Abstract
The prehistoric peopling of East Asia by modern humans remains controversial with respect to early population migrations. Here, we present a systematic sampling and genetic screening of an East Asian–specific Y-chromosome haplogroup (O3-M122) in 2,332 individuals from diverse East Asian populations. Our results indicate that the O3-M122 lineage is dominant in East Asian populations, with an average frequency of 44.3%. The microsatellite data show that the O3-M122 haplotypes in southern East Asia are more diverse than those in northern East Asia, suggesting a southern origin of the O3-M122 mutation. It was estimated that the early northward migration of the O3-M122 lineages in East Asia occurred ∼25,000–30,000 years ago, consistent with the fossil records of modern humans in East Asia.
Introduction
The Y chromosome is a powerful tool for reconstruction of human population history (Jobling and Tyler-Smith 2003). The geographic distribution of genetic variations on the Y chromosome—which contains information about the subsequent colonization, differentiations, and migrations overlaid on recent population ranges—provides clues about prehistoric migrations (Underhill et al. 2001). The combination of biallelic (SNPs, low mutation rate) and microsatellite (STRs, high mutation rate) markers located at the nonrecombinant region of the Y chromosome have been widely used to infer early human history (Kayser et al. 2000; Nachman and Crowell 2000; Underhill et al. 2001; Y Chromosome Consortium 2002; Jobling and Tyler-Smith 2003; Dupuy et al. 2004). It was also shown that the estimates of divergence time that were based on Y-chromosome STR (Y-STR) variations under the background of Y-chromosome SNPs are more accurate than the STR-only estimates (Ramakrishnan and Mountain 2004). This strategy has been used recently in tracing and dating prehistoric migrations of European populations (Di Giacomo et al. 2004; Rootsi et al. 2004; Semino et al. 2004).
With a large number of indigenous populations and a complex human-inhabitation history, the peopling of East Asia remains unclear with respect to patterns of the early population migrations. The major dispute among researchers is rooted in different interpretations of early migration history that are based on the observed genetic divergence between northern (NEAS) and southern (SEAS) East Asian populations (Zhao et al. 1986; Weng and Yan 1989; Chu et al. 1998; Du et al. 1998; Piazza 1998; Su et al. 1999, 2000a, 2000b; Ding et al. 2000; Jin and Su 2000; Capelli et al. 2001; Karafet et al. 2001). Our previous study on Y-chromosome variations indicated that SEAS are more polymorphic than are NEAS (Su et al. 1999; Jin and Su 2000). However, using a similar set of Y-chromosome biallelic markers, Karafet et al. (2001) argued that there was a lack of genetic divergence between SEAS and NEAS and therefore disapproved of the hypothesis of early northward migration in East Asia. Data from mtDNA also indicated discrepancy on this issue. Two previous mtDNA studies supported the southern origin of modern humans in East Asia (Ballinger et al. 1992; Yao et al. 2002), whereas another study proposed that the genetic divergence between SEAS and NEAS might be due only to isolation by distance and that, therefore, a northern origin is still a possible explanation (Ding et al. 2000).
It should be noted that when we discuss the origin and migration of human populations, a time period—which part of the human-population history is under scrutiny—should be clearly defined. Recent population movement and admixture could wipe out or significantly diminish the original genetic signatures of early population movements. Therefore, to extract information for modern human origin and early population movements that happened before the Neolithic period, population-specific markers, such as SNP markers on the Y chromosome, become useful for the study of regional population movements (Jobling and Tyler-Smith 2003). At the same time, recent gene flow between distantly related populations can also be identified and removed in an analysis based on population specificity. Hence, in this sense, extreme caution should be exercised in selection of genetic markers in the study of the origin and early migrations of a continental population, because genetic variations introduced through recent gene flow could create false interpretations, as in two previous studies (Ding et al. 2000; Karafet et al. 2001). The same logic also applies to the selection of populations; ethnic populations with long histories of inhabitation in a region are always preferred for inferring early population histories.
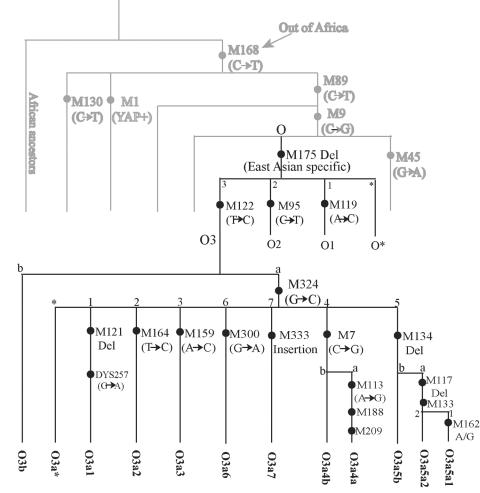
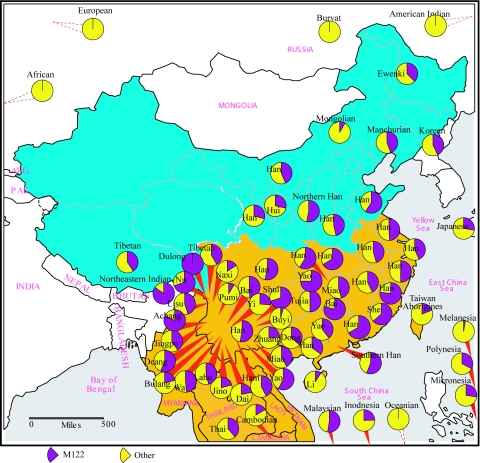
In East Asian populations, there are three regionally distributed (East Asian–specific) Y-chromosome haplogroups under the M175 lineage (fig. 1)—O3-M122, O2-M95, and O1-M119—together accounting for 57% of the Y chromosomes in East Asian populations (table 1). The O3-M122 has the highest frequency (41.8% on average) (fig. 2) in East Asians, especially in Han Chinese (52.06% in northern Han and 53.72% in southern Han) (table 1), and it is absent outside East Asia. Previous studies have shown that O2-M95 and O1-M119 are prevalent in SEAS and probably originated in the south (Su et al. 1999, 2000a; Wen et al. 2004a, 2004b) (table 1). Therefore, tracing the origin of O3-M122 became critical for a full understanding of the origin and early migrations of modern East Asians.
