Abstract
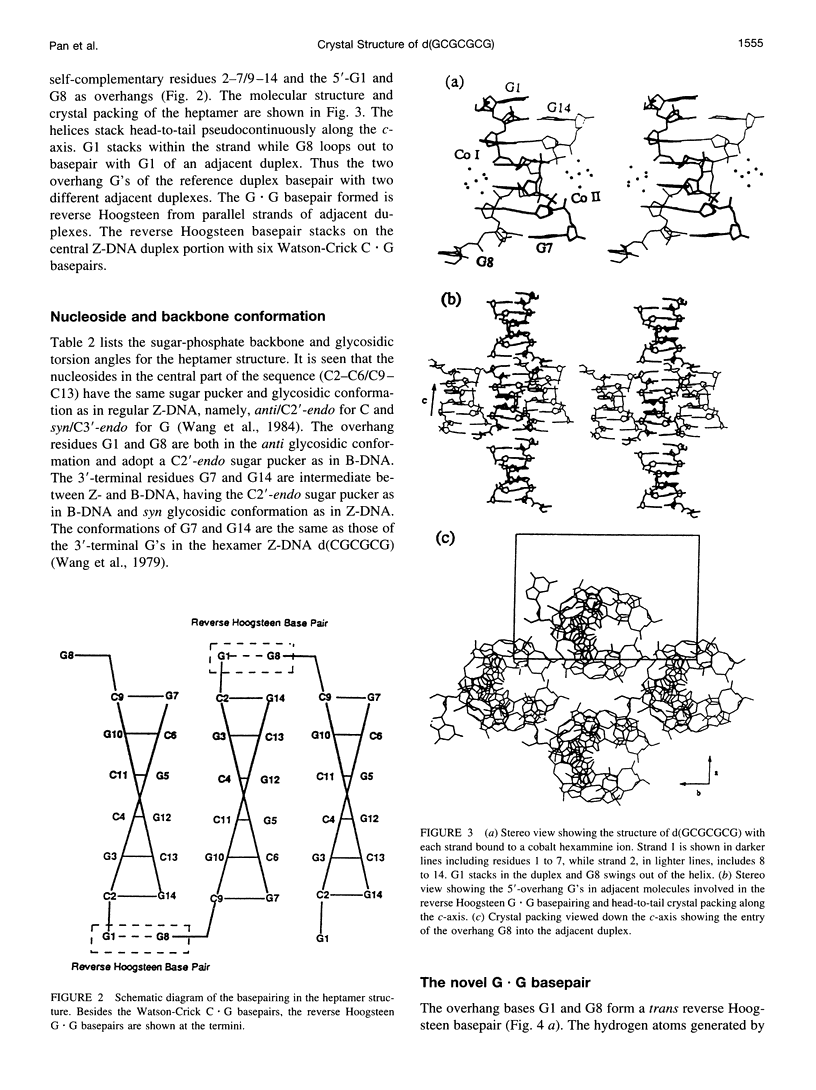
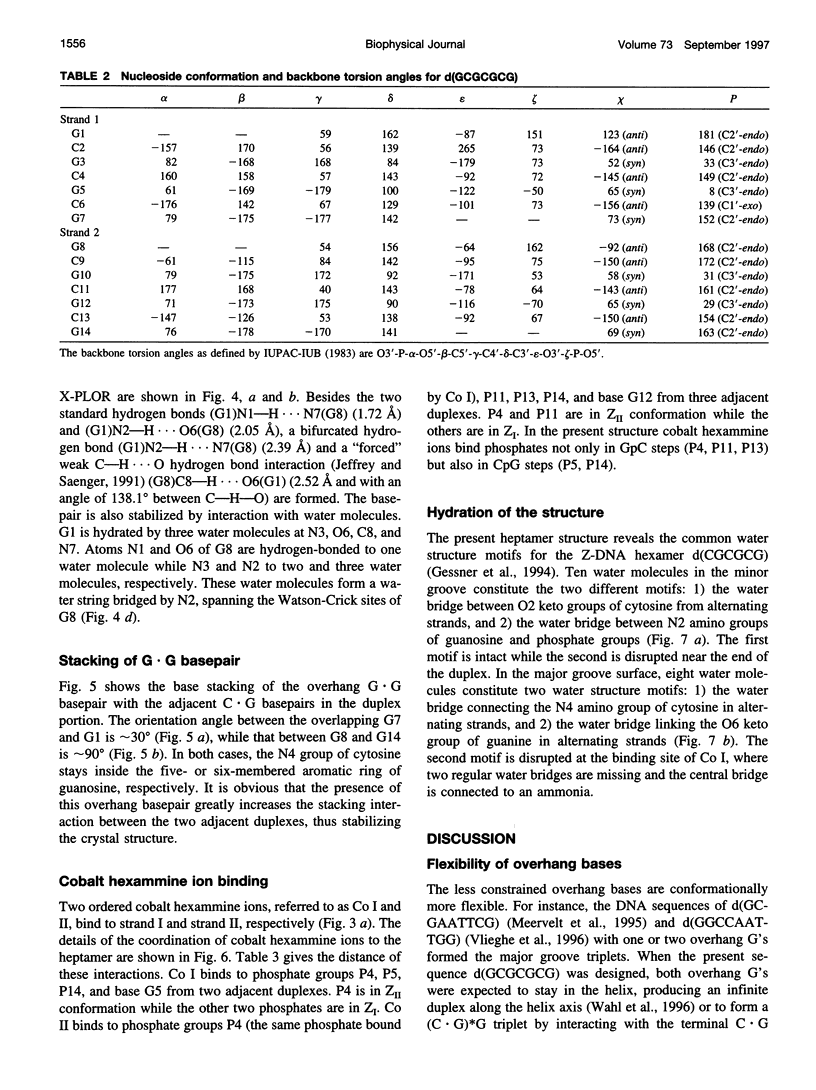
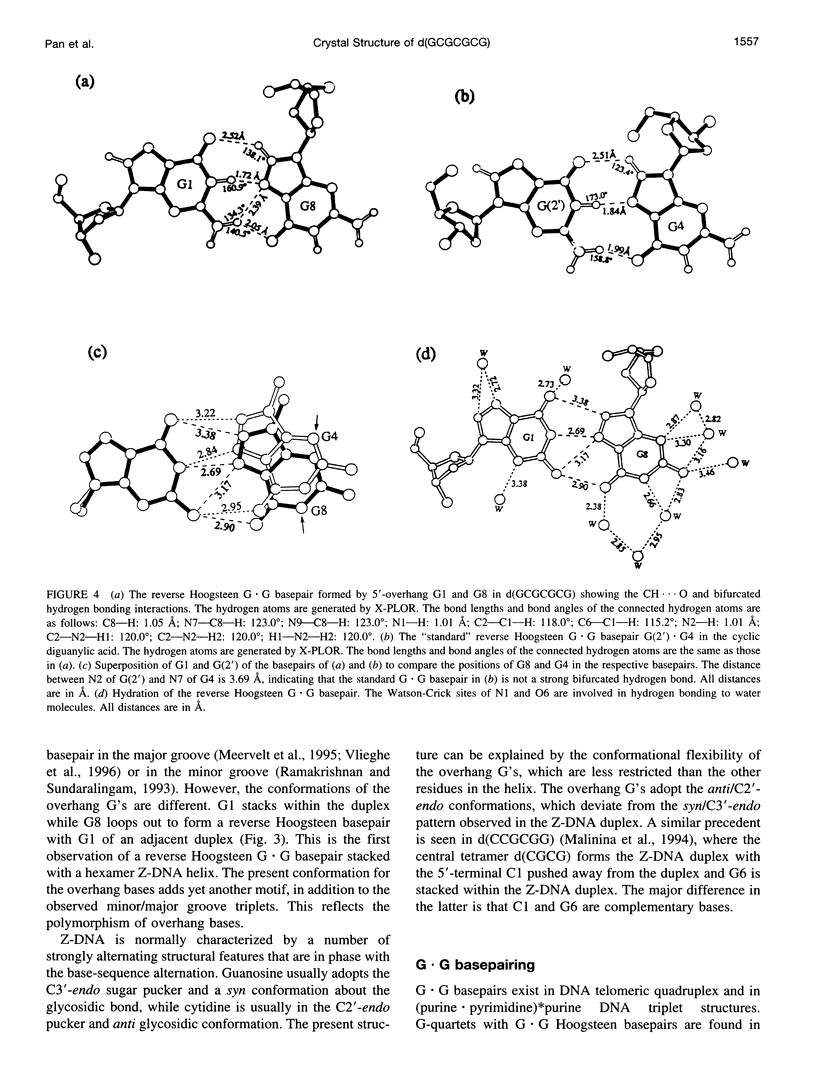
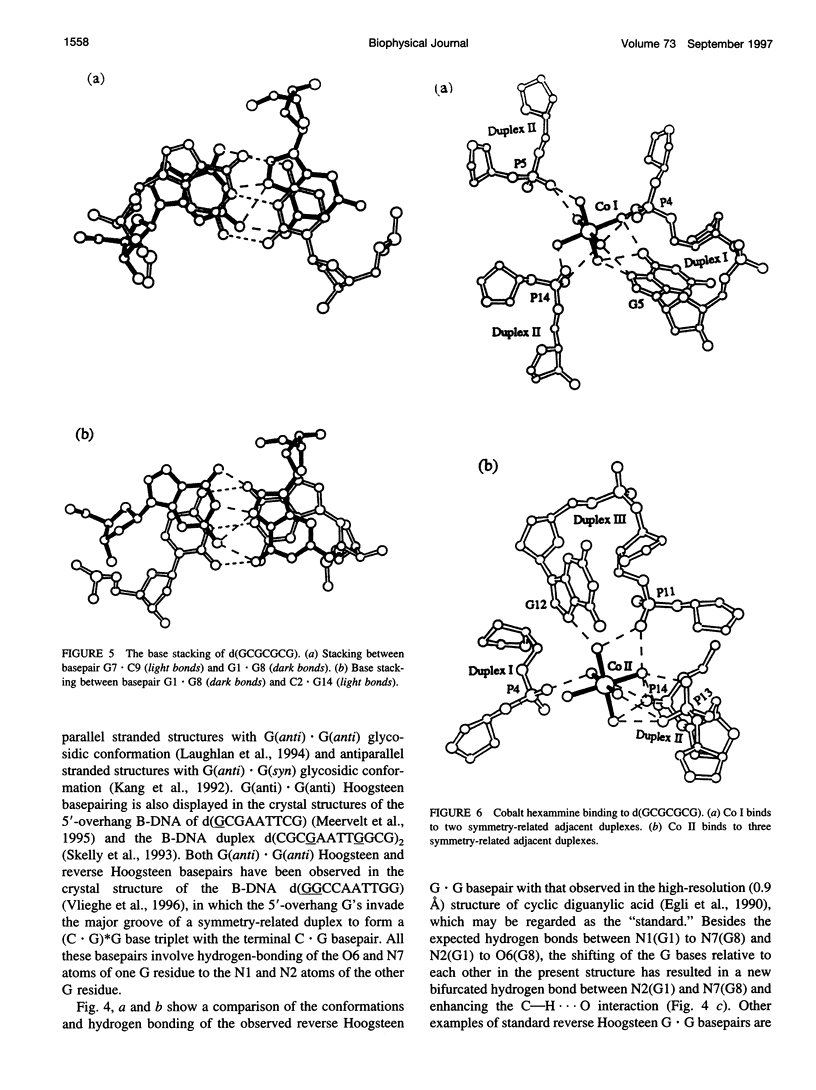
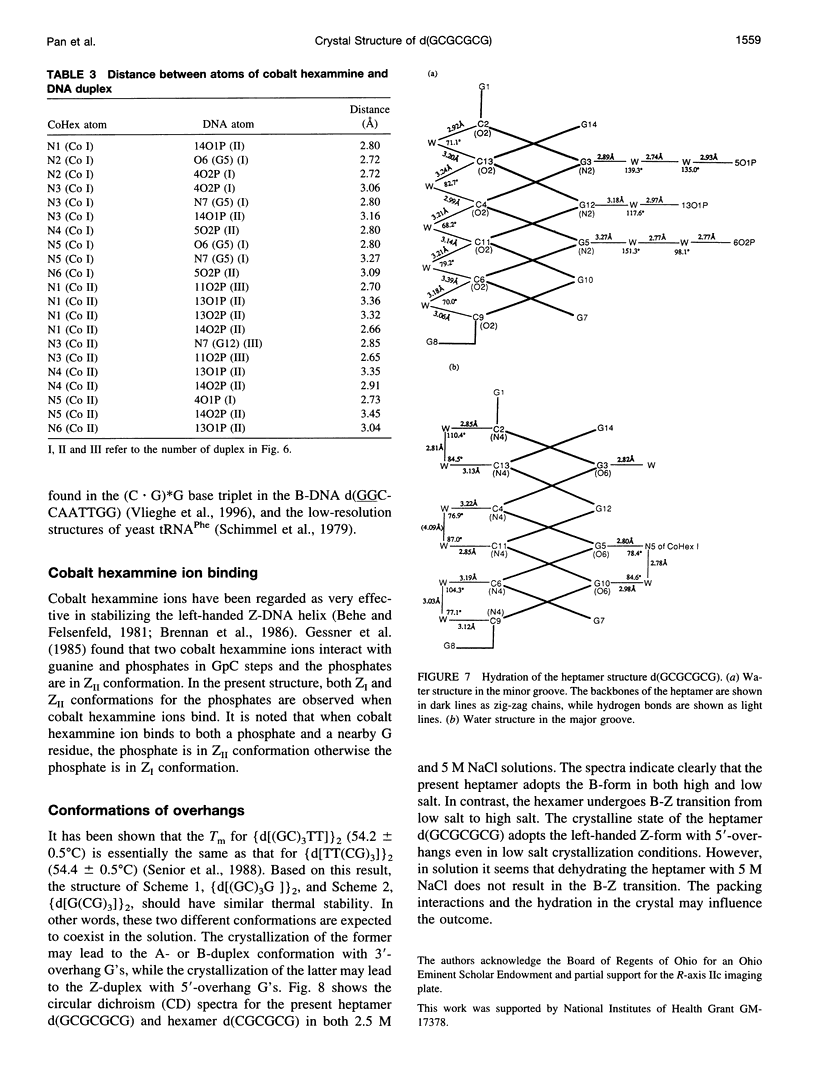
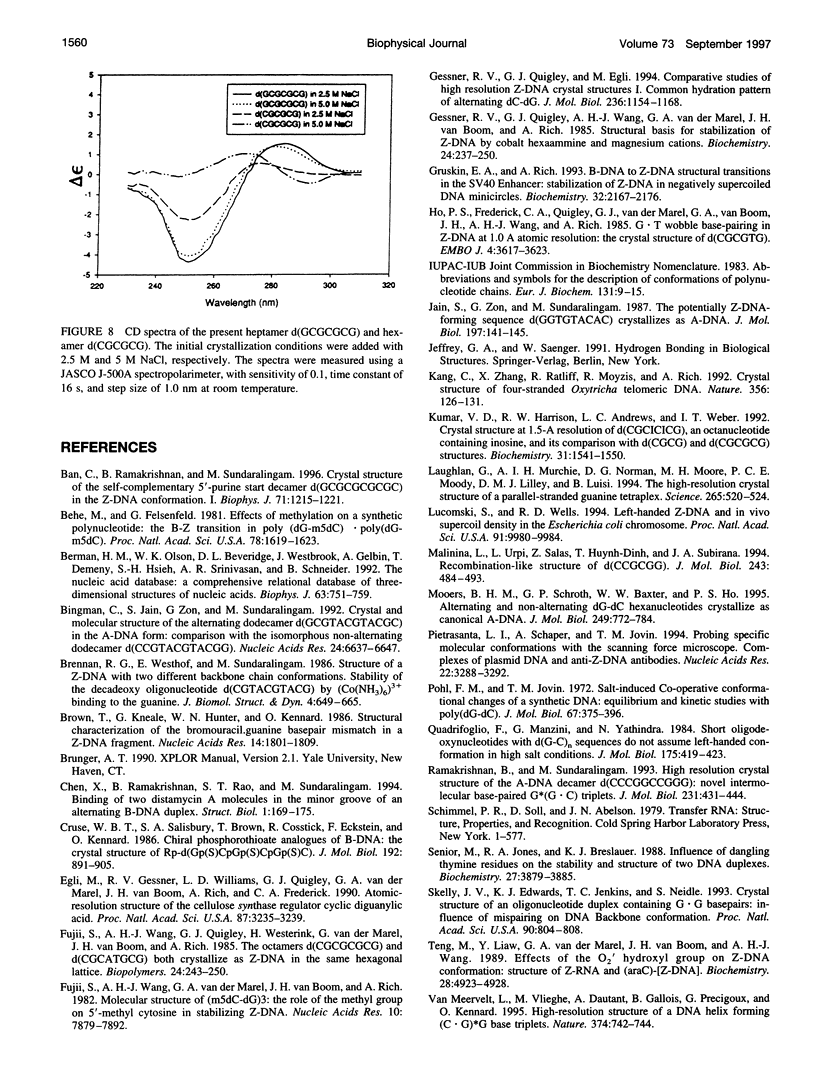
The crystal structure of the DNA heptamer d(GCGCGCG) has been solved at 1.65 A resolution by the molecular replacement method and refined to an R-value of 0.184 for 3598 reflections. The heptamer forms a Z-DNA d(CGCGCG)2 with 5'-overhang G residues instead of an A-DNA d(GCGCGC)2 with 3'-overhang G residues. The overhang G residues from parallel strands of two adjacent duplexes form a trans reverse Hoogsteen G x G basepair that stacks on the six Z-DNA basepairs to produce a pseudocontinuous helix. The reverse Hoogsteen G x G basepair is unusual in that the displacement of one G base relative to the other allows them to participate in a bifurcated (G1)N2 . . . N7(G8) and an enhanced (G8)C8-H . . . O6(G1) hydrogen bond, in addition to the two usual hydrogen bonds. The 5'-overhang G residues are anti and C2'-endo while the 3'-terminal G residues are syn and C2'-endo. The conformations of both G residues are different from the syn/C3'-endo for the guanosine in a standard Z-DNA. The two cobalt hexammine ions bind to the phosphate groups in both GpC and CpG steps in Z(I) and Z(II) conformations. The water structure motif is similar to the other Z-DNA structures.