Abstract
Specificity in the cellular immune system is controlled and regulated by the T cell antigen receptor (TCR), which specifically recognizes peptide/major histocompatibility complex (MHC) molecules. In recent years many cancer-associated MHC-restricted peptides have been isolated and because of their highly restricted fine specificity, they are desirable targets for novel approaches in immunotherapy. Antibodies that would recognize tumor-associated MHC–peptide complexes with the same specificity as the TCR would be valuable reagents for studying antigen presentation by tumor cells, for visualizing MHC–peptide complexes on cells, and eventually for monitoring the expression of specific complexes during immunotherapy. To generate molecules with such a unique fine specificity, we selected a large nonimmune repertoire of phage Fab antibodies on recombinant HLA-A2 complexed with three common antigenic T cell, HLA-A2-restricted epitopes derived from the melanoma differentiation antigen gp100. We were able to isolate a surprisingly large panel of human recombinant Fab antibodies that exhibit a characteristic TCR-like binding specificity to each of the three gp100-derived epitopes, yet unlike TCRs, they did so with an affinity in the nanomolar range. These TCR-like antibodies recognize the native MHC–peptide complex expressed on the surface of antigen-presenting cells. Moreover, they can detect the specific MHC–peptide complexes on the surface of melanoma tumor cells. These results demonstrate the ability to isolate high-affinity human recombinant antibodies with the antigen-specific, MHC-restricted specificity of T cells, and this ability was demonstrated for three different epitopes of the same melanoma-derived antigen.
In recent years, major advances in tumor immunology have led to an increased understanding of the immune responses against tumors. Especially with melanoma, it is now well established that human melanoma and other tumor cells express antigens that are recognized by cytotoxic T lymphocytes (CTLs) derived from cancer patients (1–3). The cascade of molecular recognition events associated with these tumor-associated immune responses involves the expression of specific peptides in complex with MHC class I molecules on the cancer cells (1–3). For example, human melanomas express tumor-associated peptides that are presented to the immune system in a complex with class I HLA-A2 molecules (4, 5). Although there is strong experimental evidence demonstrating the presence of these antigens on a variety of tumors, they are apparently unable to elicit a strong enough anti-tumor immune response (6).
Therefore, many modern cancer immunotherapy approaches are now designed to induce and enhance T cell reactivity against these tumor antigens (7–11). Tumor-specific MHC–peptide complexes present on the surface of tumor cells may also offer a unique and specific target for an antibody-based therapeutic approach. To develop such a strategy, targeting moieties such as recombinant antibodies that will specifically recognize peptide–MHC complexes must be isolated.
The recent advent of MHC–peptide tetramers has provided a new tool for studying antigen-specific T cell populations in health and disease, even when they are very rare, by monitoring tetramer–T cell binding by means of flow cytometry (12–14). However, to date, there are very few tools available to detect, visualize, count, and study antigen (MHC–peptide) presentation. Indeed, several studies demonstrated that the inability of the patient's immune system to elicit an effective immune response against the tumor is often due to poor antigen presentation (8, 9). Antibodies with T cell antigen receptor (TCR)-like specificity could enable measuring the antigen presentation capabilities of such tumor- or antigen-presenting cells (APCs)—for example, by direct visualization of the specific MHC–peptide complex on the cell surface. Attempts to use recombinant soluble TCRs for this purpose have largely failed because of their inherent low affinity for their target as well as their instability as recombinant-engineered molecules (15). Therefore, TCR-like antibodies would serve as a valuable tool to obtain precise information about the presence, expression pattern, and distribution of the MHC–peptide complex, on the tumor cell surface, on tumor metastases, in lymphoid organs, and on professional APCs.
Antibodies that specifically recognize class I MHC–peptide complexes have already been used in murine systems to study antigen presentation, to localize and quantify APCs displaying a T cell epitope, or as a targeting tool (16–25). Here, we have isolated a large panel of high-affinity human recombinant Fab antibodies endowed with the antigen-specific, MHC-restricted specificity of T cells. These antibodies recognize three common HLA-A2-restricted epitopes of the human melanoma differentiation antigen gp100.
We show that this panel of antibodies recognizes HLA-A2 molecules only when displaying the specific peptide against which they were selected; they do not bind HLA-A2 molecules complexed with other gp100-derived epitopes or with other HLA-A2-restricted control peptides. Hence, they exhibit a TCR-like restriction. Moreover, these antibodies have been used to directly visualize the specific HLA-A2/gp100 epitopes on APCs as well as on the surface of melanoma tumor cells by flow cytometry.
Materials and Methods
Production of Biotinylated scMHC–Peptide Complexes.
scMHC–peptide complexes were produced by in vitro refolding of inclusion bodies produced in Escherichia coli as described (26). Correctly folded MHC–peptide complexes were isolated and purified by anion-exchange Q-Sepharose chromatography (Amersham Pharmacia) and subjected to biotinylation by the BirA ligase as described (26).
Selection of Phage-Antibodies on Biotinylated Complexes.
A large human Fab library containing 3.7 × 1010 different Fab clones was used for the selection (27). Selection on biotinylated complexes was performed as described (28). Briefly, phages were subsequently incubated for 1 h with decreasing amounts of biotinylated scMHC–peptide complexes (500 nM for the first round and 100 nM for the following rounds). Streptavidin magnetic beads were added, and the mixture was incubated for 15 min with continuous rotation. A magnetic force was applied to pull down phages bound to biotinylated complexes. Phages were eluted by 1 ml of triethylamine (100 mM). The elution mixture was neutralized by the addition of 100 μl of Tris⋅HCl (1 M, pH 7.4) and used to infect E. coli TG1 cells (OD600 = 0.5) for 30 min at 37°C.
Expression and Purification of Soluble Recombinant Fab Antibodies.
Soluble Fabs were purified from the periplasmic fraction of BL21 cells by using a hexahistidine tag fused to the CH1 domain of the Fabs as described (27). The homogeneity and purity of the purified Fabs was determined by analysis on nonreduced and reduced SDS/PAGE.
