Abstract
The devastating genetic disorder Cockayne syndrome (CS) arises from mutations in the CSA and CSB genes. CS is characterized by progressive multisystem degeneration and is classified as a segmental premature-aging syndrome. The CS complementation group B (CSB) protein is at the interface of transcription and DNA repair and is involved in transcription-coupled and global genome–DNA repair, as well as in general transcription. Recent structure-function studies indicate a process-dependent variation in the molecular mechanism employed by CSB and provide a starting ground for a description of the mechanisms and their interplay.
Introduction
Patients with Cockayne syndrome (CS [MIM 133540 and MIM 216400]) are characterized by traits reminiscent of normal aging, such as systemic growth failure, neurological degeneration, and cataracts; CS has thus been classified as a segmental premature-aging syndrome. The majority of CS cases are caused by defects in the CS complementation group B (CSB) protein, which is also the factor we focus on in this review article. CS can be described as “a transcription- and DNA repair–deficiency syndrome.”
Cellular DNA is exposed to various endogenous and exogenous damaging agents. The inflicted lesions are normally repaired by the different pathways of DNA repair. However, not all lesions are removed, and it has been suggested that a gradual accumulation of DNA lesions during the cellular and organismal lifetime contributes to the normal aging process. Among the major DNA repair pathways are nucleotide-excision repair (NER) and base-excision repair (BER), both responsible for excising and repairing different lesions in DNA (reviewed by Friedberg et al. [1995]). NER is effective on bulky adducts and intrastrand cross links, whereas BER recognizes single-strand breaks and minor base lesions, such as oxidative modifications and methylations.
NER can be divided in global genome repair (GGR) and transcription-coupled repair (TCR), depending on the primary damage-recognition step. TCR is the preferential repair of the transcribed strand of active genes, where lesion-induced stalling of the transcribing RNA polymerase serves as a signal for recruitment of factors responsible for NER (Bohr et al. 1985; Mellon et al. 1987). GGR is, as implied by the name, the repair of lesions situated in all regions of the genome, and the lesion recognition is mediated by a combination of the factors XPC/hHR23B (MIM 278720 and MIM 600062), replication protein A (RPA [MIM 179835]), and XPA (MIM 278700). Many NER factors have been discovered through studies of the seven complementation groups of the genetic disorder xeroderma pigmentosum (XP [MIM 278700]) and are thus named “XPA,” “XPB” (MIM 133510), “XPC,” “XPD” (MIM 278730), “XPE” (MIM 278740), “XPF” (MIM 133520), and “XPG” (MIM 133530). Apart from XPC/hHR23B and XPE, which are specific to global genome NER (GG-NER), all NER factors are common to TCR and GGR. XPC, XPA, and RPA are responsible for recognizing and ascertaining the DNA lesion and provide binding sites for additional NER factors. The XPB and XPD helicase subunits of TFIIH (MIM 189972) unwind the DNA around the lesion, and the two structure-specific endonucleases XPG and XPF/ERCC1 (MIM 126380) make single-strand incisions 3′ and 5′ to the DNA lesion, respectively. An ∼28-base patch of DNA that contains the damaged site is thus excised, and the repair pathway is completed by DNA resynthesis by use of the undamaged complementary strand as template.
In BER, damaged bases are recognized by various DNA glycosylases, each with specific yet overlapping lesion targets. The DNA glycosylase is responsible for removing its cognate aberrant base by hydrolysis of the N-glycosylic bond. This step is succeeded by cleavage of the DNA backbone and removal of the remaining deoxyribose moiety by an endonuclease activity, pertaining either to the DNA glycosylase or to APE1 (MIM 107748). Finally, a resynthesis step of 1–7 bases, depending on the subpathway employed, restores the integrity of DNA.
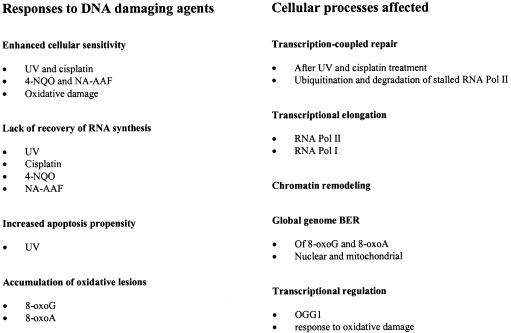
The CSB protein seems to be involved in several different processes, including transcription by RNA polymerase I (MIM 602000), II (MIM 180660), and possibly III (MIM 606007), transcription-coupled NER, and BER of some types of oxidative damage in nuclei and mitochondria (summarized in fig. 1). Apparently, CSB employs different mechanisms of action, depending on the functional context. The involvement of CSB in transcription, TCR, and BER might be simultaneous; yet, some kind of interregulation, depending on cellular status, is suspected to be a functional advantage of a common key factor in these processes. It could be through posttranslational modifications of CSB and changes in function and localization of its interaction partners. The many roles of CSB provide an explanation for the multisystem manifestations of the CS phenotype (see below) and provide rich ground for the influence of genetic background, which explains the phenotypic diversity of patients with CS.
Figure 1.
Summary of CSB cellular phenotypes: the responses of CSB cells to various DNA damaging agents and the underlying cellular processes affected.
CS: Clinical Manifestations
CS is a rare genetic disorder with autosomal recessive inheritance. It is characterized by growth failure and multisystem progressive degeneration (Cockayne 1936; Nance and Berry 1992). More than 180 cases of CS have been reported from different parts of the world, with no apparent overrepresentation in any specific population (Nance and Berry 1992; Colella et al. 1999; Mahmoud et al. 2002). Different genotypes have been found to underlie CS. Of patients with CS, ∼80% have mutations in the CSB gene, whereas the remaining patients carry mutated CSA (MIM 216400) alleles. Mutations in the XPB, XPD, or XPG genes from XP group B, D, and G, respectively, are detected in patients with a combined XP/CS phenotype (reviewed by de Boer and Hoeijmakers [2000] and Rapin et al. [2000]).
Patients with CS display variable phenotypes with respect to age at onset, symptoms displayed, and severity of the defects. However, signs of poor growth and neurological abnormalities are required for diagnosis. Growth failure is evident in height and weight, since few patients with CS exceed 115 cm in height and 20 kg in weight. Usually, weight is more affected than height, which leads to the term “cachectic dwarfism.” Neurological abnormalities are caused by progressive neurodegeneration and include delayed psychomotor development, mental retardation—measured with formal intelligence testing—and microcephaly in most patients. Gait ataxia, which indicates cerebellar dysfunction, also may be evident. Neuropathological examinations have shown multifocal patchy demyelination in the cerebral and cerebellar cortex, dilated ventricles, and calcium deposits in basal ganglia and cerebral cortex (Itoh et al. 1999; Brooks 2002). Loss of neurons was seen to a moderate degree in the vicinity of calcium deposits but was most evident throughout the cerebellum (Itoh et al. 1999). Thus, the neurological defects in CS seem to be associated with widespread demyelination of the CNS and neuronal loss in the cerebellum. Changes in glutamate transport and accumulation of oxidative products in the globus pallidus have also been reported (Hayashi et al. 2001). In addition, peripheral nerve defects have been detected as reduced velocity of nerve conduction, sensorineural hearing loss, and changes in muscle innervations (Nance and Berry 1992; Rapin et al. 2000; Lindenbaum et al. 2001).
Many patients display structural eye abnormalities, such as cataracts and pigmentary degeneration of the retina, and ciliary body defects (Dollfus et al. 2003). Another very prominent symptom in patients with CS is cutaneous photosensitivity manifested as an unusually high ultraviolet (UV) sensitivity, with resulting pigmentation abnormalities and general atrophy of sun-exposed skin (Nance and Berry 1992; Rapin et al. 2000). This is not associated with an increased risk of skin cancer. Reduced amounts of subcutaneous fat are often seen and contribute to the characteristic facial appearance of patients with CS, often called “birdlike face”: enophthalmia (sunken eyes), a beaked nose, and a narrow mouth (Nance and Berry 1992; Dollfus et al. 2003). Patients with CS also display dental caries and skeletal abnormalities such as kyphosis. The progressive degeneration seen in individuals with CS does not affect immune function and is not associated with an elevated cancer risk. The life expectancy is 12.5 years, but the individual age at death spans the spectrum from neonatal to (in one patient) age 55 years. The most common cause of death, pneumonia, is probably a result of general atrophy and cachexia.
As mentioned above, CS can be caused by mutations in five different genes, with the majority of cases attributable to CSB. Mutations in CSB, on the other hand, have also been associated with two other phenotypes, cerebro-oculo-facio-skeletal syndrome (MIM 214150) (Meira et al. 2000) and the DeSanctis-Cacchione severe neurological form of XP (MIM 278800) (Colella et al. 2000). Nine patients have so far been reported with the combined phenotype XP/CS: three with XPB/CS, two with XPD/CS, and four with XPG/CS (Rapin et al. 2000). The patients with XPD/CS and XPG/CS display a severe CS phenotype, whereas the patients with XPB/CS have very mild symptoms. Furthermore, presumably identical mutations in genes that cause CS can result in different phenotypes (Colella et al. 2000; Mahmoud et al. 2002). Thus, the genotype-phenotype relationships of these disorders and the involved genes are complex.
CSB Cellular Characteristics
Mammalian CSB studies are typically performed using hamster, mouse, or human cells. Aspects of the CSB cellular phenotype are shown in figure 1. Some rodent cells, including Chinese hamster ovary (CHO) cells, have very limited GGR of UV-induced cyclobutane pyrimidine dimers (CPD) compared with human cells. CSB studies in hamster cells are mainly performed using the UV61 cell line, which is defective in the hamster CSB homologue and which was originally used to clone the human gene for CSB (Troelstra et al. 1992). UV61 cells have been used for studies in structure-function relationships (Brosh et al. 1999; Sunesen et al. 2000, 2002) and apoptosis (Balajee et al. 2000; Proietti De Santis et al. 2001, 2002). Since the construction of a csb−/− mouse in 1997 (van der Horst et al. 1997), studies of liver cells, mouse embryonic fibroblasts (MEF), keratinocytes, and embryonic stem (ES) cells from isogenic mice have been undertaken (Osterod et al. 2002; Stevnsner et al. 2002; de Waard et al. 2003). CSB knockout mice display a phenotype resembling that of patients with CS, with mild growth-related and neurological defects and a surprising predisposition to skin cancer (van der Horst et al. 1997). CSB studies in human cells have been conducted with primary and SV40-transformed fibroblasts and with primary lymphoblastoid cells derived from patients with CS. The most frequently used cells are primary or SV40-transformed cells from patient CS1AN. The SV40-transformed cell line, CS1AN.S3.G2, has CSB alleles with A→T transversions at nucleotide 1088 that result in a stop codon at position 337 of the CSB protein (Troelstra et al. 1992). Studies using these cells must take into account the SV40-induced increased chromosomal instability and lack of p53 (MIM 191170) activity.
Hamster UV61 cells, csb−/− MEFs from Csb knockout mice, and primary as well as transformed CSB fibroblasts are hypersensitive to UV light (Wade and Chu 1979; Troelstra et al. 1992; van der Horst et al. 1997; Sunesen et al. 2000). UV irradiation induces primarily two types of lesions in DNA: CPDs, mainly removed by TCR, and pyrimidine 6-4 pyrimidone photoproducts, predominantly removed by GGR. Transfection of UV61 and CS1AN.S3.G2 cells with wild-type (WT) CSB cDNA restores the UV sensitivity to a WT level (Troelstra et al. 1992; Orren et al. 1996; Brosh et al. 1999). UV doses of 5–10 J/m2 result in a significant reduction in survival of CSB cells compared with WT cells. A hypersensitivity toward 4-nitroquinoline-1-oxide (4-NQO) has been shown for primary and SV40-transformed CS1AN fibroblasts (Wade and Chu 1979; Tuo et al. 2001; Muftuoglu et al. 2002). 4-NQO induces alkali-labile single-strand DNA (ssDNA) breaks and bulky adducts repaired without strand bias (Snyderwine and Bohr 1992). Another DNA-damaging agent, N-acetoxy-2-acetylaminofluorene (NA-AAF), inducing the lesion N-(deoxyguanosine-8-yl)-2-acetylaminofluorene (dG-C8-AAF) repaired by GGR (Tang et al. 1989), effects a reduced survival in primary CS1AN cells compared with WT cells (Wade and Chu 1979; van Oosterwijk et al. 1998). This hypersensitivity has also been shown in UV61 cells (Sunesen et al. 2000).
Ionizing radiation (IR) induces ssDNA breaks, double-strand DNA (dsDNA) breaks, and oxidative base damage. Primary and transformed CS1AN cells exposed to γ radiation have an ∼50% reduction in clonogenic survival compared with WT cells (Leadon and Cooper 1993; Tuo et al. 2001). The effect of IR on CSB-deficient cells has been ascribed to induced oxidative damage. However, Selzer et al. (2002) report no major difference in survival after H2O2, when comparing CS1AN.S3.G2 and WT cells (Selzer et al. 2002). Results of a recent study investigating the effect of IR on the clonogenic survival of MEFs, ES cells, and keratinocytes from csb knockout mice show a marked reduction in survival of csb−/− MEFs only, compared with WT cells (de Waard et al. 2003). The csb−/− MEFs were also found to be hypersensitive to paraquat (which generates superoxide) and H2O2 (causative of oxidative damage to DNA). de Waard et al. (2003) have therefore suggested that the hypersensitivity to IR seen in csb−/− MEFs is due to oxidative base damage (de Waard et al. 2003).
A characteristic cellular phenotype of CSB cells is the deficient recovery of RNA synthesis after exposure to UV (Troelstra et al. 1992; van der Horst et al. 1997). RNA synthesis in normal cells is transiently inhibited following UV exposure, but, unlike CSB cells, they recover to 90% of pretreatment levels within 90 min (Mayne 1984). The defect in recovery of RNA synthesis after UV irradiation is thought to reflect a defect in TCR (Troelstra et al. 1992). Primary CS1AN fibroblasts are also deficient in recovery of RNA synthesis after treatment with paraoxon/NA-AAF compared with WT cells (van Oosterwijk et al. 1998), which is surprising, since the inflicted damage is repaired by GGR. The same situation applies to 4-NQO in UV61 cells (Brosh et al. 1999).
