Abstract
Genomic analysis of Emesis [Fabricius], 1807 (Lepidoptera: Riodinidae Grote, 1895) reveals species richness higher than anticipated. As a result, one subgenus, 22 species, and one subspecies are proposed as new (type species or type localities in parenthesis): Diogenia Grishin, new subgenus (Emesis diogenia Prittwitz, 1865), Emesis (Emesis) aerunda Grishin, new species (Peru: Rio Pachitea, Monte Alegre), Emesis (Emesis) bartica Grishin, new species (Guyana: Cuyuni-Mazaruni), Emesis (Emesis) fatimellina Grishin, new species (Brazil: Santa Catarina), Emesis (Emesis) panamella Grishin, new species (Panama: Darién), Emesis (Mandania) mandora Grishin, new species (Ecuador: Santo Domingo), Emesis (Mandania) manduza Grishin, new species (Peru: Cuzco), Emesis (Tenedia) nimia Grishin, new species (Panama: Chiriquí), Emesis (Tenedia) faria Grishin, new species (Mexico: Tamaulipas), Emesis (Tenedia) leona Grishin, new species (Mexico: Nuevo León), Emesis (Tenedia) subangularis Grishin, new species (Argentina: Salta), Emesis (Tenedia) alisada Grishin, new species (Peru: Piura), Emesis (Tenedia) flecta Grishin, new species (Bolivia: La Paz), Emesis (Poeasia) sonorensis Grishin, new species (Mexico: Sonora), Emesis (Brimia) apagada Grishin, new species (Peru: Madre de Dios), Emesis (Brimia) boliviana Grishin, new species (Bolivia: La Paz), Emesis (Aphacitis) aurichica Grishin, new species (Mexico: Chiapas), Emesis (Aphacitis) auripana Grishin, new species (Panama: Darién), Emesis (Aphacitis) pruinapicalis Grishin, new species (Panama: Darién), Emesis (Aphacitis) furvescens Grishin, new species (Panama: Darién), Emesis (Aphacitis) pallescens Grishin, new species (Panama: Panamá), Emesis (Aphacitis) andigna Grishin, new species (Peru: Cuzco), Emesis (Aphacitis) luxata Grishin, new species (Brazil: São Paulo), and Emesis (Mandania) russula sudesta Grishin, new subspecies (Brazil: Paraná). The following five taxa are species (not subspecies): Emesis (Emesis) cronina Schaus, 1928, reinstated status (not Emesis (Emesis) cereus (Linnaeus, 1767)), Emesis (Emesis) nobilata Stichel, 1910, new status (not Emesis (Emesis) fatimella Westwood, 1851), Emesis (Tenedia) tristis Stichel, 1929, reinstated status (not Emesis (Tenedia) lupina Godman and Salvin, 1886), Emesis (Tenedia) paphia R. Felder, 1869, reinstated status (not Emesis (Tenedia) cypria C. Felder and R. Felder, 1861), and Emesis (Aphacitis) parvissima Kaye, 1921, new status (not Emesis (Aphacitis) lucinda (Cramer, 1775). Emesis tenedia ab. fasciata E. Strand, 1916, an unavailable name, is a synonym of Emesis (Tenedia) tenedia C. Felder and R. Felder, 1861, not Emesis (Tenedia) lupina Godman and Salvin, 1886. Lectotypes are designated for five species (type localities in parenthesis): Emesis russula Stichel, 1910 (Bolivia: La Paz), Emesis tenedia C. Felder and R. Felder, 1861 (Venezuela), Emesis poeas Godman, 1901 (Mexico: Guerrero, Acapulco), Emesis castigata Stichel, 1910 (Peru: Pozuzo), and Emesis condigna Stichel, 1925 (Brazil: Pará). Finally, an updated synonymic list of Emesis is provided. The list covers seven valid subgenera and 71 valid species with 14 additional subspecies.
Keywords: Cryptic species, biodiversity, metalmark butterflies, genomics, speciation, nomenclature, taxonomy
Introduction
Describing a new species signifies its official birth in our system of knowledge and is the first step in bringing it into the world. After the description, a species can be enjoyed and studied by everyone, from nature photographers and avocational naturalists to conservationists and a diverse group of researchers, such as taxonomists, ecologists, or evolutionary and molecular biologists. Cataloging species and their localities becomes increasingly important for conservation and restoration efforts. Without a name and awareness of a species’ distinctness and uniqueness, it is impossible to evaluate and value its existence and well-being.
Traditionally, biodiversity has been studied by careful comparative analysis of phenotypes coupled with observations on ecology and behavior whenever possible. Phenotype, including ecological preferences, is encoded by genotype. The inclusion of genotypic characters in the analysis, even on a limited basis, has been revolutionizing biodiversity research. The COI barcodes introduced two decades ago became a standard tool aiding species discovery and identification (Hebert et al. 2003, 2004). Even limited to 658 base pairs, this tiny segment of mitochondrial DNA has been instrumental in producing hypotheses to be tested by the analysis of phenotypes (Burns and Janzen 2005; Burns et al. 2007, 2008).
We have been applying a genomic approach to the characterization of butterflies (Zhang et al. 2022a, c; Pavulaan et al. 2023; Zhang et al. 2023a–d). A large-scale whole genome sequencing of butterfly specimens from across their distribution reveals patterns of diversification that were either not observed or underappreciated before (Núñez et al. 2022; Robbins et al. 2022; Zhang et al. 2022b; Zhang et al. 2023a–d). The results uncover a tapestry of genetic diversity that may not be obvious from phenotypes. In contrast to the COI barcoding that tracks species by a single short mitochondrial gene marker, the genomic approach is more comprehensive and reliable because organisms are represented by their nuclear genomes. Considering all genes enables us to track the entire organism rather than several of its characters.
Examining phylogenetic trees, we form hypotheses about new species. We corroborate these hypotheses by the phenotypic analysis of wing patterns and genitalia. However, our primary evidence for the species distinction is genomic. This is because the species delineation based on phenotypes is frequently subjective and speculative, sometimes without the means to identify these species confidently. Visual assessment of phenotypes is particularly challenging in species-rich groups, where even minute phenotypic differences may signify speciation. Nevertheless, careful phenotypic analysis has been successful in revealing species richness in difficult groups of butterflies. For instance, an unexpected species richness in Detritivora J. Hall and Harvey, 2002 was discovered by a comprehensive morphological analysis (Hall and Harvey 2002; Harvey and Hall 2002).
We delineate species by a combination of several criteria: (1) genetic differentiation in the Z chromosome measured by Fst (>0.20 usually corresponds to distinct species) and gene exchange Gmin (<0.05 for distinct species) (Cong et al. 2019a); (2) COI barcode difference (typically >2% for distinct species) (Hebert et al. 2003) and its correlation with phenotypic differences (Lukhtanov et al. 2016); and (3) the prominence of species-level clades (Zhang et al. 2022c), taking into account genetic differentiation within and between such clades. However, COI barcodes (together with mitochondria) frequently introgress between species (Bachtrog et al. 2006; Cong et al. 2017a), and some distinct species may possess highly similar or identical barcodes (Burns et al. 2008; Zhang et al. 2023b), which we also find in Emesis [Fabricius], 1807. See the “Species, subspecies, and genomics” section in Zhang et al. (2022a) for further discussion.
In this work, we continue our exploration of the tribe Emesidini Seraphim, Freitas and Kaminski, 2018 (Riodinidae). Previously, we focused on the higher classification and proposed a new genus, four new subgenera, and refined the synonymy of the group (Zhang et al. 2019b). One of the highlights is the finding that Apodemia C. Felder and R. Felder, 1865 (type species Lemonias mormo C. Felder and R. Felder, 1859) is also present in South America as Roeberella Strand, 1932 (type species Lemonias calvus Staudinger, 1887), which is a subgenus of Apodemia, and Plesioarida Trujano and García, 2018 (type species Apodemia walkeri Godman and Salvin, 1886) is its junior subjective synonym (Zhang et al. 2020). Significant new progress has been made recently regarding Emesis [Fabricius], 1807 (type species Hesperia ovidius Fabricius, 1793, a junior subjective synonym of Papilio cereus Linnaeus, 1767), with additional refinement of the classification (Trujano-Ortega et al. 2020), several new species described (Costa et al. 2021; Gallardo et al. 2021; Callaghan et al. 2024), and Mexican distribution records compiled and analyzed (Trujano-Ortega et al. 2021).
Here, we focus on the species level in Emesis and uncover surprising, at times cryptic, richness nearly doubling the count of valid species in the genus. This study is only an early step in advancing our knowledge about the hidden species diversity in Emesis. Even in a limited sample of specimens sequenced thus far, we found one new subgenus, 22 new species, and one new subspecies. After more detailed analysis and a more comprehensive sequencing effort, we expect these numbers to increase, and a more complete picture of diversification in Emesis will emerge.
Materials and Methods
Traditionally, new species have been discovered by means of visual comparisons of facies and genitalia, sometimes complemented with field observations about their life histories and ecology. Recently, we have been using a genomic screen approach to species discovery, i.e., to detect new taxa or test hypotheses about new species suspected from phenotypic inspection using genomic sequence comparison (Zhang et al. 2023a). First, we obtain whole genome shotgun sequences of many representative specimens of (nearly) all known species across their ranges, including phenotypically unusual specimens, using our previously established experimental protocols (Li et al. 2019; Zhang et al. 2019a). Typically, a leg of a dry pinned specimen from a collection (see the list of collections below) is used for DNA extraction. Specimens of any age are amenable to our protocol (Cong et al. 2021). Second, these genomic datasets composed of 150 bp (or less) DNA segments are subjected to computational analysis to identify and assemble (i.e., stitch together) protein-coding regions using DIAMOND (Buchfink et al. 2015) aided by a reference set of all proteins encoded in a previously assembled genome of Calephelis nemesis (W. H. Edwards, 1871) (Cong et al. 2017b). This procedure results in a master-slave alignment of all these regions (i.e., coding regions in each specimen are aligned to the reference). These alignments, which are too large (about 18 million positions in the nuclear genome) for time-efficient phylogenetic analysis, are randomly subsampled for best-sequenced positions (by codon) to be used in the construction of phylogenetic trees as described previously (Zhang et al. 2022b). The number of such positions depends on the alignment and is given in figure legends for each tree. Third, we construct phylogenetic trees using IQ-TREE v1.6.12 under the GTR+GAMMA model (Nguyen et al. 2015) from these randomly sampled positions in nuclear (autosomes and Z chromosome separately) and all protein-coding positions in mitochondrial genomes, and ultrafast bootstrap (Minh et al. 2013) is used to estimate statistical significance of tree branches. These three trees are visualized using FigTree (Rambaut 2018) and visually compared to each other.
Inspecting the genomic-level trees, we look for confident clades close to the leaves that visually appear like combs (i.e., star subtrees). Such clades typically correspond to distinct species characterized by prominent genetic differentiation from other species (Zhang et al. 2022a, c). Preference is given to the Z chromosome trees because most of the genes important in speciation (pheromone production, wing pattern control, differences between sexes) are encoded by this chromosome, which, in addition, is more resistant to introgression (Pazhenkova and Lukhtanov 2021). We also illustrate mitochondrial genome trees. Although prone to introgression, mitochondrial DNA is inherited as a single locus and frequently does not vary strongly within species but differs between species. Differences between species visually stand out in phylogenetic trees inferred from mitogenomes. The COI barcode, which is extensively used for species identification and discovery (Hebert et al. 2003), is located in the mitogenome. In many instances, only a single specimen of a species is available, and we compare its genetic distance from others with distances between specimens of the same species, using both nuclear and mitochondrial DNA.
The next step is to confidently assign available names to the clades representing species. In many instances, the assignment is supported by primary type specimens that we sequenced and included in the trees: the species represented by the clade receives the name of the type of the oldest valid name in this clade. If there are no valid names, available names in the clade serve as the basis for naming (and resurrection from synonymy). In the absence of sequenced primary types, identifications are made by the traditional phenotype-based method: comparing facies and genitalia with those of extant primary type specimens or, if types could not be found, with original descriptions while taking type localities into account. The clades or genetically differentiated branches (when only one specimen is available) that cannot be assigned available names represent potential new species and become the focus of this study. Specimens from these clades are scrutinized for their phenotypes, and genitalia are dissected to learn about morphological differences from known species.
In addition to phenotypic diagnosis (terminology per Callaghan et al. 2024), we provide diagnostic DNA characters, both in the nuclear genome and, when such characters exist, in the COI barcode. DNA characters are found in nuclear protein-coding regions using our previously developed procedure (see SI Appendix to Li et al. 2019). The logic behind the character selection was detailed in Cong et al. (2019b). The character states are provided in species diagnoses as abbreviations. E.g., cne728.44.1:G672C means position 672 in exon 1 of gene 44 from scaffold 728 of the Calephelis nemesis (W. H. Edwards, 1871) reference genome (Cong et al. 2017b) is C, changed from G in the ancestor. When characters are given for the sister clade of the diagnosed taxon, the following notation is used: aly5294.20.2:A548A (not C), which means that position 548 in exon 2 of gene 20 on scaffold 5294 is occupied by the ancestral base pair A, which was changed to C in the sister clade (so it is not C in the diagnosed taxon). The same notation is used for COI barcode characters but without a prefix ending with ‘:’. The sequences of exons from the reference genome with the positions used as character states highlighted in green are given in the supplemental file (Zhang et al. 2024a). Providing a link to these DNA sequences from this publication ensures that the numbers given in the diagnoses can be readily associated with actual sequences. Whole genome shotgun datasets we obtained and used in this work are available from the NCBI database (https://www.ncbi.nlm.nih.gov) as BioProject PRJNA1150334, and BioSample entries of the project contain the locality and other collection data of the sequenced specimens shown in the trees. Additionally, specimen data are summarized in Table S1 of the supplemental file (Zhang et al. 2024a). COI barcode sequences have been deposited in GenBank with accessions PQ203545–PQ203570. All new names have been registered with ZooBank.
Spread specimens were photographed with the Nikon 800 camera through the 105 mm Nikkor macro lens in NEF (raw) format, converted to TIF format using DxO with color-correction options adjusted to match 24 patch ColorChecker, and edited in Adobe Photoshop CS4 to standardize the background. Imperfections in specimens, such as scale damage, pinholes, and wing tears, were not digitally removed. Genitalia were prepared after DNA extraction from abdomens (see SI Appendix to Li et al. 2019), which were subsequently soaked in 10% KOH either overnight at room T (if it was convenient to take a break from work) or at 65°C for 15–60 min (depending on the size and abdomen softness after the soak) and then dissected under a binocular microscope. Genitalia were placed in glycerin and photographed using AmScope system H800-96S-18M3 (0.7–5× zoom monocular microscope on a table stand with LED ring light and USB 18.0 MP digital camera) in 3–5 focus slices, which were edited to brighten the background and merged using Adobe Photoshop CS4, and further assembled into plates. Genitalia were stored in glycerin in small vials pinned by each specimen.
The specimens were examined and sampled for sequencing in the following collections (abbreviations, which are not necessarily acronyms of the current names of these institutions, are given in parentheses and used in Table S1 of the supplemental file (Zhang et al. 2024a)): American Museum of Natural History, New York, NY, USA (AMNH), Natural History Museum, London, UK (BMNH), Colorado State University Collection, Fort Collins, CO, USA (CSUC), Field Museum of Natural History, Chicago, IL, USA (FMNH), Museum für Naturkunde, Berlin, Germany (MFNB), McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA (MGCL), Muséum National d’Histoire Naturelle, Paris, France (MNHP), Museum für Tierkunde, Dresden, Germany (MTD), Texas A&M University Insect Collection, College Station, TX, USA (TAMU), Biodiversity Center, University of Texas at Austin, Austin, TX, USA (TMMC), and National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM). Type status abbreviations are: HT holotype, LT lectotype, ST syntype, PT paratype, and PLT paralectotype.
Results and Discussion
Genomic analysis of Emesis species across their ranges reveals 23 distinct unnamed phylogenetic lineages described below as species and one as a subspecies. These taxa are genotypically unique lineages separated from other similar lineages. Some of these new species are possibly allopatric with their closest relatives, but the transition from one species to another in genotype is abrupt, without detected intermediates. A particular rationale for each species distinction is given in their “Definition and diagnosis” section.
For each new species and subspecies, phenotypic characters are given to differentiate it from its close relatives. Phenotypic diagnoses, coupled with localities, will facilitate the identification of these species from photographs. However, many new species are proposed here from a single specimen, and their phenotypic variation is yet unknown. Therefore, until this variation is explored further and if confident identification is required, it should rely on genomic sequencing. Diagnostic DNA characters in the nuclear genome and (when they exist) the COI barcode are listed as abbreviations. The COI barcode sequence itself is provided, although it may not be diagnostic for all species and specimens, as discussed in each such case.
Phylogenetic trees with the holotypes of all new taxa included show their position relative to previously known taxa. Photographs of the dorsal and ventral side of the holotype and, in most cases, photographs of genitalia are given. Additional photographs of these species and their relatives can be found on the Butterflies of America website (Warren et al. 2024). Descriptions of new taxa are accompanied by lectotype designations and taxonomic adjustments, such as the elevation of subspecies to species if supported by genomic analysis. Phylogenetic trees are shown in Fig. 1–6, specimen photographs in Fig. 7–80 (dorsal and ventral sides are denoted by odd and even figure numbers, respectively, except 42 and 44, which show dorsal), and genitalia images in Fig. 81–132 (left lateral and ventral views are denoted by odd and even numbers, respectively, except 87, 91, 95, and 101, which show ventral).
Figure 1.

Phylogenetic trees of Emesis (Emesis) species inferred from protein-coding regions in a) the nuclear genome (autosomes), based on 4,040,316 positions, b) the Z chromosome, based on 247,785 positions, and c) the mitochondrial genome: E. cereus (purple), E. cronina stat. rest. (green), E. aerunda sp. n. (orange), E. orichalceus (olive), E. bartica sp. n. (aquamarine), E. aerigera (brown), E. nobilata stat. nov. (cyan), E. panamella sp. n. (magenta), E. fatimella (blue), and E. fatimellina sp. n. (red). Ultrafast bootstrap (Minh et al. 2013) values are shown at nodes. For each specimen, the name adopted in this work is given first, and a comment about the previous taxonomic placement (if different) is shown in square brackets, supplemented with the DNA Sample number, type status (see Materials and Methods for abbreviations), general locality, and year of collection. See Table S1 in the supplemental file (Zhang et al. 2024a) or NCBI database entries for additional data about these specimens. Synonyms are given in parentheses preceded by “=”. The type status refers to this synonym if the synonym name is provided. The same notations are used throughout this work in other figures showing phylogenetic trees.
Figure 6.

Phylogenetic trees of Emesis inferred from protein-coding regions of a) the nuclear genome (autosomes), based on 4,149,375 positions, b) the Z chromosome, based on 296,487 positions, and c) the mitochondrial genome. New taxa proposed in this work are labeled in red and those with taxonomic changes such as subspecies-to-species status (changes indicated in brackets) are labeled in blue. Subgenera are labeled by their corresponding clades. See Fig. 1 caption for other notations.
Figures 7–26.

Holotypes (unless indicated) of Emesis (Emesis) and Emesis (Mandania), data in text. In Fig. 7–80, dorsal and ventral sides are denoted by odd and even numbers, respectively (except 42 and 44, which are dorsal); type status and sex are shown in each view, F indicates flipped image (left-right inverted). 7–8) E. (E.) aerunda sp. n. 9–10) E. (E.) orichalceus non-type specimen NVG-18045D03. 11–12) E. (E.) aerigera syntype NVG-18054D07. 13–14) E. (E.) bartica sp. n. 15–16) E. (E.) fatimellina sp. n. 17–18) E. (E.) panamella sp. n. 19–22) E. (M.) russula sudesta ssp. n.: 21–22) Paratype NVG-18045H11. 23–24) E. (M.) mandora sp. n. 25–26) E. (M.) manduza sp. n.
Figures 71–80.

Holotypes (unless indicated) of Emesis (Aphacitis), and data in text. 71–72) E. (A.) pallescens sp. n. 73–78) E. (A.) andigna sp. n.: 75–76) Paratype NVG-18052G01 and 77–78) paratype NVG-18039E12. 79–80) E. (A.) luxata sp. n.
Figures 81–106.

Genitalia of holotypes (unless indicated) of Emesis (Emesis), Emesis (Mandania), and Emesis (Tenedia), data in text. In Fig. 81–132, left lateral and ventral views are denoted by odd and even numbers, respectively (except 87, 91, 95, and 101, which are ventral); complete genitalia are illustrated, sometimes with parts removed and shown separately (as stated); for females, only the sternite VII (“genital plate”) (88, 92, 96, 102) is shown to scale and complete genitalia are at half the size, as indicated by the smaller scale bar. 81–82) E. (E.) fatimellina sp. n. 83–84) E. (E.) panamella sp. n. 85–88) E. (M.) russula sudesta ssp. n. paratypes: 85–86) NVG-18045H11 and 87–88) NVG-18044F07. 89–90) E. (M.) russula russula non-type specimen NVG-18044D11. 91–92) E. (M.) mandora sp. n. 93–94) E. (M.) manduza sp. n. 95–96) E. (T.) nimia sp. n. 97–98) E. (T.) faria sp. n. 99–102) E. (T.) leona sp. n.: 101–102) Paratype NVG-10598. 103–104) E. (T.) subangularis sp. n. 105–106) E. (T.) angularis non-type specimen NVG-23115A11.
