Abstract
Proteins that can be reversibly photoswitched between a fluorescent and a nonfluorescent state bear enormous potential in diverse fields, such as data storage, in vivo protein tracking, and subdiffraction resolution light microscopy. However, these proteins could hitherto not live up to their full potential because the molecular switching mechanism is not resolved. Here, we clarify the molecular photoswitching mechanism of asFP595, a green fluorescent protein (GFP)-like protein that can be transferred from a nonfluorescent “off” to a fluorescent “on” state and back again, by green and blue light, respectively. To this end, we establish reversible photoswitching of fluorescence in whole protein crystals and show that the switching kinetics in the crystal is identical with that in solution. Subsequent x-ray analysis demonstrated that upon the absorption of a green photon, the chromophore isomerizes from a trans (off) to a cis (on) state. Molecular dynamics calculations suggest that isomerization occurs through a bottom hula twist mechanism with concomitant rotation of both bonds of the chromophoric methine ring bridge. This insight into the switching mechanism should facilitate the targeted design of photoswitchable proteins. Reversible photoswitching of the protein chromophore system within intact crystals also constitutes a step toward the use of fluorescent proteins in three-dimensional data recording.
Keywords: photoisomerization, asCP, photochromism, optical bistability, asulCP
Fluorescent proteins have been widely used as genetically encodable tags to monitor protein localizations and dynamics in live cells (1–3). Recently, novel GFP-like fluorescent proteins have been discovered (4–6) that can be reversibly photoswitched between a fluorescent (on) and nonfluorescent (off) state, that is, they are optically bistable and fluorescent. This feature is remarkable, because the reversible photoswitching occurring in photochromic organic compounds, such as in fulgides and diarylethenes, is usually not accompanied by fluorescence (7). Therefore, not surprisingly, these proteins hold great promise in many areas of science reaching out far beyond their prominent use as triggerable protein markers in live cells. For example, the reversible photoswitching of fluorescent markers should provide nanoscale resolution in fluorescence microscopy by using lenses and regular illumination, which was hardly conceivable only a few years ago (8–10). As fluorescence can be sensitively read out from a bulky crystal, the prospect of erasable three-dimensional data storage is equally intriguing.
The GFP-like protein asFP595 (asCP or asulCP) from the sea anemone Anemonia sulcata is such a protein: It can be transferred by green light from a nonfluorescent off into a fluorescent on state from which it reverts back eventually, but this transition can also be promptly stimulated by gentle irradiation with blue light (6). The “on-off” cycle can be repeated many times. However, with its low quantum yield (<0.001, ref. 6) and comparatively slow switching kinetics, the photochromic properties of asFP595 are far from being optimal. To systematically exploit the potential of switchable proteins and to approach a targeted improvement of their properties, a detailed molecular understanding of the switching process is essential. Although first insights based on the spectroscopic analysis of asFP595 mutants hinted at a cis-trans isomerization of the chromophore (11), the precise switching mechanism has remained indefinite.
We now report on protein crystals (of the variant asFP595-A143S) that can be photoswitched reversibly and display the same switching properties as the proteins in solution. Crystal structures of both the on and the off chromophoric states were obtained. The structural information enabled us to perform molecular dynamics (MD) simulations of potential pathways of the conformational change of the chromophore, a conjugated 2-iminomethyl-5-(4-hydroxybenzylidene)imidazolinone system formed by the tripeptide M63-Y64-G65. Force probe calculations combined with free dynamics simulations suggest that the chromophore undergoes a trans-cis-isomerization through a bottom hula twist (HTbot) mechanism.
Materials and Methods
Protein Purification. The plasmid pQE30-asFP595 was a generous gift from K. A. Lukyanov (Russian Academy of Sciences, Moscow). The mutants asFP595-S158V and asFP595-A143S were generated by site-directed PCR mutagenesis by using the QuikChange mutagenesis kit (Stratagene). The proteins were expressed in Escherichia coli BL21CodonPlus (Stratagene), purified by Ni-NTA affinity chromatography and subsequent size exclusion chromatography according to standard procedures. The purified proteins were concentrated by ultrafiltration and taken up in 20 mM Tris·HCl, pH 7.5/120 mM NaCl to ≈28 mg/ml for crystallization.