Results
Recombinant Single-Chain MHC–Peptide Complexes with Three Melanoma-Derived gp100, HLA-A2-Restricted Peptides.
gp100 is a melanocyte lineage-specific membrane glycoprotein consisting of 661 aa that is expressed in most melanoma cells (29). This protein is recognized by many HLA-A2-restricted melanoma-reactive tumor-infiltrating lymphocytes (TILs) that have been isolated from melanoma patients (29, 30). Five T cell epitopes have been identified in gp100; three of them are common immunogenic epitopes recognized by CTLs derived from different patients (31, 32): G9-154 (KTWGQYWQV), G9-209 (ITDQVPFSV), and G9-280 (YLEPGPVTA). Recombinant MHC–peptide complexes that display the three gp100-derived epitopes were generated by using a scMHC construct that has been described (26, 33). In this construct, the extracellular domains of HLA-A2 are connected into a single-chain molecule with β2-microglobulin by using a 15-amino acid flexible linker. The scMHC–peptide complexes were produced by in vitro refolding of inclusion bodies, from bacterial cultures transformed with the scMHC construct, in the presence of each of the three gp100-derived peptides. Recombinant scMHC–peptide complexes generated by this strategy have been previously characterized in detail for their biochemical, biophysical, and biological properties and were found to be functional (26, 33).
Selection of Recombinant Antibodies with TCR-Like Specificity to Three Common T Cell Epitopes of the Melanoma Antigen gp100.
To enable efficient selection, scMHC–peptide complexes were biotinylated by using a BirA sequence tag that was engineered at the C terminus of the HLA-A2 gene for site-specific biotinylation as previously described (12, 26). A large naive repertoire of 3.7 × 1010 human recombinant Fab fragments (27), which was first depleted of anti-streptavidin antibodies, was used for the subsequent panning in solution on soluble recombinant scMHC–peptide complexes containing each of the three gp100-derived T cell epitopes. A 1,000- to 2,500-fold enrichment in phage titer was observed after three rounds of panning by using each of the three different gp100-derived peptide-MHC complexes (Table 1).
Table 1.
Selection of recombinant Fab antibodies with TCR-like specificity
| Cycle no. | Phage input (I)* | Phage output (O)* | Ratio (O/I) | Enrichment | Pan-MHC binders† | TCR-like binders‡ | No. of patterns§ |
|---|---|---|---|---|---|---|---|
| G9-209/HLA-A2 | |||||||
| 1 | 6 × 1013 | 4 × 106 | 7 × 10−8 | — | |||
| 2 | 9 × 1012 | 6 × 107 | 4 × 10−6 | 15 | 24/94 (25%) | 6 /94 (6%) | 1 |
| 3 | 6 × 1012 | 1 × 1010 | 2 × 10−3 | 2,500 | 62/94 (66%) | 20 /94 (21%) | 4 |
| G9-280/HLA-A2 | |||||||
| 1 | 6 × 1013 | 6 × 106 | 1 × 10−7 | — | |||
| 2 | 6 × 1012 | 2 × 107 | 3 × 10−6 | 3 | 16/94 (17%) | 9 /94 (9%) | 2 |
| 3 | 1 × 1013 | 8 × 109 | 8 × 10−4 | 1,300 | 63/94 (67%) | 15 /94 (16%) | 3 |
| G9-154/HLA-A2 | |||||||
| 1 | 7 × 1012 | 1 × 106 | 1 × 10−7 | — | |||
| 2 | 5 × 1013 | 8 × 106 | 2 × 10−7 | 8 | 14/90 (16%) | 4 /90 (4%) | 4 |
| 3 | 7 × 1013 | 3 × 109 | 4 × 10−5 | 3,000 | 72/90 (80%) | 24 /90 (27%) | 10 |
Phage colony-forming units determined by titration on E. coli TG1 cells before and after each round of selection.
Fabs that bind as phage-antibody to scMHC-HLA-A2 complexed to at least two different peptides.
Fabs that react only with specific scMHC-HLA-A2/peptide complex to which they were selected.
The number of different TCR-like Fabs determined by DNA fingerprint analysis.
An ELISA with phage particles was performed to determine phage specificity on biotinylated recombinant scMHC–peptide complexes immobilized to BSA-biotin-streptavidin-coated immunoplates. About 70–90% of randomly selected phages from the third round of panning on each complex reacted with the corresponding MHC–peptide complex (Table 1). The precise specificity of the selected phage antibodies was determined by a differential ELISA on wells coated with scMHC-HLA-A2 complexes containing either the specific gp100-derived peptide or control HLA-A2-restricted peptides including the two other gp100-derived epitopes (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org).
Two types of Fab phage clones were observed after these specificity assays. One type bound to the HLA-A2/peptide complex without peptide specificity (“Pan-MHC binders” in Table 1) and the second bound to the HLA-A2 complex in a peptide-specific manner (termed as “TCR-like binders” in Table 1). We assayed these specific clones and revealed the following specificity results: for the G9-154 epitope, 24 of the 90 clones analyzed (27%) reacted specifically with the HLA-A2-G9-154 complex but not with complexes containing the gp100-derived peptides G9-280, G9-209 (data not shown), nor with human T lymphotrophic virus type 1 (HTLV-1) TAX or melanoma MART1-containing scMHC complexes (Table 1; see Fig. 5A). The diversity within the selected TCR-like Fabs was assessed by DNA fingerprinting analysis; 10 different antibodies with TCR-like specificity were selected. For the G9-209 epitope, 20 of the 94 clones analyzed (21%) reacted specifically with the HLA-A2-G9-209 complex but not with control complexes (Table 1; see Fig. 5B). These positive clones yielded four different fingerprint patterns. Finally, the panning on HLA-A2 complexes containing the G9-280 epitope resulted in 15 of the 94 specific peptide-restricted clones (16%) (Table 1; see Fig. 5C), exhibiting three different fingerprint patterns. This unexpectedly high frequency of these TCR-like binders, representing 16–27% of the phage clones binding to the MHC–peptide complex (Table 1), is very surprising indeed. Further, for all three HLA-A2–gp100 peptide complexes screened, we isolated several of such Fab antibodies displaying a TCR-like binding pattern, and in all three cases, one particular clone dominated the population after three rounds of selection (at a frequency of 30–50%).
Characterization of Recombinant Soluble Fab Antibodies with TCR-Like Specificity.
We have selected two to four Fab clones for each HLA-A2–gp100-derived complex that exhibited the most specific peptide-dependent and TCR-like binding pattern as analyzed by the phage ELISAs. These Fab fragments were produced in a soluble form in E. coli TG1 or BL21 cells and were purified by immobilized metal ion affinity chromatography (IMAC). Yields were 2–4 mg of pure material from 1 liter of bacterial culture. SDS/PAGE analysis revealed a homogenous and pure population of Fabs with the expected molecular size.
