Abstract
Lufenuron is an insect growth regulator insecticide mainly used for the control of the cat flea. To understand mechanisms of resistance to lufenuron, we have characterized lufenuron resistance in a natural population of Drosophila melanogaster. In this study we have used precise genetic mapping to identify a mechanism of lufenuron resistance: the overexpression of the cytochrome P450 gene Cyp12a4. Cyp12a4 is predicted to encode a mitochondrial cytochrome P450 enzyme. Expression of Cyp12a4 in D. melanogaster third-instar larvae was detected in the midgut and Malpighian tubules of both lufenuron-resistant and wild-type strains. The level of Cyp12a4 expression in the midgut is higher in the lufenuron-resistant strain than in wild-type strains. Driving the expression of Cyp12a4 in the midgut and Malpighian tubules by using the GAL4/UAS gene expression system results in lufenuron resistance, but it does not result in resistance to three other insecticide classes. Transgenic expression of Cyp12a4 in a ubiquitous expression pattern results in late embryonic lethality, suggesting that high-level ectopic expression of Cyp12a4 is detrimental to development.
Keywords: cytochrome P450, insecticide resistance, insect growth regulator, genetic mopping
Insecticide resistance is an important example of natural selection (1). Resistance is predominantly mediated either by changes in the metabolism of the insecticide or by changes in the sensitivity of insecticide targets (2). Insecticide resistance mediated by target site modification is well documented for most commonly used insecticides (3–7). Molecular mechanisms of resistance due to increased insecticide metabolism by cytochrome P450s, esterases, and glutathione S-transferases are less well understood. Resistance is commonly found to be the result of up-regulation of single or multiple members of these enzyme families (8–13); however, resistance due to structural changes in enzymes has also been documented (14–16).
Lufenuron is an insect growth regulator insecticide, mainly used in the control of the cat flea, Ctenocephalides felis. It is active against larval developmental stages, causing cuticular lesions resulting from the disruption of chitin synthesis (17). Lufenuron is also active against Diptera, including Drosophila melanogaster (18). To understand possible resistance mechanisms to lufenuron, we have used D. melanogaster as a model. Lufenuron resistance in D. melanogaster was originally reported in populations from two widely separated locations in the United States (19). As it was not expected that D. melanogaster had been in contact with lufenuron, it was postulated that this resistance resulted from cross-resistance that had evolved to an earlier, widely used insecticide (19). Subsequently, it was found that resistance to lufenuron was caused by the up-regulation of the cytochrome P450 gene Cyp6g1, which confers resistance to numerous insecticide classes, and that Cyp6g1-mediated resistance is widespread (8, 20).
Here we describe the discovery of a mechanism of lufenuron resistance in a natural population of D. melanogaster collected in Victoria, Australia. We used a genetic mapping approach to identify a 30-kb region on the right arm of chromosome III containing a lufenuron-resistance locus. Within this 30-kb region resides a cluster of two cytochrome P450 genes, Cyp12a5 and Cyp12a4. We found Cyp12a4 to be overexpressed in third-instar larvae of the lufenuron-resistant strain. We describe the expression pattern of Cyp12a4 in third-instar larvae of resistant and susceptible D. melanogaster strains. We show that transgenic expression of Cyp12a4 in the midgut and Malpighian tubules of larvae confers lufenuron resistance.
Materials and Methods
Strains. The lufenuron-resistant D. melanogaster strain NB16 was collected from an apple orchard in Wandin, Victoria, Australia, in 1996 and maintained in the laboratory under standard conditions. NB16 was not selected for insecticide resistance as part of routine maintenance. Strains used in mapping and GAL4 expression and laboratory reference strains were obtained from the Bloomington Drosophila Stock Center at Indiana University, unless otherwise indicated.
Resistance Bioassays. Drosophila were tested for lufenuron resistance by using a feeding assay (21). Survival to adulthood from five replicates of 100 first-instar larvae per vial, for at least five different lufenuron concentrations, was used to generate dosage mortality responses. Natural mortality in control vials was taken into account when generating dosage mortality curves with the priprobitnm program (M. Sakuma, Kyoto University, Kyoto) using a natural regression model (22). Resistance bioassays using dicyclanil, nitenpyram, and diazinon were performed by using standard techniques (23, 24).
