Abstract
DNA microarrays provide an opportunity to combine the principles of signature-tagged mutagenesis (STM) with microarray technology to identify potentially important bacterial virulence genes. The scope of DNA microarrays allows for less laborious screening on a much larger scale than possible by STM alone. We have adapted a microarray-based transposon tracking strategy for use with a Salmonella enterica serovar Typhimurium cDNA microarray in order to identify genes important for survival and replication in RAW 264.7 mouse macrophage-like cells or in the spleens of BALB/cJ mice. A 50,000-CFU transposon library of S. enterica serovar Typhimurium strain SL1344 was serially passaged in cultured macrophages or intraperitoneally inoculated into BALB/cJ mice. The bacterial genomic DNA was isolated and processed for analysis on the microarray. The novel application of this approach to identify mutants unable to survive in cultured cells resulted in the identification of components of Salmonella pathogenicity island 2 (SPI2), which is known to be critical for intracellular survival and replication. In addition, array results indicated that a number of SPI1-associated genes, currently not associated with intracellular survival, are negatively selected. However, of the SPI1-associated mutants individually tested for intracellular survival, only a sirA mutant exhibited reduced numbers relative to those of wild-type bacteria. Of the mutants unable to survive in mice, significant proportions are either components of the SPI2 pathogenicity island or involved in lipopolysaccharide synthesis. This observation is in agreement with results obtained in the original S. enterica serovar Typhimurium STM screen, illustrating the utility of this approach for the high-throughput identification of virulence factors important for survival in the host.
The Enterobacteriaceae family consists of several closely related genera, including Salmonella, which are responsible for a variety of diseases. Salmonella enterica consists of a large number of highly related serovars that are responsible for a variety of diseases in a wide range of host organisms. S. enterica is organized into several subspecies, and it is the subspecies I serovars that are primarily associated with pathogenesis in warm-blooded hosts. S. enterica serovar Typhimurium (hereafter referred to as serovar Typhimurium) is one of the leading causes of food-borne gastroenteritis in humans. In susceptible strains of mice, serovar Typhimurium can cause a systemic disease that mimics the human-specific infection typhoid fever, caused by S. enterica serovar Typhi. In resistant strains of mice that carry the Nramp1Gly169 (also known as Slc11a1) allele, a persistent long-term infection can be established similar to that seen in human typhoid carriers (45). Much of our understanding of the systemic diseases that Salmonella serovars can cause has been obtained by utilizing a number of inbred mouse lines of varying susceptibility to infection and disease (reviewed in references 39 and 56). As a complement to this model, there are a number of established cell culture systems of murine and nonmurine origin that have been invaluable to the understanding of Salmonella pathogenesis (reviewed in references 21 and 59), including macrophages and epithelial cell lines.
Serovar Typhimurium possesses a number of virulence determinants that facilitate its ability to colonize, invade the intestinal epithelia, and spread systemically. In addition to the repertoire of fimbriae that facilitate the attachment of the bacteria to host cells, tracts of horizontally acquired DNA known as pathogenicity islands also play a role in disease (20, 55). Two important islands, Salmonella pathogenicity island 1 (SPI1) (23) and SPI2 (52, 62), encode the components of type III secretions systems that form molecular syringes for the translocation of bacterial effector proteins into the host cell cytosol. These effectors directly or indirectly subvert or manipulate normal cellular processes to the benefit of the bacterium (reviewed in references 25, 41, and 73). SPI1 is expressed during the intestinal phase of the infection cycle, and its effector molecules actively induce bacterial entry into both phagocytic and nonphagocytic cells of the intestinal mucosa. The SPI1 protein SipB induces cell toxicity and cell death, resulting in the release of proinflammatory cytokines and a local inflammatory response (34, 46). Macrophages and other phagocytic cells subsequently ingest the bacteria into a specialized vacuole. SPI2 expression is activated within the phagosome of macrophage cells, and the translocated effectors serve to modify the intracellular environment to create a niche within which the bacteria can replicate. Ironically, these macrophages then serve as the transport system for bacteria to gain access to and colonize the liver and spleen via the reticuloendothelial system. Mutants of SPI2 are defective for replication in both macrophages and epithelial cells and are also attenuated for disease in the mouse (11, 19; reviewed in references 27 and 73).
The sequencing of S. enterica genomes, including serovars Typhimurium and Typhi (14, 42, 53), has greatly increased our understanding of the factors contributing to their evolution and the acquisition of genetic information that facilitates their pathogenesis. However, as with most organisms sequenced to date, there is still a significant portion of each genome for which no function or phenotype has been ascribed. A number of techniques have been developed in order to identify the genes that contribute to survival of an organism under conditions of selection or stress, such as those seen during the course of infection (26). One example, genetic footprinting, involves placing a transposon-mutagenized library under a selection condition (65). PCR is then performed using gene-specific primers in conjunction with a transposon-specific primer to amplify a portion of the gene of interest from the pool. Disappearance of the PCR product from the selected library suggests a role for the gene of interest in survival under that selection condition. Another example of a transposon-mediated approach is signature-tagged mutagenesis (STM) (31). In STM, each transposon insertion possesses a unique signature tag that can be amplified using a universal set of primers. The individually maintained mutants are pooled together and placed under a selection condition, and the primers are used to amplify the unique tags. This pool of PCR products is then hybridized to a membrane where each feature corresponds to a specific mutant. Based upon the strength of the hybridization signal, mutants that are diminished after selection can be identified. STM has been applied to a wide variety of bacteria and has also been used to identify serovar Typhimurium genes that are differentially required for pathogenesis in different hosts (47, 70). Most recently, approaches that allow for the utilization of microarrays for transposon detection have been developed, facilitating the large-scale identification of conditionally essential genes. Salama et al. utilized a combination of transposon-specific primers and random primers to PCR amplify the region of the genome adjacent to each transposon insertion in order to identify essential genes in Helicobacter pylori (58). Badarinarayana et al. and Sassetti et al. both employed a T7 transcriptional promoter engineered on the end of the transposon cassette in order to generate an RNA probe for the sequence adjacent to each insertion (4, 60), an approach that has recently been modified for use in a screen for Escherichia coli genes important for growth in minimal medium (74).
Here, we describe the adaptation of the T7-based negative selection strategy developed by Badarinarayana et al. and Sassetti et al. (4, 60) to identify serovar Typhimurium genes that are important for survival in RAW 264.7 macrophage-like cells and BALB/cJ mice.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth conditions, and generation of the transposon-mutagenized library.
All bacterial strains were grown and maintained at 37°C in Luria-Bertani (LB) medium with aeration or on LB agar plates with the appropriate antibiotics at the following concentrations, unless otherwise specified: 50 μg/ml streptomycin, 30 μg/ml kanamycin, 100 μg/ml ampicillin, and 10 μg/ml chloramphenicol. Serovar Typhimurium strain SL1344 was used throughout this study and is streptomycin resistant (64). pJA1 is an oriRγ-based suicide plasmid maintained in the presence of ampicillin and is a kind gift from Vasudeo Badarinarayana. It contains a mini-Tn10 transposase under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible lacIq/Ptac promoter system, IS10 inverted repeated sequences flanking a transposable kanamycin resistance cassette with an adjacent T7 transcriptional promoter, and the RP4 plasmid mob region that confers capability of conjugative transfer (1, 4). pJA1 was transformed into Escherichia coli SM10 λpir, a strain that contains the trans-acting factors necessary for mobilization of mob-containing plasmids.
Cultures (1.5 ml) of SL1344 and pJA1/SM10 λpir grown overnight were rinsed with 10 mM MgSO4, combined in a low volume, and then pooled in the center of an LB plate upon which 10 μl of 0.1 M IPTG had been spread. The plate was incubated overnight at 37°C and then scraped into 10 mM MgSO4, and dilutions were plated on LB agar plates containing 200 μg/ml streptomycin and 30 μg/ml kanamycin to select for successful transposition events. A library of 50,000 colonies was scraped off the plates and stored in 15% glycerol at −80°C. Cultures for infections grown overnight were inoculated from the glycerol stock and grown with aeration.
