Abstract
Previously, antibodies were raised against a nuclear envelope-enriched fraction of yeast, and the essential gene NNF1 was cloned by reverse genetics. Here it is shown that the conditional nnf1-17 mutant has decreased stability of a minichromosome in addition to mitotic spindle defects. I have identified the novel essential genes DSN1, DSN3, and NSL1 through genetic interactions with nnf1-17. Dsn3p was found to be equivalent to the kinetochore protein Mtw1p. By indirect immunofluorescence, all four proteins, Nnf1p, Mtw1p, Dsn1p, and Nsl1p, colocalize and are found in the region of the spindle poles. Based on the colocalization of these four proteins, the minichromosome instability and the spindle defects seen in nnf1 mutants, I propose that Nnf1p is part of a new group of proteins necessary for chromosome segregation.
The process of mitotic cell division ensures that chromosomes are faithfully duplicated and equally segregated between mother and daughter cells, as the absence of genes or their presence in irregular numbers is typically lethal. The microtubule (MT) cytoskeleton and its associated structures are responsible for orchestrating chromosome segregation and maintaining genetic continuity. In many eukaryotic cells, chromosomal movements during mitosis are conserved and are mediated by three organelles: the bipolar mitotic spindle, the kinetochores (centromere DNA and associated proteins), and the centrosomes (MT organizing centers). These common features of chromosome segregation can be observed microscopically, as aided by fluorescence in situ hybridization studies of individual chromosomes in fixed cells (20) and green fluorescent protein (GFP)-tagged chromosomes in living cells (23, 46, 49, 66, 67). Separation of sister chromatids is initiated at and progresses from the centromeric regions where the kinetochores mediate the attachment of sister chromatids to the spindle MTs. In Saccharomyces cerevisiae, the MTs are nucleated by the spindle pole body (SPB), the yeast equivalent of the centrosome. The SPB is a large laminar structure that is embedded in the nuclear membrane throughout the cell cycle and nucleates both cytoplasmic and nuclear MTs (5, 6). One array of nuclear MTs interdigitate and span from pole to pole, while a second array is captured by the kinetochores. Chromosome segregation requires the shortening of kinetochore MTs (anaphase A) and the elongation of pole-to-pole MTs (anaphase B). In budding yeast, the most dramatic movement of chromatin is concomitant with spindle elongation, although anaphase A has also been observed (21, 49, 67). Sister centromeres that are proximally marked with GFP have been shown to align along a spindle equator, separate, and oscillate between the spindle poles before anaphase B spindle elongation (49). Following the end of mitosis and during much of the cell cycle, the centromeres are clustered near the SPBs (21, 33).
Mutations in components of the mitotic spindle, the SPB, kinetochores, MT-associated motor proteins, and various regulatory enzymes may result in aneuploidy. In one set of mutants, chromosome segregation is asymmetric, with chromosomal DNA segregating to only one pole of a bipolar spindle. Mutants in this group include kinetochore proteins of the Ndc80p complex (76), Ndc10p (16), and the protein kinase Ipl1p, which localizes to the mitotic spindle and kinetochores (2, 9, 23, 37). Alternatively, duplication of the SPB can terminate at a particular stage, resulting in a monopolar spindle and diploidization, as seen in the mps2-1 or ndc1-1 mutants (70, 78, 79). In other chromosome missegregation mutants, chromosome loss is seemingly random. In certain tub4 and spc110 mutants, which are defective in γ-tubulin and the SPB component Spc110p, respectively, the SPBs are assembled but the spindle is compromised during mitosis (36, 61, 64, 68, 69). Failure in chromosome segregation and spindle defects are also seen in duo1 and dam1 mutants, as might be expected since Duo1p and Dam1p are found along the mitotic spindle and at the kinetochores in wild-type cells (10, 23, 24, 34). These mutants demonstrate that deficient SPBs, kinetochores, or spindles can lead to disomy of individual chromosomes or diploidization of the entire genome within a single nucleus.
In S. cerevisiae, nuclear division occurs along the bud axis, and hence positioning of the mitotic spindle through the narrow bud neck is critical to ensure an equal distribution of DNA between the mother and daughter cells. Dynamic contacts between the cytoplasmic MTs and the cell cortex are chiefly responsible for nuclear migration and orientation of the mitotic spindle (8, 57). Mutants that can complete spindle extension but are defective in nuclear positioning are characterized by populations of anucleate daughter cells and binucleate mother cells. These mutants are typically defective in cytoplasmic MT proteins or have perturbations in the SPB. For example, detachment of the cytoplasmic MTs from the SPBs through loss of either of the outer plaque proteins Cnm67p and Spc72p leads to multinucleated cells (4, 60). Therefore, the observation of binucleate cells may indicate a SPB defect.
In an earlier study, we raised antibodies against a nuclear envelope-enriched fraction of yeast and subsequently cloned the essential gene NNF1 (necessary for nuclear function 1) from a yeast expression library (56). Nnf1p is a small coiled-coil protein of 201 amino acids with no homology to any known proteins. Cells carrying mutations in NNF1 exhibit primarily short mitotic spindles and, to a lesser extent, aberrant cytoplasmic MTs and defects in nuclear migration. Nnf1p is a protein of low abundance and cannot be detected by Western blotting unless it is overexpressed. When cells overexpressing NNF1 are fractionated, Nnf1p is found predominantly in the nuclear fraction and can be extracted only in the presence of 8 M urea or guanidine hydrochloride. These studies have verified that Nnf1p is a nuclear protein but have not elucidated the nature of its function. In this work, I present the results of two extensive genetic screens that were initiated with the conditional nnf1-17 allele. I show that Nnf1p is important for chromosome segregation and identify three functionally related proteins.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Yeast cell culture and genetic manipulations, including mating, diploid isolation, sporulation, and tetrad analysis, were performed as described previously (52, 58). Yeast strains (Table 1) are derivatives of W303, except for CH1305 (42) and ABY112 (4). Media were prepared according to standard recipes (3, 58, 74). Sectoring assays were performed on low-adenine (5 mg/liter) synthetic medium.
TABLE 1.
Yeast strains used in this study
| Strain(s)a | Genotype |
|---|---|
| W961-5A | MATaleu2-3,112 trp1-1 ura3-1 ade2-1 HIS3+can1-100 |
| GEY145 | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/TRP1+ura3-1/ura3-1 ade2-ade2-1 HIS3+/his3-11,15 can1-100/can1-100 NNF1::LEU2/NNF1::LEU2 |
| GEY146 | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/TRP1+ura3-1/ura3-1 ade2-/ade2-1 HIS3+/his3-11,15 can1-100/can1-100 nnf1-17::LEU2/nnf1-17::LEU2 |
| CH1305 | MATalys2 leu2 ura3 ade2 ade3 can1 |
| ABY112 | MATa/MATα leu2-3,112/leu2-3,112 trp1-289/trp1-289 ura3-52/ura3-52 his3Δ1/his3Δ1CNM67::GFP(S65T)-kanMX6/CNM67::GFP(S65T)-kanMX6 |
| CY6 | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-/ade2-1 his3-11,15 his3-11,15 can1-100/can1-100 |
| GEY160 | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-/ade2-1 ade3Δ/ade3Δ his3-11,15/his3-11,15 can1-100/can1-100 |
| GEY122 | MATα leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 NNF1::LEU2 |
| GEY138 | MATα leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 nnf1-17::LEU2 |
| GEY110 | MATaleu2-3,112 trp1-1 ura3-1 ade2-1 HIS3+can1-100 NNF1-myc6::LEU2 |
| GEY111 | MATaleu2-3,112 trp1-1 ura3-1 ade2-1 HIS3+can1-100 NNF1-myc6::LEU2 CNM67::GFP(S65T)-kanMX6 |
| GEY165 | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 ade3Δ/ade3Δ his3-11,15/his3-11,15 can1-100/can1-100 nnf1-17::LEU2/nnf1-17::LEU2 |
| YSLP1 | MATalys2 leu2 trp1 ura3 ade2 ade3 can1 nnf1-17::LEU2 |
| YSL6 | MATalys2 leu2 trp1 ura3 ade2 ade3 can1 nsl1-6 nnf1-17::LEU2 |
| YSL8 | MATalys2 leu2 trp1 ura3 ade2 ade3 can1 nsl1-8 nnf1-17::LEU2 |
| YSL68 | MATalys2 leu2 trp1 ura3 ade2 ade3 can1 nsl-68 nnf1-17::LEU2 |
| YSL29 | MATalys2 leu2 trp1 ura3 ade2 ade3 can1 nsl2-29(dsn3-29) nnf1-17::LEU2 |
| GEY170 to 172 | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-/ade2-1 his3-11,15 his3-11,15 can1-100/can1-100 dsn1::HIS3/DSN1 |
| GEY190 to 191 | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 ade3Δ/ade3Δ his3-11,15/his3-11,15 can1-100/can1-100 nsl1::LEU2/NSL1 |
| GEY210 to 212 | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-/ade2-1 ade3Δ/ade3Δ his3-11,15 his3-11,15 can1-100/can1-100 nsl2(dsn3)::LEU2/NSL2(DSN3) |
| GEY176 | MATα leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 dsn1::HIS3 [pGE36; DSN1-GFP] |
| GEY206 | MATaleu2-3,112 trp1-1 ura3-1 ade2-1 ade3Δ his3-11,15 can1-100 nsl1::LEU2 [pGE55; NSL1-GFP] |
| GEY216 | MATaleu2-3,112 trp1-1 ura3-1 ade2-1 ade3Δ his3-11,15 can1-100 nsl2(dsn3)::LEU2 [pGE74; DSN3(NSL2)-GFP] |
Strains with a GEY prefix are in a W303 background.
Plasmid constructions.
