Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that can play crucial regulatory roles in eukaryotes by targeting mRNAs for silencing. To test whether miRNAs play roles in the regulation of wood development in tree species, we isolated small RNAs from the developing xylem of Populus trichocarpa stems and cloned 22 miRNAs. They are the founding members of 21 miRNA gene families for 48 miRNA sequences, represented by 98 loci in the Populus genome. A majority of these miRNAs were predicted to target developmental- and stress/defense-related genes and possible functions associated with the biosynthesis of cell wall metabolites. Of the 21 P. trichocarpa miRNA families, 11 have sequence conservation in Arabidopsis thaliana but exhibited species-specific developmental expression patterns, suggesting that even conserved miRNAs may have different regulatory roles in different species. Most unexpectedly, the remaining 10 miRNAs, for which 17 predicted targets were experimentally validated in vivo, are absent from the Arabidopsis genome, suggesting possible roles in tree-specific processes. In fact, the expression of a majority of the cloned miRNAs was upregulated or downregulated in woody stems in a manner consistent with tree-specific corrective growth against tension and compression stresses, two constant mechanical loads in trees. Our results show that plant miRNAs can be induced by mechanical stress and may function in one of the most critical defense systems for structural and mechanical fitness.
INTRODUCTION
MicroRNAs (miRNAs) are a class of gene products of ∼21 nucleotides in length that are derived from primary miRNAs transcribed from miRNA loci (Lee et al., 2002). In plants, the primary miRNAs are cleaved by Dicer-like 1 (DCL1) (Xie et al., 2004) and possibly other proteins in the nucleus to produce an ∼60- to 300-nucleotide hairpin structure known as the miRNA precursor. The miRNA precursor may be transported out of the nucleus by HASTY or retained in the nucleus and further processed by DCL1 to release the stem portion of the hairpin as an miRNA:miRNA* duplex (for reviews, see Bartel, 2004; Kidner and Martienssen, 2005). The duplex, which comprises a mature miRNA of ∼21 nucleotides and a similarly sized miRNA* fragment on the opposing arm of the miRNA precursor, is then presumably unwound by a helicase, releasing the single-stranded mature miRNA. The miRNA then enters the RNA-induced silencing complex (for review, see Bartel, 2004) and guides the complex to identify target messages for posttranscriptional gene silencing through direct target cleavage (Llave et al., 2002a; Rhoades et al., 2002; Emery et al., 2003; Kasschau et al., 2003; Palatnik et al., 2003; Tang et al., 2003; Achard et al., 2004; Juarez et al., 2004; Kidner and Martienssen, 2004; Laufs et al., 2004; Mallory et al., 2004a, 2004b; McHale and Koning, 2004; Baker et al., 2005) or, in a few cases, for translational repression (Aukerman and Sakai, 2003; Chen, 2004).
So far, a large number of miRNAs have been found in various plant species (Llave et al., 2002b; Park et al., 2002; Reinhart et al., 2002; Palatnik et al., 2003; Bonnet et al., 2004; Floyd and Bowman, 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004a, 2004b; Adai et al., 2005; Bedell et al., 2005). The original efforts, focusing on molecular cloning supplemented with computation, resulted in the identification of >90 Arabidopsis thaliana miRNAs, which were systematically classified into 33 miRNA gene families (Reinhart et al., 2002; Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). However, the argument that cloning may exclude underexpressed or tissue- or development-specific miRNAs has shifted identification efforts toward computational analyses, which have resulted in a considerably larger number of identified Arabidopsis miRNAs (Bonnet et al., 2004; Wang et al., 2004b; Adai et al., 2005). Interestingly, using the cloning approach, Sunkar and Zhu (2004) identified a significant number of miRNAs from Arabidopsis grown under abiotic stress conditions that have been missed by computation. A majority of these abiotic stress-responsive miRNAs are not conserved in rice (Oryza sativa), suggesting the existence of other nonconserved miRNAs and that nonconserved miRNAs are critical to regulation associated with specific growth and developmental processes.
As for predicting miRNA targets, computation has been the most effective approach for plant species (Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004). Approximately 120 targets were predicted for the 33 classified Arabidopsis miRNA families (Reinhart et al., 2002; Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). Of the predicted targets of the first 14 Arabidopsis miRNA families, 70% are transcription factor genes (Rhoades et al., 2002), representing one of the most important discoveries associated with plant miRNA function. These transcription factor genes include those encoding SQUAMOSA PROMOTER BINDING PROTEIN-LIKE, MYB, NAC, HD-Zip, and SCARECROW-LIKE proteins that have known or putative roles in plant development. However, as more nonconserved miRNAs are being identified, the diversity of target genes also expands beyond the dominating transcription factor genes to include classes associated with other metabolic and cellular processes (Eckardt, 2004; Sunkar and Zhu, 2004). The large number of nonconserved miRNAs from Arabidopsis under abiotic stress (Sunkar and Zhu, 2004) is an important discovery providing direct evidence for the involvement of miRNAs in specialized plant processes. This observation lends support to the notion that miRNA networks may also modulate species-specific processes such as wood formation in trees.
Long-term deposition in trees of secondary xylem, or wood, provides strength to support the tremendous crown structures. In addition, trees constantly develop specialized woody tissues, termed “reaction wood,” to correct inclined branch and stem growth as a result of mechanical stimuli/loads generated by, for example, wind and gravity (Sinnott, 1952; Barnett, 1981; Timell, 1986). This corrective growth is a result of coordinately enhanced development of strength-contributing cells and wall metabolites (Wardrop and Davies, 1964; Scurfield, 1973; Barnett, 1981; Timell, 1986) and is one of the most critical defense systems in the long-term growth of woody species. However, little is known about the underlying genetic regulation of this remarkable defense system or of wood formation in general. To gain a better understanding of these tree-specific processes that likely require the coordinated regulation of many genes, we investigated the miRNA networks in stem xylem tissues of the Nisqually-1 clone of Populus trichocarpa.
RESULTS
Cloning of miRNAs from Developing Secondary Xylem of P. trichocarpa Stems
Small RNAs with sizes of 16 to 36 nucleotides were gel purified from total RNA isolated from developing secondary xylem of P. trichocarpa stems. RNAs were end modified and ligated with adaptors for unidirectional polarity, RT-PCR amplified, concatamerized, and cloned into a library as described by Lau et al. (2001), Lagos-Quintana et al. (2002), and Elbashir et al. (2001). A total of 379 individual clones were isolated and sequenced, yielding 898 sequences. BLAST analyses against the GenBank and the P. trichocarpa genome databases showed that 809 of these sequences, which were not further analyzed, correspond to known noncoding rRNAs, tRNAs, and small nuclear RNAs and to those associated with retrotransposons or transposons. The 89 remaining sequences, of which 30 are located in various protein-coding genes and 59 in the intergenic regions, were subjected to detailed characterization. The Populus genome fragments (∼600 nucleotides) surrounding these small RNA sequences were used to predict the secondary structure using the mfold program (Zuker, 2003). No hairpin structure could be identified for 66 of the 89 RNAs analyzed, and as such, these 66 small RNAs could not be considered as miRNAs. They could, however, be produced after the cleavage of double-stranded RNAs following processes similar to the biogenesis of small interfering RNAs (siRNAs) (Bartel, 2004) and therefore were considered as endogenous putative siRNAs (see Supplemental Table 1 online).
The remaining 23 unique sequences were found capable of forming stable stem-loop structures in their precursor sequences (Figure 1; see Supplemental Table 2 online). One of them is miR160*, which we do not consider a miRNA. The other 22 are likely miRNAs (denoted as ptr-miRNAs, Tables 1and 2), of which 11 (belonging to 10 families) are either identical or highly similar in sequence to the reported Arabidopsis miRNAs (Park et al., 2002; Reinhart et al., 2002; Rhoades et al., 2002; Palatnik et al., 2003; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). The other 11 are novel miRNAs.
Figure 1.
Predicted Stem-Loop Structures of Precursors Containing the Newly Cloned miRNA Sequence (Red).
Table 1.
