Abstract
Bruton's tyrosine kinase (Btk) acts downstream of phosphoinositide 3-kinase (PI3K) in a pathway required for B cell receptor (BCR)-dependent proliferation. We used DNA microarrays to determine what fraction of genes this pathway influences and to investigate whether PI3K and Btk mediate distinct gene regulation events. As complete loss-of-function mutations in PI3K and Btk alter B cell subpopulations and may cause compensatory changes in gene expression, we used B cells with partial loss of function in either PI3K or Btk. Only about 5% of the BCR-dependent gene expression changes were significantly affected by reduced PI3K or Btk. The results indicate that PI3K and Btk share target genes, and that PI3K influences additional genes independently of Btk. These data are consistent with PI3K acting through Btk and other effectors to regulate expression of a critical subset of BCR target genes that determine effective entry into the cell cycle.
Phosphoinositide 3-kinase (PI3K) and Bruton's tyrosine kinase (Btk) are two critical signaling components activated after B cell receptor (BCR) stimulation (1). The term PI3K refers to a family of lipid kinases that phosphorylate phosphatidylinositol (PtdIns) and its derivatives on the 3′-hydroxyl of the inositol head group. The resulting 3′-phosphorylated phosphoinositides act as second messengers to recruit specific proteins to cellular membranes (2). Blocking PI3K activation by treatment with specific inhibitors or by disruption of the PI3K p85α gene in mice abolishes BCR-dependent proliferation (3–5). Btk is a Tec family tyrosine kinase whose activation is crucial for generation of a sustained increase in intracellular Ca2+ concentration, a required event for B cell activation (6). Mutations in Btk block B cell proliferation and are the cause of the B cell deficiencies X-linked agammaglobulinemia in humans and X-linked immunodeficiency (Xid) in mice (7–10).
There is good evidence that Btk activation is PI3K-dependent. The pleckstrin homology (PH) domain of Btk binds selectively to the PI3K product PtdIns(3,4,5)P3 and is required for membrane recruitment and activation of Btk by membrane-associated Src-family kinases (11–13). Binding of PtdIns(3,4,5)P3 to the PH domain also contributes directly to Btk activation by relieving intramolecular inhibition of the kinase domain (14). A point mutation in the Btk PH domain (R28C) that abrogates PtdIns(3,4,5)P3 binding is the genetic lesion in Xid and in some patients with X-linked agammaglobulinemia (10, 15).
Analysis of Btk-deficient B cells has indicated that Btk function is required for up-regulation of cyclin D2, cyclin E, and Bcl-XL (16, 17). Although the induction of these genes has not been assessed in PI3K-deficient B cells, PI3K has been shown to regulate expression of cyclins and Bcl-XL downstream of cytokine receptors (18–20). In those systems, the serine/threonine kinase Akt/PKB is a critical link between PI3K and gene expression. Akt is also activated in a PI3K-dependent manner after BCR engagement (21–23) and represents a likely Btk-independent pathway for gene regulation. It is also possible that Btk participates in PI3K-independent signaling pathways. A number of proteins have been reported to associate with various domains of Btk (reviewed in ref. 6).
We wished to test the hypothesis that PI3K and Btk regulate B cell function through both common and distinct pathways. DNA microarray hybridization provides a detailed and global view of the gene expression differences that ultimately determine cellular responses. Here we present an analysis of global gene expression changes at two time points after BCR stimulation of B cells with normal or diminished PI3K or Btk function.
Materials and Methods
Mice.
The generation of Balb/Xid(BtkTg) (Tg, transgenic) mice was described previously (24). Animals used in this study had been backcrossed 8–14 times to BALB/c. Balb/Xid(BtkTg) and BALB/c controls were used at 8–16 weeks of age. Procedures were approved by institutional review boards at the University of California, Los Angeles and at the University of California, Irvine.
B Cell Purification.
Spleens were removed and processed in groups of five. Single cell suspensions were prepared and red blood cells lysed. T cells were depleted with anti-Thy1.2 (Sigma) and rabbit Low-Tox complement (Accurate, San Diego, CA). Live cells were collected over a gradient of Lympholyte-M (Accurate). Macrophages, dendritic cells, and remaining T cells were then depleted by using magnetic beads coated with anti-CD43 followed by separation on MACS-CS columns (Miltenyi Biotec, Auburn, CA). Cell purity was checked by flow cytometry after staining with anti-IgM-FITC, anti-Thy1.2-PE, and anti-B220-CyChrome (BD Biosciences, San Diego, CA). The percentage of cells expressing both B220 and IgM was 98–99% for all experiments. All B cells of a given genotype were pooled before separation into treatment groups.
Proliferation Assay.
Purified B cells were suspended at 2 × 106/ml in B cell medium (RPMI medium 1640 containing 10% heat-inactivated FCS, 5 mM Hepes, pH 7.5, 2 mM l-glutamine, 1× penicillin–streptomycin, and 50 μM 2-mercaptoethanol). Cells were pretreated for 15 min at 37°C with either 3 μM LY294002 or diluent (0.1% EtOH), then seeded in 96-well flat-bottom plates at 1 × 106/ml in a total volume of 100 μl B cell medium in the absence or presence of 10 μg/ml of anti-IgM (F(ab′)2, Jackson Immunoresearch). After 48 h, 1μCi of 3H-thymidine in 50 μl B cell medium was added. Sixteen hours later, plates were harvested onto filters and counted with a BetaPlate system (Wallac/Perkin-Elmer, Gaithersburg, MD).