Negative selection in RAW 264.7 mouse macrophage-like cells and BALB/cJ mice.
RAW 264.7 cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Invitrogen Life Technologies) and used for infections at passage 20 or less. Three independent experiments of three serial passages each were performed in the macrophages. Two experiments were conducted at a multiplicity of infection (MOI) of 10, while the third was carried out at an MOI of 1. For each of the three experiments, cultures of the transposon library grown overnight in LB containing streptomycin and kanamycin were opsonized in 15% normal mouse serum (Sigma-Aldrich) and phosphate-buffered saline (PBS) at 37°C for 15 min. The bacteria were pelleted and resuspended in PBS and used to infect a total of 5 × 106 macrophage cells at an MOI of 10 or 5 × 107 cells at an MOI of 1 for 60 min. Cells were rinsed in PBS and treated with DMEM-FBS containing 100 μg/ml gentamicin for 2 h and then maintained in 10 μg/ml gentamicin for the duration of the passage. Parallel wells of DMEM-FBS were also inoculated as medium controls. After 12 h, macrophages were lysed with 1% Triton X-100, and the bacteria were harvested by centrifugation and inoculated into 25 ml LB containing streptomycin and kanamycin for 12 h of growth at 37°C with aeration. Medium-passaged bacteria were similarly treated with Triton X-100, pelleted, and grown in LB containing streptomycin and kanamycin. The output from each passage was used for the next round of infection following opsonization, and the MOI was kept constant for all three passages of an experiment. Bacterial cells from the third passage for both the macrophage output and medium control were harvested for genomic DNA extraction.
For each of the three independent passages of the library through mice, five female BALB/cJ mice (Jackson Laboratory), 7 to 9 weeks old, were each injected intraperitoneally with 5 × 105 CFU bacteria from the library. Mice were sacrificed 48 h after infection, when they started to demonstrate severe symptoms of illness. Spleens were harvested, homogenized, and combined into 300 ml LB with streptomycin and kanamycin for overnight outgrowth. Bacterial cell pellets of the input inoculum and the mouse-passaged outputs were harvested for genomic DNA extraction.
Genomic DNA extraction, microarray sample processing, and microarray hybridizations.
Genomic DNA was isolated and purified from bacterial pellets by phenol-chloroform extraction. DNA was digested with HinP1I (New England Biolabs Inc.) and processed as previously described except that the Y-linker annealing and PCR steps were eliminated (4). A total of 47,159 HinP1I recognition sites are present within the LT2 genome sequence at a mean frequency of every 103 bases. The purified, digested DNA was then used directly as the template for in vitro transcription with the MEGAscript T7 transcription kit (Ambion Inc.) following the manufacturer's protocol except that the reaction volumes were doubled, 2 μg of digested DNA was used, and the reaction was allowed to proceed for 12 to 16 h. cDNA synthesis and amino-allyl dUTP incorporation were performed on the RNA using Superscript II(−) (Invitrogen Life Technologies) in a standard reverse-transcription reaction with random hexamers as primers (as detailed at http://cmgm.stanford.edu/pbrown/protocols). The Cy5 and Cy3 dyes were conjugated to amino-allyl dUTP nucleotides in the selected libraries and controls (either the medium-passaged macrophage control or the mouse library input), respectively. The serovar Typhimurium DNA microarray and the hybridization protocol used have also been described previously (9). At least one microarray was hybridized for each biological replicate. All raw data sets are freely available for download from the Stanford Microarray Database (http://genome-www5.stanford.edu/) (63).
Data analysis.
Normalized data were downloaded from the Stanford Microarray Database according to the mean log2 Cy5/Cy3 (logRAT2N). Filters for feature quality, including a Cy3 net mean intensity of ≥150 and regression correlation of >0.6, were also applied. In addition, features (spots) missing values for ≥40% of the arrays were removed. The final RAW 264.7 data set included seven arrays representing all the biological replicates and consisted of a total of 5,044 features. These features correspond to 3,657 genes and 45 intergenic regions, as many genes are represented by more than one feature on the array. This data set is referred to as the macrophage microarray data set throughout the text. The BALB/cJ data set included five arrays with 3,473 genes and 36 intergenic regions represented by a total of 3,508 features and also represents all the biological replicates. The data sets were analyzed with the Significant Analysis of Microarrays (SAM) program, v. 1.21 (72), using the one-class analysis option to identify features that consistently deviated from logRAT2N = 0 across all the arrays. (The lists of statistically significant genes, also known as the SGLs or significant genes lists, have been made available for download at http://falkow.stanford.edu/whatwedo/supplementarydata/ as noted in the text.) The lexical analysis program LACK, v. 3.1, was used to assess the significance of the enrichment of specific categories of genes in the SAM data sets (38).
Generation of defined gene deletions.
Mutant strains are listed in Table 1. Gene deletions were performed using a PCR-mediated λ Red recombinase system developed by Datsenko and Wanner (12). Primers for deleting each open reading frame in serovar Typhimurium strain LT2 were designed using a Perl script (available upon request) and are available at http://falkow.stanford.edu/whatwedo/index.html. Primers were designed manually for the nonpolar deletions. All deletions were constructed in the LT2 background, verified by PCR, and transduced into SL1344 using standard P22 phage transduction methods. P22 transducing phage for the SPI2 effector mutants, a kind gift from Stephanie M. Brandt (7), was transduced into the appropriate SL1344 background prior to use.
TABLE 1.
Strains and mutants used in this study
| Straina | Genotypeb | Source or reference |
|---|---|---|
| SL1344 | hisG xyl rpsL | 64 |
| CK188 | SL1344 ΔhisG::rpsM-luxDCABE-cat | This work |
| CK113 | SL1344 ΔssrAB::ahp | 37 |
| KC182 | SL1344 ΔssrAB::ahp ΔhisG::rpsM-luxDCABE-cat | This work |
| BJ66 | SL1344 orgA::Tn5lacZY | 36 |
| KC228 | SL1344 orgA::Tn5lacZY ΔhisG::rpsM-luxDCABE-cat | This work |
| KC183 | SL1344 ΔsseB::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC184 | SL1344 ΔsseC::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC185 | SL1344 ΔsseE::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC186 | SL1344 ΔsseF::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC187 | SL1344 ΔsseG::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC188 | SL1344 ΔsseI::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC189 | SL1344 ΔsseJ::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC190 | SL1344 ΔssaB::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC191 | SL1344 ΔsifA::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC192 | SL1344 ΔsifB::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC193 | SL1344 ΔslrP::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC194 | SL1344 ΔsspH2::ahp ΔhisG::rpsM-luxDCABE-cat | This workc |
| KC223 | SL1344 ΔbarA::ahp ΔhisG::rpsM-luxDCABE-cat | This work |
| KC211 | SL1344 ΔsirA::ahp (polar) ΔhisG::rpsM-luxDCABE-cat | This work |
| KC229 | SL1344 ΔsirA::FRT (nonpolar) ΔhisG::rpsM-luxDCABE-cat | This work |
| KC210 | SL1344 ΔuvrC::ahp ΔhisG::rpsM-luxDCABE-cat | This work |
| KC213 | SL1344 ΔhilA::ahp (polar) ΔhisG::rpsM-luxDCABE-cat | This work |
| KC230 | SL1344 ΔhilA::FRT (nonpolar) ΔhisG::rpsM-luxDCABE-cat | This work |
All strains designated “KC” were generated by P22-mediated transduction of the mutant from the original strain into either SL1344 or CK188. In the case of the light-producing nonpolar mutants, ΔhisG::rpsM-luxDCABE-cat was transduced into each nonpolar mutant strain.
FRT is the FLP Recognition Target. In the case of these genotype designations, it refers to the FRT lesion that is left behind after the drug marker is removed by the FLP recombinase.