Balajee et al. (1997) have shown a 50% reduction in transcription in intact and permeabilized CSB fibroblasts and lymphoblasts. This defect is ascribed to a defect in elongation of transcription. A recent study implicates CSB in rRNA synthesis by RNA polymerase I: recombinant CSB stimulates rRNA synthesis 10-fold in vitro, and CSB cells have an apparently reduced rRNA synthesis rate in vivo, which is associated with a reduced growth rate (Bradsher et al. 2002).
Analyses of differential gene expression in CSB and WT cells by use of an array technique have recently been reported. Selzer et al. (2002) compared expression profiles in CS1AN.S3.G2 fibroblasts transfected with vector alone or vector carrying the CSB WT allele, but they did not find any significant change (Selzer et al. 2002). Kyng et al. (2003) compared the expression profile of 6,912 genes after H2O2-inflicted oxidative stress for CSB-deficient and CSB-rescued SV40-transformed fibroblasts (Kyng et al. 2003). CSB-deficient cells, in response to H2O2, had a differential expression of 122 genes, compared with WT rescued cells. The down-regulated genes in the CSB-deficient cells encompassed genes encoding uracil-DNA glycosylase (UNG [MIM 191525]), heat-shock proteins, cell-cycle–related proteins, and ribosomal proteins (Kyng et al. 2003).
Another characteristic cellular feature of CSB cells is their predisposition to undergo apoptosis in response to UV treatment (Ljungman and Zhang 1996). Induction of apoptosis after UV or cisplatin treatment is associated with an accumulation of active p53 and inhibition of total RNA synthesis (Ljungman and Zhang 1996; Ljungman et al. 1999). Specific inhibition of RNA polymerase II by various agents leads to the same response (Ljungman et al. 1999). Balajee et al. (2000) showed that a mutated CSB gene is directly responsible for the propensity to UV-induced apoptosis in CSB cells (Balajee et al. 2000). Inhibition of replication by use of aphidicolin does not induce apoptosis in the normal hamster AA8 cells, whereas transcription inhibition by α-amanitin does (Balajee et al. 2000). CSB has been suggested to exert an antiapoptotic effect after UV treatment by preventing blockage of RNA Pol II transcription by UV-induced DNA damage (Ljungman et al. 1999; Balajee et al. 2000). The tumor suppressor protein p53 is induced at lower UV doses in CSB cells than in normal human fibroblasts, and the induction lasts longer (Balajee et al. 2000). It is possible that UV-induced apoptosis can be independent of p53 (Spivak et al. 2003). This is also suggested by the increased propensity to UV-induced apoptosis in UV61 cells, which have a mutated p53. Reports indicate that progression into and/or through S phase after UV treatment is necessary to induce apoptosis (Proietti De Santis et al. 2001, 2002; McKay et al. 2002). At low UV doses, CSB cells, owing to their impaired TCR, retain more damage than normal cells and are predisposed to undergo apoptosis after S phase entry. At high UV doses, the damage to CSB cells could be so devastating that entry into S phase is inhibited and apoptosis reduced compared with normal cells that, have a functional TCR, and can repair some of the damage and proceed into S phase and programmed cell death (McKay et al. 2002).
Studies by Tuo et al. (2001) show that CSB-deficient human cells accumulate oxidative damage, compared with WT cells, after treatment with γ radiation. They find significant accumulation of the oxidative purine modifications 8-hydroxy-7,8-dihydroguanine (8-oxoG) (Tuo et al. 2001) and 8-hydroxy-7,8-dihydroadenine (8-oxoA) (Tuo et al. 2002b). These studies detect no significant difference in accumulation of endogenous oxidative damage in the CSB-proficient and -deficient fibroblasts tested (Tuo et al. 2001, 2002b). However, csb−/−/ogg1 (8-oxoguanine DNA glycosylase 1 or ogg1−/− [MIM 601982]) double-knockout mice accumulate, with age, higher levels of formamidopyrimidine DNA-glycosylase (FPG)–sensitive sites—representing 8-oxoG lesions and abasic sites—in hepatocytes, splenocytes, and kidney cells than ogg1−/− mice, which indicates an influence of CSB on accumulation of endogenous oxidative damage (Osterod et al. 2002).
CSB-Influenced Processes
Venema et al. (1990) first showed that CS cells have a defect in TCR of UV-induced DNA damage (Venema et al. 1990); later, the hamster gene ercc6 (MIM 133540), encoding the homologue of human CSB, was cloned on the basis of its ability to complement the TCR defect (Troelstra et al. 1992). Using a nucleotide resolution approach, Tu et al. (1997) showed that CSB-deficient cells have a defect in repair of the transcribed strand of the JUN gene (MIM 165160) downstream from +20—a fast repair around the transcription initiation site—and a reduced repair of the promoter region (Tu et al. 1997).
TCR is thought to remove transcription-blocking lesions in actively transcribed genes, possibly by a recruitment of repair factors to the DNA lesion after removal of the stalled RNA polymerase (Svejstrup 2002). On the basis of the lack of recovery of RNA synthesis in CSB cells after 4-NQO and NA-AAF treatment, in spite of the inflicted lesions being repaired by GGR, an alternative model has been proposed. In this model, CSB acts as a transcription-repair-uncoupling factor, possibly by helping TFIIH switch from a repair to a transcription mode. However, a recent study by Hoogstraten et al. (2002) showed that TFIIH moves freely and rapidly between transcription and repair in vivo (Hoogstraten et al. 2002). Also, UV-induced transcription inhibition is reported not to correlate with any lack of TFIIH in nuclear extracts (Rockx et al. 2000).
The importance of preferential repair of actively expressed genes is suggested by the conservation of the TCR pathway from Escherichia coli and yeast to man (van Gool et al. 1997b). In E. coli, the transcription-repair coupling factor (TRCF) that is known as “the Mfd protein” can dissociate transcription complexes blocked by DNA lesions and can recruit DNA repair proteins. In Saccaromyces cerevisiae, TCR is divided into two subpathways mediated by either the CSB homologue Rad26 or the RNA Pol II subunit Rpb9 and is interregulated by Rpb4 (Li and Smerdon 2002). Results suggesting a role of TCR in the repair of thymine glycol (TG) and 8-oxoG have been published (Leadon and Cooper 1993; Cooper et al. 1997; Le Page et al. 2000), but there are some concerns over the validity of these results, since a number of these articles have been retracted (Gowen et al. 2003). TCR and strand-specific repair were not observed in the repair of 8-oxoG lesions in endogenous genes of CHO cells (Thorslund et al. 2002). TCR of UV-induced lesions was initially observed in endogenous genes, and, if TCR of 8-oxoG lesions cannot be detected in endogenous genes in a similar setting, it may suggest that 8-oxoG is repaired without TCR. Whereas UV lesions are known to block transcription, it is uncertain whether TG and 8-oxoG lesions do the same (Le Page et al. 2000; Spivak et al. 2003). Thus, if transcription arrest is a signal for TCR, this signal may not exist in the repair of 8-oxoG. In addition to CSB, several proteins appear to be involved in TC-NER: CSA, RNA Pol II, XPA-binding protein 2 (XAB2), and NER proteins (Henning et al. 1995; Nakatsu et al. 2000; Svejstrup 2002). Controversy exists as to a possible role of the mismatch-repair proteins hMSH2 (MIM 120435) and hMLH1 (MIM 120436) in TC-NER (Mellon et al. 1996; Rochette et al. 2002; Leadon and Avrutskaya 2003).
As mentioned above, a characteristic of CSB cells is the reduced general RNA synthesis. The effect on transcription has been measured using methods specifically detecting elongation, and CSB is suggested to function as an elongation factor for RNA Pol II on undamaged DNA (Balajee et al. 1997; Selby and Sancar 1997a; Tantin et al. 1997). CSB has been shown (i) to promote RNA Pol II transcription past natural pause sites in vitro (Selby and Sancar 1997a; Tantin et al. 1997); (ii) to promote transcription of genes encoding highly structured RNAs, such as the U1 (MIM 180680) and U2 (MIM 180690) small nuclear RNAs and 5S rRNA [MIM 180420] (Yu et al. 2000); and (iii) to stimulate rRNA synthesis in vitro and in vivo (Bradsher et al. 2002). This implicates CSB in transcription by RNA Pol I and II and, indirectly through the effect on 5S genes, also in RNA Pol III transcription. It is paradoxical that CSB counteracts transcript shortening by elongation factor TFIIS (MIM 601425) (Selby and Sancar 1997b), which normally is a step in rescuing backtracked, arrested elongation complexes. Perhaps the action of TFIIS and CSB in promoting elongation is somewhat redundant. The Mfd protein of E. coli has recently been found to rescue arrested transcription complexes from their typical backtracked position, possibly by rewinding DNA upstream of the complex and thereby pushing the complex forward (Park et al. 2002).
UV—or cisplatin—treatment, which induces Pol II–blocking lesions that are removed by TCR, also induces ubiquitination of RNA Pol II in normal fibroblasts but not in CSA or CSB fibroblasts (Bregman et al. 1996). The ubiquitinated Pol II is in a hyperphosphorylated state (Ratner et al. 1998). The C-terminal domain (CTD) of the Pol II large subunit changes phosphorylation state during different stages of transcription, going from a hypophosphorylated state, Pol IIa, at promoter binding and transcription initiation to a hyperphosphorylated state, Pol IIo, upon promoter escape and elongation. The implication of CSB in the ubiquitination of Pol II is supported by the preferential interaction of CSB with elongating or stalled RNA Pol II (Tantin et al. 1997). After UV-induced ubiquitination of the Pol II large subunit, the level of this polypeptide is reduced. This reduction is inhibited by proteasome inhibitors, indicating that ubiquitination targets the large subunit for proteasomal degradation (Ratner et al. 1998). The reduction in the Pol II large subunit level after UV treatment is not seen in CSA or CSB cells (McKay et al. 2001). Also, a correlation between ubiquitination and blockage of transcription in vitro by α-amanitin has been found (Lee et al. 2002a). Together, these results have led to a model in which an elongating RNA Pol IIo that is stalled by a CPD or cisplatin adduct can be ubiquitinated and proteasomally degraded; this is somehow affected by CSA and CSB (Svejstrup 2003). This proteasome-mediated degradation is independent of p53 and appears important for recovery of RNA synthesis but not for TCR (McKay et al. 2001).
By analogy to the Mfd protein of E. coli, CSB has been suggested to directly dissociate stalled RNA Pol II. However, using recombinant CSB in an in vitro transcription assay, Selby and Sancar (1997b) found that CSB could not dissociate the ternary complex of DNA, mRNA, and stalled RNA Pol IIo (Selby and Sancar 1997b). This discrepancy has been proposed to be a result of differences between prokaryotes and eukaryotes in average transcript length, leading to a difference in propensity to abort an incomplete transcript due to DNA lesions. Eukaryotes might invest more resources in DNA repair and resumption of transcription when confronted with RNA Pol II blocks than do prokaryotes. It could also be an artifact of the in vitro system or the fact that recombinant-purified CSB is used (Svejstrup 2003). However, another factor, human transcription release factor 2 (HuF2), has been shown to dissociate RNA Pol I and II stalled at CPDs in vitro (Hara et al. 1999). This dissociation was not affected by addition of recombinant CSB (Hara et al. 1999). This observation suggests some functional similarity between HuF2 and TRCF and makes the incorporation of a rapid dissociation activity of stalled RNA polymerases important in models describing the effect of CSB on stalled RNA Pol II in TCR in humans (Hara et al. 1999).
CSB has also been suggested to remodel the DNA-Pol II interface to allow DNA repair or to promote bypass of some types of damage (Svejstrup 2003). This has been suggested from the findings that (i) the Mfd protein is an ATP-dependent DNA translocase that can move arrested RNA polymerase forward (Park et al. 2002), (ii) the CSB-related RSC enzyme from the SNF2-like family is a DNA translocase, and (iii) CSB can make a damage-stalled Pol II add an extra nucleotide (Selby and Sancar 1997a; Svejstrup 2003).
The yeast homologue of CSB, Rad26, is found in a complex with a factor called “Def1” (Woudstra et al. 2002). Rad26 protects RNA Pol II from degradation after DNA damage, whereas Def1 is necessary for ubiquitin-mediated degradation of stalled RNA Pol II (Woudstra et al. 2002). These factors apparently exert opposing effects on the choice between repair and transcript abortion after DNA damage in yeast. A recent hypothesis proposes the existence of a similar complex in human cells with CSB and an as-yet-unknown Def1-like factor (Svejstrup 2003). CSB could promote either translesion transcription or ubiquitin-mediated degradation of RNA polymerase followed by NER, depending on the cause of polymerase stalling. If neither approach is successful, another activity, such as HuF2, could provoke removal of RNA Pol II by dissociation, thereby preventing apoptosis.
As a member of the SNF2-like family, CSB is related to the human Brg1 (MIM 603254), hBrm (MIM 600014), and the Drosophila ISWI, which are all capable of chromatin remodeling. This activity is implicated in changing the structure of chromatin during transcription regulation and was suggested to be relevant in DNA repair as well (Citterio et al. 2000). The chromatin remodeling activity of CSB has been investigated in vitro, and CSB was shown to remodel mononucleosomes and the chromatin structure on a chromatinized plasmid (Citterio et al. 2000). This was not accomplished through complete dissociation of the histone octamer from DNA nor by octamer transfer from the mononucleosome to free DNA; however, CSB was found to interact with core histones through their N-terminal tails and to induce negative supercoiling in a naked singly nicked plasmid (Citterio et al. 2000). In support of these results, a study of S. cerevisiae found, on deletion of rad26, the expression of genes normally repressed by transposon δ elements (Gregory and Sweder 2001). These δ elements are thought to repress expression of certain genes by affecting local chromatin structure, and the result of the study therefore could implicate Rad26 in chromatin remodeling (Gregory and Sweder 2001).