Figures 107–132.

Genitalia of holotypes (unless indicated) of Emesis (Tenedia), Emesis (Poeasia), Emesis (Aphacitis) and Emesis (Brimia), data in text. 107–108) E. (T.) alisada sp. n. 109–110) E. (T.) flecta sp. n. 111–112) E. (P.) sonorensis sp. n. 113–114) E. (P.) poeas non-type specimen NVG-23111B11. 115–116) E. (B.) apagada sp. n. 117–118) E. (B.) boliviana sp. n. 119–120) E. (A.) aurichica sp. n. 121–122) E. (A.) auripana sp. n. 123–124) E. (A.) pruinapicalis sp. n. 125–126) E. (A.) furvescens sp. n., aedeagus separated, shown right below the number 125. 127–128) E. (A.) pallescens sp. n. 129–130) E. (A.) andigna sp. n. 131–132) E. (A.) luxata sp. n. paratype NVG-23114H12.
In proposing new names, we refrain from using patronyms, partly because they may be more challenging to remember and associate with corresponding species. The new names are derived either from the names of species’ close relatives (making the name longer for southern counterparts), a descriptive phenotypic feature, or the type locality. We hope such names will integrate easily with the existing classification and be more straightforward to learn.
Emesis (Emesis) cronina Schaus, 1928 is a species distinct from Emesis (Emesis) cereus (Linnaeus, 1767)
Genomic analysis reveals that Emesis cronina Schaus, 1928 (type locality in Paraguay: Sapucaí, syntypes sequenced as NVG-18045C08 and NVG-18045C09) currently regarded as a subspecies of Emesis (Emesis) cereus (Linnaeus, 1767) (type locality originally given as “Indiis”, likely in Suriname) (Callaghan and Lamas 2004) is genetically differentiated from it at the species level (Fig. 1), e.g., their COI barcodes differ by 2.7% (18 bp). In the presence of recognizable phenotypic differences—specimens from Paraguay are smaller, typically with reduced silvery streaks and vestigial subapical pale spot by the forewing costa, which is weaker curved compared to E. cereus—we propose to treat Emesis (Emesis) cronina Schaus, 1928, reinstated status, as a species-level taxon.
Emesis (Emesis) aerunda Grishin, new species
http://zoobank.org/7ADDFF04-F4DC-4379-AE2E-59B99593C46E
Definition and diagnosis.
Genomic analysis of Emesis [Fabricius], 1807 reveals that a specimen from Peru (Fig. 1 orange) is genetically differentiated from its sister Emesis (Emesis) orichalceus Stichel, 1916 (type locality in Bolivia, syntype sequenced as NVG-18043E06) (Fig. 1 olive-colored clade) at the species level, e.g., their COI barcodes differ by 2.1% (14 bp). Therefore, this specimen represents a new species. This new species is phenotypically similar to E. orichalceus and Emesis neemias Hewitson, 1872 (type locality in Brazil) and differs from its relatives by postdiscal and submarginal bands of metallic crescents on hindwing being farther from each other, less orange-red and more purplish ventral side of wings with more prominent pale ray near the anal margin of hindwing, and typically larger and more diffuse tornal dark spots on ventral side of both wings. In addition to the holotype of the new species (Fig. 7–8), we also illustrate a typical male of E. orichalceus (NVG-18045D03, USNMENT 01466452 Bolivia: La Paz Province, San Lorenzo Valley, Rio San Lorenzo, 800 m, GPS −15.8056, −67.4908, Brian Harris leg. [USNM]) (Fig. 9–10). Due to unexplored phenotypic variation in the new species, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne1775.9.1:T306C, cne1775.9.1:A795G, cne403.3.3:A120G, cne403.3.3:T135C, cne4207.3.2:C32G, cne253.1.13:G171G (not A), cne10789.3.8:A150A (not G), cne2851.12.1:C104C (not A), cne2851.12.1:A123A (not G), cne13070.7.1:G1255G (not A), and COI barcode: T34A, A130T, T133C, 169C, T484T.
Barcode sequence of the holotype.
Sample NVG-18052F04, GenBank PQ203545, 658 base pairs:
AACATTATATTTTATCTTTGGAATTTGAGCAGGAATAGTAGGAACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACCTCAGGCTCATTAATTGGAGATGACCAAATTTATAATACTATTGTAACTGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCCATTATAATTGGAGGATTTGGTAATTGATTAGTACCTTTAATACTTGGAGCACCAGATATAGCATTTCCACGTATAAATAATATAAGATTTTGATTATTACCTCCTTCTTTATTTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCAAATATTGCTCATGGAGGATCTTCTGTTGATTTAGCTATTTTTTCATTACATTTAGCTGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACTATTATTAATATACGAATTAATAATTTATCTTTTGATCAAATACCATTATTTGTTTGATCAGTAGGAATTACAGCTCTTTTATTATTATTATCTTTACCTGTATTAGCTGGAGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCATTTTTTGATCCTGCTGGAGGAGGAGACCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ currently deposited in the Museum für Naturkunde, Berlin, Germany (MFNB), illustrated in Fig. 7–8, bears the following four rectangular labels (1st green, last red, others white; 2nd handwritten, others printed): [ Mt. Alegre, Rio | Pachitea O. Peru | G.Tessmann ], [ DNA sample ID: | NVG-18052F04 | c/o Nick V. Grishin ], [ neemias Hew. ], and [ HOLOTYPE ♂ | Emesis (Emesis) | aerunda Grishin ].
Type locality.
Peru: Rio Pachitea, Monte Alegre. This is also the type locality of Pseudophaloe tessmanni Hering, 1925 (Erebidae: Arctiinae), Hylesia natex Draudt, 1929 (Saturniidae), and Synargis flavicauda Grishin, 2023 (Riodinidae), among others.
Etymology.
In Latin, aeruginosus means brassy or verdigris-colored, and unda means wave. The name is given for the metallic green wavy pattern of this species: aeru[ginosus] + [u]nda and is treated as a feminine noun in apposition.
Distribution.
Currently known only from the holotype collected in central Peru.
Emesis (Emesis) bartica Grishin, new species
http://zoobank.org/5C122118-275F-4682-A81C-F499174B98C3
Definition and diagnosis.
Genomic analysis of Emesis [Fabricius], 1807 reveals that a specimen from Guyana (Fig. 1 aquamarine) is genetically differentiated from its sister Emesis (Emesis) aerigera (Stichel, 1910) (type locality in Brazil: Sao Paulo, syntype sequenced as NVG-18054D07) (Fig. 1 brown) at the species level, e.g., their COI barcodes differ by 4.0% (26 bp). Therefore, this specimen represents a new species. This new species is phenotypically similar to E. aerigera and differs from it by narrower metallic bands with less interconnected and aligned spots, a metallic postdiscal spot in the forewing cell M1-M2 being stronger offset basad from the band, and forewings with slightly less hooked apex. In addition to the holotype of the new species (Fig. 13–14), we also illustrate a syntype, a male, of E. aerigera (NVG-18054D07 Brazil: Sao Paulo, Casa Branca, 1890, Garbe leg. [MFNB]) (Fig. 11–12). Furthermore, see iNaturalist observation 160938035 of E. aerigera female from Brazil: Santa Catarina for comparison (iNaturalist 2024). Due to unexplored phenotypic variation in this species and males still unknown, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne2800.4.2:T142A, cne6221.18.5:T129C, cne6221.18.5:A144G, cne15953.4.2:T15A, cne15953.4.2:A51G, cne2337.3.4:A45A (not T), cne2337.3.4:T48T (not C), cne7747.1.14:C116C (not G), cne7747.1.14:C120C (not G), cne4614.6.1:C264C (not T), and COI barcode: T49T, T103C, A238A, T373C, T442C, T553C.
Barcode sequence of the holotype.
Sample NVG-18048H03, GenBank PQ203546, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCTGGTATAGTAGGTACATCTTTAAGTTTATTAATTCGTATAGAATTAGGAACTTCTGGTTCTTTAATTGGAGACGATCAAATTTATAATACTATTGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGGTTAGTTCCTCTTATATTAGGAGCCCCTGATATAGCATTCCCACGTATAAATAATATAAGATTTTGATTATTACCCCCATCCTTATTTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCTCATGGAGGCTCTTCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCTATTTTAGGAGCAATTAACTTTATTACAACTATTATTAATATACGAATTAATAATATATCTTTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACTGCTCTTTTATTATTACTATCTCTTCCCGTATTAGCAGGAGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGACCCAGCAGGTGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 13–14, bears the following four printed rectangular labels, three white: [ Bartica | Bartica District | British Guiana ], [ DNA sample ID: | NVG-18048H03 | c/o Nick V. Grishin ], [ USNMENT | {QR Code} | 01466572 ], and one red [ HOLOTYPE ♀ | Emesis (Emesis) | bartica Grishin ]. The holotype lacks its abdomen.
Type locality.
Guyana: Cuyuni-Mazaruni Region, Bartica.
Etymology.
The name is given for the type locality and is a feminine noun in apposition. Furthermore, the name Bartica comes from an Amerindian word, possibly Arawakan or Cariban, that means “red earth,” and seems suitable for this reddish-colored species.
Distribution.
Currently known only from the holotype collected in Guyana.
Emesis (Emesis) nobilata Stichel, 1910 is a species distinct from Emesis (Emesis) fatimella Westwood, 1851
Genomic analysis reveals that Emesis fatima nobilata Stichel, 1910 (type locality in Costa Rica, syntype sequenced as NVG-18052D01) currently regarded as a subspecies of Emesis (Emesis) fatimella Westwood, 1851 (type locality in Suriname and Brazil: Amazonas) (Callaghan and Lamas 2004) is genetically differentiated from it at the species level (Fig. 1), e.g., their COI barcodes differ by 2.9% (19 bp). In the presence of recognizable phenotypic differences—males of E. fatimella nobilata have darker ground color and larger, more diffuse spotting compared to the nominate subspecies—we propose to treat Emesis (Emesis) nobilata Stichel, 1910, new status, as a species-level taxon.
Emesis (Emesis) fatimellina Grishin, new species
http://zoobank.org/FAE56D8B-3EF1-4E43-B997-CB21188D67C7
Definition and diagnosis.
Genomic analysis of Emesis [Fabricius], 1807 reveals that two specimens from Southeast Brazil and South Brazil (Fig. 1 red) form a clade sister to both Emesis (Emesis) nobilata Stichel, 1910, new status (type locality in Costa Rica) and Emesis (Emesis) fatimella Westwood, 1851 (type locality in Suriname and Brazil: Amazonas) and are genetically differentiated from their relatives (Fig. 1 cyan, blue, and magenta) at the species level, e.g., their COI barcodes differ by 4.1%–4.6% (27–30 bp). Therefore, these specimens represent a new species. This new species is phenotypically similar to E. nobilata and E. fatimella and differs from its relatives by the ground color that is paler and more orange than in E. nobilata, but yellower (rather than orange) beneath compared even to E. fatimella, sharper defined and less diffuse dark markings on the dorsal side, and generally smaller submarginal brown spots on the ventral side; these spots are not all the same size and the size difference among them appears more pronounced than in other species. In male genitalia (Fig. 81–82), the lower valval projection is not developed, the upper projection is claw-like with the inner broad tooth, aedeagus terminally with several stronger sclerotized broad teeth around the posterior margin. Due to unexplored phenotypic variation in this species, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne11524.2.14:G153A, cne877.3.6:T267C, cne764.2.12:C144T, cne764.2.12:C154A, cne6813.1.17:G103T, and COI barcode: A1T, T169A, T232C, A433T, T526C, T646C.
Barcode sequence of the holotype.
Sample NVG-18044H01, GenBank PQ203547, 658 base pairs:
TACATTATATTTTATTTTTGGTATTTGAGCCGGAATAGTAGGAACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGATCTTTAATTGGCGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGCGGATTTGGTAATTGATTAGTACCTTTAATATTAGGAGCTCCTGATATAGCCTTCCCACGTATAAATAATATAAGATTTTGATTATTACCTCCATCTTTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCTCATGGTGGATCTTCTGTAGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCTTCTATTTTAGGTGCTATTAATTTTATTACTACTATTATTAACATACGAATTAATAATATATCATTTGATCAAATACCATTATTTGTTTGATCAGTAGGAATTACCGCTCTTTTATTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCATTTTTTGATCCAGCTGGTGGTGGAGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 15–16, bears the following six printed (text in italics handwritten) rectangular labels, five white: [ Brasil:Santa Catarina | Joinville: 10–200 m | 26 Feb 1991 | Leg. H. Miers ], [ DNA sample ID: | NVG-18044H01 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114G04 | c/o Nick V. Grishin ], [ genitalia | NVG240817-09 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466402 ], and one red [ HOLOTYPE ♂ | Emesis (Emesis) | fatimellina Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratype: 1♂: NVG-18052G08 Brazil: Rio de Janeiro, Teresópolis, coll. H. Stichel number 3305 [MFNB].
Type locality.
Brazil: Santa Catarina, Joinville.
Etymology.
The name is formed from the name of its South American relative, E. fatimella, which is made longer for this more southern species and is treated as a feminine noun in apposition.
Distribution.
Southeast and South Brazil.
Emesis (Emesis) panamella Grishin, new species
http://zoobank.org/02586855-7FA6-4F8F-AAC5-15D46FF43F03
Definition and diagnosis.
A more detailed comparison of Emesis (Emesis) fatimella Westwood, 1851 (type locality in Suriname and Brazil: Amazonas) relatives reveals that in addition to the most genetically divergent species, Emesis (Emesis) fatimellina, new species, a specimen from Panama (Fig. 1 magenta) is not tightly clustered with either Emesis (Emesis) nobilata Stichel, 1910, new status (type locality in Costa Rica) (Fig. 1 cyan) or E. fatimella (Fig. 1 blue) and instead is genetically differentiated from them at the species level, e.g., their COI barcodes differ by 2.4% (16 bp) from E. nobilata and by 3.2% (21 bp) from E. fatimella. Therefore, this specimen represents a new species. This new species is phenotypically similar to E. nobilata and E. fatimella and differs from them by the color of both sides of wings being brighter orange than in E. nobilata, and by more deep orange (rather than yellower) colors compared to E. fatimella, approximately the same color of dorsal and ventral side of wings (ventral is typically yellower than dorsal in E. fatimella), and usually sharper defined and less diffuse submarginal spots on forewing, and whiter scales on thorax, basal half of legs and abdomen beneath. In male genitalia (Fig. 83–84), the lower valval projection is much smaller, directed inward, the upper projection is strongly elongated, longer than falces, claw-like with the inner broad tooth, aedeagus is narrower and longer. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne2539.10.4:T57C, cne33461.1.2:A87G, cne7180.6.10:A120G, cne7180.6.10:T174A, cne65262.1.1:A648G, cne254622.4.2:G30G (not A), cne254622.4.2:A42A (not G), cne3597.4.3:A73A (not G), cne3597.4.3:C74C (not A), cne3597.4.3:C85C (not T), and COI barcode: A85A, T361C, A379C, T533C, A604C.
Barcode sequence of the holotype.
Sample NVG-18044G07, GenBank PQ203548, 658 base pairs:
AACATTATATTTTATTTTTGGTATTTGAGCAGGAATAGTAGGAACATCATTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAGTACCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGTATAAATAATATAAGATTTTGATTATTACCTCCATCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCAAATATCGCTCATGGCGGATCTTCCGTAGATTTAGCTATTTTTTCCTTACATTTAGCTGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACTATTATTAACATACGAATTAATAATATATCATTTGATCAAATACCTTTATTTGTATGATCTGTAGGAATTACTGCTCTTCTATTATTATTATCTCTACCCGTATTAGCAGGAGCTATTACCATATTATTAACAGATCGTAATTTAAATACCTCATTCTTTGATCCAGCTGGTGGTGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 17–18, bears the following six printed (text in italics handwritten) rectangular labels, five white: [ PANAMA:DARIEN | Cana (Cerro Pirre) | 1000m | 7°56’N 77°43’W | 31 I 1984 | leg. G.B.Small ], [ DNA sample ID: | NVG-18044G07 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114G05 | c/o Nick V. Grishin ], [ genitalia | NVG240817-10 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466396 ], and one red [ HOLOTYPE ♂ | Emesis (Emesis) | panamella Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection.
Type locality.
Panama: Darién Province, Cana, Cerro Pirre, elevation 1000 m, approx. GPS 7.9333, −77.7167.
Etymology.
The name is a fusion of the type locality country name with the name of a relative from South America: Pana[ma[ + [fati]mella. The name is treated as a feminine noun in apposition.
Distribution.
Currently known only from the holotype collected in eastern Panama.
Lectotype designation for Emesis russula Stichel, 1910
Emesis russula Stichel, 1910 was described from three specimens: two males from Bolivia (Stichel collection Nos. 924 and 3207) and one female from Brazil (Stichel collection No. 584, “S. Leopoldo” in the original description and “S. Leopoldina” on the label of this syntype) (Stichel 1910). Genomic sequencing of male and female syntypes (No. 924 as NVG-18052C11 and No. 584 as NVG-18052C11) reveals that while they both belong to the clade of specimens we initially identified as E. russula, the Bolivian syntype forms a subclade with another Bolivian specimen we sequenced. The Brazilian syntype belongs to a different subclade that includes specimens from southeastern regions of South America (Fig. 2). The two subclades signify two major groups of E. russula populations: the Andean one exemplified by two Bolivian specimens (Fig. 2 green) and the one from the plains and hills of southeastern South America (Fig 2 brown). These two major groups correspond to two different genetically differentiated subspecies, and the type series of E. russula is polytypic. Because the original description of E. russula centers around Bolivian males and only mentions the Brazilian female as being similar to males but with slightly broader wings (Stichel 1910), and the males with their locality are listed first; the Bolivian males better represent Stichel’s concept of E. russula.
Figure 2.
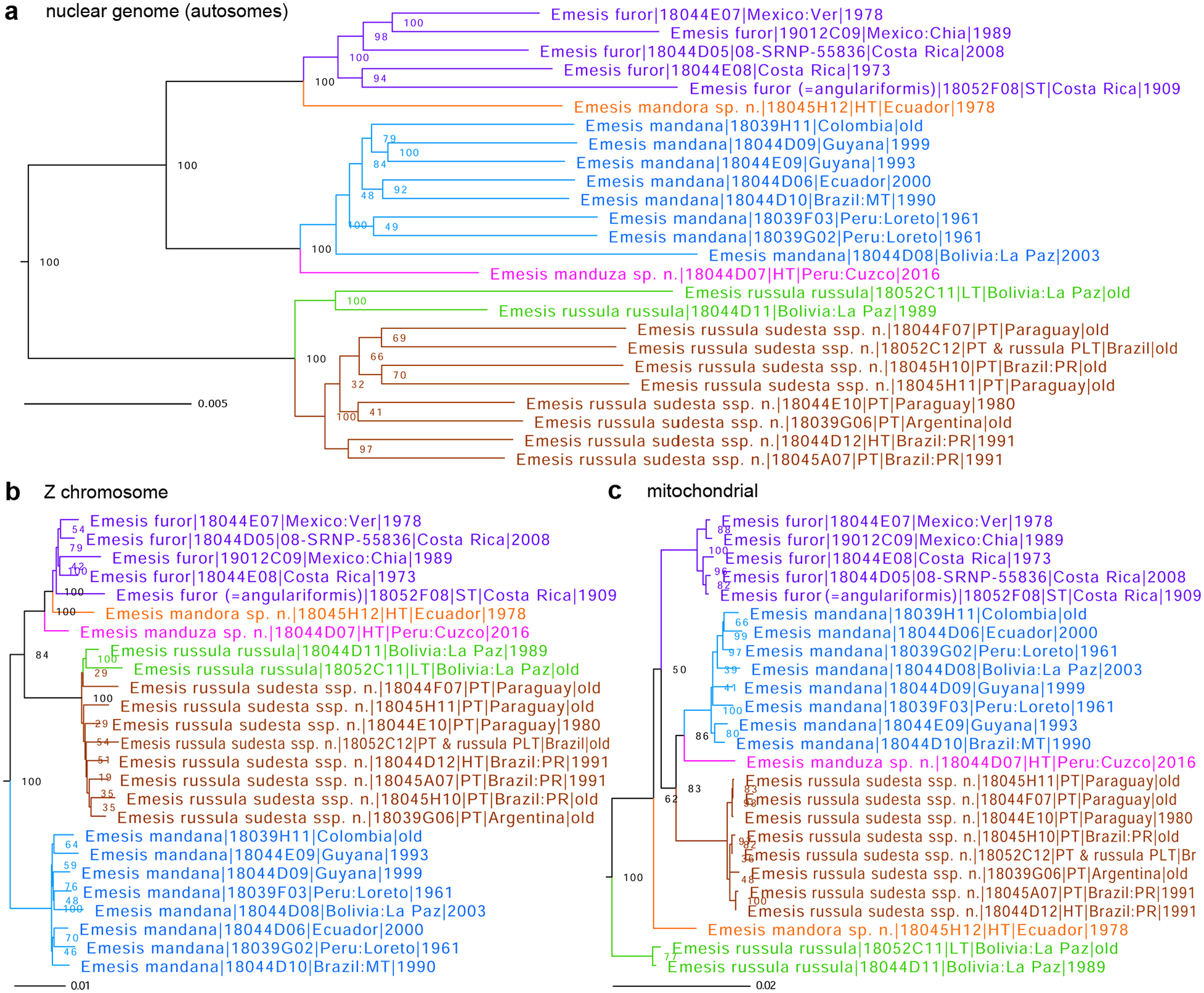
Phylogenetic trees of Emesis (Mandania) species inferred from protein-coding regions in a) the nuclear genome (autosomes), based on 7,989,090 positions, b) the Z chromosome, based on 327,183 positions, and c) the mitochondrial genome: E. furor (purple), E. mandora sp. n. (orange), E. mandana (blue), E. manduza sp. n. (magenta), E. russula russula (green), and E. russula sudesta ssp. n. (brown). See Fig. 1 caption for other notations.
