Abstract
Molecular phylogenies challenge the view that bats belong to the superordinal group Archonta, which also includes primates, tree shrews, and flying lemurs. Some molecular studies also challenge microbat monophyly and instead support an alliance between megabats and representative rhinolophoid microbats from the families Rhinolophidae (horseshoe bats, Old World leaf-nosed bats) and Megadermatidae (false vampire bats). Another molecular study ostensibly contradicts these results and supports traditional microbat monophyly, inclusive of representative rhinolophoids from the family Nycteridae (slit-faced bats). Resolution of the microbat paraphyly/monophyly issue is essential for reconstructing the temporal sequence and deployment of morphological character state changes associated with flight and echolocation in bats. If microbats are paraphyletic, then laryngeal echolocation either evolved more than once in different microbats or was lost in megabats after evolving in the ancestor of all living bats. To examine these issues, we used a 7.1-kb nuclear data set for nine outgroups and twenty bats, including representatives of all rhinolophoid families. Phylogenetic analyses and statistical tests rejected both Archonta and microbat monophyly. Instead, bats are in the superorder Laurasiatheria and microbats are paraphyletic. Further, the superfamily Rhinolophoidea is polyphyletic. The rhinolophoid families Rhinolophidae and Megadermatidae belong to the suborder Yinpterochiroptera along with rhinopomatids and megabats. The rhinolophoid family Nycteridae belongs to the suborder Yangochiroptera along with vespertilionoids, noctilionoids, and emballonuroids. These results resolve the apparent conflict between previous molecular studies that sampled different rhinolophoid families. An important implication of rhinolophoid polyphyly is independent evolution of key anatomical innovations associated with the nasal-emission of echolocation pulses.
Keywords: bats‖Chiroptera‖echolocation‖Mammalia‖phylogeny
The recent history of bat systematics is rife with controversies. Pettigrew and colleagues (1, 2) challenged the systematics community with the “flying primate hypothesis,” which associates megabats with primates and dermopterans rather than with microbats. Recent morphological and molecular studies disagree with Pettigrew's hypothesis and support traditional bat monophyly (3–5). At the interordinal level, the conventional view based on morphology is that bats group in Archonta with dermopterans, primates, and scandentians (6, 7). Molecular studies reject this hypothesis and instead place bats in the superordinal clade Laurasiatheria, which also includes Carnivora, Cetartiodactyla, Eulipotyphla, Perissodactyla, and Pholidota (8–10).
Within Chiroptera, morphology supports the monophyly of living microbats, all of which possess complex laryngeal echolocation systems (11, 12). Recent DNA studies disagree with morphology and suggest that microbats are paraphyletic (13–16), with megadermatid (Macroderma, Megaderma) and rhinolophid (Rhinolophus, Hipposideros) representatives of the superfamily Rhinolophoidea associating with megabats instead of with other microbats (13–15); The name Yinpterochiroptera is suggested for this clade (table 1 in ref. 15). This result is ostensibly contradicted by other molecular studies. Murphy et al. (10) included a nycterid (Nycteris) rhinolophoid in their study and found robust support for microbat monophyly. Liu et al. (17) recovered microbat paraphyly in their molecular supertree, but with emballonurids rather than rhinolophoids as the sister-taxon to megabats.
Resolution of these conflicts is a prerequisite for understanding the evolution of echolocation and flight in mammals. If Chiroptera and Microchiroptera are both monophyletic, then flight evolved before echolocation and both features evolved only once in the evolutionary history of bats. If microbats are paraphyletic, then laryngeal echolocation either evolved twice in extant microbats or was lost in megabats after evolving in the common ancestor of Chiroptera. An implication of the latter scenario (i.e., loss of echolocation in megabats) is that the “flight-first,” “echolocation-first,” and “tandem-evolution” hypotheses of flight and echolocation all remain viable (15).
To date, DNA sequence studies arguing for microbat paraphyly have included only eight bats (14–15). All microbat superfamilies [sensu Simmons and Geisler (18), table 1] were represented in these studies, but potential systematic biases deriving from limited taxon sampling remain untested. We investigated the effects of increased taxon sampling on the microbat paraphyly hypothesis by using a 7.1-kb nuclear data set that included segments of protein-coding regions for twenty bats and nine outgroups.
Methods
Sequences and Taxa.
New protein-coding sequences for portions of A2AB (α-2B adrenergic receptor gene, 1.3 kb), exon 11 of BRCA1 (breast cancer susceptibility gene, 2.8 kb), RAG1 (recombination activating gene 1, 1.1 kb), RAG2 (recombination activating gene 2, 0.8 kb), and exon 28 of vWF (von Willebrand factor gene, 1.2 kb) were obtained as described (9, 14). Additional sequences are from Teeling et al. (14), Madsen et al. (9), and Springer et al. (15). Following Simmons and Geisler's (18) classification of bats (Table 1), our data set included four pteropodids (Cynopterus, Nyctimene, Pteropus, Rousettus), one rhinopomatid (Rhinopoma), two rhinolophids (Rhinolophus, Hipposideros), one megadermatid (Megaderma), two nycterids (Nycteris), two emballonurids (Emballonura, Taphozous), one noctilionid (Noctilio), one molossid (Tadarida,), one natalid (Natalus), two vespertilionids (Myotis, Rhogeessa), one antrozoid (Antrozous), and two phyllostomids (Desmodus, Tonatia). Outgroup taxa included one rodent, two euarchontans, and six laurasiatherians (9, 10). The A2AB sequences were aligned using CLUSTAL W (19). All other nuclear genes were aligned using EYEBALL sequence editor (20). The repeat regions in BRCA1 and A2AB were removed because of alignment ambiguity (9). GenBank accession numbers for all sequences included in this study, voucher information for bat samples, and DNA sequence alignments, are given in the supporting information, which is published on the PNAS web site, www.pnas.org.
