfI-EcoRI-EPO was excised from pLM116 using XbaI/ClaI and ligated into the compatible sites (NheI/ClaI) of pLM120, thereby resulting in PhCMV-ONIC2-4bp-ONIC1-EPO-pA | This work |
| pLM126 | ONIC-NheI-SbfI-EcoRI-EPO was excised from pLM116 using XbaI/ClaI and ligated into the compatible sites (NheI/ClaI) of pLM121, thereby resulting in PhCMV-ONIC2-6bp-ONIC1-EPO-pA | This work |
| pLM127 | ONIC-NheI-SbfI-EcoRI-EPO was excised from pLM116 using XbaI/ClaI and ligated into the compatible sites (NheI/ClaI) of pLM122, thereby resulting in PhCMV-ONIC2-8bp-ONIC1-EPO-pA | This work |
| pLM128 | ONIC-NheI-SbfI-EcoRI-EPO was excised from pLM116 using XbaI/ClaI and ligated into the compatible sites (NheI/ClaI) of pLM123, thereby resulting in PhCMV-ONIC2-10bp-ONIC1-EPO-pA | This work |
| pLM129 | PhCMV-ONIC2-0bp-ONIC1-NheI was excised from pLM118 using ScaI/SbfI and ligated into the corresponding sites (ScaI/SbfI) of pLM105, thereby resulting in PhCMV-ONIC2-0bp-ONIC1-NheI-SbfI-4bp-PhCMVmin-SEAP-pA | This work |
| pLM130 | PhCMV-ONIC2-2bp-ONIC1 was excised from pLM124 using ScaI/SbfI and ligated into the corresponding sites (ScaI/SbfI) of pLM105, thereby resulting in PhCMV-ONIC2-2bp-ONIC1-NheI-SbfI-4bp-PhCMVmin-SEAP-pA | This work |
| pLM131 | PhCMV-ONIC2-4bp-ONIC1 was excised from pLM125 using ScaI/SbfI and ligated into the corresponding sites (ScaI/SbfI) of pLM105, thereby resulting in PhCMV-ONIC2-4bp-ONIC1-NheI-SbfI-4bp-PhCMVmin-SEAP-pA | This work |
| pLM132 | PhCMV-ONIC2-6bp-ONIC1 was excised from pLM126 using ScaI/SbfI and ligated into the corresponding sites (ScaI/SbfI) of pLM105, thereby resulting in PhCMV-ONIC2-6bp-ONIC1-NheI-SbfI-4bp-PhCMVmin-SEAP-pA | This work |
| pLM133 | PhCMV-ONIC2-8bp-ONIC1 was excised from pLM127 using ScaI/SbfI and ligated into the corresponding sites (ScaI/SbfI) of pLM105, thereby resulting in PhCMV-ONIC2-8bp-ONIC1-NheI-SbfI-4bp-PhCMVmin-SEAP-pA | This work |
| pLM134 | PhCMV-ONIC2-10bp-ONIC1 was excised from pLM128 using ScaI/SbfI and ligated into the corresponding sites (ScaI/SbfI) of pLM105, thereby resulting in PhCMV-ONIC2-10bp-ONIC1-NheI-SbfI-4bp-PhCMVmin-SEAP-pA | This work |
| pLM135 | Vector encoding a PNIC2a-driven SEAP expression unit (PNIC2a-SEAP-pA; PNIC2a, ONIC2-0bp-ONIC1-NheI-SbfI-4bp-PhCMVmin). PhCMV was excised from pLM129 using AatII/AatII and the pLM129 backbone was self-ligated | This work |
| pLM136 | Vector encoding a PNIC2b-driven SEAP expression unit (PNIC2b-SEAP-pA; PNIC2b, ONIC2-2bp-ONIC1-NheI-SbfI-4bp-PhCMVmin). PhCMV was excised from pLM130 using AatII/AatII and the pLM130 backbone was self-ligated | This work |
| pLM137 | Vector encoding a PNIC2c-driven SEAP expression unit (PNIC2c-SEAP-pA; PNIC2c, ONIC2-4bp-ONIC1-NheI-SbfI-4bp-PhCMVmin). PhCMV was excised from pLM131 using AatII/AatII and the pLM131 backbone was self-ligated | This work |
| pLM138 | Vector encoding a PNIC2d-driven SEAP expression unit (PNIC2d-SEAP-pA; PNIC2d, ONIC2-6bp-ONIC1-NheI-SbfI-4bp-PhCMVmin). PhCMV was excised from pLM132 using AatII/AatII and the pLM132 backbone was self-ligated | This work |
| pLM139 | Vector encoding a PNIC2e-driven SEAP expression unit (PNIC2e-SEAP-pA; PNIC2e, ONIC2-8bp-ONIC1-NheI-SbfI-4bp-PhCMVmin). PhCMV was excised from pLM133 using AatII/AatII and the pLM133 backbone was self-ligated | This work |
| pLM140 | Vector encoding a PNIC2f-driven SEAP expression unit (PNIC2f-SEAP-pA; PNIC2f, ONIC2-10bp-ONIC1-NheI-SbfI-4bp-PhCMVmin). PhCMV was excised from pLM134 using AatII/AatII and the pLM134 backbone was self-ligated | This work |
| pLM141 | PNIC2d (ONIC2-6bp-ONIC1-NheI-SbfI-4bp-PhCMVmin) was excised from pLM132 (AscI/EcoRI) and ligated into the corresponding sites (AscI/EcoRI) of pBM105 (5′LTR-ψ+-oriSV40-cPPT-RRE-PNIC2d-SEAP-3′LTRΔU3; PNIC2d, ONIC2-6bp-ONIC1-NheI-SbfI-4bp-PhCMVmin) | This work |
| pLM145 | PhCMVmin-VEGF121 was excised from pBM104 using SbfI/SspI and ligated into the corresponding sites (SbfI/SspI) of pLM120, thereby resulting in PNIC3-VEGF121-pA (PNIC3, ONIC-4bp-NheI-SbfI-PhCMVmin) | This work |
| pLM146 | Lentiviral expression vector encoding a PNIC3-driven VEGF121 expression unit (5′LTR-ψ+-oriSV40-cPPT-RRE-PNIC3-VEGF121-3′LTRΔU3; PNIC3, ONIC-4bp-NheI-SbfI-PhCMVmin). PNIC3–VEGF121 was excised from pLM145 using AscI/MluI and ligated into the corresponding sites (AscI/MluI) of pBM104 | This work |
| pMF111 | Vector encoding a PhCMV*−1-driven SEAP expression unit (PhCMV*−1-SEAP-pA) | (69) |
| pMF391 | Lentiviral ET1 expression vector (5′LTR-ψ+-oriSV40-PPT-RRE- PhEF1α-ET1-3′LTRΔU3) | (70) |
| pRevTRE | Oncoretroviral expression vector containing a tetracycline-responsive expression unit | Clontech, Palo Alto, CA |
| Pseap2-Control | Constitutive PSV40-driven SEAP expression vector | Clontech, Palo Alto, CA |
| pWW35 | Constitutive ET1 expression vector (PSV40-ET1-pA) | (10) |
| pWW42 | Constitutive ET2 expression vector (PSV40-ET2-pA) | (10) |
| pWW64 | Constitutive ET4 expression vector (PSV40-ET4-pA) | (10) |
| pWW139 | EPO expression vector | (18) |
3′LTRΔU3, enhancer-free 3′ long terminal repeat; 5′LTR, 5′ long terminal repeat; 6HNic, 6-hydroxy-nicotine; cPPT, central polypurine tract; E2F4, human transcription factor, transactivation domain of the human E2F4; EPO, erythropoetin; ET1, macrolide-dependent transactivator (MphR(A)-VP16); ET2, macrolide-dependent transactivator (MphR(A)-p65); ET4, macrolide-dependent transactivator (MphR(A)-E2F4); ETR, operator module specific for MphR(A); EYFP, enhanced yellow fluorescent protein; HdnoR, repressor of the A.nicotinovorans pAO1 6HNic oxidase gene; MphR(A), E.coli-derived repressor of the macrolide resistance gene mphA; NF-κB, human transcription factor; NT1, 6HNic-dependent transactivator (HdnoR-VP16); NT2, 6HNic-dependent transactivator (HdnoR-p65); NT3, 6HNic-dependent transactivator (HdnoR-E2F4); ONIC, HdnoR-specific operator; ONIC1/2, ONIC numbering in tandem operator configurations; oriSV40, origin of replication of the SV40; p65, transactivation domain of NF-κB; pA; SV40-derived polyadenylation site; PETR2, macrolide-responsive promoter (ETR-PhCMVmin); PETR5-9, macrolide-responsive promoters containing different spacers between ETR and PhCMVmin; PhCMV, promoter of the human cytomegalovirus immediate early promoter; PhCMVmin, minimal PhCMV; PhCMV*−1, tetracycline-responsive promoter; PhEF1α, promoter of the human elongation factor 1 alpha; PNIC1a-f, 6HNic-responsive promoters containing different spacers between ONIC and PhCMVmin; PNIC2a-f, 6HNic-responsive promoters containing tandem ONIC operators with different inter-ONIC spacing but fixed spacing relative to PhCMVmin; PNIC3, 6HNic-responsive promoter with extended spacing between ONIC and PhCMVmin; PPIR8, streptogramin-dependent promoter; PSV40, constitutive SV40 promoter; RRE, rev response element; SEAP, human placental secreted alkaline phosphatase; VEGF121, human vascular endothelial growth factor 121; VP16, H.simplex virus-derived transactivation domain; ψ+, extended lentiviral packaging signal.
