Abstract
Evolution of an oxygenic atmosphere required primordial life to accommodate the toxicity associated with reactive oxygen species. We have characterized an archaeal antioxidant from the hyperthermophilic acidophile Sulfolobus solfataricus. The amino acid sequence of this ≈22-kDa protein shares little sequence similarity with proteins with known function. However, the protein shares high sequence similarity with hypothetical proteins in other archaeal and bacterial genomes. Nine of these hypothetical proteins form a monophyletic cluster within the broad superfamily of ferritin-like diiron-carboxylate proteins. Higher order structural predictions and image reconstructions indicate that the S. solfataricus protein is structurally related to a class of DNA-binding protein from starved cells (Dps). The recombinant protein self assembles into a hollow dodecameric protein cage having tetrahedral symmetry (SsDps). The outer shell diameter is ≈10 nm, and the interior diameter is ≈5 nm. Dps proteins have been shown to protect nucleic acids by physically shielding DNA against oxidative damage and by consuming constituents involved in Fenton chemistry. In vitro, the assembled archaeal protein efficiently uses H2O2 to oxidize Fe(II) to Fe(III) and stores the oxide as a mineral core on the interior surface of the protein cage. The ssdps gene is up-regulated in S. solfataricus cultures grown in iron-depleted media and upon H2O2 stress, but is not induced by other stresses. SsDps-mediated reduction of hydrogen peroxide and possible DNA-binding capabilities of this archaeal Dps protein are mechanisms by which S. solfataricus mitigates oxidative damage.
Keywords: ferritin, iron, oxidative stress, biomineralization, hyperthermophile
The evolution of oxygenic photosynthesis marks the dawn of oxidative stress and represents one of the greatest selective pressures imposed on primordial life. The association of molecular oxygen with abundant ferrous iron pools created two major biological consequences. First, life dependent on the redox properties of Fe(II) would have to contend with its oxidation and precipitation as Fe(III). Second, life would have to contend with the toxicity of reactive oxygen species generated by the partial reduction of dioxygen by ferrous iron. Iron metabolism and oxidative stress are thus intimately interwoven, with the toxicity of one being dependent on the other. The significance of these selective pressures remains evident in the genomes of modern organisms where complex and overlapping defense mechanisms protect against oxygen toxicity and iron stress (1–3).
Oxidative stress is a universal phenomenon experienced by both aerobic and anaerobic organisms from all three domains of life (4, 5). Although the significance of oxidative stress in biology is well established, the processes by which intracellular reactive oxygen species (ROS) are generated continue to be recognized. One pathway generally recognized as a significant source of ROS is the partial reduction of molecular oxygen by the autoxidation of flavoproteins (5, 6). This process generates a mixture of superoxide (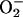 ) and hydrogen peroxide (H2O2). The strong anionic charge of superoxide limits its reactivity with electron-rich molecules such as nucleic acids and amino acids (5, 7). However, metal clusters within proteins are highly susceptible to superoxide. For example, Fe(II) ions at the catalytic center of 4Fe–4S-containing enzymes are electrostatically attractive targets (8–10). Oxidized clusters are unstable and degrade, resulting in the release of iron and the inactivation of the enzyme. The released iron is available to react with hydrogen peroxides through the well characterized Fenton reaction (described below) to produce hydroxyl radicals (HO•), the most toxic of all ROS (11). Hydroxyl radicals indiscriminately oxidize most biomolecules at nearly diffusion-limiting rates.
) and hydrogen peroxide (H2O2). The strong anionic charge of superoxide limits its reactivity with electron-rich molecules such as nucleic acids and amino acids (5, 7). However, metal clusters within proteins are highly susceptible to superoxide. For example, Fe(II) ions at the catalytic center of 4Fe–4S-containing enzymes are electrostatically attractive targets (8–10). Oxidized clusters are unstable and degrade, resulting in the release of iron and the inactivation of the enzyme. The released iron is available to react with hydrogen peroxides through the well characterized Fenton reaction (described below) to produce hydroxyl radicals (HO•), the most toxic of all ROS (11). Hydroxyl radicals indiscriminately oxidize most biomolecules at nearly diffusion-limiting rates.
The identification of superoxide dismutase (SOD) provided the first indication of active mechanisms designed to specifically minimize the effects of oxidative stress (12). This enzyme catalyzes the disproportionation of superoxide (Eq. 1):
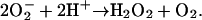 |
[1] |
SOD genes are ubiquitous in the genomes of aerobic organisms, and homologous genes are found in the genomes of some anaerobes. The apparent absence of a recognizable SOD gene in the genomes of some anaerobes may be a reflection of this enzyme's generation of toxic oxygen (13). A second mechanism, originally identified in Pyrococcus furiosus has been described for superoxide detoxification (4, 13). This mechanism utilizes a mononuclear iron-containing enzyme, super oxide reductase (SOR), for the efficient reduction of superoxide to H2O2 without an O2 byproduct (Eq. 2):
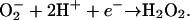 |
[2] |
H2O2 is a powerful oxidant produced by both superoxide reductase and superoxide dismutase reactions. Potential targets of H2O2 oxidation include 4Fe–4S clusters and the sulfur atoms of cysteine and methionine residues, with potentially lethal consequences (5). More importantly, the interaction of free Fe(II) with hydrogen peroxide is known to efficiently generate HO• through the Fenton reaction (Eq. 3) (11, 14):
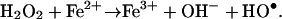 |
[3] |
Peroxide detoxification and iron metabolism are important components in mitigating oxidative damage. Other enzymes, such as catalase, affecting peroxide detoxification have been characterized. Although these enzymes efficiently remove H2O2, they do little to minimize the toxicity of free iron.
