Abstract
We have sequenced six overlapping clones from a library of bacterial artificial chromosome (BAC) clones derived from a laboratory strain of the mosquito, Anopheles gambiae, the major vector of human malaria in Africa. The resulting uninterrupted 528-kb sequence is from the 8C region of the mosquito 2R chromosome, at or very near the major refractoriness locus associated with melanotic encapsulation of parasites. This sequence represents the first extensive view of the mosquito genome structure encompassing 48 genes. Genomic comparison reveals that the majority of the orthologues are found in six microsyntenic clusters in Drosophila melanogaster. A BAC clone that is wholly contained within this region demonstrates the existence of a remarkable degree of local polymorphism in this species, which may prove important for its population structure and vectorial capacity.
Anopheles gambiae is the most important vector of Plasmodium falciparum malaria in Africa, where almost 90% of the world's malaria-specific mortality occurs. Historically, malaria in many parts of the world has been controlled with two public health interventions, antimalarial drugs like chloroquine and insecticides like DDT. Chloroquine resistance, which first appeared in Africa in the late 1970s, has now spread throughout virtually all of sub-Saharan Africa, and resistance to alternative drugs like Fansidar and Mefloquine has begun to appear. Although malaria control programs based on residual spray insecticides have seen only limited use over much of Africa, control programs using pyrethroid insecticide-impregnated bed nets are now being implemented in many African countries. Resistance to pyrethroid insecticides has now been recorded in A. gambiae populations from both West and East Africa, threatening to undermine even these programs.
The decreasing efficacy of insecticides has led to the hope that study of vector biology will facilitate the development of new strategies for vector-targeted malaria control. In this broad context, the biological interaction between the parasite and its vector is of special interest. In the Plasmodium-refractory L3-5 strain of A. gambiae, the mosquito is able to kill the ookinete stage of the parasite by encapsulating it in a proteinaceous envelope cross-linked with melanin, as the parasite completes its transit through the mosquito midgut epithelium (1–3). Genetic analysis has ascribed melanotic encapsulation to the concerted action of three quantitative trait loci, Pen1, Pen2, and Pen3 (4, 5). Pen1 is the major and most essential locus and is very close to a microsatellite marker H175 from which it has not been separated recombinationally, in contrast to the nearest flanking markers H290 and ND3B6. Thus, Pen1 has been assigned with confidence to the polytene chromosomal region 8C–8D where all three microsatellite markers map, a region spanning ≈1.5 Mb of DNA.
With the ultimate goal of positionally cloning this gene, we have used clones from two bacterial artificial chromosome (BAC) libraries produced with genomic DNA from the PEST strain of A. gambiae (Y.S.H., X.W., and F.H.C., unpublished data) to sequence a 528-kb region of DNA from the 8C region, including both the H175 and H290 markers. This sequence has identified candidate genes that can be assessed in the future by finer-scale genetic analysis coupled with functional testing by transgenesis. This work affords the opportunity to compare the organization of the A. gambiae genome with that of another dipteran insect, Drosophila melanogaster, at the sequence level. The analysis of this region has served as a testing ground for the whole-genome sequence assembly and annotation (R. Holt, personal communication). It has also revealed an unexpected phenomenon, unusually high sequence variation in localized chromosomal regions, which may have important implications for the population structure and evolution of this important vector species.
Methods
Construction and Sequencing of a BAC Contig Spanning the Pen1 Locus.
Two BAC genomic DNA libraries (Y.S.H., X.W., and F.H.C., unpublished data) from the A. gambiae PEST laboratory strain (6) were used in these studies. The 8C–D region of ovarian polytene chromosomes was microdissected, amplified (7), and used to probe a filter (http://www.genomesystems.com) displaying all of the BAC clones in one library. Positive clones were isolated and subjected to in situ hybridization to ovarian nurse cell polytene chromosomes from PEST mosquitoes (8) to exclude false positive or potentially chimeric clones. The ends of most clones had been sequenced for about 600–800 bp each (http://www.genoscope.cns.fr and www.tigr.org), allowing use of the sequence-tagged connector strategy (9) to construct the minimal tiling path. We used PCR analysis of successively less complex pools of BAC clone DNA to isolate BAC 11N17 containing marker H175. This BAC was sequenced completely, then two minimally flanking BACs were selected by sequence comparison with the BAC ends database (http://www.genoscope.cns.fr). This process was repeated in both directions, and BAC overlaps were confirmed by PCR and restriction analysis. PCR primers were designed by primer 3 (10).
