Abstract
8-oxoguanine (8-oxoG), ring-opened purines (formamidopyrimidines or Fapys), and other oxidized DNA base lesions generated by reactive oxygen species are often mutagenic and toxic, and have been implicated in the etiology of many diseases, including cancer, and in aging. Repair of these lesions in all organisms occurs primarily via the DNA base excision repair pathway, initiated with their excision by DNA glycosylase/AP lyases, which are of two classes. One class utilizes an internal Lys residue as the active site nucleophile, and includes Escherichia coli Nth and both known mammalian DNA glycosylase/AP lyases, namely, OGG1 and NTH1. E. coli MutM and its paralog Nei, which comprise the second class, use N-terminal Pro as the active site. Here, we report the presence of two human orthologs of E. coli mutM nei genes in the human genome database, and characterize one of their products. Based on the substrate preference, we have named it NEH1 (Nei homolog). The 44-kDa, wild-type recombinant NEH1, purified to homogeneity from E. coli, excises Fapys from damaged DNA, and oxidized pyrimidines and 8-oxoG from oligodeoxynucleotides. Inactivation of the enzyme because of either deletion of N-terminal Pro or Histag fusion at the N terminus supports the role of N-terminal Pro as its active site. The tissue-specific levels of NEH1 and OGG1 mRNAs are distinct, and S phase-specific increase in NEH1 at both RNA and protein levels suggests that NEH1 is involved in replication-associated repair of oxidized bases.
Reactive oxygen species
(ROS), including the O −
radical, H2O2, and the
OH⋅ radical, may be the most important genotoxic agents because of
their ubiquitous and continuous production as by-products of
respiration (1). They are also generated in mammalian cells during the
inflammatory response, and have been implicated in the etiology of a
variety of pathophysiologies ranging from cancer to rheumatoid
arthritis, cardiovascular diseases, and Alzheimer's disease, and also
in aging (2–4). Genotoxicity of ROS results from their reaction with
DNA to produce a plethora of DNA base lesions as well as strand breaks
(5). All of these lesions except the double-strand breaks are repaired
primarily via the base excision repair (BER) pathway in both bacteria
and eukaryotes (6). BER is initiated with excision of oxidatively
damaged bases by several DNA glycosylase/AP lyases. These
glycosylases have broad substrate ranges, commensurate with the need to
excise lesions of widely different chemical structures by using a
limited number of enzymes. However, these enzymes have preferred
substrates. Thus 8-oxoguanine (8-oxoG), an abundant and arguably the
most critical mutagenic lesion, is repaired primarily by 8-oxoG-DNA
glycosylase (OGG1) in mammalian cells and by MutM/Fpg in
Escherichia coli. Formamidopyrimidines (Fapys), derived from
purines by ROS, are more abundant than 8-oxoG in normal mammalian DNA
(7). Although not extensively investigated, Fapys, which are likely to
inhibit replication (8) and may be mutagenic, are also excised by OGG1
and other DNA glycosylases. A number of pyrimidine lesions, including
5,6 dihydrouracil (DHU) and 5-hydroxyuracil (5-OHU), generated from
cytosine by ionizing radiation and ROS, are excised primarily by
E. coli endonuclease III (Nth) and its mammalian ortholog
NTH1, and also by E. coli endonuclease VIII (Nei; refs. 5,
9, and 10). DNA glycosylase/AP lyases can be divided into two
classes. The two mammalian enzymes, specific for oxidized base lesions
and identified so far, namely, NTH1 and OGG1, belong to the E.
coli Nth class, based on similar structural features and reaction
mechanism (11). These enzymes use an internal Lys residue as the active
site nucleophile, and carry out β elimination (after excising the
damaged base), to cleave the DNA strand, and generate 3′ phospho
α,β-unsaturated aldehyde and 5′ phosphate termini (11, 12). Only
prokaryotic MutM/Fpg and its paralog Nei comprise the second class,
and use N-terminal Pro as the active site nucleophile. These enzymes
carry out βδ-elimination and cleave the DNA strand at the site of
lesion to generate 3′ phosphate and 5′ phosphate termini (13). Both
classes of enzymes form covalent Schiff base intermediates with the
lesion-containing DNA strand, both before and after its cleavage, and
these unstable intermediates can be converted into stable “trapped
complexes” by reduction with NaCNBH3. With
oligo substrates, the mobility of these complexes in SDS/PAGE depends
on the relative size of the enzyme.
−
radical, H2O2, and the
OH⋅ radical, may be the most important genotoxic agents because of
their ubiquitous and continuous production as by-products of
respiration (1). They are also generated in mammalian cells during the
inflammatory response, and have been implicated in the etiology of a
variety of pathophysiologies ranging from cancer to rheumatoid
arthritis, cardiovascular diseases, and Alzheimer's disease, and also
in aging (2–4). Genotoxicity of ROS results from their reaction with
DNA to produce a plethora of DNA base lesions as well as strand breaks
(5). All of these lesions except the double-strand breaks are repaired
primarily via the base excision repair (BER) pathway in both bacteria
and eukaryotes (6). BER is initiated with excision of oxidatively
damaged bases by several DNA glycosylase/AP lyases. These
glycosylases have broad substrate ranges, commensurate with the need to
excise lesions of widely different chemical structures by using a
limited number of enzymes. However, these enzymes have preferred
substrates. Thus 8-oxoguanine (8-oxoG), an abundant and arguably the
most critical mutagenic lesion, is repaired primarily by 8-oxoG-DNA
glycosylase (OGG1) in mammalian cells and by MutM/Fpg in
Escherichia coli. Formamidopyrimidines (Fapys), derived from
purines by ROS, are more abundant than 8-oxoG in normal mammalian DNA
(7). Although not extensively investigated, Fapys, which are likely to
inhibit replication (8) and may be mutagenic, are also excised by OGG1
and other DNA glycosylases. A number of pyrimidine lesions, including
5,6 dihydrouracil (DHU) and 5-hydroxyuracil (5-OHU), generated from
cytosine by ionizing radiation and ROS, are excised primarily by
E. coli endonuclease III (Nth) and its mammalian ortholog
NTH1, and also by E. coli endonuclease VIII (Nei; refs. 5,
9, and 10). DNA glycosylase/AP lyases can be divided into two
classes. The two mammalian enzymes, specific for oxidized base lesions
and identified so far, namely, NTH1 and OGG1, belong to the E.
