Abstract
Oxidative stress, resulting from an imbalance in the accumulation and removal of reactive oxygen species such as hydrogen peroxide (H2O2), is a challenge faced by all aerobic organisms. In plants, exposure to various abiotic and biotic stresses results in accumulation of H2O2 and oxidative stress. Increasing evidence indicates that H2O2 functions as a stress signal in plants, mediating adaptive responses to various stresses. To analyze cellular responses to H2O2, we have undertaken a large-scale analysis of the Arabidopsis transcriptome during oxidative stress. Using cDNA microarray technology, we identified 175 non-redundant expressed sequence tags that are regulated by H2O2. Of these, 113 are induced and 62 are repressed by H2O2. A substantial proportion of these expressed sequence tags have predicted functions in cell rescue and defense processes. RNA-blot analyses of selected genes were used to verify the microarray data and extend them to demonstrate that other stresses such as wilting, UV irradiation, and elicitor challenge also induce the expression of many of these genes, both independently of, and, in some cases, via H2O2.
Oxidative stress, arising from an imbalance in the generation and removal of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2), is a challenge faced by all aerobic organisms (Finkel and Holbrook, 2000). Although ROS were originally considered to be detrimental to cells, it is now widely recognized that redox regulation involving ROS is a key factor modulating cellular activities (Allen and Tresini, 2000; Dat et al., 2000).
Increasing evidence indicates that H2O2 functions as a signaling molecule in plants. H2O2 generation during the oxidative burst is one of the earliest cellular responses to potential pathogens and elicitor molecules (Lamb and Dixon, 1997). H2O2 induces the expression of defense-related genes such as GST, encoding glutathione S-transferase, and PAL, encoding Phe ammonia lyase (Levine et al., 1994; Desikan et al., 1998a; Grant et al., 2000). H2O2 also activates mitogen-activated protein kinases (MAPKs), conserved signaling kinases that modulate gene expression and transduce cellular responses to extracellular stimuli (Desikan et al., 1999; Grant et al., 2000; Kovtun et al., 2000; Samuel et al., 2000). Furthermore, several studies indicate that H2O2 is a key factor mediating programmed cell death (PCD) in response to pathogens, elicitors, and hormones (Tenhaken et al., 1995; Levine et al., 1996; Desikan et al., 1998a; Mittler et al., 1999; Solomon et al., 1999; Bethke and Jones, 2001). In addition, two recent studies have shown that H2O2 is synthesized in response to exogenous abscisic acid (ABA), and that H2O2 mediates, at least in part, ABA responses including stomatal closure and gene expression (Guan et al., 2000; Pei et al., 2000).
There are several possible sources of H2O2 in plants, and a number of abiotic and biotic stress stimuli induce H2O2 generation and thereby oxidative stress. Potential enzymatic sources include a plasma membrane-located NADPH oxidase (Desikan et al., 1998b; Keller et al., 1998; Torres et al., 1998), cell wall-bound peroxidases, and amine oxidase (Bolwell and Wojtaszek, 1997). Other sources of ROS include electron transport processes in chloroplasts and mitochondria, and photooxidative stress in peroxisomes (Noctor and Foyer, 1998; Dat et al., 2000). In addition to pathogen challenge, other stimuli that induce H2O2 synthesis and oxidative stress include drought stress and ABA, itself synthesized following loss of turgor (see Dat et al., 2000; Guan et al., 2000; Pei et al., 2000); low and high temperatures (Prasad et al., 1994; Foyer et al., 1997; Dat et al., 1998); excess excitation energy (Karpinski et al., 1999); UV irradiation (A-H-Mackerness et al., 1999); and ozone (Langebartels et al., 2000). A number of similarities can be seen in the cellular responses to these stresses, suggesting that H2O2 could be a common factor regulating various signaling pathways (Neill et al., 1999). For example, it is clear that there are overlapping spectra of genes induced by stresses such as ozone, UV, and pathogen challenge (Langebartels et al., 2000). Furthermore, the phenomenon of cross tolerance, in which exposure to one stress can induce tolerance to other stresses, is one in which H2O2 is likely to play a pivotal role (Bowler and Fluhr, 2000). Identification of genes and proteins regulated by H2O2 is thus an important step toward treatments that might confer tolerance of multiple stresses.
It is already known that H2O2 can induce the expression of genes involved in antioxidant defense (Levine et al., 1994; Karpinski et al., 1999; Morita et al., 1999; Mullineaux et al., 2000). In addition, recent work has shown that H2O2 induces the expression of genes required for peroxisome biogenesis (Lopez-Huertas et al., 2000); peroxisomes are organelles of direct importance for antioxidant defense. In a previous report, we utilized differential mRNA display to identify H2O2-regulated genes in Arabidopsis suspension cultures (Desikan et al., 2000). Here, we use cDNA microarray technology to carry out a transcriptomic analysis of oxidative stress-regulated genes in Arabidopsis. We identify H2O2-regulated genes and use RNA-blot analyses of some of the genes to demonstrate that their expression is also modulated by other stimuli that involve oxidative stress. A substantial proportion of these genes have predicted functions in cell rescue and defense responses, cell signaling, and transcription, implying that H2O2 does have multiple roles in plant responses to stress.
RESULTS
To obtain a global picture of gene expression during oxidative stress, Arabidopsis cultures were exposed to 20 mm H2O2 for 1.5 and 3 h, cells harvested, and mRNA from control and H2O2-treated cells (pooled from 1.5- and 3-h treatments) used as probes for the Arabidopsis Functional Genomics Consortium (AFGC) Cycle 1 microarray. This concentration and the two time points were chosen based on earlier studies (Desikan et al., 1998a, 2000). Arabidopsis cultures have a very high H2O2 scavenging capacity (Desikan et al., 1998a). At the cell density typical of 7-d-old cultures (approximately 0.2 g fresh weight cells mL−1), the half-life of exogenous 20 mm H2O2 is 2 min, and after only 5 min, no H2O2 is detectable (Desikan et al., 1998a). This means that the concentration of H2O2 at its cellular site(s) of action is very much lower than 20 mm. We used two time points because previous work indicates that increased expression of some genes in response to H2O2 is transient (Desikan et al., 1998b, 2000). Thus, the choice of 1.5 and 3 h was an attempt to capture both rapid and longer term changes in gene expression.
Microarray Analysis
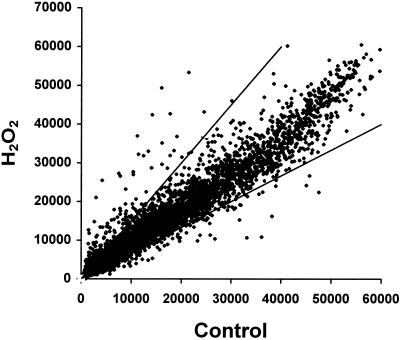
A global representation of the changes in expression of all the expressed sequence tags (ESTs) on the microarray is depicted in Figure 1. For the vast majority of transcripts, expression appeared unchanged with H2O2 treatment (Fig. 1). Using the selection criteria outlined in “Materials and Methods,” and accounting for any ESTs that corresponded to the same GenBank hit (Schaffer et al., 2001), we were able to identify 175 nonredundant ESTs with a change in expression greater than 1.5-fold in response to H2O2 (Fig. 1). Of these, expression of 113 was up-regulated and 62 down-regulated. The AFGC microarray (Cycle 1 experiments) contains approximately 11,000 ESTs, and the redundancy on this AFGC array is approximately 25% (Schaffer et al., 2001); thus, approximately 30% of the genome is represented on the array.
Figure 1.
Scatter plot of signal intensities for all ESTs on the microarray. Normalized channel intensities for each clone on the microarray are plotted with signals from control and H2O2-treated cells on the x and y axes, respectively. The diagonal lines represent 1.5-fold induction/repression ratio cutoffs.
The utility of the microarray system for identifying H2O2-responsive genes is illustrated by the demonstration that ESTs corresponding to the genes PAL1, GST6, and a stress-related protein, previously found to be H2O2 responsive in Arabidopsis suspension cultures (Desikan et al., 1998a, 2000) were among those identified via microarray. Expression data and sequence homologies (based on EST description and BLAST homologies) for H2O2-responsive ESTs are shown in Table I. ESTs that have an average induction greater than 5-fold include those encoding a heat shock protein, a zinc finger protein, calmodulin, and an ethylene-responsive transcription factor (Table I). We also identified ESTs with transcripts of low abundance (i.e. those with low channel intensities post-hybridization) that were H2O2 inducible (Table I). These included ESTs encoding potential signaling proteins such as a heat shock transcription factor, a His kinase, and a protein Tyr phosphatase.
Table I.