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ban C., Ramakrishnan B., Sundaralingam M. Crystal structure of the self-complementary 5'-purine start decamer d(GCGCGCGCGC) in the Z-DNA conformation. I. Biophys J. 1996 Sep;71(3):1215–1221. doi: 10.1016/S0006-3495(96)79350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Olson W. K., Beveridge D. L., Westbrook J., Gelbin A., Demeny T., Hsieh S. H., Srinivasan A. R., Schneider B. The nucleic acid database. A comprehensive relational database of three-dimensional structures of nucleic acids. Biophys J. 1992 Sep;63(3):751–759. doi: 10.1016/S0006-3495(92)81649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingman C., Jain S., Zon G., Sundaralingam M. Crystal and molecular structure of the alternating dodecamer d(GCGTACGTACGC) in the A-DNA form: comparison with the isomorphous non-alternating dodecamer d(CCGTACGTACGG). Nucleic Acids Res. 1992 Dec 25;20(24):6637–6647. doi: 10.1093/nar/20.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Westhof E., Sundaralingam M. Structure of a Z-DNA with two different backbone chain conformations. Stabilization of the decadeoxyoligonucleotide d(CGTACGTACG) by [Co(NH3)6]3+ binding to the guanine. J Biomol Struct Dyn. 1986 Feb;3(4):649–665. doi: 10.1080/07391102.1986.10508453. [DOI] [PubMed] [Google Scholar]
