Abstract
The ability of Pseudomonas syringae pv. tomato DC3000 to parasitize tomato and Arabidopsis thaliana depends on genes activated by the HrpL alternative sigma factor. To support various functional genomic analyses of DC3000, and specifically, to identify genes involved in pathogenesis, we developed a draft sequence of DC3000 and used an iterative process involving computational and gene expression techniques to identify virulence-implicated genes downstream of HrpL-responsive promoters. Hypersensitive response and pathogenicity (Hrp) promoters are known to control genes encoding the Hrp (type III protein secretion) machinery and a few type III effector proteins in DC3000. This process involved (i) identification of 9 new virulence-implicated genes in the Hrp regulon by miniTn5gus mutagenesis, (ii) development of a hidden Markov model (HMM) trained with known and transposon-identified Hrp promoter sequences, (iii) HMM identification of promoters upstream of 12 additional virulence-implicated genes, and (iv) microarray and RNA blot analyses of the HrpL-dependent expression of a representative subset of these DC3000 genes. We found that the Hrp regulon encodes candidates for 4 additional type III secretion machinery accessory factors, homologs of the effector proteins HopPsyA, AvrPpiB1 (2 copies), AvrPpiC2, AvrPphD (2 copies), AvrPphE, AvrPphF, and AvrXv3, and genes associated with the production or metabolism of virulence factors unrelated to the Hrp type III secretion system, including syringomycin synthetase (SyrE), Nɛ-(indole-3-acetyl)-l-lysine synthetase (IaaL), and a subsidiary regulon controlling coronatine production. Additional candidate effector genes, hopPtoA2, hopPtoB2, and an avrRps4 homolog, were preceded by Hrp promoter-like sequences, but these had HMM expectation values of relatively low significance and were not detectably activated by HrpL.
Pseudomonas syringae pv. tomato DC3000 has emerged as an important model organism in molecular plant pathology because of its genetic tractability, its pathogenicity on both tomato and Arabidopsis thaliana, and its ability to deliver virulence effector proteins into host cells via a type III protein secretion system. To support worldwide functional genomic investigations of P. s. tomato DC3000, we are determining the complete sequence of the DC3000 genome. A draft sequence is now available (http://www.tigr.org/tdb/mdb/mdbinprogress.html), and we have initiated a genomewide investigation of potential virulence factors with a search for virulence-related genes in the same regulon as the type III secretion system.
Virulence effector proteins delivered to or into host cells by type III secretion systems are key factors in the pathogenicity of many bacteria, including animal pathogens in the genera Salmonella, Yersinia, Shigella, and Escherichia, and plant pathogens in the genera Pseudomonas, Erwinia, Xanthomonas, Ralstonia, and Pantoea (1). In plant pathogens, the type III secretion machinery is referred to as the hypersensitive response and pathogenicity (Hrp) system because secretion mutants typically lose their ability to elicit the defense-associated hypersensitive response in nonhost plants and to grow parasitically or be pathogenic in host plants (2). These phenotypes demonstrate the importance of the Hrp system in bacterium–plant interactions, and global identification of effectors will be important for understanding the pathogenesis of bacteria that use type III secretion systems. Unfortunately, several factors have hindered searches for type III effector genes. (i) Effectors are often redundant, with mutants having only subtle phenotypes. (ii) With few exceptions (3), motifs that can identify proteins as substrates for type III secretion have not been recognized (4). (iii) Many effectors show no similarity to known proteins. (iv) Some pathogens have multiple type III secretion systems, which deliver different sets of effectors (5). Thus, a complete inventory of type III effector genes is lacking for any pathogen, although it seems that pathogens such as Salmonella may have many such genes (6).
Plant pathogen type III effector proteins are mostly designated Avr (avirulence) or Hop (Hrp-dependent outer proteins), depending on whether their primary phenotype involves plant reaction or secretion behavior. Many effectors were initially discovered through their ability to betray the pathogen to the host R (resistance) gene surveillance system, thereby rendering the pathogen avirulent on a test plant (7). Over 25 effector genes have been identified by Avr or Hop phenotypes in various P. syringae pathovars and races (8, 9). The encoded effectors seem to determine both basic pathogenicity and host range, but the number of such proteins produced by any single strain has not been systematically investigated. P. s. tomato DC3000 is known to carry at least three avr genes, avrPto (10), avrPtoB (Y.-J. Kim and G.B.M., unpublished data), and avrE (11), with the latter being in the Hrp pathogenicity island along with five other poorly characterized candidate effector genes (9, 11).
Promoter-based screens provide a potentially facile method for identifying Hrp effector genes in P. syringae for several reasons. All known P. syringae effector genes are associated with “Hrp box” promoters and seem to be in the Hrp regulon along with the type III secretion machinery genes. Furthermore, P. syringae strains have only one type III secretion system and regulon (excluding the flagellar system), and Hrp promoters have been shown to be activated by the HrpL alternative sigma factor, a member of the extracytoplasmic factor (ECF) family of sigma factors (12).
The P. syringae Hrp system is activated in apoplast-mimicking minimal medium by a regulatory cascade that includes HrpR and HrpS (12–14), which are σ54 enhancer-binding proteins in the NtrC family, and HrpL, which has a σ54-dependent promoter and is expressed in an RpoN-dependent manner (15). Overexpression of HrpV represses hrp gene expression (16), as does deletion of the hrpA (Hrp pilus) gene (17). Little is known about other factors that act upstream of HrpR/S; however, constitutive expression of HrpL activates hrp gene expression in all known genetic backgrounds and culture conditions.
