Abstract
Electropermeabilization is one of the nonviral methods successfully used to transfer genes into living cells in vitro and in vivo. Although this approach shows promise in the field of gene therapy, very little is known about the basic processes supporting DNA transfer. The present investigation studies this process at the single-cell level by using digitized fluorescence microscopy. Permeabilization is a prerequisite for gene transfer. Its assay by propidium-iodide (PI) penetration shows that it occurs at the sides of the cell membrane facing the two electrodes, whereas fluorescently labeled plasmids only interact with the electropermeabilized side of the cell facing the cathode. The plasmid interaction with the electropermeabilized part of the cell surface results in the formation of localized aggregates. These membrane-associated spots are formed only when pulses with a longer duration than a critical value are applied. These complexes are formed within 1 s after the pulses and cannot be destroyed by pulses of reversed polarities. They remain at the membrane level up to 10 min after pulsing. Although freely accessible to DNA dye (TOTO-1) 1 min after the pulses, they are fully protected when the addition takes place 10 min after. They diffuse in the cytoplasm 30 min after pulses and are present around the nucleus 24 h later.
Viral vectors still are considered to be the most efficient method for gene transfer, but often they are associated with host inflammatory and immune responses (1). Therefore, nonviral methods of gene delivery have been developed recently. These methods are chemical or physical. Among physical methods, electropermeabilization, also named electroporation (see ref. 2 for review), a method based on the application of electric field pulses to cells was introduced in the early 1980s (3). Electropermeabilization indeed can be used to introduce a large variety of molecules into many different cells (4–6). For the last 10 years, medical applications of this method have been developed successfully. A local antitumoral drug delivery to patients (electrochemotherapy) is under clinical trial (7). Transdermal drug delivery is obtained in vivo (8). Electropermeabilization has been used also to transfer DNA in vivo into the skin, liver, melanoma, and skeletal muscle cells (9–14). Many theoretical models have been proposed in an attempt to explain the mechanisms of electroporation and gene transfer (15–18), but no clear evidence has been obtained on how the DNA molecules interact with the electropermeabilized cell plasma membrane at the single-cell level. Indeed, several models have been proposed in the case of mammalian cells where the plasmid crosses the membrane (i) because of the existence of long-lived “electropores” (3, 17, 18), (ii) after a binding step at the cell surface and then electrodiffusion through the electropores (17), (iii) during application of the electric pulses caused by electrophoretic forces associated with the external field (19, 20), or (iv) by adsorption by sphingosine-DNA interactions, insertion, and passage of DNA through a hydrophilic percolated porous zone (16). In such models, plasmid DNA could enter the cell either after the application of electric pulses (i and ii) or only during pulse application (iii and iv). However, experimental results have been reported that lead to the conclusion that plasmids had to be present during electropulsation but crossed the electropulsed membrane in the minute after it. No gene transfer was detected with a postpulse DNA addition. These results were obtained on bacteria, yeast, and mammalian cells (21–23). We propose a model in which only the localized part of the cell membrane brought to the permeabilized state by the external field is competent for the transfer (24). It is a complex process, in which an anchoring step connecting the plasmid to the electropermeabilized membrane takes place during the pulse, followed by a postpulse transfer into the cytoplasm (23). All investigations published up to now were performed on cell population. Gene transfer was always evaluated by the associated gene expression.
With the recent development of fluorescence microscopy digitized imaging, it is now possible to work at the single-cell level and to follow the localization of fluorescently tagged plasmids. In the present study, this technology is used to give new insights into the process supporting the electrically mediated DNA transfer in mammalian cells.
Experimental Procedures
Cells.
Chinese hamster ovary cells were used. The wild-type Chinese hamster ovary clone (Toronto strain) was selected for its ability to grow in suspension or plated on Petri dishes or a 22 × 22-mm microscope glass coverslip. They were grown as described (25). Their ability to grow on a support after being maintained in suspension is direct evidence of their viability.
DNA Staining.
A 4.7-kb plasmid (pEGFP-C1, CLONTECH) carrying the green fluorescent protein (GFP) gene controlled by the cytomegalovirus promoter was stained stoichiometrically with the DNA intercalating dye TOTO-1 (Molecular Probes; ref. 26). The plasmid was stained with 2.3 × 10−4 M dye at a DNA concentration of 1 μg/μl for 60 min on ice. This concentration yields an average base pair to dye ratio of 5. Even if the labeling is not covalent, the equilibrium is dramatically in favor of the linked form (26). Plasmids were prepared from Escherichia coli transfected cells by using Maxiprep DNA purification system (Qiagen, Chatsworth, CA).
A version of pGeneGrip (Gene Therapy Systems, San Diego, CA) was used also. pGFP/Rhod is a rhodamine-labeled vector that codes for an enhanced GFP.
Electropermeabilization Apparatus.