Microarray Hybridization.
For unstimulated samples (three independent preparations), purified B cells were immediately pelleted, washed with PBS, and snap-frozen. For the stimulated samples (two independent preparations), cells were treated with LY294002 or EtOH and incubated with anti-IgM exactly as for the proliferation assays, except that volumes were scaled up for the larger cell numbers used (5–60 × 106). At 2 or 12 h after initiation of culture, dishes were placed on ice and cells transferred to centrifuge tubes, washed in PBS, and snap-frozen. PolyA-enriched RNA was prepared by using either the OligoTex Direct mRNA Kit (Qiagen, Valencia, CA) or the MicroPolyA-Pure Kit (Ambion, Austin, TX). Labeled cRNA target for microarray hybridization was prepared according to Affymetrix recommendations. cRNA was fragmented and hybridized sequentially to Mu11K subA, Mu11K subB, U74Bv2, and U74Cv2 chips (Affymetrix, Santa Clara, CA). Hybridization, washing, and scanning were performed by staff at the Beckman Center (Stanford University). The Mu11K subA and subB arrays together carried 13,027 probe sets corresponding to ≈11,000 unique genes (some represented more than once with different probes). The U74Bv2 and U74Cv2 arrays carried 24,279 probe sets representing expressed sequence tags that correspond to an unknown number of unique genes. Data from all the arrays will be made available at mbb.bio.uci.edu/fruman/.
Data Analysis.
Detailed descriptions of data analysis tools and statistical methods are published as supporting information on the PNAS web site (www.pnas.org).
Quantitative Real-Time PCR (Q-PCR).
Separate groups of mice were used to prepare purified B cell RNA. Contaminating genomic DNA was removed by treatment with RNase-free DNase (Promega, Madison, WI). First-strand cDNA was synthesized by using a kit (Promega) and oligo-dT priming. Genes of interest were detected by using primers that had been optimized to generate a single amplicon of 75–200 nucleotides. Real-time PCR was performed by using an iCycler (Bio-Rad) and SyBr green detection. Reactions were performed in 96-well plates with optical sealing tape (Bio-Rad) and contained 20 μl total volume and the following components: 50 mM Tris, pH 8.3/3 mM MgCl2/0.5 mg/ml BSA/200 μM each dNTP/0.33× SyBr green (Molecular Probes)/0.5 units HotStarTaq (Qiagen), primers at the optimal concentration, and cDNA template or water. Mouse β-actin primers were used in parallel for each run. Primer sequences are published as supporting information on the PNAS web site. The relative expression level of the gene of interest in different cDNA samples was determined as follows. For each primer pair, the threshold cycle for detection over a range of cDNA dilutions was plotted to generate a standard curve for extrapolating unknowns. The relative expression of the gene of interest was determined by calculating the ratio of the extrapolated concentration of that gene to the extrapolated concentration of β-actin.
Results
Experimental Design.
Complete loss of Btk function (Xid mice or Btk gene-targeted mice) or PI3K function (p85α gene-targeted mice) disrupts B cell differentiation, altering the percentages of B cell subpopulations in the spleen (4, 5, 25). Deletion of p85α results in up-regulated expression of the related protein p85β (4, 26, 27), and other compensatory changes in gene expression may also occur. Moreover, many fewer B cells can be recovered from Btk- or PI3K-deficient mice, making it difficult to isolate enough mRNA for microarray analysis. We used the following approaches to obtain cells that were closely matched both developmentally and for genetic context.
We took advantage of the finding that reduced gene dosage of Btk affects B cell proliferation but not development (24). In Xid mice hemizygous for a Btk transgene, B cells express functional Btk at ≈25% of wild-type levels. To minimize genetic background differences, we used Xid(BtkTg) mice that had been extensively backcrossed with BALB/c mice. Balb/Xid(BtkTg) mice possess normal numbers of splenic B cells with no apparent defects in differentiation compared with BALB/c controls (24). Nevertheless, B cell proliferation in response to BCR crosslinking was reduced to about 25% of wild-type levels, and humoral immune responses to T-independent antigens were greatly diminished (Fig. 1 and ref. 24).
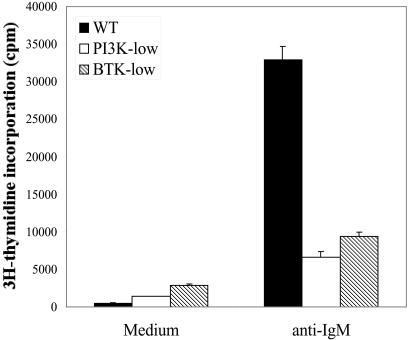
Figure 1.
Reduced proliferation in PI3K- and Btk-low B cells. Highly purified B cells were stimulated with anti-IgM (F(ab′)2, 10 μg/ml) in triplicate wells for 60 hr, with 3H-thymidine added during the last 12 hr. Cells were harvested and incorporated radioactivity counted.
