Abstract
The calmodulin-dependent adenylate cyclase domain (Cya) of the Bordetella pertussis cyclolysin was used as a reporter protein to study the direct translocation of the Xanthomonas effector protein, AvrBs2, into the plant host cell. Adenylate cyclase activity (production of cAMP) depends on the presence of eukaryotic plant calmodulin and is only active after translocation from the prokaryotic cell into the eukaryotic plant cell. Here, we show that infection of pepper plants by Xanthomonas campestris pv. vesicatoria strains expressing the AvrBs2:Cya fusion protein results in detectable increases of cAMP levels in plant cells as early as 3 h after inoculation. Adenylate cyclase activity was shown to be type III secretion-dependent as the Xanthomonas hrp mutations, hrcV or hrpF, failed to produce detectable levels of cAMP in infected pepper plants. Furthermore, the N-terminal secretion and translocation signals of AvrBs2 were shown to be required for activity of the fusion protein in the plant. A single genomic copy of the avrBs2:cya fusion gene expressed under the control of the wild-type avrBs2 promoter was used to compare the effect of a susceptible and resistant plant interaction on the kinetics of effector protein delivery. Implications of these results and additional applications of this reporter construct are discussed.
The type III secretion system (TTSS) is a protein translocation system conserved in many Gram-negative pathogenic bacteria that infect plants and animals (1–3). The main function of the TTSS is to deliver bacterial effector proteins into the eukaryotic host cell to mimic, suppress, or modulate host defense signaling pathways and to cause disease. In the case of plants, a surveillance system encoded in part by plant resistance genes has evolved to specifically recognize these virulence effector proteins and to trigger host defense signaling pathways that result in pathogen inhibition. However, definitive evidence that effector proteins are translocated from phytopathogenic bacteria into the plant cell is currently lacking. Indirect evidence that effector proteins are translocated to the plant host is supported by the data that transient expression of these genes in host cells is sufficient to elicit a genetically defined hypersensitive cell death-resistance response (4–9). In addition, initiation of plant disease or plant resistance depends on a functional (TTSS) mechanism in these bacteria (10, 11), suggesting that bacterial effector proteins are translocated from the pathogen into the plant cytosol. Finally, that the majority of NBS/LRR disease-resistance proteins are thought to be cytosolic or associated with the plasma membrane is consistent with this hypothesis.
In plant pathogens, the TTSS is encoded by the clustered hrp (hypersensitive reaction and pathogenicity) genes (12). The homologies between the TTSS of mammalian and plant pathogens suggest that phytopathogenic bacteria translocate proteins into host cells by a process similar to that of animal pathogens. Indeed, the phytopathogen Xanthomonas campestris pv. vesicatoria (Xcv) is able to secrete type III effector proteins of the mammalian pathogen Yersinia enterocolitica (13). Plant cells, unlike animal cells, have a cell wall, however, and one might expect specific differences between pathogen secretion systems. Along these lines, a recent report discusses the role of pili in the in vitro protein secretion of phytopathogenic bacterial effector proteins (14).
In vitro evidence exists for type III effector protein secretion by the phytopathogenic bacteria Xcv, (13, 15), Pseudomonas syringae (16, 17), Ralstonia solanacearum (18), and Erwinia amylovora (19). Furthermore, the P. syringae AvrRpt2 effector protein was shown to be cleaved at the N terminus during bacterial infection and only occurred in bacteria that contained a functional TTSS. The proteolytic processing was demonstrated not to occur outside the host, suggesting that cleavage most likely occurred after translocation into the cytosol of the plant cell (16). Direct evidence for translocation into plant cells is lacking (10, 15), as it has been problematic to target reporter proteins through the TTSS (16, 20). In contrast to plant pathogens, type III-secreted effector proteins of the mammalian pathogens Y. enterocolitica (21, 22) and Salmonella dublin (23) have been detected inside host cells by immunofluorescence (24) as well as by detection of reporter fusions (21, 25).
In this article, we present biochemical evidence that supports the notion that phytopathogenic bacteria can deliver effector proteins directly into the plant host cell. We constructed a chimeric protein between the calmodulin-dependent adenylate cyclase domain (cya) of Bordetella pertussis cyclolysin (21) and the Xcv effector protein AvrBs2. Adenylate cyclase fusions have been used to demonstrate translocation of type III-secreted bacterial effector proteins of Yersinia, Salmonella, and Chlamydia into mammalian cells (21, 23, 26). Activity of this reporter enzyme depends on eukaryotic calmodulin and therefore the Cya fusion is only active after translocation from the bacteria into eukaryotic host cells (21). The use of this reporter strategy will also allow us to analyze further the TTSS in phytopathogenic bacteria in a more sensitive and precise manner.
Materials and Methods
Bacterial Strains.
The strains used in this study were Escherichia coli DH5α, Xcv strain 85-10 (contains avrBs2 and avrBs1) (27), 85-10 hrpG* (85*) (85-10 with a mutation for constitutive hrp gene expression) (13), 85*ΔhrcV (85* with a type III secretion-defective mutation) (15, 28), 85*ΔhrpF (85* with a mutation in hrpF) (29), and Xcv strain GM98-38 (lacks avrBs2 activity) (30). Xcv strains were grown at 28°C on nutrient yeast glucose agar containing 100 μg/ml of rifampicin. E. coli strain DH5α was grown at 37°C on Luria agar.
