Abstract
Trichromatic color vision in humans results from the combination of red, green, and blue photopigment opsins. Although color vision genes have been the targets of active molecular and psychophysical research on color vision abnormalities, little is known about patterns of normal genetic variation in these genes among global human populations. The current study presents nucleotide sequence analyses and tests of neutrality for a 5.5-kb region of the X-linked long-wave “red” opsin gene (OPN1LW) in 236 individuals from ethnically diverse human populations. Our analysis of the recombination landscape across OPN1LW reveals an unusual haplotype structure associated with amino acid replacement variation in exon 3 that is consistent with gene conversion. Compared with the absence of OPN1LW amino acid replacement fixation since divergence from chimpanzee, the human population exhibits a significant excess of high-frequency OPN1LW replacements. Our results suggest that subtle changes in L-cone opsin wavelength absorption may have been adaptive during human evolution.
Introduction
Trichromatic color vision in humans is made possible by three genes that code for red, green, and blue light-sensitive cone pigments (Nathans et al. 1986). Retinal photoreceptor cells that contain either short (S)–, middle (M)–, or long (L)–wavelength cone pigments have spectral sensitivities that enable light absorption and color vision. These visual pigments are small membrane-bound G-protein–coupled photoreceptors (fig. 1), which contain an 11-cis retinal chromophore surrounded by an “opsin” protein that has seven α-helical barrels connected by loops (Sharpe et al. 1999) and that has wavelength absorption maxima (λmax) at either 420 (blue, S), 530 (green, M), or 560 (red, L) nm in the visible light spectrum. The phenotypic expression of mutations in the genes that code for these photopigments has long been studied in human patients because of the mutations' association with color blindness, which occurs in ∼8% of the male population (Sharpe et al. 1999).
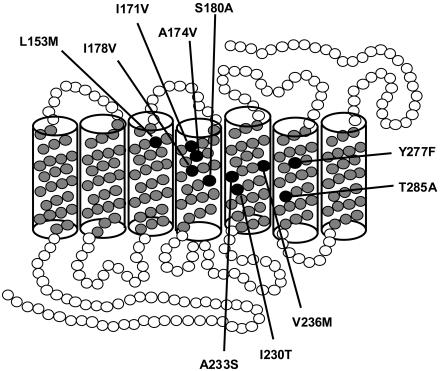
Figure 1.
Diagram of the opsin pigment with the seven α-helical barrels that form a ring around a light-trapping chromophore. Each circle on the polypeptide chain represents 1 of the 364 amino acids that form the L-cone opsin pigments. The Y277F and T285A variants distinguish L- and M-cone pigments’ λmax. Eight polymorphic residues were found in this study (table 3).
Human S-cone opsins show only 43% amino acid identity with M- and L-cone opsins, whereas the M- and L-cone opsins are 96% identical (in 364 amino acids), suggesting that the chromosome 7–linked S-cone opsin gene and X-chromosome–linked L/M-cone opsin genes diverged ∼300–500 million years ago (MYA) (Nathans et al. 1986). The L- (OPN1LW [MIM 303900]) and M-cone (OPN1MW) opsins are probably the result of a tandem duplication ∼30–40 MYA, because only a single X-linked opsin gene is found in most New World primates, whereas Old World primates (including humans) possess both the OPN1LW and OPN1MW genes (Sharpe et al. 1999). OPN1LW and OPN1MW genes are separated by only 24 kb; therefore, frequent unequal recombination and gene conversion between these genes often result in deletions and duplications in this gene array that can cause color blindness (Hayashi et al. 1999). Because of this frequent exchange, X chromosomes have around one to five OPN1MW copies, whereas individuals rarely have more than one OPN1LW copy (Sjoberg et al. 1998; Sharpe et al. 1999; Carroll et al. 2002).
The OPN1LW and OPN1MW genes have six exons and are 15 and 13.7 kb in length, respectively, differing by the presence of Alu repeats in intron 1 of OPN1LW (Jorgensen et al. 1990). Exons 1 and 6 are identical in nucleotide sequence for OPN1LW and OPN1MW genes, but exons 2–5, which code for α-helices that contact the chromophore, contain several amino acid replacement variants that greatly influence λmax (Merbs and Nathans 1992; Asenjo et al. 1994; Sharpe et al. 1998). Three amino acid changes account for the majority of the variation in λmax, an observation that remains consistent even across distantly related taxa (Yokoyama and Radlwimmer 2001). These changes are a Ser180Ala variant (∼5-nm difference) coded by exon 3 and Tyr277Phe (∼7 nm) and Thr285Ala (∼14 nm) variants coded by exon 5.
All expressed L-cone opsins have the Tyr277 and Thr285 residues, whereas expressed M-cone opsins have the Phe277 and Ala285 residues (Deeb et al. 1992). However, OPN1LW and OPN1MW genes also share four amino acid replacement polymorphisms in exon 3, including the Ser180Ala variant (Merbs and Nathans 1992; Winderickx et al. 1992, 1993; Asenjo et al. 1994; Sharpe et al. 1998). Humans and other primates also share several L- and M-cone opsin amino acid polymorphisms; however, human OPN1LW and OPN1MW introns are more closely related to each other than they are to their homologs in other primates (Shyue et al. 1994; Zhou and Li 1996; Boissinot et al. 1998). Therefore, because of gene conversion, introns have become historically homogenized between OPN1LW and OPN1MW, yet purifying selection has maintained specific amino acid differences (i.e., 277 and 285) between L- and M-cone opsins.
Despite the several hundreds of studies on the functional and genetic basis of color vision variation, most have focused on genetic variation among color-blind individuals. There have been a few studies of coding-region variation in European-derived populations (Winderickx et al. 1993; Sharpe et al. 1998), yet there has never been a study of nucleotide diversity in both coding and noncoding regions of the color vision genes across a random sample of geographically diverse ethnic groups. Although M-cone variants are rare and exhibit no measurable effect on λmax, L-cone variants are common in number (eight) and in frequency (several >20%), cause λmax to shift ∼1–5 nm, and have been linked to differences in color vision perception for humans and other primates (Winderickx et al. 1992; Jameson et al. 2001; Regan et al. 2001).
Although previous studies attempted to estimate the adaptive value of L-cone variants with functional analyses (Winderickx et al. 1992; Jameson et al. 2001), these variants are typically discounted because they result in subtle effects on λmax, compared with other residues (i.e., 277 and 285). However, molecular evolutionary genetic analyses can elucidate the adaptive value of SNPs by inferring their historical impact on the underlying haplotype structure. In the current study, we characterized DNA sequence diversity, linkage disequilibrium (LD), and recombination patterns for a 5.5-kb region of OPN1LW in a global sample of 236 humans. In comparing polymorphism and fixation between humans and chimpanzees along coding and noncoding regions of OPN1LW, we apply population genetic and statistical analyses to detect how natural selection and gene conversion have historically shaped diversity at this gene. An understanding of the molecular evolution of color vision will have broad implications for many disciplines, including medical and physiological research that focuses on how sensory perception has evolved in humans and other primates.