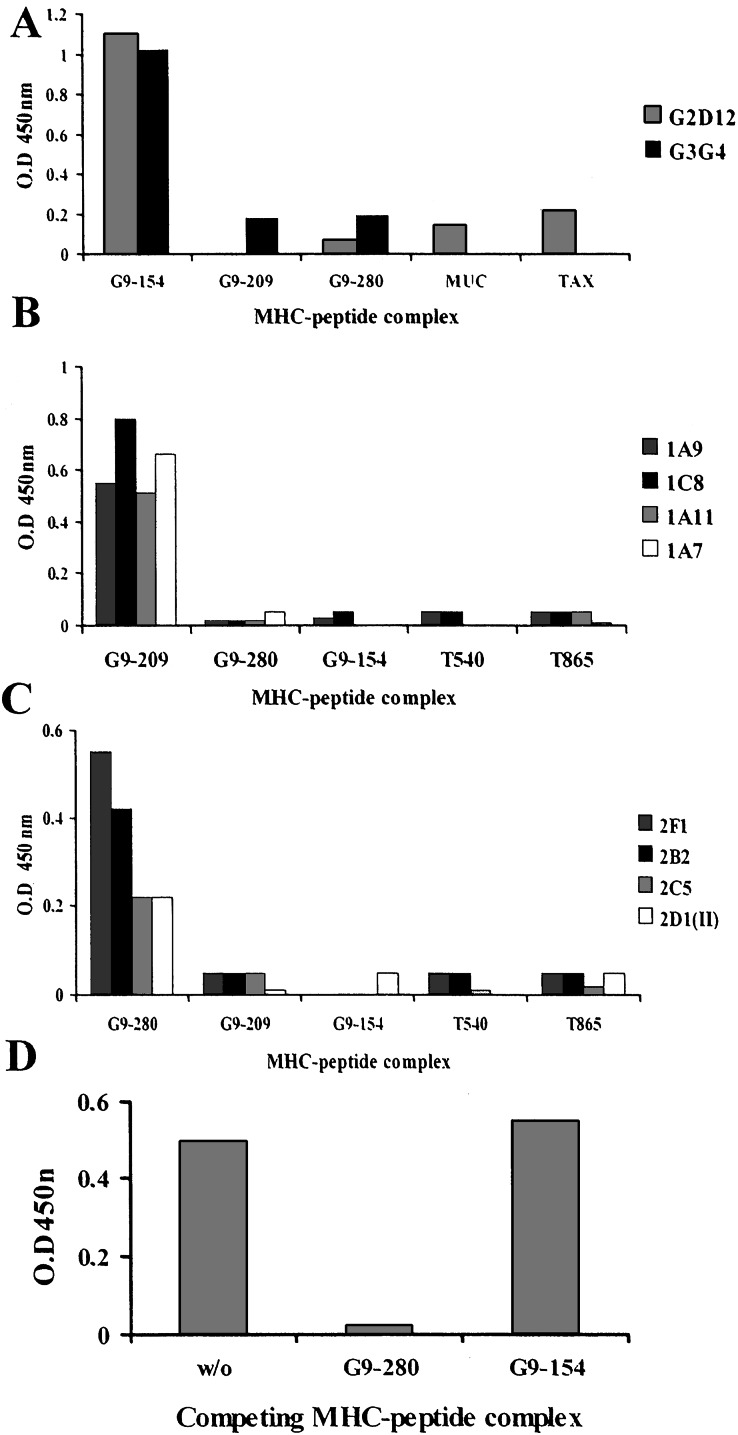
The binding specificity of these purified Fab fragments was determined by ELISAs on biotinylated MHC–peptide complexes immobilized to wells through BSA-biotin-streptavidin. The correct folding of the bound complexes and their stability during the binding assays were determined by their ability to react with the conformational specific monoclonal antibody W6/32, which binds HLA complexes only when folded correctly and when it contains peptide (data not shown). When we used soluble purified Fabs, these ELISAs revealed a very specific recognition pattern (Fig. 1). Two Fab clones, G2D12 and G3G4, selected to bind the G9-154 HLA-A2 complex, bound only to the specific complex but not to complexes displaying the G9-209 or G9-280 peptides, nor to HLA-A2 complexes containing a MUC1-derived peptide or the HTLV-1-derived TAX peptide (Fig. 1A).
Figure 1.
Binding in ELISA of soluble purified Fabs to recombinant scMHC-HLA-A2-peptide complexes. Binding of soluble purified Fab clones specific for the gp100-derived epitopes G9-154 (A), G9-209 (B), and G9-280 (C) to immobilized scMHC-HLA-A2-peptide complexes is indicated. Shown are the specificities of several Fab clones to the gp100-derived epitopes for which they were selected, but not to the indicated control MHC–peptide complexes containing other gp100 and telomerase-derived HLA-A2-restricted epitopes. (D) Competition of MHC–peptide complexes for binding of Fab 2F1 to immobolized G9-280–HLA-A2 complexes; W/O, without competitior.
Fab clones specific for the G9-209 HLA-A2 complex recognized only this complex, but not the two other gp100-derived peptides in the same context, nor the two telomerase-derived HLA-A2 complexes (Fig. 1B). Finally, the HLA-A2-G9-280-specific Fab clones recognized only their G9-280-derived complexes and no other MHC–peptide complexes (Fig. 1C). The Fab antibodies did not recognize any of five to seven other HLA-A2-peptide complexes, the peptide alone, empty HLA-A2 molecules (which are difficult to produce because they are unstable in the absence of a peptide), and neither streptavidin nor other protein antigens (data not shown). Thus, these antigen-specific Fab fragments exhibited binding characteristics and the fine specificity of a TCR-like molecule.
The ELISA binding specificity results were confirmed in competition experiments, in which excess specific and control soluble scMHC–peptide complexes were present in solution and competed for binding to the immobilized complex. As expected, competition was observed with the specific soluble MHC–peptide complex but not with control complexes. An example for this type of assay is shown in Fig. 1D, in which soluble G9-280-containing HLA-A2 but not G9-154/HLA-A2 complexes in solution competed and inhibited the binding of Fab 2F1 to the immobilized G9-280/HLA-A2 complexes.
