Abstract
Under solution conditions where the native state is destabilized, the largely helical polypeptide hormone insulin readily aggregates to form amyloid fibrils with a characteristic cross-β structure. However, there is a lack of information relating the 4.8 Å β-strand repeat to the higher order assembly of amyloid fibrils. We have used cryo-electron microscopy (EM), combining single particle analysis and helical reconstruction, to characterize these fibrils and to study the three-dimensional (3D) arrangement of their component protofilaments. Low-resolution 3D structures of fibrils containing 2, 4, and 6 protofilaments reveal a characteristic, compact shape of the insulin protofilament. Considerations of protofilament packing indicate that the cross-β ribbon is composed of relatively flat β-sheets rather than being the highly twisted, β-coil structure previously suggested by analysis of globular protein folds. Comparison of the various fibril structures suggests that very small, local changes in β-sheet twist are important in establishing the long-range coiling of the protofilaments into fibrils of diverse morphology.
Under conditions that destabilize the native state, proteins can self-aggregate into insoluble, fibrillar assemblies (1–3). In the form of amyloid fibrils or fibril precursors, the proteins not only lack their original biological function but also may be harmful to organisms, causing pathologies such as Alzheimer's and prion diseases. Although amyloid precursor proteins do not share any sequence or structural homology, amyloid fibrils are typically unbranched, protease-resistant filaments approximately 100 Å in diameter and composed of ≈20–35 Å wide protofilaments, which are sometimes arranged around an electron lucent core (4, 5). Recently, several nonpathological proteins and short peptides have been shown to self-assemble into amyloid-like fibrils (6–9), leading to the suggestion that amyloid formation is a generic property of polypeptide chains (3, 10).
The overall morphology of amyloid aggregates depends on the conditions in which fibrillogenesis takes place, and different fibril morphologies are often observed in the same preparation (7, 11–14). Variable structures also are seen in ex vivo fibrils extracted from amyloidotic tissue (4, 15). The morphological variation seems to be caused by fibrils with a variable number and arrangement of protofilaments. X-ray fiber diffraction studies reveal a characteristic cross-β structure with β-strands of the precursor protein arranged perpendicular to, and ribbon-like β-sheets parallel to, the fibril axis (2, 16, 17). The β-strand repeat has also been directly visualized by cryo-EM (8). However, there is a lack of three-dimensional (3D) structural information on how the 4.8 Å β-strand repeat relates to the overall fibril assembly.
The polypeptide hormone insulin has a mainly helical native structure, with its two polypeptide chains linked by two inter-chain and one intra-chain disulfide bonds (18). In vitro, insulin is readily converted to an inactive fibrillar form by incubation at high insulin concentrations, low pH and high temperatures (19, 20). The early studies of Waugh demonstrated that no chemical modification was involved in the assembly, and described nucleation and growth steps (19). In the late 1960's, the characteristic 4.8 Å cross-β x-ray fiber diffraction of pathological amyloid was recognized (4, 16), and the cross-β structure of insulin fibrils was reported by Burke and Rougvie in 1972 (21). Amyloid deposits containing intact insulin molecules, including the disulfide bridges, have been reported in a patient with insulin-dependent diabetes undergoing treatment by injection of porcine insulin (22). At pH 2, mass spectrometry and hydrogen exchange measurements reveal that insulin forms soluble assemblies of up to 12 molecules in equilibrium with monomers and smaller oligomers (23, 24). At elevated temperatures, these species further assemble into larger, irreversible aggregates and ultimately fibrils. Studies using Fourier transform infrared spectroscopy (FTIR) and circular dichroism (CD) spectroscopy indicate that the initial aggregates retain their predominantly helical structure, but that there is a subsequent conversion to β-sheet structure as well organized fibrils become visible in negative stain EM images (14). EM and atomic force microscopy studies of other amyloid proteins have indicated similar assembly pathways (25–28).
The aim of the work presented here has been to obtain 3D reconstructions by cryo-EM of insulin fibrils with different morphologies, to gain insight into their common underlying structure and the ways that protofilaments can interact to form fibrils. A compact shape of the insulin protofilament has been observed and may be the result of the constraints of disulfide bridges upon rearrangement of the native conformation into the fibrillar form. Comparison of the various 3D fibril structures suggests some general principles of amyloid fibril assembly.
Materials and Methods
Sample Preparation.
Bovine insulin (2 mM; Sigma) was dissolved in water adjusted to pH 2 with HCl, heated to 60°C for 2 days, and then left incubating at room temperature. This procedure yielded fibrils with a predominantly compact morphology. Fibrils also were prepared at 2 mM insulin and pH 2.3, but the insulin solution was heated to 70°C for 1 h and then fast-frozen in liquid nitrogen, followed by incubation at room temperature. These samples contained a variety of morphologies, several of which were analyzed. Samples were imaged at intervals between 1 month and 1 year.
Microscopy and Image Processing.
