Abstract
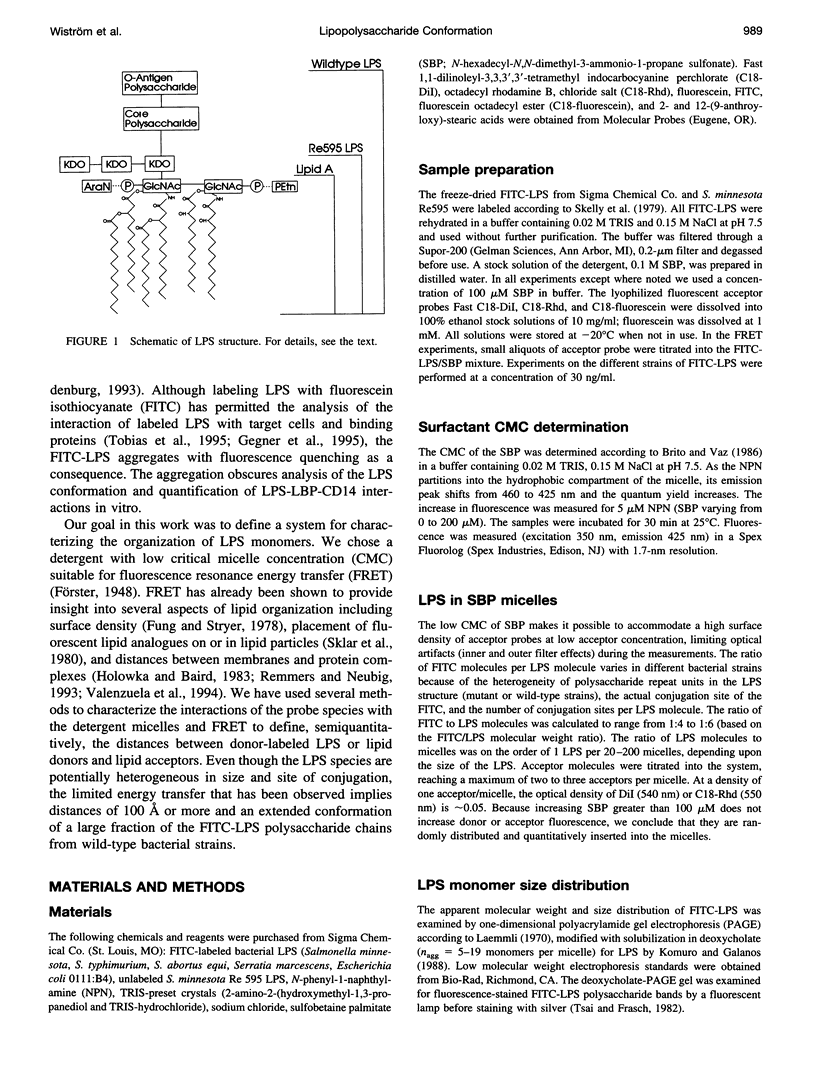
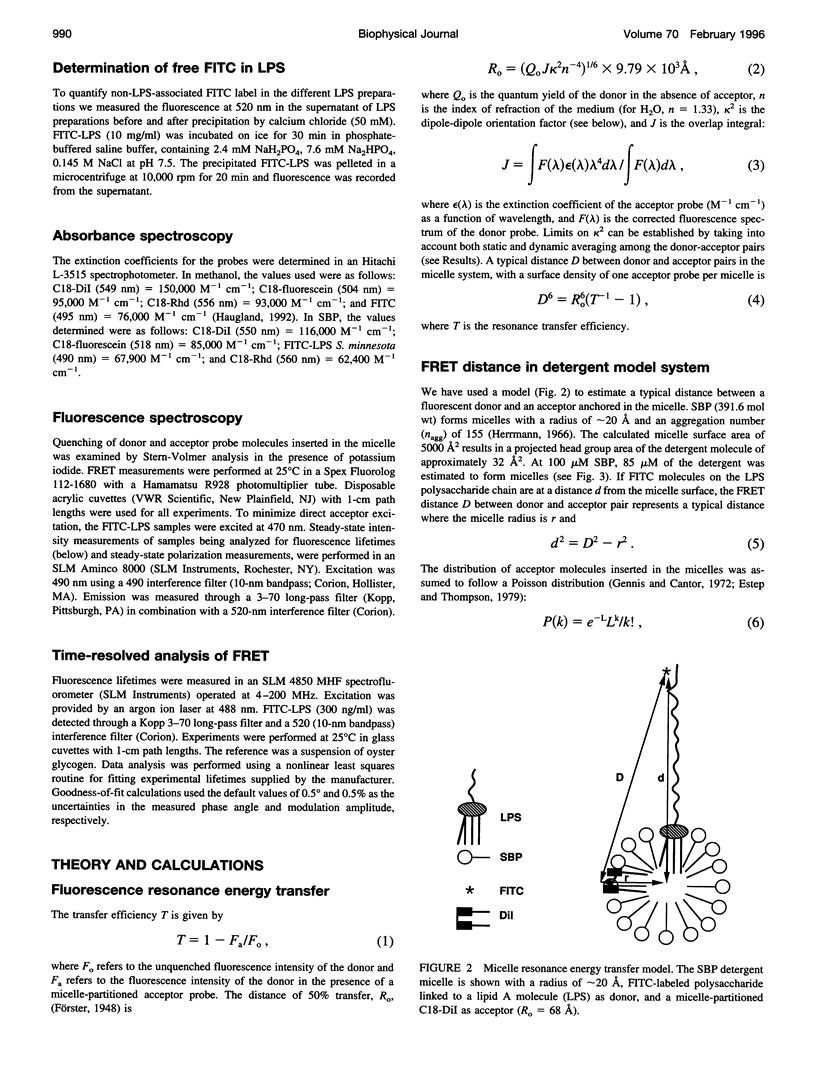
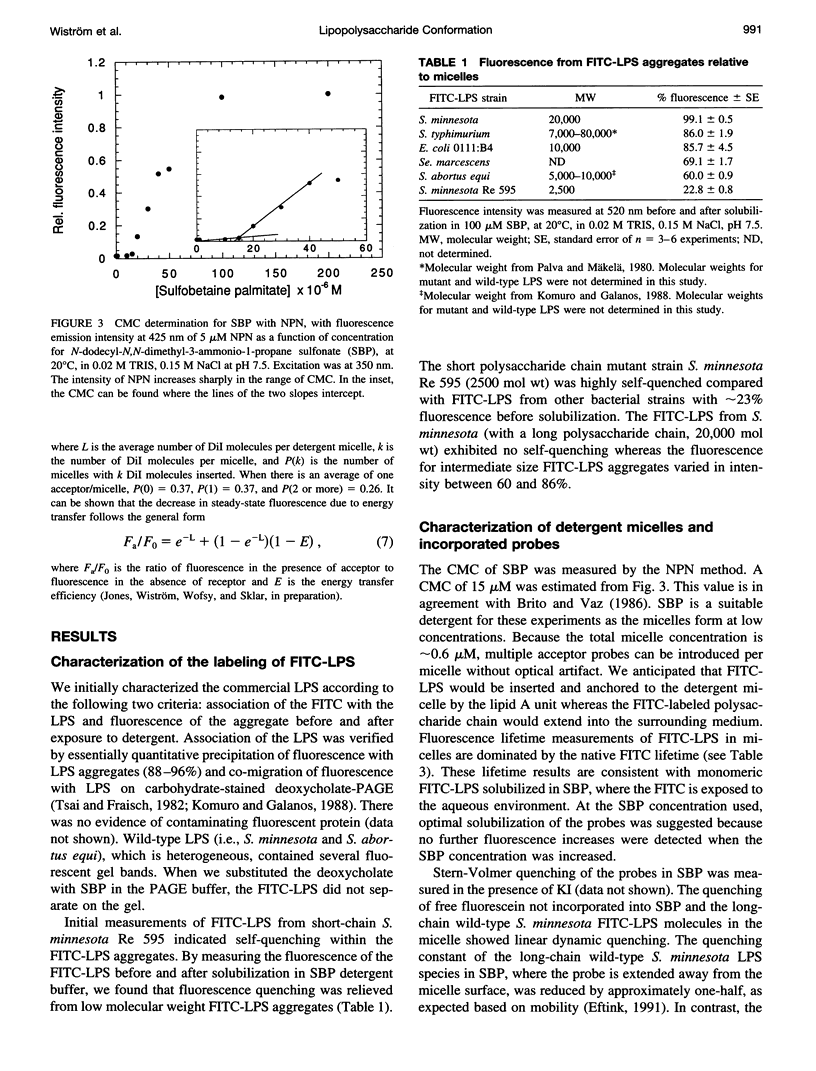
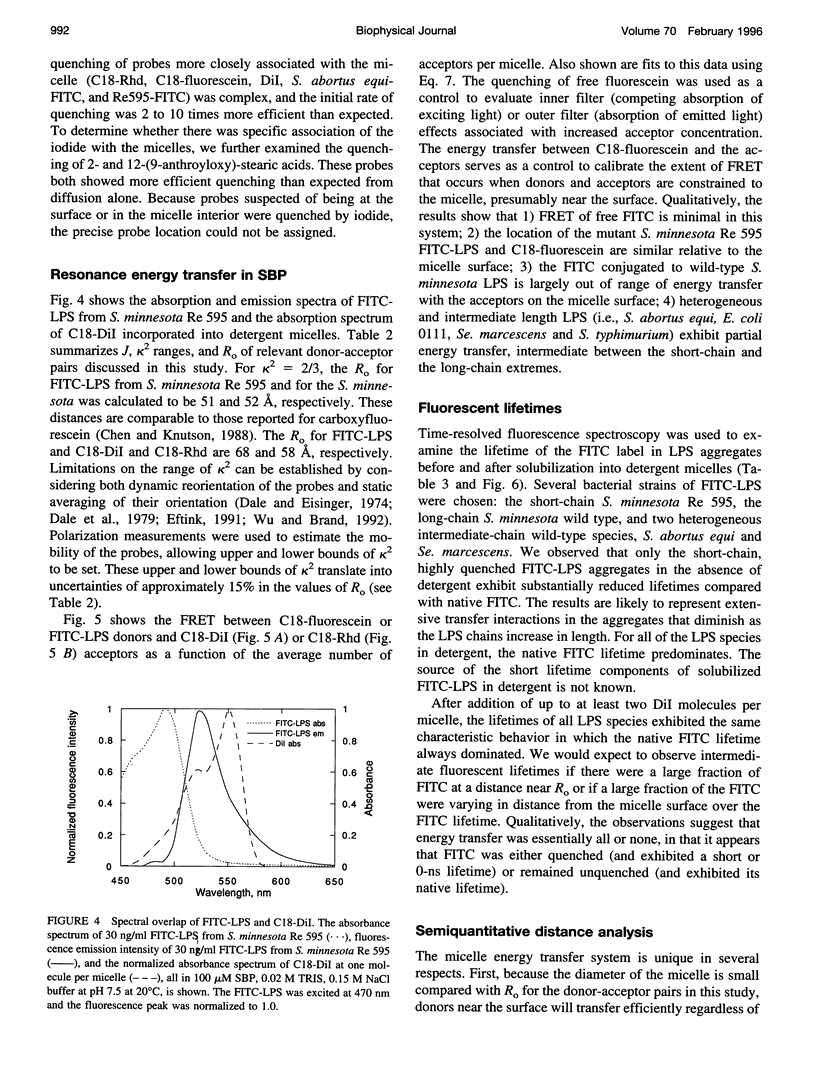
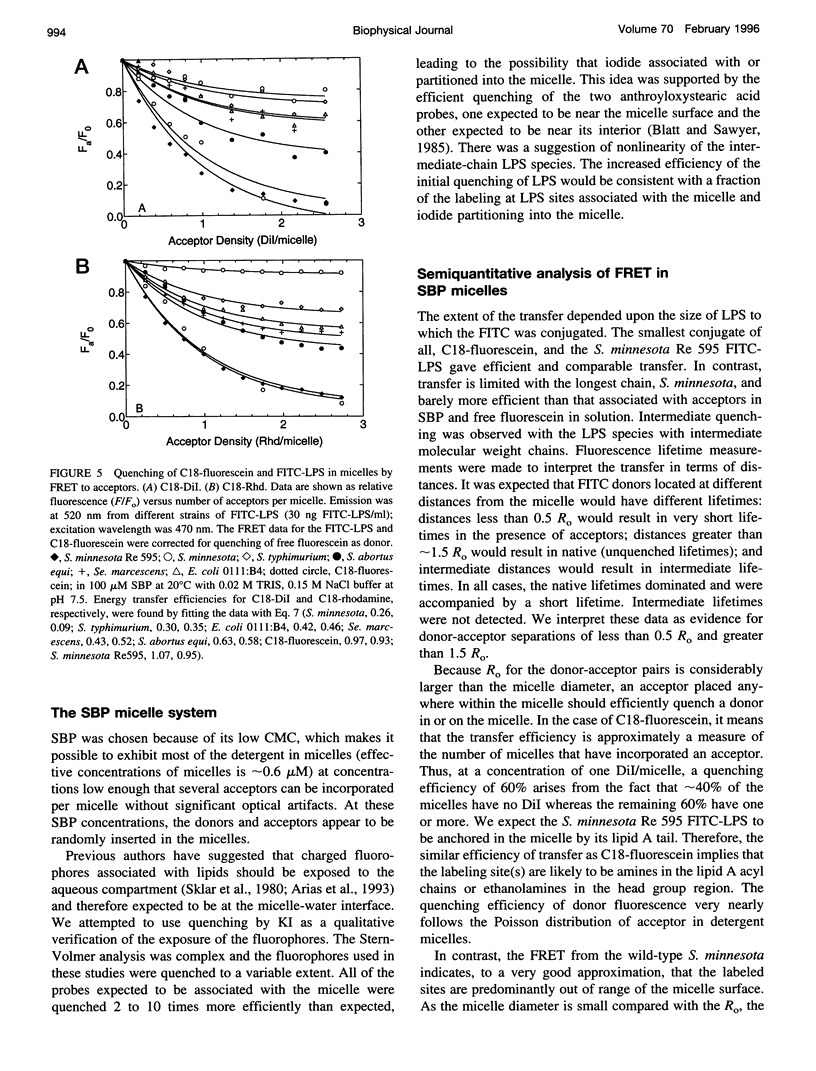
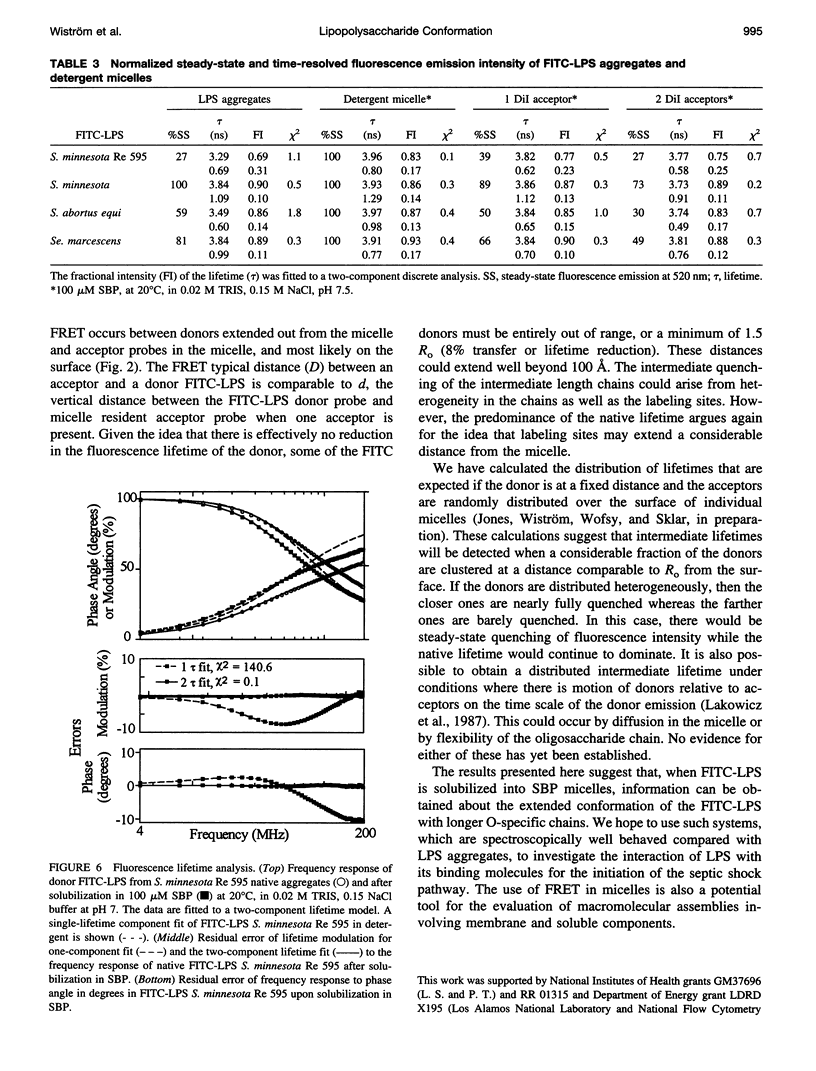
Bacterial endotoxins or lipopolysaccharides (LPS), cell wall components of gram-negative bacteria, are involved in septic shock. LPS consists of a lipid A tail attached to core and O-antigen polysaccharides, but little is known about the supramolecular structure of LPS in blood. We have developed an approach to locate donor and acceptor probes in sulfobetaine palmitate detergent micelles using steady-state and time-resolved fluorescence resonance energy transfer. C18-fluorescein and several LPS species of varying molecular weight labeled with fluorescein isothiocyanate (FITC-LPS) were the donor probes. Acceptor probes were 1,1-dilinoleyl-3,3,3',3'-tetramethyl indocarbocyanine perchlorate (Fast C18-Dil, Ro approximately 68 A), and octadecyl B rhodamine chloride (C18-Rhd, Ro approximately 58 A). With either acceptor, the transfer was of similar high efficiency when FITC-LPS Salmonella minnesota Re 595 (2,500 mol wt, lacking both core and O-antigen) or C18-fluorescein were the fluorescent donor probes. Thus, the donor FITC-LPS with short polysaccharide chain S. minnesota Re 595 and the control donor C18-fluorescein appear to be close to the micelle surface. The transfer efficiency decreased as the molecular weight of the LPS increased. Separation distances between the longest FITC-LPS, S. minnesota (20,000 mol wt, with a long O-antigen), and the micelle were estimated to be 1.5 Ro or more (approximately 100 A), consistent with an extended conformation for the longer O-antigen polysaccharide chain in the detergent.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arias H. R., Valenzuela C. F., Johnson D. A. Quinacrine and ethidium bind to different loci on the Torpedo acetylcholine receptor. Biochemistry. 1993 Jun 22;32(24):6237–6242. doi: 10.1021/bi00075a017. [DOI] [PubMed] [Google Scholar]