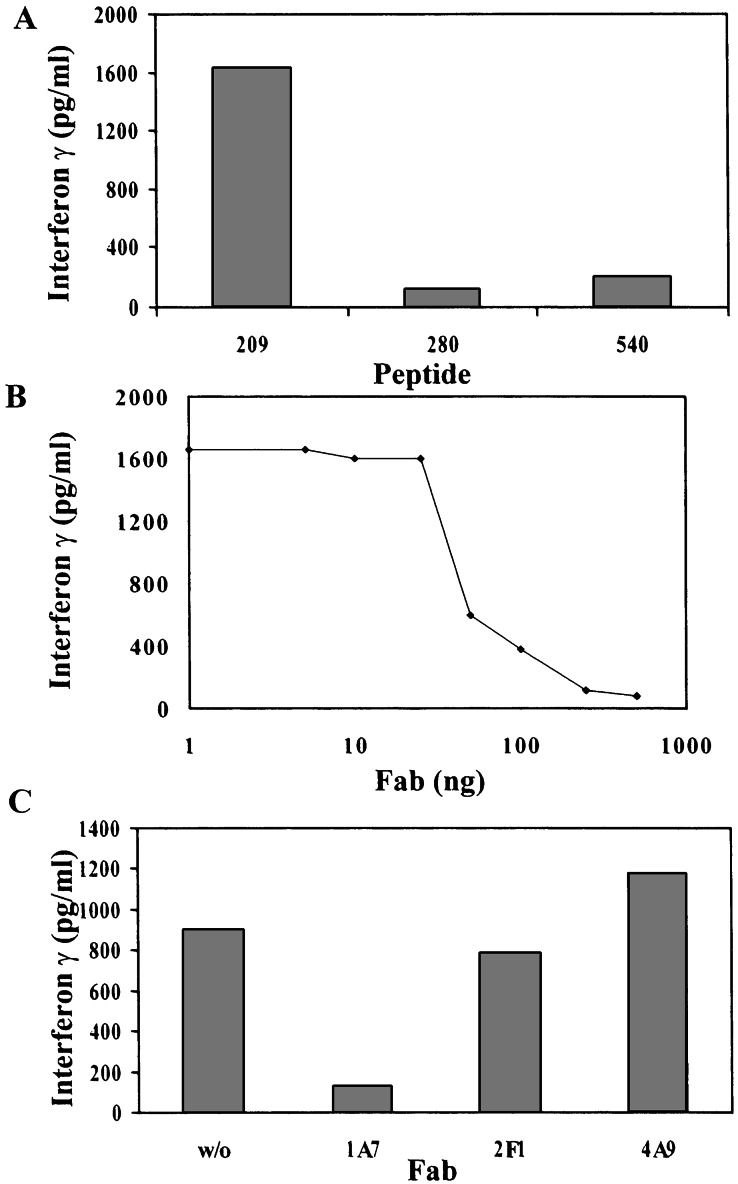
Further proof for the specificity of the TCR-like Fab antibodies isolated in this study was obtained in T cell stimulation/inhibition assays (Fig. 2). The HLA-A2-restricted, G9-209-specific CTL clone R6C12 was stimulated in the presence of APCs loaded with the G9-209 peptide but not with control HLA-A2-restricted peptides (Fig. 2A). In the inhibition assays the G9-209-specific Fab 1A7 was able to inhibit the release of IFN-γ from G9-209-specific CTL, R6C12, whereas a control G9-280-specific or telomerase-specific Fab did not inhibit peptide-specific CTL stimulation (Fig. 2 B and C).
Figure 2.
Specific inhibition of peptide-specific, MHC-restricted T cell activation with TCR-like Fab. T2 cells were pulsed with peptide as indicated and incubated with the G9-209-specific HLA-A-restricted CTL clone R6C12 in the presence of various concentrations of Fab 1A7 or control Fabs as indicated. T cell stimulation was measured by the release of IFN-γ to the culture medium. IFN-γ was determined by a double sandwich ELISA. (A) Stimulation of R6C12 CTLs with the G9-209 peptide but not control peptides. (B) Inhibition of T cell response with Fab 1A7 (C) Inhibition of T cell response is specific to Fab 1A7 but not to control Fabs 2F1 or 4A9.
Next, the affinity binding properties of the TCR-like soluble Fabs were determined by using a competition binding assay in which binding of 125I-labeled Fab competed with increasing concentrations of unlabeled Fab fragment. The apparent binding affinity of three Fabs, each of them specific for one of the three gp100-derived T cell epitopes, was measured to be 15 to 30 nM (see Fig. 6, which is published as supporting information on the PNAS web site). These results underscore our success in isolating high-affinity Fab antibodies with TCR-like specificity from a large nonimmune phage-displayed repertoire of antibodies.
Binding of Fab Antibodies to APCs Displaying the gp100-Derived Epitopes.
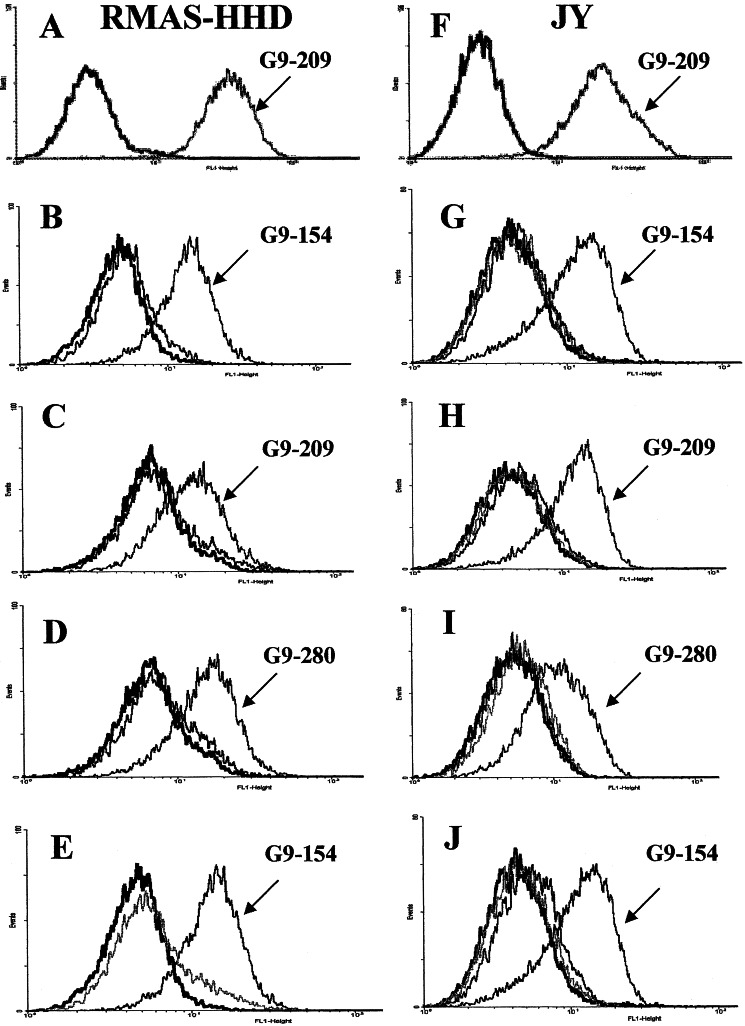
To demonstrate that the isolated soluble Fab antibodies can bind the specific MHC–peptide complex not only in its recombinant soluble form but also in the native form as expressed on the cell surface, we used two APC systems. The first consists of the murine TAP2-deficient RMA-S cells transfected with the human HLA-A2 gene in a single-chain form (HLA-A2.1/Db-β2-microglobulin single chain) (RMA-S-HHD cells). gp100-derived or control peptides were loaded on the RMA-S-HHD cells and the ability of the selected Fab antibodies to bind to peptide-loaded cells was monitored by fluorescence-activated cell sorter (FACS). Peptide-induced MHC stabilization of the TAP2 mutant RMA-S-HHD cells was determined by analyzing the reactivity of anti-HLA-A2 monoclonal antibody BB7.2 with peptide-loaded and unloaded cells by using a fluorescence-activated cell sorter (FACS) (Fig. 3A). Fab G2D12, which recognized the G9-154-containing HLA-A2 complex, reacted with RMA-S-HHD cells loaded with the G9154 peptide but not with cells loaded with the G9-280 peptide (Fig. 3B) or control cells not loaded with peptide. Similarly the G9-209-HLA-A2-specific Fab antibody 1A7 recognized RMA-S-HHD cells loaded with G9-209 peptide but not at all cells loaded with G9-154 peptide (Fig. 3C). Similar results were observed in FACS analysis of the G9-280-specific Fab antibody 2F1 (Fig. 3D). The Fab antibodies were analyzed on RMAS-HHD cells loaded with five different control HLA-A2-restricted peptides including cross-reaction studies among the gp100-derived peptides, and similar specificity results were observed. Moreover, RMAS-HHD cells loaded with the G9-154 epitope reacted with Fab G2D12 directed toward the G9-154-containing complex but not with Fabs 1A7 and 2F1 recognizing HLA-A2 in complex with the G9-209 or G9-280 epitopes, respectively (Fig. 3E).