coli Nth class, based on similar structural features and reaction
mechanism (11). These enzymes use an internal Lys residue as the active
site nucleophile, and carry out β elimination (after excising the
damaged base), to cleave the DNA strand, and generate 3′ phospho
α,β-unsaturated aldehyde and 5′ phosphate termini (11, 12). Only
prokaryotic MutM/Fpg and its paralog Nei comprise the second class,
and use N-terminal Pro as the active site nucleophile. These enzymes
carry out βδ-elimination and cleave the DNA strand at the site of
lesion to generate 3′ phosphate and 5′ phosphate termini (13). Both
classes of enzymes form covalent Schiff base intermediates with the
lesion-containing DNA strand, both before and after its cleavage, and
these unstable intermediates can be converted into stable “trapped
complexes” by reduction with NaCNBH3. With
oligo substrates, the mobility of these complexes in SDS/PAGE depends
on the relative size of the enzyme.
MutM and human OGG1 (hOGG1) have no obvious structural similarity despite their common substrate preference for excising 8-oxoG from an 8-oxoG⋅C pair. Their activity toward Fapys is more complex. Whereas MutM cleaves both G-derived Fapy (FapyG) and A-derived Fapy (FapyA), human OGG1 (hOGG1) excises only FapyG (14–16). The teleological reason for evolution of several DNA glycosylases with overlapping substrate range is not clear. It is possible that they have distinct roles in in vivo repair. For example, oxidized base lesions have now been shown to be preferentially repaired in the transcribed strand in mammalian cells (17, 18). This “transcription-coupled repair” (TCR) was discovered for DNA bulky adducts, which are repaired via the nucleotide excision repair (NER) pathway in which XPG functions as an essential endonuclease (18, 19). TCR was also observed for thymine glycol (Tg) in human cells, a substrate of Nth, Nei, and human NTH1 that requires the presence of the XPG protein but not its endonuclease activity (20). Thus TCR of Tg is likely to use the base excision repair and not the NER pathway. Evidence for TCR of 8-oxoG in human cells was elegantly provided by Le Page et al. (21). Accumulation of 8-oxoG in DNA extracted from Ogg1 (−/−) mouse cells and tissues indicates that OGG1 is the major repair enzyme for 8-oxoG (22, 23). However, complete absence of 8-oxoG removal from the nontranscribed strand but proficient and nearly normal removal of 8-oxoG from the transcribed sequences in Ogg1 (−/−) cells indicated TCR of 8-oxoG in mouse cells as well (24). These results are consistent with either the presence of a distinct DNA glycosylase that functions primarily in TCR, or removal of this lesion during TCR by an altogether different mechanism. Although TCR of Fapys has not been demonstrated, it is likely that Fapys, like 8-oxoG and Tg, are also subject to TCR.
We identified and partially characterized a second OGG (OGG2), in HeLa cells. It excises 8-oxoG preferentially from 8-oxoG⋅G or 8-oxoG⋅A pairs (12). In contrast, the major substrate for both OGG1 and MutM is the 8-oxoG⋅C pair (12, 25). We showed subsequently that E. coli Nei also has OGG activity and its substrate preference mirrors that of human OGG2 (26). These observations led us to propose that OGG2 preferentially excises 8-oxoG when it is misincorporated in the nascent strand opposite A (12). Similar replication-associated repair (RAR) also occurs for normal bases, e.g., A or G, when misincorporated opposite 8-oxoG. Such repair is mediated by MYH in human cells, an ortholog of E. coli MutY (27, 28).
Searching human genomic databases, we identified two candidate DNA glycosylases with sequence homology to MutM and Nei. In this report, we describe characterization of one of these DNA glycosylases, and discuss its potential role in repair of oxidatively damaged bases via RAR and TCR processes.
Materials and Methods
Human Genome Database Analysis and Molecular Cloning of NEH.
The NCBI and Celera Genomic Databases were searched by using the blastp program to identify human cDNA clones with significant homology to E. coli MutM or Nei. Two putative clones of human genes along with their mouse homologs were identified that we tentatively named NEH1 and NEH2 (Nei Homolog). The cDNA for human NEH1 (hNEH1; accession no. AAH10876), obtained from Research Genetics (Huntsville, AL), was inserted between the NdeI and XhoI sites of pRSETB plasmid, and its identity was confirmed by sequence analysis. Predicting that N-terminal Pro is the catalytic site (after cleavage of the initiator Met residue), we constructed expression plasmids encoding the Pro 1 deletion mutant of NEH1, and its N-terminal Histag fusion by using PCR-mediated site-specific mutagenesis.
Purification of NEH1.