The samples were checked by negative staining (2% uranyl acetate) on carbon-coated grids. Unidirectional shadowing of samples blotted and dried onto carbon-coated grids was carried out with platinum/carbon in a Bal-Tec Med 020 coating system. For cryo-EM, 5 μl samples were incubated for about half a minute on perforated carbon grids, which then were blotted with filter paper and plunged into liquid ethane. The micrographs were taken at minimum dose on Agfa Scientia EM film in a JEOL 1200 EX with a tungsten filament at 120 kV. The micrographs were digitized at a pixel size of 3.33 Å on the specimen by using a Leafscan 45 linear charge-coupled device scanner (Ilford, Cheshire, U.K.) and then averaged to 6.67 Å per pixel. The software packages SUPRIM V.5.2 (29), SPIDER V.4.8 (30), and IMAGIC-5 (31) were run on either Silicon Graphics or DEC Alpha workstations.
Because of variations in the pitch of the fibrils examined here, we used a combination of single particle and helical reconstruction methods, as described (7). The problem of variable pitch was considered in earlier work by Crowther (32), who interpolated fiber images to a constant pitch. Details of the procedures used here are given in the supporting information, which is published on the PNAS web site, www.pnas.org, and in ref. 33. Briefly, for the compact fibrils, a map at ≈25 Å resolution was generated from a class average of 80 helical repeats obtained by alignment and classification of a data set of 747 repeats. For the other fibril morphologies, only individual examples of any particular morphology and spacing could be found. Therefore, only the repeats in one fibril could be averaged. Such fibrils were first interpolated to a straight axis by using PHOELIX (34), and then the Fourier filtered image was generated from the visible layer lines. This averaged image was backprojected to produce a continuous helical structure. Protofilaments from the different fibril cross sections were translationally and rotationally aligned to determine their average and variance. The mass of the protofilament section was estimated by determining the number of pixels above a density threshold (mean plus 2 or 3 SDs) and by calculating the corresponding volume, assuming a protein density of 0.81 Da per Å (3) and a β-strand separation of 4.8 Å. This calculation gives 0.54–0.64 insulin molecules per 4.8 Å of height, suggesting that a molecule of insulin occupies two β-strand layers along the fiber axis, which is consistent with a 30 × 40 Å protofilament cross section.
Results
Morphological Variability of Insulin Fibrils.
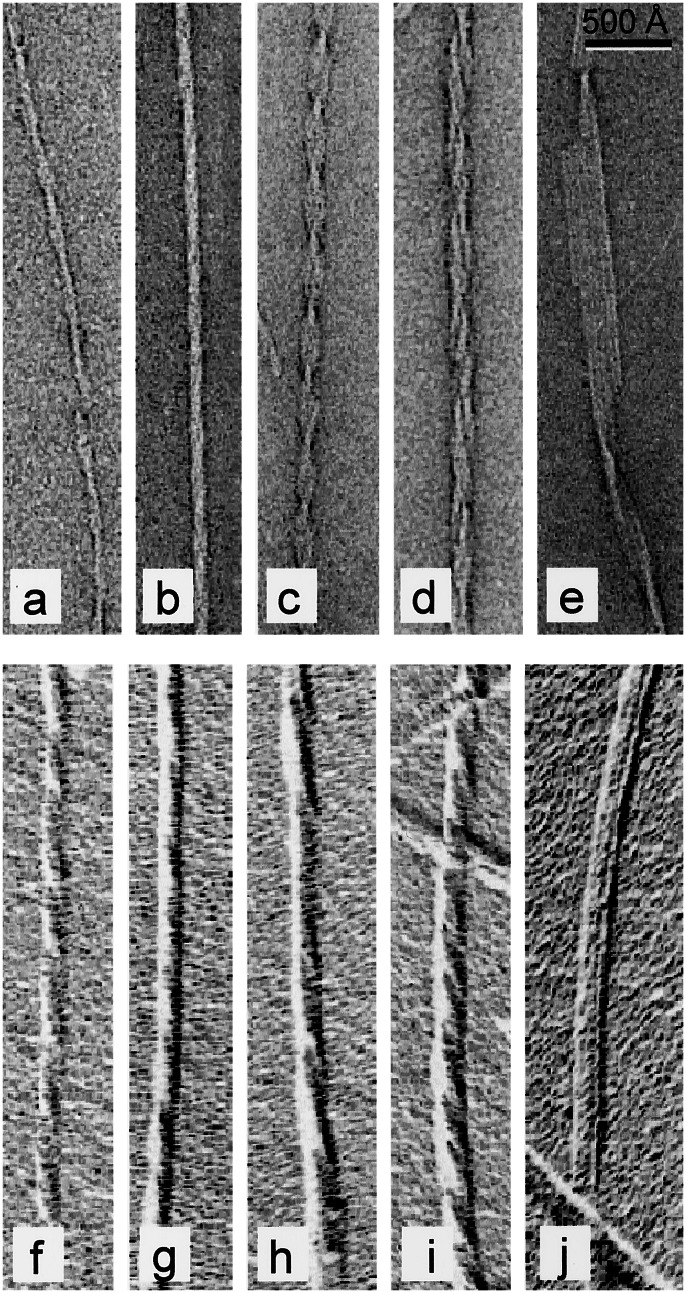
Fibril morphologies found in the two samples studied included a thin fibril (Fig. 1a) and a thicker, compact fibril with a faintly discernable periodicity (Fig. 1b). In other fibrils, there are clearly repeating variations in width, indicative of a helical arrangement of protofilaments (Fig. 1c). We also observed many twisting ribbon-like structures of different width and pitch (Fig. 1d) and flat ribbons of parallel protofilaments (Fig. 1e). The same morphologies were identified in samples imaged by metal shadowing to provide surface relief and thus identify the handedness of the twisted structures (Fig. 1 f–j). All of the intact filaments with a clearly visible twist were found to be left handed (Fig. 1 g–j). All of the different morphologies were found at all incubation times examined.
