Abstract
Although some mitochondrial, X chromosome, and autosomal sequence diversity data are available for our closest relatives, Pan troglodytes and Pan paniscus, data from the nonrecombining portion of the Y chromosome (NRY) are more limited. We examined ≈3 kb of NRY DNA from 101 chimpanzees, seven bonobos, and 42 humans to investigate: (i) relative levels of intraspecific diversity; (ii) the degree of paternal lineage sorting among species and subspecies of the genus Pan; and (iii) the date of the chimpanzee/bonobo divergence. We identified 10 informative sequence-tagged sites associated with 23 polymorphisms on the NRY from the genus Pan. Nucleotide diversity was significantly higher on the NRY of chimpanzees and bonobos than on the human NRY. Similar to mtDNA, but unlike X-linked and autosomal loci, lineages defined by mutations on the NRY were not shared among subspecies of P. troglodytes. Comparisons with mtDNA ND2 sequences from some of the same individuals revealed a larger female versus male effective population size for chimpanzees. The NRY-based divergence time between chimpanzees and bonobos was estimated at ≈1.8 million years ago. In contrast to human populations who appear to have had a low effective size and a recent origin with subsequent population growth, some taxa within the genus Pan may be characterized by large populations of relatively constant size, more ancient origins, and high levels of subdivision.
A great deal has been learned about the evolutionary history of humans and great apes through comparisons of homologous DNA sequences. Humans are thought to be most closely related to chimpanzees (Pan troglodytes) and bonobos (Pan paniscus), with whom we share ≈99% of our genomic sequence (1). Much is also being learned about levels of genetic diversity within humans through comparisons of allelic variation at different loci in the human genome (2). Studies of intraspecific diversity in our closest living relatives, however, have lagged far behind those of humans. Comparisons of levels of intraspecific variability have important implications for understanding the processes involved in speciation and clarifying the demographic history of contemporary populations of humans and great apes. Because chimpanzees and bonobos have such a poor fossil record, genetic data can play a major role in elucidating the evolutionary history of the genus Pan.
Currently, there are comparative human-chimpanzee sequence data for loci representing three of the four compartments of the genome: the autosomes, the X chromosome, and mtDNA. Comparisons of mitochondrial sequence variation in chimpanzees, bonobos, and humans are consistent in showing 2- to 4-fold higher levels of diversity in both Pan species than in humans (3–6). Studies of autosomal and X-chromosome loci have generally shown higher levels of nucleotide variation in Pan, with some exceptions. In a study of a ≈1-kb region of the HOXB6 gene from 105 humans, 45 chimpanzees, and 19 bonobos, Deinard and Kidd (7) found more than three times as much nucleotide diversity in both species of Pan than in humans. Adams et al. (8) found that chimpanzees have greater diversity than humans at two of the three highly polymorphic MHC class I genes (HLA-B and HLA-C). However, the third locus was less diverse in chimpanzees, which led to the suggestion that HLA-A diversity patterns were influenced by differential pathogen-mediated selection in the human and chimpanzee lines. An investigation of X-chromosome sequence variation in Homo and Pan indicated that chimpanzees have twice the diversity found in humans, whereas bonobos have less diversity than humans (9). Results of these and other studies have led to the suggestion of a smaller effective population size for humans, perhaps as the result of a severe bottleneck followed by an expansion (7–11). On the other hand, based on their finding of lower levels of autosomal short tandem repeat diversity in P. troglodytes compared with humans and based on the earlier protein studies of King and Wilson (12), Wise et al. (5) have argued that nuclear diversity in chimpanzees reflects a lower effective population size, and that reduced mtDNA diversity in humans may be explained by directional and/or background selection.
Although studies of human nonrecombining portion of the Y chromosome (NRY) sequence variation have intensified over the last few years (13–17), comparative data from the genus Pan have been very limited. Hammer (13) sequenced 2.6-kb within the YAP region on the long arm of the NRY from four chimpanzees and found no variation. Burrows and Ryder (18) sequenced the 729-bp third intron of ZFY in a chimpanzee and a bonobo and found only a single base substitution. They suggested that the lack of intraspecific variation in this intron in humans and great apes is caused by selection acting at the ZFY gene or at a linked site on the NRY. Deinhard and Kidd (19) examined a 940-bp region of the pseudoautosomal boundary region and found three haplotypes in 19 chimpanzees and two haplotypes in 11 bonobos.
Motivated by the need for more comprehensive surveys of NRY diversity in the genus Pan, we examined a total of 2,787 bp on the NRY of 101 chimpanzees, seven bonobos, and 42 humans. The purpose of this survey was 2-fold. First, we wanted to test levels of Homo–Pan diversity from the perspective of the fourth compartment of the genome (i.e., the NRY). Under a model of larger chimpanzee/bonobo effective population sizes, we would expect to find higher levels of NRY diversity in populations of Pan compared with humans. On the other hand, if selection was limiting variation on the NRY, we might expect to find low levels of nucleotide diversity on the NRY in populations of Homo and Pan species. Second, we wanted to evaluate whether different male versus female dispersal patterns are responsible for contrasting patterns of lineage sorting in mitochondrial and nuclear DNA lineages. The latter observations stem from research begun by Morin et al. (20) on mtDNA variation in the three currently recognized subspecies of chimpanzees: P. t. troglodytes, P. t. schweinfurthii, and P. t. verus. They demonstrated that the three chimpanzee subspecies are characterized by distinct clades of mtDNA hypervariable region I (HVI) haplotypes (3, 20). In contrast, haplotypes based on sequences from a 10-kb region on Xq13.3 (9), and the autosomal genes APOB and HOXB6 (19, 21), do not show distinct clades defining chimpanzee subspecies, and lineages are often shared between subspecies. By surveying variation on the NRY in these subspecies, we wanted to test whether the phylogenetic structure indicated by mtDNA data reflects a model in which there has been ongoing male gene flow or a model where subspecies have formed recently without subsequent gene flow.