Genetic Constructs.
For protein expression experiments, the broad host range vector pDD62 was used (15). Plasmids were mobilized from E. coli into Xcv by triparental mating with standard methods (15). The plasmid pDD62(avrBs2:HA) (15) has wild-type avrBs2 activity and was used as a positive control strain. To construct pDD62 (avrBs2:cya), plasmid 81533B (pUC118 with avrBs2) (31) was modified by replacing the translational stop site at codon 714 with a BamHI site. AvrBs2 was then cloned as a BamHI fragment into the BglII site of pMS107 (cya) (21), in-frame with cya. The avrBs2:cya fusion was then cloned as an XhoI/SacI fragment into the XhoI/SacI sites of pDD62, yielding pDD62 (avrBs2:cya). To make the C-terminal deletion constructs pDD62 (avrBs21-97:cya), pDD62 (avrBs21-58:cya), pDD62 (avrBs21-41:cya), pDD62 (avrBs21-28:cya), and pDD62 (avrBs21-18:cya), we recloned cya from pMS107 (cya) cut with BglII and SacI, and cloned into the BamHI/SacI sites of pUC19. A HindIII/SacI cya fragment from pUC19(cya) was used to replace the avrRpt280-255 HindIII/SacI fragments of pDD62 (avrBs21-97+avrRpt280-255), pDD62 (avrBs21-58+avrRpt280-255), pDD62 (avrBs21-41+avrRpt280-255), pDD62 (avrBs21-28+avrRpt280-255), and pDD62 (avrBs21-18+avrRpt280-255) (10). pDD62 (avrBs218-714:cya) was constructed by replacing the N-terminal BamHI and HindIII fragment of pDD62 (avrBs2:cya) with the N-terminal BamHI and HindIII fragments of pDD62 (avrBs218-714+avrRpt280-255) (15).
For chromosomal gene exchanges, vector pLVC18 (DNA Plant Technology, Oakland, CA) was used. pLVC18 is a derivative of pBR322 with a 1.8-Kb mob+ fragment of pRSF1010 cloned into the PvuII site. To fuse Cya to the chromosomal copy of avrBs2, we cloned the EcoRI fragment from pDD62 (avrBs2:cya) (codons 74–714) fused to cya, into the EcoRI site of pLVC18, and the vector was conjugated into Xvc strains 85-10 and 85* by triparental mating. Single crossover events were selected on 10 μg/ml of tetracycline. Immunoblot analysis verified that strains 85-10(avrBs2:cya) and 85*(avrBs2:cya) expressed the expected 126-kDa fusion protein (data not shown).
To construct pTOPO(hrpF), a 2,565-bp fragment (beginning 140 bp N-terminal to the start site) was PCR-amplified from Xcv 85-10 genomic DNA with XbaI restriction sites on each end and cloned into pCR 2.1-Topo (Invitrogen). pSVP61 (hrpF) was made by cloning the 2,565-Kb XbaI hrpF fragment (C,T partially filled in) into the pSVP61 HindIII site (G,A partially filled in) behind the Lac promoter.
To construct the 85*ΔhrpF mutant, a 700-bp EcoRI-XhoI fragment of the hrpF ORF was cloned from pSVP61(hrpF) into the EcoRI and XhoI sites of the pBlueKS(+) vector pBKS+hrpF(0.7). The hrpF(0.7) fragment was cut back out with KpnI and XbaI, and cloned into the KpnI and XbaI sites of pLVC18L to make pLVC18LhrpF(0.7). Vector pLVC18L is vector pLVC18 with an additional polylinker from the puc19-polylinker (sites EcoRI to PstI). pLVC18LhrpF(0.7) was conjugated into strain 85*. Single crossover events between chromosomal DNA and vector hrpF 700-bp EcoRI-XhoI piece of DNA resulted in disruption of the chromosomal hrpF gene and were selected by tetracycline resistance. The selected colonies were confirmed to have a hrp(−) phenotype. To confirm that the hrp− phenotype resulted from the disruption of hrpF, pSVP61 (hrpF) was conjugated into the 85*ΔhrpF mutant and tested for complementation to wild-type activity.
Plant Growth and Inoculation.
Pepper cultivars ECW-0 (bs2/bs2) and the nearly isogenic line ECW-20R (Bs2/Bs2) (32) were grown in a greenhouse. The upper fully expanded leaves of 2-month-old plants were hand-infiltrated with a 108 colony-forming unit per ml bacterial suspension with a 1-ml syringe. After infiltrations, plants were kept in continuous low light, and leaf samples were harvested at various times and stored at −80°C for cAMP and protein analyses.
Cya Activity Assay and cAMP Extraction.