Cell culture and transfection
Chinese hamster ovary cells (CHO-K1, ATCC CCL 61) were cultivated in standard medium: FMX-8 medium (Cell Culture Technologies, Zurich, Switzerland) supplemented with 10% fetal calf serum (FCS) (Pan Biotech GmbH, Aidenbach, Germany; catalog no. 3302-P231902, lot no. P231902). Human embryonic kidney cells transgenic for simian virus 40 (SV40) large T antigen [HEK293-T (33)] were cultivated in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FCS. All cell lines were cultivated at 37°C in a 5% CO2-containing humidified atmosphere. CHO-K1 cells were transfected using an optimized calcium phosphate-based protocol that resulted in standard transfection efficiencies of 35 ± 5%. In brief, 40 000 CHO-K1 cells were seeded per well of a 24-well plate and cultivated overnight. Aliquots containing 6 μg of plasmid DNA (for cotransfections equal amounts of each plasmid were used) were diluted in 60 μl of 250 mM CaCl2 and precipitated following drop-wise addition of 60 μl phosphate solution for 20 s (50 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4, pH 7.1). Following further incubation for 10 s, 2 ml FMX-8 containing 2% FCS were added. The culture medium was replaced by the DNA-precipitate-containing medium and incubated for 5 h prior to a glycerol shock for 30 s (FMX-8 medium supplemented with 15% glycerol and 2% FCS). After a single washing step using standard medium, cells were cultivated for analysis in the presence or absence of regulating 6HNic (50 μg/ml, unless stated otherwise). Forty-eight hours post glycerol shock, reporter protein expression was profiled.
Lentiviral particle production and transduction
For production of replication-incompetent self-inactivating HIV-1-derived lentiviral particles, HEK293-T cells were co-transfected following an optimized calcium phosphate-based protocol. In brief, 200 000 HEK293-T cells were seeded per well of a 6-well plate and cultivated overnight in 2 ml 10% FCS-containing DMEM. For each transfection, 1 μg pLTR-G [encoding the pseudotyping envelope protein VSV-G of the vesicular stomatitis virus (34)], 1 μg pCD/NL-BH* [helper construct (35)] and 1 μg of the desired transgene-encoding lentiviral expression vector were diluted in 100 μl of 250 mM CaCl2. The DNA mixture was added drop-wise to 100 μl phosphate solution (100 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4, pH 7.1), incubated for 15 min to enable formation of DNA-CaPO4 precipitates, which were subsequently added to HEK293-T cultures. Five-hours post-transfection, the DNA–CaPO4 complex was removed by medium exchange and lentiviral particles were produced for another 48 h prior to collection from the supernatant by filtration through a 0.45 μm filter (PIR8FP 030/2; Schleicher & Schuell GmbH, Dassel, Germany). This protocol typically yielded lentiviral particle titers of 2 × 107 c.f.u./ml following titration on CHO-K1 cells or enzyme-linked immunosorbent assay-based p24 quantification according to the manufacturer's protocol (catalog no. 103; Immunodiagnostics Inc., Woburn, MA). In order to prevent cross-contamination of secreted proteins from production supernatants and increase overall transduction efficiency, lentiviral particles were concentrated by ultracentrifugation for 2 h at 43 000 g and 4°C (Beckman Quick-Seal centrifuge tubes; catalog no. 342413, Beckman Instruments Inc., CA). The pellets were resuspended in 10% FCS-containing DMEM to adjust viral concentrations to desired levels. Furthermore, the culture medium was exchanged 6 h post transduction to ensure that transgene expression was exclusively based on transduction. Unless stated otherwise, standard transduction experiments included infection of 24 000 target cells seeded per well of a 12-well plate with 8 × 105 c.f.u. lentiviral particles (4 ng of p24).
Quantification of reporter protein production
Product proteins were quantified in cell culture supernatants 48 h after transduction. Human placental secreted alkaline phosphatase (SEAP) production was assessed using a chemiluminescence-based assay (Roche Diagnostics AG, Rotkreuz, Switzerland). Human vascular endothelial growth factor 121 (VEGF121) production was quantified using the human VEGF-specific DuoSet ELISA System (R&D Systems, Minneapolis, MO) according to the manufacturer's protocol.
In vivo methods I—mice
CHO-K1 cells engineered for NICE-controlled SEAP expression by cotransfection of pLM82 (PSV40-NT1-pA) and pLM104 (PNIC1b-SEAP-pA) were encapsulated in coherent alginate-poly-(L-lysine)-alginate beads (200 cells/capsule, 2 × 106 cells/mouse) as described previously (10) and implanted intraperitoneally into female OF1 mice (oncins France souche 1; Iffa-Credo, Lyon, France). At 1 h after capsule implantation, 6HNic was administered by intraperitoneal injection at doses ranging from 0 to 100 mg/kg. 6HNic was formulated for in vivo administration by dilution of stock solutions to appropriate concentrations using physiological salt solution [0.9% (w/v); Laboratoire Aguettant, Lyon, France]. Control mice harbored encapsulated wild-type CHO-K1 cells. At 72 h after 6HNic administration, the mice were killed for blood collection and quantification of SEAP serum levels using microtainer SST tubes (Beckton Dickinson, Plymouth, UK) according to the manufacturer's protocol. All experiments involving mice were approved by the French Ministry of Agriculture and Fishery (Paris, France) and performed by M. D. El-Baba at the Institut Universitaire de Technologie, IUTA, F-69622 Villeurbanne Cedex, France.
In vivo methods II—transduction of chicken embryos
The shell-free cultivation protocols of Djonov and co-workers (36) were used for all experiments involving chicken embryos. Brown Leghorn eggs were opened after 3 days incubation at 37°C and their contents were carefully poured into 80 mm plastic Petri dishes. The chicken embryos were incubated at 37°C in a humidified atmosphere. On embryonic day 9, pLM146- (100 μl in DMEM, 8.5 × 106 c.f.u., 40 ng p24) and pLM103- (90 μl in DMEM, 6.4 × 105 c.f.u., 3 ng p24) derived lentiviral particles were co-applied locally on top of the growing chorioallantoic membrane (CAM) together with 0.5 μl (0.5 nM in DMEM) CellTracker orange CMTMR (catalog no. C-2927; Molecular Probes Inc., Eugene, OR) to tag the transduction site. In order to modulate heterologous VEGF121 expression, different concentrations of 6HNic (0, 0.1, 1 and 50 μg/ml) were administered 1 h post transduction. On embryonic day 12, the CAMs were examined by in vivo fluorescence microscopy following intravenous injection of 100 μl 2.5% fluoresceine isothiocyanate dextran (FITC) (2 000 000; Sigma Chemicals, St Louis, MO) (37). FITC-stained CAM blood vessels were visualized at 50× and 100× magnifications using a Leica DM-RB fluorescence microscope equipped with a Leica digital fluorescence camera DC300 FX (Leica Microsystems AG, Heerbrugg, Switzerland) and a XF114 filter (Omega Optical Inc., Brattleboro, VT).
