Abstract
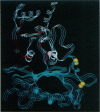
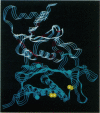
Light-dependent reduction of target disulfides on certain chloroplast enzymes results in a change in activity. We have modeled the tertiary structure of four of these enzymes, namely NADP-linked glyceraldehyde-3-P dehydrogenase, NADP-linked malate dehydrogenase, sedoheptulose bisphosphatase, and fructose bisphosphatase. Models are based on x-ray crystal structures from non-plant species. Each of these enzymes consists of two domains connected by a hinge. Modeling suggests that oxidation of two crucial cysteines to cystine would restrict motion around the hinge in the two dehydrogenases and influence the conformation of the active site. The cysteine residues in the two phosphatases are located in a region known to be sensitive to allosteric modifiers and to be involved in mediating structural changes in mammalian and microbial fructose bisphosphatases. Apparently, the same region is involved in covalent modification of phosphatase activity in the chloroplast.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Biesecker G., Harris J. I., Thierry J. C., Walker J. E., Wonacott A. J. Sequence and structure of D-glyceraldehyde 3-phosphate dehydrogenase from Bacillus stearothermophilus. Nature. 1977 Mar 24;266(5600):328–333. doi: 10.1038/266328a0. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Bradshaw R. A., Banaszak L. J. Structure of porcine heart cytoplasmic malate dehydrogenase: combining X-ray diffraction and chemical sequence data in structural studies. Biochemistry. 1987 May 19;26(10):2722–2734. doi: 10.1021/bi00384a011. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Rhodes G., Banaszak L. J. Refined crystal structure of cytoplasmic malate dehydrogenase at 2.5-A resolution. Biochemistry. 1989 Jul 11;28(14):6065–6081. doi: 10.1021/bi00440a051. [DOI] [PubMed] [Google Scholar]
- Brinkmann H., Cerff R., Salomon M., Soll J. Cloning and sequence analysis of cDNAs encoding the cytosolic precursors of subunits GapA and GapB of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinach. Plant Mol Biol. 1989 Jul;13(1):81–94. doi: 10.1007/BF00027337. [DOI] [PubMed] [Google Scholar]
- Crétin C., Luchetta P., Joly C., Decottignies P., Lepiniec L., Gadal P., Sallantin M., Huet J. C., Pernollet J. C. Primary structure of sorghum malate dehydrogenase (NADP) deduced from cDNA sequence. Homology with malate dehydrogenase (NAD). Eur J Biochem. 1990 Sep 11;192(2):299–303. doi: 10.1111/j.1432-1033.1990.tb19227.x. [DOI] [PubMed] [Google Scholar]
- Ferri G., Stoppini M., Meloni M. L., Zapponi M. C., Iadarola P. Chloroplast glyceraldehyde-3-phosphate dehydrogenase (NADP): amino acid sequence of the subunits from isoenzyme I and structural relationship with isoenzyme II. Biochim Biophys Acta. 1990 Oct 18;1041(1):36–42. doi: 10.1016/0167-4838(90)90119-z. [DOI] [PubMed] [Google Scholar]
- Fickenscher K., Scheibe R. Limited proteolysis of inactive tetrameric chloroplast NADP-malate dehydrogenase produces active dimers. Arch Biochem Biophys. 1988 Feb 1;260(2):771–779. doi: 10.1016/0003-9861(88)90507-3. [DOI] [PubMed] [Google Scholar]
- Goldenberg D. P. Native and non-native intermediates in the BPTI folding pathway. Trends Biochem Sci. 1992 Jul;17(7):257–261. doi: 10.1016/0968-0004(92)90405-x. [DOI] [PubMed] [Google Scholar]
- Hatch M. D., Agostino A. Bilevel disulfide group reduction in the activation of c(4) leaf nicotinamide adenine dinucleotide phosphate-malate dehydrogenase. Plant Physiol. 1992 Sep;100(1):360–366. doi: 10.1104/pp.100.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill E., Tsernoglou D., Webb L., Banaszak L. J. Polypeptide conformation of cytoplasmic malate dehydrogenase from an electron density map at 3.0 angstrom resolution. J Mol Biol. 1972 Dec 30;72(3):577–589. doi: 10.1016/0022-2836(72)90176-3. [DOI] [PubMed] [Google Scholar]
- Horsnell P. R., Raines C. A. Nucleotide sequence of a cDNA clone encoding chloroplast fructose-1,6-bisphosphatase from Arabidopsis thaliana. Plant Mol Biol. 1991 Jul;17(1):185–186. doi: 10.1007/BF00036829. [DOI] [PubMed] [Google Scholar]