A DNA-binding protein from nutrient-starved Escherichia coli cells (Dps), which protects nucleic acids against oxidative damage, has been described (15). Twelve copies of the Dps protein subunit self-assemble into an ≈10-nm cage-like structure. These structures are homologous to the multimeric assemblies formed by the iron-mineralizing family of ferritin proteins. In addition to their structural similarities, assembled Dps proteins also mineralize iron within the interior of the protein cage. However, Dps mineralization is distinct from that described for ferritins. Whereas the ferroxidase activity of ferritins preferentially use O2 as an oxidant, the ferroxidase center in Dps proteins more efficiently uses H2O2 as the oxidant according to Eq. 4 (16, 17):
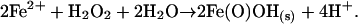 |
[4] |
In this way, Dps simultaneously eliminates the two components of the Fenton reaction [Fe(II) and H2O2] that contribute to the generation of hydroxyl radicals. This mineralization reaction, in combination with the in vivo association of DPS with DNA, represents a multifunctional approach to cellular protection against oxidative damage.
We are interested in the evolutionarily conserved mechanism by which iron-dependent life manages the paradoxical relationship between iron and oxygen. Hyperthermophilic archaea are deeply rooted in the tree of life and therefore may harbor ancient mechanisms for managing oxidative stress. Sulfolobus solfataricus is an aerobic hyperthermophilic (70–90°C) archaeon that thrives in acidic terrestrial thermal features that are commonly associated with high iron (18). We have identified a previously unrecognized protein from S. solfataricus that functions as a Dps. Here, we provide evidence supporting the role of this protein in protection against oxidative damage.
Materials and Methods
Identification of S. solfataricus Dps (SsDps). The S. solfataricus (P2) genome was queried by using the blast algorithm, with ferritin and Dps-like proteins from several sources [i.e., human, horse, bullfrog, E. coli, Listeria innocua, Archeaoglobus fulgidus, and an uncultured crenarchaeote (4B7)] (19). A putative ferritin from the uncultured crenarchaeote 4B7 was the only sequence sharing detectable similarity to any annotated protein in the S. solfataricus (P2) genome (20). The identified protein, annotated as a hypothetical protein (gi:15898865) was subjected to a threading routine and submitted to the fold recognition server 3D-pssm (www.sbg.bio.ic.ac.uk/servers/3dpssm/) (21).
Phylogenetics. Amino acid sequences of ferritin-like diiron carboxylate proteins were aligned with clustalx (22). Parsimony analysis was performed with test version 4.0b10 of paup* and bootstrapped with 10,000 resamplings (23).
Culturing of S. solfataricus. Liquid cultures of S. solfataricus (P2), were grown aerobically in a revised version of an established salt-based medium: 22.70 mM (NH4)2SO4, 2.19 mM K2HPO4·3H2O, 13.42 mM KCl, 9.33 mM glycine, 4.55 μM MnCl2·4H2O, 9.44 μM Na2B4O7·2H2O, 382.56 nM ZnSO4·7H2O, 146.64 nM CuCl2·2H2O, 61.99 nM Na2MoO4·2H2O, 59.29 nM VOSO4·5H2O, 17.79 nM CoSO4·7H2O, and 19.02 nM NiSO4·6H2O, supplemented with 0.2% tryptone (24). Iron-depleted versions of this medium were prepared by using an iron-specific chelating column, phosphorylated silica polyamine composite (D. Nielsen and E. Rosenberg, personal communication). Iron-depleted media were used directly or supplemented with FeSO4 or FeCl3. All cultures were grown in long neck Erlenmeyer flasks at 78°C in shaking oil bath incubators.
Western Analysis. Polyclonal antibodies were raised in mice against purified recombinant SsDps protein. IgG antibodies were isolated by using the ImmunoPure purification kit according to the manufacturer's protocol (Pierce). Western blots were performed by using the purified IgG. S. solfataricus cells were harvested by centrifugation and resuspended in 2× SDS gel-loading buffer (25). Lanes were loaded with approximately equal cell numbers according to OD650. S. solfataricus proteins were separated on 15% SDS-polyacrylamide gels and transferred to Hybond-ECL nitrocellulose membrane (Amersham Pharmacia Bioscience). Colorometric detection was used to identify the SsDps protein (Bio-Rad). Protein and RNA concentrations were estimated by using AlphaImager (Alpha Innotech, San Leandro, CA) imaging system (IS-2200) according to average pixel density.
Northern Analysis. Total cellular RNA was extracted from S. solfataricus (P2) cells, according to the TRI Reagent protocol (Molecular Research Center, Cincinnati). Total RNA concentrations were estimated by agarose gel electrophoresis and by spectrophotometric measurements (OD260). For Northern analysis, 0.5–2 μg of total RNA was separated by electrophoresis in a 1.5% agarose (wt/vol) formaldehyde gel and transferred to GeneScreen membranes as recommended by the manufacturer (NEN). RNA was membrane-crosslinked in a UV Stratalinker (Stratagene). Blots were probed with 32P-labeled PCR products (Ready-To-Go Labeling Kit, Amersham Pharmacia Bioscience). S. solfataricus DNA was isolated by using previously established methods and served as the template in PCRs (26).
Cloning and Expression. PCR primers were designed to amplify the dps-like gene from S. solfataricus (P2) (gi:15898865). The forward primer (5′-gggtgtacatatgcaagagaaacccc-3′) included an NdeI restriction endonuclease site (in bold) directly upstream of the start codon (underlined). The reverse primer (5′-acggatccttatttcttggaatatggagcg-3′) included the stop codon for the gene (underlined) and a BamHI site (in bold). The resulting PCR product was digested with NdeI and BamHI restriction endonucleases, purified with a PCR purification kit (Qiagen, Valencia, CA), ligated into pET-30a(+) (Novagen), and transformed into Xl-2 blue E. coli (Stratagene). The cloned sequence was confirmed by DNA sequencing (Applied Biosystems). The plasmid was subsequently transformed into BL21 E. coli for protein expression (Novagen). For protein expression, E. coli were grown and induced according to the manufacturer's protocol (Novagen). Cells were harvested by centrifugation and screened for SsDps expression by SDS/PAGE.