HPLC-based quantification of 6-hydroxy-nicotine
6HNic concentrations in 2 μl mouse urine samples were quantified by high-performance liquid chromatography (HPLC) [reversed-phase C18 column (Atlantis®C18, 4.6 × 150 mm; Waters Associates Inc.), Waters 2695 Separation Module (Waters Associates Inc.), Waters 996 Photodiode Array Detector (Waters Associates Inc.)]. Using an isocratic elution system of 0.1% TFA in water/acetonitrile (98:2 v/v) at a flow rate of 1 ml/min, the retention time of 6HNic was 7.1 min. The chromatograms were analyzed at 230 and 295 nm using Waters Empower® software. 6HNic concentrations in each sample were determined based on a peak area calibration curve generated using pure 6HNic.
Regulating 6-hydroxy-nicotine
6HNic (InterBioScreen, Moscow, Russia) was prepared as a stock solution of 100 mg/ml in water and used at indicated final concentrations.
RESULTS
Design of the 6-hydroxy-nicotine-responsive mammalian transgene regulation system
Capitalizing on machinery enabling A.nicotinovorans pAO1 to metabolize nicotine, we have designed a system called NICE. NICE-controlled transgene modulation in mammalian cells required two functionally crosstalking components: (i) an artificial transactivator (NT1) engineered by fusing A.nicotinovorans pAO1's 6-hydroxy-d-nicotine oxidase gene (6HDNO) repressor HdnoR to the generic H.simplex type 1 (HSV-1) VP16 transactivation domain (1,38) (pLM82; PSV40-NT1-pA, NT1, HdnoR-VP16) and (ii) a chimeric promoter (PNIC) assembled by cloning 6HDNO-specific operator modules (ONIC) adjacent to a minimal version of the human cytomegalovirus immediate early promoter (PhCMVmin; PNIC, ONIC-PhCMVmin) (1,39). Following cotransfection of pLM82 (PSV40-NT1-pA) and pLM83 (PNIC1a-SEAP-pA; PNIC1a, ONIC-0bp-PhCMVmin), enabling PNIC1a-driven human placental SEAP expression, into CHO-K1 grown in the absence of 6HNic, NT1 bound PNIC1a via HdnoR-ONIC interaction and initiated high-level SEAP production (48.0 ±7.3 U/l). Akin to 6HNic-mediated derepression of 6HDNO in A.nicotinovorans pAO1 NT1 adopts a binding-incompetent allosteric conformation in the presence of 6HNic, which results in disruption of the NT1-PNIC1a interaction and shutdown of SEAP production (1.3 ± 0.2 U/l) (Figure 1).
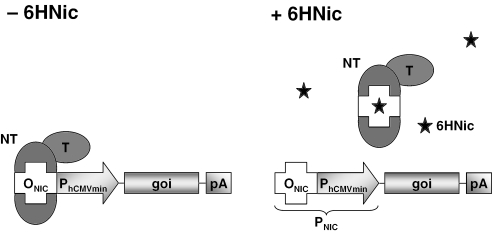
Figure 1.
Schematic representation of key components of the 6HNic (6HNic)-responsive transgene regulation system (NICE). As a binary transcription-control system, NICE consists of an artificial 6HNic-dependent transactivator (NT), assembled by fusing the A.nicotinovorans pAO1 6HNic oxidase repressor HdnoR to functional mammalian transactivation domains (T; e.g. H.simplex VP16, p65 of human NF-κB, a domain of human E2F4) and a chimeric promoter engineered by placing HdnoR-specific operator modules (ONIC) adjacent to a minimal version of the human cytomegalovirus immediate early promoter (PhCMVmin). In the absence of 6HNic (−6HNic), NT binds to PNIC via direct HdnoR-ONIC interaction and induces PhCMVmin-mediated transcription of the gene of interest (goi). However, 6HNic modifies NT's allostery such that it is no longer able to bind and induce PNIC, which results in complete transgene repression.
6HNic-responsive promoter configurations I—PNIC containing a single NT1-specific operator
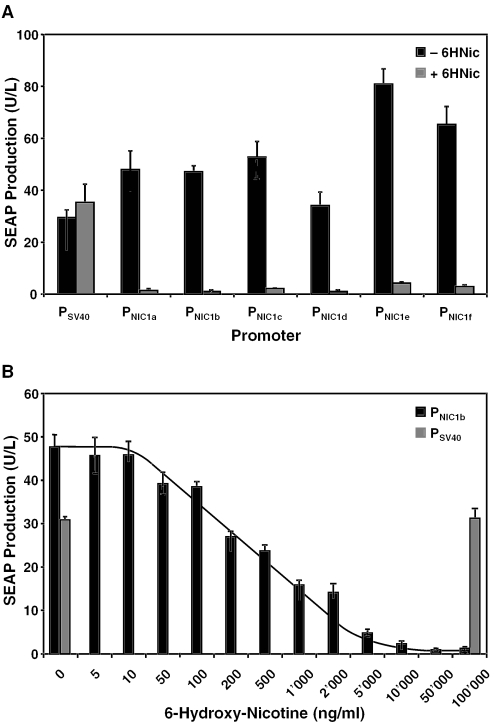
Efficient transcription-initiation of regulated promoters requires optimal crosstalk between the transcription machinery, the transactivator tethered to the cognate operator and the minimal promoter. The distance and twist between the operator module and the minimal promoter represent spatio-steric constraints for the assembly of the transcription-initiation complex and so influence overall performance of regulated promoters (40–46). With the aim of designing optimal PNIC configurations, we engineered linkers of 2 bp increments ranging from 0 to 10 bp between ONIC and PhCMVmin (PNIC1a, ONIC-0bp-PhCMVmin; PNIC1b, ONIC-2bp-PhCMVmin; PNIC1c, ONIC-4bp-PhCMVmin; PNIC1d, ONIC-6bp-PhCMVmin; PNIC1e, ONIC-8bp-PhCMVmin; PNIC1f, ONIC-10bp-PhCMVmin). PNIC1a-f-driven SEAP expression units (PNIC1a, pLM83; PNIC1b, pLM104; PNIC1c, pLM105; PNIC1d, pLM106; PNIC1e, pLM107; PNIC1f, pLM108) were cotransfected with the NT1 expression vector pLM82 (PSV40-NT1-pA) into CHO-K1 and cultivated for 48 h in the presence (50 μg/ml 6HNic) and absence of 6HNic before SEAP production was quantified. The PSV40-driven SEAP expression used as the control indicated that 6HNic showed no negative impact on host cell physiology at regulation-effective concentrations (Figure 2A). PNIC1b harboring 2 bp between ONIC and PhCMVmin showed the tightest repression of all promoter configurations while its maximum SEAP expression levels compared favorably with PSV40. Although PNIC1e and PNIC1f promoted higher SEAP production compared with PNIC1b, their leaky expression was increased as well. Direct comparison of the isogenic promoters PNIC1a and PNIC1f, which harbor ONIC and PhCMVmin on the same face of the DNA but at different distances, suggested a distance-dependent increase of both maximum as well as leaky expression (Figure 2A).
Figure 2.
Regulation performance and adjustability of 6HNic-responsive promoters (PNIC1) containing a single 6HNic-dependent transactivator (NT)-specific operator module. (A) 6HNic-responsive promoters containing a single operator module (ONIC) were engineered to contain linkers of 2 bp increments ranging from 0 to 10 bp between ONIC and the minimal promoter PhCMVmin (PNIC1a, ONIC-0bp-PhCMVmin; PNIC1b, ONIC-2bp-PhCMVmin; PNIC1c, ONIC-4bp-PhCMVmin; PNIC1d, ONIC-6bp-PhCMVmin; PNIC1e, ONIC-8bp-PhCMVmin; PNIC1f, ONIC-10bp-PhCMVmin). PNIC1a-f-driven SEAP expression units (PNIC1a-SEAP-pA [pLM83]; PNIC1b-SEAP-pA [pLM104]; PNIC1c-SEAP-pA [pLM105]; PNIC1d-SEAP-pA [pLM106]; PNIC1e-SEAP-pA [pLM107]; PNIC1f-SEAP-pA [pLM108]) were cotransfected with the NT1 expression vector pLM82 (PSV40-NT1-pA; NT1, HdnoR-VP16) into CHO-K1 and cultivated for 48 h in the presence (50 μg/ml 6HNic) and absence of 6HNic before SEAP production was quantified. SEAP production was compared with pSEAP2-Control-transfected CHO-K1 cells harboring a glycoprotein expression unit driven by the SV40 promoter (PSV40). (B) CHO-K1 transfected with pLM82 (PSV40-NT1-pA) and pLM104 (PNIC1b-SEAP-pA) were cultivated for 48 h in the presence of increasing 6HNic concentrations, which resulted in adjustable repression of the model product protein SEAP. (The line was added for clarity.)
