Abstract
Auxiliary splicing signals in introns play an important role in splice site selection, but these elements are poorly understood. We show that a subset of serine/arginine (SR)-rich proteins activate a cryptic 3′ splice site in a sense Alu repeat located in intron 4 of the human LST1 gene. Utilization of this cryptic splice site is controlled by juxtaposed Alu-derived splicing silencers and enhancers between closely linked short tandem repeats TNFd and TNFe. Systematic mutagenesis of these elements showed that AG dinucleotides that were not preceded by purine residues were critical for repressing exon inclusion of a chimeric splicing reporter. Since the splice acceptor-like sequences are present in excess in exonic splicing silencers, these signals may contribute to inhibition of a large number of pseudosites in primate genomes.
Most eukaryotic genes contain introns that are removed from pre-mRNAs by splicing. The removal of introns occurs in two sequential transesterification reactions that are catalyzed by a large ribonucleoprotein (RNP) complex termed the spliceosome (10). The formation of the spliceosome involves the stepwise assembly of snRNPs (U1, U2, U4/U6, and U5) and a large number of non-snRNP proteins on a pre-mRNA. The spliceosome assembly is characterized by multiple and relatively weak interactions that require conserved cis-acting elements in the pre-mRNA: the 5′ splice site (5′ ss), 3′ ss, polypyrimidine tract (PPT), and the branchpoint sequence (BPS). In higher eukaryotes, these consensus signals are necessary but often insufficient to define exon-intron boundaries and efficient splicing requires auxiliary cis elements that activate or repress splicing, known as exonic splicing enhancers (ESEs) and intronic splicing enhancers or exonic splicing silencers (ESSs) and intronic splicing silencers. These signals allow the genuine splice sites to be correctly recognized among a vast excess of pseudosites that have similar sequences but outnumber authentic splice sites by an order of magnitude (10, 22, 60). The auxiliary signals have been identified through mutations that alter splicing, through computational comparisons, and through selection of sequences that activate or repress splicing or bind to splicing regulatory proteins (16, 23, 61, 65, 68). Exonic sequences, which often regulate both constitutive and alternative splicing through binding of serine/arginine-rich (SR) proteins (6, 27) and are important for exon definition (3), have been studied more extensively than auxiliary signals in introns. In particular, repression of pseudosites by intronic splicing silencers has been poorly understood (22, 60).
Introns contain several classes of repetitive elements that have been shown to influence pre-mRNA splicing. Short tandem repeats (STRs), or 1- to 6-bp iterative motifs called microsatellites, were reported to affect pre-mRNA splicing of at least four human genes if located close to the 3′ or 5′ ss (1, 25, 28, 33, 51). Alu repeats, the largest family of mobile elements in the human genome (2), can be exonized by a single or two point mutations if they are in the antisense orientation (44, 47, 58) and provide a source of ready-to-use segments with a coding potential (45, 46). In yeast (Saccharomyces cerevisiae), extensive self-complementarity in intronic sequences has been shown to promote exon definition (32), suggesting that repetitive sequences could have a more general and underappreciated role in gene expression.
The LST1 gene encodes a leukocyte-specific transcript with a predominant expression in monocytes and dendritic cells (31, 52). LST1 generates at least 14 alternatively spliced variants (designated LST1/A to LST1/N) that encode transmembrane as well as soluble molecules (53). The predicted polypeptides vary with regard to the presence of the N- and C-terminal sequences and have been shown to differentially stimulate lymphocyte proliferation, suggesting that LST1 plays a role in the immune system (19, 53). Overexpression of LST1 isoforms that contain the transmembrane domain induced the formation of long filopodia and microspikes at the cell surface (52), but the exact function of the gene is not understood. LST1 is located in the major histocompatibility complex class III region centromeric of the gene for tumor necrosis factor alpha (TNF-α) in an area predicted to encode genes that have a role in mRNA processing (42). The LST1 gene contains well-known STRs that were termed TNFd and TNFe (63) and mapped close to the alternative 3′ ss of intron 4 (31).
Here, we have investigated a role of TNFd and surrounding sequences in alternative splicing of LST1. We show that an SR-induced activation of a cryptic splice acceptor located in a sense Alu repeat between TNFd and TNFe is influenced by a downstream Alu-derived segment that contains juxtaposed splicing silencers. Systematic mutagenesis of these elements in a heterologous context revealed that AG dinucleotides that were not preceded by purine residues were critical for the inclusion of Alu-derived sequences in mature transcripts. Since these acceptor-like sequences are overrepresented in ESSs, we propose that they play an important role in suppressing a large number of pseudosplice sites in the primate genomes.
MATERIALS AND METHODS
Minigene constructs and site-directed mutagenesis.
The LST1 minigene was cloned into pCR3.1 (Invitrogen) with primers 1F (5′-AACTGTTGGAGAGGGAATCTGAGA) and 1R (5′-ATTCAAAGGTCAAAAAGCCACATA) (Fig. 1A). This minigene contains TNFd allele d3 characterized by a (AG)14GG(AG)9 repeat. The allelic structure of TNFd and TNFe is shown in Table S1 in the supplemental material. The XPC construct, which was derived from the human gene deficient in xeroderma pigmentosum (group C) and contains exons 3 through 5, was cloned into EcoRV/XbaI sites of pCR3.1 with primers 5′-CGTAGATATCATCTTCCTTCCTGTGGTCTTTACAC and 5′-TAGCTCTAGACTTCCCCATCATTCCTTGCCCTTAC. The truncated XPC construct XPC-T was created with mutagenic primers xpc-din3-f (AAGAAGGCTGAGACTGTCTGGGTTGTGTGG) and xpc-din3-r (CAGACAGTCTCAGCCTTCTTTGGTAACTTG). The TH minigene, which contains exons 11 through 13 of the human gene for tyrosine hydroxylase, was cloned into EcoRI/XbaI sites of the same expression vector using primers 5′-GACTGAATTCGGCTGCCTCTCCCCATCCTTC and 5′-GATCTCTAGAAGCAGCCTAGCCCACCTGAGC. The chimeric XPC and TH minigenes were prepared by inserting Alu-derived LST1 segments 24 nucleotides (nt) downstream of the 3′ ss of XPC intron 3 and TH intron 11. An ASF/SF2 cDNA clone containing a 21-nt deletion (CGGCGGGGGTGGAGGTGGCGG) that removes a heptaglycine stretch between RNA recognition motif 1 (RRM1) and RRM2 was obtained by amplifying cDNAs prepared from HeLa cells with primers A1 (ATCCTCGAGATGTCGGGAGGTGGTGTGATT)and A2 (ATCACGCGTCAACATGGGTTCTACAAAAAG).