Expression data and sequence homologies of identified ESTs
| EST Accession No. | BLAST Hit | Putative ID | Ratio 1 | Ratio 2 | Avea |
|---|---|---|---|---|---|
| Induced genes | |||||
| Cellular organization and biogenesis | |||||
| T44253d | I39698 | Blue copper-binding protein | 4 | 2.8 | 3.4 |
| AA585751d | A84841 | Putative embryo-abundant protein | 2.7 | 2.2 | 2.4 |
| H37424d | T52410 | Blue copper-binding protein | 2.2 | 2.1 | 2.2 |
| T76263de | T05577 | Putative mitochondrial uncoupling protein | 1.8 | 2 | 1.9 |
| T45414de | T51579 | Cellulose synthase catalytic subunitb | 1.5 | 2 | 1.8 |
| H37373f | AB024029 | Outer membrane lipoprotein precursor | 1.5 | 2.2 | 1.8 |
| H36601ej | Q42589 | Lipid transfer proteinb | 1.6 | 1.6 | 1.6 |
| N96229e | T45791 | Lipid transfer proteinb | 1.5 | 1.6 | 1.6 |
| T43399jk | C84606 | ras GTP-binding proteinb | 1.9 | 1.4 | 1.6 |
| Signal transduction | |||||
| R30557d | BAB10479 | Calmodulin | 5.7 | 8.1 | 6.9 |
| H36168jk | S40469 | ATMPK3 | 2.3 | 2.4 | 2.4 |
| N96364dj | T46189 | Calcium-dependent protein kinase | 2.3 | 2.6 | 2.4 |
| T20648d | F84429 | Histidine kinasebc | 1 | 3.4 | 2.2 |
| H76238d | C84473 | Putative protein kinase | 2.3 | 1.9 | 2.1 |
| T88109dk | D96689 | Calmodulin-related protein2 | 1.5 | 2 | 1.8 |
| N37896k | AL132978 | Tyr phosphatasebc | 1.2 | 1.9 | 1.5 |
| Metabolism | |||||
| N38687de | T04731 | Cytochrome P450 | 6.4 | 2 | 4.2 |
| AA394580k | G84823 | Anthocyanin 5-aromatic acyltransferase | 2.4 | 5.2 | 3.8 |
| AA713237ejk | G84787 | PAL1 | 2.3 | 3.9 | 3.1 |
| H36318g | C84531 | Nitrite reductase | 3.7 | 1.8 | 2.7 |
| R90669jk | H84918 | GST6 | 3 | 1.9 | 2.4 |
| T04251j | A96502 | Stearoyl acyl carrier protein | 2.5 | 2.2 | 2.3 |
| R90074dj | T05315 | Pyruvate decarboxylase-1 | 1.8 | 2.8 | 2.3 |
| T45380ek | U96045 | Adenosine-5′-phosphosulfate reductase | 1.8 | 2.6 | 2.2 |
| T43668e | T48390 | Monodehydroascorbate reductase | 1.6 | 2.4 | 2 |
| N37618d | T07698 | Short-chain alcohol dehydrogenase | 1.7 | 2.3 | 2 |
| H76730d | AL078467 | Gln-dependent Asn synthetase | 1.6 | 2.3 | 1.9 |
| N96471d | AL022023 | Phospholipase-like protein | 1.6 | 2.2 | 1.9 |
| N97306k | AB026643 | Myrosinase-binding protein | 1.6 | 2.2 | 1.9 |
| T04244ej | S56653 | Myrosinase/thioglucosidaseb | 1.8 | 1.4 | 1.6 |
| Energy | |||||
| H36160j | AB022223 | Light-induced protein | 4.7 | 2.4 | 3.6 |
| Transcription | |||||
| T41722ej | X98674 | Zn finger protein Zat12 | 7.2 | 9.4 | 8.3 |
| AA042693e | T49070 | Heat shock transcription factorb | 5.3 | 9.7 | 7.5 |
| AA720262d | BAB01997 | Ethylene-responsive transcription factor | 2.6 | 7.6 | 5.1 |
| H77041j | T51833 | DREB2A | 4 | 3.3 | 3.6 |
| AA720306d | E96663 | RING Zn finger protein | 3.1 | 3.4 | 3.3 |
| ATHATL2Ad | T52079 | Zn finger protein | 2.8 | 2.9 | 2.9 |
| N65178dk | T50672 | Putative Zn finger protein | 2.4 | 2.8 | 2.6 |
| T75904d | AAC6343 | Zn finger protein (CONSTANS-like B Box)b | 3.1 | 1.8 | 2.5 |
| AA720308d | – | bHLH protein | 2.3 | 2.3 | 2.3 |
| T21879dj | AF022658 | Zn finger transcription factor | 2.7 | 1.7 | 2.2 |
| R29900fjk | AB012239 | Ethylene-responsive element binding factor (EREBP-4) | 1.7 | 2.6 | 2.2 |
| T44892j | F84792 | Zn finger protein (C2H2 type)b | 2.7 | 1.7 | 2.2 |
| N38276j | T49142 | CCR4-associated factor | 2 | 2.2 | 2.1 |
| R30043jk | AC011708 | RING Zn finger protein | 2.4 | 1.8 | 2.1 |
| T41938dk | T02684 | DNA-binding protein CCA1 | 1.9 | 2.3 | 2.1 |
| N97162d | E71444 | EREBP | 1.7 | 2.5 | 2.1 |
| R30551jk | H86320 | myb-Related transcription factor | 2.1 | 1.8 | 2 |
| H37631j | S37100 | ATAF 2 protein | 2.1 | 1.9 | 2 |
| T43796e | AF132016 | RINGH2 Zn finger protein | 1.6 | 2.2 | 1.9 |
| AA404906dj | A84682 | Zn finger/DNA binding proteinb | 1.9 | 1.4 | 1.6 |
| Protein destination and transport | |||||
| T04221d | AB022214 | 2S seed storage protein | 2.1 | 2 | 2 |
| T20525j | T48847 | Syntaxinb | 1.8 | 2 | 1.9 |
| T44252j | T04623 | 12S cruciferin seed storage proteinb | 2.1 | 1.3 | 1.7 |
| Cell rescue/defense | |||||
| AA042551d | AAC95188 | Small heat shock protein | 7.4 | 16 | 11.7 |
| R29801dj | A45508 | hsp 83 | 6.1 | 9 | 7.6 |
| AA597555h | T49264 | hsp 17 | 3.7 | 5.9 | 4.8 |
| AA650758df | T48562 | hsp | 3.3 | 4.2 | 3.7 |
| N38277dk | T05001 | Gamma glutamyl transferaseb | 3 | 2.6 | 2.8 |
| AA042089jk | AAC19273 | Similar to several small proteins inducible by various stresses | 3.1 | 2.1 | 2.6 |
| AA394361jk | BAB10761 | Salt-inducible calcium-binding protein | 3.1 | 2.1 | 2.6 |
| R29894dj | A84824 | Putative nematode resistance protein | 2.5 | 2.4 | 2.4 |
| W43132j | AC006577 | hsp20/Alpha crystallin family | 1.9 | 2.8 | 2.4 |
| N38383dk | BAB01138 | Jasmonic acid (JA)-inducible proteinb | 1.9 | 2.6 | 2.3 |
| AA042366j | G96604 | hsp (DNAJ like)b | 2.1 | 2.2 | 2.2 |
| AA597442h | AC20579 | hsp101 | 1.9 | 2 | 2 |
| AA042774f | AAD25796 | rab28b | 1.9 | 1.9 | 1.9 |
| AA720285d | U75202 | Germin-like proteinb | 1.9 | 1.9 | 1.9 |
| T04362e | T00971 | Cf2-like disease resistance proteinb | 1.9 | 1.9 | 1.9 |
| T22424jk | AJ249794 | Lipoxygenaseb | 2.1 | 1.2 | 1.7 |
| AA720182d | G96806 | Thaumatin-like proteinb | 1.5 | 1.9 | 1.7 |
| R65190dj | X98775 | Peroxidaseb | 1.9 | 1.2 | 1.6 |
| N95898d | AP000377 | Selenium-binding proteinb | 1.2 | 1.9 | 1.6 |
| AA040958d | Y12673 | Germin-like auxin-induced proteinb | 1.2 | 1.8 | 1.5 |
| Unknown/unclassified | |||||
| T88199j | T51418 | Unknown | 8.8 | 4.1 | 6.5 |
| T46413fj | B84778 | Unknown | 3.3 | 4.5 | 3.9 |
| R30050i | T05004 | Unknown | 3.7 | 3.5 | 3.6 |
| T13956dj | F96649 | Unknown | 3.9 | 2.8 | 3.4 |
| AA394803j | BAB08381 | Unknown | 3.3 | 3.5 | 3.4 |
| N65722k | AL392144 | Unknown | 3.8 | 2.6 | 3.2 |
| AA394358d | G96596 | Unknown | 2.9 | 3.3 | 3.1 |
| AA394561djk | AC002343 | Unknown | 3.5 | 2.5 | 3 |
| T88573dej | BAB10082 | Unknown | 2.5 | 3.5 | 3 |
