Abstract
By using high-density oligonucleotide arrays, we profiled gene expression in reward-related brain regions of rats that developed escalated cocaine intake after extended access to cocaine (6 h per day). Rats allowed restricted daily access to cocaine (only 1 h) that displayed a stable level of cocaine intake and cocaine naive rats were used for controls. Four analysis methods were compared: Affymetrix microarray suite 4 and microarray suite 5, which use perfect-match-minus-mismatch models, and dchip and rma, which use perfect-match-only models to generate expression values. Results were validated by RT-PCR in individual animals from an independent replication of the experiment. A small number of genes was associated with escalated cocaine intake (ESC genes). Unexpectedly, of the brain regions examined [prefrontal cortex, nucleus accumbens, septum, lateral hypothalamus (LH), amygdala, and ventral tegmental area], the LH was the most transcriptionally responsive in escalation of cocaine intake. Most of the ESC genes identified are also expressed during synaptogenesis and synaptic plasticity and include genes that code for several presynaptic and postsynaptic proteins involved in neurotransmission. These results suggest that LH intrinsic circuitry undergoes a structural reorganization during escalation of cocaine use. This remodeling of LH circuitry could contribute to the chronic deficit in reward function that has been hypothesized to drive the transition to drug addiction. Results also support the value of using multiple analysis strategies to identify the most robust changes in gene expression and to compensate for the biases that affect each strategy.
Keywords: release machinery, δ-catenin, Glu receptor-interacting protein 2, PKCγ, fractalkine
Acurrent challenge for the neuroscience of drug addiction is to understand the molecular mechanisms responsible for the development of compulsive drug use (1). Such a transition is generally associated with a pattern of escalating drug use whereby consumption increases over time and becomes increasingly difficult to control. In the present study, we investigated gene expression changes associated with drug addiction by using an animal model that demonstrates escalation of cocaine intake. In this model, drug intake gradually escalates when daily access to the drug is increased to ≥6 h [long access (LgA)] (2, 3), whereas with only 1 h of access per day [short access (ShA)], drug intake remains low and stable over time. The difference in cocaine consumption between ShA and LgA animals has been hypothesized to model the difference drawn by clinicians between controlled and compulsive drug use (2, 3). This view is supported by recent results showing that escalation in cocaine intake in LgA animals is associated with an increased motivation to seek and to take cocaine and with a persistent down-regulation of brain reward function when compared with ShA animals (3–5). Therefore, escalation of cocaine intake in LgA rats appears to replicate the behavioral and neuroadaptive changes associated with the development of addiction (1, 6). Thus the elucidation of gene expression changes in the present model may provide unique clues to the molecular mechanisms behind the reward dysfunction that drives the transition to compulsive cocaine use.
By using high-density oligonucleotide arrays, we profiled gene expression changes in several reward-related brain regions in LgA rats in comparison with ShA rats and cocaine naive controls. Results were extensively validated by RT-PCR in individual animals from an independent replication of the experiment. The results suggest that the lateral hypothalamus (LH) in particular undergoes a structural reorganization during escalation of cocaine use. This remodeling of LH circuitry could contribute to the chronic deficit in reward function that has been hypothesized to drive the transition to drug addiction (3, 7).
Materials and Methods
Behavioral Procedures. Male Wistar rats (280–340 g) were prepared with a chronic i.v. catheter and were food-restricted 5 days later and trained for 7 days to press a lever to obtain food pellets. Two days after food training, 20 rats were tested for cocaine self-administration during two consecutive phases: a screening phase (1 day) and an escalation phase (18 days). Eight more rats were exposed to the same experimental manipulations as the other rats, except that they were not exposed to cocaine. During the screening phase, the 20 rats tested for self-administration were allowed to self-administer cocaine during only 1 h on a fixed-ratio schedule where one lever press results in one cocaine infusion (250 μg per injection in a volume of 0.1 ml delivered in 4 sec), after which two balanced groups with the same mean weight and mean cocaine intake were formed. During the escalation phase, one group had access to cocaine self-administration for only 1 h per day (ShA rats) and the other group for 6 h per day (LgA rats). Four of the 20 rats allowed to self-administer cocaine were discarded from the study either because of a failure to reach the criterion for acquisition of cocaine self-administration (n = 3) (i.e., at least eight injections per h) or because of inconsistent within-session intake for several days (n = 1), leaving eight rats per group.
Brain Dissection. Forty-eight hours after the last self-administration session, drug-naive, ShA, and LgA rats (n = 8 per group) were killed in random order to obtain tissue samples from six reward-related brain regions. Animals were perfused with 10% RNAlater (Ambion, Austin, TX) in 0.1 M PBS under anesthesia by CO2 narcosis. Brains were sliced with a wire brain slicer (Research Instruments and Manufacturing, Corvallis OR) and dissected with the assistance of a rat brain atlas (8). A 14-gauge needle constructed from a spinal tap needle and equipped with a plunger to facilitate the transfer of the dissected tissue was used to collect the nucleus accumbens (NAc), the LH, the septum (SEP), and the ventral tegmental area (VTA). The medial prefrontal cortex and the amygdaloid complex were dissected free-handedly with established anatomical landmarks (8).