Figure 3.
Binding of Fab antibodies to APCs. RMAS-HHD or JY cells were loaded with the indicated HLA-A2-restricted peptides. Peptide-loaded cells were then incubated with the specific soluble purified Fab antibodies as shown. Detection of binding was with FITC-labeled anti-human Fab. (A) RMAS-HHD cells loaded with the G9-209 peptide and control unloaded cells were stained with the anti-HLA-A2 antibody BB7.2 to demonstrate the stabilization/expression of HLA-A2 complexes on the surface of peptide-loaded but not on peptide-unloaded cells. (B) RMAS-HHD cells were loaded with the G9-154 (marked) and control G9-280 peptides or control unloaded cells. Cells were stained with the G9-154-specific Fab G2D12. (C) RMAS-HHD cells were loaded with peptides G9-209-specific (marked) and G9-154 (control). Loaded and unloaded cells were stained with the G9-209-specific Fab 1A7. (D) RMAS-HHD cells were loaded with G9-280-specific (marked) and G9-209 (contol) peptides. Loaded and unloaded cells were stained with the G9-280-specific Fab 2F1. (E) RMAS-HHD cells were loaded with peptide G9-154 and incubated with Fabs G2D12 (marked), 1A7, and 2F1 specific for G9-154, G9-209, and G9-280, respectively. (F) JY cells were loaded with peptides G9-209 and stained with anti-HLA-A2 BB7.2 antibody. Controls are cells incubated with secondary anti-mouse-FITC antibody. (G) JY cells were loaded with peptides G9-154 (marked), G9-209, G9-280, or unloaded and then reacted with the G9-154/HLA-A-specific Fab G2D12. (H) JY cells were loaded with peptides G9-209 (marked), G9-154, G9-280, or unloaded and then reacted with the G9-209/HLA-A-specific Fab 1A7. (I) JY cells were loaded with peptides G9-280 (marked), G9-154, G9-209, or unloaded and then reacted with the G9-280/HLA-A-specific Fab 2F1. (J) JY cells were loaded with the peptide G9-154 and incubated with Fabs G2D12 (marked), 1A7, and 2F1 specific for G9-154, G9-209, and G9-280 in complex with HLA-A2, respectively. Control unloaded cells are represented by black traces.
The second type of APCs tested were Epstein–Barr virus (EBV)-transformed B lymphoblast JY cells, which express HLA-A2, and were incubated with the gp100-derived or control peptides. These cells express TAP (transporter associated with antigen processing), and consequently, displaying the exogenous peptide is facilitated by peptide exchange. By using this strategy, we obtained a mixture of exogenously and endogenously derived peptides presented on HLA-A2 that are displayed on the cell surface. In testing the HLA-A2/gp100-specific antibodies 1A7, 2F1, and G2D12, we found intensive staining of JY cells loaded with the specific gp100-derived peptide for which they were selected, but no binding was observed when other gp100 or control peptides were used (Fig. 3 G–I). Control antibodies recognizing a telomerase-derived peptide in complex with scMHC-HLA-A2 did not bind to the gp100-derived peptide-loaded JY cells (Fig. 3J). Furthermore, no binding was observed when these antibodies were incubated with an HLA-A2/EBVB cell line loaded with the gp100 or control peptides.
These results show that the Fab antibodies exhibit TCR-like fine specificity and can specifically recognize their corresponding native HLA-A2 complexes in situ on the surface of cells.
Binding of gp100-Specific TCR-Like Fab Antibodies to Melanoma Cells.