Table 1.
Bat classification incorporated in this paper
| Simmons & Geisler (18) |
| Order Chiroptera |
| Suborder Megachiroptera |
| Family Pteropodidae (Pteropus, Cynopterus, Nyctimene, Rousettus) |
| Suborder Microchiroptera |
| Superfamily Emballonuridae |
| Family Emballonuridae (Emballonura, Taphozous) |
| Infraorder Yinochiroptera |
| Superfamily Rhinopomatoidea |
| Family Rhinopomatidae (Rhinopoma) |
| Superfamily Rhinolophoidea |
| Family Nycteridae (Nycteris) |
| Family Megadermatidae (Megaderma) |
| Family Rhinolophidae |
| Subfamily Hipposiderinae (Hipposideros) |
| Subfamily Rhinolophinae (Rhinolophus) |
| Infraorder Yangochiroptera |
| Superfamily Noctilionoidea |
| Family Noctilionidae (Noctilio) |
| Family Phyllostomidae (Tonatia, Desmodus) |
| Superfamily Nataloidea |
| Family Natalidae (Natalus) |
| Superfamily Molossoidea |
| Family Antrozoidae (Antrozous) |
| Family Molossidae (Tadarida) |
| Superfamily Vespertilionoidea |
| Family Vespertilionidae (Rhogeessa, Myotis) |
| New classification expanded from Springer et al. (15) |
| Order Chiroptera |
| Suborder Yinpterochiroptera |
| Superfamily Pteropodoidea |
| Family Pteropodidae (Pteropus, Cynopterus, Nyctimene, Rousettus) |
| Superfamily Rhinolophoidea |
| Family Rhinopomatidae (Rhinopoma) |
| Family Megadermatidae (Megaderma) |
| Family Rhinolophidae (Rhinolophus) |
| Subfamily Rhinolophinae (Rhinolophus) |
| Subfamily Hipposiderinae (Hipposideros) |
| Suborder Yangochiroptera |
| Family Nycteridae (Nycteris) incertae sedis |
| Superfamily Emballonuroidea |
| Family Emballonuridae (Emballonura, Taphozous) |
| Superfamily Noctilionoidea |
| Family Noctilionidae (Noctilio) |
| Family Phyllostomidae (Tonatia, Desmodus) |
| Superfamily Vespertilionoidea |
| Family Natalidae (Natalus) |
| Family Vespertilionidae (Antrozous*, Rhogeessa, Myotis) |
| Family Molossidae (Tadarida) |
In agreement with Koopman (ref. 11; see supporting information) our results place Antrozous in the Vespertilionidae.
Base Composition.
A χ2 test of homogeneity (21) was used to test the assumption of base-compositional homogeneity. Hutcheon et al. (13) suggested the possibility that differences in the percentage of A + T drive the rhinolophoid–megabat alliance. To investigate this potential bias, we used a Mann–Whitney U test to compare the percentage of A + T in the Yinpterochiroptera versus other microbats (i.e., Yangochiroptera).
Phylogenetic Analyses.
Phylogenetic analyses were performed on the concatenated data set (≈7.1 kb), with and without Tonatia present. Using De Queiroz's (22) method for evaluating data set incongruence with a support/conflict criterion of 90% bootstrap support, there were no conflicting nodes (see supporting information). In contrast, the partition homogeneity test (23) with four separate partitions [A2AB; BRCA1; RAG1 + RAG2, analyzed together due to their physical linkage (24); vWF] resulted in significant data set heterogeneity. Given these mixed results and because individual genes lack resolving power, we chose to combine our data. As discussed by Lyons-Weiler et al. (25) and Teeling et al. (14), it is important to investigate the effects of removing taxa that may introduce phylogenetic artifacts. Tonatia was excluded in a subset of the analyses because models of sequence evolution used in our analyses assume base-compositional stationarity. With the inclusion of Tonatia, there was statistically significant base-compositional heterogeneity (P < 0.0001). With the removal of Tonatia, base-compositional heterogeneity was not significant for the remaining 28 taxa (P = 0.1915). With the exception of Bayesian methods, all phylogenetic analyses were performed with PAUP 4.0 (21) and included maximum likelihood (ML), minimum evolution (ME), and maximum parsimony (MP). Bayesian analyses were carried out with MRBAYES 2.0 (26).
Modeltest (27) suggested the general time reversible (GTR) model of sequence evolution with an allowance for a gamma (Γ) distribution of rates and a proportion of invariant sites (I). ML analyses were performed with the GTR + Γ + I model as well as with the less complex HKY85 model, with and without an allowance for a Γ distribution of rates. The latter models were used to assess whether results of ML analyses were sensitive to the model of sequence evolution. With the exception of ML analyses with the GTR + Γ + I model, we used heuristic searches with tree-bisection and reconnection (TBR) branch swapping. ML bootstrap analyses with the GTR + Γ + I model used nearest-neighbor interchange branch swapping because of computational demands.