In order to assess the dose-response characteristics of the NICE technology, CHO-K1 were cotransfected with pLM104 (PNIC1b-SEAP-pA) and pLM82 (PSV40-NT1-pA) and cultivated for 48 h in the presence of increasing 6HNic concentrations prior to SEAP production profiling. SEAP production gradually decreased at and beyond 50 ng/ml 6HNic until transgene expression was fully repressed at 50 μg/ml 6HNic. Control configurations including PSV40-driven SEAP expression in CHO-K1 reached identical glycoprotein production in the absence and presence of 100 μg/ml 6HNic confirming that this nicotine derivative exhibits no deleterious physiologic effects on mammalian cells at regulation-effective concentrations (Figure 2B).
6HNic-responsive promoter configurations II—PNIC containing a tandem NT1-specific operator
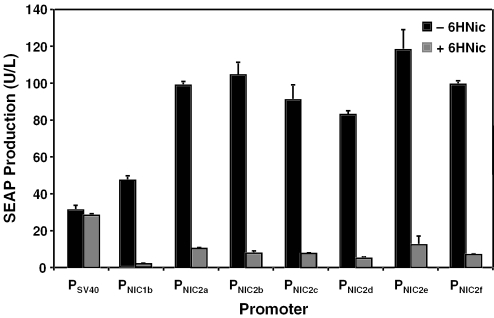
In order to increase the expression performance of PNIC1b, we cloned a second ONIC (ONIC2) module 5′ of ONIC1. PNIC2 derivatives contain a first NT1 operator (ONIC1) at an optimal distance 5′ of PhCMVmin and a second operator (ONIC2) placed 0/2/4/6/8/10 bp upstream of ONIC1 (PNIC2, ONIC2-spacer-ONIC1-NheI-SbfI-4bp-PhCMVmin). Resulting promoters (PNIC2a, ONIC2-0bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2b, ONIC2-2bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2c, ONIC2-4bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2d, ONIC2-6bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2e, ONIC2-8bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2f, ONIC2-10bp-ONIC1-NheI-SbfI-4bp-PhCMVmin) were configured for SEAP expression (pLM135, PNIC2a-SEAP-pA; pLM136, PNIC2b-SEAP-pA; pLM137, PNIC2c-SEAP-pA; pLM138, PNIC2d-SEAP-pA; pLM139, PNIC2e-SEAP-pA; pLM140, PNIC2f-SEAP-pA) and cotransfected with the NT1 expression vector pLM82 (PSV40-NT1-pA) into CHO-K1. Transfected cell populations were grown for 48 h in the presence (50 μg/ml) and absence of 6HNic prior to SEAP quantification (Figure 3). Tandem ONIC-containing modules doubled maximum SEAP expression levels compared with mono-ONIC PNIC1 promoter derivatives. However, leaky transcription in the presence of 6HNic was also increased, which compromised overall regulation performance. PNIC2d, containing 6 bp between the two ONIC modules supported optimal tightness among 2-ONIC-containing promoters while transgene expression reached 2-fold higher levels compared with PNIC1b (Figures 2 and 3). Direct correlation between the number of tandem operator modules and maximum expression levels is a common observation for transcription control modalities of the NICE type. Also, a qualitative correlation between maximum and leaky expression is frequently observed (47). Generic PNIC1 and PNIC2 promoters offer a wide portfolio of transgene regulation performance and provide a choice of specific expression/regulation characteristics, depending on the gene regulation system's mission.
Figure 3.
Regulation performance of 6HNic-responsive promoters (PNIC2) containing a twin 6HNic-dependent transactivator (NT)-specific operator module (ONIC1 and ONIC2). Whereas the distance of the most proximal ONIC1 to the minimal version of the human cytomegalovirus promoter (PhCMVmin) was kept constant, the spacing between ONIC2 and ONIC1 was increased by 2 bp increments (PNIC2a, ONIC2-0bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2b, ONIC2-2bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2c, ONIC2-4bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2d, ONIC2-6bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2e, ONIC2-8bp-ONIC1-NheI-SbfI-4bp-PhCMVmin; PNIC2f, ONIC2-10bp-ONIC1-NheI-SbfI-4bp-PhCMVmin). PNIC2a-f-driven SEAP expression units (PNIC2a-SEAP-pA [pLM135]; PNIC2b-SEAP-pA [pLM136]; PNIC2c-SEAP-pA [pLM137]; PNIC2d-SEAP-pA [pLM138]; PNIC2e-SEAP-pA [pLM139]; PNIC2f-SEAP-pA [pLM140]) were cotransfected with the NT1 expression vector pLM82 (PSV40-NT1-pA; NT1, HdnoR-VP16) into CHO-K1 and cultivated for 48 h in the presence (50 μg/ml 6HNic) and absence of 6HNic before SEAP production was quantified. SEAP production was compared with pSEAP2-Control-transfected CHO-K1 cells harboring a glycoprotein expression unit driven by the constitutive SV40 promoter (PSV40).
Engineering of different 6HNic-dependent transactivators
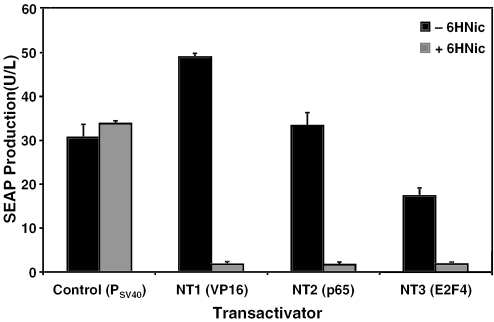
In addition to rigorous engineering of 6HNic-responsive promoters, we designed 6HNic-dependent transactivators containing alternative transactivation domains. We selected the potent transactivation domains of NF-κB (p65) and E2F4 (E2F4), which showed efficient transactivation in antibiotic-adjustable transgene regulation settings (47–51). VP16 of NT1 was replaced by p65 and E2F4 transactivation domains, which resulted in NT2 (HdnoR-p65) and NT3 (HdnoR-E2F4), respectively. The relative transactivation properties of NT1, NT2 and NT3 were assessed by cotransfection of either pLM82 (PSV40-NT1-pA), pLM101 (PSV40-NT2-pA) or pLM102 (PSV40-NT3-pA) and pLM104 (PNIC1b-SEAP-pA) into CHO-K1 followed by cultivation for 48 h in the presence and absence of 6HNic and SEAP quantification (Figure 4). Although all transactivators mediated comparable basal expression levels, their maximum transgene production levels differed significantly. NT2's transactivation efficiency was lower compared with NT1 but higher than NT3. Thus, 6HNic-dependent transactivators showed graded transcription-initiation capacity and provided a choice of different expression windows.
Figure 4.
Regulation performance of different 6HNic-dependent transactivators. A.nicotinovorans pAO1's 6HNic oxidase repressor HdnoR was fused to different transactivation domains derived from (i) H.simplex virus (VP16; NT1, HdnoR-VP16), (ii) human NF-κB (p65; NT2, HdnoR-p65) and (iii) human E2F4 (E2F4; NT3, HdnoR-E2F4). The regulation performance of NT1, NT2 and NT3 was assessed by cotransfection of either pLM82 (PSV40-NT1-pA), pLM101 (PSV40-NT2-pA) or pLM102 (PSV40-NT3-pA) and pLM104 (PNIC1b-SEAP-pA) into CHO-K1 followed by cultivation for 48 h in the presence and absence of 6HNic and SEAP quantification. SEAP production was compared with pSEAP2-Control-transfected CHO-K1 cells harboring a glycoprotein expression unit driven by the constitutive SV40 promoter (PSV40).