BAC sequencing used the randi strategy (11), based on simultaneous sequencing on both strands from pUC18 clones of both random and directed libraries. The former was generated by partial BAC digestion with Tsp509 I or Sau3A whereas the latter used complete EcoRI or HindIII digests yielding clones that served as a “scaffold” for assembling the BAC sequence and as templates for primer walking during finishing. Direct BAC DNA sequencing was used for proof sequencing (12). Plasmid sequencing was performed with the AmpliTaqFS core kit (Applied Biosystems) by using standard forward and reverse primers labeled with FITC or CY5. All reactions were analyzed on the arakis sequencing system (13). Raw sequencing data were evaluated, analyzed, and assembled with the software packages lane tracker and gene skipper (14). Gaps were covered by primer walking (15) or transposon-mediated sequencing (16).
Sequence Analysis.
The combined BAC sequence was annotated with ab initio gene prediction algorithms and alignment algorithms based on sequence similarity. The ab initio methods included GENSCAN 1.0 (17), FGENES 1.0 (V. Solovyev, unpublished data), and GENEID 1.1 (18) and were used with default parameters and D. melanogaster or human sequences as the organismal option. Similarity-based methods included blastx and blastp searches (19). The expressed sequence tag (EST) searches were run against Anopheles and Drosophila ESTs from the Gene2EST server (20). The genewise tool (21) was also used as a combined method. Protein domain analysis was performed by pfam, smart, and interpro. All analyses were viewed with the artemis graphical tool releases 3 and 4 (22). To avoid over-prediction, only genes fulfilling one or more of the following criteria were accepted: (i) prediction by at least two algorithms; (ii) prediction by one ab initio method and matches with ESTs, cDNAs, or proteins in the databases; and/or (iii) detectable expression (data not shown). tRNA genes were predicted with tRNASCAN-SE 1.21 (23). For comparative analysis of Pen1 region genes with Drosophila genes, we relied on annotations and cytological locations presented by the FlyBase/Berkeley Drosophila Genome Project GadFly annotation database, releases 1 and 2 (http://flybase.bio.indiana.edu and http://www.fruitfly.org/).
PCR Cloning and Analysis.
Gene 22J3.4 was amplified (Expand Long Template PCR system, Roche Molecular Biochemicals) from genomic DNAs extracted from pools of eight Plasmodium-susceptible 4Ar/r and eight Plasmodium-refractory L3–5 mosquitoes, respectively. Primers were designed from PEST sequences flanking the predicted gene in BAC 22J3. PCR products were cloned into the pCR2.1-TOPO vector with the TA cloning kit (Invitrogen) and sequenced by primer walking. The conceptually translated amino acid sequences of 11 (L3–5) and 6 (4Ar/r) clones were aligned by CLUSTAL X 1.81 (24), after setting aside variants occurring only once and suspected of being PCR artifacts. A bootstrapped tree was constructed with clustal x and viewed with TREETOOL 2.0.1 (25).
Results
Physical Mapping and Sequencing in the Pen1 Region.
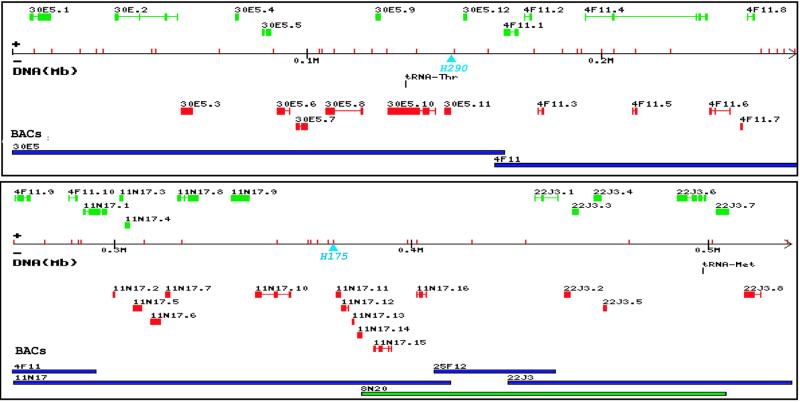
One of the A. gambiae BAC libraries was screened by PCR using primers for the two microsatellite markers H290 and H788 that delimit the Pen1 region. This 8C–D region of polytene chromosome 2 was then microdissected, PCR-amplified, and used to probe a filter containing DNA from all BAC clones. In situ hybridization to polytene chromosomes confirmed that more than 120 of the BAC clones so identified mapped uniquely to this region, and the available end-sequences from most of these BACs (http://www.genoscope.cns.fr and www.tigr.org), combined with filter hybridization experiments, permitted us to develop a contig of clones spanning the region. BAC clone 11N17 encompassing H175 (the closest marker to the Pen1 locus) was chosen to begin sequencing, and minimally overlapping end-sequenced BACs were used to extend the sequence (Fig. 1). A BAC trimming strategy (26) was used to remove about 50 kb of excess DNA from one end of clone 25F12, reducing unnecessary overlap and speeding up the sequencing. The 528-kb uninterrupted Consensus Sequence that we report here was determined from five BACs (Fig. 1). Additional sequences totaling 52 kb were determined as overlaps between these clones and showed an average sequence variation of 0.03%, well within the acceptable range of intraspecies polymorphism. In addition, we fully sequenced 8N20, a 129-kb BAC which overlapped three of the canonical clones (Fig. 1) and showed surprisingly high variation from the Consensus Sequence, as discussed below.