SsDps Purification from E. coli. Cells were pelleted from 1-liter cultures by centrifugation, resuspended in 30 ml of lysis buffer (50 mM Mes/100 mM NaCl, pH 6.5/0.002 mg/ml DNase/0.05 mg/ml RNase/1 mg/ml lysozyme), and incubated for 30 min at room temperature. The slurry was sonicated (3 × 5 min), and cellular debris was removed by centrifugation. The resulting supernatant was heated at 65°C for 10 min, cooled on ice, and then centrifuged to remove denatured proteins. The recovered supernatant was passed through a 0.2-μm filter and loaded onto a Superose 6 size exclusion column (Amersham Pharmacia Biosciences) equilibrated with Mes buffer (50 mM Mes/100 mM NaCl, pH 6.5). Elution of the protein was monitored at 260, 280, and 410 nm. Protein concentration was determined by the biuret method, and confirmed by the molar absorptivity at 280 nm of 3.00 × 104 M-1·cm-1.
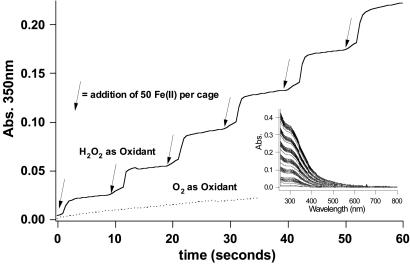
SsDps Fe(O)OH Mineralization. An amount equal to 0.2 mg of SsDps (7.66 × 10-7 mmol) was diluted with 3 ml of Mes buffer (100 mM Mes/100 mM NaCl, pH 6.5) in a standard quartz cuvette. A total of 2.25 × 10-4 mmol of deaerated Fe(II) was added in six increments (50 Fe per cage) at 10-min intervals. A half molar equivalent of hydrogen peroxide (1 H2O2:2 Fe2+) was delivered 2 min after each iron addition. A UV-Vis spectrum was recorded every 2 sec to monitor Fe(O)OH core formation by the increase in the characteristic absorbance at 350–400 nm (27).
SsDps Particle Characterization. Dynamic light scattering was performed on the assembled protein before and after mineralization to determine particle size as described (28). Particle size was independently estimated by mobility on size exclusion columns (Superose 6, Amersham Pharmacia Biosciences) and by transmission electron microscopy (Leo 912AB TEM, Oberkochen, Germany) operating at 120 keV. Samples for TEM were imaged both unstained and stained with 2% uranyl acetate. Mass of the SsDps protein was determined by electrospray mass spectrometry (LC-MS).
Electron Microscopy and 3D Image Reconstruction. Aliquots of un-mineralized SsDps (≈3 μl) were stained for 30 s with 2% uranyl acetate. Images were recorded by using a CM120 electron microscope (FEI/Philips, Eindhoven, The Netherlands) at a magnification of 60,000 ± 1% and focal pairs with a range of ≈0.5–2.5 μm underfocus. Negatives were digitized on a Zeiss SCAI flat-bed scanning densitometer (ZI/Zeiss) with a step size of 7 μm, which resulted in a pixel size of 0.117 nm on the object scale.
Image processing was performed with polar Fourier transform (PFT) methods, with a modification for tetrahedral symmetry by D. Belnap (29). Approximately 2,230 particle pairs were manually extracted by using the program x3d, and the images were scaled to the mean and SD for all images (30). The contrast transfer function (CTF) parameters for each micrograph were determined from the computed Fourier transform of the carbon film of each micrograph, and phase corrections were then applied to each particle image. A 3-nm starting model was calculated from the x-ray structure of a ferritin-like protein isolated from L. innocua (PDB ID code 1QGH) for the reference-based alignment. The x,y origin and rotational orientation of the particles were iteratively refined. For each cycle, particles that deviated by 0.5 below the average were rejected. In general, approximately one-quarter of the particles were rejected. Two independent reconstructions were computed to estimate the resolution of the refined data by Fourier shell correlation and subsequently combined. By using a correlation coefficient cut-off value of 0.5, the resolution was estimated to be 1.8 nm. The final 3D map was derived from 1,402 image pairs.
Results
SsDps Identification. Using the default parameters of blast, we were unable to identify any amino acid sequence in the S. solfataricus (P2) genome related to characterized ferritin or Dps proteins. However, a blast search using a putative ferritin identified in an uncultured crenarchaeote (AAK66802) matched an S. solfataricus (P2) sequence (gi:15898865) with 55% identity. This S. solfataricus (P2) ORF (SSO2079), annotated as a conserved hypothetical protein, codes for a predicted 188 aa with an estimated molecular mass of 21,753 Da. This sequence threads onto the four-helix bundle structure of the ferritin-like protein from L. innocua, with high confidence [position-specific scoring matrix (PSSM) E = 0.0171] (17, 31, 32). Based on this predicted secondary and tertiary structural similarity, the S. solfataricus (P2) protein was expected to assemble into a dodecameric cage similar to Dps family proteins (33).
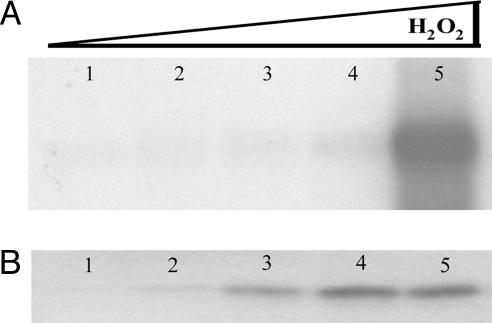
In Vivo Expression Patterns of SsDps. Northern blot analysis of RNA isolated from exponential growth phase culture of S. solfataricus (P2) identifies a single transcript corresponding to the approximate size of the ssdps gene (≈600 nt). The transcript is up-regulated in a nonlinear fashion in response to oxidative stress (Fig. 1A), with a dramatic increase observed at 30 μMH2O2. Western blots indicate a more linear accumulation of the SsDps protein in response to increasing concentrations of H2O2 (Fig. 1B). Loading controls and densitometry are available in Fig. 8, which is published as supporting information on the PNAS web site.
