Abstract
A century and a half since the time of Hewitson, we are experiencing a renaissance in species discovery fueled by whole genome sequencing. A large-scale genomic analysis of Hesperiidae Latreille, 1809 (Lepidoptera), including primary type specimens, reveals a deluge of species new to science. One hundred of them (one in a new genus) are described here from the New World (type localities are given in parenthesis): Drephalys (Drephalys) diovalis Grishin, new species (Ecuador: Napo), Euriphellus panador Grishin, new species (Ecuador: Esmeraldas), Euriphellus panamicus Grishin, new species (Panama: Panama), Cecropterus (Thorybes) viridissimus Grishin, new species (Ecuador: Zamora-Chinchipe), Cecropterus (Murgaria) dariensis Grishin, new species (Panama: Darien), Urbanus (Urbanus) mericuti Grishin, new species (Ecuador: Napo), Telegonus (Telegonus) pastus Grishin, new species (Panama: Panama), Autochton (Autochton) dora Grishin, new species (Ecuador: Pastaza), Astraptes centralis Grishin, new species (Panama: Colón), Aguna claxonica Grishin, new species (Ecuador: Napo), Aguna esmeralda Grishin, new species (Ecuador: Esmeraldas), Aguna lata Grishin, new species (Guyana), Ridens angulinea Grishin, new species (Peru: Cuzco), Pythonides lera Grishin, new species (Peru: Cuzco), Pythonides latemarginatus Grishin, new species (Panama: Panama), Gindanes variegatus Grishin, new species (Brazil: Mato Grosso), Milanion (Milanion) virga Grishin, new species (Brazil: Rondônia), Milanion (Milanion) furvus Grishin, new species (Panama: Panama), Milanion (Milanion) laricus Grishin, new species (Ecuador: Napo), Charidia ronda Grishin, new species (Brazil: Rondônia), Pseudodrephalys tinas Grishin, new species (Peru: Loreto), Pseudodrephalys argus Grishin, new species (Suriname: Para), Achlyodes calvus Grishin, new species (Brazil: Santa Catarina), Spioniades artemis Grishin, new species (Panama: Panama), Spioniades artemidoides Grishin, new species (Brazil: Santa Catarina), Myrinia orieca Grishin, new species (Ecuador: Orellana), Myrinia aragua Grishin, new species (Venezuela: Aragua), Myrinia maculosa Grishin, new species (Guatemala), Myrinia manchada Grishin, new species (Guyana), Polyctor (Fenops) lamperus Grishin, new species (Panama: Darien), Nisoniades (Nisoniades) lutum Grishin, new species (Mexico: Guerrero.), Bolla (Stolla) vena Grishin, new species (Venezuela: Aragua), Staphylus (Vulga) vula Grishin, new species (Mexico: Veracruz), Staphylus (Vulga) vulga Grishin, new species (Panama: Darien), Staphylus (Staphylus) rotundalus Grishin, new species (Ecuador: Napo), Staphylus (Staphylus) yucatanus Grishin, new species (Mexico: Quintana Roo/Yucatan), Heliopetes (Heliopetes) lana Grishin, new species (Guatemala), Canesia ella Grishin, new species (Venezuela: Barinas), Paches (Paches) loxeca Grishin, new species (Ecuador: Morona-Santiago), Clito congruens Grishin, new species (Panama: Colón), Cycloglypha corax Grishin, new species (Brazil: Rio de Janeiro), Festivia peruvia Grishin, new species (Peru: Huánuco), Decinea notata Grishin, new species (Ecuador: Napo), Pompeius fuscus Grishin, new species (Brazil: Minas Gerais), Vernia clara Grishin, new species (Panama: Chiriquí), Oligoria (Oligoria) obtena Grishin, new species (Ecuador: Napo), Thespieus mandal Grishin, new species (Brazil: Rio de Janeiro), Psoralis (Saniba) magnamacus Grishin, new species (Panama: Darien), Alychna ayonis Grishin, new species (Ecuador: Napo), Wahydra banios Grishin, new species (Ecuador: Tungurahua), Wahydra cuzcona Grishin, new species (Peru: Cuzco), Cynea (Cynea) aureofimbra Grishin, new species (Ecuador), Cynea (Nycea) quada Grishin, new species (Ecuador: Napo), Cynea (Quinta) achirae Grishin, new species (Mexico: Tamaulipas), Eutus amazonicus Grishin, new species (Peru: Madre de Dios), Eutus incus Grishin, new species (Peru: Cuzco), Eutus septemaculatus Grishin, new species (Brazil: Mato Grosso), Godmia viridicapita Grishin, new species (Ecuador: Napo), Rhomba pulla Grishin, new species (Peru: Cuzco), Niconiades victoria Grishin, new species (Mexico: Tamaulipas), Lancephallus purpurus Grishin, new genus and new species (Guyana), Mnasicles (Remella) ecua Grishin, new species (Ecuador: Pichincha), Amblyscirtes (Amblyscirtes) aeratus Grishin, new species (Mexico: Oaxaca), Amblyscirtes (Mastor) chrysoplea Grishin, new species (Mexico: Oaxaca), Amblyscirtes (Mastor) chrysomisa Grishin, new species (Mexico: Chiapas), Amblyscirtes (Flor) meridus Grishin, new species (Mexico: Veracruz), Rectava chiriquensis Grishin, new species (Panama: Chiriquí), Cobalopsis adictys Grishin, new species (Panama: Veraguas), Cymaenes melaporphyrus Grishin, new species (Mexico: San Luis Potosí), Lerema (Morys) ecuadorica Grishin, new species (Ecuador: Pichincha), Saturnus obscurior Grishin, new species (Panama: Darien), Cantha zoirodicta Grishin, new species (Peru: Madre de Dios), Cantha meiodicta Grishin, new species (Peru: Madre de Dios), Phlebodes duplex Grishin, new species (Guatemala: Cayuga), Lychnuchus (Enosis) valle Grishin, new species (Colombia: Valle), Eutychide ochoides Grishin, new species (Peru: Cuzco), Dion bora Grishin, new species (Panama: Darien), Dion occida Grishin, new species (Peru: Madre de Dios), Eprius (Eprius) veledinus Grishin, new species (Ecuador: Pichincha), Radiatus panamensis Grishin, new species (Panama: Panama), Pheraeus pulcher Grishin, new species (Peru: Madre de Dios), Callimormus rades Grishin, new species (Panama: Panama), Gubrus lubens Grishin, new species (Ecuador: Loja), Ludens labens Grishin, new species (Panama: Darien), Rigga isa Grishin, new species (Ecuador: Napo), Flaccilla lactea Grishin, new species (Peru: Cuzco), Falga athena Grishin, new species (Panama: Darien), Panoquina jay Grishin, new species (Peru: Loreto), Calpodes salianus Grishin, new species (Peru: Madre de Dios), Calpodes stingo Grishin, new species (Ecuador: Sucumbíos), Aides nobra Grishin, new species (Panama: Colón), Thracides pavo Grishin, new species (Mexico: Tabasco), Talides eluta Grishin, new species (Peru: Cuzco), Talides laeta Grishin, new species (Peru: Cuzco), Neoxeniades angustior Grishin, new species (Brazil: Rio de Janeiro), Damas zea Grishin, new species (Guyana), Tromba xantha Grishin, new species (Mexico: Veracruz), Perichares fura Grishin, new species (Ecuador: Pichincha), Carystoides (Balma) goliath Grishin, new species (Colombia: Valle), and Agathymus galeana Grishin, new species (Mexico: Nuevo Leon). Additionally, we present evidence to support 22 taxa as species (not subspecies or synonyms) and synonymize one genus and four species. Namely, the following taxa are species: Milanion pilta Evans, 1953 (not Milanion pilumnus Mabille and Boullet, 1917), Milanion latior Mabille and Boullet, 1917 (not a synonym of Milanion marciana Godman and Salvin, 1895), Charidia pilea Evans, 1953, and Charidia pocus Evans, 1953 (not Charidia lucaria (Hewitson, 1868)), Paches (Paches) gloriosus Röber, 1925 and Paches (Paches) loxana Evans, 1953 (not Paches (Paches) loxus (Westwood, 1852)), Spioniades anta Evans, 1953 (not Spioniades abbreviata (Mabille, 1888)), Decinea onasima (Hewitson, 1877) and Decinea formosus (Hayward, 1940) (not Decinea dama (Herrich-Schäffer, 1869)), Thespieus guerreronis (Dyar, 1913) (not Thespieus dalman (Latreille, [1824])), Cynea (Nycea) erebina (Möschler, 1879) and Cynea (Nycea) cleochares (Mabille, 1891) (not Cynea (Cynea) diluta (Herrich-Schäffer, 1869)), Amblyscirtes (Mastor) repta Evans, 1955 (not Amblyscirtes (Flor) florus (Godman, 1900)), Saturnus tiberius (Möschler, 1883), Saturnus conspicuus (E. Bell, 1941), Saturnus meton (Mabille, 1891), and Saturnus obscurus (E. Bell, 1941) (not Saturnus reticulata (Plötz, 1883)), Phlebodes sifax Evans, 1955 (not Phlebodes campo (E. Bell, 1947)), Eutychide ochus Godman, 1900 and Eutychide rogersi (Kaye, 1914) (not a subspecies and a synonym, respectively, of Eutychide subcordata (Herrich-Schäffer, 1869)), Falga mirabilis Evans, 1955, Falga jacta Evans, 1955, and Falga ombra Evans, 1955 (not Falga jeconia (A. Butler, 1870)); and the following taxa are junior subjective synonyms: Libra Evans, 1955 (of Phemiades Hübner, [1819]), Papilio clito Fabricius, 1787 of Milanion hemes hemes (Cramer, 1777), Pamphila hycsos Mabille, 1891 of Cynea (Nycea) erebina (Möschler, 1879), Hesperia olympia Plötz, 1882 of Eutychide subcordata (Herrich-Schäffer, 1869), and Hesperia ocrinus Plötz, 1882 of Aides aegita (Hewitson, 1866). Furthermore, we propose new combinations for genus-species: Lychnuchus (Enosis) ponka (Evans, 1955) (not Thoon Godman, 1900), and species-subspecies: Charidia pocus mayo Evans, 1953 (not Charidia lucaria (Hewitson, 1868)), Decinea onasima boliviensis (E. Bell, 1930) (not Decinea dama (Herrich-Schäffer, 1869)), Cynea (Nycea) erebina somba Evans, 1955 (not Pamphila hycsos Mabille, 1891), Saturnus tiberius suffuscus (Hayward, 1940) (not Saturnus reticulata (Plötz, 1883)), and Falga mirabilis odol Evans, 1955 (not Falga jeconia (A. Butler, 1870)). Then, Milanion pilumnus var. hemestinus Mabille and Boullet, 1917 is a junior subjective synonym of Milanion pilumnus pilumnus Mabille and Boullet, 1917, not of Milanion leucaspis (Mabille, 1878). Lectotypes are designated for nine taxa (names in original combinations below): Pellicia bromias Godman and Salvin, 1894 (Mexico: Veracruz, Atoyac), Nisoniades perforata Möschler, 1879 (Colombia), Helias ascalaphus Staudinger, 1876 (central Panama), Pamphila hycsos Mabille, 1891 (Colombia), Amblyscirtes fluonia Godman, 1900 (Mexico: Guerrero, Xocomanatlan), Mastor anubis Godman, 1900 (Mexico: Guerrero, Omiltemi), Eutychide ochus Godman, 1900 (Mexico: Veracruz, Atoyac), Cobalus subcordata Herrich-Schäffer, 1869 (Southeast Brazil), and Thracides xanthura Godman, 1901 (Panama: Chiriquí Province, Bugaba). A neotype is designated for Eudamus briccius Plötz, 1881 (Guyana: Iwokrama Forest).
Keywords: Cryptic species, biodiversity, skipper butterflies, genomics, speciation, nomenclature, taxonomy
Introduction
More than 150 years have passed since Hewitson’s papers describing dozens of new butterfly species in a single publication (Hewitson 1867, 1868). At the time of Hewitson, wing pattern differences were nearly the only criterion for species delineation in Lepidoptera. The superficial appearance of spread specimens, as judged by an author, was described in a short paragraph preceded by a newly proposed species name. The influx of new species descriptions was fueled by increased collecting efforts in species-rich and previously unexplored tropical areas of the world. Most of the commonly encountered species differing in wing patterns were described within a few decades, suggesting to some that we have cataloged nearly all butterfly species.
The situation changed with the realization that consistent genitalic differences typically signify species-level differences even without obvious wing pattern characters. This prompted a genitalia screening approach where large series of specimens were dissected in a search for new species. In Hesperiidae, this approach proposed by Godman and Salvin (1893–1899) was used on a large scale by Evans (Evans 1937, 1949, 1951, 1952, 1953, 1955), who discovered more than 1500 species and subspecies. Genitalia screening was taken to the next level by Austin and collaborators (Austin and Mielke 1998; Austin 2000, 2008) and is being used by others (Dolibaina et al. 2014, 2017; Siewert et al. 2020). For example, revisions of Phanus Hübner, [1819], Entheus Hübner, [1819], and Aguna R. Williams, 1927 by Austin and colleagues nearly doubled the number of species in each genus as a result of extensive genitalic screening.
With the advent of DNA-based methods, which are particularly suited for screening due to automation in both experimental and computational pipelines, we are experiencing the next renaissance in species discovery. The COI barcoding popularized by Hebert and colleagues (Hebert et al. 2003), the most widespread approach, appears to generally fall short if unaccompanied by phenotypic inspection (Rubinoff et al. 2006) but is productive if used with caution (Lukhtanov et al. 2016). Barcoding screens yield impressive results when applied to species rich and poorly studied insect groups (Fernandez-Triana et al. 2014, 2023; Sharkey et al. 2021).
Although expensive, whole genome screens are significantly more reliable than barcode screens for this work because the genome represents its organism (i.e., genotype determines phenotype) and is ideally suited for species delimitation, identification, and discovery. We have been applying a genomic screening approach to butterflies with the focus on Hesperiidae, thus refining their higher classification (Cong et al. 2019b; Li et al. 2019; Zhang et al. 2019b, 2019c, 2022b, 2023c, 2023d) and finding new species (Zhang et al. 2022a, 2022c, 2023b). Our overall strategy is to obtain whole genome shotgun sequences using Illumina short-read sequencing from as many butterfly specimens (leg samples) from diverse localities as possible and infer phylogenetic trees from nuclear and mitochondrial genome protein-coding genes. Inspection of these trees reveals species as tight clades of specimens, as described in the “Species, subspecies, and genomics” section of Zhang et al. (2022a). Whenever possible, we identify species from the first principles, i.e., by sequencing primary type specimens and including them in phylogenetic trees. The primary types identify the clades they fall into. If type specimens were not yet sequenced or are lost, we use traditional identification methods, starting from the original description and comparing genitalia of specimens from or near type localities.
As a result, we find many clades that are not associated with available names and represent new taxa. To test whether these clades are likely to be species, we compute genetic differentiation (as Fst) and estimate gene exchange (as Gmin) between taxa on proteins predicted to be located on the Z chromosome: Fst > 0.20 and Gmin < 0.05 typically correspond to distinct species (Cong et al. 2019a). We also compute percent difference on the COI barcode region, and a value > 2% (~13 bp) is characteristic of distinct species (Hebert et al. 2003), although some species (as delineated by genitalic differences and their biology) may exhibit smaller differences in their barcodes (Burns et al. 2008; Zhang et al. 2023b).
The degree of genetic difference between specimens of the same species is consistent: DNA differs little within species and prominently between species. Therefore, in contrast to the need for a large number of specimens to gauge phenotypic variation, a small series, even a single specimen, the holotype, is sufficient for genomic-based species delineation if genetic differentiation from its relatives is substantial. Furthermore, the genomic approach enables us not to rely on males as holotypes in placing the new species among its relatives, which was frequently necessary before because most species have been confidently identified only by male genitalia. Even a single female genetically differentiated from other species would suffice as the name bearer using the genomic approach.
After finding potential new species in genomic trees, we return to phenotypic inspection and compare wing patterns and genitalia of the proposed new species with its relatives to rationalize genomic differences phenotypically and provide phenotypic characters for species diagnosis. Such characters can usually be found, but in several instances, they should be treated with caution due to the small number of specimens sequenced. Genetic diagnoses given as DNA characters are more reliable for identifying these more cryptic species, at least before larger series of specimens are assembled and examined.
Materials and Methods
Traditionally, new species are discovered through visual comparisons of facies and genitalia, sometimes complemented with field observations about their life histories and ecology. Here, we use a genomic screen approach to species discovery: i.e., to detect new taxa or to test a hypothesis about a new species suspected from phenotypic inspection. First, we obtain whole genome shotgun sequences of many representative Hesperiidae specimens of (nearly) all known species across their ranges, including phenotypically unusual specimens, using our previously established experimental protocols (Li et al. 2019; Zhang et al. 2019a). Typically, a leg of a dry pinned specimen from a collection (see the list of collections below) is used for DNA extraction. Specimens of any age are amenable to our protocol (Cong et al. 2021). Second, these genomic datasets composed of 150 bp (or less) DNA segments are subjected to computational analysis to identify and assemble (i.e., stitch together) protein-coding regions using DIAMOND (Buchfink et al. 2015) aided by a reference set of all proteins encoded in a previously assembled genome of Cecropterus lyciades (Geyer, 1832) (Shen et al. 2017). This procedure results in a master-slave alignment of all these regions (i.e., coding regions in each specimen are aligned to the reference), and these alignments, which are too large (about 18 million positions) for time-efficient phylogenetic analysis, are randomly subsampled for 300,000 positions (by codon) to be used in the construction of phylogenetic trees as described previously (Zhang et al. 2022b). Third, we construct phylogenetic trees using IQ-tree v1.6.12 under GTR+GAMMA model (Nguyen et al. 2015) from these randomly sampled positions in nuclear (autosomes and Z chromosome separately) and mitochondrial genomes and estimate statistical significance by codon resampling from the original complete alignment. These three trees are visualized using FigTree (Rambaut 2018) and visually compared to each other.
Inspecting the genomic-level trees, we look for high confidence clades close to the leaves that visually appear like combs (i.e., star subtrees). Such clades typically correspond to distinct species characterized by prominent genetic differentiation from other species (Zhang et al. 2022a, 2022c). Preference is given to the Z chromosome trees, which are illustrated in this work, because most of the genes important in speciation (pheromone production, wing pattern control, differences between sexes) are encoded by this chromosome, which, in addition, is more resistant to introgression (Pazhenkova and Lukhtanov 2021). Additionally, we illustrate segments of mitochondrial genome trees. Although prone to introgression, mitochondrial DNA is inherited as a single locus and frequently does not vary strongly within species but differs between species. Differences between species visually stand out in phylogenetic trees inferred from mitogenomes. The COI barcode, which is extensively used for species identification and discovery (Hebert et al. 2003), is located in the mitogenome. In many instances, only a single specimen of a species is available, and we compare its genetic distance from others with distances between specimens of the same species, using both nuclear and mitochondrial DNA.
The next step is to confidently assign available names to the clades representing species. In many instances, the assignment is supported by sequenced primary type specimens that we sequenced and included in the trees: the species represented by the clade receives the name of the type of the oldest valid name in this clade. If there are no valid names, available names in the clade serve as the basis for naming (and resurrection from synonymy). In the absence of sequenced primary types, identifications are made by the traditional phenotype-based method: comparing facies and genitalia with those of extant primary type specimens or, if types could not be found, with original descriptions while taking type localities into account. The clades or genetically differentiated branches (when only one specimen is available) that cannot be assigned available names represent potential new species and become the focus of this study. Specimens from these clades are scrutinized for their phenotypes, and genitalia are dissected to learn about morphological differences from known species.
In addition to phenotypic diagnosis, we provide diagnostic DNA characters, both in the nuclear genome and, when such characters exist, in the COI barcode. DNA characters are found in nuclear protein-coding regions using our previously developed procedure (see SI Appendix to Li et al. 2019). The logic behind the character selection was detailed in Cong et al. (2019b). The character states are provided in species diagnoses as abbreviations. For example, aly728.44.1:G672C means position 672 in exon 1 of gene 44 from scaffold 728 of the Cecropterus lyciades (Geyer, 1832) (formerly in Achalarus Scudder, 1872, thus “aly”) reference genome (Shen et al. 2017) is C, changed from G in the ancestor. When characters are given for the sister clade of the diagnosed taxon, the following notation is used: aly5294.20.2:A548A (not C), which means that position 548 in exon 2 of gene 20 on scaffold 5294 is occupied by the ancestral base pair A, which was changed to C in the sister clade (so it is not C in the diagnosed taxon). The same notation is used for COI barcode characters but without a prefix ending with ‘:’. The sequences of exons from the reference genome with the positions used as character states highlighted in green are given in the supplemental file (Zhang et al. 2023a). Providing a link to these DNA sequences from this publication ensures that the numbers given in the diagnoses can be readily associated with actual sequences. Whole genome shotgun datasets we obtained and used in this work are available from the NCBI database <https://www.ncbi.nlm.nih.gov/> as BioProject PRJNA1044449, and BioSample entries of the project contain the locality and other collection data of the sequenced specimens shown in the trees. Additionally, specimen data are summarized in Table S1 of the supplemental file (Zhang et al. 2023a). COI barcode sequences have been deposited in GenBank with accessions OR835792 and OR837624–OR837723. All new names have been registered with ZooBank.
Spread specimens were photographed with a Nikon 800 camera using a 105 mm Nikkor macro lens in NEF (raw) format, converted to TIF format using DxO with color-correction options adjusted to match 24 patch ColorChecker, and edited in Adobe Photoshop CS4 to standardize the background. Imperfections in specimens, such as scale damage, pinholes, and wing tear, were not digitally removed. Genitalia were prepared after DNA extraction from abdomens, which had been soaked in 10% KOH either overnight at room T (if it was convenient to take a break from work) or at 65°C for 15–60 min (depending on the size and abdomen softness after the soak) and then dissected under a binocular microscope. Genitalia were placed in glycerin and photographed using the AmScope system H800–96S-18M3 (0.7–5× zoom monocular microscope on a table stand with LED ring light and USB 18.0MP digital camera) in 3–5 focus slices, which were edited to brighten the background and merged using Adobe Photoshop CS4, and further assembled into plates. Genitalia were stored in glycerin in small vials pinned by each specimen.
The specimens were examined and sampled for sequencing from the following collections (abbreviations, which are not necessarily acronyms of the current names of these institutions, are given in parenthesis and used in Table S1 of the supplemental file (Zhang et al. 2023a)): American Museum of Natural History, New York, NY, USA (AMNH), Academy of Natural Sciences of Drexel University, Philadelphia, PA, USA (ANSP), Natural History Museum, London, UK (BMNH), California Academy of Sciences, San Francisco, CA, USA (CAS), Carnegie Museum of Natural History, Pittsburgh, PA, USA (CMNH), Colorado State University Collection, Fort Collins, CO, USA (CSUC), Cornell University Insect Collection, Ithaca, New York, USA (CUIC), Universidade Federal do Paraná, Curitiba, Paraná, Brazil (DZUP), Field Museum of Natural History, Chicago, IL, USA (FMNH), Los Angeles County Museum of Natural History, Los Angeles, CA, USA (LACM), Mississippi Entomological Museum, Starkville, MS, USA (MEM), Museum für Naturkunde, Berlin, Germany (MFNB), McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA (MGCL), Museo del Instituto de Zoología Agrícola “Francisco Fernandez Yépez”, Universidad Central de Venezuela, Maracay, Venezuela (MIZA), Muséum National d’Histoire Naturelle, Paris, France (MNHP), Museum für Tierkunde, Dresden, Germany (MTD), Museo de Historia Natural, Lima, Peru (MUSM), Peabody Museum of Natural History, Yale University, New Haven, CT, USA (PMNH), Naturalis Biodiversity Center, Leiden, Netherlands (RMNH), Natural History Museum, Frankfurt, Germany (SMF), Texas A&M University Insect Collection, College Station, TX, USA (TAMU), Biodiversity Center, University of Texas at Austin, Austin, TX, USA (TMMC), National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), Burke Museum of Natural History and Culture, Seattle, WA, USA (UWBM), Zoological Institute and Museum Greifswald, Germany (ZIMG), Zentrum fur Biodokumentation des Saarlandes, Schiffweiler, Germany (ZfBS), Natural History Museum of Denmark, University of Copenhagen, Copenhagen, Denmark (ZMUC), Zoologische Staatssammlung München, Germany (ZSMC), and research collections of Pierre Boyer, France (PBoyer), Jim P. Brock, USA (JPBrock), Ernst Brockmann, Germany (EBrockmann), Matthew J. W. Cock, UK (MJWCock), Bill Dempwolf, USA (WRDempwolf), Bernard Hermier, French Guiana (BHermier), and Kiyoshi Maruyama, Japan (KMaruyama). Type status abbreviations are HT holotype, LT lectotype, NT neotype, ST syntype, PT paratype, and PLT paralectotype.
Results and Discussion
Genomic analysis of Hesperiidae species across their ranges reveals 100 distinct unnamed phylogenetic lineages that are described below as species. These species are genotypically unique lineages separated from other similar lineages. Many of them are possibly allopatric with their closest relatives, but the transition from one species to another in genotype is abrupt, without detected intermediates. The specific rationale for each species distinction is given below in their “Definition and diagnosis.”
Species are placed in previously published identification keys (Evans 1952, 1953, 1955), and phenotypic characters are given to differentiate it from their closest relatives. A tree with the holotype included shows its position relative to other taxa. We present photographs of the dorsal and ventral sides of the holotype and, in nearly all cases, photographs of genitalia of either the holotype or a paratype. Diagnostic DNA characters in the nuclear genome and the COI barcode (if they exist) are given as abbreviations. The COI barcode sequence itself is provided for every new species. Species descriptions are accompanied by other nomenclatural acts (designation of a neotype and lectotypes) and taxonomic adjustments necessary as supporting evidence for the new species. Phylogenetic trees are shown in Fig. 1–8, specimen photographs in Fig. 9–220, 465–467 (dorsal and ventral sides are denoted by odd and even figure numbers, respectively), and genitalia images in Fig. 221–464.
Figure 1.
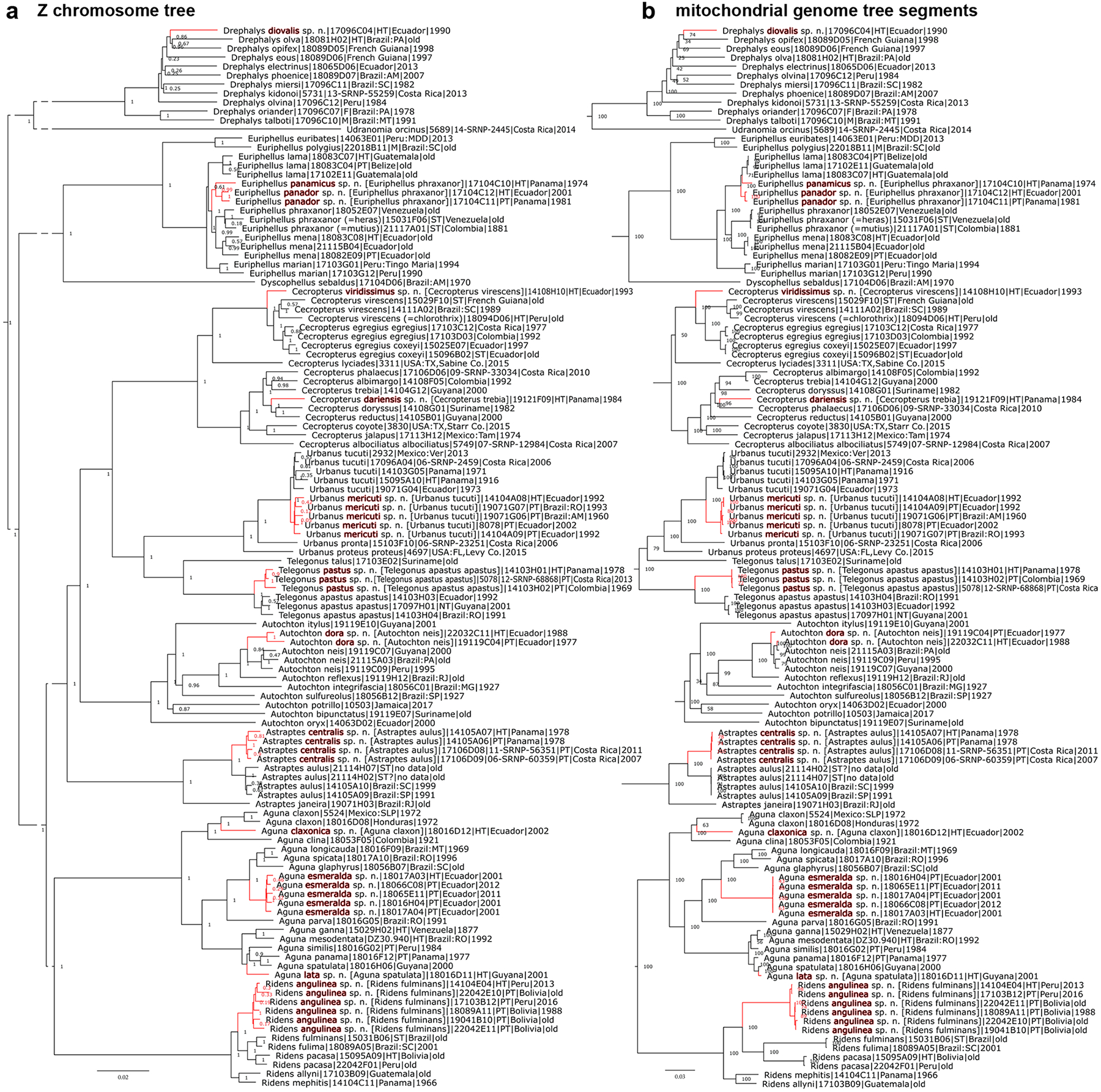
Phylogenetic trees of Eudaminae inferred from protein-coding regions in a) the Z chromosome; gaps in branches indicate places where vertical slices of the tree were removed to reduce its horizontal dimension (to allow an increase of the font size), i.e., branches with gaps, are longer than shown; and b) the mitochondrial genome (in segments): clades are cut from the tree and rearranged to match better the order of taxa in the Z chromosome tree and to save space. Statistical support values are shown by nodes. For each specimen, the name adopted in this work is given first, and a previously used name (or misidentification for the new species) is listed in square brackets (if different), supplemented with the DNA sample number, type status (see Materials and Methods for abbreviations), general locality, and year of collection. See Table S1 in the Supplemental file (Zhang et al. 2023a) or NCBI database entries for additional data about these specimens. Synonyms are given in parentheses preceded by “=”. The type status refers to this synonym if the synonym name is provided. The same notations are used throughout this work in other figures showing phylogenetic trees. Clades of new species proposed here are shown in red color, and their species epithets are highlighted in red. In other phylogenetic tree figures (e.g., Fig. 2), yellow highlight indicates taxonomic changes, e.g., transfer to a different genus (genus name highlighted) or change in species, subspecies, or synonym status (species epithet highlighted) and clades with taxonomic changes are shown in different colors. Green arrows point from the clade of the genus/species where a species/synonym was previously placed to the clade of its placement proposed in this work.
Figure 8.

Phylogenetic trees of several Hesperiini subtribes inferred from protein-coding regions in a) the Z chromosome and b) the mitochondrial genome (in segments). See Fig. 1 legend for other notations.
In the present work, we refrain from using patronyms, partly because they may be more challenging to remember and associate with corresponding species. The new names proposed here are derived either from the names of species’ close relatives (usually making the name longer for southern counterparts, a practice that we introduce as a menmonic), a descriptive phenotypic feature, or the type locality. We hope such names will integrate easily with the existing classification and be more straightforward to learn.
Subfamily Eudaminae Mabille, 1877 Tribe Entheini Grishin, 2019
Drephalys (Drephalys) diovalis Grishin, new species https://zoobank.org/23F826FB-40E4-4664-823D-C671FF1FEDEC (Fig. 1 part, 9–10, 221–222)
Definition and diagnosis.
Both genomic sequences and genitalia characters (valva with knobs distally and without a long process from the ampulla) place this species in the nominal subgenus Drephalys E. Watson, 1893. However, the yellow colors of the spots and the wing patterns show some similarity to species from the subgenus Paradrephalys Burns, 2000. The closest in appearance and the only known nominotypical dorsally yellow-patterned Drephalys with ventral hindwing pale spots instead of a band is Drephalys opifex Evans, 1952 (type locality in Suriname). The new species keys to it (B.6.8) in Evans (1952), although not precisely, because the hindwing base in the new species is ventrally orange infused with purple instead of yellow, and palpi are whiter. More specifically, it differs from all described Drephalys species by a combination of honey-yellow markings on the dorsal wing surface (cell Rs-M1 with a yellow spot), two prominent oval white spots on the purple-orange ventral hindwing, and the lack of costal fold. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1146.54.9:A585G, aly1146.54.9:A591T, aly275211.5.3:A92G, aly2284.4.3:T87C, aly2284.4.3:T148C, aly113.11.7:C105C (not T), aly113.11.7:C138C (not T), aly2363.7.5:G102G (not A), aly525.90.3:G21G (not C), aly10226.17.2:C63C (not T), and COI barcode: T268T, T274A, T475C, T487C, C536C.
Barcode sequence of the holotype.
Sample NVG-17096C04, GenBank OR837624, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACATCTTTAAGTCTTTTAATTCGAACTGAATTAGGAACCCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGTTTTTGATTACTTCCCCCATCATTAACTCTTTTAATTTCTAGAAGAATTGTAGAAAATGGAGCAGGAACTGGATGAACAGTTTATCCCCCTCTTTCATCAAATATTGCTCATCAAGGTTCTTCTGTAGATTTAGCTATTTTTTCCCTTCATTTAGCTGGTATTTCATCTATTTTAGGAGCTATTAACTTTATTACTACAATTATTAATATACGAATTAACAATTTATCATTCGATCAAATACCTTTATTTGTATGAGCTGTAGGAATTACTGCTTTATTACTTTTATTATCTTTACCTGTTTTAGCAGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAATACATCATTTTTTGATCCAGCAGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 9–10, bears the following five rectangular labels, four white: [ECUADOR Napo | Tena-Pano Rd. 600m | 27 Sept. ‘90 | D. H. Ahrenholz], [Drephalys | dumeril | Det. ♂ | S. S. Nicolay], [DNA sample ID: | NVG-17096C04 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 00894990], and one red [HOLOTYPE ♂ | Drephalys | diovalis Grishin].
Type locality.
Ecuador: Napo Province, Tena-Pano Road, elevation 600 m.
Etymology.
The name is given for the doublet of oval spots on the ventral hindwing and is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Ecuador.
Tribe Phocidini Tutt, 1906
Euriphellus panador Grishin, new species https://zoobank.org/2CF28569-39B0-4379-9C26-25992D63BFFA (Fig. 1 part, 11–12, 223–224)
Definition and diagnosis.
Among the clades representing named species of the Euriphellus phraxanor (Hewitson, 1876) group, we see a clade, sister to both E. phraxanor and Euriphellus mena (Evans, 1952) (type locality in Ecuador) that was not associated with any available name and therefore consists of new species (Fig. 1). One of these new species (see below for the second one) keys to “Dyscophellus phraxanor phraxanor” (D.4.2(b)) in Evans (1952) but differs from it and other relatives by a combination of flatter and narrower tegumen in lateral view, the sharper and terminally narrower basal tooth of harpe, ventral margin of harpe being only slightly shouldered (Fig. 224), but more so than that in the new species described next (Fig. 226), moderately defined hindwing discal spots, which are brown on the dorsal side, not hyaline, spot in cell M2-M3 offset basad from the row, comparatively (to the ventral hindwing discal yellow spots) larger forewing subapical spots, and weaker orange overscaling in the anterior part of ventral forewing (Fig. 11–12). COI barcode of this new species differs from E. phraxanor and E. mena by 4.7% (31 bp) and 4.6% (30 bp), respectively, but only 1.4% (9 bp) different from Euriphellus lama (Evans, 1952) (type locality in Guatemala), while being well differentiated from it in the Z chromosome (Fig. 1a). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly151.14.2:A75G, aly2090.1.4:A54G, aly127.43.4:G102A, aly127.43.4:A160G, aly443.22.1:C88A, and COI barcode: A181G, T259C, T364C, T376A, T553A.
Barcode sequence of the holotype.
Sample NVG-17104C12, GenBank OR837625, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATGTTAGGAACTTCTTTAAGTTTACTAATTCGAACTGAATTAGGAACTCCAGGATCTTTAATTGGAAATGATCAAATTTATAATACTATTGTTACAGCCCATGCTTTTATTATAATTTTTTTTATAGTAATGCCTATTATAATTGGGGGATTCGGAAACTGATTAGTACCATTAATATTAGGAGCCCCAGATATAGCTTTTCCACGAATAAATAATATAAGATTCTGATTACTTCCCCCTTCTTTAATATTATTAATTTCAAGAAGAATCGTTGAAAATGGAGCAGGAACAGGATGAACAGTTTATCCTCCTTTATCTGCTAACATTGCCCATCAAGGATCATCAGTTGATTTAGCAATTTTTTCTCTTCACTTAGCTGGTATTTCTTCAATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAGAAACTTATCTTTCGATCAAATACCATTATTTGTTTGAGCTGTAGGAATTACAGCTTTATTATTACTTCTCTCTTTACCAGTACTAGCAGGTGCAATTACTATATTATTAACAGACCGAAATTTTAATACATCTTTTTTTGATCCTTCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 11–12, bears the following four rectangular labels, three white: [ECUADOR: Esmeraldas | La Chaquita Exp Station | 10 Km San Lorenzo-Lita Road | 01° 13.82′S, 78° 45.95′W | 3 March 2001, 50 m | D.H. Ahrenholz leg.], [DNA sample ID: | NVG-17104C12 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 00913861], and one red [HOLOTYPE ♂ | Euriphellus | panador Grishin]. Paratype: 1♂ NVG-17104C11, USNMENT_00913860 Panama, Darien Province, Cana (Cerro Pirre), elevation 400 m, GPS 7.9333, −77.5667, 6-Jul-1981, G. B. Small leg. [USNM].
Type locality.
Ecuador: Esmeraldas Province, km 11 of San Lorenzo-Lita Road, La Chiquita Wildlife Refuge, elevation 50 m, GPS 1.23033, −78.76583.
Etymology.
The name is a fusion of this species’ known localities: Pana[ma and Ecua]dor. The name is a noun in apposition.
Distribution.
Currently known from Ecuador and eastern Panama.
Comment.
Sequencing of the Telegonus mutius Plötz, 1882 (type locality in Colombia) syntype we found in MFNB reveals that it is conspecific with a syntype of Telegonus heras Mabille, 1888 (type locality Venezuela: Porto Cabello), currently a junior subjective synonym of Euriphellus phraxanor (Hewitson, 1876) (type locality “New Granada”—likely referring to Colombia—and Panama: Chiriquí) (Fig. 1), thus confirming our previously hypothesized synonymy of T. mutius with E. phraxanor (Zhang et al. 2022b). We are currently undertaking a search for syntypes of E. phraxanor to complete this investigation.
Euriphellus panamicus Grishin, new species https://zoobank.org/F9D83659-187D-4AC5-8EAC-D1B5A599D972 (Fig. 1 part, 13–14, 225–226)
Definition and diagnosis.
Sister to previous species and differs from it by 1.8% (12 bp) in COI barcode. The previous species is either sympatric with this new species in Panama or comes close to it in distribution. Keys to “Dyscophellus phraxanor phraxanor” (D.4.2(b)) in Evans (1952) but differs from it and other relatives by a combination of more convex and wider tegumen in lateral view, terminally rounded and wider basal tooth of harpe (Fig. 226), ventral margin of harpe being even less shouldered than in E. panador new species (Fig. 224), well-defined hindwing discal spots, not hyaline (could be pale-centered), spot in cell M2-M3 nearly within the row, comparatively (to the ventral hindwing discal yellow spots) smaller forewing subapical spots, and stronger orange overscaling in the anterior part of ventral forewing (Fig. 13–14). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly671.39.2:T432C, aly887.9.1:G232A, aly102.20.9:G45T, aly272.9.2:G61A, aly272.9.2:G79A, aly2578.3.9:G222G (not T), aly2578.3.9:A230A (not G), aly2275.23.9:A72A (not G), aly4036.9.5:G321G (not A), aly27.16.1:T1497T (not C), and COI barcode: T118C, A181A, A202G, T376G, A625G.
Barcode sequence of the holotype.
Sample NVG-17104C10, GenBank OR837626, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATGTTAGGAACTTCTTTAAGTTTACTAATTCGAACTGAATTAGGAACTCCAGGATCTTTAATTGGAAATGATCAAATTTATAACACTATTGTTACAGCCCATGCTTTTATTATAATTTTTTTTATAGTAATGCCTATTATAATTGGAGGATTCGGAAACTGATTAGTGCCATTAATATTAGGAGCCCCAGATATAGCTTTTCCACGAATAAACAATATAAGATTTTGATTACTTCCCCCTTCTTTAATATTATTAATTTCAAGAAGAATCGTTGAAAATGGAGCAGGAACAGGATGAACAGTTTATCCTCCTTTATCTGCTAATATTGCTCATCAAGGATCGTCAGTTGATTTAGCAATTTTTTCTCTTCACTTAGCTGGTATTTCTTCAATTTTAGGAGCTATTAATTTTATTACAACGATTATTAATATACGAATTAGAAACTTATCTTTCGATCAAATACCATTATTTGTTTGAGCTGTAGGAATTACAGCTTTATTATTACTTCTCTCTTTACCTGTACTAGCAGGTGCAATTACTATATTATTAACAGACCGAAATTTTAATACATCTTTTTTTGATCCTTCTGGGGGAGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 13–14, bears the following four rectangular labels, three white: [Cerro Jefe 2200’ | Pma., Panama | April 10, 1974 | G B Small], [DNA sample ID: | NVG-17104C10 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 00913859], and one red [HOLOTYPE ♂ | Euriphellus | panamicus Grishin].
Type locality.
Panama: Panama Province, Cerro Jefe, elevation 2200′.
Etymology.
The name is given for the type locality and is a masculine adjective.
Distribution.
Currently known only from the type locality in central Panama.
Tribe Eudamini Mabille, 1877 Subtribe Eudamina Mabille, 1877
Cecropterus (Thorybes) viridissimus Grishin, new species https://zoobank.org/855F436E-D634-4129-AB6F-FCFB51E374E0 (Fig. 1 part, 15–16, 227–228)
Definition and diagnosis.
This new species is very similar to Cecropterus virescens (Mabille, 1877) (type locality in French Guiana) phenotypically and keys to it (C.13.27) in Evans (1952). However, in both Z chromosome and mitochondrial genome trees, it is sister to the clade that consists of both C. virescens and Cecropterus egregius (A. Butler, 1870) (type locality unknown), which are rather different-looking species. COI barcode differs from C. virescens syntype by 2.7% (18 bp). Phenotypically, it differs from most C. virescens specimens by the white border on the hindwing underside, which harbors brown overscaling and is reduced in width from vein M2 to the apex (Fig. 16), strongly humped in the middle dorsoposterior margin on the harpe, and wider separated uncus arms (Fig. 228). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2668.2.9:C109T, aly2668.2.9:C123T, aly1370.7.4:A221G, aly347.8.1:A265C, aly923.7.1:A781C, aly1313.24.3:A72A (not G), aly138.12.1:A515A (not G), aly208.4.3:T84T (not C), aly5623.1.3:C1206C (not T), aly173.13.2:G59G (not A), and COI barcode: T263C, A319G, T400T, T529C, T553C.
Barcode sequence of the holotype.
Sample NVG-14108H10, GenBank OR837627, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAATTGGAACTTCATTAAGTTTACTTATTCGAACTGAATTAGGAACTCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCTCTTATATTAGGAGCCCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGACTATTACCCCCTTCATTAACTCTTTTAATTTCAAGAAGTATTGTTGAAAATGGAGCGGGTACTGGATGAACTGTTTATCCTCCTTTATCTTCTAATATTGCCCATCAAGGAGCATCAGTAGATTTAGCAATTTTTTCTTTACATCTTGCAGGAATTTCATCTATTCTTGGAGCTATTAATTTTATTACAACTATTATTAATATACGAATTAATAATTTATCATTTGATCAAATACCATTATTTATTTGAGCTGTAGGAATTACAGCCTTATTATTATTACTTTCATTACCCGTTTTAGCTGGAGCCATTACTATATTATTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCTGCAGGTGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 15–16, bears the following three rectangular labels, two white: [ECUADOR: Zamora | 56 km Loja-Zamora | 4° 2.7′S 78° 59.2′W | 4 October 1993 | S. S. Nicolay, leg.], [DNA sample ID: | NVG-14108H10 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Cecropterus | viridissimus Grishin].
Type locality.
Ecuador: Zamora-Chinchipe Province, km 56 of Loja-Zamora road, elevation 976 m, GPS −4.0450, −78.9867.
Etymology.
In Latin, viridissimus means very green or bright green. The name is given for the extensive green coloring of this species and is a masculine adjective.
Distribution.
Only known from the holotype collected in Ecuador.
Comment.
Curiously, in the tree constructed from autosome genes, the new species is sister to C. virescens in accord with phenotypic similarities, suggesting introgression and other irregularities in the evolution of its mitochondrial genome and Z chromosome.
Cecropterus (Murgaria) dariensis Grishin, new species https://zoobank.org/269C0E17-EE28-439C-8F7C-7247A6D6CC62 (Fig. 1 part, 17–18, 229–230)
Definition and diagnosis.
Superficially somewhat resembles sympatric Cecropterus trebia (Möschler, 1879) but differs from it and other relatives by completely brown hindwing ventral surface (without white at the outer margin, but with whitish fringes) and 4 subapical hyaline spots on the forewing. It also resembles Spicauda Grishin, 2019 but differs in the hyaline spot in forewing cell M3-CuA1, being dash-like and strongly offset from the cell base and discal hyaline band. Keys (imperfectly) to “Urbanus carmelita carmelita” (C.13. 22(b)) in Evans (1952) but differs from this species currently known as Cecropterus carmelita (Herrich-Schäffer, 1869) (type locality in Brazil) by vestigial, line-like and not scalloped white outer marginal band on the hindwing underside (the fringe in mostly white from the apex to vein CuA1); differs also from the somewhat similar Cecropterus athesis (Hewitson, 1867) by longer hindwing tails and hyaline spot in forewing cell CuA2-1A+2A being better aligned with the discal band, and by the lack of a darker-brown line along the outer margin of ventral hindwing, which is replaced with a vestigial white band. In male genitalia (Fig. 229–230), most similar to Cecropterus doryssus (Swainson, 1831) (type locality in Brazil: Bahia) and relatives but differs from them by less angled harpe with rounder curve from ventral to posterior margin and with serrated dorsal margin that is more prominently expanded into a small distal lobe. This species is not cryptic and is unambiguously recognizable by its phenotype. The following base pairs are diagnostic in the nuclear genome: aly2750.3.3:G39A, aly515.2.2:G496T, aly1651.24.15:C120A, aly235.4.4:T51C, aly23605.18.5:G118A, aly1038.17.30:C61C (not A), aly1038.17.30:A62A (not C), aly383.7.2:T54T (not A), aly1042.3.1:A138A (not G), aly1651.24.15:C91C (not T), and COI barcode: T266C, T401C, T424A, A550G, T581C.
Barcode sequence of the holotype.
Sample NVG-19121F09, GenBank OR837628, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAGTTGGAACTTCATTAAGTTTACTTATTCGAACTGAATTAGGAACTCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAATCCCCCTTATATTAGGAGCCCCCGATATAGCTTTCCCCCGTATAAATAATATAAGATTTTGATTACTACCTCCATCTTTAACTCTTTTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGTACTGGATGAACAGTTTACCCCCCTTTATCATCTAATATTGCTCATCAAGGAGCATCCGTAGATTTAGCAATTTTCTCACTACATCTTGCTGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACAACTATTATTAACATACGAATTAATAATTTATCATTTGATCAAATACCATTATTTATTTGAGCTGTTGGAATTACAGCTTTACTATTATTACTTTCATTGCCTGTTTTAGCTGGAGCTATTACTATACTACTAACTGATCGAAATTTAAATACTTCTTTTTTTGACCCAGCAGGTGGGGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 17–18, bears the following four rectangular labels, three white: [PANAMA: 1000m. | Darien, Cana | 5. Jan. 1984 | Gordon Small], [DNA sample ID: | NVG-19121F09 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01602755], and one red [HOLOTYPE ♂ | Cecropterus | dariensis Grishin].
Type locality.
Panama: Darien Province, Cana, elevation 1000 m.
Etymology.
The name reflects the type locality and is a masculine adjective.
Distribution.
This species is currently known only from the holotype collected in Panama.
Urbanus (Urbanus) mericuti Grishin, new species https://zoobank.org/5C620BB9-A60D-42F8-B72B-8C832D79A9D4 (Fig. 1 part, 19–20, 231–232)
Definition and diagnosis.
Inspection of genomic trees reveals that most South American populations identified as Urbanus tucuti (R. Williams, 1927) (type locality in Panama, holotype sequenced as NVG-15095A10) are strongly differentiated genetically from U. tucuti (Fig. 1): e.g., their COI barcodes differ by 3.5% (23 bp), and therefore represent a new species. It keys to “Astraptes tucuti” (C.14.5) in Evans (1952) and differs from it by comparatively shorter harpe with a straighter dorsal margin and less acute terminal angle, the wider separation between harpe and ampulla (wider notch) (Fig. 232), uncus arms being more parallel to each other and closer together, rather than terminally diverging in dorsal view (Fig. 231), and usually absent or reduced hyaline dash in M3-CuA1 cell (Fig. 19). Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly53.2.40:C42T, aly1968.11.7:A106G, aly58.10.2:C13T, aly58.10.2:G45A, aly7098.1.5:C115A, and COI barcode: A43T, A79G, T178C, T479C, T601C.
Barcode sequence of the holotype.
Sample NVG-14104A08, GenBank OR837629, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAATTGGTACTTCTTTAAGATTACTTATTCGAACTGAATTAGGGACTCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCCCATGCATTTATTATAATTTTCTTTATAGTTATACCTATCATAATCGGAGGATTTGGTAATTGACTTGTACCTTTAATAATAGGTGCCCCTGATATAGCTTTCCCCCGTATAAATAACATAAGATTTTGATTACTACCCCCTTCCTTAACTTTATTAATTTCAAGAAGAATTGTTGAAAATGGTGCTGGTACTGGATGAACAGTTTATCCCCCCCTTTCATCTAACATTGCTCATCAAGGAGCTTCTGTTGATTTAGCAATTTTCTCTCTTCATCTTGCCGGAATTTCATCAATTCTTGGAGCTATTAATTTTATTACAACAATTATTAACATACGAATTAATAGACTAACTTTTGATCAAATACCTTTATTTGTATGAGCTGTAGGAATTACAGCATTATTATTATTACTTTCTTTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACTGATCGAAATCTAAACACATCATTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 19–20, bears the following four rectangular labels, three white: [ECUADOR: Napo Pr. | 8 km Napo-Ahuano | 1° 2.5′ S 77° 43.5′ W | 9 Nov. 1992, 480m. | S. S. Nicolay, leg.], [Astraptes | Det. fulgor | S.S. Nicolay], [DNA sample ID: | NVG-14104A08 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Urbanus mericuti | Grishin]. Paratypes: 4♂♂ in USNM: 1♂ NVG-14104A09 the same data as the holotype, except 13-Nov-1992; Ecuador 1♂ NVG-8078 Napo, Misahualli Jungle Lodge, 450 m, GPS −1.0257, −77.6570, 6–8-Jan-2002, J. P. W. Hall and M. A. Solis leg., genitalia NVG170208–63 (Fig. 231–232); Brazil: 1♂ NVG-19071G06 Amazonas, Benjamin Constant, Nov-1960, Jorge Kesselring leg.; 1♂ NVG-19071G06 Rondonia, 62 km S Ariquemes, Fazenda Rancho Grande, 165 m, GPS −10.533, −62.800, 14–25-Nov-1993, Brian Harris leg.