Materials and Methods
DNA Samples.
The three subspecies of chimpanzees (i.e., P. t. verus, P. t. troglodytes, and P. t. schweinfurthii) are found in western, central, and eastern Africa, respectively. Although physical differences have been reported among the subspecies, the phenotypic variation among individuals precludes using appearance for accurate subspecies identification (22). Most chimpanzees in this sample were exported from Africa during the 15 years before the Convention on International Trade in Endangered Species of Wild Fauna and Flora enacted in 1975 and are of unknown subspecies status. It is also typically unknown where in Africa a particular chimpanzee was captured. Pan DNA samples were obtained from blood taken during routine veterinary exams, from cheek swabs acquired by sanctuary or zoo personnel, or from purified DNA from other laboratories. Pedigree information for each individual enabled us to exclude paternally related males. The majority of the P. troglodytes samples (64 of 101) were from wild-born individuals, with 17 of known subspecies. Therefore, mtDNA HVI sequencing was performed (see below) to determine the maternal origin of each wild-born sample. Human samples were selected from the Y Chromosome Consortium cell line panel to maximize diversity. The samples included 10 Africans, 11 Asians, nine Europeans, four Oceanians, and eight Native Americans. We also examined variation in one Gorilla gorilla gorilla male.
PCR and Mutation Detection.
Sequence tagged site (STSs) on the NRY (i.e., sY15, sY19, sY65, sY67, sY74, sY84, sY85, sY123, and sY126) were amplified by using primers from Vollrath et al. (23). SMCY gene primers (U1241 5′-TGGGGATGAAGATAATAAG-3′ and L1430 5′-AGCCATTTCACCAAAACTC-3′) were designed based on the published sequence (24). Of the 16 STSs originally chosen, six were dropped from subsequent analyses because either they would not amplify in Pan (sY130), they amplified in both males and females (sY17, sY68, and sY95), or they were found to be duplicated (sY102 and sY108). Mutation detection analysis was performed by using denaturing HPLC (DHPLC) (15). PCR and DHPLC conditions for the 10 informative STSs are in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. DNA sequencing was performed by standard procedures to identify mutations that caused shifted DHPLC chromatograms. For the NRY analyses, seven individuals (one H. sapiens, one P. paniscus, one P. t. verus, two P. t. schweinfurthii, and two P. t. troglodytes) were routinely sequenced for each marker in addition to the individuals recognized as polymorphic by DHPLC analysis. No discrepancies between DHPLC and sequencing results were found. mtDNA HVI sequence data were obtained (data not shown) from the wild-born males, nine captive-born males, and the wild-born purported fathers of two captive-born males. These data were compared with published sequences (3, 25, 26) to identify subspecies status. For subsequent population genetic analyses, we obtained mtDNA ND2 sequence data from four P. t. verus, 16 P. t. troglodytes, five P. t. schweinfurthii, and four P. paniscus (all of whom were from maternally unrelated individuals), as well as from the literature (5, 27). mtDNA HVI and ND2 sequences were amplified as described in Morin et al. (3) and Wise et al. (5).
Data Analysis.
For each individual, the combination of point mutations observed across the 10 STSs is referred to as a NRY haplogroup. Two measures of nucleotide diversity were calculated for each locus: (i) π is based on the average number of nucleotide differences per site between two sequences randomly drawn from a sample; and (ii) θ (28) is based on the sample size-corrected proportion of segregating sites (29). The Jukes and Cantor correction was applied to all sequence comparisons involving interspecific variation and divergence (30). Under equilibrium conditions with respect to mutation and drift, both π and θ estimate the neutral parameters: 4Neμ for autosomal loci, 3Neμ for X-chromosome loci, and 2Neμ for NRY and mtDNA loci, where Ne is the effective population size and μ is the neutral mutation rate.
Mutation rates were calculated from the data as follows: mutation rate per site per year = k/2Tsplitl, where k is the average number of substitutions, l is the length of the sequence, and Tsplit is the time in years since the human and chimpanzee (or bonobo) divergence, which is assumed to be 6 million years ago. Long-term effective population sizes (Ne) were estimated for the NRY, mtDNA (ND2 synonymous sites), Xq13.3, and HOXB6 data using mutation rates of 1.08 × 10−9, 3.49 × 10−8, 7.42 × 10−10, and 5.62 × 10−10 per site per year, respectively. For all calculations, generation times were 15 years in chimpanzees and bonobos and 20 years in humans (31). Tajima's D statistic was calculated to test for deviations from neutral frequency distribution (32). Ratios of polymorphism to divergence were compared with expectations under a neutral model using the Hudson Kreitman Aguadé test (33). These tests take into consideration differences in Ne for the NRY/mtDNA, X chromosome, and autosomal loci and assume a sex ratio of one. The measures of diversity and tests of neutrality were performed with the program dnasp (34). An analysis of molecular variance (35) was used to determine the significance of the differences among samples, using the computer package ARLEQUIN 2.000 (36).