- Blatt E., Sawyer W. H. Depth-dependent fluorescent quenching in micelles and membranes. Biochim Biophys Acta. 1985 Jun 12;822(1):43–62. doi: 10.1016/0304-4157(85)90003-6. [DOI] [PubMed] [Google Scholar]
- Brade L., Brandenburg K., Kuhn H. M., Kusumoto S., Macher I., Rietschel E. T., Brade H. The immunogenicity and antigenicity of lipid A are influenced by its physicochemical state and environment. Infect Immun. 1987 Nov;55(11):2636–2644. doi: 10.1128/iai.55.11.2636-2644.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg K. Fourier transform infrared spectroscopy characterization of the lamellar and nonlamellar structures of free lipid A and Re lipopolysaccharides from Salmonella minnesota and Escherichia coli. Biophys J. 1993 Apr;64(4):1215–1231. doi: 10.1016/S0006-3495(93)81488-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg K., Koch M. H., Seydel U. Phase diagram of deep rough mutant lipopolysaccharide from Salmonella minnesota R595. J Struct Biol. 1992 Mar-Apr;108(2):93–106. doi: 10.1016/1047-8477(92)90010-8. [DOI] [PubMed] [Google Scholar]
- Brandenburg K., Koch M. H., Seydel U. Phase diagram of lipid A from Salmonella minnesota and Escherichia coli rough mutant lipopolysaccharide. J Struct Biol. 1990 Oct-Dec;105(1-3):11–21. doi: 10.1016/1047-8477(90)90093-r. [DOI] [PubMed] [Google Scholar]
- Brandenburg K., Mayer H., Koch M. H., Weckesser J., Rietschel E. T., Seydel U. Influence of the supramolecular structure of free lipid A on its biological activity. Eur J Biochem. 1993 Dec 1;218(2):555–563. doi: 10.1111/j.1432-1033.1993.tb18409.x. [DOI] [PubMed] [Google Scholar]