To stabilize nomenclature and define the name E. russula objectively in the light of polytypic type series, N.V.G. hereby designates the sequenced male syntype in the MFNB collection that bears the following five rectangular labels (4th handwritten, others printed, 1st red, 4th pale green, and others white): [ Typus ], [ Bolivia La Paz | Farinas | e.c.H.Stichel ], [ 924 ], [ russula | Stich. ], and [ DNA sample ID: | NVG-18052C11 | c/o Nick V. Grishin ] as the lectotype of Emesis russula Stichel, 1910. The lectotype lacks the left antenna and has a small piece of left hindwing chipped away around the middle of the outer margin. Images of this specimen are shown on the Butterflies of America website (Warren et al. 2024). The type locality of E. russula becomes Bolivia: La Paz, Farinas. The COI barcode sequence of the lectotype, sample NVG-18052C11, GenBank PQ203549, 658 base pairs is:
AACATTATATTTTATTTTTGGAATTTGAGCAGGGATAGTTGGAACTTCACTAAGATTATTAATTCGAATAGAATTAGGAACTTCAGGATCATTAATTGGTGATGATCAAATTTATAATACTATTGTTACAGCTCACGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCTCCAGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGACTTTTACCTCCATCTTTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCTTCAAATATTGCTCATGGAGGTTCTTCAGTAGATTTAGCTATTTTCTCTTTACATTTAGCAGGAATTTCTTCAATTTTAGGTGCAATTAACTTTATTACTACTATTATTAATATACGAATTAATAATATATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACAGCTCTTTTATTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACAGATCGAAATTTAAATACATCATTCTTTGATCCTGCTGGTGGTGGTGATCCTATTTTATATCAACATTTATTT
Due to the possibility of introgression and the similarity of COI barcode sequences among Emesis (Mandania) Grishin, 2019 (type species Papilio mandana Cramer, 1780), we do not expect that the COI barcode sequence would be sufficient to identify the nominate subspecies of E. russula, and nuclear genome sequences should be compared. The female syntype from Brazil represents a different subspecies of E. russula, which does not have a name and, therefore, is new, as described below.
Emesis (Mandania) russula sudesta Grishin, new subspecies
http://zoobank.org/0FDAEBAB-A780-403B-B1F8-1F4C772A15D6
Definition and diagnosis.
As discussed above, Emesis russula Stichel, 1910 (type locality in Boliv ia: La Paz, Farinas) populations partition into two subclades that we define as subspecies (Fig. 2 green and brown). Their COI barcodes differ by 1.2% (8 bp). The lectotype designation assigns the nominate subspecies to the northwestern populations of E. russula from the Andes, and the southeastern subspecies is new. This new subspecies differs from the nominotypical subspecies by more developed and darker pattern elements on the wings’ dorsal side that is somewhat redder in color, usually without purplish gloss, and the ventral side is typically with heavier reddish markings. Due to unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne5853.5.7:C70T, cne12666.1.11:T552C, cne2259.5.9:A147C, cne6241.3.5:G24A, cne4342.8.1:A349T. However, the COI barcode may not differentiate between subspecies due to introgression. We hypothesize this because, in the specimens we sequenced, the barcode of the new subspecies is more similar to Emesis (Mandania) mandana (Cramer, 1780) (type locality in Suriname) than to its closer relative E. russula russula (Fig. 2c brown to blue rather than brown to green). Therefore, these barcodes likely represent a later introgression event rather than the original barcodes of the new subspecies. It remains unclear if the introgression is complete, and these E. mandana-like barcodes are present in all specimens of the new subspecies (less likely), or if the original, E. russula-like barcodes are still present in some individuals of the new subspecies (more likely). In our current dataset, the two subspecies differ in their COI barcodes, and the COI barcode corresponding to the new subspecies is diagnosed by a combination of the following base pairs A34A, T136T, C220C, T397T, A412G, C421C. However, if some specimens of the new subspecies possess barcodes similar to those of the nominate subspecies, they may not be identifiable by barcodes.
Barcode sequence of the holotype.
Sample NVG-18044D12, GenBank PQ203550, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACTTCACTAAGATTATTAATTCGAATAGAATTAGGAACTTCAGGATCATTAATTGGTGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCCCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCTCCATCTTTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCTTCTAATATTGCTCATGGAGGTTCTTCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCGGGAATTTCCTCAATTTTAGGTGCAATTAACTTTATTACTACTATTATTAATATACGAATTAATAATATATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACAGCTCTTTTATTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACAGATCGAAATTTAAATACATCATTCTTTGATCCTGCTGGTGGTGGTGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♀ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 19–20, bears the following six rectangular labels (3rd blue, the last red, and others white; 2nd handwritten, others printed with handwritten text marked in italics): [ BRAZIL:PR | 30 km NW Ponta | Grossa, 900m | 24°57’S 50°28’W | 19 Mar 1991 | Robbins, Mielke & | Cassagrande, leg. ], [ russula ], [ JHALL | -0002 ], [ DNA sample ID: | NVG-18044D12 | c/o Nick V. Grishin ], [ USNMENT | {QR Code} | 01466367 ], and [ HOLOTYPE ♀ | Emesis (Mandania) russula | sudesta Grishin ]. Paratypes: 4♂♂ and 3♀♀: Paraguay [USNM]: 1♂ NVG-18045H11 (leg), NVG-23114G06 (abdomen), USNMENT 01466507, genitalia vial NVG240817-11 (Fig. 21–22, 85–86) and 1♀ NVG-18044F07 (leg), NVG-23114G07 (abdomen), USNMENT 01466386 from Sapucaí, old (around 1900), W. T. Foster leg., genitalia vial NVG240817-12 (Fig. 87–88) and 1♂ NVG-18044E10, USNMENT 01466377 Paraguarí Department, 25 km SE of Ybycui, Ybycui National Park, 12–24-Apr-1980, P. J. Spangler et al.; Brazil, Paraná [USNM]: 1♂ NVG-18045H10, USNMENT 01466506 Castro, old, W. Schaus collection [USNM] and 1♀ NVG-18045A07, USNMENT 01466420 the same data as the holotype; 1♀ NVG-18052C12, paralectotype of E. russula, Brazil, “S. Leopoldina” or “S. Leopoldo” [MFNB]; and 1♂ NVG-18039G06 Argentina: Buenos Aires, old, Strecker collection, No. 9477 [FMNH].
Type locality.
Brazil: Paraná, 30 km northwest of Ponta Grossa, elevation 900 m, GPS −24.950, −50.467.
Etymology.
The name russula possibly originated from the Latin word russus, which means reddish, although the color of this species is less red than that of E. mandana. In Portuguese, sudeste means southeast, and the name is given for the southeastern distribution of this subspecies compared to the nominate. The name is treated as a feminine noun in apposition.
Distribution.
Recorded from southern Brazil (e.g., Paraná), Paraguay, and northeastern Argentina.
Comment.
We conservatively propose this new taxon as a subspecies due to its small genetic differentiation, especially in the Z chromosome (Fig. 2b), and limited phenotypic distinction. To aid future comparisons, we illustrate the genitalia of the nominate E. russula male NVG-18044D11 (leg), NVG-23114G08 (abdomen), USNMENT 01466366 from Bolivia: La Paz, Chulumani, 600 m, GPS-16.400, -67.517, 27-May-1989, C. Covell leg., genitalia vial NVG240817-13 [USNM] (Fig. 89–90), which may have less robust valvae with narrower upper and lower projections. We note that considering generally small genetic differences among species of Emesis (Mandania), it is possible that future research may demonstrate that the new subspecies described here is a species-level taxon.
Five species in the subgenus Mandania Grishin, 2019
Genomic analysis of the subgenus Mandania Grishin, 2019 (type species Papilio mandana Cramer, 1780) that currently consists of three species: E. furor A. Butler and H. Druce, 1872 (type locality in Costa Rica), E. mandana (type locality in Suriname), and E. russula Stichel, 1910 (type locality in Bolivia: La Paz) reveals incongruence between the three phylogenetic trees (Fig. 2), those constructed from autosomes in the nuclear genome (Fig. 2a), the Z chromosome (Fig. 2b), and the mitochondrial genome (Fig. 2c). First, a specimen from Peru (Fig. 2 magenta) is closer related to (but distinct from) E. mandana in the autosome tree (Fig. 2a blue) while being in the clade with E. furor in the Z chromosome tree (Fig. 2b purple). Second, in the mitogenome tree, a specimen from Ecuador (Fig. 2c orange) is uniquely distinct and not grouped with E. furor as in the other two trees (Fig. 2a, b purple). Moreover, and in addition to E. russula sudesta, new subspecies, described above, the mitogenome tree reveals five distinct lineages (Fig. 2c purple, blue, magenta, orange, and green) with little genetic variability within each lineage of more than one specimen and prominent separation between the lineages. These lineages are seen in both nuclear genome trees (Fig. 2a, b) and correspond to phenotypically distinct specimens. Due to genetic and phenotypic differentiation between the lineages, incongruence of the trees suggesting non-linear evolutionary scenarios, and close clustering of specimens from different localities within each lineage (when more than one specimen was sequenced), we hypothesize that these lineages represent five distinct species: three previously known and two new. However, COI barcodes of Mandania species are rather similar, and although the species partition accordingly to the mitochondrial DNA (and thus barcode) clusters (Fig. 2c different colors), barcode differences are small, e.g., 0.8% (5 bp) between the new species from Ecuador and E. mandana, and 0.9% (6 bp) from E. furor.
Emesis (Mandania) mandora Grishin, new species
http://zoobank.org/E7455377-B43E-415E-AAAB-2A760E201C04
Definition and diagnosis.
As discussed above, a genetically and phenotypically distinct specimen from Ecuador (Fig. 2 orange) represents a new species of the subgenus Mandania Grishin, 2019. This new species is phenotypically similar to other Mandania and differs from its closest relatives by being paler and less saturated in color (i.e., plainer, less red, grayer) than E. mandana, but with a more contrasting pattern of dark spots and bands than E. furor, and broader wings than E. russula. The holotype is also notably larger than a typical Mandania (Fig. 19–26), and the size may be one of the characters for the new species. However, the size is typically variable, and without a series of specimens, it is not possible to ascertain. In female genitalia (Fig. 91–92), ductus bursae with a loop near a spherical corpus bursae, two very large (about ½ of the corpus diameter) horn-like signa at the caudal end of corpus, signum is more curved and with a larger base compared to its length, sternite VII (“genital plate”) with posterior margin shaped as a broad V less rounded on the sides and in the middle. Due to unexplored phenotypic variation and males still unknown, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne1684.6.12:G143A, cne3364.6.1:A299G, cne2551.8.1:A315G, cne2551.8.1:C327T, cne350.7.4:T726C, cne399.1.1:T225T (not G), cne2564.2.1:A56A (not T), cne6857.5.3:A318A (not G), cne6857.5.3:T375T (not A), cne1186.1.1:A119A (not T), and COI barcode: C50C, T106T, T235T, A412A, T581T, T595C.
Barcode sequence of the holotype.
Sample NVG-18045H12, GenBank PQ203551, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACTTCACTAAGATTATTAATTCGAATAGAATTAGGAACTTCAGGATCATTAATTGGTGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCCCCAGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGACTTTTACCTCCATCTTTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCTTCTAATATTGCTCATGGAGGTTCTTCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGAATTTCCTCAATTTTAGGTGCAATTAACTTTATTACTACTATTATTAATATACGAATTAATAATATATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACAGCTCTTTTATTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACAGATCGAAACTTAAATACATCATTCTTTGATCCTGCTGGTGGTGGTGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 23–24, bears the following six printed rectangular labels, five white: [ Riodinidae V-31-1978 | Emesis fatima M | SDLC,Ecuador,1800 ft ], [ DNA sample ID: | NVG-18045H12 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114G09 | c/o Nick V. Grishin ], [ genitalia | NVG240817-14 | Nick V. Grishin ], [ USNMENT | {QR Code} | 00940170 ], and one red [ HOLOTYPE ♀ | Emesis (Mandania) | mandora Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection.
Type locality.
Ecuador: Santo Domingo de los Colorados, elevation 550 m.
Etymology.
The name is a modified fusion of the name of a related species with the name of the country with the type locality: mand[an] + [Ecua]dor + a. The name is treated as a feminine noun in apposition.
Distribution.
Currently known only from the holotype collected in Ecuador.
Emesis (Mandania) manduza Grishin, new species
http://zoobank.org/9C0ED1F0-211A-4588-B534-E3292AD60323
Definition and diagnosis.
As discussed above, a genetically and phenotypically distinct specimen from Peru (Fig. 2 magenta) represents a new species of the subgenus Mandania Grishin, 2019. This new species is phenotypically similar to other Mandania and differs from its closest relatives by being darker, dorsally more maroon than orange, orange-red, or brown; in particular, the difference in darkness is more obvious towards the apex and costal margin of the dorsal hindwing and on the ventral side, towards the margins. Due to unexplored phenotypic variation in this species, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne216.12.3:T242C, cne216.12.3:T414A, cne216.12.3:A426T, cne9580.1.6:T99G, cne9580.1.6:G111T, cne5285.1.8:C60C (not T), cne5285.1.8:C81C (not T), cne5285.1.8:G114G (not A), cne6560.2.3:A489A (not T), cne6560.2.3:T504T (not C), and COI barcode: C50C, T106T, T235T, A388G, A412A, T581C, T595T.
Barcode sequence of the holotype.
Sample NVG-18044D07, GenBank PQ203552, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACTTCACTAAGATTATTAATTCGAATAGAATTAGGAACTTCAGGATCATTAATTGGTGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCCCCAGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGACTTTTACCTCCATCTTTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCTTCTAATATTGCTCATGGAGGTTCTTCAGTAGATTTGGCTATTTTTTCTTTACATTTAGCAGGAATTTCCTCAATTTTAGGTGCAATTAACTTTATTACTACTATTATTAATATACGAATTAATAATATATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACAGCTCTTTTATTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACTATATTACTAACAGATCGAAATTTAAATACATCATTCTTTGATCCTGCTGGTGGTGGTGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 25–26, bears the following six printed (text in italics handwritten) rectangular labels, five white: [ PERU:Cuzco, 1050m | Quitacalzón | Cosnipata Valley 4856 | 01-XI-2016 Kinyon ], [ DNA sample ID: | NVG-18044D07 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114G10 | c/o Nick V. Grishin ], [ genitalia | NVG240817-15 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466363 ], and one red [ HOLOTYPE ♂ | Emesis (Mandania) | manduza Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection.
Type locality.
Peru: Cuzco Department, Cosñipata Valley, Quebrada Quitacalzón, elevation 1050 m, GPS −13.0167, −71.4833.
Etymology.
The name is a modified fusion of the name of a related species with the name of the Peruvian region with the type locality: mand[an] + [c]uz[co] + a. The name is treated as a feminine noun in apposition.
Distribution.
Currently known only from the holotype collected in southern Peru.
Emesis (Tenedia) tristis Stichel, 1929 is a species distinct from Emesis (Tenedia) lupina Godman and Salvin, 1886
Genomic analysis reveals that Emesis tristis Stichel, 1929 (type locality in Mexico: Colima, syntype sequenced as NVG-18043E08) currently regarded as a junior subjective synonym of Emesis (Tenedia) lupina Godman and Salvin, 1886 (type locality in Costa Rica) (Zhang et al. 2019b), while being closely related to it, is genetically differentiated from it at the species level (Fig. 3), e.g., their COI barcodes differ by 2.3% (15 bp). In the presence of recognizable phenotypic differences—E. tristis of both sexes being darker in ground color, with less contrasting spots and bands compared to E. lupina—we propose to treat Emesis (Tenedia) tristis Stichel, 1929, reinstated status, as a species-level taxon.
Figure 3.
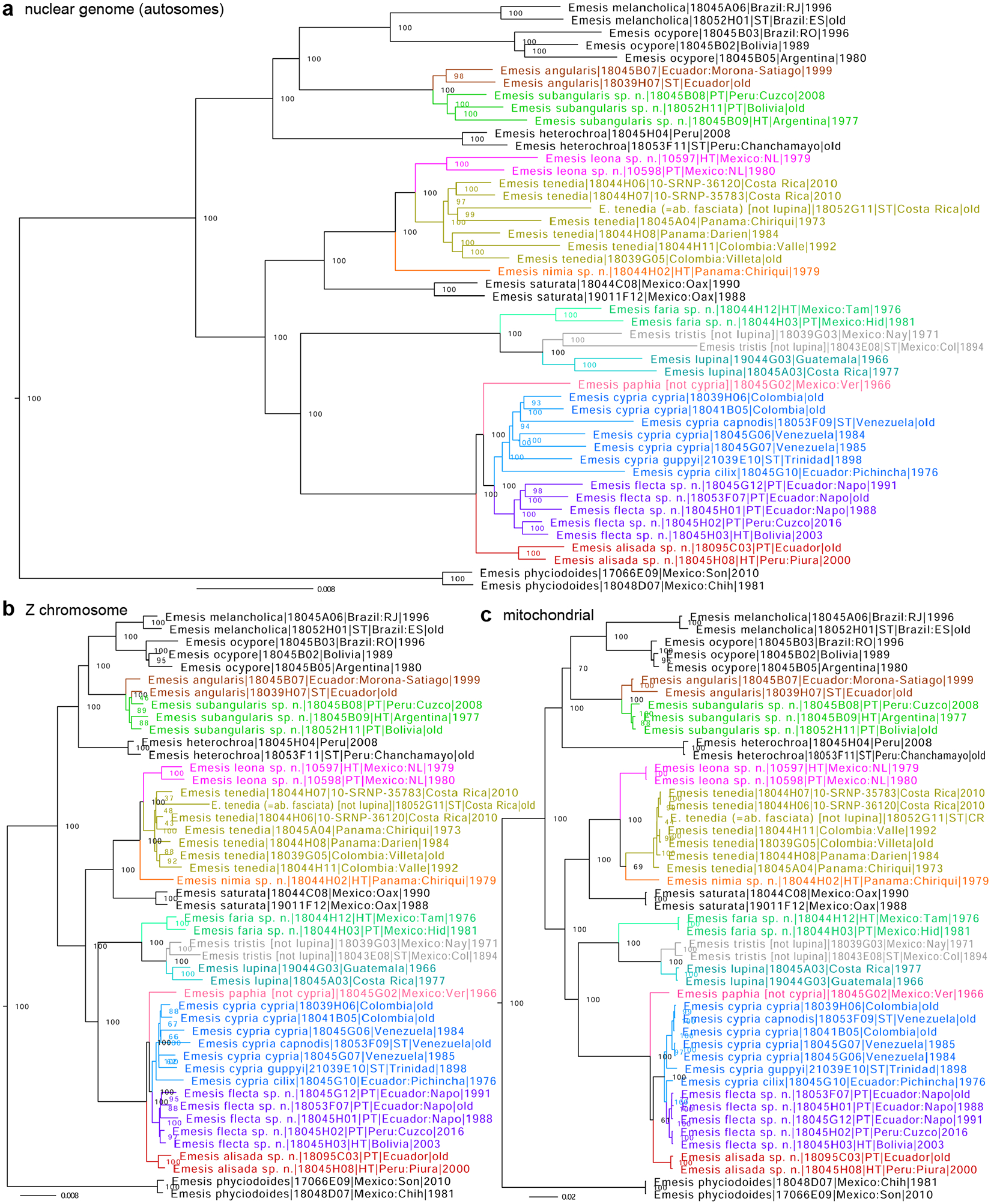
Phylogenetic trees of Emesis (Tenedia) species inferred from protein-coding regions in a) the nuclear genome (autosomes), based on 5,436,138 positions, b) the Z chromosome, based on 202,944 positions, and c) the mitochondrial genome: E. angularis (brown), E. subangularis sp. n. (green), E. leona sp. n. (magenta), E. tenedia (olive), E. nimia sp. n. (orange), E. faria sp. n. (aquamarine), E. tristis stat. rest. (gray), E. lupina (cyan), E. paphia stat. rest. (pink), E. cypria (blue), E. flecta sp. n. (purple), and E. alisada sp. n. (red). See Fig. 1 caption for other notations.
