Abstract
Isopentenyl diphosphate and dimethylallyl diphosphate serve as the universal precursors for the biosynthesis of terpenes. Although their biosynthesis by means of mevalonate has been studied in detail, a second biosynthetic pathway for their formation by means of 1-deoxy-d-xylulose 5-phosphate has been discovered only recently in plants and certain eubacteria. Earlier in vivo experiments with recombinant Escherichia coli strains showed that exogenous 1-deoxy-d-xylulose can be converted into 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate by the consecutive action of enzymes specified by the xylB and ispCDEFG genes. This article describes the transformation of exogenous [U-13C5]1-deoxy-d-xylulose into a 5:1 mixture of [U-13C5]isopentenyl diphosphate and [U-13C5]dimethylallyl diphosphate by an E. coli strain engineered for the expression of the ispH (lytB) gene in addition to recombinant xylB and ispCDEFG genes.
Keywords: biosynthesis of isoprenoids‖1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate‖isopentenyl diphosphate‖dimethylallyl diphosphate
Terpenes are one of the largest groups of natural products comprising numerous medically relevant compounds (e.g., vitamins, hormones, and antitumor agents such as Taxol) (1). Bloch, Lynen, Cornforth, and their coworkers showed that the universal terpenoid precursors, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), are biosynthesized by means of the mevalonate pathway in yeasts and animals (for review see refs. 2–5). These studies served as the basis for the development of metabolic inhibitors that are widely used for the treatment of hypercholesterolemia.
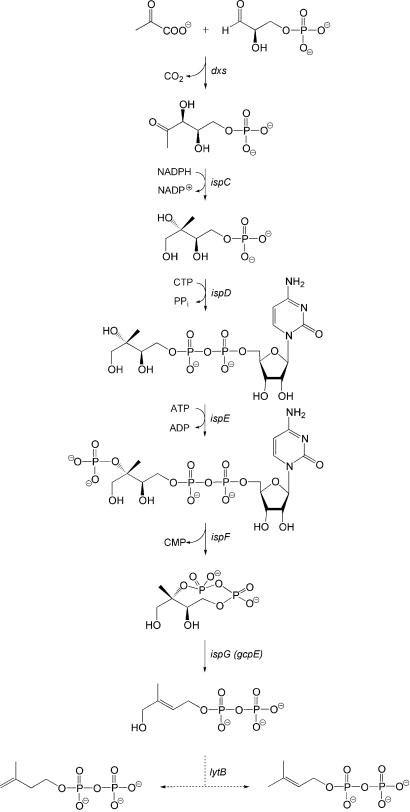
For a period of several decades, these milestone achievements completely eclipsed the existence of a second terpenoid pathway for the biosynthesis of IPP and DMAPP. Recently, however, knowledge on that pathway has been unfolding rapidly on the basis of seminal discoveries by the research groups of Arigoni and Rohmer (refs. 6–8, for review see refs. 9–12). Briefly, the mevalonate independent pathway starts from 1-deoxy-d-xylulose 5-phosphate, which is assembled from pyruvate and d-glyceraldehyde 3-phosphate (13, 14) and was already known to serve as a biosynthetic precursor of vitamins B1 (thiamine) and B6 (pyridoxal) (Fig. 1) (15–17). 1-Deoxy-d-xylulose 5-phosphate is converted into 2C-methyl-d-erythritol 4-phosphate, the first committed intermediate of the nonmevalonate pathway, by isomerization followed by a two-electron reduction step catalyzed by 2C-methyl-d-erythritol 4-phosphate synthase specified by the ispC gene (formerly designated yaeM and then dxr) (18) (Fig. 1).
Figure 1.
The nonmevalonate pathway of isoprenoid biosynthesis.
In the next step, 2C-methyl-d-erythritol 4-phosphate is converted into the 2C-methyl-d-erythritol 2,4-cyclodiphosphate by the sequential action of three enzymes specified by the ispD, ispE, and ispF genes (19–24). The last known step of the sequence, the reductive transformation of the cyclic diphosphate into 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate has been identified recently in work with a recombinant Escherichia coli strain engineered for hyperexpression of the ispG gene (previously designated gcpE) (25). Comparative genomics suggest that the protein specified by the lytB gene is involved in the conversion of 2C-methyl-d-erythritol 2,4-cyclodiphosphate into IPP and DMAPP (26). Mutagenesis studies confirmed this suggestion (27–29), but no direct evidence for the specific role of the LytB protein has been reported up to now.
The nonmevalonate pathway is present in many eubacteria (for review see refs. 9–12). In higher plants, both pathways are operative; while triterpenes including the sterol precursor, cycloartenol, are known to be produced by means of the mevalonate pathway in the cytoplasmic compartment of plant cells, a large number of other terpenes (e.g., mono- and diterpenes) are produced in the plastid compartment by means of the deoxyxylulose phosphate pathway. The picture is complicated by the exchange of certain terpene precursors between the two compartments; because of this crosstalk between the two pathways, terpenes synthesized in plastids can be derived in part from the cytoplasmically located mevalonate pathway and vice versa.
