Abstract
Certain HIV-encoded proteins modify host-cell gene expression in a manner that facilitates viral replication. These activities may contribute to low-level viral replication in nonproliferating cells. Through the use of oligonucleotide microarrays and high-throughput Western blotting we demonstrate that one of these proteins, gp120, induces the expression of cytokines, chemokines, kinases, and transcription factors associated with antigen-specific T cell activation in the absence of cellular proliferation. Examination of transcriptional changes induced by gp120 in freshly isolated peripheral blood mononuclear cells and monocyte-derived-macrophages reveals a broad and complex transcriptional program conducive to productive infection with HIV. Observations include the induction of nuclear factor of activated T cells, components of the RNA polymerase II complex including TFII D, proteins localized to the plasma membrane, including several syntaxins, and members of the Rho protein family, including Cdc 42. These observations provide evidence that envelope-mediated signaling contributes to the productive infection of HIV in suboptimally activated T cells.
HIV preferentially replicates in proliferating CD4+ T cells (1). However recent evidence suggests that, in vivo, resting and suboptimally activated T cells may serve as targets for low-level productive infection in the absence of cellular proliferation (2–5). Infection in this manner may contribute to the establishment and/or maintenance of persistent viral reservoirs that currently prevent the eradication of virus. To productively infect suboptimally activated CD4+ T cells, HIV must overcome post-entry barriers to replication (6–8).
DNA microarrays have been used to characterize the effect of HIV on target cell transcription (9, 10); in one microarray-based study, HIV Nef was shown to diminish barriers to viral replication by mimicking antigen-specific T cell proliferation signals (11, 12). It has been suggested that HIV gp120 also facilitates replication in suboptimally activated cells (12–15). Gp120 transduces near-simultaneous signals through CD4 (16), a component of the T cell receptor complex, and CCR5, a chemokine receptor (17–19). In vivo concentrations of gp120 (20, 21) fall within the range required to induce signaling in vitro (17–19, 22). To provide a more complete picture of the complex cascade of signals induced by gp120, we treated freshly isolated peripheral blood mononuclear cells (PBMCs) and monocyte-derived macrophages (MDMs) with an envelope derived from a CCR5-using virus and measured temporal changes in the levels of mRNA by using Affymetrix (Santa Clara, CA) U95A oligonucleotide microarrays that include probes encompassing ≈12,600 genes. In addition, we used a high-throughput Western blot analysis that allowed us to screen protein lysates with 800 monoclonal antibodies. The gp120 used was derived from JR-FL, a CCR5-tropic molecular clone obtained from a minimally passaged viral isolate (23). We used concentrations of envelope near or below that detected in the serum of infected patients (20).
Materials and Methods
Cells and Reagents.
PBMCs were obtained by apheresis from normal volunteers followed by ficoll-hypaque isolation. Cells were resuspended in RPMI medium 1640/10% FBS. Donor PBMCs carrying one or two CCR5 Δ32 alleles were not used. MDMs were derived from elutriated monocytes cultured for 10 days in RPMI medium 1640/10%FBS/10% pooled human AB serum supplemented with granulocyte/macrophage colony-stimulating factor (GM-CSF; 10 units/ml). Gp120 was expressed and purified as described (17, 24). Briefly, a Chinese hamster ovary (CHO) cell line expressing a recombinant gp120 derived from the HIV molecular clone JR-FL (23) was cultured in hollow-fiber cartridges (Fibercell Systems, Frederick, MD). Protein was purified in three steps, employing metal-chelating, lectin, and size-exclusion chromatography. Protein concentrations were determined by absorbance at wavelength 280 nanometers. Protein was visualized by silver-stain and determined to be greater than 97% pure and endotoxin-free (LAL assay; BioWhittaker, Walkersville, MD). A mock protein prepared in an identical manner was derived from untransfected CHO cells and used as a mock.
gp120 Treatment of PBMCs and MDMs.
Freshly isolated PBMCs and MDMs (1–5 × 107) per time point were incubated in 10% FBS/RPMI medium 1640 at 37°C during a time course ranging from 1 to 16 h at a concentration of 50 ng per 106 cells of gp120. Parallel cultures were treated with a mock protein preparation.
cRNA Preparation for Oligonucleotide Arrays.
