Abstract
During postembryonic plant development, cell division is coupled to cell growth. There is a stringent requirement to couple these processes in shoot and root meristems. As cells pass through meristems, they transit through zones with high rates of cell growth and proliferation during organogenesis. This transition implies a need for coordinate regulation of genes underpinning these two fundamental cell functions. Here, we report a mechanism for coregulation of cell division control genes and cell growth effectors. We identified a GCCCR motif necessary and sufficient for high-level cyclin CYCB1;1 expression at G2/M. This motif is overrepresented in many ribosomal protein gene promoters and is required for high-level expression of the S27 and L24 ribosomal subunit genes we examined. p33TCP20, encoded by the Arabidopsis TCP20 gene, binds to the GCCCR element in the promoters of cyclin CYCB1;1 and ribosomal protein genes in vitro and in vivo. We propose a model in which organ growth rates, and possibly shape in aerial organs, are regulated by the balance of positively and negatively acting teosinte-branched, cycloidea, PCNA factor (TCP) genes in the distal meristem boundary zone where cells become mitotically quiescent before expansion and differentiation.
Keywords: cyclin B1;1, growth control, teosinte-branched, cycloidea, PCNA factor genes
Enhanced expression of mitotic cyclin CYCB1;1 in transgenic Arabidopsis or of the D-type cyclin CYCD2;1 in tobacco has yielded plants with accelerated organ growth, without affecting overall developmental control or final organ size in determinate organs (1, 2). These results are paradoxical, because cell proliferation does not proceed without concomitant cell growth.
Plant organ growth is mediated in aggregate by three processes: cell growth, division, and expansion. Although the net outcome of cell growth and cell expansion is cell enlargement, the difference between these two is not semantic because they are driven by different processes. During cell growth, the increase in mass is a consequence of stimulated macromolecular, mostly protein, synthesis required for enhanced metabolism. In meristems and organs, cell growth is necessary to ensure the survival of cells after division and, hence, must be spatially and temporally tightly coupled with proliferation. In contrast, cell expansion predominates in postmitotic cells, accounts for the bulk of the overall size increase in developing organs, and is therefore responsible for most of the growth of the plant body. In cell expansion, mass increase is largely due to osmotically driven water uptake and the initiation of cell enlargement spatio-temporally coincides with the development and expansion of vacuoles. Despite their fundamental importance for plant growth, the genetic mechanisms governing cell growth and size are far less well understood than those regulating division and expansion.
In a shoot or root meristem, cells transit through a succession of developmental zones with different local rates of cell division. Spatial analysis of cell division patterns in meristems has revealed that stem cells, the ultimate source of all shoot and root cells, proliferate very slowly. Their immediate progeny, which form the flanks of the shoot meristem from which lateral organ primordia arise, or the initials in the root that generate the cell types of the root, proliferate slightly faster (3-5). Subsequently, in newly initiated leaf primordia and in the domain distal to the initials in the root, a rapid increase of cell division rates is observed (5-7). We term this the zone of multiplicative divisions. Finally, the rates of cell division gradually decline at the distal end of the root meristem and in a distal-proximal gradient in leaves as cells begin to differentiate (6-10). Analysis of cell morphology in these zones has revealed a gradual size increase as cells progress through these zones (4, 5, 11, 12). Specifically, no size reduction as cells transit into the zone of rapid proliferation was observed in roots (ref. 12 and J. Dubrovsky and P.D., unpublished data).
To maintain approximately constant cell sizes in meristems, the rates of cell division and cell growth must locally be identical; this implies a fundamental requirement for coordinate regulation of cell growth and division in the meristem, specifically in the zone of multiplicative divisions, where proliferation rates change rapidly. Such coordinate regulation is revealed when shoot apices are treated with oryzalin, an inhibitor of mitosis: Cells in young primordia grow more than cells in the central or peripheral zones of the meristem, revealing differential control of growth rates (4). Cell growth requires increased rates of metabolism, mediated by up-regulation of ribosome synthesis and other components involved in protein synthesis (13). Stimulated ribosome biogenesis involves coordinated gene expression mediated by all three types of RNA polymerase; however, coordination of gene expression programs for cell division and cell growth is likely to rely on RNA polymerase II-dependent transcription.
Here, we examine quantitative aspects of Arabidopsis mitotic cyclin CYCB1;1 and ribosomal protein gene L24 and S27 expression and report a DNA sequence motif necessary and sufficient for high-level expression. This motif is overrepresented in promoters of genes that function in ribosome biogenesis. We show that the protein encoded by Arabidopsis TCP20 specifically binds to the cyclin B1;1 promoter, as well as to several ribosomal protein promoters, thereby revealing mechanistic links between growth and cell cycle control in plants.