HrpL presumably interacts with Hrp box sequences, which were initially defined as 5′-GGAACCNA-N13–14CCACNNA-3′ (18–20). A 34-bp fragment cloned from the HrpZ operon promoter region of P. s. syringae 61, which encompasses this motif, possesses HrpL-dependent promoter activity in Escherichia coli (20). However, direct interaction of HrpL with Hrp box sequences and determination of essential nucleotides within Hrp box sequences by site-specific mutagenesis have not been reported. Furthermore, avr genes have been found with Hrp promoters containing variations in four positions in the canonical motif (19, 21), and we have little knowledge of the extent of the Hrp regulon beyond the Hrp secretion system and a handful of effector genes. Thus, methods for defining functional Hrp boxes and identifying them globally in a P. syringae genome would advance the search for effector genes and other factors in a regulon that is central to plant pathogenicity.
In this article, we describe a reporter transposon search for functional Hrp promoters controlling candidate virulence factors in the DC3000 genome and the use of these sequences to develop a hidden Markov model (HMM) for genomewide identification of potential Hrp promoters. We analyzed the first ORF downstream of these novel promoters for sequences with similarity to genes implicated in virulence. The HrpL-dependent expression of several of these genes was then tested by microarray and RNA blot analyses. These analyses confirmed the utility of the HMM search and revealed that the Hrp regulon includes at least 17 candidate effectors and other virulence factors apparently unrelated to the Hrp system, including the Cor (phytotoxin coronatine biosynthesis) regulon.
Materials and Methods
Strains and Media.
Conditions for routine culturing of E. coli and P. syringae have been described (22). AB medium (23) supplemented with 10 mM citrate was used for the transposon mutagenesis and gene expression analyses. Antibiotics were used at the following concentrations (μg/ml): kanamycin, 50; rifampicin, 100; chloramphenicol 20; and tetracycline, 20.
Draft Sequence of the P. s. tomato DC3000 Genome.
High molecular weight DNA (genomic and plasmid) was isolated from P. s. tomato DC3000 by using standard DNA isolation procedures (24). The DNA was mechanically sheared by nebulization, ligated to BstXI adaptors, and size-selected on an agarose gel. Two shotgun libraries, a small insert library of 2–3 kbp and a larger insert library of 8–12 kbp, were constructed and sequenced on Applied Biosystems 3700 sequencers from both ends with BigDye terminators (Perkin–Elmer). Sequencing methods have been described (25) and will be explained in more detail when the genomic sequence is closed and published. A total of 73,744 sequences were generated and then assembled by using The Institute for Genomic Research assembler program (26) into 392 assemblies representing 6.55 Mb with an overall GC content of 59%. A pseudomolecule of the assemblies (5801 version) was constructed to assist in further bioinformatic analyses. ORFs were identified by using the glimmer algorithm (27).
Reporter Transposon Screen for HrpL-Activated Genes.
The construction of the reporter transposon mutagenesis system is described elsewhere (D.E.F., R.B.A., D. Bauer, D. Tscherne, and A.C., unpublished data). In brief, the hrp/hrc cluster was deleted from DC3000 by allelic exchange of a DNA fragment containing a CmR interposon flanked by exchangeable effector locus (EEL) and conserved effector locus (CEL) sequences that border hrpK and hrpR, to produce mutant CUCPB5114 (D.E.F., A. Ramos, R. Rapp, and A.C., unpublished data). Plasmid pCPP5032 was constructed by cloning hrpL under control of the nptII promoter in pRK415 (28). This TetR plasmid is rapidly lost from P. syringae in liquid media lacking tetracycline. Vector control pCPP5031 is pRK415 containing the nptII promoter fragment but not hrpL. A miniTn5gus transposon that can generate transcriptional fusions with uidA [β-glucuronidase (Gus)] was constructed and introduced into CUCPB5114(pCPP5032) in 5 independent triparental matings and plated on AB citrate agar medium containing 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-gluc), rifampicin, kanamycin, chloramphenicol, and tetracycline. Blue colonies were transferred to 96-well microtiter plates containing LB medium supplemented with the same antibiotics. Cells from grown cultures were transferred with a multipin replicator to identical 96-well plates with or without tetracycline, and after further growth, to AB citrate, X-gluc, rifampicin, kanamycin, chloramphenicol agar plates with and without tetracycline. Transposon insertion points were then determined in colonies that were more intensely blue on agar plates containing tetracycline after 5 days of growth by sequencing of genomic templates using an Epicentre Technologies (Madison, WI) MasterPure DNA Purification kit, BigDye terminator (Perkin–Elmer) chemistries, an Applied Biosystems 3100 sequencer (at the Boyce Thompson Institute), and primers that hybridized to transposon sequences.
Bioinformatic Techniques.
Routine sequence analyses were performed with Lasergene (DNAStar, Madison, WI). The HMM training set was comprised of 51 P. syringae hrp promoter sequences extending from 1 nucleotide 5′ of the −35 motif through 3 nucleotides 3′ of the −10 motif. A training set of promoter sequences lacking the 4 nucleotides outside of the −35 and −10 motifs was also analyzed. De novo alignment of these 51 training sequences was accomplished with the sequence alignment and modeling (sam) package (Ultrasparc binary distribution 3.2) on a Sun Ultrasparc 10 running Solaris 2.7 (29). An initial generic node was specified to reduce gaps: 0.05, 0.01, 0.00; 0.95, 0.99, 1; 0.00, 0.00, 0.00; 0.25, 0.25, 0.25, 0.25; 0.25, 0.25, 0.25, 0.25. A reverse-complement alignment was generated by using the BCM Search Launcher online (http://searchlauncher.bcm.tmc.edu/). The forward and reverse sam alignments were used in building HMMs with HMMER 2.1.1 models (compiled from source for Cygwin, run on a Dell Latitude LS) (30). The resulting HMMs were scanned across the P. syringae genome pseudomolecule 5801 and the GenBank Pseudomonas aeruginosa sequence. The models were calibrated with the following flags: –no. 5,000; –mean 1,000; and –SD 200. Searches were conducted by using a model built with the null base composition of the target genomes. A PERL script was used to compare the locations of putative ORFs from TIGR and those of the Hrp promoter-like sequences identified by the HMMER searches. HMMER training sequences and output histograms for the two alternative training sets are provided in Tables 3 and 4, which are published as supporting information on the PNAS web site, www.pnas.org. To produce the Hrp promoter logo of the HMM used for searching, hmmemit was used to generate 1,000 random sequences from the search profile and aligned with hmmalign. The figure was generated with the GENIO/logo RNA/DNA and Amino Acid Sequence Logos web server (http://genio.informatik.uni-stuttgart.de/GENIO/logo/).