Figure 1.
Diagram of the 528-kb genomic sequence, presented in two segments. Predicted genes are in green (+ strand, left to right and telomere to centromere orientation) and red (− strand). tRNA genes are shown in black. The sequenced BAC clones are identified, in blue for those containing the Consensus Sequence and green for the variant 8N20 BAC. The sequence axis includes red tickmarks representing 51 microsatellites (more than 8 dinucleotide repeats each); two light blue arrowheads represent microsatellites that partially delimit the Pen1 region, of which H175 has not been separated from Pen1 recombinationally. The genes belong to the following functional categories according to the Gene Ontology classification (www.geneontology.org): 7 signal transduction, 6 metabolism, 5 transport/binding, 4 transcription/translation, 3 each structural protein and regulation, 2 nucleic acid metabolism, and 1 each cell adhesion and replication. The figure was produced with the Bio∷Graphics library from the Generic Model Organism Database (GMOD) project (http://www.gmod.org).
Annotation Features.
As described in Methods, we have used both ab initio and similarity-based gene prediction methods to annotate the Consensus Sequence. All outputs of the computational searches were reviewed manually. The analysis resulted in prediction of a total of 48 genes, of which 46 are putative protein-coding and 2 are tRNA-coding genes. Provisionally we have named the protein-coding genes sequentially, in a telomere-to-centromere order and according to the BAC in which they are fully contained (30E5.1 to .12, 4F11.1 to .10, 11N17.1 to .16, and 22J3.1 to .8; Fig. 1 and Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). Thirty-nine of these (85%) were predicted with both ab initio programs and sequence similarity to database entries of other organisms or Anopheles EST matches and 32 of these were ascribed by similarity to functional classes (see Methods and legend to Fig. 1; also see Table 1 and Fig. 5, which are published as supporting information on the PNAS web site). Seven putative genes were predicted only on the basis of the ab initio approach, but their existence is considered firm, as it was supported by reverse transcription–PCR tests of expression (not shown). The average length of protein-coding genes in this sequence (including introns but not 5′ and 3′ untranslated regions) is 1.98 kb, and their average density is one gene per 11.5 kb. The gene density along the Consensus Sequence is variable, as in the D. melanogaster genome (27). For example, the 36-kb region between 11N17.16 and 22J3.1 is predicted to be gene-free, whereas a nearby 19-kb region contains five genes (11N17.11 to .15). The average gene density in this sequence, combined with the reported A. gambiae haploid DNA content of 280 Mb, of which 61% is composed of single-copy or “unique” sequences according to reassociation kinetics (28, 29), would predict a total of approximately 14,800 genes in the A. gambiae genome, which is very similar to that reported for D. melanogaster, 13,601 (27).
Annotation of the Adh region sequence from D. melanogaster revealed a large number of genes (8%) nested within the introns of other genes (30). Because gene-finding programs do not predict this class of genes, one usually has to rely on supporting evidence obtained from cDNA and EST homologies or expression profiling. We have predicted one nested gene (4F11.5) within a 28-kb intron of the 4F11.4 gene (Fig. 1). The 4F11.4 gene product resembles transcription factor AFX from D. melanogaster (AAL28078.1) and shows a forkhead domain encoded in two exons, separated by the large intron that contains gene 4F11.5. These nested genes are transcribed from opposite DNA strands. Even though the putative 4F11.5 gene product shows no similarity to other proteins in the databases, its prediction is supported by reverse transcription–PCR expression data (not shown). Similarly, a tRNA-Thr gene is found within an intron of gene 30E5.10, in the same orientation.
Comparison of the Anopheles Consensus Sequence with D. melanogaster.
The genome of the fruit fly D. melanogaster has been sequenced, allowing detailed comparison of genome content and organization with A. gambiae. The ancestral lines of these two Diptera are thought to have split approximately 250 million years ago (31); for comparison, the separation between human and mouse is typically placed at around 100 million years ago, and the divergence of insects and vertebrates occurred more than 650 million years ago (32, 33). Only seven of the predicted Anopheles proteins showed no significant sequence similarity to proteins from any organism (analysis with blastp and default thresholds). All of the other 39 genes were similar to Drosophila sequences and 38 showed a clear strong match to a single Drosophila sequence (see Table 1). Homologous genes can be classified as orthologues (when related by a speciation event) and paralogues (when related by an intragenome duplication event). The 38 genes are considered orthologues of their Drosophila best matches, with which they cluster (rather than other members of the same gene family). However, in the absence of a complete analysis of the Anopheles genome we cannot rule out the possibility of misassigning orthologues. The average protein identity of the putative orthologues is approximately 54%.