Methods
Constructs. We made 5′ deletions of the CYCB1;1 promoter in pCDG (7) with exonuclease III (14). Constructs were ligated into the pBIB vector (15) to give p351CDG, p205CDG, p143CDG, p120CDG, and p60CDG, with the numbers indicating the 5′ deletion endpoint relative to the transcription start site. Constructs expressing CYCB1;1 were made by replacing a XhoI-SacI fragment from uidA with a XhoI-SacI fragment from the CYCB1;1 cDNA. Modification of candidate binding sites was made by inverse PCR. Gain-of-function constructs were generated on the basis of the -46 35S minimal promoter (16), and contained five or three copies each of the GCCCR or the MSA motif (17), or both, respectively. The minimal promoter, or synthetic promoters were used to express the cyclin-GUS (CGU) cassette in pCDG (7) downstream of the translation start site.
Plant Material and Transformation. Arabidopsis thaliana Col-0 was grown at 21°C with a 16-h light/8-h dark cycle. Bright Yellow 2 tobacco (BY2) cells were grown at 27°C in the dark (18). Plants and cell cultures were transformed with Arabidopsis tumefaciens GV3101 (19) as described by Clough et al. (20) and An (21), respectively. At least 100 calli were pooled for each transgenic BY2 cell culture.
Expression Analysis. For histochemical or fluorometric GUS analysis, tissues were assayed as described (7, 22). Tobacco BY2 cells were synchronized as described (7, 18). RNA was isolated with TRIzol (Invitrogen). RNA (0.5 μg) was reverse-transcribed by using oligo(dT) or gene-specific primers. PCRs (see Supporting Text, which is published as supporting information on the PNAS web site, for primer sets) all contained one primer designed to cross an intron and were performed in quadruplicate with the iCycler iQ (Bio-Rad). To normalize for cDNA loading, threshold cycle differences were obtained by subtracting the mean threshold cycle (MTC) for each gene (designated a) from the MTC of β-ATPase (23) amplification from the same cDNA sample (designated b). Relative amounts were then calculated by subtracting this value from the MTC of the sample designated to be the reference (designated c). To represent relative amounts, these logarithms were inverted by using the relationship: relative amount = 2c-(b-a).
In Vivo Footprint Analysis. In vivo footprinting was performed by using Arabidopsis suspension cells (24) or mature Arabidopsis leaves as described (25-27), using separate primer sets for upper and lower strands (see Supporting Text).
EMSA. Full-length TCP20 protein was expressed as GST-fusion in E. coli, purified by affinity chromatography, and subsequently cleaved from GST. EMSA was conducted with radiolabeled oligonucleotides corresponding to wild-type or mutated GCCCR motifs. Binding reactions were done at room temperature for 20 min in 25 μl containing 20 mM Tris·HCl (pH 8.5), 50 mM KCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 200 ng/μl BSA, 10% glycerol, 1 μg poly (dI-dC), 1 ng of labeled, probe and 0.1-1 μg of protein. For competition experiments, a 100- to 500-fold molar excess of unlabeled competitors was added 10 min before the labeled probe.
Chromatin Immunoprecipitation (ChIP). Antibodies were raised in sheep against a GST-fusion protein corresponding to amino acids 275-315 of TCP20. Antibodies were affinity purified, and 5-μl aliquots were used for ChIP experiments using extracts from young seedlings (28). See Supporting Text for primer sequences. All reactions included primers for the internal control At4. Band intensities were measured with NIH image version 1.62, and enrichment was calculated as ratio of precipitated versus input DNA after normalization to At4.
Results
Enhanced Expression of Arabidopsis CYCB1;1 Stimulates Growth. Increased expression of Arabidopsis CYCB1;1 under control of the Arabidopsis CDKA;1 promoter enhances root and shoot growth (1). CYCB1;1 expression peaks at G2/M in the cell cycle (1, 7, 29), whereas the CDKA;1 promoter directs expression at uniform levels throughout the cell cycle (30-33). This raised the possibility that enhanced growth observed in CDKA;1::CYCB1;1 transformed plants was due to an unknown CYCB1;1 function in the G1 phase of the cell cycle. To address whether CYCB1;1 was an effector of growth control pathways at G2/M, we increased expression of CYCB1;1 specifically at G2/M. Arabidopsis was transformed with constructs in which CYCB1;1 was expressed under its own promoter. In lines expressing CYCB1;1 at high levels (data not shown) due to variation of expression observed in individual transformation events, we observed enhanced shoot (data not shown) and root growth rates (Table 1, which is published as supporting information on the PNAS web site). The cell length of mature cortical cells was very similar in wild-type Col-0 and these transgenic lines (Col-0, 164.1 μm ± 1.96 μm SEM; 11D, 165.6 μm ± 1.57 μm SEM; 26A, 165.5 μm ± 1.72 μm SEM) and, therefore, we concluded that enhanced organ growth was due to increased cell production. Hence, CYCB1;1 is an effector for growth control at G2/M. Together with our previous report that CYCB1;1 expression levels limit growth (1), these results highlight the importance of quantitative control of CYCB1;1 expression for plant growth.