Xcv cells were grown on nutrient yeast glucose agar plates, resuspended to an OD600 of 2.0 in sonication buffer (3.3 mM Tris⋅HCl, pH 7.4; 0.75 M sucrose), and sonicated on ice 3 times for 30 sec (Branson sonifier at 50% power). An aliquot of 20 μl was suspended in 79 μl of reaction buffer (final concentrations: 50 mM Tris⋅HCl, pH 8; 2 mM ATP; 6 mM MgCl2; 100 μg/ml BSA; 0.12 mM CaCl) (21). As indicated, 1 μl of bovine calmodulin (Sigma, 100 mM stock) or water was added to the reaction mix. The reaction was allowed to proceed for 5 min at 30°C and was stopped by the addition of 650 μl of HClO4 (1.1 M). After centrifugation, a 670-μl aliquot of the supernatant was neutralized with 90 μl of K2CO3 (6 M). The salt was removed by pelleting in an Eppendorf microfuge for 10 min. Two 100-μl aliquots of the resulting supernatant were transferred to new tubes and dried in a heated speed-vac. Samples were stored frozen at −80°C until assayed for cAMP. The cAMP levels were measured with a cAMP enzyme immunoassay kit (Biotrak, Amersham Pharmacia Biotech) according to the manufacturer's instruction and expressed as nmol of cAMP per mg of total protein.
For the isolation of cAMP from plants (modified after ref. 33), two leaf discs (0.5 cm2 each) were collected and ground in liquid nitrogen. After evaporation of the liquid nitrogen, leaves were further ground in 325 μl of HClO4 (1.1 M). After centrifugation, the pellet was saved for protein concentration determination [a modified Lowry procedure (34)]. Then 280 μl of the supernatant was neutralized in 37.3 μl of K2CO3 (6 M). The salt was removed by pelleting in an Eppendorf microfuge for 10 min. Two 100-μl aliquots of the resulting supernatant were transferred into new tubes and dried in a heated speed-vac. Samples were stored frozen at −80°C until assayed for cAMP.
Secretion Assay.
The secretion of AvrBs2:Cya fusion protein by Xcv strains 85* pDD62 (avrBs2:cya), 85*ΔhrcV pDD62 (avrBs2:cya), and 85*ΔhrpF pDD62 (avrBs2:cya) into culture media was detected by immunoblot analysis of trichloroacetic acid (TCA) protein-precipitated culture filtrates as described (15).
SDS/PAGE Gels and Immunoblot Analysis.
Xcv cells were grown on nutrient yeast glucose agar plates and resuspended to an OD600 of 2.0 in 100 mM MgCl2. A 50-μl aliquot was mixed with 50 μl 3 times Laemmli buffer (35), boiled for 5 min, and centrifuged at 14,000 × g for 10 min. Fifteen microliter of the supernatant was loaded on 8% SDS/PAGE gels. Proteins were transferred onto nitrocellulose membranes and detected with polyclonal rabbit antisera raised against AvrBs2 (30) and NPT II (obtained from 5 Prime → 3 Prime), and monoclonal mouse antiserum against adenylate cyclase (courtesy of N. Giuso, Institut Pasteur, Paris). Membranes were incubated with peroxidase-conjugated secondary Ab and proteins were visualized by chemiluminescence.
Results
Avirulence Activity of the AvrBs2:Cya Fusion.
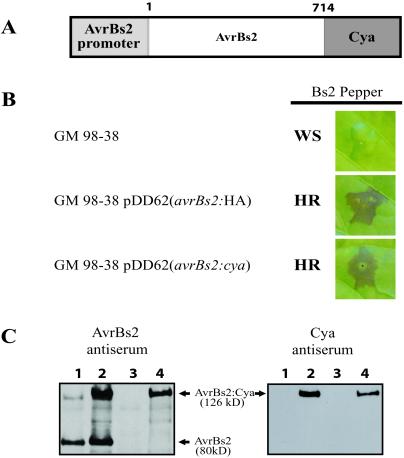
To determine whether Cya could be used as reporter protein without altering the avirulence activity of avrBs2, a translational C-terminal fusion of the Cya protein to the full-length AvrBs2 protein was constructed. The plasmid pDD62(avrBs2:cya), which encodes this chimeric protein, was conjugated into Xcv strain GM98-38 and was tested for avrBs2 activity (30). The phenotype of this exconjugant and GM98-38 pDD62 (avrBs2:HA) showed a Bs2-specific hypersensitive cell death response (HR) when inoculated on the Bs2-resistance pepper cultivar (ECW-20R) (Fig. 1A). These results indicated that the AvrBs2:Cya fusion protein fully complemented avrBs2 activity (Fig. 1B).
Figure 1.
Biological activity of the AvrBs2:Cya fusion protein. (A) Schematic of chimeric AvrBs2:Cya fusion protein construct shows the 180-bp promoter region of avrBs2 with the full-length codons (1–714) fused to Cya as a translational fusion. (B) Xcv strain GM98-38 (no avrBs2 activity) with avrBs2 fusion constructs showing avrBs2-dependent cell death on Bs2-resistant pepper (ECW-20R). Leaves were inoculated with a 5 × 108 colony-forming units per ml suspension of bacteria and then photographed 48 h p.i. WS, water-soaked lesions; HR, hypersensitive resistance. (C) Immunoblot analysis of cellular lysate from Xcv expressing AvrBs2:Cya with either α-AvrBs2 polyclonal Ab or monoclonal mouse antiserum against adenylate cyclase. Lane 1, 85* (hrpG mutant with constitutive hrp expression and a wild-type copy of avrBs2); lane 2, 85* pDD62(avrBs2:cya) (85* also expressing AvrBs2:Cya); lane 3, GM98-38 (mutant for avrBs2); lane 4, GM98-38 pDD62(avrBs2:cya).