Lectotype designation for Emesis tenedia C. Felder and R. Felder, 1861
Emesis tenedia C. Felder and R. Felder, 1861 was described from several specimens from Venezuela and Colombia (Felder and Felder 1861). A male syntype from Venezuela was, according to its label, illustrated by Godman and Salvin (1886) on plate 43, figs. 16, 17. To stabilize nomenclature, clarify the type locality, and define the name E. tenedia objectively, N.V.G. hereby designates this male syntype in the BMNH collection that bears the following nine labels (1st round, 5th T-shaped, others rectangular; 1st with red outer circle, 2nd blue, 6th green others white; 6th handwritten, others printed): ( Type | H. T. ), [ ] no text on this blue label, [ Venezuela. | Druce Coll. ], [ Coll. Kaden. ], [ Type. | Sp. figured. ], [ Emesis | tenedia. Fd.Vz | ? ], [ B.C.A.Lep.Rhop. | Emesis | tenedia, | Feld. | Godman-Salvin | Coll. 1914.—5. ], [ {QR Code} | NHMUK010430897 ], and [ MOLECULAR | 0247281291 ], as the lectotype of Emesis tenedia C. Felder and R. Felder, 1861. The lectotype’s left forewing is chipped at the outer margin by the apex, and the left hindwing is nicked in the middle of the outer margin. Images of this specimen are shown on the Butterflies of America website (Warren et al. 2024). The type locality of E. tenedia becomes Venezuela.
Emesis tenedia ab. fasciata E. Strand, 1916 is a synonym of Emesis (Tenedia) tenedia C. Felder and R. Felder, 1861, not Emesis (Tenedia) lupina Godman and Salvin, 1886
An infrasubspecific name Emesis tenedia ab. fasciata E. Strand, 1916 proposed for specimen(s) from Costa Rica is currently placed in synonymy with Emesis (Tenedia) lupina Godman and Salvin, 1886 (type locality in Costa Rica) (Callaghan and Lamas 2004). Genomic sequencing of a female labeled as a “type” (formally, infrasubspecific names do not have type specimens) of E. tenedia ab. fasciata (NVG-18052G11) reveals that it is a specimen of Emesis (Tenedia) tenedia C. Felder and R. Felder, 1861 (Fig. 3), consistently with its phenotype (a prominent postdiscal pale band in the anterior half of forewing). Images of this specimen are shown on the Butterflies of America website (Warren et al. 2024). Therefore, we return Emesis tenedia ab. fasciata E. Strand, 1916 in synonymy with Emesis tenedia C. Felder and R. Felder, 1861, as originally proposed.
Emesis (Tenedia) nimia Grishin, new species
http://zoobank.org/90A323A7-029F-4C4D-92DC-FEC247F900FD
Figures 27–48.

Holotypes (unless indicated) of Emesis (Tenedia), data in text. 27–28) E. (T.) nimia sp. n. 29–32) E. (T.) faria sp. n.: 31–32) paratype NVG-18044H03. 33–36) E. (T.) leona sp. n.: 35–36) paratype NVG-10598. 37–42) E. (T.) subangularis sp. n.: 39–40) paratype NVG-18045B0, 41–42) paratypes, right hindwing, dorsal: 41) NVG-23115A12 (outer marginal area), 42) NVG-18052H11. 43–44) E. (T.) angularis non-type specimens, right hindwing, dorsal: 43) NVG-18045B07 and 44) NVG-23115A11. 45–48) E. (T.) alisada sp. n.: 47–48) Paratype NVG-18095C03.
Definition and diagnosis.
Genomic analysis of a female Emesis specimen from Panama with angular wings and a very prominent pale postdiscal half-band on the forewing (Fig. 27–28) reveals that it is genetically unique and differentiated from all others we sequenced at the species level (Fig. 3), e.g., its COI barcode differs from the closest species Emesis (Tenedia) tenedia C. Felder and R. Felder, 1861 by 2.7% (18 bp). Therefore, it represents a new species. This new species is closest to E. tenedia in females having a sharply defined cream-yellow postdiscal band in the anterior half of the forewing, but this band is deeper yellow and with the spot in the cell M3-CuA1 about half of the width of the spot in the cell M2-M3 (usually wider in E. tenedia) and differs in generally more angular wings with a stronger hooked forewing apex. In other species, the outer margin of the forewing is less wavy, with reduced concavity near the apex and in the cell CuA1-CuA2. In female genitalia (Fig. 95–96), ductus bursae is not looped, two small (less than ⅛ of the corpus diameter) horn-like signa at the caudal end of the spherical corpus bursae, sternite VII (“genital plate”) is trapezoidal, weakly sclerotized especially at the margins, with a straight posterior margin. Due to unexplored phenotypic variation and males still unknown, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne7345.5.3:T40C, cne7345.5.3:A42T, cne3225.1.1:T168C, cne285.13.6:A126T, cne13338.1.2:A216T, cne26870.1.5:G63G (not A), cne26870.1.5:C64C (not A), cne1953.11.4:A45A (not G), cne1953.11.4:T57T (not G), cne5876.11.2:T321T (not C), and COI barcode: T92C, T121C, A238G, A334G, T475C, T523C.
Barcode sequence of the holotype.
Sample NVG-18044H02, GenBank PQ203553, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACATCTTTAAGTCTATTAATTCGAATAGAATTAGGAACTTCAGGATTTCTAATTGGTGATGATCAAATTTATAATACCATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAGTACCATTAATATTAGGAGCTCCAGATATAGCTTTCCCGCGAATAAATAACATAAGATTTTGATTATTACCCCCCTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCTGGAACAGGATGAACGGTGTACCCCCCACTTTCATCTAATATTGCCCATAGAGGCTCATCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATCTTAGGAGCAATTAATTTTATCACTACTATTATTAATATACGTATTAACAATTTATCATTTGATCAAATACCCTTATTTATTTGATCAGTAGGTATCACAGCACTTTTACTTTTATTATCTTTACCTGTATTAGCTGGAGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCATTTTTTGACCCAGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 27–28, bears the following six printed (text in italics handwritten) rectangular labels, five white: [ PANAMA:CHIRIQUI | Cerro Colorado 1450m | 8°32’N 81°47’W | 9.VIII.1979 | leg. G.B.Small ], [ DNA sample ID: | NVG-18044H02 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114G11 | c/o Nick V. Grishin ], [ genitalia | NVG240817-16 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01532799 ], and one red [ HOLOTYPE ♀ | Emesis (Tenedia) | nimia Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection.
Type locality.
Panama: Chiriquí Province, Cerro Colorado, elevation 1450 m., approx. GPS 8.533, −81.783.
Etymology.
In Latin, nimius means excessive, extreme, or exaggerated and is given for the extreme looks of this species, both in its more angular wing shape and contrasty yellow patch cut through by dark veins. The name is a feminine adjective.
Distribution.
Currently known only from the holotype collected in western Panama.
Emesis (Tenedia) faria Grishin, new species
http://zoobank.org/FC8D5649-CD80-455B-9501-79C4C20959A3
Definition and diagnosis.
Genomic analysis of two Emesis (Tenedia Grishin, 2019) (type species Emesis tenedia C. Felder and R. Felder, 1861) specimens from Mexico (Fig. 3 aquamarine) reveals that they form a clade sister to both Emesis (Tenedia) lupina Godman and Salvin, 1886 (type locality in Costa Rica) (Fig. 3 cyan) and Emesis (Tenedia) tristis Stichel, 1929, reinstated status (type locality in Mexico: Colima, syntype sequenced as NVG-18043E08) (Fig. 3 gray) and genetically differentiated from them at the species level, e.g., their COI barcodes differ by 4.6% (30 bp) from E. lupina and 5.2% (34 bp) from E. tristis. Therefore, these specimens represent a new species. This new species is similar in appearance to E. tristis and E. tenedia in its darker brown dorsal colors of males and more uniformly orange-brown ventral side with darker spots and streaks, but differs from them by slightly narrower wings than in E. tristis, straighter forewing costa and less hooked apex than in E. tenedia, better defined darker bands on dorsal forewing bordered by sharper dark-brown lines composed of curved streaks and dashes, and by brighter orange color of ventral side of wings. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne3200.4.3:A136T, cne13412.2.4:G67A, cne6684.2.15:C105T, cne9580.1.12:A234C, cne3815.7.4:A169C, and COI barcode: T50C, T56C, C271T, T463C, A541G, T571C.
Barcode sequence of the holotype.
Sample NVG-18044H12, GenBank PQ203554, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACATCTCTAAGTCTATTAATTCGAATAGAATTAGGAACTTCAGGTTCTTTAATTGGTGATGATCAAATTTATAATACTATTGTCACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGTTTTGGTAACTGATTAGTACCATTAATACTAGGAGCTCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTACCTCCCTCATTAATCTTATTAATTTCAAGAAGAATCGTAGAAAATGGAGCTGGAACAGGATGAACAGTGTACCCCCCACTTTCTTCTAATATCGCTCATGGAGGATCATCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCATCTATTTTAGGAGCAATTAATTTTATTACTACTATTATTAACATACGAATTAATAATTTATCATTTGATCAAATACCTCTTTTCATTTGATCAGTAGGTATCACAGCACTTTTACTTTTGCTATCTTTACCTGTTTTAGCTGGAGCTATCACTATACTATTAACAGATCGTAATCTAAATACATCATTTTTTGATCCTGCAGGAGGAGGAGACCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 29–30, bears the following six printed (text in italics handwritten) rectangular labels, five white: [ Tamps, Mexico | Gomez Farias | leg. E.C.Knudson | 12-X-1976 ], [ DNA sample ID: | NVG-18044H12 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114G12 | c/o Nick V. Grishin ], [ genitalia | NVG240817-17 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466413 ], and one red [ HOLOTYPE ♂ | Emesis (Tenedia) | faria Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratype: 1♂ NVG-18044H03, USNMENT 01466404 Mexico: Hidalgo, Cuesta Colorada, 21-Jul-1981, W. H. Howe leg. [USNM] (Fig. 31–32).
Type locality.
Mexico: Tamaulipas, Gomez Farías.
Etymology.
The name is formed from the name of the type locality: [Gomez ]Faria[s], and is treated as a feminine noun in apposition.
Distribution.
Eastern Mexico.
Emesis (Tenedia) leona Grishin, new species
http://zoobank.org/8EA1F0FE-925B-41E4-86F7-DECF0735B178
Definition and diagnosis.
Genomic analysis of a pair of Emesis (Tenedia Grishin, 2019) specimens from Nuevo Leon, Mexico (Fig. 3 magenta) reveals that they form a clade sister to E. tenedia (Fig. 3 olive) and are genetically differentiated from it at the species level, e.g., their COI barcodes differ by 2.0% (13 bp). Therefore, these two specimens represent a new species. This new species is phenotypically similar to E. tenedia and differs from it by males with typically less elongated and weaker hooked at the apex forewings, more uniform ground color with weaker defined postdiscal paler bands, and weaker expressed gray overscaling along the postdiscal dark narrow band; and by females with less developed pale postdiscal band on the forewing, which is frequently prominent in its anterior half in E. tenedia. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne603.4.1:A54G, cne9180.2.1:A57T, cne1600.2.7:T120G, cne498.4.1:C141T, cne498.4.1:T183C, and COI barcode: T92C, T115C, T266C, T427C, T448C.
Barcode sequence of the holotype.
Sample NVG-10597, GenBank PQ203555, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGATTTCTAATTGGTGATGATCAAATTTACAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGTAATTGATTAGTACCATTAATATTAGGAGCTCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTACTACCCCCCTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCTGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCTCATAGAGGCTCATCAGTAGATTTAGCCATTTTTTCTTTACATTTAGCTGGAATTTCTTCTATCTTAGGAGCAATTAATTTTATCACTACTATTATTAATATACGTATTAATAATTTATCATTTGATCAAATACCTTTATTTATTTGATCAGTAGGTATTACAGCACTATTACTTTTATTATCTTTACCTGTATTAGCTGGAGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCATTTTTTGATCCAGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the Texas A&M University Insect Collection, College Station, TX, USA (TAMU), illustrated in Fig. 33–34, bears the following six printed (text in italics handwritten) rectangular labels, five white: [ MEXICO: | NUEVO LEON | Cola de Caballo | (horsetail falls) ], [ coll. 24-X-1979 | Roy O. Kendall | & C. A. Kendall ], [ RIODINIDAE: | Emesis tenedia | C. & R. Felder, 1861 | det. Roy O. Kendall | ♂ M. & B. no. 543.1 ], [ DNA sample ID: | NVG-10597 | c/o Nick V. Grishin ], [ genitalia | NVG180106-14 | Nick V. Grishin ], and one red [ HOLOTYPE ♂ | Emesis (Tenedia) | leona Grishin ]. Paratype: 1♀ NVG-10598 Mexico: Nuevo Leon, 25 km WSW of Linares, 12-Nov-1980, R. O. Kendall and C. A. Kendall leg., genitalia NVG180106-15 [TAMU] (Fig. 35–36, 101–102).
Type locality.
Mexico: Nuevo León, Cola de Caballo.
Etymology.
The name is formed from the name of the state with the type locality and is treated as a feminine noun in apposition.
Distribution.
Currently known only from the Sierra Madre Oriental in Nuevo Leon, Mexico.
Emesis (Tenedia) subangularis Grishin, new species
http://zoobank.org/6FB2105E-32C6-4AAB-943F-5A519563E15D
Definition and diagnosis.
Genomic analysis of Emesis (Tenedia) angularis Hewitson, 1870 (type locality in Ecuador, a syntype sequenced as NVG-18038H07) reveals that specimens initially identified as this species and collected to the south of Ecuador are genetically differentiated from the true E. angularis at the species level in the nuclear genome (Fig. 3) and their COI barcodes are 1.2% (8 bp) different. While the COI barcode difference is not very prominent, the nuclear genomes of the two species differ, and the difference correlates with the wing shape in both sexes. Therefore, we hypothesize that the nuclear genome clade (Fig. 3 green), sister to E. angularis (Fig. 3 brown), represents a new species. This new species is similar to E. angularis and differs from it by generally less angular hindwing in males, with less developed protrusion in the middle of the outer margin of the hindwing, with less concave margin anteriad and posteriad of it and usually less contrasting submarginal spot in central hindwing cell RS-M1. The female of the new species possesses a more obtuse hindwing angle at the outer margin but a more prominently hooked forewing apex with a more concave costal margin in the middle and the outer margin by the apex (i.e., more prominently hooked forewing apex). To illustrate the wing shape difference, we show the hindwing or its part for all three males in the type series (Fig. 37, 41, 42) and two males of E. angularis from Ecuador in USNM: NVG-18045B07, USNMENT 01466432 Morona-Santiago, Nueve de Octubre, 1800 m, −2.2167, −78.2167, 10-Sep-1999, R. Robbins, R. Busby, G. Estevez, and A. Aldas leg. (Fig. 43) and NVG-23115A11 Pichincha, Baeza, 2000 m, 28-Sep-1975, S. S. Nicolay leg. (Fig. 44, 105–106). In male genitalia (Fig. 103–104), the lower and upper valval projections are more parallel to each other, at a smaller angle than in E. angularis (Fig. 105–106), and uncus is convex in the middle, without a small notch. Due to relatively unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne475.6.4:T21C, cne475.6.4:C57A, cne6005.2.1:C180T, cne3598.3.3:A107T, cne7168.1.1:C410G, and COI barcode: A40G, A88G, C361C, T421C, T646C.
Barcode sequence of the holotype.
Sample NVG-18045B09, GenBank PQ203556, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTGGGAACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGGTCTTTAATCGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCTCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTACCCCCCTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCTGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATCGCCCATGGAGGATCATCAGTAGATTTAGCTATTTTTTCCTTACATTTAGCTGGTATCTCCTCTATTTTAGGAGCAATTAATTTTATTACTACTATTATTAACATACGAATTAACAATTTATCATTTGATCAAATACCTCTTTTTATTTGATCAGTAGGTATTACAGCACTTTTACTTTTATTATCTTTACCTGTATTAGCAGGAGCTATTACTATATTATTAACAGATCGTAATTTAAACACATCATTTTTTGATCCAGCAGGAGGAGGAGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 37–38, bears the following eight rectangular labels (first three handwritten, others printed), seven white: [ ARGENTINA | Salta, 750m. | Agua Blanca rd. | to Angosta, km.30–31 ], [ 17.VI.1977 | R.C.Eisele ], [ Emesis | angularis ♂ | det. Eisele ], [ DNA sample ID: | NVG-18045B09 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H01 | c/o Nick V. Grishin ], [ genitalia | NVG240817-18 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466434 ], and one red [ HOLOTYPE ♂ | Emesis (Tenedia) | subangularis Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratypes: 2♂♂ and 1♀: Peru, Cuzco [USNM]: 1♂ NVG-23115A12 Qda. Morro Leguia, 1950–2150 m, GPS −13.133, −71.550, R. 30-Aug-1989, Robbins leg. (Fig. 41, right hindwing outer margin) and 1♀ NVG-18045B08, USNMENT 01466433 Peru: Cuzco, Qbrda Buenos Aires, Cosñipata Rd., 2400 m, 19-Nov-2008, S. Kinyon leg. (Fig. 39–40, right side of the specimen) and 1♂ NVG-18052H11 Bolivia (no detailed locality), H. Stichel collection no. 3334 [MFNB] (Fig. 42, right hindwing).
Type locality.
Argentina: Salta, west of Aguas Blancas, km 30–31 of the road to El Angosto, elevation 750 m.
Etymology.
The name of this new species is formed by adding a prefix sub- to the name of its sister species, given for the less angular shape of the hindwing, and is also made longer for this more southern species, living on the map “below” (i.e., sub-) of E. angularis. The name is a feminine adjective.
Distribution.
Currently known from southern Peru, Bolivia, and northern Argentina.
Emesis (Tenedia) paphia R. Felder, 1869 is a species distinct from Emesis (Tenedia) cypria C. Felder and R. Felder, 1861
Genomic analysis reveals that Emesis paphia R. Felder, 1869 (type locality in Mexico: Veracruz), currently regarded as a subspecies of Emesis (Tenedia) cypria C. Felder and R. Felder, 1861 (type locality in Venezuela) (Callaghan and Lamas 2004), is genetically differentiated from it at the species level (Fig. 3), e.g., their COI barcodes differ by 2.1% (14 bp). In the presence of recognizable phenotypic differences—e.g., males of E. paphia are darker in ground color and typically have slightly rounder forewings with a broader orange band with more sharply defined and less diffuse edges compared to E. cypria—we propose to treat Emesis (Tenedia) paphia R. Felder, 1869, reinstated status, as a species-level taxon.
Emesis (Tenedia) alisada Grishin, new species
http://zoobank.org/8283D573-C2FA-49BA-904B-45010AE37E36
Definition and diagnosis.
Genomic analysis reveals that a pair of specimens (Fig. 3 red) initially identified as Emesis (Tenedia) cypria C. Felder and R. Felder, 1861 forms a clade sister to all other E. cypria-like specimens, including Emesis (Tenedia) paphia R. Felder, 1869, reinstated status (type locality in Mexico: Veracruz), and therefore represents a new species, which in COI barcode differs by 1.7% (11 bp) from E. cypria and by 2.0% (13 bp) from E. paphia. This new species is most similar to and sympatric with Emesis cypria cilix Hewitson, 1870 (type locality in Ecuador: Sarayaku and Mexico), at least in Alluriquin at 700 m (Pichincha, Ecuador), e.g., the specimen NVG-18045G10 we sequenced. Phenotypically, males of the two species differ in the following ways: the discal narrow, wavy dark band on the forewing is not at the right angle towards the costal margin as in E. cypria cilix (the character mentioned in the original description), but is tilted slightly distad at costa, and is offset distad between veins M3 and CuA1 (more obvious on the ventral side), and the segment of the band in cell CuA2-1A+2A is not offset distad as strongly as in E. cypria cilix. In females, the yellow transverse band does not reach the forewing tornus. In male genitalia (Fig. 107–108), uncus is as long as tegumen, lower valval projection is less robust and stronger turned inward, the upper projection is rounder and broader in ventral view. Due to unexplored phenotypic variation in this species, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne2063.5.4:A71G, cne2024.5.4:T75G, cne14049.3.3:A19C, cne14049.3.3:A108G, cne254203.4.6:G58A, and COI barcode: C226T, T364C, T442T, T448C, T457T, C508T, A586A.
Barcode sequence of the holotype.