Qualitative and Quantitative cis-Elements in the CYCB1;1 Promoter. To identify the regulatory elements responsible for CYCB1;1 expression, we used a cyclin-GUS gene fusion (pCDG, with 1.2 kb of 5′ DNA sequence) (7), to generate 5′ promoter deletions and analyzed these in transformed Arabidopsis plants and tobacco BY-2 cells. Removal of 5′ promoter sequences up to -351 relative to the transcription start site (Fig. 1) revealed no detectable differences in expression pattern or level, when compared to pCDG (data not shown). Therefore, we focused subsequent analysis on the promoter deletions terminating 351, 205, 143, 120, and 60 bp upstream of the transcription start, respectively (Fig. 1). Histochemical analysis of 5-12 homozygous plant lines for each construct revealed similar levels of expression in plants carrying p351CDG and p205CDG when compared to the full-length promoter. However, further 5′ deletion of promoters to -143, -120, and -60 resulted in a drastic reduction of histochemical staining.
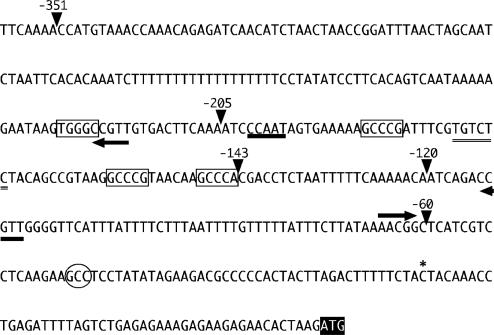
Fig. 1.
Proximal CYCB1;1 promoter. The transcription start site, indicated by an asterisk, was determined by primer extension (data not shown). Endpoints of 5′ deletions are indicated by arrowheads. Possible CCAAT and ARF-binding sites are indicated by thick and double underscore, respectively. GCCCR elements are boxed and MSA elements are indicated by arrows. The hypersensitive footprint is marked with an oval and the lettering of the start codon is inverted.
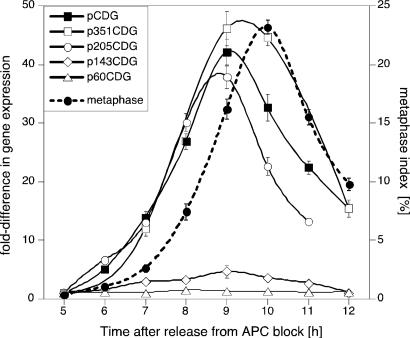
To delineate the promoter elements responsible for regulating CYCB1;1 RNA levels, we analyzed RNA in synchronized BY2 cells transformed with the aforementioned constructs. Cyclin-GUS (CGU) RNA levels were normalized to endogenous β-ATPase mRNA levels, which do not fluctuate during the cell cycle (23). Quantitative analysis in cells transformed with pCDG, p351CDG and p205CDG revealed high-level induction of CGU RNA as cells approached M phase (Fig. 2). In contrast, cells transformed with p143CDG expressed CGU RNA with 8- to 10-fold lower amplitude but still with appropriate cell cycle timing (Fig. 2). Further 5′ deletion of the promoter to -60 led to the loss of M phase induction of CGU expression (Fig. 2). We concluded that sequences required for cell cycle-specific timing were located between -143 and -60 bp and that elements required for high-magnitude expression were located between -205 and -143 bp of the CYCB1;1 promoter.
Fig. 2.
A major quantitative determinant of CYCB1;1 expression. BY2 cells were transformed with CYCB1;1 promoter 5′ deletion constructs directing expression of a chimeric cyclin-GUS (CGU) marker. RNA was isolated from BY2 cells synchronized with Aphidicolin and analyzed by real time RT-PCR. The figure shows gene expression (left axis) and metaphase index (right axis) relative to the values measured 5 h after release from Aphidicolin block. Expression of the CGU reporter in mitotic cells was significantly lower when the DNA sequence between -205 and -143 was removed from the promoter.
Identification of Quantitative Elements by in Vivo Footprinting. Inspection of the DNA sequence between -143 and -60 bp revealed two copies of the previously identified MSA element required for G2/M phase-specific timing of expression (17) (Fig. 1). We also identified a sequence weakly homologous to the CCAATCA-box and an auxin response factor (ARF)-binding site (Fig. 1). We used in vivo footprint analysis (25) to identify the binding sites of factors involved in CYCB1;1 expression. We identified two classes of footprints. In Arabidopsis suspensioncultured cells or young leaves that still contained mitotic cells (6), we observed a hypersensitive site at GCC residues at -41 to -43 relative to the transcription start site (data not shown, Fig. 1), which was not observed in older, quiescent tissues.