| AA712985e | A85024 | Unknown | 3.4 | 2.4 | 2.9 |
| T42821d | A96697 | Unknown | 2 | 3.3 | 2.7 |
| N95870de | BAB09266 | Unknown | 3.2 | 1.9 | 2.6 |
| AA041108d | F84603 | Unknown | 2.6 | 2.6 | 2.6 |
| AA712865 | – | No protein match | 2.8 | 2.2 | 2.5 |
| N37850dj | C96721 | Unknown | 2.8 | 1.6 | 2.2 |
| R30393djk | T48018 | Unknown | 2.6 | 1.8 | 2.2 |
| AA404905d | D96613 | Unknown | 2.6 | 1.7 | 2.2 |
| T44436d | C86221 | Similar to yeast (Saccharomyces cerevisiae) hypothetical protein | 2.2 | 1.9 | 2.1 |
| N97061j | AC007396 | Unknown | 2.5 | 1.7 | 2.1 |
| H76737djk | T47990 | Unknown | 2.3 | 1.9 | 2.1 |
| N96660k | AB010698 | Unknown | 1.8 | 2.4 | 2.1 |
| AA651091d | AC102562 | Unknown | 2.3 | 1.8 | 2.1 |
| R30143df | C86410 | Unknown | 2.3 | 1.6 | 2 |
| AA395824j | AB005240 | Unknown | 2.2 | 1.9 | 2 |
| T20758 | – | No protein match | 1.9 | 2.2 | 2 |
| W43585 | – | No protein match | 1.8 | 2.2 | 2 |
| H36052d | H96798 | Unknown | 2.1 | 1.7 | 1.9 |
| R30074de | D85014 | Unknown | 1.6 | 2.2 | 1.9 |
| T46201dj | AB010069 | Unknown | 2.3 | 1.6 | 1.9 |
| T46597djk | T48305 | Unknown | 2.3 | 1.6 | 1.9 |
| H37568k | AF326884 | Unknown | 2.2 | 1.7 | 1.9 |
| AA394961dj | T01480 | Unknown | 2.2 | 1.7 | 1.9 |
| AA395225d | F86200 | Unknown | 2.2 | 1.6 | 1.9 |
| AA651022k | T14084 | Unknown | 1.5 | 2.2 | 1.9 |
| R86787dej | T48437 | Unknown | 1.5 | 2.4 | 1.9 |
| T45451dj | E86490 | Unknown | 1.5 | 2.2 | 1.9 |
| N96028jk | A86350 | Unknown | 1.6 | 2 | 1.8 |
| AA042745d | AB016870 | Lipase/hydrolase-like proteinb | 1.8 | 1.3 | 1.6 |
| N96263l | AF160973 | shyc pr (Mouse/p53-inducible protein)b | 1.9 | 1 | 1.5 |
| Repressed genes | |||||
| Metabolism | |||||
| N95951 | AL080254 | Berberine bridge enzyme-like protein | 3.1 | 5.6 | 4.3 |
| T43750 | AL022140 | Pectinesterase-like protein | 2.4 | 4.4 | 3.4 |
| T88304 | AC007169 | Stearoyl-coenzyme A desaturase | 1.7 | 4.1 | 2.9 |
| N96662 | AC008046 | Putative pectinesterase | 2.4 | 2.6 | 2.5 |
| H36710 | AP001297 | Nucleotide sugar epimerase-like protein | 1.9 | 2.6 | 2.2 |
| T04189 | AC008075 | Alpha xylosidase precursor | 1.8 | 2.4 | 2.1 |
| R30147 | AC007797 | Sulfate adenylyl transferase | 1.6 | 2.6 | 2.1 |
| Cellular organization and biogenesis | |||||
| AA042610 | AC006072 | Lipid transfer protein | 2.7 | 6 | 4.3 |
| T46457 | AC004238 | Extensin homolog | 2.4 | 3.4 | 2.9 |
| Energy/chloroplast located | |||||
| N96255 | AP000423 | NADH plastoquinone oxidoreductase | 2.6 | 3.3 | 2.9 |
| T04794 | AP000423 | Chloroplast 50S ribosomal protein | 2.3 | 3.4 | 2.9 |
| N95853 | AP000423 | 30S ribosomal protein | 2.9 | 2.1 | 2.5 |
| AA042459 | AP000423 | Photosystem I chl A ribosomal protein | 1.8 | 3.1 | 2.5 |
| T76754 | AP000423 | Ribosomal protein L32 | 1.6 | 3.3 | 2.4 |
| N96876 | AC007654 | Photosystem I subunit III | 1.5 | 3.1 | 2.3 |
| W43885 | AP000423 | Photosystem I protein | 2.2 | 1.9 | 2 |
| AA712552 | AP000423 | Photosystem II reaction center protein | 1.8 | 2.1 | 2 |
| T21974 | AP000423 | Chloroplast protease-like protein | 1.7 | 2.1 | 1.9 |
| W43521 | AP000423 | Photosystem II 44-kD reaction center precursor | 1.6 | 2.2 | 1.9 |
| Transcription | |||||
| H77088 | AC006592 | Homoeodomain transcription factor | 2.1 | 2.6 | 2.3 |
| Protein distribution/destination/transport | |||||
| AA042384 | AC006922 | p-Glycoprotein | 1.9 | 2.7 | 2.3 |
| AA042644 | AT08315 | Calnexin homolog precursor | 1.7 | 2.7 | 2.2 |
| T22409 | AL035679 | Cys protease RD19A precursor | 1.7 | 2.2 | 2 |
| W43857 | P98205 | Phospholipid-transporting ATPase | 1.5 | 2.3 | 1.9 |
| R83980 | AB008267 | Cys protease RD21A precursor | 1.6 | 2 | 1.8 |
| AA597751 | B84606 | Putative ATP synthase | 1.5 | 2.2 | 1.8 |
| T22351 | AB028611 | Gamma tonoplast intrinsic protein | 1.5 | 2 | 1.8 |
| Cell rescue/defense | |||||
| W43838 | AC002339 | Receptor-like protein kinase | 2 | 2.2 | 2.1 |
| T21130 | AC007017 | Harpin-induced protein | 1.6 | 2.1 | 1.9 |
| Unknown/unclassified | |||||
| AA598175 | – | No protein match | 2.6 | 5.3 | 4 |
| AA597909 | – | No protein match | 2.9 | 4.8 | 3.8 |
| A394830 | – | No protein match | 2.9 | 3.6 | 3.2 |
| AA713102 | – | No protein match | 2.8 | 3.3 | 3 |
| N96775 | – | No protein match | 3.3 | 2.6 | 2.9 |
| AA394802 | – | No protein match | 2.6 | 3.1 | 2.9 |
| T75890 | AC011663 | Unknown | 2.3 | 3.5 | 2.9 |
| N96785 | – | No protein match | 2.5 | 2.7 | 2.6 |
| T43336 | AC069471 | Unknown | 1.9 | 3.4 | 2.6 |
| H37549 | AC010155 | Unknown | 2.1 | 2.9 | 2.5 |
| H37081 | A71406 | Unknown | 2.3 | 2.5 | 2.4 |
| AA586206 | AC011664 | Unknown | 2.1 | 2.7 | 2.4 |
| N97271 | C85084 | Unknown | 1.5 | 3.3 | 2.4 |
| T43162 | F96511 | Unknown | 2.6 | 1.8 | 2.2 |
| AA395049 | AC009853 | Unknown | 2.2 | 2.1 | 2.2 |
| H37137 | AB005246 | Unknown | 2.1 | 2.3 | 2.2 |
| N96223 | AC000104 | Ankyrin-like protein | 1.6 | 2.8 | 2.2 |
| T42914 | AL021687 | Unknown | 2.3 | 1.9 | 2.1 |
| T42914 | AL021687 | Unknown | 2.3 | 1.9 | 2.1 |
| N96632 | AC013483 | Unknown | 1.9 | 2.4 | 2.1 |
| AA042200 | T05877 | Unknown | 1.7 | 2.5 | 2.1 |
| T04212 | T10238 | Unknown | 1.5 | 2.8 | 2.1 |
| R84204 | AB007649 | Unknown | 1.5 | 2.7 | 2.1 |
| AA395457 | AL023094 | Auxin-regulated protein | 2.2 | 1.7 | 2 |
| AA598031 | C84663 | Unknown | 2.1 | 1.9 | 2 |
| AA394401 | AC013430 | Unknown | 1.9 | 2 | 2 |
| N96862 | – | No protein match | 2.2 | 1.6 | 1.9 |
| AA042489 | G84923 | Unknown | 2 | 1.9 | 1.9 |
| T42055 | – | No protein match | 1.8 | 2 | 1.9 |
| AA042412 | – | No protein match | 1.7 | 2.1 | 1.9 |
| AA042748 | – | No protein match | 1.6 | 2.3 | 1.9 |
| T45103 | D84447 | Unknown | 1.6 | 2 | 1.8 |
| H37648 | B96534 | Unknown | 1.6 | 2.1 | 1.8 |
| AA041029 | AP002048 | Unknown | 1.5 | 2 | 1.8 |
Average channel intensity ratio of H2O2-treated over control-treated cells (induced) or vice versa (repressed) from two independent experiments (ratio 1 and ratio 2).