We have identified two pairs of adjacent genes that seem to have duplicated after the divergence of the Drosophila and Anopheles lineages. One pair (11N17.5 and 11N17.6) shares 66% amino acid identity and encodes proteins highly similar to the V-ATPase subunit-α from a number of organisms, and to the immune-suppressor gene TJ6 from human (AAD04632.1) and mouse (AAA39336.1). The other recently duplicated neighboring genes (22J3.2 and 22J3.3) are transcribed from opposite strands and encode proteins that are 59% identical. They belong to the apyrase/5′-nucleotidase family of proteins that facilitate hematophagy by inhibiting aggregation of the host platelets in blood-feeding arthropods. In both cases, each of the duplicated genes is most similar to the same Drosophila gene. In contrast, our analysis identified two homeobox-containing genes (11N17.11 and 30E5.4) that were not adjacent; each showed a clear one-to-one relationship with distinct Drosophila genes (gsc and ind, respectively), indicating that both were present in the last common ancestor of the two species.
Syntenic and Microsyntenic Relationships with Drosophila.
By comparing in situ localization of putative A. gambiae and D. melanogaster orthologues on polytene chromosomes, we have reported recently that the major chromosomal arms of the mosquito and the fruit fly (five arms in each case) have retained sufficient similarity of gene content (ranging from 41% to 73%) to be recognizably homologous in terms of synteny (34). That study also suggested the existence of limited microsynteny (local conservation of gene order). To examine these questions more fully at the sequence level, we compared the chromosomal positions of putative Drosophila and Anopheles orthologues in the Pen 1 region (Fig. 2).
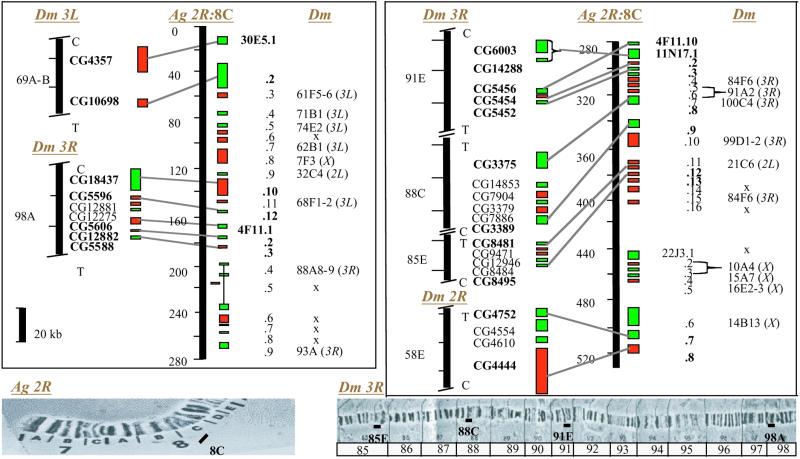
Figure 2.
Syntenic analysis, displaying all protein-coding genes of the Pen1 sequence in two large blocks, 0–280 and 280–528 kb (telomere to centromere orientation). Genes are numbered and colored as in Fig. 1. The chromosomal location of this sequence (Ag 2R:8C) is seen Lower Left. Microsyntenic Drosophila clusters are shown to the left of the mosquito sequence diagrams. Orthologues in both species are connected and shown in bold. C and T indicate the centromeric/telomeric orientation in Drosophila, and CG numbers refer to genes in the consensus fruit fly genome. Of the six microsyntenic clusters, one each maps to 3L and 2R and four to 3R in Drosophila (diagrammed at Lower Right). The Drosophila 69A-B cluster consists of two adjacent orthologues in both species, the 91E cluster shows a simple transposition, and the other four clusters are “hyphenated” by extra genes in either species. Hyphenated and nonclustered genes are not in bold. Their locations in Drosophila, if not in clusters, are indicated by individual cytogenetic locations to the right of the mosquito sequence; x represents the absence of a Drosophila orthologue. Two pairs of duplicated Anopheles genes are shown in brackets.
The analysis confirmed the homology of a chromosomal arm in the two species. The 36 mosquito genes that we are considering (counting duplicated genes only once) are all on 2R and of these 20 (55.6%) have putative orthologues in the 3R chromosomal arm of the fruit fly. Of the remainder, seven map to the fruit fly 3L, five to X, and two each to 2L and 2R. By comparison to a polynomial random distribution, this observed distribution has clear statistical significance and confirms the predominant homology of the A. gambiae 2R arm with the D. melanogaster 3R arm, which was estimated as 61.5% (34).