These deletion strains were originally constructed as previously described (7) and were transduced by P22 phage into the light-producing strain.
Macrophage survival and replication assays.
Salmonella constitutively expressing the luxCDABE genes of Photorhabdus luminescens has been previously described as strain SMB500 (8). SMB500 is the result of a transposition event using the suicide vector pTn5-EM7-Lux-Km1, in which the synthetic E7 promoter (Invitrogen Life Technologies) drives the constitutive expression of the lux operon. The insertion in SMB500 is in hha, which is reported to affect expression of SPI1 (18). We therefore reengineered the insertion construct by PCR amplifying a 400-bp region of the rpsM promoter using a 5′ oligonucleotide with an Flp recombinase target (FRT) site (boldface) and an NcoI restriction site (italics) (CCATGGGAAGTTCCTATTCTCTAGAAAGAATAGGAACTTC CGATCCCGGAGTTCATGCGT) and a 3′ oligonucleotide with an SpeI site (TCGCTGAAAATGTTAAGATCACTAGT) and cloning this fragment upstream of luxCDABE in pUT-Tn5-EM7-Lux-Km2 to yield pCK67. This vector was partially digested with PmlI and MscI to disrupt the transposase, yielding pCK68. The pCK68 plasmid was integrated into the hisG locus of CK500 (ΔhisG::FRT [37]) by expression of Flp recombinase from pCP20 (12) to yield strain CK168. The chromosomally integrated construct was transferred to additional strains via P22-mediated transduction. A chloramphenicol-marked version of the lux construct was constructed by λ Red recombinase replacement of the integrated pCK68 ahp gene with cat of pKD3 (12). The construct produces approximately 10 to 30 times more light than SMB500.
For CFU-based survival and replication assays, RAW macrophages were seeded at a density of 2.5 × 105 cells per well of a 24-well culture dish. Individual mutants from a culture grown overnight that had been opsonized in 15 to 50% normal mouse serum were used to infect the macrophages at an MOI of 10. Gentamicin-protected bacteria were harvested from the macrophages at various time points by lysis of macrophages with 1% Triton X-100 and plated for enumeration. To assess bacterial levels as a function of luxCDABE light production, RAW cells were seeded at a density of 5 × 104 cells per well in DMEM lacking phenol red in Optilux 96-well black-wall clear-bottom microtest plates (BD Falcon). Cells were infected as usual with luminescent strains, and light production was assayed using a Veritas Microplate Luminometer kindly provided by Turner BioSystems (Sunnyvale, Calif.). The raw light units obtained from luminometer readings were log10 transformed and normalized to levels at 3 h postinfection to account for variations in bacterial uptake by the macrophages. The data were additionally normalized to the average value for the wild-type replicates so that the behavior of each strain could be assessed relative to wild-type levels.
RESULTS
The negative selection strategy identified SPI2 genes as being critical for macrophage survival and replication.
Individual bacteria harboring mutations in genes that are important for survival are compromised in their ability to be maintained within a population. Upon application of selective pressure, a subset of mutants that cannot persevere under the given condition will disappear from a library of mutants. These negatively selected mutants can be detected on a DNA microarray based upon the intensity of fluorescence signal for the corresponding feature on the microarray. Utilizing this concept, a 50,000-CFU library of transposon-mutagenized SL1344 was subjected to three rounds of serial passage in RAW macrophage cells. Bacterial genomic DNA from the third passage was isolated and processed for hybridization on the microarray along with a medium-passaged control sample. Statistical analysis of the microarray data set for multiple biological and technical replicates using SAM (72) revealed an enrichment of SPI2 genes in the top-scoring negatively significant genes (supplementary Table A1).
In order to assess the significance of this enrichment and distinguish it from a random level of SPI2 appearance in the list of significant genes, we performed a lexical analysis for SPI2 genes using LACK (38). Using a negative SGL identified by SAM consisting of 395 features (<2% median false discovery rate), we determined that it was unlikely that the 51 SPI2 features we observed would appear randomly (P < 10−10). Because many of the serovar Typhimurium genes are represented by multiple features on the microarray, these 395 SGL features correspond to a total of 304 genes and intergenic regions (supplementary Table A1). The 51 SPI2 features correspond to 25 genes (and one intergenic region) that represent the pathogenicity-associated region of the SPI2 island.
With the aim of understanding the overall behavior of the genes in the entire SPI2 island (30), we extracted the corresponding features from the full microarray data set and organized them in genome order (Fig. 1A). The majority of SPI2 features have a hybridization pattern indicating that mutations in these genes result in the disappearance of these mutants from the library upon passage through macrophages. The majority of SPI2 features also appear in the SGL as negatively selected. The exceptions to this are the tetrathionate reductase (ttr) complex genes and the putative genes orf48, orf70, and orf242, all of which are within the pathogenicity island but have been shown to have a minimal contribution to systemic infection in BALB/cJ mice (28-30). They exhibit a hybridization pattern indicating that they are not selected against in macrophages (Fig. 1A; see supplementary Table A1). The genes that have a greater than threefold reduction from the library include ssaB, ssrA, ssaC, ssaV, sseA, and ssaM. Thus, the microarray results indicate that the majority of genes in SPI2, once disrupted, render the bacteria less capable of surviving in RAW macrophages.
FIG.1.
Data corresponding to (A) the SPI2 island and (B) the SPI1 island were extracted from the full macrophage microarray data set and organized by genome order. Features corresponding to sirA and barA, neither of which are in SPI1 but have been shown to regulate the island, are included at the bottom of the SPI1 image. All seven arrays, including the average for each feature, are represented. Arrays that are independent biological replicates are grouped at the top by color bars and designated by the letters A, B, and C, corresponding to experiments at an MOI of 1, an MOI of 10, and an MOI of 10, respectively. If multiple features are present for a given gene, they are grouped by the presence of a vertical bar next to the gene name. Asterisks indicate features that are present in the SAM macrophage SGL. The color scheme for change (n-fold) is indicated at the bottom. Generally, yellow indicates genes whose corresponding mutants are absent from the macrophage-selected library, while blue/black indicates their presence.
In addition to the known effector molecules encoded within SPI2, there are a number of molecules located elsewhere in the genome that are known to be coregulated with SPI2, and in some cases, translocation of these effectors has been shown to be dependent upon SPI2 (44, 73). We looked for their presence within the SGL (supplementary Table A1) and found that two of these, srfA and sifA, were negatively selected in macrophages. srfA was identified in a screen for SsrB-regulated genes, although its function inside of macrophage cells is not known (76). sifA was originally described as being responsible for the formation of membrane-containing filaments in Salmonella-infected epithelial cells (24, 67) and was shown to be regulated by SsrA. In addition, mutants not only have a macrophage replication defect but are also released into the host cytosol, thereby implicating SifA in the maintenance of the Salmonella-containing vacuole (6). Of the other effectors encoded outside of SPI2, only slrP and sspH2 are represented in the macrophage microarray data set. However, neither is present in the SGL and is therefore not predicted by the microarray to be compromised for macrophage survival or replication.
The behavior of individual SPI2 mutants in a macrophage replication assay parallels microarray predictions.
The genes that were detected in our negative-selection experiment as having been lost during passage of the library in macrophages could represent defects in entry, persistence, viability, or replication. Individual mutants with deletions in SPI2 genes that had been identified as negatively selected were tested for replication and persistence defects in RAW macrophage cells. To facilitate the rapid screening of individual mutants, we introduced them into a common SL1344 background where the Photorhabdus luminescens luxCDABE operon is expressed under the control of the serovar Typhimurium rpsM promoter. These bacteria constitutively produce light, providing a means of continuously assessing bacterial levels without disrupting infected macrophage cells or the need to perform colony counts. Also included in the macrophage assays were mutants representing genes that were not negatively selected in our analysis and would therefore be predicted to persist in macrophage cells at wild-type levels. Finally, a number of SPI2 genes that are not represented in the microarray data set were also included. Their inclusion serves as an additional control for the light-based replication assay. While we cannot make array-based predictions as to the behavior of these mutants, we can correlate our observations with published literature.