Subjects and Methods
Population Samples
The OPN1LW gene resides on the X chromosome (Xq28); therefore, a sampling of males enabled the unambiguous determination of all polymorphic sites (i.e., no heterozygous sites) and the complete haplotype phase for all individuals. Nucleotide sequence variation was surveyed from 236 men from 19 groups (table 1). Our sub-Saharan African sample (labeled as “African”) consists of 163 individuals from 11 different groups. In cases where we had very small sample sizes (often <5) for distinct ethnic groups, these groups were pooled by country of origin (e.g., Sierra Leone, Gambia, Nigeria, Cameroon, or South Africa). We also sampled 73 individuals from outside of sub-Saharan Africa (referred to as “non-African”). Finally, the same OPN1LW fragment was sequenced from six unrelated male chimpanzees (Pan troglodytes) for interspecific comparisons. This study was approved by the institutional review board at the University of Maryland, and all samples were gathered with informed consent.
Table 1.
Population-Diversity Estimates
|
Silentb |
Replacement |
|||||
| Samplea | No. of Chromosomes | No. of SNPs | πc | No. of SNPs | πc | hd |
| African: | 163 | 67 | .21 | 9 | .60 | .87 |
| West African: | 76 | 50 | .23 | 6 | .59 | .86 |
| Sierra Leone | 18 | 36 | .24 | 4 | .50 | .79 |
| Gambia | 15 | 38 | .25 | 4 | .58 | .89 |
| Nigeria | 16 | 33 | .20 | 4 | .45 | .78 |
| Cameroon | 13 | 36 | .22 | 6 | .70 | .94 |
| Bakola Pygmiese | 14 | 29 | .21 | 4 | .50 | .88 |
| East Africanf: | 71 | 45 | .19 | 9 | .62 | .88 |
| Hadza | 16 | 30 | .20 | 5 | .61 | .89 |
| Sandawe | 13 | 33 | .23 | 8 | .73 | .92 |
| Maasai | 14 | 24 | .10 | 6 | .45 | .69 |
| Turu | 13 | 27 | .22 | 6 | .74 | .94 |
| Burunge | 15 | 32 | .21 | 6 | .52 | .78 |
| Southern African (Bantu speakers) | 16 | 36 | .22 | 6 | .66 | .88 |
| Non-African: | 73 | 36 | .12 | 9 | .55 | .81 |
| Algeria (Berber) | 9 | 23 | .14 | 6 | .63 | .92 |
| Tunisia | 9 | 27 | .19 | 4 | .51 | .81 |
| Cyprus | 8 | 8 | .07 | 3 | .43 | .75 |
| Lebanon | 10 | 8 | .05 | 3 | .42 | .82 |
| Northern Europe | 10 | 20 | .14 | 7 | .84 | .84 |
| Western Russia (Zversky) | 9 | 5 | .05 | 1 | .06 | .22 |
| China (Han) | 9 | 9 | .05 | 2 | .21 | .56 |
| Papua New Guinea | 9 | 8 | .05 | 3 | .27 | .69 |
| Global | 236 | 74 | .19 | 9 | .61 | .86 |
| Chimpanzee (P. troglodytes) | 6 | 10 | .09 | 0 | .00 | 0 |
African and non-African samples are described in the “Subjects and Methods” section.
Includes coding and noncoding regions.
Average pairwise sequence differences per bp (×10-2).
Amino acid haplotype diversity estimates, weighted by sample size.
Pygmies are from Southern Cameroon.
East Africans are from Tanzania.
PCR and Sequence Determination
Primers constructed from regions of exons 2 and 5 that distinguish OPN1LW from OPN1MW genes were adapted from Winderickx et al. (1992), to amplify single OPN1LW gene copies by PCR. The region amplified is shown in figure 2, and primers can be found at the Tishkoff Web page. PCR products were prepared for sequencing with shrimp alkaline phosphatase and exonuclease I (U.S. Biochemicals). All nucleotide sequence data were obtained using the ABI Big Dye terminator kit and 3100 automated sequencer (Applied Biosystems). Sequence files were aligned using the Sequencher v. 4.0.5 program (Gene Codes).
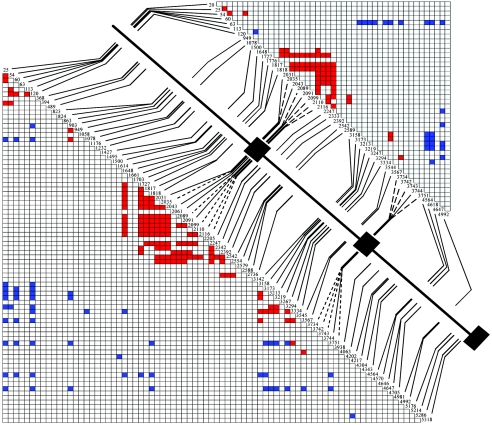
Figure 2.
Diagram of sequenced 5.5-kb OPN1LW region with LD plots. The large black boxes are exon 3 (169 bp), exon 4 (166 bp), and partial exon 5 (121 bp), which are separated by introns 2 (1,987 bp), 3 (1,467 bp), and 4 (1,554 bp). Dashed lines are replacement SNPs. Plots represent pairwise associations among the 78 variants from the 163 Africans (bottom half) and among the 46 variants from the 73 non-Africans (top half). Red and blue blocks are pairwise associations that exhibit significantly less LD and significantly more LD, respectively, than expected on the basis of the ML estimate of ρMAF (see the “Subjects and Methods” section).
As described above, the amino acid residues 277 and 285, coded by exon 5, largely determine the opsins’ λmax. In this study, we analyzed nucleotide sequence variation associated with L-cone opsin phenotypes; therefore, only nucleotide sequences that coded for Tyr277 and Thr285 were included in our analyses. Although X chromosomes rarely have more than one OPN1LW copy, in the rare cases in which heterozygous sites were observed (<5% of our sequences), these individuals were excluded to ensure analysis of single OPN1LW haplotypes.
Data Analysis
Individuals were chosen with no a priori knowledge of their color vision phenotypes; thus, our data set represents a random sample of genetic variation, at the OPN1LW locus, across geographic regions of human populations appropriate for statistical tests of neutrality. African populations typically have significantly higher levels of genetic diversity compared with other regions, which is consistent with a larger and more ancestral effective population size (Ne) for African groups and a relatively recent ancestry and smaller Ne for other regions (Tishkoff and Williams 2002; Tishkoff and Verrelli 2003a). Differences in demographic histories can result in distinct patterns of LD and haplotype structure; therefore, our African and non-African samples were analyzed separately for all statistical tests. Silent polymorphisms (this term refers, throughout the present article, to introns and synonymous sites in exons) do not alter the amino acid sequence; therefore, they are typically expected to reflect the neutral mutation rate in humans. Estimates of genetic diversity and statistical tests for exons and introns were calculated independently to determine whether replacements sites (i.e., amino acid residues) are neutrally evolving compared with silent sites.