Interestingly the mosquito genes seem to be broadly clustered within the 528-kb chromosomal sequence, depending on which Drosophila chromosomal arm bears their homologues. All 20 genes with Drosophila orthologues on 3R are clustered within only half the mosquito sequence, 265 kb. All seven genes with Drosophila orthologues on 3L are found within 150 kb; four of the five mosquito genes with Drosophila X orthologues are clustered within 50 kb; the two genes with Drosophila 2R orthologues are adjacent in the mosquito, but the two with Drosophila 2L orthologues are far apart (250 kb). This loose clustering pattern persisting in the contemporary species may reflect residual persistence of gene arrangements in the last common ancestor of Anopheles and Drosophila at the chromosomal arm level.
Residual ancestral arrangements are most clearly evident at the finer level of local clustering (microsynteny). Six local clusters with 17 genes (nearly half of the total) are identified in Fig. 2 as including some genes that are neighbors in both species. However, local cluster may be interrupted (“hyphenated”) in either species by the presence of one or more noncorresponding genes. Direct contiguity is seen in the cluster of two genes, 30E5.1 and 30E5.2, whose orthologues are also contiguous on the Drosophila 3L arm (69A-B region). The genes 22J3.7 and 22J3.8 are also adjacent but correspond to a hyphenated cluster in the Drosophila 2R arm (58E region), including two extra genes. Four clusters of mosquito 8C region genes correspond to widely separated clusters on the D. melanogaster 3R arm. Of these the 85E and 88C Drosophila clusters are hyphenated; each cluster matches two adjacent mosquito genes (11N17.12/.13 and 11N17.8/.9, respectively). The 91E Drosophila cluster encompasses the same four genes in both species but with one local transposition. Finally, the 98A cluster includes the same five genes but in a hyphenated arrangement in both species.
A Variant Sequence Segment in the Pen1 Region.
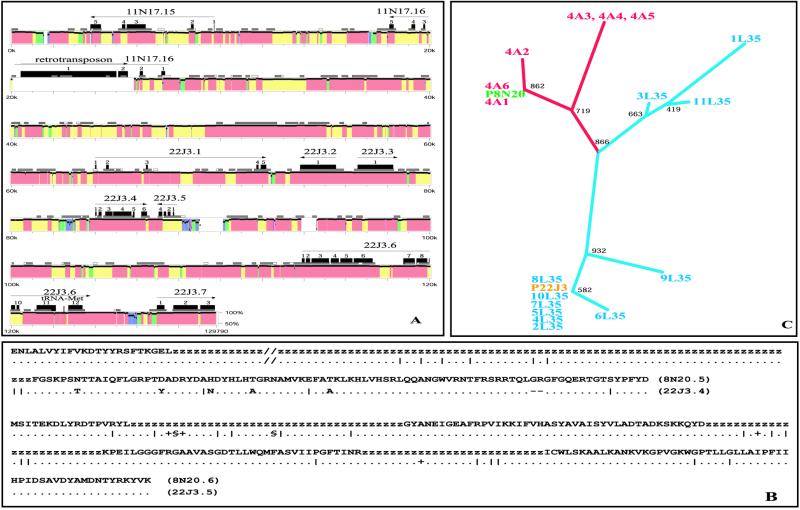
We serendipitously sequenced the 129-kb BAC 8N20, which clearly overlaps the Consensus Sequence between coordinates 382.9 and 505.8 kb, i.e., in part of 11N17, the entire trimmed 25F12 and much of 22J3 (Fig. 1). The latter three BACs show a high degree of similarity to each other, with their overlaps (21.2 kb in total) differing by only 0.02%. In strong contrast, the sequence of 8N20 differs extensively from its Consensus counterpart, by an average of 3.3% in 121.8 kb of aligned sequence. The differences are both single-base changes and short deletions/insertions (indels) and are widely distributed, more so but not exclusively in intergenic regions (Fig. 3A). For example, the three longest gene-free regions between 11N17.15 and 11N17.16 (8.3 kb in length), 11N17.16 to 22J3.1 (36.6 kb), and 22J3.5 to 22J3.6 (26 kb) show high overall divergence between the Consensus and the 8N20 sequences, but also include notable segments of high conservation (e.g., between 49.2–55 kb and 108.3–111 kb in the 8N20 sequence). On the other hand, gene-bearing segments tend to be well conserved (especially 22J3.2, .3), although many introns and some exons in several genes are also variant. Fig. 3B illustrates patterns of intragenic sequence variation in two of the genes. In the 8N20 sequence, gene 11N17.16 is interrupted by a putative retrotransposon similar to the BEL element (7511879) of D. melanogaster and is extensively diverged in sequence.