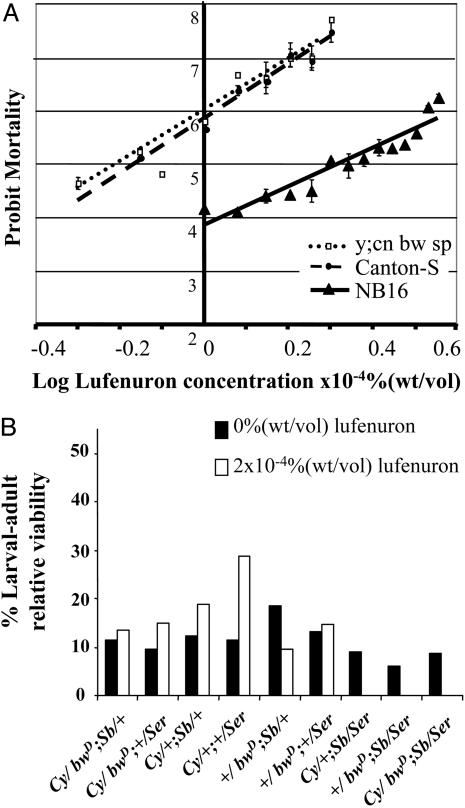
Genetic Mapping of Lufenuron Resistance. Survival from first instar to adulthood on 2 × 10–4% lufenuron was used to score for lufenuron resistance in mapping crosses. Crosses were set up at a controlled density of 100 larvae per vial. Initially, lufenuron resistance was mapped to a chromosome by using the SM5/Dp(?;2)bwD,wgSp-1 bwD; TM3, y+ Ser-1/Sb1 strain, (referred to as Cy/bwD; Sb/Ser) with the dominant markers curly wings and brown eyes on chromosome II, and stubble bristles and serrate wings on chromosome III. Reciprocal crosses of Cy/bwD; Sb/Ser × NB16 were set up. Emerging +/Cy; +/Ser males (where + represents a chromosome from NB16) were test-crossed by using Cy/bwD; Sb/Ser and reared on 2 × 10–4% lufenuron. Mapping within chromosome III was performed by a standard test cross strategy, using three different strains, ru h st ry e, red e, and gl e, each with phenotypic markers that progressively refined the location of the resistance locus. Ten molecular markers within the gl e region were also used (Table 1 and Supporting Text, which are published as supporting information on the PNAS web site).
Real-Time PCR. Total RNA was isolated from third-instar larvae by using TRIzol reagent (GIBCO/BRL). Each RNA sample was treated with RQ1 RNase-free DNase (Promega). PCR using each RNA sample as a template was used to test for the absence of DNA contamination. The RNA was deemed to be free from DNA by the absence of a PCR product after 40 cycles of PCR. Reverse transcription was then performed on 2 μg of each RNA sample in a 20-μl reaction using Superscript III Reverse Transcriptase (Invitrogen Life Technologies) and oligo(dT)20 primer, following the supplier's instructions. One microliter of a 1 in 10 dilution of cDNA was used in real-time PCR. Real-time PCR was conducted on a RotorGene-3000 (Corbett Research, Sydney) using a QuantiTect SYBR Green PCR kit (Qiagen, Valencia, CA). PCR conditions were 95°C for 10 min to activate the hot-start polymerase, followed by 50 cycles of 95°C for 20 sec, 55°C for 30 sec, and 72°C for 30 sec. Fluorescence was measured after each cycle. Relative expression of each gene was measured in reference to the housekeeping gene RpL11. PCR primers used were RpL11F (CGA TCC CTC CAT CGG TAT CT) and RpL11R (AAC CAC TTC ATG GCA TCC TC) for RPL11, Cyp12a4F (CCA ATC GTC CAG GCA ACT AT) and Cyp12a4R (TCG GGA TCT CTC AGT TCG AG) for Cyp12a4, and Cyp12a5F (ATC CTG GGG AGA CTT TCG AT) and Cyp12a5R (GCG GTT AAT GGT TTC CAA GA) for Cyp12a5. For every experiment, standard curves of target genes and RpL11 were made from a reference sample of cDNA generated from w1118, using duplicate serial dilutions with at least five different cDNA concentrations covering a 1,000-fold concentration range. Standard curves were used to quantify amounts of target and housekeeping transcripts in each sample.
Transgenic Expression of Cyp12a4. Cyp12a4 cDNA from a y; cn bw sp strain was amplified by using a high-fidelity PCR system (Roche) and the primers ORF12A4F (GTG AGC CGG AAA AGT TCT AAT C) and ORF12a4R (TTT GAC CAT GAC TGT ATA TCG C). The PCR product was cloned into pCR-BluntII-TOPO (Invitrogen Life Technologies), sequenced, and subcloned into pUAST (25). This construct was transformed into w1118 flies by using standard techniques. Independent transformed lines were made homozygous, and the inserted construct was mapped to a chromosome by using the If/CyO; MKRS/TM6b, Tb strain. Two independent, homozygous viable transformed lines, UAS-Cyp12a42 and UAS-Cyp12a43, with the construct inserted on chromosome II and III, respectively, were chosen for further analysis.
GAL4 driver strains were used to specifically drive the expression of Cyp12a4 in the F1 of crosses between UAS-Cyp12a4 and driver strains by using the GAL4/UAS system (25). y w; tub-GAL4/TM3, Sb was used to express Cyp12a4 in a ubiquitous pattern (26). The 6g1Cs-GAL4-1a and 6g1HR-GAL4-6c drivers were used to drive the expression of Cyp12a4 in the midgut and Malpighian tubules, and in the midgut, Malpighian tubules, and fat body, respectively. These drivers, constructed in the w1118 strain, contain upstream sequence of Cyp6g1 from different strains cloned upstream of GAL4. The expression pattern of GAL4 when these drivers were used was determined by using crosses to y1 w*; P{UAS-GFP::lacZ.nls}15.3 (27) followed by the detection of GFP in third-instar larvae (H.C., M.R.B., P.B., and P.D., unpublished work).