Fig. 1.
SsDps expression in response to oxidative stress. (A) Northern blot analysis of RNA extracted from exponential growth phase (OD650 ≈0.5) S. solfataricus cells. Lane 1, standard growth conditions (0 μM H2O2); lane 2, 5 μM H2O2; lane 3, 10 μM H2O2; lane 4, 20 μM H2O2; lane 5, 30 μM H2O2. Approximately 1.5 μg of total RNA was loaded in each lane. (B) Western blot analysis of total protein extract from logarithmic (OD650 ≈0.5) S. solfataricus cells. Loads are normalized according to number of cells, and lane assignments are as above. Densitometry and loading controls are available in Fig. 8.
Taking advantage of this oxidative induction, we were able to isolate ≈0.3 mg of the assembled protein from a 1-liter culture of S. solfataricus (P2) grown in media supplemented with 30 μM H2O2. No detectable amounts of this protein could be purified from S. solfataricus (P2) culture grown under standard conditions following this same protocol. These results confirmed our prior observations that neither the transcript nor the protein is readily detectable under normal growth conditions (Fig. 1).
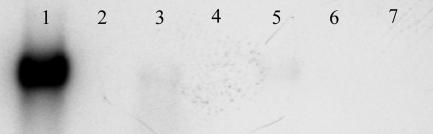
We further examined the possibility of SsDps being involved in a general stress response rather then being exclusively related to oxidative stress (Figs. 2 and 9, which is published as supporting information on the PNAS web site). Transcription of the ssdps gene is low or not detectable in late log phase cells or in exponential growth phase cells UV irradiated (300 KJ), grown in an alternative carbon source (sucrose), virus-infected [Sulfolobus spindle-shaped virus, Ragged Hills (SSV RH)], heat-shocked at 90°C, or cold-shocked at 60°C for 11 h (Fig. 2). However, consistent with the characterized expression profiles of other Dps-like proteins, ssdps is expressed under iron-limiting growth conditions (Fig. 3) (2, 3, 34). Dps proteins use ferrous iron atoms as cofactors for the two-electron reduction of H2O2. It follows then that, in the absence of iron, Dps-mediated reduction of hydrogen peroxide will be compromised, resulting in increased H2O2 stress.
Fig. 2.
Ssdps expression is specific to oxidative stress. Shown is Northern blot analysis of RNA extracted from S. sulfataticus cultures. Lane 1, cells cultured in media supplemented with 30 μMH2O2; lane 2, late log phase cells; lane 3, 300 KJ UV, which accounts for RNA degradation as shown in the load control; lane 4, sucrose as the sole carbon source; lane 5, SSV RH (Sulfolobus spindle-shaped virus, Ragged Hills)-infected; lane 6, 11 h heat shock at 90°C; lane 7, 11 h cold shock at 60°C. Approximately 0.75 μg of total RNA was loaded in each lane. Loading controls are available in Fig. 9.
Fig. 3.
SsDps expression in response to iron. Western blot analysis of total protein extract from exponential growth phase (OD650 ≈0.5) S. solfataricus cells. Loads were normalized according to number of cells. Lane 1, standard growth conditions; lane 2, iron-extracted media; lane 3, iron-extracted media supplemented with 1.25 μM Fe2SO4; lane 4, iron-extracted media supplemented with 1.25 mM Fe2SO4; lane 5, iron-extracted media supplemented with 5.0 mM Fe2SO4; lane 6, iron-extracted media supplemented with 5.0 mM Fe2Cl3; lane 7, molecular weight standard; lane 8, 2 ng of purified SsDps.
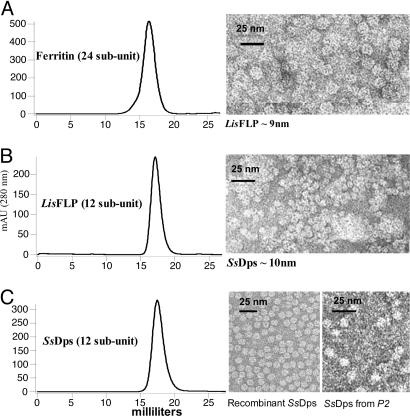
Particle Purification and Characterization. The SsDps protein expresses to high levels in E. coli. Approximately 40 mg of the recombinant protein can be purified from a 1-liter E. coli culture. This yield compares with 0.3 mg of the protein that could be purified from 1-liter cultures of S. solfataricus stressed with H2O2. Purification of the recombinant SsDps protein was simplified by taking advantage of the protein's inherent thermal stability. A 10-min incubation at 65°C was sufficient to denature a majority of the E. coli proteins, which were precipitated by a low-speed centrifugation. The recombinant protein eluted as a single peak on an analytical size exclusion column (Fig. 4). SDS/PAGE of the purified protein revealed a single band with molecular mass of ≈22 kDa (Fig. 10, which is published as supporting information on the PNAS web site). This finding was in good agreement with the deconvoluted mass of the recombinant protein (21,768.23 Da), as determined by electrospray mass spectrometry.
Fig. 4.
Size exclusion liquid chromatography elution profiles and TEM images of corresponding peaks. Retention times according to size exclusion liquid chromatography are shown for the 24-subunit horse spleen ferritin (A), the 12-subunit ferritin-like protein (Flp) from L. innocua (B), and the 12-subunit Dps-like protein from S. solfataricus (C). The L. innocua FLP and the Dps-like protein from S. solfataricus have retention times consistent with a 12-subunit, 260-kDa protein, whereas the 24-subunit ferritin from horse spleen elutes earlier. Transmission electron microscopy of each peak reveals intact cage diameters of ≈13 nm (A), ≈9 nm (B), and ≈10 nm (C), commensurate with each protein's retention time.