Conserved miRNAs between Populus and Arabidopsis
| miRNA Gene | miRNA Sequence (5′→3′) | Arm | Length (nt) | Expression | Populus EST |
|---|---|---|---|---|---|
| UUGACAGAAGAUAGAGAGCAC (1) | 21 | N | |||
| ptr-MIR156d | UUGACAGAAGAGAGUGAGCAC | 5′ | |||
| ptr-MIR156e | UUGACAGAAGAGAGUGAGCAC | 5′ or 3′ | |||
| ptr-MIR156g | UUGACAGAAGAUAGAGAGCAC | 5′ | |||
| ptr-MIR156h | UUGACAGAAGAUAGAGAGCAC | 5′ | |||
| ptr-MIR156i | UUGACAGAAGAUAGAGAGCAC | 5′ | |||
| ptr-MIR156j | UUGACAGAAGAUAGAGAGCAC | 5′ | |||
| UUUGGAUUGAAGGGAGCUCUA (5) | 20–21 | N | Yes | ||
| ptr-MIR159a | UUUGGAUUGAAGGGAGCUCUA | 3′ | |||
| ptr-MIR159b | UUUGGAUUGAAGGGAGCUCUA | 3′ | |||
| ptr-MIR159c | UUUGGAUUGAAGGGAGCUCUA | 3′ | |||
| UGCCUGGCUCCCUGUAUGCCA (3) | 21 | N | |||
| ptr-MIR160a | UGCCUGGCUCCCUGUAUGCCA | 5′ | |||
| ptr-MIR160b | UGCCUGGCUCCCUGUAUGCCA | 5′ | |||
| ptr-MIR160c | UGCCUGGCUCCCUGUAUGCCA | 5′ | |||
| ptr-MIR160d | UGCCUGGCUCCCUGUAUGCCA | 5′ | |||
| ptr-MIR160e | UGCCUGGCUCCCUGAAUGCCA | 5′ | |||
| ptr-MIR160f | UGCCUGGCUCCCUGAAUGCCA | 5′ | |||
| ptr-MIR160g | UGCCUGGCUCCCUGGAUGCCA | 5′ | |||
| ptr-MIR160h | UGCCUGGCUCCCUGCAUGCCA | 5′ | |||
| UCGAUAAACCUCUGCAUCCAG (2) | 21 | N | |||
| ptr-MIR162a | UCGAUAAACCUCUGCAUCCAG | 3′ | |||
| ptr-MIR162b | UCGAUAAACCUCUGCAUCCAG | 3′ | |||
| ptr-MIR162c | UCGAUAAACCUCUGCAUCCAG | 3′ | |||
| UGGAGAAGCAGGGCACGUGCA (3) | 21 | N | Yes | ||
| ptr-MIR164a | UGGAGAAGCAGGGCACGUGCA | 5′ | |||
| ptr-MIR164b | UGGAGAAGCAGGGCACGUGCA | 5′ or 3′ | |||
| ptr-MIR164c | UGGAGAAGCAGGGCACGUGCA | 5′ | |||
| ptr-MIR164d | UGGAGAAGCAGGGCACGUGCA | 5′ | |||
| ptr-MIR164e | UGGAGAAGCAGGGCACGUGCA | 5′ or 3′ | |||
| ptr-MIR164f | UGGAGAAGCAGGGCACAUGCU | 5′ | |||
| UCGCUUGGUGCAGGUCGGGAA (3) | 21 | N | Yes | ||
| ptr-MIR168a | UCGCUUGGUGCAGGUCGGGAA | 5′ | |||
| ptr-MIR168b | UCGCUUGGUGCAGGUCGGGAA | 5′ | |||
| ptr-MIR168c | UCGAUUGGUGCAGGCCGGGAA | 5′ | |||
| UGAUUGAGCCGUGCCAAUAUC (1) | 21 | N | |||
| ptr-MIR171a | UGAUUGAGCCGUGCCAAUAUC | 3′ | |||
| ptr-MIR171b | UGAUUGAGCCGUGCCAAUAUC | 3′ | |||
| ptr-MIR171c | AGAUUGAGCCGCGCCAAUAUC | 3′ | |||
| ptr-MIR171d | AGAUUGAGCCGCGCCAAUAUC | 3′ | |||
| ptr-MIR171e | UGAUUGAGCCGUGCCAAUAUC | 3′ | |||
| ptr-MIR171f | UGAUUGAGCCGUGCCAAUAUC | 3′ | |||
| ptr-MIR171g | UGAUUGAGCCGUGCCAAUAUC | 3′ | |||
| ptr-MIR171h | UGAUUGAGCCGUGCCAAUAUC | 3′ | |||
| ptr-MIR171i | UGAUUGAGCCGUGCCAAUAUC | 3′ | |||
| GGAAUCUUGAUGAUGCUGCAGU (1) | 22 | N | |||
| ptr-MIR172d | GGAAUCUUGAUGAUGCUGCAUC | 3′ | |||
| ptr-MIR172e | GGAAUCUUGAUGAUGCUGCAUC | 3′ | |||
| ptr-MIR172g | GGAAUCUUGAUGAUGCUGCAGC | 3′ | |||
| ptr-MIR172h | GGAAUCUUGAUGAUGCUGCAGC | 3′ | |||
| UUGGACUGAAGGGAGCUCCC (1) | 20 | N | yes | ||
| UUGGACUGAAGGGAGCUCCUUU (2) | 22 | ||||
| ptr-MIR319a | UUGGACUGAAGGGAGCUCCC | 3′ | |||
| ptr-MIR319b | UUGGACUGAAGGGAGCUCCC | 3′ | |||
| ptr-MIR319c | UUGGACUGAAGGGAGCUCCC | 3′ | |||
| ptr-MIR319d | UUGGACUGAAGGGAGCUCCC | 3′ | |||
| ptr-MIR319e | UUGGACUGAAGGGAGCUCCU | 3′ | |||
| ptr-MIR319f | UUGGACUGAAGGGAGCUCCU | 3′ | |||
| ptr-MIR319g | UUGGACUGAAGGGAGCUCCU | 3′ | |||
| ptr-MIR319h | UUGGACUGAAGGGAGCUCCU | 3′ | |||
| ptr-MIR319i | UUGGGCUGAAGGGAGCUCCC | 3′ | |||
| AUGCACUGCCUCUUCCCUGGC (3) | 20–21 | N | Yes | ||
| ptr-MIR408 | AUGCACUGCCUCUUCCCUGGC | 3′ | |||
| UUUUCCCUACUCCACCCAUCCC (3) | 22 | N | Yes | ||
| ptr-MIR472a | UUUUCCCUACUCCACCCAUCCC | 3′ | |||
| ptr-MIR472b | UUUUCCCAACUCCACCCAUCCC | 3′ |
The cloned small RNA sequences are shown in bold. Only the longest small RNA sequence in each family is shown. The frequency of cloning is indicated in parentheses after the small RNA sequence. Small RNA location in the predicted hairpin structure is specified (5′ or 3′ arm). “N” indicates that the small RNA was detected by RNA gel blotting. “Yes” indicates that precursors containing the cloned small RNA sequence have been found in the Populus EST database. nt, nucleotides.
Table 2.
Nonconserved miRNAs between Populus and Arabidopsis
| miRNA Gene | miRNA Sequence (5′→3′) | Arm | Length (nt) | Expression | Populus EST |
|---|---|---|---|---|---|
| ACUCUCCCUCAAGGCUUCCA (1) | |||||
| ptr-MIR473a | ACUCUCCCUCAAGGCUUCCA | 5′ or 3′ | 20 | N | |
| ptr-MIR473b | GCUCUCCCUCAGGGCUUCCA | 5′ | |||
| CAAAAGUUGCUGGGUUUGGCUGGG (1) | 24 | PCR | |||
| ptr-MIR474a | CAAAAGUUGCUGGGUUUGGCUGGG | 5′ | |||
| ptr-MIR474b | CAAAAGUUGUUGGGUUUGGCUGGG | 5′ | |||
| ptr-MIR474c | CAAAAGCUGUUGGGUUUGGCUGGG | 3′ | |||
| UUACAGUGCCCAUUGAUUAAG (10) | 21 | N | Yes | ||
| ptr-MIR475a | UUACAGUGCCCAUUGAUUAAG | 3′ | |||
| ptr-MIR475b | UUACAGUGCCCAUUGAUUAAG | 3′ | |||
| ptr-MIR475c | UUACAAUGUCCAUUGAUUAAG | 3′ | |||
| ptr-MIR475d | UUACAGAGUCCAUUGAUUAAG | 3′ | |||
| UAGUAAUCCUUCUUUGCAAAG (1) | 21 | N | |||
| ptr-MIR476a | UAGUAAUCCUUCUUUGCAAAG | 5′ | |||
| ptr-MIR476b | UAGUAAUUCUUCUUUGCAAAA | 5′ | |||
| ptr-MIR476c | UAGUAAUUCUUCUUUGCAAAA | 5′ | |||
| AUCUCCCUCAGAGGCUUCCAA (1) | 21 | N | |||
| ptr-MIR477a | AUCUCCCUCAGAGGCUUCCAA | 5′ | |||
| ptr-MIR477b | AUCUCCCUCAGAGGCUUCCAA | 5′ | |||
| UGACGUGUCUUCUAUUUUUAGGGA (1) | 24 | PCR | |||
| ptr-MIR478a | UGACGUGUCUUCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478b | UGACGUGUCUUCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478c | UGACGUGUCUUCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478d | UGACGUGUCUUCUAUUUUUAGGAA | 3′ | |||
| ptr-MIR478e | UGACGAGUCUUCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478f | UGACAUGUCUUCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478g | UGACAUGUCUUCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478h | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478i | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478j | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478k | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478l | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478m | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478n | UAACGUGUCUCCUAUUUUAGGGA | 3′ | |||
| ptr-MIR478o | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478p | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478q | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478r | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478s | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478t | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| ptr-MIR478u | UAACGUGUCUCCUAUUUUUAGGGA | 3′ | |||
| UGUGAUAUUGGUCCGGCUCAUC (2) | 22 | N | |||
| ptr-MIR479 | UGUGAUAUUGGUCCGGCUCAUC | 5′ | |||
| ACUACUACAUCAUUGACGUUGAAC (1) | 24 | PCR | |||
| ptr-MIR480a | ACUACUACAUCAUUGACGUUGAAC | 3′ | |||
| ptr-MIR480b | ACUACUACAUCAUUGAUGUUGAAC | 3′ | |||
| AAGACCUCACUUAACAGCUUAAGC (1) | 24 | PCR | |||
| ptr-MIR481a | AGGACCUCACUUAACAGCUUAAGC | 3′ | |||
| ptr-MIR481b | AGGACCUCACUUAACAGCUUAAGC | 5′ | |||
| ptr-MIR481c | AGGACCUCACUUAACAGCUUAAGC | 5′ | |||
| ptr-MIR481d | AAGACCUCACCUAACAGCUUAAGC | 3′ | |||
| ptr-MIR481e | AGGACCUCACCUAACAGCUUAAGC | 5′ | |||
| CCUACUCCUCCCAUUCC (1) | 17 | PCR | |||
| ptr-MIR482 | CCUACUCCUCCCAUUCC | 3′ |
The cloned small RNA sequences are shown in bold. Only the longest small RNA sequence in each family is shown. The frequency of cloning is indicated in parentheses after the small RNA sequence. Small RNA location in the predicted hairpin structure is specified (5′ or 3′ arm). Expression of the small RNAs was detected by RNA gel blot hybridization (N) or real-time PCR. “Yes” indicates that precursors containing the cloned small RNA sequence have been found in the Populus EST database. ptr-miR473a, ptr-miR478a, and ptr-miR482 miRNA sequences are conserved between rice and Populus. nt, nucleotides.