A second class of footprint was identified, which defined an element with the consensus sequence GCCCR (R = G or A) (Fig. 7, which is published as supporting information on the PNAS web site). This element was protected from dimethyl sulfate modification in actively dividing cells, as well as in cells competent to divide, but not in quiescent cells in tissues such as mature leaves. This element is repeated four times in the CYCB1;1 -351 promoter (Fig. 1). Three of the four copies of the GCCCR element are located between the -205 and -143 5′ deletion endpoints, raising the possibility that the cognate transcription factor is a major determinant of CYCB1;1 transcript abundance.
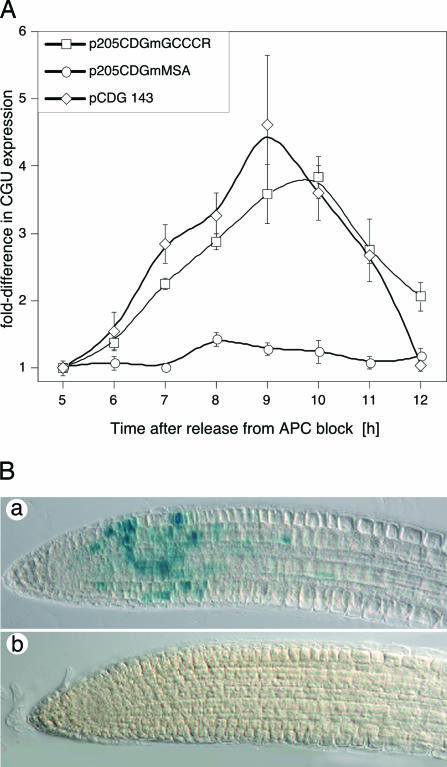
Functional Analysis of Elements in Synchronized BY2 cells and Arabidopsis Plants. To test this possibility, the three GCCCR motifs between -143 and -205 were mutated to AAATR. The corresponding construct (pCDGmGCCCR) was introduced into BY2 cells and the accumulation of CGU RNA analyzed in synchronized cells (see Fig. 3A). CGU expression from the mutant promoter peaked at 9-10 h, coincident with the peak in mitotic activity (Fig. 2) and the maximum of p205CDG expression, but the magnitude of expression was low and similar to that observed in cells transformed with the p143CDG construct. In plants transformed with the AAATR mutant, we did not detect any GUS activity by histochemical analysis (Fig. 3B), and quantitative analysis by fluorescence spectrophotometry revealed a 20-fold reduction of GUS activity in root tips of these plants when compared to the wild-type (data not shown). We concluded that the GCCCR motif was necessary for high-level CYCB1;1 expression at G2/M. We also examined whether the MSA elements (Fig. 1) were required to confer G2/M-specific expression in the context of the -205 promoter. Previous work had shown that this motif is required for G2/M phase expression in Catharanthus roseus cyclin CYM, when assayed in BY2 cells (17, 34). The AACGG core MSA motif was mutated to ACTAG or AGCTG. CGU expression in BY2 lines transformed with this construct was not cell cycle regulated (Fig. 3A), indicating that the MSA motif is necessary for G2/M-specific CYCB1;1 expression.
Fig. 3.
Elements required for CYCB1;1 expression in mitosis. The GCCCR elements within the -205 promoter were destroyed by site-directed mutagenesis and transformed into BY2 cells (A) or Arabidopsis plants (B). Mutation of the GCCCR element abrogates high-level CYCB1;1 expression, but does not affect cell cycle phase-specific expression at G2/M. However, mutation of the MSA elements within the -205 promoter quenches CYCB1;1 expression at G2/M. (A) Gene expression (left axis) relative to the values measured 5 h after release from the Aphidicolin block. (B) Histochemical analysis of GUS activity. (a) p205CDG-directed GUS activity. (b) p205CDGmGCCCR-directed GUS activity in roots. Identical results were obtained from 12 independently transformed lines.