For numerous microbial pathogens such as Enterobacteria and Mycobacterium tuberculosis, the nonmevalonate pathway is the exclusive source of terpenoids. The alternative terpenoid pathway has also been shown to be essential in Plasmodium spp., which is responsible for more than a million deaths per year (30).
Because the enzymes of the nonmevalonate pathway have no orthologs in mammalian hosts, they are attractive targets for the development of novel antibiotic and/or antiprotozoal agents. Thus, the further elucidation of the nonmevalonate pathway enzymes appears urgent to provide molecular information for drug design. This article describes the role of the ispH (formerly designated lytB) gene product in the conversion of 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate into IPP and DMAPP.
Experimental Procedures
Materials.
[U-13C5]1-deoxy-d-xylulose 5-phosphate was prepared by published procedures (31). Oligonucleotides were custom-synthesized by MWG Biotec, Ebersberg, Germany. IPP and DMAPP were purchased from Echelon, Salt Lake City, UT. Diaminobenzidine and alkaline phosphatase from bovine intestinal mucosa were purchased from Sigma. A rabbit polyclonal antiserum directed against recombinant IspH (LytB) protein was prepared by Eurogentec, Seraing, Belgium. Anti-rabbit horseradish peroxidase conjugated was purchased from Promega.
Preparation of 1-Deoxy-d-Xylulose.
Reaction mixtures containing 150 mM Tris⋅HCl (pH 9.5), 30 mM MgCl2, 32 mM [U-13C5]1-deoxy-d-xylulose 5-phosphate, and 130 μg/ml−1 alkaline phosphatase were incubated at 37°C for 16 h. The reaction mixtures were monitored by NMR spectroscopy and used for feeding experiments without further treatment.
Bacterial Strains and Plasmids.
Bacterial strains and plasmids used in this study are summarized in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristic | Ref. or source |
|---|---|---|
| E. coli | ||
| XL1-Blue | RecA1, endA1, gyrA96, thi-1, hsdR17, supE44, relA1, lac, [F′, proAB, laclqZΔM15, Tn10 (tetr)] | 33, Stratagene |
| M15(pREP4) | Lac, ara, gal, mtl, recA+, uvr+, [pREP4, lacI, kanr] | 34 |
| Plasmids | ||
| pQE30 | High copy His-Tag expression vector | Qiagen |
| pQElytB | Expression of ispH (lytB) from E. coli | This study |
| pACYC184 | Low copy cloning vector | 35, NEB |
| pACYClytB | Expression of ispH from E. coli | This study |
| pACYClytBgcpE | Expression of ispG and ispH from E. coli | This study |
| pBluescript SKII− | High copy cloning vector | Stratagene |
| pBScyclo | Expression of xylB, ispC, ispD, ispE, and ispF from E. coli | 25 |
| pBSxispC-H | Expression of xylB, ispC, ispD, ispE, ispF, ispG, and ispH from E. coli | This study |
DNA Sequence Determination.
DNA was sequenced by the automated dideoxynucleotide method (32) using a 377 Prism sequencer from Perkin–Elmer.
Protein Sequencing.
Automated Edman degradation was performed by using a PE BIO systems model 492 from Perkin–Elmer.
Cloning and Expression of the ispH Gene.
The ispH gene of E. coli (formerly designated lytB; GenBank accession no. AE000113) was amplified from bp position 5618 to 6568 by PCR using chromosomal E. coli DNA as template and the oligonucleotides lytBvo and lytBhi as primers (Table 2). The amplificate was purified, treated with the restriction enzymes BamHI and PstI, and ligated into the expression vector pQE30 (Qiagen, Hilden, Germany), which had been treated with the same enzymes. The resulting plasmid pQElytB was electrotransformed into E. coli strains XL1-Blue (Stratagene) (33) and M15(pREP4) (34), affording the recombinant strains XL1-pQElytB and M15-pQElytB, respectively.
Table 2.
Oligonucleotides used in this study
| Designation | 5′-Sequence-3′ |
|---|---|
| lytBvo | TGGAGGGGATCCATGCAGATCCTGTTGGCC |
| lytBhi | GCATTTCTGCAGAACTTAGGC |
| lytBSalIvo | GCTTGCGTCGACGAGGAGAAATTAACCATGCAGATCCTGTTGGCCACC |
| lytBEagIhi | GCTGCTCGGCCGTTAATCGACTTCACGAATATCG |
| gcpEBamHIvo | CGTACCGGATCCGAGGAGAAATTAACCATGCATAACCAGGCTCCAATTC |
| gcpESalIhi | CCCATCGTCGACTTATTTTTCAACCTGCTGAACGTC |
| gcpESacIIvo | GCGGGAGACCGCGGGAGGAGAAATTACCATGCATAACCAGGCTCCAATTCAACG |
| gcpENotIhi | AGGCTGGCGGCCGCTTAATCGACTTCACGAATATCG |
Construction of a Synthetic Operon.
Vector constructs for the in vivo utilization of 1-deoxy-d-xylulose into recombinant E. coli strains were synthesized in consecutive cloning steps as follows. The ispH gene of E. coli was amplified by PCR using chromosomal E. coli DNA as template and the oligonucleotides lytBSalIvo and lytBEagIhi as primers (Table 2). The amplificate was purified, treated with SalI and EagI, and ligated into the plasmid vector pACYC184 (35), which had been treated with the same restriction enzymes. The resulting plasmid pACYClytB was electrotransformed into E. coli strain XL1-Blue, yielding the recombinant strain XL1-pACYClytB.