Microarray Procedures. Total RNAs of regions of interest were prepared with an RNeasy miniprep kit according to manufacturer's (Qiagen, Valencia, CA) protocol. RNA was quantified by using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and quality was assessed with a 2100 Bioanalyzer (Agilent Technologies, Palo Alta, CA) and with agarose gel electrophoresis. Depending on the brain region, between 1 and 5 μg of total RNA was used to prepare double-stranded cDNA with a cDNA synthesis kit (GIBCO/BRL) for first and second strand. Biotinylated cRNA was generated from these cDNAs with a BioArray high-yield RNA transcript labeling kit (Enzo Life Sciences). It was then purified with RNeasy spin columns (Qiagen) and fragmented before hybridization (9). Hybridization mixtures were boiled at 99°C, loaded on Affymetrix Neurobiology RNU34 chips and hybridized at 45°C for 16 h. Washes were performed on a Fluidics Station with the manufacturer's (Affymetrix) recommended wash solutions, and the chips were stained with a streptavidin–phycoerythrin conjugate for fluorescence detection. After staining, chips were scanned with an Affymetrix GeneArray Scanner 2500. For the amygdaloid complex and medial prefrontal cortex, hybridizations were run in quadruplicate (four independent pools of two animals each were hybridized on four RNU34 chips); for smaller regions, such as NAc and LH, four RNU34 chips were hybridized with duplicate hybridizations of two independent pools of four animals each; for VTA and SEP, we ran three replicate hybridizations of single pools each of eight animals. Results were extensively validated by RT-PCR in brain regions of individual animals from an independent replication of the experiment as described below.
Data Analysis. Gene expression changes associated with escalated cocaine intake (ESC genes) were investigated by using four analysis strategies differing by type of image analysis software and statistical treatment of the data. microarray suite (mas)4- and mas5-generated expression values, which are derived with perfect match (PM) minus mismatch (MM) models, were analyzed with ANOVA and Fischer's post hoc test. The data were also analyzed with a nonparametric test (Kruskall–Wallis; P < 0.05), obtaining virtually the same statistical outcome. Expression values generated with dchip 1.3 (10) or rma (11, 12) software with PM-only models were analyzed with sam using the permuted unpaired two-class test (13). ESC genes were defined as genes whose expression levels in LgA rats were significantly different from control and ShA rats (ANOVA; P < 0.05, Fischer's post hoc test for mas4 and mas5 or sam, with a median number of false significant < 1 for dchip and rma), regardless of whether they were significantly different between control and ShA rats (Fig. 4 and Table 1, which is published as supporting information on the PNAS web site). Conversely, genes whose expression levels differed significantly from control in ShA and LgA but were not significantly different between ShA and LgA were defined as being associated only with cocaine self-administration (CSA genes) but not with escalation of cocaine intake. Global normalization was applied to mas4 and mas5 data. With dchip, we normalized to the invariant set of genes across all chips in the experiment (10). With rma, we subtracted the background from PM values, performed quantile normalization, and generated log2-transformed expression values. Negative expression levels occurring in mas4 (12, 14) were converted to 0.01 rather than 0 to avoid calculation artifacts, such as fold change ratios equal to infinity (15). The following three empirically defined additional filtering criteria were also applied: a minimum fold change of 1.4 for mas4 and mas5 data and 1.2 for dchip and rma, consistent with the criteria of other studies (16–19); genes labeled absent in all chips by the mas5 “detection call” algorithm were filtered out in all four analysis strategies; and genes that did not display an average expression level of at least 100 in at least one condition (after scaling to a target intensity of 250) were also excluded in mas4 and mas5. This minimum expression level filter was not applied to dchip or rma because of their lower levels of noise. These empirical filters were validated by initial RT-PCR results. Gene lists were annotated by use of National Center for Biotechnology Information EN-TREZ Nucleotide and Unigene databases and National Institutes of Health david (20). Biological themes were explored with National Institutes of Health ease (21) and the Kyoto Encyclopedia of Genes and Genomes (22).
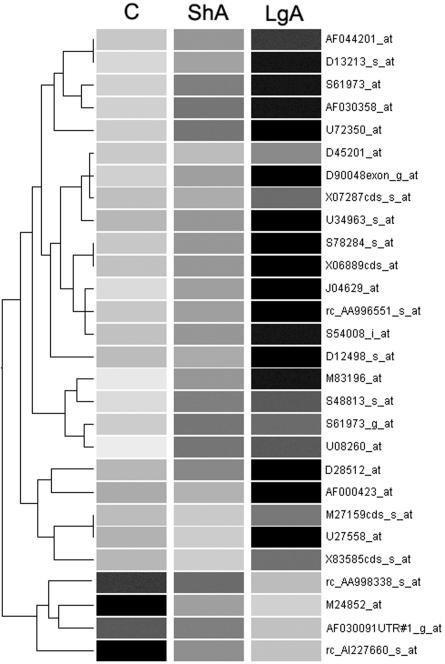
Fig. 4.
Hierarchical clustering of genes specifically associated with escalated cocaine intake (ESC genes) obtained using mas5-generated expression values. The dendrogram was generated by using genespring 4.2.1. The first node separates genes up-regulated from genes down-regulated in LgA rats relative to controls (C) and ShA rats. Darker shades of gray represent higher expression levels.
RT-PCR Validation of Microarray Results. For RT-PCR validation, an independent experiment was conducted (n = 5 in all groups) in the same manner described above (“Behavioral Procedures”). RNA was extracted from the aforementioned brain regions of individual animals, and cDNAs were obtained as described above. Sixty-five genes (represented by 78 probe sets) were tested by RT-PCR in cDNA samples from individual animals. Primers were designed with beacon designer software (PREMIER Biosoft International, Palo Alto, CA). iQ SYBR Green Supermix (Bio-Rad) was used in a 25-μl reaction volume with a MyiQ real-time PCR detection system (Bio-Rad) using 0.2-ml, 96-well, thin-wall PCR plates from Bio-Rad. The relative amounts of mRNA were normalized to β-actin with the exception of the VTA for which Na+/K+-ATPase α1-subunit gene (Atp1a1) was used because its expression proved more invariant than β-actin's in this region.