Figure 1.
The phylogenetic relationships of the O3-M122 SNPs and haplotypes
Table 1.
Distribution of the East Asian–Specific Y-Chromosome Haplogroups in Worldwide Populations
|
Percentage of Population with Lineagea |
|||||
| Population | No. of Samples | M122 | M119 | M95 | Total |
| African | 329 | ||||
| American Indian | 221 | ||||
| European | 1,046 | ||||
| Caucasus | 147 | ||||
| Russian | 243 | ||||
| Siberian | 328 | 3.05 | 3.05 | ||
| CAS | 1,231 | 4.55 | .57 | .61 | 5.28 |
| Southern Indian | 259 | .39 | 1.16 | 1.55 | |
| NEAS: | 34.65 | ||||
| Altai | 561 | 15.15 | 1.43 | 1.60 | 18.18 |
| Korean | 81 | 38.27 | 2.47 | 40.74 | |
| Japanese | 29 | 27.59 | 3.45 | 31.04 | |
| Northern Han | 413 | 52.06 | 4.36 | .72 | 57.14 |
| Hui | 74 | 22.97 | 1.35 | 24.32 | |
| Tibetan | 129 | 36.43 | .78 | 37.21 | |
| SEAS: | 72.31 | ||||
| Southern Han | 1,102 | 53.72 | 15.34 | 6.17 | 75.23 |
| Tibeto-Burman | 293 | 48.81 | 4.20 | 6.74 | 59.75 |
| Daic | 178 | 29.78 | 10.67 | 40.45 | 80.90 |
| Hmong-Mien | 249 | 51.41 | 2.41 | 16.10 | 69.92 |
| Austro-Asiatic | 63 | 11.11 | 3.18 | 50.80 | 65.09 |
| Austronesian | 555 | 26.31 | 22.34 | 16.94 | 65.59 |
| Melanesian | 113 | 2.65 | 2.65 | ||
Figure 2.
The frequency distribution of the O3-M122 haplotypes in East Asian and other continental populations. The data used were from published studies (Su et al. 1999, 2000a, 2000b; Qian et al. 2000; Semino et al. 2000; Underhill et al. 2000; Karafet et al. 2001; Lell et al. 2002; Jin et al. 2003; Wen et al. 2004a).
In the present study, through extensive sampling of East Asian populations and genotyping of Y-chromosome markers (SNPs and STRs) that define an East Asian–specific Y-chromosome haplogroup—O3-M122—we intended to delineate the origin of the O3-M122 haplogroup, to shed more light on the prehistoric migration of modern humans in East Asia.
Material and Methods
Samples
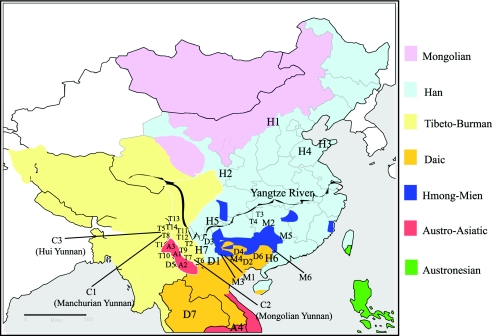
In the present study, 2,332 unrelated male samples were collected from the sites shown in figure 3, including 40 populations in East Asia. The criteria for population selection were based on the distribution of the O3-M122 haplogroups. Most of the populations sampled were from southern and southwestern China, where ∼80% of the Chinese ethnic populations live; most of them have inhabitation histories longer than 3,000 years (Wang 1994). Most of the northern ethnic populations in China, (e.g., Hui, Uygur, and Mongolian) were recently established (<1,000 years ago), with extensive admixture with Caucasian and Central Asian populations (CAS) (Wang 1994); therefore, those populations were not included in this study. The study populations were defined as “SEAS” and “NEAS” on the basis of their geographic locations. The Yangtze River was used as the geographic border to separate the SEAS and NEAS. In the SEAS, there are 14 Tibeto-Burman–speaking populations with a recorded history of migration from northern China ∼3,000 years ago (Wang 1994). The three Altai-speaking populations in Yunnan (southwestern China) were recent immigrants from northern China, <1,000 years ago (Wang 1994). The data about other populations were obtained from previous studies (Su et al. 1999, 2000a, 2000b; Qian et al. 2000; Semino et al. 2000; Underhill et al. 2000; Karafet et al. 2001; Lell et al. 2002; Jin et al. 2003; Wen et al. 2004a).