In Situ Hybridization. Cyp12a4 was amplified by PCR using Taq DNA polymerase (Promega) with the primers ORF12a4F and ORF12a4R, and cloned into pGEM-T Easy (Promega) in both orientations. The sense and antisense constructs were then linearized with SalI (Promega), transcribed with Megascript T7 polymerase (Ambion), and labeled with digoxigenin-labeled dNTP mix (Roche). The final concentration and purity of probes were determined by UV spectrophotometry and agarose gel electrophoresis. In situ hybridization was performed on dissected third-instar larvae by using standard techniques (28).
Results
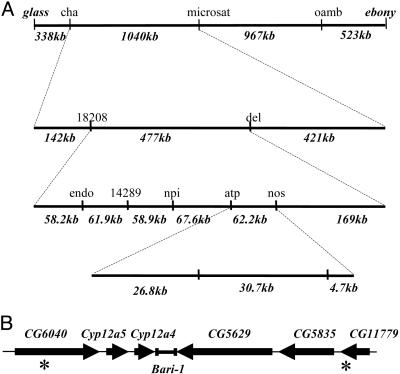
Genetic Mapping of Lufenuron Resistance. The D. melanogaster NB16 strain, collected from an apple orchard in Victoria, Australia, was found to be 3-fold resistant to lufenuron (Fig. 1A). PCR of the upstream region of Cyp6g1 was performed as in ref. 8 to confirm that NB16 does not carry the Accord allele of Cyp6g1 (data not shown). The lufenuron resistance was mapped to chromosome III (Fig. 1B). Genetic mapping within chromosome III (Fig. 7 and Table 2, which are published as supporting information on the PNAS web site) revealed a region on the right arm of chromosome III to confer lufenuron resistance. A fine-scale mapping cross, scoring recombinants between glass (91A3) and ebony (93C7-D1) for 10 molecular markers (Table 1) and lufenuron resistance was conducted. A total of 264 lufenuron-resistant recombinants (62 gl e+ and 204 gl+ e) were genotyped for molecular markers, and the region was limited to a 30-kb region at 91F3 (Fig. 2A and Fig. 8, which is published as supporting information on the PNAS web site). This region contains five genes, including the cytochrome P450 enzymes Cyp12a4 and Cyp12a5 (Fig. 2B). The other three genes have unknown functions (29). Cyp12a4 and Cyp12a5 were considered as candidates for the lufenuron-resistance locus.
Fig. 1.
Characterization of lufenuron resistance in NB16. (A) Dosage–mortality response for the lufenuron-resistant NB16 strain and lufenuron-susceptible strains Canton-S and y; cn bw sp. NB16 is 3.06 and 3.40 times more lufenuron resistant than Canton-S and y; cn bw sp, respectively. (B) Lufenuron resistance in NB16 maps to chromosome III. +/Cy; +/Ser progeny from Cy/bwD; Sb/Ser × NB16 were test-crossed to Cy/bwD; Sb/Ser and reared in the presence or absence of 2 × 10–4% lufenuron. The relative numbers of each class of progeny are shown. + represents a chromosome from NB16. Note the lack of survivors on lufenuron in progeny where both copies of chromosome III are from the mapping strain.
Fig. 2.
The location of the lufenuron resistance locus. (A) Fine-scale genetic mapping of lufenuron resistance within chromosome III. Recombinants between the genetic markers glass and ebony were progressively scored for molecular markers. Molecular distances between markers are indicated. Numbers of recombinants between each marker are presented in Fig. 8. Lufenuron resistance was mapped to a region of 30.7 kb. (B) Genes in the region of the lufenuron-resistance locus at 91F3–6. Arrows indicate direction of transcription.
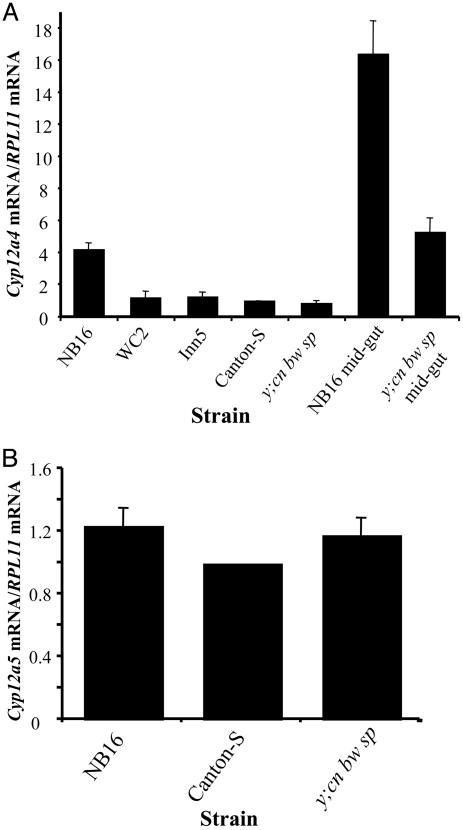
Expression Analysis and Sequencing of Candidate Genes. The expression levels of the candidate genes Cyp12a4 and Cyp12a5 were examined by real-time PCR. Approximately 4 times more Cyp12a4 transcript was detected in NB16, compared with the lufenuron-susceptible strains Canton-S and y; cn bw sp (Fig. 3A). No difference in transcript levels of Cyp12a5 were detected between NB16 and susceptible strains Canton-S and y; cn bw sp (Fig. 3B). Two strains that are lufenuron resistant because of increased expression of Cyp6g1 had levels of Cyp12a4 mRNA similar to those of lufenuron-susceptible strains (Fig. 3A). Cyp12a4 mRNA levels were assayed in a further 16 strains from natural populations known to be lufenuron resistant, with no increase in Cyp12a4 expression detected (data not shown), suggesting that this lufenuron-resistance mechanism is rare.