Times to the most recent common ancestor (TMRCAs) were estimated by using the program genetree (37, 38). TMRCAs and split times also were estimated by using a program written by R. C. Griffiths called SPLIT_TIME. Mutation rates and split times were estimated based on posterior means obtained from a Bayesian analysis of the data, which was comprised of the numbers of mutations observed in the different regions of the gene tree. A Markov Chain Monte Carlo computation was then carried out to find the mean and standard deviations of the MRCA times in the gene tree, conditional on the position of the mutations in the tree. Uniform priors for the mutation rate (2.0 × 10−6 to 5.0 × 10−6 per sequence per year) and the Homo-Pan split time (5–7 million years ago) were used. Different prior distributions (0.5 to 3.0 million years) including uniform, normal-shaped, and gamma-shaped were tried for the chimpanzee-bonobo split time. Indels were counted as mutations in the analysis and hypervariable sites were removed. In the SPLIT_TIME analyses, we also assumed that (i) the male effective sizes are 20,000 for chimpanzees and bonobos and 5,000 for humans; (ii) the Nem of all ancestral populations are 20,000; (iii) the generation times for chimpanzees, bonobos, and humans are 15 years, 15 years, and 20 years, respectively; and (iv) the generation times for all common ancestral populations were 15 years.
Results
NRY Haplogroups in the Genus Pan.
A total of 98 mutations were discovered on the Y chromosomes of 101 chimpanzees, seven bonobos, 42 humans, and one gorilla (see Table 3, which is published as supporting information on the PNAS web site). Within P. troglodytes, there were a total of 19 polymorphisms in 2,787 bp. Of these, three were insertion/deletions (indels), eight were transversions, and eight were transitions. Within P. paniscus, one transition and three transversions were identified. No polymorphic sites were found in humans.
Variation at all sites gave rise to 10 haplogroups in chimpanzees and three in bonobos. Haplogroups were named according to the subspecies in which they were found. We inferred that there were two haplogroups within P. t. verus (Ptv1 and Ptv2), which differ only by a single-base indel. Of the 77 individuals reported as P. t. verus, 47 were wild-born (46 had Ptv1 and one had Ptv2). All but one of these individuals had mtDNA HVI sequences characteristic of P. t. verus (3). The discrepant individual had NRY haplogroup Ptv1 and a HVI sequence related to that of the recently proposed subspecies, P. t. vellerosus (26). We also genotyped five Y-linked short tandem repeats (DYS388, DYS390, DYS391, DYS392, and DYS393) in a sample of 20 or more P. t. verus and found relatively high levels of diversity, confirming that most of these individuals are, indeed, from independent male lineages (data not shown).
Two captive-born males had NRY haplogroup Ptt1 (P. t. troglodytes haplogroup 1). We were able to obtain serum from the wild-born father of one of these individuals and found that his HVI sequence clustered with those of P. t. troglodytes. Haplogroups Ptt2, Ptt3, Ptt4, and Ptt5 (P. t. troglodytes haplogroups 2–5) were found in three, six, two, and one wild-born individuals, respectively. Four wild-born individuals (one from Gombe and three confiscated in Tanzania) had the Pts1 haplogroup (P. t. schweinfurthii haplogroup 1).
The final two Pan troglodytes haplogroups, Pt?1 and Pt?2, were from captive-born individuals for whom pedigree information was either inaccurate or does not provide subspecies information. Unfortunately, the wild-born fathers (or grandfathers) of these individuals are no longer living so subspecies status could not be inferred directly. Two individuals currently residing in an European zoo had the divergent NRY haplogroup Pt?1. Given the large number of P. t. verus Y chromosomes sampled here, and the larger importation of P. t. troglodytes to Europe compared with the United States, we inferred that this haplogroup is characteristic of P. t. troglodytes (although further testing is necessary to confirm this idea). As a result, subsequent analyses were performed both with and without this haplogroup as P. t. troglodytes. NRY haplogroup Pt?2 was closely related to Pts1. We inferred that this individual is P. t. schweinfurthii and included this individual as such in most analyses.
Phylogenetic Tree of NRY Haplogroups.
In the gorilla, seven of the 10 loci were successfully amplified (i.e., all except sY65, sY67, and sY84). A maximum parsimony tree was reconstructed with 87 steps (not including indels) or 99 steps (including indels), both with a consistency index of 1.0. There was only a single homoplasy: a C > G transversion and a C > T transition occurred at the same site on the lineages leading to haplogroup Pt?1 and to the common ancestor of haplogroups Pp2 and Pp3, respectively. The topology of the tree shows humans and chimpanzees to be more closely related to each other than to the gorilla (Fig. 1). A clear separation between chimpanzees and bonobos is also indicated.
Figure 1.
NRY maximum parsimony haplogroup tree. Haplogroup code names are defined in Results. Color coding refers to taxonomic status within Pan. Numbers associated with branches correspond to the number of point mutational events followed by the number of indels in parentheses. Indels were also treated as mutational events in a branch and bound search analysis (55). Dotted lines denote uncertainty associated with Pt?1 and Pt?2.
Within P. troglodytes, there were four subclades defined by six synapomorphic sites: one specific to P. t. verus, one specific to P. t. schweinfurthii, and two within P. t. troglodytes (Ptt1/2; Ptt4/5) (Fig. 1). Haplogroup Ptt3 was the most basal haplogroup within P. troglodytes (i.e., a zero-length branch attached to the base of the P. t. troglodytes clade), whereas the lineage leading to the Pt?1 haplogroup had the longest branch on the Pan portion of the tree (i.e., with six base substitutions, a single-base deletion, and a two-base insertion).