Electropulsation was operated by using a CNRS cell electropulsator (Jouan, St. Herblain, France), which delivered square-wave electric pulses. An oscilloscope (Enertec, St. Etienne, France) monitored pulse shape. A polarity inverter built in the laboratory allowed to trigger bipolar pulses. The electropulsation chamber was built by using two stainless steel parallel rods (diameter = 0.5 mm, length = 10 mm, interelectrode distance = 5 mm) put on a 22 × 22-mm microscope glass coverslip. The electrodes were connected to a voltage generator. A uniform electric field was generated. The chamber was placed on the stage of an inverted digitized videomicroscope (Leica DMIRB, Wetzlar, Germany).
Electropermeabilization.
Permeabilization of cells was performed by the application of long electric pulses required to transfer genes and load macromolecules into cells (25, 27). Cell viability was preserved when using a long pulse duration by decreasing the electric field intensity (24, 28). Penetration of PI (100 μM in a low ionic strength pulsing buffer: 10 mM phosphate/1 mM MgCl2/250 mM sucrose, pH 7.4) was used to monitor permeabilization. Cells in suspension were centrifuged for 5 min at 120 × g and resuspended in the pulsing buffer. The cell suspension (100 μl, i.e., 105 cells) was poured between the electrodes, and 2 min were allowed for cell sedimentation on the coverslip surface. Ten pulses with controlled duration at a frequency of 1 Hz were applied at a given electric field intensity at room temperature. For plated cells, the culture medium was removed and replaced by the same buffer described above.
Electrotransfection.
The cells in suspension were washed and resuspended in a labeled plasmid-containing pulsation buffer. For each assay, 100 μl of cell suspension were used corresponding to 106 cells and 4 μg of labeled plasmid. The number of copies per cell was 7.9 × 105. Ten pulses with controlled duration at a frequency of 1 Hz were applied at a given electric field intensity at room temperature. For plated cells, identical conditions were used except that 3 × 105 cells were plated on the microscope coverslip.
Microscopy.
Cells were observed with a Leica ×100, 1.3 numerical aperture oil immersion objective. The wavelengths were selected by using the Leica L4 filter block (450 nm ≤ λex ≤ 490 nm; dichromatic mirror pass band, 515 nm ≤ λem ≤ 560 nm) for the TOTO-1-labeled DNA and the Leica M2 filter block (532 nm ≤ λex ≤ 560 nm; dichromatic mirror cut-off, 590 nm ≤ λem) for the rhodamine-labeled DNA and PI-labeled cells. Images (and optical sections) were recorded with the CELLscan system (Scanalytics, Billerica, MA) fitted with a cooled charge-coupled device camera (Princeton Instruments, Trenton, NJ).
This digitizing set-up allowed quantitative localized analysis of the fluorescence emission. This was done along the cell membrane as well as radially across the cell. Plot histograms detected local increase above the background level outside of the cells. Two characteristic parameters were used: the peak intensity and the integral under the peak. Both were related directly to the number of fluorescent molecules locally present. The light haze contributed by fluorescent-labeled structures located above and below the plane of optimal focus was reassigned mathematically to its original place [EXHAUSTIVE PHOTON REASSIGNMENT software (Scanalytics, Billerica, MA)] after accurate characterization of the blurring function of the optical system.
Results
Cell Permeabilization.
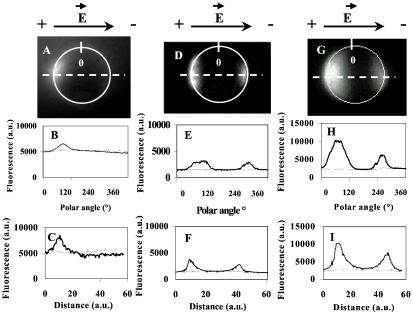
Permeabilization of Chinese hamster ovary cells was only detected for electric field values higher than thresholds (0.2 kV/cm for plated cells and to 0.4 kV/cm for cells in suspension) as observed already in many systems (29). A localized inflow of PI in the seconds after the pulses was observed at the single-cell level. Membrane permeabilization was observed at the sites of the cell membrane facing the two electrodes. When short pulses were used (0.1 ms), PI inflow was present only on the side facing the anode, showing that only that side of the cell surface was altered. When the pulse duration was increased up to 0.5 ms (or longer), permeabilization was detected on both cell sides (Fig. 1). The inflow of PI was quantified by the associated fluorescence intensity. It reflects its free diffusion across permeabilized parts of the membrane in the cytoplasm.
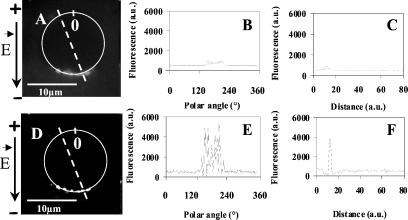
Figure 1.
