Abstract
Scavenger receptor class B type I (SR-BI) is a high-density lipoprotein (HDL) receptor that mediates the selective uptake of HDL cholesterol and cholesterol secretion into bile in the liver. Previously, we identified an SR-BI-associated protein, termed PDZK1, from rat liver membrane extracts. PDZK1 contains four PSD-95/Dlg/ZO-1 (PDZ) domains, the first of which in the N-terminal region is responsible for the association with SR-BI. PDZK1 controls hepatic SR-BI expression in a posttranscriptional fashion both in cell culture and in vivo. In this study, we demonstrated that the C-terminal region of PDZK1 is crucial for up-regulating SR-BI protein expression. Metabolic labeling experiments and phosphoamino acid analysis revealed that PDZK1 is phosphorylated at Ser residues within this region. Point-mutation analysis demonstrated that PDZK1 is phosphorylated at Ser-509. Interestingly, a mutant PDZK1, in which Ser-509 was replaced with Ala, lost the ability to up-regulate SR-BI protein. We identified Ser-509 of PDZK1 as the residue that is phosphorylated by the cAMP-dependent PKA in vitro as well as in cell culture. Ser-509 of PDZK1 in rat liver was also phosphorylated, as shown by an Ab that specifically detects phosphorylated Ser-509. Administration of glucagon to Wistar rats increased PDZK1 phosphorylation as well as hepatic SR-BI and PDZK1 expression while it decreased plasma HDL levels, indicating that PDZK1 phosphorylation is hormonally regulated. These findings suggest that phosphorylation of PDZK1 has an important role in the regulation of hepatic SR-BI expression and, thus, influences plasma HDL levels.
Keywords: glucagon, high-density lipoprotein, PKA
Plasma high-density lipoprotein (HDL) has a critical role in cholesterol metabolism, and plasma HDL concentrations are inversely related to the risk of developing cardiovascular disease (1, 2). The protective role of HDL against cardiovascular disease is commonly attributed to its ability to remove excess cholesterol from cells in the arterial wall and transport it to the liver for disposal, which is a process that is called reverse cholesterol transport (3, 4). Scavenger receptor class B type I (SR-BI) is an HDL receptor that is expressed in the liver as well as in steroidogenic tissues at high levels and mediates the selective uptake of HDL cholesterol (5, 6). Overexpression of SR-BI in murine hepatocytes results in the virtual disappearance of plasma HDL and a substantial increase in biliary cholesterol (7-10). Mice with a targeted mutation in the SR-BI gene exhibit increased plasma HDL cholesterol levels, increased HDL particle size (11, 12), and impaired biliary cholesterol secretion (13). These studies have established the concept that SR-BI determines the level of plasma HDL by taking up HDL cholesterol into the liver for transport into bile.
SR-BI contains a large extracellular domain that is anchored in the plasma membrane by transmembrane domains adjacent to short cytoplasmic N- and C-terminal regions (6). We have identified (14) an SR-BI C-terminal binding protein from rat liver membranes and named it CLAMP (C-terminal linking and modulating protein). CLAMP contains four PSD-95/Dlg/ZO-1 (PDZ) domains and associates with the C terminus of SR-BI via its N-terminal first PDZ domain (14). CLAMP is expressed mainly in the liver, kidney, and small intestine, whereas steroidogenic organs that express high levels of SR-BI, such as the adrenal grand, showed no significant CLAMP expression (15, 16). This protein has been shown to interact with a number of membrane-associated transporter proteins from different tissues, such as cMOAT/MRP2 (17), cystic fibrosis transmembrane regulator (CFTR) (16), CIC-3B (18), type IIa Na+/Pi cotransporter (Na/Pi-IIa) (19), solute carrier SLC17A1 (Na/Pi-I), Na+/H+ exchanger (NHE-3), organic cation transporter (OCTN), chloride-formate exchanger (CFEX), and urate-anion exchanger (URAT1) (20). CLAMP is also called diphor-1 (21), PDZK1 (15), CAP70 (16), and NaPi-Cap1 (19), depending on its binding partners. Here, we refer to CLAMP as PDZK1.
In recent studies, Kocher et al. showed that PDZK1 knockout mice had significantly increased plasma cholesterol levels (22) as well as dramatically reduced hepatic SR-BI protein levels and abnormally large HDL particles in plasma (23). Similar changes were observed in SR-BI knockout mice (11). These studies established that PDZK1 up-regulates hepatic SR-BI expression at the protein level in vivo. Consistent with these observations, we have demonstrated (14) that coexpression of PDZK1 and SR-BI in CHO cells results in increased SR-BI protein levels without affecting the SR-BI mRNA level.
In this study, we searched for other domains, in addition to the N-terminal first PDZ domain in PDZK1, that are required for its SR-BI up-regulating activity. We found evidence that phosphorylation of the PDZK1 C-terminal region is also involved in the up-regulation of SR-BI protein expression.
Materials and Methods
Cells. CHO-K1 cells were grown in Ham's F-12 medium, supplemented with 10% FCS, 50 units/ml penicillin, 50 mg/ml streptomycin, and 2 mM glutamine at 37°C in a humidified 5% CO2/95% air incubator. Rat hepatoma Fao cells were maintained in Coon's F-12 medium supplemented with 10% FCS, 50 units/ml penicillin, 50 mg/ml streptomycin, and 2 mM glutamine at 37°C in a humidified 10% CO2/90% air incubator.
