Abstract
Colorectal cancer is one of the most significant causes of cancer death. A genetic model for colorectal cancer has been proposed in which the sequential accumulation of mutations in specific genes, including adenomatous polyposis coli (APC), Kirsten-ras (K-ras), and p53, drives the transition from healthy colonic epithelia through increasingly dysplastic adenoma to colorectal cancer. We have characterized tumor mutation spectra in a large cohort of colorectal cancer patients. In marked contrast to the predictions of the sequential model of mutation accumulation, only 6.6% of tumors were found to contain mutations in APC, K-ras, and p53, with 38.7% of tumors containing mutations in only one of these genes. The most common combination of mutations was p53 and APC (27.1%), whereas mutations in both p53 and K-ras were extremely rare. Statistical analysis (two-sided Fisher's exact test) confirmed that mutations in K-ras and p53 co-occurred less frequently than expected by chance (P < 0.01, Fisher's exact test). This finding suggests that these mutations lie on alternate pathways of colorectal tumor development. The heterogeneous pattern of tumor mutations in our patient cohort suggests that multiple alternative genetic pathways to colorectal cancer exist and that the widely accepted genetic model of cancer development is not representative of the majority of colorectal tumors.
Colorectal cancer is one of the major causes of cancer death worldwide and in Western society is second only to lung cancer (1). Only a small proportion (between 5 and 10%) of colorectal cancer cases are attributable to familial cancer syndromes and the majority seem to arise sporadically. Colorectal cancer incidence rates vary widely in different geographical areas, with relatively low incidence in Asia, Africa, and parts of Latin America but with high incidence in “Western” countries, including Northern Europe, Australia, New Zealand, and the U.S. (2). Studies in migrant populations have demonstrated that populations moving from low- to high-risk countries rapidly acquire an increased cancer risk, suggesting that local environmental exposures influence colorectal cancer susceptibility (3). Epidemiological studies have suggested that specific components of the Western diet, including dietary fat and red meat, are risk factors in colorectal cancer pathogenesis whereas other dietary components, including fruit, vegetables, and dietary fiber, are protective (4).
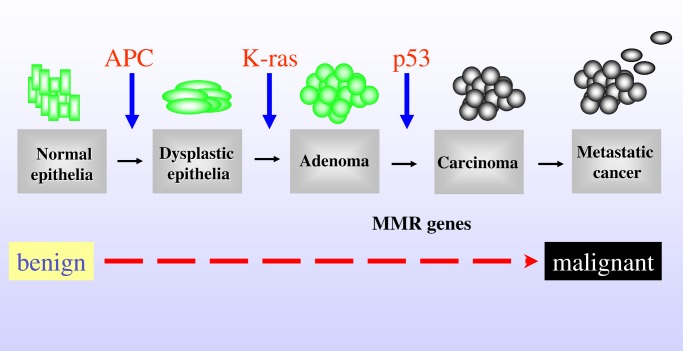
Carcinogenesis is a multistep process, which can arise from a combination of mutations in oncogenes or tumor suppressor genes or from epigenetic changes in DNA such as methylation. A genetic model describing the transition from healthy colonic epithelia through increasingly dysplastic adenoma to malignant cancer has been proposed (5) that identifies a number of key oncogenes and tumor suppressor genes, the progressive acquisition of activating or loss of function mutations in which drives the adenoma to carcinoma transition (Fig. 1). Certain of the genes identified in this pathway, including adenomatous polyposis coli (APC) and the DNA mismatch repair (MMR) genes, are the same as those mutated in familial colorectal cancer syndromes (6, 7).
Figure 1.
A model of the genetic changes required for progression from adenoma to carcinoma in the development of colorectal cancer (6). The proposed order of mutations in APC, K-ras, p53, and the DNA MMR genes is illustrated.
It has been suggested that as many as seven distinct genetic changes are required for a cell to progress from adenoma to carcinoma, and that the accumulation rather than the specific nature and temporal order of the mutations is most critical (8). Of the genes characterized to date, inactivation of the tumor suppressor genes APC and p53 and activation of the oncogene Kirsten-ras (K-ras) are thought to be particularly important determinants of tumor initiation and progression (6).
The APC gene was first localized to chromosome 5q21 by disease linkage in patients with familial adenomatous polyposis (FAP) (9). APC is a relatively large gene, encoding an 8.5-kB mRNA and 312-kD protein. The APC protein is structurally complex, with many possible protein/protein interaction sites, including binding sites for β-catenin, EB1, and axin (10). A mutation cluster region (MCR) has between identified in exon 15 in both patients with FAP and patients with sporadic colorectal cancer, located between codons 1286 and 1513 (11). Although the MCR comprises only 10% of the total APC coding sequence, it contains more than 90% of APC mutations reported in both familial and sporadic colorectal cancers (8). The majority of APC mutations in the MCR introduce a stop codon into the APC mRNA, resulting in deletion of the carboxyl-terminal functions of the protein, including β-catenin and axin binding. APC has been proposed to function as a “gatekeeper” gene, regulating the entry of epithelial cells into the adenoma-carcinoma progression (8).