Conserved miRNAs and P. trichocarpa miRNAs That Are Absent from Arabidopsis
Next, we analyzed whether these 11 novel miRNAs are also in the Arabidopsis genome. By allowing zero to three nucleotide substitutions, the sequences of these 11 ptr-miRNAs were searched against the Arabidopsis genome to identify their Arabidopsis homologs and the surrounding genomic sequences. Analysis of the ptr-miRNA sequence-containing loci in Arabidopsis by the mfold program resulted in the identification of only one distinct hairpin precursor structure (see Supplemental Table 2 online), which harbors three mismatched ptr-miR472a sequences (Table 1). Therefore, together with the ptr-miR472a, we identified 12 miRNAs (belonging to 11 families) conserved between Populus and Arabidopsis.
Using the hairpin precursor structure as a criterion, we found that, under the search algorithm with the highest (0) mismatch stringency, the remaining 10 of the 22 cloned Populus miRNAs could not be identified in the Arabidopsis genome (Table 2). This cannot be attributed to an incomplete Arabidopsis genome sequence but was thought to be the result of the high search stringency. Indeed, a majority of the known Arabidopsis and rice miRNA homologs exhibit zero to two nucleotide mismatches, and normally a nucleotide mismatch of two is the score cutoff applied to computational identification of conserved miRNA sequences across species (Jones-Rhoades and Bartel, 2004). However, lowering the search stringency progressively to allow up to three mismatches relative to the Populus miRNA sequences also led to the same conclusion that homologs to these 10 ptr-miRNAs are absent from Arabidopsis. Taken together, 12 of the 22 miRNAs cloned in this study are conserved between Arabidopsis and Populus, and the other 10 appear to be missing from Arabidopsis. Using the same algorithm, we then searched the rice genome (O. sativa ssp japonica cv Nipponbare; http://www.tigr.org/tdb/e2k1/osa1/index.shtml) and found that, among the 10 ptr-miRNAs that are absent from Arabidopsis, only three (ptr-miR473a, ptr-miR478a, and ptr-miR482) are conserved between rice and Populus (see Supplemental Table 2 online).
To look for further evidence of these 22 cloned miRNAs, we searched the Populus ESTs (http://www.ncbi.nlm.nih.gov/dbEST) for expressed sequences that would match these miRNAs. The resulting ESTs were then analyzed for their secondary structures using the mfold program. This search identified 11 ESTs (Tables 1 and 2; see Supplemental Table 3 online) that have the hairpin precursor structures. These precursors contain distinct miRNA sequences that are identical to 8 of the 22 cloned ptr-miRNAs, indicating that, in the context of expressed miRNA precursors, a significant portion of the cloned miRNAs could be considered as miRNAs. Such a verification frequency is reasonable, considering the limitations both in quantity and in sequence attributes of the available ESTs.
Identification and Sequence Analysis of ptr-miRNA Homologs and Their Gene Families
We further searched the Populus genome using PatScan for sequences similar to the cloned 22 miRNA sequences (one nucleotide substitution for ptr-miR482 and up to two nucleotides for the others), which resulted in the identification of additional candidate miRNAs. mfold analysis of the ∼600-nucleotide Populus genomic sequences centered on each of these candidates for secondary structure prediction identified 26 additional miRNA sequences, raising the number of identified ptr-miRNA sequences to 48 (Tables 1 and 2). These 48 sequences, representing members of 21 miRNA families, map to 98 genomic loci in the Populus genome (Tables 1 and 2; see Supplemental Table 2 online). Of the 21 miRNA gene families (denoted as ptr-MIRs), three are each encoded by a single copy gene, whereas each of the other 18 correspond to multiple (2 to 21) genomic loci.
In each of the ptr-MIR162 and ptr-MIR476 gene families, an identical precursor sequence is derived from two distinct loci (Tables 1 and 2; see Supplemental Table 2 online). Twenty-two distinct genomic loci were identified for the ptr-MIR478 family, of which the members ptr-MIR478i, ptr-MIR478j, ptr-MIR478k, ptr-MIR478l, ptr-MIR478m, and ptr-MIR478n are identical in sequence, as are ptr-MIR478s and ptr-MIR478t (Table 2; see Supplemental Table 2 online). However, perfect genomic matches to the cloned ptr-miR172, ptr-miR481, and one of the two ptr-miR319 sequences could not be identified in the Populus genome. Perhaps some unknown cloning artifacts have caused sequence modifications in these miRNAs. miRNA sequence alterations originating from nuclear editing of the corresponding fold-back precursors are also a possibility, as observed for human and mouse miRNA22 (Bass, 2002; Luciano et al., 2004). Nevertheless, multiple genomic matches to family members with one to two nucleotide substitutions relative to ptr-miR172, ptr-miR319, and ptr-miR481 were successfully identified (Table 1).
We also found that, in most cases, the ptr-miRNA having multiple genomic loci is present on the same arm of its fold-back precursors, as reported previously (Reinhart et al., 2002; Jones-Rhoades and Bartel, 2004; Wang et al., 2004a). However, we observed exceptions in P. trichocarpa, with a mature miRNA having multiple genomic loci on either the 5′ arm or the 3′ arm, depending upon the genomic association of the mature miRNA with its miRNA* fragments and the transcription process for miRNA precursor production. For instance, the ptr-miR156e sequence is flanked in the Populus genome by two upstream miRNA* fragments and one downstream miRNA* fragment (Table 1; see Supplemental Figure 1 and Table 2 online), each of which can pair with ptr-miR156e for a stable fold-back structure. Thus, three potential hairpin precursors might result—with the ptr-miR156e sequence residing in the 5′ arm of one of the precursors (see Supplemental Figure 1A online) or in the 3′ arm of either one of the other two (see Supplemental Figures 1B and 1C online). Similarly, the miRNA sequence in ptr-MIR473a (Figure 1, Table 2), ptr-MIR164b, or ptr-MIR164e (Table 1) is enclosed by two miRNA* fragments, allowing the production of either a 5′ arm or a 3′ arm miRNA-bearing precursor. Searching the genome sequences flanking these ptr-miRNAs confirmed that they are not siRNAs with inverted repeat sequences. Because the transcription mechanism for processing miRNA genes is unknown, the exact miRNA position in these ptr-miRNA precursors cannot be certain.
Developmental and Mechanical Stress–Responsive Expression of Populus miRNAs
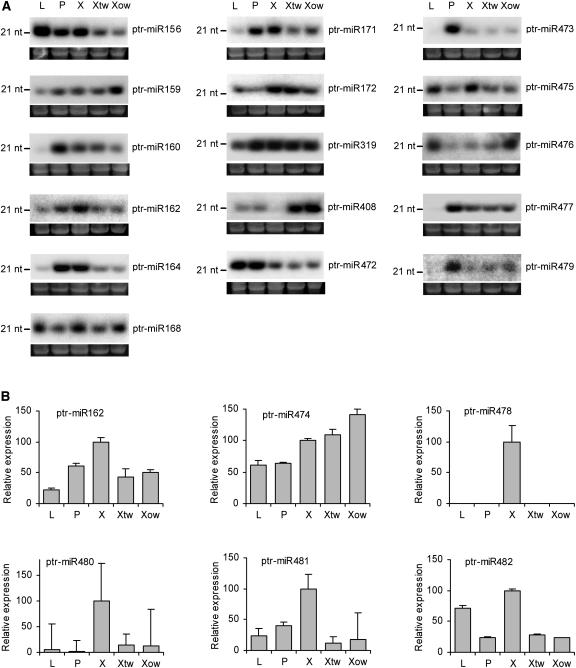
To predict the functional relevance of miRNAs to development in P. trichocarpa, we examined the ptr-miRNA transcript levels in leaves, phloem, and developing xylem by RNA gel blot analysis of the total RNA samples isolated from these tissues. Hybridization probes complementary to the cloned ptr-miRNA sequences were designed, and these probes detected 20- to 22-nucleotide RNA signals of 16 ptr-miRNAs in at least one of the tested tissue types (Figure 2A). Because members within each miRNA family share high sequence homology, the hybridization signals shown probably reflect the transcript levels of the family rather than a single member. Five ptr-miRNAs could not be detected by RNA gel blot analysis using the conditions described. Failure to detect the transcripts of cloned or computationally identified miRNAs by RNA gel blotting has been reported in previous studies (Park et al., 2002; Reinhart et al., 2002; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004a, 2004b; Adai et al., 2005). Instead, miRNAs with expression below the RNA gel blot analysis detection level were validated by PCR in Caenorhabditis elegans (Lau et al., 2001; Grad et al., 2003; Lim et al., 2003) and in plants (Jones-Rhoades and Bartel, 2004). Following the amplification principles used in these validations, we applied real-time PCR, using the RNA gel blot-positive ptr-miR162 as a reference, and verified the expression of the five ptr-miRNAs that could not be detected by RNA gel blot analysis (Figure 2B).