Sample NVG-18045H08, GenBank PQ203557, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGTTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTCACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAGTACCATTAATACTAGGAGCCCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTACCCCCCTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCTGGAACAGGATGAACAGTGTACCCCCCACTTTCCTCTAATATTGCCCATGGAGGATCCTCAGTTGATTTAGCTATTTTTTCTTTACACTTAGCAGGTATCTCTTCTATTCTAGGAGCAATTAATTTTATCACCACTATTATCAATATACGAATTAATAACTTATCATTTGATCAAATACCTCTTTTTATTTGATCAGTAGGTATTACTGCACTTTTACTTTTATTATCATTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCCTTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 45–46, bears the following six printed rectangular labels, five white: [ PERU: Piura: 3km SW | Chinchin, 1800m. | 04 42’S 79 49’W | 30 May 2000 | Robbins & Lamas Leg. ], [ DNA sample ID: | NVG-18045H08 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H02 | c/o Nick V. Grishin ], [ genitalia | NVG240817-20 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466504 ], and one red [ HOLOTYPE ♂ | Emesis (Tenedia) | alisada Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratype: 1♀ NVG-18095C03 Ecuador: “Slanos” [Los Llanos], old [MTD] (Fig. 47–48).
Type locality.
Peru: Piura Region, 3 km southwest of Chinchin, elevation 1800 m, approx. GPS −4.700, −79.817.
Etymology.
In Spanish, alisado means smoothed, flattened, or straightened. The name, treated as a feminine adjective, is given for the lack of the strong kink in the postdiscal band in males at the forewing vein CuA2.
Distribution.
Currently known from the Andes of southern Ecuador and northern Peru.
Emesis (Tenedia) flecta Grishin, new species
http://zoobank.org/66B61DB6-8554-4B5C-B20D-81086B8CB475
Figures 49–62.

Holotypes (unless indicated) of Emesis (Tenedia), Emesis (Poeasia), Emesis (Brimia), and Emesis (Aphacitis), and data in text. 49–52) E. (T.) flecta sp. n.: 51–52) Paratype NVG-18045H01. 53–54) E. (P.) sonorensis sp. n. 55–56) E. (B.) apagada sp. n. 57–58) E. (B.) boliviana sp. n. 59–62) E. (A.) aurichica sp. n.: 61–62) Paratype NVG-18095C02.
Definition and diagnosis.
Genomic sequencing reveals that several specimens from Ecuador, Peru, and Bolivia (Fig. 3 purple) form a clade sister to Emesis (Tenedia) cypria C. Felder and R. Felder, 1861 (Fig. 3 blue) which, in the nuclear genome, is genetically differentiated from it at the species level with Fst/Gmin of 0.31/0.015. In the mitochondrial genome, however, the differences are small, e.g., 0.6% (4 bp) in the COI barcode. This clade represents a new species, which is similar to E. cypria and differs from it by males with dark ventral forewing postdiscal band that is wider and stronger kinked at vein CuA2, the segment in the CuA2-1A+2A cell is stronger offset distad than in E. cypria, and the two segments in this cell do not form into an arrowhead pointing basad. In male genitalia (Fig. 109–110), uncus is shorter than tegumen, lower valval projection is more robust and slightly tilted inward, the upper projection is more pointed and narrower in ventral view. Due to the cryptic nature of this species and poorly explored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne3772.6.14:C28T, cne686.4.4:A36C, cne686.4.4:A78G, cne686.4.4:A144G, cne1307.2.3:T96C, and COI barcode (may not always distinguish this species): T364T, T442C, G586G, A628A.
Barcode sequence of the holotype.
Sample NVG-18045H03, GenBank PQ203558, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGCTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTCACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAGTACCATTAATACTAGGAGCCCCAGACATAGCTTTTCCACGAATAAATAATATAAGATTTTGATTATTACCCCCCTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCTGGAACAGGATGAACAGTGTACCCCCCACTTTCCTCTAATATTGCTCATGGAGGATCCTCAGTTGATTTAGCTATTTTTTCTTTACACTTAGCAGGTATTTCTTCTATTCTAGGAGCAATTAACTTTATTACCACTATCATCAATATACGAATTAATAACTTATCATTCGATCAAATACCTCTTTTTATCTGATCAGTAGGTATTACTGCACTTTTACTTTTATTATCATTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACGGATCGTAATTTAAATACATCCTTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 49–50, bears the following eight printed (text in italics handwritten) rectangular labels, seven white: [ BOLIVIA: La Paz Province | San Lorenzo Valley | Rio San Lorenzo 800 m. | 15°48.338’S, 67°29.447’W, 12–19 April 2003 | Brian Harris, coll. ], [ ON DAMP SOIL | BY RIVER ], [ Emesis | cypria ♂ | det. Brian Harris 2003 ], [ DNA sample ID: | NVG-18045H03 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H03 | c/o Nick V. Grishin ], [ genitalia | NVG240817-21 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466499 ], and one red [ HOLOTYPE ♂ | Emesis (Tenedia) | flecta Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratypes: 3♂♂ and 1♀: Ecuador, Napo Province: 1♂ NVG-18053F07 Santa Inés, R. Haensch S. leg, old, H. Stichel collection No. 3339 [MFNB]; 1♀ NVG-18045H01, USNMENT 01466497 4 km E Puerto Napo, 500 m, −1.050, −77.783 (could be a mistake and this is the same locality as listed for the next specimen), 6–10-Nov-1988, D. H. Ahrenholz leg. [USNM] (Fig. 51–52); and 1♂ NVG-18045G12, USNMENT 01466496 14 km E Puerto Napo, 470 m, −1.050, −77.683, 24-Sep-1991, D. H. Ahrenholz leg. [USNM]; and 1♂ NVG-18045H02, USNMENT 01466498 Peru, Cuzco, Cosñipata Valley, El Mirador, km 68, elevation 1720 m, 25-Oct-2016, S. Kinyon leg. [USNM].
Type locality.
Bolivia: La Paz Province, San Lorenzo Valley, Rio San Lorenzo, elevation 800 m, GPS −15.8056, −67.4908.
Etymology.
In Latin, flectus means bent, curved, or bowed. The name, a participle, refers to the orange band and its dark framing along the proximal margin bent near the ventral forewing tornus.
Distribution.
Ecuador, Peru, and Bolivia.
Lectotype designation for Emesis poeas Godman, 1901
Emesis poeas Godman, 1901 was “described from two males and three females from Western Mexico” (Godman 1901). A male syntype from Acapulco, Guerrero, Mexico was, according to its label, illustrated by Godman (1901) on plate 110, figs. 7, 8. To stabilize nomenclature, clarify the type locality, and define the name E. poeas objectively, N.V.G. hereby designates this male syntype in the BMNH collection that bears the following eight labels (1st round, others rectangular; 5th handwritten, others printed; 1st with red outer circle, others white): ( Type | H. T. ), [ Acapulco, | Guerrero. | Sept. H.H.Smith ], [ ♂ ], [ Sp. figured. ], [ E. poeas ♂. ], [ B.C.A.Lep.Rhop. | Emesis | poeas, | G. & S. | Godman-Salvin | Coll. 1914.—5. ], [ {QR Code} | NHMUK010430896 ], [ MOLECULAR | 0247278436 ], as the lectotype of Emesis poeas Godman, 1901. The lectotype’s left forewing is damaged at the apex, and the left hindwing is nicked twice at the outer margin towards the tornus. Images of this specimen are shown on the Butterflies of America website (Warren et al. 2024). The type locality of E. poeas becomes Mexico: Guerrero, Acapulco.
Emesis (Poeasia) sonorensis Grishin, new species
http://zoobank.org/536E5AFC-C7D0-4BE4-8020-6C3473916B3B
Figure 4.

Phylogenetic trees of Emesis (Poeasia) and Emesis (Brimia) species (top and bottom clades, respectively) inferred from protein-coding regions in a) the nuclear genome (autosomes), based on 2,139,909 positions, b) the Z chromosome, based on 401,862 positions, and c) the mitochondrial genome: E. (P.) sonorensis sp. n. (green), E. (P.) poeas (purple), E. (B.) temesa (blue, with E. temesa peruviana labeled in cyan), E. (B.) apagada sp. n. (magenta), and E. (B.) boliviana sp. n. (orange). See Fig. 1 caption for other notations.
Definition and diagnosis.
Genomic analysis reveals that specimens from Mexico identified as Emesis (Poeasia) poeas Godman, 1901 (type locality in Mexico: Guererro, lectotype sequenced as NVG-18081G07, NHMUK_010430896) partition into two clades (Fig. 4 purple and green) genetically differentiated from each other at the species level, e.g., their COI barcodes differ by 1.8% (12 bp). One clade includes the lectotype of E. poeas, while the other one, more northern in distribution, corresponds to a new species. This new species is phenotypically similar to E. poeas and differs from it by being, on average, smaller, darker, and with a less contrasting pattern, i.e., more uniformly colored with weaker defined alternating grayer and more saturated in color reddish bands, and ventrally yellower orange rather than browner as E. poeas. In male genitalia (Fig. 111–112), valvae are less robust than in E. poeas (Fig. 113–114, NVG-23111B11 Mexico: Oaxaca, Rt. 200 at Rio Huamelula, 16-Jul-1981, D. S. Bogar, J. C. Schaffner, and T. P. Friedlander leg. [CMNH]), the lower projection is stronger curved inward and terminally less rounded, the upper projection is laterally narrower and more curved dorsad; the posterior lobe in the middle of vinculum is larger, plate-like, trapezoidal, with more concave posterior margin. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne213.6.1:A360G, cne562.20.2:A522C, cne896.9.8:G42A, cne3178.4.9:C6A, cne3178.4.9:T9C, and COI barcode: T212T, C235T, T250T, A382A, T418T.
Barcode sequence of the holotype.
Sample NVG-18044G03, GenBank PQ203559, 658 base pairs:
AACATTATATTTCATTTTTGGAATTTGAGCAGGAATAGTAGGAACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGATCTTTAATTGGTGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTCTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTACCTCTTATATTAGGAGCACCTGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGATTATTACCTCCTTCATTATTTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCTTCTAATATTGCTCATAGAGGTTCTTCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTTTAGGAGCAATTAATTTTATTACTACTATTATTAATATACGTATTAATAATATATCATTTGATCAAATACCTTTATTTGTTTGATCAGTAGGAATTACAGCTCTTTTATTATTATTATCTTTACCTGTTTTAGCTGGTGCTATTACTATATTATTAACTGATCGTAATTTAAATACATCATTTTTTGACCCTGCAGGTGGTGGAGACCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 53–54, bears the following six printed rectangular labels, five white: [ Riodinidae IX-23-1987 | Emesis poeas M | Tepoca, Sonora, Mexico | Leg: John Kemner ], [ DNA sample ID: | NVG-18044G03 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H04 | c/o Nick V. Grishin ], [ genitalia | NVG240817-22 | Nick V. Grishin ], [ USNMENT | {QR Code} | 00940216 ], and one red [ HOLOTYPE ♂ | Emesis (Poeasia) | sonorensis Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratypes: 2♀♀: NVG-18044G04, USNMENT 00940173 the same data as the holotype and NVG-18052H06 “Texas” Fruhstorfer, H. Stichel collection No. 3304 [MFNB]. The locality of the last paratype is most likely in error.
Type locality.
Mexico: Sonora, Tepoca.
Etymology.
The name is formed from the name of the state with the type locality and is a feminine adjective.
Distribution.
Currently known only from Sonora in Mexico.
Two new species related to Emesis (Brimia) temesa (Hewitson, 1870)
Genomic analysis of the subgenus Brimia Grishin, 2019 (type species Emesis brimo Godman and Salvin, 1889) reveals a clade in the nuclear genome trees (Fig. 4 magenta and orange) that is sister to Emesis (Brimia) temesa (Hewitson, 1870) (type locality in Ecuador) (Fig. 4 blue and cyan) and consists of two unnamed subclades genetically differentiated from each other and E. temesa at the species level, e.g., their COI barcodes differ from each other by 2.1% (14 bp) and from E. temesa by 2.7% (18 bp) and 2.3% (15 bp). Therefore, these subclades correspond to two new species that are described next.
Emesis (Brimia) apagada Grishin, new species
http://zoobank.org/B7A63CB1-2EDC-4641-9DE0-EB5D5A0271C8
Definition and diagnosis.
This is a more northern species from the clade sister to Emesis (Brimia) temesa (Hewitson, 1870) (type locality in Ecuador) and is sympatric with it in Rondônia, Brazil. This new species is phenotypically similar to E. temesa and differs from it, including sympatric Emesis temesa peruviana (Lathy, 1904) (type locality in Peru: Junín) by the reduced red coloration of the ventral side, including much reduced red overscaling on ventral hindwing. Most notably, the forewing margin is brown (not largely reddish with a narrow brown frame towards the apex as is E. temesa) including part of the area with the submarginal row of black spots. In male genitalia (Fig. 115–116), the posterior lobe in the middle of vinculum is longer and more triangular, uncus is narrower and with a more prominent central bulge, saccus and the lower valval projection are longer, the upper valval projection is only slightly curved dorsad and outward. Due to unexplored phenotypic variation in this species, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne6505.1.20:G347A, cne6505.1.20:T366C, cne4919.1.4:T36C, cne4919.1.4:A111G, cne10177.13.11:T75C, and COI barcode: A73G, A208G, A238T, T259C, C406T, A619C.
Barcode sequence of the holotype.
Sample NVG-18045D08, GenBank PQ203560, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACATCTTTAAGTTTATTAATTCGTATGGAGTTAGGAACTTCAGGCTCTTTAATTGGAGATGATCAAATCTATAATACTATTGTAACAGCTCACGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGAAATTGATTAGTACCTTTGATATTAGGAGCCCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTCTGACTATTACCCCCATCATTATTTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCCTCTAATATTGCACATGGAGGTTCTTCAGTAGATTTAGCTATTTTTTCCTTACATTTAGCAGGAATTTCTTCAATTTTAGGAGCAATTAATTTTATTACTACAATCATTAATATACGAATTAATAATATATCATTTGACCAAATACCATTATTTGTTTGATCAGTTGGAATTACTGCTTTATTATTATTATTATCCTTACCAGTATTAGCAGGTGCCATCACTATATTATTAACTGACCGTAACTTAAATACATCCTTTTTTGACCCCGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 55–56, bears the following six printed rectangular labels, five white: [ PERU, M. de Dios, Par- | que Manu, Pakitza 340m | 11°55’48”S 71°15’18”W | 12 Oct 1991 | Leg. M. Cassagrande ], [ DNA sample ID: | NVG-18045D08 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H05 | c/o Nick V. Grishin ], [ genitalia | NVG240817-24 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466457 ], and one red [ HOLOTYPE ♂ | Emesis (Brimia) | apagada Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratype: 1♂ NVG-18045D11 USNM 01466460 Brazil: Rondonia, vic. Caucalandia, −10.533, −62.800 10-Oct-1991, J. MacDonald leg. [USNM].
Type locality.
Peru: Madre de Dios Region, Manu National Park, Pakitza, elevation 340 m, approx. GPS −11.930, −71.255.
Etymology.
In Spanish, apagado means extinguished or muted, referring to the reduced flaming coloration of the ventral side of this species. The name is treated as a feminine adjective.
Distribution.
From southeastern Peru to west-central Brazil.
Emesis (Brimia) boliviana Grishin, new species
http://zoobank.org/9D5266C2-EA39-49B7-9BE2-6652F489A469
Definition and diagnosis.
This is a more southern species from the clade sister to Emesis (Brimia) temesa (Hewitson, 1870) (type locality in Ecuador). This new species is phenotypically similar to E. temesa and Emesis (Brimia) apagada, new species (type locality in Peru: Madre de Dios), and differs from them by the hindwing being red beneath up to the postdiscal row of black spots and forewing brown margin being wider than in E. temesa, especially towards the tornus. In male genitalia (Fig. 117–118), the posterior lobe in the middle of vinculum is shorter and more trapezoidal with rounded angles and strongly concave in the middle, uncus is broader and with a smaller central bulge, saccus and the lower valval projection are shorter, the upper valval projection is stronger curved dorsad and very slightly inward. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne10024.5.4:T69A, cne520.3.3:C647T, cne520.3.3:T675C, cne1657.3.4:A66G, cne15996.2.4:T24C, cne6505.1.20:G347G (not A), cne6505.1.20:T366T (not C), cne3772.15.9:T120T (not C), cne3772.15.9:T133T (not C), cne4809.1.3:T156T (not C), and COI barcode: T88C, T163C, C586C, T595T, T634C.
Barcode sequence of the holotype.
Sample NVG-18045D10, GenBank PQ203561, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACATCTTTAAGTTTATTAATTCGTATAGAATTAGGAACTTCAGGCTCTTTAATTGGAGATGATCAAATCTATAATACTATTGTAACAGCTCACGCTTTTATTATAATTTTTTTTATAGTCATACCTATTATAATTGGTGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTACCCCCATCATTATTTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCTTCTAATATTGCACATGGAGGTTCTTCAGTAGATTTAGCTATTTTTTCCTTACACTTAGCAGGAATTTCTTCAATTTTAGGAGCAATTAATTTTATTACTACAATCATTAATATACGAATTAATAATATATCATTTGACCAAATACCATTATTTGTTTGATCAGTTGGAATTACTGCTTTATTATTATTATTATCCTTACCAGTATTAGCAGGTGCCATCACTATATTATTAACCGACCGTAATTTAAATACATCCTTTTTTGACCCAGCAGGAGGAGGAGACCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 57–58, bears the following seven printed (text in italics handwritten) rectangular labels, six white: [ BOLIVIA: La Paz Province | San Lorenzo Valley | Rio San Lorenzo 800 m. | 15°48.338’S, 67°29.447’W, 12–19 April 2003 | Brian Harris, coll. ], [ Emesis ♂ | temesa | det. Brian Harris 2003 ], [ DNA sample ID: | NVG-18045D10 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H06 | c/o Nick V. Grishin ], [ genitalia | NVG240817-25 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466459 ], and one red [ HOLOTYPE ♂ | Emesis (Brimia) | boliviana Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection.
Type locality.
Bolivia: La Paz Province, San Lorenzo Valley, Rio San Lorenzo, elevation 800 m, GPS −15.8056, −67.4908.
Etymology.
The name is formed from the name of the country with the type locality and is a feminine adjective.
Distribution.
Currently known only from the holotype collected in western Bolivia.
Emesis (Aphacitis) parvissima Kaye, 1921 is a species distinct from Emesis (Aphacitis) lucinda (Cramer, 1775)
Genomic analysis reveals that Emesis lucinda parvissima Kaye, 1921 (type locality in Trinidad, syntypes sequenced as NVG-21037B02 and NVG-21037B03) currently regarded as a junior subjective synonym of Emesis (Aphacitis) lucinda (Cramer, 1775) (type locality in Suriname) (Callaghan and Lamas 2004) is genetically differentiated from it at the species level (Fig. 5), e.g., their COI barcodes differ by 6.2% (41 bp), and therefore is not synonymous with it. Moreover, the syntypes of E. lucinda parvissima, together with another specimen from Venezuela (NVG-18044B08) that we identified as this taxon, form a clade sister to Emesis (Aphacitis) aurimna (Boisduval, 1870) (type locality in Colombia) in the nuclear genome trees (Fig. 5a, b) and distinct from it at the species level, e.g., their COI barcodes differ by 2.3% (15 bp). In the presence of recognizable phenotypic differences—females of E. lucinda parvissima are darker, with smaller white apical spots on forewing compared to E. aurimna—we propose to treat Emesis (Aphacitis) parvissima Kaye, 1921, new status, as a species-level taxon.
Figure 5.