Creation of PDZK1 Mutants. PDZK1 mutations were constructed by PCR-mediated mutagenesis. All cDNAs were cloned into pcDNA 3.1/Hygro (Invitrogen), pGEX6P-1 (Amersham Biosciences), or pShuttle-CMV (Q-BIO Gene, Carlsbad, CA) and verified by sequencing. Fusion proteins of GST were expressed in Escherichia coli and purified as described (14). The shuttle vector plasmids were inserted into pAdEasy-1 (Q-BIO Gene). Recombinant adenoviral constructs were transfected into E1-transformed human embryonic kidney (HEK293ΔE1) cells to produce viral particles. The recombinant adenoviruses were purified by cesium chloride ultracentrifugation. As controls, the LacZ virus that carries β-gal cDNA was also constructed and purified as described above.
Transfection of Plasmids and Transduction of Adenoviral Vectors into Cells. CHO-K1 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Constitutively transfected cells were selected with hygromycin. Cultured Fao cells at 70% confluence were infected with the respective adenoviruses at a rate of 5,000 particles per cell in Coon's F-12 medium containing 2% FBS for 2 h. The cultures were supplemented an equal volume of Coon's F-12 medium containing 20% FBS and incubated for an additional 4 days.
Preparation of Rat Liver Membrane Extracts. Livers obtained from male Wistar rats (200-250 g, 8 weeks old) were homogenized with a homogenizing buffer (14), and the homogenate was centrifuged at 100,000 × g for 1 h at 4°C. The resulting precipitate was suspended in a homogenizing buffer.
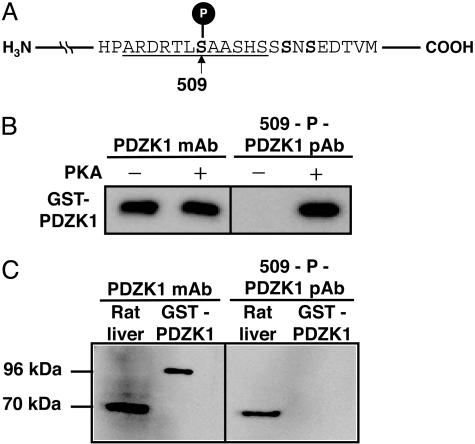
Ab Production. A synthetic phosphopeptide based on residues 503-514 of rat PDZK1 with phospho-Ser-509 was conjugated to keyhole limpet hemocyanin [KLH-CARDRTLS (PO3H2) AASHS] and injected subcutaneously into rabbits together with adjuvants. After eight booster injections at weekly intervals, anti-509-P-PDZK1 polyclonal Ab (pAb) was purified from rabbit serum by using a peptide-ligand-affinity column.
Western Blot Analysis. Proteins in the cell lysates or rat liver membrane extracts were separated by SDS/PAGE. The gels were blotted to polyvinylidene difluoride membranes (Millipore). The membranes were treated with anti-SR-BI pAb (NB400-101, Novus Biologicals, Littleton, CO), anti-PDZK1 mAb (14), or anti-509-P-PDZK1. The immunoreactive proteins were visualized by using chemiluminescence and recorded with a digital recorder (LAS-1000, Fuji Film, Tokyo). The relative amounts of proteins were quantified by using image gauge v.3.45 (Fuji Film).
Metabolic Labeling, Immunoprecipitation, and Phosphoamino Acid Analysis. CHO-KI cells expressing PDZK1 or mutants of PDZK1 were incubated for 30 min in phosphate-depleted DMEM. Cultures were then labeled in labeling media containing 0.3 mCi (1 Ci = 37 GBq) of [32P]orthophosphate for 4 h. Cells were lysed in immunoprecipitation buffer (0.5% Nonidet P-40/1% Triton X-100/10 mM Tris·HCl, pH 7.4/1 mM EDTA/150 mM NaCl/0.2 mM sodium vanadate/10 mM NaF/1 μg/ml pepstatin/1 μg/ml leupeptin/2 μg/ml aprotinin/1 mM PMSF). Lysates were immunoprecipitated with anti-PDZK1 mAb, as described in ref. 14. The immunoprecipitates were then analyzed by Western blotting and autoradiography. Phosphoamino acid maps of phosphorylated PDZK1 were generated by excising 32P-labeled PDZK1 bands from polyvinylidene difluoride membranes and eluting the proteins with 6 M HCl for 1 h at 100°C. The samples were dried in a SpeedVac concentrator (Savant) and dissolved in first-dimension buffer [2.2% (vol/vol) formic acid/7.8% (vol/vol) acetic acid, pH 1.9], each sample containing three phosphoamino-acid standards (phospho-Ser, phospho-Thr, and phospho-Tyr). Phosphoamino acids were subjected to two-dimensional electrophoresis on TLC cellulose plates (pH 1.9, 1.5 kV for 30 min; pH 3.5, 1.3 kV for 15 min), followed by ninhydrin staining. The TLC plates were scanned with a bas 2000 imaging system (Fuji Film) to detect labeled phosphoamino acids.