- Brown T., Kneale G., Hunter W. N., Kennard O. Structural characterisation of the bromouracil.guanine base pair mismatch in a Z-DNA fragment. Nucleic Acids Res. 1986 Feb 25;14(4):1801–1809. doi: 10.1093/nar/14.4.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ramakrishnan B., Rao S. T., Sundaralingam M. Binding of two distamycin A molecules in the minor groove of an alternating B-DNA duplex. Nat Struct Biol. 1994 Mar;1(3):169–175. doi: 10.1038/nsb0394-169. [DOI] [PubMed] [Google Scholar]
- Cruse W. B., Salisbury S. A., Brown T., Cosstick R., Eckstein F., Kennard O. Chiral phosphorothioate analogues of B-DNA. The crystal structure of Rp-d[Gp(S)CpGp(S)CpGp(S)C]. J Mol Biol. 1986 Dec 20;192(4):891–905. doi: 10.1016/0022-2836(86)90035-5. [DOI] [PubMed] [Google Scholar]
- Egli M., Gessner R. V., Williams L. D., Quigley G. J., van der Marel G. A., van Boom J. H., Rich A., Frederick C. A. Atomic-resolution structure of the cellulose synthase regulator cyclic diguanylic acid. Proc Natl Acad Sci U S A. 1990 Apr;87(8):3235–3239. doi: 10.1073/pnas.87.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Wang A. H., Quigley G. J., Westerink H., Van der Marel G., Van Boom J. H., Rich A. The octamers d(CGCGCGCG) and d(CGCATGCG) both crystallize as Z-DNA in the same hexagonal lattice. Biopolymers. 1985 Jan;24(1):243–250. doi: 10.1002/bip.360240118. [DOI] [PubMed] [Google Scholar]
- Fujii S., Wang A. H., van der Marel G., van Boom J. H., Rich A. Molecular structure of (m5 dC-dG)3: the role of the methyl group on 5-methyl cytosine in stabilizing Z-DNA. Nucleic Acids Res. 1982 Dec 11;10(23):7879–7892. doi: 10.1093/nar/10.23.