Development of 6HNic-adjustable lentivectors
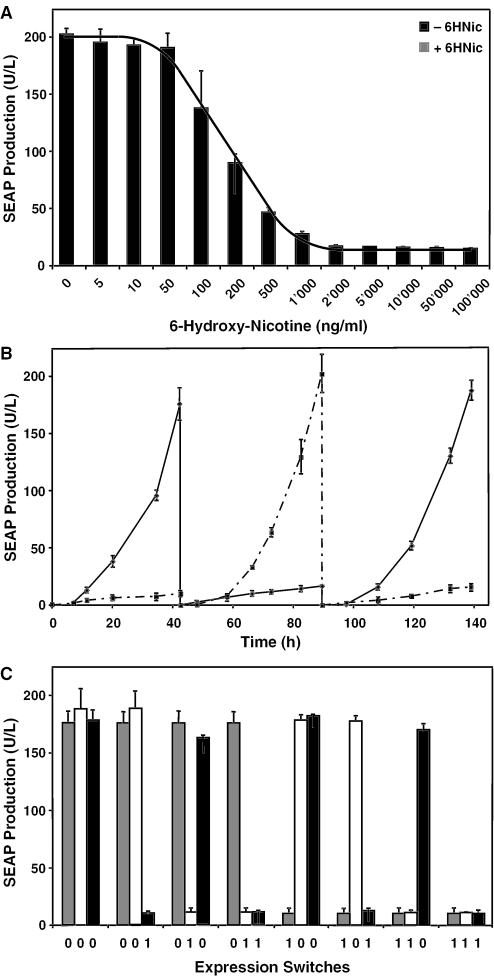
In order to enable straightforward one-step engineering of mammalian cells for NICE-controlled transgene expression, we have designed a set of HIV-1-derived self-inactivating lentiviral expression vectors exemplified by pLM141 (5′LTR-ψ+-oriSV40-PPT-RRE-PNIC2d-SEAP-3′LTRΔU3) and pLM103 (5′LTR-ψ+-oriSV40-PPT-RRE-PhEF1α-NT1-3′LTRΔU3). Cotransduction of pLM141- and pLM103-derived lentiviral particles into CHO-K1 followed by cultivation of engineered cell populations at increasing 6HNic concentrations resulted in precise dose-dependent SEAP expression fine-tuning (Figure 5A). Besides excellent adjustability, NICE-controlled SEAP production was fully reversible when followed over a 1 week period of 6HNic addition and removal alternating every 48 h. SEAP accumulation kinetics, maximum expression levels in the absence and basal expression levels in the presence of 50 μg/ml 6HNic remained reproducible following consecutive expression status switches (Figure 5B). Repeated ON–OFF or OFF–ON expression switching did neither compromise maximum nor basal expression levels.
Figure 5.
Adjustability, reversibility and expression imprinting of NICE-controlled transgene transduction using HIV-1-derived lentiviral particles. (A) 6HNic-adjustable SEAP expression of CHO-K1 cells cotransduced with pLM103- (5′LTR-ψ+-oriSV40-cPPT-RRE-PhEF1α-NT1-3′LTRΔU3) and pLM141- (5′LTR-ψ+-oriSV40-cPPT-RRE-PNIC2d-SEAP-3′LTRΔU3) derived lentiviral particles. Transduced cells were grown for 48 h in medium supplemented with increasing 6HNic concentrations prior to SEAP production profiling (The line was added for clarity). (B) Reversibility of NICE-controlled transgene transduction. Aforementioned CHO-K1 cell populations (40000 cells/ml) transduced with pLM103/141-derived lentiviral particles were cultivated in the presence and absence of 6HNic (50 μg/ml). The SEAP expression status (presence of 6HNic, OFF; absence of 6HNic, ON) was reversed and quantified on alternate days (48, 96 and 144 h) after culture medium exchanges. (C) Assessment of expression imprinting of NICE-controlled transgene transduction. CHO-K1 transduced for NICE-controlled SEAP expression were set for 48 h to high (0, −6HNic) or basal (1, +6HNic) glycoprotein production, which was then either maintained or switched twice during subsequent 48 h cultivation periods (48 h→48 h→48 h; 0→0→0, 0→0→1, 0→1→0, 0→1→1, 1→0→0, 1→0→1, 1→1→0, 1→1→1).
In parallel, we studied the impact of repressed or induced expression status imprinting on follow-up expression scenarios. CHO-K1 transduced for NICE-controlled SEAP expression were set for 48 h to high (0, −6HNic) or basal (1, +6HNic) glycoprotein production, which was either maintained or switched twice during subsequent 48 h cultivation periods (48 h→48 h→48 h; 0→0→0, 0→0→1, 0→1→0, 0→1→1, 1→0→0, 1→0→1, 1→1→0, 1→1→1). Irrespective of the cell population's previous NICE-controlled expression history, SEAP production levels in the presence or absence of 6HNic remained consistent (Figure 5C). Lentiviral implementation of NICE-controlled fine-tuning of transgene expression enabled stable, adjustable and reversible regulation characteristics as well as sustained regulation kinetics for over 6 days.
In vivo validation of NICE-adjustable transgene expression in mice and chicken embryos
For in vivo validation of NICE-adjustable transgene expression, we implanted microencapsulated CHO-K1 cells transgenic for NICE-controlled SEAP expression into coherent alginate-poly-l-lysine-alginate capsules intraperitoneally into mice (200 cells/capsule). Treated mice were exposed to different daily 6HNic doses, and serum SEAP levels were quantified 72 h after implantation. While NICE-induced SEAP production resulted in high-level concentrations of this glycoprotein in the serum of treated mice (106 ± 21 mU/l), 6HNic was unable to repress heterologous production even at concentrations of 100 mg/kg. 6HNic is a unique unphysiologic compound, which may fail to control NICE-driven gene expression due to unknown in vivo derivatization/degradation or rapid clearance from the body. Indeed, HPLC-based analysis of urine samples collected from 6HNic-treated mice showed that 90% of the administered nicotine derivative was excreted after 2 h (Figure 6). Therefore, further efforts will be required to establish NICE technology in mammals. However, the 6HNic concentration administered to mice without significant side effects remains impressive and holds promises for future applications.
Figure 6.
HPLC-based 6HNic analysis of mouse urine samples. (A) Chromatograms of a 6HNic standard (2.5 nmol, 0.45 μg; dotted line) and a urine sample collected from mice 2 h after intraperitoneal injection of 20 mg/kg 6HNic (solid line). (B) Comparative UV spectra of the peaks at 7.09 and 7.14 min [see (A)].
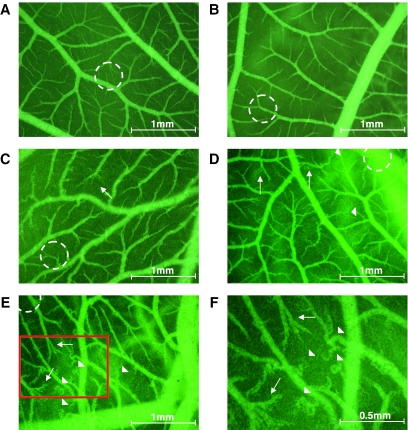
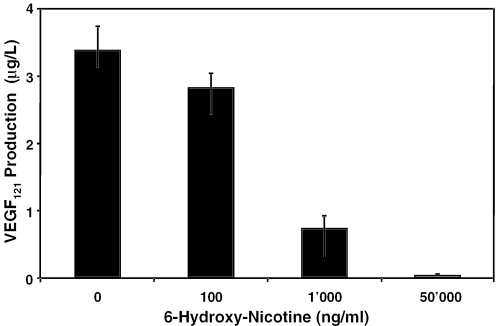
In order to alleviate possible 6HNic clearance, we cotransduced chicken embryos with lentiviral particles derived from pLM103- (5′LTR-ψ+-oriSV40-cPPT-RRE-PhEF1α-NT1-3′LTRΔU3) and pLM146- (5′LTR-ψ+-oriSV40-cPPT-RRE-PNIC3-VEGF121-3′LTRΔU3). The pLM146 lentivector harbors a PNIC3 (ONIC-4bp-NheI-SbfI-PhCMVmin)-driven human VEGF121 expression cassette to modulate the chicken embryo's vascularization in a 6HNic-adjustable manner (Figure 7). Cotransduction of these lentiviral particles into CHO-K1 followed by cultivation of infected cell populations at increasing 6HNic concentrations resulted in tight dose-dependent VEGF121 expression fine-tuning (Figure 8). Microscopic analysis of VEGF121-mediated neovascularization, as well as vessel morphology in the CAM of chicken embryos grown for 3 days at decreasing 6HNic concentrations or 6HNic-free conditions (50, 1, 0.1, 0 μg/ml, see Figure 7) showed a dose-dependent angiogenic response characterized by a general increase in blood vessel number, atypical (brush- and delta-like) endpoint patterns, irregular tortuous vessel shape and multiple vessel branching. All of those effects could be completely repressed by treating the chicken embryo with 50 μg/ml 6HNic. VEGF121-induced impact on vessel structure was confined to a 4 mm radius around the site of lentiviral particle application and could not be observed on the same CAM beyond this perimeter (Figure 7). These results confirm 6HNic-adjustable transgene transduction in vivo.