Total RNA from ≈1–5 × 107 stimulated or unstimulated cells per time point was extracted using the TRIZOL method (Life Technologies, Frederick, MD). Briefly, cell pellets containing 1 × 107 cells were lysed in 1 ml of TRIZOL and homogenized using a 1-ml syringe and a 19-gauge needle. Samples were layered with 200 μl of chloroform, inverted 15 times, and incubated on ice for 15 min. Lysates were centrifuged at 4°C for 15 min at 14,000 rpm and the aqueous phase transferred to a clean tube. RNA was precipitated with equal volumes of isopropanol for 15 min at room temperature. Precipitates were spun for 30 min at 4°C and 14,000 rpm. Pellets were washed twice with 70% cold EtOH, and dried at room temperature. RNA was resuspended in 50 μl of diethyl pyrocarbonate (DEPC) water, quantitated, and analyzed by denaturing gel electrophoresis to check purity. A total of 15 μg of RNA was used for microarray analysis. First- and second-strand DNA synthesis reactions were performed using the Superscript Choice System (Life Technologies) followed by in vitro transcription (Enzo Diagnostics) using biotin-labeled dNTPs. The resulting biotin-labeled cRNA was quantitated and analyzed for purity on a 2% agarose Tris acetate EDTA (TAE) gel. cRNA samples were then fragmented and prepared for hybridization to Affymetrix Human Genome U95A oligonucleotide arrays according to protocols specified by the manufacturer.
Western Blot Analysis.
Immunoblot analysis of proteins was carried out as described (www.translab.com/shtml). Briefly, PBMCs were treated with gp120 as described above and proteins were lysed in 10 mM Tris (pH 7.4)/1 mM Na+orthovanadate/1% SDS followed by sonication and clarified by centrifugation. Gels used were 16 × 16 cm, 5–15% gradient SDS-polyacrylamide, 1-mm-thick. A gradient system was used so a wide size range of proteins could be detected on one gel. Four hundred micrograms of protein was loaded in long well across the entire width of the gel. This translates into ≈15 μg per lane on a standard 25-well gel. The gel was then run at constant milliamps. Subsequently the gel was transferred to Immobilon-P membrane (Millipore, Bedford, MA) overnight at 200 mA. After transfer, membranes were blocked for 1 h with 5% milk. Subsequently, the membrane was inserted into a Western blotting manifold that isolates 45 channels across the membrane. In each channel, different complex antibody cocktails were added and allowed to hybridize for 1 h. Following staining, the membranes were washed and hybridized for 30 min with secondary goat anti-mouse horseradish peroxidase (HRP). All antibodies were mouse monoclonal. Membranes were washed and developed with SuperSignal West Pico (Pierce).
Statistical Determination of Significant Differential Expression.
A Significant Analysis of Microarrays (sam; ref. 25) algorithm was used to determine significant differential expression after extensive prefiltering of genes. The prefilter was established as follows. The expression value, termed average difference (Avg. Diff.), was derived using the Affymetrix software program MAS 4.0. The Avg. Diff. of genes <10 were truncated to 10. The difference between mean Avg. Diff. between comparison groups was set at >30. Mean Avg. Diff. between comparison groups was set at >1.4-fold or <−1.4-fold, and the significant difference of a Student's t test between the comparison group was set at P < 0.1. Only those genes that generated lists from sam after 5,000 randomizations and with a 90% median false discovery rate equal to zero were defined as differentially expressed.
Results
Six Hundred Genes Modulated by HIV-1 Envelope in PBMCs and MDMs.
Fresh PBMCs (four donors) were treated with gp120 for 1, 5 and 16 h, whereas MDMs (three donors) were treated for 3, 6, and 10 h. In PBMCs, >9,000 genes were expressed of the ≈12,600 genes represented on the microarray whereas in MDMs, >8,900 genes were expressed. To identify genes that were modulated in a significant manner, we used a recently developed statistical method termed sam (25). This method was designed specifically to analyze large numbers of biological responses (≈10,000) typical of microarray data. Using sam we identified ≈600 genes (≈300 for PBMCs and ≈300 for MDMs) that were significantly modulated in response to envelope treatment. From that list we extracted subsets of genes that were up-regulated to the greatest degree (Fig. 1 a and b). We report these responses at each of the time points analyzed (1, 5, and 16 h for PBMCs; 3, 6, and 10 h for MDMs). Because individual donors may modulate a given gene at different time points, we also identified genes that were significantly modulated irrespective of the time of the response (Fig. 1 a and b; all time points). Although many of the genes we identified are of unknown function or have never been associated with HIV, our analysis identified at least 23 genes that have previously been associated with HIV replication and/or envelope signaling (Table 1). This finding indicates that the system and strategy that we used was sufficiently sensitive and reliable to identify relevant responses. We chose to focus our current analysis on genes likely to influence viral replication.
Figure 1.
Induction of genes in response to gp120 treatment. A list of genes, which were determined by sam to be significantly modulated in response to JR-FL gp120, was generated. A subset of that list that includes those genes most up-regulated is listed by time point for PBMCs (a) and MDMs (b). Responses were also evaluated by sam independent of time, and a subset of those genes that were most up-regulated is reported in the list termed “all time points.” Accession number and definition are included for each gene. avg FC, the fold change relative to PBMCs or MDMs treated with a mock protein preparation. Genes are ranked by descending fold change. Results represent the average of four donors in PBMCs or three donors in MDMs.