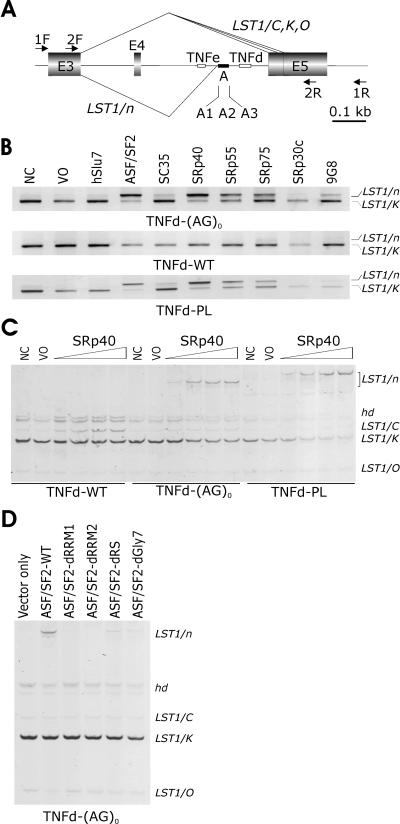
FIG. 1.
SR-dependent activation of a cryptic 3′ splice site in LST1. (A) Schematic representation of the LST1 minigene (to scale; scale units are kilobases). Exons are shown as shadowed boxes; introns are shown as lines. TNFe and TNFd are indicated by small open boxes; segment A is denoted by a closed rectangle. Arrows indicate primers. For a full genomic sequence of this region, see Fig. S1 in the supplemental material. Naturally occurring LST1 isoforms arising by alternative 3′ ss and designated LST1/C, LST1/K (53), and LST1/O (this study) are shown above the minigene, whereas cryptic 3′ ss activation generating LST1/n is shown below. The location of the alternative 3′ ss of intron 4 was as described in reference 53 (and see Fig. S1 in the supplemental material). (B) Cryptic 3′ ss activation in cells overexpressing SR proteins in constructs lacking TNFd. LST1 reporters are shown below each panel, and LST1 RNA products are shown to the right. LST1/K is a predominant isoform in peripheral blood mononuclear cells generated by alternative 3′ ss of intron 4 (53). WT, wild-type; NC and VO; no-cotransfection and vector-only controls, respec-tively. No-template controls are not shown. (C) SR-dependent upregulation of LST1/n and LST1/C. We used 0.5 μg of each reporter plasmid together with 0.1, 0.5, 1.0, and 2.0 μg of plasmids expressing SRp40. Hd, heteroduplexes. (D) Binding of ASF/SF2 to the pre-mRNA is essential for the LST1/n formation. d, deletion of ASF/SF2 domains RRM1, RRM2, and RS or a heptaglycine repeat (Gly7). LST1 isoforms described above are shown on the right side.
Mutated clones were obtained by two-step overlap extension PCR (30). A→G mutations of the predicted branchpoint (BP) adenines of XPC exon 4 and TH exon 12 were introduced with mutagenic primers 5′-ACTATTACTGGTTTTTAAAAA and 5′-CTCTGGGCTGGTGCTGCCCGGC, respectively (mutation is in bold). BPs of both exons were mapped previously in case reports of disease-causing BPS mutations that produced splicing defects (34, 39) and through our own investigation (I.V. et al., in preparation). A 48-bp polylinker (PL) sequence (ACTAGTGCGGCCGCCTGCAGGTCGACCATATGGGAGAGCTCCCAACGC) was used for TNFd replacements to generate constructs TNFd-PL (Fig. 1B). Clone TNFd-(AG)0-A>PL was prepared by replacing segment A (Fig. 1A) with the first 38 bp of PL, except for an A-to-T transversion (in bold in the PL sequence) to destroy a potential 3′ AG. All wild-type and mutated reporter constructs were sequenced using the automated ABI377 sequencer as described previously (64) to confirm intended changes and exclude clones with undesired mutations. ASF/SF2 constructs described previously (12) were a generous gift of G. Screaton (University of Oxford) and J. Cáceres (University of Edinburgh). Wild-type and mutated ASF/SF2 constructs were validated by sequencing.
Cell cultures and transfection.
Transient transfections were performed in six-well (growth area, 9.4 cm2) plates with FuGENE 6 (Roche) as described previously (41). The plating density was 1.5 × 105. When reaching ∼50% confluence, the medium was changed 2 h before adding DNA mixture, which was prepared by combining 3 μl of FuGENE and 50 μl of serum-free medium, followed by addition of plasmid DNA (1 μg, except for cotransfection experiments [see figure legends]) purified with the Wizard Plus SV Minipreps (Promega). The DNA mixture was incubated for 20 min at room temperature prior to transfections. Cells were harvested for RNA extraction 48 h posttransfection. The amounts of plasmids expressing SR proteins are shown in the figure legends.
RNA extraction and detection of RNA products.
Total RNA was extracted using Tri reagent (Sigma) according to the manufacturer's recommendation and measured on a spectrophotometer. Five hundred nanograms of RNA was reverse transcribed with primer PL2 (CCGACTAGTCGCCCAAAT) and Moloney murine leukemia virus reverse transcriptase (RT; Promega) at 42°C. RT was inactivated for 5 min at 95°C. RT-PCR was performed with vector primers PL1 (ACTCACTATAGGGAGACC) and PL2 and validated by a combination of vector and cDNA primers 2F (TCTGTCCGCCTGCCTGTGTTG) and 2R (GGATCCTCCTTGGTGCCTCTC; Fig. 1A). PCR products were separated on polyacrylamide (Fig. 1C) or agarose gels. RNA products were confirmed by sequencing as described previously (64). The relative ratios of RNA products were measured with FluorImager 595 using FluorQuant and Phoretix software packages (Nonlinear Dynamics Inc.) for at least two transfections in duplicate as described previously (41).
RESULTS
SR-mediated activation of a cryptic 3′ ss in LST1 intron 4.
TNFd and TNFe are highly polymorphic STRs located upstream of the alternative 3′ ss of LST1 intron 4 (Fig. 1A). To determine their organization, we sequenced both STRs in cell lines homozygous in the major histocompatibility complex (see Table S1 in the supplemental material). The structure of TNFd was (AG)12-16GG(AG)9-11 on the analyzed haplotypes, whereas TNFe was a simple (AG)7-9 repeat. To test if TNFd, which is located ∼70 bp upstream of the proximal splice acceptor site (Fig. 1A; and see Fig. S1 in the supplemental material), can influence alternative 3′ ss selection, we transfected minigene constructs that contained TNFd allele d3 [(AG)14GG(AG)9, minigene TNFd-WT], a TNFd deletion [TNFd-(AG)0], and a PL sequence replacing TNFd (minigene TNFd-PL) into 293T cells. Examination of the minigene splicing pattern showed that splicing reporters lacking TNFd generated RNA products identical to those observed for the wild-type minigene (data not shown), suggesting that TNFd does not influence splicing.
