Abstract
Mouse models have provided significant insights into the molecular mechanisms of tumor suppressor gene function. Here we use mouse models of prostate carcinogenesis to demonstrate that the Nkx3.1 homeobox gene undergoes epigenetic inactivation through loss of protein expression. Loss of function of Nkx3.1 in mice cooperates with loss of function of the Pten tumor suppressor gene in cancer progression. This cooperativity results in the synergistic activation of Akt (protein kinase B), a key modulator of cell growth and survival. Our findings underscore the significance of interactions between tissue-specific regulators such as Nkx3.1 and broad-spectrum tumor suppressors such as Pten in contributing to the distinct phenotypes of different cancers.
Molecular investigations of the functions of oncogenes and tumor suppressor genes have been facilitated by analysis of mouse models (1, 2). With respect to prostate carcinogenesis, mouse models potentially can overcome inherent difficulties in studying the molecular genetics of this disease in humans (3–5). Human prostate cancer is characterized by the long latency between the appearance of precursor lesions, termed prostatic intraepithelial neoplasia (PIN), which can appear in men as early as in their twenties, and the manifestation of clinically detectable carcinomas that generally arise late in life. Thus, mouse models can provide insight into the molecular mechanisms involved in prostate cancer initiation and early steps of progression, which otherwise are nearly inaccessible in humans.
We have been using mouse models to investigate the individual and collaborative roles of candidate tumor suppressor genes for prostate carcinogenesis. Our work has focused on the Nkx3.1 homeobox gene because of its restricted expression in the prostate and essential role in prostate differentiation and function (6, 7). Loss of function of Nkx3.1 in mice results in prostatic epithelial hyperplasia and dysplasia as a correlate of aging (7), and Nkx3.1 heterozygotes display a similar although less severe phenotype than homozygotes, indicating haploinsufficiency (7). The relevance of NKX3.1 for human prostate cancer has been suggested by its localization to chromosomal region 8p21 (8, 9), which undergoes loss of heterozygosity (LOH) in ≈80% of prostate cancers (10–13). Notably, 8p21 LOH represents an early event in prostate carcinogenesis, because it occurs at high frequency in PIN (11, 12), suggesting that genes within this region are involved in cancer initiation. However, the role of NKX3.1 in human prostate carcinogenesis has been unclear, because it is not mutated in prostate cancer specimens (9). Thus, although one allele of NKX3.1 is presumed to be lost at high frequency in human prostate cancer because of its localization to 8p21, the remaining allele does not undergo mutational inactivation.
Pten encodes a lipid phosphatase that functions as an inhibitor of the phosphatidylinositol 3-kinase/Akt pathway (14–17), and its essential function is evident from the early embryonic lethality of homozygous mutants (18–20). In humans, PTEN maps to chromosomal region 10q23, which undergoes LOH at relatively advanced stages in many cancers (16, 17), suggesting that genes within this region are important for progression. PTEN represents a frequent target of mutational inactivation in human cancers (16, 17), and Pten heterozygous mutant mice develop cancers or dysplasias of multiple tissues including prostate (18, 20–22).
Our current investigations demonstrate that loss of function of Nkx3.1 and Pten cooperate in prostate carcinogenesis in mice. Nkx3.1;Pten compound mutant mice display an increased incidence of high-grade PIN (HGPIN)/early carcinoma lesions, which resemble early stages of human prostate carcinogenesis. A hallmark of these lesions in mutant mice as well as human prostate cancer is the loss of Nkx3.1 protein expression without LOH. Finally, our analysis reveals the unexpected convergence of Nkx3.1 and Pten in negative regulation of Akt activity.
Experimental Procedures
The Nkx3.1 and Pten mutant mice have been described (7, 20). Analyses were performed on a hybrid 129/SvImJ and C57BL/6J strain background by using virgin males. The primary histological analysis was performed on a nonblinded basis by R.D.C.; M.M.S. independently reviewed the histological data on a blinded basis. The scheme for histopathological grading will be described elsewhere (J.-H. Park, M.J.K., C.A.-S., M.M.S., and R.D.C., unpublished data). The human prostate tumor specimens were paraffin-embedded samples retrieved from the surgical pathology files at the University of California Davis Medical Center (generously supplied by Regina Gandour-Edwards). The histological diagnosis and Gleason grade were verified by independently R.D.C. and Regina Gandour-Edwards; no relationship between NKX3.1 protein loss and Gleason grade was observed.