The amount of light produced by each gentamicin-protected strain over the course of 24 hours was monitored in infected macrophage cells. The data were normalized to both the early 3-h time point to control for the number of internalized bacteria as well as the SL1344 wild-type control for each time point in order to more readily observe deviations from the wild-type strain's growth kinetics. Microarray results had indicated that mutants in the SPI2 genes ssrA, ssrB, ssaB, sseC, sseE, and sseF were negatively selected during passage of the library in macrophages. Individual mutants for each of these genes were defective for replication by 12 h (Fig. 2A). According to the luminescence assay, the ssrAB, ssaB, and sseC mutants had the greatest macrophage defect and produced approximately 30- to 46-fold less light than the wild-type strain at 24 h. sseE and sseF mutants had more moderate phenotypes, with approximately eight- and fivefold less light production than the wild type, respectively. The macrophage defects for these mutants were confirmed using a separate CFU assay (Fig. 2B), validating the use of the lux-based approach for monitoring intracellular bacterial load. sseB is not represented on the microarray. However, its SPI2-encoded product is believed to associate with SseC and SseD to form a translocon through which effectors are secreted (50). The phenotype for the sseB mutant was comparable to that seen with the sseC mutant by both luminescence and CFU assays, and both have been shown to have equivalent levels of attenuation in RAW 264.7 macrophage cells (33). sseG is also not represented in the array data set but has been shown to have a mild macrophage replication defect similar to that of an sseF mutant (33), which is in accord with our observations. Therefore, all of the mutants in genes physically within the SPI2 locus display a macrophage defect corresponding with predictions made by both the microarray and published literature.
FIG. 2.
RAW 264.7 macrophage survival and replication assays with individual SPI2 mutants. (A) Mutants were placed in a background where the luxCDABE operon, under the control of an rpsM promoter, was constitutively expressed. The level of intracellular, gentamicin-protected bacteria was tracked by light production. Individual mutants are indicated on the x axis. The ssrA and ssrB genes were the only genes tested as a double mutant, as they are part of the same SPI2 regulatory unit. Each strain was tested in quadruplicate. The average log10 relative light units (RLU) are reported with normalization to wild-type SL1344 levels and to the levels at 3 h postinfection as described in Materials and Methods. These results are representative of multiple assays. (B) The same strains were assayed by traditional colony counts. Each strain was tested in triplicate, and the average log10 CFU/milliliter are reported after normalization to wild-type SL1344 levels and to the levels at 3 h postinfection. Open circles indicate genes that appear in the macrophage SGL as negatively selected, closed circles indicate genes that are represented in the macrophage microarray data set but did not appear in the SGL, and strains without any symbols are not represented in the microarray data set. Hours postinfection are indicated, mutants are ordered as they appear in the genome, and none of the strains displayed a growth defect in LB. Standard errors are reported, and asterisks indicate a P value of <0.01 as calculated using an unpaired t test comparing each mutant to the wild-type strain for each respective time point.
SifA, SifB, SlrP, SseI, SseJ, and SspH2 are members of a family of proteins that are encoded outside of the SPI2 island but share a translocation signal in their N termini that directs their secretion through the SPI2 apparatus (44). Of these, only sifA appeared in the SGL, although only sifA, slrP, and sspH2 are present in the microarray data set (supplementary Table A1). The luminescence and CFU results corroborate a macrophage defect for the sifA mutant with approximately 8- and 14-fold fewer light and CFU units, respectively, than the wild-type control at 24 h (Fig. 2A and B). SifA has been shown to be critical for maintaining the membrane of the Salmonella-containing vacuole, and mutants are attenuated in macrophage cells (6). While sseJ is not represented in our microarray data set, it has been reported to have a minor replication defect in macrophages and mice (22, 43, 57). We observed a slight decrease in bacterial numbers relative to the wild type in the CFU assay but not in the luminescence assay. slrP and sspH2 mutants do not show an appreciable defect by either luminescence or CFU assay. slrP was initially identified in a signature-tagged mutagenesis screen looking for bacterial genes that may play a role in differential host disease between the mouse and calf models of infection (70). However, its role in intracellular replication has not been examined. The behavior of the individual mutants in macrophages correlates well with both microarray-predicted behavior and published literature. These results validate the use of both the light-based assay to monitor intracellular levels of bacteria and the microarray-based negative selection approach.
SPI1 genes are negatively selected upon serial passage in macrophage cells.
During the course of analysis, we also observed a strong enrichment of features corresponding to SPI1 genes in the SAM SGL (supplementary Table A1). Lexical analysis revealed that the overrepresentation of SPI1 genes in the SGL was highly significant (P < 10−10). The 36 SPI1 features in the SGL correspond to 22 genes and include structural components of the secretion machinery, regulatory genes such as hilA, hilC, and hilD, and genes for effector molecules such as sipA and sipB. Additionally, the genes for the SirA/BarA two-component regulatory system, which are known to have a role in regulating SPI1 expression (3, 35, 69), also appear as negatively selected (Fig. 1B). Given the fact that the cultures were grown to stationary phase under SPI1-suppressing conditions and that complement-mediated opsonization allowed internalization without activation of bacterium-mediated invasion, the observation that disruption of SPI1 genes resulted in negative selection during passage through macrophages was unexpected. The role of SPI1 has thus far primarily been associated with invasion of host cells in the gastrointestinal tract and not believed to be critical for macrophage survival at systemic sites. Indeed, SPI1 mutants are attenuated when delivered by oral inoculation but appear to be fully virulent when assessed by 50% lethal dose (LD50) experiments after intraperitoneal challenge in BALB/cJ mice (23).
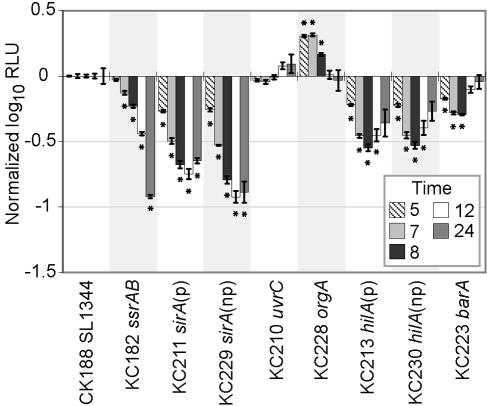
In order to assess the contribution of SPI1 genes to macrophage survival and replication, individual deletion mutants of SPI1 genes were monitored for light production during infection of RAW 264.7 cells. A sirA mutant (annotated uvrY) was defective for intracellular replication (Fig. 3). This mutant also exhibited an invasion defect (data not shown) that had been previously observed in epithelial cells (35). In order to circumvent the problem of differential invasion in measuring replication, the data were normalized to the 3-h time point in order to assess replication relative to the number of internalized bacteria. Since a mutation in sirA is likely to be polar on the downstream gene uvrC, we also generated a nonpolar sirA mutant and a uvrC mutant. The nonpolar sirA mutant still had a macrophage defect comparable to that of the polar mutant, while the uvrC mutant behaved like the wild type, indicating that the negative selection of sirA was not due to downstream effects on uvrC. A barA mutant, which encodes the cognate sensor for SirA, was also tested but did not have a replication defect. An orgA mutant, which encodes a structural component of the SPI1 secretion system, was also tested and shown not to have a macrophage defect. Polar and nonpolar mutants of hilA were tested and appeared to have a slight phenotype (Fig. 3). However, this result was variable across multiple experiments. CFU assays corroborate the sirA phenotype and also maintain that orgA, barA, and hilA mutants do not have a defect (data not shown). Other mutants, including sipA, sipB, and a deletion spanning the entire SPI1 island, were also tested for a macrophage defect and were found to have none (data not shown). We have therefore shown that despite the observation that SPI1 mutants are negatively selected upon macrophage serial passage, all SPI1 deletion mutants, with the exception of sirA, do not exhibit a macrophage replication defect when tested individually.