Results
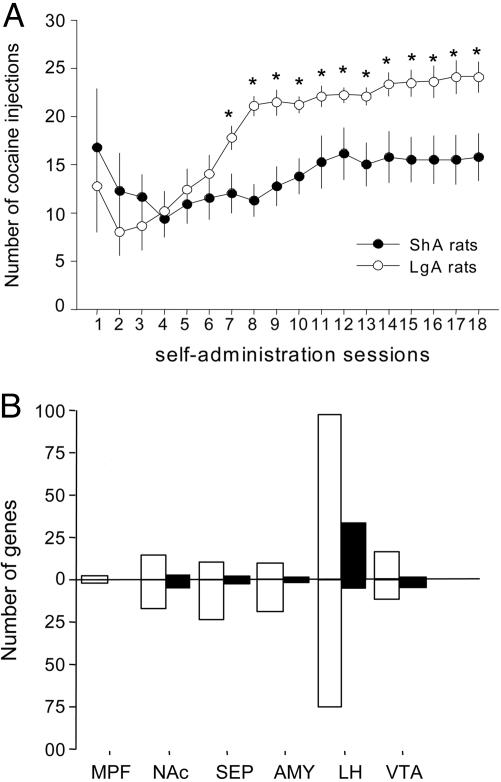
The experimental design for this study comprised three groups of rats (n = 8) with different daily access to a continuous schedule of i.v. cocaine self-administration (0.25 mg per injection): 0 h access per day (drug-naive control rats), 1 h access per day (ShA rats), or 6 h access per day (LgA rats) (see Materials and Methods). As expected, in rats allowed to self-administer cocaine, the duration of access dramatically influenced cocaine intake. Within 18 days, cocaine intake in the first hour in LgA rats rose to a level almost 2-fold greater than that observed in ShA rats, which remained stable over time (Fig. 1A). Total cocaine intake in LgA rats also increased over the same period from an initial average of 48 to an average of 126 cocaine injections over the 6-h period. Gene expression profiling was then performed for each dissected brain region by using Affymetrix rat neurobiology arrays. This array is representative of known neurotransmitter receptors, transporters, synthetic and metabolic enzymes, signal transduction proteins, and other brain-specific transcripts. To validate the microarray results, 65 genes (represented by 78 probe sets) were tested by RT-PCR in individual animals from an independent experiment conducted with the same procedure as above (Materials and Methods, Behavioral Procedures). All of the genes identified as ESC genes with all four analysis strategies were confirmed by RT-PCR as such. Of the genes identified as ESC with at least two analysis strategies, >70% were confirmed by RT-PCR. Of these genes, all of the ones that were not confirmed as ESC genes proved nevertheless to be CSA genes. Of the genes identified as ESC genes with only one of the four analysis strategies, ≈62% were confirmed as ESC genes, with ≈50% of those remaining belonging to the CSA class. For this reason, all of the genes identified with only one of the four analysis strategies were further analyzed by RT-PCR (Table 1, which is published as supporting information on the PNAS web site). These data support the value of using multiple analysis strategies both to identify the most robust changes in gene expression, and to compensate for the biases that affect each strategy, as noted by others (23, 24).
Fig. 1.
Effects of access time to cocaine self-administration on drug intake and brain gene expression levels. (A) Escalation of i.v. cocaine consumption in rats. Rats had access to cocaine for either 1 h (ShA rats, n = 8) or 6 h per day (LgA rats, n = 8). Data represent the mean (±SEM) number of cocaine injections obtained during the first hour of each daily self-administration session. *, Different from ShA rats; P < 0.05, tests of simple main effects after appropriate two-way analyses of variance. (B) Total number of probe sets per brain region that significantly changed in LgA and ShA rats compared with drug-naive control rats (CSA genes, white bars) and probe sets that significantly changed in LgA rats from ShA and drug-naive rats (ESC genes, black bars). AMY, amygaloid complex; MPF, medial prefrontal cortex.
In all of the brain regions studied, the majority of genes whose expression levels were affected by a history of cocaine self-administration were not differentially affected by the pattern of cocaine intake (stable/moderate in ShA rats vs. escalating/compulsive in LgA rats) (Fig. 1B). Only the expression of a small set of genes was differentially regulated in rats with escalated cocaine intake (Fig. 1B). Of all of the brain regions examined, the LH was the most transcriptionally responsive to cocaine intake escalation (Fig. 1B). Relative changes in expression levels from control in ShA and LgA are plotted vs. each other in Fig. 2. As shown in Table 1, ESC genes can be grouped in five functional categories: (i) genes coding for proteins involved in the regulation of neuronal growth, survival, and functional and structural plasticity; (ii) neurotransmitter receptors, synthetic and metabolic enzymes, and transducers; (iii) genes coding for proteins involved in the regulation of membrane potential, such as ion pumps and channels; (iv) genes involved in the neurotransmitter release machinery; and (v) genes involved in tissue response to insults.
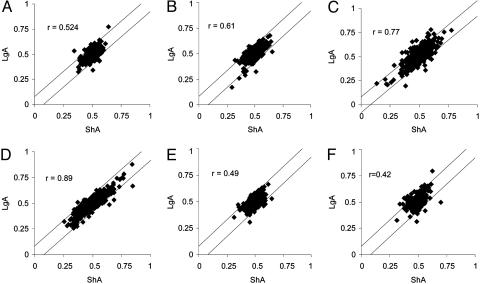
Fig. 2.
Correlation between changes in gene expression levels and patterns of cocaine intake. (A) NAc. (B) Amygdala. (C) LH. (D) Medial prefrontal cortex. (E) SEP. (F) VTA. In LgA and ShA groups, the expression levels corresponding to each probe set were normalized to control levels (drug-naive rats), as described in Materials and Methods, to obtain values ranging from 0 to 1, with 0.5 corresponding to no change from the control level. Only probe sets with average expression levels ≥100 in at least one condition were included in the regression analysis. ESC genes were defined as significantly different in LgA vs. control and LgA vs. ShA, regardless of whether they were significant in ShA vs. controls. Regression analysis showed a positive correlation between the change in expression in ShA from controls and the change in expression in LgA from controls. This correlation suggests that, in all of the brain regions considered, the majority of genes whose expression levels were affected by a history of cocaine self-administration were not differentially affected by the pattern of cocaine intake (stable/moderate in ShA rats vs. escalating/excessive in LgA rats). The diagonal lines in each graph represent the 1.4-fold cutoffs used in the analysis of mas4 and mas5 data (see Materials and Methods for details). In this plot, deviations from the center within the area defined by the diagonal lines indicate changes in ShA and LgA rats from controls (CSA genes). Points outside the area defined by the diagonal lines indicate genes that differed in LgA from ShA and control rats (ESC genes).