- Issakidis E., Decottignies P., Miginiac-Maslow M. A thioredoxin-independent fully active NADP-malate dehydrogenase obtained by site-directed mutagenesis. FEBS Lett. 1993 Apr 19;321(1):55–58. doi: 10.1016/0014-5793(93)80620-a. [DOI] [PubMed] [Google Scholar]
- Issakidis E., Miginiac-Maslow M., Decottignies P., Jacquot J. P., Crétin C., Gadal P. Site-directed mutagenesis reveals the involvement of an additional thioredoxin-dependent regulatory site in the activation of recombinant sorghum leaf NADP-malate dehydrogenase. J Biol Chem. 1992 Oct 25;267(30):21577–21583. [PubMed] [Google Scholar]
- Issakidis E., Saarinen M., Decottignies P., Jacquot J. P., Crétin C., Gadal P., Miginiac-Maslow M. Identification and characterization of the second regulatory disulfide bridge of recombinant sorghum leaf NADP-malate dehydrogenase. J Biol Chem. 1994 Feb 4;269(5):3511–3517. [PubMed] [Google Scholar]
- Jackson R. M., Sessions R. B., Holbrook J. J. A prediction of the three-dimensional structure of maize NADP(+)-dependent malate dehydrogenase which explains aspects of light-dependent regulation unique to plant enzymes. J Comput Aided Mol Des. 1992 Feb;6(1):1–18. doi: 10.1007/BF00124383. [DOI] [PubMed] [Google Scholar]
- Ke H. M., Liang J. Y., Zhang Y. P., Lipscomb W. N. Conformational transition of fructose-1,6-bisphosphatase: structure comparison between the AMP complex (T form) and the fructose 6-phosphate complex (R form). Biochemistry. 1991 May 7;30(18):4412–4420. doi: 10.1021/bi00232a007. [DOI] [PubMed] [Google Scholar]
- Ke H. M., Thorpe C. M., Seaton B. a., Lipscomb W. N., Marcus F. Structure refinement of fructose-1,6-bisphosphatase and its fructose 2,6-bisphosphate complex at 2.8 A resolution. J Mol Biol. 1990 Apr 5;212(3):513–539. doi: 10.1016/0022-2836(90)90329-k. [DOI] [PubMed] [Google Scholar]
- Liang J. Y., Zhang Y., Huang S., Lipscomb W. N. Allosteric transition of fructose-1,6-bisphosphatase. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2132–2136. doi: 10.1073/pnas.90.6.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaud M. F., Zhang D. X., Cerff R. Differential intron loss and endosymbiotic transfer of chloroplast glyceraldehyde-3-phosphate dehydrogenase genes to the nucleus. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8918–8922. doi: 10.1073/pnas.87.22.8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd J. C., Raines C. A., John U. P., Dyer T. A. The chloroplast FBPase gene of wheat: structure and expression of the promoter in photosynthetic and meristematic cells of transgenic tobacco plants. Mol Gen Genet. 1991 Feb;225(2):209–216. doi: 10.1007/BF00269850. [DOI] [PubMed] [Google Scholar]
- Luchetta P., Cretin C., Gadal P. Structure and characterization of the Sorghum vulgare gene encoding NADP-malate dehydrogenase. Gene. 1990 May 14;89(2):171–177. doi: 10.1016/0378-1119(90)90003-a. [DOI] [PubMed] [Google Scholar]
- Marcus F., Edelstein I., Reardon I., Heinrikson R. L. Complete amino acid sequence of pig kidney fructose-1,6-bisphosphatase. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7161–7165. doi: 10.1073/pnas.79.23.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus F., Harrsch P. B. Amino acid sequence of spinach chloroplast fructose-1,6-bisphosphatase. Arch Biochem Biophys. 1990 May 15;279(1):151–157. doi: 10.1016/0003-9861(90)90475-e. [DOI] [PubMed] [Google Scholar]
- Marcus F., Harrsch P. B., Moberly L., Edelstein I., Latshaw S. P. Spinach chloroplast fructose-1,6-bisphosphatase: identification of the subtilisin-sensitive region and of conserved histidines. Biochemistry. 1987 Nov 3;26(22):7029–7035. doi: 10.1021/bi00396a026. [DOI] [PubMed] [Google Scholar]
- Marcus F., Moberly L., Latshaw S. P. Comparative amino acid sequence of fructose-1,6-bisphosphatases: identification of a region unique to the light-regulated chloroplast enzyme. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5379–5383. doi: 10.1073/pnas.85.15.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Cerff R. Prokaryotic features of a nucleus-encoded enzyme. cDNA sequences for chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenases from mustard (Sinapis alba). Eur J Biochem. 1986 Sep 1;159(2):323–331. doi: 10.1111/j.1432-1033.1986.tb09871.x. [DOI] [PubMed] [Google Scholar]