All DNA and bacterial manipulations were by standard protocols (55). Plasmids are listed in Table 2. The CEN vectors pRS313 (HIS3 marker), pRS314 (TRP1 marker), and pRS316 (URA3 marker) (59) were primarily used for cloning. The CEN plasmid pCT3 contains the URA3 marker and was a gift from C. Thompson. The 2μm vector YEp352 contains the URA3 marker and was a gift from A. Tzagoloff. Three plasmids (pGE98 to pGE100) were constructed for the synthetic lethal screen. A 1.4-kb XmnI/PvuII fragment containing NNF1 and its promoter was ligated into the SmaI site of pCH1122 to create pGE98 and into the SmaI site of pRS314 to create pGE99. Plasmid pGE100 was constructed in a pRS314 vector in two steps. First, the ADE3 marker was band isolated on a 3.6-kb EagI/NheI fragment from pCH1023 and cloned into the EagI and SpeI sites of pRS314. Second, the 1.4-kb XmnI/PvuII fragment containing NNF1 and its promoter was inserted in the SmaI site of the multiple cloning site to complete pGE100.
TABLE 2.
Plasmids used in this study
| Plasmida | Relevant markers |
|---|---|
| pCH1122b | CEN, URA3, ADE3 |
| pCH1023b | CEN, LEU2, ADE3 |
| pDK243c | CEN, LEU2; ade3-2p |
| pW35d | 2μm, URA3; SSD1/SRK1 |
| pGE36 | CEN, TRP1; DSN1-GFP |
| pGE55 | CEN, TRP1; NSL1-GFP |
| pGE74 | CEN, TRP1; DSN3-GFP |
| pGE81 | CEN, URA3; NSL1; pGE180 gap-repaired SnaBI-EcoNI in yeast strain YSLP1 |
| pGE82 | CEN, URA3; nsl1-6; pGE180 gap-repaired SnaBI-EcoNI in yeast strain YSL6 |
| pGE83 | CEN, URA3; nsl1-8; pGE180 gap-repaired SnaBI-EcoNI in yeast strain YSL8 |
| pGE84 | CEN, URA3; nsl1-68; pGE180 gap-repaired SnaBI-EcoNI in yeast strain YSL68 |
| pGE89 | CEN, URA3; DSN3(NSL2); pGE170 gap-repaired MluI-SpeI in yeast strain YSLP1 |
| pGE90 | CEN, URA3; dsn3-29/nsl2-29; pGE170 gap-repaired MluI-SpeI in yeast strain YSL29 |
| pGE99 | CEN, TRP1; NNF1 on a 1.4-kb XmnI-PvuII insert |
| pGE100 | CEN, TRP1; NNF1 on a 1.4-kb XmnI-PvuII insert and ADE3 on a 3.6-kb EagI-NheI insert |
| pGE101 | CEN, URA3; NNF1 on a 1.4-kb XmnI-PvuII insert |
| pGE102 | CEN, URA3; ade3-2p |
| pGE151 | 2μm, URA3; 1.4-kb HindIII subclone from YEp24-based library; contains DSN3(NSL2) |
| pGE164 | 2μm, URA3; 2.7-kb BsaAI subclone from YEp24-based library; contains DSN1 |
| pGE165 | CEN, URA3; 2.7-kb BsaAI subclone from YEp24-based library; contains DSN1 |
| pGE170 | CEN, URA3; 3.1-kb KpnI subclone from YEp24-based library; contains DSN3(NSL2) |
| pGE171 | CEN, URA3; 1.4-kb HindIII subclone from YEp24-based library; contains DSN3(NSL2) |
| pGE180 | CEN, URA3; 2.0-kb PvuII-NruI subclone from pCT3-based library; contains NSL1 |
| pGE190 | 2μm, URA3; 3.3-kb HindIII subclone from YEp24-based library; contains SLG1 |
Plasmids are from this study unless otherwise indicated.
From C. Holm.
From D. Koshland.
From K. Tatchell.
Generation of NNF1 mutants.
Temperature-sensitive mutations of NNF1 were generated by methods described previously(43, 47). Briefly, NNF1 was amplified by mutagenic PCR, and cotransformed with gapped vector into a nnf1::URA3 strain carrying PGAL1-NNF1 in a HIS3 plasmid. Transformants were selected at 25°C on SC-His-Leu medium (containing 2% galactose as the sole carbon source) to maintain expression of NNF1. The transformation plates were then replica plated onto two sets of SC-Leu plates and incubated at 25 or 37°C. Plasmid DNA from potential temperature-sensitive nnf1 mutants was rescued into Escherichia coli, retested in the nnf1::URA3 yeast strain, and sequenced. The nnf1-17 allele analyzed here had three amino acid changes: C53S, V78D, and C121G. For integration at the NNF1 chromosomal locus by one-step gene replacement, the LEU2 selectable marker and NNF1 3′ noncoding region were added after the C-terminal ends of NNF1 and nnf1-17. The coding sequence at the 3′ ends of NNF1 and nnf1-17 was changed from ENY* to ENYWILV* (asterisks designate stop codons). Band-isolated 3.2-kb HindIII fragments were transformed into the W303 strain W961-5A for Leu+ selection. Correct integration at the NNF1 locus was verified by Southern analysis.
Selection for dosage suppressors.
High- and low-copy suppressors of the temperature-sensitive growth defect of nnf1-17 cells were selected at 35°C following transformation of strain GEY138 with S. cerevisiae genomic libraries in either 2μm (7) or CEN (71) plasmids. Cells were plated on SC-Ura medium for plasmid selection and allowed an initial recovery for 12 h at 25°C before the plates were shifted to the restrictive temperature (35°C) for 4 days. A total of 24 colonies were isolated at 35°C, corresponding to over 150,000 transformants at the permissive temperature of 25°C. Twenty of the 24 plasmids recovered from these colonies allowed growth at 35°C when retransformed into strain GEY138. Restriction digests showed that these 20 plasmids contained nine different inserts. Subclones were made in the 2μm vector YEp352 to identify the genes suppressing the nnf1-17 growth defect. Plasmids pGE164, pGE190, and pGE151 contained the smallest functional subclones for DSN1 (YIR010w on a 2.7-kb BsaAI fragment), DSN2 (YOR008c on a 3.3-kb HindIII fragment), and DSN3 (YAL034w-a on a 1.4-kb HindIII fragment), respectively. The nnf1-17 phenotype was complemented at 35°C by two distinct plasmids from the low-copy CEN library. Of 58,000 transformants, one colony grew well at 35°C, and one colony grew poorly at this temperature. Sequencing of rescued plasmids indicated that their inserts included NNF1 and SSD1-v, respectively. Plasmids pW31 and pW35, containing SSD1/SRK1 (a gift of K. Tatchell) (77) were transformed into the nnf1-17 mutant to confirm the partial suppression effect at 35°C.
Synthetic lethal screen.
The nnf1-17 ade2 ade3 strain YSLP1 was a segregant from the cross GEY138 × CH1305. For the synthetic lethal screen, strain YSLP1 carrying plasmid pGE98 (CEN/NNF1/ADE3/URA3) was grown in SC-Ura at 25°C, sonicated, plated on SC low-Ade plates, and UV irradiated to ∼10% survival. The mutagenized cells were incubated at 30°C for 7 days, when the colony color had fully developed. From 102,000 colonies that survived the UV mutagenesis, 17 nonsectoring derivatives were identified. As integration of the plasmid or mutations at the chromosomal ade3 locus could also result in a nonsectoring phenotype, the stably red strains were transformed with pGE99 (CEN/NNF1/TRP1) or empty pRS314 (CEN/TRP1) vector. Six mutants which sectored upon transformation with pGE99, but not with empty vector, were selected for cloning by complementation of the nonsectoring phenotype at 30°C.
Cloning and allelic rescue of NSL1.
The screening plasmid pGE98 (CEN/NNF1/ADE3/URA3) in the six candidate strains from the synthetic lethal screen was replaced with pGE100 (CEN/NNF1/ADE3/TRP1) by plasmid shuffling and counterselection on 5-fluoroorotic acid (5-FOA) so that a URA3/CEN yeast genomic library (71) could be used to clone the gene(s) causing synthetic lethality with nnf1-17. Ura+ transformants were selected on SC-Ura low-Ade plates at 30°C. Two sectoring colonies were identified after transformation of the library DNA: one colony from isolate YSL6 (12,000 transformants) and one colony from isolate YSL8 (14,000 transformants). Plasmid DNA from these sectored colonies was recovered and checked for the ability to confer sectoring in all six of the synthetic lethal strains. Both plasmids complemented the nonsectoring phenotype of strains YSL6 and YSL8 as well as the nonsectoring phenotype of a third strain, YSL68. The plasmids contained a common yeast genomic insert from chromosome XVI. Subcloning of this insert into pRS316 to create pGE180 reduced the complementing region to a 2.0-kb PvuII/NruI fragment that contained NSL1 (YPL233w) as the only open reading frame (ORF).
Chromosomal NSL1 and its alleles nsl1-6, nsl1-8, and nsl1-68 were recovered by gap repair of pGE180 from the parent strain YSLP1 (NSL1) and the mutant strains YSL6 (nsl1-6), YSL8 (nsl1-8), and YSL68 (nsl1-68). For the gap repair, pGE180 was digested with SnaBI and EcoNI and band isolated to remove a 920-bp fragment containing 640 bp of wild-type NSL1 and 280 bp of 5′ noncoding region. Plasmid DNA was isolated from Ura+ transformants, and gap repair was verified with an EcoRV digest to produce a 1.2-kb fragment. The gap-repaired vectors were designated pGE81, pGE82, pGE83, and pGE84 for NSL1, nsl1-6, nsl1-8, and nsl1-68, respectively (Table 2). The four gap-repaired plasmids pGE81 to pGE84 were transformed into the nnf1-17 nsl1 strains YSL6, YSL8, and YSL68 carrying pGE100 (CEN/NNF1/ADE3/TRP1). Only pGE81 with wild-type NSL1 could be plasmid shuffled with pGE100 in YSL6, YSL8, and YSL68 to restore sectoring.
Cloning and allelic rescue of NSL2.