For PI3K, we treated wild-type BALB/c splenic B cells with the specific PI3K inhibitor, LY294002. At 10 μM, this compound completely blocks BCR-mediated proliferation yet is not generally toxic, as it has no effect on proliferation triggered by the combination of anti-CD40 plus IL-4 (4). At 3 μM, LY294002 reduced proliferation to the level seen in Btk-low cells (Fig. 1). Henceforth, the term “Btk-low” will be used to denote cells from Balb/Xid(BtkTg) mice, and “PI3K-low” will refer to wild-type (BALB/c) cells treated with 3 μM LY294002. Although some Btk and PI3K function remains in these cells, quantitative differences in gene expression could be taken as evidence that these signaling proteins are rate-limiting for specific gene regulation events.
Gene Expression in Wild-Type Cells.
Highly purified (>98% B220+/IgM+) B cells were harvested directly (time t = 0) or after anti-IgM stimulation for 2 or 12 h. These times were chosen to view changes in gene expression as B cells transition from G0 into the G1 phase of the cell cycle (2 h) and as they progress through G1 (12 h) (28, 29). mRNA was prepared and used for the generation of labeled cRNA as described in Materials and Methods. Samples were hybridized to high-density oligonucleotide arrays from Affymetrix (GeneChip Mu11K) carrying 13,027 probe sets. Subsequently, the same cRNA samples were hybridized to arrays (Affymetrix U74Bv2 and U74Cv2) carrying an additional 24,279 expressed sequence tags.
First, we determined which genes changed significantly in expression from 0 to 2 h or from 0 to 12 h in wild-type cells. Analysis of the Mu11K chips showed that 1,020 genes changed expression at 2 h and 673 changed at 12 h after BCR stimulation (P < 0.01). Thus, BCR crosslinking leads to significant changes in mRNA levels for ≈5–8% of the genes queried. A complete list of genes whose mRNA levels changed significantly at 2 or 12 h is published as supporting information (Excel Spreadsheet) on the PNAS web site. Table 1 lists selected mRNAs, grouped by functional category, with fold-change values. Inspection of the gene lists revealed expected regulatory patterns, helping to validate the current experimental design and analysis. For example, cyclin D2 and cdk4 were significantly induced in wild-type cells at 2 h, consistent with previous studies (28, 29). In addition, the antiapoptotic proteins Bcl-XL, A1, and c-FLIPL were induced (16, 30, 31). Activation of lymphocytes leads to a marked increase in cell size, which would be expected to require an increase in the rate of protein production. Strikingly, nearly all of the genes involved in RNA processing or translation were up-regulated at both 2 and 12 h (see Table 3, which is published as supporting information on the PNAS web site). Hallmarks of cellular differentiation include changes in the array of surface receptors and transcription factors. Indeed, each of these categories was represented by both up- and down-regulated genes (Table 1).
Table 1.
Selected genes whose expression changes significantly after BCR stimulation, grouped by general function, with fold change indicated
| Accession no. | Name | Fold change |
|---|---|---|
| Cell cycle | ||
| M83749 | cyclin D2 | 9.3 |
| D87911 | Ki antigen | 4.1 |
| L01640 | cdk4 | 3.4 |
| X53068 | PCNA | 2.0 |
| U36799 | Rb2/p130 | −7.2 |
| Z37110 | cyclin G | 3.6 |
| Transcription | ||
| X55499 | Ig/EBP-1 | 3.2 |
| U25096 | LKLF | −4.0 |
| U33626 | Pml | −4.3 |
| M12848 | Myb | 5.7 |
| U20735 | junB | −6.7 |
| Survival | ||
| U97076 | cFLIP long | 6.2 |
| U78031 | Bcl-X long | 2.7 |
| L16462 | A1 | 1.7 |
| Surface protein | ||
| L25606 | CD86/B7-2 | 11.0 |
| M29697 | IL-7R | 7.2 |
| L16928 | CD22 | −6.6 |
| M25324 | CD62L/Mel-14 | −11.4 |
| U70139 | CCR4 | 3.9 |
| D16432 | CD63 | 3.1 |
| M59378 | TNFR-1 | −3.1 |
| AB000803 | CXCR-4 | −4.4 |
Italics denote 12-hr data.
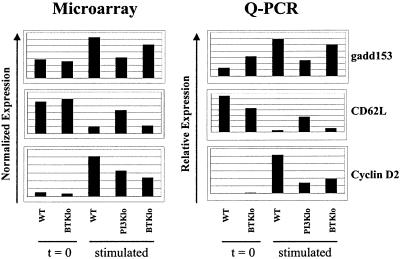
To further test the reliability of the microarray measurements, expression levels were measured in independent RNA samples by using Q-PCR (32). Of eight selected genes, each showed the expected increase or decrease in mRNA level after stimulation of wild-type cells (Fig. 2 and Fig. 5, which is published as supporting information on the PNAS web site).
Figure 2.