To test if factors known to affect RNA processing influence selection of alternative 3′ ss, we cotransfected the splicing reporters with a panel of plasmids expressing SR proteins. Coexpression of TNFd-(AG)0 and TNFd-PL, but not the wild-type construct, with ASF/SF2, SRp40, SRp55, SRp75, and, to a minor extent, 9G8 and SC35 in both 293T (Fig. 1B) and HeLa (data not shown) cells activated an upstream cryptic 3′ ss located between TNFe and TNFd. The resulting isoform was termed LST1/n (Fig. 1A, B). The LST1/n expression increased with increasing amounts of plasmids expressing SRp40 (Fig. 1C) and ASF/SF2 (data not shown) cotransfected with TNFd-(AG)0 or TNFd-PL constructs. In addition, we observed upregulation of isoform LST1/C described previously (53) and downregulation of a novel isoform designated LST1/O, consistent with promotion of proximal splicing by a subset of SR proteins (Fig. 1A to C), as described first for ASF/SF2 (40).
Since most SR proteins that produced significant amounts of LST1/n had two RRMs (ASF/SF2, SRp40, SRp55, SRp75) (27) and those lacking the second RRM (SC35, 9G8) generated little LST1/n (Fig. 1B), we tested the importance of their modular domains. We cotransfected the TNFd-(AG)0 construct with the wild-type ASF/SF2 and ASF/SF2 lacking RRM1, RRM2, the C-terminal RS domain (13), and a heptaglycine tract between RRM1 and RRM2 (see Materials and Methods). Deletion of either RRM1 or RRM2 eliminated LST1/n (Fig. 1D). In contrast, deletion of the RS domain and the intervening heptaglycine only reduced the LST1/n yield, suggesting that the SR-induced activation of cryptic 3′ ss depends on binding of SR proteins to the pre-mRNA and only partially on contacts made by the RS domain.
Identification of splicing silencers in sense Alu sequences.
Interestingly, a search for sequences homologous to the region between TNFe and TNFd showed that the SR-induced cryptic 3′ ss was located in the free left Alu monomer (FLAM) (Fig. 2A; see also Fig. S1 in the supplemental material). Alus are dimeric ∼300-nt repeats that probably originated from a fusion between the left and right Alu monomers (35). Alus in antisense orientation are amenable to exonization as they contain a strong PPT and many potential 3′ and 5′ ss, whereas exonization of sense Alus is rare (44, 57, 58). The presence of the cryptic 3′ ss in the sense Alu element (Fig. 2A) thus provided an opportunity to examine Alu-derived sequences that repress splicing.
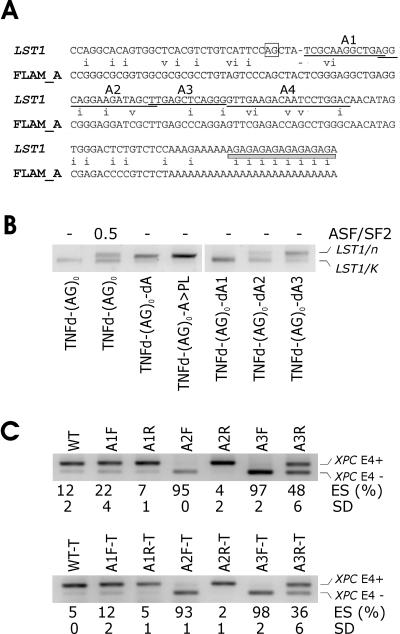
FIG. 2.
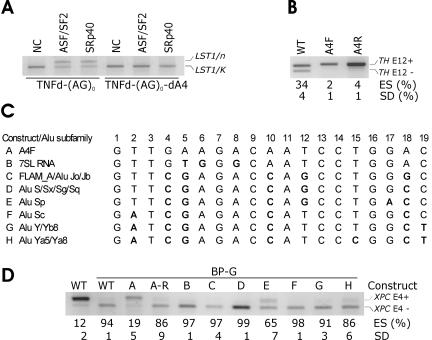
Identification of Alu-derived splicing signals that inhibit LST1/n. (A) Cryptic 3′ ss and segment A sequences are derived from Alu repeats. Sequence alignment was generated by the RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). The cryptic 3′ ss (position 33/34 in the Alu consensus sequence) (2, 37) is boxed. Deleted segments A1 (positions 40 to 51 in the Alu consensus), A2 (positions 51 to 66), A3 (positions 66 to 77), and A4 (positions 78 to 96) are underlined. The telomeric end of TNFd is shown as a gray bar. Mismatches are indicated by the letter i (transition) or v (transversion). (B) Activation of cryptic 3′ ss upon removal of segment A3 or A2. The amount of plasmid expressing ASF/SF2 is in micrograms. Mutated constructs (bottom) are described in the text. d, deletion; >, replacement. PL refers to a polylinker sequence. Expression of LST1/C and LST1/O was negligible and is not shown. Reporter constructs containing the TNFd repeat that lacked segment A were not obtained because of a detection bias against RNA isoforms with this STR (see Fig. 1B and Discussion). (C) A3 and A2 in the sense orientation promote skipping of XPC exon 4. (Upper panel) Mutated minigenes transfected into 293T cells are shown at the top. F and R, forward (sense) and reverse (antisense) orientation, respectively. ES, exon skipping as a ratio of transcripts lacking exon 4 (E4−) to the sum of E4− and E4+ transcripts. SD, standard deviation as calculated from two transfection experiments. (Lower panel) ES following transfection of the truncated (T) XPC minigene carrying identical insertions.
To map cis-elements important for the LST1/n formation, we constructed a splicing reporter lacking a 38-bp segment (termed A) located between the cryptic 3′ ss and TNFd (Fig. 1A; see also Fig. S1 in the supplemental material). Interestingly, transfection of mutated constructs and sequencing of the resulting RNAs showed that this deletion activated the same cryptic 3′ ss in the absence of transiently expressed SR proteins (Fig. 2B). Replacement of this segment with a vector sequence of identical length had the same effect (Fig. 2B). Further mapping of segment A, which was subdivided into three subregions termed A1 to A3 (Fig. 1A and 2A), showed that deletion of A2 and A3 promoted cryptic splicing, whereas deletion of A1 did not (Fig. 2B), suggesting that A3 and A2 contain splicing silencers important for the LST/n repression.