Type locality.
Ecuador: Napo Province, km 8 of Puerto Napo-Ahuano Road, elevation 480 m, GPS −1.0417, −77.725.
Etymology.
The name denotes a more southern range of this species than U. tucuti: meri[dionalis (Latin for southern) + tu]cuti. The name is a noun in apposition.
Distribution.
Currently known from the upper Amazonian region in Ecuador and Brazil.
Neotype designation for Eudamus briccius Plötz, 1881
Eudamus briccius Plötz, 1881 was described from an unstated number of specimens from South America (Plötz 1881). Translating and assembling relevant sections of Plötz’s identification key, we get for E. briccius: “Body and wing bases are mostly overscaled with shiny green or blue above, forewings with wide and complete, pale, oblique hyaline median band broken into spots starting from cell 1 (i.e., CuA2-1A+2A) and reaching the costal margin, the spot in cell 3 (i.e., M3-CuA1) is integrated into the band; no hyaline spots by the apex, hindwings without a white transverse band. Hindwings with a smooth outer margin and uncheckered brown fringes. Underside of the hindwings is green, mixed with brown. Forewing length 31 mm. From South America.”
We were not able to locate syntypes of E. briccius. At least one of the syntypes was drawn by Plötz as t[afel]. 72 (Plötz 1881), which has not been found with the rest of the original Plötz drawings. This drawing is not among Godman’s copies, now in BMNH (Godman 1907). However, we found a drawing labeled “briccius Pl. t. 72” (Fig. 467) in ZSMC, pinned in a drawer together with specimens, and styled similarly to Godman’s copies of drawings. We hypothesize that this is either another copy by a different artist or the original, giving a visual of E. briccius. Currently, E. briccius is treated as a junior subjective synonym of Telegonus (Telegonus) apastus (Cramer, 1777) (type locality in Suriname), and the drawing confirms this treatment.
Next, we proceeded with the neotype designation because there is an exceptional need to clarify both the taxonomic identity and the type locality of E. briccius. In the light of the new species proposed below, it is essential to define E. briccius objectively. Hereby, N.V.G. designates a specimen in USNM, DNA sample NVG-18115F05, USNM ENT 00179346, illustrated in Fig. 465–466, with the label data given below as the neotype of Eudamus briccius Plötz, 1881. This neotype reaffirms the current treatment of E. briccius as a junior subjective synonym of Telegonus (Telegonus) apastus (Cramer, 1777) (type locality in Suriname).
The neotype satisfies all requirements set forth by the ICZN Article 75.3, namely: 75.3.1. It is designated to clarify the taxonomic identity of E. briccius, which is necessary because additional species are present among its close relatives and to define the type locality that was only generally stated in the original description as South America; 75.3.2. The characters to differentiate this taxon from others were given in the original description (Plötz 1881), and given above, we summarize them as follows: body and wing bases shiny-green, otherwise brown-black, forewing with a single broad transverse band of hyaline spots from mid-costa to near tornus, the spot in cell M3-CuA1 is within the band; ventral hindwing green intermixed with brown; forewing length around 31 mm; 75.3.3. The neotype specimen is a male bearing three labels: [GUYANA: Iwokrama | Rainforest Res Middle | Essequibo R/Turtle Mt | 200–950’ 20–26.III.2001 | 04°43.90′N 58°43.08′W | Leg. S. Fratello et al], [DNA sample ID: | NVG-17097H01 | c/o Nick V. Grishin], [{QR Code} | USNM ENT 00179346], the neotype has a small piece of wing missing along the margin of right hindwing; 75.3.4. We carefully searched for syntypes of E. briccius in the MFNB, ZSMC, and ZIMG. We failed to find syntypes among Hesperiidae holdings in these collections and, therefore, believe that they were lost; 75.3.5. The neotype closely agrees with the original description of E. briccius in all characters, as evidenced by comparing the neotype illustrated in Fig. 465–466 with the characters for this taxon given in the original description (Plötz 1881) complemented with the drawing (Fig. 467) and listed above (75.3.2.); 75.3.6. The neotype is from Guyana: Iwokrama Forest, Essequibo River, Turtle Mountain, GPS 4.7317, −58.7180, which becomes the type locality of E. briccius, consistently with the original type locality given as “South America”; 75.3.7. The neotype is in the National Museum of Natural History, Washington, DC, USA (USNM). The COI barcode sequence of E. briccius neotype, sample NVG-17097H01, GenBank OR835792, 658 base pairs, is:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAATTGGAACTTCCTTAAGATTACTTATTCGAACTGAATTAGGAACCCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACAATTGTTACAGCTCATGCATTTATCATAATTTTTTTCATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCACTAATAATAGGAGCCCCAGATATAGCATTCCCCCGTATAAATAATATAAGATTCTGATTATTACCTCCATCTTTAACATTATTAATTTCAAGAAGAATTGTTGAAAATGGTGCTGGAACAGGATGAACAGTTTACCCCCCCCTTTCATCTAATATCGCTCATCAAGGAGCATCTGTTGACTTAGCAATTTTCTCCCTTCATCTTGCTGGTATTTCTTCAATTCTTGGAGCAATTAATTTTATTACAACAATTATTAATATACGAATTAACAATTTATCTTTTGATCAAATACCTCTATTCGTTTGAGCAGTAGGAATTACAGCATTATTATTATTACTTTCTTTACCTGTTTTAGCTGGAGCTATTACCATATTATTAACTGATCGAAATTTAAATACTTCTTTCTTTGATCCTGCTGGTGGTGGAGATCCCATTTTATATCAACATTTATTT
Telegonus (Telegonus) pastus Grishin, new species https://zoobank.org/24FB11E2-E34F-4B88-B36E-B2D2CB62BBE5 (Fig. 1 part, 21–22, 233–234)
Definition and diagnosis.
Phylogenetic trees reveal prominent genetic differentiation of northern populations identified as Telegonus apastus (Cramer, 1777) (type locality in Suriname) (Fig. 1): e.g., their COI barcodes differ by 5.9% (39 bp), and therefore they represent a new species. The new species keys to “Astraptes apastus” (C.14.13) in Evans (1952) and differs from it by shorter harpe (shorter than valva) with its distal angle less acute, more robust bulge on costa-ampulla (Fig. 234) and a hyaline spot in forewing cell CuA2-1A+2A typically more angular and extended into a “beak” towards tornus (Fig. 21 vs. 465), usually with less prominent white overscaling around this spot on ventral side (Fig. 22 vs. 466). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1038.17.34:G379A, aly125.11.2:G46A, aly499.47.2:C118A, aly1603.18.3:T185C, aly6286.2.7:T42C, and COI barcode: 49A, T91C, T226C, T382C, T571C.
Barcode sequence of the holotype.
Sample NVG-14103H01, GenBank OR837630, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAATTGGAACTTCATTAAGATTACTTATTCGAACTGAATTAGGAACCCCAGGATCCTTAATTGGAGATGATCAAATTTATAATACAATTGTTACAGCTCATGCATTTATCATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCATTAATAATAGGAGCCCCAGACATAGCATTCCCCCGTATAAATAATATAAGATTTTGATTATTACCCCCATCTCTAACATTATTAATTTCAAGAAGAATTGTTGAAAATGGTGCAGGAACAGGATGAACAGTTTATCCCCCTCTTTCATCTAATATTGCCCATCAAGGAGCATCAGTCGACTTAGCAATTTTCTCCCTTCATCTTGCCGGTATTTCCTCAATTCTTGGGGCAATCAATTTTATTACAACAATTATTAATATACGAATCAATAATTTATCATTTGATCAAATACCTTTATTTGTTTGAGCAGTAGGAATTACAGCATTATTACTATTACTTTCTTTACCTGTTTTAGCAGGAGCTATCACTATATTATTAACTGATCGAAATTTAAATACTTCTTTCTTTGACCCTGCGGGAGGGGGTGACCCAATTCTTTACCAACACTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 21–22, bears the following three rectangular labels, two white: [PANAMÁ: Panamá Prov. | Distrito de El Llano | Cordillera de San Blas | North of El Llano ca. 330 m. | VI. 1978 | Gordon B. Small: Coll.], [DNA sample ID: | NVG-14103H01 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Telegonus (Telegonus) | pastus Grishin]. Paratypes: 2♂♂ in USNM: NVG-5078, 12-SRNP-68868 Costa Rica: Area de Conservación Guanacaste, Alajuela Prov., Sector Rincon Rain Forest, Palomo, el. 96 m, GPS 10.96187, −85.28045, eclosed 09-Jan-2013; and NVG-14103H02 Colombia: Caldas, Victoria, el. 2400′, 9-Feb-1969, S. S. Nicolay leg.
Type locality.
Panama: Panama Province, Distrito de El Llano, Cordillera de San Blas, north of El Llano, elevation 330 m.
Etymology.
The name removes the negating “a” from its sister species name and is a noun in apposition.
Distribution.
From southeastern Mexico to western Colombia.
Autochton (Autochton) dora Grishin, new species https://zoobank.org/2CBF81CD-A45A-40E8-B6B9-E4E64513578D (Fig. 1 part, 23–24, 235–236)
Definition and diagnosis.
Ecuadorian specimens identified (incorrectly) as Autochton neis (Geyer, 1832) (type locality in Brazil) differ prominently from it in Z chromosome genes (Fig. 1a) and, therefore, although not differing in the COI barcode (0.15%, 1 bp), represent a new species. This new species partly keys to “Autochton neis” (C.16.6) in Evans (1952) and has somewhat similar male genitalia but differs from A. neis and other species of Autochton in a rounder distal end of harpe (near ventral margin), harpe with terminally smaller, rounded dorsal projection (Fig. 235–236), two (not three) subapical forewing spots, less irregular forewing hyaline discal band with spots more aligned with each other, and the spot in M3-CuA1 cell being minute, dot-like (Fig. 23–24). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly159.20.1:G156A, aly159.20.1:C183A, aly214.14.1:G229T, aly525.55.2:G403T, aly275215.9.3:A42C, however COI barcodes do not differentiate this species from A. neis.
Barcode sequence of the holotype.
Sample NVG-22032C11, GenBank OR837631, 658 base pairs:
AACTTTATATTTTATTTTTGGAATCTGAGCAGGATTAGTAGGTACTTCTTTAAGATTACTAATTCGAACTGAACTAGGAACACCTGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAACTGACTAGTACCCCTTATACTAGGAGCTCCAGATATAGCCTTCCCACGAATAAATAATATAAGATTCTGACTATTACCTCCATCATTAACTCTTTTAATTTCAAGTAGTATTGTAGAAAATGGAGCAGGAACTGGATGAACTGTTTATCCTCCCCTTTCTTCTAATATTGCTCATCAAGGAGCTTCAGTAGATTTAGCAATTTTTTCTCTTCATTTAGCAGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACAACTATTATTAATATACGAATTAATAATATATCTTTTGATCAAATACCTTTATTTGTATGAGCTGTAGGAATTACAGCTCTTCTTCTTTTACTTTCTTTACCAGTTCTAGCTGGAGCTATTACTATATTATTAACAGATCGAAATTTAAATACTTCATTCTTTGACCCAGCTGGGGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 23–24, bears the following three rectangular labels, two white: [ECUADOR Pastaza | Puyo-Napo Rd. | Km 25 – 1200m | 11 Nov. ‘88 | S.S. Nicolay], [DNA sample ID: | NVG-22032C11 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Autochton (Autochton) | dora Grishin]. Paratype: 1♀ NVG-19119C04, USNMENT_01602625 the same data as the holotype but collected on 10-Sep-1977.
Type locality.
Ecuador: Pastaza Province, km 25 of Puyo-Napo road, elevation 1200 m.
Etymology.
The name is for its distribution in [Ecua]dor + a and is a noun in apposition.
Distribution.
Known only from the type locality in Ecuador.
Astraptes centralis Grishin, new species https://zoobank.org/556FE76C-4D98-46DD-802A-D57449CFD0AD (Fig. 1 part, 25–26, 237–238)
Definition and diagnosis.
Phylogenetic trees reveal that Central American specimens identified as Astraptes aulus (Plötz, 1881) (type locality in Brazil, a syntype sequenced as NVG-21114H07) show prominent genetic differentiation from it (Fig. 1): e.g., their COI barcodes differ by 5% (33 bp), and therefore represent a new species. This new species keys to “Astraptes fulviluna” (C.14.14) in Evans (1952), which is currently a junior subjective synonym of A. aulus and differs from it by a straighter dorsoposterior margin of harpe (Fig. 238), less extensive green area on the dorsal forewing (Fig. 25) and not pupillated pale spot by inner margin on the ventral hindwing (Fig. 26). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1341.9.2:G264C, aly1019.14.10:G1446A, aly1497.4.1:T63C, aly887.6.4:C66G, aly594.10.7:G171A, and COI barcode: T35T, T169C, A217G, T292C, T508C.
Barcode sequence of the holotype.
Sample NVG-14105A07, GenBank OR837632, 658 base pairs:
AACTCTATATTTTATTTTTGGAATTTGAGCAGGATTAGTTGGAACTTCTTTAAGTCTTCTTATTCGAACTGAATTAGGAACACCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCCATTATAATTGGAGGATTTGGTAATTGACTAGTACCATTAATATTAGGGGCCCCTGATATAGCTTTCCCTCGAATAAATAACATAAGATTCTGATTATTACCCCCCTCTTTAACTCTCTTAATCTCAAGAAGTATCGTAGAAAATGGTGCTGGTACTGGTTGAACTGTCTATCCCCCACTTTCATCTAATATTGCCCACCAAGGAACTTCTGTTGATTTAGCAATTTTCTCCCTTCATCTTGCTGGAATTTCCTCCATTTTAGGAGCTATTAATTTTATTACAACTATTATTAATATACGAATCAATAATTTATCATTTGATCAAATACCATTATTTATCTGAGCAGTAGGAATTACTGCATTATTATTATTACTTTCTTTACCCGTACTAGCAGGAGCTATTACTATATTACTAACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCAGGTGGTGGGGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 25–26, bears the following three rectangular labels, two white: [PANAMÁ: Canal Zone | Gamboa | X.18.78 | Gordon B. Small], [DNA sample ID: | NVG-14105A07 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Astraptes | centralis Grishin]. Paratypes: 2♂♂ and 1♀ in USNM: 1♂ NVG-14105A06 the type locality, with the same data, additionally “Pipeline Road, NW of Gamboa 9° 07′N, 79° 41′W”; and Costa Rica, Area de Conservación Guanacaste, Guanacaste Prov., Sector Mundo Nuevo, Estacion La Perla, 325 m, GPS 10.76737, −85.43313: 1♂ NVG-17106D09 06-SRNP-60359 eclosed on 10-Jan-2007 and 1♀ NVG-17106D08 11-SRNP-56351 eclosed on 29-Aug-2011.
Type locality.
Panama: Colón Province, Gamboa.
Etymology.
The name is for the range of this species in Central America and is a masculine adjective.
Distribution.
Costa Rica and Panama.
Subtribe Loboclina Grishin, 2019
Aguna claxonica Grishin, new species https://zoobank.org/D5952070-7C94-498A-9066-62A9F3F52544 (Fig. 1 part, 27–28, 239–240)
Definition and diagnosis.
A specimen from Ecuador identified as Aguna claxon Evans, 1952 (type locality in Mexico: Veracruz) is genetically differentiated from it (COI barcode difference 4.9%, 32 bp) and represents a new species. This new species keys to Aguna claxon (C.5.3) in Evans (1952) and differs from it by harpe less extended dorsal, thus forming a wider gap from a longer and terminally narrower process of ampulla, harpe with a more convex dorsal margin forming into distal tooth (Fig. 240), and usually wider hyaline spot in forewing cell M3-CuA1 (Fig. 27). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1651.2.6:A45G, aly168.15.2:A94G, aly491.2.1:A394C, aly876.6.1:A81G, aly103.35.5:C187T, aly1405.20.9:A489A (not G), aly1405.20.9:C507C (not T), aly527.6.11:G81G (not T), aly4265.5.1:G63G (not A), aly2130.10.1:T75T (not C), and COI barcode: A22G, A316T, 361C, T385T, A550G.
Barcode sequence of the holotype.
Sample NVG-18016D12, GenBank OR837633, 658 base pairs:
AACTTTATATTTTATTTTTGGGATTTGAGCTGGTTTAGTTGGTACTTCTTTAAGATTACTTATTCGAACTGAATTAGGAACCCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGACTAGTACCCCTTATACTAGGAGCCCCAGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAACTCTTTTAATTTCTAGAAGTATTGTAGAAAATGGTGCAGGTACTGGATGAACTGTCTATCCCCCTCTTTCATCTAATATCGCCCACCAAGGAGCTTCTGTAGATTTAGCAATTTTCTCATTACATTTAGCAGGAATTTCTTCTATTCTTGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAATAATTTATCATTTGATCAAATATCATTATTTATTTGAGCTGTAGGAATTACAGCTTTATTATTATTACTTTCATTGCCTGTTTTAGCAGGAGCTATTACTATATTATTAACTGATCGTAATTTAAATACATCATTTTTTGACCCTGCAGGAGGGGGTGATCCTATTCTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 27–28, bears the following four rectangular labels, three white: [ECUADOR: Napo, | Misahuallí Jungle Lodge | 1° 01.54′ S, 77° 39.42′ W | 450 m, 6, 8 Jan 2002 | J.P.W. Hall & M.A. Solis], [DNA sample ID: | NVG-18016D12 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450788], and one red [HOLOTYPE ♂ | Aguna claxonica | Grishin].
Type locality.
Ecuador: Napo Province, Puerto Misahuallí, Misahuallí Amazon Lodge, elevation 450 m, GPS −1.0257, −77.6570.
Etymology.
The name is formed by adding the suffix -ica to its sister species name, making the name longer for this more southern species. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Ecuador.
Aguna esmeralda Grishin, new species https://zoobank.org/81C0476E-A9A8-4A6E-8EE9-F25C3C13130F (Fig. 1 part, 29–30, 241–242)
Definition and diagnosis.
A species related to Aguna glaphyrus (Mabille, 1888) (type locality in Brazil: Santa Catarina), Aguna longicauda Austin and O. Mielke, 1998 (type locality in Brazil: Rondônia), and Aguna spicata Austin and O. Mielke, 1998 (type locality in Brazil: Rondônia) (Fig. 1), but genetically differs from them more than they are from each other, e.g., COI barcode difference from A. glaphyrus is 5% (33 bp). Phenotypically can be identified by the contrasting tint of upperside green overscaling: yellower on the forewing and bluer on the hindwing (Fig. 29), short and stubby tails, nearly oval valva (when taken together with harpe), longer than in A. glaphyrus and A. spicata (among others), and separated from harpe only with a small notch, harpe rounded distad rather than narrowing, sacculus expanded distad. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly671.26.9:C88T, aly671.16.4:T181A, aly1468.8.8:A146G, aly1468.8.8:T156C, aly1340.1.1:C1566T, and COI barcode: A148G, A181T, TC208, T430G, T523C.
Barcode sequence of the holotype.
Sample NVG-18017A03, GenBank OR837634, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAATTGGAACTTCATTAAGATTACTTATTCGAACTGAATTAGGAACCCCCGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATGATTTTTTTTATAGTAATACCTATTATAATTGGTGGATTTGGAAATTGACTTGTACCTCTCATACTAGGAGCCCCTGATATAGCATTCCCCCGAATAAATAATATAAGATTTTGACTTTTACCCCCCTCCTTAACTCTTTTAATCTCTAGAAGCATTGTAGAAAATGGTGCAGGCACAGGATGAACAGTTTATCCCCCTCTTTCATCTAATATTGCACATCAGGGAGCTTCCGTAGATTTAGCAATTTTTTCTTTACATTTAGCAGGAATTTCCTCTATTCTGGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAATAATTTATCTTTCGATCAAATATCTTTATTCATTTGAGCAGTTGGTATCACTGCATTATTACTATTACTTTCTTTACCTGTTTTAGCAGGAGCTATTACAATATTATTAACAGATCGAAACTTAAACACTTCATTCTTTGATCCTGCAGGAGGAGGTGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 29–30, bears the following four rectangular labels, three white: [ECUADOR: Esmeraldas: | Río Chuchuví, km. 12.5 Lita- | San Lorenzo rd. 800–900m | 0° 53.01’ N 78° 30.90′ W | I.2001 I.Aldas leg.], [DNA sample ID: | NVG-18017A03 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450831], and one red [HOLOTYPE ♂ | Aguna esmeralda | Grishin]. Paratypes: 4♂♂ from Ecuador: Esmeraldas Province: 2♂♂ from the type locality: NVG-18017A04 USNMENT_01450832 the same data as the holotype, but collected in Mar-2001 [USNM]; and NVG-18066C08 Oct-2012, ex coll. M. Büche [EBrockmann]; 1♂ NVG-18016H04, USNMENT_01450823 Km 18.5 San Mateo-Pto. Libre Road, Zapatta Hilltop, 500 m, GPS 0.884500, −79.540333, 6-Mar-2001, D. H. Ahrenholz leg. [USNM]; 1♂ NVG-18065E11 Durango, 500–800 m, Mar-Jun-2011, ex coll. M. Büche [EBrockmann].
Type locality.
Ecuador: Esmeraldas Province, Río Chuchuví, km. 12.5 Lita-San Lorenzo Road, elevation 800– 900 m, GPS 0.883500, −78.515000.
Etymology.
The name is formed from the Esmeraldas Province, the type locality of this species. The name is a noun in apposition, originating from a Greek word meaning emerald, which also nicely reflects the emeraldgreen color on the dorsal hindwing of this species.
Distribution.
Currently known from northwestern Ecuador.
Aguna lata Grishin, new species https://zoobank.org/1D86CDBD-1D55-4A24-9705-84847DF9E86B (Fig. 1 part, 31–32, 243–245)
Definition and diagnosis.
Inspection of genomic trees reveals that a specimen identified as Aguna spatulata Austin and O. Mielke, 1998 (type locality in Brazil: Rondônia) is sister to the clade consisting of A. spatulata, Aguna panama Austin and O. Mielke, 1998 (type locality in Panama), and Aguna similis Austin and O. Mielke, 1998 (type locality in Brazil: Rondônia) and therefore is a distinct species. The new species differs from its closest relatives by a more angular distal margin of lamella antevaginalis, with slightly concave lobes on the sides of the notch (Fig. 243); these lobes appear truncate rather than rounded as in other species (Austin and Mielke 1998). Due to the cryptic nature of this species and unknown males, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly37338.40.1:A567G, aly1651.2.15:T844C, aly1651.2.15:T1098C, aly1146.14.5:T39A, aly318.14.14:T432C, aly806.11.5:G1380G (not A), aly383.14.5:C90C (not T), aly114.6.2:T960T (not C), aly770.33.9:A75A (not G), aly725.4.3:A123A (not G), but the COI barcode does not distinguish this species from A. spatulata.
Barcode sequence of the holotype.
Sample NVG-18016D11, GenBank OR837635, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGATTAGTAGGAACTTCTTTAAGATTATTAATTCGAACTGAATTAGGAACACCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGAAATTGACTAGTACCCCTTATACTAGGAGCTCCTGATATAGCATTCCCCCGAATAAATAATATAAGATTTTGACTTTTACCCCCTTCTTTAACCCTTTTAATTTCTAGTAGTATCGTAGAAAATGGTGCAGGCACAGGATGAACAGTTTACCCCCCACTTTCATCTAATATTGCCCATCAAGGAGCTTCAGTTGATTTAGCAATTTTCTCCTTACACTTAGCAGGAATTTCTTCTATTCTTGGAGCTATTAATTTTATCACCACAATTATTAACATACGAATTAATAATATATCATTTGATCAAATATCATTATTTATTTGAGCTGTTGGAATTACAGCTTTATTATTACTACTATCTTTACCAGTTTTAGCAGGAGCTATTACAATATTATTAACAGATCGAAATTTAAATACATCATTTTTTGATCCTGCTGGAGGGGGAGATCCTATTCTTTACCAACATTTATTC
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 31–32, bears the following four rectangular labels, three white: [GUYANA: Trop F Res | Middle Demerara R | 200–400′ | 31.I-12.II.2001| 5°9.32′N 58°41.98′W | Leg. S.Fratello et al], [DNA sample ID: | NVG-18016D11 | c/o Nick V. Grishin], [{QR Code} | USNM ENT 00234359], and one red [HOLOTYPE ♀ | Aguna | lata Grishin].
Type locality.
Guyana: Potaro-Siparuni Region, middle Demerara River, Tropenbos forest reserve, elevation 200–400′, GPS 5.1553, −58.6997.
Etymology.
The name is formed from its similar species, [spatu]lata, and is a noun in apposition.
Distribution.
Known only from the holotype collected in Guyana.
Ridens angulinea Grishin, new species https://zoobank.org/F16FA82A-8646-4B46-A2F2-7C7A8DDECC6B (Fig. 1 part, 33–34, 246–247)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Peru and Bolivia identified as Ridens fulminans (Herrich-Schäffer, 1869) (type locality not specified) are not monophyletic with it and are instead sister to several species of Ridens Evans, 1952 (Fig. 1), and therefore represent a new species. COI barcodes of this new species and a syntype of R. fulminans differ by 6.8% (45 bp). The new species keys to Ridens fulminans (C.12.7) in Evans (1952) and differs from R. fulminans and Ridens fulima Evans, 1952 (type locality in Brazil: Espírito Santo) in typically having narrower forewing hyaline spots (although some specimens have broader spots as well) and less prominent, mostly vestigial, pale discal band on the ventral hindwing (Fig. 33–34). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly116.28.11:A377G, aly116.28.11:A405G, aly2284.34.16:A61C, aly2284.34.16:A90G, aly2311.2.8:G123A, and COI barcode: T34C, T70C, A100C, T145C, T263C.
Barcode sequence of the holotype.
Sample NVG-14104E04, GenBank OR837636, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCTGGCTTAATTGGAACTTCCTTAAGATTACTTATTCGTACCGAATTAGGAATTCCCGGTTCTTTAATTGGCGATGACCAAATTTATAATACTATTGTAACAGCACATGCCTTTATCATAATCTTTTTTATAGTAATACCAATTATAATTGGTGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCCCCTGATATAGCATTCCCCCGAATAAATAATATAAGATTTTGACTATTACCCCCATCTCTTACTCTTTTAATTTCTAGAAGAATTGTAGAAAATGGTGCTGGAACAGGTTGAACAGTTTATCCCCCTCTTTCAACTAATATTGCCCATCAAGGGGCATCTGTCGACTTAGCCATTTTTTCTCTTCATTTAGCTGGAATTTCCTCAATTTTAGGAGCAATTAATTTTATTACAACAATTATTAATATGCGAATTAATAATTTATCATTTGATCAAATACCATTATTTATTTGAGCCGTAGGAATTACAGCATTATTATTATTACTTTCTTTACCTGTATTGGCAGGAGCTATTACCATATTACTTACCGATCGAAACTTAAATACTTCATTTTTTGATCCTGCAGGAGGTGGAGATCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 33–34, bears the following three rectangular labels, two white: [PERU:Cuzco 1,194m | Bridge@1194 m | Cosnipata Road 3167 | 29.I.2013 Kinyon], [DNA sample ID: | NVG-14104E04 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Ridens angulinea | Grishin]. Paratypes: 5♂♂: 1♂ NVG-17103B12 USNM_00913749 Peru: Cuzco, Cosnipata Valley, Quebrada Santa Isabel, 24-Oct-2016, S. Kinyon leg. [USNM]; 1♂ NVG-18089A11 Bolivia: Yungas Region, Caranavi Province, 50 km N of Coroico, 700–1200 m, Nov-1988, Carlos Tello leg. [EBrockmann]; 3♂♂ Bolivia: Rio Songo: NVG-19041B10 AMNH_IZC 00337748 750 m, coll. Fassl, genitalia slide G770 [AMNH]; and R. C. Williams, Jr. collection [ANSP]: NVG-22042E10 and NVG-22042E11 genitalia slide No. 316.
Type locality.
Peru: Cuzco, Cosñipata Road, bridge at 1194 m.
Etymology.
The name reflects narrower white bands and spots in many specimens of this species. It is derived from the Latin phrase “narrow lines”: angu[stis]line[is]+a. The name is a noun in apposition.
Distribution.
Southern Peru and Bolivia.
Subfamily Pyrginae Burmeister, 1878 Tribe Achlyodini Burmeister, 1878
Pythonides lera Grishin, new species https://zoobank.org/CE3F7AF8-AAA1-4496-8044-F42391762519 (Fig. 2 part, 35–36, 248–249)
Figure 2.
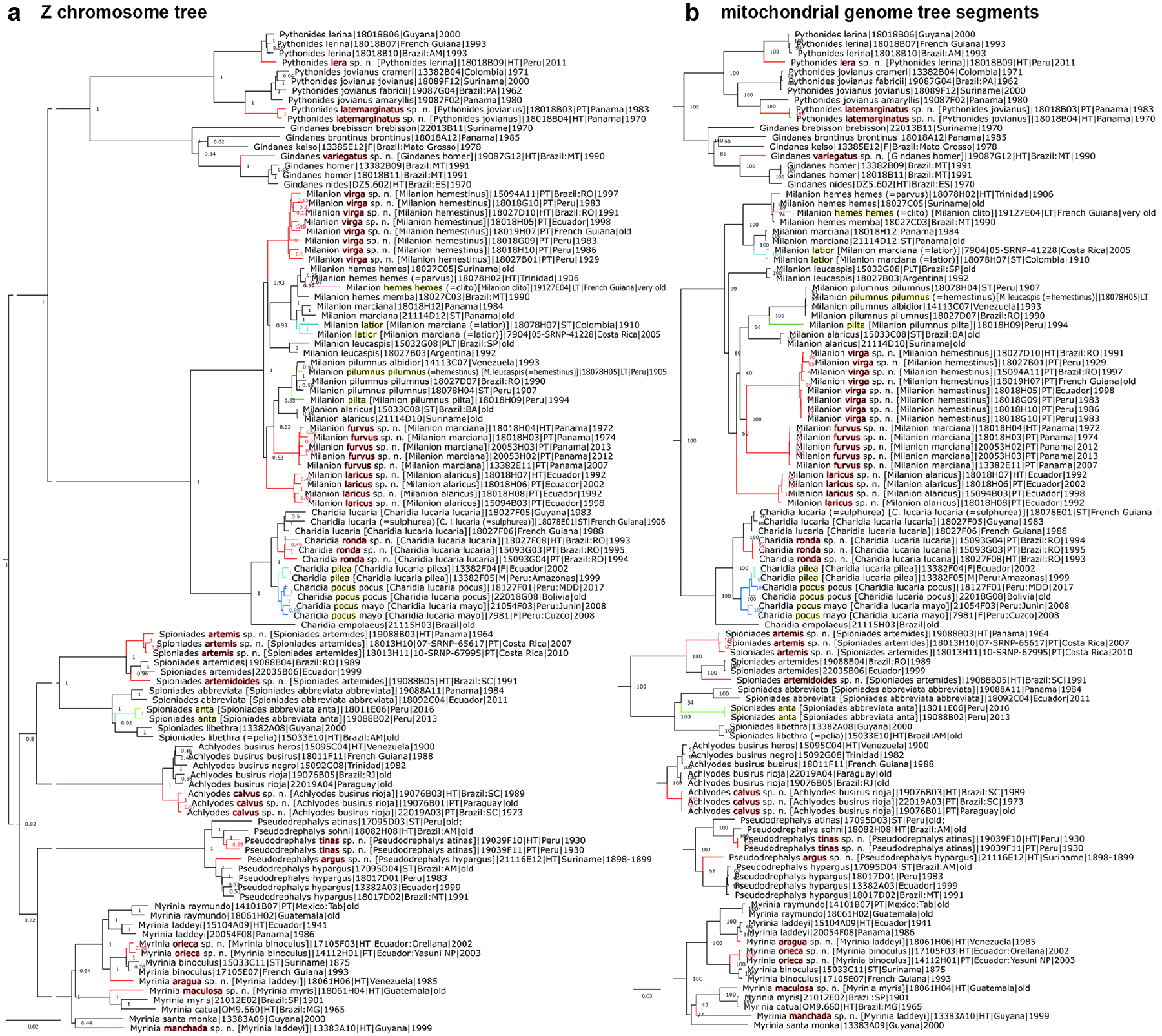
Phylogenetic trees of Achlyodini inferred from protein-coding regions in a) the Z chromosome and b) the mitochondrial genome (in segments). See Fig. 1 legend for other notations.
Definition and diagnosis.
A specimen from Peru identified as Pythonides lerina (Hewitson, 1868) (type locality in French Guiana and Brazil: Para) is genetically differentiated from it (COI barcodes differ by 5% (33 bp)) (Fig. 2) and therefore is a new species. This new species keys to P. lerina (E.41.4) in Evans (1953) and differs from it by a more elongated lower hyaline spot in the forewing discal cell, less developed network of brown “webbing” on the ventral hindwing (Fig. 35–36), and more robust harpe that protrudes dorsad of costa (249). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly393.15.7:G381C, aly393.15.7:A395T, aly531.17.6:C195T, aly82.4.5:A198T, aly173.28.2:A84G, aly345.5.1:C36C (not T), aly2116.6.1:C247C (not T), aly221.16.10:A57A (not T), aly3512.5.3:T141T (not C), aly2012.21.2:C863C (not G), and COI barcode: T79C, T133T, A190T, T280C, T536C.
Barcode sequence of the holotype.
Sample NVG-18018B09, GenBank OR837637, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCTGGTATAGTTGGAACATCTTTAAGTTTATTAATTCGAACTGAACTAGGCAACCCTGGTTCTTTAATTGGAGATGATCAAATTTATAATACCATTGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGTAATTGACTAGTACCCCTTATACTAGGAGCTCCTGATATAGCCTTTCCTCGAATAAATAATATAAGTTTTTGACTATTACCTCCTTCTCTCACATTATTAATTTCAAGAAGTATTGTAGAAAATGGAACTGGAACTGGATGAACAGTTTACCCCCCTCTTTCTTCTAATATTGCTCATCAAGGTTCTTCCGTAGATTTAGCTATTTTTTCCCTGCATTTAGCTGGAATTTCATCAATTTTAGGTGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAACCTTTCTTTTGATCAAATACCTTTATTCGTTTGAGCAGTAGGAATTACAGCATTACTTCTACTATTATCATTACCTGTTTTAGCAGGAGCTATTACTATACTATTAACTGATCGAAATTTAAATACATCATTTTTTGACCCTGCTGGAGGAGGAGACCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 35–36, bears the following four rectangular labels, three white: [PERU:Cuzco 1050m | Quitacalzone | Cosnipata Road 1530 | 5.ii.2011 Kinyon], [DNA sample ID: | NVG-18018B09 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450938], and one red [HOLOTYPE ♂ | Pythonides | lera Grishin].
Type locality.
Peru: Cuzco, Cosñipata Road, Quitacalzone, elevation 1050 m.
Etymology.
The name is formed from its sister species name ler[in]a and is a feminine noun in apposition.
Distribution.
Currently known only from the holotype collected in southern Peru.
Pythonides latemarginatus Grishin, new species https://zoobank.org/7434EACB-635C-435F-A365-2932F2749590 (Fig. 2 part, 37–38, 250–251)
Definition and diagnosis.
Specimens from Panama identified as Pythonides jovianus amaryllis Staudinger, 1876 (type locality in Panama and Colombia) are sister to all Pythonides jovianus (Stoll, 1782) and are genetically differentiated from it (Fig. 2): e.g., their COI barcodes differ by 3.5% (24 bp) from P. j. amaryllis, and therefore they represent a new species. This new species keys to P. j. amaryllis (E.41.3(a)) in Evans (1953) and differs from it by a broader brown marginal area on the hindwing and correspondingly smaller blue patch, but more extensive forewing postdiscal blue spotting, in particular having a blue streak by the inner margin and two prominent blue streaks near forewing tornus beneath (Fig. 37–38). This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly2178.10.1:C78T, aly2178.10.1:C96T, aly363.24.5:G81A, aly522.12.2:G39A, aly9588.7.2:A27G, and COI barcode: T49A, T268C, T385C, T457C, T499C.
Barcode sequence of the holotype.
Sample NVG-18018B04, GenBank OR837638, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCTGGAATAGTAGGAACATCATTAAGTTTATTAATTCGAACTGAATTAGGTAATCCCGGTTCATTAATTGGAGATGATCAAATTTATAATACTATTGTAACTGCTCATGCTTTTATTATAATTTTCTTTATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGACTAGTACCCCTTATACTAGGAGCTCCTGATATAGCATTCCCTCGAATAAATAATATAAGTTTTTGATTACTCCCCCCTTCTCTTACATTATTAATTTCAAGAAGTATTGTTGAAAATGGAACTGGAACAGGATGAACAGTATACCCCCCTCTTTCCTCTAATATTGCTCATCAAGGATCTTCTGTTGACTTAGCTATTTTTTCCTTACACCTAGCTGGTATTTCATCAATTTTAGGAGCTATTAATTTTATTACTACAATCATTAATATACGAATTAGAAATTTATCATTTGATCAAATACCCTTATTTGTTTGAGCTGTAGGAATTACAGCATTATTATTATTATTATCATTACCTGTTTTAGCCGGAGCTATTACCATATTATTAACTGACCGAAATTTAAATACATCATTTTTTGATCCAGCTGGTGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 37–38, bears the following four rectangular labels, three white: [PANAMA: Panama | Cerro Campana 2500′ | 8°41′N 79°55′W | August 1970 | leg. G.B.Small], [DNA sample ID: | NVG-18018B04 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450934], and one red [HOLOTYPE ♂ | Pythonides | latemarginatus Grishin]. Paratype: 1♂ NVG-18018B03 USNMENT_01450933 Panama: Darien, Cana, 750 m, 15-Jun-1983, G. B. Small leg. [USNM].
Type locality.
Panama: Panama Province, Cerro Campana, elevation 2500 ft, GPS 8.6833, −79.9167.
Etymology.
The name is Latin for the broad (dark) margin and is a masculine adjective.
Distribution.
Panama.
Gindanes variegatus Grishin, new species https://zoobank.org/1855A4ED-6F31-4ED4-9416-C43CDC581D57 (Fig. 2 part, 39–40, 252–253)
Definition and diagnosis.
A specimen from Mato Grosso, Brazil, identified (incorrectly) as Gindanes homer (Evans, 1953) (type locality in Brazil: Mato Grosso) is genetically differentiated from it (Fig. 2): e.g., COI barcodes differ by 4.9% (32 bp). The new species keys (incompletely) to “Pythonides homer” (E.41.10) or “Pythonides herennius herennius” (E.41.7a) in Evans (1953) and differs from them and Gindanes nides (Mielke and Casagrande, 2002) (type locality in Brazil: Espírito Santo) by a more variegated appearance, larger forewing hyaline spots, conjoined forewing cell spots, and more extensive whitish coloration of ventral hindwing that includes its anterior part: a prominent central brown spot near costa is framed by white on both sides. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly26.1.2:C18T, aly4305.19.19:G123A, aly4305.19.19:G120T, aly264.4.18:C93T, aly253.13.3:G99A, aly276558.27.2:C121C (not A), aly276558.27.2:C144C (not T), aly1158.11.3:G702G (not A), aly1281.15.3:C82C (not G), aly6954.10.6:C1206C (not T), and COI barcode: A22G, T121T, C205C, C343G, T442C.
Barcode sequence of the holotype.
Sample NVG-19087G12, GenBank OR837639, 658 base pairs:
AACTTTATATTTTATTTTTGGGATTTGGGCAGGAATAATTGGAACATCTTTAAGCCTACTAATTCGAACTGAATTAGGTAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTACCCTTAATATTAGGAGCTCCAGATATAGCTTTCCCCCGAATAAACAATATAAGATTTTGATTATTACCTCCATCTTTATTATTATTAATTTCAAGAAGTATTGTTGAAAATGGAGCAGGAACAGGATGAACTGTTTATCCGCCTCTTTCTGCTAATATTGCTCATCAAGGTTCATCTGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCCTCAATTTTAGGAGCTATTAACTTTATTACTACAATTATTAATATACGAATTAATAATCTTTCATTTGATCAAATACCTTTATTTATTTGAGCAGTGGGAATTACAGCATTACTTTTATTATTATCTCTTCCTGTTTTAGCAGGTGCTATTACTATATTATTAACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCAGGTGGGGGTGATCCAATCTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 39–40, bears the following four rectangular labels, three white: [BRASIL: Mato Grosso | Diamantino, 350–400m | Alto Rio Arinos | 14°13′S 56°12′W | 18 August 1990 | Leg. E. Furtado], [DNA sample ID: | NVG-19087G12 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01588890], and one red [HOLOTYPE ♂ | Gindanes | variegatus Grishin].
Type locality.
Brazil: Mato Grosso, Diamantino, Alto Rio Arinos, elevation 350–400 m, GPS −14.217, −56.200.
Etymology.
The name is given for the variegated appearance of this species. The name is a masculine perfect passive participle.
Distribution.
Known only from the holotype collected in Brazil: Mato Grosso.
Papilio clito Fabricius, 1787 is a junior subjective synonym of Milanion hemes hemes (Cramer, 1777)
Genomic sequencing of the lectotype of Papilio clito Fabricius, 1787 (type locality in French Guiana), currently a valid species in the genus Milanion Godman and Salvin, 1895 (type species Papilio hemes Cramer, 1777), reveals that it is conspecific with specimens we identified as Milanion hemes hemes (Cramer, 1777) (type locality in Suriname), including a specimen from Suriname (Fig. 2). Therefore, we propose that Papilio clito Fabricius, 1787 is a new junior subjective synonym of Milanion hemes hemes (Cramer, 1777).
Milanion pilta Evans, 1953 is a species distinct from Milanion pilumnus Mabille and Boullet, 1917
Proposed by Evans (1953) as a subspecies of Milanion pilumnus Mabille and Boullet, 1917 (type locality in Bolivia and Peru) from Peru: Moyabamba, M. p. pilta is genetically differentiated from it at the level typical for distinct species: e.g., their COI barcodes differ by 6.7% (44 bp) (Fig. 2). Therefore, we propose that it is a species-level taxon Milanion pilta Evans, 1953, new status.
Milanion latior Mabille and Boullet, 1917 is a species distinct from Milanion marciana Godman and Salvin, 1895
Sequencing of a syntype of Milanion marciana var. latior Mabille and Boullet, 1917 (type locality in Colombia), a taxon currently treated as a junior subjective synonym of Milanion marciana Godman and Salvin, 1895 (type locality in Panama), reveals genetic differentiation between them (Fig. 2): e.g., COI barcodes of syntypes of these taxa differ by 3% (20 bp). Therefore, we propose to treat it as a species-level taxon Milanion latior Mabille and Boullet, 1917, new status.
Milanion pilumnus var. hemestinus Mabille and Boullet, 1917 is a junior subjective synonym of Milanion pilumnus pilumnus Mabille and Boullet, 1917 not of Milanion leucaspis (Mabille, 1878)
Genomic sequencing of the lectotype of Milanion pilumnus var. hemestinus Mabille and Boullet, 1917 (type locality in Peru) reveals that it is in a clade different from Milanion leucaspis (Mabille, 1878) (type locality in Brazil), which is its current senior subjective synonym but is placed among specimens of Milanion pilumnus Mabille and Boullet, 1917 (type locality in Bolivia and Peru). COI barcodes of the M. p. hemestinus lectotype and a syntype of M. pilumnus pilumnus differ by 0.5% (3 bp). Therefore, as proposed by the original authors, M. p. hemestinus is conspecific with M. pilumnus, and we treat these two names as synonymous.
Milanion (Milanion) virga Grishin, new species https://zoobank.org/2C440233-C0FA-49D4-9496-A451B5F57764 (Fig. 2 part, 41–42, 254–255)
Definition and diagnosis.
As detailed above, Papilio clito Fabricius, 1787 and Milanion pilumnus var. hemestinus Mabille and Boullet, 1917 are junior subjective synonyms of Milanion hemes hemes (Cramer, 1777) and Milanion pilumnus Mabille and Boullet, 1917, respectively. As a result of this, and because no other available name applies to it, the species that Evans (1953) called “Milanion hemestinus” was misidentified and is new. This new species keys to (E.46.3) in Evans (1953). It differs from its relatives by a combination of the following characters: a white spot in forewing cell CuA2-1A+2A, a dorsal hindwing white band about the same width as the brown marginal area, basal half of the ventral hindwing is white with a dark ray near costa from the base to costal margin (i.e., there is an elongated white spot near the base by costa); both gnathos and uncus arms are equally wide apart, tips of uncus not turned outwards; harpe broad, terminally rounded and with a spine in the middle at the dorsal margin. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly536.107.1:A81G, aly536.107.1:C93T, aly770.13.3:T153C, aly1937.10.4:C88T, aly1937.10.4:C112A, and COI barcode: T19C, T172C, A499C, A541C, T613C.
Barcode sequence of the holotype.
Sample NVG-18027D10, GenBank OR837640, 658 base pairs:
AACTTTATATTTTATTTTCGGAATTTGAGCAGGATTAGTAGGAACTTCTTTAAGTTTATTAATTCGAACTGAATTAGGAAATCCAGGATCTTTAATTGGAGATGACCAAATTTATAATACTATTGTAACTGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCAATCATAATTGGTGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCCCCAGATATAGCTTTCCCTCGAATAAATAACATAAGATTTTGATTATTACCCCCATCTCTTATATTATTAATTTCAAGAAGTATTGTAGAAAATGGTGCAGGAACTGGATGAACAGTTTATCCACCTCTTTCAGCTAACATCGCTCATCAAGGTTCATCAGTAGATCTAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACTACTATTATTAATATACGAGTAAATAATTTATCATTTGATCAAATACCCTTATTTGTGTGAGCTGTAGGTATTACAGCTTTACTTTTACTCCTATCATTACCCGTATTAGCAGGTGCTATTACTATATTATTAACTGATCGAAATTTAAATACATCATTTTTCGACCCAGCAGGAGGAGGAGATCCAATCTTATATCAACATCTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 41–42, bears the following five rectangular labels, four white: [BRASIL:Rondonia | 62 km S Ariquemes | Fazenda Rancho | Grande, 165m | 10–32′S, 62–48′W | 29 Oct-10 Nov 1991 | Brian P. Harris], [Milanion | hemes | hemes], [DNA sample ID: | NVG-18027D10 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01465169], and one red [HOLOTYPE ♂ | Milanion | virga Grishin]. Paratypes: 5♂♂ and 2♀♀: 1♂ NVG-15094A11 the type locality, 27-Oct-1997, G. T. Austin leg. [MGCL]; 1♂ NVG-18018H05 USNMENT_01450997 Ecuador: Orellana, Yasuni Research Station, Rios Tivacuno and Tiputini, elevation 220 m, GPS −0.675000, −76.396667, 30-Oct-1998, D. H. Ahrenholz leg., genitalia NVG-22032E07 [USNM]; 1♂ NVG-18018H10 USNMENT_01465137 Peru: Madre de Dios, Tambopata Res., Rio La Torre, elevation 300 m, 6-Oct-1986, S. S. Nicolay leg., genitalia NVG-22032E02 [USNM]; 1♂ NVG-18027B01 Peru: Middle Rio Ucayali, 6-Apr-1929, H. Bassler Collection [AMNH]; 1♂ NVG-18018G09, USNMENT_01450989 Peru: 30 km SW of Pto. Maldonado, 300 m, 20-Oct-1983, S. S Nicolay leg. [USNM]; 1♀ NVG-18018G10, USNMENT_01450990 Peru: Pto. Aldonado, 290 m, 15-Oct-1983, S. S Nicolay, leg. [USNM]; 1♀ NVG-18019H07 French Guiana [AMNH].
Type locality.
Brazil: Rondônia, 62 km S Ariquemes, Fazenda Rancho Grande, elevation 165m, GPS −10.533, −62.800.
Etymology.
The name is given for the pale streak at the base by the costa of the ventral hindwing characteristic of this species. In Latin, virga means rod or streak. The name is a noun in apposition.
Distribution.
Widely distributed in tropical South America: Ecuador, Peru, French Guiana, and Brazil.
Milanion (Milanion) furvus Grishin, new species https://zoobank.org/371AD2A6-502E-44AF-B185-91A95AC388CE (Fig. 2 part, 43–44, 256–257)
Definition and diagnosis.
Several specimens from Panama identified as Milanion marciana Godman and Salvin, 1895 (type locality in Panama) are in a clade different from M. marciana and Milanion latior Mabille and Boullet, 1917 (type locality in Colombia) and are genetically differentiated from all other species in the genus (Fig. 2). Therefore, they represent a new species. This new species does not key correctly to any species, including M. marciana (E.46.5), in Evans (1953) and differs from its relatives by a combination of the following characters: white spot in forewing cell CuA2-1A+2A is small, located by the vein 1A+2A and does not reach the middle of the cell, white patch on hindwing is broader than brown border, ventral hindwing with wide brown area by costa, cell Sc+R1-RS brown with a (usually) small white spot in the middle joined with the white central area with veins darker by the dark border. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly2027.2.5:C51T, aly971.9.9:G51T, aly2811.4.1:G248A, aly2811.4.1:C249T, aly1475.2.1:G48A, and COI barcode: T133G, T157C, T358C, T514C, A607C.
Barcode sequence of the holotype.
Sample NVG-18018H04, GenBank OR837641, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAGTAGGAACCTCTTTAAGTTTATTAATTCGAACTGAATTAGGAAACCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACTGCGCATGCATTTATTATAATTTTTTTCATGGTCATACCAATTATAATTGGTGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCCCCAGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGATTATTACCCCCATCTCTTATATTATTAATTTCAAGAAGTATTGTAGAAAATGGAGCAGGAACTGGATGAACAGTTTACCCACCTCTTTCAGCTAACATTGCCCATCAAGGCTCATCAGTAGATTTAGCAATTTTTTCTTTACATTTAGCTGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACTACTATTATTAACATACGAGTAAATAATTTATCATTTGATCAAATACCTTTATTTGTATGAGCCGTAGGTATTACAGCTTTACTTTTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACTATATTATTAACTGACCGAAATTTAAACACATCCTTCTTTGACCCAGCAGGAGGAGGAGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 43–44, bears the following four rectangular labels, three white: [PANAMA: Panama | Farfan (C.Z.) | 8° 56′N, 79° 34′W | 6 Jan ‘72 | leg. S. S. Nicolay], [DNA sample ID: | NVG-18018H04 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450996], and one red [HOLOTYPE ♂ | Milanion | furvus Grishin]. Paratypes: 4♂♂ from Panama: Panama Province: 1♂ 11-BOA-13382E11 Panama City, Albrook Hotel and Area, 1–4-Aug-2007, R. H. Leuschner leg. [USNM]; 1♂ NVG-18018H03 Bayano, 23-Nov-1974, G. B. Small leg. [USNM]; 1♂ NVG-20053H02 Gamboa, Pipeline Road, 75 m, 9.127167, −79.714833, 2-Jun-2012, John R. MacDonald leg. [MEM]; 1♂ NVG-20053H03 Veracruz, ca. 50 m, GPS 8.911833, −79.565722, 2-Jul-2013, John R. MacDonald leg. [MEM].
Type locality.
Panama: Panama Province, Farfan, GPS 8.933, −79.567.
Etymology.
In Latin, furvus means dark, dusky, gloomy, or swarthy. The name, a masculine adjective, is given partly for the smaller white area and very small white spot in cell 7 on the ventral hindwing.
Distribution.
Currently known only from central Panama.
Milanion (Milanion) laricus Grishin, new species https://zoobank.org/5BD7C016-6141-4D71-BAAB-DD8F18887811 (Fig. 2 part, 45–46, 258–259)
Definition and diagnosis.
