Abstract
The kinetic properties of the three taxonomic A substates of sperm whale carbonmonoxy myoglobin in 75% glycerol/buffer are studied by flash photolysis with monitoring in the infrared stretch bands of bound CO at nu(A0) approximately 1967 cm-1, nu(A1) approximately 1947 cm-1, and nu(A3) approximately 1929 cm-1 between 60 and 300 K. Below 160 K the photodissociated CO rebinds from the heme pocket, no interconversion among the A substates is observed, and rebinding in each A substate is nonexponential in time and described by a different temperature-independent distribution of enthalpy barriers with a different preexponential. Measurements in the electronic bands, e.g., the Soret, contain contributions of all three A substates and can, therefore, be only approximately modeled with a single enthalpy distribution and a single preexponential. The bond formation step at the heme is fastest for the A0 substate, intermediate for the A1 substate, and slowest for A3. Rebinding between 200 and 300 K displays several processes, including geminate rebinding, rebinding after ligand escape to the solvent, and interconversion among the A substates. Different kinetics are measured in each of the A bands for times shorter than the characteristic time of fluctuations among the A substates. At longer times, fluctuational averaging yields the same kinetics in all three A substates. The interconversion rates between A1 and A3 are determined from the time when the scaled kinetic traces of the two substates merge. Fluctuations between A1 and A3 are much faster than those between A0 and either A1 or A3, so A1 and A3 appear as one kinetic species in the exchange with A0. The maximum-entropy method is used to extract the distribution of rate coefficients for the interconversion process A0 <--> A1 + A3 from the flash photolysis data. The temperature dependencies of the A substate interconversion processes are fitted with a non-Arrhenius expression similar to that used to describe relaxation processes in glasses. At 300 K the interconversion time for A0 <--> A1 + A3 is 10 microseconds, and extrapolation yields approximately 1 ns for A1 <--> A3. The pronounced kinetic differences imply different structural rearrangements. Crystallographic data support this conclusion: They show that formation