Table 1.
JR-FL gp120-induced expression of genes previously associated with HIV
| Induced gene | Cell type | GenBank accession no. | Ref. and/or PubMed ID no. |
|---|---|---|---|
| Naf-1 | PBMC, MDM | AJ011896 | 11, 12 |
| RANTES | PBMC, MDM | M21121 | 35 |
| MIP-1α | PBMC | D90144 | 35 |
| MIP-1β | PBMC | J04130 | 35 |
| NFATc | PBMC, MDM | U08015 | 11, 28, 29 |
| IL8 | PBMC, MDM | M28130 | 33 |
| MGSA (Gro-α) | PBMC | X54489 | 34 |
| IL10 | PBMC | U16720 | 26 |
| IL1β | PBMC | M15330 | 26 |
| TAFII-28 | PBMC | X83928 | 11 |
| c-jun | PBMC | J04111 | 10027715 |
| c-myc | PBMC, MDM | V00568 | 11208609 |
| JKN2 | MDM | U09759 | 9045910, 9403476 |
| RNA polymerase II | MDM | L37127 | 11 |
| LcK | MDM | M36881 | 8887682, 7953537 |
| STAT2 | PBMC | U18671 | 15 |
| IFN-γ | MDM | L07633 | 26 |
| TGFβ | PBMC | X02812 | 26 |
| vav | PBMC | X16316 | 10394361, 9582304 |
| p53 | MDM | X02469 | 11313714 |
| sCD44 | PBMC | U94902 | 8345206 |
| 70Kd HSP1 | MDM | M11717 | 11, 11727548 |
| NFkB | MDM | M58603 | 15, 1380488 |
Genes determined by sam analysis to be induced by JR-FL gp120 that have previously been reported to be modulated by HIV infection and/or gp120 signaling are listed along with the associated GenBank accession no. and literature citation.
The Term Cytokine Ranked Highest Among Descriptive Categories Modulated by gp120.
To assess the potential of the ≈600 differentially expressed genes to influence the replication of HIV, we used a newly developed literature-mining algorithm (D.A.H. and R.A.L., unpublished work). This algorithm utilizes a set of descriptive categories that includes all of the descriptive terms in the Gene Ontology and Proteome At-A-Glance classification systems found in the Locus-Link reports (National Center for Biotechnology Information, www.ncbi.nlm.nih.gov/locuslink). We identified descriptive categories of genes that were over-represented in our list of differentially expressed genes, relative to all 8,443 annotated genes represented on the U95A microarray. In this manner, we generated an index with corresponding significance values. Envelope-induced genes encompassed 353 descriptive categories and among those categories the term cytokine achieved the highest degree of significance and was among the most frequently associated with gp120-modulated genes (Table 2). HIV replication is strongly influenced by cytokines in vivo (1), and thus this result is highly consistent with the hypothesis that envelope-mediated cell signaling plays a role in viral replication. Other relevant terms, including cell proliferation and viral replication, also occurred with a high degree of significance (Table 2).
Table 2.
Incidence of descriptive categories associated with genes modulated by gp120
| Descriptive term | P value |
|---|---|
| Cytokine | 8.6E-06 |
| Growth factor | 3.1E-05 |
| Ligand | 1.6E-04 |
| Inhibitor or repressor | 2.7E-04 |
| Chemokine | 3.5E-04 |
| Response to bacteria | 4.0E-04 |
| Cell motility | 5.3E-04 |
| Antiviral response protein | 1.3E-03 |
| Response to biotic stimulus | 1.3E-03 |
| Chemotaxis | 1.4E-03 |
| Stress response | 1.8E-03 |
| Cell migration/motility | 4.3E-03 |
| Defense response | 9.0E-03 |
| Viral replication | 9.7E-03 |
| Cell proliferation | 2.3E-02 |
A sam-derived list of genes modulated in both PBMCs and MDMs by gp120 was analyzed for significant association with descriptive categories that distinguish this list from the rest of the genes represented on the genechip. Descriptive categories were derived from Gene Ontology and Proteome At-A-Glance classification systems. Descriptive categories that were significantly over-represented in the list that includes all of the genes modulated by HIV envelope at all time points, were ranked by P value (one-tailed Fisher Exact test). Shown are the top ten over-represented terms and five other highly represented terms relevant to the theme of viral replication.
We observed increased transcription of twelve cytokine messages (Table 3). TNF-α, IL1-β, and IL3 all enhance HIV replication (26) and all were up-regulated in PBMCs. In MDMs, four cytokines that enhance HIV replication, TNF-α, IL6, IL15, and IFN-γ, were up-regulated. Only two cytokines that suppress viral replication, IL13 and IFN-β, were induced. This pattern of cytokine induction, although complex, favors viral replication.