Immunohistochemical analysis was performed on 4% paraformaldehyde-fixed cryosections (for anti-P-Akt) or formalin-fixed paraffin sections after antigen retrieval (for all other antibodies). The antibodies used were anti-cytokeratin 14 (monoclonal, BioGenex Laboratories, San Ramon, CA), anti-CD105/endoglin (monoclonal, Dako), anti-polycytokeratins (polyclonal, CK-P, Dako), anti-Ki67 antigen (polyclonal, NovoCastra Laboratories, Newcastle, U.K.), PTEN/MMAC1 (polyclonal, Ab-2, NeoMarkers, Fremont, CA), and anti-Akt and anti-P-Akt (Ser 473) (polyclonal, Cell Signaling Technology, Beverly, MA). Anti-mouse and anti-human Nkx3.1 antisera were generated by using as antigens the full-length proteins purified from Escherichia coli by hexahistidine affinity chromatography. Immunodetection was performed by using the Vector M.O.M. kit for monoclonal antibodies, the Vector Elite ABC kit for rabbit IgG for polyclonal antisera, and a Vector NovaRED kit for substrate detection. The data in Fig. 4 were obtained with polyclonal antisera against mouse or human Nkx3.1; similar results were obtained with other anti-Nkx3.1 antisera. Ki67-labeled nuclei were quantitated as reported in ref. 7. Western blot analysis was done with extracts prepared from anterior prostates by sonication in 20 mM Hepes (pH 7.4)/450 mM NaCl/0.2 mM EDTA/0.5 mM DTT/25% glycerol and protease inhibitor and phosphatase inhibitor cocktails (Sigma p2714 and p2850, respectively). Laser-capture microdissection (LCM) of immunostained sections was performed by using a PixCell apparatus (Arcturus Engineering, Mountain View, CA).
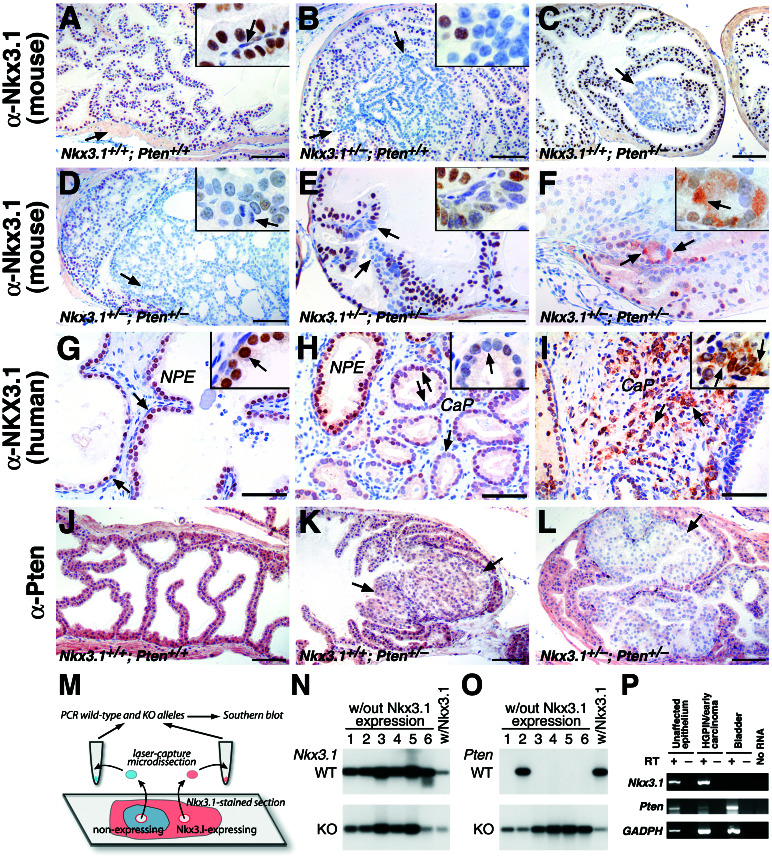
Figure 4.
Absence of Nkx3.1 protein expression in mouse and human prostate cancer. (A–F) Immunohistochemical analysis of the anterior prostate of Nkx3.1 and Pten mutants with an anti-mouse Nkx3.1 polyclonal antisera. (A) Uniform immunostaining of luminal epithelium with adjacent stroma unstained (arrow). (Inset) Nuclear staining of secretory cells and absence of staining of basal cells (arrow). (B) Absence of staining in LGPIN in a 12-month prostate. (Inset) Unstained and stained nuclei at the margin of the PIN (arrow). (C and D) Lack of staining in HGPIN in a 6-month prostate (C, arrow) and in an HGPIN/early carcinoma lesion of an 8-month prostate (D, arrow). Note the uniform staining of adjacent unaffected regions (C). (Inset) Unstained nuclei with atypia and mitotic figure (arrow). (E) Heterogeneity of staining (arrows) in a cluster of hyperplastic cells in an 8-month prostate. (Inset) Juxtaposition of stained, unstained, and lightly stained nuclei. (F) Example of cytoplasmic immunostaining (arrows and inset) at the margins of an HGPIN/early carcinoma lesion in a 12-month prostate. (G–I) Immunohistochemical analysis of human prostatectomy specimens using a polyclonal anti-human NKX3.1 antisera. (G) Immunostaining of normal prostate epithelium (NPE). Note absence of staining in basal cells (arrows) and stroma. (Inset) Nuclear staining of secretory cells (arrow). (H) Absent or heterogeneous staining in well differentiated cancer (CaP) compared with adjacent normal prostate epithelium. (Inset) Lack of staining in cancer cells (arrow). (I) Predominantly cytoplasmic immunostaining of a poorly differentiated cancer (arrows and Inset). (J–L) Immunohistochemical analysis of the anterior prostate using anti-Pten antisera. (J) Uniform staining in the epithelium and stroma. (K) Moderate reduction of Pten staining in a region of HGPIN in a 5-month prostate (arrows). (L) Absence of Pten staining in an HGPIN/early carcinoma lesion of a 12-month prostate. Note that in K and L the staining of adjacent unaffected regions is similar to wild type (J). (M–O) Analysis of allelic status of Nkx3.1 and Pten in HGPIN/early carcinoma lesions. (M) LCM was performed on Nkx3.1-immunostained sections to isolate genomic DNA from lesions (n = 20, Nkx3.1-nonexpressing) and adjacent unaffected regions (n = 8, Nkx3.1-expressing); representative data are shown. (N and O) Southern blot analysis to detect the wild-type (WT) alleles for Nkx3.1 and Pten; detection of the targeted allele (KO) serves as an internal control. (P) Reverse transcriptase–PCR analysis showing robust expression of Nkx3.1 and reduced expression of Pten in HGPIN/early carcinoma lesion of Nkx3.1+/−;Pten+/− mutant relative to the adjacent unaffected epithelium. cDNA was prepared in the absence (−) or presence (+) of reverse transcriptase (RT). Note that Nkx3.1 is not expressed in bladder; glyceraldehyde-3-phosphate dehydrogenase is the positive control.