All six of the synthetic lethal strains carrying pGE100 (CEN/NNF1/ADE3/TRP1) were also transformed with the dosage suppressors SSD1-v, SLG1, DSN1, and DSN3 subcloned into the CEN/URA3 plasmid pRS316 to ascertain whether any of the genes might be both a multicopy suppressor of the nnf1-17 allele and also have a mutation that is synthetically lethal in combination with nnf1-17. Plasmid pGE171 containing DSN3 was able to complement the nonsectoring phenotype of the mutant YSL29. To distinguish between the possibilities that DSN3(NSL2) is able to act as both a high- and low-copy suppressor of nnf1-17 and that a dsn3(nsl2) mutation is synthetically lethal with nnf1-17, the plasmid shuffle with pGE100 was repeated in synthetic lethal strain YSL29 with DSN3(NSL2) and dsn3-29(nsl2-29) from strains YSLP1 and YSL29 cloned into pRS316. To isolate DSN3(NSL2) and the dsn3-29(nsl2-29) allele, genomic DNA was prepared from the parent strain YSLP1 and the mutant strain YSL29, and a 1.7-kb region around the ORFs was PCR amplified with primers YAL560 (5′-CTATCTGGGGTCGTCATGTTGA-3′) and YAL2220 (5′-CTGCTGAACTAGTTCTCGCTCA-3′). The 1.7-kb PCR product was then restricted to 1.3 kb with MluI [which cuts 105 bp before the ATG of DSN3(NSL2)] and SpeI [which cuts 350 bp after the DSN3(NSL2) stop codon]. This 1.3-kb MluI/SpeI fragment was used to replace the same 1.3-kb MluI/SpeI sequence in pGE170, resulting in pGE89 [DSN3(NSL2) from the parent strain) and pGE90 (dsn3-29(nsl2-29) from the UV-mutagenized strain]. Plasmid pGE89 was able to complement the nonsectoring phenotype of YSL29, whereas plasmid pGE90 was not able to complement the nonsectoring phenotype of this strain. Therefore, NSL2 is identical to DSN3.
Gene disruptions.
DSN1, DSN3(NSL2), and NSL1 were disrupted singly in a W303 background by one-step gene replacement (53). Correct integrations at the DSN1, DSN3(NSL2), and NSL1 chromosomal loci were verified by PCR or Southern blotting. For each gene disruption, three independent transformants which had a single integration at the intended locus were chosen for tetrad dissection and analysis. For the dsn1::HIS3 disruption, the resulting heterozygous diploid strains were GEY170, GEY171, and GEY172. For the dsn3::LEU2 disruption, the resulting heterozygous diploid strains were GEY210, GEY211, and GEY212. For the nsl1::LEU2 disruption, the resulting heterozygous diploid strains were GEY190, GEY191, and GEY192.
Epitope tagging.
Six repeats of the Myc epitope on a 270-bp DraI/XbaI fragment were inserted at the 3′ end of NNF1. The sequence at the C terminus of NNF1 now translates as YWIKA(MEQKLISEEDLNE)6SCSP*. For integration and selection at the NNF1 chromosomal locus, the LEU2 selectable marker on a 2.0-kb XbaI fragment was ligated after the NNF1 stop codon and before the 164 bp of 3′ noncoding region. Yeast strain W961-5A was transformed with NNF1-myc6::LEU2 on a 3.5-kb HindIII fragment, and genomic DNA from Leu+ transformants was analyzed by Southern blotting. Strain GEY110 has NNF1-myc6::LEU2 successfully integrated at the NNF1 locus. Strain GEY111 was derived from GEY110 and expresses a GFP(S65T)-tagged allele of CNM67 from the CNM67 chromosomal locus. The CNM67-GFP(S65T)-kanMX6 gene fusion was PCR amplified from ABY112 genomic DNA with primers CNM1940 (5′-GAGCTGAGCCGCATTTCTCAG-3′) and CNM3200 (5′-CTTCATAAGCGGCTCGAATCACAG-3′). The PCR product was band isolated and transformed into strain GEY110. Transformants were selected on YPD-G418 plates. Correct integration at the CNM67 locus was verified by PCR analysis.
GFP(S65T) was fused to the C terminus of DSN1 in several steps. DSN1 was amplified from pGE164 by using primers YIRSacI (5′-GCCTTTGAAAGTTGAGCTCAGCAAGAG-3′) and YIREcoRV (5′-CTTTTATTTCAGATATCCAGTTTTTTACTGA-3′). Primer YIRSacI anneals 500 bp before the ATG of DSN1. Primer YIREcoRV mutates an EcoRV site at the C terminus of DSN1, which after EcoRV digestion removes the final codon (leucine) and the stop codon. The resulting 2.3-kb PCR product was digested with SacI and EcoRV, and this fragment was ligated into the SacI and EcoRV sites of pRS313. GFP(S65T) on a NotI fragment was cut from pSF-GP1 (a gift of J. Hirsch), filled in with Klenow polymerase, and ligated into the EcoRV site to create pGE35.
For GFP(S65T)-tagging of NSL1, NSL1 and its promoter were amplified by PCR from pGE180 using the primers T7 and YPL1640 (5′-GTTATTTATTCAATCCTCCTGCAGGAAG-3′). The NSL1 PCR product was digested with SnaBI and PstI and cloned into the EcoRV and PstI sites of pBS SK(−). The resulting plasmid was cut with SmaI (in the multiple cloning site region next to PstI), and GFP(S65T) on a 750-bp NotI fragment (filled with Klenow polymerase) was ligated into this SmaI site. This cloning changes the last amino acids of NSL1 from EED to QP. The entire NSL1-GFP fusion was removed from the pBS SK(−) backbone with a HindIII-XbaI digest and cloned into the HindIII and XbaI sites of pRS316, to make pGE54.
GFP(S65T) was fused to the C terminus of DSN3 by first PCR amplifying the DSN3 promoter and ORF from pGE170 with primers YAL120EcoRI (5′-TCCGCGGGAATTCCCAGAGTATGCCGTGATGTTATG-3′) and YAL1860NotI (5′-GTAACGATTGCGGCCGCCTAACACATCATCAAGTAAATCCAATTGAGG-3′). Primer YAL120EcoRI introduces an EcoRI site 900 bp before the DSN3 start codon, and primer YAL1860NotI anneals before the stop of DSN3, to remove the termination codon and add a NotI restriction site. The 1,740-bp PCR product was digested with EcoRI and NotI and cloned into the EcoRI and NotI sites of pRS316. GFP(S65T) on a 750-bp NotI fragment was then ligated into the NotI site, resulting in plasmid pGE73.
Plasmid stability assay.
The plasmid stability assay is based on greater incidence of plasmid loss under nonselective conditions in affected mutants (40, 41). Diploid strains GEY160 and GEY165 were transformed with pCT3/ade3-2p (pGE102), and single transformants were grown overnight at 25°C in SC-Ura medium. The next day, the starter cultures were diluted in SC complete medium at 30°C for ∼4 doublings before plating on SC low-Ade plates prewarmed to 30°C. Colonies were scored after 5 days at 30°C as follows: (i) half-sectored, colonies that are half pink (with red and white sectors) and half white; (ii) pink, pink colonies that have pink and white sectors, not including half-sectored colonies; (iii) white, colonies that are solid white with no sectors; and (iv) red, colonies that are dark red. To confirm that plasmid loss in the nnf1-17 mutant was due to the nnf1-17 mutation alone, the nnf1-17 diploid strain GEY165 was cotransformed with pCT3/ade3-2p (pGE102) and pRS314/NNF1 (pGE98) or pRS314/empty vector, and the NNF1 diploid strain GEY160 was cotransformed with pCT3/ade3-2p and pRS314/empty vector. Single transformants were grown overnight at 25°C in SC-Ura-Trp medium, diluted in SC-Trp medium at 30°C for ∼4 doublings, and then plated on SC-Trp low-Ade plates prewarmed to 30°C. After 5 days at 30°C, colonies were scored as described above.
Flow cytometry.
Cells were prepared for flow cytometry as described previously (27). For each sample a minimum of 25,000 cells were analyzed using a FACS Vantage flow cytometer (Becton Dickinson, San Jose, Calif.).
Fluorescence microscopy.
Indirect immunofluorescence against tubulin was carried out essentially as described previously (50). For localization of SPB antigens, short fixation times were used, as suggested previously (54). Cells carrying GFP and/or Myc6 fusions were fixed for 5 to 10 min in 3.7% formaldehyde at 25°C. Spheroplasts were mounted on polyethyleneimine-coated slides and then rinsed in −20°C methanol for 6 min followed by −20°C acetone for 30 s. Primary antibody sources and dilutions were rat anti-α-tubulin monoclonal antibody YOL1/34 at 1:200 (Harlan Sera-lab, Indianapolis, Ind.), mouse anti-c-Myc epitope monoclonal 9E10 at 1:50 (BAbCO, Richmond, Calif.), and rabbit anti-GFP at 1:30 (Clontech, Palo Alto, Calif.). Secondary antibody sources and dilutions were fluorescein isothiocyanate (FITC)-conjugated goat anti-rat immunoglobulin G (IgG) at 1:200, Cy3-conjugated goat anti-mouse IgG at 1:500, and FITC-conjugated goat anti-rabbit IgG at 1:200. All secondary antibodies were purchased from Jackson Immunoresearch, West Grove, Pa. Cells were photographed with Kodak T-MAX p3200 film at ASA 1600.
Three controls lacking primary antibody were used to make sure that there was not any crossover fluorescence between the FITC and Cy3 fluorophores. First, only faint GFP fluorescence was seen when cells from strain GEY111 were incubated with both FITC- and Cy3-conjugated secondary antibodies in the absence of primary antibodies. Second, FITC signal alone was detected when GEY111 cells were incubated with the anti-GFP polyclonal antibody as the only primary antibody and with both FITC- and Cy3-conjugated secondary antibodies. The third control experiment with the anti-Myc 9E10 monoclonal antibody as the only primary antibody and both FITC- and Cy3-conjugated secondary antibodies had to be carried out with Nnf1p-Myc6 cells from strain GEY110 because GFP fluorescence can still be seen even without amplification from the anti-GFP antibody. In summary, no crossover fluorescence between Cy3 and FITC was detected in the absence of either or both anti-Myc and anti-GFP primary antibodies.