Table 3.
Cytokines and chemokines induced by JR-FL gp120 in PBMCs and MDMs
| Accession no. | Cell type | HIV replication | Ref./PubMed ID no. | |
|---|---|---|---|---|
| Cytokine | ||||
| IL1β | M15330 | PBMC ↑ | ↑ | 26 |
| IL3 | M20137 | PBMC ↑ | ↑ | 26 |
| IL6 | X04430 | MDM ↑ | ↑ | 26 |
| IL10 | U16720 | PBMC ↑ | ↑↓ | 26 |
| IL13 precursor | U31120 | MDM ↑ | ↓ | 26 |
| IL15 precursor | AF031167 | MDM ↑ PBMC ↓ | ↑ | 9533656 |
| IFN β 2A | X04430 | MDM ↑ PBMC ↑ | ↓ | 26 |
| IFN γ | L07633 | MDM ↑ | ↑↓ | 26 |
| INF ω 1 | X58822 | MDM ↑ | ||
| TNF α | X02910 | MDM ↑ PBMC ↑ | ↑ | 26 |
| VEGF | X58822 | PBMC ↑ | ||
| VEGF B precursor | U48801 | PBMC ↓ | ||
| LIF (IL6 family) | X13967 | MDM ↑ PBMC ↓ | ↑ | 8207643 |
| Chemokine | ||||
| GRO-α | X54489 | PBMC ↑ | ↑ | 34 |
| GRO-γ | M36821 | PBMC ↑ | ||
| NAP2 | M54995 | MDM ↓ | ||
| IL8 | M28130 | MDM ↑ PBMC ↑ | ↑ | 33 |
| IP-10 | X02530 | PBMC ↓ | ||
| MIP-1β | J04130 | PBMC ↑ | ↑↓ | 35 |
| MIP-1α | D90144 | PBMC ↑ | ↑↓ | 35 |
| RANTES | M21121 | MDM ↑ PBMC ↑ | ↑↓ | 35 |
| MIP-3α | U64197 | PBMC ↑ |
Cytokines and chemokines that were differentially expressed in either PBMCs and/or MDMs at any individual time point as determined by sam analysis are listed along with their GenBank accession no. The cell type in which differential expression was detected is listed. Increased mRNA expression is denoted by ↑, and decreased mRNA expression by ↓. Effect on viral replication, based on published; literature is also listed by arrows.
Increased Expression of the Transcription Factor NFAT (Nuclear Factor of Activated T Cells).
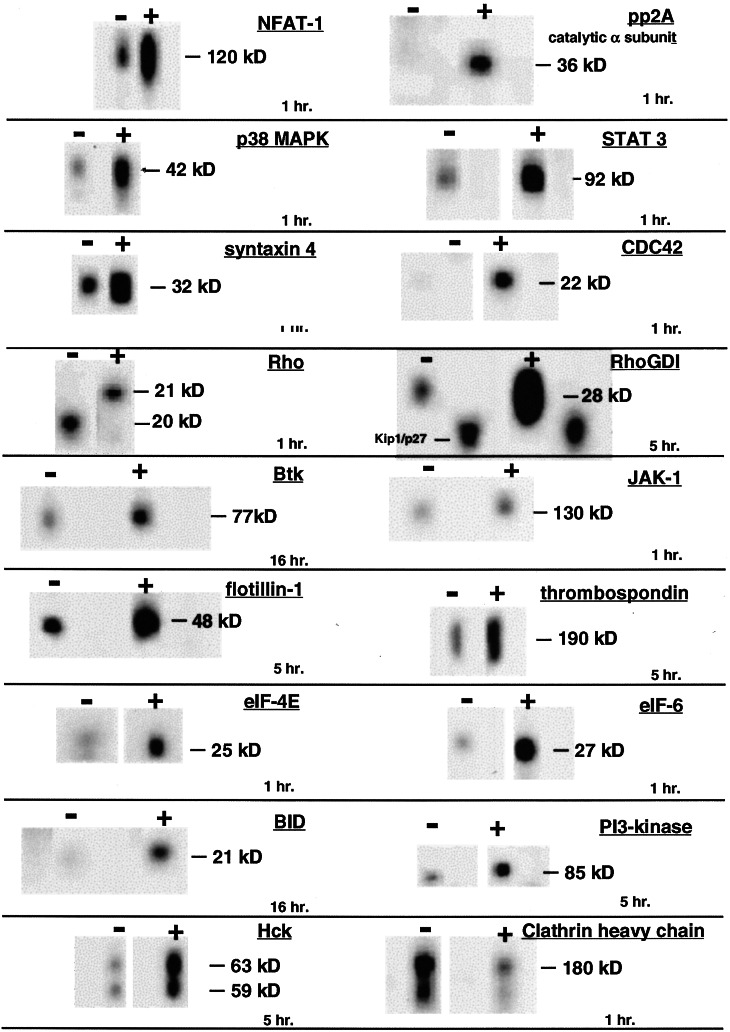
An increase in NFATc message was observed in both PBMCs and MDMs (Table 1). In addition, Western blot analysis demonstrated that gp120 induced a pronounced increase in NFAT-1 protein expression (Fig. 2). The NFAT family of transcription factors plays a central role in the transcriptional regulation of inflammatory cytokines and other genes central to immune responses (27). Two NFAT recognition elements reside in the HIV long terminal repeat (LTR) (28), and both NFAT-1 and NFATc induce the replication of HIV in primary cells (28–30). Furthermore, NFATc promotes the productive infection of HIV in the absence of CD4+ T cell proliferation (29). This occurs as a consequence of two activities: NFAT facilitates reverse-transcription of the HIV genome (29), and it strongly induces transcription from the HIV LTR (28). Because NFAT acts both directly on the HIV transcription and indirectly through increased cytokine expression, we regard its induction as particularly significant to HIV replication. Although this is the first report of gp120 inducing NFAT expression, it has previously been shown that NEF induces NFATc transcription (11). Interestingly, the α subunit of protein phosphatase 2A (PP2A), which activates NFAT (31), was also up-regulated (Fig. 2).