Two-dimensional fluorescence intensity profiles of the permeabilized areas. Cells in suspension were incubated in the presence of 100 μM PI in the pulsing buffer. Ten pulses of 0.1, 0.5, and 5 ms were applied at 0.8 kV/cm (A, D, and G). The time exposure of the camera was set at 1 s in A and 0.1 s in D and G. a.u., arbitrary units. (A, D, and G) Fluorescence images 1 s after the pulses. (B, E, and H) Fluorescence intensity profile along the cell membrane highlighted by the white line in A, D, and G. (C, F, and I) Fluorescence intensity radial profile along the dotted lines shown in A, D, and G. Thin lines represent the fluorescence detected before pulse application. Thick lines represent the fluorescence intensity after electropulsation. The black arrows on the top of the figure indicate the direction of the electric field.
Cell Electrotransfection.
No transfected cells were detected in the absence of electric field, in the absence of DNA, or when DNA was added after the pulses. Electrotransfection was detected only for electric field values leading to permeabilization. Transfection threshold values were equal to 0.2 kV/cm for plated cells and 0.4 kV/cm for cells in suspension when pulses lasting 5 ms were applied. These thresholds were the same as those for cell permeabilization.
Visualization of DNA-Membrane Interaction.
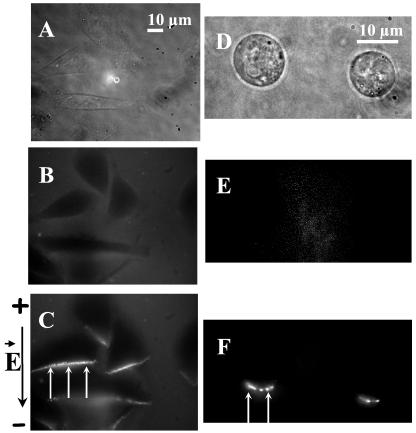
To go further into the mechanism of gene transfer, direct visualization of DNA interaction with membrane was performed at the single-cell level by using fluorescent plasmid DNA. The labeled plasmid was added to cells (Fig. 2). Cells appeared in black. No fluorescence ring caused by spontaneous adsorption of DNA to the plasma membrane was detected (Fig. 2 B and E). When a train of millisecond electric pulses known to permeabilize cells was applied, a sharp fluorescence increase appeared at the membrane level and was restricted to the side of the cell facing the cathode (Fig. 2 C and F). These observations were the same as those obtained with a rhodamine covalently labeled plasmid expressing GFP (Fig. 8). Because of its high quantum yield, the TOTO-1-labeled plasmid was used for quantification of the early events of the transfer. Postpulse addition of fluorescent DNA did not bring any enhancement of fluorescence on the cell membrane (data not shown).
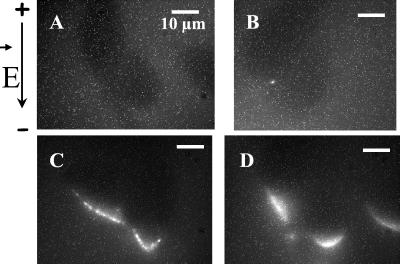
Figure 2.
Fluorescence microscopy observation of the DNA-membrane interaction areas. Plated cells or cells in suspension were incubated in presence of TOTO-1-labeled DNA (pEGFP-C1). Ten pulses of 5 ms (1 Hz) were applied at 0.6 kV/cm (A, B, and C) and 0.8 kV/cm (D, E, and F). The time exposure of the camera was set at 1 s. (A and D) Light contrast images before pulsation. (B and E) Fluorescence images before the pulsation. Cells appeared in black in labeled DNA solution. (C and F) Fluorescence images 10 s after the pulsation. Labeled DNA interacts with the permeabilized area of the membrane facing the cathode (white arrows). The black arrow on the left side indicates the direction of the electric field.
Figure 8.
Cellular traffic of fluorescent-labeled DNA. Chinese hamster ovary cells in suspension were submitted to 10 pulses of 5 ms at 0.8 kV/cm in the presence of GFP/rhodamine plasmid (8 μg/100 μl). They were observed during the postpulse incubation. The time exposure of the camera was set at 2 s. A, before pulsation; B–E, 1 sec, 10 min, 30 min, and 24 h after the pulses (rhodamine filter); F, GFP emission 24 h after the pulses (GFP filter).
When adult bovine aortic endothelial cells were used, spot formation again was observed (data not shown). The DNA-membrane interaction is not cell line-specific.
Quantification of Fluorescence.