To test how the silencers affect splicing in a heterologous context, we individually inserted A1-to-A3 subregions in both the sense and antisense orientations into the central exon of a minigene carrying exons 3 through 5 of the XPC gene. Intron 3 of the wild-type XPC minigene has a weak 3′ ss and generates an excess of exon inclusion over exon skipping, thus providing a suitable system for testing putative splicing repressors (Fig. 2C). Insertion of A2 or A3 in the sense orientation resulted in almost full exon skipping, whereas insertion of A1 had only a minor effect. In the antisense orientation, A1 and A2 constructs produced exon inclusion levels similar to those of the wild-type XPC, whereas insertion of A3 moderately decreased exon inclusion. Similar levels of exon skipping were produced by a truncated construct (termed XPC-T), in which XPC intron 3 was shortened from ∼2.1 kb to ∼450 bp to facilitate mutagenesis (Fig. 2C, lower panel). In addition to XPC, the insertion of A3 in the sense orientation into a minigene carrying exons 11 through 13 of the human TH gene, which produced a mixture of exon inclusion and exon skipping in the wild type (see further below), also led to exon repression (data not shown). Together, these results suggested that A2 and A3 contained splicing silencers derived from sense Alus.
Importance of AG dinucleotides for A3-mediated exon repression.
To determine which A3 residues are critical for splicing, we systematically mutated each A3 position in minigene XPC-T-A3F to the remaining nucleotides. This XPC-T construct contained a 12-nt segment A3 that was inserted in the sense orientation in the central XPC exon. Transfection of mutated constructs into 293T cells and examination of exon inclusion showed that, unlike any other A3 dinucleotides, each single substitution of A9G10 dramatically reduced exon skipping (Fig. 3A) as compared to the wild-type XPC-T-A3F. Similarly, mutations of A4G5 to cytosines significantly enhanced exon inclusion (Fig. 3A). In contrast, mutations at positions 3 and 6 led only to a minor decrease of exon skipping, whereas the remaining substitutions had an intermediate effect (Fig. 3A). Of 10 non-C A3 positions, cytosine replacements in at least 6 of them showed a higher exon inclusion than the remaining nucleotides. These results indicated that AG dinucleotides were core A3 residues that promoted exon repression and that cytosines, which are underrepresented in ESSs (65), were powerful enhancers of exon inclusion in most A3 positions.
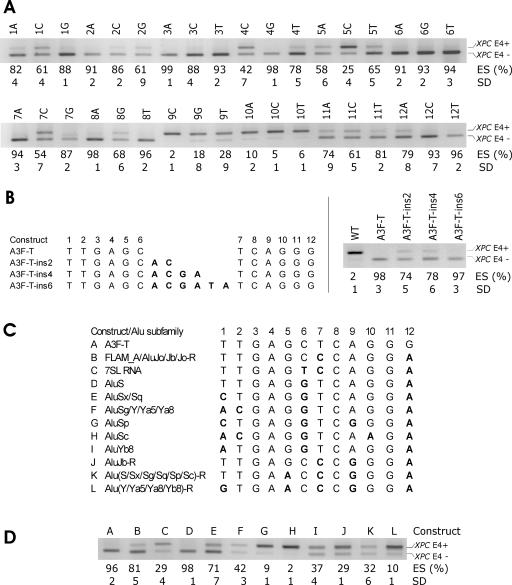
FIG. 3.
Characterization of Alu-derived splicing silencer A3. (A) Exon inclusion levels following transfection of mutated A3 in XPC exon 4. Point mutations above each panel are numbered according to the A3 position as shown in panels B and C. ES, exon skipping as a ratio of transcripts lacking exon 4 (E4−) to the sum of E4− and E4+ transcripts. SD, standard deviation of two transfection experiments. (B) The influence of AG-AG distance in segment A3 on inclusion of a heterologous exon. (Left panel) Two-, 4-, and 6-nt insertions in the XPC-T-A3F construct. The XPC exon 4 sequences flanking A3 were GTGCTGGGTG (upstream) and ACGTGAGAGA (downstream). (Right panel) Exon inclusion levels from a single transfection experiment in duplicate. (C) Alignment of LST1 A3 segment and consensus sequences of Alu subfamilies. Alu sequences were derived from the left arm (2, 37), except for those denoted by -R (the right arm of Alu). The designation of Alu subfamilies was as described previously (2, 37). Mutations are shown in bold. (D) The influence of minigene mutations in A3 on exon inclusion. Constructs A to L shown at the top correspond to mutations listed in Fig. 3C.
Since A4G5 and A9G10 were separated by only three nucleotides, their repressive function might be related to interplay between the two closely linked AGs, similar to the effect of neighboring AGs on 3′ ss selection (18, 44). To examine whether the gap between the two AGs affects exon inclusion, we extended the A4G5-to-A9G10 distance in the XPC-T-A3F minigene from 5 to 7, 9, and 11 nt as shown in Fig. 3B. Constructs extended by 2 and 4 nt resulted in minor increase of exon inclusion levels, whereas further extension eliminated the difference. Thus, extending the AG-AG distance up to 9 nt slightly weakened repressive effect on exon inclusion, although we could not exclude an inhibitory influence of short insertions per se.
Splicing activity of A3 sequences derived from Alu subfamilies.
Alu repeats are composed of J, S, and Y families that are further categorized into subfamilies based on their sequence variability (2, 37). Alignment of LST1 segment A3 and sequences representative of Alu subfamilies (Fig. 3C) showed that nucleotides 1, 6, and 12, which were not essential for exon inclusion (Fig. 3A), were in the most variable positions. To test how natural variability of A3-like Alus affects splicing, we mutated the XPC-T-A3F minigene to create constructs that corresponded to consensus sequences of Alu subfamilies (Fig. 3C). Interestingly, splicing reporters with variations in positions 9 and 10 showed the greatest reduction of exon skipping, whereas minigenes with mutations in positions 1, 6, and 12 showed no or only minor alterations. A3 segments of Alu subfamilies that did not contain A9G10 induced only minor or moderate exon skipping (splicing reporters G, H, J, K, and L in Fig. 3C and D). Thus, A3 derived from FLAM_A, the left arm of AluJo/Jb/S/Sx/Sq, and the right arm of AluJo remained strong inhibitors of exon inclusion, whereas segments representing the remaining Alu subfamilies were less efficient silencers.
Characterization of Alu-derived splicing silencer A2.
The importance of two AGs in A3 led us to examine the contribution of four AGs in segment A2 to exon skipping (Fig. 1A and 2A). We analyzed exon skipping in the truncated XPC construct, in which segment A2 was inserted in the central exon in the same position as A3. The splicing reporter (termed XPC-T-A2F) was mutated in each AG. Figure 4A shows that guanosine-to-cytosine transversions in the first two AGs markedly diminished exon skipping. The same mutation in the fourth AG also reduced exon skipping; however, mutation of the third AG showed little effect.