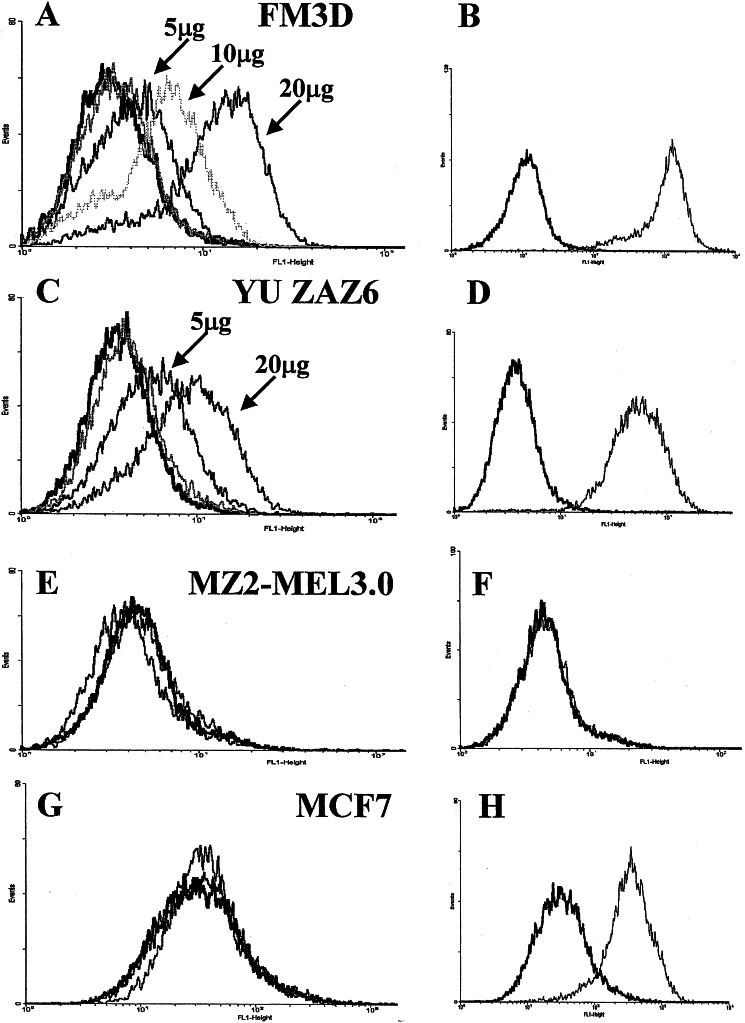
To explore whether these TCR-like Fab antibodies would bind endogenously derived MHC–peptide complexes and therefore may eventually be used to visualize these complexes on the surface of tumor cells, we performed flow cytometry analysis on HLA-A2+ melanoma tumor cell lines (Fig. 4). These cells represent the normal situation in which MHC–peptide complexes are expected to be present at a much lower density on the cell surface compared with the peptide-loaded RMAS-HHD or JY cells used above. The G9-154-specific Fab antibody G2D12 reacted very intensely in a dose-dependent manner with the HLA-A2+ gp100+ melanoma FM3D (Fig. 4 A and B) and YU ZAZ6 cells (Fig. 4 C and D), but not with the HLA-A2-melanoma MZ2-MEL3.0 cells (Fig. 4 E and F) or the HLA-A2+ breast carcinoma tumor cell line MCF7 (Fig. 4 G and H). Anti-HLA-A2 antibody BB7.2 was used to confirm HLA type expression (in addition to genomic PCR using HLA-A2-specific primers, data not shown). A control Fab antibody specific for the HTLV-1-derived HLA-A2–TAX complex did not bind to either cell line (Fig. 4 A, C, E, and G). These results demonstrate that, although the Fab antibodies are in a monovalent form, their high affinity enables efficient detection and visualization of MHC–peptide complexes on the surface of tumor cells. Hence, these TCR-like antibodies can bind to cells that express the specific MHC–peptide complex at a density most likely to be found on gp100-expressing tumor cells, APCs, and dendritic and other lymphoid cells involved in tumor antigen presentation to the immune system. Fab antibodies 1A7 and 2F1 specific to the G9-209 or G9-280 gp100-derived epitopes, respectively, also reacted with FM3D cells but with a lower intensity (data not shown). This finding may reflect differential expression of gp100-derived epitopes known as the antigenic variation phenomenon. Indeed, FM3D cells were shown to express high levels of the G9-154 epitope in comparison with the two other epitopes as revealed by their relative sensitivity to CTLs specific to the different gp100-derived epitopes in killing assays (35).
Figure 4.
Detection of MHC–peptide complexes on the surface of tumor cells. Melanoma FM3D (A) and YU ZAZ6 (C), which express HLA-A2 (B and D), as determined by reactivity with monoclonal antibody BB7.2, were stained with 5, 10, or 20 μg of Fab G2D12 specific for the melanoma gp100-derived G9-154 epitope, or with a Fab TCR-like antibody specific for the viral epitope TAX. Detection of binding was with FITC-labeled anti-human Fab. The melanoma HLA-A2- MZ2-MEL3.0 cells were not stained with G2D12 (E) or BB7.2 (F) (indication for HLA-A2−). MCF7 HLA-A2+ breast carcinoma cells were stained with BB7.2 (H) but not with Fab G2D12 or the TAX-specific Fab (G). Black traces represent cells incubated with the secondary FITC-labeled antibody.
Discussion
In this study we have demonstrated the ability to select, from a large nonimmune repertoire of human Fab fragments, a panel of antibodies directed to several T cell epitopes within a single cancer antigen, the melanoma-associated antigen gp100.
These antibodies exhibit a very specific and special binding pattern; they can bind with a peptide-specific manner to HLA-A2 complexes. Hence, these are recombinant antibodies with TCR-like specificity. In contrast to the inherent low affinity of TCRs, these molecules display the high-affinity binding characteristics of antibodies, while retaining TCR specificity. Most importantly, these recombinant antibodies specifically recognize native gp100-derived MHC–peptide complexes on the surface of cells. Interestingly, we were able to isolate a repertoire of several antibodies against each of the gp100-derived epitopes. They exhibit a very specific recognition pattern toward each of the three T cell epitopes even though they are encoded within a single cancer antigen. Until now antibodies with TCR-like specificity have been generated against murine MHC–peptide complexes by employing various immunization strategies (16, 17, 21–23). Recently the same Fab library was used to select for HLA-A1-MAGE-A1-specific binding antibodies (36). One specific clone, G8, was selected that exhibited TCR-like specificity but revealed a relatively low affinity of 250 nM. Most strikingly, here we selected several different TCR-like antibodies against each MHC–peptide complex screened, whereas all previous successful experiments reported the ability to isolate only a single antibody clone (16, 17, 36). The observation that 20–30% of the MHC–peptide binding antibodies had the fine specificity of a TCR-like molecule is nevertheless surprising, especially because they were selected from a nonimmune repertoire considered not to be biased toward such specificity. More recently we have been able to isolate from the same phage library recombinant Fabs against a large variety of MHC–peptide complexes containing other cancer-associated or viral HLA-A2-restricted peptides (28), indicating that this behavior is not gp100 or peptide related.