Crystallographic Analyses. wt asFP595 and variants could be readily crystallized overnight by sitting drop vapor diffusion with reservoirs containing 20% polyethylene glycol 3350, 0.2 M Tris·HCl (pH 7.1), and 0.3 M NaCl. After transfer into perfluoropolyether, crystals were mounted in a liquid nitrogen stream (Cryostream 700 series, Oxford Cryosystems, Oxford) for data collection at 100 K. Data were obtained on a rotating anode source (FR591, Bruker-Nonius, Karlsruhe, Germany) or on beamline BW6 (Deutsches Elektronen Synchrotron, Hamburg, Germany) and processed with the hkl package (ref. 12; see Table 1, which is published as supporting information on the PNAS web site).
The structure of wt asFP595 was solved by molecular replacement (molrep; ref. 13) by using the structure coordinates of Entacmaea quadricolor fluorescent protein 611 (14); (Protein Data Bank ID code 1UIS) and subsequently refined by standard protocols with refmac5 (13). The sequence was adjusted by manual model building in MAIN (www-bmb.ijs.si/doc/index.html). The chromophore was initially omitted, later on placed into clear difference electron density, and eventually included in the refinement (restraints from http://davapc1.bioch.dundee.ac.uk/programs/prodrg/prodrg.html). Final steps included the refinement of anisotropic temperature factors. The structures of the variants of asFP595 were solved by molecular replacement by using the refined wt structure but omitting the chromophore, which was placed manually into the difference electron densities later on. Refinement to the maximum resolutions (Table 1) was completed as outlined for the wt protein, but temperature factors were modeled isotropically.
The coordinates and structure factors of the five crystal structures (Table 1) have been deposited with the Protein Data Bank (ID codes 2A50, 2A52, 2A53, 2A54, and 2A56).
Optical Switching. Photoswitching experiments were performed with a fluorescence microscope (Leica, Wetzlar, Germany) equipped with a ×40 NA 0.6 air objective lens for imaging of protein crystals, and alternatively a ×20 NA 0.4 air objective lens for imaging protein solutions. For irradiation, a 100 W Hg lamp with a 550/40 nm excitation filter (“green light”) and a 50 W Hg lamp with a 450/40 nm excitation filter (“blue light”) were used. For kinetic measurements, fluorescence was recorded with a photomultiplier tube (HR9306-0, Hamamatsu, Hamamatsu City, Japan) by using a HQ600LP longpass detection filter (AHF Analysetechnik, Tübingen, Germany). Images were taken with a color charge-coupled device camera (DFK 3003/P, The Imaging Source, Bremen, Germany) by using the same detection filter. For the measurements, proteins and crystals were suspended in 0.2 M Tris·HCl pH 7.1/20% polyethylene glycol 3350/0.3 M NaCl.
The determination of reversion rates 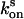 and
and 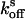 into the equilibrium was performed as follows: After complete switching into the on state with 50 W·cm–2 and 20 W·cm–2 green light for crystals and protein solutions, respectively, the reversion into the equilibrium state (
into the equilibrium was performed as follows: After complete switching into the on state with 50 W·cm–2 and 20 W·cm–2 green light for crystals and protein solutions, respectively, the reversion into the equilibrium state (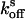 ) was followed by consecutive short (0.02 sec) measurements with 15 W·cm–2 or 6 W·cm–2 green light, respectively. For determination of the reversion rate
) was followed by consecutive short (0.02 sec) measurements with 15 W·cm–2 or 6 W·cm–2 green light, respectively. For determination of the reversion rate 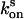 after complete switching into the off state, the proteins were quenched with blue light (3 W·cm–2 for crystals and 2 W·cm–2 for solutions), and the fluorescence recovery was analyzed by short (0.02 sec) measurements at varying time points. To avoid an influence by the short measurements on the measured
after complete switching into the off state, the proteins were quenched with blue light (3 W·cm–2 for crystals and 2 W·cm–2 for solutions), and the fluorescence recovery was analyzed by short (0.02 sec) measurements at varying time points. To avoid an influence by the short measurements on the measured 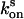 , we repeated the quenching for each recovery time point measurement. The final rate constants
, we repeated the quenching for each recovery time point measurement. The final rate constants 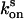 and
and 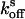 were deduced from single-exponential fits.
were deduced from single-exponential fits.