of the A0 substate involves a major change of the protein structure; the distal histidine rotates about the C(alpha)-C(beta) bond, and its imidazole sidechain swings out of the heme pocket into the solvent, whereas it remains in the heme pocket in the A1 <--> A3 interconversion. The fast A1 <--> A3 exchange is inconsistent with structural models that involve differences in the protonation between A1 and A3.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abadan Y., Chien E. Y., Chu K., Eng C. D., Nienhaus G. U., Sligar S. G. Ligand binding to heme proteins. V. Light-induced relaxation in proximal mutants L89I and H97F of carbonmonoxymyoglobin. Biophys J. 1995 Jun;68(6):2497–2504. doi: 10.1016/S0006-3495(95)80432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alben J. O., Beece D., Bowne S. F., Doster W., Eisenstein L., Frauenfelder H., Good D., McDonald J. D., Marden M. C., Moh P. P. Infrared spectroscopy of photodissociated carboxymyoglobin at low temperatures. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3744–3748. doi: 10.1073/pnas.79.12.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alben J. O., Moh P. P., Fiamingo F. G., Altschuld R. A. Cytochrome oxidase (a3) heme and copper observed by low-temperature Fourier transform infrared spectroscopy of the CO complex. Proc Natl Acad Sci U S A. 1981 Jan;78(1):234–237. doi: 10.1073/pnas.78.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Berendzen J., Bowne S. F., Frauenfelder H., Iben I. E., Sauke T. B., Shyamsunder E., Young R. D. Protein states and proteinquakes. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A., Berendzen J., Braunstein D., Cowen B. R., Frauenfelder H., Hong M. K., Iben I. E., Johnson J. B., Ormos P., Sauke T. B. Rebinding and relaxation in the myoglobin pocket. Biophys Chem. 1987 May 9;26(2-3):337–355. doi: 10.1016/0301-4622(87)80034-0. [DOI] [PubMed] [Google Scholar]
- Ansari A., Jones C. M., Henry E. R., Hofrichter J., Eaton W. A. Conformational relaxation and ligand binding in myoglobin. Biochemistry. 1994 May 3;33(17):5128–5145. doi: 10.1021/bi00183a017. [DOI] [PubMed] [Google Scholar]
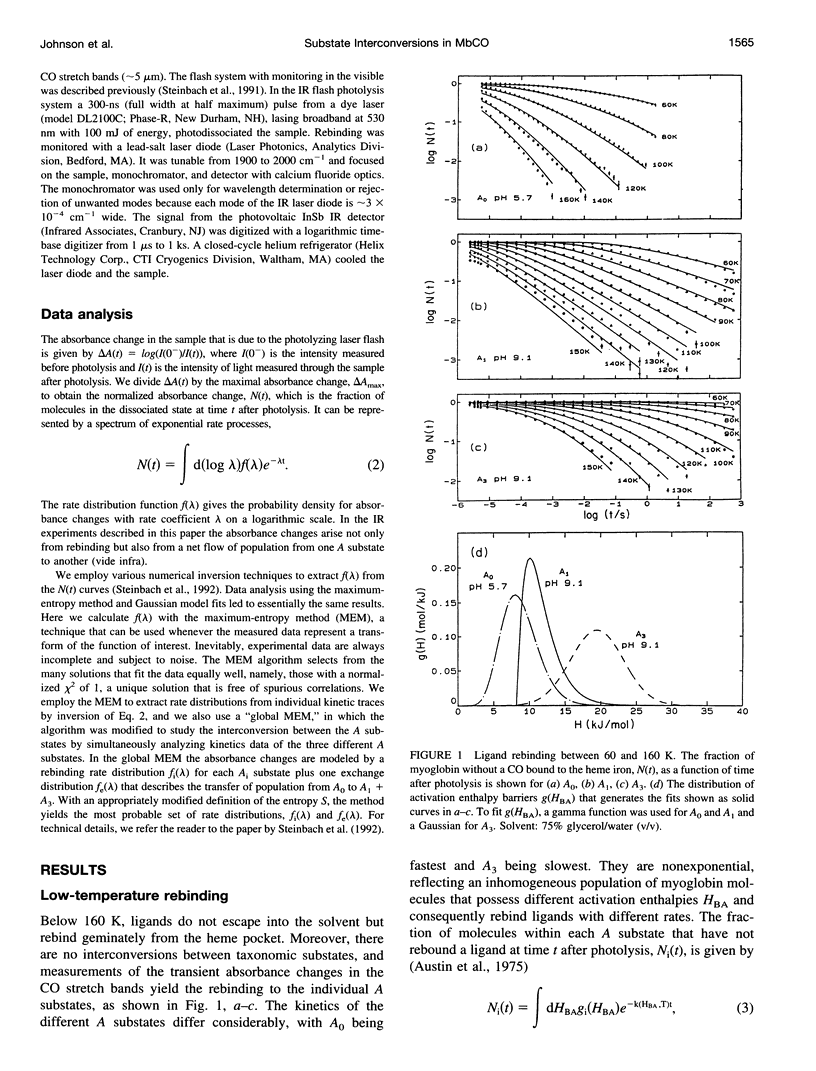
- Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., Gunsalus I. C. Dynamics of ligand binding to myoglobin. Biochemistry. 1975 Dec 2;14(24):5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S., Lambright D. G., Marden M. C., Boxer S. G. CO recombination to human myoglobin mutants in glycerol-water solutions. Biochemistry. 1993 Mar 9;32(9):2202–2212. doi: 10.1021/bi00060a011. [DOI] [PubMed] [Google Scholar]
- Berendzen J., Braunstein D. Temperature-derivative spectroscopy: a tool for protein dynamics. Proc Natl Acad Sci U S A. 1990 Jan;87(1):1–5. doi: 10.1073/pnas.87.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein D. P., Chu K., Egeberg K. D., Frauenfelder H., Mourant J. R., Nienhaus G. U., Ormos P., Sligar S. G., Springer B. A., Young R. D. Ligand binding to heme proteins: III. FTIR studies of His-E7 and Val-E11 mutants of carbonmonoxymyoglobin. Biophys J. 1993 Dec;65(6):2447–2454. doi: 10.1016/S0006-3495(93)81310-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bässler H. Viscous flow in supercooled liquids analyzed in terms of transport theory for random media with energetic disorder. Phys Rev Lett. 1987 Feb 23;58(8):767–770. doi: 10.1103/PhysRevLett.58.767. [DOI] [PubMed] [Google Scholar]
- Case D. A., Karplus M. Stereochemistry of carbon monoxide binding to myoglobin and hemoglobin. J Mol Biol. 1978 Aug 25;123(4):697–701. doi: 10.1016/0022-2836(78)90214-0. [DOI] [PubMed] [Google Scholar]
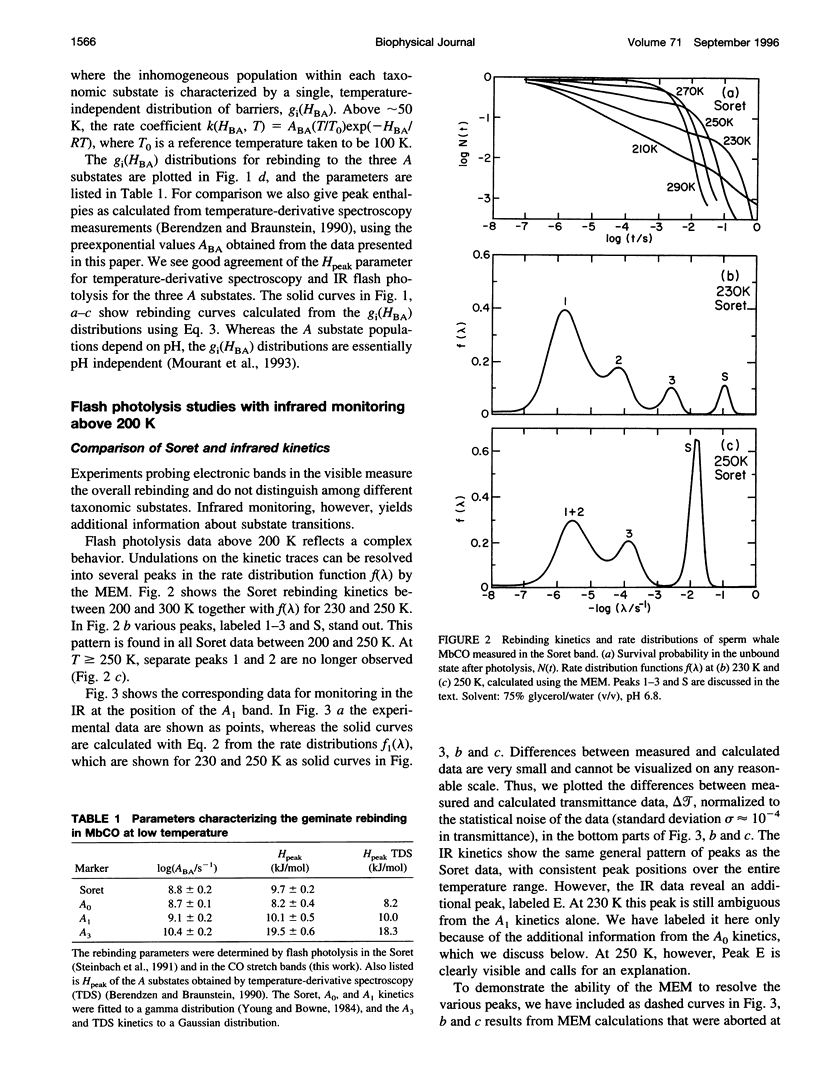
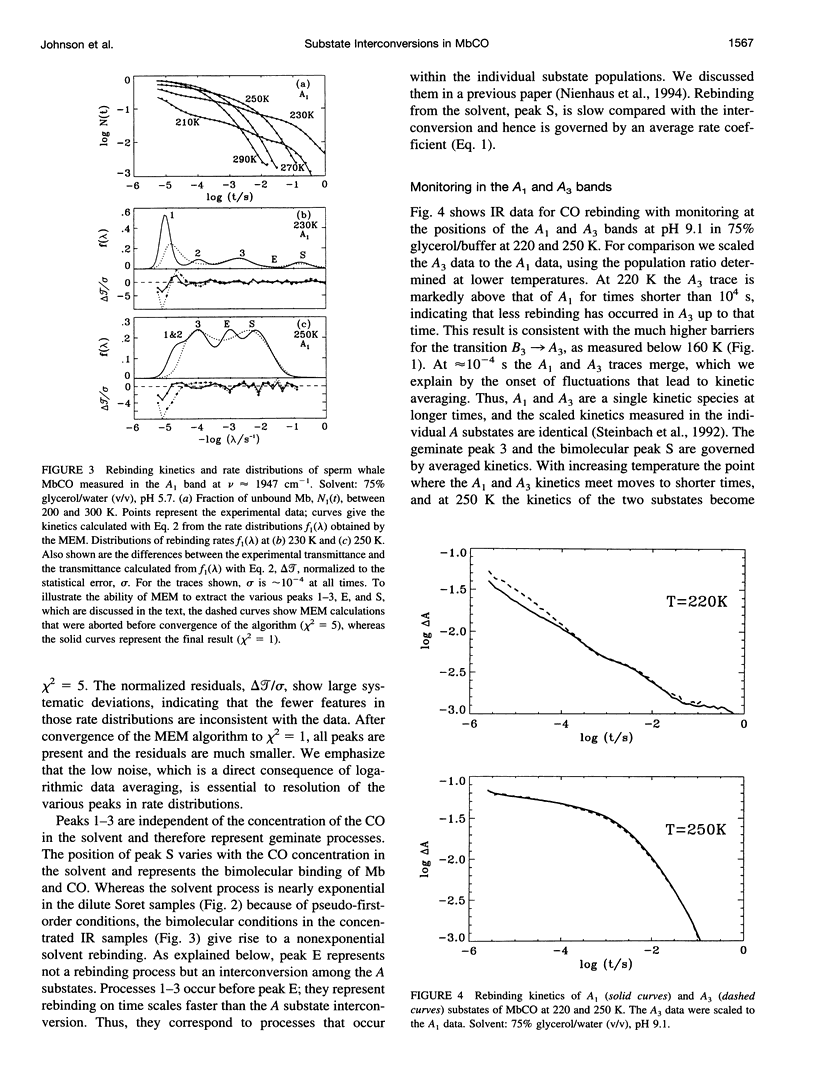
- Cheng X. D., Schoenborn B. P. Neutron diffraction study of carbonmonoxymyoglobin. J Mol Biol. 1991 Jul 20;220(2):381–399. doi: 10.1016/0022-2836(91)90020-7. [DOI] [PubMed] [Google Scholar]
- Doster W., Beece D., Bowne S. F., DiIorio E. E., Eisenstein L., Frauenfelder H., Reinisch L., Shyamsunder E., Winterhalter K. H., Yue K. T. Control and pH dependence of ligand binding to heme proteins. Biochemistry. 1982 Sep 28;21(20):4831–4839. doi: 10.1021/bi00263a001. [DOI] [PubMed] [Google Scholar]
- Doster W., Bowne S. F., Frauenfelder H., Reinisch L., Shyamsunder E. Recombination of carbon monoxide to ferrous horseradish peroxidase types A and C. J Mol Biol. 1987 Mar 20;194(2):299–312. doi: 10.1016/0022-2836(87)90377-9. [DOI] [PubMed] [Google Scholar]
- Ehrenstein D., Filiaci M., Scharf B., Engelhard M., Steinbach P. J., Nienhaus G. U. Ligand binding and protein dynamics in cupredoxins. Biochemistry. 1995 Sep 26;34(38):12170–12177. doi: 10.1021/bi00038a010. [DOI] [PubMed] [Google Scholar]