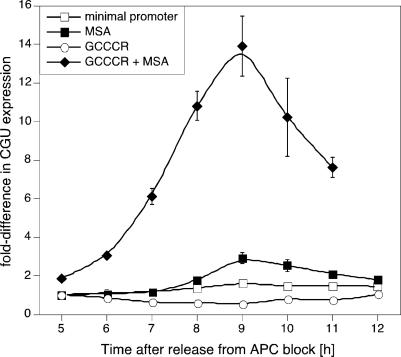
To examine whether the GCCCR motif was sufficient to confer high-level induction of CYCB1;1 expression, we generated a series of synthetic promoters on the basis of the -46 minimal 35S promoter. When expressed under control of the minimal promoter, CGU RNA did not exhibit any cell cycle regulation and addition of the GCCCR motif alone slightly suppressed the steady-state levels of CGU RNA (Fig. 4). However, when three copies of the MSA motif were added to the minimal promoter, G2/M phase regulation of CGU abundance was observed, which was strikingly enhanced, when five copies of the GCCCR motif were added 5′ to the MSA elements (Fig. 4). Thus, the GCCCR motif is necessary and sufficient to enhance G2/M expression mediated by MSA elements.
Fig. 4.
The GCCCR element is necessary and sufficient to confer high-level CYCB1;1 expression in mitosis. Oligonucleotides with the MSA element or the GCCCR element were cloned 5′ of the minimal 35S-46 promoter. The minimal promoter is not cell cycle phase-regulated, but addition of the MSA element restores expression at G2/M. Addition of the GCCCR element alone slightly suppresses expression directed by the minimal promoter. However, addition of the GCCCR element to the MSA element strongly enhances expression with appropriate timing at G2/M.
Genomic Analysis of the GCCCR Element in Arabidopsis. We next examined the distribution of GCCCR elements in promoters of Arabidopsis genes. Genes with the GCCCR element in their promoters were assigned to functional categories based on the scheme developed by the Munich Information Center for Protein Sequences (35). The frequency of each functional term in the list of genes was then compared to the frequency in the whole genome, and a P value of overrepresentation was calculated by using the hypergeometric distribution (R.A.G., L. Lejay, and G. M. Coruzzi, unpublished data). This analysis revealed that ribosomal protein, ribosome biogenesis, translation, and protein synthesis functions were significantly overrepresented (P value < 0.0001) in genes with three to four copies of the GCCCR element (Fig. 8, which is published as supporting information on the PNAS web site). Strikingly, genes with similar functions are induced when Arabidopsis plants are exposed to optimal concentrations of nitrogen and carbon, which strongly stimulate growth (R.A.G., L. Lejay, and G. M. Coruzzi, unpublished data). This finding raised the possibility that a subset of cell cycle genes, e.g., cyclins, and genes required for cell growth are coordinately regulated by shared transcription factors.
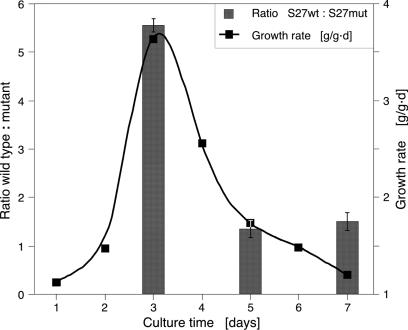
GCCCR Elements Are Required for High-Level Ribosomal Protein Expression. We then examined whether GCCCR elements in promoters of ribosomal protein (RP) genes were also required for their high-level expression. We selected two ribosomal protein genes, RPS27aB (At3g46040) and RPL24B (At3g53020), with six and five GCCCR motifs, respectively, and mutated these elements. Constructs expressing the CGU reporter under control of wild-type or mutant promoters were introduced into BY2 cells. High-level RPS27aB expression was strongly dependent on the GCCCR motif, specifically when growth rates were high during the culture cycle (Fig. 5). High-level expression of RPL24B was also dependent on GCCCR elements, but we observed less correlation with culture growth rates. We concluded that, as for CYCB1;1, high-level expression of these two RP genes depended on intact GCCCR elements.
Fig. 5.
The GCCCR element is necessary for high-level ribosomal protein gene expression. The promoter of RPS27aB was mutated to remove GCCCR motifs. Corresponding wild-type and mutant promoter-reporter constructs were introduced into BY2 cells. Growth rate was determined by measuring fresh weights of cultures. RPS27aB gene expression is more dependent on GCCCR motifs at high growth rates.
p33TCP20 Binds to the GCCCR Element in Vitro and in Vivo. Class I teosinte-branched, cycloidea, PCNA factor (TCP) transcription factors bind to GGNCCCAC consensus sites (36), and the GCCCR element identified here forms a subset of these. Therefore, we examined whether TCP20, which had been shown to bind the Arabidopsis PCNA2 promoter (37), interacts with the GCCCR elements in the CYCB1;1 promoter in EMSAs. Recombinant TCP20 (p33TCP20) binds specifically to the GCCCR elements in the CYCB1;1 promoter (Fig. 9, which is published as supporting information on the PNAS web site), and is competed by an excess of unlabelled probe but cannot be competed with mutant probes.