Figure 2.
Developmental and Mechanical Stress–Responsive Expression of Populus miRNAs.
(A) RNA gel blots of total RNA isolated from leaf (L), phloem (P), developing xylem (X), tension-stressed developing xylem (Xtw), and compression-stressed developing xylem (Xow) were probed with end-labeled oligonucleotides. The 5S rRNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls. nt, nucleotides.
(B) Relative expression of miRNAs in leaf (L), phloem (P), developing xylem (X), tension-stressed developing xylem (Xtw), and compression-stressed developing xylem (Xow) was analyzed by real-time PCR. The 5.8S rRNA was selected as a reference. The data are means of three measurements ± se.
The RNA gel blot and PCR analyses revealed that all 21 ptr-miRNA families expressed at some (similar or differential) level in developing xylem and/or phloem (Figure 2), suggesting their functional association with cambium differentiation activities. Whereas some of them, such as ptr-miR156 and ptr-miR472, also are highly expressed in leaves, the others (ptr-miR160, 164, 171, 473, 477, 478, 479, and 480), of which transcripts could barely be detected in leaves, are clearly more xylem or woody tissue specific. The tissue-specific expression patterns of various ptr-miRNAs, however, differ markedly from those of their conserved Arabidopsis counterparts (Park et al., 2002; Reinhart et al., 2002; Mallory et al., 2004a; Sunkar and Zhu, 2004). We then tested whether the cloned ptr-miRNAs are more related to tree-specific developmental processes.
The formation of tension wood (TW) and opposite wood (OW) is a developmental feature characteristic of angiosperm trees and has been thought to be part of a stress-sensing mechanism leading to increased mechanical support (Timell, 1986; Wu et al., 2000). TW can be induced easily in the upper side of a bent tree stem (Wu et al., 2000), whereas the accumulation of compression stressed xylem tissue, called OW, occurs in the stem wood opposite to TW. Collectively, these specialized woody tissues are termed “reaction wood” (Sinnott, 1952). We examined ptr-miRNA transcript levels in tension-stressed and compression-stressed xylem tissues and compared them with those in normal developing xylem tissues. Overall, the expression levels of nearly all ptr-miRNAs were altered to various extents in these stressed tissues (Figure 2). The expression patterns under mechanical stress of these ptr-miRNAs could be categorized into five major types. The expression of type A ptr-miRNA is similarly suppressed by tension and compression stresses. This is most conspicuous for ptr-miR156, 162, 164, 475, 480, and 481. By contrast, transcript levels of type B ptr-miRNA, represented by ptr-miR408, are upregulated in both tension- and compression-stressed tissues. Thus, both types of miRNAs respond toward mechanical stress, but such responses are generally not discriminatory toward tension or compression stresses, suggesting important roles for types A and B ptr-miRNAs in counteracting overall mechanical stimuli. However, type C ptr-miRNA, such as ptr-miR159, 476, and 479, exhibit preferentially upregulated expression in compression tissues. The expression of ptr-miR160 and ptr-miR172 (type D ptr-miRNA) is suppressed only in compression tissue, whereas only tension stress appears to induce an attenuated expression of ptr-miR168, the type E ptr-miRNA. These results suggest that types C, D, and E ptr-miRNAs are possibly associated with more specialized regulations that may lead to a preferential development of either TW or OW.
ptr-miRNA Target Prediction
Plant miRNAs were found to have perfect or near-perfect complementarity to their targets, allowing an effective prediction of the target sequences through computation (Rhoades et al., 2002). A sensitive procedure for identifying authentic targets, including those that would form bulged and mismatched nucleotides when basepaired with their miRNAs, was recently developed by Jones-Rhoades and Bartel (2004). It includes a penalty scoring system for mismatched patterns in the miRNA:mRNA duplexes within a 20-base sequence window, with 0 points being assigned to each complementary pair, 0.5 points to each G:U wobble, 1 point to each non-G:U wobble mismatch, and 2 points to each bulged nucleotide in either RNA strand. A score of 3.0 or less was demonstrated for the identification of authentic targets with very high confidence (Jones-Rhoades and Bartel, 2004). This scoring scheme was implemented in this study with PatScan (Dsouza et al., 1997) for identifying miRNA targets in the transcript sequences of 58,036 gene models, the complete gene model set collected from the currently annotated Populus genome (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html). The transcript sequences complementary to ptr-miRNA sequences with up to seven mismatches, three mismatches plus one insertion, and three mismatches plus one deletion were searched to accommodate all possible G:U wobble pairs and bulges but keeping the scores at 3.5 or lower. This modified procedure was then applied to the prediction of targets for all cloned ptr-miRNAs (Tables 3 and 4) with, in general, a cutoff score of ≤2.5 to further minimize the number of nonauthentic targets.
Table 3.
Potential Targets for the Conserved miRNAs
| miRNA | Predicted Function | Target Genes in P. trichocarpa |
|---|---|---|
| ptr-miR156 | SPB-like | eugene3.00160416(1), eugene3.01640028(1), fgenesh1_pg.C_LG_X001417(1.5), grail3.0010026801(1.5), grail3.0033026601(2), eugene3.00400033(2), estExt_Genewise1_v1.C_1240187(2),a estExt_Genewise1_v1.C_LG_XV2186(2) |
| Nitrate transporter | grail3.0015018701(2.5) | |
| Unknown | grail3.0008008901(1), fgenesh1_pg.C_scaffold_9189000001(2), eugene3.04930001(2.5), fgenesh1_pg.C_LG_I003208(2.5), fgenesh1_pg.C_scaffold_464000004(2.5) | |
| ptr-miR159 | MYB | eugene3.00011133(2), estExt_fgenesh1_pg_v1.C_LG_IX1359(2), eugene3.00400064(2), grail3.0100003501(2.5) |
| Asparagine synthase | fgenesh1_pg.C_LG_X001429(2.5) | |
| (1-4)-β-mannan endohydrolase | eugene3.00161335(2.5) | |
| Unknown | grail3.0043007201(0),a grail3.0016033301(1.5),a fgenesh1_pg.C_LG_I002929(2.5), eugene3.00111000(2.5), eugene3.01340043(2.5), grail3.0123001501(2.5) | |
| ptr-miR160 | Auxin responsive factor | fgenesh1_pg.C_LG_V000905(0.5), eugene3.00020832(0.5), eugene3.00280173(0.5), eugene3.00280175(0.5), estExt_Genewise1_v1.C_LG_XVI2818(0.5), fgenesh1_pm.C_LG_IX000578(1), eugene3.00660262(1), fgenesh1_pm.C_LG_VIII000107(1), estExt_fgenesh1_pm_v1.C_LG_X0730(1) |
| ptr-miR162 | DCL1 | eugene3.00021687(2) |
| Unknown | eugene3.00160687(2.5) | |
| ptr-miR164 | NAC-domain protein | fgenesh1_pg.C_LG_V000368(2), estExt_Genewise1_v1.C_LG_VII2720(2), fgenesh1_pg.C_LG_XI000904(2), fgenesh1_pg.C_LG_XV000195(2.5), eugene3.00120150(2.5) |
| Protein kinase | eugene3.00110658(2.5) | |
| ptr-miR168 | Vesicle coat protein complex COPI | eugene3.00120688(2.5), estExt_Genewise1_v1.C_LG_XV1316(2.5) |
| Unknown | grail3.0110002401(0) | |
| AGO1 | grail3.0122002801(4)b | |
| ptr-miR171 | Scarecrow-like transcription factor | fgenesh1_pm.C_LG_II000611(0.5), eugene3.00030827(0.5), eugene3.01270071(0.5), eugene3.00400008(0.5), eugene3.44860001(0.5), |
| estExt_Genewise1_v1.C_LG_II3184(0.5),c grail3.0057018101(1), grail3.0019015901(2) | ||
| ptr-miR172 | Homeotic protein APETALA2 | grail3.0019003502(0.5), eugene3.00050501(1), fgenesh1_pg.C_LG_X001967(1.5), fgenesh1_pg.C_scaffold_28000114(1.5), eugene3.00160775(1.5) |
| Unknown | estExt_fgenesh1_pg_v1.C_LG_VIII1617(1.5) | |
| ptr-miR319 | MYB | grail3.0100003501(2) |
| Unknown | eugene3.00131146(2.5), eugene3.00090112(2.5)a | |
| ptr-miR408 | Plastocyanin-like | estExt_fgenesh1_pg_v1.C_400101(1),ad eugene3.00002467(2.5)d |
| Early-responsive to dehydration-related protein | estExt_fgenesh1_pm_v1.C_LG_XI0014(3)d | |
| Unknown | fgenesh1_pg.C_LG_II001930(0)c | |
| ptr-miR472 | Putative disease resistance protein | fgenesh1_pg.C_LG_XI000133(1.5), eugene3.00110125(1.5), eugene3.06450005(1.5), eugene3.00700173(2), eugene3.03110003(2), eugene3.08360002(2), fgenesh1_pg.C_scaffold_12467000001(2.5), eugene3.00190077(2.5), fgenesh1_pg.C_scaffold_548000004(2.5), fgenesh1_pg.C_scaffold_645000003(2.5), eugene3.00030137(2.5), eugene3.00110135(2.5), eugene3.113680001(2.5), eugene3.32720001(2.5), eugene3.00770103(2.5) |
All predicted miRNA targets with scores (shown in parentheses) of 2.5 or less and 5′-RACE validated targets with scores >2.5 are listed. The putative ptr-miR168 target, AGO1 homolog, is retained with a score of 4.0. Target sites are located in the predicted ORFs unless noted specifically.