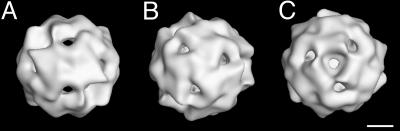
The recombinant SsDps protein self-assembles into a 12-subunit cage-like architecture. Dynamic light scattering (DLS) of the recombinant protein indicates an average diameter of 9.7 ± 0.4 nm, consistent with a 12-subunit cage-like assembly. This architecture is directly evident from negatively stained TEM (Fig. 4). SsDps image reconstruction, at 1.8 nm resolution, reveals a dodecameric protein cage with an exterior diameter of ≈100 Å and an interior cavity of ≈5 nm (Fig. 5). As a tetrahedron, the structure is composed of a trimer of dimers that assembles to form two nonequivalent three-fold (3F) environments at opposing ends of the 3F axis (Fig. 5 B and C).
Fig. 5.
3D image reconstruction of the assembled SsDps cage. Surface-shaded views of reconstructed negative-stained images displayed along the twofold (2F) axis (A), and along the two nonequivalent environments at each end of the threefold (3F) axis (B and C). (Scale bar: 2.5 nm).
SsDps Mineralization. At pH 6.5, the recombinant SsDps protein efficiently catalyzes the oxidation of Fe(II) to Fe(III) in the presence of H2O2 (Fig. 6). Substitution of O2 for H2O2 results in significantly retarded rates of Fe oxidation. This result is in contrast to Fe(II) oxidation by ferritins, which efficiently catalyze the oxidation of Fe(II) with O2. Fe oxidation reactions were monitored by absorbance of the Fe(O)OH at 350–400 nm (27). Fe(II) was added to the protein in aliquots of 50 iron atoms per protein cage and oxidized in the presence of H2O2. This iron loading process (50 Fe:25 H2O2) could be repeated six times, resulting in a theoretical load of 300 iron atoms per cage. Iron mineralized (Fe(O)OH) preparations of SsDps exhibited no change in hydrodynamic diameter before and after the mineralization reaction. Electron dense cores commensurate with the ≈5-nm interior cavity could be visualized by TEM. This finding is indicative of a spatially controlled reaction in which Fe(O)OH particles are formed within the confines of the protein cage, analogous to ferritin and Dps biomineralization.
Fig. 6.
SsDps-catalyzed mineralization of iron. SsDps efficiently uses H2O2 to oxidize iron in a stepwise progression (1H2O2:2Fe) (solid line). In contrast, O2 serves as a relatively poor oxidant of iron in this reaction (dotted line). (Inset) Iron confined within the SsDps cage remains soluble as shown in the full-spectrum.
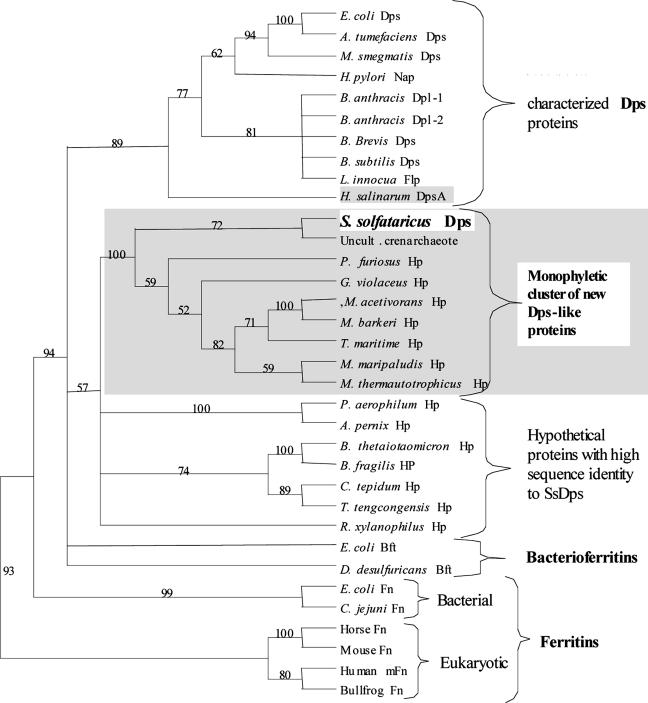
Phylogenetic Identification of a New Dps Subclass. The SsDps protein shares high sequence similarity with hypothetical proteins in 14 prokaryotic genomes (expect ≥7e-04). Nine of these sequences form a well supported monophyletic cluster outside of the currently characterized Dps clade (Fig. 7). These sequences are only distantly related to those in the currently characterized Dps clade. Despite this distant relationship, a putative Dps-like metal-binding motif (diiron-binding site) is identifiable in the primary structures of each of these new Dps-like sequences (sequence alignments are in Fig. 11, which is published as supporting information on the PNAS web site). Whereas this putative metal-binding motif is distinct from that identified in currently recognized Dps proteins, it does share similar charge and spacing characteristics with the known Dps metal-binding sites.
Fig. 7.
