Abstract
Genomic analysis of Pyrginae Burmeister, 1878 (Lepidoptera: Hesperiidae Latreille, 1809) with an emphasis on the tribes Achlyodini Burmeister, 1878 and Carcharodini Verity, 1940 reveals many inconsistencies between the resulting phylogeny and the current classification. These problems are corrected by proposing new taxa, changing the ranks of others, or synonymizing them, and transferring species between genera. As a result, five subtribes, one genus, 20 subgenera, and one species are proposed as new: Cyclosemiina Grishin, new subtribe (type genus Cyclosemia Mabille, 1878), Ilianina Grishin, new subtribe (type genus Iliana E. Bell, 1937), Nisoniadina Grishin, new subtribe (type genus Nisoniades Hübner, [1819]), Burcina Grishin, new subtribe (type genus Burca E. Bell and W. Comstock, 1948), and Pholisorina Grishin, new subtribe (type genus Pholisora Scudder, 1872), all in Carcharodini; Lirra Grishin, new genus (type species Leucochitonea limaea Hewitson, 1868) in Pythonidina Grishin, 2019; Trifa Grishin, new subgenus (type species Tagiades jacobus Plötz, 1884), Tuberna Grishin, new subgenus (type species Pythonides contubernalis Mabille, 1883), Ebona Grishin, new subgenus (type species Quadrus eboneus E. Bell, 1947), Noctis Grishin, new subgenus (type species Achlyodes accedens Mabille, 1895), and Cyrna Grishin, new subgenus (type species Achlyodes cyrna Mabille, 1895) of Quadrus Lindsey, 1925; Liddia Grishin, new subgenus (type species Helias pallida R. Felder, 1869), Minna Grishin, new subgenus (type species Achlyodes minna Evans, 1953), and Thilla Grishin, new subgenus (type species Eurypterus later Mabille, 1891) of Eantis Boisduval, 1836; Torgus Grishin, new subgenus (type species Ouleus gorgus E. Bell, 1937) of Iliana E. Bell, 1937; Fenops Grishin, new subgenus (type species Cabares enops Godman and Salvin, 1894) of Polyctor Evans, 1953; Bezus Grishin, new subgenus (type species Pellicia bessus Möschler, 1877) and Macarius Grishin, new subgenus (type species Pellicia macarius Herrich-Schäffer, 1870) of Nisoniades Hübner, [1819]; Quadralis Grishin, new subgenus (type species Pterygospidea extensa Mabille, 1891) of Gorgopas Godman and Salvin, 1894; Menuda Grishin, new subgenus (type species Nisoniades menuda Weeks, 1902) and Narycus Grishin, new subgenus (type species Pythonides narycus Mabille, 1889) of Perus Grishin, 2019; Bovaria Grishin, new subgenus (type species Achlyodes cyclops Mabille, 1876), Sebia Grishin, new subgenus (type species Nisoniades eusebius Plötz, 1884), and Stolla Grishin, new subgenus (type species Pholisora balsa E. Bell, 1937) of Bolla Mabille, 1903; Vulga Grishin, new subgenus (type species Achlyodes vulgata Möschler, 1879) and Capilla Grishin, new subgenus (type species Helias aurocapilla Staudinger, 1876, currently a junior subjective synonym of Hesperia musculus Burmeister, 1875) of Staphylus Godman and Salvin, 1896; and Quadrus (Zera) vivax Grishin, new species (type locality in Brazil: Rio de Janeiro). The following 10 are subgenera, not genera or synonyms: Ouleus Lindsey, 1925 and Zera Evans, 1953 of Quadrus Lindsey, 1925; Atarnes Godman and Salvin, 1897 and Eburuncus Grishin, 2012 of Milanion Godman and Salvin, 1895; Pachyneuria Mabille, 1888 and Austinus O. Mielke and Casagrande, 2016 of Sophista Plötz, 1879; Hemipteris Mabille, 1889 and Mictris Evans, 1955 of Pellicia Herrich-Schäffer, 1870; and Hesperopsis Dyar, 1905 and Scantilla Godman and Salvin, 1896 of Staphylus Godman and Salvin, 1896. The following 7 are species, not subspecies: Quadrus (Ebona) cristatus (Steinhauser, 1989) (not Quadrus (Ebona) negrus (Nicolay, 1980)), Quadrus (Quadrus) ophia (A. Butler, 1870) (not Quadrus (Quadrus) lugubris (R. Felder, 1869)), Quadrus (Zera) gellius (Mabille, 1903) and Quadrus (Zera) servius (Plötz, 1884) (not Quadrus (Zera) hyacinthinus (Mabille, 1877)), Mimia pazana Evans, 1953 (not Mimia phidyle (Godman and Salvin, 1894)), Polyctor (Polyctor) dagua Evans, 1953 (not Polyctor (Polyctor) polyctor (Prittwitz, 1868)), and Staphylus (Vulga) satrap Evans, 1953 (not Staphylus (Vulga) saxos Evans, 1953); and these 8 are species, not synonyms: Quadrus (Zera) menedemus (Godman and Salvin, 1894) (not Quadrus (Zera) tetrastigma (Sepp, [1847])), Pellicia (Pellicia) bilinea Mabille, 1889 (not Pellicia (Pellicia) dimidiata Herrich-Schäffer, 1870), Pellicia (Hemipteris) nema Williams and Bell, 1939 (not Pellicia (Pellicia) theon Plötz, 1882), Bolla (Bovaria) sodalis Schaus, 1913 (not Bolla (Bolla) imbras (Godman and Salvin, 1896)), Bolla (Bovaria) aplica (E. Bell, 1937) (not Bolla (Sebia) eusebius (Plötz, 1884)), Bolla (Sebia) chilpancingo (E. Bell, 1937) (not Bolla (Bolla) subapicatus (Schaus, 1902)), and Bolla (Stolla) madrea (R. Williams and E. Bell, 1940) and Bolla (Stolla) hazelae (Hayward, 1940) (not Bolla (Stolla) zorilla (Plötz, 1886)). The following 2 are junior subjective synonyms: Achlyodes erisichthon Plötz, 1884 of Quadrus (Zera) servius (Plötz, 1884) (not a subspecies of Quadrus (Zera) tetrastigma (Sepp, [1847]) and Staphylus subapicatus Schaus, 1902 of Bolla (Bolla) imbras (Godman and Salvin, 1896). Furthermore, we propose the following additional new genus-species combination: Gindanes homer (Evans, 1953), Gindanes nides (O. Mielke and Casagrande, 2002), Gindanes maraca (O. Mielke and Casagrande, 1992), Gindanes jenmorrisae (Shuey and Ramírez. 2022), Gindanes tullia (Evans, 1953), Gindanes herennius (Geyer, [1838]), Gindanes proxenus (Godman and Salvin, 1895), Gindanes parallelus (Mabille, 1898), Gindanes braga (Evans, 1953), Gindanes hampa (Evans, 1953), Gindanes rosa (Steinhauser, 1989), Gindanes neivai (Hayward, 1940), Gindanes mundo (H. Freeman, 1979), Gindanes eminus (E. Bell, 1934), Quadrus (Trifa) francesius Freeman, 1969, Quadrus (Trifa) ineptus (Draudt, 1922), Quadrus (Trifa) jacobus (Plötz, 1884), Quadrus (Tuberna) lancea (Hewitson, 1868), Quadrus (Ebona) pescada (E. Bell, 1956), Lirra pteras (Godman and Salvin, 1895), and Lirra limaea (Hewitson, 1868) (not Pythonides Hübner, 1819); Quadrus (Cyrna) zora (Evans, 1953) (not Bolla Mabille, 1903); Eantis later (Mabille, 1891) and Eantis haber (Mabille, 1891) (not Aethilla Hewitson, 1868); Iliana (Torgus) gorgus (E. Bell, 1937) and Iliana (Torgus) taurus (Evans, 1953) (not Eantis Boisduval, 1836); Bolla (Stolla) evemerus (Godman and Salvin, 1896), Bolla (Stolla) chlora (Evans, 1953), Bolla (Stolla) astra (R. Williams and E. Bell, 1940), Bolla (Stolla) balsa (E. Bell, 1937), Bolla (Stolla) tridentis (Steinhauser, 1989), Bolla (Stolla) esmeraldus (L. Miller, 1966), Bolla (Stolla) chlorocephala (Latreille, [1824]), and Bolla (Stolla) incanus (E. Bell, 1932) (not Staphylus Godman and Salvin, 1896). Finally, lectotypes are designated for Achlyodes servius Plötz, 1884 (type locality in Brazil: Rio de Janeiro), Pellicia theon Plötz, 1882 (type locality in South America), and Nisoniades eusebius Plötz, 1884 (type locality in Central America). Unless stated otherwise, all subgenera, species, subspecies, and synonyms of mentioned genera and species are transferred with their parent taxa, and others remain as previously classified.
Keywords: taxonomy, classification, genomics, phylogeny, biodiversity
Introduction
This work is a continuation of our efforts to improve the taxonomic classification of Hesperiidae Latreille, 1809 using large-scale DNA sequencing (Cong et al. 2019b; Li et al. 2019; Zhang et al. 2019a, 2019b, 2022b, 2023c, 2023d) and it complements the studies of others (Warren et al. 2008, 2009; Fan et al. 2016; Sahoo et al. 2016, 2017; Toussaint et al. 2018; Huang et al. 2019; Toussaint et al. 2019, 2021a, 2021b, 2022). Continuing with the whole genome shotgun sequencing of Hesperiidae, we focus on two tribes in the subfamily Pyrginae Burmeister, 1878, namely Achlyodini Burmeister, 1878 and Carcharodini Verity, 1940. These two tribes appear particularly problematic taxonomically, revealing that many taxa in the current classification are non-monophyletic.
In classification decisions, we strongly rely on nuclear genomic trees and use morphological considerations as secondary evidence to rationalize the results. This is because, contrary to a small set of gene markers, genomes offer a comprehensive picture of an organism richer than the morphology of adults typically used to classify butterflies. Genomes encode life histories, habitat and mating preferences, and food sources. All this information is present in the genomic sequence. While we lack the knowledge to extract it and predict phenotypes, we can use a genetic equivalent of this information in an aggregate to deduce phylogenetically sound taxonomic classification. We do so by selecting random codons from all protein-coding genes. This process yields a dataset that mirrors the natural balance of this information as found in the organism’s genome.
We see that phylogenetic trees based on nuclear genomes are particularly suited to address the questions of higher classification and offer guidance on discretizing continuous evolutionary processes of speciation and extinction, fitting the clades to taxonomic ranks. Genome-based phylogenies frequently reveal levels, i.e., timepoints when diversification occurred in several lineages independently (Zhang et al. 2021, 2023b). These synchronized diversifications result from geological events affecting all major lineages at the same time, offering an opportunity to match taxonomic ranks (we utilize tribe, subtribe, genus, and subgenus) to the levels in genome-based phylogenies, which leads to a more objective and consistent classification tied to both genetic differentiation and paleontological history. We attempt to define secondary levels (subtribes and subgenera) in the classification whenever possible if we see noteworthy clustering of genera or species into groups of relatives prominently differentiated from other such groups. We find these subdivisions useful in taxonomic lists, even if for no other reason than to place closer relatives close to each other in alphabetically sorted hierarchical lists.
Species are delineated by a combination of criteria that include genetic differentiation in the Z chromosome measured by Fst (>0.20 usually corresponds to distinct species) and gene exchange Gmin (<0.05 for distinct species) (Cong et al. 2019a), COI barcode difference (typically >2% for distinct species) (Hebert et al. 2003) and its correlation with phenotypic differences (Lukhtanov et al. 2016), and the prominence of species-level clades (Zhang et al. 2022c). However, COI barcodes (together with mitochondria) can be transfer between species through occasional hybridization (Bachtrog et al. 2006; Cong et al. 2017), and some distinct species may possess highly similar or identical barcodes (Burns et al. 2008; Zhang et al. 2023a). See the “Species, subspecies, and genomics” section in Zhang et al. (2022a) for further discussion.
Following the logic outlined above, we are reclassifying two tribes in the subfamily Pyrginae Burmeister, 1878 based on the phylogeny inferred from whole genome shotgun datasets. The nuclear genomic tree we obtain is strongly supported, and the taxa we propose are confidently monophyletic. Moreover, their taxonomic ranks are consistent because they are tied to levels in the phylogenetic tree.
Materials and Methods
Methods employed in this study are the same as detailed in our previous publications (Li et al. 2019; Zhang et al. 2019a, 2022b; Cong et al. 2021). First, we obtain whole genome shotgun sequence datasets from butterfly specimens using our previously established experimental protocols (Li et al. 2019; Zhang et al. 2019a). Typically, a leg of a dry pinned specimen is used for DNA extraction. Our protocol is non-destructive, and legs are preserved. Specimens of any age are amenable to this protocol (Cong et al. 2021). We do not rely on the amplification of specific genes or segments, and every extracted DNA piece is sequenced. Therefore, the protocol succeeds with very old specimens, in which DNA may be fragmented into 30–50 bp segments. Second, these genomic datasets composed of 150 bp (or less) DNA segments are subjected to computational analysis to identify and assemble (i.e., stitch together) protein-coding regions using DIAMOND (Buchfink et al. 2015) and a reference set of all proteins encoded in a previously assembled reference genome of Cecropterus lyciades (Geyer, 1832) (Shen et al. 2017). This procedure results in a master-slave alignment of all these regions (i.e., coding regions in each specimen are aligned to the reference). These alignments (about 18 million positions) are too large for time-efficient phylogenetic analysis. They are randomly subsampled for 300,000 positions (by codon) that are used to infer phylogenetic trees as described previously (Zhang et al. 2022b). Third, we construct phylogenetic trees using IQ-tree v1.6.12 under the GTR+GAMMA model (Nguyen et al. 2015) from these randomly sampled positions in the nuclear genome (300 thousand positions) and estimate statistical significance by standard codon resampling from the original complete alignment (18 million positions). The trees are visualized, manipulated, and colored to prepare illustrations using FigTree (Rambaut 2018).
The current taxonomic classification was overlaid on the trees to find non-monophyletic taxa and clades corresponding to taxa without names. Taxa we define are monophyletic groups in nuclear genome trees that correspond to prominent clades. By “prominent,” we mean tree branches strongly supported statistically (typically by 100% of replicates) and usually longer than neighboring branches. The length of a branch is proportional to the number of base-pair substitutions along the branch. Not only are longer branches better supported statistically, but the larger number of genetic changes along them likely leads to more pronounced phenotypic changes that should be reflected in some morphological characters, not necessarily in adults, but could be in immature stages or other aspects of the phenotype. Nevertheless, due to highly non-linear relationships between the number of genetic changes and visually drastic phenotypic differences (Zhang et al. 2019a), there are short tree branches that correspond to visually recognizable taxa, and each case should be considered individually. It is unclear, however, if some drastic phenotypic change in adult appearance that was caused by a small number of genetic changes (maybe even a single inversion of a genomic segment) should be grounds for the erection of a separate taxon for this lineage because all other characters, e.g., those of caterpillars, would remain rather similar to the relatives of this lineage. Generally, we prefer to avoid monotypic (or nearly monotypic, i.e., consisting of very close relatives) higher taxa unless they cannot be confidently assigned to other taxa of the same rank or show prominent genetic differentiation from them at the level of the tree that corresponds to their rank. Furthermore, currently employed taxonomy is considered, and currently used names and their taxonomic ranks serve, on average, as a reference point to define levels in the trees and new taxa.
All taxa in this work are listed in their suggested phylogenetic order that agrees with phylogenetic trees augmented with the considerations of phenotypic similarity so that more similar species are closer to each other in the list (under the constraints of the trees). In addition to phenotypic diagnoses, we provide diagnostic DNA characters, both in the nuclear genome and the COI barcode. DNA characters are found in nuclear protein-coding regions using our previously developed procedure (see SI Appendix to Li et al. 2019). The logic behind the character selection was detailed in Cong et al. (2019b). The character states are provided in species diagnoses as abbreviations. E.g., aly728.44.1:G672C means position 672 in exon 1 of gene 44 from scaffold 728 of the Cecropterus lyciades (Geyer, 1832) (formerly in Achalarus Scudder, 1872, thus “aly”) reference genome (Shen et al. 2017) is C, changed from G in the ancestor. When characters are given for the sister clade of the diagnosed taxon, the following notation is used: aly5294.20.2:A548A (not C), which means that position 548 in exon 2 of gene 20 on scaffold 5294 is occupied by the ancestral base pair A, which was changed to C in the sister clade (so it is not C in the diagnosed taxon). The same notation is used for COI barcode characters but without a prefix ending with ‘:’. The sequences of exons from the reference genome with the positions used as character states highlighted in green are given in the supplemental file (Zhang et al. 2023d). Linking to the DNA sequences accessible from this publication ensures that DNA characters given in the diagnoses can be readily associated with actual sequences. General locality and collection year for specimens are given in the tree figures, and detailed data are provided in Table S1 of the supplemental file (Zhang et al. 2023d). COI barcode sequences have been deposited in GenBank with accessions OR665721–OR665734 and OR721875–OR721877. All new names have been registered with ZooBank, and registration numbers are provided for each.