Settings for the GTR + Γ + I model of sequence evolution derived from Modeltest (26) were as follows: 29 taxa; R-matrix = (1.0951, 4.2318, 0.7229, 1.1534, 4.9283, 1.000); base frequencies = (0.2741, 0.2482, 0.2534, 0.2243); proportion of invariant sites = 0.1445; and shape parameter (α) of gamma distribution = 1.0585. For 28 taxa, settings were as follows: R-matrix = (1.0864, 4.2203, 0.6975, 1.1243, 4.9002, 1.000); base frequencies = (0.2772, 0.2458, 0.2504, 0.2267); proportion of invariant sites = 0.1463; and shape parameter of gamma distribution = 1.0382. For ML analyses with the HKY 85 model the transition to transversion ratios (ts/tv), estimated from the best maximum parsimony tree, were as follows: 29 taxa (2.33 w/Γ, 2.17 w/o Γ); 28 taxa (2.14 w/Γ, 2.29 w/o Γ). The α values for HKY85 analyses with a Γ distribution were 0.71 for 29 taxa and 0.69 for 28 taxa. In all ML analyses, starting trees were obtained via neighbor joining. Because of computational demands, all ML analyses (both HKY85 and GTR) that incorporated a Γ distribution of rates used the following taxonomic constraint: (((Tonatia, Desmodus), (Myotis, Antrozous, Rhogeessa), (Hipposideros, Rhinolophus), (Nycteris thebaica, Nycteris grandis), (Cynopterus, Pteropus, Nyctimene, Rousettus), Tadarida, Megaderma, Rhinopoma, Natalus, Noctilio, Taphozous, Emballonura), (cow, whale), pangolin, mole, cat, horse, human, mouse, flying lemur). All constrained clades received 100% bootstrap support from MP and ME analyses and are well supported by other studies (10, 12, 28).
ME analyses were performed with (i) ML distances corrected according to the GTR + Γ + I model of evolution and (ii) logdet distances. In MP analyses, nucleotide positions were unweighted and gaps were coded as missing data. Branch-swapping was TBR for both ME and MP. Starting trees were obtained by means of neighbor-joining for ME analyses. In MP analyses, we used stepwise addition with ten randomized input orders. Bootstrap analyses included 100 replications with ML and 500 replications with ME and MP.
MRBAYES (26) calculates Bayesian posterior probabilities by using a Metropolis-coupled, Markov chain Monte Carlo (MCMCMC) sampling approach. Bayesian analyses used the GTR + Γ + I model of sequence evolution. Starting trees were random, phylogenetic constraints were not used, four simultaneous Markov chains were run for 200,000 generations, burn-in values were set at 30,000 generations (based on empirical evaluation of likelihood values), and trees were sampled every 20 generations.
Statistical Tests.
Goldman et al. (29) suggested that statistical hypothesis testing in phylogenetics is inappropriate when the optimal tree(s) is compared a posteriori with other trees. We deem it appropriate to use the Kishino and Hasegawa (30) test in this paper because we compared a priori hypotheses. Microbat monophyly, megabats + rhinolophoids, and megabats + emballonurids have all been suggested before and independent of our sequence data (13, 17, 31). Similarly, rhinolophoid monophyly (28, 31) and rhinolophoid polyphyly, the latter with nycterids associating with yangochiropteran microbats rather than with other rhinolophoids,‡‡ have been suggested.
Monte Carlo Simulations.
Monte Carlo simulations, otherwise known as parametric bootstrapping or SOWH tests (29, 33), were used to determine whether Yinpterochiroptera and rhinolophoid nonmonophyly are the result of systematic biases and/or taxonomic sampling. Tonatia was excluded from simulations because of nonstationarity. To generate the simulated data sets, we first determined the best likelihood trees, including branch lengths, by using the constraints of microbat monophyly and rhinolophoid monophyly, respectively. One hundred simulated data sets (length = 7063 bp) were generated for each tree by using seq-gen (34) under the GTR + Γ + I model of sequence evolution, using the parameters described above for the 28-taxon data set. Simulated data sets were analyzed using ML to determine the differences in −ln likelihood between the best trees with and without microbat monophyly, and with and without rhinolophoid monophyly, respectively. We compared the results from our actual data set with the distribution of tree length differences from our simulated data sets (35).
Character Mapping.
Simmons and Geisler (18) reported 195 morphological characters for all 18 chiropteran families and two archontan outgroups (Dermoptera and Scandentia). MACCLADE (36) was used to map characters of interest in the context of the molecular tree presented in Fig. 1.
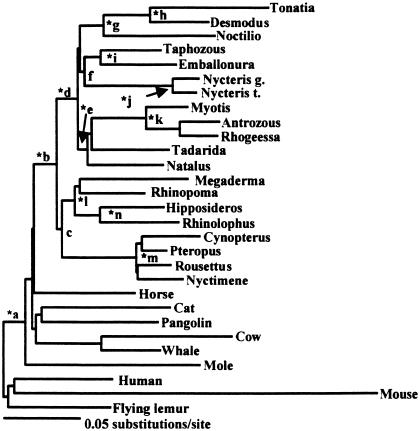
Figure 1.
The maximum likelihood tree (−ln likelihood = 62500.75) for the concatenated data set under the GTR + Γ + I model of sequence evolution. Asterisks indicate clades that were supported at or above the 90% bootstrap level in all analyses. Lowercase letters refer to clades that are discussed in the text.
Results
Base Composition.