Preferential repair of DNA associated with the nuclear matrix after UV irradiation (Mullenders et al. 1988) and recruitment of different factors to the nuclear matrix after UV irradiation is inhibited in CSB fibroblasts. After UV irradiation, the proliferating cell nuclear antigen (PCNA [MIM 176740]) forms an insoluble complex with nuclear substructures in nondividing cells. This occurs at a rate that is twofold lower in UV61 cells than in normal hamster cells, and this defect can be rescued by transfection with the WT human CSB gene (Balajee et al. 1998). PCNA can form a trimeric sliding clamp, which is loaded onto DNA by replication factor C (MIM 102579) and is involved in replication by polymerases δ and ɛ in NER resynthesis, long-patch BER, postreplication repair, and mismatch repair. Balajee et al. (1999) also investigated the induction of the PCNA complex in human cells in response to UV and H2O2 (Balajee et al. 1999). They found a ninefold induction of PCNA complex in normal cells after UV irradiation, compared with fourfold induction in CSB fibroblasts and close to none in XP-A cells, which corroborates the data obtained with hamster cells (Balajee et al. 1999). H2O2-induced oxidative damage gave an increase in PCNA, in the detergent-insoluble fraction, of fivefold in WT cells (Balajee et al. 1999). The induction was 1.7-fold lower in CSB cells (Balajee et al. 1999). Another factor with a CSB-affected translocation to the nuclear matrix after DNA damage is CSA (Kamiuchi et al. 2002). Translocation of CSA is CSB-dependent; is induced by UV irradiation, cisplatin, and H2O2 treatment; and results in colocalization of CSA with RNA Pol IIo (Kamiuchi et al. 2002). It is not induced by dimethyl sulfate and is independent of XPA and XPC, which indicates the TCR-specific nature of the response (Kamiuchi et al. 2002). However, that study found that most of the CSB was not present in the nuclear matrix fraction after damage (Kamiuchi et al. 2002). Thus, the role of CSB in the nuclear matrix is not yet understood. However, it could be related to the chromatin-remodeling activity of CSB.
The reduced PCNA complex formation in CSB cells after H2O2 treatment indicates a defect in the response to oxidative damage (Balajee et al. 1999). Dianov et al. (1999) made the first demonstration of a global genome–DNA repair defect in CS cells, showing that the incision of an 8-oxoG–containing oligonucleotide was 40%–50% reduced in whole-cell extracts from CS1AN and primary CSB fibroblasts compared with normal cells (Dianov et al. 1999). The 8-oxoG incision could be increased 60% in CSB-deficient cells by transfection with the CSB gene (Dianov et al. 1999). This in vitro incision activity is thought to reflect global genome BER (GG-BER), since the oligonucleotide is not transcribed and is unlikely to have chromatin structure (Tuo et al. 2001). UV61 transfectants without the CSB gene have 40% of the 8-oxoG incision in whole-cell extracts, compared with cells carrying a WT CSB gene copy (Sunesen et al. 2002). Using transfectants of CS1AN.S3.G2, Tuo et al. (2001) also found a reduction in 8-oxoG incision, in whole-cell extracts of CSB-deficient cells, to 33% of levels found in CSB-proficient transfectants (Tuo et al. 2001). This correlated with a reduced cellular resistance to γ irradiation (Tuo et al. 2001). Further support of a role for CSB in the processing of 8-oxoG lesions comes from the finding that antibody-mediated depletion of CSB from whole-cell extracts leads to a 25% reduction in 8-oxoG incision, which can be restored by the addition of recombinant CSB (Tuo et al. 2002a). Also, the electrophoretic mobility-shift assay (EMSA) protein-binding capacity to an 8-oxoG–containing oligonucleotide seen in whole-cell extracts from CSB-proficient cells was abrogated in whole-cell extracts from cells lacking CSB (Tuo et al. 2002a).
Apart from the in vitro incision assay, repair of oxidative damage has also been measured in vivo by the removal of FPG-sensitive sites after damage induction with light-activated photosensitizers, such as acridine orange, methylene blue, and RO19-8022, primarily introducing 8-oxoG. FPG comes from E. coli and excises 8-oxoG and formamidopyrimidine, followed by DNA strand cleavage. Sunesen et al. (2002) showed that the removal of FPG-sensitive sites in an active dihydrofolate reductase (DHFR [MIM 126060]) gene is dependent on a functional CSB gene in UV61 transfectants (Sunesen et al. 2002). This dependence on CSB was also shown, in human CS1AN cells, by Stevnsner et al. (2002), in a study in which expression of WT CSB increased the repair rate of FPG-sensitive sites (Stevnsner et al. 2002). Generally, the 8-oxoG incision and removal of FPG-sensitive sites have provided corroborating results. The incision of 8-oxoG has also recently been investigated in mitochondrial extracts from both CS1AN fibroblasts and mouse csb−/− liver cells (Stevnsner et al. 2002). Stevnsner et al. (2002) found a reduced activity in both mutant extracts, compared with those from WT cells, which indicates a role for CSB in mitochondrial repair of oxidative damage (Stevnsner et al. 2002).
The influence of CSB on BER of other types of oxidative damage has been investigated. CSB has not been found to have any effect on the incision of a TG-containing oligonucleotide in whole-cell extracts (Balajee et al. 1999; Selzer et al. 2002) or in mitochondrial extracts (Stevnsner et al. 2002). However, Tuo et al. (2002b) have recently found a 30% reduction in incision activity of 8-oxoA:T– and 8-oxoA:C–containing oligonucleotides by whole-cell extracts from CSB-deficient human fibroblasts, compared with WT cells (Tuo et al. 2002b). A recent article by Jensen et al. (2003) reported that 8-oxoA:C is incised in mitochondria by a splice-form of the enzyme OGG1 (Jensen et al. 2003). OGG1 also incises the lesion in the nucleus (Jensen et al. 2003) and is known to be responsible for incision of the majority of 8-oxoG:C in mammals. Human CSB-deficient cells have been shown to contain a reduced level of OGG1 mRNA (Dianov et al. 1999; Tuo et al. 2002a) and protein (Tuo et al. 2002a) compared with WT cells. OGG1 exists in a nuclear and a mitochondrial (mtOGG1) isoform, and the mtOGG1 protein level is also significantly reduced in whole-cell extracts and mitochondrial extracts from CSB-deficient human cells (Stevnsner et al. 2002). A direct physical and functional interaction between CSB and OGG1, however, has been difficult to prove by use of purified components (see below), but they are likely to be in the same protein complex (Tuo et al. 2002a).
The general role of CSB in the removal of 8-oxoG and 8-oxoA and the significance for the CS phenotype is suggested by a recent study of primary fibroblasts from 11 patients with CS, 4 belonging to the CSB complementation group and 7 so far unassigned (Tuo et al. 2003). They all show a reduced incision activity compared with WT whole-cell extracts (Tuo et al. 2003).
Proteins Interacting with CSB
Many proteins have been investigated for interactions with CSB, often with contradicting results. These interaction studies are reviewed below; the positive interactions are listed in table 1.
Table 1.
Proteins Interacting or in Complex with CSB
| Protein Factor | Cellular Function | References |
| CSA | TCR | Henning et al. 1995 |
| XPB and XPD of TFIIH | Transcription by Pol I and II, NER | Selby and Sancar 1997b; Bradsher et al. 2002 |
| XPG | NER, stimulation of BER | Iyer et al. 1996; Bradsher et al. 2002 |
| XAB2 | TCR | Nakatsu et al. 2000 |
| RNA Pol II | Transcription | Selby and Sancar 1997a; Tantin et al. 1997; van Gool et al. 1997; Bradsher et al. 2002 |
| RNA Pol I | Transcription | Bradsher et al. 2002 |
| p44 and p62 of TFIIH | Transcription by Pol I and II, NER | Bradsher et al. 2002 |
| p53 | Cell cycle control, G1-arrest, NER, and apoptosis | Wang et al. 1995; Yu et al. 2000 |
| Histone 2A, 2B, 3, and 4 | Chromatin component | Citterio et al. 2000 |
| p34 of TFIIE | Transcription by RNA Pol II | Selby and Sancar 1997b |
| OGG1 | BER of 8-oxoG and 8-oxoA | Tuo et al. 2002a |
| XPA | NER | Selby and Sancar 1997b |
The CSA gene is mutated in patients with CS complementation group A (CSA). Henning et al. (1995) cloned the gene in 1995 by complementation of the UV sensitivity in CSA cells (Henning et al. 1995). The gene is located at 5q12-q31 and encodes a protein of 46 kDa with five WD40-repeats (Henning et al. 1995). Proteins in the WD-repeat family are functionally diverse—with roles in signal transduction, RNA processing, general transcription, and cell division—but they have a high structural similarity (reviewed by Smith et al. [1999]). The only solved crystal structure is of the Gβ subunit of heterotrimeric G proteins. It has a barrellike shape made up of repeating units of four-stranded β sheets. CSA-deficient cells share many cellular characteristics with CSB-deficient cells and have inhibited recovery of RNA synthesis and TCR after UV irradiation (van Gool et al. 1997a). CSA interacts with XAB2 (see below) in vitro and in vivo (Nakatsu et al. 2000) and with the p44 subunit (MIM 601748) of TFIIH in vitro (Henning et al. 1995). A recent report has also shown an interaction with the N-terminal domain of topoisomerase I by use of phage display (Trzcinska et al. 2002). CSA has been found in a complex with DDB1 (MIM 600045), Cul4A (MIM 603137), Roc1 (MIM 603814), and CSN (Groisman et al. 2003). This complex has ubiquitin ligase activity, regulated in a UV-dependent manner (Groisman et al. 2003). No interaction with p62 (MIM 189972), XPD, XPB from TFIIH (Henning et al. 1995), or XPG (van Gool et al. 1997a) has been found.
Conflicting reports exist regarding a putative interaction between CSA and CSB. Henning et al. (1995) found an interaction using coimmunoprecipitation (co-IP) of in vitro translated proteins, and they reported a corroborating result from a yeast two-hybrid study (Henning et al. 1995). However, no association has been found using co-IP in whole-cell extracts, gel filtration, or copurification (Tantin et al. 1997; van Gool et al. 1997a); CSB and CSA do not colocalize in nuclear matrix preparations when immunofluorescence is used; and CSB can be isolated in a large complex devoid of CSA (Bradsher et al. 2002). Though implying the lack of a strong direct interaction, these results do not rule out the possibility of weak, transient, possibly damage-induced interactions. As mentioned above, CSA is translocated to the nuclear matrix after damage in a CSB-dependent manner, where it colocalizes with RNA Pol IIo (Kamiuchi et al. 2002). Also, the CSA-containing complex binds RNA Pol IIo after UV irradiation (Groisman et al. 2003).
The XPB gene, located at 2q21, encodes an 89-kDa polypeptide. Mutations in this gene result in the XPB/CS complex or in the transcription disorder trichothiodystrophy (TTD [MIM 601675]) (reviewed by de Boer and Hoeijmakers [2000]). The XPB protein is a 3′–5′ helicase and one of nine subunits of TFIIH. XPB functions as a part of TFIIH in transcription initiation of RNA Pol I and II and in the unwinding step of NER. Through use of a green fluorescent protein tag, XPB has recently been shown to be homogeneously distributed throughout the nucleoplasm, with particular enrichment in nucleoli (Hoogstraten et al. 2002). The nucleolar localization was abrogated by UV irradiation. XPB interacts with the TFIIH subunits XPD, p62, p44, and p34, as well as with XPG (Iyer et al. 1996). XPB and CSB have been shown to interact by use of a pull-down with CSB-GST fusion protein (Selby and Sancar 1997b). An interaction is also indicated by the ability of an XPB-containing factor to supershift a complex of elongating RNA Pol II:DNA:RNA:CSB only when it contains CSB (Tantin 1998). However, Gool et al. (1997a) found XPB and CSB fractionating differently on chromatography and saw no co-IP of the two (van Gool et al. 1997a). Contrary to this, a recent study reports XPB colocalizing with CSB and RNA Pol I in nuclear matrix preparations and identifies XPB in a CSB-containing complex immunoprecipitated at physiological salt concentration (Bradsher et al. 2002).
XPD is a helicase like XPB but with the opposite directionality, going 5′→3′. It is also a subunit of TFIIH and participates in the same processes as mentioned above for XPB. The gene is located at 19q13.2-q13.3 and, when mutated, gives rise to one of three disorders: XP, TTD, or XPD/CS. Like XPB, it interacts with some of the other subunits of TFIIH, namely p62, p44, and p34 (Iyer et al. 1996). It can be coimmunopurified with XPG in vitro (Iyer et al. 1996). XPD has been reported not to copurify with CSB (van Gool et al. 1997a). However, XPD can be found in the same large CSB-containing complex described for XPB (Bradsher et al. 2002). This complex could replace TFIIH in transcription and incision assays in vitro. It was suggested that mutations in XPB and XPD, which both can result in a partial CS phenotype, could destabilize this complex and thereby affect the function of CSB (Bradsher et al. 2002).