For MT depolymerizations, cultures of haploid yeast strains (GEY110, GEY176, GEY206, and GEY216) expressing either Nnf1p-Myc6, Dsn1p-GFP, Dsn3p-GFP, or Nsl1p-GFP from their endogenous promoters were split and either treated with 17 μg of nocodazole (Sigma Chemical Co.) per ml from a 1.5-mg/ml stock in dimethyl sulfoxide (DMSO) or mock treated with DMSO for 2.5 h at 30°C. Cells were fixed and prepared for indirect immunofluorescence, as described above, using either antitubulin antibodies or anti-Myc or anti-GFP antibodies. For the nocodazole experiment, cells were imaged with a Leica fluorescence microscope and a CCD camera (Princeton Instruments, Inc.).
RESULTS
The nnf1-17 mutant is defective in nuclear division.
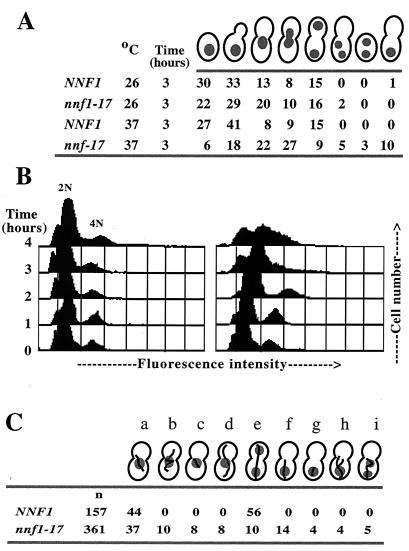
NNF1 is essential for viability. To determine whether Nnf1p may have a specific role in the cell division cycle and to analyze the onset of the nnf1-17 defect, phenotypes of logarithmically growing nnf1-17 cells were examined after a shift to the nonpermissive temperature of 37°C. In wild-type strains, large buds are indicative of cells at the G2/M border (reviewed in reference 51). After 3 h at 37°C, large-budded cells comprised more than 70% of the nnf1-17 population, compared to ∼30% of the NNF1 population (Fig. 1A). The majority of these nnf1-17 cells were uninucleate with the nucleus positioned at the bud neck. In a small percentage of nnf1-17 cells, nuclear division was completed in the mother cell, resulting in binucleate cells. To estimate DNA content and look for aneuploidy, flow cytometry was carried out on asynchronous cultures of NNF1 and nnf1-17 cells. At time zero (Fig. 1B) homozygous diploid NNF1 and nnf1-17 cells generated two peaks, corresponding to 2N and 4N DNA contents. After 3 h at 37°C, these peaks are maintained in the wild-type population, whereas in the nnf1-17 population, these two peaks merge and broaden, indicating cells with a wide range of DNA content. Thus, it appears that the nnf1-17 mutant fails to arrest at the G2/M border. The nnf1-17 mutation leads to a mitotic defect, as DNA does not segregate evenly in the mutant.
FIG. 1.
nnf1-17 cells have a mitotic defect. (A) Nuclear distribution (percent occurrence) in nnf1-17 cells. Wild-type (GEY145) or nnf1-17 (GEY146) cells were shifted from 25 to 37°C at time zero. After 3 h at 37°C, cells (n > 200 for each time point) were fixed and stained with DAPI. Anucleate cells were not observed, perhaps due to either cell lysis or difficulty in detection. (B) The DNA content of logarithmically growing NNF1 (GEY145) and nnf1-17 (GEY146) cells was analyzed by flow cytometry. (C) MT morphologies in large-budded nnf1-17 cells (percent occurrence) after 3 h at 37°C. Wild-type (GEY122) or nnf1-17 (GEY138) cells were shifted from 25 to 37°C at time zero. MTs were visualized by indirect immunofluorescence with antitubulin antibody. Cells were scored as follows: a, short spindle at or inserted in the bud neck; b, short, misoriented spindle; c, short spindle with missing cytoplasmic MTs; d, short spindle with elongated cytoplasmic MTs; e, extended spindle with divided nucleus; f, anaphase occurring in the mother cell; g. anaphase occurring in the mother cell with missing cytoplasmic MTs; h. anaphase occurring in the mother cell with elongated cytoplasmic MTs; and i, binucleated mother cell.
MT morphologies in large-budded cells were examined after 3 h at 37°C (Fig. 1C). The mitotic spindle was short in most (∼85%) large-budded nnf1-17 cells (columns a to d and f to h in Fig. 1C). These short spindles were sometimes misoriented with respect to the mother-daughter axis (column b in Fig. 1C). In a smaller percentage of the nnf1-17 population, the cytoplasmic MTs were also affected, in that they were either elongated (columns d and h) or missing altogether (columns c and g). The nnf1-17 mutation does not cause a global defect in MT function, because the mutant is not hypersensitive or resistant to benomyl and displays no karyogamy defect (data not shown). Therefore, Nnf1p is required for mitotic spindle elongation and orientation.
nnf1-17 cells display plasmid instability.
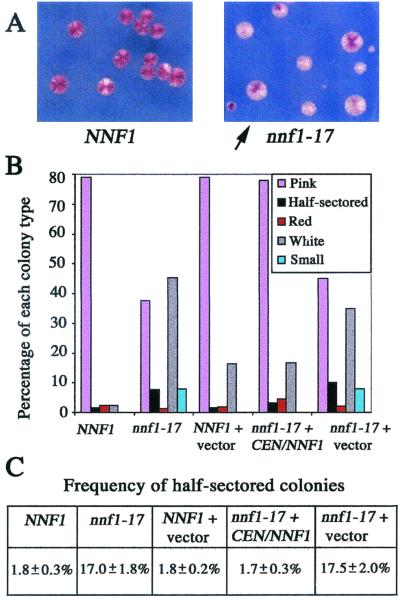
If mitotic spindle function is the primary nnf1-17 defect, then there may be a partial spindle defect in nnf1-17 cells under semipermissive growth conditions. A centromere-plasmid stability assay was used to detect mitotic segregation defects (40, 41). NNF1 ade2 ade3 and nnf1-17 ade2 ade3 strains carrying the CEN/ade3-2p plasmid pGE102 were grown under nonselective conditions at 30°C (Fig. 2A and B). The rate of plasmid loss can be determined from the number of colonies that are half pink (one copy of pGE102) and half white (zero copies of pGE102) due to plasmid loss during the first mitotic division upon plating. For NNF1 cells, the CEN plasmid loss rate was 1.8% ± 0.3% per generation; for nnf1-17 cells, the rate was 17.0% ± 1.8% (see Materials and Methods) (Fig. 2C). The presence of an NNF1 plasmid had no effect on wild-type plasmid stability but complemented the nnf1-17 defect. Therefore, the nnf1-17 mutation causes CEN plasmid instability under semipermissive growth conditions. These observations support the hypothesis that Nnf1p is required for mitotic spindle function.
FIG. 2.
nnf1-17 cells show unstable transmission of a centromeric plasmid. Assays were performed as described in Materials and Methods. (A) The nnf1-17 mutant (GEY165) rapidly loses a centromeric ade3-2p plasmid (pGE102) when grown under nonselective conditions. The arrow points to a half-sectored colony. A wild-type strain (GEY160) carrying pGE102 is shown for comparison. (B) Percentages of each colony type were scored for >800 colonies of the following strains carrying pGE102: NNF1, GEY160; nnf1-17, GEY165; NNF1 + vector, GEY160 + pRS314; nnf1-17 + CEN/NNF1, GEY165 + pGE99; and nnf1-17 + vector, GEY165 + pRS314. Small colonies were ∼1 mm in diameter with irregular margins and thus could not be scored for half-sectoring. (C) The frequency of half-sectored colonies was calculated for each transformant as the number of half-sectored colonies divided by the total number of colonies with one copy of pCEN/ade3-2p (pGE102) upon plating. Errors are standard deviations of the means for the four or five transformants. Approximately 200 colonies were scored for each transformant.
Isolation and characterization of suppressors of nnf1-17.
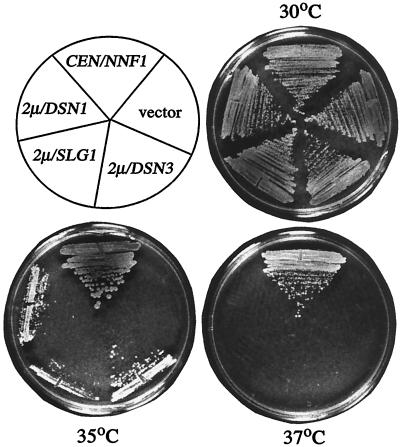
Dosage suppressors of nnf1-17 were isolated by selection for growth at the minimum restrictive temperature (35°C). In addition to NNF1 itself, four other genes were identified: SSD1(SRK1), SLG1(WSC1), and the newly identified genes DSN1 and DSN3 (dosage suppressor of NNF1). Only NNF1 in either a CEN or 2μm vector was able to restore wild-type growth to the nnf1-17 strain at 37°C. High-copy DSN1, SLG1, and DSN3 allowed growth at 35°C to various degrees (Fig. 3).
FIG. 3.
Growth of an nnf1-17 mutant with various suppressors at 30, 35, and 37°C. The nnf1-17 strain GEY138 was transformed to Ura+ at 25°C with pGE101 (CEN/NNF1), pRS316 (empty vector), pGE164 (2μm/DSN1), pGE190 (2μm/SLG1), or pGE151 (2μm/DSN3). The transformed strains were streaked on the same minimal SC-Ura plates and incubated for 2 days at the indicated temperatures.
The DSN1 protein is 576 amino acids and has a predicted molecular mass of 66 kDa and a predicted pI of 5.2. A match to a consensus bipartite nuclear localization sequence is found at residues 414 to 430; a potential EF-hand calcium binding site is found at residues 398 to 410. Expression of DSN1 peaks during G2 (62). Dsn1p has no significant overall similarity to any known protein in the available databases.