Estimates of locus-specific diversity, based on the number of segregating sites (S) and corrected by sample size, were calculated using Watterson’s θ (Watterson 1975). This was compared with an estimate based on the average number of pairwise differences per base pair among all alleles (π). These two estimates of the parameter θ=3Neμ (for X-linked genes, where μ is the mutation rate) were calculated for silent and replacement sites across populations and gene regions, by use of the Rozas and Rozas (1999) DnaSP v. 4.0 program (available at the Rozas Web page). The estimates S and π are expected to be equal under a model of neutrality, which can be assessed using Tajima’s (1989) D test. Significant positive D values may indicate an excess of high-frequency SNPs consistent with balancing selection, whereas significant negative D values indicate an excess of low-frequency SNPs consistent with recent directional selection.
There is compelling evidence from archeological and genetic studies, including nucleotide sequence analyses (see reviews by Tishkoff and Williams [2002] and by Tishkoff and Verrelli [2003a]), that the human population rapidly expanded in size ∼50–200 thousand years ago (KYA) from an Ne of ∼10,000 individuals. Tajima’s D test assumes a constant historical population size; therefore, test results may be consistent with demographic influences such as population structure (positive D) or expansion (negative D). Tajima’s D test also assumes no recombination in the sample of nucleotide sequences; however, this is an invalid assumption for genes that have a history of gene conversion, which can greatly affect this statistical test (Ardlie et al. 2001). We used Hudson’s (2002) ms program (available at the Hudson Web page) to generate 10,000 genealogical trees simulating exponential population expansion, with generation times of 20 years, at periods of 50, 100, or 200 KYA from historical Ne, of 0.0001%, 0.001%, or 0.005% of the current size of 109. We also incorporated the effects of recombination into these simulations by varying the recombination parameter ρ=3Ner (for X-linked genes, where r is the recombination rate per nucleotide site) at intervals of 0 (no recombination) to 100, for each run. These simulated distributions of Tajima’s D values under different recombination parameters and population expansion times and sizes serve as a more appropriate model of human evolution with which to test our observed Tajima’s D values for statistical significance.
We calculated FST (by use of DnaSP) as a relative estimate of population differentiation for the major defined groups of Africans and non-Africans (with estimates weighted by sample size), and we tested these estimates for statistical significance by use of simulations from the permtest program of Hudson (2000) that is implemented in DnaSP. We also compared the frequencies of single amino acid polymorphisms between the major geographic regions of sampled Africans and non-Africans, with standard χ2 tests.
A simple neutral model predicts that drift and mutation rate determine the level of nucleotide variation that accumulates within and between species; therefore, the relative amount of within-species polymorphism should reflect the amount of between-species fixation under neutrality (McDonald and Kreitman 1991). Genetic diversity for human OPN1LW was compared with fixation between human and chimpanzee OPN1LW sequences to test this hypothesis. Although introns and exons are expected to have different levels of constraint and inferred mutation rates under neutrality, we still expect that the ratios of polymorphism to fixation will be similar across these regions (J. H. McDonald, personal communication). For example, if we observe more replacement polymorphism than silent-site polymorphism, we would then expect proportionately more replacement fixation than silent-site fixation. The Hudson, Kreitman, and Aguade (HKA) test (Hudson et al. 1987) was applied to test whether levels of variation across coding regions were consistent with that of noncoding regions. We also tested whether OPN1LW replacement polymorphisms were consistent with neutrality by comparing the ratio of replacement polymorphism to fixation with the ratio of silent-site polymorphism to fixation (in introns and exons) using a Fisher’s exact test of independence that was described by McDonald and Kreitman (1991).
Although population genetic analyses traditionally test for significant LD utilizing different summary statistics (i.e., D′ or r2), a more appropriate theoretical method would take into consideration the underlying recombination rate (ρ) across the genomic region being tested (Hudson 2001). Although this has historically been impractical, maximum-likelihood (ML) methods for estimating ρ have recently become less computationally intensive. We used the LDhat program of McVean et al. (2002) (available at the McVean Web page)—which uses the approximate-likelihood method of Hudson (2001)—to estimate ρ (ρMAF) across the 5.5-kb region. A permutation analysis is then conducted to determine whether pairwise comparisons among SNPs exhibit significantly more or less LD, given the SNP allele frequencies and underlying gene-specific estimate of ρMAF. We then employed the “hotspotter” program of Li and Stephens (2003) (available at the Stephens Web page), which also generates a gene-specific estimate of ρ (ρLS) by use of a ML method, but, in contrast to the LDhat algorithm, tests for varying recombination rates across the OPN1LW region. This algorithm tests whether a model that includes a recombination “hotspot” has a significantly higher likelihood (MLHOT) in explaining the haplotype structure than does a model that ignores variation in recombination rates (MLNOT).
Results
OPN1LW Population Variation
A diagram of the 5.5-kb region of the OPN1LW gene that was surveyed in this study is shown in figure 2. There are 456 bp in exons 3–5 and 5,008 bp in introns 2–4 that result in a total of 5,118 effectively neutral silent sites. In our global sample of 236 chromosomes, we find 85 variants in the 5.5-kb region, which includes two 2-bp in/dels in intron 3. Of the 83 SNPs, there are 9 replacement and 5 silent SNPs in exons, all of which occur in exons 3 and 4 (table 2). The nine replacements result in eight changes to the amino acid sequence, because two replacement SNPs in exon 4 are completely linked (nucleotides 3742–3743 in fig. 2; numbers starting with the first base pair of intron 2) and together result in an Ala233Ser variant.
Table 2.
Gene-Region Diversity Estimates
|
Total |
Silentb |
Replacement |
|||||||
| Samplea | No. of Sites | No. of SNPs | πc | No. of SNPs | πc | Dd | No. of SNPs | πc | Dd |
| Africans (n=163): | 5,464 | 76 | .24 | 67 | .21 | −.15 | 9 | .60 | .71e |
| Intron 2 | 1,987 | 28 | .25 | 28 | .25 | −.03 | … | … | … |
| Exon 3 | 169 | 9 | 1.46 | 4 | 2.29 | .48 | 5 | 1.24 | 1.61e |
| Intron 3 | 1,467 | 17 | .27 | 17 | .27 | 1.31e | … | … | … |
| Exon 4 | 166 | 5 | .29 | 1 | .03 | −.99 | 4 | .40 | −.46 |
| Intron 4 | 1,554 | 17 | .07 | 17 | .07 | −1.50 | … | … | … |
| Exon 5 | 121 | 0 | 0 | 0 | 0 | … | 0 | 0 | … |
| Coding region | 456 | 14 | .66 | 5 | .84 | .06 | 9 | .60 | .71e |
| Non-Africans (n=73): | 5,464 | 45 | .15 | 36 | .12 | −.48 | 9 | .55 | .07 |
| Intron 2 | 1,987 | 15 | .10 | 15 | .10 | −1.01 | … | … | … |
| Exon 3 | 169 | 8 | 1.28 | 3 | 1.91 | .38 | 5 | 1.12 | .88e |
| Intron 3 | 1,467 | 13 | .19 | 13 | .19 | .15 | … | … | … |
| Exon 4 | 166 | 5 | .29 | 1 | .13 | −.93 | 4 | .40 | −.77 |
| Intron 4 | 1,554 | 4 | .04 | 4 | .04 | −.37 | … | … | … |
| Exon 5 | 121 | 0 | 0 | 0 | 0 | … | 0 | 0 | … |
| Coding region | 456 | 13 | .59 | 4 | .74 | −.06 | 9 | .55 | .07 |
See table 1 for population samples. n = no. of chromosomes.