Figure 3.
The geographic distribution of language families/subfamilies in East Asia (Wang 1994) and the sampling sites for the present study. The population labels correspond with those in table 2.
Y-Chromosome Markers and Genotyping
We typed 15 Y-chromosome SNPs—including M122, M119, and M95—that represent the three major East Asian–specific lineages, and M134, M117, M162, M7, M113, M159, M121, M164 (Underhill et al. 2001; Y Chromosome Consortium 2002), M324, M300, M333 (P. Shen, A. E. Hirsh, T. Kivisild, B. Do, S. Song, R. Sung, V. Chou, H. Tang, L. Zhivotovsky, P. A. Underhill, L. L. Cavalli-Sforza, M. W. Feldman, P. J. Oefner, unpublished data), and DYS257 (Hammer et al. 1998), which are derived SNPs under the background of M122. The haplotypes were named according to the Y Chromosome Consortium (2002). Both PCR-RFLP and sequencing were used for genotyping (Su et al. 2000b). The phylogenetic relationship of the Y-SNPs is shown in figure 1. We initially typed M122, M119, and M95 in the 2,332 samples, in which 1,032 O3-M122 lineages were observed and were subjected to further typing of the other 12 SNPs and 8 STRs (DYS19/394, DYS388, DYS389 I, DYS389 II, DYS390, DYS391, DYS392, and DYS393). Among them, 854 samples generated complete sets of allele counting for all the SNPs and STRs (see detailed listing in our PDF supplement [online only]).
Data Analysis
The Y-chromosome SNP data were used to analyze the frequency distribution of the markers across the geographic regions by generation of contour maps (Golden Software Surfer 7.0). The multidimensional scaling (MDS) was used to analyze the relationship among populations with Arlequin 2.0 (Rst genetic distance matrix) (Schneider et al. 1998) and SPSS11.0 (MDS plot) software, with use of the SNP data.
The divergence times between the subclades of O3-M122 were estimated using the STR data by use of the SNP-STR coalescence method (Zhivotovsky 2001; Rootsi et al. 2004; Semino et al. 2004). The average mutation rate of the Y-STRs used was 0.00069 (Zhivotovsky et al. 2004). We also performed AMOVA analysis using the STR data with Arlequin 2.0 software to dissect population structure. The networks of the Y-STR haplotypes of major O3-M122 subhaplogroups were constructed using the program NETWORK4.1.0.6 (Fluxus). With use of the Arlequin 2.0 program (Schneider et al. 1998), we recalculated the gene diversity of SEAS and NEAS on the basis of the data reported by Karafet et al. (2001), and the NEAS with known history of recent admixture were removed from the calculation.
Results
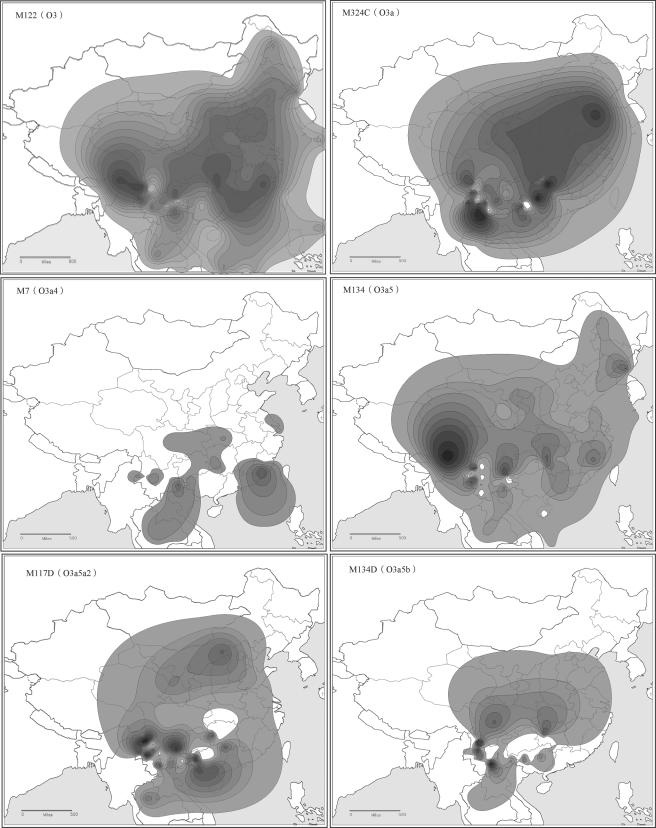
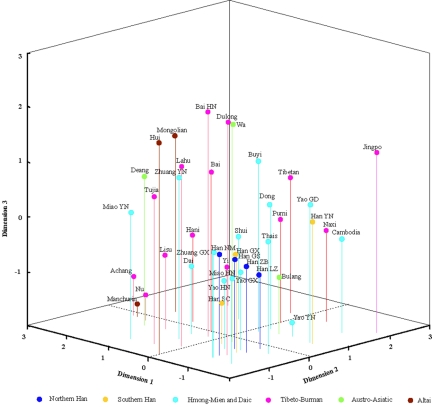
Among the 2,332 samples typed, 1,032 (44.3%) were O3-M122, which is consistent with previous reports of the dominant occurrence of M122 in East Asian populations (table 1) (Su et al. 1999, 2000a, 2000b; Karafet et al. 2001). There are 12 haplotypes within the O3-M122 haplogroup (fig. 1). We observed seven of them; their distribution is shown in table 2. The dominant O3-M122 subhaplogroups are O3-M134 (which includes O3a5a2 and O3a5b), O3-M7 (which includes O3a4b), and O3-M324 (which includes O3a*). Haplotype O3a1 (defined by M121 and DYS257) was observed only in two individuals (one from Shandong Han in northern China and the other from Cambodia). Haplotype O3a2 (defined by M164) was observed only in one Cambodian. We did not observe O3a3 (M159), O3a4a (M113), O3a5a1 (M162), O3a6 (M300), or O3a7 (M333), which were originally identified in SEAS samples in the initial screening panel of both