Several specimens from Ecuador identified as Milanion alaricus (Plötz, 1884) (type locality in Brazil: Bahia) were not closely related to the syntype of M. alaricus (Fig. 2) and formed a clade distinct from all other species of the genus. Therefore, they represent a new species. This new species keys (incompletely) to M. alaricus (E.46.6) in Evans (1953) and differs from its relatives by a vestigial (or lacking) white spot in forewing cell CuA2-1A+2A, spot at the base of cell M3-CuA1 is larger, more triangular than rounded (as in M. alaricus) with its distal part protruding distad of the spot in the cell CuA1-CuA2, ventral hindwing area by costa in basal two-thirds is not white, but is pale-brown, not the same dark color as the brown marginal wide border. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1313.22.1:G201A, aly923.23.1:T1497C, aly15192.3.7:G100A, aly1405.11.14:C72T, aly2095.5.1:C135T, and COI barcode: T133A, T301C, A577G, A592G, A628T.
Barcode sequence of the holotype.
Sample NVG-18018H07, GenBank OR837642, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAGTAGGAACTTCTTTAAGTTTATTAATTCGAACTGAATTAGGAAACCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACTGCACATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCCCCAGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTACCCCCATCTCTTATATTATTAATTTCAAGAAGCATTGTGGAAAATGGTGCAGGAACTGGATGAACAGTTTATCCCCCCCTTTCAGCCAATATTGCCCACCAAGGTTCATCAGTAGATTTAGCAATCTTTTCTTTACATTTAGCTGGAATTTCTTCAATTCTAGGAGCTATTAATTTTATCACCACTATTATTAACATACGAGTAAATAACTTATCATTTGATCAAATACCATTATTTGTATGAGCAGTAGGAATTACTGCACTACTTTTATTGTTATCTTTACCTGTATTAGCGGGTGCTATTACTATGTTATTAACTGATCGGAATTTAAATACATCATTTTTTGACCCAGCAGGAGGTGGAGATCCAATTTTATATCAACATCTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 45–46, bears the following four rectangular labels, three white: [ECUADOR: Napo Pr. | Jatun Sacha Biol St. | 1° 4.0′S 77° 37.0′W | 13 Nov. 1992. 450m. | S. S. Nicolay, leg.], [DNA sample ID: | NVG-18018H07 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450999], and one red [HOLOTYPE ♂ | Milanion | laricus Grishin]. Paratypes: 3♂♂ from Ecuador: 1♂ NVG-18018H08, USNMENT_01465135 the same data as the holotype but collected on 7-Nov-1992; 1♂ NVG-15094B03, Napo, Yasuni Research Station, Rios Tivacuno and Tiputini, 250 m, GPS −0.633, −76.600, 22-Oct-1998, J. D. Turner leg. [MGCL]; 1♂ NVG-18018H06, USNMENT_01450998 Sucumbios, Cerro Lumbaqui Norte, 800–950 m, GPS 0.028333, −77.320333, 18–22-Aug-2002, J. P. W. Hall and M. A. Solis leg. [USNM].
Type locality.
Ecuador: Napo Province, Jatun Sacha Biological Reserve, elevation 450 m, GPS −1.0667, −77.6167.
Etymology.
The name alaricus is derived from Alarīks (king of all) with the prefix “alla-” (all, everybody, entire) suggests a kingly, royal, or noble identity. Laricus (i.e., not alaricus) is a noun in apposition and a shorter name for this more northern species.
Distribution.
Known only from northeastern Ecuador.
Charidia pilea Evans, 1953, and Charidia pocus Evans, 1953 (with Charidia lucaria mayo Evans, 1953 as its subspecies) are species distinct from Charidia lucaria (Hewitson, 1868)
Genomic sequencing of taxa currently treated as subspecies of Charidia lucaria (Hewitson, 1868) (type locality in French Guiana) reveals prominent genetic differentiation between some of them (Fig. 2): e.g., COI barcodes of C. lucaria lucaria differ from Charidia lucaria pilea Evans, 1953 (type locality in Colombia) and Charidia lucaria pocus Evans, 1953 (type locality in Peru) by 2.3% (15 bp) and 2.3% (15 bp), respectively. However, despite an obvious phenotypic difference in males having ventral hindwing largely white, Charidia lucaria mayo Evans, 1953 is not strongly differentiated genetically from C. l. pocus. However, we find an irregularity in the mitochondrial genome: C. l. pocus is not distinguishable from C. l. pilea, probably due to introgression from it, but C. l. mayo might have kept its original mitochondrial DNA differing by 1.2% (8 bp) in the COI barcode. Due to genetic and phenotypic differences, we propose a new status of a species for Charidia pilea Evans, 1953 and Charidia pocus Evans, 1953, and a new combination Charidia pocus mayo Evans, 1953 (type locality in Peru). Moreover, we found another species-level taxon in this complex that is described next.
Charidia ronda Grishin, new species https://zoobank.org/B46214EB-61ED-4174-8AAA-AF92820DF9BF (Fig. 2 part, 47–48, 260–261)
Definition and diagnosis.
Sister to Charidia lucaria (Hewitson, 1868) (type locality in French Guiana) and keys to it (E.48.1(b)) in Evans (1953) but is genetically differentiated from it in nuclear genome at the level more typical for species-level taxa (Fig. 2), with the COI barcode distance of 1.5% (10 bp). Differs from its relatives by the following combination of characters: males with more extensive orange-yellow scaling on ventral side, e.g., forewing tornal spots larger, hindwing overscaling more prominent towards tornus (Fig. 48) (yellow overscaling is equally extensive at the end of the discal cell and the hindwing base in more boldly patterned C. lucaria specimens), and costa-ampulla strongly humped (Fig 261); females with wide yellowish area on ventral hindwing with broadly brown costal margin and somewhat paler than the brown ground color subapical band on ventral forewing. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly283.5.1:T359C, aly283.5.1:G429A, aly502.6.3:C325T, aly1937.18.2:C52A, aly2202.3.1:A55T, and COI barcode: A73G, T421C, T460C, C529A, A550G.
Barcode sequence of the holotype.
Sample NVG-18027F08, GenBank OR837643, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACTTCTTTAAGTTTATTAATTCGAACTGAGCTAGGAAATCCAGGATCTTTAATTGGAAATGATCAAATTTATAATACTATTGTTACTGCTCATGCATTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGACTAGTACCCCTTATACTAGGAAGTCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTATTACCCCCATCTCTTATATTATTAATTTCAAGAAGTATTGTAGAAAATGGAGCAGGAACTGGATGAACAGTTTATCCCCCTCTTTCTGCTAATATTGCTCATCAAGGTTCATCTGTAGACTTAGCAATTTTTTCATTACATTTAGCTGGAATTTCCTCAATTTTAGGAGCTATTAATTTTATTACCACTATTATCAATATACGAGTAAATAATCTATCATTTGATCAAATACCATTATTTGTATGAGCTGTAGGAATTACAGCATTACTTTTATTACTATCATTGCCTGTATTAGCAGGAGCTATTACTATATTATTAACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 47–48, bears the following five rectangular labels, four white: [BRASIL: Rondonia | 6km S Cacaulandia | Rio Pardo | 21 November 1993 | Brian Harris], [Charidia | lucaria], [DNA sample ID: | NVG-18027F08 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01465190], and one red [HOLOTYPE ♂ | Charidia | ronda Grishin]. Paratypes:1♂ and 1♀ from Brazil: Rondonia [MGCL]: 1♂ NVG-15093G03 Cacaulandia, Fazenda Rancho Grande, 12-Nov-1995, D. and J. Lindsley leg.; 1♀ NVG-15093G04 5 km S of Cacaulandia, linea C-10 at Rio Pardo off B-65, 7-Jul-1994, O. Gomes leg.
Type locality.
Brazil: Rondônia, 6 km S of Cacaulandia, Rio Pardo.
Etymology.
The name is given for the type locality in Rond[oni]a and is a noun in apposition.
Distribution.
Currently known only from the area around the type locality in Rondônia, Brazil.
Pseudodrephalys tinas Grishin, new species https://zoobank.org/37160735-2E27-4AE4-9B65-DBD0DD58B076 (Fig. 2 part, 49–52, 262–263)
Definition and diagnosis.
Genomic sequencing reveals that two specimens from Iquitos, Peru, identified as Pseudodrephalys atinas (Mabille, 1888) (type locality in Peru) are sister to Pseudodrephalys sohni Burns, 1999 (type locality in Brazil: Amazonas) with P. atinas being a more distant species (Fig. 2): COI barcodes differ by 4.7% (31 bp) from P. atinas syntype and 1.8% (12 bp) from P. sohni holotype. Due to genetic differentiation, the two Peruvian specimens represent a new species, which is distinguished from its relatives by a narrower valva with a larger cleft between the harpe and ampulla than in P. sohni and a much narrower harpe with a smaller cleft separating it from ampulla compared with P. atinas. In facies, males differ from P. sohni by the ventral hindwing white band being narrower and interrupted in the middle of the cell CuA2-1A+2A, thus leaving an isolated spot near the anal fold; from P. atinas by this band reaching Sc+R1 (instead of ending at Rs); and from both P. sohni and P. atinas by the postdiscal pale lavender line on ventral hindwing being vestigial. Females differ from P. atinas by a strongly concave border of the basal yellow-orange basal area of ventral hindwing and from P. sohni by not being similar to males in wing pattern (e.g., having a round ventral hindwing white spot instead of a band). Due to the somewhat cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly671.6.3:T102C, aly6954.5.63:C66T, aly207.8.7:A150G, aly4456.10.1:A194G, aly1709.2.6:C87G, and COI barcode: A28G, A100T, T361C, T535C, 616C.
Barcode sequence of the holotype.
Sample NVG-19039F10, GenBank OR837644, 658 base pairs:
AACTTTATACTTTATTTTTGGAATTTGGGCAGGAATAGTAGGTACTTCTTTAAGATTATTAATTCGTACTGAATTAGGAAATCCAGGATCTTTAATTGGTGATGATCAAATTTATAATACTATTGTTACAGCTCACGCTTTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGAGGATTTGGAAATTGATTAGTACCTCTGATACTAGGAGCTCCTGATATAGCATTCCCACGAATAAATAATATAAGATTTTGACTACTTCCCCCCTCTTTATTGTTATTAATTTCAAGAAGTATTGTAGAAAATGGTGCTGGAACTGGATGAACTGTATATCCTCCTCTTTCTTCTAATATCGCCCATCAAGGAGCATCAGTAGATTTAGCAATTTTTTCATTACATTTAGCAGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAATAATCTTTCCTTTGATCAATTACCCTTATTCGTATGAGCTGTTGGTATCACAGCTTTACTCTTATTACTCTCTTTACCAGTATTAGCTGGAGCTATTACTATACTATTAACTGATCGAAATTTAAATACATCTTTTTTTGACCCTGCGGGAGGAGGAGATCCAATCTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the American Museum of Natural History, New York, NY, USA (AMNH), illustrated in Fig. 49–50, bears the following six rectangular labels, five white: [Iquitos, Peru | XII-15 1930], [G646], [DNA sample ID: | NVG-19039F10 | c/o Nick V. Grishin], [Leg removed on | 20-Feb-2019 by | N. V Grishin for | DNA extraction], [{QR Code} | AMNH_IZC 00337700] and one red [HOLOTYPE ♂ | Pseudodrephalys | tinas Grishin]. Paratype: 1♀: NVG-19039F11, AMNH_IZC 00337701, the same data as the holotype (Fig. 51–52).
Type locality.
Peru: Loreto Region, Iquitos.
Etymology.
The name is formed by removing the negating prefix “a” from atinas. In Latin, tinus means small or tiny. This species has a smaller ampulla. The name is a noun in apposition.
Distribution.
Currently known only from Northern Peru.
Pseudodrephalys argus Grishin, new species https://zoobank.org/D90B1BBA-F8B9-4EC3-8188-EC8E8667C4C1 (Fig. 2 part, 53–54)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Suriname identified as Pseudodrephalys hypargus (Mabille, 1891) (type locality in Brazil: Amazonas) is genetically differentiated from a series of P. hypargus that included a syntype (Fig. 2), e.g., their COI barcodes differ by 2.1% (14 bp), and therefore represents a new species. This new species keys to “Drephalys hypargus” (B.6.13) in Evans (1952) and differs from it by being generally paler, has a paler streak along dorsal hindwing vein 1A+2A, ventral dark hindwing border is rounder and without a tooth-like contour in cell CuA2-1A+2A. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly4105.1.1:T783C, aly54.9.3:C21A, aly2178.10.1:A52G, aly345.5.4:G72C, aly2284.13.13:G63C, aly423.15.5:A24A (not T), aly499.4.2:C81C (not T), aly15220.6.2:C129C (not A), aly151.8.3:A126A (not G), aly925.30.8:C24C (not T), and COI barcode: T97C, T220T, T277A, 322G, G622A.
Barcode sequence of the holotype.
Sample NVG-21116E12, GenBank OR837645, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACTTCTTTAAGATTATTAATTCGTACTGAATTAGGTAATCCAGGATCTTTAATCGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCTCCTGATATAGCATTTCCACGAATAAATAATATAAGATTTTGACTTCTTCCTCCTTCATTATTATTATTAATTTCAAGAAGTATTGTAGAAAATGGTGCTGGGACAGGATGAACTGTTTACCCTCCTCTTTCTTCTAATATTGCTCATCAAGGAGCATCTGTAGATTTAGCAATTTTTTCATTACATTTAGCAGGAATTTCATCTATTTTAGGAGCCATTAATTTTATTACAACAATTATTAATATACGAATTAATAATCTTTCTTTTGATCAATTACCTTTATTTGTATGAGCTGTTGGAATTACAGCTTTACTTTTATTACTTTCTTTACCAGTATTAGCTGGAGCTATTACCATATTATTAACTGATCGAAATTTAAATACATCTTTTTTTGACCCTGCAGGAGGAGGAGATCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♀ deposited in the Museum für Naturkunde, Berlin, Germany (MFNB), illustrated in Fig. 53–54, bears the following five rectangular labels, four white: [Bersaba | Surinam | 1898—9 Michls.], [Coll. | Staudinger], [] (empty label), [DNA sample ID: | NVG-21116E12 | c/o Nick V. Grishin], and one red [HOLOTYPE ♀ | Pseudodrephalys | argus Grishin].
Type locality.
Suriname: Para District, Bersaba.
Etymology.
The name is formed by removing hyp from hypargus. In Greek, the prefix hyp[o] means low, under, beneath, down, or less than normal. We removed the prefix, making this species higher. In Greek, argos means bright or white. The holotype is indeed brighter and whiter than hypargus. However, it is not clear whether this trait is general for the species or an individual variation. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Suriname.
Achlyodes calvus Grishin, new species https://zoobank.org/D5824D70-DF3A-42A2-9730-6B719506E741 (Fig. 2 part, 55–56, 266–267)
Definition and diagnosis.
Phylogenetic analysis reveals that several specimens from Southern Brazil and Paraguay identified as Achlyodes busirus rioja Evans, 1953 (type locality in Brazil: Rio de Janeiro) are genetically differentiated from all valid subspecies of Achlyodes busirus (Cramer, 1779) (type locality in Suriname), including sympatric A. b. rioja (Fig. 2): e.g., their COI barcodes differ by 3% (20 bp), and therefore represent a new species. This new species keys to A. b. rioja (F.2.1(c)) in Evans (1953) and differs from it by the absence of androconia patches (“brands” in Evans) at the base of the forewing beneath and at the humeral lobe on hindwing above (the two patches are contacting each other in spread specimens). Evans (1953) mentioned this character as a variation in A. b. rioja, but genomic analysis reveals that their absence corresponds to a distinct sympatric genetically differentiated species. Moreover, male genitalia are diagnostic (Fig. 266–267) and differ from sympatric A. b. rioja (Fig. 264–265) in the larger process of ampulla and more strongly upturned harpe with truncate, instead of rounded, distal end. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1260.10.2:A69T, aly151.22.21:G41A, aly1349.5.5:G30T, aly2844.10.4:C132T, aly84.50.2:A45G, and COI barcode: 49A, A169G, T250C, T346C, T472C.
Barcode sequence of the holotype.
Sample NVG-19076B03, GenBank OR837646, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCCGGAATAATTGGAACTTCACTAAGTATATTAATTCGAACTGAACTAGGAAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGAGGATTTGGAAATTGATTAGTACCCCTTATATTAGGAGCCCCTGATATAGCTTTTCCCCGAATAAATAACATAAGATTTTGATTATTACCCCCCTCTTTAATATTATTAATTTCAAGAAGAATTGTTGAAAATGGAGCTGGAACAGGATGAACAGTTTATCCCCCCCTTTCATCTAATATTGCTCACCAAGGATCATCTGTAGATTTAGCTATTTTTTCCCTACATTTAGCAGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATCATAAATCTTTCTTTTGATCAAATACCTCTTTTTGTATGAGCTGTAGGAATTACAGCTTTACTTTTATTACTTTCCTTACCAGTACTAGCTGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 55–56, bears the following four rectangular labels, three white: [BRAZIL:Sta Catarina | Sao Bento do Sul | 26°17′ S 49°25′W | 21.II.1989 | 850m, leg. Rank], [DNA sample ID: | NVG-19076B03 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01588644], and one red [HOLOTYPE ♂ | Achlyodes | calvus Grishin]. Paratypes: 2♂♂: NVG-22019A03 from near the type locality, Rio Vermelho, elevation 600–850 m, 24-Feb-1973, Rank leg. [ZSMC] and NVG-19076B01, USNMENT_01588642 Paraguay [USNM].
Type locality.
Brazil: Santa Catarina, São Bento do Sul, elevation 850 m, GPS −26.283333, −49.416667.
Etymology.
In Latin, calvus means hairless, bold, naked, or bare. It is given to this species for the lack of hair tufts. The name is an adjective.
Distribution.
Southern Brazil and Paraguay.
Spioniades artemis Grishin, new species https://zoobank.org/994E86A1-7BEF-42D2-807F-8DC7623058E6 (Fig. 2 part, 57–58, 268–270)
Definition and diagnosis.
Phylogenetic analysis reveals that Central American specimens identified as Spioniades artemides (Stoll, 1782) (type locality in Suriname) are genetically differentiated from it (Fig. 2): e.g., their COI barcodes differ by 5.5% (36 bp), and therefore represent a new species. This new species keys to S. artemides (E.14.2) in Evans (1953) and differs from it and another new species described below by a combination of the following characters: a more regular, straight, and diffuse border between discal brown and postdiscal white area on the hindwing, lack of pale diffuse spot(s) towards inner margin and tornus of the ventral forewing, nearly evenly convex (not angled at vein CuA1) forewing outer margin (Fig. 57–58), more robust uncus in lateral view, narrower in dorsal view, deeper divided harpe with more concave distal margin in lateral view and protruding less inward in dorsal view, more concave in the middle, wavy costa with a narrower knob-like process that protrudes deeper inward (Fig. 268–270). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly378.30.2:A75G, aly1838.60.2:A36G, aly1838.60.2:A126G, aly294.15.14:C81T, aly274.33.1:G1047A, and COI barcode: T50C, T127A, T529C, A541G, T547C.
Barcode sequence of the holotype.
Sample NVG-19088B03, GenBank OR837647, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACTTCTCTAAGTTTATTAATTCGAACTGAATTAGGAAATCCTGGAGCTCTTATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATGCTAGGAGCCCCTGATATAGCATTCCCTCGAATAAATAATATAAGATTCTGATTATTACCTCCATCTTTAATATTACTAATTTCTAGAAGAATCGTTGAAAATGGAGCAGGTACTGGATGAACAGTTTACCCCCCTCTTTCAGCTAATATCGCACACCAAGGATCTTCAGTTGATTTAGCAATTTTTTCTTTACATCTTGCTGGAATTTCCTCTATTTTAGGAGCTATTAATTTTATTTCTACAATTATTAATATACGAATTAGAAATCTTTCATTTGATCAAATACCATTATTTGTTTGAGCTGTAGGAATTACTGCCTTACTTTTATTGTTATCCTTACCAGTATTAGCAGGTGCTATTACTATACTTTTAACTGATCGAAATCTTAATACATCATTTTTTGATCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 57–58, bears the following four rectangular labels, three white: [Madden For. | Panama C.Z. | IV-4–64], [DNA sample ID: | NVG-19088B03 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01588916], and one red [HOLOTYPE ♂ | Spioniades | artemis Grishin]. Paratypes: 1♂ and 1♀: Costa Rica: Area de Conservación Guanacaste, Alajuela Prov.: 1♂ NVG-18013H11, 10-SRNP-67995 Sector Rincon Rain Forest, Palomo, 96 m, 10.96187, −85.28045, eclosed on 04-Nov-2010; and 1♀ NVG-18013H10, 07-SRNP-65617 Brasilia, Piedrona, 340 m, 11.01618, −85.35902, eclosed on 13-Oct-2007.
Type locality.
Panama: Panama Province, Madden Road Forest Preserve.
Etymology.
The name formed from its sister species, artemides, made shorter for this more northern species. The name is a noun in apposition.
Distribution.
Costa Rica and Panama.
Spioniades artemidoides Grishin, new species https://zoobank.org/85986CDE-04A2-42E3-BD64-178DD5DB5583 (Fig. 2 part, 59–60, 271–273)
Definition and diagnosis.
Phylogenetic analysis reveals that a specimen from Southern Brazil identified as Spioniades artemides (Stoll, 1782) (type locality in Suriname) is genetically differentiated from it (Fig. 2): e.g., their COI barcodes differ by 5.0% (33 bp), and therefore represent a new species. This new species keys to S. artemides (E.14.2) in Evans (1953) and differs from it and S. artemis new species by the following combination of characters: the irregular, wavy, and sharp border between discal brown and postdiscal white area on the hindwing, frequent expression of pale diffuse spot(s) towards inner margin and tornus of the ventral forewing (Fig. 59–60), less robust uncus in lateral view, broader in dorsal view, shallowly divided harpe with slightly concave distal margin in lateral view and protruding deeper inward in dorsal view, nearly straight with a broader knob-like process that protrudes less inward (Fig. 271–273). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly251.4.1:G89A, aly6370.13.3:C61T, aly15220.10.2:C66T, aly2012.51.1:T768G, aly336.2.3:A52T, aly4333.9.1:C60C (not T), aly1113.5.5:T45T (not A), aly1456.5.1:T1015T (not A), aly1456.5.1:T1044T (not C), aly1139.42.1:C66C (not T), and COI barcode: T70C, T91A, T178C, T202C, T226C, A379G.
Barcode sequence of the holotype.
Sample NVG-19088B05, GenBank OR837648, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACTTCCTTAAGTTTACTAATTCGAACCGAATTAGGAAATCCTGGAGCACTTATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATCGGAGGATTTGGAAATTGATTAATCCCATTAATGCTAGGGGCCCCTGACATAGCATTTCCTCGAATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAATACTTTTGATTTCTAGTAGTATTGTTGAAAATGGGGCAGGTACTGGTTGAACAGTTTATCCCCCTCTTTCAGCTAATATTGCACATCAAGGATCTTCGGTTGATTTAGCAATTTTTTCTTTACATCTTGCTGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTTCTACAATTATTAATATACGAATTAGAAATCTTTCATTTGATCAAATACCATTATTTGTTTGAGCTGTTGGAATTACTGCTTTACTTTTATTATTATCTTTACCAGTATTAGCTGGTGCTATTACTATACTTTTAACTGACCGAAATCTTAATACATCATTTTTTGATCCTGCTGGAGGGGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 59–60, bears the following four rectangular labels, three white: [Brasil:Santa Catarina | Joinville: 10–200 m | 16 Feb. 1991 | Leg. H. Miers], [DNA sample ID: | NVG-19088B05 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01588918], and one red [HOLOTYPE ♂ | Spioniades | artemidoides Grishin].
Type locality.
Brazil: Santa Catarina, Joinville.
Etymology.
The name is formed from its sister species name and is a noun in apposition. From north to south, short to long, we get artemis, artemides, and artemidoides.
Distribution.
Currently known only from Southern Brazil.
Spioniades anta Evans, 1953 is a species distinct from Spioniades abbreviata (Mabille, 1888)
Genomic sequencing of Spioniades abbreviata anta Evans, 1953 (type locality in Bolivia) reveals that it is not monophyletic with Spioniades abbreviata (Mabille, 1888) (type locality Panama: Chiriquí) in the Z chromosome tree (Fig. 2a) and the two taxa are strongly differentiated genetically, e.g., their COI barcodes differ by 5.6% (37 bp). In addition, their male genitalia and wing shapes differ (Evans 1953). Therefore, we propose that Spioniades anta Evans, 1953, new status, is a species-level taxon.
Myrinia orieca Grishin, new species https://zoobank.org/E284FB0F-A886-47D0-B039-E684CA8EF130 (Fig. 2 part, 61–62, 274–276)
Definition and diagnosis.
Phylogenetic analysis reveals that specimens from eastern Ecuador identified as Myrinia binoculus (Möschler, 1877) (type locality in Suriname, syntype sequenced as NVG-15033C11) are genetically differentiated from it in both nuclear and mitochondrial genomes (Fig. 2): e.g., their COI barcodes differ by 1.8% (12 bp), and therefore represent a new species. This new species keys to M. binoculus ((E.24.1) in Evans (1953) and differs from it by more elongated eyespot on the dorsal forewing, wider dark-brown forewing margins, better defined and more contrasting from the ground color bands (discal, postdiscal, and submarginal) on the ventral hindwing, and a smaller dark ventral hindwing tornal spot (Fig. 61–62). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2178.55.2:C48T, aly2178.55.2:C63T, aly1084.13.12:C93T, aly276558.43.4:A408G, aly4645.9.3:T103C, and COI barcode: A43G, A79G, T97C, A166G, A184G, A586G.
Barcode sequence of the holotype.
Sample NVG-17105F03, GenBank OR837649, 658 base pairs:
TACTTTATACTTTATTTTTGGAATTTGAGCAGGAATAGTAGGGACATCCTTAAGTTTATTAATTCGTACTGAATTAGGGAATCCAGGATCATTAATCGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCAATTATAATTGGAGGGTTTGGAAATTGACTTGTTCCATTAATATTAGGTGCCCCAGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGACTTTTACCTCCATCATTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACTGTTTACCCTCCTTTATCTGCTAATATTGCTCATCAAGGATCCTCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCTGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGTATTAATAATCTTTCATTCGATCAAATACCTTTATTTGTATGAGCAGTAGGAATTACAGCTTTATTATTACTATTATCTTTACCTGTATTAGCAGGAGCAATTACTATACTTTTAACGGATCGAAATTTAAATACATCATTTTTTGATCCTGCAGGTGGGGGAGATCCTATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 61–62, bears the following four rectangular printed labels, three white: [ECUADOR: Orellana, | Tiputini Biodiversity Station, | Lower Río Tiputini, | 0 37′55″ S, 76 08′39″ W | 200 m, 10–15 Aug 2002 | J.P.W. Hall & M.A. Solis], [DNA sample ID: | NVG-17105F03 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 00894950], and one red [HOLOTYPE ♂ | Myrinia | orieca Grishin]. Paratype: 1♂ NVG-14112H01 Ecuador: Napo, Yasuní National Park, 20-Sep–4-Oct-2003, C. Bordelon and E. Knudson, the Texas Lepidoptera Survey collection [MGCL].
Type locality.
Ecuador: Orellana Province, Lower Río Tiputini, Tiputini Biodiversity Station, elevation 200 m, GPS −0.6319, −76.1442.
Etymology.
This species comes from the Ori[ente (eastern region) in]Ec[u]a[dor]. The name is a Latinized feminine adjective.
Distribution.
Currently known only from the Amazonian region in eastern Ecuador.
Myrinia aragua Grishin, new species https://zoobank.org/055C6BE9-3405-428D-9BD1-B20AFA4C3F5D (Fig. 2 part, 63–64, 277–278)
Definition and diagnosis.
Inspection of the Z chromosome tree reveals that a specimen from Venezuela identified as Myrinia laddeyi (E. Bell, 1942) (type locality in Ecuador) is not monophyletic with it and instead is sister to Myrinia binoculus (Möschler, 1877) (type locality in Suriname) (Fig. 2a), thus representing a new species. Curiously, the mitochondrial DNA of this new species is much closer to M. laddeyi than to any other species (Fig. 2b): COI barcode of the holotype of M. laddeyi differs from the new species by 1% (7 bp), suggesting either introgression or hybrid origin of this species. The new species keys to Myrinia binoculus (E.24.1) in Evans (1953), which includes M. laddeyi that Evans misidentified and Myrinia raymundo H. Freeman, 1979 (type locality in Mexico, Tabasco) described later and differs from them by the following combination of characters: paler, especially on dorsal hindwing, where the marginal pale band is wider; forewing eyespot rounder, not elongated; ventral hindwing without a dark spot at tornus. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2012.66.4:G225A, aly1222.4.1:A99G, aly890.3.2:C130T, aly887.6.1:C142T, aly1838.60.9:G111A, aly2130.12.1:C162C (not A), aly2130.12.1:A179A (not C), aly2130.12.1:C183C (not T), aly85.3.11:C106C (not T), aly4645.9.9:C156C (not T), but COI barcode is not different from M. laddeyi.
Barcode sequence of the holotype.
Sample NVG-18061H06, GenBank OR837650, 658 base pairs:
TACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACATCTTTAAGTTTATTAATCCGTACTGAATTAGGAAATCCAGGATCGTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGACTTGTTCCATTAATACTAGGTGCTCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCTCCATCATTAATACTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACAGGGTGAACTGTTTACCCTCCTTTATCTGCTAATATTGCCCATCAAGGATCTTCTGTAGATTTAGCTATTTTTTCTTTACATTTAGCTGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGTATTAATAACCTTTCATTTGATCAAATACCTTTATTTGTATGAGCAGTAGGAATTACAGCTCTATTATTATTATTATCTTTACCTGTTTTAGCAGGAGCAATTACAATACTTTTAACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCAGGAGGAGGAGATCCTATTCTTTATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 63–64, bears the following four rectangular labels, three white: [VENEZUELA-ARAGUA | Rancho Grande 1100m | 28 May ‘85 | S. S. Nicolay], [DNA sample ID: | NVG-18061H06 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01466907], and one red [HOLOTYPE ♀ | Myrinia | aragua Grishin].
Type locality.
Venezuela: Aragua, Rancho Grande, elevation 1100 m.
Etymology.
The name is given for the type locality and is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Aragua, Venezuela.
Myrinia maculosa Grishin, new species https://zoobank.org/9BB52D48-C7A3-4940-97B0-FC88016521D3 (Fig. 2 part, 65–66, 279–281)
Definition and diagnosis.
Phylogenetic analysis reveals that a specimen from Guatemala identified as Myrinia myris (Mabille, 1898) (type locality in Brazil: Santa Catarina) is genetically differentiated from it (Fig. 2): e.g., their COI barcodes differ by 3.2% (21 bp), and therefore represents a new species. The new species keys to M. myris (E.24.2) in Evans (1953) and differs from it by more contrasting wing patterns: dark bands and spots stand out on paler ground color stronger than in M. myris, e.g., forewing subapical dark spots. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly528.33.1:T420A, aly707.3.1:G46A, aly315.8.2:G129C, aly536.208.9:A36G, aly4645.9.3:T87A, aly728.25.1:T72T (not A), aly1475.3.2:G1077G (not A), aly600.8.1:G81G (not A), aly1720.13.9:C206C (not A), aly1350.21.3:T54T (not C), and COI barcode: A208G, A238A, T287C, A298T, T601C.
Barcode sequence of the holotype.
Sample NVG-18061H04, GenBank OR837651, 658 base pairs:
AACTTTATATTTTATTTTCGGAATTTGAGCAGGAATAGTTGGAACATCTTTAAGTTTATTAATTCGTACTGAATTAGGTAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGTGGATTTGGTAATTGATTAGTACCTTTGATATTAGGAGCTCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGACTTTTACCTCCATCTTTAATATTACTAATTTCAAGTAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACTGTTTATCCTCCTTTATCAGCTAATATTGCTCATCAAGGATCTTCTGTAGATTTAGCTATTTTTTCTCTACATTTAGCAGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGTATTAATAACTTATCATTTGATCAAATACCATTATTTGTATGAGCAGTAGGAATTACAGCTTTATTATTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACTATACTTTTAACAGATCGAAATTTAAACACATCTTTTTTTGATCCAGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 65–66, bears the following four rectangular labels, three white: [fromLThiel | SSebastian | Retalhuleu | Guatemala], [DNA sample ID: | NVG-18061H04 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01466905], and one red [HOLOTYPE ♂ | Myrinia | maculosa Grishin].
Type locality.\
Guatemala: Retalhuleu department, San Sebastián.
Etymology.
In Latin, maculosa means spotted, speckled, dappled, mottled, or variegated. It reflects a more contrasting and spottier wing pattern compared to its closest relatives. The name is an adjective.
Distribution.
Currently known only from the holotype collected in Guatemala.
Myrinia manchada Grishin, new species https://zoobank.org/F341D06C-4741-4C54-ABCE-A7BD83C5C264 (Fig. 2 part, 67–68, 282–284)
Definition and diagnosis.
Sequencing of the holotype of Myrinia laddeyi (E. Bell, 1942) (type locality in Ecuador) revealed that it is a species related to Myrinia binoculus (Möschler, 1877) (type locality in Suriname) and not the species Evans (1953) identified as M. laddeyi (Fig. 2). Therefore, the species that Evans misidentified as M. laddeyi does not have an available name and is new. In USNM, we found a specimen that keys to E.24.4 in Evans (1953) and is distinguished from its relatives, including the true M. laddeyi, by larger size (forewing typically longer than 21 mm), more segments in nudum (more than 25 segments), the lack of eyespot on the ventral forewing, large dark tornal spot on the ventral hindwing, short and rounded harpe of the right valva, style of the left valva longer than harpe. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly798.33.50:A117G, aly2149.2.8:G45A, aly38.5.1:T141C, aly767.7.2:T78C, aly767.7.2:T468A, aly651.4.5:T106T (not C), aly536.173.2:C78C (not T), aly17.12.4:C112C (not T), aly17.12.4:A336A (not G), aly1022.2.1:G96G (not A), and COI barcode: T82C, 277C, T364A, T352C, T386C.
Barcode sequence of the holotype.
Sample 11-BOA-13383A10, GenBank OR837652, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACATCTTTAAGTTTATTAATTCGAACTGAATTAGGTAACCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGACTTGTACCATTAATATTAGGAGCCCCAGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGATTATTACCCCCCTCCTTAATATTATTAATCTCCAGAAGAATTGTAGAAAATGGAGCAGGAACTGGATGAACTGTTTATCCCCCTTTATCCGCTAATATTGCACACCAAGGATCTTCAGTAGACCTAGCTATTTTTTCTCTACATTTAGCTGGAATTTCATCTATTTTAGGAGCTATCAATTTTATTACAACAATTATTAATATACGTATTAATAATCTTTCATTTGATCAAATACCATTATTCGTTTGAGCTGTAGGAATCACAGCTTTATTATTATTATTATCTTTACCTGTATTAGCAGGTGCTATTACTATACTTCTAACAGATCGAAATTTAAATACATCTTTTTTTGATCCTGCTGGAGGAGGAGATCCTATTCTTTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 67–68, bears the following five rectangular labels, four white: [GUYANA: Region 8 | Potaro River nr. Tukeit | 250′ – 1000′ | 18–23 Mar 1999 | leg. S. Fratello], [DNA sample ID: | 11-BOA-13383A10 | c/o Nick V. Grishin], [DNA sample ID: | NVG-22034H05 | c/o Nick V. Grishin], [{QR Code}| USNM ENT 00232398], and one red [HOLOTYPE ♂ | Myrinia | manchada Grishin]. Paratype: 1♂: NVG-23026F10, EL83681 French Guiana, Saint-Laurent-du-Maroni, 1905, E. Le Moult leg. [MNHP].
Type locality.
Guyana: Potaro-Siparuni Region, Potaro River nr. Tukeit.
Etymology.
In Spanish, manchada means stained, spotted, spotty, or smudgy. The name is given for the large tornal spot on the ventral hindwing and is a noun in apposition.
Distribution.
The Guianas.
Tribe Carcharodini Verity, 1940
Polyctor (Fenops) lamperus Grishin, new species https://zoobank.org/CF3D0B2D-8ED2-4D3F-AD93-03621C2AFC9B (Fig. 3 part, 69–70, 285–287)
Figure 3.
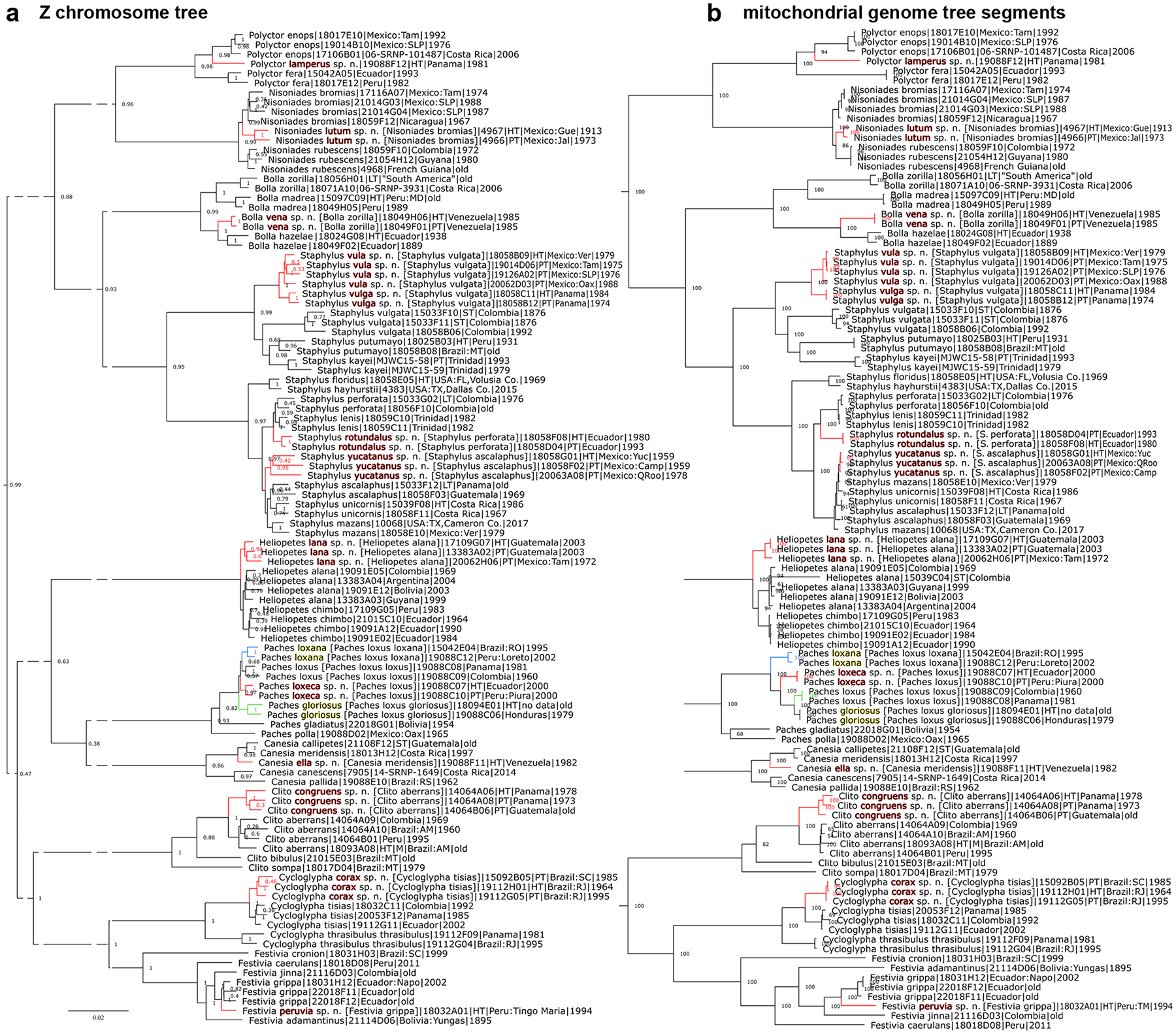
Phylogenetic trees of remaining Pyrginae (excluding Achlyodini) inferred from protein-coding regions in a) the Z chromosome and b) the mitochondrial genome (in segments). See Fig. 1 legend for other notations.
Definition and diagnosis.
This new species is sister to Polyctor enops (Godman and Salvin, 1894) (type locality in Mexico: Veracruz and Honduras) and differs from it by 2.3% (15 bp) in COI barcode and is recognizable phenotypically by paler wing ground color, larger forewing hyaline spots and mostly white ventral hindwing (marginally brown towards tornus), in particular by costa and apex, which are brown in P. enops and Polyctor fera (Weeks, 1901) (distal half of hindwing can be white). This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly85.29.3:A228G, aly214.4.1:C375T, aly3268.8.1:T900C, aly72.21.1:G933A, aly1022.3.1:G150A, aly315.8.6:G63G (not A), aly570.6.1:G57G (not A), aly1841.5.20:T96T (not C), aly1656.40.1:C575C (not A), aly1656.40.1:A1038A (not G), and COI barcode: T50C, A100T, G353A, 445C, T529C.
Barcode sequence of the holotype.
Sample NVG-19088F12, GenBank OR837653, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGATCAGGAATAGTAGGAACATCACTAAGTTTAATTATTCGATCTGAATTAGGTATACCTGGTTCATTAATTGGTAATGATCAAATTTATAATACAATTGTAACTGCTCATGCTTTTATTATAATTTTCTTCATAGTTATACCCATTATAATTGGAGGATTCGGAAATTGATTAGTACCCCTTATATTAGGAGCTCCTGATATAGCTTTTCCCCGAATAAATAATATAAGATTTTGATTATTACCTCCATCTTTAACTTTATTAATTTCAAGAAGTATTGTTGAAAATGGTGCTGGAACAGGATGAACAGTTTATCCTCCTTTATCTACTAATATTGCTCATCAAGGTTCTTCTGTAGATTTAGCAATTTTCTCTTTACATTTAGCAGGTATTTCTTCAATTTTAGGTGCTATTAACTTCATTACAACTATTATTAATATACGAATTAATAATATATCTTTTGATCAAATACCTCTTTTTATTTGAGCTGTTGGAATTACAGCCTTACTTTTATTATTATCTTTACCAGTTTTAGCAGGAGCTATTACAATATTATTAACTGATCGTAATTTAAATACATCTTTTTTTGATCCTTCAGGAGGAGGAGATCCTATTCTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 69–70, bears the following four rectangular labels, three white: [PANAMA: Darien | Cana 400m | 9.VII.1981 | Leg. G. B. Small | on mud], [DNA sample ID: | NVG-19088F12 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01588973], and one red [HOLOTYPE ♂ | Polyctor | lamperus Grishin].
Type locality.
Panama: Darien Province, Cana, elevation 400 m.
Etymology.
In Greek, λαμπερός (lamperós) means shiny bright. The name reflects the species’ bright colors compared to its relatives and is a noun in apposition.
Distribution.
Currently known only from the holotype collected in eastern Panama.
Lectotype designation for Pellicia bromias Godman and Salvin, 1894
Pellicia bromias Godman and Salvin, 1894 (type locality in Mexico, Guatemala, Costa Rica, Panama), currently in the genus Nisoniades Hübner, [1819] (type species Papilio bromius Stoll, 1787, which is a junior subjective synonym of Nisoniades mimas (Cramer, 1775)) was treated as a valid species, sister to Nisoniades rubescens (Möschler, 1877) (type locality in Suriname) by Zhang et al. (2020). To define Nisoniades bromias objectively due to likely polytypic type series, N.V.G. hereby designates a male syntype in the Natural History Museum, London, UK that according to its label was illustrated by Godman and Salvin (1893–1899) and bears the following seven rectangular (except the first one, which is round and with a red circle on the upper side) white labels: (Type) and on the other side (H | 707), [Atoyac, | Vera Cruz. | May. H.H.S.], [♂], [Sp. figured.], [B.C.A.Lep.Rhop. | Pellicia | bromias, | Mab.], [Godman-Salvin | Coll. 1912.—23.], and [{QR Code} | BMNH(E) 1669504], as the lectotype of Pellicia bromias Godman and Salvin, 1894. The type locality of Nisoniades bromias becomes Mexico: Veracruz, Atoyac.
Nisoniades (Nisoniades) lutum Grishin, new species https://zoobank.org/8CA203F6-1E1A-4B01-9D9E-FC3A9F5F0AA4 (Fig. 3 part, 71–72, 288–290)
Definition and diagnosis.
Genomic trees reveal that specimens from western Mexico identified as Nisoniades bromias Godman and Salvin, 1894 (type locality in Mexico: Veracruz) form a distinct clade that is not even monophyletic with N. bromias in the mitochondrial genome tree (albeit with weaker statistical support of 86%) (Fig. 3). Due to genetic differentiation, amplified by the genetic similarity within N. bromias and its lower evolutionary rate (comparatively shorter branches in Fig. 3), these western populations represent a new species. In the COI barcode, the new species differs from N. bromias and Nisoniades rubescens (Möschler, 1877) by 1.1% (7 bp) and 2.0% (13 bp), respectively. The new species keys to N. rubescens (E.19.5) in Evans (1953) and differs from its relatives by paler-brown wings with more contrasty and restricted dark-brown areas that form bands rather than ground color (Fig. 71–72), broader ampulla of the right valva, and less sharply curved inward ampulla of the left valva (Fig. 288–290). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly60.18.10:G18T, aly60.18.10:G78A, aly2165.8.15:G39C, aly2303.1.7:A31T, aly1222.34.1:G57A, and COI barcode: T82C, T292C, T328C, T619C, A628G.
Barcode sequence of the holotype.
Sample NVG-4967, GenBank OR837654, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGATCCGGAATAGTAGGAACATCATTAAGCTTACTAATTCGATCTGAATTAGGTAACCCAGGATTTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCCTTTATTATAATTTTCTTTATAGTAATACCCATTATAATTGGAGGATTCGGAAATTGATTAGTACCCCTTATATTAGGAGCTCCTGATATAGCTTTTCCCCGAATAAATAACATAAGATTTTGATTACTTCCTCCTTCTATTACTTTATTAATCTCAAGTAGTATTGTAGAAAATGGAGCTGGAACAGGCTGAACTGTGTATCCACCTTTATCATCTAATATTGCTCACCAAGGCTCATCTGTTGATTTAGCAATTTTTTCATTACATTTAGCTGGAATTTCATCTATTTTAGGTGCTATTAATTTTATTACTACTATTATCAATATACGAATTAATAATTTATCACTTGATCAAATACCTTTATTTGTTTGAGCCGTAGGAATTACAGCATTACTTTTATTATTATCTTTACCAGTTTTAGCTGGAGCTATCACAATATTATTAACTGATCGAAATTTAAATACATCTTTTTTTGATCCCGCAGGAGGGGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 71–72, bears the following seven rectangular labels, six white: [Sierra de | GuerreroMex], [Jan. | ‘13], [RMuller | Collector], [3682], [DNA sample ID: | NVG-4967 | c/o Nick V. Grishin], [genitalia: | NVG151101–18 | Nick V. Grishin], and one red [HOLOTYPE ♂ | Nisoniades | lutum Grishin]. Paratype: 1♂: NVG-4966 Mexico: Jalisco, Mismaloya area, Puerto Vallarta, 15–31-Oct-1973, P. M. Aubry leg., genitalia NVG151101–17 [USNM].
Type locality.
Mexico: Guerrero.
Etymology.
In Greek, βρωμιάς (bromias) means dirt or dirty, and lutum is dirt in Latin, used here as the name for the sister species of N. bromias. The name is a noun in apposition.
Distribution.
Western Mexico, currently confirmed from Jalisco and Guerrero.
Bolla (Stolla) vena Grishin, new species https://zoobank.org/F6F25879-BE84-46BB-9450-0F9A35F55461 (Fig. 3 part, 73–74, 291–292)
Definition and diagnosis.
In addition to the three species in the Bolla zorilla (Plötz, 1886) (type locality in Panama) complex, genomic trees reveal a fourth (and new) species from Venezuela (Fig. 3): it differs from its sister Bolla hazelae (Hayward, 1940) (type locality Ecuador: Mapoto) by 4.0% (26 bp). This new species keys to B. zorilla (E.31.21) in Evans (1953) and differs from its relatives by the following combination of characters: hindwing with a more scalloped outer margin, convex in the middle and noticeably concave towards tornus, a hyaline subapical spot in cell R5-M1 is smaller than the spot in cell R3-R4 (spots subequal in other species) (Fig. 73–74), ampulla with a thin style directed dorsad, harpe broader in lateral view, less extended distad, with a more prominent expansion directed dorsad, dorsal margin between the dorsal-most points of harpe and costa deeper concave (with a style in the middle) (Fig. 291–292). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1281.15.8:C123T, aly813.2.4:G44C, aly727.20.3:C69T, aly1329.6.5:G105A, aly1329.6.5:G108T, and COI barcode: A202T, C235T, T334G, T535C, A622G.
Barcode sequence of the holotype.
Sample NVG-18049H06, GenBank OR837655, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGATCTGGTATAGTAGGAACTTCTTTAAGAATACTTATTCGTTCAGAATTAGGAACCCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTCATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGACTTTTACCCCCTTCTTTAATACTTTTAATTTCTAGAAGTGTAGTAGAAAATGGAGCAGGTACAGGATGAACGGTTTACCCCCCCCTTTCAGCTAATATTGCTCATCAAGGATCATCTGTAGATTTAGCTATTTTTTCCCTTCATTTAGCAGGTATTTCTTCAATTTTAGGGGCAATTAATTTTATCACAACTATTATTAATATACGAATTAACAACTTATCATTTGATCAAATACCTTTATTTGTATGAGCAGTTGGTATCACCGCTTTACTCTTATTATTATCTTTACCTGTATTAGCAGGAGCTATTACAATACTTCTAACAGATCGAAATTTAAATACTTCATTCTTTGACCCTGCGGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 73–74, bears the following six rectangular labels, five white: [VENEZUELA-ARAGUA | Rancho Grande 1100m | 18 May ‘85 | S. S. Nicolay], [Bolla ♂ | zorilla | Det. | S.S. Nicolay], [Bolla | zorilla ♂ | (Ploetz) | det. H. A. Freeman], [DNA sample ID: | NVG-18049H06 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01466668], and one red [HOLOTYPE ♂ | Bolla vena | Grishin]. Paratype: 1♀ NVG-18049F01, USNMENT_01466639 the same data as the holotype, but collected on 7-Jun-1985.
Type locality.
Venezuela: Aragua, Rancho Grande, elevation 1100 m.
Etymology.
The name is given for Ven[ezuel]a and is a noun in apposition.
Distribution.
Venezuela.
Staphylus (Vulga) vula Grishin, new species https://zoobank.org/1E5D2A3B-D751-4144-A814-9F916BD25F36 (Fig. 3 part, 75–76, 293–294)
Definition and diagnosis.
Phylogenetic analysis that included syntypes of Staphylus vulgata (Möschler, 1879) (type locality in Colombia) reveals that Mexican specimens identified as this species are strongly differentiated genetically from and are not even monophyletic with it (Fig. 3), e.g., their COI barcodes differ by 5.9% (39 bp), and therefore represent a new species. This new species keys to S. vulgata (E.32.6(a)) in Evans (1953) and differs from its relatives by the following combination of characters: costa of valva less convex, valva narrower in lateral view, expansion of ampulla directed distad and reaches about the middle of harpe, its terminal spines shorter, harpe semi-triangular, distally rounded, with slightly concave dorsoposterior margin (Fig. 293–294). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1146.51.1:T1173C, aly536.133.3:T30C, aly1244.6.1:C819T, aly2811.1.8:T52C, aly2811.1.8:C53A, and COI barcode: T50C, T118C, T205C, T463C, T479C.