The K-ras gene, located on the short arm of chromosome 12, encodes a 21-kD protein (p21ras) involved in G protein-mediated signal transduction. K-ras has constitutive GTPase activity, which is lost when the gene is mutated, most commonly at codons 12, 13, and 61 (12). Mutations in K-ras lead to increased and unregulated cellular proliferation and malignant transformation.
Mutations in p53 are proposed to be relatively late events in the development of colorectal tumors, with the loss of p53-mediated pathways of apoptosis considered to be an important determinant of progression from adenoma to malignant tumor. The p53 gene, localized on the short arm of chromosome 17, is mutated in up to 70% of colorectal cancers (13). p53 functions as a transcription factor, exerting cell cycle control by binding to specific recognition sequences in variety of genes including p21, Bax, and Bcl-2 in response to DNA damage or other cellular stress (14). Constitutive p53 levels are tightly regulated by interaction with the E3 ubiquitin ligase MDM2, itself induced by p53, which targets p53 for ubiquitination and subsequent degradation (15).
Based on the hypothesis that mutation of specific target genes may be important molecular mechanisms that influence the pathogenesis of colorectal cancer, as part of a wider study to investigate the relationship among diet, genetic polymorphism, and colorectal cancer susceptibility, we have analyzed the mutation spectra in three key genes, APC, K-ras, and p53 in more than 100 patients with colorectal cancer.
Materials and Methods
Patient Recruitment.
Patients (age range 45–80; median age 67; 64 males and 42 females) undergoing surgery for colorectal cancer at Ninewells Hospital, Dundee, or Perth Royal Infirmary were invited to participate in the study. All patients were white, had preoperative pathological conformation of diagnosis, and had no history of previous cancer or diverticular disease. Patient details, including tumor location and Dukes' staging, are given in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. All tumors were classified by the standard Dukes' staging system where Dukes' A tumors were confined to the bowel wall, Dukes' B tumors extended locally beyond the bowel, and Dukes' C tumors involved lymph nodes. All patients were asked to complete a detailed Food Frequency Questionnaire. The study was approved by the Tayside Committee for Medical Research Ethics.
Sample Preparation.
Resection specimens were brought fresh from theatre to pathology. Normal and tumor tissue samples were selected by an experienced pathologist (F.A.C.) and stored in liquid nitrogen before analysis. Genomic DNA for mutation analysis was extracted from each tumor tissue with the Wizard Genomic DNA Purification kit (Promega) according to the manufacturer's instructions.
Mutation Detection.
Denaturing high-performance liquid chromatography (HPLC) “WAVE” analysis.
Because of the number and complexity of mutations in p53 and APC, the transgenomic WAVE denaturing HPLC system was used as a “prescreen” to identify samples containing mutations in these genes. WAVE analysis is based on the temperature-dependent differences in column-retention time of PCR products generated from homoduplex (wild-type) and heteroduplex (mutated) DNA, resulting in the presence of distorted or additional peaks when mutations are present.** The presence of homozygous mutations or mutations in samples that had undergone allelic loss was confirmed by mixing each test sample with a known wild-type DNA control. Assay conditions (acetonitrile gradient and temperature profile) were optimized for WAVE analysis of each PCR fragment with a bank of samples previously characterized by DNA sequencing. All samples were denatured and cooled slowly to room temperature before WAVE analysis to maximize heteroduplex formation.
(i) p53.
Oligonucleotide primers were designed to amplify the entire p53 coding region in eight fragments (see Fig. 5 and Table 3, which are published as supporting information on the PNAS web site). Mutation detection gradients (see Table 4, which is published as supporting information on the PNAS web site) were optimized for WAVE analysis as described above.
(ii) APC.
The MCR (codons 1028–1712) was analyzed (8). Oligonucleotide primers were designed to amplify a 2-kb region of exon 15 of APC, in 3 overlapping PCR fragments, which were optimized for WAVE analysis as described previously. WAVE analyses of fragments APC2 and APC3 were found to contain a particularly high number of base pair changes, the majority of which were the result of the presence of characterized polymorphisms (16). WAVE analysis was therefore used to screen only fragment APC1—mutations in fragments APC2 and APC3 were identified by direct sequencing.
(iii) K-ras.
Mutations in codons 12, 13, and 61 of K-ras were identified by direct sequencing as described in the next paragraph.
Direct Sequencing.