Examples of genes involved in structural and functional plasticity that were induced in the LH of LgA rats with escalated drug intake include δ-catenin, microtubule-associated protein (MAP)-1a and the fibroblast growth factor receptor (Table 1). δ-Catenin is a postsynaptic density neuron-specific catenin expressed at high level in the cortex, basal ganglia, and midbrain catecholaminergic nuclei (see the symatlas database of the Genomics Institute of the Novartis Research Foundation at http://symatlas.gnf.org/SymAtlas), and it is involved in dendritic branching (25, 26). The absence of δ-catenin leads to mental retardation in humans and mutant mice (25, 26). It is also involved in the organization of postsynaptic density where it interacts with scaffolding proteins involved in the targeting and trafficking of ionotropic NMDA and metabotropic glutamate receptors (Fig. 3 and Table 1) (27, 28). Additionally, the α1, α3, and β2 isoforms of Na+/K+-ATPase, which are induced during peripheral nerve regeneration (29), were also induced in the LH. The α3 isoform of Na+/K+-ATPase has been linked to bipolar disorder (30). Decreases in MAP-2 and neurofilament light chain in the NAc (Table 1) could be the result of trophic changes in these regions, as suggested by Nestler and colleagues (31). Increased expression of apoptotic and antiapoptotic gene products, such as Bcl-2 and related genes, was observed in various regions (Table 1).
Fig. 3.
Coordinated presynaptic and postsynaptic gene expression changes in the LH after cocaine intake escalation. The levels of the mRNAs for several synaptic proteins were altered in the LH of cocaine-escalating rats. ESC genes are indicated in bold within yellow boxes. The diagram shows their presynaptic and postsynaptic distributions and some of their possible interactions. In some cases, e.g., Map1a and PKCγ, the proteins could be present presynaptically and postsynaptically. Changes in elements of the release machinery were usually increases and could reflect changes in protein content or the result of increased synaptic contacts. SNAP-25, synaptosome-associated protein-25; ERK, extracellular signal-regulated kinase; ANIA-6, activity and neurotransmitter-induced early gene 6; VAMP, vesicle-associated membrane protein; Nf1, neurofibromin; PSD-95, postsynaptic density-95; CamKII, Ca2+/calmodulin protein kinase II; mGlur, metabotropic GluR; NMDAR, NMDA receptor. The differential regulation of several genes related to structural plasticity in the LH of animals with escalated cocaine intake but not in rats with stable cocaine intake suggests that a remodeling of LH circuitry involving synaptogenesis and neuritogenesis contributes to the transition to cocaine addiction.
Among the genes in the neurotransmitter receptors, enzymes, and signal transduction group, the NMDA receptor subunit 2D (NR2D) was increased in rats with escalated levels of cocaine intake. This subunit is predominantly expressed during development and confers slow channel kinetics to the NMDA receptors (32–34). The slow deactivation of the embryonic subunits is believed to lower the temporal threshold for coincidence detection favoring synaptic strengthening during development (32–34). Extrasynaptically located NR2D receptors have been demonstrated (35, 36). It has been proposed that such extrasynaptic NR2D receptors mediate glutamate trophic actions rather than contribute to neural transmission (34). Thus, the increased expression of the embryonic NR2D subunit in the LH of cocaine-escalating rats could also be a hallmark of plastic structural rearrangements.
The PDZ (postsynaptic density-95/Drosophila disk large-tumor suppressor zona occludens 1) domain-containing glutamate receptor (GluR)-interacting protein (GRIP)2 was also increased in the LH (Table 1). GRIP2 binds to the C terminus of the GluR2 and GluR3 subunits of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors and is enriched in synaptic plasma membrane and postsynaptic density fractions, and its expression relatively late in development parallels the expression of AMPA receptors (37, 38). The complex formed by GluR2 and GRIP1 (a protein highly homologous to GRIP2 expressed at earlier developmental stages) has been shown to be capable of directly interacting with kinesin heavy chains in dendrites, acting as motors for AMPA receptors (39). MAP-1a was also induced in the LH of LgA rats (Table 1). This interaction has been proposed to contribute to AMPA receptors targeting to synapses (40). Interestingly, GRIP1 has also been shown to bind to MAP-1a and MAP-1b in the yeast two-hybrid assay, an interaction that could play a role in AMPA receptors' dynamics (40). Other proteins involved in trafficking of glutamatergic receptors and synaptic plasticity, such as Homer (41) and postsynaptic density-95 (42–44), have been shown to be involved in cocaine's actions in other brain regions and models of cocaine exposure, suggesting that cocaine-induced adaptations affect multiple genes involved in glutamatergic receptor trafficking.
The GTPase-activating protein neurofibromin was induced in the LH of LgA rats. Neurofibromin is involved in the regulation of the activity of the low-molecular-weight G protein Ras (45, 46) and cAMP levels (46). Neurofibromin deficiency is associated with impaired synaptic plasticity and cognitive impairments (46, 47). Increased neurofibromin is expected to result in reduced ERK activation (47) and higher cAMP levels (48). The G protein β-subunit 1 (GNB1; also known as rGβ1), was found to be down-regulated in the LH of escalating rats. GNB1 is enriched in the brain, is up-regulated by cocaine and amphetamine in the shell region of the NAc, and it is required for behavioral sensitization induced by repeated administration of psychostimulants (49). GNB1 mediates the fast voltage-dependent inhibition of N-type Ca2+ channels produced by many G protein-coupled receptors (50). Therefore, decreased GNB1 could result in greater neurotransmitter release. GNB1 also inhibits adenylyl cyclase and reduces cAMP levels (51). Therefore, increased neurofibromin and decreased GNB1 could synergize in increasing accumulation of cAMP. Interestingly, GNB1 was increased in the present study in the SEP of LgA rats (Table 1) and in the VTA of ShA and LgA rats (data not shown).
