Abstract
We constructed two site-specifically modified nucleosomes containing an intrastrand cis-{Pt(NH3)2}2+ 1,3-d(GpTpG) cross-link, similar to one formed by the anticancer drugs carboplatin and cisplatin on DNA, and investigated their structures by hydroxyl radical footprinting and exonuclease III digestion. Hydroxyl radical footprinting demonstrated that the presence of the platinum cross-link selects out a specific rotational setting of DNA on the histone octamer core in each of two reconstituted nucleosomes in which the platinum positions differ by half a DNA helical turn. The {Pt(NH3)2}2+ cross-link is situated in a structurally similar location, with the undamaged strand projecting outward, forcing the DNA to adopt opposite rotational settings in its wrapping around the histone octamer in the two nucleosomes. Enzymatic digestion by exonuclease III of the nucleosome substrates revealed that the platinum cross-link affects the translational positioning of the DNA, forcing it into an asymmetric arrangement with respect to the core histone proteins. We suggest that these phasing phenomena may be central to the recognition and processing of platinum-DNA adducts in cancer cells treated with these drugs and possibly may be common to other DNA damaging events.
Keywords: cancer, carboplatin, chromatin, DNA repair, DNA structure
The anticancer drugs cisplatin and carboplatin form intrastrand cross-links on DNA that evoke their antineoplastic activity by inhibiting DNA and RNA polymerases while escaping resistance mechanisms including nucleotide excision repair (NER) (1). In the cell, recognition of the platinum lesion by the repair machinery in the context of chromatin may require exposure of the damaged DNA duplex to the exterior of the nucleosome.
The nucleosome is the fundamental building block of chromatin. Nature has therefore constructed it in such a manner as to lack a preferred positional setting for most sequences of the double-stranded DNA that wraps around the central core of eight histone proteins (2, 3). The rotational setting of DNA on the surface of the histone octamer defines which nucleotides are directed toward the solvent and which are occluded by the histones.
Determining where platinum-DNA cross-links are located within the nucleosome represents an important step in understanding their cellular recognition and processing. A key question is whether the platinum adduct or the DNA sequence determines the rotational setting of a DNA segment on the nucleosome and therefore how the platinum lesion is presented to the replication, transcription, and repair machinery of the cell. To answer this question, we prepared two nucleosomes in which a 1,3-d(GpTpG) cross-link was incorporated site-specifically near the center of the DNA molecule. The sites of platination were placed one-half turn of the DNA helix apart in the two nucleosomal DNA sequences. We then used high-resolution footprinting and exonuclease mapping experiments to reveal the rotational and translational settings of the platinated DNA in the two nucleosomes and the structural environment of the platinum cross-link in the context of the nucleosome.
Materials and Methods
Cisplatin was obtained as a gift from Engelhard. Reagents for DNA synthesis were purchased from Glen Research. T4 polynucleotide kinase, T4 DNA ligase, and exonuclease III were procured from New England Biolabs. [γ-32P]ATP was obtained from Perkin-Elmer. All other reagents were purchased from either Sigma or Mallinckrodt.
Synthesis of Platinated Oligonucleotides. Oligonucleotides were synthesized by using an Applied Biosystems DNA synthesizer (Model 392) and purified by conventional methods. The two platinated oligonucleotides (strands B; see Fig. 1), in which the platinum atom forms a 1,3-cross-link between the two guanine bases of the GTG sequence, were prepared, purified, and characterized as described in ref. 4.
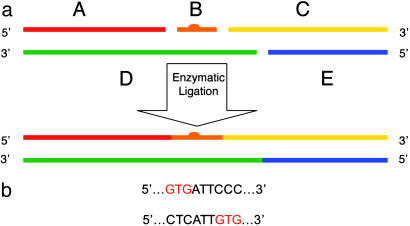
Fig. 1.
Strategy for synthesizing site-specifically platinated DNA molecules. Five synthetic oligonucleotides, one bearing a 1,3-d(GpTpG) intrastrand platinum-DNA cross-link (fragment B), were annealed and ligated as shown. The result is a 146-bp DNA molecule. (a) Scheme for constructing a 146-bp DNA duplex. (b) Sites of platination (GTG) in fragment B for the two nucleosomes studied. The remainder of the sequence, identical in the two nucleosomes, is as follows (platinated strand): 5′-ATCAATATCCACCTGCAGATTCTACCAAAAGTGTATTTGGAAACTGCTCCATCAAAAGGCATGTTCACCXXXATTCYYYTCAACATCGGAAAACTACCTCGTCAAAGGTTTATGTGAAAACCATCTTAGACGTCCACCTATAACTA-3′. The bases that differ between the two sequences used in this study are indicated by X and Y. For sequence 1, XXX = GTG and YYY = CCC. For sequence 2, YYY = GTG and XXX = CTC.