Figure 7.
In vivo microscopy of angiogenic response in the CAM of 12-day-old chicken embryos 72 h post cotransduction with lentiviral particles derived from pLM103- (5′LTR-ψ+-oriSV40-cPPT-RRE-PhEF1α-NT1-3′LTRΔU3) and pLM146- (5′LTR-ψ+-oriSV40-cPPT-RRE-PNIC3-VEGF121-3′LTRΔU3). Following administration of 50 μg/ml 6HNic 1 h post transduction, NICE-controlled VEGF121 production was completely repressed (A) and microvascular growth compared with mock-transduced 6HNic-treated control embryos (B). Transduced cultures treated with decreasing 6HNic concentrations showed an increasing dose-dependent angiogenic response with atypical (brush- and delta-like) endpoint patterns (arrows) and irregular tortuous vessel shape (arrowhead) within a perimeter of 4 mm of the transduction site (dashed circle). [(C) 1 μg/ml 6HNic; (D) 0.1 μg/ml 6HNic; (E) no 6HNic; see Figure 8 for VEGF121 expression profiles.] (F) Detail representation of the red-framed part of D.
Figure 8.
Dose–response characteristics of PNIC3-driven VEGF121 expression engineered into a lentiviral expression configuration. CHO-K1 cells were cotransduced with pLM103 lentiviral particles derived from (5′LTR-ψ+-oriSV40-cPPT-RRE-PhEF1α-NT1-3′LTRΔU3)- and pLM146 (5′LTR-ψ+-oriSV40-cPPT-RRE-PNIC3-VEGF121-3′LTRΔU3) and grown for 48 h in medium supplemented with increasing 6HNic concentrations before VEGF121 production was quantified.
DISCUSSION
Long appreciated in basic sciences for their power to reveal gene-function correlations, heterologous transgene control systems have gathered momentum and now stand on the eve of therapeutic and pilot production implementation (20,52). Currently available transcription control modalities capitalize on the generic design principle of the pioneering TET system: (i) clinically licensed tetracycline as inducer, (ii) bacterial repressor fused to mammalian cell-compatible transactivation domain as transactivator and (iii) transactivator-specific promoter assembled by fusing transactivator-specific operator modules to a minimal eukaryotic promoter. Besides their relationship to the TET configuration alternative transgene control systems differ significantly in the regulating small molecule (clinically licensed small-molecule drugs [antibiotics (7,9–11), immunosuppressive agents, (12), hormones and hormone agonists, (13–15), type-2 diabetes drug, (19,53), clinically inert compounds (17,18), temperature (16) and gaseous acetaldehyde (20)]), the origin of the transactivator [prokaryotic origin (7,9–11,16–18,20) (http://www.qbiogene.com/products/gene-expression/qmateslideshow/index.htm), mammalian origin (13,14,19,53)] and their promoter configurations [tandem operator modules, minimal promoter origin, relative promoter-operator spacing; see (40) as well as (52) for a non-limiting overview]. Despite the portfolio of different transgene regulation systems, there is nothing like the best control modality. All systems are associated with pros and cons:
clinically licensed inducers are intuitively better suited for therapeutic applications, although ongoing administration of small-molecule drugs is prone to side effects,
prokaryotic-derived promoter-transactivator systems operate in the absence of pleiotropies, yet may elicit immune responses,
eukaryotic-derived promoter-transactivator systems are compatible with the immune system, yet may interfere with endogenous regulatory networks.
The demands for transgene control modalities are as different as the systems themselves: while the gene therapy and tissue engineering communities prefer endogenous regulation systems adjusted by clinically licensed small-molecule drugs, biopharmaceutical manufacturing representatives favor heterologous prokaryotic-derived systems fine-tuned by clinically inert compounds.
Based on the components of A.nicotinovorans pAO1's nicotine mobilization machinery, we have designed a novel gene control system to adjust transgene expression in response to the water-soluble nicotine derivative 6HNic. The NICE technology mediated high-level expression, tight repression, precise expression fine-tuning and reversible transcription control in the absence of compromising imprinting originating from previous switching history. All of those performance characteristics have been confirmed in a variety of expression configurations in mammalian cells, by lentivirus-mediated transduction as well as in chicken embryos. As a closed system, chicken embryos supported adjustable angiogenic responses in vivo. However, 6HNic was unable to modulate SEAP expression in mice, suggesting that its high water solubility promoted rapid renal clearance and resulted in control-incompatible pharmacokinetics in whole animals.
Starting with a generic design concept, we have improved the NICE technology by (i) varying ONIC-PhCMVmin spacing, (ii) engineering tandem ONIC repeats and modifying the inter-ONIC distance and by (iii) swapping the transactivation domains fused to HdnoR. In order to selectively improve the regulation performance of hybrid NICE promoters, we focused on optimizing the spatio-steric relation between promoter, transactivator and transcription-initiation complex. The importance of rotational alignment of cis-acting elements on overall promoter performance has been investigated in several systems. The synthesis of those studies suggested appropriate helical phasing to be required for optimal promoter activity. Non-limiting examples included the promoter of the human serine protease B gene (54), the promoter of the Escherichia coli araBAD operon (55) and the promoter of the human HLA-DR gene (56). Furthermore, helical periodicity has been observed for promoter-enhancer crosstalk in SV40 and HIV-1 viruses (57,58). As for heterologous gene regulation systems, Weber and co-workers (47) have recently exemplified that the spacing and torsion angle between the operator and minimal promoters had dramatic impact on the overall regulation performance. This observation could be confirmed for the NICE system. Increasing the ONIC-PhCMVmin distance by 2 bp increments resulted in a promoter portfolio with graded response characteristics.
It is commonly accepted that transactivator-mediated transcription-initiation increases with the number of cognate operator modules (59,60). Similarly, NICE promoters harboring tandem operator modules showed 2- to 3-fold higher transgene expression compared with mono ONIC-containing promoters. The net increase of the maximum expression levels was a function of the inter-ONIC distance. The relative spacing between different operator modules has long been recognized as an essential factor for promoter performance (46,61–65). For example, analysis of beta interferon (IFN-β) gene activation showed that deletion, or rearrangement of any one of the enhancer modules compromises transcription performance (66). Also, variation of the spacing between individual activator binding sites within the TCR α enhancer modulated enhancer activity in a helical phasing-dependent manner (67). We have modified the inter-ONIC spacing by 2 bp increments while leaving the tandem operator-minimal promoter distance invariant. Optimal spacing resulted in up to 3-fold increased maximum expression levels. However, basal expression was higher as well.
Since the transactivator interfaces with promoter and transcription-initiation machinery it has a decisive impact on the overall regulation and expression performance of transgene control modalities. We have therefore equipped HdnoR with different viral and human transactivation domains, which mediated graded response characteristics in CHO-K1.
The NICE optimization studies have resulted in a portfolio of different 6HNic-responsive promoter/transactivator configurations, which show superior key characteristics including tight repression and maximum expression levels. The choice of different promoter/transactivator combinations enables unmatched adaptation of NICE-controlled transgene expression to specific needs: tight repression is best satisfied by mono-ONIC-containing promoters while promoters with tandem ONIC modules support high-level transgene expression. Our rigorous NICE analysis has exemplified its potential for sophisticated cell engineering related to basic and applied research applications.