SEAS and NEAS (Shen et al. 2000; Underhill et al. 2000; P. Shen, A. E. Hirsh, T. Kivisild, B. Do, S. Song, R. Sung, V. Chou, H. Tang, L. Zhivotovsky, P. A. Underhill, L. L. Cavalli-Sforza, M. W. Feldman, P. J. Oefner, unpublished data). The contour maps in figure 4 demonstrate the geographic distribution of the major O3-M122 subhaplogroups in East Asia. In general, the distribution of the O3-M122 haplotypes did not show distinctive divergence between southern and northern populations, with all the major subhaplogroups shared between them—except for O3-M7, which was observed only in the southern populations and therefore indicates a recent common ancestry of the O3-M122 lineage in East Asia. Using the STR data, we calculated the gene diversities; no significant differences were observed between SEAS and NEAS or among different language groups (data not shown). The AMOVA analysis did not show significant between-group divergence either (data not shown). However, the MDS analysis showed that the NEAS are closely related by clustering together, whereas the SEAS showed relatively loose connections with larger variance, indicating that SEAS are genetically more polymorphic than are NEAS (fig. 5). It should be noted that the difference in genetic variance between NEAS and SEAS could be due to the sampling-density discrepancy. However, our previous studies showed that northern Han populations are relatively homogenous, with similar Y-chromosome haplotype distributions (Ke et al. 2001a, 2001b). In addition, the four northern Han populations sampled in the present study covered different geographic regions in northern China. Therefore, the genetic variance observed probably reflected the true genetic background of the northern populations in China. In the MDS map, the Hmong-Mien populations were clustered closely with Han populations, which reflects the recorded history of admixture (Wang 1994).
Table 2.
Distribution of the O3-M122 Haplotypes in the East Asian Populations Studied[Note]
|
No. of Samples of Lineage |
|||||||||||
| Population | Label | Language | No. of Samples | M122 | O3b | O3a* | O3a1 | O3a2 | O3a4b | O3a5b | O3a5a2 |
| NEAS: | |||||||||||
| Han Inner Mongolian | H1 | Han | 60 | 29 | 1 | 11 | 2 | 10 | |||
| Han Gansu | H2 | Han | 60 | 22 | 8 | 3 | 5 | ||||
| Han Laizhou | H3 | Han | 86 | 49 | 1 | 31 | 5 | 2 | |||
| Han Zibo | H4 | Han | 98 | 61 | 2 | 18 | 1 | 6 | 8 | ||
| SEAS: | |||||||||||
| Han Sichuan | H5 | Han | 64 | 38 | 3 | 10 | 2 | 13 | 3 | ||
| Han Guangxi | H6 | Han | 39 | 15 | 4 | 1 | 3 | ||||
| Han Yunnan | H7 | Han | 81 | 37 | 1 | 16 | 3 | 1 | 16 | ||
| Achang | T1 | Tibeto-Burman | 40 | 33 | 30 | 3 | |||||
| Bai | T2 | Tibeto-Burman | 80 | 38 | 2 | 12 | 11 | 3 | 10 | ||
| Bai Hunan | T3 | Tibeto-Burman | 38 | 31 | 12 | 2 | 16 | 1 | |||
| Tujia | T4 | Tibeto-Burman | 100 | 54 | 14 | 2 | 4 | 7 | |||
| Nu | T5 | Tibeto-Burman | 50 | 35 | 2 | 2 | 31 | ||||
| Hani | T6 | Tibeto-Burman | 41 | 17 | 1 | 6 | 3 | 7 | |||
| Lahuo | T7 | Tibeto-Burman | 88 | 38 | 2 | 19 | 8 | 5 | 4 | ||
| Lisu | T8 | Tibeto-Burman | 49 | 23 | 2 | 17 | 1 | 3 | |||
| Yi | T9 | Tibeto-Burman | 47 | 12 | 1 | 4 | 6 | 1 | |||
| Jingpo | T10 | Tibeto-Burman | 17 | 4 | 1 | 1 | 1 | ||||
| Pumi | T11 | Tibeto-Burman | 47 | 4 | 1 | 3 | |||||
| Naxi | T12 | Tibeto-Burman | 87 | 12 | 5 | 4 | 3 | ||||
| Tibetan Yunnan | T13 | Tibeto-Burman | 50 | 22 | 5 | 6 | 11 | ||||
| Dulong | T14 | Tibeto-Burman | 28 | 28 | 19 | 9 | |||||
| Zhuang Yunnan | D1 | Daic | 47 | 15 | 2 | 6 | 1 | 5 | 1 | ||