Preparation of 5′-Radiolabeled, Site-Specifically Platinated DNA Molecules. The synthesis of a DNA duplex containing a single site of platination was performed by enzymatic ligation of complementary oligonucleotides, as described in refs. 5 and 6. In brief, an equimolar amount of 5′-32P-radiolabeled strand A (see Fig. 1 a) was combined with the site-specifically cisplatin-modified oligonucleotide (strand B) and the three other oligonucleotide strands. The five strands were annealed and then incubated with T4 DNA ligase at 16°C for 16 h. The resulting ligation products were separated by denaturing PAGE and subsequently isolated by nondenaturing PAGE. Unmodified DNA molecules were synthesized and purified in the same manner as their platinated counterparts.
Nucleosome Reconstitution. The 5′-32P-radiolabeled DNA molecule was assembled into a nucleosome in a manner similar to that in previous reports, except that glycerol was excluded from the nucleosome assembly buffer (4, 7) so as not to interfere with the hydroxyl radical footprinting reaction. Further details of nucleosome characterization are provided in Figs. 8 and 9, which are published as supporting information on the PNAS web site.
Hydroxyl Radical Footprinting. Hydroxyl radical footprinting of nucleosomes was carried out according to published methods (8, 9), omitting the step involving sucrose gradient centrifugation. About 10 fmol of 5′-32P-radiolabeled mononucleosomes were treated with 0.6% H2O2, 1 mM Fe(II)/2 mM EDTA, and 2 mM sodium ascorbate. The footprinting reaction was quenched by addition of glycerol to 5%. The reaction mixture was electrophoresed on a 5% native polyacrylamide gel to separate the nucleosome from residual free DNA. The band containing the radiolabeled nucleosome was excised. The DNA was eluted from the gel slice, purified, and resolved on an 8% denaturing polyacrylamide gel. The presence of a bridging platinum atom in the hydroxyl radical cleavage products made it impossible to appreciate the footprinting pattern around the platination site. Therefore, the cleavage products were treated with 0.2 M NaCN (pH 10.4) at 45°C for 12 h to remove the DNA-platinum cross-links before denaturing PAGE analysis (Fig. 2). This procedure significantly clarified the footprint pattern.
Fig. 2.
Denaturing PAGE analysis of hydroxyl radical footprinting products. In these experiments the platinated DNA strand was radiolabeled at the 5′ end. Hydroxyl radical cleavage products were separated on an 8% denaturing polyacrylamide gel. Lane 1, sequence 1 nucleosome; lane 2, platinated sequence 1 nucleosome; lane 3, sequence 2 nucleosome; lane 4, platinated sequence 2 nucleosome; lane 5, sequence 1 nucleosome, treated with NaCN; lane 6, platinated sequence 1 nucleosome, treated with NaCN; lane 7, sequence 2 nucleosome, treated with NaCN; lane 8, platinated sequence 2 nucleosome, treated with NaCN; lane 9, Maxam–Gilbert G-reaction.
Exonuclease III Mapping of Nucleosome Boundaries. Both free and nucleosomal DNA samples were characterized by determining their exonuclease III digestion stop sites. For each of the nucleosome and free DNA samples, an equal amount of 5′-radiolabeled DNA (≈10 fmol) was treated with 0.5 units of exonuclease III in New England Biolabs Buffer 1 (10 mM bis-Tris propane-HCl/10 mM MgCl2/10 mM DTT, pH 7.0) at 25°C for 2 min. The reaction was quenched by the addition of proteinase K to 2.5 mg/ml and SDS to 0.6%. The DNA digestion products were purified from residual protein by phenol-chloroform extraction and resolved by 8% denaturing PAGE.
Excision Repair Assay. A cell-free extract prepared from CHO AA8 cells was used to measure damage excision (4). Substrate DNA (1.5 to 3 fmol), either free or nucleosomal, was mixed with 50 μg of the cell-free extract at 30°C in 25 μl of excision repair buffer (32 mM Hepes-KOH, pH 7.9/64 mM KCl/6.4 mM MgCl2/0.24 mM EDTA/0.8 mM DTT/2 mM ATP/0.2 mg/ml BSA/5.5% glycerol/4.8% sucrose). The reaction mixture was incubated at 30°C for 3 h. Reaction products were purified by phenol-chloroform extraction and analyzed by denaturing PAGE (8% polyacrylamide/1× TBE). The extent of excision was determined by measuring the levels of radioactivity in the bands of excised products (24–32 nt range) and unexcised substrate, using a PhosphorImager and associated imagequant software (Molecular Dynamics).