Because plasmid-encoded expression of hNEH1 was low in mutM nei E. coli DE884, we introduced the recombinant plasmid into E. coli BL21 (DE3) codon plus (Stratagene) for large-scale purification of the enzyme after its induction with 0.2 mM isopropyl β-thiogalactoside at 16°C for 16 h. We purified wild type (WT) NEH1 from the sonicated extract of log-phase bacteria in a series of steps starting with the removal of nucleic acids by Polymin P, followed by enzyme precipitation with 60% saturation ammonium sulfate. After dissolving the precipitate in a minimum volume of buffer A (25 mM Tris⋅HCl, pH 7.5/10% glycerol/1 mM DTT/0.1 mM EDTA) containing 150 mM NaCl, and chromatography in a Superdex 75 column, fractions containing NEH1 were adjusted to 50 mM NaCl, and then applied to a 5 ml HiTrap Q and SP columns (Amersham Pharmacia) connected in tandem. After a wash with the starting buffer, the Q column was disconnected, and the proteins were eluted from the SP column at 500 mM NaCl with a 100-ml linear gradient of 50 mM to 1 M NaCl. The final step involved chromatography on a 1-ml Mono S column, and NEH1 was eluted without detectable impurity at 500 mM NaCl. The active fractions were frozen in liquid nitrogen and stored at −80°C. The final yield of the protein was about 5 mg per liter of culture.
Enzymatic Assay of NEH1 by GC-MS Analysis.
The release of modified bases by NEH1 from irradiated calf thymus DNA was assayed as described earlier (29). In short, 100 μg of irradiated calf thymus DNA was incubated with or without the enzyme at 37°C for 30 min in 100 μl of 50 mM phosphate buffer (pH 7.4), containing 100 mM KCl, 1 mM EDTA, and 0.1 mM DTT. Aliquots of stable isotope-labeled analogs of modified bases were added, and the samples were treated and analyzed by GC/MS as previously described (29).
Incision Assay with Duplex Oligo Substrates.
We used the standard procedure for assaying DNA glycosylase/AP lyase activity of NEH1 with 32P-labeled duplex oligo substrates. A 31-mer oligo containing either 8-oxoG or DHU at position 16 and 32P-labeled at the 5′ terminus (12) was annealed with the complementary strand, which contained either A, G, C, or T opposite the lesion site. In a different experiment, we used a 32-mer oligo containing 5-OHU in the sequence, 5′-TTCCAGACTGTCCTTCGTXACTTTCCTCTCAA-3′, where X is 5-OHU and the complementary strand had G opposite this lesion. The 32P-labeled duplex oligo (4 pmol) was incubated with 0.5 pmol NEH1 at 37°C in 20 μl mixture containing 20 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 0.1 mM EDTA, and 100 μg/ml BSA for 20 min, unless otherwise stated. After terminating the reaction with a stop buffer, the substrates and the cleaved products were separated by denaturing gel electrophoresis in 15% polyacrylamide containing 7 M urea in 90 mM Tris-borate (pH 8.3) and 2 mM EDTA and detected by PhosphorImager (Molecular Dynamics) analysis. NEH1-cleaved products were identified by comparing with those generated by E. coli Nth, Nei, and MutM.
Analysis of Trapped Complexes of NEH1.
The 32P-labeled duplex oligo substrate (100 fmol) was incubated with 1 μg crude extract in 20 μl assay buffer in the presence of 25 mM NaCNBH3 at 37°C for 30 min, and the trapped complexes were separated by SDS/PAGE (12% polyacrylamide; ref. 12).
Cell Synchronization and Analysis of Cell Cycle.
Primary human diploid MRC5 fibroblasts were synchronized by serum starvation for 120 h, and then stimulated to proliferate by adding FBS (10%) to the Eagle's minimal essential medium (EMEM) culture medium. The cells were harvested at various times; the aliquots were used for cell cycle analysis, and for preparing lysates for blot analysis of NEH1 mRNA and protein (30).
Blot Analysis for NEH1.
Nitrocellulose membranes containing 2 μg poly(A)+ RNA from various human tissues (CLONTECH) or 50 μg total RNA from synchronized MRC5 cells were probed with 32P-labeled NEH1 cDNA or an 18s ribosomal RNA gene probe following standard protocol (31). Forty micrograms total cell lysate proteins were used for immunoblot analysis with anti-NEH1 or anti-tubulin antibody (Santa Cruz Biotechnology) by using the enhanced chemiluminescence system (Amersham Pharmacia; ref. 12).
Other Reagents.
E. coli MutM, Nei and Nth, and human OGG1 and NTH1, used as references in this study, were purified to apparent homogeneity (26, 32, 33). Purified hNEH1 was used for raising polyclonal antibodies in rabbits (Alpha Diagnostic, San Antonio, TX).
Results
Identification of Mammalian Orthologs of E. coli MutM/Nei.
Fig. 1 shows comparative sequence homology of hNEH1 and hNEH2, with E. coli MutM and Nei as analyzed by clustalw. Although the overall identity between the human NEH1 and MutM is only 14.6% (57 identical residues of 390 residues in NEH1, and 14.9% between NEH1 and Nei), key residues, in particular the N-terminal PE (L/G) PE motif, are completely conserved in NEH1, along with an internal Lys residue (Lys-56 in MutM; ref. 34). Furthermore, NEH1 shares a potential helix-2-turn-helix (H2TH) motif with MutM and Nei (35). However, unlike in MutM, the absence of conserved cysteines suggests the absence of a Zn-finger motif in NEH1. It is thus expected that Pro-1 (after proteolytic cleavage of the initiator Met) is the active site in NEH1, which, like MutM/Nei, should carry out βδ elimination. Several coding sequences for hNEH1, deposited in GenBank, show two polymorphic sites in nonconserved regions, with Ser or Asn at position 147 and Arg or Lys at position 242. It is not known whether NEH1 activity is affected by polymorphic substitution. Our clone (accession no. AAH10876) contains Asn and Arg at positions 147 and 242 respectively. In any event, we decided to investigate the DNA glycosylase activity of NEH1.
Figure 1.