Results
Loss of Function of Nkx3.1 and Pten Cooperate in Prostate Cancer Progression.
To examine whether loss of function of Nkx3.1 and Pten collaborate in prostate carcinogenesis, we intercrossed compound heterozygotes (Nkx3.1+/−;Pten+/−) in a mixed C57BL/6J-129/SvJ strain background to produce cohort groups comprised of all six viable genotypes (Fig. 1). Interestingly, our comparison of the prostatic phenotype of Nkx3.1 and Pten single mutant mice (Nkx3.1−/−;Pten+/+, Nkx3.1+/−;Pten+/+, and Nkx3.1+/+;Pten+/−) at 6 months of age revealed notable histological differences. At this age, the Pten mutant prostates displayed localized regions of severely dysplastic epithelium, unlike Nkx3.1 mutants in which the prostatic epithelium was more broadly hyperplastic but less severely dysplastic (Fig. 1 C–F, I, and J).
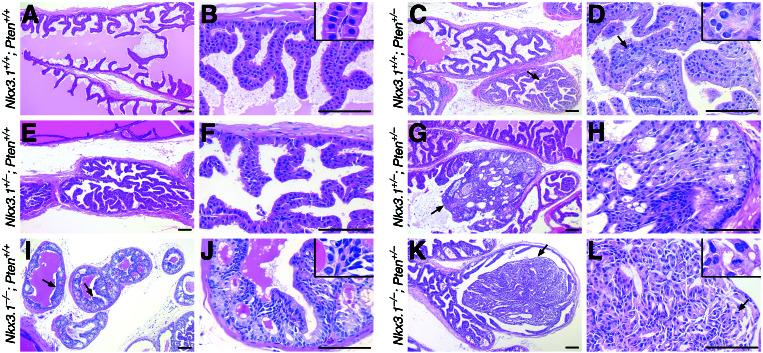
Figure 1.
HGPIN/early carcinoma lesions in Nkx3.1;Pten compound mutant prostates. Hematoxylin and eosin staining of anterior prostates of Nkx3.1;Pten compound mutants at 6 months is shown. (A and B) Well differentiated columnar epithelium. (Inset) High-power view. (C and D) Localized region of severely dysplastic epithelium (arrows) surrounded by well differentiated columnar epithelium. (Inset) Example of nuclear atypia. (E and F) Moderately hyperplastic epithelium. (G and H) HGPIN/early carcinoma lesion (arrow) surrounded by relatively unaffected epithelium. (I and J) Extensive low-grade PIN (LGPIN, arrows). (Inset) Example of nuclear atypia. (K and L) HGPIN/early carcinoma lesion surrounded by relatively unaffected epithelium. (Inset) Atypical nuclei with mitotic figure. (Scale bars, 100 microns.)
Our analysis of the compound mutants revealed that loss of function of Nkx3.1 and Pten displayed striking cooperativity in the anterior and dorsolateral prostatic lobes by 6 months of age (Figs. 1 and 2). In particular, Nkx3.1+/−;Pten+/− and Nkx3.1−/−;Pten+/− mice developed extensive multifocal lesions comprised of poorly differentiated cells with prominent and multiple nucleoli, an increased nuclear/cytoplasmic ratio, and frequent mitotic figures (Fig. 1 G, H, K, and L). These lesions were readily discernible as light-dense regions within the normally transparent prostatic ducts, usually filled the affected ducts, and were highly vascularized (Fig. 3 A–D, I, and J). Among the histopathological features that distinguished these lesions from the relatively normal (“unaffected”) adjacent epithelium were a marked elevation of wide-spectrum cytokeratins and an absence of basal epithelium (Fig. 3 E–H). In addition, the lesions displayed a high proliferative index (≈15%) relative to the adjacent unaffected epithelium, as indicated by the prevalence of mitotic figures and the abundance of Ki67-labeled nuclei (Figs. 1L and 3 K and L). Based on their undifferentiated cytology, microvascularization, and high proliferative index, we have defined these multifocal lesions as HGPIN/early carcinoma in accordance with terminology used to describe human prostate cancer precursors (23).