MD. All simulations used the 1.3-Å crystal structure of wt asFP595 described in this paper (PDB ID code 2A50) and were carried out with the gromacs 3.2.1 simulation suite (15) by using the OPLS force field (16). A monomer of the protein was solvated in a box of TIP4P water molecules at physiological salt concentration. An ensemble with constant particle number N, pressure p, and temperature T was simulated at T = 300 K and P = 1 atm (1 atm = 101.3 kPa) by using periodic boundary conditions, the Particle-Mesh-Ewald method (17), and an integration time step of 2 fs. After energy minimization, the system was equilibrated for 100 ps. All four possible trans-cis isomerizations of the chromophore were induced by force probe simulations (18), shifting the respective dihedral potentials. For the chromophore force field and for the details of the simulations, see Table 2 and Supporting Methods, which are published as supporting information on the PNAS web site.
Results and Discussion
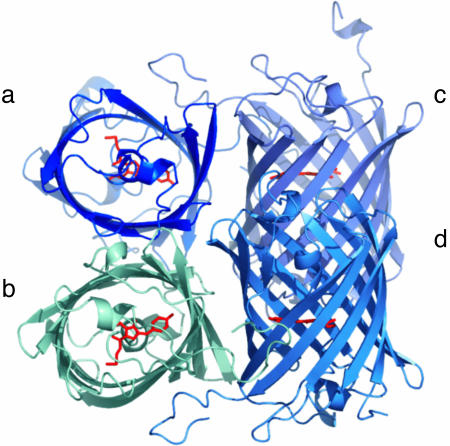
Overall Structure of asFP595. The photochromic protein asFP595 can be reversibly switched between a nonfluorescent off and a fluorescent on state by irradiation with green and blue light, respectively. In asFP595 the on state quickly and spontaneously relaxes back to the off state. Hence, in a nonirradiated solution, asFP595 is found predominantly in the off state. After crystallization, we determined the off-state asFP595 structure at 1.3-Å resolution (Fig. 1). The structure is similar to the dark state structure of KFP1 (5), a mutant of asFP595 that has recently been reported at 2.1 Å resolution (19) (Cα root mean square deviation ≈ 0.33 Å; see also Table 3, which is published as supporting information on the PNAS web site). As in all GFP-like proteins, the asFP595 chromophore resides in a helical segment, which is enclosed in an 11-stranded β-barrel (20–23). The tripeptide M63-Y64-G65 rearranges to a chromophoric, conjugated 2-iminomethyl-5-(4-hydroxybenzylidene)imidazolinone (MYG) system, in agreement with previous biochemical analysis of proteolytic peptides (24). Unlike most other GFP-like proteins, the protein backbone is broken between C62 and the chromophore. Nevertheless, the former M63 Cα and backbone nitrogen atoms are in plane with the imidazolinone ring and, thus, part of the conjugated system. The imino group expands the conjugated system of MYG, likely accounting for the shift of the absorption maximum toward a longer wavelength (572 nm) as compared with that of GFP (470 nm). The MYG chromophore of the off state asFP595 exclusively adopts the trans conformation.
Fig. 1.
Overall structure of asFP595. A schematic ribbon representation of the quaternary tetrameric structure of asFP595 shows the four molecules in different colors and the chromophores highlighted in red.
Because of the very low quantum efficiency of asFP595 and the fast spontaneous relaxation of the on state, we refrained from determining the on state structure of asFP595 directly but concentrated on the variant asFP595-A143S.