The mutations in p53 and in APC1 detected by WAVE analysis were confirmed by bidirectional direct sequencing (Li-Cor long Readir 4200 DNA Sequencer, MWG Biotech, Ebersberg, Germany). Direct sequencing was also used to identify mutations in APC2 and APC3 and in codons 12, 13, and 61 of the K-ras gene. All PCR products for direct sequencing were amplified with M13-tagged primers (see Table 5, which is published as supporting information on the PNAS web site) and were purified by polyethylene glycol precipitation before sequencing. Fragments were simultaneously bidirectionally sequenced with fluorescently labeled M13-tagged primers by using a Thermosequenase DYEnamic Direct Cycle Sequencing kit (Amersham Pharmacia Biotech), according to the manufacturer's instructions. All sequences obtained were aligned with previously published sequences [National Center for Biotechnology Information (NCBI) GenBank accession nos.: M74088 (APC), M54968 (K-ras), and X54156 (p53)] for each of the target genes; the presence and nature of each mutation was confirmed by repeat PCR and sequencing.
hMSH-2/hMLH-1 Immunohistochemistry.
Four micrometer sections were cut from paraffin blocks chosen to include both tumor and normal mucosa. The sections were processed in a standard immunohistochemical procedure with microwave antigen retrieval and a horseradish peroxidase-labeled streptavidin detection system. The primary Abs (hMSH-2, Calbiochem; hMLH-1, Cambridge Bioscience, Cambridge, U.K.) were applied at a 1/100 dilution (17). All sections showed nuclear staining in the internal control (normal) mucosa.
Results
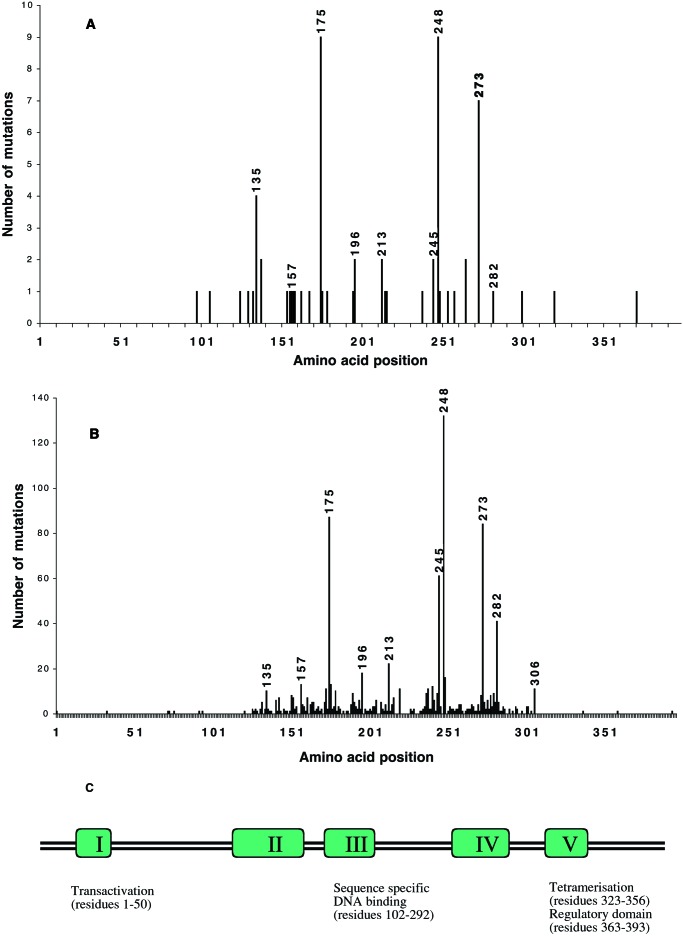
Of the genes studied, p53 was found to be most frequently mutated, with 65/106 (61.3%) of the tumors analyzed containing a p53 mutation (Tables 1 and 2). This finding is in agreement with previous studies in colorectal cancer, which report frequencies between 50 and 70% (13). Consistent with literature reports, the majority of the base pair changes in p53 were in a “hotspot” region between exons 5 and 8, a core functional domain responsible for DNA binding (Fig. 2). The International Agency for Research on Cancer (IARC) p53 database identifies codons 175, 245, 248, and 273 as particularly frequently mutated in colorectal cancer (Fig. 2B) (18). These somatically mutated hotspot codons may identify functionally important amino acid residues, which provide a selective growth advantage to the tumor cell. Fig. 2 A and B shows a comparison of the p53 mutations identified in our patients with colorectal cancer with those reported in the IARC p53 database. To ensure that an appropriate comparison was made and particularly to exclude the possibility that there may be geographical differences that may influence the number and nature of p53 mutations, only database entries relating to solid tumors (i.e., not cell lines) from white individuals were compared. The hotspot codons identified in our study were almost identical to those described in the IARC data set. These data, together with previous reports, confirm that the majority of p53 mutations lie within the central conserved DNA-binding region of the protein and justify the approaches adopted by previous studies that screened for mutations only in this restricted area of the protein. It should be noted, however, that we did find additional changes throughout this region, e.g., between amino acid residues 92 and 112. Consistent with previous literature reports (19), the majority (47.1%) of p53 mutations in our patient group were C to T transitions at CpG dinucleotides.