Figure 1.
Gallery of negatively stained (a–e) and shadowed (f–j) insulin fibrils, showing the diversity of fibril structures. The protofilament substructure is visible in e. The shadowing shows that all of the fibrils are left handed (g–j). (Bar = 500 Å.)
Cryo-EM and 3D Reconstruction of Insulin Fibrils.
The sample incubated after a heating and a freezing step (see Materials and Methods) yielded a great diversity of fibrils with different structures and spacings, so that different fibrils could not be averaged. In the other sample, which was only exposed to elevated temperature, a single type of compact fibril was abundant, and many fibrils were averaged for structure determination. Four fibril structures are presented, in order of increasing diameter. The compact fibril is the second of these. As we found for SH3 amyloid fibrils (7), the diffraction patterns contained substantially higher resolution information in the direction perpendicular to the fibril axis than along it. The intensities of the layer lines extended laterally to the first minimum of the contrast transfer function, a resolution cut-off around 22 Å. In all cases, the pitch was quite variable, with the distance between crossovers varying up to 20–30%.
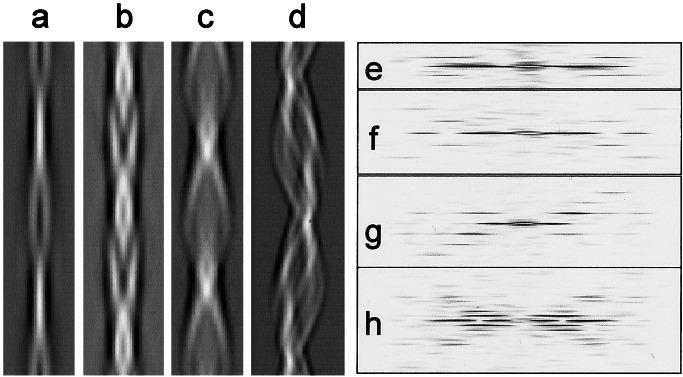
In Fig. 2, a reprojected image and a diffraction pattern are shown for each of the four fibril structures studied here in detail. The thin fibril (Fig. 2 a and e), similar to the one in Fig. 1a, showed only a single layer line at a spacing of ≈525 Å. The finding that the layer line indexed to a Bessel order of 2 (determined by the positions of amplitude peaks on the layer line and their phase relationships) implies that the fibril is a double helix.
Figure 2.
Reprojected images and calculated diffraction patterns of the helical repeat from cryo-EM images of four different insulin amyloid fibril types. (a and e) Filtered image and diffraction pattern of the smallest diameter fibril. (b and f) For the compact fibril. (c and g) A larger fibril. (d and h) A twisted ribbon. The images are reprojected from the 3D maps in Fig. 3. Distances between crossovers (layer line spacings) are: a, 525 Å; b, 355 Å; c, 426 Å; d, 940 Å. Note that a–c are double helices, so that their layer line spacings correspond to one half of a complete turn.
The diffraction patterns of the abundant compact fibrils (Figs. 1b and 2b) interpolated onto a straight axis contain two layer lines (Fig. 2f). The layer line spacing (≈355 Å) corresponds to the distance between crossovers observed in the EM images. The reprojected image has mirror symmetry (Fig. 2b), suggesting that the filaments have an even number of protofilaments, in agreement with the determination of Bessel orders 2 and 4 for the first and second layer lines, respectively. The image presents clear polarity and shows that the width varies little along the projection, accounting for the compact appearance of this fibril type. The fibril polarity results from the deviation of its cross section from mirror symmetry (see, for example, Fig. 3f).
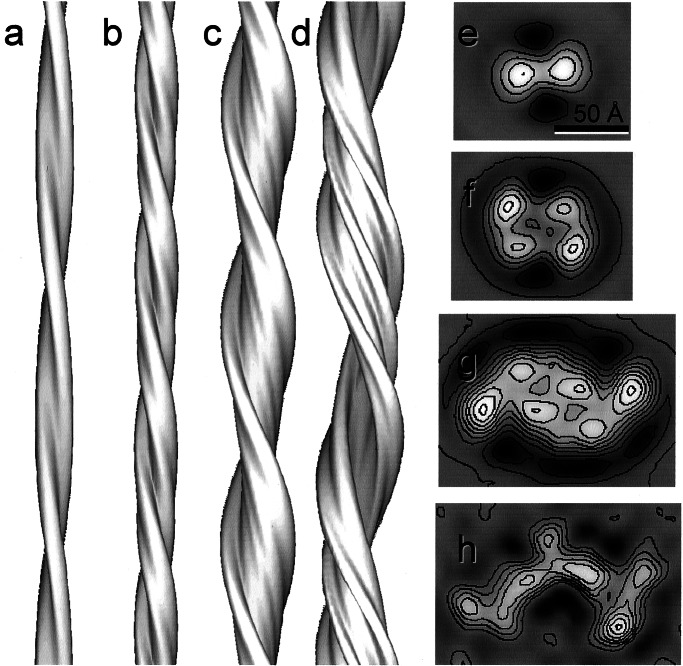
Figure 3.