XPG is the third gene in which mutations can give rise to an XP/CS phenotype (reviewed by de Boer and Hoeijmakers [2000]). It is located at 13q33 and encodes a DNA endonuclease responsible for the incision 3′ to the DNA lesion in NER. XPG mutations leading to XPG/CS have, in all reported cases, resulted in severely truncated proteins (Nouspikel et al. 1997), whereas patient XP125LO with XPG has a full-length protein with a mutation that abolishes its endonuclease activity. Klungland et al. (1999) found that XPG can stimulate the incision activity of the DNA glycosylase Nth1 (MIM 602656) involved in BER of oxidative damage in vitro (Klungland et al. 1999). This effect is intact with a full-length mutant XPG lacking endonuclease activity, indicating that the XPG stimulation of BER is intact in patients with XPG (Klungland et al. 1999). The authors suggest that a full-length XPG is necessary for this function and that this contributes to the difference in the XPG and XPG/CS phenotypes. Interactions between XPG and XPB, XPD, p62, and p44 have been demonstrated (Iyer et al. 1996). XPG has been shown to interact with CSB through use of in vitro co-IP (Iyer et al. 1996), but they do not copurify or coimmunoprecipitate from HeLa whole-cell extracts (van Gool et al. 1997a). However, XPG participates in a large CSB-containing complex (Bradsher et al. 2002). Large truncating mutations in XPG, as seen in patients with XPG/CS, could potentially destabilize this complex, as suggested for XPB and XPD.
XPA is involved in the early steps of NER, but its exact role is not yet clear (Reardon and Sancar 2002). It is thought to help recognize the damaged DNA and to be necessary for both GG-NER and TC-NER (reviewed by de Boer and Hoeijmakers [2000]). XPA interacts with other NER proteins, including TFIIH, XPF/ERCC1, and RPA. Used as bait in a yeast two-hybrid screen, XPA was found to interact with a previously unknown protein, XAB2 (see below) (Nakatsu et al. 2000). An interaction between CSB and XPA could theoretically provide the link between TCR and a classical, early NER factor. However, this has been tested by two different methods, with contradicting results (Selby and Sancar 1997b; van Gool et al. 1997a).
XAB2 is a tetratricopeptide repeat protein homologous to SYF1 from S. cerevisiae and to predicted ORFs from several other organisms (Nakatsu et al. 2000). The functions of the homologous proteins are unknown, but XAB2 is apparently involved in TCR and transcription. Microinjection of anti-XAB2 antisera into WT and XP-C fibroblasts resulted in reduction in recovery of RNA synthesis after UV irradiation, in unscheduled DNA repair synthesis after UV irradiation, and in reduction in normal transcription (Nakatsu et al. 2000). XAB2 interacts with XPA, CSA, and a fraction of RNA Pol II (Nakatsu et al. 2000). XAB2 can be immunoprecipitated from whole-cell extracts of CSB cells with a stable expression of HA-His6–double-tagged CSB by use of anti-HA antibody, and it cofractionates with CSB and RNA Pol II (Nakatsu et al. 2000). Tetratricopeptide repeat motifs are arranged into a right-handed superhelix and are proposed to simultaneously interact with many proteins. XAB2 could serve as a bridging protein among the transcription, TCR, and the NER machinery (Nakatsu et al. 2000).
As suggested by its role in transcription and TCR, CSB has been found to interact with RNA Pol II in vitro and to reside in a common, large complex (Selby and Sancar 1997a; van Gool et al. 1997a; Bradsher et al. 2002). CSB interacts with both Pol IIa and IIo, with a preference for Pol IIo (Selby and Sancar 1997a; Tantin et al. 1997). As mentioned above, CSB has also been implicated in transcription by RNA Pol I and was found to reside in a complex with RPA116 (from RNA Pol I) and TAF68 (TIF-1B) but lacking RPB1 (RNA Pol II) and TAFII 250 (MIM 313650) (Bradsher et al. 2002). Explaining this discrepancy, CSB was in complex with RNA Pol I when purified using 150 mM KCl but with RNA Pol II when using 50 mM KCl (Bradsher et al. 2002). It is possible that the salt concentration–dependent variation of CSB-complex association reflects the difference in osmotic concentration between the nucleolus and the regions of RNA Pol II transcription. The effect of CSB on the metaphase stability of the 5S rRNA locus indicates a role in RNA Pol III transcription and the possibility of a direct interaction (Yu et al. 2000).
The CSB-containing complex isolated at physiological salt conditions also contains the p44 and p62 subunits of TFIIH (Bradsher et al. 2002). Both of these interact with XPB, XPD, and XPG (Iyer et al. 1996), whereas only p44 interacts with CSA (Henning et al. 1995). CSB has been reported to interact with the p34 subunit of TFIIE (MIM 189962) (Selby and Sancar 1997b), the tumor suppressor protein p53 (Wang et al. 1995; Yu et al. 2000), and the N-terminal tail of core histones (Citterio et al. 2000). Recently, p53 was reported to be a chromatin-accessibility factor for GG-NER (Rubbi and Milner 2003). The chromatin relaxation is probably initiated by the blocking of a transcribing RNA polymerase but is independent of TCR and CSB function. A CSB interaction with OGG1 not mediated by DNA was found using a pull-down from whole-cell extracts (Tuo et al. 2002a). However, this could not be confirmed by a yeast two-hybrid assay or by EMSA super shift, indicating other factors might mediate the interaction (Tuo et al. 2002a).
Several proteins have been tested for an interaction with CSB, with negative results. These include p56 of TFIIE, TFIIF, RPA, XPC/hHR23B, and XPF/ERCC1 (Selby and Sancar 1997b; van Gool et al. 1997a). In summary, CSB interacts physically with CSA, RNA Pol II, XAB2, histones, TFIIE, and p53 and can exist in complex with XPA, XPB, XPD, XPG, RNA Pol I, the TFIIH subunits p44 and p62, and possibly OGG1. These relations are listed in table 1. However, many of the protein interactions described here are observations of CSB binding to other proteins and, although these physical interactions are of interest, it is important to search for functional interactions.
Biochemical Characteristics of CSB
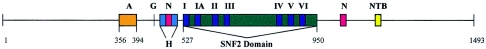
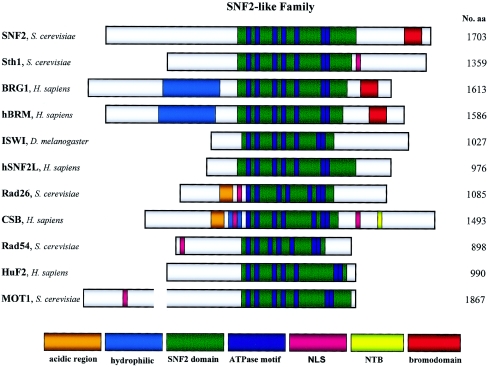
In 1992, the human homologue of the hamster ercc6 gene was cloned by Troelstra et al. (1992). Because of alternative polyadenylation, the expression of ercc6 results in two mRNAs, of 5 kb and 7.5 kb. The longest ORF encodes a predicted protein sequence of 1,493 amino acids, which, through complementation studies, was found to be the protein responsible for the phenotype of CS. CSB has a molecular weight of 168 kDa and contains several conserved motifs (Troelstra et al. 1992) (fig. 2). Among these are the seven consecutive ATPase motifs I, Ia, II, III, IV, V, and VI from amino acids 527–950. These motifs form a nucleotide-binding fold and are conserved among three superfamilies of RNA and DNA helicases, which places CSB in superfamily 2. The region of CSB encompassing these motifs is highly homologous to proteins of the SNF2-like family, such as SWI2/SNF2 (MIM 600014), Sth1, hBrm, BRG1, ISWI, MOT1 (or TAF-172 [MIM 605191]), SRCAP, RAD54 (MIM 603615), RAD16, RAD5 (MIM 607266), and HuF2 (fig. 3). Thus, CSB is placed in the ERCC6 subfamily of the SNF2-like family, together with its yeast homologue Rad26 (reviewed by Pazin and Kadonaga [1997]).
Figure 2.
Predicted motifs of CSB. A = acidic domain. G = glycine-rich stretch. H = hydrophilic region. N = bipartite NLS. I–VI = ATPase motifs located in the SNF2 domain conserved among the members of the SNF2-like family. NTB = putative NTB motif.
Figure 3.
Selected members of the SNF2-like family. Examples of family members from S. cerevisiae, Drosophila melanogaster, and humans (Homo sapiens) are listed. Conserved regions and functionally important motifs are indicated.
As shown in figure 2, CSB has an acidic region extending N-terminal to the ATPase motifs from amino acids 356–394, encompassing 60% acidic residues and an acidic stretch of 10 consecutive amino acids (Troelstra et al. 1992). Between these two motifs is a glycine-rich stretch of seven residues and a highly hydrophilic region with >60% charged amino acids. C-terminal to the ATPase motifs is a second putative nucleotide-binding (NTB) domain. The protein contains a bipartite nuclear-localization signal (NLS) with two predicted casein kinase II (CKII [MIM 115440]) phosphorylation sites nearby (Troelstra et al. 1992). Christiansen et al. (2003) recently reported that CSB contains several additional putative CKII sites, multiple phosphorylation sites for protein kinase C, and two sites recognizable by tyrosine kinases (Christiansen et al. 2003).
Owing to its relatedness to RNA and DNA helicases, CSB has been tested for DNA-helicase activity. When a strand-displacement assay was used in vitro, no helicase activity was found, nor did the addition of CSA to the reaction have any effect (Selby and Sancar 1997b). In corroboration of this, a later study found no CSB-helicase activity on a variety of different DNA substrates (Citterio et al. 1998). Indeed, no member of the SNF2-like family has thus far exhibited helicase activity (Pazin and Kadonaga 1997). It remains to be established whether the lack of measurable helicase activity could be due to the absence of a crucial accessory protein or cofactor. However, CSB exhibits DNA-dependent ATPase activity (Selby and Sancar 1997b; Citterio et al. 1998; Christiansen et al. 2003). It is highly stimulated by the presence of DNA, with a markedly larger effect of dsDNA than ssDNA (Tantin et al. 1997; Citterio et al. 1998; Christiansen et al. 2003). Comparing the effects of different dsDNA structures, Christiansen et al. (2003) recently found a 60% increase in ATPase activity when using dsDNA cofactors with a bubble or loop (corresponding to the open DNA structure during NER and transcription arrest), compared with using closed dsDNA (Christiansen et al. 2003). A dsDNA with a larger bubble, corresponding to what is found during transcription elongation, had no additional effect, compared with closed dsDNA, nor did loose structures with dsDNA-to-ssDNA transitions (Christiansen et al. 2003). These data corroborate the role for CSB in coupling NER to arrested transcription complexes in TCR. Damaged DNA used as cofactor did not result in any significant change in the ATPase activity of CSB (Christiansen et al. 2003), indicating that CSB is not a damage-recognition factor on its own. The presence of XPA or p53 did not affect the ATP hydrolysis (Christiansen et al. 2003); however, the addition of CSA to CSB and dsDNA was reported by Tantin et al. (1997) to increase the ATPase activity. Data on the effect of the various DNA cofactors are all in vitro results, through use of naked DNA. However, there is no detectable difference in the stimulatory effect of naked versus chromatinized DNA (Citterio et al. 1998).
CSB is capable of binding dsDNA in vitro in the absence of ATP and the nonhydrolyzable ATPγS (Tantin et al. 1997). DNA binding is actually inhibited in the presence of ATPγS and ADP (Christiansen et al. 2003). Thus, it is not necessary for CSB to bind ATP in order to bind DNA, but DNA binding is stimulated by certain protein conformations. These conformations are probably compatible with binding and hydrolyzing ATP but not with binding the structurally different and nonhydrolyzable ATPγS or the hydrolysis end-product ADP.
Through use of immunofluorescence and CS1AN.S3.G2 cells stably transfected with enhanced cyano-fluorescent protein–tagged CSB, CSB localizes to nuclei (van Gool et al. 1997a; Christiansen et al. 2003). This is in agreement with the prediction of a bipartite nuclear-localization signal in the amino acid sequence of CSB. During metaphase, CSB colocalizes with microtubuli of the mitotic spindle rather than with chromatin (van Gool et al. 1997a). Recently, it has been found that TCR is localized to specific chromosomal domains corresponding to the distribution of the human transcriptome, CpG islands, and regions of hyperacetylated histones (Surralles et al. 2002). This suggests that CSB might also be concentrated in certain subnuclear regions. Indeed, Bradsher et al. (2002) have recently reported that CSB localizes to nucleoplasmic foci and accumulates in nucleoli of normal human fibroblasts (Bradsher et al. 2002). In nuclear matrix preparations, CSB is detected in the nucleolus together with RNA Pol I and the XPB subunit of TFIIH (Bradsher et al. 2002). The localization of CSB is dynamically dependent on the RNA polymerase activity: when RNA Pol II is inhibited, the nucleoplasmic CSB signal weakens and, on treatment with actinomycin D, CSB is dispersed from nucleoli (Bradsher et al. 2002). Unlike what has been reported for CSA, no relocalization of CSB after UV irradiation was reported by van Gool et al. (1997a).
CSB has not been reported to localize to mitochondria, and no mitochondrial target sequences have been identified in the protein sequence. However, these signals are variable and not always easily recognizable, and CSB may be present in levels below the present detection limit. The effect of CSB on GG-BER– and OGG1-mediated incision (see above) in mitochondrial preparations naturally leads to the proposition of a fraction of the CSB, perhaps the alternatively polyadenylated form, localizing to the mitochondria, where these processes also take place.
A recent study by Christiansen et al. (2003) shows that CSB can be phosphorylated in vitro by CKII and in whole-cell extracts from human fibroblasts (Christiansen et al. 2003). CSB is also phosphorylated in vivo; this can be reversed by UV irradiation (Christiansen et al. 2003). This indicates a phosphorylation-dependent regulation of CSB activity after DNA damage, corroborated by the finding that dephosphorylation of CSB by protein phosphatase 1 (MIM 176875) increases the ATPase activity by 38% (Christiansen et al. 2003). So far, no other type of posttranslational modification has been reported for CSB. However, the recent discovery associating CSA with ubiquitin-ligase activity prompts speculations as to the role of ubiquitin in TCR. A general role for ubiquitin and the related SUMO proteins in the regulation of DNA repair is emerging with their differential effect on PCNA in postreplicative repair (Pickart 2002).
Structure-Function Relationships
Several functional studies have been conducted using various CSB mutants. The emerging structure-function relationships are discussed below and are summarized in table 2.