| Zhuang Guangxi | D2 | Daic | 39 | 5 | 1 | 2 | 1 | ||||
| Buyi | D3 | Daic | 48 | 3 | 1 | 1 | 1 | ||||
| Shui | D4 | Daic | 40 | 28 | 6 | 3 | 19 | ||||
| Dai | D5 | Daic | 132 | 29 | 2 | 13 | 5 | 1 | 1 | ||
| Thais | D6 | Daic | 60 | 21 | 3 | 7 | 7 | 4 | |||
| Miao Yunnan | M1 | Hmong-Mien | 48 | 21 | 1 | 8 | 8 | 2 | 2 | ||
| Miao Hunan | M2 | Hmong-Mien | 105 | 48 | 1 | 9 | 2 | 1 | 3 | ||
| Yao Yunnan | M3 | Hmong-Mien | 90 | 50 | 9 | 10 | 20 | 6 | |||
| Yao Guangxi | M4 | Hmong-Mien | 225 | 104 | 2 | 30 | 6 | 7 | 25 | ||
| Yao Hunan | M5 | Hmong-Mien | 20 | 15 | 3 | 1 | 1 | 4 | |||
| Yao Guangdong | M6 | Hmong-Mien | 37 | 24 | 5 | 9 | 1 | 5 | |||
| Wa | A1 | Austro-Asiatic | 31 | 15 | 1 | 6 | 3 | 5 | |||
| Bulang | A2 | Austro-Asiatic | 28 | 6 | 2 | 3 | 1 | ||||
| Deang | A3 | Austro-Asiatic | 16 | 9 | 2 | 2 | 5 | ||||
| Cambodian | A4 | Austro-Asiatic | 14 | 2 | 1 | 1 | |||||
| Manchurian Yunnan | C1 | Altai | 41 | 24 | 1 | 2 | 21 | ||||
| Monngolian Yunnan | C2 | Altai | 46 | 8 | 4 | 2 | 2 | ||||
| Hui Yunnan | C3 | Altai | 15 | 3 | 2 | 1 | |||||
Note.— The geographic locations of the sampling sites are shown in figure 3. A total of 854 samples generated complete sets of both SNP and STR data; therefore, the sums of the sublineages in some populations are smaller than the total O3-M122 samples obtained.
Figure 4.
The contour maps of the Y-haplotype–frequency distribution. The data used to construct the contour maps of O3-M122, O3a-M324, O3a5-M134, and O3a4-M7 were from published studies (Su et al. 1999, 2000a, 2000b; Qian et al. 2000; Wen et al. 2004a) and the present study. The data used to construct the contour maps of O3a* and O3a5a2 (M117) were from the present study.
Figure 5.
The map of multidimensional scaling analysis based on the O3-M122 SNP haplotype distribution (table 2) of the 40 populations studied. Three dimensions were used in construction of the MDS map.
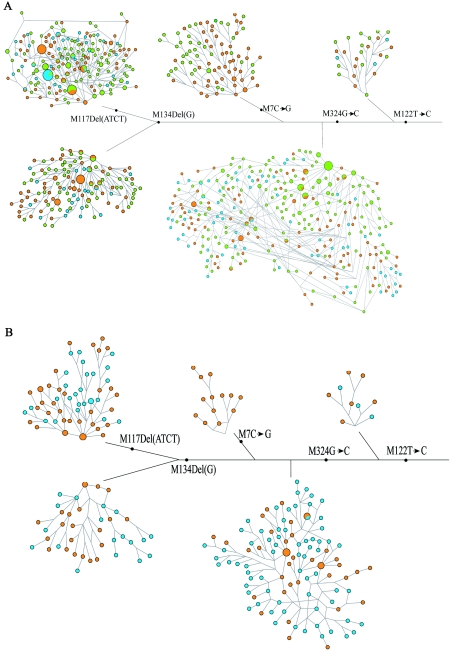
Assuming the O3-M122 lineage has a common origin in East Asia, we next sought to answer the question of where this lineage originally occurred. We conducted a detailed analysis by constructing a network of both the SNP and STR haplotype data. Figure 6A shows that there was a lot of similar STR evolution after the emergence of O3-M122, and many shared STR haplotypes were observed between northern and southern populations, again confirmation of the recent common ancestry of the M122 lineage in East Asia. It has been well documented that the Tibeto-Burman populations living in southwestern China were originally, during the late Neolithic period, from the north, but they have been under extensive influence from the southern ethnic groups, including Daic- and Austro-Asiatic–speaking populations (Wang 1994; Wen et al. 2004b). In addition, the Hmong-Mien populations have a recorded history of admixture with Han populations, although they are often considered southern populations (Wang 1994; Wen et al. 2005). The southern Han populations were recent northern immigrants because of the expansion of Han culture in the past several thousand years (Wang 1994). To remove the influence of relatively recent population admixture, we constructed the STR network excluding the Tibeto-Burman, Altaic, Hmong-Mien, and southern Han populations. The rebuilt network (fig. 6B) shows that most of the major STR haplotypes occurred in southern populations (Daic and Austro-Asiatic). We argue that both the MDS analysis and the STR network support a southern origin of the O3-M122 lineage in East Asia (fig. 6B).
Figure 6.