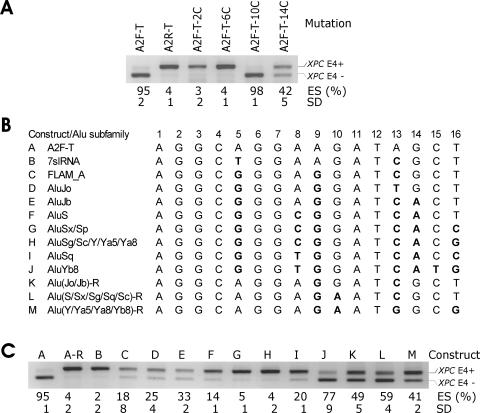
FIG. 4.
Characterization of splicing silencer A2. (A) The influence of A2 mutations in four AGs on exon inclusion. The splicing reporter constructs are shown at the top. Guanine-to-cytosine mutations are in positions shown in panel B. ES, exon skipping as a ratio of transcripts lacking exon 4 (E4−) to the sum of E4− and E4+ transcripts. SD, standard deviation of two transfection experiments. (B) Alignment of LST1 segment A2 with consensus sequences of Alu subfamilies. Mutations (in bold) that corresponded to sequence variations in the subfamilies were introduced in segment A2 inserted in the XPC-T construct in the sense orientation. The designation of Alu subfamilies was as described previously (2, 37). (C) Influence of A2 variants representing Alu subfamilies on inclusion of XPC exon 4 in mRNA. Constructs A to M shown at the top correspond to mutations listed in Fig. 4B. A-R, constructs with segment A2 inserted in the antisense orientation.
To compare the influence of A2 segments that were representative of Alu subfamilies, we prepared a series of mutated clones B to M (Fig. 4B), transfected them into 293T cells, and examined exon inclusion. Interestingly, exon inclusion levels correlated with the number of intact AGs (r = 0.66). Correlation was higher (r = 0.85) if minigene J, which contains a mutation in position 15 important for splicing, was disregarded. The presence of three remaining AGs (clones K to M) generated about equal mixtures of exon inclusion and skipping, whereas the presence of only one AG was associated with low levels of skipping, except for AluYb8. Constructs containing two AGs (C to E, Fig. 4C) produced intermediate levels. Together, most A2 sequences representative of the left arm of Alu showed only a minor increase of exon skipping, whereas those that were characteristic of the right arm were more active.
Identification of a downstream splicing enhancer in sense Alu.
Comparison of the newly exonized sequences in LST1/n with octamer (68)- and RESCUE (23)-derived enhancers revealed a cluster of putative exonic splicing enhancers (designated A4; see Tables S2 and S3 in the supplemental material) located immediately downstream of segment A3 (Fig. 2A). We deleted the last 16 nt from segment A4 in the LST1 TNFd-(AG)0 minigene, leaving the first 3 nt in the construct to maintain the flanking sequence of A3 intact. Interestingly, transfection of this minigene into cells transiently expressing ASF/SF2 or SRp40 did not induce LST1/n (Fig. 5A), indicating that this element was critical for SR-induced activation of the cryptic 3′ ss. Insertion of A4 in both orientations into exon 12 of the TH minigene markedly increased exon inclusion (Fig. 5B), and insertion of both sense and antisense A4 segments into XPC exon 4 virtually eliminated exon skipping (data not shown). Since there was no significant difference in exon inclusion between the sense and antisense orientations of A4 in either minigene, we compared the strength of both inserts further in an XPC construct with weakened 3′ ss. This construct was created by mutating the XPC-T minigene in the BP adenosine that was colocalized previously with a disease-causing mutation in a patient with xeroderma pigmentosum (39). The A→G mutation in predicted BP in the wild-type XPC-T minigene increased exon skipping to over 90% (Fig. 5D). Insertion of A4F into the XPC-T construct with the BP A→G mutation restored exon 4 inclusion to a level comparable to the wild-type XPC-T minigene, whereas A4R showed no effect, indicating that A4F is a stronger splicing enhancer than A4R. Similarly, insertion of A4F into the TH minigene carrying an A→G transition of the predicted exon 12 BP (34, 41) rescued splicing (data not shown). Together, these results suggested that A4 contains a splicing enhancer derived from a sense Alu that promotes exon inclusion in a heterologous context.
FIG. 5.
Identification and characterization of Alu-derived splicing enhancer A4. (A) Deletion of A4 in TNFd-(AG)0 constructs eliminated the expression of SRp40- and ASF/SF2-induced LST1/n. We used 1 μg of plasmids expressing the two SR proteins and 0.5 μg of the reporter plasmid. NC, no-cotransfection controls. No-template controls are not shown. (B) Insertion of A4 into the TH minigene in both sense and antisense orientations resulted in full exon inclusion. The insertions were made in TH exon 12. ES, exon skipping as a ratio of transcripts lacking exon 12 (E12−) to the sum of E12− and E12+ transcripts. WT, wild type. SD, standard deviation as calculated from two transfection experiments. (C) Alignment of LST1 segment A4 with consensus sequences of Alu subfamilies. Mutations (in bold) that corresponded to sequence variations in the subfamilies were introduced in segment A4 inserted in the XPC-T construct in the sense orientation. The designation of Alu subfamilies was as described previously (2, 37). (D) Splicing of the XPC-T minigene containing the branchpoint A→G mutation could be rescued by A4F but not by A4 sequences representing most Alu subfamilies. BP-G, constructs containing the adenine-to-guanine mutation in the predicted branchpoint of XPC exon 4. Constructs A to H shown at the top correspond to mutations listed in panel C. A-R, construct with segment A4 inserted in the antisense orientation.
Finally, to determine the extent to which sequence variability in segment A4 in Alu subfamilies influences splicing, we transfected the XPC-T minigene with the BP A→G transition that was further mutated in A4 as shown in Fig. 5C. In contrast to A4F, which rescued splicing to a level comparable to that of the wild type (Fig. 5D), most A4 sequences derived from other Alu subfamilies did not generate significant exon inclusion, with AluSp-derived A4 exhibiting a moderate effect.
DISCUSSION
We have shown the first examples of splicing silencers and enhancers in sense Alu repeats, providing additional evidence for splicing regulatory sequences in these mobile elements, first reported for antisense Alus (22, 60). Alus are primate specific and are present in hundreds of thousands of copies in the human genome, suggesting that splicing silencers identified in this study contribute to repression of a large number of pseudoacceptor sites and that these repeats may provide a substantial reservoir of sequences that inhibit splicing. Since the Alu-derived splicing silencers inhibited utilization of a cryptic 3′ ss located in the same repeat, they function to eliminate Alu exonization.