Nucleotide Diversity.
Two measures of nucleotide diversity were used to compare levels of NRY variation in chimpanzees, bonobos, and humans (Table 1). With respect to both measures, chimpanzees and bonobos are considerably more diverse than humans. Despite very different sample sizes, estimates of nucleotide diversity in chimpanzees and bonobos were similar. Within chimpanzees, P. t. troglodytes exhibited the most variation, followed by P. t. schweinfurthii, and P. t. verus. This rank order was the same when the two individuals with divergent haplogroup Pt?1 were not included in the analysis. The estimate of the male long-term effective population size (Nem) for chimpanzees was 21,000, whereas the bonobo estimate was 24,000. In comparison, the female long-term effective population sizes (Nef), estimated by using synonymous sites from the mtDNA ND2 gene, were higher (≈41,000 and 28,000 for chimpanzees and bonobos, respectively). These estimates should be considered with caution as they assume that chimpanzees form a randomly mating population (see below).
Table 1.
NRY, mtDNA, and X chromosome diversity statistics for chimpanzees, bonobos, and humans
| n | s | Indels | π, % | θ, %* | Ne† | Tajima's D, P value | Reference | |
|---|---|---|---|---|---|---|---|---|
| NRY | ||||||||
| P.t.s. (+Pt?2) | 6 | 2 | 0 | 0.024 | 0.032 | 7,000 | −1.13 (>0.10) | This study |
| P.t.s. (−Pt?2) | 5 | 0 | 0 | 0.000 | 0.000 | This study | ||
| P.t.t. (+Pt?1) | 18 | 12 | 2 | 0.092 | 0.105 | 28,000 | −0.42 (>0.10) | This study |
| P.t.t. (−Pt?1) | 16 | 4 | 0 | 0.051 | 0.043 | 16,000 | 0.57 (>0.10) | This study |
| P.t.v.‡ | 77 | 0 | 1 | 0.001 | 0.007 | 300 | −1.06 (>0.10) | This study |
| P. troglodytes | 101 | 16 | 3 | 0.067 | 0.111 | 21,000 | −1.12 (>0.10) | This study |
| P. paniscus | 7 | 4 | 0 | 0.079 | 0.059 | 24,000 | 1.63 (>0.10) | This study |
| H. sapiens | 42 | 0 | 0 | 0.000 | 0.000 | This study | ||
| mtDNA ND2 (synonymous sites)§ | ||||||||
| P.t.s. | 6 | 5 | 0 | 0.718 | 0.837 | 7,000 | −0.63 (>0.10) | This study; 5 |
| P.t.t. | 17 | 13 | 0 | 1.200 | 1.489 | 12,000 | −1.19 (>0.10) | This study; 5 |
| P.t.v. | 22 | 25 | 0 | 2.410 | 2.697 | 23,000 | −0.63 (>0.10) | This study; 5 |
| P. troglodytes | 45 | 52 | 0 | 4.320 | 4.613 | 41,000 | −0.31 (>0.10) | This study; 5 |
| P. paniscus | 5 | 13 | 0 | 2.879 | 2.365 | 28,000 | 1.39 (>0.10) | This study; 51 |
| H. sapiens | 73 | 33 | 0 | 0.609 | 2.565 | 4,000 | −2.44 (<0.01) | 5; 27 |
| Xq13.3 | ||||||||
| P.t.t. | 12 | 64 | 4 | 0.175 | 0.209 | 52,000 | −0.75 (>0.10) | 9 |
| P.t.v. | 17 | 24 | 2 | 0.050 | 0.070 | 15,000 | −1.15 (>0.10) | 9 |
| P. troglodytes | 30 | 85 | 6 | 0.131 | 0.211 | 39,000 | −1.47 (>0.10) | 9 |
| P. paniscus | 5 | 5 | 1 | 0.022 | 0.024 | 7,000 | −0.56 (>0.10) | 9 |
| H. sapiens | 69 | 33 | 0 | 0.033 | 0.068 | 7,000 | −1.63 (>0.05) | 56 |
| HOXB6¶ | ||||||||
| P.t.s. | 4 | 0 | 0 | 0.000 | 0.000 | 21 | ||
| P.t.t. | 19 | 4 | 0 | 0.094 | 0.112 | 28,000 | −0.48 (>0.10) | 21 |
| P.t.v. | 58 | 5 | 0 | 0.105 | 0.106 | 31,000 | −0.02 (>0.10) | 21 |
| P. troglodytes | 89 | 8 | 0 | 0.176 | 0.155 | 52,000 | 0.33 (>0.10) | 21 |
| P. paniscus | 36 | 6 | 0 | 0.175 | 0.142 | 52,000 | 0.64 (>0.10) | 21 |
| H. sapiens | 210 | 4 | 0 | 0.060 | 0.066 | 13,000 | −0.16 (>0.10) | 21 |
n is the number of chromosomes; s is the number of segregating sites.
Calculated from s.
Calculated using π.
P.t.v. was calculated with the indel represented as a base substitution.
Tajima's D was based on all sites.
Chimpanzee data do not include recombinant haplogroup Trog G.
Coalescence and Divergence Time Analyses.