Barcode sequence of the holotype.
Sample NVG-18058B09, GenBank OR837656, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATAGTAGGAACTTCTCTAAGTATTCTTATTCGATCAGAATTAGGAACCCCTGGATCTTTAATTGGAGATGATCAAATTTATAACACTATTGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGACTTGTACCCCTTATATTAGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTACCCCCATCCTTAATACTTCTAATTTCAAGTAGCATTGTAGAAAATGGAGCAGGAACTGGATGAACTGTATATCCCCCACTTTCAGCCAATATTGCCCATCAAGGTTCATCTGTAGATTTAGCTATTTTTTCCCTTCATTTAGCTGGAATTTCTTCCATTTTAGGAGCAATTAACTTCATCACAACTATTATTAACATACGAATTAATAATCTATCATTTGATCAAATACCTTTATTTGTATGAGCTGTAGGAATCACAGCATTACTTTTACTCCTATCTTTACCAGTATTAGCTGGAGCTATTACTATACTTTTAACTGATCGAAATCTTAATACATCATTTTTTGATCCCGCTGGAGGAGGTGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 75–76, bears the following five rectangular labels, four white: [7 mi N Santa | Comapan, Ver., | Mex. V-22–1979 | J.R.Powers,Collr.], [Staphylus | vulagata ♂ | (Moeschler) | det. H. A. Freeman], [DNA sample ID: | NVG-18058B09 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01466683], and one red [HOLOTYPE ♂ | Staphylus | vula Grishin]. Paratypes: 7♂♂ from Mexico, Roy O. Kendall and C. A. Kendall leg., at TAMU, unless stated otherwise: 1♂ NVG-20062D03 Oaxaca, Pluma Hidalgo, 18-Apr-1988, J. Kemner leg., genitalia [TMMC]; 1♂ NVG-19014D06 Tamaulipas, Ciudad Mante, Los Arcos Ct., larval foodplant Achyranthes aspera L., 11-Jan-1975; 2♂♂ Tamaulipas, Rancho Pico de Oro, vic. Of Los Kikos: NVG-19126A03 21-Dec-1972 and 19126A05 6-Feb-1974; 1♂ NVG-19126A04 Tamaulipas, El Nacimiento, Rio Mante, 23-Jan-1974; 2♂♂ San Luis Potosi, El Naranjo: NVG-19014D03 1-Mar-1976 and NVG-19126A02 13-Feb-1976.
Type locality.
Mexico: Veracruz, 7 mi north of Santa Comapan.
Etymology.
The name is formed from the species epithet of S. vulgata (in Latin, vulgate means common). By significantly shortening the name, it refers to this most northern species of the S. vulgata complex. The name is a noun in apposition.
Distribution.
Widely distributed in Mexico: recorded from Tamaulipas, San Luis Potosi, Veracruz and Oaxaca.
Comment.
Reared on Achyranthes aspera L. by Roy O. Kendall and C. A. Kendall.
Staphylus (Vulga) vulga Grishin, new species https://zoobank.org/1ECE61F4-A057-4903-8706-D6CD468A8B87 (Fig. 3 part, 77–78, 295–297)
Definition and diagnosis.
Phylogenetic analysis that included syntypes of Staphylus vulgata (Möschler, 1879) (type locality in Colombia) reveals that Panamanian specimens identified as this species are strongly differentiated genetically from and are not even monophyletic with it (Fig. 3), e.g., their COI barcodes differ by 5.9% (39 bp). These specimens are sister to Staphylus vula new species but differ genetically from it (e.g., COI barcodes differ by 1.5%, 10 bp), and, therefore, represent a new species. This new species keys to S. vulgata (E.32.6(a)) in Evans (1953) and differs from its relatives by the following combination of characters: costa of valva strongly convex, nearly angled at the dorsal-most point, valva broader in lateral view, expansion of ampulla directed distad and slightly ventrad towards its end, and reaches about the middle of harpe, its terminal spines longer, harpe semi-triangular, distally rounded, with concave near its base dorsoposterior margin (Fig. 295–297). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly361.16.7:C229T, aly887.42.4:A63G, aly116.34.5:G392A, aly727.17.5:G94T, aly728.49.1:T1077C, and COI barcode: A40G, T50T, 85A, A202A, 400T.
Barcode sequence of the holotype.
Sample NVG-18058C11, GenBank OR837657, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATAGTGGGAACTTCTTTAAGTATTCTTATTCGATCAGAATTAGGAACCCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGACTTGTACCTCTTATATTAGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTACCCCCATCCTTAATACTTTTAATTTCAAGTAGCATTGTAGAAAATGGAGCAGGAACTGGATGAACTGTATATCCCCCACTTTCAGCCAATATTGCCCATCAAGGTTCATCTGTAGATTTAGCTATTTTTTCTCTTCATTTAGCTGGAATTTCTTCCATTTTAGGAGCAATTAACTTCATCACAACTATTATTAATATACGAATTAATAATTTATCATTTGATCAAATACCTTTATTTGTATGAGCTGTAGGAATTACAGCATTACTTTTACTCCTATCTTTACCAGTATTAGCTGGAGCTATTACTATACTTTTAACTGATCGAAATCTTAATACATCATTTTTTGATCCCGCTGGAGGAGGTGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 77–78, bears the following five rectangular labels, four white: [PANAMA:500m. | Darien, Cana | 3 Jan. 1984 | Gordon Small], [Staphylus ♂ | caribbea (Williams | and Bell) | det. H. A. Freeman], [DNA sample ID: | NVG-18058C11 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01466697], and one red [HOLOTYPE ♂ | Staphylus | vulga Grishin]. Paratype: 1♂ NVG-18058B12, USNMENT_01466686 Panama: Panama, Bayano, “8°[likely 9°]03′N 78°40′W”, 26-Oct-1974, G. B. Small leg., genitalia vial H-2198 by H. A. Freeman.
Type locality.
Panama: Darien Province, Cana, elevation 500 m.
Etymology.
The name is formed from the species epithet of S. vulgata (in Latin, vulgate means common). By slightly shortening the name, it refers to this species in the middle of the range of the S. vulgata complex. The name is a noun in apposition.
Distribution.
Recorded from eastern Panama.
Lectotype designation for Nisoniades perforata Möschler, 1879
Genomic sequencing of Nisoniades perforata Möschler, 1879 (type locality in Colombia and Panama) syntypes reveals that the type series is polytypic. To define this species objectively, N.V.G. hereby designates a male syntype in the Museum für Naturkunde, Berlin, Germany, that bears the following ten rectangular labels (the first three are red, green, and purple, respectively; others are white): [Lectotypus], [Columbia | Stdr. 76.], [Origin], [Type. | Verhdlg. zool. | bot. Gesellschft. | 1878. p. 223. | no= 32.], [Coll. Möschl.], [Hayhursti | Edw. Perforata Msch.], [Haÿhursti | Edw.], [GEN.PEP., | MIELKE | 1996], [{OR Code} http://coll.mfn-berlin.de/u/ | 80a649], [DNA sample ID: | NVG-15033G02 | c/o Nick V. Grishin], as the lectotype of Nisoniades perforata Möschler, 1879, currently in the genus Staphylus Godman and Salvin, 1896. The type locality of Staphylus perforata becomes Colombia.
Staphylus (Staphylus) rotundalus Grishin, new species https://zoobank.org/0568D95D-9F75-4538-823C-07E64A7D7CD0 (Fig. 3 part, 79–80, 298–299)
Definition and diagnosis.
Phylogenetic analysis reveals that specimens from Ecuador identified as Staphylus perforata (Möschler, 1879) (type locality in Colombia, lectotype sequenced as NVG-15033G02) are not monophyletic with it and instead are sister to both S. perforata and Staphylus lenis Steinhauser, 1989 (type locality in Trinidad) and genetically differentiated from them (Fig. 3), e.g., their COI barcodes differ by 1% (7 bp). Provided low mitochondrial DNA differentiation in these species (COI barcodes of S. perforata and S. lenis differ by only 1 base pair), the Ecuadorian specimens represent a new species. This new species keys to “Staphylus ascaphalus” [sic!] (E.32.20(d)) in Evans (1953) and differs from its relatives by the following combination of characters: rounder wings, most visible as more convex forewing outer margin, prominent forewing hyaline spots, especially the one in cell CuA1-CuA2, valva elongated with broad dorsally directed expansion between valva and harpe, rounded along its dorsal margin and armed with two large socketed bristles at the distal margin, and one medium bristle at its base, plus many smaller spines on harpe (Fig. 298–299). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1916.7.3:A81T, aly2928.14.8:T107C, aly103.40.5:C95A, aly9349.5.1:A206C, aly1249.14.3:C204A, and COI barcode: A34G, A217G, T298A, A511A, T601C.
Barcode sequence of the holotype.
Sample NVG-18058F08, GenBank OR837658, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGATCTGGGATAGTAGGAACTTCTTTAAGTATTCTTATTCGTTCTGAATTAGGAACTCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGAAATTGACTTGTTCCTCTTATATTAGGGGCCCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAATACTTTTAATTTCAAGAAGAATCGTAGAAAATGGAGCAGGTACTGGATGAACAGTTTATCCCCCCCTTTCAGCTAACATTGCCCATCAAGGTTCTTCTGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCTATTTTAGGAGCAATTAATTTTATTACAACTATTATCAATATACGAATTAATAATTTATCCTTTGATCAAATACCTTTATTTGTTTGAGCAGTTGGAATTACAGCATTATTATTACTTTTATCTTTACCAGTATTAGCAGGTGCTATTACTATACTTTTAACAGATCGAAATCTTAACACATCATTTTTTGATCCTGCTGGTGGAGGAGATCCTATTTTATATCAACATTTATTC
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 79–80, bears the following six rectangular labels, five white: [Rio Jondachi Napo | Ecuador 1000m | 8 July ‘80 | S. S. Nicolay], [Staphylus | m. ♂ | ascalaphus | Det. Staud. | S.S. Nicolay], [Staphylus | perforatus ♂ | (Moeschler) | det. H.A. Freeman], [DNA sample ID: | NVG-18058F08 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01466730], and one red [HOLOTYPE ♂ | Staphylus | rotundalus Grishin]. Paratype: 1♂ NVG-18058D04, USNMENT_01466702 1 km E of Pto. Napo, 490 m, GPS −1.04, −77.7867, 10-Oct-1993, S. S. Nicolay leg. [USNM].
Type locality.
Ecuador: Napo Province, Río Jondachi.
Etymology.
In Latin, rotund means round, and ala means wings. The name is given to describe the rounder than usual wing shape and is a noun in apposition.
Distribution.
Currently known from Ecuador.
Lectotype designation for Helias ascalaphus Staudinger, 1876
Helias ascalaphus Staudinger, 1876, the type species of a valid genus Staphylus Godman and Salvin, 1896, was described from several specimens from two localities in Panama. To define this species objectively with a single specimen, N.V.G. hereby designates a male syntype in the Museum für Naturkunde, Berlin, Germany, that bears the following eight rectangular labels (the first two are red and purple, respectively, others are white): [Lectotypus], [Origin], [Panama | Ribbe], [ascalaphus], [Ascalaphus | Stdgr.], [GEN.PEP., | MIELKE | 1996], [{OR Code} http://coll.mfn-berlin.de/u/ | 908619], [DNA sample ID: | NVG-15033F12 | c/o Nick V. Grishin], as the lectotype of Helias ascalaphus Staudinger, 1876. Because some of H. ascalaphus paralectotypes are labeled “Chiriqui” in a similar style as “Panama”, we suggest that the locality label “Panama” is more detailed than a country and refers to central Panama around Panama City. Therefore, the type locality of Staphylus ascalaphus is in central Panama.
Staphylus (Staphylus) yucatanus Grishin, new species https://zoobank.org/E2DA1C62-273A-4640-8B7D-FE16B5FBE315 (Fig. 3 part, 81–82, 300–301)
Definition and diagnosis.
Phylogenetic analysis reveals that specimens from the Yucatan peninsula in Mexico identified as Staphylus ascalaphus (Staudinger, 1876) (type locality in Panama: Panama, lectotype sequenced as NVG-15033F12) or Staphylus lenis Steinhauser, 1989 (type locality in Trinidad) are not monophyletic with either S. ascalaphus or S. lenis and instead form a separate clade that is sister to the clade of three species: S. ascalaphus, Staphylus unicornis Steinhauser and Austin, 1993 (type locality in Costa Rica, holotype sequenced as NVG-15039F08), and Staphylus mazans (Reakirt, [1867]) (type locality in Mexico: Veracruz) and is genetically differentiated from them (Fig. 3). Therefore, the Yucatecan specimens represent a new species. The four species (the new one, S. ascalaphus, S. unicornis and S. mazans) are closely related to each other and possess nearly identical DNA barcodes (0–5 bp difference) but are considered distinct due to differences in genitalia (Steinhauser and Austin 1993). This new species keys to “Staphylus ascaphalus” [sic!] (E.32.20(d)) in Evans (1953) and differs from it and other relatives by the following combination of characters: valva without prominent expansion directed dorsad between valva and harpe, but with a small (shorter than bristles), plate-like triangular, tooth-shaped lobe armed with three socketed bristles at its base, harpe more elongated than in relatives, terminally rounded, valva near the base of harpe and harpe towards dorsal margin armed on the outer surface with two to three dozen of long (length of harpe), medium-sized (~½ of long), and short (~¼ of long) socketed bristles (Fig. 300–301). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly419.1.7:G960A, aly1222.1.15:G494A, aly276558.8.2:C64T, aly1651.33.11:T222A, aly2555.6.3:G64C, and COI barcode: T67C, A160G, C238C.
Barcode sequence of the holotype.
Sample NVG-18058G01, GenBank OR837659, 658 base pairs:
AACTTTATATTTTATCTTTGGTATTTGATCTGGAATAGTAGGAACTTCTTTAAGTATTCTTATTCGCTCTGAATTAGGAACTCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGAGGTTTTGGAAATTGACTTGTTCCTCTTATATTAGGAGCCCCTGATATAGCTTTTCCCCGAATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAATACTTTTAATTTCAAGTAGAATCGTAGAAAATGGAGCAGGTACTGGATGAACAGTTTATCCCCCCCTTTCAGCTAACATTGCCCATCAAGGTTCTTCTGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCTATTTTAGGAGCAATTAATTTTATTACAACTATTATCAATATACGAATTAATAATTTATCCTTTGATCAAATACCCTTATTTGTTTGAGCAGTTGGAATTACAGCATTATTATTACTTTTATCTTTACCAGTATTAGCAGGTGCTATTACTATACTTTTAACAGATCGAAATCTTAATACATCATTTTTTGATCCTGCTGGTGGAGGAGATCCTATTTTATATCAACATTTATTC
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 81–82, bears the following seven rectangular labels, six white: [Near Xcan, Quin- | tana Roo – Yuca- | tan border, | MEXICO | May 19–23, 1959 | T.C. Emmel], [♂ genitalia | slide/ # | H169 | Prep. S.S. Nicolay], [Staphylus ♂ | m.ascalaphus | Det. Stgr. | S. S. Nicolay], [Staphylus | lenis ♂ | Steinhauser | det. H.A. Freeman], [DNA sample ID: | NVG-18058G01 | c/o Nick V. Grishin], [USNMENT| {QR Code} | 01466735], and one red [HOLOTYPE ♂ | Staphylus | yucatanus Grishin]. Paratypes: 2♂♂ from Mexico: NVG-18058F02, USNMENT_01466724 Campeche, 40 km E of Escarcega, 3–4-May-1959, T. C. Emmel leg. [USNM]; and NVG-20063A08 Qintana Roo, Nuevo X-Can 25-Nov-1978 ECW leg., genitalia H-1271 H. A. Freeman (Fig. 300–301) [CSUC].
Type locality.
Mexico: Quintana Roo–Yucatan border, near X-Can.
Etymology.
The name is for the distribution in the Yucatan peninsula and is a noun in apposition.
Distribution.
Currently known only from the Yucatan peninsula in Mexico.
Tribe Pyrgini Burmeister, 1878
Heliopetes (Heliopetes) lana Grishin, new species https://zoobank.org/2E6992E1-9CAE-4FF2-AD84-B73CB2C57F00 (Fig. 3 part, 83–84, 302–303)
Definition and diagnosis.
Phylogenetic analysis reveals that specimens from North America identified as Heliopetes alana (Reakirt, 1868) (type locality in Colombia, syntype sequenced as NVG-15039C04, sequence completeness insufficient to add it to the Z chromosome tree) are not monophyletic with it and instead are sister to both H. alana and Heliopetes chimbo Evans, 1953 (type locality in Ecuador), exhibiting notable genetic differentiation (Fig. 3): e.g., COI barcodes of a specimen from Guatemala and Colombia differ by 2% (14 bp). All available names in synonymy with H. alana have type localities in South America. Therefore, the North American populations are a new species. This new species keys to H. alana (G.2.12) in Evans (1953) and differs from its relatives by the following combination of characters: typically whiter (i.e., less yellow), especially on ventral hindwing (Fig. 84, but due to extensive variation hardly identifiable by facies), rounder in dorsal view anterior margin of tegumen, more extended harpe and, correspondingly, longer expansion of ampulla (Fig. 302–303). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1079.16.3:T70C, aly173.27.12:C85T, aly173.27.12:A144G, aly276665.11.2:T224G, aly276665.11.2:A302T, and COI barcode: A229G, C235T, A289A, T397C, A477G, T557C.
Barcode sequence of the holotype.
Sample NVG-17109G07, GenBank OR837660, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGTACTTCTTTAAGTTTATTAATTCGAACTGAATTAGGAAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCCCATGCTTTTATTATAATTTTTTTCATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCCCCAGATATGGCATTTCCTCGTATAAATAATATAAGATTTTGACTTTTACCCCCATCCCTAACATTATTAATTTCAAGAAGTGTAGTAGAAAATGGAGCAGGAACTGGTTGAACAGTTTACCCCCCTCTCTCGGCTAATATTGCCCATCAAGGATCTTCTGTTGATTTAGCTATTTTCTCTTTACATTTAGCTGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACAACTATTATTAATATACGTATTAGAAGTATATCATTTGATCAAATACCTTTATTTGTATGAGCGGTAGGAATTACTGCTTTATTACTACTATTATCATTACCTGTTCTAGCAGGTGCCATTACAATATTATTAACAGATCGAAATTTAAATACATCATTCTTCGATCCTGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTC
Type material.
Holotype: ♂ deposited in the Los Angeles County Museum of Natural History, Los Angeles, CA, USA (LACM), illustrated in Fig. 83–84, bears the following three rectangular labels, two white: [GUATEMALA. Peten District | Finca Ixobel S of Poptun | 16° 18’ 14” N: 89° 25’ 20” W | 5–10 JUNE 2003: 1700 ft. | Ron Leuschner. coll.], [DNA sample ID: | NVG-17109G07 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Heliopetes | lana Grishin]. Paratypes: 2♂♂: 11-BOA-13383A02 the same data as the holotype [USNM] and NVG-20062H06 Mexico: Tamaulipas, 1–4 km N of Gomez Farias, 350 m, 17-Aug-1972, C. J. Durden leg. (genitalia Fig. 302–303) [TMMC].
Type locality.
Guatemala: Peten District, Finca Ixobel S of Poptún, elevation 1700 ft, GPS 16.3039, −89.4222.
Etymology.
The name removes a negating “a” from the name of its more southern sister species H. alana and is a feminine noun in apposition.
Distribution.
This species is currently known from Mexico and Guatemala but is expected from other Central American countries.
Canesia ella Grishin, new species https://zoobank.org/A4979CC1-C793-4277-808B-45D5388EE312 (Fig. 3 part, 85–86, 304–306)
Definition and diagnosis.
Phylogenetic analysis reveals that a more brightly colored than typical for the genus specimen of Canesia Grishin, 2019 (type species Leucochitonea canescens R. Felder, 1869) is sister to both Canesia meridensis (Godman and Salvin, 1895) (type locality in Costa Rica) and Canesia callipetes (Godman and Salvin, 1895) (type locality in Mexico, Guatemala, and Colombia) and therefore is a new species. This new species keys to “Carrhenes callipetes callipetes” (E.51.3(a)) in Evans (1953) and differs from its relatives by being more vividly and vibrantly colored, more prominent dark spots, and the lack of whitish scales beneath: ventral side of wings is hay-yellow with well-developed brown spots and brown overscaling in the posterior half. This darker overscaling is not present in either C. meridensis or C. callipetes. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1651.28.14:C132T, aly1651.28.14:C147T, aly747.2.1:A480C, aly747.2.1:T513C, aly671.50.9:T53C, aly412.9.2:A201A (not G), aly636.8.1:A84A (not G), aly6841.59.1:A39A (not G), aly5719.1.9:A24A (not C), aly1155.1.7:G112G (not T), and COI barcode: A40G, T82C, T85C, T439C, A508G.
Barcode sequence of the holotype.
Sample NVG-19088F11, GenBank OR837661, 658 base pairs:
AACTTTATATTTTATTTTTGGAATCTGAGCAGGAATAGTGGGTACTTCCTTAAGTTTATTAATTCGAACTGAATTAGGAAACCCCGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCCCCAGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGATTATTACCACCTTCATTAACATTATTAATTTCTAGAAGTATTGTAGAAAACGGAGCAGGAACTGGATGAACAGTTTACCCCCCTCTCTCAGCTAATATCGCTCATCAAGGATCTTCTGTTGATTTAGCTATTTTTTCTTTACATTTAGCTGGAATTTCATCAATTTTAGGAGCTATCAATTTTATTACTACAATTATTAATATGCGAATTAGAAATTTATCTTTTGATCAAATACCATTATTTGTGTGAGCAGTAGGTATTACTGCATTATTATTATTATTATCATTACCAGTATTAGCTGGAGCAATTACTATATTATTAACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCTGGTGGAGGAGACCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 85–86, bears the following four rectangular labels, three white: [VENEZUELA:Barinas; | 27.4kmNW.Barinitas | 17 March 1982 | G.F. & J.F. Hevel], [DNA sample ID: | NVG-19088F11 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01588972], and one red [HOLOTYPE ♂ | Canesia | ella Grishin].
Type locality.
Venezuela: Barinas, 27.4 km NW of Barinitas.
Etymology.
The name is for the type locality in [Venezu]el(l)a. Also, הלא (ela) in Hebrew is Goddess. This probably is the most colorful and beautiful species of Canesia. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Venezuela.
Paches (Paches) gloriosus Röber, 1925 and Paches (Paches) loxana Evans, 1953 are species distinct from Paches (Paches) loxus (Westwood, 1852)
Genomic sequencing reveals that Paches gloriosus Röber, 1925 (type locality not given, holotype sequenced as NVG-18094E01) and Paches loxus loxana Evans, 1953 (type locality in Bolivia) currently regarded as subspecies of Paches loxus (Westwood, 1852) (type locality not given, possibly in Venezuela) are genetically differentiated from each other at the level characteristic of species (Fig. 3): e.g., COI barcodes of P. loxus and P. l. loxana differ by 3% (20 bp). Curiously, despite the strong genetic differentiation of P. gloriosus in the Z chromosome from all other taxa in this complex (Fig. 3a), it is more similar to P. loxus in the mitochondrial genome (Fig. 3b): COI barcode difference of 0.9% (6 bp). Due to phenotypic differences accompanied by genetic differentiation, we reinstate Paches (Paches) gloriosus Röber, 1925 as a species and propose a new status of species for Paches (Paches) loxana Evans, 1953.
Paches (Paches) loxeca Grishin, new species https://zoobank.org/D34A1B66-34BC-4424-A3DF-73B6D72B695C (Fig. 3 part, 87–88, 307–309)
Definition and diagnosis.
Phylogenetic analysis of the Paches loxus (Westwood, 1852) complex reveals a fourth (and new) species in addition to P. loxus, Paches gloriosus Röber, 1925 and Paches loxana Evans, 1953 that stands out in its genetic differentiation (Fig. 3): e.g., its COI barcodes differ from those of P. loxus, P. gloriosus, and P. loxana by 2.4% (16 bp), 2.7% (18 bp), and 2.9% (19 bp), respectively. This new species keys to Paches loxus loxus or “Paches loxus loxana” (E.43.1(b) or (c)) in Evans (1953) and differs from its relatives by the lack of median brown band (or its remnants) on dorsal hindwing in males and narrower brown hindwing margin (like P. loxus, and P. gloriosus, the band typically developed in P. loxana), more extensive dark discal area on forewing in males (like P. loxana and P. gloriosus, forewing mostly blue in P. loxus), white distal two-third of the ventral hindwing in females (like P. loxus and different from other species). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly536.116.5:T25C, aly2578.2.1:A51T, aly1079.4.3:C141T, aly1222.17.1:T51C, aly1489.18.2:C55T, and COI barcode: T97T, A202G, A214G, T274C, T412C, G620T.
Barcode sequence of the holotype.
Sample NVG-19088C07, GenBank OR837662, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGTACCTCATTAAGTTTATTAATTCGAACAGAACTAGGAAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCCATTATAATTGGAGGATTTGGAAATTGATTAGTGCCACTAATATTGGGAGCCCCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGAATATTACCCCCCTCATTAACCCTTTTAATTTCTAGAAGTATTGTGGAAAATGGAGCTGGAACCGGATGAACAGTTTACCCCCCGCTTTCATCAAATATTGCTCATCAAGGTTCTTCAGTTGATTTAGCTATTTTTTCTCTTCACTTAGCCGGTATTTCATCAATTTTAGGAGCTATTAATTTTATTACCACAATTATTAATATACGTATTATAAATTTATCTTTTGATCAAATACCTCTATTTGTTTGAGCAGTAGGAATTACTGCATTATTACTATTATTATCTTTACCTGTTTTAGCGGGAGCTATTACAATATTATTAACAGATCGAAATTTAAATACATCATTTTTTGACCCCTCTGGAGGTGGAGATCCAATCTTATATCAACACTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 87–88, bears the following four rectangular labels, three white: [ECUADOR: Morona-Santiago | 15 km S Gualaquiza, 800 m | 3° 27.6′S, 78° 33.1′W | 15 September 2000 | D H Ahrenholz, leg.], [DNA sample ID: | NVG-19088C07 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01588932], and one red [HOLOTYPE ♂ | Paches (Paches) | loxeca Grishin]. Paratype: 1♀: NVG-19088C10 USNMENT_01588935 Peru: Piura, 3 km W Canchaque, 1300 m, −5.366667, −79.616667, 4-Jun-2000, R. Robbins and G Lamas, leg., genitalia NVG-22032F04 [USNM].
Type locality.
Ecuador: Morona-Santiago, 15 km S of Gualaquiza, elevation 800 m, GPS −3.460000, −78.551667.
Etymology.
The name is formed as lox[us from]Ec[u]a[dor] and is a noun in apposition.
Distribution.
Southern Ecuador and northern Peru.
Tribe Erynnini Brues and Carpenter, 1932
Clito congruens Grishin, new species https://zoobank.org/1F3053D3-130E-4382-894F-8C5A3EEE0D70 (Fig. 3 part, 89–90, 310–311)
Definition and diagnosis.
Phylogenetic analysis of specimens from Panama and Guatemala identified as Clito aberrans (Draudt, 1924) (type locality in Brazil: Amazonas, holotype sequenced as NVG-18093A08) reveals their genetic differentiation at the species level (Fig. 3): e.g., COI barcodes differ by 2% (13 bp). This species does not have an available name and is, therefore, new. This new species keys to “Clito clito” (which is C. aberrans) (E.52.3) in Evans (1953) and differs from its relatives by narrower lobe of ampulla that is twisted perpendicular to the plane of valva and extends inward, closely approaching the dorsal end of harpe with its distal margin; both this lobe over its entire surface and harpe distally are serrated (Fig. 310–311). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2388.3.1:G181A, aly6940.3.8:G84C, aly1689.4.2:T51C, aly1779.17.11:A168G, aly151.30.3:A30G, and COI barcode: T49C, A79A, T85T, C235C, T284T, T653C.
Barcode sequence of the holotype.
Sample NVG-14064A06, GenBank OR837663, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGATCAGGAATAGTGGGAACTTCCTTAAGTATATTAATTCGAACTGAATTAGGAAATCCTGGATCTTTAATTGGAGATGACCAAATTTATAATACTATTGTTACAGCTCATGCTTTCATTATAATTTTCTTTATAGTAATACCAATTATAATCGGAGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGAGCCCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGATTATTACCCCCTTCTTTAATATTATTAATTTCAAGTAGTATTGTAGAAAATGGTGCAGGAACAGGGTGAACTGTTTACCCCCCTTTATCTGCTAATATTGCCCATCAAGGATCTTCTGTAGATTTAGCTATTTTCTCATTACACTTAGCTGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACTACTATTATTAATATACGTGTTAGAAATTTATCATTTGATCAAATACCTTTATTTGTATGAGCAGTAGGTATTACTGCACTATTATTATTATTATCATTACCTGTTTTAGCTGGAGCTATTACAATACTTTTAACAGATCGAAATTTAAATACATCTTTTTTTGATCCAGCAGGAGGAGGAGATCCTATTTTATATCAACATCTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 89–90, bears the following three rectangular labels, two white: [PANAMA:CANAL ZONE | Gatun | 9° 17′W 79° 57′W | 10.XII.1978 | leg. G.B.Small], [DNA sample ID: | NVG-14064A06 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Clito congruens | Grishin]. Paratypes: 1♂ and 2♀♀ in [USNM]: 1♂ NVG-14064A08 Panama: Colon, 1000′, 6-Jan-1973, G. B. Small leg.; 2♀♀ Guatemala: Cayuga, around 1900, Schaus and Barnes collection: NVG-14064B06 May, genitalia No. X-6371 J. M. Burns 2006 and NVG-14064B07 April.
Type locality.
Panama: Colón Province, Gatún, GPS 9.2833, −79.9500.
Etymology.
In Latin, aberrans means wandering, straying, or deviating. The word congruens means agreeing, according, or consistent and is an antonym of aberrans. The name is a present active participle in the nominative case.
Distribution.
Currently known from Guatemala and Panama.
Cycloglypha corax Grishin, new species https://zoobank.org/D267A571-CE78-45DD-B5D4-1C5BB65E9556 (Fig. 3 part, 91–92, 312–314)
Definition and diagnosis.
Phylogenetic analysis of specimens from Southeast and Southern Brazil identified as Cycloglypha tisias (Godman and Salvin, 1896) (type locality in Costa Rica, Panama, and Brazil: Amazon Valley) reveals their genetic differentiation (Fig. 3): e.g., COI barcode difference of 2% (13 bp), and suggests that they are a new species. This new species keys to C. tisias (F.9.2) in Evans (1953) and differs from its relatives by typically stronger outlined and less diffuse postdiscal and subapical brown spots inside yellower posterior area of ventral hindwing (Fig. 92), dorsally directed tooth on right harpe narrower, harpe distal end stronger bent dorsad, and right valva with bigger and more robust, rhomboid-shaped process on ampulla (Fig. 312–314). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly3241.4.3:T126C, aly10226.17.12:C156T, aly1872.5.1:A228G, aly1405.12.15:C54T, aly1283.1.11:A26C, and COI barcode: T187C, A211G, A217G, T235T, T394C, A550G.
Barcode sequence of the holotype.
Sample NVG-19112H01, GenBank OR837664, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACTTCTTTAAGTTTATTAATTCGAACTGAATTAGGAAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACAATTGTTACAGCTCATGCTTTTATTATAATTTTTTTCATAGTTATACCAATTATAATCGGAGGATTCGGAAATTGATTAGTACCTTTAATGTTAGGGGCTCCTGATATAGCATTTCCACGAATAAATAATATAAGATTTTGATTATTACCCCCTTCATTAATATTATTAATTTCAAGAAGAATCGTAGAAAATGGAGCAGGTACAGGTTGAACAGTTTACCCCCCTCTTTCAGCTAATATTGCTCATCAAGGTTCTTCAGTAGATTTAGCTATCTTTTCATTACATTTAGCTGGAATTTCCTCAATTCTTGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAAAAACTTATCATTTGATCAAATACCTTTATTTGTTTGAGCAGTAGGTATTACAGCTTTACTTTTATTACTTTCTTTGCCTGTATTAGCAGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAATACATCTTTTTTTGATCCAGCTGGAGGAGGTGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 91–92, bears the following five rectangular labels, four white: [RIO de JAN | GUA, BRAZIL |13 Aug 64], [Collection of | Bryant Mather], [DNA sample ID: | NVG-19112H01 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01602089], and one red [HOLOTYPE ♂ | Cycloglypha | corax Grishin]. Paratypes: 2♂♂ from Brazil: NVG-19112G05, USNMENT_01602081 Rio de Janeiro, Itatiaia National Park, 800 m, GPS −22.450, −44.617, 22-Feb-1995, A. Caldas and students leg. [USNM]; and NVG-15092B05 Santa Catarina, Fazenda Alpina, nr. Joinville, 25-Feb-1985, J. Y. Miller leg. [MGCL].
Type locality.
Brazil: Rio de Janeiro, Guadalupe.
Etymology.
Tisias and Corax were founders of ancient Greek rhetoric, and Corax was Tisias’s teacher. Although Corax and Tisias might have been the same person (Wikipedia contributors 2023), C. corax is a species distinct from but sister to C. tisias. The name is a noun in apposition.
Distribution.
Southeast and Southern Brazil.
Festivia peruvia Grishin, new species https://zoobank.org/15FFE992-AC82-4640-864B-5C549EB970CD (Fig. 3 part, 93–94, 315–317)
Definition and diagnosis.
Genomic analysis reveals that a female from Tingo Maria, Peru, identified as Festivia grippa (Evans, 1953) (type locality in eastern Ecuador), is genetically differentiated from it (Fig. 4), e.g., COI barcode difference of 2.3% (15 bp) and because no published names apply to it, represents a new species. This new species keys to “Sostrata grippa” (E.42.5) in Evans (1953) and differs from its relatives by a combination of the following characters: forewing discal cell with one large upper hyaline spot (lower spot absent), the two segments of the hyaline spot in forewing cell CuA1-CuA2 are connected to each other at their bases on both dorsal and ventral sides, ventral forewing blue basal overscaling broader, extends from costa to cover discal cell, cell CuA2-1A+2A with a pale spot at 1/3 from its base, ventral hindwing with an apical blue spot and the dark brown streak in cell Sc+R1-RS is smaller and separated from vein Sc+R1 by blue (Fig. 93–94). Due to the cryptic nature of this species and unexplored phenotypic variation, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2700.10.9:G42A, aly300.20.2:G126A, aly116.12.4:G66T, aly2578.2.1:A22C, aly536.138.7:A319C, aly1260.2.1:T124T (not C), aly10226.27.3:A51A (not T), aly4523.3.2:C153C (not T), aly235.8.17:T150T (not C), aly10235.5.16:C81C (not T), and COI barcode: T139C, T287C, T319A, 514T, A526T, T619C.
Figure 4.

Phylogenetic trees of Hesperiina inferred from protein-coding regions in a) the Z chromosome and b) the mitochondrial genome (in segments). See Fig. 1 legend for other notations.
Barcode sequence of the holotype.
Sample NVG-18032A01, GenBank OR837665, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACCTCACTAAGAATATTAATTCGAACTGAATTAGGAAACCCCGGATCTTTAATTGGAGATGATCAAATTTATAACACTATTGTTACAGCTCATGCCTTTATTATAATTTTTTTCATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTCCCACTTATACTAGGAGCCCCTGATATAGCATTCCCCCGAATAAATAATATAAGATTTTGACTTTTACCCCCCTCTTTAATACTGCTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGTACTGGATGAACTGTTTACCCCCCTCTTTCTGCTAATATTGCTCACCAGGGCTCTTCTGTAGATTTAGCTATTTTTTCATTACATTTAGCTGGAATTTCATCAATTCTTGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAGAAATTTATCTTTTGATCAAATACCTTTATTTGTTTGAGCTGTAGGAATTACTGCATTATTATTATTACTTTCACTACCAGTATTGGCTGGTGCTATTACTATACTATTAACAGATCGAAATTTAAATACTTCCTTTTTTGATCCCGCAGGAGGAGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♀ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 93–94, bears the following four rectangular printed labels, three white: [PERU: Huanuco | Tingo Maria, 800 m. | May–June, 1994], [DNA sample ID: | NVG-18032A01 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01466114], and one red [HOLOTYPE ♀ | Festivia | peruvia Grishin].
Type locality.
Peru: Huánuco, Tingo Maria, elevation 800 m.
Etymology.
The name derives from the country of the type locality and is a feminine adjective.
Distribution.
Currently known only from the holotype collected in Peru.
Subfamily Hesperiinae Latreille, 1809 Tribe Hesperiini Latreille, 1809 Subtribe Hesperiina Latreille, 1809
Libra Evans, 1955 is a junior subjective synonym of Phemiades Hübner, [1819]
Genomic sequencing of representative species, including the type species of both genera, reveals that Libra Evans, 1955 (type species Augiades aligula Schaus, 1902) originates within Phemiades Hübner, [1819] (type species Phemiades pseudophineus de Jong, 1983) and is not monophyletic in the Z chromosome tree (Fig. 4). The two genera are genetically close to each other: e.g., COI barcodes of their type species differ by 7% (46 bp). Therefore, we propose that Libra Evans, 1955 is a junior subjective synonym of Phemiades Hübner, [1819].
Decinea onasima (Hewitson, 1877) with Cobalus boliviensis E. Bell, 1930 as its subspecies and Decinea formosus (Hayward, 1940) are species distinct from Decinea dama (Herrich-Schäffer, 1869)
Genomic analysis of primary type specimens of the following four taxa: Hesperia onasima Hewitson, 1877 (type locality in Brazil: Rio de Janeiro, syntype sequenced as NVG-18043C12), Cobalus boliviensis E. Bell, 1930 (type locality in Bolivia, holotype sequenced as NVG-18025E07), Cobalus formosus Hayward, 1940 (type locality in Ecuador, holotype sequenced as NVG-18025H07) and Cobalus dama Herrich-Schäffer, 1869 (type locality not specified, syntypes sequenced as NVG-15036H04 and NVG-18042G04), the former three of which are currently regarded as junior subjective synonyms of the latter, placed in the genus Decinea Evans, 1955 (type species Hesperia decinea Hewitson, 1876), reveals that they represent three genetically differentiated non-sister lineages (Fig. 4). COI barcodes of D. dama differ from either H. onasima or C. formosus by 7.9% (52 bp). However, H. onasima and C. boliviensis are more closely related to each other (Fig. 4). Therefore, we propose to reinstate Decinea onasima (Hewitson, 1877) and Decinea formosus (Hayward, 1940) as species-level taxa and, for the time being, pending further studies, treat Decinea onasima boliviensis (E. Bell, 1930), new combination, as a subspecies.
Decinea notata Grishin, new species https://zoobank.org/D0018528-567A-43E6-8476-17D4BD2DFE8E (Fig. 4 part, 95–96, 318–319)
Definition and diagnosis.
Phylogenetic analysis of specimens from Ecuador identified as Decinea lydora lyco (Mabille, 1878) (type locality in Peru) reveals that they are not monophyletic, and the former specimens represent a most prominently differentiated new species without close relatives (Fig. 4). COI barcodes of the new species differ from D. lydora lyco and D. dama by 8.5% (56 bp) and 10% (66 bp), respectively. The new species keys to “D. neroides lyco” (currently D. lydora lyco) (L.11.5(b)) in Evans (1955) but differs from it by the abdomen being white beneath (not brown) and forewing with three well-defined subapical dots in a row. These dots are smaller, and some may be missing in Decinea lydora (Plötz, 1882) (type locality in Venezuela) subspecies and junior subjective synonyms. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly499.50.4:G45C, aly1656.33.2:A51T, aly291.13.2:G39T, aly291.13.2:G66A, aly2631.9.19:G72A, and COI barcode: T121C, T145C, T250C, T436C, T589C, T461C.
Barcode sequence of the holotype.
Sample NVG-18118B04, GenBank OR837666, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCCTTAAGTTTACTAATTCGTACAGAATTAGGTAGACCTGGATCTTTAATTGGAGATGATCAAATTTATAATACCATCGTAACAGCTCATGCTTTTATCATAATTTTTTTCATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCCTTAATATTAGGAGCTCCTGATATAGCCTTCCCTCGAATAAATAACATAAGATTTTGAATACTACCCCCTTCTTTAACCTTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACTGGTTGAACAGTTTACCCCCCTTTATCATCAAATATTGCTCACCAAGGATCTTCAGTTGATTTAGCTATTTTTTCTCTTCATTTAGCTGGTATTTCTTCTATTTTAGGAGCCATTAATTTTATTACTACAATTATTAACATACGAATTAAAAATCTATCATTTGATCAAATACCACTATTTGTTTGATCAGTAGGAATTACAGCTTTATTATTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACTATACTTCTTACTGACCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGTGGAGGAGACCCTATTCTTTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 95–96, bears the following five rectangular labels, four white: [ECUADOR:Napo Prov | 9 km E Puerto Napo | 1° 03′S 77° 44′W | 600m 20 Sept 1990 | S S Nicolay], [♂ genitalia | slide/vial # | H1076 | Prep. S.S. Nicolay], [DNA sample ID: | NVG-18118B04 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01531768], and one red [HOLOTYPE ♂ | Decinea | notata Grishin]. Paratype: 1♂ NVG-18091E07 Ecuador: Morona-Santiago, Mendez, 800 m, GPS −2.42, −78.20, 10-Nov-2012, J.-C. Petit leg. [EBrockmann].
Type locality.
Ecuador: Napo Province, 9 km E of Puerto Napo, elevation 600 m, GPS −1.05, −77.73.
Etymology.
In Latin, notatus means marked, written, signified, noted, or distinguished. The name reflects the prominent subapical dots and a white abdomen below. It also reflects the prominent phylogenetic position separate from all others. The name is a feminine perfect passive participle in the nominative singular.
Distribution.
Ecuador.
Pompeius fuscus Grishin, new species https://zoobank.org/A3D335E7-A11D-4361-B8C1-A9BD1924462E (Fig. 4 part, 97–98, 320–321)
Definition and diagnosis.
Phylogenetic analysis of specimens from Brazil identified as Pompeius amblyspila (Mabille, 1898) (type locality in Bolivia) reveals that they are not monophyletic with and strongly differentiated genetically from it (Fig. 4): e.g., their COI barcodes differ by 4.2% (28 bp) and therefore represent a new species. Inspection of (and sequencing of two, Fig. 4) of primary type specimens of taxa placed as junior subjective synonyms of P. amblyspila confirms their synonymy and distinctness of the new species from Brazil. This new species keys to P. amblyspila (M.15.4) in Evans (1955) but differs from it and other relatives by rounder wings, narrower stigma, pale spots in forewing cells CuA1-CuA2 and M3-CuA1 being rounder and placed farther distad from stigma, hindwing pale spots smaller and in a more even arc closer to the margin, and veins ventrally with pale overscaling towards outer margin. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly320.4.1:T75C, aly1838.7.1:T387C, aly1838.7.1:T2038C, aly1651.4.1:A404G, aly525.109.4:G81A, and COI barcode: T19C, T139C, T340C, T376A, T472C, T541C.
Barcode sequence of the holotype.
Sample NVG-19093D05, GenBank OR837667, 658 base pairs:
AACTTTATATTTTATTTTCGGTATTTGAGCAGGAATATTAGGAACTTCTTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCTGGTTCTTTAATTGGAGATGATCAAATTTTTAATACTATTGTTACAGCTCATGCCTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCTCCAGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGAATATTACCACCATCATTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACTGGATGAACAGTTTACCCCCCTTTATCTTCTAATATTGCTCACCAAGGATCATCTGTTGATTTAGCAATTTTTTCCTTACATTTAGCTGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATCAAAAATATATCATTTGATCAAATACCATTATTTGTTTGATCTGTTGGAATTACAGCTTTATTATTACTCTTATCTTTACCTGTTTTAGCTGGAGCTATTACTATACTTCTTACAGATCGAAATTTAAATACTTCTTTTTTTGATCCAGCAGGAGGAGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 97–98, bears the following four rectangular labels, three white: [BRAZIL:MG 1250m | Serra do Cipo | 19°16′S 43°35′W | 17 Apr 1991 | Robbins & Becker], [DNA sample ID: | NVG-19093D05 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01589476], and one red [HOLOTYPE ♂ | Pompeius | fuscus Grishin]. Paratypes: 2♂♂ from Brazil in USNM: NVG-18115B11 the same data as the holotype, except 18-Apr-1991 and NVG-19093D03 Mato Grosso, Diamantino, Alto Rio Arinos, 350–400 m, GPS −14.2167, −56.2000, 7-Feb-1986, E. Furtado.
Type locality.
Brazil: Minas Gerais, Serra do Cipo, elevation 1250 m, GPS −19.2667, −43.5833.
Etymology.
In Latin, fuscus means dark, muffled, dingy, brown, dusky, or tawny. Moreover, the pattern of paler yellow-brown spots and overscaling along veins in this species reminds us of the pattern observed in Anthrenus fuscus Olivier, 1789 (Coleoptera). The name is a masculine adjective.
Distribution.
Recorded from the states of Mato Grosso and Minas Gerais in Brazil.
Vernia clara Grishin, new species https://zoobank.org/D20A8C16-E197-4F52-806F-01921EE16C07 (Fig. 4 part, 99–100, 322–323)
Definition and diagnosis.
Genomic analysis of specimens from Mexico and Panama identified as Vernia dares (Plötz, 1883) (type locality not specified) reveals that they differ significantly from the South American V. dares (Fig. 4): e.g., COI barcodes differ by 5.8% (38 bp) and, therefore, this complex consists of at least two species. Although we were not able to locate primary type specimens, according to the original description (Plötz 1883) and the illustration in Draudt (1921–1924), which probably is a copy of an unpublished drawing by Plötz, V. dares is of a darker phenotype that corresponds to South American specimens. Therefore, North and Central American specimens that are characterized by more extensive yellow-orange coloration represent a new species. This new species keys to “Pompeius dares” (M.15.5) in Evans (1955), but differs from it by larger yellow-orange spots, more developed yellow-orange overscaling, and less contrasting dark spots on the ventral hindwing. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2487.24.2:T675G, aly2487.24.2:T765C, aly536.138.7:T78C, aly82.18.3:T303C, aly82.18.3:G321A, and COI barcode: T5C, 31C, T46C, T193C, A283T, T536C.
Barcode sequence of the holotype.
Sample NVG-22035A08, GenBank OR837668, 658 base pairs:
AACTCTATATTTTATTTTCGGTATTTGAGCCGGTATATTAGGAACCTCTCTAAGTTTATTAATTCGAACAGAATTAGGTAATCCTGGTTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGGTTTGGAAACTGATTAGTCCCATTAATATTAGGAGCCCCTGACATAGCTTTCCCACGAATAAATAACATAAGATTTTGAATATTACCCCCATCTTTAACTCTTTTAATCTCAAGTAGAATTGTAGAAAATGGTGCAGGAACAGGTTGAACTGTTTATCCTCCTTTATCATCAAATATTGCTCACCAAGGATCTTCTGTTGATTTAGCAATTTTTTCTCTTCATTTAGCTGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAAAAACTTATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACAGCTTTATTACTACTTTTATCTTTACCTGTTTTAGCTGGAGCTATTACAATATTACTTACTGATCGAAATTTAAACACTTCATTTTTTGATCCTGCTGGAGGAGGAGATCCAATTCTTTACCAACATCTTTTC
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 99–100, bears the following three rectangular labels, two white: [Panama:Chiriqui | Santa Cruz | 10.VIII.1975 | G.B. Small], [DNA sample ID: | NVG-22035A08 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Vernia | clara Grishin]. Paratypes: 2♂♂ and 3♀♀: Mexico: 1♂ NVG-22102C01, CASENT 8566973 Nayarit, 20 air mi NE of San Blas, 23-Dec-1971, C. D. MacNeill leg. [CAS]; 1♀ NVG-18021D01 San Luis Potosi, Valles, 10-Jun-1966, H. A. Freeman leg. [AMNH]; Chiapas: 1♂ NVG-18115C04, USNMENT_01531564 35 mi W of Tuxtla Gutierrez, 16-Aug-1972, G. F. and S. Hevel leg. [USNM]; 1♀ NVG-21044A02 3 mi S of Simojovel, 3000′, 22-Jun-1989, J. Kemner leg., genitalia SRS-4469 [MGCL] and Panama: 1♀ NVG-18115C05, USNMENT_01531565 Panama, Cerro Campana, 2000′, GPS 8.6833, −79.9167, 9-Mar-1963, G. B. Small leg. [USNM].
Type locality.
Panama: Chiriquí Province, Santa Cruz.
Etymology.
The name clara, which is Latin for bright and clear, is given for the brighter colors of this species. The name is an adjective.
Distribution.
From Mexico to Panama.
Oligoria (Oligoria) obtena Grishin, new species https://zoobank.org/74FCABDE-324D-4C60-A191-BD4C1E8F1B3B (Fig. 4 part, 101–102, 324–327)
Definition and diagnosis.
Phylogenetic analysis of specimens from Ecuador identified as Oligoria lucifer (Hübner, [1831]) (type locality in Suriname) reveals that they are not monophyletic with it and instead are sister to both Oligoria maculata (W. H. Edwards, 1865) (type locality in USA: Louisiana) and Oligoria percosius (Godman, 1900) (type locality in Mexico, Guatemala, and Panama), being genetically differentiated from them (Fig. 4): e.g., COI barcodes of Ecuadorian specimens differ from O. maculata and O. percosius by 5.3% (35bp) and 4.6% (30 bp), respectively, and therefore they represent a new species. This new species keys to “Decinea lucifer” (L.11.8) in Evans (1955) but differs from its relatives by a small spot and reduced pale overscaling around it in ventral forewing cell CuA2-1A+2A. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly3446.8.5:C48T, aly3446.8.5:T87C, aly127.87.2:C51T, aly127.87.2:G75A, aly903.2.14:T860C, and COI barcode: A166G, T277A, T530C, T553A, A628T.
Barcode sequence of the holotype.
Sample NVG-18118B05, GenBank OR837669, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCTTTAAGATTATTAATTCGTACAGAATTAGGTAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGAGGATTCGGAAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGAATGTTACCCCCCTCATTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCAGGAACTGGTTGAACAGTTTATCCTCCTTTATCTTCTAATATTGCCCACCAAGGATCTTCTGTTGATTTAGCAATTTTTTCCCTTCATTTAGCTGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAAAAATTTATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGTATTACTGCTCTATTATTACTTTTATCTTTACCAGTTTTAGCTGGAGCTATTACTATACTACTTACTGATCGAAATCTTAATACCTCATTTTTTGATCCAGCAGGAGGTGGTGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 101–102, bears the following five rectangular labels, four white: [ECUADOR: Napo Prov | 4 km Tena-Pano Rd | 1° 02′S, 77 ° 50′W | 28 Sep 1990 600 m | DH Ahrenholz leg | SS Nicolay curator], [Decinea | lucifer ♂ | Det. Hbn | S.S. Nicolay], [DNA sample ID: | NVG-18118B05 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01531769], and one red [HOLOTYPE ♂ | Oligoria (Oligoria) | obtena Grishin]. Paratype: 1♂ NVG-18118B07, USNMENT_01531771 Ecuador: Napo, Jatun Sacha Biological Reserve, GPS −1.0667, −77.6000, 30-Sep-1991, D. H. Ahrenholz leg., genitalia H1096 (Fig. 324–325) [USNM].
Type locality.
Ecuador: Napo Province, km 4 of Tena-Pano Rd., elevation 600 m, GPS −1.033, −77.833.
Etymology.
In Latin, obtenebro means darken, make dark, obscure, or conceal. The name is given for the reduced pale scaling near the ventral forewing tornus compared to its congeners and is a noun in apposition.
Distribution.