Results
Design and Construction of Specifically Platinated Nucleosomes. Whereas some DNA sequences have a high affinity for the histone octamer and form nucleosomes that are well positioned in rotation and translation, most genomic DNA sequences exhibit only moderate affinity and ability to position a nucleosome precisely (10). For our experiments, we chose to study a DNA sequence that does not form a high-affinity, well positioned nucleosome so as to investigate the effect of platination on nucleosome structure by a DNA sequence that is representative of bulk genomic DNA. We based our DNA sequence on one-half of the palindromic duplex used to obtain the first high-resolution x-ray crystal structure of the nucleosome (11). Because the palindromic nature of this sequence would make it difficult to ligate the constituent oligonucleotide fragments together, which is required to construct our site-specifically damaged probes (Fig. 1), we made modifications to produce the ones that we used in this study. The first 67 nucleotides of our sequence are identical to that used for the x-ray structural study, and the last 60 nucleotides are the complement of that sequence. Our DNA was designed so that only one GTG site occurs in the central region, to allow for site-specific platination.
We constructed two 146-bp DNA molecules by ligating five synthetic oligonucleotides together for each (Fig. 1a). Only five base pairs in the central segment (Fig. 1b) differed between the two constructs. A single binding site for cisplatin, GTG, was placed near the middle of this segment and was shifted by six base pairs between the two constructs. The remainder of the DNA sequence in each construct was identical. The central segment was platinated before ligation so as to place a single, site-specific platinum 1,3-d(GpTpG) intrastrand cross-link near the center of the 146-bp DNA molecule. The 5′ end of the platinated strand in the 146-bp DNA molecule was radiolabeled with 32P. We then used standard reconstitution conditions to form a nucleosome on each radiolabeled, site-specifically platinated DNA molecule (4). Control nucleosomes were formed with the same two DNA sequences that had not been subjected to platination. We used hydroxyl radical footprinting (12) to characterize the rotational setting of each of these four DNA molecules when incorporated into a nucleosome (Fig. 2). Further details about these nucleosomes and their characterization are available in Supporting Text, which is published as supporting information on the PNAS web site.
Footprinting and Exonuclease III Digestion Analysis of Nucleosome Structure. Hydroxyl radical cleavage patterns of the two input DNA sequences not in complex with a nucleosome are very similar to each other (Fig. 3A), as expected from their near identity in sequence. The hydroxyl radical footprints of the two control nucleosomes, formed with unplatinated DNA (Fig. 3B), show weakly periodic cleavage patterns that generally are in phase with one another. Such patterns indicate that these two closely related sequences do not form well positioned nucleosomes.
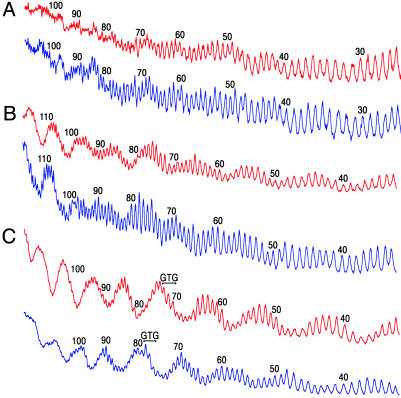
Fig. 3.
Hydroxyl radical cleavage patterns of DNA and nucleosomes. For these experiments, the DNA strand having the platination site (GTG) at the center was radiolabeled on the 5′ end. Footprints plotted in red are those of substrates containing DNA with sequence 1. Plots shown in blue are those of samples containing DNA with sequence 2. (A) Cleavage pattern of free DNA. (B) Hydroxyl radical footprints of unplatinated nucleosomes. (C) Footprints of platinated nucleosomes.
Further evidence supporting this point is provided by exonuclease III digestion experiments on platinated and unplatinated nucleosomes (Fig. 4). Nonplatinated sequences 1 and 2 do not exhibit well defined stop sites, as demonstrated by the extended shoulder on the peak in the trace of sequence 1 (red) and the apparently fused peaks in the pattern for sequence 2 (green). The lack of a well defined stop site is indicative of a heterogeneous population of DNA translational positions on the nucleosomes in these samples. Moreover, most of the DNA in the unmodified nucleosomes is not digested by exonuclease III (Fig. 4, lanes 7 and 9), indicating that the 146-bp DNA molecule is more or less symmetrically disposed around the nucleosome dyad. In contrast, platinated sequence 1 nucleosome (orange) and platinated sequence 2 nucleosome (blue) exhibit sharp peaks corresponding to exonuclease III stop sites. Well defined exonuclease III stops are characteristic of nucleosomes in which the DNA is translationally well positioned.