Immunoblot analysis of proteins from whole-cell lysates from the two Xcv strains, 85* and GM98-38 expressing AvrBs2:Cya, were probed with either the AvrBs2 polyclonal Ab or monoclonal adenylate cyclase antisera (Fig. 1C). As shown in Fig. 1C, only the Xcv strains that express avrBs2:cya contain the expected 126-kDa hybridizing band with both antisera (lanes 2 and 4). The Xcv strain 85* (lane 1) has a genomic wild-type copy of avrBs2 and has the appropriate 80-kDa hybridizing band when probed with the AvrBs2 polyclonal Ab. The 80-kD band is missing in strain GM 98-38 (lane 3), which lacks the genomic copy of avrBs2.
Type III-Dependent Secretion of AvrBs2:Cya Fusion Protein.
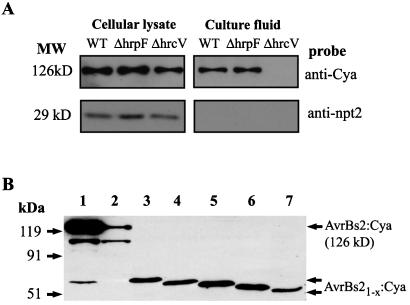
Once it was established that the AvrBs2:Cya fusions were capable of eliciting a Bs2-specific resistance reaction, we tested whether these proteins were secreted in vitro in a TTSS-dependent manner. To this end, we used the Xcv 85* strain containing the AvrBs2:Cya fusion and the previously characterized hrp mutation, hrcV. Immunoblot analyses revealed that AvrBs2:Cya fusion protein was present in whole-cell lysates from strains of Xcv 85* and 85*ΔhrcV (Fig. 2A). As predicted, Xcv 85* secreted the AvrBs2:Cya fusion protein into the culture media, whereas the negative control strain, 85*ΔhrcV, failed to secrete the protein into the culture medium (Fig. 2A).
Figure 2.
Type III-dependent secretion of AvrBs2:Cya protein from Xcv. (A) Immunoblot analysis of AvrBs2:Cya protein and NPT II protein in cellular lysates and proteins secreted into culture fluids isolated from Xcv strains expressing AvrBs2:Cya protein (126 kD) and NPT II protein (29-kDa nonsecreted control). Lane WT, wild-type 85* secretion strain; lane ΔhrpF, strain 85* with an hrpF mutation; lane ΔhrcV, strain 85* with a TTSS mutation. (B) Immunoblot analysis of AvrBs2 translocation domains with Cya fusion reporter. Equally loaded bacterial lysates of Xcv strain 85* expressing AvrBs2 or various AvrBs2 deletions as translational fusions with Cya. Lane 1, AvrBs21-714:Cya; lane 2, AvrBs218-714:Cya; lane 3, AvrBs21-98:Cya; lane 4, AvrBs21-58:Cya; lane 5, AvrBs21-41:Cya; lane 6, AvrBs21-28:Cya; lane 7, AvrBs21-18:Cya. Immunoblot was probed with monoclonal mouse antiserum against adenylate cyclase.
Furthermore, we constructed an additional control strain 85*ΔhrpF, which contained the avrBs2:cya fusion. The inclusion of this strain served as an important control as it is unable to elicit a Bs2-specific HR response when inoculated into pepper plants (data not shown), but is still capable of secretion (Fig. 2A). A plasmid-borne copy of the wild-type hrpF gene pSVP61(hrpF) was introduced into 85*ΔhrpF and the strain was complemented for HR-inducing activity (data not shown).
Calmodulin-Dependent Adenylate Cyclase Activity of the AvrBs2:Cya Fusion Protein Expressed in Bacteria.
Because the AvrBs2:Cya fusion should only be active in the presence of plant calmodulin, we determined whether the AvrBs2:Cya fusion protein was biochemically active with respect to production of cAMP when expressed in bacteria. Adenylate cyclase activity in total bacterial protein extracts was assayed to test the enzymatic activity of the AvrBs2:Cya fusion protein and to demonstrate that it was possible to activate the enzyme by the addition of eukaryotic calmodulin in vitro. Bacterial extracts containing the AvrBs2:Cya fusion protein had negligible cAMP activity in the absence of calmodulin, whereas addition of exogenous calmodulin resulted in significant increases in cAMP levels (Table 1). The calmodulin-dependent increase in in vitro Cya activity was comparable to that reported in Yersinia strains expressing a YopE:Cya fusion (21), although the total amount of in vitro Cya activity in Xcv was slightly lower. These experiments confirmed the eukaryotic calmodulin-dependent nature of the activity of the fusion protein expressed in Xcv and the measurement of the in planta translocation of the AvrBs2:Cya fusion protein.