In Vitro Phosphorylation of PDZK1 by PKA. We incubated 1 μg of recombinant GST-fused wild-type PDZK1 protein or GST-fused mutant PDZK1 protein, in which Ser-509 was replaced with Ala (S509A), with 5 units of cAMP-dependent PKA (Sigma) in phosphorylation buffer (50 mM Tris·HCl, pH 7.5/2 mM EDTA/7 mM MgCl2/0.1 mM DTT/1 mM PMSF). We then added [γ-32P]ATP (6 μCi), and the mixture was incubated at 30°C for 30 min. The reaction was stopped by boiling for 5 min in SDS sample buffer. The samples were subjected to SDS/PAGE and then stained with Coomassie brilliant blue (CBB). The gels were scanned with bas 2000 to detect labeled PDZK1.
Phosphorylation of PDZK1 by PKA in Cell Culture. CHO-K1 cells stably expressing PDZK1 were treated with 10 μM forskolin (a PKA inducer) or 10 μM H-89 (a PKA inhibitor) and then labeled in labeling media containing 0.3 mCi of [32P]orthophosphate for 4 h. Cells were lysed and immunoprecipitated with PDZK1. The immunoprecipitates were separated by SDS/PAGE. The gel was stained with CBB and scanned with bas 2000 to detect labeled PDZK1.
Glucagon Administration. Male Wistar rats (200-250 g, 8 weeks old) were used. Animals were kept under standardized conditions with free access to water and chow. The light-cycle was from 7 a.m. to 7 p.m. Rats were injected s.c. under light ether anesthesia at 10 a.m. and 4 p.m. with 400 μg (115 nmol) of glucagon or a vehicle on each of 2 consecutive days. The blood was removed by venipuncture of the orbital sinus at 12 a.m., which was 2 h after the last injection. Then, rats were immediately killed, and livers were removed. Plasma was separated by low-speed centrifugation for 10 min at 4°C. Plasma total cholesterol and HDL cholesterol were prepared and assayed by using the cholesterol test kit (Wako Pure Chemical, Osaka).
Quantitative RT-PCR. Total RNA of rat liver was isolated with Isogen reagent (Nippon Gene, Toyama, Japan). Total RNA (1 μg) was reverse-transcribed in the presence of poly(dT) sequences in a total volume of 10 μl. We used 1 μl of this mixture as template in the quantitative RT-PCR. Quantitative RT-PCR reactions were performed by using the Prism 7000 sequence-detection system (Applied Biosystems). The following PCR primers were used: β-actin, 5-CCTTCTACAATGAGCTGCGTGT-3 (forward) and 5-TGGGGTGTTGAAGGTCTCAAAC-3 (reverse); SR-BI, 5-TTCTGGTGCCCATCATTTACC-3 (forward) and 5-AGCCCTTTTTACTACCACTCCAAA-3 (reverse); and CLAMP, 5-TTGAAGTGAATGGAGAAAATGTAG-3 (forward) and 5-TGATACGGCTTCCTGACTTTGTC-3 (reverse). Results were normalized to β-actin data.
Results
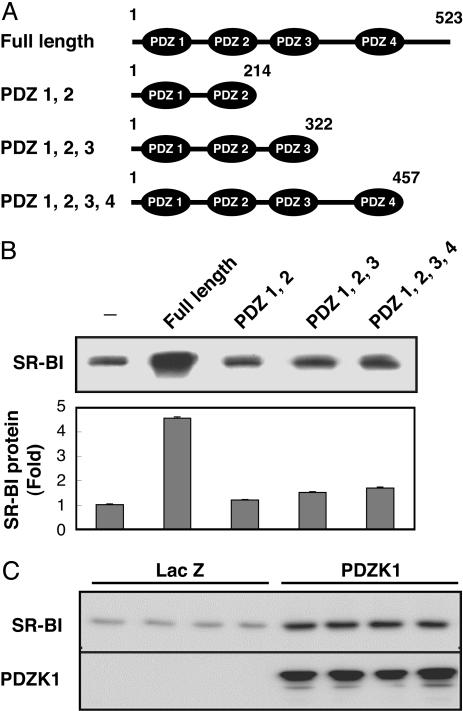
The PDZK1 C-Terminal Domain Is Required for Up-Regulation of SR-BI Expression. CHO-K1 cells expressing various deletion mutants of PDZK1 were established (Fig. 1A) and transiently transfected with SR-BI. SR-BI protein expression increased 4- to 5-fold in the cells coexpressing SR-BI and full-length PDZK1 (Fig. 1B). We also examined the effect of PDZK1 on SR-BI expression using rat hepatoma Fao cells, which intrinsically express SR-BI but not PDZK1. Expression of PDZK1 in Fao cells was achieved by infection with an adenovirus vector. The same degree of up-regulation of SR-BI protein was observed in this hepatoma cell line (Fig. 1C). However, none of the PDZK1 deletion mutants lacking the C-terminal region were able to up-regulate the SR-BI protein (Fig. 1B). Even the mutant lacking only 66 aa at the C terminus was less effective than the full-length PDZK1. These results suggest that PDZK1 is capable of up-regulating the SR-BI protein and that a region within the C-terminal 66 aa is indispensable for the up-regulation.
Fig. 1.
The C-terminal region of PDZK1 is required for SR-BI up-regulation. (A) Schematic representation of various deletion mutants of PDZK1. (B) Western blot analysis of SR-BI from CHO-K1 cells constitutively expressing various deletion mutants of PDZK1 and transiently transfected with SR-BI. The graph represents relative quantities of SR-BI proteins in these Western blot analyses. (C) Western blot analysis of SR-BI and PDZK1 from cell lysates of Fao cells transduced with the recombinant adenovirus construct of PDZK1. LacZ is a control. The data represent at least three independent experiments.