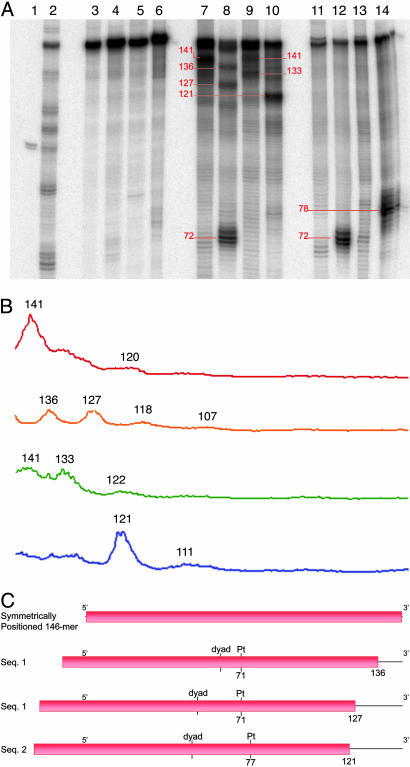
Fig. 4.
Exonuclease III analysis of native and platinated nucleosomes. All four nucleosome samples, and the corresponding free DNA, were treated with exonuclease III to assess the positioning of the DNA on the histone core. (A) Denaturing PAGE analysis (8% polyacrylamide) of exonuclease III digestion products. This analysis reveals several stop sites in the nucleosome samples (lanes 7–10) that are not present in the undigested nucleosome samples (lanes 3–6) or in the digestion products of the analogous free DNA substrates (lanes 11–14). The lengths of each of the major digestion products are indicated in red. Lane 1, 100-nt DNA molecular weight standard; lane 2, Maxam–Gilbert G-reaction; lane 3, undigested sequence 1 nucleosome; lane 4, undigested platinated sequence 1 nucleosome; lane 5, undigested sequence 2 nucleosome; lane 6, undigested platinated sequence 2 nucleosome; lane 7, sequence 1 nucleosome; lane 8, platinated sequence 1 nucleosome; lane 9, sequence 2 nucleosome; lane 10, platinated sequence 2 nucleosome; lane 11, sequence 1 DNA; lane 12, platinated sequence 1 DNA; lane 13, sequence 2 DNA; lane 14, platinated sequence 2 free DNA. (B) Plots of the intensities of bands resulting from exonuclease III digestion of nucleosome samples. The intense band from intact DNA at the top of each lane is omitted from the traces. The approximate lengths of the major digestion products are indicated above each peak. Red trace, sequence 1 nucleosome (lane 7); orange trace, platinated sequence 1 nucleosome (lane 8); green trace, sequence 2 nucleosome (lane 9); blue trace, platinated sequence 2 nucleosome (lane 10). (C) Schematic representation of the translational positioning of the 146-mers on the histone octamer. Exonuclease III digestion of the platinated nucleosomes reveals that the 146-mer DNAs in these substrates are positioned asymmetrically with respect to the histone core. The platinum damage site is not located in the same translational position on the histone octamer for the two different platinated sequences. The diagram illustrates the relative positioning of the 146-mer substrate within each platinated nucleosome based on the locations of its major exonuclease III stoppage sites. The platinated sequence 1 nucleosomes showed two major bands in the exonuclease III digestion pattern, both of which are illustrated in this representation. The pink rectangles denote the position of the histone octamer, and the black lines indicate the locations of the 146-mers.
The hydroxyl radical footprints of the two platinated nucleosomes are remarkably different from those of the unplatinated nucleosomes. A highly modulated cleavage pattern with an ≈10-nt periodicity is apparent for each platinated nucleosome (Fig. 3C), indicating that platination enforces a defined rotational setting on the DNA as it wraps around the histone octamer. Most striking is the observation that the footprints of the two platinated DNAs are almost precisely out of phase with each other. Because the sites of platination are six base pairs apart in the two nucleosomes, it is apparent that the platinum cross-link dominates the rotational setting of the DNA sequence in the nucleosome. The platinum lesion resides in a structurally similar position in each nucleosome, as judged by the footprinting pattern, whereas identical DNA sequences in the two nucleosomes occupy different positions relative to the surface of the histone octamer.
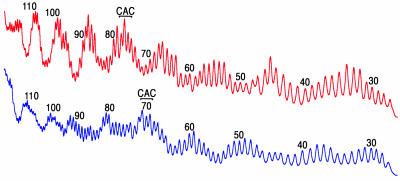
Although the platinum cross-link occupies structurally comparable locations in the two nucleosomes we studied, the cis-diammineplatinum(II) moiety itself is not situated at the most solvent-accessible site on the surface of the core particle (Fig. 3C). Hydroxyl radical footprinting revealed that position 73–74 in one platinated nucleosome, and position 80 in the other, is most exposed to radical attack. In each nucleosome, the center of the platinum cross-link, the T residue in the GTG sequence, is situated 2–3 nt away from the most exposed site, at position 71 in one nucleosome and at position 77 in the other. On the unplatinated strand, however, the complementary CAC sequence is found at the most solvent-exposed portion of the duplex (Fig. 5).