Table 1.
Calmodulin-dependent adenylate cyclase activity of AvrBs2:Cya fusion protein expressed in vitro and in planta
| Xcv strain | AvrBs2 protein† |
in vitro Cya activity cAMP§ (nmol/mg total proteins)
|
in planta Cya activity cAMP§ (nmol/mg total proteins)
|
|
|---|---|---|---|---|
| − Calmodulin | + Calmodulin | Time (24 h p.i.) | ||
| Mature protein | ||||
| 85* | 1-714-HA‡ | 0.6 ± 0.6¶ | 0.6 ± 0.4 | 0.02 ± 0.006 |
| 85* | 1-714-Cya | 0.9 ± 1.1 | 917 ± 196 | 38.0 ± 0.9 |
| 85*ΔhrpF | 1-714-Cya | 0.6 ± 0.6 | 1,340 ± 60 | 0.02 ± 0.002 |
| 85*ΔhrcV | 1-714-Cya | 0.9 ± 1.0 | 941 ± 23 | 0.03 ± 0.002 |
| N-terminal deletions | ||||
| 85* | 18-714-Cya | 340 | 0.4 ± 0.2 | |
| C-terminal deletions | ||||
| 85* | 1-98-Cya | 580 | 43.9 ± 1.6 | |
| 85* | 1-58-Cya | 529 | 38.6 ± 1.2 | |
| 85* | 1-41-Cya | 850 | 5.7 ± 3.2 | |
| 85* | 1-28-Cya | 910 | 0.9 ± 0.19 | |
| 85* | 1-18-Cya | 334 | 0.3 ± 0.17 | |
Codon numbering is based on predicted protein for ORF1 (30).
HA, hemagglutinin epitope.
Adenylate cyclase activity in vitro (nmol cAMP per mg total protein) in Xcv-sonicated cellular lysates with and without exogenous calmodulin. Adenylate cyclase activity in planta (nmol cAMP per mg total protein) pepper (ECW) inoculated with Xcv and sampled 24 h p.i. All AvrBs2 fusion proteins are expressed in plasmid pDD62.
Each measurement has three replications that are averaged with SDs represented.
Translocation of the AvrBs2:Cya Fusion Protein into the Plant Cell.
To determine whether AvrBs2:Cya is translocated into the plant cytosol and reacts with plant-produced calmodulin, we inoculated pepper plants with Xcv strains expressing the AvrBs2:Cya fusion protein. We predicted that if the fusion protein was translocated into the plant cytosol, Cya would be active in the presence of plant-produced calmodulin and increased levels of adenylate cyclase activity could be measured. Table 1 depicts a large increase in cAMP levels in plants (adenylate cyclase activity in planta) 24 h after inoculation with Xcv stain 85* carrying pDD62 (avrBs2:cya) compared with a control strain with no AvrBs2:Cya. In addition, there was no increase in adenylate cyclase activity in planta with either Xcv strain 85*ΔhrcV, a secretion-defective strain, or 85*ΔhrpF, a translocation-defective mutant. The lack of Cya activity in 85*ΔhrpF is especially significant as this result rules out the possibility that the reporter protein can be first secreted and then delivered to the plant cell.
Analysis of the AvrBs2 Translocation Domains on the Delivery of Cya to Plant Cells.
To compare the quantitative sensitivity of the Cya reporter to the published AvrRpt2 reporter protein (15), we exchanged the AvrRpt2 reporter with Cya on a number of AvrBs2 deletions that define the in planta secretion and translocation domains of AvrBs2 (15). Immunoblot analyses of whole-cell lysates from these Xcv strains showed the expected sizes of these fusion proteins (Fig. 2B). As shown in Table 1, the calmodulin-dependent in vitro Cya activity measurements of bacterial extracts expressing these fusion proteins was reduced compared with the expression of full-length AvrBs2:Cya fusion protein.
To measure increases of in planta Cya activity of the various constructs, leaf samples were taken 24 h after inoculation and assayed for cAMP levels. Table 1 shows the results of the inoculation of pepper plants with Xcv strains expressing AvrBs21-98:Cya and AvrBs21-58:Cya fusions. These constructs resulted in increased in planta cAMP accumulation (similar to the expression level of full-length AvrBs2:Cya) in plants and were comparable to previously reported experiments for the translocation of the AvrRpt2 reporter. Infections of pepper plants with Xcv strains expressing progressively smaller truncated proteins, AvrBs21-41:Cya, AvrBs21-28:Cya, and AvrBs21-18:Cya resulted in significantly less cAMP accumulation in plants. The AvrBs21-18:Cya and AvrBs21-28:Cya fusions were inactive in planta, but the AvrBs21-41:Cya fusion had low activity (Table 1). As reported (15), these three C-terminal deletions, when fused to the AvrRpt280-255 reporter, were not able to translocate high enough levels to elicit the AvrRpt2-dependent cell death. Thus, the AvrBs2:Cya reporter protein seems to be a more sensitive assay in the case of AvrBs21-41:Cya fusion.