Includes coding and noncoding regions.
Average pairwise sequence differences per bp (×10-2).
Tajima’s (1989) D statistic.
P<.01 under models of demographic growth and recombination (see the “Subjects and Methods” section).
Africans have twice as much silent-site diversity (π) as do non-Africans (tables 1 and 2), and our global FST of 0.16 (P<.0001) is comparable to other nucleotide studies (Tishkoff and Verrelli 2003a). We find significant differentiation between Africans and non-Africans (FST=0.10; P<.0001), yet there is also significant differentiation among the 8 African groups (FST=0.06; P<.0001) and among the 11 non-African groups (FST=0.28; P<.0001). In addition, Africans and non-Africans possess the same nine replacement SNPs; however, these two samples show significant frequency differences for several amino acid variants (table 3).
Table 3.
L-Cone Opsin Amino Acid Frequencies
|
Frequency in |
|||
| Varianta | Africans(n=163) | Non-Africans(n=73) | χ2 Testb |
| Exon 3: | |||
| Leu153Met | .22 | .21 | NS |
| Ile171Val | .71 | .90 | P<.001 |
| Ala174Val | .24 | .21 | NS |
| Ile178Val | .06 | .03 | NS |
| Ser180Ala | .08 | .30 | P<.0001 |
| Exon 4: | |||
| Ile230Thr | .01 | .05 | NS |
| Ala233Ser | .01 | .05 | NS |
| Val236Met | .58 | .78 | P<.01 |
Frequencies are shown for the derived allele for each amino acid variant (e.g., frequency of the Met153 allele is reported for the Leu153Met variant).
Statistical significance of difference in frequency between geographic groups, each with n chromosomes. NS = not significant.
Neutrality Tests
OPN1LW is X-linked; therefore, diversity estimates need to be adjusted (multiplied by 4/3) for comparisons with autosomal genes. The adjusted estimate for OPN1LW (πadj=0.31%) is more than three times the average diversity for autosomal and X-linked human genes (π=0.10%) (Nachman 2001; Tishkoff and Verrelli 2003a). However, silent-site fixation at OPN1LW (0.9%) is similar to other comparisons between humans and chimpanzees (1.1%) (Przeworski et al. 2000; Nachman 2001), which suggests that silent sites reflect the typical neutral fixation rate. Our HKA analyses find that the ratio of polymorphism/fixation was significantly greater in exons versus introns for Africans (14/0 vs. 62/45; P=.015) and non-Africans (13/0 vs. 32/45; P=.001). In addition, our significant McDonald-Kreitman tests (table 4) indicate that the number of replacement SNPs segregating in Africans and non-Africans is unexpectedly high, given the absence of fixed replacements between human and chimpanzee OPN1LW.
Table 4.
McDonald-Kreitman Tests[Note]
|
No. of Differences |
|||
| Samplea | Silent | Replacement | Fisher’s Exact Test(P) |
| Africans (n=163): | |||
| Polymorphic | 67 | 9 | |
| Fixed | 45 | 0 | .014 |
| Non-Africans (n=73): | |||
| Polymorphic | 36 | 9 | |
| Fixed | 45 | 0 | .002 |
| Global (n=236): | |||
| Polymorphic | 74 | 9 | |
| Fixed | 45 | 0 | .026 |
Note.— Nos. of SNPs and fixations found at silent and replacement sites are contrasted with a Fisher’s exact test (see the “Subjects and Methods” section).
See table 1 for population samples. n = no. of chromosomes.
Our Tajima’s D tests indicate that frequency distributions tend to be skewed toward rare variants (i.e., negative D values) (table 2), a result typically observed at many human genes and consistent with a model of historical population expansion. However, when we simulated the allelic frequency distribution under models of varying population growth, we observed significant positive D values for exon 3 replacements and intron 3 in Africans, as well as for exon 3 replacements in non-Africans (all P<.01). In fact, our simulations indicate that a neutral model could explain the observed replacement frequency distributions only if the human population had expanded >200 KYA from an Ne >50,000. However, this time period and size of expansion are much older and larger, respectively, than predicted by the majority of genetic, morphological, and archaeological studies.
LD and Haplotype Structure
Our analysis of recombination finds gene-specific ML estimates of ρMAF=95 and ρLS=144 for Africans and ρMAF=100 and ρLS=153 for non-Africans. Although estimates for non-Africans are larger than those for Africans, this difference is not statistically significant and probably reflects the high variance associated with estimates of ρ>100 (McVean et al. 2002). Figure 2 shows the results of the LDhat permutation analysis that tests for significantly high or low LD among SNPs, given the underlying recombination estimate for the gene region (ρMAF). In light of the high estimates of ρ for Africans (ρMAF=95) and non-Africans (ρMAF=100), it is not surprising that <2% of the pairwise comparisons among SNPs exhibit significant LD, all of which are sporadically found among introns 2, 3, and 4. However, in spite of these high estimates of ρMAF, we find that a number of pairwise comparisons exhibit significantly less LD than expected. Of particular interest is the observation that these pairwise comparisons are clustered around exon 3 in Africans and non-Africans, suggesting that this “cluster” is not population specific. When we applied the Li and Stephens (2003) hotspotter algorithm to test our data for a localized region with elevated ρ, we found that the interval spanning SNPs 1817–2342 has an estimated ρ that is 20 and 5 times greater in Africans and non-Africans, respectively, than any other interval of the gene (P<.05). In fact, compared with a model that ignores recombination rate heterogeneity (MLNOT), a model that assumes a recombination hotspot (MLHOT) more likely explains the African (9×1019 times more likely; P<.05) and non-African (148 times more likely; P<.05) haplotype structures.