The expression of various presynaptic proteins found at excitatory synapses was affected (generally increased) in the LH of LgA rats (Fig. 3 and Table 1), including proteins involved in vesicle docking and neurotransmitter release, such as Rab-3A, synaptotagmin-3 and -11, syntaxin-1, and some potential regulator of their function, PKCγ (Fig. 3 and Table 1). The presynaptic protein synaptophysin, which is increased by plasticity of excitatory synapses (52), was also induced in the LH (Fig. 3 and Table 1). Synapsin 1 was also induced; however, it did not differ in ShA and LgA and was therefore classified as a CSA gene. These changes could reflect altered levels of presynaptic proteins or increased numbers of synapses in the LH. Synaptotagmin-3 binds phospholipids in a Ca2+-dependent manner, whereas synaptotagmin-11 is one of two members of the synaptotagmin family with reduced Ca2+ sensitivity and that has been proposed to be induced as an adaptive mechanism at times of excessive neuronal stimulation (53). The Bcl-2-related protein Bcl-XL was also increased in the LH. As indicated above, this increase could reflect apoptotic pressure in this brain region; however, Bcl-XL is also enriched in mitochondria of presynaptic terminals and has been shown to increase synaptic transmission (54).
Alterations in the expression of different K+ channels suggest changes in cellular excitability. Particularly in cocaine-escalating rats, the expressions of Kir4.1 (also known as KCNJ10) and Kv1.6 (also known as KCNA6) were increased in the LH, whereas Kv4.2 (also known as KCND2) was decreased in the SEP. These changes in the LH would be expected to result in reduced excitability and could be a counteradaptation to decreased GNB1 and increased neurofibromin, which would be expected to lead to cAMP accumulation and reduced activity of G protein-coupled inwardly rectifying K+ channels, similarly to what has been shown for N-type Ca2+ channels (50).
The transcript for the chemokine fractalkine was also up-regulated in the LH of rats with escalating cocaine use. Fractalkine is a chemokine predominantly expressed in the brain and is believed to be part of a response mechanism to excitotoxic neuronal injuries (55). Fractalkine also reduces glutamate neurotransmission, and its induction could be a response to chronic activation of glutamate-mediated excitatory neurotransmission (55, 56). Also worth noticing is that fractalkine has been found to be up-regulated in the brain tissue from patients with HIV-1 encephalitis (57) and to potently induce the migration of monocytes across endothelial cells (58). Although fractalkine is neuroprotective to cultured neurons exposed to HIV, it has also been proposed to contribute to HIV invasion of the CNS (58). Therefore, increased fractalkine induction by cocaine intake could contribute to the pathogenesis of HIV invasion of the CNS in human cocaine addicts.
Discussion
The goal of the present study was to identify genes specifically associated with escalated and compulsive cocaine intake (ESC genes) by using a microarray-based transcriptional approach. Four different strategies were used to analyze the gene expression data obtained. RT-PCR of individual animal samples from an independent replication of the experiment was used to validate the genes identified. The main finding of the present study is that the expression levels of only a small fraction of genes changed specifically in association with drug intake escalation (ESC genes). ESC genes varied across different brain regions, suggesting that different components of the reward circuitry undergo specific adaptations during escalation of cocaine self-administration. The most dramatic changes were observed in the LH. This observation points to a previously underappreciated importance of this brain area in the development of drug addiction.
Data Analysis Considerations. Multiple strategies for the analysis of microarray data have been devised. However, optimal protocols for gene expression analysis in specific experimental settings remain to be defined. In the present study, we have analyzed expression levels obtained with two versions of Affymetrix analysis software and with sam analysis of dchip- and rma-generated PM-only expression values. The mas4 and mas5 software use a PM-minus-MM model to subtract nonspecific binding from expression values, subtract background, and normalize the data by globally scaling to an arbitrary target intensity; these programs also have additive signal algorithms (12, 14). The mas5 output incorporates an anti-log of robust average to transform negative expression levels, which occur with and are a limitation of mas4 (12, 14). Conversely, dchip normalizes individual probes across arrays to a computed “invariant set” of genes, determined by ranking and iteratively comparing the PM probes across all of the arrays in the experiment or across experiments (10, 59). The arrays in the experiment are then scaled to the median array in the experiment. Signal values are generated with a multiplicative algorithm called Model-Based Expression Indices (10, 59). rma uses a robust average of log2-transformed background-corrected PM intensities combined with a quantile normalization method (11, 12). The use of PM-only expression values in the latter two methods as opposed to the PM-minus-MM model used in the mas algorithms is motivated by the observation that some MM probes can be unreliable indicators of nonspecific binding (10). The comparison of four microarray analysis strategies performed here suggests that all of the analysis strategies used have validity and supports the usefulness of using multiple analysis strategies to identify the most robust changes in gene expression, as noted by others (23, 24), and to compensate for the biases that affect each strategy (24).
Implications of the Remodeling of LH Circuitry in Cocaine Addiction. The LH is a heterogeneous hypothalamic region whose function and neuronal organization are still poorly understood (60). LH comprises intrinsic neuronal populations and circuitry that are bidirectionally connected by the medial forebrain bundle to key reward-related forebrain regions, such as the NAc. The LH is a brain region that is highly rewarding when electrically stimulated (61, 62). Intracranial self-stimulation (ICSS) in the LH is potentiated by drugs of abuse, including cocaine, amphetamine, nomifensine, morphine, heroin, and nicotine (63–69). Conversely, decreased ICSS reward in the LH is seen in drug-dependent animals during withdrawal from cocaine, alcohol, amphetamine, nicotine, and morphine (65, 70–73) and is highly correlated with the development of compulsive cocaine self-administration (4).