Table 2.
Structure-Function Relationships of CSB[Note]
|
Motif |
|||||||||
| Function | Acidic | I | IA | II | III | IV | V | VI | NTB |
| ATPase activity | ND | − | ND | − | ND | ND | + | + | ND |
| ATP binding | ND | ND | ND | ++++ | ND | ND | ++ | ++ | ND |
| DNA binding | ND | ND | ND | +++ | ND | ND | +++ | +++ | ND |
| Gene-specific repair | +++ | ND | ND | − | ND | ND | ND | ND | ND |
| Chromatin remodeling | ND | − | ND | ND | ND | ND | ND | ND | ND |
| 8-oxoG incision | +++ | ND | +++ | + | +++ | ND | ++ | (+) | +++ |
| 8-oxoA incision | ND | ND | ND | ND | ND | ND | ND | − | ND |
| hOGG1 expression | ND | ND | ND | +++ | ND | ND | ND | − | ND |
| RRSa after UV irradiation | ND | + | ++ | − | +(+) | ND | − | − | +++ |
| RRSa after 4-NQO | ND | ND | ND | − | ND | ND | ND | ND | ND |
| UV-induced apoptosis | +++ | ND | +(+) | − | +(+) | ND | − | − | +++ |
| UV sensitivity | +++ | ND | +(+) | − | +(+) | ND | + | + | +++ |
| 4-NQO sensitivity | ND | ND | ++ | − | ++ | ND | + | + | ++(+) |
| 8-oxoG accumulation | ND | ND | ND | ND | ND | ND | ND | − | ND |
| 8-oxoA accumulation | ND | ND | ND | ND | ND | ND | ND | − | ND |
| γ-radiation sensitivity | ND | ND | +++ | ND | +++ | ND | ++ | − | +++ |
Note.— Selected functional characteristics of the CSB protein carrying mutations in various protein motifs. Levels of function: ++++ = better than WT; +++ = WT; ++ = weak deficiency; + = strong deficiency; − = complete deficiency. ND = no data. Plus signs (+) in parentheses indicate a level of function intermediate between the level indicated by the total number of plus signs for a given function and the level below.
RRS = recovery of RNA synthesis.
Of the seven ATPase motifs, motifs I and II have also been called “Walker A” and “Walker B,” respectively, because they contain consensus sequences originally found by Walker in a family of NTP-binding proteins. They are therefore thought to be involved in ATP binding by CSB and in CSB-mediated hydrolysis (Hall and Matson 1999). Mutant proteins with point mutations of highly conserved residues in motifs I and II are devoid of ATPase activity in vitro (Citterio et al. 1998; Christiansen et al. 2003). Mutation of motif I and the resulting lack of ATPase activity leads to a partial defect in recovery of RNA synthesis after UV treatment, indicating an inhibition of TCR (Citterio et al. 1998). A functional motif I is also necessary for chromatin remodeling in vitro but not for the induction of topological change in naked DNA (Citterio et al. 1998). Comparison with crystal structures of proteins that belong to SF1 and SF2 suggests an interaction between the invariant lysine of the signature amino acid sequence GSGKS Walker A of motif I and the ATP β-phosphate. It is possible that this helps stabilize the transition state during hydrolysis (Hall and Matson 1999).
Several functional studies have been conducted using the E646Q CSB motif II mutant. The altered glutamate is a part of the conserved amino acid sequence DEXH involved in Mg2+ binding and necessary for ATP hydrolysis (Hall and Matson 1999). The CSB E646Q mutant cannot complement the UV sensitivity, the inhibited recovery of RNA synthesis after UV treatment, or the increased UV-induced apoptosis of UV61 and CS1AN.S3.G2 cells (Brosh et al. 1999; Balajee et al. 2000; Selzer et al. 2002). Thus, it appears defective in TCR of UV-induced damage. This is corroborated by a defect in gene-specific repair of CPDs in the DHFR gene of E646Q-transfected UV61 cells (Brosh et al. 1999). This mutant was also unable to complement the sensitivity toward and the defect in recovery of RNA synthesis after 4-NQO (Brosh et al. 1999). Since 4-NQO–induced damage is repaired by GGR, this indicates that the mutant is defective in reactivation of RNA synthesis after damage, possibly owing to a lack of reactivation of RNA polymerases. The role of motif II and the ATPase activity in the processing of oxidative damage has also been investigated. Sunesen et al. (2002) reported a reduced 8-oxoG incision in whole-cell extracts from UV61 cells transfected with CSB E646Q, compared with WT transfectants (Sunesen et al. 2002). Also, the rate of removal of FPG-sensitive sites in the DHFR gene was reduced in the mutant transfectants (Sunesen et al. 2002). However, Selzer et al. (2002) did not see any effect of E646Q on H2O2 sensitivity or 8-oxoG incision in transfected CS1AN.S3.G2 cells (Selzer et al. 2002). Stevnsner et al. (2002) have recently investigated the role of motif II in mitochondrial repair of oxidative damage in human cells. The integrity of this motif and the ATPase activity has no influence on the repair of 8-oxoG in vitro and in vivo, nor does it affect the level of mtOGG1 protein (Stevnsner et al. 2002). In a recent study, Kyng et al. (2003), using CS1AN.S3.G2 fibroblasts, reported a transcriptional response of the E646Q mutant after exposure to H2O2, that corresponds to what is seen for a CSB-null mutant, with a reduced, delayed induction of the response and changed expression of 122 genes, compared with what is seen for the WT (Kyng et al. 2003). Thus, this mutation might have an effect on the cellular response to oxidative damage, and it seems to be primarily nuclear. Recently, Christiansen et al. (2003) did not find any change in DNA binding of the CSB motif II mutant in vitro (Christiansen et al. 2003). This confirms the primary involvement of motif II in ATP binding and hydrolysis and not in nucleic-acid binding.
Studies of CSB-deficient cells transfected with CSB mutant proteins with point mutations in either ATPase motif Ia or III have revealed almost identical phenotypes, suggesting a common functional role for these motifs. Both motifs can only partially complement the increased sensitivity toward UV and 4-NQO of CSB-deficient cells (Tuo et al. 2001; Muftuoglu et al. 2002). This malfunction is also reflected in a partial defect in the modulation of UV-induced apoptosis and in a reduced recovery of RNA synthesis after UV irradiation (Muftuoglu et al. 2002). The CSB mutant deviates neither from the WT level in sensitivity toward γ irradiation nor in 8-oxoG incision (Tuo et al. 2001), indicating that, whatever their normal role is, the conservation of important residues in motifs Ia and III is not important for the function of CSB in GG-BER. Studies of homologous proteins suggest the involvement of motifs Ia and III in energy transduction between the ATPase site and the nucleic acid–binding site (Hall and Matson 1999). However, future studies of the ATPase activity and DNA-binding capacity of these mutants are necessary to further clarify their exact role.
The ATPase motifs V and VI of CSB have also been studied using point mutations of invariant residues and stable CS1AN transfectants. These mutants exhibit a similar, almost complete, inhibition of ATPase activity and reduced ATP binding in vitro (Christiansen et al. 2003). They are also equally sensitive to 4-NQO and UV treatment and have comparable defects in recovery of RNA synthesis and apoptosis induction after UV irradiation (Tuo et al. 2001; Muftuoglu et al. 2002). These results indicate a necessary contribution of motifs V and VI to TCR and RNA synthesis resumption, a role shared by motif I and II mutants and thereby probably attributable to the common effect on ATPase activity. However, motif V and VI mutants show differences in their response to oxidative damage. They both show increased γ irradiation sensitivity and reduced 8-oxoG incision, but although these defects in motif VI mutants are comparable to what is seen for CSB null mutants, motif V mutants are at an intermediary level (Tuo et al. 2001). Further studies of the involvement of CSB motif VI mutants in BER of oxidative damage have been conducted by Tuo et al. (2001). CS1AN transfectants that carry motif VI mutants accumulate 8-oxoG and 8-oxoA in genomic DNA after γ irradiation to the same extent as does a CSB-null mutant (Tuo et al. 2001, 2002b). Also, the mutants exhibit a twofold reduction in 8-oxoA:C and 8-oxoA:T incision compared with WT (Tuo et al. 2002b). These defects correlate with a reduction in the cellular level of OGG1 transcript and protein (Tuo et al. 2002a), suggesting a causative relationship between a failure to induce the expression of OGG1 and an inhibited BER of 8-oxoG and 8-oxoA. The structure of related proteins in the helicase superfamilies propose that motif V binds ssDNA directly (Hall and Matson 1999). This has not been tested, but CSB motif V mutants bind to dsDNA in vitro as efficiently as WT CSB (Christiansen et al. 2003).
The effect of CSB motif VI mutants on DNA binding is not detectable on dsDNA in vitro (Christiansen et al. 2003) but could possibly be evident on ssDNA, as suggested for motif V. As mentioned above, the CSB ATPase activity is stimulated to a much higher degree by dsDNA than by ssDNA, apparently contradicting ssDNA binding by motif V and maybe VI. However, as mentioned above, Christiansen et al. (2003) have recently found that the stimulatory effect is even higher for dsDNA containing a small bubble or loop, suggesting that some parts of CSB contact dsDNA—whereas others contact ssDNA—and placing CSB at the ssDNA-to-dsDNA transition of the structure. A putative candidate for the dsDNA binding is the as-yet-uninvestigated motif IV, which, from the structure of the SF2 member NS3, is known to contact the DNA backbone directly (Hall and Matson 1999).
The members of the SNF2-like family are highly homologous over the entire region that contains the seven ATPase motifs, named “the SNF2 domain” by Eisen et al. (1995). Therefore, the motifs and structures giving them their functional specificity are expected to lie outside this domain. In CSB, one of the most evident domains outside the ATPase region is the acidic region. Different types of acidic domain mutants have been established and investigated (Brosh et al. 1999; Sunesen et al. 2000). A mutant with a complete deletion of the 39 amino acids composing the acidic domain showed full complementation of UV survival, gene-specific CPD repair, UV-induced apoptosis propensity, PCNA relocation after UV irradiation, and NA-AAF survival of UV61 cells (Sunesen et al. 2000). A recent article from Sunesen et al. (2002) further shows that the acidic domain is superfluous in BER of oxidative damage in vivo and in vitro in UV61 cells (Sunesen et al. 2002). Thus, so far, no role has been assigned to the acidic domain. However, all published studies on this domain have been performed in UV61 cells, and the possibility for interspecies differences exists. Acidic domains in other proteins have been implicated in transcriptional activation and in protein-protein interactions, and an investigation of the transcriptional response after oxidative stress in CSB acidic domain deletion mutants could be interesting.
The putative NTB box C-terminal to the ATPase domain contains a Walker A GXGKT amino acid motif, and a mutant with a substitution of the highly conserved lysine CSBK1137Q has been established (Tuo et al. 2001). This mutant is phenotypically very close to WT CSB in all parameters tested, including cellular sensitivities, recovery of RNA synthesis, and 8-oxoG incision (Tuo et al. 2001; Muftuoglu et al. 2002). Thus, no function has so far been assigned to this motif.
So far, there have been no reported structure-function experiments involving the putative NLS sequences, the glycine-rich stretch, the hydrophilic segment, or the ATPase motif IV. It is most likely that the bipartite NLS signal is necessary for the nuclear localization of CSB. This is corroborated by the fact that mutations in other parts of CSB—namely motifs II, V, and VI—did not affect the localization of CSB (Christiansen et al. 2003). The glycine-rich stretch could give this region of the protein a high conformational flexibility and may be a flexible joining of two major protein domains. This would subdivide CSB into an N-terminal third and a C-terminal two-thirds. A subdivision with a flexible hinge domain is reminiscent of what is seen in transcription factors, with separate DNA-binding and transcription-activating or -repressing domains. It is interesting that no function has yet been assigned to any part of this N-terminal third and that hardly any mutations in this part of the gene have been found in the investigated patients with CSB (Mallery et al. 1998). This can imply one of two things: either this region is practically dispensable and mutations located here do not lead to CS or any other overt phenotype, or mutations in this region lead to severe inviable phenotypes and are therefore never presented in any patient.
The ATPase activity of motif Ia and III mutants has not been assayed. However, it has been suggested that these motifs mediate the internal energy transduction of CSB between ATP hydrolysis and nucleic-acid binding. Combining this with their defects in TCR-related parameters and WT response to oxidative damage suggests that a separation of the ATPase activity and the DNA binding inhibits TCR but not CSBs function in BER. The interrelation between energy production and DNA binding is likely to be necessary to mediate changes in protein-DNA interactions. Thus, CSB might work through fundamentally different mechanisms in mediating its function in TCR and GG-BER. It is possible that DNA binding and protein interactions are sufficient for transcriptional activation and stimulation of various factors involved in BER and factors induced by oxidative stress. The role of motifs V and VI in BER could be due to their effect on nucleic-acid binding and general stabilization of the protein structure rather than caused by the effect on ATP hydrolysis.