Reversible Switching of asFP595-A143S Crystals. This variant has >12 times the fluorescence quantum yield as compared with that of the wt asFP595 protein (6). In a shaded solution, asFP595-A143S spontaneously reaches equilibrium between the off and the on state, exhibiting a certain level of fluorescence (that can be fully quenched by irradiation with blue light). The fluorescence at equilibrium amounts to ≈10% of the fluorescence induced by the strongest possible photoactivation. Hence, contrary to the wt asFP595, a significant fraction resides in the on state already. Besides, the spontaneous relaxation from the on to the equilibrium state is ≈35 times slower (≈400 sec) than that of the wt asFP595 (≈12 sec).
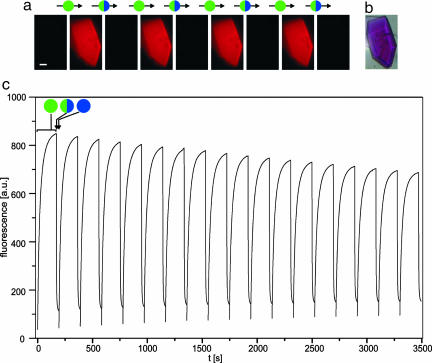
The equilibrated protein solution yielded faintly fluorescent crystals. Irradiation with green light in a standard fluorescence microscope converted these crystals into brightly fluorescent ones that were readily quenched by irradiation with blue light as well (Fig. 2). In fact, photoswitching of asFP595-A143S crystals was reversible for >30 cycles (Fig. 2c). The slight increase in the baseline seen in Fig. 2c indicates the occurrence of a small fraction of fluorophores in a permanently fluorescent state upon several switching cycles. We assume that the molecular mechanism resulting in a permanently fluorescent state is different from the reversible photoswitching described herein.
Fig. 2.
Reversible photoswitching of a protein crystal under a fluorescence microscope. (a) asFP595-A143S crystals in the equilibrium state are barely fluorescent. Irradiation with green light (550 ± 20 nm, 50 W·cm–2, 170 sec) yields brightly fluorescent crystals. The fluorescence is quenched by gentle irradiation with blue light (450 ± 20 nm, 3 W·cm–2, 24 sec); the quenching also takes place while simultaneously irradiating with green light (required to survey the fluorescence). (Scale bar: 20 μm.) (b) Brightfield image of the asFP595-A143S crystal. (c)On/off cycles of the asFP595-A143S crystal fluorescence by using the same intensities as in a. Excitation sequence: green light, 170 sec; green plus blue light, 24 sec; blue light, 1 sec. Fluorescence was recorded during the first and the second section of the sequence.
After on or off switching, the crystals exhibited slow spontaneous relaxation to the equilibrium state with rate constants of 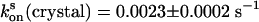 and
and 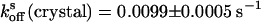 , respectively. The recovery kinetics of asFP595-A143S in solution was as follows:
, respectively. The recovery kinetics of asFP595-A143S in solution was as follows: 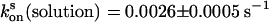 and
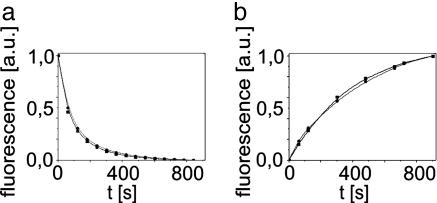
and 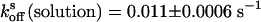 (Fig. 3). Comparison shows that the rates do not change upon the capture of asFP595-A143S within the crystal lattice, leading us to conclude that the crystal form does not impose significant constraints on the conversion kinetics or the switching mechanism.
(Fig. 3). Comparison shows that the rates do not change upon the capture of asFP595-A143S within the crystal lattice, leading us to conclude that the crystal form does not impose significant constraints on the conversion kinetics or the switching mechanism.
Fig. 3.
Switching kinetics of asFP595-A143S are the same in solution and in crystals. asFP595-A143S crystals (dotted) and asFP595-A143S in solution (solid) were fully switched on (a) or off (b) by green light or by blue light, respectively. Shown is the subsequent return of the fluorescence to the equilibrium level.