The ispG gene of E. coli (GenBank accession no. AE000338) was amplified from bp position 372 to 1204 by PCR using chromosomal E. coli DNA as template and the oligonucleotides gcpEBamHIvo and gcpESalIhi as primers (Table 2). The amplificate was purified, treated with BamHI and SalI, and ligated into the plasmid vector pACYClytB, which had been treated with the same restriction enzymes. The resulting plasmid pACYClytBgcpE was electrotransformed into E. coli strain XL1-Blue, yielding the recombinant strain XL1-pACYClytBgcpE.
The artificial gcpE, lytB operon of the plasmid pACYClytBgcpE was amplified by PCR and the oligonucleotides gcpESacIIvo and lytBNotI as primers (Table 2). The amplificate was purified, treated with SacII and NotI, and ligated into the plasmid construct pBScyclo expressing the xylB and the ispCDEF genes (25), which had been treated with the same restriction enzymes. The resulting plasmid pBSxispC-H was electrotransformed into E. coli strain XL1-Blue, affording the recombinant strain XL1-pBSxispC-H.
Isotope Incorporation Studies.
LB medium (0.2 l containing 36 mg ampicillin) was inoculated with recombinant E. coli XL1-Blue cells carrying plasmids overexpressing xylB in conjunction with genes of the nonmevalonate pathway (Table 1). The cells were grown with shaking at 37°C overnight. At an OD (600 nm) of 1.4, 24 ml of a solution containing 1.1 M lithium lactate and 6.7 mM [U-13C5]1-deoxy-d-xylulose, pH 7.0, was added continuously within 2 h. Aliquots of 40 ml were retrieved at intervals of 30 min. The cells were harvested, centrifuged for 10 min at 5,000 rpm and 4°C, washed with 0.9% NaCl, and centrifuged. The cells were resuspended in 600 μl of 10 mM NaF/D2O in 50% (vol/vol) methanol-d4, cooled on ice, and subjected to ultrasonic treatment. The suspension was centrifuged at 15,000 rpm for 15 min. To avoid degradation during work-up, the supernatants were subjected to NMR analysis without further purification.
NMR Spectroscopy.
1H, 13C, and 31P NMR spectra were recorded by using an AVANCE DRX 500 spectrometer from Bruker Instruments, Karlsruhe, Germany. Two-dimensional heteronuclear multiple quantum correlation (HMQC) and HMQC- total correlation spectroscopy (TOCSY) experiments were performed by using standard Bruker software (XWINNMR 3.0). The duration of TOCSY transfer was 60 ms in HMQC-TOCSY experiments.
Purification of Recombinant IspH (LytB) Protein.
The E. coli strain M15-pQElytB was grown in LB broth containing 180 mg ampicillin and 50 mg kanamycin sulfate per liter. Cultures were incubated at 37°C with shaking. At an OD of 0.7 (600 nm), isopropyl β-d-thiogalactoside was added to a final concentration of 2 mM, and the culture was incubated for 5 h. The cells were harvested by centrifugation, washed with 0.9% (g/vol) sodium chloride, and stored at −20°C.
Frozen cell mass (8 g) was thawed in 80 ml of 50 mM potassium phosphate, pH 7.0, containing 0.5 M sodium chloride and 20 mM imidazole hydrochloride. The suspension was subjected to ultrasonic treatment and centrifuged. The supernatant was applied to a column of Ni-chelating Sepharose FF (2.0 × 5 cm, Amersham Pharmacia), which had been equilibrated with 50 mM potassium phosphate, pH 7.0, containing 0.5 M sodium chloride and 20 mM imidazole (flow rate, 2 ml⋅min−1). The column was washed with 150 ml of 50 mM potassium phosphate, pH 7.0, containing 0.5 M sodium chloride and 20 mM imidazole, and was then developed with a gradient of 20–300 mM imidazole in 240 ml of 20 mM potassium phosphate, pH 7.0. Fractions were combined and dialyzed overnight against 50 mM potassium phosphate, pH 7.0. The solution was concentrated by ultrafiltration. The protein solution was loaded on top of a Superdex 200 column (2.6 × 60), which had been equilibrated with 50 mM potassium phosphate, pH 7.0 at a flow rate of 3 ml⋅min−1. The retention volume of IspH protein was 238 ml. Fractions were combined and stored at −20°C.
PAGE.
SDS/PAGE was performed with the SE 250 Mighty small II electrophoresis system from Amersham Pharmacia at a constant current of 20 mA per gel, using 5% acrylamide stacking gels and 15% acrylamide separating gels (36). Gels were stained with 0.25% (wt/vol) Coomassie R-250 in 50% methanol/acetic acid/water (46:10:46; vol/vol/vol) and destained in methanol/acetic acid/water (30:10:70; vol/vol/vol).
Western Blotting.