The rich array of gene expression changes observed in the LH suggests that the reward deficit seen in rats with compulsive cocaine use could result from a remodeling of the LH intrinsic circuitry, and not only from changes in medial forebrain bundle fibers. For instance, the expression of several synaptic proteins was increased in the LH (Fig. 3 and Table 1) and could reflect a structural reorganization of LH intrinsic circuitry characterized by changes in protein content and/or increased synaptic contacts as seen in some forms of plasticity at excitatory synapses (52). This interpretation is in agreement with the previous observation of the ability of cocaine to increase the density of dendritic spines and synaptic contacts in other brain regions (74).
The role of LH-intrinsic neurons in ICSS reward is also supported by classic studies showing that ICSS in the LH is preserved by unilateral removal of all telencephalic structure and associated ipsilateral precollicular transection (75) and that ICSS is impaired by lesions of LH-intrinsic neurons that spare transhypothalamic aminergic fibers (76, 77). Future studies will be required to elucidate the specific role of LH intrinsic neurons in the reward deficit associated with compulsive cocaine self-administration.
Experimental Design Considerations. Several methodological issues should be considered in interpreting the present data. Increased access to cocaine self-administration results in increasing the levels of operant experience and of drug exposure, both of which could affect the behavioral and molecular changes observed here. Although plausible, a role of operant experience in cocaine intake escalation is unlikely. In fact, extended exposure to cocaine without changes in operant experience is sufficient to induce escalated levels of cocaine self-administration (78). Additionally, gene expression changes were assessed after a period of cocaine withdrawal (48 h) when cocaine has been eliminated from the body and transient gene expression changes are likely to have subsided. It is widely believed that protracted gene expression changes induced by chronic drug self-administration are key in craving and dependence (1). Therefore, changes in brain gene expression at this withdrawal time point are likely to reflect longer-lasting adaptations that may underlie the maintenance of escalated cocaine use. Future studies will be needed to extend these findings to longer withdrawal periods to assess the dynamics and persistence of these neuroadaptations.
In conclusion, the present multianalysis study reports a list of strong candidate genes specifically associated with the development of compulsive cocaine intake. The differential regulation of several genes related to structural plasticity in the LH of animals with escalated cocaine intake but not in rats with stable cocaine intake suggests that a remodeling of LH circuitry involving synaptogenesis and neuritogenesis contributes to the transition to cocaine addiction. Future studies need to focus more on this understudied brain region by specifically targeting the genes identified in this study.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants DA13821, DA017208, and DA004398. D.L. and V.R.-C. were partially supported by Training Grant AA007456 (Neuropsycopharmacology–Multidisciplinary training), S.H.A. was supported by Centre National de la Recherche Scientifique, and V.R.-S. was supported by Italian Ministry of Health Commission for the Monitoring and Control of Doping Grant N. 2002-18.
Abbreviations: AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; GluR, glutamate receptor; GNB1, G protein β-subunit 1; GRIP, GluR-interacting protein; ICSS, intracranial self-stimulation; LgA, long access; LH, lateral hypothalamus; MAP, microtubule-associated protein; mas, microarray suite; MM, mismatch; NAc, nucleus accumbens; PM, perfect match; SEP, septum; ShA, short access; VTA, ventral tegmental area.
Data deposition: The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (NCBI GEO accession no. GSE 3016).
References
- 1.Koob, G. F., Sanna, P. P. & Bloom, F. E. (1998) Neuron 21, 467-476. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, S. H. & Koob, G. F. (1998) Science 282, 298-300. [DOI] [PubMed] [Google Scholar]
- 3.Ahmed, S. H., Kenny, P. J., Koob, G. F. & Markou, A. (2002) Nat. Neurosci. 5, 625-626. [DOI] [PubMed] [Google Scholar]
- 4.Vanderschuren, L. J. & Everitt, B. J. (2004) Science 305, 1017-1019. [DOI] [PubMed] [Google Scholar]
- 5.Paterson, N. E. & Markou, A. (2003) NeuroReport 14, 2229-2232. [DOI] [PubMed] [Google Scholar]
- 6.Koob, G. F. & Le Moal, M. (2001) Neuropsychopharmacology 24, 97-129. [DOI] [PubMed] [Google Scholar]
- 7.Koob, G. F., Ahmed, S. H., Boutrel, B., Chen, S. A., Kenny, P. J., Markou, A., O'Dell, L. E., Parsons, L. H. & Sanna, P. P. (2004) Neurosci. Biobehav. Rev. 27, 739-749. [DOI] [PubMed] [Google Scholar]
- 8.Paxinos, G. & Watson, C. (1998) The Rat Brain in Stereotaxic Coordinates (Academic, San Diego), 4th Ed.
- 9.Hacia, J. G., Brody, L. C., Chee, M. S., Fodor, S. P. & Collins, F. S. (1996) Nat. Genet. 14, 441-447. [DOI] [PubMed] [Google Scholar]
- 10.Li, C. & Wong, W. (2001) Genome Biol. 92, 1-11. [Google Scholar]
- 11.Bolstad, B. M., Irizarry, R. A., Astrand, M. & Speed, T. P. (2003) Bioinformatics 19, 185-193. [DOI] [PubMed] [Google Scholar]
- 12.Irizarry, R. A., Bolstad, B. M., Collin, F., Cope, L. M., Hobbs, B. & Speed, T. P. (2003) Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tusher, V. G., Tibshirani, R. & Chu, G. (2001) Proc. Natl. Acad. Sci. USA 98, 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajagopalan, D. (2003) Bioinformatics 19, 1469-1476. [DOI] [PubMed] [Google Scholar]
- 15.Silico Genetics (2001) genespring Basic Instruction Manual (Silico Genetics, Redwood City, CA).