RNA Preparation.
Total RNA was isolated from bacterial cells grown at room temperature in AB medium (23) supplemented with 10 mM sodium citrate and containing tetracycline to select for the plasmids pCPP5031 or pCPP5032. Overnight cultures were transferred to fresh medium and adjusted to OD600 = 0.4. After 6 h of growth, 30-ml cultures were centrifuged at 4,000 rpm for 10 min at room temperature, and total RNA was extracted (31). Genomic DNA was removed by digestion with RQ1 RNase-free DNase (Promega). The RNA was evaluated on denaturing agarose gels, and the concentration was determined by absorbance at 260 nm.
Microarray Analysis.
ORF-specific DNA fragments were amplified by PCR from DC3000 genomic DNA and printed onto amine-coated slides from Cell Associates (Houston). Each DNA sample was printed three times on each slide with a BioRobotics (Boston) Microgrid II Arrayer by using MicroSpot 2500 split pins. Slides were blocked according to the recommended protocol from Cell Associates. Of total RNA, 50–100 μg was used to synthesize the cDNA probes for microarray analysis. RNA was mixed with 3 μg of random hexamers (Invitrogen) in a total volume of 15 μl and incubated at 65°C for 10 min. Reactions were then placed on ice for 2 min, to which were added 3 μl of 1 mM FluoroLink Cy3- or Cy5-dUTP (Amersham Biosciences, Piscataway, NJ), 3 μl of 0.1 M DTT, 6 μl of 5X first-strand buffer, 0.6 μl of 50X dNTPs mix (25 mM dATP, dCTP, dGTP/10 mM dTTP), and 2 μl of Superscript II (GIBCO/BRL). Reactions were incubated at room temperature for 10 min, followed by 42°C for 110 min. RNA was hydrolyzed by adding 1.5 μl of 1 M NaOH at 65°C for 10 min followed by neutralizing with 1.5 μl of 1 M HCl. cDNA probes were purified by using a PCR purification kit (Qiagen, Valencia, CA) and were resuspended in 20 μl of hybridization buffer [5× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), 0.1% SDS, and 25% formamide]. Denatured probes (99°C, 2 min) were hybridized to slides at 60°C overnight in hybridization cassettes (Corning), after which slides were washed twice with 2× SSC, 0.1% SDS (60°C, 5 min), once with 2× SSC (room temperature, 5 min), and once with 0.2× SSC (room temperature, 5 min).
Microarray images were visualized by using a ScanArray 5000 (Packard), using laser and PMT settings of 100 and 90, respectively. Images were overlaid and quantified by using IMAGENE 4.1 software (BioDiscovery; Marina Del Rey, CA). Ratio data were extracted by using GENESIGHT 2.1 software (BioDiscovery). For these analyses, local background for each spot was corrected, and signals lower than 50 were flagged and eliminated. After flooring low signals to the value of 100, ratios of the overlaid images were calculated for individual spots. We used 16S rRNA, which was expressed to similar levels in both tested strains based on RNA blots, to normalize the data. Finally, all of the replicated data were combined, and mean ratio data and SDs were calculated for each ORF.
RNA Blots.
RNA blot analyses were performed as described (32). Of each RNA sample, 25 μg was resolved on 1.2% formaldehyde-agarose gels and transferred to Nylon membranes (Hybond-N+) by capillary blotting using 20× SSC. RNA was bound to the membrane by UV cross-linking. Probes were generated by PCR amplification from genomic DNA, using ORF-specific primers, and labeled with 32P-dATP by random priming with a DECAprime II kit (Ambion). Hybridization was performed in 5× SSC, 50% formamide, 0.1% sodium-lauroylsarcosine, 0.02% SDS, and 2% blocking reagent (Roche Molecular Biochemicals) at 42°C overnight. Membranes were then washed twice with 2× SSC/0.1% SDS for 15 min, twice with 1× SSC/0.1% SDS for 15 min, and once with 0.1× SSC/0.1% SDS for 15 min before exposure on a phosphor screen. Signals were detected and evaluated by using a Storm system (Molecular Dynamics).
Results
Reporter Transposon Screen for HrpL-Activated Genes.
To identify additional HrpL-dependent promoters in P. s. tomato DC3000, we used a mutant lacking the hrp/hrc gene cluster, an unstable plasmid constitutively expressing hrpL, and miniTn5gus, a reporter transposon that can generate transcriptional fusions expressing βglucuronidase. The 25-kb hrp/hrc cluster was deleted from DC3000 to remove hrpL and many genes already known to be expressed in a HrpL-dependent manner. The Δhrp/hrc mutant, CUCPB5114, harboring PnptII-hrpL plasmid pCPP5032, was mutagenized with miniTn5gus and then cultured under conditions in which replicate arrays of mutant colonies either retained (through selection for tetracycline resistance) or lost pCPP5032. Insertion sites in miniTn5gus mutants that exhibited HrpL-dependent β-glucuronidase expression were determined by sequence analyses of genomic templates with a primer that hybridized to the transposon.