Target site located in the predicted 3′ untranslated region.
Target site located in the predicted intron.
Target site located in the predicted 5′ untranslated region.
5′-RACE validated targets.
Table 4.
Potential Targets for the Nonconserved miRNAs
| miRNA | Predicted Function | Target Genes in P. trichocarpa |
|---|---|---|
| ptr-miR473 | UV-B–resistant protein (UVR8) | estExt_Genewise1_v1.C_LG_VII1592(2) |
| GRAS domain–containing protein | fgenesh1_pm.C_LG_XII000350(3),a fgenesh1_pm.C_LG_XV000218(3) | |
| ptr-miR474 | PPR | fgenesh1_pg.C_scaffold_165000036(2.5)a |
| Protein kinase | fgenesh1_pg.C_LG_XII000479(2.5) | |
| Kinesin | eugene3.03440011(3),a eugene3.08910001(3)a | |
| Leucine-rich repeat | eugene3.00700255(3)a | |
| ptr-miR475 | PPR | fgenesh1_pg.C_scaffold_1064000001(1),ab eugene3.00062011(1) (2.5),ab eugene3.00190210(1),ab fgenesh1_pg.C_scaffold_2400000001(1) (2.5),a,b eugene3.00061747(1),ab eugene3.01250075(1)(2.5), eugene3.20170001(1), estExt_Genewise1_v1.C_400113(1.5), eugene3.00002232(2), eugene3.00040809(2),b eugene3.00061748(2) (2.5),b eugene3.00130327(2), eugene3.00160204(2),b eugene3.00640083(2), eugene3.03980005(2),b eugene3.00040299(2.5), eugene3.00070793(2.5),b eugene3.00110033(2.5), eugene3.00130309(2.5), eugene3.00130325(2.5), eugene3.00130326(2.5), eugene3.01240090(2.5), eugene3.127570001(2.5), fgenesh1_pm.C_LG_XVIII000087(2.5)b |
| ptr-miR476 | PPR | eugene3.00131192(0.5),a eugene3.00131188(1), eugene3.00131190(1), eugene3.00160204(1),b eugene3.03980005(1),b fgenesh1_pm.C_LG_XVIII000087(1),b eugene3.00061748(1.5),b eugene3.00062011(1.5),b eugene3.00190210(1.5),b eugene3.15500003(1.5), fgenesh1_pg.C_LG_IV000814(1.5), fgenesh1_pg.C_scaffold_1064000001(1.5),b fgenesh1_pg.C_scaffold_2400000001(1.5),b eugene3.00040809(2),b eugene3.00061747(2),b eugene3.00140626(2), eugene3.00012769(2.5), eugene3.00070793(2.5),b eugene3.00131194(2.5), eugene3.00160470(2.5) |
| ptr-miR477 | GRAS domain–containing protein | fgenesh1_pm.C_LG_XII000350(1.5),a fgenesh1_pm.C_LG_XV000218(1.5) |
| NAC-domain protein | eugene3.00150047(2.5) | |
| Zinc finger protein | grail3.0116008302(2.5) | |
| ptr-miR478 | Organic anion transporter-like protein | estExt_fgenesh1_pg_v1.C_LG_XI1072(3)a |
| ptr-miR479 | Unknown | eugene3.00031416(3.5)a |
| ptr-miR480 | Proton-dependent oligopeptide transport family protein | fgenesh1_pg.C_LG_XIX000707(3)a |
| ptr-miR482c | Putative disease resistance protein | grail3.0140004801(1.5),a fgenesh1_pg.C_LG_XIX000055(1.5),a eugene3.02400005 (2.5)a |
All predicted miRNA targets with scores (in parentheses) of 2.5 or less and 5′-RACE validated targets with scores >2.5 are listed. All target sites listed are located in the predicted ORFs.
5′-RACE validated target.
Eleven predicted targets with multiple complementary sites for ptr-miR475 and ptr-miR476.
Eighty-seven targets with score <2.5 were predicted for ptr-miR482, and only those confirmed by 5′-RACE are listed.
A total of 82 targets were identified for the conserved ptr-miRNAs (Table 3). As expected, most of these targets were similar or related to the previously validated plant miRNA targets. Of these predicted targets, ∼49% are associated with the members of the transcription factor gene families with known or predicted functions, mostly in development, and ∼29% are related to metabolic and cellular processes and stress/defense functions (Table 3). We also identified a significant number (18 or ∼22%) of the predicted targets with unknown functions, suggesting possible new roles for conserved miRNAs in tree species. Overall, the range of miRNA targets in Populus appears to be broader than in Arabidopsis.
Approximately 130 targets were identified for the ptr-miRNAs that are not conserved in Arabidopsis (Table 4). All of these target sites are located in open reading frames (ORFs). Only three of these predicted targets are transcription factor genes, which were not previously identified and belong to the GRAS (GIBBERELLIN-INSENSITIVE [GAI], REPRESSOR of GAI [RGA], SCARECROW [SCR]) and NAC-domain protein families. Of these 130 targets, >70% appear to be involved in the defense response and other cellular processes, whereas ∼26% encode pentatricopeptide repeat proteins (PPRs). These predictions are consistent with the current belief that transcription factor genes may not be the dominant miRNA targets as previously thought (Eckardt, 2004; Sunkar and Zhu, 2004). It is interesting to note that although ptr-miR473a, ptr-miR478a, and ptr-miR482 (Table 2) have conserved miRNA sequences in rice, the predicted target sequences of these ptr-miRNAs and their rice counterparts are not conserved. The association of Populus miRNAs with generally more and distinct targets may further suggest more diverse miRNA functions in tree than in herbaceous species. Nevertheless, validation of predicted targets is necessary for verifying whether the small RNAs are miRNAs and for establishing functional evidence for the newly identified miRNAs.
Experimental Validation of Predicted Populus miRNA Targets
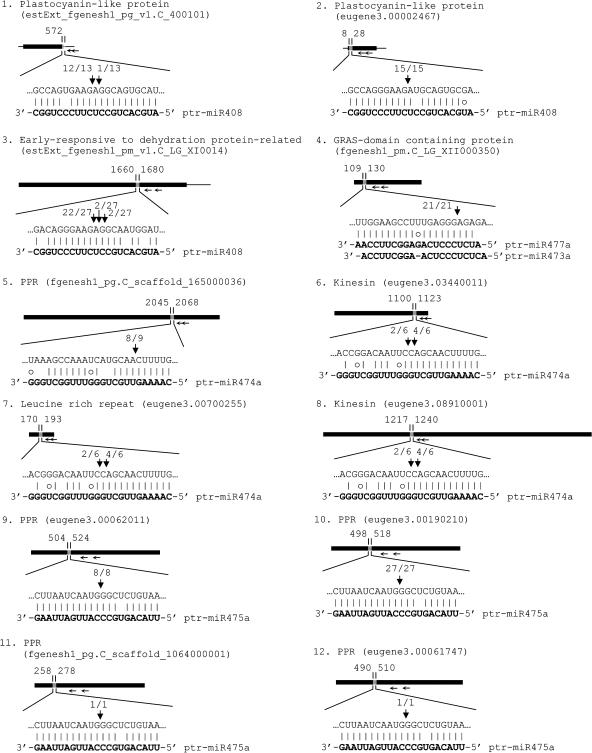
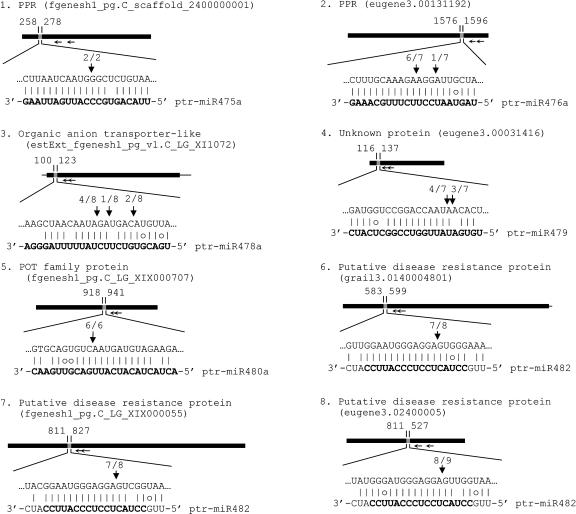
The predominant mode of miRNA-mediated gene silencing in plants is believed to be the direct cleavage of target mRNAs by binding to coding segments with near-perfect complementarity (Llave et al., 2002b; Reinhart et al., 2002; Carrington and Ambros, 2003; Tang et al., 2003). A modified 5′-rapid amplification of cDNA ends (RACE) can readily detect this cleavage and has been used to validate 48 predicted miRNA targets for 18 of the 33 Arabidopsis miRNA families (Jones-Rhoades and Bartel, 2004). To verify whether the cloned ptr-miRNAs can mediate the cleavage of their predicted targets in vivo, we isolated RNAs from the developing secondary xylem and phloem of P. trichocarpa and performed 5′-RACE on three predicted targets for ptr-miR408, a conserved miRNA between Populus and Arabidopsis, and 17 targets representing those for 9 of the 10 ptr-miRNAs that are not conserved in Arabidopsis (Figures 3 and 4). The 5′-RACE products revealed that the three mRNAs are indeed the targets of ptr-miR408 (Figure 3). Similarly, sequencing of the 5′-RACE products of the targets for the nonconserved ptr-miRNA indicated that all cleavage sites are mapped to the miRNA complementary sequences, as seen for other miRNA targets (Llave et al., 2002b; Aukerman and Sakai, 2003; Kasschau et al., 2003; Palatnik et al., 2003; Xie et al., 2003; Jones-Rhoades and Bartel, 2004; Mallory et al., 2004a, 2004b; Vazquez et al., 2004a), validating miRNA target cleavage for all nine tested nonconserved ptr-miRNAs (Figures 3 and 4). This brings the number of validated plant miRNA targets to 68. In addition, the overall target cleavage results in Populus suggest that whereas some plant miRNAs mediate cleavage predominantly at positions around 10 nucleotides from the 5′ end of the miRNA, some might mediate the cleavage mainly at other locations. However, the possibility that some of these small RNAs are transacting siRNAs (Parizotto et al., 2004; Peragine et al., 2004; Vazquez et al., 2004b) cannot be ruled out. Further study of the variability in target cleavage sites may help add new insights into small RNA-mediated gene regulation in plants.