The DSN3 protein is 289 amino acids and has a predicted molecular mass of 33 kDa and a predicted pI of 5.1. DSN3 was found to be equivalent to MTW1, a gene identified in S. cerevisiae through its homology to the Schizosaccharomyces pombe gene mis12 (17, 18). In budding yeast, Mtw1p-GFP is closely situated near the SPBs as seen by fluorescence microscopy, Mtw1p coimmunoprecipitates with centromere DNA, and the temperature sensitive mtw1-1 mutant exhibits unequal chromosome segregation (18). Recently Mtw1p was reported to be present in a highly enriched preparation of spindle poles, as detected by matrix-assisted laser desorption-ionization mass spectrometric analysis (76).
SLG1 and SSD1 are both nonessential genes. Slg1p is a plasma membrane protein that is part of the Pkc1p pathway (19, 31, 44, 73). Overexpression of SLG1 may maintain viability in the nnf1-17 background through the heat shock response and cell integrity signaling or through its effects on the cell cycle and SPB duplication (29, 35, 65). Ssd1p is a cytoplasmic protein which may control RNA metabolism by affecting RNA stability (72). Various wild-type yeast laboratory strains have polymorphisms of SSD1. The dominant SSD1-v allele, present in the wild-type strain S288C, has been found in single or multiple copies to partially suppress growth defects associated with diverse mutations (13, 14, 38, 63). In this study nnf1-17 was present in a W303 background, and the wild-type strain W303 has the recessive ssd1-d allele.
Identification of mutations that are lethal in a nnf1-17 strain.
A synthetic lethal screen was also used to identify gene products that may act in conjunction with Nnf1p. These clones were called NSL genes (NNF1 synthetic lethal). An ade2 ade3 colony sectoring assay (1, 42) was carried out at 30°C. A total of 102,000 mutagenized colonies of strain YSLP1 (ade2 ade3 nnf1-17) carrying an ADE3/NNF1 plasmid were screened to find isolates that had a stable red, nonsectoring phenotype. Four isolates (YSL6, YSL8, YSL29, and YSL68) were chosen for cloning by complementation of the nonsectoring phenotype.
Cloning of NSL1.
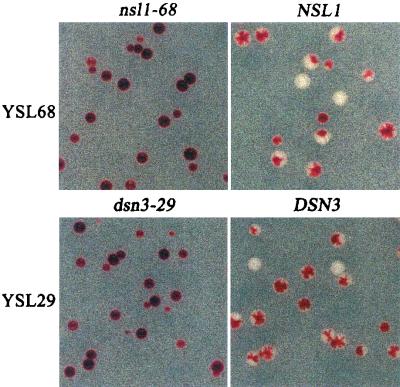
Plasmids carrying YPL233w and small flanking regions complemented the nsl1 mutants YSL6, YSL8, and YSL68. To verify that mutants YSL6, YSL8, YSL68 carry nsl1 mutations, the alleles nsl1-6, nsl1-8, nsl1-68, and NSL1 were isolated by gap repair of YPL233w from strains YSL6, YSL8, YSL68, and YSLP1, respectively. Colony sectoring assays indicated that the retrieved NSL1 allele complemented mutants YSL6, -8, and -68, whereas the retrieved nsl1-6, -8, and -68 did not (Fig. 4). Therefore, YPL233w is NSL1, and it encodes a novel protein of 216 amino acids with a predicted molecular mass of 25 kDa and a predicted pI of 4.7. No significant sequence similarity was found between Nsl1p and proteins in current databases.
FIG. 4.
Complementation of strain YSL68 by NSL1 and of strain YSL29 by DSN3(MTW1). The synthetic lethal mutant YSL68 carrying plasmid pGE100 (CEN/TRP1/ADE3/NNF1) was transformed with either pGE84 (CEN/URA3/nsl1-68) or pGE81 (CEN/URA3/NSL1). The synthetic lethal mutant YSL29 carrying pGE100 was transformed with either pGE90 (CEN/URA3/dsn3-29) or pGE89 (CEN/URA3/DSN3). Transformants were selected on SC-Ura low-Ade plates at 30°C for 5 days. Wild-type NSL1 and DSN3(MTW1) restore the sectoring phenotype to strains YSL68 and YSL29, respectively. Comparable results were obtained with plasmid pGE82 in strain YSL6 and plasmid pGE83 in strain YSL8.
The UV mutagenesis might have produced nsl1 mutations conferring a distinct growth defect. However, no growth defects were observed when the nsl1-6, nsl1-8, and nsl1-68 alleles were placed in a nsl1::LEU2 mutant strain. Hence, nsl1-6, nsl1-8, and nsl1-68 probably represent only a partial loss of Nsl1p function.
Mutations in DSN3(MTW1) are synthetically lethal in combination with nnf1-17.
I reasoned that the same genes might be isolated through nnf1-17 synthetic lethality and dosage suppression. Therefore low-copy plasmids carrying each dosage suppressor were tested for complementation of all YSL mutants. DSN3(MTW1) complemented the nonsectoring phenotype of mutant YSL29.
To establish that a mutation in DSN3 was synthetically lethal in combination with nnf1-17 and that DSN3 was not just a low-copy suppressor of the mutations in YSL29, chromosomal dsn3-29 and DSN3 were recovered from the mutant strain YSL29 and the parent strain YSLP1, respectively, and a plasmid shuffle experiment similar to the one performed with the nsl1 isolates was carried out (see also Materials and Methods). DSN3 (in plasmid pGE89) fully complemented the nonsectoring phenotype of the YSL29 mutant, but dsn3-29 (in plasmid pGE90) did not (Fig. 4). Therefore, YAL034w-a displays both synthetic lethality and dosage suppression with nnf1-17. The dsn3-29 allele does not confer a growth defect, since no temperature sensitivity was observed in a dsn3::LEU2 strain carrying dsn3-29 in a CEN plasmid as the only source of Dsn3p.
DSN1, DSN3(MTW1), and NSL1 are essential genes.
Wild-type diploid strains (CY6 or GEY160) were transformed with dsn1::HIS3, dsn3::LEU2, or nsl1::LEU2 constructs, and for each disruption, three independent transformants that showed a single integration at the DSN1, DSN3, or NSL1 locus were chosen for tetrad analysis. Dissection of the dsn1::HIS3, dsn3::LEU2, or nsl1::LEU2 heterozygotes resulted in poor spore viability (28 to 40%) and a number of His+ or Leu+ progeny (Tables 3 to 5). Southern analysis showed that all three His+ spores from the dsn1::HIS3 heterozygote still maintained a wild-type copy of DSN1, and these spores were not able to mate with MATa or MATα tester strains (Table 3). Therefore, these His+ spores may be diploids or disomes. Similar results were obtained for the DSN3 and NSL1 disruptions (Tables 4 and 5). Low spore viability and disome or diploid progeny appear to be part of the mutant phenotypes, as sporulation and dissection of the CY6 and GEY160 parent strains resulted in 95 and 93% viability, respectively. None of these spores was a nonmater. By inference, the His+ or Leu+ spores recovered from the dsn1::HIS3, dsn3::LEU2, or nsl1::LEU2 heterozygotes are the result of nuclear or chromosome missegregation.
TABLE 3.
Tetrad data from DSN1 disruption
| Strain(s) (genotype) | % Spore viability | DSN1 (His− spores) | DSN1/dsn1::HIS3 (His+ spores)a | dsn1::HIS3 + pCEN/ URA3/DSN1 (His+ Ura+ spores)b |
|---|---|---|---|---|
| CY6 (DSN1/DSN1) | 95 | 19 | NAc | NA |
| GEY170, GEY171, GEY172 (dsn1::HIS3/DSN1) | 28 | 19 | 3 | NA |
| GEY170, GEY171, GEY172 + pGE165 (dsn1::HIS3/DSN1 + pCEN/URA3/DSN1) | 43 | 17 | 6 | 6 |
All nine of the His+ spores were nonmating and still had a wild-type copy of DSN1 which was detected by Southern analysis, as described in Materials and Methods.
All six of the His+ Ura+ spores were not able to lose pCEN/URA3/DSN1 on 5-FOA plates.
NA, not applicable.
TABLE 5.
Tetrad data from NSL1 disruption
| Strain(s) (genotype) | % Spore viability | NSL1 (Leu− spores) | NSL1/nsl1::LEU2 (Leu+ spores)a | nsl1::LEU2 + pCEN/ URA3/NSL1 (Leu+ Ura+ spores)b |
|---|---|---|---|---|
| GEY160 (NSL1/NSL1) | 93 | 41 | NAc | NA |
| GEY190, GEY191, GEY192 (nsl1::LEU2/NSL1) | 30 | 18 | 5 | NA |
| GEY190, GEY191, GEY192 + pGE180 (nsl1::LEU2/NSL1 + pCEN/URA3/NSL1) | 32 | 27 | 0 | 8 |
Three out of the five Leu+ spores were nonmating. All five of the Leu+ spores still had a wild-type copy of NSL1 which was detected by PCR, as described in Materials and Methods.
All eight of the Leu+ Ura+ spores were not able to lose pCEN/URA3/NSL1 on 5-FOA plates.
NA, not applicable.
TABLE 4.
Tetrad data from DSN3 disruption
| Strain(s) (genotype) | % Spore viability | DSN3 (Leu− spores) | DSN3/dsn3::LEU2 (Leu+ spores)a | dsn3::LEU2 + pCEN/ URA3/DSN3 (Leu+ Ura+ spores)b |
|---|---|---|---|---|
| GEY160 (DSN3/DSN3) | 93 | 41 | NAc | NA |
| GEY210, GEY211, GEY212 (dsn3::LEU2/DSN3) | 40 | 35 | 4 | NA |
| GEY210, GEY211, GEY212 + pGE171 (dsn3::LEU2/DSN3 + pCEN/URA3/DSN3) | 60 | 43 | 5 | 19 |
Six out of the nine Leu+ spores were nonmating. All nine of the Leu+ spores still had a wild-type copy of DSN3 which was detected by PCR, as described in Materials and Methods.
All 19 of the Leu+ Ura+ spores were not able to lose pCEN/URA3/DSN3 on 5-FOA plates.
NA, not applicable.