FIG. 3.
Individual SPI1-associated mutants that constitutively produce light were assayed for replication defects in RAW 264.7 macrophage cells as described in the legend of Fig. 2. Each strain was tested in quadruplicate, and the average log10 relative light units (RLU) are reported after normalization to wild-type SL1344 levels and levels at 3 h postinfection. The ssrAB strain was included as a control. These results are representative of multiple assays. “p” indicates polar mutants, while “np” indicates nonpolar mutants. Standard errors are reported, and asterisks indicate significance (P < 0.01) as calculated using an unpaired t test comparing each mutant to the wild-type strain for each respective time point. Hours postinfection are indicated, and none of the strains displayed a growth defect in LB.
Salmonella-specific genes are enriched in the RAW 264.7 SGL.
We were interested in the composition of the genes that were enriched in the SGL. If certain classes of genes appeared enriched in the SGL, this could provide some insight into the intracellular environment or particular needs of the bacteria. We used two approaches to analyze the composition of the RAW 264.7 SGL: we organized the genes according to whether or not they were conserved in other members of the Enterobacteriaceae family, and we also utilized the Clusters of Orthologous Groups (COGs) database designations (68) available for the serovar Typhimurium LT2 genes.
Comparison of Enterobacteriaceae genomes facilitates the identification of genes that are unique to each genus and are therefore potentially important in determining the specifics of pathogenesis of each respective organism. We associated all of the features in both the SGL and the microarray data set with the expected distribution across members of the Enterobacteriaceae, including multiple S. enterica serovars, S. bongori, Klebsiella pneumoniae, E. coli K-12, and E. coli O157:H7, as determined by both sequence analysis and microarray genomic comparison (42). Each serovar Typhimurium gene is defined by the presence or absence of a homolog in the other genomes, and genes with similar patterns of presence or absence are placed in the same category. By comparing the relative size of each category in both the SGL (Fig. 4A), and the microarray data set (Fig. 4B), we could assess whether or not specific classes of genes were being negatively selected. The distribution of the microarray data set was compared to the distribution within the LT2 genome and observed to be nearly identical, indicating even representation of all classes of genes on our microarray (data not shown). One of the greatest differences between the SGL and microarray data set is the category that represents genes that are shared across all of the Enterobacteriaceae genomes. This category comprises 53% of the microarray data set (Fig. 4B) but represents a much smaller proportion of the SGL at 29% (Fig. 4A). Much of the Enterobacteriaceae category consists of genes involved in common functions such as biosynthesis, respiration, and macromolecule synthesis (supplementary Table A1). There is a concomitant increase in size of some of the Salmonella-specific categories in the SGL relative to the microarray data set. The S. enterica category represents 12% of the SGL compared to 2% of the array data set and includes SPI2, which is known to be present only in the S. enterica serovars and is associated with intracellular survival and replication (51). SPI1 genes, the acquisition of which evolutionarily distinguishes Salmonella from the other Enterobacteriaceae and which is important during the intestinal phase of disease (5), can be found in the category representing genes found in some or all Salmonella species that was enriched from 19% in the array data set to 41% in the SGL. Interestingly, the categories representing genes found only in subspecies 1 and genes found only in serovar Typhimurium do not change in proportion between the SGL and microarray data set. Taken together, these observations indicate that the loci which alter systemic invasion and survival are conserved across most or all Salmonella species but not across the Enterobacteriaceae.
FIG. 4.
The distribution of S. enterica serovar Typhimurium LT2 genes across the Enterobacteriaceae and how this distribution in (A) the macrophage SGL compares to (B) the complete macrophage array data set of 5,044 features. The features in the SGL and the macrophage data set were associated with various categories as defined by McClelland et al. according to whether or not a homolog for each LT2 gene was present in various Enterobacteriaceae genomes including Klebsiella pneumoniae, E. coli K-12, E. coli O257:H7, S. bongori, and multiple S. enterica serovars (42). The “unknown” category comprised a negligible portion of the genes and was removed. In addition, the original “some or all Salmonella only,” “some Salmonella only,” and “all Salmonella only” categories were combined into the single “some or all Salmonella only” category for simplification. This category represents serovar Typhimurium LT2 genes that are present in most if not all the Salmonella serovars represented. The “all nine genomes” category represents genes that are conserved across all the Enterobacteriaceae sampled. The “other distributions” category encompasses genes that may have a varied distribution, being present in some of the non-Salmonella Enterobacteriaceae but not others. The remaining categories are self-explanatory. The key indicates color scheme and category association, and the percent values indicate the proportion of either the SGL or array data set that is comprised by each category.
The COG database was originally developed to facilitate the characterization of genes according to the presence of orthologs in other bacterial genomes. We organized the features in the SGL and the microarray data set according to the functional class of their corresponding genes and assessed the differences in proportions of the COG categories between the two as well as the genes comprising each functional class in the SGL (supplementary Table A1). When the COG comparison was made between the SGL and the array data set, the “not in COGs” category, which represents proteins without sufficient homology upon which to base functional predictions, had the greatest difference, representing 33% of the SGL and only 15% of the array data set (supplementary Table A1). This observation indicates that many of the genes that are important for survival in macrophages encode proteins with little homology to other genes in the COG database, which complements our observation that the SGL is enriched for genes unique to serovar Typhimurium or to Salmonella. Of the 127 features that fall within this category in the SGL, 29 correspond to SPI2, 16 correspond to SPI1, several others correspond to various prophage genes, and many more correspond to genes of putative function (http://falkow.stanford.edu/whatwedo/index.html). The remainders of the SPI1 and SPI2 features are distributed throughout other COG categories including “intracellular trafficking and secretion,” “signal transduction,” and “cell motility,” all of which are proportionately enriched in the SGL relative to the array data set.
There are other categories whose proportions do not change significantly in the SGL. These include genes involved in basic bacterial functions including amino acid, nucleotide, and carbohydrate metabolism; energy production; translation; and DNA replication and repair. Within the SGL “energy production and conversion” functional group, the mutants in genes encoding the subunits of cytochrome d terminal oxidase, cydA and cydB, and a membrane-bound ATP synthase F0 component, atpE and atpF, are negatively selected (supplementary Table A1). In a study to probe the contribution of respiratory proton translocators to virulence in serovars Typhimurium, Gallinarum, and Dublin in both mice and chicks, it was found that mutants in cydA were moderately attenuated and that mutants in atpB and atpH, which encode components of the F0 and F1 ATPases, respectively, were strongly attenuated with regard to their ability to kill both hosts. Moreover, atpB was shown to have a replication defect in mouse J774.1 cells (71). Also included within this category are genes in the putative operon STM0854-0859. STM0854-0859 includes components of an electron transport protein, dehydrogenases, and a transcriptional regulator. This mini pathogenicity island has recently been shown to be induced in RAW 264.7 macrophages (17). Salmonella's ability to modify its respiration according to changing environmental conditions could therefore play a critical role in survival in the intracellular conditions of the macrophage.
Passage of the library in BALB/cJ mice reveals that SPI2 and lipopolysaccharide (LPS) mutants are compromised in their ability to colonize the spleen.
Given the successful application of this approach to in vivo screens for Mycobacterium tuberculosis genes important for survival in mice (61), we were interested in examining its utility for screening serovar Typhimurium genes important for establishing an infection in mice. We infected BALB/cJ mice intraperitoneally with the library and recovered bacteria from the spleen 2 days postinfection, similar to the conditions used in the original STM screen that identified the SPI2 locus (31, 62). Three independent sets of five mice each were inoculated, and the composition of bacteria from the spleens was compared to the initial inoculum.