Quantitative Changes in Early Type III Translocation Kinetics.
The development of a sensitive biochemical reporter to assay translocation of bacterial effector proteins to plant cells prompted us to investigate the early translocation kinetics of AvrBs2:Cya. To carry out these experiments, we initially characterized Xcv strains expressing different levels of AvrBs2:Cya fusion protein based on whether the gene was expressed in an HrpG* or wild-type background and whether the gene was expressed from a plasmid or as a single chromosomal copy. Immunoblot analyses of three Xcv strains with different expression levels of the AvrBs2:Cya fusion protein all contained the predicted 126-kDa-size fusion protein (data not shown). Xcv 85* pDD62(avrBs2:cya) contained the highest level of protein expression and Xcv 85*(avrBs2:cya) expressed an intermediate level, whereas 85-10 (avrBs2:cya) expressed the lowest level (data not shown). Furthermore, the levels of protein expression correlated with the level of adenylate cyclase activity of these strains in the in vitro cAMP assay (Table 2).
Table 2.
Effect of gene copy no. on adenylate ayclase activity in vitro (nmol cAMP per mg total proteins) in Xcv-sonicated cellular lysates with and without exogenous calmodulin
| Xcv strain | Plasmid construct |
in vitro Cya activity cAMP (nmol/mg total proteins)
|
|
|---|---|---|---|
| − Calmodulin | + Calmodulin | ||
| 85* | pDD62 (avrBs2:HA) | 0.6 ± 0.6† | 0.6 ± 0.4 |
| 85* | pDD62(avrBs2:cya) | 0.9 ± 1.1 | 917 ± 196 |
| 85*(avrBs2:cya) | — | 0.7 ± 1.0 | 120 ± 27 |
| 85-10(avrBs2:cya) | — | 0.9 ± 1.0 | 11.4 ± 4.3 |
HA, hemagglutinin.
Each measurement has three replications that are averaged with SDs represented.
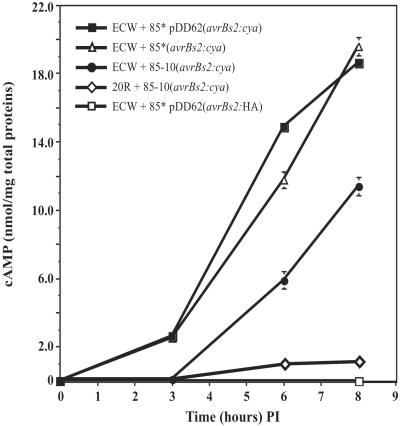
To determine the kinetics of AvrBs2:Cya translocation in the early stages of plant infection, Cya activity was assayed over an 8-h period at 0, 3, 6, and 8 h post inoculation (p.i.). The kinetic analysis was limited to 8 h p.i. to minimize the effects of in planta bacterial growth and to precede the visual symptoms associated with hypersensitive cell death. At the 8-h time point there was no significant increase in bacterial growth in any strains used. A comparison between the 85*(avrBs2:cya) and 85*pDD62 (avrBs2:cya) strains revealed that the in planta Cya activity can be detected as early as 3 h p.i. and rapidly increases to a high level within 8 h (Fig. 3). Although there is a significant difference in the calmodulin-reconstituted in vitro activity between these two strains, there seems to be little effect on whether the reporter protein is expressed from a single chromosomal copy [85*(avrBs2:cya)] or a from a multiple-copy plasmid [85*pDD62 (avrBs2:cya)]. Finally, it is also apparent that the 85-10(avrBs2:cya) strain, which lacks the HrpG* mutation, is delayed in its ability to translocate the reporter protein, as in planta activity can only be slightly detected after 6 h p.i. However, these levels may more closely reflect natural infections and the time it takes to synthesize the secretion apparatus encoded by the Hrp loci. Thus, we used this strain to compare the effect of resistant host genotype on effector protein delivery.
Figure 3.
Kinetic analysis of in planta adenylate cyclase activity for different strains of Xcv inoculated into pepper plants. Leaf samples (1 cm2) were taken at times 0, 3, 6, and 8 h p.i. with a 5 × 108 cell per ml suspension of bacteria. Each point represents three replications that are averaged with SDs represented by error bars.
Xcv strain 85-10(avrBs2:cya) was inoculated on the resistant pepper ECW-20R (Bs2/Bs2) to observe the effect of an incompatible host–pathogen interaction on the delivery of AvrBs2:Cya to plant cells. Interestingly, the translocation of AvrBs2:Cya is significantly inhibited in the ECW-20R pepper plants at 8 h p.i (Fig. 3).
Discussion
The work described in this article provides biochemical evidence for TTSS-dependent translocation of a bacterial Avr effector protein from a phytopathogenic bacterium into the cytosol of plant cells. We fused the calmodulin-activated adenylate cyclase domain (Cya) of B. pertussis cyclolysin (21) to AvrBs2 and expressed this construct either from the bacterial chromosome or in trans from a broad host-range vector. Inside bacteria, no calmodulin is available and Cya has little activity (Table 1). However, after translocation of AvrBs2:Cya into plant cells, the eukaryotic calmodulin activates the adenylate cyclase, and cAMP levels inside the plants increase significantly.