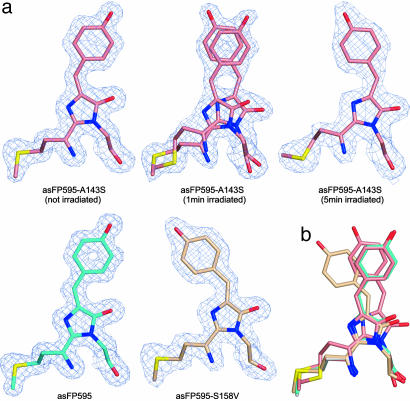
Structure of asFP595-A143S in the On and the Off State. Crystals of asFP595-A143S in dark equilibrium are barely fluorescent. Hence, we determined the equilibrium state asFP595-A143S crystal structure at 1.45 Å resolution. We found it to be very similar to the off state structure of asFP595 (root mean square deviation ≈ 0.23 Å). The obtained 2Fo – Fc electron density is consistent with the chromophore that is predominantly in a trans conformation (Fig. 4a; see also Fig. 6, which is published as supporting information on the PNAS web site). Scrutinizing difference Fourier maps reveals slight deviations from their asFP595 counterpart accounting for a fraction of asFP595-A143S chromophores in a cis conformation. We attribute this fraction of cis-conformation MYG to the ≈10% on state chromophores pertinent to the asFP595-A143S crystals at the equilibrium.
Fig. 4.
Trans and cis conformation of the MYG chromophore in the off and the on state. Views of the MYG chromophores after least-squares superpositioning of all Cα atoms. (a) MYG chromophores of asFP595 variants as indicated in the figure. Atoms are color-coded by atom type (carbon wt asFP595, cyan; carbon asFP595-A143S, salmon; carbon asFP595-S158V, beige; oxygen, red; nitrogen, blue). Final 2Fo – Fc electron densities around the chromophores are contoured at the 1σ level. For asFP595-A143S (irradiated for 1 min) both isomerization states are depicted, because both structures were observed in a single crystal with equal proportions. (b) Overlay of the four chromophore structures. Color-coding as in a.
Next, we solved the atomic structure of asFP595-A143S in the on state. Crystals were irradiated at room temperature with green light for ≈1 min and ≈5 min, respectively. The slow relaxation of on state asFP595-A143S crystals back to the off state facilitated the capture of on state crystals by flash freezing in liquid nitrogen. The structure of on state asFP595-A143S is unchanged (root mean square deviation ≈ 0.22 Å) with regard to that in the untreated asFP595-A143S crystals, with the exception of the ratio of occurrence of the two conformational states. Whereas in briefly irradiated (≈1 min) crystals ≈50% of MYG moieties adopted a cis conformation, in the intensely irradiated (≈5 min) crystals, virtually all chromophores were found in its cis counterpart (Figs. 4a and 6). This finding provides direct evidence that the on state is given by the cis conformation of the chromophore in agreement with a hypothesis in ref. 11.
In the cis conformation, the MYG imidazolinone ring is rotated by ≈20° about the Cα atom of the former G65 compared with the transconformation. Furthermore, the hydroxy group of the p-hydroxyphenyl ring shifted its position because of a rotation of the ring by ≈60°. Nevertheless, due to the isomerization about the methine bridge, the p-hydroxyphenyl rings of the cis and the trans conformations remain almost congruent in both forms (Fig. 4b). The cis conformer in asFP595-A143S is stabilized by an additional hydrogen bond of the MYG p-hydroxyphenyl ring to S143 (Fig. 7, which is published as supporting information on the PNAS web site). This stabilization cannot form in wt asFP595, likely explaining the shift of the equilibrium toward the on state.
Structure of asFP595-S158V. To further confirm that the cis conformation represents the on state of the chromophore, we solved the structure of the permanently fluorescent variant asFP595-S158V (11) at 1.7-Å resolution. Crystals of asFP595-S158V are brightly fluorescent but not switchable. The x-ray structure shows that the chromophore of asFP595-S158V exclusively adopts a cis conformation, in which the MYG imidazolinone ring is almost congruent to imidazolinone ring of the off state asFP595-A143S. However, the p-hydroxyphenyl ring is noticeably displaced (Fig. 4). In fact, because of steric constraints imposed by V158, the cis chromophores of asFP595-A143S and asFP595-S158V are differently positioned relative to the respective protein environments (Figs. 4 and 7). We conclude that in the different asFP595 variants, fluorescence is maintained irrespective of the small changes in the location of the cis conformer with respect to the protein matrix.