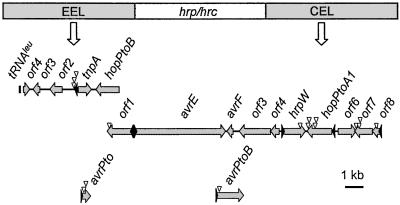
Of 5,184 mutants examined, 71 were identified as more intensely blue on plates containing tetracycline. Among these, 3 insertions were in pCPP5032, 53 were downstream of 18 different Hrp boxes, and 15 were unlinked to Hrp boxes but located in the DC3000 genome. MiniTn5gus insertions were observed downstream of 7 of the 8 Hrp boxes previously reported outside of the hrp/hrc cluster in DC3000, with multiple independent insertions in 4 of them (Fig. 1). Further analysis of the mutants revealed several novel genes that were linked to Hrp boxes and predicted to encode products homologous to known virulence-implicated factors: HopPsyA, AvrPpiC2, AvrPphE, pectin lyase (Ppr), Nɛ-(indole-3-acetyl)-l-lysine synthase (IaaL), CEL ORF1, and HrpA (Table 1). Additional insertions involved two virulence-implicated genes unlinked to a Hrp promoter—the coronatine biosynthesis genes cfa1 and cfa6 (hereafter, cfa1Pto and avrPphEPto, etc., to designate these P. s. tomato homologs). Finally, HrpL-activated insertions occurred in four ORFs that are downstream of Hrp boxes and predicted proteins with no homology to known virulence factors (see supporting information).
Figure 1.
Physical map of miniTn5gus insertions in the P. s. tomato CUCPB5114 Hrp pathogenicity island EEL and CEL regions and in avrPtoDC3000 and AvrPtoB. Transposon insertions were identified on the basis of the higher β-glucuronidase activity in CUCPB5114∷miniTn5gus(pCPP5032) mutants grown in media containing tetracycline, which prevents loss of the PnptII-hrpL plasmid. Arrowheads upstream of ORFs denote Hrp promoters. Independent Tn5gus insertions are indicated by triangles.
Table 1.
Virulence-implicated P. s. tomato DC3000 ORFs identified by HrpL-dependent expression following miniTn5gus mutagenesis
| Designation* | GenBank accession no.** | Amino acid %identity‡‡ | blastpP value | Tn hits†† |
|---|---|---|---|---|
| Known ORFs | ||||
| EEL ORF2 | AF232004 | NA | NA | 3 |
| CEL ORF1 | AF232004 | NA | NA | 1 |
| HrpW | AF232004 | NA | NA | 1 |
| HopPtoA1† | AF232004 | NA | NA | 4 |
| CEL ORF7 | AF232004 | NA | NA | 3 |
| CEL ORF8 | AF232004 | NA | NA | 1 |
| AvrPtoDC3000‡ | L20425 | 97 | 3e-79 | 2 |
| AvrPtoB | AF141883 | 55 | 1e-140 | 1 |
| Newly found ORFs | ||||
| AvrPphEPto | U16817 | 67 | 1e-117 | 1 |
| AvrPpiC2Pto | AJ277496 | 99 | 1e-149 | 1 |
| HopPsyAPto§ | L14926 | 52 | 9e-93 | 2 |
| Pectin lyase-related | AF027868 | 43 | 1e-54 | 1 |
| IaaLPto¶ | M35373 | 91 | 0 | 1 |
| CEL ORF1-related | AE005994 | 40 | 4e-72 | 1 |
| HrpA-related‖ | AF232004 | 67 | 5e-07 | 2 |
| Cfa1Pto | JC5745 | 98 | 2e-31 | 1 |
| Cfa6Pto | AF098795 | 89 | 0 | 1 |
Nucleotide sequences of ORFs and 5′ regions are provided in supporting information.
HopPtoA1 (CEL ORF5) is secreted in an Hrp-dependent manner (J. Badel and A.C., unpublished data).
The subscript is used to distinguish this allele from that of P. s. tomato JL1065.
HopPsyAPto is given in the table, although the putative chaperone gene is promoter proximal.
The miniTn5gus insertion was in iaaLPto; the upstream promoter-proximal ORF encoded a putative MATE efflux transport and binding protein.
The C-terminal 43 amino acids of HrpA and the HrpA-related protein are highly similar.
GenBank accession no. AF232004 is for DC3000 sequences, all others are for homologs originally found in another P. syringae pathovar or, in the case of AvrPto, another strain of P. s. tomato.
Amino acid identity of newly found ORF to most similar protein in GenBank, based on blastp using filter for low-complexity regions; NA, not applicable.
Tn hits represent independent insertions of miniTn5gus.
Development of a HMM to Efficiently Identify Potential Hrp Promoters in P. syringae Genomes.
Analysis of the new transposon-identified Hrp promoter sequences revealed novel variations in the canonical sequence for avrPphEPto (−35 GGCACC) and iaaLPto (−10 TCACnnA). The existence of multiple variations in functional Hrp boxes prompted us to use an HMM to find additional Hrp promoters in the DC3000 genome (Fig. 2). The training set was comprised of the −35, spacer, and −10 regions and bordering nucleotides from 33 promoter regions located upstream of hrp/avr genes previously reported to be functional in P. syringae and from 18 promoter regions identified in DC3000 by miniTn5gus mutagenesis. The ORFs immediately downstream of all candidate Hrp promoters were analyzed by blast searches for any ORFs that had similarity to known virulence-implicated factors. This analysis yielded a number of additional candidate virulence factors: CorR (a two-component regulator of coronatine biosynthesis) and homologs of AvrPphD (2 copies), AvrPphF, AvrPpiB1 (2 copies), AvrRps4, AvrXv3, HopPtoA, HopPtoB, HrpW, and SyrE (syringomycin synthetase) (Table 2). Independently, all putative ORFs in the current assembly of the genome were identified with glimmer (27) and searched with blastp. Also, the sequences of published phytopathogen Avr/Hop proteins were used to search the DC3000 genome with blastx. Neither analysis yielded additional avr/hop homologs.