The failure to recover haploid dsn1::HIS3, dsn3::LEU2, or nsl1::LEU2 spores suggested that DSN1, DSN3, and NSL1 are essential for spore germination and perhaps viability. To verify this conclusion, these dsn1::HIS3, dsn3::LEU2, or nsl1::LEU2 heterozygous diploids carrying CEN/URA3 plasmids with DSN1, DSN3, and NSL1, respectively, were tested for the ability to produce meiotic segregants lacking the plasmid (Tables 3 to 5). All of the His+ or Leu+ spores either were unable to grow on 5-FOA medium (which selects against Ura+ and thus against plasmid loss) or still maintained a wild-type copy of the gene targeted for disruption, as detected by PCR analysis. To conclude, DSN1, DSN3, and NSL1 are essential genes.
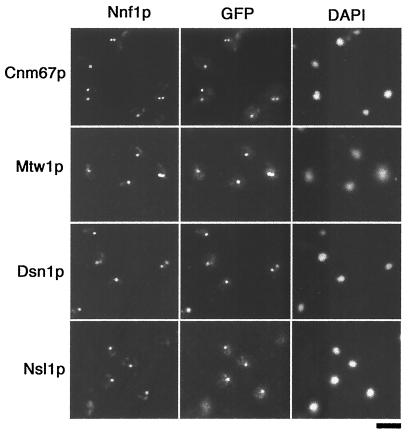
Nnf1p, Mtw1p, Dsn1p. and Nsl1p colocalize and are found near the spindle poles.
Epitope-tagged Nnf1p-Myc6 expressed from the NNF1 locus (strain GEY110) was used for subcellular localization studies. Indirect immunofluorescence showed that Nnf1p-Myc6 is present at the nuclear periphery as a single dot in unbudded cells and as side-by-side dots in small-budded cells. In large-budded cells, Nnf1p appears at opposite ends of the dividing nucleus. This localization pattern is typical of spindle pole body proteins (see, e.g., reference 75) and certain kinetochore proteins that transiently cluster at the spindle poles during various stages of mitosis (10, 16, 32, 33, 45, 75, 76).
Double-label immunofluorescence with Nnf1p-Myc6 and an SPB marker, GFP-tagged Cnm67p (4, 75), was undertaken to verify spindle pole proximity of Nnf1p (strain GEY111) (Fig. 5). Several controls indicated an absence of crossover fluorescence between the GFP and Myc detection systems (see Materials and Methods). In cells where both Nnf1p-Myc6 and Cnm67p-GFP were visible, the fluorescent dots were closely positioned (in 54 of 54 cells scored), implying that Nnf1p resides near the spindle poles. This Nnf1p-Myc6 staining pattern was visible throughout the cell cycle, and no MT staining was observed.
FIG. 5.
Localization of Nnf1p, Mtw1p, Dsn1p, and Nsl1p to the region of the spindle poles. First row, localization of Nnf1p and the SPB component Cnm67p in strain GEY111. For colocalization of Nnf1p with Mtw1p, Dsn1p, and Nsl1p, strain GEY110 was transformed with pGE74, pGE36, or pGE55, respectively. NNF1-myc6 and CNM67-GFP are integrated in place of wild-type NNF1 or CNM67 in a haploid strain and are expressed from the endogenous promoters. The MTW1-GFP, DSN1-GFP, and NSL1-GFP fusions are expressed from their own promoters in CEN plasmids. For the indirect double-label immunofluorescence, cells were short fixed first in formaldehyde and then in methanol and acetone. The Myc6 epitope was detected with the monoclonal antibody 9E10 and a Cy3-conjugated goat anti-mouse secondary antibody. GFP fluorescence was enhanced with a rabbit anti-GFP antibody and an FITC-labeled goat anti-rabbit secondary antibody. Bar, 5 μm.
Fusions between GFP(S65T) and the C-terminal ends of DSN1, MTW1, and NSL1 were constructed to allow visualization of Dsn1p, Mtw1p, or Nsl1p in both living and fixed cells. All three fusions were expressed under control of endogenous promoters from CEN plasmids and were able to rescue the respective disruptions in plasmid shuffle experiments. The same localization patterns that are characteristic of SPBs or some kinetochore proteins were seen for Dsn1p-GFP, Mtw1p-GFP, and Nsl1p-GFP. Single cells showed one fluorescent dot which was seen to be nuclear when the cells were grown in the presence of DAPI (4′,6′-diamidino-2-phenylindole). Two fluorescent dots were seen in budding cells. The dots were adjacent in small-budded cells but were separated in large-budded cells. For colocalization with Nnf1p, Dsn1p-GFP, Mtw1p-GFP, and Nsl1p-GFP were transformed separately into the strain GEY110, which stably expresses Nnf1p-Myc6 from the NNF1 locus. Double-label immunofluorescence was performed with monoclonal anti-c-Myc and polyclonal anti-GFP antibodies. Each of the Dsn1p-GFP, Mtw1p-GFP, and Nsl1p-GFP signals was found to colocalize with the signal obtained for Nnf1p-Myc6 (Fig. 5). By single-plane immunofluorescence, Nnf1p localizes with both the SPB protein Cnm67p and the known kinetochore protein Mtw1p. However, at this resolution, it cannot be determined whether Nnf1p, Dsn1p, and Nsl1p are present at the SPBs and kinetochores, since Mtw1p has been shown to accumulate near the SPBs (18, 76). Labeling of cytoplasmic MTs or the mitotic spindle was not observed for Nnf1p, Dsn1p, Mtw1p, and Nsl1p.
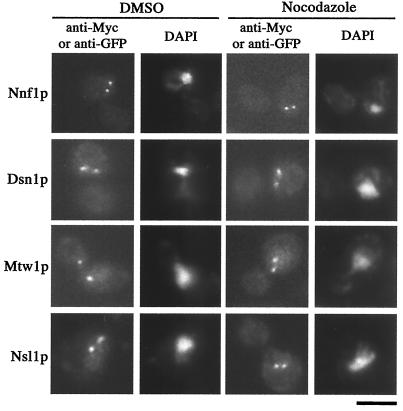
To determine whether the Nnf1p-Myc6, Dsn1p-GFP, Mtw1p-GFP, and Nsl1p-GFP localizations could be maintained in the absence of MTs, cultures of haploid yeast strains expressing these fusion proteins were treated with nocodazole to depolymerize MTs or were mock treated with DMSO. After depolymerization, antitubulin immunofluorescence revealed that MT arrays were lost in more than 93% of the cells (data not shown), indicative of effective nocodazole treatment (30). As shown (Fig. 6), the characteristic dot staining patterns were still present for the Nnf1p-Myc6, Dsn1p-GFP, Mtw1p-GFP, and Nsl1p-GFP proteins. Nocodazole treatment did not vary the number of cells showing Nnf1p-Myc6, Dsn1p-GFP, Mtw1p-GFP, and Nsl1p-GFP staining (>85% for both nocodazole and DMSO treatments), and staining intensities were comparable in treated and untreated cultures. In large-budded cells with a single nucleus, the percentage of cells with only one visible dot was higher in nocodazole-treated Nnf1p-Myc6, Dsn1p-GFP, Mtw1p-GFP, and Nsl1p-GFP cultures (∼40 to 50%; n > 90) than in the untreated cultures (∼3%; n > 100). This increase in the single-dot staining pattern may reflect either duplicated but unseparated SPBs or sister chromatids (15, 30, 66). In short, Nnf1p, Dsn1p, Mtw1p, and Nsl1p staining did not diminish with nocodazole treatment and spindle collapse.
FIG. 6.
The localization of Nnf1p, Dsn1p, Mtw1p, and Nsl1p does not depend on MTs. Cultures of haploid yeast expressing either NNF1-myc6 (strain GEY110), DSN1-GFP (strain GEY176), MTW1-GFP (strain GEY216), or NSL1-GFP (strain GEY206) were split and with treated either 17 μg of nocodazole per ml or mock treated with DMSO for 2.5 h. Cells were fixed and prepared for indirect immunofluorescence using either antitubulin antibodies (not shown) or anti-Myc or anti-GFP antibodies. Since MT arrays were lost in nocodazole-treated cells, Nnf1p, Dsn1p, Mtw1p, and Nsl1p localizations are independent of MTs (n > 90 for each strain). Each fusion was expressed from its endogenous promoter, and localizations were not dependent on untagged versions of these proteins as only tagged copies were present in each strain. Bar, 5 μm.
DISCUSSION
We previously identified Nnf1p as a protein associated with the nuclear envelope in S. cerevisiae. In this study, I show that Nnf1p is needed for genetic stability. I performed two genetic screens and identified novel proteins that have genetic interactions with NNF1. MTW1, a gene encoding a kinetochore protein, was isolated from both screens. Mutations in MTW1 enhance the temperature-sensitive nnf1-17 allele, and overexpression of MTW1 suppresses the nnf1-17 phenotype. Use of the temperature-sensitive nnf1-17 mutant has enabled me to propose that Nnf1p is necessary for chromosome segregation. Loss of Nnf1p function leads to abnormal MTs, an accumulation of large-budded cells, altered DNA content, and decreased plasmid stability. Moreover, the genetically related proteins Mtw1p, Dsn1p, and Nsl1p share the same subcellular localization as Nnf1p. Although Nnf1p, Mtw1p, and Nsl1p are similar in size, no significant sequence similarities were found between any two of these proteins. These four proteins must have unique functions, because each is essential for cell viability.
Previously (56), we described two additional phenotypes observed in the nnf1-17 mutant. First, a small percentage of nnf1-17 cells (∼15%) had a slight nuclear accumulation of poly(A)+ RNA after 3 h at 35°C. This nuclear accumulation of poly(A)+ RNA is of uncertain significance and may simply be part of the terminal phenotype due to its late onset and low penetrance. Second, in some nnf1-17 cells there were changes in the nucleolus as detected by immunofluorescence against a nucleolar antigen. The changes in nucleolar morphology may be related to the phenomenon that the nucleolus in wild-type cells is often found opposite to the SPBs (80). Conceivably, the loss of a crescent-shaped nucleolus may be due to chromosome loss, since nnf1-17 cells have a wide range of DNA content and nucleolar structure is determined at least in part by ribosomal DNA. The genetic screens reported here did not uncover any links to the nucleolus or mRNA export.