There was no evidence for high A + T content in the pteropodids and rhinolophoids relative to other bat taxa. Pteropodids (49.7–50.0%), emballonurids (48.9–51.0%), and the natalid (49.8%) shared the highest percentages of A + T. Non-nycterid rhinolophoids (43.3–48.9%), vespertilionids (46.5–47.5%), and the phyllostomids (44.2%) had among the lowest percentages (see Fig. 3, which is published as supporting information on the PNAS web site). A Mann–Whitney nonparametric U test indicated no difference in the median percentage of A + T between the Yinpterochiroptera and Yangochiroptera (P = 0.4328).
Phylogenetic Analyses.
The concatenated data set, with or without Tonatia, provided robust bootstrap for Laurasiatheria (range = 93–100%; mean = 99%) and the posterior probability for this clade was 1.000 (Fig. 1, node a; Table 2). Bootstrap support for Archonta was 0% in all analyses and the posterior probability for this clade was 0.000 (Table 2). The monophyly of Chiroptera (Fig. 1, node b) received 100% bootstrap support in all analyses that did not constrain this clade, and the posterior probability for Chiroptera was 1.000.
Table 2.
Bootstrap support and posterior probabilities for selected phylogenetic hypotheses
| No. of taxa | Bootstrap support percentages
|
Bayesian posterior probabilities | |||||
|---|---|---|---|---|---|---|---|
| MP | ME logdet | ME-ML (GTR + Γ + I) | ML (HKY) | ML (HKY + Γ) | ML (GTR + Γ + I) | ||
| Laurasiatheria (node a, Fig. 1) versus Archonta | |||||||
| 29 | 95/0 | 100/0 | 100/0 | 100/0 | 100/0 | 100/0 | 1.000/0.000 |
| 28 | 93/0 | 97/0 | 100/0 | 100/0 | 100/0 | 100/0 | 1.000/0.000 |
| Yinpterochiroptera (node c, Fig. 1) versus microbat monophlyly | |||||||
| 29 | 80/10 | 94/6 | 75/24 | 97/1 | 99/0 | 93/0 | 1.000/0.000 |
| 28 | 82/13 | 91/7 | 88/12 | 95/0 | 99/1 | 93/1 | 1.000/0.000 |
| Rhinolophoid polyphyly versus rhinolophoid monophyly | |||||||
| 29 | 100/0 | 100/0 | 100/0 | 100/0 | 100/0 | 100/0 | 1.000/0.000 |
| 28 | 100/0 | 100/0 | 100/0 | 100/0 | 100/0 | 100/0 | 1.000/0.000 |
| Vespertilionids + molossids + natalids (node e, Fig. 1) | |||||||
| 29 | 99 | 100 | 99 | 100 | 100 | 100 | 1.000 |
| 28 | 98 | 97 | 100 | 100 | 100 | 100 | 1.000 |
| Rhinopoma + non-nycterid rhinolophoids (node l, Fig. 1) | |||||||
| 29 | 100 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 28 | 100 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| Noctilio + Phyllostomidae (node g, Fig. 1) | |||||||
| 29 | 100 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| 28 | 100 | 100 | 100 | 100 | 100 | 100 | 1.000 |
| Nycterids + emballonurids (node f, Fig. 1) | |||||||
| 29 | 73 | 89 | 75 | 98 | 96 | 98 | 1.000 |
| 28 | 89 | 78 | 77 | 100 | 94 | 100 | 1.000 |
ME logdet, minimum evolution with logdet distances; ME-ML, minimum evolution with ML-corrected distances; GTR, general time reversible model; Γ, gamma distribution of rates; I, allowance for invariant sites; HKY, Hasegawa–Kishino–Yano model of sequence evolution. All analyses were performed on concatenated data sets that included segments of A2AB, BRCA1, RAG1, RAG2, and vWF. Analyses were performed with Tonatia (29 taxa) and without Tonatia (28 taxa) because of base-compositional heterogeneity that results from the inclusion of sequences for this taxon.
Microbat monophyly was not well supported (range = 0–24%; mean = 6%; posterior probability = 0.000; Table 2). Instead, there was a well supported basal split between two groups of bats—Yinpterochiroptera and Yangochiroptera. Yinpterochiroptera (Fig. 1, node c; Table 2) includes Pteropodidae and the microbat families Rhinolophidae, Megadermatidae, and Rhinopomatidae; bootstrap support for this clade ranged from 80–99% (mean = 90%) and the posterior probability for this clade was 1.000. Yangochiroptera (Fig. 1, node d) includes all of the remaining microbats and was supported at the 100% level in all bootstrap analyses and with a posterior probability of 1.000. This phylogenetic arrangement for Chiroptera renders microbats paraphyletic and indicates that rhinolophoids are not monophyletic.
Analyses with 28 and 29 taxa never supported rhinolophoid monophyly (mean bootstrap support = 0%; posterior probability = 0.000; Table 2). Additional support for grouping nycterids in Yangochiroptera derives from a 15-bp deletion in BRCA1 (see Fig. 4, which is published as supporting information on the PNAS web site) that occurs in nycterids and all yangochiropterans; this deletion is absent in all yinpterochiropteran and outgroup taxa.