Table 1.
Mutation frequencies in Dundee patients with colorectal cancer
| Gene | Left-sided (n = 88) | Right-sided (n = 18) | All tumors (n = 106) | Colon tumors (n = 65) | Rectal tumors (n = 41) |
|---|---|---|---|---|---|
| p53 | 56/88 (63.6%) | 9/18 (50%) | 65/106 (61.3%) | 44/65 (67.7%) | 21/41 (51.2%) |
| APC | 49/88 (55.7%) | 11/18 (61.1%) | 60/106 (56.%) | 41/65 (63.1%) | 19/41 (46.3%) |
| K-ras | 24/88 (27.3%) | 5/18 (27.7%) | 29/106 (27.4%) | 13/65 (20.0%) | 16/41 (39.0%) |
Figure 2.
Spectrum of p53 mutations in colorectal cancer. (A) p53 mutations in Dundee patients with colorectal cancer and (B) p53 mutations in all white colorectal solid tumors in the IARC p53 mutation database (18). The amino acid positions of the most frequently mutated hotspot codons are highlighted. (C) The position of each of the hotspot codons relative to the conserved regions and functional domains of p53 is illustrated.
APC mutations were found in approximately 60% of the samples studied, in agreement with previous literature reports (20, 21). The vast majority of APC mutations were frame shifts, introducing a premature stop codon (Table 2). Of the two APC mutations that were not frame shifts (patients 1004 and 1073), one (patient 1004) was found in conjunction with an additional frame shift mutation.
K-ras was the least frequently mutated of the genes studied, with approximately 30% of tumors affected. This finding was in good agreement with the frequency of 37.7% reported in a recent metaanalysis of K-ras mutation frequencies in colorectal cancer (22). Consistent with literature reports, the majority of K-ras mutations were found in codon 12 (Table 2), with a smaller number of nucleotide substitutions in codon 13. No mutations were found in codon 61 in this patient group. In contrast to p53 and APC, K-ras mutations were significantly more common in rectal (39.0%) than in colon tumors (20.0%, P = 0.044), suggesting different etiologies for these cancers. The majority of K-ras mutations were base pair transitions, occurring predominantly at the second bases of codons 12 and 13. In agreement with previous studies (22), all of the codon 13 mutations were G → A transitions, whereas both G → A and G → C transitions and G → T transversions were found in codon 12.
There were no significant differences in mutation rates between males and females for either APC (55.6% vs. 59.5%), K-ras (28.6% vs. 26.2%), or p53 (61.9% vs. 59.5%, respectively).
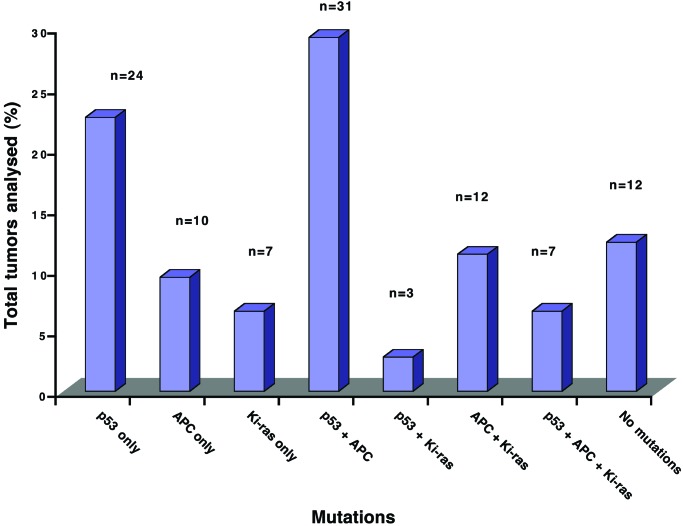
The proposed sequence of genetic changes leading to the development of colorectal cancer involves mutations in all three genes analyzed, i.e., APC, K-ras, and p53. However, in our study a significant percentage of the tumors contained mutations in only one of these genes, p53 (24.1%), APC (12.0%), or K-ras (8.3%). Only 7 tumors (6.0% of all tumors analyzed) contained alterations in all three genes (Fig. 3), whereas 12 tumors (11.3%) contained no mutations. The most common combination of mutations was p53 and APC (27.1%), whereas mutations in both p53 and K-ras were extremely rare. Statistical analysis confirmed that mutations in K-ras and p53 co-occurred less frequently than expected by chance (P < 0.01, Fisher's exact test). This result suggests that these mutations lie on alternate pathways of colorectal cancer development.