As mentioned above, the members of the SNF2-like protein family share ATPase motifs located in the conserved SNF2 domain. However, none of these proteins exhibits helicase activity, unlike most of the other members of SF1, SF2, and SF3. A function shared by several of the SNF2-like family members is their ability to modulate protein-DNA interactions in an ATP-dependent manner (reviewed by Pazin and Kadonaga [1997]). SWI2/SNF2, BRG1, hBrm, Sth1, and ISWI are the catalytic subunits of different multiprotein complexes, which all have the ability to modulate chromatin structure when hydrolyzing ATP (Travers 1999). As reported for CSB, these proteins are capable of catalyzing nucleosome remodeling in vitro without the presence of the rest of their respective complex (Travers 1999; Citterio et al. 2000). However, their modulating abilities are changed when encompassed in different complexes, as is exemplified by the different characteristics displayed by the NURF, CHRAC (MIM 607268), and ACF complexes, which all contain ISWI (Travers 1999). It is possible that the nucleosome-remodeling activity of CSB is also modulated by different protein interactions as well as by its incorporation into large complexes. Whereas the nucleosome-remodeling activity of ISWI, hBrm, BRG1, Sth1, and SWI2/SNF2 is implicated in changes in gene expression, it has been suggested that the CSB-mediated change in chromatin structure makes DNA lesions accessible to repair (Citterio et al. 2000). The ATP-dependent nature of nucleosome remodeling suggests that the process is necessary for the function of CSB in TCR but not for its role in BER of oxidative lesions. Protein-DNA interactions are also affected by the family members MOT1 and HuF2. These factors displace the TATA box-binding protein (TBP [MIM 600075]) from the TATA box and stalled RNA Pol I and II from CPD-containing DNA, respectively (Auble and Steggerda 1999; Hara et al. 1999). It has been suggested by Citterio et al. (2000) that CSB could use its ATPase activity to weaken and change the interaction between stalled RNA Pol II and DNA (Citterio et al. 2000). This could allow repair of a polymerase-blocking lesion and/or resumption of elongation.
Some SNF2-related proteins, such as MOT1 and Sth1, have a general effect on transcription, whereas others target specific genes or cofactors. SRCAP is a coactivator for the CREB-binding protein (CBP [MIM 600140]) at a subset of the CBP-influenced genes (Johnston et al. 1999), and the SWI/SNF complex targets and activates specific genes through local nucleosome remodeling. CSB has been suggested to have a role in general transcription as a positive elongation factor, but it has also recently been implicated in specific transcription regulation after oxidative stress (Kyng et al. 2003), and the cellular CSB status influences the transcript and protein level of OGG1 (Tuo et al. 2002a). It is not known how CSB mediates these gene-specific effects, but it appears to be in an ATP-independent manner. CSB nucleosome remodeling is therefore most probably not involved.
Little is known about the mechanisms of action of SNF2 family members. A DNA-tracking model resulting in dissociation of all attached proteins has been tested and rejected for MOT1 (Auble and Steggerda 1999). Rather, MOT1 appears to be distributively recruited to TBP bound to DNA (Auble and Steggerda 1999). SWI/SNF is also recruited to certain promoters through protein interactions. The interaction of CSB with elongating RNA Pol II and the lack of visible redistribution after DNA damage could suggest that CSB is constitutively associated with transcribing polymerases. The results from MOT1 suggest that the SNF2 region is unfit to track DNA and that CSB may bind to only the hyperphosphorylated CTD of RNA Pol II and not to DNA during elongation. However, when the polymerase is stalled by RNA secondary structure, natural pause sites, or a DNA lesion, the polymerase-DNA interface changes, possibly allowing CSB to interact with the polymerase and to bind DNA at the same time. The DNA-mediated stimulation of its ATPase activity could lead to ATP hydrolysis, and the resulting conformational change could then change the interaction between the polymerase and DNA, with CSB as the connecting lever. The same mechanism could account for nucleosome-remodeling activity employment at stalled transcription complexes. The changed histone-DNA interactions could allow DNA repair and/or transcription resumption by opening up the chromatin, making possible NER and transition from the smaller stalling bubble to the larger elongation bubble.
The ISWI-containing CHRAC complex associates with a topoisomerase II (MIM 126430) dimer that is thought to be implicated in the nucleosome-remodeling activity (Travers 1999). Interaction with topoisomerases could be a means for CSB to change the DNA twist and thereby facilitate changes in DNA-histone interactions. RAD5, the SNF2-family member involved in postreplication repair, is part of a complex which polyubiquitinates monoubiquitinated PCNA, resulting in error-free DNA repair (Pickart 2002).
A Tentative Model for the Function of CSB in Humans
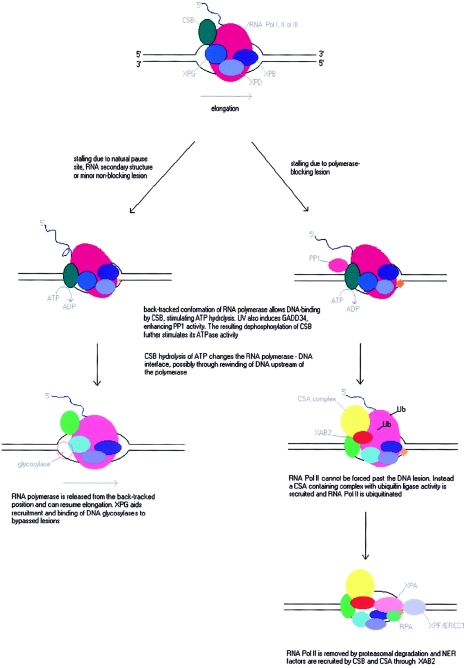
On the basis of data reviewed above and models previously suggested for the actions of CSB, a tentative and speculative model is developed and explained below (fig. 4).
Figure 4.
Tentative model for CSB function in transcription and TCR. (See text for further explanation.)
CSB is implicated in RNA Pol I-, II-, and possibly III-dependent transcription. It is suggested to stimulate elongation when an RNA polymerase is paused at natural pause sites or by strong RNA secondary structure. CSB is also important in preventing UV-induced apoptosis, most likely through a pivotal role in TC-NER. The necessity of CSB in TC-NER results in increased sensitivity of CSB-deficient cells toward damaging agents that introduce bulky lesions and adducts capable of blocking elongating RNA polymerases. A part of the role of CSB in TC-NER is the removal of RNA polymerase II by ubiquitination and proteasomal degradation of the large subunit. This degradation may be necessary for the cellular recovery of RNA synthesis after polymerase-blocking damage to DNA, whether or not the inflicted lesions are normally removed by TCR or GGR. Even lesions normally removed by GGR, which can be introduced in DNA by 4-NQO and NA-AAF, can be expected to block some elongating polymerases before being removed. The release of these polymerases, partly through proteasomal degradation, can be assumed to be important for allowing repair and the resumption of RNA synthesis.
CSB can be isolated in large complexes with either RNA polymerase I or II. It appears to specifically associate with the elongating hyperphosphorylated form of RNA polymerase II. This, combined with the apparent lack of redistribution of CSB after damage induction, suggests that CSB is constitutively associated with transcribing RNA polymerases. The SNF2-like domain appears incapable of DNA tracking, as shown for MOT1, so CSB most probably does not interact with DNA during elongation. However, when the polymerase is paused or stalled, it backtracks, and its interface with DNA changes. This could allow CSB to bind DNA, which would stimulate its ATPase activity. Christiansen et al. (2003) show that the ATPase activity of CSB is most strongly stimulated by dsDNA with a small bubble or loop reminiscent of a stalled transcription bubble (Christiansen et al. 2003). Thus, CSB can be speculated to bind DNA at the dsDNA-to-ssDNA transition of the stalled transcription bubble, possibly behind the RNA polymerase. The ATPase activity is necessary for the role of CSB in TCR and is increased by dephosphorylation of CSB in response to UV irradiation. The energy released from ATP hydrolysis is most likely used to change the RNA polymerase-DNA interface. This might be achieved through conformational changes in CSB, which, owing to the fixation of CSB to DNA and its simultaneous interaction with RNA polymerase, propagates to change the DNA interaction of the polymerase. As suggested for Mfd, CSB might pull the DNA through the RNA polymerase, thereby pushing it forward (Park et al. 2002). The pulling effect could be achieved by DNA rewinding upstream of the RNA polymerase (Park et al. 2002), an interesting putative function for a protein originally supposed to have helicase activity. The forward pushing of RNA polymerase can be speculated to release it from the backtracked position and thus to resume elongating, if the stalling were due to nonblocking causes. However, if the forward movement is blocked by, for instance, a CPD, it can be suspected to result in either a return to the backtracked position or a changed conformation of the RNA polymerase. This prolonged backtracking or a changed conformation could serve as a signal for TCR. Under the supposition that the RNA polymerase is close to the lesion and hindering repair, a natural beginning of the TCR would be the recruitment of a ubiquitinating activity, such as the CSA-containing complex. This would, through degradation, remove the RNA polymerase and allow CSB—still bound to DNA and possibly keeping the DNA open—to recruit NER factors. The recruitment might be mediated through the interaction of CSB and CSA with XAB2, which also interacts with the damaged DNA-binding early NER factor, XPA. A lack of CSB or CSA hinders the ubiquitination of blocked RNA polymerase. It is possible that a factor, such as HuF2, can displace the RNA polymerase from DNA. However, if the RNA polymerase remains stalled, it will result in apoptosis, either through a p53-mediated pathway or through S phase entry and progression.
The discovery of DNA polymerases capable of translesion synthesis (Friedberg and Gerlach 1999) has prompted speculations regarding the putative existence of a related function in RNA polymerases. This, as described above, has also been suggested by the translocase activity of both the Mfd and the RSC complexes. Also, a study of yeast shows that Rad26 promotes transcription through methyl methanesulfonate–damaged bases, normally removed by BER (Lee et al. 2002b). It can be speculated that CSB is involved in stimulating transcription through minor base lesions, such as 8-oxoA and 8-oxoG, by use of the same mechanism suggested for naturally paused, backtracked RNA polymerases. Preventing prolonged polymerase stalling would increase the cellular resistance to oxidative damage.
The role of XPB, XPD, and XPG in this model, in relation to the XP/CS phenotype, is also speculative. Their involvement in the TC-NER reaction is no different from that of GG-NER, which is deficient in patients with classical XP with mutated XPD or XPG, and thus cannot explain the CS-related symptoms. The isolation of these three factors in a large complex with CSB and RNA Pol I at physiological salt concentrations suggests that they may be constitutively associated with transcribing RNA Pol I. It is possible that the putative translesion-stimulating effect of CSB past oxidatively modified bases is specific for RNA Pol I. This might be an advantage for the high level of rRNA synthesis necessary for cell growth and function. Also, the possibility of incorporating mismatched bases during translesion transcription might be less devastating in rRNA than in mRNA, given that rRNA consists of both structural and “coding” bases. Thus, a lack of this translesion activity would sensitize a cell to oxidative damage, affecting most strongly the rRNA synthesis and, therefore, cellular growth. The specific roles of XPB, XPD, and XPG in this complex can also only be speculated on. As discussed above, the endonuclease activity of XPG appears not to be important for the XPG/CS phenotype. It is possible that the role of XPG in the RNA Pol I complex is participation in stabilizing protein interactions. However, XPG is also reported to stimulate BER by stimulating the incision activity of the Nth1 glycosylase. This might be mediated by a recruitment of DNA glycosylases by XPG to the translesion synthesis site. The role of XPB and XPD in this putative CSB function could be the use of their helicase activity to open the minor bubble of a stalled transcription complex to the larger transcription-elongation bubble. By opening the dsDNA in front of the RNA polymerase, XPB and XPD would support the forward-translocating effect of the CSB DNA-rewinding behind the polymerase.
CSB also has a role in transcription regulation after oxidative stress and has an effect on GG-BER. This might be mediated by entirely different mechanisms than those described above and most likely does not require ATPase activity. It is possible that a free fraction of CSB is modified in response to oxidative stress and thereby activated to induce transcription of a specific set of genes important for the stress response. The acidic region of CSB could function as an N-terminal activation domain separated from the DNA-binding domain by the glycine-rich stretch. The specificity of activation could be mediated by protein interactions with other transcription factors. Thus, CSB would function as a coactivator, as reported for the SNF2-like factor SRCAP. By inducing transcription of genes encoding DNA glycosylases such as OGG1 and UNG, CSB indirectly stimulates GG-BER.
All CSB-influenced pathways are affected to a common end by a deficient CSB protein: stalled RNA polymerases. This inhibits transcription and leads to apoptosis. Thus, cellularity and cellular growth are both inhibited, providing a possible explanation for the low cancer risk in patients with CS. Over time, a high cell-death rate and inhibited growth can be expected to affect nondividing tissues significantly and to empty the stem cell pools of the body, resulting in reduced growth, functional defects, and general atrophy in the patients.
Perspectives
The multisystem character of CS and the complexity of the genotype-phenotype relationship suggest that the underlying gene products are involved in basal cellular processes. Through the participation of CSB in several direct and functional protein interactions, the CS phenotype becomes sensitive to genetic variation affecting concentration, activity, and structure of its many interaction partners. The recent finding that CSB is involved not only in transcription and TCR by RNA Pol II but also in transcription by RNA Pol I adds to its general basal role in transcription. In yeast, RNA Pol I–transcribed genes are subject to TCR (Conconi et al. 2002). The implication of the CSB homologue Rad26 has not yet been investigated. Apart from RNA Pol I and II, CSB has indirectly been implicated in transcription by RNA Pol III. Together, these results suggest a general role for CSB in transcription and TCR by RNA Pol I, II, and III. Such generality could be mediated by the interaction of CSB with common subunits of the RNA polymerases. A further extrapolation of these speculations leads to the question of CSB involvement in mitochondrial transcription.
The role of CSB in TCR has typically been studied using exogenous DNA-damaging agents, such as UV and cisplatin. However, UV irradiation does not penetrate the skull and therefore cannot be causative of the neurological symptoms of CS. Candidates for natural endogenous DNA lesions possibly repaired by TCR are the cyclo-purines—cyclo-dA and cyclo-dG—which can be induced by free radicals and are capable of blocking RNA Pol II transcription (Brooks et al. 2000). The role of CSB in GG-BER could also explain some of the subcutaneous symptoms of CS.