Surface representation of 3D maps and contoured density cross sections of the four insulin fibril structures shown in Fig. 2. (a and e) Structure of the fibril with a pair of protofilaments twisting around each other. (b and f) The four-protofilament compact fibril. (c and g) The six-protofilament fibril. (d and h) The twisted ribbon. The protofilaments are well resolved in the first three structures, but are less clear in the twisted ribbon. (e–h, bar = 50 Å.)
The thicker, helical filament (Fig. 2c), similar to the one in Fig. 1c, presents a diffraction pattern with three layer lines at a spacing around 426 Å (Fig. 2g), corresponding to the distance between crossovers. The near and far sides of the pattern are asymmetric, probably owing to noise or distortion. Fig. 2d shows the reprojected image of a filament with a twisting ribbon morphology similar to that seen in Fig. 1d. Its diffraction pattern contains five clear layer lines with a spacing around 940 Å (Fig. 2h).
The 3D map and cross section of a thin fibril of the type shown in Fig. 1a reveal two protofilaments twisting around each other (Fig. 3 a and e). The 3D structure of the compact filament at 25 Å resolution was obtained by averaging ≈80 helical repeats (Fig. 3b). It is a left-handed double helix with four peaks of high electron density in cross section (Fig. 3f). Although two of the peaks have a higher density than the other two, their overall size is very similar and, therefore, each peak can be attributed to a single protofilament of approximate dimensions 30 × 40 Å. Fig. 3c shows a reconstruction of a fibril containing six protofilaments. The cross section (Fig. 3g) shows six protofilaments of similar size but unequal density. The 3D map of the twisted ribbon is shown in Fig. 3d. Although the curved shape of the fibril section is well resolved, the number of protofilaments cannot be unambiguously determined, because they do not contain equivalent features (Fig. 3h). The three regions of higher density each might consist of a pair of protofilaments.
The resolution of the reconstructions from single filaments (Fig. 3 a, c, and d) was estimated as 30–40 Å by the lateral extent of the layer lines. Because the protofilament cross section seems to be common to all of the reconstructions, its features should be enhanced by averaging.
Insulin Amyloid Fibrils Share a Common Protofilament Structure.
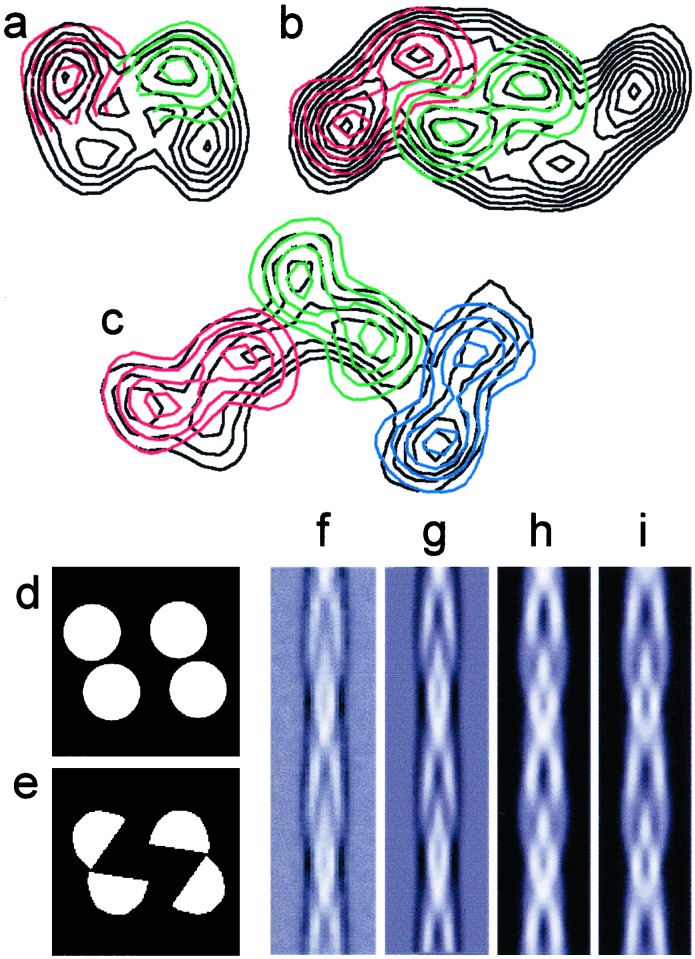
To compare the protofilaments in the different fibrils, pairs of maps were superimposed and manually aligned to find the best fit (Fig. 4 a–c). For convenience, the cross section of the fibril containing the pair of protofilaments was used as the reference. In general, the protofilament shapes, sizes, and packing agree well in the different maps. To examine whether the different protofilaments in the three well defined fibrils (excluding the ribbon) have common features, the set of six nonsymmetry-related protofilaments were aligned by cross-correlation and averaged. In addition, a variance map was calculated to assess the consistency of the average features. The average and variance of the protofilament densities and also the comparison with the SH3 protofilament are shown in the supporting information. The average protofilament is about 30 × 40 Å, very similar to the core of the SH3 protofilament (7).