Examination of chimeric splicing reporters that contained the newly identified inhibitory sequences highlighted the importance of AG dinucleotides in splice site repression. AGs are commonly found in splicing silencer elements of both mammalian (17, 21, 26, 38, 49) and viral (4, 54, 59, 62, 70) pre-mRNAs and are also common in groups B and D to G of ESSs that have been recently identified through comprehensive fluorescence-activated screens (FAS) (65). Although they were neither over- nor underrepresented among FAS ESSs (65), only 16 of 85 (18.8%) informative AGs found in 133 nonrepetitive FAS ESS decamers were preceded by a purine, whereas the majority of the remaining AGs had uracil in this position (67/69; 97.1%). Similarly, only three of 79 (3.8%) informative FAS ESS AGs were preceded by purine dinucleotides, whereas 38/79 (48.1%) FAS ESS AGs were preceded by pyrimidine dinucleotides (P < 10−4, assuming independence).
The occurrence of nucleotides that precede AGs in FAS ESSs (T67>A13>G3>C2) is similar to a C>T>A>G hierarchy observed for the vertebrate 3′ ss (48, 56, 67), except for a depletion of cytosines in this position among FAS ESSs. The efficiency of CAG 3′ ss in in vitro splicing reactions was comparable to that of UAGs (56), raising speculation that a higher proportion of cytosines versus uracils in position −3 of authentic 3′ ss might be linked to a silencer effect of UAG pseudoacceptors. In addition to FAS ESS, UAGs predominate also in other ESSs (4, 21, 36, 38, 54, 59, 66), although winner decamers identified by selection experiments contained cytosines in this position (22). Similarly, our inspection of previously identified ESSs (reviewed in reference 69) showed that AGs preceded by dipurines were extremely rare, whereas RRAGs dominated purine-rich ESEs. This is consistent with the observed lack of efficient exonization in our splicing reporter mutated in the third AG of A2, which was preceded by two purines (Fig. 4A). In addition, A3 mutations 3T and 8T that introduced uracils in front of the critical AG dinucleotides were associated with almost full exon skipping, whereas an A3 2G mutation, which creates a dipurine in front of the same AG, increased exon inclusion (Fig. 3A).
The importance of AGs in repression of 3′ ss selection is also supported by rare cases of disease-causing splicing mutations that create new AGs between BPS and authentic 3′ AG. Although they usually create de novo acceptor sites, as first described for HBB (11), at least several well-documented reports (7, 8, 24, 29) showed that these AGs suppressed authentic acceptor sites, while activating a cryptic 3′ ss further upstream of the BPS (I. Vořechovský et al., in preparation).
Together, the above observations suggest that AGs represent important motifs in the RNA splicing code that governs 3′ ss selection. As for the splice-like sequences proposed to negatively regulate authentic 5′ ss (15, 21, 22, 55, 60, 65), inhibitory pseudoacceptors identified in this study may act in a similar manner, most likely by recruiting components of the spliceosome to inappropriate places where they compete for interactions with genuine 3′ ss. These signals are likely to be partially recognized by splicing factors such as U2 snRNP (36), but may not assemble fully functional splicing complexes (50). The results of our initial experiment, in which the A4G5-A9G10 distance was modified in segment A3 (Fig. 3B), were not inconsistent with the notion that complexes assembled at the authentic 3′ ss and A3 AGs share common spliceosomal components and similar AG-AG measuring mechanisms.
Segment A3 (Fig. 2B and 3B) contains an AGGG motif that was previously shown to interact with several trans-acting factors, including hnRNP A1 (9) and the H family of hnRNPs (5, 14). Interestingly, our inspection of FAS ESS sequences showed that guanosine was the most frequent nucleotide in a position that immediately followed FAS ESS AGs, whereas cytosines and adenines were underrepresented (G44>T33>A2>C1). In addition, all AGs in FAS ESS decamers that were obtained twice in independent transfections were followed by guanine. GG dinucleotides were also overrepresented in two positions that followed FAS ESS AGs (14 among 74 AGs, versus the ∼5 expected; P < 0.001). However, the AGGG motif alone did not appear to explain all the observed effects, since the A-to-G mutation in position 6 of A1 (Fig. 2A), which creates an AGGG signature, increased exon skipping only from ∼12 to ∼30% (data not shown). Our systematic mutagenesis showed (Fig. 3A) that AG dinucleotides in this motif were critical, whereas the second and third guanines in the G triplet were less important for exon repression. Thus, future studies should determine if these trans-acting factors differentially interact with RNAs transcribed from our mutated constructs.
The apparent lack of LST1/n in the wild-type construct (Fig. 1B) could be due to a silencing effect of TNFd on cryptic 3′ ss activation or impaired amplification and/or reverse transcription of repeat-containing transcripts. To address the latter hypothesis, we examined the amplification yield of plasmids with and without TNFd. The yield of PCR products was slightly lower for TNFd-containing plasmids than for those lacking the repeat (43). However, separate transcription of the two plasmids in vitro and repeated treatment with DNase I followed by RT-PCR of mixed cDNAs showed a substantial decrease of signal intensity from TNFd-containing transcript, indicating that the detection bias toward templates lacking TNFd was introduced mainly by the RT step (43; data not shown). The insertion of TNFd into a PY7 construct, which is spliced efficiently in vitro (20), did not reduce in vitro splicing as compared to the wild-type PY7 in 30- to 90-min reactions (43; data not shown). However, this construct has a very strong PPT (20), which is a central splicing recognition element of human introns, and a putative weak silencing effect of TNFd may not be revealed. Thus, rigorous evidence for a direct TNFd role in splicing inhibition will require further studies. TNFd is not a perfect AG repeat since it has two AGGG tetramers both in the middle and at the 3′ end (see Fig. S1 in the supplemental material) that may recruit trans-acting factors that directly contact A3 in the pre-mRNA or may prevent accessibility of the cryptic 3′ ss through secondary structure.
Finally, antisense Alus are amenable to exonization as they contain a strong PPT and many potential 3′ and 5′ ss, whereas exonization of sense Alus is less common due to a lack of strong PPT and a lower number of both 3′ and 5′ ss (44, 57, 58). Since antisense Alus contain sequences that inhibit (22, 60) and enhance (H. Lei, I. Day, and I. Vořechovský, submitted for publication) Alu inclusion in mature transcripts, Alu-derived silencers identified in this study may contribute to a low exonization potential of sense Alus. It will be interesting to test this hypothesis in future studies by modifying splicing consensus signals of naturally occurring sense Alus in human introns.
Supplementary Material
Acknowledgments
This study was supported by grants from the European Commission and the Wellcome Trust, the Karolinska Institute, and the University of Southampton School of Medicine.