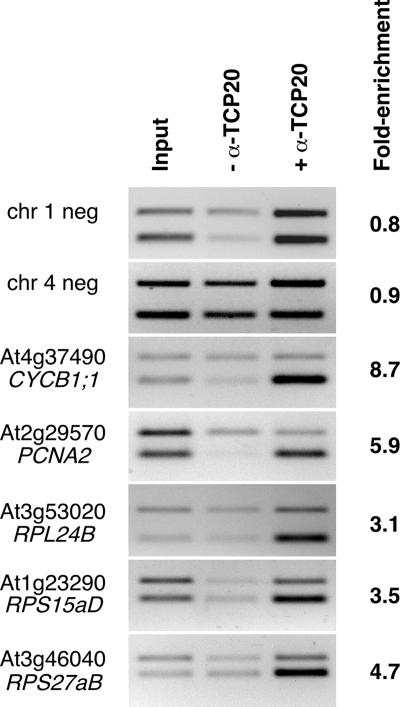
We then used chromatin immunoprecipitation assays to examine whether p33TCP20 binds in vivo to the CYCB1;1 and promoters of other genes involved in executing growth programs with GCCCR motifs. Antibodies raised in sheep were used to precipitate p33TCP20 cross-linked with DNA isolated from 12-day-old Arabidopsis seedlings. Subsequently, primers specific for CYCB1;1, PCNA2, RPL24B, RPS15aD, and RPS27aB were used to determine whether these genes are p33TCP20 targets (Fig. 6). As controls, we examined p33TCP20 binding to two genomic fragments on chromosome 1 and 4, which were >2 or >1.4 kb distant from any GCCCR motif, respectively. The negative controls showed no enrichment of templates recognized by the anti-p33TCP20 antibody, whereas CYCB1;1 and PCNA2 were specifically enriched by 8.7and 5.9-fold, respectively (Fig. 6). DNA fragments corresponding to the ribosomal proteins were also enriched (Fig. 6). We conclude that p33TCP20 coregulates the expression of a suite of cell cycle control and ribosomal protein genes.
Fig. 6.
p33TCP20 coregulates cell cycle and ribosomal protein genes. Chromatin cross-linked to DNA isolated from Arabidopsis seedlings was immunoprecipitated with anti-p33TCP20 antibodies. PCRs were performed with input DNA (Left), DNA precipitated without addition of anti-p33TCP20 antibodies (Center), and DNA immunoprecipitated with anti-p33TCP20 antibodies (Right). All reactions were performed in the presence of an internal control used to normalize reactions (top band), corresponding to sequences within At5g03545 (At4). Enrichment was calculated after normalizing input and immunoprecipitated reactions and comparing their ratios. Gene identifiers are shown at left.
Discussion
We report the first mechanistic links between the regulation of cell growth and division in plants. The Arabidopsis TCP20 gene product, p33TCP20, binds in vivo to cognate GCCCR elements in the promoters of CYCB1;1, of PCNA2, as well as the RPL24B, RPS15aD, and RPS27aB ribosomal protein genes we examined. GCCCR elements are required for high-level expression of the cyclin and ribosomal protein genes we examined.
Almost half (40%) of all Arabidopsis RP genes carry several clustered GCCCR motifs in their proximal promoters, suggesting that TCP-gene mediated transcriptional regulation of RP gene expression is likely to significantly contribute to the regulation of ribosome biogenesis. Most RP genes in Arabidopsis comprise small gene families. It is striking that, with two exceptions, GCCCR motifs are present in the promoters of only one member per RP gene family, suggesting that RP gene duplication has enabled their promoters to be differentially regulated. Moreover, the different temporal expression patterns of genes with GCCCR motifs (for example, PCNA2 expression peaks in S phase, whereas the maximum of CYCB1;1 expression is in G2/M; ref. 32) suggests that the cognate TCP factors are likely to interact with different specific transcription factors to mediate high expression levels. However, we were not able to demonstrate in vivo p33TCP20 binding to all promoter sequences with GCCCR motifs. For example, Arabidopsis cyclin CYCA3;4 (At1g47230) and the transcription factor E2Fc (At1g47870) did not show enrichment in ChIP assays (data not shown). It is possible that p33TCP20 does not bind to these promoters in vivo, that these factors are not coexpressed in the same cells, or alternatively, that the epitope recognized by the anti-p33TCP20 antibody was occluded in the cross-linked tissue samples.