CSB has been studied at phenotypic, cellular, subcellular, and biochemical levels. Recently, several structure-function studies have aided in integrating the knowledge of these different levels. However, the question of the control of CSB itself is as yet unresolved. Given the involvement of CSB in several different processes, it can be suspected that the participation of CSB in each process is somehow interregulated in response to DNA damage, development, cellular differentiation, and aging. Also, the expression of CSB could vary during development, differentiation, and aging and between different tissues. It could be interesting to establish a transcriptional profile of CSB and to study it in comparison with profiles of CSA, XPB, XPD, and XPG. Also, a comparison of the promoters of these CS-causative genes might suggest common transcriptional regulators. The interregulation of CSB among different cellular processes is most likely of a posttranslational nature. CSB is differentially phosphorylated in response to UV irradiation, and other posttranslational modifications might also be involved. CSB could also be regulated, at the protein level, through changes in quaternary structure and/or through changes in protein-interaction partners. To investigate this, it would be interesting to map the interaction domains of CSB with the different interaction partners and to see if the different areas overlap with each other or with a putative additional CSB-binding site. Such studies, and structure-function studies in general, would be profoundly aided by having the CSB tertiary structure solved.
An understanding of the processes and factors controlling expression, activity, and choice of pathway by CSB in response to different cellular stages and conditions will help explain the CS phenotype. It will also provide specific factors to pay attention to in comparing the genetic backgrounds of patients with CS. It could be especially rewarding to compare, using microarray or probing for specific key factors, the CS-affected patients with identical mutations in CSB who display different symptoms with respect to genetic background. A further characterization of the several functions of CSB and the underlying mechanisms employed will also contribute to the understanding of general processes of DNA metabolism.
Acknowledgments
We thank Tina Thorslund for critical comments on the manuscript. T.S. was supported, in part, by The Velux Foundation, The Danish Research Council (grant 22-02-0397), and The Danish Centre for Molecular Gerontology.
Appendix A: Abbreviations
4-NQO: 4-nitroquinoline-1-oxide
8-oxoA: 8-hydroxy-7,8-dihydroadenine
8-oxoG:8-hydroxy-7,8-dihydroguanine
6-4 PP: pyrimidine 6-4 pyrimidone photoproduct
BER: base excision repair
CBP: CREB-binding protein
CHO: Chinese hamster ovary
CKII: casein kinase II
CO-IP: coimmunoprecipitation
CPD: cyclobutane pyrimidine dimer
COFS: cerebro-oculo-facio-skeletal syndrome
CS: Cockayne syndrome
CSA: Cockayne syndrome complementation group A
CSB: Cockayne syndrome complementation group B
CTD: C-terminal domain
dG-C8-AAF: (deoxyguanosine-8-yl)-2-acetylaminofluorene
DHFR: dihydrofolate reductase
dsDNA: double-strand DNA
EMSA: electrophoretic mobility-shift assay
ES: embryonic stem
FPG: formamidopyrimidine glycosylase
GG-BER: global genome base excision repair
GG-NER: global genome nucleotide excision repair
GGR: global genome repair
HuF2: human transcription release factor 2
IR: ionizing radiation
MEF: mouse embryonic fibroblast
NA-AAF: N-acetoxy-2-acetylaminofluorene
NER: nucleotide excision repair
NLS: nuclear-localization signal
NTB: nucleotide-binding
OGG1: 8-oxoguanine glycosylase
PCNA: proliferating cell nuclear antigen
RPA: replication protein A
ssDNA: single-strand DNA
TBP: TATA box-binding protein
TCR: transcription coupled repair
TG: thymine glycol
TRCF: transcription-repair coupling factor
TTD:, trichothiodystrophy
UNG: uracil-DNA glycosylase
UV: ultraviolet
WT: wild type
XAB2: XPA-binding protein 2
XP: xeroderma pigmentosum
XPB: xeroderma pigmentosum complementation group B
XPD: xeroderma pigmentosum complementation group D
XPG: Xeroderma pigmentosum complementation group G
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CS, CSB, XPC/hHR23B, RPA, XPA, XP, XPB, XPD, XPE, XPF/ERCC1, XPG, TFIIH, APE1, RNA polymerase I, RNA polymerase II, RNA polymerase III, CSA, XP/CS, cerebro-oculo-facio-skeletal syndrome, DeSanctis-Cacchione severe neurological form of XP, XPB/CS, XPD/CS, XPG/CS, p53, UNG, OGG1, ERCC6, JUN, hMSH2, hMLH1, U1, U2, 5S rRNA, TFIIS, Brg1, hBrm, PCNA, replication factor C, DHFR, p44 subunit of TFIIH, DDB1, Cul4A, Roc1, CSN, p62, TTD, Nth1, TAFII 250, TFIIE, SWI2/SNF2, TAF-172, RAD54, RAD5, CKII, protein phosphatase 1, CHRAC, TBP, CBP, and topoisomerase II)
References
- Auble DT, Steggerda SM (1999) Testing for DNA tracking by MOT1, a SNF2/SWI2 protein family member. Mol Cell Biol 19:412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee AS, Dianova I, Bohr VA (1999) Oxidative damage-induced PCNA complex formation is efficient in xeroderma pigmentosum group A but reduced in Cockayne syndrome group B cells. Nucleic Acids Res 27:4476–4482 10.1093/nar/27.22.4476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA (1997) Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci USA 94:4306–4311 10.1073/pnas.94.9.4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee AS, May A, Dianova I, Bohr VA (1998) Efficient PCNA complex formation is dependent upon both transcription coupled repair and genome overall repair. Mutat Res 409:135–146 [DOI] [PubMed] [Google Scholar]
- Balajee AS, Proietti De Santis L, Brosh RM Jr, Selzer R, Bohr VA (2000) Role of the ATPase domain of the Cockayne syndrome group B protein in UV induced apoptosis. Oncogene 19:477–489 10.1038/sj.onc.1203372 [DOI] [PubMed] [Google Scholar]
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC (1985) DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell 40:359–369 [DOI] [PubMed] [Google Scholar]
- Bradsher J, Auriol J, Proietti de Santis L, Iben S, Vonesch JL, Grummt I, Egly JM (2002) CSB is a component of RNA pol I transcription. Mol Cell 10:819–829 [DOI] [PubMed] [Google Scholar]
- Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL (1996) UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA 93:11586–11590 10.1073/pnas.93.21.11586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks PJ (2002) DNA repair in neural cells: basic science and clinical implications. Mutat Res 509:93–108 [DOI] [PubMed] [Google Scholar]
- Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH (2000) The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem 275:22355–22362 10.1074/jbc.M002259200 [DOI] [PubMed] [Google Scholar]
- Brosh RM Jr, Balajee AS, Selzer RR, Sunesen M, Proietti De Santis L, Bohr VA (1999) The ATPase domain but not the acidic region of Cockayne syndrome group B gene product is essential for DNA repair. Mol Biol Cell 10:3583–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen M, Stevnsner T, Modin C, Martensen PM, Brosh RM Jr, Bohr VA (2003) Functional consequences of mutations in the conserved SF2 motifs and post-translational phosphorylation of the CSB protein. Nucleic Acids Res 31:963–973 10.1093/nar/gkg164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W (1998) Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem 273:11844–11851 10.1074/jbc.273.19.11844 [DOI] [PubMed] [Google Scholar]
- Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W (2000) ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol 20:7643–7653 10.1128/MCB.20.20.7643-7653.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockayne EA (1936) Dwarfism with retinal atrophy and deafness. Arch Dis Child 11:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Nardo T, Botta E, Lehmann AR, Stefanini M (2000) Identical mutations in the CSB gene associated with either Cockayne syndrome or the DeSanctis-Cacchione variant of xeroderma pigmentosum. Hum Mol Genet 9:1171–1175 10.1093/hmg/9.8.1171 [DOI] [PubMed] [Google Scholar]
- Colella S, Nardo T, Mallery D, Borrone C, Ricci R, Ruffa G, Lehmann AR, Stefanini M (1999) Alterations in the CSB gene in three Italian patients with the severe form of Cockayne syndrome (CS) but without clinical photosensitivity. Hum Mol Genet 8:935–941 10.1093/hmg/8.5.935 [DOI] [PubMed] [Google Scholar]
- Conconi A, Bespalov VA, Smerdon MJ (2002) Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast. Proc Natl Acad Sci USA 99:649–654 10.1073/pnas.022373099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper PK, Nouspikel T, Clarkson SG, Leadon SA (1997) Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science 275:990–993 10.1126/science.275.5302.990 [DOI] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH (2000) Nucleotide excision repair and human syndromes. Carcinogenesis 21:453–460 10.1093/carcin/21.3.453 [DOI] [PubMed] [Google Scholar]
- de Waard H, de Wit J, Gorgels TG, van den Aardweg G, Andressoo JO, Vermeij M, van Steeg H, Hoeijmakers JH, van der Horst GT (2003) Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2:13–25 [DOI] [PubMed] [Google Scholar]
- Dianov G, Bischoff C, Sunesen M, Bohr VA (1999) Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res 27:1365–1368 10.1093/nar/27.5.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollfus H, Porto F, Caussade P, Speeg-Schatz C, Sahel J, Grosshans E, Flament J, Sarasin A (2003) Ocular manifestations in the inherited DNA repair disorders. Surv Ophthalmol 48:107–122 10.1016/S0039-6257(02)00400-9 [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23:2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Gerlach VL (1999) Novel DNA polymerases offer clues to the molecular basis of mutagenesis. Cell 98:413–416 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W (1995) DNA repair and mutagenesis. American Society for Microbiology, Washington DC [Google Scholar]
- Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA (2003) Retraction. Science 300:1657 10.1126/science.300.5626.1657b [DOI] [PubMed] [Google Scholar]
- Gregory SM, Sweder KS (2001) Deletion of the CSB homolog, RAD26, yields Spt(-) strains with proficient transcription-coupled repair. Nucleic Acids Res 29:3080–3086 10.1093/nar/29.14.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y (2003) The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357–367 [DOI] [PubMed] [Google Scholar]
- Hall MC, Matson SW (1999) Helicase motifs: the engine that powers DNA unwinding. Mol Microbiol 34:867–877 10.1046/j.1365-2958.1999.01659.x [DOI] [PubMed] [Google Scholar]
- Hara R, Selby CP, Liu M, Price DH, Sancar A (1999) Human transcription release factor 2 dissociates RNA polymerases I and II stalled at a cyclobutane thymine dimer. J Biol Chem 274:24779–24786 10.1074/jbc.274.35.24779 [DOI] [PubMed] [Google Scholar]
- Hayashi M, Itoh M, Araki S, Kumada S, Shioda K, Tamagawa K, Mizutani T, Morimatsu Y, Minagawa M, Oda M (2001) Oxidative stress and disturbed glutamate transport in hereditary nucleotide repair disorders. J Neuropathol Exp Neurol 60:350–356 [DOI] [PubMed] [Google Scholar]
- Henning KA, Li L, Iyer N, McDaniel LD, Reagan MS, Legerski R, Schultz RA, Stefanini M, Lehmann AR, Mayne LV, Freidberg EC (1995) The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82:555–564 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D, Nigg AL, Heath H, Mullenders LH, van Driel R, Hoeijmakers JH, Vermeulen W, Houtsmuller AB (2002) Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol Cell 10:1163–1174 [DOI] [PubMed] [Google Scholar]
- Itoh M, Hayashi M, Shioda K, Minagawa M, Isa F, Tamagawa K, Morimatsu Y, Oda M (1999) Neurodegeneration in hereditary nucleotide repair disorders. Brain Dev 21:326–333 10.1016/S0387-7604(99)00033-9 [DOI] [PubMed] [Google Scholar]
- Iyer N, Reagan MS, Wu KJ, Canagarajah B, Friedberg EC (1996) Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry 35:2157–2167 10.1021/bi9524124 [DOI] [PubMed] [Google Scholar]
- Jensen A, Calvayrac G, Karahalil B, Bohr VA, Stevnsner T (2003) Mammalian 8-oxoguanine DNA glycosylase 1 incises 8-oxoadenine opposite cytosine in nuclei and mitochondria, while a different glycosylase incises 8-oxoadenine opposite guanine in nuclei. J Biol Chem 278:19541–19548 10.1074/jbc.M301504200 [DOI] [PubMed] [Google Scholar]
- Johnston H, Kneer J, Chackalaparampil I, Yaciuk P, Chrivia J (1999) Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J Biol Chem 274:16370–16376 10.1074/jbc.274.23.16370 [DOI] [PubMed] [Google Scholar]
- Kamiuchi S, Saijo M, Citterio E, de Jager M, Hoeijmakers JH, Tanaka K (2002) Translocation of Cockayne syndrome group A protein to the nuclear matrix: possible relevance to transcription-coupled DNA repair. Proc Natl Acad Sci USA 99:201–206 10.1073/pnas.012473199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland A, Hoss M, Gunz D, Constantinou A, Clarkson SG, Doetsch PW, Bolton PH, Wood RD, Lindahl T (1999) Base excision repair of oxidative DNA damage activated by XPG protein. Mol Cell 3:33–42 [DOI] [PubMed] [Google Scholar]
- Kyng KJ, May A, Brosh RM Jr, Cheng WH, Chen C, Becker KG, Bohr VA (2003) The transcriptional response after oxidative stress is defective in Cockayne syndrome group B cells. Oncogene 22:1135–1149 10.1038/sj.onc.1206187 [DOI] [PubMed] [Google Scholar]
- Le Page F, Kwoh EE, Avrutskaya A, Gentil A, Leadon SA, Sarasin A, Cooper PK (2000) Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101:159–171 [DOI] [PubMed] [Google Scholar]
- Leadon SA, Avrutskaya AV (2003) Retraction of: Differential involvement of the human mismatch repair proteins, hMLH1 and hMSH2, in transcription-coupled repair. Cancer Res 63:3846 [PubMed] [Google Scholar]