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessner R. V., Quigley G. J., Egli M. Comparative studies of high resolution Z-DNA crystal structures. Part 1: Common hydration patterns of alternating dC-dG. J Mol Biol. 1994 Mar 4;236(4):1154–1168. doi: 10.1016/0022-2836(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Gessner R. V., Quigley G. J., Wang A. H., van der Marel G. A., van Boom J. H., Rich A. Structural basis for stabilization of Z-DNA by cobalt hexaammine and magnesium cations. Biochemistry. 1985 Jan 15;24(2):237–240. doi: 10.1021/bi00323a001. [DOI] [PubMed] [Google Scholar]
- Gruskin E. A., Rich A. B-DNA to Z-DNA structural transitions in the SV40 enhancer: stabilization of Z-DNA in negatively supercoiled DNA minicircles. Biochemistry. 1993 Mar 9;32(9):2167–2176. doi: 10.1021/bi00060a007. [DOI] [PubMed] [Google Scholar]
- Ho P. S., Frederick C. A., Quigley G. J., van der Marel G. A., van Boom J. H., Wang A. H., Rich A. G.T wobble base-pairing in Z-DNA at 1.0 A atomic resolution: the crystal structure of d(CGCGTG). EMBO J. 1985 Dec 16;4(13A):3617–3623. doi: 10.1002/j.1460-2075.1985.tb04125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Zon G., Sundaralingam M. The potentially Z-DNA-forming sequence d(GTGTACAC) crystallizes as A-DNA. J Mol Biol. 1987 Sep 5;197(1):141–145. doi: 10.1016/0022-2836(87)90616-4. [DOI] [PubMed] [Google Scholar]