Currently known only from the Napo Province in Ecuador.
Thespieus guerreronis (Dyar, 1913) is a species distinct from Thespieus dalman (Latreille, [1824])
Genomic sequencing of Thespieus dalmani [sic] guerreronis Dyar, 1913 (type locality in Mexico: Guerrero, syntypes sequenced as NVG-18113F03 and NVG-18113F04), currently a junior subjective synonym of Thespieus dalman (Latreille, [1824]) (type locality in Brazil, lectotype sequenced as NVG-18078F01) reveals prominent genetic differentiation between these two taxa (Fig. 4): e.g., their COI barcodes differ by 4.3% (28 bp). Therefore, we propose to treat Thespieus guerreronis Dyar, 1913, new status, as a species.
Thespieus mandal Grishin, new species https://zoobank.org/BA7D773F-A815-4251-B153-DC7B3CAC5B13 (Fig. 4 part, 103–104, 328–329)
Definition and diagnosis.
Phylogenetic analysis of a specimen from Rio de Janeiro, Brazil, identified as Thespieus dalman (Latreille, [1824]) (type locality in Brazil, lectotype sequenced as NVG-18078F01) reveals its strong genetic differentiation (Fig. 4): e.g., COI barcode difference of 3.8% (25 bp) from T. dalman, and therefore it represents a new species. This new species keys to T. dalman (O.7.4) in Evans (1955) but differs from it by a generally narrower hyaline spot in the forewing discal cell but wider spot in the hindwing cell CuA1-CuA2, which is framed by a smaller and paler brown spot at the base of this cell on the ventral side, which has no brown discal dash in cell Sc+R1-RS, and exhibits paler basal area in cell 1A+2A-3A (Fig. 103–104), harpe wider, protrudes dorsad farther than ampulla, more concave along the distal margin, saccus stronger bowed ventrad (Fig. 328–329). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly168.8.1:G313A, aly2517.1.2:T285C, aly3905.3.3:A99C, aly971.11.1:A75G, aly3268.8.1:T840C, aly10226.17.4:A204A (not C), aly320.37.1:G189G (not A), aly1656.14.1:C411C (not T), aly594.20.16:C67C (not A), aly1651.3.5:A72A (not T), and COI barcode: T50C, T133C, T340C, T352C, A379C, T640C.
Barcode sequence of the holotype.
Sample NVG-18012A11, GenBank OR837670, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCACTAAGATTACTAATTCGTACAGAATTAGGTAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGAATATTACCTCCCTCTTTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACTGGATGAACAGTTTACCCCCCCTTATCCTCTAATATTGCTCATCAAGGATCTTCCGTAGATTTAGCAATTTTCTCACTTCATTTAGCTGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAAAAATTTATCATTTGATCAAATACCTTTATTTGTATGATCTGTAGGTATTACAGCTTTATTATTACTTTTATCTTTACCTGTATTAGCTGGTGCTATTACCATATTATTAACAGATCGAAATTTAAATACTTCATTCTTTGACCCTGCAGGAGGAGGAGATCCAATCTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 103–104, bears the following six rectangular labels, five white: [BRAZIL, RJ, P. N. de | Itatiaia, 800m | 22°27′S, 44°37′W | 7Apr 1995 | Diversity Project UERJ], [Thespieus | dalman], [no transect], [DNA sample ID: | NVG-18012A11 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450264], and one red [HOLOTYPE ♂ | Thespieus | mandal Grishin].
Type locality.
Brazil: Rio de Janeiro, Itatiaia National Park, elevation 800 m, GPS −22.450, −44.617.
Etymology.
The name reverses syllables in its sister species, T. dalman. The name is a noun in apposition.
Distribution.
Southeast Brazil.
Subtribe Moncina A. Warren, 2008
Psoralis (Saniba) magnamacus Grishin, new species https://zoobank.org/EE8B803C-DB04-4B78-9732-F18EE4B7007A (Fig. 5 part, 105–106, 330–333)
Figure 5.

Phylogenetic trees of Moncina (part 1) inferred from protein-coding regions in a) the Z chromosome and b) the mitochondrial genome (in segments). See Fig. 1 legend for other notations.
Definition and diagnosis.
A Hesperiidae specimen from Panama stood out due to its unusual appearance, with large hyaline spots on the forewing. Genomic sequencing placed it in subgenus Saniba Mielke and Casagrande, 2003 (type species Hesperia sabina Plötz, 1882) of the genus Psoralis Mabille, 1904 (type species Psoralis sabaeus Mabille, 1904, which is a junior subjective synonym of Pamphila idee Weeks, 1901) (Fig. 5), an affinity not immediately apparent for this recognizably new species. In the pattern of forewing, this new species resembles Psoralis (Saniba) laska (Evans, 1955) (type locality in Brazil: Mato Grosso) but the hyaline spots are larger, e.g., a spot occupies more than a third of the cell M3-CuA1, and about a third of the cell CuA1-CuA2. Only two, not three, subapical hyaline spots are present in the holotype, those in cells R5-M1 and R4-R5. The ventral side is rusty-brown, not variegated, and much more uniformly colored than in P. laska: cream-colored area occupies more than a third of forewing cell CuA2-1A+2A, hindwing with traces of postdiscal pale spots. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1038.7.10:T45A, aly1038.7.10:A129G, aly322.26.7:T472G, aly1139.29.1:T21C, aly1139.29.1:T297A, aly3712.7.1:T57T (not C), aly3712.7.1:T117T (not C), aly390.23.4:T282T (not C), aly390.23.4:C397C (not T), aly1656.10.5:C136C (not T), and COI barcode: T46C, T202C, T212C, 352A, T533C, T568A.
Barcode sequence of the holotype.
Sample NVG-19023H12, GenBank OR837671, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACCTCATTAAGTTTATTAATTCGAACAGAATTAGGTAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATCGGAGGATTTGGAAATTGATTAGTCCCTTTAATACTAGGAGCTCCTGATATAGCATTTCCACGAATAAATAATATAAGATTTTGAATATTACCCCCTTCATTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGTTGAACAGTATACCCCCCTTTATCATCTAATATTGCTCACCAAGGAGCTTCTGTTGATTTAGCAATTTTTTCATTACATTTAGCAGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATCACTACAATTATTAATATACGAATTAGAAATTTATCATTTGATCAAATACCTTTATTTGTTTGATCTGTTGGTATTACAGCTTTACTACTTCTTTTATCTTTACCTGTTTTAGCAGGTGCAATTACAATACTACTTACAGATCGAAATCTAAATACTTCATTCTTTGATCCAGCTGGAGGAGGAGATCCTATTCTTTACCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 105–106, bears the following four rectangular labels, three white: [PANAMA: Darien | Cana 1200m | 22.IX.1982 | Leg. G. B. Small], [DNA sample ID: | NVG-19023H12 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532887], and one red [HOLOTYPE ♀ | Psoralis (Saniba) | magnamacus Grishin].
Type locality.
Panama: Darien Province, Cana, elevation 1200 m.
Etymology.
The name is given for the big macules on the forewing. In Latin, magnae maculae means large spots. The name is a noun in apposition.
Distribution.
This species is known only from the holotype collected in Panama.
Alychna ayonis Grishin, new species https://zoobank.org/050EC67D-FC69-4005-B871-A861C7D74835 (Fig. 5 part, 107–108, 334–335)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Ecuador and Peru identified as Alychna exclamationis (Mabille, 1898) (type locality in Bolivia, lectotype sequenced as NVG-18042G08) show prominent genetic differentiation from it while being its sister (Fig. 5): e.g., their COI barcodes differ by 1.5% (10 bp) and, therefore, represent a new species, more distantly related to Alychna zenus (E. Bell, 1942) (type locality in Ecuador) (COI barcodes differ by 3.5%, 23 bp). This new species keys to “Psoralis exclamationis” (J.43.3) in Evans (1955) but differs from it by narrower stigma, narrower white spots and streaks on forewing, a semi-circle of discal pale dots on ventral hindwing (Fig. 107–108), undivided uncus, harpe strongly upcurved and protruding beyond ampulla dorsad, terminally rounded in lateral view and plate-like in dorsal view, costa-ampulla with a bump closer to ampulla, concave on both sides of the bump (Fig. 334–335). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly420.62.5:G73T, aly420.62.5:G84A, aly2041.8.2:A195C, aly2012.47.1:G1269C, aly25.10.1:G48C, and COI barcode: A85T, T163A, T169A, T205A, A553C, A631G.
Barcode sequence of the holotype.
Sample NVG-22035H07, GenBank OR837672, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGTATACTAGGAACTTCATTAAGTTTATTAATTCGTACAGAATTAGGAAATCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATATTACCCCCTTCCTTAATATTATTAATTTCAAGAAGAATTGTTGAAAATGGTGCAGGTACTGGATGAACTGTTTATCCCCCCCTTTCTTCTAATATCGCACACCAAGGATCATCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCTGGGATCTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAATATATCCTTTGATCAAATACCTTTATTTGTATGATCTGTAGGAATTACAGCTTTATTATTACTTTTATCTTTACCCGTATTAGCTGGAGCTATTACAATACTTTTAACTGATCGAAACTTAAATACTTCTTTTTTTGATCCAGCTGGAGGAGGGGACCCTATCTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 107–108, bears the following three rectangular labels, two white: [ECUADOR Napo | Papallacta 2800m | 23 Sept. ‘87 | S. S. Nicolay], [DNA sample ID: | NVG-22035H07 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Alychna | ayonis Grishin]. Paratype: 1♂ NVG-19021H01 the same data as the holotype but collected on 17-Sep-1987, genitalia H962 by S. S. Nicolay [USNM].
Type locality.
Ecuador: Napo Province, Papallacta, elevation 2800 m.
Etymology.
The name is given for the “i” on the forewing. A close relative of this species is named exclamati-onis, probably for the exclamation mark pattern on the forewing. In the new species, the exclamation mark is thinner and resembles the letter “i” [ay]. The name is treated as a noun in apposition.
Distribution.
Currently known only from the type locality in Napo, Ecuador.
Wahydra banios Grishin, new species https://zoobank.org/2BB146AF-6818-4929-AAB0-CE36EBEC299B (Fig. 5 part, 109–110, 336–338)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Ecuador identified as Wahydra subhebetis Steinhauser, 1991 (type locality in Colombia, holotype sequenced as NVG-15039G08) shows prominent genetic differentiation from it (Fig. 5): e.g., their COI barcodes differ by 2.4% (16 bp), and therefore represents a new species. This new species phenotypically is very similar to W. subhebetis but differs from it by an even narrower and longer harpe separated from the ampulla by a larger notch, harpe projecting dorsad and terminally narrowing to a serrated and rounded point, larger gnathos (Fig. 336–338). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1042.30.1:G732A, aly1042.30.1:G747C, aly727.26.22:G33A, aly727.26.22:A38G, aly208.41.1:T54C, aly822.30.12:T1050T (not A), aly822.30.12:A1055A (not G), aly536.122.6:G99G (not C), aly536.122.6:C105C (not T), aly1018.14.4:C54C (not T), and COI barcode: T5C, A100G, A166G, 337A, T460C.
Barcode sequence of the holotype.
Sample NVG-7990, GenBank OR837673, 658 base pairs:
AACTCTATATTTTATTTTTGGTATTTGAGCAGGAATGTTAGGAACTTCCCTAAGTTTACTTATTCGTACAGAATTAGGTAATCCAGGATCCTTAATTGGCGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTCTTCATAGTTATGCCAATTATAATTGGAGGATTCGGAAATTGATTAGTTCCTTTAATATTAGGAGCACCTGATATAGCCTTCCCACGTATAAATAATATAAGATTTTGAATATTACCTCCTTCCTTAATATTATTAATTTCTAGAAGAATTGTAGAAAATGGTGCAGGGACTGGTTGAACAGTATATCCCCCCCTTTCATCTAATATTGCACATCAAGGATCATCTGTTGATTTAGCAATTTTTTCTCTTCATTTAGCAGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATCAATATACGAATTAAAAATATATCATTTGATCAAATACCTTTATTTGTATGATCAGTAGGAATTACAGCTTTACTACTACTTTTATCTTTACCAGTATTAGCTGGAGCTATTACAATACTTCTAACTGATCGAAATTTAAATACATCTTTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 109–110, bears the following five rectangular labels, four white: [ECUADOR: Tungurahua: | Baños 3000 m. | XI.2001 I.Aldas leg.], [DNA sample ID: | NVG-7990 | c/o Nick V. Grishin], [genitalia| NVG170207–75 | Nick V. Grishin], [USNMENT | {QR Code} | 01321830], and one red [HOLOTYPE ♂ | Wahydra | banios Grishin].
Type locality.
Ecuador: Tungurahua Province, Baños, elevation 3000 m.
Etymology.
The name is given for the type locality and is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Baños, Ecuador.
Wahydra cuzcona Grishin, new species https://zoobank.org/5C35C09C-FEAE-43D3-A1FD-874FF72F0803 (Fig. 5 part, 111–112, 339–341)
Definition and diagnosis.
Phylogenetic trees reveal that a unique-looking specimen of Wahydra Steinhauser, 1991 (type species Pamphila kenava Butler, 1870) from Cuzco, Peru, belongs to the W. kenava group but does not show any clear affinities to a particular species, and therefore is new. This new species is genetically distant from all others; e.g., its COI barcode differs from that of W. kenava (type locality in Venezuela) by 7.3% (48 bp). This new species is diagnosed by an unique (for Wahydra) wing pattern: forewing orange spots are narrower, the cell CuA2-1A+2A is with one small elongated spot, situated along vein 1A+2A, the spot in cell CuA1-CuA2 is rhomboidal and the spot in cell M3-CuA1 is separated from it by dark vein and shifted distad, overlapping with it by less than half of the width, the spot in cell R5-M1 is the smallest, slightly elongated; hindwing with a cluster of four spots in the subapical area, separated by dark veins: the spot between veins M1 and M3 is the largest, about a third of wing’s length, other spots are less than half of this spot in length, those in cells RS-M1 and M3-CuA1 are aligned to its distal margin (to follow the curvature of the outer wing margin) and the spot in the cell CuA1-CuA2 is aligned to the center of the M1-M3 spot, thus shifted basad from the spot in cell M3-CuA1; ventral side of wings is similar to Wahydra thisbe (Hayward, 1942) (type locality in Ecuador) in lacking the paler ray anteriad of the dark wing section by inner margin, but the forewing yellow spots are smaller. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly3176.7.1:A170G, aly770.23.1:A79G, aly770.23.1:A93G, aly276634.3.2:C125A, aly536.138.1:T18C, aly159.13.1:A378A (not G), aly276634.3.2:C140C (not T), aly50.26.1:C33C (not G), aly587.20.1:T1440T (not A), aly499.36.22:G111G (not A), and COI barcode: T38C, T106C, T127C, T259C, T349C, A514C.
Barcode sequence of the holotype.
Sample NVG-7989, GenBank OR837674, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGGGCAGGAATACTAGGAACTTCTTTAAGTTTATTAATTCGTACAGAATTAGGTAATCCAGGATCTTTAATTGGAGATGACCAAATTTATAATACTATTGTCACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGTGCACCTGATATAGCTTTTCCACGAATAAATAATATAAGATTCTGAATATTACCTCCTTCCTTAATACTTCTAATTTCTAGTAGAGTCGTAGAAAATGGTGCAGGGACTGGTTGAACAGTTTACCCCCCCCTCTCATCTAATATTGCACATCAAGGAGCATCTGTCGATTTAGCAATTTTTTCTCTTCATTTAGCAGGAATTTCCTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATATATCATTTGATCAAATACCCTTATTTGTATGATCCGTAGGAATTACAGCTTTACTTTTACTTTTATCTTTACCAGTATTAGCTGGAGCTATTACAATACTTTTAACAGATCGAAATTTAAATACATCTTTTTTTGATCCAGCAGGAGGAGGAGATCCAATCCTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 111–112, bears the following five rectangular labels, four white: [PERU: Cusco | Qda. Morro Leguia | 13°08′ 71°33′ | 29 Aug 1989, 2150m | Leg. R. Robbins], [DNA sample ID: | NVG-7989 | c/o Nick V. Grishin], [genitalia| NVG170207–74 | Nick V. Grishin], [USNMENT | {QR Code} | 01321829], and one red [HOLOTYPE ♂ | Wahydra | cuzcona Grishin].
Type locality.
Peru: Cuzco Region, Cosñipata Valley, Quebrada Moro Leguia, elevation 2150 m, GPS −13.133, −71.550.
Etymology.
The name is given for the type locality and is a noun in apposition.
Distribution.
Currently known only from the holotype collected in the Cosñipata Valley, Peru.
Cynea (Cynea) aureofimbra Grishin, new species https://zoobank.org/E2C5918D-5713-48C4-B171-FACD197E03B7 (Fig. 5 part, 113–114, 342–344)
Definition and diagnosis.
Phylogenetic analysis reveals that an unspotted brown specimen of Cynea Evans, 1955 (type species Hesperia cynea Hewitson, 1876) with unique orange fringes is sister to a group of species closely related to Cynea cynea (type locality in Venezuela) but is not genetically close to any of these species (Fig. 5): e.g., its COI barcode differs from that of C. cynea by 3.8% (25 bp), and therefore is a new species. This new species is recognizable by its uniformly dark brown wings on both sides, the forewing tornal area ventrally only very slightly paler than the rest of the wing due to sparse overscaling and without spots, and orange fringes on both wings (except apical areas). This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly5294.26.2:A30G, aly5294.26.2:T183A, aly638.8.1:A318G, aly18826.6.2:A171G, aly18826.6.2:T180G, aly12063.12.6:A81A (not G), aly8211.10.1:A693A (not G), aly3277.16.1:C533C (not T), aly3277.16.1:C1022C (not T), aly37338.4.2:A48A (not C), and COI barcode: A81C, G200A, A211G, T358C, 484C, A514T.
Barcode sequence of the holotype.
Sample NVG-18119D08, GenBank OR837675, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACTTCATTAAGATTATTAATTCGTACAGAATTAGGTACCCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAATTCCTTTAATGTTAGGAGCCCCTGATATAGCCTTCCCTCGAATAAATAACATAAGATTTTGAATACTTCCCCCCTCATTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACTGGATGAACAGTTTATCCTCCTTTATCTTCTAACATTGCTCACCAAGGATCCTCAGTTGATTTAGCAATTTTTTCTCTTCATTTAGCAGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATTACAACAATCATTAATATACGAATTAAAAATTTAACCTTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACTGCTCTTTTATTACTTTTATCTTTACCAGTATTAGCTGGAGCTATTACAATACTTTTAACAGATCGAAATTTAAATACTTCTTTTTTTGATCCTGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 113–114, bears the following six rectangular labels, five white: [Old Sto. Domingo | Rd. 1800 m ECUADOR | 7 Oct. ‘73 | S.S. Nicolay], [♂ genitalia | slide/vial # | H600 | Prep. S.S. Nicolay], [Cynea sps. | nr. megalops | Det. | S.S. Nicolay], [DNA sample ID: | NVG-18119D08 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01531869], and one red [HOLOTYPE ♂ | Cynea aureofimbra | Grishin].
Type locality.
Ecuador: Pichincha/Santo Domingo de los Tsáchilas Province, Old Santo Domingo road, elevation 1800 m.
Etymology.
In Latin, aurea fimbria means golden fringe. The name reflects the golden-orange fringe on the wings of this species and is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Ecuador.
Cynea (Nycea) erebina (Möschler, 1879) and Cynea (Nycea) cleochares (Mabille, 1891) are species distinct from Cynea (Cynea) diluta (Herrich-Schäffer, 1869)
Phylogenetic analysis of Cynea Evans, 1955 (type species Hesperia cynea Hewitson, 1876) reveals that Carystus erebina Möschler, 1879 (type locality in Colombia, holotype sequenced as NVG-21118B08) and Pamphila cleochares Mabille, 1891 (type locality Venezuela: Valera, syntype sequenced as NVG-15036G08) currently regarded as junior subjective synonyms of Cynea (Cynea) diluta (Herrich-Schäffer, 1869) (type locality not specified) are in a clade (and subgenus) different from a specimen from Guyana identified as C. diluta and the holotype of Proteides osembo Möschler, 1883 (type locality in Suriname), currently treated as a junior subjective synonym of C. diluta (Fig. 5) (Zhang et al. 2022b). The two taxa C. erebina and P. cleochares belong to the subgenus Nycea Grishin, 2022 (type species Pamphila hycsos Mabille, 1891), differ from each other by 3.5% (23 bp) in COI barcodes and differ from other species of Nycea (Fig. 5). Therefore, we reinstate them as species: Cynea (Nycea) erebina (Möschler, 1879) and Cynea (Nycea) cleochares (Mabille, 1891).
Pamphila hycsos Mabille, 1891 is a junior subjective synonym of Cynea (Nycea) erebina (Möschler, 1879), with lectotype designation
Genomic sequencing of the primary type specimens of Carystus erebina Möschler, 1879 (holotype sequenced as NVG-21118B08) and Pamphila hycsos Mabille, 1891 (syntype sequenced as NVG-18043B08), both from Colombia, reveals their genetic similarity (COI barcodes differ by 0.3%, 2 bp). The two type specimens are phenotypically similar and are from the same country of origin. Therefore, we propose that Pamphila hycsos Mabille, 1891, is a new junior subjective synonym of Cynea (Nycea) erebina (Möschler, 1879). As a result, Cynea (Nycea) erebina somba Evans, 1955 is a new combination, originally described and maintained as Cynea hycsos somba Evans, 1955. To stabilize these treatments, N.V.G. hereby designates a male syntype in the Museum für Naturkunde, Berlin, Germany, that bears the following nine rectangular labels (the first three are purple, blue, and red, respectively, others are white): [Origin], [Columb], [Pa. hycsos | ♂ Mab.], [Coll. Sommer], [Coll. | Staudinger], [Hÿcsos | Mab.], [GEN.PEP., | MIELKE | 1996], [{OR Code} http://coll.mfn-berlin.de/u/ | 44a043], [DNA sample ID: | NVG-18043B08 | c/o Nick V. Grishin], as the lectotype of Pamphila hycsos Mabille, 1891. The Sommer collection was bought by Staudinger in 1873 (before the description of P. hycsos), and the red label appears to be in Mabille handwriting, thus providing additional evidence that this specimen is a syntype.
Cynea (Nycea) quada Grishin, new species https://zoobank.org/6A719582-3B76-4FD4-9C4C-FEC6DF85C27E (Fig. 5 part, 115–116, 345–347)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Ecuador identified as Cynea (Cynea) diluta (Herrich-Schäffer, 1869) (type locality not specified) or Cynea bistrigula (Herrich-Schäffer, 1869) (type locality not specified) is more related to Cynea (Nycea) erebina (Möschler, 1879), but shows prominent genetic differentiation from the latter species (Fig. 5): e.g., their COI barcodes differ by 2.6% (17 bp), and therefore the Ecuadorian specimen represents a new species. This new species keys to C. popla Evans, 1955 (type locality in Trinidad) (L.7.8) in Evans (1955) and differs from its relatives by narrower forewing spots, four yellowish dots on the ventral hindwing: one in the middle and three in the postdiscal area (Fig. 115–116), more concave sides of uncus in dorsal view, arms not separated but with a shallow notch between them, their distal margin relatively flat and angled on the sides, gnathos arms thinner, shorter, strongly sclerotized, sacculus distally with a couple of small teeth, harpe terminally rounded, continues into a rounded thumb-like finely serrated process directed anterodorsad, situated inner from ampulla and covered with it in outer lateral view (Fig. 345–347). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly653.3.7:G45A, aly798.8.27:G99A, aly1651.42.3:T36A, aly525.64.4:C90T, aly127.90.1:T201C, aly390.26.4:G93G (not A), aly4265.5.1:T96T (not C), aly531.44.1:C1237C (not T), aly2011.20.4:G135G (not A), aly18826.6.2:A102A (not G), and COI barcode: A100C, A253G, T268C, 325C, T500C.
Barcode sequence of the holotype.
Sample NVG-18119D03, GenBank OR837676, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGTATATTAGGAACTTCCTTAAGATTATTAATTCGTACAGAATTAGGTAACCCAGGATCATTAATTGGCGATGATCAAATTTATAATACTATTGTAACAGCTCATGCCTTTATTATAATTTTTTTTATAGTTATACCTATTATAATCGGAGGATTTGGTAATTGATTAGTACCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCCCGAATAAATAACATGAGATTTTGAATACTCCCACCATCTTTAATACTACTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACCGGTTGAACAGTTTACCCCCCTCTTTCATCTAATATTGCCCATCAAGGAAGTTCAGTTGATTTAGCAATTTTTTCCCTTCATTTAGCGGGAATTTCTTCTATTTTAGGGGCTATCAATTTTATTACAACAATTATTAATATACGAATCAATAATATATCATTTGATCAAATATCCCTATTTATTTGATCAGTAGGTATTACAGCACTTTTATTACTTTTATCCTTACCAGTATTAGCAGGAGCTATTACTATACTTTTAACAGATCGAAATTTAAATACTTCTTTTTTTGATCCTGCAGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 115–116, bears the following six rectangular labels, five white: [ECUADOR: Napo Prov | 4 km Tena-Pano Rd. | 1° 02′S 77° 50′W | 600m 27 Sep 1990 | S S Nicolay leg], [♂ genitalia | slide/vial # | H1068 | Prep. S.S. Nicolay], [Cynea ♂ | bistrigula | Det. H-S. | S.S. Nicolay], [DNA sample ID: | NVG-18119D03 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01531867], and one red [HOLOTYPE ♂ | Cynea quada | Grishin].
Type locality.
Ecuador: Napo Province, km 4 of Tena-Pano Rd., elevation 600 m, GPS −1.033, −77.833.
Etymology.
The name is for the four points on each ventral hindwing and four major spots on both forewings taken together and also for the type locality in Ecuador. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Napo, Ecuador.
Cynea (Quinta) achirae Grishin, new species https://zoobank.org/1E962C5C-976E-4C07-9D15-B8FBD3EE63FF (Fig. 5 part, 117–118, 348–349)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Mexico identified as Cynea (Quinta) cannae (Herrich-Schäffer, 1869) (type locality not specified, possibly in Venezuela, lectotype sequenced as NVG-15035D04) show prominent genetic differentiation from it in the Z chromosome (Fig. 5) (although COI barcodes differ only by 0.8%, 5 bp), and therefore represent a new species. This new species keys to “Quinta cannae” (L.6.1) in Evans (1955) but differs from it by usually less contrasting patterns of wings venter (Fig. 118), shorter and directed dorsad (not dorsoposteriad) process at the base of harpe, overlapping the process of ampulla for its entire width, but not projecting much dorsad of it in lateral view (Fig. 349). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1011.4.6:C72T, aly37338.16.1:C549T, aly37338.16.1:C573T, aly390.11.3:C108G, aly2612.8.8:T199C, and COI barcode: 352C, T367C, A586C.
Barcode sequence of the holotype.
Sample NVG-19012H03, GenBank OR837677, 658 base pairs:
AACTTTATATTTTATTTTCGGAATTTGAGCAGGAATATTAGGAACTTCTTTAAGATTATTAATTCGTACAGAATTAGGAAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATCATAATTGGAGGATTTGGTAATTGATTAGTTCCTCTTATATTAGGAGCCCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTCTGAATACTTCCCCCATCTTTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGTACAGGCTGAACAGTATATCCCCCTCTTTCCTCTAATATTGCTCACCAAGGAAGATCTGTTGACTTAGCTATTTTTTCTCTTCATTTAGCTGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACAACAATTATCAATATACGAATTAGTAATATATCATTTGATCAAATACCTTTATTTGTATGATCTGTAGGTATTACAGCTCTTTTATTATTATTATCCTTACCTGTATTAGCTGGAGCAATTACTATATTATTAACCGATCGAAATTTAAATACCTCATTTTTTGACCCTGCAGGAGGAGGTGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the Texas A&M University Insect Collection, College Station, TX, USA (TAMU), illustrated in Fig. 117–118, bears the following seven rectangular labels, six white: [MEXICO: | Tamaulipas, | Ciudad Mante | Los Arcos Ct.], [ex pupa | 7 Feb 1974 | Roy O. Kendall | & C. A. Kendall], [CHROMOSOMES: | Kendall Log- | H-201 -M | N=301, 315], [Larva Foodplant: | CANNACEAE | Canna indica | Linnaeus | (foliage)], [HESPERIIDAE, | Hesperiinae: | Quinta cannae | (Herr.-Schaffer, 1869) | ♂ det. R. O. Kendall | [M. & B. No. 138.2], [DNA sample ID: | NVG-19012H03 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Cynea achirae | Grishin]. Paratypes: 3♂♂ from Mexico: 1♂ NVG-22035B12 Cuernavaca, Jun-1906, genitalia vial NVG230216–44 (Fig. 348–349) [USNM] and 2♂♂ Sonora, Alamos, J. P. Brock leg., ex larvae on Canna indica L.: NVG-21056B01 eclosed Sep-1994; NVG-21056B02 eclosed 8-Oct-1993.
Type locality.
Mexico: Tamaulipas, Ciudad Mante, Los Arcos Ct.
Etymology.
Achira is one of the common names for Canna indica L., a frequent larval foodplant of this species. The name is treated as a noun in apposition.
Distribution.
Mexico.
Eutus amazonicus Grishin, new species https://zoobank.org/FE50E403-652D-4E02-8EB3-6EAA7C30B600 (Fig. 5 part, 119–120, 350–352)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from the Amazonian region that were challenging to place belong to the genus Eutus Grishin, 2022 (type species Cobalus rastaca Schaus, 1902) and are the closest to Eutus yesta (Evans, 1955) (type locality in Peru: Inambari) or its relative (Fig. 5) but are phenotypically distinct and therefore represent a new species. Identified by the following combination of characters: wings are narrower than in E. yesta, forewing is with a contrasting black wedge-shaped brand at the base of forewing cell CuA1-CuA2, hyaline spot shaped as an angle bracket distad of the brand, three round spots of decreasing size in cells M3-CuA1, R5-M1, and R4-R5, and a small spot at the upper side of forewing discal cell; ventral forewing is with a large (about half of wing length) cream area near tornus, ventral hindwing with more or less apparent yellowish spots and a trace of a pale ray in the cell CuA2-1A+2A. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1656.16.2:A48C, aly1656.16.2:A63G, aly4456.8.2:C72T, aly517.17.2:C372G, aly517.17.2:G516A, and COI barcode: A22G, T25C, T103C, T157C, T304C, A586C.
Barcode sequence of the holotype.
Sample NVG-19121G08, GenBank OR837678, 658 base pairs:
AACTTTATATTTTATTTTTGGGATCTGAGCAGGAATATTAGGAACTTCCTTAAGTTTATTAATTCGTACTGAATTAGGAAATCCGGGTTCTTTAATTGGAGACGATCAAATTTATAACACTATCGTTACAGCACATGCTTTTATTATAATTTTTTTCATAGTTATACCTATTATAATTGGTGGATTTGGAAATTGACTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATATTACCCCCTTCTTTATTTTTATTAATTTCAAGAAGAATCGTAGAAAATGGAGCAGGAACAGGATGAACAGTATACCCTCCTTTATCTTCTAACATTGCCCACCAAGGATCTTCTGTTGATTTAGCAATTTTTTCTCTACATTTAGCAGGAATTTCATCCATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAATATATCATTTGACCAAATACCTTTATTTGTATGATCTGTAGGTATTACCGCTTTATTACTACTCTTATCTTTACCTGTATTAGCTGGAGCTATTACTATACTTTTAACCGATCGAAATTTAAATACCTCATTCTTTGATCCTGCTGGAGGAGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 119–120, bears the following six rectangular labels, five white: [PERU Madre De Dios | Rio La Torre 300m | Tambopata Res. | 5 Oct. ‘86 | S.S. Nicolay], [♂ genitalia | slide/vial # | H956 | Prep. S.S. Nicolay], [Thoon ♂ | Det. ponka | S.S. Nicolay], [DNA sample ID: | NVG-19121G08 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01602763], and one red [HOLOTYPE ♂ | Eutus amazonicus | Grishin]. Paratypes: 1♂ NVG-22023D03, H20815 French Guiana, Montagne Favard, GPS 4.500, −52.050, 18-Sep-2003, B. Hermier leg. [BHermier] and 1♀: NVG-21048D10 62 km S of Ariquemes, linha C-10, 5 km S of Cacaulandia, 8-Oct-1994 O. Gomes leg. [MGCL].
Type locality.
Peru: Madre de Dios, Tambopata National Reserve, Rio La Torre, elevation 300 m.
Etymology.
The name is for the Amazonian distribution of this species and is a noun in apposition.
Distribution.
The Amazonian region: recorded from Peru, French Guiana, and Brazil.
Eutus incus Grishin, new species https://zoobank.org/9ACA4D61-7A97-46AC-A262-E1DBD30EC6D2 (Fig. 5 part, 121–122, 353–354)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from the Cosñipata Valley in Peru that was superficially similar to Eutus amazonicus new species is genetically differentiated from it (Fig. 5): e.g., their COI barcodes differ by 6.5% (43 bp) and, therefore, represents a new species. This new species is similar to Eutus amazonicus new species and differs from it by having a larger brand at the base of forewing cell CuA1-CuA2, larger hyaline spots, smaller ventral hindwing tornal pale area, and larger (although still small, dot-like) yellowish spots on ventral hindwing. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly85.15.2:A144C, aly349.15.2:A54G, aly1468.20.2:T507C, aly18826.15.1:T129C, aly1432.18.2:A42G, aly1656.16.2:A48A (not C), aly1656.16.2:A63A (not G), aly4456.8.2:C72C (not T), aly425.14.6:C90C (not T), aly517.17.2:C372C (not G), and COI barcode: T10C, A34G, T112C, T223A, T499C.
Barcode sequence of the holotype.
Sample NVG-19023C07, GenBank OR837679, 658 base pairs:
AACTTTATACTTTATTTTTGGAATTTGAGCAGGGATATTAGGAACTTCTTTAAGTTTATTAATTCGTACTGAATTAGGAAATCCAGGCTCTTTAATTGGAGATGATCAAATCTATAATACTATTGTTACAGCACATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGGGCCCCAGACATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATACTACCTCCTTCTTTATTTTTATTAATCTCAAGAAGAATTGTAGAAAATGGAGCAGGTACAGGATGAACAGTTTACCCCCCTCTATCTTCTAATATTGCCCACCAAGGATCCTCTGTTGATTTAGCAATTTTTTCCTTACATTTAGCAGGAATTTCATCCATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAATATATCATTTGATCAAATACCCTTATTTGTTTGATCTGTAGGTATTACTGCTTTATTATTACTTTTATCTTTGCCCGTATTAGCTGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 121–122, bears the following four rectangular labels, three white: [PERU: Cuzco: Cosñipata Valley | Quebrada Quitacalzón 1,050m. | 13° 01′ 13″S, 71° 29′ 50″W | 12 August 2009 Brian Harris], [DNA sample ID: | NVG-19023C07 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532830], and one red [HOLOTYPE ♂ | Eutus incus | Grishin].
Type locality.
Peru: Cuzco Region, Cosñipata Valley, Quebrada Quitacalzón, elevation 1050 m, GPS −13.020278, −71.497222.
Etymology.
The name is for the type locality in Cuzco, the center of the Inca Empire. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Cosñipata Valley, Peru.
Eutus septemaculatus Grishin, new species https://zoobank.org/3F5D4627-A738-4C6B-9DB4-687DFCA5FE1D (Fig. 5 part, 123–124, 355–356)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Mato Grosso Brazil with wing pattern similarities to Eutus mubevensis (E. Bell, 1932) (type locality in Paraguay) shows prominent genetic differentiation from it (Fig. 5): e.g., their COI barcodes differ by 9.1% (60 bp) and, therefore, represents a new species. This new species keys to “Decinea mubevensis” (L.11.9) in Evans (1955) but differs from it (males) by rounder hindwings, larger hyaline forewing spots, and brighter ventral forewing tornal area (Fig. 123–124), harpe more robust, not shorter than valva, and ampulla process more massive, only slightly shorter than costa (Fig. 355–356). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly18826.14.1:C60G, aly1295.8.3:A42T, aly207.9.6:T195C, aly207.9.6:A312G, aly5294.34.2:A116G, aly490.2.1:T435T (not C), aly490.2.1:A501A (not T), aly531.12.1:T103T (not A), aly1146.43.2:T93T (not C), aly2127.7.4:T726T (not C), and COI barcode: T50C, A101G, T346A, T382A, A421C.
Barcode sequence of the holotype.
Sample NVG-19021H05, GenBank OR837680, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAAGAGGTATATTAGGAACTTCTCTAAGTTTATTAATTCGTACAGAATTAGGTAATCCTGGATCTTTAATTGGAAATGACCAAATTTATAATACTATCGTAACAGCTCACGCTTTCATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCACCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATATTACCCCCTTCTTTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCTGGAACAGGATGAACAGTATATCCCCCACTTTCTTCTAATATTGCCCATCAAGGATCTTCTGTAGATTTAGCAATTTTCTCCCTTCATTTAGCTGGTATTTCCTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATATATCCTTTGATCAAATACCATTATTTGTTTGATCGGTAGGAATTACAGCTCTTTTATTACTTTTATCTTTACCAGTATTAGCAGGAGCAATTACTATATTATTAACAGATCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGAGGAGGAGACCCTATTTTATATCAACATCTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 123–124, bears the following five rectangular labels, three white: [BRAZIL MT | Cuiba/Santarem | Sinope-km 500 | 13 July 1979 | leg. S.S. Nicolay], [DNA sample ID: | NVG-19021H05 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532718], and one red [HOLOTYPE ♂ | Eutus septemaculatus | Grishin].
Type locality.
Brazil: Mato Grosso, km 500 of Cuiabá–Santarém highway, Sinop.
Etymology.
The name is for seven spots on the forewing. In Latin, septem means seven, and macula means spot. The name is a masculine perfect passive participle in the nominative singular.
Distribution.
Currently known only from the holotype collected in Mato Grosso, Brazil.
Godmia viridicapita Grishin, new species https://zoobank.org/6255941E-1A19-4ED0-810E-A334F5C57211 (Fig. 5 part, 125–126, 357–358)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Ecuador identified as Godmia chlorocephala (Godman, 1900) (type locality in Panama) shows prominent genetic differentiation from it (Fig. 5): e.g., their COI barcodes differ by 4.7% (31 bp), and therefore represents a new species. This new species keys to “Onophas chlorocephala” (J.51.3) in Evans (1955) but differs from it by less elongated upper part of the brand, both upper and lower segments, and even weaker developed forewing pale spots (essentially none visible in the holotype) (Fig. 125–126), uncus arms thin, bowed, terminally converging, space between them is drop-shaped in dorsal view, harpe is not separated from valva, together they form semi-rectangular shape in lateral view, gnathos with flattened, blade-like overlapping arms, harpe with a semi-circular ridge on the inner surface, saccus is the same length as uncus (Fig. 357–358). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1405.6.4:A75G, aly1405.6.4:T78C, aly15220.1.1:G864T, aly525.64.4:C108T, aly525.64.4:G162A, aly5773.2.5:T315T (not C), aly1175.3.8:G72G (not C), aly1175.3.8:G93G (not A), aly1772.7.1:G714G (not A), aly5294.37.11:G897G (not A), and COI barcode: A64G, T67C, T103C, T403C, A559T.
Barcode sequence of the holotype.
Sample NVG-19023F02, GenBank OR837681, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACATCTCTTAGTTTATTGATTCGCTCAGAATTAGGAAATCCAGGATCTTTAATTGGAGACGATCAAATTTATAACACTATTGTAACTGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCCTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATATTACCACCTTCTTTAATATTATTAATTTCAAGAAGAATCGTAGAAAATGGTGCTGGAACTGGATGAACAGTTTATCCCCCCCTTTCATCTAATATTGCCCATCAAGGATCATCTGTTGATCTAGCAATTTTTTCTCTCCACTTAGCAGGTATTTCATCAATCTTAGGGGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATTTATCTTTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACAGCACTATTATTACTACTATCTTTACCTGTACTTGCAGGAGCTATTACTATATTATTAACAGATCGAAATTTAAATACTTCATTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 125–126, bears the following five rectangular labels, four white: [Cotundo NAPO | ECUADOR 800m | 3 Oct ‘76 | S. S. Nicolay], [Onophas ♂ | chlorocephala | Det. God. | S.S. Nicolay], [DNA sample ID: | NVG-19023F02 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532855], and one red [HOLOTYPE ♂ | Godmia viridicapita | Grishin].
Type locality.
Ecuador: Napo Province, Cotundo, elevation 800 m.
Etymology.
The name is Latin from the Greek name of its sister species, G. chlorocephala (i.e., green-headed). In Latin, viridi capite is green head. The name is a feminine adjective.
Distribution.
Only known from the holotype collected in northcentral Ecuador.
Rhomba pulla Grishin, new species https://zoobank.org/8AE2A86B-97B8-4871-B55F-994B301AA720 (Fig. 5 part, 127–128, 359–360)
Definition and diagnosis.
Phylogenetic trees reveal that a nearly unmarked Hesperiidae specimen from San Pedro Lodge in Peru is closely related to Rhomba gertschi (E. Bell, 1937) (type locality in Panama) while showing prominent genetic differentiation from it (Fig. 5): e.g., their COI barcodes differ by 4.3% (28 bp) and, therefore, represents a new species. This species is similar to R. gertschi in the shape of brands (although the brand segments are bulkier), spotting of the ventral hindwing, and morphology of genitalia, but differs from it by the lack of hyaline spots, which gives it a very different appearance. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly728.22.1:A207G, aly2163.5.1:A138C, aly2163.5.1:T169G, aly736.5.4:A39G, aly736.5.4:T87C, aly1585.9.2:G97G (not T), aly2011.4.1:A84A (not G), aly2011.4.1:T90T (not C), aly85.11.3:G162G (not A), aly3312.7.2:T120T (not A), and COI barcode: T16C, C235T, A241T, T406C, A433G, T457C.
Barcode sequence of the holotype.
Sample NVG-19023E08, GenBank OR837682, 658 base pairs:
AACTTTATATTTTATCTTTGGTATTTGAGCAGGAATATTAGGAACTTCTTTAAGTTTATTAATTCGTACAGAATTAGGTAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGTTTTGGAAATTGATTAGTACCATTAATATTAGGGGCCCCAGATATAGCCTTTCCACGTATAAATAATATAAGATTTTGAATATTACCACCTTCATTAATATTACTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACTGGTTGAACAGTTTATCCACCTTTATCCTCTAATATCGCTCATCAAGGATCTTCTGTAGATTTAGCAATTTTTTCCCTTCACTTAGCAGGAATTTCATCAATTTTAGGGGCAATTAATTTTATTACAACAATCATTAATATACGAATTAGAAATTTATCATTTGATCAAATACCTTTATTTATTTGATCAGTAGGTATTACAGCACTTTTATTACTTTTATCTTTACCTGTTCTAGCTGGTGCTATTACTATACTTTTAACAGATCGAAATTTAAATACTTCATTTTTTGATCCAGCTGGTGGAGGTGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 127–128, bears the following five rectangular labels, four white: [PERU:Cuzco, 1450m. | San Pedro Lodge | Cosnipata Rd 548 | 9-XI-2008 Kinyon], [DNA sample ID: | NVG-19023E08 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532850], and one red [HOLOTYPE ♂ | Rhomba | pulla Grishin].
Type locality.
Peru: Cuzco Region, Cosñipata Road, San Pedro Lodge, elevation 1450 m.
Etymology.
In Latin, pullus means blackish or dark colored. The name is given for this darker (thus far) nearly unspotted species of the genus and is a feminine adjective.
Distribution.
Currently known only from the holotype collected in the Cosñipata Valley, Peru.
Niconiades victoria Grishin, new species https://zoobank.org/216DED72-3642-44BA-B553-22AD2C42EFF6 (Fig. 5 part, 129–130, 361–363)
Definition and diagnosis.
Genomic analysis of specimens we identified as Niconiades nikko Hayward, 1948 (type locality Argentina: Misiones) reveals their partitioning into two clades genetically differentiated in the Z chromosome (Fig. 5a) but not in the mitogenome (Fig. 5b). Due to prominent genetic differentiation in the nuclear genome, the two clades represent species-level taxa, and the North American species is new. This new species keys to N. nikko (O.11.6) in Evans (1955) and differs from it by typically weaker defined whitish band on the ventral hindwing and more extensive greenish-yellow overscaling (Fig. 130). Due to the cryptic nature of this species and significant wing pattern variability, most reliable identification is achieved by DNA and a combination of the following nuclear genomic base pairs is diagnostic: aly366.12.1:A1164G, aly383.17.7:C1401T, aly383.17.7:G1869A, aly1651.5.1:T1086C, aly694.1.1:A441C. There are no consistent differences between the new species and N. nikko in COI barcodes.
Barcode sequence of the holotype.
Sample NVG-19013C03, GenBank OR837683, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGGGCTGGAATATTAGGAACTTCCCTCAGTTTATTAATTCGTACAGAATTAGGTAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTACCTTTAATATTAGGAGCACCTGATATGGCTTTCCCCCGAATAAATAATATAAGATTTTGAATATTACCCCCTTCTTTAATATTATTAATCTCAAGAAGAATTGTAGAAAATGGTGCTGGAACTGGATGAACAGTTTACCCCCCTCTATCCTCTAATATTGCCCACCAAGGATCATCAGTTGATTTAGCAATTTTCTCTCTTCATCTAGCCGGAATTTCCTCAATTCTAGGAGCAATTAATTTTATTACTACAATCATTAATATACGAATTAGTAATATATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGTATTACAGCATTATTATTACTTTTATCTTTACCAGTTTTAGCAGGGGCTATTACTATACTTCTTACAGATCGAAATTTAAACACTTCATTTTTTGATCCTGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the Texas A&M University Insect Collection, College Station, TX, USA (TAMU), illustrated in Fig. 129–130, bears the following seven rectangular labels: six white [MEXICO: | Tamaulipas | Rancho Pico de Oro | vic. of Los Kikos], [ex larva | 26 Dec 1974 | Roy O. Kendall | and C. A. Kendall], [Larval foodplant: | GRAMINEAE | Bambusa vulgaris | Schrad. | (foliage -)], [Niconiades ♂ | nikko | Hayward | det.H.A.Freeman], [HESPERIIDAE: | Hesperiinae: | Niconiades nikko | ♂ Hayward, 1948 | Det. R. O. Kendall | [M. & B. No. 264.3]], [DNA sample ID: | NVG-19013C03 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Niconiades | victoria Grishin]. Paratypes: 2♂ and 1♀: 1♀ NVG-18012C11, USNMENT_ 01450288 Mexico: San Luis Potosi, Cd. Valles, 15-Oct-1976, E. C. Knudson leg., genitalia X-3289 J. M. Burns 1992 [USNM], 1♂ NVG-20057B04 Mexico: Chiapas (no other data), genitalia vial NVG231118–03 [TMMC] (Fig. 361–363), and 1♂ NVG-18012C12, USNMENT_01450289 Honduras, San Pedro Sula, 24-Dec-1971, Robert D. Lehman leg., genitalia X-3249 J. M. Burns 1992 [USNM].
Type locality.
Mexico: Tamaulipas, vic. Los Kikos, Rancho Pico de Oro.
Etymology.
In Greek, νίκη (nike) is the word for “victory” which sounds similar to the name N. nikko, the sister species. In Latin, victory is victoria, the word used as this species’ epithet. The name is a noun in apposition.
Distribution.
From eastern Mexico to Guatemala, at least.
Lancephallus purpurus Grishin, new genus and new species https://zoobank.org/3AD6C438-4774-4FE6-9020-54F6A4A6D92E https://zoobank.org/7BACF558-7208-4EFE-AF4C-923515E25AE5 (Fig. 5 part, 131–134, 364–365)
Definition of the new genus.
Phylogenetic analysis places specimens that agree with Evans’ unpublished (curation in BMNH only) concept of males of Phlebodes confixa (A. Butler, 1877) (type locality in Brazil: Amazonas) as a unique lineage sister to Vettius Godman, 1901 (type species Papilio phyllus Cramer, 1777) (Fig. 5). This lineage is prominently separated in trees from Vettius, stronger than Vettius species within the genus, and therefore represents a new genus. This new genus is diagnosed by unique male genitalia, in particular, by the shape of the aedeagus with extended caudal end, slightly curved and terminating in a sharp point and valva with a spike-like projection from the ampulla and rounded harpe, uncus arms strongly divergent, gnathos arms convergent. Males have a bi-partite brand with a longer section along cubitus between the origins of veins CuA1 and CuA2 and a dash below it, below vein CuA2. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly2178.10.1:G66A, aly2178.10.1:T109C, aly1260.24.1:G228A, aly1019.12.6:C66T, aly1019.12.6:G78A.
Type species.
Lancephallus purpurus Grishin, new species.
Species included in the genus.
Only the type species.
Parent taxon for the genus.
Subtribe Moncina A. Warren, 2008
Definition of the new species.
While we have not sequenced true P. confixa, females of Lancephallus that were associated with males by DNA sequencing (and specimens in copula in BMNH) do not resemble the only known P. confixa syntype, a female, in having sharply defined pale spots on the ventral hindwing, which has a strong purple sheen and slightly paler veins. Therefore, this species of Lancephallus is new. In wing patterns, it is uniquely different from all of its closest relatives in Vettius, its sister genus. The only similarly patterned species formerly associated with Vettius was Phlebodes fuldai (E. Bell, 1930) (type locality in Colombia), now transferred to its rightful genus Phlebodes Hübner, [1819] (type species Papilio pertinax Stoll, [1781]) (Zhang et al. 2022b), which Lancephallus resembles. However, Lancephallus and Phlebodes are not closely related to each other (Fig. 5, 6). This new species is identified by chestnut-brown wings with small and narrow (in males) yellowish forewing spots in cells CuA1-CuA2 and M3-CuA1, and in some specimens, one to three subapical dots, spots are larger and paler in females who may have a spot in upper discal cell beneath; ventrally with strong purple sheen, especially on hindwing that has variously developed postdiscal pale dots and a dot in the center, veins are paler. In DNA, a combination of the following base pairs is diagnostic in the COI barcode: T38C, A184T, A208T, A334C, A532C.
Figure 6.

Phylogenetic trees of Moncina (part 2) inferred from protein-coding regions in a) the Z chromosome and b) the mitochondrial genome. See Fig. 1 legend for other notations.
Barcode sequence of the holotype.
Sample NVG-19021F03, GenBank OR837684, 658 base pairs:
AACTTTATATTTTATTTTTGGAATCTGAGCAGGAATACTAGGAACTTCTTTAAGTTTATTAATTCGTTCAGAATTAGGAAATCCAGGTTCATTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGAAATTGATTAGTACCTCTTATATTAGGAGCCCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTCTGAATACTTCCCCCCTCATTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACTGGATGAACCGTTTACCCCCCTCTTTCTTCTAATATTGCTCATCAAGGATCTTCAGTAGATTTAGCTATCTTTTCCCTTCATTTAGCAGGAATTTCATCTATTCTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATTTATCTTTTGATCAAATACCTTTATTTGTATGATCAGTAGGAATTACTGCACTCTTATTACTTTTATCTTTACCTGTATTAGCAGGAGCTATTACTATACTTTTAACAGATCGAAATTTAAATACTTCATTTTTTGATCCAGCAGGAGGAGGAGATCCTATTCTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 131–132, bears the following five rectangular labels, four white: [GUYANA: Cuyuni R, | Kamaria Falls 100′ | 30.XI.–5.XII.2000 | 6° 24′N 58° 54.6′W | Leg. S. Fratello et al] (GPS given as “546′W”), [DNA sample ID: | NVG-19021F03 | c/o Nick V. Grishin], [{QR Code} | USNM ENT 00275117], and one red [HOLOTYPE ♂ | Lancephallus | purpurus Grishin]. Paratypes: 5♂♂ and 2♀♀: 1♂ Colombia/Venezuela border, Orinoco River, Maipures, Dec-1898, Cherrie leg. [BMNH]; 1♂ Venezuela, Suapure, 2-Mat-1899, Klages leg. [BMNH]; 2♂♂ NVG-18098F02, H15049 and (not sequenced) H15046 French Guiana, Bagne des Annamites, GPS 4.8333, −52.5167, H. Crampette leg., 12-Jul-1998 [BHermier], 1♂ and 1♀ in copula, Guyana 1962–3 [BMNH]; 1♀ NVG-19024B11, USNM ENT 00275122 Guyana: Acarai Mts., Sipu River, elevation 900 ft, GPS 1.4183, −58.9533, 24-Oct-12-Nov-2000, S. Fratello et al. leg. [USNM] (Fig. 133–134).