- Leadon SA, Cooper PK (1993) Preferential repair of ionizing radiation-induced damage in the transcribed strand of an active human gene is defective in Cockayne syndrome. Proc Natl Acad Sci USA 90:10499–10503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB, Wang D, Lippard SJ, Sharp PA (2002a) Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc Natl Acad Sci USA 99:4239–4244 10.1073/pnas.072068399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Yu SL, Prakash L, Prakash S (2002b) Yeast RAD26, a homolog of the human CSB gene, functions independently of nucleotide excision repair and base excision repair in promoting transcription through damaged bases. Mol Cell Biol 22:4383–4389 10.1128/MCB.22.12.4383-4389.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Smerdon MJ (2002) Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. Embo J 21:5921–5929 10.1093/emboj/cdf589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbaum Y, Dickson D, Rosenbaum P, Kraemer K, Robbins I, Rapin I (2001) Xeroderma pigmentosum/Cockayne syndrome complex: first neuropathological study and review of eight other cases. Eur J Paediatr Neurol 5:225–242 10.1053/ejpn.2001.0523 [DOI] [PubMed] [Google Scholar]
- Ljungman M, Zhang F (1996) Blockage of RNA polymerase as a possible trigger for u.v. light-induced apoptosis. Oncogene 13:823–831 [PubMed] [Google Scholar]
- Ljungman M, Zhang F, Chen F, Rainbow AJ, McKay BC (1999) Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene 18:583–592 10.1038/sj.onc.1202356 [DOI] [PubMed] [Google Scholar]
- Mahmoud AA, Yousef GM, Al-Hifzi I, Diamandis EP (2002) Cockayne syndrome in three sisters with varying clinical presentation. Am J Med Genet 111:81–85 10.1002/ajmg.10492 [DOI] [PubMed] [Google Scholar]
- Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M, Lehmann AR (1998) Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am J Hum Genet 62:77–85 10.1086/301686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayne LV (1984) Inhibitors of DNA synthesis (aphidicolin and araC/HU) prevent the recovery of RNA synthesis after UV-irradiation. Mutat Res 131:187–191 [DOI] [PubMed] [Google Scholar]
- McKay BC, Becerril C, Spronck JC, Ljungman M (2002) Ultraviolet light-induced apoptosis is associated with S-phase in primary human fibroblasts. DNA Repair (Amst) 1:811–820 [DOI] [PubMed] [Google Scholar]
- McKay BC, Chen F, Clarke ST, Wiggin HE, Harley LM, Ljungman M (2001) UV light-induced degradation of RNA polymerase II is dependent on the Cockayne’s syndrome A and B proteins but not p53 or MLH1. Mutat Res 485:93–105 [DOI] [PubMed] [Google Scholar]
- Meira LB, Graham JM Jr, Greenberg CR, Busch DB, Doughty ATB, Ziffer DW, Coleman DM, Savre-Train I, Friedberg EC (2000) Manitoba aboriginal kindred with original cerebro-oculo-facio-skeletal syndrome has a mutation in the Cockayne syndrome group B (CSB) gene. Am J Hum Genet 66:1221–1228 10.1086/302867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Rajpal DK, Koi M, Boland CR, Champe GN (1996) Transcription-coupled repair deficiency and mutations in human mismatch repair genes. Science 272:557–560 [DOI] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt PC (1987) Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241–249 [DOI] [PubMed] [Google Scholar]
- Muftuoglu M, Selzer R, Tuo J, Brosh RM Jr, Bohr VA (2002) Phenotypic consequences of mutations in the conserved motifs of the putative helicase domain of the human Cockayne syndrome group B gene. Gene 283:27–40 10.1016/S0378-1119(01)00870-8 [DOI] [PubMed] [Google Scholar]
- Mullenders LH, van Kesteren van Leeuwen AC, van Zeeland AA, Natarajan AT (1988) Nuclear matrix associated DNA is preferentially repaired in normal human fibroblasts, exposed to a low dose of ultraviolet light but not in Cockayne’s syndrome fibroblasts. Nucleic Acids Res 16:10607–10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, Kamiuchi S, Yeo JP, Khaw MC, Saijo M, Kodo N, Matsuda T, Hoeijmakers JH, Tanaka K (2000) XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J Biol Chem 275:34931–34937 10.1074/jbc.M004936200 [DOI] [PubMed] [Google Scholar]
- Nance MA, Berry SA (1992) Cockayne syndrome: review of 140 cases. Am J Med Genet 42:68–84 [DOI] [PubMed] [Google Scholar]
- Nouspikel T, Lalle P, Leadon SA, Cooper PK, Clarkson SG (1997) A common mutational pattern in Cockayne syndrome patients from xeroderma pigmentosum group G: implications for a second XPG function. Proc Natl Acad Sci USA 94:3116–3121 10.1073/pnas.94.7.3116 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Orren DK, Dianov GL, Bohr VA (1996) The human CSB (ERCC6) gene corrects the transcription-coupled repair defect in the CHO cell mutant UV61. Nucleic Acids Res 24:3317–3322 10.1093/nar/24.17.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterod M, Larsen E, Le Page F, Hengstler JG, Van Der Horst GT, Boiteux S, Klungland A, Epe B (2002) A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene 21:8232–8239 10.1038/sj.onc.1206027 [DOI] [PubMed] [Google Scholar]
- Park JS, Marr MT, Roberts JW (2002) E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109:757–767 [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT (1997) SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell 88:737–740 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2002) DNA repair: right on target with ubiquitin. Nature 419:120–121 10.1038/419120a [DOI] [PubMed] [Google Scholar]
- Proietti De Santis L, Garcia CL, Balajee AS, Brea Calvo GT, Bassi L, Palitti F (2001) Transcription coupled repair deficiency results in increased chromosomal aberrations and apoptotic death in the UV61 cell line, the Chinese hamster homologue of Cockayne’s syndrome B. Mutat Res 485:121–132 [DOI] [PubMed] [Google Scholar]
- Proietti De Santis L, Garcia CL, Balajee AS, Latini P, Pichierri P, Nikaido O, Stefanini M, Palitti F (2002) Transcription coupled repair efficiency determines the cell cycle progression and apoptosis after UV exposure in hamster cells. DNA Repair (Amst) 1:209–223 [DOI] [PubMed] [Google Scholar]
- Rapin I, Lindenbaum Y, Dickson DW, Kraemer KH, Robbins JH (2000) Cockayne syndrome and xeroderma pigmentosum. Neurology 55:1442–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner JN, Balasubramanian B, Corden J, Warren SL, Bregman DB (1998) Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II: implications for transcription-coupled DNA repair. J Biol Chem 273:5184–5189 10.1074/jbc.273.9.5184 [DOI] [PubMed] [Google Scholar]
- Reardon JT, Sancar A (2002) Molecular anatomy of the human excision nuclease assembled at sites of DNA damage. Mol Cell Biol 22:5938–5945 10.1128/MCB.22.16.5938-5945.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochette PJ, Bastien N, McKay BC, Therrien JP, Drobetsky EA, Drouin R (2002) Human cells bearing homozygous mutations in the DNA mismatch repair genes hMLH1 or hMSH2 are fully proficient in transcription-coupled nucleotide excision repair. Oncogene 21:5743–5752 10.1038/sj.onc.1205641 [DOI] [PubMed] [Google Scholar]
- Rockx DA, Mason R, van Hoffen A, Barton MC, Citterio E, Bregman DB, van Zeeland AA, Vrieling H, Mullenders LH (2000) UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc Natl Acad Sci USA 97:10503–10508 10.1073/pnas.180169797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J (2003) p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. Embo J 22:975–986 10.1093/emboj/cdg082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A (1997a) Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA 94:11205–11209 10.1073/pnas.94.21.11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (1997b) Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem 272:1885–1890 10.1074/jbc.272.3.1885 [DOI] [PubMed] [Google Scholar]
- Selzer RR, Nyaga S, Tuo J, May A, Muftuoglu M, Christiansen M, Citterio E, Brosh RM Jr, Bohr VA (2002) Differential requirement for the ATPase domain of the Cockayne syndrome group B gene in the processing of UV-induced DNA damage and 8-oxoguanine lesions in human cells. Nucleic Acids Res 30:782–793 10.1093/nar/30.3.782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24:181–185 10.1016/S0968-0004(99)01384-5 [DOI] [PubMed] [Google Scholar]
- Snyderwine EG, Bohr VA (1992) Gene- and strand-specific damage and repair in Chinese hamster ovary cells treated with 4-nitroquinoline 1-oxide. Cancer Res 52:4183–4189 [PubMed] [Google Scholar]
- Spivak G, Lloyd RS, Sweder KS (2003) Workshop on DNA Repair and Related DNA Transactions, a conference report. DNA Repair (Amst) 2:235–242 [DOI] [PubMed] [Google Scholar]
- Stevnsner T, Nyaga S, de Souza-Pinto NC, van der Horst GT, Gorgels TG, Hogue BA, Thorslund T, Bohr VA (2002) Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene 21:8675–8682 10.1038/sj.onc.1205994 [DOI] [PubMed] [Google Scholar]
- Sunesen M, Selzer RR, Brosh RM Jr, Balajee AS, Stevnsner T, Bohr VA (2000) Molecular characterization of an acidic region deletion mutant of Cockayne syndrome group B protein. Nucleic Acids Res 28:3151–3159 10.1093/nar/28.16.3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunesen M, Stevnsner T, Brosh RM Jr, Dianov GL, Bohr VA (2002) Global genome repair of 8-oxoG in hamster cells requires a functional CSB gene product. Oncogene 21:3571–3578 10.1038/sj.onc.1205443 [DOI] [PubMed] [Google Scholar]
- Surralles J, Ramirez MJ, Marcos R, Natarajan AT, Mullenders LH (2002) Clusters of transcription-coupled repair in the human genome. Proc Natl Acad Sci USA 99:10571–10574 10.1073/pnas.162278199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup JQ (2002) Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol 3:21–29 10.1038/nrm703 [DOI] [PubMed] [Google Scholar]
- ——— (2003) Rescue of arrested RNA polymerase II complexes. J Cell Sci 116:447–451 10.1242/jcs.00271 [DOI] [PubMed] [Google Scholar]
- Tang MS, Bohr VA, Zhang XS, Pierce J, Hanawalt PC (1989) Quantification of aminofluorene adduct formation and repair in defined DNA sequences in mammalian cells using the UVRABC nuclease. J Biol Chem 264:14455–14462 [PubMed] [Google Scholar]
- Tantin D (1998) RNA polymerase II elongation complexes containing the Cockayne syndrome group B protein interact with a molecular complex containing the transcription factor IIH components xeroderma pigmentosum B and p62. J Biol Chem 273:27794–27799 10.1074/jbc.273.43.27794 [DOI] [PubMed] [Google Scholar]
- Tantin D, Kansal A, Carey M (1997) Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol 17:6803–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, Sunesen M, Bohr VA, Stevnsner T (2002) Repair of 8-oxoG is slower in endogenous nuclear genes than in mitochondrial DNA and is without strand bias. DNA Repair (Amst) 1:261–273 [DOI] [PubMed] [Google Scholar]
- Travers A (1999) An engine for nucleosome remodeling. Cell 96:311–314 [DOI] [PubMed] [Google Scholar]
- Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH (1992) ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell 71:939–953 [DOI] [PubMed] [Google Scholar]
- Trzcinska AM, Girstun A, Piekielko A, Kowalska-Loth B, Staron K (2002) Potential protein partners for the N-terminal domain of human topoisomerase I revealed by phage display. Mol Biol Rep 29:347–352 10.1023/A:1021237021338 [DOI] [PubMed] [Google Scholar]
- Tu Y, Bates S, Pfeifer GP (1997) Sequence-specific and domain-specific DNA repair in xeroderma pigmentosum and Cockayne syndrome cells. J Biol Chem 272:20747–20755 10.1074/jbc.272.33.20747 [DOI] [PubMed] [Google Scholar]
- Tuo J, Chen C, Zeng X, Christiansen M, Bohr VA (2002a) Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst) 1:913–927 [DOI] [PubMed] [Google Scholar]
- Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M (2003) Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. Faseb J 17:668–674 10.1096/fj.02-0851com [DOI] [PubMed] [Google Scholar]
- Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA (2002b) The Cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem 277:30832–30837 10.1074/jbc.M204814200 [DOI] [PubMed] [Google Scholar]
- Tuo J, Muftuoglu M, Chen C, Jaruga P, Selzer RR, Brosh RM Jr, Rodriguez H, Dizdaroglu M, Bohr VA (2001) The Cockayne syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J Biol Chem 276:45772–45779 10.1074/jbc.M107888200 [DOI] [PubMed] [Google Scholar]
- van der Horst GT, van Steeg H, Berg RJ, van Gool AJ, de Wit J, Weeda G, Morreau H, Beems RB, van Kreijl CF, de Gruijl FR, Bootsma D, Hoeijmakers JH (1997) Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell 89:425–435 [DOI] [PubMed] [Google Scholar]
- van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D, Hoeijmakers JH (1997a) The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. Embo J 16:5955–5965 10.1093/emboj/16.19.5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool AJ, van der Horst GT, Citterio E, Hoeijmakers JH (1997b) Cockayne syndrome: defective repair of transcription? Embo J 16:4155–4162 10.1093/emboj/16.14.4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterwijk MF, Filon R, de Groot AJ, van Zeeland AA, Mullenders LH (1998) Lack of transcription-coupled repair of acetylaminofluorene DNA adducts in human fibroblasts contrasts their efficient inhibition of transcription. J Biol Chem 273:13599–13604 10.1074/jbc.273.22.13599 [DOI] [PubMed] [Google Scholar]
- Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV (1990) The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci USA 87:4707–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MH, Chu EH (1979) Effects of DNA damaging agents on cultured fibroblasts derived from patients with Cockayne syndrome. Mutat Res 59:49–60 [DOI] [PubMed] [Google Scholar]
- Wang XW, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly JM, Wang Z, et al (1995) p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet 10:188–195 [DOI] [PubMed] [Google Scholar]
- Woudstra EC, Gilbert C, Fellows J, Jansen L, Brouwer J, Erdjument-Bromage H, Tempst P, Svejstrup JQ (2002) A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415:929–933 10.1038/415929a [DOI] [PubMed] [Google Scholar]
- Yu A, Fan HY, Liao D, Bailey AD, Weiner AM (2000) Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol Cell 5:801–810 10.1016/S1097-2765(00)80320-2 [DOI] [PubMed] [Google Scholar]