Low-abundance genes with channel intensity between 300 and 1,000.
Low expressed ESTs whose expression was confirmed by RNA dot blot analysis.
d–k Potential binding sites present in 1.1-kb upstream sequence of genomic sequences for ESTs, for redox-sensitive transcription factors (identified using PLACE database; H2O2-repressed genes not analyzed).
MYBCORE, CNGTTR (for all animals and AtMyb1/2).
MYBPLANT, MACCWAMC (all plant Mybs).
MYB2AT, TAACTG (AtMyb2).
MYBPZM, CCWACC (maize myb).
MYBST1, GGATA (potato mybst1).
MYBGAHV, TAACAAA (GA-regulated myb).
Ocs (AS1 element) binding site, TGACG.
AP1/3 binding site, TGAGTCA.
No upstream sequences identified in the genome.
Functional Classification of Genes
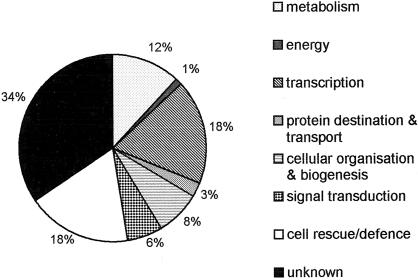
To determine whether H2O2 regulates the differential expression of only particular classes of genes, a functional classification of all the H2O2-induced ESTs was performed (Table I, Fig. 2). This was achieved using BLAST homology searches of all the ESTs, with potential functions then being categorized as in the Munich Information Center for Protein Sequences Arabidopsis database (see “Materials and Methods”). The broad spectrum of gene functions depicted in Figure 2 is similar to that predicted for the entire Arabidopsis genome (The Arabidopsis Genome Initiative, 2000). However, as might be expected, the representation of ESTs involved in cell rescue/defense responses is increased. Twenty of the H2O2-induced ESTs encode proteins with potential functions in transcription, indicating that the subsequent expression of further genes is likely at later time points. A third of the H2O2-induced ESTs are unknown, similar to the proportion of unknown genes in the entire genome.
Figure 2.
Functional distribution of oxidative stress-induced genes. The top BLASTX hits of the 113 ESTs were classified according to the functional organization of the Arabidopsis genome (The Arabidopsis Genome Initiative, 2000).
Expression Analyses of Selected Genes
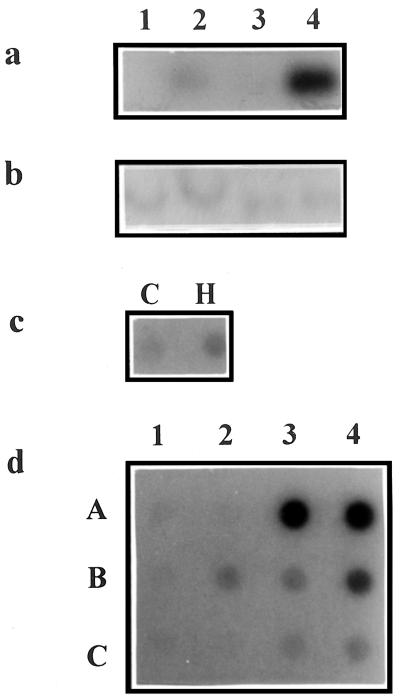
To validate and extend the microarray data, the expression of 14 ESTs was monitored by RNA-blot analyses (Fig. 3, Table II). This was to verify the effects of H2O2 on the expression of ESTs with high and low abundance transcripts, and to investigate the effects of other stress stimuli such as wilting, UV irradiation, and elicitation. Representative northern and dot blots are shown in Figure 3. Induction by H2O2 of ESTs encoding calmodulin (Fig. 3a) and a heat shock protein (Fig. 3d) is evident.
Figure 3.
RNA blot analyses of selected H2O2-induced genes. a, Northern analysis of a calmodulin (accession no. R30557): lanes 1 and 2, total RNA from control and H2O2 (20 mm, 1.5 and 3 h pooled)-treated cells; lanes 3 and 4, mRNA from control and H2O2-treated cells, respectively. b, Control northern blot: RNA used in a was hybridized to EF1α cDNA to confirm equal loadings. c, RNA dot-blot analysis of calmodulin in control (C) and H2O2 (500 μm, H)-treated cells at 0.02 g fresh weight cells mL−1. d, RNA-blot analyses of an hsp (accession no. AA042551): row A, time course analysis; lane 1, control; lanes 2 through 4, cells treated with H2O2 (20 mm) for 0.5 , 1.5, and 3 h. Row B, Dose response; lane 1, control; lanes 2–4, cells treated with 5, 10, and 20 mm H2O2 for 1.5 and 3 h (pooled RNA). Row C, Expression in rosette leaves: lane 1, leaves mock infiltrated with water; lanes 2 through 4, leaves vacuum infiltrated with 20 mm H2O2 for 0.5, 1.5, and 3 h.
Table II.
Expression analysis of selected H2O2-induced genes via RNA blots
| EST Accession No. (Gene) | Treatmenta
|
||||||
|---|---|---|---|---|---|---|---|
| H2O2 (cells) | H2O2 (leaves) | Wilt | Wilt + AsA | UV | UV + AsA | Harpin | |
| R30557 (Calmodulin) | 2.6 | 8 | 2.6 | 1.3 | 2.5 | 2 | 1.5 |
| N37896 (Tyr phosphatase)b | 1.6 | 1.5 | 1.9 | 2.5 | 2 | 1.5 | – |
| AA042551 (small hsp) | 14 | 2.8 | 7.5 | 6.5 | 3 | 2.4 | 2 |
| H77041 (DREB2A) | 2.3 | 1.6 | 1.8 | 0.9 | 2 | 1.9 | 2.5 |
| N65549 (disease resistance protein) | 1.9 | 2.1 | 2.2 | 2.7 | 2.4 | 0.9 | 1.5 |
| H36168 (ATMPK3) | 2.2 | 2.5 | 1.6 | 0.6 | 8.2 | 7.8 | 1.2 |
| AA720262 (ethylene-responsive transcription factor [TF]) | 7.4 | 1.8 | 1.9 | 2.3 | 5.1 | 2 | – |
| T20525 (syntaxin)b | 3 | 3 | – | – | 3 | 2.7 | 7 |
| R30551 (myb-related TF) | 2 | – | 2.3 | 5.2 | 3 | 1.7 | – |
| H76238 (protein kinase) | 1.8 | 1.7 | 1.8 | 2.7 | 1.7 | 0.4 | 2.9 |
| T44253 (blue Cu-binding protein) | 1.5 | 5.7 | 1.9 | 1.8 | 4 | 5.2 | 1.9 |
| T41722 (Zn finger protein) | 9.2 | 2.6 | 3.5 | 1 | 3 | 1.9 | 3.8 |
| T20648 (His kinase)b | 1.5 | 1.5 | 1.4 | 1 | 1.5 | 1.5 | – |
| N97306 (myrosinase-binding protein) | 3.8 | – | 1.5 | 1.7 | 3.5 | 0.8 | – |
Figures in table represent fold-induction over appropriate controls (either water or ascorbic acid [AsA] treatment alone).
Low abundance genes identified on the microarray.
RNA-blot analysis confirmed that the expression of all the 14 ESTs was H2O2 responsive. Although the absolute values of fold induction were not identical to those on the array (Tables I and II), a similar trend was observed. Moreover, the H2O2 inducibility of three “low abundance” genes from the array, encoding a potential His kinase, a protein Tyr phosphatase, and a syntaxin, was confirmed by the dot-blot analysis, demonstrating the veracity of the microarray data. The effects of H2O2 on the expression of 14 ESTs was determined in rosette leaves, which, like suspension cultures, also have a high H2O2-scavenging capacity (data not shown). The expression of most of the ESTs was increased by H2O2 treatment, demonstrating that differentiated tissues responded similarly to H2O2 in terms of gene expression (Table II). The H2O2 sensitivity of some of the ESTs was also assessed in cell culture using various lower doses of H2O2. Lower density cell cultures have considerably reduced H2O2-scavenging capacity, such that exogenous H2O2 is correspondingly much longer lived. For example, at a culture density of approximately 0.02 g fresh weight cells mL−1, the half-life of 20 mm H2O2 is approximately 60 min (at 0.2 g fresh weight cells mL−1, the half-life of 20 mm H2O2 is approximately 2 min). We found that at this culture density, the expression of ESTs encoding calmodulin (Fig. 3c), DREB2A, and a protein kinase was induced by exposure to 500 μm H2O2, and that the expression of ESTs encoding a His kinase, a protein Tyr phosphatase, a blue copper-binding protein, and a disease resistance protein was induced by H2O2 at concentrations down to 200 μm (data not shown). These data demonstrate that Arabidopsis cells can perceive and respond to low concentrations of exogenous H2O2.