TCP20 is a class I TCP gene, and its closest paralogs in Arabidopsis are TCP6 and -11, which also bind to the GCCCR motif present in the CYCB1;1 promoter in vitro (data not shown). These three genes, and possibly additional class I genes, may be functionally partially redundant, because the TCP20 knockout mutant has no obvious growth phenotype (C.L. and P.D., unpublished data). Class I TCP genes positively regulate gene expression (38), whereas class II TCP genes, such as Arabidopsis TCP2 and -4 or Antirrhinum majus CYCLOIDEA, DICHOTOMA, and CINCINNATA negatively regulate proliferation (8, 39-41). Precise spatio-temporal regulation of class II gene RNA accumulation is critical for leaf growth and morphogenetic development (8, 39). Interestingly, the DNA sequences recognized by class I (GGNCCCAC) and class II (GGNCCC) TCP genes are not mutually exclusive (36). Strikingly, we found that, in promoters with three or four GCCCR motifs (Fig. 8), these are much more likely to be nested within a GGNCCC motif than expected by chance alone (P < 0.0001). This finding raises the attractive possibility that both classes of TCP genes share target genes.
We propose a model in which class I TCP factors mediate the marked stimulation of cell growth and division required for elevated cell production rates in young lateral primordia in shoots (6) or in the multiplicative division zone of the root meristem (7). This elevation would be followed by concerted suppression of cell growth and division by class II TCP genes as cells exit the multiplicative zone. Thus, organ growth is proposed to be regulated by the balance of antagonistic activities of class I and II TCP genes. In this model, the crucial variable underpinning organ growth is the population size of dividing cells within the zone of multiplicative divisions. Indeed, kinematic analysis of root growth reveals that, although there is no evidence for shortening cell cycle duration when root growth accelerates, there is clear evidence that the multiplicative division zone expands distally as root organ growth accelerates (9, 42). This finding highlights the importance of controlling the exit from this zone for overall organ growth rate in roots and organ shape and morphology in shoots. It will be interesting to test this model by examining the balance of TCP gene activities in this boundary zone.
Supplementary Material
Acknowledgments
We thank Ratha You, Tal Haimovitch-Gal, Hang Phung, Agnoula Gourzoulidou, Simone Baumann, and Kristin Höwing for outstanding support during the course of the project; Daniel Schubert for help with the ChIP analysis; and all members of the P.D. laboratory for discussions. A.C.-C. was supported by National Science Foundation Postdoctoral Fellowship BIR-9510821 and Grant MCB-000-3328. T.P. was supported by an Erwin Schrödinger Auslandsstipendium (J1907-Bio) of the Fondszur Förderung der Wissenschaftlichen Forschung and European Molecular Biology Organization Short-Term Fellowship ASTF 9427; we thank Marie-Edith Chabouté, Strasbourg, for hosting T.P. during the Short-Term Fellowship. This work was supported by U.S. Department of Agriculture Grant 95-37304-2228 and Biotechnology and Biological Sciences Research Council Grant G13698 (to P.D.).
Author contributions: C.L., T.P., A.C.-C., and P.D. designed research; C.L., T.P., A.C.-C., and P.D. performed research; R.A.G. contributed new reagents/analytic tools; C.L., T.P., A.C.-C., R.A.G., and P.D. analyzed data; and P.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TCP, teosinte-branched, cycloidea, PCNA factor; ChIP, chromatin immunoprecipitation; RP, ribosomal protein.
References
- 1.Doerner, P., Jorgensen, J. E., You, R., Steppuhn, J. & Lamb, C. (1996) Nature 380, 520-523. [DOI] [PubMed] [Google Scholar]
- 2.Cockcroft, C. E., den Boer, B. G., Healy, J. M. & Murray, J. A. (2000) Nature 405, 575-579. [DOI] [PubMed] [Google Scholar]
- 3.Laufs, P., Grandjean, O., Jonak, C., Kieu, K. & Traas, J. (1998) Plant Cell 10, 1375-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grandjean, O., Vernoux, T., Laufs, P., Belcram, K., Mizukami, Y. & Traas, J. (2004) Plant Cell 16, 74-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reddy, G. V., Heisler, M. G., Ehrhardt, D. W. & Meyerowitz, E. M. (2004) Development (Cambridge, U.K.) 131, 4225-4237. [DOI] [PubMed] [Google Scholar]
- 6.Donnelly, P. M., Bonetta, D., Tsukaya, H., Dengler, R. E. & Dengler, N. G. (1999) Dev. Biol. 215, 407-419. [DOI] [PubMed] [Google Scholar]
- 7.Colon-Carmona, A., You, R., Haimovitch-Gal, T. & Doerner, P. (1999) Plant J. 20, 503-508. [DOI] [PubMed] [Google Scholar]
- 8.Nath, U., Crawford, B. C., Carpenter, R. & Coen, E. (2003) Science 299, 1404-1407. [DOI] [PubMed] [Google Scholar]
- 9.Beemster, G. & Baskin, T. (1998) Plant Physiol. 116, 1515-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov, V. B., Dobrochaev, A. E. & Baskin, T. I. (2002) J. Plant Growth Regul. 21, 60-67. [DOI] [PubMed] [Google Scholar]
- 11.Kwiatkowska, D. & Dumais, J. (2003) J. Exp. Bot. 54, 1585-1596. [DOI] [PubMed] [Google Scholar]
- 12.Dolan, L., Janmaat, K., Willemsen, V., Linstead, P. J., Poethig, S., Roberts, K. & Scheres, B. (1993) Development (Cambridge, U.K.) 119, 71-84. [DOI] [PubMed] [Google Scholar]
- 13.Conlon, I. & Raff, M. (1999) Cell 96, 235-244. [DOI] [PubMed] [Google Scholar]
- 14.Ausubel, F. M., Brent, R., Kingston, R. E., More, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (1987) Current Protocols in Molecular Biology (Wiley-Interscience, New York).