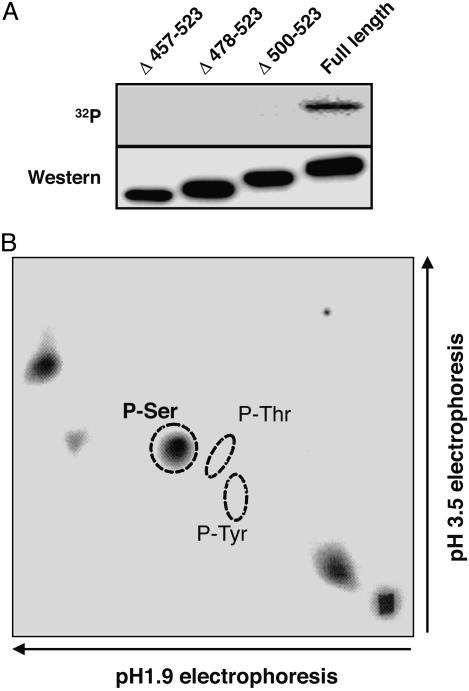
PDZK1 Is Phosphorylated at the Ser Residue in the C-Terminal Region. PDZK1 possesses multiple potential phosphorylation sites within the C-terminal 66 aa, as indicated by the NetPhos database (available at www.cbs.dtu.dk/services/NetPhos). To examine whether the C-terminal region of PDZK1 is phosphorylated in cells, we constructed three C-terminal deletion mutants (namely, Δ457-523, Δ478-523, and Δ500-523), corresponding to deletions of the indicated amino acids. The C-terminal deletion mutants and full-length PDZK1 were transfected into CHO-KI cells and metabolically labeled with [32P]orthophosphate. The expressed proteins were immunoprecipitated with anti-PDZK1 mAb. Western blotting with the anti-PDZK1 mAb revealed that similar levels of wild-type and mutant PDZK1 proteins were expressed (Fig. 2A). A 32P-labeled protein band comigrating with PDZK1 was detected in the CHO cell extract, indicating that PDZK1 is a phosphorylated protein. However, all of the tested C-terminal deletion mutants resulted in significant reduction of 32P-labeling. These data demonstrate that the phosphorylation of PDZK1 occurs in a region within the C-terminal 66 aa.
Fig. 2.
PDZK1 is a phosphorylated protein. (A) Autoradiography (Upper) and the Western blot analysis of PDZK1 (Lower). CHO-KI cells were transiently transfected with an expression plasmid of C-terminal deletion mutants of PDZK1 (Δ457-523, Δ478-523, and Δ500-523) and incubated with [32P]orthophosphate. (B) Phosphoamino acid analysis of 32P-labeled PDZK1. Circles indicate where the phosphoamino acid standards migrated. P-Ser, phospho-Ser; P-Thr, phospho-Thr; P-Tyr, phospho-Tyr.
We then analyzed phosphoamino acids of PDZK1 labeled with [32P]orthophosphate. As shown in Fig. 2B, PDZK1 was phosphorylated on Ser residues but not on Thr or Tyr residues.
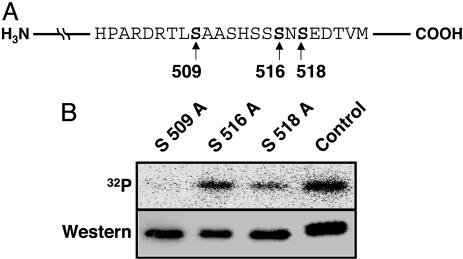
Ser-509 of PDZK1 Is Phosphorylated. Based on the database search for potential phosphorylation sites within the C-terminal 66 aa, we chose three potential sites of Ser phosphorylation (namely, Ser-509, Ser-516, and Ser-518). To determine whether these Ser residues were phosphorylated, we constructed respective Ala substitution mutants as shown in Fig. 3A. Transfection with these mutants into CHO-KI cells and metabolic labeling with 32P revealed that S509A was no longer phosphorylated in transfected cells, whereas S516A and S518A were phosphorylated (Fig. 3B), thus implicating Ser-509 as a critical site of PDZK1 phosphorylation.
Fig. 3.
Ser-509 of PDZK1 is phosphorylated. (A) Schematic representation of potential sites of Ser phosphorylation in the PDZK1 C-terminal region. (B) Autoradiography (Upper) and the Western blot analysis of PDZK1 (Lower). CHO-KI cells were transiently transfected with various single-point mutants of PDZK1 (S509A, S516A, and S518A) and labeled with [32P]orthophosphate.