Validation of selected microarray results by Q-PCR. (Left) Normalized expression values from microarray experiments were averaged and plotted for each of the indicated conditions. (Right) mRNA samples isolated from cells treated as for the microarray experiments were reverse-transcribed; then, relative quantitation of genes of interest was performed by using real-time detection of PCR amplification in the presence of SyBr green, as described in Materials and Methods. The “stimulated” time points were 2 hr for gadd153 and cyclin D2, 12 hr for CD62L.
Role of PI3K and Btk in Gene Expression.
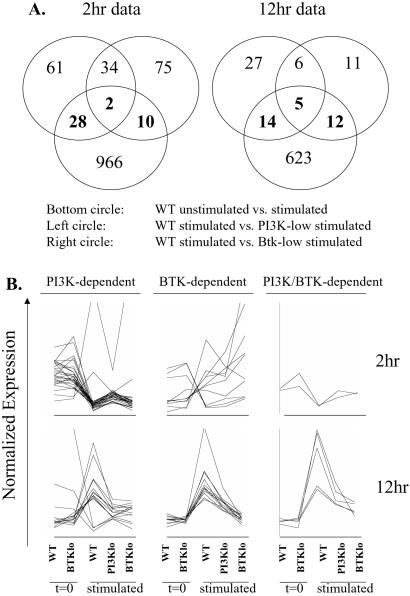
Next, we determined which genes showed significantly different expression at each time point in stimulated wild-type cells vs. either PI3K- or Btk-low (P < 0.01). The resulting gene lists were then plotted in a Venn diagram that also contained the list of genes from the wild type unstimulated to stimulated comparison (Fig. 3A). The overlap of the three groups, in the center of the diagrams, represents BCR target genes that are regulated by both PI3K and Btk. To the left and right of the center are areas of overlap corresponding to genes whose regulation is either PI3K- or Btk-dependent. Some of the genes in these categories have likely roles in cell cycle regulation (Table 2). Reduced induction of cyclin D2 in Btk-low cells may be sufficient to explain their diminished proliferation, as B cells lacking cyclin D2 proliferate poorly in response to BCR engagement (33). Failure to adequately repress the retinoblastoma family member Rb2/p130, TIS (topoisomerase inhibitor-suppressed) or GAP-III could affect cell cycle progression in the PI3K-low cells. The PI3K/Btk-dependent up-regulation of nucleolin could be required for RNA processing events important for cell growth.
Figure 3.
(A) Venn diagram analysis of gene list overlap. Genes showing significant differences between the stimulated conditions (P<0.01) were identified by using CyberT statistical analysis of normalized expression data. These groups (Upper Left and Upper Right) were plotted along with the group of genes regulated by BCR stimulation of wild-type cells (Lower). The numbers in the center represent genes that are PI3K- and Btk-dependent; numbers left of center, PI3K-dependent; numbers right of center, Btk-dependent. Numbers within the bottom circle represent genes regulated in wild-type cells independently of PI3K and Btk. (B) Graphical view of gene categories revealed by statistical analysis followed by Venn diagram. By using genespring software (Silicon Genetics, Redwood City, CA), expression values were normalized to plot all on the same scale. Each line represents a single gene across the five indicated conditions.
Table 2.
Annotated genes determined to be significantly regulated by PI3K, Btk, or both
| Accession no. | Name |
|---|---|
| 2 hr, PI3K-dependent | |
| X51834 | Osteopontin |
| X67083 | gadd153/chop-10 |
| D63679 | Decay accelerating factor |
| J03168 | Irf-2 |
| U36799 | Rb2/p130 |
| X86405 | CB2 cannabinoid receptor |
| AF005885 | Cyclin G2 |
| U20238 | GAP III |
| U10871 | MAP kinase |
| U69488 | G7e |
| M16465 | Calpactin light chain |
| M25324 | CD62L/Mel-14 |
| J03857 | B29 |
| D63902 | Estrogen-responsive finger protein |
| U15635 | Mg11 |
| X15643 | Beta-2-adrenergic receptor |
| M89956 | Calcium binding protein pp52 |
| D86344 | Topoisomerase inhibitor suppressed |
| 2 hr, Btk-dependent | |
| AA051121 | GU4 nucleic-binding homolog |
| M65027 | gp49 |
| W33838 | Asn-tRNA synthetase homolog |
| M83749 | cyclin D2 |
| J05261 | Protective protein Mo54 |
| 2 hr, PI3K/Btk-dependent | |
| AA238081 | Similar to C1R complement precursor |
| 12 hr, PI3K-dependent | |
| AA050852 | NM23 |
| W34756 | Uridine kinase homolog |
| Y11666 | Hexokinase II |
| AA146127 | Ran/TC4 homolog |
| AA123463 | PHAPII homolog |
| W59723 | Pre-B cell enhancing factor homolog |
| AA137580 | Heat shock cognate protein homolog |
| AA002883 | Mitochond. elong. factor G homolog |
| AA096793 | ADP/ATP translocase 2 homolog |
| M25324 | CD62L/Mel-14 |
| X06143 | CD2 |
| D63679 | Decay accelerating factor |
| 12 hr, Btk-dependent | |
| U25691 | Lymphocyte-specific helicase |
| X62154 | P1 |
| AA138121 | Multifunctional tRNA synthetase hom. |
| X04097 | RP2 |
| AA068712 | PKC inhibitor 1 homolog |
| W75484 | hnRNP A1 homolog |
| M25149 | Tum-P91A |
| X95591 | C1D |
| 12 hr, PI3K/Btk-dependent | |
| M63445 | NAD-dependent MTHF dehydrogenase |
| X07699 | Nucleolin |
| X70100 | mal1 lipid-binding protein |
| Z31557 | Chaperonin containing TCP-1, ζ subunit |
Italics denote genes down-regulated in wild-type cells.