- Elber R., Karplus M. Multiple conformational states of proteins: a molecular dynamics analysis of myoglobin. Science. 1987 Jan 16;235(4786):318–321. doi: 10.1126/science.3798113. [DOI] [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983 Nov;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Parak F., Young R. D. Conformational substates in proteins. Annu Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Petsko G. A., Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979 Aug 16;280(5723):558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Sligar S. G., Wolynes P. G. The energy landscapes and motions of proteins. Science. 1991 Dec 13;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
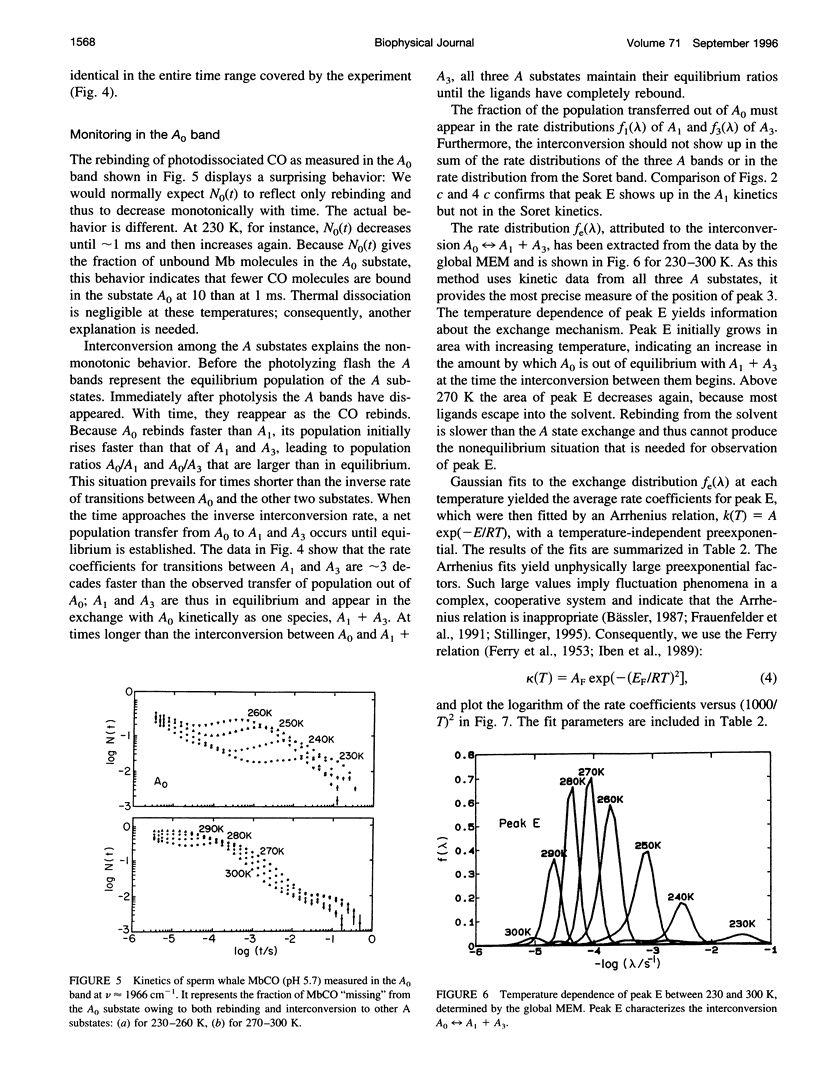
- Fuchsman W. H., Appleby C. A. CO and O2 complexes of soybean leghemoglobins: pH effects upon infrared and visible spectra. Comparisons with CO and O2 complexes of myoglobin and hemoglobin. Biochemistry. 1979 Apr 3;18(7):1309–1321. doi: 10.1021/bi00574a030. [DOI] [PubMed] [Google Scholar]
- Henry E. R. Molecular dynamics simulations of heme reorientational motions in myoglobin. Biophys J. 1993 Mar;64(3):869–885. doi: 10.1016/S0006-3495(93)81447-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeycutt J. D., Thirumalai D. Metastability of the folded states of globular proteins. Proc Natl Acad Sci U S A. 1990 May;87(9):3526–3529. doi: 10.1073/pnas.87.9.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M. K., Braunstein D., Cowen B. R., Frauenfelder H., Iben I. E., Mourant J. R., Ormos P., Scholl R., Schulte A., Steinbach P. J. Conformational substates and motions in myoglobin. External influences on structure and dynamics. Biophys J. 1990 Aug;58(2):429–436. doi: 10.1016/S0006-3495(90)82388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvidt A., Nielsen S. O. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- Iben IE, Braunstein D, Doster W, Frauenfelder H, Hong MK, Johnson JB, Luck S, Ormos P, Schulte A, Steinbach PJ. Glassy behavior of a protein. Phys Rev Lett. 1989 Apr 17;62(16):1916–1919. doi: 10.1103/PhysRevLett.62.1916. [DOI] [PubMed] [Google Scholar]