The specimens were examined and sampled for sequencing in the following collections (abbreviations, which are not necessarily acronyms of the current names of these institutions, are given in parenthesis and used in Table S1 of the supplemental file (Zhang et al. 2023d)): American Museum of Natural History, New York, NY, USA (AMNH), Natural History Museum, London, UK (BMNH), Carnegie Museum of Natural History, Pittsburgh, PA, USA (CMNH), Colorado State University Collection, Fort Collins, CO, USA (CSUC), Los Angeles County Museum of Natural History, Los Angeles, CA, USA (LACM), Museum of Comparative Zoology, Harvard University, Cambridge, MA, USA (MCZ), Mississippi Entomological Museum, Starkville, MS, USA (MEM), Museum für Naturkunde, Berlin, Germany (MFNB), McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA (MGCL), Muséum National d’Histoire Naturelle, Paris, France (MNHP), Museo de Historia Natural, Lima, Peru (MUSM), Texas A&M University Insect Collection, College Station, TX, USA (TAMU), Biodiversity Center, University of Texas at Austin, Austin, TX, USA (TMMC), Bohart Museum of Entomology, University of California, Davis, CA, USA (UCDC), National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM), University of Texas Southwestern, freezers of the Grishin lab, Dallas, TX, USA (UTSW), Zoologische Staatssammlung München, Germany (ZSMC), and research collections of Ernst Brockmann, Germany (EBrockmann), Matthew J. W. Cock, UK (MJWCock), Bill Dempwolf, USA (WRDempwolf), Bernard Hermier, French Guiana (BHermier), and the late James A. Scott, USA (JAScott).
Results and Discussion
Tribe Achlyodini Burmeister, 1878, subtribe Pythonidina Grishin, 2019
Genomic analysis of the subtribe Pythonidina Grishin, 2019 reveals a large number of inconsistencies within the current classification (Fig. 1). Out of traditionally used genera, only Gindanes Godman and Salvin, 1895 (type species Gindanes panaetius Godman and Salvin, 1895) (Fig. 1 cyan) was monophyletic (but also included a species from a different genus), while species of other genera, such as Pythonides Hübner, 1819 (type species Papilio jovianus Stoll, 1782) (Fig. 1 red), Ouleus Lindsey, 1925 (type species Achlyodes fridericus Geyer, 1832) (Fig. 1 blue), Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) (Fig. 1 green), and Zera Evans, 1953 (type species Achlyodes zera Butler, 1870) (Fig. 1 magenta) were scattered among several clades. The tree reveals two major levels (genus and subgenus) in the clade containing all these species. We observe five prominent clades at the genus level (Fig. 1) that represent genera Livida Grishin, 2019 (type species Pythonides assecla Mabille, 1883), Pythonides, Gindanes, Quadrus, and the unnamed genus described below. To restore the monophyly of all taxa and to provide an internally consistent classification of this subtribe, we propose taxonomic changes detailed below.
Figure 1.
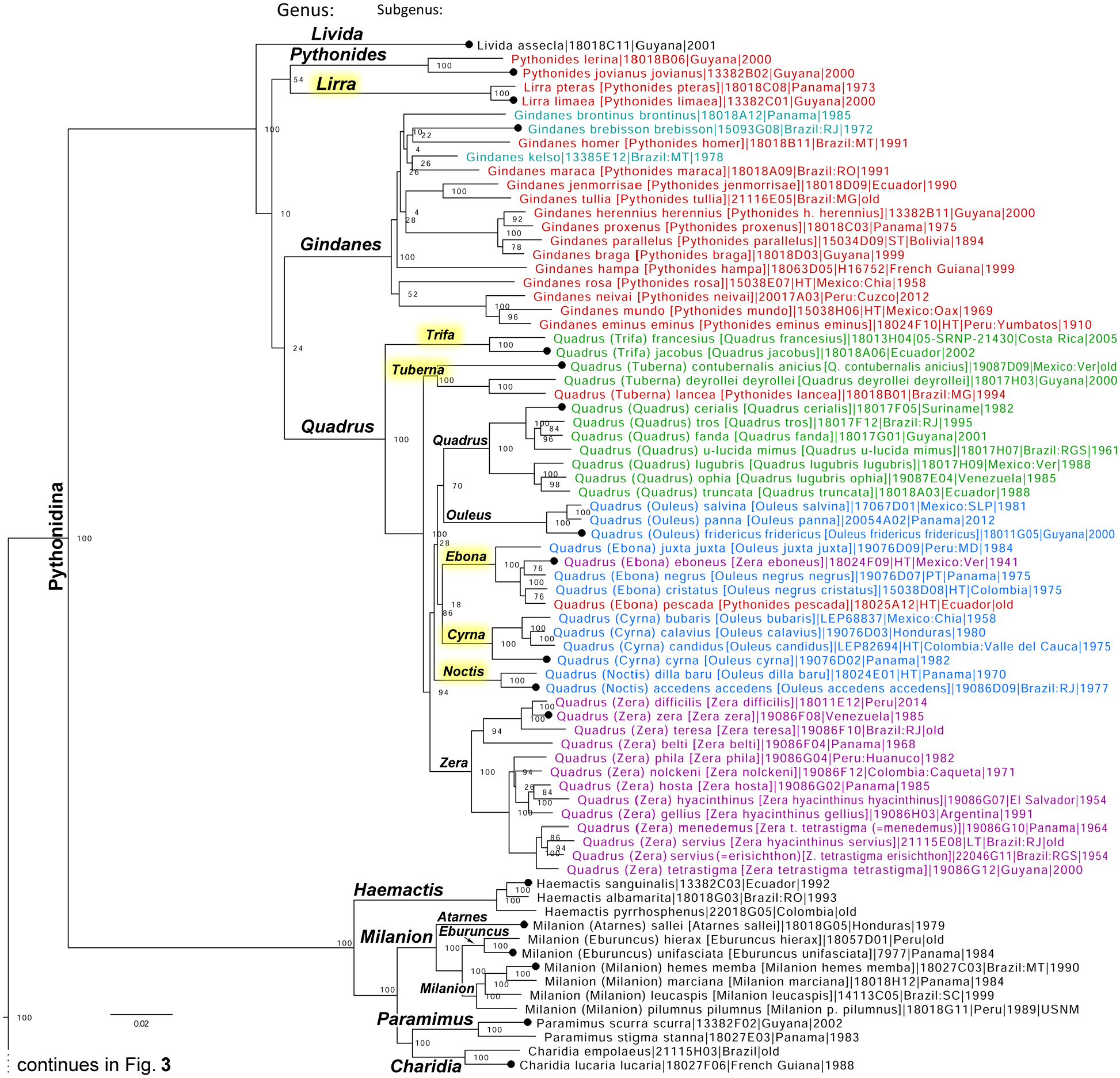
The nuclear genome-based phylogeny of Achlyodini (continued in Fig. 3). Only the segment of the tree corresponding to the subtribe Pythonidina is shown. The bottom segment of the tree (subtribe Achlyodinia) is replaced with four dots and is shown in Fig. 3. The tree is constructed from protein-coding regions in autosomes. The tree is rooted with Carcharodus alceae (NVG-18087F02, GenBank barcode OR665734), not displayed. Statistical support values are shown by nodes. For each specimen, its name adopted in this work is given first, and a previously used name is listed in square brackets (if different), supplemented with the DNA sample number, type status (HT holotype, LT lectotype, ST syntype, T type, PT paratype, and PLT paralectotype), general locality and collection year (“old” means collection year known, but likely before 1950, mostly around the turn of 20th century). See Table S1 in the Supplemental file (Zhang et al. 2023d) for additional data about these specimens. Synonyms are given in parentheses preceded by “=”. The type status refers to this synonym if the synonym name is provided. Names of genera (larger font, more prominent tree branches, on the left) and subgenera (smaller font, less prominent tree branches, on the right) are shown by the clades corresponding to these taxa. New genus-group names are highlighted in yellow. Branches leading to the type species (or their closest sequenced relatives) of valid genus-group names are marked with large black dots and refer to the subgenus (in parenthesis) given in the corresponding name, or the genus if the subgenus is not specified. The same notations are used in Fig. 3–6. Names of taxa that before this work were placed in genera Pythonides (red), Gindanes (cyan), Quadrus (green), Ouleus (blue), and Zera (purple) are shown in color to illustrate rampant violations of monophyly in these taxa, corrected here.
Ouleus Lindsey, 1925 and Zera Evans, 1953 are subgenera of Quadrus Lindsey, 1925
The genome-based phylogeny reveals that Ouleus Lindsey, 1925 (type species Achlyodes fridericus Geyer, 1832) and Zera Evans, 1953 (type species Achlyodes zera Butler, 1870) currently treated as genera, in addition to being polyphyletic are at a subgenus level (second from the root major level) (Fig. 1). Therefore, we propose that Ouleus Lindsey, 1925, new status, and Zera Evans, 1953, new status, are subgenera of Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) and restore their monophyly as detailed below. We note that Ouleus and Quadrus were proposed in the same work issued on the same date, and we give precedence to the name Quadrus acting as First Revisers (ICZN Code Art. 24.2.2.) (ICZN 1999).
Taxonomic rearrangement of Pythonides Hübner, 1819
Inspection of the genome-based phylogeny reveals that Pythonides Hübner, 1819 (type species Papilio jovianus Stoll, 1782) as currently circumscribed (Fig. 1 red) is not monophyletic with its species dispersed among three genera: Gindanes Godman and Salvin, 1895 (type species Gindanes panaetius Godman and Salvin, 1895), Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782), and a new genus described below. Clades corresponding to these genera receive 100% statistical support, and to restore the monophyly of Pythonides, we transfer all of its species, but two that remain in Pythonides (P. lerina (Hewitson, 1868) and P. jovianus (Stoll, 1782)), in these other genera to form the following new combinations: Gindanes homer (Evans, 1953), Gindanes nides (O. Mielke and Casagrande, 2002), Gindanes maraca (O. Mielke and Casagrande, 1992), Gindanes jenmorrisae (Shuey and Ramírez. 2022), Gindanes tullia (Evans, 1953), Gindanes herennius (Geyer, [1838]), Gindanes proxenus (Godman and Salvin, 1895), Gindanes parallelus (Mabille, 1898), Gindanes braga (Evans, 1953), Gindanes hampa (Evans, 1953), Gindanes rosa (Steinhauser, 1989), Gindanes neivai (Hayward, 1940), Gindanes mundo (H. Freeman, 1979), Gindanes eminus (E. Bell, 1934), Quadrus lancea (Hewitson, 1868), Quadrus pescada (E. Bell, 1956), and the remaining two in the new genus described below.
Lirra Grishin, new genus
http://zoobank.org/C0278D7F-5AF8-4C68-8306-35A67EA4AC52
Type species. Leucochitonea limaea Hewitson, 1868.
Definition. Currently in Pythonides Hübner, 1819 (type species Papilio jovianus Stoll, 1782), two closely related sister species Ate pteras Godman and Salvin, 1895 (type locality Panama: Chiriqui) and Leucochitonea limaea Hewitson, 1868 (type locality in French Guiana) are genetically distant from it and statistical support for their monophyly with species included here in Pythonides is not strong in the genomic tree (54%) (Fig. 1). The clade formed by these two species corresponds to a genus-level taxonomic category based on its position in the phylogeny and comparison to established genera-level clades. Therefore, we propose that this clade corresponds to a new genus that keys to E.41.11 in Evans (1953) and is distinguished from its relatives by the following characters: hind tibiae with one upper spur and with a tuft in males, forewing without hyaline spots, dorsal hindwing broadly blue at the margin, ventral hindwing without black overscaling of veins; uncus undivided, semi-triangular, gradually narrowing towards its end, aedeagus straight, not downturned or upturned, harpe approximately the same length as valva, narrowing distad, upturned and ending in a broad tooth pointing dorsad, costa slightly convex, ampulla with long process over harpe reaching its half. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly531.8.1:T654C, aly536.138.5:A1345C, aly1603.75.1:C49G, aly74.2.4:A627G, aly1656.25.1:A333G.
Etymology. The name is a feminine noun in the nominative singular formed as a modified fusion of species names: Li[maea]+[pte]{r}ra[s].
Species included. Ate pteras Godman and Salvin, 1895 and Leucochitonea limaea Hewitson, 1868.
Parent taxon. Subtribe Pythonidina Grishin, 2019.
Trifa Grishin, new subgenus
http://zoobank.org/80AA9B97-38A1-4668-9A65-1B2D621F6DEA
Type species. Tagiades jacobus Plötz, 1884.
Definition. The newly expanded genus Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) can be divided into eight clades at the subgenus level in the tree (Fig. 1). In addition to the nominotypical subgenus, two of these clades correspond to traditional subgenera, previously treated as genera: Ouleus Lindsey, 1925 (type species Achlyodes fridericus Geyer, 1832) and Zera Evans, 1953 (type species Achlyodes zera Butler, 1870), and five clades represent new subgenera. One of these new subgenera forms a clade sister to the rest and is stronger differentiated from them genetically. It keys to E.39.1 (in part, as a synonym of cerealis [sic], a synonymy probably based on a misidentification) or E.13.2 in Evans (1953) and is distinguished from its relatives by the following characters: dorsal hindwing with dark-brown base and two bands (postdiscal and submarginal), ventral hindwing mostly blue except submarginal brown area, forewing with one or two dash-like (not collected into a “C”) hyaline spots in discal cell; valva with a process from mid-costa, not from the ampulla area; harpe longer than valva, with strongly convex distal margin, narrowing to a point. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly3312.4.6:C55A, aly84.97.14:C160T, aly84.97.14:G171T, aly276558.35.2:T64A, aly276558.35.2:C65G.
Etymology. The name is a feminine noun in the nominative singular, inspired by the three dark bands on dorsal hindwings of these species: Tri-fa[scia].
Species included. Pythonides ineptus Draudt, 1922, Quadrus francesius Freeman, 1969, and Tagiades jacobus Plötz, 1884.
Parent taxon. Genus Quadrus Lindsey, 1925.
Tuberna Grishin, new subgenus
http://zoobank.org/7C63B146-7D14-42AC-B8EC-071C103D39D4
Type species. Pythonides contubernalis Mabille, 1883.
Definition. The newly expanded genus Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) can be divided into eight clades at the subgenus level in the tree (Fig. 1). In addition to the nominotypical subgenus, two of these clades correspond to traditional subgenera, previously treated as genera: Ouleus Lindsey, 1925 (type species Achlyodes fridericus Geyer, 1832) and Zera Evans, 1953 (type species Achlyodes zera Butler, 1870), and five clades represent new subgenera. One of these new subgenera is sister to all others except Trifa new subgenus and keys to or E.39.4a or E.41.1 in Evans (1953) and is distinguished from its relatives by the following characters: dorsal hindwing either with 2 bright shiny blue bands, or primarily blue with dark veins and central white streak; uncus undivided, stem-shaped, rounded at its end, with parallel sides in dorsal view, ampulla with projection (or a hump), harpe longer than valva, either with concave dorsal margin or rounded, nearly oval, merged with valva and ampulla with a small hump. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly2116.6.1:T200C, aly2116.6.1:C326T, aly1475.25.1:A1840C, aly1475.25.1:T1200A, aly1838.49.3:G435T.
Etymology. The name is a feminine noun in the nominative singular, formed from the type species name: [con] Tuberna[lis].
Species included. Pythonides contubernalis Mabille, 1883, Pythonides deyrollei Mabille, 1877, and Leucochitonea lancea Hewitson, 1868.
Parent taxon. Genus Quadrus Lindsey, 1925.
Ebona Grishin, new subgenus
http://zoobank.org/8A1637CD-1681-42BC-AE8A-04FF28148C42
Type species. Quadrus eboneus E. Bell, 1947.
Definition. The newly expanded genus Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) can be divided into eight clades at the subgenus level in the tree (Fig. 1). In addition to the nominotypical subgenus, two of these clades correspond to traditional subgenera, previously treated as genera: Ouleus Lindsey, 1925 (type species Achlyodes fridericus Geyer, 1832) and Zera Evans, 1953 (type species Achlyodes zera Butler, 1870), and five clades represent new subgenera. One of these new subgenera brings together species dispersed among three genera (Ouleus, Zera, and Pythonides). It keys to E.37.3a in Evans (1953) and is distinguished from its relatives by mostly dark wings without blue, hind-tibiae with one pair of spurs, tapered to a point uncus tip, and stout ampulla process. In DNA, a combination of the following base nuclear genomic pairs is diagnostic: aly770.31.1:C235A, aly770.31.1:A588G, aly6311.5.2:A42G, aly6311.5.2:A60G, aly1370.7.2:T139G.