- Brito R. M., Vaz W. L. Determination of the critical micelle concentration of surfactants using the fluorescent probe N-phenyl-1-naphthylamine. Anal Biochem. 1986 Feb 1;152(2):250–255. doi: 10.1016/0003-2697(86)90406-9. [DOI] [PubMed] [Google Scholar]
- Chen R. F., Knutson J. R. Mechanism of fluorescence concentration quenching of carboxyfluorescein in liposomes: energy transfer to nonfluorescent dimers. Anal Biochem. 1988 Jul;172(1):61–77. doi: 10.1016/0003-2697(88)90412-5. [DOI] [PubMed] [Google Scholar]
- Dale R. E., Eisinger J., Blumberg W. E. The orientational freedom of molecular probes. The orientation factor in intramolecular energy transfer. Biophys J. 1979 May;26(2):161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftink M. R. Fluorescence techniques for studying protein structure. Methods Biochem Anal. 1991;35:127–205. doi: 10.1002/9780470110560.ch3. [DOI] [PubMed] [Google Scholar]
- Estep T. N., Thompson T. E. Energy transfer in lipid bilayers. Biophys J. 1979 May;26(2):195–207. doi: 10.1016/S0006-3495(79)85244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung B. K., Stryer L. Surface density determination in membranes by fluorescence energy transfer. Biochemistry. 1978 Nov 28;17(24):5241–5248. doi: 10.1021/bi00617a025. [DOI] [PubMed] [Google Scholar]
- Galanos C., Lüderitz O. Electrodialysis of lipopolysaccharides and their conversion to uniform salt forms. Eur J Biochem. 1975 Jun;54(2):603–610. doi: 10.1111/j.1432-1033.1975.tb04172.x. [DOI] [PubMed] [Google Scholar]