MT defects are the most prominent cytological defect seen in nnf1-17 cells, and pleiotropic MT defects are observed in large-budded nnf1-17 cells at the nonpermissive temperature. The range of MT defects observed in the nnf1-17 mutant are in line with those morphologies seen in mutants lacking spindle function. In the nnf1-17 mutant, both cytoplasmic and spindle MTs are aberrant, although not always in the same cell. However, a particular MT pattern is not always diagnostic for the role of a protein in a specific cellular substructure, perhaps due to the delicate stoichiometry of many components of the cytoskeleton. For example, mutations in the SPB outer plaque protein Spc72p result mainly in loss of cytoplasmic MTs and binucleate cells, but some spc72 mutants also exhibit spindle defects (11, 39, 60). After shifting a synchronized nnf1-17 population to the restrictive temperature (56), short, thick mitotic spindles in nuclei at the bud necks were observed at early time points (1 h after the shift). Binucleate cells and cells with elongated cytoplasmic MTs were not observed until later time points (3 h after the shift). Therefore, one possibility is that spindle elongation is the primary MT defect in nnf1-17 cells, with a nuclear migration defect occurring subsequently due to perturbations in the nuclear envelope or in tubulin pools.
MTs are essential for mitotic chromosome segregation, and so mutants with MT defects may also have higher frequencies of chromosome or plasmid loss. To test this, nnf1-17 cells were assayed for plasmid stability and were found to lose a marked plasmid at a 10-fold-higher frequency than the wild-type strain. Simple loss could be due to either MT defects or, possibly, errors in DNA replication or repair (26). If the plasmid loss phenotype is accompanied by an increase in mitotic recombination, this could imply that the mutant has DNA lesions and that the wild-type protein has a role in DNA metabolism. Plasmid loss accompanied by nondisjunction only might support a role in the MT cytoskeleton, although DNA replication defects can also prevent sister chromatid separation (25, 48). Neither recombination nor nondisjunction was tested in nnf1-17 cells in an attempt to distinguish between these possibilities. However, flow cytometry analysis of DNA content in nnf1-17 cells at the restrictive temperature and colocalization of Nnf1p and Mtw1p strongly suggest that the defect in the nnf1-17 mutant is in MT or spindle function.
Mtw1p has recently been characterized in a separate study, and the cytological behavior of an mtw1-1 mutant is consistent with defects observed in the nnf1-17 cells. The mtw1-1 mutant exhibits unstable transmission of a CEN plasmid at a semirestrictive temperature, as shown by the colony color assay (18). Synchronous cultures of mtw1-1 cells accumulate as large-budded cells with short spindles at early time points, while at later time points, some binucleate cells are seen (18), as was similarly observed in a synchronous population of nnf1-17 cells (56). Goshima and Yanagida (18) also found that in unbudded cells, Mtw1p-GFP was seen as a single dot at the nuclear periphery, whereas two Mtw1p-GFP dots were seen near the SPBs in budding cells. Mtw1p-GFP fluorescence was lost in an ndc10-1 mutant, and from Ndc10p and Mtw1p colocalization studies, these two proteins share the same kinetochore localization but Ndc10p is found additionally along the mitotic spindle (16, 18). The combined use of Mtw1p-GFP, Tub4p-GFP (γ-tubulin), and GFP-tagged chromosomes has facilitated the observations that yeast sister chromatids separate early in the cell cycle and are situated near the SPBs and that sister arms remain connected until spindle elongation (18). Further support of this phenomenon has come from He et al. (22), who have used Mtw1p-GFP as a kinetochore marker to show that sister chromatids undergo transient separations during metaphase and that centromeric chromatin has an elastic quality.
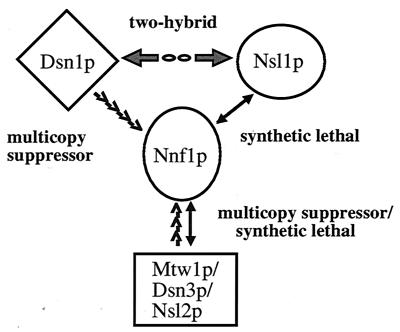
A variety of different relationships among Nnf1p, Dsn1p, Mtw1p, and Nsl1p can be envisioned. In a straightforward model, Nnf1p, Dsn1p, Mtw1p, and Nsl1p exist in a subcomplex. In this model, DSN1 and MTW1 are interaction suppressors of nnf1-17. An excess of Dsn1p and Mtw1p at the nonpermissive temperature may prevent degradation of nnf1-17 protein by sequestering the unstable monomer. Similarly, increases in the concentration of Dsn1p or Mtw1p may stabilize a structure already formed with nnf1-17 protein by maintaining an assembled complex through mass action. At lower temperatures, the Nnf1p subcomplex would be functional with the nnf1-17 protein, but partial loss of function in either Mtw1p or Nsl1p would destabilize the Nnf1p assembly and the cells would be inviable.
Thus, a future question to be addressed is whether Nnf1p, Dsn1p, Mtw1p, and Nsl1p are present in a subcomplex. I have also found that Dsn1p and Nsl1p interact in the two-hybrid system (G. Euskirchen, unpublished results). Although the mechanism of the Dsn1p and Nsl1p interaction remains to be determined, this two-hybrid result establishes another link between the genes cloned from the suppressor and synthetic lethal analysis starting with the nnf1-17 mutant (Fig. 7). The disomy or diploidization that occurs when either DSN1, MTW1, or NSL1 is disrupted implies that these proteins are required in a precise stoichiometry for chromosome segregation and suggests that these loci might be haploinsufficient. Interestingly, haploinsufficient phenotypes are not often observed. In yeast, loss of diploid dosage of the SPB duplication gene, NDC1, results in aneuploidy (12). Chromosome disorders in heterozygous dsn1, mtw1, and nsl1 diploids are consistent with the plasmid loss phenotype observed in the nnf1-17 mutant and with the strong spindle defects observed in nnf1-17 cells. The most direct explanation is that these four proteins are needed to maintain chromosome number and act together in spindle function.
FIG. 7.
Summary of interactions among NNF1, DSN1, MTW1, and NSL1. DSN1 and MTW1(DSN3) were isolated as high-copy suppressors of the nnf1-17 growth defect at 35°C. Mutations in NSL1 or MTW1(NSL2) are synthetically lethal with nnf1-17. Finally, Dsn1p and Nsl1p interact in the two-hybrid system (G. Euskirchen, unpublished data).
An extension of this model is that an Nnf1p complex is peripherally associated with the SPB. This model derives from the biochemical identification of Nnf1p from a nuclear envelope preparation (56); the localization patterns of Nnf1p, Cnm67p, Dsn1p, Mtw1p, and Nsl1p; the copurification of Mtw1p with a highly enriched spindle pole preparation (76); and the genetic interactions among NNF1, MTW1, DSN1, and NSL1 (Fig. 7). In S. cerevisiae, comparable patterns of proximal SPB localization are seen with the kinetochore proteins Ndc10p (16), Cse4p (45), Ctf19p (28), Ndc80p (75, 76), and Slk19p (81). Cytological observations that centromeres transiently cluster near the SPBs further support this hypothesis (21, 22, 33, 67). A kinetochore association has been found for Spc24p and Spc25p (32, 75), proteins that were originally identified as SPB components, both biochemically and by immunoelectron microscopy (76). The essential proteins identified in this study are well positioned to link data from various chromosome segregation studies.
Acknowledgments
Foremost, I am very grateful to Teri Mélèse and Aaron Mitchell for their generous support and critical review throughout the course of this work. The CNM67-GFP fusion was a gift from the Philippsen laboratory. Other strains and plasmids were freely given by Marion Carlson, Craig Thompson, Rodney Rothstein, Kelly Tatchell, Connie Holm, Elaine Yeh, Jeanne Hirsch, Alex Tzagoloff, and Charles diComo. I thank Rocco Carbone for help with flow cytometric analysis. Thanks go to Mike Rout, Elaine Yeh, Mike Snyder, Florian Schärer, Catherine Yan, Xiaoyin Shan, Zhixiong Xue, Anuj Kumar, Mike Smith, Jackie Vogel, and members of the Mitchell laboratory for providing suggestions and discussions.
This work was supported by a grant from the National Institutes of Health (GM44901) to Teri Mélèse. Flow cytometry was carried out with support from the Yale Cancer Center Flow Cytometry Shared Resource, Public Health Service grant CA-16359 from the National Cancer Institute. I was supported by GAANN fellowships (P200A50013-97 and P200A80111-99) from the Department of Education.