Figure 3.
Distribution of APC, K-ras, and p53 mutations in patients with colorectal cancer. The presence of mutations in APC, K-ras, and p53 was determined in a cohort of patients with colorectal cancer (n = 106) by a combination of WAVE denaturing HPLC analysis and direct sequencing, as described in Materials and Methods. The percentage of tumors with each combination of mutations is illustrated.
We failed to detect alterations in p53, APC, or K-ras in 12 (11.3%) of the patients studied, although these individuals had a histologically confirmed diagnosis of colorectal cancer. We therefore used immunohistochemical analysis to study the expression of the DNA repair genes hMLH1 and hMSH2 (17), of which loss of expression is a marker for microsatellite instability resulting from defects in DNA MMR pathways. Although all of the tumors studied showed were positive for hMLH2, 5 of the 106 tumors analyzed (4.7%) were negative for hMLH1 (data not shown). Three of these 5 tumors (patients 1011, 1034, and 1261) had wild-type APC, K-ras, and p53; one (patient 1001) had mutations in both APC and p53; and one (patient 1153) had a gene-inactivating mutation in APC. The association between loss of hMLH1 expression and wild-type genotype at the three genes evaluated is significant (P = 0.01, Fisher's exact text), and loss of expression of MMR genes may therefore rationalize tumor formation in a small proportion of the tumors in our series.
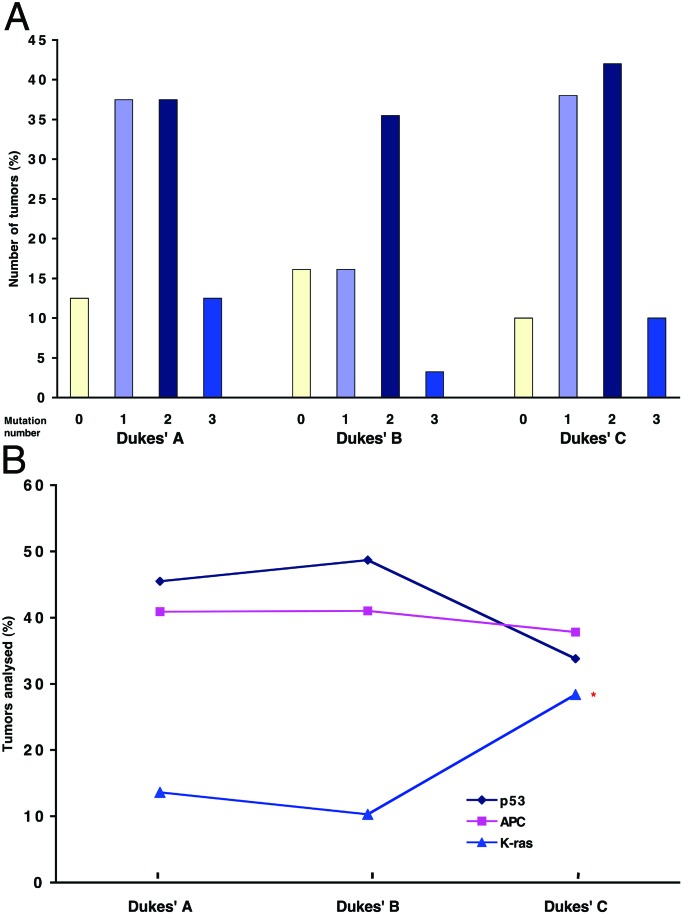
We further investigated whether the accumulation of multiple mutations was attributable to differences in Dukes' stages (23). There was no significant correlation among Dukes' stages and the number of tumor mutations (correlation coefficient = 0.07) (Fig. 4A). Interestingly, analysis of the relative contribution of APC, K-ras, and p53 to the overall mutation burden in early and advanced tumors demonstrated a statistically significant increase in the K-ras mutation frequency in Dukes' C tumors (Fig. 4B), suggesting that altered K-ras function is an important determinant of tumor progression.
Figure 4.
Distribution of APC, K-ras, and p53 mutations according to Dukes' stages. The presence of mutations in APC, K-ras, and p53 was determined in a cohort of patients with colorectal cancer (n = 106) by a combination of WAVE denaturing HPLC analysis and direct sequencing, as described in Materials and Methods. (A) Tumors were categorized by the number of mutations and further subdivided according to Dukes' stages. (B) Tumors were categorized according to Dukes' stages and further subdivided by the presence of mutations in APC, K-ras, and p53. *, P = 0.032, comparing Dukes' B and Dukes' C.