Figure 4.
Superposition of cross sections from the different fibril morphologies suggests the presence of a common protofilament structure. (a) The protofilaments fit well when compared individually between the two- and four-protofilament fibrils. (b) The pair of protofilaments fits well in size with pairs of protofilaments in the six-protofilament fibril. (c) The ribbon could hold six-protofilaments although their boundaries are not clear. (d and e) Two possible, simple models for the cross section of the compact, four-protofilament fibril. (f) Average of the repeats extracted from cryo-EM images of the compact fibrils and aligned as single particles. (g) Reprojection of the 3D map of the compact fibril. (h and i) Reprojections of the 3D models whose cross sections are shown in d and e, respectively.
Fig. 4 d and e show cross sections of two simple models of the compact, four-protofilament fibril. An average of extracted and aligned repeats of the cryo-EM images of this fibril type (Fig. 4f) is compared with reprojections of the 3D map (Fig. 4g) and of the two models (Fig. 4 h and i).
Discussion
A Model of the Molecular Packing in Insulin Amyloid.
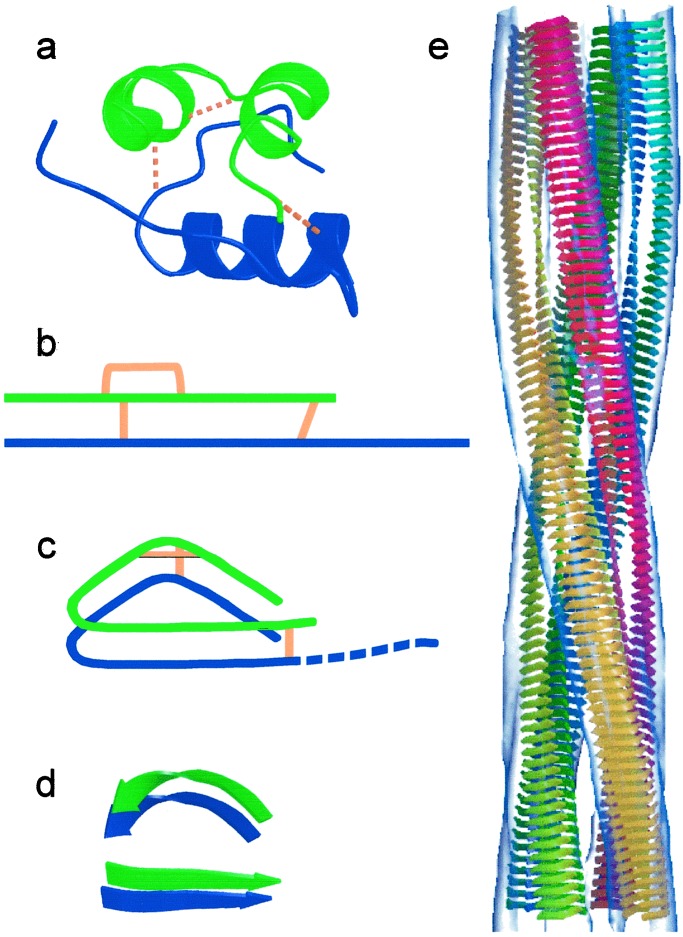
The α-helical structure of native insulin must unfold completely to form the amyloid cross-β structure. However, mass spectrometry of insulin molecules retrieved from amyloid fibrils by alkali extraction confirms that the disulfide bonds of the native hormone are retained in the amyloid form (35), providing substantial constraints to refolding. A simple model for the chain packing is, however, consistent with these constraints. The inter-chain disulfide bonds require the strands to run in parallel, consistent with FTIR analysis (14). The intra-chain disulfide bond introduces some curvature into the strand, so that the protofilament has a compact cross-sectional shape. Schematic representations of the topology in native insulin and of a possible arrangement of the polypeptide chain in the amyloid protofilament are shown in Fig. 5 a–c. It is possible to unwind all of the α-helices and wrap the chains into segments of parallel β-helix. The density is consistent with the presence of two β-sheets within each protofilament, as found in the SH3 domain fibrils, although the intra-chain disulfide bond may induce a bulge in one of the sheets. Comparison of insulin and SH3 protofilaments suggests that the character of the polypeptide forming the amyloid fibril can influence β-sheet packing and thus protofilament shape, although the basic protofilament structure remains similar.
Figure 5.
(a) Insulin structure showing the three native disulfide bonds. A chain, green; B chain, blue; disulfide bonds, gold. (b) Topology diagram of insulin color coded as in a. (c) Possible topology for the amyloid protofilament. Orientations of the termini and disulfide bonds within the curved structure are arbitrary. The C terminus of chain B (dashed) is not required for amyloid fibril formation (see ref. 43). (d) β-strand model of a protofilament. Each chain is shown in two segments, a straight and a curved β-strand (PDB accession no. 1umu, residues 93–100). Each insulin molecule would occupy two layers, connected by the interchain disulfide bonds. (e) A possible β-strand model docked into the EM density of the compact fibril (transparent gray surface). The four protofilaments are colored separately.