Within Yangochiroptera, there was strong support for an association of vespertilionids, molossids, and natalids (97–100%; mean = 99%; posterior probability = 1.000; Fig. 1, node e; Table 2). Also in clade d, Noctilio grouped with the phyllostomid microbats; this association received 100% bootstrap support and the posterior probability was 1.000 (Fig. 1, node g; Table 2). Nycterids and emballonurids associated together (range of bootstrap support = 73–100%; mean bootstrap support = 88%; posterior probability = 1.000; Fig. 1, node f; Table 2). Phyllostomid (Fig. 1, node h), emballonurid (Fig. 1, node i), nycterid (Fig. 1, node j), and vespertillionid (Fig. 1, node k) monophyly, respectively, were all supported at the 100% bootstrap level in all analyses and had posterior probabilities of 1.000.
Within Yinpterochiroptera (Fig. 1, node c), Rhinopoma grouped with the non-nycterid rhinolophoids with 100% bootstrap support and this arrangement had a posterior probability of 1.000 (Fig. 1, node l). Pteropodid monophyly (Fig. 1, node m) and Rhinolophinae + Hipposiderinae (Fig. 1, node n) were both supported at the 100% bootstrap level and had posterior probabilities of 1.000 in all analyses.
Statistical Tests.
Kishino-Hasewaga (ML) tests rejected the monophyly of microbats in favor of Yinpterochiroptera (P = 0.0374, 29 taxa; P = 0.0448, 28 taxa; see Table 6, which is published as supporting information on the PNAS web site). Liu et al.'s (17) hypothesis of emballonurids + megabats was also rejected (see Table 6), both in comparison to Yinpterochiroptera (P < 0.0001) and microbat monophyly (P < 0.0001). The monophyly of the Rhinolophoidea was rejected in favor of the association of the Nycteridae with Yangochiroptera (P < 0.0001; see Table 6).
Monte Carlo Simulations.
The results of Monte Carlo simulations provide no support for the possibility that systematic biases in ML are responsible for the failure of molecular data to recover microbat and rhinolophoid monophyly, respectively. Instead, the results of Monte Carlo simulations provide statistical support for rejecting these hypotheses. In both cases, the decrease in ln likelihood associated with the actual data set are statistically significant and fall outside of the “failure distribution” associated with systematic biases. In simulations that assumed microbat monophyly, likelihood correctly recovered this clade in 90/100 simulations. When microbat monophyly was not recovered among the best trees, the decrease in ln likelihood associated with forcing microbat monophyly never exceeded three. With the actual data set, the decrease in ln likelihood was 18 (see Table 6). When rhinolophoid monophyly was assumed, likelihood analyses correctly recovered this clade in all simulations.
Discussion
Laurasiatheria, Archonta, and Volitantia.
Archonta was originally proposed by Gregory (37) to include “menotyphlan” insectivores (i.e., tree shrews and elephant shrews), flying lemurs, primates, and bats. Morphologists subsequently removed elephant shrews from Archonta, leaving the arboreal orders Primates, Scandentia, Dermoptera, and Chiroptera (6). Even before the explosion of DNA data, the validity or “naturalness” of this superordinal group had been under scrutiny (38). Primates, tree shrews, and flying lemurs share derived features of the astragalocalcaneal complex. However, tarsal features uniting bats with other archontans are absent (39). Similarly, albumin and transferrin immunological data place tree shrews, flying lemurs, and primates into a clade to the exclusion of bats (40).
Given the absence of tarsal modifications that unite bats with other archontans, the primary rationale for including bats in Archonta is the suite of novel features that bats share with flying lemurs (37, 39, 41). Bats and flying lemurs share a patagium with two unique features not seen in other gliding mammals: a humeropatagialis muscle and interdigital patagia on the manus (41). Bats and flying lemurs also have a tendon locking mechanism (TLM) on their feet that allows them to hang without continuous contraction of the flexor muscles (41). Also, megabats (not microbats) and flying lemurs share features such as peculiar primate-like retinal projections to the superior colliculus (2). The assumption that the patagial and TLM features are synapomorphies and not homoplastic characters, as well as the similarities between flying lemurs and megabats, are the basis for the concept of Volitantia. According to this hypothesis, Chiroptera and Dermoptera are united together as sister taxa within Archonta. If Dermoptera and Chiroptera share a most recent common ancestor, the megabat condition must be primitive within Chiroptera. However, this is not apparent from the fossil record. The oldest known bat fossil (50 million years old), Icaronycteris index, is a microbat (18).
Molecular data do not support a sister-group relationship between bats and flying lemurs, nor do they provide any support for Archonta. Instead, bats are placed in Laurasiatheria (8). This result is corroborated by two recent molecular studies based on 19 different nuclear genes and three mitochondrial genes for representatives of all placental orders (9, 10). According to these findings, the living placental orders are divided into four groups: Xenarthra, Afrotheria, Laurasiatheria, and Euarchonta-Glires. Bats group in Laurasiatheria and are not the sister-taxon to Dermoptera. Similarily, complete mitochondrial genome studies provide high support for a clade resembling Laurasiatheria and inclusive of bats (42–44).
The dissociation of bats and flying lemurs on molecular trees mandates that presumed synapomorphies of Volitantia evolved independently in these two groups. Even morphological data challenge the Volitantia hypothesis. Beard (45) argued that the patagium evolved independently in bats and flying lemurs because early Tertiary members of Dermoptera lack this feature. Thiele et al. (46) contested the claim that megabats, primates, and flying lemurs share the same unique retinal projections. Chiroptera and Dermoptera share the unique ratchet TLM that allows members of these orders to hang with minimal energy expenditure (41). However, the dermopteran TLM differs from that of bats in having cartilage nodules and a more complex pattern of tendon division and insertion (41). We suggest that these differences reflect the independence of this evolutionary innovation.