Etymology. The name is a feminine noun in the nominative singular, given for the dark appearance of these species, with “o” to derive from the type species name and to avoid homonymy with Ebena Schumacher, 1817 (in Mollusca); from the genus Ebenus, commonly known as ebony plants. Ebony wood is known for its dark, brown, and often nearly black colors.
Species included. Pythonides juxta Bell, 1934, Quadrus eboneus E. Bell, 1947, Ouleus negrus Nicolay, 1980, Ouleus negrus cristatus Steinhauser, 1989 (see below), and Pythonides pescada Bell, 1956.
Parent taxon. Genus Quadrus Lindsey, 1925.
Quadrus (Ebona) cristatus (Steinhauser, 1989) is a species distinct from Quadrus (Ebona) negrus (Nicolay, 1980)
The genomic tree reveals that the holotype of Ouleus negrus cristatus Steinhauser, 1989 (type locality Colombia: Valle del Cauca, Rio Anchicayá, sequenced as NVG-15038D08, GenBank barcode OR665721) and a topotypical paratype of Ouleus negrus Nicolay, 1980 (type locality Panama: Veraguas Prov., Santa Fe, sequenced as NVG-19076D07, GenBank barcode OR665722) are not monophyletic and belong to different, albeit closely related, clades in the subgenus Ebona new subgenus of Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) (Fig. 1). Furthermore, they are genetically differentiated from each other, e.g., COI barcode difference of 4.3% (28 bp). Therefore, we propose that Quadrus (Ebona) cristatus (Steinhauser, 1989), new status, is a species distinct from Quadrus (Ebona) negrus (Nicolay, 1980). We note that Q. cristatus is closely related to (conspecific with?) Pythonides pescada E. Bell, 1956 (type locality in Ecuador: Río Pescado) (Fig. 1) and is phenotypically similar to it in having a distal part of ventral hindwing white. However, O. negrus is closely related to (but distinct from!) Zera eboneus E. Bell, 1947 (type locality in Mexico: Veracruz) and is phenotypically similar to it in largely uniform dark-brown, unspotted appearance on both sides of wings. These close relationships were overlooked, likely because species in the two pairs were incorrectly placed in different genera before (Ouleus and Pythonides, and Ouleus and Zera). If they were classified correctly, each pair’s more recently proposed species might have never been described due to phenotypic similarities with its previously named counterpart. Analysis of additional specimens, including their genomic sequencing, may lead to conclusion that Q. pescada and Q. cristatus are conspecific because they are more similar to each other genetically (0.3%, 2 bp COI barcode difference) than O. eboneus to O. negrus, the latter pair being sufficiently different from each other for us to be confident about their specie-level status (COI barcode difference of 2.6%, 17 bp).
Noctis Grishin, new subgenus
http://zoobank.org/628D6182-4F0F-4953-83CB-7781E5AFBC99
Type species. Achlyodes accedens Mabille, 1895.
Definition. The newly expanded genus Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) can be divided into eight clades at the subgenus level in the tree (Fig. 1). In addition to the nominotypical subgenus, two of these clades correspond to traditional subgenera, previously treated as genera: Ouleus Lindsey, 1925 (type species Achlyodes fridericus Geyer, 1832) and Zera Evans, 1953 (type species Achlyodes zera Butler, 1870), and five clades represent new subgenera. One of these new subgenera keys to E.37.6a in Evans (1953) and is distinguished from its relatives by mostly dark wings without blue, uncus tip with a small stalked triangle, and slender ampulla process. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly1139.36.2:G59A, aly1838.6.2:A63G, aly529.46.1:A354G, aly451.31.2:T72A, aly2487.16.4:A87G.
Etymology. The name is a masculine noun in the nominative singular, given for the black appearance of these species: noctis is Latin for “of the night” or “nighttime.”
Species included. Ouleus dilla Evans, 1953 and Achlyodes accedens Mabille, 1895.
Parent taxon. Genus Quadrus Lindsey, 1925.
Cyrna Grishin, new subgenus
http://zoobank.org/1E1041B3-4CDC-4C7C-ACF3-522D4ED3233A
Type species. Achlyodes cyrna Mabille, 1895.
Definition. The newly expanded genus Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) can be divided into eight clades at the subgenus level in the tree (Fig. 1). In addition to the nominotypical subgenus, two of these clades correspond to traditional subgenera, previously treated as genera: Ouleus Lindsey, 1925 (type species Achlyodes fridericus Geyer, 1832) and Zera Evans, 1953 (type species Achlyodes zera Butler, 1870), and five clades represent new subgenera. One of these new subgenera keys to E.37.1c in Evans (1953) and is distinguished from its relatives by mostly dark wings without blue, no tibial tuft in males, hind-tibiae with two pairs of spurs, and uncus tip tapered to a point. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly84.28.1:T2136A, aly84.28.1:G2577A, aly203.14.2:T24C, aly203.14.2:T54C, aly1656.40.1:A1276G.
Etymology. The name is a feminine noun in the nominative singular, tautonymous with the type species name.
Species included. Achlyodes bubaris Godman and Salvin, 1895, Achlyodes calavius Godman and Salvin, 1895, Ouleus candidus Steinhauser, 1989, Bolla zora Evans, 1953 (see below), and Achlyodes cyrna Mabille, 1895.
Parent taxon. Genus Quadrus Lindsey, 1925.
Quadrus (Cyrna) zora (Evans, 1953), new combination
Inspection of the holotype of Bolla zora Evans, 1953 (type locality in Ecuador), which is a female, reveals that it does not belong to Bolla Mabille, 1903 (type species Bolla pullata Mabille, 1903, treated as a junior subjective synonym of Staphylus imbras Godman and Salvin, 1896), but instead is a species of Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) similar to Quadrus (Cyrna) calavius Godman and Salvin, 1895. On the dark-brown forewing, it possesses three subapical white dots in an arc, the middle dot smallest (the shape, size, and location of these dots suggest Quadrus), and two faint bands from costa to inner margin, postdiscal and subapical. Therefore, we propose a new combination Quadrus (Cyrna) zora (Evans, 1953).
Quadrus (Quadrus) ophia (A. Butler, 1870) is a species distinct from Quadrus (Quadrus) lugubris (R. Felder, 1869)
The genomic tree reveals that Achlyodes ophia Butler, 1870 (type locality in Venezuela), currently treated as a subspecies of Quadrus lugubris (R. Felder, 1869) (type locality in Mexico: Veracruz), is not monophyletic with it and instead is sister to Quadrus truncata (Hewitson, 1870) (type locality in Ecuador) (Fig. 1). Moreover, genetic differentiation between Q. lugubris ophia and Q. lugubris lugubris is at the level characteristic of distinct species, e.g., their COI barcodes differ by 3.5% (23 bp). Therefore, we propose that Quadrus (Quadrus) ophia (A. Butler, 1870), restored status, is a species distinct from Quadrus (Quadrus) lugubris (R. Felder, 1869).
Lectotype designation for Achlyodes servius Plötz, 1884
Achlyodes servius Plötz, 1884 (type locality in Brazil), treated above as a valid species in the subgenus Zera Evans, 1953 (type species Achlyodes zera Butler, 1870) of the genus Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782), was described from an unstated number of specimens and the name “servius” was attributed to a letter or a manuscript by Herrich-Schäffer (Plötz 1884). A detailed search in the MFNB collection yielded two specimens we believe are syntypes of A. servius, sequenced as NVG-21115E08 (Fig. 2b) and NVG-21115E10. The first one, a male, agrees with the original description and bears an identification label crediting Herrich-Schäffer for the manuscript name “sergius” (similar enough to “servius” to assume the difference resulting from some trivial error), as mentioned by Plötz in the original description. The second specimen, a female, bears a label in Plötz’s handwriting, “Brasilia / Nov Frib.” but has two hyaline spots by the forewing apex, not three as mentioned in the original description (it was probably not the specimen drawn, and Plötz wrote descriptions from his drawings). The two specimens were near each other in the same drawer, were collected in the Brazilian state of Rio de Janeiro, and are conspecific judging from our genomic sequencing results (Fig. 2a). Because one of these specimens was labeled by Plötz himself, and the other one bears a label attributing its name to the manuscript by Herrich-Schäffer (as in the original description of A. servius) and also bears a specimen number label (5908) identifying it as a part of Hesperiidae collection inspected (and at times references in publications using these numbers) by Plötz, these specimens are syntypes. To stabilize nomenclature, N.V.G. hereby designates the syntype in the MFNB collection, a male shown in Fig. 2b, bearing three labels (2nd green, others white): [5908], [Sergius | HSch. ms | Rio v. Lgsdf], [DNA sample ID: | NVG-21115E08 | c/o Nick V. Grishin], as the lectotype of Achlyodes servius Plötz, 1884. The lectotype was missing an abdomen when inspected. According to its large green label in the handwriting of Carl Heinrich Hopffer (1810–1876), who was the curator of the collection in Berlin until his death, this specimen was collected in Brazil: Rio de Janeiro by Georg Heinrich von Langsdorff (1774–1852) and identified by the name “sergius” in an unpublished manuscript by Herrich-Schäffer. The number on the first label, “5908”, is the entry in the collection Catalog by Hopffer. In addition to confirming the information repeated on the green label, the Catalog reveals that there were three specimens with this number. All these details increase our confidence that the lectotype was indeed a syntype. The COI barcode sequence of A. servius lectotype, sample NVG-21115E08, GenBank OR665723, 658 base pairs, is: AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACTTCATTAAGTTTATTAATTCGAACTGAATTAGGAAATCCTGGATCCTTAATTGGAGATGATCAAATTTATAATACTATTGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGGAATTGACTAGTACCCCTTATACTAGGAGCCCCTGATATAGCTTTTCCTCGAATAAATAATATAAGTTTTTGATTATTACCTCCCTCTTTAATATTATTAATTTCAAGTAGTGTTGTAGAAAATGGAGCCGGTACAGGTTGAACCGTATACCCTCCTCTTTCTGCTAATATTGCTCATCAAGGCTCATCAGTAGATTTAGCAATCTTTTCTCTTCATTTAGCAGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAATAATCTTTCCTTAGATCAAATACCCCTATTTGTTTGATCTGTTGGAATTACAGCATTACTTTTACTATTATCTTTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACTGACCGAAATTTAAATACATCATTTTTTGATCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Figure 2.

Quadrus (Zera) nuclear genome-based phylogeny and type specimens. a) The nuclear genome-based phylogeny of selected species: Q. vivax sp. n. (red), Q. hyacinthinus (purple), Q. hosta (cyan), Q. servius stat. rest. (blue and magenta, with its synonym Achlyodes erisichthon labeled in blue) rooted with the remaining Zera species (not shown; see Fig. 1). Specimens shown below are highlighted in yellow; see Fig. 1 for other notations. b) Lectotype of Quadrus (Zera) servius designated herein, with its labels shown above on the right. c) Holotype of Quadrus (Zera) vivax sp. n. Specimens are in dorsal (left) and ventral (right) views, data in text. The scale bar at the bottom refers to specimens; the scale bar at the top refers to labels reduced by 1/3 compared to specimens.
Quadrus (Zera) gellius (Mabille, 1903) and Quadrus (Zera) servius (Plötz, 1884) are species distinct from Quadrus (Zera) hyacinthinus (Mabille, 1877)
Genomic sequencing of the lectotype of Achlyodes servius Plötz, 1884 (type locality in Brazil, sequenced as NVG-21115E08) from “Rio” in MFNB currently treated as a subspecies of Quadrus (Zera) hyacinthinus (Mabille, 1877) (type locality not specified, likely in Central America) reveals that it is in a clade different from it and instead is closer to Quadrus (Zera) tetrastigma (Sepp, [1847]) (type locality in Suriname) while being genetically differentiated from the latter as well (Fig. 1, 2a), e.g., COI barcode difference of 0.9% (6 bp) while showing consistent separation in the Z chromosome with Fst/Gmin of 0.51/0.002. Furthermore, Pythonides gellius Mabille, 1903 (type locality in “Ecuador”, likely in SE South America), currently another subspecies of Q. hyacinthinus, while being closer related to it than A. servius, is also non-monophyletic with it and shows notable genetic differentiation from it (Fig. 1, 2a), e.g., COI barcode difference of 3.8% (25 bp). Therefore, we propose that Quadrus (Zera) gellius (Mabille, 1903), restored status, and Quadrus (Zera) servius (Plötz, 1884), restored status, are species distinct from Quadrus (Zera) hyacinthinus (Mabille, 1877) and from Quadrus (Zera) tetrastigma (Sepp, [1847]).
Quadrus (Zera) vivax Grishin, new species
http://zoobank.org/6B5F54D9-0706-4CDC-9811-05E1A9414155
Definition and diagnosis. Evans misidentified (at least some) Achlyodes servius Plötz, 1884 (type locality in Brazil), which he treated as a subspecies of Zera hyacinthinus (Mabille, 1877) (type locality not specified, likely in Central America). Above, we placed Zera Evans, 1953 (type species Achlyodes zera Butler, 1870) in the genus Quadrus Lindsey, 1925 (type species Papilio cerialis Stoll, 1782) as a subgenus. Also, as shown above, A. servius is not closely related to Q. hyacinthinus but instead is a distinct species closer to Quadrus (Zera) tetrastigma (Sepp, [1847]) (type locality in Suriname). However, the species that keys to Evans’ “Zera hyacinthinus servius” and indeed is closer related to Q. hyacinthinus does not have a name and is described here. It is a species-level taxon, not a subspecies of Q. hyacinthinus, due to genetic differentiation (Fig. 2a), e.g., their COI barcodes differ by 2% (13 bp) in the presence of consistent phenotypic differences described below. Furthermore, while this species is in the same clade as Q. hyacinthinus, it is sister to Quadrus (Zera) hosta (Evans, 1953) (type locality in Costa Rica), differing from it by 3.8% (25 bp) in the COI barcode. This new species keys to E.38.5b in Evans (1953) and is distinguished from its relatives by the following combination of characters: dorsal forewing without a hyaline spot in cell CuA1-CuA2, dark central area extending neither to the wing base nor the end of discal cell, wing base violaceous, dark area in discal cell more extensive than in relatives and without prominent, continuous pale bar crossing it, but with one or two small, pale, sometimes narrowly connected spots; ventral forewing with a conspicuous tawny spot by the apex and tornal area broadly tawny, could be extending to and reaching costal margin, mostly unmarked, but sometimes with submarginal dark spot(s); dark discal band absent or vestigial on dorsal hindwing; tornal half of ventral hindwing bluish-white, contrasting with the tawny area and not gradually merging into it as in Q. servius.
Barcode sequence of the holotype. Sample NVG-19086H02, GenBank OR721875, 658 base pairs: AACCTTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACTTCTTTAAGTTTGTTAATTCGAACTGAATTAGGAAATCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATCGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCAATTATAATTGGAGGATTTGGAAATTGACTAGTACCCCTTATACTAGGAGCACCTGATATAGCTTTCCCCCGAATAAATAATATAAGTTTTTGGTTATTACCCCCTTCTTTAATATTATTAATCTCAAGTAGTATTGTAGAAAATGGAGCTGGTACAGGTTGAACAGTTTACCCACCTCTTTCAGCTAATATTGCCCATCAAGGATCATCTGTAGATTTAGCAATTTTTTCTCTTCATTTAGCAGGAATTTCTTCAATTCTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAGTCAATAATCTTTCCTTAGATCAAATACCCCTTTTTGTTTGATCCGTAGGAATTACAGCATTACTTTTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACTATACTATTAACTGATCGAAATTTAAATACATCATTTTTTGATCCTGCTGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material. Holotype: ♂ currently deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA [USNM], illustrated in Fig. 2c, bears four printed (date handwritten) labels: three white [BRAZIL: RJ | Petropolis 900m | 6 Aug.’78 | S. S. Nicolay], [DNA sample ID: | NVG-19086H02 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01588802] and one red [HOLOTYPE ♂ | Quadrus (Zera) | vivax Grishin]. Paratypes: 1♂ 1♀ from Brazil: 1♂ Rio de Janeiro, Teresópolis, elevation 1000 m, GPS −22.4500, −42.9833, 17-Feb-1995, Robbins and Caldas leg. (NVG-19086H09, USNMENT 01588809) [USNM]; 1♀ Santa Catarina, Sao Bento do Sul, Feb-1984, Rank leg., from D. and J. Jenkins collection (NVG-15093C04) [MGCL].
Type locality. Brazil: Rio de Janeiro, Petrópolis.
Etymology. In Latin, vivax means lively or vibrant, given for the contrasty and more vivid appearance of this species compared to its relatives: ventral side of wings with more conspicuous tawny patches and spots and brighter, whiter, better defined pale area on the hindwing. The name is an adjective.