Figure 2.
A sequence logo representing the Hrp promoter HMM used for searching the P. s. tomato DC3000 genome (51). The vertical axis is information content in bits. The height of a nucleotide reflects its representation in the sequence at that position. Nucleotides are displayed upside-down in positions where they occur less frequently than 25%. The expected frequency of nucleotides is listed below each position. This figure represents only the expected nucleotide frequencies at each position, but not the underlying transition and insertion/deletion probabilities. Positions of relaxed insertion probabilities are indicated with an “i.”
Table 2.
Microarray analysis of the HrpL-dependent expression of virulence-implicated ORFs that were previously shown by miniTn5gus to be HrpL-activated or were newly identified by HMM searching of the P. s. tomato DC3000 genome for Hrp promoter-like sequences
| Designation* | GenBank accession no.‡ | Amino acid %identity | blastpP value | HMM E value | Microarray signal ratio§ |
|---|---|---|---|---|---|
| Known ORFs | |||||
| AvrPtoDC3000 | See Table 1 | NA | NA | 2.0e-8 | 158 ± 57 |
| AvrPtoB | ‘ | ‘ | ‘ | 7.7e-8 | 26 ± 13 |
| CEL ORF7 | ‘ | ‘ | ‘ | 5.7e-6 | 4 ± 2 |
| HrpW | ‘ | ‘ | ‘ | 6.1e-6 | 5 ± 4 |
| HopPsyAPto | ‘ | ‘ | ‘ | 1.0e-5 | 11 ± 9 |
| Pectin lyase-related | ‘ | ‘ | ‘ | 2.1e-5 | 17 ± 9 |
| IaaL | ‘ | ‘ | ‘ | 2.9e-5 | 31 ± 17 |
| HrpA-related | ‘ | ‘ | ‘ | 4.5e-5 | 5 ± 1 |
| AvrPphEPto | ‘ | ‘ | ‘ | 2.5e-4 | 5 ± 2 |
| Cfa1Pto | ‘ | ‘ | ‘ | NA | 29 ± 11 |
| Cfa6Pto | ‘ | ‘ | ‘ | NA | 28 ± 7 |
| ORFs newly found by HMM | |||||
| SyrEPto | AF047828 | 43 | 0 | 9.8e-7 | NT |
| AvrPphFPto | AF231452 | 51 | 3e-36 | 1.7e-6 | 3 ± 2 |
| HrpW-related | AF232004 | 44 | 2e-4 | 1.9e-6 | 14 ± 7 |
| AvrPphD1Pto | AJ277494 | 89 | 0 | 1.9e-6 | 30 ± 17 |
| AvrXv3Pto | AF190120 | 27 | 7e-12 | 3.4e-6 | ND |
| AvrPpiB11Pto | X84843 | 100 | 1e-152 | 7.8e-6 | 11 ± 9 |
| AvrPpiB12Pto | X84843 | 100 | 1e-150 | 7.8e-6 | 10 ± 6 |
| AvrPphD2Pto | AJ277494 | 53 | 2e-44 | 3.0e-5 | 27 ± 11 |
| HopPtoA2 | AF232004 | 74 | 1e-177 | 2.8e-4 | ND |
| HopPtoB2† | AF232004 | 2.6e-3 | ND | ||
| aa 147–369 | 28 | 5e-10 | |||
| aa 498–735 | 29 | 2e-16 | |||
| AvrRps4Pto | L43559 | 72 | 2e-44 | 2.5e-2 | ND |
| CorRPto | AF400252 | 82 | 2e-72 | 6.6e-2 | ND |
| Reference genes | |||||
| 16S rRNA | 1 | ||||
| 23S rRNA | 1 | ||||
Nucleotide sequences of ORFs and 5′ regions are provided in supporting information.
HopPtoB1 (EEL ORF1) is secreted in an Hrp-dependent manner (T. Petnicki-Ocwieja and J.R.A., unpublished data); HopPtoB2 has duplicated regions of homology with HopPtoB1.
GenBank accession no. AF232004 is for DC3000 sequences; all others are for homologs originally found in other bacteria.
Microarray signal is the mean ratio and SD from 3 replicates of 2 independent experiments, calculated as described in Materials and Methods. AvrPpiB11Pto and AvrPpiB12Pto are 100% identical, therefore their signals cannot be distinguished. AvrPphD1Pto and AvrPphD2Pto are 62% identical. NT, not tested; ND, not detected.
Microarray Analysis of the Expression of Representative HrpL-Activated Genes.
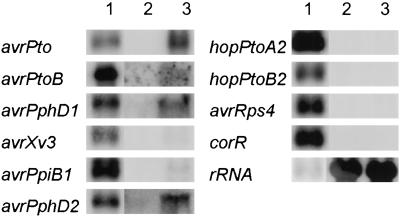
To test the ability of HMM expectation (E) values to predict functional Hrp promoters, we performed microarray analyses of 9 representative miniTn5gus-mutated ORFs and 11 representative virulence-implicated ORFs that were newly found by the hmmer program. ORF-specific primers were used to print the arrays and random primers to generate labeled cDNAs from Δhrp/hrc mutant CUCPB5114 cells carrying either PnptII-hrpL plasmid pCPP5032 or vector control pCPP5031. To corroborate the microarray results, RNA blotting was performed on 10 ORFs from similarly grown cultures (Fig. 3). The microarray experiments were in qualitative agreement with both the RNA blot and previous miniTn5gus expression experiments. These data indicate that Hrp promoter candidates with E values smaller (more significant) than 1e-4 are expressed at levels detected by our methods. However, within this group there was no apparent relationship between the magnitude of the E value and the level of expression. Furthermore, 1 of the 16 ORFs with an E value substantially lower than this threshold, AvrXv3 (4e-6), was expressed at a level that was detected only by RNA blot analysis (Table 2 and Fig. 3), indicating that significant E values do not always predict strong expression.