The TMRCA for the NRY was estimated for each species by using two different approaches. Using the genetree program with the assumptions of constant population sizes and panmixia, we estimated TMRCAs (±SD) of 660,000 (±190,000) years and 800,000 (±320,000) years for chimpanzees and bonobos, respectively (Table 1). Neither the mutation rate nor the human-chimpanzee/bonobo split times were sensitive to the different priors used in the Bayesian analyses (whereas the chimpanzee-bonobo split time depended to a greater degree on different priors). Posterior means and SDs for the mutation rate, Homo-Pan, and chimpanzee-bonobo split times were 3.0 × 10−6 (±0.7 × 10−6) per sequence per year, 5.9 (±0.6) million years, and 1.7 (±0.7) million years, respectively. Using parameters similar to these (i.e., 3.3 × 10−6/sequence per year, 5.8 million years, and 1.5 million years, respectively) in a Markov Chain Monte Carlo computation produced the best fit of the observed numbers of mutations on different regions of the gene tree (Fig. 2). The TMRCAs (±SD) of chimpanzees, bonobos, and humans were estimated to be 720,000 (±350,000) years, 500,000 (±270,000) years, and 190,000 (±100,000) years, respectively (Fig. 2). The divergence time for chimpanzees and bonobos was estimated at 1.8 (±0.3) million years and that for the Homo-Pan split was 6.0 (±0.2) million years. When the Pt?1 haplogroup was omitted from the analysis, the chimpanzee TMRCA decreased to 610,000 (±300,000) years.
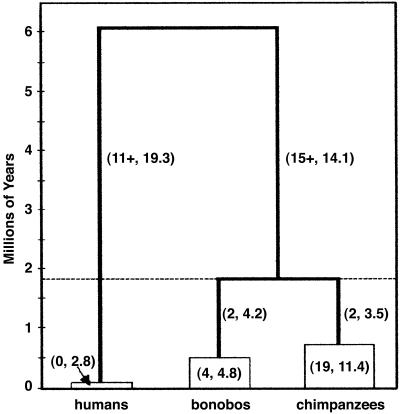
Figure 2.
SPLIT_TIME tree for humans, bonobos, and chimpanzees. The (observed, expected) numbers of mutations are shown on each branch. The total number of expected mutations is random and is not conditional on the total number of observed mutations. Observed mutations on the branches leading to Homo and Pan include a + symbol referring to eight mutations, which could not be assigned to one particular branch. Numbers within boxes correspond to the (observed, expected) numbers of mutations within each taxonomic group. The dotted line indicates the chimpanzee-bonobo divergence time (1.8 million years) estimated with the Pt?1 haplogroup. When the Pt?1 haplogroup was excluded from the analysis, the divergence time remained the same; however, the chimpanzee TMRCA was 610,000 ± 300,000 years and the observed and expected numbers of mutations were 11 and 10.4, respectively.
Tests of Neutrality.
Tajima's D statistic (32) was slightly negative in most cases pertaining to P. troglodytes (e.g., all chimpanzees considered together or by subspecies), whereas it was slightly positive in the case of P. paniscus. However, in no case was the test statistically significant (Table 1). When the two divergent Pt?1 individuals were removed from the analysis, Tajima's D changed to slightly positive for P. t. troglodytes and slightly negative for chimpanzees as a whole, reflecting the rare and divergent nature of this haplogroup. This finding highlights the fact that there is subdivision within P. t. troglodytes, and that the assumptions of these tests were likely violated. The only statistically significant Tajima's D statistic in our analyses was at the human ND2 locus.
Hudson Kreitman Aguadé tests were performed by comparing our NRY data from all three species with mtDNA ND2 data presented here and from published data (5, 27), and with data from the literature for the Xq13.3. locus (9, 39) and the HOXB6 locus (19) (data not shown). Of the 18 comparisons involving the NRY, only the Homo-Pan comparison with ND2 was statistically significant. Nevertheless, the four other comparisons involving the human NRY (i.e., with respect to the X and autosomal loci) approached statistical significance (i.e., 0.05 < P < 0.10). Excess variation at human ND2 may have been partly responsible for the patterns of statistical significance in the HKA test with the other loci. Finally, three comparisons were statistically significant and showed reduced variation at the bonobo Xq13.3 locus in comparison with the NRY, ND2, and HOXB6 systems.
Nucleotide Divergence between Subspecies and Species.
Pairwise NRY sequence comparisons (data not shown) indicated lower levels of divergence between each pair of subspecies (mean = 0.16%) than between the two species of Pan (0.25%). A similar pattern held for ND2 (among subspecies mean = 4.9% versus 12.6% between species). According to the NRY evidence, the greatest subspecies divergence was between P. t. schweinfurthii and P. t verus (0.18%), followed by P. t. troglodytes and P. t. verus (0.16%). For all other loci, the level of sequence divergence was higher between P. t. troglodytes and P. t. verus, and according to the nuclear loci (Xq13.3 and HOXB6), this level of divergence was greater or equal to the level of sequence divergence between the two species of Pan (data not shown).
Analysis of Molecular Variance.
ΦST measures the extent of genetic differentiation among subpopulations. For the NRY data, the overall ΦST for chimpanzees was 0.88. These results are similar to the mtDNA ND2 data in chimpanzees (ΦST = 0.72). In contrast, previous analyses in humans show worldwide Fst and ΦST values for autosomal and NRY genetic sources of ≈0.10 and 0.36, respectively (40, 41).