Distribution. Southeast and South Brazil.
Achlyodes erisichthon Plötz, 1884 is a junior subjective synonym of Quadrus (Zera) servius (Plötz, 1884), not a subspecies of Quadrus (Zera) tetrastigma (Sepp, [1847])
Genomic comparison of the lectotype of Achlyodes servius Plötz, 1884 (type locality in Brazil, sequenced as NVG-21115E08) from “Rio” in MFNB (white ventral hindwing by inner margin) with specimens identified by us as Quadrus (Zera) tetrastigma erisichthon (Plötz, 1884) (type locality not stated, syntypes not located, specimens from SE Brazil with ventral hindwing largely pale-orange, yellower by inner margin but not white) reveals the lack of genetic differentiation between them (Fig. 1, 2a), i.e., their COI barcodes are identical. Therefore, these taxa are different color morphs of the same species, and we propose that Achlyodes erisichthon Plötz, 1884 is a junior subjective new synonym of Quadrus (Zera) servius (Plötz, 1884), not a subspecies of Quadrus (Zera) tetrastigma (Sepp, [1847]). We note that A. servius and A. erisichthon were proposed in the same work (on the same page) issued on the same date, and we give precedence to the name A. servius acting as First Revisers (ICZN Code Art. 24.2.2.).
Furthermore, in the absence of extant type specimens, the identity of A. erisichthon remains somewhat uncertain, and we use this name for the taxon identified as such by Evans (1953). Judging from the original description (Plötz 1884) and copies (in BMNH and USNM) of the unpublished and lost illustration t[afel]. 982 by Plötz (Godman 1907), we believe that Evans identified this species correctly. Moreover, due to the extensive tawny coloration of the ventral forewing, prominent forewing subapical spots, and other wing pattern details as illustrated in the drawings, A. erisichthon is not conspecific with Q. tetrastigma, a conclusion that would hold even if Evans misidentified A. erisichthon. However, two characters of the A. erisichthon drawing reveal an unusual specimen, i.e., four—instead of the usual three—forewing subapical spots (also mentioned in the original description) and the absence of a dark cross-bar in the forewing discal cell, which is mostly violaceous—instead of dark-brown in the middle—cast certain doubt on the identification of this taxon and even its attribution to Quadrus (Zera).
Nevertheless, out of all Hesperiidae species known to us worldwide (the type locality of A. erisichthon was not stated), Quadrus (Zera) is the best match to the drawing due to this violaceous coloration, s-shaped placement of the forewing subapical spots, and other details of wing pattern, in particular, on the ventral side. While we have seen Quadrus (Zera) specimens with a vestigial 4th subapical forewing spot, none was suitable for the neotype of A. erisichthon because these specimens did not agree with the drawing and the original description in other characters. In its largely tawny hindwing, A. erisichthon is also similar to Quadrus (Zera) gellius (Mabille, 1903) (type locality in “Ecuador”, likely in SE South America). However, the hindwing is more uniformly tawny in the latter species, but the original description of A. erisichthon specifically mentions greenish-gray overscaling by the inner margin (Plötz 1884). This overscaling is typical of Q. servius form lacking extensive white area on the ventral hindwing and is the manifestation of this white overscaling, but much reduced.
Finally, Plötz studied Q. servius specimens from SE Brazil (at least four, see above), proposing this name for the white-overscaled form. It is likely that he also encountered the tawny hindwing form from the same area and, additionally noticing four (not three) subapical spots on an unusual specimen, considered it to be a closely related but distinct species he called A. erisichthon, which he placed in the key next to Q. servius and, therefore, these two species were likely quite close to each other in appearance. We suggest the synonymy proposed above for all these reasons, and will be looking for a specimen suitable for A. erisichthon neotype designation, keeping in mind that it could be not yet re-discovered species different from all others currently known.
Quadrus (Zera) menedemus (Godman and Salvin, 1894) is a valid species, not a synonym of Quadrus (Zera) tetrastigma (Sepp, [1847])
The genomic tree reveals that Pythonides menedemus Godman and Salvin, 1894 (type locality in Panama: Chiriqui) currently treated as a junior subjective synonym of Quadrus (Zera) tetrastigma (Sepp, [1847]) (type locality in Suriname) is not monophyletic with it and instead is sister to Quadrus (Zera) servius (Plötz, 1884) (type locality in Brazil) (Fig. 1, 2a). Moreover, genetic differentiation between P. menedemus and Q. tetrastigma is at the level characteristic of distinct species, e.g., their COI barcodes differ by 1.8% (12 bp). Therefore, we propose that Quadrus (Zera) menedemus (Godman and Salvin, 1894), restored status, is a valid species, not a synonym of Quadrus (Zera) tetrastigma (Sepp, [1847]). We note that while the name Q. menedemus is defined by its extant syntypes, the identity of Q. tetrastigma could be questioned, because it is identified from the original description and illustrations only; its syntypes likely lost. However, this name has been stably applied to the populations in the Guianas, and we do not have a compelling reason to challenge this interretation. Currently, there is no exceptional need to define Q. tetrastigma by a neotype.
Atarnes Godman and Salvin, 1897 and Eburuncus Grishin, 2012 are subgenera of Milanion Godman and Salvin, 1895
Inspection of the genomic tree reveals close clustering of the three genera: Atarnes Godman and Salvin, 1897 (type species Leucochitonea sallei C. Felder and R. Felder, 1867), Eburuncus Grishin, 2012 (type species Leucochitonea unifasciata C. Felder and R. Felder, 1867), and Milanion Godman and Salvin, 1895 (type species Papilio hemes Cramer, 1777) that taken together are prominently distinct from their relatives (Fig. 1). The tree taxa share a number of phenotypical features and some species in them were misclassified before (Grishin 2012). Therefore, to simplify the taxonomic classification and reflect all these similarities, we propose to treat Atarnes Godman and Salvin, 1897, new status, and Eburuncus Grishin, 2012, new status, as subgenera of Milanion Godman and Salvin, 1895.
Tribe Achlyodini Burmeister, 1878, subtribe Achlyodina Burmeister, 1878
Eurypterus later Mabille, 1891 and Eurypterus haber Mabille, 1891 belong to the genus Eantis Boisduval, 1836, not Aethilla Hewitson, 1868
Eurypterus later Mabille, 1891 (type locality likely in Peru) and Eurypterus haber Mabille, 1891 (type locality probably in Peru) are currently in the genus Aethilla Hewitson, 1868 (type species Aethilla eleusinia Hewitson, 1868), because of their general appearance and visual similarities with Aethilla. However, the genomic tree reveals that they are not monophyletic with Aethilla, which is sister to Achlyodes Hübner, 1819 (type species Papilio busirus Cramer, 1779), but instead originate within Eantis Boisduval, 1836 (type species Urbanus thraso Hübner, 1807) (Fig. 3). Therefore, we transfer them from Aethilla to Eantis forming new combinations: Eantis later (Mabille, 1891) and Eantis haber (Mabille, 1891).
Figure 3.
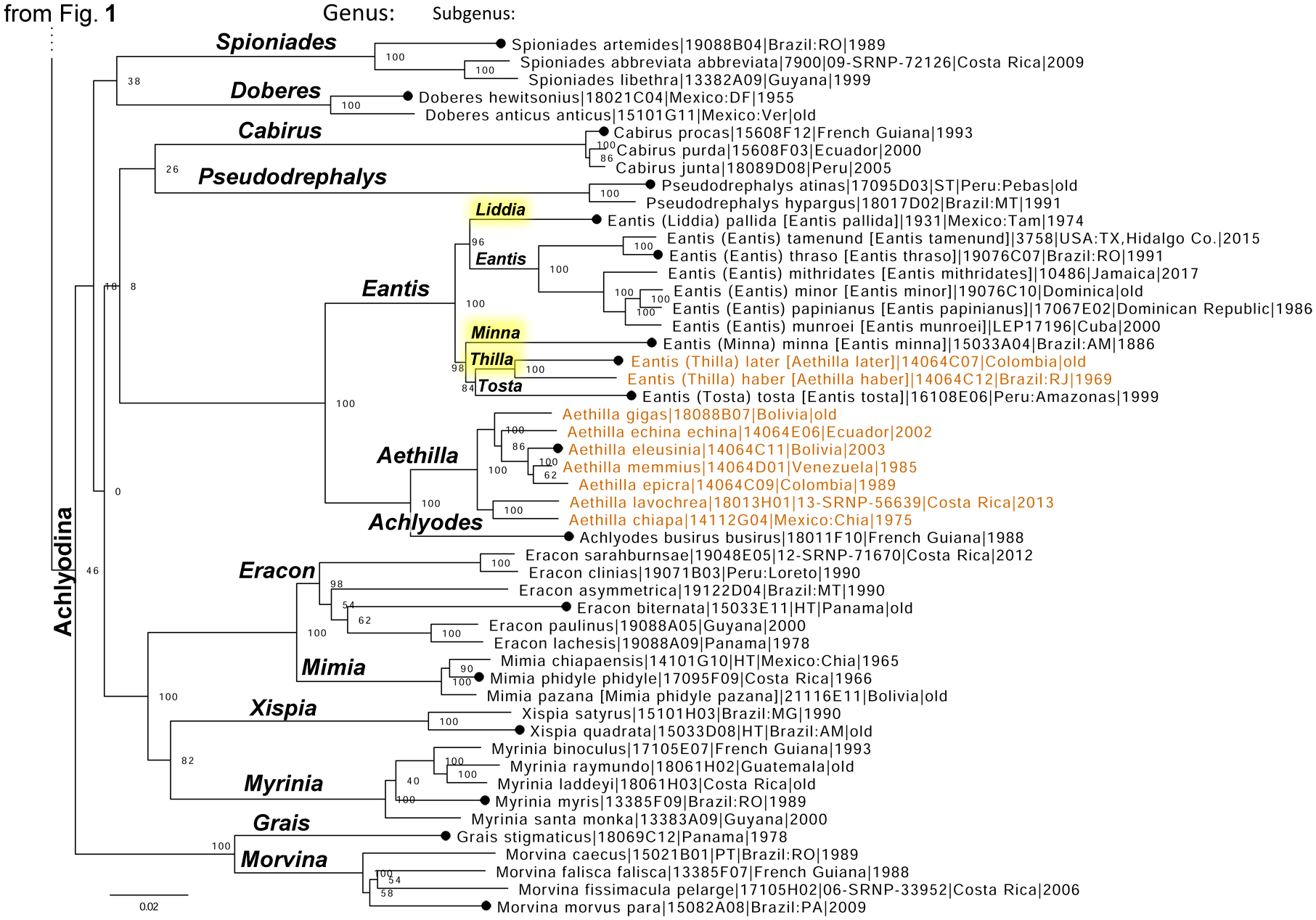
The nuclear genome-based phylogeny of Achlyodini (continued from Fig. 1). A segment corresponding to the subtribe Achlyodinia is shown. The top segment of the tree (subtribe Pythonidina) is replaced with four dots and is shown in Fig. 1. See Fig. 1 for notations. The names of taxa that before this work were placed in the genus Aethilla are shown in orange to illustrate the lack of monophyly.
Liddia Grishin, new subgenus
http://zoobank.org/BEF8C41C-DD78-426C-BB39-91BDA3210A75
Type species. Helias pallida R. Felder, 1869.
Definition. The genomic tree shows that the genus Eantis Boisduval, 1836 (type species Urbanus thraso Hübner, 1807) splits into five lineages at the subgenus level, and we regard these lineages as representing five subgenera (Fig. 3). Out of these subgenera, in addition to the nominotypical, only one has a name: Tosta Evans, 1953 (type species Tosta tosta Evans, 1953), while three others are new. One of these new subgenera keys to F.2.4 in Evans (1953) and is distinguished from its relatives by the following characters: forewing only slightly falcate, wings paler brown above; valvae asymmetric, left with a process from the ampulla, right without, harpe with convex dorsal margin without a process at its base by ampulla, aedeagus not strongly bent in the middle. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly1249.23.2:T82G, aly322.13.2:A64C, aly2612.12.4:A232C, aly2612.12.4:G284C, aly619.2.4:A36T.
Etymology. The name is a feminine noun in the nominative singular, formed from the type species name: [pal] Lid{di}a.
Species included. Only the type species.
Parent taxon. Genus Eantis Boisduval, 1836.
Minna Grishin, new subgenus
http://zoobank.org/BAE6E4C1-284A-401C-99CD-2F51DB4A382B
Type species. Achlyodes minna Evans, 1953.
Definition. The genomic tree shows that the genus Eantis Boisduval, 1836 (type species Urbanus thraso Hübner, 1807) splits into five lineages at the subgenus level, and we regard these lineages as representing five subgenera (Fig. 3). Out of these subgenera, in addition to the nominotypical, only one has a name: Tosta Evans, 1953 (type species Tosta tosta Evans, 1953), while three others are new. One of these new subgenera keys to F.2.3. in Evans (1953) and is distinguished from its relatives by the following characters: forewing only slightly falcate, wings dark-brown above; process from ampulla is longer and thinner than in others, and there is no process from the base of harpe near ampulla, aedeagus not strongly bent in the middle. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly4333.9.1:C37T, aly822.26.1:T192C, aly127.21.4:T198G, aly1313.7.1:G322T, aly481.7.1:A380C.
Etymology. The name is a feminine noun in the nominative singular, a tautonym of the type species name.
Species included. Only the type species.
Parent taxon. Genus Eantis Boisduval, 1836.
Thilla Grishin, new subgenus
http://zoobank.org/ADF9A522-9D1D-466B-8A62-65A40F9E70F2
Type species. Eurypterus later Mabille, 1891.
Definition. The genomic tree shows that the genus Eantis Boisduval, 1836 (type species Urbanus thraso Hübner, 1807) splits into five lineages at the subgenus level, and we regard these lineages as representing five subgenera (Fig. 3). Out of these subgenera, in addition to the nominotypical, only one has a name: Tosta Evans, 1953 (type species Tosta tosta Evans, 1953), while three others are new. One of these new subgenera consists of species previously placed in Aethilla Hewitson, 1868 (type species Aethilla eleusinia Hewitson, 1868) and keys to F.1.8 or F.1.4a (in part, as incorrect synonymy) in Evans (1953) and is distinguished from its relatives by the following characters: forewing more pointed or even slightly falcate at the apex, hindwing less produced or more rounded at the tornus; ampulla with a long terminally serrated process directed dorsoposterad and extending over harpe, harpe with process-like expansion at its base by ampulla. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly577.14.5:A210G, aly728.5.4:G78A, aly461.13.3:T129C, aly2012.59.3:A90G, aly2012.59.3:T100C.
Etymology. The name is a feminine noun in the nominative singular, formed from the previous genus name for these species: [Ae]Thilla.
Species included. Eurypterus later Mabille, 1891 and Eurypterus haber Mabille, 1891.
Parent taxon. Genus Eantis Boisduval, 1836.
Mimia pazana Evans, 1953 is a species distinct from Mimia phidyle (Godman and Salvin, 1894)
The genomic tree reveals that Mimia phidyle (Godman and Salvin, 1894) (type locality in Panama: Chiriqui) is paraphyletic with respect to Mimia chiapaensis Freeman, 1969 (type locality in Mexico: Chiapas), with Mimia phidyle pazana Evans,1953 (type locality in Bolivia) being sister to the clade of the former two taxa (Fig. 3). The two subspecies of M. phidyle are genetically differentiated at the level typical for distinct, and not even very closely related, species (e.g., their COI barcodes differ by 4.9%, 32 bp) and are phenotypically different (Evans 1953). Therefore, we propose that Mimia pazana Evans,1953, new status, is a species distinct from Mimia phidyle (Godman and Salvin, 1894).
Tribe Carcharodini Verity, 1940
Genomic analysis of the tribe Carcharodini Verity, 1940 reveals six prominent clades at the subtribe tree level (Fig. 4–6). Therefore, we partition the tribe into six subtribes corresponding to these clades. Only one of these subtribes, the nominotypical, has a name, and others are new. These five subtribes are described below.
Figure 4.

The nuclear genome-based phylogeny of Carcharodini, first segment (continues in Fig. 5, 6). See Fig. 1 for notations.
Figure 6.

The nuclear genome-based phylogeny of Carcharodini, the last segment (continues from Fig. 4 and 5) showing Carcharodina. Different genera are shown in different colors. See Fig. 1 for notations. The red asterisk on a tree branch denotes the arrival of Carcharodina in the Old World from the New World. See the discussion about Muschampia (a junior name) vs. Sloperia (should have priority) in the text.
Cyclosemiina Grishin, new subtribe
http://zoobank.org/C9F6823E-F97F-4C0B-8C9A-BF1E09CC49BE
Type genus. Cyclosemia Mabille, 1878.