The Venn diagrams (Fig. 3A) suggest that PI3K and Btk have both shared and independent influences on gene expression. However, as illustrated in Fig. 3B, many of the genes classified as either PI3K- or Btk-dependent showed similar expression in PI3K- and Btk-low cells, which is explained by replicate measurements for those two conditions having similar means but different variances. Thus, all genes whose up-regulation depended statistically on Btk appeared to be influenced by PI3K as well. On the other hand, there were clear examples of genes whose induction or repression was affected primarily by PI3K. Q-PCR results supported these conclusions (Fig. 2). Notably, although the Btk dependence of cyclin D2 induction (P = 0.0097) was confirmed, the effect in PI3K-low was found to be equivalent (Fig. 2). On the other hand, the greater influence of PI3K on gadd153 up-regulation (P = 0.0056) and CD62L down-regulation (P = 0.0016) was verified by Q-PCR (Fig. 2).
Overall, less than 1% (2 at 2 h, 5 at 12 h) of the gene expression changes were significantly affected by reduced signaling via PI3K and Btk. Less than 3% of the genes were regulated by PI3K alone (28 at 2 h, 14 at 12 h), and less than 2% by Btk alone (10 at 2 h, 12 at 12 h). Consequently, statistical analysis indicates that about 95% of the gene expression changes were not altered significantly in PI3K- or Btk-low cells. This may reflect the use of B cells with only partial loss of PI3K or Btk function and proliferation. To gain a broader view of the role of PI3K and Btk in regulation of gene expression, we clustered genes on the basis of their similarity in expression across different conditions. Several clustering algorithms (34) were tested, and K-means clustering (see supporting information on the PNAS web site) was found to yield the most informative output. Only those genes whose expression changed in wild-type cells were included in this analysis.
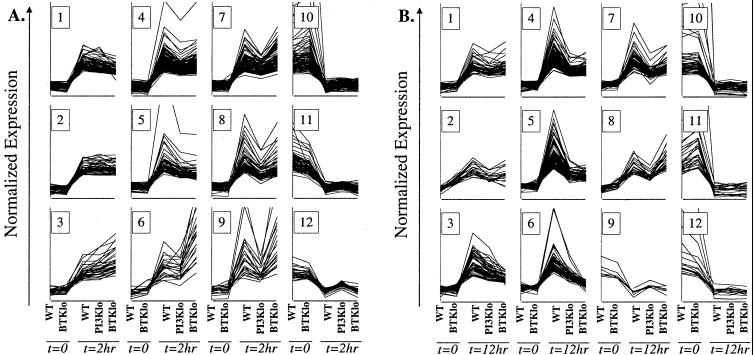
Graphical views of the clusters (Fig. 4) confirmed that many of the gene expression changes after BCR stimulation were largely independent of both PI3K and Btk (2 h clusters 1–5, 10 and 11 and 12 h clusters 1,2, 10 and 11). However, clustering also suggested that PI3K and Btk contribute at least partially to a larger fraction of the gene expression program than was suggested by statistical analysis. Induction of some genes was regulated strongly by both PI3K and Btk. This was especially apparent at 12 h, where clusters 4 and 5 showed considerably less induction in both PI3K-low and Btk-low (Fig. 4). In addition, there were clearly genes whose regulation depended primarily on PI3K, as their induction/repression was attenuated in PI3K- but not Btk-low cells. At 2 h, clusters 7, 8, and 9 showed PI3K-dependent induction, and cluster 12 showed PI3K-dependent repression. At 12 h, clusters 7 and 8 showed PI3K-dependent induction, and cluster 9 showed PI3K-dependent repression. It was less clear whether some clusters represent genes primarily dependent on Btk. Cluster 6 at 12 h included genes whose up-regulation appeared more attenuated in Btk-low than in PI3K-low. At 2 h, cluster 6 showed several genes that were superinduced in Btk-low cells relative to wild-type or PI3K-low, a pattern that can also be recognized in the statistically Btk-dependent gene list (Fig. 3B). Overall, clustering analysis supports the conclusions that PI3K signals independently of Btk, but that PI3K-independent Btk signaling has at most a modest contribution to gene expression. It is worth noting that nearly all genes showed similar basal expression in wild-type and Btk-low cells, helping to confirm that the starting cell populations were closely matched.
Figure 4.