Protein samples dissolved in 3% SDS containing 3% mercaptoethanol and 0.1% bromophenol blue were subjected to SDS/PAGE as described above. The proteins were electro-transferred to Immobilon-P transfer membranes by using the Transblot SD from Bio-Rad with a constant current of 2 mA⋅cm−2 gel for 1 h (37). A rabbit polyclonal antiserum directed against IspH (LytB) protein from E. coli (diluted 1:8,000) was used as the first antibody. Anti-rabbit IgG (Fc) conjugated to horseradish peroxidase was used as second antibody. Blots were developed with diaminobenzidine and H2O2 (38).
Radial Immunodiffusion.
Experiments were performed by the procedure of Mancini et al. (39). Gels contained 50 mM Tris⋅HCl (pH 7.6), 0.7% agarose (Serva), 0.03% sodium azide, and 90 μl/ml anti-IspH serum containing 150 mM sodium chloride. Wells with diameters of 4 mm were loaded with samples of 20 μl. The plates were incubated in a humid chamber at room temperature. Recombinant IspH protein was used as standard.
Results
The ispH (lytB) gene of E. coli was placed under the control of a T5 promoter and lac operator in the hyperexpression plasmid pQE30. The ORF was preceded by a synthetic DNA segment specifying the amino acid motif MRGSHHHHHHGS to expedite the purification of the recombinant protein by metal chelating affinity chromatography. A recombinant E. coli strain carrying that plasmid directed the synthesis of a peptide with a relative mass of ≈37 kDa as judged from SDS/PAGE. The recombinant protein accounted for ≈5.6% of soluble cell protein as judged by single radial immunodiffusion.
The recombinant protein was purified to apparent homogeneity by affinity chromatography on a Ni-chelating column. The N-terminal sequence obtained by partial Edman degradation matched the prediction based on the DNA sequence of the expression plasmid. Electrospray MS indicated a relative mass of 36,161 ± 40 Da in agreement with the predicted mass of 36,173 Da. The recombinant IspH (LytB) protein was used to prepare a rabbit antiserum, which was subsequently used to monitor IspH (LytB) protein in recombinant strains designed for hyperexpression of isoprenoid biosynthesis genes.
We have shown earlier that the hyperexpression of d-xylulokinase specified by the xylB gene enables E. coli to convert exogenous 1-deoxy-d-xylulose into 1-deoxy-d-xylulose 5-phosphate with high efficacy (25) and that strains expressing the xylB gene in conjunction with ispCDEFG genes convert exogenous 1-deoxy-d-xylulose into a mixture of 2C-methyl-d-erythritol 2,4-cyclodiphosphate and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (25) (Fig. 1).
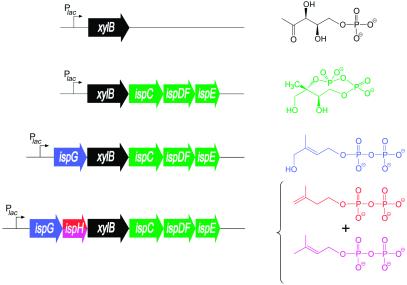
Elaborating on this strategy, we have now constructed a synthetic operon directing the expression of the ispH gene (formerly designated lytB) together with the xylB and ispCDEFG genes (Fig. 2). The synthetic operon was placed under the control of a lac promoter and operator in the plasmid pBSXispC-H (Table 2). Cell extracts of an E. coli strain harboring that plasmid were shown to contain ≈1.5% of IspH protein based on total cell protein as shown by immunoanalysis. The levels of IspH protein in E. coli strains expressing the xylB and ispCDEFG genes (25) (Fig. 2) were below the detection level (<80 ng = 0.5% based on cell protein).
Figure 2.
Expression vectors comprising synthetic operons of xylB together with genes of the nonmevalonate pathway used for the in vivo production of intermediates in the nonmevalonate pathway. 13C-labeled compounds detected in cell extracts obtained from E. coli cells overexpressing the indexed genes and supplied with [U-13C5]1-deoxy-d-xylulose are indicated. Genes and their respective products are shown in the same colors.
Recombinant E. coli strains carrying one of the plasmids shown in Fig. 2 were supplemented with [U-13C5]1-deoxy-d-xylulose and cultivated as described under Experimental Procedures. The cells were harvested and disrupted by ultrasonic treatment. The crude supernatants were analyzed by 13C NMR spectroscopy without any pretreatment.
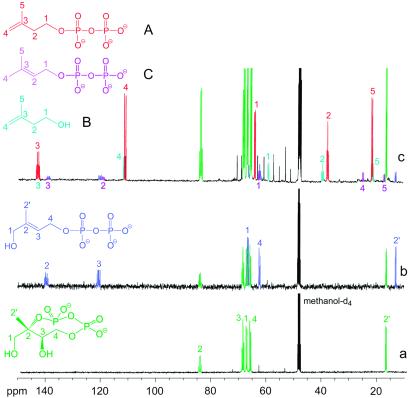
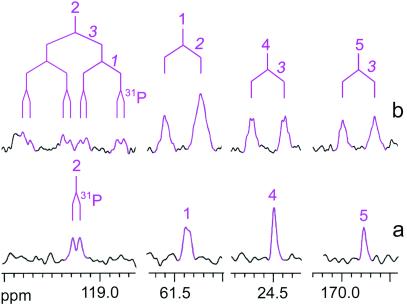
All spectra in Fig. 3 display a set of five multiplets shown in green, which is characteristic of 2C-methyl-d-erythritol 2,4-cyclodiphosphate, the product of the IspF protein (Fig. 2). A second set of five signals shown in blue in Fig. 3 B and C represents the product of IspG protein, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate, which has been described in our previous study (25).
