Abstract
We compared putative molecular markers of virulence (vacA, cagA, and iceA) of Helicobacter pylori strains isolated from 52 adult duodenal ulcer patients from West Bengal, India, with those of H. pylori strains isolated from 48 adult healthy volunteers from the same region. On the basis of genotyping by PCR, we conclude that the H. pylori strains isolated from the two study groups were indistinguishable and that there are geographic variations in the association of certain putative H. pylori virulence genes with clinical status.
Helicobacter pylori infection is associated with duodenal ulcer (DU) or gastric ulcer, gastritis, and gastric adenocarcinoma (5). Although more than half of the population worldwide acquires H. pylori infection early in life, probably from their parents and older siblings (5), only about 10% suffer from overt disease, which mostly occurs in the later part of life, while large portions of the H. pylori-infected population remain asymptomatic carriers or have chronic gastritis (15). Therefore, it appears that both pathogenic and nonpathogenic strains of H. pylori coexist, as has been observed for several other bacteria, and those associated with disease may be endowed with a specific genotype (12).
Some strains of H. pylori can produce vacuoles in various cell lines due to the cytotoxic effect of the vacuolating cytotoxin, which comprises a signal sequence, a midregion, and a C-terminal end, coded by the vacA gene. The signal sequence in vacA is either s1 or s2, and the midregion of vacA can also be divided into m1 and m2. Strains carrying the s1ml mosaic combination of the vacA gene exhibit higher levels of cytotoxic activity than s1m2 strains, while s2m2 strains do not secrete the vacuolating cytotoxin (10). Furthermore, in Western countries, H. pylori strains which carry the s1m1 mosaic combination of vacA also usually carry a 40-kb pathogenicity island, whose marker gene is cagA (which encodes cytotoxin-associated gene product A). Strains that carry this pathogenicity island are associated with disease significantly more often than strains that do not carry this pathogenicity island (4). iceA1, an allele whose sequence has a high degree of homology to the sequence of nlaIIIR (which encodes a CATG-specific restriction endonuclease [NlaIII] in Neisseria lactamica), is replaced by iceA2 in some strains. The promoters of hpyIM, a CATG-specific methylase gene, which exists immediately downstream of iceA, may differ between iceA1 and iceA2 strains. It is not clear, however, how strains carrying iceA1 can produce higher levels of the proinflammatory cytokine interleukin 8 in the gastric mucosa and are more often associated with DU in the West than strains carrying iceA2 (17).
Most of the studies described above were performed with strains from symptomatic individuals, while the control groups in those studies comprised asymptomatic carriers (3, 6), which does not necessarily mean that they were healthy volunteers. To our knowledge very few studies that have compared H. pylori strains from patients with H. pylori-associated disease with strains from healthy volunteers have been described in the literature. In India, virtually no study on the genotypic status of H. pylori strains in relation to clinical outcome has been conducted except for our earlier study of the genotypes of strains isolated from DU patients and of DNA extracted from the gastric juice or from strains cultured from patients with gastritis (11). This lack of information was the impetus for this study.
Biopsy specimens were obtained as described earlier (11) from 52 adult Bengali patients of both sexes with a diagnosis of DU on the basis of endoscopic examination of the stomach and duodenum and 48 adult Bengali healthy volunteers (HVs) of both sexes who had no gastritis or dyspeptic syndromes like abdominal pain. The DU patients were recruited from patients seeking care for gastroduodenal disease at the Hospital of the Institute of Post Graduate Medical Education and Research in Calcutta, India, while the HVs were recruited on request from among a variety of individuals, including medical students and semiskilled and unskilled laborers. Written informed consent was obtained from all the individuals according to the recommendations of the Ethical Committee of the Institute of Post Graduate Medical Education and Research. One biopsy specimen from each patient was used for an in-house rapid urease test, while a second biopsy specimen was transported in 1 ml of brucella broth (Difco, Detroit, Mich.) containing 25% glycerol under cold conditions for culture at the Helicobacter unit of the National Institute of Cholera and Enteric Diseases. H. pylori strains were isolated on brain heart infusion (BHI; Difco) agar plates supplemented with 7% sheep blood, 0.4% IsoVitale X (BBL), and Dent (Oxoid, Basingstoke, England) in an atmosphere of 10% CO2-5% O2-85% N2 in a double gas incubator (Heraeus Instruments, Hanau, Germany) and were identified on the basis of their typical morphologies and positivities by urease, oxidase, and catalase tests and by subsequent gene-specific tests. The H. pylori cells that grew from a biopsy specimen on the primary culture plate were collected as a pooled population and were preserved in sterile BHI broth with 20% glycerol at −70°C.