Graphical view of clusters of genes whose expression changes significantly in wild-type B cells 2 or 12 hr after BCR stimulation. The graphs are presented in the same format as in Fig. 3B. Each line represents a gene found to change significantly in expression (P<0.01) between WT t = 0 and WT t = 2 hr (A) or WT t = 12 hr (B). K-means clustering was done to identify genes with similar expression patterns, by using a Pearson correlation. The number of clusters was set arbitrarily at 10. One of the 12-hr clusters contained a single gene and is not shown. For both 2 and 12 hr, all down-regulated genes fell into a single cluster, which was further clustered into three (2-hr) or four (12-hr) subsets. The 2- and 12-hr clusters are numbered arbitrarily 1–12 for ease of identification in the text.
Selected genes from the cluster plots were analyzed by Q-PCR. The results suggested that clustering is more useful for identifying global patterns of gene regulation than for making definitive lists of target genes. Although Q-PCR confirmed that induction of Gfi-1 was PI3K-dependent, the PI3K/Btk dependence of Enx-1 was not reproduced, nor was the Btk dependence of glutamic acid decarboxylase (Fig. 5, which is published as supporting information on the PNAS web site). Stat4, with an apparent Btk-dependent pattern, showed lower expression in stimulated Btk-low cells compared with stimulated wild-type or PI3K-low (Fig. 5). However, the slightly lower basal expression of Stat4 in Btk-low suggests that the fold induction was similar.
Discussion
We have used DNA arrays to compare gene expression in normal and signaling-deficient B cells. Our results support the following conclusions. First, BCR stimulation triggers significant changes (P < 0.01) in the expression of less than 10% of cellular genes at the time points studied. Some of these genes encode proteins with established roles in control of cell growth/survival (Table 1). An important goal is to determine which of the other genes and expressed sequence tags whose mRNA levels are altered after BCR stimulation play an important role in B cell proliferation.
Second, reduced PI3K or Btk function has a statistically significant effect on only about 5% of this gene expression program. However, by looking at clustering results (Fig. 4) instead of statistical analysis, it is apparent that signaling via PI3K and Btk does contribute at least partially to a larger fraction of the gene expression changes. Nevertheless, the prevalence of PI3K/Btk-independent genes suggests that other signals emanating from the BCR can act alone to alter gene expression. Overall, the data indicate that PI3K and Btk provide a measured input, rather than an all-or-none effect, on expression of different genes.
A third conclusion is that PI3K clearly influences gene expression by pathways not involving Btk (Figs. 3 and 4). Another likely PI3K effector in B cells is the serine/threonine kinase Akt/PKB. PI3K signaling through Akt/PKB has been shown to enhance E2F function in T cells (18) and to contribute to NF-κB activation in certain systems (35, 36). The PI3K/Atk pathway also inhibits activity of forkhead family transcription factors in various cell types (37). The striking cluster of PI3K-dependent down-regulated genes observed at 2 h (Fig. 3B) may be transcriptional targets of forkhead factors.
Finally, few genes appear to be strongly regulated by Btk independently of PI3K. Using Venn diagrams to represent the overlap in gene lists generated by CyberT analysis, we identified groups of genes at 2 and 12 h that were statistically affected by only Btk-low (Fig. 3A). However, graphical views of these lists suggest that most of these genes were also affected to some degree by reduced PI3K function (Fig. 3B). Using a clustering approach, we found few genes whose induction or repression was strongly influenced by Btk alone (Fig. 4). Independent measurement of mRNA levels by Q-PCR failed to confirm that induction of three selected genes was both Btk-dependent and PI3K-independent (Figs. 2 and 5, supporting information on the PNAS web site). These data support the model that the function of Btk downstream of the BCR depends primarily on PI3K. This interpretation is consistent with the finding that a Btk-null allele produces a phenotype nearly indistinguishable from Xid mice, in which only the ability of Btk to interact with PtdIns-3,4,5-P3 is compromised (25). A unique role for Btk in opposing BCR signaling is suggested by the observation that some genes induced in wild-type and PI3K-low were more strongly up-regulated in Btk-low cells (Figs. 3 and 4). A PI3K-independent inhibitory function of Btk could be mediated through one of many proteins reported to interact with Btk (6).
Phosphatase 2B (calcineurin) is a potential effector of Ca2+ signaling downstream of PI3K/Btk because of its Ca2+-dependent dephosphorylation of nuclear factor of activated T cells (NFAT). However, microarray analysis of B cells treated with the calcineurin inhibitor FK506 (38) defined a set of calcineurin-dependent genes quite different from the PI3K/Btk-dependent genes determined in the present study. Some of the differences could be because of measurement of gene expression 1 h after BCR engagement by Glynne et al. (38), compared with 2 h in the present study. However, we did observe significant overlap between the genes induced or repressed 1 and 2 h after BCR engagement of wild-type cells (unpublished data). It is possible that residual signaling in PI3K- and Btk-low cells is sufficient to activate calcineurin but not other pathways. Other potential downstream targets of PI3K/Btk signaling include Ca2+-dependent PKC isoforms. In support of this possibility, the phenotype of PKCβ-deficient B cells is quite similar to p85α knockout or Xid B cells (39). In addition, treatment of Xid B cells with phorbol ester restores proliferation driven by anti-IgM plus IL-4 (17).