- Gallay P., Heumann D., Le Roy D., Barras C., Glauser M. P. Lipopolysaccharide-binding protein as a major plasma protein responsible for endotoxemic shock. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):9935–9938. doi: 10.1073/pnas.90.21.9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegner J. A., Ulevitch R. J., Tobias P. S. Lipopolysaccharide (LPS) signal transduction and clearance. Dual roles for LPS binding protein and membrane CD14. J Biol Chem. 1995 Mar 10;270(10):5320–5325. doi: 10.1074/jbc.270.10.5320. [DOI] [PubMed] [Google Scholar]
- Gennis R. B., Cantor C. R. Use of nonspecific dye labeling for singlet energy-transfer measurements in complex systems. A simple model. Biochemistry. 1972 Jun 20;11(13):2509–2517. doi: 10.1021/bi00763a020. [DOI] [PubMed] [Google Scholar]
- Holst O., Müller-Loennies S., Lindner B., Brade H. Chemical structure of the lipid A of Escherichia coli J-5. Eur J Biochem. 1993 Jun 15;214(3):695–701. doi: 10.1111/j.1432-1033.1993.tb17970.x. [DOI] [PubMed] [Google Scholar]
- Kastowsky M., Gutberlet T., Bradaczek H. Comparison of X-ray powder-diffraction data of various bacterial lipopolysaccharide structures with theoretical model conformations. Eur J Biochem. 1993 Oct 15;217(2):771–779. doi: 10.1111/j.1432-1033.1993.tb18305.x. [DOI] [PubMed] [Google Scholar]
- Kastowsky M., Gutberlet T., Bradaczek H. Molecular modelling of the three-dimensional structure and conformational flexibility of bacterial lipopolysaccharide. J Bacteriol. 1992 Jul;174(14):4798–4806. doi: 10.1128/jb.174.14.4798-4806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro T., Galanos C. Analysis of Salmonella lipopolysaccharides by sodium deoxycholate-polyacrylamide gel electrophoresis. J Chromatogr. 1988 Oct 26;450(3):381–387. doi: 10.1016/s0021-9673(01)83593-7. [DOI] [PubMed] [Google Scholar]
- Komuro T., Nakazawa R. Detection of low molecular size lipopolysaccharide contaminated in dialysates used for hemodialysis therapy with polyacrylamide gel electrophoresis in the presence of sodium deoxycholate. Int J Artif Organs. 1993 May;16(5):245–248. [PubMed] [Google Scholar]