A possible schematic model for the β-strand arrangement and protofilament assembly for the compact, four-protofilament form of insulin amyloid is shown in Fig. 5 d and e. Volume calculations suggest that each insulin molecule occupies two β-strand layers (see Materials and Methods). For simplicity, all chains are shown with parallel packing, although alternate molecules could be antiparallel. The connectivity between the two sections of strand within each protofilament is unknown, but open rings of such β-structures are sufficient to account for the observed protofilament density. This model is an interpretation consistent with the available data, but other models are, of course, not excluded.
β-Sheet Twist, Protofilament Packing, and Amyloid Assembly.
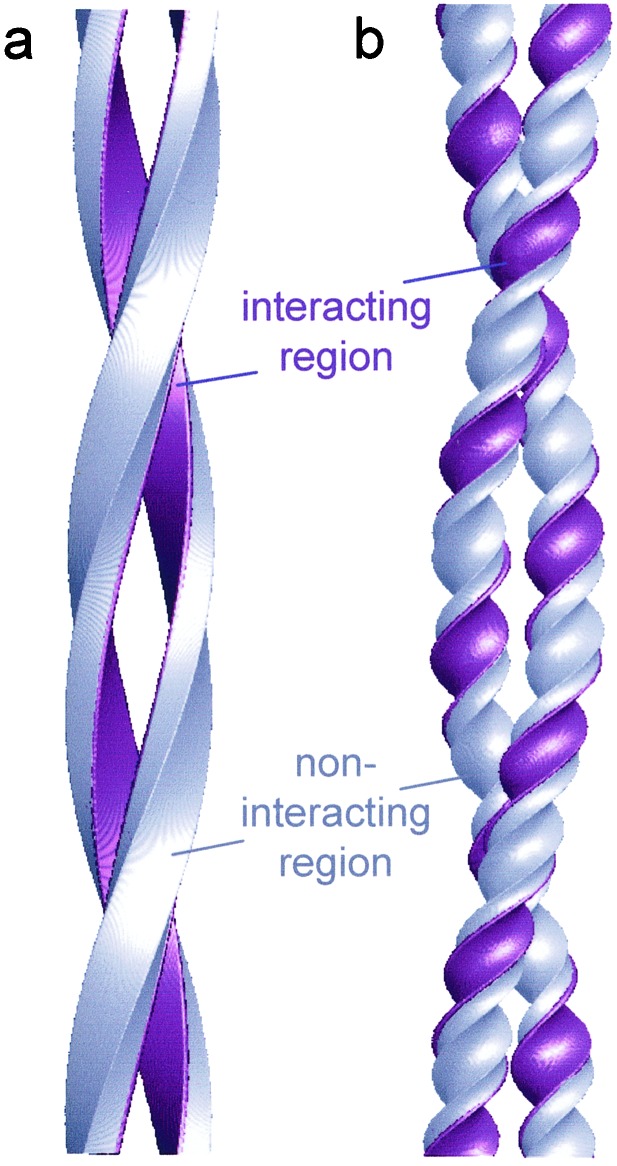
All of the amyloid fibrils for which the hand has been determined have been reported to have a left-handed helical twist (12, 13, 28, 36). [The hand of SH3 fibrils was initially not determined, and was arbitrarily shown as right-handed (7), but was subsequently shown to be left-handed (37)]. The twist between β-strands in β-sheets is also left-handed and, therefore, the twist of the protofilaments must be left-handed as well. This, together with the existence of a region of the protofilament free of interactions with other protofilaments, adds a new constraint to the molecular nature of the protofilament. In order for the protofilaments to pack together in a consistent interface, the protofilament twist must follow the overall twist of the fibril (Fig. 6a). If the twist between the β-strands of insulin molecules in the protofilament were of shorter pitch than the twist of the whole fibril, as in the β-coil protofilament model of transthyretin (2, 17), the regions of interaction between protofilaments would rotate progressively around the protofilament (Fig. 6b). When the twist of the protofilament accompanies the twist of the fibril, the regions of interaction between protofilaments are constant. This constraint permits the presence of noninteracting regions in which loops and motifs outside the cross-β core are always on surfaces free of contacts to other protofilaments, and thus do not disrupt the protofilament packing (Fig. 6a). This finding is in agreement with the average and variance of the protofilament sections (Fig. 7, which is published as supporting information on the PNAS web site), suggesting that interactions between protofilaments are indeed restricted to one surface, as required by the latter model.
Figure 6.
Models for protofilament packing. (a) A twisted pair of rectangular protofilaments in which an interactive surface is colored purple. The protofilament twist accompanies the filament twist. (b) A supercoiled pair of protofilaments in which the regions involved in packing interactions rotate around each protofilament. In the correlated twist model a, interacting regions would be fixed relative to the cross-β structure, and other regions could accommodate large loops and/or folded domains that would not interfere with protofilament packing. Similar models can be constructed with more than two protofilaments, in which the cross section rotates as a rigid unit in the helical structure. Keeping the cross section fixed means that all packing contacts can be preserved in the helical fibril. This is the case for the four-protofilament model in Fig. 5e.