Figure 3.
Sequence variants. (A) Percent identity plot (PIP) analysis of the displayed BAC 8N20 (jagged black line) vs. the Consensus Sequence. Percent sequence identity is shown by colored blocks in pink (>98%), yellow (95–98%), green (90–95%), blue (80–90%), and purple (10–80%). Blocks in white between 1.6–1.7 kb, 88.9–90.1 kb, and 93.8–94.6 kb represent sequences present only in 8N20; the block between 20.1 and 25.8 kb is a retrotransposon that is only present in 8N20. Exons of the indicated genes are shown in black numbered boxes (short gray and white boxes represent CpG islands). (B) Examples of sequence variations with each letter corresponding to 3 nucleotides (amino acids or z for introns). The top sequence is exons 5 and 6 of (8N20.5/22J3.4) interrupted by an intron, and the bottom sequence is the four exons of (8N20.6/22J3.5) interrupted by introns. In each case, the aligned (8N20.5/22J3.4) and (8N20.6/22J3.5) sequences are shown by dots if identical. Coding differences are indicated as amino acid changes, whereas silent changes are indicated by tickmarks, + or §, depending on whether they involve one, two, or three DNA changes in the same triplet, respectively. (C) Phylogenetic tree of conceptually translated confirmed alleles of the complete gene 22J3.4 from susceptible 4Ar/r (red), refractory L3–5 (blue), and PEST laboratory strains. The PEST strains are from BAC 22J3 (Consensus Sequence, yellow) and BAC 8N20 (Variant, green). Bootstrap values are associated with the tree nodes.
Three potential explanations for the variant 8N20 clone can be advanced: that it represents a duplicated chromosomal region, that it is a chance contamination by DNA from a different mosquito species, and that it represents high localized polymorphism in A. gambiae. In situ hybridizations to polytene chromosomes showed that 8N20 is located in a single, not very dense polytene band near the telomeric end of subdivision 8C, as are clones 11N17, 25F12, and 22J3. BACs 30E5 and 4F11 hybridize to an adjacent band in 8C (http://konops.imbb.forth.gr/AnoDB/Cytomap/). These results strongly argue against the duplication hypothesis. To exclude the contamination hypothesis, we used two additional A. gambiae strains to clone by PCR and sequence one of the polymorphic genes, 22J3.4 (which has a Drosophila orthologue, the amiloride-sensitive Na+ channel gene, CG4805). These strains, 4Ar/r and L3–5, are susceptible and refractory (melanotically encapsulating) to Plasmodium, respectively, and are unrelated to the PEST strain from which both the 8N20 and the Consensus Sequence have been derived. Six clones from 4Ar/r and 11 clones from L3–5 were recovered. Their sequences and the corresponding PEST sequences derived from BACs 22J3 and 8N20 were conceptually translated to amino acid sequences, aligned, and a phylogenetic tree constructed with clustal x (24). The tree (Fig. 3C) showed interstrain differences and a certain level of polymorphism within both 4Ar/r and L3–5. Importantly, the version of the gene present in BAC 8N20 clustered with the 4Ar/r sequences, and the canonical 22J3 version clustered with most of the L3–5 sequences. Thus, the 8N20 and 22J3 versions of this putative Na+ channel gene have each been found in two different laboratory strains, effectively excluding the contamination hypothesis and arguing strongly that major local sequence variations exist in the A. gambiae genome.
Discussion
Determination of the 528-kb genomic sequence from the Pen1 region has yielded a preview into the structure of the A. gambiae genome. Comparison with the D. melanogaster genome identified 38 of the 46 protein-coding mosquito genes as putative orthologues of specific fruit fly genes, emphasizing the value of comparing these two insect genomes. Thus, for annotating the A. gambiae genome and for experimentally focusing future functional analysis on interesting candidate genes, the D. melanogaster sequence will serve as an invaluable guide.
Identification of the putative orthologues will become more secure soon, once both genomes are almost fully sequenced and annotated. Aside from the degree of sequence similarity, identification of orthologues will be greatly facilitated by their genomic context, whenever putative orthologues are found in microsyntenic clusters. Orthology can be challenged by postulating that the last common ancestor of these species had two adjacent paralogous genes, of which one was lost in the mosquito and the other in the fruit fly. Ocam's razor will have to be applied in the absence of any supporting evidence for such a hypothesis.