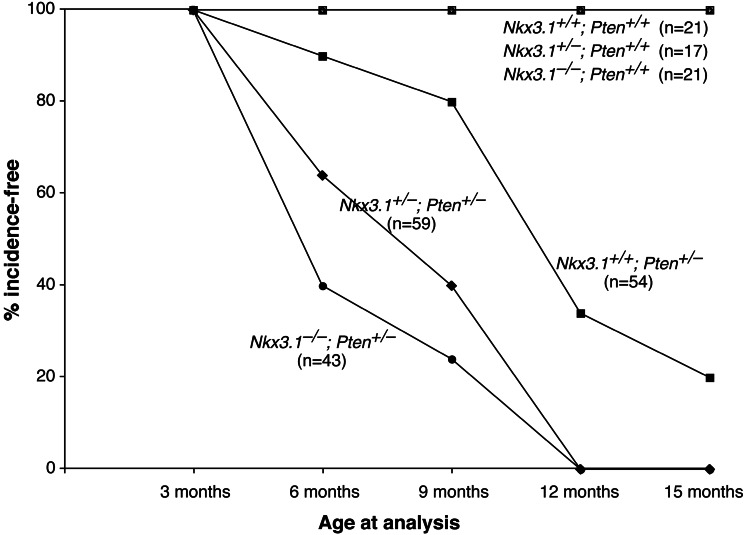
Figure 2.
Loss of function of Nkx3.1 and Pten cooperate in prostate cancer progression. A graphical representation of the percentage of mice lacking HGPIN/early carcinoma lesions in the dorsolateral prostate at the indicated ages is shown. Incidence corresponds to the occurrence of HGPIN/early carcinoma lesions, which is defined by using the histopathological and morphological criteria described in the text. The percentage incidence-free was calculated by dividing the number of unaffected mice by the total number of mice analyzed for each age group.
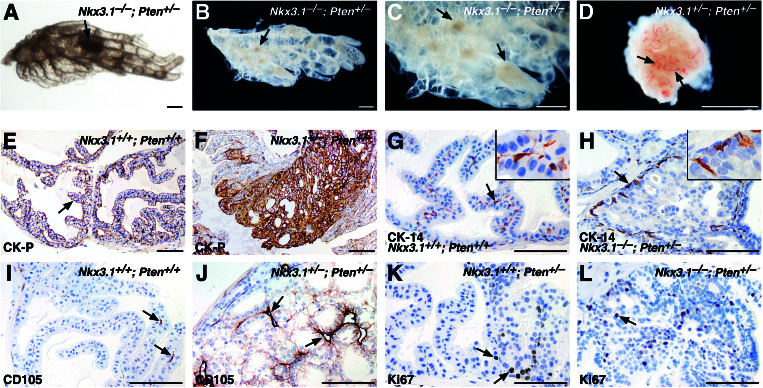
Figure 3.
Analysis of HGPIN/early carcinoma lesions. (A–D) Bright-field (A) and dark-field (B–D) images of anterior prostates at 6 months. (A–C) Whole mounts show light dense masses corresponding to the HGPIN/early carcinoma lesions (arrows). (D) Microdissected lesion containing numerous blood vessels (arrows). (Scale bars, 500 microns.) (E–L) Immunohistochemical analysis of the anterior prostate at 6 months. (E and F) Robust staining of wide-spectrum cytokeratins (CK-P) in HGPIN/early carcinoma lesions. (G and H) Immunodetection with anti-CK14 (arrows) shows an absence of basal cells in lesion. (Insets) High-power views. (I and J) Anti-CD105 (endoglin) shows increased microvascularization (J, arrows) of lesions. (K and L) Ki67 antibody shows increased cellular proliferation in lesions. (Scale bars, 100 microns.)
The cooperativity of loss of function of Nkx3.1 and Pten in prostate carcinogenesis was apparent from the increased incidence of HGPIN/early carcinoma lesions in the Nkx3.1−/−;Pten+/− and Nkx3.1+/−;Pten+/− prostates relative to Nkx3.1+/+;Pten+/− prostates (Fig. 2). Notably, at 6 months of age these lesions were observed in 60% of the Nkx3.1−/−;Pten+/− prostates and 36% of the Nkx3.1+/−;Pten+/− prostates, in contrast to only 10% of Nkx3.1+/+;Pten+/− prostates. By 12 months, all the Nkx3.1−/−;Pten+/− and Nkx3.1+/−;Pten+/− prostates displayed lesions, whereas 34% of Nkx3.1+/+;Pten+/− prostates remained lesion-free. When found, however, the HGPIN/early carcinoma lesions of the Nkx3.1+/+;Pten+/− prostates were indistinguishable morphologically and immunohistochemically from those of the Nkx3.1;Pten compound mutants. Therefore, the cooperativity of Nkx3.1 and Pten loss of function reflects the decreased latency of HGPIN/early carcinoma lesion formation in compound mutants. In contrast, we observed no increase in the occurrence of LGPIN in compound mutants (data not shown), suggesting that cooperativity of Nkx3.1 and Pten loss of function occurs at the level of cancer progression.