Figure 3.
13C NMR spectra of crude extracts obtained from the feeding of [U-13C5]1-deoxy-d-xylulose to recombinant cells of E. coli overexpressing (a) the xylB, ispC-F genes, (b) the xylB, ispC-ispG genes, and (c) the xylB, ispC-ispH genes.
In addition to these features, the spectrum in Fig. 3C shows a series of novel multiplets, which are displayed in more detail in Figs. 4 and 5. The complex 13C13C coupling patterns indicate that these novel metabolites are universally 13C-labeled and must therefore have been derived from the proffered [U-13C5]1-deoxy-d-xylulose. On the basis of 13C13C coupling constants, 1H13C correlation experiments and chemical shift comparison with reference compounds, these novel compounds can be identified as IPP (metabolite A, shown in red), isopentenol (metabolite B, shown in light blue), and DMAPP (metabolite C, shown in magenta). A detailed description of the NMR data follows.
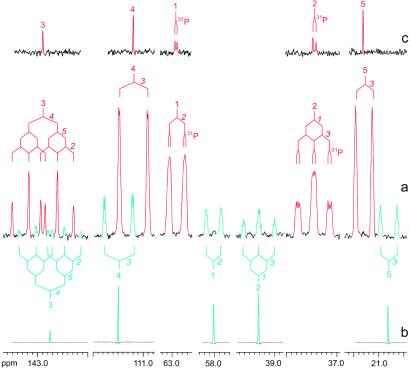
Figure 4.
13C NMR signals of IPP (in red) and isopentenol (in light blue) (a) of the crude extract obtained from the feeding of [U-13C5]1-deoxy-d-xylulose to recombinant cells of E. coli overexpressing the xylB, ispC-H genes (cf. Fig. 3) (b and c) of reference samples with natural 13C abundances.
Figure 5.
13C NMR signals of DMAPP (in magenta) (b) of the crude extract obtained from the feeding of [U-13C5]1-deoxy-d-xylulose to recombinant cells of E. coli overexpressing the xylB, ispC-H genes (cf. Fig. 3) (a) of a reference sample with natural 13C abundance.
Fig. 4 shows the expanded signals of metabolites A and B. Three 13C NMR signals (21.6, 63.0, and 111.2 ppm) of metabolite A are doublets, indicating that each of the three carbon atoms is connected to a single 13C-labeled neighbor. One signal (37.4 ppm) displays a pseudotriplet signature, indicating a 13C atom with two adjacent 13C atoms, and one signal (142.9 ppm) is a doublet of pseudotriplets, indicating a 13C atom with three directly connected 13C atoms. The large coupling constant of 71 Hz observed for the signals at 111.2 and 142.9 ppm is typical for olefinic carbon atoms. Long-range 13C13C coupling is responsible for fine splitting or apparent signal broadening for the signals at 37.4, 63.0, and 111.2 ppm of the universally 13C-labeled metabolite A.
HMQC and HMQC-TOCSY experiments revealed the 1H NMR chemical shifts as well as the complete 13C and 1H spin systems of metabolite A. Specifically, the 13C NMR signal at 111.2 ppm is correlated to a 1H NMR signal at 4.73 ppm. The signal at 142.9 ppm gave no 1H13C correlation and represents a quaternary carbon. The 13C signals at 63.0, 37.4, and 21.6 ppm show correlations to 1H signals at 4.00, 2.31, and 1.68 ppm, respectively. As shown by HMQC-TOCSY experiments, the proton signals at 2.31 and 4.00 were 1H-coupled, whereas the signals at 4.73 and 1.68 ppm appeared as singlets in the HMQC-TOCSY experiment. All of these features identify metabolite A as an isopentene derivative.
The 13C and 1H chemical shifts of metabolite A agree with those of authentic, unlabeled IPP. Carbon atoms 1 and 2 of biosynthetic IPP as well as the synthetic reference sample show 13C31P coupling. The assignment of metabolite A as IPP is also well in line with 31P NMR spectra, indicating the presence of an organic diphosphate derivative in significant amounts (Table 3).
Table 3.