Fig. 5.
Hydroxyl radical footprints of platinated nucleosomes. For these experiments, the nonplatinated DNA strand was radiolabeled on the 5′ end. Red trace, footprint of the nucleosome reconstituted with DNA of sequence 1; blue trace, footprint of the nucleosome reconstituted with DNA of sequence 2.
NER of a Platinated Nucleosome. In previous studies it was discovered that nucleosomes inhibit the NER of UV- (13, 14), AAF- (15), and platinum-damaged (4) DNA. In particular, the constraints imposed by the nucleosome structure reduce the efficiency of NER in substrates containing a single, site-specifically located 1,3-d(GpTpG) platinum-DNA cross-link by 10-fold when compared with that of free DNA samples (4). To investigate the effect of a structurally well defined platinum lesion on NER, we performed repair experiments on platinated sequence 1 nucleosomes (Fig. 6). We find that NER of this platinated nucleosome is reduced to ≈10% of the level observed for platinated free DNA, entirely consistent with our experiments on other nucleosomes (4).
Fig. 6.
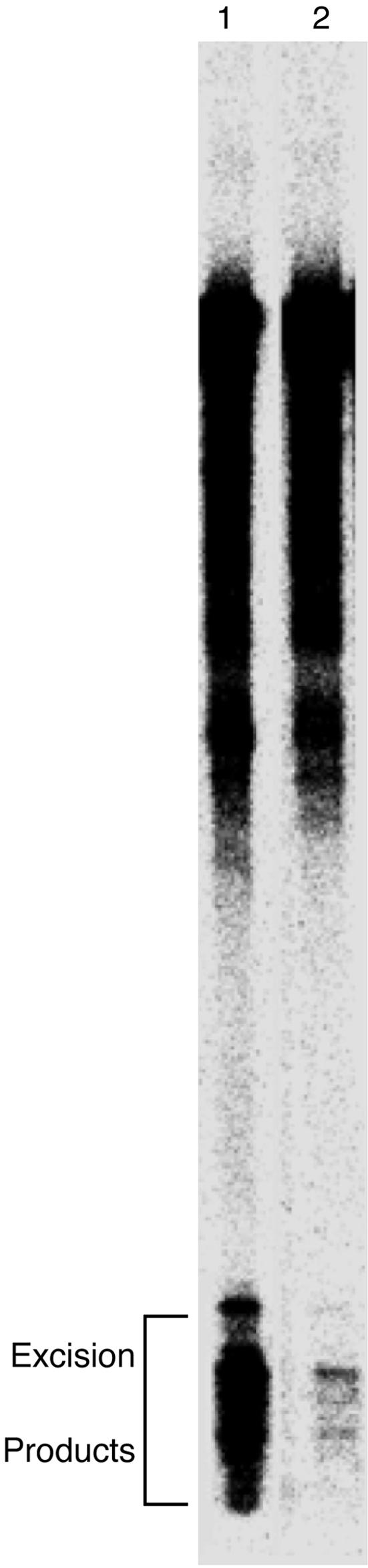
Denaturing PAGE analysis of 146-mer repair products. After treatment with cell-free extract, the 146-mer repair substrates were resolved by denaturing PAGE. Consistent with our previous results with 199-mer probes (4), the presence of the histone core inhibited repair in the nucleosome substrates by 10-fold over the efficiency of the analogous free DNA substrates. Lane 1, sequence 1 free DNA; lane 2, sequence 1 nucleosome. The bracket indicates the bands corresponding to the excised fragments containing a platinum-DNA cross-link (6). The “smear” of radioactivity extending from the full-length DNA bands results from nuclease activity present in the cell-free extract.
Discussion
Influence of Damage on the Rotational Positioning of DNA in Nucleosomes. We demonstrate here that a 1,3-d(GpTpG) platinum-DNA cross-link has a dominant effect on the rotational positioning of DNA in a nucleosome, overcoming any inherent preference of the DNA sequence that we studied. In the case of the cis-syn cyclobutane thymine dimer (CTD), the most prevalent UV-induced DNA photoproduct, a previous experiment (16) showed that the presence of this photolesion influenced the rotational setting of DNA in a bulk population of nucleosomes. More recently, a CTD was incorporated site-specifically into a nucleosome (17) by similar methods to those that we employ here. Hydroxyl radical footprinting showed that the CTD-lesioned DNA adopts a highly preferred rotational setting in the nucleosome, in which the CTD lesion is directed toward solvent. However, the DNA sequence that was used to form the CTD nucleosome contained strong nucleosome positioning elements to ensure that the DNA adopted a single rotational and translational setting that was designed to direct the CTD to the exterior of the nucleosome. Our experiments, in contrast, test two different platination sites in a DNA sequence not designed to form a conformationally homogeneous nucleosome, and we observe a dominating effect of the platinum lesion on the structure of the nucleosome.