Aside from the development of these HGPIN/early carcinoma lesions, the compound mutants displayed no additional phenotypes compared with the single mutants. In particular, the compound mutants displayed a similar survival profile to the Pten single mutants (data not shown), which generally succumb to lymphomas and other nonprostate tumors (18, 20, 22). These observations are consistent with the prostate-specific phenotype of Nkx3.1, which does not have an impact on survival rate, and emphasize the prostate specificity of the cooperativity between loss of function of Nkx3.1 and Pten.
Absence of Nkx3.1 Protein Expression in Mouse and Human Prostate Cancer.
Because the HGPIN/early carcinoma lesions occurred frequently in Nkx3.1+/−;Pten+/− compound mutants, which are heterozygous for Nkx3.1, we examined the status of Nkx3.1 expression in these lesions (Fig. 4). Strikingly, Nkx3.1 immunostaining was invariably absent from lesions of the compound heterozygotes (100%; n = 25; Fig. 4D) but displayed robust nuclear staining in the unaffected regions adjacent to the lesions. In some cases, we observed mislocalization of Nkx3.1 protein to the cytoplasm (Fig. 4F), which may provide an alternative means for inactivating Nkx3.1 function. In contrast to alterations of Nkx3.1 protein, Nkx3.1 mRNA was readily detected in these HGPIN/early carcinoma lesions as well as the adjacent unaffected epithelium by reverse transcriptase–PCR (Fig. 4P).
More generally, we have observed the absence of Nkx3.1 protein expression in regions of atypical hyperplasia, LGPIN, and HGPIN occurring in both single and compound mutant mice (n = 20). For example, Nkx3.1 protein was absent in HGPIN regions in Pten single mutants, which are genotypically wild type for Nkx3.1 (Fig. 4C). Loss of Nkx3.1 protein was common also in LGPIN of Nkx3.1 heterozygotes, which are wild type for Pten (Fig. 4B). Finally, we have observed the absence of Nkx3.1 immunostaining in small clusters of atypical hyperplastic cells in both Nkx3.1 heterozygotes and Nkx3.1;Pten compound heterozygotes (Fig. 4E), suggesting that Nkx3.1 protein loss may precede formation of PIN during cancer progression.
In a parallel analysis of human prostate cancer, we observed that NKX3.1 protein expression was reduced significantly or absent (56 or 26%, respectively; n = 27) in a majority of cancer specimens, with an occasional shift from nuclear to cytoplasmic subcellular localization (11%; n = 27; Fig. 4 G–I). These findings are in accordance with a recent report demonstrating the frequent reduction or absence of NKX3.1 protein expression in a large-scale tissue array analysis of human PIN and prostate cancer specimens (24). Thus, loss and/or mislocalization of NKX3.1 protein expression is characteristic of prostate carcinogenesis in the mouse model as well as in human cancer.
Pten but Not Nkx3.1 Undergoes Allelic Loss in HGPIN/Early Carcinoma Lesions of Compound Mutants.
To examine the status of the wild-type Nkx3.1 and Pten alleles in the HGPIN/early carcinoma lesions of compound heterozygotes, we performed LCM on Nkx3.1-immunostained sections to recover genomic DNA from lesions (Nkx3.1-nonexpressing) and adjacent unaffected regions (Nkx3.1-expressing controls; Fig. 4M). In all cases analyzed, the wild-type Nkx3.1 allele was retained (n = 20); moreover, no mutations were detected in the Nkx3.1 coding region (Fig. 4N and data not shown).
In contrast, Pten sustained allelic loss in 9 of 10 lesions from Nkx3.1;Pten compound heterozygotes (Fig. 4O); this was accompanied by a loss of Pten protein expression, which was apparent both by immunohistochemistry and Western blotting (Figs. 4L and 5A). Interestingly, Pten did not undergo allelic loss in adjacent unaffected regions, and Pten protein expression was reduced but not absent in regions of HGPIN in Pten single mutants (Fig. 4 K, L, and O). Thus, our findings using LCM demonstrate that Pten undergoes LOH within HGPIN/early carcinoma lesions and help to reconcile discrepancies in the literature regarding the allelic status of Pten in mouse models (21, 22, 25).
Figure 5.