Discussion
Chimpanzees and bonobos have greater NRY diversity than humans. For example, in this study the seven bonobos tested have three haplogroups compared with a single haplogroup found in the 42 humans. Although we found no polymorphism in humans, in a separate study of eight of the same STSs in 21 humans, Underhill et al. (15) found five polymorphic sites. By comparison, we discovered 14 polymorphic sites (including two indels) in 101 chimpanzees, and four polymorphic sites in seven bonobos in these same eight STSs. If we take the average level of human nucleotide heterozygosity reported for four genes totaling more than 63 kb of noncoding DNA on the NRY (π = 0.011%) (16), there is 6–7 times more variation on the NRY of chimpanzees and bonobos than on the human NRY. As has been observed with other genetic systems, NRY nucleotide diversity within any single chimpanzee subspecies appears to be the same as (or greater) than the diversity within the entire human species (5, 9, 19).
The contrast between chimpanzees and humans is also apparent in their respective effective population sizes. In chimpanzees, Nem is ≈21,000 whereas the Nef calculated from mtDNA ND2 data are 41,000. Kaessmann et al. (9) estimated the chimpanzee Ne to be 35,000 based on X chromosome data (assuming a 20-year generation time and a separation time of the chimpanzee and human lineages of 5 million years). Our re-estimate of Kaessmann et al.'s Ne value was based on a 15-year generation time and a 6 million-year separation time. This new Ne was 39,000 (Table 1). In humans, estimates of male effective population size range from 4,900 to 7,500 (13, 42) with similar estimates in females of 3,500 to 7,500 (11, 43, 44). We estimated a human female long-term effective population size of 4,400 by using synonymous sites from the mtDNA ND2 gene. In general, if males have a higher variance in reproductive success and/or higher premating mortality rates, we would expect Ne for males to be smaller than that for females. According to these data, that is, indeed, the case for chimpanzees.
Our results, as well as analyses of single gene regions on mtDNA (3), the X chromosome (9), and the autosomes (8, 19) all support greater diversity in Pan. These findings stand in direct contrast to the multilocus data of Wise et al. (45), who claimed that human autosomal short tandem repeat heterozygosity was significantly greater than that of chimpanzees. This finding led to a discrepancy in mtDNA/nuclear genome diversity ratios: humans appeared to have lower levels of mtDNA diversity whereas chimpanzees appeared to have lower levels of nuclear diversity. Possible explanations for this discrepancy include: selection on mtDNA combined with a lower Ne for chimpanzees, ascertainment bias (but see ref. 46), a higher average genomewide microsatellite mutation rate in humans, and/or sampling artifacts. More surveys of diversity at additional loci in chimpanzees are necessary to resolve this debate. This need for more surveys is particularly true because the effects of selection can alter levels of diversity in a locus- and species-specific manner. For example, the statistically significant lower levels of nucleotide variability at Xq13.3 in P. paniscus may result from a recent selective sweep near this region in this species.
Tajima's test (32) did not show significant departures from neutrality at the NRY, Xq13.3, and HOXB6 loci, although Tajima's D values tended to be somewhat negative for these loci in chimpanzees (Table 1). These results must be considered in light of the fact that there is significant subdivision among chimpanzee subspecies, especially for the haploid compartments of the genome, and that poor sampling of subdivided populations can lead to negative Tajima's D values. Goldberg and Ruvolo (47) proposed that their negative Tajima's D value for HVI in P. t. schweinfurthii represented a signature of a Pleistocene expansion. Our negative Tajima's D value for the P. t. schweinfurthii NRY data (Table 1) may lend support to this hypothesis. We also found a statistically significant negative Tajima's D value for HVI (data not shown) in our sample of P. t. schweinfurthii. Such an expansion signature was not detected in mtDNA HVI sequences in the other subspecies of chimpanzee or bonobos. Interestingly, we did not observe a statistically significant negative Tajima's D value at the ND2 locus in P. t. schweinfurthii.
The autosomal sequence data do not show distinct clades defining chimpanzee subspecies. On the other hand, neither mtDNA nor NRY haplogroups are shared among subspecies. The lack of sharing of NRY lineages across subspecies is not consistent with a model of continued male gene flow and female philopatry, as seen in some species of primates (48). Rather, these results suggest that sharing of biparentally inherited gene lineages among subspecies is the result of the short evolutionary time since the subspecies have diverged from each other (49). Chimpanzee subspecies should be considered as populations in the process of differentiation, a process that could ultimately lead to speciation (as was the case when bonobos diverged from the P. troglodytes lineage). The estimated divergence time of chimpanzees and bonobos from the NRY data of ≈1.8 million years is intermediate between estimates based on X chromosome data (930,000 years) (9) and those from β globin and some mtDNA data, which suggest a divergence time closer to 2.5 million years ago (50–52). Our estimate from the NRY data is similar to the estimate based on mtDNA restriction mapping (1.3 million years), and this date may correspond with the formation of the Congo River that currently separates the two species (53).
In contrast to studies of humans in which the most recent common ancestor for the NRY is only about 50,000 to 134,000 years old (42, 54), the NRY TMRCAs for chimpanzees and bonobos are both more than 500,000 years. Furthermore, nucleotide diversity values for all P. troglodytes subspecies are ≥ the corresponding values for H. sapiens. In sum, our results suggest that chimpanzees and bonobos have had a different demographic history compared with humans. Although humans have experienced a relatively low long-term effective population size, a recent common ancestry, and a recent size expansion, chimpanzees and bonobos appear to have had a relatively large effective population size, a deeper time depth, and a constant population size (with perhaps the exception of P. t. schweinfurthii). In essence, Homo and Pan may represent the opposite ends of the spectrum of replacement versus continuity, respectively.