Discussion
A model, proposed to describe the accumulation of genetic changes necessary to drive the transition from adenoma to carcinoma in the development of colorectal cancer, has become generally accepted as a paradigm for the genetic basis of colorectal cancer development (6, 8). Fundamental to this model is the concept that progression from normal epithelium to carcinoma is accompanied by the accumulation of mutations in a number of key genes that contribute to the maintenance of healthy colonic epithelia, the regulation of cell cycle control, and the inhibition of apoptosis.
Our data, however, suggest that the existing model will only rationalize tumor formation in an extremely small number of colorectal cancers. Indeed, in view of the relatively low frequency of tumors with mutations in all three genes, APC, K-ras, and p53, it is feasible that, where multiple mutations do exist in the same tumor, their occurrence is a chance event and does not represent a synergistic evolutionary pathway. The model described by Fearon and Vogelstein (6) was based on the analysis of a relatively limited number of colorectal tumors, where more than 90% of tumors had allelic losses with, presumably, a corresponding loss of function of at least two of the four genes studied (p53, APC, K-ras, and DCC). We did not consider DCC in the present study but have described a comprehensive analysis of mutations in p53, APC, and K-ras in 106 colorectal tumors where only 50% of tumors contained multiple mutations and less than 7% of tumors contained mutations in all three genes. In confirmation of these findings, we have also analyzed a second cohort of 63 tumors, where only 2% of tumors analyzed contained mutations in all three genes. More than one-third of our tumors contained mutations in only one of p53, APC, or K-ras, demonstrating that the progressive accumulation of multiple mutations in these genes is not a prerequisite for tumor development. Our mutation frequencies for the individual genes was entirely consistent with previous literature reports in colorectal cancer (13, 20–22), although many previous studies considered relatively small patient groups and the majority studied only a single gene in isolation.
The current study provides a number of insights into the mechanisms of colorectal tumor development. First, K-ras mutations were significantly more common in rectal than in colon tumors, indicating differences in the pathways of carcinogenesis in these tissues. Second, p53 and K-ras mutations were rarely found together in the same tumor, suggesting different genetic pathways leading to tumor formation. The significant overrepresentation of K-ras mutations in Dukes' C tumors suggests that K-ras activation may be an important determinant of tumor progression. In a minority of tumors, mutations were found in both p53 and K-ras. Although the combination of mutant p53 and K-ras is likely to have an effect on tumor phenotype, the low number of tumors containing mutations in both genes suggests that this phenotype may have arisen by chance.
Consistent with other studies, we have found that a significant number of tumors do not seem to have lost APC tumor suppressor gene function, or at least do not contain mutations within the APC exon 15 MCR, suggesting that APC-driven alterations in colonic crypt architecture are not an absolute requirement for the initiation of dysplasia. Tumors retaining APC integrity must have alternative mechanisms of tumor initiation. Our data suggest that mutations in K-ras, p53, or other events can also independently initiate the adenoma to carcinoma transition (24). These observations are supported by mutation analysis in patients with ulcerative colitis, an inflammatory bowel disease that confers a significantly increased risk of colorectal cancer (25). APC mutations are relatively rare (<10%) in patients with ulcerative colitis, whereas p53 mutations are common, suggesting that loss of cell cycle apoptotic control mechanisms through mutation in p53 may be one mechanism to explain the observed increase in colorectal cancer risk (26).
The effects of mutations in p53 are likely to be codon-specific, with certain amino acid changes having a more significant effect on p53 function than others. For example, several of the hotspot codons identified in both the IARC database and in our own patients (codons 248 and 273) have been localized within the DNA-binding domain of the protein, a region which influences protein–p53 interactions and is therefore a critical determinant of p53 sequence-specific transactivation functions (27). Other hotspots (e.g., codon 175) disrupt protein conformation, whereas various insertion and deletion mutations lead to loss of protein function through frame shifts and the generation of premature stop codons.
Previous studies have reported K-ras mutation rates to be higher in left-sided colorectal cancers (28). This finding has been attributed to the increased exposure of the left-sided bowel lumen to ingested carcinogens and mutagens. No differences in K-ras frequencies in left- and right-sided tumors were found in this study, although the number of right-sided tumors was small. There is also evidence that K-ras mutations may be lost through selection as cells progress from adenoma to carcinoma, with some studies reporting K-ras mutation frequencies that are higher in patients with adenoma compared with colorectal cancer (29). Although we have not studied adenomas here, our finding that K-ras mutations were significantly overrepresented in Dukes' C tumors is not consistent with this observation.