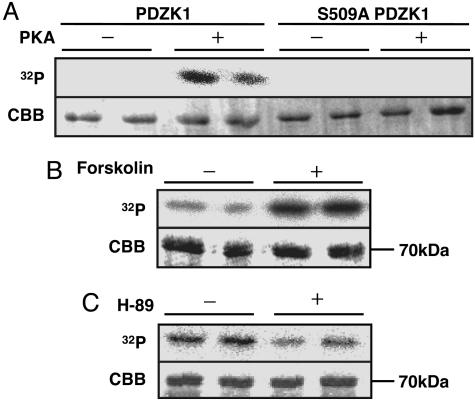
PDZK1 Ser-509 Is Phosphorylated by PKA in Vitro and in Cell Culture. According to ref. 24, Ser-509 was found to be a potential PKA target Ser. To examine whether Ser-509 can be directly phosphorylated by PKA, we incubated the recombinant PDZK1 with the purified catalytic subunit of PKA in the presence of [γ-32P]ATP. We found that PKA phosphorylated PDZK1 in vitro, whereas PKA did not phosphorylate the S509A mutant to any extent (Fig. 4A). Next, we treated CHO-KI cells that constitutively express PDZK1 with the PKA inducer forskolin or the PKA inhibitor H-89 in the presence of [32P]orthophosphate. Endogenous PDZK1 was then immunoprecipitated and subjected to CBB staining and autoradiography. The CBB-stained ≈70-kDa band was found to be PDZK1, as judged from the fact that this ≈70-kDa band in SDS/PAGE (Fig. 4 B and C Lower) reacted with anti-PDZK1 Ab by Western blot analysis and could not be detected by performing the same procedures with parental CHO-K1 cells (data not shown). Phosphorylation levels of PDZK1 were significantly increased by forskolin (Fig. 4B) and reduced by H-89 (Fig. 4C). These in vitro and cell-culture experiments demonstrated that PDZK1 Ser-509 is phosphorylated by PKA.
Fig. 4.
PKA phosphorylates PDZK1. (A) Recombinant GST-PDZK1 or GST-S509A mutant fused protein expressed by E. coli was incubated with or without PKA in the presence of [γ-32P]ATP. (B and C) Metabolic labeling with [32P]orthophosphate of CHO-K1 cells constitutively expressing PDZK1 were performed in the absence or presence of 10 μM forskolin (B) or 10 μM H-89 (C). Autoradiography (Upper) and CBB staining (Lower) are shown. The data represent at least three independent experiments.
PDZK1 Expressed in Liver Is Phosphorylated. Although mutagenesis studies using CHO-K1 cells showed the importance of the Ser site for PDZK1 phosphorylation, it was not clear whether Ser-509 was phosphorylated in vivo. To answer this question, we established specific Abs against synthetic peptides corresponding to amino acids 503-514 of rat PDZK1 in which Ser-509 was phosphorylated (Fig. 5A). By using a phospho-specific Ab (anti-509-P-PDZK1) that was developed to detect Ser-509 phosphorylation, a prominent immunoreactive band was detected in the recombinant PDZK1 when the recombinant PDZK1 was incubated with PKA, whereas no anti-509-P-PDZK1 immunoreactivity was detected when the recombinant PDZK1 was incubated without PKA (Fig. 5B). In a Western blot analysis (Fig. 5C), anti-509-P-PDZK1 detected PDZK1 in rat liver extract but not the recombinant PDZK1 produced by E. coli. These results showed that Ser-509 of PDZK1 expressed in rat liver was phosphorylated.
Fig. 5.
PDZK1 expressed in the liver is phosphorylated per se.(A) Preparation of Abs against phospho-Ser-509. The underlined phosphorylated peptide was synthesized for immunization. (B) The recombinant PDZK1 expressed by E. coli was incubated with or without PKA and then subjected to Western blotting using anti-PDZK1 mAb (Left), anti-509-P-PDZK1 pAb (Right). (C) Western blot analysis of PDZK1 from rat liver membrane extract (70 kDa) and the recombinant GST-PDZK1 fused protein (96 kDa) using anti-PDZK1 mAb (Left) and anti-509-P-PDZK1 pAb (Right).
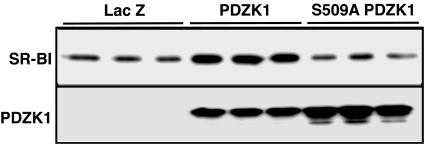
SR-BI Protein Level Is Affected by the PDZK1 Phosphorylation State. Because we found that PDZK1 is phosphorylated at position Ser-509, we examined next whether PDZK1 phosphorylation could regulate SR-BI protein expression. Wild-type PDZK1 and the PDZK1 S509A mutant were overexpressed in rat hepatoma Fao cells by using recombinant adenovirus vectors (Fig. 6). The amount of SR-BI protein increased ≈4-fold in Fao cells transduced with an adenovirus vector harboring PDZK1 (Ad-PDZK1), as compared with Ad-LacZ. However, transduction of Fao cells with Ad-PDZK1 S509A mutant did not affect the amount of SR-BI protein. These results suggested that PDZK1 phosphorylation of Ser-509 is one of the key factors regulating SR-BI protein levels.
Fig. 6.
SR-BI protein level is regulated by PDZK1 through phosphorylation. Western blot analysis of SR-BI (Upper) and PDZK1 (Lower) from cell lysates of Fao cells transduced with the recombinant adenovirus of PDZK1 or S509A PDZK1. LacZ is a control. The results of this figure are representative of at least three independent experiments.