We thank C. W. J. Smith, University of Cambridge; G. Screaton, University of Oxford; J. Cáceres, Medical Research Council Human Genetics Unit, Edinburgh; J. Královičová, University of Southampton; M. Boutros, DKFZ; S. Marsh and N. Mayor, Anthony Nolan Research Institute, London; and A. Krainer, Cold Spring Harbor Laboratory, for reagents and/or helpful discussions.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arrisi-Mercado, P., M. Romano, A. F. Muro, and F. E. Baralle. 2004. An exonic splicing enhancer offsets the atypical GU-rich 3′ splice site of human apolipoprotein A-II exon 3. J. Biol. Chem. 279:39331-39339. [DOI] [PubMed] [Google Scholar]
- 2.Batzer, M. A., and P. L. Deininger. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3:370-379. [DOI] [PubMed] [Google Scholar]
- 3.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 270:2411-2414. [DOI] [PubMed] [Google Scholar]
- 4.Bilodeau, P. S., J. K. Domsic, A. Mayeda, A. R. Krainer, and C. M. Stoltzfus. 2001. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 75:8487-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchette, M., and B. Chabot. 1999. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 18:1939-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 7.Brand, K., K. A. Dugi, J. D. Brunzell, D. N. Nevin, and S. Santamarina-Fojo. 1996. A novel A→G mutation in intron I of the hepatic lipase gene leads to alternative splicing resulting in enzyme deficiency. J. Lipid Res. 37:1213-1223. [PubMed] [Google Scholar]
- 8.Bruggenwirth, H. T., A. L. Boehmer, S. Ramnarain, M. C. Verleun-Mooijman, D. P. Satijn, J. Trapman, J. A. Grootegoed, and A. O. Brinkmann. 1997. Molecular analysis of the androgen-receptor gene in a family with receptor-positive partial androgen insensitivity: an unusual type of intronic mutation. Am. J. Hum. Genet. 61:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burd, C. G., and G. Dreyfuss. 1994. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 13:1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burge, C. B., T. Tuschl, and P. A. Sharp. 1999. Splicing of precursors to mRNAs by the spliceosome, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 11.Busslinger, M., N. Moschonas, and R. A. Flavell. 1981. Beta+ thalassemia: aberrant splicing results from a single point mutation in an intron. Cell 27:289-298. [DOI] [PubMed] [Google Scholar]
- 12.Cáceres, J. F., T. Misteli, G. R. Screaton, D. L. Spector, and A. R. Krainer. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cáceres, J. F., G. R. Screaton, and A. R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caputi, M., and A. M. Zahler. 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 276:43850-43859. [DOI] [PubMed] [Google Scholar]
- 15.Carothers, A. M., G. Urlaub, D. Grunberger, and L. A. Chasin. 1993. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol. Cell. Biol. 13:5085-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3:285-298. [DOI] [PubMed] [Google Scholar]
- 17.Chew, S. L., L. Baginsky, and I. C. Eperon. 2000. An exonic splicing silencer in the testes-specific DNA ligase III beta exon. Nucleic Acids Res. 28:402-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chua, K., and R. Reed. 2001. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 21:1509-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Baey, A., B. Fellerhoff, S. Maier, S. Martinozzi, U. Weidle, and E. H. Weiss. 1997. Complex expression pattern of the TNF region gene LST1 through differential regulation, initiation, and alternative splicing. Genomics 45:591-600. [DOI] [PubMed] [Google Scholar]
- 20.Deirdre, A., J. Scadden, and C. W. Smith. 1995. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 14:3236-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Del Gatto, F., and R. Breathnach. 1995. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 15:4825-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fairbrother, W. G., and L. A. Chasin. 2000. Human genomic sequences that inhibit splicing. Mol. Cell. Biol. 20:6816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairbrother, W. G., R. F. Yeh, P. A. Sharp, and C. B. Burge. 2002. Predictive identification of exonic splicing enhancers in human genes. Science 297:1007-1013. [DOI] [PubMed] [Google Scholar]
- 24.Fujimaru, M., A. Tanaka, K. Choeh, N. Wakamatsu, H. Sakuraba, and G. Isshiki. 1998. Two mutations remote from an exon/intron junction in the beta-hexosaminidase beta-subunit gene affect 3′-splice site selection and cause Sandhoff disease. Hum. Genet. 103:462-469. [DOI] [PubMed] [Google Scholar]
- 25.Gabellini, N. 2001. A polymorphic GT repeat from the human cardiac Na+Ca2+ exchanger intron 2 activates splicing. Eur. J. Biochem. 268:1076-1083. [DOI] [PubMed] [Google Scholar]
- 26.Graham, I. R., M. Hamshere, and I. C. Eperon. 1992. Alternative splicing of a human α-tropomyosin muscle-specific exon: identification of determining sequences. Mol. Cell. Biol. 12:3872-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graveley, B. R., K. J. Hertel, and T. Maniatis. 1999. SR proteins are ‘locators’ of the RNA splicing machinery. Curr. Biol. 9:R6-R7. [DOI] [PubMed] [Google Scholar]
- 28.Hefferon, T. W., J. D. Groman, C. E. Yurk, and G. R. Cutting. 2004. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc. Natl. Acad. Sci. USA 101:3504-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higashi, Y., A. Tanae, H. Inoue, T. Hiromasa, and Y. Fujii-Kuriyama. 1988. Aberrant splicing and missense mutations cause steroid 21-hydroxylase [P-450(C21)] deficiency in humans: possible gene conversion products. Proc. Natl. Acad. Sci. USA 85:7486-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 31.Holzinger, I., A. de Baey, G. Messer, G. Kick, H. Zwierzina, and E. H. Weiss. 1995. Cloning and genomic characterization of LST1: a new gene in the human TNF region. Immunogenetics 42:315-322. [DOI] [PubMed] [Google Scholar]
- 32.Howe, K. J., and M. Ares, Jr. 1997. Intron self-complementarity enforces exon inclusion in a yeast pre-mRNA. Proc. Natl. Acad. Sci. USA 94:12467-12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hui, J., K. Stangl, W. S. Lane, and A. Bindereif. 2003. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat. Struct. Biol. 10:33-37. [DOI] [PubMed] [Google Scholar]
- 34.Janssen, R. J., R. A. Wevers, M. Haussler, J. A. Luyten, G. C. Steenbergen-Spanjers, G. F. Hoffmann, T. Nagatsu, and L. P. Van den Heuvel. 2000. A branch site mutation leading to aberrant splicing of the human tyrosine hydroxylase gene in a child with a severe extrapyramidal movement disorder. Ann. Hum. Genet. 64:375-382. [DOI] [PubMed] [Google Scholar]
- 35.Jurka, J., and E. Zuckerkandl. 1991. Free left arms as precursor molecules in the evolution of Alu sequences. J. Mol. Evol. 33:49-56. [DOI] [PubMed] [Google Scholar]
- 36.Kan, J. L., and M. R. Green. 1999. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 13:462-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapitonov, V., and J. Jurka. 1996. The age of Alu subfamilies. J. Mol. Evol. 42:59-65. [DOI] [PubMed] [Google Scholar]
- 38.Kashima, T., and J. L. Manley. 2003. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 34:460-463. [DOI] [PubMed] [Google Scholar]
- 39.Khan, S. G., A. Metin, E. Gozukara, H. Inui, T. Shahlavi, V. Muniz-Medina, C. C. Baker, T. Ueda, J. R. Aiken, T. D. Schneider, and K. H. Kraemer. 2003. Two essential splice lariat branchpoint sequences in one intron in a xeroderma pigmentosum DNA repair gene: mutations result in reduced XPC mRNA levels that correlate with cancer risk. Hum. Mol. Genet. 13:343-352. [DOI] [PubMed] [Google Scholar]
- 40.Krainer, A. R., G. C. Conway, and D. Kozak. 1990. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell 62:35-42. [DOI] [PubMed] [Google Scholar]
- 41.Královičová, J., S. Houngninou-Molango, A. Krämer, and I. Vořechovský. 2004. Branch sites haplotypes that control alternative splicing. Hum. Mol. Genet. 13:3189-3202. [DOI] [PubMed] [Google Scholar]
- 42.Lehner, B., J. I. Semple, S. E. Brown, D. Counsell, R. D. Campbell, and C. M. Sanderson. 2004. Analysis of a high-throughput yeast two-hybrid system and its use to predict the function of intracellular proteins encoded within the human MHC class III region. Genomics 83:153-167. [DOI] [PubMed] [Google Scholar]
- 43.Lei, H. 2004. Ph.D. thesis. Karolinska Institute, Stockholm, Sweden.