An alternative pathway of tumor development is characterized by microsatellite instability resulting from defects in DNA MMR pathways (30). Defects in DNA MMR are responsible for the familial syndrome HNPCC, an autosomal dominant disease which accounts for up to 5% of colorectal cancer cases (31). In these cases, multiple polyps are found in many tissues including stomach, ovary, and bladder as well as the gastrointestinal tract (32). HNPCC arises from mutations in one of several DNA MMR genes (hMSH2, hMLH1, hPMS1, hPMS2, hMSH3, or hMSH6), all of which regulate the fidelity of DNA synthesis during replication. Although microsatellite instability frequencies of up to 15% have been reported in sporadic colorectal cancers, more recent reports, in agreement with our data, suggest that only 1–2% of sporadic colorectal tumors arise from defects in MMR pathways (33).
The presence of mutations in APC, K-ras, and p53 has all been identified as prognostic indicators of tumor behavior and survival in patients with colorectal cancer, although the findings of all studies are not consistent. In the majority of colorectal cancers, “outcome” is likely to be determined by the skill of the surgeon, although the presence of specific tumor mutations may be an important identifier of “high-risk” individuals, with particularly aggressive or rapidly growing tumors. Our data specifically associate K-ras mutation with Dukes' C tumors, consistent with previous literature reports that K-ras mutation, in particular the glycine to valine substitution at codon 12, is associated with poorer prognosis and time to relapse (22).
The heterogeneous nature of the mutations in APC, K-ras, and p53 in tumors analyzed in the current study raises a number of questions—Do tumors with multiple mutations grow faster/become more aggressive/have a worse prognosis that tumors with only a single mutation? Can specific mutation patterns identify individuals at particularly high risk of progression or relapse? Is it possible that targeted dietary intervention may influence disease progression and/or relapse or that the appropriate dietary regime can influence susceptibility to colorectal cancer in high-risk individuals? In support of this hypothesis, there is some evidence that K-ras mutations are more common in patients with colorectal cancer with diets with a high red meat content, whereas p53 mutations are less common in patients with a high vegetable intake.‡‡ To investigate whether specific dietary and lifestyle choices influence mutation burden in individual patients, we are currently analyzing data from detailed Food Frequency Questionnaires completed by all our cancer patients (unpublished data). In an ongoing comprehensive analysis of drug-metabolizing enzyme polymorphisms in a matched case-control study of these plus additional patients with colorectal cancer, we hope to relate individual dietary choice and patient genotype to the presence of specific tumor mutations.
Various gene therapy approaches have recently been proposed, for example, to “rescue” mutant p53 phenotype in colorectal tumors, including the design of synthetic peptides and small molecule drugs that have successfully restored p53 function in model systems (34, 35). Although these approaches are exciting and may indeed lead to the development of novel therapeutic interventions in the treatment of colorectal and other cancers, our data suggest that future therapies must be targeted to individual patients, based on a detailed understanding of individual genetic background and the nature of the mutations present in individual tumors.
Supplementary Material
Acknowledgments
We thank all of the research nurses who were responsible for patient recruitment and sample collection. We acknowledge financial support from the Food Standards Agency (Contracts TO1003, TO1004, and TO1005). G.S. acknowledges additional financial support from Medical Research Council Grant G0000281.
Abbreviations
- MCR
mutation cluster region
- MMR
mismatch repair
Footnotes
Freedman, A. N., Michalek, A. M., Muro, K., Mettlin, C. J., Asirwatham, J. E., Brooks, J. S., Petrelli, N. J., Caporaso, N. E. & Hamilton, S. R. (1997) Proc. Am. Assoc. Cancer Res. 38, 457A (abstr.).
This paper was submitted directly (Track II) to the PNAS office.
Jin, L., Underhill, P. A., Oefner, P. J. & Cavalli-Sforza, L. L. (1995) Am. J. Hum. Genet. 57, 26A (abstr.).