Figure 2.
Western blot analysis of gp120-treated PBMCs. Freshly isolated PBMCs were treated with JR-FL gp120 for 1, 5, or 16 h. Lysates were analyzed by Western blot analysis with a collection of ≈800 mAbs or sera (www.translab.com/TOC.shtml). Selected results are displayed.
Relevant Transcription Factors and Kinases Induced by HIV-1 Envelope.
Other transcription factors and protein kinases, including c-jun, JNK, MEK, p38 MAPK, STAT, and JAK, all of which participate in antigen-specific T cell activation (14, 15, 32), were induced (Figs. 1 and 2, Table 1). We observed increased transcription of two components of the transcription-elongation factor complex, TFII H and TFII D (Fig. 1, Table 1). We also observed a large increase in the transcription of RNA polymerase II (Fig. 1, Table 1). In addition to NFATc, NEF also induces RNA polymerase II, TFII D, and a third component of the transcription-elongation complex, CDK9 (11). Because envelope-mediated signaling occurs at a very early step in HIV infection of target cells, and NEF is among the first proteins expressed, we suggest that gp120 and NEF may act synergistically in driving transcription from the HIV LTR in suboptimally activated cells.
gp120 Induced Five Chemokines and Modulates Chemokine-Related Signal Transduction.
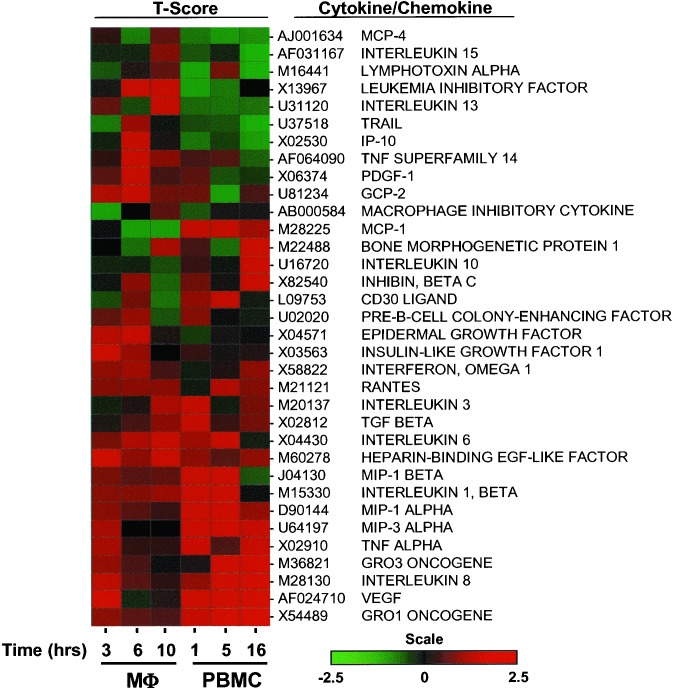
Five chemokines, IL8, GRO-α, GRO-γ, MIP1α, MIP1β, and RANTES, were up-regulated in response to gp120 (Figs. 1 and Fig. 3, Tables 1 and 3). IL8 and GRO-α enhance HIV replication (33, 34). GRO-γ is closely related to GRO-α and is also likely to enhance viral replication. Although MIP1α, MIP-1β, and RANTES are generally viewed as inhibitory chemokines, they only block entry of R5 viruses at very high concentrations (35). It is of note these same chemokines enhance viral replication at postentry steps of the viral life cycle (35–37). A common paradigm in gene expression analysis holds that genes that are regulated similarly function in a coordinated manner (38). In this regard, cluster analysis of genes included in the descriptive category “cytokine/chemokine” illustrate the simultaneous induction of Gro-α, IL-8, MIP-1α, and MIP1-β, which are all up-regulated within 5 h of treatment with envelope (Fig. 3).