Microbat Monophyly Versus Microbat Paraphyly.
Pettigrew's flying primate hypothesis stimulated a gestalt of scientific studies aimed at examining the question of chiropteran monophyly (1). Both hard- and soft-character morphological studies and molecular studies unanimously support the monophyly of Chiroptera (3, 5, 10, 12, 31, 44).
Initial investigations into chiropteran monophyly revealed that microbats may be paraphyletic, with members of the superfamily Rhinolophoidea grouping with the megabats as opposed to other microbats (13, 47). This finding was viewed with suspicion because all microbats use a sophisticated form of laryngeal echolocation not used by megabats. Further, cladistic analyses of morphological characters indicate that the suborder is monophyletic (18, 28, 31). In view of the morphological evidence, Hutcheon et al. (13) suggested that high levels of adenine and thymine in rhinolophoids and megabats may be responsible for the alliance of these taxa in DNA hybridization studies.
Teeling et al. (14) addressed the question of microbat monophyly by using four different nuclear genes (vWF, RAG1, RAG2, BRCA1) and three mitochondrial genes (12S-tRNA valine-16S) for eight bat species and four diverse outgroups. Although providing robust support for Yinpterochiroptera, the results of Teeling et al. (14) may have been affected by taxon representation. Springer et al. (15) added A2AB to the Teeling et al. (14) data set but did not expand taxonomic representation. The present results address the issue of taxonomic sampling by including twelve additional bats (representing five different families) and five additional outgroups. Even with increased taxon sampling, support for Yinpterochiroptera is high and microbats remain paraphyletic.
As with the DNA hybridization data, base compositional biases are an important consideration in evaluating the Yinpterochiroptera hypothesis. Our findings indicate that the percentage of A + T is not significantly different in Yinpterochiroptera and Yangochiroptera. Also in support of Yinpterochiroptera, Monte Carlo simulations refute the possibility that microbat paraphyly is a result of systematic biases associated with ML. It thus appears that Yinpterochiroptera and microbat paraphyly are not the result of poor taxon representation, base compositional biases, or systematic errors associated with phylogenetic methodology.
Morphological evidence does not agree with molecular evidence. Nevertheless, the Yinpterochiroptera hypothesis is not without support from morphology. All yinpterochiropteran families are characterized by two lower incisors on each side of the jaw; this character is convergently present in Tomopeatinae and Mormoopidae within Yangochiroptera.
The inability of morphological data to recover microbat paraphyly may be affected by outgroup choice. A recent comprehensive morphological examination of interfamilial relationships within Chiroptera (18) only used Scandentia and Dermoptera as outgroups. The authors (18) noted that ideal outgroups should comprise the sister-taxa to the ingroup because homoplasy may increase with time. Molecular data indicate that other laurasiatherians are bats' closest relatives. In particular, the placement of bats in Laurasiatheria rather than Archonta may affect the polarity of characters associated with vision. The well developed visual system in pteropodids, for example, is derived relative to most other laurasiatherians; in contrast, the pteropodid visual system is primitive among bats if Archonta is monophyletic.
Convergent Evolution of a Key Innovation in Rhinolophoid Echolocation.
In apparent conflict with previous molecular studies (13–15, 47), Murphy et al. (10) found that microbat monophyly was well supported in an analysis that included Nycteris thebaica as the sole representative of the superfamily Rhinolophoidea. Our results resolve this apparent conflict: microbats are paraphyletic but the superfamily Rhinolophoidea is not monophyletic. The family Nycteridae, which is traditionally grouped in the superfamily Rhinolophoidea (11, 18) along with (minimally) megadermatids and rhinolophids (18), instead has closer affinities with yangochiropteran microbats. Evidence for including Nycteridae in Yangochiroptera, rather than Rhinolophoidea, includes a 15-bp deletion in BRCA1 (see supporting information). Another version of microbat paraphyly was suggested by Liu et al. (17), who found that emballonurids grouped with megabats on their molecular supertree. Our analyses firmly reject this possibility. Also, fundamental errors in the supertree approach used by Liu et al. (17) cast doubt on their results (48).
The nonmonophyly of rhinolophoids, like microbat paraphyly, is in striking contrast to traditional taxonomy. Phylogenetic studies based on morphology provide strong support for the association of Nycteridae, Megadermatidae, and Rhinolophidae in the superfamily Rhinolophoidea (11, 12, 18, 31). All rhinolophoids, including Nycteridae, emit their echolocation calls through their nasal passages (49). Further, rhinolophoids have been placed in the infraorder Yinochiroptera along with the Craseonycteridae, Rhinopomatidae, and (variably) Emballonuridae (11, 12, 18, 31). All members have premaxillaries that are moveable in relation to the maxillaries (11, 12, 18).
From a molecular perspective, both rhinolophoid and yinochiropteran monophyly are untenable. The deployment of rhinolophoids into Yinpterochiroptera and Yangochiroptera implies that features of the chiropteran skull associated with the nasal emission of echolocation have more complex evolutionary histories than previously believed.
