Abstract
Genomic sequencing of worldwide butterfly fauna followed by phylogenetic analysis of protein-coding genes informs butterfly classification throughout the taxonomic hierarchy, from families to species. As a rule, we attribute the same taxonomic rank to more prominent clades of comparable divergence (i.e., at the same level in the tree). For species delimitation, we use criteria based on relative genetic differentiation and the extent of gene exchange between populations. We analyze the current taxonomic classification of butterflies in the light of genomic phylogenies and encounter clades that correspond to yet unnamed taxa. As a result, 11 tribes, 33 subtribes, 2 genera, 11 subgenera, and 12 species are proposed as new. New tribes are: in family Nymphalidae Rafinesque, 1815: in subfamily Heliconiinae Swainson, 1822: Vindulini Grishin, trib. n. (type genus Vindula Hemming, 1934) and Algiini Grishin, trib. n. (type genus Algia Herrich-Schäffer, 1864), Lebadeini Grishin, trib. n. (type genus Lebadea C. Felder, 1861, in subfamily Limenitidinae Behr, 1864), and Amnosiini Grishin, trib. n. (type genus Amnosia E. Doubleday, 1849, in subfamily Pseudergolinae Jordan, 1898) and in family Lycaenidae [Leach], [1815]: in subfamily Aphnaeinae Distant, 1884: Cigaritini Grishin, trib. n. (type genus Cigaritis Donzel, 1848) and Axiocersini Grishin, trib. n. (type genus Axiocerses Hübner, [1819]) and in subfamily Theclinae: Drinini Grishin, trib. n. (type genus Drina Nicéville, 1890), Hypochrysopini Grishin, trib. n. (type genus Hypochrysops C. Felder & R. Felder, 1860), Jalmenini Grishin, trib. n. (type genus Jalmenus Hübner, 1818), Pseudalmenini Grishin, trib. n. (type genus Pseudalmenus H. H. Druce, 1902), and Rapalini Grishin, trib. n. (type genus Rapala F. Moore, 1881). New subtribes are: in Papilionidae Latreille, [1802], Meandrusina Grishin, subtrib. n. (type genus Meandrusa F. Moore, 1888, in Papilionini Latreille, [1802]); in Pieridae Swainson, 1820: Gandacina Grishin, subtrib. n. (type genus Gandaca F. Moore, 1906, in Coliadini Swainson, 1821), Hebomoiina Grishin, subtrib. n. (type genus Hebomoia Hübner, [1819], in Anthocharidini Scudder, 1889), and Pseudopierina Grishin, subtrib. n. (type genus Pseudopieris Godman & Salvin, 1890, in Dismorphiini Schatz, 1886); in Nymphalidae: Lachnopterina Grishin, subtrib. n. (type genus Lachnoptera E. Doubleday, 1847, in Algiini Grishin, trib. n.), in Vagrantini Pinratana & Eliot, 1996: Terinosina Grishin, subtrib. n. (type genus Terinos Boisduval, 1836) and Smerinina Grishin, subtrib. n. (type genus Smerina Hewitson, 1874), in Adoliadini Doubleday, 1845: Evenaina Grishin, subtrib. n. (type genus Evena Westwood, 1850) and Pseudathymina Grishin, subtrib. n. (type genus Pseudathyma Staudinger, 1891), and Kumothalina Grishin, subtrib. n. (type genus Kumothales Overlaet, 1940, in Cymothoini Dhungel & Wahlberg, 2018); in Riodinidae Grote, 1895 (1827): Teratophthalmina Grishin, subtrib. n. (type genus Teratophthalma Stichel, 1909, in Mesosemiini Bates, 1859), Argyrogrammanina Grishin, subtrib. n. (type genus Argyrogrammana Strand, 1932, in Symmachiini Reuter, 1896), in the tribe Calydnini Seraphim, Freitas & Kaminski, 2018: Echenaidina Grishin, subtrib. n. (type genus Echenais Hübner, [1819]) and Echydnina Grishin, subtrib. n. (type genus Echydna J. Hall, 2002), and Cariina Grishin, subtrib. n. (type genus Caria Hübner, 1823, in Riodinini Grote, 1895 (1827); in Lycaenidae: Megalopalpina Grishin, subtrib. n. (type genus Megalopalpus Röber, 1886, in Miletini Reuter, 1896), Pseudaletidina Grishin, subtrib. n. (type genus Pseudaletis H. H. Druce, 1888, in Cigaritini Grishin, trib. n.), in Aphnaeini Distant, 1884: Aloeidina Grishin, subtrib. n. (type genus Aloeides Hübner, [1819]) and Phasisina Grishin, subtrib. n. (type genus Phasis Hübner, [1819]), Pilodeudorigina Grishin, subtrib. n. (type genus Pilodeudorix H. H. Druce, 1891, in Rapalini Grishin, trib. n.), Hemiolaina Grishin, subtrib. n. (type genus Hemiolaus Aurivillius, 1922, in Oxylidini Eliot, 1973), Cupidopsina Grishin, subtrib. n. (type genus Cupidopsis Karsch, 1895, in Hypotheclini Eliot, 1973), and in tribe Polyommatini Swainson, 1827: Theclinesthina Grishin, subtrib. n. (type genus Theclinesthes Röber, 1891), Azanina Grishin, subtrib. n. (type genus Azanus F. Moore, 1881), Unina Grishin, subtrib. n. (type genus Una Nicéville, 1890), Ionolycina Grishin, subtrib. n. (type genus Ionolyce Toxopeus, 1929), Pithecopina Grishin, subtrib. n. (type genus Pithecops Horsfield, 1828), Zizulina Grishin, subtrib. n. (type genus Zizula Chapman, 1910), Jamidina Grishin, subtrib. n. (type genus Jamides Hübner, [1819]), Fameganina Grishin, subtrib. n. (type genus Famegana Eliot, 1973), Oboroniina Grishin, subtrib. n. (type genus Oboronia Karsch, 1893), and Uranothaumatina Grishin, subtrib. n. (type genus Uranothauma Butler, 1895); and in Hesperiidae Latreille, 1809, Cupithina Grishin, subtrib. n. (type genus Cupitha F. Moore, 1884, in Astictopterini Swinhoe, 1912). The new genera are Balenga Grishin, gen. n. (type species Proteides balenge Holla nd, 1891, in Gretnini Grishin, 2019) and Tulsia Grishin, gen. n. (type species Parnara tulsi Nicéville, 1884, in Baorini Doherty, 1886). New subgenera are: in Pieridae, Lirinia Grishin, subgen. n. (type species Terias lirina H. Bates, 1861, in genus Pyrisitia A. Butler, 1870); in Nymphalidae: Hyperanartia Grishin, subgen. n. (type species Vanessa dione Latreille, [1813], in genus Hypanartia Hübner, [1821]) and Paranartia Grishin, subgen. n. (type species Hypanartia hippomene Hübner, [1823], in genus Vanessa [Fabricius], 1807); in Lycaenidae, Auricirrus Grishin, subgen. n. (type species Papilio thysbe Linnaeus, 1764, in genus Chrysoritis Butler, 1898); and in Hesperiidae: Isocleros Grishin, subgen. n. (type species Pamphila (?) mackenii Trimen, 1868, in genus Acleros Mabille, 1885), Mesna Grishin, subgen. n. (type species Parnara leucophaea Holland, 1894, in genus Fresna Evans, 1937), Lippina Grishin, subgen. n. (type species Carystus telesinus Mabille, 1878, in genus Xanthoneura Eliot, 1978), Ganda Grishin, subgen. n. (type species Zophopetes ganda Evans, 1937, in genus Leona Evans, 1937), Zarida Grishin, subgen. n. (type species Hesperia lacida Hewitson, 1876, in genus Gretna Evans, 1937), and in genus Gegenes Hübner, 1819: Flanga Grishin, subgen. n. (type species Parnara perobscura H. H. Druce, 1912), and Havea Grishin, subgen. n. (type species Hesperia havei Boisduval, 1833). New species are: in Nymphalidae: Microtia elvira Grishin, sp. n. (type locality in the USA: AZ, Pima/Santa Cruz Cos.) and Cyllopsis brocki sp. n. (type locality in Mexico: Sonora, Yécora), in Riodinidae, Argyrogrammana astuta Grishin, sp. n. (type locality in Peru: Madre de Dios), in Hesperiidae: Cecropterus (Thorybes) rockiensis Grishin, sp. n. (type locality in USA: CO, Jefferson Co.), Cecropterus (Thorybes) floridianus Grishin, sp. n. (type locality in USA: FL, Volusia Co.), Cecropterus (Thorybes) oaxacensis Grishin, sp. n. (type locality in Mexico: Oaxaca), Nascus (Bron) lux Grishin, sp. n. (type locality in Brazil: Amapá), Cogia chiagua Grishin, sp. n. (type locality in Guatemala), Celotes sabinus Grishin, sp. n. (type locality in USA: AZ, Pima Co.), Acleros togo Grishin, sp. n. (type locality in Togo), Ceratrichia notata Grishin, sp. n. (type locality in Central African Republic), and Semalea malawi Grishin, sp. n. (type locality in Malawi). Furthermore, we elevate a tribe to subfamily, resurrect 3 tribes, 3 subtribes, 5 genera (and confirm 1), 3 subgenera, change the rank of 5 currently recognized tribes to subtribes (and confirm 1), 10 genera to subgenera, synonymize 4 genera, and present evidence to support 21 taxa as species instead of subspecies and 3 taxa as subspecies instead of synonyms. Namely, we reinstate Liphyrinae Doherty, 1889 as a subfamily (was a tribe of Miletinae Reuter, 1896), treat the following as tribes: Leptidiini Grote, 1897 (in Dismorphiinae Schatz, 1886) and in Theclinae Swainson, 1830: Surendrini Koçak & Seven, 1997 (not a subtribe of Arhopalini Bingham, 1907) and Myrinini Toxopeus, 1929 (not a synonym of Amblypodiini Doherty, 1886), and subtribes: Callidryina Kirby, 1896 and Gonepterygina Verity, 1920 (in Coliadini Swainson, 1821), Abrotina Hemming, 1960 and Bebeariina Hemming, 1960 (in Adoliadini Doubleday, 1845), and Sarotina Bridges, 1988 (in Helicopini Stichel, 1928). We confirm Libytheana Michener, 1943 as a valid genus, not a junior subjective synonym of Prolibythea Scudder, 1889 and resurrect from synonymy the following genera: Pseudanaphaeis Bernardi, 1953 (not Belenois Hübner, [1819]), Charmion Nicéville, 1894 (not Celaenorrhinus Hübner, [1819]), Sape Mabille, 1891 (not Sarangesa F. Moore, [1881]), Milena Evans, 1912 (not Caltoris Swinhoe, 1893) and subgenera: Neofieldia Özdikmen, 2008 and Bassaris Hübner, [1821] of Vanessa [Fabricius], 1807 and Spindasis Wallengren, 1857 of Cigaritis Donzel, 1848. We change the status of the following taxa from tribes to subtribes: Nathalina Bálint, 2022 and Kricogonina Bálint, 2022 of Euremini Grote, 1898, Tarakina Eliot, 1973 of Spalgini, 1929, Horagina Swinhoe, 1910 and Loxurina Swinhoe, 1910 of Cheritrini Swinhoe, 1910, and Niphandina Sibatani & Ito, 1942 of Polyommatini; from genus to subgenus: Leucidia E. Doubleday, 1847 of Abaeis Hübner, [1819], Cesa Seven, 1997 of Crudaria Wallengren, 1875, Paralycaeides Nabokov, 1945 of Itylos Draudt, 1921, Eldoradina Balletto, 1993 of Nabokovia Hemming, 1960, Pyrrhochalcia Mabille, 1904 of Coeliades Hübner, 1818, Paracleros Berger, 1978 of Acleros Mabille, 1885, Mopala Evans, 1937 of Leona Evans, 1937, Afrogegenes Jong & Coutsis, 2017 and Torbenlarsenia Kemal & Koçak, 2020 of Gegenes Hübner, 1819, and Zenonoida Fan & Chiba, 2016 of Zenonia Evans, 1935; and from genus to junior subjective synonym: Vansomerenia Heath, 1997 of Chloroselas Butler, 1886, Ceratricula Larsen, 2013 of Paronymus Aurivillius, 1925, Xanthodisca Aurivillius, 1925 of Semalea Holland, 1896, Perrotia Oberthür, 1916 of Galerga Mabille, 1898. The following taxa are species, not subspecies or synonyms: Pyrisitia mayobanex (M. Bates, 1939), stat. nov. and Pyrisitia memulus (A. Butler, 1871), stat. rest. (not Pyrisitia dina (Poey, 1832)), Abaeis gratiosa (E. Doubleday, 1847), stat. rest. and Abaeis angulata (Wallengren, 1860), stat. rest. (not Abaeis arbela (Geyer, 1832)), Teriocolias doris (Röber, 1909), stat. rest. (not Teriocolias deva (E. Doubleday, 1847)), Vanessa madegassorum (Aurivillius, 1899), stat. nov. (not Vanessa hippomene (Hübner, 1823)), Eresia (Anthanassa) seminole Skinner, 1911, stat. rev. (not Eresia (Anthanassa) texana (W. H. Edwards, 1863)), Cecropterus (Thorybes) albosuffusa (H. Freeman, 1943), stat. nov. and Cecropterus (Thorybes) indistinctus (Austin & J. Emmel, 1998), stat. nov. (not Cecropterus (Thorybes) pylades (Scudder, 1870)), Cogia hiska Evans, 1953, stat. nov. (not Cogia hippalus (W. H. Edwards, 1882)), Cogia moschus (W. H. Edwards, 1882), stat. rest. (not Cogia caicus (Herrich-Schäffer, 1869)), Pholisora albicirrus Glassberg 2023, stat. nov. (not Pholisora catullus (Fabricius, 1793)), Acleros (Isocleros) instabilis Mabille, 1889, stat. rest. and Acleros (Isocleros) olaus (Plötz, 1884), stat. rest. (not Acleros (Isocleros) mackenii (Trimen, 1868)), Fresna (Mesna) bassa (Lindsey & L. Miller, 1965), stat. nov., comb. nov. (not Meza leucophaea (Holland, 1894)), Paronymus volta (L. Miller, 1971), stat. nov., comb. nov. (not Meza cybeutes (Holland, 1894)), Paronymus indeterminabilis (Strand, 1912), stat. rest., comb. nov. and Paronymus congdoni (Larsen, 2013), stat. nov., comb. nov. (not Ceratricula semilutea (Mabille, 1891)), Semalea corvinus (Mabille, 1890), stat. rest. (not Semalea sextilis (Plötz, 1886)), Xanthoneura patmapana (Fruhstorfer, 1911), stat. nov. (not Xanthoneura corissa (Hewitson, 1876)), Gretna capra Evans, 1937, stat. nov. (not Gretna carmen Evans, 1937), and Lerodea dysaules Godman, 1900, stat. rest. (not a synonym of Lerodea arabus (W. H. Edwards, 1882)), while Vernia verna sequoyah (H. Freeman, 1942), stat. rest. (not a synonym of Vernia verna W. H. Edwards, 1862) and Euphyes vestris osceola (Lintner, 1878), stat. rev. (not a synonym of Euphyes vestris vestris (Boisduval, 1852)) are subspecies. In addition, we propose new genus-species combinations: Pyrisitia amelia (Poey, [1852]), Pyrisitia lirina (H. Bates, 1861), Abaeis paulina (H. Bates, 1861), Abaeis xantochlora (Kollar, 1850), Abaeis fabiola (C. Felder & R. Felder, 1861), Abaeis tupuntenem (Lichy, 1976), and Abaeis adamsi (Lathy, 1898) (not Eurema Hübner, [1819]), Shijimia potanini (Alphéraky, 1889) (not Tongeia Tutt, 1908), Fresna (Mesna) larea (Neave, 1910), Fresna (Mesna) leucophaea (Holland, 1894), Fresna (Mesna) mabea (Holland, 1894), Paronymus banda (Evans, 1937), Paronymus cybeutes (Holland, 1894), Paronymus elba (Evans, 1937), Paronymus gardineri (Collins & Larsen, 2008), Paronymus indusiate (Mabille, 1891), and Paronymus mabillei (Holland, 1893) (not Meza Hemming, 1939), Paronymus punctata (Holland, 1896) (not Ceratrichia Butler, 1870), Galerga ariel (Mabille, 1878) (not Xanthodisca Aurivillius, 1925), Isoteinon anomoeus (Plötz, 1879), Isoteinon bruno (Evans, 1937), Isoteinon inornatus (Trimen, 1864), and Isoteinon punctulata (A. Butler, 1895) (not Astictopterus C. Felder & R. Felder, 1860), Borbo gemella (Mabille, 1884) (not Torbenlarsenia Kemal & Koçak, 2020), and Gegenes (Torbenlarsenia) cottrelli (Larsen, 2013), Gegenes (Torbenlarsenia) fallax (Gaede, 1916), Gegenes (Torbenlarsenia) fanta (Evans, 1937), Gegenes (Torbenlarsenia) micans (Holland, 1896), and Gegenes (Torbenlarsenia) sirena (Evans, 1937) (not Borbo Evans, 1949); and a new species-subspecies combination Cogia hiska hester Evans, 1953 (not Cogia hippalus). Acleros nyassicola Strand, 1921 is a junior subjective synonym of Acleros (Isocleros) olaus (Plötz, 1884), not of Acleros (Isocleros) mackenii (Trimen, 1868). We transferred the tribes Oxylidini Eliot, 1973, Remelanini Eliot, 1973, and Hypolycaenini Swinhoe, 1910 from Theclinae Swainson, 1831 to Polyommatinae Swainson, 1827. We conclude that [No genus] osibius Draudt, 1924 is an unavailable name. The lectotype is designated for Eudamus caicus Herrich-Schäffer, 1869 (type locality likely in Mexico: Oaxaca, as deduced by sequence comparison), and the neotype is designated for Telemiades solon Plötz, 1882 (type locality becomes Brazil: Bahia). The type localities of Ceratrichia punctata Holland, 1896 and Osmodes staudingeri Holland, 1896 are Sierra Leone: Freetown and Cameroon: Efoulan, respectively, as determined from the locality labels of primary type specimens. Finally, we provide taxonomic lists for Euremini (to worldwide subgenera and American species) and Lycaenidae (to subtribes).
Additional keywords: taxonomy, classification, genomics, phylogeny, biodiversity
INTRODUCTION, CONCEPTS, AND METHODS
This work continues a series of studies derived from genomic sequencing of butterflies and uses the same principles and methods (Cong et al. 2019a, b; Li et al. 2019; Zhang et al. 2019a–d; Cong et al. 2020; Zhang et al. 2020; Cong et al. 2021; Zhang et al. 2021; Robbins et al. 2022; Zhang et al. 2022b, c; Zhang et al. 2023c, e). The goal is to improve butterfly classification through genomic data analyses. The approach is that of screening. Specimens of various taxa throughout the butterfly phylogeny worldwide are selected. The emphasis is placed on species recorded from the United States, and specimens across the range are taken for each species. Most specimens come from collections, both museum and private (see acknowledgments section for their list); specimen age varies from ~250 years to recently collected. Whenever possible, we sequence primary type specimens to have an objective reference for the name (Zhang et al. 2022a). Primarily, legs are used for DNA extraction. Our protocol is non-destructive, and legs are preserved. Extracted DNA is fragmented (unless a specimen is old and its DNA is already short) and sequenced at 150 bp on Illumina next-generation sequencing platform. We do not rely on the amplification of specific genes or segments, and every extracted DNA piece is sequenced. Therefore, the protocol succeeds with very old specimens, in which DNA may be fragmented into 30–50 bp segments.
Sequence data (i.e., sequence segments of 150 bp or shorter) of each specimen are used to assemble exons of protein-coding genes as guided by a reference genome available for the phylogenetically closest species. These protein-coding genes are used for phylogeny reconstruction. Three trees are constructed using IQtree v1.6.12 under the GTR+GAMMA model (Nguyen et al. 2015): from autosomes in the nuclear genome, the gene predicted to be in the Z chromosome, and the mitochondrial genome. To decrease computational load, 100,000 codons (resulting in 300,000 base pairs, about 2% of the total) are randomly selected from the entire dataset to use for the nuclear trees. Statistical support for branches is estimated from 100 replicates of 10,000 codons each, sampled from the total set of codons, and trees are constructed from each replicate. Statistical support value (from 0 to 100) is simply the number of replicates with a particular bipartition identical to the one in the 100,000-codon tree. For further methods details, see our previous publications (Li et al. 2019; Zhang et al. 2022b).
The resulting trees were visualized, rotated, and colored in FigTree (Rambaut 2018). The current taxonomic classification was overlaid on the trees to find non-monophyletic taxa and clades corresponding to taxa without names. Genomic trees frequently reveal “levels,” i.e., timepoints when diversification occurred in several lineages independently (Zhang et al. 2021). These “synchronized” diversifications result from geological events affecting all major lineages at the same time, offering an opportunity to match taxonomic ranks (tribe, subtribe, genus, subgenus) to the levels in genomic trees, which leads to a more objective and internally consistent classification tied to both genetic differentiation and paleontological history. In classification decisions, we strongly rely on genomic trees and use morphological considerations as secondary evidence to rationalize the results. This is because, contrary to a small set of gene markers, genomes offer a comprehensive picture of an organism richer than the morphology of adults typically used to classify butterflies. Genomes encode life histories, habitat and mating preferences, and food sources. All this information is present in the genomic sequence. While we lack the knowledge to extract it and predict phenotypes, we can use a genetic equivalent of this information in an aggregate, in a way that the organism balances it in its genome (we select random codons from all protein-coding genes), to deduce phylogenetically sound taxonomic classification.
Taxa we define are monophyletic groups in nuclear genome trees that correspond to prominent clades. By “prominent,” we mean tree branches strongly supported statistically (typically by 100% of replicates) and usually longer than neighboring branches. The length of a branch is proportional to the number of base-pair substitutions along the branch. Not only are longer branches better supported statistically, but the larger number of genetic changes along them likely leads to more pronounced phenotypic changes that should be reflected in some morphological characters, not necessarily in adults, but could be in immature stages or other aspects of the phenotype. Nevertheless, due to highly non-linear relationships between the number of genetic changes and visually drastic phenotypic differences (Zhang et al. 2019a), there are short tree branches that correspond to visually recognizable taxa, and each case should be considered individually. It is unclear, however, if some drastic phenotypic change in adult appearance that was caused by a small number of genetic changes (maybe even a single inversion of a genomic segment) should be grounds for erection of a separate taxon for this lineage because all other characters, e.g., those of caterpillars, would remain rather similar to the relatives of this lineage. Generally, we prefer to avoid monotypic (or nearly monotypic, i.e., consisting of very close relatives) taxa unless they cannot be confidently assigned to other taxa of the same rank or show prominent genetic differentiation from them at the level of the tree that corresponds to their rank. Furthermore, currently employed taxonomy is considered, and currently used names and their taxonomic ranks serve, on average, as a reference point to define levels in the trees and new taxa.
Tribes are defined as a prominent level in genomic trees between subfamilies and genera that supports most tribes as they are currently circumscribed in each subfamily. Subtribes correspond to a notable tree level between tribes and genera. Genera are defined as the most prominent level in genomic trees between tribes and species that largely corresponds to the current classification into genera. Subgenera correspond to a rather prominent level between genera and species. We attempt to define secondary levels (subtribes and subgenera) in the classification whenever possible if we see noteworthy clustering of genera or species into groups of relatives prominently differentiated from other such groups. We find these subdivisions useful in taxonomic lists, even if for no other reason than to place closer relatives close to each other in alphabetically sorted hierarchical lists.
Species are delineated by a combination of criteria that include genetic differentiation in the Z chromosome measured by Fst (>0.20 usually corresponds to distinct species) and gene exchange Gmin (<0.05 for distinct species) (Cong et al. 2019a), COI barcode difference (typically >2% for distinct species) (Hebert et al. 2003) and its correlation with phenotypic differences (Lukhtanov et al. 2016), and the prominence of species-level clades (Zhang et al. 2022c). However, COI barcodes (together with mitochondria) frequently introgress between species (Bachtrog et al. 2006; Cong et al. 2017a), and some distinct species may possess highly similar or identical barcodes (Burns et al. 2008; Zhang et al. 2023a). See the “Species, subspecies, and genomics” section in Zhang et al. (2022a) for further discussion.
Sections below are arranged in the taxonomic order deduced from genome-scale phylogeny complemented by phenotypic considerations. For most new taxa, in addition to brief phenotypic diagnoses (genitalia terminology follows Carneiro et al. (2013), we use “harpe” for Evans’ “cuiller” (Evans 1949)) frequently accompanied by references that discuss and illustrate morphological characters in greater detail, we provide diagnostic DNA characters in the nuclear genome and/or (when meaningful) in the COI barcode. DNA characters are found in nuclear protein-coding regions using our previously developed procedure (see SI Appendix to Li et al. 2019). The logic behind the character selection was described in Cong et al. (2019b) and is aimed at finding more robust characters likely to stand when additional specimens and species are sequenced.
The character states are given in species diagnoses as abbreviations for one of the six reference genomes: Pterourus glaucus (Linnaeus, 1758) (pg) (Cong et al. 2015), Pieris rapae (Linnaeus, 1758) (pra) (Shen et al. 2016), Heliconius melpomene (Linnaeus, 1758) (hm) (Davey et al. 2016), Calephelis nemesis (W. H. Edwards, 1871) (cne) (Cong et al. 2017b), Calycopis cecrops (Fabricius, 1793) (cce) (Cong et al. 2016), or Cecropterus lyciades (Geyer, 1832) (aly, because this species was formerly in the genus Achalarus Scudder, 1872) (Shen et al. 2017). E.g., aly728.44.1:G672C means position 672 in exon 1 of gene 44 from scaffold 728 of the Cecropterus lyciades (Geyer, 1832) (aly) reference genome (Shen et al. 2017) is C, changed from G in the ancestor. When characters are given for the sister clade of the diagnosed taxon, the following notation is used: aly5294.20.2:A548A (not C), which means that position 548 in exon 2 of gene 20 on scaffold 5294 is occupied by the ancestral base pair A, which was changed to C in the sister clade (so it is not C in the diagnosed taxon). The same notation is used for COI barcode characters but without a prefix ending with ‘:’. The sequences of exons from the reference genome with the positions used as character states highlighted in green are in the supplemental file deposited at < https://osf.io/7tnqy/ >. This link to the DNA sequences accessible from this publication ensures that DNA characters given in the diagnoses can be readily associated with actual sequences.
Whole genome shotgun datasets we obtained and used in this work are available from the NCBI database < https://www.ncbi.nlm.nih.gov/ > as BioProject PRJNA1022154 and BioSample entries of the project contain the locality and other collection data of the sequenced specimens shown in the trees. For each specimen in tree figures, the following information is provided (separated by “|”): taxon name with comments in square brackets, DNA sample code, type status, general locality, and year of collection (“old” if not dated and likely collected 100–150 years ago). Type status abbreviations are: HT holotype, LT lectotype, ST syntype, T type (could be ST, LT, paralectotype, or HT, status not investigated), PT paratype; and if a synonym name is given (in parenthesis, preceded by “=”, and in addition by “‡” for unavailable names), type status refers to the synonym. COI barcode sequences reported here have been deposited in GenBank with accessions OR578710–OR578722 and OR589636–OR589640. Several photographs shown in this work were taken from iNaturalist (2023). Links to observations by observation number reported in figure legends are < https://www.inaturalist.org/observations/xxx >, where xxx is the observation number. Abbreviations or acronyms for collections are listed in the acknowledgments section.
Family Papilionidae Latreille, [1802]
Meandrusina Grishin, new subtribe http://zoobank.org/72E085A2-EA04-4D25-8124-04A86D17F26B
Type genus.
Meandrusa F. Moore, 1888.
Definition.
Meandrusa (type species Papilio evan E. Doubleday, 1845 currently treated as a subspecies of Papilio payeni Boisduval, 1836) was placed in the subtribe Teinopalpini Grote, 1899, but our genomic trees show that it is not monophyletic with Teinopalpus Hope, 1843 (type species Teinopalpus imperialis Hope, 1843) and instead is sister to Papilionini Latreille, [1802] with 100% support (Fig. 1). Genetic differentiation of Meandrusa from Papilionini is approximately the same as of Battus Scopoli, 1777 (type species Papilio polydamas Linnaeus, 1758) from other Troidini Talbot, 1939 (comparable distance from the root, Fig. 1). Therefore, we consider that Meandrusa belongs to the tribe Papilionini. However, the tree shows that Meandrusa is prominently separated from the rest of Papilionini and, therefore, belongs to a distinct subtribe analogously to Battus. This subtribe does not have a name and is proposed as new. As detailed by Miller (1987) for Meandrusa, the new subtribe is characterized by bifid tarsal claws and differs from the rest of Papilionini by strongly incurved middle discocellular vein on the forewing, shorter forewing discal cell (less than half of the wing), and scaled tibiae and tarsi, and from Teinopalpini by scaled antennae and prodiscrimen (equivalent to the prosternum of other insects) with a spine. Visually, species in the new subtribe are recognized by the distinct shape of curved forewings (especially in males) with produced and somewhat hooked apex narrowing to a point (less prominently in Meandrusa sciron (Leech, 1890)). A combination of the following nuclear genomic base pairs is diagnostic: pgl2854.3.1: A400G, pgl827.10.7:G122A, pgl827.10.7:G138A, pgl1898.30.3:G202A, pgl1397.11.1: G1138A.
Fig. 1.

The phylogenetic tree of selected Papilionidae inferred from protein-coding regions of the nuclear genome (autosomes). Family-group names are shown above or below the corresponding branches. A new subtribe proposed in this work is shown in red. Names of other subtribes are shown in black, and names of subfamilies and tribes are shown in shades of gray.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Papilionini Latreille, [1802].
Family Pieridae Swainson, 1820
Lirinia Grishin, new subgenus http://zoobank.org/A59BC65A-CEE7-4380-8D40-877A90B74CFD
Type species.
Terias lirina H. Bates, 1861.
Definition.
Our genomic tree reveals that the T. lirina (type locality in Brazil: Pará) lineage is monophyletic with neither Abaeis Hübner, [1819] (type species Papilio nicippe Cramer, 1779) nor Eurema Hübner, [1819] (type species Papilio delia Cramer, 1780, a junior homonym, valid name for this species is Pieris daira Godart, 1819), and instead is a confident sister to all other Pyrisitia A. Butler, 1870 (type species Papilio proterpia Fabricius, 1775), but is genetically differentiated from them at the level of a subgenus (Fig. 2). Therefore, we transfer T. lirina to Pyrisitia forming a new combination Pyrisitia lirina (H. Bates, 1861) and propose that its lineage represents a distinct subgenus of Pyrisitia. This new subgenus differs from its relatives by the characters given and illustrated for Eurema furtadoi Casagrande & O. Mielke, 1979 in its original description (Casagrade and Mielke 1979). In brief, wings rounded, mostly white with black forewing apex, reminiscent of Abaeis albula (Cramer, 1775) (type locality in Suriname), with which it was lumped in the past (Klots 1929), but with very different genitalia: in males, uncus broader, with two shot side processes (absent in A. albula); saccus shorter, about the same length as tegumen with uncus; aedeagus bow-shaped, broader and shorter, twice as long as saccus; valva shaped as a half-circle, harpe with robust ventral tooth and much smaller vestigial dorsal tooth; in females, corpus bursae smaller with much smaller signum and a small bubble-shaped appendix (instead of the appendix as long as corpus). A combination of the following nuclear genomic base pairs is diagnostic: pse1181.9.1: G68A, pse988.17.1:A57G, pse6193.9.1:G135T, pse5030.21.1:A392T, pse5030.21.1:T376G, pse907.3.2: A270C, pse1899.1.7:G805A, pse1899.1.7:T806C, pse2087.5.1:C260T, pse102.3.4:C1026G.
Fig. 2.

The phylogenetic tree of Euremini inferred from protein-coding regions of the nuclear genome (autosomes). Levels in the tree that correspond to the taxonomic hierarchy are marked above as Tribes, Subtribes, Genera, and Subgenera. The translucent vertical lime bar denotes the level approximately corresponding to genera. Family-group names (roman font) and genus-group names (genera in bold italics and subgenera in italics) are shown by corresponding branches. Different genera of Euremini are shown in different colors. Numbers in cyan on a yellow background followed by “Mya” placed on the right of several tree nodes indicate the approximate ages of these nodes in million years (rounded) as estimated by Kawahara et al. (2023); smaller numbers are statistical support values of the tree branches (in %, most are 100% implying high confidence). The red asterisk denotes Euremini’s arrival in the Old World from the New World, and the genus Terias originated as a result.
Etymology.
The name is a feminine noun in the nominative singular formed from the type species name.
Species included.
Only the type species.
Parent taxon.
Genus Pyrisitia A. Butler, 1870.
Pyrisitia mayobanex (M. Bates, 1939) and Pyrisitia memulus (A. Butler, 1871) are species-level taxa
As we previously concluded, Pyrisitia westwoodii (Boisduval, 1836) (type locality in Mexico) is a distinct species, not a subspecies of Pyrisitia dina (Poey, 1832) (type locality in Cuba) (Zhang et al. 2021). Sequencing of additional specimens that included the nominal P. dina (Fig. 3 blue) confirms this conclusion, also confirming Pyrisitia parvumbra (Kaye, 1925) (type locality in Jamaica) (Fig. 3 magenta) as a distinct species (Turner and Turland 2017): Fst/Gmin between P. parvumbra and P. dina dina of 0.59/0.002 and the COI barcode difference of 2.7% (18 bp). Furthermore, we find prominent genetic differentiation between P. dina and the taxon originally proposed as Eurema helios mayobanex M. Bates, 1939 (type locality in Haiti): Fst/Gmin of 0.39/0.00, COI difference of 1.8% (12 bp). Therefore, we propose that Pyrisitia mayobanex (M. Bates, 1939), stat. nov. is a species-level taxon. Inspecting the genomic trees, we see that Terias memulus Butler, 1871 (type locality in Haiti), currently regarded as a subspecies of Pyrisitia leuce (Boisduval, 1836) (type locality in Brazil: Rio Grande do Sul), is most strongly differentiated from it genetically: Fst/Gmin 0.60/0.00 and the COI barcode difference of 7.3% (48 bp) (Fig. 3 orange), while other P. leuce subspecies cluster closely together in the tree (Fig. 3 violet). Therefore, we reinstate Pyrisitia memulus (A. Butler, 1871), stat. rest. as a species.
Fig. 3.