Phylogenetic analysis of the ferritin-like diiron-carboxylate superfamily. The Dps protein from S. solfataricus is more closely related to a group of hypothetical proteins than it is to other characterized members of the ferritin-like diiron-carboxylate superfamily. H. salinarum DpsA (shaded) and S. solfataricus Dps are currently the only examples of archaeal Dps proteins; all other characterized Dps proteins are bacterial. Database gene identification numbers are as follows: E. coli Dps (Escherichia coli) gi:16128780, A. tumefaciens Dps (Agrobacterium tumefaciens) gi:15889746, M. smegmatis Dps (Mycobacterium smegmatis) gi:17887432, H. pylori Nap (Helicobacter pylori) gi:15611298, B. anthracis Dpl-1 (Bacillus anthracis) gi: 21730371, B. anthracis Dpl-2 (Bacillus anthracis) gi: 21730378, B. Brevis Dps (Bacillus brevis) gi:31615600, B. subtilis Dps (Bacillus subtilis) gi:16080351, L. innocua Flp (Listeria innocua) gi:16800011, H. salinarum DpsA (Halobacterium salinarum sp. NRC-1) gi:15791220, S. solfataricus Dps (Sulfolobus solfataricus) gi:15898865, Uncult. crenarchaeote (uncultured crenarchaeote non-heme iron-containing protein) gi:14548145, P. furiosus Hp (Pyrococcus furiosus) gi:18977565, G. violaceus Hp (Gloeobacter violaceus) gi:37523861, M. acetivorans Hp (Methanosarcina acetivorans) gi:20091704, M. barkeri Hp (Methanosarcina barkeri) gi:48837814, T. maritima Hp (Thermotoga maritima) gi: 15644274, M. maripaludis Hp (Methanococcus maripaludis) gi:45358735, M. thermautotrophicus Hp (Methanothermobacter thermautotrophicus) gi:7482214, P. aerophilum Hp (Pyrobaculum aerophilum) gi:18313528, A. pernix Hp (Aeropyrum pernix) gi:14601419, B. thetaiotaomicron Hp (Bacteroides thetaiotaomicron) gi:29349231, B. fragilis HP (Bacteroides fragilis) gi:53714735, C. tepidum Hp (Chlorobium tepidum) gi:21674150, T. tengcongensis Hp (Thermoanaerobacter tengcongensis) gi:20808605, R. xylanophilus Hp (Rubrobacter xylanophilus) gi:46106146, E. coli Bft (Escherichia coli) gi:16131215, D. desulfuricans Bft (Desulfovibrio desulfuricans) gi:14326006, E. coli Fn (Escherichia coli) gi:16129855, C. jejuni Fn (Campylobacter jejuni) gi:15791972, Horse Fn (Equus caballus L-chain ferritin) gi:406209, Mouse Fn (Mus musculus L-chain ferritin) gi:55154579, Human mFn (Homo sapiens mitochondrial ferritin) gi:29126241, and Bullfrog Fn (Rana catesbeiana) gi:85895. Protein abbreviations are as follows: Hp, hypothetical protein; Dps, DNA-binding protein from nutrient-starved cells; Bft, bacterioferritin; Fn, ferritin; mFn, mitochondrial ferritin; Dpl, Dps-like protein; Flp, ferritin-like protein; and Nap, neutrophil-activating protein. Numbers at branching nodes are bootsrap values from 10,000 resamplings.
Dps proteins are widely distributed across the bacterial domain; however, before this study, only one other archaeal Dps had been described (DpsA) (35). Interestingly, our phylogenetic analysis indicates that the DpsA protein from Halobacterium salinarum is more closely related to members of the previously identified Dps subclass than it is to the new, predominantly archaeal, Dps subclass identified in this study.
Dps Surface Charge and DNA-binding Domains. Surface charge determined by zeta potential measurements of the SsDps assembly indicate that the cage retains a net negative surface charge at pH values >4.1. This overall net negative surface charge is consistent with all other characterized Dps proteins irrespective of their DNA-binding character. Although the DNA-binding domain in Dps proteins are not well defined, basic residues in either the N-terminal or C-terminal regions have been implicated. For example, extension of the N-terminal E. coli Dps subunit or in the C-terminal extension of the Mycobacterium smegmatis Dps subunit have been implicated in DNA binding (33, 36, 37). These suggestions have recently been corroborated by N-terminal deletion mutants of E. coli Dps that do not bind DNA (38). Multiple sequence alignments identify basic residues in extended N-terminal domains of both the SsDps and in the Dps-like protein from H. salinarum, suggesting their role in DNA association (35, 39, 40). A short C-terminal extension of the SsDps protein also contains three basic amino acids that may also play a role in DNA binding (Fig. 11). Further investigations of DNA binding by archaeal Dps's will be important for appreciating the significance of Dps proteins in Archaea.
Discussion
The evolution of oxygenic photosynthesis marks the dawn of oxidative stress and represents one of the greatest selective pressures imposed on primordial life. The evolution of a single protein capable of managing the paradoxical relationship between iron and oxygen may represent an important ancestral component of the antioxidant defense system. We have identified and characterized a Dps-like protein from the hyperthermophilic archaeon S. solfataricus (SsDps). Although the primary structure of this protein is only distantly related to characterized Dps proteins, the SsDps protein does assemble into dodecameric cage-like structures that efficiently oxidize Fe(II) to Fe(III) using H2O2 as an oxidant. In all previously characterized Dps proteins, this redox reaction is mediated by a unique diiron-binding motif (Fig. 11) (16, 33, 36, 37, 41). This motif has been implicated in coordinating the two-electron reduction of H2O2. This mechanism has been shown to avoid the production of toxic hydroxyl radicals generated through the Fenton reaction (Eq. 4) (16). A putative Dps diiron-binding motif is identifiable in the primary structure of each new Dps sequence. Whereas this putative metal-binding motif is distinct from that described in previously recognized Dps proteins, it does share similar characteristics with known Dps metal-binding sites, including charge and motif spacing.
In exponential growth, the primary role of previously characterized Dps proteins is to minimize H2O2 stress. In vivo expression patterns of SsDps are consistent with this protein's role in mitigating oxidative damage. SsDps seems to be exclusively up-regulated in exponential growth phase cultures of S. solfataricus in response to H2O2 stress and does not function in a general stress response. However, SsDps is up-regulated in exponential growth phase cultures of S. solfataricus under iron-limited conditions. Expression of this protein under iron-limiting conditions seems contradictory, given the well established role of iron in oxidative stress. However, Dps proteins use two ferrous iron ions as cofactors for the two-electron reduction of H2O2. In the absence of ferrous iron, Dps-mediated reduction of hydrogen peroxide is compromised, resulting in the continued accumulation of H2O2. This increase in H2O2 likely induces the up-regulation of the SsDps protein. Although other cellular mechanisms are present to minimize H2O2 stress (42–46), Dps proteins play a proven role in mitigating oxidative damage. Dps knockouts have been shown to be more sensitive to H2O2-mediated oxidative damage during exponential growth (47). Whereas dps expression is generally considered specific to oxidative stress during exponential growth, Dps proteins have been shown to effectively protect against a broad spectrum of different stresses while in stationary phase (48). SsDps is also likely to confer protection against a broader range of environmental stress during stationary phase; however, growth of S. solfataricus is limited by the accumulation of inorganic ions and thus complicates studies aimed at understanding S. solfataricus physiologies under true nutrient limitation (49). Nevertheless, it is clear that the oxidative stress response in S. solfataricus is coupled to both H2O2 and iron levels through the expression of ssdps.