Reduced PI3K/Btk function causes immunodeficiency, and enhanced PI3K signaling leads to systemic autoimmunity (40). The severity of the X-linked agammaglobulinemia and Xid phenotypes vary among individual humans or strains of mice (41, 42). A candidate gene approach has revealed several modifiers of the Xid phenotype in mice, including the p85α subunit of PI3K (43). Comparative gene expression profiling similar to that presented in this work will be a valuable tool to define whether these modifiers act through Btk-dependent, Btk-independent, or overlapping pathways. This information will increase our understanding of and define potential therapeutic targets for diseases associated with dysregulation of Btk and PI3K signaling pathways.
Supplementary Material
Acknowledgments
We thank Hajir Dadgostar and Genhong Cheng for advice on microarray experiments, Wes Hatfield for advice on statistical analysis, Chris Carpenter for critical reading of the manuscript, James Johnson and Fiona Willis for assistance with mouse husbandry and genotyping, and Shirley Quan for administrative support. D.A.F. and A.B.S. were supported in part by Special Fellowships from the Leukemia and Lymphoma Society. A.B.S. is a Southwestern Medical Foundation Scholar in Biomedical Research. O.N.W. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- Btk
Bruton's tyrosine kinase
- PI3K
phosphoinositide 3-kinase
- BCR
B cell receptor
- PtdIns
phosphatidylinositol
- Xid
X-linked immunodeficiency
- Tg
transgenic
- Q-PCR
quantitative real-time PCR
References
- 1.Fruman D A, Satterthwaite A B, Witte O N. Immunity. 2000;13:1–3. doi: 10.1016/s1074-7613(00)00002-9. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B, Waterfield M D. Exp Cell Res. 1999;253:239–254. doi: 10.1006/excr.1999.4701. [DOI] [PubMed] [Google Scholar]
- 3.Aagaard-Tillery K M, Jelinek D F. J Immunol. 1996;156:4543–4554. [PubMed] [Google Scholar]
- 4.Fruman D A, Snapper S B, Yballe C M, Davidson L, Yu J Y, Alt F W, Cantley L C. Science. 1999;283:393–397. doi: 10.1126/science.283.5400.393. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki H, Terauchi Y, Fujiwara M, Aizawa S, Yazaki Y, Kadowaki T, Koyasu S. Science. 1999;283:390–392. doi: 10.1126/science.283.5400.390. [DOI] [PubMed] [Google Scholar]
- 6.Satterthwaite A B, Li Z, Witte O N. Semin Immunol. 1998;10:309–316. doi: 10.1006/smim.1998.0123. [DOI] [PubMed] [Google Scholar]
- 7.Thomas J D, Sideras P, Smith C I, Vorechovsky I, Chapman V, Paul W E. Science. 1993;261:355–358. doi: 10.1126/science.8332900. [DOI] [PubMed] [Google Scholar]
- 8.Tsukada S, Saffran D C, Rawlings D J, Parolini O, Allen R C, Klisak I, Sparkes R S, Kubagawa H, Mohandas T, Quan S, et al. Cell. 1993;72:279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 9.Vetrie D, Vorechovsky I, Sideras P, Holland J, Davies A, Flinter F, Hammarstrom L, Kinnon C, Levinsky R, Bobrow M, et al. Nature (London) 1993;361:226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 10.Rawlings D J, Saffran D C, Tsukada S, Largaespada D A, Grimaldi J C, Cohen L, Mohr R N, Bazan J F, Howard M, Copeland N G, et al. Science. 1993;261:358–361. doi: 10.1126/science.8332901. [DOI] [PubMed] [Google Scholar]
- 11.Salim K, Bottomley M J, Querfurth E, Zvelebil M J, Gout I, Scaife R, Margolis R L, Gigg R, Smith C I, Driscoll P C, et al. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- 12.Rameh L E, Arvidsson A, Carraway K L, III, Couvillon A D, Rathbun G, Crompton A, VanRenterghem B, Czech M P, Ravichandran K S, Burakoff S J, et al. J Biol Chem. 1997;272:22059–22066. doi: 10.1074/jbc.272.35.22059. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Wahl M I, Eguinoa A, Stephens L R, Hawkins P T, Witte O N. Proc Natl Acad Sci USA. 1997;94:13820–13825. doi: 10.1073/pnas.94.25.13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito K, Scharenberg A M, Kinet J P. J Biol Chem. 2001;276:16201–16206. doi: 10.1074/jbc.M100873200. [DOI] [PubMed] [Google Scholar]
- 15.Vihinen M, Mattsson P T, Smith C I. Front Biosci. 2000;5:D917–D928. doi: 10.2741/vihinen. [DOI] [PubMed] [Google Scholar]
- 16.Anderson J S, Teutsch M, Dong Z, Wortis H H. Proc Natl Acad Sci USA. 1996;93:10966–10971. doi: 10.1073/pnas.93.20.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forssell J, Nilsson A, Sideras P. Scand J Immunol. 1999;49:155–161. doi: 10.1046/j.1365-3083.1999.00483.x. [DOI] [PubMed] [Google Scholar]