Chromosomal DNA from bacterial pellets was prepared from confluent growth on BHI agar plate cultures by the cetyltrimethylammonium bromide extraction method (2). PCRs for detection of the vacA and iceA alleles were carried out by using the primers and PCR conditions described previously (11). We used the primers described by Yang et al. (19) to amplify the 208-bp cagA-specific products, and the reaction was carried out in 25-μl reaction volume containing 10 ng of genomic DNA, 25 pmol of each primer, each deoxynucleoside triphosphate (Takara) at a concentration of 0.25 mM, 1 U of Taq DNA polymerase (Takara), and 1.5 mM MgCl2 in a standard PCR buffer (Takara). All the products were amplified under the following conditions: 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 35 cycles in a Perkin-Elmer 9700 thermal cycler. Positive and negative controls were included in each assay.
All 100 biopsy specimens included in this study gave a positive result by the rapid urease test. On the basis of PCRs with specific primers for alternative alleles of vacA s1 and s2, vacA m1 and m2, and iceA1 and iceA2, mixed genotype patterns were observed for isolates in cultures of specimens from 15 of the 100 subjects (7 were from DU patients and 8 were from HVs), indicating the prevalence of mixed infections. These isolates were eliminated from the final analysis. A total of 85 strains (45 from DU patients and 40 from HVs) whose genotypes did not indicate the presence of a mixed infection were further analyzed in this study.
We found that strains of H. pylori carrying the s1 allele of vacA, cagA, and iceA1 predominate in West Bengal, India (Table 1). Previous descriptions of the vacuolating activities of strains with different vacA subtypes demonstrate that, in vitro, s1 strains secrete larger amounts of cytotoxin than s2 strains, which are known to be less virulent since they have a 12-amino-acid type s2 hydrophilic amino-terminal extension that results in the less efficient formation of membrane channels compared to the efficiency of membrane channel formation by s1 strains (9, 10). Recently, De Gusmao et al. (6) demonstrated a strong association between the presence of the s1 allele and the presence of DUs in Brazilian children. In contrast, our findings demonstrate the high frequency of occurrence of the s1 allele among adult HVs (Table 1). Therefore, there is a clear geographic difference in the prevalence of a particular genotype among patients with DUs and HVs.
TABLE 1.
Genotypes of H. pylori strains from patients with DUs and HVs
| Genotype | No. (%) of strains from:
|
|
|---|---|---|
| DU patients (n = 45) | HVs (n = 40) | |
| s1 | 44 (97.8) | 39 (97.5) |
| s2 | 1 (2.2) | 1 (2.5) |
| m1 | 29 (64.4) | 26 (65) |
| m2 | 16 (35.6) | 14 (35) |
| cagA positive | 43 (95.6) | 38 (95) |
| cagA negative | 2 (4.4) | 2 (5) |
| iceA1 | 26 (57.8) | 25 (62.5) |
| iceA2 | 15 (33.3) | 13 (32.5) |
| iceA negative | 4 (8.9) | 2 (5) |
De Gusmao et al. (6) also observed an association between the presence of the m1 allele of the vacA gene and the presence of peptic ulcers in children. The toxigenic role of m1 was established by experiments that showed that the product of vacA m1 can produce vacuoles, which are acidic in nature, in HeLa cells (1, 14). m2 strains, on the contrary, were thought to be the less toxigenic, as culture supernatants of m2 strains cannot produce vacuoles in HeLa cells, mainly because the toxin does not bind to the HeLa cell membrane (10). However, subsequent studies have shown that the product of the m2 allele also can produce vacuoles in RK13 cells (13). In our study, we have found that the profiles of the midregion of the vacA gene in strains isolated from DU patients and HVs were almost the same (Table 1).
In this study cagA-positive H. pylori strains were found to predominate (95.3%) over cagA-negative strains in West Bengal. This virulence marker was observed at almost equal frequencies in strains from DU patients and HVs (Table 1), indicating that the presence of the cagA gene cannot be considered a key virulence factor for determination of the clinical status of the host, as has been reported for strains from other geographic regions (16). Moreover, in this study we have found that iceA1 and iceA2 were almost equally distributed among strains from DU patients and HVs. This result demonstrates that, unlike in the West, iceA1 is not associated with disease in West Bengal and is analogous to an earlier observation made in Japan (7).