It is of interest to recognize that sequences related to the SsDps protein are not limited to hyperthermophilic Archaea. In fact, blast analysis indicates that SsDps-like sequences are well distributed across the spectrum of phylogenetically diverse prokaryotes. This finding suggests that SsDps-like sequences are not evolutionarily maintained, solely as a consequence of protracting thermal stability. A case in point is the identification of a SsDps-like sequence (85% similar) in the genome of a mesophilic bacteria (Gloeobacter violaceus PCC 7421) (50). Although the amino acid sequence of this Dps-like protein is highly conserved, the nucleotide sequence is not. The low G plus C content of the ssdps gene (40%) is typical of genes originating from hyperthermophiles and is consistent with that of S. solfataricus (P2) genome. In contrast, the relatively high G plus C content (60%) of the dps-like gene from G. violaceus (gi:37523861) is consistent with that of the overall G plus C content of the genome. This finding suggests that the distribution of this newly identified dps gene is not a result of a recent horizontal gene transfer event.
This newly identified Dps protein fits within the broad ferritin-like diiron-carboxylate protein superfamily (Fig. 7). Other members of this superfamily include other Dps proteins, ferritins, and bacterioferritins, which form three distinct subclasses within the superfamily (51). All proteins in this superfamily assemble into multimeric cage-like structures that functionally sequester iron. However, unlike the 24-subunit ferritins and bacterioferritins that function to sequester iron when concentrations are high, Dps proteins are 12-subunit assemblies that functionally sequester iron only as a consequence of reducing H2O2. Identification of an amino acid sequence in the S. solfataricus genome that self-assembles into a dodecameric cage and functions to mitigate oxidative damage has allowed us to confidently assign Dps-like function to genes from nine different organisms whose function was previously unknown. The identification of a previously uncharacterized subclass of Dps genes provides further insights into the evolution and diversity of biochemical adaptations to oxidative stress.
Supplementary Material
Acknowledgments
We thank S. Brumfield, L. Liepold, Z. Varpness, D. Nielsen, and M. Lavin for invaluable technical assistance. This investigation was supported by Office of Naval Research Grant N00014-04-1-0673, National Science Foundation Grant MCB 01322156, and National Institutes of Health Grants GM54076 and GM 066087, and was supported through the National Aeronautics and Space Administration support of the Thermal Biology Institute's Life in Extreme Environments Program (NAG5-8807).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Dps, DNA-binding protein from nutrient-starved cells; TEM, transmission electron microscopy; SsDps, Sulfolobus solfataricus Dps.
References
- 1.Touati, D., Jacques, M., Tardat, B., Bouchard, L. & Despied, S. (1995) J. Bacteriol. 177, 2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, L., James, L. P. & Helmann, J. D. (1993) J. Bacteriol. 175, 5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, L. & Helmann, J. D. (1995) Mol. Microbiol. 18, 295-300. [DOI] [PubMed] [Google Scholar]
- 4.Jenney, F. E., Jr., Verhagen, M. F. J. M., Cui, X. Y. & Adams, M. W. W. (1999) Science 286, 306-309. [DOI] [PubMed] [Google Scholar]
- 5.Imlay, J. A. (2003) Annu. Rev. Microbiol. 57, 395-418. [DOI] [PubMed] [Google Scholar]
- 6.Massey, V. (1994) J. Biol. Chem. 269, 22459-22462. [PubMed] [Google Scholar]
- 7.Sawyer, D. T. & Valentine, J. S. (1981) Acc. Chem. Res. 14, 393-400. [Google Scholar]
- 8.Liochev, S. I. & Fridovich, I. (1994) Free Radical Biol. Med. 16, 29-33. [DOI] [PubMed] [Google Scholar]
- 9.Liochev, S. I. (1996) Free Radical Res. 25, 369-384. [DOI] [PubMed] [Google Scholar]
- 10.Flint, D. H., Tuminello, J. F. & Emptage, M. H. (1993) J. Biol. Chem. 268, 22369-22376. [PubMed] [Google Scholar]
- 11.Sawyer, D. T., Sobkowiak, A. & Matsushita, T. (1996) Acc. Chem. Res. 29, 409-416. [Google Scholar]
- 12.McCord, J. M. & Fridovic, I. (1969) J. Biol. Chem. 244, 6049-6055. [PubMed] [Google Scholar]
- 13.Adams, M. W. W., Jenney, F. E., Jr., Clay, M. D. & Johnson, M. K. (2002) J. Biol. Inorg. Chem. 7, 647-652. [DOI] [PubMed] [Google Scholar]
- 14.Kehrer, J. P. (2000) Toxicology 149, 43-50. [DOI] [PubMed] [Google Scholar]
- 15.Almiron, M., Link, A. J., Furlong, D. & Kolter, R. (1992) Genes Dev. 6, 2646-2654. [DOI] [PubMed] [Google Scholar]
- 16.Zhao, G., Ceci, P., Ilari, A., Giangiacomo, L., Laue, T. M., Chiancone, E. & Chasteen, N. D. (2002) J. Biol. Chem. 277, 27689-27696. [DOI] [PubMed] [Google Scholar]
- 17.Su, M., Cavallo, S., Stefanini, S., Chiancone, E. & Chasteen, N. D. (2005) Biochemistry 44, 5572-5578. [DOI] [PubMed] [Google Scholar]
- 18.She, Q., Singh, R. K., Confalonieri, F., Zivanovic, Y., Allard, G., Awayez, M. J., Chan-Weiher, C. C.-Y., Clausen, I. G., Curtis, B. A., De Moors, A., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J. H., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein, J. L., Marsh, T. L., Wu, K. Y., Shizuya, H. & DeLong, E. F. (1996) J. Bacteriol. 178, 591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelley, L. A., MacCallum, R. M. & Sternberg, M. J. E. (2000) J. Mol. Biol. 299, 499-520. [DOI] [PubMed] [Google Scholar]
- 22.Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G. & Gibson, T. J. (1998) Trends Biochem. Sci. 23, 403-405. [DOI] [PubMed] [Google Scholar]
- 23.Swofford, D. L. (2001) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b10.