Phylogenetic trees of Pyrisitia dina relatives inferred from protein-coding regions of a) the nuclear (autosomes) and b) the mitochondrial genomes. Different species are shown in different colors: P. westwoodii (green), P. dina (blue), P. mayobanex stat. nov. (red), P. parvumbra (magenta), P. memulus stat. rest. (orange), and P. leuce (violet).
Abaeis gratiosa (E. Doubleday, 1847) and Abaeis angulata (Wallengren, 1860) are species-level taxa, not subspecies of Abaeis arbela (Geyer, 1832)
Genomic sequencing of taxa treated as subspecies of Eurema arbela Geyer, 1832 (type locality in “Java”, possibly southern Brazil) in Lamas (2004), currently in the genus Abaeis Hübner, [1819] (type species Papilio nicippe Cramer, 1779) (Zhang et al. 2019b), confirms that Abaeis boisduvaliana (C. Felder & R. Felder, 1865) (type locality in Mexico) is a distinct species due to its genetic differentiation from A. arbela (Fig. 4 green and red). E.g., its COI barcode is 3.3% (22 bp) different. Moreover, we find that Abaeis gratiosa (E. Doubleday, 1847), stat. rest. (type locality in Venezuela) and Abaeis angulata (Wallengren, 1860), stat. rest. (type locality not given, likely in Ecuador) are prominently differentiated genetically from A. arbela, A. boisduvaliana, and each other both in nuclear and mitochondrial genomes (Fig. 4) and, therefore, are best treated as distinct species. Fst/Gmin/COI difference for the pairs of closest (in nuclear genome) relatives are: A. gratiosa and A. arbela: 0.21/0.01/3.5% (23 bp) and A. angulata and A. boisduvaliana: 0.64/0.001/3.6% (24 bp).
Fig. 4.
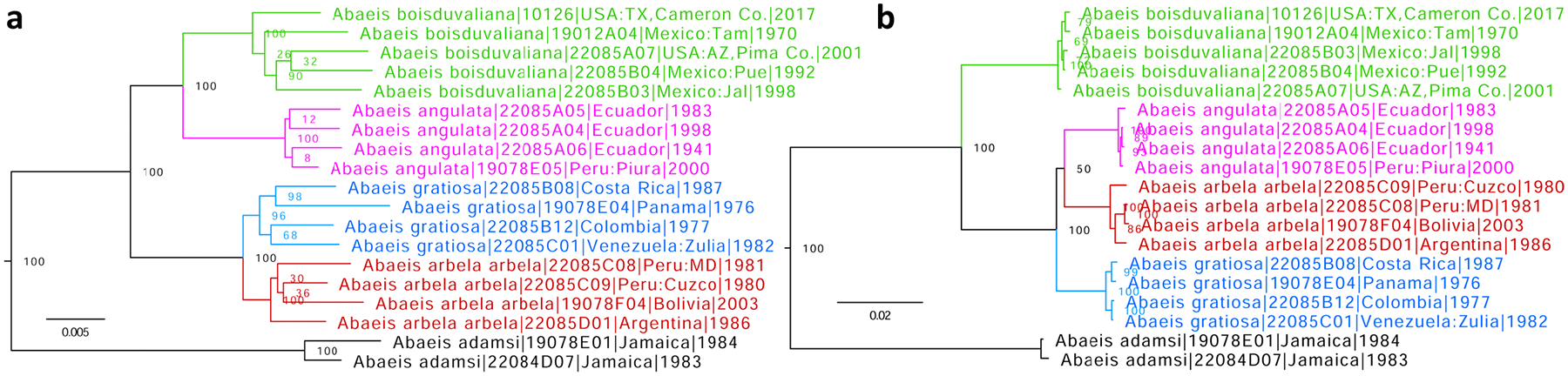
Phylogenetic trees of Abaeis arbela species group (rooted with Abaeis adamsi comb. nov.) inferred from protein-coding regions of a) the nuclear (autosomes) and b) the mitochondrial genomes. Different species are shown in different colors: A. boisduvaliana (green), A. angulata (magenta), A. gratiosa (blue), and A. arbela (red).
Classification of the tribe Euremini Grote, 1898
The phylogenetic tree of the tribe Euremini Grote, 1898 (type genus Eurema Hübner, [1819]) inferred from protein-coding regions in the nuclear genome (autosomes only) reveals strong statistical support (100% for most branches) and noticeable tree levels (resulting from coinciding diversifications in different clades at about the same distance from the root) to aid its higher classification (Fig. 2). The level closest to the origin of the tribe consists of three prominent clades that we treat as subtribes: Nathalina Bálint, 2022, stat. nov. (type genus Nathalis Boisduval, 1836), Kricogonina Bálint, 2022, stat. nov. (type genus Kricogonia Reakirt, 1864) and the nominotypical, Euremina. The former two were originally proposed as tribes, but the early split of the subfamily Coliadinae Swainson, 1821 (type genus Colias [Fabricius], 1807) into two prominent clades argues for treating these clades as tribes (Euremini and Coliadini) rather than dividing them further. Therefore, further divisions would correspond to the subtribal level. The monotypic genus Prestonia Schaus, 1920 (type species Prestonia clarki Schaus, 1920) placed in Euremini by Zhang et al. (2021) is sister to Kricogonia and diverged from it approximately 27 million years ago (Mya), as estimated in Kawahara et al. (2023), which is close to the split of the subtribe Euremina into two clades (25 Mya). Due to this similar level of genetic differentiation between Kricogonia and Prestonia and between the two first clades of Euremina, Prestonia is placed in the subtribe Kricogonina rather than in a subtribe of its own. Such classification emphasizes the sister relationship of the two genera (Kricogonia and Prestonia) rather than the distinction between them.
While there is little doubt that the five known species in Nathalina and Kricogonina are best classified into the three genera corresponding to the most prominent clades in the tree (Fig. 2), the taxonomy of the subtribe Euremina is more complex. Traditionally, the entire subtribe has been treated as a single genus Eurema Hübner, [1819] (type species Papilio delia Cramer, 1780, a junior homonym, valid name for this species is Pieris daira Godart, 1819) (Klots 1933). However, early DNA work suggested strong genetic differentiation within the subtribe (Pollock et al. 1998), formalized by Opler and Warren (2002), who partitioned the US species into three genera: Eurema, Pyrisitia A. Butler, 1870 (type species Papilio proterpia Fabricius, 1775), and Abaeis Hübner, [1819] (type species Papilio nicippe Cramer, 1779). This split has been largely followed (Lamas 2004; Pelham 2008), although some species have been reassigned between these genera (Zhang et al. 2019b). Our work (Zhang et al. 2019c; Zhang et al. 2019b) and more recent publications (Kawahara et al. 2023) support the notion of strong genetic differentiation within Euremina and date its diversification to approximately 25 Mya (Fig. 2). This level of differentiation and age are too large for placing all Euremina in the single genus Eurema.
The subtribe Euremina splits into two prominent clades at approximately 25 Mya, as estimated by Kawahara et al. (2023). These two clades can be taken to represent genera, and the subtribe can be divided into two genera: Eurema and Abaeis. However, even this level of genetic differentiation and age would be larger than for most genera of butterflies (Talavera et al. 2012; Li et al. 2019; Zhang et al. 2019a, c), and the ages between 15 and 20 Mya would be more consistent with the genus level. Even if absolute values of age estimates are not particularly accurate due to various errors, their relative values and the fact they are all estimated mainly with the same method and on similar datasets argue for the validity of such comparisons of estimated ages between nodes. For these reasons, we regard the two clades of Euremina as “sections”: the Eurema section and the Abaeis section, and take the next level in the tree to define genera.
The next tree level, dating to approximately 20 Mya, consists of five clades, one of which is entirely from the Old Word, the only Old Word group in the entire tribe Euremina (Fig. 2 marked with red asterisk). We propose that these five clades represent genera in Euremina: Terias W. Swainson, 1821 (type species Papilio hecabe Linnaeus, 1758) and Eurema with its sister Pyrisitia A. Butler, 1870 (type species Papilio proterpia Fabricius, 1775) belong to the Eurema section, and Abaeis with Teriocolias Röber, 1909 (type species Terias atinas Hewitson, 1874, a junior subjective synonym of Terias zelia Lucas, 1852) belong to the Abaeis section. This partitioning into genera is biogeographically significant because it reflects the invasion of the only Euremini lineage from the New World, where the tribe likely originated, into the Old World, giving rise to the genus Terias, followed by its extensive diversification. Genera of Euremina defined this way are analogous to Codatractus Lindsey, 1921 vs. Lobocla Moore, 1884, Heliopetes Billberg, 1820 vs. Pyrgus Hübner, [1819], and Oarisma Scudder, 1872 vs. Thymelicus Hübner, [1819] (in the latter two, pairs some species from the Old World genus returned to the New World at a later time) and correspond to similar geological time-frame of the mid-Miocene climatic optimum characterized by elevated biotic movement from America to Asia (Jiang et al. 2019).
The phylogenetic tree (Fig. 2) guides the assignment of species to genera, and we restore the monophyly by proposing new genus-species combinations: Pyrisitia amelia (Poey, [1852]), comb. nov., Pyrisitia lirina (H. Bates, 1861), comb. nov., Abaeis paulina (H. Bates, 1861), comb. nov., Abaeis xantochlora (Kollar, 1850), comb. nov., Abaeis fabiola (C. Felder & R. Felder, 1861), comb. nov., Abaeis tupuntenem (Lichy, 1976), comb. nov., Abaeis adamsi (Lathy, 1898), comb. nov., Abaeis brephos (Hübner, [1809]), comb. nov., and Abaeis elvina (Godart, 1819), comb. nov. The latter two combinations reflect our treatment of Leucidia E. Doubleday, 1847, stat. nov. (type species Pieris elvina Godart, 1819) as a subgenus of Abaeis despite its unique phenotype. Such treatment results in a more internally consistent classification because an Abaeis that includes Leucidia corresponds to a more prominent clade in the tree, and the Leucidia clade has split from the rest of Abaeis at the tree level corresponding to subgenera (Fig. 2). This level allows us to define five subgenera in Euremina in addition to the five genera (Fig. 2 and listed below). Finally, we note that Teriocolias doris (Röber, 1909), stat. rest. (type locality in Bolivia), currently regarded as a subspecies of Teriocolias deva (E. Doubleday, 1847) (type locality in French Guiana), is genetically very distant from it: e.g., COI barcodes differ by 7.6% (50 bp) and is a distinct species.
Below is a proposed classification of the tribe Euremini. Only available genus-group names are listed; subspecies names are not given and can be found on the Butterflies of America website (Warren et al. 2023), and the Old Word species are not provided (all belong to the genus Terias, which consists only of Old World species). Type genus (for family-group names) or type species (for genus-group names) names are given in parenthesis; synonyms are preceded by = and all but subjective synonyms also by ‡ with a valid name of this species following the colon. New taxa and status changes are shown in red font.
Tribe Euremini Grote, 1898 (Eurema Hübner, [1819]) Subtribe Nathalina Bálint, 2022, stat. nov. (Nathalis Boisduval, 1836) Genus Nathalis Boisduval, 1836 (Nathalis iole Boisduval, 1836) Nathalis iole Boisduval, 1836 Nathalis plauta E. Doubleday, 1847 Subtribe Kricogonina Bálint, 2022, stat. nov. (Kricogonia Reakirt, 1864) Genus Kricogonia Reakirt, 1863 (Colias lyside Godart, 1819) Kricogonia lyside (Godart, 1819) Kricogonia cabrerai Ramsden, 1920 Genus Prestonia Schaus, 1920 (Prestonia clarki Schaus, 1920) Prestonia clarki Schaus, 1920 Subtribe Euremina Grote, 1898 (Eurema Hübner, [1819]) Eurema section Genus Terias W. Swainson, 1821 (Papilio hecabe Linnaeus, 1758) Consists of all Old World species of Euremini Subgenus Maiva Grose-Smith & W.F. Kirby, 1893 (=M. sulphurea Gr-Sm. & Kirby: Papilio brigitta Stoll, 1780) =Kibreeta F. Moore, 1906 (=‡Papilio libythea Fabricius, 1798: Terias brigitta rubella Wallace, 1867) =Nirmula F. Moore, 1906 (=Terias venata F. Moore, 1858: Terias laeta Boisduval, 1836) Subgenus Terias W. Swainson, 1821 (Papilio hecabe Linnaeus, 1758) Genus Eurema Hübner, [1819] (=‡Papilio delia Cramer, 1780: Pieris daira Godart, 1819) Eurema priddyi (Lathy, 1898) Eurema lucina (Poey, [1852]) Eurema daira (Godart, 1819) Eurema elathea (Cramer, 1777) Eurema nigrocincta Dognin, 1889 Eurema agave (Cramer, 1775) Eurema phiale (Cramer, 1775) Genus Pyrisitia A. Butler, 1870 (Papilio proterpia Fabricius, 1775) Subgenus Pyrisitia A. Butler, 1870 (Papilio proterpia Fabricius, 1775) Pyrisitia proterpia (Fabricius, 1775) Pyrisitia westwoodii (Boisduval, 1836) Pyrisitia dina (Poey, 1832) Pyrisitia mayobanex (M. Bates, 1939), stat. nov. Pyrisitia parvumbra (Kaye, 1925) Pyrisitia memulus (A. Butler, 1871), stat. rest. Pyrisitia leuce (Boisduval, 1836) Pyrisitia larae (Herrich-Schäffer, 1862) Pyrisitia venusta (Boisduval, 1836) Pyrisitia chamberlaini (A. Butler, 1898) Pyrisitia nise (Cramer, 1775) Pyrisitia lisa (Boisduval & Le Conte, [1830]) Pyrisitia euterpiformis (Munroe, 1947) Pyrisitia amelia (Poey, [1852]), comb. nov. Pyrisitia portoricensis (Dewitz, 1877) Pyrisitia pyro (Godart, 1819) Pyrisitia messalina (Fabricius, 1787) Subgenus Lirinia Grishin, subgen. n. Pyrisitia lirina (H. Bates, 1861), comb. nov. Abaeis section Genus Abaeis Hübner, [1819] (Papilio nicippe Cramer, 1779) Subgenus Leucidia E. Doubleday, 1847 (Pieris elvina Godart, 1819), stat. nov. Abaeis brephos (Hübner, [1809]), comb. nov. Abaeis elvina (Godart, 1819), comb. nov. Subgenus Lucidia Lacordaire, 1833 (Papilio albula Cramer, 1775) Abaeis albula (Cramer, 1775) Subgenus Sphaenogona Butler, 1870 (Terias bogotana C. & R. Felder, 1861: a ssp. of T. mexicana Boisduval) Abaeis paulina (H. Bates, 1861), comb. nov. Abaeis xantochlora (Kollar, 1850), comb. nov. Abaeis fabiola (C. Felder & R. Felder, 1861), comb. nov. Abaeis tupuntenem (Lichy, 1976), comb. nov. Abaeis salome (C. Felder & R. Felder, 1861) Abaeis mexicana (Boisduval, 1836) Abaeis boisduvaliana (C. Felder & R. Felder, 1865) Abaeis angulata (Wallengren, 1860), stat. rest. Abaeis gratiosa (E. Doubleday, 1847), stat. rest. Abaeis arbela (Geyer, 1832) Abaeis adamsi (Lathy, 1898), comb. nov. Subgenus Abaeis Hübner, [1819] (Papilio nicippe Cramer, 1779) Abaeis nicippe (Cramer, 1779) Abaeis nicippiformis (Munroe, 1947) Genus Teriocolias Röber, 1909 (=Terias atinas Hewitson, 1874: Terias zelia Lucas, 1852) Teriocolias deva (E. Doubleday, 1847) Teriocolias doris (Röber, 1909), stat. rest. Teriocolias zelia (Lucas, 1852) Teriocolias reticulata (A. Butler, 1871)
Subtribes in Coliadini Swainson, 1821
The genomic tree reveals four prominent clades in the tribe Coliadini Swainson, 1821 that are at approximately the same distance from the root (Fig. 5). We propose treating these clades as subtribes. Three of these subtribes have names: the nominotypical one, Callidryina Kirby, 1896, stat. nov., and Gonepterygina Verity, 1920, stat. nov. The name Callidryina is formed from the genus Callidryas Boisduval & Le Conte, 1830 (type species Papilio eubule Linnaeus, 1767), which is currently treated as a junior subjective synonym of Phoebis Hübner, [1819] (type species Phoebis cypris Hübner, [1819], which is an unjustified emendation of Papilio cipris Cramer, 1777, which is a junior subjective synonym of Papilio argante Fabricius, 1775). Callidryina consists of two closely related genera, Phoebis (that includes Rhabdodryas Godman & Salvin, 1889) and Aphrissa Butler, 1873. In addition to the type genus Gonepteryx Leach, 1815, we place Dercas E. Doubleday, 1847 in Gonepterygina. The fourth subtribe does not have a name. It is described below.
Fig. 5.

The phylogenetic tree of selected Pieridae inferred from protein-coding regions of the nuclear genome (autosomes). Family-group names are shown by corresponding branches, or an arrow points to the branch. Names, clades, and species of new subtribes proposed in this work are shown in color, except that red indicates a reinstated genus Pseudanaphaeis (not a synonym of Belenois). Names of other subtribes are shown in black, and names of subfamilies and tribes are shown in shades of gray. Note the extreme variation of base-pair substitution rate in Pieridae, where DNA of some taxa (e.g., Gonepteryx) changes at least two times slower than others (e.g., Belenois), as evidenced by the distance from the root to these leaves. Therefore, no vertical bars or lines can demarcate tribes and subtribes in this tree, where branch length corresponds to the estimated number of base pair changes along the branch.
Gandacina Grishin, new subtribe http://zoobank.org/EF448D21-E251-4ABA-9A22-22EBD93A8303
Type genus.
Gandaca F. Moore, 1906.
Definition.
Gandaca (type species Terias harina Horsfield, 1829) is sister to Gonepterygina Verity, 1920, stat. nov. but is more distant from the members of this subtribe both genetically and phenotypically. Furthermore, the statistical support for Gonepterygina to include Gandaca is lower than for most other clades in the tree (88%) (Fig. 5). Therefore, the clade with Gandaca is defined as a subtribe. This new subtribe is diagnosed by the following characters: in male genitalia, uncus pointed dorsad at the base, with dorsal margin strongly arched and finely serrated, uncus shorter than in relatives and broader towards its distal end in dorsal view; in female genitalia, corpus bursae nearly spherical with a ring-shaped signum at its base and an appendix that is larger than the corpus itself; in facies, is more similar to Eurema Hübner, [1819] and relatives (tribe Euremini Grote, 1898) than to Gonepterygina: lemon-yellow wings without central spots, hindwings plain or with dark margin by the apex and forewings with dark apex continuing into thin marginal border. See Kaur et al. (2022) for illustrations. A combination of the following nuclear genomic base pairs is diagnostic: pse19182.2.2:A3375G, pse1982.1.2:A51T, pse200.39.1:T1209C, pse1378. 26.1:G839A, pse200.29.1:C1534G.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Coliadini Swainson, 1821.
Pseudanaphaeis Bernardi, 1953 is a genus distinct from Belenois Hübner, [1819]
Currently considered a junior subjective synonym of Belenois Hübner, [1819] (type species Papilio calypso Drury, 1773, sequenced as NVG-19068G08), Pseudanaphaeis Bernardi, 1953 (type species Pieris gidica Godart, 1819, sequenced as NVG-19068G12), is not monophyletic with it, and instead is sister to Dixeia Talbot, 1932 (type species Pieris charina Boisduval, 1836, sequenced as NVG-19069F05) (Fig. 5). Extensive genetic differentiation between Dixeia and Belenois precludes from considering them congeneric. Therefore, we propose to treat Pseudanaphaeis Bernardi, 1953, stat. rest. as a distinct genus.
Hebomoiina Grishin, new subtribe http://zoobank.org/71C79ADE-4BBF-4CC8-86B6-331BCD38863B
Type genus.
Hebomoia Hübner, [1819].
Definition.
Hebomoia (type species Papilio glaucippe Linnaeus, 1758) belongs to the tribe Anthocharidini Scudder, 1889, but is genetically differentiated from its other genera (Fig. 5). Phenotypically, species in this lineage are characterized by a more robust appearance that the rest of the tribe. Therefore, we propose that the clade with Hebomoia corresponds to a subtribe. This new subtribe is diagnosed by a bifurcate uncus and bifurcate valva, forewing with five radial veins, two of which and M1 (not stalked with R) originate at the discal cell, larger size (forewing longer than 40 mm), and pointed broadly orange apex of the forewing. See Klots (1933) for additional discussion and illustrations of these characters given for Hebomoia. A combination of the following nuclear genomic base pairs is diagnostic: pse123.37.1:A2228G, pse123.37.1:T1285A, pse9809.2.1:G1812A, pse657.5.2:A89G, pse5906.8.2:G3017A.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Anthocharidini Scudder, 1889.
Tribes and subtribes in Dismorphiinae Schatz, 1886
The subfamily Dismorphiinae Schatz, 1886 splits into two prominent clades that we propose to treat as tribes: the nominotypical and Leptidiini Grote,1897, stat. rev., which is monotypic (Fig. 5). The two tribes are well-defined morphologically (Klots 1933) and biogeographically, with Leptidiini being the Old World tribe and Dismorphiini restricted to the New World.
Pseudopierina Grishin, new subtribe http://zoobank.org/25317F65-DA92-4FD1-88BB-ED275C6B46A1
Type genus.
Pseudopieris Godman & Salvin, 1890.
Definition.
Pseudopieris (type species Pieris nehemia Boisduval, 1836) is sister to and is stronger differentiated genetically from the rest of Dismorphiini Schatz, 1886 (Fig. 5). Therefore, combined with phenotypic differences, we propose that the lineage with Pseudopieris corresponds to a subtribe. This new subtribe is distinguished from the rest of Dismorphiini (and Dismorphiinae, for that matter) by much broader wings that are more like in Pieris Schrank, 1801, rather than the elongated wings of Dismorphiinae, the last abdominal segment with rounded lobes and a cleft, and M1 stalked with R stem on the forewing. See Klots (1933) for details and illustrations given for Pseudopieris. A combination of the following base pairs in the nuclear genome is diagnostic: pse7986.9.2:A4048G, pse19182.2.2:T3830C, pse165.20.1:A803T, pse578.3.2:C31A, pse578.3.2:A12G.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Dismorphiini Schatz, 1886.
Family Nymphalidae Rafinesque, 1815
Libytheana Michener, 1943 is a genus distinct from Prolibythea Scudder, 1889
Considered synonyms in some publications (Kawahara 2009; Sohn et al. 2012), fossil Prolibythea Scudder, 1889 (type species Prolibythea vagabunda Scudder, 1889) and contemporary Libytheana Michener, 1943 (type species Libythea bachmanii Kirtland, 1851, which is regarded as a subspecies of Papilio carinenta Cramer, 1777), both American, are separated by at least 30 million years according to the age estimate of the fossil as late Priabonian (Sohn et al. 2012). The typical age of congeners is not larger than 20 million years. Furthermore, the divergence between Libytheana and the Old World genus Libythea [Fabricius], 1807 (type species Papilio celtis Laicharting, 1782) was dated to about 12 Mya (Kawahara et al. 2023), which is more recent than 30 Mya. Thus, it is most likely that Prolibythea lived before the divergence between Libytheana and Libythea. Therefore, if Libytheana and Libythea are treated as separate genera, then Prolibythea is not congeneric with Libytheana to avoid paraphyly.
The tribe Vagrantini Pinratana & Eliot, 1996 as currently defined is paraphyletic
Currently, the tribe Vagrantini Pinratana & Eliot, 1996 consists of ten genera: Vagrans Hemming, 1934, Cupha Billberg, 1820, Phalanta Horsfield, 1829, Smerina Hewitson, 1874, Terinos Boisduval, 1836, Algia Herrich-Schäffer, 1864), Algiachroa Parsons, 1989, Cirrochroa E. Doubleday, 1847, Lachnoptera E. Doubleday, 1847, and Vindula Hemming, 1934 (Wahlberg 2019). However, our genomic tree reveals that Vagrantini, defined to include all ten genera, is paraphyletic with respect to Argynnini Swainson, 1833 with the highest support (Fig. 6). The first five genera listed above form a clade sister to Argynnini. This clade includes Vagrans, which is the type genus of Vagrantini. Therefore, to restore monophyly, we restrict Vagrantini to include only these five genera: Vagrans, Cupha, Phalanta, Smerina, and Terinos. The remaining five genera previously included in Vagrantini form a clade sister to both Vagrantini and Argynnini (Fig. 6) and, therefore, belong to other tribes. No published family-group names have been formed from any of these five genera; hence, these other tribes are new. They are described below.
Fig. 6.

The phylogenetic tree of selected Nymphalidae inferred from protein-coding regions of the nuclear genome (autosomes). Sequences of the three samples with numbers starting with “SAMN” were taken from the alignment provided in Supplementary materials to Kawahara et al. (2023). Family-group names are shown above or below the corresponding branches. Names, clades, and species of new tribes and subtribes proposed in this work are shown in color. Names of other subtribes are shown in black, and names of subfamilies and tribes are shown in shades of gray. The translucent vertical lime bar denotes a level in the tree approximately corresponding to subtribes; however, due to variation in base pair substitution rates (i.e., not all tree leaves are at the same level), the correspondence may not be exact. E.g., DNA changes in Argynnini are slower than in most other groups (many Argynnini leaves are closer to the left compared to most others); hence, the level is shifted to the left, which also preserves the originally proposed (Simonsen 2006) division into two and not three subtribes.
Vindulini Grishin, new tribe http://zoobank.org/151AC163-E036-49BE-A49C-48D7B9F0108F
Type genus.
Vindula Hemming, 1934.
Definition.
Vindula (type species Papilio arsinoe Cramer, 1777) constitutes a lineage sister to four other genera that were previously included in Vagrantini Pinratana & Eliot, 1996 but did not belong to this tribe (see above). This lineage diverged from these other genera at about the same level as (if not earlier than) Vagrantini from Argynnini Swainson, 1833, and therefore corresponds to a tribe (Fig. 6). This new tribe is distinguished from its relatives by sclerotized subpapillary glands in females and forked humeral vein (Penz and Peggie 2003). A combination of the following nuclear genomic base pairs is diagnostic: hm2013347-RA.4:T162C, hm2013540-RA.5:G265A, hm2015146-RA.7:G80A, hm2015146-RA.7:G79A, hm2013347-RA.4:A220C.
Genera included.
Only the type genus.
Parent Taxon.
Subfamily Heliconiinae Swainson, 1822.
Algiini Grishin, new tribe http://zoobank.org/AE49C6E7-8E51-4E4E-9947-151B37A467E8
Type genus.
Algia Herrich-Schäffer, 1864.
Definition.
This tribe corresponds to the second major subclade in the clade that is sister to both Argynnini Swainson, 1833 and Vagrantini Pinratana & Eliot, 1996. This subclade is sister to Vindulini trib. n., diverging from it at about the same level as (if not earlier than) Argynnini from Vagrantini (Fig. 6). Due to this prominent genetic differentiation, it is defined as a tribe. This new tribe is distinguished from its relatives by unsclerotized subpapillary glands in females, forked humeral vein, and/or smooth eyes and undifferentiated androconial scales, or by an oval patch of androconial scales in males in the apical area of dorsal hindwing covering 1/7–1/6 of its surface (Penz and Peggie 2003). A combination of the following nuclear genomic base pairs is diagnostic: hm2000037-RA.1:C698G, hm2008858-RA.12: T1021C, hm2008858-RA.12:C1022T, hm2016824-RA.4:C154A, hm2017493-RA.1:A2480G.
Genera included.
The type genus (i.e., Algia Herrich-Schäffer, 1864), Algiachroa Parsons, 1989, Cirrochroa E. Doubleday, 1847, and Lachnoptera E. Doubleday, 1847.
Parent Taxon.
Subfamily Heliconiinae Swainson, 1822.
Lachnopterina Grishin, new subtribe http://zoobank.org/8BBABF1F-145B-4A67-94F5-F498130FB857
Type genus.
Lachnoptera E. Doubleday, 1847.
Definition.
Lachnoptera (type species Papilio iole Fabricius, 1781) forms a lineage that splits from all other Algiini trib. n. at the tree level of subtribes (Fig. 6), therefore representing a subtribe. This new subtribe is distinguished from its relatives by unsclerotized subpapillary glands in females (Penz and Peggie 2003) and an oval patch of androconial scales in males in the apical area of dorsal hindwing covering 1/7–1/6 of its surface. A combination of the following nuclear genomic base pairs is diagnostic: hm2008958-RA.5:T118G, hm2008958-RA.5:G119T, hm2006642-RA.2:A2275C, hm2006706-RA.1:A709T, hm2006706-RA.1:A748T, hm2015589-RA.1:A1852A (not C), hm2014529-RA.4:A139A (not G), hm2012118-RA.6:A79A (not G), hm2016492-RA.5:A1813A (not G), hm2016492-RA.5:C1831C (not A).
Genera included.
Only the type genus.
Parent Taxon.
Tribe Algiini Grishin, trib. n.
Terinosina Grishin, new subtribe http://zoobank.org/31DFAAA5-EA63-431B-AC9D-BF686549FEC8
Type genus.
Terinos Boisduval, 1836.
Definition.
Terinos (type species Terinos clarissa Boisduval, 1836) forms a deep-diverging lineage in the tribe Vagrantini Pinratana & Eliot, 1996 at about the tree level corresponding to subtribes (Fig. 6) and, therefore, represents a subtribe. This new subtribe is distinguished from its relatives by a combination of the following characters: larval head with scoli, hindwing cell closed, a fold across the forewing between R and M veins, vein R2 arising from discal cell, vein R4 arising at about the end of R2, humeral vein simple and straight, gnathos arms not ventrally fused, valva with a long (about 2/3 of valva length) projection off its base inside (Penz and Peggie 2003). A combination of the following base pairs in the nuclear genome is diagnostic: hm2007153-RA.3:A305G, hm2014625-RA.4:T887A, hm2012596-RA.1: C853G, hm2011273-RA.1:T484G, hm2011273-RA.1:G486A.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Vagrantini Pinratana & Eliot, 1996.
Smerinina Grishin, new subtribe http://zoobank.org/06DF6620-9B17-4522-9E4B-BD5342AC8DF5
Type genus.
Smerina Hewitson, 1874.
Definition.
Smerina (type species Smerina vindonissa Hewitson, 1874) forms a deep-diverging lineage in the tribe Vagrantini Pinratana & Eliot, 1996 at about the tree level corresponding to subtribes (Fig. 6) and therefore represents a subtribe. This new subtribe is distinguished from its relatives by a combination of the following characters: papilla anales moderately retracted (not deeply) inside the body, aedeagus not broadened at the tip in ventral view, costa of valva with one spiny process (Penz and Peggie 2003), forewing apex more produced and hindwing margin evenly curved (not wavy, no tails). A combination of the following base pairs in the nuclear genome is diagnostic: hm2015462-RA.1:T360C, hm2015689-RA. 5:T57C, hm2009280-RA.3:T94G, hm2010526-RA.2:A67G, hm2017391-RA.1:A139C.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Vagrantini Pinratana & Eliot, 1996.
Lebadeini Grishin, new tribe http://zoobank.org/F26F931E-9141-4329-92B6-6E86A9F369E9
Type genus.
Lebadea C. Felder, 1861.
Definition.
Lebadea (type species Limenitis ismene E. Doubleday, 1848, which is a junior subjective synonym of Papilio martha Fabricius, 1787), currently in Neptini Newman, 1870 (Dhungel and Wahlberg 2018; Wahlberg 2019) is not monophyletic with it and instead is placed as sister to the clade of Chalingini Hemming, 1960 and Limenitidini Behr, 1864 but with weak support; therefore, it is distinct from them (Fig. 6). Thus, this lineage, currently consisting only of Lebadea, represents a tribe that does not have a name. This new tribe is diagnosed by a combination of the following characters: tegumen and uncus are smaller than typical for Limenitidini, uncus is more gracile, thus more similar to Neptis [Fabricius], 1807, and differs from Neptis in having a well-defined and projecting anteriad lobe (not just a hump) on the dorsal side of the segment A2 in the pupa (Willmott 2003). A combination of the following nuclear genomic base pairs is diagnostic: hm2005025-RA.3:C373A, hm2006832-RA.2:C97A, hm2004700-RA.1:C590A, hm2004700-RA.1:G351T, hm2005025-RA.3:G319A.
Genera included.
Only the type genus.
Parent Taxon.
Subfamily Limenitidinae Behr, 1864.
Comment.
Using morphological analysis, Willmott (2003) has placed Lebadea in the “Limenitis group” of genera away from Neptis, which seems to agree more with our genomic results.
Subtribes in Adoliadini Doubleday, 1845
Currently, no subtribes are in use for Adoliadini (Wahlberg 2019). However, our genomic tree reveals five prominent clades in this tribe (Fig. 6), confirming the results reported by Dhungel and Wahlberg (2018). We propose to treat these clades as subtribes. This subtribal arrangement will bring additional order to the species-rich tribe Adoliadini. Three of these subtribes have names: the nominotypical one, Abrotina Hemming, 1960, stat. nov., and Bebeariina Hemming, 1960, stat. nov. (the last two were originally proposed as tribes), and two do not. They are described below.
Evenaina Grishin, new subtribe http://zoobank.org/AB51B17D-4CB5-4909-8C7C-C76C81E84C4B
Type genus.
Evena Westwood, 1850.
Definition.
Evena (type species Papilio crithea Drury, 1773) forms a prominent phylogenetic lineage within Adoliadini Doubleday, 1845 on par with other subtribes (Fig. 6) and therefore represents a new subtribe. The new subtribe is diagnosed by genitalia and venation as described in detail and illustrated by Chermock (1950) for the genus Catuna W. F. Kirby, 1871 (a junior objective synonym of Evena). In brief, uniquely long and narrow saccus nearly as long as valva and R1 vein arising before the end of the discal cell, then fusing with Sc for some distance and diverging to meet costal margin are diagnostic. Furthermore, the subtribe is recognized by the unique appearance of its species, somewhat resembling Heliconiinae Swainson, 1822: with elongated forewings and shorter, rounded hindwings, spider-web forewing pattern, and a pale frequently triangular area across the hindwing toward the apex, hidden from view when the butterfly is sitting. A combination of the following nuclear genomic base pairs is diagnostic: hm2005164-RA.2:C119T, hm2017194-RA.1:C92T, hm2017194-RA.1:G298A, hm2005515-RA.6:C97G, hm2016751-RA.4:C44T, hm2007706-RA.6:G759G (not T), hm2009397-RA.1:A367A (not T), hm2020285-RA.1:C553C (not A), hm2020285-RA.1:A554A (not G), hm2004293-RA.6:A149A (not G).
Genera included.
Only the type genus.
Parent Taxon.
Tribe Adoliadini Doubleday, 1845.
Comment.
The name for the subtribe is formed by taking the entire name of the type genus as a root to avoid homonymy with Evenina Faynel & Grishin, 2022 (type genus Evenus Hübner, [1819], in Eumaeini E. Doubleday, 1847).
Pseudathymina Grishin, new subtribe http://zoobank.org/55581E3A-18C6-492F-BD1A-3B3F205896AF
Type genus.
Pseudathyma Staudinger, 1891.
Definition.
Pseudathyma (type species Pseudacraea sibyllina Staudinger, 1890) forms a prominent phylogenetic lineage within Adoliadini Doubleday, 1845 on par with other subtribes (Fig. 6) and, therefore, represents a new subtribe. This new subtribe is diagnosed by open discal cells of both wings, R2 that originates slightly beyond, instead of before, the end of the discal cell, and the absence of the anal lobe on the hindwing, per Chermock (1950), who gave these characters for Pseudathyma. In wing patterns, members of this subtribe are more similar to Neptis [Fabricius], 1807 in having four generally pale areas on the forewing (by the middle of the inner margin, in the discal area distad of the discal cell, by the apex, and in the discal cell) than to most Adoliadini. A combination of the following nuclear genomic base pairs is diagnostic: hm2007185-RA.1:A1149G, hm2007185-RA.1:C1150T, hm2017262-RA.1: A935G, hm2018054-RA.1:T155C, hm2017807-RA.2:A68T.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Adoliadini Doubleday, 1845.
Kumothalina Grishin, new subtribe http://zoobank.org/101F0041-F957-4CD5-92F7-079A780A043D
Type genus.
Kumothales Overlaet, 1940.
Definition.
Wahlberg et al. (2020) placed Kumothales (type species Kumothales inexpectata Overlaet, 1940) in the tribe Cymothoini Dhungel & Wahlberg, 2018. Our analysis confirms this conclusion and previously published phylogenies (Wahlberg et al. 2020; Kawahara et al. 2023) and places the Kumothales lineage as sister to all other Cymothoini that diverged from them before the divergence of Adoliadini Doubleday, 1845 into subtribes (Fig. 6). This substantial genetic differentiation of Kumothales is also the reason for the difficulty in finding the place for this genus in taxonomic hierarchy without DNA analysis. Therefore, this lineage represents a subtribe. This new subtribe is distinguished from its relatives by the details of wing venation as described for Kumothales by Overlaet (1940) and a combination of the following characters: wings less rounded, forewing apex lobed, hindwing margin wavy, wings without bands, with a unique submarginal wavy pattern consisting of dark inverted deep U with a sharp tooth (narrow V) inserted into it in every cell. A combination of the following nuclear genomic base pairs is diagnostic: hm2008200-RA.1:A517C, hm2006358-RA.1:C1151A, hm2009464-RA.1:C128T, hm2010867-RA.7:G38C, hm2012380-RA.2:A48G, hm2012713-RA.1:T428T (not C), hm2012713-RA.1:G946G (not A), hm2007718-RA.2:C205C (not A), hm2006845-RA.2:T1311T (not A), hm2006845-RA.2:A1314A (not G).
Genera included.
Only the type genus.
Parent Taxon.
Tribe Cymothoini Dhungel & Wahlberg, 2018.
Amnosiini Grishin, new tribe http://zoobank.org/AD32BD0A-6755-4650-AB24-3FD7A43DFA06
Type genus.
Amnosia E. Doubleday, 1849.
Definition.
Amnosia (type species Amnosia decora E. Doubleday, 1849) belongs to the subfamily Pseudergolinae Jordan, 1898, but is more distant from and sister to the rest of the subfamily (Fig. 6). Genetic differentiation of the Amnosia lineage from other Pseudergolinae is at the level of a tribe. Therefore, we propose that the Amnosia lineage corresponds to a tribe. This new tribe is diagnosed by a combination of the following characters: wings mostly dark in males, forewing with a pale stripe from mid-costa to tornus, and hindwing with two pairs of larger submarginal eyespots beneath that are better defined than in other Pseudergolinae. A combination of the following nuclear genomic base pairs is diagnostic: hm2002290-RA.3:A49G, hm2013678-RA.4:A234C, hm2013678-RA.4:G246A, hm2013678-RA.4:T294C, hm2006306-RA.3:G107C, hm2013835-RA.2:G178G (not A), hm2014102-RA.2:C115C (not T), hm2014102-RA.2:A117A (not T), hm2009568-RA.1:T184T (not A), hm2009568-RA.1:C185C (not A).
Genera included.
Only the type genus.
Parent Taxon.
Subfamily Pseudergolinae Jordan, 1898.
Hyperanartia Grishin, new subgenus http://zoobank.org/D264019C-2106-4F29-9F1F-7830321FAD68
Type species.
Vanessa dione Latreille, [1813].
Definition.
The genus Hypanartia Hübner, [1821] (type species Hypanartia demonica Hübner, [1821], which is a junior subjective synonym of Papilio lethe Fabricius, 1793) has been divided into two species groups: the paullus group (includes the type species of Hypanartia) and the dione group (Willmott et al. 2001; Llorente et al. 2023). Our genomic tree shows this split (Fig. 7) and is consistent with the cladogram constructed using morphological characters (Willmott et al. 2001). Here, the division of Hypanartia into two clades is formalized, and the new subgenus is proposed to encompass the dione group. This new subgenus is distinguished from the nominotypical subgenus by male genitalia: nearly triangular in lateral view valvae, separated at the base in ventral view; vinculum broader near the base of succus in lateral view; saccus with narrower anterior part (at least in lateral view); and gnathos continuously sclerotized, joined. See Willmott, Hall, and Lamas (2001) for additional information and illustrations. A combination of the following nuclear genomic base pairs is diagnostic: hm2021257-RA.1:A261G, hm2002154-RA.32:T1114A, hm2013826-RA.2:G702A, hm2006214-RA.3:T888C, hm2005917-RA.1:C3222G.
Fig. 7.

The phylogenetic tree of Nymphalis relatives inferred from protein-coding regions of the nuclear genome (autosomes). Genus-group names are shown above or below corresponding branches. Names of genera are shown on the left and in a larger font than names of subgenera. Names, clades, and species of new subgenera proposed in this work are shown in color, except that magenta indicates a new status of Vanessa madegassorum (species, not a subspecies of Vanessa hippomene).
Etymology.
The Latin prefix hypo- means “below”, “beneath”, and sometimes “less than”. Species of the new subgenus are clearly more than that, and this prefix is replaced with hyper- (i.e., “above”, “high”, “beyond”, “excessive”) for an exaggerated look of some of these butterflies: typically with more angular wings, longer tails, and bolder white spots. The name is a feminine noun in the nominative singular.
Species included.
The type species (i.e., Vanessa dione Latreille, [1813]), Hypanartia celestia Lamas, Willmott & J. Hall, 2001, Eurema charon Hewitson, 1878, Hypanartia christophori Jasinski, 1998, Hypanartia cinderella Lamas, Willmott & J. Hall, 2001, Hypanartia fassli Willmott, J. Hall & Lamas, 2001, Eurema kefersteini Doubleday, [1847], Eurema lindigii C. Felder & R. Felder, 1862, Hypanartia splendida Rothschild, 1903, and Hypanartia trimaculata Willmott, J. Hall & Lamas, 2001 (including their subspecies and synonyms).
Parent taxon.
Genus Hypanartia Hübner, [1821].
Comments.
In their genetic differentiation (Fig. 7), the two subgenera of Hypanartia are approximately the same as some subgenera in Nymphalis Kluk, 1780 (type species Papilio polychloros Linnaeus, 1758). Therefore, they are phylogenetically equivalent to the two subgenera Nymphalis and Aglais Dalman, 1816 (type species Papilio urticae Linnaeus, 1758) (i.e., the same level in the tree), and are stronger differentiated genetically than Nymphalis from Polygonia Hübner, [1819] (type species Papilio c-aureum Linnaeus, 1758).
Vanessa madegassorum (Aurivillius, 1899) is a species distinct from Vanessa hippomene (Hübner, 1823)
Genomic sequencing reveals notable genetic differentiation between the nominotypical Vanessa hippomene (Hübner, 1823) (type locality not given, deduced by wing shape and patterns of a specimen shown in the original illustration to be in South Africa) and Vanessa hippomene madegassorum (Aurivillius, 1899) (type locality in Madagascar) (Fig. 7). The COI barcodes of the two taxa differ by 2.9% (19 bp). Phenotypically, V. h. madegassorum is characterized by more prominently scalloped (even toothed at veins) wing margins, a longer and thinner major hindwing tail at the end of vein M3, a second, shorter tail at the end of vein CuA2 (absent in the nominotypical V. hippomene), orange (rather than yellower) forewing band, green scaling by the forewing apex beneath, and reduced pale scaling and spot by mid-costa on the ventral hindwing (Fig. 8). Taken together, these observations suggest that Vanessa madegassorum (Aurivillius, 1899), stat. nov. is a species distinct from Vanessa hippomene (Hübner, 1823). Vanessa madegassorum stat. nov. is known only from Madagascar, with the last reported record from 1976 (Lees et al. 2003), and may be of conservation concern, if not already extinct. The COI barcode sequence of V. madegassorum stat. nov., sample NVG-19122B12, GenBank accession OR578710, 658 base pairs, is:
TACTTTATATTTTATTTTCGGAATTTGAGCAGGAATAGTTGGAACTTCACTTAGTTTATTAATTCGAACTGAATTAGGAAATCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACAATTGTTACAGCTCATGCTTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGAGGTTTTGGTAATTGATTAATTCCACTTATATTAGGAGCCCCTGATATAGCTTTTCCACGTATAAATAATATAAGATTTTGACTTTTACCCCCTTCATTAATATTATTAATTTCTAGTAGAATTGTTGAAAATGGAGCAGGAACAGGATGAACAGTTTACCCCCCACTTTCATCTAATATTGCTCATAGAGGATCTTCTGTAGATCTAGCAATTTTTTCATTACATTTAGCTGGAATTTCCTCTATTTTAGGAGCAATTAATTTTATTACTACTATTATTAATATACGAATTAATAGAATATCTTTTGATCAAATACCTTTATTTGTTTGAGCTGTAGGTATTACAGCTTTACTTTTATTAATCTCTCTTCCTGTTTTAGCTGGAGCTATTACTATACTTCTAACAGATCGAAATATTAATACATCATTTTTTGATCCTGCGGGAGGAGGAGACCCAATTCTTTATCAACATTTATTT
Fig. 8.

Vanessa madegassorum stat. nov. male (dorsal: left, ventral: right) in USNM collection sequenced as NVG-19122B12 with its labels. All images are to scale.
Subgenera in Vanessa [Fabricius], 1807
The genus Vanessa [Fabricius], 1807 (type species Papilio atalanta Linnaeus, 1758) has been divided into five species groups: the atalanta group (includes the type species of Vanessa), the cardui group (Papilio cardui Linnaeus, 1758 is the type species of Cynthia [Fabricius], 1807), the carye group (Hamadryas carye Hübner, 1812 is the type species of Neofieldia Özdikmen, 2008), the itea group (Papilio itea Fabricius, 1775 is the type species of Bassaris Hübner, [1821]), and the hippomene group (Wahlberg and Rubinoff 2011). Because four of these groups are characterized by notable genetic differentiation (Fig. 7), we propose to treat the two names as subgenera: Neofieldia Özdikmen, 2008, stat. rest. and Bassaris Hübner, [1821], stat. rev. and leave Cynthia as a junior subjective synonym of Vanessa. The hippomene species group does not have a name. It is described below.
Paranartia Grishin, new subgenus http://zoobank.org/5760367D-44F8-48AF-B592-2B1AEE4CC755
Type species.
Hypanartia hippomene Hübner, [1823].
Definition.
Comprises the hippomene species group in Vanessa [Fabricius], 1807, as proposed by Wahlberg and Rubinoff (2011), who inferred a comprehensive phylogeny of Vanessa and its relatives and discovered the phylogenetic position of the hippomene group within Vanessa. This group is unusual because its constituent species have been previously placed in Antanartia Rothschild & Jordan, 1903 (type species Papilio delius Drury, 1782) due to phenotypic similarities. We treat the hippomene group as a new subgenus (Fig. 7). This subgenus is distinguished from Antanartia by genitalia: in males, aedeagus without spines and projections (does not end in a “barb” like a fish hook end structure) and valva without projections off costa, which is nearly straight; and in females, with developed signa in corpus bursae (Howarth 1966), and from Vanessa by a sharp tooth-like tail at the vein M3 on hindwing. A combination of the following nuclear genomic base pairs is diagnostic: hm2005743-RA.6:T48C, hm2002542-RA.3:A237G, hm2017019-RA.1:T160C, hm2002154-RA.32:A1116G, hm2021745-RA.5: C862A.
Etymology.
The prefix para- means “beside”, “beyond”, or “similar to”. Species of the new subgenus are similar to and were previously placed in Antanartia, and the prefix para- is fused with the latter genus name to form the new name. The name is a feminine noun in the nominative singular.
Species included.
The type species (i.e., Hypanartia hippomene Hübner, [1823]), Antanartia dimorphica Howarth, 1966, and Hypanartia hippomene var. madegassorum Aurivillius, 1899, stat. nov. (including their subspecies and synonyms).
Parent taxon.
Genus Vanessa [Fabricius], 1807.
Comments.
The four subgenera of Vanessa are more genetically distinct from each other than some subgenera within Nymphalis Kluk, 1780 (type species Papilio polychloros Linnaeus, 1758) (Fig. 7), and several of such Nymphalis subgenera are commonly treated as genera, e.g., Polygonia Hübner, [1819] (type species Papilio c-aureum Linnaeus, 1758).
Eresia (Anthanassa) seminole Skinner, 1911 is a species distinct from Eresia (Anthanassa) texana (W. H. Edwards, 1863)
Currently in the subgenus Anthanassa Scudder, 1875 (of Eresia Boisduval, 1836) and treated as a subspecies of its type species Melitaea texana W. H. Edwards, 1863 (type locality USA: Texas, probably Comal Co., New Braunfels), Eresia texana seminole Skinner, 1911 (type locality USA: Georgia, Decatur Co., Bainbridge), is genetically differentiated from it (Fig. 9 blue vs. red), showing Fst/Gmin in the Z chromosome of 0.39/0.01 and COI barcode difference of 3.6% (24 bp). Therefore, in agreement with Calhoun (1997), we regard Eresia (Anthanassa) seminole Skinner, 1911, stat. rev. as a species-level taxon.
Fig. 9.

Nuclear genome tree (autosomes) of Eresia texana (blue) and Eresia seminole stat. rest. (red). Primary types are in magenta.
Microtia elvira Grishin, new species http://zoobank.org/17FC3D08-DF04-487B-A1F1-F536C01E624F (Figs. 10 part, 11, 12a, b)
Fig. 10.

Phylogenetic trees of Microtia inferred from protein-coding regions of a) nuclear (autosomes) and b) mitochondrial genomes: Microtia elvira sp. n. (red), M. elva (blue with M. e. horni labeled in teal), M. perse (green), and M. elada (violet).
Fig. 11.

Holotype of Microtia elvira sp. n. in dorsal (left) and ventral (right) views, data in text.
Fig. 12.