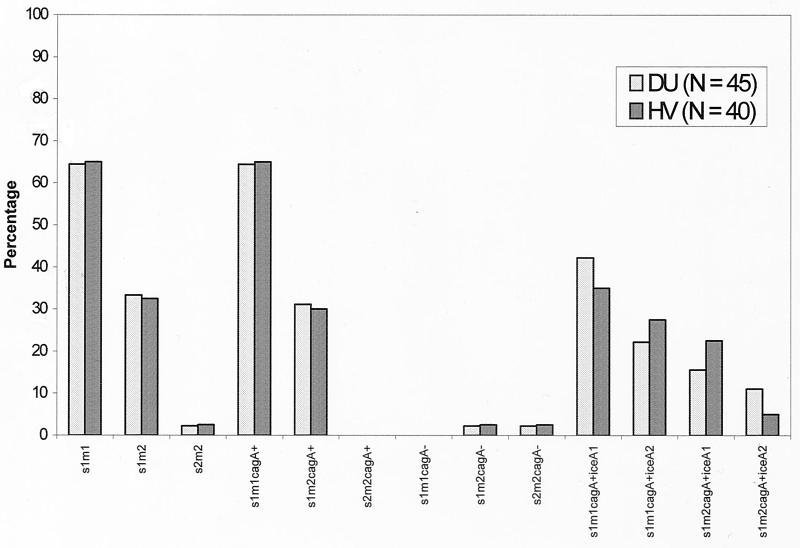
In an attempt to find associations between the presence of the combination of the cagA gene and the vacA and iceA alleles and disease outcome by using H. pylori strains from different countries (Korea, Japan, the United States, and Colombia), Yamaoka et al. (18) have concluded that the presence of neither iceA nor particular combinations of iceA, vacA, and cagA was helpful in predicting a patient's disease status. From earlier studies, it is evident that the genotype varies among H. pylori strains isolated from different geographic regions (5, 8), which means that strains from patients with overt H. pylori-associated disease and from healthy individuals of the same geographic region should be evaluated for disease-specific genotypes. Importantly, toxin gene expression by H. pylori is inconsistent with the general phenomenon observed for several pathogenic bacteria, that is, that expression of key toxin genes is strictly associated with disease. Therefore, our aim was to find the most probable H. pylori genotype associated with disease and its distinctiveness from the genotypes of H. pylori strains isolated from healthy individuals in West Bengal. We found that s1m1 cagA iceA1 was the dominant genotype in West Bengal and that the prevalence of this genotype was higher than those of the other allelic combinations in both the DU patient and HV populations (Fig. 1). Statistical analysis by the chi-square test and Fisher's exact test, with significance set at a P value of <0.05, further confirmed the absence of significant differences in the genotypes of strains isolated from DU patients and HVs (data not shown). We conclude that the strains isolated from the two study groups were almost indistinguishable and that no association could be made between the presence of these virulence marker genes or their combinations and the clinical status of H. pylori-infected individuals in West Bengal.
FIG. 1.
Combinations of vacA, cagA, and iceA genotypes of H. pylori strains isolated from DU patients and HVs. +, positive for the gene; −, negative for the gene.
Acknowledgments
This work was supported by a grant (grant BT/MB/VAP/3/2/98) from the Department of Biotechnology, Government of India (no. BT/MB/VAP/3/2/98), and by grants from the U.S. Public Health Service (AI 38166, AI49161, DK53727, and P30DK52574).
REFERENCES
- 1.Atherton, J. C., P. Cao, R. M. J. Peek, M. K. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1993.. Current protocols in molecular biology. Greene Publishing and Wiley-Interscience, New York, N.Y.