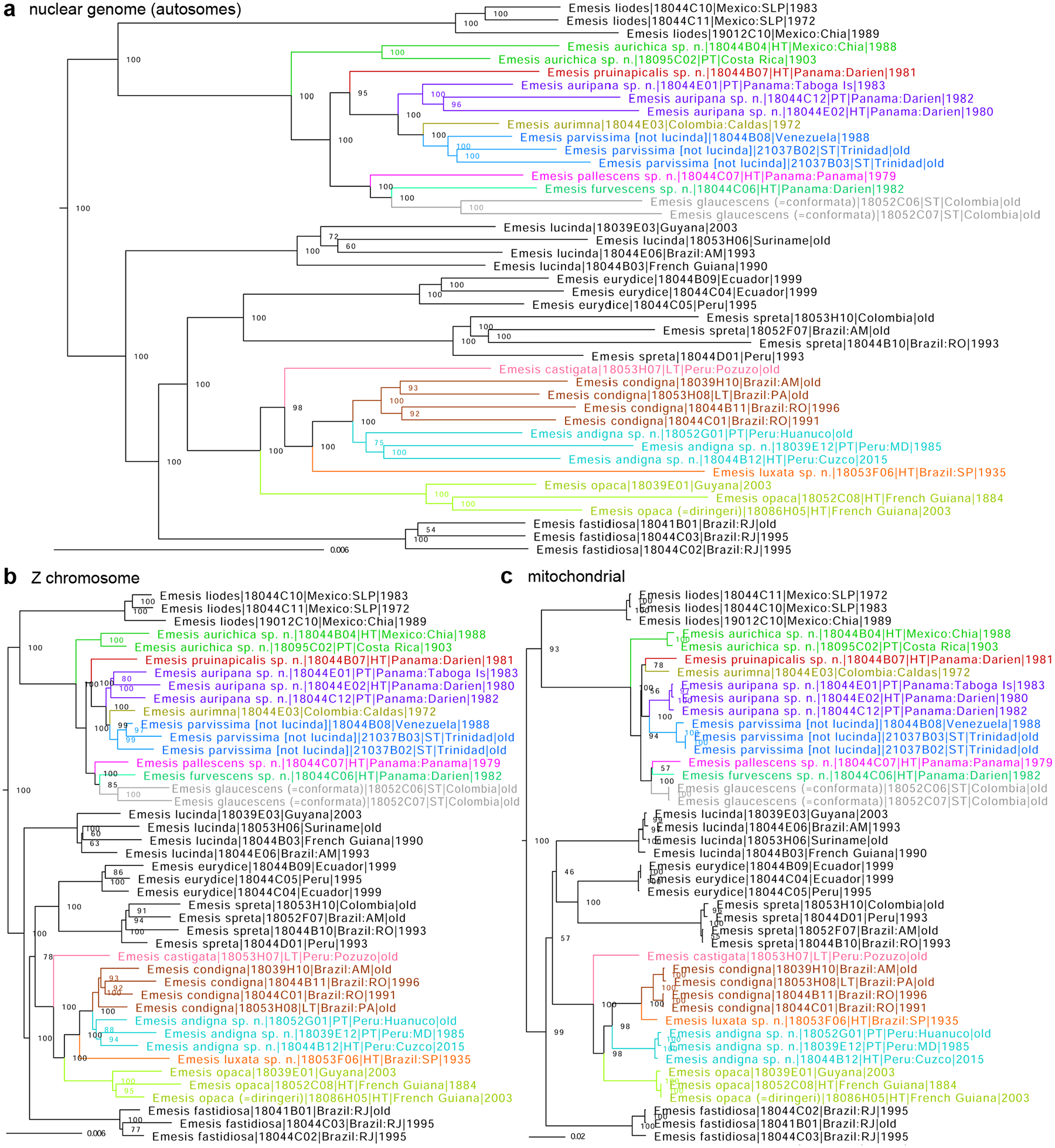
Phylogenetic trees of Emesis (Aphacitis) species inferred from protein-coding regions in a) the nuclear genome (autosomes), based on 3,692,175 positions, b) the Z chromosome, based on 315,075 positions, and c) the mitochondrial genome: E. aurichica sp. n. (green), E. pruinapicalis sp. n. (red), E. auripana sp. n. (purple), E. aurimna (olive), E. parvissima stat. nov. (blue), E. pallescens sp. n. (magenta), E. furvescens sp. n. (aquamarine), E. glaucescens (gray), E. castigata (pink), E. condigna (brown), E. andigna sp. n. (cyan), E. luxata sp. n. (orange), and E. opaca (lime). See Fig. 1 caption for other notations.
Five new species related to Emesis (Aphacitis) aurimna (Boisduval, 1870) from Central America
Genomic analysis of the subgenus Aphacitis Hübner, [1819] (type species Papilio dyndima Cramer, [1780], a junior homonym, current name applied to this species is Papilio lucinda Cramer, [1775]) reveals that the clade with Emesis (Aphacitis) aurimna (Boisduval, 1870) (type locality in Colombia) consists of eight phylogenetic lineages with genetic differentiation between them at the species level (Fig. 5 green, red, olive, purple, blue, magenta, aquamarine, and gray). In addition to E. aurimna, this clade includes two known species: Emesis (Aphacitis) parvissima Kaye, 1921, new status (type locality in Trinidad) discussed above, differing by 2.3% (15 bp) in the COI barcode from E. aurimna and Emesis (Aphacitis) glaucescens Talbot, 1929 (type locality in Colombia) differing by 1.7% (11 bp) from E. aurimna (while not being closer related to it even in the mitochondrial genome tree, Fig. 5c, thus the smaller barcode distance is a result of some anomaly) and by 2.7% (18 bp) from E. parvissima. The other five species with approximately the same genetic differences from the known ones do not have names and are new. Specimens of these species that we sequenced were collected in Central America, from Southern Mexico to Panama, and several of these species are sympatric in eastern Panama. These five species are described next, and their COI barcode distances are given in individual species accounts.
Emesis (Aphacitis) aurichica Grishin, new species
http://zoobank.org/D421A535-0198-4C66-9E64-5E983B48351F
Definition and diagnosis.
This is the northernmost species of the five revealed by genomic sequencing of the subgenus Aphacitis Hübner, [1819], as detailed above. This new species (Fig. 5 green) is sister to all other seven species in the clade with Emesis (Aphacitis) aurimna (Boisduval, 1870) (type locality in Colombia) and most differentiated genetically from others, e.g., its COI barcode differs by 3.2% (21 bp) from E. aurimna. This new species is phenotypically similar to others in the E. aurimna clade and differs from its relatives by a combination of the following characters in female: white forewing subapical area is less extensive and stronger separated from the wing margins by the ground brown color, both above and beneath, the ventral side is with submarginal inverted crescents connected with each other, not clearly separated as in E. aurimna, and dark web pattern is generally more extensive ventrally. Males have a stronger postdiscal dark band from costa to tornus on dorsal forewing, i.e., the postdiscal band visually merges into the submarginal band instead of bending basad as in other species; this bent portion from vein M1 to the inner margin is present but is usually not as prominent as the submarginal branch; subapical forewing area is paler, but not strongly frosted with white scales, and ventral side is brighter orange compared to other species. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne16034.1.2:A125G, cne16034.1.2:A735C, cne6734.6.2:A57G, cne6734.6.2:C78T, cne2296.1.3:A34G, and COI barcode: T226C, A229G, T250C, T550C, T574C.
Barcode sequence of the holotype.
Sample NVG-18044B04, GenBank PQ203562, 658 base pairs:
AACATTATACTTTATTTTTGGAATTTGATCAGGGATAGTCGGCACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACCTCAGGTTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATCGGAGGATTTGGTAACTGATTAGTTCCATTAATACTAGGAGCACCTGACATGGCTTTCCCACGAATAAATAACATAAGATTTTGACTTTTACCACCATCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCCCATGGAGGAGCTTCAGTTGATTTAGCTATCTTTTCCCTTCATTTAGCTGGTATTTCATCTATTTTAGGAGCAATTAATTTTATCACAACAATCATTAATATACGTATTAACAATATGTCATTTGATCAAATACCATTATTTGTCTGATCTGTTGGAATTACAGCCCTTTTACTTTTATTGTCTCTCCCAGTTTTAGCTGGAGCTATTACCATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 59–60, bears the following six printed rectangular labels, five white: [ Riodinidae VIII-2-1988 | Emesis lucinda M | Agua Azul, CHIS, Mexico ], [ DNA sample ID: | NVG-18044B04 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H07 | c/o Nick V. Grishin ], [ genitalia | NVG240817–26 | Nick V. Grishin ], [ USNMENT | {QR Code} | 00940155 ], and one red [ HOLOTYPE ♂ | Emesis (Aphacitis) | aurichica Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratype: 1♀: NVG-18095C02 Costa Rica (no detailed locality), 1903 [MTD] (Fig. 61–62).
Type locality.
Mexico: Chiapas, Agua Azul.
Etymology.
The name for this E. aurimna relative from Chiapas and Costa Rica is formed as a fusion: auri[mna] + Chi[apas] + [Costa Ri]ca and is treated as a feminine noun in apposition.
Distribution.
Currently known from southern Mexico (Chiapas) and Costa Rica.
Emesis (Aphacitis) auripana Grishin, new species
http://zoobank.org/C51B6181-7DA5-4330-AC99-2AEFBA530032
Figures 63–70.

Holotypes (unless indicated) of Emesis (Aphacitis), and data in text. 63–66) E. (A.) auripana sp. n.: 65–66) Paratype NVG-18044E01. 67–68) E. (A.) pruinapicalis sp. n. 69–70) E. (A.) furvescens sp. n.
Definition and diagnosis.
This new species (Fig. 5 purple) is sister to both Emesis (Aphacitis) aurimna (Boisduval, 1870) (type locality in Colombia) and Emesis (Aphacitis) parvissima Kaye, 1921, new status (type locality in Trinidad) in the nuclear genome trees, and therefore is distinct from either of them. In the COI barcodes, the difference is 2% (13 bp) from E. aurimna and 2.1% (14 bp) from E. parvissima. This new species is phenotypically similar to others in the E. aurimna clade and differs from its relatives by a combination of the following characters in female: white forewing subapical area is less extensive (but not as small as in E. parvissima), consists of smaller spots and not as prominently extending along veins towards the apex as in E. aurimna, and the apical separate white spot is smaller. Ventral hindwing is more similar to E. aurimna and E. parvissima in pattern, with submarginal inverted crescents separated from each other, with smaller spots there, white not prominently extending along veins towards apex, and the very apical separate spot is smaller as well. In males, white subapical frosting on the dorsal forewing is present (not vestigial as in E. parvissima) but less extensive than in E. aurimna; ventrally, wings are slightly yellower and weaker patterned. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne4410.1.5:G204A, cne4410.1.5:G210A, cne2706.1.64:A165G, cne793.4.4:A96T, cne793.4.4:A165G, and COI barcode: A61A, T88C, T130T, T247T, T610C.
Barcode sequence of the holotype.
Sample NVG-18044E02, GenBank PQ203563, 658 base pairs:
AACATTATACTTTATTTTTGGAATTTGATCAGGAATAGTCGGCACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACCTCAGGCTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAACTGATTAGTTCCATTAATATTAGGAGCACCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCACCATCATTAATTTTATTAATTTCAAGAAGAGTTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCCCATGGAGGAGCCTCAGTTGATTTAGCTATTTTTTCCCTTCATTTAGCTGGTATTTCATCTATTTTGGGAGCAATTAATTTTATCACAACAATCATTAATATACGTATTAATAATATGTCATTTGATCAAATACCATTATTTGTCTGATCTGTTGGAATTACAGCTCTTTTACTTTTATTATCTCTTCCAGTTTTAGCCGGAGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCTTTCTTTGACCCTGCTGGGGGAGGAGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 63–64, bears the following six printed rectangular labels, five white: [ PANAMA: Darién | Canglón | 14.ix.1980 | leg. G. B. Small ], [ DNA sample ID: | NVG-18044E02 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H08 | c/o Nick V. Grishin ], [ genitalia | NVG240817-27 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466369 ], and one red [ HOLOTYPE ♂ | Emesis (Aphacitis) | auripana Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratypes: 1♂ and 1♀ from Panama [USNM]: 1♂ NVG-18044C12 USNMENT 01466357 Darién, Cana, 400 m, 20-Sep-1982, G. B. Small leg., genitalia #2003–46, Donald J. Harvey; 1♀ NVG-18044E01, USNMENT 01466368, Panamá, Taboga Island, 1-Nov-1983, J. F. G. Clarke (Fig. 65–66).
Type locality.
Panama: Darién Province, Canglón.
Etymology.
The name for this E. aurimna relative from Panama is formed as a fusion: auri[mna] + Pana[ma] and is treated as a feminine noun in apposition.
Distribution.
Central and eastern Panama.
Emesis (Aphacitis) pruinapicalis Grishin, new species
http://zoobank.org/0F94E2B7-C12F-4BE9-A643-B94414C23302
Definition and diagnosis.
This new species (Fig. 5 red) is sister to the clade that consists of Emesis (Aphacitis) aurimna (Boisduval, 1870) (type locality in Colombia), Emesis (Aphacitis) parvissima Kaye, 1921, new status (type locality in Trinidad), and Emesis (Aphacitis) auripana, new species (type locality in Panama: Darién), in the nuclear genome trees, and therefore is distinct from all others. In the COI barcodes, the difference is 2.1% (14 bp) from E. aurimna, 2.6% (17 bp) from E. parvissima, and 2.0% (13 bp) from E. auripana. This new species is phenotypically similar to others in the E. aurimna clade and differs from its relatives by a combination of the following characters in male (female is unknown): dorsal forewing heavily frosted with white scales in the apical quarter, frosting reaches vein CuA2 along the outer margin, white scales present in the postdiscal area between costa and vein M2, separated from the subapical white overscaling by a darker olive-brown inverted-L-shaped postdiscal band, submarginal dark spots are not developed, giving this species a more “frosted” appearance; ventral side orange, yellower in the middle of cells and darker along the veins and dark bands and spots, mostly orange along out wing margins, with dark spots towards forewing tornus and at the hindwing apex. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne23605.2.8:C216T, cne3560.4.6:C232T, cne6180.2.5:A192G, cne471.1.1:A802C, cne2676.3.1:T487C, cne426.6.1:A630A (not G), cne2343.2.13:C183C (not A), cne2343.2.13:T537T (not C), cne12832.2.1:A218A (not T), cne5563.4.3:G22G (not C), and COI barcode: T10T, T197C, A289G, A316G, A466G, T595C.
Barcode sequence of the holotype.
Sample NVG-18044B07, GenBank PQ203564, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGATCAGGAATAGTCGGCACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACCTCAGGTTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAACTGACTAGTTCCATTAATATTAGGAGCACCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCACCATCATTAATTTTATTGATTTCAAGAAGAATTGTAGAAAATGGGGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCCCATGGAGGAGCCTCAGTTGATTTAGCTATTTTTTCCCTTCATTTAGCTGGTATTTCATCTATTTTAGGAGCAATTAATTTTATCACAACAATCATTAATATGCGTATTAATAATATGTCATTTGATCAAATACCATTATTTGTTTGATCTGTTGGAATTACAGCTCTTTTACTTTTATTATCTCTTCCAGTTTTAGCTGGAGCTATTACTATATTATTAACAGATCGTAACTTAAATACATCTTTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 67–68, bears the following six printed (text in italics handwritten) rectangular labels, five white: [ PANAMA:DARIEN | Cana (Cerro Pirre) | 400m | 7°56’N 77°43’W | 26 VII 1981 | leg. G.B.Small ], [ DNA sample ID: | NVG-18044B07 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H09 | c/o Nick V. Grishin ], [ genitalia | NVG240817-28 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466341 ], and one red [ HOLOTYPE ♂ | Emesis (Aphacitis) | pruinapicalis Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection.
Type locality.
Panama: Darién Province, Cana, Cerro Pirre, elevation 400 m, approx. GPS 7.9333, −77.7167.
Etymology.
In Latin, pruina means frost, and apicalis means apical or related to the apex. The name is given for the frosted apical part of the forewing and is a feminine adjective.
Distribution.
Currently known only from the holotype collected in eastern Panama.
Emesis (Aphacitis) furvescens Grishin, new species
http://zoobank.org/4DAF6790-C417-46F3-A92D-AC27F794DD20
Definition and diagnosis.
This new species (Fig. 5 aquamarine) is sister to Emesis (Aphacitis) glaucescens Talbot, 1929 (type locality in Colombia: Montepa) in the nuclear genome tree and genetically differentiated from it at the species level. Although their COI barcodes do not differ strongly, by 1.4% (9 bp), this difference is coupled with nuclear genome differentiation and phenotypic differences. This new species is phenotypically most similar to E. glaucescens and differs from it by a combination of the following characters in male (female is unknown): darker on both sides of wings, with reduced pale bluish-white frosting towards dorsal forewing apex, with dark submarginal spots in the frosted area, ventral side of wings with dark-brown margins, orange-yellow spots within this dark-brown border are lacking or vestigial, forewing submarginal area is orange and only slightly yellower than the ground color (not pale-yellow as in E. glaucescens). Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne2149.1.2:C63G, cne2149.1.2:A90T, cne392.2.10:C100T, cne392.2.10:G145A, cne3775.6.11:G87A, cne10177.2.4:T46T (not C), cne10177.2.4:T42T (not C), cne865.2.5:C159C (not T), cne13674.5.7:G70G (not A), cne13674.5.7:A75A (not T), and COI barcode: 133T, T397C, T463C, 481G, 616T.
Barcode sequence of the holotype.
Sample NVG-18044C06, GenBank PQ203565, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGATCAGGAATAGTCGGCACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACCTCAGGTTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAACTGATTAGTTCCATTAATATTAGGAGCACCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCACCATCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCCCATGGAGGAGCCTCAGTTGATTTAGCTATTTTCTCCCTTCATTTAGCTGGTATCTCATCTATTTTAGGAGCAATTAATTTTATCACAACAATCATTAACATACGTATTAATAATATGTCATTTGATCAAATACCATTATTTGTTTGATCTGTTGGAATTACAGCTCTTTTACTTTTATTGTCTCTTCCAGTTTTAGCCGGAGCTATTACCATACTATTAACAGATCGTAATTTAAATACATCTTTTTTTGATCCTGCTGGAGGAGGAGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 69–70, bears the following five printed (text in italics handwritten) rectangular labels, four white: [ Panama:Darien | Cana 1000 m. | 4.IX.1982 | G.B. Small ], [ Genitalia Dissection | #2003 – 47 | Donald J. Harvey ], [ DNA sample ID: | NVG-18044C06 | c/o Nick V. Grishin ], [ USNMENT | {QR Code} | 01466352 ], and one red [ HOLOTYPE ♂ | Emesis (Aphacitis) furvescens Grishin ].
Type locality.
Panama: Darién Province, Cana, elevation 1000 m.
Etymology.
In Latin, furvus means dark or dusky, and the name is formed similarly to the name of its relative E. glaucescens to mean becoming darker, given for the reduced bluish-white overscaling at the dorsal forewing apex and darker colors of the ventral side. The name is a participle.
Distribution.
Currently known only from the holotype collected in eastern Panama.
Emesis (Aphacitis) pallescens Grishin, new species
http://zoobank.org/619F1F8C-33EA-4B25-936D-6B2628D6B3EC
Definition and diagnosis.
This new species (Fig. 5 magenta) is sister to both Emesis (Aphacitis) glaucescens Talbot, 1929 (type locality in Colombia: Montepa) and Emesis (Aphacitis) furvescens, new species (type locality in Panama: Darién) and genetically differentiated from them at the species level, e.g., their COI barcodes differ by 1.2% (8 bp, moderate, but accompanied by the nuclear genome differentiation and phenotypic differences) from E. glaucescens and 2.0% (13 bp) from E. furvescens. This new species is phenotypically most similar to E. glaucescens and E. furvescens in the general wing pattern of males (female unknown), e.g., all three species have a nearly solid-brown outer border (with pale spots in some species) on the ventral side of all wings and developed pale overscaling in the subapical area of the dorsal forewing, but differs from others by being paler overall, particularly in the ground color of the dorsal side and reduced white subapical frosting, thus is more similar in appearance of the dorsal side to Emesis (Aphacitis) aurimna (Boisduval, 1870) (type locality in Colombia). Ventrally with reduced brown bands and lines compared to E. glaucescens and E. furvescens, and more orange in the submarginal area of the forewing than E. glaucescens. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne213.7.2:G664T, cne213.7.2:A679T, cne1255.2.6:G267A, cne1255.2.6:A271G, cne1255.2.6:T298G, cne953.8.8:T93T (not C), cne953.8.8:G108G (not A), cne2149.1.2:C63C (not G), cne2149.1.2:A90A (not T), cne6935.6.4:A218A (not C), and COI barcode: G200G, T355A, T490C, T514C, A526C.
Barcode sequence of the holotype.
Sample NVG-18044C07, GenBank PQ203566, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGATCAGGAATAGTCGGTACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACCTCAGGTTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAACTGATTAGTTCCATTAATATTAGGAGCACCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCACCATCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCAAATATTGCCCATGGAGGAGCCTCAGTTGATTTAGCTATTTTTTCCCTTCATTTAGCTGGTATCTCATCTATTTTAGGAGCAATTAATTTTATCACAACAATCATTAATATACGTATTAATAATATATCATTTGACCAAATACCATTATTTGTTTGATCCGTTGGAATTACCGCTCTTTTACTTTTACTGTCTCTTCCAGTTTTAGCCGGAGCTATTACCATATTATTAACAGATCGTAATTTAAATACATCTTTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 71–72, bears the following six printed rectangular labels, five white: [ PANAMA:Panama | Cerro Jefe 900m | 9°14’N 79°22’W | 1 April 1979 | leg. G. B. Small ], [ DNA sample ID: | NVG-18044C07 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H10 | c/o Nick V. Grishin ], [ genitalia | NVG240817-29 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466353 ], and one red [ HOLOTYPE ♂ | Emesis (Aphacitis) | pallescens Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection.
Type locality.
Panama: Panamá Province, Cerro Jefe, elevation 900 m, GPS 9.233, −79.367.
Etymology.
In Latin, pallescens means becoming pale or turning pale. The refers to the paler tones of this species compared to its relatives, uses the same ending -escens, as other species in this group, and is a participle.
Distribution.
Currently known only from the holotype collected in central Panama.