The networks of Y-STR haplotypes under the background of the Y SNPs. A, Network constructed with use of all populations. Populations shown in blue are northern Han. Populations shown in red are southern populations (Daic, Hmong-Mien, and Austro-Asiatic). Populations shown in green are southern Han and Tibeto-Burmans. B, Network constructed with southern Han, Altaic, Tibeto-Burman, and Hmong-Mien populations excluded to remove the influence of recent population admixture. Blue represents northern Han. Red represents Daic and Austro-Asiatic populations. The sizes of the dots are proportional to the haplotype frequencies, and the dots with multiple colors represent haplotypes shared among populations.
It should be noted that the lack of genetic divergence, in view of the gene diversity between the southern and northern O3-M122 lineages, indicates that the O3-M122 lineages were probably dominant in the population involved in the initial northward migration; therefore, no obvious bottleneck occurred for the O3-M122 lineage, in contrast with the skewed distribution of the O2-M95 and O1-M119 lineages (Su et al. 1999; Wen et al. 2004b). However, recent gene flows due to the expansion of Han culture could also have contributed partly to the homogeneity of the O3-M122 lineage, although Daic and Austro-Asiatic populations had much less influence from Han populations than did the Tibeto-Burman and Hmong-Mien populations (Wang 1994; Su et al. 2000b; Wen et al. 2004b).
On the basis of the STR data, we estimated the ages of the major O3-M122 subhaplogroups by use of the coalescence method developed by Zhivotovsky (2001). Table 3 lists the age estimations; all the subhaplogroups have a history older than the Neolithic time, with a range of 25,000–30,000 years ago.
Table 3.
Estimated Divergence Times of the O3-M122 Sublineages, with Use of Y-STR Data[Note]
|
Divergence Time(years ago) |
|||
| O3-M122Sublineage | Upper Limit | Lower Limit | Mean ± SE |
| O3a_M324G | 38,579 | 21,053 | 29,816±1,161 |
| O3a5_M134Del | 33,799 | 16,915 | 25,357±1,592 |
| O3a4_M7G | 36,759 | 19,875 | 28,317±4,030 |
| O3a5a2_M117Del | 37,398 | 22,217 | 29,807±1,613 |
Note.— The divergence times were estimated in accordance with the method of TD (Zhivotovsky 2001; Zhivotovsky et al. 2004). The upper limit was determined with the assumption that V0=0 (Rootsi et al. 2004). The lower limit was determined with the assumption that V0=Va, where Va is the value of within-population variance in the ancestral populations.
Discussion
As we described above, the distribution of the O3-M122 haplotypes in East Asian populations supports a southern origin. The age estimation is consistent with the fossil records unearthed in East Asia, where no modern human fossils aged >40,000 years have been found (Wu et al. 1995; Su et al. 1999; Jin and Su 2000). Interestingly, all the O3-M122 sublineages seem to diverge during a relatively short period of time (25,000–30,000 years ago), an implication that an ancient population expansion in southern East Asia might have occurred that could have triggered the initial northward migration. Hence, we argue that the time of the initial northward migration should be close to the estimated divergence times of the O3-M122 sublineages, a hypothesis that is also supported by the earliest fossil records of modern humans unearthed in northern China (27,000–39,000 years ago) (Wu et al. 1995; Jin and Su 2000).
Ding et al. (2000) point out that the southern areas are heavily populated, whereas the northern areas are sparsely populated. Consequently, between-region migration accompanied by high rates of genetic drift and lineage loss in northern groups could account for an asymmetry in lineage composition, which indicates that a northern origin could not be ruled out (Ding et al. 2000). However, in the work by Ding et al. (2000), the southern populations studied are the Tibeto-Burman populations, whose recorded history of recent northern origin, therefore, would blur the south-north divergence that was observed in multiple genetic systems (Zhao et al. 1986; Weng and Yan 1989; Chu et al. 1998; Du et al. 1998; Piazza 1998; Su et al. 1999, 2000a, 2000b; Ding et al. 2000; Jin and Su 2000; Capelli et al. 2001; Karafet et al. 2001). On the other hand, in the past 5,000 years, the major population migration in East Asia is from north to south, because of the expansion of Han culture (Cavalli-Sforza et al. 1994; Wang 1994), which was demonstrated by our recent study (Wen et al. 2004a). When we discuss the earliest migration of modern humans in East Asia before the Neolithic time, our data on O3-M122 polymorphisms revealed that southern populations are probably the ancestral populations. Data from the other two major East Asian–specific haplogroups (O1-M119 and O2-M95) also supported a southern origin of northern populations. These two lineages are prevalent in SEAS (table 1) but rare in NEAS (Wen et al. 2004b).