Reinforcing our previous postulate of chromosome arm homologies between the mosquito and the fruit fly, which was based on comparative in situ hybridizations to polytene chromosomes of a random set of putative orthologues (34), the present study indicates at the level of contiguous sequence that the 8C region of the mosquito (2R) predominantly corresponds to the fruit fly 3R chromosome arm. A close analysis indicates that extensive reshuffling is superimposed on this residual chromosomal arm homology. In summary, 13 Pen1 region genes have orthologues in four small, widely separated clusters in the Drosophila 3R chromosomal arm, 7 have matches in other parts of the same arm, and 16 in four different chromosomal arms. A striking pattern is localized microsynteny: six small gene clusters encompassing a total of 17 mosquito genes are represented in both species, but the clusters are often “hyphenated” by genes which have been added or removed in one of the species. We have also detected two cases of gene duplication in A. gambiae.
The other major conclusion of this study is that an unusually high degree of polymorphism exists in at least one specific section of the mosquito genome. Since this work was completed, a similar conclusion has been reached in the whole-genome sequencing effort (R. Holt, personal communication). A. gambiae (sensu strictu) has been distinguished into several different ecotypes on the basis of floating chromosomal inversions, whose prevalence varies geographically and seasonally, suggesting that they may “lock in” specific allelic combinations important for malaria transmission (35, 36). More recently, a small number of molecular markers have also suggested the existence of flexibility and even mosaicism in the A. gambiae genome (37, 38). The high level of polymorphism that we detected between BAC 8N20 and the Consensus Sequence is reminiscent of the human MHC locus, which encompasses at least 200 genes, most of which are highly polymorphic and have roles in immune responses (39). It remains to be determined how long this mosquito polymorphic DNA region is, and whether it may have resulted from a small (undetected) chromosomal inversion or from retroelement action (40). An intriguing possibility arising from Fig. 3C is that this extensive polymorphism may be correlated with mosquito refractoriness to the parasite. It should be noted that PEST strain mosquitoes exhibit both susceptible and melanotic refractory phenotypes.
Do any of the genes that we have described correspond to Pen1? This seems likely, as the sequence includes the closely linked marker H175, but genetic analysis of Pen1 is incomplete. Candidate genes would include at least the duplicated (V-ATPase subunit-α/mouse immune-suppressor TJ6 homologues 11N17.5/.6; the 11N17.16 gene, which is disrupted in 8N20; or indeed any of the genes encompassed in the variant 8N20 sequence. Refined genetic analysis, additional studies on sequence variations, and ultimately experimental analysis of gene function will be necessary to answer this question definitively.
Supplementary Material
Acknowledgments
We thank Kathleen Merz of Notre Dame for administrative support, James Hogan for help with BAC clone mapping, members of the Kafatos laboratory for discussions, and LION Biosciences AG for assistance in sequencing. L.Q.T. was supported by National Institutes of Health Predoctoral Training Grant T32-AI07030 (to F.H.C.). Financial support for this research was provided by the European Molecular Biology Laboratory and National Institute of Allergy and Infectious Diseases Grants R01-AI44273 and U01-AI48846 (to F.H.C.).
Abbreviations
- BAC
bacterial artificial chromosome
- EST
expressed sequence tag
Footnotes
References
- 1.Collins F H, Sakai R K, Vernick K D, Paskewitz S, Seeley D C, Miller L H, Collins W E, Campbell C C, Gwadz R W. Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 2.Paskewitz S M, Brown M R, Lea A O, Collins F H. J Parasitol. 1988;74:432–439. [PubMed] [Google Scholar]
- 3.Paskewitz S M, Brown M R, Collins F H, Lea A O. J Parasitol. 1989;75:594–600. [PubMed] [Google Scholar]
- 4.Zheng L, Cornel A J, Wang R, Erfle H, Voss H, Ansorge W, Kafatos F C, Collins F H. Science. 1997;276:425–428. doi: 10.1126/science.276.5311.425. [DOI] [PubMed] [Google Scholar]
- 5.Collins FH, Zheng L, Paskewitz S M, Kafatos F C. Ann Trop Med Parasitol. 1997;91:517–521. doi: 10.1080/00034989760888. [DOI] [PubMed] [Google Scholar]