NMR data of IPP and DMAPP
| Position | Chemical shifts, ppm
|
Coupling constants, Hz
|
Correlation pattern
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1H* | 13C* | 31P† | JPC | JHH | JPP | JPH | JCC‡ | HMQC‡ | HMQC-TOCSY‡ | |
| IPP (metabolite A) | ||||||||||
| 1 | 4.00 | 63.0 | 4.9 | 6.6 | 6.6 | 34 | 1 | 1, 2 | ||
| 2 | 2.31 | 37.4 | 8.0 | 6.7 | 40, 40 | 2 | 2, 1 | |||
| 3 | 142.9 | 71, 41, 41 | ||||||||
| 4 | 4.73 | 111.2 | 71 | 4 | ||||||
| 5 | 1.68 | 21.6 | 41 | 5 | ||||||
| P | −7.8 | nd | ||||||||
| P | −11.9 | 19.5 | ||||||||
| DMAPP (metabolite C) | ||||||||||
| 1 | 4.45 | 61.4 | 3.6 | 6.6 | 6.6 | 47 | 1 | 1, 2 | ||
| 2 | 5.43 | 119.2 | 9.0 | 7.2 | 75, 48 | 2 | 2, 1 | |||
| 3 | 139.0 | nd | ||||||||
| 4 | 1.75 | 24.5 | 42 | 4 | ||||||
| 5 | 1.71 | 16.7 | 41 | 5 | ||||||
| P | −9.1 | 21.7 | ||||||||
| P | −6.4 | 21.5 | ||||||||
nd, not determined.
Referenced to external trimethylsilylpropane sulfonate.
Referenced to external 85% orthophosphoric acid.
Observed with [U-13C5]DMAPP.
The coupling and correlation patterns of metabolite B as gleaned from one- and two-dimensional (HMQC and HMQC-TOCSY) NMR spectra are closely similar to those of metabolite A (IPP). The 13C13C and 13C1H connectivity patterns of metabolite B and IPP are identical, and the 1H and 13C chemical shift values of metabolite B are identical with those of isopentenol. The absence of 13C31P coupling agrees with the identification of metabolite B as [U-13C5]isopentenol, which may have been formed by dephosphorylation of IPP.
The structure of metabolite C (Figs. 3 and 5) was assigned by the same approach. The 13C coupling patterns (three doublets, one double doublet, one multiplet) suggested that the compound is an isopentane derivative. The chemical shifts observed for the double-doublet (119.2 ppm) and the multiplet (139.0 ppm) show that a carbon–carbon double bond connects C-2 (coupled to two adjacent 13C atoms) and C-3 (coupled to three adjacent 13C atoms) of the molecule. The signal at 119.2 ppm showed fine splitting caused by 13C31P coupling. The signals at 61.4, 24.5, and 16.7 ppm were broadened because of long-range 13C13C couplings. The 1H NMR chemical shifts revealed by HMQC and HMQC-TOCSY experiments show two singlets at 1.75 and 1.71 ppm and a spin system comprising signals at 5.43 and 4.45 ppm. In conjunction with the chemical shifts, this correlation pattern shows that metabolite C is a dimethylallyl derivative.
The 13C and 1H NMR chemical shifts of an authentic sample of DMAPP were identical to the chemical shifts of the signals attributed to metabolite C (Fig. 5).
By evaluation of the NMR signal integrals, it can be estimated that the concentrations of 2C-methyl-d-erythritol 2,4-cyclodiphosphate, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate, IPP, and DMAPP are ≈25, 1, 5, and 1 mM, respectively. The concentration of isopentenol increased during the period of feeding and NMR experiments probably caused by degradation of IPP.
Discussion
Earlier in vivo studies had shown that IspG protein catalyzes the reductive ring opening of 2C-methyl-d-erythritol 2,4-cyclodiphosphate to 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate (25). We have now extended this approach to include a recombinant IspH protein and were able to detect the formation of a 5:1 mixture of IPP and DMAPP upon feeding of 13C-labeled 1-deoxy-d-xylulose to an E. coli strain engineered for the overexpression of the ispH (lytB) gene in addition to recombinant xylB and ispCDEFG genes.
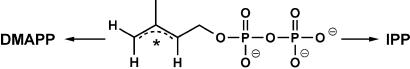
In the mevalonate pathway IPP and DMAPP are generated sequentially, the latter arising from the former in an equilibration reaction catalyzed by an isomerase that exploits a protonation-deprotonation mechanism (40–42). In the present case sequential formation of the two compounds seems unlikely for a variety of reasons. (i) The IspH protein does not display isomerase activity (27, 43). (ii) The idi gene of E. coli encodes an IPP isomerase (44). Its catalytic activity, however, is 20-fold lower than the one of the yeast enzyme (45) and does not seem to contribute significantly to terpene biosynthesis; moreover, the idi gene has been shown to be dispensable in this bacterium (44). (iii) In contrast to the ispH gene the idi gene is not present in the genome of many bacteria operating on the mevalonate independent pathway (12, 26, 27). It seems therefore more realistic to conclude that the IspH protein is capable of generating both IPP and DMAPP along parallel ways starting from 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate as a single precursor. Accordingly, this protein can be identified as the responsible agent for the branching in the nonmevalonate pathway, which had been postulated earlier to account for the anomalous results observed in feeding experiments with deuterated substrates both with E. coli (46, 47) and Eucalyptus globulus (48). Independent genetic evidence for the existence of such a branching has been provided for the machinery of E. coli (43). Elucidation of the role of the protein encoded by the ispH gene fills the last gap in the sequence of enzymatic steps of the pathway that leads from pyruvate and d-glyceraldehyde 3-phosphate to IPP and DMAPP.