Figure 3.
Experimental Validation of the Predicted mRNA Targets for the Newly Cloned ptr-miR408, 473a, 474a, 475a, and 477a.
The mRNA cleavage sites were determined by modified 5′ RNA ligase-mediated RACE. Heavy black lines represent ORFs. The lines flanking gray regions represent nontranslated regions (if known) and miRNA complementary sites, with the nucleotide positions within the ORF indicated. The mRNA sequence of each complementary site from 5′ to 3′ and the cloned miRNA sequence (bold) from 3′ to 5′ are shown in the expanded regions. Watson-Crick pairing (vertical dashes) and G:U wobble pairing (circles) are indicated. Vertical arrows indicate the 5′ termini of miRNA-guided cleavage products, as identified by 5′-RACE, with the frequency of clones shown. Positions of the nesting and the nested primers used for 5′-RACE are indicated by horizontal arrows. For targets eugene3.03440011, eugene3.00700255, and eugene3.08910001 predicted for ptr-miR474a, 5′-RACE was performed using the same set of primers, yielding products with the identical sequence for which origin could not be certain. Therefore, at least one of them is an authentic target for ptr-miR474a.
Figure 4.
Experimental Validation of the Predicted mRNA Targets for the Newly Cloned ptr-miR475a, 476a, 478a, 479, 480a, and 482.
The ptr-miR475a targets shown in this figure and in Figure 3 were validated using the same set of degenerate primers. The partial flanking sequence of ptr-miR482 in the precursor is also shown.
DISCUSSION
Many Populus miRNAs Have No Sequence Conservation in Arabidopsis
We report here the cloning of 22 miRNAs from the developing secondary xylem of P. trichocarpa, of which 10 are absent from the Arabidopsis genome. Seven of these 10 miRNAs also have no apparent sequence conservation in rice. These findings may seem inconsistent with early reports suggesting that plant miRNAs are conserved (Reinhart et al., 2002; Bartel and Bartel, 2003). Indeed, a majority of the first 16 Arabidopsis miRNA gene families identified are conserved. They were all cloned from normal growth Arabidopsis (Reinhart et al., 2002; Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004) and therefore might represent those normally abundant miRNAs. As the number of identified Arabidopsis miRNAs increases to include those that are difficult to clone or normally underexpressed, such as those associated with particular growth conditions, the presence of a large fraction of nonconserved miRNAs also becomes obvious in Arabidopsis.
Of the 33 systematically classified miRNA gene families in Arabidopsis (Reinhart et al., 2002; Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004), 15 have no apparent conservation among rice miRNAs. Eleven of these 15 nonconserved miRNA families were cloned only from Arabidopsis grown under various abiotic stress conditions (Sunkar and Zhu, 2004). These results may suggest that a majority of the conserved miRNAs might be necessary for mediating normal growth and developmental processes that are common to plants. However, the previous results, together with our findings, suggest that a significant fraction of the nonconserved miRNAs could be part of the regulatory networks associated with specific growth conditions or developmental processes. The absence of some Populus miRNAs from genomes of herbaceous species further indicates the presence of tree species–specific miRNAs. More miRNAs need to be identified under different growth and development conditions and from species across genera to test these hypotheses.
ptr-miRNAs Exhibit Tissue-Specific Expression Patterns That Are Different from Their Arabidopsis Homologs
Despite the existence of sequence conservation between Populus and Arabidopsis miRNAs, a large number of these homologous miRNAs exhibit contrasting tissue-specific expression patterns. For example, ptr-miR156 has the most conspicuous expression in leaves, and its transcripts also accumulate abundantly in the stem organs (xylem and phloem), whereas the expression of Arabidopsis miR156 in leaves and stems is negligible (Reinhart et al., 2002). ptr-miR159 is highly expressed in stem xylem but weakly in leaves (Figure 2), whereas its Arabidopsis homolog was not detectable in stems but instead was found abundantly expressed in leaves (Park et al., 2002). The expression of ptr-miR164 in Populus stem phloem and xylem is significantly higher than that in leaves, but such preferential expression was not observed for the Arabidopsis miR164 (Mallory et al., 2004a). The stem-specific (xylem and phloem) expression of ptr-miR171 was not observed for its Arabidopsis homolog, which exhibits essentially no expression in stems and seedlings (Reinhart et al., 2002). ptr-miR408 is essentially absent from stem xylem (Figure 2), but the Arabidopsis miR408 exhibits strong expression in stems (Sunkar and Zhu, 2004). Furthermore, the expression of many ptr-miRNAs could be induced in the developing xylem of stems receiving mechanical stress (Figure 2), suggesting possible regulatory roles for these miRNAs in the most critical defense systems for structural and mechanical fitness in trees. Long-term structural and mechanical fitness and associated persistent wood growth are normally not required by Arabidopsis. All of these examples suggest that miRNA sequence conservation between plant species may not indicate conserved miRNA functions.
Targets Were Experimentally Validated for Nonconserved ptr-miRNAs
The detection in developing xylem or phloem tissues of mRNA fragments validates in vivo the miRNA-mediated target cleavage for ptr-miRNAs that are absent from Arabidopsis, establishing functional evidence for nine newly identified plant miRNAs. ptr-miR473 and ptr-miR477, two homologous but distinct miRNAs exhibiting contrasting tissue-specific expression patterns (Figure 2), target a member of the GRAS gene family, suggesting that distinct miRNAs may have similar functions in a tissue-specific manner. Sequence alignment indicated that this GRAS domain–containing protein is clustered in an array of SCARECROW-like (SCL) family members of putative transcription factors (data not shown). SCR or SHOOT GRAVITROPISM 1, the founding member of GRAS, is known for its role in regulating asymmetric cell divisions and elongations to generate the cell lineages in shoot and root in response to gravitational force (Di Laurenzio et al., 1996; Tasaka et al., 1999; Helariutta et al., 2000; Nakajima et al., 2001). Because gravitropism represents a major mechanical force triggering tension and compression wood development in trees (Timell, 1986), ptr-miR473 and ptr-miR477 together with a conserved miRNA (miR171, Table 3) with validated SCL targets (Llave et al., 2002a; Kasschau, et al., 2003), may regulate the formation of specialized woody tissue in trees. Likewise, ptr-miR477, also predicted to target a NAC-domain mRNA, may represent a member for NAC-domain gene regulation (miR164) in trees affecting organ separation during development (Laufs et al., 2004; Mallory et al., 2004a).
Except for ptr-miR473 and ptr-miR477, all other seven new ptr-miRNAs appear to have less involvement in developmental patterning. ptr-miR474, ptr-miR475, and ptr-miR476 target seven PPRs (Figures 3 and 4), which belong to one of the largest plant protein families that are believed to be constitutively involved in the posttranscriptional regulation of organelle gene expression and RNA processing (Small and Peeters, 2000; Lurin et al., 2004). This is consistent with the expression at various levels of ptr-miR475 and ptr-miR476 in all tissues examined in this study (Figure 2). Interestingly, 11 PPRs retained by search scores of ≤2.5 were predicted to be the common targets for ptr-miR475 and ptr-miR476, and each of these PPRs contains multiple complementary sites that are unique to either ptr-miR475 or ptr-miR476 (Table 4). Although PPRs were targets predicted for Arabidopsis miR400 (Sunkar and Zhu, 2004), the multiple target sites in multiple PPRs have not been reported previously. Our finding may suggest the involvement of more tree-specific miRNA functions in coordinated or redundant pathways in controlling the class of genes involved in important gene and RNA processing.
ptr-miR482 targets three validated mRNAs encoding putative disease resistance proteins, which have not been validated for Arabidopsis. In addition, a large number of the related targets also were predicted for this, as well as a new conserved miRNA, ptr-miR472 (Table 3), further revealing possible roles for miRNAs in complex defense pathways. The large number of disease resistance genes targeted by tree-specific miRNAs may be important defense networks in trees for their long-term adaptation to constant pathogenic environments for survival.