Figure 3.
Hierarchical cluster analysis of the genes included in the category “Cytokine/Chemokine” that are induced by gp120. Genes included in this that were significantly differentially expressed in macrophages (three donors; Mφ) or PBMCs (four donors) following treatment with gp120 were clustered (spotfire software package, Spotfire Inc., Somerville, MA; hierarchical algorithm) according to t score values derived after performing 5,000 random permutations of the expression data by using a paired test comparing treated versus untreated samples (sam software package). Changes in gene expression relative to untreated PBMCs and MDMs are indicated by a color scale in which the color red indicates up-regulation of transcription and green indicates down-regulation. Changes in gene expression for three different time points in both MDMs and PBMCs are represented.
Intracellular factors related to chemokine signaling and cell trafficking were also induced. We observed a marked increase in the expression of both RhoGDI and Cdc42, and an apparent modification of Rho (Fig. 2). Rho and Cdc42 are members of the Rho family of small guanine nucleotide-binding proteins that participate in lymphocyte trafficking by controlling the formation of focal adhesion complexes (39). We and others have previously demonstrated that gp120 induces the phosphorylation of FAK (40) and Pyk-2 (18), two components of focal adhesion complexes. We suggest that gp120-mediated induction of chemokines has a net effect that favors viral replication directly through the stimulation of target cells and indirectly through the recruitment of target cells to sites of active viral replication.
gp120-Mediated Modulation of a Group of Genes Involved in Membrane Fusion.
Several genes associated with membrane fusion were induced by gp120. We found the increased expression of syntaxin-4 by gp120 within the first hour after treatment to be intriguing (Fig. 2). Induction of other syntaxin family members was observed at the level of mRNA. Syntaxin-7 was induced in PBMCs, whereas syntaxins 6 and 11 were induced in MDMs (data not shown). Syntaxins are members of a family of integral membrane proteins referred to as SNAREs (41). These proteins mediate membrane fusion in a compartment-specific manner. They play a central role in the fusion of intracellular vesicles with the plasma membrane (41). It has been suggested that syntaxins participate in the controlled exocytosis of cytokines from intracellular vesicles on T cell activation (42), a process consistent with gp120-induced cytokine and chemokine secretion. Of note, syntaxin-mediated membrane fusion is mechanistically similar to HIV-envelope-mediated fusion (43). Finally we note the induction of flotillin-1 (Fig. 2), a protein that is enriched in lipid rafts (44), which are membrane structures that facilitate fusion of HIV with plasma membranes of target cells (45).
Discussion
HIV-1 pathogenesis is regulated by a complex interaction between viral and cellular factors. A better understanding of these complex interactions will aid in the development of new classes of therapeutic agents designed to inhibit HIV-1 replication. We have demonstrated that CCR5-specific HIV gp120 induces the transcription and expression of factors that provide a conducive environment for HIV replication in resting or suboptimally activated PBMCs.
We used high-density oligonucleotide microarrays and high-throughput Western blotting to characterize the complex response of PBMCs and MDMs to gp120-mediated signaling. By microarray analysis, ≈300 genes were reproducibly modulated in both PBMCs and MDMs. This represents ≈3% of the total number of expressed messages detected by the Affymetrix U95A chip in our system. Changes in the expression of these genes may reflect both direct and indirect responses to envelope signaling. Presently we cannot distinguish between CD4-mediated versus CCR5-mediated signals.
The rapid induction of cytokines and chemokines at early time points is likely to initiate a cascade of responses. Using a literature-mining algorithm, we found genes related to cytokines and chemokines among the most highly represented groups of genes responding to gp120, an observation that underscores the potential impact that envelope-mediated signaling can impose on the transcriptional and translational programs of the human immune system. HIV replication in vivo is controlled by cytokines and chemokines, and the majority of those induced by gp120 enhance viral replication. In addition, gp120-induced dysregulation of cytokines and chemokines may contribute to immune dysfunction and HIV pathogenesis (1). Cytokines and chemokines that were modulated by gp120 were further analyzed by a cluster analysis program. By grouping genes that were similarly regulated in time, we observed the coordinated induction of a set of chemokines, all of which are reported to enhance viral replication. In addition, these chemokines may further enhance infection by recruiting lymphocytes to sites of viral replication
Intracellular factors including kinases and transcription factors associated with antigen-specific T cell activation were also induced. We observed an increase in the expression of NFATc and NFAT-1, factors that may be critically important in promoting viral replication. These observations support the hypothesis that signal transduction by the viral envelope contributes to the replication of HIV in resting or suboptimally activated cells. Of note, it has been suggested that such activity may contribute to the preferential transmission of R5 tropic HIV (32) and our data are consistent with this hypothesis. Interestingly, other viral gene products, notably NEF, also appear to promote the expression of factors favorable to viral replication (11). We suggest that these activities of gp120 and NEF increase the susceptibility of target cells to productive infection and may contribute to the low-level replication of HIV at sites set apart from activated T cells, thus avoiding immune surveillance. Replication in this manner may contribute to the establishment and maintenance of viral reservoirs, the persistence of which presents an obstacle to highly active antiretroviral therapy (HAART)-mediated eradication of HIV.