Type locality.
Guyana: Cuyuni-Mazaruni Region, Cuyuni River, Kamaria Falls, approx. GPS 6.40, −58.91.
Etymology.
In Latin, lancea means spear, lance, or pike. The name is given for the spear-shaped aedeagus (phallus), and the species epithet is for the extensive purple sheen on the ventral side of the wings. The name of the genus is a masculine noun in the nominative singular, and the species name is a masculine adjective.
Distribution.
Venezuela, Guyana, and French Guiana.
Mnasicles (Remella) ecua Grishin, new species https://zoobank.org/E5849270-FF1F-4A4A-B099-B9097FABABDF (Fig. 6 part, 135–136, 366–367)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Ecuador identified as Mnasicles (Remella) remus (Fabricius, 1798) (type locality in “Caiena”) shows prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 4.6% (30 bp), and therefore represents a new species. This new species keys to “Moeris remus” (J.33.1) in Evans (1955) but differs from it by more developed pale area at the apex of ventral forewing (Fig. 136), not strong spots as in Mnasicles (Remella) rita (Evans, 1955) (type locality in Guatemala) and relatives, but more prominent than in most M. remus, uncus smaller, terminally knob-like, strongly humped in lateral view, tegumen short, not much longer than vinculum width, harpe shorter, with less developed tooth on the dorsal side, costa convex in the middle and slightly concave before ampulla that is separated from harpe by a narrower groove (Fig. 366–367). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly216.58.1:T366A, aly216.58.1:A492C, aly1456.2.1:T15C, aly1139.65.24:T108A, aly1139.65.24:A117C, aly2954.3.1:T525T (not A), aly2954.3.1:C800C (not T), aly127.87.1:T24T (not G), aly127.87.1:A177A (not G), aly11700.1.14:A59A (not T), and COI barcode: T5T, A37G, A100G, T316G, T463C.
Barcode sequence of the holotype.
Sample NVG-19017B02, GenBank OR837685, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACTTCTTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAACACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTCATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTTCCACGAATAAATAATATAAGATTTTGAATACTTCCTCCCTCTTTAATACTTTTAATTTCAAGAAGAATCGTAGAAAATGGGGCAGGTACTGGTTGAACAGTTTATCCTCCTTTATCTTCTAATATTGCTCATCAAGGTTCATCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCAGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAACATACGAGTAAGAAACTTATCATTTGATCAAATACCTTTATTTGTATGATCAGTAGGTATTACTGCTTTATTATTACTTTTATCTTTACCTGTATTAGCAGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAATACTTCATTTTTTGACCCTGCTGGAGGAGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 135–136, bears the following four rectangular labels, three white: [ECUADOR-Pich. | Napac-1000m | 12 Sept. 1976 | leg.S.S.Nicolay], [DNA sample ID: | NVG-19017B02 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532380], and one red [HOLOTYPE ♂ | Mnasicles (Remella) | ecua Grishin].
Type locality.
Ecuador: Pichincha Province, Río Napac, elevation 1000 m.
Etymology.
The name is given for Ecua[dor], the type locality of this species, and is a noun in apposition.
Distribution.
Only known from the holotype collected in the Andean region of northern Ecuador.
Lectotype designation for Amblyscirtes fluonia Godman, 1900
Amblyscirtes fluonia Godman, 1900 was described from a series of specimens collected at different localities in Mexico: Guerrero and Jalisco. To define the type locality more precisely, N.V.G. hereby designates a male syntype in the Natural History Museum, London, UK, that, according to its label was illustrated by Godman (1899–1901) and bears the following nine rectangular (except the first two, which are round and with a red circle on the upper side) white labels: (Type) and on the other side (H | 2162), (Type | H. T.), [Xucumanatlan, | Guerrero, | 7000 ft. | July. H.H.Smith.], [♂], [Sp. figured.], [B.C.A.Lep.Rhop. | Amblyscirtes | fluonia, | Godm.], [Godman-Salvin | Coll. 1913.—2.], [{QR Code} | NHMUK 012824161], and [MOLECULAR | 0247279852], as the lectotype of Amblyscirtes fluonia Godman, 1900. This specimen was sequenced as NVG-18082H01. The type locality of A. (Amblyscirtes) fluonia becomes Mexico: Guerrero, Xocomanatlan, elevation 7000 ft, approximate GPS 17.550, −99.617.
Amblyscirtes (Amblyscirtes) aeratus Grishin, new species https://zoobank.org/02F9C1E1-2242-4E8B-9276-C8F3614C6FD2 (Fig. 6 part, 137–140, 368–369)
Definition and diagnosis.
Phylogenetic trees reveal that a number of specimens identified as Amblyscirtes (Amblyscirtes) fluonia Godman, 1900 (type locality in Mexico: Guerrero) show prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 5.6% (37 bp). These specimens are not even monophyletic with A. fluonia and therefore belong to a new species. This new species keys to A. fluonia (N.2.7) in Evans (1955) but differs from the true A. fluonia (Fig. 141–144) by the lack of pebbly variegated appearance on the ventral side of wings, in particular on the hindwing and the submarginal area of forewing. In wing pattern, it resembles Amblyscirtes aenus W. H. Edwards, 1878 (type locality in USA: Colorado) more than A. fluonia, but the spots are more diffuse and less defined, e.g., forewing apical spots are smaller and yellower, and ventral hindwing postdiscal band is composed of diffuse and interconnected blotches rather than smaller and separated spots. In DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: aly522.2.4:T99A, aly522.2.4:C124T, aly587.21.2:G153A, aly587.21.2:T156C, aly669.9.1:T201A, and COI barcode: T205C, T206C, A295T, T397C, T418C, A565G.
Barcode sequence of the holotype.
Sample NVG-18063E02, GenBank OR837686, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGTATATTAGGAACTTCTTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGAAATTGATTAGTCCCCCTTATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATACTTCCCCCTTCTTTATTACTTTTAATTTCTAGAAGAATTGTAGAAAATGGTGCAGGAACAGGATGAACAGTTTATCCCCCTCTTTCTTCTAATATTGCACATCAAGGATCATCAGTTGATTTAGCAATTTTCTCTCTTCATTTAGCAGGAATCTCTTCTATTCTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAGTTAGAAATTTAATATTTGATCAAATACCTTTATTTGTTTGATCAGTAGGTATTACTGCTTTATTATTACTTTTATCTTTACCAGTTCTAGCTGGGGCTATTACTATACTTCTTACAGATCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGAGGAGGGGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 137–138, bears the following four rectangular labels, three white: [MEXICO: OAXACA | El Vado-San Sebastian | 5500–6500 ft | c 16° 53’ N 96° 53′ W | 27-Jun-1992 J. Kemner], [DNA sample ID: | NVG-18063E02 | c/o Nick V. Grishin], [USNMENT | {QR Code} 01466204] and one red [HOLOTYPE ♂ | Amblyscirtes | aeratus Grishin]. Paratypes: Mexico: 1♂ NVG-19042F06 Colima, Jun-1918, C. C. Hoffmann leg., AMNH_IZC 00337883 [AMNH] and 1♀: NVG-19122A02 Oaxaca, La Canada, 2 mi S Dominguillo, ca. 2500 ft, GPS 17.6500, −96.9167, 8,10-Aug-1991, J. Kemner leg. [USNM] (Fig. 139–140).
Type locality.
Mexico: Oaxaca, El Vado–San Sebastián, elevation 5500–6500 ft, GPS 16.8833, −96.8833.
Etymology.
Like aenus, the name aeratus in Latin means made or covered with brass or bronze. Fluonia means a flow, and the pebbly appearance is like a flow on the wings, which is not present in this species. The name is an adjective.
Distribution.
At least southwestern Mexico, known from Colima and Oaxaca.
Lectotype designation for Mastor anubis Godman, 1900
Mastor anubis Godman, 1900, currently in the genus Amblyscirtes Scudder, 1872 (type species Hesperia vialis W. H. Edwards, 1862) was described from a series of specimens collected at different localities in Mexico: Guerrero and Veracruz. Due to the polytypic type series and to define the type locality more precisely, N.V.G. hereby designates a male syntype in the Natural History Museum, London, UK, that, according to its label, was illustrated by Godman (1899–1901) and bears the following nine rectangular (except the first two, which are round and with a red circle on the upper side) white labels: (Type) and on the other side (H | 2164), (Type | H. T.), [Omilteme, | Guerrero, | 8000 ft. | July. H.H.Smith.], [♂], [Sp. figured.], [B.C.A.Lep.Rhop. | Mastor | anubis, | Godm.], [Godman-Salvin | Coll. 1914.—5.], [{QR Code} | NHMUK 012824162], and [MOLECULAR | 0247281638], as the lectotype of Mastor anubis Godman, 1900. This specimen was sequenced as NVG-18082H02. The type locality of Amblyscirtes (Mastor) anubis becomes Mexico: Guerrero, Omiltemi, elevation 8000 ft. The name Anubis is associated with “dark” as it was an ancient Egyptian god of the dead. Fittingly, the lectotype designation makes A. anubis the darkest species (darkest fringes) among its close relatives.
Amblyscirtes (Mastor) chrysoplea Grishin, new species https://zoobank.org/81EE321D-0271-4A7C-A136-ED06491AB494 (Fig. 6 part, 145–146, 370–371)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Oaxaca, Mexico, identified as Amblyscirtes anubis Godman, 1900 (type locality in Mexico: Guerrero), show prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 5.8% (38 bp), and therefore represent a new species. This new species keys to A. anubis (N.2.21) in Evans (1955) but differs from it in having bright-orange forewing fringes, hindwing fringes typically with some orange scales, and ventral hindwing essentially unspotted (better defined, rounder ventral hindwing spots are typical for A. anubis). Due to the cryptic nature of this species, most reliable identification is achieved by DNA, and a combination of the following base pairs is diagnostic in the nuclear genome: aly216.64.14:G57A, aly216.64.14:A87G, aly103.15.43:C102T, aly3125.2.3:G201A, aly6398.11.1:G1269A, and COI barcode: 88T, A199G, T277C, T358C, T487C, A643G.
Barcode sequence of the holotype.
Sample NVG-18012F09, GenBank OR837687, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATACTAGGAACTTCATTAAGATTATTAATTCGTACAGAATTAGGAAACCCTGGTTCATTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTGGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGGATAAATAATATAAGATTTTGAATACTTCCCCCATCCCTAATACTTTTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGATGAACAGTATACCCCCCTTTATCATCTAACATTGCCCATCAAGGTTCATCTGTTGATTTAGCTATTTTTTCCCTTCATTTAGCTGGAATTTCCTCAATTTTAGGAGCAATTAATTTTATTACCACAATTATTAATATACGAGTTAGAAATATATCATTCGATCAAATACCCCTATTTGTTTGATCTGTAGGTATTACTGCTTTATTATTACTTTTATCTTTACCTGTATTAGCAGGTGCTATCACAATACTCCTCACTGATCGAAATTTAAATACTTCCTTTTTTGATCCTGCTGGAGGAGGGGATCCAATTCTGTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 145–146, bears the following four rectangular labels, three white: [MEXICO: OAXACA | Putla-Tlaxiaco | 5000–7000 ft | 5–7-VIII-1992 | J. Kemner], [DNA sample ID: | NVG-18012F09 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450312], and one red [HOLOTYPE ♂ | Amblyscirtes (Mastor) | chrysoplea Grishin]. Paratypes: 2♂♂ Mexico: Oaxaca, Sierra Madre del Sur, J. Kemner leg.: NVG-18063G12, USNMENT_01466218 El Guajolote, 7700′, 14-Jun-1989, genitalia X-2924 J.M.Burns 1990 [USNM] and NVG-21014G07 Sierra Madre del Sur, La Soledad–Buena Vista, 5000′, 14-Apr-1990 [CMNH].
Type locality.
Mexico: Oaxaca, Putla–Tlaxiaco, elevation 5000–7000 ft.
Etymology.
The name is formed from the Greek χρυσός (chrysós), meaning gold and πλέον (pléon), meaning most. The name is given for the orange fringes not only on the forewing but also partly on the hindwing, making this species the most orange-colored member of the A. anubis group. The name is a noun in apposition.
Distribution.
Mexico: Oaxaca.
Amblyscirtes (Mastor) chrysomisa Grishin, new species https://zoobank.org/713888C4-2DD4-4996-B0B7-05226B4462D0 (Fig. 6 part, 147–148, 372–374)
Definition and diagnosis.
Phylogenetic trees reveal that a number of specimens identified as Amblyscirtes anubis Godman, 1900 (type locality in Mexico: Guerrero) show prominent genetic differentiation from it and from their sister species Amblyscirtes chrysoplea new species (Fig. 6): e.g., their COI barcodes differ from the latter species by 4% (26 bp), and therefore represent a new species. This new species keys to A. anubis (N.2.21) in Evans (1955) but differs from its closest relatives in having yellow-orange forewing fringes, dark-brown hindwing fringes, and ventral hindwing essentially unspotted (better defined, rounder ventral hindwing spots are typical for A. anubis). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly390.21.1:C54T, aly2011.1.3:C213T, aly2487.17.5:T57A, aly2275.11.10:G75C, aly1158.9.10:C114T, and COI barcode: 88C, T103C, 268C, T349G, T386C, T553G.
Barcode sequence of the holotype.
Sample NVG-18063H02, GenBank OR837688, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATACTAGGAACTTCATTAAGATTATTAATTCGTACAGAATTAGGAAACCCTGGCTCATTAATTGGAGACGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTTTAATATTAGGGGCCCCTGATATAGCTTTCCCACGGATAAATAATATAAGATTTTGAATACTCCCCCCATCTCTAATACTTTTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGATGAACAGTGTACCCCCCTCTGTCATCCAATATTGCCCATCAAGGATCATCTGTTGATCTAGCTATTTTTTCCCTTCATTTAGCAGGAATTTCTTCAATTTTAGGAGCAATTAATTTTATTACTACAATTATTAATATACGAGTTAGAAACATATCGTTTGATCAAATACCCCTATTTGTTTGATCTGTAGGTATTACTGCTTTATTATTACTTTTATCTTTACCGGTATTAGCAGGTGCTATCACAATACTCCTTACCGATCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGAGGAGGGGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 147–148, bears the following four rectangular labels, three white: [MEXICO: CHIAPAS | nr. Navenchauc | c. 8000 ft 4-VII-1992 | J. Kemner & A. Vasquez], [DNA sample ID: | NVG-18063H02 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01466220], and one red [HOLOTYPE ♀ | Amblyscirtes (Mastor) | chrysomisa Grishin]. Paratypes: 4♂♂ and 1♀ from Mexico: 1♂ NVG-18063G10, USNMENT_01466216 no data, Collection Wm Schaus, old, genitalia X-2556 J.M.Burns 1988 [USNM]; 1♀ NVG-18063G11, USNMENT_01466217 Veracruz, Xalapa, old [USNM]; 1♂ NVG-19043A01, AMNH_IZC 00337914 Mexico: Hidalgo, Apulco, Apr-1952, T. Escalante leg. [AMNH]; 1♂ NVG-18063H01, USNMENT_01466219 Chiapas, nr. Navenchauc, c. 8000 ft, 4-Jul-1992, J. Kemner and A. Vasquez [USNM]; 1♂ NVG-19043A02, AMNH_IZC 00337915 Chiapas, Ochuc, Rancho San Ramon, 7-Jul-1975, Peter Hubbell leg. [AMNH].
Type locality.
Mexico: Chiapas, nr. Navenchauc, elevation about 8000 ft.
Etymology.
The name is formed from the Greek χρυσός (chrysós), meaning gold, and μισα (misa), meaning half. It is given for the forewings with orange fringes and hindwings with dark fringes. The name is a noun in apposition.
Distribution.
Southeastern Mexico.
Amblyscirtes (Mastor) repta Evans, 1955 is a species distinct from Amblyscirtes (Flor) florus (Godman, 1900)
Genomic sequencing of the holotype of Repens repta Evans, 1955 (type locality Mexico: Jalisco, Guadelajara) currently a junior subjective synonym of Amblyscirtes (Flor) florus (Godman, 1900) (type locality in Mexico: Nayarit, holotype sequenced as NVG-18083E07) reveals that these two taxa are not monophyletic and R. repta is sister to the clade with Amblyscirtes (Mastor) anubis Godman, 1900, the type species of the subgenus Mastor Godman, 1900 (Fig. 6). Therefore, we reinstate R. repta as a valid species and transfer it to the subgenus Mastor: Amblyscirtes (Mastor) repta Evans, 1955.
Amblyscirtes (Flor) meridus Grishin, new species https://zoobank.org/9BBDAEE2-FF8A-4F93-A33D-7C95FDCC5BE6 (Fig. 6 part, 149–150, 375–376)
Definition and diagnosis.
Phylogenetic trees reveal that a number of specimens from southeastern Mexico identified as Amblyscirtes florus (Godman, 1900) (type locality in Mexico: Nayarit, holotype sequenced as NVG-18083E07) show prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 4.1% (27 bp), and therefore represent a new species. This new species keys to A. florus (N.2.20) in Evans (1955) but differs from it by uncheckered fringes, less developed ventral white overscaling, and frequently better developed blotchy postdiscal pale band on ventral hindwing. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2195.3.17:C141T, aly252.16.2:T159C, aly1259.29.5:C66A, aly536.215.2:T210C, aly3268.16.2:A96G, and COI barcode: T118C, A166G, T193C, T304C, 373C.
Barcode sequence of the holotype.
Sample NVG-19023D09, GenBank OR837689, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCGTTAAGATTATTAATTCGTACTGAATTAGGAAATCCTGGATCTTTAATTGGAGATGACCAAATTTATAACACTATTGTAACAGCTCATGCCTTTATTATAATTTTCTTTATAGTTATGCCTATTATAATTGGAGGTTTTGGAAACTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGAATATTACCTCCTTCATTAATATTATTAATTTCAAGAAGAATCGTGGAAAATGGTGCAGGTACTGGATGAACAGTTTATCCCCCCCTTTCATCAAATATTGCACATCAAGGCTCATCTGTTGATTTAGCTATTTTTTCCCTTCATTTAGCTGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATATATCATTTGATCAAATACCCTTATTTGTTTGATCAGTAGGTATTACTGCTTTATTACTACTTTTATCTTTACCTGTTTTAGCAGGAGCTATTACTATACTTCTTACAGATCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 149–150, bears the following seven rectangular labels, six white: [Paso San Juan | V. Cruz.], [♂], [B.C.A.Lep.Rhop. | Eutychide | asema | Mab.], [Collection | W. Schaus], [DNA sample ID: | NVG-19023D09 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532841], and one red [HOLOTYPE ♂ | Amblyscirtes (Flor) | meridus Grishin]. Paratypes: 4♂♂ from Mexico, H. A. Freeman leg. [AMNH]: NVG-18021A08 Tamaulipas, 15 mi S of Llera de Canales, 27-Jul-1966; NVG-18021A09 Tamaulipas, Monte, 22-Jul-1964, genitalia H-137; NVG-19042H11, AMNH_IZC 00337912 Tamaulipas, Victoria, 16-Aug-1962; NVG-19042H09, AMNH_IZC 00337910 Veracruz, Catemaco, 10-Aug-1967.
Type locality.
Mexico: Veracruz, Paso San Juan.
Etymology.
In Latin, meridiem stands for midday and southeastern and signifies more eastern distribution of this species than its sister A. florus. The name is an adjective.
Distribution.
Southeastern Mexico.
Rectava chiriquensis Grishin, new species https://zoobank.org/B38D4A96-FE18-495F-9423-AB1EC92348CD (Fig. 6 part, 151–152, 377–379)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Panama identified as Rectava sobrinus Schaus, 1902 (type locality Brazil: Rio de Janeiro) shows prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 3.8% (25 bp), and therefore represents a new species. This new species keys to “Papias sobrinus” (J.36.2) in Evans (1955) but differs from similar-looking species by more expressed pale overscaling on somewhat darker ventral wing distal areas, smaller but visible pale spots between veins in most cells on ventral hindwing (Fig. 152), harpe shorter, its ventral margin rounder and less extended in lateral view, dorsal margin with a tooth projecting dorsad and separated from ampulla, costa only slightly concave (Fig. 377–379). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly499.36.11:A168G, aly2680.6.3:A79T, aly2680.6.3:C80A, aly113.6.1:G3177C, aly357.1.11:A96G, aly7186.4.1:A787A (not T), aly173.79.3:C261C (not T), aly88.15.4:C31C (not T), aly423.11.2:G129G (not A), aly529.34.2:A18A (not G), and COI barcode: T38C, T49C, A286G, T499A, T574C.
Barcode sequence of the holotype.
Sample NVG-19019H03, GenBank OR837690, 658 base pairs:
AACTTTATATTTTATTTTCGGAATTTGAGCCGGAATACTAGGTACATCCTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCATTCCCACGAATAAATAATATAAGATTCTGAATACTTCCCCCTTCCTTAATATTGTTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGCACTGGTTGAACTGTTTATCCCCCCCTTTCTTCTAATATTGCACATCAAGGAGCTTCAGTCGATCTAGCAATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTTTAGGAGCTATTAACTTTATCACCACAATTATTAATATACGAATTATAAATTTATCATTTGATCAAATACCATTATTTGTTTGATCAGTTGGAATTACAGCTTTATTATTACTTTTATCTTTACCTGTATTAGCTGGTGCTATTACCATACTCTTAACTGATCGAAATTTAAATACTTCTTTTTTTGATCCTGCCGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 151–152, bears the following six rectangular labels, five white: [Volcan | Chiriqui, Panama | 18 April ‘73 | S. S. Nicolay], [♂ genitalia | /vial # | H569 | Prep. S.S. Nicolay], [Papias ♂ | sobrinus | Schs | DET. BY S.S.NICOLAY], [DNA sample ID: | NVG-19019H03 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532625], and one red [HOLOTYPE ♂ | Rectava chiriquensis | Grishin].
Type locality.
Panama: Chiriquí Province, Volcán.
Etymology.
The name is given for the type locality and is a feminine adjective.
Distribution.
Currently known only from the holotype collected in Chiriquí Province, Panama. This is the only Rectava species known from Central America.
Cobalopsis adictys Grishin, new species https://zoobank.org/CBF4DB85-179A-4240-912C-8B7186AFB049 (Fig. 6 part, 153–154, 380–381)
Definition and diagnosis.
Phylogenetic trees reveal that a number of specimens identified as Cobalopsis dictys (Godman, 1900) (type locality in Mexico: Veracruz, Guatemala, Costa Rica, Panama, illustrated specimen from Guatemala) show prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 3.3% (22 bp), and therefore represent a new species. This new species keys to “Papias dictys” (J.36.6) in Evans (1955) but differs from it by comparatively darker ventral forewing and hindwing area by inner margin, vestigial or absent forewing apical spot (Fig. 153–154), smaller tooth on ventral margin of harpe, distal margin of harpe stronger projecting dorsad, and more angled connection between tegumen and uncus in lateral view (Fig. 381). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1905.5.2:A148G, aly260.3.2:A28T, aly260.3.2:T51C, aly451.31.3:C84T, aly318.41.3:G78A, and COI barcode: A100T, T112C, C343T, T445C, T536C.
Barcode sequence of the holotype.
Sample NVG-19018H05, GenBank OR837691, 658 base pairs:
AACTTTATACTTCATCTTTGGAATTTGAGCCGGAATATTAGGAACTTCTTTAAGATTATTAATTCGAACAGAATTAGGTAATCCTGGATCTTTAATTGGTGATGATCAAATCTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCCCTAATACTTGGTGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGTTTTTGAATACTACCACCTTCCTTAATATTACTAATTTCAAGAAGAATTGTAGAAAATGGGGCAGGTACTGGTTGAACTGTATACCCTCCTCTTTCTTCTAATATTGCACATCAAGGAGCATCTGTTGATTTAGCAATTTTTTCTCTTCATTTAGCTGGTATTTCTTCTATTCTAGGAGCTATTAATTTCATTACTACAATTATTAATATACGAATTAAAAATTTATCTTTTGATCAAATACCTTTATTTGTCTGATCAGTTGGAATTACAGCTTTACTACTACTATTATCTTTACCTGTACTAGCAGGAGCTATTACTATACTTCTTACTGATCGAAATTTAAATACTTCTTTTTTTGATCCTGCAGGAGGAGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 153–154, bears the following four rectangular labels, three white: [PANAMA:VERAGUAS | Sante Fe | 8°31′N 81°05′W | 12.IX.1981 | leg. G.B.Small], [DNA sample ID: | NVG-19018H05 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532536], and one red [HOLOTYPE ♂ | Cobalopsis | adictys Grishin]. Paratypes: 2♂♂ from Costa Rica: Area de Conservación Guanacaste, Guanacaste Prov., Sector Pitilla [USNM]: NVG-18064B08, 11-SRNP-33187 Sendero Nacho, 710 m, GPS 10.9845, −85.4248, eclosed 17-Nov-2011; NVG-18064C02, 11-SRNP-30141 Sendero Laguna 680 m GPS 10.9888, −85.4234, eclosed 3-Mar-2011.
Type locality.
Panama: Veraguas Province, Santa Fe, GPS 8.5167, −81.0833.
Etymology.
The name is derived by combining dictys with the negating prefix “a”. As a mnemonic, a longer name indicates it is a more southern species than C. dictys. The name is a noun in apposition.
Distribution.
Costa Rica and Panama.
Cymaenes melaporphyrus Grishin, new species https://zoobank.org/48D8BE5E-3F74-4EA4-948F-F9D60BB59FFC (Fig. 6 part, 155–156, 382–383)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Mexico and Guatemala identified as Cymaenes laureolus (Schaus, 1913) (type locality in Costa Rica, holotype sequenced as NVG-15109B05) are not monophyletic with it and show prominent genetic differentiation from it in the Z chromosome (Fig. 6), although their COI barcodes differ by only 0.9% (6 bp), and therefore represent a new species. This new species keys to Cymaenes laureolus laureolus (J.27.6(a)) in Evans (1955) but differs from it by better developed white subapical spots on forewing and more prominent pale areas on ventral hindwing that are tinted with purplish brown rather than plain brown (Fig. 155–156), valva is broader, more square, and teeth on the harpe are more prominent (Fig. 382–383). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1249.14.4:C96T, aly1249.14.4:A147G, aly527.6.11:A63G, aly594.19.2:C72T, aly3936.3.3:C88T, and does not consistently differ from its close relatives in COI barcode.
Barcode sequence of the holotype.
Sample NVG-7249, GenBank OR837692, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCCGGAATATTAGGAACATCTTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCAGGTTCTCTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCCCTAATATTAGGAGCTCCTGATATAGCATTTCCACGAATAAATAATATAAGATTTTGAATATTGCCTCCTTCTTTAATACTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACTGGTTGAACTGTTTATCCTCCTCTTTCTTCTAATATTGCTCACCAAGGAGCTTCAGTTGATTTAGCAATTTTTTCTCTTCATTTAGCAGGTATTTCTTCTATTCTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAGTTAGAAATTTATCATTTGATCAAATACCCCTATTTGTTTGATCAGTTGGAATTACAGCTTTATTATTACTTTTATCTTTACCTGTATTAGCAGGTGCTATTACTATACTTTTAACTGATCGAAATTTAAATACTTCATTTTTTGACCCTGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 155–156, bears the following six rectangular labels, five white: [Mex: S.L.P. La Mera Ceiba |13 July 1990 - el. 700′ | John Kemner], [Cymaenes ♂ | laureolus | Schaus | Det. H.A. Freeman], [DNA sample ID: | NVG-7249 | c/o Nick V. Grishin], [genitalia | NVG161005–76 | Nick V. Grishin], [USNMENT | {QR Code} | 01321097], and one red [HOLOTYPE ♂ | Cymaenes | melaporphyrus Grishin]. Paratypes: 2♂♂ AMNH: NVG-15111G01 Mexico: Veracruz, Coatepec, Jun-1917; NVG-15111G05 Guatemala: Alta Verapaz, Municipio San Cristobal Verapaz, Baleu, >1350 m, 14-Jun-1966, E. C. Welling leg.
Type locality.
Mexico: San Luis Potosí, La Mera Ceiba, elevation 700 ft.
Etymology.
In Greek, μελανό (melano) means dark, and πορφύρα (porphyra) means purple. The name is given for the purplish sheen of the ventral hindwing in this dark-brown species and is an adjective.
Distribution.
Currently known from Mexico and Guatemala.
Lerema (Morys) ecuadorica Grishin, new species https://zoobank.org/0047F48A-FECF-40CC-82C0-43631FD3A5CE (Fig. 6 part, 157–158, 384–385)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Ecuador identified as Lerema (Morys) micythus (Godman, 1900) (type locality in Mexico: Guerrero and Tabasco, and Costa Rica, illustrated specimen from Tabasco) is not monophyletic with it and shows prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 2.3% (15 bp) and, therefore, represents a new species. This new species keys to “Morys compta micythus” (J.40.2(a)) in Evans (1955) but differs from it and other relatives by wings unspotted on both sides (Fig. 157–158) and harpe massively expanded posteriad, somewhat ax-shaped, its dorsal tooth longer and narrower, smoothly merged with harpe without separation between them (Fig. 384–385). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly18312.9.17:A27T, aly18312.9.17:C39T, aly6841.19.2:A449G, aly4778.7.5:A84C, aly37338.27.1:G351A, aly1042.2.2:C27C (not T), aly1042.8.5:C138C (not G), aly1042.8.5:C151C (not T), aly29.8.12:C153C (not A), aly536.180.3:T69T (not C), and COI barcode: A100G, T169C, T304C, C355T, T616C.
Barcode sequence of the holotype.
Sample NVG-19021F08, GenBank OR837693, 658 base pairs:
AACTTTATATTTTATTTTCGGAATTTGAGCAGGAATATTAGGAACTTCTTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCAGGATCTTTAATTGGGGATGACCAAATTTATAATACTATTGTTACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCCATTATAATTGGAGGATTTGGTAATTGATTAGTTCCATTAATATTAGGAGCACCTGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGAATACTTCCTCCTTCCTTAATATTATTAATTTCAAGAAGAATCGTAGAAAATGGAGCAGGAACAGGATGAACTGTTTATCCTCCTTTATCTTCTAATATTGCTCATCAAGGAGCTTCAGTTGATTTAGCAATTTTTTCTCTTCATTTAGCTGGTATTTCTTCTATTTTAGGTGCTATTAATTTTATTACTACAATTATTAATATACGAGTAAGAAATTTATCTTTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACTGCATTATTATTACTTTTATCATTACCTGTATTAGCAGGTGCTATTACTATACTTTTAACTGATCGAAATTTAAATACTTCATTTTTTGACCCTGCCGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 157–158, bears the following four rectangular labels, three white: [ECUADOR, Pichincha | Prov.: Tinalandia | 2200′ 18–20 Apr 1990 | coll J. W. Brown], [DNA sample ID: | NVG-19021F08 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532698], and one red [HOLOTYPE ♂ | Lerema (Morys) | ecuadorica Grishin].
Type locality.
Ecuador: Pichincha Province, Tinalandia, elevation 2200 ft.
Etymology.
The name is given for the type locality. The name is a feminine adjective.
Distribution.
Currently known only from the holotype collected in Pichincha Province, Ecuador.
Saturnus tiberius (Möschler, 1883) (with Phlebodes tiberius form suffuscus Hayward, 1940 as its subspecies), Saturnus meton (Mabille, 1891), Saturnus conspicuus (E. Bell, 1941), and Saturnus obscurus (E. Bell, 1941) are species distinct from Saturnus reticulata (Plötz, 1883)
Genomic sequencing taxa currently treated as subspecies of Saturnus reticulata (Plötz, 1883) (type locality in Venezuela and Panama, syntype sequenced as NVG-18043D11), namely: Apaustus tiberius Möschler, 1883 (type locality in Suriname, holotype sequenced as NVG-15035F10), Phamphila [sic] meton Mabille, 1891 (type locality in Brazil: Amazonas, syntype sequenced as NVG-15035F11), Phlebodes tiberius form suffuscus Hayward, 1940 (type locality in Ecuador), Phlebodes tiberius race obscurus Bell, 1941 (type locality Panama: Chiriquí, holotype sequenced as NVG-18026E04), Phlebodes tiberius race conspicuus Bell, 1941 (type locality in Brazil: Santa Catarina, holotype sequenced as NVG-18025G02) reveals that all but P. t. suffuscus are genetically differentiated from each other in the Z chromosome at the level characteristic of species (Fig. 6), and the latter taxon is closely related to A. tiberius. Curiously, the mitochondrial genome (Fig. 6b) does not follow the evolutionary path of the nuclear genome (Fig. 6a), likely due to gene exchange and introgression. Therefore, the COI barcodes of some of these taxa do not differ, while others (Saturnus reticulata) are not monophyletic in mitogenomes. The southernmost P. t. conspicuus exhibits the largest divergence in COI barcodes: 5.5% (36 bp) (likely due to more restricted gene exchange with other taxa, mitochondrial genome remained unaffected), while not being the most divergent taxon in the nuclear genome (Fig. 6a). This example illustrates that it is not always possible to rely on mitochondrial DNA in general and on COI barcodes in particular in taxonomic and phylogenetic studies. As suggested by nuclear genome analysis and supported by phenotypic differences, we reinstate the following taxa as species: Saturnus tiberius (Möschler, 1883) and Saturnus meton (Mabille, 1891), and propose a new status of species for Saturnus conspicuus (E. Bell, 1941), and Saturnus obscurus (E. Bell, 1941). We leave one taxon as a subspecies but reinstate its original species-subspecies combination: Saturnus tiberius suffuscus (Hayward, 1940). As a result, S. reticulata becomes monotypic.
Saturnus obscurior Grishin, new species https://zoobank.org/6DBB3DEA-776E-42D3-8940-41664259C043 (Fig. 6 part, 159–160, 386–387)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Costa Rica and Panama identified as Saturnus obscurus (E. Bell, 1941) (type locality in Panama, holotype sequenced as NVG-18026E04) show prominent genetic differentiation from it in the Z chromosome (Fig. 6), although their COI barcodes differ by only 1.2% (8 bp), and therefore represent a new species. This new species keys to “Saturnus tiberius obscurus” (L.1.4(a)) in Evans (1955) but differs from it by being darker overall, e.g., ventral hindwing in males lacks the large yellow patch and is only tinted in yellow: in the middle, along the veins and as a series of postdiscal spots; forewing yellow spots are more widely separated from each other. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly499.34.3:T132C, aly1838.37.3:A57G, aly2011.20.4:C183T, aly1113.2.10:C121T, aly1603.31.2:C163A, and COI barcode: T193T, A217A, 235T, A607C.
Barcode sequence of the holotype.
Sample NVG-19024A12, GenBank OR837694, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATACTAGGAACATCTTTAAGTTTATTAATTCGAACAGAATTAGGAAATCCAGGTTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACTGCCCATGCATTTATTATAATTTTTTTCATAGTAATACCAATCATAATTGGAGGTTTTGGTAATTGATTAGTACCATTAATATTAGGAGCCCCTGATATAGCTTTTCCTCGAATAAATAACATAAGATTTTGAATACTTCCCCCTTCATTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCTGGTACTGGTTGAACAGTTTACCCCCCTCTTTCTGCTAATATTGCACATCAAGGAGCATCTGTAGATTTAGCAATTTTTTCACTTCATTTAGCAGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAATTTATCATTTGATCAAATACCCTTATTTGTATGATCAGTTGGTATTACTGCATTACTTTTACTTTTATCCTTACCTGTTTTAGCAGGGGCTATCACTATACTCTTAACTGATCGAAATCTTAATACATCCTTTTTTGATCCAGCAGGAGGAGGAGACCCAATCCTCTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 159–160, bears the following four rectangular labels, three white: [PANAMA: Darien | Cana 400m | 27.VI.1981 | Leg. G. B. Small], [DNA sample ID: | NVG-19024A12 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532899], and one red [HOLOTYPE ♂ | Saturnus | obscurior Grishin]. Paratypes: 2♂♂ [USNM]: NVG-17106B04, 12-SRNP-1268 Costa Rica, Area de Conservación Guanacaste, Alajuela Prov., Sector San Cristobal, Sendero Huerta, 527 m, GPS 10.9305, −85.3722 eclosed 26-Apr-2012; NVG-19024B01, USNMENT_01532900 Panama: Panama Province, Madden Forest Preserve, 6-Feb-1968, S. S. Nicolay leg., genitalia H337 prep. S. S. Nicolay.
Type locality.
Panama: Darien Province, Cana, elevation 400 m.
Etymology.
In Latin, obscurior means darker, dimmer, duskier, or more obscure. The name is given for the darker appearance of this species due to the loss of paler coloration. The name is a comparative adjective treated as a noun in apposition.
Distribution.
Costa Rica and Panama.
Cantha zoirodicta Grishin, new species https://zoobank.org/7874E2A2-2FE3-45BD-BA94-F363C90CB2CE (Fig. 6 part, 161–162, 388–390)
Definition and diagnosis.
Phylogenetic trees reveal that two specimens identified as Cantha zara (E. Bell, 1941) (type locality in Bolivia, holotype sequenced as NVG-18022E02) show prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 6.5% (43 bp), and therefore represent a new species. This new species keys to “Cantha celeus zara” (I.9.1(b)) in Evans (1955) but differs from it by rounder wings, browner (not reddish) ground color, and yellow framing at outer margins of both wings beneath, in addition to yellow veins. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly2178.10.1:C102T, aly363.37.1:A909G, aly363.37.1:T3771G, aly1139.50.8:T279G, aly1139.50.8:T372G, and COI barcode: 67G, T74C, A88T, T508C, A535G, A565G.
Barcode sequence of the holotype.
Sample NVG-8016, GenBank OR837695, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACATCATTAAGTTTATTAATTCGGACAGAACTAGGAAATCCTGGTTCTCTTATTGGAGATGATCAAATTTATAATACCATTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGAAATTGATTAGTACCCCTAATATTAGGGGCCCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATATTACCCCCTTCTTTAATATTATTAATTTCAAGAAGAATCGTAGAAAATGGTGCAGGAACTGGATGAACTGTTTATCCTCCTCTTTCATCTAATATTGCCCATCAAGGAGCATCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCAGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAATAATATATCATTTGATCAAATACCTTTATTTGTCTGATCAGTTGGTATTACCGCATTATTGTTACTTTTATCTTTACCGGTATTAGCTGGGGCTATTACTATACTTTTAACGGATCGAAATTTAAATACATCATTTTTTGATCCTGCTGGAGGAGGGGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 161–162, bears the following seven rectangular labels, six white: [PERU 300m | 30 Km S. W. | Pto. Maldonado | 8 May ‘84 | S. S. Nicolay], [♂ genitalia | slide/vial # | H864 | Prep. S.S. Nicolay], [Cantha ♂ | celeus | Det. zara Bell | S.S. Nicolay], [DNA sample ID: | NVG-8016 | c/o Nick V. Grishin], [genitalia | NVG170208–01 | Nick V. Grishin], [USNMENT | {QR Code} | 01321856], and one red [HOLOTYPE ♂ | Cantha zoirodicta | Grishin]. Paratype: 1♂ NVG-21046E03 Brazil: Rondonia, 62 km S of Ariquemes, linha C-10, 5 km S of Cacaulandia, 25-Apr-1995, G. T. Austin leg., genitalia GTA-8707 [MGCL].
Type locality.
Peru: Madre de Dios Region, 30 km SW of Puerto Maldonado, elevation 300 m.
Etymology.
The name is a compound word of Greek ζωηρός (zoirós) meaning lively, vivid, or vibrant, and δικτυωτός (diktyotós) meaning reticulated. The name reflects the bright reticulated pattern of this species and is a noun in apposition.
Distribution.
Amazonian Peru and Brazil.
Cantha meiodicta Grishin, new species https://zoobank.org/7756C17B-E4A5-42D2-B809-ECD97BC3EA77 (Fig. 6 part, 163–164, 391–393)
Definition and diagnosis.
Phylogenetic trees reveal that several specimens identified as Phlebodes eteocla (Plötz, 1882) or Phlebodes virgo Evans, 1955 are not closely related to these species and instead are conspecific, belong to Cantha Evans, 1955 (type species Cantha celeus calva Evans, 1955), and are a distant sister to Cantha zoirodicta new species (Fig. 6): e.g., their COI barcodes differ by 5.9% (39 bp), and therefore represent a new species. This new species is similar to C. zoirodicta new species but the hindwings are more pointed at the tornus, the amount of yellow framing of margins and veins beneath is reduced, and yellow spots are smaller, especially on the dorsal hindwing, which is completely unspotted in the holotype. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1935.7.3:G30C, aly1935.7.3:G87A, aly1497.7.1:A1020T, aly103.32.1:T650A, aly103.32.1:A681G, and COI barcode: A34T, A67T, T124C, A184G, A256T, T517C.
Barcode sequence of the holotype.
Sample NVG-19024C02, GenBank OR837696, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGTATATTAGGAACATCATTAAGTTTATTAATTCGTACTGAATTAGGAAATCCTGGATCTCTTATTGGAGATGATCAAATTTATAATACTATCGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGGTTTGGGAATTGATTAGTACCTTTAATATTAGGAGCCCCTGATATAGCTTTTCCTCGAATAAATAACATAAGTTTTTGAATATTACCTCCTTCTTTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACTGGATGAACTGTTTATCCTCCTCTTTCTTCTAATATTGCTCACCAAGGTTCATCTGTTGATCTAGCAATTTTTTCTTTACATTTAGCAGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAATAATATATCATTCGACCAAATACCTTTATTTGTTTGATCAGTCGGAATTACTGCTTTATTATTACTTTTATCTTTACCTGTATTAGCAGGAGCTATTACTATACTTTTAACGGATCGAAATTTAAATACATCATTTTTTGATCCTGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 163–164, bears the following seven rectangular labels, six white: [PERU 300m | 30 Km S. W. | Pto. Maldonado | 8 May ‘84 | S. S. Nicolay], [♂ genitalia | slide/vial # | H880 | Prep. S.S. Nicolay], [Phlebodes | eteocla ♂ | Det. | S.S. Nicolay], [Parphorus sp. n.], [DNA sample ID: | NVG-19024C02 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532910], and one red [HOLOTYPE ♂ | Cantha meiodicta | Grishin]. Paratypes: 2♂♂ NVG-19019B09, USNMENT_01532561 the same data as the holotype but 17-Oct-1983 and not dissected and NVG-21048C09 Brazil: Rondônia, 62 km S of Ariquemes, linha C-10, 5 km S of Cacaulandia, 10-Jul-1993, O. Gomes leg., genitalia GTA-3649 [MGCL].
Type locality.
Peru: Madre de Dios Region, 30 km SW of Puerto Maldonado, elevation 300 m.
Etymology.
The name is a compound word of Greek μειώ (meió) meaning reduce, decrease, or lessen, and δικτυωτός (diktyotós) meaning reticulated. The name points to less contrasting lattice patterns compared to the previous species and is a noun in apposition.
Distribution.
Amazonian Peru and Brazil.
Phlebodes sifax Evans, 1955 is a species distinct from Phlebodes campo (E. Bell, 1947)
Genomic sequencing of Phlebodes campo sifax Evans, 1955 (type locality in Brazil: Amazonas) and Phlebodes campo campo (E. Bell, 1947) (type locality in Brazil: Rio de Janeiro, holotype sequenced as NVG-18025F03) reveals prominent genetic differentiation between them (Fig. 6): e.g., their COI barcodes differ by 4.1% (27 bp). Therefore, we propose that the former is a species-level taxon Phlebodes sifax Evans, 1955, new status.
Phlebodes duplex Grishin, new species https://zoobank.org/0AEAD528-03AB-4BD0-B8C1-D78CD4EFCB98 (Fig. 6 part, 165–166, 394–395)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Guatemala identified as Phlebodes campo (E. Bell, 1947) (type locality Brazil: Rio de Janeiro, holotype sequenced as NVG-18025F03) show prominent genetic differentiation from it (Fig. 6): e.g., their COI barcodes differ by 4% (26 bp), and therefore represent a new species. This new species keys to Phlebodes campo campo (L.2.3(b)) in Evans (1955) but differs from its relatives by having a doublet of (merged) pale spots (rather than one spot or a large cream-colored area) in the forewing cell CuA2-1A+2A beneath and yellow framing of veins less developed than in Phlebodes sifax. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1139.40.3:T144A, aly2659.10.1:A136G, aly2659.10.1:C137A, aly2659.10.1:G139A, aly2659.10.1:A140G, and COI barcode: T91T, T106C, C529C, A532A, T619T.
Barcode sequence of the holotype.
Sample NVG-19024B10, GenBank OR837697, 658 base pairs:
AACTTTATACTTTATTTTCGGAATTTGAGCAGGAATATTAGGAACATCTTTAAGACTACTAATTCGTACAGAATTAGGAAATCCAGGATCTTTAATTGGAGATGACCAAATTTATAACACTATTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCTCGTATAAATAATATAAGATTTTGAATACTTCCCCCTTCTTTAATATTACTAATCTCAAGAAGAATCGTAGAAAATGGAGCAGGAACTGGTTGAACTGTTTATCCCCCCCTTTCCTCTAATATCGCCCATCAAGGATCTTCTGTTGATTTAGCAATTTTTTCTCTTCACTTAGCAGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAATTTATCATTCGATCAAATACCTTTATTTGTTTGATCAGTAGGTATTACAGCCTTATTATTACTTTTATCTTTACCTGTATTAGCTGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGTGGAGGAGATCCAATTCTCTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 165–166, bears the following six rectangular labels, five white: [Cayuga | Guat], [Sept.], [Schaus and | Barnes | coll], [DNA sample ID: | NVG-19024B10 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532907], and one red [HOLOTYPE ♂ | Phlebodes | duplex Grishin]. Paratype: 1♂ NVG-21048C01 Guatemala: Peten, Parque Nacional Tikal, 31-May-1992, N. M. Haddad leg., genitalia GTA-3240 [MGCL].
Type locality.
Guatemala: Cayuga.
Etymology.
In Latin, duplex means twofold, double, dual, both, thick, or strong. The name is given for the double (diffuse) spot in the ventral forewing cell CuA2-1A+2A and is a noun in apposition.
Distribution.
Currently known only from Guatemala.
Lychnuchus (Enosis) ponka (Evans, 1955), new combination
Genomic sequencing reveals that Thoon ponka Evans, 1955 (type locality in Brazil: Para) is in a different clade from Thoon Godman, 1900 (type species Proteides modius Mabille, 1889), and instead originates within the subgenus Enosis Mabille, 1889 (type species Enosis dognini Mabille, 1889) of Lychnuchus Hübner, [1831] (type species Lychnuchus olenus Hübner, [1831], which is a junior subjective synonym of Hesperia celsus Fabricius, 1793) (Fig. 7). Therefore, we propose a new combination Lychnuchus (Enosis) ponka (Evans, 1955).
Figure 7.
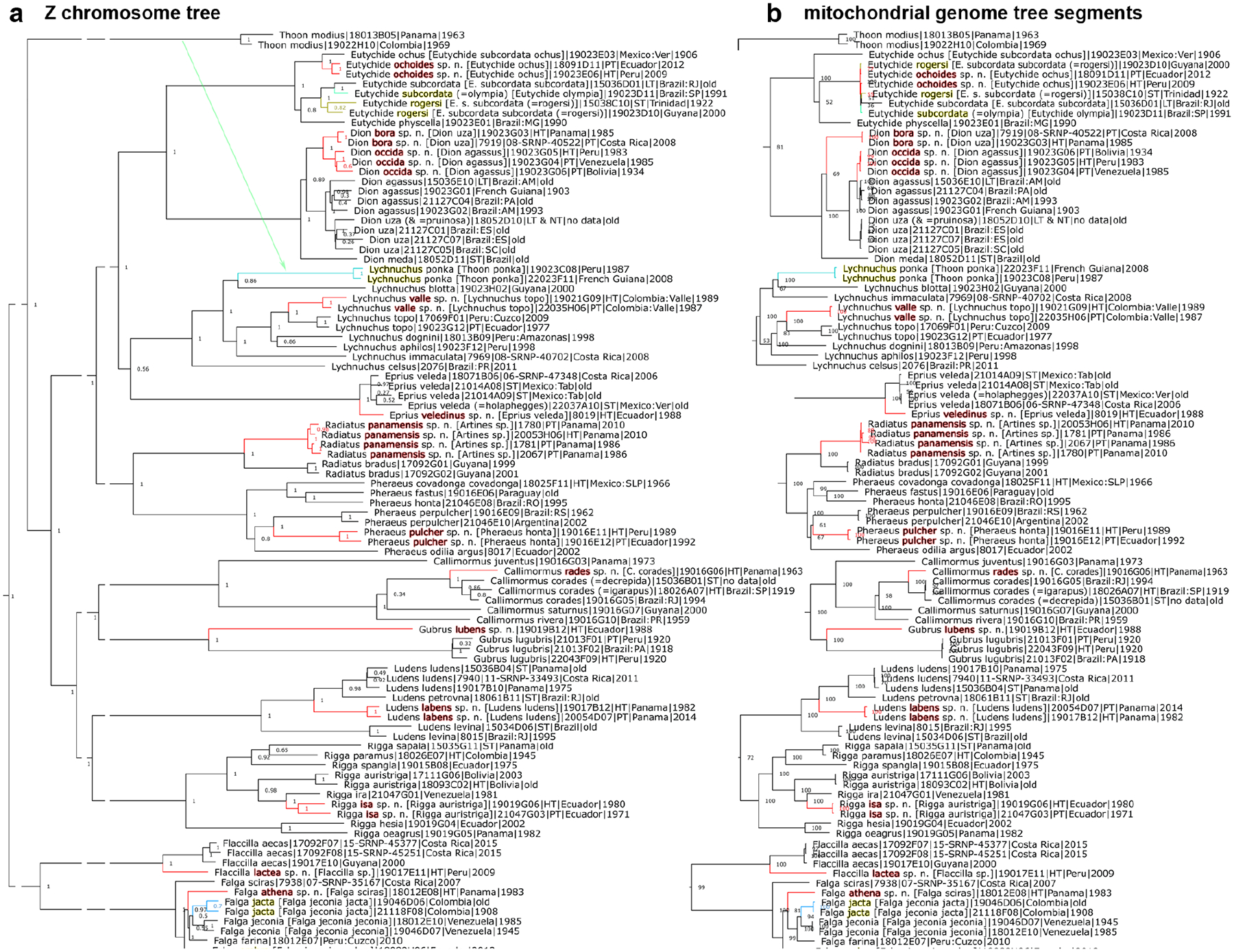
Phylogenetic trees of Moncina (part 3) and Flagina inferred from protein-coding regions in a) the Z chromosome and b) the mitochondrial genome (in segments). See Fig. 1 legend for other notations.
Lychnuchus (Enosis) valle Grishin, new species https://zoobank.org/97BEB2D4-9C1E-42D1-82F5-A1E7102CFF7F (Fig. 7 part, 167–168, 396–398)
Definition and diagnosis.