Such a correlated twist model is also in good agreement with the SH3 (7) and full-length Sup35 fibrils (27). It is also supported by observations on amyloid-like α-synuclein fibrils that can be labeled all along their length by antibodies directed against the C-terminal region of the protein, but only at one end by N-terminally directed antibodies (38, 39). In these cases, a particular domain or epitope is always oriented the same way relative to the fibril. Other amyloid models based on x-ray fiber diffraction data also propose that the β-strand twist accompanies the twisting of the fibrils (40). The observed variability in long-range repeat and morphology of amyloid fibrils follows readily from this correlated twist model, considering that there are about 200 β-strands per long-range repeat. Changes in the inter-strand angle of even a fraction of a degree will significantly change the long-range twist and, thus, the repeat length. The angles observed so far are in the range 1.5–2.5° between successive strands, corresponding to full-pitch repeats of ≈1200–700 Å, respectively.
General Principles of Protofilament Assembly into Amyloid Fibrils.
Although the fundamental cross-β structure is common to all amyloid fibrils, the packing of β-sheets into protofilaments must vary to some extent according to the constraints of the constituent polypeptide. The protofilaments themselves assemble into pairs or larger groupings by coiling around each other with a long-range twist arising from the small angle between their constituent β-strands. The left-handed twist and packing between strands is propagated for particular higher order assemblies throughout the fibril structure. Such apparently cooperative assembly could provide a structural basis for the multiple strains of prions, that are associated with distinct pathological states (41, 42). It is possible that different strains could arise from different fibril morphologies that nucleate further assembly of the same morphological type.
Overall, the present work has revealed a consistent size and compact shape for the protofilaments in amyloid fibrils and provided insight into the structural principles of the assembly of amyloid states. General conclusions are that the β-sheets in amyloid protofilaments must accompany the overall fibril twist (and thus be nearly flat), in contrast to earlier models based on a β-coil structure (17). Although different amyloid protofilaments appear similar in size, the protein sequence can influence the sheet packing into protofilaments, for example, because of the constraints of preexisting disulfide bonds. Finally, a common protofilament unit may assemble into fibrils with extremely variable packing, but particular protofilament interactions can be approximately maintained along the length of a given fibril. This finding suggests that the protofilaments adopt a series of inter-chain interactions during their assembly process that can be propagated during fibrillar growth. Moreover, it provides a graphic illustration of the long-range interactions inherent in the structure of amyloid fibrils.
Supplementary Material
Acknowledgments
We thank Jesus Zurdo, Tony Clarke, Guy Schoehn, Stephen Fuller, and Elena Orlova for helpful discussions. We thank Shaoxia Chen and Juliet Munn for EM support, Richard Westlake and David Houldershaw for computing support, and Brent Gowen for help with shadowing. We thank the Wellcome Trust for their support. H.R.S. is a member of the Bloomsbury Centre for Structural Biology. The Oxford Centre for Molecular Sciences is supported by the Biotechnology and Biological Sciences Research Council, Engineering and Physical Sciences Research Council, and Medical Research Council.
Abbreviations
- EM
electron microscopy
- 3D
three-dimensional
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pepys M B. In: The Oxford Textbook of Medicine. 3rd Ed. Weatherall D J, Ledingham J G G, Warell D A, editors. Vol. 2. Oxford: Oxford Univ. Press; 1996. pp. 1512–1524. [Google Scholar]
- 2.Sunde M, Serpell L C, Bartlam M, Fraser P E, Pepys M B, Blake C F. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 3.Dobson C M. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 4.Cohen A S, Shirahama T, Skinner M. In: Electron Microscopy of Proteins. Harris J R, editor. Vol. 3. London: Academic; 1982. pp. 165–205. [Google Scholar]
- 5.Serpell L C, Sunde M, Benson M D, Tennent G A, Pepys M B, Fraser P E. J Mol Biol. 2000;300:1033–1039. doi: 10.1006/jmbi.2000.3908. [DOI] [PubMed] [Google Scholar]
- 6.Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, Dobson C M. Proc Natl Acad Sci USA. 1999;96:3590–3594. doi: 10.1073/pnas.96.7.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiménez J L, Guijarro J I, Orlova E, Zurdo J, Dobson C M, Sunde M, Saibil H R. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serpell L C, Smith J M. J Mol Biol. 2000;299:225–231. doi: 10.1006/jmbi.2000.3650. [DOI] [PubMed] [Google Scholar]