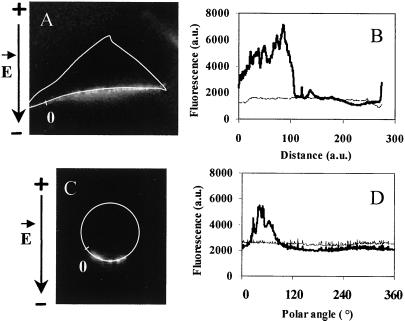
In Fig. 3, TOTO fluorescence intensity profiles were measured along the membrane on the cathode side of the cell before and after electric pulse application in the case of the plated cell (Fig. 3A) and the cell in suspension (Fig. 3C). Before electropulsation, basal fluorescence level was the same right around the membrane (thin line). One second after electropulsation, fluorescence sharply increased. It was not found to be homogeneous in the membrane as shown by the peaks of fluorescence (Fig. 3 B and D). In our experimental conditions, the local increase in fluorescence caused by pulsation varied from 1.2- to 3.2-fold. It was only present at the membrane level. No diffusion of DNA into the cytoplasm was detected (Fig. 4C).
Figure 3.
Fluorescence intensity profiles at the membrane level. Plated cells or cells in suspension were incubated in the presence of TOTO-1-labeled DNA (pEGFP-C1). Ten pulses of 5 ms (1 Hz) were applied at 0.6 kV/cm (A) and 0.8 kV/cm (C). The time exposure of the camera was set at 1 s. (A and C) Fluorescence images 10 s after the pulsation. On these images, the membrane was drawn in white. (B and D) Fluorescence intensity profiles of the permeabilized areas were determined by quantifying the intensity values of the fluorescence at the membrane level along the white line drawn in A and C. a.u., arbitrary units. The thick line represents the fluorescence intensity at the membrane level of the A and C images. The thin line represents the level before pulsation. The black arrows on the left side indicate the direction of the electric field.
Figure 4.
Visualization of the labeled DNA with the electropermeabilized membrane after exhaustive photon reassignment. Cells in suspension were incubated with TOTO-1-labeled DNA (pEGFP-C1). Ten pulses of 5 ms (1 Hz) were applied at 0.8 kV/cm. The time exposure of the camera was set at 1 s per image. The optical sections were separated by 0.25 μm on the z axis. (A) Raw data image of the 25th plane after acquisition of 50 planes on the z axis. (B) Fluorescence profile along the membrane on image A (full line). (C) Fluorescence radial profile on image A (dotted line). (D) Restored image by exhaustive photon reassignment of the 25th plane. (E) Fluorescence profile along the membrane on image D (full line). (F) Fluorescence radial profile on image D (dotted line). The black arrows on the left side indicate the direction of the electric field.
Because the DNA–membrane complex formed blurred pictures with standard fluorescence microscopy, we deconvolved it by using the exhaustive photon reassignment method. The method allows one to obtain a deblurred quantitative visualization of fluorescence patterns. Deblurring clearly showed that the complexes were not distributed homogeneously in the permeabilized areas facing the cathode (Fig. 4 A, B, D, and E). The associated increase in fluorescence was found to be 8–9-fold instead of the 1.2–3.2-fold estimated previously. Fluorescence clearly is not homogeneous but is present into membrane-associated “spots” (Fig. 4 C and F). The size of these spots varies from 0.1 to 0.5 μm.
This increase in fluorescence can be explained by the accumulation of the labeled DNA at the cell membrane surface, which was permeabilized. A fair level of agreement was found with cells in suspension between the part of the membrane that was permeabilized on the cathode size, as shown by the PI emission, and the one in which the DNA–membrane complexes were found (Fig. 4B).
Effect of the Electric Field.
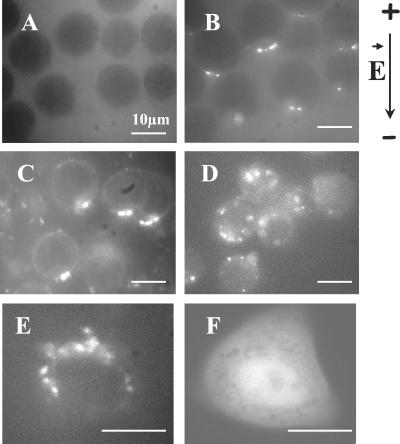
As shown for plated cells in Fig. 5, this increase in fluorescence appeared when cells were pulsed above 0.5 kV/cm, i.e., above an electric field value associated with the permeabilization of a cell on both the anode and cathode sites.
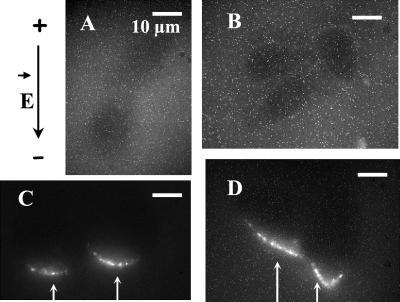
Figure 5.