Sequence alignment of hNEH1 and hNEH2 with E. coli MutM/Nei. Identical residues are underlined. Box A contains catalytic N-terminal Pro; box B, helix-2-turn-helix motif; box C, Zn-finger motif. Potential nuclear localization signal (NLS; by psort) in NEH1 is underlined. N-terminal Met is cleaved after synthesis.
Chromosomal Map and Predicted Size of NEH1 and NEH2 of Humans and Mice.
Table 1 summarizes the information gleaned from the genomic database on chromosomal maps of NEH1 and NEH2 in the human and mouse genomes. No information is yet available about the potential linkage of these genes to any known disease. HNEH1 and hNEH2, with predicted molecular masses of 44 kDa and 36 kDa, respectively, are larger than the 30-kDa MutM or Nei, but are comparable in size to other mammalian DNA glycosylases.
Table 1.
Protein size and chromosomal location of human and mouse NEHs
| Size of human gene | Species | Protein size, kDa | Chromosomal location |
|---|---|---|---|
| NEH1 | Human | 43.7 | 15q 25 |
| Mouse | 9a5.3 | ||
| NEH2 | Human | 36.8 | 4q35 |
| Mouse | 8b.1.2 |
Detection of DNA Glycosylase Activity of Recombinant NEH1 by Trapping Analysis.
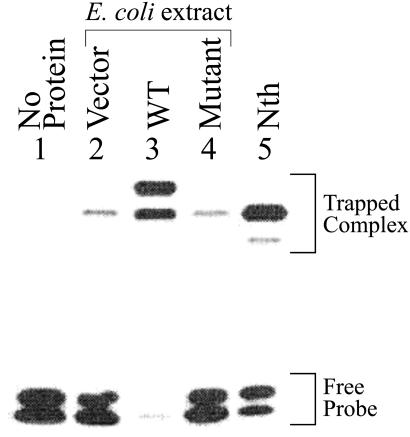
We tested DNA glycosylase/AP lyase activity of WT NEH1 and its Pro-1 deletion mutant in crude extracts of mutM nei E. coli harboring NEH1 expression plasmids. Fig. 2 shows SDS/PAGE of 32P-labeled trapped complexes generated by using extracts of mutM nei E. coli with or without expression of hNEH1. Because the mobility of such complexes reflects the size of the DNA glycosylase when the same oligo substrate is used (12), it is evident that, in control E. coli lacking MutM and Nei, only endogenous Nth formed a major trapped complex with the uncleaved oligo (lane 2). A minor species of smaller size was observed with the Nth standard, which corresponds to its complex with the cleaved oligo (lane 5). The WT NEH1 similarly forms two trapped complexes with uncleaved and cleaved oligos, respectively (lane 3). Coincidentally, the mobility of the NEH1 complex with the cleaved oligo was similar to that of the uncleaved oligo complex of Nth. Lane 4 shows that no NEH1-specific trapped complex was formed with the Pro-1 deletion mutant even though its level in the E. coli lysate was comparable to that of the WT protein as indicated by Western analysis (data not shown). Furthermore, a significant increase in strand incision activity with the DHU⋅A pair was observed in the sonicate of mutM nei E. coli expressing WT NEH1, but not the Pro-1 mutant, over low basal level (because of endogenous Nth) present in the extract of cells harboring the empty vector (data not shown). The requirement of N-terminal Pro for activity was further confirmed by the observation that recombinant NEH1 with N-terminal Histag fusion was inactive (data not shown). Taken together, these results show that recombinant NEH1 is active as a DNA glycosylase/AP lyase with the DHU oligo substrate, and that Pro-1 is likely to be its active site.
Figure 2.
Trapping analysis of recombinant NEH1 in extracts of mutM nei E. coli. Lane 1, no protein; lanes 2–4, trapping assay with lysates (1 μg) of E. coli containing empty vector, WT NEH1, and Pro 1 deletion mutant, after incubation with 0.1 pmol of DHU⋅A-containing duplex oligo; lane 5, trapped complex of purified Nth (5 ng) used as a reference with 0.1 pmol of DHU⋅A. The positions of trapped complexes and free DNA are indicated.
Purification and Properties of hNEH1.
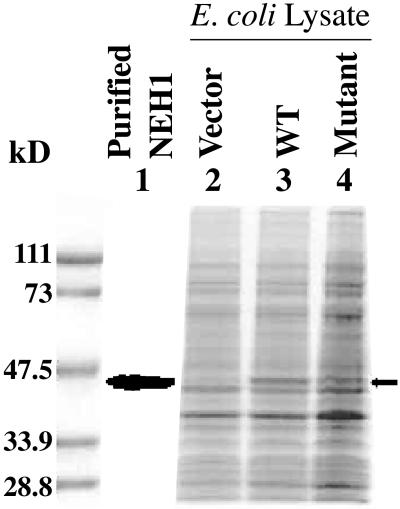
SDS/PAGE of extracts of E. coli codon plus harboring WT NEH1 and its Pro 1 mutant expression plasmids showed significant levels of expression of these proteins, most of which was present in the soluble form (Fig. 3, lanes 2–4). The WT NEH1 was then purified to apparent homogeneity by using conventional steps (Fig. 3, lane 1). The N-terminal sequence of this protein, Pro-Glu-Gly-Pro-Glu-Leu-His-Leu-Ala-Ser, matched perfectly with its predicted sequence (after cleavage of the initiator Met residue). Its molecular weight (43,633 as determined by MS analysis) was close to the predicted value of 43,582.
Figure 3.
Expression and purification of NEH1. Lysates (15 μg) of E. coli expressing NEH1 were analyzed by SDS/PAGE and Coomassie blue staining. The Bio-Rad protein markers are shown on the left. Purified WT NEH1 (lane 1); lanes 2–4, lysates of E. coli with empty vector, WT, and Pro-1 mutant NEH1 expression plasmid, respectively. The position of NEH1 is indicated by arrow.