Microtia elvira sp. n. (a, b) and Microtia elva (c, d) iNaturalist observations: USA: AZ, Santa Cruz Co.: a) 144668453 Montosa Canyon, GPS 31.6727, −110.9406, 19-Aug-2016 © jmbearce; b) 141491956 Atascosa Mts., California Gulch, GPS 31.4220, −111.2404, 3-Oct-2014 © Ken Kertell; and Mexico: Nuevo Leon, Monterrey: c) 4292755 Parque la Estanzuela, GPS 25.5507, −100.2707, 7-Oct-2016 © Roberto González; d) 171257635 Guadalupe, Contry Sol, GPS 25.6505, −100.2648, 5-Jul-2023 © Rodolfo Salinas Villarreal. Arrows in b) and d) point at the legs to draw attention to their color difference. Images are color-corrected and rotated. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/
Definition and diagnosis.
Genomic sequencing of Microtia H. Bates, 1864 (type species Microtia elva H. Bates, 1864) specimens reveals a deep split within its type species (Fig. 10). The western clade (Fig. 10 red) is profoundly differentiated from the eastern clade (Fig. 10 blue) with the Z chromosome Fst/Gmin of 0.60/0.006 and COI barcode difference of 4.7% (31 bp), which is more than expected from phenotypic similarity. This scenario parallels that of Microtia perse (W. H. Edwards, 1882) (type locality in USA: AZ, Graham Co.) vs. Microtia elada (Hewitson, 1868) (type locality in Mexico) (Fig. 10 green vs. violet). Therefore, the two clades represent distinct species. Specimens from around the type locality (Guatemala) belong to the eastern clade. Curiously, Microtia elva horni Rebel, 1906 (type locality in Mexico: Oaxaca) and its junior subjective synonym Microtia elva form draudti Röber, [1914] (type locality in Mexico, “Coatepec” on the label of a syntype) (Fig. 10 teal colored), which are also in the eastern clade but are visually rather distinct in their much broader orange-yellow markings and nearly half of dorsal hindwing orange-yellow, are not particularly different genetically from the nominotypical M. elva. Thus, the western clade does not have available names associated with it and represents a new species. This new species is most similar to M. elva, with which it was previously combined. Distinguished from M. elva by largely yellow or orange-yellow tibiae and frequently other leg parts (legs in M. elva are back). Additionally, characterized by narrower orange-yellow bands and thinner, bar-like (particularly in males) orange-yellow mark by the middle of the inner forewing margin (Figs. 11, 12a). In M. elva, this bar is more rounded, larger, and can be nearly triangular and much broader at the base, widening both distad and basad (Fig. 12c). The outer edge of the hindwing discal band (on both dorsal and ventral sides) is more angled in the middle (Figs. 11, 12b), instead of straighter and more rounded edge in M. elva (Fig. 11d). Due to variation in colors and patterns, most confident identification is provided by DNA. The following combination of characters is diagnostic in the nuclear genome: hm2014195-RA.2:C190T, hm2014195-RA.2:T201A, hm2016592-RA.17:G261A, hm2017493-RA.1:G240A, hm2008057-RA.6:T171C and COI barcode: G38A, C238C, 421C, A583T, T637C.
Barcode sequence of the holotype: Sample NVG-22096H10, GenBank OR578711, 658 base pairs:
TACTTTATATTTTATTTTTGGAATTTGAGCAGGAATAATTGGAACATCTTTAAGACTTTTAATTCGAACTGAATTAGGAAACCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGTAATTGATTAATTCCATTAATATTAGGAGCTCCTGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGACTACTACCCCCATCACTTATATTATTAATTTCTAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTGTACCCCCCACTTTCTTCTAATATTGCTCATAGAGGATCATCTGTTGATTTAGCAATTTTTTCACTACATCTAGCAGGAATTTCCTCAATTCTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAATAATATATCATTTGATCAAATACCTTTATTTGTTTGAGCAGTTGGTATTACAGCTCTTTTATTATTATTATCTTTACCAGTATTAGCAGGAGCTATTACTATACTCCTTACTGATCGAAATATTAATACATCATTTTTTGACCCAGCTGGAGGAGGGGATCCCATTTTATATCAACATCTATTT
Type material.
Holotype: ♂ deposited in the Los Angeles County Museum of Natural History, Los Angeles, CA, USA [LACM], illustrated in Fig. 11, bears three printed (number “12” handwritten) labels: two white [October 12, 1954 | Madera Canyon, Santa Rita Mt’s. | Southern Arizona. | W. Rees & H. Reid], [DNA sample ID: | NVG-22096H10 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Microtia elvira | Grishin]. Paratypes: 5♂♂ 2♀♀: 1♂ USA, Arizona, Santa Cruz Co., Sycamore canyon, GPS 31.4213, −111.1942, 25-Sep-2015, Brian Banker leg. (NVG-21091A03); others at LACM, Mexico: Sonora: 2♂♂ Rio Cuchujaqui, 8 rd. mi E of Alamos, el. 1000′, 30-Aug-1976, J. P. & K. E. Donahue leg. (NVG-22096G07 & G08); 1♀ Alamos, 25-Jul-7-Aug-1953, Fred S. Truxal leg. (NVG-22096H08); 1♂ Sinaloa, 27.8 km S Culiacán, 30-Aug-1976, C. D. George & R. K. Snelling leg. (NVG-22097A04); 1♂ 1♀ Nayarit, 5–10 mi N of Tepic, 2500′−3000′, 13-Dec-1946 (NVG-22097A03 & A01).
Type locality.
USA: Arizona, Pima/Santa Cruz Cos., Santa Rita Mountains, Madera Canyon.
Etymology.
The meaning of the name Elvira is typically associated with traits such as truthful, trustworthy, or pure, and also noble or elf-like. Formed from the name of its sister species, elva, it signifies our high confidence that this is a “trustworthy” species, i.e., strongly differentiated from M. elva but is elva- or elf-like. The name of this western counterpart of M. elva is longer to mean that it takes a long day from sunrise in the east to sunset in the west. The name is a feminine noun in apposition.
English name.
Elfoid.
Distribution.
Southeastern Arizona and western Mexico (confirmed from Sonora, Sinaloa, and Nayarit).
Comment.
Both M. elvira sp. n. and M. elva occur in the USA. Microtia elva strays into the lower Rio Grande Valley of Texas, e.g., a male from Brownsville in Cameron County collected by William W. & Nadine McGuire on 20-Jul-1971 [TAMU] that we sequenced as NVG-10626 (Fig. 10).
Cyllopsis brocki Grishin, new species http://zoobank.org/3EE47FD0-31EF-4CE0-BE21-5DC240F1FFEC (Figs. 13 part, 14–16)
Fig. 13.

Phylogenetic trees of selected Cyllopsis species inferred from protein-coding regions of a) the nuclear genome (autosomes), b) the Z chromosome, and c) the mitochondrial genome: C. brocki sp. n. (red) and its sister group (blue).
Fig. 14.

Holotype of Cyllopsis brocki sp. n. in dorsal (left) and ventral (right) views, data in text.
Fig. 16.

Genitalia of Cyllopsis brocki sp. n. holotype (data in text) in left lateral (top left), dorsal (right) and left ventrolateral (bottom left) views.
Definition and diagnosis.
Inspection of nuclear genomic trees reveals that a single specimen from Mexico, Sonora, initially identified as “Cyllopsis pertepida”, is confidently placed as sister to the clade of three species: Cyllopsis gemma (Hübner, 1808), Cyllopsis pertepida (Dyar, 1912), and Cyllopsis pyracmon (A. Butler, 1867) (Fig. 13a, b, red and blue), and therefore represents a species distinct from them. In the mitochondrial genome tree, which has lower statistical support, this species is also in the same clade with the three others but is sister to C. gemma (Fig. 13c). Because all other described species of Cyllopsis R. Felder, 1869 (type species Cyllopsis hedemanni R. Felder, 1869) belong to other clades (Fig. 13) and are phenotypically different (Miller 1974), this species is new. It differs from its relatives by a combination of larger size (forewing length about 20 mm, while typically less than 18 mm in C. gemma), a large androconial patch (cut by veins) in the discal area of the dorsal forewing (absent in C. gemma), postdiscal brown line strongly toothed towards the margin at vein M1 and not reaching the costal margin (as in C. gemma and some C. pertepida, but not in C. pyracmon), more extensive and coarse mottling on the ventral side of wings, especially in the basal part of the hindwing (as in some C. pyracmon but not in other species), and reduced rusty overscaling dorsally: wings appear more brown than reddish (Figs. 14, 15). In male genitalia (Fig. 16), diagnosed by broader valva with more convex costa-ampulla, valva more distinctively narrowing into harpe in lateral view, harpe with broader and more angular distal end in dorsal view shaped like a triangular plate rather than ending in a point. In DNA, a combination of the following characters is diagnostic in the nuclear genome: hm2006469-RA.2:C61T, hm2003322-RA.2:G69A, hm2003322-RA.2:T106C, hm2009012-RA.1:T408C, hm2009012-RA.1:G885T, hm2009732-RA.2:G72G (not A), hm2009355-RA.1:A261A (not G), hm2009355-RA.1:G270G (not A), hm2013443-RA.4:T606T (not C), hm2013443-RA.4:T702T (not A) and COI barcode: T235C, T283C, T379A, T451A, T544C, A631G. Due to pronounced and frequently seasonal wing pattern variation in Cyllopsis that is unexplored in the new species, definitive identification is possible by genitalia or DNA.
Fig. 15.

Cyllopsis brocki sp. n. (possible), iNaturalist observation 142409312 Mexico: Sonora Yécora, GPS 28.3783, −108.8370, 3-Sep-2018, © jmbearce. Brightened and color-corrected. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/
Barcode sequence of the holotype: Sample NVG-20112B02, GenBank OR578712, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGTATAGTTGGAACCTCCCTTAGTCTTATTATTCGAATAGAATTAGGTAATCCAGGATTTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGTGGATTTGGGAACTGATTAGTTCCCCTTATATTAGGGGCCCCCGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGACTACTTCCCCCCTCATTAGTCCTTTTAATTTCAAGAAGTATTGTAGAAAATGGAGCTGGRACAGGATGAACAGTTTACCCCCCTCTTTCTTCTAATATTGCCCACAGAGGATCTTCAGTCGATTTAGCAATTTTTTCTTTACATTTAGCTGGGATTTCATCAATTTTAGGAGCTATTAATTTTATTACAACAATTATTAACATACGAATTAATAATATAACGTATGATCAAATACCTCTATTTGTTTGAGCAGTTGGAATTACAGCATTATTATTATTACTCTCTTTACCAGTATTAGCTGGAGCTATTACTATACTTTTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCTGCTGGAGGAGGGGACCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA [MGCL], illustrated in Fig. 14, bears four printed labels: three white [31 July 1984 | Trinidad-Yecora Rd. | 1–5 mi. e. Trinidad | Mine, Son., Mexico | leg. Jim P. Brock], [DNA sample ID: | NVG-20112B02| c/o Nick V. Grishin], [genitalia vial | NVG230917–02 | Nick V. Grishin], and one red [HOLOTYPE ♂ | Cyllopsis brocki | Grishin].
Type locality.
Mexico: Sonora, Yécora, E of Santa Rosa, Trinidad-Yecora Rd., 3–5mi E of Trinidad mine.
Etymology.
The name honors Jim P. Brock, the collector of the holotype and one of the finest and most knowledgeable Lepidopterists with a sixth sense for butterflies and caterpillars, finding them effortlessly (or so it seems) where others fail. Jim’s significant contributions to butterfly knowledge delivered through his many books, presentations, and nature tours can only be matched by his contagious excitement, passion for sharing his expertise, and unsurpassed kindness. We deeply appreciate Jim’s extensive support of our projects throughout the years. The name is a singular noun in the genitive case.
Distribution.
Known only from the holotype collected in Mexico: Sonora.
Comment.
The type locality of this new species is near the type locality of Amblyscirtes brocki H. Freeman, 1992 (Hesperiidae), with its two paratypes collected on the same date and at approximately the same place as the holotype of Cyllopsis brocki sp. n.
Family Riodinidae Grote, 1895 (1827)
Teratophthalmina Grishin, new subtribe http://zoobank.org/8C350BD8-8094-472C-9C17-70A9C2023D20
Type genus.
Teratophthalma Stichel, 1909.
Definition.
Currently placed in the subtribe Eunogyrina Grishin, 2021, Teratophthalma (type species Mesosemia phelina C. Felder & R. Felder, 1862) is indeed sister to its type genus Eunogyra Westwood, 1851, but is rather distant from it both genetically (Fig. 17) and phenotypically, and therefore represents a subtribe of its own. The description and diagnostic characters of this new subtribe are as those given for Teratophthalma on pages 76–77 (illustrated in Fig. 11) by Stichel (1910). In brief, the subtribe belongs to Mesosemiini (see Hall (2003) for Teratophthalma) and is diagnosed by the following combination of characters: wings without multiple narrow bands, eyespots at the end of the forewing discal cell but not along wing margins; genitalic valvae short (as long as tegumen) and triangular, simple with pointed apex, pedicel unsclerotized in the middle or ventrally split (or both). A combination of the following nuclear genomic base pairs is diagnostic: cne1792.4.2:T805C, cne11317.11.9:A512G, cne12569.1.1:T2145C, cne599.10.1:A4814T, cne1398.1.1:C838G.
Fig. 17.

The phylogenetic tree of selected Riodinidae inferred from protein-coding regions of the nuclear genome (autosomes). Family-group names are shown above or below the corresponding branches. Names, clades, and species of new subtribes proposed in this work are shown in color. Names of other subtribes are shown in black, and names of subfamilies and tribes are shown in shades of gray. Note the relatively constant rate of DNA change in Riodininae (i.e., molecular clock), as evidenced by approximately the same distance from the root to each leaf. The translucent vertical lime bar denotes a level in the tree approximately corresponding to subtribes.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Mesosemiini Bates, 1859.
Argyrogrammanina Grishin, new subtribe http://zoobank.org/FA7881EE-0382-4796-9C6B-A10B4C08B0A1
Type genus.
Argyrogrammana Strand, 1932.
Definition.
Argyrogrammana (type species Erycina stilbe Godart, 1824) is prominently differentiated genetically from the rest of Symmachiini Reuter, 1896 (Fig. 17), and we propose that the lineage containing Argyrogrammana is a subtribe. This new subtribe differs from other Symmachiini by a thin submarginal line of (sometimes fused) metallic (golden or silvery-blue) spots on both wings above and beneath and a dark medial stripe across the eyes (Hall and Willmott 1996). A combination of the following nuclear genomic base pairs is diagnostic: cne3461.2.10:G397A, cne2040.8.2:A100G, cne3461.2.10:G382A, cne2478.7.10:C607A, cne6674.8.2:G397C.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Symmachiini Reuter, 1896.
Argyrogrammana astuta Grishin, new species http://zoobank.org/C3B104C6-18F0-4FAE-8506-DD345ED63531 (Figs. 18, 19)
Fig. 18.

Holotype of Argyrogrammana astuta sp. n. in dorsal (left) and ventral (right) views, data in text.
Fig. 19.

Argyrogrammana astuta sp. n. iNaturalist observations from Peru: Madre de Dios, Tambopata: a) 175878841 GPS −12.6060, −69.0324, 27-Jul-2023 © danielblanco521; b) 67719625 Tambopata, GPS −12.0467, −69.6766, 29-Sep-2017, © Ken Kertell; c) 94229584 ventral of b) © David Geale. Images are color-corrected and rotated. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/
Definition and diagnosis.
Genomic sequencing of the holotype of Argyrogrammana praestigiosa (Stichel, 1929) (type locality not specified, likely the Guianas) (in MFNB, NVG-18077D12) reveals that a sequenced specimen identified as A. praestigiosa from southeastern Peru (illustrated in Fig. 11 in Hall et al. (2023)) is genetically distant from it with COI barcodes differing by 2.6% (17 bp). In the presence of phenotypic differences, such as those in wing patterns discussed in detail by Hall et al. (2023), the observed genetic differentiation suggests that the specimen from Peru is not A. praestigiosa but a distinct species. This species is new, and its males (female is unknown) differ from superficially most similar A. praestigiosa by the characters described for “west Amazonian males” in Hall et al. (2023), such as more extensive orange coloration of dorsal wings, i.e., basal area of forewing with three orange bands and in some specimens an orange marginal streak near tornus, hindwing with more orange scaling by its apex, e.g., an orange band may cut through the brown patch to reach the costa (the apical area is mostly brown in A. praestigiosa). Beneath, brown scaling is less extensive, pale bands are wider, with more orange scales in them (particularly toward wing margins), and with sharper edges, especially towards the outer margin; submarginal dark spots are more even, e.g., on hindwing spots in cells M1-M2 and M2-M3 are not particularly larger than others (they are larger and more pointed in A. praestigiosa). The dorsal side of the abdomen with a brown central spot at the base of each segment (entirely orange in A. praestigiosa). Other species of Argyrogrammana are more distant and different, e.g., the next closest species is A. glaucopis (H. Bates, 1868), which is characterized by much less extensive orange coloration and additional blue spots on the forewing, and A. caerulea J. Hall, 2023 has even more extensive blue forewing patches.
Barcode sequence of the holotype: Sample NVG-19029F11, GenBank OR578713, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCTGGAATAATTGGAACTTCTTTAAGTTTATTAATTCGTATAGAATTAGGTAATCCAAACTCATTAATTGGTAATGACCAAATTTATAATACAATTGTAACTGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGGTTTGGAAATTGATTAATTCCTTTAATATTAGGGGCTCCTGATATAGCATTTCCACGAATAAATAACATAAGTTTTTGATTATTACCCCCCTCCTTAATTCTTTTAATTTCAAGAAGTATTGTTGAAAATGGAGCAGGAACAGGATGAACAGTTTATCCCCCTCTTTCTTCTAATATTGCTCATAGAGGCTCATCCGTTGATTTAGCCATTTTTTCTCTTCATTTAGCTGGGATTTCTTCCATTCTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGTATTAATAATATAGCTTTTGATCAAATACCTTTATTTGTTTGATCTGTAGGAATTACAGCTCTTCTTTTATTATTATCATTACCAGTTTTAGCTGGAGCTATTACCATATTATTAACAGATCGTAATTTAAATACATCATTTTTTGATCCTGCTGGAGGTGGAGATCCTATTTTATACCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Washington, DC, USA [USNM], illustrated in Fig. 18 (and Fig. 11 in Hall et al. (2023) left-right inverted—i.e., mirror image of the specimen—and with imperfections edited out), bears four printed labels: three white [PERU, Madre de Dios | Tambopata Reserve | 12° 50’S 69° 17’W, 300m | 28 Oct 1991 | Leg. R. Robbins], [DNA sample ID: | NVG-19029F11 | c/o Nick V. Grishin], [USNMENT | {QR Code} | 01544363], and one red [HOLOTYPE ♂ | Argyrogrammana | astuta Grishin].
Type locality.
Peru: Madre de Dios, Tambopata National Reserve, elevation 300 m, GPS −12.83, −69.28.
Etymology.
In Latin, astutus is astute, cunning, clever, sly, and artful, and praestigiosus is deceptive and misleading. This new species was skillfully hiding among the deceitful A. praestigiosa until genomic sequencing revealed its distinction. The name is a feminine adjective.
Distribution.
This species is genetically confirmed only from the holotype collected in southeastern Peru; however, it is expected at least in the southwestern Amazonian Basin (Fig. 19).
Comments.
Hall et al. (2023) described in detail wing pattern differences between populations of A. praestigiosa, concluding that they represent intraspecific variation due to the lack of apparent differences in genitalia and citing phenotypic intermediates. Argyrogrammana astuta sp. n. corresponds to the “west Amazonian” A. praestigiosa of Hall et al. (2023). It is possible that A. praestigiosa is even more deceitful, and additional variation described by Hall et al. (2023) may refer to additional species in this complex.
Subtribes in Helicopini Stichel, 1928
Inspection of the genomic tree reveals that Sarota Westwood, 1851 (type species Papilio chrysus Stoll, 1782), which is confidently placed in the tribe Helicopini Stichel, 1928 (Fig. 17), diverges from other members of the tribe at the tree level corresponding to subtribes (left of the lime bar in Fig. 17). Therefore, this lineage represents a valid subtribe. As a result, we propose to split Helicopini into two subtribes: the nominotypical and Sarotina Bridges, 1988, stat. rest.
Echenaidina Grishin, new subtribe http://zoobank.org/3BAD20C2-92C3-4F44-A713-583D7B3932E2
Type genus.
Echenais Hübner, [1819].
Definition.
Two subgenera, Echenais (type species Lemonias alphaea Hübner, 1808, which is a junior subjective synonym of Papilio thelephus Cramer, 1775) and Imelda Hewitson, 1870 (type species Imelda glaucosmia Hewitson, 1870), show prominent genetic differentiation from other taxa in the tribe Calydnini Seraphim, Freitas & Kaminski, 2018 and, therefore, constitute a subtribe (Fig. 17). This new subtribe differs from other Calydnini by a submarginal (not marginal) continuous yellow, blue, or metallic line or band on dorsal sides of both wings and/or largely white, yellow, orange, or blue (but not mostly black or dark brown) hindwing above, or orange patch in the middle of the hindwing. A combination of the following nuclear genomic base pairs is diagnostic: cne2872.1.1:A361C, cne12666.5.7:A497G, cne2222.10.1:A431T, cne2483.2.1:A1667G, cne254274.1.1:T198C.
Genera included.
Only the type genus (including subgenus Imelda Hewitson, 1870).
Parent Taxon.
Tribe Calydnini Seraphim, Freitas & Kaminski, 2018.
Echydnina Grishin, new subtribe http://zoobank.org/7E98DF54-98E3-41D7-BC19-3528A548CD96
Type genus.
Echydna J. Hall, 2002.
Definition.
Echydna (type species Calydna chaseba Hewitson, 1854) is genetically differentiated from other Calydnini genera at the tree level similar to that of Echenaidina subtrib. n. and therefore represents a distinct subtribe (Fig. 17). Diagnostic characters for this subtribe are as those given in detail and illustrated by Hall (2002) for Echydna. In brief, it is diagnosed by a combination of setose eyes and black frons; in male genitalia, unique cornuti of three elements: a narrow unspined plate anteriad of an unsclerotized sack with small spines and sclerotized ovoid with larger spines; and in female genitalia, small signa without surface sculpturing. A combination of the following nuclear genomic base pairs is diagnostic: cne2156.9.5:A3355G, cne5645.4.3:G360A, cne16213.4.1:A205G, cne16034.2.1:T643G, cne5785.9.3:G454A.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Calydnini Seraphim, Freitas & Kaminski, 2018.
Cariina Grishin, new subtribe http://zoobank.org/6D9F6CE6-E393-4484-8124-963E2C60DE55
Type genus.
Caria Hübner, 1823.
Definition.
The tribe Riodinini Grote, 1895 (1827) splits into two prominent clades: the larger includes the type genus, and the smaller consists of genera not previously used for family-group names (Fig. 17). We propose that these two clades represent two subtribes, one of which is new. This new subtribe is diagnosed by a combination of the following characters: eyes bare, palpi short, not visible above, both sides of all wings either with submarginal row of metallic (silver, green) spots usually connected into a band, or dark and with yellow bands or spots, or pale veins. In Barbicornis, hindwings unique, shovel-shaped with a tail in the middle nearly equal to the hindwing length; in Chamaelimnas, forewings elongated and with a yellow band, either diagonal (from mid-costa to tornus) or longitudinal (from base towards outer margin); in Caria and Seco, forewing costal margin typically at least slightly convex. Male genitalia with terminally bilobed valvae; in Chamaelimnas, boomerang-shaped, ventral lobe vestigial. Best identified by DNA, and a combination of the following nuclear genomic base pairs is diagnostic: cne1302.2.1:A1473G, cne1302.2.1:A2614C, cne1143.5.1:A883G, cne1143.5.1:A884C, cne8241.4.4:A67C.
Genera included.
The type genus (Caria Hübner, 1823), Barbicornis Godart, 1824, Chamaelimnas C. Felder & R. Felder, 1865, and Seco J. Hall & Harvey, 2002.
Parent Taxon.
Tribe Riodinini Grote, 1895 (1827).
Family Lycaenidae [Leach], [1815]
Liphyrinae Doherty, 1889 is a subfamily
Currently treated as a tribe within subfamily Miletinae Reuter, 1896, Liphyrini Doherty, 1889 show prominent genetic differentiation from the rest of Miletinae and are at the level in the genomic tree that corresponds to subfamilies (Fig. 20). Therefore, we return this morphologically unique group to the status of a subfamily as originally proposed: Liphyrinae Doherty, 1889, stat. rest.
Fig. 20.

The phylogenetic tree of selected Lycaenidae (Polyommatinae are shown in Fig. 22 below) inferred from protein-coding regions of the nuclear genome (autosomes). Family-group names are shown above or below the corresponding branches. Not all subtribes are shown and/or labeled (see the subtribal classification list of Lycaenidae below). Names, clades, and species of new tribes and subtribes proposed in this work are shown in color. Names of other subtribes are shown in black, and names of subfamilies and tribes are shown in shades of gray.
Megalopalpina Grishin, new subtribe http://zoobank.org/2A22AFFB-346D-4DCF-B256-C59DA7FCEFB1
Type genus.
Megalopalpus Röber, 1886.
Definition.
Megalopalpus Röber, 1886 (type species Megalopalpus simplex Röber, 1886) forms a lineage sister to all other Miletini Reuter, 1896 and is more prominently separated from them than they are from each other (Figs. 20, 21). Therefore, we propose that this lineage represents a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for the Megalopalpus section by Eliot (1973): uncus with tegumen plates triangular, with a unique for this group broad lobe-like triangular ventrally-pointed process, falces strongly curved, males lack secondary sexual characters, hindwing with precostal vein. A combination of the following nuclear genomic base pairs is diagnostic: cce64.1.4:G152C, cce5960.3.7:A41T, cce7884.1.3:A388A (not C), cce28693.2.4:G109G (not A), cce9880.2.2:A83A (not C), cce49.4.1:C595C (not T), cce16970.13.7:T187T (not G).
Fig. 21.