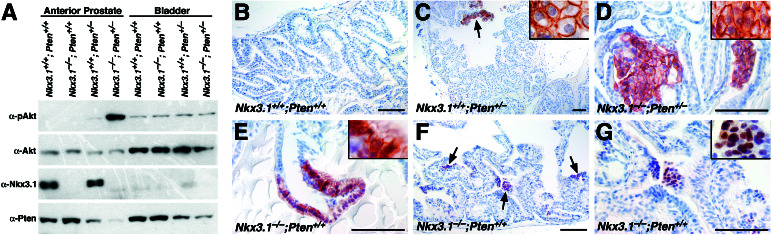
Synergistic activation of Akt in Nkx3.1;Pten compound mutant prostates. (A) Western blot analysis shows high levels of activated Akt in protein extracts from the anterior prostate but not the bladder of Nkx3.1;Pten compound mutants at 8 months. Protein lysates (20 μg) were resolved by SDS/PAGE and probed with antisera to detect P-Akt, total Akt, Pten, or Nkx3.1. Note the reduced levels of Pten in the Nkx3.1−/−;Pten+/− compound mutants relative to the Pten single mutants. (B–G) Immunohistochemical analysis of P-Akt in the anterior prostates of Nkx3.1 and Pten mutants. (B) Absence of staining in wild-type mice. (C) Staining in a cluster of hyperplastic cells (arrows and Inset). (D) Robust staining of HGPIN/early carcinoma lesion (Inset). (E–G) P-Akt immunostaining in clusters of cells in the Nkx3.1−/− prostates shows nuclear and cell surface staining (E, Inset) and primarily nuclear staining (G, Inset).
Synergistic Activation of Akt in Nkx3.1; Pten Compound Mutant Mice.
Because Pten is known to inhibit activation of the Akt kinase (refs. 26–28 and reviewed in refs. 17, 29, and 30), we examined the expression levels and distribution of activated Akt in single and compound mutant prostates by using an antibody specific for the activated (phosphorylated) form (P-Akt; Fig. 5). By Western blot analysis, we observed a synergistic increase in P-Akt in the Nkx3.1−/−;Pten+/− prostates relative to wild-type or single mutants, although the level of total Akt protein was equivalent in all genotypes (n = 9 cohorts; Fig. 5A). Moreover, Akt activation was restricted to the prostate and was not observed in tissues that do not express Nkx3.1 such as the bladder (Fig. 5A). Elevated P-Akt was detected in compound mutants as early as 2 months of age (data not shown), which precedes lesion formation (Fig. 2).
Immunohistochemical analysis revealed robust P-Akt staining in the HGPIN/early carcinoma lesions of Nkx3.1;Pten mice (n = 30; Fig. 5D). These P-Akt-positive regions correspond to those lacking Pten immunostaining and have undergone Pten LOH (Fig. 4 L and O and data not shown). In addition, we detected P-Akt staining in PIN regions of Pten single mutants as well as in small clusters of hyperplastic cells in both Pten single and Nkx3.1;Pten compound mutants (Fig. 5C and data not shown).
We also observed P-Akt staining in the prostatic epithelium of Nkx3.1 single mutants, suggesting that loss of function of Nkx3.1 can affect Akt activation in the context of wild-type Pten function (n = 13; Fig. 5 E–G and data not shown). P-Akt was relatively restricted in its distribution and detected typically in isolated clusters of cells within regions of LGPIN in Nkx3.1+/− and Nkx3.1−/− mice. Although P-Akt-positive clusters were prevalent in the prostate, they were not found in non-Nkx3.1-expressing tissues such as bladder and intestine, and Pten protein expression was unaffected in the Nkx3.1 mutant prostates (data not shown). Although we occasionally detected P-Akt staining associated with the cell surface in Nkx3.1 mutant prostates, we more frequently observed nuclear distribution of phospho-Akt (Fig. 5 E–G). In summary, our findings demonstrate synergistic activation of Akt in compound mutant prostates, suggesting a potential molecular basis for the observed cooperativity of Nkx3.1 and Pten loss of function.
Discussion
Until recently, the validity of the mouse as a model for human prostate cancer has been controversial because of the anatomical and histological differences between mouse and human prostate and the absence of spontaneous prostate cancer in mice (3–5). Our findings demonstrate the utility of mutant mouse models for recapitulating early stages of human prostate carcinogenesis and for providing novel mechanistic insights into this process.
Several lines of evidence implicate NKX3.1 as a tumor suppressor gene, the loss of function of which represents a critical step in prostate cancer initiation. First, NKX3.1 displays tumor suppressor activities in cell culture and in nude mice (M.J.K., M.M.S., and C.A.-S., unpublished data). Second, Nkx3.1 mutant mice develop PIN, paralleling the predicted consequences of chromosome 8p21 LOH in human prostate carcinogenesis. Finally, the NKX3.1 locus is contained within a minimal deletion interval (≈1,500 kb) of human chromosome 8p21 that has been defined by allelotyping studies of prostate carcinomas (C. Vocke and M. Emmert-Buck, personal communication).
Furthermore, the epigenetic inactivation of NKX3.1 function through loss of protein expression is a hallmark of prostate cancer in humans and mutant mouse models (ref. 24 and M.J.K., M.M.S., and C.A.-S., unpublished observations). Notably, this loss of protein expression occurs without an accompanying loss of mRNA expression or mutational inactivation of the NKX3.1 locus (refs. 9, 31, and 32 and this work). One possible mechanism for the loss of NKX3.1 protein is altered translational or posttranslational control, potentially involving the unusually long (≈4-kb) NKX3.1 3′ untranslated region (6); an alternative possibility is deregulated intracellular transport and/or degradation, which would account for the cytoplasmic localization of NKX3.1 protein. Regardless of the mechanism, the observed absence of Nkx3.1 protein provides an explanation for the haploinsufficient phenotype of Nkx3.1 heterozygotes and reconciles a crucial role for NKX3.1 in human prostate cancer with the failure to detect inactivating mutations.