Figure 3.
RNA blot analysis of HrpL-dependent expression of representative virulence-implicated genes. Each well was loaded with 25 μg of total RNA isolated from CUCPB5114 cultures carrying either vector control pCPP5031 or PnptII-hrpL plasmid pCPP5032 (lanes 2 and 3, respectively). PCR-amplified internal fragments were used as probes; lane 1 in each case contains PCR product of the corresponding probe. AvrPpiB11Pto and AvrPpiB12Pto are 100% identical, therefore their signals cannot be distinguished.
Global Analyses of Hrp Promoter-Like Sequences in DC3000 and P. aeruginosa.
The current draft assembly of the DC3000 genome contains 48 Hrp promoter sequences with E values lower than 1e-4. To test the specificity of the HMM, we searched the genome of P. aeruginosa PAO1, which is another member of the fluorescent pseudomonad group of gamma Proteobacteria with a relatively high GC content (66.6%), but which lacks a Hrp secretion system (33). Only one Hrp promoter-like sequence with an E value below 1e-4 occurred in the PAO1 genome. The HMM search of DC3000 seemed to be comprehensive to the extent that it identified all of the Hrp promoters that were previously known or shown to be functional. Furthermore, all of the candidate effector genes independently identified by blast searches of the DC3000 genome, with the exception of hopPtoA2, hopPtoB2, avrPphEPto, and avrRps4Pto, are downstream of Hrp promoters with HMM values below 1e-4 (Table 2). corR also has a potential Hrp promoter with a marginal E value, and it is possible that Hrp promoter sequences in this group function at a low level or perhaps only in planta. In this context, it is noteworthy that the DC3000 genome has 78 candidate Hrp promoters with E values less than 1e-3 and 212 with E values less than 1e-2.
Discussion
By using an iterative process involving computational and gene expression data, we have developed an initial inventory of P. s. tomato DC3000 candidate type III secretion effector proteins, the presumed prime agents of host metabolic subversion. Our analyses have revealed that the Hrp regulon, the primary regulon known to be expressed during infection, seems to control at least 48 genes and a subsidiary regulon directing phytotoxin production. The iterative process focused on Hrp promoters in DC3000 and featured microarray experiments that tested the activity of novel Hrp promoters and demonstrated the validity of this approach for genomewide transcriptional profiling in DC3000. Our findings suggest that the P. syringae Hrp regulon is more complex than expected and encompasses more than type III secretion system genes and effector genes.
Identifying Hrp promoters in a genomic sequence is complicated by sequence degeneracy, with the known functional Hrp promoters possessing 6 variations in the −10 and −35 motifs and spacer regions possessing different lengths. HMMs are useful for characterizing heterogeneous DNA sequences (34), identifying promoter elements (35), and have been used in a similarly iterative manner in transcriptional profiling of Bacillus subtilis sporulation genes (36). The HMM we developed, which used 4 nucleotides flanking the −10 and −35 motifs, seemed to be both specific and comprehensive in finding strongly expressed Hrp promoters controlling virulence-related genes (Fig. 2). An alternative HMM, lacking these four nucleotides, had greatly diminished specificity (Table 4). Expanded profiling of DC3000 gene expression should enable further refinement of our HMM, which will be particularly useful in analyzing draft sequences of other strains of P. syringae as they become available.
Our global analysis of Hrp promoters in DC3000 is a pioneering step in addressing the question of how many type III effectors are involved in the pathogenicity of any single bacterial strain. In this study, we limited our analysis to candidate effector proteins that are similar to known Avr/Hop proteins. We also searched the genome for homologs of known avr/hop genes regardless of 5′ sequences. Our finding that all of the avr/hop genes identified in DC3000 are preceded by Hrp promoter-like sequences validates our approach of identifying additional effectors on the basis of their promoters. However, we cannot eliminate the possibility that novel, constitutively expressed effectors exist, which would be missed by our approach.
The global search for DC3000 ORFs that are similar to known Avr/Hop proteins yielded AvrXv3Pto, AvrPtoB, and the AvrPphD families as the only candidate effectors shared with Xanthomonas spp. (37) (Y.-J. Kim and G.B.M., unpublished data). Notably missing were members of the AvrBs2 and AvrBs3 families, which are widespread in Xanthomonas spp., or any members of the AvrRxv/YopJ family, which are found in genera as diverse as Salmonella, Yersinia, Xanthomonas, Erwinia, and Rhizobium, and have also been reported in another strain of P. syringae (i.e., P. s. syringae B728a) (1, 9). However, it is important to note that further searches after closure and annotation of the DC3000 genome may yield additional homologs of known effectors. In addition, genomic projects with other pathogens will enlarge the set of candidate effector genes available for comparison.
The majority of P. syringae avr genes that have been cloned on the basis of Avr phenotype have come from three pathovars that parasitize legumes—glycinea, phaseolicola, and pisi. P. s. tomato has a different host range and diverges from these other pathovars in rRNA comparisons (38). Nevertheless, of the 15 avr gene families found in these legume-attacking pathovars, 6 are also found in DC3000. This finding suggests the existence of a core set of P. syringae effectors in addition to those in the Hrp pathogenicity island CEL.