MD Simulations of the Switching Mechanism. Starting from its off state, we calculated by MD the energetically most favorable and, hence, most probable conformation of the on state of asFP595. The resulting asFP595 on state is indeed very similar to that found by the crystal structure of the on state of the asFP595-A143S variant. The similarity between the two indicates that the A143S mutation has only minor influences on the structure of the protein in the on state. These simulations also revealed that the N-terminal chromophore residues have to rearrange upon isomerization, suggesting that the C62-M63 backbone break facilitates photoswitching (Fig. 8, which is published as supporting information on the PNAS web site). This result points to the fact that the observed break of the protein backbone is an essential element of the unique switching characteristics of asFP595.
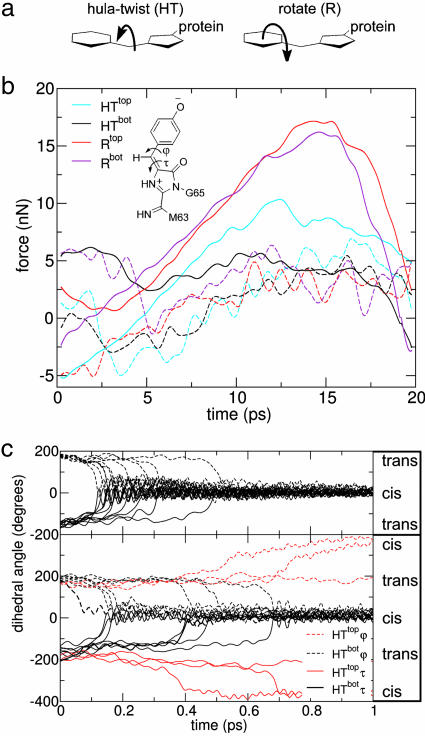
Next, we simulated by MD the two conceivable trans-cis photoisomerization pathways, namely the hula twist (HT) and the rotation (R) mechanism (Fig. 5). The HT mechanism (25) involves concurrent rotation around both dihedral angles τ and ϕ (Fig. 5 a and b Inset). In this case, both chromophore rings stay largely in place, and only the methine group linking the two rings moves. Rotation around τ only (R mechanism) results in a space-consuming rotation of the p-hydroxyphenyl ring, thus involving possible sterical hindrance by the protein matrix.
Fig. 5.
MD simulations of the switching mechanism in asFP595. (a) The two possible isomerization mechanisms. (b) Forces due to the protein matrix opposing chromophore isomerization were calculated by nonequilibrium force-probe MD simulations; 10 trajectories were averaged for the four possible pathways (solid lines). For the rotate mechanism, the MYG p-hydroxyphenyl ring can either rotate toward the initially coplanar H197 (Rtop) or toward the other side (Rbot). Likewise, during HT isomerization, the bridging methine group can move along a top (HTtop) or bottom (HTbot) pathway. Control simulations of the chromophore in water show similar forces for the R and HT mechanisms (dashed curves). (Inset) MYG chromophore and relevant torsion angles. (c) Spontaneous trans-cis isomerization during free excited state MD simulations for the chromophore within the protein matrix (Upper) and in water (Lower), monitored through the dihedral angles τ (dashed curves) and ϕ (solid curves). The protein favors the HTbot mechanism, with both dihedral angles rotating simultaneously within a narrow time frame. Simulation of protein-free isomerization of the chromophore in water also follows HT; both directions (HTtop and HTbot) are observed in this case.
For enforced fast isomerizations, strong forces of the protein matrix opposing the R and HT pathways are seen (Fig. 5b). HT is favored, because all favorable hydrophobic interactions are maintained, in particular the π stacking of MYG and H197. R is opposed by larger forces that arise from the disruption of hydrophobic chromophore–protein interactions (mainly to T60 and H197, see Fig. 9, which is published as supporting information on the PNAS web site). Irrespective of the mechanism, the hydrogen bonding network remains largely intact.
