Abstract
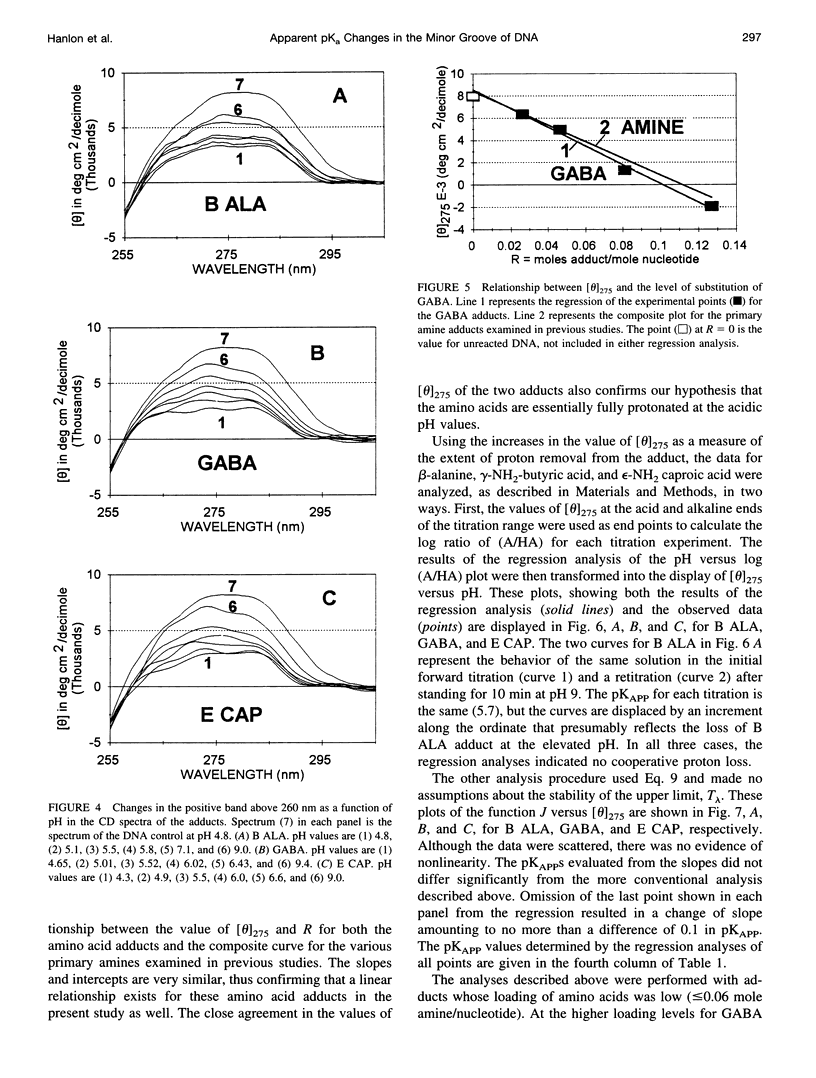
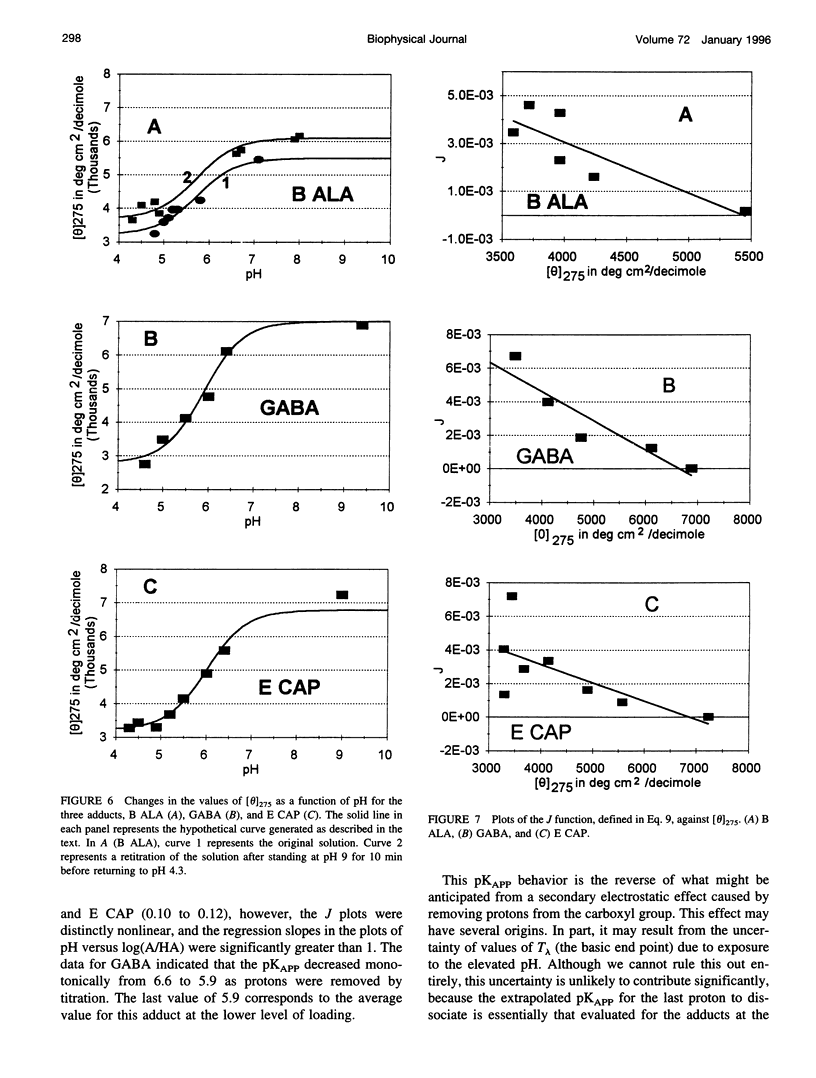
Poisson-Boltzmann calculations by Pack and co-workers suggest the presence of regions of increased hydrogen ion density in the grooves of DNA. As an experimental test of this prediction, we have attached proton-sensitive probes, with variable linker lengths, to random-sequence DNA at G sites in the minor groove. The amino groups of beta-alanine, gamma-aminobutyric acid (GABA), and epsilon-aminocaproic acid have been coupled at pH 5, via a formaldehyde link, to the exocyclic amino group of guanine, utilizing a reaction that has been extensively investigated by Hanlon and co-workers. The resulting adducts at pH 5 retained duplex B form but exhibited typical circular dichroism (CD) changes previously shown to be correlated with the presence of a net positive charge in the minor groove. Increases in the solvent pH reversed the CD spectral changes in a manner suggesting deprotonation of the carboxylic acid group of the adduct. These data were used to calculate an apparent pK(a) for the COOH. The pK(a) was increased by 2.4 units for beta-alanine, by 1.7 units for GABA, and by 1.5 units for epsilon-amino caproic acid, relative to their values in the free amino acid. This agrees well with Poisson-Boltzmann calculations and the energy minimization of the structures of the adducts that place the carboxyl groups in acidic domains whose hydrogen ion density is approximately 2 orders of magnitude greater than that of bulk solvent.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antosiewicz J., McCammon J. A., Gilson M. K. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994 May 6;238(3):415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- Bashford D., Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992 Mar 20;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- Bashford D., Karplus M. pKa's of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990 Nov 6;29(44):10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Pheiffer B. H., Zimmerman S. B., Hanlon S. Conformational characteristics of deoxyribonucleic acid-butylamine complexes with C-type circular dichroism spectra. 1. An X-ray fiber diffraction study. Biochemistry. 1983 Sep 27;22(20):4746–4751. doi: 10.1021/bi00289a020. [DOI] [PubMed] [Google Scholar]