- Kang C., Zhang X., Ratliff R., Moyzis R., Rich A. Crystal structure of four-stranded Oxytricha telomeric DNA. Nature. 1992 Mar 12;356(6365):126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- Kumar V. D., Harrison R. W., Andrews L. C., Weber I. T. Crystal structure at 1.5-A resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures. Biochemistry. 1992 Feb 11;31(5):1541–1550. doi: 10.1021/bi00120a035. [DOI] [PubMed] [Google Scholar]
- Laughlan G., Murchie A. I., Norman D. G., Moore M. H., Moody P. C., Lilley D. M., Luisi B. The high-resolution crystal structure of a parallel-stranded guanine tetraplex. Science. 1994 Jul 22;265(5171):520–524. doi: 10.1126/science.8036494. [DOI] [PubMed] [Google Scholar]
- Lukomski S., Wells R. D. Left-handed Z-DNA and in vivo supercoil density in the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):9980–9984. doi: 10.1073/pnas.91.21.9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinina L., Urpí L., Salas X., Huynh-Dinh T., Subirana J. A. Recombination-like structure of d(CCGCGG). J Mol Biol. 1994 Oct 28;243(3):484–493. doi: 10.1006/jmbi.1994.1674. [DOI] [PubMed] [Google Scholar]
- Mooers B. H., Schroth G. P., Baxter W. W., Ho P. S. Alternating and non-alternating dG-dC hexanucleotides crystallize as canonical A-DNA. J Mol Biol. 1995 Jun 16;249(4):772–784. doi: 10.1006/jmbi.1995.0336. [DOI] [PubMed] [Google Scholar]