Discussion
Previous analyses of the OPN1LW gene had revealed several replacement SNPs in color-normal and color-blind individuals; however, a population genetic analysis has never been conducted on a random sample of OPN1LW nucleotide sequences. As with other studies, we have discovered nine replacement SNPs in our global sample of 236 human chromosomes; however, we have also revealed an additional 74 silent SNPs at OPN1LW. With this new data set, our analysis of coding and noncoding regions reveals an unusual haplotype structure associated with replacement variation in exon 3 that is consistent with gene conversion at OPN1LW in Africans and non-Africans. Additionally, by combining intra- and interspecific comparisons, we demonstrate that L-cone opsin amino acid variation is not consistent with a neutral model of molecular evolution.
Gene Conversion and OPN1LW Variation
Compared with estimates for other genes, including others in the Xq28 region where OPN1LW is found (Ardlie et al. 2001; Frisse et al. 2001; Hudson 2001; Przeworski and Wall 2001; Reich et al. 2002), we find an unusually high level of recombination across the 5.5-kb region. More specifically, we find a significantly elevated ρ in an interval of ∼530 bp that is centered on the 169 bp of exon 3. These results support a previous study that suggested gene conversion with OPN1MW in exon 3 results in shared variation (Winderickx et al. 1993). Several studies have proposed that the small “hotspots” of recombination that are commonly dispersed throughout the genome are a result of gene conversion. In fact, Crawford et al. (2004) have recently estimated that the hotspot frequency may be one in every 63 kb in the human genome. Because recombination probably affects the haplotype structure on a larger scale, the unusual haplotype blocks found over smaller localized regions that mimic recombination hotspots are more consistent with gene conversion (Ardlie et al. 2001; Frisse et al. 2001; Przeworski and Wall 2001; Reich et al. 2002; Li and Stephens 2003; Wall et al. 2003; Wall 2004; Ptak et al. 2004). Although the hotspotter algorithm was developed to identify “recombination hotspots,” simulations of nucleotide sequence data with varying levels of gene conversion did not lead to the false identification of hotspots when they are not present in the data (Crawford et al. 2004).
A major consideration is that these algorithms do not take into account the impact of natural selection. In fact, evidence from our data set as well as others' (Shyue et al. 1994; Zhou and Li 1996; Hayashi et al. 1999) strongly suggests that gene conversion occurs quite frequently across the entire OPN1LW gene region. Gene conversion may increase LD on a local scale because of the insertion of sequence “tracks” as small as 50 bp (Petes 2001); however, in the “hotspot” interval that includes exon 3, we find a significant lack of LD and the presence of all four gametic types for some pairs of SNPs separated by <20 bp. Therefore, it is unclear whether this pattern is the result of gene conversion alone or whether it is due to the combined effects of gene conversion and natural selection, which we discuss below.
Theoretical studies have shown that, relative to a single-gene model, polymorphism may be elevated at duplicated genes due to gene conversion (Innan 2002, 2003a, 2003b), consistent with our observation of elevated diversity at the OPN1LW locus. Genetic drift at duplicated gene copies occurs more slowly than at single genes because variation “migrates” between gene copies as a result of gene conversion. Nonetheless, variation will eventually become fixed due to drift, and, in fact, even balancing selection cannot maintain adaptive alleles indefinitely (Takahata et al. 1992). Innan (2002, 2003b) has shown that typical models of molecular evolution and specific interlocus comparisons (i.e., HKA statistical tests) are invalid because of the higher expectation of variation at duplicated genes than at single genes. However, our HKA tests of coding and noncoding regions are valid, given that these regions have been subject to the same historical duplication event. In addition, although gene conversion may retard the level of fixation between duplicated copies, gene conversion is not expected to significantly alter the level of neutral fixation between humans and chimpanzees (Innan 2002, 2003b). As expected, in the face of gene conversion, we find that silent-site fixation has historically occurred between humans and chimpanzees across all introns. On the contrary, given the absence of fixed differences in exons, the number of SNPs in these regions is not expected under the neutral model.
Multiple Signatures of Selection
Recent work indicates that primates have higher mutation rates in exons than in introns because of a greater number of CpG dinucleotides in coding regions (Subramanian and Kumar 2003). However, an excess of CpG dinucleotides cannot account for the excess of SNPs at OPN1LW, since a higher proportion of CpG island SNPs occur in introns (24/69) than in exons (3/14). If gene conversion does inflate polymorphism, then it is possible that the overall excess of exon SNPs, as indicated by our HKA analyses, is a result of the detected “recombination hotspot” centered on exon 3, given that 9 of the 14 coding-region SNPs are found in this exon. Although this may be true, our McDonald-Kreitman tests are consistent with an excess of only replacement SNPs. Gene conversion may inflate the overall variation at a locus; however, it is unclear why gene conversion would inflate the number of replacement SNPs and not that of silent SNPs.
A significant excess of replacement SNPs is typically explained by either balancing selection or “slightly” deleterious selection. If purifying selection against replacement SNPs is weak because they are only slightly deleterious, then these SNPs may remain polymorphic at low frequencies, but weak purifying selection keeps them from reaching high frequencies and fixation (Ohta 1992; Fay et al. 2001). This theory is in accordance with numerous human genes that possess replacement SNPs that are very rare in frequency (Nachman et al. 1996; Nielsen and Weinreich 1999). However, derived replacements at OPN1LW are found at frequencies as high as 22%–90% (table 3). Although this suggests that they are not rare in frequency, it is unclear if this frequency distribution is consistent with a slightly deleterious selection model.
One way to examine the frequency distributions of SNPs is with Tajima’s D test. When we simulated potential scenarios of human population growth, our Tajima’s D analyses show that replacement SNPs in exon 3 are found at significantly high frequencies in Africans and non-Africans (table 2). Intron 3 of Africans shows a significant result as well; however, other tests indicate that this region is evolving neutrally, so it is possible that this pattern is simply a result of historical hitchhiking with exon 3. Simulations of population growth were similarly employed by Wooding et al. (2004) in demonstrating that amino acid polymorphisms at the PTC gene were significantly high in frequency and were consistent with selection.
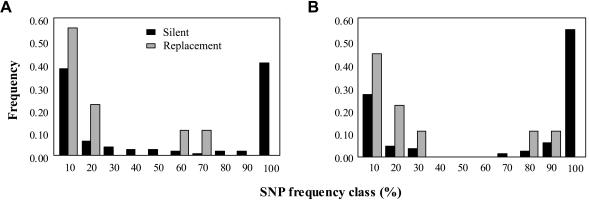
A second way to analyze amino acid frequency distributions is to compare them with the frequency distributions of silent SNPs, which are expected to reflect neutrality (Akashi 1999). Here, we employ a Mann-Whitney U (MWU) test (Akashi and Schaeffer 1997) that combines the Tajima (1989) and McDonald-Kreitman (1991) tests but which is statistically more powerful for rejecting a slightly deleterious model (Akashi 1999). If replacement polymorphisms are slightly deleterious, then we may expect them to be found at significantly rarer frequencies than silent SNPs. For our MWU test, we first compared chimpanzee and human sequences to determine the derived state at each human SNP. Frequency distributions for silent and replacement changes were created with frequency bins (fig. 3), where the 100% frequency bin represents the “fixed” class and all other bins are SNP classes. Figure 3 shows that the overall pattern of replacement SNPs in non-Africans may be consistent with a model of weak purifying selection, whereas the excess of replacement SNPs in Africans is not consistent with this model.