- Chen C., Kilkuskie R., Hanlon S. Circular dichroism spectral properties of covalent complexes of deoxyribonucleic acid and n-butylamine. Biochemistry. 1981 Aug 18;20(17):4987–4995. doi: 10.1021/bi00520a027. [DOI] [PubMed] [Google Scholar]
- Chen C., Ringquist S., Hanlon S. Covalent attachment of an alkylamine prevents the B to Z transition in poly(dG-dC). Biochemistry. 1987 Dec 15;26(25):8213–8221. doi: 10.1021/bi00399a029. [DOI] [PubMed] [Google Scholar]
- Fish S. R., Chen C. Y., Thomas G. J., Jr, Hanlon S. Conformational characteristics of deoxyribonucleic acid-butylamine complexes with C-type circular dichroism spectra. 2. A Raman spectroscopic study. Biochemistry. 1983 Sep 27;22(20):4751–4756. doi: 10.1021/bi00289a021. [DOI] [PubMed] [Google Scholar]
- Jayaram B., Sharp K. A., Honig B. The electrostatic potential of B-DNA. Biopolymers. 1989 May;28(5):975–993. doi: 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- Kilkuskie R., Wood N., Ringquist S., Shinn R., Hanlon S. Effects of charge modification on the helical period of duplex DNA. Biochemistry. 1988 Jun 14;27(12):4377–4386. doi: 10.1021/bi00412a026. [DOI] [PubMed] [Google Scholar]
- Klein B. J., Pack G. R. Calculations of the spatial distribution of charge density in the environment of DNA. Biopolymers. 1983 Nov;22(11):2331–2352. doi: 10.1002/bip.360221103. [DOI] [PubMed] [Google Scholar]
- Lamm G., Pack G. R. Acidic domains around nucleic acids. Proc Natl Acad Sci U S A. 1990 Nov;87(22):9033–9036. doi: 10.1073/pnas.87.22.9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm G., Wong L., Pack G. R. Monte Carlo and Poisson-Boltzmann calculations of the fraction of counterions bound to DNA. Biopolymers. 1994 Feb;34(2):227–237. doi: 10.1002/bip.360340209. [DOI] [PubMed] [Google Scholar]
- Le Bret M., Zimm B. H. Monte Carlo determination of the distribution of ions about a cylindrical polyelectrolyte. Biopolymers. 1984 Feb;23(2):271–285. doi: 10.1002/bip.360230208. [DOI] [PubMed] [Google Scholar]
- Maibenco D., Tang P., Shinn R., Hanlon S. Base and conformational specificity of an amine modification of DNA. Biopolymers. 1989 Feb;28(2):549–571. doi: 10.1002/bip.360280203. [DOI] [PubMed] [Google Scholar]