Plasmid-DNA accumulation on the cell membrane as a function of the electric field intensity. Plated cells were incubated in the presence of TOTO-1-labeled DNA (pEGFP-C1). Ten pulses of 5 ms (1 Hz) were applied at 0.3 (A), 0.5 (B), 0.7 (C), and 0.9 kV/cm (D). The time exposure of the camera was set at 1 s. The black arrow on the left side indicates the direction of the electric field.
The plot histograms (not shown) indicated that the peak values as well as the integral depended greatly on the field strength. As shown in Fig. 5, no increase in fluorescence was detected up to 0.5 kV/cm. For higher field strengths, the peak intensity increased with the field intensity, whereas a widening of the peak brought a strong increase in the integral.
Effect of Pulse Duration.
No effect of pulsation on fluorescence was observed when the pulse duration was less than 1 ms (Fig. 6 A and B). Above 1 ms, fluorescent-plasmid accumulation was observed at the plasma-membrane level (Fig. 6 C and D).
Figure 6.
Plasmid-DNA accumulation on the cell membrane as a function of the pulse duration. Plated cells were incubated in the presence of TOTO-1-labeled DNA (pEGFP-C1). Ten pulses were applied at 0.7 kV/cm of 100 μs (A), 500 μs (B), 1 ms (C), and 5 ms (D) at 1-Hz frequency. Cells were observed 1 s after the pulsation. The time exposure of the camera was set at 1 s. The black arrow on the left side indicates the direction of the electric field.
The plot histograms (not shown) indicated that the peak value as well as the integral depended on pulse duration. No increase in fluorescence was detected when T was smaller than 1 ms. Higher pulse durations led to a sharp increase in peak intensity, and a slight broadening was present, resulting in an increase in the integral.
Inversion of the Electrode Polarity.
Stability of the interaction between plasmid molecules and the electropermeabilized membrane was shown by inverting the polarity of the electrodes between each pulse (Fig. 7). Indeed, if the increase in fluorescence was only caused by DNA accumulation under electrophoretic forces, then inversion of the polarity would lead to the disappearance of the fluorescence by removing the plasmid from the membrane to the bulk. As shown in Fig. 7B, DNA fluorescence was observed at both sides of the cells facing the electrodes. The fluorescence increase was not caused by the simple DNA accumulation on the permeabilized cell surface but was associated with a stable interaction between DNA and the permeabilized membrane.
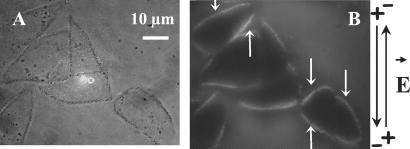
Figure 7.
Plasmid-DNA accumulation on the cell membrane with polarity inversion. Plated cells were incubated in the presence of TOTO-1-labeled DNA (pEGFP-C1). Eight pulses of 5 ms (1 Hz) were applied at 0.6 kV/cm with inversion of the polarity between each pulse. The time exposure of the camera was fixed at 1 s. (A) Light contrast images before the pulsation. The cell membrane is underlined to show the cell edge. (B) Fluorescence images 1 s after pulsation. White arrows indicate where the labeled DNA interacted with the areas of the membrane facing the two electrodes. The black arrows on the right side indicate the direction of the electric field.
Lifetime and Localization of Fluorescent DNA Spots.
The field-induced spots were still observed at the membrane level 10 min after the pulses. Electropermeabilized membrane had resealed at that time, as shown by the lack of PI penetration when added 10 min after (data not shown). Fluorescent spots were observed if DNA fluorescent dye (such as TOTO-1) was added 1 s after pulsing the cell-plasmid mixture. But if the DNA fluorescent dye addition was done 10 min after pulses, i.e., when the membrane has resealed, spots were not observed. DNA in the spots therefore was accessible to external dye only when the membrane was in an electropermeabilized state.
The long term migration of DNA within the cell was observed by using the covalently rhodamine-labeled DNA. Up to 10 min after pulses, the spots were localized at the membrane level on a restricted area (Fig. 8 A–C). After a 30-min postpulse incubation at 37°C, spots were always present, but they were distributed more randomly in the cytoplasm. Because a diffuse background of fluorescence was detected in the cytoplasm, DNA was apparently present in a free form and not any more limited to the spots (Fig. 8D). Twenty-four hours later, gene expression was fully active, GFP was detected everywhere in the cell (Fig. 8F), and spots were always present but localized around the nuclear envelope (Fig. 8E). No significant change in the spot size was detected (Fig. 8 B–E).