In our studies of compound mutant mice, we have observed that the loss of function of Nkx3.1 and Pten cooperates in prostate cancer progression, as shown by the increased incidence of HGPIN/early carcinoma lesions in the Nkx3.1;Pten compound mutants relative to the Pten single mutants. These HGPIN/early carcinoma lesions can be distinguished from the surrounding epithelium by their (i) high mitotic index, (ii) increased microvessel density, (iii) absence of the basal epithelial layer, (iv) altered wide-spectrum cytokeratin expression, (v) absence of Nkx3.1 protein expression without corresponding LOH or loss of Nkx3.1 mRNA expression, (vi) Pten LOH and corresponding absence of Pten protein expression, and (vii) elevated P-Akt.
Despite the extensive morphological changes observed in Nkx3.1;Pten compound mutant prostates, these mice rarely develop invasive prostatic adenocarcinoma. Notably, these findings contrast with the reported consequences of loss of function of Pten and the cyclin-dependent kinase inhibitor p27KIP1 (21). However, we have found that Pten;p27KIP1 compound mutants as well as Nkx3.1;Pten;p27KIP1 triple mutants develop HGPIN/early carcinoma lesions resembling those of Nkx3.1;Pten compound mutants with respect to their histopathological and immunohistochemical features (M.J.K., M.M.S., C.A.-S., unpublished observations).
These findings highlight the significance and potential synergies of Nkx3.1, Pten, and p27KIP1 in prostate cancer progression. Although the cumulative data from human studies indicate that loss of function of NKX3.1 corresponds to an initiation event, whereas loss of function of PTEN and p27KIP1 correspond to progression events, a limitation of these mutant mouse models is their inability to provide insight into the sequential order of events. Thus, although these studies have elucidated genetic components of a prostate cancer progression pathway, future studies using inducible targeting strategies or similar approaches will be necessary to explore the physiological sequence of events.
The synergistic activation of Akt in Nkx3.1;Pten compound mutant prostates suggests that deregulation of Akt activity is a critical event in prostate carcinogenesis, consistent with the recent observation of elevated phospho-Akt levels in human PIN (33). Interestingly, we have observed nonuniform activation of Akt in the prostatic epithelium of Nkx3.1 single mutants, suggesting that Nkx3.1 loss of function also affects Akt activation, albeit indirectly. Moreover, Nkx3.1 seems to affect the nuclear-cytoplasmic distribution of Akt protein (Fig. 5 and M.J.K., M.M.S., and C.A.-S., unpublished observations), which is noteworthy because Akt is activated at the cell membrane and subsequently translocated to the nucleus, where it has been proposed to phosphorylate regulatory targets (34–36).
In conclusion, we have shown that collaboration between a tissue-specific modulator of prostatic epithelial differentiation and a broad-spectrum tumor suppressor can contribute to cancer progression. These observations raise the possibility that the apparent tissue selectivity of broad-spectrum tumor suppressors (1, 2) may be generated through their synergy with tissue-specific genes to affect common signaling pathways such as what we have observed for Pten and Nkx3.1 for Akt activation in the prostate. Thus, we propose that these collaborative interactions contribute to the distinguishing features of prostate carcinoma and that similar interactions may generally explain the tissue-specific phenotypes of cancers.
Acknowledgments
We are indebted to Dr. Regina Gandour-Edwards for the human prostate cancer specimens. We acknowledge April Graham and Judy E. Walls for excellent technical assistance. We thank David Ornstein and Michael Emmert-Buck for assistance with LCM and discussions. We are grateful to David Berman, Diego Castrillon, Ron DePinho, Simon Hayward, and Danny Reinberg for comments on the manuscript. This work was supported by National Cancer Institute Grants UO1 CA84294 (to C.A.-S., M.M.S., and R.D.C) and CA76501 (to C.A.-S. and M.M.S.), National Institute of Diabetes and Digestive and Kidney Diseases Grant DK60887 (to M.M.S.), U.S. Army Prostate Cancer Research Program Grants DAMD17-00-1-0091 (to C.A.-S.) and DAMD17-98-1-8532 (to M.M.S.), and University of California Breast Cancer Research Program Grant 5JB-0014 (to R.D.C.).