- Jewsbury P., Kitagawa T. The distal residue-CO interaction in carbonmonoxy myoglobins: a molecular dynamics study of two distal histidine tautomers. Biophys J. 1994 Dec;67(6):2236–2250. doi: 10.1016/S0006-3495(94)80708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczera K., Kuriyan J., Karplus M. Temperature dependence of the structure and dynamics of myoglobin. A simulation approach. J Mol Biol. 1990 May 20;213(2):351–373. doi: 10.1016/S0022-2836(05)80196-2. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., Wilz S., Karplus M., Petsko G. A. X-ray structure and refinement of carbon-monoxy (Fe II)-myoglobin at 1.5 A resolution. J Mol Biol. 1986 Nov 5;192(1):133–154. doi: 10.1016/0022-2836(86)90470-5. [DOI] [PubMed] [Google Scholar]
- Lambright D. G., Balasubramanian S., Boxer S. G. Dynamics of protein relaxation in site-specific mutants of human myoglobin. Biochemistry. 1993 Sep 28;32(38):10116–10124. doi: 10.1021/bi00089a030. [DOI] [PubMed] [Google Scholar]
- Lambright D. G., Balasubramanian S., Boxer S. G. Ligand and proton exchange dynamics in recombinant human myoglobin mutants. J Mol Biol. 1989 May 5;207(1):289–299. doi: 10.1016/0022-2836(89)90456-7. [DOI] [PubMed] [Google Scholar]
- Lecomte J. T., La Mar G. N. 1H NMR study of labile proton exchange in the heme cavity as a probe for the potential ligand entry channel in myoglobin. Biochemistry. 1985 Dec 3;24(25):7388–7395. doi: 10.1021/bi00346a054. [DOI] [PubMed] [Google Scholar]
- Li T., Quillin M. L., Phillips G. N., Jr, Olson J. S. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994 Feb 15;33(6):1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- Lim M., Jackson T. A., Anfinrud P. A. Binding of CO to myoglobin from a heme pocket docking site to form nearly linear Fe-C-O. Science. 1995 Aug 18;269(5226):962–966. doi: 10.1126/science.7638619. [DOI] [PubMed] [Google Scholar]
- Makinen M. W., Houtchens R. A., Caughey W. S. Structure of carboxymyoglobin in crystals and in solution. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6042–6046. doi: 10.1073/pnas.76.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell J. C., Caughey W. S. An infrared study of NO bonding to heme B and hemoglobin A. Evidence for inositol hexaphosphate induced cleavage of proximal histidine to iron bonds. Biochemistry. 1976 Jan 27;15(2):388–396. doi: 10.1021/bi00647a023. [DOI] [PubMed] [Google Scholar]