- 9.Fändrich M, Fletcher M A, Dobson C M. Nature (London) 2001;410:165–166. doi: 10.1038/35065514. [DOI] [PubMed] [Google Scholar]
- 10.Dobson C M. Philos Trans R Soc London B. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fraser P E, Nguyen J T, Surewicz W K, Kirschner D A. Biophys J. 1991;60:1190–1201. doi: 10.1016/S0006-3495(91)82154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bauer H H, Aebi U, Häner M, Hermann R, Müller M, Arvinte T, Merkle H P. J Struct Biol. 1995;115:1–15. doi: 10.1006/jsbi.1995.1024. [DOI] [PubMed] [Google Scholar]
- 13.Goldsbury C S, Cooper G J S, Goldie K N, Müller S A, Saafi E L, Gruijters W T M, Misur M P, Engel A, Aebi U. J Struct Biol. 1997;119:17–27. doi: 10.1006/jsbi.1997.3858. [DOI] [PubMed] [Google Scholar]
- 14.Bouchard M, Zurdo J, Nettleton E J, Dobson C M, Robinson C V. Protein Sci. 2000;9:1969–1967. doi: 10.1110/ps.9.10.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiménez J L, Tennent G, Pepys M, Saibil H R. J Mol Biol. 2001;311:241–247. doi: 10.1006/jmbi.2001.4863. [DOI] [PubMed] [Google Scholar]
- 16.Eanes E D, Glenner G G. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 17.Blake C, Serpell L C. Structure (London) 1996;4:989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 18.Blundell T L, Cutfield J F, Cutfield S M, Dodson E J, Dodson G G, Hodgkin D C, Mercola D A, Vijayan M, et al. Nature (London) 1971;231:506–511. doi: 10.1038/231506a0. [DOI] [PubMed] [Google Scholar]
- 19.Waugh D F. J Am Chem Soc. 1946;68:247–250. [Google Scholar]
- 20.Nielsen L, Frokjaer S, Carpenter J F, Brange J. J Pharm Sci. 2001;90:29–37. doi: 10.1002/1520-6017(200101)90:1<29::aid-jps4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Burke M J, Rougvie M. Biochemistry. 1972;11:2435–2439. doi: 10.1021/bi00763a008. [DOI] [PubMed] [Google Scholar]
- 22.Dische F E, Wernstedt C, Westermark G T, Westermark P, Pepys M B, Rennie J A, Gilbey S G, Watkins P J. Diabetologia. 1988;31:158–161. doi: 10.1007/BF00276849. [DOI] [PubMed] [Google Scholar]
- 23.Nettleton E J, Tito P, Sunde M, Bouchard M, Dobson C M, Robinson C V. Biophys J. 2000;79:1053–1065. doi: 10.1016/S0006-3495(00)76359-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tito P, Nettleton E J, Robinson C V. J Mol Biol. 2000;303:267–278. doi: 10.1006/jmbi.2000.4142. [DOI] [PubMed] [Google Scholar]
- 25.Jarrett J T, Lansbury P T. Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 26.Colon W, Kelly J W. Biochemistry. 1992;31:8654–8660. doi: 10.1021/bi00151a036. [DOI] [PubMed] [Google Scholar]
- 27.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J, Lindquist S. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 28.Ionescu-Zanetti C, Khurana R, Gillespie J R, Petrick J S, Trabachino L C, Minert L J, Carter S A, Fink A L. Proc Natl Acad Sci USA. 1999;96:13175–13179. doi: 10.1073/pnas.96.23.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schroeter J P, Bretaudiere J P. J Struct Biol. 1996;116:131–137. doi: 10.1006/jsbi.1996.0021. [DOI] [PubMed] [Google Scholar]
- 30.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 31.van Heel M, Harauz G, Orlova E, Schmidt R, Schatz M. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 32.Crowther R A, Wischik C. EMBO J. 1985;4:3661–3665. doi: 10.1002/j.1460-2075.1985.tb04132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiménez J L. Ph.D. thesis. Univ. of London, London: Birkbeck College; 2000. [Google Scholar]
- 34.Whittaker M, Carragher B, Milligan R A. Ultramicroscopy. 1995;58:245–260. doi: 10.1016/0304-3991(95)00057-8. [DOI] [PubMed] [Google Scholar]
- 35.Nettleton E J. Ph.D. thesis. Oxford: Oxford University; 1998. [Google Scholar]
- 36.Harper J D, Lieber C M, Lansbury P T. Chem Biol. 1997;4:951–959. doi: 10.1016/s1074-5521(97)90303-3. [DOI] [PubMed] [Google Scholar]
- 37.Chamberlain A K, MacPhee C E, Zurdo J, Morozova-Roche L A, Hill H A, Dobson C M, Davis J J. Biophys J. 2000;79:3282–3293. doi: 10.1016/S0006-3495(00)76560-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crowther R A, Daniel S E, Goedert M. Neurosci Lett. 2000;292:128–130. doi: 10.1016/s0304-3940(00)01440-3. [DOI] [PubMed] [Google Scholar]
- 39.Serpell L C, Berriman J, Jakes R, Goedert M, Crowther R A. Proc Natl Acad Sci USA. 2001;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inouye H, Kirschner D A. Ciba Found Symp. 1996;199:22–35. doi: 10.1002/9780470514924.ch3. [DOI] [PubMed] [Google Scholar]
- 41.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chien P, Weissman J S. Nature (London) 2001;410:223–226. doi: 10.1038/35065632. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen L, Frokjaer S, Brange J, Uversky V N, Fink A L. Biochemistry. 2001;40:8397–8409. doi: 10.1021/bi0105983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.