DNA Glycosylase Activity of hNEH1 with Irradiated Natural Duplex DNA Substrate.
By using GC/MS, we examined the substrate range of NEH1 with irradiated DNA, which contained a wide variety of modified bases (29). Irradiated DNA used for these experiments contained FapyA, FapyG, and 8-oxoG at levels of 508 ± 37, 1825 ± 180, and 915 ± 55 lesions per 106 DNA bases, respectively. MutM/Fpg, used as the control, excised FapyA and FapyG, as well as 8-oxoG, from the same DNA, as expected (14, 16). Surprisingly, only FapyA and FapyG were found to be excised by hNEH1 (Fig. 4). The release of 173 ± 10, 721 ± 5, and 839 ± 1 of FapyA, FapyG, and 8-oxoG per 106 DNA bases, respectively, per microgram of 30-kDa MutM under the same conditions indicates that NEH1 excises FapyA and FapyG about 2- and 6-fold less efficiently than MutM on an equimolar basis.
Figure 4.
Excision of FapyA (□) and FapyG (○) by hNEH1 from irradiated DNA. Other details are described in Materials and Methods.
Analysis of NEH1 Activity with Duplex Oligo Substrates.
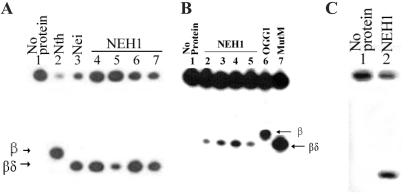
The sensitivity of NEH1 assay with irradiated DNA substrate containing low levels of multiple lesions is much less than with oligo substrates containing specific lesions. Furthermore, the effect of complementary base on the activity of DNA glycosylases could not be investigated in natural DNA. We, therefore, examined the activity of NEH1 for 8-oxoG and DHU present in 31-mer duplex oligos that had identical sequence, except for the lesion at position 16 (12, 26). For measuring relative reaction rates with various substrates, we assayed with excess substrates in the linear kinetic range. Fig. 5 shows that, under these conditions, NEH1 incised both DHU- and 8-oxoG-containing oligos at the lesion site, although its activity was higher for the pyrimidine lesion. The effect of the opposite base on NEH1 activity was also investigated. Incision of the DHU-containing strand was similar with A, C, or G opposite the lesion, and is reduced when T is present (Fig. 5A). The effect of different base pairs on hNEH1 incision activity for 8-oxoG was more pronounced (Fig. 5B). The enzyme was most active with the 8-oxoG⋅G pair (lane 4) followed by the 8-oxoG⋅T pair (lane 3) and had weak activity with 8-oxoG⋅C (lane 5) and 8-oxoG⋅A pairs (lane 2). Similar studies showed that hOGG1 was most active with the 8-oxoG⋅C pair, and the activity was barely detectable with the 8-oxoG⋅A pair (12, 36). NEH1 was also active in incising 5-OHU-strand from a duplex oligo when the lesion is paired with G (Fig. 5C). Thus, NEH1 mimics Nei more closely than MutM with respect to substrate preferences (26).
Figure 5.
Substrate specificity and AP lyase activity of purified hNEH1. (A) Purified NEH1 (0.5 pmol) was incubated with 5′ 32P-labeled DHU-containing duplex oligo (4 pmol) with A, T, G, and C in the complementary strand (lanes 4–7) opposite DHU. Lanes 2 and 3, Incision activity of Nth and Nei, respectively, with DHU⋅A oligo. (B) Incision activity of NEH1 with 8-oxoG⋅A-, 8-oxoG⋅T-, 8-oxoG⋅G-, and 8-oxoG⋅C-containing oligos, respectively (lanes 2–5). Incision activity of 8-oxoG⋅C with hOGG1 and MutM (lanes 6 and 7). Positions of β and βδ-elimination products are indicated. (C) Cleavage of 5-OHU-containing oligo (100 fmol) by NEH1. Lane 1, No enzyme; lane 2, 1 pmol NEH1.
E. coli MutM and Nei differ from other oxidized base lesion-specific DNA glycosylase/AP lyases because they generate 3′ phosphate termini after βδ elimination (13, 26). A comparison of the mobility of NEH1 digestion products with those of OGG1, and E. coli MutM, Nei, and Nth from 8-oxoG- or DHU-containing oligo substrate of the same sequences shows that NEH1 carries out βδ elimination like Nei and MutM, and not β elimination like Nth and OGG1 (Fig. 5 A and B).
Expression of NEH1 and OGG1 mRNAs in Human Tissues.
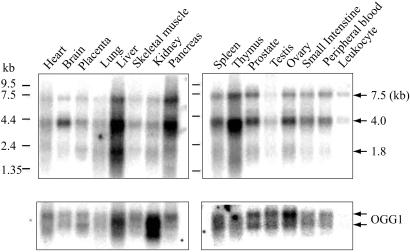
We compared the levels of NEH1 and OGG1 mRNAs in human tissues (Fig. 6). Three species of NEH1 mRNA (≈7.5, 4.0, and 1.8 kb) were observed. Because the hNEH1 gene is 8.1 kb long, it is likely that the largest RNA species corresponds to its primary transcript, and that the 4.0-kb species is a processing intermediate whereas the 1.8-kb species corresponds to mature NEH1 mRNA. We do not know the reason why hNEH1 mRNA was processed poorly in various tissue samples, although 7.5- and 4-kb precursor RNAs were not detected in the total RNA of in vitro cultured cells (see below). In any case, we observed the highest expression of the NEH1 gene in the liver, pancreas, and thymus; moderate expression in the brain, spleen, prostate, and ovary; and a relatively low level in the testis and leukocytes (Fig. 6).