Acknowledgments
We thank Audrey Kinter for helpful discussions throughout the course of this study. We also thank Guy Baratte, Ann Mastradone, John Cadwell, and Jennifer Stevenson for technical assistance.
Abbreviations
- PBMCs
peripheral blood mononuclear cells
- MDMs
monocyte-derived macrophages
References
- 1.Fauci A S. Nature (London) 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 2.Eckstein D A, Penn M L, Korin Y D, Scripture-Adams D D, Zack J A, Kreisberg J F, Roederer M, Sherman M P, Chin P S, Goldsmith M A. Immunity. 2001;15:671–682. doi: 10.1016/s1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Z, Schuler T, Zupancic M, Wietgrefe S, Staskus K A, Reimann K A, Reinhart T A, Rogan M, Cavert W, Miller C J, et al. Science. 1999;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 4.Blaak H, van't Wout A B, Brouwer M, Hooibrink B, Hovenkamp E, Schuitemaker H. Proc Natl Acad Sci USA. 2000;97:1269–1274. doi: 10.1073/pnas.97.3.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostrowski M A, Chun T W, Justement S J, Motola I, Spinelli M A, Adelsberger J, Ehler L A, Mizell S B, Hallahan C W, Fauci A S. J Virol. 1999;73:6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zack J A, Cann A J, Lugo J P, Chen I S. Science. 1988;240:1026–1029. doi: 10.1126/science.2835813. [DOI] [PubMed] [Google Scholar]
- 7.Zack J A, Haislip A M, Krogstad P, Chen I S. J Virol. 1992;66:1717–1725. doi: 10.1128/jvi.66.3.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky M I, Haggerty S, Dempsey M P, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. Nature (London) 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbeil J, Sheeter D, Genini D, Rought S, Leoni L, Du P, Ferguson M, Masys D R, Welsh J B, Fink J L, et al. Genome Res. 2001;11:1198–1204. doi: 10.1101/gr.180201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiss G K, Bumgarner R E, An M C, Agy M B, van 't Wout A B, Hammersmark E, Carter V S, Upchurch D, Mullins J I, Katze M G. Virology. 2000;266:8–16. doi: 10.1006/viro.1999.0044. [DOI] [PubMed] [Google Scholar]
- 11.Simmons A, Aluvihare V, McMichael A. Immunity. 2001;14:763–777. doi: 10.1016/s1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 12.Arendt C W, Littman D R. Genome Biol. 2001;2:1030. doi: 10.1186/gb-2001-2-11-reviews1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kornfeld H, Cruikshank W W, Pyle S W, Berman J S, Center D M. Nature (London) 1988;335:445–448. doi: 10.1038/335445a0. [DOI] [PubMed] [Google Scholar]
- 14.Popik W, Pitha P M. Virology. 2000;276:1–6. doi: 10.1006/viro.2000.0581. [DOI] [PubMed] [Google Scholar]
- 15.Stantchev T S, Broder C C. Cytokine Growth Factor Rev. 2001;12:219–243. doi: 10.1016/s1359-6101(00)00033-2. [DOI] [PubMed] [Google Scholar]
- 16.Hivroz C, Mazerolles F, Soula M, Fagard R, Graton S, Meloche S, Sekaly R P, Fischer A. Eur J Immunol. 1993;23:600–607. doi: 10.1002/eji.1830230303. [DOI] [PubMed] [Google Scholar]
- 17.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Nature (London) 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 18.Davis C B, Dikic I, Unutmaz D, Hill C M, Arthos J, Siani M A, Thompson D A, Schlessinger J, Littman D R. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arthos J, Rubbert A, Rabin R L, Cicala C, Machado E, Wildt K, Hanbach M, Steenbeke T D, Swofford R, Farber J M, Fauci A S. J Virol. 2000;74:6418–6424. doi: 10.1128/jvi.74.14.6418-6424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oh S K, Cruikshank W W, Raina J, Blanchard G C, Adler W H, Walker J, Kornfeld H. J Acquired Immune Defic Syndr. 1992;5:251–256. [PubMed] [Google Scholar]