Discussion
Our results are direct observations of the early events leading to gene electrotransfer in mammalian cells. When no DNA was present, PI (or TOTO-1) fluorescence gave access to the electropermeabilization events. As we had observed already in populations (30) or during the very first millisecond after or during the pulse (31, 32), the field strength controlled the part of the cell surface brought to the permeabilized state. Our results clearly showed the critical role of pulse duration under our experimental conditions in which 10 successive pulses were applied. An increase in PI inflow was present with the increase in pulse duration. This had been observed already either in population or at the single-cell level. But the new aspect was that although with short pulses (0.1 ms) permeabilization affected only one side of the cell surface as predicted (29), the two opposite sides facing the electrodes were observed to give free exchange of PI when pulse duration was longer than 0.5 ms. This result suggested that the resting potential difference did not contribute anymore (29, 33). Differences in cell membrane behavior when treated by short or long pulses were noticed previously (34).
Fluorescent plasmids allowed us to monitor the interaction of nucleic acids with field-treated cells. Field strength was observed to have a critical role. The cell membrane must be permeabilized as shown by the PI assay for plasmid-membrane interaction to occur. Plasmids interacted with the cell surface side where they were accumulated by the field-associated electrophoretic drag as predicted previously (24). No free plasmid diffusion into the cytoplasm was detected as suggested elsewhere (19). No plasmid membrane interaction occurred if the nucleic acids were added after electropermeabilizing cells as proposed in refs. 2 and 3. A complex between the plasmid and the cell surface was present only when the pulse duration was at least 1 ms, whereas 0.5 ms was enough to trigger permeabilization on the cathode side where accumulation was present (Fig. 5). Pulse duration under the low field conditions played a critical role in the formation of the plasmid–cell complex as observed for in vitro gene transfer and expression (27, 28). Plasmids interacted with the membrane by forming localized spots, but electrophoretic DNA accumulation is not enough to bring spot formation. Negatively charged DNA molecules migrate when submitted to an electric field (24, 35). Migration of the DNA molecules is given by the following equation: L = μ × E × T, in which μ is the electrophoretic mobility 1.5 × 10−4 cm2/V (35), E is the electric field intensity, and T is the cumulated pulse duration. When 10 pulses of 1 ms were applied, fluorescent spots appeared for electric field values above 0.7 kV/cm. The corresponding value of L is 10.5 μm. Making the hypothesis that only electrophoretic forces are involved, such a value (10.5 μm) can be obtained by applying 10 pulses of 5 ms at 0.14 kV/cm. In fact, this pulsing condition did not lead to the formation of fluorescent spots on the cathode side. These observations are consistent with a process in which plasmids interact with the electropermeabilized part of the cell surface caused by their interfacial electrophoretic accumulation. This interpretation is supported further by the observation that the DNA content, shown by the local fluorescence emission and its spot size, is under the control of the field strength and pulse duration.
Pulse duration is a key factor in the formation of these spots on electropermeabilized membranes. It acted on the electrophoretic accumulation of the plasmid DNA (24). It controlled the density of defects in the electropermeabilized part of the cell surface as shown by the increase in the PI inflow. Finally, it was the key parameter for the step in which plasmid DNA was forced against the membrane, where a high conductance was present when the field was applied (36). These contributions of the pulse duration to the plasmid-membrane interaction has been illustrated already by a complex dependence of the gene expression on the pulse duration (24).
A piece of information provided by this study is that plasmid DNA was trapped in the electropermeabilized membrane (Fig. 2). This conclusion is supported by the observation that the inversion of the electric field polarity did not lead to a release of the fluorescent spots from the cell membrane (Fig. 5).
Spots are observed only in the electropermeabilized part of the membrane. They are formed within 1 s and can resist a field-polarity inversion. DNA within the spot remains stainable by external DNA fluorescent dye (such as TOTO-1) as long as the membrane is in the permeabilized state. This information is evidence that spots did not result from the dye-labeling of DNA but from DNA alone. Spots still are present 10 min after pulsation, when the electropermeabilized membrane has resealed to its native state, but therefore DNA remains protected against the external addition of DNA dye. These observations are in agreement with previous results showing that DNA remains accessible to external DNase up to 1 min after electric pulse application and then becomes protected (6).
The structure of the spots remains to be explained. Vesicle formation has been described by results on the electropulsation of a lecithin liposome-plasmid suspension (37). Macrovesicles were formed by the addition of macromolecules to prepulsed cells (38, 39). Our observations support a model in which the field pulse induces a complex between plasmids and cell membrane. Spots can result from DNA aggregates that are strongly associated to the membrane and internalized when membrane-resealing occurs. Because intercalating dyes cannot interact with compacted DNA, DNA aggregation is present in the spots but with no DNA compaction.