- 6.Mukabayire O, Besansky N J. Chromosoma. 1996;104:585–595. doi: 10.1007/BF00352298. [DOI] [PubMed] [Google Scholar]
- 7.Saunders R D, Ashburner M, Coulson D, Glover D M, Kafatos F C, Louis C, Modolell J, Rimmington G A, Savakis C, Sidin-Kiamos I. Parassitologia (Rome) 1993;35:99–102. [PubMed] [Google Scholar]
- 8.Kumar V, Collins F H. Insect Mol Biol. 1994;3:41–47. doi: 10.1111/j.1365-2583.1994.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 9.Rowen L, Mahairas G, Hood L. Science. 1997;278:605–607. doi: 10.1126/science.278.5338.605. [DOI] [PubMed] [Google Scholar]
- 10.Rozen S, Skaletsky H. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 11.Voss H, Schwager C, Wiemann S, Zimmermann J, Stegemann J, Erfle H, Voie A M, Drzonek H, Ansorge W. J Biotechnol. 1995;41:121–129. doi: 10.1016/0168-1656(95)00007-d. [DOI] [PubMed] [Google Scholar]
- 12.Benes V, Kilger C, Voss H, Paabo S, Ansorge W. BioTechniques. 1997;23:98–100. doi: 10.2144/97231bm21. [DOI] [PubMed] [Google Scholar]
- 13.Wiemann S, Stegemann J, Grothues D, Bosch A, Estivill X, Schwager C, Zimmermann J, Voss H, Ansorge W. Anal Biochem. 1995;224:117–121. doi: 10.1006/abio.1995.1015. [DOI] [PubMed] [Google Scholar]
- 14.Schwager C, Wiemann S, Ansorge W. Genome Dig. 1995;2:8–9. [Google Scholar]
- 15.Voss H, Wiemann S, Grothues D, Sensen C, Zimmermann J, Schwager C, Stegemann J, Erfle H, Rupp T, Ansorge W. BioTechniques. 1993;15:714–721. [PubMed] [Google Scholar]
- 16.Strathmann M, Hamilton B A, Mayeda C A, Simon M I, Meyerowitz E M, Palazzolo M J. Proc Natl Acad Sci USA. 1991;88:1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 18.Parra G, Blanco E, Guigo R. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gemund C, Ramu C, Altenberg-Greulich B, Gibson T J. Nucleic Acids Res. 2001;29:1272–1277. doi: 10.1093/nar/29.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birney E, Durbin R. Genome Res. 2000;10:547–548. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M A, Barrell B. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 23.Lowe T M, Eddy S R. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen G J, Overbeek R, Larsen N, Marsh T L, McCaughey M J, Maciukenas M A, Kuan W M, Macke T J, Xing Y, Woese C R. Nucleic Acids Res. 1992;20:2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill F, Benes V, Thomasova D, Stewart A F, Kafatos F C, Ansorge W. Genomics. 2000;64:111–113. doi: 10.1006/geno.1999.6106. [DOI] [PubMed] [Google Scholar]
- 27.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 28.Besansky N J, Powell J R. J Med Entomol. 1992;29:125–128. doi: 10.1093/jmedent/29.1.125. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman S L, Subramaiian G M, Collins F H, Venter JC. Nature (London) 2002;415:702–709. doi: 10.1038/415702a. [DOI] [PubMed] [Google Scholar]
- 30.Ashburner M, Misra S, Roote J, Lewis S E, Blazej R, Davis T, Doyle C, Galle R, George R, Harris N, et al. Genetics. 1999;153:179–219. doi: 10.1093/genetics/153.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeates D K, Wiegmann B M. Annu Rev Entomol. 1999;44:397–428. doi: 10.1146/annurev.ento.44.1.397. [DOI] [PubMed] [Google Scholar]
- 32.Nei M, Xu P, Glazko G. Proc Natl Acad Sci USA. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayala F J, Rzhetsky A, Ayala F J. Proc Natl Acad Sci USA. 1998;95:606–611. doi: 10.1073/pnas.95.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolshakov V N, Topalis P, Blass C, Kokoza E, della Torre A, Kafatos F C, Louis C. Genome Res. 2002;12:57–66. doi: 10.1101/gr.196101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coluzzi M. Bull W H O. 1985;62:107–113. [PMC free article] [PubMed] [Google Scholar]
- 36.Toure Y T, Petrarca V, Traore S F, Coulibaly A, Maiga H M, Sankare O, Sow M, Di Deco M A, Coluzzi M. Parassitologia (Rome) 1998;40:477–511. [PubMed] [Google Scholar]
- 37.Mukabayire O, Caridi J, Wang X, Toure Y T, Coluzzi M, Besansky N J. Insect Mol Biol. 2001;10:33–46. doi: 10.1046/j.1365-2583.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- 38.Favia G, Lanfrancotti A, Spanos L, Siden-Kiamos I, Louis C. Insect Mol Biol. 2001;10:19–23. doi: 10.1046/j.1365-2583.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- 39.Bodmer J G, Marsh S G, Albert E D, Bodmer W F, Bontrop R E, Dupont B, Erlich H A, Hansen J A, Mach B, Mayr W R, et al. Hum Immunol. 2001;62:419–468. doi: 10.1016/s0198-8859(01)00229-4. [DOI] [PubMed] [Google Scholar]
- 40.Charlesworth B, Sniegowski P, Stephan W. Nature (London) 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.