Supplementary Material
Acknowledgments
We thank the following institutions for samples: Centre International de Recherches Medicales, the Jane Goodall Institute, Kitwe Point Sanctuary, the Language Research Center at the Georgia State University, New Iberia Research Center, Primate Foundation of Arizona, Yerkes Primate Center, Southwest Foundation for Biomedical Research, Cincinnati Zoo, Detroit Zoo, Lincoln Park Zoo, Riverside Zoo, Sunset Zoo, Welsh Mountain Zoo, and the Center for Reproduction of Endangered Species at the Zoological Society of San Diego. We thank Jean Wickings, Christian Roos, Randy Fulk, and Rick Lukens for information about the origin of samples and Tasha Altheide, Roxane Bonner, Felisa Blackmer, and Cecil Lewis for laboratory assistance. This research has been supported by a grant from the Wenner-Gren Foundation for Anthropological Research, a University of Arizona Small grant, grants from the National Science Foundation (BCS-9816508 and BCS-0073871) (to A.C.S.), and Grant GM-53566 from the National Institute of General Medical Sciences (to M.F.H.). A.C.S. conducted some of this research at the University of Arizona while supported by a postdoctoral fellowship (National Research Service Award) from the National Institutes of Health. Samples were imported under Convention on International Trade in Endangered Species of Wild Fauna and Flora Permit 99US013176/9.
Abbreviations
- NRY
nonrecombining portion of the Y chromosome
- HVI
hypervariable region I
- STS
sequence tagged site
- DHPLC
denaturing HPLC
- TMRCA
time to the most recent common ancestor
Footnotes
References
- 1.Varki A. Genome Res. 2000;10:1065–1070. doi: 10.1101/gr.10.8.1065. [DOI] [PubMed] [Google Scholar]
- 2.Przeworski M, Hudson R R, Di Rienzo A. Trends Genet. 2000;16:296–302. doi: 10.1016/s0168-9525(00)02030-8. [DOI] [PubMed] [Google Scholar]
- 3.Morin P A, Moore J J, Chakraborty R, Jin L, Goodall J, Woodruff D S. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- 4.Nachman M W, Brown W M, Stoneking M, Aquadro C F. Genetics. 1996;142:953–963. doi: 10.1093/genetics/142.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise C A, Sraml M A, Easteal S. Genetics. 1998;148:409–421. doi: 10.1093/genetics/148.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gagneux P, Wills C, Gerloff U, Tautz D, Morin P A, Boesch C, Fruth B, Hohmann G, Ryder O A, Woodruff D S. Proc Natl Acad Sci USA. 1999;96:5077–5082. doi: 10.1073/pnas.96.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deinard A, Kidd K. J Hum Evol. 1999;36:687–703. doi: 10.1006/jhev.1999.0298. [DOI] [PubMed] [Google Scholar]
- 8.Adams E J, Cooper S, Thomson G, Parham P. Immunogenetics. 2000;51:410–424. doi: 10.1007/s002510050639. [DOI] [PubMed] [Google Scholar]
- 9.Kaessmann H, Wiebe V, Pääbo S. Science. 1999;286:1159–1162. doi: 10.1126/science.286.5442.1159. [DOI] [PubMed] [Google Scholar]
- 10.Di Rienzo A, Wilson A C. Proc Natl Acad Sci USA. 1991;88:1597–1601. doi: 10.1073/pnas.88.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers A, Harpending H. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 12.King M-C, Wilson A C. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 13.Hammer M F. Nature (London) 1995;378:376–378. doi: 10.1038/378376a0. [DOI] [PubMed] [Google Scholar]
- 14.Whitfield L S, Sulston J E, Goodfellow P N. Nature (London) 1995;378:379–380. doi: 10.1038/378379a0. [DOI] [PubMed] [Google Scholar]
- 15.Underhill P A, Jin L, Lin A A, Mehdi S Q, Jenkins T, Vollrath D, Davis R W, Cavalli-Sforza L L, Oefner P J. Genome Res. 1997;7:996–1005. doi: 10.1101/gr.7.10.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen P, Wang F, Underhill P A, Franco C, Yang W-H, Roxas A, Sung R, Lin A A, Hyman R W, Vollrath D, et al. Proc Natl Acad Sci USA. 2000;97:7354–7359. doi: 10.1073/pnas.97.13.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Underhill P, Shen P, Lin A A, Jin L, Passarino G, Yang W H, Kauffman E, Bonee-Tamir B, Bertranpetit J, Francalacci P, et al. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 18.Burrows W, Ryder O A. Nature (London) 1997;385:125–126. doi: 10.1038/385125a0. [DOI] [PubMed] [Google Scholar]
- 19.Deinard A S, Kidd K. Am J Phys Anthropol. 2000;111:25–44. doi: 10.1002/(SICI)1096-8644(200001)111:1<25::AID-AJPA3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 20.Morin P A, Moore J J, Woodruff D S. Proc R Soc London Ser B. 1992;249:293–297. doi: 10.1098/rspb.1992.0117. [DOI] [PubMed] [Google Scholar]
- 21.Deinard A. Ph.D. dissertation. New Haven, CT: Yale University; 1997. [Google Scholar]