- 18.Brennan P, Babbage J W, Burgering B M, Groner B, Reif K, Cantrell D A. Immunity. 1997;7:679–689. doi: 10.1016/s1074-7613(00)80388-x. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez-Garcia A, Merida I, Martinez A C, Carrera A C. J Biol Chem. 1997;272:10220–10226. doi: 10.1074/jbc.272.15.10220. [DOI] [PubMed] [Google Scholar]
- 20.Leverrier Y, Thomas J, Mathieu A L, Low W, Blanquier B, Marvel J. Cell Death Differ. 1999;6:290–296. doi: 10.1038/sj.cdd.4400492. [DOI] [PubMed] [Google Scholar]
- 21.Aman M J, Lamkin T D, Okada H, Kurosaki T, Ravichandran K S. J Biol Chem. 1998;273:33922–33928. doi: 10.1074/jbc.273.51.33922. [DOI] [PubMed] [Google Scholar]
- 22.Astoul E, Watton S, Cantrell D. J Cell Biol. 1999;145:1511–1520. doi: 10.1083/jcb.145.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold M R, Scheid M P, Santos L, Dang-Lawson M, Roth R A, Matsuuchi L, Duronio V, Krebs D L. J Immunol. 1999;163:1894–1905. [PubMed] [Google Scholar]
- 24.Satterthwaite A B, Cheroutre H, Khan W N, Sideras P, Witte O N. Proc Natl Acad Sci USA. 1997;94:13152–13157. doi: 10.1073/pnas.94.24.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan W N, Alt F W, Gerstein R M, Malynn B A, Larsson I, Rathbun G, Davidson L, Muller S, Kantor A B, Herzenberg L A, et al. Immunity. 1995;3:283–299. doi: 10.1016/1074-7613(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 26.Fruman D A, Mauvais-Jarvis F, Pollard D A, Yballe C M, Brazil D, Bronson R T, Kahn C R, Cantley L C. Nat Genet. 2000;26:379–382. doi: 10.1038/81715. [DOI] [PubMed] [Google Scholar]
- 27.Lu-Kuo J M, Fruman D A, Joyal D M, Cantley L C, Katz H R. J Biol Chem. 2000;275:6022–6029. doi: 10.1074/jbc.275.8.6022. [DOI] [PubMed] [Google Scholar]
- 28.Carman J A, Wechsler-Reya R J, Monroe J G. J Immunol. 1996;156:4562–4569. [PubMed] [Google Scholar]
- 29.Solvason N, Wu W W, Kabra N, Wu X, Lees E, Howard M C. J Exp Med. 1996;184:407–417. doi: 10.1084/jem.184.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grumont R J, Rourke I J, Gerondakis S. Genes Dev. 1999;13:400–411. doi: 10.1101/gad.13.4.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Lobito A A, Shen F, Hornung F, Winoto A, Lenardo M J. Eur J Immunol. 2000;30:155–163. doi: 10.1002/1521-4141(200001)30:1<155::AID-IMMU155>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 32.Bustin S A. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 33.Solvason N, Wu W W, Parry D, Mahony D, Lam E W, Glassford J, Klaus G G, Sicinski P, Weinberg R, Liu Y J, Howard M, Lees E. Int Immunol. 2000;12:631–638. doi: 10.1093/intimm/12.5.631. [DOI] [PubMed] [Google Scholar]
- 34.Sherlock G. Curr Opin Immunol. 2000;12:201–205. doi: 10.1016/s0952-7915(99)00074-6. [DOI] [PubMed] [Google Scholar]
- 35.Kane L P, Shapiro V S, Stokoe D, Weiss A. Curr Biol. 1999;9:601–604. doi: 10.1016/s0960-9822(99)80265-6. [DOI] [PubMed] [Google Scholar]
- 36.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. Nature (London) 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 37.Kops G J, Burgering B M. J Mol Med. 1999;77:656–665. doi: 10.1007/s001099900050. [DOI] [PubMed] [Google Scholar]
- 38.Glynne R, Akkaraju S, Healy J I, Rayner J, Goodnow C C, Mack D H. Nature (London) 2000;403:672–676. doi: 10.1038/35001102. [DOI] [PubMed] [Google Scholar]
- 39.Leitges M, Schmedt C, Guinamard R, Davoust J, Schaal S, Stabel S, Tarakhovsky A. Science. 1996;273:788–791. doi: 10.1126/science.273.5276.788. [DOI] [PubMed] [Google Scholar]
- 40.Di Cristofano A, Kotsi P, Peng Y F, Cordon-Cardo C, Elkon K B, Pandolfi P P. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- 41.Bona C, Mond J J, Paul W E. J Exp Med. 1980;151:224–234. doi: 10.1084/jem.151.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bykowsky M J, Haire R N, Ohta Y, Tang H, Sung S S, Veksler E S, Greene J M, Fu S M, Litman G W, Sullivan K E. Am J Hum Genet. 1996;58:477–483. [PMC free article] [PubMed] [Google Scholar]
- 43.Satterthwaite A B, Willis F, Kanchanastit P, Fruman D, Cantley L C, Helgason C D, Humphries R K, Lowell C A, Simon M, Leitges M, et al. Proc Natl Acad Sci USA. 2000;97:6687–6692. doi: 10.1073/pnas.110146697. . (First Published May 30, 2000; 10.1073/pnas.110146697) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.