One parameter that must be taken into account is the confinement of DNA on a two-dimensional surface, because it is transiently induced by the electrophoretic forces in the present case. It has been shown that such an organization can induce the formation of a very clustered form of DNA (40). Studies on model membrane systems showed that DNA–lipid complexes could be formed under different conditions bringing DNA interfacial accumulation (41, 42). The local interfacial high concentration of DNA induced an electrostatic segregation of the cationic lipids and a compaction of the outer layer (16). The resulting mechanical stress resulted in formation of DNA containing vesicle-like structures. The electrophoretic accumulation of DNA against the permeabilized membrane may act by the associated increase in asymmetry of surface charges between the two sides of the cell membrane. This may help in the vesiculation that was observed on giant liposomes (43). All these studies have shown that vesicle formation was associated with the occurrence of a plasmid interfacial accumulation. A similar effect appeared to be present in our observations. Spots were formed only when pulses with long duration were applied, a condition in which the plasmid electrophoretic accumulation was large (24), but the pulse duration might be involved in the creation in a competent state of the outer layer of the cell membrane. The longer the pulse duration, the more competent the membrane would be to trap the DNA.
Because of the good correlation between visualization of DNA accumulation at the membrane and gene expression, these results are consistent with a multistep process of DNA transfer across the membrane: (i) when an electric field is applied, the plasma membrane is permeabilized on the sides of the cell facing the electrodes, and negatively charged DNAs migrate electrophoretically toward the plasma membrane on the cathode side, (ii) the accumulated plasmids interact with the membrane, (iii) plasmids are “trapped” in spots strongly associated with the plasma membrane, and (iv) a translocation of those plasmids to the cytoplasmic side of the plasma membrane occurs within a few minutes. Then DNA, either as spots or in a freed form, slowly diffuses toward the nucleus.
Acknowledgments
We thank Claire Millot for her help in cell culture and Christine Vossen, Cécile Faurie, and Bruno Gabriel for discussion. The English of the manuscript was checked by Mr. Robb. This work has been supported by Association Française Contre les Myopathies (to M.-P.R.), the Région Midi Pyrénées (to J.T.), and the European Union project “Cliniporator.”
Abbreviations
- PI
propidium iodide
- GFP
green fluorescent protein
References
- 1.Crystal R. Science. 1995;270:404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 2.Tsong T. Biophys J. 1991;60:297–306. doi: 10.1016/S0006-3495(91)82054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann E, Schaefer-Ridder M, Wang Y, Hofschneider P H. EMBO J. 1982;1:841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potter H. Methods in Enzymology. Vol. 217. New York: Academic; 1993. [DOI] [PubMed] [Google Scholar]
- 5.Orlowski S, Mir L M. Biochim Biophys Acta. 1993;1154:51–63. doi: 10.1016/0304-4157(93)90016-h. [DOI] [PubMed] [Google Scholar]
- 6.Eynard N, Rols M, Ganeva V, Galutzo B, Sabri N, Teissié J. Bioelectrochem Bioenerg. 1997;44:103–110. [Google Scholar]
- 7.Mir L M, Glass L F, Sersa G, Teissié J, Domenge C, Miklavcic D, Jaroszeski M J, Orlowski S, Reintgen D S, Rudolf Z, et al. Br J Cancer. 1998;77:2336–2342. doi: 10.1038/bjc.1998.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prausnitz M R, Bose V G, Langer R, Weaver J C. Proc Natl Acad Sci USA. 1993;90:10504–10508. doi: 10.1073/pnas.90.22.10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Titomirov A, Sukarev S, Kistanova E. Biochim Biophys Acta. 1991;1088:131–134. doi: 10.1016/0167-4781(91)90162-f. [DOI] [PubMed] [Google Scholar]
- 10.Heller R, Jaroszeski M, Atkin A, Moradpour D, Gilbert R, Wands J, Nicolau C. FEBS Lett. 1996;389:225–228. doi: 10.1016/0014-5793(96)00590-x. [DOI] [PubMed] [Google Scholar]
- 11.Rols M-P, Delteil C, Golzio M, Dumond P, Cros S, Teissié J. Nat Biotechnol. 1998;16:168–171. doi: 10.1038/nbt0298-168. [DOI] [PubMed] [Google Scholar]
- 12.Aihara H, Miyazaki J-i. Nat Biotechnol. 1998;16:867–870. doi: 10.1038/nbt0998-867. [DOI] [PubMed] [Google Scholar]