Phylogenetic trees reveal that two females from eastern Colombia identified as Lychnuchus (Enosis) topo (Nicolay, 1980) (type locality in Ecuador) show prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 5.8% (38 bp), and therefore represent a new species. This new species differs from L. topo, which is known only from males, by vestigial or lacking pale dot on the ventral hindwing and darker tornal area of the ventral forewing. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly320.2.13:T495C, aly1603.33.3:G145A, aly770.42.5:A69T, aly2096.2.3:G151C, aly3370.5.2:C48A, and COI barcode: T10C, T64C, T451C, A535T, T574C.
Barcode sequence of the holotype.
Sample NVG-19021G09, GenBank OR837698, 658 base pairs:
AACTTTATACTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACTTCTTTAAGATTATTAATCCGTACAGAACTAGGTAATCCAGGTTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCCCATGCTTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTTTAATATTAGGAGCACCTGATATAGCTTTCCCACGAATAAATAACATAAGATTTTGAATATTACCTCCCTCTTTAATGTTATTGACTTCAAGAAGTATCGTAGAAAATGGTGCAGGTACAGGATGAACAGTTTATCCTCCTTTATCTTCTAATATTGCTCATCAAGGTTCTTCTGTTGACTTAGCTATTTTTTCCTTACATTTAGCAGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACCACAATTATTAATATACGAATTAAAAATATATCATTTGATCAAATACCTTTATTTGTTTGATCAGTAGGTATTACAGCCTTACTTTTACTTTTATCTTTACCTGTTTTAGCTGGAGCTATTACCATACTTCTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCAGCCGGAGGAGGAGACCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 167–168, bears the following five rectangular labels, four white: [Jan.29, 1989 | Carret del Mar | to Dapa Rd., 1800 m | Valle, Colombia | J. B. Sullivan], [Genit. Vial | SRS-3248], [DNA sample ID: | NVG-19021G09 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532704], and one red [HOLOTYPE ♀ | Lychnuchus (Enosis) | valle Grishin]. Paratype: 1♀ NVG-22035H06 Colombia: Valle, San Antonio, above Cali, 1900 m, 2-Feb-1987, J. B. Sullivan leg., genitalia SRS-3224 [USNM].
Type locality.
Colombia: Valle, Carret del Mar to Dapa Rd., elevation 1800 m.
Etymology.
The name is for the type locality and is a noun in apposition.
Distribution.
Eastern Colombia.
Lectotype designation for Eutychide ochus Godman, 1900
Eutychide ochus Godman, 1900, currently a subspecies of Eutychide subcordata (Herrich-Schäffer, 1869), was described from a series of specimens collected at multiple localities: Mexico: Veracruz, Nicaragua, and Brazil: Amazonas. Due to the polytypic type series and to define the type locality more precisely, N.V.G. hereby designates a male syntype in the Natural History Museum, London, UK, that according to its label, was illustrated by Godman (1899–1901) and bears the following 11 rectangular (except the first three, which are round and with a red circle on the upper side, and the 9th which is round and yellow) white labels: (Type), (Type), (Type | H. T.), [Atoyac, | Vera Cruz. | May. H.H.S.], [♂], [Sp. figured.], [B.C.A.Lep.Rhop. | Eutychide | ochus, | Godm.], [Godman-Salvin | Coll. 1912.—2.], (13), [16 { genitalia glued to this label }], and [PHOTO | AA], as the lectotype of Eutychide ochus Godman, 1900. The type locality of E. ochus becomes Mexico: Veracruz, Atoyac.
Hesperia olympia Plötz, 1882 is a junior subjective synonym of Eutychide subcordata (Herrich-Schäffer, 1869) and a lectotype designation
Genomic sequencing of a syntype of Cobalus subcordata Herrich-Schäffer, 1869 (type locality not specified, probably Brazil: Rio de Janeiro per Plötz (1882)) reveals that Evans (1955) misidentified this species. This specimen is a syntype because it agrees with the original description, is from the Herrich-Schäffer collection, and bears a label in likely Herrich-Schäffer’s handwriting “subcordata | m”, where ‘m’ stands for ‘mihi’ (Latin for ‘of me’), placed after a species name as an attribution of the new species to the writer. This notation was common over a century ago, instead of the author’s name being written directly. This ‘m’ confirms that the label was written by Herrich-Schäffer and offers additional evidence that this specimen is a syntype.
To define this species objectively, N.V.G. hereby designates the sequenced male syntype in the Museum für Naturkunde, Berlin, Germany, that bears the following ten rectangular labels (the first and the fourth are purple, others are white): [Origin.], [Co subcordata | m], [Coll. H.—Sch.], [syn– t. | volesus Mab.], [912.], [Coll. | Staudinger], [subcordata | H. Sch.], [Caryst. | subcordata | HS.], [{QR Code} http://coll.mfn-berlin.de/u/ | 449f81], and [DNA sample ID: | NVG-15036D01 | c/o Nick V. Grishin] as the lectotype of Cobalus subcordata Herrich-Schäffer, 1869, currently in the genus Eutychide Godman, 1900 (type species Hesperia physcella Hewitson, 1866). While the lectotype lacks a locality label, the genomic tree places the lectotype with a specimen from Southeast Brazil, thus suggesting that the type locality of Eutychide subcordata is in Southeast Brazil, in agreement with Plötz (1882), who stated that the type locality was “Rio”.
According to genomic comparison (Fig. 7) and phenotypic characters, the lectotype of Eutychide subcordata is conspecific with Hesperia olympia Plötz, 1882 (type locality in Brazil) as the latter species was identified by Draudt (1921–1924), who already used the combination Cobalus subcordata form olympia, and later treated as a species by Evans (1955), who misidentified E. subcordata. Therefore, we propose that Hesperia olympia Plötz, 1882 is a new junior subjective synonym of Eutychide subcordata (Herrich-Schäffer, 1869).
Eutychide ochus Godman, 1900 and Eutychide rogersi (Kaye, 1914) are valid species-level taxa
Placed as a subspecies of Eutychide subcordata (Herrich-Schäffer, 1869) (type locality likely in Brazil: Rio de Janeiro) by Evans (1955), Eutychide ochus Godman, 1900 (type locality in Mexico: Veracruz) is not conspecific with the former because Evans misidentified E. subcordata, which instead, as shown above, is conspecific with and a senior name for Hesperia olympia Plötz, 1882 (type locality in Brazil). Therefore, the senior name for the species Evans misidentified as E. subcordata is E. ochus, and a senior name for the subspecies Evans misidentified as E. subcordata subcordata is Cobalopsis rogersi Kaye, 1914 (type locality in Trinidad, syntype sequenced as NVG-15038C10) (Fig. 7). However, Evans (1955) mentioned male genitalia differences between his subspecies of “E. subcordata” and genomic sequencing suggests that these “subspecies” are species-level taxa. Therefore, we reinstate these two taxa as species: Eutychide ochus Godman, 1900 and Eutychide rogersi (Kaye, 1914).
Eutychide ochoides Grishin, new species https://zoobank.org/972EB90E-A40C-45F1-8ED0-F796092C18B9 (Fig. 7 part, 169–170, 399–401)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Ecuador and Peru identified as Eutychide ochus Godman, 1900 (type locality in Mexico: Veracruz) show prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 7.3% (48 bp), and therefore represent a new species. This new species keys to “Eutychide subcordata ochus” (J.50.3(a)) in Evans (1955) but differs from it by more contrasty hindwing with submarginal area paler than discal area and typically larger subapical forewing spots of about equal size. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly5.7.1:T1986A, aly5.7.1:A2066G, aly1249.21.19:T39C, aly1036.5.4:C96T, aly851.2.8:C43T, and COI barcode: T92C, 157C, T287C, 346A, T436C, T568A.
Barcode sequence of the holotype.
Sample NVG-19023E06, GenBank OR837699, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCTTTAAGATTATTAATTCGAACAGAATTAGGAAATCCTGGTTCACTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTCATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCTTTAATATTAGGAGCACCTGATATAGCTTTCCCACGAATAAACAATATAAGATTTTGAATATTACCCCCTTCATTAATATTACTAATTTCTAGAAGAATTGTTGAAAATGGTGCAGGTACAGGATGAACAGTTTACCCCCCACTTTCATCTAATATTGCTCACCAAGGTTCTTCAGTTGATCTAGCAATTTTCTCCCTACACTTAGCAGGAATTTCTTCTATTTTAGGAGCCATTAACTTTATTACTACAATTATTAATATACGAATTAGAAATTTATCATTTGATCAAATACCTTTATTTGTATGATCTGTAGGTATTACAGCCCTATTATTATTATTATCATTACCTGTATTAGCAGGTGCAATTACAATACTTTTAACCGACCGAAATTTAAATACTTCTTTTTTTGATCCAGCTGGAGGAGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 169–170, bears the following four rectangular labels, three white: [PERU: Cuzco:Cosñipata Valley | San Pedro vicinity 1,373m. | 13°03′30″S, 71°32′64″W | 11 August 2009 Brian Harris], [DNA sample ID: | NVG-19023E06 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532848], and one red [HOLOTYPE ♂ | Eutychide | ochoides Grishin]. Paratype: 1♂ NVG-18091D11 Ecuador: Morona-Santiago, Mendez, 800 m, GPS −2.42, − 78.20, 9-Nov-2012, J.-C. Petit leg., [EBrockmann].
Type locality.
Peru: Cuzco, Cosñipata Valley, near San Pedro, elevation 1373 m, GPS −13.0583, −71.5483.
Etymology.
The name reflects a similarity to E. ochus. It is longer to signify a more southern distribution. The name is an adjective.
Distribution.
Ecuador and Peru.
Dion bora Grishin, new species https://zoobank.org/75DE2A48-50F7-4D25-8E27-F1F95CCD0CE4 (Fig. 7 part, 171–172, 402–403)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Costa Rica and Panama identified as Dion uza (Hewitson, 1877) (type locality not specified, lectotype sequenced as NVG-18052D10) show prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 3.6% (24 bp), and therefore represent a new species. This new species keys to “Enosis pruinosa pruinosa” (K.4.3(a)) in Evans (1955), which was misidentified by Evans, see Zhang et al. (2022b), and differs from its relatives by a combination of the following characters: ventral hindwing with broader blue metallic overscaling in posterior half and partly along forewing outer margin, similar to D. uza, but hindwing discal cyan-blue spots are more prominent on the blue ground color (Fig. 172) and less developed than in another new species described below, tube-like arched upcurved process from near the base of harpe is thinner and longer, harpe is broader, with two humps on its dorsal margin, costa straighter, less concave (Fig. 403). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly6954.4.5:G69A, aly6954.4.5:C91T, aly527.12.3:T74C, aly103.13.2:C60T, aly103.13.2:A78G, and COI barcode: T49C, T91A, T325C, A553G, C539T, T601C.
Barcode sequence of the holotype.
Sample NVG-19023G03, GenBank OR837700, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCCTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCTGGCTCATTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTCTAATATTAGGAGCACCTGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGAATACTACCCCCTTCCCTTATACTATTAACTTTTAGAAGAATTGTAGAAAATGGTGCAGGTACCGGATGAACAGTTTACCCCCCCCTTTCTTCTAATATTGCCCATCAAGGTTCTTCAGTTGATTTAGCAATTTTTTCATTACATTTAGCAGGAATTTCTTCTATTTTAGGTGCTATTAATTTTATTACAACAATTATTAATATACGAATTAAAAATTTATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGTATTACAGCTTTATTATTATTATTATCTTTACCGGTATTAGCAGGAGCTATTACAATACTTCTCACTGATCGAAATTTAAACACTTCTTTTTTTGATCCAGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 171–172, bears the following four rectangular labels, three white: [PANAMA: Darien | Rio Tuira at | Rio Pucuro | 16–17 Feb. 1985 | J. Louton], [DNA sample ID: | NVG-19023G03 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532867], and one red [HOLOTYPE ♂ | Dion bora | Grishin]. Paratype: ♂ NVG-7919, 08-SRNP-40522 Costa Rica: Area de Conservación Guanacaste, Alajuela Prov., Sector Rincon Rain Forest, Laureles, 95 m, GPS 10.9332, −85.25335, genitalia NVG170207–04 [USNM].
Type locality.
Panama: Darien Province, Rio Tuira at Rio Pucuro.
Etymology.
The name is formed from the Latin borealis, meaning northern. It signifies it is the northernmost species within the D. uza group. The name is a noun in apposition.
Distribution.
El Salvador to Panama.
Dion occida Grishin, new species https://zoobank.org/0458BF11-8AEB-4AF2-8B02-A4F6DF108B58 (Fig. 7 part, 173–174, 404–406)
Definition and diagnosis.
Phylogenetic trees reveal that several specimens identified as Dion agassus (Mabille, 1891) (type locality type locality Brazil: Amazonas, Massauary, lectotype sequenced as NVG-15036E10) show prominent genetic differentiation from it in the Z chromosome and are sister to Dion bora new species (Fig. 7), but their COI barcodes differ from D. bora by 4.1% (27 bp) while being closer to sympatric D. agassus, probably due to introgression: 1.1% (7 bp). Therefore, these specimens represent a new species that keys to “Enosis pruinosa agassus” (K.4.3(a)) in Evans (1955) and differs from its relatives by a combination of the following characters: ventral hindwing with comparatively reduced blue metallic overscaling and on forewing replaced by purple tint overscaling, similar to D. agassus, but hindwing discal cyan-blue spots are most prominent on the blue ground color, and blue overscaling is framing the inner margin (Fig. 174), the tube-like arched upcurved process from near the base of harpe is thicker and shorter, harpe is narrower, rounder, with a straight or convex dorsal margin that bears a small hump directed partly inward and anteriad, costa more concave (Fig. 405–406). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly87.2.2:C44T, aly1259.30.6:C96T, aly2130.12.1:C174T, aly349.2.5:A264G, aly2130.9.3:C120T, and COI barcode: T59C, T322G, T373C, T533C, T616T.
Barcode sequence of the holotype.
Sample NVG-19023G05, GenBank OR837701, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCTTTAAGTTTACTAATTCGAACAGAATTAGGTAATCCTGGCTCTTTAATTGGAGACGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTCATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTCCCTCTAATACTAGGAGCACCTGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGAATACTACCACCCTCACTTATACTATTAACTTTTAGTAGAATTGTAGAAAATGGAGCAGGGACTGGATGAACAGTTTACCCCCCTCTTTCTTCTAATATTGCTCATCAAGGCTCTTCAGTTGATTTAGCAATTTTTTCATTACATTTAGCAGGAATTTCTTCTATTTTAGGCGCTATTAATTTTATTACAACAATTATTAATATACGAATTAAAAACTTATCATTTGATCAAATACCTTTATTTGTGTGATCTGTAGGTATTACAGCCTTACTGTTACTACTATCTTTACCAGTATTAGCAGGAGCTATTACAATACTTCTCACTGATCGAAATTTAAATACTTCTTTTTTTGATCCAGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 173–174, bears the following six rectangular labels, five white: [PERU 300m | 30 Km S. W. | Pro. Maldonado | 27 Oct. ‘83 | S. S. Nicolay], [♂ genitalia | slide/vial # | H790 | Prep. S.S. Nicolay], [Enosis ♂ | pruinosa | Det. pruinosa | S.S. Nicolay], [DNA sample ID: | NVG-19023G05 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532869], and one red [HOLOTYPE ♂ | Dion occida | Grishin]. Paratypes: 2♂♂ [USNM]: NVG-19023G04, USNMENT_01532868 Venezuela: Amazonas, Cerro de la Neblina basecamp, 140 m, GPS 0.8333, −66.1667, 10–20-Feb-1985, P. J. and P. M. Spangler, R. A. Faitoute, W. E. Steiner leg. and NVG-19023G06 with a single label “17o46–55. S. Lat. 63-5-34 Long.”, which we interpreted as Bolivia: Santa Cruz department, Santa Cruz de la Sierra, GPS −17.7819, −63.0928.
Type locality.
Peru: Madre de Dios Region, 30 km SW of Puerto Maldonado, elevation 300 m.
Etymology.
In Latin, occidentalis means western. The name signifies the westernmost species of the D. uza group and is a noun in apposition.
Distribution.
Currently known from Venezuela, Peru, and Bolivia.
Eprius (Eprius) veledinus Grishin, new species https://zoobank.org/C135FD94-6DC9-4C82-9C2E-8A62D34DA640 (Fig. 7 part, 175–176, 407–409)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Ecuador identified as Eprius veleda (Godman, 1901) (type locality in Mexico: Veracruz and Tabasco, Guatemala, Honduras, and Panama, syntypes sequenced as NVG-21014A08 and NVG-21014A09) shows prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 2.7% (18 bp), and therefore represents a new species. This new species keys to “Epeus veleda veleda”(J.5.(a)) in Evans (1955) but differs from the true E. veleda by less extensive white overscaling on the abdomen beneath, which is present in the form of two stripes rather than the entire ventral side white with a narrow dark stripe in the middle (Fig. 176) and harpe more expanded dorsad into a small lobe (Fig. 409). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly276665.5.1:A864G, aly276665.5.1:A912G, aly2790.11.3:T489G, aly2487.46.1:T39G, aly2487.46.1:T45C, aly770.26.3:A73A (not C), aly860.3.1:T690T (not C), aly860.3.1:C723C (not T), aly3241.2.5:G289G (not A), aly86.14.6:C90C (not T), and COI barcode: T25C, G34C, T100G, T479C, T583C.
Barcode sequence of the holotype.
Sample NVG-8019, GenBank OR837702, 658 base pairs:
AACCTTATATTTTATTTTTGGTATCTGAGCTGGCATACTAGGAACATCCTTAAGATTATTAATTCGTACAGAATTAGGTAATCCTGGATCTTTAATTGGGGATGATCAAATTTATAATACTATTGTAACAGCTCACGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGTTTTGGTAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGGATACTACCACCTTCTTTAATATTATTAATCTCAAGAAGAATTGTTGAAAATGGTGCTGGAACTGGATGAACTGTTTATCCCCCCCTTTCATCTAATATTGCTCACCAAGGTTCTTCTGTAGATTTAGCAATTTTTTCTTTACATTTAGCAGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATCTATCATTTGATCAAATACCTTTATTTGTTTGATCTGTAGGTATTACAGCTTTATTGTTATTATTATCTTTACCTGTATTAGCTGGGGCTATTACAATATTACTCACTGATCGAAATTTAAATACTTCATTTTTTGACCCAGCAGGAGGAGGAGACCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 175–176, bears the following five rectangular labels, four white: [ECUADOR Pichincha | Alluriquin 700m | 26 May ‘88 | S. S. Nicolay], [DNA sample ID: | NVG-8019 | c/o Nick V. Grishin], [genitalia | NVG170208–04 | Nick V. Grishin], [USNMENT | {QR Code} | 01321859], and one red [HOLOTYPE ♂ | Eprius (Eprius) | veledinus Grishin].
Type locality.
Ecuador: Pichincha Province, Alluriquin, elevation 700 m.
Etymology.
The name is formed from the name of its northern sister species (E. veleda), which is made longer to indicate southern origin. The name is an adjective.
Distribution.
Currently known only from the holotype collected in Ecuador.
Radiatus panamensis Grishin, new species https://zoobank.org/FF987ADB-7F20-4972-9BFE-C5B24701E914 (Fig. 7 part, 177–178, 410–412)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Panama that someone associated with Artines Godman, 1901 (type species Hesperia aquilina Plötz, 1882) are sister to South American Radiatus bradus O. Mielke, 1968 (type locality in Brazil: Distrito Federal) while being genetically differentiated from it (Fig. 7): e.g., their COI barcodes differ by 4.6% (30 bp), and therefore represent a new species of Radiatus O. Mielke, 1968 (type species R. bradus). This new species differs from its only close relative R. bradus by the presence of forewing discal and sometimes subapical spots, longer, dash-like postdiscal spots between veins on ventral hindwing, less prominent pale area by dorsal forewing inner margin (Fig. 177–178), more extended harpe, longer process of ampulla, longer saccus, narrower undivided uncus, wider separated gnathos arms, and stronger bent aedeagus (Fig. 410–412). This species is not cryptic. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly318.14.8:A156T, aly361.13.3:G40C, aly2379.4.3:T30C, aly2011.1.3:C117T, aly2011.1.3:C192G, and COI barcode: A70G, A130T, T212C, T337C, T799A.
Barcode sequence of the holotype.
Sample NVG-20053H06, GenBank OR837703, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACTTCATTAAGTTTATTAATTCGAACGGAATTAGGTAACCCAGGCTCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACTGCTCACGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCTTTAATACTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAACATAAGATTTTGAATATTACCACCTTCTTTAATATTATTAATTTCAAGAAGAATTGTTGAAAATGGTGCAGGTACTGGATGAACTGTCTATCCCCCCCTTTCTTCTAATATTGCTCACCAAGGATCTTCAGTTGACTTAGCAATTTTTTCATTACATTTAGCAGGTATTTCTTCAATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAATTTATCATTTGATCAAATACCATTATTTGTTTGATCTGTAGGAATTACAGCATTATTACTTCTTTTATCTTTACCTGTTTTAGCTGGAGCTATTACAATACTTCTTACAGACCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGAGGAGGGGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the Mississippi Entomological Museum, Starkville, MS, USA (MEM), illustrated in Fig. 177–178, bears the following three rectangular labels, two white: [Panama: Panama | Cerro Jefe | “elfin forest” | ca. 950 m. | N 09° 13′ 27.1″ | W 079° 22′ 32.9″ | July 31, 2010 | J. R. MacDonald], [DNA sample ID: | NVG-20053H06 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Radiatus panamensis | Grishin]. Paratypes: 2♂♂ and 1♀ from Panama, John R. MacDonald leg. in MEM: 1♂ NVG-2067 Panama: Cocle, El Valle, 800–850 m, 7-Jan-1986, John R. MacDonald leg., genitalia vial no. RAA 0212; 1♂ NVG-1781 Cocle, El Valle, 800–850 m, 7-Jan-1986, John R. MacDonald leg.; 1♀ NVG-1780 Panama, Cerro Jefe, ca. 950 m, GPS 9.22419, −79.37581, 31-Jul-2010, John R. MacDonald leg.
Type locality.
Panama: Panama Province, Cerro Jefe, elevation ca. 950 m, GPS 9.22419, −79.37581
Etymology.
The name is given for the type locality and is an adjective.
Distribution.
Known only from Panama but may be present in western Colombia.
Comment.
A significant extension of the range of this formerly monotypic genus known only from South America.
Pheraeus pulcher Grishin, new species https://zoobank.org/309A87AC-194F-4AC8-BA2A-4E489BA69D46 (Fig. 7 part, 179–180, 413–414)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Ecuador and Peru that look superficially similar to Pheraeus honta Evans, 1955 (type locality in Peru) are not monophyletic with it and show prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 5.5% (36 bp), and therefore represent a new species. This new species is a distant sister to Pheraeus perpulcher (Hayward, 1934) (type locality in Argentina), while not being very similar to it in ventral wing patterns and differs by 6.1% (40 bp) in the COI barcode. This new species keys (incompletely and as the best overall compromise) to P. honta (L.11.1.) in Evans (1955) but has an orange tinted with brown at the tip tuft of long scales near the inner hindwing margin above. The holotype was misidentified as Pheraeus rumba Evans, 1955 (type locality in Brazil) but differs from it by the lack of yellow and hyaline spots in forewing cells M1-M2 and M2-M3 and the lack of submarginal brown “shading” of ventral hindwing postmedian spots: these white spots are encircled by relatively well-defined blackish frames and the submarginal area is of orange-yellow ground color, similar to P. honta. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly536.17.10:C39T, aly166.5.1:C267T, aly166.5.1:G282A, aly594.8.2:G80C, aly1454.4.1:A246G, and COI barcode: T38C, A100G, T121C, C220A, T548C.
Barcode sequence of the holotype.
Sample NVG-19016E11, GenBank OR837704, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATACTAGGAACTTCATTAAGTTTATTAATTCGAACTGAATTAGGAAACCCAGGTTCTTTAATTGGGGATGATCAAATTTATAATACCATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATCGGAGGATTTGGTAATTGATTAGTTCCTCTTATATTAGGTGCACCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATATTACCTCCTTCTTTAATATTATTAATCTCAAGAAGAATTGTTGAAAATGGTGCAGGAACTGGATGAACAGTTTACCCTCCTTTATCTTCTAATATTGCCCATCAAGGTTCTTCAGTTGATTTAGCAATTTTTTCTTTACATTTAGCAGGAATTTCATCAATTTTAGGAGCTATCAATTTTATTACAACAATTATTAATATACGAGTTAAAAACTTATCTTTTGATCAAATACCTTTATTTGTATGATCTGTAGGTATTACAGCATTACTATTACTTCTATCTCTACCTGTTTTAGCAGGAGCTATTACTATACTTCTTACTGATCGAAATTTAAATACTTCATTTTTTGATCCAGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 179–180, bears the following five rectangular labels, four white: [PERU Madre De Dios | Rio La Torre 300m | Tambopata Res. | 27 Sept. ‘89 | S. S. Nicolay], [Pheraeus | rumba ♂ | Det. | S.S. Nicolay], [DNA sample ID: | NVG-19016E11 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532334], and one red [HOLOTYPE ♂ | Pheraeus | pulcher Grishin]. Paratype: 1♀ Ecuador: Napo Province, Jatun Sacha Biological Reserve, elevation 450 m, GPS −1.0667, −77.6167, 7-Nov-1992, S. S. Nicolay leg. [USNM].
Type locality.
Peru: Madre de Dios, Tambopata National Reserve, Rio La Torre, elevation 300 m.
Etymology.
In Latin, perpulcher means very beautiful. Even after removing the prefix “per”, we still have a beautiful species. A shorter name signifies a more northern distribution of this species. The name is a noun in apposition.
Distribution.
The upper Amazonian region, known from Ecuador and Peru.
Callimormus rades Grishin, new species https://zoobank.org/6B68354B-2C44-4669-8A87-E093C704272A (Fig. 7 part, 181–182, 415–417)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Panama identified as Callimormus corades (C. Felder, 1862) (type locality in Brazil: Rio de Janeiro) shows prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 3% (20 bp) and, therefore, represents a new species, provided that we sequenced primary type specimens of taxa treated as synonyms of Callimormus corades and show that they are distinct from the new species (Fig. 7). This new species keys to C. corades (J.2.7) in Evans (1955) but differs from it by discal spot in ventral hindwing cell Sc+R1-RS slightly more offset basad, rounder discal spots on forewing, and generally more prominent yellow overscaling of veins on the ventral side, more evident as a yellow bar at the end of discocellular vein (Fig. 181–182). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2041.16.10:C33T, aly159.20.1:C99G, aly814.17.1:G105A, aly4007.2.5:G48A, aly1631.11.9:T43G, aly50.38.2:G177G (not C), aly686.17.2:G54G (not A), aly60.25.7:C69C (not A), aly26.25.5:C138C (not T), aly6847.3.5:C54C (not T), and COI barcode: T212C, 271A, C334T, T337A, T574C.
Barcode sequence of the holotype.
Sample NVG-19016G06, GenBank OR837705, 658 base pairs:
AACTTTATATTTTATTTTCGGAATTTGAGCAGGAATAATTGGTACTTCATTAAGTTTATTAATTCGTACAGAATTAGGTAATCCAGGTTCTTTAATTGGAGATGATCAAATTTATAATACCATTGTTACTGCTCATGCCTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGTAATTGATTAGTTCCCTTAATACTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTCTGAATACTTCCACCATCATTAACACTTTTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGTACTGGTTGAACTGTATACCCCCCTCTTTCTTCTAATATTGCTCATCAAGGATCTTCAGTTGATTTAGCAATTTTTTCATTACATCTTGCAGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTACAACTATTATTAATATACGAATTAAAAATCTTTCTTTTGATCAAATACCTTTATTTGTATGATCAGTTGGTATTACAGCTTTACTTCTACTTTTATCTTTACCAGTTTTAGCTGGAGCTATTACCATACTTTTAACAGATCGTAATCTTAATACTTCATTTTTTGATCCTGCAGGAGGAGGAGACCCAATTCTTTATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 181–182, bears the following five rectangular labels, four white: [Farfan, | Panama C. Z. | 17 Feb. ‘63 | S. S. Nicolay], [Callimormus | corades | Felder | DET.BY S.S.NICOLAY], [DNA sample ID: | NVG-19016G06 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532350], and one red [HOLOTYPE ♀ | Callimormus | rades Grishin].
Type locality.
Panama: Panama Province, Farfan.
Etymology.
The name is formed from its sister species name (C. corades). By shortening the name, we specify a more northern distribution of this species. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Panama.
Gubrus lubens Grishin, new species https://zoobank.org/AEEFAD08-1F9A-406F-8AD7-AD386BBA716D (Fig. 7 part, 183–184, 418–420)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Loja, Ecuador, placed in the USNM collection among possible new species of “Vehilius” is a distant sister of Gubrus lugubris (Lindsey, 1925) (type locality in Peru) that was previously in the genus Vehilius Godman, 1900 (type species Cobalus illudens Mabille, 1891, a subspecies of Pamphila stictomenes Butler, 1877) (Fig. 7): e.g., their COI barcodes differ by 10.8% (71 bp), a large divergence likely caused by an elevated evolutionary rate in this lineage and its sister genus Callimormus Scudder, 1872 (type species Callimormus juventus Scudder, 1872), and therefore represents a new species in the previously monotypic genus Gubrus. This new species is distinct in its appearance and is recognized among small brown Hesperiidae by rounder hindwing, without produced tornus in males, uniformly colored wings without paler veins and on the forewing with three pale-yellow spots in a line near the bases of forewing cells CuA1-CuA2, M3-CuA1, and M2-M3 and a small paler area instead of a developed spot in cell R5-M1; fringes are paler and indistinctly checkered. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly349.39.4:A117G, aly824.21.1:T348C, aly824.21.1:G351C, aly770.22.2:T84G, aly770.22.2:T120C, aly536.107.1:A129A (not C), aly536.107.1:G135G (not A), aly595.10.1:G801G (not A), aly9855.3.1:C140C (not T), aly1405.20.4:T18T (not C), and COI barcode: T22G, A79C, A379G, A529C, T619A.
Barcode sequence of the holotype.
Sample NVG-19019B12, GenBank OR837706, 658 base pairs:
AACTTTATATTTTATTTTTGGGATTTGAGCAGGAATATTAGGAACTTCATTAAGATTATTAATTCGTACAGAATTAGGCAATCCTGGATCTTTAATTGGGGATGACCAAATTTATAATACTATTGTTACAGCCCATGCTTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGAGGTTTTGGTAATTGATTAGTCCCTTTAATACTAGGAGCCCCTGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGAATATTGCCCCCCTCACTAATATTATTAATTTCTAGAAGAATTGTAGAAAATGGTGCAGGAACAGGTTGAACAGTTTATCCCCCTCTTTCTTCTAATATTGCCCATCAAGGTTCATCGGTTGATTTAGCAATTTTTTCCCTTCATTTAGCAGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACTACTATCATTAACATACGAATTAAAAATCTTTCATTTGATCAAATACCTTTATTTGTTTGATCAGTAGGTATTACAGCCTTACTTTTACTTTTATCTTTACCAGTATTAGCTGGAGCTATTACTATACTTCTAACAGATCGAAATTTAAATACTTCTTTTTTTGATCCAGCTGGAGGAGGTGACCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 183–184, bears the following six rectangular labels, five white: [ECUADOR Loja | Loja-Catamayo Rd. | Km 28 1800m | 16 May ‘88 | S. S. Nicolay], [♂ genitalia | slide/vial # | H989 | Prep. S.S. Nicolay], [Vehilius | n. sps. | Det. | S.S. Nicolay], [DNA sample ID: | NVG-19019B12 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532564], and one red [HOLOTYPE ♂ | Gubrus lubens | Grishin].
Type locality.
Ecuador: Loja Province, km 28 of Loja–Catamayo road.
Etymology.
In Latin, lugubris means sad, and lubens means happy, merry, joyful, willing, ready, or eager. This new species is happy to be the second species of this genus and is eager to be described here. The name is a masculine adjective.
Distribution.
Currently known only from the holotype collected in Ecuador.
Ludens labens Grishin, new species https://zoobank.org/7B83772E-AFB1-4A3F-995A-363407AD85FC (Fig. 7 part, 185–186, 421–422)
Definition and diagnosis.
Phylogenetic trees reveal that several specimens from Panama identified as Ludens ludens (Mabille, 1891) (type locality in Panama, syntype sequenced as NVG-15036B04) are not monophyletic with it and show prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 8.4% (55 bp), and therefore represent a new species. This new species is sister to the species pair L. ludens and Ludens petrovna (Schaus, 1902) (type species Brazil: Rio de Janeiro) (Fig. 7). This new species keys to Ludens ludens (J.7) that included L. petrovna as its synonym in Evans (1955) but differs from these species by the lack of postdiscal spots that connect yellow veins on the ventral hindwing (Fig. 186), longer harpe than in L. ludens that is more similar to L. petrovna, shorter (compared to the length of costa) “thumb”-like expansion of the ampulla, and more concave ventral margin of harpe in lateral view (Fig. 422). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly216.51.3:T66C, aly890.73.2:C99T, aly890.73.2:G111A, aly600.14.2:G80A, aly1493.4.5:C60T, and COI barcode: T118A, T163A, C346A, A391T, A565G.
Barcode sequence of the holotype.
Sample NVG-19017B12, GenBank OR837707, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATATTAGGTACATCATTAAGAATATTAATTCGTACTGAATTAGGTAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAACACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATGGTAATACCAATTATAATTGGCGGATTCGGTAATTGATTAGTCCCATTAATATTAGGAGCCCCAGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGAATATTACCCCCATCATTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGATGAACTGTATATCCCCCACTTTCATCCAATATTGCACATCAAGGATCCTCTGTTGATTTAGCTATTTTCTCCCTTCATTTAGCAGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATCACCACTATTATTAATATACGAATTAAAAACCTATCTTTCGATCAAATACCTTTATTTGTATGATCTGTAGGTATTACAGCTTTATTATTATTATTATCATTACCTGTTCTAGCTGGGGCCATTACTATACTTTTAACAGATCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGAGGGGGGGACCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 185–186, bears the following four rectangular labels, three white: [PANAMA:Darien | Cana 1200m | 7°56′N 77°43′W | 22 September 1982 | leg. G.B.Small], [DNA sample ID: | NVG-19017B12 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532848], and one red [HOLOTYPE ♂ | Ludens | labens Grishin]. Paratype: 1♂ NVG-20054D07 Panama: Panama Province, Cerro Jefe, elevation ca. 880 m, GPS 9.2242, −79.3758, 17-Feb-2014, J. R. MacDonald leg. [MEM].
Type locality.
Panama: Darien Province, Cana, elevation 1200 m, GPS 7.9333, −77.7167.
Etymology.
In Latin, ludens means playful. An antonym for play is work. In Latin, labor means work, and the name derived from this word, similarly to ludens, is labens, which actually means slipping, gliding, or flowing. The name is treated as a noun in apposition.
Distribution.
Panama.
Rigga isa Grishin, new species https://zoobank.org/82DE66CF-AF62-4C68-9D1F-3ABCEBCFD07E (Fig. 7 part, 187–188, 423–424)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Ecuador similar in appearance to Rigga auristriga (Draudt, 1923) (type locality in Bolivia, holotype sequenced as NVG-18093C02) are not monophyletic with it and show prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 5.5% (36 bp), and therefore represent a new species. This new species is sister to Rigga ira (A. Butler, 1870) (type locality not given, possibly in Venezuela) and differs from it by 3.2% (21 bp) in the COI barcode. The hindwing of the new species is uniformly colored above, lacking yellow rays of R. ira, ventral hindwing veins are overscaled with approximately uniform thickness (except M2, which is weaker), and there is no appearance of a ray along the radius and vein M1 as in R. ira. The new species differs from R. auristriga, which frequently has similar uniform overscaling of veins, by wider forewing spots and wider lower section of stigma. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly2284.14.4:G85C, aly4778.18.1:C946T, aly4778.18.1:T1398C, aly1937.17.47:C39A, aly1019.14.1:A84T, and COI barcode: A100G, C343A, T397C, T463C, T556C.
Barcode sequence of the holotype.
Sample NVG-19019G06, GenBank OR837708, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGTATATTAGGAACTTCTTTAAGTATACTAATTCGAACAGAATTAGGTAACCCAGGATCTTTAATTGGGGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTCCCCCTTATACTAGGAGCCCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATGCTTCCCCCTTCTTTAACACTTTTAATTTCTAGAAGAATTGTTGAAAATGGAGCAGGTACTGGTTGAACAGTTTACCCACCTCTTTCTTCTAATATTGCCCACCAAGGTTCTTCTGTTGATTTAGCAATTTTCTCCCTTCATTTAGCAGGTATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAACATACGAGTTAGAAATTTATCATTTGATCAAATACCTTTATTTGTTTGATCAGTAGGTATTACAGCATTATTATTACTTTTATCTTTACCTGTCTTAGCAGGAGCTATTACTATACTTCTTACAGATCGAAATTTAAATACTTCTTTTTTTGACCCTGCTGGAGGAGGAGACCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 187–188, bears the following five rectangular labels, four white: [ECUADOR Napo | Baeza 2000m | 6 July ‘80 | S. S. Nicolay], [Parphorus | hesia ♂ | Det. Hew. | S.S. Nicolay], [DNA sample ID: | NVG-19019G06 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532617], and one red [HOLOTYPE ♂ | Rigga isa | Grishin]. Paratype: 1♀ NVG-21047G03 Ecuador: Napo, El Chaco, 1500 m, Nov-1971, R. de Lafebre leg. [MGCL].
Type locality.
Ecuador: Napo Province, Baeza, elevation 2000 m.
Etymology.
The name is formed from the Greek ίσος (isos), meaning equal. It reflects the equal overscaling of nearly all veins and signifies a more uniform appearance than R. ira and other congeners. The name is a noun in apposition.
Distribution.
Ecuador.
Subtribe Falgina Grishin, 2019
Flaccilla lactea Grishin, new species https://zoobank.org/CB051F7E-A99E-4DE6-B233-993276D1F393 (Fig. 7 part, 189–190, 425–426)
Definition and diagnosis.
Sequencing of an unusual specimen of Flaccilla Godman, 1901 (type species Papilio aecas Stoll, 1781) from Peru with largely cream-colored ventral hindwing confirms its expected prominent genetic differentiation from F. aecas (type locality in Surinam) (Fig. 7): e.g., their COI barcodes differ by 7% (46 bp), and therefore it represents a new species. This new species (incompletely) keys to “Aecas aecas” (J.13.1) in Evans (1955) but differs from it by the lack of purple sheen on the ventral side and hindwing cream in color with discal brown spots and brown marginal spots nearly fused into a band. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly123.8.1:A51T, aly123.8.1:T72G, aly1779.16.4:C84T, aly1779.16.4:G111A, aly13170.2.2:A27G, aly1139.48.20:T97T (not C), aly1139.48.20:G102G (not A), aly499.35.1:G419G (not A), aly2096.12.3:A137A (not C), aly2096.12.3:T155T (not G), and COI barcode: A35T, T142C, A241T, 484C, T580C.
Barcode sequence of the holotype.
Sample NVG-19017E11, GenBank OR837709, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGATTATTAGGAACTTCATTAAGATTATTAATTCGGACAGAACTAGGAAACCCAGGTTCTTTAATCGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTCATTATAATTTTTTTTATAGTAATACCTATTATAATCGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGAGCTCCTGACATAGCCTTCCCACGTATAAATAATATAAGATTTTGAATACTACCCCCATCATTAACTTTATTAATTTCAAGAAGAATTGTAGAAAATGGGGCAGGAACAGGATGAACTGTTTATCCGCCCCTTTCCTCTAATATTGCCCATCAAGGTTCTTCTGTTGATTTAGCAATTTTTTCATTACATTTAGCTGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATCACTACAATTATTAATATACGAATTAAAAATATATCCTTTGATCAAATACCATTATTTGTTTGATCTGTAGGAATTACAGCATTATTATTACTTTTATCTTTACCTGTTTTAGCTGGAGCTATTACCATACTCCTTACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCAGGAGGAGGAGACCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 189–190, bears the following five rectangular labels, four white: [PERU: Cuzco: Cosñipata Valley | Quebrada Quitacalzón 1,050m. | 13° 01′ 13″S, 71° 29′ 50″W | 12 August 2009 Brian Harris], [Flaccilla sp. n.], [DNA sample ID: | NVG-19017E11 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01532369], and one red [HOLOTYPE ♂ | Flaccilla | lactea Grishin].
Type locality.
Peru: Cuzco Region, Cosñipata Valley, Quebrada Quitacalzón, elevation 1050 m, GPS −13.020278, −71.497222.
Etymology.
In Latin, lacteus means milky. The name is given for the milky-colored hindwing and is a feminine adjective.
Distribution.
Currently known only from the holotype collected in the Cosñipata Valley, Peru.
Falga mirabilis Evans, 1955 (with Falga jeconia odol Evans, 1955 as its subspecies), Falga jacta Evans, 1955, and Falga ombra Evans, 1955 are species distinct from Falga jeconia (A. Butler, 1870)
Genomic sequencing taxa currently treated as subspecies of Falga jeconia (A. Butler, 1870) (type locality in Venezuela), namely: Falga mirabilis Mabille, 1898 (type locality in Bolivia), Falga jeconia jacta Evans, 1955 (type locality in Colombia), and Falga jeconia ombra Evans, 1955 (type locality in Ecuador) show substantial genetic differentiation from it and each other (Fig. 7): e.g., the smallest barcode difference among them is 2.1% (14 bp), between F. jeconia and F. j. jacta, with the next being 2.6% (17 bp) between F. mirabilis and F. j. ombra. Therefore, we propose a new status of species for Falga mirabilis Evans, 1955, Falga jacta Evans, 1955, and Falga ombra Evans, 1955. The phylogenetic tree reveals that Falga jeconia odol Evans, 1955 (type locality in southern Peru) is closely related to Falga mirabilis (Fig. 7). Therefore, we propose to treat the former as a subspecies of the latter: Falga mirabilis odol Evans, 1955, new combination.
Falga athena Grishin, new species https://zoobank.org/E9BA59DB-2467-42BA-9C5F-BEB41F8CF500 (Fig. 7 part, 191–192, 427–429)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Panama resembling a darker Falga sciras Godman, 1901 (type locality in Honduras) is not monophyletic with it and shows prominent genetic differentiation from it (Fig. 7): e.g., their COI barcodes differ by 5.9% (39 bp). This specimen also differs from all other species of Falga Mabille, 1898 (type species Carystus jeconia Butler, 1870) and, therefore, represents a new species. This new species (incompletely) keys to Falga jeconia jeconia (I.1.2(b)) in Evans (1955). It differs from F. sciras by lacking a discal brown spot at the end of discal cell on the ventral hindwing and orange spots on the dorsal forewing not reaching the wing base. It differs from other species by orange, rather than yellow or yellow-orange spots and patches on the dorsal side and brown around the tornus on the ventral hindwing. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly2423.6.2:G57A, aly1838.37.5:T96C, aly666.26.4:A100C, aly542.3.12:C99T, aly1651.8.4:A153T, aly2548.20.5:C27C (not T), aly725.21.1:A114A (not G), aly208.47.15:T281T (not C), aly7689.2.1:T36T (not C), aly11945.4.1:T2088T (not C), and COI barcode: T250C, T352C, T355A, T530C, A565T.
Barcode sequence of the holotype.
Sample NVG-18012E08, GenBank OR837710, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACTTCTTTAAGTATATTAATTCGTACTGAATTAGGAAATCCAGGATTTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAACATAAGATTTTGAATATTGCCTCCTTCCCTAACATTATTAATTTCAAGAAGAATTGTTGAAAATGGTGCAGGAACTGGATGAACAGTTTATCCCCCTCTTTCCTCAAATATTGCTCATCAAGGATCTTCTGTTGATTTAGCAATTTTTTCTCTTCATTTAGCTGGAATTTCTTCTATTCTTGGAGCTATTAACTTTATTACTACAATTATTAATATACGAATTAAAAATAGATCATTTGATCAAATACCATTATTTGTATGATCTGTGGGAATTACAGCACTTTTATTACTTTTATCTTTACCTGTATTAGCAGGTGCTATTACTATACTTCTTACAGATCGAAATCTTAATACATCTTTTTTTGATCCTGCAGGAGGAGGGGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 191–192, bears the following four rectangular labels, three white: [PANAMA: Darien | Cana 1550m | 23.III.1983 | leg. G.B.Small], [DNA sample ID: | NVG-18012E08 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450279], and one red [HOLOTYPE ♀ | Falga athena | Grishin].
Type locality.
Panama: Darien Province, Cana, elevation 1550 m.
Etymology.
Sciras is the surname of Athena, an Olympian goddess. The name athena signifies that the new species is closest in wing pattern to F. sciras. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in eastern Panama. Subtribe Calpodina A. Clark, 1948
Panoquina jay Grishin, new species https://zoobank.org/B6EEC7CF-A0A4-4AB2-BB1D-14F036CB0D20 (Fig. 8 part, 193–194, 430–431)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Peru with a general appearance of Panoquina bola Bell, 1942 (type locality in Bolivia) shows prominent genetic differentiation from all named relatives without strong affinity to any of them (Fig. 8): e.g., their COI barcodes differ by 6.5% (43 bp) and, therefore, represents a new species. This new species keys to P. bola (O.2.9) in Evans (1955) but differs from it and other relatives by broader ventral hindwing discal band of uniform width and extending into the cell CuA2-1A+2A on chestnut ground color and a smaller forewing spot in the cell M2-M3. Due to the lack of additional specimens and unknown phenotypic variation, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1041.32.2:T54C, aly1019.32.2:A169C, aly221.7.14:A63T, aly10226.12.4:A54G, aly971.2.12:C129G, aly481.23.3:C109C (not A), aly481.23.3:T117T (not C), aly927.1.16:C177C (not T), aly171.7.2:C69C (not T), aly531.29.1:A138A (not G), and COI barcode: T49C, A67G, A251T, A433G, A550G.
Barcode sequence of the holotype.
Sample NVG-8064, GenBank OR837711, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGTATATTAGGTACTTCCTTAAGTTTATTAATTCGGACAGAATTAGGAAATCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACCATCGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTTTAATATTAGGAGCTCCTGATATAGCATTCCCTCGTATAAATAATTTAAGATTTTGAATACTACCTCCTTCTTTAACTTTATTAATTTCAAGAAGTATTGTAGAAAATGGAGCAGGTACAGGTTGAACTGTTTATCCCCCCCTTTCTTCTAATATTGCTCATCAAGGAGCTTCTGTTGATTTAGCAATTTTTTCCCTTCATTTAGCAGGTATTTCATCAATCTTAGGGGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATTTATCATTTGATCAAATACCTTTATTTGTATGATCCGTTGGAATTACAGCCTTATTATTACTTTTATCTTTGCCAGTTTTAGCTGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAATACTTCATTTTTTGACCCTGCTGGAGGGGGGGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 193–194, bears the following five rectangular labels, four white: [PERU: Loreto Prov. | Rio Amazonas, 200m | Explorama Lodge | 50 mi E Iquitos | 12–16 Sept. 1990 | Brian P. Harris], [DNA sample ID: | NVG-8064 | c/o Nick V. Grishin], [genitalia | NVG170208–49 | Nick V. Grishin], [USNMENT | {QR Code} | 01321904], and one red [HOLOTYPE ♂ | Panoquina | jay Grishin].
Type locality.
Peru: Loreto Province, 50 mi E Iquitos, Amazon River, Explorama Lodge, elevation 200 m.
Etymology.
The name reflects the white band on the ventral hindwing, which reminds us of the letter J (jay). The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in northeastern Peru.
Calpodes salianus Grishin, new species https://zoobank.org/9916050C-DBCE-4EDA-A05C-6BB74C2BD7B3 (Fig. 8 part, 195–196, 432–434)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Peru identified as Calpodes salius (Cramer, 1775) (type locality in Suriname) is not monophyletic with and shows prominent genetic differentiation from it (Fig. 8): e.g., their COI barcodes differ by 5.2% (34 bp), and therefore represents a new species. This new species keys to “Saliana salius” (O.14.17) in Evans (1955) but differs from it by longer costal shoulder of the discal cell spot that has an appearance of a separate spot shifted distad (at the base too) but joined with the lower spot (instead of a single spot with a flatter base and irregular outer margin), and the lack of ash-gray overscaling at the base of ventral hindwing (this area is maroon-colored instead with violet sheen) (Fig. 195–196), ampulla more protruding dorsad, and more convex costa near ampulla (Fig. 434). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly276558.9.2:A84G, aly276558.9.2:A150G, aly2258.12.5:G486C, aly272.25.11:T84C, aly1660.4.4:C157T, aly638.8.1:G327G (not A), aly638.8.1:A369A (not G), aly827.13.1:C42C (not G), aly827.13.1:C75C (not T), aly525.2.6:G186G (not A), and COI barcode: T19C, A44T, T121C, A175G, T596C.
Barcode sequence of the holotype.
Sample NVG-18112H11, GenBank OR837712, 658 base pairs:
AACTTTATATTTTATTTTCGGTATTTGAGCAGGAATATTAGGTTCTTCATTAAGTTTATTAATTCGTACAGAATTAGGTAATCCTGGTTCATTAATTGGAGATGACCAAATTTATAATACCATTGTTACAGCTCACGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATGATTGGAGGATTTGGAAATTGATTAGTACCATTAATATTAGGAGCCCCAGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGAATACTCCCCCCTTCATTAACTTTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGTTGAACAGTTTATCCCCCCCTTTCAGCTAATATCGCCCATCAAGGATCTTCAGTTGATCTAGCAATTTTTTCTTTACATTTAGCAGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAATTTAATATTTGACCAAATACCATTATTTGTTTGATCTGTAGGAATTACAGCATTATTATTACTATTATCATTACCAGTTTTAGCAGGAGCTATTACAATACTTCTTACTGACCGAAATCTAAATACATCTTTCTTTGACCCTGCAGGAGGAGGTGACCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 195–196, bears the following four rectangular labels, three white: [PERU 300m | 30 Km S.W. | Pto. Maldonado | 26 Oct. ‘83 | S. S. Nicolay], [DNA sample ID: | NVG-18112H11 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01531430], and one red [HOLOTYPE ♂ | Calpodes | salianus Grishin].
Type locality.
Peru: Madre de Dios Region, 30 km SW of Puerto Maldonado, elevation 300 m.
Etymology.
The name reflects its somewhat similar appearance to C. salius. Made longer, the name signifies it is a more southern relative. The name is treated as a noun in apposition.
Distribution.
Currently known only from the holotype collected in the Amazonian region of Peru.
Calpodes stingo Grishin, new species https://zoobank.org/C1F403E7-718A-427C-9882-2C56D6D2B124 (Fig. 8 part, 197–198, 435–437)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Ecuador identified as Calpodes placens (A. Butler, 1874) (type locality Colombia: Bogota) shows prominent genetic differentiation from it (Fig. 8): e.g., their COI barcodes differ by 3.6% (24 bp), and therefore represents a new species. This new species keys to “Saliana placens” (O.14.9) in Evans (1955) but differs from it by much reduced orange overscaling between the pale base and brown tornus of ventral hindwing with that area being more brown than pale or orange (in C. placens, pale basal color intrudes into the brown area and is framed with yellow and orange), as well as darker and more restricted rusty overscaling on forewing above. Due to the lack of additional specimens and unknown phenotypic variation, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1603.82.16:T51C, aly1603.82.16:A54G, aly84.57.9:A48T, aly671.7.4:T54C, aly619.9.1:T48C, aly4305.15.6:T303T (not C), aly1041.22.3:G133G (not A), aly2103.6.1:A392A (not G), aly144.20.2:T57T (not A), aly18882.2.3:G48G (not T), and COI barcode: A34C, A58G, A373T, 220C, T653C.
Barcode sequence of the holotype.