Because the experiments reported in this article were carried out in vivo the cofactors of the reaction catalyzed by the IspH protein remain unidentified and detailed reaction mechanisms can be proposed only with qualification. The simplest general explanation for the observed branching is provided by assuming the formation of a bidentate reactive intermediate of the type illustrated in Fig. 6, in which * is a reference to the unknown number of electrons (2 to 4) in the delocalized allyl system. Results from the Rohmer group on the mevalonate-independent biosynthesis of bacteriohopans in Zymomonas mobilis (49) suggest the involvement of NADPH as a reducing agent in the last step of the reaction sequence that leads to IPP, and this, in turn, could be taken to imply the formation of a cationic intermediate in the reaction catalyzed by the IspH protein. The available evidence, however, does not conclusively rule out alternative pathways involving radical or anionic intermediates and the development of an appropriate in vitro system is clearly needed to settle the remaining mechanistic issues.
Figure 6.
A generalized structure for the hypothetical intermediate(s) at the branching point of the IspH-mediated reaction. * is related to the unknown number of electrons (2 to 4) in the delocalized allyl system.
Acknowledgments
We thank Fritz Wendling and Ingrid Obersteiner for skillful assistance and Angelika Werner for expert help with the preparation of the manuscript. We thank the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Hans-Fischer Gesellschaft for support. Financial support by Novartis International AG, Basel (to D.A.) is gratefully acknowledged.
Abbreviations
- IPP
isopentenyl diphosphate
- DMAPP
dimethylallyl diphosphate
- HMQC
heteronuclear multiple quantum correlation
- TOCSY
total correlation spectroscopy
Note Added in Proof.
In a paper published after submission of this manuscript (50) M. Hintz et al. have reported a significant accumulation in lytB- (now ispH-) deficient mutants of E. coli of a compound subsequently identified as 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY062212).
References
- 1.Sacchettini J C, Poulter C D. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 2.Qureshi N, Porter J W. In: Biosynthesis of Isoprenoid Compounds. Porter J W, Spurgeon S L, editors. Vol. 1. New York: Wiley; 1981. pp. 47–94. [Google Scholar]
- 3.Bloch K. Steroids. 1992;57:378–382. doi: 10.1016/0039-128x(92)90081-j. [DOI] [PubMed] [Google Scholar]
- 4.Bach T J. Lipids. 1995;30:191–202. doi: 10.1007/BF02537822. [DOI] [PubMed] [Google Scholar]
- 5.Bochar D A, Friesen J A, Stauffacher C V, Rodwell V W. In: Comprehensive Natural Product Chemistry. Cane D, editor. Vol. 2. Oxford: Pergamon; 1999. pp. 15–44. [Google Scholar]
- 6.Rohmer M, Knani M, Simonin P, Sutter B, Sahm H. Biochem J. 1993;295:517–524. doi: 10.1042/bj2950517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broers S T J. Thesis. Zürich: Eidgenössische Technische Hochschule; 1994. [Google Scholar]
- 8.Schwarz M K. Thesis. Zürich: Eidgenössische Technische Hochschule; 1994. [Google Scholar]
- 9.Eisenreich W, Schwarz M, Cartayrade A, Arigoni D, Zenk M H, Bacher A. Chem Biol. 1998;5:R221–R233. doi: 10.1016/s1074-5521(98)90002-3. [DOI] [PubMed] [Google Scholar]
- 10.Rohmer M. In: Comprehensive Natural Product Chemistry. Cane D, editor. Vol. 2. Oxford: Pergamon; 1999. pp. 45–68. [Google Scholar]
- 11.Schwarz M, Arigoni D. In: Comprehensive Natural Product Chemistry. Cane D, editor. Vol. 2. Oxford: Pergamon; 1999. pp. 367–399. [Google Scholar]
- 12.Rohdich F, Kis K, Bacher A, Eisenreich W. Curr Opin Chem Biol. 2001;5:535–540. doi: 10.1016/s1367-5931(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 13.Sprenger G A, Schörken U, Wiegert T, Grolle S, deGraaf A A, Taylor S V, Begley T P, Bringer-Meyer S, Sahm H. Proc Natl Acad Sci USA. 1997;94:12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lois L M, Campos N, Putra S R, Danielsen K, Rohmer M, Boronat A. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White R H. Biochemistry. 1978;17:3833–3840. doi: 10.1021/bi00611a024. [DOI] [PubMed] [Google Scholar]
- 16.David S, Estramareix B, Fischer J-C, Thèrisod M. J Am Chem Soc. 1981;103:7341–7342. [Google Scholar]
- 17.Hill R E, Sayer B G, Spenser J D. J Am Chem Soc. 1989;111:1916–1917. [Google Scholar]
- 18.Takahashi S, Kuzuyama T, Watanabe H, Seto H. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk M H. Proc Natl Acad Sci USA. 1999;96:11758–11763. doi: 10.1073/pnas.96.21.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuzuyama T, Takagi M, Kaneda K, Dairi T, Seto H. Tetrahedron Lett. 2000;41:703–706. [Google Scholar]
- 21.Lüttgen H, Rohdich F, Herz S, Wungsintaweekul J, Hecht S, Schuhr C A, Fellermeier M, Sagner S, Zenk M H, Bacher A, Eisenreich W. Proc Natl Acad Sci USA. 2000;97:1062–1067. doi: 10.1073/pnas.97.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuzuyama T, Takagi M, Kaneda K, Watanabe H, Dairi T, Seto H. Tetrahderon Lett. 2000;41:2925–2928. [Google Scholar]
- 23.Herz S, Wungsintaweekul J, Schuhr C A, Hecht S, Lüttgen H, Sagner S, Fellermeier M, Eisenreich W, Zenk M H, Bacher A, Rohdich F. Proc Natl Acad Sci USA. 2000;97:2486–2490. doi: 10.1073/pnas.040554697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takagi M, Kuzuyama T, Kaneda K, Watanabe H, Dairi T, Seto H. Tetrahedron Lett. 2000;41:3395–3398. [Google Scholar]
- 25.Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Proc Natl Acad Sci USA. 2001;98:14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adam, P., Eisenreich, W., Fellermeier, M., Hecht, S., Rohdich, F., Schuhr, C. A., Wungsintaweekul, J., Zenk, M. H. & Bacher, A. (2000) German Patent Appl. 10027821.3.