Translocation of AvrBs2 depends on the TTSS of Xcv. A mutant deficient in hrcV, an inner membrane protein of the type III secretion protein complex (29), did not secrete AvrBs2:Cya, and pepper infections with this strain did not result in increased cAMP levels in the plant. In accordance with this result, an N-terminal truncation of AvrBs2:Cya lacking the previously identified type III translocation signal (15) of AvrBs2 (AvrBs218-714:Cya) did not result in increased cAMP levels when assayed in planta (Table 1).
To ensure that accumulation of cAMP in plants depends not only on secretion of AvrBs2:cya into the plant apoplast but also on translocation into plant cells, we used a bacterial mutant that lacks hrpF, a putative translocator of the type III export mechanism (29). The mutant bacteria secreted the fusion protein into induction medium but failed to increase cAMP levels in plant cells, suggesting that it was inhibited in type III translocation.
In addition, we fused the adenylate cyclase reporter to C-terminal truncations of AvrBs2 that lacked the defined translocation signal sequence between amino acids 41 and 58 (15). The AvrBs21-41:Cya fusion construct resulted in reduced adenylate cyclase activity in plant cells, whereas the AvrBs21-58:Cya fusion was similar to a full-length AvrBs2 fusion (Table 1). These results suggest that AvrBs2 has two N-terminal secretion and translocation signals: one within the first 18 amino acids, and a second signal between amino acids 41 and 58. The first signal is absolutely required for secretion (15) and the second signal enhances translocation. The existence of two translocation signals has also been observed in the type III-secreted Yersinia effector YopE. The first YopE signal is located in the 15 N-terminal amino acids. However, there is controversy as to whether it resides in the mRNA (36) or in the protein (37). The second signal is located between amino acid residues 15–100 and requires binding of the chaperone SycE for protein stability and type III secretion (38).
Our experiments showed that cAMP accumulation was significantly lower in plants that are resistant to AvrBs2-expressing Xcv strains compared with susceptible cultivars. There was no difference in bacterial growth on resistant or susceptible plants during the duration of the experiment. An HR-related general protein breakdown may reduce the amount of the fusion protein, although Boyes et al. (39) have shown that a general protein breakdown only occurs well after the visible HR-related tissue collapse. Indeed, preliminary results suggest that the decrease in the adenylate cyclase activity of AvrBs2:Cya is specifically related to the resistance response mediated by the AvrBs2 effector protein and the Bs2 plant-resistance gene product. This decrease does not seem to involve a specific AvrBs2:Cya breakdown, and it is therefore possible that AvrBs2-induced plant resistance mechanisms interfere directly with type III-dependent translocation of effector proteins into plant cells. As our results indicate, such interference seems to occur within hours of infection of resistant pepper cultivars. Alternatively, it is also possible that calmodulin levels may rapidly decrease in a Bs2-specified resistance response or that the AvrBs2:Cya fusion protein may be sequestered into complexes that are not mutually accessible.
The adenylate cyclase reporter protein will have wide utility to establish further early events after pathogen infection. Preliminary results in our laboratory have demonstrated that chimeric fusions of Cya with the translocation signals of either AvrRpt2 and AvrRpm1 result in accumulation of cAMP in planta when delivered from P. syringae (M. Mudgett, D.D., and B.J.S., unpublished observations). Finally, fusions to other effector proteins will be instrumental to dissect specific and common responses of the numerous resistance pathways that are mediated by bacterial Avr genes and the corresponding plant-resistance genes.
Acknowledgments
We thank Christina Morales for technical assistance in the PCR-cloning of hrpF. We are thankful to Dr. Nicole Giuso for the generous gift of adenylate cyclase antibody. We also thank Dr. Ulla Bonas for use of the various published hrp strains of Xcv. We thank Mary Beth Mudgett, Stephen Chisholm, Mike Axtell, Brad Day, Todd Leister, and other members of the Staskawicz lab for helpful suggestions and critical reading of the manuscript. This work was supported by the Department of Energy and the Torrey Mesa Research Institute, Syngenta Research and Technology, San Diego.
Abbreviations
- TTSS
the type III secretion system
- HR
hypersensitive cell death response
- p.i.