The phylogenetic tree of Miletinae and Aphnaeinae inferred from protein-coding regions of the nuclear genome (autosomes). Names are shown by corresponding branches: family-group in roman font and subgenera in italics. Names proposed in this work are marked with red asterisks. Names, clades, and species of taxa referring to this figure from the text are colored: Tarakina stat. nov. (green), Spindasis stat. rest. (magenta), Vansomerenia syn. nov. (orange), and Auricirrus subgen. n. (blue). Other names of subtribes are shown in black, and names of subfamilies and tribes are shown in shades of gray.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Miletini Reuter, 1896.
Tarakina Eliot, 1973 is a subtribe
Currently treated as a tribe in the subfamily Miletinae Reuter, 1896, Tarakini Eliot, 1973 is sister to Spalgini Toxopeus, 1929 and shows less extensive genetic differentiation from them than typical for the tribal level (Fig. 21). The Taraka Doherty, 1889 lineage has split from Spalgini at the same tree level as Megalopalpina subtrib. n. has split from Miletina Reuter, 1896. Therefore, we place Tarakina Eliot, 1973, stat. rev. as a subtribe in the tribe Spalgini, 1929.
Cesa Seven, 1997 is a subgenus of Crudaria Wallengren, 1875
The monotypic genus Cesa Seven, 1997 (type species Spindasis waggae Sharpe, 1898) is closely related to Crudaria Wallengren, 1875 (type species Arhopala ? leroma Wallengren, 1857): the COI barcodes of the type and the only species of Cesa (GenBank JN286116) and the type species of Crudaria (NVG-22012H12) differ by 5.5% (36 bp). For comparison, the COI barcode difference between the type species of sister genera (according to Boyle et al. (2015)) Crudaria and Cigaritis Donzel, 1848 (type species Cigaritis zohra Donzel, 1848, NVG-22012H05) is 9.6% (63 bp), which is nearly twice as large and does not preclude genus-level genetic differentiation between Crudaria and Cigaritis. Therefore, Cesa does not represent a significant deviation from Crudaria to warrant its monotypic status as a genus and may be placed in this genus with its close relatives. Acknowledging some morphological distinction of Spindasis waggae, we propose to treat Cesa Seven, 1997, stat. nov. as a subgenus of Crudaria Wallengren, 1875.
Vansomerenia Heath, 1997 is a junior subjective synonym of Chloroselas Butler, 1886
The monotypic genus Vansomerenia Heath, 1997 (type species Desmolycaena rogersi Riley, 1932) originates within the genus Chloroselas Butler, 1886 (type species Chloroselas esmeralda Butler, 1886), rendering it paraphyletic (Fig. 21). The difference between their COI barcodes is 6.2% (41 bp), which is typical for congeners. To restore monophyly and place close relatives in a single genus, we propose that Vansomerenia Heath, 1997, syn. nov. is a junior subjective synonym of Chloroselas Butler, 1886.
Auricirrus Grishin, new subgenus http://zoobank.org/B527E473-8675-489E-B979-EAFF56996F4B
Type species.
Papilio thysbe Linnaeus, 1764.
Definition.
African genus Chrysoritis Butler, 1898 (type species Zeritis oreas Trimen, 1891) has been divided into two clades: the eastern (chrysaor clade) and the western (includes the thysbe clade) (Talavera et al. 2020) (Fig. 21). Strong genetic differentiation between these clades warrants their recognition at least as subgenera: estimated divergence time between them is about 17 Mya (Talavera et al. 2020), comparable to that of some genera. Among all Chrysoritis species, the type species of available genus-group names belong to the eastern clade (Talavera et al. 2020): Zeritis oreas Trimen, 1891 (of Chrysoritis), Zeritis lycegenes Trimen, 1874, which is a subspecies of Zeritis lyncurium Trimen, 1868 (of Poecilmitis Butler, 1899), Zeritis phosphor Trimen, 1864 (of Bowkeria Quickelberge, 1972), and Phasis dicksoni Gabriel, 1947 (of Oxychaeta Tite & Dickson, 1973). This eastern clade represents the nominotypical subgenus. However, the western clade remains without a name and represents a new subgenus. This subgenus differs from the nominal by the absence of tibial spicules (Heath 1997), produced (but not tailed) hindwing tornus and/or blue or violet scaling on dorsal surface of wings in some species (lacking in the nominotypical subgenus), and in species with more rounded tornus, spots on ventral forewing—especially at the end of discal cell—are with more extensive silvery scaling. A combination of the following nuclear genomic base pairs is diagnostic: cce3229.3.2:A633G, cce2784.3.3:C85G, cce2423.1.2:A2767T, cce7537.1.5:C57T, cce5689.1.7:A744G.
Etymology.
The name Chrysoritis may have been derived from the Greek “chryso” (gold) and “ritis” (hair or curl). Their Latin equivalents ar “aurum” (gold) and “cirrus” (lock or curl of hair), connected with a vowel i to form the name of the subgenus. The name is a masculine noun in the nominative singular.
Species included.
The type species (i.e., Papilio thysbe Linnaeus, 1764) and 41 others, names given in their original combinations: Poecilmitis adonis Pennington, 1962, Poecilmitis turneri amatola Dickson & McMaster, 1967, Chrysoritis adonis aridimontis Heath & Pringle, 2007, Poecilmitis aridus Pennington, 1953, Poecilmitis azurius Swanepoel, 1975, Poecilmitis beaufortia Dickson, 1966, Poecilmitis beulah Quickelberge, 1966, Poecilmitis blencathrae Heath & Ball, 1992, Poecilmitis braueri Pennington, 1967, Poecilmitis thysbe brooksi Riley, 1938, Zeritis chrysantas Trimen, 1868, Poecilmitis (Poecilmitis) daphne Dickson, 1975, Poecilmitis endymion Pennington, 1962, Zeritis felthami Trimen, 1904, Poecilmitis irene Pennington, 1968, Poecilmitis lyndseyae Henning, 1979, Poecilmitis lysander Pennington, 1962, Poecilmitis mithras Pringle, 1994, Phasis thysbe var. nigricans Aurivillius, 1924, Poecilmitis orientalis Swanepoel, 1976, Papilio palmus Stoll, 1781, Poecilmitis pan Pennington, 1962, Poecilmitis pelion Pennington, 1953, Poecilmitis penningtoni Riley, 1938, Poecilmitis perseus Henning, 1977, Poecilmitis plutus Pennington, 1967, Poecilmitis pyramus Pennington, 1953, Zeritis pyroeis Trimen, 1864, Poecilmitis rileyi Dickson, 1966, Poecilmitis stepheni Dickson, 1978, Poecilmitis swanepoeli Dickson, 1965, Poecilmitis thysbe trimeni Riley, 1938, Poecilmitis turneri Riley, 1938, Poecilmitis uranus Pennington, 1962, Poecilmitis violescens Dickson, 1971, Poecilmitis whitei Dickson, 1994, Poecilmitis williami Heath, 1997, Poecilmitis wykehami Dickson, 1980, Papilio zeuxo Linnaeus, 1764, Phasis zeuxo zonarius Riley, 1938, Poecilmitis nigricans zwartbergae Dickson, 1982 (including their subspecies and synonyms). Species taxonomy follows Heath (2023) and Williams (2023c).
Parent taxon.
Genus Chrysoritis Butler, 1898.
Spindasis Wallengren, 1857 is a subgenus of Cigaritis Donzel, 1848
Phylogenetic analysis of Aphnaeinae Distant, 1884 revealed that the genus Cigaritis Donzel, 1848 (type species Cigaritis zohra Donzel, 1848) splits into two prominent clades: mostly Oriental (hindwing tornal lobe not well developed or lacking, at least one hindwing tail short, < 2 mm) and African (hindwing tornal lobe well developed, both hindwing tails long) (Riley 1925) with genetic differentiation between them similar to that of the two subgenera in Chrysoritis Butler, 1898 (type species Zeritis oreas Trimen, 1891) (Boyle et al. 2015) (Fig. 21). Therefore, the two clades of Cigaritis correspond to subgenera. The (mostly) Oriental clade is the nominal subgenus because the type species of Cigaritis (from NW Africa) belongs to this clade. Two additional genus-group names have type species currently placed in Cigaritis: Spindasis Wallengren, 1857 (type species Spindasis masilikazi Wallengren, 1857, which is a junior subjective synonym of Aphnaeus natalensis Westwood, [1851]) and Apharitis Riley, 1925 (type species Polyommatus epargyros Eversmann, 1855). The latter is in the Asian clade and, therefore, remains a junior subjective synonym of Cigaritis. The former belongs to the African clade and, therefore, this clade corresponds to the subgenus Spindasis Wallengren, 1857, stat. rest., which we propose to treat as a valid subgenus instead of a synonym.
Tribes in Aphnaeinae Distant, 1884
Inspection of the genomic tree reveals three strongly supported clades in the subfamily Aphnaeinae Distant, 1884 that we define as tribes (Figs. 20, 21): nominotypical, “Apharitini” Chou et al., 1994, and the third one that does not have a name and is described below. The name “Apharitini” was published in Chou et al. (1994) in an informal way without description, definition, or references to them (fails ICZN Art. 13.) and is a nomen nudum. The name for this tribe that satisfies the requirements of the ICZN Code is proposed below. We define three tribes and not two because the phylogenetic affinities of the unnamed tribes with the nominotypical one are not particularly strong (86%), and combining their clades may not result in a monophyletic taxon if this topology is incorrect. The three clades are the same in the recently published large-scale tree (Kawahara et al. 2023), but the topology is different in a tree based on a more limited set of gene markers (but a larger set of taxa) (Boyle et al. 2015).
Cigaritini Grishin, new tribe http://zoobank.org/E44D9831-7137-4120-86CB-700DBCC8D752
Type genus.
Cigaritis Donzel, 1848.
Definition.
The clade with Cigaritis (type species Cigaritis zohra Donzel, 1848) and related genera is genetically differentiated from other clades and is at the tree level of a tribe (Figs. 20, 21). This new tribe is morphologically diverse and can be distinguished from other members of the subfamily Aphnaeinae Distant, 1884 by the shape of uncus, which is highly variable, but nevertheless bilobed or divided, with centrally concave distal margin (see Stempffer (1967) for illustrations and more detailed descriptions): in Lipaphnaeus Aurivillius, 1916, subtriangular uncus is deeply divided with narrow, tooth-like pointed lobes; in Chloroselas Butler, 1886, the lobes are small and subtriangular uncus is with a central notch separating finely serrated lobes; in Cigaritis Donzel, 1848, uncus is broader than long, nearly rectangular or trapezoidal in dorsal view (sometimes nearly vestigial), lobes widely separated, uncus mostly concave (may be with irregularities) between the lobes; in Chrysoritis Butler, 1898 and Crudaria Wallengren, 1875, uncus is similar, trapezoidal or heart-shaped, but less broad, with rounded lobes and convex margin between them; in Pseudaletis H. H. Druce, 1888, uncus is divided for nearly its entire length with finger-like terminally rounded lobes. In other Aphnaeinae tribes, uncus in undivided and not strongly bilobed: typically, from almost square to broadly rectangular, with nearly flat (with some irregularities) distal margin, or dome-shaped with convex distal margin, which is only slightly concave in Trimenia Tite & Dickson, 1973 (type species Zeritis wallengrenii Trimen, 1887), thus resembling Chrysoritis, but uncus is narrower and without clearly defined lobes expanded laterally (as in Chrysoritis and Crudaria). Furthermore, Trimenia has saccus, which is not developed in Chrysoritis. A combination of the following nuclear genomic base pairs is diagnostic: cce332.12.5:A61T, cce1232.20.1:A596G, cce1351.40.3:A349T, cce5072.9.1:A67C, cce2368.14.7:G95T.
Genera included.
The type genus (i.e., Cigaritis Donzel, 1848), Chloroselas Butler, 1886, Chrysoritis Butler, 1898, Crudaria Wallengren, 1875, Lipaphnaeus Aurivillius, 1916, and Pseudaletis H. H. Druce, 1888, including their subgenera and synonyms (e.g., Cesa Seven, 1997, Vansomerenia Heath, 1997, and Apharitis Riley, 1925).
Parent Taxon.
Subfamily Aphnaeinae Distant, 1884.
Comment.
This tribe corresponds to “Apharitini” of Chou et al. (1994), published without description, definition, or references to them: a nomen nudum (fails ICZN Art. 13.). Apharitis Riley, 1925 (type species Polyommatus epargyros Eversmann, 1855) is a junior subjective synonym of Cigaritis.
Pseudaletidina Grishin, new subtribe http://zoobank.org/D8A065B1-F0CF-4A7F-BADA-F06FDEECA919
Type genus.
Pseudaletis H. H. Druce, 1888.
Definition.
Pseudaletis (type species Pseudaletis agrippina H. H. Druce, 1888) is in the lineage that is sister to all other Cigaritini Grishin, trib. n. and is genetically differentiated from them at the subtribal level (Figs. 20, 21). Therefore, we propose to treat this lineage as a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for the Pseudaletis section by Eliot (1973): palpi short—much less than half of the head length—covered in appressed scales, proboscis very short (but functional), forewing appears disproportionately large comparatively to hindwing; uncus divided for nearly its entire length with finger-like terminally rounded lobes, falces rudimentary, pointed processes inflexibly fused to tegumen; female abdomen with a prominent tuft of specialized scales, which are spoon-shaped with long “handles”. A combination of the following nuclear genomic base pairs is diagnostic: cce1853.23.8:T1384A, cce133.4.1:A287G, cce4260.4.1:G34A, cce1367.9.3:G1252A, cce17940.6.2:C71A.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Cigaritini Grishin, trib. n.
Axiocersini Grishin, new tribe http://zoobank.org/1BBFE9EC-AB8B-4709-A250-4DA082ED2B4E
Type genus.
Axiocerses Hübner, [1819].
Definition.
Axiocerses (type species Papilio perion Stoll, 1782, which is a junior subjective synonym of Papilio harpax Fabricius, 1775) and Zeritis Boisduval, 1836 (type species Zeritis neriene Boisduval, 1836) are confident sisters in the genomic tree (Figs. 20, 21), consistently with similarities in their morphology reported previously (Stempffer 1967). The two genera form a clade that originates early in the radiation of the subfamily Aphnaeinae Distant, 1884, and, therefore, it corresponds to a tribe. This new tribe is distinguished from the relatives by a combination of the following characters: uncus very broad and short, distal margin straight or convex, lobes nearly triangular or rounded, falces thick at the base, then strongly angled with free branch long and slender, ventral side with apophysis, tegumen with uncus hood-shaped, tegumen with convex anterior margin; palpi short, not extending or slightly extending beyond the fronts, 2nd segment of palpi with long scales and hairs; tarsus unsegmented, with spines below; forewing with only 10 veins (Stempffer 1967; Henning and Henning 1996). The similarity in uncus, falces, and tegumen unifies Axiocerses and Zeritis Boisduval, 1836 (Stempffer 1967). A combination of the following nuclear genomic base pairs is diagnostic: cce243.9.9:A1417C, cce15587.11.3:G103A, cce127.4.3:A149T, cce980.22.7:C902T, cce2423.1.2:T2315C.
Genera included.
The type genus (i.e., Axiocerses Hübner, [1819]) and Zeritis Boisduval, 1836.
Parent Taxon.
Subfamily Aphnaeinae Distant, 1884.
Aloeidina Grishin, new subtribe http://zoobank.org/A32E1EE2-A06D-4100-AFD9-3214F38A2C93
Type genus.
Aloeides Hübner, [1819].
Definition.
Within Aphnaeini Distant, 1884, the clade of several genera that includes Aloeides (type species Papilio pierus Cramer, 1779) corresponds to the subtribal level in the tree (Figs. 20, 21). This new subtribe is distinguished from its relatives by a combination of these characters, as discussed and illustrated by Stempffer (1967), Tite & Dickson (1973), and Eliot (1973): foreleg and midleg tibiae with apical spurs, palpi smooth, with equal length scales (in several species with some scattered long ribbon-shaped blunt scales), or with uniquely long and bristly white scales (in Erikssonia Trimen, 1891), forewing vein R4+5 originates at or beyond the junction of the discocellular vein and vein M1, vein R2 originates next to the origin of vein R4+5. A combination of the following nuclear genomic base pairs is diagnostic: cce5760.10.2:C484T, cce2790.13.2:T63C, cce2790.13.2:A183G, cce1354.3.7:A61T, cce1354.3.7:T1903A.
Genera included.
The type genus (i.e., Aloeides Hübner, [1819]), Argyraspodes Tite & Dickson, 1973, Erikssonia Trimen, 1891, and Trimenia Tite & Dickson, 1973.
Parent Taxon.
Tribe Aphnaeini Distant, 1884.
Phasisina Grishin, new subtribe http://zoobank.org/B6D8239D-DAF0-4865-9F3E-18677ABDEA6F
Type genus.
Phasis Hübner, [1819].
Definition.
Within Aphnaeini Distant, 1884, the clade of several genera that includes Phasis (type species Papilio salmoneus Stoll, 1781, which is a junior subjective synonym of Papilio thero Linnaeus, 1764) corresponds to the subtribal level in the tree (Figs. 20, 21). This new subtribe is distinguished from its relatives by a combination of the following characters, as discussed and illustrated by Tite & Dickson (1973): tibia of all legs without apical spurs, forewing vein M2 originates much closer to the origin of vein M1 than to the origin of vein M3, vein R2 originates at a distance from vein R4+5, size larger with forewing longer than 16 mm. A combination of the following nuclear genomic base pairs is diagnostic: cce288.3.2:A79C, cce33.3.3:A234G, cce2859.6.1:T729C, cce511.6.1:A54G, cce14215.5.1:T850C, cce33280.1.7:T944T (not A), cce3467.4.4:A374A (not G), cce7428.5.1:G156G (not A), cce4435.12.1:C616C (not A), cce1239.1.3:T235T (not G).
Genera included.
The type genus (i.e., Phasis Hübner, [1819]) and Tylopaedia Tite & Dickson, 1973.
Parent Taxon.
Tribe Aphnaeini Distant, 1884.
Surendrini Koçak & Seven, 1997 is a tribe
Currently placed in the tribe Arhopalini Bingham, 1907, Surendrina Koçak & Seven, 1997 is not monophyletic with it and instead is sister to Theclini Swainson, 1830 (Fig. 20). It is conceivable to place Surendrina in Theclini, but the two groups are more distinct from each other than subtribes and are at the tree level of tribes. Therefore, we propose to treat the former taxon as a tribe Surendrini Koçak & Seven, 1997, stat. nov.
Drinini Grishin, new tribe http://zoobank.org/2001E28A-4C8C-45D9-B228-E34D99B26E85
Type genus.
Drina Nicéville, 1890.
Definition.
Currently in Loxurini Swinhoe, 1910, Drina (type species Myrina donina Hewitson, 1865) is not monophyletic with it and is not closely related to that tribe, instead forming a separate lineage originating early in the diversification of Theclinae Swainson, 1831 (Fig. 20). Therefore, we propose that this lineage corresponds to a tribe. This new tribe is diagnosed by a combination of the following characters, as given for the Drina section by Eliot (1973): male genitalia with significantly reduced (nearly vestigial) tegumen (not wider than valva in lateral view), uncus, and falces, and long and narrow vinculum; forewing with 11 veins, the three M veins are nearly at the same distance from each other, hindwing with a single tail at vein CuA2. A combination of the following nuclear genomic base pairs is diagnostic: cce305.14.6:G130A, cce305.14.6:T131C, cce1105.7.5:G93A, cce144.8.2:G223A, cce204.17.10:C49A, cce3081.9.2:G97G (not T), cce2598.5.13:G484G (not C), cce2598.5.13:C485C (not A), cce948.2.2:T73T (not A), cce948.2.2: A139A (not G).
Genera included.
Only the type genus.
Parent Taxon.
Subfamily Theclinae Swainson, 1831.
Hypochrysopini Grishin, new tribe http://zoobank.org/71317481-C0FE-4839-97E2-341444327DD7
Type genus.
Hypochrysops C. Felder & R. Felder, 1860.
Definition.
Currently placed in Luciini Waterhouse & Lyell, 1914, Hypochrysops (type species Papilio polycletus Linnaeus, 1758) and related genera are not monophyletic with it and instead form a distinct clade within Theclinae Swainson, 1831 not confidently associated with any other subtribe (Fig. 20). Therefore, this clade represents a subtribe. This new tribe is diagnosed by a combination of the following characters, as given for the Hypochrysops section by Eliot (1973): uncus not produced, rounded, falces prominent, strongly curved, juxta absent or vestigial, aedeagus very thick, just slightly narrower than valva, valva rhomboidal, saccus not developed; ventral wing surface typically with obsolete or distorted patterns with metallic silver or green spots and bands and red blotches. A combination of the following nuclear genomic base pairs is diagnostic: cce3313.6.1:G1784C, cce3313.6.1:G2131C, cce67043.1.6:C55G, cce67043.1.6:A56T, cce1184.15.22:G142C.
Genera included.
The type genus (i.e., Hypochrysops C. Felder & R. Felder, 1860), Philiris Röber, 1891, and Titea Eliot, 1973.
Parent Taxon.
Subfamily Theclinae Swainson, 1831.
Jalmenini Grishin, new tribe http://zoobank.org/D58D0D0A-382D-4E6D-B879-75D3DE7A3670
Type genus.
Jalmenus Hübner, 1818.
Definition.
Currently in Zesiusinae Swinhoe, 1912, Jalmenus (type species Jalmenus evagoras Hübner, 1818, which is a junior homonym of Papilio evagoras Donovan, 1805) is far removed from the genus Zesius Hübner, [1819] (type species Zesius chrysomallus Hübner, 1823) in the genomic tree, and instead forms a distinct lineage within Theclinae Swainson, 1831 not confidently associated with other genera (Fig. 20). Therefore, this lineage represents a tribe. This new tribe is diagnosed by a combination of the following characters, as given for the Jalmenus section (excluding Pseudalmenus H. H. Druce, 1902) by Eliot (1973): uncus and tegumen narrower in lateral view, longer than in relatives, ventral side not expanded, falces smaller, tegumen not expanded anteriad, valva without costal process, simple and rounded; palpi with 3rd segment very long, 2nd segment with bristle-like scales. A combination of the following nuclear genomic base pairs is diagnostic: cce9330.11.2:G69A, cce7486.3.3:T58G, cce9657.10.14:T3166C, cce10680.1.1:T28C, cce4529.2.2:A67G.
Genera included.
Only the type genus.
Parent Taxon.
Subfamily Theclinae Swainson, 1831.
Pseudalmenini Grishin, new tribe http://zoobank.org/BB6B6C59-EAD7-42B4-B7E6-CA95E94A419D
Type genus.
Pseudalmenus H. H. Druce, 1902.
Definition.
In the genomic tree, Pseudalmenus (type species Ialmenus myrsilus Westwood, 1851, which is a subspecies of Thecla chlorinda Blanchard, 1848) is a weakly supported (54%) sister of Jalmenus Hübner, 1818, and therefore may not be monophyletic with it (Fig. 20). Hence, we propose that this lineage represents a tribe. This new tribe was included in the Jalmenus section by Eliot (1973) and is diagnosed by a combination of the following characters: 3rd segment of palpi shorter than in Jalmenini trib. n., 2nd segment hairy; uncus with ventral portion expanded, protruding posteriad of the dorsal margin, falces long and strongly curved, tegumen well-developed, expanded anteriad, valva with the curved costal process giving it a crab claw-like appearance. A combination of the following nuclear genomic base pairs is diagnostic: cce10780.3.1:G247A, cce1122.11.2:C1079G, cce11670.2.2:C750T, cce24738.4.8:A1793G, cce10780.2.2:A2954G.
Genera included.
Only the type genus.
Parent Taxon.
Subfamily Theclinae Swainson, 1831.
Myrinini Toxopeus, 1929 is a tribe
Currently in Amblypodiini Doherty, 1886, close relatives Myrina [Fabricius], 1807 (type species Papilio alcides Cramer, 1776) and Iraota F. Moore, 1881 (type species Hesperia maecenas Fabricius, 1793) are distantly related to Amblypodia Horsfield, 1829 (type species Thecla narada Horsfield, 1828). The clade of these three genera is not strongly supported (42%) in our tree (Fig. 20), and their union is not monophyletic in a global phylogeny of butterflies (Kawahara et al. 2023). Therefore, we propose a status of a tribe for Myrinini Toxopeus, 1929, stat. nov., which consists of two genera (Myrina and Iraota).
Horagina Swinhoe, 1910 and Loxurina Swinhoe, 1910 are subtribes of Cheritrini Swinhoe, 1910
Currently regarded as distinct tribes, Cheritrini Swinhoe, 1910, Horagini Swinhoe, 1910, and Loxurini Swinhoe, 1910 (Eliot 1973) are closely related to each other and are at the tree level of subtribes (Fig. 20). Being combined, all three constitute one tribe. The priority of these names could not be determined because they were proposed (as subfamilies) on the same page of the same work (i.e., issued on the same date). As the first revisers, we give priority to Cheritrini Swinhoe, 1910, because this group includes more genera and species than the other two. As a result, we propose that Horagina Swinhoe, 1910, stat. nov. and Loxurina Swinhoe, 1910, stat. nov. are subtribes of Cheritrini Swinhoe, 1910.
Rapalini Grishin, new tribe http://zoobank.org/6B76C64C-1C57-4CC8-BBA2-925D4AFBCF7D
Type genus.
Rapala F. Moore, 1881.
Definition.
Currently, in Deudorigini Doherty, 1886, several genera, including Rapala (type species Thecla varuna Horsfield, 1829), form a clade that is not confidently monophyletic with it: only 26% partitions in the genomic tree support their grouping with Deudorigini. Low support values indicate a more distant relationship and a possibility of incomplete lineage sorting or gene exchange around the time of origin of these clades. The clade consisting of Rapala (mostly Oriental) and three Afrotropical genera closely related to Pilodeudorix H. H. Druce, 1891 (type species Pilodeudorix barbatus H. H. Druce, 1891) is most confidently supported (100%) and corresponds to the level of a tribe in the tree (Fig. 20). This new tribe differs from relatives by male genitalia having conjoined valvae that evenly taper to narrow rounded or pointed apices, uncus and tegumen broad, two lobes of uncus with a concave margin between them, hood-shaped; secondary sexual characters in males: an oval brand of small androconia near the base of cell Sc+R1-RS (at least in non-African species), and typically a hair tuft on ventral forewing near inner margin to complement the brand, sometimes with other brands, including those on abdomen; hindwing tailed, forewing veins R4+5 and M1 originate separately, although may be very narrowly separated at their origins (Eliot 1973). Most confidently distinguished by DNA and a combination of the following base pairs in the nuclear genome is diagnostic: cce303273.1.1:G197A, cce1093.2.1:A4672C, cce3516.7.1:A173T, cce12299.7.3:G134A, cce4160.2.2:A207G.
Genera included.
The type genus (i.e., Rapala F. Moore, 1881), Pilodeudorix H. H. Druce, 1891, Hypomyrina H. H. Druce, 1891, and Paradeudorix Libert, 2004.
Parent Taxon.
Subfamily Theclinae Swainson, 1831.
Pilodeudorigina Grishin, new subtribe http://zoobank.org/5826F9A8-6834-487C-B207-DC883CE19307
Type genus.
Pilodeudorix H. H. Druce, 1891.
Definition.
Afrotropical clade of Rapalini trib. n. that includes Pilodeudorix (type species Pilodeudorix barbatus H. H. Druce, 1891, which is a junior subjective synonym of Sithon camerona Plötz, 1880) is genetically differentiated from non-Arican (mostly Oriental) species at the tree level of a subtribe (Fig. 20). This new subtribe is diagnosed by a combination of the following characters: abdomen frequently with scent brands and hindwings with hair tufts; aedeagus shorter and less gracile that in the nominotypical tribe, frequently expanded terminally, vesica with many small cornuti and usually with large cuneus; basally fused valvae with a rectangular gap between them in many species, in others, each valva with a process, hook-like in ventral view, uncus lobes are frequently less separated from each other than in the nominotypical subtribe (Stempffer 1967). Best distinguished by DNA and a combination of the following nuclear genomic base pairs is diagnostic: cce303351.9.14:A88G, cce303351.9.14:A89G, cce14215.8.1:C143T, cce881.1.6:A79G, cce462.24.2:C89G.
Genera included.
The type genus (i.e., Pilodeudorix H. H. Druce, 1891), Hypomyrina H. H. Druce, 1891, and Paradeudorix Libert, 2004.
Parent Taxon.
Tribe Rapalini Grishin, trib. n.
Oxylidini Eliot, 1973, Remelanini Eliot, 1973, and Hypolycaenini Swinhoe, 1910 belong to Polyommatinae Swainson, 1827, not to Theclinae Swainson, 1831
Traditionally placed among hairstreaks (Theclinae Swainson, 1831) due to their appearance (frequently with very long hindwing tails), species from the tribes Oxylidini Eliot, 1973, Remelanini Eliot, 1973, and Hypolycaenini Swinhoe, 1910 are confidently placed as a sister clade to “Blues” and are not monophyletic with the subfamily Theclinae (Figs. 20, 22). Therefore, they do not belong to Theclinae. Their genetic differentiation from the clade that consists of traditional Polyommatinae Swainson, 1827 (tribes Lycaenesthini Toxopeus, 1929, Hypotheclini Eliot, 1973, and Polyommatini Swainson, 1827) is smaller than that for other subfamilies of Lycaenidae. Insufficient differentiation argues against treating Oxylidini, Remelanini, and Hypolycaenini as a subfamily of their own. Therefore, we place them in the subfamily Polyommatinae. We see that the three subfamilies, Lycaeninae, Theclinae, and Polyommatinae, are closely related to each other (Figs. 20, 22), as expected and evidenced by morphology (Eliot 1973). The genetic distinction of Lycaeninae from the two other subfamilies is more pronounced, and the tree branches separating them are more prominent (Fig. 20). However, Theclinae and Polyommatinae are less distinct from each other, and “blues” originate from within “hairstreaks”, not next to them, like butterflies originate from within moths, birds from within reptiles, and tetrapods from within fishes. Furthermore, due to their close relationship, it is conceivable to unify Theclinae and Polyommatinae into one subfamily (Polyommatinae). We are not taking this step here and continue with the traditional tri-subfamily arrangement for now, simply rearranging tribes among Theclinae and Polyommatinae to restore monophyly. See our proposed subtribal classification of Lycaenidae below.
Fig. 22.
The phylogenetic tree of selected Polyommatinae (other Lycaenidae are shown in Fig. 20) inferred from protein-coding regions of the nuclear genome (autosomes). Family-group names are shown above or below the corresponding branches. Not all subtribes are shown and/or labeled (see the subtribal classification list of Lycaenidae below). Names, clades, and species of new subtribes proposed in this work are shown in color. Names of other subtribes are shown in black, and names of subfamilies and tribes are shown in shades of gray.
Hemiolaina Grishin, new subtribe http://zoobank.org/45FFC7EB-6CFA-49A6-A688-EC98E721BA62
Type genus.
Hemiolaus Aurivillius, 1922.
Definition.
Hemiolaus (type species Jolaus caeculus Hopffer, 1855) forms a lineage sister to other Oxylidini Eliot, 1973, but is genetically differentiated from them at the subtribal level, and therefore represents a subtribe (Fig. 22). This new subtribe is diagnosed by a combination of the following characters, as given for the Hemiolaus section by Eliot (1973): juxta enlarged, longer than half of valva, shaped as the end of a nail puller with a deep cleft; ventral hindwing of males with a scent patch beneath a hair brush; antennal club short, nudum confined to the club, about 32 segments, palpi with 3rd segment slightly less than half of the 2nd, tarsus of male foreleg terminally tapered and downturned. A combination of the following nuclear genomic base pairs is diagnostic: cce54422.3.2:C373T, cce993.15.2:A922T, cce303329.2.7:A2399C, cce2502.15.1:A320C, cce5483.2.11:T370A.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Oxylidini Eliot, 1973.
Cupidopsina Grishin, new subtribe http://zoobank.org/8AFD9453-3743-4ECB-BA87-4DAC0A3297FB
Type genus.
Cupidopsis Karsch, 1895.
Definition.
Confidently placed in the tribe Hypotheclini Eliot, 1973, the lineage with Cupidopsis (type species Lycaena jobates Hopffer, 1855) is genetically differentiated from the Hypothecla Semper, 1890 lineage at the tree level of subtribes (Fig. 22) and therefore represents a subtribe. This new subtribe is diagnosed by a combination of the following characters as given for the Cupidopsis section by Eliot (1973): only 10 veins on the forewing, secondary sexual characters absent, aedeagus with developed coecum and ductus entrance on dorsal side, saccus smaller than in relatives, nearly vestigial, tegumen with uncus comparatively massive, the same length as valva in lateral view. A combination of the following nuclear genomic base pairs is diagnostic: cce9377.2.3:A136T, cce4053.12.2:A3235C, cce332.21.2:G117T, cce199. 20.2:A244C, cce3203.9.2:A172C.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Hypotheclini Eliot, 1973.
Comments.
A family-group name formed from the same genus was proposed as a nomen nudum (fails ICZN Art. 13.) by Koçak (1996). The genomic tree demonstrates that this subtribe (as other Hypotheclini) belongs to Polyommatinae Swainson, 1827 (Figs. 20, 22), contrary to the hypothesis of Stradomsky (2016), who nevertheless correctly associated Cupidopsis with Hypothecla.
Niphandina Sibatani & Ito, 1942 is a subtribe
In agreement with Stradomsky (2016), we find that Niphandini Sibatani & Ito, 1942 clusters closely with Polyommatini Swainson, 1827, being at the tree level with subtribes (Fig. 22). Therefore, we confirm its treatment as a subtribe Niphandina Sibatani & Ito, 1942, stat. conf.
Theclinesthina Grishin, new subtribe http://zoobank.org/32E65131-DDFB-4EBC-BE37-4F42E8F5F6D3
Type genus.
Theclinesthes Röber, 1891.
Definition.
The clade with Theclinesthes (type species Plebeius (Theclinesthes) eremicola Röber, 1891, which is a junior subjective synonym of Nacaduba miskini gaura Doherty, 1891) originates near the base of Polyommatini Swainson, 1827 (Fig. 22) and therefore represents a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for the Theclinesthes section by Eliot (1973) and Stradomsky (2016): uncus lobes and (vestigial) falces directed ventrad, vinculum in lateral view much broader than in relatives, as broad as valva, aedeagus basally bulbous and apically tapered, ductus enters at anterior end, valva constricted in the middle. A combination of the following nuclear genomic base pairs is diagnostic: cce2737.15.2:A259G, cce3516.7.8:A98T, cce10374.3.2:A67C, cce2234.8.11:G1960A, cce2399.18.8:A86G.
Genera included.
The type genus (i.e., Theclinesthes Röber, 1891), Neolucia G. Waterhouse & Turner, 1905, and Sahulana Hirowatari, 1992.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was published in a pioneering study by Stradomsky (2016) without explicitly indicating that the name was intentionally new (fails ICZN Art. 16.1.) and not stating what the type genus of this taxon was (fails ICZN Art. 16.2.). This name for the subtribe already discovered by Stradomsky and confirmed by our genomic analysis is simply formalized here to comply with the ICZN Code (ICZN [International Commission on Zoological Nomenclature] 1999).
Azanina Grishin, new subtribe http://zoobank.org/DF55B888-23ED-4B2E-94F9-B1B6685CBDA3
Type genus.
Azanus F. Moore, 1881.
Definition.
Azanus (type species Papilio ubaldus Stoll, 1782) is confidently placed in a clade of Polyommatini Swainson, 1827 containing several subtribes, not confidently grouping with any of them (Fig. 22). Therefore, the lineage with Azanus represents a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for the Azanus section by Eliot (1973): male genitalia elongated and flattened, uncus narrowly divided into separate lobes, forewings veins SC and R1 come together and then diverge, androconia of two unusual types: nearly rectangular scales with concave bases and long padded scales, eyes hairy, ventral forewing with a dark streak below SC vein. A combination of the following nuclear genomic base pairs is diagnostic: cce9549.2.2:A815G, cce302383.7.1:T1153A, cce302383.7.1:C1154G, cce2510.1.2:A686G, cce178.15.6:A356G.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was proposed as a nomen nudum (fails ICZN Art. 13.) by Koçak (1996) and then published in a pioneering study by Stradomsky (2016) without explicitly indicating that the name was intentionally new (fails ICZN Art. 16.1.) and not stating what the type genus of this taxon was (fails ICZN Art. 16.2.). This name for the subtribe already discovered by Koçak and Stradomsky and confirmed by our genomic analysis is simply formalized here to comply with the ICZN Code (ICZN [International Commission on Zoological Nomenclature] 1999).
Unina Grishin, new subtribe http://zoobank.org/2FD670E6-5ECD-404E-A1F7-842AEF97335C
Type genus.
Una Nicéville, 1890.
Definition.
Una (type species Zizera ? usta Distant, 1886) is confidently placed in the clade of Polyommatini Swainson, 1827 with several subtribes, not confidently grouping with any of them (Fig. 22). Therefore, the lineage with Ionolyce represents a subtribe. This new subtribe is a union of the Una and Petrelaea sections of Eliot (1973), who listed characters for them, and is diagnosed as follows: male genitalia elongated and appear flattened, with gracile and long aedeagus and prominent saccus (absent in close relatives); forewings with 11 veins, veins SC and R1 fuse at least for some distance. A combination of the following nuclear genomic base pairs is diagnostic: cce10730.5.6:A179T, cce7658.4.3:A184G, cce4822.3.3:A98T, cce5018.4.1:A78G, cce870.7.1:G452A.
Genera included.
The type genus (i.e., Una Nicéville, 1890), Orthomiella Nicéville, 1890, Petrelaea Toxopeus, 1929, and Pseudonacaduba Stempffer, 1942.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was proposed as a nomen nudum (fails ICZN Art. 13.) by Koçak & Seven (1997).
Ionolycina Grishin, new subtribe http://zoobank.org/77081BED-FF23-424A-98B3-541E04BA53E5
Type genus.
Ionolyce Toxopeus, 1929.
Definition.
Ionolyce (type species Ionolyce helicon javanica Toxopeus, 1929) is confidently placed in the clade of Polyommatini Swainson, 1827 with several subtribes, not confidently grouping with any of them (Fig. 22). Therefore, the lineage with Ionolyce represents a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for Ionolyce by Tite (1963): cornuti in aedeagus are large and spine-like, ribs in androconial scales are ribbon-like with nodular irregularities mainly in the posterior third of the scale; fused part of veins SC and R1 is typically longer than in relatives, and the free end of vein SC is faint. A combination of the following nuclear genomic base pairs is diagnostic: cce437.15.1:A182G, cce303334.4.2:C106G, cce3111.1.6:A86G, cce3368.2.2:T241A, cce29649.12.1:A151G.
Genera included.
The type genus (i.e., Ionolyce Toxopeus, 1929) and Paraduba Bethune-Baker, 1906.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Pithecopina Grishin, new subtribe http://zoobank.org/3E5E4A9A-64A9-4AFC-85DD-81B13BE53FBF
Type genus.
Pithecops Horsfield, 1828.
Definition.
The clade with Pithecops (type species Pithecops hylax corax Fruhstorfer, 1919, which is a subspecies of Pithecops corvus Fruhstorfer, 1919) is confidently placed as sister to the “crown group” of Polyommatini Swainson, 1827 that undergoes extensive diversification (Fig. 22), and we include it in the “crown group” despite its unusual wing patterns. The Pithecops clade does not have close relatives within Polyommatini and, therefore, represents a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for the Pithecops section by Eliot (1973): male genitalia elongated and appear flattened, uncus broadly divided nearly to its base, secondary sexual characters absent, veins SC and R1 fuse at least for some distance, eyes not hairy, palpi hairy. A combination of the following nuclear genomic base pairs is diagnostic: cce59502.1.2:A118G, cce1246.19.3:A71G, cce9990.6.1:G740A, cce1317.1.1:C772T, cce3516.4.2:G83T.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was published in a pioneering study by Stradomsky (2016) without explicitly indicating that the name was intentionally new (fails ICZN Art. 16.1.) and not stating what the type genus of this taxon was (fails ICZN Art. 16.2.). This name for the taxon already discovered by Stradomsky and confirmed by our genomic analysis is simply formalized here to comply with the ICZN Code (ICZN [International Commission on Zoological Nomenclature] 1999).
Zizulina Grishin, new subtribe http://zoobank.org/A230CCD1-B083-4025-8B78-A31A89478932
Type genus.
Zizula Chapman, 1910.
Definition.
The lineage with Zizula (type species Lycaena gaika Trimen, 1862, which is a junior subjective synonym of Papilio hylax Fabricius, 1775) is a confident sister to Brephidiina Stempffer, 1957, but is at the tree level that corresponds to subtribes in the “crown group” of Polyommatini Swainson, 1827 (Fig. 22) and therefore represents a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for the Zizula section by Eliot (1973): male genitalia unusual, aedeagus stout and terminally divided into two processes of about half of its length, dorsal and ventral, together resembling a beak, valva with a rod-like process of about the same length as genitalia and long bristles twice of valval length, veins SC and R1 fused towards costa, secondary sexual characters absent. A combination of the following nuclear genomic base pairs is diagnostic: cce993.15.2:A161G, cce993.15.2:T162C, cce811.10.3:A2207G, cce811.10.3:T2197C, cce1162.15.2:T151A.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was proposed as a nomen nudum (fails ICZN Art. 13.) by Koçak (1996) and then published in a pioneering study by Stradomsky (2016) without explicitly indicating that the name was intentionally new (fails ICZN Art. 16.1.) and not stating what the type genus of this taxon was (fails ICZN Art. 16.2.). This name for the subtribe already discovered by Koçak and Stradomsky and confirmed by our genomic analysis is simply formalized here to comply with the ICZN Code (ICZN [International Commission on Zoological Nomenclature] 1999).
Jamidina Grishin, new subtribe http://zoobank.org/F3E26669-CDF0-4EFC-9539-F88A3D47B1F7
Type genus.
Jamides Hübner, [1819].
Definition.
The lineage with Jamides (type species Papilio bochus Stoll, 1782) is a confident sister to Celastrinina Tutt, 1907, but splits from the latter at the tree level of subtribes (Fig. 22), thus representing a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for the Jamides section by Eliot (1973): male genitalia with vinculum lacking cephalad expansion, uncus directed ventrad, falces very long (nearly half of valva length), aedeagus longer than valva (usually by a third or more), valva deeply bilobed; forewing veins SC and R1 are not fused for any distance, but connected with a short cross-vein. A combination of the following nuclear genomic base pairs is diagnostic: cce23510.3.2:T93C, cce23510.3.2:A432G, cce1568.2.4:A5519G, cce1568.2.4:A6856C, cce10823.2.3:T258C.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was proposed as a nomen nudum (fails ICZN Art. 13.) by Koçak (1996) and then published in a pioneering study by Stradomsky (2016) without explicitly indicating that the name was intentionally new (fails ICZN Art. 16.1.) and not stating what the type genus of this taxon was (fails ICZN Art. 16.2.). This name for the subtribe already discovered by Koçak and Stradomsky and confirmed by our genomic analysis is simply formalized here to comply with the ICZN Code (ICZN [International Commission on Zoological Nomenclature] 1999).
Fameganina Grishin, new subtribe http://zoobank.org/07D24435-1EAD-4C80-9350-0A1FA6003972
Type genus.
Famegana Eliot, 1973.
Definition.
The lineage with Famegana (type species Lycaena alsulus Herrich-Schäffer, 1869, which is a junior subjective synonym of Lycaena nisa Wallace, 1866) is sister to Zizeeriina Chapman, 1910 with moderate confidence (Fig. 22). Because the confidence of this grouping is not the highest and because this lineage originates at the tree level of subtribes, it represents a subtribe. This new subtribe is diagnosed by a combination of the following characters, as given for the Famegana section by Eliot (1973): in male genitalia, tegumen and uncus bulky, falces stout and nearly rigidly connected, uncus lobes terminally pointed and slightly downturned; veins SC and R1 touch each other over a short distance, eyes not hairy, palpi with bristles. A combination of the following nuclear genomic base pairs is diagnostic: cce1088.12.3:A100T, cce1088.12.3:G102A, cce18.45.4:G1256T, cce18.45.4:G1255A, cce10490.1.2:T1933A.
Genera included.
Only the type genus.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was proposed as a nomen nudum (fails ICZN Art. 13.) by Koçak & Seven (1997) and then published in a pioneering study by Stradomsky (2016) without explicitly indicating that the name was intentionally new (fails ICZN Art. 16.1.) and not stating what the type genus of this taxon was (fails ICZN Art. 16.2.). This name for the subtribe already discovered by Koçak& Seven and Stradomsky and confirmed by our genomic analysis is simply formalized here to comply with the ICZN Code (ICZN [International Commission on Zoological Nomenclature] 1999).
Oboroniina Grishin, new subtribe http://zoobank.org/FB96541D-672A-41A6-9C47-2EDAADEC0D60
Type genus.
Oboronia Karsch, 1893.
Definition.
The clade with Oboronia (type species Oboronia staudingeri Hemming, 1960, which is a junior subjective synonym of Plebeius punctatus Dewitz, 1879) is placed within the “crown group” of Polyommatini Swainson, 1827 without strongly supported phylogenetic affinity to any subtribe (Fig. 22) and therefore represents a subtribe of its own. This new subtribe is diagnosed by a combination of the following characters, as given for the Euchrysops section by Eliot (1973): male genitalia with long falces, vinculum broadening in the middle in lateral view, long and narrow valva, massive rod-shaped aedeagus with anterior ductus entrance; veins SC and R1 not fused. A combination of the following nuclear genomic base pairs is diagnostic: cce349.2.1:C166A, cce349.2.1:A167T, cce935.8.2:A66G, cce2073.8.1:A230G, cce178.15.6:A190G.
Genera included.
The type genus (i.e., Oboronia Karsch, 1893), Euchrysops Butler, 1900, Lepidochrysops Hedicke, 1923, Orachrysops Vári, 1986, and Thermoniphas Karsch, 1895.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was published in a pioneering study by Stradomsky (2016) without explicitly indicating that the name was intentionally new (fails ICZN Art. 16.1.) and not stating what the type genus of this taxon was (fails ICZN Art. 16.2.). This name for the subtribe already discovered by Stradomsky and confirmed by our genomic analysis is simply formalized here to comply with the ICZN Code (ICZN [International Commission on Zoological Nomenclature] 1999).
Uranothaumatina Grishin, new subtribe http://zoobank.org/57D2B8F3-36FA-4457-88B2-FC99DADC2EF8
Type genus.
Uranothauma Butler, 1895.
Definition.
The lineage with Uranothauma (type species Uranothauma crawshayi Butler, 1895) is confidently placed as sister to Scolitantidina Tutt, 1907 (Fig. 22), but it was not traditionally included in the latter subtribe. Additionally, because it originates at the tree level corresponding to subtribes, it represents a subtribe. This new subtribe is a union of the Uranothauma and Phlyaria sections of Eliot (1973), who listed characters for them, and is diagnosed as follows (see also Stradomsky (2016) for genitalia illustrations): in male genitalia, saccus absent, falces developed, as long as tegumen with uncus, vinculum expanded in the middle in lateral view, aedeagus shorter than valva, rod-shaped with ductus entrance dorso-cephalad, valva elongated, undivided; veins SC and R1 touch each other or fuse at least for some distance, eyes hairy, palpi hairy or bristly. A combination of the following nuclear genomic base pairs is diagnostic: cce993.29.2:A515G, cce103.22.12:A47G, cce462.35.1:G193T, cce912.1.1:C56A, cce1162.12.1:G739A.
Genera included.
The type genus (i.e., Uranothauma Butler, 1895) and Phlyaria Karsch, 1895.
Parent Taxon.
Tribe Polyommatini Swainson, 1827.
Comment.
The same name was published in a pioneering study by Stradomsky (2016) without explicitly indicating that the name was intentionally new (fails ICZN Art. 16.1.) and not stating what the type genus of this taxon was (fails ICZN Art. 16.2.). This name for the subtribe already discovered by Stradomsky and confirmed by our genomic analysis is simply formalized here to comply with the ICZN Code (ICZN [International Commission on Zoological Nomenclature] 1999).
Higher classification of Lycaenidae to the subtribal level
Based on our genome-scale phylogeny (Figs. 20–22) complemented with other studies (Talavera et al. 2012; Boyle et al. 2015; Robbins et al. 2022; Boyle et al. 2023; Kawahara et al. 2023), we propose the following provisional classification of Lycaenidae into subfamilies, tribes, and subtribes. We partition the family into eight subfamilies. If no tribes and subtribes are listed for a subfamily, we consider that subfamily to be monotypic. If no subtribes are listed for a tribe, we consider that tribe to be monotypic. The type genus name for each taxon is given in parentheses. New taxa and status changes are shown in red font. Synonymy is not provided.
Family Lycaenidae [Leach], [1815] (Lycaena [Fabricius], 1807)
Subfamily Curetinae Distant, 1884 (Curetis Hübner, [1819])
Subfamily Liphyrinae Doherty, 1889 (Liphyra Westwood, [1864]), stat. rest.
Subfamily Miletinae Reuter, 1896 (Miletus Hübner, [1819])
Tribe Lachnocnemini Clench, 1955 (Lachnocnema Trimen, 1887)
Tribe Miletini Reuter, 1896 (Miletus Hübner, [1819])
Subtribe Miletina Reuter, 1896 (Miletus Hübner, [1819])
Subtribe Megalopalpina Grishin, subtrib. n. (Megalopalpus Röber, 1886)
Tribe Spalgini Toxopeus, 1929 (Spalgis F. Moore, 1879)
Subtribe Spalgina Toxopeus, 1929 (Spalgis F. Moore, 1879)
Subtribe Tarakina Eliot, 1973, stat. rev. (Taraka Doherty, 1889)
Subfamily Poritiinae Doherty, 1886 (Poritia F. Moore, [1866])
Tribe Liptenini Röber, 1892 (Liptena Westwood, [1851])
Subtribe Durbaniina Clench, 1955 (Durbania Trimen, 1862)
Subtribe Pentilina Aurivillius, 1914 (Pentila Westwood, [1851])
Subtribe Liptenina Röber, 1892 (Liptena Westwood, [1851]); includes Mimacraeina Stempffer, 1961
Subtribe Iridanina Clench, 1965 (Iridana Aurivillius, 1920)
Subtribe Epitolina Jackson, 1962 (Epitola Westwood, [1851])
Subtribe Cooksoniina Sáfián, Boyle & Pierce, 2023 (Cooksonia H. H. Druce, 1905)
Tribe Poritiini Doherty, 1886 (Poritia F. Moore, [1866])
Subfamily Aphnaeinae Distant, 1884 (Aphnaeus Hübner, [1819]); not monophyletic with Theclinae!
Tribe Axiocersini Grishin, trib. n. (Axiocerses Hübner, [1819])
Tribe Cigaritini Grishin, trib. n. (Cigaritis Donzel, 1848)
Subtribe Pseudaletidina Grishin, subtrib. n. (Pseudaletis H. H. Druce, 1888)
Subtribe Cigaritina Grishin (Cigaritis Donzel, 1848)
Tribe Aphnaeini Distant, 1884 (Aphnaeus Hübner, [1819])
Subtribe Aloeidina Grishin, subtrib. n. (Aloeides Hübner, [1819])
Subtribe Phasisina Grishin, subtrib. n. (Phasis Hübner, [1819])
Subtribe Aphnaeina Distant, 1884 (Aphnaeus Hübner, [1819])
Subfamily Lycaeninae [Leach], [1815] (Lycaena [Fabricius], 1807)
Subfamily Theclinae Swainson, 1830 (Thecla [Fabricius], 1807)
Tribe Theclini Swainson, 1830 (Thecla [Fabricius], 1807)
Tribe Surendrini Koçak & Seven, 1997, stat. nov. (Surendra F. Moore, 1879)
Tribe Arhopalini Bingham, 1907 (Arhopala Boisduval, 1832)
Tribe Drinini Grishin, trib. n. (Drina Nicéville, 1890)
Tribe Candalidini Eliot, 1973 (Candalides Hübner, [1819])
Tribe Hypochrysopini Grishin, trib. n. (Hypochrysops C. Felder & R. Felder, 1860)
Tribe Luciini Waterhouse & Lyell, 1914 (Lucia W. Swainson, 1833)
Tribe Ogyrini Waterhouse & Lyell, 1914 (Ogyris Angas, 1847)
Tribe Jalmenini Grishin, trib. n. (Jalmenus Hübner, 1818)
Tribe Pseudalmenini Grishin, trib. n. (Pseudalmenus H. H. Druce, 1902)
Tribe Amblypodiini Doherty, 1886 (Amblypodia Horsfield, 1829)
Tribe Myrinini Toxopeus, 1929, stat. nov. (Myrina [Fabricius], 1807)
Tribe Cheritrini Swinhoe, 1910 (Cheritra F. Moore, 1881)
Subtribe Cheritrina Swinhoe, 1910 (Cheritra F. Moore, 1881)
Subtribe Horagina Swinhoe, 1910, stat. nov. (Horaga F. Moore, 1881)
Subtribe Loxurina Swinhoe, 1910, stat. nov. (Loxura Horsfield, [1829])
Tribe Iolaini Riley, 1958 (Iolaus Hübner, [1819])
Tribe Catapaecilmatini Eliot, 1973 (Catapaecilma Butler, 1879)
Tribe Zesiusini Swinhoe, 1912 (Zesius Hübner, [1819])
Tribe Tomarini Eliot, 1973 (Tomares Rambur, 1840)
Tribe Rapalini Grishin, trib. n. (Rapala F. Moore, 1881)
Subtribe Rapalina Grishin (Rapala F. Moore, 1881)
Subtribe Pilodeudorigina Grishin, subtrib. n. (Pilodeudorix H. H. Druce, 1891)
Tribe Deudorigini Doherty, 1886 (Deudorix Hewitson, 1863)
Tribe Eumaeini Doubleday, 1847 (Eumaeus Hübner, [1819])
Subtribe Eumaeina Doubleday, 1847 (Eumaeus Hübner, [1819])
Subtribe Rhammina Prieto & Busby, 2022 (Rhamma K. Johnson, 1992)
Subtribe Timaetina Busby & Prieto, 2022 (Timaeta K. Johnson, Kruse & Kroenlein, 1997)
Subtribe Atlidina Martins & Duarte, 2022 (Atlides Hübner, [1819])
Subtribe Evenina Faynel & Grishin, 2022 (Evenus Hübner, [1819])
Subtribe Jantheclina Robbins & Faynel, 2022 (Janthecla Robbins & Venables, 1991)
Subtribe Paiwarriina Lamas & Robbins,2022 (Paiwarria Kaye, 1904)
Subtribe Cupatheclina Lamas & Grishin, 2022 (Cupathecla Bálint, 2005)
Subtribe Parrhasiina Busby & Robbins, 2022 (Parrhasius Hübner, [1819])
Subtribe Ipideclina Martins & Grishin, 2022 (Ipidecla Dyar, 1916)
Subtribe Calycopidina Duarte & Robbins, 2010 (Calycopis Scudder, 1876)
Subtribe Strymonina Tutt, 1907 (Strymon Hübner, 1818)
Subtribe Strephonotina K. Johnson, Austin, Le Crom & Salazar, 1997 (Strephonota K. Johnson et al., 1997)
Subtribe Trichonidina Duarte & Faynel, 2022 (Trichonis Hewitson, 1865)
Subtribe Callophryidina Tutt, 1907 (Callophrys Billberg, 1820)
Subfamily Polyommatinae Swainson, 1827 (Polyommatus Latreille, 1804)
Tribe Oxylidini Eliot, 1973 (Oxylides Hübner, [1819])
Subtribe Oxylidina Eliot, 1973 (Oxylides Hübner, [1819])
Subtribe Hemiolaina Grishin, subtrib. n. (Hemiolaus Aurivillius, 1922)
Tribe Remelanini Eliot, 1973 (Remelana F. Moore, 1884)
Tribe Hypolycaenini Swinhoe, 1910 (Hypolycaena C. Felder & R. Felder, 1862)
Tribe Lycaenesthini Toxopeus, 1929 (Lycaenesthes F. Moore, 1866)
Tribe Hypotheclini Eliot, 1973 (Hypothecla G. Semper, 1890)
Subtribe Hypotheclina Eliot, 1973 (Hypothecla G. Semper, 1890)
Subtribe Cupidopsina Grishin, subtrib. n. (Cupidopsis Karsch, 1895)
Tribe Polyommatini Swainson, 1827 (Polyommatus Latreille, 1804)
Subtribe Niphandina Sibatani & Ito, 1942, stat. conf. (Niphanda F. Moore, 1875)
Subtribe Theclinesthina Grishin, subtrib. n. (Theclinesthes Röber, 1891)
Subtribe Azanina Grishin, subtrib. n. (Azanus F. Moore, 1881)
Subtribe Unina Grishin, subtrib. n. (Una Nicéville, 1890)
Subtribe Danina Koçak & Seven, 1997 (Danis [Fabricius], 1807)
Subtribe Ionolycina Grishin, subtrib. n. (Ionolyce Toxopeus, 1929)
Subtribe Pithecopina Grishin, subtrib. n. (Pithecops Horsfield, 1828)
Subtribe Zizulina Grishin, subtrib. n. (Zizula Chapman, 1910)
Subtribe Brephidiina Stempffer, 1957 (Brephidium Scudder, 1876)
Subtribe Celastrinina Tutt, 1907 (Celastrina Tutt, 1906)
Subtribe Jamidina Grishin, subtrib. n. (Jamides Hübner, [1819])
Subtribe Zizeeriina Chapman, 1910 (Zizeeria Chapman, 1910)
Subtribe Fameganina Grishin, subtrib. n. (Famegana Eliot, 1973)
Subtribe Catochrysopina Toxopeus, 1929 (Catochrysops Boisduval, 1832)
Subtribe Oboroniina Grishin, subtrib. n. (Oboronia Karsch, 1893)
Subtribe Castaliina Distant, 1884 (Castalius Hübner, [1819])
Subtribe Scolitantidina Tutt, 1907 (Scolitantides Hübner, [1819])
Subtribe Uranothaumatina Grishin, subtrib. n. (Uranothauma Butler, 1895)
Subtribe Actizerina Koçak, 1996 (Actizera Chapman, 1910)
Subtribe Leptotina Wagener, 1995 (Leptotes Scudder, 1876)
Subtribe Lampidina Tutt, 1907 (Lampides Hübner, [1819])
Subtribe Everina Tutt, 1907 (Everes Hübner, [1819])
Subtribe Polyommatina Swainson, 1827 (Polyommatus Latreille, 1804)
Our classification of Polyommatini Swainson, 1827 compared to that by Stradomsky (2016)
In his pioneering work based on limited DNA sequence data and careful genitalic comparison, Stradomsky (2016) proposed to partition the tribe Polyommatini into 22 subtribes. Stradomsky’s classification fares well compared to our genomic results (see above), and his study is impressive in its insight. Out of our 23 subtribes, 20 match Stradomsky’s in names, and one, Celastrinina Tutt, 1907, was referred to by its junior subjective synonym, Lycaenopsina Swinhoe, 1910. The remaining two subtribes, Ionolycina subtrib. n. and Unina subtrib. n., were placed by Stradomsky into his Danina Koçak & Seven, 1997 and “Azanina,” respectively. Una Nicéville, 1890 cannot belong to “Azanina,” because it is in the clade with Danis [Fabricius], 1807 and not with Azanus F. Moore, 1881 (see Fig. 22 and Kawahara et al. (2023)). While it is indeed possible to unify these four groups into a single subtribe, Danina, genetic differentiation within this subtribe would be larger than in other subtribes in Polyommatini (Fig. 22). Therefore, dividing the group into four subtribes brings the classification in line with the genetic differentiation within other Polyommatini subtribes. Finally, Stradomsky’s “Cacyreina” is merged into our Lampidina Tutt, 1907 due to genetic similarities, and Stradomsky’s phylogeny based on a small number of gene markers is poorly supported around these taxa. We put names of subtribes used by Stradomsky in quotes because they were proposed somewhat informally: authorship of previously published names was not indicated, and for the names we could not find in previous publications, Stradomsky did not explicitly indicate that they were intentionally new (fails ICZN Art. 16.1.) and did not specify what their type genera were (fails ICZN Art. 16.2.). Moreover, it remains unclear from the text of his article which names Stradomsky considered new in the sense of the ICZN Code (if any) and if he intended to propose new ICZN-compliant names in his work.
Paralycaeides Nabokov, 1945 is a subgenus of Itylos Draudt, 1921 and Eldoradina Balletto, 1993 of Nabokovia Hemming, 1960
Inspection of genomic trees reveals that two pairs of genera: (1) Itylos Draudt, 1921 (type species Cupido speciosa Staudinger, 1894, which is a junior subjective synonym of Lycaena titicaca Weymer, 1890) and Paralycaeides Nabokov, 1945 (type species Itylos inconspicua Draudt, 1921) and (2) Nabokovia Hemming, 1960 (type species Thecla faga Dognin, 1895 and Eldoradina Balletto, 1993 (type species Nabokovia (Eldoradina) cyanea Balletto, 1993) are closely related to each other in each pair (Fig. 23): COI barcode difference of 5.9%–6% (39–40 bp) in each pair. Furthermore, at least one genus in each pair consists of a small number of species. Therefore, we propose to treat the junior name in each pair as a subgenus name: Paralycaeides Nabokov, 1945 of Itylos Draudt, 1921 and Eldoradina Balletto, 1993 of Nabokovia Hemming, 1960.
Fig. 23.