The skull of adult microchiropterans has two distinct morphotypes corresponding to the mode of echolocation pulse emission—oral or nasal. This dichotomy reflects developmental constraints associated with the co-opting of the nasopharynx as an acoustical organ in nasal-emitting species (49, 50). Pedersen suggested that the shift from the ancestral developmental path (oral axis) to the derived developmental path (nasal axis) is a “key innovation” in bat echolocation. The skull of oral-emitting bats exhibits a more linear shape, whereas the nasal-emitting skull is distinctive and is bent into a more angular shape to accommodate the position of the hard palate (51). This difference is established in utero as a result of ontogenetic shifts in the spatial accommodation of facial structures around the nasopharynx or orophyranx (49). Although rhinolophoids are restricted to the Old World, nasal emission is not restricted to the Old World and the independent evolution of this key innovation also occurs in members of the exclusively New World family Phyllostomidae. Molecular evidence discussed in this paper supports the view that convergent evolution of this key innovation has also occurred in the nycterid skull versus other rhinolophoid skulls. In support of this hypothesis, the resonating chambers characteristic of nycterids differ from other Old World nasal emitters as they are found outside of the bony nasal cavity (50).
Despite morphological evidence for rhinolophoid monophyly, there are also derived morphological features that exclude the Nycteridae from Rhinolophoidea and even from Yinochiroptera and Yinpterochiroptera. Pubic nipples are present in few bat taxa and may function as additional holdfasts for the young. Simmons (32) examined 1,723 individuals representing 206 species in 83 genera and found that pubic nipples were only present in the yinochiropteran families Rhinolophidae, Megadermatidae, Craseonycteridae, and Rhinopomatidae, but not in Nycteridae. The first costal cartilage is ossified and fused to the manubrium and to the first rib, forming a wing-like lateral process of the manubrium in all yinochiropteran families. In all other bat families, including Nycteridae, the first costal cartilage is not fused to either the manubrium or the first rib (18). Finally, all yinpterochiropteran families except Nycteridae are characterized by the presence of two lower incisors (18).
Supplementary Material
Acknowledgments
We thank Dr. David Jacobs, Dr. Peter Taylor, and Dr. Jon Russ for DNA samples. This work was sponsored by National Science Foundation Grant DEB-9903810 (to M.S.S.) and the Training and Mobility of Researchers program of the European Commission (M.J.S. and W.W.d.J.).
Abbreviations
- ML
maximum likelihood
- ME
maximum evolution
- MP
maximum parsimony
- GTR
general time reversible
Footnotes
References
- 1.Pettigrew J D. Science. 1986;231:1304–1306. doi: 10.1126/science.3945827. [DOI] [PubMed] [Google Scholar]
- 2.Pettigrew J D, Jamieson B G W, Robson S K, Hall L S, McAnally K I, Cooper H M. Philos Trans R Soc London B. 1989;325:489–559. doi: 10.1098/rstb.1989.0102. [DOI] [PubMed] [Google Scholar]
- 3.Simmons N B. Am Mus Novit. 1994;3103:1–54. [Google Scholar]
- 4.Kirsch J A W, Flannery T F, Springer M S, Lapointe F-J. Aust J Zool. 1995;43:395–428. [Google Scholar]
- 5.Van Den Bussche R, Baker R J, Huelsenbeck J P, Hillis D M. Mol Phylogenet Evol. 1998;13:408–416. doi: 10.1006/mpev.1998.0531. [DOI] [PubMed] [Google Scholar]
- 6.Novacek M J. Nature (London) 1992;356:121–125. doi: 10.1038/356121a0. [DOI] [PubMed] [Google Scholar]
- 7.Shoshani J, McKenna M C. Mol Phylogenet Evol. 1998;9:572–584. doi: 10.1006/mpev.1998.0520. [DOI] [PubMed] [Google Scholar]
- 8.Waddell P J, Okada N, Hasegawa M. Syst Biol. 1999;48:1–5. [PubMed] [Google Scholar]
- 9.Madsen O, Scally M, Douady C J, Kao D J, DeBry R W, Adkins R, Amrine H, Stanhope M J, deJong W W, Springer M S. Nature (London) 2001;409:614–617. doi: 10.1038/35054544. [DOI] [PubMed] [Google Scholar]
- 10.Murphy W J, Eizirk E, Johnson W E, Zhang Y P, Ryder O A, O'Brien S J. Nature (London) 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 11.Koopman K F. Handb Zool. 1994;8:1–217. [Google Scholar]
- 12.Simmons N. In: Bats: Phylogeny, Morphology, Echolocation and Conservation. Kunz T H, Racey P A, editors. Washington, DC: Smithsonian Inst. Press; 1998. pp. 3–26. [Google Scholar]
- 13.Hutcheon J M, Kirsh J A W, Pettigrew J D. Philosos Trans R Soc London B. 1998;353:607–617. doi: 10.1098/rstb.1998.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teeling E C, Scally M, Kao D J, Romagnoli M L, Springer M S, Stanhope M J. Nature (London) 2000;403:188–192. doi: 10.1038/35003188. [DOI] [PubMed] [Google Scholar]