Definition. This new subtribe corresponds to one of the six major clades in the tribe Carcharodini Verity, 1940 (Fig. 4–6). It keys to E.27 in Evans (1953) and is diagnosed by the following combination of characters: uncus undivided, gracile, valvae symmetrical, typically with a style; palpi with short 2nd and long 3rd segments, males without costal fold, dorsal forewing at the end of discal cell with an eyespot, usually bi-pupiled, and both wings with dark bands from costa to inner margin. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly1294.6.1:T204G, aly1264.5.2:G47C, aly1264.5.2:A130C, aly838.12.1:T458A, aly838.12.1:A427C..
Genera included. Only the type genus.
Parent taxon. Tribe Carcharodini Verity, 1940.
Ilianina Grishin, new subtribe
http://zoobank.org/AE693F6C-DD1E-4F25-9D8B-15E1AC97A09A
Type genus. Iliana E. Bell, 1937.
Definition. This new subtribe corresponds to one of the six major clades in the tribe Carcharodini Verity, 1940 (Fig. 4–6). It keys to E.16.3a, E.13.6a, or F.7.2, 3, or 4 in Evans (1953) and is diagnosed by the following combination of characters: both wings are semi-triangular in males; uncus frequently with flanges at its base (one on each side) either divided (arms are usually widely separated) or aedeagus unusually flared, trumpet-shaped and juxta large (longer than a third of aedeagus), or aedeagus very thin (narrower than uncus in lateral view); ampulla modified with lobes or processes, and/or expanded in many species, harpe either narrow and upturned, wrapping around distal margin of valva, or broad and protruding posteriad, diamond-shaped to nearly triangular with serrated dorsoposterior margin. Most reliably identified by DNA, and a combination of the following nuclear genomic base pairs is diagnostic: aly276665.25.1:G295A, aly594.12.19:T108A, aly594.12.19:C131T, aly3241.2.5:C191G, aly3241.2.5:C235G.
Genera included. Cornuphallus Austin, 1997, Iliana E. Bell, 1937, and Tiana Grishin, 2019.
Parent taxon. Tribe Carcharodini Verity, 1940.
Nisoniadina Grishin, new subtribe
http://zoobank.org/06559DF6-FAB5-41D4-B86D-397F658ED67F
Type genus. Nisoniades Hübner, [1819].
Definition. This new subtribe corresponds to one of the six major clades in the tribe Carcharodini Verity, 1940 (Fig. 4–6). Approximately corresponds to the Nisoniades sub-group of Evans (1953) (with several exceptions, but generally agrees with his description) and is diagnosed by the following combination of characters: antennae usually not shorter than half of costa, apiculus hooked and gracile, shorter than the rest of the club; forewing without lower median veinlet, hindwing inner margin produced and usually longer than forewing costal margin, dorsal hindwing in males frequently with a tuft of scales at the base of cell Sc+R1-RS; uncus undivided, aedeagus usually bent and wider at the bent, valvae asymmetrical, broad, oval to nearly circular, harpe narrow, frequently framing the valva or slightly protruding distad. Most reliably identified by DNA, and a combination of the following nuclear genomic base pairs is diagnostic: aly1259.30.2:T226A, aly127.55.5:G122C, aly9181.1.1:A400T, aly9181.1.1:G401C, aly2101.3.2:T247A.
Genera included. Arteurotia A. Butler and H. Druce, 1872, Conognathus C. Felder and R. Felder, 1862, Ocella Evans, 1953, Sophista Plötz, 1879 (includes Austinus O. Mielke and Casagrande, 2016 and Pachyneuria Mabille, 1888, see below), Polyctor Evans, 1953, Pellicia Herrich-Schäffer, 1870 (includes Mictris Evans, 1955, see below), Nisoniades Hübner, [1819], Viola Evans, 1953, and Viuria Grishin, 2019.
Parent taxon. Tribe Carcharodini Verity, 1940.
Burcina Grishin, new subtribe
http://zoobank.org/4E6C387B-AD5F-41C2-BBD3-7DEC415A315C
Type genus. Burca E. Bell and W. Comstock, 1948.
Definition. This new subtribe corresponds to one of the six major clades in the tribe Carcharodini Verity, 1940 (Fig. 4–6). It keys to E.30 in Evans (1953) and is diagnosed by the following combination of characters: unusual for the subfamily secondary sexual characters in males: either a costal fold that covers a long hair-pencil, or brands in the middle of forewing, no tibial tufts; uncus undivided, valvae mostly symmetrical, ampulla modified, frequently with process(es), protrudes beyond harpe posteriad; palpi short and porrect, forewing not produced, hindwing quadrate, angled at apex and vein M3, and rounded at tornus. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly481.18.1:G1591A, aly318.41.3:A82C, aly671.39.1:C130A, aly671.39.1:A1039G, aly291.11.14:C85A.
Genera included. Only the type genus.
Parent taxon. Tribe Carcharodini Verity, 1940.
Pholisorina Grishin, new subtribe
http://zoobank.org/9FEFDDB5-1BE4-4377-861F-B40047521F9C
Type genus. Pholisora Scudder, 1872.
Definition. This new subtribe corresponds to one of the six major clades in the tribe Carcharodini Verity, 1940 (Fig. 4–6). Approximately corresponds to the Staphylus sub-group of Evans (1953) (with many exceptions, but generally agrees with his description) and is diagnosed by the following combination of characters: antennae usually not shorter than half of costa, apiculus obtuse, shorter than the rest of the club; forewing without lower median veinlet, hindwing quadrate, longest at vein CuA1, inner margin not produced and approximately the same length as forewing costal margin, no tufts of scales on hindwing; uncus typically undivided and thinning toward distal end, aedeagus narrower, not bent, valvae almost always symmetrical (if asymmetrical then elongated and rectangular, not rounded), valva mostly rectangular, harpe protruding distad, variable in shape from narrow, almost needle-like to broad and wide, nearly the same length and height as valva. Most reliably identified by DNA, and combination of the following nuclear genomic base pairs is diagnostic: aly1139.42.2:C42T, aly1139.42.2:G63A, aly1107.9.6:G214A, aly1107.9.6:G215A, aly1107.9.6:C922A.
Genera included. Gorgopas Godman and Salvin, 1894, Incisus Grishin, 2019, Clytius Grishin, 2019, Perus Grishin, 2019, Bolla Mabille, 1903, Staphylus Godman and Salvin, 1896 (includes Hesperopsis Dyar, 1905 as a subgenus, see below), and Pholisora Scudder, 1872.
Parent taxon. Tribe Carcharodini Verity, 1940.
Torgus Grishin, new subgenus
http://zoobank.org/324B2041-EFAA-41BE-9403-70E49EF66BB9
Type species. Ouleus gorgus E. Bell, 1937.
Definition. Instead of originating within the genus Eantis Boisduval, 1836 (type species Urbanus thraso Hübner, 1807) as previously assumed, members of this new subgenus form a lineage sister to all other Iliana E. Bell, 1937 (type species Iliana romulus Bell, 1937), but are separated from them by prominent genetic differentiation at the subgenus level (Fig. 4). Therefore, they are placed in Iliana and a new subgenus is proposed for them. It keys to F.7.2 or F.7.5. in Evans (1953) and is distinguished from its relatives by the following characters: tegumen with longer side processes, harpe forked, not simply directed dorsal as in the subgenus Iliana, and ampulla not expanded. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly315.4.2:A178C, aly1249.27.3:C34A, aly1249.27.3:A35C, aly806.12.1:C2122T, aly806.12.1:A1482G.
Etymology. The name is a masculine noun in the nominative singular, a fusion of the names of the two species placed in the genus: T[aurus]+[g]orgus.
Species included. Ouleus gorgus E. Bell, 1937 and Tosta taurus Evans, 1953, for which we therefore propose new genus-species combinations: Iliana (Torgus) gorgus and Iliana (Torgus) taurus.
Parent taxon. Genus Iliana E. Bell, 1937.
Fenops Grishin, new subgenus
http://zoobank.org/FF4C0769-80D2-4BCC-8906-DA6D7B852FC6
Type species. Cabares enops Godman and Salvin, 1894.
Definition. The genomic tree reveals that the genus Polyctor Evans, 1953 (type species Pirgus [sic!] polyctor Prittwitz, 1868) splits into two prominent clades at the tree level of subgenera (Fig. 4). A subgenus represented by one of the clades does not have a name. Species of this new subgenus key to E.18.2a in Evans (1953) and are distinguished from the relatives by the following characters: absent white bands and spots on dorsal side of wings, hindwing outer margin strongly humped or angled in the middle, terminal projection of harpe on the right valva strongly upturned and reaches costa. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly7032.3.4:A46C, aly798.5.3:T760G, aly798.5.3:A794C, aly16.3.1:T537A, aly16.3.1:T825C.
Etymology. The name is a masculine noun in the nominative singular, a fusion of the names of the two species placed in the genus: Fe[ra]+[e]nops.
Species included. Cabares enops Godman and Salvin, 1894, Achlyodes fera Weeks, 1901, and Polyctor tensa Evans, 1953.
Parent taxon. Genus Polyctor Evans, 1953.
Polyctor (Polyctor) dagua Evans, 1953 is a species distinct from Polyctor (Polyctor) polyctor (Prittwitz, 1868)
Sequencing of the holotype of Polyctor (Polyctor) polyctor (Prittwitz, 1868) (type locality in Brazil: Rio de Janeiro) and its comparison with present-day specimens reveals distinction of Polyctor polyctor dagua Evans, 1953 (type locality Colombia: Dagua), phenotypically unique taxon in its darker aspect and yellower white bands. E.g., the Z chromosome Fst / Gmin between them is 0.39/0.00. Therefore, we propose that Polyctor (Polyctor) dagua Evans, 1953 new status, is a species distinct from Polyctor (Polyctor) polyctor (Prittwitz, 1868).
Pachyneuria Mabille, 1888 and Austinus O. Mielke and Casagrande, 2016 are subgenera of Sophista Plötz, 1879
Genomic tree places three current recognized genera, Sophista Plötz, 1879 (type species Thracides aristoteles Westwood, 1852), Pachyneuria Mabille, 1888 (type species Pachyneuria obscura Mabille, 1888) and Austinus O. Mielke and Casagrande, 2016 (type species Echelatus heros Mabille and Boullet, 1917) at the subgenus level (Fig. 4). E.g., the COI barcode difference of Pachyneuria obscura and Austinus heros from Sophista aristoteles is 7.2% (48 bp) and 6.4% (4 bp), respectively. Furthermore, all these species possess similar wing shape and male genitalia in the shape of valvae rounder than in their relatives, harpe folded along the inner surface of the right valva, and lanceolate in dorsal view undivided uncus. Differences between these three genera are confined mainly to wing patterns. Therefore, we propose that Pachyneuria Mabille, 1888 and Austinus O. Mielke and Casagrande, 2016 are subgenera of Sophista Plötz, 1879.
Pellicia (Pellicia) bilinea Mabille, 1889 is a species distinct from Pellicia (Pellicia) dimidiata Herrich-Schäffer, 1870
Genomic sequencing of Pellicia bilinea Mabille, 1889 (type locality in Panama: Chiriqui) syntype (NVG-15032E01, GenBank barcode OR665724) and a possible syntype of Pellicia dimidiata Herrich-Schäffer, 1870 (type locality Mexico and La Guaira, [Venezuela]), names currently regarded as subjective synonyms, and their comparison with additional specimens suggests that they belong to distinct species (Fig. 4): e.g., their Fst/Gmin/COI differences are 0.31/0.00/2.3% (15 bp). The only possible syntype of P. dimidiata we found (NVG-15032E10 in MFNB, GenBank barcode OR665725) was from Venezuela: La Guaira, from the Weymer’s collection, with identification label in Plötz’s handwriting “Pellicia (157.) / Dimidiata HS.” This is likely the specimen illustrated by Plötz in his unpublished drawings as t[afel]. 199 (Godman 1907) that also corresponds to an unnecessary replacement name Pellicia corinna Plötz, 1882, as mentioned by Godman (1907) and by Weymer on another label of this specimen: “Corinna Plötz i. l. / Dimidiata HS / non Feld. / Laguayra”. The remark “non Feld.” refers to Carterocephalus dimidiatus C. Felder and R. Felder, 1867, currently a valid species of Dalla Mabille, 1904 (type species Cyclopides eryonas Hewitson, 1877), a possible reason behind the replacement name P. corinna. The taxonomic identity of this syntype is consistent with the current application of the name P. dimidiata. However, exercising caution, we do not yet designate this specimen as a lectotype (or neotype if it is not a syntype). We will continue searching for Mexican syntypes and further investigating Weymer’s specimen. We suspect that the identification label by Plötz was the earliest label on this specimen, also accompanied by a small label “H S / 66 / Weymer”, probably meaning that this might have been a Herrich-Schäffer specimen in Weymer’s possession. The identification label by Weymer was added later when Plötz decided (incorrectly) that the name needed a replacement and suggested the name corinna, yet unpublished (therefore “i. l.”, for “in litteris”, on this label). For these reasons, it is possible that this specimen is sufficiently old and may indeed be a syntype of P. dimidiata as labeled. Regardless of its status as a syntype, South American populations represent a species distinct from the Central American species. Therefore, we propose that Pellicia (Pellicia) bilinea Mabille, 1889, restored status, is a species distinct from Pellicia (Pellicia) dimidiata Herrich-Schäffer, 1870, assuming that P. dimidiata is a South American species, pending further research. We note that only P. bilinea Mabille, 1889 was recorded from the US (Fig. 4).
Hemipteris Mabille, 1889 and Mictris Evans, 1955 are subgenera of Pellicia Herrich-Schäffer, 1870
The genomic tree reveals that Pellicia Herrich-Schäffer, 1870 (type species Pellicia dimidiata Herrich-Schäffer, 1870) as currently circumscribed is paraphyletic with respect to Mictris Evans, 1955 (type species Mycteris caerula Mabille, 1877) with the highest statistical support (Fig. 4). The third prominent clade in the group corresponds to an available name Hemipteris Mabille, 1889 (type species Hemipteris fumida Mabille, 1889, a junior subjective synonym of Pellicia tyana Plötz, 1882), currently a junior subjective synonym of Pellicia. Because the diversification of the three clades (Pellicia, Mictris, and Hemipteris) corresponds to a subgenus level in the tree, and all these species are similar in appearance, it seems that placing them into a single genus is a better choice for restoring monophyly instead of elevating Hemipteris to genus. Therefore, we propose that Hemipteris Mabille, 1889, restored status, and Mictris Evans, 1955, new status, are subgenera of Pellicia Herrich-Schäffer, 1870. As a result, only Pellicia bilinea Mabille, 1889, restored status, Pellicia dimidiata Herrich-Schäffer, 1870 and Pellicia theon Plötz, 1882 (as judged by their primary type specimens) belong to the nominotypical subgenus (Fig. 4). All other species currently placed in Pellicia belong to the subgenus Hemipteris. The subgenus Mictris is monotypic.