Influence of Platination on Translational Positioning of DNA in Nucleosomes. Digestion of a nucleosome by exonuclease III provides insight into the translational positioning of DNA on the histone octamer. Although equal amounts of radioactive material were used in each of our exonuclease III digestions (see Fig. 4), bands corresponding to undigested nucleosomal DNA that has not been modified with platinum (Fig. 4A, lanes 7 and 9) are more intense than those for the platinated samples (Fig. 4A, lanes 8 and 10). The lack of significant exonuclease III digestion of undamaged nucleosomes is evidence that the 146-bp DNA molecule in these nucleosomes is positioned symmetrically with respect to the histone core. In contrast, the presence of strong pauses in the digestion patterns of the platinated nucleosomes (Fig. 4A, lanes 8 and 10), corresponding to the removal of DNA that extends beyond the histone octamer core, indicates that the DNA is asymmetrically positioned in these nucleosomes. These observations indicate that the platinum adduct affects the translational positioning of the damaged DNA on the histone core, in addition to controlling the rotational setting. Although the platinum cross-link induces changes in the translational positioning of the DNA in these nucleosomes, the adduct does not reside in the same translational location on the histone octamer in the two substrates. Fig. 4C depicts the relative positioning of the 146-mers with respect to the histone octamer core. These results indicate that, unlike the rotational positioning phenomenon, the platinum adduct does not impose a translational preference on the DNA.
Structural Insights into the Repair of Platinum Lesions. We also learned through our footprinting experiments that the undamaged DNA strand, not the strand bearing the DNA-platinum adduct itself, across from the damage occupies the most solvent-accessible site of the nucleosome (Fig. 5). Because of the position of the platinum cross-link within the nucleosome, the histone core presents a physical barrier that may allow the platinum-DNA adduct to elude the repair machinery. Our results provide structural insight into the inhibition of the repair of platinum damage in a nucleosome (4) and highlight the importance of remodeling proteins (13, 18) in providing access to platinum-damaged DNA.
In addition, our results suggest the possibility of strong interactions, perhaps through hydrogen bonds, between the 1,3-d(GpTpG) platinum-DNA adduct and the histone core, because both platinated nucleosomes are well positioned and the platinum damage resides in structurally similar sites in both. Specific interactions might involve the ammine ligands of the cis-diammineplatinum(II) group or heteroatoms of the extruded thymine base (see below) (19). Such interactions would contribute another level of complexity to the repair-shielding phenomenon, whereby cellular proteins block excision repair of a DNA adduct by forming a specific complex at the site of the lesion (1), by concealing the platinum damage from the NER recognition apparatus and thereby inhibiting repair in these substrates. These hypotheses remain to be tested by an x-ray structure determination of a site-specifically platinated nucleosome.
Our results may also indicate that access to the undamaged strand is an important component of damage recognition in nucleosomes. A single-stranded DNA binding protein, replication protein A (RPA), binds specifically to cisplatin-damaged DNA (20). A subsequent NMR study showed that, in the presence of the repair protein xeroderma pigmentosum complementation group A (XPA), RPA binds specifically to the undamaged strand of a DNA duplex containing a CTD lesion (21). Because the undamaged strand is the most exposed to solvent in our nucleosomes, it is conceivable that its recognition by RPA is facilitated within platinated nucleosomes.
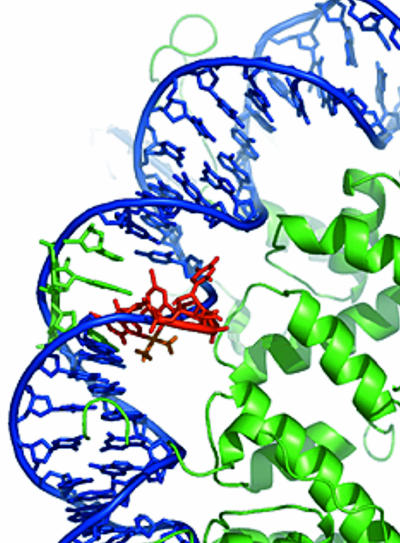
Modeling the Platinum Cross-Link into a Nucleosome. The structure of the Pt-GTG intrastrand cross-link in an 11-bp oligonucleotide duplex has been determined by NMR spectroscopy (19). Two geometric features are of interest in the present context. The most profound effect on DNA structure resulting from the Pt-GTG cross-link is expulsion of the central T base from the base stack into the solution, to accommodate the cross-link that connects the two adjacent guanines. The DNA duplex is bent around the cross-link toward the major groove by ≈30°. In Fig. 7 we present a model in which the NMR structure of the 1,3-d(GpTpG) platinum-DNA cross-link (19) and the crystal structure of the nucleosome core particle (11, 22) are superimposed. The setting of the DNA with respect to the surface of the histone octamer was chosen to be in accord with our hydroxyl radical footprinting results. This modeling exercise indicates that the Pt-GTG cross-link can be accommodated in a nucleosome at the site we detect by our footprinting experiments.