- Labischinski H., Barnickel G., Bradaczek H., Naumann D., Rietschel E. T., Giesbrecht P. High state of order of isolated bacterial lipopolysaccharide and its possible contribution to the permeation barrier property of the outer membrane. J Bacteriol. 1985 Apr;162(1):9–20. doi: 10.1128/jb.162.1.9-20.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Cherek H., Gryczynski I., Joshi N., Johnson M. L. Analysis of fluorescence decay kinetics measured in the frequency domain using distributions of decay times. Biophys Chem. 1987 Oct;28(1):35–50. doi: 10.1016/0301-4622(87)80073-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. D., Kravchenko V., Kirkland T. N., Han J., Mackman N., Moriarty A., Leturcq D., Tobias P. S., Ulevitch R. J. Glycosyl-phosphatidylinositol-anchored or integral membrane forms of CD14 mediate identical cellular responses to endotoxin. Proc Natl Acad Sci U S A. 1993 Nov 1;90(21):9930–9934. doi: 10.1073/pnas.90.21.9930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B., Van Alphen L. Molecular architecture and functioning of the outer membrane of Escherichia coli and other gram-negative bacteria. Biochim Biophys Acta. 1983 Mar 21;737(1):51–115. doi: 10.1016/0304-4157(83)90014-x. [DOI] [PubMed] [Google Scholar]
- Lüderitz T., Brandenburg K., Seydel U., Roth A., Galanos C., Rietschel E. T. Structural and physicochemical requirements of endotoxins for the activation of arachidonic acid metabolism in mouse peritoneal macrophages in vitro. Eur J Biochem. 1989 Jan 15;179(1):11–16. doi: 10.1111/j.1432-1033.1989.tb14514.x. [DOI] [PubMed] [Google Scholar]