Lectotype designation for Pellicia theon Plötz, 1882
Pellicia theon Plötz, 1882 (type locality in South America) currently treated as a valid species in its original genus was described from unstated number of specimens (Plötz 1882). A single syntype is curated as such in the MFNB collection. It agrees with the original description and is likely the specimen illustrated by Plötz in his unpublished drawings as t[afel]. 200 (Godman 1907), copies in BMNH and USNM, because the details of the illustration match the details of the specimen. For instance, it appears that a glue patch in the middle of ventral hindwing along vein CuA2 applied to secure the tear in the syntype is depicted as a pair of gray streaks. There are no natural markings at this spot in Pellicia. To stabilize nomenclature, N.V.G. hereby designates the syntype in the MFNB collection, a male bearing nine labels (1st red, others white): [typus], [53 |Weymer], [Pellicia (156i.) | Theon Pl.], [Hesperia], [26:17], [Coll. Weymer], [Theon Plötz i l. | Amer. Mer.], [DNA sample ID: | NVG-15032E09 | c/o Nick V. Grishin], and [{QR Code} http://coll.mfn-berlin.de/u/ | 940b9e], as the lectotype of Pellicia theon Plötz, 1882. The third label, matching Plötz’s handwriting, adds to our confidence that the lectotype was indeed one of the syntypes. The fifth label is the number (genus:species) of P. theon in the Mabille catalog (1903). The COI barcode sequence of P. theon lectotype, sample NVG-15032E09, GenBank OR721877, 658 base pairs, is: AACTTTATATTTTATTTTTGGTATTTGATCTGGAATAGTAGGAACATCATTAAGTTTAATTATTCGATCCGAATTAGGTACCCCTAGATCTTTTATTGGAGATGATCAAATTTATAATACCATTGTAACAGCTCATGCCTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTCGGAAATTGATTAGTACCCCTTATATTAGGAGCTCCTGATATAGCTTTTCCCCGAATAAATAATATAAGATTTTGACTTTTACCTCCTTCTATTACTTTACTAATTTCAAGAAGTTTTGTAGAAAATGGTGTTGGTACAGGTTGAACTGTTTATCCCCCTTTATCTGCTAATATTGCTCACCAAGGTTCTTCTGTAGATTTAGCAATTTTTTCTTTACATTTAGCTGGTATTTCATCTATTTTAGGTGCTATTAATTTTATTACAACCATTATTAATATACGTATTAACAATTTATTATTTGATCAAATACCTTTATTTATTTGAGCTGTTGGAATTACAGCTTTACTTTTATTATTATCTCTACCAGTTTTAGCTGGAGCTATTACCATACTATTAACTGATCGAAATTTAAATACATCTTTTTTTGACCCTGCGGGAGGAGGAGATCCAATTTTATATCAACATTTATTT
Pellicia (Hemipteris) nema R. Williams and E. Bell, 1939 is a species distinct from Pellicia (Pellicia) theon Plötz, 1882
Genomic sequencing of the holotype of Pellicia nema Williams and Bell (type locality in Brazil: Mato Grosso, sequenced as NVG-15097B11, GenBank barcode OR721876) and the lectotype of Pellicia theon Plötz, 1882 (type locality in South America, sequenced as NVG-15032E09, GenBank barcode OR721877) currently considered conspecific reveals that they belong to different subgenera, Hemipteris Mabille, 1889 (type species Hemipteris fumida Mabille, 1889, a junior subjective synonym of Pellicia tyana Plötz, 1882) and Pellicia Herrich-Schäffer, 1870 (type species Pellicia dimidiata Herrich-Schäffer, 1870), respectively (Fig. 4). Not finding other species with older names that are justifiably conspecific with either of the two, we propose that Pellicia (Hemipteris) nema Williams and Bell, 1939, restored status, is a species distinct from Pellicia (Pellicia) theon Plötz, 1882. We hypothesize that the two species were confused with each other because Evans (1953) misidentified P. theon. What he identified as P. theon was probably P. nema. As identified by its lectotype, labeled as “theon Pl.” in Plötz’s writing, P. theon is a species closely related to Pellicia dimidiata Herrich-Schäffer, 1870 (type locality Mexico and La Guaira [Venezuela]) (Fig. 4). Evans treated the former within his concept of the latter species, but their COI barcodes differ by 1.4% (9 bp).
Bezus Grishin, new subgenus
http://zoobank.org/F70C2876-F6A2-4633-8938-B52917E75CF9
Type species. Pellicia bessus Möschler, 1877.
Definition. The genus Nisoniades Hübner, 1819 (type species Papilio bromius Stoll, 1787, a junior subjective synonym of Papilio mimas Cramer, 1775) splits into three prominent clades at the subgenus level (Fig. 4). Because no available genus-group names have been associated with Nisoniades, two of three subgenera are new. This new subgenus keys to E.19.1c in Evans (1953) and is distinguished from its relatives by the following characters: vein RS on ventral hindwing in males does not run along vein M1 but diverges from it at its origin, which is around the middle between the base and end of the discal cell, and is swollen until the end of cell, not much beyond, and harpe of right valva is not divided. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly84.3.1:T55C, aly84.3.1:C101T, aly37338.21.1:A1627T, aly37338.21.1:G1861T, aly3486.1.4:T485A.
Etymology. The name is a masculine noun in the nominative singular formed from the type species name to keep it short and avoid a homonym.
Species included. Nisoniades godma Evans, 1953, Nisoniades panama Evans, 1953, Nisoniades suprapanama Steinhauser, 1989, Nisoniades benda Evans, 1953, Pellicia bessus Möschler, 1877, Nisoniades brazia Evans, 1953, Pellicia hecale Hayward, 1940, Nisoniades cauca Evans, 1953, Nisoniades remo Evans, 1953, Pellicia guianae R. Williams and E. Bell, 1939, Pellicia montana R. Williams and E. Bell, 1939, Nisoniades coca Steinhauser, 1989, and Nisoniades lata Steinhauser, 1989.
Parent taxon. Genus Nisoniades Hübner, [1819].
Macarius Grishin, new subgenus
http://zoobank.org/3C0E9973-A2BA-4F62-8B65-C9DA18692468
Type species. Pellicia macarius Herrich-Schäffer, 1870.
Definition. The genus Nisoniades Hübner, 1819 (type species Papilio bromius Stoll, 1787, a junior subjective synonym of Papilio mimas Cramer, 1775) splits into three prominent clades at the subgenus level (Fig. 4). Because no available genus-group names have been associated with Nisoniades, two of three subgenera are new. This new subgenus keys to E.19.3, E19.4, or E.19.12a in Evans (1953) and is distinguished from its relatives by the following characters: vein RS on ventral hindwing in males usually runs parallel to vein M1 from its origin to the end of the swollen portion, if divergent then right harpe is bifid or vein RS origin is closer to the end of cell than to the wing base and dorsal hindwing tuft in males is inconspicuous, harpe of right valva is frequently bifid, hindwing is frequently quadrate and angled at vein CuA1, harper of left valva frequently does not extend past ampulla. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly6517.5.2:T1476C, aly6517.5.2:G1699T, aly4975.2.1:C43T, aly4975.2.1:T73G, aly23605.23.23:A145G.
Etymology. The name is a masculine noun in the nominative singular, tautonymous with the type species name.
Species included. Pellicia laurentina R. Williams and E. Bell, 1939, Nisoniades evansi Steinhauser, 1989, Nisoniades supra Steinhauser, 1989, Pellicia maura Mabille and Boullet, 1917, Pellicia hora Hayward, 1939, Pellicia criton Mabille, 1898, Pellicia hesperia Hayward, 1939, Arteurotia castolus Hewitson, 1878, Pellicia haywardi R. Williams and E. Bell, 1939, Pellicia rimana E. Bell, 1942, Pellicia macarius Herrich-Schäffer, 1870, and Achlyodes nyctineme A. Butler, 1877.
Parent taxon. Genus Nisoniades Hübner, [1819].
Quadralis Grishin, new subgenus
http://zoobank.org/C859F739-AC31-4F2E-9A2A-14778E4FC67B
Type species. Pterygospidea extensa Mabille, 1891.
Definition. Sister to the rest of the genus Gorgopas Godman and Salvin, 1894 (type species Achlyodes viridiceps Butler & H. Druce, 1872, which is currently regarded as a junior subjective synonym of Pellicia chlorocephala Herrich-Schäffer, 1870) and sufficiently distinct from it (Fig. 5), resembling in appearance some species of Polyctor Evans, 1953 (type species Pirgus [sic!] polyctor Prittwitz, 1868). Keys to E.18.4 in Evans (1953) and distinguished from its relatives by nearly square hindwing due to a blunt tooth at the vein CuA1 and hyaline spots in forewing cells between veins M1 to 1A+2A. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly770.25.10:A48C, aly451.25.1:G1816A, aly3686.9.6:G137C, aly1656.12.3:G757C, aly5196.9.2:C832T.
Figure 5.

The nuclear genome-based phylogeny of Carcharodini, middle segment (continues in 6, see Fig. 4 for the first segment) showing Pholisorina subtrib. n. Species formerly placed in Staphylus are labeled in red. Asterisk marks a species with a dedicated study (Lemes et al. 2023) that already assigned it to Bolla. See Fig. 1 for notations.
Etymology. The name is a masculine noun (to agree in gender with the names of species in this subgenus) in the nominative singular, given for the square-shaped hindwing: in Latin, quadratum is square, and alis is wing.
Species included. Only the type species.
Parent taxon. Genus Gorgopas Godman and Salvin, 1894.
Menuda Grishin, new subgenus
http://zoobank.org/61EF142F-EE57-49AF-A475-041DBC620ED4
Type species. Nisoniades menuda Weeks, 1902.
Definition. The genus Perus Grishin, 2019 (type species Pholisora cordillerae Lindsey, 1925) splits into three prominent lineages at the subgenus level in the tree (Fig. 5), two of which correspond to currently monotypic and unnamed subgenera. One of these new subgenera keys to E.32.16 in Evans (1953) and is distinguished from its relatives by undivided and narrow, nearly needle-shaped and somewhat upturned in lateral view uncus without projections at its base and harpe nearly as long as valva, with a beak-like tooth on its dorsal margin and projection off ampulla reaching the “beak”. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly686.40.2:T38C, aly686.40.2:T99C, aly671.7.4:T75C, aly2811.5.5:A489G, aly171.6.1:G5424C.
Etymology. The name is a feminine noun in the nominative singular, tautonymous with the type species name.
Species included. Only the type species.
Parent taxon. Genus Perus Grishin, 2019.
Narycus Grishin, new subgenus
http://zoobank.org/7D5FD1DE-9860-48E2-AB25-D0B6F00C7CAB
Type species. Pythonides narycus Mabille, 1889.
Definition. The genus Perus Grishin, 2019 (type species Pholisora cordillerae Lindsey, 1925) splits into three prominent lineages at the subgenus level in the tree (Fig. 5), two of which correspond to currently monotypic and unnamed subgenera. One of these new subgenera keys to E.37.8 in Evans (1953) and is distinguished from its relatives by undivided and semi-triangular in dorsal view, narrowing to a point, and downturned in lateral view uncus without projections at its base and harpe only slightly shorter than valva, with a broad beak-like tooth on its dorsal margin projecting dorsad beyond costa-ampulla in lateral view. In DNA, a combination of the following nuclear genomic base pairs is diagnostic: aly736.3.1:G100T, aly736.3.1:C101T, aly214.18.3:A37C, aly214.18.3:C38T, aly173.28.7:A73T.
Etymology. The name is a masculine noun in the nominative singular, tautonymous with the type species name.
Species included. Only the type species.
Parent taxon. Genus Perus Grishin, 2019.
Bovaria Grishin, new subgenus
http://zoobank.org/86413A71-3528-4B5A-BB4D-9C7C43667518
Type species. Achlyodes cyclops Mabille, 1876.
Definition. The nuclear genomic tree reveals that Bolla Mabille, 1903 (type species Bolla pullata Mabille, 1903, treated as a junior subjective synonym of Staphylus imbras Godman and Salvin, 1896) splits into four clades at the subgenus level (Fig. 5). The subgenera corresponding to three of these clades do not have available names and are new. One of these new subgenera keys to E.31.1b or E.31.18a in Evans (1953) and is distinguished from its relatives by the following characters: at least around tornus of ventral hindwing with gray or ocherous overscaling, distal third of ventral forewing paler than the rest, if not then forewing cell CuA1-CuA2 with a prominent hyaline spot (at least in females) and/or wings ocherous-brown with conspicuous darker spots somewhat connected into bands, antenna nudum with about 13 segments; tegumen in dorsal view rounded, wider at the base, without processes near uncus (but could be with small “knobs”), uncus lanceolate to narrowly triangular, valva more square than rectangular, or rounded on the sides, ampulla with a bulge or a bulbous process, harpe usually extended posteriad, could be claw-like and upturned or relatively short and overlapping with valva and with inner dorsal margin serrated and/or expanded. Confident identification is provided by DNA, and a combination of the following base pairs is diagnostic: in the nuclear genome: aly537.31.1:G611A, aly1139.42.5:C79A, aly1139.42.5:A27T, aly84.37.1:T1180C, aly84.37.1:T1395C, and in the COI barcode: A181A, 263C, A265T, A424A, 592A, T652T, T653T.
Etymology. The name is a feminine noun in the nominative singular, unifying a number of Bolla species with variable superficial appearance: Bo[lla] + varia[ble].
Species included. Bolla cybele Evans, 1953, Staphylus cylindus Godman and Salvin, 1896, Bolla sonda Evans, 1953, Achlyodes cyclops Mabille, 1877, Pholisora aplica E. Bell, 1937, restored status (see below), Bolla sodalis Schaus, 1913, restored status (see below), Bolla solitaria Steinhauser, 1991, Staphylus saletas Godman and Salvin, 1896, Pholisora tepeca E. Bell, 1942, and Bolla fenestra Steinhauser, 1991.
Parent taxon. Genus Bolla Mabille, 1903.
Sebia Grishin, new subgenus
http://zoobank.org/D31D04CF-D4BF-4D85-BE43-D68B7FB57421
Type species. Nisoniades eusebius Plötz, 1884.
Definition. The nuclear genomic tree reveals that Bolla Mabille, 1903 (type species Bolla pullata Mabille, 1903, treated as a junior subjective synonym of Staphylus imbras Godman and Salvin, 1896) splits into four clades at the subgenus level (Fig. 5). The subgenera corresponding to three of these clades do not have available names and are new. One of these new subgenera keys to E.31.10 or 11c (except 18a) in Evans (1953) and is distinguished from its relatives by the following characters: at least around tornus of ventral hindwing with gray or ocherous overscaling, distal third of ventral forewing not paler than the rest, forewing typically broader, more pointed at the apex, discal cell without white or hyaline spots, antenna nudum with about 13 segments; tegumen in dorsal view notched, but not lobed, in the middle on its sides, with projections at the base of uncus in some species, uncus bulbous at the base, narrowing to a sharp or more rounded point, valva approximately rectangular (at least more so than in relatives), ampulla small, indistinct, harpe usually long, could also be rectangular and not shorted than valva, bilobed, or short and upturned, in which case tegumen with short processes at the base of uncus. Confident identification is provided by DNA, and a combination of the following base pairs is diagnostic: in the nuclear genome: aly594.10.2:A96T, aly594.10.2:T117G, aly84.89.2:C467G, aly84.89.2:T480C, aly84.99.2:A925T, and in the COI barcode: A40T, C81(mostly T), 82R, T652C, or if 652T then T653C.
Etymology. The name is a masculine noun in the nominative singular, formed from the type species name: [eu] Sebi[us] + a.
Species included. Staphylus evippe Godman and Salvin, 1896, Staphylus orsines Godman and Salvin, 1896, Nisoniades eusebius Plötz, 1884, Pholisora chilpancingo E. Bell, 1937, restored status (see below), Bolla tetra guerra Evans, 1953, Bolla tetra oriza Evans, 1953, Staphylus brennus Godman and Salvin, 1896, Bolla antha Evans, 1953, Bolla dorsolaciniae Steinhauser, 1989, Hesperia giselus Mabille, 1883, and Pholisora giselus race boliviensis E. Bell, 1937.
Parent taxon. Genus Bolla Mabille, 1903.
Stolla Grishin, new subgenus
http://zoobank.org/CA87FEC7-A5E2-413D-96D0-9E0CE260713B
Type species. Pholisora balsa E. Bell, 1937.
Definition. The nuclear genomic tree reveals that Bolla Mabille, 1903 (type species Bolla pullata Mabille, 1903, treated as a junior subjective synonym of Staphylus imbras Godman and Salvin, 1896) splits into four clades at the subgenus level (Fig. 5). The subgenera corresponding to three of these clades do not have available names and are new. One of these new subgenera includes many species previously placed in Staphylus Godman and Salvin, 1896 (type species Helias ascalaphus Staudinger, 1876). Species in this subgenus key to E.31.21, E.32.1b, 9, 27, 32, or 37 in Evans (1953) and are distinguished from its relatives by the following characters: ventral hindwing even around tornus frequently without gray or ocherous overscaling, distal third of ventral forewing not paler than the rest, forewing usually narrower, more rounded at the apex, discal cell with white or hyaline spot(s) in some species, antenna nudum with 10–11 segments in most species; tegumen in dorsal view not notched and not lobed in the middle on its sides, and could be with projections (sometimes long) at the base of uncus, uncus undivided, terminally narrowing, usually separated from tegumen by a notch (or a concavity) in lateral view, harpe may be elongated, extending posteriad, or broader and upturned, ampulla with a process, projection, or a hump. Confident identification is provided by DNA, and a combination of the following base pairs is diagnostic: in the nuclear genome: aly291.13.2:C57T, aly1720.4.1:A1080G, aly1720.4.1:A837G, aly1720.4.1:A1147C, aly728.40.3:T120C, and in the COI barcode: C81C, 82Y, T284C, A286T, A298A, T652T, T653T.
Etymology. The name is a feminine noun in the nominative singular, meaning that the genus includes a large number of species that were transferred from Staphylus to Bolla: St[aphylus] + [B]olla.
Species included. Antigonus zorilla Plötz, 1886, Pholisora madrea R. Williams and E. Bell, 1940, restored status (see below), Pholisora hazelae Hayward, 1940, restored status (see below), Staphylus evemerus Godman and Salvin, 1896, Staphylus chlora Evans, 1953, Pholisora astra R. Williams and E. Bell, 1940, Pholisora balsa E. Bell, 1937, Staphylus tridentis Steinhauser, 1989, Staphylus esmeraldus L. Miller, 1966, Hesperia chlorocephala Latreille, [1824], and Staphylus incanus E. Bell, 1932.
Parent taxon. Genus Bolla Mabille, 1903.