Lectotype designation for Emesis lucinda castigata Stichel, 1910
Emesis lucinda castigata Stichel, 1910 was described from several males and females from Peru: Pozuzo and Bolivia: La Paz (Stichel 1910). Originally described as a subspecies, it is currently recognized as a separate species (Callaghan and Lamas 2004), as confirmed by our genomic analysis (Fig. 5). We sequenced a male bearing a green identification label by Stichel, which was likely the header label in the collection, later placed on the first specimen in the column, the specimen with the smallest Stichel collection number in the type series (No. 3255). As such, this specimen in excellent condition properly represents the concept of Stichel’s name. To stabilize nomenclature and define the name E. castigata objectively in the light of possibly polytypic type series from different localities, N.V.G. hereby designates the sequenced male syntype in the MFNB collection that bears the following five rectangular labels (4th handwritten, others printed, 1st red, 4th pale green, and others white): [ Typus ], [ Süd Peru | Pozuzo | e.c.H.Stichel ], [ 3255 ], [ castigata | Stich. ], and [ DNA sample ID: | NVG-18053H07 | c/o Nick V. Grishin ] as the lectotype of Emesis castigata Stichel, 1910. The lectotype has pinholes in the middle of the discal cell of the right forewing and some damage to the fringe by the apex of the left forewing. Images of this specimen are shown on the Butterflies of America website (Warren et al. 2024). The type locality of E. castigata becomes Peru: Pozuzo. The COI barcode sequence of the lectotype, sample NVG-18053H07, GenBank PQ203567, 658 base pairs is:
AACATTATATTTTATTTTTGGAATTTGATCAGGAATAGTCGGTACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGTTCTTTAATTGGTGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCCATTATAATTGGCGGATTTGGTAATTGATTAGTTCCATTAATATTAGGAGCCCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCCCCTTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCCAATATTGCCCATGGAGGAGCTTCAGTTGATTTAGCTATTTTTTCCCTTCATTTAGCTGGAATTTCATCTATTTTAGGAGCAATTAATTTTATTACTACAATTATTAATATACGTATTAATAATTTAACATTTGATCAAATACCATTATTTGTCTGATCTGTTGGAATTACAGCTCTTTTACTTTTATTGTCTCTTCCAGTTTTAGCGGGAGCTATTACTATATTATTAACAGACCGTAATTTAAATACATCATTTTTTGACCCTGCCGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Lectotype designation for Emesis lucinda condigna Stichel, 1925
Emesis lucinda condigna Stichel, 1925 was described from a series of two males and two females (Stichel 1925). Originally described as a subspecies, it is currently recognized as a separate species (Callaghan and Lamas 2004), as confirmed by our genomic analysis (Fig. 5). We sequenced a male syntype that bears a green identification label by Stichel, which was likely the header label in the collection, later placed on the first specimen in the column. The specimen is in excellent condition and agrees with the original description. To stabilize nomenclature and define the name E. condigna objectively, N.V.G. hereby designates the sequenced male syntype in the MFNB collection that bears the following five rectangular labels (4th handwritten, others printed with handwritten “II” on the 2nd label, 1st red, 4th pale green, and others white): [ Typus ], [ Amazonas II | (Santarem) | e.c.H.Stichel ], [ 238 ], [ condigna | Stich. ], and [ DNA sample ID: | NVG-18053H08 | c/o Nick V. Grishin ] as the lectotype of Emesis condigna Stichel, 1925. The lectotype has a slightly damaged tornus of the right hindwing and has lost a small portion of the fringe in the middle of the hindwing outer margin. Images of this specimen are shown on the Butterflies of America website (Warren et al. 2024). The type locality of E. condigna becomes Brazil: Pará, Santarém. The COI barcode sequence of the lectotype, sample NVG-18053H08, GenBank PQ203568, 658 base pairs is:
AACCTTATATTTTATTTTTGGAATTTGATCAGGAATAGTAGGTACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGTTCTTTAATTGGTGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAGTCCCATTAATATTAGGAGCTCCTGACATAGCTTTCCCACGAATAAATAACATAAGATTTTGACTTTTACCCCCCTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAACATTGCCCATGGAGGAGCTTCAGTTGATTTAGCTATTTTTTCTCTCCATTTAGCTGGAATTTCATCTATTTTAGGAGCAATTAATTTTATTACTACAATTATTAACATACGTATTAATAATTTAGCATTTGATCAAATACCATTATTTGTTTGATCTGTTGGAATTACGGCTCTTTTACTTTTATTATCTCTACCAGTTTTAGCGGGAGCTATTACTATATTATTAACAGATCGTAATTTAAATACATCATTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Emesis (Aphacitis) andigna Grishin, new species
http://zoobank.org/DD77E3C7-D040-4B57-9C97-F0FDEABB56AA
Definition and diagnosis.
Genomic analysis of specimens initially identified as Emesis (Aphacitis) condigna Stichel, 1925 (type locality Brazil: Pará, Santarém, lectotype sequenced as NVG-18053H08) reveals that they partition into two clades genetically differentiated from each other at the species level (Fig. 5 brown and cyan), e.g., their COI barcodes differ by 2.3% (15 bp). One clade includes the lectotype of E. condigna, thus representing this species, and the other clade corresponds to a new species. This new species is phenotypically most similar to E. condigna in having a prominent pale spot in the middle near the costal margin of the dorsal hindwing and differs from it by the discal dark dash in the dorsal hindwing cell M3-CuA1 being somewhat offset distad from the line of connected dashes in anterior cells (the dash is aligned with others in the lectotype of E. condigna), and generally paler ventral side of wings, which is more heavily marked with dark framing, bands, and dashes in E. condigna. Additionally, it differs from closely related Emesis (Aphacitis) castigata Stichel, 1910 (Peru: Pozuzo, lectotype sequenced as NVG-18053H07) by darker forewing apical area beneath, including the apex itself, which is more orange in E. castigata, and possesses a better-defined pale spot in the middle of the costal area of dorsal hindwing. In male genitalia (Fig. 129–130), falces are narrower, cornutus is more robust; in ventral view, the lower valval projection is narrower at the base and the upper projection is less rounded, more square-shaped. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne6600.5.10:T61C, cne16584.1.1:T63C, cne16584.1.1:G93C, cne50888.1.8:A36G, cne50888.1.8:A63T, and COI barcode: 250T, 358T, G482A, T581C, T634C.
Barcode sequence of the holotype.
Sample NVG-18044B12, GenBank PQ203569, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGATCAGGAATAGTCGGTACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGTTCTTTAATTGGTGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGTAATTGATTAGTTCCATTAATATTAGGAGCCCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCCCCCTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAATATTGCCCATGGAGGAGCTTCAGTTGATTTAGCTATTTTTTCTCTTCATTTAGCTGGAATTTCATCTATTTTAGGAGCAATTAATTTTATTACTACAATTATTAATATACGTATTAATAATTTAACATTTGATCAAATACCATTATTTGTTTGATCTGTTGGAATTACAGCTCTTTTACTTTTATTATCTCTTCCAGTTTTAGCAGGAGCTATTACTATATTACTAACAGATCGTAATTTAAATACATCATTTTTTGACCCTGCTGGAGGAGGAGACCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 73–74, bears the following six printed (text in italics handwritten) rectangular labels, five white: [ PERU:Cuzco 540 m. | Villa Carmen | Pilcopata 4264 | 29-IV-2015 Kinyon ], [ DNA sample ID: | NVG-18044B12 | c/o Nick V. Grishin ], [ DNA sample ID: | NVG-23114H11 | c/o Nick V. Grishin ], [ genitalia | NVG240817-30 | Nick V. Grishin ], [ USNMENT | {QR Code} | 01466346 ], and one red [ HOLOTYPE ♂ | Emesis (Aphacitis) | andigna Grishin ]. The first NVG number corresponds to a sampled leg, while the second refers to DNA extraction from the abdomen, followed by genitalia dissection. Paratypes: 2♂♂ from Peru: NVG-18052G01 Monte Alegre, Rio Pachitea, old, G. Tessmann leg. [MFNB] (Fig. 75–76) and NVG-18039E12 Puerto Maldonado, forest across Tambopata River from Explorer’s Inn Nature Reserve, 900’, 28-Aug-1985, J. Mix collection No. 13921 [FMNH] (Fig. 77–78).
Type locality.
Peru: Cuzco, Villa Carmen, Pilcopata, elevation 540 m.
Etymology.
A species from or near the Andes, closely related to E. condigna (means deserved, appropriate in Latin): And[es] + [cond]igna. The name is treated as a feminine adjective.
Distribution.
Currently known only from Peru.
Emesis (Aphacitis) luxata Grishin, new species
http://zoobank.org/F4979856-9A3D-46CC-A772-2DBD9A72D87C
Definition and diagnosis.
Genomic analysis of a specimen from Southeast Brazil (Fig. 5 orange) that was identified in the collection as Emesis (Aphacitis) fastidiosa Ménétriés, 1855 (type locality in Brazil), probably due to locality, reveals that it is not closely related to this species and instead is sister to both Emesis (Aphacitis) condigna Stichel, 1925 (type locality Brazil: Para, Santarem, lectotype sequenced as NVG-18053H08) and Emesis (Aphacitis) andigna, new species (type locality in Peru: Cuzco), thus representing a new species due to that and genetic differentiation from others, e.g., its COI barcode differs by 1.2% (8 bp, low probably due to introgression) from E. condigna and by 2.6% (17 bp) from E. andigna. This new species is phenotypically most similar to E. condigna, E. andigna, and Emesis (Aphacitis) opaca Stichel, 1910 (type locality in French Guiana) and differs from them and other in the subgenus Aphacitis Hübner, [1819] by a combination of the following characters: dorsal hindwing with a weakly defined pale spot in the middle near the costal margin (absent in E. opaca), the discal dark dash in cell M3-CuA1 on dorsal hindwing that is strongly offset distad, as in Emesis (Aphacitis) fastidiosa Ménétriés, 1855 (type locality in Brazil) (the dash is mostly aligned with the anterior part of the discal band in E. condigna and weakly offset in E. andigna), more washed-out less distinct dark dashes on dorsal side, apex of forewing with significant dark overscaling beneath, not mostly orange as in Emesis (Aphacitis) castigata Stichel, 1910 (Peru: Pozuzo, lectotype sequenced as NVG-18053H07). Males of the new species differ from the sympatric E. fastidiosa by rounder forewing outer margin, less produced forewing apex darker on both sides, smaller and sharper defined submarginal inverted lunules on ventral hindwing, and a discal bar in cell Sc+R1-RS nearly aligned with the bar in cell RS-M1 (the latter is offset distad from the former in E. fastidiosa). In male genitalia (Fig. 131–132), the lower and upper valval projections are smaller and weaker separated from each other, rounded, knob-like. Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: cne5787.2.15:G27C, cne11414.2.6:A69G, cne11414.2.6:T18C, cne4293.8.1:A13C, cne4293.8.1:A22T, cne2256.1.1:A1045A (not T), cne3201.5.1:A64A (not G), cne3201.5.1:C66C (not T), cne43189.1.2:G462G (not A), cne939.13.9:C182C (not T), and COI barcode: T181C, G200A, T463C, T542C, A586G.
Barcode sequence of the holotype.
Sample NVG-18053F06, GenBank PQ203570, 658 base pairs:
AACCTTATATTTTATTTTTGGAATTTGATCAGGAATAGTAGGTACATCTTTAAGTTTATTAATTCGAATAGAATTAGGAACTTCAGGTTCTTTAATTGGTGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGCGGATTTGGTAATTGATTAATCCCATTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAACATAAGATTTTGACTTTTACCCCCCTCATTAATTTTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCATCTAACATTGCCCATGGAGGAGCTTCAGTTGATTTAGCTATTTTTTCTCTCCATTTAGCTGGAATTTCATCTATTTTAGGAGCAATTAATTTTATTACTACAATCATTAACATACGTATTAATAATTTAGCATTTGATCAAATACCATTATTTGTTTGATCTGTTGGAATTACAGCTCTTTTACTTTTACTATCTCTTCCAGTTTTAGCGGGAGCTATTACTATATTATTAACGGATCGTAATTTAAATACATCATTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the Museum für Naturkunde, Berlin, Germany (MFNB), illustrated in Fig. 79–80, bears the following four rectangular labels (1st green, last red and others white; 2nd handwritten, others printed): [ Brazil, Sao Paulo | Aracatuba | Oct. 1935 | ex coll. Arnold Schultze ], [ Aracatuba | Oct-35 | Aze ] (the last line is illegible and appears to be a short signature), [ DNA sample ID: | NVG-18053F06 | c/o Nick V. Grishin ], and [ HOLOTYPE ♂ | Emesis (Aphacitis) | luxata Grishin ]. Paratype: 1♂ NVG-23114H12 Brazil: São Paulo, 17 km W of Teodoro Sampaio, 600 m, GPS −22.517, −52.200 (GPS does not exactly correspond to the stated locality), 16-Mar-1991, R. Robbins, O. Mielke, and M. Casagrande leg., genitalia vial NVG240817-31 (Fig. 131–132) [USNM].
Type locality.
Brazil: São Paulo, Araçatuba.
Etymology.
In Latin, luxatus means dislocated and is given for the central spot in the hindwing discal band strongly offset distad in this species. Also, lux is Latin for light, fitting this brightly colored species. The name is a participle.
Distribution.
Currently known only from Southeast Brazil.
Diogenia Grishin, new subgenus
http://zoobank.org/A49E23F7-2EA1-4A4A-BDB8-2E38D326C3E5
Type species.
Emesis diogenia Prittwitz, 1865.
Definition.
In our first attempt to rationalize the genetic diversity of Emesis [Fabricius], 1807, we took a conservative approach and minimized the number of new subgenus names. As a result, we combined two phenotypically distinct clades in a subgenus that already had an available name: Aphacitis Hübner, [1819]. However, due to the recognizability of these two clades by observers in the field and the rather prominent genetic differentiation between them, each clade represents a subgenus of its own (Fig. 6). The clade consisting of closer relatives of Emesis lucinda (Cramer, 1775) (type locality in Suriname) corresponds to the subgenus Aphacitis. Its species are recognizable by their large size, bright orange ground color of ventral side in males (paler to pale yellow and nearly white in females) with a flipped-L, concave on the sides, brown postdiscal forewing band in both sexes and usually a large nearly white apical forewing spot in females. The second clade does not have an available name. Callaghan et al. (2024) demonstrated the utility of this clade as a taxonomic unit when they discussed species in this clade separately from Aphacitis, mentioning phenotypic similarity between them. Thus, this clade corresponds to a new subgenus. This new subgenus differs from its relatives by a combination of the following characters: the ventral side is mostly orange (females can be yellow or pale yellow) or with large orange bands and patches on wings, without a strong flipped-L postdiscal brown band on forewing and females are without a large, white forewing apical spot, submarginal spots on ventral hindwing usually well-developed and frequently larger near the apex and tornus, forewing postdiscal spots are mostly joined in a band composed of inverted crescents; aedeagus is shorter and stronger curved. In DNA, a combination of the following characters is diagnostic in the nuclear genome: cne5008.7.3:A87G, cne5008.7.3:C124A, cne7265.2.1:T165C, cne7265.2.1:T168C, cne7265.2.1:C40A, and in COI barcode: T49A, A130T, 517T, T538A.
Etymology.
The name is tautonymous with the type species name and is a feminine noun in the nominative singular.
Species included.
The type species (i.e., Emesis diogenia Prittwitz, 1865), Emesis vulpina Godman and Salvin, 1886, Emesis tegula Godman and Salvin, 1886, Emesis heteroclita Stichel, 1929, and Emesis hypoaithos Callaghan, Trujano-Ortega, and Ríos-Málaver, 2024.
Parent taxon.
Genus Emesis [Fabricius], 1807.
A taxonomic list of Emesis
Here, we refine our previous taxonomic list of Emesis (Zhang et al. 2019b), to accommodate new data, those sequenced by us (Fig. 6) and published by others (Trujano-Ortega et al. 2020; Costa et al. 2021; Trujano-Ortega et al. 2021; Callaghan et al. 2024), and correct mistakes. Specifically, the list is adjusted to reflect the discovery of Trujano-Ortega et al. (2020) that Apodemia planeca R. de la Maza and J. de la Maza, 2017 (type locality in Mexico: Michoacán) belongs to Emesis, and we follow the identification of Emesis tegula Godman and Salvin, 1886 (type locality in Mexico: Yucatan, Guatemala, Nicaragua, and Panama) detailed and illustrated by Callaghan et al. (2024).
We attempt ordering Emesis species to maximize the phenotypic similarity and geographic proximity of the list neighbors but without disrupting phylogenetic orders given in genomic trees (Fig. 1–6): i.e., a strongly supported clade in the trees is a continuous segment in the list. However, the evolution and diversification of Emesis are riddled with complexities due to the confidently supported incongruence of the trees (Fig. 6). While closer to the leaves, the topology is nearly always identical in different trees, the deeper branching such as between different subgenera differs. For instance, while Emesis (Emesis) is sister to Emesis (Tenedia) in the autosome tree with 100% fast bootstrap support (Fig. 6a), Emesis (Emesis) is sister to Emesis (Mandania) in the Z chromosome tree with the same highest confidence (Fig. 6b). These differences may be due to evolutionary events like incomplete lineage sorting or introgression during the diversification of Emesis into subgenera, and its investigation is outside the scope of this study. However, the incongruence creates challenges in ordering species phylogenetically according to these trees. For the sake of simplicity, here we take the Z chromosome tree as a guide because the phylogeny of the sex chromosomes typically correlates better with the species phylogeny (Fontaine et al. 2015).
The order of species is proposed based on the following considerations. First, we start the list with species with strongly curved forewing costa and pale subapical spot due to their similarity to the genus Curvie Grishin, 2019 (type species Symmachia emesia Hewitson, 1867) and the possibility of having these Emesis species and Curvie next to each other in the overall list of Riodinidae. Second, we end with large species that belong to the subgenus Aphacitis and look most different from the rest. Only one order of subgenera if Emesis (Emesis) is placed first and Emesis (Aphacitis) is placed last is implied by the Z chromosome tree (Fig. 6b). Third, within each subgenus, we follow the Z chromosome phylogeny and rotate the clades to agree better with phenotypic similarity and generally place more northern species first, ordering taxa from north to south where possible and contradicts neither phylogeny nor phenotypic similarity. We note that this order of subgenera, species groups, and species is tentative, and improvements to it are much desired.
The list is formatted as follows. Names treated as synonyms (including genera and names of type species that are regarded as synonyms) are preceded by “=”: not followed by daggers are junior subjective synonyms; † junior objective synonyms; ‡ junior homonyms and unavailable names (such as nomina nuda); “preocc.” indicates preoccupied, followed by the author and date. Species-group synonyms are given with their original genus. Type species for genera and subgenera are listed. For type species regarded as synonyms, valid names are shown in parenthesis. The general area of the type locality (originally published or deduced; the country name and frequently its major subdivision; erroneous or uninformative published localities are shown in quotes) follows “;” after each species-group name. Taxonomic changes are explained after the name: stat. nov. (status novus, new status, not previously published), stat. rest. (status restitutus, status reinstated to that proposed in the original description), and pos. rest. (positio restituta, placement of a synonym reinstated to that originally proposed). The former status and the former placement are given in remarks that follow the | symbol. Genomic sequencing has not been done for several taxa (as noted in remarks); their placement in the list follows phenotypic considerations and, therefore, is hypothetical. The list covers 7 valid subgenera (1 new) and 71 valid species (22 new and 4 elevated from subspecies) with 14 additional subspecies (1 new). New taxa and the category of taxonomic change are shown in bold font.