The effects of other stress stimuli on expression of the 14 selected ESTs was also assessed. Wilting results in oxidative stress, and ABA, synthesized in response to drought stress, induced H2O2 synthesis in both Arabidopsis guard cells (Pei et al., 2000) and maize (Zea mays) plants and cell cultures (Guan et al., 2000). Moreover, H2O2 partly mediated the effects of ABA on both gene expression and stomatal closure (Guan et al., 2000; Pei et al., 2000). As a consequence, the effects of wilting, in the absence and presence of the antioxidant ascorbic acid, were determined. Plants were subjected to rapid dehydration and then RNA isolated after 2 h of incubation. It can be seen in Table II that this wilting treatment induced the expression of H2O2-responsive ESTs. However, the effects of dehydration on the expression of some of these ESTs was partly mediated by H2O2. Pretreatment with the antioxidant ascorbic acid reduced the wilt-induced expression of ESTs encoding calmodulin, DREB2A transcription factor, the MAP kinase ATMPK3, and a zinc finger protein.
UV irradiation is another abiotic stimulus that causes oxidative stress in plants and induces genes that are responsive to oxidative stress (A-H-Mackerness et al., 1999, 2001). Thus, we determined the expression of the H2O2-responsive genes in plants that had been exposed to UV irradiation. All of the ESTs analyzed were induced by UV (Table II). Pretreatment with ascorbic acid reduced the effects of UV on the expression of several ESTs (disease resistance protein, ethylene-responsive transcription factor, myb-related transcription factor, protein kinase, and myrosinase-binding protein), suggesting that for these genes the effects of UV were partly mediated by H2O2.
H2O2 generation is a rapid response to potential pathogens and microbial elicitors (Lamb and Dixon, 1997), and involved in the activation of various signaling pathways (Bolwell, 1999). As a consequence, we determined the effects of harpin, a bacterial elicitor previously shown to induce H2O2 production and defense responses in Arabidopsis cells (Desikan et al., 1996, 1998a, 1999). Harpin induced the expression of many of the oxidative stress-responsive genes in suspension cultures (Table II), suggesting a potential role for these genes in host defense.
DISCUSSION
H2O2 is now widely recognized as a key signaling molecule in all eukaryotes, including plants. Generation of H2O2 occurs under a diverse range of conditions, and it appears likely that H2O2 accumulation in specific tissues, and in the appropriate quantities, is of benefit to plants and can mediate cross tolerance toward other stresses (Bolwell, 1999; Bowler and Fluhr, 2000). H2O2 is intimately involved in plant defense responses, affecting both gene expression and the activation of proteins such as MAP kinases, which in turn function as regulators of transcription (Desikan et al., 1998a, 1999, 2000; Mittler et al., 1999; Kovtun et al., 2000). It is clear, then, that identification of all the changes in gene expression regulated by oxidative stress is of considerable interest. However, a global analysis of the effects of H2O2 on the transcriptome of any one plant species has not yet been described.
The development of publicly funded and accessible resources such as the AFGC microarray facility means that it has become possible to undertake large-scale studies at the genome level. We accessed the AFGC microarray (Cycle 1) to identify H2O2-responsive genes expressed in Arabidopsis suspension cultures. Our data were obtained using the normalized mean intensity ratios derived from two independent experiments. We chose a minimum threshold figure of 1.5-fold change in intensity ratio; however, most of the ESTs that we identified were 2-fold or more induced. It is possible that small changes in gene expression could result in larger changes in protein levels, especially within specific subcellular compartments. Proteomic analysis will be required to correlate changes in protein and RNA content. It is important, however, that we utilized RNA-blot analysis to verify the microarray data: 14 ESTs identified as being H2O2 responsive (some of which are 1.5-fold induced) via microarray were confirmed as H2O2 responsive, including those that were classed as “low abundance” genes on the microarray. Our data indicate that 1% to 2% of the genes represented on the array (taking into account redundancy) is affected by oxidative stress imposed by H2O2. This figure is comparable with those determined for oxidative stress in yeast (Godon et al., 1998; Gasch et al., 2000) and pathogen challenge for plant cells (Durrant et al., 2000; Maleck et al., 2000). It should be noted that the AFGC microarray used here is estimated to represent only about 30% of the Arabidopsis genome, depending on redundancy. Moreover, we used RNA from undifferentiated suspension cultures as the hybridization probes. However, suspension cultures do represent excellent model systems (e.g. McCabe and Leaver, 2000), and many of the genes analyzed by RNA blots were found to be similarly H2O2 responsive in rosette leaves (Table II). Although this study was restricted to an analysis of ESTs that are responsive to H2O2, it does identify those genes necessary to form the basis of further studies using gene-specific sequences to analyze the expression and function of the genes that are sensitive to H2O2.
Of the 175 genes identified as being H2O2 responsive, most do not have an obvious direct role in oxidative stress. However, roles in other abiotic and biotic stresses and developmental processes that might be linked to oxidative stress could explain their sensitivity to H2O2. The genes that were sensitive to H2O2 have a range of potential functions based on their sequence homologies (Table I, Fig. 2). Some of these genes are discussed in more detail below.
Several ESTs encoding heat shock proteins were induced by H2O2. Heat stress stimulates H2O2 generation in plants (Foyer et al., 1997; Dat et al., 1998). Moreover, heat shock proteins are involved in enhancing survival following oxidative stress in yeast, animals, and plants (Banzet et al., 1998; Godon et al., 1998; Finkel and Holbrook, 2000). Thus, the induction of genes encoding heat shock proteins and a heat shock transcription factor by H2O2 may lead to increased tolerance of further oxidative stress, as in tomato (Lycopersicon esculentum) cells (Banzet et al., 1998), as well as contributing to tolerance of other stresses such as pathogen challenge (Vallelian-Bindschedler et al., 1998) or high temperatures (Dat et al., 1998). It is interesting that one of the heat shock protein genes was also induced independently by wilting, UV irradiation, and elicitation (Table II), demonstrating signaling cross talk.
A calmodulin gene was also strongly induced by H2O2. Calmodulin is a calcium-binding protein that may well have a pivotal role in stress tolerance. Intracellular calcium concentrations increase in response to oxidative stress (Price et al., 1994), and calcium influx is required for the activation of ROS generation (Schwacke and Hager, 1992; Baker et al., 1993; Desikan et al., 1997; Harding et al., 1997). Furthermore, NADPH oxidase, a potential ROS-generating enzyme, contains EF hand calcium-binding motifs (Desikan et al., 1998b; Keller et al., 1998), and at least one of the NADPH oxidase genes is induced by H2O2 (Desikan et al., 1998b). These observations suggest that H2O2 induction of a calmodulin might, at least in part, be regulating the activity of this enzyme. Moreover, a calmodulin has been shown to mediate between calcium and ROS generation in tobacco (Nicotiana tabacum) cells undergoing the hypersensitive response (HR). Calmodulin is a regulator of NAD kinase, which generates NADPH for NADPH oxidase activity (Harding et al., 1997). Thus, a significant amount of cross talk occurs between ROS and calcium, and both these signaling molecules mediate cross tolerance to a variety of stresses (Bowler and Fluhr, 2000).
One of the genes identified via microarray analysis as being expressed at low levels but H2O2 responsive was that encoding a protein Tyr phosphatase. Protein Tyr phosphatases are important signaling enzymes that regulate protein phosphorylation events in all eukaryotes (Walton and Dixon, 1993; Fauman and Saper, 1996), particularly the inactivation of MAPK cascades (Luan, 1998). Oxidative stress activates MAPK cascades not only in plants (Desikan et al., 1999; Kovtun et al., 2000), but also in animals (Fialkow et al., 1994; Irani et al., 1997), where Tyr phosphatases have been identified as a primary target for H2O2 (Wu et al., 1998). The Tyr phosphatase identified as H2O2 inducible in the present study was also induced by wilting and UV irradiation. An Arabidopsis protein Tyr phosphatase has previously been identified that is transcriptionally regulated by environmental stresses such as cold and salt stress (Xu et al., 1998).
Among the genes induced by H2O2 was one encoding a blue copper-binding protein. Such proteins might function to sequester copper, a potentially toxic element that is also an essential cellular catalyst for redox reactions (Himelbau and Amasino, 2000). This gene was also induced by wilting, UV, and harpin (Table II), and its expression up-regulated in senescent leaves (data not shown). Genes encoding blue copper-binding proteins have been shown previously to respond to abiotic stresses such as drought and ozone (Cho, 1997; Langebartels et al., 2000), and expression increased during senescence, in which copper sequestration is an important event (Himelbau and Amasino, 2000). A regulatory role for ROS such as H2O2 has been implicated during senescence (Pastori and del Rio, 1997), and we have already shown that H2O2 induces the expression of a senescence-related gene (Desikan et al., 2000). Thus, it is not surprising that there are genes that are induced by both oxidative stress and senescence.