- Nikaido H., Vaara M. Molecular basis of bacterial outer membrane permeability. Microbiol Rev. 1985 Mar;49(1):1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva E. T., Mäkelä P. H. Lipopolysaccharide heterogeneity in Salmonella typhimurium analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis. Eur J Biochem. 1980;107(1):137–143. doi: 10.1111/j.1432-1033.1980.tb04634.x. [DOI] [PubMed] [Google Scholar]
- Peterson A. A., McGroarty E. J. High-molecular-weight components in lipopolysaccharides of Salmonella typhimurium, Salmonella minnesota, and Escherichia coli. J Bacteriol. 1985 May;162(2):738–745. doi: 10.1128/jb.162.2.738-745.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugin J., Schürer-Maly C. C., Leturcq D., Moriarty A., Ulevitch R. J., Tobias P. S. Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipopolysaccharide-binding protein and soluble CD14. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2744–2748. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi N., Mascagni P., Ribi E., Takayama K. Monophosphoryl lipid A obtained from lipopolysaccharides of Salmonella minnesota R595. Purification of the dimethyl derivative by high performance liquid chromatography and complete structural determination. J Biol Chem. 1985 May 10;260(9):5271–5278. [PubMed] [Google Scholar]
- Raetz C. R. Biochemistry of endotoxins. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- Raetz C. R., Ulevitch R. J., Wright S. D., Sibley C. H., Ding A., Nathan C. F. Gram-negative endotoxin: an extraordinary lipid with profound effects on eukaryotic signal transduction. FASEB J. 1991 Sep;5(12):2652–2660. doi: 10.1096/fasebj.5.12.1916089. [DOI] [PubMed] [Google Scholar]
- Remmers A. E., Neubig R. R. Resonance energy transfer between guanine nucleotide binding protein subunits and membrane lipids. Biochemistry. 1993 Mar 9;32(9):2409–2414. doi: 10.1021/bi00060a036. [DOI] [PubMed] [Google Scholar]