Fig. 3.
Level of Cyp12a5 (A) and Cyp12a4 (B) mRNA in third-instar larvae of NB16 compared with susceptible strains Canton-S and y; cn bw sp, as determined by real-time PCR (±SEM, n = 3). All levels are relative to the susceptible strain Canton-S. Also shown are the mRNA levels of other lufenuron-resistant strains, WC2 and Inn5 (23). Compared with Canton-S, 4.2 times more Cyp12a4 mRNA is detected in NB16. Cyp12a4 mRNA levels were also measured in midguts from third-instar larvae, with 3.1 times more Cyp12a4 detected in the midgut of NB16 than in that of y; cn bw sp.
Given that lufenuron resistance in the NB16 strain maps to the 91F3 region containing Cyp12a4 and Cyp12a5, we would expect to find the resistance-associated mutation somewhere in this region. We would also predict that this mutation is a cis-acting mutation, resulting in an increase of Cyp12a4 mRNA. The 91F region was uncovered in lufenuron-resistant flies from the NB16 strain by using deficiency lines Df5597 (Df(3R)Dl-KX23, e/TM3, Ser) and Df2411 (Df(3R)Dl-M2/TM6C, Sb). A total of 7,041 bp of sequence encompassing a region 796 bp upstream of Cyp12a5 to 2,068 bp downstream of Cyp12a4 was compared between NB16 (GenBank database accession DQ026292) and y; cn bw sp. No sequence polymorphisms between NB16 and y; cn bw sp were detected in the 451-bp intergenic region between Cyp12a4 and Cyp12a5. Two coding region polymorphisms within Cyp12a4, neither leading to amino acid substitutions, and a 33-bp deletion downstream of Cyp12a4 were detected. The 33-bp deletion was internal to a 1,707-bp Bari-1 element, and it encompasses the translation start site of the Bari-1 transposase (CAA47913) (30). This Bari-1 element is inserted in the 3′ untranslated region of Cyp12a4. The same polymorphism was subsequently found in other strains that are not lufenuron resistant, suggesting that the mutation is not the resistance-associated mutation. No sequence differences in the 796 bp directly upstream of Cyp12a5 were detected. Several sequence polymorphisms in Cyp12a5 were detected between NB16 and y; cn bw sp, the most significant being a 2-bp deletion in exon 2 of Cyp12a5. This deletion causes a premature termination codon in Cyp12a5, predicted to result in a truncated protein of 77 aa. This mutation was confirmed by sequencing Cyp12a5 from cDNA. This mutation was not detectable in a further 64 natural populations collected from various locations in Australia (data not shown). Analysis of polymorphisms between NB16 and y; cn bw sp was conducted by using p-match and match (31) from the TRANSFAC web site (www.gene-regulation.com/). This analysis failed to reveal any differences in putative transcriptional factor binding sites.
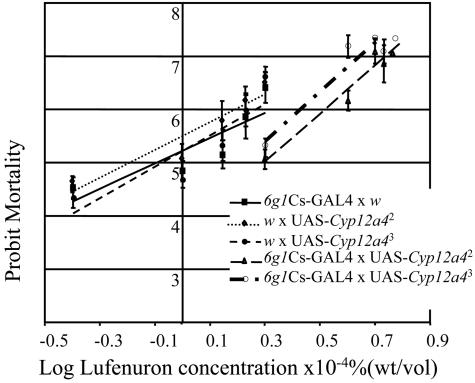
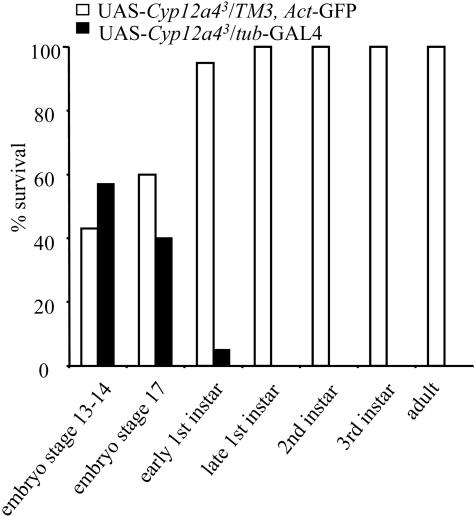
Transgenic Expression of Cyp12a4. To confirm that Cyp12a4 over-expression is responsible for lufenuron resistance, the Cyp12a4 gene was overexpressed by using the Gal4/UAS system (25). Different driver strains were used to drive Cyp12a4 expression in a tissue-specific manner. Driving the expression of Cyp12a4 in the larval midgut and Malpighian tubules by using the 6g1Cs-GAL4 driver resulted in 1.5- 1.8-fold resistance to lufenuron (Fig. 4). Driving expression of Cyp12a4 in the midgut, fat body, and Malpighian tubules by using the 6g1HR-GAL4 driver resulted in 1.5- to 1.7-fold lufenuron resistance (data not shown). The NB16 strain shows 3-fold resistance to lufenuron. Direct comparison between these resistance levels is difficult, as it is not known if the expression patterns and levels