The expression of genes encoding a mitochondrial uncoupling protein, pyruvate decarboxylase, and a myb-related transcription factor were induced by H2O2. Mitochondrial uncoupling proteins are key factors regulating ATP synthesis and generation of ROS in mitochondria, this redox balance affecting the longevity of organisms (Finkel and Holbrook, 2000). Moreover, a gene encoding such a protein was found to be highly up-regulated in mammalian cells induced to undergo PCD (Voehringer et al., 2000). Pyruvate decarboxylase catalyzes the decarboxylation of pyruvate to acetaldehyde and CO2 during ethanolic fermentation as a result of oxygen deprivation. Pyruvate decarboxylase was found to be induced during oxygen deprivation stress in rice (Oryza sativa) seedlings (Minhas and Grover, 1999), and plants expressing a bacterial pyruvate decarboxylase showed enhanced levels of cell death in response to pathogen challenge (Tadege et al., 1998), suggesting that sugar metabolism is a crucial activity during the HR and other stresses. Myb genes represent a large gene family in Arabidopsis (Kranz et al., 1998) and a myb oncogene homolog has been implicated as a critical regulator of the HR cell death pathway (Daniel et al., 1999). Moreover, myb transcription factors possess conserved amino acid motifs that are redox sensitive (Myrset et al., 1993). H2O2 induces PCD in Arabidopsis and other species (Levine et al., 1994; Desikan et al., 1998a; Mittler et al., 1999; Solomon et al., 1999); consequently, the expression of potential PCD-related genes following H2O2 treatment might be expected.
Some of the H2O2-sensitive genes could also be involved in plant hormone signaling. For example, a gene encoding a syntaxin was identified as H2O2 responsive by both microarray and RNA-blot analyses. Syntaxins are docking proteins involved in vesicle trafficking, and a role in the hormonal control of guard cell ion channels has been demonstrated for an ABA-inducible syntaxin in tobacco (Leyman et al., 1999). Because both elicitors and ABA induce H2O2 production in guard cells (Lee et al., 1999; Pei et al., 2000), it could be that induction of a syntaxin by H2O2 is involved in regulating guard cell functioning. Genes encoding myrosinase binding proteins and JA-inducible proteins were shown to be H2O2 responsive on the microarray. Myrosinases are enzymes involved in the degradation of glucosinolates, and a myrosinase-binding protein was found to be induced by both wounding and dehydration (Reymond et al., 2000). We also found that wilting induced the expression of a gene encoding a myrosinase-binding protein. Levels of JA and ROS increase with water stress, which might lead to the induction of such genes.
Various genes encoding transcription factors were induced by H2O2, suggesting that these transcription factors mediate further downstream H2O2 responses, and that several other genes are likely to be induced at later times. Transcription factors have been reported to be rapidly induced during defense responses (Rushton and Somssich, 1998; Durrant et al., 2000). Among the transcription factors induced by H2O2, EREBP and DREB2A are important ones that regulate gene expression during various stresses (Liu et al., 1998, Riechmann and Meyerowitz, 1998). EREBP was also induced during Cf-9: Avr9 interactions in tobacco (Durrant et al., 2000). Other transcription factors induced by H2O2 include a myb-related TF, several zinc finger proteins, and a heat shock transcription factor. Zinc finger proteins have wide-ranging functions and several types exist in plants (Takatsuji, 1999). The involvement of zinc finger transcription factors in stress responses has been reported. For example, during barley-powdery mildew interactions, a zinc finger protein was identified as a key mediator of R gene-induced resistance responses such as H2O2 generation (Shirasu et al., 1999). Furthermore, during Avr-9: Cf-9 interactions in tomato, a gene encoding a zinc finger protein was induced (Durrant et al., 2000), and other stresses such as UV, high salinity, ozone, and wounding also induce this class of genes (Takatsuji, 1999).
Exogenous H2O2 not only activated gene expression, but also repressed the expression of some genes (Table I). Oxidative stress represses several genes in animals (Morel and Barouki, 1999), and on our array, the expression of 62 genes was down-regulated. Many of these encode proteins of unknown function. It is interesting to note that genes encoding a receptor protein kinase and Cys proteases were repressed by H2O2.
The microarray analysis has identified a number of ESTs regulated by oxidative stress that are of potential importance to diverse stress responses. Coregulation of these genes by various stresses supports the hypothesis that H2O2 mediates cross tolerance (Bowler and Fluhr, 2000). However, it is likely that the exact mechanism and levels of expression of individual genes is dependent on cell type and the specific stress stimulus. Coordinated expression of several genes in response to a specific stimulus can be achieved via the interaction of transcription factors with cis-elements common to the promoter regions of those genes. For example, the WRKY binding site was identified in the promoter region of all 26 genes making up the “pathogen regulon” in Arabidopsis (Maleck et al., 2000). Analysis of the 1.1-kb 5′-upstream region of all the oxidative stress-induced genes did not reveal the presence of a known binding site common to them all. However, we identified 5′-upstream regions in the H2O2-induced genes that are potential binding sites for redox-sensitive transcription factors (see Table I). These included binding sites for myb (Myrset et al., 1993; Yang and Klessig, 1996), Ocs/AS-1-like elements (that are present in SA- and auxin-induced genes; Qin et al., 1994) and AP-1 (Abate et al., 1990). Such redox-sensitive motifs have previously been identified in H2O2-induced genes such as tcI7 (Etienne et al., 2000) and GST6 (Chen et al., 1996). However, our identification of potential binding sites for redox-sensitive transcription factors is as yet merely an observation. Confirmation of functional significance will require promoter analysis.
The degree of induction or repression of individual genes by specific stimuli will clearly depend on a complex interaction of all the components of the transcription apparatus with all the regulatory sequences associated with the gene. The promoters of several of the oxidative stress-responsive genes identified here also contain known binding sites for WRKY (Maleck et al., 2000) and bZIP transcription factors (Kim et al., 1997). Moreover, it is interesting that expression of the MAP kinase ATMPK3 is induced by oxidative stress, that oxidative stress also activates the Arabidopsis MAP kinases ATMPK3 and ATMPK6, and that such activation can itself mediate the induction of oxidative stress-responsive genes (Desikan et al., 1999; Kovtun et al., 2000).
Our data demonstrate that H2O2 can modulate the expression of a subset of genes within the Arabidopsis genome. Furthermore, it is also clear from other studies that H2O2 can alter the activity of cellular proteins. The mechanisms by which these changes are effected remain to be elucidated. It is possible that in some cases H2O2 can interact directly with target proteins; for example, by oxidizing Cys residues and thereby altering protein conformation (Wu et al., 1998; Morel and Barouki, 1999). In addition, it may be that plant cells contain redox sensors that detect and respond to signals such as H2O2. In this context, the induction of a gene encoding a potential hybrid His kinase is of particular interest. His kinases and two component signal transduction systems are well represented in the Arabidopsis genome and have already been shown to modulate cellular responses to ethylene, cytokinin, and possibly osmotic stress (Urao et al., 2000; Inoue et al., 2001). His kinases are also important sensory enzymes in yeast, in which the osmo-sensing SLN1-SSK1 system has been particularly well characterized (Maeda et al., 1994). Here, the His kinase signaling module is connected to a MAPK system, such that activation of the HOG1 MAPK is regulated by osmotic stress. Recent work has shown that the SLN1 His kinase-HOG1 MAPK signaling system also functions as an H2O2 sensor in yeast (Singh, 2000). This finding, along with the fact that H2O2 activates the Arabidopsis MAPK ATMPK6 (Kovtun et al., 2000), a MAPK with high sequence homology to HOG1, coupled with the observation that H2O2 induces the expression of a His kinase, suggests strongly that this His kinase may also function as an H2O2 sensor in plants.
MATERIALS AND METHODS
Plant Treatments
Suspension cultures of Arabidopsis (var Landsberg erecta) were maintained as described by Desikan et al. (1996). Plants were grown in a controlled environment growth cabinet with an 8-h photoperiod at 20°C or, for UV treatment (var Columbia), as described previously (A-H-Mackerness et al., 1999). Seven-day-old cultures were exposed to H2O2 (20 mm) and harpin (2 μg mL−1) as described (Desikan et al., 1998a). Rosette leaves were vacuum infiltrated with a solution of 20 mm H2O2; control leaves were vacuum infiltrated with distilled water. Drought stress was imposed by wilting leaves in a stream of cool air until a 10% loss in fresh weight was achieved, followed by incubation in a sealed plastic bag for 2 h. Plants were sprayed prior to wilting with a 10 mm solution of ascorbic acid; controls involved spraying the plants with water. Plants (var Columbia) were exposed to UV irradiation, with or without ascorbate pretreatment, as described by A-H-Mackerness et al. (1999).