When the SAM-generated SGL of 188 BALB/cJ features was analyzed, we observed an enrichment of features that could be organized into three categories (supplementary Table A2). The first class of genes included 23 features that corresponded to the SPI2 island, the significance of which was confirmed by lexical analysis (Table 2). These 23 features consisted primarily of SPI2 secretion apparatus structural components and did not include any of the effector molecules that are encoded elsewhere in the genome. The second group of features that appeared enriched corresponded to genes that are involved in LPS biosynthesis (supplementary Table A2). The appearance of these nine genes in the SGL is also significant upon lexical analysis (Table 2). In agreement with our results, both of these categories of genes were also enriched in the serovar Typhimurium STM screen (31, 62). We also noticed the presence of a few SPI1 features. However, lexical analysis resulted in a P value of 0.059, indicating that the appearance of these genes may be stochastic (Table 2). When the BALB/cJ SGL was compared to the macrophage SGL, 32 features were present in both gene lists, of which 23 corresponded to genes in SPI2. Other genes present in both SGLs include the putative mouse virulence gene mviM, a putative periplasmic protein, STM1621, and a putative cytoplasmic protein, STM3180 (http://falkow.stanford.edu/whatwedo/index.html).
TABLE 2.
Lexical analysis of BALB/cJ SGL
| Category | No. of hitsa | Genesb | P valuec |
|---|---|---|---|
| SPI2 | 23 | ssrA, ssaB, ssaC (4), ssaD (4), ssaE (2), sseA, ssaI, ssaJ, ssaK, ssaM, ssaV, ssaN, ssaQ, ssaS, ssaT, ssaU | <1 × 10−10 |
| SPI1 | 5 | orgA, prgI, prgH (2), spaP | 0.059 |
| LPS | 9 | rfbX, rfbF, rfbC (3), rfbB, rfaK, rfaB, rfaG | <7.1 × 10−5 |
Number of features that correspond to the category in an SGL size of 188.
The parenthetical value indicates the number of features corresponding to that gene that appear in the SGL.
P value calculated by LACK binomial analysis and a data set size of 3,508 features.
DISCUSSION
In this work, we have generated a 50,000-CFU-complexity transposon library of serovar Typhimurium and were able to demonstrate that we can detect the negative selection of SPI2 mutants in the mouse. Out of the top 188 features in our SGL, 23 (12% of the SGL) corresponded to mutants in SPI2. In addition, we also identified LPS mutants as being compromised for survival in the mouse, corresponding with the known role of LPS in serovar Typhimurium virulence (20). In the original STM screen to identify mutants attenuated for establishing an infection in BALB/cJ mice, 1,152 clones were screened in 12 pools of 96 mutants each. This led to the identification of 43 attenuated mutants, 19 of which had never been characterized and showed either no known homology or homology to known bacterial virulence determinants. Of these, 16 (1.4% of the original library and 37% of the attenuated mutants) were eventually mapped to what is now known as SPI2 (31, 62). Additionally, five of the original STM insertions localized to genes involved in LPS biosynthesis. We have therefore demonstrated that use of the microarray-based negative selection approach is comparable to another established technique. The microarray-based approach requires considerably less labor, thereby greatly facilitating the execution of comprehensive screens.
We have also demonstrated the utility of a microarray-based negative selection approach in the identification of genes important for serovar Typhimurium's survival and replication in RAW 264.7 macrophage-like cells. The enrichment of Salmonella-specific genes in the SGL, as determined by the Enterobacteriaceae distribution and COG distribution analyses, indicates that the genetic contribution to intracellular survival and replication involves factors that are either sparsely distributed in bacterial genera or specifically adapted by Salmonella. Furthermore, the observation that the enriched genes are shared across Salmonella species, in particular, S. enterica, and not specific to just serovar Typhimurium or subspecies 1 suggests that with regard to the experimental conditions we used, serovar Typhimurium's ability to survive within RAW macrophage cells does not involve genes that are specific to serovars associated with disease in warm-blooded hosts. Instead, it is the genes that Salmonella as a genus possesses which facilitate intracellular survival and replication. A significant number of the genes identified are within SPI2, corresponding with the established role of this pathogenicity island in macrophage survival (73). In addition, we have verified the array-predicted intracellular growth defect of some of the genes both physically within SPI2 and encoded outside of the island but whose protein products have been shown to be secreted by the SPI2 apparatus. This verification was performed using both the traditional CFU assay as well as a luxCDABE-based assay to monitor intracellular replication of individual deletion mutants. In the cases where genes were not predicted by the array to be compromised for macrophage survival, such as with slrP and sspH2, no replication defect was seen in the single infection experiments. In situations where a replication defect was predicted by the array data, as in the case of ssrAB, sifA, ssaB, sseC, sseE, and sseF, intracellular bacteria did not reach the same levels as the wild-type strain, thereby corroborating the microarray results.
We have observed that the luminescence and CFU results do not always perfectly mimic one another. For example, while both assays confirmed that the sifA and ssrAB mutants have a replication defect, the magnitudes of the defects for the two mutants are different between the assays (Fig. 2). One possible explanation may have to do with the specific microenvironment that the mutants are in and the subsequent effects on luciferase activity. For example, a sifA mutant is unable to maintain the integrity of its vacuole and is consequently released into the cytosol of the cell. However, an ssrA mutant is able to maintain its association with vacuolar markers and is therefore still membrane bound (6). Luciferase synthesis places an increased metabolic burden on the bacteria as it requires and competes for many of the same components required by other processes within the organism. The localized levels of various metabolic substrates and oxygen concentration and even the growth rate, growth phase, and density of the bacteria will therefore impact luminescence (49). Thus, while the CFU assay assesses viability of a mutant, luminescence is likely to be a measure of both viability and metabolism. Regardless of the differences between the two assays, the luminescence assay provides a means of simultaneously evaluating replication and survival of hundreds of mutants under multiple conditions and with high time resolution (unpublished work). Once the behavior of a particular mutant has been characterized using the luminescent system, CFU assays can be also performed to corroborate and complement the findings.
We looked for Salmonella mutants that are defective for macrophage survival and/or replication by serially passaging our mutant library through macrophages. Serial passage can select against multiple types of serovar Typhimurium mutants. Among the possibilities are mutants with a generalized susceptibility to killing during the course of opsonization and infection due to compromised membrane integrity. Mutants could be less capable of resisting the initial onslaught of cellular antibacterial factors. In addition, mutants that cannot properly conduct vacuole biogenesis will not be able to replicate as well as competing strains and will disappear from the population over the course of multiple rounds of selection. As a result, care must be taken to ascertain where the vulnerability of a given mutant lies. The compelling observation of SPI1 genes being negatively selected during the course of macrophage serial passage is an example of when there are multiple possibilities as to why certain mutants appear to be selected against. Unlike the SPI2 results, macrophage replication assays with individual SPI1 mutants do not completely reciprocate the array predictions (Fig. 3). With the exception of the sirA mutant, all SPI1 mutants tested, including a complete deletion of the island, did not have a replication defect. One technical explanation for this inconsistency is that the SPI1 mutants are invasion defective despite the fact that experiments were carried out under invasion-independent conditions. Although the bacteria were opsonized prior to macrophage infection, SPI1 mutants simply may not be as capable of being internalized and become reduced in representation in the library over the course of the experiment. Another parameter that may amplify this effect is that each mutant is but a member of a very large and complex population during the selection process but is present in a homogenous population during macrophage replication assays. Competition may therefore amplify subtle defects in a population, while these deficiencies in fitness could go undetected when mutants are tested individually. This competition model is already widely implemented by microbiologists in the form of competitive index experiments.