- Moore J. N., Hansen P. A., Hochstrasser R. M. Iron-carbonyl bond geometries of carboxymyoglobin and carboxyhemoglobin in solution determined by picosecond time-resolved infrared spectroscopy. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5062–5066. doi: 10.1073/pnas.85.14.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikis D., Champion P. M., Springer B. A., Sligar S. G. Resonance raman investigations of site-directed mutants of myoglobin: effects of distal histidine replacement. Biochemistry. 1989 May 30;28(11):4791–4800. doi: 10.1021/bi00437a041. [DOI] [PubMed] [Google Scholar]
- Mourant J. R., Braunstein D. P., Chu K., Frauenfelder H., Nienhaus G. U., Ormos P., Young R. D. Ligand binding to heme proteins: II. Transitions in the heme pocket of myoglobin. Biophys J. 1993 Oct;65(4):1496–1507. doi: 10.1016/S0006-3495(93)81218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nar H., Messerschmidt A., Huber R., van de Kamp M., Canters G. W. Crystal structure analysis of oxidized Pseudomonas aeruginosa azurin at pH 5.5 and pH 9.0. A pH-induced conformational transition involves a peptide bond flip. J Mol Biol. 1991 Oct 5;221(3):765–772. doi: 10.1016/0022-2836(91)80173-r. [DOI] [PubMed] [Google Scholar]
- Nienhaus G. U., Mourant J. R., Frauenfelder H. Spectroscopic evidence for conformational relaxation in myoglobin. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2902–2906. doi: 10.1073/pnas.89.7.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormos P., Braunstein D., Frauenfelder H., Hong M. K., Lin S. L., Sauke T. B., Young R. D. Orientation of carbon monoxide and structure-function relationship in carbonmonoxymyoglobin. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8492–8496. doi: 10.1073/pnas.85.22.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osapay K., Theriault Y., Wright P. E., Case D. A. Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints. J Mol Biol. 1994 Nov 25;244(2):183–197. doi: 10.1006/jmbi.1994.1718. [DOI] [PubMed] [Google Scholar]
- Park K. D., Guo K. M., Adebodun F., Chiu M. L., Sligar S. G., Oldfield E. Distal and proximal ligand interactions in heme proteins: correlations between C-O and Fe-C vibrational frequencies, oxygen-17 and carbon-13 nuclear magnetic resonance chemical shifts, and oxygen-17 nuclear quadrupole coupling constants in C17O- and 13CO-labeled species. Biochemistry. 1991 Mar 5;30(9):2333–2347. doi: 10.1021/bi00223a007. [DOI] [PubMed] [Google Scholar]