- Putkey J. A., Dotson D. G., Mouawad P. Formation of inter- and intramolecular disulfide bonds can activate cardiac troponin C. J Biol Chem. 1993 Apr 5;268(10):6827–6830. [PubMed] [Google Scholar]
- Quigley F., Brinkmann H., Martin W. F., Cerff R. Strong functional GC pressure in a light-regulated maize gene encoding subunit GAPA of chloroplast glyceraldehyde-3-phosphate dehydrogenase: implications for the evolution of GAPA pseudogenes. J Mol Evol. 1989 Nov;29(5):412–421. doi: 10.1007/BF02602911. [DOI] [PubMed] [Google Scholar]
- Quigley F., Martin W. F., Cerff R. Intron conservation across the prokaryote-eukaryote boundary: structure of the nuclear gene for chloroplast glyceraldehyde-3-phosphate dehydrogenase from maize. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2672–2676. doi: 10.1073/pnas.85.8.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines C. A., Lloyd J. C., Longstaff M., Bradley D., Dyer T. Chloroplast fructose-1,6-bisphosphatase: the product of a mosaic gene. Nucleic Acids Res. 1988 Aug 25;16(16):7931–7942. doi: 10.1093/nar/16.16.7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines C. A., Lloyd J. C., Willingham N. M., Potts S., Dyer T. A. cDNA and gene sequences of wheat chloroplast sedoheptulose-1,7-bisphosphatase reveal homology with fructose-1,6-bisphosphatases. Eur J Biochem. 1992 May 1;205(3):1053–1059. doi: 10.1111/j.1432-1033.1992.tb16873.x. [DOI] [PubMed] [Google Scholar]
- Reng W., Riessland R., Scheibe R., Jaenicke R. Cloning, site-specific mutagenesis, expression and characterization of full-length chloroplast NADP-malate dehydrogenase from Pisum sativum. Eur J Biochem. 1993 Oct 1;217(1):189–197. doi: 10.1111/j.1432-1033.1993.tb18233.x. [DOI] [PubMed] [Google Scholar]
- Scheibe R., Kampfenkel K., Wessels R., Tripier D. Primary structure and analysis of the location of the regulatory disulfide bond of pea chloroplast NADP-malate dehydrogenase. Biochim Biophys Acta. 1991 Jan 8;1076(1):1–8. doi: 10.1016/0167-4838(91)90212-i. [DOI] [PubMed] [Google Scholar]
- Scheibe R. Redox-modulation of chloroplast enzymes : a common principle for individual control. Plant Physiol. 1991 May;96(1):1–3. doi: 10.1104/pp.96.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih M. C., Heinrich P., Goodman H. M. Cloning and chromosomal mapping of nuclear genes encoding chloroplast and cytosolic glyceraldehyde-3-phosphate-dehydrogenase from Arabidopsis thaliana. Gene. 1991 Aug 15;104(2):133–138. doi: 10.1016/0378-1119(91)90242-4. [DOI] [PubMed] [Google Scholar]
- Shih M. C., Lazar G., Goodman H. M. Evidence in favor of the symbiotic origin of chloroplasts: primary structure and evolution of tobacco glyceraldehyde-3-phosphate dehydrogenases. Cell. 1986 Oct 10;47(1):73–80. doi: 10.1016/0092-8674(86)90367-3. [DOI] [PubMed] [Google Scholar]
- Skarzyński T., Moody P. C., Wonacott A. J. Structure of holo-glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus at 1.8 A resolution. J Mol Biol. 1987 Jan 5;193(1):171–187. doi: 10.1016/0022-2836(87)90635-8. [DOI] [PubMed] [Google Scholar]
- Skarzyński T., Wonacott A. J. Coenzyme-induced conformational changes in glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus. J Mol Biol. 1988 Oct 20;203(4):1097–1118. doi: 10.1016/0022-2836(88)90130-1. [DOI] [PubMed] [Google Scholar]
- Weissman J. S., Kim P. S. The pro region of BPTI facilitates folding. Cell. 1992 Nov 27;71(5):841–851. doi: 10.1016/0092-8674(92)90559-u. [DOI] [PubMed] [Google Scholar]
- White J. L., Hackert M. L., Buehner M., Adams M. J., Ford G. C., Lentz P. J., Jr, Smiley I. E., Steindel S. J., Rossmann M. G. A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes. J Mol Biol. 1976 Apr 25;102(4):759–779. doi: 10.1016/0022-2836(76)90290-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liang J. Y., Huang S., Ke H., Lipscomb W. N. Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase. Biochemistry. 1993 Feb 23;32(7):1844–1857. doi: 10.1021/bi00058a019. [DOI] [PubMed] [Google Scholar]