References
- 1.Boyle P, Lagman J. Br Med J. 2000;321:805–808. doi: 10.1136/bmj.321.7264.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkin D M, Muir C S, Whelan S L, Gao J T, Ferlay J, Powell J. Cancer Incidence in Five Continents. Lyon, France: Intl. Agency Res. Cancer; 1992. [Google Scholar]
- 3.McMichael A J, Giles G G. Cancer Res. 1988;48:751–756. [PubMed] [Google Scholar]
- 4.Potter J D, editor. World Cancer Research Fund (WCRF) Panel. Food, Nutrition and the Prevention of Cancer: A Global Perspective. Washington, DC: WCRF/Am. Inst. Cancer Res.; 1997. [Google Scholar]
- 5.Vogelstein B, Fearon E R, Hamilton S R, Kern S E, Preisinger A C, Leppert M, Nakamura Y, White R, Smits A M, Bos J L. N Engl J Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 6.Fearon E R, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 7.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 8.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 9.Bodmer W F, Bailey C J, Bodmer J, Bussey H, Ellis A, Gorman P, Lucibello F C, Murday V A, Rider S H, Scambler P. Nature (London) 1987;328:614–616. doi: 10.1038/328614a0. [DOI] [PubMed] [Google Scholar]
- 10.Rubinfeld B, Albert I, Porfiri E, Munemitsu S, Polakis P. Cancer Res. 1997;57:4624–4630. [PubMed] [Google Scholar]
- 11.Nakamura Y. Adv Cancer Res. 1993;62:65–87. doi: 10.1016/s0065-230x(08)60315-2. [DOI] [PubMed] [Google Scholar]
- 12.Bos J L. Cancer Res. 1989;49:4682–4686. [PubMed] [Google Scholar]
- 13.Baker S, J, Preisinger A C, Jessup J M, Paraskeva C, Markowitz S, Willson J K, Hamilton S, Vogelstein B. Cancer Res. 1990;50:7717–7722. [PubMed] [Google Scholar]
- 14.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 15.May P, May E. Oncogene. 1999;18:7621–7636. doi: 10.1038/sj.onc.1203285. [DOI] [PubMed] [Google Scholar]
- 16.Lamlum H, Al Tassan N, Jaeger E, Frayling I, Sieber O, Reza F B, Eckert M, Rowan A, Barclay E, Atkin W, et al. Hum Mol Genet. 2000;9:2215–2221. doi: 10.1093/oxfordjournals.hmg.a018912. [DOI] [PubMed] [Google Scholar]
- 17.Cawkwell L, Gray S, Murgatroyd H, Sutherland F, Haine L, Longfellow M, O'Loughlin S, Cross D, Kronborg O, Fenger C, et al. Gut. 1999;45:409–415. doi: 10.1136/gut.45.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hainaut P, Hollstein M. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 19.Harris C C, Hollstein M. N Engl J Med. 1993;329:1318–1327. doi: 10.1056/NEJM199310283291807. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi Y, Nagase H, Ando H, Horii A, Ichii S, Nakatsuru S, Aoki T, Miki Y, Mori T, Nakamura Y. Hum Mol Genet. 1992;1:229–233. doi: 10.1093/hmg/1.4.229. [DOI] [PubMed] [Google Scholar]
- 21.Powell S M, Zilz N, Beazer-Barclay Y, Bryan T M, Hamilton S R, Thibodeau S N, Vogelstein B, Kinzler K W. Nature (London) 1992;359:235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 22.Andreyev H J N, Norman A R, Cunningham D, Oates J R, Clarke P A. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 23.Dukes C E. J Pathol Bacteriol. 1932;35:323–332. [Google Scholar]
- 24.Soengas M S, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman J, G, Gerald W L, Lazebnik Y A, et al. Nature (London) 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 25.Potter J D. J Natl Cancer Inst. 1999;91:916–932. doi: 10.1093/jnci/91.11.916. [DOI] [PubMed] [Google Scholar]
- 26.Brentnall T A, Crispin D A, Rabinovitch P S, Haggit R C, Rubin C E, Stevens A C, Burmer G C. Gastroenterology. 1994;107:369–378. doi: 10.1016/0016-5085(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 27.van Oijen M G, Slootweg P J. Clin Cancer Res. 2000;6:2138–2145. [PubMed] [Google Scholar]
- 28.Toribara N W, Sleisenger M H. N Engl J Med. 1995;332:861–867. doi: 10.1056/NEJM199503303321306. [DOI] [PubMed] [Google Scholar]
- 29.Pretlow T P, Brasitus T A, Fulton N C, Cheyer C, Kaplan E L. J Natl Cancer Inst. 1993;85:2004–2007. doi: 10.1093/jnci/85.24.2004. [DOI] [PubMed] [Google Scholar]
- 30.Peltomaki P, de la Chapelle A. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/s0065-230x(08)60097-4. [DOI] [PubMed] [Google Scholar]
- 31.Huang J, Papadopoulos N, McKinley A J, Farrington S M, Curtis L J, Wyllie A H, Zheng S, Willson J K, Markowitz S D, Morin P, et al. Proc Natl Acad Sci USA. 1996;93:9049–9054. doi: 10.1073/pnas.93.17.9049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch P M, Lynch H T. Colon Cancer Genetics. New York: Van Nostrand Rheinhold; 1985. [Google Scholar]
- 33.Evans D G, Walsh S, Jeacock J, Robinson C, Hadfield L, Davies D R, Kingston R. Br J Surg. 1997;84:1281–1285. [PubMed] [Google Scholar]
- 34.Selivanova G, Ryabchenko L, Jansson E, Iotsova V, Wiman K G. Mol Cell Biol. 1999;19:3395–3402. doi: 10.1128/mcb.19.5.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foster B A, Coffey H A, Morin M J, Rastinejad F. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.