Abbreviations
- PIN
prostatic intraepithelial neoplasia
- LOH
loss of heterozygosity
- HGPIN
high-grade PIN
- LGPIN
low-grade PIN
- LCM
laser-capture microdissection
References
- 1.Jacks T. Annu Rev Genet. 1996;30:603–636. doi: 10.1146/annurev.genet.30.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Macleod K F, Jacks T. J Pathol. 1999;187:43–60. doi: 10.1002/(SICI)1096-9896(199901)187:1<43::AID-PATH246>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 3.Abate-Shen C, Shen M M. Genes Dev. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 4.Matusik R, Masumori M, Thomas T, Case T, Paul M, Kasper S, Shappell S B. In: Transgenics in Endocrinology. Matzuk M, Brown C, Kumar T, editors. Totowa, NJ: Humana; 2002. pp. 401–425. [Google Scholar]
- 5.Green J E, Greenberg N M, Ashendel C L, Barrett J C, Boone C, Getzenberg R H, Henkin J, Matusik R, Janus T J, Scher H I. Prostate. 1998;36:59–63. doi: 10.1002/(sici)1097-0045(19980615)36:1<59::aid-pros11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 6.Sciavolino P J, Abrams E W, Yang L, Austenberg L P, Shen M M, Abate-Shen C. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia-Gaur R, Donjacour A A, Sciavolino P J, Kim M, Desai N, Young P, Norton C R, Gridley T, Cardiff R D, Cunha G R, et al. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He W W, Sciavolino P J, Wing J, Augustus M, Hudson P, Meissner P S, Curtis R T, Shell B K, Bostwick D G, Tindall D J, et al. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- 9.Voeller H J, Augustus M, Madike V, Bova G S, Carter K C, Gelmann E P. Cancer Res. 1997;57:4455–4459. [PubMed] [Google Scholar]
- 10.Bergerheim U S, Kunimi K, Collins V P, Ekman P. Genes Chromosomes Cancer. 1991;3:215–220. doi: 10.1002/gcc.2870030308. [DOI] [PubMed] [Google Scholar]
- 11.Emmert-Buck M R, Vocke C D, Pozzatti R O, Duray P H, Jennings S B, Florence C D, Zhuang Z, Bostwick D G, Liotta L A, Linehan W M. Cancer Res. 1995;55:2959–2962. [PubMed] [Google Scholar]
- 12.Haggman M J, Wojno K J, Pearsall C P, Macoska J A. Urology. 1997;50:643–647. doi: 10.1016/S0090-4295(97)00304-X. [DOI] [PubMed] [Google Scholar]
- 13.Vocke C D, Pozzatti R O, Bostwick D G, Florence C D, Jennings S B, Strup S E, Duray P H, Liotta L A, Emmert-Buck M R, Linehan W M. Cancer Res. 1996;56:2411–2416. [PubMed] [Google Scholar]
- 14.Maehama T, Dixon J E. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 15.Myers M P, Pass I, Batty I H, Van der Kaay J, Stolarov J P, Hemmings B A, Wigler M H, Downes C P, Tonks N K. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantley L C, Neel B G. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Cristofano A, Pandolfi P P. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 18.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi P P. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki A, de la Pompa J L, Stambolic V, Elia A J, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, Fukumoto M, Mak T W. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 20.Podsypanina K, Ellenson L H, Nemes A, Gu J, Tamura M, Yamada K M, Cordon-Cardo C, Catoretti G, Fisher P E, Parsons R. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi P P. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 22.Stambolic V, Tsao M S, Macpherson D, Suzuki A, Chapman W B, Mak T W. Cancer Res. 2000;60:3605–3611. [PubMed] [Google Scholar]
- 23.Bostwick D G, Brawer M K. Cancer. 1987;59:788–794. doi: 10.1002/1097-0142(19870215)59:4<788::aid-cncr2820590421>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 24.Bowen C, Bubendorf L, Voeller H J, Slack R, Willi N, Sauter G, Gasser T C, Koivisto P, Lack E E, Kononen J, Kallioniemi O P, Gelmann E P. Cancer Res. 2000;60:6111–6115. [PubMed] [Google Scholar]
- 25.Kwabi-Addo B, Giri D, Schmidt K, Podsypanina K, Parsons R, Greenberg N, Ittmann M. Proc Natl Acad Sci USA. 2001;98:11563–11568. doi: 10.1073/pnas.201167798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stambolic V, Suzuki A, de la Pompa J L, Brothers G M, Mirtsos C, Sasaki T, Ruland J, Penninger J M, Siderovski D P, Mak T W. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Senechal K, Neshat M S, Whang Y E, Sawyers C L. Proc Natl Acad Sci USA. 1998;95:15587–15591. doi: 10.1073/pnas.95.26.15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H, Lesche R, Li D M, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, Wu H. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coffer P J, Jin J, Woodgett J R. Biochem J. 1998;335:1–13. doi: 10.1042/bj3350001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta S R, Brunet A, Greenberg M E. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 31.Ornstein D K, Cinquanta M, Weiler S, Duray P H, Emmert-Buck M R, Vocke C D, Linehan W M, Ferretti J A. J Urol. 2001;165:1329–1334. [PubMed] [Google Scholar]
- 32.Xu L L, Srikantan V, Sesterhenn I A, Augustus M, Dean R, Moul J W, Carter K C, Srivastava S. J Urol. 2000;163:972–979. [PubMed] [Google Scholar]
- 33.Paweletz C P, Charboneau L, Bichsel V E, Simone N L, Chen T, Gillespie J W, Emmert-Buck M R, Roth M J, Petricoin I E, Liotta L A. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 34.Meier R, Alessi D R, Cron P, Andjelkovic M, Hemmings B A. J Biol Chem. 1997;272:30491–30497. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- 35.Andjelkovic M, Alessi D R, Meier R, Fernandez A, Lamb N J, Frech M, Cron P, Cohen P, Lucocq J M, Hemmings B A. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 36.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]