To also consider intramolecular properties of the chromophore, we carried out free dynamics simulations, including the relevant internal chromophore energy barriers opposing the τ and the ϕ rotations. To this end, we adopted high-level quantum mechanical computations on the chemically very similar chromophore of GFP (26). When starting the simulation from the first electronically excited energy level of the asFP595 off state, the bridging methine group spontaneously rotated toward H197 within 100–500 fs through a bottom hula twist (HTbot) (see Fig. 5c; see also Movie 1, which is published as supporting information on the PNAS web site). To test the robustness of this semiquantitative description, 30 simulations with different barrier heights were carried out (see Supporting Methods and Fig. 10, which is published as supporting information on the PNAS web site). Taken together, these simulations showed that both the intrinsic chromophore properties and the overall protein conformation designate HTbot as the predominant isomerization mechanism in asFP595.
Conclusion and Perspectives
Accomplishing reversible photoswitching both in solution and in a protein crystal enabled us to synergistically combine optical spectroscopy, x-ray crystallography, and MD simulations to unravel the molecular mechanism of reversible photoswitching of a bistable fluorescent protein. Our data indicate that the key event in asFP595 is a HTbot mechanism resulting in a trans-cis isomerization of the chromophore. The chromophore on state is attributed to the cis conformation.
We note that the isomerization might also be accompanied by a proton transfer, as it is known for several GFP variants (27). In the cis conformation, the chromophoric p-hydroxyphenyl group is likely to be in equilibrium between a (nonfluorescent) protonated and a (fluorescent) nonprotonated form (11).
Recently, other highly intriguing photoswitchable proteins have been described in ref. 4. Hence, it will be enlightening to see the generality of this mechanism. Because a methine group linking two rings appears to be common to all GFP-like fluorophores (28), it might well prove that the mechanism described herein is a common mechanism for reversible protein photoswitching. The spectroscopic properties of asFP595 are currently far from optimal for most aspired applications. The structure and mechanism reported herein should open up the road for specific modifications of switchable fluorescent proteins to facilitate applications, including three dimensional data optical memories, protein tracking and live cell imaging at the nanoscale.
Supplementary Material
Acknowledgments
We thank Dr. K. A. Lukyanov for providing us with the plasmid pQE30-asFP595 and Dr. Gerrit Groenhof for fruitful discussions. L.V.S. and F.G. were supported by the Boehringer Ingelheim Fonds.
Author contributions: M.C.W., H.G., S.W.H., and S.J. designed research; M.A., M.C.W., A.C.S., F.G., L.V.S., S.T., and G.W. performed research; M.A., M.C.W., A.C.S., F.G., L.V.S., C.E., H.G., S.W.H., and S.J. analyzed data; and M.C.W., F.G., L.V.S., H.G., S.W.H., and S.J. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office. J.W.S. is a guest editor invited by the Editorial Board.
Abbreviations: HTbot: bottom hula twist, MD: molecular dynamics; MYG, 2-iminomethyl-5-(4-hydroxybenzylidene)imidazolinone.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2A50, 2A52, 2A53, 2A54, and 2A56).
References
- 1.Lippincott-Schwartz, J., Altan-Bonnet, N. & Patterson, G. H. (2003) Nat. Cell Biol. S7–S14. [PubMed]
- 2.Miyawaki, A., Sawano, A. & Kogure, T. (2003) Nat. Cell Biol. S1–S7. [PubMed]
- 3.Tsien, R. Y. (1998) Annu. Rev. Biochem. 67, 509–544. [DOI] [PubMed] [Google Scholar]
- 4.Ando, R., Mizuno, H. & Miyawaki, A. (2004) Science 306, 1370–1373. [DOI] [PubMed] [Google Scholar]
- 5.Chudakov, D. M., Belousov, V. V., Zaraisky, A. G., Novoselov, V. V., Staroverov, D. B., Zorov, D. B., Lukyanov, S. & Lukyanov, K. A. (2003) Nat. Biotechnol. 21, 191–194. [DOI] [PubMed] [Google Scholar]
- 6.Lukyanov, K. A., Fradkov, A. F., Gurskaya, N. G., Matz, M. V., Labas, Y. A., Savitsky, A. P., Markelov, M. L., Zaraisky, A. G., Zhao, X. N., Fang, Y., et al. (2000) J. Biol. Chem. 275, 25879–25882. [DOI] [PubMed] [Google Scholar]
- 7.Crano, J. C. & Guglielmetti, R. J., eds. (1999) Organic Photochromic and Thermochromic Compounds: Main Photochromic Families (Plenum, New York), Vol. 1.