Genus Emesis [Fabricius], 1807; type species =Hesperia ovidius (E. cereus)
Subgenus Emesis [Fabricius], 1807; type species =Hesperia ovidius (E. cereus)
=Polystichtis Hübner, [1819]; type species E. cereus
=†Tapina Billberg, 1820; type species =Hesperia ovidius (E. cereus)
=‡Polystichthis Agassiz, 1847; type species E. cereus | unjustified emendation
=Nelone Boisduval, 1870; type species =Papilio fatima (E. cereus)
Emesis cereus (Linnaeus, 1767); “Indiis”: likely Suriname
=‡Papilio caeneus Linnaeus, 1767; “Indiis”: likely Suriname | preocc. by Linnaeus, 1758
=Papilio fatima Cramer, 1780; Suriname
=Hesperia ovidius Fabricius, 1793; “Indiis”: likely Suriname
=Polystichtis cerea Hübner, [1819]; “Indiis”: likely Suriname | emended
Emesis cronina Schaus, 1928, stat. rest.; Paraguay | was a subspecies of E. cereus
Emesis neemias Hewitson, 1872; Brazil
Emesis neemias cayennensis Gallard, 2017; French Guiana
Emesis neemias neemias Hewitson, 1872; Brazil
Emesis aerunda Grishin, sp. n.; Peru
Emesis orichalceus Stichel, 1916; Bolivia
Emesis malik Callaghan, Costa, Trujano-Ortega and M. Benmesbah, 2021; Venezuela
Emesis bartica Grishin, sp. n.; Guyana
Emesis aerigera (Stichel, 1910); Brazil (SP)
Emesis lacrines Hewitson, 1870; Nicaragua
Emesis nobilata Stichel, 1910, stat. nov.; Costa Rica | was a subspecies of E. fatimella
Emesis panamella Grishin, sp. n.; Panama
Emesis fatimella Westwood, 1851; Surinam, Brazil (AM)
Emesis fatimellina Grishin, sp. n.; Brazil (SC)
Subgenus Mandania Grishin, 2019; type species E. mandana
Emesis furor A. Butler and H. Druce, 1872; Costa Rica
=Emesis mandana var. angulariformis E. Strand, 1916; Costa Rica
Emesis mandora Grishin, sp. n.; Ecuador
Emesis manduza Grishin, sp. n.; Peru: Cuzco
Emesis mandana (Cramer, 1780); Suriname
=‡Papilio flegia Fabricius, 1787; French Guiana | preocc. by Cramer, 1779
=Papilio polymenus Fabricius, 1793; Suriname
=Papilio arminius Fabricius, 1793; not specified, likely the Guianas
=Erycina ops Latreille [1813]; not specified, likely the Guianas
=Polystichtis mandane Hübner, [1819]; Suriname | emended
Emesis russula Stichel, 1910; Bolivia
Emesis russula russula Stichel, 1910; Bolivia
Emesis russula sudesta Grishin, ssp. n.; Brazil (PR)
Subgenus Tenedia Grishin, 2019; type species E. tenedia
Emesis melancholica Stichel, 1916; Brazil (ES)
Emesis ocypore (Geyer, 1837); “Africa”: likely Amazonian
Emesis ocypore aethalia H. Bates, 1868; Colombia
=Emesis olivae Butler and H. Druce, 1872; Costa Rica
Emesis ocypore ocypore (Geyer, 1837); “Africa”: likely Amazonian
=Emesis samius A. Seitz, 1916; Peru
Emesis ocypore zelotes Hewitson, 1872; Brazil, likely SE or S
Emesis angularis Hewitson, 1870; Ecuador
Emesis subangularis Grishin, sp. n.; Argentina
Emesis sinuatus Hewitson, 1877; Ecuador | no DNA work, placement tentative
Emesis heterochroa Hopffer, 1874; Peru, Bolivia
Emesis leona Grishin, sp. n.; Mexico (NL)
Emesis tenedia C. Felder and R. Felder, 1861; Venezuela
Emesis tenedia tenedia C. Felder and R. Felder, 1861; Venezuela
=‡Emesis tenedia ab. fasciata E. Strand, 1916, pos. rest.; Costa Rica | was a synonym of E. lupina
Emesis tenedia guyanensis Gallard, 2017; French Guiana
Emesis nimia Grishin, sp. n.; Panama: Chiriquí
Emesis toltec Reakirt, 1866; Mexico (Ver) | taxonomic identity unclear
Emesis saturata Godman and Salvin, 1886; Mexico (Oax)
Emesis faria Grishin, sp. n.; Mexico (Tam)
Emesis tristis Stichel, 1929, stat. rest.; Mexico (Col) | was a synonym of E. lupina
Emesis lupina Godman and Salvin, 1886; Costa Rica | identification tentative
Emesis pacis Callaghan, Trujano-Ortega, and Ríos-Málaver, 2024; Colombia | no DNA work
Emesis paphia R. Felder, 1869; stat. rest.; Mexico (Ver) | was a subspecies of E. cypria
Emesis cypria C. Felder and R. Felder, 1861; Venezuela
Emesis cypria cypria C. Felder and R. Felder, 1861; Venezuela
Emesis cypria capnodis Stichel, 1911; Venezuela
Emesis cypria guppyi Kaye, 1904; Trinidad, Colombia
Emesis cypria cilix Hewitson, 1870; Ecuador
Emesis flecta Grishin, sp. n.; Bolivia
Emesis alisada Grishin, sp. n.; Peru: Piura
Emesis phyciodoides (W. Barnes and Benjamin, 1924); USA (AZ)
Emesis planeca (R. de la Maza and J. de la Maza, 2017); Mexico (Mich) | no genomic work
Subgenus Poeasia Grishin, 2019; type species E. poeas
Emesis sonorensis Grishin, sp. n.; Mexico (Son)
Emesis poeas Godman, 1901; Mexico (Gue)
Subgenus Brimia Grishin, 2019; type species E. brimo
Emesis brimo Godman and Salvin, 1889; Panama
Emesis brimo vimena Schaus, 1928; Guatemala, Panama
Emesis brimo brimo Godman and Salvin, 1889; Panama
Emesis brimo insulicola Vargas and Salazar, 2018; Colombia
Emesis progne (Godman, 1903); Bolivia, Peru
Emesis temesa (Hewitson, 1870); Ecuador
Emesis temesa temesa (Hewitson, 1870); Ecuador
=Symmachia emesina Staudinger, [1887]; Peru: Loreto
Emesis temesa peruviana (Lathy, 1904); Peru: Junín
Emesis apagada Grishin, sp. n.; Peru: Madre de Dios
Emesis boliviana Grishin, sp. n.; Bolivia
Emesis satema (Schaus, 1902); Brazil (RJ)
Subgenus Diogenia Grishin, subgen. n.; type species E. diogenia | was included in Aphacitis
Emesis vulpina Godman and Salvin, 1886; Mexico (Ver)
Emesis tegula Godman and Salvin, 1886; Mexico (Yuc), Guatemala, Nicaragua, Panama
Emesis eleanorae Gallardo and Grishin, 2021; Honduras
Emesis diogenia Prittwitz, 1865; Brazil (RJ)
=Emesis aurelia Bates, 1867; Brazil (MA)
=Emesis tenedia ravidula Stichel, 1910; Paraguay
Emesis hypoaithos Callaghan, Trujano-Ortega, and Ríos-Málaver, 2024; Colombia
Emesis hypoaithos hypoaithos Callaghan, Trujano-Ortega, and Ríos-Málaver, 2024; Colombia
Emesis hypoaithos ochros Callaghan, Trujano-Ortega, and Ríos-Málaver, 2024; Colombia | no DNA work
Emesis heteroclita Stichel, 1929; Peru
Emesis heteroclita heteroclita Stichel, 1929; Peru
Emesis heteroclita vicaria Le Cerf, 1958; “Amazon”
Emesis heteroclita adelpha Le Cerf, 1958; Bolivia
Subgenus Aphacitis Hübner, [1819]; type species =‡Papilio dyndima (E. lucinda)
=Nimula Blanchard, 1840; type species E. lucinda
Emesis liodes Godman and Salvin, 1886; Mexico (Yuc)
Emesis aurichica Grishin, sp. n.; Mexico (Chis)
Emesis pruinapicalis Grishin, sp. n.; Panama: Darién
Emesis auripana Grishin, sp. n.; Panama: Darién
Emesis aurimna (Boisduval, 1870); Colombia
Emesis parvissima Kaye, 1921, stat. nov.; Trinidad | was a synonym of E. lucinda
Emesis pallescens Grishin, sp. n.; Panama: Panama
Emesis furvescens Grishin, sp. n.; Panama: Darién
Emesis glaucescens Talbot, 1929; Colombia: Montepa
=Emesis lucinda conformata Stichel, 1929; Colombia: Rio Dagua
Emesis lucinda (Cramer, 1775); Suriname
=‡Papilio lucinde Cramer, 1775; Suriname | incorrect original spelling
=‡Papilio dyndima Cramer, 1780; Suriname | preocc. by Cramer, 1775
=Papilio lassus Fabricius, 1787; French Guiana
Emesis eurydice Godman, 1903; Ecuador
Emesis spreta H. Bates, 1868; Brazil (AM)
Emesis castigata Stichel, 1910; Peru: Pozuzo
Emesis condigna Stichel, 1925; Brazil (PA)
Emesis andigna Grishin, sp. n.; Peru: Cuzco
Emesis luxata Grishin, sp. n.; Brazil (SP)
Emesis opaca Stichel, 1910; French Guiana
=Emesis castigata diringeri Gallard 2008; French Guiana
Emesis fastidiosa Ménétriés, 1855; Brazil
=Nelone godartii Boisduval, 1870; Brazil
=‡Emesis lucinda ab. albida A. Seitz, 1916; Brazil (BA) | infrasubspecific
Supplementary Material
Table S1. Data for sequenced specimens: 178 Emesis and two Apodemia available at https://osf.io/d48aj/ (Zhang et al. 2024a).
Acknowledgments
We acknowledge Ping Chen and Ming Tang for their excellent technical assistance. We are grateful to David Grimaldi and Courtney Richenbacher (AMNH: American Museum of Natural History, New York, NY, USA), Blanca Huertas, David Lees, and Geoff Martin (BMNH: Natural History Museum, London, UK), the late Paul Opler, Chuck Harp, and the late Boris Kondratieff (CSUC: Colorado State University Collection, Fort Collins, CO, USA), Crystal Maier and Rebekah Baquiran (FMNH: Field Museum of Natural History, Chicago, IL, USA), Théo Léger, Viola Richter, and Wolfram Mey (MFNB: Museum für Naturkunde, Berlin, Germany), Andrei Sourakov, Andrew D. Warren, Debbie Matthews-Lott, Riley J. Gott, and Keith R. Willmott (MGCL: McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, Gainesville, FL, USA), Rodolphe Rougerie (MNHP: Muséum National d’Histoire Naturelle, Paris, France), Matthias Nuss and Manuela Bartel (MTD: Museum für Tierkunde, Dresden, Germany), Mario Cupello, Edward G. Riley, Karen Wright, and John Oswald (TAMU: Texas A&M University Insect Collection, College Station, TX, USA), Alex Wild (TMMC: University of Texas Biodiversity Center, Austin, TX, USA), Robert K. Robbins, John M. Burns, and Brian Harris (USNM: National Museum of Natural History, Smithsonian Institution, Washington, DC, USA) for granting access to the collections under their care, loan of specimens, and stimulating discussions; to Curtis Callaghan and Gerardo Lamas for discussions and helpful suggestions, to Ezequiel O. Núñez Bustos for clarifying locality data, to Ernst Brockmann for the help with deciphering handwriting on specimen labels, and to James K. Adams and David Wright for critical review of the manuscript and many helpful comments and corrections. We are indebted to the U. S. National Park Service for the research permit BIBE-2008-SCI-0044, Big Bend (Raymond Skiles). We acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources. This study was supported in part by the HHMI Investigator funds and by grants from the National Institutes of Health GM127390 and the Welch Foundation I-1505 to N.V.G.
Contributor Information
Jing Zhang, Eugene McDermott Center for Human Growth and Development and Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-8816 USA.
Qian Cong, Eugene McDermott Center for Human Growth and Development and Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-8816 USA.
Jinhui Shen, Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-9050 USA.
Leina Song, Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-9050 USA.
Nick V. Grishin, Departments of Biophysics and Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-9050 USA
Literature Cited
- Bachtrog D, Thornton K, Clark A, Andolfatto P. 2006. Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution 60(2): 292–302. [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nature Methods 12(1): 59–60. [DOI] [PubMed] [Google Scholar]
- Burns JM, Janzen DH. 2005. Pan-neotropical genus Venada (Hesperiidae: Pyrginae) is not monotypic: Four new species occur on one volcano in the Area de Conservación Guanacaste, Costa Rica. Journal of the Lepidopterists’ Society 59(1): 19–34. [Google Scholar]
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN. 2007. DNA barcodes of closely related (but morphologically and ecologically distinct) species of skipper butterflies (Hesperiidae) can differ by only one to three nucleotides. Journal of the Lepidopterists’ Society 61(3): 138–153. [Google Scholar]
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN. 2008. DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proceedings of the National Academy of Sciences of the United States of America 105(17): 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan C, Trujano-Ortega M, Ríos-Málaver C. 2024. Two new species of Emesis Fabricius, 1807 from northwestern South and Central America (Lepidoptera: Riodinidae). Zootaxa 5443(3): 406–416. [DOI] [PubMed] [Google Scholar]
- Callaghan CJ, Lamas G. 2004. Riodinidae, p. 141–170. In: Lamas G (ed.). Checklist: Part 4A. Hesperioidea - Papilionoidea. Association for Tropical Lepidoptera; Scientific Publishers; Gainesville. 439 p. [Google Scholar]
- Cong Q, Shen J, Borek D, Robbins RK, Opler PA, Otwinowski Z, Grishin NV. 2017a. When COI barcodes deceive: complete genomes reveal introgression in hairstreaks. Proceedings of the Royal Society B: Biological Sciences 284(1848): 1–9. 10.1098/rspb.2016.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Li W, Borek D, Otwinowski Z, Grishin NV. 2017b. The first complete genomes of metalmarks and the classification of butterfly families. Genomics 109: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Zhang J, Li W, Kinch LN, Calhoun JV, Warren AD, Grishin NV. 2021. Genomics reveals the origins of historical specimens. Molecular Biology and Evolution 38(5): 2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Zhang J, Grishin NV. 2019a. Genomic determinants of speciation [Preprint]. bioRxiv BIORXIV/2019/837666. Available at https://www.biorxiv.org/content/10.1101/837666v1 (Last accessed August 2024.)
- Cong Q, Zhang J, Shen J, Grishin NV. 2019b. Fifty new genera of Hesperiidae (Lepidoptera). Insecta Mundi 0731: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Viloria AL, Callaghan C, Trujano-Ortega M, Neild AFE, Benmesbah M, Attal S, Grishin NV. 2021. Lepidoptera del Pantepui. Parte XI Nuevos Riodinidae (Riodininae), Pieridae (Dismorphiinae) y Nymphalidae (Satyrinae). Antenor 8(1): 2–28. [Google Scholar]
- Felder C, Felder R. 1861. Lepidoptera nova Columbiae. Wiener Entomologische Monatschrift 5: (3): 72–87, (4): 97–111. [Google Scholar]
- Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani E, Mitchell SN, Wu YC, Smith HA, Love RR, Lawniczak MK, Slotman MA, Emrich SJ, Hahn MW, Besansky NJ. 2015. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347(6217): 1–6. 10.1126/science.1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo RJ, Zhang J, Cong Q, Shen J, Grishin NV. 2021. A uniquely patterned new species of Emesis from Honduras (Riodinidae). Tropical Lepidoptera Research 31(1): 53–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godman FD. 1901. Lepidoptera-Rhopalocera. 2(167): 701–740, pl. 110–111. In: Godman FD, Salvin O (eds.). Biologia Centrali-Americana. Insecta. Dulau & Co., Bernard Quaritch; London. 782 p. [Google Scholar]
- Godman FD, Salvin O. 1886. Biologia Centrali-Americana. Insecta. Lepidoptera-Rhopalocera. 1(44): 401–440, pl. 42–44. Dulau & Co., Bernard Quaritch; London. 487 p. [Google Scholar]
- Hall JPW, Harvey DJ. 2002. A phylogenetic review of Charis and Calephelis (Lepidoptera: Riodinidae). Annals of the Entomological Society of America 95(4): 407–421. [Google Scholar]
- Harvey DJ, Hall JPW. 2002. Phylogenetic revision of the Charis cleonus complex (Lepidoptera: Riodinidae). Systematic Entomology 27(4): 265–300. [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270(1512): 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PD, Penton EH, Burns JM, Janzen DH, Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America 101(41): 14812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- iNaturalist. 2024. A Community for Naturalists – iNaturalist. Available at https://www.inaturalist.org (Last accessed August 2024.)
- Li W, Cong Q, Shen J, Zhang J, Hallwachs W, Janzen DH, Grishin NV. 2019. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proceedings of the National Academy of Sciences of the United States of America 116(13): 6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Sourakov A, Zakharov E. 2016. DNA barcodes as a tool in biodiversity research: testing pre-existing taxonomic hypotheses in Delphic Apollo butterflies (Lepidoptera, Papilionidae). Systematics and Biodiversity 14: 599–613. [Google Scholar]
- Minh BQ, Nguyen MA, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30(5): 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez R, Willmott KR, Álvarez Y, Genaro JA, Pérez-Asso AR, Quejereta M, Turner T, Miller JY, Brévignon C, Lamas G, Hausmann A. 2022. Integrative taxonomy clarifies species limits in the hitherto monotypic passion-vine butterfly genera Agraulis and Dryas (Lepidoptera, Nymphalidae, Heliconiinae). Systematic Entomology 47(1): 152–178. [Google Scholar]
- Pavulaan H, Patterson R, Grishin NV. 2023. Reassessment of Amblyscirtes hegon (Hesperiidae) as a complex of four distinct species revealed by genomic analysis. The Taxonomic Report of the International Lepidoptera Survey 11(5): 1–38.36817548 [Google Scholar]
- Pazhenkova EA, Lukhtanov VA. 2021. Genomic introgression from a distant congener in the Levant fritillary butterfly, Melitaea acentria. Molecular Ecology 30(19): 4819–4832. [DOI] [PubMed] [Google Scholar]
- Rambaut A 2018. FigTree, version 1.4.4. Available at http://tree.bio.ed.ac.uk/software/figtree/ (Last accessed August 2024.)
- Robbins RK, Cong Q, Zhang J, Shen J, Busby RC, Faynel C, Duarte M, Martins ARP, Prieto C, Lamas G, Grishin NV. 2022. Genomics-based higher classification of the species-rich hairstreaks (Lepidoptera: Lycaenidae: Eumaeini). Systematic Entomology 47(3): 445–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stichel H 1910. Vorarbeiten zu einer Revision der Riodinidae Grote (Erycinidae Swains.) (Lep. Rhop.). Berliner entomologische Zeitschrift 55(1/2): 9–103. [Google Scholar]
- Stichel H 1925. Beiträge zur Kenntnis der Riodinidenfauna Südamerikas. VII. Zeitschrift für wissenschaftliche Insektenbiologie 20: (1): 14–18, (2): 19–23, (3): 53–56, (4): 84–93. [Google Scholar]
- Trujano-Ortega M, Callaghan CJ, Arellano-Covarrubias A, Luis-Martinez A, Avalos-Hernandez O, Llorente-Bousquets J. 2021. Geographical distribution of Emesis Fabricius (Lepidoptera: Riodinidae) in Mexico: Updated checklist and temporal patterns. Zootaxa 4964(3): 401–442. [DOI] [PubMed] [Google Scholar]
- Trujano-Ortega M, Callaghan CJ, GarcIa VUO, Luis-MartInez A, Avalos-HernAndez O, Llorente-Bousquets J. 2020. Emesis planeca n. comb. (Lepidoptera: Riodinidae): a new combination revealed by molecular evidence with a description of its morphological variation. Zootaxa 4853(2): 218–234. [DOI] [PubMed] [Google Scholar]
- Warren AD, Davis KJ, Stangeland EM, Pelham JP, Willmott KR, Grishin NV. 2024. Illustrated Lists of American Butterflies [9-III-2024]. Available at https://www.butterfliesofamerica.com/ (Last accessed August 2024.)
- Zhang J, Cong Q, Burns JM, Grishin NV. 2022a. Checking the checkered taxonomy of Plötz’s checkered skippers (Hesperiidae: Pyrgini). The Taxonomic Report of the International Lepidoptera Survey 10(5): 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Grishin NV. 2023a. Descriptions of one hundred new species of Hesperiidae. Insecta Mundi 1026: 1–115. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Grishin NV. 2023b. Thirteen new species of butterflies (Lepidoptera: Hesperiidae) from Texas. Insecta Mundi 0969: 1–58. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Brockmann E, Grishin NV. 2019a. Genomes reveal drastic and recurrent phenotypic divergence in firetip skipper butterflies (Hesperiidae: Pyrrhopyginae). Proceedings of the Royal Society B: Biological Sciences 286(1903): 1–6. 10.1098/rspb.2019.0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Grishin NV. 2022b. Taxonomic changes suggested by the genomic analysis of Hesperiidae (Lepidoptera). Insecta Mundi 0921: 1–135. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, Grishin NV. 2020. Genomic evidence suggests further changes of butterfly names. The Taxonomic Report of the International Lepidoptera Survey 8(7): 1–40. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2022c. Genomic DNA sequencing reveals two new North American species of Staphylus (Hesperiidae: Pyrginae: Carcharodini). The Taxonomic Report of the International Lepidoptera Survey 10(4): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2023c. Butterfly classification and species discovery using genomics. The Taxonomic Report of the International Lepidoptera Survey 11(3): 1–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2023d. Genomic analysis reveals new species and subspecies of butterflies. The Taxonomic Report of the International Lepidoptera Survey 11(6): 1–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2024a. Supplementary information to “Genomic analysis reveals hidden species diversity in Emesis Fabricius (Lepidoptera: Riodinidae).” Table S1 and DNA sequences of exons with diagnostic characters. Available at https://osf.io/d48aj/ (Last accessed September 2024.) [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2024b. Taxonomic advances driven by the genomic analysis of butterflies. The Taxonomic Report of the International Lepidoptera Survey 11(7): 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shen J, Cong Q, Grishin NV. 2019b. Genomic analysis of the tribe Emesidini (Lepidoptera: Riodinidae). Zootaxa 4668(4): 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Data for sequenced specimens: 178 Emesis and two Apodemia available at https://osf.io/d48aj/ (Zhang et al. 2024a).