- 15.Springer M S, Teeling E C, Madsen O, Stanhope M J, de Jong W W. Proc Natl Acad Sci USA. 2001;98:6241–6246. doi: 10.1073/pnas.111551998. . (First Published May 15, 2001; 10.1073/pnas.111551998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker R J, Longmire J L, Maltbie M, Hamilton M J, Van Den Bussche R. Syst Biol. 1997;46:579–589. doi: 10.1093/sysbio/46.4.579. [DOI] [PubMed] [Google Scholar]
- 17.Liu F-G R, Miyamoto M M, Freire N P, Ong P Q, Tennant M R, Young T S, Gugel K F. Science. 2001;291:1786–1789. doi: 10.1126/science.1056346. [DOI] [PubMed] [Google Scholar]
- 18.Simmons N B, Geisler J H. Bull Am Mus Nat Hist. 1998;235:1–182. [Google Scholar]
- 19.Thompson J D, Higgins G D, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cabot E L, Beckenbach A T. Comput Appl Biosci. 1989;5:233–234. doi: 10.1093/bioinformatics/5.3.233. [DOI] [PubMed] [Google Scholar]
- 21.Swofford D L. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 1998. , Version 4. [Google Scholar]
- 22.de Queiroz A. Syst Biol. 1993;42:368–372. [Google Scholar]
- 23.Farris J S, Källersjö M K, Kluge A G, Bult C. Cladistics. 1994;10:315–319. [Google Scholar]
- 24.Oettinger M A. Trends Genet. 1992;8:412–416. doi: 10.1016/0168-9525(92)90323-v. [DOI] [PubMed] [Google Scholar]
- 25.Lyons-Weiler J, Hoelzer G A, Tausch R J. Mol Biol Evol. 1996;13:749–757. doi: 10.1093/oxfordjournals.molbev.a025635. [DOI] [PubMed] [Google Scholar]
- 26.Huelsenbek J P. MRBAYES: Bayesian Inference of Phylogeny. Rochester, NY: Department of Biology, University of Rochester; 2000. [Google Scholar]
- 27.Posada D, Crandall K A. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 28.Koopman K F. Bat Res News. 1983;24:1–2. [Google Scholar]
- 29.Goldman N, Anderson J P, Rodrigo A G. Syst Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- 30.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 31.Simmons N B. In: Ontogeny, Functional Ecology, and Evolution of Bats. Adams R A, Pedersen S C, editors. London: Cambridge Univ. Press; 2000. pp. 9–58. [Google Scholar]
- 32.Simmons N B. Am Mus Novit. 1993;3077:1–37. [Google Scholar]
- 33.Huelsenbeck J P, Hillis D M, Jones R. In: Molecular Zoology. Ferraris J D, Palumbi S R, editors. New York: Wiley–Liss; 1996. pp. 19–42. [Google Scholar]
- 34.Rambaut A, Grassly N C. Comput Appl Biosci. 1997;13:303–306. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- 35.Hillis D M, Mable B K, Moritz C. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 515–543. [Google Scholar]
- 36.Maddison W P, Maddison D R. MACLADE: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 37.Gregory W K. Bull Amer Mus Nat Hist. 1910;27:1–524. [Google Scholar]
- 38.MacPhee R D E. In: Primates and Their Relatives in Phylogenetic Perspective. MacPhee R D E, editor. New York: Plenum Press; 1993. pp. 363–373. [Google Scholar]
- 39.Szalay F S, Drawhorn G. In: Comparative Biology and Evolutionary Relationships of Tree Shrews. Luckett W P, editor. New York: Plenum; 1980. pp. 133–170. [Google Scholar]
- 40.Cronin J E, Sarich V M. In: Comparative Biology and Evolutionary Relationships of Tree Shrews. Luckett W P, editor. New York: Plenum Press; 1980. pp. 293–312. [Google Scholar]
- 41.Simmons N B, Quinn T H. J Mamm Evol. 1994;2:231–254. [Google Scholar]
- 42.Pumo D E, Finamore P E, Franek W R, Philips C J, Tarzami S, Balzarano D. J Mol Evol. 1998;47:709–717. doi: 10.1007/pl00006430. [DOI] [PubMed] [Google Scholar]
- 43.Mouchaty S K, Gullberg A, Janke A, Arnason U. Mol Biol Evol. 2000;17:60–67. doi: 10.1093/oxfordjournals.molbev.a026238. [DOI] [PubMed] [Google Scholar]
- 44.Nikaido M, Harada M, Cao Y, Hasegawa M, Okada N. J Mol Evol. 2000;51:318–328. doi: 10.1007/s002390010094. [DOI] [PubMed] [Google Scholar]
- 45.Beard K C. In: Mammal Phylogeny: Placentals. Szalay F S, Novacek M J, McKenna M C, editors. New York: Springer–Verlag; 1993. pp. 129–150. [Google Scholar]
- 46.Thiele A, Vogelsang M, Hoffman K-P. J Comp Neurol. 1991;314:671–683. doi: 10.1002/cne.903140404. [DOI] [PubMed] [Google Scholar]
- 47.Porter C A, Goodman M, Stanhope M. Mol Biol Evol. 1996;5:89–101. doi: 10.1006/mpev.1996.0008. [DOI] [PubMed] [Google Scholar]
- 48.Springer M S, de Jong W W. Science. 2001;291:1709–1711. doi: 10.1126/science.1059434. [DOI] [PubMed] [Google Scholar]
- 49.Pederson S C. J Morphol. 1993;218:85–98. doi: 10.1002/jmor.1052180107. [DOI] [PubMed] [Google Scholar]
- 50.Pederson S. J Morphol. 1995;225:107–123. doi: 10.1002/jmor.1052250109. [DOI] [PubMed] [Google Scholar]
- 51.Pedersen S C. In: Ontogeny, Functional Ecology, and Evolution of Bats. Adams R A, Pedersen S C, editors. London: Cambridge Univ. Press; 2000. pp. 174–213. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.