- Pietrasanta L. I., Schaper A., Jovin T. M. Probing specific molecular conformations with the scanning force microscope. Complexes of plasmid DNA and anti-Z-DNA antibodies. Nucleic Acids Res. 1994 Aug 25;22(16):3288–3292. doi: 10.1093/nar/22.16.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Yathindra N. Short oligodeoxynucleotides with d(G-C)n sequence do not assume left-handed conformation in high salt conditions. J Mol Biol. 1984 May 25;175(3):419–423. doi: 10.1016/0022-2836(84)90358-9. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B., Sundaralingam M. High resolution crystal structure of the A-DNA decamer d(CCCGGCCGGG). Novel intermolecular base-paired G*(G.C) triplets. J Mol Biol. 1993 May 20;231(2):431–444. doi: 10.1006/jmbi.1993.1292. [DOI] [PubMed] [Google Scholar]
- Senior M., Jones R. A., Breslauer K. J. Influence of dangling thymidine residues on the stability and structure of two DNA duplexes. Biochemistry. 1988 May 17;27(10):3879–3885. doi: 10.1021/bi00410a053. [DOI] [PubMed] [Google Scholar]
- Skelly J. V., Edwards K. J., Jenkins T. C., Neidle S. Crystal structure of an oligonucleotide duplex containing G.G base pairs: influence of mispairing on DNA backbone conformation. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):804–808. doi: 10.1073/pnas.90.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng M. K., Liaw Y. C., van der Marel G. A., van Boom J. H., Wang A. H. Effects of the O2' hydroxyl group on Z-DNA conformation: structure of Z-RNA and (araC)-[Z-DNA]. Biochemistry. 1989 Jun 13;28(12):4923–4928. doi: 10.1021/bi00438a001. [DOI] [PubMed] [Google Scholar]