- 15.Becker, D. (1990) Nucleic Acids Res. 18, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benfey, P. N., Ren, L. & Chua, N. H. (1990) EMBO J. 9, 1685-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, M., Iwase, M., Kodama, H., Lavisse, P., Komamine, A., Nishihama, R., Machida, Y. & Watanabe, A. (1998) Plant Cell 10, 331-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagata, T., Nemoto, Y. & Hasezawa, S. (1992) Int. Rev. Cytol. 132, 1-29. [Google Scholar]
- 19.Koncz, C. & Schell, J. (1986) Mol. Gen. Genet. 204, 383-396. [Google Scholar]
- 20.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 21.An, G. (1985) Plant Physiol. 1985, 568-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jefferson, R. A. (1987) Plant Mol. Biol. Rep. 5, 387-405. [Google Scholar]
- 23.Boutry, M. & Chua, N. H. (1985) EMBO J. 4, 2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May, M. J. & Leaver, C. J. (1993) Plant Physiol. 103, 621-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller, P. R. & Wold, B. (1989) Science 246, 780-786. [DOI] [PubMed] [Google Scholar]
- 26.Hornstra, I. K. & Yang, T. P. (1993) Anal. Biochem. 213, 179-193. [DOI] [PubMed] [Google Scholar]
- 27.Chaboute, M. E., Clement, B., Sekine, M., Philipps, G. & Chaubet-Gigot, N. (2000) Plant Cell 12, 1987-2000. [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, H., Tang, W., Zhu, C. & Perry, S. E. (2002) Plant J. 32, 831-843. [DOI] [PubMed] [Google Scholar]
- 29.Shaul, O., Mironov, V., Burssens, S., Van Montagu, M. & Inze, D. (1996) Proc. Natl. Acad. Sci. USA 93, 4868-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez, M. C., Jørgensen, J.-E., Lawton, M. A., Lamb, C. J. & Doerner, P. W. (1992) Proc. Natl. Acad. Sci. USA 89, 7360-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemerly, A. S., Ferreira, P., Engler, J. D. A., Montagu, M. V., Engler, G. & Inzé, D. (1993) Plant Cell 5, 1711-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menges, M., Hennig, L., Gruissem, W. & Murray, J. A. (2002) J. Biol. Chem. 277, 41987-42002. [DOI] [PubMed] [Google Scholar]
- 33.Menges, M., Hennig, L., Gruissem, W. & Murray, J. A. (2003) Plant Mol. Biol. 53, 423-442. [DOI] [PubMed] [Google Scholar]
- 34.Ito, M., Araki, S., Matsunaga, S., Itoh, T., Nishihama, R., Machida, Y., Doonan, J. H. & Watanabe, A. (2001) Plant Cell 13, 1891-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frishman, D., Mokrejs, M., Kosykh, D., Kastenmuller, G., Kolesov, G., Zubrzycki, I., Gruber, C., Geier, B., Kaps, A., Albermann, K., et al. (2003) Nucleic Acids Res. 31, 207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosugi, S. & Ohashi, Y. (2002) Plant J. 30, 337-348. [DOI] [PubMed] [Google Scholar]
- 37.Tremousaygue, D., Garnier, L., Bardet, C., Dabos, P., Herve, C. & Lescure, B. (2003) Plant J 33, 957-966. [DOI] [PubMed] [Google Scholar]
- 38.Kosugi, S. & Ohashi, Y. (1997) Plant Cell 9, 1607-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C. & Weigel, D. (2003) Nature 425, 257-263. [DOI] [PubMed] [Google Scholar]
- 40.Luo, D., Carpenter, R., Vincent, C., Copsey, L. & Coen, E. (1996) Nature 383, 794-799. [DOI] [PubMed] [Google Scholar]
- 41.Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J. & Coen, E. (1999) Cell 99, 367-376. [DOI] [PubMed] [Google Scholar]
- 42.Baskin, T. I. (2000) Plant Mol. Biol. 43, 545-554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.