Nuclear genome tree (autosomes): genus Itylos (red, subgenus Paralycaeides in magenta) and genus Nabokovia (blue, subgenus Eldoradina in violet color).
Shijimia Matsumura, 1919 is confirmed as a valid genus with Shijimia potanini Alphéraky, 1889, new combination
Genomic sequencing and analysis of representatives of the subtribe Everina Tutt, 1907 (type genus Everes Hübner, [1819])) reveals that Shijimia Matsumura, 1919 (type species Lycaena moorei Leech, 1889) may not be monophyletic with Cupido Schrank, 1801 (type species Papilio minimus Fuessly, 1775) and instead may be sister to Bothrinia Chapman, 1909 (type species Cyaniris chennellii de Nicéville, [1884]) (Fig. 24), confirming that it is meaningful to treat Shijimia as a valid genus. We sequenced one of the two syntypes of Everes umbriel Doherty, 1889 (type locality Tenasserim Valley) currently treated as a junior subjective synonym (or subspecies) of Lycaena potanini Alphéraky, 1889 (type locality in China: Gansu Province) in the genus Tongeia Tutt, 1908 (type species Lycaena fischeri Eversmann, 1843), and found that it is not monophyletic with Tongeia fischeri, but is closely related to Shijimia Matsumura, 1919 (type species Lycaena moorei Leech, 1889) (Fig. 24). Therefore, we propose a new combination: Shijimia potanini (Alphéraky, 1889), comb. nov.
Fig. 24.

Nuclear genome tree (autosomes) of Everina representatives: genera Cupido (green), Shijimia (red), Bothrinia (violet), and Tongeia (blue).
Family Hesperiidae Latreille, 1809
Pyrrhochalcia Mabille, 1904 is a subgenus of Coeliades Hübner, 1818
Although immediately distinguishable by its much larger size, black wings with metallic-green shine (and broad green streaks in females) above, and shiny cyan-green-olive hindwings beneath, Pyrrhochalcia Mabille, 1904 (type and the only species Papilio iphis Drury, 1773) was recognized as a close relative of Coeliades Hübner, 1818 (type species Papilio forestan Stoll, 1782) by Evans (1937) and placed next to Pyrrhiades Lindsey & Miller, 1965 (type species Papilio lucagus Cramer, 1777)—currently within Coeliades—by Chiba (2009) in his key, differing by the relative positions of vein origins on hindwing. Moreover, some species of Coeliades are patterned similarly, although without extensive green coloration on ventral hindwing: C. lucagus (Cramer, 1777) and C. aeschylus (Plötz, 1884) (both formerly in Pyrrhiades). Our genomic tree and recently published phylogeny (Toussaint et al. 2021) support this close relationship between Pyrrhochalcia and Coeliades by placing the former as a close sister of the latter without prominent separation between them (Fig. 25). Genetic differentiation within Coeliades even after including Pyrrhochalcia is approximately the same as that within the genus Choaspes F. Moore, 1881 (type species Hesperia (Thymele) benjaminii Guérin-Méneville, 1843) (Fig. 25 blue vs. green). Therefore, we propose that Pyrrhochalcia Mabille, 1904, stat. nov. is a subgenus of Coeliades Hübner, 1818.
Fig. 25.

Nuclear genome tree (autosomes): genus Coeliades (blue, with subgenus Pyrrhochalcia labeled in violet) and genus Choaspes (green).
Cecropterus (Thorybes) nevada (Scudder, 1872) is restricted to OR, CA, and NV and Cecropterus (Thorybes) dobra (Evans, 1952) is found eastward in the US
Genomic sequencing of specimens from the northeastern part of the range of the Cecropterus (Thorybes) mexicana species group that consists of three species: Cecropterus (Thorybes) nevada (Scudder, 1872), Cecropterus (Thorybes) dobra (Evans, 1952), and Cecropterus (Thorybes) mexicana (Herrich-Schäffer, 1869) reveals that all of them are C. dobra, which is genetically uniform throughout its range (Fig. 26). Therefore, C. nevada is restricted to the westernmost part of the USA (Oregon, California, and Nevada) and is separated from C. dobra by a wide gap in its distribution, according to iNaturalist observations (2023) (Fig. 26).
Fig. 26.

Cecropterus (Thorybes) dobra (blue) and Cecropterus (Thorybes) nevada (red): a) a distribution map made on the basis of iNaturalist (2023) observations: iNaturalist maps for C. dobra and C. nevada were superimposed and merged in Photoshop and re-colored (CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/), the map does not refer to sequenced specimens, records were not checked and some may be misidentifications; b) nuclear genome tree (autosomes) showing that specimens we sequenced from several localities in Utah, Colorado, New Mexico, and Arizona are C. dobra.
Cecropterus albosuffusa (H. Freeman, 1943) and Cecropterus indistinctus (Austin & J. Emmel, 1998) are species distinct from Cecropterus pylades (Scudder, 1870)
Genomic comparison of Cecropterus Herrich-Schäffer, 1869 (type species Cecrops zarex Hübner, 1818) reveals close relationship between Cecropterus (Thorybes) pylades (Scudder, 1870) (type locality in USA: Massachusetts) and Cecropterus (Thorybes) drusius (W. H. Edwards, [1884]) (type locality in southern Arizona) that are unified by the presence of costal fold in males (Evans 1955). We regard them as the C. pylades species group (Fig. 27). Genomic trees (the Z chromosome and the mitogenome) confirm that C. drusius with its distinctive genitalia (Fig. 30q) is sister to all others but reveal that the entity currently treated as a single species C. pylades consists of several genetically differentiated clades (Fig. 27). The Z chromosome (Fig. 27a) and the mitochondrial genome (Fig. 27b) trees show identical strongly supported topology between the clades (but not within clades, where the subtrees are comb-like due to genetic similarity and gene exchange) and placement of specimens into them, thus increasing the confidence in the results due to this uncommon congruence between nuclear and mitochondrial trees.
Fig. 27.

Phylogenetic trees of Cecropterus (Thorybes) pylades species group inferred from protein-coding regions of a) the Z chromosome (best for species delimitation) and b) the mitochondrial genome. Different species are shown in different colors: C. floridianus sp. n. (red), C. pylades (green), C. indistinctus stat. nov. (cyan), C. rockiensis sp. n. (purple), C. albosuffusa stat. nov. (blue), C. oaxacensis sp. n. (orange), and C. drusius (black). Primary type specimens are labeled in magenta (except that C. oaxacensis is left in orange as the sole representative of its species).
Fig. 30.

Male genitalia of Cecropterus (Thorybes) pylades species group: a–d) C. floridianus sp. n. from USA, FL: a) NVG-22101A01, paratype, genitalia slide J. W. T. 25–34, data in text [CAS]; b) holotype, data in text; c) and d) from genitalia slides G11 and G14, respectively, by E. L. Bell, only valvae shown [AMNH]; e–i) C. pylades from USA: e–g) left valva in situ, N. V. Grishin, leg.: e) TX, Wise Co., LBJ National Grassland, 4-May-2008 and f, g) OK: Murray Co., 5 mi S of Davis, 23-Apr-2008; h) NVG-9288, AR, Montgomery Co., Ouachita National Forest, 6.5 air mi WNW of Oden, 6-Jul-2017, N. V. Grishin leg.; i) NJ, from genitalia slide G13 by E. L. Bell, only valvae shown [AMNH]; j) C. indistinctus stat. nov., USA: CA, San Diego Co., El Cajon, 28-Apr-1934, Ray Hulbirt leg., genitalia slide J. W. T. 25–36 by J. W. Tilden [CAS]; k, l) C. rockiensis sp. n. paratypes from USA: k) NVG-11465 from AZ: Apache Co. and l) NVG-8970 from NM: Otero Co., data in text; m–o) C. albosuffusa stat. nov.: m, n) Mexico [TMMC]: m) NVG-22056A08, Coahuila, Zaragosa, C. J. Durden leg., 11-Apr-1976, genitalia vial NVG-22056A09 and n) NVG-22056A04, Nuevo Leon: Galeana, Cerro Potosi, C. J. Durden leg., 25-Jul-1981, genitalia vial NVG-22056A05; o) USA: AZ, Santa Rita Mts., 10-Jul-1941, J. W. Tilden leg. and genitalia slide J. W. T. 25–13 [CAS]; p) C. oaxacensis sp. n. holotype from Mexico: Oaxaca, data in text; q) C. drusius USA: AZ, from genitalia slide G47 by E. L. Bell, aedeagus (left) and valva (right) [AMNH]. Panels b, h, k, l, n, p) show complete genitalia in dorsal (above) and left lateral (below) views, and m) only left lateral view. Panels a, j, o) assemble genitalia parts from slides to display aedeagus (if detached, top left), detached valva (left), and the rest of the genital capsule in lateral view (right). All images are to scale.
The extent of genetic differentiation suggests that the clades within C. pylades represent distinct species (Cong et al. 2019a). We find that Cecropterus pylades albosuffusa (H. Freeman, 1943) (type locality USA: Texas, Ft. Davis) forms a clade distant from the nominotypical C. pylades with Fst/Gmin of 0.39/0.001 and COI barcode difference of 2.1% (14 bp). Similarly, the westernmost Cecropterus pylades indistinctus (Austin & J. Emmel, 1998) (type locality in USA: California: San Diego Co.) is separated from the nominotypical C. pylades with Fst/Gmin of 0.41/0.002 and COI barcode difference of 1.7% (11 bp). Therefore, we propose treating Cecropterus (Thorybes) albosuffusa (H. Freeman, 1943), stat. nov. and Cecropterus (Thorybes) indistinctus (Austin & J. Emmel, 1998), stat. nov. as species-level taxa, not subspecies of C. pylades. Hence, no subspecies are recognized in the C. pylades species group. Furthermore, three additional clades with strong genetic differentiation do not have names and represent new species. These three new species are described below. Finally, we note that the speciation scenario in the “crown” subgroup of the C. pylades group consisting of four USA species: Floridian, eastern, west-coastal, and central (Fig. 27 red, green, cyan, and purple) parallels that of the Erynnis brizo (Boisduval & Le Conte, [1837]) species group as laid out by Burns (2020).
Cecropterus (Thorybes) rockiensis Grishin, new species http://zoobank.org/7CC2D85B-497D-4A81-9486-82434CCBE143 (Figs. 27 part, 28, 29 part, 30k, l)
Fig. 28.

Holotype of Cecropterus (Thorybes) rockiensis sp. n. in dorsal (left) and ventral (right) views, data in text.
Fig. 29.

A map of sequenced specimens from the Cecropterus (Thorybes) pylades group. Different species are shown in different symbols of different colors: C. floridianus sp. n. (larger red circles), C. pylades (smaller green circles), C. indistinctus stat. nov. (cyan diamonds), C. rockiensis sp. n. (purple squares), C. albosuffusa stat. nov. (smaller dark blue downturned triangles), and C. oaxacensis sp. n. (larger orange upturned triangle) and labeled on the map near their type localities. Type localities for valid names are indicated by tiny circles placed inside symbols, or (if no specimens from these localities were sequenced) by a small circle framed with the color of the taxon (for C. pylades only).
Definition and diagnosis.
Both the Z chromosome and the mitogenome trees reveal partitioning of western US populations previously assigned to Cecropterus (Thorybes) pylades (Scudder, 1870) (type locality in USA: Massachusetts) into two clades: Cecropterus (Thorybes) indistinctus (Austin & J. Emmel, 1998), stat. nov. (type locality in the USA: California: San Diego Co., holotype sequenced as NVG-17109A08) and a clade consisting of specimens from and around the Rocky Mountains region in the US, not associated with any available names (Fig. 27 cyan and purple). The Fst/Gmin between the two clades are 0.37/0.004, and their COI barcodes differ by 1.1% (7 bp). Therefore, the Rocky Mountains clade represents a species-level taxon. This new species is generally similar in appearance to its closest relative, C. indistinctus, in the following combination of characters: stronger checkered fringes (especially on the forewing), ventral wing surface distally paler gray-brown, but not strongly overscaled with white, on average smaller hyaline spots, and less rounded forewings; and differs from it by more prominently checkered fringes, paler ground color, stronger expressed darker framing around (or in place of) hyaline forewing spots, better defined ventral hindwing bands, typically larger hyaline spots and specimen size. In male genitalia, uncus arms are thicker and not as widely separated as in C. (Thorybes) albosuffusa (H. Freeman, 1943), stat. nov. (type locality USA: Texas, Ft. Davis), more angled than rounded at the base between them; similar to C. pylades and C. indistinctus and differ from them by somewhat wider separation of uncus arms, rather parallel than slightly converging distad and usually longer compared to tegumen than in the other two species, a notch between ampulla and harpe is typically shallower and wider, harpe is longer, more upcurved. Definitive identification is provided by DNA, and a combination of the following characters is diagnostic in nuclear genome: aly5196.9.2:C915T, aly536.8.1:C750T, aly1259.10.2:A1122G, aly276561.5.1:G2469A, aly5021.6.4:A1940G and in the COI barcode: A217G, T460C, T596C, A637A.
Barcode sequence of the holotype: Sample NVG-22099H03, GenBank OR578714, 658 base pairs:
AACTTTATATTTTATTTTCGGAATTTGAGCAGGATTAATTGGAACTTCTTTAAGTTTACTTATTCGAACTGAATTAGGAACTCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTCACAGCTCATGCTTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAATTCCCCTTATACTAGGGGCTCCTGACATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTACCTCCATCTTTAACTCTTTTAATTTCAAGAAGTATTGTTGAAAATGGAGCAGGTACTGGATGAACTATTTACCCCCCTTTATCTTCTAATATTGCTCATCAAGGAGCTTCAGTAGATTTAGCAATTTTTTCTTTACATCTTGCTGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACAACTATTATCAATATACGAATTAATAATTTATCATTTGATCAAATACCATTATTTATTTGAGCTGTTGGAATTACAGCTTTATTACTTTTACTTTCATTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACTGATCGAAACCTAAATACTTCATTTTTTGATCCAGCAGGTGGAGGAGATCCAATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the California Academy of Sciences, San Francisco, CA, USA [CAS], illustrated in Fig. 28, bears six printed (other text but “COLO” on the first label is handwritten) labels: five white [COLO. V-24–79 | Jefferson Co. | Clear Cr. Cyn.], [Ray E. Stanford | collector], [Collection of | C.D.MacNeill], [DNA sample ID: | NVG-22099H03 | c/o Nick V. Grishin], [{QR Code} CASENT | 8566837], and one red [HOLOTYPE ♂ | Cecropterus (Thorybes) | rockiensis Grishin]. Paratypes: 20♂♂ 7♀♀: 1♀ Montana, Yellowstone Co., Billings, 5-Jun-1948, Neil Euting leg. (NVG-22099H04, CASENT8566838) [CAS]; 2♂♂ Nebraska, Sioux Co., Monroe Cyn., 6 mi. N of Harrison, 25-Jun-1983, S. M. Spomer leg. (NVG-22064H01 & H02); Utah: Salt Lake Co.: 1♂ Mill Creek Canyon, 28-Jun-1967, C. J. Callaghan leg. (NVG-22099H07, CASENT8566841) [CAS]; Salt Lake, City Creek Canyon: 1♂ 15-Jun-1930, Lloyd M. Martin leg. (NVG-22094A09) [LACM]; 1♂ 9-Jun-1946, L. I. Hewes leg. (NVG-22099H08, CASENT 8566842) [CAS]; Utah Co.: 1♂ Provo Canyon, 14-Jun-1947, William A. Hammer leg. (NVG-22099G12, CASENT8566834) [CAS]; 1♂ Mt. Timpanogos, 5.6 mi W of Jct. Hwy 189 & 92 on 92, 22-Jul-1946, C. D. MacNeill leg. (NVG-22099H09, CASENT8566843) [CAS]; 1♂ Beaver Co., East Fork Baker Canyon, SH153, Tushar Mts, N side of Beaver Canyon, larva collected on 1-Jul-2022, eclosed 7-Dec-2022, Todd Stout leg. (NVG-22068D07); 1♀ Washington Co., 11-May-1977, D. F. Shillingburg leg. (NVG-22099H06, CASENT8566840) [CAS]; 1♀ Zion National Park, 15-Jun-1928, T. Craig leg. (NVG-22099H10, CASENT8566844) [CAS]; San Juan Co.: 1♀ La Sal Mts., Pack Creek day use area, 31-May-2016, Robb Hannawacker leg. (NVG-20045E07); 1♂ Abajo Mts., Indian Creek trail, el. 6400′−7400′, 8-May-2020, Robb Hannawacker leg. (NVG-20045E08); Colorado: 1♂ Garfield Co., Glenwood Cyn, el. 6200′, 8-Jun-1977, Ray E. Stanford leg. (NVG-22099H02, CASENT8566836) [CAS]; 1♂ Eagle Co., Fryingpan River, 10-Jun-1976, Ray E. Stanford leg. (NVG-22099H05, CASENT8566839) [CAS]; Boulder Co.: 1♀ Lefthand Canyon, 16-May-1954, Donald Eff leg. (NVG-22099H01, CASENT8566835) [CAS]; 1♂ Flagstaff Mtn., 15-Jun-1953, Donald Eff leg. (NVG-22094A08) [LACM]; New Mexico: 1♂ Sandoval Co., Cibola National Forest, SH165 4.2 mi S of Placitas, GPS 35.2502, −106.4104, 14-May-2017, Qian Cong, Jing Zhang & Nick V. Grishin leg. (NVG-8800); 1♂ 1♀ Lincoln Co.,1.5 mi E of Capitan Gap Rd N water course, el.7000′, 10-May-1981, J. McCaffrey leg. (NVG-15099H08 & H09) [FMNH]; 2♂♂ Otero Co., Lincoln National Forest, La Luz Canyon Rd., 4.8 air mi NE of High Rolls, GPS 32.9992, −105.7759, 21-May-2017, Qian Cong, Jing Zhang & Nick V. Grishin leg. (NVG-8970, Fig. 30l & NVG-9057); Arizona: Coconino Co.: 1♂ 1♀ S rim of Grand Cyn, 10-Jun-1942, J. S. Garth leg. (NVG-22094A02 & A03) [LACM]; 1♂ Lockett Meadow, GPS 35.3605, −111.6208, 24-May-2021, Brian Banker leg. (NVG-21089E12); 1♂ Apache Co., Greens Peak area, Pipeline Spring at Fs117, GPS 34.1413, −109.5809, 25-May-2018, Jing Zhang & Nick V. Grishin leg. (NVG-11465, Fig. 30k); 1♂ Graham Co., Adam’s Flat, GPS 32.6508, −109.8125, 23-Apr-2009, Mark Walker leg. (NVG-18037D10).
Type locality.
USA: Colorado, Jefferson Co., Clear Creek Canyon.
Etymology.
The name is given for the general area of the distribution of this species, which is in and around the Rocky Mountains (the Rockies), by fusing the Latin suffix -ensis (meaning “from place” or “of place”) with the word “Rockies”. The name is a masculine adjective in the nominative case.
English name.
Rocky Mountains Cloudywing.
Distribution.
Across the Rocky Mountains and neighboring states: confirmed from Montana, Nebraska, Utah, Colorado, New Mexico, and Arizona.
Cecropterus (Thorybes) floridianus Grishin, new species http://zoobank.org/EA537AE1-E4FB-4C72-8615-D9BE2DE7336A (Figs. 27 & 29 parts, 30a–d, 31)
Fig. 31.

Holotype of Cecropterus (Thorybes) floridianus sp. n. in dorsal (left) and ventral (right) views, data in text.
Definition and diagnosis.
Both the Z chromosome and the mitogenome trees reveal that eastern US populations previously assigned to Cecropterus (Thorybes) pylades (Scudder, 1870) (type locality in USA: Massachusetts) are not monophyletic, and populations from Florida form a clade sister to three species combined (C. pylades, C. indistinctus, and C. rockiensis sp. n.) (Fig. 27). Genetic differentiation between C. pylades and the Floridian populations is notable: Fst/Gmin between the two clades are 0.24/0.003 and the COI barcode difference is 2.3%–2.4% (15–16 bp). Therefore, the Floridian clade represents a distinct species. This new species is similar in appearance to C. pylades in darker ground color, weaker checkered fringes (especially on forewing), weaker developed marginal pale overscaling on wings beneath, larger size, and rounder wings; and differs from it by darker appearance and generally smaller hyaline spots. In the male genitalia, most similar to C. pylades, e.g., uncus arms slightly converge distad, but valva is usually broader, the central bump on its costa is typically less pronounced (in lateral view), harpe is relatively shorter, broader, and straighter, less angled along the ventral margin, and uncus arms are usually shorter compared to tegumen (Fig. 30a–d). Definitive identification is provided by DNA and a combination of the following characters is diagnostic in nuclear genome: aly1409.4.2:G1779A, aly383.20.2:T1131A, aly3507.12.1:G3947C, aly383.21.1:A1654G, aly1409.4.2:A1477C and in COI barcode: T91A, T232C, T355C, T478C, T514A.
Barcode sequence of the holotype: Sample NVG-22032A09, GenBank OR578715, 658 base pairs:
AACTTTATATTTTATTTTCGGAATTTGAGCAGGATTAATTGGAACTTCTTTAAGTTTACTTATTCGAACTGAATTAGGAACTCCAGGATCATTAATTGGAGATGACCAAATTTATAATACTATTGTCACAGCTCATGCTTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAATTCCTCTTATACTAGGAGCTCCTGATATAGCCTTTCCTCGTATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAACTCTTTTAATTTCAAGAAGTATTGTTGAAAATGGAGCAGGTACTGGATGAACTGTTTATCCCCCATTATCTTCCAATATTGCTCACCAAGGAGCTTCAGTAGATTTAGCAATTTTTTCTTTACATCTTGCAGGAATTTCTTCAATTTTAGGAGCTATTAATTTTATTACAACTATTATTAATATACGAATTAATAACTTATCATTTGATCAAATACCATTATTTATTTGAGCAGTTGGAATTACAGCTTTATTACTTTTACTTTCACTACCTGTTTTAGCTGGAGCTATTACTATATTATTAACTGATCGAAATTTAAATACTTCATTTTTTGATCCAGCAGGTGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA [USNM], illustrated in Fig. 31, bears four printed labels: three white [USA:FLA: Volusia Co. | New Smyrna Beach | 3 April 1969 | Leg. G. Rawson], [DNA sample ID: | NVG-22032A09 | c/o Nick V. Grishin], [genitalia vial | NVG230917–01 | Nick V. Grishin], and one red [HOLOTYPE ♂ | Cecropterus (Thorybes) | floridianus Grishin]. Paratypes: 4♂♂ 3♀♀ from USA, Florida: 1♂ Franklin Co., USH98 0.4 mi. S of junction with Co. Rd. 370, 3 air mi N of Alligator Point, GPS 29.9378, −84.3907, 29-Mar-1988, J. M. Burns leg., (NVG-22032A08) [USNM]; 1♀ Dixie Co., 16 km S Steinhatchee, end of Rt. 361, 4-Apr-1988, Scott W. Gross leg. (NVG-22032A07) [USNM]; 1♂ Levy Co., Cedar Key, 25-Jul-1963, C. J. Durden leg. (NVG-20062C06) [TMMC]; 1♀ Alachua Co., Gainesville, 20-Apr-1973, E. C. Knudson leg. (NVG-22032A10) [USNM]; 1♂ Marion Co., Ocala National Forest nr. Juniper Springs, 15-May-1988, Scott W. Gross leg. (NVG-22032A12) [USNM]; 1♀ Volusia Co., New Smyrna Beach, G. W. Rawson leg. 10-Mar-1969 (NVG-22032A11) [USNM]; 1♂ Martin Co., Port Sewall, 16-Mar-1939, genitalia 25–34 J. W. Tilden (NVG-22101A01, Fig. 30a) [CAS].
Type locality.
USA: Florida, Volusia Co., New Smyrna Beach.
Etymology.
The name for this Floridian species is formed by adding “-us” and is a masculine adjective.
English name.
Florida Cloudywing.
Distribution.
Confirmed only from the USA: Florida, but is likely to be found at least in Georgia.
Cecropterus (Thorybes) oaxacensis Grishin, new species http://zoobank.org/5638F005-DC60-47A2-BB6F-6D9B0650F399 (Figs. 27 & 29 parts, 30p, 32, 33)
Fig. 32.

Holotype of Cecropterus (Thorybes) oaxacensis sp. n. pinned through its side (to be spread later), data in text.
Fig. 33.

Cecropterus (Thorybes) oaxacensis sp. n. female, iNaturalist observation 110602850 from Mexico: Oaxaca, San Miguel Tequixtepec, 17-Jun-2014, © John Kemner; photographs show the same individual. Images are color-corrected, and the rightmost is rotated approximately 90° clockwise. CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/
Definition and diagnosis.
Inspection of genomic trees reveals that a single specimen from Mexico, Oaxaca, initially identified by us as “Cecropterus albosuffusa” (curated with “Cecropterus drusius” in the collection), is not placed among Cecropterus (Thorybes) albosuffusa (H. Freeman, 1943) (type locality USA: Texas, Ft. Davis) specimens we sequenced from across the range (Fig. 27). This specimen is sister to all analyzed C. albosuffusa, some from Puebla and DF in Mexico. This consistent placement of the specimen in the trees constructed from protein-coding regions in autosomes, Z chromosome, and mitochondrial genome, where all sequenced C. albosuffusa specimens across the range from Arizona and Texas to Pueblo cluster closely together, and its distinction in the COI barcode of 1.2% (8 bp, while C. albosuffusa did not show variation in the barcode) suggest that it represents a distinct species. This new species is diagnosed by white hindwing fringe from vein M2 to tornus, similar to Cecropterus (Thorybes) drusius (W. H. Edwards, [1884]) (type locality in southern Arizona) but has C. albosuffusa-like genitalia with rounded (Fig. 30p), not claw-shaped (Fig. 30q) harpe, rounder hindwings in males (in C. drusius males, hindwings are slightly extended at tornus, almost lobed), broad paler “frosty” submarginal area on ventral hindwing, which C. drusius typically lacks (some specimens with narrower pale overscaling), and broader forewing subapical spots in a straight line, each spot is nearly square. In male genitalia (Fig. 30p), it is most similar to C. albosuffusa (Fig. 30m–o) but differs in thicker and shorter relative to tegumen uncus arms, broader and more rounded in lateral view tegumen, and broader, rounder and more upturned harpe. In DNA, a combination of the following characters is diagnostic in nuclear genome: aly638.13.2:A120C, aly638.13.2:C165T, aly4592.3.6:T120C, aly4592.3.6:A159T, aly727.34.3:C417T, aly596.8.8:G120G (not A), aly596.8.8:G618G (not A), aly767.17.3:A273A (not G), aly767.17.3:T941T (not G), aly1432.13.4:C50C (not T) and COI barcode: A181G, A421A, T574C, C595C, T604C.
Barcode sequence of the holotype: Sample NVG-19125B09, GenBank OR578716, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGATTAATTGGAACTTCTTTAAGTTTACTTATTCGAACTGAATTAGGAACTCCAGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTCACAGCTCATGCTTTTATTATAATTTTCTTTATAGTTATACCTATTATAATTGGGGGATTTGGAAATTGATTAATTCCTCTTATATTAGGAGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTATTACCCCCATCTTTAACTCTTTTAATTTCAAGAAGTATTGTTGAAAACGGAGCAGGTACTGGATGAACTGTTTATCCCCCTTTATCTTCTAATATTGCTCATCAAGGAGCTTCAGTAGATTTAGCAATTTTTTCTTTACATCTTGCAGGAATTTCATCAATTTTAGGAGCTATTAATTTTATTACAACTATTATTAATATACGAATTAATAATTTATCATTTGATCAAATACCATTATTTATTTGAGCTGTTGGAATTACAGCCTTATTACTTTTACTTTCATTACCTGTTTTAGCTGGAGCTATTACCATATTATTAACTGATCGAAACTTAAATACCTCATTTTTTGATCCTGCAGGTGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the University of Texas Biodiversity Center collection, Austin, TX, USA [TMMC], illustrated in Fig. 32, bears four printed labels: three white [OA.Tlalixtac.002 | 5 mi N Oaxaca | KemnerJ 88138A02], [DNA sample ID: | NVG-19125B09 | c/o Nick V. Grishin], [DNA sample ID: | NVG-22056A03 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Cecropterus (Thorybes) | oaxacensis Grishin], collected by John Kemner on 17-May-1988 (i.e., “88138”: day 138 of 1988).
Type locality.
Mexico: Oaxaca, Tlalixtac de Cabrera, Hwy 175, ca. 5 mi north of Oaxaca City.
Etymology.
The name is given for the type locality and is a masculine adjective in the nominative case.
English name.
Oaxaca Cloudywing.
Distribution.
Known only from Mexico: Oaxaca. Specimens from phenotypically similar (but mostly darker-fringed) populations to the north (e.g., Puebla and Ciudad de México) are Cecropterus (Thorybes) albosuffusa (H. Freeman, 1943) as identified by genomic sequencing (Fig. 27).
Telemiades solon Plötz, 1882 is a junior subjective synonym of Nascus (Bron) broteas (Cramer, 1780)
The name Telemiades solon Plötz, 1882 (type locality in South America), currently applied to a valid species of Nascus Watson, 1893 (type species Papilio phocus Cramer, 1777), was proposed in a key with a specimen number 4874 in Berlin collection (MFNB) mentioned and the drawing “t. 154” referenced (Plötz 1882). Neither the specimen nor the illustration or its copy could be found. Godman (1907) skipped the numbers t. 149 to t. 159 in his analysis of unpublished illustrations of American Hesperiidae by Plötz, and none of these drawings were copied. The fate of the original Plötz’s illustrations is still unknown (Zhang et al. 2023d). A search of the Hesperiidae holdings in MFNB did not yield the specimen with the number 4874. However, the catalog of the old Hesperiidae collection in MFNB handwritten by Hopffer (Zhang et al. 2023d) has a record for No. 4874: two specimens collected in “Rio” [Brazil: Rio de Janeiro]. Initially, they were listed as “sp.” (i.e., unidentified), but subsequently, “sp.” was crossed out, and “Pherenice Hew.” was written instead. Eudamus pherenice Hewitson, 1867 (type locality in Brazil) is currently regarded as a junior subjective synonym of Nascus (Nascus) phocus (type locality in Suriname). Thus, the appearance of these two specimens No. 4874 was likely that of Nascus, even if they were misidentified by Hopffer (i.e., they were not pherenice but some other similar species), and thus, if they were T. solon, consistent with the placement of T. solon in Plötz’s key among species currently in Nascus. Therefore, it is possible that the specimen(s) referred to by Plötz as “4874” was (were) listed in the Hopffer catalog for that entry and collected in Southeast Brazil (Rio de Janeiro), a detail not given in the original description of T. solon.
Evans (1952) applied the name “Nascus solon” to an Amazonian species not known from Southeast Brazil. This discrepancy found after an inspection of the Berlin collection catalog prompted us to study the original description of T. solon more closely. We concluded that Evans misidentified T. solon because the species Evans identified as “N. solon” does not agree with the original description of T. solon. Most significantly, Plötz mentions a brown “hair pencil” (a tuft of long hair-like scales) at the base of cell 1b [i.e., 1A+2A-3A] on ventral hindwing in T. solon, but this tuft is pale, mostly yellow, in Evans’ “N. solon.” Then, in T. solon, pale spots in the middle of the forewing are close together and “only separated by veins” (Plötz 1882); but in “N. solon”, the spot in cell M3-CuA1 does not reach the cell origin, which is filled with the ground color (yellow-brown) for at least half of the width of the pale spot in cell CuA1-CuA2 along vein CuA1. Furthermore, in T. solon, the hyaline spot in forewing cell M1-M2 is small, a pale spot by the costa in the middle of the forewing is mentioned only as “beneath, costal margin is also spotted with pale”, and the ventral hindwing is with brown “crossbar in cell 7 [Sc+R1-RS] and in the discal cell”. In contrast, in “N. solon”, the spot in the forewing cell M1-M2 is nearly the same size as the spot in the cell R5-M1, forewing is with a well-developed hyaline spot by mid-costa, which is seen from the dorsal side as well (hyaline!), hindwing is with weakly developed or missing cental brown spot in the discal cell, and the spot in the cell Sc+R1-RS is small, not a “crossbar”. Therefore, Evans’ “N. solon” is not conspecific with the true T. solon.
Next, we searched for specimens that agree with the original description of T. solon. The specimen we found to match the description closely was No. 4865 in MFNB. This specimen was previously curated as a type of Netrocoryne seneca Plötz, 1882 (type locality Brazil) because No. 4865 was mentioned by Plötz (1882) in the original description of N. seneca. However, this specimen is a pseudotype because it agrees neither with the original description nor with a copy of Plötz’s unpublished illustration of N. seneca (Zhang et al. 2023d). It is possible that due to some mistakes in referencing the MFNB specimen numbers, this specimen No. 4865 was instead (or in addition) a syntype of T. solon. However, we do not have defendable evidence to support this hypothesis. Therefore, we proceeded with the neotype designation because there is an exceptional need to clarify both the taxonomic identity and the type locality of T. solon. This taxon has been misidentified by Evans (1952), who applied this name to a species that does not agree with the original description of T. solon. This mistake creates inconsistencies in the literature and the potential for further destabilization of nomenclature due to the existence of additional species in this group unless the name T. solon is objectively defined by the neotype. Therefore, N.V.G. designates the specimen No. 4865 in MFNB illustrated in Fig. 1a–c in Zhang et al. (2023d) (DNA sample NVG-15031F11) as the neotype of Telemiades solon Plötz, 1882.
Our neotype of T. solon satisfies all requirements set forth by the ICZN Article 75.3, namely: 75.3.1. It is designated to clarify the taxonomic identity of Telemiades solon Plötz, 1882, which has been misinterpreted and attributed to a different species that does not agree with the original description of T. solon, and to detail its type locality that was given only generally (as “South America”) in the original description; 75.3.2. The characters to differentiate this taxon from others were specified by Plötz (1882), and we regard them as follows: forewing with a hyaline spot in cell CuA2-1A+2A, hyaline spots in the middle of the forewing are crowded together and separated only by veins, hyaline spots in cells M1-M2 and M2-M3 are small, and the former is merged with the apical spots that together form an oval-shaped hyaline patch separated by veins, a hyaline spot in cell R2-R3 is merged with this patch and not offset basad, the hyaline spot is absent by the costal margin near its middle, but the costal margin with a pale spot in the middle beneath; ground color of wings olive-brown, the base of ventral hindwing and most of ventral hindwing are clay-yellow, ventral hindwing with brown crossbars in cell Sc+R1-RS and the discal cell, and, in addition to the broadly brown outer margin area, with a postdiscal brown band and a brown tuft of hair-like scales at the base of cell 1A+2A-3A; 75.3.3. The neotype specimen is a male bearing eight labels (1st red, 3rd bluish-greenish, the last orange, and others white): [typus], [4865], [Bahia Sello], [GEN.PREP., | MIELKE | 1996], [seneca | Pl. | type], [DNA sample ID: | NVG-15031F11 | c/o Nick V. Grishin], [{QR Code} http://coll.mfn-berlin.de/u/ | 940b3c], [not a type specimen of | Netrocoryne seneca | Plötz, 1882 | determined by Zhang, | Cong et al. 2023] and illustrated in Fig. 1a–c (without the last label, which was added later) in Zhang et al. (2023d); the neotype has a chipped off tornus on both hindwings; 75.3.4. We searched for syntypes of T. solon in the MFNB collection because the original description specified specimen(s) with the number 4874 in Berlin. While there was an entry in the old collection catalog with No. 4874 listing two specimens, we could not find them among Hesperiidae holdings, and therefore, we believe that syntypes were lost; 75.3.5. The neotype closely agrees with the original description of T. solon in all (but one) characters, as evidenced by comparing the neotype illustrated in Fig. 1a–c in Zhang et al. (2023d) with the characters for this taxon given in the original description (Plötz 1882) and listed above (75.3.2.); the only discrepancy is that palpi are nearly white beneath, not clay-yellow (could have been discolored) as stated by Plötz (1882); 75.3.6. The neotype is from Brazil: Bahia, which is within the original type locality given as “South America”; 75.3.7. The neotype is in the collection of the Museum für Naturkunde, Berlin, Germany (MFNB).
Genomic sequencing confirms the phenotypic assessment of the T. solon neotype as a specimen of Nascus (Bron) broteas (Cramer, 1780) (type locality in Suriname) (Zhang et al. 2023d) because it groups with specimens of the latter species in the tree (Fig. 34). Notably, Draudt (1922) has already placed T. solon within his Nascus cous (Möschler, 1879) (listing Nascus eugamon Godman & Salvin, 1893 as a synonym of the latter), and expressed an opinion that these may be males of N. broteas, which was then known only by females. Therefore, as a result of the neotype designation, Telemiades solon Plötz, 1882, syn. nov. becomes a junior subjective synonym of Nascus (Bron) broteas (Cramer, 1780). The COI barcode sequence of T. solon neotype, sample NVG-15031F11, GenBank OR578717, 658 base pairs, is:
AACATTATATTTTATTTTTGGAATTTGAGCTGGAATAATTGGAACTTCTCTTAGATTACTAATTCGAACTGAATTAGGAACCCCCGGATCTTTAATTGGAGATGATCAAATTTATAATACTATCGTAACAGCTCATGCTTTCATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCTCTTATACTAGGAGCTCCTGATATAGCATTTCCACGAATAAATAACATAAGATTTTGATTATTACCTCCATCATTAACATTATTAATTTCAAGAAGAATTGTCGAAAATGGTGCTGGTACTGGATGAACAGTTTACCCCCCTTTATCAGCAAATATTGCTCACCAAGGTTCTTCCGTAGATTTAGCAATCTTTTCTTTACATTTAGCTGGAATTTCTTCTATTTTAGGAGCTATTAACTTTATTACAACAATTATTAATATACGAATTAGAAATTTATCTTTTGATCAAATACCATTATTTATTTGAGCTGTAGGAATTACAGCATTATTATTACTACTTTCTTTACCTGTTTTAGCAGGAGCTATTACTATATTACTTACTGATCGAAATTTAAATACATCTTTCTTTGACCCAGCTGGAGGAGGAGATCCTATTCTTTATCAACATTTATTT
Fig. 34.

The phylogenetic tree of Nascus (Bron) inferred from nuclear genome protein-coding regions (autosomes): N. (B.) corilla stat. nov. (blue), N. (B.) lux sp. n. (red), and N. (B.) broteas (black). Primary type specimens are labeled in magenta.
Nascus (Bron) corilla Evans, 1952 is a species-level taxon
Nascus solon corilla Evans, 1952 (type locality in Venezuela) was proposed as a subspecies of a species that Evans misidentified as “N. solon”. We argue above that the true Telemiades solon Plötz, 1882 (type locality in Brazil: Bahia) is conspecific with Nascus broteas (Cramer, 1780) (type locality in Suriname). Because a species that Evans considered to be “N. solon” is distinct from N. broteas, it is not conspecific with the true T. solon and should not be identified by this name. Because no other available name applies to Evans’ “N. solon”, Nascus corilla Evans, 1952, stat. nov. becomes a species-level taxon.
Nascus (Bron) lux Grishin, new species http://zoobank.org/4B4097CE-A43D-4ABB-A5E5-1EAE4FD5C049 (Figs. 34 part, 35)
Fig. 35.