The unexpectedly high frequency of these antibodies and our ability to isolate several different antibodies directed to either complex is even more surprising in view of previous reports, in which the use of immunized or naive phage libraries resulted in only a single antibody clone (16, 17, 36).
It would have been possible that one particular antibody family or antibody V-gene segment would have an intrinsic propensity to bind HLA-A2 molecules, and that the high frequency could be explained by a high abundance of such antibodies in the nonimmune library. However, the sequences of the selected clones are derived from many different antibody families and germ-line segments, without any biases visible in the complementarity-determining regions either (data not shown). The high frequency and high affinity for some of the antibodies isolated here suggest that these molecules may well be present at a high frequency in the antibody repertoires from the B cell donors of the phage library, but an in vivo role for such antibodies remains unclear.
Whatever the eventual reason for this high frequency of antibodies to MHC–peptides may be, it appears that this phage-based approach can be successfully applied to isolate recombinant antibodies with TCR-like specificity to a large variety of MHC–peptide complexes. Thus, it may now become possible to dissect the role of antigens in various pathological conditions such as cancer, viral infections, and autoimmune disease, not only at the level of the T cell by using MHC tetramers, but also at the level of the APC and diseased cell, by using antibodies of the type described in this paper.
Recombinant antibodies with TCR-like specificity, such as have been selected and characterized in this study, represent a valuable tool in molecular immunology for several major fields of research: (i) for studying antigen presentation in cancer (ii), for developing new immunotherapy targeting molecules, and (iii) for studying structure–function relationships in TCR–peptide–MHC interactions. They should be very useful for the study and analysis of antigen presentation on tumor cells by determining the expression of specific tumor-related MHC–peptide complexes on the surface of tumor cells, metastases, APCs, and lymphoid cells. Such antibodies are expected to be particularly useful for determining the alterations in MHC–peptide complex expression on antigen-presenting cells before, during, and after vaccination protocols with peptides, APCs loaded with tumor cell extracts, or dendritic–tumor cell hybrids (7, 10, 11).
To the best of our knowledge, the molecules described here are the first examples of high-affinity human antibodies directed against the most frequent HLA haplotype, HLA-A2, complexed with cancer peptides. Therefore there are now opportunities to use these very specific molecules, which recognize a very specific human tumor antigen, as candidates for targeting moieties by using various antibody-based immunotherapeutic approaches. This includes the use of these antibodies to construct recombinant immunotoxins (37), fusions with cytokine molecules (38), for bispecific antibody therapy (39), or immunogene therapy (40).
Another interesting aspect for the use of these TCR-like Fab antibodies is for structure–function studies of MHC–peptide–TCR interactions. By mutating particular residues in the specific peptide and testing the influence of these mutations on the binding of the Fab antibodies and peptide-specific T cell clones it may be possible to obtain important data on structure–function relationship and the different nature of recognition between the TCR-like Fabs and the native TCR.
The most important question with respect to immunodiagnostic and immunotherapeutic applications of TCR-like Fabs relates to the low density and turnover of the specific epitope on the target cell surface. It remains to be determined what the density is of the gp100-derived complexes on cancer cells, APCs, and other cells involved in antigen presentation. To improve the sensitivity and targeting capabilities of these TCR-like antibody molecules, two antibody engineering approaches can be used: the first increases the affinity of the parental antibodies by affinity maturation strategies without altering their TCR-like fine specificity (41, 42), and the second increases the avidity of these recombinant monovalent molecules by making them multivalent. Combining these strategies may well result in improved second-generation antibody molecules that will be sensitive enough and specific for immunotherapeutic approaches as well as for studying the interaction of tumor cells and the human immune system. The advent in recent years of the application of tetrameric arrays of class I MHC–peptide complexes now enables us to detect and study rare populations of antigen-specific T cells (12). Our approach produces antibody molecules that enable phenotypic analysis of antigen (MHC–peptide) presentation, the other side of the coin to MHC–peptide–TCR interactions. Combining these two approaches will significantly enhance our ability to understand immune responses in health as well as under various pathological conditions such as cancer and viral infections, and also when applied to class II MHC molecules and autoimmune diseases. The effectiveness and feasibility of this approach, as presented in this study, makes it realistic to generate, in a generic form, antibodies directed toward a large variety of specific MHC–peptide complexes.
Supplementary Material
Acknowledgments
We thank Drs. Steven Rosenberg and Mark Dudley, Surgery Branch, National Cancer Institute, National Institutes of Health, for the R6C12 CTL clone. This work was supported in part by a research grant from the Israel Science Foundation administered by the Israel National Academy for Sciences and Humanities, Jerusalem, and a Research Career Development Award by the Israel Cancer Research Foundation, New York. C.J.C. was supported by a short-term fellowship administered by the European Molecular Biology Organization.