Lectotype designation for Nisoniades eusebius Plötz, 1884
Nisoniades eusebius Plötz, 1884 (type locality in Central America), currently a valid species in the genus Bolla Mabille, 1903 (type species Bolla pullata Mabille, 1903, treated as a junior subjective synonym of Staphylus imbras Godman and Salvin, 1896) and the type species of the subgenus Sebia, new subgenus, was described from an unstated number of specimens (Plötz 1884). One specimen is curated in the MFNB collection as a syntype of N. eusebius. It agrees with the original description and was sequenced as NVG-15033G09. To stabilize nomenclature, N.V.G. hereby designates the syntype in the MFNB collection, a male bearing eight labels (1st red, others white): [typus], [Centr Amer.], [Parna Plötz | n. 151 best. v. Plötz.], [Eusebius Plötz | Plötz taf 1053], [Coll. Weymer], [Eusebius Plötz i. l. | Parna Plötz i. l. | olim | Amer centr.], [DNA sample ID: | NVG-15033G09 | c/o Nick V. Grishin], [{QR Code} http://coll.mfn-berlin.de/u/ | 80a659] as the lectotype of Nisoniades eusebius Plötz, 1884. According to its labels, this specimen was identified by Plötz (“best[immt]. v[on]. Plötz”) as “parna”, which was initially suggested (“olim”, Latin for ‘formerly’, ‘once’, or ‘formerly known as’), and unpublished, the name for this species that was later changed to “Eusebius”, and this label with the name was added to the specimen prior to publication (“i[n] l[itteris]”). Furthermore, per the dedicated label, this specimen is from Central America, exactly as stated in the original description. These data on the labels increase our confidence in this specimen, originally from the Weymer collection, being a syntype. The COI barcode sequence of N. eusebius lectotype, sample NVG-15033G09, GenBank OR665726, 658 base pairs, is: AACTTTATACTTTATTTTTGGTATTTGATCTGGAATAGTTGGAACTTCTTTAAGTATTCTTATTCGTTCTGAACTAGGAATGCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGAAATTGATTGGTGCCCCTTATATTAGGAGCTCCTGATATAGCTTTTCCTCGAATAAATAATATAAGATTTTGACTTTTACCTCCTTCTCTAATATTATTAATTTCTAGTAGTATTGTAGAAAGTGGGGCAGGTACAGGATGAACAGTATATCCCCCACTTTCAGCTAATATTGCCCATCAAGGTTCTTCTGTAGATTTAGCTATTTTTTCTCTTCATTTAGCTGGTATTTCTTCAATTTTAGGTGCTATTAATTTCATTACAACTATTATCAATATACGAATTAATAACTTATCCTTTGATCAAATACCTTTATTTGTTTGAGCAGTAGGTATTACTGCATTACTTTTATTATTATCTTTACCAGTATTAGCAGGAGCTATTACTATACTTCTAACTGATCGTAATTTAAATACATCATTCTTTGATCCAGCAGGTGGAGGAGATCCTATTTTATACCAACATCTATTT
Bolla (Bovaria) sodalis (Schaus, 1913) is a species distinct from Bolla (Bolla) imbras (Godman and Salvin, 1896)
Evans (1953) apparently misinterpreted Bolla sodalis Schaus, 1913 (type locality Costa Rica: El Alto) in treating it as a junior subjective synonym of Bolla (Bolla) imbras (Godman and Salvin, 1896) (type locality in Mexico: Veracruz and Tabasco, and Guatemala). We suggest Evan’s mistake, because the original description and illustration of B. sodalis show a species that is “Nearest [to] B. cylindus G. & S.” (Schaus 1913), if not for any other reason than the pattern of forewing hyaline spots, which B. imbras lacks. Genomic sequencing of a syntype of Bolla sodalis (NVG-18061C04, GenBank barcode OR665727) places it away from B. imbras (currently a valid name for the type species of Bolla Mabille, 1903) and indeed in the subgenus Bovaria new subgenus Consideraing only species with names more senior than B. sodalis, it is sister to the clade of Bolla cylindus Godman and Salvin, 1896) (type locality in Mexico: Veracruz and Costa Rica) and Bolla cyclops (Mabille, 1877) (type locality in Guatemala and Colombia) (Fig. 5). Therefore, we propose that Bolla (Bovaria) sodalis (Schaus, 1913), restored status, is a species distinct from Bolla (Bolla) imbras (Godman and Salvin, 1896).
Bolla (Bovaria) aplica (E. Bell, 1937) is a species distinct from Bolla (Sebia) eusebius (Plötz, 1884)
Genomic sequencing of the holotype of Pholisora aplica E. Bell, 1937 (type locality in Mexico: Guerrero, NVG-18024D09, GenBank barcode OR665728) and the lectotype of Nisoniades eusebius Plötz, 1884 (type locality in Central America, NVG-15033G09) regarded by Evans (1953) as conspecific in the genus Bolla Mabille, 1903 (type species Bolla pullata Mabille, 1903, treated as a junior subjective synonym of Staphylus imbras Godman and Salvin, 1896) reveals that they belong to different subgenera: Bovaria new subgenus (type species Achlyodes cyclops Mabille, 1876) and Sebia new subgenus (type species Nisoniades eusebius Plötz, 1884), respectively (Fig. 5). This problem arose because Evans (1953) misidentified N. eusebius, e.g., its syntype has costal fold, but according to Evans’ key, his “eusebius” lacks costal fold. The phylogenetic tree shows that N. eusebius is sister to Bolla (Sebia) orsines (Godman and Salvin, 1896) (type locality in Mexico: Jalisco) and P. aplica is sister to Bolla (Bovaria) sodalis (Schaus, 1913) (type locality Costa Rica: El Alto), restored status, Thus, although we have not sequenced specimens identified by Evans as “eusebius”, we hypothesize that they were B. sodalis. We find that while P. aplica is closely related to B. sodalis, they show some genetic differentiation, e.g., their COI barcodes differ by 0.6% (4 bp) and come from different biogeographic areas. Therefore, to attract attention to the problem and due to confusion in the literature, we propose that Bolla (Bovaria) aplica (E. Bell, 1937), restored status, is a species-level taxon distinct from both Bolla (Sebia) eusebius (Plötz, 1884) (definitively) and Bolla (Bovaria) sodalis (Schaus, 1913) (tentatively), and leave it for future studies to determine whether it is best treated as a subspecies of the latter.
Bolla (Sebia) chilpancingo (E. Bell, 1937) is a species distinct from Bolla (Bolla) subapicatus (Schaus, 1902); the latter is a junior subjective synonym of Bolla (Bolla) imbras (Godman and Salvin, 1896)
Genomic sequencing of the holotype of Pholisora chilpancingo Bell, 1937 (type locality in Mexico: Guerrero, NVG-18024E12, GenBank barcode OR665729) and a syntype of Staphylus subapicatus Schaus, 1902 (type locality in Mexico: Veracruz, NVG-18061C07, GenBank barcode OR665730) regarded by Evans (1953) as conspecific in the genus Bolla Mabille, 1903 (type species Bolla pullata Mabille, 1903, treated as a junior subjective synonym of Staphylus imbras Godman and Salvin, 1896) reveals that they belong to different subgenera: Sebia new subgenus (type species Nisoniades eusebius Plötz, 1884) and the nominotypical subgenus, respectively (Fig. 5). This problem arose because Evans (1953) misidentified S. subapicatus. Indeed, P. chilpancingo forms a distinct lineage in the tree and keys to the “subapicatus” of Evans. However, S. subapicatus falls within the genetic variation of Bolla (Bolla) imbras (Godman and Salvin, 1896) (type locality in Mexico: Veracruz and Tabasco, and Guatemala) (Fig. 5), e.g., the COI barcodes of the specimens we sequenced differ by 1 bp. Therefore, we propose that Bolla (Sebia) chilpancingo (E. Bell, 1937), restored status, is a valid species distinct from Bolla (Bolla) subapicatus (Schaus, 1902), but the latter taxon is a new junior subjective synonym of Bolla (Bolla) imbras (Godman and Salvin, 1896).
Bolla (Stolla) madrea (R. Williams and E. Bell, 1940) and Bolla (Stolla) hazelae (Hayward, 1940) are species distinct from Bolla (Stolla) zorilla (Plötz, 1886)
Genomic sequencing of the holotypes of Pholisora madrea R. Williams and E. Bell, 1940 (type locality in Peru: Madre de Dios, NVG-15097C09, GenBank barcode OR665731) and Pholisora hazelae Hayward, 1940 (type locality in Ecuador, NVG-18024G08, GenBank barcode OR665732) and a syntype of Antigonus zorilla Plötz, 1886 (type locality in Panama, NVG-18056H01, GenBank barcode OR665733), currently treated as conspecific in the genus Bolla Mabille, 1903 (type species Bolla pullata Mabille, 1903, considered a junior subjective synonym of Staphylus imbras Godman and Salvin, 1896) reveals that while all three are in the same clade, they are genetically differentiated at the level not unprecedented even for subgenera (Fig. 5), e.g., COI barcode differences are: 8.2% (54 bp) for P. madrea and P. hazelae, 7.1% (47 bp) for P. madrea and A. zorilla, and 7.6% (50 bp) for P. hazelae and A. zorilla. Therefore, we propose that Bolla (Stolla) madrea (R. Williams and E. Bell, 1940), restored status, and Bolla (Stolla) hazelae (Hayward, 1940), restored status, are species distinct from Bolla (Stolla) zorilla (Plötz, 1886).
Hesperopsis Dyar, 1905 is a subgenus of Staphylus Godman and Salvin, 1896
The phylogenetic tree did not reveal a prominent separation between Staphylus Godman and Salvin, 1896 (type species Helias ascalaphus Staudinger, 1876) and Hesperopsis Dyar, 1905 (type species Thanaos alpheus W. H. Edwards, 1876), quite the opposite, it illustrates their closeness (Fig. 5), which is equally reflected in the similarity of their wing shapes and patterns. Staphylus and Hesperopsis are closer related than some species of Bolla to each other. While Staphylus is a large genus, Hesperopsis is a small group of three North American species. Including them in Staphylus is not expected to cause confusion but would highlight their close relationships. Therefore, we proposed to treat Hesperopsis Dyar, 1905 as a subgenus of Staphylus Godman and Salvin, 1896.
Scantilla Godman and Salvin, 1896 is a subgenus of Staphylus Godman and Salvin, 1896
The nuclear genomic tree reveals that Staphylus Godman and Salvin, 1896 (type species Helias ascalaphus Staudinger, 1876) splits into five clades at the subgenus level (Fig. 5). The subgenus Hesperopsis Dyar, 1905 (type species Thanaos alpheus W. H. Edwards, 1876) is sister to all others. The next prominent clade is sister to the clade with the type species of Staphylus and contains the type species of the available genus-group name Scantilla Godman and Salvin, 1896, Scantilla opites Godman and Salvin, 1896. Therefore, this clade corresponds to the subgenus Scantilla, new status, resurrected from synonymy. In addition to the type species, the following species belong to this subgenus: Pholisora cartagoa R. Williams & E. Bell, 1940, Tagiades vincula Plötz, 1886, Pholisora ceos W. H. Edwards, 1882, and Staphylus ecos Grishin, 2022. Curiously, the two subgenera (Hesperopsis and Scantilla) are characterized by a slower substitution rate than others: tree distance from the last common ancestor of Staphylus to the leaves is smaller for species in these two subgenera than for others.
Vulga Grishin, new subgenus
http://zoobank.org/DA9638E7-54AF-4B18-B4B8-D6E275D4977B
Type species. Achlyodes vulgata Möschler, 1879.
Definition. The nuclear genomic tree reveals that Staphylus Godman and Salvin, 1896 (type species Helias ascalaphus Staudinger, 1876) splits into five clades at the subgenus level (Fig. 5). The subgenera corresponding to two of these clades, which are sister to each other, do not have available names and are new. One of these new subgenera keys to E.32.5c, 7, or 9c in Evans (1953) and is distinguished from its relatives by the following characters: spots on wings rather inconspicuous, head without green scales but is golden in some species; ampulla of valva frequently expanded, overlaps harpe from inside (if not then harpe and valva of approximately equal size, and ampulla expansion reaches the end of harpe), vinculum brushes absent, uncus undivided, narrowing to a point, lanceolate and semi-triangular towards the tip, typically broader than in relatives, and not strongly concave in lateral view, tegumen typically not broader than uncus. Confident identification is provided by DNA, and a combination of the following base pairs is diagnostic: in the nuclear genome: aly3824.12.4:A540G, aly3824.12.4:A595C, aly259.26.2:T1260A, aly259.26.2:A590C, aly390.10.14:G2931A and the COI barcode: T29G, A241A, A334T, T337A, T346A, T412T, T578C, A580T.
Etymology. The name is a feminine noun in the nominative singular formed from the type species name: Vulga[ta].
Species included. Achlyodes vulgata Möschler, 1879, Pholisora melaina Hayward, 1947, Staphylus kayei Cock, 1996, Staphylus putumayo sambo Evans, 1953, Pholisora putumayo E. Bell, 1937, Staphylus saxos Evans, 1953, Staphylus saxos satrap Evans, 1953 (see below), Staphylus tingo Steinhauser, 1989, Nisoniades oeta Plötz, 1884, and Staphylus punctiseparatus Hayward, 1933.
Parent taxon. Genus Staphylus Godman and Salvin, 1896.
Staphylus (Vulga) satrap Evans, 1953 is a species distinct from Staphylus (Vulga) saxos Evans, 1953
Described as a subspecies of Staphylus saxos Evans, 1953 (type locality in Colombia: Cali), S. s. satrap Evans, 1953 (type locality in Bolivia) is not monophyletic with it and is genetically differentiated from it at the level characteristic of distinct species (Fig. 5), e.g., their COI barcodes differ by 4.4% (29 bp). Moreover, Evans (1953) mentions and illustrates differences in genitalia that suggest species level of these taxa. Therefore, we propose that Staphylus (Vulga) satrap Evans, 1953, new status, is a species distinct from Staphylus (Vulga) saxos Evans, 1953, and the latter becomes monotypic.
Capilla Grishin, new subgenus
http://zoobank.org/942D19CE-7B9C-47E7-9650-1721B9A18AB4
Type species. Helias aurocapilla Staudinger, 1876, currently a junior subjective synonym of Hesperia musculus Burmeister, 1875.
Definition. The nuclear genomic tree reveals that Staphylus Godman and Salvin, 1896 (type species Helias ascalaphus Staudinger, 1876) splits into five clades at the subgenus level (Fig. 5). The subgenera corresponding to two of these clades, which are sister to each other, do not have available names and are new. One of these new subgenera is phenotypically diverse, keying to E.32.3, 8, 10a, 11a, 13d, 19, 21, or 22 in Evans (1953) and is distinguished from its relatives by the following characters: wings typically narrower than in relatives, several species with more extensive white/hyaline forewing spots; the only subgenus that includes species with divided, rounded (except the very tip), or bulbous at the tip uncus, but uncus undivided and narrow, nearly needle-like in some species, and not strongly concave in lateral view, tegumen usually broader than uncus, vinculum relatively straight in lateral view, harpe mostly broad, with rounded or truncate distal margin, overlays ampulla from the inside (at least in species with lanceolate uncus), ampulla frequently expanded and projects to the end of harper in some species, aedeagus frequently shorter and broader than in relatives. Confident identification is provided by DNA, and a combination of the following base pairs is diagnostic: in the nuclear genome: aly1405.20.35:T489C, aly1405.20.35:T444C, aly1405.20.35:G452C, aly1370.7.2:A2191G, aly1370.7.2:T804G, and the COI barcode T29(mostly T), A130Y, A241(mostly T), A256T, T412(mostly A), T578(mostly T), A580(mostly A).
Etymology. The name is a feminine noun in the nominative singular, formed from the type species name: [auro] Capilla.
Species included. Pholisora lizeri Hayward, 1938, Pholisora caribbea R. Williams and E. Bell, 1940, Pholisora buena R. Williams and E. Bell, 1940, Staphylus melius Steinhauser, 1989, Hesperia musculus Burmeister, 1875, Pholisora imperspicua Hayward, 1940, Pholisora corumba R. Williams and E. Bell, 1940, Helias ascalon Staudinger, 1876, Staphylus eryx Evans, 1953, Hesperia melangon Mabille, 1883, Nisoniades tucumanus Plötz, 1884, Staphylus perna Evans, 1953, Staphylus insignis O. Mielke, 1980, Helias tyro Mabille, 1878, and Pholisora azteca Scudder, 1872.
Parent taxon. Genus Staphylus Godman and Salvin, 1896.