- 16.Daniels, G. M. & Buck, K. J. (2002) Genes Brain Behav. 1, 35-45. [DOI] [PubMed] [Google Scholar]
- 17.Pass, H. I., Liu, Z., Wali, A., Bueno, R., Land, S., Lott, D., Siddiq, F., Lonardo, F., Carbone, M. & Draghici, S. (2004) Clin. Cancer Res. 10, 849-859. [DOI] [PubMed] [Google Scholar]
- 18.Lockhart, D. J. & Barlow, C. (2001) Nat. Rev. Neurosci. 2, 63-68. [DOI] [PubMed] [Google Scholar]
- 19.Lee, C. K., Klopp, R. G., Weindruch, R. & Prolla, T. A. (1999) Science 285, 1390-1393. [DOI] [PubMed] [Google Scholar]
- 20.Dennis, G., Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R60. [PubMed] [Google Scholar]
- 21.Hosack, D. A., Dennis, G., Jr., Sherman, B. T., Lane, H. C. & Lempicki, R. A. (2003) Genome Biol. 4, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H. & Kanehisa, M. (1999) Nucleic Acids Res. 27, 29-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabakoff, B., Bhave, S. V. & Hoffman, P. L. (2003) J. Neurosci. 23, 4491-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J., Spletter, M. L. & Johnson, J. A. (2005) Physiol. Genomics 21, 43-58. [DOI] [PubMed] [Google Scholar]
- 25.Medina, M., Marinescu, R. C., Overhauser, J. & Kosik, K. S. (2000) Genomics 63, 157-164. [DOI] [PubMed] [Google Scholar]
- 26.Israely, I., Costa, R. M., Xie, C. W., Silva, A. J., Kosik, K. S. & Liu, X. (2004) Curr. Biol. 14, 1657-1663. [DOI] [PubMed] [Google Scholar]
- 27.Jones, S. B., Lanford, G. W., Chen, Y. H., Moribito, M., Kim, K. & Lu, Q. (2002) Neuroscience 115, 1009-1021. [DOI] [PubMed] [Google Scholar]
- 28.Kim, K., Sirota, A., Chen, Y. H., Jones, S. B., Dudek, R., Lanford, G. W., Thakore, C. & Lu, Q. (2002) Exp. Cell Res. 275, 171-184. [DOI] [PubMed] [Google Scholar]
- 29.Kawai, H., Yasuda, H., Terada, M., Omatsu-Kanbe, M. & Kikkawa, R. (1997) J. Neurochem. 69, 330-339. [DOI] [PubMed] [Google Scholar]
- 30.Mynett-Johnson, L., Murphy, V., McCormack, J., Shields, D. C., Claffey, E., Manley, P. & McKeon, P. (1998) Biol. Psychiatry 44, 47-51. [DOI] [PubMed] [Google Scholar]
- 31.Beitner-Johnson, D., Guitart, X. & Nestler, E. (1992) J. Neurosci. 12, 2165-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cull-Candy, S., Brickley, S. & Farrant, M. (2001) Curr. Opin. Neurobiol. 11, 327-335. [DOI] [PubMed] [Google Scholar]
- 33.Monyer, H., Burnashev, N., Laurie, D. J., Sakmann, B. & Seeburg, P. H. (1994) Neuron 12, 529-540. [DOI] [PubMed] [Google Scholar]
- 34.Vicini, S. & Rumbaugh, G. (2000) J. Physiol. 525, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misra, C., Brickley, S. G., Farrant, M. & Cull-Candy, S. G. (2000) J. Physiol. 524, 147-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misra, C., Brickley, S. G., Wyllie, D. J. & Cull-Candy, S. G. (2000) J. Physiol. 525, 299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyszynski, M., Valtschanoff, J. G., Naisbitt, S., Dunah, A. W., Kim, E., Standaert, D. G., Weinberg, R. & Sheng, M. (1999) J. Neurosci. 19, 6528-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong, H., Zhang, P., Song, I., Petralia, R. S., Liao, D. & Huganir, R. L. (1999) J. Neurosci. 19, 6930-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Setou, M., Seog, D. H., Tanaka, Y., Kanai, Y., Takei, Y., Kawagishi, M. & Hirokawa, N. (2002) Nature 417, 83-87. [DOI] [PubMed] [Google Scholar]
- 40.Seog, D. H. (2004) Biosci. Biotechnol. Biochem. 68, 1808-1810. [DOI] [PubMed] [Google Scholar]
- 41.Kalivas, P. W., McFarland, K., Bowers, S., Szumlinski, K., Xi, Z. X. & Baker, D. (2003) Ann. N.Y. Acad. Sci. 1003, 169-175. [DOI] [PubMed] [Google Scholar]
- 42.Elkins, R. L., Orr, T. E., Rausch, J. L., Fei, Y. J., Carl, G. F., Hobbs, S. H., Buccafusco, J. J. & Edwards, G. L. (2003) Ann. N.Y. Acad. Sci. 1003, 386-390. [DOI] [PubMed] [Google Scholar]
- 43.Sanna, P. P. & Koob, G. F. (2004) Nat. Med. 10, 340-341. [DOI] [PubMed] [Google Scholar]
- 44.Yao, W. D., Gainetdinov, R. R., Arbuckle, M. I., Sotnikova, T. D., Cyr, M., Beaulieu, J. M., Torres, G. E., Grant, S. G. & Caron, M. G. (2004) Neuron 41, 625-638. [DOI] [PubMed] [Google Scholar]
- 45.Cichowski, K. & Jacks, T. (2001) Cell 104, 593-604. [DOI] [PubMed] [Google Scholar]
- 46.Arun, D. & Gutmann, D. H. (2004) Curr. Opin. Neurol. 17, 101-105. [DOI] [PubMed] [Google Scholar]