Abbreviations
- TCR
T cell antigen receptor
- CTL
cytotoxic T lymphocyte
- scMHC
single-chain MHC
- APC
antigen-presenting cell
- HTLV-1
human T lymphotrophic virus type 1
References
- 1.Boon T, van der Bruggen P. J Exp Med. 1996;183:725–729. doi: 10.1084/jem.183.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenberg S A. Nature (London) 2001;411:380–384. doi: 10.1038/35077246. [DOI] [PubMed] [Google Scholar]
- 3.Renkvist N, Castelli C, Robbins P F, Parmiani G. Cancer Immunol Immunother. 2001;50:3–15. doi: 10.1007/s002620000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anichini A, Maccalli C, Mortarini R, Salvi S, Mazzocchi A, Squarcina P, Herlyn M, Parmiani G. J Exp Med. 1993;177:989–998. doi: 10.1084/jem.177.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coulie P G, Brichard V, Van Pel A, Wolfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora J P. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivoltini L, Loftus D J, Squarcina P, Castelli C, Rini F, Arienti F, Belli F, Marincola F M, Geisler C, Borsatti A, et al. Crit Rev Immunol. 1998;18:55–63. doi: 10.1615/critrevimmunol.v18.i1-2.70. [DOI] [PubMed] [Google Scholar]
- 7.Offringa R, van der Burg S H, Ossendorp F, Toes R E, Melief C J. Curr Opin Immunol. 2000;12:576–582. doi: 10.1016/s0952-7915(00)00145-x. [DOI] [PubMed] [Google Scholar]
- 8.Restifo N P, Esquivel F, Kawakami Y, Yewdell J W, Mule J J, Rosenberg S A, Bennink J R. J Exp Med. 1993;177:265–272. doi: 10.1084/jem.177.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seliger B, Maeurer M J, Ferrone S. Immunol Today. 2000;21:455–464. doi: 10.1016/s0167-5699(00)01692-3. [DOI] [PubMed] [Google Scholar]
- 10.Esche C, Shurin M R, Lotze M T. Curr Opin Mol Ther. 1999;1:72–81. [PubMed] [Google Scholar]
- 11.Kugler A, Stuhler G, Walden P, Zoller G, Zobywalski A, Brossart P, Trefzer U, Ullrich S, Muller C A, Becker V, et al. Nat Med. 2000;6:332–336. doi: 10.1038/73193. [DOI] [PubMed] [Google Scholar]
- 12.Altman J D, Moss P A, Goulder P J, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 13.Lee P P, Yee C, Savage P A, Fong L, Brockstedt D, Weber J S, Johnson D, Swetter S, Thompson J, Greenberg P D, et al. Nat Med. 1999;5:677–685. doi: 10.1038/9525. [DOI] [PubMed] [Google Scholar]
- 14.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, et al. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 15.Wulfing C, Plückthun A. J Mol Biol. 1994;242:655–669. doi: 10.1006/jmbi.1994.1615. [DOI] [PubMed] [Google Scholar]
- 16.Andersen P S, Stryhn A, Hansen B E, Fugger L, Engberg J, Buus S. Proc Natl Acad Sci USA. 1996;93:1820–1824. doi: 10.1073/pnas.93.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porgador A, Yewdell J W, Deng Y, Bennink J R, Germain R N. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 18.Day P M, Yewdell J W, Porgador A, Germain R N, Bennink J R. Proc Natl Acad Sci USA. 1997;94:8064–8069. doi: 10.1073/pnas.94.15.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong G, Reis e Sousa C, Germain R N. Proc Natl Acad Sci USA. 1997;94:13856–13861. doi: 10.1073/pnas.94.25.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong G, Sousa C R, Germain R N. J Exp Med. 1997;186:673–682. doi: 10.1084/jem.186.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dadaglio G, Nelson C A, Deck M B, Petzold S J, Unanue E R. Immunity. 1997;6:727–738. doi: 10.1016/s1074-7613(00)80448-3. [DOI] [PubMed] [Google Scholar]
- 22.Murphy D B, Lo D, Rath S, Brinster R L, Flavell R A, Slanetz A, Janeway C A., Jr Nature (London) 1989;338:765–768. doi: 10.1038/338765a0. [DOI] [PubMed] [Google Scholar]
- 23.Aharoni R, Teitelbaum D, Arnon R, Puri J. Nature (London) 1991;351:147–150. doi: 10.1038/351147a0. [DOI] [PubMed] [Google Scholar]
- 24.Krogsgaard M, Wucherpfennig K W, Canella B, Hansen B E, Svejgaard A, Pyrdol J, Ditzel H, Raine C, Engberg J, Fugger L. J Exp Med. 2000;191:1395–1412. doi: 10.1084/jem.191.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reiter Y, Di Carlo A, Fugger L, Engberg J, Pastan I. Proc Natl Acad Sci USA. 1997;94:4631–4636. doi: 10.1073/pnas.94.9.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denkberg G, Cohen C J, Segal D, Kirkin A F, Reiter Y. Eur J Immunol. 2000;30:3522–3532. doi: 10.1002/1521-4141(2000012)30:12<3522::AID-IMMU3522>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 27.de Haard H J, van Neer N, Reurs A, Hufton S E, Roovers R C, Henderikx P, de Bruine A P, Arends J W, Hoogenboom H R. J Biol Chem. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 28.Lev A, Denkberg G, Cohen C J, Tzukerman M, Skorecki K L, Chames P, Hoogenboom H R, Reiter Y. Cancer Res. 2002;62:3184–3194. [PubMed] [Google Scholar]
- 29.Kawakami Y, Eliyahu S, Delgado C H, Robbins P F, Sakaguchi K, Appella E, Yannelli J R, Adema G J, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakker A B, Schreurs M W, de Boer A J, Kawakami Y, Rosenberg S A, Adema G J, Figdor C G. J Exp Med. 1994;179:1005–1009. doi: 10.1084/jem.179.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawakami Y, Eliyahu S, Jennings C, Sakaguchi K, Kang X, Southwood S, Robbins P F, Sette A, Appella E, Rosenberg S A. J Immunol. 1995;154:3961–3968. [PubMed] [Google Scholar]
- 32.Cox A L, Skipper J, Chen Y, Henderson R A, Darrow T L, Shabanowitz J, Engelhard V H, Hunt D F, Slingluff C L., Jr Science. 1994;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 33.Denkberg G, Cohen C J, Reiter Y. J Immunol. 2001;167:270–276. doi: 10.4049/jimmunol.167.1.270. [DOI] [PubMed] [Google Scholar]
- 34.Dudley M E, Ngo L T, Westwood J, Wunderlich J R, Rosenberg S A. Cancer J. 2000;6:69–77. [PubMed] [Google Scholar]
- 35.Kirkin A F, Petersen T R, Olsen A C, Li L, thor Straten P, Zeuthen J. Cancer Immunol Immunother. 1995;41:71–81. doi: 10.1007/BF01527402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chames P, Hufton S E, Coulie P G, Uchanska-Ziegler B, Hoogenboom H R. Proc Natl Acad Sci USA. 2000;97:7969–7974. doi: 10.1073/pnas.97.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastan I. Biochim Biophys Acta. 1997;1333:C1–C6. doi: 10.1016/s0304-419x(97)00021-8. [DOI] [PubMed] [Google Scholar]
- 38.Lode H N, Reisfeld R A. Immunol Res. 2000;21:279–288. doi: 10.1385/IR:21:2-3:279. [DOI] [PubMed] [Google Scholar]
- 39.Withoff S, Helfrich W, de Leij L F, Molema G. Curr Opin Mol Ther. 2001;3:53–62. [PubMed] [Google Scholar]
- 40.Willemsen R A, Debets R, Hart E, Hoogenboom H R, Bolhuis R L, Chames P. Gene Ther. 2001;8:1601–1608. doi: 10.1038/sj.gt.3301570. [DOI] [PubMed] [Google Scholar]
- 41.Chowdhury P S, Pastan I. Nat Biotechnol. 1999;17:568–572. doi: 10.1038/9872. [DOI] [PubMed] [Google Scholar]
- 42. Chames, P., Willemsen, R. A., Rojas, G., Dieckmann, D., Rem, L., Schuler, G., Bolhuis, R. L. & Hoogenboom, H. R. (2002) J. Immunol. 169, in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.