of Cyp12a4 are identical between NB16 and the transgenic strains. It is also possible that the genetic background of the strains is contributing a level of lufenuron resistance. Driving expression of Cyp12a4 in the midgut, fat body, and Malpighian tubules by using the 6g1HR-GAL4 driver did not result in resistance to other insecticides, including diazinon, nitenpyram, and dicyclanil (Fig. 9, which is published as supporting information on the PNAS web site). These three insecticides differ in terms of both their structure and mode of action. Diazinon is an organophosphorus chemical and exerts its lethal effect on acetylcholinesterase; nitenpyram is a neonicotinoid and targets the nicotinic acetylcholine receptor; and dicyclanil is an insect growth regulator insecticide with an undetermined mode of action. It is noteworthy that in these transgenic overexpression experiments, the genetic background is consistent (all strains were constructed in a w1118 background), and therefore lufenuron resistance is due to Cyp12a4 overexpression. The expression of Cyp12a4 in a ubiquitous pattern in the y w; tub-GAL4/TM3, Sb strain (26) results in late embryonic lethality (Fig. 5).
Fig. 4.
Dosage–mortality response for transgenic flies overexpressing Cyp12a4 in the midgut and Malpighian tubules by using the 6g1Cs-GAL4 driver, compared with controls, reared on increasing concentrations of lufenuron. Between 1.5-(UAS-Cyp12a43) and 1.8-(UAS-Cyp12a42) fold lufenuron resistance is seen in flies overexpressing Cyp12a4 in the midgut and Malpighian tubules, compared with flies not overexpressing Cyp12a4.
Fig. 5.
Percentage survival at different life cycle stages for individuals overexpressing Cyp12a4 in a ubiquitous expression pattern by using the tubulin-GAL4 driver (UAS-Cyp12a43/tub-GAL4), compared with flies not over-expressing Cyp12a4 (UAS-Cyp12a43/TM3, Act-GFP). The presence/absence of GFP was used to determine overexpression of Cyp12a4. Overexpression of Cyp12a4 results in early first-instar lethality.
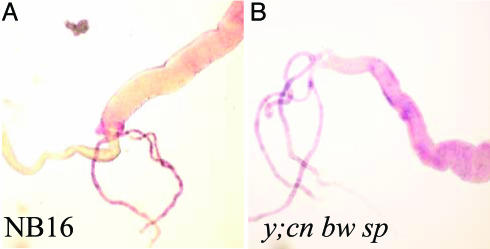
Native Expression Pattern of Cyp12a4. In situ hybridization to third-instar larvae was carried out to determine which tissues express Cyp12a4. Expression of Cyp12a4 in the y; cn bw sp strain was detected predominantly in the midgut and Malpighian tubules (Fig. 6A). In situ hybridization of Cyp12a4 in third-instar larvae of the NB16 strain showed the same expression pattern as seen in y; cn bw sp (Fig. 6B). There was no detectable signal in controls when a Cyp12a4 sense probe was used (data not shown). Real-time PCR was used to compare expression levels of Cyp12a4 in dissected midguts of NB16 and y; cn bw sp. Cyp12a4 levels were 3.1 times higher in the midgut of NB16 than in y; cn bw sp, indicating overexpression of Cyp12a4 in the midgut (Fig. 3A). Midgut samples were enriched for Cyp12a4 compared with whole third-instar larvae, (3.9 times for NB16 and 6.2 times for y; cn bw sp). This observation indicates that Cyp12a4 is expressed at higher levels in the midgut compared with other tissues, confirming in situ hybridization results (Fig. 6). It is possible that Cyp12a4 is also expressed in other tissues in NB16, and expression in these tissues was not detectable by in situ hybridization. The level of enrichment for Cyp12a4 in the NB16 midgut is lower than for y; cn bw sp.
Fig. 6.
In situ hybridization of Cyp12a4 in third-instar larvae of the lufenuron-resistant strain NB16 (A) and the susceptible control y; cn bw sp (B). Expression of Cyp12a4 is detected in the midgut and Malpighian tubules in both strains.