- 24.Zillig, W., Kletzin, A., Schleper, C., Holz, I., Janekovic, D., Hain, J., Lanzendorfer, M. & Kristjansson, J. K. (1994) Syst. Appl. Microbiol. 16, 609-628. [Google Scholar]
- 25.Maniatis, T., Fritsch, E. F. & Shambrook, J. (1982) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 26.Stedman, K. M., Schleper, C., Rumpf, E. & Zillig, W. (1999) Genetics 152, 1397-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornell, R. M. & Schwertmann, U. (2003) The Iron Oxides (Wiley-VCH Verlag, Weinheim, Germany).
- 28.Allen, M., Willits, D., Young, M. & Douglas, T. (2003) Inorg. Chem. 42, 6300-6305. [DOI] [PubMed] [Google Scholar]
- 29.Baker, T. S. & Cheng, R. H. (1996) J. Struct. Biol. 116, 120-130. [DOI] [PubMed] [Google Scholar]
- 30.Conway, J. F., Cheng, N., Zlotnick, A., Wingfield, P. T., Stahl, S. J. & Steven, A. C. (1997) Nature 386, 91-94. [DOI] [PubMed] [Google Scholar]
- 31.Ilari, A., Stefanini, S., Chiancone, E. & Tsernoglou, D. (2000) Nat. Struct. Biol. 7, 38-43. [DOI] [PubMed] [Google Scholar]
- 32.Bozzi, M., Mignogna, G., Stefanini, S., Barra, D., Longhi, C., Valenti, P. & Chiancone, E. (1997) J. Biol. Chem. 272, 3259-3265. [DOI] [PubMed] [Google Scholar]
- 33.Grant, R. A., Filman, D. J., Finkel, S. E., Kolter, R. & Hogle, J. M. (1998) Nat. Struct. Biol. 5, 294-303. [DOI] [PubMed] [Google Scholar]
- 34.Polidoro, M., De Biase, D., Montagnini, B., Guarrera, L., Cavallo, S., Valenti, P., Stefanini, S. & Chiancone, E. (2002) Gene 296, 121-128. [DOI] [PubMed] [Google Scholar]
- 35.Zeth, K., Offermann, S., Essen, L.-O. & Oesterhelt, D. (2004) Proc. Natl. Acad. Sci. USA 101, 13780-13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy, S., Gupta, S., Das, S., Sekar, K., Chatterji, D. & Vijayan, M. (2004) J. Mol. Biol. 339, 1103-1113. [DOI] [PubMed] [Google Scholar]
- 37.Ren, B., Tibbelin, G., Kajino, T., Asami, O. & Ladenstein, R. (2003) J. Mol. Biol. 329, 467-477. [DOI] [PubMed] [Google Scholar]
- 38.Ceci, P., Cellai, S., Falvo, E., Rivetti, C., Rossi, G. L. & Chiancone, E. (2004) Nucleic Acids Res. 32, 5935-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reindel, S., Schmidt, C. L., Anemuller, S. & Matzanke, B. F. (2002) Biochem. Soc. Trans. 30, 713-715. [DOI] [PubMed] [Google Scholar]
- 40.Reindel, S., Anemuller, S., Sawaryn, A. & Matzanke, B. F. (2002) Biochim. Biophys. Acta 1598, 140-146. [DOI] [PubMed] [Google Scholar]
- 41.Kauko, A., Haataja, S., Pulliainen, A. T., Finne, J. & Papageorgiou, A. C. (2004) J. Mol. Biol. 338, 547-558. [DOI] [PubMed] [Google Scholar]
- 42.Poole, L. B. (2005) Arch. Biochem. Biophys. 433, 240-254. [DOI] [PubMed] [Google Scholar]
- 43.Kengen, S. W. M., Bikker, F. J., Hagen, W. R., de Vos, W. M. & van der Oost, J. (2001) Extremophiles 5, 323-332. [DOI] [PubMed] [Google Scholar]
- 44.Mongkolsuk, S. & Helmann, J. D. (2002) Mol. Microbiol. 45, 9-15. [DOI] [PubMed] [Google Scholar]
- 45.Pedone, E., Bartolucci, S. & Fiorentino, G. (2004) Front. Biosci. 9, 2909-2926. [DOI] [PubMed] [Google Scholar]
- 46.Amo, T., Atomi, H. & Imanaka, T. (2002) J. Bacteriol. 184, 3305-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueshima, J., Shoji, M., Ratnayake, D. B., Abe, K., Yoshida, S., Yamamoto, K. & Nakayama, K. (2003) Infect. Immun. 71, 1170-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nair, S. & Finkel, S. E. (2004) J. Bacteriol. 186, 4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park, C. B. & Lee, S. B. (1999) J. Biosci. Bioeng. 87, 315-319. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura, Y., Kaneko, T., Sato, S., Mimuro, M., Miyashita, H., Tsuchiya, T., Sasamoto, S., Watanabe, A., Kawashima, K., Kishida, Y., et al. (2003) DNA Res. 10, 137-145. [DOI] [PubMed] [Google Scholar]
- 51.Grossman, M. J., Hinton, S. M., Minak-Bernero, V., Slaughter, C. & Stiefel, E. I. (1992) Proc. Natl. Acad. Sci. USA 89, 2419-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.