- 13.Mir L M, Bureau M F, Rangara R, Schwartz B, Scherman D. C R Acad Sci Ser III. 1998;321:893–899. doi: 10.1016/s0764-4469(99)80003-1. [DOI] [PubMed] [Google Scholar]
- 14.Muramatsu T, Nakamura A, Park H M. Int J Mol Med. 1998;1:55–62. doi: 10.3892/ijmm.1.1.55. [DOI] [PubMed] [Google Scholar]
- 15.Klenchin V. Biol Membr (Russia) 1994;7:1–16. [Google Scholar]
- 16.Hristova N I, Tsoneva I, Neumann E. FEBS Lett. 1997;415:81–86. doi: 10.1016/s0014-5793(97)01097-1. [DOI] [PubMed] [Google Scholar]
- 17.Xie T, Tsong T. Biophys J. 1993;65:1684–1689. doi: 10.1016/S0006-3495(93)81208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Gennes P G. Proc Natl Acad Sci USA. 1999;96:7262–7264. doi: 10.1073/pnas.96.13.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klenchin V A, Sukharev S I, Serov S M, Chernomordik L V, Chizmadzhev Yu A. Biophys J. 1991;60:804–811. doi: 10.1016/S0006-3495(91)82115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sukharev S I, Klenchin V A, Serov S M, Chernomordik L V, Chizmadzhev Yu A. Biophys J. 1992;63:1320–1327. doi: 10.1016/S0006-3495(92)81709-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eynard N, Sixou S, Duran N, Teissié J. Eur J Biochem. 1992;209:431–436. doi: 10.1111/j.1432-1033.1992.tb17306.x. [DOI] [PubMed] [Google Scholar]
- 22.Ganeva V, Galutzov B, Teissié J. Biochem Biophys Res Commun. 1995;214:825–832. doi: 10.1006/bbrc.1995.2361. [DOI] [PubMed] [Google Scholar]
- 23.Golzio M, Mora M P, Raynaud C, Delteil C, Teissié J, Rols M-P. Biophys J. 1998;74:3015–3022. doi: 10.1016/S0006-3495(98)78009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolf H, Rols M-P, Boldt E, Neumann E, Teissié J. Biophys J. 1994;66:524–531. doi: 10.1016/s0006-3495(94)80805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rols M-P, Coulet D, Teissié J. Eur J Biochem. 1992;206:115–121. doi: 10.1111/j.1432-1033.1992.tb16908.x. [DOI] [PubMed] [Google Scholar]
- 26.Rye H, Yue S, Wemmer D, Quesada M, Haugland R, Mathies R, Glazer A. Nucleic Acids Res. 1992;20:1803–1812. doi: 10.1093/nar/20.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hui S. Methods Mol Biol. 1995;55:29–40. doi: 10.1385/0-89603-328-7:29. [DOI] [PubMed] [Google Scholar]
- 28.Rols M-P, Teissié J. Biophys J. 1998;75:1415–1423. doi: 10.1016/S0006-3495(98)74060-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teissié J, Rols M-P. Biophys J. 1993;65:409–413. doi: 10.1016/S0006-3495(93)81052-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rols M-P, Teissié J. Biophys J. 1990;58:1089–1098. doi: 10.1016/S0006-3495(90)82451-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabriel B, Teissié J. Biophys J. 1997;73:2630–2637. doi: 10.1016/S0006-3495(97)78292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabriel B, Teissié J. Biophys J. 1999;76:2158–2165. doi: 10.1016/S0006-3495(99)77370-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tekle E, Astumian R D, Chock P B. Proc Natl Acad Sci USA. 1994;91:11512–11516. doi: 10.1073/pnas.91.24.11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akinlaja J, Sachs F. Biophys J. 1998;75:247–254. doi: 10.1016/S0006-3495(98)77511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann E, Werner E, Sprafke A, Krüger K. In: Colloid and Molecular Electro-Optics. Jennings B a S., editor. Bristol, U.K.: Institute of Physics; 1992. [Google Scholar]
- 36.Hibino M, Itoh H, Kinosita K., Jr Biophys J. 1993;64:1789–1800. doi: 10.1016/S0006-3495(93)81550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chernomordik L V, Sokolov A V, Budker V G. Biochim Biophys Acta. 1990;1024:179–183. doi: 10.1016/0005-2736(90)90222-a. [DOI] [PubMed] [Google Scholar]
- 38.Glogauer M, Lee W, McCulloch C A. Exp Cell Res. 1993;208:232–240. doi: 10.1006/excr.1993.1242. [DOI] [PubMed] [Google Scholar]
- 39.Rols M-P, Femenia P, Teissié J. Biochem Biophys Res Commun. 1995;208:26–35. doi: 10.1006/bbrc.1995.1300. [DOI] [PubMed] [Google Scholar]
- 40.Koltover I, Wagner K, Safinya C R. Proc Natl Acad Sci USA. 2000;97:14046–14051. doi: 10.1073/pnas.97.26.14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarahovsky T S, Khusainova R S, Gorelov A V, Nicolaeva T I, Deev A A, Dawson A K, Ivanitsky G R. FEBS Lett. 1996;390:133–136. doi: 10.1016/0014-5793(96)00643-6. [DOI] [PubMed] [Google Scholar]
- 42.Angelova M I, Tsoneva I. Chem Phys Lipids. 1999;101:123–137. doi: 10.1016/s0009-3084(99)00060-2. [DOI] [PubMed] [Google Scholar]
- 43.Mathivet L, Cribier S, Devaux P F. Biophys J. 1996;70:1112–1121. doi: 10.1016/S0006-3495(96)79693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]