- 22.Reyonds V, Luscombe G. Folia Primatol. 1971;5:80–87. [Google Scholar]
- 23.Vollrath D, Foote S, Hilton A, Brown L G, Beer-Romero P, Bogan J S, Page D C. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- 24.Kent-First M G, Maffitt M, Muallem A, Brisco P, Shultz J, Ekenberg S, Agulnik A I, Agulnik I, Shramm D, Bavister B, et al. Nat Genet. 1996;14:128–129. doi: 10.1038/ng1096-128. [DOI] [PubMed] [Google Scholar]
- 25.Goldberg T L, Ruvolo M. Biol J Linnean Soc. 1997;61:301–324. [Google Scholar]
- 26.Gonder M K, Oates J F, Disotell T R, Forstner M R J, Morales J C, Melnick D J. Nature (London) 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- 27.Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Nature (London) 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 28.Watterson G A. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 29.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 30.Jukes T H, Cantor C R. In: Mammalian Protein Metabolism. Munro H N, editor. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 31.Takahata N. Mol Biol Evol. 1993;10:2–22. doi: 10.1093/oxfordjournals.molbev.a039995. [DOI] [PubMed] [Google Scholar]
- 32.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudson R R, Kreitman M, Aguadé M. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rozas J, Rozas R. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 35.Excoffier L, Smouse P E, Quattro J M. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider S, Roessli D, Excoffier L. arlequin, A Software for Population Genetics Data. Geneva: University of Geneva; 2000. [Google Scholar]
- 37.Bahlo M, Griffiths R C. Theor Popul Biol. 2000;57:79–95. doi: 10.1006/tpbi.1999.1447. [DOI] [PubMed] [Google Scholar]
- 38.Griffiths R C, Tavaré S. Theor Popul Biol. 1994;46:131–159. [Google Scholar]
- 39.Kaessmann H, Heissig F, von Haeseler A, Pääbo S. Nat Genet. 1999;22:78–81. doi: 10.1038/8785. [DOI] [PubMed] [Google Scholar]
- 40.Relethford J H, Harpending H C. Am J Phys Anthropol. 1994;95:249–270. doi: 10.1002/ajpa.1330950302. [DOI] [PubMed] [Google Scholar]
- 41.Hammer M F, Karafet T M, Redd A J, Jarjanazi H, Santachiara-Benerecetti A S, Soodyall H, Zegura S L. Mol Biol Evol. 2001;18:189–203. doi: 10.1093/oxfordjournals.molbev.a003906. [DOI] [PubMed] [Google Scholar]
- 42.Thomson R, Pritchard J K, Shen P, Oefner P J, Feldman M W. Proc Natl Acad Sci USA. 2000;97:7360–7365. doi: 10.1073/pnas.97.13.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kocher T D, Wilson A C. In: Evolution of Life. Osawa S, Honjo T, editors. Tokyo: Springer; 1991. pp. 391–413. [Google Scholar]
- 44.Ruvolo M, Zehr S, von Dornum M, Pan D, Chang B, Lin J. Mol Biol Evol. 1993;10:1115–1135. doi: 10.1093/oxfordjournals.molbev.a040068. [DOI] [PubMed] [Google Scholar]
- 45.Wise C A, Sraml M, Rubinsztein D C, Easteal S. Mol Biol Evol. 1997;14:707–716. doi: 10.1093/oxfordjournals.molbev.a025810. [DOI] [PubMed] [Google Scholar]
- 46.Cooper G, Rubinsztein D C, Amos W. Hum Mol Genet. 1998;7:1425–1429. doi: 10.1093/hmg/7.9.1425. [DOI] [PubMed] [Google Scholar]
- 47.Goldberg T L, Ruvolo M. Mol Biol Evol. 1997;14:976–984. doi: 10.1093/oxfordjournals.molbev.a025841. [DOI] [PubMed] [Google Scholar]
- 48.Melnick D J, Hoeltzer G A. Evol Anthropol. 1993;2:2–10. [Google Scholar]
- 49.Tajima F. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesole G, Sbisá E, Preparata G, Saccone C. Mol Biol Evol. 1992;9:587–598. doi: 10.1093/oxfordjournals.molbev.a040747. [DOI] [PubMed] [Google Scholar]
- 51.Horai S, Satta Y, Hayasaka K, Kondo R, Inoue T, Ishida T, Hayashi S, Takahata N. J Mol Evol. 1992;35:32–43. doi: 10.1007/BF00160258. [DOI] [PubMed] [Google Scholar]
- 52.Bailey W J, Hayasaka K, Skinner C G, Kehoe S, Sieu L C, Slightom J L, Goodman M. Mol Phylogenet Evol. 1992;1:97–135. doi: 10.1016/1055-7903(92)90024-b. [DOI] [PubMed] [Google Scholar]
- 53.Ferris S D, Brown W M, Davidson W S, Wilson A C. Proc Natl Acad Sci USA. 1981;78:6319–6323. doi: 10.1073/pnas.78.10.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karafet T M, Zegura S L, Posukh O, Osipova L, Bergen A, Long J, Goldman D, Klitz W, Harihara S, de Knijff P, et al. Am J Hum Genet. 1999;64:817–831. doi: 10.1086/302282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swofford D. paup*, Phylogenetic Analysis Using Parsimony. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- 56.Kaessmann H, Wiebe V, Weiss G, Pääbo S. Nat Genet. 2001;27:155–156. doi: 10.1038/84773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.