Karafet et al. (2001) found that the CAS, the NEAS, and the SEAS shared some Y haplotypes, and NEAS were more polymorphic than SEAS. However, when we looked into the detailed haplotype distribution described by Karafet et al. (2001), we found that 9 of the 28 haplogroups observed are derived from M175, which comprise 30.5% (230/754) and 79.1% (398/503) of the Y chromosomes of northern and southern East Asian males studied, respectively. It has been shown that M175 is specific to East Asians (Underhill et al. 2000, 2001). Although SEAS were found to have fewer haplogroups than NEAS, the pattern is the opposite within the M175 lineage (haplogroups 28–36 in the study by Karafet et al. [2001]), where SEAS are more polymorphic than NEAS. In SEAS, the haplogroups derived from M175 show higher gene diversity than for NEAS (0.86 vs. 0.67), and their distribution in the NEAS is highly skewed (table 1 in the study by Karafet et al. 2001). On the other hand, it would be misleading to interpret data from populations with a known history of recent population admixture (<1,000 years ago [Wang 1994]). For example, Hui and Uygurs, studied by Karafet et al. (2001), are recently established NEAS with various degrees of admixture with CAS. Mongolians have constant genetic exchange with CAS and North Asian (Siberian) populations. Consequently, the claimed larger number of Y-chromosome haplogroups observed in NEAS by Karafet et al. (2001) is in fact a false impression due to recent population admixture. This pattern was clearly reflected by frequent occurrence of haplogroups derived from M45 (haplogroups 37–45, European specific) (Semino et al. 2000) and M89 (haplogroups 20–24, highly prevalent in Europe, the Middle East, Central Asia, and the Indian subcontinent [Semino et al. 2000; Ramana et al. 2001; Wells et al. 2001]) lineages in NEAS, whereas they are almost absent in SEAS (Su et al. 1999). When the genetic influence of recent gene flow—that is, the M45 and M89 haplogroups—are removed from the analysis, the distribution of East Asian–specific haplogroups in the study by Karafet et al. (2001) indicates that SEAS are ancestral to NEAS.
On the basis of the published data and data presented in the present study, we propose that there might be two major reasons that led to the current genetic divergence between SEAS and NEAS. The first is due to the initial founder effect of the early northward migration and subsequent geographic isolation, reflected by the distribution of the O1-M119 and O2-M95 lineages (Su et al. 1999; Wen et al. 2004b). The second is probably due to the relatively recent population admixture in NEAS that introduced Caucasian and Central/Northern Asian Y chromosomes (Jin and Su 2000; Karafet et al. 2001).
In summary, our data about the East Asian–specific haplogroup O3-M122 indicates a southern origin of the O3-M122 lineage, therefore supporting the hypothesized southern origin of modern humans in East Asia. The initial prehistoric northward migration was estimated at 25,000–30,000 years ago.
Supplementary Material
Acknowledgments
We are thankful to the technical help of Xiao-na Fan and Yi-Chuan Yu. This study was supported by grants from the Chinese Academy of Sciences (KSCX2-SW-121), the National Natural Science Foundation of China (30370755 and 30440018), the Natural Science Foundation of Yunnan Province of China, and the National 973 Project of China (2006CB701506).
Web Resource
The URL for data presented herein is as follows:
References
- Ballinger SW, Schurr TG, Torroni A, Gan YY, Hodge JA, Hassan K, Chen KH, Wallace DC (1992) Southeast Asian mitochondrial DNA analysis reveals genetic continuity of ancient Mongoloid migrations. Genetics 130:139–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli C, Wilson JF, Richards M, Stumpf MPH, Gratrix F, Oppenheimer S, Underhill P, Pascali VL, Ko T-M, Goldstein DB (2001) A predominantly indigenous paternal heritage for the Austronesian-speaking peoples of insular Southeast Asia and Oceania. Am J Hum Genet 68:432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton [Google Scholar]
- Chu JY, Huang W, Kuang SQ, Wang JM, Xu JJ, Chu ZT, Yang ZQ, Lin KQ, Li P, Wu M, Geng ZC, Tan CC, Du RF, Jin L (1998) Genetic relationship of populations in China. Proc Natl Acad Sci USA 95:11763–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo F, Luca F, Popa LO, Akar N, Anagnou N, Banyko J, Brdicka R, et al (2004) Y chromosomal haplogroup J as a signature of the post-Neolithic colonization of Europe. Hum Genet 115:357–371 [DOI] [PubMed] [Google Scholar]
- Ding YC, Wooding S, Harpending HC, Chi HC, Li HP, Fu YX, Pang JF, Yao YG, Yu JG, Moyzis R, Zhang Y (2000) Population structure and history in East Asia. Proc Natl Acad Sci USA 97:14003–14006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du RF, Xiao CJ, Cavalli-Sforza LL (1998) Genetic distances between Chinese groups calculated on gene frequencies of 38 loci. Sci China Series C 83–89 [DOI] [PubMed] [Google Scholar]
- Dupuy BM, Stenersen M, Egeland T, Olaisen B (2004) Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum Mutat 23:117–124 [DOI] [PubMed] [Google Scholar]
- Hammer MF, Karafet T, Rasanayagam A, Wood ET, Altheide TK, Jenkins T, Griffiths RC, Templeton AR, Zegura SL (1998) Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol Biol Evol 15:427–441 [DOI] [PubMed] [Google Scholar]
- Jin HJ, Kwak KD, Hammer MF, Nakahori Y, Shinka T, Lee JW, Jin F, Jia X, Tyler-Smith C, Kim W (2003) Y-chromosomal DNA haplogroups and their implications for the dual origins of the Koreans. Hum Genet 114:27–35 [DOI] [PubMed] [Google Scholar]
- Jin L, Su B (2000) Natives or immigrants: modern human origin in East Asia. Nat Rev Genet 1:126–133 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 [DOI] [PubMed] [Google Scholar]
- Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF (2001) Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet 69:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Krüger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Su B, Li H, Chen L, Qi C, Guo X, Huang W, Jin J, Lu D, Jin L (2001a) Y chromosome evidence for no independent origin of modern human in China. Chinese Sci Bul 46:935–937 [Google Scholar]
- Ke Y, Su B, Xiao C, Chen H, Huang W, Chen Z, Chu J, Tan J, Jin L, Lu D (2001b) Y-chromosome haplotype distribution in Han Chinese populations and modern human origin in East Asians. Sci China Series C 44:225–232 [DOI] [PubMed] [Google Scholar]
- Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC (2002) The dual origin and Siberian affinities of Native American Y chromosomes. Am J Hum Genet 70:192–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156:297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza A (1998) Towards a genetic history of China. Nature 395:636–639 [DOI] [PubMed] [Google Scholar]
- Qian Y, Qian B, Su B, Yu J, Ke Y, Chu Z, Shi L, Lu D, Chu J, Jin L (2000) Multiple origins of Tibetan Y chromosomes. Hum Genet 106:453–454 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan U, Mountain JL (2004) Precision and accuracy of divergence time estimates from STR and SNPSTR variation. Mol Biol Evol 21:1960–1971 [DOI] [PubMed] [Google Scholar]
- Ramana GV, Su B, Jin L, Singh L, Wang N, Underhill P, Chakraborty R (2001) Y-chromosome SNP haplotypes suggest evidence of gene flow among caste, tribe, and the migrant Siddi populations of Andhra Pradesh, South India. Eur J Hum Genet 9:695–700 [DOI] [PubMed] [Google Scholar]
- Rootsi S, Magri C, Kivisild T, Benuzzi G, Help H, Bermisheva M, Kutuev I, et al (2004) Phylogeography of Y-chromosome haplogroup I reveals distinct domains of prehistoric gene flow in Europe. Am J Hum Genet 75:128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Kueffer JM, Roessli D, Excoffier L (1998) Arlequin: a software for population genetic analysis, Genetics and Biometry Laboratory, University of Geneva, Geneva [Google Scholar]
- Semino O, Magri C, Benuzzi G, Lin AA, Al-Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner PJ, Zhivotovsky LA, King R, Torroni A, Cavalli-Sforza LL, Underhill PA, Santachiara-Benerecetti AS (2004) Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 74:1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 [DOI] [PubMed] [Google Scholar]
- Shen P, Wang F, Underhill PA, Franco C, Yang WH, Roxas A, Sung R, Lin AA, Hyman RW, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ (2000) Population genetic implications from sequence variation in four Y chromosome genes. Proc Natl Acad Sci USA 97:7354–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Jin L, Underhill P, Martinson J, Saha N, McGarvey ST, Shriver MD, Chu J, Oefner P, Chakraborty R, Deka R (2000a) Polynesian origins: insights from the Y chromosome. Proc Natl Acad Sci USA 97:8225–8228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, Jin L (2000b) Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet 107:582–590 [DOI] [PubMed] [Google Scholar]
- Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, Huang W, Shen D, Lu D, Luo J, Chu J, Tan J, Shen P, Davis R, Cavalli-Sforza L, Chakraborty R, Xiong M, Du R, Oefner P, Chen Z, Jin L (1999) Y-chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am J Hum Genet 65:1718–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Passarino G, Lin AA, Shen P, Mirazon Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL (2001) The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann Hum Genet 65:43–62 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 [DOI] [PubMed] [Google Scholar]
- Wang ZH (1994) [History of nationalities in China.] China Social Science Press, Beijing [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, et al (2001) The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA 98:10244–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Li H, Gao S, Mao X, Gao Y, Li F, Zhang F, He Y, Dong Y, Zhang Y, Huang W, Jin J, Xiao C, Lu D, Chakraborty R, Su B, Deka R, Jin L (2005) Genetic structure of Hmong-Mien speaking populations in East Asia as revealed by mtDNA lineages. Mol Biol Evol 22:725–734 [DOI] [PubMed] [Google Scholar]
- Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, Qian J, Tan J, Jin J, Huang W, Deka R, Su B, Chakraborty R, Jin L (2004a) Genetic evidence supports demic diffusion of Han culture. Nature 431:302–305 [DOI] [PubMed] [Google Scholar]
- Wen B, Xie X, Gao S, Li H, Shi H, Song X, Qian T, Xiao C, Jin J, Su B, Lu D, Chakraborty R, Jin L (2004b) Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am J Hum Genet 74:856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng ZL, Yan YD (1989) [Analysis of the genetic structure of human populations in China.] Acta Anthropol Sin 8:261–268 [Google Scholar]
- Wu HC, Poirier FE, Wu XZ (1995) Human evolution in China: a metric description of the fossils and a review of the sites. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- Yao Y-G, Kong Q-P, Bandelt H-J, Kivisild T, Zhang Y-P (2002) Phylogeographic differentiation of mitochondrial DNA in Han Chinese. Am J Hum Genet 70:635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao TM, Zhang GL, Zhu YM, Zheng SQ, Lui DY, Chen Q, Zhang X (1986) [The distribution of immunoglobulin Gm allotypes in forty Chinese populations.] Acta Anthropol Sin 6:1–8 [Google Scholar]
- Zhivotovsky LA (2001) Estimating divergence time with the use of microsatellite genetic distances: impacts of population growth and gene flow. Mol Biol Evol 18:700–709 [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioğlu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, Chambers GK, Herrera RJ, Yong KK, Gresham D, Tournev I, Feldman MW, Kalaydjieva L (2004) The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 74:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.