Sample NVG-18112H02, GenBank OR837713, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGCATATTAGGTACTTCATTAAGTTTGTTAATTCGTACTGAATTAGGTAACCCTGGTTCATTAATTGGAGATGACCAAATTTATAATACTATTGTTACAGCTCATGCCTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCATTAATATTAGGTGCCCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGAATACTCCCCCCTTCATTAACTTTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGTTGAACGGTTTACCCCCCCCTTTCATCCAATATTGCCCACCAAGGTTCATCTGTTGATTTAGCAATTTTTTCTTTACATTTAGCAGGAATCTCATCAATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATTTAATATTTGATCAAATACCATTATTTGTTTGATCTGTAGGAATTACAGCATTATTATTACTTTTATCATTACCTGTTTTAGCAGGAGCTATTACTATATTACTTACTGATCGAAATTTAAATACATCTTTTTTTGATCCTGCAGGAGGAGGTGATCCTATTTTATATCAACATCTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 197–198, bears the following four rectangular labels, three white: [ECUADOR: Sucumbíos, | Cerro Lumbaquí Norte, | 0° 01′70″ N, 77° 19′22″ W | 800–950 m, 18–22 Aug 2002 | J.P.W. Hall & M.A. Solis], [DNA sample ID: | NVG-18112H02 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01531422], and one red [HOLOTYPE ♂ | Calpodes | stingo Grishin].
Type locality.
Ecuador: Sucumbíos Province, Cerro Lumbaquí Norte, elevation 800–950 m, approx. GPS 0.0283, −77.3203
Etymology.
In Latin, placens means pleasing, and stinguō means to put out or extinguish. The name stingo is given to this species with a “pleasing” orange streak removed from the ventral hindwing. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in north-central Ecuador.
Subtribe Carystina Mabille, 1878
Hesperia ocrinus Plötz, 1882 is a junior subjective synonym of Aides aegita (Hewitson, 1866)
Genomic sequencing of a single syntype of Hesperia ocrinus Plötz, 1882 (type locality in Colombia, NVG-15035A10) in MFNB, currently a valid species in the genus Aides Billberg, 1820 (type species Papilio epitus Stoll, 1781, a homonym, the valid name for this species is Aides duma argyrina Cowan, 1970) reveals that it is Aides aegita (Hewitson, 1866) (type locality in Brazil: Para) (Fig. 8). Indeed, inspection of the H. ocrinus syntype, a male, reveals brands on the wings that are characteristic of A. aegita and its junior subjective synonyms. Therefore, we propose to treat Hesperia ocrinus Plötz, 1882 as a junior subjective synonym of Aides aegita (Hewitson, 1866). Hesperia ocrinus was misidentified by Evans (1955), and the species without brands that Evans called “Aides ocrinus” does not have a name and is described next as new.
Aides nobra Grishin, new species https://zoobank.org/3466088D-C59F-48E0-BEE9-4231E4D349A7 (Fig. 8 part, 199–200, 438–439)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Panama identified as Aides ocrinus show prominent genetic differentiation from a syntype of Hesperia ocrinus Plötz, 1882 (type locality in Colombia, NVG-15035A10, see above for its synonymization) (Fig. 8): e.g., their COI barcodes differ by 3.6% (24 bp), and therefore represent a new species. This new species keys to “Aides ocrinus” (O.12.6) in Evans (1955), which he misidentified, and specimens he identified as “A. ocrinus” are probably this species. Differs from its relatives by the lack of brands in males, redder (instead of more olive in color) ventral wing coloration, and harpe with broad and rounded end. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1222.14.21:A112T, aly1222.14.21:A228G, aly7999.4.21:G189A, aly19602.6.1:G288A, aly19602.6.1:G574T, and COI barcode: T10C, T154C, T171C, T367C, A628T.
Barcode sequence of the holotype.
Sample NVG-18012E05, GenBank OR837714, 658 base pairs:
AACTTTATACTTTATTTTTGGAATTTGAGCAGGTATATTAGGAACTTCCTTAAGTTTATTAATTCGAACAGAATTAGGTAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTTACTGCTCATGCTTTTATTATAATTTTCTTTATAGTAATACCTACTATAATTGGAGGATTTGGAAATTGACTAGTCCCTCTAATACTAGGAGCCCCTGATATAGCATTTCCTCGAATAAATAATATAAGATTTTGATTATTACCTCCCTCACTTACTTTACTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACTGGATGAACAGTTTATCCCCCTCTTTCATCTAATATTGCTCACCAAGGATCATCAGTTGATTTAGCAATTTTTTCTCTTCACTTAGCAGGTATTTCATCAATTTTAGGAGCAATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATTTATCTTTTGACCAAATACCTTTATTTGTATGATCTGTAGGAATTACAGCTTTATTATTACTTTTATCATTACCAGTATTAGCCGGAGCCATTACAATACTCCTTACTGATCGAAATTTAAATACTTCATTTTTTGATCCAGCTGGAGGTGGTGATCCAATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 199–200, bears the following four rectangular labels, three white: [COCO SOLO | PANAMA CANAL ZONE | 1 APRIL 1944 | W. M. WAGNER, JR.], [DNA sample ID: | NVG-18012E05 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450333], and one red [HOLOTYPE ♂ | Aides nobra | Grishin]. Paratypes: 3♂♂ and 3♀♀ from Panama, Canal Zone: 1♂ NVG-18012E04, USNMENT_01450332 the same data as the holotype but collected on 30-Mar-1944; 1♂ Gulf of Panama nr. Cape Mala, 30-Sep-1024 [BMNH]; 1♂ Pedro Miguel, G. Tryhane leg. [BMNH]; 1♀ 1-Feb-1913, A. Hall leg. [BMNH]; 2♀♀ [AMNH]: NVG-18021C10 no other data than “Panama”; and NVG-18021C11 May-1911, collection F. E. Watson.
Type locality.
Panama: Colón Province, Cativá [formerly Coco Solo].
Etymology.
The name reflects the lack of brands: no + bra[nds] and is a noun in apposition.
Distribution.
Currently known only from central Panama.
Thracides pavo Grishin, new species https://zoobank.org/14FC64FB-D37D-43E1-8764-08D112459C19 (Fig. 8 part, 201–202, 440–441)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Mexico identified as Thracides phidon (Cramer, 1779) (type locality in Suriname) show prominent genetic differentiation from it in the Z chromosome (Fig. 8) and, therefore, represent a new species. Curiously, despite the prominent differentiation in the Z chromosome, their COI barcodes differ by only 1.1% (7 bp). This new species keys to T. phidon (O.15.7) in Evans (1955) but differs from it by typically longer segments of stigma, by lacking or vestigial subapical white spots on ventral forewing (many T. phidon specimens have these spots), especially in males, yellower and more extensive overscaling at the forewing apex beneath, less extensive blue overscaling on dorsal hindwings (Fig. 201–202), longer saccus, and broader rounded distal end of harpe (Fig. 441). Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly38.8.4:C87A, aly423.2.7:A162T, aly214.15.4:G34A, aly1074.2.19:G51A, aly1074.2.19:C66T, and COI barcode: T5A, T118T, 127A, T442T, A508G, T533T.
Barcode sequence of the holotype.
Sample NVG-18114B07, GenBank OR837715, 658 base pairs:
AACTCTATATTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACATCTTTAAGATTATTAATTCGAACTGAATTAGGTAACCCAGGATCTTTAATTGGAGATGATCAAATCTATAATACTATTGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCTTTAATATTAGGAGCTCCCGATATAGCTTTCCCCCGAATAAATAATATAAGTTTTTGAATGCTACCCCCTTCATTAACTCTTTTAATTTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGATGAACAGTTTACCCCCCTTTATCCTCTAATATTGCTCATCAAGGATCTTCAGTAGATTTAGCCATTTTTTCTCTTCATTTAGCCGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATTTATCTTTCGATCAAATACCTTTATTTGTGTGATCAGTAGGAATTACAGCTTTATTATTACTTTTATCTCTACCAGTATTAGCAGGTGCTATTACTATACTTCTTACAGATCGAAATTTAAATACATCTTTTTTTGATCCTGCTGGAGGAGGAGATCCTATTCTTTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 201–202, bears the following seven rectangular labels, six white: [Teapa | Tabasco | Mexico], [March | 1913], [RMuller | Collector], [3708], [DNA sample ID: | NVG-18114B07 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01531463], and one red [HOLOTYPE ♂ | Thracides | pavo Grishin]. Paratypes: 2♀♀: Mexico: San Luis Potosi, Sierra Madre O., El Salto Falls, Roy O. Kendall and C. A. Kendall leg., larval foodplant: Heliconia latispatha Benth. [TAMU]: NVG-19013C04 4-Feb-1976 and NVG-19013C05 14-Feb-1975.
Type locality.
Mexico: Tabasco, Teapa.
Etymology.
In Latin, pavo means peacock. The name is given for the colorful wings of this species and is a noun in apposition.
Distribution.
Currently recorded from Tabasco and San Luis Potosi in Mexico.
Talides eluta Grishin, new species https://zoobank.org/900BB006-C1DB-4CC9-ADD3-B2393A6B4539 (Fig. 8 part, 203–204, 442–443)
Definition and diagnosis.
Phylogenetic trees reveal that several specimens of Talides Hübner, [1819] (type species Talides sinois Hübner, [1819]) from Peru and Brazil show prominent genetic differentiation from named species of this genus (Fig. 8): e.g., their COI barcodes differ from that of Talides alternata E. Bell, 1941 (type locality in Brazil: Santa Catarina, holotype sequenced as NVG-18025D05) by 5.6% (37 bp), and therefore represent a new species. This new species keys to T. alternata hispa (K.13.3(b)) in Evans (1955) but differs from it by less rounded wings and less contrasting patterns of ventral side, shorter and not as bright orange fringes, straighter and shorter upper segment of stigma, and a more elongated and thinner apical spot in the cell R5-M1. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly1244.8.2:G99A, aly927.2.7:G49T, aly927.2.7:G128A, aly596.8.4:C10A, aly35002.7.1:C96T, and COI barcode: T247C, T373C, T436A, T482A, A622G.
Barcode sequence of the holotype.
Sample NVG-18012D01, GenBank OR837716, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACTTCTTTAAGATTATTAATTCGAACAGAATTAGGTAATCCAGGATTTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTTCCTCTTATACTTGGAGCCCCTGATATAGCTTTCCCCCGAATAAACAATATAAGATTCTGAATACTTCCCCCTTCTTTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCTGGTACAGGATGAACTGTATATCCCCCTCTTTCAGCTAATATTGCTCATCAAGGCTCATCTGTTGATTTAGCAATTTTTTCCCTTCATTTAGCAGGAATTTCTTCTATTTTAGGAGCAATTAACTTTATTACAACAATTATTAATATACGAATTAGAAATTTAATATTTGATCAAATACCTCTATTTGTATGATCTGTGGGAATTACAGCTTTATTATTATTATTATCTTTACCTGTTTTAGCAGGAGCTATTACTATACTTCTTACTGATCGTAATTTAAATACTTCATTTTTTGACCCTGCGGGTGGAGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 203–204, bears the following four rectangular labels, three white: [PERU:Cuzco 540 m. | Villa Carmen | Pilcopata 4057| 03-V-2015 Kinyon], [DNA sample ID: | NVG-18012D01 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450290], and one red [HOLOTYPE ♂ | Talides | eluta Grishin]. Paratypes: 3♂♂: 1♂ NVG-21046B04 Brazil: Rondônia, 5 km S Cacaulandia, linha C-10 (at Rio Pardo) off B-65, 18-Apr-1995, O. Gomes leg., genitalia GTA-7238 [MGCL]; 2♂♂: Peru: Madre de Dios, Alto Madre de Dios at Pantiacolla Lodge, 400 m, GPS −12.6500, −71.2167 W. Dempwolf leg. [WRDempwolf]: NVG-18125D05, WRD 14975 4-Nov-2017; and NVG-18125D07, WRD 14981 7-Nov-2017.
Type locality.
Peru: Cuzco, Pilcopata, Villa Carmen Biological Reserve, elevation 540 m.
Etymology.
In Latin, elutus means washed out. The name is given to signify the reduced coloration and duller appearance of this species compared to the new species described next. The name is an adjective.
Distribution.
Currently known from Peru (Cuzco and Madre de Dios) and Brazil (Rondônia).
Talides laeta Grishin, new species https://zoobank.org/61A18EF8-BC80-4CF3-9143-26179E53CECA (Fig. 8 part, 205–206, 444–445)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Peru identified as Talides alternata E. Bell, 1941 (type locality in Brazil: Santa Catarina, holotype sequenced as NVG-18025D05) show prominent genetic differentiation from it and other species in the genus (Fig. 8): e.g., their COI barcodes differ by 6.4% (42 bp) and from Talides eluta new species by 4.9% (32 bp), and therefore represent a new species. This new species keys to T. alternata alternata (K.13.3(a)) in Evans (1955) but differs from it by a broader, nearly square-shaped yellow spot in the forewing discal cell and the broader area of ventral forewing with submarginal pale overscaling. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly536.106.3:C159T, aly444.1.12:C24T, aly444.1.12:G45A, aly444.1.12:G138A, aly971.9.9:C61T, and COI barcode: A34T, A67G, T133A, C271A, 499C.
Barcode sequence of the holotype.
Sample NVG-18111H05, GenBank OR837717, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGTATATTAGGAACTTCATTAAGATTATTAATTCGGACAGAATTAGGTAATCCAGGATTTTTAATCGGAGATGATCAAATCTATAATACTATTGTAACAGCACATGCTTTTATTATAATTTTTTTTATAGTAATACCAATTATAATTGGAGGATTTGGAAATTGATTAGTTCCTCTTATACTTGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGAATACTTCCACCCTCTTTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCTGGTACAGGATGAACTGTATACCCCCCTCTTTCAGCTAATATTGCCCATCAAGGTTCCTCTGTTGATTTAGCAATTTTTTCACTTCATTTAGCAGGAATTTCTTCTATTTTGGGAGCTATTAATTTTATTACAACAATTATTAATATACGAATTAGAAATTTATTATTTGATCAAATACCCCTATTTGTATGATCTGTAGGAATTACAGCTTTATTGTTATTATTATCTTTACCTGTTTTAGCAGGAGCTATTACAATACTTCTTACAGATCGAAACTTAAATACTTCATTTTTTGATCCTGCAGGTGGAGGAGATCCTATCTTATACCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 205–206, bears the following four rectangular labels, three white: [PERU:Cuzco, 1375m | San Pedro Lodge | Cosnipata Valley 3756 | 21.IX.2014 Kinyon], [DNA sample ID: | NVG-18111H05 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01531341], and one red [HOLOTYPE ♂ | Talides laeta | Grishin]. Paratypes: 2♂♂ NVG-18124A01, WRD 14984 at the type locality, GPS −13.05, −71.55, 1-Nov-2017, W. Dempwolf leg. [WRDempwolf]and NVG-20017B02, 2019–043 Peru: Madre de Dios, Amazonia Lodge, 491m, 24-Oct-2013, M. McInnis leg. [MUSM].
Type locality.
Peru: Cuzco Region, Cosñipata Road, San Pedro Lodge, elevation 1375 m.
Etymology.
In Latin, laetus means colorful, joyful, glad, pleasing, or vivid. The name is given to signify this species is more brightly colored than the previous one (Talides eluta new species). The name is an adjective.
Distribution.
Currently known from Peru.
Neoxeniades angustior Grishin, new species https://zoobank.org/AD9B121F-E94F-4090-A0FF-339601F78275 (Fig. 8 part, 207–210, 446–450)
Definition and diagnosis.
Phylogenetic trees reveal that some specimens identified as Neoxeniades ethoda (Hewitson, 1866) (type locality in Brazil: Rio de Janeiro) show prominent genetic differentiation from it (Fig. 8): e.g., their COI barcodes differ by 4% (26 bp), and therefore represent a new species. This new species keys to “Xeniades ethoda” (O.13.4) in Evans (1955) but differs from it by smaller forewing hyaline yellow spots: e.g., the spot in cell CuA1-CuA2 does not overlap with the discal cell spot, the presence of subapical forewing hyaline spots (small in some males), a differently shaped hindwing white band: gets very thin, intermittent, or disappears towards vein Sc+R1-RS, and more olive in color in contrast to redder N. ethoda. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly318.10.1:A2253G, aly318.10.1:T2289C, aly133.4.1:C446T, aly133.4.1:G456A, aly159.20.1:C63G, and COI barcode: T46C, T56C, T133C, T340C, T364C, 500C.
Barcode sequence of the holotype.
Sample NVG-22035D03, GenBank OR837718, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATACTAGGAACCTCTTTAAGACTATTAATTCGTACAGAATTAGGAAATCCTGGATTTTTAATTGGAAATGATCAAATTTATAATACCATTGTAACAGCCCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTCGGAAATTGATTAGTTCCATTAATATTAGGTGCTCCTGATATAGCATTTCCTCGATTAAATAATATAAGATTTTGATTATTACCACCTTCATTAATATTATTAATTTCAAGAAGAATTGTAGAAAATGGAGCTGGAACAGGTTGAACTGTTTACCCACCTCTTTCTTCTAATATTGCCCATCAAGGATCTTCTGTAGATTTAGCAATTTTTTCCCTTCATTTAGCAGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACCATTATTAATATACGAATTAAAAATTTATCATTTGATCAAATACCTCTATTTGTTTGATCTGTAGGAATTACAGCTTTACTTTTATTATTATCTCTCCCAGTATTAGCAGGAGCAATTACAATACTTCTTACTGATCGTAATCTTAATACTTCATTTTTTGATCCTTCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 207–208, bears the following four rectangular labels, three white: [BRAZIL, RJ, RJ | P. N. da Tijuca | Morro de Dona Marta | 22°56.7′S, 43°11.4′W | 7 May 1994, 362 m | Leg. Robbins/Caldas], [Territorial | Behavior | Time: 0715], [DNA sample ID: | NVG-22035D03 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Neoxeniades | angustior Grishin]. Paratypes: 1♂ and 2♀♀: Brazil: Rio de Janeiro, old (around 1900) [FMNH]: 1♂ NVG-18039C02 and 1♀ NVG-18039C03; and 1♀ NVG-21119A09, #5243 Brazil, prior to 1876 [MFNB] (Fig. 209–210).
Type locality.
Brazil: Rio de Janeiro, Tijuca National Park, Morro de Dona Marta, elevation 362 m, GPS −22.945, −43.190.
Etymology.
In Latin, angustior means narrower, limited, or constructed. The name is given to signify both its wing pattern and distribution and is treated as a noun in apposition.
Distribution.
Currently only known from around Rio de Janeiro in Brazil.
Damas zea Grishin, new species https://zoobank.org/60CBF52F-A78C-4AA8-8C6C-5E47E7638F4D (Fig. 8 part, 211–212, 451–453)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen from Guyana that is similar in appearance to Damas cervelina Orellana and Costa 2019 (type locality in Venezuela) shows prominent genetic differentiation from it (Fig. 8): e.g., their COI barcodes differ by 4.1% (27 bp), and therefore represents a new species. This new species differs from its sister D. cervelina by forewing spots being rounder and subapical spots lacking. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly393.18.11:A61G, aly860.2.2:G81T, aly1772.7.1:T522G, aly1772.7.1:T732A, aly1405.1.1:T303C, aly499.36.2:C202C (not A), aly499.36.2:A240A (not G), aly16540.2.1:C225C (not T), aly16540.2.1:A289A (not T), aly2700.18.3:G117G (not A), and COI barcode: T133C, T136C, T337C, T406C, A562G.
Barcode sequence of the holotype.
Sample NVG-8006, GenBank OR837719, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGATTATTAGGAACTTCTTTAAGTATATTAATTCGAACAGAATTAGGAAATCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACAATTGTTACAGCCCACGCCTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTACCTTTAATATTAGGTGCTCCTGATATAGCTTTCCCTCGAATAAATAATATAAGATTTTGAATATTACCTCCATCTTTAATCTTATTAATTTCAAGAAGAATCGTAGAAACTGGAGCAGGAACTGGTTGAACTGTCTATCCCCCTCTTTCCTCCAATATTGCTCACCAAGGAGCTTCAGTAGATTTAGCTATTTTTTCCTTACACTTAGCAGGAATTTCTTCCATTTTAGGAGCAATTAATTTTATTACCACAATCATTAATATACGTGTAAGAAATTTATCTTTTGATCAAATACCTTTATTTGTATGATCAGTAGGAATTACAGCCCTATTATTACTTTTATCTTTACCAGTTTTAGCGGGAGCTATTACTATATTACTTACTGATCGAAATCTTAATACTTCTTTTTTTGACCCAGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 211–212, bears the following six rectangular labels, five white: [Guyana: Region 7 | Mt. Ayanganna | 4500’ - 5500’ | 5°24.1′N 59°57.4′W | 13 – 18 Apr 1999 | leg. S. Fratello, R. Hanner, | W. Prince, R. Williams.], [{QR Code} | USNM ENT 00232408], [DNA sample ID: | NVG-8006 | c/o Nick V. Grishin], [genitalia | NVG170207–91 | Nick V. Grishin], [USNMENT | {QR Code} | 01321846], and one red [HOLOTYPE ♀ | Damas zea | Grishin].
Type locality.
Guyana: Cuyuni-Mazaruni Region, Mt. Ayanganna, elevation 4500–5500 ft, GPS 5.4017, −59.9567.
Etymology.
The name reflects wing pattern similarity to the Old World species Unkana mytheca (Hewitson, 1877), which is the type species of the genus Zea Distant, 1886, currently regarded as a junior subjective synonym of the genus Unkana Distant, 1886. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Guyana.
Lectotype designation for Thracides xanthura Godman, 1901
Thracides xanthura Godman, 1901, currently a species of Tromba Evans, 1955 (type species Tromba tromba Evans, 1955), was described from a series of specimens collected at multiple localities: Belize, Honduras, Panama, and Colombia. Due to the polytypic type series and to define the type locality more precisely, N.V.G. hereby designates a male syntype in the Natural History Museum, London, UK, that according to its label, was illustrated by Godman (1899–1901), and bears the following five rectangular white labels: [Bugaba, | 800–1,000 ft. | Champion.], [Sp. figured.], [B.C.A.Lep.Rhop. | Thracides | xanthura, | Godm.], [Godman-Salvin | Coll. 1914.—5.], [{QR Code} | BMNH(E) 1669777], and a genitalia mini-slide number 111 that is pinned with the labels, as the lectotype of Thracides xanthura Godman, 1901. The lectotype had scales of left wings removed to study venation. The type locality of Thracides xanthura becomes Panama: Chiriquí Province, Bugaba.
Tromba xantha Grishin, new species https://zoobank.org/8A465187-542D-43AA-B5DA-BF3B8DB19614 (Fig. 8 part, 213–214, 454–455)
Definition and diagnosis.
Phylogenetic trees reveal that a number of specimens identified as Tromba xanthura (Godman, 1901) (type locality in Panama) show prominent genetic differentiation from it (Fig. 8): e.g., their COI barcodes differ by 3.6% (24 bp), and therefore represent a new species. This new species keys to T. xanthura (K.14.2) in Evans (1955) but differs from it by male genitalia with deeper separation of harpe from ampulla, which is only slightly notched in T. xanthura, but in some specimens of the new species (including the holotype, Fig. 454–455) is separated by a groove, more prominent serrations towards distal end of the dorsal margin of harpe, and less bulky ventral margin on harpe near its fusion with the body of valva. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly2672.7.10:C117T, aly1295.7.1:A156T, aly133.6.8:G260A, aly4592.2.4:G73A, aly4592.2.4:A74T, and COI barcode: 49G, 238C, T340C, T367C, T640C.
Barcode sequence of the holotype.
Sample NVG-5054, GenBank OR837720, 658 base pairs:
AACTTTATATTTTATTTTCGGTATTTGAGCAGGAATATTAGGAACATCGTTAAGATTATTAATTCGAACTGAATTAGGTAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACCATTGTAACAGCTCATGCCTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGTTTTGGAAATTGATTAGTACCATTAATATTAGGAGCACCAGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGAATATTACCTCCTTCATTAACTTTATTAATCTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGATGAACTGTTTACCCCCCTTTATCTTCTAATATTGCTCACCAAGGATCTTCAGTAGATTTAGCAATTTTTTCTCTTCATTTAGCAGGTATTTCTTCTATTTTAGGAGCAATTAATTTTATTACTACAATTATTAATATACGAATTATAAACTTATCTTTTGACCAAATACCTTTATTTGTTTGATCTGTAGGAATTACAGCTTTATTATTATTATTATCTTTACCTGTATTAGCTGGAGCTATTACAATATTACTTACTGATCGAAATTTAAATACTTCTTTTTTTGATCCTGCAGGAGGTGGAGATCCAATCTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 213–214, bears the following five rectangular labels, four white: [Paso San Juan, | V. Cruz.], [Thracides | xanthura | Godm. | comp. type.], [DNA sample ID: | NVG-5054 | c/o Nick V. Grishin], [genitalia | NVG151102–09 | Nick V. Grishin], and one red [HOLOTYPE ♂ | Tromba xantha | Grishin]. Paratypes: 3♂♂: 1♂ NVG-17111B07 Mexico: Guerrero, Ixtapa, 5-Mar-1985, Benjamin Landing leg. [LACM]; 1♂ NVG-5055 Honduras: 18 km W of La Ceiba, 17-Apr-1980 Robert D. Lehman leg., genitalia NVG151102–10 [USNM]; and 1♂ NVG-5056 Nicaragua: San Marcos, old specimen ca. 1900, Coll. Baker, genitalia NVG151102–11 [USNM].
Type locality.
Mexico: Veracruz, Paso de San Juan.
Etymology.
The name is derived from T. xanthura, the southern counterpart of this species. It has been shortened to indicate this is a more northern species. The name is a feminine adjective.
Distribution.
From Mexico to Nicaragua.
Tribe Pericharini Grishin, 2019 Subtribe Pericharina Grishin, 2019
Perichares fura Grishin, new species https://zoobank.org/2FD4A73B-7AC8-4B32-BC27-CB0F8F5FE674 (Fig. 8 part, 215–216, 456–458)
Definition and diagnosis.
Phylogenetic trees reveal that specimens from Ecuador identified as Perichares furcata (Mabille, 1891) (type locality in Brazil: São Paulo) are not monophyletic with it and show prominent genetic differentiation from it (Fig. 8): e.g., their COI barcodes differ by 6.7% (44 bp), and therefore represent a new species. This new species is a distant sister to Perichares lotus (A. Butler, 1870) (type locality in Venezuela), differing from it by 4.9% (32 bp) in COI barcode, but has remarkably different ventral wing pattern, being nearly identical to Perichares furcata instead. This new species keys to “Alera furcata” (K.32.3) in Evans (1955) but differs from it by a brown spot in the humeral area of ventral hindwing that is missing in P. furcata and a more diffuse boundary between the darker discal forewing area and paler apex, particularly towards costa that is rather sharp in P. furcata. Due to the cryptic nature of this species, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly525.25.7:G450T, aly15656.2.3:G461A, aly275209.10.4:C66G, aly173.49.2:C33T, aly2012.16.4:T135A, and COI barcode: T55A, T241C, T250C, T394C, T484C.
Barcode sequence of the holotype.
Sample NVG-18014F08, GenBank OR837721, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGAATATTAGGAACATCTCTAAGATTATTAATTCGTACTGAATTAGGAAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTTACTGCCCATGCTTTTATTATAATTTTTTTTATAGTTATACCCATTATAATTGGAGGATTCGGAAATTGACTTGTCCCTCTTATATTAGGAGCTCCTGATATAGCTTTTCCTCGCATAAATAACATAAGATTTTGAATATTACCCCCATCATTAACTCTTTTAATTTCAAGAAGAATCGTTGAAAACGGTGCTGGAACTGGATGAACAGTTTACCCCCCACTTTCATCTAATATTGCCCATCAAGGATCTTCAGTTGATTTAGCAATCTTTTCCCTTCATTTAGCAGGAATTTCCTCTATTCTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTATAAATTTATCCTTTGACCAAATACCTTTATTTGTTTGATCAGTAGGTATTACAGCCTTATTATTACTACTATCTTTACCAGTATTAGCAGGAGCTATTACAATACTTCTTACAGATCGAAATTTAAATACCTCATTTTTTGATCCTGCAGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♀ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 215–216, bears the following five rectangular labels, four white: [Alluriquin 700 m | PICHINCHA ECUADOR | 14 Sept. ‘76 | S. S. Nicolay], [Perichares | lotus ♀ | Det. Btlr. | S.S. Nicolay], [DNA sample ID: | NVG-18014F08 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450684], and one red [HOLOTYPE ♀ | Perichares | fura Grishin]. Paratype: 1♀ NVG-18014F07, USNMENT_01450683 Ecuador: Pichincha, 10 mi E of Santo Domingo de los Colovados, Tinalandia Grounds/Trails, 16–21-Apr-1984, Brian Harris leg. [USNM].
Type locality.
Ecuador: Pichincha, Alluriquin, elevation 700 m.
Etymology.
The name reflects a strong superficial similarity to Perichares furcata (Mabille, 1891) but does not reflect a close relationship. The name is a non-Latinized noun in apposition.
Distribution.
Currently known only from Pichincha Province, Ecuador.
Tribe Megathymini Comstock and Comstock, 1895 Subtribe Carystoidina Grishin, 2022
Carystoides (Balma) goliath Grishin, new species https://zoobank.org/A2EA2056-49E6-4315-A42C-A9FF87A5C1A3 (Fig. 8 part, 217–218, 459–464)
Definition and diagnosis.
Phylogenetic trees reveal that a unique in appearance specimen from Colombia belongs to the subgenus Balma Grishin, 2022 (type species Carystoides balza Evans, 1955) of the genus Carystoides Godman, 1901 (type species Hesperia basoches Latreille, [1824]) and differs prominently from both species of the subgenus: C. balza (type locality in Ecuador) and Carystoides maroma Möschler, 1877 (type locality in Suriname) (Fig. 8): by 8.1% (52 bp) and 5.5% (36 bp) in COI barcodes, respectively, and therefore represents a new species. This new species differs from all others by a pale yellow spot at the base of forewing cell CuA1-CuA2 above and a unique for a male arrangement of three forewing hyaline spots with the spot in cell CuA1-CuA2 basad of the spot in the discal cell without overlapping it. This species is not cryptic and is diagnosed reliably by phenotype. In DNA, a combination of the following base pairs is diagnostic in the nuclear genome: aly1146.46.3:T206C, aly1139.84.1:A2445G, aly923.1.5:T72C, aly215.21.1:A1036C, aly215.21.1:A1078C, aly85.38.8:G72G (not T), aly85.38.8:A81A (not T), aly128.26.2:A160A (not C), aly4778.18.1:A394A (not C), aly1282.12.2:T204T (not C), and COI barcode: A37G, T115C, T142C, T487C, A577G, T637C.
Barcode sequence of the holotype.
Sample NVG-18012D11, GenBank OR837722, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATGTTAGGAACATCATTAAGTTTAATAATTCGTACAGAATTAGGAAATCCTGGATCTTTAATTGGAGATGACCAAATTTACAATACTATTGTCACAGCCCATGCTTTCATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGACTTGTACCTTTAATACTAGGAGCTCCTGATATAGCATTCCCTCGAATAAATAATATAAGTTTTTGAATACTACCCCCCTCATTAACTTTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCTGGTACAGGATGAACTGTCTATCCCCCCCTTTCATCTAATGTAGCTCACCAAGGATCATCTGTAGATTTAGCTATTTTTTCTTTACATTTAGCAGGTATTTCTTCAATTTTAGGAGCTATTAATTTTATTACTACTATTATTAATATACGAATTAAAAATTTATCATTCGACCAAATACCTTTATTTATTTGATCTGTAGGAATTACTGCTTTATTATTATTATTATCTTTACCTGTTTTAGCGGGAGCTATTACTATGCTTCTTACTGATCGAAATTTAAATACCTCTTTTTTTGACCCAGCGGGAGGAGGAGACCCCATTCTTTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), illustrated in Fig. 217–218, bears the following five rectangular labels, four white: [FEB. 1 1989 | Anchicaya, 650m | Valle, Colombia | J. Bolling Sullivan], [Genit. Vial | SRS-3255], [DNA sample ID: | NVG-18012D11 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01450298], and one red [HOLOTYPE ♂ | Carystoides (Balma) | goliath Grishin].
Type locality.
Colombia: Valle, Anchicaya, elevation 650 m.
Etymology.
The name is given for the large size of this species and is a noun in apposition.
Distribution.
Colombia.
Subtribe Megathymini Comstock and Comstock, 1895
Agathymus galeana Grishin, new species https://zoobank.org/1705C581-B49C-4679-9A6C-B1ACDD11F9CB (Fig. 8 part, 219–220)
Definition and diagnosis.
Phylogenetic trees reveal that a specimen of Agathymus H. Freeman, 1959 (type species Megathymus neumoegeni W. H. Edwards, 1882) from Nuevo Leon Mexico is not closely related to any of the known species of the genus and is a distant sister to both Agathymus estelleae (D. Stallings and Turner, 1958) (type locality in Mexico: Nuevo Leon) and Agathymus remingtoni (D. Stallings and J. Turner, 1958) (type locality in Mexico: Hidalgo) (Fig. 8): COI barcode differences from them are 6.2% (41 bp), and 5.3% (35 bp), respectively, while being phenotypically dissimilar to them. This new species is more similar to Agathymus hoffmanni (H. Freeman, 1952) (type locality in Mexico: “Valle de Mexico”), among others, in its orange coloration and pattern of spots but differs from it and other species by rounder wings, uniformly gray ventral hindwing without any spots and blotches, relatively narrow dorsal hindwing orange postdiscal band contrasting with broader orange spots on forewing overscaled with orange at its base, and orange along the cubitus in forewing discal cell. Due to the lack of additional specimens and unknown phenotypic variation, most reliable identification is achieved by DNA and a combination of the following base pairs is diagnostic in the nuclear genome: aly5773.3.3:C118T, aly5773.3.3:T428C, aly2874.12.6:C66T, aly3850.2.2:T84A, aly2041.8.3:T41G, aly2012.46.3:A132A (not G), aly5729.2.6:A854A (not C), aly536.163.5:T18T (not G), aly798.8.13:G48G (not C), aly16.30.9:A241A (not G), and COI barcode: A37G, T59A, T136C, T346C, 581C.
Barcode sequence of the holotype.
Sample NVG-1895, GenBank OR837723, 658 base pairs:
TACTTTATATTTTATTTTTGGTATTTGAGCAGGTATGTTAGGAACATCTTTAAGATTAATAATTCGTACTGAATTAGGAAATCCAGGATCATTAATTGGAGATGATCAAATTTATAATACCATTGTAACAGCTCACGCTTTTATTATAATTTTTTTCATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTACCTTTAATATTAGGAGCACCTGATATAGCTTTTCCCCGTATAAATAACATAAGATTTTGAATATTACCCCCCTCATTAATATTACTAATCTCAAGAAGAATTGTAGAAAATGGTGCAGGAACAGGATGAACTGTATACCCTCCCCTATCTTCTAATATTGCACATCAAGGTTCTTCAGTTGATTTAGCAATTTTTTCCCTTCATTTAGCAGGTATCTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATTTATCATTTGATCAAATACCTCTATTTGTATGATCTGTTGGAATTACTGCTTTATTATTATTATTATCTCTACCTGTTTTAGCTGGAGCTATTACTATACTTCTAACAGATCGAAATTTAAATACTTCTTTTTTTGACCCTGCTGGAGGAGGAGATCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♀ deposited in the Texas A&M University Insect Collection, College Station, TX, USA (TAMU), illustrated in Fig. 219–220, bears the following six rectangular labels, five white: [MEXICO: | Nuevo Leon | ± 40 Km. WSW | Linares], [♂], [coll. | 21-IX-1977 | Roy O. Kendall | & C. A. Kendall], [DNA sample ID: | NVG-1895 | c/o Nick V. Grishin], [NVG140104–44], and one red [HOLOTYPE ♀ | Agathymus | galeana Grishin].
Type locality.
Mexico: Nuevo Leon, ca 40 km WSW of Linares.
Etymology.
The name is given for the type locality of this species near Galeana in Mexico and is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Nuevo Leon, Mexico.
Supplementary Material
Acknowledgments
We acknowledge Jinhui Shen, Leina Song, Ping Chen, and Ming Tang for their excellent technical assistance. We are grateful to David Grimaldi and Courtney Richenbacher (AMNH: American Museum of Natural History, New York, NY, USA), Jason Weintraub (ANSP: Academy of Natural Sciences of Drexel University, Philadelphia, PA, USA), Blanca Huertas, David Lees, and Geoff Martin (BMNH: Natural History Museum, London, UK), Chris Grinter, Denise Montelongo, David Bettman, Vince F. Lee, and the late Norm Penny (CAS: California Academy of Sciences, San Francisco, CA, USA), Jim Fetzner, Bob Androw, Vanessa Verdecia, Cat Giles, and the late John Rawlins (CMNH: Carnegie Museum of Natural History, Pittsburgh, PA, USA), the late Paul A. Opler, Chuck Harp, and the late Boris Kondratieff (CSUC: Colorado State University Collection, Fort Collins, CO, USA), Jason Dombroskie (CUIC: Cornell University Insect Collection, Ithaca, New York, USA), Olaf. H. H. Mielke and Mirna M. Casagrande (DZUP: Universidade Federal do Paraná, Curitiba, Paraná, Brazil), Crystal Maier and Rebekah Baquiran (FMNH: Field Museum of Natural History, Chicago, IL, USA), Weiping Xie and Giar-Ann Kung (LACM: Los Angeles County Museum of Natural History, Los Angeles, CA, USA), John R. MacDonald and Richard L. Brown (MEM: Mississippi Entomological Museum, Starkville, MS, USA), Théo Léger, Christoph L. Häuser, Wolfram Mey, and Viola Richter (MFNB: Museum für Naturkunde, Berlin, Germany), Andrei Sourakov, Andrew D. Warren, Debbie Matthews-Lott, Riley J. Gott, and Keith R. Willmott (MGCL: McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA), José “Pepe” Clavijo, Quintin Arias, and Mauro Costa (MIZA: Museo del Instituto de Zoología Agrícola, Universidad Central de Venezuela, Maracay, Venezuela), Rodolphe Rougerie (MNHP: Muséum National d’Histoire Naturelle, Paris, France), Matthias Nuss and Manuela Bartel (MTD: Museum für Tierkunde, Dresden, Germany), Gerardo Lamas (MUSM: Museo de Historia Natural, Lima, Peru), Larry F. Gall (PMNH: Peabody Museum of Natural History, Yale University, New Haven, CT, USA), Rob de Vos (RMNH: Naturalis Biodiversity Center, Leiden, Netherlands), Wolfgang A. Nässig (SMF: Natural History Museum, Frankfurt, Germany), Edward G. Riley, Karen Wright, and John Oswald (TAMU: Texas A&M University Insect Collection, College Station, TX, USA), Alex Wild (TMMC: Biodiversity Center, University of Texas at Austin, Austin, TX, USA), Robert K. Robbins, John M. Burns, and Brian Harris (USNM: National Museum of Natural History, Smithsonian Institution, Washington, DC, USA), Jonathan Pelham (UWBM: Burke Museum of Natural History and Culture, Seattle, WA, USA), Peter Michalik and Lara Lopardo (ZIMG: Zoological Institute and Museum Greifswald, Germany), Ernst Brockmann (ZfBS: Zentrum fur Biodokumentation des Saarlandes, Schiffweiler, Germany), Thomas Pape (ZMUC: Natural History Museum of Denmark, University of Copenhagen, Copenhagen, Denmark), and Axel Hausmann, Andreas Segerer, and Ulf Buchsbaum (ZSMC: Zoologische Staatssammlung München, Germany), for granting access to or sampling specimens in the collections under their care and for stimulating discussions; to Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the research permit 08-02Rev; to U. S. National Park Service for the research permits: Big Bend (Raymond Skiles) for BIBE-2004-SCI-0011 and Yellowstone (Erik Oberg and Annie Carlson) for YELL-2017-SCI-7076; to the National Environment and Planning Agency of Jamaica for the permission to collect specimens; to Pierre Boyer, Jim P. Brock, Ernst Brockmann, Matthew J. W. Cock, Bill Dempwolf, Bernard Hermier, the late Edward C. Knudson, John R. MacDonald, and Kiyoshi Maruyama for specimens and leg samples, to Ernst Brockmann for help with sampling specimens for DNA in several German collections, to Gerardo Lamas and Bernard Hermier for fruitful discussions, comments, critiques and suggestions. We are indebted to James K. Adams, Bernard Hermier, Brian G. Scholtens, and David Wright for their critical reviews of the manuscript. We acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources. The study was supported in part by the HHMI Investigator funds and grants (to N.V.G.) from the National Institutes of Health (GM127390) and the Welch Foundation (I-1505).
Footnotes
ZooBank registration. urn:lsid:zoobank.org:pub:ACDF923B-906D-460E-9707-259E0ECDBCA8
Contributor Information
Jing Zhang, Eugene McDermott Center for Human Growth and Development and Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-8816 USA.
Qian Cong, Eugene McDermott Center for Human Growth and Development and Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-8816 USA.
Nick V. Grishin, Departments of Biophysics and Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-9050 USA
Literature Cited
- Austin GT. 2000. Hesperiidae of Rondônia, Brazil: “Antigonus” genus group (Pyrginae), with taxonomic comments and descriptions of new species from Brazil and Guatemala. Journal of the Lepidopterists’ Society 54(1): 1–28. [Google Scholar]
- Austin GT. 2008. Hesperiidae of Rondônia, Brazil: Taxonomic comments on ‘night’ skippers, with descriptions of new genera and species (Lepidoptera: Eudaminae). Insecta Mundi 29: 1–36. [Google Scholar]
- Austin GT, Mielke OHH. 1998. Hesperiidae of Rondônia, Brazil: Aguna Williams (Pyrginae), with a partial revision and descriptions of new species from Panama, Ecuador, and Brazil. Revista brasileira de Zoologia 14(4): 889–965. [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nature Methods 12(1): 59–60. [DOI] [PubMed] [Google Scholar]
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PD. 2008. DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proceedings of the National Academy of Sciences of the United States of America 105(17): 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Zhang J, Li W, Kinch LN, Calhoun JV, Warren AD, Grishin NV. 2021. Genomics reveals the origins of historical specimens. Molecular Biology and Evolution 38(5): 2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Zhang J, Grishin NV. 2019a. Genomic determinants of speciation. bioRxiv BIORXIV/2019/837666. Available at 10.1101/837666 (Last accessed November 2023.) [DOI] [Google Scholar]
- Cong Q, Zhang J, Shen J, Grishin NV. 2019b. Fifty new genera of Hesperiidae (Lepidoptera). Insecta Mundi 0731: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolibaina DR, Mielke OH, Casagrande MM. 2014. Taxonomic revision of Cumbre Evans, 1955 (Hesperiidae: Hesperiinae: Moncini), with the description of two new species. Zootaxa 3841(1): 47–66. [DOI] [PubMed] [Google Scholar]
- Dolibaina DR, Mielke OHH, Casagrande MM. 2017. Taxonomy of Rufocumbre gen. nov., a new Moncini skipper genus (Lepidoptera: Hesperiidae: Hesperiinae). Zootaxa 4365(2): 196–216. [DOI] [PubMed] [Google Scholar]
- Draudt MWK. 1921–1924. B. Grypocera, breitköpfige Tagfalter. p. 833–1011, 1046–1139, pl. 113B, 160–193. In: Seitz A (Ed.). Die Gross-Schmetterlinge der Erde. Alfred Kernen; Stuttgart. 1141 p. [Google Scholar]
- Evans WH. 1937. A catalogue of the African Hesperiidae indicating the classification and nomenclature adopted in the British Museum. The Trustees of the British Museum (Natural History); London. xii + 212 p., 30 pl. [Google Scholar]
- Evans WH. 1949. A catalogue of the Hesperiidae from Europe, Asia, and Australia in the British Museum (Natural History). The Trustees of the British Museum (Natural History); London. xix + 502 p., 53 pl. [Google Scholar]
- Evans WH. 1951. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part I. Introduction and Group A Pyrrhopyginae. The Trustees of the British Museum (Natural History); London. x + 92 p., pl. 1–9. [Google Scholar]
- Evans WH. 1952. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part II. Pyrginae. Section I. The Trustees of the British Museum (Natural History); London. v + 178 p., pl. 10–25. [Google Scholar]
- Evans WH. 1953. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part III. Pyrginae. Section 2. The Trustees of the British Museum (Natural History); London. v + 246 p., pl. 26–53. [Google Scholar]
- Evans WH. 1955. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part IV. Hesperiinae and Megathyminae. The Trustees of the British Museum (Natural History); London. v + 499 p., pl. 54–88. [Google Scholar]
- Fernandez-Triana JL, Shimbori EM, Whitfield JB, Penteado-Dias AM, Shaw SR, Boudreault C, Sones J, Perez K, Brown A, Manjunath R, Burns JM, Hebert PDN, Smith MA, Hallwachs W, Janzen DH. 2023. A revision of the parasitoid wasp genus Alphomelon Mason with the description of 30 new species (Hymenoptera, Braconidae). ZooKeys 1175: 5–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Triana JL, Whitfield JB, Rodriguez JJ, Smith MA, Janzen DH, Hallwachs WD, Hajibabaei M, Burns JM, Solis MA, Brown J, Cardinal S, Goulet H, Hebert PD. 2014. Review of Apanteles sensu stricto (Hymenoptera, Braconidae, Microgastrinae) from Area de Conservacion Guanacaste, northwestern Costa Rica, with keys to all described species from Mesoamerica. ZooKeys 383: 1–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godman FD. 1899–1901. Hesperiidae. p. 457–782. In: Godman FD, Salvin O. Biologia Centrali-Americana. Insecta. Lepidoptera-Rhopalocera. Vol 2. Dulau & Co., Bernard Quaritch; London. 782 p. [Google Scholar]
- Godman FD. 1907. Notes on the American species of Hesperiidae described by Plötz. Annals and Magazine of Natural History (7)20(16): 132–155. [Google Scholar]
- Godman FD, Salvin O. 1893–1899. Biologia Centrali-Americana. Insecta. Lepidoptera-Rhopalocera. Vol 2. Dulau & Co., Bernard Quaritch; London. 782 p. [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270(1512): 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitson WC. 1867. Descriptions of one hundred new species of Hesperiidae 1. John Van Voorst; London. ii+25 p. [Google Scholar]
- Hewitson WC. 1868. Descriptions of one hundred new species of Hesperiidae 2. John Van Voorst; London. p. 25–56. [Google Scholar]
- Li W, Cong Q, Shen J, Zhang J, Hallwachs W, Janzen DH, Grishin NV. 2019. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proceedings of the National Academy of Sciences of the United States of America 116(13): 6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Sourakov A, Zakharov E. 2016. DNA barcodes as a tool in biodiversity research: testing pre-existing taxonomic hypotheses in Delphic Apollo butterflies (Lepidoptera, Papilionidae). Systematics and Biodiversity 14: 599–613. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhenkova EA, Lukhtanov VA. 2021. Genomic introgression from a distant congener in the Levant fritillary butterfly, Melitaea acentria. Molecular Ecology 30(19): 4819–4832. [DOI] [PubMed] [Google Scholar]
- Plötz C 1881. Die Hesperiinen-Gattung Eudamus und ihre Arten. Stettiner entomologische Zeitung 42(10/12): 500–504; 43(1/3): 87–101. [Google Scholar]
- Plötz C 1882. Die Hesperiinen-Gattung Hesperia Aut. und ihre Arten. Stettiner entomologische Zeitung 44(1/3): 26–64. [Google Scholar]
- Plötz C 1883. Die Hesperiinen-Gattung Hesperia Aut. und ihre Arten. Stettiner entomologische Zeitung 44(4/6): 195–233. [Google Scholar]
- Rambaut A 2018. FigTree, version 1.4.4. Available at http://tree.bio.ed.ac.uk/software/figtree/ (Last accessed November 2023.)
- Rubinoff D, Cameron S, Will K. 2006. A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. Journal of Heredity 97(6): 581–594. [DOI] [PubMed] [Google Scholar]
- Sharkey MJ, Janzen DH, Hallwachs W, Chapman EG, Smith MA, Dapkey T, Brown A, Ratnasingham S, Naik S, Manjunath R, Perez K, Milton M, Hebert P, Shaw SR, Kittel RN, Solis MA, Metz MA, Goldstein PZ, Brown JW, Quicke DLJ, van Achterberg C, Brown BV, Burns JM. 2021. Minimalist revision and description of 403 new species in 11 subfamilies of Costa Rican braconid parasitoid wasps, including host records for 219 species. ZooKeys 1013: 1–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Cong Q, Borek D, Otwinowski Z, Grishin NV. 2017. Complete genome of Achalarus lyciades, the first representative of the Eudaminae subfamily of skippers. Current Genomics 18(4): 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewert RR, Mielke OHH, Casagrande MM. 2020. Taxonomic revision of the Neotropical genus Telemiades Hubner, [1819] (Lepidoptera: Hesperiidae: Eudaminae), with descriptions of fourteen new species. Zootaxa 4721(1): 1–111. [DOI] [PubMed] [Google Scholar]
- Steinhauser SR, Austin GT. 1993. New species of Hesperiidae from Costa Rica. Tropical Lepidoptera 4(Suppl. 2): 12–20. [Google Scholar]
- Wikipedia contributors. 2023. Corax of Syracuse. Wikipedia, The Free Encyclopedia. August 2, 2023, 07:50 UTC. Available at https://en.wikipedia.org/w/index.php?title=Corax_of_Syracuse&oldid=1168349841 (Last accessed November 2023.) [Google Scholar]
- Zhang J, Cong Q, Burns JM, Grishin NV. 2022a. Checking the checkered taxonomy of Plötz’s checkered skippers (Hesperiidae: Pyrgini). The Taxonomic Report of the International Lepidoptera Survey 10(5): 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Grishin NV. 2023a. Supplementary information to “Descriptions of one hundred new species of Hesperiidae.” Table S1 and DNA sequences of exons with diagnostic characters. Available at https://osf.io/hq7g4/ (Last accessed December 2023.) [Google Scholar]
- Zhang J, Cong Q, Grishin NV. 2023b. Thirteen new species of butterflies (Lepidoptera: Hesperiidae) from Texas. Insecta Mundi 0969: 1–58. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Brockmann E, Grishin NV. 2019a. Genomes reveal drastic and recurrent phenotypic divergence in firetip skipper butterflies (Hesperiidae: Pyrrhopyginae). Proceedings of the Royal Society B: Biological Sciences 286(1903): 20190609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Brockmann E, Grishin NV. 2019b. Three new subfamilies of skipper butterflies (Lepidoptera, Hesperiidae). ZooKeys 861: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Grishin NV. 2022b. Taxonomic changes suggested by the genomic analysis of Hesperiidae (Lepidoptera). Insecta Mundi 0921: 1–135. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, Grishin NV. 2020. Genomic evidence suggests further changes of butterfly names. The Taxonomic Report of the International Lepidoptera Survey 8(7): 1–40. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2022c. Genomic DNA sequencing reveals two new North American species of Staphylus (Hesperiidae: Pyrginae: Carcharodini). The Taxonomic Report of the International Lepidoptera Survey 10(4): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2023c. Butterfly classification and species discovery using genomics. The Taxonomic Report of the International Lepidoptera Survey 11(3): 1–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dolibaina DR, Cong Q, Shen J, Song L, Mielke CGC, Casagrade MM, Mielke OHH, Grishin NV. 2023d. Taxonomic notes on Neotropical Hesperiidae (Lepidoptera). Zootaxa 5271(1): 91–114. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shen J, Cong Q, Grishin NV. 2019c. Genomic analysis of the tribe Emesidini (Lepidoptera: Riodinidae). Zootaxa 4668(4): 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