Figure 6.
Tissue-specific expression of NEH1. The blots containing 2 μg poly(A)+ RNA purified from various human tissues (CLONTECH) were probed sequentially for Northern analysis with 32P-labeled cDNAs of hNEH1 (Upper) and hOGG1 (Lower).
When the same membranes were probed for OGG1 expression, two mRNA species were detected in all of the tissues, as observed earlier (36). More importantly, the patterns of expression for OGG1 and NEH1 were different. For example, the brain has low expression of OGG1 but high expression level of NEH1. Assuming that the levels of NEH1 and OGG1 polypeptides reflect their mRNA levels, it is possible that these two enzymes serve as each other's back-up system, but that their levels may also reflect the need to use distinct types of repair mechanisms such as RAR and TCR in various tissues.
S Phase-Specific Expression of hNEH1.
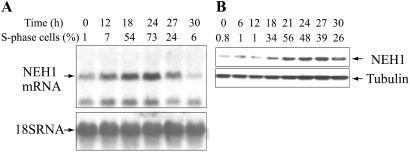
Cell cycle dependence of NEH1 expression was examined by both Northern analysis with total RNA isolated from synchronized human diploid fibroblasts and Western analysis with whole cell lysates. Fig. 7A shows that the NEH1 mRNA level reached its maximum at 24 h after release from serum starvation, paralleling the fraction of cells in the S phase. About a 4-fold increase over the level in G0/G1 cells was observed, which declined at 27 h. The level of 18s ribosomal RNA, used as an internal control, did not change significantly with the cell cycle (Fig. 7A).
Figure 7.
S phase-specific activation of NEH1. (A) Northern analysis of NEH1 mRNA, and (B) Western analysis of NEH1 polypeptide in synchronized MRC5 cells collected at various times after serum addition. The percentage of S phase cells in these different experiments is shown.
It is interesting to note that only 1.8-kb mRNA species were observed in MRC5 cells, along with a smaller RNA species whose origin is not clear. The absence of the 7.5- and 4-kb RNA species suggests that NEH1 mRNA is efficiently processed in these cells.
In a separate experiment, Western analysis of the level of the NEH1 polypeptide in synchronized cells showed a similar increase of the enzyme level during the S phase, reaching a maximum that was at 27 h about 6-fold higher than that in G0/G1 cells (Fig. 7B). Interestingly, the protein level did not go down as rapidly as the RNA level in cells beyond the S phase. These results showing S phase-specific activation of NEH1 suggest a linkage of this enzyme to DNA replication.
Discussion
Identification and cloning of hNEH1 and hNEH2, the first mammalian orthologs of E. coli Nei and MutM that carry out βδ elimination, became possible because of the human genome project, for several reasons. First, the cellular abundance of these enzymes, particularly in non-S phase cells, may be too low for their detection in cell extracts on the basis of activity. Second, 8-oxoG⋅C pair-containing DNA or oligos, commonly used for assaying DNA glycosylases for oxidized bases, are poor substrates, at least for NEH1, whereas FapyA and FapyG, the major substrates of NEH1, are not routinely used as substrates. Although hNEH1 has significant homology with both MutM and Nei in the catalytic domain (Fig. 1), we named it NEH (Nei Homolog) based on its substrate preference. NEH1, unlike MutM/Fpg but like Nei, has very weak 8-oxoG⋅C-specific OGG activity, the hallmark of MutM, and stronger activity with the 8-oxoG⋅G pair. Furthermore, it resembles Nei, and not MutM, in its stronger excision activity for pyrimidine lesions than for 8-oxoG. Although we have tentatively named the second candidate enzyme NEH2, its definitive naming should depend on its substrate preference, once determined. Interestingly, NEH1 acts like MutM in excising both FapyA and FapyG in natural DNA, whereas hOGG1 cleaves only FapyG (14, 16). Among all of the lesions in irradiated natural DNA, only FapyA and FapyG were found to be substrates of NEH1, and this is the only human enzyme identified so far that excises FapyA. Interestingly, FapyA is repaired by E. coli Nth but not its human ortholog NTH1 (29, 37). It appears that multiple glycosylases have evolved to excise Fapys because of their abundance in the genomes of both normal and oxidatively stressed cells (7).
We had identified and partially purified an enzyme from HeLa cells with an apparent molecular mass of 36 kDa, which we named OGG2 (12). Both OGG2 and NEH1 prefer the 8-oxoG⋅G pair as a substrate. However, apart from the size difference between OGG2 and NEH1, the fact that OGG2 carried out β elimination, whereas NEH1 has βδ elimination activity, indicates that OGG2 is not identical to NEH1. It is thus evident that the hOGG2 gene would not have been detected in the search that led to the discovery of hNEH1.
NEH1 has no activity toward 2-hydroxyadenine, 1-N6 ethenoadenine, 3-N4 ethenocytosine, hypoxanthine, and xanthine (data not shown). As expected, NEH1 efficiently cleaves AP site in DNA by functioning as an AP lyase (data not shown). 8-oxoG is present only as 8-oxoG⋅C pairs in nonreplicating DNA. Thus, our inability to observe significant excision of 8-oxoG from natural DNA is likely to be due to the weak activity of NEH1 toward an 8-oxoG⋅C pair. A similar lack of activity toward pyrimidine lesions in natural DNA (whereas a robust activity was observed with synthetic oligos) may be due to the limiting lesion concentration and competition among substrates in natural DNA, whereas excess substrates were used to compare reaction rates with oligos.