- 47.Costa, R. M., Federov, N. B., Kogan, J. H., Murphy, G. G., Stern, J., Ohno, M., Kucherlapati, R., Jacks, T. & Silva, A. J. (2002) Nature 415, 526-530. [DOI] [PubMed] [Google Scholar]
- 48.Tong, J., Hannan, F., Zhu, Y., Bernards, A. & Zhong, Y. (2002) Nat. Neurosci. 5, 95-96. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X. B., Funada, M., Imai, Y., Revay, R. S., Ujike, H., Vandenbergh, D. J. & Uhl, G. R. (1997) J. Neurosci. 17, 5993-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia, D. E., Li, B., Garcia-Ferreiro, R. E., Hernandez-Ochoa, E. O., Yan, K., Gautam, N., Catterall, W. A., Mackie, K. & Hille, B. (1998) J. Neurosci. 18, 9163-9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McIntire, W. E., MacCleery, G. & Garrison, J. C. (2001) J. Biol. Chem. 276, 15801-15809. [DOI] [PubMed] [Google Scholar]
- 52.Antonova, I., Arancio, O., Trillat, A. C., Wang, H. G., Zablow, L., Udo, H., Kandel, E. R. & Hawkins, R. D. (2001) Science 294, 1547-1550. [DOI] [PubMed] [Google Scholar]
- 53.von Poser, C., Ichtchenko, K., Shao, X., Rizo, J. & Sudhof, T. C. (1997) J. Biol. Chem. 272, 14314-14319. [DOI] [PubMed] [Google Scholar]
- 54.Jonas, E. A., Hoit, D., Hickman, J. A., Brandt, T. A., Polster, B. M., Fannjiang, Y., McCarthy, E., Montanez, M. K., Hardwick, J. M. & Kaczmarek, L. K. (2003) J. Neurosci. 23, 8423-8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chapman, G. A., Moores, K., Harrison, D., Campbell, C. A., Stewart, B. R. & Strijbos, P. J. (2000) J. Neurosci. 20, RC87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sims, K. D., Straff, D. J. & Robinson, M. B. (2000) J. Biol. Chem. 275, 5228-5237. [DOI] [PubMed] [Google Scholar]
- 57.Tong, N., Perry, S. W., Zhang, Q., James, H. J., Guo, H., Brooks, A., Bal, H., Kinnear, S. A., Fine, S., Epstein, L. G., Dairaghi, D., et al. (2000) J. Immunol. 164, 1333-1339. [DOI] [PubMed] [Google Scholar]
- 58.Umehara, H., Bloom, E. T., Okazaki, T., Nagano, Y., Yoshie, O. & Imai, T. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 34-40. [DOI] [PubMed] [Google Scholar]
- 59.Li, C. & Wong, W. H. (2001) Proc. Natl. Acad. Sci. USA 98, 31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swanson, L. W. (2000) Brain Res. 886, 113-164. [DOI] [PubMed] [Google Scholar]
- 61.Olds, J. & Milner, P. (1954) J. Comp. Physiol. Psychol. 47, 419-427. [DOI] [PubMed] [Google Scholar]
- 62.Milner, P. M. (1991) Can. J. Psychol. 45, 1-36. [DOI] [PubMed] [Google Scholar]
- 63.Wise, R. A. (1996) Annu. Rev. Neurosci. 19, 319-340. [DOI] [PubMed] [Google Scholar]
- 64.You, Z. B., Chen, Y. Q. & Wise, R. A. (2001) Neuroscience 107, 629-639. [DOI] [PubMed] [Google Scholar]
- 65.Markou, A. & Koob, G. F. (1991) Neuropsychopharmacology 4, 17-26. [PubMed] [Google Scholar]
- 66.Kenny, P. J., Koob, G. F. & Markou, A. (2003) Behav. Neurosci. 117, 1103-1107. [DOI] [PubMed] [Google Scholar]
- 67.Markou, A. & Koob, G. F. (1992) Physiol. Behav. 51, 111-119. [DOI] [PubMed] [Google Scholar]
- 68.Lin, D., Koob, G. F. & Markou, A. (2000) Pharmacol. Biochem. Behav. 65, 407-417. [DOI] [PubMed] [Google Scholar]
- 69.Bauco, P. & Wise, R. A. (1994) J. Pharmacol. Exp. Ther. 271, 294-301. [PubMed] [Google Scholar]
- 70.Schulteis, G., Markou, A., Cole, M. & Koob, G. F. (1995) Proc. Natl. Acad. Sci. USA 92, 5880-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paterson, N. E., Myers, C. & Markou, A. (2000) Psychopharmacology 152, 440-446. [DOI] [PubMed] [Google Scholar]
- 72.Epping-Jordan, M. P., Watkins, S. S., Koob, G. F. & Markou, A. (1998) Nature 393, 76-79. [DOI] [PubMed] [Google Scholar]
- 73.Schulteis, G., Markou, A., Gold, L. H., Stinus, L. & Koob, G. F. (1994) J. Pharmacol. Exp. Ther. 271, 1391-1398. [PubMed] [Google Scholar]
- 74.Robinson, T. E. & Kolb, B. (1999) Eur. J. Neurosci. 11, 1598-1604. [DOI] [PubMed] [Google Scholar]
- 75.Huston, J. P., Ornstein, K. & Lehner, R. (1982) Brain Res. 245, 187-191. [DOI] [PubMed] [Google Scholar]
- 76.Velley, L., Chaminade, C., Roy, M. T., Kempf, E. & Cardo, B. (1983) Brain Res. 268, 79-86. [DOI] [PubMed] [Google Scholar]
- 77.Velley, L. (1986) Behav. Brain Res. 22, 141-152. [DOI] [PubMed] [Google Scholar]
- 78.Ahmed, S. H., Lin, D., Koob, G. F. & Parsons, L. H. (2003) J. Neurochem. 86, 102-113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.