- Paul J., Smith M. L., Paul K. G. The vibrational bands of carbon monoxide bound to hemes or metal surfaces. Biochim Biophys Acta. 1985 Dec 20;832(3):257–264. doi: 10.1016/0167-4838(85)90258-4. [DOI] [PubMed] [Google Scholar]
- Potter W. T., Hazzard J. H., Choc M. G., Tucker M. P., Caughey W. S. Infrared spectra of carbonyl hemoglobins: characterization of dynamic heme pocket conformers. Biochemistry. 1990 Jul 3;29(26):6283–6295. doi: 10.1021/bi00478a025. [DOI] [PubMed] [Google Scholar]
- Quillin M. L., Arduini R. M., Olson J. S., Phillips G. N., Jr High-resolution crystal structures of distal histidine mutants of sperm whale myoglobin. J Mol Biol. 1993 Nov 5;234(1):140–155. doi: 10.1006/jmbi.1993.1569. [DOI] [PubMed] [Google Scholar]
- Ramsden J., Spiro T. G. Resonance Raman evidence that distal histidine protonation removes the steric hindrance to upright binding of carbon monoxide by myoglobin. Biochemistry. 1989 Apr 18;28(8):3125–3128. doi: 10.1021/bi00434a001. [DOI] [PubMed] [Google Scholar]
- Shimada H., Caughey W. S. Dynamic protein structures. Effects of pH on conformer stabilities at the ligand-binding site of bovine heart myoglobin carbonyl. J Biol Chem. 1982 Oct 25;257(20):11893–11900. [PubMed] [Google Scholar]
- Steinbach P. J., Ansari A., Berendzen J., Braunstein D., Chu K., Cowen B. R., Ehrenstein D., Frauenfelder H., Johnson J. B., Lamb D. C. Ligand binding to heme proteins: connection between dynamics and function. Biochemistry. 1991 Apr 23;30(16):3988–4001. doi: 10.1021/bi00230a026. [DOI] [PubMed] [Google Scholar]
- Steinbach P. J., Chu K., Frauenfelder H., Johnson J. B., Lamb D. C., Nienhaus G. U., Sauke T. B., Young R. D. Determination of rate distributions from kinetic experiments. Biophys J. 1992 Jan;61(1):235–245. doi: 10.1016/S0006-3495(92)81830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillinger F. H. A topographic view of supercooled liquids and glass formation. Science. 1995 Mar 31;267(5206):1935–1939. doi: 10.1126/science.267.5206.1935. [DOI] [PubMed] [Google Scholar]