The larger size of hNEH1 compared with its ortholog Nei follows the pattern of other DNA glycosylases in that the mammalian ortholog is larger than the prokaryotic enzyme. It is likely that the nonhomologous C-terminal sequence in hNEH1 is involved in noncatalytic functions of the enzyme, e.g., in interaction with other proteins and/or in nuclear translocation. The C-terminal region of NEH1 contains several putative nuclear localization signal (NLS) sequences (Fig. 1). Splice variants of hOGG1 are targeted to the nucleus or mitochondria (38). The mitochondrial target sequence (MTS) is usually present at the N terminus, which is cleaved off during transit through the mitochondrial membrane (39). The presence of the active site Pro at the start of the coding sequence suggests that hNEH1 is not localized in the mitochondria.
Potential Involvement of NEH1 in TCR and RAR.
As pointed out in the introduction, preferential repair of 8-oxoG located in the transcribed sequences does not involve OGG1. It is possible that NEH1 (and NEH2) are responsible for TCR of 8-oxoG. Although the observed activity of NEH1 for 8-oxoG and oxidized purines is rather low, this result may not reflect the in vivo situation where the activity could be significantly enhanced in the presence of other proteins, e.g., XPG, which is involved in TCR (17, 40). Furthermore, although not tested, Fapys may also be subject to TCR by using NEHs in this process.
NEH1 may also be involved in replication-associated repair of oxidatively damaged bases. We proposed that RAR is necessary to prevent misincorporation of damaged bases in the nascent strand during DNA replication (12). In view of the broad substrate specificity of NEH1, at least with oligo substrates, it is possible that NEH1 preferentially excises 8-oxoG, as well as pyrimidine lesions, when incorporated in the nascent strand. We have shown earlier that both human OGG2 and E. coli Nei preferentially incise 8-oxoG-strand when 8-oxoG is paired with G and A (12, 26). The present study shows that NEH1 is also more active with the 8-oxoG⋅G pair than with the 8-oxoG⋅C pair. Electrophoretic gel mobility-shift assay (EMSA) showed distinct patterns of affinities of hOGG1 and hNEH1 for different 8-oxoG base pairs. OGG1 bound much more tightly to the 8-oxoG⋅C pair than to the 8-oxoG⋅A pair, whereas NEH1 preferred 8-oxoG⋅G and 8-oxoG⋅A pairs (data not shown). 8-oxoG in syn conformation was shown to mispair with A during in vitro replication (41). However, it can also pair with G as a 6-enolate-8-keto tautomer (42). Whether 8-oxoG⋅G pairing occurs during DNA replication in vivo, which would lead to GC → CG transversion mutation, is not known. Such mutations have, in fact, been found in oxidatively stressed E. coli and mammalian cells (43, 44). That these mutations in E. coli are prevented by the presence of Nei suggests that Nei in E. coli (45), and possibly NEH1 in mammalian cells, are responsible for repair of 8-oxoG (and other oxidized bases) in the nascent strand. We have observed that the level of hNEH1 polypeptide is highest in the S phase, with 6- to 7-fold enhancement over that in G0/G1-phase. In contrast, the level of hOGG1 is not significantly affected by the cell cycle (46). These results further support our prediction that NEH1 is involved in RAR. RAR of oxidized bases requires targeting of the DNA glycosylase to the nascent strand, which is presumably mediated by its interaction with replication-associated proteins. Identification of the interacting partners of hNEH1 should be an important topic for future studies.
Acknowledgments
We thank Eric Wilmore and Julie Lock for excellent technical assistance, and Alex Kurosky and Stephen Smith of the National Institute on Environmental Health Sciences (NIEHS) Center for protein sequence analysis, David Konkel for editing the manuscript, and Wanda Smith for expert secretarial assistance. We also thank Rabindra Roy for a gift of human NTH1. Certain commercial equipment or materials are identified in this paper to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose. This research was supported by U.S. Public Health Service Grants CA81063 (to S.M.), CA90860 (to Y.W.K.), and CA 84461 (to I.B.), and NIEHS Center Grant ES06676.
Abbreviations
- DHU
dihydrouracil
- 8-oxoG
8-oxoguanine
- Fapy
formamidopyrimidine
- 5-OHU
5-hydroxyuracil
- hOGG1
human 8-oxoguanine-DNA glycosylase 1
- NEH
Nei homolog
- RAR
replication-associated repair
- TCR
transcription-coupled repair
- ROS
reactive oxygen species
- WT
wild type
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AAH10876 (NEH1) and XL055431 (NEH2)].