- 44.Lev-Maor, G., R. Sorek, N. Shomron, and G. Ast. 2003. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300:1288-1291. [DOI] [PubMed] [Google Scholar]
- 45.Makalowski, W. 2003. Genomics. Not junk after all. Science 300:1246-1247. [DOI] [PubMed] [Google Scholar]
- 46.Makalowski, W., G. A. Mitchell, and D. Labuda. 1994. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 10:188-193. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell, G. A., D. Labuda, G. Fontaine, J. M. Saudubray, J. P. Bonnefont, S. Lyonnet, L. C. Brody, G. Steel, C. Obie, and D. Valle. 1991. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc. Natl. Acad. Sci. USA 88:815-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mount, S. M. 1982. A catalogue of splice junction sequences. Nucleic Acids Res. 10:459-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muro, A. F., M. Caputi, R. Pariyarath, F. Pagani, E. Buratti, and F. E. Baralle. 1999. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 19:2657-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemeroff, M. E., U. Utans, A. Krämer, and R. M. Krug. 1992. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol. Cell. Biol. 12:962-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niksic, M., M. Romano, E. Buratti, F. Pagani, and F. E. Baralle. 1999. Functional analysis of cis-acting elements regulating the alternative splicing of human CFTR exon 9. Hum. Mol. Genet. 8:2339-2349. [DOI] [PubMed] [Google Scholar]
- 52.Raghunathan, A., R. Sivakamasundari, J. Wolenski, R. Poddar, and S. M. Weissman. 2001. Functional analysis of B144/LST1: a gene in the tumor necrosis factor cluster that induces formation of long filopodia in eukaryotic cells. Exp. Cell Res. 268:230-244. [DOI] [PubMed] [Google Scholar]
- 53.Rollinger-Holzinger, I., B. Eibl, M. Pauly, U. Griesser, F. Hentges, B. Auer, G. Pall, P. Schratzberger, D. Niederwieser, E. H. Weiss, and H. Zwierzina. 2000. LST1: a gene with extensive alternative splicing and immunomodulatory function. J. Immunol. 164:3169-3176. [DOI] [PubMed] [Google Scholar]
- 54.Si, Z., B. A. Amendt, and C. M. Stoltzfus. 1997. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 25:861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siebel, C. W., L. D. Fresco, and D. C. Rio. 1992. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev. 6:1386-1401. [DOI] [PubMed] [Google Scholar]
- 56.Smith, C. W. J., T. T. Chu, and B. Nadal-Ginard. 1993. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 13:4939-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorek, R., G. Ast, and D. Graur. 2002. Alu-containing exons are alternatively spliced. Genome Res. 12:1060-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sorek, R., G. Lev-Maor, M. Reznik, T. Dagan, F. Belinky, D. Graur, and G. Ast. 2004. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in Alu exons. Mol. Cell 14:221-231. [DOI] [PubMed] [Google Scholar]
- 59.Staffa, A., and A. Cochrane. 1995. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol. 15:4597-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun, H., and L. A. Chasin. 2000. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 20:6414-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tacke, R., and J. L. Manley. 1999. Determinants of SR protein specificity. Curr. Opin. Cell Biol. 11:358-362. [DOI] [PubMed] [Google Scholar]
- 62.Tomonaga, K., T. Kobayashi, B. J. Lee, M. Watanabe, W. Kamitani, and K. Ikuta. 2000. Identification of alternative splicing and negative splicing activity of a nonsegmented negative-strand RNA virus, Borna disease virus. Proc. Natl. Acad. Sci. USA 97:12788-12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Udalova, I. A., S. A. Nedospasov, G. C. Webb, D. D. Chaplin, and R. L. Turetskaya. 1993. Highly informative typing of the human TNF locus using six adjacent polymorphic markers. Genomics 16:180-186. [DOI] [PubMed] [Google Scholar]
- 64.Vořechovský, I., L. Luo, M. J. Dyer, D. Catovsky, P. L. Amlot, J. C. Yaxley, L. Foroni, L. Hammarstrom, A. D. Webster, and M. A. Yuille. 1997. Clustering of missense mutations in the ataxia-telangiectasia gene in a sporadic T-cell leukaemia. Nat. Genet. 17:96-99. [DOI] [PubMed] [Google Scholar]
- 65.Wang, Z., M. E. Rolish, G. Yeo, V. Tung, M. Mawson, and C. B. Burge. 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119:831-845. [DOI] [PubMed] [Google Scholar]
- 66.Wentz, M. P., B. E. Moore, M. W. Cloyd, S. M. Berget, and L. A. Donehower. 1997. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type 1 induces dominant aberrant splicing and arrests virus production. J. Virol. 71:8542-8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeo, G., S. Hoon, B. Venkatesh, and C. B. Burge. 2004. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl. Acad. Sci. USA 101:15000-15005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, X. H., and L. A. Chasin. 2004. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 18:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng, Z. M. 2004. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11:278-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng, Z. M., M. Huynen, and C. C. Baker. 1998. A pyrimidine-rich exonic splicing suppressor binds multiple RNA splicing factors and inhibits spliceosome assembly. Proc. Natl. Acad. Sci. USA 95:14088-14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.