Holotype of Nascus lux sp. n. in dorsal (left) and ventral (right) views, data in text.
Definition and diagnosis.
Inspection of genomic trees reveals that Amazonian populations, which Evans (1952) considered to be conspecific with Nascus corilla Evans, 1952, stat. nov. (type locality in Venezuela) and misidentified as Telemiades solon Plötz, 1882 (type locality in Brazil: Bahia) as determined above, are genetically differentiated from N. corilla with Fst/Gmin of 0.48/0.003 (although their COI barcodes do not reveal differences but in 1 or 2 base pairs) and therefore represent a distinct species (Fig. 34). This species does not have a name. This new species keys to “Nascus solon solon” D.5.3(b) in Evans (1952) and is distinguished from its closest relative, N. corilla, stat. nov. (see above), by pale-yellow to orange-yellow patches of scales, variable in their expression, dividing the brown outer marginal area on the ventral hindwing (that is solid dark brown in N. corilla) into narrow postdiscal and broad submarginal bands in males, and usually having five (not four) subapical hyaline spots in females; and from other species of Nascus by the following combination of characters: forewing apical spot in cell R2-R3 is in line with others, not offset basad; yellow or pale brown tuft of long scales at the base of cell 1A+2A-3A on ventral hindwing; palpi beneath and cheeks are white; prominent hyaline spot in the middle of forewing by costal margin in males; forewing typically larger than 28 mm in males and 30 mm in females. A combination of the following nuclear genomic characters is diagnostic: aly525.35.1:C54T, aly214.21.1:C303T, aly214.21.1:G312A, aly26.5.3:C294T, aly50.27.2:A51G.
Barcode sequence of the holotype: Sample NVG-21012D07, GenBank OR578718, 658 base pairs:
AACTTTATATTTTATTTTTGGAATTTGAGCTGGAATAATTGGAACTTCTCTTAGATTATTAATTCGAACTGAATTAGGCACCCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACAATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTCGGAAATTGACTAGTACCTCTTATATTAGGAGCTCCTGATATAGCATTTCCACGAATAAATAATATAAGATTTTGATTATTACCTCCATCATTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCTGGTACTGGTTGAACAGTTTACCCCCCTTTATCAGCAAATATTGCTCACCAAGGATCTTCTGTAGATTTAGCAATTTTTTCATTACATTTAGCAGGAATTTCCTCAATTTTAGGAGCTATTAACTTTATTACAACAATTATTAATATACGAATTAGAAATTTATCTTTTGATCAAATACCATTATTTATTTGAGCTGTAGGTATTACAGCTTTATTATTATTACTTTCTTTACCTGTTTTAGCAGGAGCTATTACTATATTACTTACTGATCGAAATTTAAATACATCTTTTTTTGATCCTGCAGGAGGAGGTGATCCAATTCTTTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the Carnegie Museum of Natural History, Pittsburgh, PA, USA [CMNH], illustrated in Fig. 35, bears four printed (last two digits of the year handwritten) labels: three white [Uassa Island, | Uassa Swamp. | S. M. Klages, C. M. Acc. 6176], [May | 1918], [DNA sample ID: | NVG-21012D07 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Nascus (Bron) | lux Grishin]. Paratypes: 4♂♂, one from each locality Peru: Huanuco, Tingo Maria, 800m, May-Jun-1994, genitalia X-5174 J. M. Burns 2002 (NVG-17103E04, USNMENT00913776); Brazil: Rondônia, 62 km S Ariquemes, Fazenda Rancho Grande, elevation 165m, GPS −10.53, −63.80, 27-Aug-8-Sep-1994, Ron Leuschner leg. (NVG-17103G06, USNMENT00913805); Amazonas, Maues, Rio Preto, 15–25-Nov-2007 (NVG-18088H10); and French Guiana: Roura, Coralie, GPS 4.5083, −52.3750, L. Sénécaux & A. Docquin leg. 24-Jul-1992 (NVG-18098E10, H3956).
Type locality.
Brazil: Amapá, Rio Uaçá region (“Uassa Swamp”), which is by the border between Brazil and French Guyana.
Etymology.
In Latin, lux means “light” and refers to the lighter (i.e., paler) overall appearance of this sunny species, light—instead of brown—tuft of scales on the hindwing (a beam of light), and a sprinkle of light scales between the postdiscal and submarginal brown bands on the ventral hindwing (a phenotypic character that typically separates this species from its closest relative). The name is a noun in apposition.
Distribution.
Generally, in and around the Amazonian region in South America. This species has been recorded from French Guiana, Peru, Ecuador, and Brazil (Amapá, Amazonas, Rondônia).
Comment.
This name is proposed for Evans’ concept of “Nascus solon solon” (Evans misidentified Telemiades solon Plötz, 1882), and due to genetic differentiation, the two taxa Evans considered to be subspecies: “Nascus solon corilla” and “Nascus solon solon” are distinct species: Nascus corilla and Nascus lux sp. n., respectively.
Cogia hiska Evans, 1953 (with its subspecies Cogia hippalus hester Evans, 1953) is a species distinct from Cogia hippalus (W. H. Edwards, 1882)
Genomic sequencing of Cogia hippalus (W. H. Edwards, 1882) (type locality USA: AZ, Pima Co. Tucson, lectotype sequenced as NVG-15101B12) specimens across the range reveals their partitioning into two clades with genetic differentiation at the species level: Fst/Gmin and COI barcode difference of 0.49/0.004/2.9% (19 bp) (Fig. 36). Therefore, the two clades represent two distinct species. One clade contains specimens from the northwestern part of the range northeast to South Texas, USA, and south to Oaxaca, Mexico: the nominotypical C. hippalus and its close relative Cogia hippalus peninsularis L. Miller & MacNeill, 1969 (type locality Mexico: Baja California Sur, Arroyo San Bartolo, holotype sequenced as NVG-15095C07). The other clade consists of specimens from the southeastern part of the range from Mexico: Tamaulipas to Venezuela: Cogia hippalus hiska Evans, 1953 (type locality Costa Rica: Puerto Carrillo) and Cogia hippalus hester Evans, 1953 (type locality Venezuela: Merida). Our methods of analysis do not find species-level genetic differences between C. h. hiska and C. h. hester. The names hiska and hester were proposed in the same work issued on the same date. As the first revisers, we give precedence to hiska due to its larger distribution. Thus, we propose that Cogia hiska Evans, 1953, stat. nov. is a species and Cogia hiska hester Evans, 1953, comb. nov. is its subspecies.
Fig. 36.

Phylogenetic trees of selected Cogia inferred from protein-coding regions of a) the nuclear (autosomes) and b) the mitochondrial genomes. Different species are shown in different colors: C. hippalus (blue with its subspecies C. h. peninsularis labeled in violet), C. hiska (red, with its subspecies C. h. hester labeled in magenta), and C. outis (black).
Lectotype designation for Eudamus caicus Herrich-Schäffer, 1869 with the type locality likely in Mexico: Oaxaca
Eudamus caicus Herrich-Schäffer, 1869 (type locality Tropical America, possibly into southern USA), currently a valid species in the genus Cogia Butler, 1870 (type species Cogia hassan Butler, 1870) was described from an unstated number of specimens without details about their localities (Herrich-Schäffer 1869). One specimen without a locality label is curated in the MFNB collection as a syntype of E. caicus. We determine that this specimen is indeed a syntype because it agrees with the original description and matches closely in wing patterns a specimen BMNH(E) 1236437 (a male, in BMNH) that Godman selected as the most similar in his collection to the unpublished drawing by Plötz of Eudamus schaefferi Plötz, 1881 (a replacement name for E. caicus Herrich-Schäffer, 1869, erroneously deemed preoccupied), and labeled as “Compared with | Plotz’s drawing of | schaefferi, | Plötz | caicus, | HS.” Godman placed such similar-styled labels on specimens that, in his opinion, closely resembled the originals of Plötz’s drawings that he inspected (Godman 1907), and we use this specimen as a “proxy” for the drawing. We note that E. schaefferi (Plötz’s t[afel]. 129, which is E. caicus H.-S.) was not mentioned in Godman’s publication (1907), which might be because that work dealt with American species, but the locality for E. schaefferi was not explicitly stated in the original description and, due to the lack of locality labels on the syntype, likely not appearing on the Plötz’s drawing numbered 129. This Godman’s specimen from “N. Sonora, | Mexico” is actually Cogia caicus moschus (W. H. Edwards, 1882), but is similar in wing pattern to the nominotypical C. caicus; its photographs are shown on the Butterflies of America website (Warren et al. 2023). To stabilize nomenclature, N.V.G. hereby designates the syntype in the MFNB collection, a female with the following seven printed labels, the 1st purple and others white: [Origin], [Coll. H.—Sch.], [Daunus Cr.], [Caicus | HS. | Achlyodes], [Caicus | H-Sch.], [{QR Code} http://coll.mfn-berlin.de/u/ | 940b5d], and [DNA sample ID: | NVG-15032A02 | c/o Nick V. Grishin] as the lectotype of Eudamus caicus Herrich-Schäffer, 1869. Genomic trees place the lectotype among specimens from Mexico, Oaxaca (Fig. 37), suggesting that it was collected there because it is genetically within this metapopulation. Therefore, we infer that the type locality of Cogia caicus (Herrich-Schäffer, 1869) may be in Oaxaca and will test this hypothesis further by sequencing specimens from other localities.
Fig. 37.

Phylogenetic trees of selected Cogia species inferred from protein-coding regions of a) the nuclear genome (autosomes), b) the Z chromosome, and c) the mitochondrial genome. Different species are shown in different colors: C. moschus stat. rest. (blue), C. caicus (green), and C. chiagua sp. n. (red). Primary type specimens are labeled in magenta.
Cogia moschus (W. H. Edwards, 1882) is a species distinct from Cogia caicus (Herrich-Schäffer, 1869)
We find the genetic differentiation between Cogia caicus (Herrich-Schäffer, 1869) (type locality likely in Mexico: Oaxaca, as deduced above, lectotype sequenced as NVG-15032A02, in MFNB) and Cogia caicus moschus (W. H. Edwards, 1882) (type locality in USA: AZ, Graham Co., lectotype sequenced as NVG-15097H06, in CMNH) to be of a magnitude suggestive of distinct species: Fst/Gmin/COI differences are 0.50/0.003/0.9% (6 bp) (Fig. 37). Therefore, we propose that Cogia moschus (W. H. Edwards, 1882), stat. rest. is a species-level taxon.
Cogia chiagua Grishin, new species http://zoobank.org/00E80EF8-25B0-4A36-97FC-E6157CD96DE5 (Figs. 37 part, 38, 39a)
Fig. 38.

Holotype of Cogia chiagua sp. n. in dorsal (left) and ventral (right) views, data in text.
Fig. 39.

Male genitalia of Cogia in lateral view (left valva separated and rotated 180°). a) C. chiagua sp. n. paratype, mini-slide 153, data in text. b) C. moschus stat. rest., Mexico: N Sonora, Morrison leg., BMNH(E) 1236437, mini-slide 151. Photographs by N. V. G. © The Trustees of the Natural History Museum London and are made available under Creative Commons License 4.0 (https://creativecommons.org/licenses/by/4.0/).
Definition and diagnosis.
In addition to Cogia moschus (W. H. Edwards, 1882) (type locality in USA: AZ, Graham Co.) and Cogia caicus (Herrich-Schäffer, 1869) (type locality likely in Mexico: Oaxaca), genomic trees reveal a third lineage of similar genetic differentiation (Fig. 37 red) with Fst/Gmin/COI differences from the former and the latter, respectively: 0.49/0.009/0.9% (6 bp) and 0.4/0.006/0.5% (3 bp) that represents a species. This new species differs from its two close relatives by being darker overall, smaller (or lacking) hyaline spots, and darker palpi beneath. Male genitalia are variable, but the harpe typically is broader, rounder, and dorsally with more robust teeth on a shorter ridge near the ampulla; aedeagus with a smaller number of cornuti (Fig. 39). A combination of the following characters in the COI barcode is diagnostic: A40A, C343C, T349T, T386T, A481G, T556A.
Barcode sequence of the holotype: Sample NVG-22079A01, GenBank OR578719, 658 base pairs:
AACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTTGGAACTTCTTTAAGTTTATTAATTCGTACTGAATTAGGAACTCCAGGATCATTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCTCATGCTTTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTTCCCTTAATATTAGGAGCTCCTGATATAGCTTTTCCTCGTATAAATAATATAAGATTTTGATTACTTCCCCCATCATTAACATTATTAATTTCTAGAAGTATTGTAGAAAATGGTGCTGGTACTGGATGAACAGTATATCCCCCCCTTTCATCAAATATTGCACATCAAGGTTCATCAGTAGATTTAGCTATTTTTTCTTTACATTTAGCTGGTATTTCATCTATTTTAGGTGCTATTAATTTTATTACTACAATTATTAATATACGAATTAGAAATTTGTCATTTGATCAAATACCTTTATTTGTATGAGCAGTAGGAATTACAGCTTTATTACTTTTACTTTCTTTACCTGTATTAGCTGGTGCTATTACTATACTTTTAACAGATCGAAATCTTAATACATCATTTTTTGATCCTGCTGGAGGAGGAGACCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA [MGCL], illustrated in Fig. 38, bears five printed labels: four white [Antigua, Sacatepequez | Guatemala | September 16, 1993 | D.L.Lindsley], [Cogia Butler | caicus (Herrich-Schäffer) | caicus (Herrich-Schäffer)], [D.L. Lindsley colln. | MGCL Accession | # 2008–20], [DNA sample ID: | NVG-22079A01 | c/o Nick V. Grishin], and one red [HOLOTYPE ♂ | Cogia chiagua | Grishin]. Paratypes: 3♂♂, 1♀: 1♂ Mexico, Chiapas, Comitán, Laguna Chamula, 7100’, 13-May-1987, C.J. Durden leg. (NVG-19124H05) [TMMC] and Guatemala: 1♂ Panajachel, 2-Jun-1968, Beals leg. (NVG-17109F09) [LACM] and Duenas, G.C. Champion leg. [BMNH]: 1♂ (BMNH(E) 1236437, genitalia mini-slide 153, Fig. 39a) and 1♀.
Type locality.
Guatemala: Sacatepéquez Department, Antigua.
Etymology.
The name is a fusion of Chia[pas] + Gua[temala] for the known distribution of this species and is a feminine noun in apposition.
Distribution.
Currently known from Mexico: Chiapas and from Guatemala.
Pholisora albicirrus Glassberg, 2023 is a species distinct from Pholisora catullus (Fabricius, 1793)
We find the genetic differentiation between Pholisora catullus (Fabricius, 1793) (type locality “Indiis,” probably eastern USA) and Pholisora catullus albicirrus Glassberg 2023 (type locality in USA: AZ, Santa Rita Mts.) to be of a magnitude suggestive of distinct species: Fst/COI differences are 0.46/2.3% (15 bp) (Fig. 40), similar to that for Pholisora crestar J. Scott & Davenport, 2017 (type locality in USA: CA, Tulare Co.) (Zhang et al. 2020). Therefore, we propose that Pholisora albicirrus Glassberg 2023, stat. nov. is a species-level taxon in agreement with phenotypic differences mentioned in the original description (Glassberg 2023), which, from our experience with other Hesperiidae, also suggest species, and not subspecies, status.
Fig. 40.

Phylogenetic trees of Pholisora species recorded from the USA inferred from protein-coding regions of a) the Z chromosome (best for species delimitation) and b) the mitochondrial genome. Different species are shown in different colors: P. catullus (blue), P. crestar (violet), and P. albicirrus stat. nov. (red).
Celotes sabinus Grishin, new species http://zoobank.org/085EAF93-91A9-4F81-886C-0E77073C8B0E (Figs. 41 part, 42, 43a)
Fig. 41.
Phylogenetic trees of Celotes inferred from protein-coding regions of a) the nuclear genome (autosomes) and b) the Z chromosome. Different species are shown in different colors: C. sabinus sp. n. (red), C. nessus (blue), and C. spurcus (green). Primary type specimens are labeled in magenta.
Fig. 42.

Holotype of Celotes sabinus sp. n. in dorsal (left) and ventral (right) views, data in text.
Fig. 43.
Male genitalia of Celotes from USA, in lateral view. a) Celotes sabinus sp. n. paratype, slide J. W. T. 25–15, AZ: Baboquivari Mts., Brown Canyon, 21-Mar-1938, DNA sample NVG-22104H08. b) Celotes nessus, slide 25–16, TX: George West, 12-Jun-1940, DNA sample NVG-22104H10. Specimens are in CAS, collected and genitalia prepared by J. W. Tilden. Genital capsule is shown on the right, separated valva is on the left with aedeagus above it. Aedeagus of C. sabinus is shown in ventrolateral view as mounted on the slide.
Definition and diagnosis.
Inspection of genomic trees reveals that Celotes nessus (W. H. Edwards, 1877) (type locality USA: Texas, Bexar Co., San Antonio; lectotype sequenced as NVG-15097F12) as currently circumscribed (Fig. 41 blue and red) is not monophyletic, and populations from the eastern part of the range (Fig. 41 blue) are sister to Celotes spurcus A. Warren, Steinhauser, Hernandez-Mejía & Grishin, 2008 (type locality in Mexico: Querétaro) (Fig. 41 green). While species, in general, do not have to be monophyletic, populations currently identified as C. nessus from the western part of the range (Fig. 41 red) are genetically differentiated from the eastern populations (which include nominotypical) at the level characteristic of distinct species, with Z chromosome Fst/Gmin of 0.32/0.008 and COI barcode difference of ~2.0% (13 bp, but barcodes introgress between these species). Therefore, these western populations are a species distinct from C. nessus. This western species does not have a name because the only two junior synonyms of C. nessus: Spilothyrus notabilis Strecker, [1878] (type locality in the USA: Texas, vicinity of New Braunfels and San Antonio; two syntypes sequenced as NVG-15039C06 and NVG-15039C07) and Carcharodus radiatus Plötz, 1884 (type locality in USA: Texas, syntypes not located, attributed to a species by locality), are conspecific with the eastern species. This new species is distinguished from C. spurcus by approximately two times shorter process of valva (from the base of ampulla), which is similar in length to C. nessus, and from C. nessus by terminally broader harpe, wider separation between harpe and ampulla (broader gap between them), wider process of valva, broader valva narrowing less towards vinculum, and two small teeth on aedeagus shaft: by its bend and halfway between the bend and distal end (Fig. 43). A combination of the following nuclear genomic characters is diagnostic: aly2284.30.1:G1323A, aly40182.1.2:C154A, aly40182.1.2:C171T, aly1445.3.1:G246A, aly1445.3.1:A195G.
Barcode sequence of the holotype: Sample NVG-22105A02, GenBank OR578720, 658 base pairs:
AACTTTATATTTCATTTTTGGAATTTGAGCAGGCATAGTAGGTACTTCTCTAAGTTTATTAATTCGAACTGAATTAGGAAATCCAGGATCTCTAATTGGGGATGATCAAATTTATAATACTATTGTAACAGCACATGCCTTCATTATAATTTTTTTTATGGTAATGCCTATTATAATTGGAGGATTTGGAAATTGATTAGTACCTTTAATACTAGGAGCTCCTGATATAGCATTCCCACGTATAAATAATATAAGATTTTGATTATTACCTCCTTCTTTAACACTTCTTATTTCAAGAAGTATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTTTACCCCCCTCTATCATCTAATATTGCTCATCAAGGTTCATCTGTTGACTTAGCTATCTTTTCTTTACATCTAGCAGGAATTTCATCAATCTTAGGAGCAATTAACTTCATTACAACTATTATTAATATACGAATTAGAAATTTATCATTTGATCAAATACCTTTATTCGTATGAGCTGTAGGAATTACAGCATTACTTTTATTATTATCTTTACCTGTTTTAGCTGGAGCTATTACAATATTATTAACTGATCGAAATTTAAATACATCTTTCTTTGATCCTGCTGGAGGAGGAGATCCAATTTTATATCAACACTTATTT
Type material.
Holotype: ♂ deposited in the California Academy of Sciences, San Francisco, CA, USA [CAS], illustrated in Fig. 42, bears seven printed labels (date on the first label handwritten): six white [Sabino Cany. | Santa Catalina Mts. | Pima Co. Arizona | 1. IV. 60], [collected by | Kilian Roever], [Collection of | J. W. Tilden], [JAMES W. TILDEN | COLLECTION – 1985 | Gift to the California | Academy of Sciences], [DNA sample ID: | NVG-22105A02 | c/o Nick V. Grishin], [{QR Code} CASENT | 8568344], and one red [HOLOTYPE ♂ | Celotes sabinus | Grishin]. Paratypes: 13♂♂, 4♀♀: USA, Arizona: Mohave Co., 1♀ Hualapai Mts., lower el., 16-Apr-2008, Ken Davenport leg. (NVG-20065A08, CSU_ENT1024696) [CSUC]; 1♂ nr. Wickieup, 30-Mar-1972, J. W. Tilden leg. (NVG-22104H11, CASENT8568341) [CAS]; 1♀ Yavapai Co., 8 mi SW of Prescott, 21-Apr-1976, James W. “Bill” Tilden leg. (NVG-22105A03, CASENT8568345) [CAS]; 1♂ Maricopa Co., Camp Creek on Cave Creek Rd., 12 mi NE of Jct. of Cave Creek and Scottsdale Rds., 8-Apr-1968, J. A. Miller leg., genitalia NVG140320–91 (NVG-2250) [TAMU]; 1♂ Gila Co., Sevenmile wash, 22-Aug-1960, P. A. Opler leg. (NVG-22104H12, CASENT 8568342) [CAS]; 1♂ Pinal Co., Coronado National Forest, Santa Catalina Mts., Peppersauce Canyon, 26-Mar-2017, Q. Cong, J. Zhang, and N. V. Grishin leg. (NVG-8304); Pima Co.: 1♂ Baboquivari Mts., Brown Canyon, 21-Mar-1938, J. W. Tilden leg., genitalia J.W.T. 25–15 (NVG-22104H08, CASENT8568338) [CAS] (Fig. 43a); 1♂ Santa Catalina Mts., Molino Basin, 7-Sep-1951, C. D. MacNeill leg. (NVG-22105A01, CASENT8568343) [CAS]; Santa Cruz Co.: 1♂ Pena Blanca Canyon, 11-Jul-1981, Jim P. Brock leg. (NVG-2115); 2♂♂ Walker Canyon nr. Pena Blanca Lake, 16-Aug-1972, John Hafernik leg., genitalia NVG140320–92 & −93 (NVG-2251 & NVG-2252) [TAMU]; 1♂ Pajarito Mtns., California Gulch, 30-Mar-2016, N. Grishin leg. (NVG-5997); 1♂ Patagonia, 24-Mar-1938, J. W. Tilden leg. (NVG-22104H09, CASENT8568339) [CAS]; 1♀ 3.5mi SW of Patagonia, 6-Aug-1978, Jim P. Brock leg. (NVG-2116); Mexico: Sonora: 1♂ 16 mi E of Tecoripa, 15-Mar-1984, Jim P. Brock leg. (NVG-2114); 1♀ 8 mi W of Rio Yaqui, 16-Mar-1984, Jim P. Brock leg. (NVG-2113); 1♂ ca. 8 mi NE of Bavispe, elevation ca. 4000’, 25-Mar-1998, Richard W. Holland leg. (NVG-20066B02, CSU_ENT1024701) [CSUC].
Type locality.
USA: Arizona, Pima Co., Santa Catalina Mountains, Sabino Canyon.
Etymology.
The name is formed from the type locality of this species, Sabino Canyon, a place familiar to many naturalists, which is just northeast of Tucson at the foothills of the Santa Catalina Mountains in southeastern Arizona, where this species is common. The name is a masculine adjective.
English name.
Arizona streaky-skipper.
Distribution.
Currently known from USA: Arizona and Mexico: Sonora and is likely present in southwestern New Mexico.
Comment.
We were surprised to learn that C. sabinus sp. n. is more distant from C. nessus than morphologically distinct C. spurcus, which is rather close to C. nessus genetically.
Charmion Nicéville, 1894 is a genus distinct from Celaenorrhinus Hübner, [1819]
Genomic tree reveals that Hesperia ficulnea Hewitson, 1868, the type species of the genus Charmion Nicéville, 1894 currently regarded as a junior subjective synonym of the genus Celaenorrhinus Hübner, [1819] (type species Papilio eligius Stoll, 1781) is not monophyletic with it (Fig. 44 red). We find that Bettonula Libert & Larsen, 2014 (type species Celaenorrhinus bettoni Butler, 1902) renders Celaenorrhinus paraphyletic. Therefore, to restore the monophyly, we propose that Charmion Nicéville, 1894, stat. rest. is a genus distinct from Celaenorrhinus Hübner, [1819].
Fig. 44.

Nuclear genome tree (autosomes): Charmion (red) and Celaenorrhinus (blue), Sape (magenta) and Sarangesa (green).
Sape Mabille, 1891 is a genus distinct from Sarangesa F. Moore, [1881]
Genomic tree reveals that Sape lucidella Mabille, 1891, the type species of the genus Sape Mabille, 1891 currently treated as a junior subjective synonym of the genus Sarangesa F. Moore, [1881] (type species Sarangesa albicilia F. Moore, [1881]) is not monophyletic with it (Fig. 44 magenta). We find that Eretis Mabille, 1891 (type species Eretis melania Mabille, 1891) renders Sarangesa paraphyletic. Therefore, to restore the monophyly, we propose that Sape Mabille, 1891, stat. rest. is a genus distinct from Sarangesa F. Moore, [1881].
Cupithina Grishin, new subtribe http://zoobank.org/905E8110-34EC-4CB3-A0C0-3F9002414D3E
Type genus.
Cupitha F. Moore, 1884.
Definition.
The tribe Astictopterini Swinhoe, 1912 splits into two clades containing approximately the same number of genera, each supported by 100% of partitions (see Fig. 1 in Zhang et al. (2023b) and Figs. 45, 46). Although neither clade is very prominent (branches separating them are not particularly long), both are most strongly supported. Therefore, to bring order to the genera in Astictopterini, we divide the tribe into two subtribes. The clade with the type species of the tribe contains generally larger species with more robust bodies and corresponds to the nominotypical subtribe. The second clade, with Cupitha (type species Cupitha tympanifera F. Moore, 1884, which is a junior subjective synonym of Pamphila purreea Moore, 1877), consists of smaller and frequently brighter patterned species and corresponds to the subtribe without an available name. This new and mostly Afrotropical subtribe is distinguished from its relatives by a combination of the following characters (Evans 1937; Evans 1949): palpi either porrect with convergent 3rd segment or erect with thin and erect 3rd segment; antennal club with pointed apiculus, either obtuse or hooked; caudal angle of hindwing discal cell not bent up: median vein and vein M3 collinear, end of cell relatively straight, and vein 3A usually shorter than vein CuA2; thorax not shaggy beneath; coxae and tibiae of hindlegs not densely fringed; valvae symmetrical, uncus typically narrowing to a point, ovate, terminal part narrow, frequently needle-like, uncus usually not expanded terminally and not hourglass-shaped. Most confidently identified by DNA and a combination of the following nuclear genomic base pairs is diagnostic: aly1775.3.2:A50G, aly27.16.1:A638C, aly27.16.1:C644G, aly1603.20.3:G76C, aly1651.28.7:A167G.
Fig. 45.

The phylogenetic tree of selected Ceratrichiini and Astictopterini inferred from protein-coding regions of the nuclear genome (autosomes). Levels in the tree that approximately correspond to the taxonomic hierarchy are marked above as Tribes, Subtribes, Genera, and Subgenera. Family-group names (roman font) and relevant genus-group names (genera in bold italics and subgenera in italics) are shown by corresponding branches. New species are in green, clades of new subgenera are in magenta, labels of taxa previously in Meza are in red, and others are colored as follows: elevated to species (blue), transferred between genera (cyan; green arrows show the transfer), and included into other genera as subgenera or synonyms (violet).
Fig. 46.

The phylogenetic tree of selected Ceratrichiini and Astictopterini inferred from protein-coding regions of the mitochondrial genome. See Fig. 45 for notations.
Genera included.
The type genus (i.e., Cupitha F. Moore, 1884), Acada Evans, 1937, Acleros Mabille, 1885, Andronymus Holland, 1896, Caenides Holland, 1896, Ceratricula Larsen, 2013, Fresna Evans, 1937, Gorgyra Holland, 1896, Gyrogra Lindsey & Miller, 1965, Hollandus Larsen & Collins, 2015, Hypoleucis Mabille, 1891, Melphina Evans, 1937, Melphinyet Larsen, 2012, Noctulana Larsen, 2012, Osmodes Holland, 1892, Osphantes Holland, 1896, Paracleros Berger, 1978 (see below), Paronymus Aurivillius, 1925, Parosmodes Holland, 1896, Platylesches Holland, 1896, Rhabdomantis Holland, 1896, Semalea Holland, 1896, Teniorhinus Holland, 1892, Xanthodisca Aurivillius, 1925 (see below), Xanthonymus Grishin, 2019, and Zographetus Watson, 1893. All other genera placed in Astictopterini by Zhang et al. (2023b) belong to the nominotypical subtribe Astictopterina.
Parent Taxon.
Tribe Astictopterini Swinhoe, 1912.
Paracleros Berger, 1978 is a subgenus of Acleros Mabille, 1885
Acleros Mabille, 1885 (type species Cyclopides leucopyga Mabille, 1877) and Paracleros Berger, 1978 (type species Acleros biguttulus Mabille, 1889) are two phenotypically similar sister genera, which, taken together, are prominently separated from others (Figs. 45, 46). COI barcode difference between their type species is 9.1% (60 bp), which is borderline for distinct genera in many butterfly groups. Moreover, Acleros, as currently circumscribed, is not a genetically prominent group and splits into two such groups in the nuclear genome tree (Fig. 45): the nominotypical Acleros and the other one, which is proposed as a new subgenus below. In the mitochondrial genome tree, Acleros is not even monophyletic, albeit with a weaker support (Fig. 46). On the one hand, we see that Acleros and Paracleros taken together form a prominent clade. On the other hand, Acleros and Paracleros are not prominently separated from each other, and there is a third clade of nearly the same rank. This clade is currently included in Acleros but is sister to Paracleros in the mitochondrial genome tree. Neither of these three groups consists of a large number of species. For all these reasons, we propose to treat Paracleros Berger, 1978, stat. nov. as a subgenus of Acleros Mabille, 1885.
Isocleros Grishin, new subgenus http://zoobank.org/1109E9E2-43A8-422D-BF56-967608087687
Type species.
Pamphila (?) mackenii Trimen, 1868.
Definition.
As discussed in the previous section, Acleros Mabille, 1885 (type species Cyclopides leucopyga Mabille, 1877) that includes Paracleros Berger, 1978 (type species Acleros biguttulus Mabille, 1889) splits into three prominent clades: the nominotypical Acleros, Paracleros, and the third clade not associated with any available genus-group names (Figs. 45, 46). The third clade represents a new subgenus that keys to 41.B.(a) (excluding a2) or 41.B.(b)(b1) in Evans (1937) and is distinguished from its relatives by darker distal half of ventral hindwing or if hindwing is more uniform in color then dorsal forewing is with broadly white inner margin; tegumen with uncus are narrow in lateral view, uncus in dorsal view is either bulb-shaped or continuously narrows to a point. A combination of the following nuclear genomic base pairs is diagnostic: aly2627.4.1:T63A, aly2627.4.1:C69T, aly116.29.1:T2049A, aly116.29.1:G2814A, aly9673.4.1:A480G.
Etymology.
The name of the other subgenus was formed by adding the prefix par- (i.e., besides, alongside) to the genus name. Similarly, we fuse the prefix iso- (i.e., similar, equal, the same) to the genus name. The name is a masculine noun in the nominative singular.
Species included.
The type species (i.e., Pamphila (?) mackenii Trimen, 1868), Acleros bibundica Strand, 1913, Acleros instabilis Mabille, 1889, Apaustus olaus Plötz, 1884, Acleros ploetzi Mabille, 1889, and a new species described below.
Parent taxon.
Genus Acleros Mabille, 1885.
Acleros (Isocleros) instabilis Mabille, 1889 and Acleros (Isocleros) olaus (Plötz, 1884) are species distinct from Acleros (Isocleros) mackenii (Trimen, 1868)
Inspection of genomic trees (Figs. 45, 46) and analysis of genetic differentiation reveals that Acleros instabilis Mabille, 1889 (type locality in Tanzania) and Apaustus olaus Plötz, 1884 (type locality in Congo, syntype sequenced as NVG-18073C07) currently treated as subspecies of Acleros (Isocleros) mackenii (Trimen, 1868), (type locality in South Africa, possible syntype sequenced as NVG-20126G11) are genetically differentiated from it with Fst/COI barcode difference of 0.23/4% (26 bp) and 0.49/2.6% (17 bp), respectively, and from each other with 0.39/4.7% (31 bp). Therefore, we propose to reinstate these two taxa as species: Acleros (Isocleros) instabilis Mabille, 1889, stat. rest. and Acleros (Isocleros) olaus (Plötz, 1884), stat. rest.
Acleros nyassicola Strand, 1921 is a junior subjective synonym of Acleros (Isocleros) olaus (Plötz, 1884), not of Acleros (Isocleros) mackenii (Trimen, 1868)
In the genomic trees, the holotype of Acleros nyassicola Strand, 1921 (type locality in Malawi, sequenced as NVG-20082G06, in SDEI) reveals that it is not conspecific with Acleros (Isocleros) mackenii (Trimen, 1868), (type locality in South Africa, possible syntype sequenced as NVG-20126G11) and instead groups with Acleros (Isocleros) olaus (Plötz, 1884), stat. rest. (type locality in Congo, syntype sequenced as NVG-18073C07) (Figs. 45, 46). Therefore, we propose to treat Acleros nyassicola Strand, 1921 as a junior subjective synonym of Acleros (Isocleros) olaus (Plötz, 1884).
Acleros (Isocleros) togo Grishin, new species http://zoobank.org/10245D25-D8E1-4342-A2AA-0104A185EA97 (Figs. 45–46 part, 47)
Fig. 47.
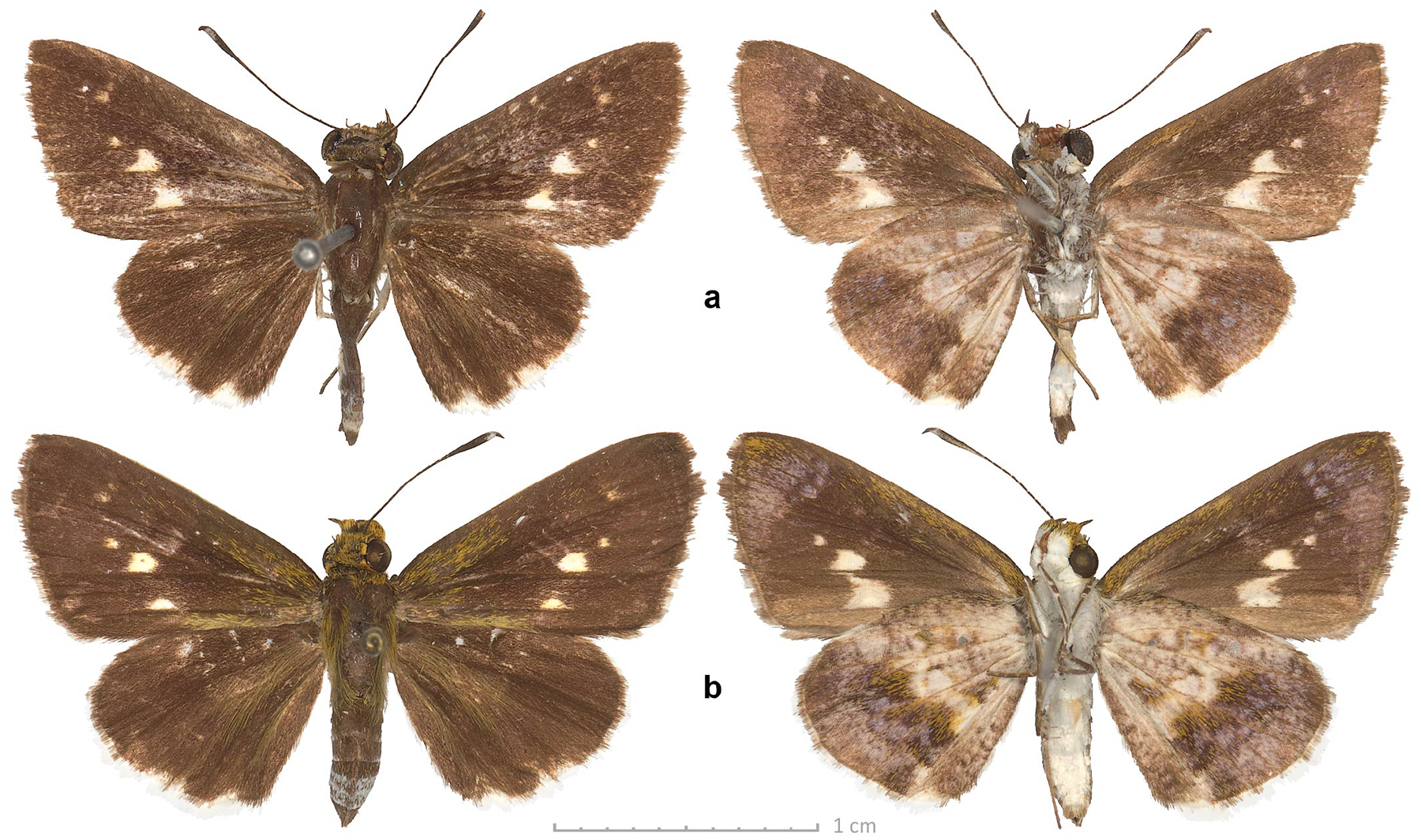
Holotype ♂ (a) and paratype ♀ (b) of Acleros togo sp. n. in dorsal (left) and ventral (right) views, data in text.
Definition and diagnosis.
Genomic trees reveal that two specimens from Western Africa identified as Acleros olaus (Plötz, 1884), stat. rest. (type locality in Congo, syntype sequenced as NVG-18073C07) are not monophyletic with A. olaus and form a clade approximately equidistant from it and Acleros mackenii (Trimen, 1868) (type locality in South Africa, possible syntype sequenced as NVG-20126G11) with Acleros instabilis Mabille, 1889, stat. rest. (type locality in Tanzania) (Figs. 45, 46), suggesting that these two specimens belong to a distinct species. Its genetic differentiation measured by Fst/COI barcode difference is: 0.31/5.5% (36 bp) from A. olaus, 0.31/4.7% (31 bp) from A. mackenii, and 0.41/5.3% (35 bp from A. instabilis, which is substantial and provides strong support for these two specimens as representatives of a new species. This species is distinguished from its relatives by the following combination of characters: forewing subapical spots are present, yellowish above and purplish beneath (not white in the specimens of the type series), diffuse, nearly in line (spot by costa is not strongly offset distad from others), other forewing spots are crisper, with sharper defined edges, the spot in cell CuA2-1A+2A on ventral forewing is with equally sharp edges (not more diffuse and disappearing towards the outer margin), forewing spot in cell CuA1-CuA2 is closer to the spot in cell CuA2-1A+2A than in A. olaus. Due to variability in phenotype, this rather cryptic species is confidently identified by DNA: in the nuclear genome: aly164.2.2:C73A, aly173.7.6:A182T, aly8937.5.1:A305G, aly1260.26.1:A247G, aly24.4.2:G63T and in the COI barcode: T38A, T91C, T376A, G380A, A514C, T571C.
Barcode sequence of the holotype: Sample NVG-22017G12, GenBank OR589638, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGTATAATAGGATCATCTTTAAGATTATTAATTCGTACAGAATTAGGTAACCCTGGATCCTTAATCGGAGATGATCAAATTTATAATACAATTGTTACAGCTCATGCTTTTATTATAATTTTTTTTATAGTAATACCTATTATAATTGGAGGATTTGGTAATTGATTAGTTCCTCTTATATTAGGAGCTCCTGATATAGCTTTCCCCCGAATAAATAATATAAGATTTTGAATACTTCCCCCATCTTTAACATTATTAATTTCAAGAAGAATTGTAGAAAATGGTGCTGGAACTGGCTGAACTGTTTACCCCCCTCTTTCTTCTAATATTGCACATCAAGGATCATCAATTGATTTAGCTATTTTTTCTTTACATTTAGCCGGAATTTCATCTATTTTAGGAGCTATTAATTTTATTACAACTATTATTAATATACGAATTAAAAATATATCATTTGATCAATTACCTTTATTTATTTGATCCGTAGGTATTACTGCTTTACTTTTACTTTTATCTCTACCTGTTTTAGCAGGTGCTATCACAATACTTCTTACTGACCGAAACTTAAATACTTCTTTTTTCGATCCTGCCGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the Zoologische Staatssammlung München, Germany [ZSMC], illustrated in Fig. 47a, bears six labels: 2nd blue, last red, and others white [43390], [Togo | Misahöhe | 1893 | E. Baumann S.], [363 | 7.XII.93 | A. olaus], [olaus | Plötz], [DNA sample ID: | NVG-22017G12 | c/o Nick V. Grishin], and [HOLOTYPE ♂ | Acleros (Isocleros) | togo Grishin]. Paratype: ♀ Liberia: Monrovia, Firestone Plantation, Dec-1946, Harry A. Beatty leg. (NVG-22047H11) [CUIC] (Fig. 47b).
Type locality.
Togo: Misahöhe.
Etymology.
The name is given for the country with the type locality. The name is a noun in apposition.
Distribution.
Western Africa, recorded from Togo and Liberia.
Taxonomic rearrangement of species currently in the genus Meza Hemming, 1939
The genus Meza Hemming, 1939 (type species Hesperia meza Hewitson, 1877) currently consists of 10 species. Genomic trees reveal that the genus is not monophyletic (Figs. 45, 46) and partitions according to the three sections of the identification key provided by Evans (1937): (i) no secondary sexual characters; (ii) male dorsal hindwing with a hair tuft that overlays bases of veins CuA1 and CuA2 and (iii) hair tuft enters a pouch either at the end of discal cell or along the median vein. The type species M. meza—section (i), no tuft—belongs to the tribe Ceratrichiini Grishin, 2019 and other species (with tuft) belong to two clades in the tribe Astictopterini Swinhoe, 1912. Section (ii) consists of three species and is sister to the genus Fresna Evans, 1937 (type species Hesperia netopha Hewitson, 1878). Although this clade is prominent, it is at the tree level of a subgenus (Figs. 45, 46). We include these species in Fresna, forming the following new combinations: Fresna larea (Neave, 1910), comb. nov., Fresna leucophaea (Holland, 1894), comb. nov., and Fresna mabea (Holland, 1894), comb. nov. All the remaining species are from section (iii) and are dispersed within the clade corresponding to the genus Paronymus Aurivillius, 1925 (type species Hesperia ligora Hewitson, 1876) (Figs. 45, 46). We assign them to this genus, forming new combinations: Paronymus banda (Evans, 1937), comb. nov., Paronymus cybeutes (Holland, 1894), comb. nov., Paronymus elba (Evans, 1937), comb. nov., Paronymus gardineri (Collins & Larsen, 2008), comb. nov., Paronymus indusiate (Mabille, 1891), comb. nov., Paronymus mabillei (Holland, 1893), comb. nov. As a result, Meza becomes monotypic to consist of only the type species M. meza, while other species currently in Meza are transferred to Fresna or Paronymus.
Mesna Grishin, new subgenus http://zoobank.org/A3DDA72C-3E34-42E8-82FF-457715B8E65E
Type species.
Parnara leucophaea Holland, 1894.
Definition.
As discussed above, Meza Hemming, 1939 (type species Hesperia meza Hewitson, 1877) was not monophyletic, and some of its species formed a clade sister to Fresna Evans, 1937 (type species Hesperia netopha Hewitson, 1878) (Figs. 45, 46). To restore the monophyly, we placed these species in Fresna. However, they are genetically and morphologically distinct from other Fresna and constitute a new subgenus. Species of this new subgenus key to 44.B. in Evans (1937) and are distinguished from similar-looking species (e.g., Meza and Paronymus) by the following characters: male dorsal hindwing with a hair tuft that overlays bases of veins CuA1 and CuA2 (does not enter a pouch either at the end of discal cell or along the median vein as in Paronymus; Meza lacks the tuft) and uncus narrowly squared at the tip in dorsal view, not terminating in a sharp point (as in Paronymus), and not bulb-shaped (as in Meza). A combination of the following nuclear genomic base pairs is diagnostic: aly1249.14.7:T1966C, aly1249.14.7:C2004T, aly536.106.2:A3984G, aly1656.5.1:A135C, aly8937.7.1:A198C.
Etymology.
The name indicates that these species started in the genus Me[za] and ended in [Fre]sna. The name is a feminine noun in the nominative singular.
Species included.
The type species (i.e., Parnara leucophaea Holland, 1894), Meza leucophaea bassa Lindsey & L. Miller, 1965 (see below), Parnara larea Neave, 1910, and Parnara mabea Holland, 1894.
Parent taxon.
Genus Fresna Evans, 1937.
Fresna (Mesna) bassa (Lindsey & L. Miller, 1965) is a species distinct from Fresna (Mesna) leucophaea (Holland, 1894)
Genetic differentiation between Meza leucophaea bassa Lindsey & L. Miller, 1965 (type locality in Liberia, holotype sequenced as NVG-20124H07) that we placed in the genus Fresna Evans, 1937 (type species Hesperia netopha Hewitson, 1878) and Fresna leucophaea leucophaea (Holland, 1894) (type locality in Gabon, syntypes sequenced as NVG-21099F07 and NVG-21099F08) is notable, e.g., COI barcode differ by 3.3% (22 bp) and the two taxa form separate clades in genomic trees (Figs. 45, 46). Therefore, we propose that Fresna (Mesna) bassa (Lindsey & L. Miller, 1965), stat. nov. is a species distinct from Fresna (Mesna) leucophaea (Holland, 1894).
Paronymus volta (L. Miller, 1971) is a species distinct from Paronymus cybeutes (Holland, 1894)
Genetic differentiation as measured by Fst/COI barcode difference between Meza cybeutes volta Miller, 1971 (type locality in Ghana, holotype sequenced as NVG-21043D12) that we placed in the genus Paronymus Aurivillius, 1925 (type species Hesperia ligora Hewitson, 1876) and Paronymus leucophaea leucophaea (Holland, 1894) (type locality in Gabon, syntype sequenced as NVG-21099G06) is: 0.43/3.8% (25 bp) and the two taxa form separate clades in genomic trees (Figs. 45, 46). Therefore, we propose that Paronymus volta (L. Miller, 1971), stat. nov. is a species distinct from Paronymus cybeutes (Holland, 1894). Paronymus cybeutes pallida (Holland, 1896) (type locality in Congo) does not differ prominently from the nominotypical subspecies, and we retain it as a subspecies of P. cybeutes.
Ceratricula Larsen, 2013 is a junior subjective synonym of Paronymus Aurivillius, 1925
The genus Ceratricula Larsen, 2013 (type species Ceratrichia semilutea Mabille, 1891) forms a tight clade within Paronymus Aurivillius, 1925 (type species Hesperia ligora Hewitson, 1876) that is a confident sister to Paronymus xanthias (Mabille, 1891) (type locality in Nigeria) (Figs. 45, 46) rendering Paronymus paraphyletic. To restore the monophyly, we propose that Ceratricula Larsen, 2013, syn. nov. is a junior subjective synonym of Paronymus Aurivillius, 1925.
Paronymus punctata (Holland, 1896), new combination; its type locality is Sierra Leone: Freetown
The name Ceratrichia punctata (type locality in “Tropical West Africa”) was published by Holland (1896) in synonymy with Ceratrichia phocion (Fabricius, 1781) (type locality in South Africa) and attributed to Mabille as a “MS. name”, with the holotype specimen (“another male … has been designated as the type”) from the Staudinger collection, now in MFNB. A short description was provided: “primaries more spotted than is quite usual.” Evans (1937) used the name “Ceratrichia punctata Holland” as valid, attributing it to Holland’s publication and citing the same “type” specimen in Berlin. Therefore, under Articles 11.6.1 and 50.7 of the ICZN Code (1999), the name Ceratrichia punctata Holland 1896 is currently both available and valid and is based on the holotype in Berlin. N.V.G. found the holotype, which bears a purple “Origin.” label, an identification label in Mabille’s handwriting, “C. punctata / Mab. ♂”, and an identification label in Holland’s handwriting, “Ceratrichia / phocion, / ♂”, among several others. The locality label on the holotype is “Frtn. / Pr.” meaning that it was collected by Preuss in Freetown, Serra Leone. German botanist Paul Rudolph Preuss collected in Sierra Leone in 1886–1888 (Barnhart 1965), giving a possible range of dates when the holotype was captured.
Genomic sequencing of the holotype (NVG-18073A05) places it as sister to Paronymus semilutea (Mabille, 1891) (type locality in Nigeria, syntype sequenced as NVG-21118G05) (Figs. 45, 46), being distinct from it due to genetic differentiation, e.g., COI barcode difference of 2.1% (14 bp). Therefore, we propose Paronymus punctata (Holland, 1896), comb. nov. The holotype of P. punctata is not conspecific with the species Evans (1937) identified as C. punctata: e.g., the holotype has ventral forewing nearly uniformly brown without pale areas by the inner margin characteristic of Evans’ C. punctata; smaller forewing and ventral hindwing spots, and brownish tuft of hair-like scales by the base of dorsal hindwing. Evans’ C. punctata lacks androconia and is a species of Ceratrichia Butler, 1870 (type species Papilio nothus Fabricius, 1787); it does not have a name. This new species is described below.
Ceratrichia notata Grishin, new species http://zoobank.org/3BD674EC-B100-4309-8181-393A29D4EE03
Definition and diagnosis.
As shown above, Evans misidentified Paronymus punctata Holland, 1896, comb. nov. The distinctive species he considered “Ceratrichia punctata” does not have a name and is new. This new species keys to 31.A.(b)(b2)(a3) in Evans (1937) and is identified by a combination of brown dorsal forewing with white dots, single (upper) dot in discal cell, ventrally with pale-yellow area by inner margin; most of dorsal hindwing is yellow with costal third brown, ventrally pale yellow, not white, broadly brown at the outer margin and with submarginal dots encircled with brown, such dot in the discal cell, and (frequently replaced with brown spots) 3–4 additional dots around.
Type material.
Holotype: ♂ deposited in the African Butterfly Research Institute, Nairobi, Kenya [ABRI], collected in Central African Republic: Zomea, Oct-1982, ABRI-2019–2412, and its photographs can be found in Williams (2023a), as the male “Ceratrichia punctata” (misidentification). Paratypes: 2♂♂ 2♀♀: 1♀ with the same data as the holotype but Sep-1996 and ABRI-2019–2413; others in BMNH: 1♂ 1♀ from Cameroon and 1♂ from Angola.
Type locality.
Central African Republic: Zomea.
Etymology.
This species was previously misidentified as “punctata”, which means “dotted” in Latin. A Latin synonym of “punctata” is “notata”, which is adopted as the name of this species. The name “notata” can also be translated as “noted”, and this species is noted for its larger white dots and bright colors and for the confusion about its name due to misidentification that genomic analysis of primary type specimens helped to resolve. The name is a feminine adjective.
Distribution.
Western Africa, recorded from Cameroon, Central African Republic, and Angola.
Paronymus indeterminabilis (Strand, 1912) and Paronymus congdoni (Larsen, 2013) are species distinct from Paronymus semilutea (Mabille, 1891)
Genomic trees reveal that sister taxa Ceratrichia indeterminabilis Strand, 1912 (type locality Equatorial Guinea [not Cameroon!]: Benito [river] area, Monte Alen, syntypes sequenced as NVG-18073A07 and NVG-18073A08) and Ceratricula semilutea congdoni Larsen, 2013 (type locality in Uganda) prior to this publication treated as subspecies of Ceratricula semilutea (Mabille, 1891) (type locality Nigeria: Lagos, syntype sequenced as NVG-21118G05) that we placed in Paronymus Aurivillius, 1925 (type species Hesperia ligora Hewitson, 1876) (see above), are not monophyletic with C. semilutea (Figs. 45, 46) and are well differentiated from it genetically with COI barcode difference of 3.0% (20 bp) and 3.6% (24 bp), respectively, and from each other of 3.3% (22 bp). Therefore, we propose to treat them as species-level taxa: Paronymus indeterminabilis (Strand, 1912), stat. rest. and Paronymus congdoni (Larsen, 2013), stat. nov.
Semalea corvinus (Mabille, 1890) is a species distinct from Semalea sextilis (Plötz, 1886)
Genomic sequencing of Cobalus corvinus Mabille, 1890 (type locality in Sierra Leone, type sequenced as NVG-18075A05) currently treated as a junior subjective synonym of Semalea sextilis (Plötz, 1886) (type locality in Ghana) reveals that it is not monophyletic with it and instead originates deep in the radiation of Semalea Holland, 1896 (type species Hesperia pulvina Plötz, 1879) (Figs. 45, 46). Therefore, we reinstate Semalea corvinus (Mabille, 1890), stat. rest. as a species-level taxon.
Xanthodisca Aurivillius, 1925 is a junior subjective synonym of Semalea Holland, 1896
Genomic trees reveal that Semalea Holland, 1896 (type species Hesperia pulvina Plötz, 1879) in paraphyletic with respect to Xanthodisca Aurivillius, 1925 (type species Astictopterus vibius Hewitson, 1878) (Figs. 45, 46). Restoring monophyly, we propose that Xanthodisca Aurivillius, 1925, syn. nov. is a junior subjective synonym of Semalea Holland, 1896 due to genetic similarity.
Semalea malawi Grishin, new species http://zoobank.org/A1AA303B-9ABF-4F7C-B84B-2ABD765583DF (Figs. 45–46 part, 48)
Fig. 48.