Acknowledgments
The authors thank Roderich Brandsch for providing pH6EX3-HdnoR. This work was supported by the Swiss National Science Foundation (grant no. 631-065946) as well as the Swiss State Secretariat for Education and Research within EC Framework 6. Funding to pay the Open Access publication charges for this article was provided by ETH Zurich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sandu C., Chiribau C.B., Brandsch R. Characterization of HdnoR, the transcriptional repressor of the 6-hydroxy-d-nicotine oxidase gene of Arthrobacter nicotinovorans pAO1, and its DNA-binding activity in response to l- and d-nicotine derivatives. J. Biol. Chem. 2003;278:51307–51315. doi: 10.1074/jbc.M307797200. [DOI] [PubMed] [Google Scholar]
- 2.Schmid A., Dordick J.S., Hauer B., Kiener A., Wubbolts M., Witholt B. Industrial biocatalysis today and tomorrow. Nature. 2001;409:258–268. doi: 10.1038/35051736. [DOI] [PubMed] [Google Scholar]
- 3.Brenik W., Rudhard H. US4343318. Patent. 1982
- 4.Gravely L.E., Geiss V.L., Gregory C.F. US4557280. United States Patent. 1985
- 5.Yildiz D. Nicotine, its metabolism and an overview of its biological effects. Toxicon. 2004;43:619–632. doi: 10.1016/j.toxicon.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Hillen W., Klock G., Kaffenberger I., Wray L.V., Reznikoff W.S. Purification of the TET repressor and TET operator from the transposon Tn10 and characterization of their interaction. J. Biol. Chem. 1982;257:6605–6613. [PubMed] [Google Scholar]
- 7.Gossen M., Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gossen M., Freundlieb S., Bender G., Muller G., Hillen W., Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 9.Fussenegger M., Morris R.P., Fux C., Rimann M., von Stockar B., Thompson C.J., Bailey J.E. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 2000;18:1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 10.Weber W., Fux C., Daoud-el Baba M., Keller B., Weber C.C., Kramer B.P., Heinzen C., Aubel D., Bailey J.E., Fussenegger M. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 2002;20:901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 11.Zhao H.F., Boyd J., Jolicoeur N., Shen S.H. A coumermycin/novobiocin-regulated gene expression system. Hum. Gene Ther. 2003;14:1619–1629. doi: 10.1089/104303403322542266. [DOI] [PubMed] [Google Scholar]
- 12.Rivera V.M., Clackson T., Natesan S., Pollock R., Amara J.F., Keenan T., Magari S.R., Phillips T., Courage N.L., Cerasoli F., Jr, et al. A humanized system for pharmacologic control of gene expression. Nature Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 13.Braselmann S., Graninger P., Busslinger M. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. Proc. Natl Acad. Sci. USA. 1993;90:1657–1661. doi: 10.1073/pnas.90.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beerli R.R., Schopfer U., Dreier B., Barbas C.F., III Chemically regulated zinc finger transcription factors. J. Biol. Chem. 2000;275:32617–32627. doi: 10.1074/jbc.M005108200. [DOI] [PubMed] [Google Scholar]
- 15.No D., Yao T.P., Evans R.M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber W., Marty R.R., Link N., Ehrbar M., Keller B., Weber C.C., Zisch A.H., Heinzen C., Djonov V., Fussenegger M. Conditional human VEGF-mediated vascularization in chicken embryos using a novel temperature-inducible gene regulation (TIGR) system. Nucleic Acids Res. 2003;31:e69. doi: 10.1093/nar/gng069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber W., Schoenmakers R., Spielmann M., El-Baba M.D., Folcher M., Keller B., Weber C.C., Link N., van de Wetering P., Heinzen C., et al. Streptomyces-derived quorum-sensing systems engineered for adjustable transgene expression in mammalian cells and mice. Nucleic Acids Res. 2003;31:e71. doi: 10.1093/nar/gng071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber W., Malphettes L., de Jesus M., Schoenmakers R., El-Baba M.D., Spielmann M., Keller B., Weber C.C., van de Wetering P., Aubel D., et al. Engineered Streptomyces quorum-sensing components enable inducible siRNA-mediated translation control in mammalian cells and adjustable transcription control in mice. J. Gene Med. 2004;7:518–525. doi: 10.1002/jgm.682. [DOI] [PubMed] [Google Scholar]
- 19.Tascou S., Sorensen T.K., Glenat V., Wang M., Lakich M.M., Darteil R., Vigne E., Thuillier V. Stringent rosiglitazone-dependent gene switch in muscle cells without effect on myogenic differentiation. Mol. Ther. 2004;9:637–649. doi: 10.1016/j.ymthe.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Weber W., Rimann M., Spielmann M., Keller B., Daoud-El Baba M., Aubel D., Weber C.C., Fussenegger M. Gas-inducible transgene expression in mammalian cells and mice. Nat. Biotechnol. 2004;22:1440–1444. doi: 10.1038/nbt1021. [DOI] [PubMed] [Google Scholar]
- 21.Beck C., Uramoto H., Boren J., Akyurek L.M. Tissue-specific targeting for cardiovascular gene transfer. Potential vectors and future challenges. Curr. Gene Ther. 2004;4:457–467. doi: 10.2174/1566523043346138. [DOI] [PubMed] [Google Scholar]
- 22.Aubel D., Morris R., Lennon B., Rimann M., Kaufmann H., Folcher M., Bailey J.E., Thompson C.J., Fussenegger M. Design of a novel mammalian screening system for the detection of bioavailable, non-cytotoxic streptogramin antibiotics. J. Antibiot. (Tokyo) 2001;54:44–55. doi: 10.7164/antibiotics.54.44. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Nicolini V., Fux C., Fussenegger M. A novel mammalian cell-based approach for the discovery of anticancer drugs with reduced cytotoxicity on non-dividing cells. Invest. New Drugs. 2004;22:253–262. doi: 10.1023/B:DRUG.0000026251.00854.77. [DOI] [PubMed] [Google Scholar]
- 24.Umaña P., Jean-Mairet J., Bailey J.E. Tetracycline-regulated overexpression of glycosyltransferases in Chinese hamster ovary cells. Biotechnol. Bioeng. 1999;65:542–549. [PubMed] [Google Scholar]
- 25.Fussenegger M., Schlatter S., Datwyler D., Mazur X., Bailey J.E. Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nat. Biotechnol. 1998;16:468–472. doi: 10.1038/nbt0598-468. [DOI] [PubMed] [Google Scholar]
- 26.Malleret G., Haditsch U., Genoux D., Jones M.W., Bliss T.V., Vanhoose A.M., Weitlauf C., Kandel E.R., Winder D.G., Mansuy I.M. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- 27.Cohlan S.Q. Tetracycline staining of teeth. Teratology. 1977;15:127–129. doi: 10.1002/tera.1420150117. [DOI] [PubMed] [Google Scholar]
- 28.Lautermann J., Dehne N., Schacht J., Jahnke K. [Aminoglycoside- and cisplatin-ototoxicity: from basic science to clinics.] Laryngorhinootologie. 2004;83:317–323. doi: 10.1055/s-2004-814280. [DOI] [PubMed] [Google Scholar]
- 29.Sartor O., Cutler G.B., Jr Mifepristone: treatment of Cushing's syndrome. Clin. Obstet Gynecol. 1996;39:506–510. doi: 10.1097/00003081-199606000-00024. [DOI] [PubMed] [Google Scholar]
- 30.Kramer B.P., Weber W., Fussenegger M. Artificial regulatory networks and cascades for discrete multilevel transgene control in mammalian cells. Biotechnol. Bioeng. 2003;83:810–820. doi: 10.1002/bit.10731. [DOI] [PubMed] [Google Scholar]
- 31.Kramer B.P., Viretta A.U., Daoud-El-Baba M., Aubel D., Weber W., Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 32.Kramer B.P., Fischer C., Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 33.Mitta B., Rimann M., Ehrengruber M.U., Ehrbar M., Djonov V., Kelm J., Fussenegger M. Advanced modular self-inactivating lentiviral expression vectors for multigene interventions in mammalian cells and in vivo transduction. Nucleic Acids Res. 2002;30:e113. doi: 10.1093/nar/gnf112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiser J., Harmison G., Kluepfel-Stahl S., Brady R.O., Karlsson S., Schubert M. Transduction of nondividing cells using pseudotyped defective high-titer HIV type 1 particles. Proc. Natl Acad. Sci. USA. 1996;93:15266–15271. doi: 10.1073/pnas.93.26.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mochizuki H., Schwartz J.P., Tanaka K., Brady R.O., Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J. Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djonov V., Schmid M., Tschanz S.A., Burri P.H. Intussusceptive angiogenesis: its role in embryonic vascular network formation. Circ. Res. 2000;86:286–292. doi: 10.1161/01.res.86.3.286. [DOI] [PubMed] [Google Scholar]