Fig. 7.
Model showing the location of the Pt-GTG cross-link in the nucleosome. Three base pairs containing the platinum-DNA adduct, adapted from the NMR structure of a platinated 11-mer (19), were superimposed on the structure of the nucleosome (22). The CAC trinucleotide of the platinum adduct NMR structure was modeled into a solvent-exposed position, in accord with our footprinting data. The histone octamer is shown as a green ribbon. DNA from the nucleosome x-ray structure (22) is blue. The GTG-Pt trinucleotide is red. The complementary CAC trinucleotide is green. The cis-{Pt(NH3)2}2+ group is orange. This image was generated by pymol (www.pymol.org).
Although the 1,2-intrastrand d(GpG) cross-link has received much attention as the major adduct formed by the antitumor drug cis-diamminedichloroplatinum(II) (cisplatin) (23), we note that the intrastrand 1,3-d(GpXpG) cross-link is the most abundant lesion produced by the related drug carboplatin [cis-diammine(1,1-cyclobutanedicarboxylato)platinum(II)] (24). The only difference between cisplatin and carboplatin is the leaving group; the ultimate DNA-binding moiety, cis-diammineplatinum(II), is the same for both drugs. Previous x-ray and NMR investigations have produced a wealth of information about the structure of the 1,2-intrastrand d(GpG) cisplatin cross-link. Although a range of values was determined in these studies, the bulk of the work showed that a cisplatin 1,2-d(GpG) cross-link results in a substantially larger DNA bend angle, ranging from ≈60° (25) to ≈80° (26), compared with the 1,3-d(GpTpG) cross-link that we studied here. Additionally, the 1,2-d(GpG) intrastrand cross-link lacks the extruded thymine base that we suggest may be an important component of the DNA positioning phenomenon revealed by this study. To accommodate ≈80 bp of DNA in a single superhelical turn around the histone octamer, the DNA must bend by an average of 45° per helical turn of the DNA (27). The ≈30° bend induced at the site of the 1,3-d(GpTpG) cross-link (19) is therefore expected to contribute to the accommodation of the modified duplex in a nucleosome. However, a detailed study of the conformation of DNA in the nucleosome (27) found that the real picture is not so simple, because the actual curvature of nucleosomal DNA is twice that necessary to form the nucleosome superhelix. Local alternation of DNA conformation accounts for the excess curvature, indicating that there exist microenvironments of unusual DNA structure in the nucleosome. These structural features of nucleosomal DNA may influence the preference of the Pt-GTG cross-link to occupy the site we observe in our experiments. It remains to be determined how the more extensively kinked 1,2-d(GpG) platinum cross-link will be accommodated within nucleosomes.
Implications for Platinum Binding to Chromatin in Vivo. In cells, the platinum drug initially binds to DNA that is already assembled into chromatin. It is now widely appreciated, however, that chromatin remodeling can give rise to “naked” DNA, sections of the genome not bound to nucleosomes, which must eventually be reassembled into chromatin, at which point the platinum lesion would most likely exert its influence on the rotational setting. Moreover, in recent work (28) the nucleosome itself was determined to be a dynamic structure, with DNA wrapping and unwrapping from the histone octamer on a time scale of tens to hundreds of milliseconds. We therefore propose from our present findings that, although the platinum adduct may initially form on DNA in a nucleosome having a particular rotational setting, the nucleosome will rearrange to accommodate the structural influence of the platinum cross-link discovered here.
Supplementary Material
Acknowledgments
We thank Jan Reedijk (Leiden University, Leiden, The Netherlands) for providing the coordinates of the Pt-GTG oligonucleotide structure, and Karolin Luger (Colorado State University, Fort Collins) for the clones that we used to express the histones for nucleosome reconstitution. This work was supported by National Cancer Institute Grant CA-34992 (to S.J.L.) and National Institute of General Medical Sciences Grant GM 41930 (to T.D.T.). A.J.D. was supported in part by resources from the Massachusetts Institute of Technology Class of 1973 UROP Fund.
Author contributions: S.J.L., T.D.T., D.W., and A.J.D. designed research; A.J.D., D.W., and Q.W. performed research; T.D.T., S.J.L., A.J.D., D.W., and Q.W. analyzed data; and T.D.T., S.J.L., and A.J.D. wrote the paper.
Abbreviations: CTD, cyclobutane thymine dimer; NER, nucleotide excision repair.