Holotype of Semalea malawi sp. n. in dorsal (left) and ventral (right) views, data in text.
Definition and diagnosis.
The mitochondrial gaenome tree reveals that a specimen from Malawi identified as Semalea vibius (Hewitson, 1878), comb. nov. (type locality in Gabon) is sister to both S. vibius and Semalea rega (Mabille, 1889), comb. nov. (type locality in Sierra Leone) (Fig. 46), suggesting that it is a third species distinct from them. We sequenced primary type specimens of all four names associated with S. rega and confirmed their synonymy (Figs. 45, 46). Specimens from western Africa (Cameroon, Congo) serve as references for S. vibius. Therefore, the third species is new. In the COI barcode, it differs from S. vibius by 1.1% (7 bp), which is larger than the difference between S. vibius and S. rega of 0.6% (4 bp). Despite this moderate difference in the barcode, Fst/Gmin of 0.35/0.008 for S. vibius and S. rega indicate that they are distinct species, in agreement with phenotypic distinction. However, we could not compute these statistics for the new species because it is known from a single specimen (at least two specimens are needed). This new species is distinguished from its relatives by the lack of subapical spots on the forewing, larger forewing orange patch that reaches closer to the outer margin, palpi beneath and cheeks more orange than yellow in color, and largely brown ventral hindwing, which is unspotted, and sparsely overscaled with orange. Due to variability in phenotype, confidently identified by DNA: in the nuclear genome: aly281.6.2:A130G, aly281.6.2:C131T, aly1249.8.1:T514C, aly1603.31.1:A370C, aly37338.23.1:C264T, aly54.4.1:T1461T (not A), aly6648.1.2:A161A (not C), aly5294.20.2:T630T (not C), aly386.8.2:A506A (not T), aly393.3.1:A492A (not G) and in the COI barcode: T88C, T118C, T139A, T202C, T547T, T610T, T646T.
Barcode sequence of the holotype: Sample NVG-19043B12, GenBank OR589640, 658 base pairs:
AACTTTATATTTTATTTTTGGTATTTGAGCAGGTATATTAGGAACATCTTTAAGTTTATTAATTCGAACTGAATTAGGTAATCCTGGCTCATTAATTGGAGATGATCAAATTTATAACACAATTGTAACAGCTCATGCATTTATTATAATTTTTTTTATAGTTATACCTATTATAATTGGAGGATTTGGAAATTGATTAGTCCCTTTAATATTAGGAGCTCCTGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGATTACTTCCCCCCTCCCTTACCTTATTAATCTCAAGAAGAATTGTAGAAAATGGAGCTGGAACTGGATGAACTGTCTATCCCCCCCTTTCATCTAATATTGCTCACCAAGGTTCTTCTGTTGATTTAGCAATCTTTTCTTTACATTTAGCAGGAATTTCTTCTATTTTAGGAGCTATTAATTTTATTACTACAATTATTAATATACGAATTAAAAATTTATCTTTTGATCAACTACCTTTATTTGTTTGATCTGTTGGTATTACTGCTTTACTTCTTCTTCTTTCTTTACCTGTTTTAGCTGGAGCTATTACAATATTATTAACTGATCGTAATCTTAATACTTCTTTTTTTGACCCTGCTGGAGGAGGAGACCCTATTCTTTATCAACATTTATTT
Type material.
Holotype: ♂ deposited in the American Museum of Natural History, New York, NY, USA [AMNH], illustrated in Fig. 48, bears five labels: four white [11-viii-40. | ♂ | Vizara | 2600’. | Nyasaland | R. C. Wood], [vibius | vibius | ♂], [DNA sample ID: | NVG-19043B12 | c/o Nick V. Grishin], [{QR Code} | AMNH_IZC 00337937], and one red [HOLOTYPE ♂ | Semalea malawi | Grishin].
Type locality.
Malawi: ca. 9 mi E of Nkhata Bay, Vizara Rubber Plantation, elevation 2600’.
Etymology.
The name is given for the country with the type locality. The name is a noun in apposition.
Distribution.
Currently known only from the holotype collected in Malawi.
The type locality of Osmodes staudingeri Holland, 1896 is Cameroon: Efoulan
The type locality of Osmodes staudingeri Holland, 1896, currently treated as a junior subjective synonym of Semalea rega (Mabille, 1889), comb. nov. (type locality in Sierra Leone) was given in the original description as “Valley of the Ogové” (Holland 1896), which is in Gabon, frequently spelled as Ogooué River Valley. This species was described from the female holotype in Holland’s collection, which is in CMNH: “Type [not types] in my collection” and “I do not know the male of this species. The solitary female in my collection …,” which was illustrated (Holland 1896). N.V.G. found a female in CMNH with the following five labels, white, but the 4th (without any text) red: [Efulen, | Kamerun, | A. I. Good | C. M. Acc. 4454], [Osmodes | staudingeri | ♀ type Holl.], [unknown | to Mabille], [], [DNA sample ID: | NVG-20125A08 | c/o Nick V. Grishin]. The female matches the original description perfectly and agrees with the illustration rather closely. Provided that it carries its identification label (2nd label) in Holland’s handwriting, it is the holotype of O. staudingeri. However, the locality label does not mention “Valley of the Ogové,” but points to Cameroon: Efoulan (in current spelling). This is a more likely locality because this species is not otherwise known from Gabon but recorded from Cameroon (Williams 2023b). Therefore, we hypothesize that the locality in the original description is erroneous, and the locality label on the holotype points to its provenance. Hence, the type locality of O. staudingeri is Cameroon: Efoulan, and genomic sequencing of the holotype confirms this taxon as a junior subjective synonym of S. rega (syntype in MFNB sequenced as NVG-18073A03) (Figs. 45, 46).
Xanthoneura patmapana (Fruhstorfer, 1911) is a species distinct from Xanthoneura corissa (Hewitson, 1876)
Two phenotypically different subspecies, Xanthoneura corissa corissa (Hewitson, 1876) (type locality in Kalimantan) and Xanthoneura corissa patmapana (Fruhstorfer, 1911) (type locality in Java), are genetically differentiated (Figs. 45, 46) and differ by 4.1% (27 bp) in their barcodes. Therefore, we propose treating Xanthoneura patmapana (Fruhstorfer, 1911), stat. nov. as a species distinct from Xanthoneura corissa (Hewitson, 1876).
Lippina Grishin, new subgenus http://zoobank.org/22B4014B-4B5D-45D4-8283-6C65583887B7
Type species.
Carystus telesinus Mabille, 1878.
Definition.
Genomic trees reveal that while Xanthoneura Eliot, 1978 (type species Hesperia corissa Hewitson, 1876) is monophyletic (and we keep it as a single genus), it deeply splits into two clades: the nominotypical and unnamed that represents a new subgenus. Members of this new subgenus key to J.10.23. in Evans (1949) and are distinguished from their relatives by the 3rd segment of palpi stout and bent forward, ventral hindwing largely unmarked (or with a central spot), veins not yellower, forewing with a spot in cell M2-M3 right above the spot in cell M3-CuA1, these two spots together give an appearance of a single larger spot; uncus in dorsal view much narrower than long, narrowing in the middle, divided, arms small knob-like, not widely separated. A combination of the following nuclear genomic base pairs is diagnostic: aly577.16.2:T174C, aly577.16.2:T198A, aly577.16.2:A216G, aly1591.7.3:A333C, aly1222.40.2:A15G.
Etymology.
The species of this subgenus are from [Phi]lippin[es]+a. The name is a feminine noun in the nominative singular.
Species included.
The type species (i.e., Carystus telesinus Mabille, 1878) and Xanthoneura obscurior de Jong & Treadaway, 2007.
Parent taxon.
Genus Xanthoneura Eliot, 1978.
Perrotia Oberthür, 1916 is a junior subjective synonym of Galerga Mabille, 1898
Genomic trees reveal a tight clade of Malagasy species sister to a Malagasy genus Fulda Evans, 1937 (type species Hesperia coroller Boisduval, 1833) (Fig. 45, 46). Currently, these species are placed in two genera: Perrotia Oberthür, 1916 (type species Perrotia albiplaga Oberthür, 1916) and Galerga Mabille, 1898 (type species Galerga hyposticta Mabille, 1898). However, the two genera are genetically close, e.g., the COI barcode difference between their type species is 7.3% (48 bp), and not distinctly differentiated from each other. Therefore, we propose to treat Perrotia Oberthür, 1916, syn. nov. is a junior subjective synonym of Galerga Mabille, 1898.
Galerga ariel (Mabille, 1878), new combination
Currently in Xanthodisca Aurivillius, 1925 (type species Astictopterus vibius Hewitson, 1878), Pamphila ariel Mabille, 1878 (type locality in Madagascar) is not monophyletic with it and instead originates deep within Galerga Mabille, 1898 (type species Galerga hyposticta Mabille, 1898) (Figs. 45, 46), where we place it as Galerga ariel (Mabille, 1878), comb. nov.
Afrotropical species of Astictopterus C. Felder & R. Felder, 1890 belong to Isoteinon C. Felder & R. Felder, 1862
Genomic trees reveal that Afrotropical species currently placed in Astictopterus C. Felder & R. Felder, 1860 (type species Astictopterus jama C. Felder & R. Felder, 1860) are not monophyletic with it and instead originate within Isoteinon C. Felder & R. Felder, 1862 (type species Isoteinon lamprospilus C. Felder & R. Felder, 1862) (Fig. 45, 46). Therefore, to restore monophyly, we transfer these species from Astictopterus to Isoteinon, forming the following new combinations: Isoteinon anomoeus (Plötz, 1879), comb. nov. (type locality in Ghana), Isoteinon bruno (Evans, 1937), comb. nov. (type locality in Tanzania), Isoteinon inornatus (Trimen, 1864), comb. nov. (type locality in South Africa), and Isoteinon punctulata (A. Butler, 1895), comb. nov. (type locality in Tanzania).
Mopala Evans, 1937 is a subgenus of Leona Evans, 1937
The monotypic genus Mopala Evans, 1937 (type species Ismene ? orma Plötz, 1879), which stands out by the large white patch on ventral hindwing and otherwise unspotted wings, is genetically close to Leona Evans, 1937 (type species Hesperia leonora Plötz, 1879) in our genomic trees (Fig. 45, 46), while they both are distant from other genera that are their closest relatives. Male genitalia in both genera are similar in the shape of uncus and valva, and all these species could be accommodated within one genus. Because Mopala and Leona were described in the same work published on the same date, as the first revisers, we give priority to Leona due to it being a larger genus, and propose that Mopala Evans, 1937 stat. nov. is a subgenus of Leona Evans, 1937.
Ganda Grishin, new subgenus http://zoobank.org/9A4C7BA6-2C42-47E6-B619-E54A20FF850E
Type species.
Zophopetes ganda Evans, 1937.
Definition.
Described by Evans (1937) in the genus Zophopetes Mabille, 1904 (type species Pamphila dysmephila Trimen, 1868) and kept in it since, Z. ganda (type locality in Ivory Coast) is not monophyletic with Zophopetes and instead is in the same clade with Leona Evans, 1937 (type species Hesperia leonora Plötz, 1879) and its subgenus Mopala Evans, 1937 (type species Ismene ? orma Plötz, 1879): sister to the subgenus Leona in the nuclear genome tree (Fig. 45) and sister to both Leona and Mopala, but less confidently, in the mitochondrial genome tree (Fig. 46). In either case, it is closely related to them both and yet genetically differentiated from them at approximately the same level as they are from each other. Therefore, similarly to Mopala, the lineage with Z. ganda represents a subgenus of Leona. This new subgenus keys to 53.A. in Evans (1937) and is distinguished from its relatives by males with a brand (from the base of vein CuA1 to near vein 1A+2A) and an apical spot on dorsal forewing (both absent in Zophopetes), ventral hindwing uniformly pale-brown with small dark-brown-framed spots (not like Leona and Mopala); upturned harpe, uncus in dorsal view narrower than in Zophopetes, especially in the middle, with smaller knob-shaped arms. A combination of the following nuclear genomic base pairs is diagnostic: aly1038.19.1:T165C, aly1405.10.1:T456C, aly925.11.3:A69G, aly587.17.1:T205A, aly2127.3.3:A24G, aly1838.61.1:T618T (not C), aly1838.61.1:T675T (not A), aly4305.26.4:C31C (not G), aly27.16.1:C354C (not A), aly614.16.1:A1098A (not G).
Etymology.
The name is a feminine noun in the nominative singular, a tautonym of the type species name.
Species included.
Only the type species.
Parent taxon.
Genus Leona Evans, 1937.
Balenga Grishin, new genus http://zoobank.org/21C6F5FF-4E85-4D88-AD3B-D010B1F11F7B
Type species.
Proteides balenge Holland, 1891.
Definition.
Currently, Gretnini Grishin, 2019 is a monotypic tribe consisting of a single genus Gretna Evans, 1937 (type species Hesperia cylinda Hewitson, 1876). Inspection of genomic trees reveals that Grenta balenge (Holland, 1891) (type locality in Gabon) is strongly differentiated genetically from the rest of the genus (e.g., COI barcode difference of 10.9%, 72 bp), at least to the same extent as Isma Distant, 1886 (type species Isma obscura Distant, 1886) from Iambrix Watson, 1893 (type species Nisoniades salsala F. Moore, 1866) or Hyarotis F. Moore, 1881 (type species Papilio adrastus Stoll, 1780) from Quedara Swinhoe, 1919 (type species Quedara comoplea Swinhoe, 1919) and is at the tree level corresponding to genera. Therefore, the lineage with G. balenge represents a genus-level taxon (Fig. 49). This new genus keys to 57.B. in Evans (1937) and is distinguished from its relatives by the subapical forewing spot in cell R5-M1 being offset distad from others, uncus with processes at its base directed sideways (one on each side, no processes in Gretna), and deeper separation between harpe and ampulla that is more expanded than in Gretna. A combination of the following COI barcode base pairs is diagnostic: T206C, A494T, A520C, A562C, A586T.
Fig. 49.

Phylogenetic trees of selected Gretnini species and ingroups inferred from protein-coding regions of a) the nuclear (autosomes) and b) the mitochondrial genomes. Different genera are shown in different colors: Gretna (blue with subgenus Zarida subgen. n. in violet and Gretna capra stat. nov. with Gretna carmen in orange), Balenga gen. n., Isma (cyan), Idmon (magenta), Iambrix (green), Hyarotis (olive), and Quedara (brown).
Etymology.
The name is a feminine noun in the nominative singular, formed from the type species name.
Species included.
Only the type species.
Parent taxon.
Tribe Gretnini Grishin, 2019.
Zarida Grishin, new subgenus http://zoobank.org/699B2792-5716-45FF-B4D9-477760251BD8
Type species.
Hesperia lacida Hewitson, 1876.
Definition.
Within a smaller genus Gretna Evans, 1937 (type species Hesperia cylinda Hewitson, 1876) due to removal of Balenga balenge Holland, 1891, comb. nov. (type locality in Gabon), we note the genetic differentiation of a clade sister to that with the type species (Fig. 49). This clade diverges from other Gretna at least at the tree level corresponding to subgenera (if not genera), i.e., approximately the same as Isma Distant, 1886 (type species Isma obscura Distant, 1886) from Idmon Nicéville, 1895 (type species Baoris unicolor Distant, 1886, a junior homonym, available name for this species is Iambrix distanti Shepard, 1937) (Fig. 49), and we regard it as representing a subgenus. This new subgenus keys to 53.A.(a)(b1) in Evans (1937) and is distinguished from its relatives by an undivided and much narrower uncus, almost spike-shaped in dorsal view; better developed (and hyaline) spot in the middle of dorsal hindwing, prominent spot in forewing cell CuA2-1A+2A, and clearly defined whitish bands or area on ventral hindwing. A combination of the following COI barcode base pairs is diagnostic: T56C, T154C, T379A, T553A, T556A.
Etymology.
The name is a feminine noun in the nominative singular and is a fusion of species names in this subgenus: Zar[emba]+[lac]ida.
Species included.
The type species (i.e., Hesperia lacida Hewitson, 1876) and Telesto zaremba Plötz, 1884.
Parent taxon.
Genus Gretna Evans, 1937.
Gretna capra Evans, 1937 is a species distinct from Gretna carmen Evans, 1937
Originally proposed as a subspecies, Gretna carmen capra Evans, 1937 (type locality in Kenya) is not monophyletic with Gretna carmen Evans, 1937 (type locality in Cameroon) in genomic trees (Fig. 49) and is strongly differentiated from it genetically, e.g., COI barcode difference of 3.6% (24 bp). Therefore, we propose that Gretna capra Evans, 1937, stat. nov. is a species distinct from Gretna carmen Evans, 1937, as hinted by Larsen (1991).
Milena Evans, 1912 is a genus distinct from Caltoris Swinhoe, 1893
Genomic trees reveal that Caltoris Swinhoe, 1893 (type species Hesperia kumara F. Moore, 1878) as currently circumscribed is not monophyletic, and phylogenetic positions of some species differ between trees not being strongly supported in any of them (Fig. 50). One of such lineages is Caltoris plebeia (Nicéville, 1887) (type locality in India): it is sister to the clade of Caltoris with its type species in the Z chromosome tree (with weak support: 46% partitions), and sister to Prusiana Evans, 1937 (type species Pamphila prusias C. Felder, 1861) in the nuclear genome tree from autosomes (58% of partitions) and mitogenome (70% of partitions), indicating complex evolutionary history of this lineage. Recognizing the uniqueness of C. plebeia, it has been chosen as the type species of the available genus-group name Milena Evans, 1912, which is currently regarded as a synonym of Caltoris. Presently, we have decided to keep Prusiana as a genus (instead of placing it as a subgenus in Caltoris) due to its genetic differentiation and phenotypic differences. Therefore, to ensure monophyly of all the taxa, we propose that Milena Evans, 1912, stat. rest. is a genus distinct from Caltoris Swinhoe, 1893.
Fig. 50.

Phylogenetic trees of selected Baorini species inferred from protein-coding regions of a, b) the nuclear genome (autosomes), b) proportionally rescaled to bring leaves to the same level, c) the Z chromosome, and d) the mitochondrial genome. Parnara is genetically removed from the rest, and the corresponding tree branches were truncated (as indicated by dots) to save space. The translucent vertical lime bar in (b) denotes the level approximately corresponding to genera. Shown by branches they refer to, names of genera are in bold italics, and subgenera are in italics: Gegenes (blue with Havea subgen. n. red and Flanga subgen. n. aquamarine), Borbo (olive), Baoris (gray), Tulsia gen. n. (magenta), Caltoris (violet), Milena stat. rest. (bright green), Prusiana (brown), Zenonia (cyan with subgenus Zenonoida stat. nov. orange), and Zinaida (purple).
Tulsia Grishin, new genus http://zoobank.org/52B76DE3-029E-4623-B87F-578013E5C53F
Type species.
Parnara tulsi Nicéville, 1884.
Definition.
Genomic trees reveal that the lineage of Caltoris tulsi (Nicéville, 1884) (type locality in India) originates in rapid radiation and is not strongly grouped with any other single genus. Complemented with its strong genetic differentiation from others (Fig. 50), it represents a genus-level taxon. This new genus keys to M.7.12. in Evans (1949) and is distinguished from its relatives by forewing without tuft of scales beneath and spots in discal cell, ventral hindwing in its basal half and ventral forewing along costal margin overscaled with pale purplish (not ocherous) scales; uncus rounded at its distal end, only slightly knobbed on the sides, ampulla expanded and pointed dorsad (not rounded at its apex). A combination of the following nuclear genomic base pairs is diagnostic: aly2643.5.1:T192C, aly2613.4.2:G2377C, aly1877.13.1:A892T, aly1877.13.1:T938C, aly1877.13.1:A1195T.
Etymology.
The name is a feminine noun in the nominative singular formed from the type species name.
Species included.
Only the type species.
Parent taxon.
Tribe Baorini Doherty, 1886.
Afrogegenes Jong & Coutsis, 2017 and Torbenlarsenia Kemal & Koçak, 2020 are subgenera of Gegenes Hübner, 1819
Nuclear genome trees point to a strongly supported clade that is sister to Borbo Evans, 1949 (type species Hesperia borbonica Boisduval, 1833) and includes type species of three genera: Gegenes Hübner, 1819 (type species Papilio pumilio Hoffmansegg, 1804), Afrogegenes Jong & Coutsis, 2017 (type species Hesperia hottentota Latreille, 1824), and Torbenlarsenia Kemal & Koçak, 2020 (type species Hesperia holtzi Plötz, 1882) in addition to several species currently in Borbo Evans, 1949 (type species Hesperia borbonica Boisduval, 1833) (Fig. 50). However, within this clade, strongly supported major subclades are absent, and Torbenlarsenia as currently circumscribed (Fan et al. 2016) is not monophyletic. Moreover, Torbenlarsenia gemella (Mabille, 1884) (type locality in Madagascar) is not in this clade but belongs to Borbo, where we transfer it as Borbo gemella (Mabille, 1884), stat. rev. This whole clade with Gegenes is at the genus level in the tree and splits into five confident subclades, three of which represent named genus-group taxa: we propose to treat Afrogegenes Jong & Coutsis, 2017, stat. rev. and Torbenlarsenia Kemal & Koçak, 2020, stat. rev. as subgenera of Gegenes Hübner, 1819. The remaining two subclades do not have names and are described as new subgenera below. Finally, to restore the monophyly of Borbo, we transfer several species to the subgenus Torbenlarsenia, forming new combinations: Gegenes (Torbenlarsenia) cottrelli (Larsen, 2013), comb. nov., Gegenes (Torbenlarsenia) fallax (Gaede, 1916), comb. nov., Gegenes (Torbenlarsenia) fanta (Evans, 1937), comb. nov., Gegenes (Torbenlarsenia) micans (Holland, 1896), comb. nov., and Gegenes (Torbenlarsenia) sirena (Evans, 1937), comb. nov.
Flanga Grishin, new subgenus http://zoobank.org/894F0986-7046-4A95-8D78-731C61C881A0
Type species.
Parnara perobscura H. H. Druce, 1912.
Definition.
As discussed above and shown in the trees (Fig. 50), the newly expanded genus Gegenes Hübner, 1819 (type species Papilio pumilio Hoffmansegg, 1804) can be partitioned into five subgenera. The species previously called Torbenlarsenia perobscura (H. H. Druce, 1912) (type locality in Ghana) is not monophyletic with Hesperia holtzi Plötz, 1882 (type locality in Angola), which is the type species of Torbenlarsenia Kemal & Koçak, 2020, and belongs to a distinct clade that we define as one of the five subgenera. It does not have a name. This new subgenus keys to 68.B.(b)(a1)(a2)(a3) in Evans (1937) and is distinguished from its relatives by long, finger-like flanges at the base of uncus and thinner, more terminally rounded tooth of harpe projecting anteriad. A combination of the following nuclear genomic base pairs is diagnostic: aly151.39.1:A351T, aly1139.20.2:A245C, aly1412.8.1:G430C, aly1412.8.1:C431A, aly1349.7.9:G58T.
Etymology.
The name is a feminine noun in the nominative singular and refers to long flanges from the base of uncus that identify this subgenus.
Species included.
The type species (i.e., Parnara perobscura H. H. Druce, 1912) and Pamphila detecta Trimen, 1893.
Parent taxon.
Genus Gegenes Hübner, 1819.
Havea Grishin, new subgenus http://zoobank.org/4816F043-591A-46E9-B19C-5C9761CD7AD9
Type species.
Hesperia havei Boisduval, 1833.
Definition.
As discussed above and shown in the trees (Fig. 50), the newly expanded genus Gegenes Hübner, 1819 (type species Papilio pumilio Hoffmansegg, 1804) can be partitioned into five subgenera. The lineage with the species currently called Borbo havei (Boisduval, 1833) (type locality in Madagascar) corresponds to the last one (others discussed above). This new subgenus keys to 68.B.(b)(b1)(c2) in Evans (1937) and is distinguished from its relatives by narrower uncus terminally rounded in dorsal view with approximately parallel sides (not concave, and not gradually narrowing distad) and ampulla expanded distad and rounded, nearly reaching the distal end of harpe, which is with a sharp, tooth-like process directed anterodorsad at its base. A combination of the following nuclear genomic base pairs is diagnostic: aly1952.2.12:C46T, aly1952.2.12:T48A, aly310.6.1:G1542C, aly4196.3.1:T1023C, aly4196.3.1:G1039T, aly127.59.2:T289T (not C), aly6209.2.1:T1095T (not G), aly813.4.5:A855A (not C), aly275211.12.5:A60A (not G), aly7600.1.6:T103T (not A).
Etymology.
The name is a feminine noun in the nominative singular formed from the type species name.
Species included.
Only the type species.
Parent taxon.
Genus Gegenes Hübner, 1819.
Zenonoida Fan & Chiba, 2016 is a subgenus of Zenonia Evans, 1935
Genomic trees reveal that genetic differentiation between Zenonia Evans, 1935 (type species Pamphila zeno Trimen, 1864) and Zenonoida Fan & Chiba, 2016 (type species Hesperia eltola Hewitson, 1869) is smaller than that within their sister genus Zinaida Evans, 1937 (type species Parnara nascens Leech, 1893) and places the former two at the subgenus tree level (Fig. 50). COI barcodes of their type species differ by 8.8% (58 bp). Furthermore, these genera are phenotypically close, and neither of them includes a large number of species. Therefore, we propose that Zenonoida Fan & Chiba, 2016, stat. nov. is a subgenus of Zenonia Evans, 1935.
On the distribution of Euphyes vestris (Boisduval, 1852) and Euphyes kiowah (Reakirt, 1866)
On the basis of genetic differences, Euphyes kiowah (Reakirt, 1866) (type locality in the USA: Colorado, Rocky Mountains) was regarded by Zhang et al. (2022b) as a species distinct from Euphyes vestris (Boisduval, 1852) (type locality in USA: California, Plumas Co.). Here, we investigate the distribution of these two species using genomic sequencing and analysis. Specimens from across the range were selected for sequencing (Fig. 51), and genomic trees are shown in Fig. 52. Both autosome and Z chromosome trees confirm prominent genetic differentiation between the two species (E. vestris and E. kiowah). In the US, Euphyes kiowah is confined to the south-central area, confirmed from Colorado, Arizona, New Mexico, and Texas (excluding northeastern, eastern, and coastal regions) (Fig. 51 squares), and E. vestris occurs everywhere else (Fig. 51 circles). Interestingly, the entire central Texas (Edwards Plateau region) is inhabited by E. kiowah, but E. vestris is found immediately to the south of it in more coastal regions. The eastern boundary between these two parapatric species is relatively straight in the northwest-to-southeast direction and likely goes through northeastern Colorado, Oklahoma, and Texas panhandles and towards the Texas coast, where the boundary turns southwest to circumvent coastal areas. Studies of specimens along this boundary may bring insights into the speciation process and isolation mechanisms of these two species. On the western side, E. kiowah and E. vestris are separated by the Great Basin, in which neither species seems to be present. As a result, we see that the distribution of E. vestris engulfs that of E. kiowah, the latter being restricted to more arid areas.
Fig. 51.
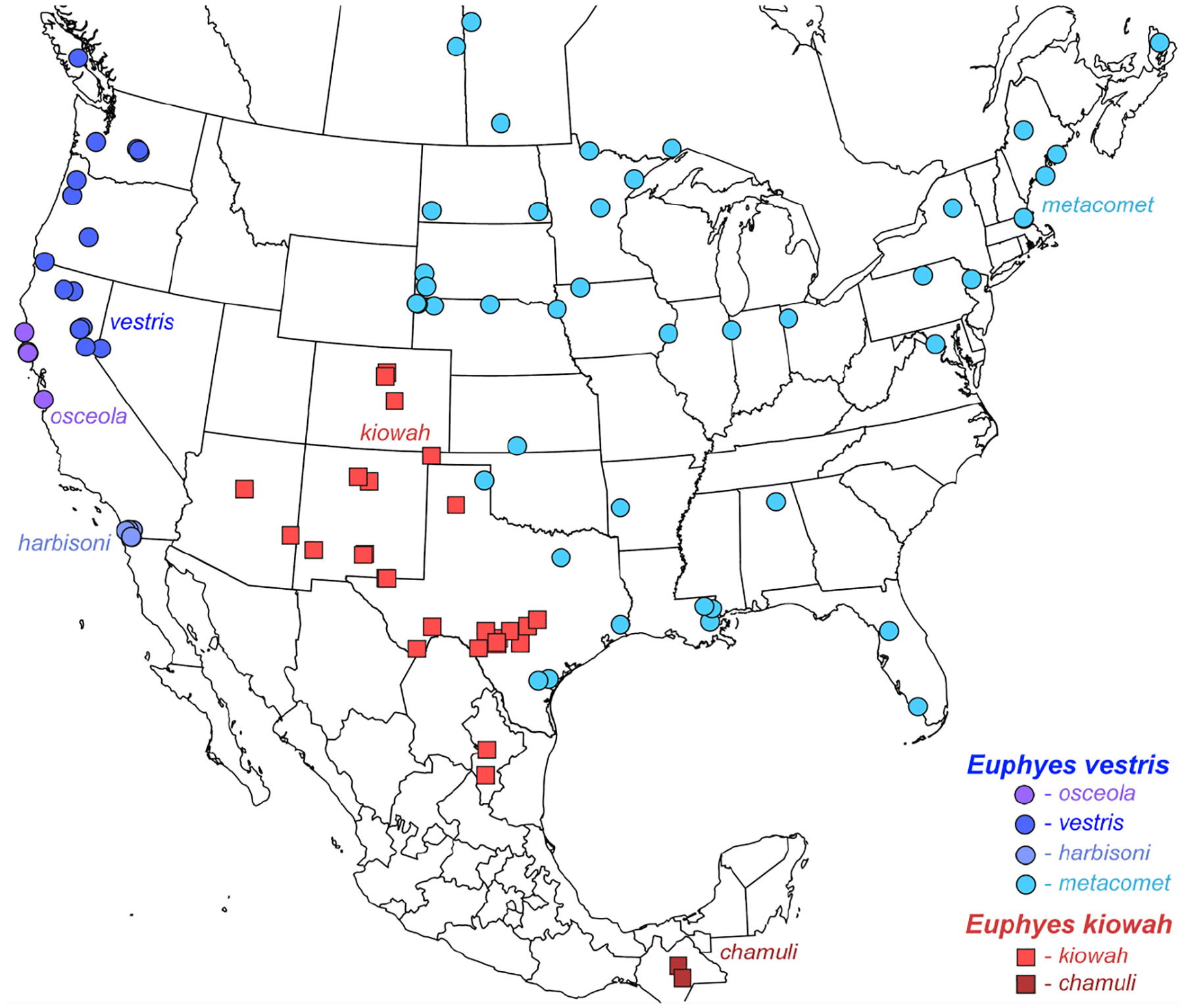
A map of sequenced specimens: Euphyes vestris (circles, purple and shades of blue) and Euphyes kiowah (squares, shades of red) with their subspecies: E. vestris osceola (purple), E. vestris vestris (darker blue), E. vestris harbisoni (paler blue), E. vestris metacomet (sky blue), E. kiowah kiowah (red), and E. kiowah chamuli (maroon).
Fig. 52.
Phylogenetic trees of Euphyes vestris (purple and shades of blue) and Euphyes kiowah (red) inferred from protein-coding regions of a) the nuclear genome (autosomes) and b) the Z chromosome. Different subspecies are shown in different shades of colors and labeled in smaller font by their clades or regions (if not monophyletic) in the tree.
Euphyes vestris osceola (Lintner, 1878) is a valid subspecies
Currently placed as a junior subjective synonym of Euphyes vestris vestris (Boisduval, 1852) (type locality in the USA: California, Plumas Co.), Pamphila osceola Lintner, 1878 (type locality in the USA: California, Mendocino Co.) shows genetic differentiation from it and forms a distinct clade in genomic trees (Fig. 52). Due to this genetic differentiation, coastal north-central California populations of E. vestris are best treated as a separate subspecies, even more distant genetically from the nominotypical subspecies than southern Californian Euphyes vestris harbisoni J. Brown & McGuire, 1983 (type locality in USA: California, San Diego Co.) (Fig. 52). Therefore, in agreement with Emmel et al. (1998), we consider Euphyes vestris osceola (Lintner, 1878), stat. rev. to be a valid subspecies.
Vernia verna sequoyah (H. Freeman, 1942) is a valid subspecies
Polites verna sequoyah H. Freeman, 1942 (type locality USA: Arkansas, Little Rock) was placed by Evans (1955) as a junior subjective synonym of Pamphila verna W. H. Edwards, 1862 (type locality in USA: Mercer Co.), the type species of and currently in the genus Vernia Grishin, 2019. The genomic tree reveals that sequenced specimens of Vernia verna partition into two clades (Fig. 53). The clades are characterized by close genetic similarity of specimens within each clade and a certain level of genetic differentiation between the clades. Therefore, populations assigned to these clades can be treated as two distinct subspecies. The neotype of V. verna and a syntype of Pamphila pottawattomie Worthington, 1880 from “N. Ind.” (type locality in USA: IL, Cook Co., and IN, Lake Co.) belong to one clade, and the holotype of P. v. sequoyah belongs to the other. We propose that the two clades represent two subspecies and reinstate Vernia verna sequoyah (H. Freeman, 1942), stat. rest. as a valid subspecies. Moreover, due to genetic differentiation, future studies of additional specimens may determine that it is a species-level taxon.
Fig. 53.