Our analyses revealed a striking apparent redundancy among the candidate effector protein genes hopPtoA, hopPtoB, avrPphDPto, and avrPpiB1Pto, as well as in three Hrp-related factors that may play a role in type III protein translocation across bacterial and plant cell walls. CEL ORF1 is a homolog of the E. coli MltD peptidoglycan hydrolase (9) and was found here to have a Hrp-induced paralog. In addition, we found an ORF with homology to the C terminus of the HrpA pilus subunit (39) and two novel proteins with features of harpins. The function of harpins, which are secreted in relative abundance by the Hrp system, is unclear. However, the P. syringae HrpZ protein can associate with plant cell walls (40), and HrpW has a pectate lyase domain that binds pectate (41), which suggests that harpins may promote effector translocation through an interaction with pectic polymers controlling wall porosity (42). Harpins may also function through direct interaction with the host plasma membrane (43). Harpins are characteristically glycine-rich cysteine-lacking proteins, and lack N-terminal signal peptides that would target them to the general secretory pathway. We have identified here two Hrp-induced proteins that share these properties. The HrpW-related protein contains a region with similarity to the harpin domain of HrpW. The pectin lyase-related protein shows strongest homology with a B. subtilis pectin lyase; however, unlike Ppr, the DC3000 homolog lacks cysteine and a classical N-terminal signal peptide, and like HrpW, it has a long N-terminal region (280 amino acids) showing no similarity to any known enzymes (44).
All of the candidate effector genes we analyzed seem to be expressed in a HrpL-dependent manner except for avrRps4Pto, hopPtoA2, and hopPtoB2 (avrXv3Pto was HrpL-activated, but relatively poorly). avrRps4 was cloned originally from Pseudomonas syringae pisi and renders recombinant DC3000 avirulent on most Arabidopsis accessions (45), and avrXv3 is from an Xanthomonas campestris pv. vesicatoria race that is avirulent on tomato carrying the Xv3 R gene (46). We may explore the possibility that poor expression of these two avr genes in DC3000 is a factor in the virulence of DC3000 on Arabidopsis and tomato carrying the cognate R genes.
The Hrp regulon also includes virulence-implicated factors that may act independently of the Hrp secretion system. These are involved in the biosynthesis or metabolism of the phytotoxins coronatine and syringomycin and the phytohormone indole acetic acid. However, the significance of the syrE syringomycin synthetase gene is unclear because P. s. tomato is reported not to produce this phytotoxin (47). Coronatine is an important virulence factor in DC3000 and may act early in pathogenesis when Hrp effectors are being delivered (48). We have observed that coronatine biosynthesis genes, which lack Hrp promoters and are activated by CorR in P. s. glycinea (49), are expressed in a HrpL-dependent manner in DC3000. However, the corR gene in DC3000 is downstream of a Hrp promoter-like sequence with an E value of relatively low significance and is not expressed at detectable levels in response to HrpL. Despite the uncertain role of CorR, it seems that the Hrp secretion system and coronatine production are coregulated and that HrpL is a master regulator of at least one subsidiary virulence regulon.
Another finding of our global analysis of Hrp promoter sequences is their frequent association with mobile genetic elements. P. syringae effector genes are commonly associated with such elements (50), and here we found transposase sequences downstream of several Hrp promoters, including the avrPphEPto promoter (Table 3). We also noted that the active exchangeable effector locus ORF2 Hrp promoter is more closely linked to the tnpA gene than to ORF2. These observations suggest that HrpL activation of transposases during infection could trigger genomic rearrangements, and one consequence of this could be the recruitment of new genes into the Hrp regulon through transposition of “portable” Hrp promoters.
A more complete picture of the HrpL regulon, the effector protein inventory, and other virulence systems are expected with the closure and annotation of the P. s. tomato DC3000 genome, the development of ORF-specific whole-genome microarrays, measurements of gene expression in planta, and tests for the secretion of candidate effector proteins. The tools we have developed here reveal the complexity of the Hrp regulon and provide a founding inventory of DC3000 Hrp effector genes. Moreover, these methods can now be applied to other virulence-related regulons to systematically reveal the web of differentially expressed genes underlying pathogenesis.
Supplementary Material
Acknowledgments
We thank Kristi Berry, Nadia Fedorova, Tamara Feldblyum, Dan Haft, Hoda Khouri, William Nelson, Jeremy Peterson, Dan Russell, Bao Tran, Lowell Umayam, Teresa Utterback, and Susan Van Aken for their efforts in the sequencing, closure, and bioinformatic analyses of the unfinished P. s. tomato DC3000 genome; and Niels Mache for providing the GENIO/logo web server. We thank the National Science Foundation Plant Genome Research Program for supporting this work through Cooperative Agreement DBI-0077622. We also acknowledge support from a Natural Sciences and Engineering Research Council of Canada Fellowship (to R.B.A.), United States Department of Agriculture/National Research Initiative Competitive Grants Program Postdoctoral Fellowship 98-35303-6662 (to D.E.F.), and from Specific Cooperative Agreement 1907-21000-008-03S to the Cornell Theory Center from the United States Department of Agriculture-Agricultural Research Service (to D.J.S.). Part of this research was conducted by using the resources of the Cornell Theory Center, which receives funding from Cornell University, New York State, federal agencies, and corporate partners.
Abbreviations
- Hrp
hypersensitive response and pathogenicity
- Avr
avirulence
- Hop
Hrp-dependent outer protein
- CEL
conserved effector locus
- HMM
hidden Markov model
- E
expectation (value)
- EEL
exchangeable effector locus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. J03681, J03682, L11336, M15194, M21965, L20425, Z21715, X84843, L14926, I16119, AF232006, M22219, AJ251482, X67808, AJ22647, M86401, AJ277494–AJ277496, AF231453, PSJ224433, L41863, and AF268940).