Figure 3.
Silent and replacement SNP frequency distributions. A, 163 Africans (Mann-Whitney U=1.85 [not significant; df ∞]). B, 73 non-Africans (Mann-Whitney U=2.93 [P<.01; df ∞]). The X-axis shows frequency bins; the “100%” bin refers to fixations between chimpanzee and human OPN1LW. The Y-axis is the relative proportion of the different frequency bins found on the X-axis (e.g., 40% of the silent SNPs in Africans are each found at ∼10% frequency, whereas 57% of the replacement SNPs in Africans are each found at ∼10% frequency).
Whereas it is possible to reject a null model of neutrality at OPNILW, it is more difficult to test specific alternative models of selection. Nonetheless, our statistically significant McDonald-Kreitman, Tajima, and MWU test results for replacement SNPs in Africans are consistent with a model of balancing selection (i.e., maintenance of high-frequency alleles), whereas the overall pattern of replacement SNPs in non-Africans may be consistent with weak purifying selection. However, we may interpret these results as a reflection of the differential selection pressures across exons. For example, it is clear from the rare frequencies of SNPs in exon 4 and the absence of SNPs in exon 5 that purifying selection removes replacements that cause large λmax changes. On the contrary, the high frequencies of replacement SNPs in exon 3 that have subtle effects on λmax may be a result of balancing selection in Africans as well as non-Africans. In fact, this selection model may also explain the detected “recombination hotspot” that is centered on the 169 bp of exon 3. If gene conversion events generate combinations of replacement SNPs that are adaptive, then these conversion events will be maintained more often than those that result in neutral variation or, more importantly, deleterious variation. Therefore, it may be the case that selection—and not a “recombination hotspot”—is responsible for the high haplotype diversity around exon 3.
Without further characterizing the nucleotide diversity at OPN1MW, we cannot rule out the possibility that balancing selection operates at OPN1MW as well. However, it is highly unlikely that selection at OPN1MW alone explains the different patterns of variation and divergence associated with silent and replacement sites at OPN1LW. Previous studies of this gene region show that OPN1MW has only four replacement SNPs, all of which are shared with OPN1LW in exon 3, are reportedly rare in frequency, and show no effect on M-cone opsin λmax (Merbs and Nathans 1992; Winderickx et al. 1992, 1993; Asenjo et al. 1994; Sharpe et al. 1998). It is clear that signatures of selection at OPN1LW are complex and may be consistent with balancing selection at exon 3 and purifying selection at exons 4 and 5. Further analyses focused on these specific regions in OPN1LW and OPN1MW will reveal how selection and gene conversion have influenced the evolution of color vision phenotypes.
Balancing selection and gene conversion may also explain the observation that many primates and humans share the same OPN1LW replacement SNPs (Deeb et al. 1994; Boissinot et al. 1998). If this were true, we might expect an excess of linked silent-site diversity (Slatkin 2000; Navarro and Barton 2002) and genealogies with branch lengths that are longer (i.e., “older”) relative to a neutral model, as is seen at MHC-HLA (Takahata et al. 1992; Garrigan and Hedrick 2003). However, this expectation may not be similar across genomic regions because of recombination and gene conversion rate heterogeneity (Przeworski and Wall 2001). The unique haplotype structure associated with exon 3 and the fact that SNPs have become fixed in introns but not in exons demonstrates that replacement and neutral SNPs have historically become uncoupled from each other by gene conversion and recombination. Therefore, a significant excess of linked neutral variation is not expected, and coalescence analyses will not result in “old” age estimates for haplotypes.
Finally, non-Africans commonly show greater LD than do Africans, owing to the larger Ne of the latter (Tishkoff and Verrelli 2003a), yet this is not seen in our data set. However, over short chromosomal distances, gene conversion and selection may play a large role in the decay of LD and may produce a similar haplotype structure across Africans and non-Africans (Frisse et al. 2001; Reich et al. 2002). Demographic influences are also seen in our analyses of geographic variation. Several replacement SNPs exhibit significant geographic variation, most notably the 22% difference for the Ser180Ala variant that shifts λmax ∼5 nm. Because silent SNPs also exhibit geographic variation, the geographic variation observed for replacement SNPs between Africans and non-Africans may be consistent with a model of genetic drift and reduced gene flow. With additional global sampling of OPN1LW haplotypes, we can determine whether specific replacement SNP combinations may have been adaptive in different geographic regions.
Selection for Color Discrimination?
In vitro analyses have shown that OPN1LW replacement SNPs significantly shift λmax into the “red-orange” visual spectrum (Asenjo et al. 1994; Sharpe et al. 1998; Carroll et al. 2002). As a result of random X inactivation, red and red-orange pigments are expressed in females heterozygous for different OPN1LW haplotypes and enable discrimination among colors in the red-orange spectrum, compared with expression in homozygous individuals (Jameson et al. 2001). Many New World primate species are dichromatic; however, species that have a single X-linked opsin gene often segregate L- and M-like alleles that enable heterozygous females to be trichromatic. These different alleles have routinely been argued to be balanced polymorphisms (Mollon et al. 1984; Surridge et al. 2003), because they enable discrimination among colored fruits, insects, and background foliage (Dominy and Lucas 2001; Regan et al. 2001; Lucas et al. 2003). Additionally, transgenically constructed mice heterozygous for L-cone opsin variation display significant differences in the chromatic sensitivities of retinal ganglion cells (Smallwood et al. 2003). As with other primates, L-cone color vision variation may have been adaptive during the evolution of humans, who, until 10 KYA, thrived from hunting-gathering activities, with females playing the primary role in gathering. Although not all replacements may result in detectable λmax shifts, there is convincing evidence that other aspects of photoreceptor function may be altered, such as the efficiency with which the photoreceptor absorbs light (Neitz et al. 1999). Because we find that populations possess high L-cone haplotype diversity (table 1), future functional studies should focus on haplotypes (Carroll et al. 2002)—and not single SNPs—when determining L-cone opsin λmax.
As a final comment, models that incorporate gene conversion and selection may explain LD hotspots in some genomic regions and therefore will have important implications for “tagged” SNPs and mapping studies, like the HapMap project, that rely on patterns of LD remaining somewhat consistent across chromosomal regions and populations (Clark 2003; Tishkoff and Verrelli 2003b; Wall and Pritchard 2003). For example, gene conversion at OPN1LW may create localized low LD within the long-range high LD in Xq28 that has been generated by recent selection for malarial resistance at G6PD (Tishkoff et al. 2001; Sabeti et al. 2002; Saunders et al. 2002; Verrelli et al. 2002). In addition, gene conversion may be elevated in regions in which duplicated gene copies are still somewhat similar. Therefore, if gene duplication has played as large a role in human evolution as is predicted, then fine-scale DNA sequence analyses will be important for determining how gene conversion generates complex phenotypes like color vision.