REFERENCES
- 1.Bender, A., and J. R. Pringle. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biggins, S., F. F. Severin, N. Bhalla, I. Sassoon, A. A. Hyman, and A. W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke, J. D., J. Trueheart, G. Natsoulis, and G. R. Fink. 1987. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154:164-175. [DOI] [PubMed] [Google Scholar]
- 4.Brachat, A., J. V. Kilmartin, A. Wach, and P. Philippsen. 1998. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol. Biol. Cell 9:977-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byers, B., and L. Goetsch. 1974. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp. Quant. Biol. 38:123-131. [DOI] [PubMed] [Google Scholar]
- 6.Byers, B., and L. Goetsch. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson, M., and D. Botstein. 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28:145-154. [DOI] [PubMed] [Google Scholar]
- 8.Carminati, J. L., and T. Stearns. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, C. S. M., and D. Botstein. 1993. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics 135:677-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheeseman. I. M., M. Enquist-Newman, T. Müller-Reichert, D. G. Drubin, and G. Barnes. 2001. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J. Cell Biol. 15:197-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, X. P., H. Yi, and T. C. Huffaker. 1998. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J. Cell Biol. 141:1169-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chial, H. J., T. H. Giddings, E. A. Siewert, M. A. Hoyt, and M. Winey. 1999. Altered dosage of the Saccharomyces cerevisiae spindle pole body duplication gene, NDC1, leads to aneuploidy and polyploidy. Proc. Natl. Acad. Sci. USA 96:10200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiannilkulchai, N., A. Moenne, A. Sentenac, and C. Mann. 1992. Biochemical and genetic dissection of the Saccharomyces cerevisiae RNA polymerase C53 subunit through the analysis of a mitochondrially mis-sorted mutant construct. J. Biol. Chem. 267:23099-23107. [PubMed] [Google Scholar]
- 14.Evans, D. R., and M. J. Stark. 1997. Mutations in the Saccharomyces cerevisiae type 2A protein phosphatase catalytic subunit reveal roles in cell wall integrity, actin cytoskeleton organization and mitosis. Genetics 145:227-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraschini, R., E. Formenti, G. Lucchini, and S. Piatti. 1999. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol. 145:979-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goh., P. Y., and J. V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goshima, G., S. Saitoh, and M. Yanagida. 1999. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 13:1664-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell 100:619-633. [DOI] [PubMed] [Google Scholar]
- 19.Gray, J. V., J. P. Ogas, Y. Kamada, M. Stone, D. E. Levin, and I. Herskowitz. 1997. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 16:4924-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guacci, V., E. Hogan, and D. Koshland. 1994. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 125:517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guacci, V., E. Hogan, and D. Koshland. 1997. Centromere position in budding yeast: evidence for anaphase A. Mol. Biol. Cell 8:957-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, X., S. Asthana, and P. K. Sorger. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell 101:763-775. [DOI] [PubMed] [Google Scholar]
- 23.He, X., D. R. Rines, C. W. Espelin, and P. K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106:195-206. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann, C., I. M. Cheeseman, B. L. Goode, K. L. McDonald, G. Barnes, and D. G. Drubin. 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143:1029-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoyt, M. A., T. Stearns, and D. Botstein. 1990. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol. Cell. Biol. 10:223-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huffaker, T. C., M. A. Hoyt, and D. Botstein. 1987. Genetic analysis of the yeast cytoskeleton. Annu. Rev. Genet. 21:259-284. [DOI] [PubMed] [Google Scholar]
- 27.Hutter, K. J., and H. E. Eipel. 1979. Microbial determinations by flow cytometry. J. Gen. Microbiol. 113:369-375. [DOI] [PubMed] [Google Scholar]
- 28.Hyland, K. M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and the mitotic spindle. J. Cell Biol. 145:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanovska, I., and M. D. Rose. 2000. SLG1 plays a role during G1 in the decision to enter or exit the cell cycle. Mol. Gen. Genet. 262:1147-1156. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs, C. W., A. E. M. Adams, P. J. Szanislo, and J. R. Pringle. 1988. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 107:1409-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacoby, J. J., S. M. Nilius, and J. J. Heinisch. 1998. A screen for upstream components of the yeast protein kinase C signal transduction pathway identifies the product of the SLG1 gene. Mol. Gen. Genet. 258:148-155. [DOI] [PubMed] [Google Scholar]
- 32.Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, A. Shevchenko, M. M. Magiera, C. Schramm, and E. Schiebel. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin, Q., J. Fuchs, and J. Loidl. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113:1903-1912. [DOI] [PubMed] [Google Scholar]
- 34.Jones, M. H., J. B. Bachant, A. R. Castillo, T. H. Giddings, and M. Winey. 1999. Yeast Dam1p is required to maintain spindle integrity during mitosis and interacts with the Mps1p kinase. Mol. Biol. Cell 10:2377-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khalfan, W., I. Ivanovska, and M. D. Rose. 2000. Functional interaction between the PKC1 pathway and CDC31 network of SPB duplication genes. Genetics 155:1543-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilmartin, J. V., and P. Y. Goh. 1996. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 15:4592-4602. [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, J., J. Kang, and C. S. M. Chan. 1999. Sli15 associates with the Ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, Y. J., L. Francisco, G. C. Chen, E. Marcotte, and C. S. Chan. 1994. Control of cellular morphogenesis by the Ip12/Bem2 GTPase-activating protein: possible role of protein phosphorylation. J. Cell Biol. 127:1381-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knop, M., and E. Schiebel. 1998. Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J. 17:3952-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koshland, D., and P. Hieter. 1987. Visual assay for chromosome ploidy. Methods Enzymol. 155:351-372. [DOI] [PubMed] [Google Scholar]
- 41.Koshland, D., J. C. Kent, and L. H. Hartwell. 1985. Genetic analysis of the mitotic transmission of minichromosomes. Cell 40:393-403. [DOI] [PubMed] [Google Scholar]
- 42.Kranz, J. E., and C. Holm. 1990. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc. Natl. Acad. Sci. USA 87:6629-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung, D. W., E. Chen, and D. V. Goeddel. 1989. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11-15. [Google Scholar]
- 44.Lodder, A. L., T. K. Lee, and R. Ballester. 1999. Characterization of the Wsc1 protein, a putative receptor in the stress response of Saccharomyces cerevisiae. Genetics 152:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 46.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 47.Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8:79-82. [DOI] [PubMed] [Google Scholar]
- 48.Palmer, R. E., E. Hogan, and D. Koshland. 1990. Mitotic transmission of artificial chromosomes in cdc mutants of the yeast Saccharomyces cerevisiae. Genetics 125:763-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pearson, C. G., P. S. Maddox, E. D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 51.Pringle, J. R., and L. H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle, p. 97-142. In N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 52.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:203-211. [DOI] [PubMed] [Google Scholar]
- 54.Rout, M. P., and J. V. Kilmartin. 1990. Components of the yeast spindle and spindle pole body. J. Cell Biol. 111:1913-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Shan, X., Z. Xue, G. Euskirchen, and T. Mélèse. 1997. NNF1 is an essential yeast gene required for proper spindle orientation, nucleolar and nuclear envelope structure and mRNA export. J. Cell Sci. 110:1615-1624. [DOI] [PubMed] [Google Scholar]
- 57.Shaw, S. L., E. Yeah, P. Maddox, E. D. Salmon, and K. Bloom. 1997. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 139:985-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sherman, F., J. B. Hicks, and G. R. Fink. 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 59.Sikorski, R., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soues, S., and I. R. Adams. 1998. SPC72: a spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. J. Cell Sci. 111:2809-2818. [DOI] [PubMed] [Google Scholar]
- 61.Spang, A., K. Grein, and E. Schiebel. 1996. The spacer protein Spc110p targets calmodulin to the central plaque of the yeast spindle pole body. J. Cell Sci. 109:2229-2237. [DOI] [PubMed] [Google Scholar]
- 62.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stettler, S., N. Chiannilkulchai, S. Hermann-Le Denmat, D. Lalo, F. Lacroute, A. Sentenac, and P. Thuriaux. 1993. A general suppressor of RNA polymerase I, II and III mutations in Saccharomyces cerevisiae. Mol. Gen. Genet. 239:169-176. [DOI] [PubMed] [Google Scholar]
- 64.Stirling, D. A., T. R. Rayner, A. R. Prescott, and M. J. Stark. 1996. Mutations which block the binding of calmodulin to Spc110p cause multiple mitotic defects. J. Cell Sci. 109:1297-1310. [DOI] [PubMed] [Google Scholar]
- 65.Stirling, D. A., and M. J. R. Stark. 2000. Mutations in SPC110, encoding the yeast spindle pole body calmodulin binding protein, cause defects in cell integrity as well as spindle formation. Biochim. Biophys. Acta 1499:85-100. [DOI] [PubMed] [Google Scholar]
- 66.Straight, A. F., A. S. Belmont, C. C. Robinett, and A. W. Murray. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599-1608. [DOI] [PubMed] [Google Scholar]
- 67.Straight, A. F., W. F. Marshall, J. W. Sedat, and A. W. Murray. 1997. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277:574-578. [DOI] [PubMed] [Google Scholar]
- 68.Sundberg, H. A., L. Goetsch, B. Byers, and T. N. Davis. 1996. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J. Cell Biol. 133:111-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sundberg, H. A., and T. N. Davis. 1997. A mutational analysis identifies three functional regions of the spindle pole component Spc110p in Saccharomyces cerevisiae. Mol. Biol. Cell 8:2575-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas, J. H., and D. Botstein. 1986. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell 44:65-76. [DOI] [PubMed] [Google Scholar]
- 71.Thompson, C. M., A. J. Koleske, D. M. Chao, and R. A. Young. 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73:1361-1375. [DOI] [PubMed] [Google Scholar]
- 72.Uesono, Y., A. Toh-e, and Y. Kikuchi. 1997. Ssd1p of Saccharomyces cerevisiae associates with RNA. J. Biol. Chem. 272:16103-16109. [DOI] [PubMed] [Google Scholar]
- 73.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wach, A., A. Brachat, C. Rebischung, S. Steiner, K. Pokorni, S. te Heesen, and P. Philippsen. 1998. PCR-based gene targeting in Saccharomyces cerevisiae. Methods Microbiol. 26:67-81. [Google Scholar]
- 75.Wigge, P. A., O. N. Jensen, S. Holme, S. Soues, M. Mann, and J. V. Kilmartin. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson, R. B., A. A. Brenner, T. B. White, M. J. Engler, J. P. Gaughran, and K. Tatchell. 1991. The Saccharomyces cerevisiae SRK1 gene, a suppressor of bcy1 and ins1, may be involved in protein phosphatase function. Mol. Cell. Biol. 11:3369-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winey, M., L. Goetsch, P. Baum, and B. Byers. 1991. MPS1 and MPS2: novel yeast genes defining distinct steps of yeast spindle pole body duplication. J. Cell Biol. 114:745-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Winey, M., M. A. Hoyt, C. Chan, L. Goetsch, D. Botstein, and B. Byers. 1993. NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol. 122:743-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang, C. H., E. J. Lambie, J. Hardin, J. Craft, and M. Snyder. 1989. Higher order structure is present in the yeast nucleus: autoantibody probes demonstrate that the nucleolus lies opposite the spindle pole body. Chromosoma 98:123-128. [DOI] [PubMed] [Google Scholar]
- 81.Zeng, X., J. A. Kahana, P. A. Silver, M. K. Morphew, J. R. McIntosh, I. T. Fitch, J. Carbon, and W. S. Saunders. 1999. Slk19p is a centromere protein that functions to stabilize mitotic spindles. J. Cell Biol. 146:415-425. [DOI] [PMC free article] [PubMed] [Google Scholar]