Discussion
We have identified a mechanism of lufenuron resistance in a natural population of D. melanogaster, the up-regulation of the cytochrome P450 gene Cyp12a4. This is the second D. melanogaster cytochrome P450 gene identified capable of conferring lufenuron resistance, the first being Cyp6g1 (8). The presence of lufenuron resistance in natural populations of D. melanogaster is unexpected, because D. melanogaster is unlikely to have been in contact with lufenuron (32). It is postulated that lufenuron resistance is the result of selection by another chemical (32). The widespread high frequency of insecticide resistance due to the overexpression of Cyp6g1 is postulated to be the result of cross-resistance to any of a number of other chemicals to which D. melanogaster is exposed (8, 20). Lufenuron resistance by means of Cyp12a4 overexpression, however, is not at a high frequency in natural populations, possibly because it has not been selected for. Cross-resistance to other insecticides was not detected in strains overexpressing Cyp12a4 (Fig. 9). The fact that we detected lufenuron resistance due to Cyp12a4 overexpression in one population highlights the likelihood that sufficient variation is present in natural populations, allowing insecticide resistance by metabolic means to be selected for. This possibility has implications for the evolution of insecticide resistance by metabolic mechanisms in pest species routinely treated with insecticides.
For both Cyp12a4 and Cyp6g1, it is an increase in the expression level that results in lufenuron resistance. Increases in cytochrome P450 expression have also been implicated in insecticide resistance in other species (9, 33–36). Few studies, however, have actually confirmed that the overexpression of an identified cytochrome P450 actually causes the insecticide resistance (8, 37). This overexpression is currently achievable by using transgenic D. melanogaster, as in this study, or by using heterologous expression systems (38).
Of the cytochrome P450 enzymes positively identified as having a role in insecticide resistance, most are microsomal. Based on sequence homology, CYP12A4 is thought to be a mitochondrial cytochrome P450. Early studies with mammalian cytochrome P450s suggested that those found in microsomes were involved in xenobiotic metabolism, whereas the primary function of those located in mitochondria was hormone metabolism. An insect exception to this is the mitochondrial cytochrome P450, CYP12A1, from the house fly, Musca domestica (39). Cyp12a1 expression is increased in an insecticide-resistant strain of M. domestica. The increased metabolism of insecticides by CYP12A1 purified in a bacterial heterologous expression system is also seen, suggesting a role for this mitochondrial P450 in insecticide metabolism (39). Our study has identified a role of a related mitochondrial cytochrome P450, Cyp12a4, in insecticide resistance. As recently suggested, the subcellular location of a cytochrome P450 does not necessarily indicate its physiological role (38).
Driving the expression of Cyp12a4 in a ubiquitous pattern in transgenic D. melanogaster by using a tubulin driver and the GAL4/UAS system resulted in late embryonic lethality (Fig. 5). This result does not necessarily suggest that CYP12A4 has a native role in the metabolism of endogenous compounds, but rather that the misexpression of Cyp12a4 in this case can lead to either a depletion of an essential endogenous substrate or a build-up of a toxic metabolite. The native function of CYP12A4, if any, is not known. Another important implication of this experiment is that it suggests that resistance due to changes in the expression pattern of Cyp12a4, and possibly other cytochrome P450 genes, may carry a fitness cost, implying there may be a tradeoff for metabolic resistance mechanisms.
Cyp12a4 mRNA was detected in the midgut and Malpighian tubules of third-instar larvae. Generally, the expression patterns of cytochrome P450 genes are not well characterized. Cyp6a2 expression has been localized to the midgut, fat body, and Malpighian tubules in adult D. melanogaster (40). Also, a recent gene expression study using microarrays generally found cytochrome P450 genes to be expressed predominantly in the midgut of late third-instar larvae and not in the salivary glands, epidermis, central nervous system, or wing disk (41). Clearly, it is difficult to make generalizations about the expression patterns of genes when such a large, functionally diverse gene family is involved. The diversity of endogenous and exogenous substrates of different cytochrome P450s is large, and the number of functions cytochrome P450s provide to the organism is also presumed to be large. However, the midgut and Malpighian tubules are tissues likely to be involved in the detoxification of foreign compounds, consistent with the notion that Cyp12a4 has a role in this process.
A 1.7-kb Bari-1 transposable element, a Tc-1-like element, is inserted within the 3′ untranslated region of Cyp12a4, 23 bp distal to the stop codon, in the lufenuron-resistant strain NB16. This Bari-1 insertion is also present in y; cn bw sp and 30 of 30 Australian natural populations we studied (data not shown). A previous study using in situ hybridization to polytene chromosomes has shown the Bari-1 element present in this same position in all 7 of 7 populations collected from different locations in the world (30). There are a growing number of examples of transposable element sequences affecting the function and expression of neighboring genes (42, 43). The fact that all strains studied carry a Bari-1 insert downstream of Cyp12a4, and not only the lufenuron-resistant NB16 strain, suggests that although the Bari-1 element insertion may be important, it is unlikely to cause the difference in Cyp12a4 expression between the NB16 and y; cn bw sp strains. A number of polymorphisms were detected upstream of Cyp12a4. It is possible that one of these polymorphisms is the resistance-associated mutation. The mutation is likely to be cis-acting, as resistance maps to the same region as the gene overexpressed. However, the possibility that the 2-bp deletion within the coding region of the upstream gene Cyp12a5 is the resistance-associated mutation, and the mutation is transacting, cannot be excluded.