- 8.Hell, S. W., Jakobs, S. & Kastrup, L. (2003) Appl. Phys. A Materials Sci. Processing 77, 859–860. [Google Scholar]
- 9.Hell, S. W. (2003) Nat. Biotechnol. 21, 1347–1355. [DOI] [PubMed] [Google Scholar]
- 10.Hell, S. W., Dyba, M. & Jakobs, S. (2004) Curr. Opin. Neurobiol. 14, 599–609. [DOI] [PubMed] [Google Scholar]
- 11.Chudakov, D. M., Feofanov, A. V., Mudriku, N. N., Lukyanov, S. & Lukyanov, K. A. (2003) J. Biol. Chem. 278, 7215–7219. [DOI] [PubMed] [Google Scholar]
- 12.Otwinowski, Z. & Minor, W. (1996) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 13.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 14.Petersen, J., Wilmann, P. G., Beddoe, T., Oakley, A. J., Devenish, R. J., Prescott, M. & Rossjohn, J. (2003) J. Biol. Chem. 278, 44626–44631. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl, E., Hess, B. & van der Spoel, D. (2001) J. Mol. Model. 7, 306–317. [Google Scholar]
- 16.Jorgensen, W. L. & Tirado-Rives, J. (1988) J. Am. Chem. Soc. 110, 1657–1666. [DOI] [PubMed] [Google Scholar]
- 17.Darden, T., York, D. & Pedersen, L. (1993) J. Chem. Phys. 98, 10089–10092. [Google Scholar]
- 18.Grubmüller, H., Heymann, B. & Tavan, P. (1996) Science 271, 997–999. [DOI] [PubMed] [Google Scholar]
- 19.Wilmann, P. G., Petersen, J., Devenish, R. J., Prescott, M. & Rossjohn, J. (2005) J. Biol. Chem. 280, 2401–2404. [DOI] [PubMed] [Google Scholar]
- 20.Örmo, M., Cubitt, A. B., Kallio, K., Gross, L. A., Tsien, R. Y. & Remington, S. J. (1996) Science 273, 1392–1395. [DOI] [PubMed] [Google Scholar]
- 21.Wall, M. A., Socolich, M. & Ranganathan, R. (2000) Nat. Struct. Biol. 7, 1133–1138. [DOI] [PubMed] [Google Scholar]
- 22.Yang, F., Moss, L. G. & Phillips, G. N., Jr. (1996) Nat. Biotechnol. 14, 1246–1251. [DOI] [PubMed] [Google Scholar]
- 23.Yarbrough, D., Wachter, R. M., Kallio, K., Matz, M. V. & Remington, S. J. (2001) Proc. Natl. Acad. Sci. USA. 98, 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zagranichny, V. E., Rudenko, N. V., Gorokhovatsky, A. Y., Zakharov, M. V., Balashova, T. A. & Arseniev, A. S. (2004) Biochemistry 43, 13598–13603. [DOI] [PubMed] [Google Scholar]
- 25.Liu, S. H. & Brown, D. T. (1986) Acc. Chem. Res. 19, 42–48. [Google Scholar]
- 26.Martin, M. E., Negri, F. & Olivucci, M. (2004) J. Am. Chem. Soc. 126, 5452–5464. [DOI] [PubMed] [Google Scholar]
- 27.Chattoraj, M., King, B. A., Bublitz, G. U. & Boxer, S. G. (1996) Proc. Natl. Acad. Sci. USA 93, 8362–8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labas, Y. A., Gurskaya, N. G., Yanushevich, Y. G., Fradkov, A. F., Lukyanov, K. A., Lukyanov, S. A. & Matz, M. V. (2002) Proc. Natl. Acad. Sci. USA 99, 4256–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.