Microarray Analysis
Total RNA and mRNA were purified from control and H2O2-treated cells as described by Desikan et al. (1998a). RNA labeling and microarray hybridizations were performed by AFGC on the two biological replicate samples (representing independent experiments) using two duplicate slides with reverse labeling (Michigan State University, East Lansing; Schaffer et al., 2001). Microarray data analysis and identification of H2O2-regulated genes were performed using the Stanford Microarray Database where the data are publicly available (http://genome-www4.stanford.edu/MicroArray/SMD). The criteria used for selection of the genes were based on: (a) normalized channel intensities >1,000, with greater than a 1.5-fold increase in mRNA abundance; and (b) for genes categorized as low abundance, those with channel intensities between 300 and 1,000 (see AFGC Web site), and intensity ratios of >1.5. Normalization was based on the average of the natural log of the ratio of channel intensities (based on threshold values greater than 1.5 times background channel intensities) after background subtraction (see Stanford Microarray Database Web site).
RNA-Blot Analysis
Total RNA was dot blotted onto nylon membranes. Total RNA and mRNA were fractionated by denaturing agarose gel electrophoresis and transferred to nylon membranes (Desikan et al., 1998a). Blots were then hybridized with 32P-labeled cDNA probes as described by Desikan et al. (1998a). ESTs were obtained from Arabidopsis Biological Resource Center (Ohio State University, Columbus) and cDNA probes prepared using PCR-generated inserts or restriction-digested plasmids as templates, as described by Desikan et al. (1998a). Hybridization signals were quantified using a scanning densitometer (Shimadzu, Kyoto), and normalized using hybridization signals from a constitutive EF-1 αcDNA probe (http://www.afgc.stanford.edu).
Bioinformatics
Sequence homologies of ESTs were analyzed using the BLASTX program (http://www.Arabidopsis.org/blast). Functional classification of H2O2-induced genes was according to the functional organization of the Arabidopsis genome (http://mips.gsf.de/proj/thal/db/index.html). Promoter analysis was performed using the PLACE software (http://www.dna.affrc.go.jp/htdocs/PLACE/signalscan. html).
ACKNOWLEDGMENTS
We would like to thank Vernadette Simon and Ellen Wisman (Michigan State University, East Lansing) for their helpful advice.
LITERATURE CITED
- Abate C, Patel L, Rauscher FJ, Curran T. Redox regulation of fos and jun DNA binding activity in vitro. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- A-H-Mackerness S, John CF, Jordan B, Thomas B. Early signaling components in ultraviolet-B responses: distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett. 2001;489:237–242. doi: 10.1016/s0014-5793(01)02103-2. [DOI] [PubMed] [Google Scholar]
- A-H-Mackerness S, Surplus SL, Blake P, John CF, Buchanan-Wollaston V, Jordan BR, Thomas B. Ultraviolet-B induced stress and changes in gene expression in Arabidopsis thaliana: role of signaling pathways controlled by jasmonic acid, ethylene and reactive oxygen species. Plant Cell Environ. 1999;22:1413–1423. [Google Scholar]
- Allen RG, Tresini M. Oxidative stress and gene regulation. Free Radic Biol Med. 2000;28:463–499. doi: 10.1016/s0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- Baker CJ, Orlandi EW, Mock NM. Harpin, an elicitor of the hypersensitive response in tobacco caused by Erwinia amylovora, elicits active oxygen production in suspension cells. Plant Physiol. 1993;102:1341–1344. doi: 10.1104/pp.102.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzet N, Richaud C, Deveaux Y, Kazmier M, Gagnon J, Triantaphylides Accumulation of small heat shock proteins, including mitochondrial HSP22, induced by oxidative stress and adaptive response in tomato cells. Plant J. 1998;13:519–527. doi: 10.1046/j.1365-313x.1998.00056.x. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Jones RL. Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J. 2001;25:19–29. doi: 10.1046/j.1365-313x.2001.00930.x. [DOI] [PubMed] [Google Scholar]
- Bolwell GP. Role of active oxygen species and NO in plant defense responses. Curr Opin Plant Biol. 1999;2:287–294. doi: 10.1016/S1369-5266(99)80051-X. [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Wojtaszek P. Mechanisms for the generation of reactive oxygen species in plant defense: a broad perspective. Physiol Mol Plant Pathol. 1997;51:347–366. [Google Scholar]
- Bowler C, Fluhr R. The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci. 2000;5:241–245. doi: 10.1016/s1360-1385(00)01628-9. [DOI] [PubMed] [Google Scholar]
- Chen W, Chao G, Singh KB. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 1996;10:955–966. doi: 10.1046/j.1365-313x.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- Cho B-H. Characterization of fatty acid desaturase, glutathione S-transferase and blue copper binding protein genes in Arabidopsis thaliana. J Plant Biol. 1997;40:170–175. [Google Scholar]
- Daniel X, Lacomme C, Morel J-B, Roby D. A novel myb oncogene homologue in Arabidopsis thalianarelated to hypersensitive cell death. Plant J. 1999;20:57–66. doi: 10.1046/j.1365-313x.1999.00578.x. [DOI] [PubMed] [Google Scholar]
- Dat JF, Lopez-Delgado H, Foyer CH, Scott IM. Parallel changes in H2O2and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol. 1998;116:1351–1357. doi: 10.1104/pp.116.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat J, Vandenbeele S, Vranova E, Van Montagu M, Inze D, Van Breusegm F. Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci. 2000;57:779–795. doi: 10.1007/s000180050041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Hancock JT, Coffey MJ, Neill SJ. Generation of active oxygen in elicited cells of Arabidopsis thalianais mediated by a NADPH oxidase-like enzyme. FEBS Lett. 1996;382:213–217. doi: 10.1016/0014-5793(96)00177-9. [DOI] [PubMed] [Google Scholar]
- Desikan R, Burnett EC, Hancock JT, Neill SJ. Harpin and hydrogen peroxide induce the expression of a homologue of gp91-phox in Arabidopsis thalianasuspension cultures. J Exp Bot. 1998b;49:1767–1771. [Google Scholar]
- Desikan R, Clarke A, Hancock JT, Neill SJ. H2O2 activates a MAP kinase-like enzyme in Arabidopsis thalianasuspension cultures. J Exp Bot. 1999;50:1863–1866. [Google Scholar]
- Desikan R, Neill SJ, Hancock JT. Generation of active oxygen in Arabidopsis thaliana. Phyton. 1997;37:65–70. [Google Scholar]
- Desikan R, Neill SJ, Hancock JT. Hydrogen peroxide-induced gene expression in Arabidopsis thaliana. Free Radic Biol Med. 2000;28:773–778. doi: 10.1016/s0891-5849(00)00157-x. [DOI] [PubMed] [Google Scholar]
- Desikan R, Reynolds A, Hancock JT, Neill SJ. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on gene expression in Arabidopsissuspension cultures. Biochem J. 1998a;330:115–120. doi: 10.1042/bj3300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound-response gene expression profiles. Plant Cell. 2000;12:963–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne P, Petitot A-S, Houot V, Blein J-P, Suty L. Induction of tcI 7, a gene encoding a β subunit of proteasome, in tobacco plants treated with elicitin, salicylic acid or hydrogen peroxide. FEBS Lett. 2000;466:213–218. doi: 10.1016/s0014-5793(00)01084-x. [DOI] [PubMed] [Google Scholar]
- Fauman EB, Saper MA. Structure and function of the protein tyrosine phosphatases. Trends Biochem Sci. 1996;21:413–417. doi: 10.1016/s0968-0004(96)10059-1. [DOI] [PubMed] [Google Scholar]
- Fialkow L, Chan CK, Rotin D, Grinstein S, Downey GP. Activation of the mitogen-activated protein kinase signalling pathway in neutrophils. J Biol Chem. 1994;269:31234–31242. [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Lopez-Delgado H, Dat JF, Scott IM. Hydrogen peroxide and glutathione-associated mechanisms of acclimatory stress tolerance and signaling. Physiol Plant. 1997;100:241–254. [Google Scholar]
- Gasch P, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godon C, Lagniel G, Lee J, Buhler J-M, Kieffer S, Perrot M, Boucherie H, Toledano MB, Labarre J. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- Grant JJ, Yun B-W, Loake GJ. Oxidative burst and cognate redox signalling reported by luciferase imaging: identification of a signal network that functions independently of ethylene, SA and Me-JA but is dependent on MAPKK activity. Plant J. 2000;24:569–582. doi: 10.1046/j.1365-313x.2000.00902.x. [DOI] [PubMed] [Google Scholar]