- 21.Jones M V, Bell J E, Nath A. AIDS. 2000;14:2709–2713. doi: 10.1097/00002030-200012010-00010. [DOI] [PubMed] [Google Scholar]
- 22.Del Corno M, Liu Q H, Schols D, de Clercq E, Gessani S, Freedman B D, Collman R G. Blood. 2001;98:2909–2916. doi: 10.1182/blood.v98.10.2909. [DOI] [PubMed] [Google Scholar]
- 23.Koyanagi Y, Miles S, Mitsuyasu R T, Merrill J E, Vinters H V, Chen I S. Science. 1987;236:819–822. doi: 10.1126/science.3646751. [DOI] [PubMed] [Google Scholar]
- 24.Mossman S P, Bex F, Berglund P, Arthos J, O'Neil S P, Riley D, Maul D H, Bruck C, Momin P, Burny A, et al. J Virol. 1996;70:1953–1960. doi: 10.1128/jvi.70.3.1953-1960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tusher V G, Tibshirani R, Chu G. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal B B, Puri R K. Human Cytokines: Their Role in Disease and Therapy. Cambridge, MA: Blackwell Science; 1998. [Google Scholar]
- 27.Rao A, Luo C, Hogan P G. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 28.Cron R Q, Bartz S R, Clausell A, Bort S J, Klebanoff S J, Lewis D B. Clin Immunol. 2000;94:179–191. doi: 10.1006/clim.1999.4831. [DOI] [PubMed] [Google Scholar]
- 29.Kinoshita S, Chen B K, Kaneshima H, Nolan G P. Cell. 1998;95:595–604. doi: 10.1016/s0092-8674(00)81630-x. [DOI] [PubMed] [Google Scholar]
- 30.Bassuk A G, Anandappa R T, Leiden J M. J Virol. 1997;71:3563–3573. doi: 10.1128/jvi.71.5.3563-3573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Handschumacher R E. Biochem Biophys Res Commun. 1996;219:96–99. doi: 10.1006/bbrc.1996.0187. [DOI] [PubMed] [Google Scholar]
- 32.Popik W, Pitha P M. J Virol. 2000;74:2558–2566. doi: 10.1128/jvi.74.6.2558-2566.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane B R, Lore K, Bock P J, Andersson J, Coffey M J, Strieter R M, Markovitz D M. J Virol. 2001;75:8195–8202. doi: 10.1128/JVI.75.17.8195-8202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lane B R, Strieter R M, Coffey M J, Markovitz D M. J Virol. 2001;75:5812–5822. doi: 10.1128/JVI.75.13.5812-5822.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kinter A, Arthos J, Cicala C, Fauci A S. Immunol Rev. 2000;177:88–98. doi: 10.1034/j.1600-065x.2000.17708.x. [DOI] [PubMed] [Google Scholar]
- 36.Kelly M D, Naif H M, Adams S L, Cunningham A L, Lloyd A R. J Immunol. 1998;160:3091–3095. [PubMed] [Google Scholar]
- 37.Kinter A L, Ostrowski M, Goletti D, Oliva A, Weissman D, Gantt K, Hardy E, Jackson R, Ehler L, Fauci A S. Proc Natl Acad Sci USA. 1996;93:14076–14081. doi: 10.1073/pnas.93.24.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quackenbush J. Nat Rev Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 39.Parsons J T, Martin K H, Slack J K, Taylor J M, Weed S A. Oncogene. 2000;19:5606–5613. doi: 10.1038/sj.onc.1203877. [DOI] [PubMed] [Google Scholar]
- 40.Cicala C, Arthos J, Ruiz M, Vaccarezza M, Rubbert A, Riva A, Wildt K, Cohen O, Fauci A S. J Immunol. 1999;163:420–426. [PubMed] [Google Scholar]
- 41.Gerst J E. Cell Mol Life Sci. 1999;55:707–734. doi: 10.1007/s000180050328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shukla A, Berglund L, Nielsen L P, Nielsen S, Hoffmann H J, Dahl R. Respir Med. 2001;95:773–780. doi: 10.1053/rmed.2001.1167. [DOI] [PubMed] [Google Scholar]
- 43.Bentz J, Mittal A. Cell Biol Int. 2000;24:819–838. doi: 10.1006/cbir.2000.0632. [DOI] [PubMed] [Google Scholar]
- 44.Dermine J F, Duclos S, Garin J, St-Louis F, Rea S, Parton R G, Desjardins M. J Biol Chem. 2001;276:18507–18512. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
- 45.Liao Z, Cimakasky L M, Hampton R, Nguyen D H, Hildreth J E. AIDS Res Hum Retroviruses. 2001;17:1009–1019. doi: 10.1089/088922201300343690. [DOI] [PubMed] [Google Scholar]