Composition of Carcharodina Verity, 1940 with comments on nomenclature
The subtribe Carcharodina Verity, 1940 consists of the following genera: Noctuana E. Bell, 1937, Windia H. Freeman, 1969, Carcharodus Hübner, [1819], Muschampia Tutt, 1906 (the senior name for this genus is Sloperia Tutt, 1906, see below), Favria Tutt, 1906, Gomalia Moore, 1879, Agyllia Grishin, 2019, Ernsta Grishin, 2019, and Spialia Swinhoe, 1912 (Fig. 6). We phylogenetically arrange genera in the subtribe to start with the New World representatives, because they would follow all the rest of Carcharodini that are from the New World, thus not disrupting the geographic arrangement. The ordering ends with the strongly checkered in wing pattern members of the subtribe, such as Spialia, because the next tribe to follow in the taxonomic list would be Pyrginae Burmeister, 1878. To link the two tribes by phenotypic similarity of their constituent species, Pyrginae could start from the Old World checkered skippers in the genus Pyrgus Hübner, [1819]), which are superficially similar to Spialia and many Muschampia, and are the only representatives of Pyrginae in the Old Word. Furthermore, genera in the two pairs (Favria, Muschampia) and (Carcharodus, Muschampia) best be next to each other due to phenotypic similarity between some species in each pair, and the phylogeny additionally constrains the pairing (Favria, Gomalia). We choose to place Carcharodus first due to some wing pattern similarity with Windia, and because this order results in a continuous placement of African members (in Gomalia, Agyllia, and Ernsta), ending with mostly Palearctic Spialia to follow with Palearctic and similar-looking Pyrgus.
In agreement with the ICZN Code, the name Sloperia Tutt, 1906 takes precedence over Muschampia Tutt, 1906, as selected by the first reviser (Warren 1926) and should be the correct name to use instead of Muschampia, as pointed out by Hemming (1967). However, judging by the literature (Wiemers et al. 2018; Wiemers et al. 2020) and discussions with colleagues, the prevailing usage of Muschampia is preferred by most of them. We do not have a solution to this problem, and it joins the list of similar problems when there is a disagreement between the articles of the ICZN Code and the community opinion (e.g., gender agreement ICZN Code articles vs. the community of Lepidopterists is an example of a more significant, but similar in spirit, issue). We see the advantages of both treatments in this specific case. Indeed, Muschampia has been a widely used name familiar to many people, and it seems that the community opinion should outweigh the priority of a semi-random decision made by the first reviser. Conversely, the rules are universal, clearly spelled out, and long-term, but the community opinions are subject to change, especially with generational changes. Furthermore, the name Sloperia is shorter and may be easier for newcomers to learn, fitting the array of other names in the subtribe, such as Spialia, Gomalia, and Favria. Whatever the decision, it is not a question to be solved by scientific tools. It is possible that after some period of deliberation, the community might embrace Sloperia (it is not a bad option at all), or the community would be more ready to make a stronger case for ICZN to issue an opinion. However, whichever path is taken, we strongly oppose solving this (and other) nomenclatural problem by fiddling with taxonomy just so that the name Muschampia is still in use (e.g., with the sole purpose of accommodating the use of Muschampia, treating both Muschampia and Sloperia as valid genera).
Supplementary Material
Acknowledgments
We acknowledge Ping Chen and Ming Tang for their excellent technical assistance. We are grateful to David Grimaldi and Courtney Richenbacher (AMNH: American Museum of Natural History, New York, NY, USA), Blanca Huertas, David Lees, and Geoff Martin (BMNH: Natural History Museum, London, UK), Jim Fetzner, Bob Androw, Vanessa Verdecia, Cat Giles, and the late John Rawlins (CMNH: Carnegie Museum of Natural History, Pittsburgh, PA, USA), the late Paul Opler, Chuck Harp, and the late Boris Kondratieff (CSUC: Colorado State University Collection, Fort Collins, CO, USA), Weiping Xie and Giar-Ann Kung (LACM: Los Angeles County Museum of Natural History, Los Angeles, CA, USA), Naomi Pierce, Phil Perkins, and Rachel Hawkins (MCZ: Museum of Comparative Zoology, Harvard University, Cambridge, MA, USA), John R. MacDonald and Richard L. Brown (MEM: Mississippi Entomological Museum, Starkville, MS, USA), Théo Léger, Viola Richter, and Wolfram Mey (MFNB: Museum für Naturkunde, Berlin, Germany), Andrei Sourakov, Andrew D. Warren, Debbie Matthews-Lott, Riley J. Gott, and Keith R. Willmott (MGCL: McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA), Rodolphe Rougerie (MNHP: Muséum National d’Histoire Naturelle, Paris, France), Gerardo Lamas (MUSM: Museo de Historia Natural, Lima, Peru), Edward G. Riley, Karen Wright, and John Oswald (TAMU: Texas A&M University Insect Collection, College Station, TX, USA), Alex Wild (TMMC: University of Texas Biodiversity Center, Austin, TX, USA), Jeff Smith and Lynn Kimsey (UCDC: Bohart Museum of Entomology, University of California, Davis, CA, USA), Robert K. Robbins, John M. Burns, and Brian Harris (USNM: National Museum of Natural History, Smithsonian Institution, Washington, DC, USA), and Axel Hausmann, Andreas Segerer, and Ulf Buchsbaum (ZSMC: Zoologische Staatssammlung München, Germany), for granting access to the collections under their care, sampling specimens, and stimulating discussions; to Ernst Brockmann, Matthew J. W. Cock, Bill R. Dempwolf, Bernard Hermier, the late Edward C. Knudson (specimens now in MGCL), the late James A. Scott, and Mark Walker for specimens and leg samples, to Gerardo Lamas and Jonathan P. Pelham for discussions, advice, helpful suggestions, patiently answering numerous questions, and sharing their research files, and to Bernard Hermier, John A. Shuey, and J. Bruce Walsh for critical reviews of the manuscript and many helpful corrections. We are indebted to the California Department of Fish and Game for collecting permit SC13645, Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the research permit 08-02Rev, to U. S. National Park Service for the research permits: Big Bend (Raymond Skiles) for BIBE-2004-SCI-0011 and Yellowstone (Erik Oberg and Annie Carlson) for YELL-2017-SCI-7076, and to the National Environment & Planning Agency of Jamaica for the permission to collect specimens. We acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources. This study was supported in part by the HHMI Investigator funds and by grants from the National Institutes of Health GM127390 and the Welch Foundation I-1505 to N.V.G.
Footnotes
ZooBank registration. http://zoobank.org/B9AFA1A9-8664-4F31-B4D9-ACF7780C7CC6
Contributor Information
Jing Zhang, Eugene McDermott Center for Human Growth and Development and Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-8816 USA.
Qian Cong, Eugene McDermott Center for Human Growth and Development and Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-8816 USA.
Jinhui Shen, Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-9050 USA.
Leina Song, Department of Biophysics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-9050 USA.
Nick V. Grishin, Departments of Biophysics and Biochemistry, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX, 75390-9050 USA
Literature Cited
- Bachtrog D, Thornton K, Clark A, Andolfatto P. 2006. Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution 60(2): 292–302. [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nature Methods 12(1): 59–60. [DOI] [PubMed] [Google Scholar]
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PD. 2008. DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proceedings of the National Academy of Sciences of the United States of America 105(17): 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Borek D, Robbins RK, Opler PA, Otwinowski Z, Grishin NV. 2017. When COI barcodes deceive: complete genomes reveal introgression in hairstreaks. Proceedings of the Royal Society B: Biological Sciences 284(1848): 20161735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Zhang J, Li W, Kinch LN, Calhoun JV, Warren AD, Grishin NV. 2021. Genomics reveals the origins of historical specimens. Molecular Biology and Evolution 38(5): 2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Zhang J, Grishin NV. 2019a. Genomic determinants of speciation [Preprint]. bioRxiv BIORXIV/2019/837666. Available at https://www.biorxiv.org/content/10.1101/837666v1 (Last accessed November 2023.) [Google Scholar]
- Cong Q, Zhang J, Shen J, Grishin NV. 2019b. Fifty new genera of Hesperiidae (Lepidoptera). Insecta Mundi 0731: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WH. 1953. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part III. Pyrginae. Section 2. The Trustees of the British Museum (Natural History); London. v + 246 p., pls. 26–53. [Google Scholar]
- Fan X, Chiba H, Huang Z, Fei W, Wang M, Safian S. 2016. Clarification of the phylogenetic framework of the tribe Baorini (Lepidoptera: Hesperiidae: Hesperiinae) inferred from multiple gene sequences. PLoS One 11(7): e0156861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godman FD. 1907. Notes on the American species of Hesperiidae described by Plötz. Annals and Magazine of Natural History (7) 20(16): 132–155. [Google Scholar]
- Grishin NV. 2012. Uncus shaped akin to elephant tusks defines a new genus for two very different-in-appearance Neotropical skippers (Hesperiidae: Pyrginae). The Journal of Research on the Lepidoptera 45: 101–112. [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270(1512): 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemming AF. 1967. The generic names of the butterflies and their type-species (Lepidoptera: Rhopalocera). Bulletin of the British Museum (Natural History). Entomology Suppl. 9: 1–509. [Google Scholar]
- Huang Z-F, Chiba H, Jin J, Kizhakke AG, Wang M, Kunte K, Fan X-L. 2019. A multilocus phylogenetic framework of the tribe Aeromachini (Lepidoptera: Hesperiidae: Hesperiinae), with implications for taxonomy and biogeography. Systematic Entomology 44(1): 163–178. [Google Scholar]
- ICZN [International Commission on Zoological Nomenclature]. 1999. International code of zoological nomenclature. Fourth edition. International Trust for Zoological Nomenclature; London. xxx + 306 p. [Google Scholar]
- Lemes JRA, Siewert RR, Mielke OHH, Casagrade MM, Warren AD. 2023. Taxonomic and distributional notes on Bolla tepeca (Bell, 1942), new combination (Lepidoptera: Hesperiidae: Pyrginae). Tropical Lepidoptera Research 33(2): 102–110. [Google Scholar]
- Li W, Cong Q, Shen J, Zhang J, Hallwachs W, Janzen DH, Grishin NV. 2019. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proceedings of the National Academy of Sciences of the United States of America 116(13): 6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Sourakov A, Zakharov E. 2016. DNA barcodes as a tool in biodiversity research: testing pre-existing taxonomic hypotheses in Delphic Apollo butterflies (Lepidoptera, Papilionidae). Systematics and Biodiversity 14: 599–613. [Google Scholar]
- Mabille P 1903. Lepidoptera Rhopalocera. Fam. Hesperidae. Genera Insectorum 17a: 1–78. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plötz C 1882. Einige Hesperiinen-Gattungen und deren Arten. Berliner entomologische Zeitschrift 26(2): 253–266. [Google Scholar]
- Plötz C 1884. Die Hesperiinen-Gruppe der Achlyoden. Jahrbücher des nassauischen Vereins für Naturkunde 37: 1–55. [Google Scholar]
- Rambaut A 2018. FigTree, version 1.4.4. Available at http://tree.bio.ed.ac.uk/software/figtree/ (Last accessed November 2023.)
- Sahoo RK, Warren AD, Collins SC, Kodandaramaiah U. 2017. Hostplant change and paleoclimatic events explain diversification shifts in skipper butterflies (Family: Hesperiidae). BMC Evolutionary Biology 17(1): 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo RK, Warren AD, Wahlberg N, Brower AV, Lukhtanov VA, Kodandaramaiah U. 2016. Ten genes and two topologies: an exploration of higher relationships in skipper butterflies (Hesperiidae). PeerJ 4: e2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaus W 1913. New species of Rhopalocera from Costa Rica. Proceedings of the Zoological Society of London 1913(3): 339–367. [Google Scholar]
- Shen J, Cong Q, Borek D, Otwinowski Z, Grishin NV. 2017. Complete genome of Achalarus lyciades, the first representative of the Eudaminae subfamily of skippers. Current Genomics 18(4): 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint EFA, Braby MF, Müller CJ, Petrie EA, Kawahara AY. 2022. Molecular phylogeny, systematics and generic classification of the butterfly subfamily Trapezitinae (Lepidoptera: Papilionoidea: Hesperiidae). Zoological Journal of the Linnean Society 195: 1407–1421. [Google Scholar]
- Toussaint EFA, Breinholt JW, Earl C, Warren AD, Brower AVZ, Yago M, Dexter KM, Espeland M, Pierce NE, Lohman DJ, Kawahara AY. 2018. Anchored phylogenomics illuminates the skipper butterfly tree of life. BMC Evolutionary Biology 18(1): 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint EFA, Chiba H, Yago M, Dexter KM, Warren AD, Storer C, Lohman DJ, Kawahara AY. 2021a. Afrotropics on the wing: phylogenomics and historical biogeography of awl and policeman skippers. Systematic Entomology 46(1): 172–185. [Google Scholar]
- Toussaint EFA, Ellis EA, Gott RJ, Warren AD, Dexter KM, Storer C, Lohman DJ, Kawahara AY. 2021b. Historical biogeography of Heteropterinae skippers via Beringian and post-Tethyan corridors. Zoologica Scripta 50(1): 100–111. [Google Scholar]
- Toussaint EFA, Vila R, Yago M, Chiba H, Warren AD, Aduse-Poku K, Storer C, Dexter KM, Maruyama K, Lohman DJ, Kawahara AY. 2019. Out of the Orient: Post-Tethyan transoceanic and trans-Arabian routes fostered the spread of Baorini skippers in the Afrotropics. Systematic Entomology 44(4): 926–938. [Google Scholar]
- Warren AD, Ogawa JR, Brower AVZ. 2008. Phylogenetic relationships of subfamilies and circumscription of tribes in the family Hesperiidae (Lepidoptera: Hesperioidea). Cladistics 24(5): 642–676. [Google Scholar]
- Warren AD, Ogawa JR, Brower AVZ. 2009. Revised classification of the family Hesperiidae (Lepidoptera: Hesperioidea) based on combined molecular and morphological data. Systematic Entomology 34(3): 467–523. [Google Scholar]
- Warren BCS. 1926. Monograph of the tribe Hesperiidi (European species) with revised classification of the subfamily Hesperiinae (Palaearctic species) based on the genital armature of the males. Transactions of the entomological Society of London 74: 1–170. [Google Scholar]
- Wiemers M, Balletto E, Dincă V, Fric ZF, Lamas G, Lukhtanov V, Munguira ML, van Swaay CAM, Vila R, Vliegenthart A, Wahlberg N, Verovnik R. 2018. An updated checklist of the European Butterflies (Lepidoptera, Papilionoidea). ZooKeys 811: 9–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers M, Chazot N, Wheat CW, Schweiger O, Wahlberg N. 2020. A complete time-calibrated multi-gene phylogeny of the European butterflies. ZooKeys 938: 97–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Burns JM, Grishin NV. 2022a. Checking the checkered taxonomy of Plötz’s checkered skippers (Hesperiidae: Pyrgini). The Taxonomic Report of the International Lepidoptera Survey 10(5): 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Grishin NV. 2023a. Thirteen new species of butterflies (Lepidoptera: Hesperiidae) from Texas. Insecta Mundi 0969: 1–58. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Brockmann E, Grishin NV. 2019a. Genomes reveal drastic and recurrent phenotypic divergence in firetip skipper butterflies (Hesperiidae: Pyrrhopyginae). Proceedings of the Royal Society B: Biological Sciences 286(1903): 20190609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Brockmann E, Grishin NV. 2019b. Three new subfamilies of skipper butterflies (Lepidoptera, Hesperiidae). ZooKeys 861: 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Grishin NV. 2022b. Taxonomic changes suggested by the genomic analysis of Hesperiidae (Lepidoptera). Insecta Mundi 0921: 1–135. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, Grishin NV. 2021. Genomics-guided refinement of butterfly taxonomy. The Taxonomic Report of the International Lepidoptera Survey 9(3): 1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2022c. Genomic DNA sequencing reveals two new North American species of Staphylus (Hesperiidae: Pyrginae: Carcharodini). The Taxonomic Report of the International Lepidoptera Survey 10(4): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2023b. Butterfly classification and species discovery using genomics. The Taxonomic Report of the International Lepidoptera Survey 11(3): 1–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2023c. A taxonomic list of the Old World genera in the subfamily Hesperiinae (Hesperiidae) arranged into tribes. The Taxonomic Report of the International Lepidoptera Survey 11(2): 1–6.36817548 [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Grishin NV. 2023d. Supplementary information to “Genomics-based taxonomic rearrangement of Achlyodini and Carcharodini (Lepidoptera: Hesperiidae: Pyrginae)”. Table S1, Table S2, and DNA sequences of exons with diagnostic characters. Available at https://osf.io//8be5h/ (Last accessed November 2023.) [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dolibaina DR, Cong Q, Shen J, Song L, Mielke CGC, Casagrade MM, Mielke OHH, Grishin NV. 2023d. Taxonomic notes on Neotropical Hesperiidae (Lepidoptera). Zootaxa 5271(1): 91–114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