Glucagon Increases PDZK1 Phosphorylation as Well as Hepatic SR-BI Expression. Next, we investigated physiological conditions regulating PDZK1 phosphorylation in rat liver. Reports (25-28) have indicated that the administration of glucagon to laboratory animals as well as humans decreases plasma HDL cholesterol levels. To our knowledge, no information is available regarding the effects of glucagon on hepatic SR-BI in vivo. The actions of glucagon are mediated by the glucagon receptor linked to a heterotrimeric G-protein complex, leading to increased cellular levels of cAMP and activation of PKA. We hypothesized that the reduction of plasma levels of HDL by glucagon is due to the up-regulation of SR-BI protein expression through PDZK1 phosphorylation by PKA.
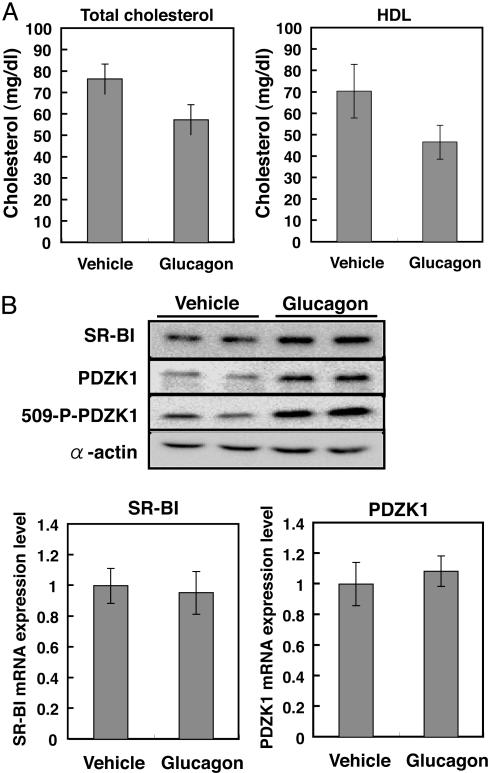
To determine the influence of glucagon on hepatic SR-BI expression and PDZK1 phosphorylation, rats were injected twice daily with 400 μg of glucagon or a vehicle for 2 days before they were killed. Total plasma cholesterol levels and plasma HDL cholesterol levels were decreased significantly, as reported in ref. 28 (Fig. 7A). Upon glucagon treatment, the phosphorylated form of PDZK1 was increased by 3.6 ± 0.2, whereas SR-BI and PDZK1 protein levels were increased by 1.8 ± 0.3 and 1.8 ± 0.4, respectively. Glucagon treatment did not affect SR-BI and PDZK1 mRNA levels (Fig. 7C), indicating the posttranscriptional regulation of hepatic SR-BI and PDZK1 expression by this hormone. These results suggested that PDZK1 phosphorylation and protein expression were hormonally regulated, and that up-regulation of hepatic SR-BI expression by glucagon is at least partly due to elevation of PDZK1 phosphorylation and/or its protein expression.
Fig. 7.
Glucagon increases PDZK1 phosphorylation and hepatic SR-BI expression. Five rats were injected twice daily with 400 μg of glucagon or a vehicle for 2 days. Rats were killed 2 h after the last injection. (A) Total and HDL cholesterol levels. (B) Western blot analysis of SR-BI, PDZK1, phosphorylated PDZK1, and α-actin from rat liver homogenates of two representative animals. (C) Quantitative analysis of SR-BI and PDZK1 transcripts. Total RNA purified from rat liver was subjected to real time RT-PCR for measuring SR-BI and PDZK1 transcripts, as described in Materials and Methods.
Discussion
We previously showed that coexpression of PDZK1 and SR-BI in CHO cells resulted in up-regulation of SR-BI protein expression without affecting the SR-BI mRNA level (14). Moreover, Kocher et al. (23) demonstrated that there is a 95% reduction in SR-BI protein expression in the livers of PDZK1-deficient mice compared with wild-type controls. These in vitro and in vivo data indicate that PDZK1 controls hepatic expression of SR-BI in a posttranscriptional fashion. In this study, we demonstrated that the phosphorylation of Ser-509 in the C-terminal region of PDZK1 is required for SR-BI up-regulation in cultured hepatoma cells. By using a phospho-specific Ab developed to detect Ser-509 phosphorylation, we showed that Ser-509 of hepatic PDZK1 is phosphorylated.
Three lines of evidence suggest that Ser-509 of PDZK1 is phosphorylated by PKA. First, the sequence around Ser-509 matches the potential phosphorylation site of PKA. Second, the recombinant PDZK1 is phosphorylated by PKA in vitro. Third, the phosphorylation level of PDZK1 in cultured cells is increased by a PKA activator and reduced by a PKA inhibitor. It has been demonstrated that the dual-specific A kinase-anchoring protein 2 (D-AKAP2), a member of the PKA-anchoring protein family (AKAP) binds to the fourth PDZ domain of PDZK1 in the kidney (20, 29), which is close to the Ser-509 position. D-AKAP2 is also expressed in the liver (30). It remains to be determined whether it interacts with PDZK1 in the liver.
The sequence around Ser-509 of PDZK1 also matches the consensus phosphorylation motif for p70 S6 kinase (p70S6K) (24). Interestingly, the C-terminal region of p70S6K contains a sequence capable of binding to a PDZ domain. Indeed, p70S6K has been reported (31) to interact with a PDZ domain-containing protein, called Neurabin, that exists in neuronal cells. In preliminary experiments, we found that p70S6K can phosphorylate Ser-509 in PDZK1 in vitro. Stimulation of hepatoma cells with insulin is known to activate p70S6K (32). Therefore, it is plausible that diverse extracellular events can lead to PDZK1 phosphorylation, thus controlling SR-BI expression levels.