In summary, this study describes a mechanism of lufenuron resistance in a natural population of D. melanogaster: the over-expression of Cyp12a4. This is the second cytochrome P450 gene from D. melanogaster that can confer resistance to lufenuron, the other being overexpression of Cyp6g1 (8). The frequency of lufenuron resistance by means of Cyp12a4 overexpression is low in natural populations, largely because of the lack of widespread exposure of D. melanogaster to lufenuron, and the apparent inability of Cyp12a4 overexpression to confer resistance to other insecticide classes. This study was possible only because of the capacity to positionally clone and manipulate genes in the model insect system, D. melanogaster. The study highlights the important role some individual members of the cytochrome P450 multigene family play, and have the potential to play, in insecticide resistance.
Supplementary Material
Acknowledgments
We thank Julie Thompson for technical assistance with initial screening for lufenuron resistance, Chris Lumb for help with Drosophila transformation, Fiona Pyke for providing the NB16 strain, Novartis Animal Health Australasia Pty. Ltd. for providing insecticides, and Charles Robin for discussion. We acknowledge two anonymous reviewers foruseful comments. This work was supported by grants from the Australian Research Council through its funding of the Special Research Centre CESAR (Centre for Environmental Stress and Adaptation Research), and an Australian Research Council Linkage–Australian Postdoctoral Fellowship (Commonwealth Scientific and Industrial Research Organisation) (to P.J.D.).
Author contributions: M.R.B., H.C., J.A.M., P.B., and P.J.D. designed research; M.R.B., H.C., L.M., S.R., W.W., M.O., and P.J.D. performed research; M.R.B., L.M., P.B., and P.J.D. analyzed data; and P.J.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. DQ026292).
References
- 1.Roush, R. T. & McKenzie, J. A. (1987) Annu. Rev. Entomol. 32, 361–380. [DOI] [PubMed] [Google Scholar]
- 2.Wilson, T. G. (2001) Annu. Rev. Entomol. 46, 545–571. [DOI] [PubMed] [Google Scholar]
- 3.ffrench-Constant, R. H., Anthony, N., Aronstein, K., Rocheleau, T. & Stilwell, G. (2000) Annu. Rev. Entomol. 45, 449–466. [DOI] [PubMed] [Google Scholar]
- 4.Williamson, M. S., Martinez-Torres, D., Hick, C. A. & Devonshire, A. L. (1996) Mol. Gen. Genet. 252, 51–60. [DOI] [PubMed] [Google Scholar]
- 5.Pittendrigh, B., Reenan, R., ffrench-Constant, R. H. & Ganetzky, B. (1997) Mol. Gen. Genet. 256, 602–610. [DOI] [PubMed] [Google Scholar]
- 6.Weill, M., Malcolm, C., Chandre, F., Mogensen, K., Berthomieu, A., Marquine, M. & Raymond, M. (2004) Insect Mol. Biol. 13, 1–7. [DOI] [PubMed] [Google Scholar]
- 7.Fournier, D., Mutero, A., Pralavorio, M. & Bride, J. M. (1993) Chem. Biol. Interact. 87, 233–238. [DOI] [PubMed] [Google Scholar]
- 8.Daborn, P. J., Yen, J. L., Bogwitz, M. R., Le Goff, G., Feil, E., Jeffers, S., Tijet, N., Perry, T., Heckel, D., Batterham, P., et al. (2002) Science 297, 2253–2256. [DOI] [PubMed] [Google Scholar]
- 9.Carino, F. A., Koener, J. F., Plapp, F. W., Jr., & Feyereisen, R. (1994) Insect Biochem. Mol. Biol. 24, 411–418. [DOI] [PubMed] [Google Scholar]
- 10.Le Goff, G., Boundy, S., Daborn, P. J., Yen, J. L., Sofer, L., Lind, R., Sabourault, C., Madi-Ravazzi, L. & ffrench-Constant, R. H. (2003) Insect Biochem. Mol. Biol. 33, 701–708. [DOI] [PubMed] [Google Scholar]
- 11.Field, L. M., Devonshire, A. L. & Forde, B. G. (1988) Biochem. J. 251, 309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mouches, C., Pasteur, N., Berge, J. B., Hyrien, O., Raymond, M., de Saint Vincent, B. R., de Silvestri, M. & Georghiou, G. P. (1986) Science 233, 778–780. [DOI] [PubMed] [Google Scholar]
- 13.Enayati, A. A., Ranson, H. & Hemingway, J. (2005) Insect Mol. Biol. 14, 3–8. [DOI] [PubMed] [Google Scholar]
- 14.Berge, J. B., Feyereisen, R. & Amichot, M. (1998) Philos. Trans. R. Soc. London B 353, 1701–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amichot, M., Tares, S., Brun-Barale, A., Arthaud, L., Bride, J. M. & Berge, J. B. (2004) Eur. J. Biochem. 271, 1250–1257. [DOI] [PubMed] [Google Scholar]
- 16.Campbell, P. M., Newcomb, R. D., Russell, R. J. & Oakeshott, J. G. (1998) Insect Biochem. Mol. Biol. 28, 139–150. [DOI] [PubMed] [Google Scholar]
- 17.Dean, S. R., Meola, R. W., Meola, S. M., Sittertz-Bhatkar, H. & Schenker, R. (1998) J. Med. Entomol. 35, 720–724. [DOI] [PubMed] [Google Scholar]
- 18.Wilson, T. G. & Cryan, J. R. (1997) J. Exp. Zool. 278, 37–44. [DOI] [PubMed] [Google Scholar]
- 19.Wilson, T. G. & Cryan, J. R. (1996) in Molecular Genetics and Evolution of Pesticide Resistance, ed. Brown, T. (Am. Chem. Soc., Washington, DC), pp. 141–148.