- Manning G. S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978 May;11(2):179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Misra V. K., Honig B. On the magnitude of the electrostatic contribution to ligand-DNA interactions. Proc Natl Acad Sci U S A. 1995 May 9;92(10):4691–4695. doi: 10.1073/pnas.92.10.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordenskiöld L., Chang D. K., Anderson C. F., Record M. T., Jr 23Na NMR relaxation study of the effects of conformation and base composition on the interactions of counterions with double-helical DNA. Biochemistry. 1984 Sep 11;23(19):4309–4317. doi: 10.1021/bi00314a009. [DOI] [PubMed] [Google Scholar]
- Oberoi H., Allewell N. M. Multigrid solution of the nonlinear Poisson-Boltzmann equation and calculation of titration curves. Biophys J. 1993 Jul;65(1):48–55. doi: 10.1016/S0006-3495(93)81032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack G. R., Garrett G. A., Wong L., Lamm G. The effect of a variable dielectric coefficient and finite ion size on Poisson-Boltzmann calculations of DNA-electrolyte systems. Biophys J. 1993 Oct;65(4):1363–1370. doi: 10.1016/S0006-3495(93)81187-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack G. R., Klein B. J. Generalized Poisson-Boltzmann calculation of the distribution of electrolyte ions around the B- and Z-conformers of DNA. Biopolymers. 1984 Dec;23(12):2801–2823. doi: 10.1002/bip.360231208. [DOI] [PubMed] [Google Scholar]
- Pack G. R., Prasad C. V., Salafsky J. S., Wong L. Calculations on the effect of methylation on the electrostatic stability of the B- and Z-conformers of DNA. Biopolymers. 1986 Sep;25(9):1697–1715. doi: 10.1002/bip.360250912. [DOI] [PubMed] [Google Scholar]
- Pack G. R., Wong L., Prasad C. V. Counterion distribution around DNA: variation with conformation and sequence. Nucleic Acids Res. 1986 Feb 11;14(3):1479–1493. doi: 10.1093/nar/14.3.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellman J. A. Electrical double layer, zeta potential, and electrophoretic charge of double-stranded DNA. Biopolymers. 1977 Jul;16(7):1415–1434. doi: 10.1002/bip.1977.360160704. [DOI] [PubMed] [Google Scholar]
- Wilson R. W., Rau D. C., Bloomfield V. A. Comparison of polyelectrolyte theories of the binding of cations to DNA. Biophys J. 1980 May;30(2):317–325. doi: 10.1016/S0006-3495(80)85097-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A. S., Gunner M. R., Sampogna R., Sharp K., Honig B. On the calculation of pKas in proteins. Proteins. 1993 Mar;15(3):252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]