References
- 1.Grisham M B, McCord J M. In: Physiology of Oxygen Radicals. Taylor A E, Matalon S, Ward P A, editors. Baltimore: Waverly Press; 1986. pp. 1–18. [Google Scholar]
- 2.Ames B N, Shigenaga M K, Hagan T M. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gotz M E, Kunig G, Riederer P, Youdim M B. Pharmacol Ther. 1994;63:37–122. doi: 10.1016/0163-7258(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 4.Lovell M A, Xie C, Markesbery W R. Brain Res. 2000;855:116–123. doi: 10.1016/s0006-8993(99)02335-5. [DOI] [PubMed] [Google Scholar]
- 5.Breen A P, Murphy J A. Free Rad Biol Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 6.Krokan H E, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. FEBS Lett. 2000;476:73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 7.Jaruga P, Speina E, Gackowski D, Tudek B, Olinski R. Nucleic Acids Res. 2000;28:E16. doi: 10.1093/nar/28.6.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Connor T R, Boiteux S, Laval J. Nucleic Acids Res. 1988;16:5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kow Y W, Wallace S S. Biochemistry. 1987;26:8200–8206. doi: 10.1021/bi00399a027. [DOI] [PubMed] [Google Scholar]
- 10.Jiang D, Hatahet Z, Blaisdell J O, Melamede R J, Wallace S S. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nash H M, Bruner S D, Scharer O D, Kawate T, Addona T A, Spooner E, Lane W S, Verdine G L. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 12.Hazra T K, Izumi T, Maidt L, Floyd R A, Mitra S. Nucleic Acids Res. 1998;26:5116–5122. doi: 10.1093/nar/26.22.5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zharkov D O, Rieger R A, Iden C R, Grollman A P. J Biol Chem. 1997;272:5335–5341. doi: 10.1074/jbc.272.8.5335. [DOI] [PubMed] [Google Scholar]
- 14.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 15.Dherin C, Radicella J P, Dizdaroglu M, Boiteux S. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karakaya A, Jaruga P, Bohr V A, Grollman A P, Dizdaroglu M. Nucleic Acids Res. 1997;25:474–479. doi: 10.1093/nar/25.3.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leadon S A. Cold Spring Harbor Symp Quant Biol. 2000;65:561–566. doi: 10.1101/sqb.2000.65.561. [DOI] [PubMed] [Google Scholar]
- 18.Tsutakawa S E, Cooper P K. Cold Spring Harbor Symp Quant Biol. 2000;65:201–215. doi: 10.1101/sqb.2000.65.201. [DOI] [PubMed] [Google Scholar]
- 19.Mellon I, Bohr V A, Smith C A, Hanawalt P C. Proc Natl Acad Sci USA. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- 21.Le Page F, Kwoh E E, Avrutskaya A, Gentil A, Leadon S A, Sarasin A, Cooper P K. Cell. 2000;101:159–171. doi: 10.1016/s0092-8674(00)80827-2. [DOI] [PubMed] [Google Scholar]
- 22.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes D E. Proc Natl Acad Sci USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, et al. Proc Natl Acad Sci USA. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Page F, Klungland A, Barnes D E, Sarasin A, Boiteux S. Proc Natl Acad Sci USA. 2000;97:8397–8402. doi: 10.1073/pnas.140137297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tchou J, Kasai H, Shibutani S, Chung M-H, Laval J, Grollman A P, Nishimura S. Proc Natl Acad Sci USA. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazra T K, Izumi T, Venkataraman R, Kow Y W, Dizdaroglu M, Mitra S. J Biol Chem. 2000;275:27762–27767. doi: 10.1074/jbc.M004052200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q M, Ishikawa N, Nakahara T, Yonei S. Nucleic Acids Res. 1998;26:4669–4675. doi: 10.1093/nar/26.20.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boldogh I, Milligan D, Lee M S, Bassett H, Lloyd R S, McCullough A K. Nucleic Acids Res. 2001;29:2802–2809. doi: 10.1093/nar/29.13.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dizdaroglu M, Bauche C, Rodriguez H, Laval J. Biochemistry. 2000;39:5586–5592. doi: 10.1021/bi9927787. [DOI] [PubMed] [Google Scholar]
- 30.Bresnahan W A, Boldogh I, Ma T, Albrecht T, Thompson E A. Cell Growth Differ. 1996;7:1283–1290. [PubMed] [Google Scholar]
- 31.Ramana C V, Boldogh I, Izumi T, Mitra S. Proc Natl Acad Sci USA. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill J W, Hazra T K, Izumi T, Mitra S. Nucleic Acids Res. 2001;29:430–438. doi: 10.1093/nar/29.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ikeda S, Biswas T, Roy R, Izumi T, Boldogh I, Kurosky A, Sarker A H, Seki S, Mitra S. J Biol Chem. 1998;273:21585–21593. doi: 10.1074/jbc.273.34.21585. [DOI] [PubMed] [Google Scholar]
- 34.Sidorkina O M, Laval J. Nucleic Acids Res. 1998;26:5351–5357. doi: 10.1093/nar/26.23.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. EMBO J. 2000;19:3857–3869. doi: 10.1093/emboj/19.15.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radicella J P, Dherin C, Desmaze C, Fox M S, Boiteux S. Proc Natl Acad Sci USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dizdaroglu M, Karahalil B, Senturker S, Buckley T J, Roldan-Arjona T. Biochemistry. 1999;38:243–246. doi: 10.1021/bi9819071. [DOI] [PubMed] [Google Scholar]
- 38.Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Mol Biol Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lithgow T. FEBS Lett. 2000;476:22–26. doi: 10.1016/s0014-5793(00)01663-x. [DOI] [PubMed] [Google Scholar]
- 40.Klungland A, Hoss M, Gunz D, Constantinou A, Clarkson S G, Doetsch P W, Bolton P H, Wood R D, Lindahl T. Mol Cell. 1999;3:33–42. doi: 10.1016/s1097-2765(00)80172-0. [DOI] [PubMed] [Google Scholar]
- 41.Grollman A P, Moriya M. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 42.Culp S J, Cho B P, Kadlubar F F, Evans F E. Chem Res Toxicol. 1989;2:416–422. doi: 10.1021/tx00012a010. [DOI] [PubMed] [Google Scholar]
- 43.McBride T J, Preston B D, Loeb L A. Biochemistry. 1991;30:207–213. doi: 10.1021/bi00215a030. [DOI] [PubMed] [Google Scholar]
- 44.de Oliveira R C, Ribeiro D T, Nigro R G, Di Mascio P, Menck C F. Nucleic Acids Res. 1992;20:4319–4323. doi: 10.1093/nar/20.16.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumoto Y, Zhang Q M, Takao M, Yasui A, Yonei S. Nucleic Acids Res. 2001;29:1975–1981. doi: 10.1093/nar/29.9.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dhenaut A, Boiteux S, Radicella J P. Mutat Res. 2000;461:109–118. doi: 10.1016/s0921-8777(00)00042-2. [DOI] [PubMed] [Google Scholar]