- 3.Carattoli, A., C. Pezzella, A. Pietroiusti, A. Galante, P. Pezzotti, and I. Luzzi. 2000. Cytotoxin-associated gene A and vacuolating cytotoxin A in human isolates of Helicobacter pylori and their association with the clinical status of ulcer disease. Eur. J. Gastroenterol. Hepatol. 12:1207-1213. [DOI] [PubMed] [Google Scholar]
- 4.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covacci, A., J. L. Telford, G. D. Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 6.De Gusmao, V. R., E. N. Mendes, D. M. De Magalhaes Queiroz, G. A. Rocha, A. M. C. Rocha, A. A. R. Ashour, and A. S. T. Carvalho. 2000. vacA genotypes in Helicobacter pylori strains isolated from children with and without duodenal ulcer in Brazil. J. Clin. Microbiol. 38:2853-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito, Y., T. Azuma, S. Ito, H. Suto, H. Miyaji, Y. Yamazaki, T. Kato, Y. Kohli, Y. Keida, and M. Kuriyama. 2000. Sequence analysis and clinical significance of the iceA gene from Helicobacter pylori strains in Japan. J. Clin. Microbiol. 38:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kersulyte, D., A. K. Mukhopadhyay, B. Velapatino, W. Su, Z. Pan, C. Garcia, V. Hernandez, Y. Valdez, R. S. Mistry, R. H. Gilman, Y. Yuan, H. Gao, T. Alarcon, M. Lopez-Brea, G. B. Nair, A. Chowdhury, S. Datta, M. Shirai, T. Nakazawa, R. Ally, I. Segal, B. C. Y. Wong, S. K. Lam, F. Olfat, T. Boren, L. Engstrand, O. Torres, R. Schneider, J. E. Thomas, S. Czinn, and D. E. Berg. 2000. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 182:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClain, M. S., P. Cao, H. Iwamoto, A. D. Vinion-Dubiel, G. Szabo, Z. Shao, and T. L. Cover. 2001. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J. Bacteriol. 183:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montecucco, C., E. Papini. M. de Bernard, J. L. Telford, and R. Rappuoli. 1999. Helicobacter pylori vacuolating cytotoxin and associated pathogenic factors, p. 264-283. In J. E. Alouf and J. H. Freer (ed.), The comparative source book of bacterial protein toxins, 2nd ed. Academic Press, San Diego, Calif.
- 11.Mukhopadhyay, A. K., D. Kersulyte, J. Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, S. Chowdhury, A. Santra, S. K. Bhattacharya, T. Azuma, G. B. Nair, and D. E. Berg. 2000. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Occhialini, A., A. Marais, R. Alm, F. Garcia, R. Sierra, and F. Megraud. 2000. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect. Immun. 68:6240-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pagliaccia, C., M. de Bernard, P. Lupetti, X. Ji, D. Burroni, T. L. Cover, E. Papini, R. Rappuoli, J. L. Telford, and J. M. Reyrat. 1998. The m2 form of the Helicobacter pylori cytotoxin has cell type specific vacuolating activity. Proc. Natl. Acad. Sci. USA 95:10212-10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papini, E., E. Gottardi, B. Satin, M. de Bernard, J. Telford, P. Massari, R. Rappuoli, S. B. Sato, and C. Montecucco. 1996. The vacuolar ATPase proton pump on intracellular vacuoles induced by Helicobacter pylori. J. Med. Microbiol. 44:1-6. [DOI] [PubMed] [Google Scholar]
- 15.Telford, J. L., A. Covacci, P. Ghiara, C. Montecucco, and R. Rappuoli. 1994. Unravelling the pathogenic role of Helicobacter pylori in peptic ulcer: potential new therapies and vaccines. Trends Biotechnol. 12:420-426. [DOI] [PubMed] [Google Scholar]
- 16.van Doorn, L. J., C. Figueiredo, R. Sanna, A. Plaisier, P. Schneeberger, W. de Boer, and W. Quint. 1998. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 115:58-66. [DOI] [PubMed] [Google Scholar]
- 17.Xu, Q., and M. J. Blaser. 2001. Promoters of the CATG-specific methyltransferase gene hpyIM differ between iceA1 and iceA2 Helicobacter pylori strains. J. Bacteriol. 183:3875-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaoka, Y., T. Kodama, O. Gutierrez, J. G. Kim, K. Kashima, and D. Y. Graham. 1999. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J. Clin. Microbiol. 37:2274-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, J.-C., T.-H. Wang, H.-J. Wang, C.-H. Kuo, J.-T. Wang, and W.-C. Wang. 1997. Genetic analysis of the cytotoxin-associated gene and the vacuolating toxin gene in Helicobacter pylori strains isolated from Taiwanese patients. Am. J. Gastroenterol. 92:1316-1321. [PubMed] [Google Scholar]