- Tian W. D., Sage J. T., Champion P. M. Investigations of ligand association and dissociation rates in the "open" and "closed" states of myoglobin. J Mol Biol. 1993 Sep 5;233(1):155–166. doi: 10.1006/jmbi.1993.1491. [DOI] [PubMed] [Google Scholar]
- Tian WD, Sage JT, Srajer V, V, Champion PM. Relaxation dynamics of myoglobin in solution. Phys Rev Lett. 1992 Jan 20;68(3):408–411. doi: 10.1103/PhysRevLett.68.408. [DOI] [PubMed] [Google Scholar]
- Tsubaki M., Hiwatashi A., Ichikawa Y. Effects of cholesterol and adrenodoxin binding on the heme moiety of cytochrome P-450scc: a resonance Raman study. Biochemistry. 1986 Jun 17;25(12):3563–3569. doi: 10.1021/bi00360a014. [DOI] [PubMed] [Google Scholar]
- Tsubaki M., Ichikawa Y. Resonance Raman detection of a v(Fe-CO) stretching frequency in cytochrome P-450scc from bovine adrenocortical mitochondria. Biochim Biophys Acta. 1985 Mar 1;827(3):268–274. doi: 10.1016/0167-4838(85)90211-0. [DOI] [PubMed] [Google Scholar]
- Uno T., Nishimura Y., Tsuboi M., Makino R., Iizuka T., Ishimura Y. Two types of conformers with distinct Fe-C-O configuration in the ferrous CO complex of horseradish peroxidase. Resonance Raman and infarared spectroscopic studies with native and deuteroheme-substituted enzymes. J Biol Chem. 1987 Apr 5;262(10):4549–4556. [PubMed] [Google Scholar]
- Wagner G., Wüthrich K. Observation of internal motility of proteins by nuclear magnetic resonance in solution. Methods Enzymol. 1986;131:307–326. doi: 10.1016/0076-6879(86)31047-4. [DOI] [PubMed] [Google Scholar]
- Wilbur D. J., Allerhand A. Titration behavior and tautomeric states of individual histidine residues of myoglobins. Application of natural abundance carbon 13 nuclear magnetic resonance spectroscopy. J Biol Chem. 1977 Jul 25;252(14):4968–4975. [PubMed] [Google Scholar]
- Woodward C., Simon I., Tüchsen E. Hydrogen exchange and the dynamic structure of proteins. Mol Cell Biochem. 1982 Oct 29;48(3):135–160. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]
- Yang F., Phillips G. N., Jr Crystal structures of CO-, deoxy- and met-myoglobins at various pH values. J Mol Biol. 1996 Mar 8;256(4):762–774. doi: 10.1006/jmbi.1996.0123. [DOI] [PubMed] [Google Scholar]
- Zhu L., Sage J. T., Rigos A. A., Morikis D., Champion P. M. Conformational interconversion in protein crystals. J Mol Biol. 1992 Mar 5;224(1):207–215. doi: 10.1016/0022-2836(92)90584-7. [DOI] [PubMed] [Google Scholar]