Acknowledgments
We thank A. Lane, T. Jenkins, M. Newport, G. Argyropoulos, G. Destro-Bisol, A. Froment, E. Chouery, V. Delague, A. Gessain, A. Helal, J. Loiselet, A. Marbane, A. Salem, G. Lefranc, A. Drousiotou, and M. Stoneking, for providing DNA samples; N. Li and M. Stephens, for advice on interpretation of the hotspotter program results; and J. H. McDonald, S. Mount, R. Payne, A. G. Clark, and two anonymous reviewers, for critical review of the manuscript. This work was funded by a Burroughs Wellcome Fund Career Award, a David and Lucile Packard Career Award, National Science Foundation grant BCS-9905396 (to S.A.T.), and National Science Foundation Integrative Graduate Education and Research Traineeships grant BCS-9987590 (to S.A.T. and B.C.V.).
Electronic-Database Information
The URLs for data presented herein are as follows:
- Hudson Lab at the University of Chicago, http://home.uchicago.edu/~rhudson1/ (for the ms program)
- McVean Lab at the University of Oxford, http://www.stats.ox.ac.uk/~mcvean/ (for the LDhat program)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for OPN1LW) [PubMed]
- Rozas Lab at the Universitat de Barcelona, http://www.ub.es/molevol/julio/ (for the DnaSP program)
- Stephens Lab at the University of Washington, http://www.stat.washington.edu/stephens/ (for the hotspotter program)
- Tishkoff Lab at the University of Maryland, http://www.life.umd.edu/biology/tishkofflab/ (for OPN1LW primers)
References
- Akashi H (1999) Inferring the fitness effects of DNA mutations from polymorphism and divergence data: statistical power to detect directional selection under stationarity and free recombination. Genetics 151:221–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi H, Schaeffer SW (1997) Natural selection and the frequency distributions of “silent” DNA polymorphism in Drosophila. Genetics 146:295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie K, Liu-Cordero SN, Eberle MA, Daly M, Barrett J, Winchester E, Lander ES, Kruglyak L (2001) Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am J Hum Genet 69:582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo AB, Rim J, Oprian DD (1994) Molecular determinants of human red/green color discrimination. Neuron 12:1131–1138 10.1016/0896-6273(94)90320-4 [DOI] [PubMed] [Google Scholar]
- Boissinot S, Tan Y, Shyue SK, Schneider H, Sampaio I, Neiswanger K, Hewett-Emmett D, Li WH (1998) Origins and antiquity of X-linked triallelic color vision systems in New World monkeys. Proc Natl Acad Sci USA 95:13749–13754 10.1073/pnas.95.23.13749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J, Neitz J, Neitz M (2002) Estimates of L:M cone ratio from ERG flicker photometry and genetics. J Vis 2:531–542 [DOI] [PubMed] [Google Scholar]
- Clark AG (2003) Finding genes underlying risk of complex disease by linkage disequilibrium mapping. Curr Opin Genet Dev 13:296–302 10.1016/S0959-437X(03)00056-X [DOI] [PubMed] [Google Scholar]
- Crawford DC, Bhangale T, Li N, Hellenthal G, Rieder MJ, Nickerson DA, Stephens M (2004) Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet 36:700–706 15184900 [DOI] [PubMed] [Google Scholar]
- Deeb SS, Jorgensen AL, Battisti L, Iwasaki L, Motulsky AG (1994) Sequence divergence of the red and green visual pigments in great apes and humans. Proc Natl Acad Sci USA 91:7262–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb SS, Lindsey DT, Hibiya Y, Sanocki E, Winderickx J, Teller DY, Motulsky AG (1992) Genotype-phenotype relationships in human red/green color-vision defects: molecular and psychophysical studies. Am J Hum Genet 51:687–700 [PMC free article] [PubMed] [Google Scholar]
- Dominy NJ, Lucas PW (2001) Ecological importance of trichromatic vision to primates. Nature 410:363–366 10.1038/35066567 [DOI] [PubMed] [Google Scholar]
- Fay JC, Wyckoff GJ, Wu C-I (2001) Positive and negative selection on the human genome. Genetics 158:1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisse L, Hudson RR, Bartoszewicz A, Wall JD, Donfack J, Di Rienzo A (2001) Gene conversion and different population histories may explain the contrast between polymorphism and linkage disequilibrium levels. Am J Hum Genet 69:831–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, Hedrick PW (2003) Perspective: detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57:1707–1722 [DOI] [PubMed] [Google Scholar]
- Hayashi T, Motulsky AG, Deeb SS (1999) Position of a ‘green-red’ hybrid gene in the visual pigment array determines colour-vision phenotype. Nat Genet 22:90–93 10.1038/8798 [DOI] [PubMed] [Google Scholar]
- Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155:2011–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2002) Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18:337–338 10.1093/bioinformatics/18.2.337 [DOI] [PubMed] [Google Scholar]
- ——— (2001) Two-locus sampling distributions and their application. Genetics 159:1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H (2002) A method for estimating the mutation, gene conversion and recombination parameters in small multigene families. Genetics 161:865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2003a) A two-locus gene conversion model with selection and its application to the human RHCE and RHD genes. Proc Natl Acad Sci USA 100:8793–8798 10.1073/pnas.1031592100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2003b) The coalescent and infinite-site model of a small multigene family. Genetics 163:803–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson KA, Highnote SM, Wasserman LM (2001) Richer color experience in observers with multiple photopigment opsin genes. Psychon Bull Rev 8:244–261 [DOI] [PubMed] [Google Scholar]
- Jorgensen AL, Deeb SS, Motulsky AG (1990) Molecular genetics of X chromosome-linked color vision among populations of African and Japanese ancestry: high frequency of a shortened red pigment gene among Afro-Americans. Proc Natl Acad Sci USA 87:6512–6516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M (2003) Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165:2213–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas PW, Dominy NJ, Riba-Hernandez P, Stoner KE, Yamashita N, Loria-Calderon E, Petersen-Pereira W, Rojas-Duran Y, Salas-Pena R, Solis-Madrigal S, Osorio D, Darvell BW (2003) Evolution and function of routine trichromatic vision in primates. Evolution 57:2636–2643 [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M (1991) Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654 10.1038/351652a0 [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P (2002) A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160:1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merbs SL, Nathans J (1992) Absorption spectra of human cone pigments. Nature 356:433–435 10.1038/356433a0 [DOI] [PubMed] [Google Scholar]
- Mollon JD, Bowmaker JK, Jacobs GH (1984) Variations of color-vision in a New World primate can be explained by polymorphism of retinal photopigments. Proc R Soc Lond B Biol Sci 222:373–399 [DOI] [PubMed] [Google Scholar]
- Nachman MW (2001) Single nucleotide polymorphisms and recombination rate in humans. Trends Genet 17:481–485 10.1016/S0168-9525(01)02409-X [DOI] [PubMed] [Google Scholar]