post inoculation
References
- 1.Anderson D M, Derrick E F, Collmer A, Schneewind O. Proc Natl Acad Sci USA. 1999;96:12839–12843. doi: 10.1073/pnas.96.22.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galán J E, Collmer A. Science. 1999;284:1322–1327. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis G R, Van Gijsegem F. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 4.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leister R T, Ausubel F M, Katagiri F. Proc Natl Acad Sci USA. 1996;93:15497–15502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 7.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 8.Bonas U, Van den Ackerveken G. Plant J. 1997;12:1–7. doi: 10.1046/j.1365-313x.1997.12010001.x. [DOI] [PubMed] [Google Scholar]
- 9.McNellis T W, Mudgett M B, Li K, Aoyama T, Horvath D, Chua N-H, Staskawicz B J. Plant J. 1998;14:247–257. doi: 10.1046/j.1365-313x.1998.00106.x. [DOI] [PubMed] [Google Scholar]
- 10.Kjemtrup S, Nimchuk Z, Dangl J L. Curr Opin Microbiol. 2000;3:73–78. doi: 10.1016/s1369-5274(99)00054-5. [DOI] [PubMed] [Google Scholar]
- 11.Mudgett M B, Staskawicz B J. Curr Opin Microbiol. 1998;1:109–114. doi: 10.1016/s1369-5274(98)80150-1. [DOI] [PubMed] [Google Scholar]
- 12.Bonas U, Schulte R, Fenselau S, Minsavage G V, Staskawicz B. Mol Plant–Microbe Interact. 1991;4:81–88. [Google Scholar]
- 13.Rossier O, Wengelnik K, Hahn K, Bonas U. Proc Natl Acad Sci USA. 1999;96:9368–9373. doi: 10.1073/pnas.96.16.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Q, He S-Y. Science. 2001;294:2556–2558. doi: 10.1126/science.1066397. [DOI] [PubMed] [Google Scholar]
- 15.Mudgett M B, Chesnokova O, Dahlbeck D, Clark E T, Rossier O, Bonas U, Staskawicz B J. Proc Natl Acad Sci USA. 2000;97:13324–13329. doi: 10.1073/pnas.230450797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mudgett M B, Staskawicz B J. Mol Microbiol. 1999;32:927–941. doi: 10.1046/j.1365-2958.1999.01403.x. [DOI] [PubMed] [Google Scholar]
- 17.Van Dijk K, Fouts D E, Rehm D E, Hill A H, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Gijsegem F, Vasse J, Camus J-C, Marenda M, Boucher C. Mol Microbiol. 2000;36:249–260. doi: 10.1046/j.1365-2958.2000.01851.x. [DOI] [PubMed] [Google Scholar]
- 19.Wei Z, Kim J F, Beer S V. Mol Plant–Microbe Interact. 2000;13:1251–1262. doi: 10.1094/MPMI.2000.13.11.1251. [DOI] [PubMed] [Google Scholar]
- 20.Collmer A, Badel J L, Charkowski A O, Deng W-D, Fouts D E, Ramos A R, Rehm A H, Anderson D M, Schneewind O, van Dijk K, Alfano J R. Proc Natl Acad Sci USA. 2000;97:8770–8777. doi: 10.1073/pnas.97.16.8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sory M P, Cornelis G R. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 22.Sory M-P, Boland A, Lambermont I, Cornelis G R. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones M A, Wood M W, Mullan P B, Watson P R, Wallis T S, Galyov E E. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosqvist R, Magnusson K-E, Wolf-Watz H. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobi C A, Roggenkamp A, Rakin A, Zumbihl R, Leitritz L, Heesemann J. Mol Microbiol. 1998;30:865–882. doi: 10.1046/j.1365-2958.1998.01128.x. [DOI] [PubMed] [Google Scholar]
- 26.Subtil A, Parsot C, Dautry-Varsat A. Mol Microbiol. 2001;39:792–800. doi: 10.1046/j.1365-2958.2001.02272.x. [DOI] [PubMed] [Google Scholar]
- 27.Bonas U, Stall R E, Staskawicz B. Mol Gen Genet. 1989;218:127–136. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- 28.Wengelnik K, Rossier O, Bonas U. J Bacteriol. 1999;181:6828–6831. doi: 10.1128/jb.181.21.6828-6831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossier O, Van den Ackerveken G, Bonas U. Mol Microbiol. 2000;38:828–838. doi: 10.1046/j.1365-2958.2000.02173.x. [DOI] [PubMed] [Google Scholar]
- 30.Gassmann W, Dahlbeck D, Chesnokova O, Minsavage G V, Jones J B, Staskawicz B J. J Bacteriol. 2000;182:7053–7059. doi: 10.1128/jb.182.24.7053-7059.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swords K M M, Dahlbeck D, Kearney B, Roy M, Staskawicz B J. J Bacteriol. 1996;178:4661–4669. doi: 10.1128/jb.178.15.4661-4669.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minsavage G V, Dahlbeck D, Whalen M C, Kearney B. Mol Plant–Microbe Interact. 1990;3:41–47. [Google Scholar]
- 33.Legendre L, Derckel J P, Wrisez F, Correze C, Audran J C, Haye B, Lambert B. Phytochemistry. 1997;44:769–774. [Google Scholar]
- 34.Bailey J L. Techniques in Protein Chemistry. New York: Elsevier; 1967. [Google Scholar]
- 35.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Cheng L W, Schneewind O. J Biol Chem. 1999;274:22102–22108. doi: 10.1074/jbc.274.31.22102. [DOI] [PubMed] [Google Scholar]
- 37.Lloyd S A, Norman M, Rosqvist R, Wolf-Watz H. Mol Microbiol. 2001;39:520–531. doi: 10.1046/j.1365-2958.2001.02271.x. [DOI] [PubMed] [Google Scholar]
- 38.Cheng L W, Anderson D M, Schneewind O. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 39.Boyes D C, Nam J, Dangl J L. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]