- Rietschel E. T., Kirikae T., Schade F. U., Mamat U., Schmidt G., Loppnow H., Ulmer A. J., Zähringer U., Seydel U., Di Padova F. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994 Feb;8(2):217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- Schumann R. R., Leong S. R., Flaggs G. W., Gray P. W., Wright S. D., Mathison J. C., Tobias P. S., Ulevitch R. J. Structure and function of lipopolysaccharide binding protein. Science. 1990 Sep 21;249(4975):1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- Seydel U., Brandenburg K., Koch M. H., Rietschel E. T. Supramolecular structure of lipopolysaccharide and free lipid A under physiological conditions as determined by synchrotron small-angle X-ray diffraction. Eur J Biochem. 1989 Dec 8;186(1-2):325–332. doi: 10.1111/j.1432-1033.1989.tb15212.x. [DOI] [PubMed] [Google Scholar]
- Skelly R. R., Munkenbeck P., Morrison D. C. Stimulation of T-independent antibody responses by hapten-lipopolysaccharides without repeating polymeric structure. Infect Immun. 1979 Feb;23(2):287–293. doi: 10.1128/iai.23.2.287-293.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar L. A., Doody M. C., Gotto A. M., Jr, Pownall H. J. Serum lipoprotein structure: resonance energy transfer localization of fluorescent lipid probes. Biochemistry. 1980 Apr 1;19(7):1294–1301. doi: 10.1021/bi00548a005. [DOI] [PubMed] [Google Scholar]
- Tobias P. S., Soldau K., Gegner J. A., Mintz D., Ulevitch R. J. Lipopolysaccharide binding protein-mediated complexation of lipopolysaccharide with soluble CD14. J Biol Chem. 1995 May 5;270(18):10482–10488. doi: 10.1074/jbc.270.18.10482. [DOI] [PubMed] [Google Scholar]
- Tobias P. S., Soldau K., Ulevitch R. J. Identification of a lipid A binding site in the acute phase reactant lipopolysaccharide binding protein. J Biol Chem. 1989 Jun 25;264(18):10867–10871. [PubMed] [Google Scholar]