- Nachman MW, Brown WM, Stoneking M, Aquadro CF (1996) Nonneutral mitochondrial DNA variation in humans and chimpanzees. Genetics 142:953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans J, Thomas D, Hogness DS (1986) Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science 232:193–202 [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton NH (2002) The effects of multilocus balancing selection on neutral variability. Genetics 161:849–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neitz J, Neitz M, He JC, Shevell SK (1999) Trichromatic color vision with only two spectrally distinct photopigments. Nat Neurosci 2:884–888 10.1038/13185 [DOI] [PubMed] [Google Scholar]
- Nielsen R, Weinreich DM (1999) The age of nonsynonymous and synonymous mutations in animal mtDNA and implications for the mildly deleterious theory. Genetics 153:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T (1992) The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst 23:263–286 10.1146/annurev.es.23.110192.001403 [DOI] [Google Scholar]
- Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2:360–369 10.1038/35072078 [DOI] [PubMed] [Google Scholar]
- Przeworski M, Hudson RR, Di Rienzo A (2000) Adjusting the focus on human variation. Trends Genet 16:296–302 10.1016/S0168-9525(00)02030-8 [DOI] [PubMed] [Google Scholar]
- Przeworski M, Wall JD (2001) Why is there so little intragenic linkage disequilibrium in humans? Genet Res 77:143–151 10.1017/S0016672301004967 [DOI] [PubMed] [Google Scholar]
- Ptak SE, Voelpel K, Przeworski M (2004) Insights into recombination from patterns of linkage disequilibrium in humans. Genetics 167:387–397 15166163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan BC, Julliot C, Simmen B, Vienot F, Charles-Dominique P, Mollon JD (2001) Fruits, foliage and the evolution of primate colour vision. Philos Trans R Soc Lond Biol Sci 356:229–283 11316480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Schaffner SF, Daly MJ, McVean G, Mullikin JC, Higgins JM, Richter DJ, Lander ES, Altshuler D (2002) Human genome sequence variation and the influence of gene history, mutation and recombination. Nat Genet 32:135–142 10.1038/ng947 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 10.1093/bioinformatics/15.2.174 [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837 10.1038/nature01140 [DOI] [PubMed] [Google Scholar]
- Saunders MA, Hammer MF, Nachman MW (2002) Nucleotide variability at G6PD and the signature of malarial selection in humans. Genetics 162:1849–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe LT, Stockman A, Jagle H, Knau H, Klausen G, Reitner A, Nathans J (1998) Red, green, and red-green hybrid pigments in the human retina: correlations between deduced protein sequences and psychophysically measured spectral sensitivities. J Neurosci 18:10053–10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe LT, Stockman A, Jagle H, Nathans J (1999) Opsin genes, cone photopigments, color vision, and color blindness. In: Gegenfurtner KR, Sharpe LT (eds) Color vision: from genes to perception. Cambridge University Press, New York, pp 3–51 [Google Scholar]
- Shyue SK, Li L, Chang BH, Li WH (1994) Intronic gene conversion in the evolution of human X-linked color vision genes. Mol Biol Evol 11:548–551 [DOI] [PubMed] [Google Scholar]
- Sjoberg SA, Neitz M, Balding SD, Neitz J (1998) L-cone pigment genes expressed in normal colour vision. Vision Res 38:3213–3219 10.1016/S0042-6989(97)00367-2 [DOI] [PubMed] [Google Scholar]
- Slatkin M (2000) Balancing selection at closely linked, overdominant loci in a finite population. Genetics 154:1367–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood PM, Olveczky BP, Williams GL, Jacobs GH, Reese BE, Meister M, Nathans J (2003) Genetically engineered mice with an additional class of cone photoreceptors: implications for the evolution of color vision. Proc Natl Acad Sci USA 100:11706–11711 10.1073/pnas.1934712100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Kumar S (2003) Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res 13:838–844 10.1101/gr.1152803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surridge AK, Osorio D, Mundy NI (2003) Evolution and selection of trichromatic vision in primates. Trends Ecol Evol 18:198–205 10.1016/S0169-5347(03)00012-0 [DOI] [Google Scholar]
- Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Satta Y, Klein J (1992) Polymorphism and balancing selection at major histocompatibility complex loci. Genetics 130:925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, Piro A, Stoneking M, Tagarelli A, Tagarelli G, Touma EH, Williams SM, Clark AG (2001) Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science 293:455–462 10.1126/science.1061573 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Verrelli BC (2003a) Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet 4:293–340 10.1146/annurev.genom.4.070802.110226 [DOI] [PubMed] [Google Scholar]
- ——— (2003b) Role of evolutionary history on haplotype block structure in the human genome: implications for disease mapping. Curr Opin Genet Dev 13:569–575 10.1016/j.gde.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM (2002) Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 3:611–621 [DOI] [PubMed] [Google Scholar]
- Verrelli BC, McDonald JH, Argyropoulos G, Destro-Bisol G, Froment A, Drousiotou A, Lefranc G, Helal AN, Loiselet J, Tishkoff SA (2002) Evidence for balancing selection from nucleotide sequence analyses of human G6PD. Am J Hum Genet 71:1112–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD (2004) Close look at gene conversion hot spots. Nat Genet 36:114–115 10.1038/ng0204-114 [DOI] [PubMed] [Google Scholar]
- Wall JD, Frisse LA, Hudson RR, Di Rienzo A (2003) Comparative linkage-disequilibrium analysis of the β-globin hotspot in primates. Am J Hum Genet 73:1330–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Pritchard JK (2003) Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet 4:587–597 10.1038/nrg1123 [DOI] [PubMed] [Google Scholar]
- Watterson GA (1975) On the number of segregating sites in genetic models without recombination. Theor Popul Biol 7:256–276 [DOI] [PubMed] [Google Scholar]
- Winderickx J, Battisti L, Hibiya Y, Motulsky AG, Deeb SS (1993) Haplotype diversity in the human red and green opsin genes: evidence for frequent sequence exchange in exon 3. Hum Mol Genet 2:1413–1421 [DOI] [PubMed] [Google Scholar]
- Winderickx J, Lindsey DT, Sanocki E, Teller DY, Motulsky AG, Deeb SS (1992) Polymorphism in red photopigment underlies variation in colour matching. Nature 356:431–433 10.1038/356431a0 [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim U-k, Bamshad MJ, Larsen J, Jorde LB, Drayna D (2004) Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet 74:637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB (2001) The molecular genetics and evolution of red and green color vision in vertebrates. Genetics 158:1697–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YH, Li WH (1996) Gene conversion and natural selection in the evolution of X-linked color vision genes in higher primates. Mol Biol Evol 13:780–783 [DOI] [PubMed] [Google Scholar]