Phylogenetic trees inferred from protein-coding regions in: a) the Z chromosome and b) the mitochondrial genome with Vernia verna verna (blue) and V. verna sequoyah (red). Primary type specimens are labeled in magenta. The sequence of SAMN18587413 is taken from the alignment provided in Kawahara et al. (2023).
Lerodea dysaules Godman, 1900 is a valid species distinct from Lerodea arabus (W. H. Edwards, 1882)
Originally proposed as a species and treated (with some reservations) as such by Evans (1955), Lerodea dysaules Godman, 1900 (type locality Mexico: Guerrero, Venta de Zopilote) was later synonymized with Lerodea arabus (W. H. Edwards, 1882) (type locality USA: Arizona, Pima Co., Sabino Canyon), because no appreciable differences in genitalia were observed, and wing pattern differences given by Evans (1955) do not hold up in a larger series of specimens (Warren and Mielke 2005). Genomic sequencing of L. arabus from across its range reveals a deep split into two clades (Fig. 54a, b), unexpected from phenotypic assessment. The two clades are most strongly differentiated genetically with Fst/Gmin statistics 0.70/0.00 and COI barcode difference of 8.2% (54 bp), typical for species from different subgenera and remarkably large for close relatives. Therefore, the two clades represent two distinct species.
Fig. 54.
Lerodea arabus (blue) and Lerodea dysaules stat. rest. (red): phylogenetic trees inferred from protein-coding regions in: a) the nuclear genome (autosomes) and b) the mitochondrial genome; c) a distribution map made from iNaturalist (2023) observations: some points in the iNaturalist map L. arabus were re-colored in Photoshop and the two points corresponding to sequenced specimens from Mexico: Sonora were added (CC BY-NC 4.0 https://creativecommons.org/licenses/by-nc/4.0/), other points do not refer to sequenced specimens, records were not checked, and some may be misidentifications.
The distribution of L. arabus as recorded on iNaturalist (2023) is disjoint, showing a cluster of observations in southeastern Arizona and the major range from Sinaloa – Durango – Coahuila – Nuevo Leon – south Texas southwards (Fig. 54c). These two disjoint areas in the distribution correspond perfectly to the two distinct species revealed by genomic sequencing (plus specimens from both clades in Sonora, Mexico where no iNaturalist observations existed) (Fig. 54). Sequenced specimens from near the type locality of L. arabus provide the name for the southeastern Arizona and Sonora (except southernmost) cluster (Fig. 54 blue). The type locality of Lerodea dysaules falls well within the major range, and the sequenced specimen from Guerrero belongs to the second species along with other specimens across the major range (Fig. 54 red), providing an available name for this species. Therefore, we reinstate Lerodea dysaules Godman, 1900, stat. rest. as a species-level taxon. In conclusion, L. arabus is the northwestern species, and L. dysaules is the southeastern species. The two species may meet in southern Sonora, Mexico, and the westernmost record of L. dysaules is from Alamos, Sonora (sequenced as NVG-21056A02) (Fig. 54 red). Curiously, genomic sequencing suggests that populations from Mexico Baja California Sur are L. arabus (Fig. 54).
As for the phenotypic assessment, both species are variable in wing patterns, and if identification is possible, it should be done within each pattern form, comparing specimens with similar patterns between species. Generally, ventral overscaling in L. dysaules is more mottled in appearance, especially in a form with a well-developed brown discal patch on the ventral hindwing; this patch is smaller, not extending towards the apex but turning towards mid-costa and is bordered by more diffuse small pale spots (if the spots are present at all), these spots are also diffuse and frequently connected into a band even if the central brown patch is absent. In L. arabus, pale ventral overscaling is more uniform, without apparent mottling, the hindwing brown patch (if expressed) is larger and frequently extending towards the apex (at least the area between the patch and the apex is darker than the central submarginal area), giving the patch a more triangular appearance; if postdiscal small pale spots are expressed, their edges are better defined, and the spots are more contrasty on the background, better separated from each other, especially in specimens lacking the patch, and the spot in cell Sc+R1-RS may be particularly prominent. To aid in the identification of these species, we provide COI barcodes of specimens from near the type localities:
Lerodea arabus from USA: AZ, Pima Co. [LACM], sample NVG-22094F10, GenBank OR578721:
AACTTTATATTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACATCATTAAGTCTTTTAATTCGAACAGAATTAGGTAATCCTGGATCTTTAATTGGAGATGATCAAATTTACAATACTATTGTGACAGCTCACGCTTTTATTATAATTTTTTTCATGGTAATACCTATTATAATTGGTGGATTTGGTAATTGATTAGTTCCTCTAATATTAGGTGCCCCAGATATAGCTTTCCCACGAATAAATAATATAAGATTTTGAATACTGCCACCTTCCTTAATATTATTAATTTCAAGTAGAATTGTAGAAAATGGTGCAGGAACAGGTTGAACTGTATATCCTCCTCTTTCTTCTAATATTGCCCACCAAGGAGCCTCAGTTGACTTAGCGATTTTTTCATTACATTTAGCAGGTATTTCATCTATTTTAGGAGCTATTAACTTTATTACTACTATTATTAATATACGAATTAAAAATTTATCATTTGATCAAATACCTTTATTTGTTTGATCAGTAGGAATTACAGCATTATTATTACTTTTATCTCTACCTGTTTTAGCAGGTGCTATTACCATACTTTTAACTGACCGAAATTTAAACACTTCATTTTTTGATCCTGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
Lerodea dysaules from Mexico: Guerrero [MGCL], sample NVG-21085D09, GenBank OR578722:
AACTTTATACTTTATTTTTGGAATTTGAGCAGGAATATTAGGAACATCTTTAAGTCTTTTAATTCGAACAGAATTAGGCAATCCTGGATCTTTAATTGGAGATGATCAAATTTATAATACTATTGTAACAGCCCATGCCTTTATTATAATTTTTTTTATAGTTATGCCTATTATAATTGGAGGATTTGGTAATTGACTAGTTCCTTTAATATTAGGAGCACCTGACATAGCATTCCCACGAATAAATAATATAAGATTTTGAATACTACCACCTTCTCTAATATTATTAATCTCAAGTAGAATTGTAGAAAATGGTGCAGGTACAGGTTGAACAGTATATCCCCCTCTTTCATCTAATATTGCACATCAAGGAGCCTCAGTTGACCTTGCAATTTTTTCTCTTCATTTAGCTGGTATTTCATCCATTTTAGGAGCTATTAATTTTATTACTACTATTATTAACATACGAATTAAAAATTTATCATTCGATCAAATACCTTTATTTGTCTGATCTGTAGGAATTACAGCATTATTATTACTTTTATCTTTACCTGTCTTAGCAGGAGCTATTACTATACTTTTAACCGATCGAAACCTTAATACTTCATTCTTTGATCCTGCTGGAGGAGGAGATCCTATTTTATATCAACATTTATTT
[No genus] osibius Draudt, 1924 is an unavailable name
The name “osibius” was published by Draudt (1921–1924) below a specimen illustration on the plate 113B entitled “AGRIAS-ERYNNIS”, row c image 4 from the left, out of 7. No other mention of the name is found in this work. The species illustrated first (i.e., top left: row a image [1]) on plate 113B was placed in the genus Agrias, and the species illustrated last (i.e., bottom right: row c image [7], current name Hesperia colorado (Scudder, 1874)) was placed by Draudt (1923b) in the genus Erynnis. The species illustrated next to last (row c, images [5] and [6], current name Turesis complanula (Herrich-Schäffer, 1869), misidentified as “lucasi”) was placed by Draudt (1923a) in the genus Turesis. Only two genera are mentioned in the title of this plate (Agrias and Erynnis), but at least three are illustrated. Therefore, it remains unclear which genus “osibius” was placed in (Agrias, Turesis [not mentioned on the plate], Erynnis, or some other genus). Mielke (1993) listed “osibius” (as “osybius”) in combination with the genus Turesis Godman, 1901 (type species Hesperia lucas Fabricius, 1793) and treated it as a valid species (Mielke 2004; Mielke 2005). However, the name “osibius” by Draudt is unavailable because it was not proposed “in unambiguous combination with a generic name” as demonstrated above (fails ICZN Code Art. 11.9.3) and therefore cannot be used as a valid name for any species. Finally, we were not able to unambiguously determine the identity of the specimen illustrated by Draudt (1921–1924) (among others, it might have been Rhinthon Godman, 1900 or Niconiades Hübner, [1821]), but it does not seem to belong to Turesis due to details in its wing pattern, such as a postdiscal arc of increasing in size pale spots on ventral hindwing, that are not characteristic of Turesis.
Supplementary Material
ACKNOWLEDGMENTS
This study is dedicated to the memory of Paul A. Opler, our close collaborator on genomic projects. We are most grateful to Paul and his wife Evi for participating in these projects, sharing knowledge, discussions, insights, critiques, and numerous specimens they collected for genomic research, which we used in this and future studies. We acknowledge Ping Chen and Ming Tang for their excellent technical assistance. We are grateful to David Grimaldi and Courtney Richenbacher (AMNH: American Museum of Natural History, New York, NY, USA), Jason Weintraub (ANSP: The Academy of Natural Sciences of Drexel University, Philadelphia, PA, USA), Blanca Huertas, David Lees, and Geoff Martin (BMNH: Natural History Museum, London, UK), Chris Grinter, Denise Montelongo, David Bettman, Vince Lee, and the late Norm Penny (CAS: California Academy of Sciences, San Francisco, CA, USA), Jim Fetzner, Bob Androw, Vanessa Verdecia, Cat Giles, and the late John Rawlins (CMNH: Carnegie Museum of Natural History, Pittsburgh, PA, USA), the late Paul Opler, Chuck Harp, and the late Boris Kondratieff (CSUC: Colorado State University Collection, Fort Collins, CO, USA), Jason Dombroskie (CUIC: Cornell University Insect Collection, Ithaca, New York, USA), Crystal Maier and Rebekah Baquiran (FMNH: Field Museum of Natural History, Chicago, IL, USA), Weiping Xie and Giar-Ann Kung (LACM: Los Angeles County Museum of Natural History, Los Angeles, CA, USA), Théo Léger, Viola Richter, and Wolfram Mey (MFNB: Museum für Naturkunde, Berlin, Germany), Andrei Sourakov, Andrew D. Warren, Debbie Matthews-Lott, Riley J. Gott, and Keith R. Willmott (MGCL: McGuire Center for Lepidoptera and Biodiversity, Gainesville, FL, USA), Rodolphe Rougerie (MNHP: Muséum National d’Histoire Naturelle, Paris, France), Matthias Nuss and Manuela Bartel (MTD: Museum für Tierkunde, Dresden, Germany), Gerardo Lamas (MUSM: Museo de Historia Natural, Lima, Peru), Larry F. Gall (PMNH: Peabody Museum of Natural History, Yale University, New Haven, CT, USA), Martin Wiemers and Christian Kutzscher (SDEI: Senckenberg Deutsches Entomologisches Institut, Müncheberg, Germany), Wolfgang A. Nässig (SMF: Natural History Museum, Frankfurt, Germany), Edward G. Riley, Karen Wright, and John Oswald (TAMU: Texas A&M University Insect Collection, College Station, TX, USA), Alex Wild (TMMC: University of Texas Biodiversity Center, Austin, TX, USA), Jeff Smith and Lynn Kimsey (UCDC: Bohart Museum of Entomology, University of California, Davis, CA, USA), Robert K. Robbins, John M. Burns, and Brian Harris (USNM: National Museum of Natural History, Smithsonian Institution, Washington, DC, USA), Jonathan P. Pelham (UWBM: Burke Museum of Natural History and Culture, Seattle, WA, USA), and Axel Hausmann, Andreas Segerer, and Ulf Buchsbaum (ZSMC: Zoologische Staatssammlung München, Germany), for granting access to the collections under their care, sampling specimens, and stimulating discussions; to Brian Banker, Gian C. Bozano, Jim P. Brock, Ernst Brockmann, Jack S. Carter, Bill R. Dempwolf, Mike Fisher, Robb Hannawacker, Bernard Hermier, James McDermott, the late James A. Scott, Kojiro Shiraiwa, Steve M. Spomer, and Mark Walker for specimens and leg samples, to Gerardo Lamas and Jonathan P. Pelham for discussions, advice, helpful suggestions, patiently answering numerous questions, and sharing their research files, and to Bernard Hermier and Gerardo Lamas for critical review of the manuscript and many helpful corrections. We are indebted to the California Department of Fish and Game for collecting permit SC13645, Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the research permit 08-02Rev, to U. S. National Park Service for the research permits: Big Bend (Raymond Skiles) for BIBE-2004-SCI-0011 and Yellowstone (Erik Oberg and Annie Carlson) for YELL-2017-SCI-7076, and to the National Environment & Planning Agency of Jamaica for the permission to collect specimens. Please note that photographs from iNaturalist (2023) reproduced in this work and photographs ©The Trustees of the Natural History Museum, London are made available under Creative Commons License 4.0 (https://creativecommons.org/licenses/by/4.0/), which means in particular that when using the images you must give appropriate credit and provide a link to the license. We acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources. This study was supported in part by the HHMI Investigator funds and by grants from the National Institutes of Health GM127390 and the Welch Foundation I-1505 to N.V.G.
Footnotes
ZooBank registration: http://zoobank.org/9CAA87E5-B1FE-4948-BD3B-EFAA9B95575D
LITERATURE CITED
- Bachtrog D, Thornton K, Clark A, and Andolfatto P. 2006. Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution 60(2): 292–302. [PubMed] [Google Scholar]
- Barnhart JH 1965. Biographical Notes Upon Botanists, vol. 3. G. K. Hall and Co.; Boston, MA. 545 pp [Google Scholar]
- Boyle JH, Espeland M, Sáfián S, Ducarme R, Gardiner AJ, Coleman JW, Heath A, Fisher S, Collins SC, Martins DJ, Aduse-Poku K, Libert M, Dankowicz E, Kawahara AY, Lohman DJ, and Pierce NE. 2023. Phylogeny of the Poritiinae (Lepidoptera: Lycaenidae), butterflies with ant associations and unusual lichenivorous diets. Systematic Entomology 48(3): 422–433. [Google Scholar]
- Boyle JH, Kaliszewska ZA, Espeland M, Suderman TR, Fleming J, Heath A, and Pierce NE. 2015. Phylogeny of the Aphnaeinae: myrmecophilous African butterflies with carnivorous and herbivorous life histories. Systematic Entomology 40(1): 169–182. [Google Scholar]
- Burns JM 2020. Taxonomic status of a Florida differentiate in the Erynnis brizo species group: classical evidence (Lepidoptera: Hesperiidae: Pyrginae). Proceedings of the Entomological Society of Washington 122(1): 25–41. [Google Scholar]
- Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, and Hebert PD. 2008. DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proceedings of the National Academy of Sciences of the United States of America 105(17): 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun JV 1997. Updated list of the butterflies and skippers of Florida (Lepidoptera: Papilionoidea and Hesperioidea). Holarctic Lepidoptera 4: 39–50. [Google Scholar]
- Carneiro E, Mielke OHH, and Casagrade MM. 2013. Thorax and abdomen morphology of some Neotropical Hesperiidae (Lepidoptera). Insecta Mundi 0327: 1–47. [Google Scholar]
- Casagrade MM, and Mielke OHH. 1979. Eurema (Eurema) furtadoi sp. n. de Mato Grosso, Brasil (Lepidoptera, Pieridae). Dusenia (Curitiba) 11(3): 137–142. [Google Scholar]
- Chermock RL 1950. A Generic Revision of the Limenitini of the World. The American Midland Naturalist 43(3): 513–569. [Google Scholar]
- Chiba H 2009. A revision of the subfamily Coeliadinae (Lepidoptera: Hesperiidae). Bulletin of the Kitakyushu Museum of Natural History and Human History, Series A (Natural History) 7: 1–102. [Google Scholar]
- Chou I (Editor) 1994. Monographia Rhopalocerorum Sinensium [in Chinese]. Henan Scientific and Technological Publishing House; Zhengzhou, China. viii + 854 pp. [Google Scholar]
- Cong Q, Borek D, Otwinowski Z, and Grishin NV. 2015. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Reports 10(6): 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Borek D, Robbins RK, Opler PA, Otwinowski Z, and Grishin NV. 2017a. When COI barcodes deceive: complete genomes reveal introgression in hairstreaks. Proceedings of the Royal Society B: Biological Sciences 284(1848): 20161735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Borek D, Robbins RK, Otwinowski Z, and Grishin NV. 2016. Complete genomes of hairstreak butterflies, their speciation, and nucleo-mitochondrial incongruence. Scientific Reports 6: 24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Li W, Borek D, Otwinowski Z, and Grishin NV. 2017b. The first complete genomes of metalmarks and the classification of butterfly families. Genomics 109: 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Shen J, Zhang J, Li W, Kinch LN, Calhoun JV, Warren AD, and Grishin NV. 2021. Genomics reveals the origins of historical specimens. Molecular Biology and Evolution 38(5): 2166–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Zhang J, and Grishin NV. 2019a. Genomic determinants of speciation. bioRxiv BIORXIV/2019/837666. [Google Scholar]
- Cong Q, Zhang J, Shen J, Cao X, Brevignon C, and Grishin NV. 2020. Speciation in North American Junonia from a genomic perspective. Systematic Entomology 45(4): 803–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Q, Zhang J, Shen J, and Grishin NV. 2019b. Fifty new genera of Hesperiidae (Lepidoptera). Insecta Mundi 0731: 1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey JW, Chouteau M, Barker SL, Maroja L, Baxter SW, Simpson F, Merrill RM, Joron M, Mallet J, Dasmahapatra KK, and Jiggins CD. 2016. Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 (Bethesda) 6(3): 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhungel B, and Wahlberg N. 2018. Molecular systematics of the subfamily Limenitidinae (Lepidoptera: Nymphalidae). PeerJ 6: e4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draudt MWK 1921–1924. B. Grypocera, breitköpfige Tagfalter. In: Seitz A(Ed.). Die Gross-Schmetterlinge der Erde. Alfred Kernen; Stuttgart, pp. 833–1011, 1046–1139, pls 113B, 160–193. [Google Scholar]
- Draudt MWK 1922. B. Grypocera, breitköpfige Tagfalter. In: Seitz A (Ed.). Die Gross-Schmetterlinge der Erde. Alfred Kernen; Stuttgart, pp. 873–880. [Google Scholar]
- Draudt MWK 1923a. B. Grypocera, breitköpfige Tagfalter. In: Seitz A(Ed.). Die Gross-Schmetterlinge der Erde. Alfred Kernen; Stuttgart, pp. 953–992. [Google Scholar]
- Draudt MWK 1923b. B. Grypocera, breitköpfige Tagfalter. In: Seitz A (Ed.). Die Gross-Schmetterlinge der Erde. Alfred Kernen; Stuttgart, pp. 913–952. [Google Scholar]
- Eliot JN 1973. The higher classification of the Lycaenidae (Lepidoptera): a tentative Arrangement. Bulletin of the British Museum of Natural History (Entomology) 28(6): 373–505. [Google Scholar]
- Emmel JF, Emmel TC, and Mattoon SO. 1998. The types of California butterflies named by Jean Alphonse Boisduval: Designation of lectotypes and a neotype, and fixation of type localities. In: Emmel TC(Ed.). Systematics of Western North American Butterflies. Mariposa Press; Gainesville, pp. 3–76. [Google Scholar]
- Evans WH 1937. A catalogue of the African Hesperiidae indicating the classification and nomenclature adopted in the British Museum. The Trustees of the British Museum (Natural History); London. xii + 212 pp., 30 pls. [Google Scholar]
- Evans WH 1949. A catalogue of the Hesperiidae from Europe, Asia, and Australia in the British Museum (Natural History). The Trustees of the British Museum (Natural History); London. xix + 502 pp., 53 pls. [Google Scholar]
- Evans WH 1952. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part II. Pyrginae. Section I. The Trustees of the British Museum (Natural History); London. v + 178 pp., pls. 10–25. [Google Scholar]
- Evans WH 1955. A catalogue of the American Hesperiidae indicating the classification and nomenclature adopted in the British Museum (Natural History). Part IV. Hesperiinae and Megathyminae. The Trustees of the British Museum (Natural History); London. v + 499 pp., pls. 54–88. [Google Scholar]
- Fan X, Chiba H, Huang Z, Fei W, Wang M, and Safian S. 2016. Clarification of the phylogenetic framework of the tribe Baorini (Lepidoptera: Hesperiidae: Hesperiinae) inferred from multiple gene sequences. PLoS One 11(7): e0156861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassberg J 2023. Description of a new subspecies of common sootywing, ‘white-tufted’ common sootywing, Pholisora catullus albicirrus new subspecies. American Butterflies 31(3): 36–39. [Google Scholar]
- Godman FD 1907. Notes on the American species of Hesperiidae described by Plötz. Annals and Magazine of Natural History (7)20(16): 132–155. [Google Scholar]
- Hall JP, Willmott KR, and Ahrenholz DH. 2023. A review of the Argyrogrammana amalfreda group fauna of the western Amazon (Lepidoptera: Riodinidae: Symmachiini). Tropical Lepidoptera Research 33(Suppl. 1): 61–85. [Google Scholar]
- Hall JPW 2002. A phylogenetic revision of Calydna and relatives (Lepidoptera: Riodinidae). Insect Systematics & Evolution 33: 185–237. [Google Scholar]
- Hall JPW 2003. Phylogenetic reassessment of the five forewing radial-veined tribes of Riodininae (Lepidoptera: Riodinidae). Systematic Entomology 28(1): 23–38. [Google Scholar]
- Hall JPW, and Willmott KR. 1996. Notes on the genus Argyrogrammana, part 2, with one new species (Lepidoptera: Riodinidae). Tropical Lepidoptera 7: 71–80. [Google Scholar]
- Heath A 1997. A review of African genera of the tribe Aphnaeini (Lepidoptera: Lycaenidae). Metamorphosis 8(Suppl. 2): 1–60. [Google Scholar]
- Heath A, Pringle EL, and Quek S-P. 2023. Chrysoritis Butler (Papilionoidea: Lycaenidae: Aphnaeinae) – Part III: An integrative taxonomic revision. Metamorphosis 33: 11–28. [Google Scholar]
- Hebert PD, Cywinska A, Ball SL, and deWaard JR. 2003. Biological identifications through DNA barcodes. Proceedings of the Royal Society B: Biological Sciences 270(1512): 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning SF, and Henning GA. 1996. A review of the genus Axiocerces Hübner (Lepidoptera: Lycaenidae), with descriptions of nine new species and three new subspecies. Metamorphosis 7(Suppl. 1): 1–48. [Google Scholar]
- Herrich-Schäffer GAW 1869. Prodromus systematis lepidopterorum. Versuch einer systematischen Anordnung der Schmetterlinge. Correspondenz-Blatt des zoologisch-mineralogischen Vereines in Regensburg 23(12): 184–204. [Google Scholar]
- Holland WJ 1896. A preliminary revision and synonymic catalogue of the Hesperiidae of Africa and the adjacent islands, with descriptions of some apparently new species. Proceedings of The Zoological Society of London 1896: 2–107. [Google Scholar]
- Howarth TG 1966. Revisional notes on the genus Antanartia (Lepidoptera: Nymphalidae). Bulletin of the British Museum of Natural History 18: 21–43. [Google Scholar]
- ICZN [International Commission on Zoological Nomenclature]. 1999. International code of zoological nomenclature. Fourth edition. International Trust for Zoological Nomenclature; London. xxx + 306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- iNaturalist. 2023. A Community for Naturalists – iNaturalist. Available from https://www.inaturalist.org. Accessed 25-Aug-2022.
- Jiang D, Klaus S, Zhang YP, Hillis DM, and Li JT. 2019. Asymmetric biotic interchange across the Bering land bridge between Eurasia and North America. National Science Review 6(4): 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur M, Kirti JS, and Sidhu AK. 2022. Taxonomic study of the Genus Gandaca Moore (Lepidoptera: Pieridae) from India. Ecology, Environment and Conservation Journal 28(May Suppl. Issue): S572–S578. [Google Scholar]
- Kawahara AY 2009. Phylogeny of snout butterflies (Lepidoptera: Nymphalidae: Libytheinae): combining evidence from the morphology of extant, fossil, and recently extinct taxa. Cladistics 25: 263–278. [DOI] [PubMed] [Google Scholar]
- Kawahara AY, Storer C, Carvalho APS, Plotkin DM, Condamine FL, Braga MP, Ellis EA, St Laurent RA, Li X, Barve V, Cai L, Earl C, Frandsen PB, Owens HL, Valencia-Montoya WA, Aduse-Poku K, Toussaint EFA, Dexter KM, Doleck T, Markee A, Messcher R, Nguyen YL, Badon JAT, Benitez HA, Braby MF, Buenavente PAC, Chan WP, Collins SC, Rabideau Childers RA, Dankowicz E, Eastwood R, Fric ZF, Gott RJ, Hall JPW, Hallwachs W, Hardy NB, Sipe RLH, Heath A, Hinolan JD, Homziak NT, Hsu YF, Inayoshi Y, Itliong MGA, Janzen DH, Kitching IJ, Kunte K, Lamas G, Landis MJ, Larsen EA, Larsen TB, Leong JV, Lukhtanov V, Maier CA, Martinez JI, Martins DJ, Maruyama K, Maunsell SC, Mega NO, Monastyrskii A, Morais ABB, Muller CJ, Naive MAK, Nielsen G, Padron PS, Peggie D, Romanowski HP, Safian S, Saito M, Schroder S, Shirey V, Soltis D, Soltis P, Sourakov A, Talavera G, Vila R, Vlasanek P, Wang H, Warren AD, Willmott KR, Yago M, Jetz W, Jarzyna MA, Breinholt JW, Espeland M, Ries L, Guralnick RP, Pierce NE, and Lohman DJ. 2023. A global phylogeny of butterflies reveals their evolutionary history, ancestral hosts and biogeographic origins. Nature Ecology & Evolution 7(6): 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klots AB 1929. A revision of the genus Eurema Hübner (Lepidoptera, Pieridae). Part II: New World species, taxonomy and synonymy. Entomologica americana; 9(3): 99–163. [Google Scholar]
- Klots AB 1933. A generic classification of the Pieridae (Lepidoptera) together with a study of the male genitalia. Entomologica Americana 12: 139–242. [Google Scholar]
- Koçak AÖ 1996. Güney batı asya Polyommatinae taksonlarının listesi ve bazı nomenklatür notları (Lepidoptera, Lycaenidae). Centre for Entomological Studies Ankara Miscellaneous Papers 30/33(1–32). [Google Scholar]
- Koçak AÖ, and Seven S. 1997. Negros Papilionoidea, Hesperioidea faunası ve taksonomisi üzerine araştırmalar (Lepidoptera) (Filipin entomolojik ekspedisyonu sonuçları-IV). Priamus 9(2/3): 57–114. [Google Scholar]
- Lamas G 2004. Pieridae, pp. 99–117. In: Lamas G (Ed.). Checklist: Part 4A. Hesperioidea - Papilionoidea. In: Heppner JB(Ed.), Atlas of Neotropical Lepidoptera. Volume 5A. Association for Tropical Lepidoptera; Scientific Publishers; Gainesville. [Google Scholar]
- Larsen TB 1991. The Butterflies of Kenya and their Natural History. Oxford University Press; Oxford. 490 pp [Google Scholar]
- Lees DC, Kremen C, and Raharitsimba H. 2003. Classification, diversity and endemism of the butterflies (Papilionoidea and Hesperioidea): a revised species checklist. In: Goodman SM and Benstead JP (Eds.). The Natural History of Madagascar. University of Chicago Press; Chicago, pp. 762–793. [Google Scholar]
- Li W, Cong Q, Shen J, Zhang J, Hallwachs W, Janzen DH, and Grishin NV. 2019. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proceedings of the National Academy of Sciences of the United States of America 116(13): 6232–6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente JE, Nieves S, and Flores A. 2023. Exochorion in the tribe Nymphalini (Lepidoptera: Nymphalidae): the genus Hypanartia Hübner, [1821] and comparison with related genera. Zootaxa 5330(2): 151–200. [DOI] [PubMed] [Google Scholar]
- Lukhtanov VA, Sourakov A, and Zakharov E. 2016. DNA barcodes as a tool in biodiversity research: testing pre-existing taxonomic hypotheses in Delphic Apollo butterflies (Lepidoptera, Papilionidae). Systematics and Biodiversity 14: 599–613. [Google Scholar]
- Mielke OHH 1993. Sobre os tipos de Hesperiidae (Lepidoptera) neotropicais descritos por M. Draudt. Revista Brasileira de Entomologica 37: 611–638. [Google Scholar]
- Mielke OHH 2004. Hesperioidea. In: Lamas G(Ed.). Checklist: Part 4A. Hesperioidea - Papilionoidea. Association for Tropical Lepidoptera; Scientific Publishers; Gainesville. [Google Scholar]
- Mielke OHH 2005. Catalogue of the American Hesperioidea: Hesperiidae (Lepidoptera). Sociedade Brasileira de Zoologia; Curitiba, Paraná, Brazil. xiii + 1536 pp. [Google Scholar]
- Miller JS 1987. Phylogenetic studies in the Papilioninae (Lepidoptera, Papilionidae). Bulletin of the American Museum of Natural History 186(4): 365–512. [Google Scholar]
- Miller LD 1974. Revision of the Euptychiini (Satyridae) 2. Cyllopsis R. Felder. Bulletin of the Allyn Museum 20: 1–98. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, and Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opler PA, and Warren AD. 2002. Butterflies of North America. 2. Scientific names list for butterfly species of North America, north of Mexico. Colorado State University, C. P. Gillette Museum of Arthropod Diversity; Fort Collins. [2] + 79 pp. [Google Scholar]
- Overlaet FG 1940. Formes nouvelles ou peu connues de Nymphalides africains. Revue de Zoologie et de Botanique Africaine 33: 150–172. [Google Scholar]
- Pelham JP 2008. Catalogue of the Butterflies of the United States and Canada. Journal of Research on the Lepidoptera 40: 1–658. [Google Scholar]
- Penz CM, and Peggie D. 2003. Phylogenetic relationships among Heliconiinae genera based on morphology (Lepidoptera: Nymphalidae). Systematic Entomology 28(4): 451–479. [Google Scholar]
- Plötz C 1882. Einige Hesperiinen-Gattungen und deren Arten. Berliner entomologische Zeitschrift 26(1): 71–82. [Google Scholar]
- Pollock DD, Watt WB, Rashbrook VK, and Iyengar EV. 1998. Molecular phylogeny for Colias butterflies and their relatives (Lepidoptera: Pieridae). Annals of the Entomological Society of America 91(5): 524–531. [Google Scholar]
- Rambaut A 2018. FigTree, version 1.4.4 http://tree.bio.ed.ac.uk/software/figtree/.
- Riley ND 1925. The species usually referred to the Genus Cigaritis Boisd. (Lycaenidae). Novitates Zoologicae 32: 70–95. [Google Scholar]
- Robbins RK, Cong Q, Zhang J, Shen J, Busby RC, Faynel C, Duarte M, Martins ARP, Prieto C, Lamas G, and Grishin NV. 2022. Genomics-based higher classification of the species-rich hairstreaks (Lepidoptera: Lycaenidae: Eumaeini). Systematic Entomology 47(3): 445–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Cong Q, Borek D, Otwinowski Z, and Grishin NV. 2017. Complete genome of Achalarus lyciades, the first representative of the Eudaminae subfamily of skippers. Current Genomics 18(4): 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Cong Q, Kinch LN, Borek D, Otwinowski Z, and Grishin NV. 2016. Complete genome of Pieris rapae, a resilient alien, a cabbage pest, and a source of anti-cancer proteins. F1000Research 5: 2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen TJ 2006. Fritillary phylogeny, classification and larval hostplants: reconstructed mainly on the basis of male and female genitalic morphology (Lepidoptera: Nymphalidae: Argynnini). Biological Journal of the Linnean Society 89: 627–673. [Google Scholar]
- Sohn J-C, Labandeira CC, Davis DR, and Mitter C. 2012. An annotated catalog of fossil and subfossil Lepidoptera (Insecta: Holometabola) of the world. Zootaxa 3286(1): 1–132. [Google Scholar]
- Stempffer H 1967. The genera of the African Lycaenidae (Lepidoptera: Rhopalocera). Bulletin of the British Museum (Natural History) Entomology Suppl. 10: 3–322. [Google Scholar]
- Stichel H 1910. Fam. Riodinidae. Allgemeines - Subfam. Riodininae. Genera Insectorum 112A: 1–238. [Google Scholar]
- Stradomsky BV 2016. A molecular phylogeny of subfamily Polyommatinae (Lepidoptera: Lycaenidae). Caucasian Entomological Bulletin 12(1): 145–156. [Google Scholar]
- Talavera G, Kaliszewska ZA, Heath A, and Pierce NE. 2020. Recent diversification of Chrysoritis butterflies in the South African Cape (Lepidoptera: Lycaenidae). Molecular Phylogenetics and Evolution 148: 106817:1–10. [DOI] [PubMed] [Google Scholar]
- Talavera G, Lukhtanov VA, Pierce NE, and Vila R. 2012. Establishing criteria for higher-level classification using molecular data: the systematics of Polyommatus blue butterflies (Lepidoptera, Lycaenidae). Cladistics 29: 166–192. [DOI] [PubMed] [Google Scholar]
- Tite GE 1963. A synonymic list of the genus Nacaduba and allied genera (Lepidoptera: Lycaenidae). Bulletin of the British Museum (Natural History) Entomology 13(4): 69–116. [Google Scholar]
- Tite GE, and Dickson CGC. 1973. The genus Aloides and allied genera (Lepidoptera: Lycaenidae). Bulletin of the British Museum (Natural History) Entomology 29(5): 227–280. [Google Scholar]
- Toussaint EFA, Chiba H, Yago M, Dexter KM, Warren AD, Storer C, Lohman DJ, and Kawahara AY. 2021. Afrotropics on the wing: phylogenomics and historical biogeography of awl and policeman skippers. Systematic Entomology 46(1): 172–185. [Google Scholar]
- Turner T, and Turland V. 2017. Discovering Jamaican butterflies and their relationships around the Caribbean. Caribbean Wildlife Publications; Safety Harbor, FL. 492 [Google Scholar]
- Wahlberg N 2019. The higher classification of Nymphalidae. <http://www.nymphalidae.net/Nymphalidae/Classification/Higher_class.htm> Accessed on 16 Aug 2023.
- Wahlberg N, Maresova J, Murillo-Ramos L, Collins S, and Wu L-W. 2020. The phylogenetic positions of Bhagadatta Moore, 1898, Kumothales Overlaet, 1940 and Harmilla Aurivillius, 1892 (Lepidoptera, Nymphalidae, Limenitidinae) based on molecular data. Nota Lepidopterologica 43: 167–171. [Google Scholar]
- Wahlberg N, and Rubinoff D. 2011. Vagility across Vanessa (Lepidoptera: Nymphalidae): mobility in butterfly species does not inhibit the formation and persistence of isolated sister taxa. Systematic Entomology 36: 362–370. [Google Scholar]
- Warren AD, Davis KJ, Stangeland EM, Pelham JP, Willmott KR, and Grishin NV. 2023. Illustrated Lists of American Butterflies. [23-VI-2023] <https://www.butterfliesofamerica.com/>.
- Warren AD, and Mielke OHH. 2005. Synonymic notes on North and Central American Lerodea Scudder, 1872 and Corticea Evans, 1955 (Lepidoptera, Hesperiidae, Hesperiinae). Revista Brasileira de Zoologia 22(1): 285–291. [Google Scholar]
- Williams M 2023a. Afrotropical butterflies: 051aa Genus Ceratrichia Butler, 1870, updated 29 December 2020, <http://www.lepsocafrica.org/?p=publications&s=atb>, accessed on 20 August 2023. [Google Scholar]
- Williams M 2023b. Afrotropical butterflies: 070 Genus Xanthodisca Aurivillius, [1925], updated 9 Feb 2021, <http://www.lepsocafrica.org/?p=publications&s=atb>, accessed on 20 August 2023. [Google Scholar]
- Williams M 2023c. Afrotropical butterflies: 286 Genus Chrysoritis Butler, [1897], updated 7 May 2023, <http://www.lepsocafrica.org/?p=publications&s=atb>, accessed on 20 August 2023. [Google Scholar]
- Willmott KR 2003. Cladistic analysis of the Neotropical butterfly genus Adelpha (Lepidoptera: Nymphalidae), with comments on the subtribal classification of Limenitidini. Systematic Entomology 28(3): 279–322. [Google Scholar]
- Willmott KR, Hall JPW, and Lamas G. 2001. Systematics of Hypanartia (Lepidoptera: Nymphalidae: Nymphalinae), with a test for geographical speciation mechanisms in the Andes. Systematic Entomology 26(4): 369–399. [Google Scholar]
- Zhang J, Cong Q, Burns JM, and Grishin NV. 2022a. Checking the checkered taxonomy of Plötz’s checkered skippers (Hesperiidae: Pyrgini). The Taxonomic Report of the International Lepidoptera Survey 10(5): 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, and Grishin NV. 2023a. Thirteen new species of butterflies (Lepidoptera: Hesperiidae) from Texas. Insecta Mundi 0969: 1–58. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Brockmann E, and Grishin NV. 2019a. Genomes reveal drastic and recurrent phenotypic divergence in firetip skipper butterflies (Hesperiidae: Pyrrhopyginae). Proceedings of the Royal Society B: Biological Sciences 286(1903): 20190609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, and Grishin NV. 2022b. Taxonomic changes suggested by the genomic analysis of Hesperiidae (Lepidoptera). Insecta Mundi 0921: 1–135. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2019b. Changes to North American butterfly names. The Taxonomic Report of the International Lepidoptera Survey 8(2): 1–11. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2019c. Genomics of a complete butterfly continent. bioRxiv BIORXIV/2019/829887. [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2020. Genomic evidence suggests further changes of butterfly names. The Taxonomic Report of the International Lepidoptera Survey 8(7): 1–40. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Opler PA, and Grishin NV. 2021. Genomics-guided refinement of butterfly taxonomy. The Taxonomic Report of the International Lepidoptera Survey 9(3): 1–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, and Grishin NV. 2022c. Genomic DNA sequencing reveals two new North American species of Staphylus (Hesperiidae: Pyrginae: Carcharodini). The Taxonomic Report of the International Lepidoptera Survey 10(4): 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, and Grishin NV. 2023b. A taxonomic list of the Old World genera in the subfamily Hesperiinae (Hesperiidae) arranged into tribes. The Taxonomic Report of the International Lepidoptera Survey 11(2): 1–6.36817548 [Google Scholar]
- Zhang J, Cong Q, Shen J, Song L, Opler PA, and Grishin NV. 2023c. Additional taxonomic refinements suggested by genomic analysis of butterflies. The Taxonomic Report of the International Lepidoptera Survey 11(1): 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cong Q, Song L, Shen J, Léger T, Lamas G, Mielke OHH, and Grishin NV. 2023d. Resolving inconsistencies between Plötz’s descriptions and presumed type specimens of some Hesperiidae (Lepidoptera). Deutsche Entomologische Zeitschrift 70(1): 159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dolibaina DR, Cong Q, Shen J, Song L, Mielke CGC, Casagrade MM, Mielke OHH, and Grishin NV. 2023e. Taxonomic notes on Neotropical Hesperiidae (Lepidoptera). Zootaxa 5271(1): 091–114. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shen J, Cong Q, and Grishin NV. 2019d. Genomic analysis of the tribe Emesidini (Lepidoptera: Riodinidae). Zootaxa 4668(4): 475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