Another intriguing interpretation of the data is that SPI1 does play a role in macrophage survival and replication. While the current understanding of serovar Typhimurium pathogenicity islands generally restricts the contributions of SPI1 to invasion and SPI2 to intracellular replication, there is evidence that points to some overlap in function. For example, SPI2 secretion apparatus mutants have deficiencies in inducing RAW 264.7 cytotoxicity and invasion of HEp2 cells, the latter of which was associated with the inability of the cells to secrete SipC in vitro (32). Subsequent to this, it was shown that the in vitro expression of SPI1 genes, including hilA, prgK, and sipC, was reduced in SPI2 mutants (13), and Zaharik et al. observed increased expression of a hilA::lacZYA transcriptional fusion upon infection of Nramp1Asp169 macrophage cells (77). By artificially inducing the uptake of SPI1 invA mutants into epithelial cells, either by coinfection with wild-type serovar Typhimurium or by providing the Yersinia pseudotuberculosis invasin (inv) gene in trans, Steele-Mortimer et al. were able to show that replication did not occur even though the mutants were found inside of cells at appreciable levels (66). Furthermore, vacuole biogenesis under inv-mediated conditions proceeded in a manner distinct from that of wild-type infections, indicating that the method of invasion greatly impacts downstream development of the Salmonella-containing vacuole. Recently, Drecktrah et al. have provided evidence that the SPI1 effector SopB persists in macrophage cells many hours postinvasion and may function to posttranscriptionally regulate macrophage-inducible nitrous oxide synthase levels (15). With regard to the function of SPI1 in the host beyond the intestinal wall, Murray and Lee have shown that a Δspi1 strain was recovered from all tissues at a level comparable to that of the wild type from both competition and single-strain experiments, whereas hilA regulatory and invG secretion apparatus mutants were recovered at lower levels. In addition, the Δspi1 strain did not have an intestinal colonization defect, although it did have a higher oral LD50 than the wild type (48). Their interpretation of these observations is that some SPI1 factor that is independent of hilA regulation and SPI1 secretion stimulated clearance of the bacteria by the host. So while Δspi1 is less capable of infecting mice, possibly accounting for its higher LD50, once it reaches systemic sites, the SPI1-encoded factor is absent and cannot stimulate clearance, allowing the bacteria to achieve wild-type levels. Our observations, in combination with these data, suggest that the role of SPI1 in pathogenesis may extend beyond the initial invasion step.
We were intrigued by the observation that a sirA mutant was compromised for macrophage survival and replication. BarA/SirA is a two-component regulatory system with orthologs in a variety of bacteria. In E. coli, UvrY (the SirA response regulator homolog) and BarA have been shown to be important for the transition between glycolysis and gluconeogenesis, most likely by regulating CsrA/B, which is itself critical for regulation of carbon utilization (54). In Pseudomonas and Erwinia species as well as Vibrio cholera, SirA orthologs have been shown to play a role in secondary metabolism, virulence, and motility (10, 16, 75). In serovar Typhimurium, sirA was originally identified in a screen for mutants that affected the transcription of the SPI1 prgHIJK operon (35), and phosphorylated SirA has been shown to directly interact with the hilA, hilC, and csrB promoters (69). Additionally, production of flagella in serovar Typhimurium is also regulated by BarA/SirA, primarily by its effect on expression of the flhDC operon and also via CsrA/B (69). BarA/SirA is therefore part of the complex signaling network in serovar Typhimurium, one function of which is to modulate the level of hilA expression and subsequently the level of SPI1 gene expression, possibly through CsrA/B (2, 3).
While BarA in Salmonella has been shown to be the sensor kinase of SirA (69) and mutation of either barA or sirA leads to inefficient invasion of HEp-2 epithelial cells (3, 35), a BarA-independent pathway for SirA activation has been observed during oral infection of mice (40). This observation was attributed to the presence of intestinal short-chain fatty acids, in particular, acetate, which can be converted by bacterial enzymes into acetyl-phosphate. Acetyl-phosphate is in turn capable of directly phosphorylating sensor kinases to activate two-component systems. While the contribution of SirA to epithelial cell invasion and virulence upon oral infection in mice has been reported, the role it plays in macrophage survival and replication has remained unexplored. Our data suggest that SirA may play an additional regulatory role within RAW macrophage cells promoting intracellular survival and replication of serovar Typhimurium. Whether this is due to regulatory effects on SPI1 or through alternative pleiotropic effects remains to be determined. In addition, given our observation that a barA mutant did not have a macrophage defect, a BarA-independent pathway for SirA activation could also be present in macrophage cells. It should be noted that acetyl-phosphate has previously been shown to have no influence on the induction of SPI2 gene expression in either medium or RAW 264.7 macrophage cells (37). However, this does not obviate another route of BarA-independent activation of SirA that ultimately contributes to intracellular survival and replication.
In summary, we have been able to apply the microarray-based negative selection approach to successfully identify serovar Typhimurium virulence determinant mutants that are compromised for survival under both in vivo and in vitro conditions. The potential application of this negative selection approach to a myriad of systems is readily apparent. The selection conditions are also innumerable, and differential genetic requirements in multiple cell types, various host genetic backgrounds, over a time course, and even tissue distribution could feasibly be assessed. This technique may be particularly beneficial in cases where characterization of the molecular pathogenesis of a bacterium is still in its early stages, provided a genome sequence, microarray, and the ability to perform random mutagenesis are available.
Acknowledgments
We gratefully acknowledge Christopher Sassetti and Vasudeo Badarinarayana for their advice in adapting the microarray-based negative selection approach. We also thank Lucinda J. Thompson, Denise M. Monack, and Elizabeth A. Joyce for critical reading of the manuscript.
This work was supported by the Ellison Medical Foundation, grant AI26195 from the National Institutes of Health, and Digestive Disease Center grant DK56339. C.C.K. was supported by a Howard Hughes Medical Institute predoctoral fellowship and a Stanford graduate fellowship. K.C. was supported by a National Science Foundation graduate research fellowship.
Editor: A. D. O'Brien
REFERENCES
- 1.Alexeyev, M. F., and I. N. Shokolenko. 1995. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery into the chromosome of gram-negative bacteria. Gene 160:59-62. [DOI] [PubMed] [Google Scholar]
- 2.Altier, C., M. Suyemoto, and S. D. Lawhon. 2000. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect. Immun. 68:6790-6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altier, C., M. Suyemoto, A. I. Ruiz, K. D. Burnham, and R. Maurer. 2000. Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol. Microbiol. 35:635-646. [DOI] [PubMed] [Google Scholar]
- 4.Badarinarayana, V., P. W. Estep III, J. Shendure, J. Edwards, S. Tavazoie, F. Lam, and G. M. Church. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol. 19:1060-1065. [DOI] [PubMed] [Google Scholar]
- 5.Baumler, A. J., R. M. Tsolis, T. A. Ficht, and L. G. Adams. 1998. Evolution of host adaptation in Salmonella enterica. Infect. Immun. 66:4579-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandt, S. M., M. S. Dionne, R. S. Khush, L. N. Pham, T. J. Vigdal, and D. S. Schneider. 2004. Secreted bacterial effectors and host-produced Eiger/TNF drive death in a Salmonella-infected fruit fly. PLoS Biol. 2:e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns-Guydish, S. M., I. N. Olomu, H. Zhao, R. J. Wong, D. K. Stevenson, and C. H. Contag. 2005. Monitoring age-related susceptibility of young mice to oral Salmonella enterica serovar Typhimurium infection using an in vivo murine model. Pediatr. Res. 58:153-158. [DOI] [PubMed] [Google Scholar]
- 9.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185:553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee, A., Y. Cui, H. Yang, A. Collmer, J. R. Alfano, and A. K. Chatterjee. 2003. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant-Microbe Interact. 16:1106-1117. [DOI] [PubMed] [Google Scholar]