References
- 1.Jamieson, E. R. & Lippard, S. J. (1999) Chem. Rev. 99, 2467–2498. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg, R. D. & Lorch, Y. (1999) Cell 98, 285–294. [DOI] [PubMed] [Google Scholar]
- 3.Caserta, M., Verdone, L. & Di Mauro, E. (2002) ChemBioChem 3, 1172–1182. [DOI] [PubMed] [Google Scholar]
- 4.Wang, D., Hara, R., Singh, G., Sancar, A. & Lippard, S. J. (2003) Biochemistry 42, 6747–6753. [DOI] [PubMed] [Google Scholar]
- 5.Huang, J. C., Zamble, D. B., Reardon, J. T., Lippard, S. J. & Sancar, A. (1994) Proc. Natl. Acad. Sci. USA 91, 10394–10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zamble, D. B., Mu, D., Reardon, J. T., Sancar, A. & Lippard, S. J. (1996) Biochemistry 35, 10004–10013. [DOI] [PubMed] [Google Scholar]
- 7.Steger, D. J., Eberharter, A., John, S., Grant, P. A. & Workman, J. L. (1998) Proc. Natl. Acad. Sci. USA 95, 12924–12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tullius, T. D., Dombroski, B. A., Churchill, M. E. A. & Kam, L. (1987) Methods Enzymol. 155, 537–558. [DOI] [PubMed] [Google Scholar]
- 9.Hayes, J. J., Clark, D. J. & Wolffe, A. P. (1991) Proc. Natl. Acad. Sci. USA 88, 6829–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowary, P. T. & Widom, J. (1997) Proc. Natl. Acad. Sci. USA 94, 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luger, K., Maeder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. (1997) Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 12.Hayes, J. J., Tullius, T. D. & Wolffe, A. P. (1990) Proc. Natl. Acad. Sci. USA 87, 7405–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hara, R., Mo, J. & Sancar, A. (2000) Mol. Cell. Biol. 20, 9173–9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosmoski, J. V., Ackerman, E. J. & Smerdon, M. J. (2001) Proc. Natl. Acad. Sci. USA 98, 10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara, R. & Sancar, A. (2002) Mol. Cell. Biol. 22, 6779–6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suquet, C. & Smerdon, M. J. (1993) J. Biol. Chem. 268, 23755–23757. [PubMed] [Google Scholar]
- 17.Kosmoski, J. V. & Smerdon, M. J. (1999) Biochemistry 38, 9485–9494. [DOI] [PubMed] [Google Scholar]
- 18.Ura, K., Araki, M., Saeki, H., Masutani, C., Ito, T., Iwai, S., Mizukoshi, T., Kaneda, Y. & Hanaoka, F. (2001) EMBO J. 20, 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teuben, J.-M., Bauer, C., Wang, A. H.-J. & Reedijk, J. (1999) Biochemistry 38, 12305–12312. [DOI] [PubMed] [Google Scholar]
- 20.Clugston, C. K., McLaughlin, K., Kenny, M. K. & Brown, R. (1992) Cancer Res. 52, 6375–6379. [PubMed] [Google Scholar]
- 21.Lee, J.-H., Park, C.-J., Arunkumar, A. I., Chazin, W. J. & Choi, B.-S. (2003) Nucleic Acids Res. 31, 4747–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davey, C. A., Sargent, D. F., Luger, K., Maeder, A. W. & Richmond, T. J. (2002) J. Mol. Biol. 319, 1097–1113. [DOI] [PubMed] [Google Scholar]
- 23.Fichtinger-Schepman, A. M. J., van der Veer, J. L., den Hartog, J. H. J., Lohman, P. H. M. & Reedijk, J. (1985) Biochemistry 24, 707–713. [DOI] [PubMed] [Google Scholar]
- 24.Blommaert, F. A., van Dijk-Knijnenburg, H. C., Dijt, F. J., den Engelse, L., Baan, R. A., Berends, F. & Fichtinger-Schepman, A. M. (1995) Biochemistry 34, 8474–8480. [DOI] [PubMed] [Google Scholar]
- 25.Ohndorf, U.-M., Rould, M. A., He, Q., Pabo, C. O. & Lippard, S. J. (1999) Nature 399, 708–712. [DOI] [PubMed] [Google Scholar]
- 26.Dunham, S. U., Dunham, S. U., Turner, C. J. & Lippard, S. J. (1998) J. Am. Chem. Soc. 120, 5395–5406. [Google Scholar]
- 27.Richmond, T. J. & Davey, C. A. (2003) Nature 423, 145–150. [DOI] [PubMed] [Google Scholar]
- 28.Li, G., Levitus, M., Bustamante, C. & Widom, J. (2005) Nat. Struct. Mol. Biol. 12, 46–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.