References
- 1.Galán J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 2.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miao E A, Miller S I. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lloyd S A, Norman M, Rosqvist R, Wolf-Watz H. Mol Microbiol. 2001;39:520–532. doi: 10.1046/j.1365-2958.2001.02271.x. [DOI] [PubMed] [Google Scholar]
- 5.Cornelis G R, Van Gijsegem F. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 6.Worley M J, Ching K H L, Heffron F. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]
- 7.Keen N T. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 8.Vivian A, Arnold D L. J Plant Pathol. 2000;82:163–178. doi: 10.1046/j.1364-3703.2000.00019.x. [DOI] [PubMed] [Google Scholar]
- 9.Alfano J R, Charkowski A O, Deng W-L, Badel J L, Petnicki-Ocwieja T, van Dijk K, Collmer A. Proc Natl Acad Sci USA. 2000;97:4856–4861. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ronald P C, Salmeron J M, Carland F M, Staskawicz B J. J Bacteriol. 1992;174:1604–1611. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorang J M, Keen N T. Mol Plant–Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm C, Aufsatz W, Panopoulos N J. Mol Microbiol. 1995;15:155–165. doi: 10.1111/j.1365-2958.1995.tb02230.x. [DOI] [PubMed] [Google Scholar]
- 14.Hutcheson S W, Bretz J, Sussan T, Jin S, Pak K. J Bacteriol. 2001;183:5589–5598. doi: 10.1128/JB.183.19.5589-5598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrickson E L, Guevera P, Ausubel F M. J Bacteriol. 2000;182:3508–3516. doi: 10.1128/jb.182.12.3508-3516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preston G, Deng W-L, Huang H-C, Collmer A. J Bacteriol. 1998;180:4532–4537. doi: 10.1128/jb.180.17.4532-4537.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei W, Plovanich-Jones A, Deng W-L, Collmer A, Huang H-C, He S Y. Proc Natl Acad Sci USA. 2000;97:2247–2252. doi: 10.1073/pnas.040570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen H, Keen N T. J Bacteriol. 1993;175:5916–5924. doi: 10.1128/jb.175.18.5916-5924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Innes R W, Bent A F, Kunkel B N, Bisgrove S R, Staskawicz B J. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao Y, Hutcheson S. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mansfield J, Jenner C, Hockenhull R, Bennett M A, Stewart R. Mol Plant–Microbe Interact. 1994;7:726–739. doi: 10.1094/mpmi-7-0726. [DOI] [PubMed] [Google Scholar]
- 22.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chilton M-D, Currier T, Farrand S, Benddich A J, Gordon M P, Nester E W. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausubel F M, Brent R, Kingston R, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 25.Tettelin H, Nelson K E, Paulsen I T, Eisen J A, Read T D, Peterson S, Heidelberg J, DeBoy R T, Haft D H, Dodson R J, et al. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 26.Sutton G G, White O, Adams M D, Kerlavage A R. Genome Seq Technol. 1995;1:9–19. [Google Scholar]
- 27.Salzberg S L, Delcher A L, Kasif S, White O. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 29.Hughey R, Krogh A. Comput Appl Biosci. 1996;12:95–107. doi: 10.1093/bioinformatics/12.2.95. [DOI] [PubMed] [Google Scholar]
- 30.Eddy S R. Curr Opin Struct Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 31.Aiba H, Adhya S, de Crombrugghe B. J Biol Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 33.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, et al. Nature (London) 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 34.Churchill G A. Bull Math Biol. 1989;51:79–94. doi: 10.1007/BF02458837. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen A G, Baldi P, Brunak S, Chauvin Y. Proc Int Conf Intell Syst Mol Biol. 1996;4:182–191. [PubMed] [Google Scholar]
- 36.Fawcett P, Eichenberger P, Losick R, Youngman P. Proc Natl Acad Sci USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noel L, Thieme F, Nennstiel D, Bonas U. Mol Microbiol. 2001;41:1271–1281. doi: 10.1046/j.1365-2958.2001.02567.x. [DOI] [PubMed] [Google Scholar]
- 38.Manceau C, Horvais A. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoyos M E, Stanley C M, He S Y, Pike S, Pu X-A, Novacky A. Mol Plant–Microbe Interact. 1996;9:608–616. [Google Scholar]
- 41.Charkowski A O, Alfano J R, Preston G, Yuan J, He S Y, Collmer A. J Bacteriol. 1998;180:5211–5217. doi: 10.1128/jb.180.19.5211-5217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baron-Epel O, Gharyal P K, Schindler M. Planta. 1988;175:389–395. doi: 10.1007/BF00396345. [DOI] [PubMed] [Google Scholar]
- 43.Lee J, Klusener B, Tsiamis G, Stevens C, Neyt C, Tampakaki A P, Panopoulos N J, Noller J, Weiler E W, Cornelis G R, et al. Proc Natl Acad Sci USA. 2001;98:289–294. doi: 10.1073/pnas.011265298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto T, Kawasaki H, Sakai T. FEBS Lett. 1996;398:269–273. doi: 10.1016/s0014-5793(96)01257-4. [DOI] [PubMed] [Google Scholar]
- 45.Hinsch M, Staskawicz B. Mol Plant–Microbe Interact. 1996;9:55–61. doi: 10.1094/mpmi-9-0055. [DOI] [PubMed] [Google Scholar]
- 46.Astua-Monge G, Minsavage G V, Stall R E, Davis M J, Bonas U, Jones J B. Mol Plant–Microbe Interact. 2000;13:911–921. doi: 10.1094/MPMI.2000.13.9.911. [DOI] [PubMed] [Google Scholar]
- 47.Volksch B, Weingart H. J Basic Microbiol. 1998;38:135–145. [PubMed] [Google Scholar]
- 48.Mittal S, Davis K R. Mol Plant–Microbe Interact. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- 49.Bender C L, Alarcon-Chaidez F, Gross D C. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J F, Charkowski A O, Alfano J R, Collmer A, Beer S V. Mol Plant–Microbe Interact. 1998;11:1247–1252. [Google Scholar]
- 51.Schneider T D, Stephens R M. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.