- Van Meervelt L., Vlieghe D., Dautant A., Gallois B., Précigoux G., Kennard O. High-resolution structure of a DNA helix forming (C.G)*G base triplets. Nature. 1995 Apr 20;374(6524):742–744. doi: 10.1038/374742a0. [DOI] [PubMed] [Google Scholar]
- Vlieghe D., Van Meervelt L., Dautant A., Gallois B., Précigoux G., Kennard O. Parallel and antiparallel (G.GC)2 triple helix fragments in a crystal structure. Science. 1996 Sep 20;273(5282):1702–1705. doi: 10.1126/science.273.5282.1702. [DOI] [PubMed] [Google Scholar]
- Wahl M. C., Rao S. T., Sundaralingam M. The structure of r(UUCGCG) has a 5'-UU-overhang exhibiting Hoogsteen-like trans U.U base pairs. Nat Struct Biol. 1996 Jan;3(1):24–31. doi: 10.1038/nsb0196-24. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Hakoshima T., van der Marel G., van Boom J. H., Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: the crystal structure of d(m5CGTAm5CG). Cell. 1984 May;37(1):321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Yoon C., Privé G. G., Goodsell D. S., Dickerson R. E. Structure of an alternating-B DNA helix and its relationship to A-tract DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6332–6336. doi: 10.1073/pnas.85.17.6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Lockshin C., Herbert A., Winter E., Rich A. Zuotin, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO J. 1992 Oct;11(10):3787–3796. doi: 10.1002/j.1460-2075.1992.tb05464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