- 27.Cunningham F X, Jr, Lafond T P, Grantt E. J Bacteriol. 2001;182:5841–5848. doi: 10.1128/jb.182.20.5841-5848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altincicek B, Kollas A, Eberl M, Wiesner J, Sanderbrand S, Hintz M, Beck F, Jomaa H. FEBS Lett. 2001;499:37–40. doi: 10.1016/s0014-5793(01)02516-9. [DOI] [PubMed] [Google Scholar]
- 29.McAteer S, Coulson A, McLennan N, Master M. J Bacteriol. 2001;183:7403–7407. doi: 10.1128/JB.183.24.7403-7407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jomaa H, Wiesner J, Sanderbrand S, Altinicicek B, Weidemeyer C, Hintz M, Türbachova I, Eberl M, Zeidler J, Lichtenthaler H K, et al. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 31.Hecht S, Kis K, Eisenreich W, Amslinger S, Wungsintaweekul J, Herz S, Rohdich F, Bacher A. J Org Chem. 2001;66:3948–3952. doi: 10.1021/jo0100300. [DOI] [PubMed] [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. Biotechnology. 1992;24:104–108. [PubMed] [Google Scholar]
- 33.Bullock W O, Fernandez J M, Short J M. BioTechniques. 1978;5:376–379. [Google Scholar]
- 34.Zamenhof P J, Villarejo M. J Bacteriol. 1972;110:171–178. doi: 10.1128/jb.110.1.171-178.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hinzpeter M. In: Methods of Immunological Analysis. Massyeff R F, Albert V H, Steines N A, editors. Vol. 1. Weinheim, Germany: VCH; 1993. pp. 529–535. [Google Scholar]
- 39.Mancini G, Carbonara A O, Heremans J F. Immunochemistry. 1965;2:235–254. doi: 10.1016/0019-2791(65)90004-2. [DOI] [PubMed] [Google Scholar]
- 40.Reardon J E, Abeles R H. Biochemistry. 1986;25:5609–5616. doi: 10.1021/bi00367a040. [DOI] [PubMed] [Google Scholar]
- 41.Muehlbacher M, Poulter C D. Biochemistry. 1988;27:7315–7328. doi: 10.1021/bi00419a021. [DOI] [PubMed] [Google Scholar]
- 42.Poulter C D, Muehlbacher M, Davis D R. J Am Chem Soc. 1989;101:3740–3742. [Google Scholar]
- 43.Rodriguez-Concepcion M, Campos N, Maria Lois L, Maldonado C, Hoeffler J-F, Grosdemange-Billiard C, Rohmer M, Boronat A. FEBS Lett. 2000;473:328–332. doi: 10.1016/s0014-5793(00)01552-0. [DOI] [PubMed] [Google Scholar]
- 44.Hahn F M, Hurlburt P A, Poulter C D. J Bacteriol. 1999;181:4499–4504. doi: 10.1128/jb.181.15.4499-4504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Street I P, Poulter C D. Biochemistry. 1990;29:7531–7538. doi: 10.1021/bi00484a023. [DOI] [PubMed] [Google Scholar]
- 46.Giner J-L, Jaun B, Arigoni D. Chem Commun. 1998;17:1857–1858. [Google Scholar]
- 47.Charon L, Hoeffler J F, Pale-Grosdemange C, Losi L M, Campos N, Boronat A, Rohmer M. Biochem J. 2000;346:737–742. [PMC free article] [PubMed] [Google Scholar]
- 48.Rieder C, Jaun B, Arigoni D. Helv Chim Acta. 2000;83:2504–2513. [Google Scholar]
- 49.Charon L, Pale-Grosdemange C, Rohmer M. Tetrahedron Lett. 1999;40:7231–7234. [Google Scholar]
- 50.Hintz M, Reichenberg A, Altincicek B, Bahr U, Gschwind R M, Kollas A K, Beck E, Wiesner J, Eberl M, Jomaa H. FEBS Lett. 2001;509:317–322. doi: 10.1016/s0014-5793(01)03191-x. [DOI] [PubMed] [Google Scholar]