The phosphorylation level of PDZK1 was reduced by approximately one half when Ala was substituted for Ser-518 in 32P-incorporation experiments (Fig. 3B). Therefore, it is possible that PDZK1 is also phosphorylated at position Ser-518. However, the Ser-509-to-Ala substitution leads to complete suppression of phosphorylation, which indicates that Ser-518 is phosphorylated after Ser-509 is phosphorylated. Whether the PDZK1 C-terminal region has other phosphorylation sites besides the Ser-509 location remains to be determined.
Glucagon, being a major regulator of plasma glucose concentration (33), has an important role in carbohydrate metabolism. The effects of glucagon are mediated by the cAMP second-messenger system, including PKA. Glucagon also influences cholesterol metabolism. Conditions with increased glucagon levels are associated with reduced plasma cholesterol levels (34, 35), and the administration of glucagon to laboratory animals as well as humans decreases plasma cholesterol levels (25-28). However, the physiological role of this phenomenon remains unclear. Glucagon has been shown to increase the number of hepatic low-density lipoprotein (LDL) receptors and concomitantly decrease plasma cholesterol and apoprotein B levels (28). According to ref. 28, the plasma HDL cholesterol level was decreased also by glucagons treatment, as we observed in this study (Fig. 7A). Our results show that glucagon increases expression of the HDL receptor SR-BI in the liver. Overexpression of hepatic SR-BI leads to reduced plasma HDL levels (7, 8), which supports the hypothesis that increased SR-BI expression is responsible for reduced plasma HDL levels. Furthermore, glucagon was shown to enhance cholesterol uptake into bile in rats (36). HDL cholesterol is known to be a major substrate source for bile acid production in both rats and humans (37). Overexpression of SR-BI markedly promotes the hepatic uptake of HDL cholesterol, thus facilitating the secretion of cholesterol into bile (7, 10). One explanation for our results is that glucagon administration to rats leads to enhanced PDZK1 phosphorylation via PKA, which subsequently leads to enhanced SR-BI protein expression, regulated at a posttranscriptional level. However, it cannot yet be proven that the phosphorylation of PDZK1 leads to the described SR-BI elevation because glucagon treatment leads not only to an elevation of SR-BI but simultaneously to a 2-fold elevation of PDZK1, respectively. Use of mouse models such as PDZK1 knockout mice and SR-BI knockout mice, as well as transgenic animals expressing only the PDZK1 mutant where Ser-509 is substituted by Ala, would bring us further toward solving this problem.
In conclusion, we demonstrated in this study that phosphorylation of the PDZK1 C-terminal region is crucial for upregulation of SR-BI protein expression and that PDZK1 phosphorylation is hormonally regulated. A major question that remains to be answered is how the phosphorylation of the PDZK1 C-terminal region is involved in the up-regulation of SR-BI levels.
Abbreviations: HDL, high-density lipoprotein; PDZ, PSD-95/Dlg/ZO-1; CLAMP, C-terminal linking and modulating protein; pAb, polyclonal Ab; CBB, Coomassie brilliant blue; SR-BI, scavenger receptor class B type I.
References
- 1.Gordon, D. J. & Rifkind, B. M. (1989) N. Engl. J. Med. 321, 1311-1316. [DOI] [PubMed] [Google Scholar]
- 2.Barter, P. J. & Rye, K. A. (1996) Atherosclerosis 121, 1-12. [DOI] [PubMed] [Google Scholar]
- 3.Fielding, C. J. & Fielding, P. E. (1995) J. Lipid Res. 36, 211-228. [PubMed] [Google Scholar]
- 4.Wang, M. & Briggs, M. R. (2004) Chem. Rev. 104, 119-137. [DOI] [PubMed] [Google Scholar]
- 5.Trigatti, B., Rigotti, A. & Krieger, M. (2000) Curr. Opin. Lipidol. 11, 123-131. [DOI] [PubMed] [Google Scholar]
- 6.Rigotti, A., Miettinen, H. E. & Krieger, M. (2003) Endocr. Rev. 24, 357-387. [DOI] [PubMed] [Google Scholar]
- 7.Kozarsky, K. F., Donahee, M. H., Rigotti, A., Iqbal, S. N., Edelman, E. R. & Krieger, M. (1997) Nature 387, 414-417. [DOI] [PubMed] [Google Scholar]
- 8.Wang, N., Arai, T., Ji, Y., Rinninger, F. & Tall, A. R. (1998) J. Biol. Chem. 273, 32920-32926. [DOI] [PubMed] [Google Scholar]