- Tsai C. M., Frasch C. E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982 Jan 1;119(1):115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Valenzuela C. F., Weign P., Yguerabide J., Johnson D. A. Transverse distance between the membrane and the agonist binding sites on the Torpedo acetylcholine receptor: a fluorescence study. Biophys J. 1994 Mar;66(3 Pt 1):674–682. doi: 10.1016/s0006-3495(94)80841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenweber H. W., Seydel U., Lindner B., Lüderitz O., Rietschel E. T. Nature and location of amide-bound (R)-3-acyloxyacyl groups in lipid A of lipopolysaccharides from various gram-negative bacteria. Eur J Biochem. 1984 Dec 3;145(2):265–272. doi: 10.1111/j.1432-1033.1984.tb08547.x. [DOI] [PubMed] [Google Scholar]
- Wright S. D., Ramos R. A., Tobias P. S., Ulevitch R. J., Mathison J. C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990 Sep 21;249(4975):1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- Wu P., Brand L. Orientation factor in steady-state and time-resolved resonance energy transfer measurements. Biochemistry. 1992 Sep 1;31(34):7939–7947. doi: 10.1021/bi00149a027. [DOI] [PubMed] [Google Scholar]
- Yeh H. Y., Jacobs D. M. Characterization of lipopolysaccharide fractions and their interactions with cells and model membranes. J Bacteriol. 1992 Jan;174(1):336–341. doi: 10.1128/jb.174.1.336-341.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alphen L., Verkleij A., Burnell E., Lugtenberg B. 31P nuclear magnetic resonance and freeze-fracture electron microscopy studies on Escherichia coli. II. Lipopolysaccharide and lipopolysaccharide-phospholipid complexes. Biochim Biophys Acta. 1980 Apr 24;597(3):502–517. doi: 10.1016/0005-2736(80)90223-0. [DOI] [PubMed] [Google Scholar]