A validated target for ptr-miR408, a plastocyanin-like protein, is interesting because this class of proteins is believed to mediate lignin polymerization (Nersissian et al., 1998). The near absence of ptr-miR408 transcripts in developing xylem, the major lignification tissue, and the moderate expression of ptr-miR408 in low lignin containing tissues, phloem and leaf (Figure 2), support the role of ptr-miR408 in coordinating lignin deposition in different tissues. ptr-miR473 is predicted to target a UV-B–resistant gene (UVR8) (Table 4), which is believed to positively regulate phenylpropanoid metabolism associated with cinnamate 4-hydroxylase for the production of cell wall polyphenolics, such as lignin and flavonoids (Jin et al., 2000; Kliebenstein et al., 2002). The (1→4)-β-mannan endohydrolase target predicted for ptr-miR159 suggests roles for miRNAs in regulating cell wall polysaccharide biosynthesis. The biosynthesis of cellulose and hemicelluloses has been extensively studied in Arabidopsis, but their regulation remains largely unknown (Richmond and Somerville, 2000; Taylor et al., 2003; Dhugga et al., 2004). Nothing is known about the genes and genetic regulation of hemicellulose biosynthesis in tree species. miRNAs may be useful, novel tools for investigating the regulation of the biosynthesis of these polysaccharides and lignin, the major cell wall components.
miRNAs Are Possibly Associated with the Formation of Specialized Wood
Although the three major woody cell wall components, lignin, cellulose, and hemicelluloses, have distinct biosynthetic pathways, the spatiotemporal deposition of all these components can be profoundly altered by mechanical stress. The alteration, creating the specialized reaction wood (TW and OW), however, leaves the total mass of these components in wood more or less unchanged (Scurfield, 1973; Fujii et al., 1982; Timell, 1986; Hu et al., 1999). In TW, decreased lignin and hemicellulose deposition is compensated for by an increase in cellulose, whereas more lignin and a less amount of polysaccharides are found in OW (Scurfield, 1973; Fujii et al., 1982; Timell, 1986; Hu et al., 1999; Wu et al., 2000). Concomitantly, the number of vessel elements decreases in TW as more cambial fusiform initials are diverted into fiber cell development, while the opposite is true in OW (Wardrop and Davies, 1964; Esau, 1965; Scurfield, 1973). Thus, crosstalk between the biosynthesis of cell wall components and cell type developments for their compensatory regulation in wood formation has been suggested as a tree-specific adaptation to mechanical stress (Scurfield, 1973; Hu et al., 1999).
The possible association of miRNAs with cell/organ development and the biosynthesis of cell wall components together with mechanical stress–inducible expression of miRNAs suggest a model that entails a cascade of miRNA regulation that could be part of the crosstalk in assembling reaction wood. Several miRNAs cloned in this work, especially those exhibiting noticeable responses to mechanical stress (Figure 2), may be part of such a cascade. For instance, the drastically upregulated ptr-miR408 expression in TW and OW may suggest a regulatory role in suppressing lignin deposition in certain tissues/zones of reaction wood. By contrast, the diminished expression of ptr-miR164 could be responsible for upregulated cell types and/or cell wall components, consistent with the fact that the cleavage of its targets (Table 3) may negatively affect the proliferation and development of certain cells and organs (Aida et al., 1997; Xie et al., 2000; Laufs et al., 2004; Mallory et al., 2004a). Reduced ptr-miR171 expression in mechanically stressed woody tissues may have similar effects because its target genes, SCLs, are known to regulate cell division and elongation to produce organ cell lineages as a positive response to gravitropism (Di Laurenzio et al., 1996; Tasaka et al., 1999; Helariutta et al., 2000; Nakajima et al., 2001), a major mechanical stress for reaction wood induction (Timell, 1986). The ptr-miRNAs with either similarly altered expression in both TW and OW or with preferentially altered expression in TW or OW may be candidate regulators contributing to control or feedback control affecting a set of coordinated pathways leading to the formation of reaction wood. Stable phenotypes associated with the downregulation or upregulation of these candidate ptr-miRNAs (Lu et al., 2004) in transgenic P. trichocarpa will be rich resources for examining these proposed genetic regulations for reaction wood deposition and for wood formation in general.
Studies using reference species, such as Arabidopsis, have created tremendous insights into plant miRNA functions. The utility of this knowledge, however, can increase dramatically as additional knowledge of miRNAs from other genomes becomes available. For example, comparative genomics led us to identify the more tree-specific miRNAs that are not in the Arabidopsis and rice genomes. Even conserved ptr-miRNAs have either distinct or more diverse target messages as compared with their Arabidopsis counterparts. Thus, comparative miRNA studies may help increase our understanding, at the regulatory level, of the events that give rise to new species or to the emergence of specific traits. In particular, our results on ptr-miRNAs together with the knowledge of the Populus, Arabidopsis, and rice genomes are useful for guiding further studies of the regulatory mechanisms of more species-specific traits, such as wood formation.
METHODS
Plant Materials
Leaf, phloem, and developing xylem tissue were collected from 1-year-old, greenhouse-grown Populus trichocarpa (clone Nisqually-1), ground in liquid nitrogen, and used immediately for RNA isolation or stored in liquid nitrogen until use. Tension- and compression-stressed developing xylem were induced by bending the tree stem into an arch for 4 d, and developing secondary xylem tissues from the upper (TW) and lower (OW) portions of the bent segment were collected directly into liquid nitrogen for RNA isolation.
Small RNA Isolation
Total RNA was isolated by the CTAB method (Chang et al., 1993). Cloning of miRNAs was performed as described by Elbashir et al. (2001), Lau et al. (2001), and Lagos-Quintana et al. (2002). Briefly, total RNA was separated on a denaturing 12% polyacrylamide gel. The 16- to 36-nucleotide band was excised, and the recovered RNA was dephosphorylated by alkaline phophatase. A 5′-phosphorylated 3′ adaptor oligonucleotide (5′-pCTGTAGGCACCATTCATCACx-3′; p, phosphate; x, 3′-amino-modifier C-7) was then ligated to the dephosphorylated RNA. The ligation product was separated from the nonligated RNA on a denaturing 12% polyacrylamide gel, and the ligated RNA was recovered from the gel and 5′ phosphorylated. A 5′ adaptor oligonucleotide (5′-ATGTCGTGaggcacctgaaa-3′, RNA/DNA version, lowercase RNA) containing hydroxyl groups at both 5′ and 3′ ends was then ligated to the phosphorylated ligation product. The new ligation product was gel purified and eluted from the gel slice. Reverse transcription was performed using an RT primer (5′-GATGAATGGTGCCTAC-3′), followed by PCR with a 5′ primer (5′-GTCGTGAGGCACCTGAAA-3′) and a 3′ primer (5′-GATGAATGGTGCCTACAG-3′). The PCR product was then digested with BanI and concatamerized using T4 DNA ligase. Concatamers with sizes >500 bp were separated on an agarose gel and recovered from the gel slice. The unpaired ends were filled by incubation with Taq polymerase, and the DNA product was directly ligated into the pCR2.1-TOPO vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA). A total of 379 individual clones were isolated and sequenced, yielding 898 unique sequences. BLAST analyses against the GenBank (http://www.ncbi.nlm.nih.gov/BLAST) and the P. trichocarpa genome assembly (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html) showed that 809 of these sequences, which were not analyzed, correspond to known noncoding rRNAs, tRNAs, and small nuclear RNAs and to those associated with retrotransposons or transposons. The 89 remaining sequences were further characterized.
Prediction of Stem-Loop Structures
The cloned small RNAs or their related sequences (one to three mismatches) were aligned with the Populus draft genome assembly (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html), Populus ESTs (http://www.ncbi.nlm.nih.gov/dbEST), Arabidopsis thaliana genome annotation version 5.0 (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/), and Oryza sativa ssp japonica cv Nipponbare genome assembly release 3 (http://www.tigr.org/tdb/e2k1/osa1/index.shtml) using PatScan (Dsouza et al., 1997). Secondary structures were predicted by the mfold program (www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi) using the default parameters (Zuker, 2003). In each case, only the lowest energy structure was selected for manual inspection, as described by Reinhart et al. (2002). Small RNA sequences were folded with flanking sequences in five contexts: (1) 300 bp upstream and 20 bp downstream, (2) 150 bp upstream and 20 bp downstream; (3) 150 bp upstream and 150 downstream, (4) 20 bp upstream and 150 downstream, and (5) 20 bp upstream and 300 bp downstream.
Gel Blot Analysis of miRNA Expression
RNA gel blot hybridization of small RNAs was performed essentially as described by Hutvágner et al. (2001), with minor modifications. Total RNA (30 μg) was denatured for 10 min at 65 to 70°C, separated on a 12% polyacrylamide/8 M urea gel (Amersham, Piscataway, NJ) in a Protean II apparatus (Bio-Rad, Hercules, CA), and electroblotted onto Hybond-N+ membrane (Amersham) by Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Bio-Rad). After UV cross-linking and air drying, blots were prehybridized in ULTRAhyb-Oligo hybridization buffer (Ambion, Austin, TX) and hybridized with [γ-32P]ATP-labeled DNA oligonucleotides complementary to small RNA sequences. The hybridization was performed overnight in ULTRAhyb-Oligo buffer at 37°C. Blots were washed twice with washing buffer consisting of 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.5% SDS at 37°C for 0.5 h. Signals were visualized by autoradiography on x-ray film at −80°C.
Real-Time PCR Validation of miRNA Expression
Total RNA was isolated using plant RNA purification reagent (Invitrogen) and treated with RNase-free DNase I (Promega, Madison, WI). Poly(A) tails were then added to the 3′ end of the RNAs by poly(A) polymerase (Ambion), and the polyadenylated RNAs were reverse transcribed by SuperScript II reverse transcriptase (Invitrogen) with the oligo(dT) 3′-RACE adaptor (Ambion). To amplify the miRNA from the reverse-transcribed cDNAs, we used the miRNA sequence as the forward primer and the 3′-RACE outer primer (Ambion) as the reverse primer. The 5.8S rRNA was selected as the endogenous reference. Real-time PCR was conducted with the Applied Biosystems 7900HT sequence detection system (Foster City, CA). For each reaction, 1 μL of diluted cDNA (equivalent to ∼100 pg of total RNA) was mixed with 12.5 μL of 2× SYBR green PCR master mix (Applied Biosystems) and 5 pmol each of the forward and the reverse primers in a final volume of 25 μL. The conditions for PCR amplification were as follows: 45 cycles at 95°C for 15 s and 60°C for 1 min. The amplification was followed by a thermal denaturing step to generate dissociation curves for verifying amplification specificity.