- 9.Ueda, Y., Royer, L., Gong, E., Zhang, J., Cooper, P. N., Francone, O. & Rubin, E. M. (1999) J. Biol. Chem. 274, 7165-7171. [DOI] [PubMed] [Google Scholar]
- 10.Ji, Y., Wang, N., Ramakrishnan, R., Sehayek, E., Huszar, D., Breslow, J. L. & Tall, A. R. (1999) J. Biol. Chem. 274, 33398-33402. [DOI] [PubMed] [Google Scholar]
- 11.Rigotti, A., Trigatti, B. L., Penman, M., Rayburn, H., Herz, J. & Krieger, M. (1997) Proc. Natl. Acad. Sci. USA 94, 12610-12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varban, M. L., Rinninger, F., Wang, N., Fairchild-Huntress, V., Dunmore, J. H., Fang, Q., Gosselin, M. L., Dixon, K. L., Deeds, J. D., Acton, S. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 4619-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mardones, P., Quinones, V., Amigo, L., Moreno, M., Miquel, J. F., Schwarz, M., Miettinen, H. E., Trigatti, B., Krieger, M., VanPatten, S., et al. (2001) J. Lipid Res. 42, 170-180. [PubMed] [Google Scholar]
- 14.Ikemoto, M., Arai, H., Feng, D., Tanaka, K., Aoki, J., Dohmae, N., Takio, K., Adachi, H., Tsujimoto, M. & Inoue, K. (2000) Proc. Natl. Acad. Sci. USA 97, 6538-6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kocher, O., Comella, N., Tognazzi, K. & Brown, L. F. (1998) Lab. Invest. 78, 117-125. [PubMed] [Google Scholar]
- 16.Wang, S., Yue, H., Derin, R. B., Guggino, W. B. & Li, M. (2000) Cell 103, 169-179. [DOI] [PubMed] [Google Scholar]
- 17.Kocher, O., Comella, N., Gilchrist, A., Pal, R., Tognazzi, K., Brown, L. F. & Knoll, J. H. (1999) Lab. Invest. 79, 1161-1170. [PubMed] [Google Scholar]
- 18.Gentzsch, M., Cui, L., Mengos, A., Chang, X. B., Chen, J. H. & Riordan, J. R. (2003) J. Biol. Chem. 278, 6440-6449. [DOI] [PubMed] [Google Scholar]
- 19.Gisler, S. M., Stagljar, I., Traebert, M., Bacic, D., Biber, J. & Murer, H. (2001) J. Biol. Chem. 276, 9206-9213. [DOI] [PubMed] [Google Scholar]
- 20.Gisler, S. M., Pribanic, S., Bacic, D., Forrer, P., Gantenbein, A., Sabourin, L. A., Tsuji, A., Zhao, Z. S., Manser, E., Biber, J. & Murer, H. (2003) Kidney Int. 64, 1733-1745. [DOI] [PubMed] [Google Scholar]
- 21.Custer, M., Spindler, B., Verrey, F., Murer, H. & Biber, J. (1997) Am. J. Physiol. 273, F801-F806. [DOI] [PubMed] [Google Scholar]
- 22.Kocher, O., Pal, R., Roberts, M., Cirovic, C. & Gilchrist, A. (2003) Mol. Cell. Biol. 23, 1175-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kocher, O., Yesilaltay, A., Cirovic, C., Pal, R., Rigotti, A. & Krieger, M. (2003) J. Biol. Chem. 278, 52820-52825. [DOI] [PubMed] [Google Scholar]
- 24.Pinna, L. A. & Ruzzene, M. (1996) Biochim. Biophys. Acta 1314, 191-225. [DOI] [PubMed] [Google Scholar]
- 25.Amatuzio, D. S., Grande, F. & Wada, S. (1962) Metabolism 11, 1240-1249. [PubMed] [Google Scholar]
- 26.Eaton, R. P. (1973) J. Lipid Res. 14, 312-318. [PubMed] [Google Scholar]
- 27.Guettet, C., Rostaqui, N., Mathe, D., Lecuyer, B., Navarro, N. & Jacotot, B. (1991) Lipids 26, 451-458. [DOI] [PubMed] [Google Scholar]
- 28.Rudling, M. & Angelin, B. (1993) J. Clin. Invest. 91, 2796-2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gisler, S. M., Madjdpour, C., Bacic, D., Pribanic, S., Taylor, S. S., Biber, J. & Murer, H. (2003) Kidney Int. 64, 1746-1754. [DOI] [PubMed] [Google Scholar]
- 30.Huang, L. J., Durick, K., Weiner, J. A., Chun, J. & Taylor, S. S. (1997) Proc. Natl. Acad. Sci. USA 94, 11184-11189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burnett, P. E., Blackshaw, S., Lai, M. M., Qureshi, I. A., Burnett, A. F., Sabatini, D. M. & Snyder, S. H. (1998) Proc. Natl. Acad. Sci. USA 95, 8351-8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemenoff, R. A., Gunsalus, J. R. & Avruch, J. (1986) Arch. Biochem. Biophys. 245, 196-203. [DOI] [PubMed] [Google Scholar]
- 33.Unger, R. H. (1983) Diabetes 32, 575-583. [DOI] [PubMed] [Google Scholar]
- 34.Vaughan, G. M., Becker, R. A., Unger, R. H., Ziegler, M. G., Siler-Khodr, T. M., Pruitt, B. A., Jr., & Mason, A. D., Jr. (1985) Metabolism 34, 637-641. [DOI] [PubMed] [Google Scholar]
- 35.Knapp, M. L., al-Sheibani, S. & Riches, P. G. (1991) Clin. Chem. 37, 2093-2101. [PubMed] [Google Scholar]
- 36.Guettet, C., Mathe, D., Riottot, M. & Lutton, C. (1988) Biochim. Biophys. Acta 963, 215-223. [DOI] [PubMed] [Google Scholar]
- 37.Botham, K. M. & Bravo, E. (1995) Prog. Lipid Res. 34, 71-97. [DOI] [PubMed] [Google Scholar]