- 11.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deiwick, J., T. Nikolaus, J. E. Shea, C. Gleeson, D. W. Holden, and M. Hensel. 1998. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J. Bacteriol. 180:4775-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, W., S. R. Liou, G. Plunkett III, G. F. Mayhew, D. J. Rose, V. Burland, V. Kodoyianni, D. C. Schwartz, and F. R. Blattner. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drecktrah, D., L. A. Knodler, K. Galbraith, and O. Steele-Mortimer. 2005. The Salmonella SPI1 effector SopB stimulates nitric oxide production long after invasion. Cell. Microbiol. 7:105-113. [DOI] [PubMed] [Google Scholar]
- 16.Eriksson, A. R., R. A. Andersson, M. Pirhonen, and E. T. Palva. 1998. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 11:743-752. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 18.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fierer, J., and D. G. Guiney. 2001. Diverse virulence traits underlying different clinical outcomes of Salmonella infection. J. Clin. Investig. 107:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finlay, B. B., and J. H. Brumell. 2000. Salmonella interactions with host cells: in vitro to in vivo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman, J. A., M. E. Ohl, and S. I. Miller. 2003. The Salmonella enterica serovar Typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infect. Immun. 71:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galan, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-del Portillo, F., M. B. Zwick, K. Y. Leung, and B. B. Finlay. 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. USA 90:10544-10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 26.Hayes, F. 2003. Transposon-based strategies for microbial functional genomics and proteomics. Annu. Rev. Genet. 37:3-29. [DOI] [PubMed] [Google Scholar]
- 27.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 28.Hensel, M., A. P. Hinsley, T. Nikolaus, G. Sawers, and B. C. Berks. 1999. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32:275-287. [DOI] [PubMed] [Google Scholar]
- 29.Hensel, M., T. Nikolaus, and C. Egelseer. 1999. Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol. Microbiol. 31:489-498. [DOI] [PubMed] [Google Scholar]
- 30.Hensel, M., J. E. Shea, A. J. Baumler, C. Gleeson, F. Blattner, and D. W. Holden. 1997. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J. Bacteriol. 179:1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 32.Hensel, M., J. E. Shea, B. Raupach, D. Monack, S. Falkow, C. Gleeson, T. Kubo, and D. W. Holden. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol. Microbiol. 24:155-167. [DOI] [PubMed] [Google Scholar]
- 33.Hensel, M., J. E. Shea, S. R. Waterman, R. Mundy, T. Nikolaus, G. Banks, A. Vazquez-Torres, C. Gleeson, F. C. Fang, and D. W. Holden. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol. Microbiol. 30:163-174. [DOI] [PubMed] [Google Scholar]
- 34.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 96:2396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 36.Jones, B. D., and S. Falkow. 1994. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect. Immun. 62:3745-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, C. C., and S. Falkow. 2004. Delineation of upstream signaling events in the Salmonella pathogenicity island 2 transcriptional activation pathway. J. Bacteriol. 186:4694-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, C. C., and S. Falkow. 2003. Significance analysis of lexical bias in microarray data. BMC Bioinformatics 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam-Yuk-Tseung, S., and P. Gros. 2003. Genetic control of susceptibility to bacterial infections in mouse models. Cell. Microbiol. 5:299-313. [DOI] [PubMed] [Google Scholar]
- 40.Lawhon, S. D., R. Maurer, M. Suyemoto, and C. Altier. 2002. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46:1451-1464. [DOI] [PubMed] [Google Scholar]
- 41.Lostroh, C. P., and C. A. Lee. 2001. The Salmonella pathogenicity island-1 type III secretion system. Microbes Infect. 3:1281-1291. [DOI] [PubMed] [Google Scholar]
- 42.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 43.Miao, E. A., M. Brittnacher, A. Haraga, R. L. Jeng, M. D. Welch, and S. I. Miller. 2003. Salmonella effectors translocated across the vacuolar membrane interact with the actin cytoskeleton. Mol. Microbiol. 48:401-415. [DOI] [PubMed] [Google Scholar]
- 44.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monack, D. M., D. M. Bouley, and S. Falkow. 2004. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J. Exp. Med. 199:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monack, D. M., D. Hersh, N. Ghori, D. Bouley, A. Zychlinsky, and S. Falkow. 2000. Salmonella exploits caspase-1 to colonize Peyer's patches in a murine typhoid model. J. Exp. Med. 192:249-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 48.Murray, R. A., and C. A. Lee. 2000. Invasion genes are not required for Salmonella enterica serovar Typhimurium to breach the intestinal epithelium: evidence that Salmonella pathogenicity island 1 has alternative functions during infection. Infect. Immun. 68:5050-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neilson, J. W., S. A. Pierce, and R. M. Maier. 1999. Factors influencing expression of luxCDABE and nah genes in Pseudomonas putida RB1353(NAH7, pUTK9) in dynamic systems. Appl. Environ. Microbiol. 65:3473-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nikolaus, T., J. Deiwick, C. Rappl, J. A. Freeman, W. Schroder, S. I. Miller, and M. Hensel. 2001. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 183:6036-6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ochman, H., and E. A. Groisman. 1996. Distribution of pathogenicity islands in Salmonella spp. Infect. Immun. 64:5410-5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 93:7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 54.Pernestig, A. K., D. Georgellis, T. Romeo, K. Suzuki, H. Tomenius, S. Normark, and O. Melefors. 2003. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J. Bacteriol. 185:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Porwollik, S., and M. McClelland. 2003. Lateral gene transfer in Salmonella. Microbes Infect. 5:977-989. [DOI] [PubMed] [Google Scholar]
- 56.Roy, M. F., and D. Malo. 2002. Genetic regulation of host responses to Salmonella infection in mice. Genes Immun. 3:381-393. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz-Albert, J., X. J. Yu, C. R. Beuzon, A. N. Blakey, E. E. Galyov, and D. W. Holden. 2002. Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44:645-661. [DOI] [PubMed] [Google Scholar]
- 58.Salama, N. R., B. Shepherd, and S. Falkow. 2004. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J. Bacteriol. 186:7926-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santos, R. L., S. Zhang, R. M. Tsolis, R. A. Kingsley, L. G. Adams, and A. J. Baumler. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335-1344. [DOI] [PubMed] [Google Scholar]
- 60.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith, B. P., M. Reina-Guerra, S. K. Hoiseth, B. A. Stocker, F. Habasha, E. Johnson, and F. Merritt. 1984. Aromatic-dependent Salmonella typhimurium as modified live vaccines for calves. Am. J. Vet. Res. 45:59-66. [PubMed] [Google Scholar]
- 65.Smith, V., D. Botstein, and P. O. Brown. 1995. Genetic footprinting: a genomic strategy for determining a gene's function given its sequence. Proc. Natl. Acad. Sci. USA 92:6479-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steele-Mortimer, O., J. H. Brumell, L. A. Knodler, S. Meresse, A. Lopez, and B. B. Finlay. 2002. The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell. Microbiol. 4:43-54. [DOI] [PubMed] [Google Scholar]
- 67.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia-del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 68.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 69.Teplitski, M., R. I. Goodier, and B. M. Ahmer. 2003. Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J. Bacteriol. 185:7257-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Turner, A. K., L. Z. Barber, P. Wigley, S. Muhammad, M. A. Jones, M. A. Lovell, S. Hulme, and P. A. Barrow. 2003. Contribution of proton-translocating proteins to the virulence of Salmonella enterica serovars Typhimurium, Gallinarum, and Dublin in chickens and mice. Infect. Immun. 71:3392-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 74.Winterberg, K. M., J. Luecke, A. S. Bruegl, and W. S. Reznikoff. 2005. Phenotypic screening of Escherichia coli K-12 Tn5 insertion libraries, using whole-genome oligonucleotide microarrays. Appl. Environ. Microbiol. 71:451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong, S. M., P. A. Carroll, L. G. Rahme, F. M. Ausubel, and S. B. Calderwood. 1998. Modulation of expression of the ToxR regulon in Vibrio cholerae by a member of the two-component family of response regulators. Infect. Immun. 66:5854-5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36:749-761. [DOI] [PubMed] [Google Scholar]
- 77.Zaharik, M. L., V. L. Cullen, A. M. Fung, S. J. Libby, S. L. K. Choy, B. Coburn, D. G. Kehres, M. E. Maguire, F. C. Fang, and B. B. Finlay. 2004. The Salmonella enterica serovar Typhimurium divalent cation transport systems MntH and SitABCD are essential for virulence in an Nramp1G169 murine typhoid model. Infect. Immun. 72:5522-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]