Modified 5′ RNA Ligase-Mediated RACE for Mapping of mRNA Cleavage Sites
Aliquots of total RNA, isolated from developing xylem and phloem of 1-year-old P. trichocarpa plants by the CTAB method (Chang et al., 1993), were mixed. Poly(A)+ RNA was enriched from 100 μg of mixed total RNA using the Absolutely mRNA purification kit (Stratagene, La Jolla, CA). The 5′-RACE was conducted using the GeneRacer kit (Invitrogen) as described by Vazquez et al. (2004a). PCR amplifications were performed using the GeneRacer 5′ primer and the nesting gene-specific primers (see Supplemental Table 4 online). Nested PCR amplifications were performed using the GeneRacer 5′ nested primer and the nested gene-specific nested primers (see Supplemental Table 4 online). The amplification products were gel purified, cloned, and sequenced.
Prediction of miRNA Targets
Targets of the ptr-miRNAs were predicted from 58,036 gene models of the current P. trichocarpa draft genome assembly (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html). PatScan (Dsouza et al., 1997) was first used to identify the predicted transcripts that contain sequence complementary to any of the cloned miRNAs with mismatches and bulges, and then the scoring scheme developed by Jones-Rhode and Bartel (2004) to screen for authentic miRNA targets was applied. For ptr-miR482, the sequence was extended to 20 bases at either the 5′- or 3′-end based on the predicted precursor sequence before target prediction. Functions of the predicted targets were assigned manually based on either the eukaryotic orthologous group annotation of the Populus genome or the function of the best hit from the BLAST homology search (Altschul et al., 1997) against the Arabidopsis annotation version 5.0 peptide sequence database.
Sequence data from this article have been deposited with the miRNA Registry (http://www.sanger.ac.uk/Software/Rfam/mirna/) with the following numbers: ptr-miR472, ptr-miR473, ptr-miR474, ptr-miR475, ptr-miR476, ptr-miR477, ptr-miR478, ptr-miR479, ptr-miR480, ptr-miR481, and ptr-miR482.
Supplementary Material
Acknowledgments
We are grateful for the sequence information produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov). This work was supported by a grant from the U.S. Department of Energy Division of Energy Biosciences (DE-FG02-03ER15442) to V.L.C.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Vincent L. Chiang (vincent_chiang@ncsu.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.033456.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. [DOI] [PubMed] [Google Scholar]
- Adai, A., Johnson, C., Mlotshwa, S., Archer-Evans, S., Manocha, V., Vance, V., and Sundaresan, V. (2005). Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 15 78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., Fukaki, H., Fujiswa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the CUP-SHAPED COTYLEDON mutant. Plant Cell 9 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, C.C., Sieber, P., Wellmer, F., and Meyerowitz, E.M. (2005). The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 15 303–315. [DOI] [PubMed] [Google Scholar]
- Barnett, J.R. (1981). Secondary xylem cell development. In Xylem Cell Development, J.R. Barnett, ed (Tunbridge Wells, UK: Castle House Publications), pp. 47–95.
- Bartel, B., and Bartel, D.P. (2003). MicroRNAs: At the root of plant development. Plant Physiol. 132 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Bass, B.L. (2002). RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell, J.A., et al. (2005). Sorghum genome sequencing by methylation filtration. PLoS Biol. 3 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet, E., Wuyts, J., Rouze, P., and Van de Peer, Y. (2004). Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 101 11511–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301 336–338. [DOI] [PubMed] [Google Scholar]
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga, K.S., Barreiro, R., Whitten, B., Stecca, K., Hazebroek, J., Randhawa, G.S., Dolan, M., Kinney, A.J., Tomes, D., Nichols, S., and Anderson, P. (2004). Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 303 363–366. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio, L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86 423–433. [DOI] [PubMed] [Google Scholar]
- Dsouza, M., Larsen, N., and Overbeek, R. (1997). Searching for patterns in genomic data. Trends Genet. 13 497–498. [DOI] [PubMed] [Google Scholar]
- Eckardt, N.A. (2004). Small RNA on the move. Plant Cell 16 1951–1954. [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2001). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Esau, K. (1965). Plant Anatomy, 2nd ed. (New York: John Wiley & Sons).
- Floyd, S.K., and Bowman, J.L. (2004). Gene regulation: Ancient microRNA target sequences in plants. Nature 428 485–486. [DOI] [PubMed] [Google Scholar]
- Fujii, M., Azuma, J., Tanaka, F., Kato, A., and Koshijima, T. (1982). Studies on hemicelluloses in tension wood. I. Chemical composition of tension, opposite and side woods of Japanese beech (Fagus crenata Blume). Wood Res. 68 8–21. [Google Scholar]
- Grad, Y., Aach, J., Hayes, G.D., Reinhart, B.J., Church, G.M., Ruvkun, G., and Kim, J. (2003). Computational and experimental identification of C. elegans microRNAs. Mol. Cell 11 1253–1263. [DOI] [PubMed] [Google Scholar]
- Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., Hauser, M.-T., and Benfey, P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101 555–567. [DOI] [PubMed] [Google Scholar]
- Hu, W.-J., Lung, J., Harding, S.A., Popko, J.L., Ralph, J., Stokke, D.D., Tsai, C.-J., and Chiang, V.L. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17 808–812. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., McLachlan, J., Pasquinelli, A.D., Bálint, É., Tuschl, T., and Zamore, P.D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293 834–838. [DOI] [PubMed] [Google Scholar]
- Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., Tonelli, C., Weisshaar, B., and Martin, C. (2000). Transcriptional repression by AtMyB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., and Bartel, D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14 787–799. [DOI] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev. Cell 4 205–217. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 81–84. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2005). The developmental role of microRNA in plants. Curr. Opin. Plant Biol. 8 38–44. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Lim, J.E., Landry, L.G., and Last, R.L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12 735–739. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294 858–862. [DOI] [PubMed] [Google Scholar]
- Laufs, P., Peaucelle, A., Morin, H., and Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131 4311–4322. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. (2002). MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Rhoades, M.W., Burge, C.B., and Bartel, D.P. (2003). The microRNAs of Caenorhabditis elegans. Genes Dev. 17 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. (2002. b). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002. a). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lu, S., Shi, R., Tsao, C.-C., Yi, X., Li, L., and Chiang, V.L. (2004). RNA silencing in plants by the expression of siRNA duplexes. Nucleic Acids Res. 32 e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano, E.J., Mirsky, H., Vendetti, N.J., and Maas, S. (2004). RNA editing of a miRNA precursor. RNA 10 1174–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. (2004. a). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004. b). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHale, N.A., and Koning, R.E. (2004). MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell 16 1730–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima, K., Sena, G., Nawy, T., and Benfey, P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413 307–311. [DOI] [PubMed] [Google Scholar]
- Nersissian, A.M., Immoos, C., Hill, M.G., Hart, P.J., Williams, G., Herrmann, R.G., and Valentine, J.S. (1998). Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: Plant-specific mononuclear blue copper proteins. Protein Sci. 7 1915–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. [DOI] [PubMed] [Google Scholar]
- Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C., and Voinnet, O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H.L., and Poethig, R.S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18 2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110 513–520. [DOI] [PubMed] [Google Scholar]
- Richmond, T.A., and Somerville, C.R. (2000). The cellulose synthase superfamily. Plant Physiol. 124 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scurfield, G. (1973). Reaction wood: Its structure and function. Science 179 647–655. [DOI] [PubMed] [Google Scholar]
- Sinnott, E.W. (1952). Reaction wood and the regulation of tree form. Am. J. Bot. 39 69–78. [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif: A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25 46–47. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.-K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaka, M., Kato, T., and Fukaki, H. (1999). The endodermis and shoot gravitropism. Trends Plant Sci. 4 103–107. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Howells, R.M., Huttly, A.K., Vickers, K., and Turner, S.R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100 1450–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timell, T.E. (1986). Compression Wood in Gymnosperms. (Berlin: Springer Verlag).
- Vazquez, F., Gasciolli, V., Crété, P., and Vaucheret, H. (2004. a). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14 346–351. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.-L., Bartel, D.P., and Crete, P. (2004. b). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16 69–79. [DOI] [PubMed] [Google Scholar]
- Wang, J.-F., Zhou, H., Chen, Y.-Q., Luo, Q.-J., and Qu, L.-H. (2004. a). Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res. 32 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.J., Reyes, J.L., Chua, N.-H., and Gaasterland, T. (2004. b). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5 R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardrop, A.B., and Davies, G.W. (1964). The structure of reaction wood: The structure and differentiation of compression wood. Aust. J. Bot. 12 24–38. [Google Scholar]
- Wu, L., Joshi, C.P., and Chiang, V.L. (2000). A xylem-specific cellulose synthase gene from aspen (Populus tremuloides) is responsive to mechanical stress. Plant J. 22 495–502. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Frugis, G., Clogan, D., and Chua, N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2 e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., Kasschau, K.D., and Carrington, J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13 784–789. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- miRNA and miRNA targets were independently annotated by Matthew Jones-Rhoades (Whitehead Institute for Biomedical Research; http://web.wi.mit.edu/bartel/pub/) as part of the assembly and annotation efforts of the International Populus Genome Consortium (http://www.ornl.gov.sci/ipgc/).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.