- Guan LM, Zhao J, Scandalios JG. Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2is the likely intermediary signaling molecule for the response. Plant J. 2000;22:87–95. doi: 10.1046/j.1365-313x.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- Harding SA, Oh S-H, Roberts DM. Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 1997;16:1137–1144. doi: 10.1093/emboj/16.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelbau E, Amasino RM. Delivering copper within plant cells. Curr Opin Plant Biol. 2000;3:205–210. [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Irani K, Yong X, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ. Mitogenic signaling mediated by oxidants in Ras transformed fibroblasts. Science. 1997;275:1649–1652. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- Karpinski S, Reynolds H, Karpinska B, Wingsle G, Creissen G, Mullineaux P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284:654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- Keller T, Damude HG, Werner D, Doerner P, Dixon RA, Lamb C. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+binding motifs. Plant Cell. 1998;10:255–266. doi: 10.1105/tpc.10.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Chung HJ, Thomas TL. Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3promoter using a modified yeast one-hybrid system. Plant J. 1997;11:1237–1251. doi: 10.1046/j.1365-313x.1997.11061237.x. [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu W-L, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97:2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, Jin H, Levya A, Meissner RC, Petroni K, Urzainqui A, Bevan M, Martin C. Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998;16:263–276. doi: 10.1046/j.1365-313x.1998.00278.x. [DOI] [PubMed] [Google Scholar]
- Lamb C, Dixon RA. The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- Langebartels C, Schraudner M, Heller W, Ernst D, Sandermann H. Oxidative stress and defense reactions in plants exposed to air pollutants and UV-B radiation. In: Inze D, Van Montagu M, editors. Oxidative Stress in Plants. London: Harwood Academic Publishers; 2000. pp. 105–135. [Google Scholar]
- Lee S, Choi H, Suh S, Du I-S, Oh K-Y, Choi EJ, Schroder Taylor AT, Low PS, Lee Y. Oligogalacturonic acid and chitosan reducs stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis. Plant Physiol. 1999;121:147–152. doi: 10.1104/pp.121.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Pennell RI, Alvarez ME, Palmer R, Lamb C. Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr Biol. 1996;6:427–437. doi: 10.1016/s0960-9822(02)00510-9. [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. H2O2from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Leyman B, Geelen D, Qunitero FJ, Blatt MR. A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science. 1999;283:537–540. doi: 10.1126/science.283.5401.537. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Huertas E, Charlton WL, Johnson B, Graham IA, Baker A. Stress induces peroxisome biogenesis genes. EMBO J. 2000;19:6770–6777. doi: 10.1093/emboj/19.24.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. Protein phosphatases and signaling cascades in higher plants. Trends Plant Sci. 1998;3:271–275. [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA, Dangl JL, Dietrich RA. The transcriptome of Arabidopsis thalianaduring systemic acquired resistance. Nat Genet. 2000;26:403–410. doi: 10.1038/82521. [DOI] [PubMed] [Google Scholar]
- McCabe PF, Leaver CJ. Programmed cell death in cell cultures. Plant Mol Biol. 2000;44:359–368. doi: 10.1023/a:1026500810877. [DOI] [PubMed] [Google Scholar]
- Minhas D, Grover A. Transcript levels of genes encoding various glycolytic and fermentation enzymes change in response to abiotic stress. Plant Sci. 1999;146:41–51. [Google Scholar]
- Mittler R, Herr EH, Orvar BL, Van Camp W, Willekens H, Inze D, Ellis BE. Transgenic tobacco plants with reduced capability to detoxify reactive oxygen intermediates are hyperresponsive to pathogen infection. Proc Natl Acad Sci USA. 1999;96:14165–14170. doi: 10.1073/pnas.96.24.14165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel Y, Barouki R. Repression of gene expression by oxidative stress. Biochem J. 1999;342:481–496. [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kaminaka H, Masumura T, Tanaka K. Induction of rice cytosolic ascorbate peroxidase mRNA by oxidative stress: the involvement of hydrogen peroxide in oxidative stress signalling. Plant Cell Physiol. 1999;40:417–422. [Google Scholar]
- Mullineaux P, Ball L, Escobar C, Karpinska B, Creissen G, Karpinski S. Are diverse signalling pathways integrated in the regulation of Arabidopsis antioxidant defense gene expression in response to excess excitation energy? Philos Trans R Soc Lond. 2000;355:1531–1540. doi: 10.1098/rstb.2000.0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrset AH, Bostad A, Jamin N, Lirsac P-N, Toma F, Gabrielsen OS. DNA and redox state induced conformational changes in the DNA-binding domain of the Myb oncoprotein. EMBO J. 1993;12:4625–4633. doi: 10.1002/j.1460-2075.1993.tb06151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hancock JT. H2O2signaling in plant cells. In: Clavert C, Smallwood M, Bowles D, editors. Plant Responses to Environmental Stress. Oxford: BIOS Scientific Publishers; 1999. pp. 59–63. [Google Scholar]
- Noctor G, Foyer CH. Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- Pastori GM, del Rio LA. Natural senescence of pea leaves: an activated oxygen-mediated function for peroxisomes. Plant Physiol. 1997;113:411–418. doi: 10.1104/pp.113.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- Prasad TK, Anderson MD, Martin BA, Stewart CR. Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell. 1994;6:65–74. doi: 10.1105/tpc.6.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AH, Taylor A, Ripley SJ, Griffiths A, Trewavas AJ, Knight MR. Oxidative signals in tobacco increase cytosolic calcium. Plant Cell. 1994;6:1301–1310. doi: 10.1105/tpc.6.9.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XF, Holuigue L, Horvath DM, Chua NH. Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell. 1994;6:863–874. doi: 10.1105/tpc.6.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Somssich IE. Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol. 1998;1:311–315. doi: 10.1016/1369-5266(88)80052-9. [DOI] [PubMed] [Google Scholar]
- Samuel MA, Miles GP, Ellis BE. Ozone treatment rapidly activates MAP kinase signalling in plants. Plant J. 2000;22:367–376. doi: 10.1046/j.1365-313x.2000.00741.x. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Hager A. Fungal elicitors induce a transient release of active oxygen species from cultured spruce cells that is dependent on Ca2+and protein kinase activity. Planta. 1992;187:136–141. doi: 10.1007/BF00201635. [DOI] [PubMed] [Google Scholar]
- Shirasu K, Lahaye T, Tan M-W, Zhou F, Azevedo C, Schulze-Lefert P. A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell. 1999;99:355–366. doi: 10.1016/s0092-8674(00)81522-6. [DOI] [PubMed] [Google Scholar]
- Singh KK. The Saccharomyces cerevisiaeSLN1P-SSK1P two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic Biol Med. 2000;29:1043–1050. doi: 10.1016/s0891-5849(00)00432-9. [DOI] [PubMed] [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell. 1999;11:431–443. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M, Bucher M, Stahl W, Suter M, Dupuis I, Kuhlemeier C. Activation of plant defense responses and sugar efflux by expression of pyruvate decarboxylase in potato leaves. Plant J. 1998;16:661–671. [Google Scholar]
- Takatsuji H. Zinc finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol. 1999;39:1073–1078. doi: 10.1023/a:1006184519697. [DOI] [PubMed] [Google Scholar]
- Tenhaken R, Levine A, Brisson LF, Dixon RA, Lamb C. Function of the oxidative burst in hypersensitive disease resistance. Proc Natl Acad Sci USA. 1995;92:4158–4163. doi: 10.1073/pnas.92.10.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Torres MA, Onuchi H, Hamada S, Machida C, Hammond-Kosack KE, Jones JDG. Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox) Plant J. 1998;14:365–370. doi: 10.1046/j.1365-313x.1998.00136.x. [DOI] [PubMed] [Google Scholar]
- Urao T, Yamaguchi-Shinozaki K, Shinozaki K. Two-component systems in plant signal transduction. Trends Plant Sci. 2000;5:67–74. doi: 10.1016/s1360-1385(99)01542-3. [DOI] [PubMed] [Google Scholar]
- Vallelian-Bindschedler L, Schweizer P, Mosinger E, Metraux J-P. Heat-induced resistance to powdery mildew (Blumeria graminis f.sp. hordei) is associated with a burst of active oxygen species. Physiol Mol Plant Pathol. 1998;52:185–199. [Google Scholar]
- Voehringer DW, Hirschberg DL, Xiao J, Lu Q, Roederer M, Lock CB, Herzenberg LA, Steinman L, Herzenberg LA. Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis. Proc Natl Acad Sci USA. 2000;97:2680–2685. doi: 10.1073/pnas.97.6.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton KM, Dixon JE. Protein tyrosine phosphatases. Annu Rev Biochem. 1993;62:101–120. doi: 10.1146/annurev.bi.62.070193.000533. [DOI] [PubMed] [Google Scholar]
- Wu Y, Kwon K-S, Rhee SG. Probing cellular targets of H2O2with fluorescein-conjugated iodoacetamide and antibodies to fluorescein. FEBS Lett. 1998;440:111–115. doi: 10.1016/s0014-5793(98)01415-x. [DOI] [PubMed] [Google Scholar]
- Xu Q, Hui-Hua F, Gupta R, Luan S. Molecular characterization of a tyrosine-specific protein phosphatase encoded by a stress-responsive gene in Arabidopsis. Plant Cell. 1998;10:849–857. doi: 10.1105/tpc.10.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Klessig DF. Isolation and characterization of a tobacco mosaic virus-inducible myboncogene homolog from tobacco. Proc Natl Acad Sci USA. 1996;93:14972–14977. doi: 10.1073/pnas.93.25.14972. [DOI] [PMC free article] [PubMed] [Google Scholar]