- 20.Catania, F., Kauer, M. O., Daborn, P. J., Yen, J. L., ffrench-Constant, R. H. & Schlotterer, C. (2004) Mol. Ecol. 13, 2491–2504. [DOI] [PubMed] [Google Scholar]
- 21.Magoc, L., Yen, J. L., Hill-Williams, A., McKenzie, J. A., Batterham, P. & Daborn, P. J. (2005) Pestic. Biochem. Physiol. 81, 129–135. [Google Scholar]
- 22.Sakuma, M. (1998) Appl. Entomol. Zool. 33, 339–347. [Google Scholar]
- 23.Pyke, F. M., Bogwitz, M. R., Perry, T., Monk, A., Batterham, P. & McKenzie, J. A. (2004) Genetica 121, 13–24. [DOI] [PubMed] [Google Scholar]
- 24.Daborn, P., Boundy, S., Yen, J., Pittendrigh, B. & ffrench-Constant, R. (2001) Mol. Genet. Genomics 266, 556–563. [DOI] [PubMed] [Google Scholar]
- 25.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 26.Lee, T. & Luo, L. (1999) Neuron 22, 451–461. [DOI] [PubMed] [Google Scholar]
- 27.Shiga, Y., Tanaka-Matakatsu, M. & Hayashi, S. (1996) Dev. Growth Differ. 38, 99–106. [Google Scholar]
- 28.Tautz, D. & Pfeifle, C. (1989) Chromosoma 98, 81–85. [DOI] [PubMed] [Google Scholar]
- 29.Drysdale, R. A., Crosby, M. A., Gelbart, W., Campbell, K., Emmert, D., Matthews, B., Russo, S., Schroeder, A., Smutniak, F., Zhang, P., et al. (2005) Nucleic Acids Res. 33, D390–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caizzi, R., Caggese, C. & Pimpinelli, S. (1993) Genetics 133, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kel, A. E., Gossling, E., Reuter, I., Cheremushkin, E., Kel-Margoulis, O. V. & Wingender, E. (2003) Nucleic Acids Res. 31, 3576–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, T. G. & Cain, J. W. (1997) J. Econ. Entomol. 90, 1131–1136. [DOI] [PubMed] [Google Scholar]
- 33.Tomita, T. & Scott, J. G. (1995) Insect Biochem. Mol. Biol. 25, 275–283. [DOI] [PubMed] [Google Scholar]
- 34.Pridgeon, J. W., Zhang, L. & Liu, N. (2003) Gene 314, 157–163. [DOI] [PubMed] [Google Scholar]
- 35.Sabourault, C., Guzov, V. M., Koener, J. F., Claudianos, C., Plapp, F. W., Jr., & Feyereisen, R. (2001) Insect Mol. Biol. 10, 609–618. [DOI] [PubMed] [Google Scholar]
- 36.Scharf, M. E., Parimi, S., Meinke, L. J., Chandler, L. D. & Siegfried, B. D. (2001) Insect Mol. Biol. 10, 139–146. [DOI] [PubMed] [Google Scholar]
- 37.Dunkov, B. C., Guzov, V. M., Mocelin, G., Shotkoski, F., Brun, A., Amichot, M., ffrench-Constant, R. H. & Feyereisen, R. (1997) DNA Cell Biol. 16, 1345–1356. [DOI] [PubMed] [Google Scholar]
- 38.Feyereisen, R. (2005) in Comprehensive Molecular Insect Science, eds. Gilbert, L. I., Iatrou, K. & Gill, S. S. (Elsevier), Vol. 4, pp. 1–77. [Google Scholar]
- 39.Guzov, V. M., Unnithan, G. C., Chernogolov, A. A. & Feyereisen, R. (1998) Arch. Biochem. Biophys. 359, 231–240. [DOI] [PubMed] [Google Scholar]
- 40.Brun, A., Cuany, A., Le Mouel, T., Berge, J. & Amichot, M. (1996) Insect Biochem. Mol. Biol. 26, 697–703. [DOI] [PubMed] [Google Scholar]
- 41.Li, T. R. & White, K. P. (2003) Dev. Cell 5, 59–72. [DOI] [PubMed] [Google Scholar]
- 42.Mozer, B. A. & Benzer, S. (1994) Development (Cambridge, U.K.) 120, 1049–1058. [DOI] [PubMed] [Google Scholar]
- 43.Franchini, L. F., Ganko, E. W. & McDonald, J. F. (2004) Mol. Biol. Evol. 21, 1323–1331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.