- 37.Djonov V.G., Galli A.B., Burri P.H. Intussusceptive arborization contributes to vascular tree formation in the chick chorio-allantoic membrane. Anat. Embryol. (Berl.) 2000;202:347–357. doi: 10.1007/s004290000126. [DOI] [PubMed] [Google Scholar]
- 38.Herrera F.J., Triezenberg S.J. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 2004;78:9689–9696. doi: 10.1128/JVI.78.18.9689-9696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boshart M., Weber F., Jahn G., Dorsch-Hasler K., Fleckenstein B., Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 40.Weber W., Fussenegger M. Artificial mammalian gene regulation networks-novel approaches for gene therapy and bioengineering. J. Biotechnol. 2002;98:161–187. doi: 10.1016/s0168-1656(02)00130-x. [DOI] [PubMed] [Google Scholar]
- 41.Semsey S., Geanacopoulos M., Lewis D.E., Adhya S. Operator-bound GalR dimers close DNA loops by direct interaction: tetramerization and inducer binding. EMBO J. 2002;21:4349–4356. doi: 10.1093/emboj/cdf431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sathya G., Li W., Klinge C.M., Anolik J.H., Hilf R., Bambara R.A. Effects of multiple estrogen responsive elements, their spacing, and location on estrogen response of reporter genes. Mol. Endocrinol. 1997;11:1994–2003. doi: 10.1210/mend.11.13.0039. [DOI] [PubMed] [Google Scholar]
- 43.Schreiter E.R., Sintchak M.D., Guo Y., Chivers P.T., Sauer R.T., Drennan C.L. Crystal structure of the nickel-responsive transcription factor NikR. Nature Struct. Biol. 2003;10:794–799. doi: 10.1038/nsb985. [DOI] [PubMed] [Google Scholar]
- 44.Parkhill J., Brown N.L. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 1990;18:5157–5162. doi: 10.1093/nar/18.17.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown N.L., Stoyanov J.V., Kidd S.P., Hobman J.L. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 46.Lewis D.E., Adhya S. In vitro repression of the gal promoters by GalR and HU depends on the proper helical phasing of the two operators. J. Biol. Chem. 2002;277:2498–2504. doi: 10.1074/jbc.M108456200. [DOI] [PubMed] [Google Scholar]
- 47.Weber W., Kramer B.P., Fux C., Keller B., Fussenegger M. Novel promoter/transactivator configurations for macrolide- and streptogramin-responsive transgene expression in mammalian cells. J. Gene Med. 2002;4:676–686. doi: 10.1002/jgm.314. [DOI] [PubMed] [Google Scholar]
- 48.Schmitz M.L., Stelzer G., Altmann H., Meisterernst M., Baeuerle P.A. Interaction of the COOH-terminal transactivation domain of p65 NF-kappa B with TATA-binding protein, transcription factor IIB, and coactivators. J. Biol. Chem. 1995;270:7219–7226. doi: 10.1074/jbc.270.13.7219. [DOI] [PubMed] [Google Scholar]
- 49.Urlinger S., Helbl V., Guthmann J., Pook E., Grimm S., Hillen W. The p65 domain from NF-kappaB is an efficient human activator in the tetracycline-regulatable gene expression system. Gene. 2000;247:103–110. doi: 10.1016/s0378-1119(00)00112-8. [DOI] [PubMed] [Google Scholar]
- 50.Akagi K., Kanai M., Saya H., Kozu T., Berns A. A novel tetracycline-dependent transactivator with E2F4 transcriptional activation domain. Nucleic Acids Res. 2001;29:E23. doi: 10.1093/nar/29.4.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baron U., Gossen M., Bujard H. Tetracycline-controlled transcription in eukaryotes: novel transactivators with graded transactivation potential. Nucleic Acids Res. 1997;25:2723–2729. doi: 10.1093/nar/25.14.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber W., Fussenegger M. Inducible gene expression in mammalian cells and mice. Methods Mol. Biol. 2004;267:451–466. doi: 10.1385/1-59259-774-2:451. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y., O'Malley B.W., Jr, Tsai S.Y., O'Malley B.W. A regulatory system for use in gene transfer. Proc. Natl Acad. Sci. USA. 1994;91:8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanson R.D., Grisolano J.L., Ley T.J. Consensus AP-1 and CRE motifs upstream from the human cytotoxic serine protease B (CSP-B/CGL-1) gene synergize to activate transcription. Blood. 1993;82:2749–2757. [PubMed] [Google Scholar]
- 55.Dunn T.M., Hahn S., Ogden S., Schleif R.F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc. Natl Acad. Sci. USA. 1984;81:5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vilen B.J., Cogswell J.P., Ting J.P. Stereospecific alignment of the X and Y elements is required for major histocompatibility complex class II DRA promoter function. Mol. Cell Biol. 1991;11:2406–2415. doi: 10.1128/mcb.11.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi K., Vigneron M., Matthes H., Wildeman A., Zenke M., Chambon P. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986;319:121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- 58.Rao E., Dang W., Tian G., Sen R. A three-protein-DNA complex on a B cell-specific domain of the immunoglobulin mu heavy chain gene enhancer. J. Biol. Chem. 1997;272:6722–6732. doi: 10.1074/jbc.272.10.6722. [DOI] [PubMed] [Google Scholar]
- 59.Ellwood K., Huang W., Johnson R., Carey M. Multiple layers of cooperativity regulate enhanceosome-responsive RNA polymerase II transcription complex assembly. Mol. Cell Biol. 1999;19:2613–2623. doi: 10.1128/mcb.19.4.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J., Ellwood K., Lehman A., Carey M.F., She Z.S. A mathematical model for synergistic eukaryotic gene activation. J. Mol. Biol. 1999;286:315–325. doi: 10.1006/jmbi.1998.2489. [DOI] [PubMed] [Google Scholar]
- 61.Hebner C., Lasanen J., Battle S., Aiyar A. The spacing between adjacent binding sites in the family of repeats affects the functions of Epstein-Barr nuclear antigen 1 in transcription activation and stable plasmid maintenance. Virology. 2003;311:263–274. doi: 10.1016/s0042-6822(03)00122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nikolajczyk B.S., Nelsen B., Sen R. Precise alignment of sites required for mu enhancer activation in B cells. Mol. Cell Biol. 1996;16:4544–4554. doi: 10.1128/mcb.16.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong G., Maser E. Regulation of the steroid-inducible 3alpha-hydroxysteroid dehydrogenase/carbonyl reductase gene in Comamonas testosteroni. J. Biol. Chem. 2001;276:9961–9970. doi: 10.1074/jbc.M010962200. [DOI] [PubMed] [Google Scholar]
- 64.Chi T., Lieberman P., Ellwood K., Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- 65.Lederer H., Tovar K., Baer G., May R.P., Hillen W., Heumann H. The quaternary structure of Tet repressors bound to the Tn10-encoded tet gene control region determined by neutron solution scattering. EMBO J. 1989;8:1257–1263. doi: 10.1002/j.1460-2075.1989.tb03499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merika M., Williams A.J., Chen G., Collins T., Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 67.Giese K., Kingsley C., Kirshner J.R., Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein–protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 68.Mitta B., Weber C.C., Rimann M., Fussenegger M. Design and in vivo characterization of self-inactivating human and non-human lentiviral expression vectors engineered for streptogramin-adjustable transgene expression. Nucleic Acids Res. 2004;32:e106. doi: 10.1093/nar/gnh104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fussenegger M., Moser S., Mazur X., Bailey J.E. Autoregulated multicistronic expression vectors provide one-step cloning of regulated product gene expression in mammalian cells. Biotechnol. Prog. 1997;13:733–740. doi: 10.1021/bp970108r. [DOI] [PubMed] [Google Scholar]
- 70.Weber W., Marty R.R., Keller B., Rimann M., Kramer B.P., Fussenegger M. Versatile macrolide-responsive mammalian expression vectors for multiregulated multigene metabolic engineering. Biotechnol. Bioeng. 2002;80:691–705. doi: 10.1002/bit.10461. [DOI] [PubMed] [Google Scholar]